
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 22 May 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1158325
This article is part of the Research TopicMining, Development and Utilization of Genetic Resources Related to Economic Traits of Aquatic Animals Based on Omics TechnologyView all 5 articles
 Tingting Li1†
Tingting Li1† Yihan Zhang1†
Yihan Zhang1† Shiqi Liu1
Shiqi Liu1 Moli Li1
Moli Li1 Ruixing Yao1
Ruixing Yao1 Senyu Niu1
Senyu Niu1 Jingyao Yuan1
Jingyao Yuan1 Huizhen Wang1,2*
Huizhen Wang1,2* Jingjie Hu1,3
Jingjie Hu1,3 Zhenmin Bao1,2,3
Zhenmin Bao1,2,3 Xiaoli Hu1,2*
Xiaoli Hu1,2*Carotenoids are essential nutrients for humans and animals, and carotenoid content has become an important trait to evaluate the nutritional value of many cultured animals. Marine animals provide humans with diverse carotenoids, and developing carotenoid-enriched varieties has been the focus of marine animal breeding. Understanding the molecular mechanism of carotenoid deposition could benefit marine animal breeding for carotenoid content improvement. In the present study, transcriptomic analysis of adductor muscle was performed between Yesso scallop (Patinopecten yessoensis) with white muscle (WM) and carotenoid-enriched orange muscle (OM). A total of 683 differentially expressed genes (DEGs) were identified, with 302 and 381 genes being up- and down-regulated in OM scallop. Gene co-expression network analysis identified four carotenoid accumulation−related modules, including three up-regulated modules and one down-regulated module. The genes in up-regulated modules mainly participate in the pathways of translation and transcription (MEgreen), immune system (MElightyellow), and lipid metabolism (MEpink), while the down-regulated module is mainly enriched with genes involved in various metabolic pathways (MEturquoise). As the causal gene responsible for muscle coloration in scallop, PyBCO-like 1 is the hub gene of MEturquoise and showed strong connectivity with NR2F1A, a transcriptional factor involved in the regulation of retinoic acid. In addition, the up-regulated DEGs, including WDR3, RPP29, TBL3, RIOK2, and NOB1 from “ribosome biogenesis”, HSP70s and HSP702Bs from “antigen processing and presentation”, and ACOX1 from “PPAR signaling pathway” were identified as hub genes, indicating the potential regulatory role of these genes and pathways in response to carotenoid accumulation. Our data contribute to a deeper understanding of the regulatory and response mechanisms of carotenoid accumulation in marine animals.
Carotenoids are a diverse group of colorful pigments that are essential for all photosynthetic organisms and most non-photosynthetic prokaryotes (Sandmann, 2015). Carotenoids and their derivatives have various biological functions in animal growth, development, and reproduction. They also act as effective photo-protectors against UV radiation, antioxidants, and enhancers of immunity (Maoka, 2020). Furthermore, they are important compounds for human nutrition and health due to their significant antioxidant function and as precursors for vitamin A biosynthesis (Miao and Wang, 2013). As almost all animals cannot synthesize carotenoids de novo and must obtain them from food, carotenoids have long been considered essential nutrients for humans (Menotti et al., 1999).
As one of the most widespread aquatic animals, many mollusks, such as scallops, mussels, oysters, clams, and abalones, provide humans with high-quality protein, lipids as well as carotenoids. More than 60 carotenoid compounds have been discovered in mollusks (Vershinin, 1996), and some carotenoids, such as astaxanthin and pectenolone, provided by mollusks are often not available to terrestrial organisms. Thus, carotenoid-based coloration has become an important phenotypic trait in aquatic breeding for improving their nutritional value.
Scallops are a popular cultured shellfish, prized for their large and delicious adductor muscles. In recent years, a color variant of adductor muscle was first found in Yesso scallops (Patinopecten yessoensis) (Li et al., 2010). After breeding and farming this variant, a new variety named “Haida golden scallop” possessing orange adductor muscle was developed (Figure 1). Compared with the normal scallop, the muscle of the “Haida golden scallop” contained higher carotenoid content. Subsequently, similar carotenoid-based color variants were found in some other scallop species, such as noble scallop (Chlamys nobilis) (Liu et al., 2015) and bay scallop (Argopecten irradians irradians) (Song and Wang, 2019). In addition to commercial value, these variants also provided excellent materials for researching the molecular mechanism underlying carotenoid accumulation in marine animals. Recently, the function of candidate genes of carotenoid pigmentation in scallop muscle has been reported. In noble scallops, SRB-like-3 was discovered to specifically express in orange scallops and its down-regulation remarkably decreased blood carotenoids in noble scallops (Liu et al., 2015). In bay scallops, RNA interference of VPS 29 resulted in the pectenolone reduction in adductor muscles (Song and Wang, 2019). In Yesso scallop, we confirmed that PyBCO-like 1 encoding carotenoid oxygenase was the causal gene for carotenoid coloration in the adductor muscle (Li et al., 2019). In addition, it has been shown that the scallop with carotenoid-enriched orange muscle showed advantages in growth and stress resistance, which may be affected by carotenoid accumulation (Zhang et al., 2018). However, it is not clear which pathways and key regulatory genes are affected by carotenoid accumulation in scallops.
