
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 08 August 2023
Sec. Marine Molecular Biology and Ecology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1152647
This article is part of the Research TopicMarine Omics in a Changing Ocean: Modelling Molecular Pathways and Networks to Understand Species Acclimation and AdaptationView all 10 articles
 Yaamini R. Venkataraman1‡
Yaamini R. Venkataraman1‡ Amanda Shore2‡
Amanda Shore2‡ Sukanya Dayal3†
Sukanya Dayal3† James Sanghyun Lee4†
James Sanghyun Lee4† Mahsa Alidoost Salimi5
Mahsa Alidoost Salimi5 Grace Crandall6
Grace Crandall6 Malina M. Loeher7
Malina M. Loeher7 Mark Stoops8
Mark Stoops8 Megan Swanger6
Megan Swanger6 Morgan E. Eisenlord9
Morgan E. Eisenlord9 Kathryn L. Van Alstyne10
Kathryn L. Van Alstyne10 Mark D. Fast11
Mark D. Fast11 Colleen A. Burge3,12*†§
Colleen A. Burge3,12*†§ Maya L. Groner11,13§
Maya L. Groner11,13§Introduction: Seagrass meadows serve as an integral component of coastal ecosystems but are declining rapidly due to numerous anthropogenic stressors including climate change. Eelgrass wasting disease, caused by opportunistic Labyrinthula spp., is an increasing concern with rising seawater temperature. To better understand the host-pathogen interaction, we paired whole organism physiological assays with dual transcriptomic analysis of the infected host and parasite.
Methods: Eelgrass (Zostera marina) shoots were placed in one of two temperature treatments, 11° C or 18° C, acclimated for 10 days, and exposed to a waterborne inoculation containing infectious Labyrinthula zosterae (Lz) or sterile seawater. At two- and five-days post-exposure, pathogen load, visible disease signs, whole leaf phenolic content, and both host- and pathogen- transcriptomes were characterized.
Results: Two days after exposure, more than 90% of plants had visible lesions and Lz DNA was detectable in 100% percent of sampled plants in the Lz exposed treatment. Concentrations of total phenolic compounds were lower after 5 days of combined exposure to warmer temperatures and Lz, but were unaffected in other treatments. Concentrations of condensed tannins were not affected by Lz or temperature, and did not change over time. Analysis of the eelgrass transcriptome revealed 540 differentially expressed genes in response to Lz exposure, but not temperature. Lz-exposed plants had gene expression patterns consistent with increased defense responses through altered regulation of phytohormone biosynthesis, stress response, and immune function pathways. Analysis of the pathogen transcriptome revealed up-regulation of genes potentially involved in breakdown of host defense, chemotaxis, phagocytosis, and metabolism.
Discussion: The lack of a significant temperature signal was unexpected but suggests a more pronounced physiological response to Lz infection as compared to temperature. Pre-acclimation of eelgrass plants to the temperature treatments may have contributed to the limited physiological responses to temperature. Collectively, these data characterize a widespread physiological response to pathogen attack and demonstrate the value of paired transcriptomics to understand infections in a host-pathogen system.
Despite occupying less than 0.1% of the ocean floor (primarily in estuaries and embayments), seagrasses provide numerous ecosystem services, including shoreline stabilization and protection from erosion, nutrient cycling, sediment retention, reduction of pathogens in the water column, and provision of critical nursery habitat for numerous invertebrate and fish species, many of which are of economic importance to the seafood industry (Nordlund et al., 2016; Lamb et al., 2017). Seagrass meadows are threatened by a wide range of global and local stressors associated with anthropogenic activity such as coastal development, increasing sea surface temperature, rising sea levels, sediment and nutrient runoff and disease (Orth et al., 2006; Sullivan et al., 2013). Consequently, over the past century, global seagrass beds have experienced approximately 19% loss in cover (Dunic et al., 2021).
Zostera marina, the most widely distributed seagrass species in the northern hemisphere (Short et al., 2007), is susceptible to infection with the protist Labyrinthula zosterae (Lz), which causes eelgrass wasting disease (EWD). While chronic Lz infections are nearly ubiquitous in distribution, acute epidemics of wasting disease can cause declines in eelgrass populations, most notably in the 1930s when Atlantic eelgrass populations were nearly eliminated along the coasts of North America and Europe (Short et al., 1987). Lz is a pathogenic Labyrinthulomycete and can be transmitted through direct contact between healthy and infected blades within an eelgrass bed (Muehlstein, 1992) or through the water column (Groner et al., 2014; Groner et al., 2018). Upon infection, Lz targets chloroplasts of eelgrass leaves, compromising photosynthesis, and ultimately causing necrosis of eelgrass leaves, and in some cases, entire plants (Muehlstein, 1992). Non-lethal impacts of EWD include reductions in growth and below-ground storage of carbohydrates, which may impact seasonal resilience of eelgrass meadows (Graham et al., 2021). Strains of Lz vary in virulence, raising questions about mechanisms of virulence employed by this pathogen and potential costs associated with those mechanisms (Martin et al., 2016).
Eelgrass has a robust defense system which includes proteins and surface-associated metabolites like fatty acids and phenolics, p-coumaric acid, rosmarinic acid, and zosteric acid (Papazian et al., 2019). These compounds play a wide-range of roles including but not limited to signaling molecules, antioxidant activity, free radical scavenging activity, and regulators of auxin transport (Ma et al., 2016). Phenolic compounds which are produced in response to stress and pathogen presence can be also considered as defense-related secondary metabolites. Indeed, some phenolic compounds have been shown to inhibit growth of Lz in vitro (Vergeer and Develi, 1997) and concentrations of total phenols (% dry mass) are increased in diseased plants in the field (Groner et al., 2016). In terrestrial plants, phytohormones including abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA) play significant roles in regulating defense responses against a great number of biotic and abiotic factors (Thaler et al., 2004; Tamaoki et al., 2013), however these hormones have not been well explored in marine plants such as eelgrass.
As with many marine diseases, EWD is sensitive to temperature (Tracy et al., 2019). Both EWD prevalence and severity are correlated with sea surface temperatures in the Salish Sea (WA, USA), and in the Isles of Scilly (UK), (Bull et al., 2012; Groner et al., 2021). Increased temperatures enhance the growth-reducing effects of EWD on eelgrass (Bull et al., 2012). These changes are likely driven by both the host and pathogen. Lz is also sensitive to temperature, with faster in vitro growth documented at 18°C compared to 11°C for a strain isolated from the Salish Sea (WA, USA) (Dawkins et al., 2018); similarly, increased disease severity was noted in eelgrass exposed to Lz and held at 18˚C compared to 11˚C (Agnew et al., 2022). Recent studies have correlated heatwaves to substantial seagrass die-offs (Strydom et al., 2020; Groner et al., 2021), reduced restoration success (Aoki et al., 2020), and changes in fatty acid composition (Beca-Carretero et al., 2018), showing that seagrasses are sensitive to warming conditions.
To better understand the factors that contribute to Lz infection and provide insight on mechanisms of virulence and host defense, we characterized the production of defensive compounds in Z. marina and the transcriptional responses of both Z. marina and Lz following a controlled Lz challenge conducted at ambient and warm seawater temperatures. We hypothesize that both increased temperature and Lz infection will alter expression of genes related to immune function and stress response in Z. marina. More specifically, we hypothesize that phenolic and hormonal gene pathways will be enriched in response to presence of Lz, but we expected these responses may be reduced at warmer temperatures that could be stressful to Z. marina. Finally, we hypothesize that Lz will exhibit a change in gene expression in response to temperature, reflecting the association between EWD and warmer seawater temperatures found in nature (Brakel et al., 2019; Groner et al., 2021).
Similarly sized eelgrass ramets were collected from False Bay, San Juan Island, WA (48.550° N, 123.008° W) on 29 July 2015. Epiphytes, grazers and damaged leaves were removed from ramets and they were held in flow-through 14°C, 0.2 μm filter-sterilized seawater at Friday Harbor Labs, WA, USA.
Eelgrass ramets were placed in one of two temperature treatments (target temperatures 11° or 18°C, actual temperatures ranged ± 1°C from target) on 3 Aug 2015 and allowed to acclimate to these treatments for 10 days. These temperatures represent typical summer sea surface temperatures found locally along coastal eelgrass beds at high and low tides, respectively. A constant exposure to 18°C would be unusually warm in this region. Each treatment was replicated eight times for a total of 16 experimental units. Each replicate unit contained four ramets. Experimental units were 4-L containers with 3500 mL of 0.2 μm filter-sterilized seawater. Experimental units were split into groups of eight that were clustered in a common cooler. Each cooler received flow-thru 0.2 µm filtered and UV-irradiated seawater. Seawater temperatures were continuously monitored and adjusted using Honeywell UDA2182 pH controller and Honeywell Durafet III electrodes. After flowing into the cooler, water was then piped into the 8 experimental units. Water temperatures were maintained by circulating it through chilled water set 1°C below target temperatures and, if necessary, heating it with aquatic heaters placed in the coolers (i.e. O’Donnell et al., 2013). Two temperature loggers (iButtons, Whitewater, WI) set to record every 30 min were placed in each cooler for independent confirmation of temperature. Full-spectrum lights were kept to a 12: 12 hr light: dark schedule with a mean light output of 161 ± 3 (mean ± SE) µmol/m2/sec PAR below the water surface. To reduce algal blooms, which could clog plumbing and block light, tanks were treated daily with 0.67 ppm (final concentration) of Germanium dioxide (GeO2; Markham and Hagmeier, 1982).
A 36-hr incubation with the pathogen inoculant was conducted after 10 days of temperature acclimation (Figure 1). Immediately prior to inoculation, all ramets were photographed, weighed, labelled with uniquely colored zip-ties, and pin-pricked at the sheath for later measurements of growth. All ramets within a replicate were placed in Whirlpaks with 98 mL of sterile seawater. Control treatments were inoculated with 2 mL of sterile seawater. Lz treatments were inoculated with 2 mL of inoculant for a final concentration of 2.5 x105 cells per mL (additional details in Supplementary Material). After adding the inoculate, the Whirlpaks were closed and floated in treatment containers to maintain target temperatures. 36 hours later, ramets were removed from Whirlpaks and placed back into treatment containers. The Lz strain used in this experiment, 8.16.D, has been used in several previous experiments, (e.g., Groner et al., 2014; Groner et al., 2018) and was isolated in 2011 from non-flowering adult Zostera marina shoots that were collected at Picnic Cove, Shaw Island, 48° 33.942’ N, 122° 55.448’ W).
