
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 February 2023
Sec. Aquatic Physiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1141231
To reveal the biosynthetic pathway of long-chain polyunsaturated fatty acids (LC-PUFA) in Ruditapes philippinarum, herein, two fatty acid desaturases (Fads, including one Δ5 Fad and one Δ6 Fad-like) and three elongases of very long-chain fatty acids (Elovls, including one Elovl2/5 and two Elovl4-like) genes were firstly cloned from this bivalve and their tissue distributions were examined. Results showed that the newly cloned Fads and Elovls contained the corresponding conserved functional domains and clustered closely with their orthologs, respectively. Meanwhile, they were expressed significantly higher in the digestive glands and intestine. Subsequently, to further understand the LC-PUFA biosynthesis in R. philippinarum, the effects of dietary LC-PUFA on Fad and Elovl expressions and the fatty acid (FA) profile in this bivalve were investigated by feeding with three microalgae varied in LC-PUFA compositions [including Chlorella sp. (rich in 18:2n-6 and 18:3n-3), Chaetoceros calcitrans (rich in eicosapentaenoic acid, EPA), and Isochrysis galbana (rich in docosahexaenoic acid, DHA]. Results showed that, throughout the experiment, the expressions of Fad and Elovl were significantly up-regulated in the visceral mass (digestive glands and intestine) of R. philippinarum fed with Chlorella sp., while no significant changes or slightly decreases were observed in those fed with I. galbana. Furthermore, in those fed with C. calcitrans, the expressions of Fad were not significantly changed, whereas the expressions of Elovl were firstly up-regulated but then restored to its initial level at the end of experiment. These results suggested that R. philippinarum could modulate Fad and Elovl expressions to adapt to the dietary LC-PUFA composition. The FA analysis showed that a significantly higher amount of DHA and EPA was found in the R. philippinarum fed with I. galbana and C. calcitrans, respectively, which reflected well of the dietary FA. However, the R. philippinarum fed with Chlorella sp. exhibited a significant decrease of 18:2n-6 and 18:3n-3 but with a significant increase of their products such as 20:3n-6 and 22:5n-3, indicating that R. philippinarum had a certain capacity for LC-PUFA biosynthesis. Collectively, this study provided valuable insights into the biosynthesis of LC-PUFA in R. philippinarum.
Omega-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFAs), particularly eicosapentaenoic acid (20:5n-3, EPA) and docosahexaenoic acid (22:6n-3, DHA), are well considered as physiologically essential nutrients that have beneficial effects on human health (Djuricic and Calder, 2021). They play crucial roles, especially in neurological development, inflammatory response, and mitigating several diseases (Sijben and Calder, 2007; Gould et al., 2013; Marventano et al., 2015). In general, humans can’t endogenously biosynthesize LC-PUFAs at a sufficient rate to satisfy their physiological requirements and thus must obtain them from diets (Hussein et al., 2005). However, it is not in question that there is a significant gap between the demand and supply of LC-PUFAs, which has prompted an interest in exploring alternative sources of them (Tocher et al., 2019). Marine molluscs, including bivalves, are rich in LC-PUFAs, making them a potential supplier of dietary LC-PUFAs for humans (Tan et al., 2020).
LC-PUFAs can be obtained directly from the diet or endogenously biosynthesized from the dietary essential 18-C PUFA precursors linoleic acid (18:2n-6, LA) and α-linolenic acid (18:3n-3, ALA) via a series of desaturation and elongation steps catalyzed by fatty acid desaturases (Fads) and elongases of very long-chain fatty acid (Elovls) (Zhang et al., 2016). Fads are key enzymes responsible for the introduction of a new double bond between defined carbons of fatty acyl chains, while Elovls catalyze the condensation reaction as the rate-limiting step, resulting in the two-carbon elongation of pre-existing fatty acids (FA) (Miyazaki and Ntambi, 2008). It has been shown that marine molluscs possess genes for enzymes responsible for producing LC-PUFAs (Surm et al., 2015; Kabeya et al., 2018; Monroig and Kabeya, 2018). Typically, Fads with Δ5/6 desaturation activities are generally regarded as vital enzymes in the biosynthesis of LC-PUFA. More specifically, a pioneering investigation on the common Octopus vulgaris revealed that this species has a Fad gene with Δ5 desaturation activity, allowing it to produce ARA and EPA from 20:3n-6 and 20:4n-3, respectively (Monroig et al., 2012b). Subsequently, similar Δ5 Fads have been cloned and characterized in a variety of molluscs, including gastropods of Haliotis discus hannai (Li et al., 2013), cephalopods of Sepia officinalis (Monroig et al., 2016), and bivalves of Chlamys nobilis (Liu et al., 2014a) and Sinonovacula constricta (Ran et al., 2018). Notably, Ran et al. (2018) further cloned and functionally characterized a Δ6 Fads that exhibited desaturation activities toward LA and ALA, respectively, which was the first Δ6 Fad in marine molluscs. At present, eight members of the Elovl family (Elovl1-8) have been identified in vertebrates, of which only Elovl2, Elovl4, Elovl5, and Elovl8 have shown the ability to elongate LC-PUFAs (Guillou et al., 2010; Li et al., 2020; Sun et al., 2021). In general, Elovl2/5 exhibits high elongation activity against C18 and C20 FA, while Elovl4 preferentially elongates C ≥ 22 FA. Thus far, two distinct types of elongases have been identified from marine molluscs involved in LC-PUFA synthesis, i.e., Elovl2/5 of Octopus vulgaris, Sepia officinalis, Chlamys nobilis, Crassostrea angulate, and S. constricta (Monroig et al., 2012a; Liu et al., 2013; Zhang et al., 2018; Ran et al., 2019); as well as Elovl4 of C. nobilis, O. vulgaris, and S. constricta (Liu et al., 2014b; Monroig et al., 2017; Ran et al., 2019). These studies provided direct molecular evidence that marine molluscs, to some extent, possess the ability to biosynthesize LC-PUFAs (like EPA or DHA) from precursors. However, the complex and variable marine environment, in particular the availability of LC-PUFA-rich diets, has a great effect on their lipid metabolism and FA profiles (Caers et al., 1999; Lee et al., 2018; Zhukova, 2019).
The manila clam, Ruditapes philippinarum, is an economically important bivalve species that is largely distributed along the coastal beaches of China, South Korea, and Japan. The global aquaculture production of this species was over 4,028,000 tons, with a value of US$ 6.7 billion in 2019 (FAO, 2021). In addition, R. philippinarum is an excellent source of health-promoting omega-3 (or n-3) LC-PUFAs, particularly DHA, which makes up approximately 15% (3.5 μg/mg) of the total FA (Fernández-reiriz et al., 2017). Previous studies have revealed that the dietary lipid plays a critical role in the growth and FA composition of R. philippinarum spat (Caers et al., 1999; Fernandez-Reiriz et al., 2006). Meanwhile, in recent years, it has been found that the expression of genes involved in LC-PUFA synthesis can be regulated by dietary lipids in aquatic animals (Monroig and Kabeya, 2018; Monroig et al., 2018). Briefly, the expression of Fad and Elovl appears to be upregulated in animals when fed with a LA or ALA-rich diet, whereas it is suppressed when fed with a DHA or EPA-rich diet. However, to our knowledge, the composition of Fad and Elovl involved in LC-PUFA biosynthesis in R. philippinarum and the dietary effect on their expressions still remain unclear.
Therefore, in the present study, we aimed to lay a foundation for understanding the biosynthesis and dietary requirement of LC-PUFAs in R. philippinarum. Firstly, the Fad and Elovl of R. philippinarum were cloned, and their tissue distributions were examined. Subsequently, the effects of dietary LC-PUFAs on Fad and Elovl expressions and FA profiles in this bivalve were investigated by feeding with three microalgae varied in LC-PUFA compositions [including Chlorella sp. (rich in LA and ALA), Chaetoceros calcitrans (rich in EPA), and Isochrysis galbana (rich in DHA).
The studies involving animals were reviewed and approved by the Ningbo University Laboratory Animal Center under permit number no. SYXK (ZHE2008-0110).
The candidate Fad and Elovl sequences were firstly retrieved from our transcriptome assembly data of R. philippinarum (unpublished) based on functional annotations. Subsequently, the existence of three diagnostic histidine boxes (H***H, H**HH, and QIEHH) and an N-terminal cytochrome b5 domain containing the heme-binding motif HPGG conserved in front-end desaturases was used to select all hits of potential Fad sequences (Hashimoto et al., 2008). Likewise, the presumed Elovl sequences were selected based on the presence of one histidine box (H**HH) and a diagnostic “Q” (glutamine) in position-5 from the H**HH, which highly conserved in all members of PUFA elongase (Hashimoto et al., 2008).
To confirm the selected Fad and Elovl sequences including homologs of Δ5 Fad (Rp_fad5), Δ6 Fad (Rp_fad6), Elovl2/5 (Rp_elovl2/5), and Elovl4 (Rp_elovl4a, Rp_elovl4b), the polymerase chain reaction (PCR) was conducted with gene-specific primers (Table S1) designed by premier primer 5 and a cDNA template of a mixture of cDNA from gill and muscle. Firstly, tissues including gill and muscle were sampled from R. philippinarum and used to extract total RNA using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan). Following that, 1 μg of RNA was reverse transcribed into cDNA using the PrimeScript RT-PCR Kit according to the manufacturer’s instructions. The PCR amplification procedure was as follows: an initial denaturing step at 98°C for 2 min, followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 15 s, and extension at 68°C for 90 s (MightyAmp™ DNA Polymerase Ver. 3, TaKaRa). The amplified PCR products were purified on a 1% agarose gel (Gel Extraction Kit, omega), then ligated into pMD18-T vector (TaKaRa), and sequenced (Hzykang, Hangzhou, China). As a result, the full-length open reading frames (ORF) of R. philippinarum Fad and Elovl were obtained.
The deduced amino acid (aa) sequences of the cloned R. philippinarum Fad and Elovl were aligned with those of a representative marine mollusc, S. constricta, the only marine mollusc known to have all of the Fad (Ran et al., 2018) and Elovl (Ran et al., 2019) crucial for LC-PUFAs production via the Sprecher pathway (Sprecher, 2000), by using ClustalX 1.83, respectively. The phylogenetic trees were constructed using the maximum-likelihood method with the MEGA 7.0 package, on the basis of deduced aa sequences of Fad or Elovl from R. philippinarum and other representative organisms, with confidence in the resulting phylogenetic tree branch topology measured by bootstrapping through 1,000 iterations.
The tissue distribution of R. philippinarum Fad and Elovl was determined by quantitative real-time PCR (qRT-PCR). Tissues including the exhalent siphon, inhalent siphon, gill, labial palps, digestive glands, intestine, foot muscle, and mantle were sampled from three individuals (39.51 ± 0.23 mm × 26.39 ± 0.41 mm, shell length × shell width) and pooled together as one sample, respectively. Each sample was performed in triplicate. Total RNA was extracted from R. philippinarum samples by using the MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan). RNA quality and concentration were determined using a Thermo Scientific NanoDrop One spectrophotometer (Thermo Fisher Scientific, USA). Subsequently, one μg of total RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix (Perfect Real Time, TaKaRa) following the manufacturer’s protocol. The qRT-PCR was carried out in a quantitative thermal cycler (LongGene Q2000A, Hangzhou) using SYBR® Premix Ex Taq™ II (Perfect Real Time, TaKaRa) and specific q-primers as shown in Table S1. The qRT-PCR procedure was conducted with an initial denaturation step at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. To determine that a single product is present in each reaction, a melting curve with 1°C increments from 65°C to 90°C was performed. The relative expression of R. philippinarum Fad and Elovl was calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001), and β-actin was selected as the reference gene.
