- 1Key Laboratory of South China Sea Fishery Resources Exploitation and Utilization, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2College of Marine Sciences, Hainan University, Haikou, Hainan, China
- 3College of Aqua-life Science and Technology, Shanghai Ocean University, Shanghai, China
- 4Tianjin Agricultural University, Tianjin, China
Antibiotic resistance represents a global health crisis for humans, animals, and the environment. However, few studies address the abundance and distribution of the environmental bacterial antibiotic resistance associated with farmed fish during the early breeding stages and their relationship with aquaculture environment. In this study, culture-dependent methods and gene chip technology were respectively used to identify and detect cultivable heterotrophic antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) of water samples from 20 sea bass-rearing ponds in spring in Zhuhai, China. Meanwhile, the relationships among ARGs, ARB, and water nutrients were elucidated. The results showed that bacterial resistance to erythromycin and sulfamethoxazole with trimethoprim was generally high (mean 48.15% and 18.07%, respectively), whereas resistance to rifampicin, florfenicol, ciprofloxacin, and enrofloxacin was generally low (mean 5.46%, 2.16%, 1.43%, and 0.16%, respectively). Acinetobacter sp. (42.31%) and Pseudomonas sp. (25.74%) were the dominant ARB, and most cultivable ARB were opportunistic pathogens. The abundance of sul family genes was higher than that of other tested ARGs. ARGs and ARB were mainly affected by NO3– and PO43–, with PO43– generally positively correlated, whereas NO3– was negatively correlated, with ARGs and ARB. Thus, recommendations for the control of antibiotic resistance risk can be made by understanding the resistance profile of the aquaculture environment.
1 Introduction
Antibiotics are widely used to prevent and treat bacterial infections. In China, the total production of antibiotics has kept increasing and was up to 223,000 tons in 2020, of which half was used for animal farming and the other half for human medical care (https://www.chinabgao.com/k/kangshengsu/66064.html). Antibiotic-resistant microorganisms can emerge after the long-term use of antibiotics. These antibiotics can enter aquatic systems, such as rivers and lakes, causing selective pressure on bacteria and leading to the enrichment of antibiotic-resistant bacteria (ARB) and the evolution of antibiotic-resistance genes (ARGs) (Muniesa et al., 2013; Hu et al., 2018; Su et al., 2018; Xiang et al., 2018; Su et al., 2019). By 2020, the antibiotic resistance in aquacultural systems in China had exceeded 50% (https://m.yicai.com/news/101235766.html). The aquaculture environment is ideal for acquiring and disseminating ARB and ARGs. Given the economic importance of the aquaculture industry, research interest in this area has increased (Taylor et al., 2011; Marti et al., 2014).
High-density farming, overfeeding, climate change, and human activity all lead to disease outbreaks (Mao et al., 2020), and there has been an increasing trend in the occurrence of bacterial antibiotic resistance in the aquacultural industry. Wu et al. (2019) found that the abundance of ARGs during the fishing period was significantly higher than that during the rearing period in aquaculture cages in both Guangdong and Hainan. Yuan et al. (2019) reported an increasing abundance of ARGs with increased rearing density and pond age in aquacultural ponds in Hangzhou Bay. Therefore, investigating antibiotic resistance profiles in breeding ponds during the early breeding stage can guide the proper use of antibiotics during the breeding season, thus avoiding the overuse of antibiotics and accumulation of antibiotic resistance. It has also been reported that bacteria in the breeding ponds, such as Acinetobacter sp. and Aeromonas sp., usually contain a large number of ARGs and are often the main pathogenic bacteria during the breeding process, causing surface bleeding, rotten tails, and visceral damage to fish (Ye et al., 2021). The highest levels of antibiotic resistance in these pathogens were found against penicillin, macrolides, sulfonamides, and tetracyclines (Gai et al., 2016). Therefore, identifying ARB is conducive to the antibiotic control of specific bacterial pathogens.
Sea bass had an output of nearly 200,000 tons in 2021 (Liu et al., 2022), providing a large amount of high-quality protein for humans. The Doumen District of Zhuhai City is the main breeding area for sea bass in China, and its output accounts for about 70% of production in Guangdong Province and 50% of Chinese sea bass production, mainly using pond aquaculture. Our team found that Edwardsiella sp., an important pathogen of sea bass in Zhuhai, is resistant to erythromycin (ERY), rifampicin (RIF), and sulfamethoxazole+trimethoprim (S×T), etc. (Mao et al., 2020). Moreover, the multidrug resistance index of the tested 16 antibiotics reaches 0.423 (Mao et al., 2020). These results suggest that serious antibiotic resistance happens in the sea bass farms; thus, there is an urgent need for intensive study of its overall antibiotic resistance in order to guide the use of antibiotics during breeding. In this study, 20 sea bass culture ponds in Doumen District were randomly selected to investigate the content of ARB with resistance to ERY, RIF, florfenicol (FFC), enrofloxacin (ENR), ciprofloxacin (CIP), and S×T and the presence of 76 ARGs, including eight mobile genetic elements (MGEs), during the spring breeding season. In addition, the isolated resistant bacteria were identified by the 16S rRNA sequence, and the inorganic nutrients from the water (NH4+, NO3–, NO2–, and PO43–) were analyzed. Finally, the relationship between the ARB, ARGs, and environmental factors was elucidated. The results of this study will be helpful for guiding the use of antibiotics during sea bass aquaculture, especially in Zhuhai, China, and could provide a theoretical basis for antibiotic resistance control via water quality regulation.
2 Materials and methods
2.1 Sample sites and sample collection
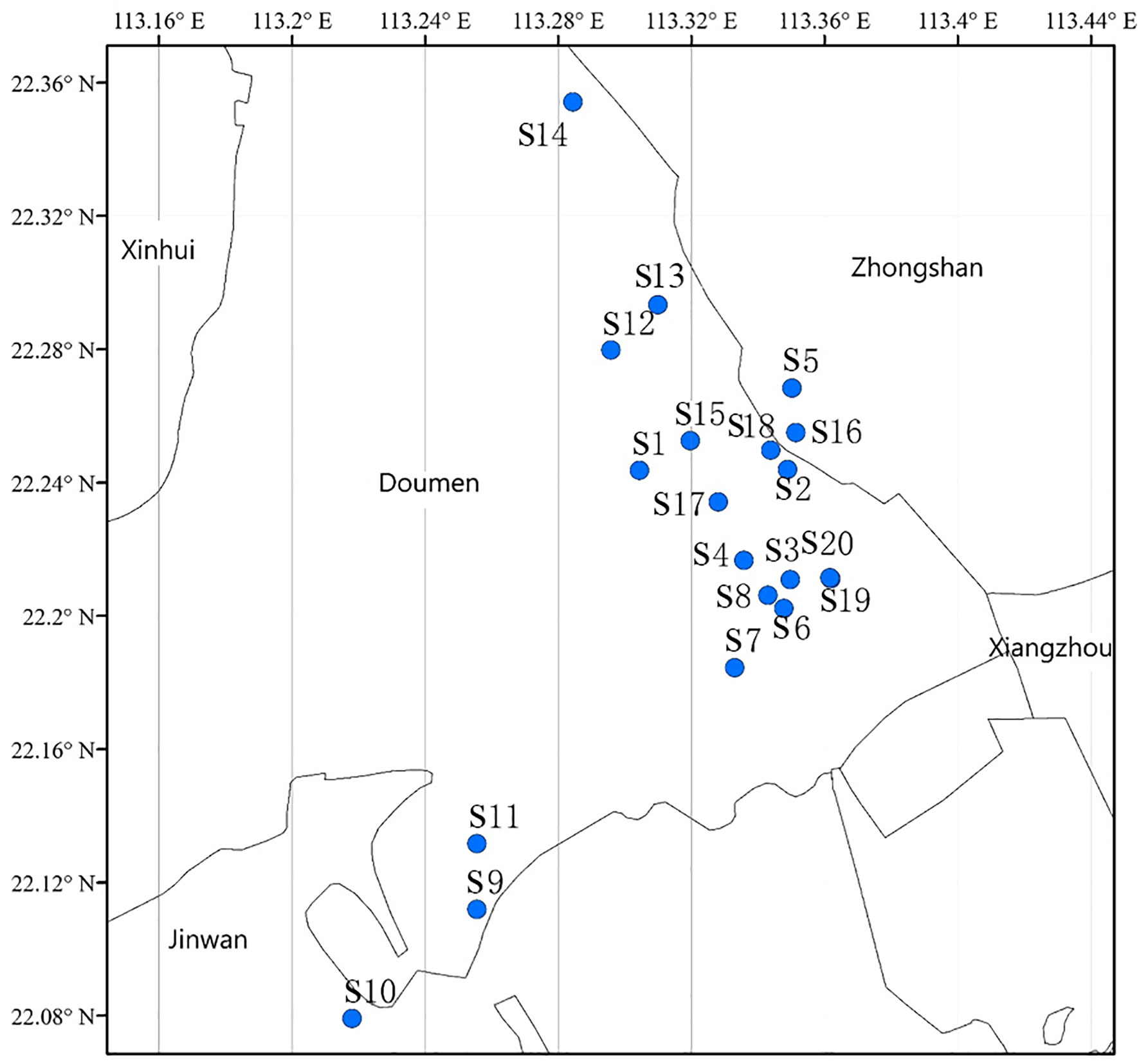
A total of 20 sea bass breeding ponds (Figure 1) were randomly selected along the sea bass culture area of Doumen District, Zhuhai City, Guangdong Province, China, to collect pond water. These ponds measured 3000-9000 m2 and contained cultured fish with the weight of 50-100 g and density of 20-40 fish/m2. All the samples were collected on 10th March, 2021. The water sampler of a 3 L capacity plexiglass container and sterilized polypropylene sampling bottles were rinsed three times with pond water before sampling. Subsequently, for each pond, the waters were collected from the middle of the four sides of the pond at 3 m from the shore and 0.5 m to 1 m below the water surface. For each side, 250 mL of water was collected, and the water from the four sides was mixed into one sample. Finally, 1 L of water was sampled in a pond and then poured into a sample bottle. All the samples were taken to the laboratory immediately for subsequent analysis.
