
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 05 May 2023
Sec. Marine Biology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1149490
This article is part of the Research TopicBiological Rhythms of Aquatic Organisms in a Changing ClimateView all 9 articles
The study aimed to gain a deeper understanding of the daily fluctuations in gene expression at a transcript level in the coral Acropora digitifera and create a comprehensive map of the biological processes that occur under natural environmental conditions. The coral is a key organism in marine ecosystems, and understanding its physiology and the adaptation mechanisms it uses to cope with daily environmental changes is vital for its survival and the preservation of coral reefs. The study’s results showed that certain genes in the coral exhibit specific patterns of expression at different times of the day. These genes play critical roles in regulating a wide range of physiological and behavioral processes, such as metabolism, development, and DNA damage repair. During the day, the coral expends energy on growth and development, and these genes are actively involved in these processes. On the other hand, at night, the coral’s focus shifts toward repair and recovery. The genes that are active during this period are involved in processes like DNA repair, hypoxia response, and transcription. This is a crucial time for the coral, as it’s exposed to a range of environmental stressors that can damage its DNA and impact its overall health. In conclusion, this study provides valuable insights into the cyclic regulatory processes that help the coral adapt to daily external variations and sustain its physiology. It highlights the importance of understanding the daily rhythms of gene expression in marine organisms and the role they play in maintaining the health of coral reefs. This research can be used to develop strategies to preserve coral reefs and mitigate the effects of environmental changes on coral physiology.
Life on Earth in many biological systems is associated with rhythmical events with frequencies ranging from seconds to several years; a predictable change and periodicity that corresponds tightly to environmental cycles caused by the Earth’s rotation (Dunlap, 1999). In approximately all organisms on Earth, a mechanism of molecular timing has emerged to meet the necessity of anticipating these cyclical changes. The adaptation of an organism’s biological rhythms to environmental cues over circadian (24-hour) and circalunar (29.5 days) periods that are vital for survival, reproduction, and fitness and is generally conserved in most living organisms (Aschoff, 1960; Tessmar-Raible et al., 2011; Torres et al., 2018; Andreatta and Tessmar-Raible, 2020; Kaiser and Neumann, 2021). The biological clock regulates homeostasis by orchestrating the temporal organization of physiological processes, which is advantageous for most organisms as day-to-day rhythms of gene expression ensure that biological functions are engaged at the most appropriate time of day (Aschoff, 1960; Herzog, 2007).
Many reef-building corals reside in tropical marine waters and live in symbiosis with dinoflagellate algae (Symbiodiniaceae) that provide most of the coral’s energy requirements (Muscatine and Cernichiari, 1969; LaJeunesse et al., 2018). In tropical marine waters, reef-building corals show large diurnal changes in their physiology between day and night to cope with the transition from obtaining energy from their photosynthetic counterparts symbiotic algae during the day to zooplankton predation and respiration during the night (Falkowski et al., 1984). Corals are exposed to large variations in their environment (temperature, oxygen, nutrients, photoperiodic light, spectra, etc.), resulting in time-dependent stress to which they are exposed daily. During the day, they are subjected to high light and UV radiation (Levy et al., 2003; Ben-Zvi et al., 2019) and excessive production of reactive oxygen species (ROS) during photosynthesis (Lesser, 1996; Ayalon et al., 2019) while at night they are exposed to hypoxic conditions and a decrease in tissue pH (Gutner-Hoch et al., 2016; Maor-Landaw and Levy, 2016). In responding to the environmental conditions, corals show diurnal behavioral patterns such as tentacles expansion and contraction and tissue inflaming (Levy et al., 2001), calcification (Goreau and Goreau, 1959), and skeletal growth (Risk and Pearce, 1992). Like in other organisms, in symbiotic corals, many aspects of physiology and development are governed by the endogenous biological clock creating temporal gene expression patterns (transcript level) characterized by daily oscillations (Levy et al., 2003; Levy et al., 2011; Reitzel et al., 2013; Sorek et al., 2014). These genes usually show peaks of expression around the time at which their function is vital to regulate downstream processes (Aschoff, 1960; Dunlap, 1999; Duffield et al., 2002).
Corals also carry environmental sensing networks like GPCRs (Bhattacharyya, 2016; Rosenberg et al., 2017) that coordinate the host animal responses of temperature, light, and pH, together with a variety of stress-related pathways that determine the fate of corals under environmental stress (Bhattacharyya, 2016). The daily biological processes of corals, synchronized with the environment, are part of the evolutionary suite of strategies that have allowed them to adapt and thrive in an ever-changing world for hundreds of millions of years. To date, most studies on corals have centered on the transition between day and night from the perspective of behavior, physiology, and gene expression. As a result, there is little data regarding coral biorhythms and circadian processes which are influenced by environmental stimuli and daily changes in extrinsic conditions (Oldach et al., 2017; Neder et al., 2022). In this work, we aimed to identify the daily biological processes that occur under environmental conditions in the coral Acropora digitifera. We conducted a 24-hour experiment, collecting samples of coral nubbins at high-resolution two-hour intervals. We discovered cyclic regulatory processes that help to sustain the coral’s physiology as it undergoes adaptation to daily external variations. Our findings could elucidate what environmental cues corals respond to and adjust physiology, behavior, and biochemistry accordingly.
On March 3rd, 2017, five Acropora digitifera colonies (measuring >50 cm in diameter) were collected from Sesoko Island, Okinawa, Japan (26°38028″ N, 127°51035″ E) at a three-meter depth. Coral colonies were kept in direct flow-through seawater, pumped directly from the reef flat, and acclimatized for 72 hours before sampling. Coral tanks were placed outside and subjected to the natural diel cycle and moon phase. Sampling started on March 6 at 20:00, during the first quarter moon, and continued for 24 hours with two-hour intervals (n=13 sampling points). At each sampling point, from each colony, a nubbin (4-5 cm long) was cut, placed in aluminum foil, dipped in liquid nitrogen, and then transferred to -80°C freezers. Light (DEFI2-L; JFE Advantech Co. Ltd., Japan) and temperature (HOBO Pro V2; Onset Computer Corporation, USA) loggers were placed in the coral tanks taking measurements at five-minute intervals. At each sampling coral nubbins were taken from different parts of the colony, trying to avoid sampling from the same proximate area.
Total RNA was extracted using TRIzol reagent (Invitrogen) and a modified version of the manufacturer’s protocol that included an additional chloroform extraction and magnesium chloride precipitation overnight (Rosenberg et al., 2019).
