- 1Department of Marine Science, Incheon National University, Incheon, Republic of Korea
- 2Research Institute of Basic Sciences, Incheon National University, Incheon, Republic of Korea
- 3Department of Ecology and Evolutionary Biology, University of Connecticut, Stamford, CT, United States
Introduction: Heat stress is one of the main threats for the seaweed aquaculture, leading to the decay of the blades and causing serious economic losses. A newly developed biostimulant, Sargassum horneri extract (SHE), can enhance the heat-tolerance in Neopyropia yezoensis, which can be applied in the seaweed aquaculture. However, the molecular mechanism underlying this acquired heat-tolerance is still unclear.
Method: In this study, we examined the transcriptional regulation of N. yezoensis under heat stress after the SHE pre-treatment by performing the RNA-Seq on illumina Novaseq platform and differentially expressed gene analysis.
Result and discussion: Our results show that, under heat stress, the SHE pre-treatment can alleviate the suppression of genes involved in normal cell activities, such as ribosome biogenesis, photosynthesis, respiration and cell cycle. On the other hand, SHE pre-treatment can also induce some key genes involved in the heat-tolerance. We also found that the combination of SHE pre-treatment and heat stress can induce some genes related to epigenetic modification, suggesting that SHE may facilitate the formation of heat stress memory in N. yezoensis. Our study provides information for understanding the molecular mechanism of heat-tolerance in N. yezoensis and for promoting the application of seaweed-derived biostimulants in the seaweed aquaculture.
1 Introduction
Neopyropia yezoensis is the commercial seaweed species that has been widely cultivated in Asian countries. In 2019, Korea was the world’s second-largest N. yezoensis producer behind China, with an annual production of 606,873 t (FAO, 2020). The cultivation of N. yezoensis in Korea has a long history, which started between 1623 and 1649 (Chung, 1937; Bae, 1991; Sohn, 1996). In the past few centuries, there has been much progress to maximize production in N. yezoensis aquaculture, such as the improvement of cultivation methods and selective breeding of high production strains (Hwang and Park, 2020). However, seaweed aquaculture is being threatened by increases in ocean temperature due to global warming (Kim et al., 2007; Tang et al., 2011; Duarte et al., 2017). According to the prediction of IPCC, the direct CO2 emission from human activities will double or even triple by 2050, resulting in global mean surface temperature increases in 2100 from 3.7°C to 4.8°C compared to pre-industrial levels (Grubb et al., 2022). As a cold-water species, N. yezoensis has a narrow range of optimal growth temperature at around 10°C (Yamamoto et al., 1991; Redmond et al., 2014). Increasing temperatures can reduce the production and quality of seaweed. For instance, elevating temperature from 10°C to 20°C has decreased growth rates, phycoerythrin content and nitrogen removal rates in the species, Neopyropia leucosticta, Porphyra linearis and Porphyra umbilicalis (Kim et al., 2007). In the past decade, several heat-tolerant strains of N. yezoensis have been developed for the Neopyropia aquaculture (Ding et al., 2016; Shin et al., 2018). However, there are several findings that domestication resulted in the reduction in genetic diversity of the cultivated seaweed population (Voisin et al., 2005; Niwa and Aruga, 2006).
Seaweed-derived biostimulants are crude extracts of seaweeds containing variety of growth regulators and hormones, which have been widely used in agriculture for decades to enhance the growth and resistance to biotic and abiotic stresses in crops (Hurtado and Critchley, 2018). Recently, it has been found that seaweed-derived biostimulants can enhance the heat-tolerance in cultivated seaweed species. For instance, pre-treatment with a commercial biostimulant, Ascophyllum nodosum (Phaeophyceae) marine plant extract powder (AMPEP, Acadian Seaplants, LLC.), significantly increased the survival and growth rates of two kelp species Saccharina latissima and Saccharina angustissima under a sub-lethal temperature of 18°C (Umanzor et al., 2020). Enhancement effects of AMPEP on stress resistance have also been reported in red alga Kappaphycus alvarezii and Eucheumatopsis isiformis (Hurtado and Critchley, 2018). Similarly, the other commercial biostimulant, Kelpak® extracted from Ecklonia maxima, can also increase the heat-tolerance in S. latissima and S. angustissima (Umanzor et al., 2019). In addition to commercial biostimulants, some new biostimulants have been developed and tested in seaweed cultivation. Our previous study tested the effects of extraction methods on the activity of biostimulants from the bloom-forming brown alga, Sargassum horneri, and found the biostimulant extracted using boiling and autoclaving methods significantly enhanced the heat-tolerance of N. yezoensis gametophyte at 20°C (Han et al., 2022). The results suggest S. horneri extract has potential application in seaweed aquaculture. However, the molecular mechanism of heat-tolerance induced by seaweed-derived biostimulants is still unclear in seaweed.
Several studies have been conducted to explore the molecular mechanisms of heat-tolerance in seaweeds (van Oppen and Coleman, 2022). Wang et al. compared the transcriptomic regulation in heat-tolerant and heat-sensitive strains of red alga, Neoporphyra haitanensis, revealing that energy metabolism, antioxidant systems, and phosphatidylinositol signal transduction played important role in its heat-tolerance (Wang et al., 2018). Recently, the overexpression of a N. haitanensis RING-type ubiquitin ligase gene, PhCUL4, significantly enhanced the heat-tolerance in Chlamydomonas reinhardtii (Wang et al., 2021). Additionally, a N. yezoensis gene (PyHRG1), encoding a novel secretory protein, was downregulated in heat-tolerant mutant Gy500, suggesting that it might be important for the heat-tolerance (Han et al., 2021). However, the research of heat-tolerance mechanism in seaweeds is still in the early stage. In this study, we explored the transcriptional mechanism of heat-tolerance induced by Sargassum horneri extract (SHE) in N. yezoensis. Our results will increase our knowledge about the molecular mechanism of heat-tolerance in seaweed and provide evidence that SHE can be applied to enhance the heat-tolerance in the N. yezoensis aquaculture.
2 Materials and methods
2.1 Seaweed material
The N. yezoensis strain used in this study was NY-HN-ST1 which was collected at a commercial seaweed farm at Haenam, Korea (34°57′N, 126°60′E) in December 2020. This strain has been artificially selected with no evidence of heat tolerance. It has been isolated and vegetatively propagated under optimal growth conditions in the Marine Ecology and Green Aquaculture (MEGA) Laboratory, Incheon National University. The optimal growth conditions for gametophytes were 10°C, 90 ± 10 μmol m−2 s−1 photosynthetically active radiation (PAR) provided by cool white, fluorescent bulbs, 12:12 L:D, and 30 psu of salinity. von Stosch enriched (VSE) artificial seawater was used as culture medium (Ott, 1966).
2.2 Preparation of biostimulants
Thalli of Sargassum horneri was collected at Jumunjin, Gangwon, Korea (37°90′N, 128°83′E) in June, 2020 and cleaned using tap water to remove sand, epiphytes, meio- and macrofauna. The thalli were mature and productive organs were observed. The clean thalli were then dried under 60°C and ground finely into powder using MM400 ball mill (Retsch, Germany). The biostimulant was prepared using the autoclaving method described in our previous study (Han et al., 2022). In brief, 8 g seaweed powder was added into 800 mL distilled water and stirred. The mixture was autoclaved at 121°C for 15 min. After cooling down, the mixture was centrifuged and the supernatant was obtained as the stock solution of SHE with the concentration of 10 g L-1. The stock solution of SHE was stored at 4°C.