2.2 Sample preparation
For the sample from each pond, 500 mL of water was mixed with 1.25 mL of 20% Tween-80 and shaken vigorously for 10 min before ARB screen. Approximately 200 mL of water was filtered through a 0.22-μm mixed cellulose ester membrane filter (PALL, New York, NY, USA) for DNA extraction. The membrane filters were immediately placed into 2 mL sterile polyethylene tubes containing ethanol at 4°C until DNA extraction. In addition, 50 mL of water was filtered through a 47-mm diameter GF/F glass microfiber filter (Whatman, UK) and stored at –20°C before nutrient analysis (NO2–, NO3–, NH4+, and PO43–) with the Cleverchem Anna Random Access Analyser (DeChem-Tech, Germany).
2.3 Screening for ARB
Six antibiotics—ERY, RIF, FFC, ENR, CIP, and S×T—which are classed as macrolides, rifamycins, chloramphenicols, fluoroquinolones, and sulfonamides, respectively, were selected to screen for ARB. According to the guidance on drug use in aquaculture published by the Ministry of Agriculture and Rural Affairs of the People’s Republic of China in 2020 (www.yyj.moa.gov.cn/gzdt/202010/t20201012_6354054.htm), in 2021—the year before our sampling year—FFC, ENR, and S×T were to be approved for use in aquaculture, whereas ERY, RIF, and CIP were not. The concentrations of antibiotics used (16 ug/mL ERY; 8 ug/mL RIF; 64 ug/mL FFC; 8 ug/mL ENR; 8 ug/mL CIP; and 152/8 ug/mL S×T) were twice the minimum inhibitory concentrations (MICs) recommended by the CLSI guidelines (Clinical and Laboratory Standards Institute, 2016), since the ARB in this study were screened on plates rather than in liquid media.
In detail, 20 mL of the shaken water (treated by Tween-80) was diluted tenfold and 100-fold with normal saline. Then, 100 μL of each dilution was directly plated and screened on nutrient agar plates, each containing an antibiotic at the concentrations detailed above. Three plates were prepared for each dilution, and nutrient agar plates without antibiotics were used as a control to count the total number of culturable bacteria. Colony-forming units (CFU) per mL (CFU/mL) were calculated after incubation for 24 h at 28°C.
2.4 Identification of ARB
The isolates that grew on the nutrient agar plates containing antibiotics were identified as ARB. Six scattered ponds were selected along the geographical distribution map (Figure 1) to identify the ARB by 16S rRNA sequence analysis. A total of 614 ARB strains, with 99, 75, 120, 100, 120, and 100 ARB strains, were randomly selected from six ponds (S4, S8, S9, S14, S18, S20) with different antibiotics. The primers of 8F (5′AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used for 16S rRNA amplification, and standard polymerase chain reactions (PCR) were performed as described previously (Wang et al., 2016). PCR products were verified with 1% agarose gels and sequenced with the next-generation sequencing platform of Beijing Ruiboxingke Biotechnology Co., Ltd. The sequence data were compared to the NCBI Nucleotide Sequence Database by using BLAST.
2.5 DNA extraction and ARGs analysis
The water total DNA was extracted using a HiPure water DNA Kit (Magen, Guangzhou, China) according to the manufacturer’s protocol. Extraction yield and the quality of DNA were verified by 1% agarose gel electrophoresis and spectrophotometry analysis (NanoPhotometer, IMPLEN, Germany). The DNA was stored at −20°C before use.
The 16S rRNA was selected as the reference gene. In total, 76 target ARGs (Table S1) in several categories (including tetracycline, sulfonamide, multidrug, MLSB, β-lactamase, and aminoglycoside) were analyzed using the SmartChip qPCR system (WaferGen Biosystems, Inc., Fremont, CA, USA) by Microanaly (An’hui) Gene Technologies Co. First, the 16S rRNA was amplified to ensure template quality. Those templates with cycle threshold (Ct) values of the 16S rRNA gene (14~17) were used for the ARG analysis. Each sample was treated using the following conditions: 10 min at 95°C (initial denaturation), 40 cycles of denaturation at 95°C for 30 s, and annealing at 60°C for 30 s with a volume of 100 nL (Chen et al., 2016; Zhao et al., 2019). Each PCR was repeated three times. Three control actions for each primer pair were set using sterile water as the template, which does not amplify or for which the Ct value was >31. The data were analyzed with qPCR quantitative software. The gene copy number was determined by Ct. At least two technical replicates were amplified with Ct <31; the average Ct was then calculated and used to determine the ARG copy number. The ARG copy numbers were normalized to 16S rRNA copies, and the ARG concentration units were gene copies per 16S rRNA gene.
2.6 Data analysis
A geographical distribution map was drawn using ArcGIS 10.2 (Jz et al., 2021). The bacterial resistance rate was calculated using Equation 1: (number of bacteria grown on plates with one antibiotic/total number of bacteria grown without antibiotics) (1). The proportion of ARB over six ponds (S4, S8, S9, S14, S18, S20) at the genus level was determined using Equation 2: (the number of ARB of a specific genus/the total number of ARB) (2). The Shannon–Weiner Index of ARB at the genus level in different ponds or resistant to different antibiotics was calculated using Equation 3: H = ∑(Pi)(ln Pi) (3). Where Pi = the number of ARB of a specific genus from one pond or resistant to one antibiotic/the total number of ARB from the same pond or resistant to the same antibiotic. The relative ratios of each ARG over 20 ponds (S1-S20) was calculated using Equation 4: (the average abundance of each ARG over the 20 ponds/the sum of the average abundance of all the ARGs over the 20 ponds) (4). The redundancy analysis (RDA) of nutrients, ARB abundance, and ARGs abundance were performed and plotted using Canoco 4.5 (Wang et al., 2020). All other images were drawn with Origin 2018 software.
3 Results
3.1 Viable bacterial counts and bacterial resistance rates
The quantity of culturable bacteria at the different sampling sites ranged from 1.5×103 to 6.7×104 cfu/mL. The average bacterial resistance rates of the six tested antibiotics over the 20 ponds were 48.15% (ERY), 18.07% (S×T), 5.46% (RIF), FFC (2.16%), 1.43% (CIP), and 0.16% (ENR), respectively (Figure 2).