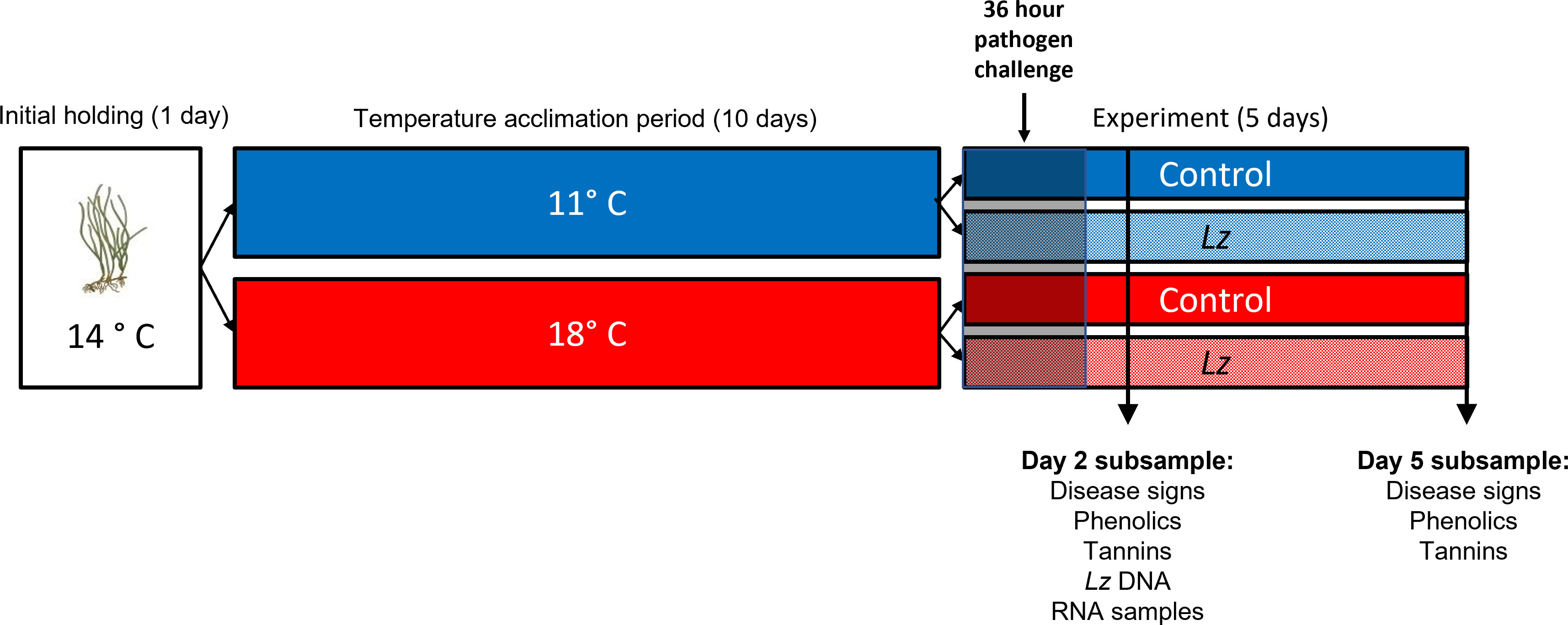
Figure 1 Timeline of experimental treatments and sampling. After collection and cleaning of plants to remove grazers, epiphytes and diseased tissue, eelgrass ramets were held at 14°C for 1d and then transferred in groups of 4 ramets to 11°C or 18°C for a 10d acclimation period. Each group of 4 ramets was replicated 8 times during the acclimation period. After that, the groups were equally divided into control and Lz – exposed treatments at each of the two temperatures. Lz or sham exposures occurred for 36 hr. Subsampling of plants occurred at days 2 and 5 post-Lz exposure.
Forty-eight hours after inoculation (experiment day 13), a random selection of half of the individuals (48 total) from each experimental unit were removed for sampling (Figure 1). Each of those individuals was photographed and weighed. Then the roots and shoots were frozen in liquid nitrogen for transcriptomic sampling. The oldest leaf was split lengthwise into two pieces. One half was frozen in liquid nitrogen for transcriptomic analysis, and the other was stored in 70% molecular grade ethanol at room temperature for subsequent measurement of Lz load using quantitative PCR (qPCR) as described in Groner et al. (2018). The second oldest leaf was frozen in liquid nitrogen for measurement of total phenolic compounds and condensed tannins. Five days after inoculation, the experiment was ended because the plants had severe disease signs. At this point, the remaining ramets were photographed and sampled (as described above) for measurement of visual disease signs (e.g., lesions with a distinct black border, sensu Groner et al., 2014) as well as phenol and tannin concentrations (Figure 1).
Total phenolic compounds were measured in 96 well microplates using the methods described in Groner et al. (2018). Briefly, sampled eelgrass leaves were frozen (-20°C) and sent on dry ice to the Shannon Point Marine Center for analysis. Frozen tissues were lyophilized and ground to a fine powder in an SPEX mixer/mill (SPEX, Metuchen, NJ). Ground tissue (9-10 mg) was extracted overnight in 1 mL of HPLC-grade methanol, then diluted in ANSI Type I water (19 parts water to 1 part extract). Forty µL of 40% Folin-Ciocalteu’s Reagent (Sigma F9252) was added to each 100 µL aliquot (n=3 per sample) of each of the diluted extracts, mixed for 5 min prior to the addition of 100 µL of 2N sodium carbonate. Samples were shaken for another 30 min in a 50°C chamber and then the absorbance of each cell was read at 765 nm. Concentrations were standardized using native standards from Z. marina collected from Ship Harbor WA, using caffeic acid as a secondary standard (Groner et al., 2016). The remaining methanolic extracts were used for measurements of condensed tannins, which were conducted with a sulfuric acid method (Bate-Smith and Rasper, 1969; Nitao et al., 2001) that was modified for use in a microplate reader. Condensed tannins (proanthocyanidins) were cleaved by the addition of 43% sulfuric acid in methanol (250 µL/well containing 100 µL of a methanol extract) for 30 min at 50°C for. Tannin cleavage produces red-colored chromophores (anthocyanins), which were then quantified spectrophotometrically at 550 nm using a Biotek Synergy multiplate reader. Cyanidin (Cayman Chemical, purity >98%) was used as a standard. All assessments were run in triplicate.
Samples (mean dry weight of leaf material = 7 mg) were preserved in 70% ethanol until DNA extractions were performed using a Qiagen DNeasy Plant Mini Kit following the manufacturer’s instructions. Prior to DNA extractions, samples were removed from ethanol, patted dry, weighed and then transferred into a clean 1.5ml micro-centrifuge tube with one 3mm tungsten carbide bead (Qiagen) per tube. Samples were then placed on dry ice for 10-15 minutes and lysed using a Qiagen TissueLyser at a speed of 25 Hz for 5 minutes.
Presence of Lz DNA was detected and quantified using qPCR detection using primers and probe targeting the Lz ITS region (Bockelmann et al., 2013); Laby_ITS_Taq_f (5’- TTG AAC GTA ACA TTC GAC TTT CGT- 3’) and the reverse primer Laby_ITS_Taq_r (5’ –ACG CAT GAA GCG GTC TTC TT -3’) were used, in addition to the TaqMan probe Laby_ITS_probe (5’ FAM- TGG ACG AGT GTG TTT TG –MGB-NFQ 3’). qPCR reaction conditions and the standard curve (serial dilution between 1.37-1.37*10^4 cells) were as described by Groner et al., 2018 on run on the Applied Biosystems 7500 Fast Real-Time PCR System. For each qPCR plate, we aimed for R2 = 0.999 and an efficiency of 90-110%; the mean reaction efficiency was 105.5% +/- 1.5% (cross three total plates). We used a cut-off of 1.37 cells as our limit of detection (amplification of 1.37 cells was achieved in all reactions in each cell curve).
Independent and interactive effects of temperature and Lz exposure on concentrations of phenols, concentrations of tannins, presence of suspected disease signs and log10 transformed Lz DNA concentrations in leaf tissues were quantified for the time points when these measurements were taken (R v. 4.1.1). Linear regression was used to quantify treatment effects on total phenolic compounds, condensed tannins, and Lz DNA concentrations, while logistic regression was used to quantify treatment effects on the presence of disease signs (package lme4, Bates et al., 2015). In all models, the 4L container replicate was included as a random effect.
Total RNA from four samples per treatment was extracted following the Qiagen RNeasy plant kit protocol; samples were chosen based on both quality of RNA and qPCR results. DNA was removed from extracted RNA using the Turbo DNA-free treatment according to the manufacturer’s instructions (Ambion Inc., The Life Technologies Corporation™, Grand Island, NY). Removal of DNA was confirmed by using RNA (1 μl) as template in a qPCR targeting 18s ribosomal DNA as previously described (Burge and Friedman, 2012). RNA concentrations were quantified using the NanoDrop® ND-1000 (NanoDrop Technologies, Wilmington, DE). Samples were shipped to McGill University and Genome Quebec for library preparation and sequencing. RNA quality was assessed prior to library preparation using an Agilent Bioanalyzer. The Illumina TruSeq stranded cDNA kit was used for library preparation and barcoding; quality assessment of libraries indicated that one sample was not of high enough quality for sequencing. The remaining samples were distributed across two sequencing lanes on an Illumina HiSeq 4000 platform (2x150 bp paired end reads).
Prior to de novo assembly, reads were quality screened (FastQC Version 0.11.4, Babraham Bioinformatics), and sequences were screened for remaining Illumina adapter sequences (Trimmomatic, Bolger et al., 2014). De novo assembly was carried out using reads from all fifteen samples using Trinity (Grabherr et al., 2011; Haas et al., 2013, Trinity RNASeq Github). Following transcriptome assembly, reads for each sample were matched back to gene isoforms.
A multi-step annotation process was used to annotate the transcriptome in order to separate isoform consensus sequences (or contigs) matching to Z. marina and those non-matching contigs (“non-host”). First, a database of annotations was created from the Z. marina genome (GCA_001185155.1) using a BLASTx search of the UniProtKB/Swiss-Prot database. The BLASTx algorithm (Altschul et al., 1990) was used to search the Z. marina database with a threshold E-value of 0.00001; each contig matching as Z. marina was filtered out. Similarly, the blastx algorithm was used to search the Swiss-Prot database with a threshold E-value of 0.00001 to form the non-Zostera annotation for subsequent analysis. Finally, to determine which non-host contigs have significant similarity to Labyrinthula and other closely related ‘slime-mold’ species, the blast algorithm was used to match non-host contigs against the recently added Labyrinthula sp. Ha genome (NCBI Accession # GCA_015227615.1) and also against the genomes of two other, better annotated genomes, a thraustochytrid, Hondaea fermentalgiana (NCBI Accession # GCA_002897355.1) and the amoeba model organism, Dictyostelium discoideum (NCBI Accession # GCF_000004695.1).
Gene ontology (GO) information was used to annotate both Z. marina and non-Zostera transcriptome data separately. Gene ontology IDs and associated GO Slim terms for biological processes, cellular components, and molecular function categories were downloaded from the UniProtKB/Swiss-Prot database. Swiss-Prot identifiers from BLASTx output were joined to gene ontology terms. Since assembly and annotation were conducted at the isoform level, transcriptome data was streamlined to conduct differential expression analysis on genes. Annotated sequences were sorted by contig name and e-value; contig names included a contig, gene, and isoform identifier. The isoform entry with the lowest e-value was retained for each gene. The final annotated dataset for both Z. marina and non-Zostera transcriptomes included only one entry per gene. Annotated genes were merged with count data to prepare for differential expression analysis. The data is deposited in the NCBI database in the SRA as Bioproject (PRJNA990835).
Statistical differences in count data were conducted for Z. marina and non-Zostera annotated genes using EdgeR (Robinson et al., 2010; McCarthy et al., 2012; Chen et al., 2016). Briefly, we utilized EdgeR to filter and normalize the count data based on the library size and examine samples for outliers using MDS plots. Next, a multi-factorial model including estimated dispersion (i.e. estimate of biological variation) was used with multiple contrasts to calculate fold change, log CPM (counts per million), and raw and Benjamini-Hochberg adjusted p-values to correct for False Discovery Rate.