2.3 Experimental design
Gametophytes of N. yezoensis were selected from cultivation stock and divided into four groups: control (C10), SHE pretreatment (S10), high temperature stress (C20) and SHE pretreatment + high temperature stress (S20). Gametophytes of S10 and S20 groups were transferred into VSE seawater in 500 mL flasks with SHE added to a final concentration of 1 g L-1. The pretreatment lasted for 10 days at 10°C. At the same time, gametophytes of C10 and C20 groups were cultured in 500 mL flask with VSE seawater at 10°C for 10 days. After pretreatment, gametophytes of each group were rinsed several times and transferred into new 500 mL flasks with VSE seawater. Each group had triplicates and 0.1 g gametophytes were used in each replicate. C10 and S10 groups were cultured at 10°C. C20 and S20 groups were transferred to 20°C. The experimental treatment lasted for 5 days. Then, samples were dried with paper towel and frozen in liquid nitrogen. Frozen samples were stored at -80°C for RNA extraction.
2.4 RNA extraction and sequencing
Total RNA was extracted using RNeasy® Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. The quality and quantity of RNA samples were examined using agarose gel electrophoresis, NanoDrop™ Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, USA) and the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The sequencing libraries were constructed following the protocol of Illumina TruSeq Stranded mRNA Library construction and sequenced using Illumina NovaSeq platform with 100 bp pair-end reads.
2.5 RNA-seq data analysis
The quality of sequencing raw reads was checked using FastQC v0.11.9 (Andrews, 2010). Then, the low quality (Phred score < 33), short reads (<50 nucleotides) and sequencing adapters were removed using Trimmomatic v0.39 (Bolger et al., 2014). The clean reads were mapped to the reference genome of N. yezoensis (Wang et al., 2020) using HISAT2 v2.2.1 (Kim et al., 2015). The genome annotations were combined with newly predicted transcripts by StringTie v2.1.2 (Kim et al., 2015) to form the final transcriptome set. Reads were remapped to the final transcriptome and quantified by StringTie. The count matrix of transcripts was extracted by a python script named prepDE.py (http://ccb.jhu.edu/software/stringtie/dl/prepDE.py) and used as input file of DESeq2 R package (Love et al., 2014) to identify differentially expressed genes (DEGs) with a threshold of |log2(fold change)| > 1 and FDR significance score (padj) < 0.05.
The transcriptome annotation was performed using Blast v2.12.0 against NCBI non-redundant (Nr) protein database (2022-7-17) and Swissprot protein database (2022-02) with an e-value cut-off of 10-5. The annotation with Kyoto Encyclopedia of Genes and Genomes (KEGG) database was performed on the online KEGG Automatic Annotation Server (http://www.genome.jp/kegg/kaas/). The Blast2GO software (Conesa et al., 2005) was used for the Gene Ontology (GO) annotation. The KEGG and GO enrichment of DEGs were performed using TBtools (Chen et al., 2020) with a adjust p-value cut-off of 0.05. The heatmap was drawn using ComplexHeatmap R package (Gu, 2022).
3 Result
3.1 Assembled transcriptome and differentially expressed genes
A total of 1.08 × 109 clean reads with an average length of 100 bp were generated from the RNA sequencing performed by Illumina NovaSeq platform. After assembly, a transcriptome of 23,663 transcripts were obtained, including 11,113 novel transcripts predicted by StringTie. Among those transcripts, 16,772 transcripts were annotated by at least one database and the overall annotation ratio of obtained transcriptome was 70.8%. In total, the differentially expressed gene (DEG) analysis identified 6,999 DEGs among three treatment conditions compared to control, in which 3,754 DEGs were up-regulated and 3,245 DEGs were down-regulated. The C20 group had the most DEGs in three conditions with 2,841 up-regulated and 2,383 down-regulated genes, followed by S20 group with 2,287 up-regulated and 2,040 down-regulated genes. S10 group induced the least DEGs in three conditions with 740 up-regulated and 718 down-regulated genes. The comparison analysis showed that 132 up-regulated and 183 down-regulated genes were identified in all three conditions (Figure 1A). Most of DEGs in C20 and S20 group were same, including 1,753 up-regulated and 1,393 down-regulated genes (Figure 1A). Only a few DEGs were common in S10 group and the other two groups under 20 °C (Figure 1A).
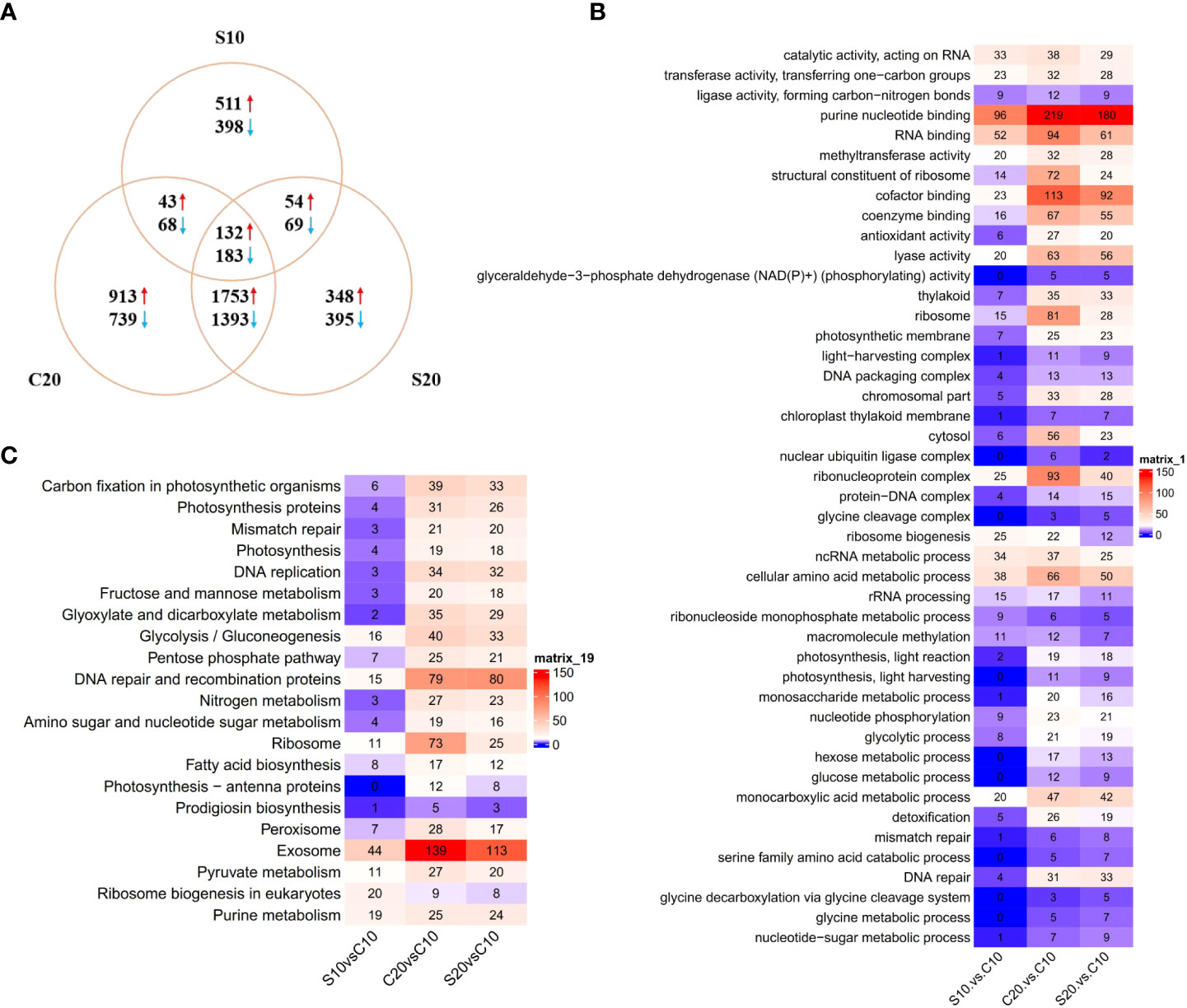
Figure 1 (A) Venn diagram showing number of DEGs found in S10, C20 and S20 groups. (B) Heatmap of GO enrichment results in S10, C20 and S20 groups. (C) Heatmap of KEGG enrichment results in S10, C20 and S20 groups.
