
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 10 May 2023
Sec. Aquatic Microbiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1163189
 Xiaohua Zhang1*†
Xiaohua Zhang1*† Shuang Cheng1†
Shuang Cheng1† Zhengquan Gao1
Zhengquan Gao1 Yulin Cui1
Yulin Cui1 Qingshou Yao1
Qingshou Yao1 Jiayang Qin1
Jiayang Qin1 Xiangyong Liu1
Xiangyong Liu1 Senjie Lin2
Senjie Lin2Phosphoesters are a dominant component of marine dissolved organic phosphorus (DOP) and an important source of the phosphorus nutrient for phytoplankton, but the molecular mechanisms of their utilization by phytoplankton are divergent and poorly understood. In this study, we used the model diatom Phaeodactylum tricornutum to investigate and compare the utilization mechanisms of two different phosphoesters, adenosine triphosphate (ATP) and glucose-6-phosphate (G6P). We found that ATP and G6P can both be efficiently used to support the growth of P. tricornutum. Cells grown on ATP or G6P showed lower pigment contents and photosynthetic rates but higher cellular lipids relative to those grown on NaH2PO4 (DIP). Surprisingly, in neither the ATP nor the G6P group were significant increases in whole-cell alkaline phosphatase (AP) activity detected, suggesting that utilization of both DOPs was not reliant on extracellular AP. Yet, ATP-grown cultures released DIP into the medium (i.e., ATP hydrolyzed extracellularly) whereas G6P-grown cultures did not. Furthermore, transcriptomic and RT-qPCR results showed that the gene encoding 5’ nucleotidase (5NT) in the ATP group and PhoD in the G6P group was upregulated. These results indicated that different pathways are involved in the use of these two DOPs, with ATP being hydrolyzed extracellularly likely by 5NT (PHATRDRAFT_44177) to release DIP for uptake, and G6P being directly absorbed and hydrolyzed intracellularly likely by PhoD (PHATRDRAFT_45757). Nevertheless, P. tricornutum under ATP and G6P conditions showed more similar transcriptomic profiles to each other than either compared to DIP-grown cultures, indicating similar metabolic functions of these two DOPs. These findings demonstrate that despite the high similarity in transcriptomic response to ATP and G6P conditions, the utilization mechanisms of these phosphoesters in the same species can be totally different, and the lack of AP activity does not necessarily signal the absence of DIP deficiency or the absence of DOP utilization.
Phosphorus (P) is an essential nutrient for all organisms, is a central component of key biochemical molecules, such as nucleic acids and phospholipids, and plays a critical role in many metabolic processes (Dyhrman et al., 2007; Paytan and McLaughlin, 2007). Available P is present in marine water in the form of dissolved inorganic P (DIP) or dissolved organic P (DOP) (Lin et al., 2016). DIP is considered the most preferred form as it can be directly utilized by phytoplankton, but it is often present at low or limiting concentrations in many marine systems (Tyrrell, 1999; Moore et al., 2013). When algae suffer from DIP deprivation, the ability to utilize DOP becomes an important competitive strategy for phytoplankton survival (Dyhrman and Ruttenberg, 2006). Therefore, DOP bioavailability is a potential driving factor affecting growth and species composition of phytoplankton.
DOP in the ocean consists of phosphoesters and phosphonates. Phosphoesters with C-O-P bonds are generally considered to be more available to phytoplankton in the ocean, accounting for the majority (80%–85%) of DOP (Young and Ingall, 2010). Numerous studies have shown that many phosphoesters, such as ATP and glucose-6-phosphate (G6P), could be utilized by phytoplankton (Wang et al., 2011; Richardson and Corcoran, 2015; Diaz et al., 2018). It is generally believed that phytoplankton possess enzymes such as alkaline phosphatase (AP) to hydrolyze DOP to phosphate (Lin et al., 2016). However, the mechanism of DOP utilization may widely vary among different species. For instance, ATP is extracellularly degraded by 5’ nucleotidase (5NT) to produce DIP before being taken up by the dinoflagellate Karenia mikimotoi (Luo et al., 2017), whereas ATP is directly taken up by Prorocentrum shikokuense (formerly P. donghaiense) without hydrolysis and directly involved in cellular processes (Li et al., 2015). The utilization of ATP by the diatom Skeletonema costatum also involves the extracellular hydrolysis of ATP, but via a different enzyme than AP or 5NT (Zhang X et al., 2020). Moreover, phytoplankton need to modulate metabolic processes to adapt to the DOP condition as the source of P. For instance, both K. mikimotoi and S. costatum modulate intracellular purine metabolism to accommodate ATP utilization despite adopting different utilization mechanisms (Luo et al., 2017; Zhang X et al., 2020). Phytic acid as the sole phosphorus source for Phaeodactylum tricornutum causes changes in cellular processes such as inositol phosphate metabolism and photosynthesis (Li et al., 2022). In the face of complex and diverse DOPs in natural marine ecosystems, the physiological responses and molecular mechanisms of phytoplankton using specific phosphoesters need to be further investigated.
Diatoms are an abundant and ubiquitous group of phytoplankton, contributing to approximately 40% of annual primary production in the marine ecosystem (Falkowski et al., 1998). P. tricornutum is a model diatom due to the availability of genome sequence and the ease of cultivation (Bowler et al., 2008). The mechanism of phosphate homeostasis in P. tricornutum has been extensively studied, and genes related to phosphorus metabolism were identified (Yang et al., 2014; Feng et al., 2015; Cruz de Carvalho et al., 2016). The first algal coordinate regulator of nitrogen–phosphorus stoichiometric homeostasis was identified and characterized in P. tricornutum (You et al., 2022). Therefore, P. tricornutum is a good model species for studying the molecular mechanism of DOP utilization in diatoms. Recently, the bioavailability and utilization mechanisms of phytic acid and 2-aminoethylphosphonate have been conducted successively in P. tricornutum (Li et al., 2022; Shu et al., 2022).
In this study, we explored and compared the physiological and transcriptomic responses in P. tricornutum to ATP and G6P, two typical phosphoesters widespread in marine areas. RNA-sequencing was conducted to identify differentially expressed genes (DEGs) of ATP- and G6P-grown cultures relative to DIP-grown cultures and explore potential shifts of key metabolic processes involved in ATP and G6P utilization, with the major aim to reveal molecular mechanisms of ATP and G6P utilization in P. tricornutum.
P. tricornutum was isolated from the coastal surface water in the Yellow Sea of China and preserved in the laboratory. Complex antibiotics (50 μg ml−1 kanamycin, 100 μg ml−1 ampicillin, and 50 μg ml−1 streptomycin) were added to the medium during algal culture to prevent bacterial contamination. Prior to experiments, the cultures were checked microscopically with the 4’,6-diamidino-2-phenylindole (DAPI) stain and incubated on LB medium to verify the absence of bacterial contamination (Shishlyannikov et al., 2011). Axenic cultures of P. tricornutum cells were maintained in artificial seawater, which was configured according to Berges et al. (2004), supplemented with f/2 medium without Na2SiO3.9H2O. Cells of P. tricornutum in the logarithmic growth phase were collected, washed with P-free medium, and transferred into 500-ml liquid cultures in 1-L conical flasks with sterilized artificial seawater enriched with f/2 medium (without Na2SiO3.9H2O and NaH2PO4). Then, the cultures were divided into four phosphorus nutrient conditions, as follows: the DIP group contained 36 μM NaH2PO4, the ATP group contained 12 μM ATP (=36 μM P), the G6P group contained 36 μM G6P, and the P-depleted group was set up without any additional P source. Each group contained three biological replicates. All cultures were incubated in a laboratory incubator (Ningbo, Jiangnan, China) at 20 ± 0.5°C under a 12 h:12 h light:dark cycle with an irradiance of 100 µmol photons m−2 s−1.
Samples were collected from each culture at regular times daily to monitor the growth curve and the DIP and DOP concentrations in the cultures. The algal cells were fixed with Lugol’s solution and placed in a 0.1-ml plankton counting chamber (Dengxun, Xiamen, China) and counted using a light microscope. Based on the growth curve, the logarithmic growth period of algal cells was divided into the early logarithmic phase (days 1 to 3) and the late logarithmic phase (days 3 to 8), and the specific growth rates were calculated separately. The specific growth rate (µ) was calculated using . , where N2 and N1 represent the algae cell number at time t2 and t1, respectively (Milligan and Harrison, 2000).
For P measurement, samples collected from cultures were filtered through a 0.22-μm PES. The DIP concentration of the filtrate was measured according to P molybdenum blue spectrophotometry (Murphy and Riley, 1962). The total P (TP) concentration was analyzed following the method of Jeffries et al. (1979). The sample was digested with potassium persulfate (K2S2O8) to convert all P to orthophosphate.