One and a half μg RNA from each sample (used three biological replicates from each sampling point n=39) was sent for sequencing. RNA samples were prepared using the Illumina TruSeq RNA Library Preparation Kit v2, according to the manufacturer’s protocol. Libraries from each sampling point were run on lanes of an Illumina NextSeq 550 machine using the multiplexing strategy of the TruSeq V2.5 protocol. The protocol starts with poly-A selection that results in RNA selection. Single-end reads were obtained for each sample; the reads were 75 bases long. All sequencing libraries were trimmed using TrimGalore version 0.4.0 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) to remove adapters, primers, and low-quality bases.
The sequenced reads were aligned to Acropora digitifera cDNA sequences downloaded from the Okinawa Institute of Science and Technology (OIST) website (https://marinegenomics.oist.jp/gallery) (Shinzato et al., 2011), according to default settings of the Bowtie2 program (v2.2.5) (Langmead and Salzberg, 2012). Using the published transcriptome and aligning the reads, accordingly, removed all Symbiodinium contamination. Previously published annotations were used (OIST and ZoophyteBase).
To find genes that have higher variance in their expression, we implemented the method described in Macosko, et al., 2015 (Macosko et al., 2015). The mean and a dispersion measure (variance/mean) were calculated for each gene across all 39 samples, and genes were placed into 20 bins based on their average expression. Within each bin, we then z-normalized the dispersion measure of all genes within the bin, to identify outlier genes whose expression values were highly variable even when compared to genes with similar average expression. A z-score cutoff of 1.7 to identify highly variable genes was used. These highly variable genes were assigned to clusters by their expression over the time course using a Dirichlet process Gaussian process model implemented in DP_GP_cluster (McDowell et al., 2018). The MPEAR criterion was used to assess optimal clustering. These methods allowed us to identify periodic signals in the time-series data throughout the day.
We have divided the daily 24-hour day into four quarters representing the morning (06:00 am-10:00 am), afternoon (12:00 pm-16:00 pm), evening (18:00 pm-22:00 pm), and night (00:00 am-04:00 am). Based on the variance analysis and clustering method we generated gene lists presenting transcript levels at each quarter. We then used the “GoTermMapper” of Princeton University (go.princeton.edu/cgi-bin/GOTermMapper) to map the granular GO annotations for genes in a list to a set of broader, high-level GO parent terms. This allowed us to simply bin the gene list to a static set of ancestors GO terms and further identify, together with the “GoTermFinder” (go.princeton.edu/cgi-bin/GOTermFinder), the enriched biological processes at each quarter of the day.
Temperature measurements in the coral tanks showed that the highest temperatures occurred between midday and sundown (22.89°C at 13:40), and the lowest temperatures occurred between 04:00 am and mid-morning local time (19.9°C at 6:20). Previous studies showed similar pH fluctuations in the water i.e., elevated pH during the day and lower pH during the night (Middelboe and Hansen, 2007; Hofmann et al., 2011). These changes in temperature and pH are coupled with solar irradiation, with the highest temperature, pH and sunlight during midday and the lowest occurring before sunrise. For the 39 samples, RNA-seq libraries of 75-bps single end had an average of 28,721,884 reads. There were 23,029 reads with at least 10 copies per sample and 18,749 reads with at least 100 copies per sample. An average of 12,476,062 reads (43.5%) per sample were mapped to the Acropora digitifera transcriptome (Supplementary Table Sequence statistics). Based on the variance analysis and clustering method and identification of genes having a synexpression we were able to identify periodic signals in the time-series data along the day. Next, we divided the daily 24-hour day into four quarters representing morning (06:00 am-10:00 am), afternoon (12:00 pm-16:00 pm), evening (18:00 pm-22:00 pm), and night (00:00 am-04:00 am) times. Based on the expression patterns we generated gene lists of 1,454 differentially expressed genes. These highly variable genes were assigned to clusters by their expression over the time course using a Dirichlet process Gaussian process model which generated 67 gene clusters (Supplementary Table Gene annotation).
There were 48 out of 1,454 genes that showed a single peak expression during the morning (Figure 1). GO term analysis revealed that ~53% of these genes cluster to the GO term “Transport”- mainly ion transport including calcium, chloride, bicarbonate, lipid, and cholesterol. 41% of these genes cluster to the Go term “signal transduction”- mainly GPCR and Wnt signaling. Additionally, 35% of these genes cluster to the GO term “cellular protein modification process mainly protein ubiquitination and phosphorylation.
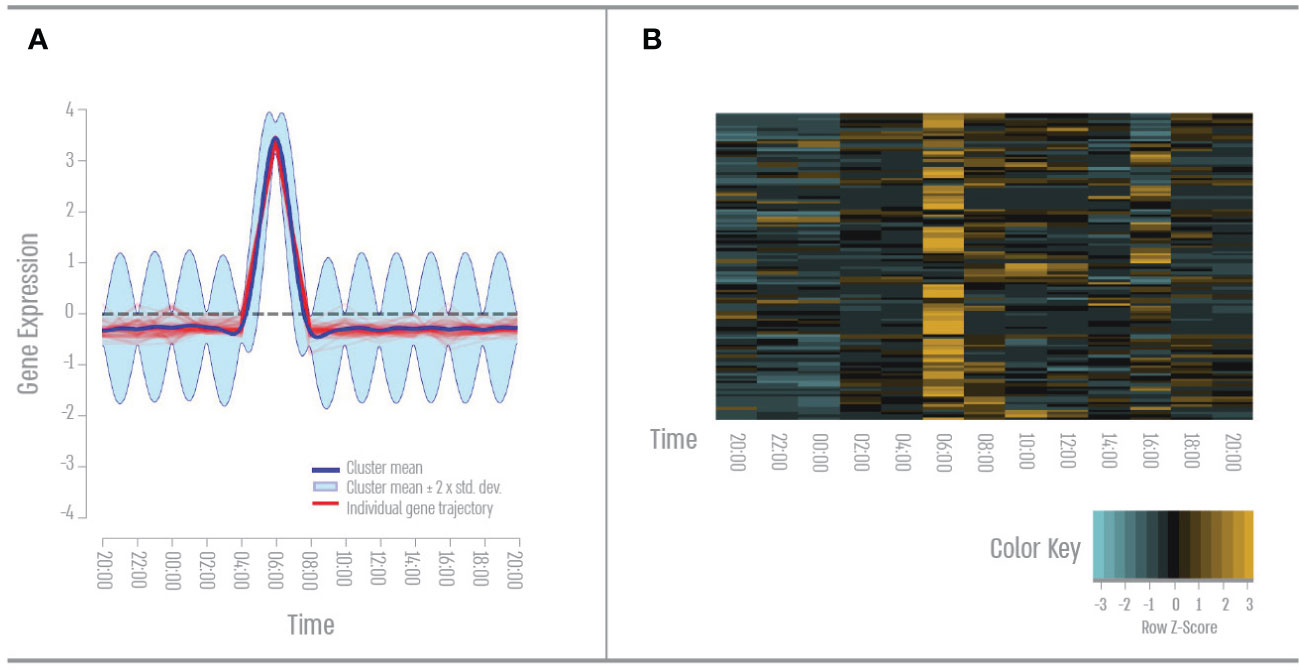
Figure 1 First quatre- Early morning gene expression pattern. (A) Expression pattern results from the DP_GP clustering (cluster 28). (B) Heatmap of morning cluster gene expression across all time points based on Z-score. The color scale indicates individual gene expression.