3.2 GO and KEGG enrichment analysis of DEGs
According to the result of GO enrichment analysis, 45 terms were enriched by DEGs in at least one conditions, including 12 terms in “molecular function”, 12 terms in “cellular component” and 21 terms in “biological process” (Figure 1B; Table S1). Among those GO terms, 6 terms in “molecular function” were enriched by DEGs in S10 group and some of them were related to nucleic acid binding and metabolism, such as “RNA binding”, “purine nucleotide binding” and “catalytic activity, acting on RNA”. Additionally, “methyltransferase activity” was also enriched in S10 group. The 10 terms of “biological process” enriched in S10 group were related to ribosome biogenesis, noncoding RNA (ncRNA) metabolism, amino acid metabolism and macromolecule methylation. Similar to the result of KEGG analysis, C20 and S20 groups had many enriched GO terms in common (Figure 1B). In addition to those GO terms, several GO terms were only enriched in C20 group, including “hexose metabolic process”, “glucose metabolic process”, “detoxification”, “antioxidant activity”, “monosaccharide biosynthetic process”, “ribosome” and “nuclear ubiquitin ligase complex” (Figure 1B). Moreover, several GO terms related to fatty acid metabolism and DNA repair were only enriched in S20 group, such as “serine family amino acid catabolic process”, “glycine metabolic process”, “DNA repair” and “mismatch repair” (Figure 1B).
The KEGG enrichment analysis showed that 21 terms were significantly enriched by identified DEGs in at least one condition (Figure 1C; Table S2). In most of those terms, S10 group had much less DEGs than other two groups. However, in the term “ribosome biogenesis in eukaryotes”, S10 group had more DEGs enriched that C20 and S20 groups. Additionally, the term “purine metabolism” was also enriched by DEGs in S10 group. The enriched KEEG terms were similar in C20 and S20 groups. Nevertheless, several KEGG terms were only enriched by DEGs in C20 condition, such as “ribosome”, “fatty acid biosynthesis”, “photosynthesis - antenna proteins”, “prodigiosin biosynthesis” and “peroxisome”. Furthermore, the term “DNA repair and recombination proteins” was only enriched by DEGs in S20 condition.
3.3 Comparative analysis of DEGs in three conditions
According to the result of KEGG and GO enrichment analysis, the expression of DEGs in those enriched terms were compared in three different conditions. For instance, comparing DEGs in S10 group with those in C20 and S20 group explores the effect of SHE on the transcriptional regulation of N. yezoensis under different temperature. Furthermore, the comparison of DEGs in C20 and S20 groups indicates the molecular mechanism of heat-tolerance induced by SHE.
3.3.1 Ribosome related DEGs
In S10 group, 12 genes were identified as ribosome related DEGs and most of them were up-regulated compared to control (Figure 2; Table S3). Among those DEGs, 3 up-regulated genes were involved in the regulation of ribosome biogenesis and a gene annotated as 60S ribosomal protein was significantly down-regulated compared to control. In C20 group, 72 genes encoding ribosomal protein were identified as DEGs, in which 69 genes were significantly down-regulated compared to control (Figure 2; Table S3). Similar to C20 group, the expression of genes encoding ribosomal protein were also significantly repressed in S20 group. However, only 23 ribosomal protein genes were significantly down-regulated in S20 group compared to control, which is much less than those in C20 groups. Additionally, several ribosomal protein genes were only significantly down-regulated in S20 group compared to control, including 3 genes encoding 40S ribosomal protein S6e and a gene encoding 60S ribosomal protein L35.

Figure 2 Heatmap showing the expression of DEGs related to (A) ribosome, (B) amino acid metabolism, (C) photosynthesis, (D) respiration and (E) cell cycle in S10, C20 and S20 groups. Due to some extremely high and low values, the expression data was scaled in each row to better illustrate the trend of gene expression in different groups. The symbols ## were used to distinguish different genes with same annotation in the heatmap.
3.3.2 Photosynthesis related DEGs
High temperature significantly inhibited the expression of photosynthesis related genes. Four genes encoding chlorophyll a-b binding protein were significantly down-regulated in C20 and S20 groups compared to control (Figure 2; Table S3). High temperature also repressed the expression of genes related to chlorophyll biosynthesis, such as genes of protochlorophyllide reductase, magnesium-chelatase subunit and plastid geranylgeranyl reductase (Figure 2; Table S3). The protochlorophyllide reductase gene (py10059.t1) showed no significant down-regulation in S20 group. Furthermore, six genes encoding light harvesting protein were identified as DEGs. Among those DEGs, five genes were significantly down-regulated in both C20 and S20 groups and one gene was only up-regulated in C20 group compared to control (Figure 2; Table S3). Similarly, six genes related to photosystem II were significantly down-regulated in both C20 and S20 groups and one gene encoding photosystem II reaction center protein PsbM was significantly up-regulated in all three treatment conditions compared to control (Figure 2; Table S3). Additionally, 11 genes of the phycobilisome linker polypeptide were significantly down-regulated in C20 groups and most of those genes were also significantly down-regulated in S20 group. However, two genes encoding phycobilisome linker polypeptides (Novel.5350.1 and py02220.t1) were not significantly down-regulated in S20 group compared to control (Figure 2; Table S3).
3.3.3 Respiration related DEGs
For respiration, 18 genes were identified as DEGs in this study. Most of those genes were involved in oxidative phosphorylation. For instance, three genes were down-regulated and a gene was up-regulated in both C20 and S20 groups, which encode subunit protein of NADH dehydrogenase complex (Figure 2; Table S3). A NADH dehydrogenase isoform 1 gene was only significantly up-regulated in S10 group with a Log2Foldchange value of 5.39. The expression of two NADH-ubiquinone oxidoreductase genes and one ubiquinone oxidoreductase gene were repressed in both C20 and S20 groups. However, the other ubiquinone oxidoreductase gene was only significantly down-regulated in C20 group (Figure 2; Table S3). Similarly, three genes encoding subunit protein of mitochondrial ATP synthase were only significantly down-regulated in C20 group compared to control (Figure 2; Table S3). Furthermore, three genes related to cytochrome c oxidase were identified as DEGs with different expression patterns. Among those genes, one gene was significantly down-regulated and one gene was significantly up-regulated in both C20 and S20 groups, which encode subunit protein of cytochrome c oxidase. One gene involved in assembly of cytochrome c oxidase and a succinate dehydrogenase assembly factor gene were only significantly up-regulated in C20 group (Figure 2; Table S3). Additionally, two genes related to pyruvate metabolism were significantly down-regulated in both C20 and S20 groups. One of them, the gene encoding pyruvate kinase, was significantly up-regulated in S10 group compared to control (Figure 2; Table S3).
3.3.4 Cell cycle related DEGs
In total, 23 genes related to cell cycle were identified as DEGs in at least one conditions (Figure 2; Table S3). Among those genes, 3 genes encoding cyclin protein were significantly down-regulated and a cyclin gene was significantly up-regulated in S10 group compared to control (Figure 2; Table S3). In C20 group, 4 up-regulated and 4 down-regulated cyclin genes were identified and a gene encoding cyclin-dependent kinase C-1 (CDKC1) was significantly up-regulated compared to control (Figure 2; Table S3). Other cell cycle related genes were also significantly up-regulated in C20 group compared to control, such as genes encoding DP-E2F-like 1 transcription factor, transcription factor-like protein DPB, retinoblastoma-related protein (RBR), anaphase-promoting complex subunit 10 protein, condensin-2 complex subunit G2 protein, kinetochore protein Nuf2, mitotic arrest deficient 2 protein and separin-like protein (Figure 2; Table S3). The expression patterns of cyclin and CDKC1 genes in S20 group were similar to those in C20 group (Figure 2; Table S3). However, for other cell cycle related genes, only genes encoding kinetochore protein Nuf2, mitotic arrest deficient 2 protein and separin-like protein were significantly up-regulated in S20 group compared to control (Figure 2; Table S3). Additionally, a gene encoding mago-nashi domain-containing protein was highly up-regulated (Log2Foldchange = 13.75) in S20 group compared to control (Figure 2; Table S3).