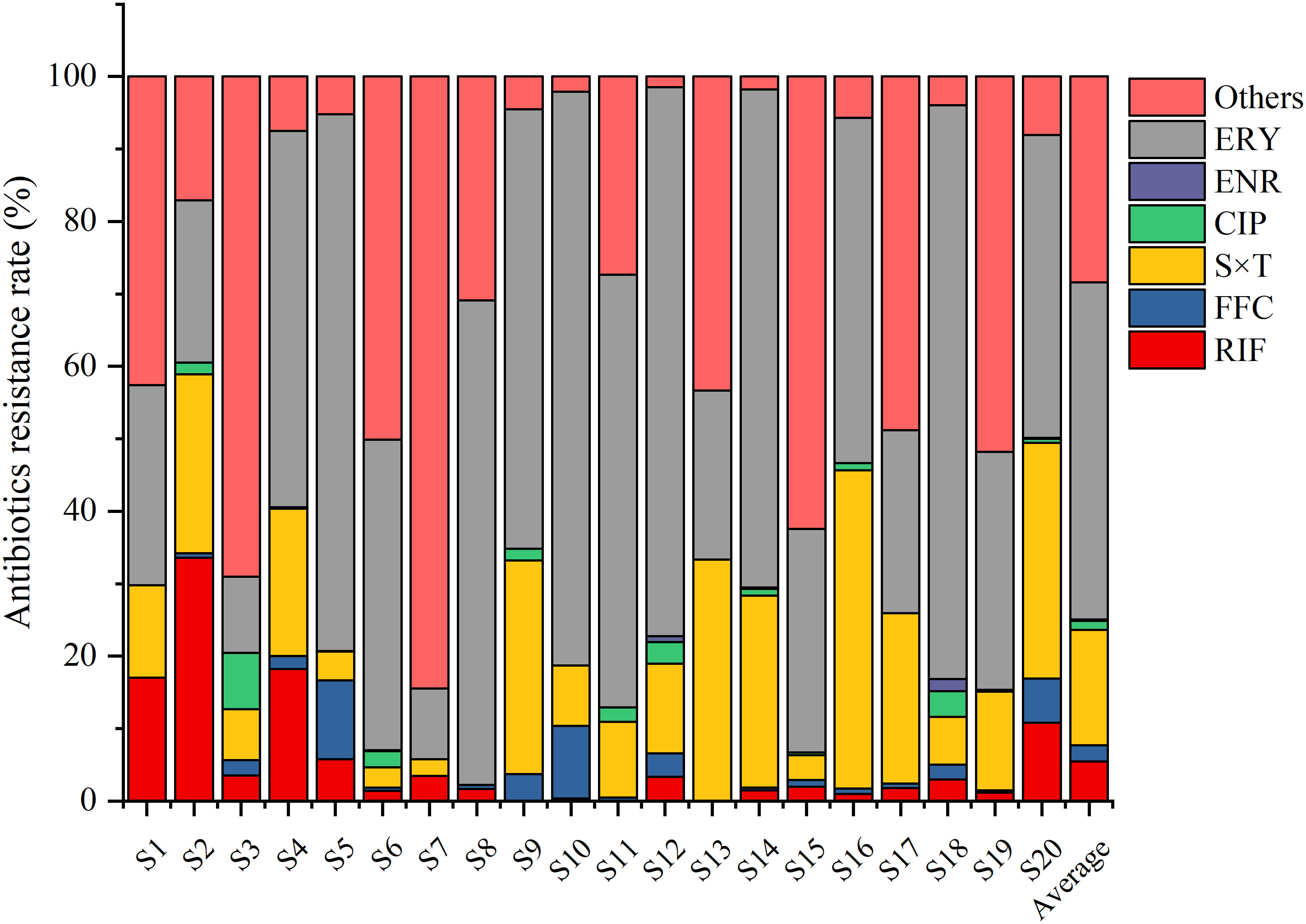
Figure 2 Bacterial resistance rates to rifampin (RIF), florfenicol (FFC), sulfamethoxazole+trimethoprim (S×T), ciprofloxacin (CIP), enrofloxacin (ENR), and erythromycin (ERY) in different sea bass ponds. The bacterial resistance rates were defined as the number of bacteria grown on plates with one antibiotic/total number of bacteria grown without antibiotics.
The resistance rate to ERY was highest especially in S18 (79.2%), S10 (79.2%), S12 (75.7%), S5 (74.1%), and S14 (68.8%). The resistance rate to S×T was the second highest, especially in S16 (43.9%), S13 (33.3%), S20 (32.6%), S9 (29.5%), and S14 (26.5%). However, the resistance rates to the other four antibiotics were relatively low in most of the other sampling sites (Figure 2).
3.2 Distribution and diversity of ARB
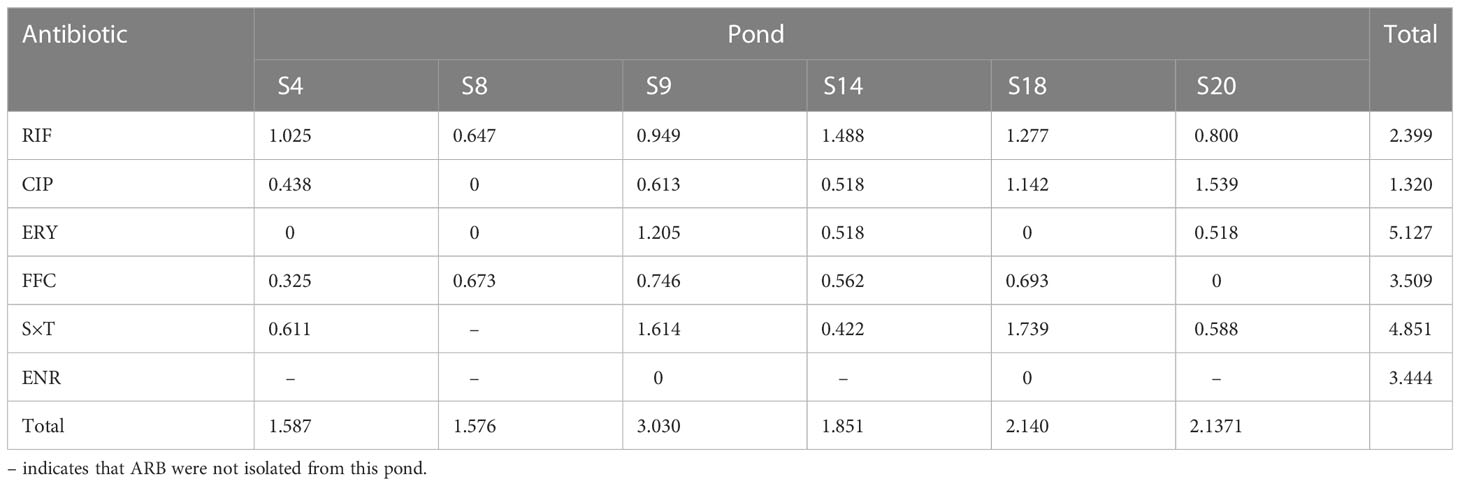
In total, 614 strains were randomly isolated from the water samples of the six ponds (S4, S8, S9, S14, S18, and S20). The Shannon–Weiner Index is often used to represent the diversity of species in the environment. The diversity of the ARB was shown in Table 1. The indices of resistance to ERY (5.127) and S×T (4.851) were relatively high, especially in S9, suggesting that there is a higher diversity of bacteria resistant to these two antibiotics compared with the other antibiotics. The Shannon–Weiner Index was the highest in S9 (3.030), especially in bacteria resistant to ERY and S×T. Thus, S9 had the highest bacterial diversity among the six ponds.
Based on the identification of 16S rRNA, 19 genera of ARB were determined, with Acinetobacter sp. (42.34%), Pseudomonas sp. (25.73%), Shewanella sp. (11.73%), Aeromonas sp. (6.51%), Citrobacter sp., (3.09%), Comamonas sp. (2.77%), Rheinheimera sp. (1.95%), and Brevundimonas sp. (1.79%) accounting for more than 1%, with a sum content of 95.93% (Figure 3). In addition, their level of antibiotic resistance was relatively high, especially that of Acinetobacter sp., Pseudomonas sp., Shewanella sp., and Aeromonas sp., with varying degrees of resistance to at least four of the six tested antibiotics (Figure 4). For example, Acinetobacter sp. showed strong resistance to RIF, ERY, FFC, and S×T, whereas Pseudomonas sp. was highly resistant to RIF, CIP, FFC, and ENR. Shewanella sp. showed resistance to RIF, CIP, FFC, ERY, and S×T and was particularly resistant to RIF. Aeromonas sp. showed resistance to all the antibiotics except ENR and ERY.
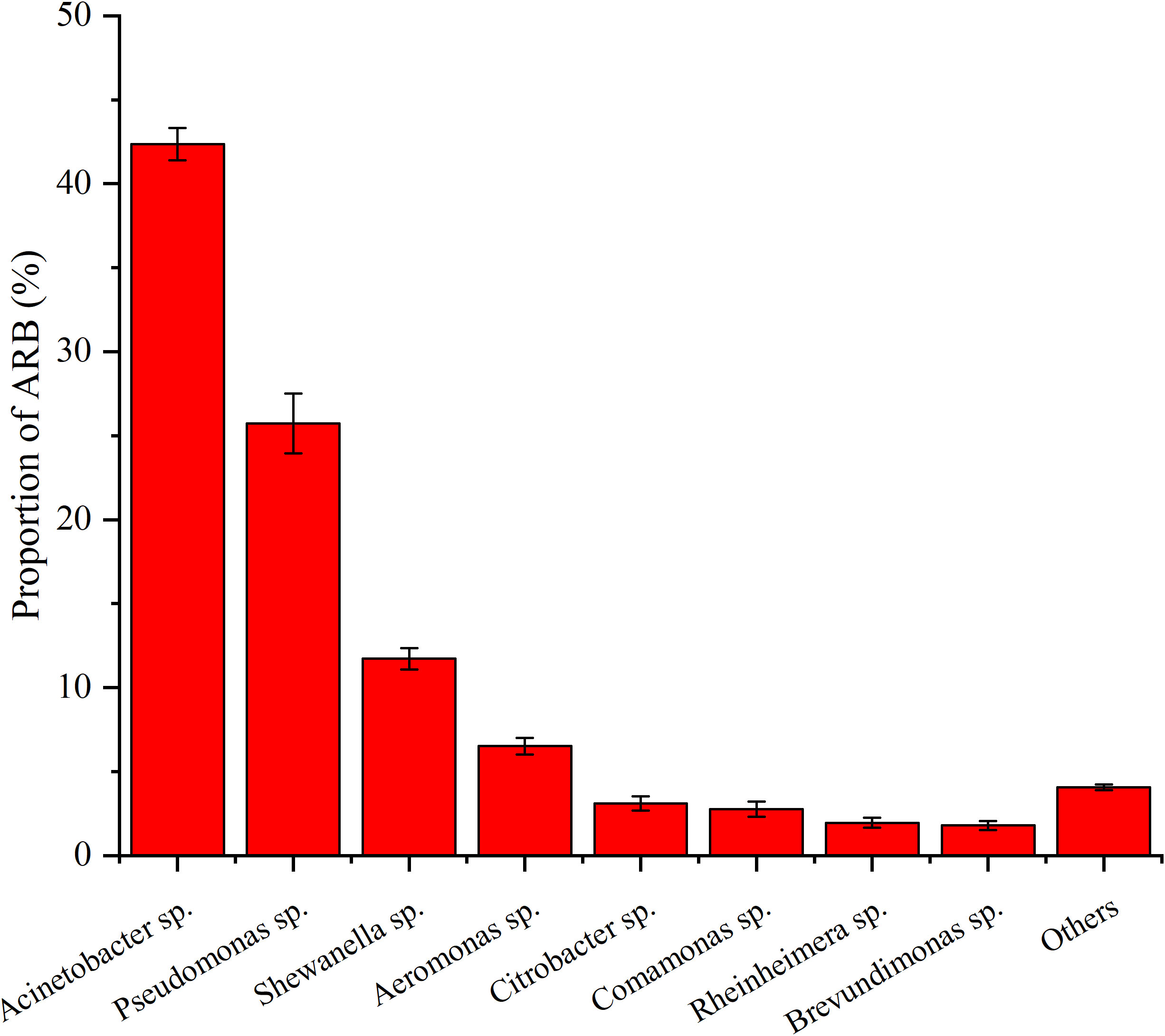
Figure 3 Proportion of different ARB over sea bass farming area of S4, S8, S9, S14, S18, and S20. Values show means and 1 standard error of mean (mean ± SEM, n = 6).