The microalgae used in this study, including Chlorella sp., C. calcitrans and I. galbana, were provided by the microalgal culture laboratory at Ningbo University. The cultivation of microalgae was conducted according to the methods described by Ran et al. (2020) with little modification. Specifically, the seawater of 23 psu (practical salinity unit) was used as the culture medium, which was obtained by adding fresh water and sea salt (Bosskas, Shanghai, China). The salinity was measured by using a refraction salinity meter (BK8180, BOKLES FIREMATE CO., LTD., Taiwan, China). The nutrient solution for microalgae cultivation was the NMB3 medium, which was composed of KNO3 (100 mg/L), KH2PO4 (10 mg/L), MnSO4·H2O (2.5mg/L), FeSO4·7H2O (2.5 mg/L), EDTA-Na2 (10 mg/L), vitamin B1 (6 μg/L), and vitamin B12 (0.05 μg/L) (Yang et al., 2016). In addition, sodium metasilicate (20 mg/L) was added as a silica source for the culture of the diatom C. calcitrans. The detailed cultivation steps were as follows. Microalgae was initially grown in 5-L flasks at 25 ± 1°C under continuous illumination (160-180 μmol photons/m2/s) provided by cool white fluorescent tubes for about 1 week. Microalgae growing in the exponential phase were then transferred to 50-L barrels with hypochlorite-sterilized seawater, and continuous aeration was supplied to support the massive growth. The microalgae growing in stationary phase were harvested and used to feed the R. philippinarum. Meanwhile, part of them were collected by centrifuging (13000×g, 4°C) and stored at -80°C for further FA analysis.
The rearing seawater (25 psu, 23 ± 0.5°C) was first filtered by 75-μm nylon cribrose silk and further hypochlorite-sterilized before use. Adult R. philippinarum with an average size of 38.14 ± 1.29 mm × 26.92 ± 1.24 mm (shell length × shell width) were purchased from a local clam farm in Ningbo, China. The clams were washed gently with seawater to clean the dirt on the shell surface, then starved for 24 h prior to the subsequent feeding experiment to eliminate the effect of feces and residual diets. A total of 270 individuals were randomly allocated to nine aquariums (40 × 40 × 30 cm, length × width × height), 30 clams per aquarium. The clams were fed either Chlorella sp., C. calcitrans or I. galbana with a concentration of 3~4 ×105 cells/mL twice daily (7:30 AM and 8:30 PM) to ensure they were satiated. Each treatment was performed in triplicate. The experiments were carried out in an air-conditioned room (23 °C) and lasted for a week. During this period, the seawater was refreshed twice daily (just before the feeding time), and continuous aeration was supplied. According to the qRT-PCR results of tissue distribution of R. philippinarum Fad and Elovl (Figure 1), the intestine and digestive gland (named as visceral mass) with significantly higher expression of those genes were sampled at 0, 3, 6, 12, 24, 48, 72 h and 7 d after the first feeding for detecting the changes of gene expression. Moreover, the muscular tissues (siphons and foot) and visceral masses were also sampled at 0 h and 7 d, respectively, for analyzing the changes in FA composition. At each sampling timepoint, the visceral mass of three individuals from each aquarium was pooled together as one sample, and each sample was in triplicate. The samples were flash frozen in liquid nitrogen and immediately stored at -80 °C for further analyses.
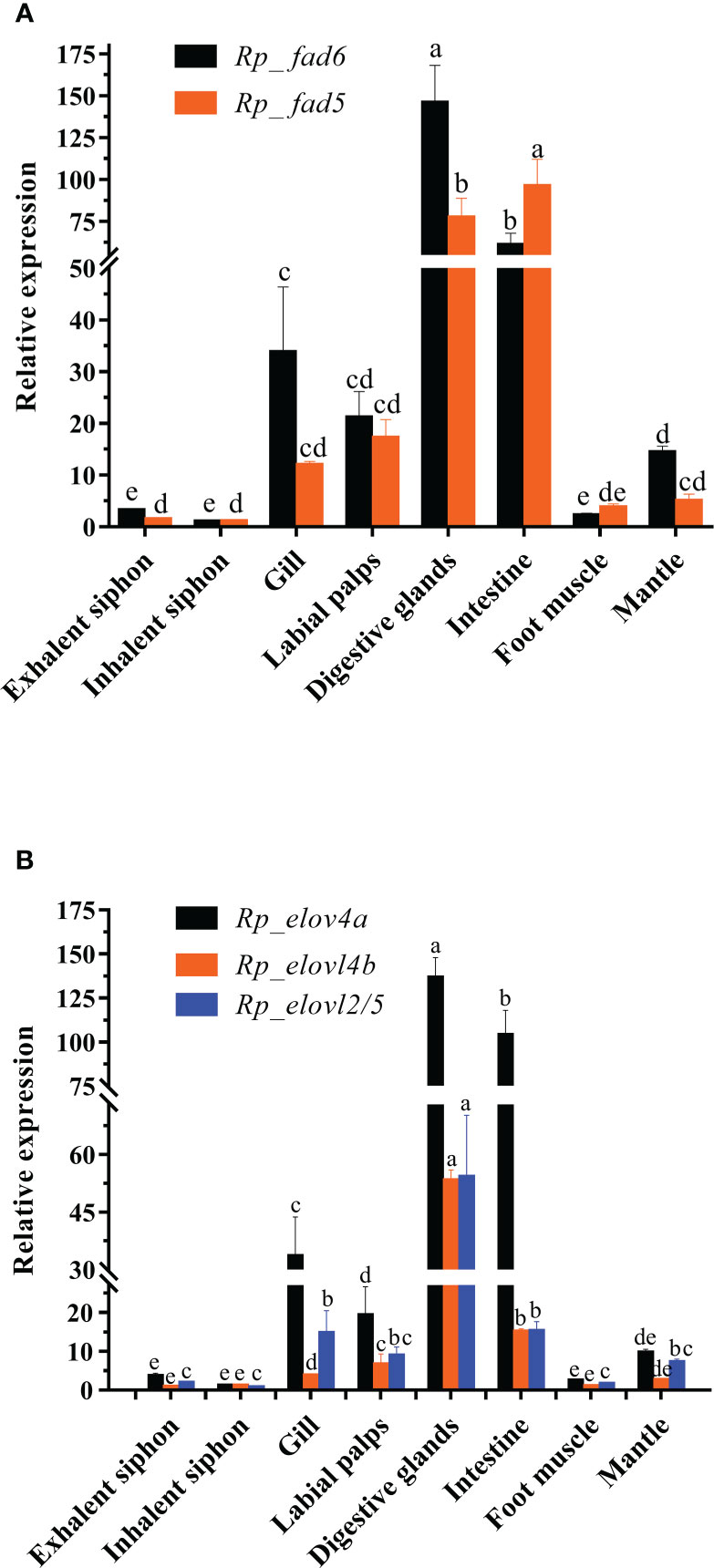
Figure 1 Tissue distributions of R. philippinarum Fads (A) and Elovls (B). Relative expression of those genes was measured by qRT-PCR, normalized by β-actin, and calibrated by that of inhalent siphon, respectively. Values sharing the common letters on the top of the same color columns are not significantly different (P ≥ 0.05).
The FA composition of the above-mentioned corresponding samples was determined using the method described by Wang et al. (2022). Briefly, an appropriate amount (30 mg) of the freeze-dried samples was first accurately weighed and then transferred into a 10 mL screw-cap glass tube along with 1 mL n-hexane, 1.5 mL of a mixed solution of methanol and acetyl chloride (Vmethanol: Vacetyl chloride = 10: 1), and 15 μL nonadecanoic acid (as an internal standard, 1 μg/μL). Followed by, the glass tube was vortexed vigorously for 30 s, and then tighten with a screw-cap and incubated in a water bath at 60 °C for 2 h. Followed by, the glass tube was equilibrated to room temperature, and then added with 2.5 mL of 6% K2CO3 and 1 mL of n-hexane. After vortexed for 30 s, the glass tube was subjected to centrifugation at 3,000 rpm for 10 min. Finally, the supernatant containing the fatty acid methyl esters (FAMEs) was collected and filtered by an organic phase filter membrane (0.22 μm) into a 2-mL screw-capped sample bottle.
The FAMEs obtained-above were further subjected to a GC-MS platform of Agilent 8890A-5977B equipped with a CD-2560 capillary column (100 m× 0.25 mm× 0.2 μm, CNW) and an auto-sampler (7963A). Briefly, high-purity helium was supplied as the carrier gas at a constant flow rate of 0.8 mL/min. The injector temperature was set at 250°C. The sample was injected with a volume of 1 μL at a split ratio of 1:5. After injection, the oven temperature was first held at 140°C for 5 min, and then raised to 240°C at a rate of 4°C/min and kept for 20 min. The solvent shutoff time was set to 13.7 min. The mass spectrometer was operated in electron compact mode with an electron energy of 70 eV. The ion source temperature and quadrupole temperature were set at 230°C and 150°C, respectively. The mass spectrometer scans from m/z 50 to m/z 600.
The FAs were identified by combining the relative retention times of commercial standards and the mass spectrometry databases (NIST14.L and Wiley7). The percentage composition of FA (%) was calculated with the formula: 100% × [area of specific FA/area of total FA] while the content composition (μg/mg) of FA was calculated with the formula [(Minternal standard × area of specific FA)/(area of internal standard × Msample)] Minternal standard = 15 μg, Msample = 30 mg.
Statistical analyses of gene expression and FA composition were tested by one-way analysis of variance (ANOVA), followed by Tukey’s honestly significant difference test using SPSS software (IBM SPSS Statistics 25.0 software, USA). Data were represented as mean ± SD, and a P value < 0.05 was considered statistically significant.
The detailed sequence information of Rp_fad5, Rp_fad6, Rp_elovl2/5, Rp_elovl4a and Rp_elovl4b had been deposited in the GenBank database with the accession numbers of OP779675-OP779679, respectively. In brief, the ORF of the putative Rp_fad5 was 1,311 bp encoding a protein of 436 aa, while the ORF of the putative Rp_fad6 was 1,317 bp encoding a protein of 438 aa. The aa sequences of Rp_fad5 and Rp_fad6 shared 68% and 76% homology with that of S. constricta Δ5 Fad and Δ6 Fad, respectively. Meanwhile, all R. philippinarum Fads contained the three histidine boxes (H***H, H**HH, and QIEHH) and an N-terminal cytochrome b5 domain with the heme-binding motif (HPGG) conserved in typical front-end Fads (Figure 2). The ORF of the putative Rp_elovl2/5 was 975 bp, encoding a polypeptide of 324 aa, and shared 67% identity with S. constricta Elovl2/5. The ORFs of Rp_elovl4a and Rp_elovl4b were 885 bp and 888 bp, encoding polypeptides of 294 aa and 295 aa, respectively. Notably, the aa sequences of Rp_elovl4a and Rp_elovl4b showed a high identity (89.15%), with the main different aa region between them being highlighted with a bold line square in Figure 3. When compared to S. constricta Elovls with Elovl4 activity, the aa sequences of Rp_elovl4a and Rp_elovl4b were 78-85% identical to S. constricta Elovl4a and Elovl4b, while 39.7-44.7% identical to S. constricta Elovlc. In addition, all R. philippinarum Elovls contained the histidine box (H**HH) and “Q,” which are conserved in the Elovl family involved in LC-PUFA elongation (Figure 3).