3.3.5 Amino acid metabolism related DEGs
SHE pre-treatment induced up-regulation of genes involved in biosynthesis pathways of several amino acid under 10°C. Three genes in arginine biosynthesis pathway, arginosuccinate synthetase, carbamoyl-phosphate synthase and glutamine-dependent carbamoyl-phosphate synthase genes, were only up-regulated in S10 group compared to control (Figure 2; Table S3). Similarly, several genes involved in biosynthesis pathways of serine, isoleucine, proline, tryptophan, asparagine and histidine were also induced in the S10 group (Figure 2; Table S3). High temperature also affected the expression of genes related to amino acid metabolism. A gene encoding reactive intermediate deaminase A involved in isoleucine biosynthesis and a tryptophan synthase gene were significantly down-regulated in C20 group compared to control (Figure 2; Table S3). Furthermore, a gene encoding serine acetyltransferase was identified as up-regulated genes in C20 group (Figure 2; Table S3). Additionally, genes involved in the biosynthesis of aspartate group amino acid were also differentially expressed in C20 and S20 group. For instance, a gene encoding B-type asparagine synthetase was significantly down-regulated in C20 group (Figure 2; Table S3). The expression of genes encoding aspartate-semialdehyde dehydrogenase was also repressed under 20°C (Figure 2; Table S3). On the other hand, a L-asparaginase gene was significantly up-regulated in C20 group compared to control (Figure 2; Table S3).
3.3.6 Epigenetic modification related DEGs
Genes related to epigenetic modification were also found differentially expressed in three conditions. For instance, 5 genes encoding histone deacetylase were differentially expressed in C20 group compared to control, including three down-regulated genes and two up-regulated genes. One of them was also significantly down-regulated in the S10 group (Table 1). Furthermore, genes encoding methyltransferase were mainly induced by SHE pretreatment combined with high temperature. Three histone methyltransferase genes and two DNA methyltransferase genes were significantly up-regulated in the S20 group compared to control (Table 1). Among those genes, a histone methyltransferase gene was also significantly up-regulated in C20 group. Additionally, one gene encoding DNA methyltransferase associated protein was significantly up-regulated in both C20 and S20 groups compared to control (Table 1).
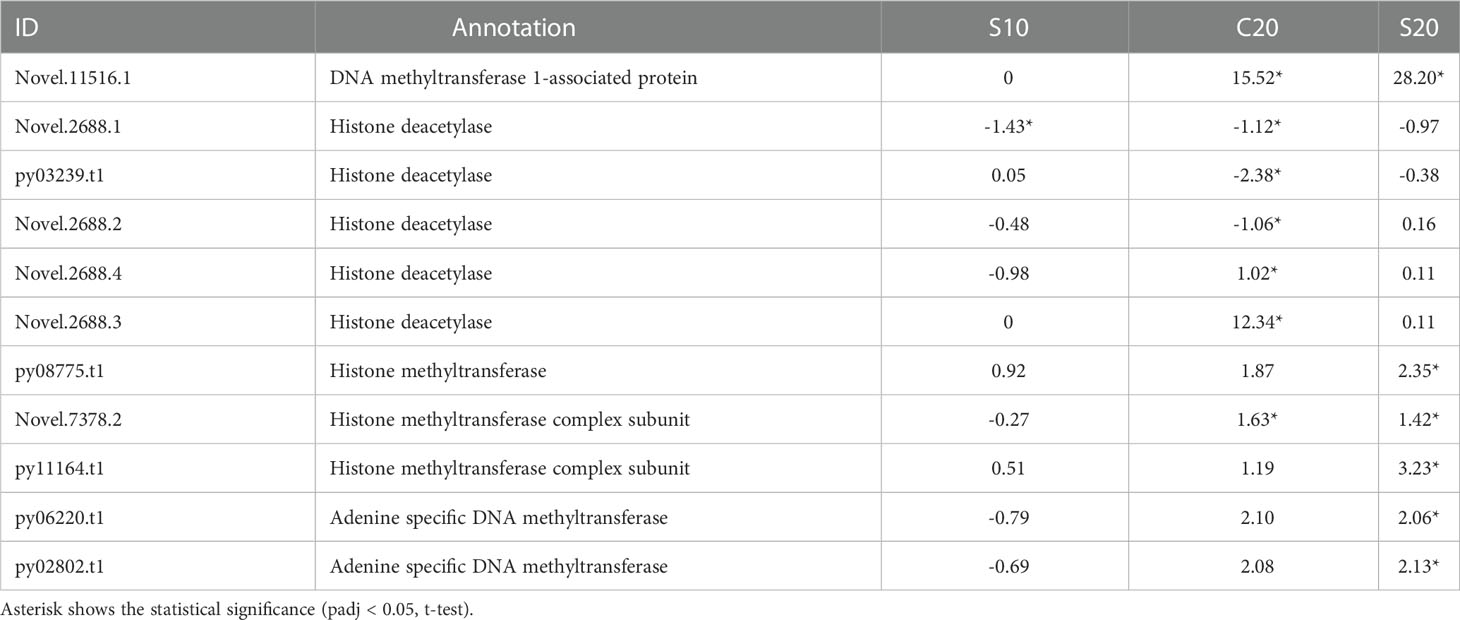
Table 1 Expression (Log2Foldchange value) of DEGs related to epigenetic modification under different conditions.