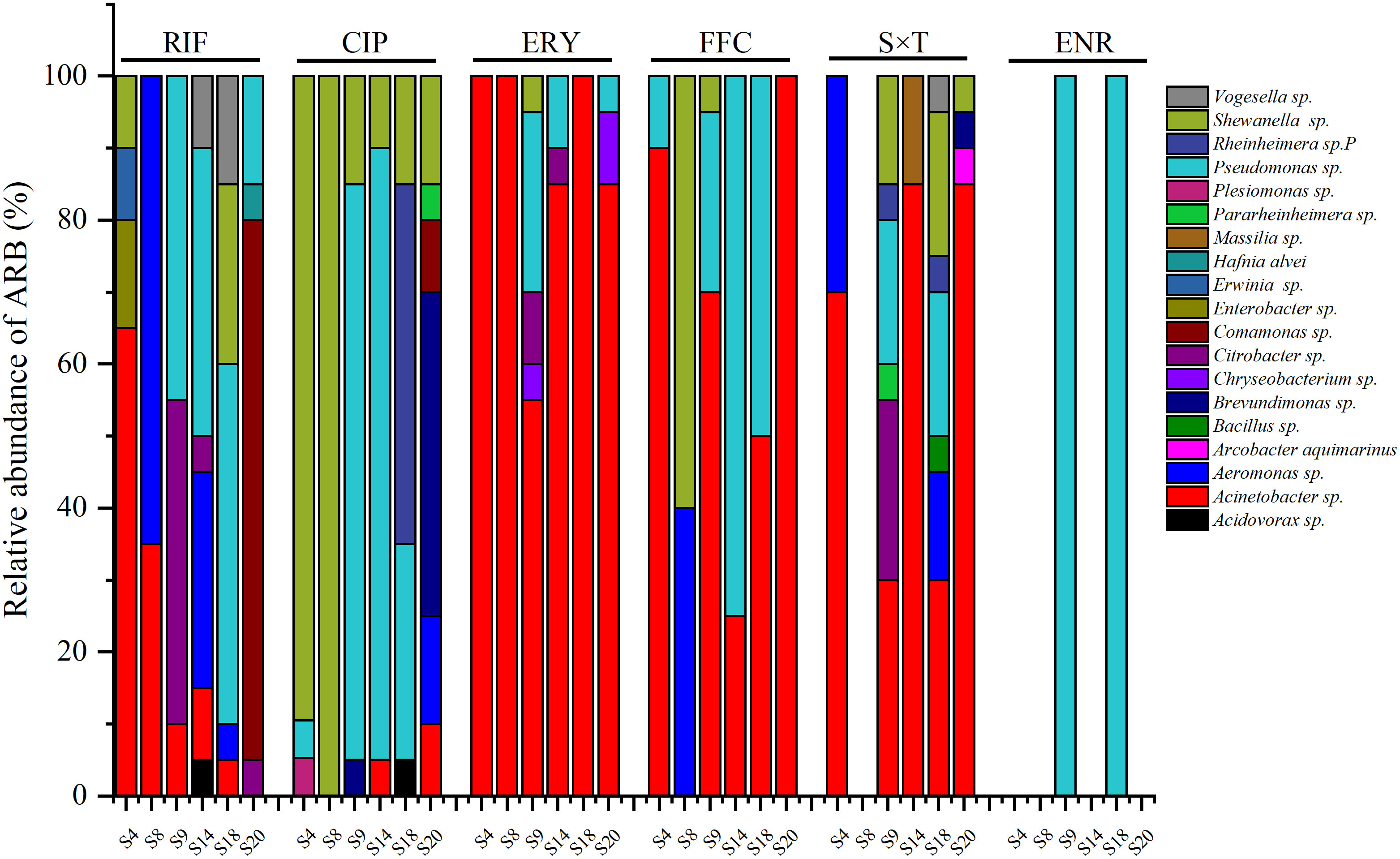
Figure 4 ARB resist to different antibiotics from different sea bass ponds (CIP, ciprofloxacin; ERY, erythromycin; FFC, florfenicol; RIF, rifampicin; S×T, sulfamethoxazole × trimethoprim; ENR, enrofloxacin).
For each pond, the ARB resistant to RIF, ERY, FFC, S×T, and ENR were generally Acinetobacter sp. and/or Pseudomonas sp. (Figure 4). However, except for Acinetobacter sp. and/or Pseudomonas sp., Aeromonas sp. accounted for 65% of ARB resistant to RIF in S8, Citrobacter sp. accounted for 45% of ARB resistant to RIF in S9, and Comamonas sp. accounted for 75% of ARB resistant to RIF in S20; no S×T-resistant bacteria were found in S8; the ARB resistant to FFC in S8 were Shewanella sp. (60%) and Aeromonas sp. (40%). The ARB resistant to CIP were mainly Shewanella sp. in S4 (90%) and S8 (100%), Pseudomonas sp. in S9 (80%) and S14 (85%), Rheinheimera sp. (50%) and Pseudomonas sp. (30%) in S20, and Brevundimonas sp. (45%) in S20. Pseudomonas sp. was the only bacteria resistant to ENR and was isolated in S9 and S18.
3.3 Occurrence and abundance of ARGs
In total, 40 of the 76 tested ARGs were detected by SmartChip qPCR, with a concentration range of 0–0.72 copies per 16S rRNA (Figure 5A). According to the average abundance over the 20 ponds, tnpA-04 was the most abundant ARG, followed by sul1, sul2, ereA, tnpA-05, dfrA1, qacEdelta1-02, intI-1(clinic), mphA-02, tetG-01, ermF, and qacEdelta1-01, with the relative abundance ranges of 0–0.72, 0–0.38, 0–0.18, 0–0.27, 0–0.31, 0–0.26, 0–0.18, 0–0.09, 0–0.06, 0–0.02, 0–0.05, and 0–0.04 copies per 16S rRNA gene, respectively (Figure 5A). The relative ratios (the average abundance of each ARG over the 20 ponds/the sum of the average abundance of all the ARGs over the 20 ponds) of these ARGs were 25.81%, 17.85%, 11.64%, 11.43%, 6.42%, 6.30% 5.52%, 3.17%, 3.09%, 2.63%, 2.60%, and 1.76%, respectively, with a sum ratio of 98.23% (Figure 5B). In addition, no aminoglycoside family ARGs were detected, and only blaCMY of the β-lactamase family was detected. Overall, the sul family genes (sul1, sul2, and drfA1) had the highest ratio (35.79%), followed by the transposase family genes (tnpA-04 and tnpA-05; 32.22%), MLSB (ereA, mphA-02, and ermF; 17.12%), multidrug family genes (qacEdelta1-01 and qacEdelta1-02; 7.29%), and tet families genes (tetG-01; 2.63%) (Figure 5B).