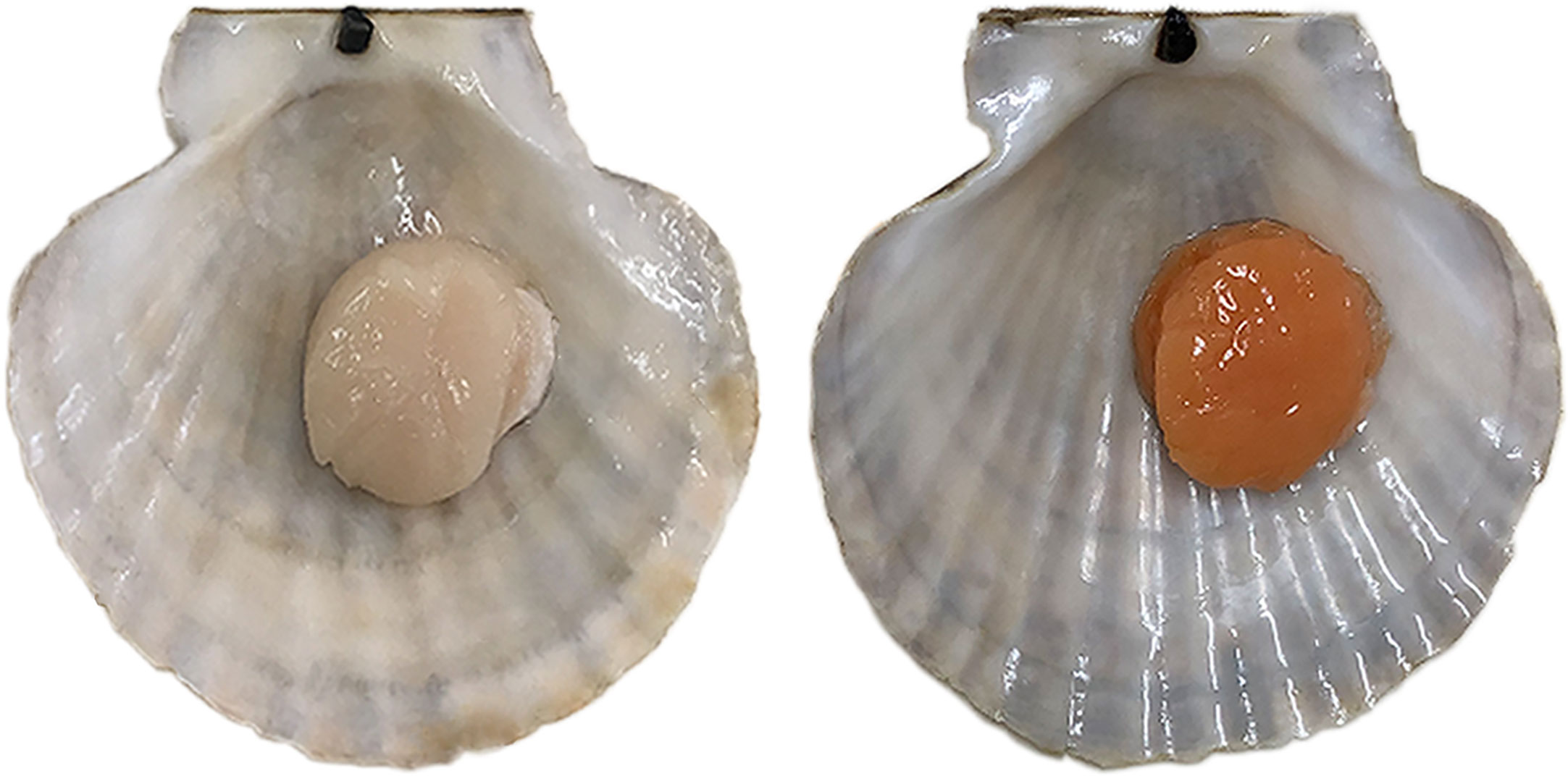
Figure 1 Color variation of the adductor muscle in Yesso scallop. Normal Yesso scallop with white muscle (WM scallop, left) and Haida golden scallop with orange muscle (OM scallop, right).
In this study, we performed a comparative transcriptome analysis of the adductor muscle between normal Yesso scallop and “Haida golden scallop”, and identified the key module and hub genes involved in regulating and responding to carotenoid accumulation through weighted gene co-expression network analysis (WGCNA). The results will provide new insights into the regulation of carotenoid accumulation and establish a foundation for carotenoid content improvement in scallops.
Normal Yesso scallops with white adductor muscle (WM) and “Haida golden scallop” with orange adductor muscle (OM) used in this study were obtained from the hatchery of Zhangzidao Group Co., Ltd. (Dalian, China), and all experimental procedures were approved by the Animal Care and Use Committee of Ocean University of China. The expression profile information was obtained from our previous study (Li et al., 2019). Concisely, total mRNA was extracted from the adductor muscles of 16 WM and 16 OM scallop individuals using the conventional guanidinium isothiocyanate method (Hu et al., 2006). RNA-seq libraries were constructed individually using the NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB, USA) and then sequenced on the Illumina HiSeq-2000 platform. For data analysis, high-quality (HQ) reads were mapped to the genome of Yesso scallop (Wang et al., 2017) using STAR aligner (v2.4.1d) (Dobin et al., 2012). Gene expression levels in terms of RPKM (Reads Per Kilobase per Million mapped reads) were estimated by HTseq and custom Perl scripts (Anders et al., 2014).
Differences in expression levels of adductor muscle between WM and OM scallops were analyzed using R (version 4.1.0) package EdgeR (v 3.36.0) (Robinson et al., 2010). Genes with a Benjamini and Hochberg corrected FDR < 0.05 and |log2 fold-change (FC)| > 1 were considered as differentially expressed genes (DEGs) (Benjamini and Hochberg, 1995).
A signed co-expression network of scallop adductor muscle was constructed using the R package weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath, 2008). Genes with RPKM > 0.5 in at least three of 32 adductor muscle samples were retained for network construction. All samples were firstly clustered to exclude any possible obvious outliers. And then, the soft threshold of 10 was chosen based on the criterion of approximate scale-free topology (model fitting index R2 = 0.8), and the co-expression modules were identified using the cutreeDynamic method, with the following parameters: detectCutHeight = 0.995, minModuleSize = 50, and deepSplit = F. The gene modules corresponding to the branch cut-off of the gene tree were labeled in different colors, and unassigned genes were labeled grey.
To identify the responsive modules of carotenoid accumulation, an overrepresentation analysis of DEGs between WM and OM scallops was performed for each module by a hypergeometric test, and modules with p value < 0.05 were considered as carotenoid accumulation−related modules. The hub genes in a given module were determined by the intramodular connectivity (Kwithin), which indicates a gene’s connection strength to other genes in the specified module. Therefore, the top 20% of genes with the highest Kwithin were defined as hub genes. Co-expression patterns of the top 50 hub genes were visualized using the heatmap. The networks were visualized using Cytoscape (version 3.9.1) (Shannon et al., 2003).
To analyze the biological functions of all DEGs and the genes in each module, gene ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using EnrichPipeline (Huang et al., 2009). Significantly enriched GO terms and KEGG pathways were defined by a hypergeometric test and a threshold of p value of less than 0.05.
To better understand the molecular mechanism of carotenoid deposition of P. yessoensis, we compared gene expression of adductor muscles between WM and OM scallops. A total of 32 libraries were constructed and named as follows: Mus-W1 to 16 and Mus-O1 to 16. After quality control of raw data, a total of 205,386,198 and 222,167,059 HQ reads were obtained from WM and OM scallops, respectively. The HQ reads were mapped to the reference genome with unique mapping rates ranging from 76.7% to 86.55% (Table S1). The correlation coefficients between the biological replicates of the same scallops were higher than 0.85 (Table S2), indicating good reproducibility of the biological repeats.
Compared with WM scallops, a total of 683 genes were significantly differentially expressed in the adductor muscles of OM scallops (|log2 (FC)| > 1 and FDR < 0.05), of which 302 were up-regulated and 381 genes showed down-regulation (Figure 2A; Table S3). We then examined the DEGs possibly involved in carotenoid deposition. Among carotenoid accumulation-related genes, PyBCO-like 1, encoding carotenoid oxygenase, showed the most significant down-regulated expression in OM scallops, which is consistent with our previous study (Li et al., 2019). We also found the genes encoding retinoic acid receptor (RXR), fatty acid-binding protein (FABP), very low-density lipoprotein receptor (VLDLR), and low-density lipoprotein receptor-related protein 1B (LRP1B) were differentially expressed between WM and OM scallops (Figure 2B).