We tested for statistical differences using several contrasts followed by the primary questions of interest, respectively examining the relationships of the pathogen-by-temperature interaction, temperature alone, and pathogen exposure alone:
1) What genes are differentially expressed between Lz-exposed shoots at elevated temperatures? At ambient temperatures?
2) What genes are differentially expressed between elevated and ambient temperatures, irrespective of Lz-exposure?
3) What genes are differentially expressed between Lz-exposed and control shoots, irrespective of temperature?
Very few genes were differentially expressed in questions 1 and 2 (25 genes and 0 genes, respectively). Thus, for the remainder of the analysis we focused on the effect of Lz exposure alone. Differentially expressed genes were pooled into host and pathogen genes. Genes were considered differentially expressed based on FDR adjusted p-values from edgeR (p < 0.05).
To identify groups of Z. marina genes with similar expression patterns (eigengenes) and associate these patterns with experimental conditions, a Weighted Gene Co-expression Network Analysis (WGCNA) was conducted using the WGCNA package in R (version 1.69; Langfelder and Horvath, 2008). The WGCNA was only conducted for Z. marina genes, as transcriptome sequencing produced a high magnitude of information for Z. marina that was difficult to parse without additional analysis. After identifying significant modules, overrepresented biological process and molecular function GOterms were identified using the GO-MWU enrichment method (Wright et al., 2015). More details on WGCNA and GO-MWU analyses are available in the Supplemental Material.
The inoculations were successful in transmitting Lz to the exposed plants. Two days after exposure, Lz concentrations in eelgrass tissue were 1.1 orders of magnitude greater in the 18°C treatment as compared to the 11°C treatment (with concentrations of 4.2 ± 4.3 x104 and 3.7 ± 3.6 x103 (mean ± SE) Lz cells/mg of eelgrass tissue, respectively) (Figure 2). These pathogen concentrations were significantly higher than the control treatments, where no Lz was detected using qPCR (t value = 12.3, p < 0.0001) No other factors affected Lz abundance in our linear regression (all p > 0.05).
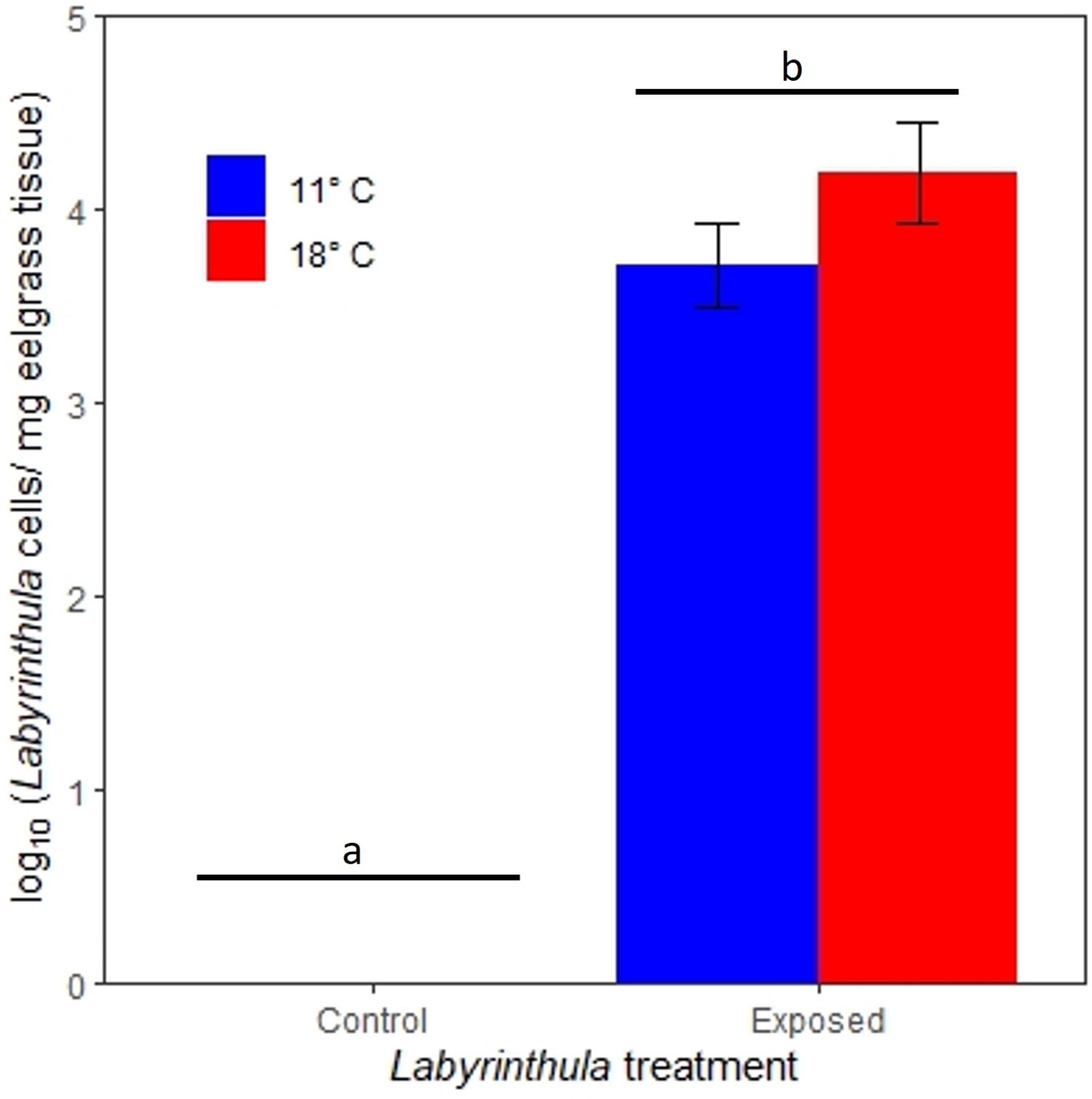
Figure 2 Lz cell equivalents (per mg eelgrass tissue) from qPCR analysis of eelgrass leaf tissue 2 days post-inoculation. Bars indicate standard errors. Letters and bars indicate significantly different treatments.
Visible disease signs were recorded in all treatments (Figure 3). Pathogen exposure was a significant predictor of disease signs on day 2 and a marginally significant predictor of disease signs on day 5. On day 2, the disease signs were recorded in 93.7 ± 8.8% (mean +/- SE) of the exposed plants and 28.8 ± 19.7% of the control plants (t = 2.1, p = 0.04). Neither temperature nor the temperature by exposure interaction were predictors of visual disease signs. These values increased on day 5 where disease signs were recorded in 100 ± 0% (mean +/- SE) of the exposed plants and 37.5 ± 17.7% of the control plants (t = 1.99, p = 0.07).
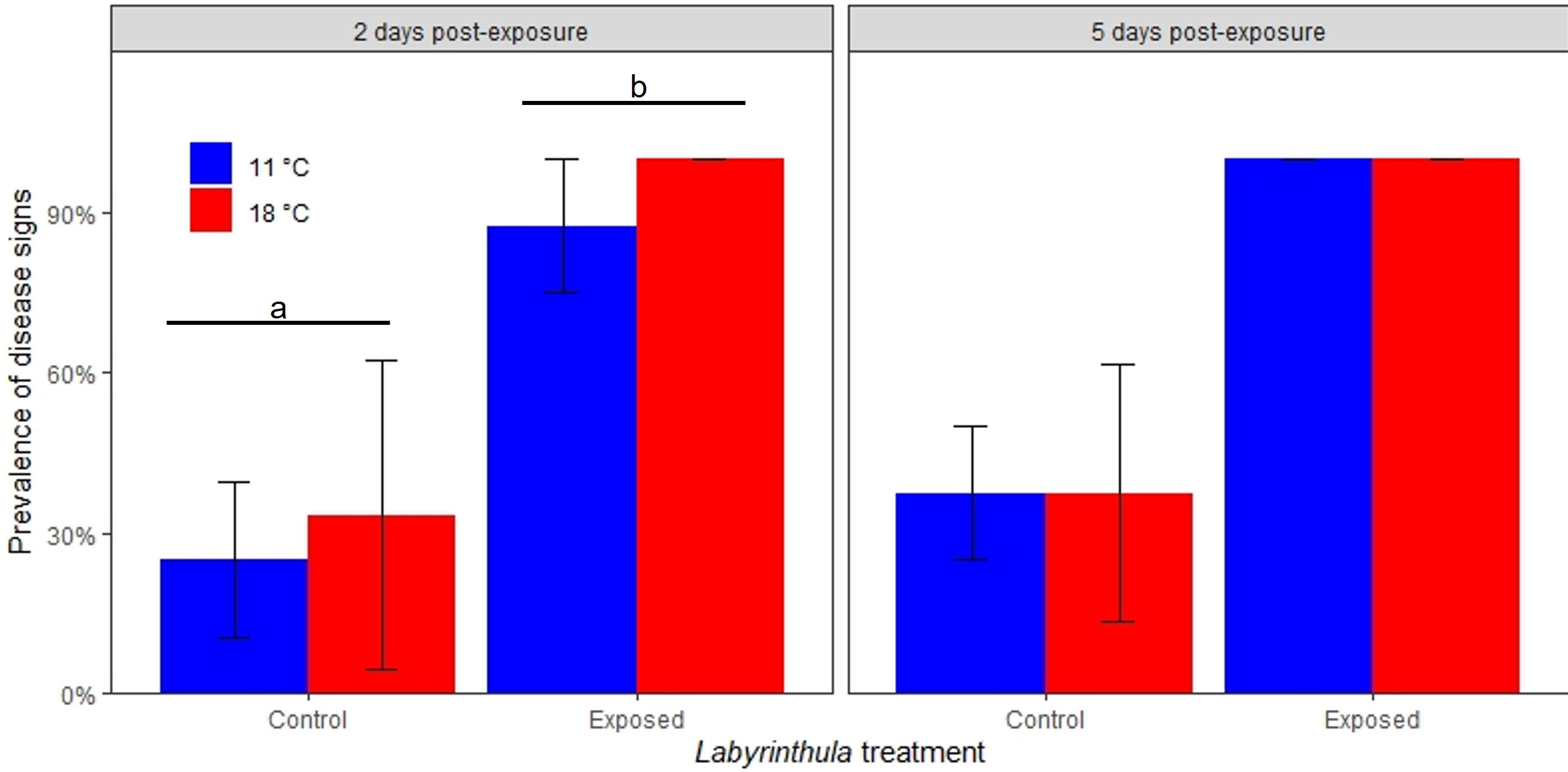
Figure 3 Prevalence of observed disease signs on eelgrass blades 2 and 5 days post-inoculation. Error bars indicate ± 1 standard error of the replicate mean. Letters and bars indicate significantly different treatments.
The concentrations of total phenolic compounds and condensed tannins were not impacted by temperature or Lz treatments on day 2 (Figure 4). By day 5, total phenolic compounds were significantly lower in the warmer exposed treatment, where they were 0.70 ± 0.17% of dry mass as opposed to ranging between 1.15 and 1.39% of dry mass in the other three treatments (t value = -3.298, p < 0.001 for the temperature by exposed interaction). There was no significant impact of temperature or Lz exposure on the concentration of condensed tannins in eelgrass leaves on day 5.