The intracellular and extracellular bulk alkaline phosphatase activity (APA) was measured daily by using 4-nitrophenyl phosphate (pNPP) as a substrate during incubation (Lin et al., 2012; Zhang T et al., 2020). Cell pellets from each group were collected daily and the pellet was lysed in 1 ml of Tris buffer (1 M, pH 9.0). pNPP at a final concentration of 1 mM was added to each sample in the dark and incubated at 20°C for 2 h. Samples were placed on ice to stop further enzymatic reactions and were prepared for enzyme activity detection. The culture medium supernatant was filtered with a 0.22-μm PES and added to pNPP with a final concentration of 1 mM for extracellular enzyme activity detection. OD measurements were performed using a microplate spectrophotometer (Readmax 1900, Flash Spectrum, Shanghai, China) at 405 nm. Single-cell APA was measured by labeling with ELF® 97 phosphatase substrate (Invitrogen, Carlsbad, CA, USA) with a final concentration of 0.25 mM (Gonzalez-Gil et al., 1998). Fluorescence measurements were performed using a laser scanning confocal microscope (LSM880, ZEISS, Jena, Germany), with the excitation wavelength set at 365 nm and the emission wavelength set at 445 nm.
Cells from each culture were collected and chlorophyll was extracted with 90% acetone at 4°C for 24 h in darkness. The chlorophyll (Chl) a and c and carotenoid concentrations were determined by spectrophotometry and calculated according to the equations of Ritchie (2006) and Li et al. (2022): Chl a (μg/ml) = 11.4902 × (A664 − A750) − 0.4504 × (A630 − A750), Chl c (μg/ml) = −3.4041 × (A664 − A750)+22.6792 × (A630 − A750), Carotenoid (μg/ml) = 7.6 × [(A480 − A750) − 1.49 × (A510 − A750)]. The chlorophyll fluorescence parameters of the photosynthetic efficiency of PSII (Fv/Fm) were obtained with a Fluorpen AP110/C (Photon Systems Instrument, Drasov, Czech Republic). For data measurement, samples were taken for dark adaptation for 15 min, and Fv/Fm was measured using the quantum yield protocol.
Fluorometric detection using Nile Red determined the intracellular relative lipid content (Wu et al., 2014). Approximately 1 × 106 cells were collected and mixed with Nile Red (Solarbio, Beijing, China) solution with a final concentration of 1 μg ml−1. Following the staining of the algal cell suspensions with Nile Red, fluorescence was measured using a SpectraMax M2 fluorescence spectrophotometer (Molecular Devices, San Jose, CA, USA) at 480-nm excitation and 570-nm emission wavelengths. Nile Red-free algal cultures and Nile Red-supplemented seawater medium were used as autofluorescence and seawater fluorescence controls, respectively.
For each sample, algae cells were collected on the fifth day by centrifugation at 4,000 × g for 15 min at 20°C and stored at −80°C until further analysis. Total RNA was extracted using a Trizol RNA extraction kit (Invitrogen, Carlsbad, CA, USA). RNA concentration and quality were analyzed using NanoDrop ND-2000 (Thermo Scientific, Wilmington, DE, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Each group had three biological replicates, and nine transcriptome libraries were constructed. Transcriptome libraries were generated using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina ® (NEB, Ipswich, MA, USA), and library quality was assessed on the Agilent Bioanalyzer 2100 system. Finally, those libraries were sequenced on an Illumina Hiseq platform and 150-bp paired-end reads were generated. High-quality clean reads were obtained after filtering out reads containing adapters and ploy-N, and low-quality reads from the raw reads. All clean reads were submitted to the Sequence Read Archive database (BioProject ID: PRJNA699705).
The reference genome of P. tricornutum was downloaded from the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/?term=phaeodactylum+tricornutum). Then, the clean reads were mapped to the P. tricornutum reference genome by using HISAT2 software. The gene expression level was quantified using HTSeq software. DEGs between two groups were selected with |log2 FoldChange| > 1 and p-value< 0.05 by using the DESeq2 software.
Reverse transcription-qPCR (RT-qPCR) was used to quantify the expression levels of selected DEGs in different treatment groups. Total RNA was isolated and transcribed into cDNA using the PrimeScript™ RT Kit with gDNA eraser (Takara, Dalian, China). The transcriptome dataset was screened for genes that were non-responsive to the P source alteration. Based on the screening results, TATA box binding protein (TBP) and ribosomal protein small subunit 30S (RPS) were selected as candidate reference genes. The web-based program RefFinder (http://blooge.cn/RefFinder/), which is a comprehensive platform integrating GeNorm, NormFinder, and BestKeeper programs, provides an overall ranking for TBP and RPS stability. Accordingly, the RPS was selected as the reference gene to normalize the target gene expression in each sample. Eight genes were selected for RT-qPCR. Gene-specific primers (Supplementary Table S1) were designed based on the sequences obtained from transcriptomic data. RT-qPCR was performed on a CFX connect qPCR system (Bio-Rad Laboratories, Singapore) by using a SYBR Green PCR kit (Takara, Dalian, China). Standard curves were generated, and the PCR amplification efficiency for all primers was between 96.3% and 110%, and the correlation coefficient was between 0.986 and 0.999. Relative gene expression was calculated by the 2−ΔΔCt method.
We used SPSS software (version 25.0, IBM, USA) for one-way analysis of variance (ANOVA) to evaluate the statistical significance of the differences among the obtained data in this study. Statistical significance was determined at the level of p< 0.05.
P. tricornutum cell density significantly increased in both the DIP and DOP (ATP and G6P) groups during the experimental period, whereas the growth in the P-depleted group was clearly depressed (Figure 1A). Population growth under ATP and G6P conditions showed trends similar to those of the DIP groups throughout the experiment. The cultures reached the maximum cell density on day 9, at 14.5 × 105 cells ml−1 in the DIP group, 12.9 × 105 cells ml−1 in the ATP group, and 12 × 105 cells ml−1 in the G6P group. During the early logarithmic growth period (from day 1 to day 3), the specific growth rate (μ) in the DIP, ATP, G6P, and P-depleted groups were 0.94, 0.93, 0.93, and 0.56 day−1, respectively. There was no significant difference in the specific growth rate of the early logarithmic growth period between the DIP and DOP groups (Figure 1B). During the late logarithmic growth period (from day 3 to day 8), the specific growth rate (μ) in the DIP, ATP, G6P, and P-depleted groups were 0.14, 0.12, 0.11, and 0.08 day−1, respectively. The specific growth rate of the late logarithmic growth phase was significantly higher in the DIP group than in ATP and G6P groups (Figure 1B).