We found 120 genes that had a single peak expression during the afternoon (Figure 2). GO term analysis revealed that 50% of these genes cluster to the GO term “signal transduction”- mainly GPCR (rhodopsin) in response to stimulus, hormones or growth factors, apoptotic pathways in response to DNA damage such as JNK, MAPK, and NF kappa B pathways. 35% of these genes cluster to the Go term “anatomical structure development”- mainly nervous system development and ECM organization. 32% of these genes cluster to the GO term “response to stress “- mainly inflammatory response, DNA damage, and innate immune response.
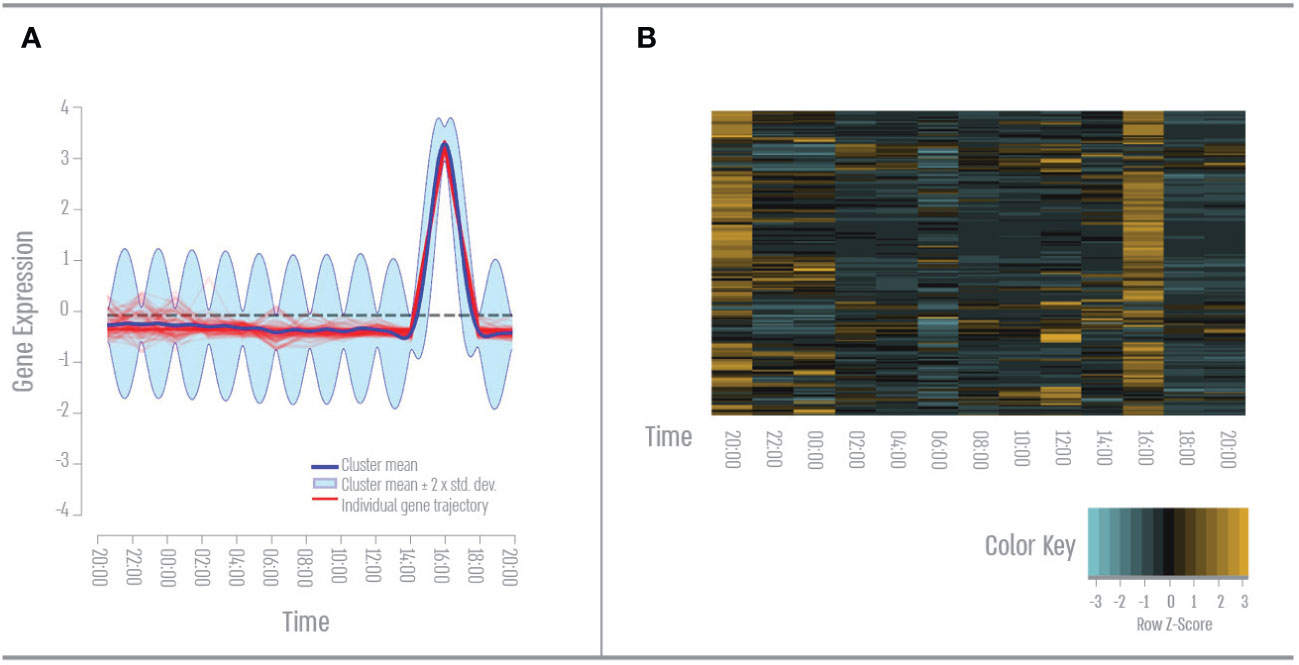
Figure 2 Second quarter – Mid-day gene expression pattern. (A) Expression pattern results from the DP_GP clustering (cluster 2). (B) Heatmap of afternoon cluster gene expression across all time points based on Z-score. The color scale indicates individual gene expression.
We found 111 genes that had a single peak expression during the evening (Figure 3). GO term analysis revealed that 46% of these genes cluster to the GO term “signal transduction”- mainly apoptotic pathways such as toll-like receptor signaling and NF kappa B. 35% of these genes cluster to the Go term “response to stress”- mainly oxidative stress and DNA damage. And 33% of these genes cluster to the GO term “cellular nitrogen compounds metabolic process “- mainly DNA repair and RNA transcription.
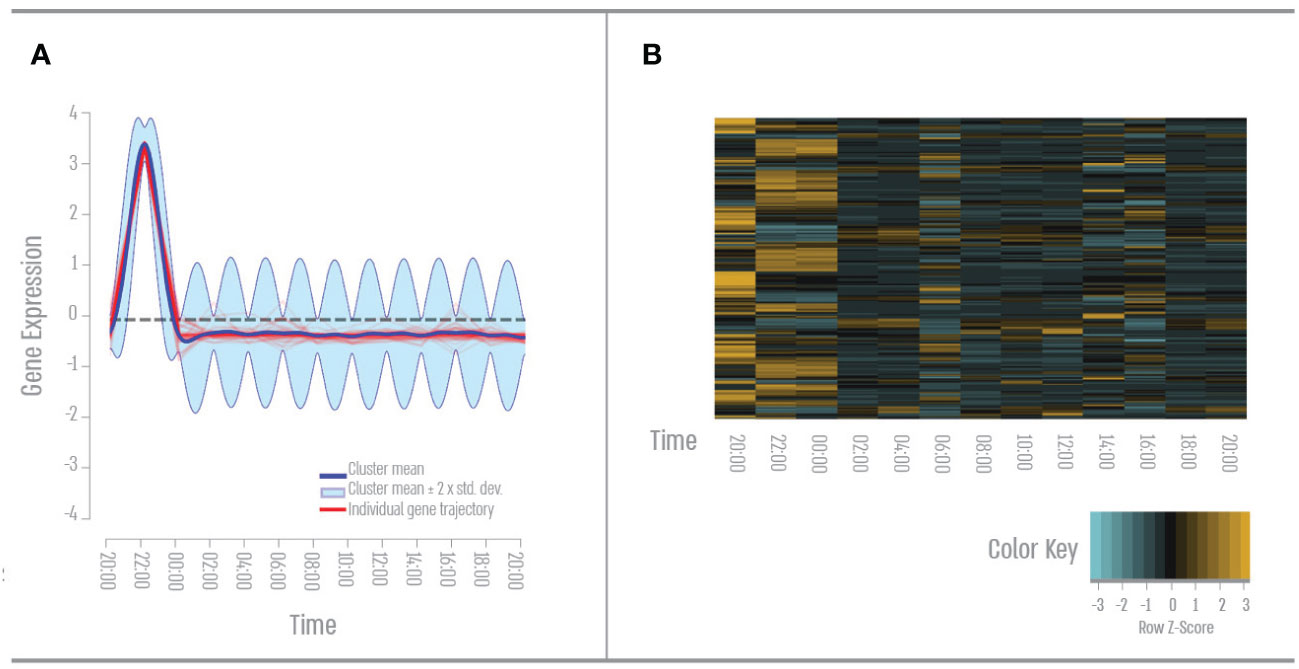
Figure 3 Third quarter – Nightfall gene expression pattern. (A) Expression pattern results from the DP_GP clustering (cluster 21). (B) Heatmap of evening cluster gene expression across all time points based on Z-score. The color scale indicates individual gene expression.