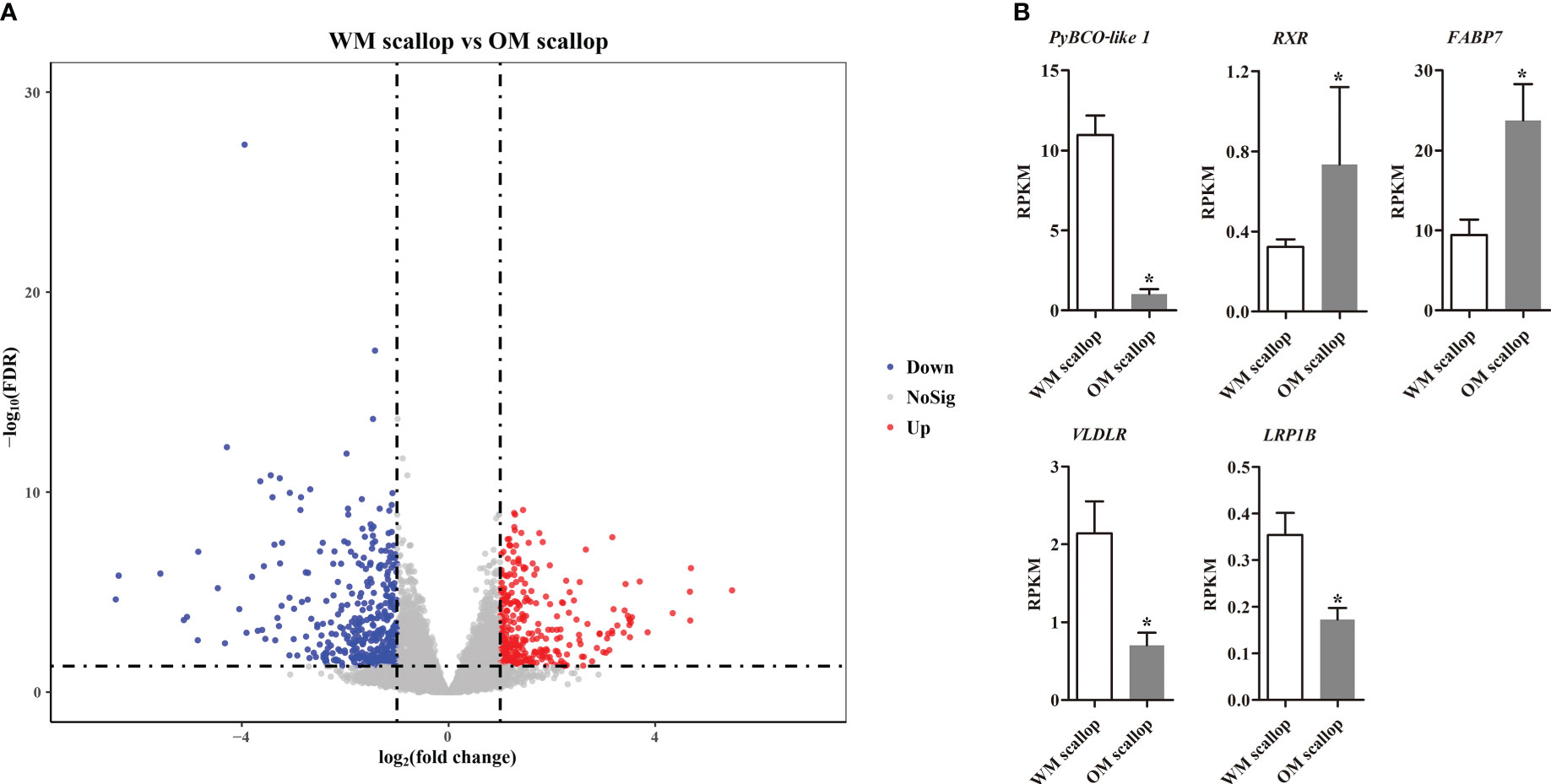
Figure 2 (A) Volcano plot for differentially expressed genes (DEGs) of adductor muscle between WM and OM scallops. The red and blue dots denote up-regulated and down-regulated genes in OM scallop. (B) Comparison of the expression levels (represented by RPKM values) of carotenoid accumulation-related genes in adductor muscle between WM and OM scallops. * indicates the significantly regulated DEGs with |log2(FC)| > 1 and FDR < 0.05.
To understand the potential functions of DEGs, GO term and KEGG pathway enrichment analysis was performed (Table S4). The GO terms of three main categories at level 4 are shown in Figure 3A. GO terms analyses showed that up-regulated DEGs were mainly significantly enriched in the cellular component (CC) related to ribosome (“preribosome”, “ribonuclease P complex”, “small nucleolar ribonucleoprotein complex”, etc.); the biological processes (BP) involving in “small molecule biosynthetic process” and “cellular macromolecule localization”; the molecular function (MF) related to transcription and translation (“RNA polymerase activity”, “ribonuclease activity”, “translation initiation factor binding”), and various enzyme activity (“catalase activity”, “transferase activity, transferring one-carbon groups”, “hydrolase activity, acting on ester bonds”). For down-regulated DEGs, “small molecule biosynthetic process” was the most significantly enriched GO term, followed by several metabolic processes (“organophosphate metabolic process”, “nitrogen cycle metabolic process”, and “urea metabolic process”) in BP category; Several enzyme activities related to lyase and hydrolase (“intramolecular lyase activity” and “hydrolase activity, acting on acid phosphorus-nitrogen bonds”) were significantly enriched in MF category, while “collagen-containing extracellular matrix” is the only significant term in CC category.
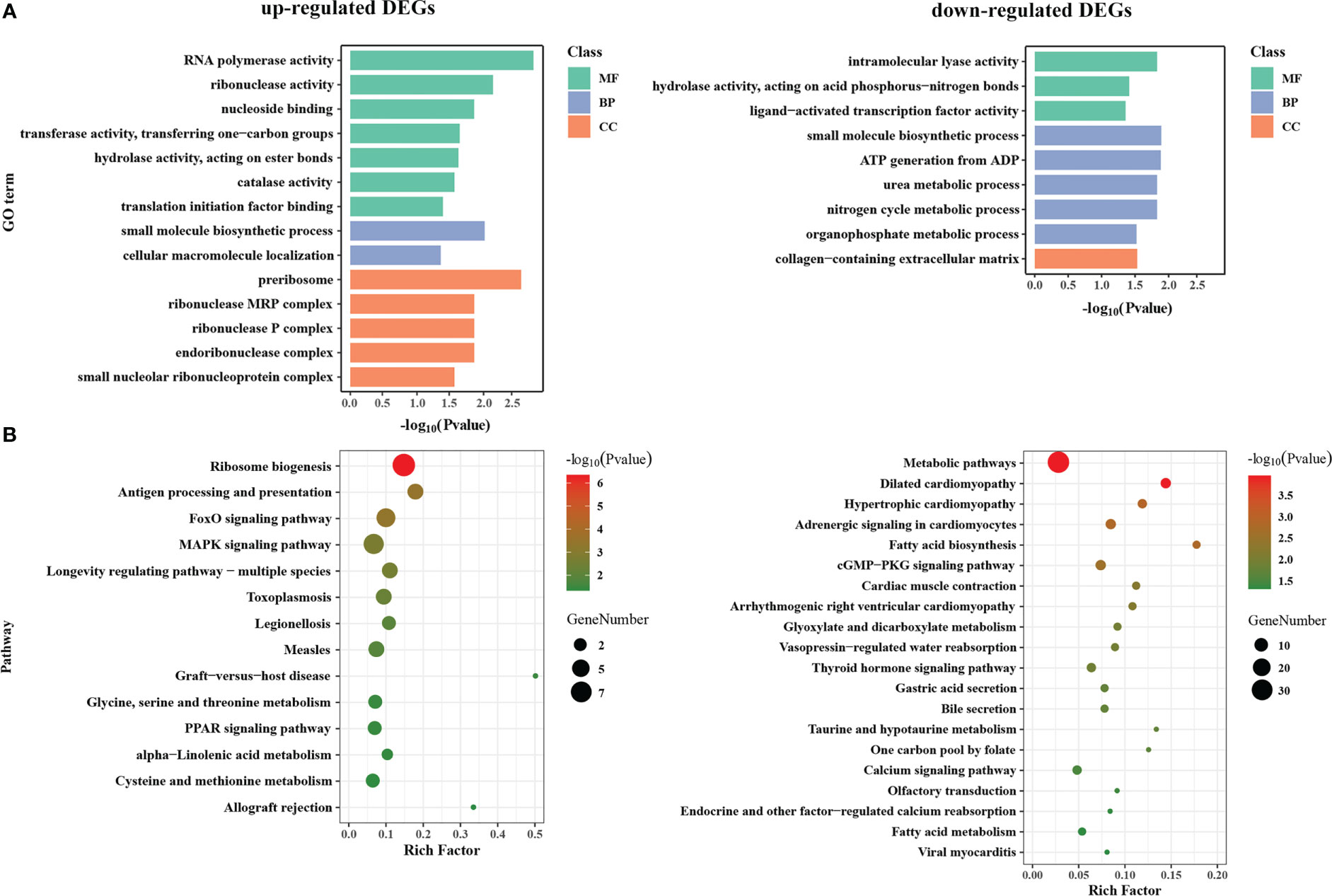
Figure 3 GO function and KEGG pathway enrichment analysis of DEGs between WM and OM scallops. (A) GO function enrichment analysis of up-regulated and down-regulated DEGs. Green, blue, and orange represent categories of molecular function (MF), biological process (BP), and cellular component (CC), respectively. (B) KEGG pathway enrichment plot of up-regulated and down-regulated DEGs.