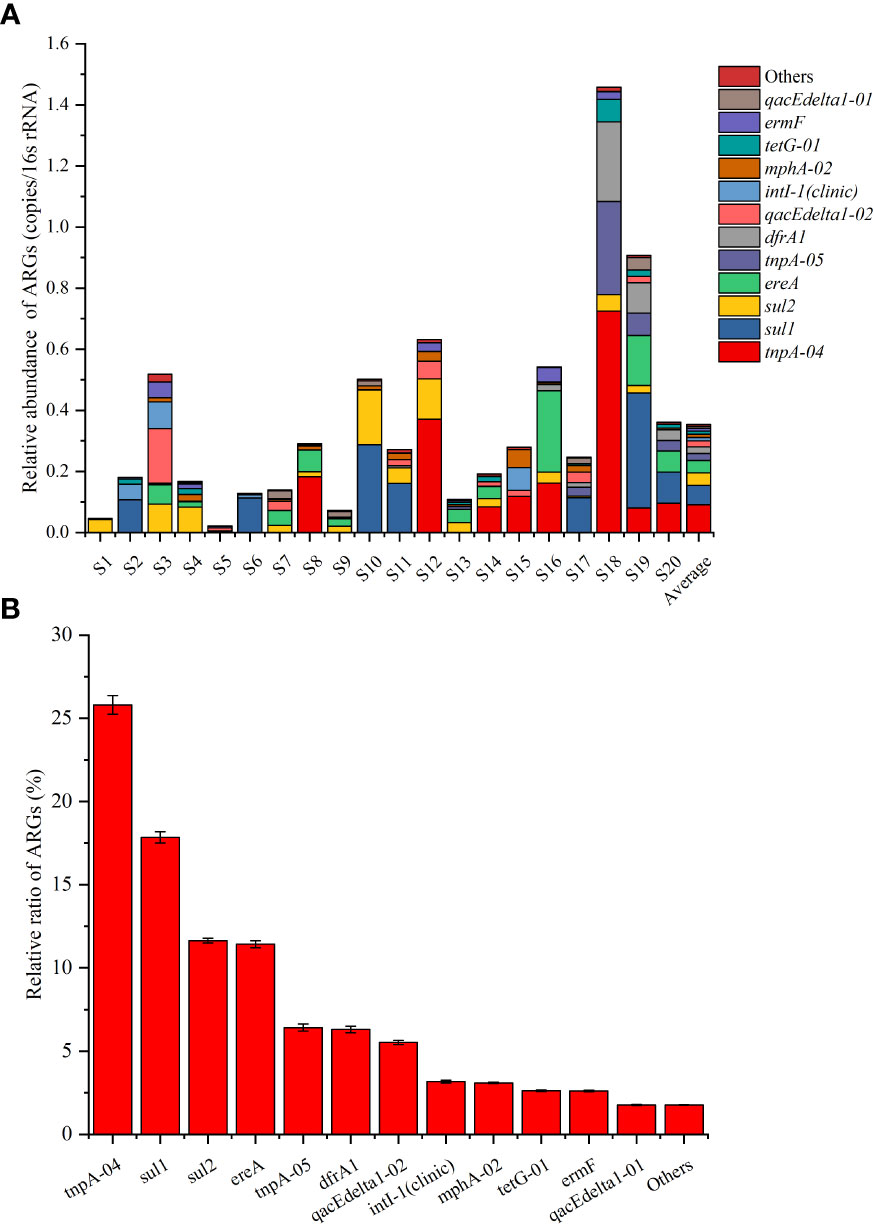
Figure 5 Relative abundance of top 12 ARGs in different sea bass ponds (A) and the relative ratio of the top 12 ARGs over the 20 ponds (S1–S20). Values show means and 1 standard error of mean (mean ± SEM, n = 20) (B).
3.4 Relationships among ARGs, ARB, and water nutrients
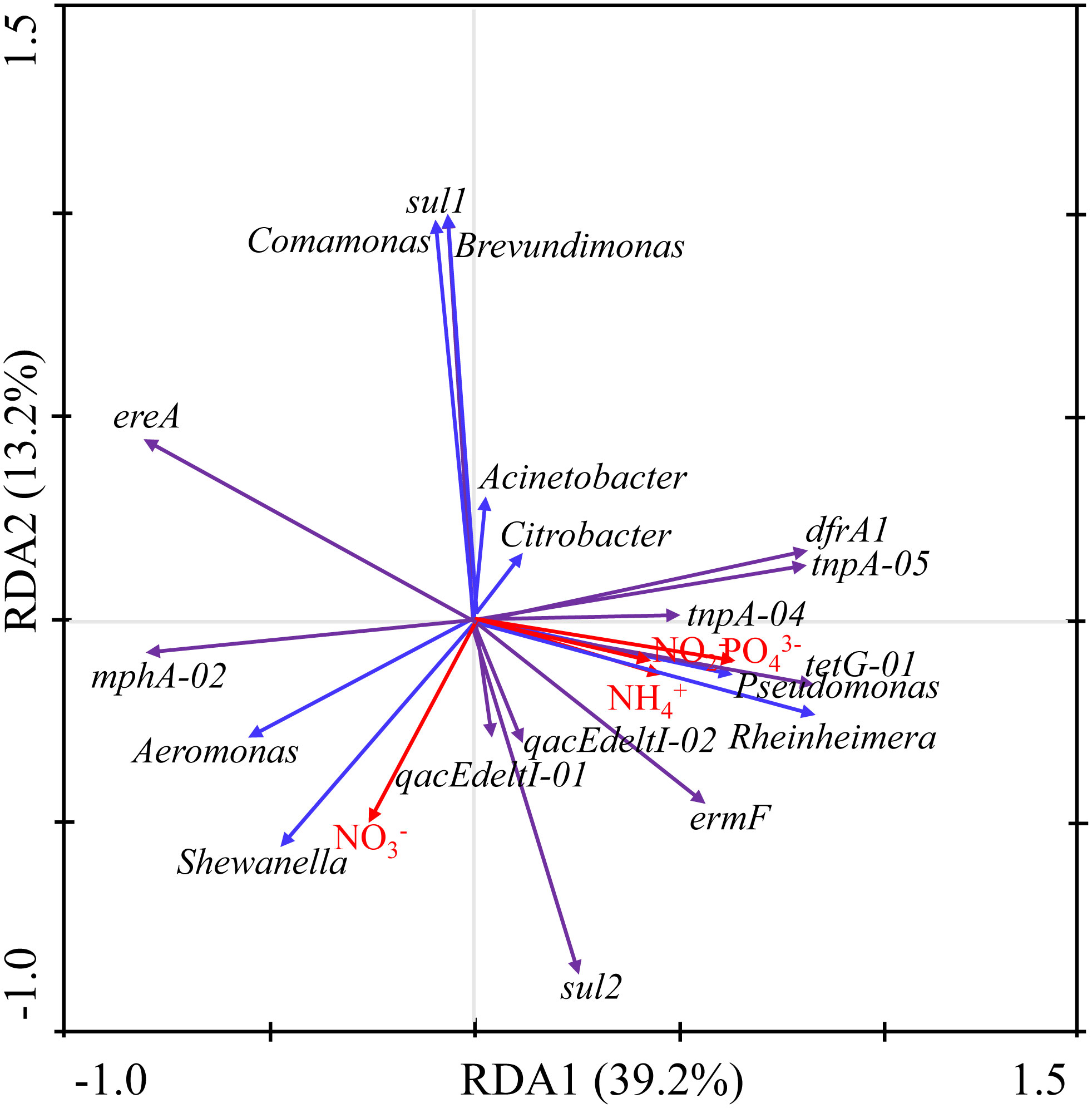
RDA analysis was conducted among the top eight ARB genera (Acinetobacter sp., Pseudomonas sp., Shewanella sp., Aeromonas sp., Citrobacter sp., Comamonas sp., Rheinheimera sp., and Brevundimonas sp.) with an average proportion exceeding 1% and a total content exceeding 95%, the top 12 ARGs [tnpA-04, sul1, sul2, ereA, tnpA-05, dfrA1, qacEdelta1-02, intI-1(clinic), mphA-02, tetG-01, ermF, and qacEdelta1-01] with an average ratio exceeding 1%, and the total ratio exceeding 95%, and water nutrients (NO3–, NO2–, PO43–, and NH4+) (Table S2). RDA1 and RDA2 determined 39.2% and 13.2% of the total variation, respectively, with a total variation of 52.4% (Figure 6).
The ARB and ARGs were mainly affected by PO43– and NO3–. PO43– was generally positive correlated with most ARGs (total relative ratio of 65.86%), except ereA, sul1, and mphA-02 (total relative ratio of 32.37%). In addition, PO43– was positively correlated with Pseudomonas sp. and Rheinheimera sp. (total proportion of 27.69%). Moreover, NO3– was generally negatively correlated with ARGs (including sul1, drfA1, tnpA-04, tnpA-05, and tetG-01 with a total relative ratio of 59.00%) and ARB (including Acinetobacter sp., Citrobacter sp., Comamonas sp., Brevundimonas sp., Pseudomonas sp., and Rheinheimera sp. with a total proportion of 77.69%). However, NO3– was positively correlated with the ARGs of qacEdelta1-01, qacEdelta1-02, sul2, and mphA-02 with a total relative ratio of 22.02% and with the Shewanella sp. and Aeromonas sp. ARB with a total proportion of 18.24% (Figure 6). Sul1 was significantly positively correlated with Comamonas sp. and Brevundimonas sp. DfrA1, tnpA-05, tnpA-04, tetG-01, and ermF were positively associated with Pseudomonas sp. and Rheinheimera sp. MphA-02 was positively associated with Aeromonas sp. and Shewanella sp. (Figure 6).