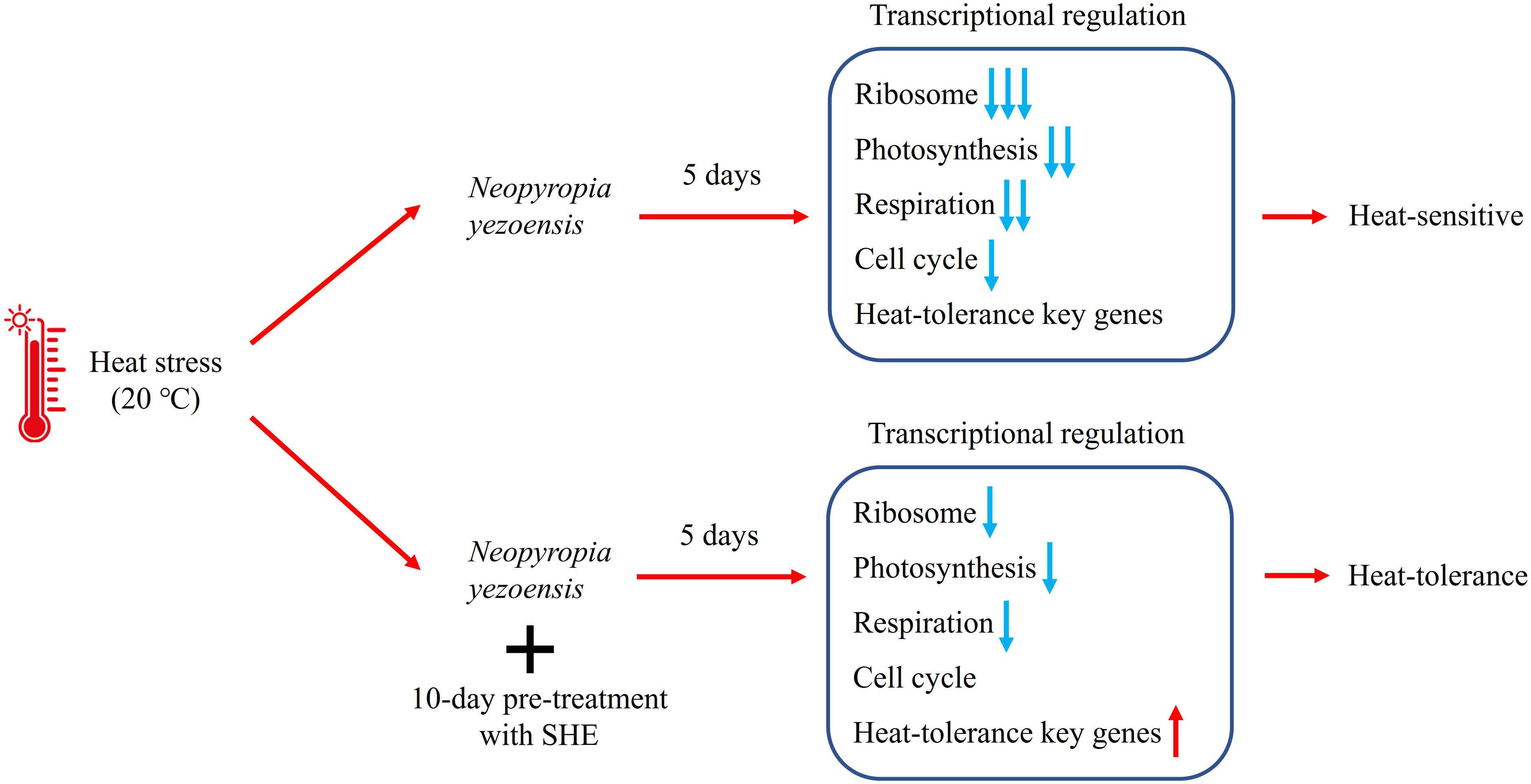
Figure 3 Scheme summarized the possible mechanisms of the different responses in Neopyropia yezoensis with or without SHE pre-treatment under heat stress. Blue arrows: down-regulation of genes; Red arrows: up-regulation of genes. Symbol +: Pretreatment of SHE.
3.3.7 Other key genes for heat-tolerance
In S20 group, approximately 20% of DEGs (866 out of 4,327 genes) were different with those in the C20 group. In addition to enrichment analysis, the key genes for heat-tolerance could also be identified in the different DEGs between two groups by comparative analysis. Among those genes, two genes encoding histidine kinase were only significantly up-regulated in S20 group (Table 2). A ferritin gene and a gene encoding iron uptake system component protein were induced by SHE pretreatment under 10°C and 20°C (Table 2). Furthermore, a spermidine synthase gene and a gene encoding carotenoid cleavage dioxygenase were also significantly up-regulated in S10 and S20 groups (Table 2).
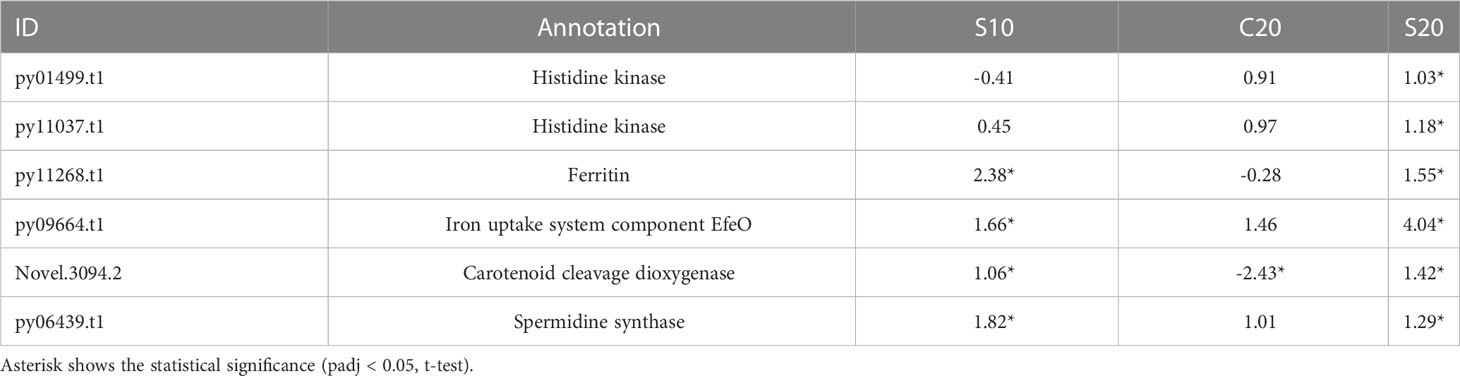
Table 2 Expression (Log2Foldchange value) of key genes for heat-tolerance under different conditions.
4 Discussion
Several studies have been conducted to explore the transcriptional responses under high temperature in N. yezoensis and its closely related cogenerors, N. tenera and N. haitanensis (Choi et al., 2013; Sun et al., 2015; Wang et al., 2018). The previous studies also revealed some molecular mechanisms of heat-tolerance in Neopyropia using comparative transcriptomic analysis of heat-tolerant and sensitive N. haitanensis strains (Wang et al., 2018). However, the molecular mechanism of heat-tolerance in N. yezoensis is still largely unknown. Our previous study reported that pre-treatment with SHE, a biostimulant extracted from Sargassum horneri, enhanced the heat-tolerance of N. yezoensis (Han et al., 2022). In the present study, we analyzed the transcriptomic regulation of heat-tolerance in N. yezoensis induced by SHE, which provided more information about heat-tolerance in N. yezoensis and the function of seaweed based biostimulants.
4.1 SHE pre-treatment alleviated negative impacts of heat stress
Heat stress inhibits the growth of algae by affecting the activities of protein production, photosynthesis and respiration (Sun et al., 2015; Fan et al., 2018). Previous studies showed that heat stress repressed the protein production by reducing the expression of ribosomal protein genes in Neoporphyra haitanensis and Dunaliella bardawil (Shi et al., 2017; Liang et al., 2020). Similar result was also found in this study. Heat stress significantly repressed the expression of 69 genes encoding ribosomal protein in N. yezoensis. However, in the group pre-treated with SHE, only 23 ribosomal protein genes were down-regulated under heat stress. This result showed that SHE pretreatment could relieve inhibition of gene expression related to protein biosynthesis induced by heat stress. Liang et al. (2021) also reported that less down-regulated ribosomal protein genes were observed in the heat-tolerant cotton strain than the heat-sensitive strain.
Photosynthesis is very sensitive to heat stress which inhibits the expression of photosynthesis related genes in seaweed (Sun et al., 2015; Fan et al., 2018; Wang et al., 2018). Transcriptomic analysis showed that genes involved in both photosystem I (PSI) and II (PSII) of N. yezoensis were significantly down-regulated under 24°C (Fan et al., 2018). In this study, genes involved in PSII were significantly down-regulated under 20°C, while no significant change was observed in PSI related genes. It has been reported that PSI activity is more stable to high temperature than PSII (Havaux, 1992). This result suggests that, as the sub-lethal temperature of N. yezoensis, 20°C did not induce a strong inhibition on photosynthesis compared to 24°C. After the pre-treatment of SHE, most of the down-regulated photosynthetic genes had higher log2foldchange value compared to those in group with only heat stress. Among down-regulated photosynthetic genes, one gene encoding protochlorophyllide reductase, which is the enzyme for chlorophyllide biosynthesis (Jespersen et al., 2016), was not significantly down-regulated under heat stress after the SHE pre-treatment. In addition, SHE pre-treatment also relieved the inhibition on the expression of two genes encoding phycobilisome linker polypeptide under heat stress. Phycobilisome is the major light harvesting complex in N. yezoensis. The function of phycobilisome linker polypeptide is stabilizing the structure of phycobilisomes, which optimizes the light absorbance and energy transfer (Liu et al., 2005). It has also been reported that phycobilisome linker polypeptides were decreased in a wild type strain and over-expressed in heat-tolerant strain of N. yezoensis in response to heat stress (Lee et al., 2019). This result suggests that SHE pre-treatment also reduced the negative effects on photosynthesis in N. yezoensis induced by heat stress.