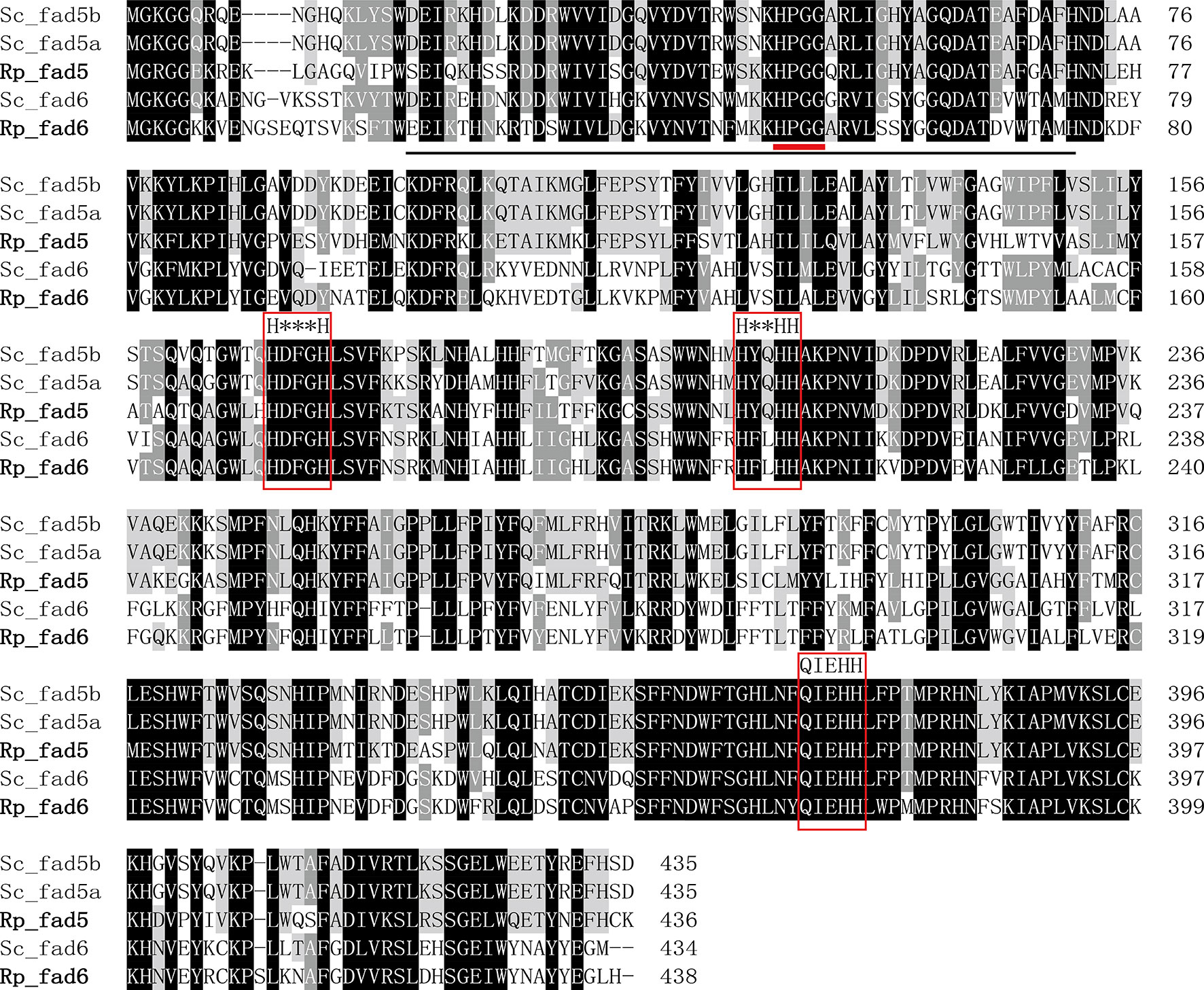
Figure 2 Alignment of the deduced amino acid (aa) sequences of Rp_fad5 and Rp_fad6 with Δ5 Fads (Sc_fad5a, AWC67984.1 and Sc_fad5b, AWC67985.1) and Δ6 Fad (Sc_fad6, AWC67986.1) from S. constricta by using ClustalX 1.83. The cytochrome-b5 like domain is solid-underlined, the heme-binding motif of HPGG is denoted with a short bold red line, and the three histidine boxes (H***H, H**HH and QIEHH) are highlighted with red frames.
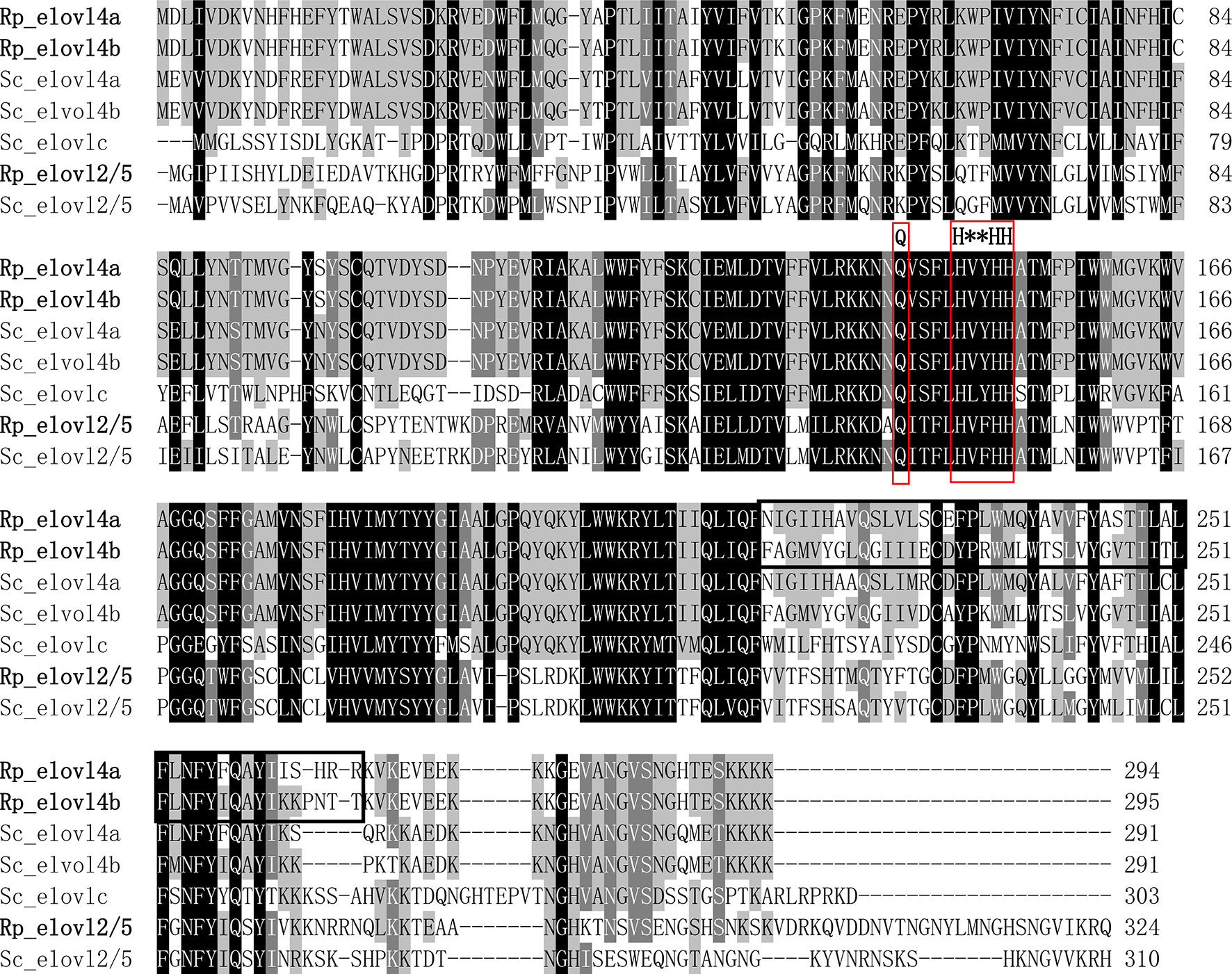
Figure 3 Alignment of the deduced aa sequences of Rp_elovl2/5, Rp_elovl4a and Rp_elovl4b and with Elovl2/5 (Sc_elovl2/5, QCF41170.1), Elovl4 (Sc_elovl4a, QCF41171.1; Sc_elovl4b, QCF41172.1) and a novel Elovl with Elovl4 activity (Sc_elovlc, QCF41173.1) from S. constricta by using ClustalX 1.83. The histidine box (H**HH) and diagnostic “Q” conserved in LC-PUFA elongase are highlighted with red frames. The different aa domains between Rp_elovl4a and Rp_elovl4b are indicated with a bold line square.
To further confirm the putative functions of R. philippinarum Fad and Elovl and reveal their evolutionary relationships with their homologies, respectively, the phylogenetic analysis was conducted. In terms of Fads, the phylogenetic tree was generally clustered into three groups (Figure 4). Briefly, Rp_fad6 was clustered together with the functionally characterized Δ6 Fad of S. constricta and Fad-like genes of other marine invertebrates (not functionally characterized, denoted with *); Rp_fad5 was clustered together with the functionally characterized Δ5 Fad of S. constricta and some other marine invertebrates, while the Δ5/6 Fads of teleosts and mammals were grouped together (Figure 4). When it comes to Elovls, the phylogenetic tree was also generally clustered into three groups (Figure 5). Briefly, Rp_elvol2/5 was grouped together with the functionally characterized Elovl2/5 from invertebrates including S. constricta; the Elovl2 and Elovl5 of vertebrates were clustered together, while Rp_elvol4a and Rp_elvol4b were clustered together with Elovl4 from both vertebrates and invertebrates and more closed to those of invertebrates.
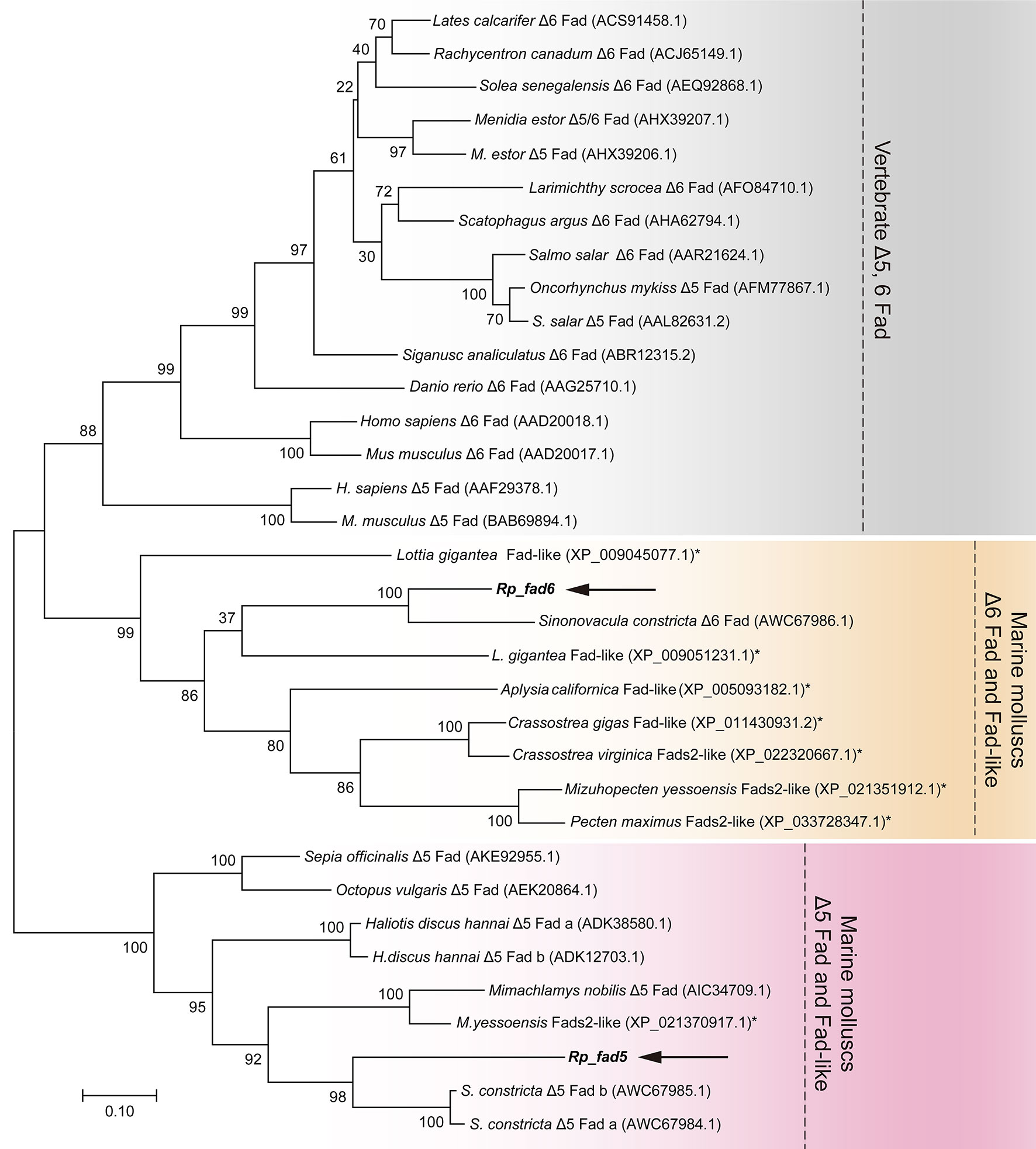
Figure 4 Phylogenetic tree comparing the deduced aa sequences of Rp_ fad5 and Rp_ fad6 with their orthologues from representative vertebrates and invertebrates. The tree was constructed using the maximum likelihood method by MEGA 7.0. The horizontal branch length is proportional to the aa substitution rate per site. The numbers represent the frequencies (%) with which the tree topology presented was replicated after 1,000 iterations. An asterisk marks Fad genes of invertebrates that have not been functionally characterized to date.