The enrichment results of KEGG were roughly corresponding to GO enrichment (Figure 3B). KEGG pathway analyses revealed that the up-regulated DEGs mainly participate in translation (“ribosome biogenesis”), signal transduction (“MAPK signaling pathway” and “FoxO signaling pathway”), immune system (“antigen processing and presentation”, “graft-versus-host disease”, etc.), and lipid metabolism and regulation (“PPAR signaling pathway”, “alpha-linolenic acid metabolism”). By contrast, down-regulated DEGs were mainly enriched in the various pathways of metabolism, including “metabolic pathways”, “glyoxylate and dicarboxylate metabolism”, “taurine and hypotaurine metabolism”, “one carbon pool by folate”, “fatty acid biosynthesis” and “fatty acid metabolism”, as well as the pathways of signal transduction (“cGMP-PKG signaling pathway”, “calcium signaling pathway”, and “thyroid hormone signaling pathway”) and organismal systems (“cardiac muscle contraction”, “gastric acid secretion”, “endocrine and other factor-regulated calcium reabsorption”, etc.).
A gene co-expression network was constructed from 32 adductor muscle transcriptomes to screen the carotenoid accumulation-related modules. Based on the approximation of scale-free topology (R2 > 0.8) and gene mean connectivity, 12,535 genes including all 683 DEGs were assigned to 20 modules, with module sizes ranging from 81 to 2872 genes (Figure 4A). Overrepresentation analysis revealed that four modules including MEturquoise (p value = 1.48E-17), MEgreen (p value = 3.46E-3), MElightyellow (p value = 0.01), and MEpink (p value = 0.02) were significantly enriched with DEGs (Figure 4B; Table S5). These modules are considered as carotenoid accumulation-related modules. Interestingly, we found all DEGs in MEturquoise (252/252) were down-regulated in OM scallops, while almost all the DEGs in MEgreen (84/84), MElightyellow (12/12), and MEpink (61/62) were up-regulated in OM scallop. Further analyses of the expression of top 50 hub genes also corroborate this result (Figure 5A). The top 50 hub genes of MEturquoise module are highly expressed in WM scallops, while the hub genes of MEgreen, MElightyellow, and MEpink showed higher expression in OM scallops. Thus, MEturquoise is considered as a down-regulated module, and MEgreen, MElightyellow, and MEpink are up-regulated modules.
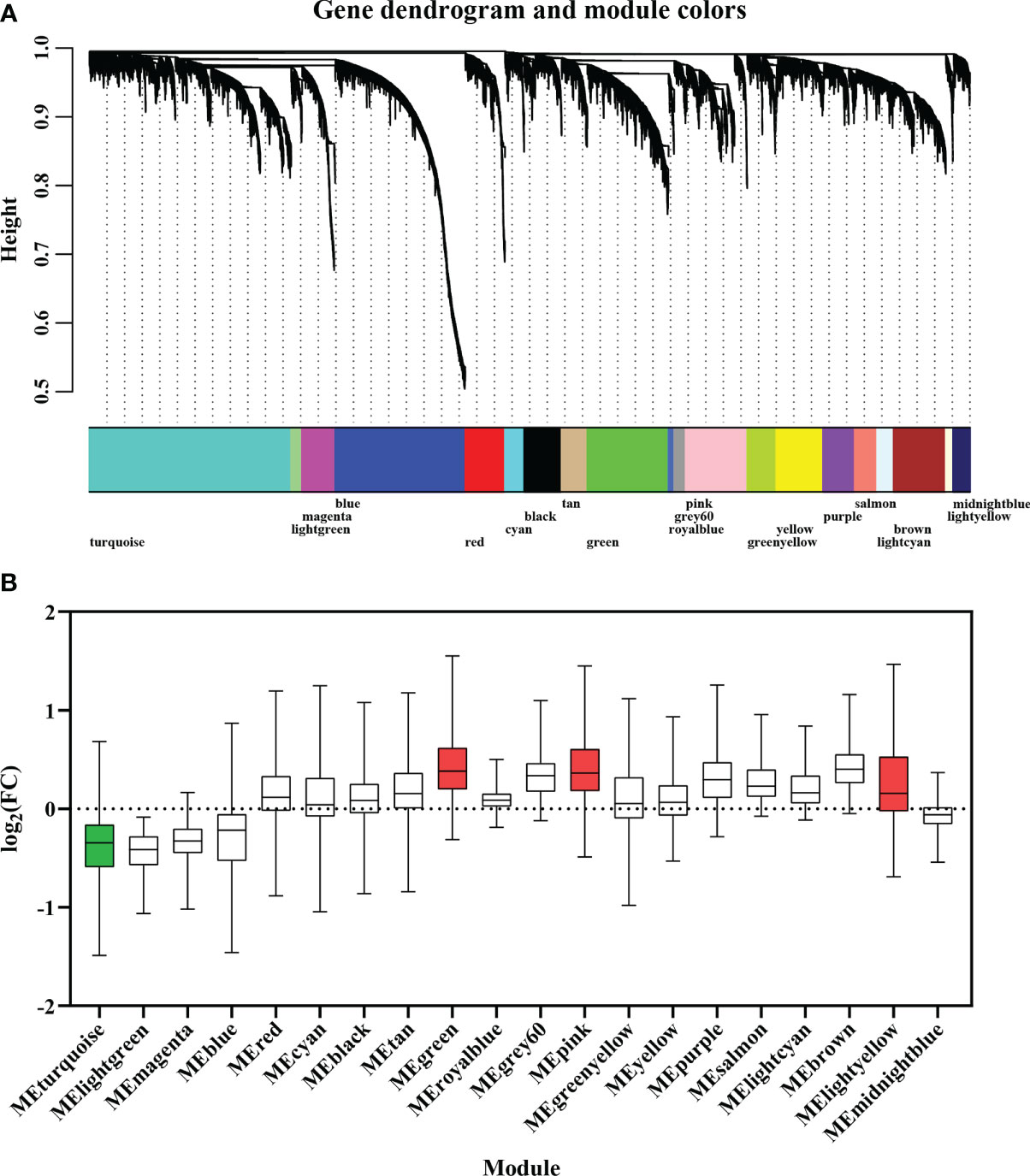
Figure 4 (A) Weighted gene co-expression network analysis (WGCNA) of 32 adductor muscle transcriptomes. Modules of co-expressed genes are labeled with different colors. (B) The boxplot of DEGs for 20 modules based on the fold change of their expression levels. The green and red colors of box represent down-regulated and up-regulated modules, respectively.