We found 61 genes with a single peak expression during the night (Figure 4). GO term analysis revealed that 40% of these genes cluster to the GO term “signal transduction”- mainly GPCR and Rho pathways in response to hormones and apoptotic pathways. 38% of these genes cluster to the Go term “response to stress”- mainly DNA repair and hypoxia. 35% of these genes cluster to the GO term “cell differentiation “- mainly neurogenesis.
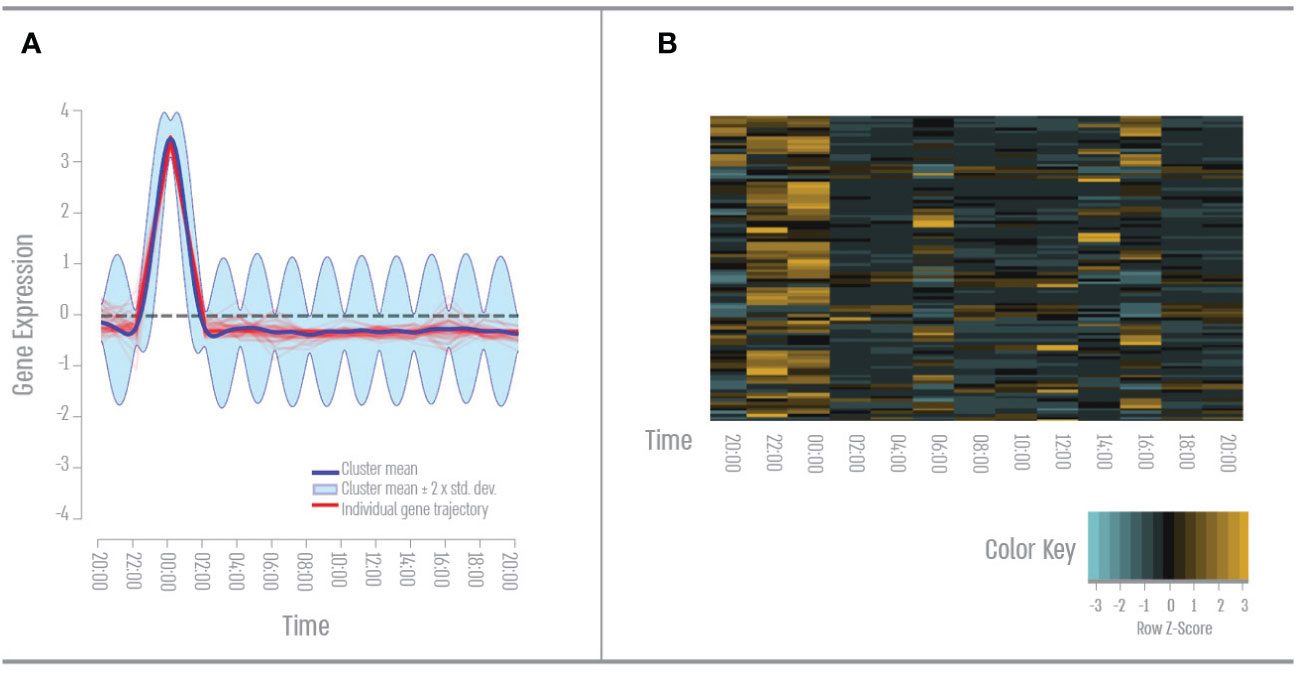
Figure 4 Nighttime gene expression pattern. (A) Expression pattern results from the DP_GP clustering (cluster 19). (B) Heatmap of nighttime cluster gene expression across all time points based on Z-score. The color scale indicates individual gene expression.
We found 19 genes that have twice-a-day expression peaks, once during the afternoon (16:00) and once during the night (00:00) (Figures 5A, B). GO term analysis revealed that 38% of these genes cluster to the GO term “biosynthetic process”- mainly RNA transcription. 38% of these genes cluster to the Go term “signal transduction”- mainly in response to stimulus, Ras, and TLR pathways. 35% of these genes cluster to the GO term “cellular nitrogen compounds metabolic process “- mainly RNA stabilization and processing. We identified a second set of genes clustering to two peak expressions a day, once during the evening (22:00) and once during the night (00:00) (Figures 5C, D). GO term analysis revealed that 44% of these genes cluster to the GO term “cell differentiation”- mainly nervous system development. 44% of these genes cluster to the Go term “signal transduction”- mainly Notch and Wnt signaling. 27% of these genes cluster to the GO term “small molecule metabolic process “- mainly amino acid metabolism.