Similar phenomenon has also been observed in the expression of genes related to respiration. Under the heat stress, the activity of respiration can be inhibited due to the thermo-sensitivity of the mitochondrial electron transport chain (Belhadj Slimen et al., 2014). In the green alga Dunaliella bardawil, genes encoding key enzymes in the mitochondrial electron transport chain were significantly down-regulated in response to heat stress (Liang et al., 2020). Our study also shows that genes involved in mitochondrial electron transport chain, such as genes encoding NADH dehydrogenase, cytochrome c oxidase, ubiquinone oxidoreductase and ATP synthase, were significantly down-regulated in N. yezoensis under heat stress. No significant repression on the expression of ATP synthase genes was observed under heat stress after the SHE pre-treatment. Similar results have also been reported in the other red alga, Gracilariopsis lemaneiformis. In that study, samples with heat-tolerance induced by an elicitor, chitooligosaccharides, had higher expression level of genes involved in mitochondrial electron transport chain than those in heat-sensitive samples in response to heat stress (Zhang et al., 2020).
Additionally, cell cycle related genes were also regulated by heat stress. In this study, a gene encoding E2F transcription factor and a gene encoding transcription factor-like protein DPB were highly up-regulated in response to heat stress. The E2F transcription factor activates the expression of genes related to mitosis by combining with DP protein to form a E2F/DP heterodimer (del Pozo et al., 2006). However, it has been reported that the transcription factor E2Fc combined with DPB protein can negatively regulated E2F target genes by competing for the E2F-binding sites, restricting the cell division (Ma et al., 2014). Given the fact that the growth rate of N. yezoensis was significantly decreased under 20°C (Han et al., 2022), the co-expression of E2F and DPB genes could be one of the reasons that suppressed the growth rate of N. yezoensis under heat stress. After the SHE pre-treatment, no significant up-regulation of these two genes was observed under 20°C. The SHE pre-treatment also significantly increased the growth rate in N. yezoensis compared to the group without pre-treatment under 20°C (Han et al., 2022). Moreover, a gene encoding retinoblastoma-related (RBR) protein was also significantly up-regulated in response to heat stress. Retinoblastoma-related protein can also inhibit the cell cycle by binding with the E2F protein (Pokora et al., 2022). Under heat stress, SHE pre-treatment decreased the expression of RBR protein gene compared to the untreated group, suggesting that SHE treatment could also reduce the inhibition of gene expression related to cell cycle induced by heat stress. Figure 3 summarized the gene regulation in N. yezoensis of SHE pretreatment group and control under heat stress and showed that SHE pretreatment can alleviate negative impacts of heat stress.
4.2 SHE pre-treatment induced genes related to heat-tolerance
In addition to reducing the negative effect of heat stress, SHE pre-treatment also induced some key genes related to heat-tolerance (Figure 3). Two genes encoding histidine kinase were significantly up-regulated under heat stress after SHE pre-treatment, showing a higher expression level than those genes C20 group. Histidine kinase can act as thermo-sensor (Albanesi et al., 2009) and the deficiency of histidine kinase reduced the long-term thermotolerance in Synechocystis sp. (Červený et al., 2015). Another key gene related to heat-tolerance is ferritin, an iron containing protein widespread in all cell types (Theil, 1987). The overexpression of a ferritin gene, TaFER, significantly enhanced the heat-tolerance in both wheat and Arabidopsis (Zang et al., 2017). In this study, a ferritin gene was only induced by SHE pre-treatment instead of heat stress, suggesting that this gene might also play a key role in the heat-tolerance in N. yezoensis. On the other hand, a gene encoding protein component in iron uptake system was also highly up-regulated under heat stress after the SHE pre-treatment, which could provide sufficient iron for the increasing biosynthesis of ferritin. Similarly, one gene encoding carotenoid cleavage dioxygenase was only significantly up-regulated after the SHE pre-treatment. Carotenoid cleavage dioxygenase has been reported participating in the biosynthesis pathways of abscisic acid and strigolactone (Schwartz et al., 2003; Stirk and Van Staden, 2014). Both of the two phytohormone can enhance the thermotolerance in land plants (Gong et al., 1998; Ahammed et al., 2021). Furthermore, it was reported that spermidine can enhance the heat-tolerance in Pinellia ternata (Ma et al., 2020). Similarly, the spermidine treatment can also enhance the heat-tolerance in a red alga species, Gracilariopsis lemaneiformis (Liu et al., 2022). In this study, we also observed the significant up-regulation of gene encoding spermidine synthase induced by SHE pre-treatment, suggesting that this gene may also contribute to the heat-tolerance in N. yezoensis. Notably, the SHE pre-treatment up-regulated genes involved in the biosynthesis pathways of several amino acids under 10°C, such as asparagine, tryptophan, proline and cysteine. Some of those genes are also related to stress resistance. For instance, previous studies reported that asparagine synthetase plays an important role in salinity tolerance (Rashmi et al., 2019) and defense responses to pathogens (Hwang et al., 2011). Also, the overexpression of serine acetyltransferase gene increased the resistance to oxidative stress in tobacco (Blaszczyk et al., 1999). These results suggest that SHE can probably protect N. yezoensis from other stresses.
4.3 SHE pre-treatment may facilitate the formation of heat stress memory
Seaweed can form stress memory through epigenetic changes, including DNA methylation, histone modification and non-coding RNA (Jueterbock et al., 2021). Epigenetic changes can maintain genomic plasticity, allowing the rapid adaptation to stresses without changes in DNA sequences (Henderson and Jacobsen, 2007). A previous study showed that high temperature induced changes in DNA methylation in Neoporphyra haitanensis (Yu et al., 2018). DNA methyltransferase is the key enzyme involved in the DNA methylation in plants (Lyko, 2018). In this study, two genes encoding DNA methyltransferase were significantly up-regulated under heat stress after the SHE pre-treatment. These two genes were also up-regulated in the group only treated with high temperature. However, the up-regulation of these two genes was not statistically significant due to the large standard deviation of gene expression level between different biological replicates. Similar results have been found in other genes related to epigenetic modification. For instance, three genes encoding histone methyltransferase were significantly up-regulated under heat stress after SHE pre-treatment. One of them was also significantly up-regulated in the group only treated with high temperature. No significant up-regulation of those genes was observed after SHE pre-treatment under 10°C. These results suggest that the epigenetic modification may not be induced by SHE pre-treatment or heat stress alone. The SHE pre-treatment may facilitate the formation of heat stress memory in N. yezoensis, which will reduce the amount of SHE used in the open water seaweed aquaculture.
5 Conclusion
In this study, we analyzed the molecular mechanism of heat-tolerance induced by a seaweed-derived biostimulant, the Sargassum horneri extract. Our results showed that SHE pre-treatment can not only alleviate negative impacts of heat stress, but also induce key genes related to heat-tolerance that has never been reported in seaweed before. Additionally, we found that SHE combined with heat stress can induce some genes related to epigenetic modification. These results provide critical information to understand the molecular mechanism underlying the heat-tolerance induced by SHE, promoting the application of this novel biostimulant in seaweed aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The Illumina sequence reads generated during the present study are available in the NCBI SRA database under the BioProject ID: PRJNA894950.