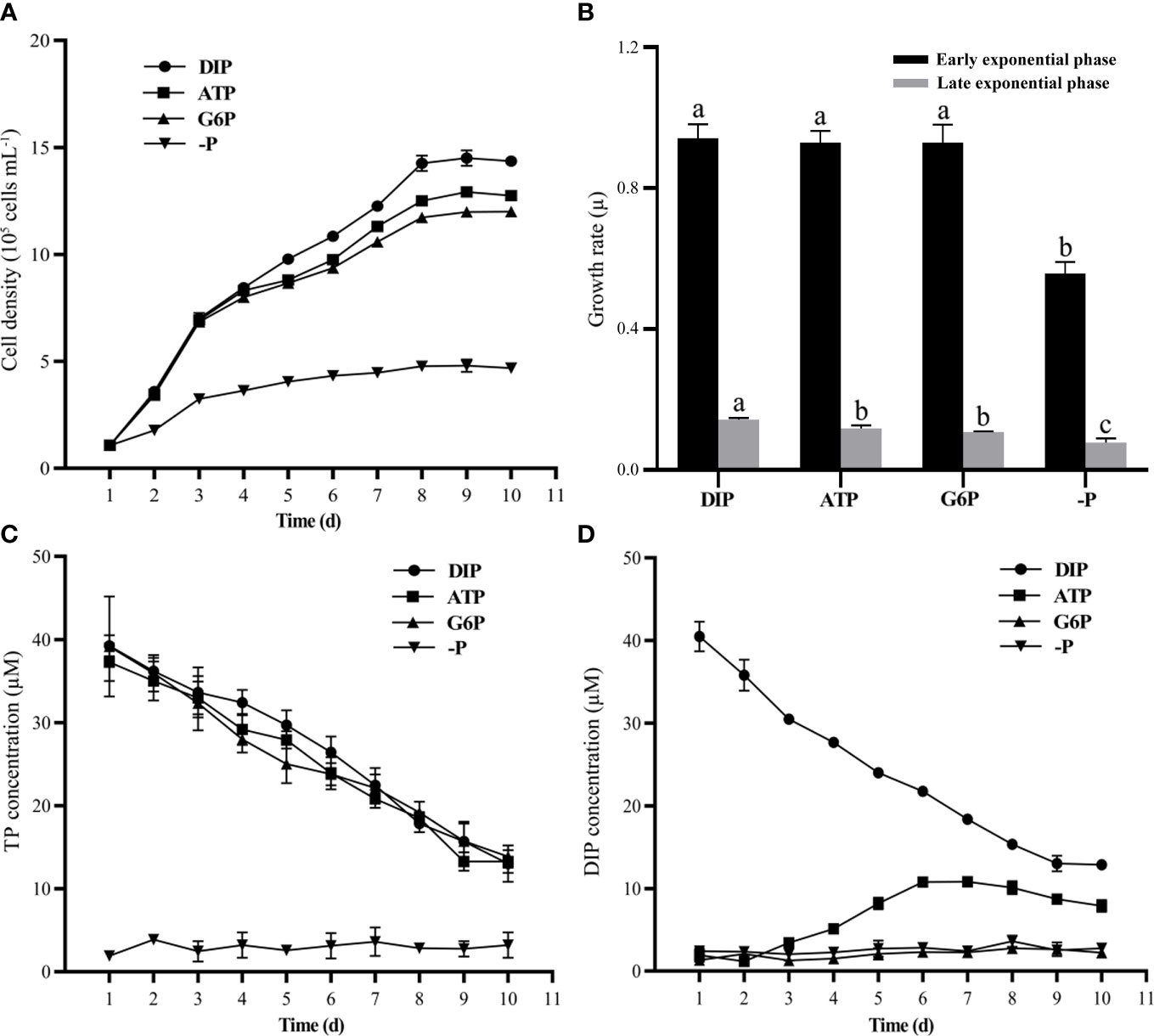
Figure 1 Physiological responses of P. tricornutum to DIP (NaH2PO4), ATP, glucose-6-phosphate (G6P), and P-depleted (–P) culture conditions. (A) Cell density. (B) Specific growth rate. (C) Total dissolved phosphorus (TP) and (D) DIP concentration in the culture medium. Different letters at the top of the columns in (B) indicate significant differences (ANOVA, p< 0.05). Values are mean ± SD, n = 3.
The TP concentration decreased significantly in the DIP, ATP, and G6P groups within 10 days of the experiment (Figure 1C), indicating that P sources were consumed in these three groups, whereas the TP concentration was barely detectable in the P-depleted group (Figure 1C). The DIP concentration in the DIP group decreased to 12.9 μM after 10 days of cultivation but showed different trends between the two DOP groups (Figure 1D). In the ATP cultures, DIP concentration in the medium started to increase on day 3 and reached the highest level of 10.8 μM on day 7. In contrast, DIP concentration in the G6P culture medium was very low throughout the experiment, similar to that in the P-depleted group (Figure 1D).
As shown in Figures 2A, B, intracellular and extracellular APA increased rapidly only in the P-depleted group and was barely detectable in the other three groups during the incubation. ELF (Enzyme-Labeled Fluorescence) 97 phosphatase substrate fluorescent labeling of single cells showed similar results, with AP-derived fluorescence detectable only in P-depleted cells (Figure 2C).
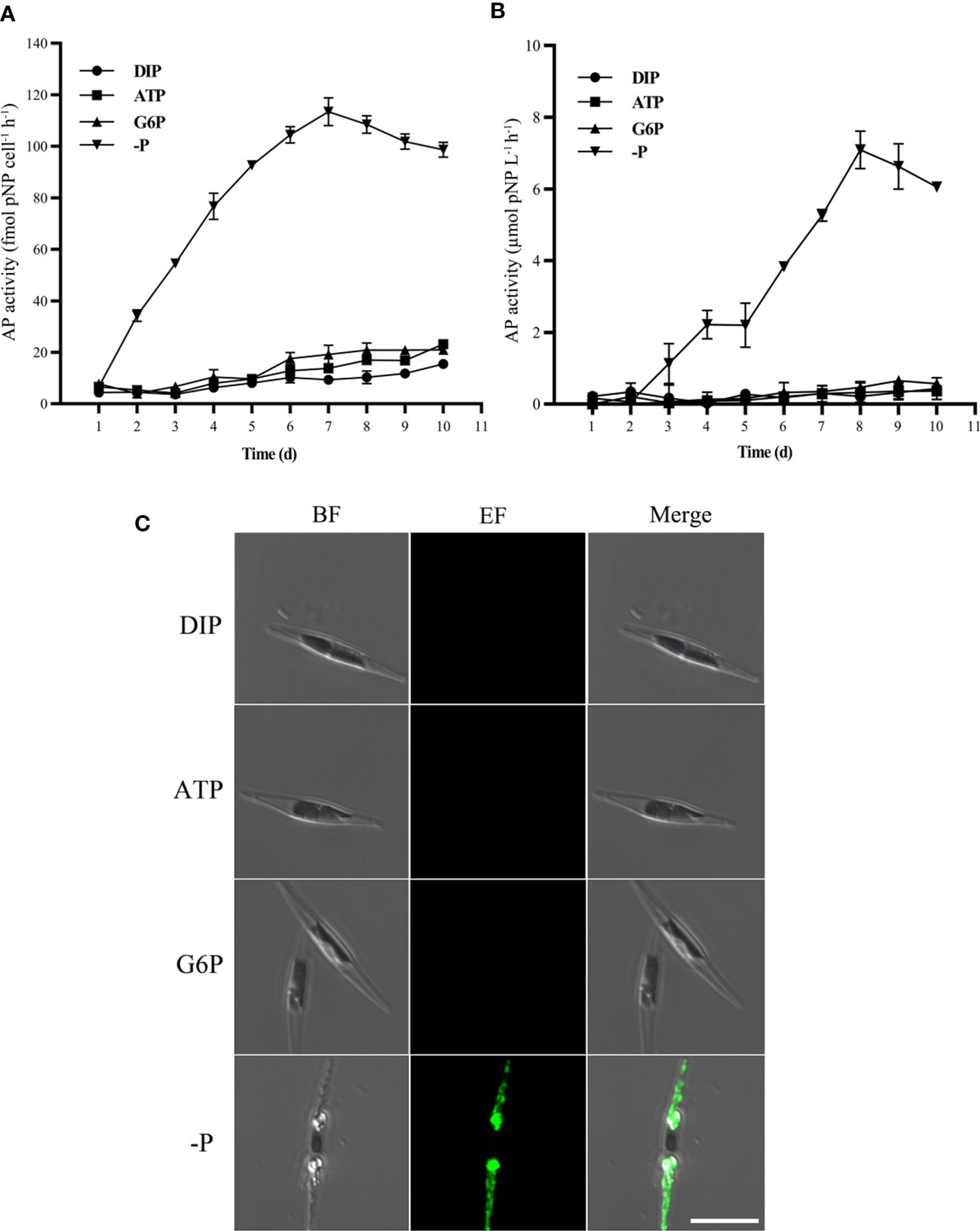
Figure 2 Alkaline phosphatase activity (APA). Bulk APA (A) inside the algal cells and (B) in the culture medium. (C) Microscopic observation of APA in the single algal cell by ELF fluorescence labeling. From left to right are bright-field (BF), ELF fluorescence (EF), and merger of bright-field and fluorescence images, respectively. Scale bar = 10 μm.
As shown in Figures 3A, B, the final Chl a and Chl c contents in the DIP group were 3.0% and 4.6% higher than that in the ATP group (albeit not significant p > 0.05), and 5.4% and 15.5% higher than that in G6P groups (p< 0.05), respectively. Meanwhile, the final carotenoid contents were 4.4% and 8.9% higher in the DIP than in the ATP and G6P groups, respectively (p< 0.05 in both comparisons) (Figure 3C). Similarly, the photosynthetic capacity of photosystem II (Fv/Fm) in the ATP and G6P groups showed different degrees of decline relative to the DIP group (Figure 3D). Fv/Fm in the ATP group decreased significantly on day 7 (p< 0.05), Fv/Fm in the G6P group decreased significantly on days 5, 7, and 9 (p< 0.05).