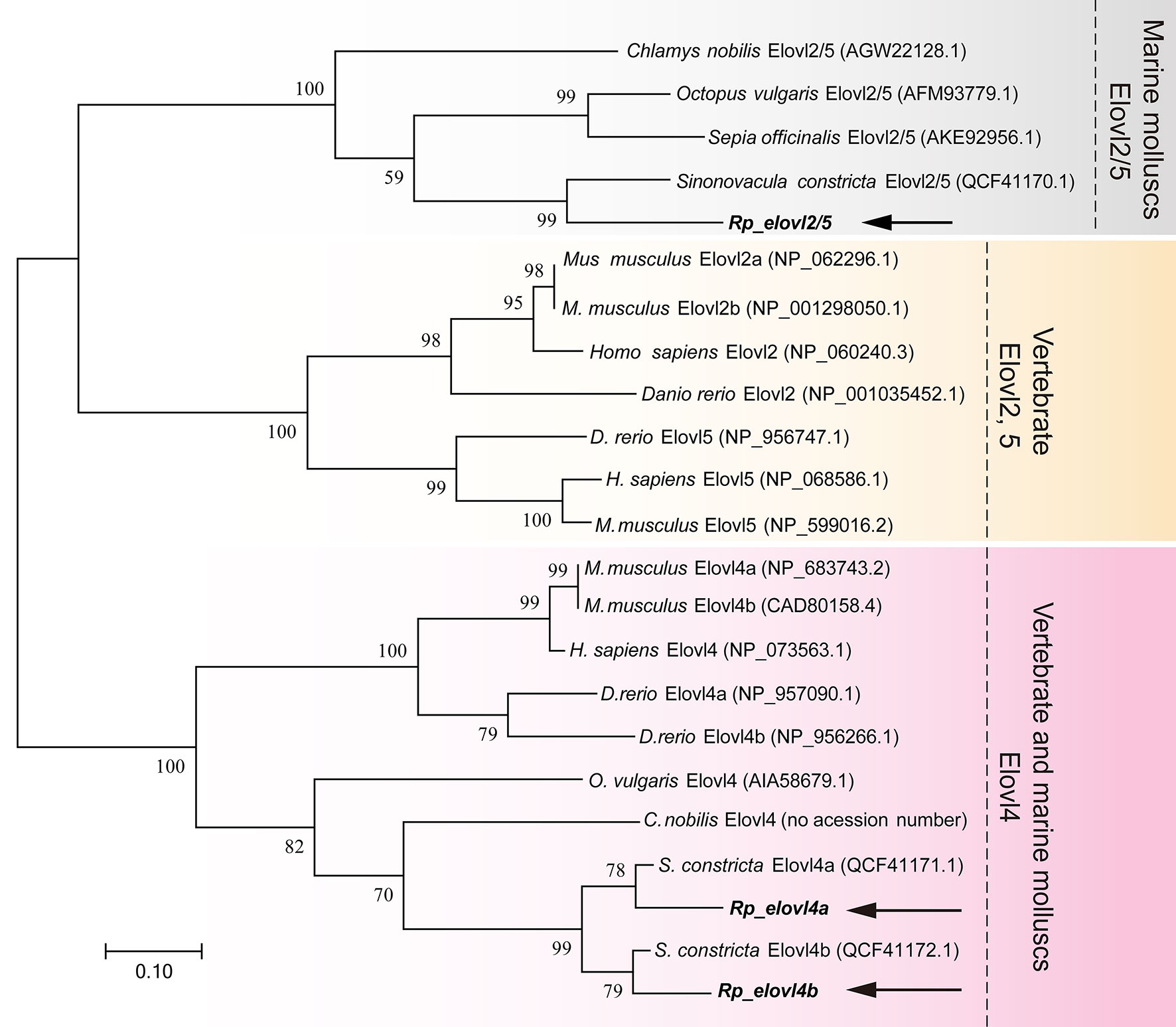
Figure 5 Phylogenetic tree comparing the deduced aa sequences of Rp_elovl2/5 and Rp_elovl4a/b with their orthologues from representative vertebrates and invertebrates. The tree was constructed using the maximum likelihood method with MEGA7.0. The horizontal branch length is proportional to aa substitution rate per site. The numbers represent the frequencies (%) with which the tree topology presented was replicated after 1,000 iterations.
To indicate the potential key tissues executing LC-PUFA biosynthesis in this clam, the tissue distributions of R. philippinarum Fad and Elovl were analyzed by qRT-PCR. The results showed that the transcripts of R. philippinarum Fad and Elovl were detected in all tissues analyzed (Figure 1). Specifically, the expression of Rp_fad6 was detected highest in digestive glands, followed by the intestine, gill, labial palps and mantle, with relatively low expression in other tissues. The expression of Rp_fad5 in the intestine was found significantly higher than that in the digestive glands, which was further significantly higher than that in the other six tissues, respectively. In terms of Elovls, Rp_elovl4a was expressed highest in the digestive glands, followed by the intestine, gill and labial palps. Similarly, Rp_elovl4b showed the highest expression in the digestive glands, followed by the intestine, labial palps, and gills. While the expression of Rp_elovl2/5 in the digestive glands was significantly higher than that in the other seven tissues, respectively.
In the present study, three microalgae including Chlorella sp., C. calcitrans and I. galbana, with the distinct LC-PUFA composition (Tables S2, S3), were selected as the microalgae diets. Specifically, as shown in Figure 6, the Chlorella sp. was characterized by significantly higher LA and ALA, each containing 14.07 ± 0.01% and 14.96 ± 0.09%, respectively, but without C>20 LC-PUFAs; C. calcitrans was characterized by significantly higher EPA (10.09 ± 0.16%) but with trace amounts of DHA (0.64 ± 0.03%), while I. galbana was characterized by significantly higher DHA (10.99 ± 0.11%).
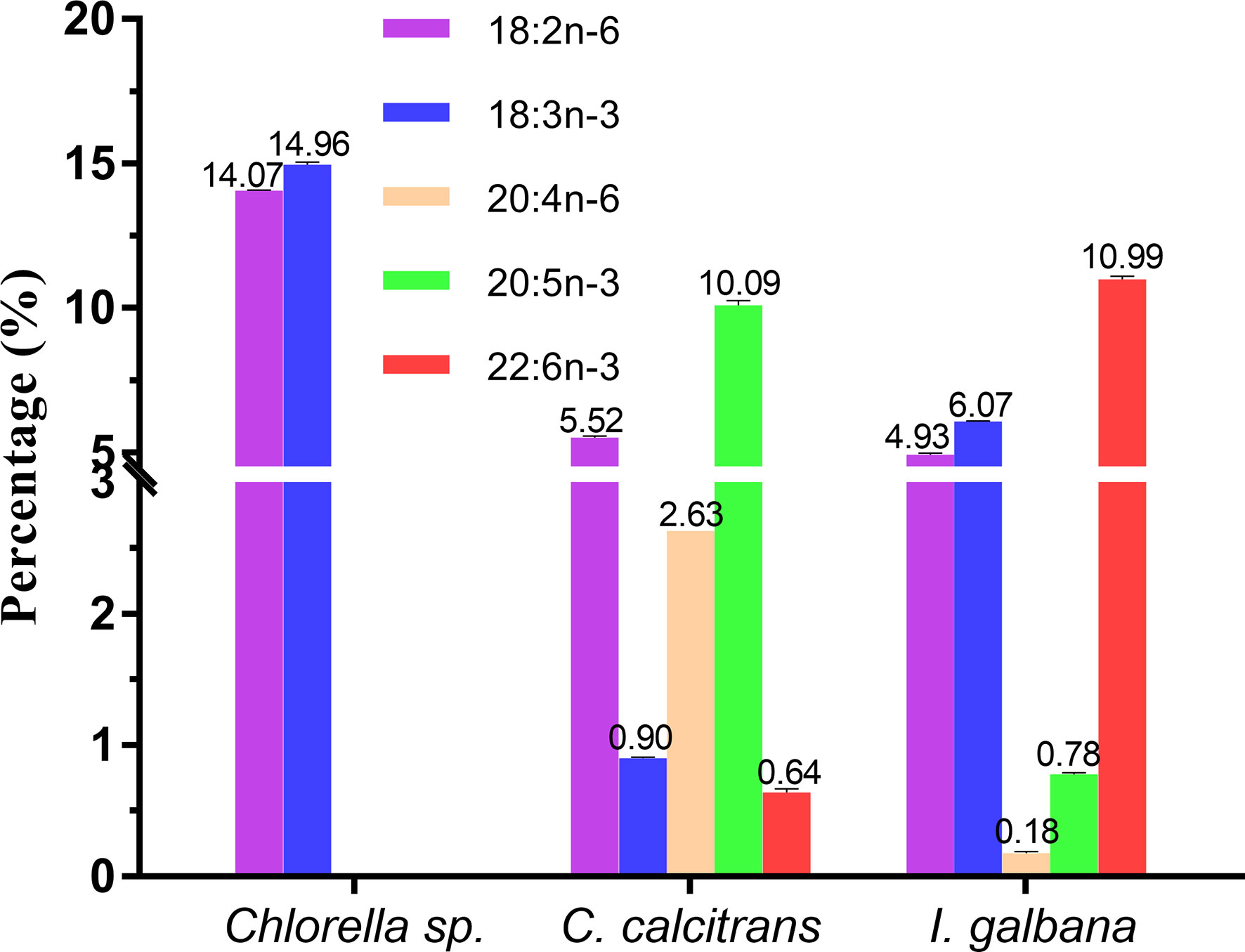
Figure 6 Representative fatty acids composition (%) of Chlorella sp., Chaetoceros calcitrans and Isochrysis galbana.
The feeding effects of the three microalgae diets on the expressions of R. philippinarum Fad and Elovl are shown in Figure 7. Generally, at most of the sampling timepoint, the expression of all R. philippinarum Fad and Elovl in visceral mass of R. philippinarum when fed with Chlorella sp. were significantly higher than those fed with C. calcitrans, which were further significantly higher than those fed with I. galbana. Specifically, during the entire experiment, the expression of all the Fad and Elovl in visceral mass of R. philippinarum fed with Chlorella sp. peaked at 48 h, and then gradually decreased, but at 7 d, the expression of most genes was still higher than those at initial stage; the expression of all the Fad and Elovl in visceral mass of R. philippinarum fed with C. calcitrans peaked at 12 h and were maintained until 48 h, then decreased, but at 7 d, the expression of most genes was significantly lower than those at the initial stage; the expression of all the Fad and Elovl in visceral mass of R. philippinarum fed with I. galbana had no significant changes or even decreased slightly compared with those at the initial stage.