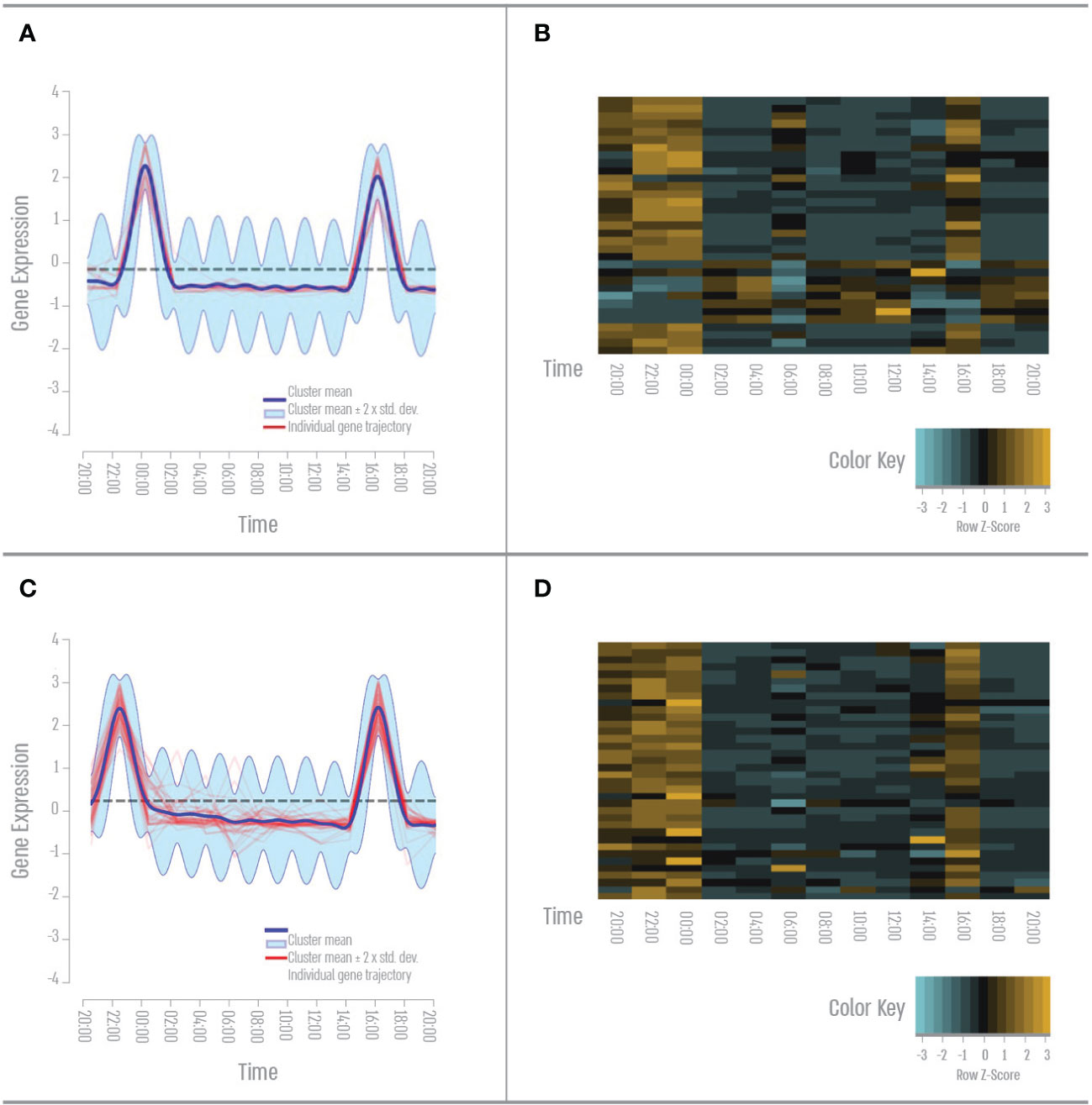
Figure 5 Bidaily gene expression pattern. (A) Afternoon and nighttime expression pattern result from the DP_GP clustering (cluster 61). (B) Heatmap of the afternoon and nighttime cluster gene expression across all time points based on Z-score. (C) Evening and nighttime Expression pattern result from the DP_GP clustering (cluster 52). (D) Heatmap of the evening and nighttime cluster gene expression across all time points based on Z-score. The color scale indicates individual gene expression.
Coral physiology and behavior have previously been shown to oscillate between day and night (Risk and Pearce, 1992; Levy et al., 2001; Levy et al., 2003; Moya, 2006; Reitzel et al., 2013; Gutner-Hoch et al., 2016; Oldach et al., 2017; Neder et al., 2022), but the cellular response linking external environmental conditions to internal gene expression throughout the day is relatively vague. The transcriptomic differences throughout the day reflect the response of the coral to irradiance, oxygen, and nutrient availability. In A. digitifera, genes that were significantly different between the various quarters of the day were involved in a range of processes (Figure 6), including signal transduction (mainly GPCR), mediation of stress responses and immunity, lipid metabolism, ion transport. In the morning, when light intensity is lower, as well as temperature and pH levels (Middelboe and Hansen, 2007; Hurd et al., 2011), we found an enhancement of biological processes involved in ion transport and metabolism. A recent review of the transcriptomic responses of marine metazoans to ocean acidification found a change in metabolic processes and ion transport and suggested a trade-off between ion homeostasis and other processes such as calcification (Strader et al., 2020). Low aragonite saturation in the early morning (Zoccola et al., 2015; Glazier et al., 2020) may be the reason for the upregulation of ion transport processes that we observed to increase pH and transport of bicarbonate to increase carbonate ion concentration in coral tissue. This trade-off with a decrease in calcification rates is also evident in the increase in genes involved in the Wnt signaling process, which has previously been shown to increase with the decrease in the biomineralization process (Kaniewska et al., 2015) (Figures 1, 6).