4 Discussion
During the breeding process, antibiotic resistance continues to increase as breeding progresses (Wu et al., 2019; Yuan et al., 2019). Here, our census of antibiotic resistance profiles (ARB and ARGs) in breeding ponds during the early stage of breeding will help to guide the use of antibiotics during the breeding process, improving the efficiency of disease prevention and control, reducing the probability of antibiotic abuse, and finally attenuating antibacterial resistance.
4.1 Distribution of ARB in sea bass ponds in spring
In total, 614 ARB strains were isolated and identified; they were most resistant to ERY (48.15%) and moderately resistant to S×T (18.07%) and RIF (5.46%), while they were less sensitive to FFC (2.16%), CIP (1.3%), and ENR (0.15%). Given that the use of ERY, RIF, and CIP has been prohibited in China since 2020, our results indicate the significant effect of the forbidden use of RIF and CIP. Although there was still 48.15% resistance to ERY, this was significantly lower than that reported in a previous study (>90%) (Mao et al., 2020).
Among the ARB, Acinetobacter sp. accounted for nearly half of species (42.34%), followed by Pseudomonas sp., Shewanella sp., Aeromonas sp., Citrobacter sp., Comamonas sp., Rheinheimera sp., and Brevundimonas sp., with a relative abundance of more than 1%. Most of them, including Acinetobacter sp., Pseudomonas sp., Shewanella sp., Aeromonas sp., Citrobacter sp., and Brevundimonas sp., are conditional pathogens (Zong, 2011; Zhang et al., 2014; Khalil et al., 2021). The Aeromonas sp., Pseudomonas sp., and Citrobacter sp. have also been found in Nile tilapia and grey mullet fish farms in Egypt, fish farms in East Malaysia, and other fish ponds in China with similar abundance (Xiong et al., 2015; Lihan et al., 2020; Sherif et al., 2021). However, here the Acinetobacter sp. (42.34%) was greatly higher than that in other farms (Lihan et al., 2020). Furthermore, other genera, especially the conditional pathogens of Citrobacter sp. and Brevundimonas sp., are rare found in other aquaculture farms. Such pathogens can cause aquaculture diseases under suitable conditions. For example, a highly virulent strain of Aeromonas sp. causes motile Aeromonas septicemia (MAS) in Chinese sucker fish (Li et al., 2021). Huang et al. (2020) reported Acinetobacter venetianus as a potential bacterial pathogen of red leg disease in whiteleg shrimp, while Pseudomonas anguilliseptica is reported to have caused disease outbreaks in four cultured seabream (Sparus aurata) farms in different localities in Egypt in 2016 (Fadel et al., 2018). These results highlight the importance of monitoring ARB in the early stage of sea bass culture ponds to guide the use of antibiotics.
In previous studies, Hoa et al. (2011) isolated sulfamethoxazole-resistant strains from Vietnamese fish ponds and found Acinetobacter sp. to be the most abundant sulfamethoxazole-resistant bacteria (35%). Gao et al. (2012) found that Acinetobacter sp. showed resistance to sulfamethoxazole and tetracycline in aquaculture establishments in Tianjin, China. In addition, Wu et al. (2019) found Alteromonas sp. (32.72%), Vibrio sp. (24.77%), and Acinetobacter sp. (11.62%) to be the major ARB in marine fish cage-culture areas in Hainan, China, with most of the ARB being opportunistic pathogens. Zago et al. (2020) reported Shewanella algae in Italian aquaculture farms to be reservoirs of ARGs, while Devarajan et al. (2017) found that >50% of multidrug resistant bacteria reported in India are Pseudomonas sp. Their results suggest that Acinetobacter sp., Shewanella sp., and Pseudomonas sp. are common ARB in aquaculture environments and that sulfonamide-resistant bacteria are widely distributed, which is consistent with the results of our study.
4.2 Distribution of ARGs in sea bass
In total, 40 of the 76 tested ARGs were detected by chip-qPCR with a concentration range of 0–0.72 copies per 16s rRNA. TnpA-04 was the most abundant ARG, followed by sul1, sul2, ereA, tnpA-05, dfrA1, qacEdelta1-02, intI-1(clinic), mphA-02, tetG-01, ermF, and qacEdelta1-01, belonging to the transposase, sul, MLSB, multidrug, and tet gene families. These types of ARGs are common in the environment and have been frequently reported in previous studies (Cheng et al., 2013; Su et al., 2014; Zhang et al., 2017). Most of the ARGs have been found in the sea bass ponds in Zhuhai, from May to December in 2018 (Deng et al., 2022). However, the abundance here (0-0.72 copies per 16S rRNA) is higher than that in 2018 (0-0.038 copies per 16S rRNA), suggesting the use of antibiotics increases the accumulation and spread of drug resistance. In addition, the abundance of ARGs, especially the frequently detected sul1 and sul2, is at a level similar to that in the freshwater aquaculture ponds in China (Wang et al., 2021). However, qnrA, qnrB, qnrS, tetA-01, tetC-01, and tetH, which were detected in the freshwater aquaculture ponds in China with the median abundance of 10−3 to 10−1 copies per 16S rRNA (Wang et al., 2021), have not been found here. These results indicate that aquaculture models, environmental factors, and biotic factors could be affecting ARGs (Wang et al., 2021), as the sea bass ponds are brackish ponds. Therefore, various usages of antibiotics and feed, environmental conditions, management methods, and water nutrition of the sea bass ponds probably contribute to different microbial communities and ARGs succession (Wang et al., 2021).
The ARG content in our sample ponds suggests that mainly sulfonamide antibiotics are used to treat aquaculture diseases during sea bass farming, which is consistent with the ARB results. In addition, the high detection rate of the mobile genetic elements of tnpA-04, tnpA-05, and intI-1 (clinic) showed that a large number of ARGs have been transferred horizontally by transposase and integrase. For example, sul1, sul2, and dfrA1 are located on class 1 integrons (Girija et al., 2019; Chaturvedi et al., 2020) and encode antibiotic-resistant variants of dihydropteroate synthase in gram-negative bacteria (Skold, 2000). Sul1, sul2, and sul3 were found to be ubiquitous in estuaries in China and elsewhere, such as Mexico and South Africa (Rodas-Suarez et al., 2006; Suzuki et al., 2015). Moreover, the high detection rate of ereA and ermF being responsible for ERY resistance is consistent with the results of the high distribution of ERY (48.15%) ARB, thus indicating that the prohibition of ERY needs to be continued. Here, only the tetG gene of the 18 tested tetracycline genes were detected, which is significantly lower than in other studies (Shen et al., 2019), suggesting the significant effects of the ban of tetracyclines such as oxytetracycline in aquaculture since 2012 and the “Drug Reduction in Aquaculture” initiative since 2020 in China (http://www.yyj.moa.gov.cn/gzdt/202004/t20200401_6340570.htm). In addition, low detection and nearly no detection of ARGs belonging to (fluoro)quinolone and beta-lactamase indicated the great effect of the forbidden use of corresponding antibiotics, such as CIP and penicillin (Shen et al., 2019) (www.yyj.moa.gov.cn/gzdt/202010/t20201012_6354054.htm). Together with our results, the abundance of sul appears to be relatively high in multiple aquaculture environments. However, the prohibition of antibiotics has played significant role in controlling the spread of drug resistance.
4.3 Relationships among ARGs, ARB, and water nutrients
The main environmental factors affecting the content of the ARB and ARGs were found to be PO43– and NO3–. Generally, PO43– was positively correlated with most ARGs and ARB, whereas NO3– showed the opposite condition, which is consistent with previous studies (Liu et al., 2015; Deng et al., 2022). Aquaculture activities can lead to nutrient accumulation in water (Farmaki et al., 2014), which is conducive to the growth of ARB, leading eventually to the accumulation of ARGs in the environment. For example, Whooley et al. (1983) found that the phosphate limitation decreased the growth rate of P. aeruginosa. In addition, Shewanella sp., Aeromonas sp., Pseudomonas sp., and Acinetobacter sp. can induce heterotrophic nitrification and aerobic denitrification; thus, they have important roles in nitrogen cycling in the environment (Zhang et al., 2011; Yao et al., 2013; Chen et al., 2014; Guo et al., 2014; Chen and Wang, 2015). Wang et al. (2020) found that NO3– had a negative relationship with Proteobacteria (Shewanella sp., Aeromonas sp., Pseudomonas sp., and Acinetobacter sp.). This is partly inconsistent with our results, in which NO3– showed a strong positive relationship with Shewanella sp. and Aeromonas sp. while showing a weak negative positive relationship with Pseudomonas sp. and Acinetobacter sp. The difference with other findings probably comes from the taxa’s response to NO3– under different environmental conditions, as these bacteria probably show differences in the denitrification capabilities and the resulting products. For example, the Aeromonas sp. strain HN-02 obtained 0.90 mg L−1 h−1 of ammonia removal rate even at 5°C, and the rate reached up to 4.19 at its optimal growth temperature 30°C (Chen et al., 2014). Moreover, it showed great performance of ammonia removal below 20 g L−1 of salinity (Chen et al., 2014). In addition, a Pseudomonas sp. YZN-001 can utilize not only nitrate and nitrite but also ammonium, and it has been shown to possess the capability of fully removing as much as 275.08 mg L−1 NO3−–N and 171.40 mg L−1 NO2−–N under aerobic conditions (Zhang et al., 2011).