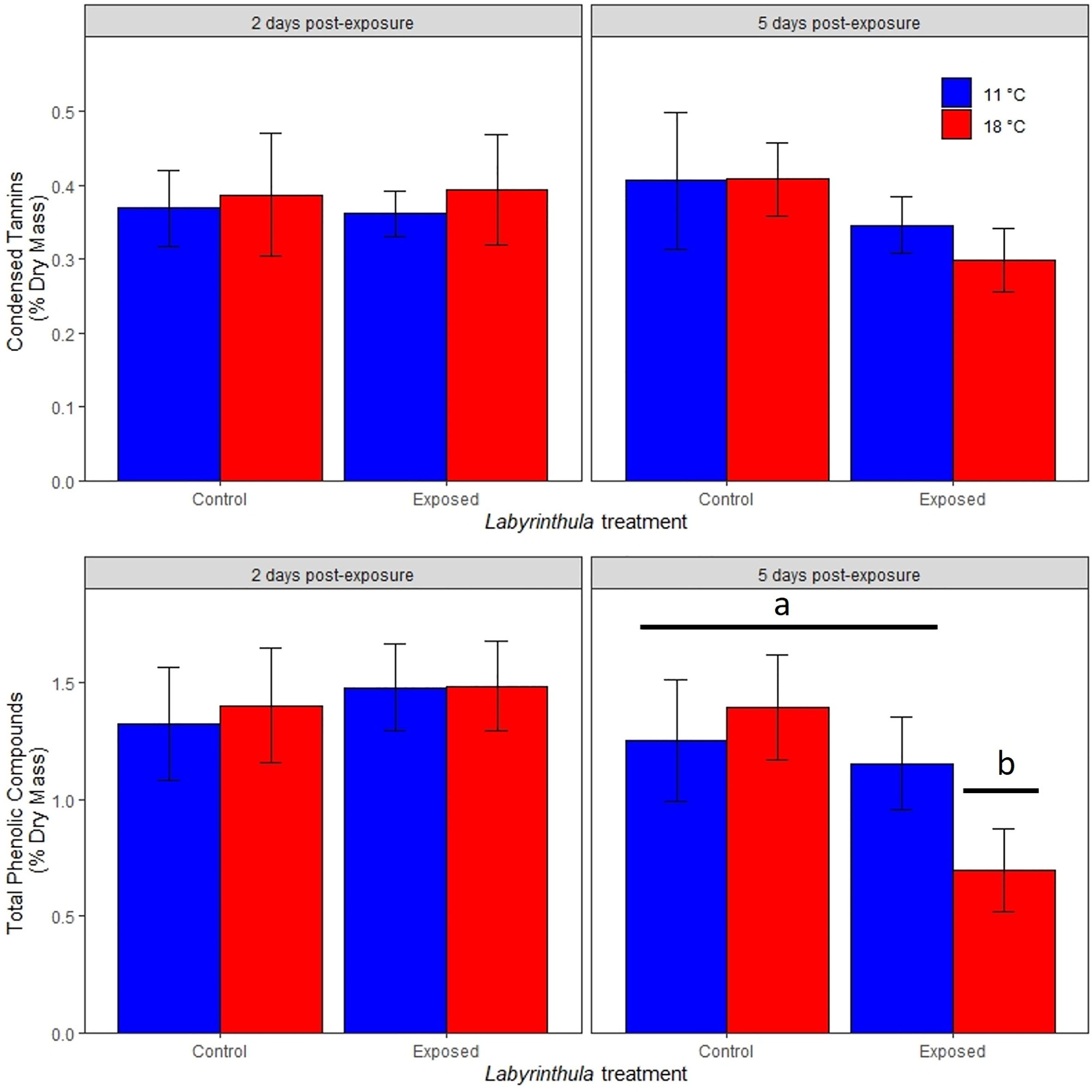
Figure 4 Effects of temperature and Lz treatment on condensed tannins and total phenolic compounds (as a percentage of eelgrass dry mass) 2 and 5 days post-inoculation. Data are means (± 1 SE). Letters and bars indicate significantly different treatments.
Trinity de novo assembly yielded 1,065,804 isoforms (767,296 potential genes) with an average read length of 654.71 bp (median of 349) and an N50 value of 1075. BLAST searches of the de novo assembly yielded 281,600 contigs matching the Z. marina genome. 784,204 contigs did not match the Z. marina genome and thus were categorized as ‘non-host’ contigs.
Of the 3,096 contigs identified as host genes and annotated, 540 genes were differentially expressed, with 338 genes with significantly higher expression levels in exposed blades and 202 genes with lower expression levels (Supplementary Table 1). The WGCNA identified thirteen module eigengenes comprised of genes with similar expression patterns for 3,096 Z. marina contigs (Figure 5A; Supplementary Table 2). Of these eigengenes, four - named “brown”, “yellow”, “blue”, and “magenta” - were significantly associated with Lz exposure (Figure 5B). These modules, labeled as color categories, are described in detail below.
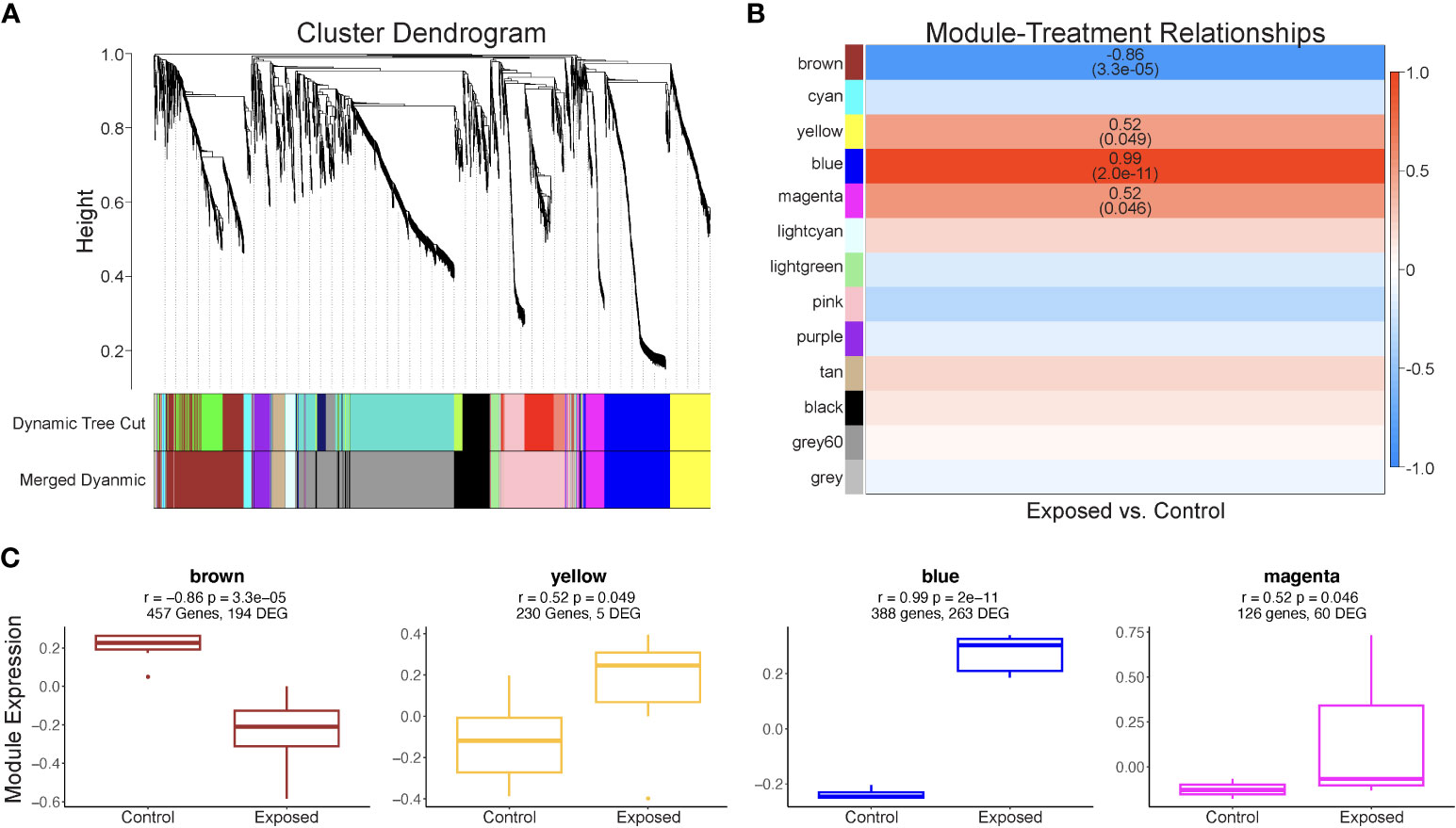
Figure 5 Results of Z. marina WCGNA analysis. (A) Cluster dendrogram of 3,096 genes used in WGCNA analysis. Modules were identified (Dynamic Tree Cut), then merged based on a cut height of 0.34 (Merged dynamic). All modules contain at least 30 genes. (B) Significant correlations between module eigengenes (rows) and Lz infection status (column). The R2 value is provided for each significant correlation, with P-value in parentheses. Positive correlations are represented by red boxes, while negative correlations are represented by blue boxes, with the intensity of the color increasing with correlation strength. (C) Expression of module eigengenes significantly correlated Lz exposure for exposed and control eelgrass.
The module eigengene “brown” was negatively correlated with Lz exposure, indicating lower module expression when eelgrass was exposed to Lz. (r2 = -0.86, p= 3.3 x 10-5) (Figure 5B) The module contained 194 differentially expressed genes (Figure 5C; Supplementary Table 2). Seven biological processes were significantly enriched, including response to fungus (GO:0009620), response to salt stress (GO:0009651), transposition (GO:0032197;GO:0032196), and cell wall organization or biogenesis (GO:0071555;GO:0045229;GO:0071554) (Table 1). Genes were also involved in responses to abscisic acid, jasmonic acid, and wounding (Supplementary Table 2). Additionally, 17 molecular functions were enriched in this module, including transferase activity, transferring glygosyl groups (GO:0016757), xenobiotic transmembrane transporter activity (GO:0042910), and transcription regulator activity (GO:0003700;GO:0140110) (Table 1).