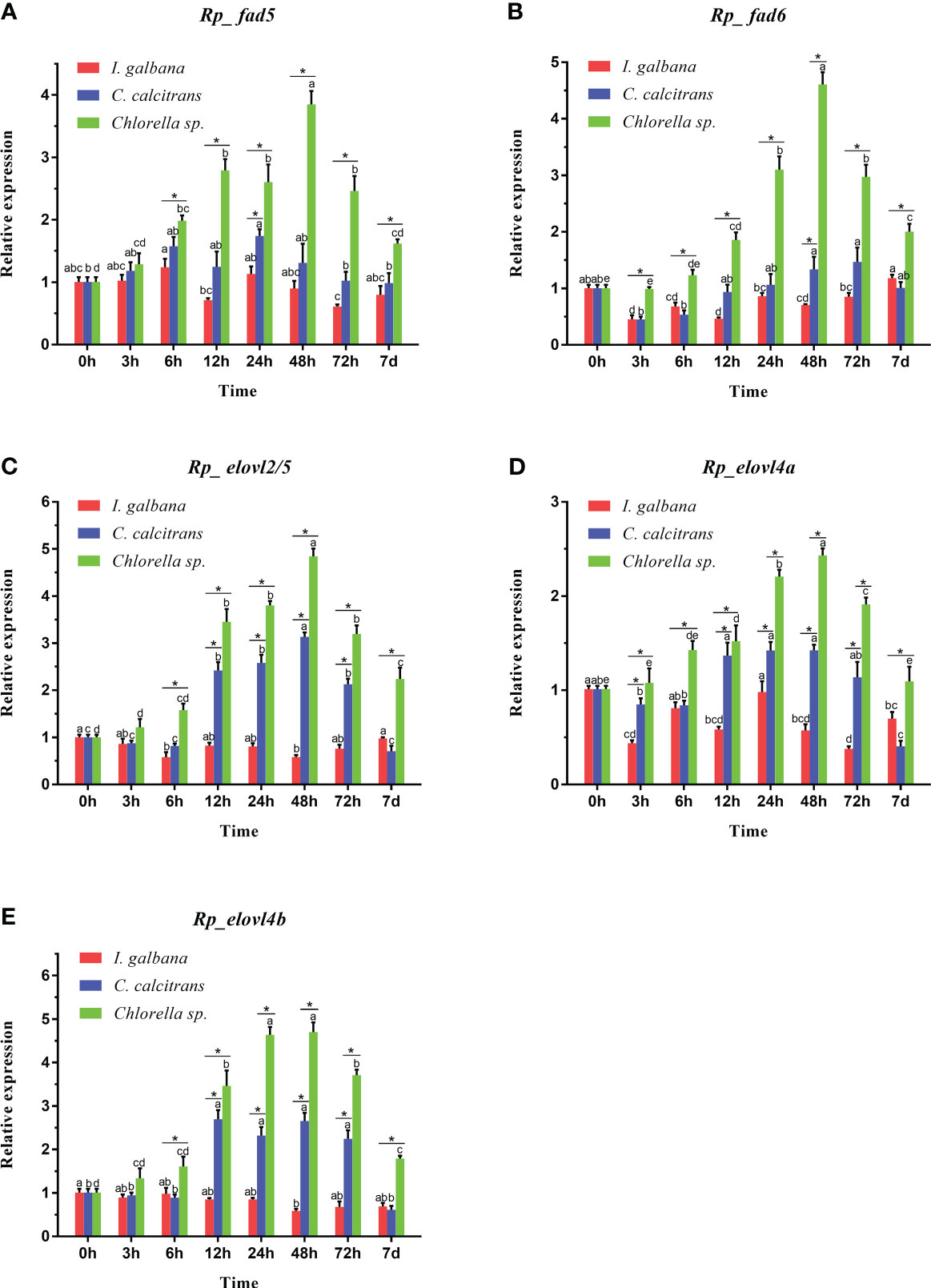
Figure 7 Expression patterns of Fad (A, B) and Elovl (C-E) in the visceral mass of R. philippinarum when fed with microalgae diets varied in LC-PUFA composition. Values sharing the common letters on the top of the same color columns are not significantly different (P ≥ 0.05). In addition, values at the same sampling timepoint sharing an asterisk are significantly different (P < 0.05).
As shown in Tables 1–4, there were significant differences in FA composition of the visceral mass and muscular tissues in R. philippinarum when fed with different microalgae diets before and after the experiment, respectively.
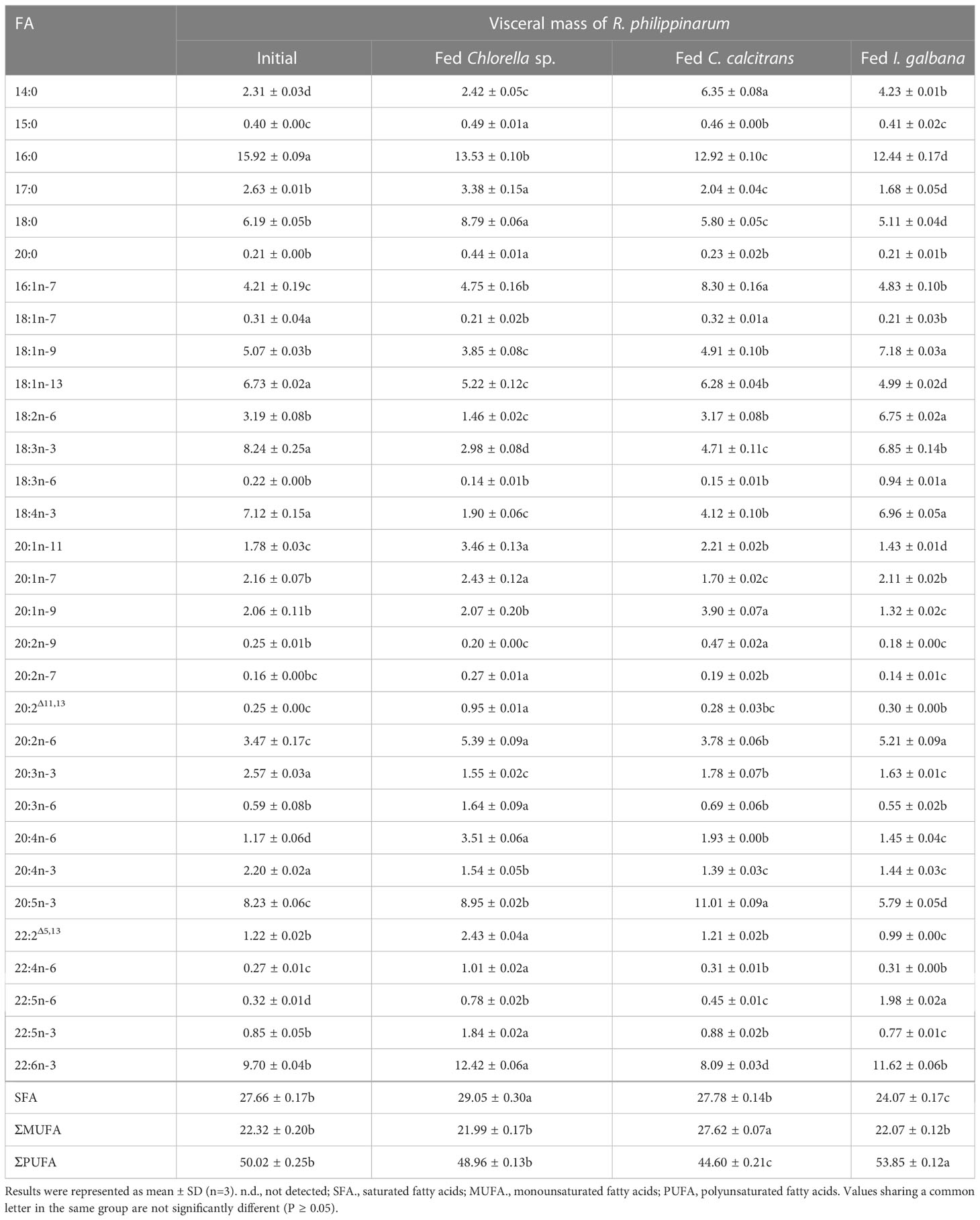
Table 1 Fatty acids composition (%) in visceral mass of R. philippinarum fed with microalgae diets varied in LC-PUFA composition before and after the experiment.
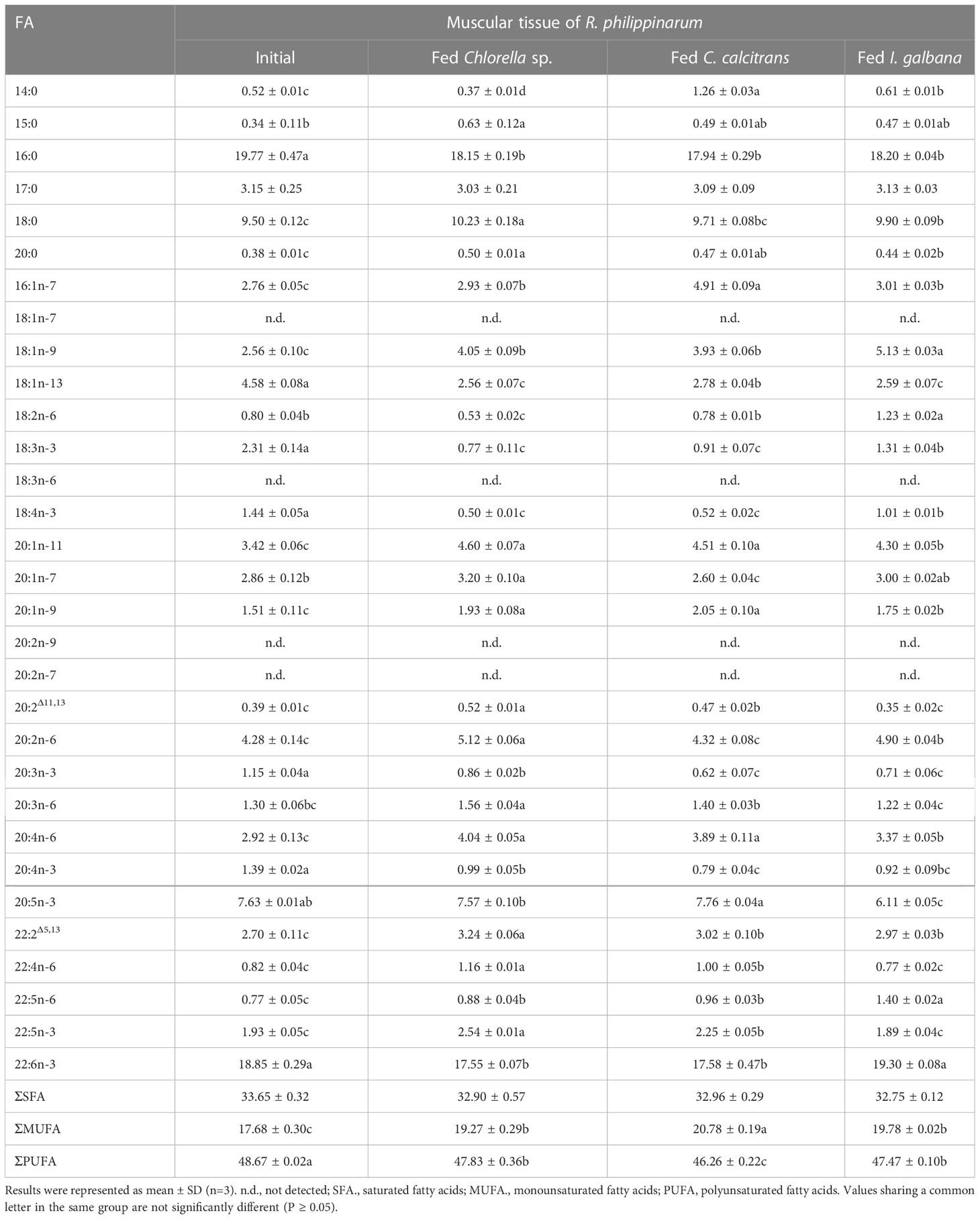
Table 2 Fatty acids composition (%) in muscular tissue of R. philippinarum fed with microalgae diets varied in LC-PUFA composition before and after the experiment.