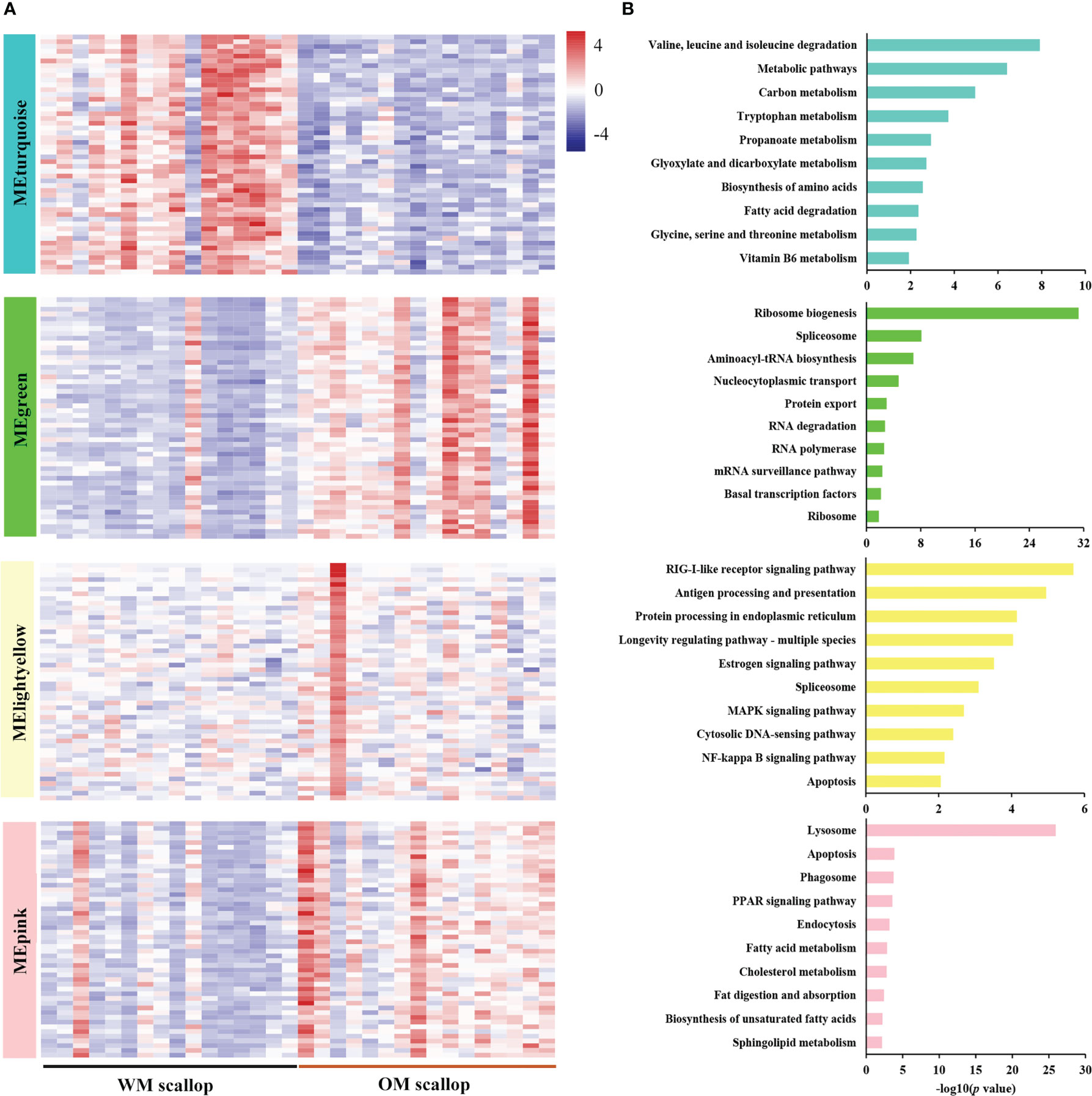
Figure 5 (A) Heatmap visualization of top 50 hub genes in carotenoid accumulation-related modules, including one down-regulated module (MEturquoise) and three up-regulated modules (MEgreen, MElightyellow, and MEpink). (B) KEGG enrichment analysis of genes in carotenoid accumulation-related modules (MEturquoise, MEgreen, MElightyellow, and MEpink).
Enrichment analysis was performed for genes in carotenoid accumulation-related modules to elucidate their potential biological roles. The down-regulated module MEturquoise was enriched with genes participating in metabolic pathways, such as amino acid metabolism (“valine, leucine and isoleucine degradation”), lipid metabolism (“fatty acid degradation”), carbohydrate metabolism (“glyoxylate and dicarboxylate metabolism”), metabolism of cofactors and vitamins (“vitamin B6 metabolism”), etc (Figure 5B). PyBCO-like 1, the key enzyme involved in carotenoid cleavage, was included in the top 20% of hub genes and showed lower expression in OM scallop than that in WM scallop. Similar expression patterns are also detected for the hub genes encoding transcriptional factors (TFs), including NR2F1A (nuclear receptor subfamily 2 group F member 1-A), EPAS1 (endothelial PAS domain-containing protein 1), and SCRT1 (Transcriptional repressor scratch 1) (Table S3). Among all TF genes in MEturquoise, NR2F1A as the member of RXR-like family, showed the highest connectivity with PyBCO-like 1 (Figure 6A), implying a potential regulatory relationship between them.