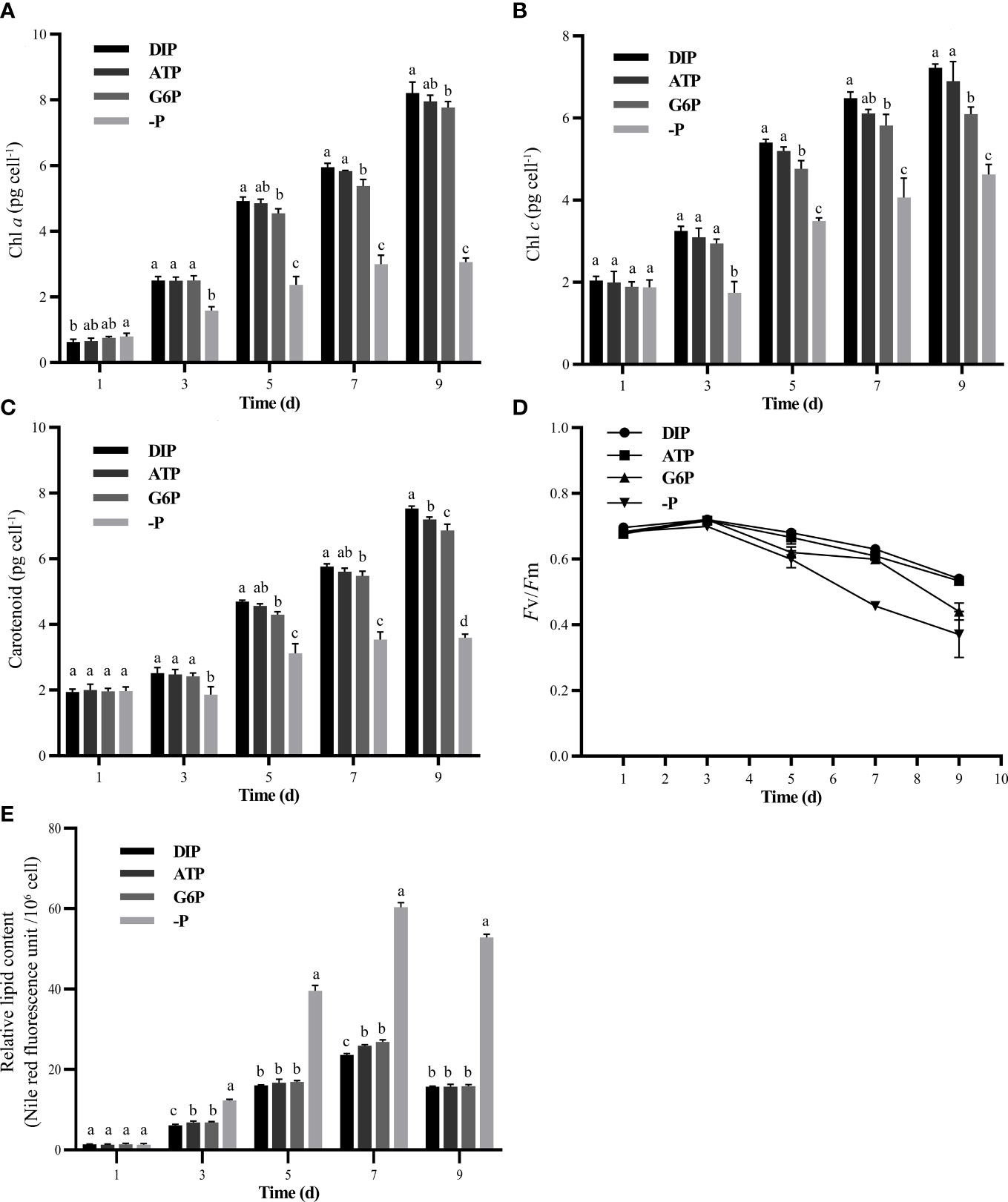
Figure 3 Photosynthetic characteristics and lipid analysis of P. tricornutum under DIP (NaH2PO4), ATP, glucose-6-phosphate (G6P), and P-depleted (–P) conditions. (A) Chlorophyll a content. (B) Chlorophyll c content. (C) Carotenoid content. (D) PS II maximum photochemical yield (Fv/Fm). (E) Relative intracellular lipid content. Different letters at the top of the column indicate significant differences (ANOVA, p< 0.05). Values are mean ± SD, n = 3.
We determined the lipid content using Nile Red and a fluorimetric spectrophotometric analysis. The relative fluorescence intensity of algae cells in the P-depleted group was consistently significantly higher than that in the DIP and DOP groups during cultivation (Figure 3E). The relative fluorescence intensity of algal cells in the ATP and G6P groups was higher than that in the DIP group, and the difference was significant on the 3rd and 7th day of the experiment (p< 0.05) (Figure 3E).
To investigate the DOP utilization mechanism and its effect on the cellular metabolism of P. tricornutum, DIP, ATP, and G6P group transcriptomes were compared by RNA-Seq. After quality filtration, an average of 91.44% of clean, high-quality reads were mapped to the reference genome (Supplementary Table S2). We performed pairwise comparisons between different groups and identified DEGs among the different groups based on the criteria of p-value< 0.05 |log2 FoldChange| > 1. A total of 632 DEGs were detected in the ATP versus DIP comparison, 537 DEGs were detected in the G6P versus DIP comparison, and 188 DEGs were detected in the ATP versus G6P comparison (Figure 4A; Supplementary File S1). There were 215 DEGs shared between ATP versus DIP and G6P versus DIP group (Figure 4B; Supplementary File S2). Hierarchical clustering analysis of DEGs found that ATP and G6P transcriptomes were more similar to each other in terms of gene expression patterns than either of these to the DIP group (Figure 4C; Supplementary File S3).
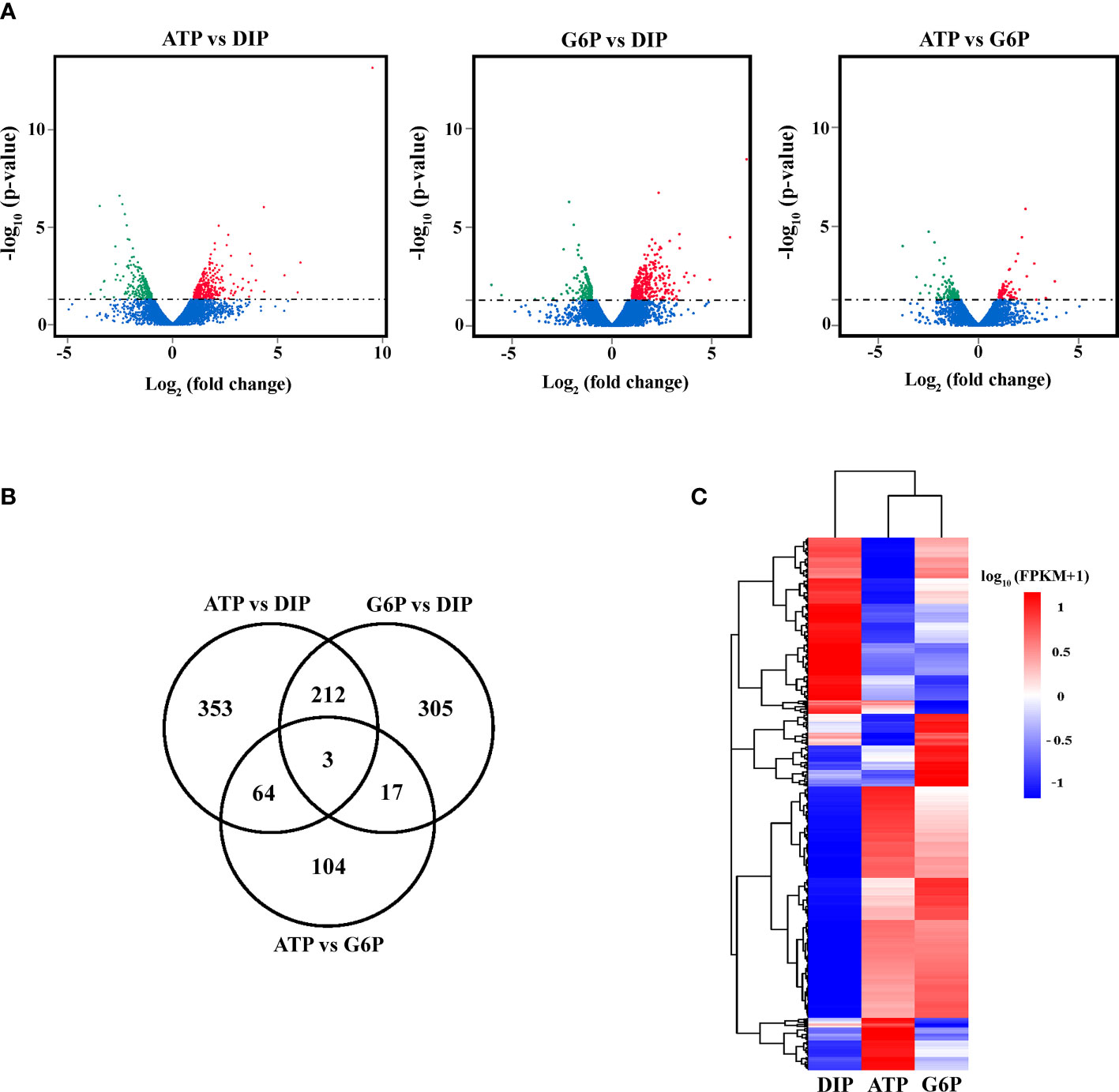
Figure 4 Comparative gene expression profiles of DIP (NaH2PO4), ATP, and glucose-6-phosphate (G6P) groups in P. tricornutum. (A) Volcano plot of differentially expressed genes (DEGs) between different phosphorus source groups. Red spots represent upregulated DEGs, green spots represent downregulated DEGs, and blue spots represent unchanged genes. (B) Venn diagram showing shared DEGs between ATP versus DIP, G6P versus DIP, and ATP versus G6P comparisons. (C) Hierarchical cluster analysis of differentially expressed transcripts between different groups. Red indicates higher-than-average abundance, while blue indicates lower-than-average abundance. The color from red to blue indicates that log10 (FPKM+1) is from large to small. FPKM indicates the expected number of fragments per kilobase of transcript sequence per million base pairs sequenced.