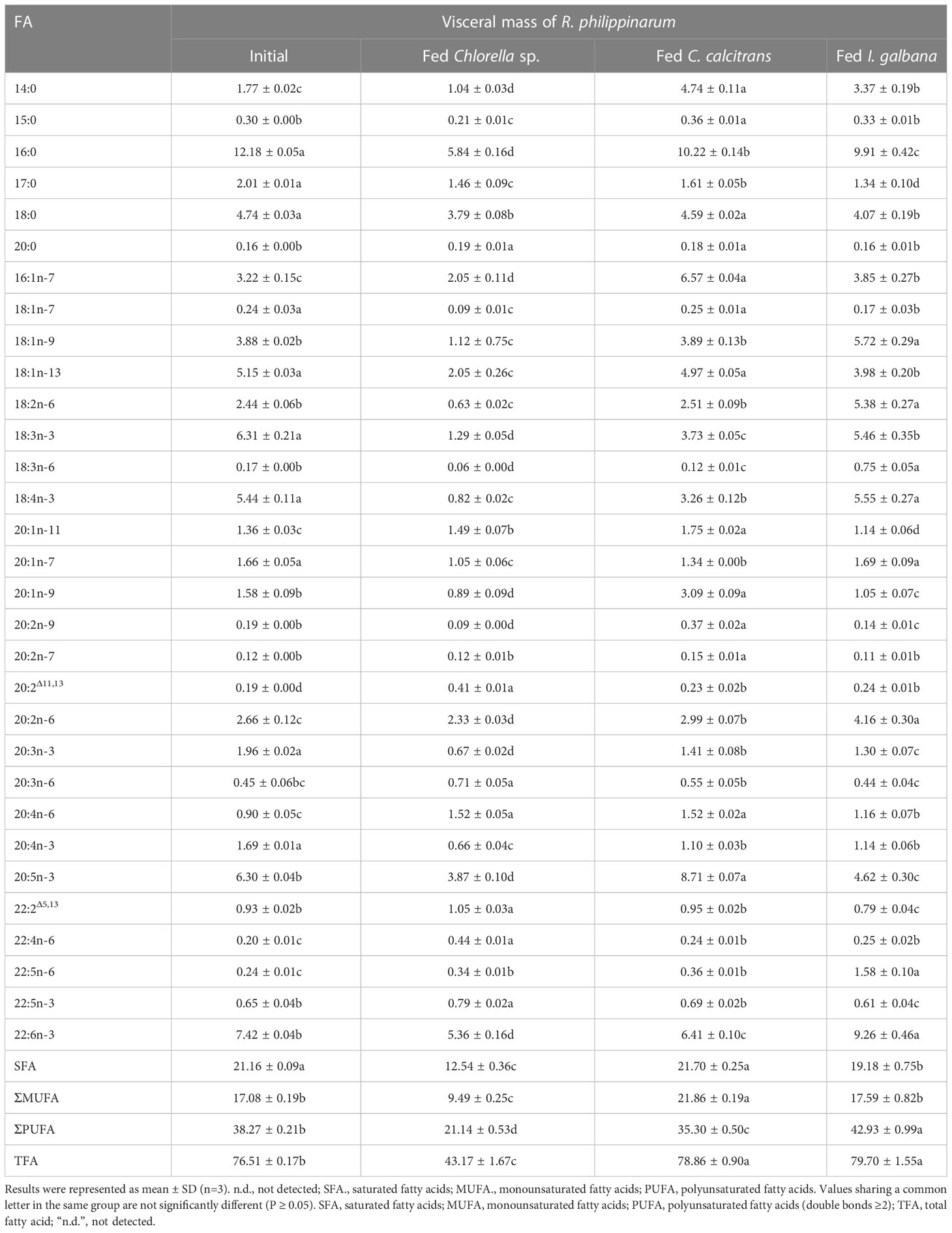
Table 3 Content compositions (μg/mg) of fatty acid in visceral mass of R. philippinarum when fed with microalgae diets varied in LC-PUFA composition before and after the experiment.
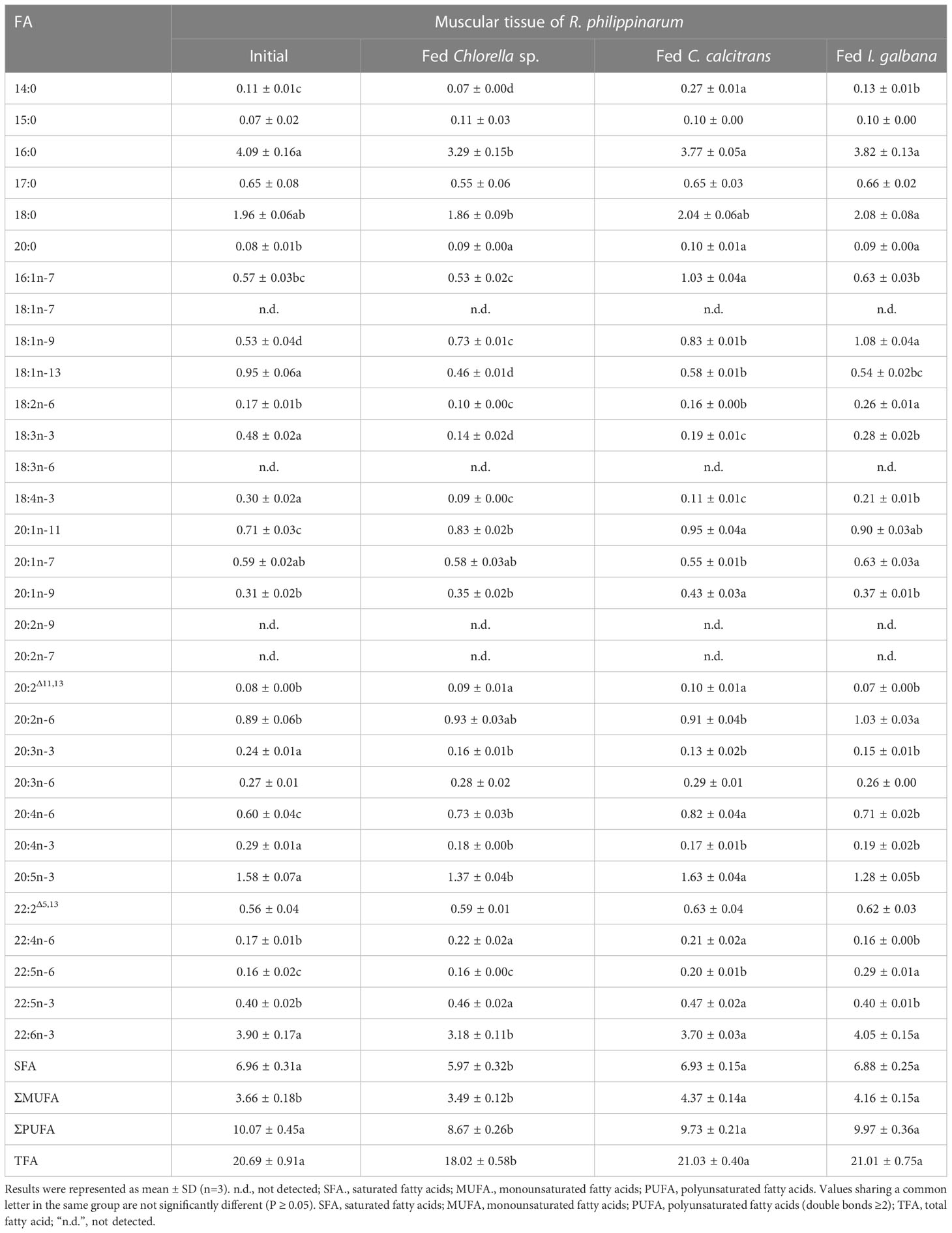
Table 4 Content compositions (μg/mg) of fatty acid in muscular tissue of R. philippinarum when fed with microalgae diets varied in LC-PUFA composition before and after the experiment.
In terms of visceral mass, the percentage composition of 14:0 in R. philippinarum fed with C. calcitrans was significantly higher than that of those fed with Chlorella sp. and I. galbana (Table 1). The 16:0 and 18:0 showed a significantly higher amount of R. philippinarum fed with Chlorella sp. than those fed with C. calcitrans and I. galbana (Table 1). The proportion of 16:1n-7 was significantly increased in R. philippinarum fed with all three microalgae diets, with the highest level in R. philippinarum fed with C. calcitrans, which was significantly higher than that of those fed with C. calcitrans and I. galbana (Table 1). Interestingly, the oleic acid (18:1n-9, OA), LA, and ALA percentages were significantly decreased in R. philippinarum fed with Chlorella sp., which were significantly lower than those fed with C. calcitrans and I. galbana (Table 1). In addition, the amount of 18:3n-6 and 18:4n-3 was decreased in R. philippinarum fed with Chlorella sp. and C. calcitrans, which was significantly lower than that in those fed with I. galbana (Tables 1, 3). The levels of 20:3n-6 and ARA were significantly increased in R. philippinarum fed with Chlorella sp. and were significantly higher than those in fed with C. calcitrans and I. galbana (Table 1). For EPA, a significantly higher level was found in R. philippinarum fed with C. calcitrans, followed by Chlorella sp., and then I. galbana (Tables 1, 3). Significantly higher percentages of 22:4n-6 and 22:5n-3 were found in R. philippinarum fed with Chlorella sp. compared to those fed with C. calcitrans and I. galbana (Table 1). Notably, a significantly higher percentage composition of DHA was found in R. philippinarum fed with Chlorella sp. (Table 1). However, the content composition of DHA was the opposite, with the lowest levels found in R. philippinarum fed with Chlorella sp. (Table 3).
In terms of muscular tissues, the percentage composition of 14:0 and 16:1n-7 in R. philippinarum fed with C. calcitrans was significantly higher than that of those fed with Chlorella sp. and I. galbana. Levels of 16:0 significantly decreased in R. philippinarum fed both experimental diets and showed no significant differences among them (Table 2). The proportion of 18:0 was significantly increased in the R. philippinarum fed with Chlorella sp., which was significantly higher than those fed with C. calcitrans and I. galbana (Table 2). Regarding the 18-C FA, the percentage composition of LA and ALA was decreased significantly in the muscular tissue of R. philippinarum fed with Chlorella sp., which was significantly lower than that of those fed with I. galbana (Table 2). The proportion of 20:3n-6 in the muscular tissue of R. philippinarum fed with Chlorella sp. was significantly increased and higher than those fed with C. calcitrans and I. galbana (Table 2). Besides, the amount of ARA was significantly increased in all three experimental diets, with a higher proportion in R. philippinarum fed with Chlorella sp. and C. calcitrans (Table 2). A significantly higher amount of EPA was found in muscular tissue of R. philippinarum fed with C. calcitrans compared to those fed with Chlorella sp. and I. galbana (Tables 2, 4). As in the visceral mass, a significantly higher percentage of 22:4n-6 and 22:5n-3 was also found in the muscular tissue of R. philippinarum fed with Chlorella sp. compared to those fed with C. calcitrans and I. galbana (Table 2). Also, the proportion of DHA in muscular tissue of R. philippinarum fed with I. galbana was significantly higher than those fed with C. calcitrans and I. galbana (Table 2).
While 16:2n-6 and 16:3n-3 had significantly higher percentage compositions in Chlorella sp. and C. calcitrans (Table S2), respectively, the two fatty acids were not detected in any of the experimental tissues of R. philippinarum. In contrast, 22:4n-6 and 22:5n-3, which were not present in the three experimental diets, showed significant increases in both visceral mass and muscle tissue in R. philippinarum fed with Chlorella sp., which were significantly higher than those fed with C. calcitrans and I. galbana (Tables 1, 2). Moreover, two non-methylene-interrupted dienoic (NMID) fatty acids, namely 20:2Δ11,13and 22:2Δ5,13, were significantly increased after the experiment in both muscular tissues and visceral masses of R. philippinarum fed with Chlorella sp., which were both deficient in all three experimental diets (Tables 1, 2). The amount of 18:4n-3 in the visceral mass of R. philippinarum fed with I. galbana rich in 18:4n-3 did not change significantly but showed a significant decrease in the muscular tissue (Tables 1, 2). The percentage composition of 20:3n-6 did not change significantly in both visceral mass and muscular tissue of R. philippinarum fed with I. galbana, while a significant increase was observed in R. philippinarum fed with Chlorella sp. (Tables 1, 2). Although there was no significant difference in the content composition of ARA in the visceral mass of R. philippinarum fed Chlorella sp. and C. calcitrans (Table 3), the content composition of ARA in muscular tissue was significantly higher in R. philippinarum fed C. calcitrans than in those fed Chlorella sp. (Table 4).