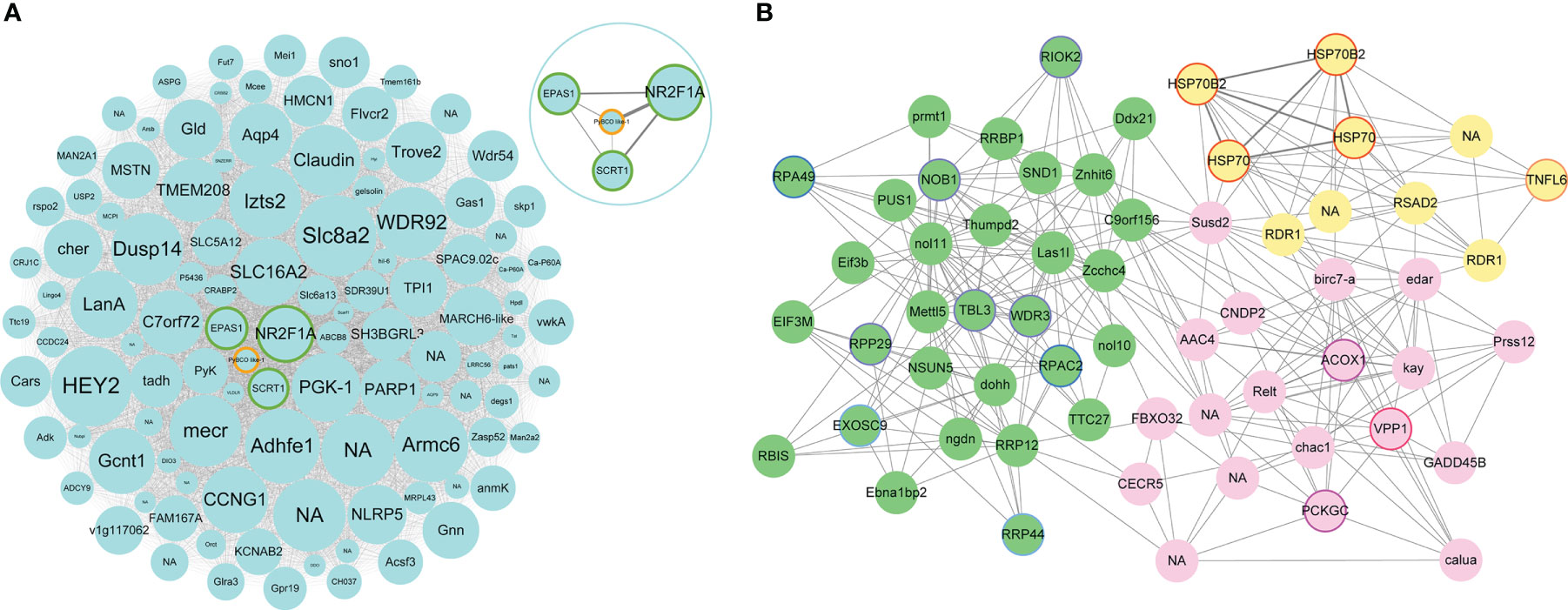
Figure 6 (A) Network visualization of hub genes with significantly different expression levels in MEturquoise. Each node represents a hub gene, which is labeled with gene name. NA represents the genes with unannotated information. The node size represents the intramodular connectivity, and larger nodes indicate higher connectivity. Green border represents hub genes encoding transcriptional factors. PyBCO-like 1 is shown in orange border. In the upper right circle is the network visualization of NR2F1A, EPAS1, SCRT1, and PyBCO-like 1 in MEturquoise. (B) Network visualization of hub genes with significantly different expression levels in up-regulated modules. Green nodes represent hub genes in MEgreen module. Purple, blue, and light blue border represents hub genes from “ribosome biogenesis”, “RNA polymerase” and “RNA degradation”, respectively. Lightyellow nodes represent hub genes in MElightyellow module. Red border represents hub genes such as HSP70s, HSP70B2s, and TNFL6. Pink nodes represent hub genes in MEpink module. Rose red and magenta border represents hub genes from “lysosome” and “PPAR signaling pathway”, respectively.
The up-regulated module MEgreen was enriched with genes involved in translation (“ribosome biogenesis”, “aminoacyl-tRNA biosynthesis”, and “nucleocytoplasmic transport”), transcription (“spliceosome” and “RNA polymerase”), folding, sorting and degradation (“protein export” and “RNA degradation”), etc (Figure 5B). Hub genes WDR3, RPP29, TBL3, RIOK2, and NOB1 from “ribosome biogenesis”, RPAC2 and RPA49 from “RNA polymerase”, RRP44 and EXOS9 from “RNA degradation” were highly expressed in OM scallops (Figure 6B; Table S3). Another up-regulated module MElightyellow is mainly enriched for the pathways of the immune system (“antigen processing and presentation” and “RIG-I-like receptor signaling pathway”), and cell growth and death (“apoptosis”) (Figure 5B). Hub genes such as HSP70 (heat shock 70 kDa protein), HSP70B2, and TNFL6 (tumor necrosis factor ligand superfamily member 6) were all up-regulated in OM scallops (Table S3). It is worth noting that two HSP70 genes and two HSP70B2 genes from “antigen processing and presentation” were included in the top 5 hub genes (Figure 6B), implying their potential regulatory roles in responding to carotenoid accumulation. MEpink is overrepresented with genes involved in transport and catabolism (“lysosome”, “apoptosis”, and “endocytosis”) and lipid metabolism (“PPAR signaling pathway”, “biosynthesis of unsaturated fatty acids”, and “fat digestion and absorption”) (Figure 5B). Hub genes including VPP1 (V-type proton ATPase 116 kDa subunit an isoform 1) from “lysosome”, ACOX1 (peroxisomal acyl-coenzyme A oxidase 1) and PCKGC (phosphoenolpyruvate carboxykinase) from “PPAR signaling pathway” were also highly expressed in OM scallop (Figure 6B; Table S3).
Carotenoids are responsible for muscle color and nutritional quality in aquatic animals, such as salmon, shrimp, and scallops. It has important economic value and social benefits to cultivate carotenoid-rich aquatic products. In recent years, research on the molecular mechanism of carotenoid accumulation in marine animals has become a focus. The color variant Yesso scallop “Haida golden scallop” used in our study provides a suitable animal model to investigate the carotenoid-related mechanism. In our previous study, we revealed the molecular mechanism underlying muscle coloration in OM scallop using multiple genomic analyses and functional verifications. Our results showed that carotenoid coloration in OM scallop muscle is recessively inherited as a Mendelian trait determined by a single gene PyBCO-like 1 (Li et al., 2019). On the basis of our previous research, we further found that some other traits such as growth, stress resistance, and amino acid content, also showed differences between WM and OM scallops. We believe that the changes of these traits are the results of carotenoid accumulation. Studying the effect of carotenoid accumulation on gene expression is helpful to understand the underlying mechanism of the changes of the traits. Therefore, the aim of this study is to investigate the response mechanisms of carotenoid accumulation in scallops by analyzing the DEGs and gene co-expression networks of WM scallops without carotenoid accumulation and OM scallops with carotenoid accumulation.