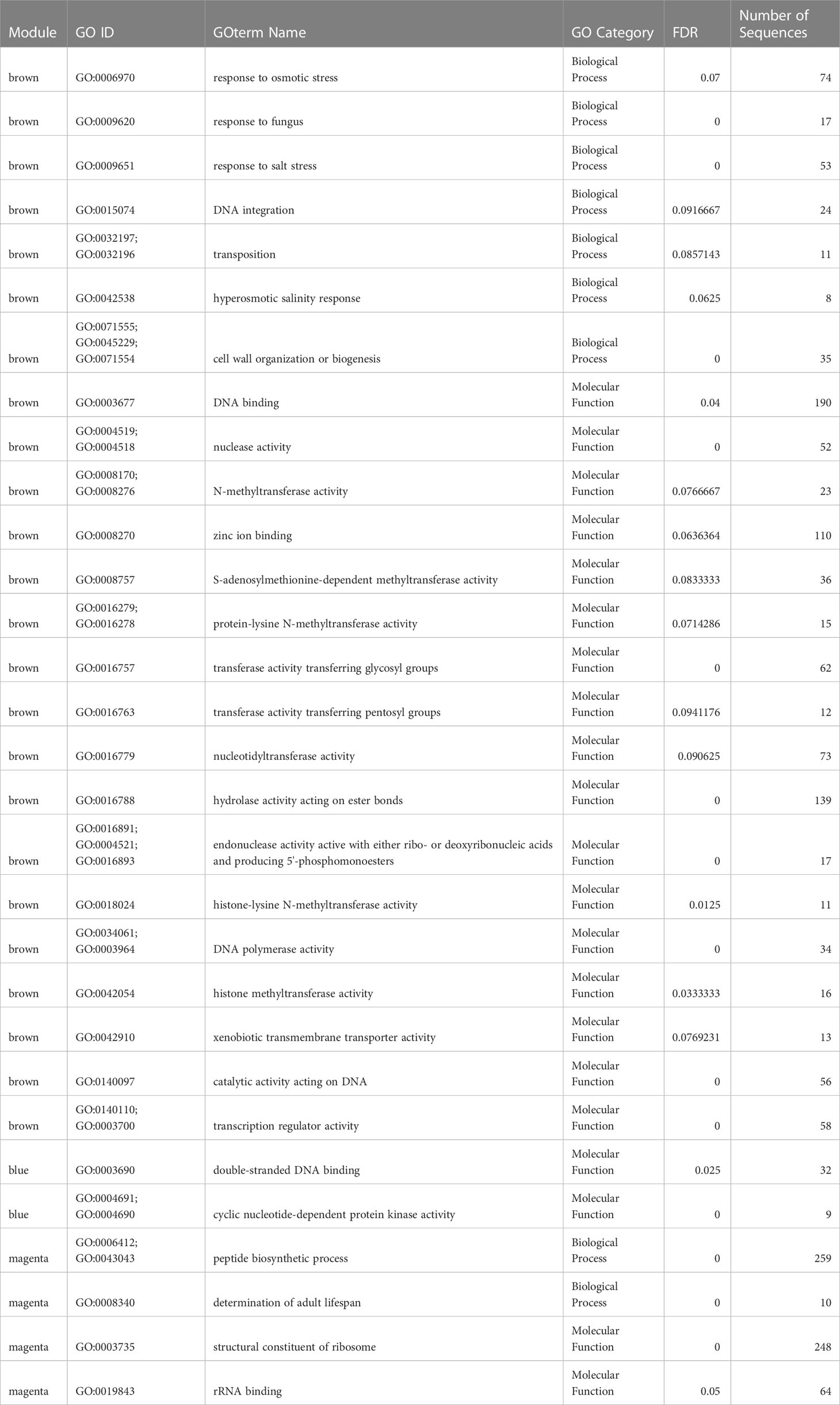
Table 1 Biological process and molecular function GOterms enriched within significant module eigengenes for Z. marina.
The module eigengene “yellow” was positively correlated with Lz exposure (r2 0.52, p= 0.049) and contained five differentially expressed genes (Figures 5B, C: Supplementary Table 2). While there were no significantly enriched GO terms in this module, common biological processes included proteolysis, cellular response to oxidative stress, response to osmotic stress, regulation of jasmonic acid metabolic process, and response to abscisic acid, and common molecular functions were binding of ATP, RNA, and metal ions (Supplementary Table 2).
The module eigengene “blue” was positively correlated with Lz exposure (r2 = 0.99, p = 2.0 x 10-11) (Figure 5B). The “blue” module contained 263 differentially expressed genes (Figure 5C; Supplementary Table 2). There were no enriched biological processes, but genes were involved in immune response, protein transport and ubiquitination, response to salt stress and oxidative stress, cell cycle, and abscisic acid-activated signaling pathway (Supplementary Table 2). Enriched molecular functions were double-stranded DNA binding (GO:0003690) and cyclic nucleotide-dependent protein kinase activity (GO:0004691;GO:0004690) (Table 1).
The module eigengene “magenta” was positively correlated with Lz exposure (r2 = 0.52, p= 0.046) (Figure 5B). This module eigengene had the most variable expression of all significant modules (Figure 5C). The “magenta” module contained 60 differentially expressed genes (Figure 5C; Supplementary Table 2). Peptide biosynthetic process (GO:0006412;GO:0043043) and determination of adult lifespan (GO:0008340), and structural constituent of ribosome (GO:0003735) and rRNA binding (GO:0019843) were significantly enriched biological process and molecular function terms in this module, respectively (Table 1). Other prominent biological processes included translation, cell redox homeostasis, and microtubule-based process (Supplementary Table 2).
Of the non-host contigs, 11.1% (87,550 out of 784,204) had significant matches to the Swiss-Prot database. Only 0.77% (5,959 out of 784,204) of non-host contigs had significant matches to the Labyrinthula sp. Ha genome. However, 42,601 non-host contigs (5.4% of total non-host contigs) had significant matches to a closely related thraustochytrid, Hondaea fermentalgiana (NCBI Accession # GCA_002897355.1). Thirty-two non-host contigs were differentially expressed; 30 up-regulated whereas two were down-regulated (Supplementary Table 3). Of the 32 differentially expressed non-host contigs, only 5 had significant similarity to the Labyrinthula sp. Ha genome; including arylsulfatase (SP_P08842), patatin-like phospholipase domain-containing protein (SP_Q8N8W4), 5-aminolevulinate synthase (SP_P08262), dynein heavy chain 10 (dhcA) (SP_Q8IVF4), and calponin-1 (SP_P`4318). In contrast, 20 differentially expressed non-host contigs had significant hits to the H. fermentalgiana genome (Supplementary Table 3). Seven non-host contigs also had homology to genes found in the well-annotated genome of the slime-mold model organism, D. discoideum, including the dynein heavy chain dhcA (DDB_G0276355), dual specificity protein kinase shkB (DDB_G0288617), adenylate cyclase sgcA (DDB_G0276269), calponin homology containing protein mp20 (DDB_G0292664), serine/threonine-protein kinase roco5 (DDB_G0294533), beta-ketoadipyl-CoA thiolase (DDB_G0269588), and regulator of G-protein signaling 21 (DDB_G0273033) (Supplementary Table 3).
Of the differentially expressed genes that displayed significant similarity to either Lz or to related species H. fermentalgiana or D. discoideum, ten up-regulated non-host contigs were linked to chemotaxis and motility (i.e. actin binding proteins, tubulin beta-chain, and microtubule formation), 11 contigs to amino acid and lipid metabolism (i.e. peptidases and fatty acid synthesis enzymes), 4 contigs to transcription/translation, 3 contigs to transporter activity (i.e. ion channel and transporter subunits), and 4 contigs to other processes including respiration (i.e. cytochrome c oxidase subunits) and production of secondary metabolites (i.e. phytoene synthase) (Supplementary Table 3). Several non-host contigs were related to phagocytosis, such as patatin-like phospholipase domain containing protein (SP_Q8N8W4), beta-ketoadipyl-CoA thiolase (SP_O26884), leucine-rich repeat serine-threonine protein kinase 2 (roco5) (SP_Q1ZXD6), ATP-dependant RNA helicase (SP_Q0DB53), hypoxanthine-guanine phosphoribosyltransferase (SP_Q72454), pepsin A-1 (SP_Q03168), and dynein heavy chain 10 (dhcA) (SP_Q8IVF4).
Exposure of eelgrass ramets to Lz resulted in rapid progression of EWD, with necrotic lesions forming within two days post-exposure and plant death occurring after five days. The temperature did not alter the pathogen load or progression of EWD in the Lz exposed plants. This may be due to the high concentration of pathogen in the exposure, which was higher than in previous studies with this Lz strain (Groner et al., 2014; Groner et al., 2018) and/or pre-acclimation of the plants to the temperature treatments prior to exposure. Analysis of the eelgrass transcriptome revealed changes in gene expression, with patterns consistent with increased defensive responses through altered regulation of genes associated with phytohormone biosynthesis, stress response, and immune function. Analysis of non-Zostera genes (likely Lz genes), revealed expression patterns suggestive of Lz disrupting host immune responses and undergoing phagocytosis. This study reveals the importance of specific pathways related to plant defense against pathogen presence, and provides molecular evidence to support previous phenotypic observations of host-pathogen interactions of eelgrass wasting disease.
The genome of Z. marina allowed us to identify 3,096 host contigs, and we detected 540 differentially expressed host genes responding to Lz infection as opposed to increased temperature (Supplementary Table 1). We found expression changes in genes and/or pathways involved with immunity such as pathogen detection, defense-related cell-cell signaling, defense-related metabolite production, and apoptosis. We also detected genes potentially involved in salt stress. The occurrence of many differentially expressed defense-response related GO terms, including genes associated with microbial detection and defense response to bacterial or fungal pathogens, indicates that Z. marina is responding to Lz infection.
Plant innate immune system pathways are highly conserved and are initiated by recognition of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) by membrane localized receptors (Choi and Klessig, 2016). Calcium signaling is a major pathway in pattern-triggered immunity, where binding of PAMPs and DAMPs to membrane receptors initiates the rapid release of calcium ions (Ca2+) by receptor like kinases and their Ca2+ dependent protein kinases and mitogen activated protein kinases (Hou et al., 2018). For example, calcium/calmodulin-dependent protein kinase (CcaMK) genes have been found to play a role in maintaining endosymbionts and in pathogen resistance in tomato, by promoting accumulation of hydrogen peroxide (Wang et al., 2015). Genes related to calcium- and protein-kinases show up eight and 43 times, respectively, in the blue module. In particular, upregulation of the calcium/calmodulin-dependent protein kinase (CcaMK) type 1 (CaM kinase I) suggests the role of Lz-related PAMP/DAMP detection and may warrant further exploration.
Pathogen detection leads to the induction of signaling pathways to induce immune and defense responses. We found that the cAMP-dependent protein kinase catalytic subunit gamma is significantly upregulated with Lz-exposure. Changes in the abundance of cAMPs can affect the activity of other signaling pathways and cellular processes, such as protein kinases and transcription factors (Świeżawska et al., 2018), demonstrating that Z. marina may respond to Lz infection by altering upstream signal transduction.
Phytohormone production in plants is important for defense-related signaling and is turned on after PAMP/DAMP detection. For example, compounds such as jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) are involved in general activation of plant defense systems against pathogens (Thaler et al., 2004; Tamaoki et al., 2013). In particular, SA is used in defense against biotic factors such as insects, mites, fungi, and bacteria and JA is used in defense pathways against saprophytic microbes like Lz (Tamaoki et al., 2013; Zhang et al., 2020). ABA is triggered in response to both abiotic and biotic stressors, and has complex interactions with both JA and SA responses to infection (Fan et al., 2009). We found GO terms for SA, JA, and ABA phytohormones [GO:0080140, GO:0009695, GO:0009753, and GO:0009751] were highly represented in the blades experimentally exposed to Lz, although these gene ontologies were not significantly enriched. Gene expression patterns in this study were indicative of phytohormone signaling via enrichment of serine/threonine-protein kinase PCRK2 gene which is involved in the resistance to bacterial pathogens and salicylate biosynthesis during pathogenic infection (Sreekanta et al., 2015). Methylesterase 1 gene, which is required during the conversion of methyl salicylate into SA was downregulated in exposed blades. These data suggest that Z. marina is altering production of these defense-related compounds. We found gene expression changes in two genes related to JA biosynthesis: chloroplastic rhomboid-like protein 11 (regulation of jasmonic acid metabolic process [GO:0080140]) and phospholipase A (defense response to fungus [GO:0050832]; jasmonic acid biosynthetic process [GO:0009695]). Phospholipase A is responsible for the release of linolenic acid from the chloroplast membrane during defensive signaling, which induces JA production (Yang et al., 2007; Mata-Pérez et al., 2015). The detection of differentially regulated genes in the JA pathway is particularly interesting, as the role of JA in seagrasses is unclear. A congener of Z. marina, Z. muelleri, lost genes encoding jasmonate methyltransferase, which converts JA into a volatile compound, methyl jasmonate, but maintain other genes along the JA pathway, including for jasmonate synthesis, and signaling (Lee et al., 2016). Although the JA-associated genes were not differentially expressed in our study, changes to expression in these genes suggests a role for the jasmonic acid pathway in responding to Lz infection.