KEGG analyses revealed that the downregulated DEGs in the ATP/DIP comparison were significantly enriched in phagosome (q-value< 0.05; Supplementary File S4). The upregulated DEGs in the G6P/DIP comparison were significantly enriched in RNA degradation, and downregulated genes were significantly enriched in ubiquitin-mediated proteolysis, phagosome, and lysine degradation (q-value< 0.05; Supplementary File S4). Further comparative analysis of the two pairs indicated that the KEGG enrichment of DEGs in the G6P group significantly overlapped with that in the ATP group. As shown in Figure 5, the co-enrichment pathways of upregulated DEGs in ATP and G6P included glycerolipid metabolism and RNA degradation, among others. The co-enrichment pathways of downregulated DEGs in ATP and G6P included photosynthesis, phagosome, and others (Figure 5; Supplementary File S4).
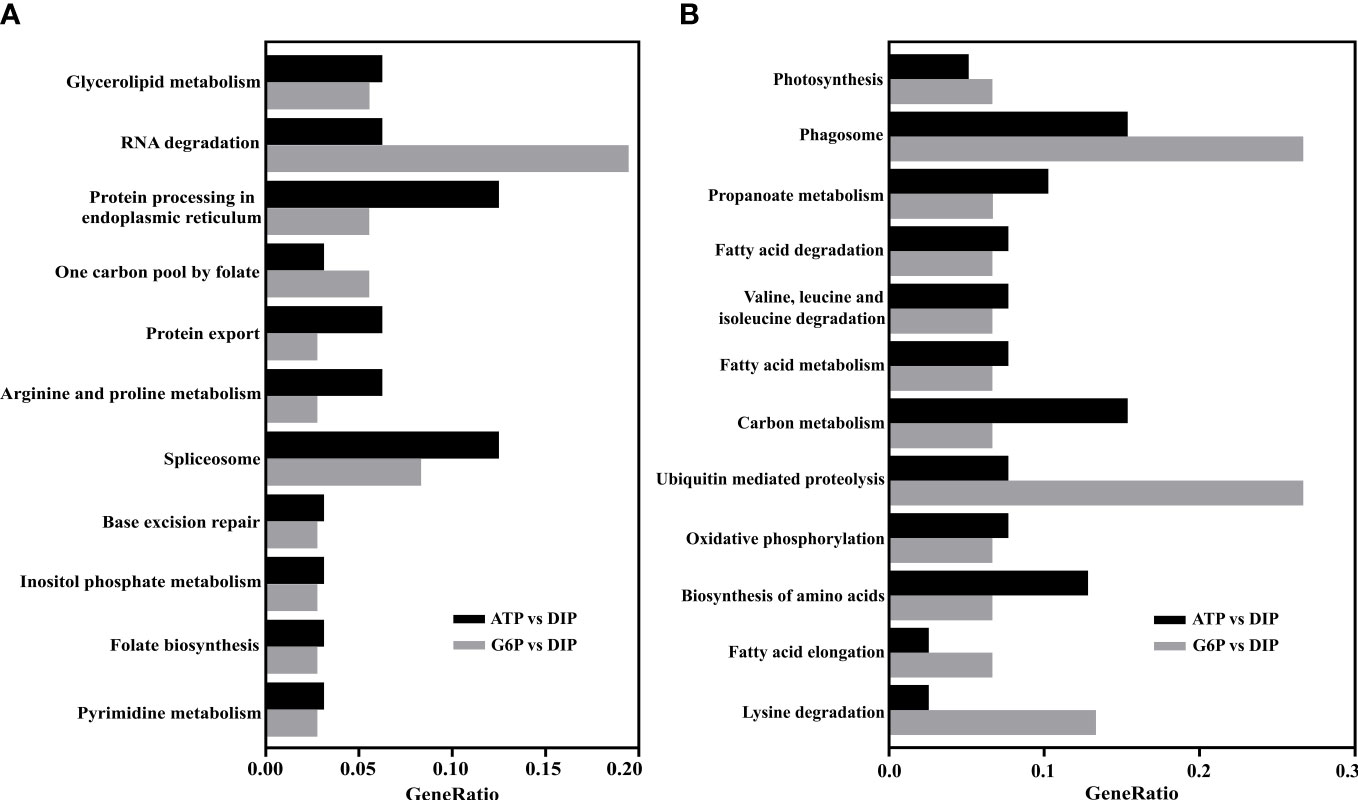
Figure 5 The overlap of the top 20 enriched KEGG pathways of (A) upregulated differentially expressed genes (DEGs) and (B) downregulated DEGs between the ATP and G6P groups.
Furthermore, we examined AP and AcP gene expression patterns in both the G6P and ATP groups compared with the DIP group (Supplementary Table S3). Consistent with the absence of detectable APA in the cultures, few upregulated AP genes were detected in the G6P and ATP groups in comparison with the DIP group, except PhoD (alkaline phosphatase, PHATDRAFT_45757), which was upregulated in the G6P group (Supplementary Table S3; Supplementary File S1; Figure 6). Moreover, 5NT (PHATRDRAFT_44177) was upregulated in the ATP group (Supplementary File S1).