Author contributions
QX, CY and JK designed the experiment QX, SH and J-SP conducted the experiment QX analyzed the data and wrote the first draft of manuscript. JK provided fund for the experiment and supervised the experiment All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A6A1A06015181) and by the Ministry of Science and ICT (2019R1F1A1059663 and 2022R1A2C1011394).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1142483/full#supplementary-material
References
Ahammed G. J., Guang Y., Yang Y., Chen J. (2021). Mechanisms of elevated CO2-induced thermotolerance in plants: the role of phytohormones. Plant Cell Rep. 40 (12), 2273–2286. doi: 10.1007/s00299-021-02751-z
Albanesi D., Martín M., Trajtenberg F., Mansilla M. C., Haouz A., Alzari P. M., et al. (2009). Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. U S A. 106 (38), 16185–16190. doi: 10.1073/pnas.0906699106
Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data, babraham bioinformatics (Cambridge, United Kingdom: Babraham Institute).
Bae S.-H. (1991). The origin and development process of laver culture industry in Korea-1. laver culture history till the end of chosun dynasty. Korean J. Fish. Aquat. Sci. 24 (3), 153–166.
Belhadj Slimen I., Najar T., Ghram A., Dabbebi H., Ben Mrad M., Abdrabbah M. (2014). Reactive oxygen species, heat stress and oxidative-induced mitochondrial damage. A review. Int. J. Hyperthermia 30 (7), 513–523. doi: 10.3109/02656736.2014.971446
Blaszczyk A., Brodzik R., Sirko A. (1999). Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J. 20 (2), 237–243. doi: 10.1046/j.1365-313x.1999.00596.x
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Červený J., Sinetova M. A., Zavřel T., Los D. A. (2015). Mechanisms of high temperature resistance of synechocystis sp. PCC 6803: an impact of histidine kinase 34. Life 5 (1), 676–699. doi: 10.3390/life5010676
Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Choi S., Hwang M. S., Im S., Kim N., Jeong W.-J., Park E.-J., et al. (2013). Transcriptome sequencing and comparative analysis of the gametophyte thalli of pyropia tenera under normal and high temperature conditions. J. Appl. Phycol. 25 (4), 1237–1246. doi: 10.1007/s10811-012-9921-2
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
del Pozo J. C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in arabidopsis. Plant Cell 18 (9), 2224–2235. doi: 10.1105/tpc.105.039651
Ding H., Zhang B., Yan X. (2016). Isolation and characterization of a heat-resistant strain with high yield of pyropia yezoensis ueda (Bangiales, rhodophyta). Aquaculture fisheries 1, 24–33. doi: 10.1016/j.aaf.2016.09.001
Duarte C. M., Wu J., Xiao X., Bruhn A., Krause-Jensen D. (2017). Can seaweed farming play a role in climate change mitigation and adaptation? Front. Mar. Sci. 4, 100. doi: 10.3389/fmars.2017.00100
Fan M., Sun X., Liao Z., Wang J., Li Y., Xu N. (2018). Comparative proteomic analysis of ulva prolifera response to high temperature stress. Proteome Sci. 16 (1), 1–22. doi: 10.1186/s12953-018-0145-5
FAO (2020). The state of world fisheries and aquaculture 2020. sustainability in action (Rome: Food and Agriculture Organization of the United Nations).
Gong M., Li Y.-J., Chen S.-Z. (1998). Abscisic acid-induced thermotolerance in maize seedlings is mediated by calcium and associated with antioxidant systems. J. Plant Physiol. 153 (3-4), 488–496. doi: 10.1016/S0176-1617(98)80179-X
Grubb M., Okereke C., Arima J., Bosetti V., Chen Y., Edmonds J., et al. (2022). “Introduction and framing,” in IPCC 2022: Climate change 2022: Mitigation of climate change. contribution of working group III to the sixth assessment report of the intergovernmental panel on climate change, IPCC. Cambridge: (Cambridge University Press)
Han S., Park J.-S., Umanzor S., Yarish C., Kim J. K. (2022). Effects of extraction methods for a new source of biostimulant from sargassum horneri on the growth of economically important red algae, neopyropia yezoensis. Sci. Rep. 12 (1), 1–11. doi: 10.1038/s41598-022-16197-0
Han N., Wi J., Im S., Lim K.-M., Lee H.-D., Jeong W.-J., et al. (2021). Downregulation of PyHRG1, encoding a novel secretory protein in the red alga pyropia yezoensis, enhances heat tolerance. Algae 36 (3), 207–217. doi: 10.4490/algae.2021.36.8.26
Havaux M. (1992). Stress tolerance of photosystem II in vivo: antagonistic effects of water, heat, and photoinhibition stresses. Plant Physiol. 100 (1), 424–432. doi: 10.1104/pp.100.1.424
Henderson I. R., Jacobsen S. E. (2007). Epigenetic inheritance in plants. Nature 447 (7143), 418–424. doi: 10.1038/nature05917
Hurtado A. Q., Critchley A. T. (2018). A review of multiple biostimulant and bioeffector benefits of AMPEP, an extract of the brown alga ascophyllum nodosum, as applied to the enhanced cultivation and micropropagation of the commercially important red algal carrageenophyte kappaphycus alvarezii and its selected cultivars. J. Appl. Phycol. 30 (5), 2859–2873. doi: 10.1007/s10811-018-1407-4
Hwang I. S., An S. H., Hwang B. K. (2011). Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67 (5), 749–762. doi: 10.1111/j.1365-313X.2011.04622.x
Hwang E. K., Park C. S. (2020). Seaweed cultivation and utilization of Korea. Algae 35 (2), 107–121. doi: 10.4490/algae.2020.35.5.15
Jespersen D., Zhang J., Huang B. (2016). Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Sci. 249, 1–12. doi: 10.1016/j.plantsci.2016.04.016
Jueterbock A., Minne A. J., Cock J. M., Coleman M. A., Wernberg T., Scheschonk L., et al. (2021). Priming of marine macrophytes for enhanced restoration success and food security in future oceans. Front. Mar. Sci. 8, 658485. doi: 10.3389/fmars.2021.658485
Kim J. K., Kraemer G. P., Neefus C. D., Chung I. K., Yarish C. (2007). Effects of temperature and ammonium on growth, pigment production and nitrogen uptake by four species of porphyra (Bangiales, rhodophyta) native to the new England coast. J. Appl. Phycol. 19 (5), 431–440. doi: 10.1007/s10811-006-9150-7
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Lee H.-J., Park E.-J., Choi J.-I. (2019). Isolation, morphological characteristics and proteomic profile analysis of thermo-tolerant pyropia yezoensis mutant in response to high-temperature stress. Ocean Sci. J. 54 (1), 65–78. doi: 10.1007/s12601-018-0060-9
Liang M.-H., Jiang J.-G., Wang L., Zhu J. (2020). Transcriptomic insights into the heat stress response of dunaliella bardawil. Enzyme Microb. Technol. 132, 109436. doi: 10.1016/j.enzmictec.2019.109436
Liang Y., Gong Z., Wang J., Zheng J., Ma Y., Min L., Li X. (2021). Nanopore-Based Comparative Transcriptome Analysis Reveals the Potential Mechanism of High-Temperature Tolerance in Cotton (Gossypium hirsutum L.). Plants 10 (11), 2517
Liu L.-N., Chen X.-L., Zhang Y.-Z., Zhou B.-C. (2005). Characterization, structure and function of linker polypeptides in phycobilisomes of cyanobacteria and red algae: an overview. Biochim. Biophys. Acta Bioenerg. 1708 (2), 133–142. doi: 10.1016/j.bbabio.2005.04.001
Liu S., Zhang J., Sun X., Xu N. (2022). Characterization of spermidine synthase (SPDS) gene and RNA–seq based identification of spermidine (SPD) and spermine (SPM) involvement in improving high temperature stress tolerance in gracilariopsis lemaneiformis (Rhodophyta). Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.939888
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
Lyko F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19 (2), 81–92. doi: 10.1038/nrg.2017.80
Ma T.-Y., Li Z.-W., Zhang S.-Y., Liang G.-T., Guo J. (2014). Identification and expression analysis of the E2F/DP genes under salt stress in medicago truncatula. Genes Genomics 36 (6), 819–828. doi: 10.1007/s13258-014-0218-5
Ma G., Zhang M., Xu J., Zhou W., Cao L. (2020). Transcriptomic analysis of short-term heat stress response in pinellia ternata provided novel insights into the improved thermotolerance by spermidine and melatonin. Ecotoxicol Environ. Saf. 202, 110877. doi: 10.1016/j.ecoenv.2020.110877
Niwa K., Aruga Y. (2006). Identification of currently cultivated porphyra species by PCR-RFLP analysis. Fish. Sci. 72 (1), 143–148. doi: 10.1111/j.1444-2906.2006.01128.x
Ott F.D. (1966). A selected listing of xenic cultures. Systematics-Ecology Program No. 72 (Mar. Biol. Lab.: Woods Hole, MA), 45.