4.4 Conclusion and aquaculture management implications
In the current study, tnpA-04, sul1, sul2, ereA, tnpA-05, dfrA1, qacEdelta1-02, intI-1(clinic), mphA-02, tetG-01, ermF, and qacEdelta-01 were prevalent ARGs in sea bass aquaculture water. In addition, numerous S×T-resistant bacteria were isolated and identified, reflecting the frequent usage of sulfonamide in sea bass disease control. The decreased ERY-resistant and low RIF- and CIP-resistant ARB suggest the effectiveness of the ban of such antibiotics. The eight most abundant ARB were Acinetobacter sp., Pseudomonas sp., Shewanella sp., Aeromonas sp., Citrobacter sp., Comamonas sp., Rheinheimera sp., and Brevundimonas sp., which could be the reservoirs of ARGs. Given that pathogenic bacteria show strong resistance to erythromycin and sulfonamide even during the early stage of sea bass culture, these antibiotics are not recommended for use during sea bass breeding. Given the low rate of bacterial resistance, quinolones, such as ciprofloxacin and enrofloxacin, could be considered for disease control during sea bass culture. RDA analysis showed that the main environmental factors affecting the content of the ARB and ARGs were PO43– and NO3–. Therefore, the abundance of some ARBs and ARGs can probably be predicted by measuring PO43– and NO3–, which would help efforts to reduce the levels of environmental ARGs and, thus, the occurrence of aquacultural diseases.
We recommend that future research should focus on the following three factors: (1) a systematic investigation of the amounts of residual antibiotics, which would confirm our hypothesis about the use of banned antibiotics for treating sea bass diseases and better guide the scientific and rational use of approved antibiotics; (2) given that this study focused on sea bass farming in a single area, resistance in different areas and species should be further investigated, and recommendations for antibiotic resistance risk in culture ponds could be made by understanding the resistance profile in the culture environment; and (3) an indoor simulating test should be conducted to confirm the effect of PO43– and NO3– on ARB and ARG proliferation and to confirm the regulatory parameters, such as the concentration of PO43– and NO3–. Such information could be used to establish reasonable ecological regulation strategies to help reduce the accumulation of ARGs and ARB in aquaculture environments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank, OQ217102-OQ217615 and OQ216893-OQ216992.
Author contributions
ZL, YD, and JF conceived and planned the experiments. ZL, YD, WS, QW, and HC carried out the experiments. All authors contributed to the interpretation of the results. ZL, YD, and JF took the lead in writing the manuscript. All authors provided critical feedback and helped to shape the research, analysis, and manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2022GH03), the Guangzhou Science and Technology Program (202201010162), the Central Public-Interest Scientific Institution Basal Research Fund, the South China Sea Fisheries Research Institute, CAFS (2021SD15), and the China Agriculture Research System (CARS-48).
Acknowledgments
We would like to thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1139641/full#supplementary-material
References
Chaturvedi P., Singh A., Chowdhary P., Pandey A., Gupta P. (2020). Occurrence of emerging sulfonamide resistance (sul1 and sul2) associated with mobile integrons-integrase (intI1 and intI2) in riverine systems. Sci. Total Environ. 751, 142217. doi: 10.1016/j.scitotenv.2020.142217
Chen Q., An X., Li H., Su J., Ma Y., Zhu Y. (2016). Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 92-93, 1–10. doi: 10.1016/j.envint.2016.03.026
Chen Y., Wang F. P. (2015). Insights on nitrate respiration by Shewanella. front. Mar. Sci. 1, 80. doi: 10.3389/fmars.2014.00080
Chen M. X., Wang W. C., Feng Y., Zhu X. H., Zhou H. Z., Tan Z. L., et al. (2014). Impact resistance of different factors on ammonia removal by heterotrophic nitrification–aerobic denitrification bacterium aeromonas sp. HN-02. Bioresour. Technol. 167, 456–461. doi: 10.1016/j.biortech.2014.06.001
Cheng W., Chen H., Su C., Yan S. (2013). Abundance and persistence of antibiotic resistance genes in livestock farms: A comprehensive investigation in eastern China. Environ. Int. 61, 1–7. doi: 10.1016/j.envint.2013.08.023
Clinical and Laboratory Standards Institute (2016). Performance standards for antimicrobial susceptibility testing. 26th ed. CLSI supplement M100S (950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA: Clinical and Laboratory Standards Institute), ISBN: ISBN 1-56238-923-8.
Deng Y. Q., Mao C., Lin Z. Y., Su W. X., Cheng C. H., Li Y., et al. (2022). Nutrients, temperature, and oxygen mediate microbial antibiotic resistance in sea bass (Lateolabrax maculatus) ponds. Sci. Total Environ. 819, 153120. doi: 10.1016/j.scitotenv.2022.153120
Devarajan N., Kohler T., Sivalingam P., Delden C. V., Mulaji C. K., Mpiana P. T., et al. (2017). Antibiotic resistant pseudomonas spp. in the aquatic environment: a prevalence study under tropical and temperate climate conditions. Water Res. 115, 256. doi: 10.1016/j.watres.2017.02.058
Fadel A., Mabrok M., Aly S. (2018). Epizootics of Pseudomonas anguilliseptica among cultured seabream (Sparus aurata) populations: Control and treatment strategies. Microb. Pathog. 121, 1–8. doi: 10.1016/j.micpath.2018.04.021
Farmaki E.G., Thomaidis N.S., Pasias I.N., Baulard C., Papaharisis L., Efstathiou C.E. (2014). Environmental impact of intensive aquaculture: Investigation on the accumulation of metals and nutrients in marine sediments of Greece. Sci. Total Environ. 485, 554–562.
Gai C., Ye W., Lu L., Yi L., Cao H. (2016). Aeromonas hydrophila: a causative agent for tail rot disease in freshwater cultured murray cod maccullochella peelii. Israeli J. aquaculture = Bamidgeh 68.
Gao P., Mao D., Luo Y., Wang L., Xu B., Xu L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46 (7), 2355–2364. doi: 10.1016/j.watres.2012.02.004
Girija A., Vijayashree P. J., Paramasivam A. (2019). Plasmid-encoded resistance to trimethoprim/sulfamethoxazole mediated by dfrA1, dfrA5, sul1 and sul2 among Acinetobacter baumannii isolated from urine samples of patients with severe urinary tract infection. J. Glob Antimicrob. Resist. 17, 145–146. doi: 10.1016/j.jgar.2019.04.001
Guo H., Chen C., Lee D. J., Wang A., Gao D., Ren N. (2014). Coupled carbon, sulfur and nitrogen cycles of mixotrophic growth of pseudomonas sp. C27 under denitrifying sulfide removal conditions. Bioresource Technol. 171, 120–126. doi: 10.1016/j.biortech.2014.08.035
Hoa P. T. P., Managaki S., Nakada N., Takada H., Shimizu A., Anh D. H., et al. (2011). Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci. Total Environ. 409 (15), 2894–2901. doi: 10.1016/j.scitotenv.2011.04.030
Hu J., Zhao F., Zhang X. X., Kan L., Mei L. (2018). Metagenomic profiling of ARGs in airborne particulate matters during a severe smog event. Sci. Total Environ. 615, 1332–1340. doi: 10.1016/j.scitotenv.2017.09.222
Huang X., Gu Y., Zhou H., Xu L., Gai C. (2020). Acinetobacter venetianus, a potential pathogen of red leg disease in freshwater-cultured whiteleg shrimp Penaeus vannamei. Aquaculture Rep. 18, 100543. doi: 10.1016/j.aqrep.2020.100543
Jz A., Cl A., Jl A., Yl A., Zh A., Mhab C., et al. (2021). Ecological assessment of current and future pogostemon cablin benth. potential planting regions in China based on MaxEnt and ArcGIS models. J. Appl. Res. Medicinal Aromatic Plants. 24, 100308. doi: 10.1016/j.jarmap.2021.100308
Khalil W., Gantois C., Lemnos L., Salle L., Salle H. (2021). Aeromonas hydrophila is a deceptive pathogen requiring reconsideration of antibiotic prophylaxis. Surg. Infect. 22 (9), 987–988. doi: 10.1089/sur.2021.125
Li F., Zhao J., Zhao Y., Liu X., Huang J., Zhang Y., et al. (2021). Pathological findings of Chinese sucker (Myxocyprinus asiaticus) infected with virulent Aeromonas hydrophila. Aquaculture Rep. 21, 100884. doi: 10.1016/j.aqrep.2021.100884
Lihan S., Jamil N. A., Azizul M., Jamian H., Kion L. N. (2020). Distribution and prevalence of antibiotic resistant bacteria in fish farms in east malaysia. Malays. J. Microbiol. 16 (4), 263–274.