The R. philippinarum is abundant in LC-PUFA, particularly DHA, providing as an excellent LC-PUFA resource for human dietary requirement. However, the potential LC-PUFA biosynthetic capability and the dietary effects on its LC-PUFA biosynthesis still remain unclear. In the present study, the critical genes of Fad and Elovl involved in LC-PUFA synthesis were firstly cloned from R. philippinarum, and then their roles in determining the FA profile in this bivalve were investigated by feeding different microalgae varied in LC-PUFA composition.
Both the newly cloned Rp_fad5 and Rp_fad6 have typical features of front-end desaturases, including three conserved histidine boxes, a heme-binding motif HPGG, and a cytochrome-like b5 structural domain (Hashimoto et al., 2008), indicating the conserved functional structural domains of Fad during evolution and a potential desaturation activity of R. philippinarum Fad. Similarly, the newly cloned Rp_elovl2/5, Rp_elovl4a and Rp_elovl4b all have a typical histidine box, and a diagnostic glutamine (Q) that is highly conserved in the Elovl protein family, suggesting their functional roles in LC-PUFA elongation. In addition, the phylogenetic tree showed that the newly cloned Rp_fad5 was grouped together with Δ5 Fads from S. constricta and other marine molluscs, indicating a potential Δ5 Fad activity of the Rp_fad5. Meanwhile, the Rp_fad6 was grouped together with Δ6 Fad of S. constricta and other unfunctionally characterized Fad from other marine molluscs, indicating a potential Δ6 Fad activity of the Rp_fad6. Similarly, the Rp_elovl2/5 and Rp_elovl4a/4b were clustered together with Elovl2/5 and Elovl4 from S. constricta and other marine molluscs, respectively, suggesting a potential corresponding Elovl2/5 and Elovl4 elongation activities of those R. philippinarum Elovl, respectively. Furthermore, the tissue distribution results showed that all R. philippinarum Fad and Elovl were expressed highest in the visceral tissues, including the intestine and digestive glands, further indicating that those genes might be functional in the elongation and desaturation of the dietary LC-PUFA precursors. Besides, the tissue distribution of R. philippinarum Fad and Elovl was consistent with that of S. constricta (Ran et al., 2018; Ran et al., 2019). Taken together, those results suggested that a complete LC-PUFA biosynthetic pathway of the “Sprecher pathway” might also exist in R. philippinarum, as was demonstrated in S. constricta (Ran et al., 2018; Ran et al., 2019). Nevertheless, the detailed functions and activities of R. philippinarum Fad and Elovl need further studies.
Based on the tissue distribution of R. philippinarum Fad and Elovl, which were expressed highest in the intestine and digestive glands, the two tissues (visceral mass) were sampled for the dietary effect on the Fad and Elovl expressions in this bivalve. Throughout the experiment, the expression of Fad and Elovl in the visceral mass of R. philippinarum fed with Chlorella sp. was significantly higher than that of those fed with I. galbana and C. calcitrans. The results might be related to the absence of LC-PUFA such as EPA and DHA in Chlorella sp., so that the R. philippinarum fed with Chlorella sp. must synthesize LC-PUFA rapidly by up-regulating Fad and Elovl expression to meet the development requirement. Meanwhile, the result might also be resulted from the high levels of LA and ALA in Chlorella sp., which provided abundant LC-PUFA precursors for elongation and desaturation. Consistently, all those genes showed no significant changes or even decreased slightly in the visceral mass of R. philippinarum fed with I. galbana, which might be due to the abundant DHA in this microalga. In contrast, the expression of Fad and Elovl in the visceral mass of R. philippinarum fed with C. calcitrans also showed an obvious increase compared to those fed with I. galbana, which might be due to the fact that C. calcitrans is rich in EPA but lacking in DHA, and DHA synthesis still must be mobilized in this bivalve when fed C. calcitrans. Meanwhile, the differences of Fad and Elovl expressions in R. philippinarum when fed I. galbana and C. calcitrans might be resulted from that DHA inhibits the expression of genes involved in the LC-PUFA synthesis but not EPA, which has been well-established in Atlantic salmon (Salmo salar) (Betancor et al., 2015; Betancor et al., 2016). Besides, those results also suggested that DHA might play a more important physiological role than EPA in R. philippinarum.
The FA composition of R. philippinarum visceral mass and muscular tissues generally mirrored that of microalgal diets. This finding is in agreement with other studies on R. philippinarum (Caers et al., 1999; Fernandez-Reiriz et al., 2006). Specifically, we found high levels of both 16:0 and 18:0 in the visceral mass and muscular tissues, and their proportions were significantly higher in R. philippinarum fed with Chlorella sp. than those fed with C. calcitrans and I. galbana, with the exception of 16:0 in the muscular tissues (Table 1 and 2). This may be related to their important cellular and metabolic functions (Legrand and Rioux, 2010) and also reflects the Chlorella sp. diet rich in 16:0 and 18:0 (Table S2, S3). A significantly higher level of 14:0 and 16:1n-7 was found in both experimental tissues of R. philippinarum fed with C. calcitrans than those fed with Chlorella sp. and I. galbana. This may be due to the fact that the amount of 14:0 and 16:1n-7 in the C. calcitrans was significantly higher than that in the Chlorella sp. and I. galbana (Tables S2, S3). Meanwhile, a significantly higher content of DHA and EPA was observed in both experimental tissues of R. philippinarum fed with I. galbana and C. calcitrans, respectively (Table 3 and 4). These results reflected well on the microalgal diets, that DHA and EPA were rich in I. galbana and C. calcitrans, respectively.
In general, the amount of 18C-FA, the main precursor of LC-PUFA biosynthesis, was significantly higher in visceral mass than in muscular tissues, which may suggest that visceral mass is the major site of LC-PUFA biosynthesis. While 16:2n-6 and 16:3n-3 had significantly higher percentage compositions in Chlorella sp. and C. calcitrans, respectively, the two FA were not detected in any of the experimental tissues of R. philippinarum. This may reflect that these two FA are not absorbed from the diet, suggesting that these two FA may not be essential for R. philippinarum. Notably, we found that R. philippinarum fed with Chlorella sp. had significantly lower levels of 18-C FA such as LA and ALA in the visceral mass than those fed with I. galbana (Tables 1– 4), despite the fact that Chlorella sp. is characterized by an abundance of LA and ALA (Figure 6). This appears to be a consequence of the deficient LC-PUFA such as EPA and DHA in the Chlorella sp., which have been considered as critical nutrients for bivalve growth and survival (Marshall et al., 2010; Pettersen et al., 2010), and the LA and ALA were rapidly consumed as LC-PUFA precursors in R. philippinarum fed with Chlorella sp., as reflected by a significant up-regulation of Fad and Elovl (Figure 7). The proportion of ARA showed an increase in the visceral mass and muscular tissues in all three dietary treatments, with the highest levels in R. philippinarum fed with Chlorella sp. (Tables 1, 2). This result indicated that ARA plays a critical role in R. philippinarum development as demonstrated by Tallima and El Ridi (2018) and R. philippinarum could selectively accumulate the ARA and might have the capacity to biosynthesize ARA with 18C-FA substrate. Although DHA was not present in the Chlorella sp. diet (Figure 6), a high proportion of DHA was found in the visceral mass of R. philippinarum fed Chlorella sp. The result indicated that R. philippinarum could synthesize DHA to a certain extent. In addition, two NMID FAs (20:2Δ11,13 and 22:2Δ5,13), 22:4n-6, and 22:5n-3 were absent from all three microalgae diets but found in both the visceral mass and muscular tissues of R. philippinarum. Besides, their percentages were significantly higher in R. philippinarum fed with Chlorella sp. than those fed with C. calcitrans and I. galbana. These results might suggest that R. philippinarum has the ability to self-synthesize FA and might point out a compensatory mechanism for the absence of EPA and DHA in Chlorella sp., as 22:5n-3 and NMID have potentially beneficial physiological effects on the organisms (Kraffe et al., 2004; Drouin et al., 2019).
From the above results, all the genes encoding Fad and Elovl required for the biosynthesis of LC-PUFA via the Sprecher pathway might be present in R. philippinarum. Their expression was significantly up-regulated in the visceral mass of R. philippinarum fed with Chlorella sp., suggesting that they might have corresponding desaturation and elongation activities and be involved in the biosynthesis of LC-PUFA in R. philippinarum with 18C-FA precursors such as LA and ALA. However, the TFA of R. philippinarum fed with Chlorella sp. was significantly lower compared to those fed with C. calcitrans and I. galbana (Tables 3, 4). These results may be due to the lower energy efficiency of the R. philippinarum in response to the Chlorella sp. diet, suggesting that the R. philippinarum might have a certain capability for LC-PUFA biosynthesis. Collectively, this study provided valuable insights into the biosynthesis of LC-PUFA in R. philippinarum, which indicated that the dietary LC-PUFA, especially EPA and DHA, were indispensable for its nutritional requirement.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank, OP779675-OP779679.
The studies involving animals were reviewed and approved by the Ningbo University Laboratory Animal Center under permit number no. SYXK (ZHE2008-0110).
KW wrote the original draft, performed the experiments, and analyzed the data. ZR designed the experiments, wrote, and revised the draft. JX and XY conceptualized and supervised the study. SW, KL, YL and HX performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (32102763), Zhejiang Major Science Project, China (2019C02057), the National Key Research and Development Program of China (2019YFD0900400), Ningbo Science and Technology Research Projects, China (202003N4124, 2019B10006), and China Agriculture Research System of MOF and MARA.
We are grateful to the microalgal culture laboratory at Ningbo University for kindly providing the microalgae used in the experiment.