Gene expression changes also indicated down-stream effects of phytohormone production. In maize roots, expression of CHC1 gene increases in response to SA or ABA, suggesting involvement of CHC1 in the SA signaling pathway in maize defense responses (Zeng et al., 2013). Upregulation of the probable clathrin heavy chain 1 gene (CHC1) ([GO:0005198]) in Lz-infected blades suggests a similar response in eelgrass. We detected gene expression changes in factors involved in stress signaling which either require ABA or interact with ABA to trigger defense responses. For example, the S-type anion channel SLAH3 is an anion channel that can be triggered by ABA during a stress response (Roelfsema et al., 2012), and was downregulated in response to Lz infection. The B3 domain-containing protein was also downregulated. This protein is a transcription factor for ABA and inhibits the production of ABA (Brady et al., 2003), so a decrease in expression of B3 could result in an increase in ABA production, indicating a greater stress response to Lz infection. One specific pathway in which ABA affects plant defense is in the callose pathway, which is involved in defense of fungal pathogens in plants. Specifically, ABA mutants displayed impaired resistance to necrotrophic fungi, and ABA addition to the infection site mimicked callose deposition and increased resistance to necrotrophic fungi (Mauch-Mani and Mauch, 2005). Therefore, it is not surprising to observe altered ABA expression in Z. marina infected with Lz, which has necrotic capabilities.
Additional immune responses in plants include the production of antimicrobial compounds and apoptosis of infected cells, the latter of which would be important for intracellular infections, such as with Lz. Upregulation of peroxisomal targeting signal 1 receptor indicates potential defenses at sites of pathogen entry. This gene is associated with concentrating antifungal glucosinolate derivatives in the peroxisome for eventual transport to sites of fungal infection entry (Bednarek et al., 2009). It would be valuable to assess whether these compounds can inhibit Lz as well. Differential expression of GDP-L-fucose synthase supports the role of apoptosis as an immune response to Lz. GDP-L-fucose plays a role in stomatal and apoptosis-related defense in Arabidopsis, indicating a similar role in Z. marina immunity (Zhang et al., 2019). Differential expression of EKC/KEOPS subunit genes, which are involved in apoptotic processes via telomere capping and elongation (Downey et al., 2006), also support the hypothesis that apoptosis is an important defense against Lz.
The production of phenolic compounds is another major immune response of plants. Many of phenolic metabolites detected in Z. marina have been implicated in Z. marina defense. These include flavonoids in sulfated and unsulfated form, as well as acids like caffeic, p-coumaric, ferulic, and zosteric, and rosmarinic acids, all of which are hydroxy, sulfoxy, or ester forms of cinnamic acid (Papazian et al., 2019). Resistance of Z. marina to Lz is hypothesized to depend upon phenolic acids, especially caffeic acid which has demonstrated activity against Lz in bioassays (Vergeer and Develi, 1997; Trevathan-Tackett et al., 2015). Lz has also been shown to induce production of total phenols in some cases (McKone and Tanner, 2009), and inhibit them in others, potentially as a mechanism to circumvent this defense. In addition, production of phenols in seagrasses are reduced at warmer temperatures (Vergeer et al., 1995). This is reflected in the reduced concentrations of phenols found five days post Lz-exposure in the warmer temperature treatment.
The shikimate pathway is the mechanism through which plants produce aromatic phenolic compounds in response to stress (Santos-Sánchez et al., 2019). Four genes - caffeic acid 3-O-methyltransferase, caffeoylshikimate esterase, 4-coumarate–CoA ligase-like 7, and cinnamoyl-CoA reductase - responsible for coding enzymes involved in the biosynthesis of different forms of cinnamic acids were differentially expressed in the exposed treatment, indicating a role for the shikimate pathway in Lz infection response. Caffeoylshikimate esterase (CSE), converts caffeoyl shikimate into caffeic acid, which has been shown to inhibit Lz growth in vitro (Vergeer and Develi, 1997; Vanholme et al., 2013). Another differentially expressed gene, caffeic acid 3-O-methyltransferase, is associated with converting caffeic acid into products associated with the lipid biosynthesis pathway. 4-coumarate-CoA is also associated with production of additional secondary compounds including isoflavonoids and furanocoumarins as phytoalexins (i.e., inhibitors of pathogen growth) (Hamberger and Hahlbrock, 2004). Cinnamoyl-CoA reductase is involved in the formation of phenolic compounds associated with the hypersensitive response (Lauvergeat et al., 2001). The hypersensitive response in plants is akin to the innate immune response in animals and is responsible for isolated apoptosis surrounding an infection in order to prevent further spread (Morel and Dangl, 1997).
Apart from individual roles in the shikimate pathway, all four differentially expressed genes in this pathway are also associated with lignin biosynthesis. The role of lignin is unclear in aquatic plants such as Z. marina. While it may be protective against microbial attack, it is typically concentrated in the slower growing rhizomes, and not in the blades (Klap et al., 2000), where gene expression was measured in this study.
In addition to genes involved in immune activation, hormone pathways, and phenolic production, the transcriptional patterns of Lz exposed plants and gene enrichment results suggest responses to environmental stress, including salt stress. Stress-related genes were significantly upregulated in the exposed treatment, indicating that Lz infection may cause damage to cells in a way that triggers this response. The mitochondrial phosphate carrier protein 3 (MPT3) is induced under high salinity conditions (Zhu et al., 2012). Increased expression of the MPT3 gene suggests that Z. marina may be experiencing salt stress as Lz breaks through host physical barriers. Similarly, the sorting nexin 1 protein regulates the process of accumulating nitrous oxide, which is an important signaling molecule in responses to abiotic stressors, including salt stress (Li et al., 2018). Higher sorting nexin 1 protein gene expression implies that Z. marina may accumulate more nitrous oxide in an effort to respond to abiotic stress. Finally, the upregulation of the calcium/calmodulin-dependent protein kinase type 1 (CaM kinase I) gene suggests changing cytosol calcium concentrations, which is a known plant response to stressors such as anoxia, increases temperature, and increased salinity (White and Broadley, 2003). Changes in expression of genes associated with stress response in Z. marina may inform our understanding of how molecular functioning is altered upon exposure to environmental changes. This information can also shed light on how Lz infection cause stressful conditions for Z. marina.
By separating host from non-host contigs, we aimed to identify genes or pathways that may contribute to the virulence of L. zosterae during experimental infection of Z. marina. We found up-regulation of genes potentially involved in breakdown of host defense, chemotaxis and phagocytosis, and metabolism. Although an unannotated genome of another species of Labyrinthula (Labyrinthula sp. Ha) is newly available, very few contigs had significant matches to the current version of the genome, and only five of the differentially expressed genes from the non-host dataset had significant BLAST hits to this genome. However, the majority of the non-host contigs had significant matches to related slime-mold species including Dictyostelium species and Hondaea fermentalgiana (a thraustochytrid closely related to the genus Labyrinthula), suggesting that the identified non-host genes were likely from Lz inoculated in the infection treatment.
Zostera species (including Z. marina) produce high concentrations of a sulfate ester, zosteric acid, as a defense mechanism that protects blades from microbial biofouling (Grignon-Dubois et al., 2012; Vilas-Boas et al., 2017). We found a 24-fold up-regulation of a non-host contig that matched to the arylsulfatase gene (GBG30795.1) of H. fermentalgiana. Arylsulfatases are enzymes that break down sulfatides and organic sulfate esters. In other protists, arylsulfatases are produced in response to sulfate deprivation (Niedermeyer et al., 1987). However, for the fungal plant pathogen, Colletotrichum gloeosporiodes, arylsulfatase expression increases during penetration of host leaf tissue after experimental inoculation (Goodwin et al., 2000). In Lz, an increase in arylsulfatase expression may facilitate attachment by breaking down the anti-biofouling zosteric acid produced by the host. However, the potential role of arylsulfatases to facilitate protist pathogen attachment and colonization has yet to be investigated.
Initial studies describing EWD observed Lz directly penetrating mesophyll cell walls, damage suggestive of feeding on plant cell organelles such as chloroplasts, and moving rapidly through the host tissues (Muehlstein, 1992; Raghukumar, 2002). In support of these microscopic observations and supporting the role of plant consumption as a mechanism of pathogenesis, we identified several up-regulated genes that are likely involved in movement, i.e. chemotaxis and cytoskeleton rearrangement and organization, as well as phagocytosis. For example, contigs homologous to shkB, sgcA and roco5 genes were all up-regulated after experimental inoculation of Lz. In D. discoideum, null mutants for shkB and sgcA have reduced chemotaxis and phagocytosis ability (Moniakis et al., 2001; Veltman and Van Haastert, 2006; Veltman et al., 2008), and null mutants for the roco5 genes have slow or no motility (Sawai et al., 2007). Six up-regulated non-host contigs were homologous to genes that are up-regulated during phagocytosis and/or have gene products which are a part of the ‘macropinocytosis proteome’ of D. discoideum (Sillo et al., 2008; Journet et al., 2012). Additionally, in D. discoideum, dhcA proteins cluster to phagosomes and promote phagolysosome fusion (Rai et al., 2016), and lysosomal aspartic protease CatD, a ubiquitous hydrolytic enzyme with a protein of homology in D. discoideum, is known to be a lysosomal pepsin in the macropinosome and is involved with phagosome maturation (Vines and King, 2019).
Genes involved in amino acid and lipid metabolism were up-regulated among the non-host contigs. M42 peptidase, a co-catalytic metallopepsidase (cytosolic enzyme) described in bacteria and archaea (reviewed by Appolaire et al., 2016), was the most expressed non-host contig. M42 may play a role in pathogenesis in bacteria by using or modifying exogenous proteins including immunological antibacterial peptides. Other peptidases were up-regulated, such as Carboxypeptidase Y and an otubain-like ubiquitin thioesterase, and may be involved with the catabolism of both large and small peptides in Lz. Ethylmalony-CoA decarboxylase, polyketide synthase, beta-ketoadipyl-CoA thiolase, and patatin-like phosphatase domain-containing protein are all involved with fatty acid synthesis and may support membrane formation and increase membrane fluidity during cell division or for the extension of ectoplasmic nets of Lz. Up-regulation of these metabolism related contigs along with the up-regulation of genes associated with transcription and translation support increased metabolism and/or cellular proliferation.
Surprisingly, no genes potentially linked to the production of zoospores were up-regulated in this study. Other mechanisms of microbial virulence, such as toxin production, iron sequestration, and host immune invasion (Finlay and Falkow, 1997), were also not up-regulated in this study. Together the non-host transcriptome suggests the possible enzymatic degradation of plant cell defenses, supports the rapid dissemination of Lz cells within host tissue, and supports the voracious consumption of plant tissue by invading Lz cells as mechanisms of virulence in seagrass wasting disease. Time course experiments, with lower exposure concentrations (sensu Bower et al., 1989) would be useful in elucidating both the genes and virulence mechanisms involved in penetration vs. pathogenesis during this disease process.