In this study, nearly 700 DEGs involved in various biological processes or pathways were identified between WM and OM scallops, implying the effects of carotenoid accumulation impacting on scallops are multifaceted. We first screened the DEGs possibly involved in carotenoid absorption, transport, and metabolism. Similar to our previous study, PyBCO-like 1 was the most significantly down-regulated gene in OM scallops among these genes (Li et al., 2019). Some other carotenoid accumulation-related genes, such as RXR, FABP7, LDLR, and LRP1B, also showed significant differences between WM and OM scallops. Given that the muscle coloration in scallop is induced by genomic variation in the upstream promoter region of PyBCO-like 1, thus the down-regulation of PyBCO-like 1 is the cause of carotenoid accumulation in muscle because of impaired carotenoid degradation. Other DEGs including some carotenoid accumulation-related genes, are not the cause of carotenoid accumulation, their expression changes are in response to carotenoid accumulation. PyBCO-like 1 was identified as the top 20% hub gene of MEturquoise, the module only enriched with down-regulated DEGs, and showed strong connectivity with NR2F1A, which was also suppressed in OM scallop. NR2F1A is the member of NR2F receptors, which are reported to bind to RAREs, and experiments in vitro have shown that they can regulate retinoic acid (RA) response genes (Tsai and Tsai, 1997). BCO is a critical cleavage enzyme for RA production, and could be regulated by the TFs (e.g. RAR/RXR) containing RAREs (Airanthi et al., 2015). Therefore, we speculate that NR2F1A may exert regulatory effects on PyBCO-like 1 by regulating RAREs-containing transcription factors. In addition, the down-regulated DEGs were significantly enriched to multiple metabolic pathways, which is consistent with the functional enrichment of MEturquoise, which is enriched with genes involved in the metabolism pathways of amino acid (e.g. “valine, leucine and isoleucine degradation”), lipid (e.g. “fatty acid degradation”), carbohydrate (e.g. “glyoxylate and dicarboxylate metabolism”), and cofactors and vitamins (e.g. “retinol metabolism”). Previous studies have shown that in carotenoid-accumulating mice constructed by silencing BCO1, a fatty liver was developed and the serum lipid levels were altered with elevated serum unesterified fatty acids (Hessel et al., 2007). Our previous research also found that the content of fatty acid, as well as amino acid and protein expression level of OM scallop are different from those of WM scallop (Zhang et al., 2014; Zhang et al., 2018). Combined with the analysis results in this study, it is suggested that the accumulation of carotenoids in scallop muscle caused by the down-regulation of PyBCO-like 1 may induce the changes in traits (e.g. amino acid and lipid content) by affecting a variety of metabolic pathways as described above.
The up-regulated DEGs were significantly enriched in transcription and translation-related processes or pathways, such as “preribosome”, “small nucleolar ribonucleoprotein complex”, “RNA polymerase activity”, “translation initiation factor binding” in GO analysis, and “ribosome biogenesis” in KEGG analysis. These functions are consistent with the enrichment results of the up-regulated module MEgreen in WGCNA. Ribosome biogenesis was the most significantly enriched pathway of MEgreen and the up-regulated DEGs WDR3, RPP29, TBL3, RIOK2, and NOB1 from this pathway were identified as hub genes, implying the important role of this pathway in response to carotenoid accumulation. Ribosome biogenesis is the process of making ribosomes, which function at the heart of the translation machinery. It is also a very tightly regulated activity and closely linked to other cellular processes, such as growth and division (Thomson et al., 2013). Previous studies have shown that marine carotenoid fucoxanthin modulated various cellular processes, including ribosome biogenesis (Guvatova et al., 2020). Similar results were observed in our study. Given carotenoid accumulation leads to the differential expression of a large number of genes, the up-regulation of the above ribosomal genes suggests that the efficiency of protein translation in ribosomes may be enhanced, thus promoting and accelerating protein synthesis in OM scallops.
Carotenoids are biologically active pigments that are well-known to enhance the defense and immunity of the vertebrate system. They also play important roles in invertebrate immunity (Tan et al., 2020). In our study, we found that the up-regulated DEGs were involved in pathways of the immune system, such as “antigen processing and presentation”. The up-regulated module MElightyellow is also enriched for the pathways of the immune system, such as “RIG-I-like receptor signaling pathway” and “antigen processing and presentation”. Many immune-related genes were identified as hub genes in this module, among which four (two HSP70s and two HSP70B2s) of the top 5 hub genes belonged to HSP70 family. HSP70 family is ubiquitous and highly conserved molecular chaperones with essential roles in various biochemical processes, such as protein folding, translocation, degradation, and regulation of cellular immune responses from extrinsic or intrinsic stimuli (Daugaard et al., 2007; Murphy, 2013). Four HSP70 genes were up-regulated in OM scallop and identified as top hub genes, suggesting that HSP70 gene may play an important regulatory role in the immune response to carotenoid accumulation.
Carotenoid accumulation also affects the expression of lipid metabolism-related genes. Carotenoids are a class of lipid-soluble molecules, and a close association between lipids and carotenoid accumulation has been reported in humans (Harrison, 2012). Our study showed that the up-reguated DEGs were enriched in lipid metabolism-related pathways, including PPAR signaling pathway. PPAR signaling pathway is a major regulator of lipid metabolism, adipogenesis, and energy homeostasis (Grygiel-Górniak, 2014). We found several members of this pathway, including ACOX1, FABP7, and PCKGC, were significantly up-regulated in OM scallop. ACOX1 is the first and rate-limiting enzyme in peroxisomal fatty acid β-oxidation of fatty acids (Reddy and Hashimoto, 2001), while FABP7 is involved in fatty acid uptake, transport, and metabolism (Owada, 2008). Studies have shown that carotenoids and their derivatives (e.g., retinoid) may exert their effect on gene expression via several similar transcriptional systems with lipid metabolism, such as RXR and PPAR (Sharoni et al., 2004). The up-regulation of these genes suggests that fatty acid accumulation and metabolism may be influenced by carotenoid accumulation. Besides, we further found ACOX1 was a key hub gene of up-regulated module MEpink, which module was also enriched for several lipid metabolism-related pathways. Previously, it was shown that ACOX1 mRNA levels were significantly elevated in BCO1 knock-out mouse (Hessel et al., 2007). Loss of ACOX1 has been shown to induce the expression of PPARs, the nuclear receptors serving as lipid sensors and regulators of lipid metabolism (Poulsen et al., 2012). Thus, it is speculated that a crosstalk between lipid homeostatic and carotenoid metabolic pathways may exist in OM scallop.
This study demonstrated the response mechanisms of carotenoid accumulation in scallops by analyzing the DEGs and gene co-expression networks of WM and OM scallops. Transcriptome analysis showed that the down-regulation of PyBCO-like 1 is the cause of carotenoid accumulation in muscle, while the expression changes of other DEGs were in response to carotenoid accumulation. Gene co-expression network analysis detected four carotenoid accumulation-related modules. The hub genes and pathways involved in translation and transcription, immune system, and lipid metabolism pathways were identified in up-regulated modules, while the down-regulated module is mainly enriched with genes involved in various metabolic pathways. These findings provide candidate genes and pathways for further studies on the effects of carotenoid accumulation on other traits in bivalves.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving animals were reviewed and approved by the Animal Care and Use 88 Committee of Ocean University of China.