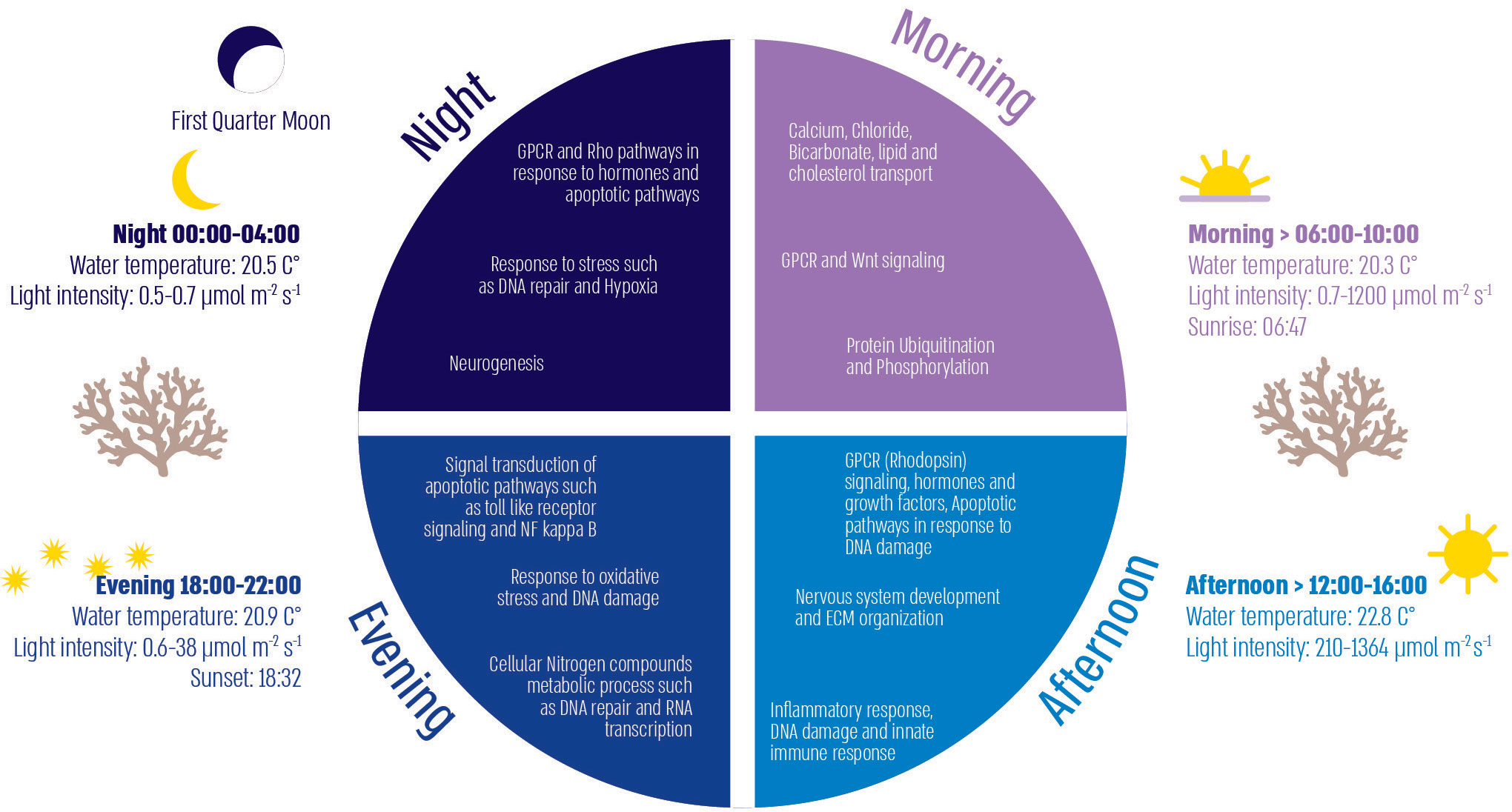
Figure 6 Illustration of diel environmental factors and cellular processes divided into the four quarters of the day.
Previous studies have shown that symbiotic corals respond to diurnal patterns of key environmental cues of temperature and light via physiological and cellular pathways (Levy et al., 2006). It is known that the net oxygen evolution rate tracks the diurnal pattern of solar energy flux, which means that in the afternoon quarter of the day, the oxygen flux and solar radiation acting on the coral host are highest (Levy et al., 2006; Sorek et al., 2014). From our results, it appears that coral biological processes in the afternoon consist mainly of light-related signal transduction, which is conspicuous by the high expression of rhodopsin GPCRs, known for their involvement in phototransduction in cnidarians (Kaniewska et al., 2015; Rosenberg et al., 2017). High solar radiation also leads to high photosynthesis performances, which might explain our results of increased development and ECM organization during the afternoon, as the coral host is known to use a high proportion of the energy supplied by photosynthesis for growth and development processes (Houlbrèque et al., 2004). Additionally, solar radiation causes DNA damage which could explain our results showing the increase in the apoptotic response to DNA damage and innate immune response activity during the afternoon (Lesser and Farrell, 2004) (Figures 2, 6).
The evening time associated with sunset brings many shifts in the transcriptomic response of corals. We found that at this time of day, corals show a response to oxidative stress due to the known hypoxic conditions resulting from the lack of photosynthesis and respiration after dusk (Shashar et al., 1993; Jones and Hoegh-Guldberg, 2001). Oxidative stress in the form of hypoxia can also induce DNA damage, this leads to the enrichment in processes involving the response to DNA damage during the evening and eventually to DNA repair and RNA transcription, both of them as part of the “transcription-coupled DNA repair”, a known mechanism that reacts to oxidative DNA damage (Møller et al., 2001; Menoni et al., 2018).
Nighttime gene expression continues to respond to the stress coupled with hypoxic tissue conditions. Additionally, we found that corals express the biological process of “signal transduction” in response to hormones, this correlates with previous studies showing that endocrine signaling in cnidarians is orders of magnitude higher during periods of darkness (Tarrant, 2005). At nighttime, we also found many genes involved in cell differentiation, specifically neurogenesis. Neurogenesis in cnidarians probably refers to the development of the nervous system and sensory cells (Galliot et al., 2009), both neurogenesis and cell proliferation are known to show higher rates during periods of darkness in most organisms (Jacobs, 2002; Grassi Zucconi et al., 2006).
When looking at the results of gene expression with two peak expressions a day we can see two trends. The first, is genes that have a peak in the late evening (22:00) and early night (00:00), with enrichment of biological processes that conform to the dark period of the day and are most likely continued across the night. The second trend, genes that have peaked during the late afternoon (16:00) and early night (00:00) are enriched with processes that are not noticeable in any quarter of the day alone. These processes involve multiple steps of the RNA life cycle, such as RNA transcription and stabilization, that are all controlled by the circadian timekeeping system. The circadian system maintains the correct period, phase, and amplitude of the rhythms of thousands of proteins that generate a wide range of rhythmic biological processes (Aschoff, 1960; Reitzel et al., 2013; Torres et al., 2018). Thus, the corals circadian gene expression is tightly regulated at several steps as we notice in their separation along the day.
The physiology of corals follows the light-dark cycle, alternating between high energy during the day and low energy at night (Levy et al., 2006; Reitzel et al., 2013; Sorek et al., 2014). External signals enable the coral’s to anticipate intake, processing, energy consumption, digestive processes, and cell repair. The temporal signals synchronize the biological clock, which orchestrates functions and behavior in a timely manner. The daily processes we observe in A. digitifera are necessary for adaptation and homeostasis. When circadian processes are perturbed, for example when corals are subjected to artificial light or anthropogenic stressors, individuals cannot anticipate and respond efficiently to the daily challenges associated with the day-night cycle (Shlesinger and Loya, 2019; Ayalon et al., 2021; Rosenberg et al., 2022). In this paper, we highlight the synchronicity between the daily environment-imposed challenges and their harmonization with the biological processes of the corals. We found that many time-dependent processes in A. digitifera are in line with those presented by Neder et al. (2022) for S. pistillata. Our analysis of gene expression profiles revealed similar synexpression patterns as Neder et al. (2022), particularly in terms of an extensive up-regulation of genes associated with reactive oxygen species, redox, metabolism, and ion transporters. We also found correlation with S. pistillata gene expression in terms of up-regulation of genes associated with tissue development, cellular movement, antioxidants, and protein synthesis. We were able to distinguish and map the processes that occur throughout the day in A. digitifera corals under baseline, natural environmental conditions. This information will serve as a valuable resource for future research to compare changes in the biological processes of corals under artificial conditions and determine the level of disturbance that allows corals to maintain homeostasis in an ever-changing ocean.