With increased outbreaks of marine disease, continued influence of anthropogenic stressors, and rapidly changing oceans contributing to significant declines in seagrass meadows across the globe, there is an increasing need to understand host-pathogen dynamics across environmental scales. Given the rapid decline of Z. marina globally coupled with active restoration, we have increased need for understanding of host-pathogen dynamics and how this might impact future restoration. This study demonstrates the value of dual host-pathogen transcriptomics for understanding physiological consequences of disease across environmental gradients. While the Lz pathogen exhibited a limited repertoire of virulence approaches, including degradation of cell walls, consumption of intracellular materials and movement within the host, the eelgrass host exhibited a wide range of responses to infection, from cascading phytohormone signals, to altered production of phenols, increased PAMP/DAMP signaling and apoptosis. As our knowledge of Lz increases, biologically realistic inoculation experiments can be used to further understand the role of changing nearshore conditions in facilitating or repressing disease in this important coastal habitat-forming species.
The RNAseq data presented in the study are deposited in the NCBI Short Read Archive repository, accession number PRJNA990835. Additional data presented in this study are available at FigShare (http://doi.org/10.6084/m9.figshare.22564528).
MG and CB obtained financial support, conceived of the experimental design, and implemented the experiment. MG led statistical analyses of phenolic and diagnostic data. YV, CB, and AS led the transcriptomics analysis, with help from all authors, and KA analyzed phenols and tannins in the samples. All authors contributed to the article and approved the submitted version.
This work was funded by a seed grant from the Canadian excellence research chair in support of aquatic epidemiology to MG, start-up funds provided to CB from the University of Maryland Baltimore County and the University of Maryland Baltimore, funding from the National Science Foundation (2109607; 1215977), as well as the University of Washington Friday Harbor Labs.
Michelle Herko, Rebecca Guenther and Connie Sullivan assisted in the execution of this experiment. N. Rivlin provided technical assistance and T. Bachvaroff bioinformatics assistance.
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1152647/full#supplementary-material
Świeżawska B., Duszyn M., Jaworski K., Szmidt-Jaworska A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9, 1428. doi: 10.3389/fpls.2018.01428
Agnew M. V., Groner M. L., Eisenlord M. E., Friedman C. S., Burge C. A. (2022). Pacific oysters are a sink and a potential source of the eelgrass pathogen, Labyrinthula zosterae. Aquacult Environ. Interact. 14, 295–307. doi: 10.3354/aei00446
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aoki L. R., McGlathery K. J., Wiberg P. L., Al-Haj A. (2020). Depth affects seagrass restoration success and resilience to marine heat wave disturbance. Estuaries Coast. 43, 316–328. doi: 10.1007/s12237-019-00685-0
Appolaire A., Colombo M., Basbous H., Gabel F., Girard E., Franzetti B. (2016). TET peptidases: a family of tetrahedral complexes conserved in prokaryotes. Biochimie 122, 188–196. doi: 10.1016/j.biochi.2015.11.001
Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting Linear Mixed-Effects Models Using lme4. J. Statistical Software. 67 (1), 1–48. doi: 10.18637/jss.v067.i01
Bate-Smith E. C., Rasper V. (1969). Tannins of grain sorghum: luteoforol (leucoluteolinidin) 3’,4,4’,5,7-pentahydroxyflavan. J. Food Sci. 34, 203–209. doi: 10.1111/j.1365-2621.1969.tb00919.x
Beca-Carretero P., Guihéneuf F., Marín-Guirao L., Bernardeau-Esteller J., García-Muñoz R., Stengel D. B., et al. (2018). Effects of an experimental heat wave on fatty acid composition in two Mediterranean seagrass species. Mar. Pollut. Bull. 134, 27–37. doi: 10.1016/j.marpolbul.2017.12.057
Bednarek P., Piślewska-Bednarek M., Svatoš A., Schneider B., Doubský J., Mansurova M., Schulze-lefert A., et al(2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Bockelmann A. C., Tams V., Ploog J., Schubert P. R., Reusch T. B. (2013). Quantitative PCR reveals strong spatial and temporal variation of the wasting disease pathogen, Labyrinthula zosterae in northern European eelgrass (Zostera marina) beds. PloS One 8, e62169. doi: 10.1371/journal.pone.0062169
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bower S. M., McLean N., Whitaker D. J. (1989). Mechanism of infection by Labyrinthuloides haliotidis (Protozoa: labyrinthomorpha), a parasite of abalone (Haliotis kamtschatkana)(Mollusca: Gastropoda). J. Invert Pathol. 53, 401–409. doi: 10.1016/0022-2011(89)90106-7
Brady S. M., Sarkar S. F., Bonetta D., McCourt P. (2003). The abscisic acid insensitive 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34, 67–75. doi: 10.1046/j.1365-313X.2003.01707.x
Brakel J., Jakobsson-Thor S., Bockelmann A. C., Reusch T. B. (2019). Modulation of the eelgrass–Labyrinthula zosterae interaction under predicted ocean warming, salinity change and light limitation. Front. Mar. Sci. 6, 268. doi: 10.3389/fmars.2019.00268
Bull J. C., Kenyon E. J., Cook K. J. (2012). Wasting disease regulates long-term population dynamics in a threatened seagrass. Oecologia 169, 135–142. doi: 10.1007/s00442-011-2187-6
Burge C.A., Friedman C.S. (2012). Quantifying Ostreid herpesvirus (OsHV-1) copies and expression during transmission. Microb Ecol. 63 (3), 596–604. doi: 10.1007/s00248-011-9937-1
Chen Y., Lun A.A.T., Smyth G.K. (2016). “From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline.” F1000Research. 5, 1438. doi: 10.12688/f1000research.8987.2
Choi H. W., Klessig D. F. (2016). DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 16, 232. doi: 10.1186/s12870-016-0921-2
Dawkins P. D., Eisenlord M. E., Yoshioka R. M., Fiorenza E., Fruchter S., Giammona F., et al. (2018). Environment, dosage, and pathogen isolate moderate virulence in eelgrass wasting disease. Dis. Aquat Org 130, 51–63. doi: 10.3354/dao03263
Downey M., Houlsworth R., Maringele L., Rollie A., Brehme M., Galicia S., et al. (2006). A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124, 1155–1168. doi: 10.1016/j.cell.2005.12.04
Dunic J. C., Brown C. J., Connolly R. M., Turschwell M. P., Cote I. M. (2021). Long-term declines and recovery of meadow area across the world’s seagrass bioregions. Global Change Biol. 27, 4096–4109. doi: 10.1111/gcb.15684
Fan J., Hill L., Crooks C., Doerner P., Lamb C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. doi: 10.1104/pp.109.137943
Finlay B. B., Falkow S. (1997). Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61, 136–169. doi: 10.1128/mmbr.61.2.136-169.1997
Goodwin P. H., Li J., Jin S. (2000). Evidence for sulfate derepression of an arylsulfatase gene of Colletotrichum gloeosporioides f. sp. malvae during infection of round-leaved mallow, Malva pusilla. Physiol. Mol. Plant Pathol. 57, 169–176. doi: 10.1006/pmpp.2000.0295
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644. doi: 10.1038/nbt.1883
Graham O. J., Aoki L. R., Stephens T., Stokes J., Dayal S., Rappazzo B., et al. (2021). Effects of seagrass wasting disease on eelgrass growth and belowground sugar in natural meadows. Front. Mar. Sci. 8, 768668. doi: 10.3389/fmars.2021.768668
Grignon-Dubois M., Rezzonico B., Alcoverro T. (2012). Regional scale patterns in seagrass defences: phenolic acid content in Zostera noltii. Estuar. Coast. shelf Sci. 114, 18–22. doi: 10.1016/j.ecss.2011.09.010
Groner M. L., Burge C. A., Couch C. S., Kim C. J., Siegmund G. F., Singhal S., et al. (2014). Host demography influences the prevalence and severity of eelgrass wasting disease. Dis. aquat org 108, 165–175. doi: 10.3354/dao02709
Groner M. L., Burge C. A., Cox R., Rivlin N. D., Turner M., Van Alstyne K. L., et al. (2018). Oysters and eelgrass: potential partners in a high pCO2 ocean. Ecology 99, 1802–1814. doi: 10.1002/ecy.2393
Groner M. L., Burge C. A., Kim C. J., Rees E., Van Alstyne K. L., Yang S., et al. (2016). Plant characteristics associated with widespread variation in eelgrass wasting disease. Dis. aqaut org 118, 159–168. doi: 10.3354/dao02962
Groner M. L., Eisenlord M. E., Yoshioka R. M., Fiorenza E. A., Dawkins P. D., Graham O. J., et al. (2021). Warming sea surface temperatures fuel summer epidemics of eelgrass wasting disease. Mar. Ecol. Prog. Ser. 679, 47–58. doi: 10.3354/meps13902
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494. doi: 10.1038/nprot.2013.084
Hamberger B., Hahlbrock K. (2004). The 4-coumarate: CoA ligase gene family in arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. PNAS 101, 2209–2214. doi: 10.1073/pnas.0307307101
Hou S., Jamieson P., He P. (2018). The cloak, dagger, and shield: proteases in plant–pathogen interactions. Biochem. J. 475, 2491–2509. doi: 10.1042/bcj20170781
Journet A., Klein G., Brugière S., Vandenbrouck Y., Chapel A., Kieffer S., et al. (2012). Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics 12, 241–245. doi: 10.1002/pmic.201100313
Klap V. A., Hemminga M. A., Boon J. J. (2000). Retention of lignin in seagrasses: angiosperms that returned to the sea. Mar. Ecol. Prog. Ser. 194, 1–11. doi: 10.3354/meps194001
Lamb J. B., Van De Water J. A., Bourne D. G., Altier C., Hein M. Y., Fiorenza E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Langfelder P., Horvath S. (2008). WGCNA: an r package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Lauvergeat V., Lacomme C., Lacombe E., Lasserre E., Roby D., Grima-Pettenati J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57, 1187–1195. doi: 10.1016/S0031-9422(01)00053-X
Lee H., Golicz A. A., Bayer P. E., Jiao Y., Tang H., Paterson A. H., et al. (2016). The genome of a southern hemisphere seagrass species (Zostera muelleri). Plant Physiol. 172, 272–283. doi: 10.1104/pp.16.00868
Li T. T., Liu W. C., Wang F. F., Ma Q. B., Lu Y. T., Yuan T. T. (2018). SORTING NEXIN 1 functions in plant salt stress tolerance through changes of NO accumulation by regulating NO synthase-like activity. Front. Plant Sci. 9, 1634. doi: 10.3389/fpls.2018.01634