XH and HW conceived and designed the study. HW, TL, and YZ performed the data analysis. TL, YZ, SL, and ML participated in the experiments. RY, SN, and JY contributed to sample collection. HW and TL wrote the original manuscript. HW and XH performed the review and editing of the manuscript. JH and ZB performed project administration. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (U2106231, 31630081), Shandong Provincial Natural Science Foundation (ZR2022QC032), and Sanya Yazhou Bay Science and Technology City (Grant No. SKJC-KJ-2019KY01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1158325/full#supplementary-material
Airanthi M., Widjajaadhi K., Lobo G. P., Golczak M., Von L. J. (2015). A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 24 (11), 3206. doi: 10.1093/hmg/ddv072
Anders S., Pyl P. T., Huber W. (2014). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 (2), 166–169. doi: 10.1093/bioinformatics/btu638
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. R. Stat. Society. Ser. B: Methodol. 57 (1), 289–300. doi: 10.2307/2346101
Daugaard M., Rohde M., Jäättelä M. (2007). The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett. 581 (19), 3702–3710. doi: 10.1016/j.febslet.2007.05.039
Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2012). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Grygiel-Górniak B. (2014). Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications - a review. Nutr. J. 13 (1), 17. doi: 10.1186/1475-2891-13-17
Guvatova Z., Dalina A., Marusich E., Pudova E., Snezhkina A., Krasnov G., et al. (2020). Protective effects of carotenoid fucoxanthin in fibroblasts cellular senescence. Mech. Ageing Dev. 189, 111260. doi: 10.1016/j.mad.2020.111260
Harrison E. H. (2012). Mechanisms involved in the intestinal absorption of dietary vitamin a and provitamin a carotenoids. Biochim. Biophys. Acta (BBA) - Mol. Cell Biol. Lipids 1821 (1), 70–77. doi: 10.1016/j.bbalip.2011.06.002
Hessel S., Eichinger A., Isken A., Amengual J., Hunzelmann S., Hoeller U., et al. (2007). CMO1 deficiency abolishes vitamin a production from β-carotene and alters lipid metabolism in mice. J. Biol. Chem. 282 (46), 33553–33561. doi: 10.1074/jbc.M706763200
Hu X., Bao Z., Hu J., Shao M., Zhang L., Bi K., et al. (2006). Cloning and characterization of tryptophan 2,3-dioxygenase gene of zhikong scallop Chlamys farreri (Jones and Preston 1904). Aquacult. Res. 37 (12), 1187–1194. doi: 10.1111/j.1365-2109.2006.01546.x
Huang D. W., Sherman B. T., Lempicki R. A. (2009). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. doi: 10.1093/nar/gkn923
Langfelder P., Horvath S. (2008). WGCNA: an r package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Li N., Hu J., Wang S., Cheng J., Hu X., Lu Z., et al. (2010). Isolation and identification of the main carotenoid pigment from the rare orange muscle of the yesso scallop. Food Chem. 118 (3), 616–619. doi: 10.1016/j.foodchem.2009.05.043
Li X., Wang S., Xun X., Zhang M., Wang S., Li H., et al. (2019). A carotenoid oxygenase is responsible for muscle coloration in scallop. Biochim. Biophys. Acta (BBA) - Mol. Cell Biol. Lipids 1864 (7), 966–975. doi: 10.1016/j.bbalip.2019.03.003
Liu H., Zheng H., Zhang H., Deng L., Liu W., Wang S., et al. (2015). A de novo transcriptome of the noble scallop, Chlamys nobilis, focusing on mining transcripts for carotenoid-based coloration. BMC Genomics 16 (1), 44. doi: 10.1186/s12864-015-1241-x
Maoka T. (2020). Carotenoids as natural functional pigments. J. Natural Medicines 74 (1), 1–16. doi: 10.1007/s11418-019-01364-x
Menotti A., Kromhout D., Blackburn H., Fidanza F., Buzina R., Nissinen A. (1999). Food intake patterns and 25-year mortality from coronary heart disease: cross-cultural correlations in the seven countries study. Eur. J. Epidemiol. 15 (6), 507–515. doi: 10.1023/A:1007529206050
Miao B., Wang X. (2013). “Provitamin a carotenoids and cancer prevention,” in Carotenoids and human health. Ed. Tanumihardjo S. A. (Totowa, NJ: Humana Press), 181–192.
Murphy M. E. (2013). The HSP70 family and cancer. Carcinogenesis 34 (6), 1181–1188. doi: 10.1093/carcin/bgt111
Owada Y. (2008). Fatty acid binding protein: localization and functional significance in the brain. Tohoku J. Exp. Med. 214 (3), 213–220. doi: 10.1620/tjem.214.213
Poulsen L., Siersbæk M., Mandrup S. (2012). PPARs: fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 23 (6), 631–639. doi: 10.1016/j.semcdb.2012.01.003
Reddy J. K., Hashimoto T. (2001). Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230. doi: 10.1146/annurev.nutr.21.1.193
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Sandmann G. (2015). “Carotenoids of biotechnological importance,” in Biotechnology of isoprenoids. Eds. Schrader J., Bohlmann J. (Cham: Springer International Publishing), 449–467.
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Sharoni Y., Danilenko M., Dubi N., Ben-Dor A., Levy J. (2004). Carotenoids and transcription. Arch. Biochem. Biophysics 430 (1), 89–96. doi: 10.1016/j.abb.2004.03.009
Song J., Wang C. (2019). Transcriptomic and proteomic analyses of genetic factors influencing adductor muscle coloration in QN orange scallops. BMC Genomics 20 (1), 363. doi: 10.1186/s12864-019-5717-y
Tan K., Zhang H., Lim L.-S., Ma H., Li S., Zheng H. (2020). Roles of carotenoids in invertebrate immunology. Front. Immunol. 10. doi: 10.3389/fimmu.2019.03041
Thomson E., Ferreira-Cerca S., Hurt E. (2013). Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 126 (Pt 21), 4815–4821. doi: 10.1242/jcs.111948
Tsai S. Y., Tsai M.-J. (1997). Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age*. Endocrine Rev. 18 (2), 229–240. doi: 10.1210/edrv.18.2.0294
Vershinin A. (1996). Carotenoids in mollusca: approaching the functions. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 113 (1), 63–71. doi: 10.1016/0305-0491(96)00104-6
Wang S., Zhang J., Jiao W., Li J., Xun X., Sun Y., et al. (2017). Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1 (5), 120. doi: 10.1038/s41559-017-0120
Zhang L., Li X., Li N., Wang S., Li X., Sun G., et al. (2018). Corralation analysis of carotenoid concentration and the growth traits of 'Haida golden scallop'. Periodical Ocean Univ. China/Zhongguo Haiyang Daxue Xuebao 48 (03), 41–47. doi: 10.16441/j.cnki.hdxb.20170188
Keywords: Patinopecten yessoensis, carotenoid accumulation, gene co-expression network analysis, PyBCO-like 1, molecular mechanism
Citation: Li T, Zhang Y, Liu S, Li M, Yao R, Niu S, Yuan J, Wang H, Hu J, Bao Z and Hu X (2023) Transcriptome and network analyses reveal key pathways and genes involved in response to carotenoid deposition in scallop muscle. Front. Mar. Sci. 10:1158325. doi: 10.3389/fmars.2023.1158325
Received: 07 February 2023; Accepted: 08 May 2023;
Published: 22 May 2023.
Edited by:
Antonella Leone, National Research Council (CNR), ItalyReviewed by:
Nick Wade, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2023 Li, Zhang, Liu, Li, Yao, Niu, Yuan, Wang, Hu, Bao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Hu, aHhsNzA3QG91Yy5lZHUuY24=; Huizhen Wang, d2FuZ2h1aXpoZW5Ab3VjLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.