Author JX is employed by Fujian Dalai Seeding Technology Co. LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1141231/full#supplementary-material
Betancor M. B., Sprague M., Sayanova O., Usher S., Campbell P. J., Napier J. A., et al. (2015). Evaluation of a high-EPA oil from transgenic camelina sativa in feeds for Atlantic salmon (Salmo salar l.): Effects on tissue fatty acid composition, histology and gene expression. Aquaculture 444, 1–12. doi: 10.1016/j.aquaculture.2015.03.020
Betancor M. B., Sprague M., Sayanova O., Usher S., Metochis C., Campbell P. J., et al. (2016). Nutritional evaluation of an EPA-DHA oil from transgenic camelina sativa in feeds for post-smolt Atlantic salmon (Salmo salar l.). PloS One 11 (7), e0159934. doi: 10.1371/journal.pone.0159934
Caers M., Coutteau P., Sorgeloos P. (1999). Dietary impact of algal and artificial diets, fed at different feeding rations, on the growth and fatty acid composition of Tapes philippinarum (L.) spat. Aquaculture 170 (3-4), 307–322. doi: 10.1016/S0044-8486(98)00410-4
Djuricic I., Calder P. C. (2021). Beneficial outcomes of omega-6 and omega-3 polyunsaturated fatty acids on human health: An update for 2021. Nutrients 13 (7), 2421. doi: 10.3390/nu13072421
Drouin G., Rioux V., Legrand P. (2019). The n-3 docosapentaenoic acid (DPA): A new player in the n-3 long chain polyunsaturated fatty acid family. Biochimie 159, 36–48. doi: 10.1016/j.biochi.2019.01.022
FAO (2021). “Fishery and Aquaculture Statistics 2019/FAO annuaire,” in FAO Yearbook. Rome: Food and Agriculture Organization of the United Nations. doi: 10.4060/cb7874t
Fernández-reiriz M. J., Irisarri J., Labarta U. (2017). Fatty acid composition of female and male clams (Ruditapes philippinarum): energy intake and temperature reliance. Aquaculture Nutr. 23 (1), 119–128. doi: 10.1111/anu.12374
Fernandez-Reiriz M. J., Labarta U., Albentosa M., Perez-Camacho A. (2006). Lipid composition of Ruditapes philippinarum spat: effect of ration and diet quality. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 144 (2), 229–237. doi: 10.1016/j.cbpb.2006.02.015
Gould J. F., Smithers L. G., Makrides M. (2013). The effect of maternal omega-3 (n-3) LCPUFA supplementation during pregnancy on early childhood cognitive and visual development: a systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 97 (3), 531–544. doi: 10.3945/ajcn.112.045781
Guillou H., Zadravec D., Martin P. G. P., Jacobsson A. (2010). The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49 (2), 186–199. doi: 10.1016/j.plipres.2009.12.002
Hashimoto K., Yoshizawa A. C., Okuda S., Kuma K., Goto S., Kanehisa M. (2008). The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J. Lipid Res. 49 (1), 183–191. doi: 10.1194/jlr.M700377-JLR200
Hussein N., Ah-Sing E., Wilkinson P., Leach C., Griffin B. A., Millward D. J. (2005). Long-chain conversion of [13C] linoleic acid and α-linolenic acid in response to marked changes in their dietary intake in men. J. Lipid Res. 46 (2), 269–280. doi: 10.1194/jlr.M400225-JLR200
Kabeya N., Fonseca M. M., Ferrier D. E. K., Navarro J. C., Bay L. K., Francis D. S., et al. (2018). Genes for de novo biosynthesis of omega-3 polyunsaturated fatty acids are widespread in animals. Sci. Adv. 4 (5), eaar6849. doi: 10.1126/sciadv.aar6849
Kraffe E., Soudant P., Marty Y. (2004). Fatty acids of serine, ethanolamine, and choline plasmalogens in some marine bivalves. Lipids 39 (1), 59–66. doi: 10.1007/s11745-004-1202-x
Lee M. C., Park J. C., Lee J. S. (2018). Effects of environmental stressors on lipid metabolism in aquatic invertebrates. Aquat Toxicol. 200, 83–92. doi: 10.1016/j.aquatox.2018.04.016
Legrand P., Rioux V. (2010). The complex and important cellular and metabolic functions of saturated fatty acids. Lipids 45 (10), 941–946. doi: 10.1007/s11745-010-3444-x
Li M., Mai K., He G., Ai Q., Zhang W., Xu W., et al. (2013). Characterization of two Δ5 fatty acyl desaturases in abalone (Haliotis discus hannai ino). Aquaculture 416-417, 48–56. doi: 10.1016/j.aquaculture.2013.08.030
Li Y., Wen Z., You C., Xie Z., Tocher D. R., Zhang Y., et al. (2020). Genome wide identification and functional characterization of two LC-PUFA biosynthesis elongase (elovl8) genes in rabbitfish (Siganus canaliculatus). Aquaculture 522 (3), 735127. doi: 10.1016/j.aquaculture.2020.735127
Liu H., Guo Z., Zheng H., Wang S., Wang Y., Liu W., et al. (2014a). Functional characterization of a Delta5-like fatty acyl desaturase and its expression during early embryogenesis in the noble scallop Chlamys nobilis reeve. Mol. Biol. Rep. 41 (11), 7437–7445. doi: 10.1007/s11033-014-3633-4
Liu H., Zhang H., Zheng H., Wang S., Guo Z., Zhang G. (2014b). PUFA biosynthesis pathway in marine scallop Chlamys nobilis reeve. J. Agric. Food Chem. 62 (51), 12384–12391. doi: 10.1021/jf504648f
Liu H., Zheng H., Wang S., Wang Y., Li S., Liu W., et al. (2013). Cloning and functional characterization of a polyunsaturated fatty acid elongase in a marine bivalve noble scallop Chlamys nobilis reeve. Aquaculture, 416–417 (3), 146-151. doi: 10.1016/j.aquaculture.2013.09.015
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marshall R., McKinley S., Pearce C. M. (2010). Effects of nutrition on larval growth and survival in bivalves. Rev. Aquaculture 2 (1), 33–55. doi: 10.1111/j.1753-5131.2010.01022.x
Marventano S., Kolacz P., Castellano S., Galvano F., Buscemi S., Mistretta A., et al. (2015). A review of recent evidence in human studies of n-3 and n-6 PUFA intake on cardiovascular disease, cancer, and depressive disorders: does the ratio really matter? Int. J. Food Sci. Nutr. 66 (6), 611–622. doi: 10.3109/09637486.2015.1077790
Miyazaki M., Ntambi J. M. (2008). “"Fatty acid desaturation and chain elongation in mammals,",” in Biochemistry of lipids, lipoproteins and membranes (Elsevier), 191–211. doi: 10.1016/B978-044453219-0.50009-X
Monroig O., de Llanos R., Varo I., Hontoria F., Tocher D. R., Puig S., et al. (2017). Biosynthesis of polyunsaturated fatty acids in Octopus vulgaris: Molecular cloning and functional characterisation of a stearoyl-CoA desaturase and an elongation of very long-chain fatty acid 4 protein. Mar. Drugs 15 (3), 82. doi: 10.3390/md15030082
Monroig Ó., Guinot D., Hontoria F., Tocher D. R., Navarro J. C. (2012a). Biosynthesis of essential fatty acids in Octopus vulgaris (Cuvie): Molecular cloning, functional characterisation and tissue distribution of a fatty acyl elongase. Aquaculture 360-361, 45–53. doi: 10.1016/j.aquaculture.2012.07.016
Monroig Ó., Hontoria F., Varó I., Tocher D. R., Navarro J. C. (2016). Investigating the essential fatty acids in the common cuttlefish Sepia officinalis (Mollusca, cephalopoda): Molecular cloning and functional characterisation of fatty acyl desaturase and elongase. Aquaculture 450, 38–47. doi: 10.1016/j.aquaculture.2015.07.003
Monroig Ó., Kabeya N. (2018). Desaturases and elongases involved in polyunsaturated fatty acid biosynthesis in aquatic invertebrates: a comprehensive review. Fisheries Sci. 84 (6), 911–928. doi: 10.1007/s12562-018-1254-x
Monroig Ó., Navarro J. C., Dick J. R., Alemany F., Tocher D. R. (2012b). Identification of a Δ5-like fatty acyl desaturase from the cephalopod Octopus vulgaris (Cuvier 1797) involved in the biosynthesis of essential fatty acids. Mar. Biotechnol. 14 (4), 411–422. doi: 10.1007/s10126-011-9423-2
Monroig O., Tocher D. R., Castro L. F. C. (2018). “Polyunsaturated fatty acid biosynthesis and metabolism in fish,” in Polyunsaturated fatty acid metabolism(Elsevier), 31-60. doi: 10.1016/B978-0-12-811230-4.00003-X
Pettersen A. K., Turchini G. M., Jahangard S., Ingram B. A., Sherman C. D. (2010). Effects of different dietary microalgae on survival, growth, settlement and fatty acid composition of blue mussel (Mytilus galloprovincialis) larvae. Aquaculture 309 (1-4), 115–124. doi: 10.1016/j.aquaculture.2010.09.024
Ran Z., Kong F., Xu J., Liao K., Xu X., Shi P., et al. (2020). Fad and elovl expressions, fatty acid compositions, and feed effects of three representative microalgae in Sinonovacula constricta (Lamarck 1818) at early developmental stages. Aquaculture 521, 735101. doi: 10.1016/j.aquaculture.2020.735101
Ran Z., Xu J., Liao K., Li S., Chen S., Yan X. (2018). Biosynthesis of polyunsaturated fatty acids in the razor clam Sinonovacula constricta: Characterization of Δ5 and Δ6 fatty acid desaturases. J. Agric. Food Chem. 66 (18), 4592–4601. doi: 10.1021/acs.jafc.8b00968
Ran Z., Xu J., Liao K., Monroig O., Navarro J. C., Oboh A., et al. (2019). Biosynthesis of long-chain polyunsaturated fatty acids in the razor clam Sinonovacula constricta: Characterization of four fatty acyl elongases and a novel desaturase capacity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864 (8), 1083–1090. doi: 10.1016/j.bbalip.2019.04.004
Sijben J. W., Calder P. C. (2007). Differential immunomodulation with long-chain n-3 PUFA in health and chronic disease. Proc. Nutr. Soc. 66 (2), 237–259. doi: 10.1017/S0029665107005472
Sprecher H. (2000). Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochem. Biophys. Acta 1486, 219–231. doi: 10.1016/s1388-1981(00)00077-9
Sun S., Wang Y., Goh P. T., Lopes-Marques M., Castro L. F. C., Monroig O., et al. (2021). Evolution and functional characteristics of the novel elovl8 that play pivotal roles in fatty acid biosynthesis. Genes (Basel) 12 (8), 1287. doi: 10.3390/genes12081287
Surm J. M., Prentis P. J., Pavasovic A. (2015). Comparative analysis and distribution of omega-3 lcPUFA biosynthesis genes in marine molluscs. PloS One 10 (8), e0136301. doi: 10.1371/journal.pone.0136301
Tallima H., El Ridi R. (2018). Arachidonic acid: physiological roles and potential health benefits–a review. J. advanced Res. 11, 33–41. doi: 10.1016/j.jare.2017.11.004
Tan K., Ma H., Li S., Zheng H. (2020). Bivalves as future source of sustainable natural omega-3 polyunsaturated fatty acids. Food Chem. 311, 125907. doi: 10.1016/j.foodchem.2019.125907
Tocher D. R., Betancor M. B., Sprague M., Olsen R. E., Napier J. A. (2019). Omega-3 long-chain polyunsaturated fatty acids, EPA and DHA: Bridging the gap between supply and demand. Nutrients 11 (1), 89. doi: 10.3390/nu11010089
Wang Y.-Y., Xu S.-M., Cao J.-Y., Wu M.-N., Lin J.-H., Zhou C.-X., et al. (2022). Co-Cultivation of isochrysis galbana and marinobacter sp. can enhance algal growth and docosahexaenoic acid production. Aquaculture 556, 738248. doi: 10.1016/j.aquaculture.2022.738248
Yang F., Chen S., Miao Z., Sheng Z., Xu J., Wan J., et al. (2016). The effect of several microalgae isolated from East China Sea on growth and survival rate of postset juveniles of razor clam, Sinonovacula constricta(Lamarck 1818). Aquaculture Nutr. 22 (4), 846–856. doi: 10.1111/anu.12310
Zhang J. Y., Kothapalli K. S., Brenna J. T. (2016). Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care 19 (2), 103–110. doi: 10.1097/MCO.0000000000000254
Zhang H., Liu H., Cheng D., Liu H., Zheng H. (2018). Molecular cloning and functional characterisation of a polyunsaturated fatty acid elongase in a marine bivalve Crassostrea angulata. J. Food Nutr. Res. 6 (2), 89–95. doi: 10.12691/jfnr-6-2-4
Keywords: Ruditapes philippinarum, fad, elovl, microalgae, fatty acids
Citation: Wu K, Ran Z, Wu S, Xie H, Li Y, Liao K, Xu J and Yan X (2023) Biosynthesis of LC-PUFA in Ruditapes philippinarum: Cloning and tissue distribution of Fad and Elovl, and effects of microalgae diets varied in LC-PUFA composition on their expressions and fatty acids profile of this bivalve. Front. Mar. Sci. 10:1141231. doi: 10.3389/fmars.2023.1141231
Received: 10 January 2023; Accepted: 10 February 2023;
Published: 21 February 2023.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaReviewed by:
Zhongming Huo, Dalian Ocean University, ChinaCopyright © 2023 Wu, Ran, Wu, Xie, Li, Liao, Xu and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoshou Ran, cmFuemhhb3Nob3VAbmJ1LmVkdS5jbg==; Jilin Xu, eHVqaWxpbkBuYnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.