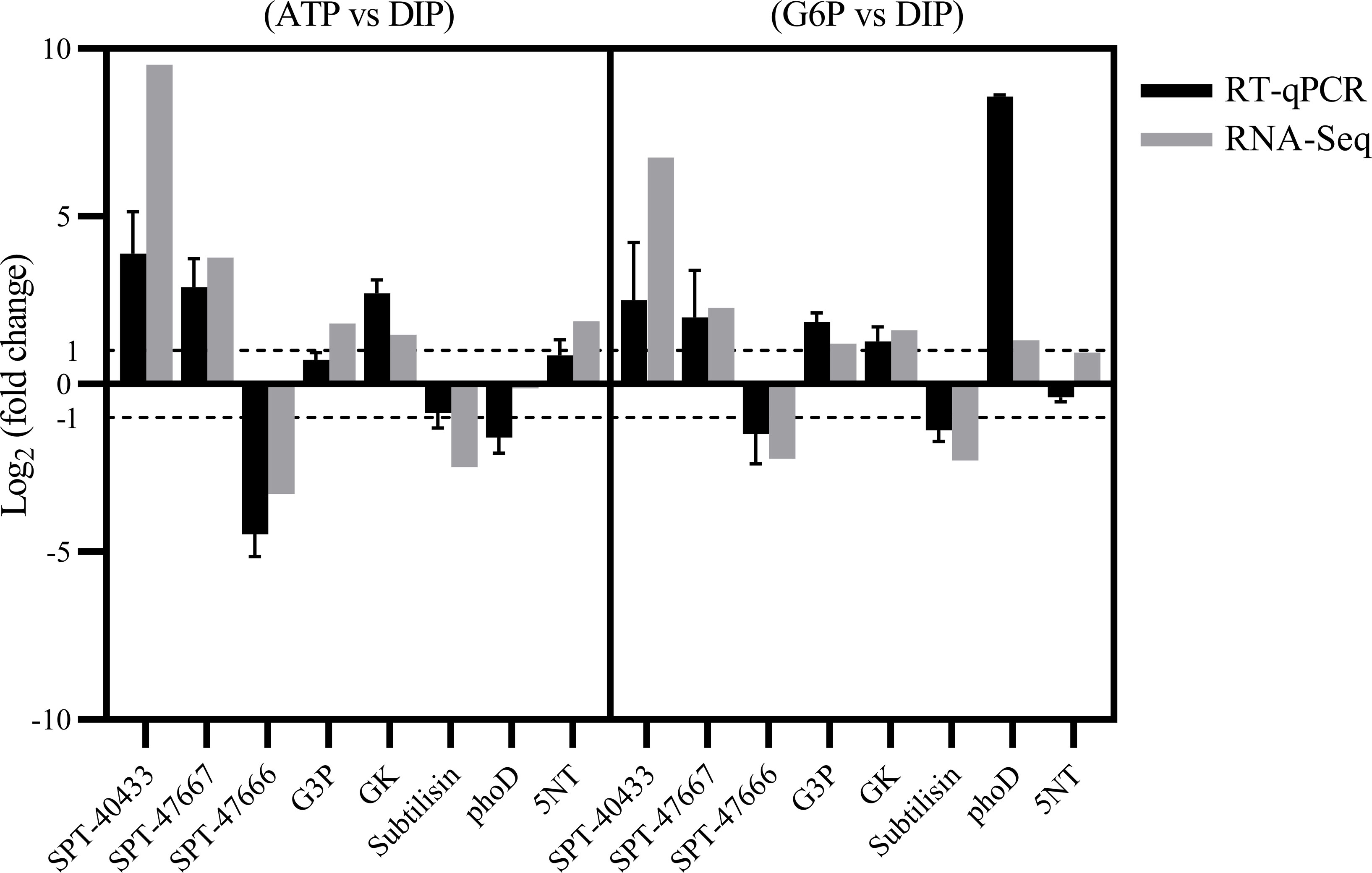
Figure 6 RT-qPCR validation of differentially expressed genes (DEGs). Ribosomal protein small subunit 30S was chosen as the internal reference gene. Gene name abbreviations: SPT-40433: Sodium-dependent phosphate transport protein (PHATRDRAFT_40433); SPT-47667: Sodium-dependent phosphate transport protein (PHATRDRAFT_47667); SPT-47666: Sodium-dependent phosphate transport protein (PHATRDRAFT_47666); Subtilisin: Subtilisin (PHATRDRAFT_50224); GK: glycerate kinase (PHATRDRAFT_56499); G3P: glycerol-3-phosphate 1-acyltransferase (PHATRDRAFT_50031); phoD: alkaline phosphatase (PHATRDRAFT_45757); 5NT: 5’ nucleotidase (PHATR_44177).
From the DEGs obtained by RNA-seq, eight DEGs were selected for RT-qPCR verification. These included three sodium-dependent phosphate transporter genes (PHATRDRAFT_40433, SPT-40433; PHATRDRAFT_47667, SPT-47667; and PHATRDRAFT_47666, SPT-47666), glycerate kinase (PHATRDRAFT_56499, GK), glycerol-3-phosphate 1-acyltransferase (PHATRDRAFT_50031, G3P), subtilisin (PHATRDRAFT_50224, Subtilisin), 5NT, and PhoD. Ribosomal protein small subunit 30S (RPS) was used as the reference gene to which expression of the eight DEGs was normalized. The expression pattern found from the RT-qPCR results was consistent with RNA-seq results, i.e., downregulation of SPT-47666 and Subtilisin and upregulation of GK, G3P, SPT-40433, and SPT-47667 in the DOP groups relative to the DIP group (Figure 6). In addition, the RT-qPCR results showed that the relative expression of phoD was upregulated in the G6P group and downregulated in the ATP group, while 5NT was upregulated in the ATP and not significantly changed in the G6P (Figure 6).
Acquiring P from organophosphorus is an effective and common mechanism for phytoplankton to cope with P deficiency in ecosystems (Dyhrman et al., 2006; Dyhrman et al., 2012; Harke et al., 2012; Lin et al., 2016). The DOP pool in the natural marine environments is composed of chemically diverse compounds, and a highly specialized and effective mechanism for a specific type of DOP might confer advantages for an algal species to outcompete others in low phosphate environments. That might define the unique ecological niche for the species. In this study, we investigated the utilization of two different DOPs, namely, ATP and G6P, by P. tricornutum. Our results showed that both ATP and G6P can be efficiently utilized to support the growth of P. tricornutum (Figure 1A). The detection of increased DIP concentration in the ATP culture medium and the lack of detectable DIP in the G6P group culture indicated that different mechanisms underlie ATP and G6P utilization in P. tricornutum.
The undetectable DIP in the G6P culture (Figure 1D) suggests that G6P was imported into algal cells directly without extracellular hydrolysis. In addition, the expression of phoD (PHATRDRAFT_45757) gene encoding a membrane-anchored intracellular phosphatase (Dell’Aquila et al., 2020; Zhang et al., 2022) was significantly upregulated in the G6P group (Supplementary Table S3; Supplementary File S1; Figure 6). This suggests that G6P might be directly transported into algal cells and then hydrolyzed intracellularly by PhoD_45757. In support of this scheme, we detected no AP activity outside the cell (Figure 2B) but upregulation of PhoD in the G6P group (Figure 6). It is expected then that intracellular AP activity would be upregulated in the G6P. However, no significant APA was detected inside the G6P-treated cells. This is possibly because phoD_45757 and other potential intracellular APs only contribute a minor fraction of the total APA, as demonstrated previously in the utilization of glycerol-P (Zhang et al., 2022). Therefore, algae might employ specific AP for certain DOP substrates, and in that case, it could be difficult to use APA to indicate active utilization of the DOPs.
In contrast, the detection of DIP release in the ATP medium (Figure 1D) clearly indicated that ATP was hydrolyzed extracellularly before uptake by the cells. Our transcriptomic analysis revealed that the expression of 5NT, but not that of AP, was upregulated in the ATP group. 5NT has been reported in P. tricornutum to be upregulated under P deficiency (Alipanah et al., 2018; Zhang et al., 2021). 5NT appears to be distinct from AP in substrate specificity and can specifically hydrolyze 5’-nucleotides, which has been reported in some eukaryotic phytoplankton and bacteria (Ammerman and Azam, 1985; Dyhrman and Palenik, 2003; Reistetter et al., 2013). For example, 5NT was reported to be responsible for ATP utilization as an extracellular hydrolase in the dinoflagellate K. mikimotoi (Luo et al., 2017) and Prochlorococcus MED4 (Reistetter et al., 2013). In this study, we detected the upregulated expression of 5NT in the ATP group, indicating that it might be involved in ATP hydrolysis and utilization in P. tricornutum. The function of 5NT involved in ATP utilization in P. tricornutum deserves further exploration, but our results demonstrate that lack of AP activity of algal cells does not necessarily mean a lack of extracellular hydrolysis of DOP.
It is clear that P. tricornutum adopts totally different mechanisms to utilize different phosphoesters. The mechanistic differentiation is not limited to this diatom, however. Previous studies have shown that the dinoflagellate K. mikimotoi also employs different mechanisms when utilizing the same DOPs as in the present study: ATP is hydrolyzed extracellularly and then absorbed (Luo et al., 2017), but G6P is taken up directly (Zhang et al., 2017). In addition, it is interesting to find that the phagosome pathway was downregulated in both the ATP and G6P groups in our study (Figure 5). Recently, P. tricornutum was reported to be able to utilize 2-aminoethylphosphonate as the sole source of P (Shu et al., 2022). Transcriptomic analysis in that study indicated that the phagosome pathway (specifically, clathrin-mediated endocytosis) was upregulated, suggesting that this phosphonate might be taken up by this alga via endocytosis. The contrast between that study and ours reported here might reflect mechanistic differentiation between utilizing phosphoesters and phosphonate, which should be further investigated in the future.
The transcriptome results were further analyzed to investigate the effect of DIP conversion to ATP or G6P as a P-nutrient on the metabolic process of P. tricornutum. The transcriptome results showed that the metabolic responses of P. tricornutum were similar under ATP and G6P conditions. It is worth mentioning that algal cells in the DIP and DOP groups have a growth delay at the late logarithmic phase (Figure 1B), probably due to CO2 restriction resulting from the absence of bubbling or shaking during culture. Co-limitation of CO2 could mask metabolic effects that might otherwise be more clearly attributable to P metabolism.