Pokora W., Tułodziecki S., Dettlaff-Pokora A., Aksmann A. (2022). Cross talk between hydrogen peroxide and nitric oxide in the unicellular green algae cell cycle: How does it work? Cells 11 (15), 2425. doi: 10.3390/cells11152425
Rashmi D., Barvkar V. T., Nadaf A., Mundhe S., Kadoo N. Y. (2019). Integrative omics analysis in pandanus odorifer (Forssk.) kuntze reveals the role of asparagine synthetase in salinity tolerance. Sci. Rep. 9 (1), 1–16. doi: 10.1038/s41598-018-37039-y
Redmond S., Green L., Yarish C., Kim J., Neefus C. (2014). New England seaweed culture handbook. Connecticut Sea Grant, Available at: http://seagrant.uconn.edu/publications/aquaculture/handbook.pdf
Schwartz S. H., Tan B. C., McCarty D. R., Welch W., Zeevaart J. A. (2003). Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim. Biophys. Acta Gen. Subj. 1619 (1), 9–14. doi: 10.1016/S0304-4165(02)00422-1
Shi J., Chen Y., Xu Y., Ji D., Chen C., Xie C. (2017). Differential proteomic analysis by iTRAQ reveals the mechanism of pyropia haitanensis responding to high temperature stress. Sci. Rep. 7 (1), 1–11. doi: 10.1038/srep44734
Shin Y. J., Min S. R., Kang D. Y., Lim J.-M., Park E.-J., Hwang M. S., et al. (2018). Characterization of high temperature-tolerant strains of pyropia yezoensis. Plant Biotechnol. Rep. 12 (5), 365–373. doi: 10.1007/s11816-018-0499-2
Stirk W. A., Van Staden J. (2014). Plant growth regulators in seaweeds: occurrence, regulation and functions. Adv. Bot. Res. Elsevier 71, 125–159. doi: 10.1016/B978-0-12-408062-1.00005-6
Sun P., Mao Y., Li G., Cao M., Kong F., Wang L., et al. (2015). Comparative transcriptome profiling of pyropia yezoensis (Ueda) MS hwang & HG choi in response to temperature stresses. BMC Genomics 16 (1), 1–16. doi: 10.1186/s12864-015-1586-1
Tang Q., Zhang J., Fang J. (2011). Shellfish and seaweed mariculture increase atmospheric CO2 absorption by coastal ecosystems. Mar. Ecol. Prog. Ser. 424, 97–104. doi: 10.3354/meps08979
Theil E. C. (1987). Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu. Rev. Biochem. 56 (1), 289–315. doi: 10.1146/annurev.bi.56.070187.001445
Umanzor S., Shin S., Marty-Rivera M., Augyte S., Yarish C., Kim J. K. (2019). Preliminary assessment on the effects of the commercial seaweed extract, AMPEP, on growth and thermal tolerance of the kelp saccharina spp. from the Northwest Atlantic. J. Appl. Phycol. 31 (6), 3823–3829. doi: 10.1007/s10811-019-01852-3
Umanzor S., Shin S., Yarish C., Augyte S., Kim J. K. (2020). Exploratory evaluation of the effects of kelpak® seaweed extract on cultivated kelp saccharina spp. exposed to sublethal and lethal temperatures. J. World Aquac Soc 51 (4), 960–969. doi: 10.1111/jwas.12687
van Oppen M. J., Coleman M. A. (2022). Advancing the protection of marine life through genomics. PloS Biol. 20 (10), e3001801. doi: 10.1371/journal.pbio.3001801
Voisin M., Engel C. R., Viard F. (2005). Differential shuffling of native genetic diversity across introduced regions in a brown alga: aquaculture vs. maritime traffic effects. Proc. Natl. Acad. Sci. U S A. 102 (15), 5432–5437. doi: 10.1073/pnas.0501754102
Wang W., Lin J., Chang J., Ji D., Xu Y., Chen C., et al. (2021). A RING type ubiquitin ligase PhCUL4 is involved in thermotolerance of pyropia haitanensis. Algal Res. 59, 102448. doi: 10.1016/j.algal.2021.102448
Wang W., Lin Y., Teng F., Ji D., Xu Y., Chen C., et al. (2018). Comparative transcriptome analysis between heat-tolerant and sensitive pyropia haitanensis strains in response to high temperature stress. Algal Res. 29, 104–112. doi: 10.1016/j.algal.2017.11.026
Wang D., Yu X., Xu K., Bi G., Cao M., Zelzion E., et al. (2020). Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 11 (1), 1–11. doi: 10.1038/s41467-020-17689-1
Yamamoto M., Watanabe Y., Kinoshita H. (1991). Effects of water temperature on the growth of red alga porphyra yezoensis form narawaensis (nori) cultivated in an outdoor raceway tank. Nippon Suisan Gakkaishi 57 (12), 2211–2217. doi: 10.2331/suisan.57.2211
Yu C., Xu K., Wang W., Xu Y., Ji D., Chen C., et al. (2018). Detection of changes in DNA methylation patterns in pyropia haitanensis under high-temperature stress using a methylation-sensitive amplified polymorphism assay. J. Appl. Phycol. 30 (3), 2091–2100. doi: 10.1007/s10811-017-1363-4
Zang X., Geng X., Wang F., Liu Z., Zhang L., Zhao Y., et al. (2017). Overexpression of wheat ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 17 (1), 1–13. doi: 10.1186/s12870-016-0958-2
Keywords: sargassum horneri extract (SHE), neopyropia yezoensis, biostimulant, heat-tolerance, transcriptome
Citation: Xing Q, Han S, Park J-S, Yarish C and Kim JK (2023) Comparative transcriptome analysis reveals the molecular mechanism of heat-tolerance in Neopyropia yezoensis induced by Sargassum horneri extract. Front. Mar. Sci. 10:1142483. doi: 10.3389/fmars.2023.1142483
Received: 11 January 2023; Accepted: 01 March 2023;
Published: 15 March 2023.
Edited by:
Shikai Liu, Ocean University of China, ChinaReviewed by:
Fanna Kong, Ocean University of China, ChinaFeng Liu, Institute of Oceanology (CAS), China
Copyright © 2023 Xing, Han, Park, Yarish and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jang K. Kim, amFuZy5raW1AaW51LmFjLmty
 Qikun Xing
Qikun Xing Sol Han
Sol Han Ji-Sook Park
Ji-Sook Park Charles Yarish
Charles Yarish Jang K. Kim
Jang K. Kim