The sequencing data reported in this study has been deposited to the Sequence Read Archive (SRA), under accession PRJNA922686: “Daily temporal homeostasis in the coral Acropora digitifera”. Correspondence and requests for materials should be addressed to YR eWFlbGlyb3NlQGdtYWlsLmNvbQ== or OL b3Jlbi5sZXZ5QGJpdS5hYy5pbA==.
Ethical review and approval was not required for the animal study because Corals are invertebrates and are not subject to the ethical committee’s approval.
YR and OL designed the research, and the experiment was carried out by YR, OL, SH, and FS. SL performed RNA sequencing. TD and YR performed data analysis. YR and OL wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
The research leading to this paper has received funding from the Israeli Science Foundation (ISF), grant number 3928 to OL.
We thank the entire staff and students of the “Tropical Biosphere Research Center” in Sesoko Island for their help and support, especially the Harii lab team members.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1149490/full#supplementary-material
Andreatta G., Tessmar-Raible K. (2020). The still dark side of the moon: molecular mechanisms of lunar-controlled rhythms and clocks. J. Mol. Biol. 432, 3525–3546. doi: 10.1016/j.jmb.2020.03.009
Aschoff J. (1960). Exogenous and endogenous components in circadian rhythms. Cold Spring Harb. Symp. Quant. Biol. 25, 11–28. doi: 10.1101/SQB.1960.025.01.004
Ayalon I., Rosenberg Y., Benichou J. I. C., Campos C. L. D., Sayco S. L. G., Nada M. A. L., et al. (2021). Coral gametogenesis collapse under artificial light pollution. Curr. Biol. 31, 413–419.e3. doi: 10.1016/j.cub.2020.10.039
Ayalon I., de Barros Marangoni L. F., Benichou J. I., Avisar D., Levy O. (2019). Red sea corals under artificial light pollution at night (ALAN) undergo oxidative stress and photosynthetic impairment. Global Change Biol. 25 (12), 4194–4207.
Ben-Zvi O., Eyal G., Loya Y. (2019). Response of fluorescence morphs of the mesophotic coral euphyllia paradivisa to ultra-violet radiation. Sci. Rep. 9 (1), 5245.
Bhattacharyya S. (2016). Inside story of group i metabotropic glutamate receptors (mGluRs). Int. J. Biochem. Cell Biol. 77, 205–212.
Duffield G. E., Best J. D., Meurers B. H., Bittner A., Loros J. J., Dunlap J. C. (2002). Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 12 (7), 551–557.
Falkowski P. G., Dubinsky Z., Muscatine L., Porter J. W. (1984). Light and the bioenergetics of a symbiotic coral. Bioscience 34 (11), 705–709.
Galliot B., Quiquand M., Ghila L., de Rosa R., Miljkovic-Licina M., Chera S. (2009). Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24. doi: 10.1016/j.ydbio.2009.05.563
Glazier A., Herrera S., Weinnig A., Kurman M., Gómez C. E., Cordes E. (2020). Regulation of ion transport and energy metabolism enables certain coral genotypes to maintain calcification under experimental ocean acidification. Mol. Ecol. 29, 1657–1673. doi: 10.1111/mec.15439
Grassi Zucconi G., Cipriani S., Balgkouranidou I., Scattoni R. (2006). ‘One night’ sleep deprivation stimulates hippocampal neurogenesis. Brain Res. Bull. 69, 375–381. doi: 10.1016/j.brainresbull.2006.01.009
Goreau T. F., Goreau N. I. (1959). The physiology of skeleton formation in corals. II. calcium deposition by hermatypic corals under various conditions in the reef. Biol. Bull. 117 (2), 239–250.
Gutner-Hoch E., Schneider K., Stolarski J., Domart-Coulon I., Yam R., Meibom A., et al. (2016). Evidence for rhythmicity pacemaker in the calcification process of scleractinian coral. Sci. Rep. 6, 20191. doi: 10.1038/srep20191
Herzog E. D. (2007). Neurons and networks in daily rhythms. Nat. Rev. Neurosci. 8, 790–802. doi: 10.1038/nrn2215
Hofmann G. E., Smith J. E., Johnson K. S., Send U., Levin L. A., Micheli F., et al. (2011). High-frequency dynamics of ocean pH: a multi-ecosystem comparison. PloS One 6, e28983. doi: 10.1371/journal.pone.0028983
Houlbrèque F., Tambutté E., Allemand D., Ferrier-Pagès C. (2004). Interactions between zooplankton feeding, photosynthesis and skeletal growth in the scleractinian coral stylophora pistillata. J. Exp. Biol. 207, 1461–1469. doi: 10.1242/jeb.00911
Hurd C. L., Cornwall C. E., Currie K., Hepburn C. D., McGraw C. M., Hunter K. A., et al. (2011). Metabolically induced pH fluctuations by some coastal calcifiers exceed projected 22nd century ocean acidification: a mechanism for differential susceptibility? Glob. Change Biol. 17, 3254–3262. doi: 10.1111/j.1365-2486.2011.02473.x
Jacobs B. L. (2002). Adult brain neurogenesis and depression. Brain. Behav. Immun. 16, 602–609. doi: 10.1016/S0889-1591(02)00015-6
Jones R. J., Hoegh-Guldberg O. (2001). Diurnal changes in the photochemical efficiency of the symbiotic dinoflagellates (Dinophyceae) of corals: photoprotection, photoinactivation and the relationship to coral bleaching. Plant Cell Environ. 24, 89–99. doi: 10.1046/j.1365-3040.2001.00648.x
Kaiser T. S., Neumann J. (2021). Circalunar clocks–old experiments for a new era. BioEssays 43, 2100074. doi: 10.1002/bies.202100074
Kaniewska P., Chan C.-K. K., Kline D., Ling E. Y. S., Rosic N., Edwards D., et al. (2015). Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PloS One 10, e0139223. doi: 10.1371/journal.pone.0139223
LaJeunesse T. C., Parkinson J. E., Gabrielson P. W., Jeong H. J., Reimer J. D., Voolstra C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28 (16), 2570–2580.
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359.
Lesser M. P. (1996). Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in ymbiotic dinoflagellates. Limnology oceanography 41 (2), 271–283.