Ma D., Li Y., Zhang J., Wang C., Qin H., Ding H., et al. (2016). Accumulation of phenolic compounds and expression profiles of phenolic acid biosynthesis-related genes in developing grains of white, purple, and red wheat. Front. Plant Sci. 7, 528. doi: 10.3389/fpls.2016.00528
Markham J. W., Hagmeier E. (1982). Observations on the effects of germanium dioxide on the growth of macro-algae and diatoms. Phycologia 21, 125–130. doi: 10.2216/i0031-8884-21-2-125.1
Martin D. L., Chiari Y., Boone E., Sherman T. D., Ross C., Wyllie-Echeverria S., et al. (2016). Functional, phylogenetic and host-geographic signatures of labyrinthula spp. provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuaries Coasts 39, 1403–1421. doi: 10.1007/s12237-016-0087-z
Mata-Pérez C., Sánchez-Calvo B., Begara-Morales J. C., Luque F., Jiménez-Ruiz J., Padilla M. N., et al. (2015). Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Front. Plant Sci. 6, 122. doi: 10.3389/fpls.2015.00122
Mauch-Mani B., Mauch F. (2005). The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. doi: 10.1016/j.pbi.2005.05.015
McCarthy D. J., Chen Y., Smyth G. K. (2012). Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McKone K. L., Tanner C. E. (2009). Role of salinity in the susceptibility of eelgrass Zostera marina to the wasting disease pathogen Labyrinthula zosterae. Mar. Ecol. Prog. Ser. 377, 123–130. doi: 10.3354/meps07860
Moniakis J., Funamoto S., Fukuzawa M., Meisenhelder J., Araki T., Abe T., et al. (2001). An SH2-domain-containing kinase negatively regulates the phosphatidylinositol-3 kinase pathway. Genes Dev. 15, 687–698. doi: 10.1101/gad.871001
Morel J. B., Dangl J. L. (1997). The hypersensitive response and the induction of cell death in plants. Cell Death differ 4, 671–683. doi: 10.1038/sj.cdd.4400309
Muehlstein L. K. (1992). The host-pathogen interaction in the wasting disease of eelgrass, Zostera marina. Can. J. Bot. 70, 2081–2088. doi: 10.1139/b92-258
Niedermeyer I., Biedlingmaier S., Schmidt A. (1987). Derepression of arylsulfatase activity by sulfate starvation in Chlorella fusca. Z. für Naturforschung C, 42 (5), 530–536, 42. doi: 10.1515/znc-1987-0507
Nitao J. K., Birr B. A., Nair M. G., Herms D. A., Mattson W. J. (2001). Rapid quantification of proanthocyanidins (condensed tannins) with a continuous flow analyzer. J. Agric. Food Chem. 49, 2207–2214. doi: 10.1021/jf001183b
Nordlund L. M., Koch E. W., Barbier E. B., Creed J. C. (2016). Seagrass ecosystem services and their variability across genera and geographical regions. PloS One 11 (10), e0163091. doi: 10.1371/journal.pone.0169942
O’Donnell M. J., George M. N., Carrington E. (2013). Mussel byssus attachment weakened by ocean acidification. Nat. clim Change 3, 587–590. doi: 10.1038/nclimate1846
Orth R. J., Carruthers T. J., Dennison W. C., Duarte C. M., Fourqurean J. W., Heck K. L., et al. (2006). A global crisis for seagrass ecosystems. Bioscience 56, 987–996. doi: 10.1641/0006-3568(2006)56[987:AGCFSE]2.0.CO;2
Papazian S., Parrot D., Burýšková B., Weinberger F., Tasdemir D. (2019). Surface chemical defence of the eelgrass Zostera marina against microbial foulers. Sci. Rep. 9, 3323. doi: 10.1038/s41598-019-39212-3
Raghukumar S. (2002). Ecology of the marine protists, the labyrinthulomycetes (Thraustochytrids and labyrinthulids). Eur. J. Protistol. 38, 127–145. doi: 10.1078/0932-4739-00832
Rai A., Pathak D., Thakur S., Singh S., Dubey A. K., Mallik R. (2016). Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell 164, 722–734. doi: 10.1016/j.cell.2015.12.054
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinform 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roelfsema M. R. G., Hedrich R., Geiger D. (2012). Anion channels: master switches of stress responses. Trends Plant Sci. 17, 221–229. doi: 10.1016/j.tplants.2012.01.009
Santos-Sánchez N. F., Salas-Coronado R., Hernández-Carlos B., Villanueva-Cañongo C. (2019). Shikimic acid pathway in biosynthesis of phenolic compounds. Plant Physiol. aspects phenolic compounds 1, 1–15. doi: 10.5772/intechopen.83815
Sawai S., Guan X. J., Kuspa A., Cox E. C. (2007). High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol. 8, 1–15. doi: 10.1186/gb-2007-8-7-r144
Short F., Carruthers T., Dennison W., Waycott M. (2007). Global seagrass distribution and diversity: a bioregional model. J. Exp. Mar. Biol. Ecol. 350, 3–20. doi: 10.1016/j.jembe.2007.06.012
Short F. T., Muehlstein L. K., Porter D. (1987). Eelgrass wasting disease: cause and recurrence of a marine epidemic. Biol. Bull. 173, 557–562. doi: 10.2307/1541701
Sillo A., Bloomfield G., Balest A., Balbo A., Pergolizzi B., Peracino B., et al. (2008). Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics 9, 1–22. doi: 10.1186/1471-2164-9-291
Sreekanta S., Bethke G., Hatsugai N., Tsuda K., Thao A., Wang L., et al. (2015). The receptor-like cytoplasmic kinase PCRK 1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol. 207, 78–90. doi: 10.1111/nph.13345
Strydom S., Murray K., Wilson S., Huntley B., Rule M., Heithaus M., et al. (2020). Too hot to handle: unprecedented seagrass death driven by marine heatwave in a world heritage area. Glob. Change Biol. 26, 3525–3538. doi: 10.1111/gcb.15065
Sullivan B. K., Sherman T. D., Damare V. S., Lilje O., Gleason F. H. (2013). Potential roles of Labyrinthula spp. in global seagrass population declines. Fungal Ecol. 6, 328–338. doi: 10.1016/j.funeco.2013.06.004
Tamaoki D., Seo S., Yamada S., Kano A., Miyamoto A., Shishido H., et al. (2013). Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal Behav. 8, e24260. doi: 10.4161/psb.24260
Thaler J. S., Owen B., Higgins V. J. (2004). The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol. 135, 530–538. doi: 10.1104/pp.104.041566
Tracy A. M., Pielmeier M. L., Yoshioka R. M., Heron S. F., Harvell C. D. (2019). Increases and decreases in marine disease reports in an era of global change. Proc. R. Soc. B 286, 20191718. doi: 10.1098/rspb.2019.1718
Trevathan-Tackett S. M., Lane A. L., Bishop N., Ross C. (2015). Metabolites derived from the tropical seagrass Thalassia testudinum are bioactive against pathogenic Labyrinthula sp. Aquat. Bot. 122, 1–8. doi: 10.1016/j.aquabot.2014.12.005
Vanholme R., Cesarino I., Rataj K., Xiao Y., Sundin L., Goeminne G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602
Veltman D. M., Keizer-Gunnik I., Van Haastert P. J. (2008). Four key signaling pathways mediating chemotaxis in Dictyostelium discoideum. J. Cell Biol. 180, 747–753. doi: 10.1083/jcb.200709180
Veltman D. M., Van Haastert P. J. (2006). Guanylyl cyclase protein and cGMP product independently control front and back of chemotaxing Dictyostelium cells. Mol. Biol. Cell 17, 3921–3929. doi: 10.1091/mbc.e06-05-0381
Vergeer L. H. T., Aarts T. L., De Groot J. D. (1995). The ‘wasting disease’and the effect of abiotic factors (light intensity, temperature, salinity) and infection with Labyrinthula zosterae on the phenolic content of Zostera marina shoots. Aquat. Bot. 52, 35–44. doi: 10.1016/0304-3770(95)00480-N
Vergeer L. H., Develi A. (1997). Phenolic acids in healthy and infected leaves of Zostera marina and their growth-limiting properties towards Labyrinthula zosterae. Aquat. Bot. 58, 65–72. doi: 10.1016/S0304-3770(96)01115-1
Vilas-Boas C., Sousa E., Pinto M., Correia-da-Silva M. (2017). An antifouling model from the sea: a review of 25 years of zosteric acid studies. Biofouling 33, 927–942. doi: 10.1080/08927014.2017.1391951
Vines J. H., King J. S. (2019). The endocytic pathways of Dictyostelium discoideum. Int. J. Dev. Biol. 63, 461–471. doi: 10.1387/ijdb.190236jk
Wang J. P., Munyampundu J. P., Xu Y. P., Cai X. Z. (2015). Phylogeny of plant calcium and calmodulin-dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 6, 1075. doi: 10.3389/fpls.2015.01075
White P. J., Broadley M. R. (2003). Calcium in plants. Ann. Bot. 9, 487–511. doi: 10.1093/aob/mcg164
Wright R. M., Aglyamova G. V., Meyer E., Matz M. V. (2015). Gene expression associated with white syndromes in a reef-building coral. Acropora hyacinthus. BMC Genomics 16, 371. doi: 10.1186/s12864-015-1540-2
Yang W., Devaiah S. P., Pan X., Isaac G., Welti R., Wang X. (2007). AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J. Biol. Chem. 282, 18116–18128. doi: 10.1074/jbc.M700405200
Zeng M. H., Liu S. H., Yang M. X., Zhang Y. J., Liang J. Y., Wan X. R., et al. (2013). Characterization of a gene encoding clathrin heavy chain in maize up-regulated by salicylic acid, abscisic acid and high boron supply. Int. J. Mol. Sci. 14, 15179–15198. doi: 10.3390/ijms140715179
Zhang L., Paasch B. C., Chen J., Day B., He S. Y. (2019). An important role of l-fucose biosynthesis and protein fucosylation genes in Arabidopsis immunity. New Phytol. 222, 981–994. doi: 10.1111/nph.15639
Zhang N., Zhou S., Yang D., Fan Z. (2020). Revealing shared and distinct genes responding to JA and SA signaling in arabidopsis by meta-analysis. Front. Plant Sci. 11, 908. doi: 10.3389/fpls.2020.00908
Keywords: Zostera marina, Labyrinthula zosterae, eelgrass wasting disease, transcriptomics, host-pathogen interactions, marine disease
Citation: Venkataraman YR, Shore A, Dayal S, Lee JS, Alidoost Salimi M, Crandall G, Loeher MM, Stoops M, Swanger M, Eisenlord ME, Van Alstyne KL, Fast MD, Burge CA and Groner ML (2023) Characterizing host-pathogen interactions between Zostera marina and Labyrinthula zosterae. Front. Mar. Sci. 10:1152647. doi: 10.3389/fmars.2023.1152647
Received: 28 January 2023; Accepted: 23 June 2023;
Published: 08 August 2023.
Edited by:
Diana Sofia Madeira, University of Aveiro, PortugalReviewed by:
Renato Crespo Pereira, Fluminense Federal University, BrazilCopyright © 2023 Venkataraman, Shore, Dayal, Lee, Alidoost Salimi, Crandall, Loeher, Stoops, Swanger, Eisenlord, Van Alstyne, Fast, Burge and Groner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Colleen A. Burge, Q29sbGVlbi5CdXJnZUB3aWxkbGlmZS5jYS5nb3Y=; Y2FidXJnZUB1Y2RhdmlzLmVkdQ==
†Present addresses: Sukanya Dayal, Department of Biology and Marine Biology, University of North Carolina Wilmington, Wilmington, NC, United States
James Sanghyun Lee, Front and Centered, Seattle, WA, United States
Colleen A. Burge, California Department of Fish & Wildlife, UC Davis Bodega Marine Laboratory, Bodega Bay, CA, United States
‡These authors have contributed equally to this work and share first authorship
§These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.