Liu X. Z., Cui L. F., Li S. M., Jiang K. Y., Yuan X. C., Yu X., et al. (2022). China Fishery statistics yearbook. China Agricultural Press, China.
Liu J., Fu B., Yang H., Zhao M., He B., Zhang X. H. (2015). Phylogenetic shifts of bacterioplankton community composition along the pearl estuary: The potential impact of hypoxia and nutrients. Front. Microbiol. 6, 64. doi: 10.3389/fmicb.2015.00064
Mao C., Su Y., Li Y., Zhang Y., Gu Q., Deng Y., et al. (2020). Analysis of the relationship between antibiotic resistance and virulence of Edwardsiella piscicida strains isolated from Lateolabrax maculatus. J. Fish. Sci. China 27 (07), 846–857. doi: 10.3724/SP.J.1118.2020.19311
Marti E., Variatza E., Balcazar J. L. (2014). The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 22 (1), 36–41. doi: 10.1016/j.tim.2013.11.001
Muniesa M., Colomer-Lluch M., Jofre J. (2013). Could bacteriophages transfer antibiotic resistance genes from environmental bacteria to human-body associated bacterial populations? Mob. Genet. Elements. 3 (4), e25847. doi: 10.4161/mge.25847
Rodas-Suarez O. R., Flores-Pedroche J. F., Betancourt-Rule J. M., Quinones-Ramirez E. I., Vazquez-Salinas C. (2006). Occurrence and antibiotic sensitivity of Listeria monocytogenes strains isolated from oysters, fish, and estuarine water. Appl. Environ. Microbiol. 72 (11), 7410–7412. doi: 10.1128/AEM.00956-06
Shen X., Jin G., Zhao Y., Shao X. (2019). Prevalence and distribution analysis of antibiotic resistance genes in a large-scale aquaculture environment. Sci. Total Environ. 711, 134626. doi: 10.1016/j.scitotenv.2019.134626
Sherif A. H., Gouda M., Darwish S., Abdelmohsin A. (2021). Prevalence of antibiotic-resistant bacteria in freshwater fish farms. Aquac. Res. 52 (5), 2036–2047. doi: 10.1111/are.15052
Skold O. (2000). Sulfonamide resistance: Mechanisms and trends. Drug Resist. Update 3 (3), 155–160. doi: 10.1054/drup.2000.0146
Su H. C., Liu Y. S., Pan C. G., Chen J., He L. Y., Ying G. G. (2018). Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 616-617, 453–461. doi: 10.1016/j.scitotenv.2017.10.318
Su H., Pan C., Ying G., Zhao J., Zhou L., Liu Y., et al. (2014). Contamination profiles of antibiotic resistance genes in the sediments at a catchment scale. Sci. Total Environ. 490, 708–714. doi: 10.1016/j.scitotenv.2014.05.060
Su Y., Wang J., Xia H., Xie B. (2019). Comparative network analysis revealing the mechanisms of antibiotic resistance genes removal by leachate recirculation under different hydraulic loadings. Sci. Total Environ. 649, 318–326. doi: 10.1016/j.scitotenv.2018.08.361
Suzuki S., Ogo M., Koike T., Takada H., Newman B. (2015). Sulfonamide and tetracycline resistance genes in total- and culturable-bacterial assemblages in south African aquatic environments. Front. Microbiol. 6, 796. doi: 10.3389/fmicb.2015.00796
Taylor N. G., Verner-Jeffreys D. W., Baker-Austin C. (2011). Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol. Evol. 26 (6), 278–284. doi: 10.1016/j.tree.2011.03.004
Wang Q., Hamilton P. B., Kang F., Zhu X., Zhang Y., Zhao H. (2020). Regional-scale investigation for microbial competition-through-environment interactions modulating antibiotic resistance. Sci. Total Environ. 734, 139341. doi: 10.1016/j.scitotenv.2020.139341
Wang C., Liu X. H., Yang Y. Y., Wang Z. (2021). Antibiotic and antibiotic resistance genes in freshwater aquaculture ponds in china: a meta-analysis and assessment. J. Clean. Prod. 329, 129719. doi: 10.1016/j.jclepro.2021.129719
Wang R., He J., Wang J. (2016). Heterotrophic bacterial abundance and diversity in the farming environment and guts of the oyster Crassostrea hongkongensis. J. Shellfish Res. 35, 343–350.
Whooley M. A., O’Callaghan J. A., Mcloughlin A. J. (1983). Effect of substrate on the regulation of exoprotease production by Pseudomonas aeruginosa ATCC 10145. Microbiology 129 (4), 981–988. doi: 10.1099/00221287-129-4-981
Wu J., Su Y., Deng Y., Guo Z., Mao C., Liu G., et al. (2019). Prevalence and distribution of antibiotic resistance in marine fish farming areas in hainan, China. Sci. Total Environ. 653, 605–611. doi: 10.1016/j.scitotenv.2018.10.251
Xiang Q., Chen Q., Zhu D., An X., Yang X., Su J., et al. (2018). Spatial and temporal distribution of antibiotic resistomes in a peri-urban area is associated significantly with anthropogenic activities. Environ. pollut. 235, 525–533. doi: 10.1016/j.envpol.2017.12.119
Xiong W., Sun Y., Zhang T., Ding X. Y., Li Y. F., Wang M. Z., et al. (2015). Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 70, 425–432. doi: 10.1007/s00248-015-0583-x
Yao S., Ni J. R., Ma T., Li C. (2013). Heterotrophic nitrification and aerobic denitrification at low temperature by a newly isolated bacterium, acinetobacter sp. HA2. Bioresour. Technol. 139, 80–86. doi: 10.1016/j.biortech.2013.03.189
Ye L., Liu G., Yao T., Lu J. (2021). Monitoring of antimicrobial resistance genes in the spotted sea bass (Lateolabrax maculatus): Association with the microbiome and its environment in aquaculture ponds. Environ. pollut. 276, 116714. doi: 10.1016/j.envpol.2021.116714
Yuan J., Ni M., Liu M., Zheng Y., Gu Z. (2019). Occurrence of antibiotics and antibiotic resistance genes in a typical estuary aquaculture region of hangzhou bay, China. Mar. pollut. Bull. 138, 376–384. doi: 10.1016/j.marpolbul.2018.11.037
Zago V., Veschetti L., Patuzzo C., Malerba G., Lleo M. M. (2020). Shewanella algae and vibrio spp. strains isolated in Italian aquaculture farms are reservoirs of antibiotic resistant genes that might constitute a risk for human health. Mar. pollut. Bull. 154, 111057. doi: 10.1016/j.marpolbul.2020.111057
Zhang Y. J., Hu H. W., Gou M., Wang J. T., Chen D., He J. Z. (2017). Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ. pollut. 231 (Pt 2), 1621–1632. doi: 10.1016/j.envpol.2017.09.074
Zhang J. B., Wu P. X., Hao B., Yu Z. N. (2011). Heterotrophic nitrification and aerobic denitrification by the bacterium pseudomonas stutzeri YZN-001. Bioresour. Technol. 102 (21), 9866–9869. doi: 10.1016/j.biortech.2011.07.118
Zhang J. T., Zhou S. M., An S. W., Chen L., Wang G. L. (2014). Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, Pseudomonas plecoglossicida. J. Fish Dis. 37 (2), 113–121. doi: 10.1111/jfd.12075
Zhao X., Li X., Li Y., Sun Y., Zhang X., Weng L., et al. (2019). Shifting interactions among bacteria, fungi and archaea enhance removal of antibiotics and antibiotic resistance genes in the soil bioelectrochemical remediation. Biotechnol. Biofuels 12 (1), 1–14. doi: 10.1186/s13068-019-1500-1
Keywords: antibiotic resistance genes, antibiotic resistant bacteria, sea bass, pond culture areas, gene chip technology
Citation: Lin Z, Deng Y, Su W, Wang Q, Chen H, Sun Y and Feng J (2023) Prevalence and distribution of antibiotic resistance in the water environment of sea bass (Lateolabrax maculatus) breeding area in spring in South China. Front. Mar. Sci. 10:1139641. doi: 10.3389/fmars.2023.1139641
Received: 07 January 2023; Accepted: 27 March 2023;
Published: 05 April 2023.
Edited by:
Ruixuan Wang, Hanshan Normal University, ChinaReviewed by:
Chui Wei Bong, University of Malaya, MalaysiaYuting Deng, Pearl River Fisheries Research Institute (CAFS), China
Biao Jiang, Zhongkai University of Agriculture and Engineering, China
Copyright © 2023 Lin, Deng, Su, Wang, Chen, Sun and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan Feng, anVhbmZlbmdAc2NzZnJpLmFjLmNu
†These authors share first authorship
 Ziyang Lin
Ziyang Lin Yiqin Deng
Yiqin Deng Wenxiao Su
Wenxiao Su Qian Wang
Qian Wang Haoxiang Chen1,3
Haoxiang Chen1,3 Yun Sun
Yun Sun Juan Feng
Juan Feng