ATP and G6P influenced the photosynthesis of P. tricornutum. Compared with the DIP group, pigment content and Fv/Fm were decreased in the ATP and G6P groups (although not significantly in the ATP group) (Figure 3). Correspondingly, several genes encoding fucoxanthin chlorophyll a/c protein were repressed in both the G6P and ATP groups compared with the DIP group. Moreover, genes involved in electron transport, such as PetJ (PHATR_44056) and PetH (PHATRDRAFT_12813) in the ATP group and PetJ in the G6P group, were also downregulated. It has also been reported that photosynthesis was inhibited when Peridinium bipes grew well in the glycerol-P culture and Micromonas pusilla thrived in the ATP culture (Whitney and Lomas, 2016; Yang et al., 2020). Cruz de Carvalho et al. (2016) suggested that there is a strict relationship between nutrient supply and the regulation of genes involved in photosynthesis. These findings suggest that DOP might serve as a source of organic carbon as well as a source of P, which triggers the downregulation of photosynthesis.
Utilization of DOPs affects the carbon metabolism pathways in P. tricornutum. The glycolysis and TCA cycle-related proteins in P. tricornutum were repressed under ATP conditions. For example, the transcriptional abundance of PYC1 (PHATRDRAFT_30519), SCS-alpha (PHATRDRAFT_42015), succinate CoA ligase (PHATRDRAFT_26921), and GapC2a (PHATRDRAFT_51128) was significantly decreased in the ATP group. The glycolysis and TCA cycle are central carbon metabolism pathways that provide not only carbon skeletons but also ATP (energy) for organisms. It is possible that some ATP or its degradation products might be transported into cells just as Karenia mikimotoi and provide energy for cells (Luo et al., 2017). The ribokase, which catalyzes D-ribose to produce D-ribo-5-phosphate, was upregulated in the G6P group. In addition, the expressions of glycerate kinase (PHATRDRAFT_56499) and glyceraldehyde 3-phosphate dehydrogenase (PHATRDRAFT_16540) were also increased in the G6P group. As an important substrate in glycolysis and oxidative pentose phosphate pathways, G6P, which was directly uptaken, could directly participate in these pathways. Thus, ATP and G6P might not only provide P for algae cells but also directly participate in intracellular carbon metabolism activities, possibly leading to intracellular carbon flux shift.
In this study, the transcript levels of glycerol-3-phosphate O-acyltransferase (PHATRDRAFT_50031) and glycerate kinase (PHATRDRAFT_56499), which are the key enzyme genes involved in the de novo triacylglycerol (TAG) synthesis pathway, displayed induced expression, whereas the transcription level of the lipid droplet-associated hydrolase gene (PHATRDRAFT_43295) was downregulated in both the ATP and G6P groups. Consistent with the transcriptome results, the lipid content in algal cells treated with ATP and G6P was increased compared with that in the DIP group but was significantly lower than that in the P-depleted group (Figure 3E). It has been reported that many microalgae synthesize and accumulate large amounts of lipids under nutrient stress conditions (Gong et al., 2013). For instance, P. tricornutum can accumulate a large amount of lipids under P limitation in previous studies (Siron et al., 1989; Alipanah et al., 2018) as well as the present study (Figure 3E). Chlorella sorokiniana was found to increase de novo triacylglycerols (TAG) biosynthesis when grown on the toxic organophosphate malathion (Nanda et al., 2019). Our results suggest that ATP and G6P conditions elicit lipid metabolism modification in P. tricornutum. Marine phytoplankton has evolved a diverse lipid remodeling mechanism in response to P situation (Cañavate et al., 2017). The effects of the types and availability of DOP on the lipid composition of algae deserve more systematic and detailed research in the future.
P. tricornutum exhibits distinct utilization mechanisms for two different DOP, namely, ATP and G6P. ATP was hydrolyzed extracellularly to produce DIP for uptake, and 5NT is most likely responsible for ATP hydrolysis. In contrast, G6P seems to be transported into P. tricornutum cells directly and then is hydrolyzed by an intracellular AP, possibly PhoD. Comparative transcriptomics analyses indicated the genome-wide expression profiles of P. tricornutum under ATP and G6P conditions were more similar to each other than either of them to those under DIP conditions. Taken together, phytoplankton like P. tricornutum might remodel metabolic programming to accommodate utilization of DOPs from the environment in a similar manner and yet adopt different specific uptake mechanisms to utilize each type of DOP. Our results also caution against interpretation of the lack of AP activity as lack of DIP deficiency or lack of DOP utilization.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XZ and XL contributed to conception and design of the study. XZ and SC performed the experiments, data analyses, and wrote the original draft of the manuscript. ZG and YC provided technological and logistic support. JQ and QY organized the database and performed statistical analysis. SL reviewed the manuscript. All authors contributed to manuscript revision, and read and approved the submitted version.
This work was supported by the Natural Science Foundation of Shandong Province of China (grant number ZR2021MC134) and the National Natural Science Foundation of China (grant number 31970086, 31972815, 42176124).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1163189/full#supplementary-material
Alipanah L., Winge P., Rohloff J., Najafi J., Brembu T., Bones A. M. (2018). Molecular adaptations to phosphorus deprivation and comparison with nitrogen deprivation responses in the diatom Phaeodactylum tricornutum. PloS One 13, e0193335. doi: 10.1371/journal.pone.0193335
Ammerman J. W., Azam F. (1985). Bacterial 5’-nucleotidase in aquatic ecosystems: a novel mechanism of phosphorus regeneration. Science 227, 1338–1340. doi: 10.1126/science.227.4692.1338
Berges J., Franklin D., Harrison P. (2004). Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 37, 1138–1145. doi: 10.1046/j.1529-8817.2001.01052.x
Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Cañavate J. P., Armada I., Hachero-Cruzado I. (2017). Interspecific variability in phosphorus-induced lipid remodelling among marine eukaryotic phytoplankton. New Phytol. 213, 700–713. doi: 10.1111/nph.14179
Cruz de Carvalho M. H., Sun H., Bowler C., Chua N. (2016). Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 210, 497–510. doi: 10.1111/nph.13787
Dell’Aquila G., Zauner S., Heimerl T., Kahnt J., Samel-Gondesen V., Runge S., et al. (2020). Mobilization and cellular distribution of phosphate in the diatom Phaeodactylum tricornutum. Front. Plant Sci. 11, 579. doi: 10.3389/fpls.2020.00579
Diaz J. M., Holland A., Sanders J. G., Bulski K., Mollett D., Chou C. W., et al. (2018). Dissolved organic phosphorus utilization by phytoplankton reveals preferential degradation of polyphosphates over phosphomonoesters. Front. Mar. Sci. 5, 380. doi: 10.3389/fmars.2018.00380
Dyhrman S. T., Ammerman J. W., Van Mooy B. (2007). Microbes and the marine phosphorus cycle. Oceanography 20, 110–116. doi: 10.5670/oceanog.2007.54
Dyhrman S. T., Chappell P. D., Haley S. T., Moffett J. W., Orchard E. D., Waterbury J. B., et al. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71. doi: 10.1038/nature04203
Dyhrman S. T., Jenkins B. D., Rynearson T. A., Saito M. A., Mercier M. L., Alexander H., et al. (2012). The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PloS One 7, e33768. doi: 10.1371/journal.pone.0033768
Dyhrman S. T., Palenik B. (2003). Characterization of ectoenzyme activity and phosphate-regulated proteins in the coccolithophorid Emiliania huxleyi. J. Plankton Res. 25, 1215–1225. doi: 10.1093/plankt/fbg086
Dyhrman S. T., Ruttenberg K. C. (2006). Presence and regulation of alkaline phosphatase activity in eukaryotic phytoplankton from the coastal ocean: implications for dissolved organic phosphorus remineralization. Limnol. Oceanogr. 51, 1381–1390. doi: 10.4319/lo.2006.51.3.1381