Lesser M. P., Farrell J. H. (2004). Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs 23, 367–377. doi: 10.1007/s00338-004-0392-z
Levy O., Achituv Y., Yacobi Y. Z., Dubinsky Z., Stambler N. (2006). Diel `tuning’ of coral metabolism: physiological responses to light cues. J. Exp. Biol. 209, 273–283. doi: 10.1242/jeb.01983
Levy O., Dubinsky Z., Achituv Y. (2003). Photobehavior of stony corals: responses to light spectra and intensity. J. Exp. Biol. 206, 4041–4049. doi: 10.1242/jeb.00622
Levy O., Mizrahi L., Chadwick-Furman N. E., Achituv Y. (2001). Factors controlling the expansion behavior of favia favus (Cnidaria: scleractinia): effects of light, flow, and planktonic prey. Biol. Bull. 200, 118–126. doi: 10.2307/1543305
Levy O., Kaniewska P., Alon S., Eisenberg E., Karako-Lampert S., Bay L. K., et al. (2011). Complex diel cycles of gene expression in coral-algal symbiosis. Science 331 (6014), 175–175.
Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161 (5), 1202–1214.
Maor-Landaw K., Levy O. (2016). Gene expression profiles during short-term heat stress; branching vs. massive scleractinian corals of the red sea. PeerJ 4, e1814.
McDowell I. C., Manandhar D., Vockley C. M., Schmid A. K., Reddy T. E., Engelhardt B. E. (2018). Clustering gene expression time series data using an infinite gaussian process mixture model. PloS Comput. Biol. 14 (1), e1005896.
Møller P., Loft S., Lundby C., Olsen N. V. (2001). Acute hypoxia and hypoxic exercise induce DNA strand breaks and oxidative DNA damage in humans. FASEB J. 15, 1181–1186. doi: 10.1096/fj.00-0703com
Menoni H., Wienholz F., Theil A. F., Janssens R. C., Lans H., Campalans A., et al. (2018). The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 46, 7747–7756. doi: 10.1093/nar/gky579
Middelboe A. L., Hansen P. J. (2007). High pH in shallow-water macroalgal habitats. Mar. Ecol. Prog. Ser. 338, 107–117. doi: 10.3354/meps338107
Moya A. (2006). Study of calcification during a daily cycle of the coral stylophora pistillata: implications for `light-enhanced calcification’. J. Exp. Biol. 209, 3413–3419. doi: 10.1242/jeb.02382
Muscatine L., Cernichiari E. (1969). Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol. Bull. 137 (3), 506–523.
Neder M., Saar R., Malik A., Antler G., Mass T. (2022). New insights on the diurnal mechanism of calcification in the stony coral, stylophora pistillata. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.745171
Oldach M. J., Workentine M., Matz M. V., Fan T.-Y., Vize P. D. (2017). Transcriptome dynamics over a lunar month in a broadcast spawning acroporid coral. Mol. Ecol. 26, 2514–2526. doi: 10.1111/mec.14043
Reitzel A. M., Tarrant A. M., Levy O. (2013). Circadian clocks in the cnidaria: environmental entrainment, molecular regulation, and organismal outputs. Integr. Comp. Biol. 53, 118–130. doi: 10.1093/icb/ict024
Risk M. J., Pearce T. H. (1992). Interference imaging of daily growth bands in massive corals. Nature 358, 572–573. doi: 10.1038/358572a0
Rosenberg Y., Doniger T., Harii S., Sinniger F., Levy O. (2017). Canonical and cellular pathways timing gamete release in acropora digitifera, Okinawa, Japan. Mol. Ecol. 26, 2698–2710. doi: 10.1111/mec.14062
Rosenberg Y., Doniger T., Levy O. (2019). Sustainability of coral reefs are affected by ecological light pollution in the gulf of Aqaba/Eilat. Commun. Biol. 2 (1), 289.
Rosenberg Y., Simon-Blecher N., Lalzar M., Yam R., Shemesh A., Alon S., et al. (2022). Urbanization comprehensively impairs biological rhythms in coral holobionts. Glob. Change Biol. 28, 3349–3364. doi: 10.1111/gcb.16144
Shashar N., Cohen Y., Loya Y. (1993). Extreme diel fluctuations of oxygen in diffusive boundary layers surrounding stony corals. Biol. Bull. 185, 455–461. doi: 10.2307/1542485
Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., et al. (2011). Using the acropora digitifera genome to understand coral responses to environmental change. Nature 476 (7360), 320–323.
Shlesinger T., Loya Y. (2019). Breakdown in spawning synchrony: a silent threat to coral persistence. Science 365, 1002–1007. doi: 10.1126/science.aax0110
Sorek M., Díaz-Almeyda E. M., Medina M., Levy O. (2014). Circadian clocks in symbiotic corals: the duet between symbiodinium algae and their coral host. Mar. Genomics 14, 47–57. doi: 10.1016/j.margen.2014.01.003
Strader M. E., Wong J. M., Hofmann G. E. (2020). Ocean acidification promotes broad transcriptomic responses in marine metazoans: a literature survey. Front. Zool. 17, 7. doi: 10.1186/s12983-020-0350-9
Tarrant A. M. (2005). Endocrine-like signaling in cnidarians: current understanding and implications for Ecophysiology1. Integr. Comp. Biol. 45, 201–214. doi: 10.1093/icb/45.1.201
Tessmar-Raible K., Raible F., Arboleda E. (2011). Another place, another timer: marine species and the rhythms of life. BioEssays 33, 165–172. doi: 10.1002/bies.201000096
Torres M., Becquet D., Franc J.-L., François-Bellan A.-M. (2018). Circadian processes in the RNA life cycle. WIREs RNA 9, e1467. doi: 10.1002/wrna.1467
Keywords: coral reefs, biological clock, gene expression, circadian, homeostasis
Citation: Rosenberg Y, Doniger T, Lampert S, Sinniger F, Harii S and Levy O (2023) Daily temporal homeostasis in the coral Acropora digitifera. Front. Mar. Sci. 10:1149490. doi: 10.3389/fmars.2023.1149490
Received: 22 January 2023; Accepted: 21 April 2023;
Published: 05 May 2023.
Edited by:
Audrey Mat, University of Vienna, AustriaReviewed by:
Tali Mass, University of Haifa, IsraelCopyright © 2023 Rosenberg, Doniger, Lampert, Sinniger, Harii and Levy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaeli Rosenberg, eWFlbGlyb3NlQGdtYWlsLmNvbQ==; Oren Levy, T3Jlbi5MZXZ5QGJpdS5hYy5pbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.