Falkowski P. G., Barber R. T., Smetacek V. V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 2, 200–207. doi: 10.1126/science.281.5374.200
Feng T., Yang Z., Zheng J., Xie Y., Li D., Murugan S. B., et al. (2015). Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom Phaeodactylum tricornutum. Sci. Rep. 5, 10373. doi: 10.1038/srep10373
Gong Y., Guo X., Wan X., Liang Z., Jiang M. (2013). Triacylglycerol accumulation and change in fatty acid content of four marine oleaginous microalgae under nutrient limitation and at different culture ages. J. Basic Microbiol. 53, 29–36. doi: 10.1002/jobm.201100487
Gonzalez-Gil S., Keafer B. A., Jovine R. V. M., Aguilera A., Lu S., Anderson D. M. (1998). Detection and quantification of alkaline phosphatase in single cells of phosphorus-starved marine phytoplankton. Mar. Ecol. Prog. Ser. 164, 21–35. doi: 10.3354/meps164021
Harke M. J., Berry D. L., Ammerman J. W., Gobler C. J. (2012). Molecular response of the bloom-forming cyanobacterium, Microcystis aeruginosa, to phosphorus limitation. Microb. Ecol. 63, 188–198. doi: 10.1007/s00248-011-9894-8
Jeffries D. S., Dieken F. P., Jones D. E. (1979). Performance of the autoclave digestion method for total phosphorus analysis. Water Res. 13, 275–279. doi: 10.1016/0043-1354(79)90206-9
Li M., Li L., Shi X., Lin L., Lin S. (2015). Effects of phosphorus deficiency and adenosine 5′-triphosphate (ATP) on growth and cell cycle of the dinoflagellate Prorocentrum donghaiense. Harmful Algae 47, 35–41. doi: 10.1016/j.hal.2015.05.013
Li J., Zhang K., Lin X., Li L., Lin S. (2022). Phytate as a phosphorus nutrient with impacts on iron stress-related gene expression for phytoplankton: insights from the diatom Phaeodactylum tricornutum. Appl. Environ. Microbiol. 88, e02097–e02021. doi: 10.1128/AEM.02097-21
Lin S., Litaker R. W., Sunda W. G. (2016). Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 52, 10–36. doi: 10.1111/jpy.12365
Lin X., Zhang H., Huang B., Lin S. (2012). Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae). Harmful Algae 17, 14–24. doi: 10.1016/j.hal.2012.02.005
Luo H., Lin X., Li L., Lin L., Zhang C., Lin S. (2017). Transcriptomic and physiological analyses of the dinoflagellate Karenia mikimotoi reveal non-alkaline phosphatase-based molecular machinery of ATP utilization. Environ. Microbiol. 19, 4506–4518. doi: 10.1111/1462-2920.13899
Milligan A. J., Harrison P. J. (2000). Effects of non-steady-state iron limitation on nitrogen assimilatory enzymes in the marine diatom Thalassiosira weissflogii (BACILLARIOPHYCEAE). J. Phycol. 36, 78–86. doi: 10.1046/j.1529-8817.2000.99013.x
Moore C. M., Mills M. M., Arrigo K. R., Berman-Frank I., Bopp L., Boyd P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Murphy J., Riley J. P. (1962). A modified single solution method for determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Nanda M., Kumar V., Fatima N., Pruthi V., Verma M., Chauhan P. K., et al. (2019). Detoxification mechanism of organophosphorus pesticide via carboxylestrase pathway that triggers de novo TAG biosynthesis in oleaginous microalgae. Aquat. Toxicol. 209, 49–55. doi: 10.1016/j.aquatox.2019.01.019
Paytan A., McLaughlin K. (2007). The oceanic phosphorus cycle. Chem. Rev. 107, 563–576. doi: 10.1021/cr0503613
Reistetter E. N., Krumhardt K., Callnan K., Roache-Johnson K., Saunders J. K., Moore L. R., et al. (2013). Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 II: gene expression. Environ. Microbiol. 15, 2129–2143. doi: 10.1111/1462-2920.12129
Richardson B., Corcoran A. A. (2015). Use of dissolved inorganic and organic phosphorus by axenic and nonaxenic clones of Karenia brevis and Karenia mikimotoi. Harmful Algae 48, 30–36. doi: 10.1016/j.hal.2015.06.005
Ritchie R. J. (2006). Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth. Res. 89, 27–41. doi: 10.1007/s11120-006-9065-9
Shishlyannikov S. M., Zakharova Y. R., Volokitina N. A., Mikhailov I. S., Petrova D. P., Likhoshway Y. V. (2011). A procedure for establishing an axenic culture of the diatom Synedra acus subsp. radians (Kütz.) skabibitsch. from lake baikal. Limnol. oceanogr. Methods. 9, 478–484. doi: 10.4319/lom.2011.9.478
Shu H., You Y., Wang H., Wang J., Li L., Ma J., et al. (2022). Transcriptomic-guided phosphonate utilization analysis unveils evidence of clathrin-mediated endocytosis and phospholipid synthesis in the model diatom, Phaeodactylum tricornutum. mSystems 7, e00563–e00522. doi: 10.1128/msystems.00563-22
Siron R., Giusti G., Berland B. (1989). Changes in the fatty acid composition of Phaeodactylum tricornutum and Dunaliella tertiolecta during growth and under phosphorus deficiency. Mar. Ecol. Prog. Ser. 55, 95–100. doi: 10.3354/meps055095
Tyrrell T. (1999). The relative influences of nitrogen and phosphorus on oceanic primary production. Nature 400, 525–531. doi: 10.1038/22941
Wang Z., Liang Y., Kang W. (2011). Utilization of dissolved organic phosphorus by different groups of phytoplankton taxa. Harmful Algae 12, 113–118. doi: 10.1016/j.hal.2011.09.005
Whitney L. P., Lomas M. W. (2016). Growth on ATP elicits a p-stress response in the picoeukaryote Micromonas pusilla. PloS One 11, e0155158. doi: 10.1371/journal.pone.0155158
Wu S., Zhang B., Huang A., Huan L., He L., Lin A., et al. (2014). Detection of intracellular neutral lipid content in the marine microalgae Prorocentrum micans and Phaeodactylum tricornutum using Nile red and BODIPY 505/515. J. Appl. Physiol. 26, 1659–1668. doi: 10.1007/s10811-013-0223-0
Yang Y., Shi J., Jia Y., Bai F., Yang S., Mi W., et al. (2020). Unveiling the impact of glycerol phosphate (DOP) in the dinoflagellate Peridinium bipes by physiological and transcriptomic analysis. Environ. Sci. Eur. 32, 1–13. doi: 10.1186/s12302-020-00317-6
Yang Z., Zheng J., Niu Y., Yang W., Liu J., Li H. (2014). Systems-level analysis of the metabolic responses of the diatom Phaeodactylum tricornutum to phosphorus stress. Environ. Microbiol. 16, 1793–1807. doi: 10.1111/1462-2920.12411
You Y., Sun X., Ma M., He J., Li L., Porto F. W., et al. (2022). Trypsin is a coordinate regulator of n and p nutrients in marine phytoplankton. Nat. Commun. 13, 4022. doi: 10.1038/s41467-022-31802-6
Young C. L., Ingall E. D. (2010). Marine dissolved organic phosphorus composition: insights from samples recovered using combined electrodialysis/reverse osmosis. Aquat. Geochem. 16, 563–574. doi: 10.1007/s10498-009-9087-y
Zhang K., Li J., Wang J., Lin X., Li L., You Y., et al. (2022). Functional differentiation and complementation of alkaline phosphatases and choreography of DOP scavenging in a marine diatom. Mol. Ecol. 31, 3389–3399. doi: 10.1111/mec.16475
Zhang X., Lin S., Liu D. (2020). Transcriptomic and physiological responses of Skeletonema costatum to ATP utilization. Environ. Microbiol. 22, 1861–1869. doi: 10.1111/1462-2920.14944
Zhang T., Lu X., Yu R., Qin M., Wei C., Hong S. (2020). Response of extracellular and intracellular alkaline phosphatase in microcystis aeruginosa to organic phosphorus. Environ. Sci. pollut. Res. Int. 27, 42304–42312. doi: 10.1007/s11356-020-09736-7
Zhang C., Luo H., Huang L., Lin S. (2017). Molecular mechanism of glucose-6-phosphate utilization in the dinoflagellate Karenia mikimotoi. Harmful Algae 67, 74–84. doi: 10.1016/j.hal.2017.06.006
Keywords: dissolved organic phosphorus, phosphoester, ATP, glucose-6-phosphate, phaeodactylum tricornutum, transcriptome
Citation: Zhang X, Cheng S, Gao Z, Cui Y, Yao Q, Qin J, Liu X and Lin S (2023) Transcriptomics and physiological analyses unveil the distinct mechanisms of ATP and glucose-6-phosphate utilization in Phaeodactylum tricornutum. Front. Mar. Sci. 10:1163189. doi: 10.3389/fmars.2023.1163189
Received: 10 February 2023; Accepted: 28 April 2023;
Published: 10 May 2023.
Edited by:
Jackie L Collier, Stony Brook University, United StatesReviewed by:
Jianrong Xia, Guangzhou University, ChinaCopyright © 2023 Zhang, Cheng, Gao, Cui, Yao, Qin, Liu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohua Zhang, eGh6aGFuZ0Biem1jLmVkdS5jbg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.