- College of Earth, Ocean, and Environment, University of Delaware, Lewes, DE, United States
Dinoflagellates are among the most toxigenic phytoplankton that cause harmful algal blooms; they can produce toxins that accumulate through the aquatic food chains to cause illness and even death in marine animals and humans. Shewanella sp. IRI-160 is a naturally-occurring bacterium that secretes a suite of algicidal compounds (collectively designated as IRI-160AA) specifically targeting dinoflagellates. Studies revealed IRI-160AA inhibited photosynthesis, while inducing cell cycle arrest, DNA damage, and reactive oxygen species (ROS) production, as well as other markers associated with programmed cell death (PCD). Recent research indicated that IRI-160AA contains ammonium and other compounds as active ingredients for its algicidal activity, while impacts by ammonium differed from the algicide with respect to photobiology. Here, transcriptomic analysis was conducted on the toxic dinoflagellate Karlodinium veneficum exposed to IRI-160AA to investigate the effects of this algicide at the molecular level. Transcriptomic analysis was also conducted on K. veneficum treated with ammonium to further differentiate its effects from IRI-160AA. Results demonstrated differential impacts by IRI-160AA and ammonium on K. veneficum at the molecular level and revealed a global response of K. veneficum to algicide exposure, supported by the enriched biological processes involved in regulating gene expression, protein activity, and morphology. Differentially expressed genes associated with stress and ROS response, DNA damage response, cell cycle checkpoint activation, and PCD were also identified in K. veneficum exposed to IRI-160AA. The potential involvement of a recovery mechanism from photodamage in K. veneficum induced by IRI-160AA was identified. Overall, results of this study further differentiated the impacts of ammonium from IRI-160AA on K. veneficum and illustrated the cellular mechanisms behind the algicidal effect. This research provided insights on algal response to bacterial derivatives in nature.
1 Introduction
Dinoflagellates are among the most abundant aquatic flagellates (reviewed by Nielsen and Kiørboe, 2015) and play important roles in both freshwater and marine ecosystems (reviewed by Hackett et al., 2004; Bi et al., 2019). The photosynthetic dinoflagellates are one of the most prominent groups of primary producers (reviewed by Bi et al., 2019). Some of them are also beneficial endosymbionts of other organisms, such as corals, sea anemones, and jellyfish (reviewed by Fukuda and Suzaki, 2015). On the other hand, dinoflagellates can form harmful algal blooms (HABs) and are among the most toxigenic HAB species (reviewed by Anderson et al., 2012). The toxins produced by these species can be transported and accumulated through the food webs, causing illness and death of fish, birds, mammals, and even human beings (reviewed by Granéli and Turner, 2006). Karlodinium veneficum is a toxic dinoflagellate that causes harmful blooms worldwide (reviewed by Farhat et al., 2022). This species can secrete polyketide toxins, referred to as karlotoxins, that are ichthyotoxic, cytotoxic, and hemolytic (reviewed by Farhat et al., 2022). Karlotoxins have caused massive fish kills worldwide (reviewed by Peng et al., 2008).
Dinoflagellates also receive attention because of their unique nuclei, which are referred to as “dinokaryon” in the core dinoflagellates (reviewed by Fukuda and Suzaki, 2015; Riaz et al., 2018; Gornik et al., 2019). Dinoflagellate genomes are extremely large (up to 80-fold of the size of a human haploid genome), and contain large amount of non-coding and highly methylated DNA. Their nuclei are characterized by an abundance of divalent cations, as well as permanently condensed chromosomes in a liquid-crystalline state (reviewed by Lin, 2011; Gornik et al., 2019).
Previous research has demonstrated a strong relationship between dinoflagellates and their surrounding and/or associated bacteria (e.g., Cruz-López and Maske, 2016; Cruz-López et al., 2018; Park et al., 2018; Matthews et al., 2020; Tarazona-Janampa et al., 2020). For instance, many dinoflagellates are reported to be vitamin B1 and B12 auxotrophs (Tang et al., 2010), and recent studies demonstrated that these vitamins could be provided by the bacterial communities (Cruz-López and Maske, 2016; Cruz-López et al., 2018). A symbiotic relationship of dinoflagellate Lingulodinium polyedra and a B vitamin producing bacterium, Dinoroseobacter shibae was also proposed, where the bacterium could supply L. polyedrum with required vitamin B1 and B12 in exchange for the algal released vitamin B7 (Cruz-López et al., 2018).
Besides the beneficial effects, many bacteria were found to inhibit the growth of dinoflagellates and/or lyse dinoflagellate cells, exhibiting algicidal activities (e.g., Hare et al., 2005; Shi et al., 2018; Dungca-Santos et al., 2019; reviewed by Coyne et al., 2022). Dungca-Santos et al. (2019), for example, isolated 48 cultivable pelagic bacteria from HAB-affected environments and tested them on the dinoflagellate Pyrodinium bahamense, demonstrating algicidal activities by 94% of the bacteria tested. As for other algal species (Meyer et al., 2017), bacteria can attach to and directly attack dinoflagellate cells (Imai and Kimura, 2008; Roth et al., 2008; reviewed by Coyne et al., 2022) or secrete active compounds to indirectly cause cell death (e.g., Pokrzywinski et al., 2012; Li et al., 2015; reviewed by Coyne et al., 2022), with the latter to be the dominant strategy (reviewed by Coyne et al., 2022).
In 2005, Hare et al. (2005) reported a gram-negative bacterium, Shewanella sp. IRI-160, isolated from the Delaware Inland Bays, Delaware, USA, that exhibited selective algicidal effects against dinoflagellates. The following research demonstrated active substances were released to the water column, and no direct interactions were required for algicidal activity; the active compounds were collectively designated as IRI-160AA (Pokrzywinski et al., 2012). Further studies on the photobiology of IRI-160AA impaired dinoflagellates revealed an inhibition of photosystem II and a disruption of the electron transport chain (Tilney et al., 2014). Morphological studies illustrated translocation of nuclei and chloroplasts in dinoflagellates exposed to IRI-160AA, along with chromosome destabilization and decondensation in these cells (Pokrzywinski et al., 2017a). Research focusing on cell cycle progression and biochemical changes in dinoflagellates demonstrated cell cycle arrest induced by IRI-160AA (Pokrzywinski et al., 2017b). DNA degradation, reactive oxygen species (ROS) production, as well as caspase 3-like protease (DEVDase) activity were also observed in these organisms, implying a programmed pathway leading to cell death (PCD) (Pokrzywinski et al., 2017b). Recent metabolomics research on K. veneficum treated with IRI-160AA demonstrated an increase of a suite of compounds in the cell pellets of this alga, including oxidative stress biomarkers, antioxidants, and compounds involved in DNA damage and PCD (Wang and Coyne, 2022).
Research conducted by Ternon et al. (2019) indicated ammonium and several amines were among the active compounds in IRI-160AA (also see Wang, 2021), and acted synergistically to increase the activity of IRI-160AA. Grasso (2018) further demonstrated the differential effects of ammonium alone vs. IRI-160AA on the photobiology of dinoflagellates. The contribution of ammonium to the activity of IRI-160AA remains unclear, although previous reports on ammonium toxicity indicated that dinoflagellates may be more sensitive than other phytoplankton groups (Collos and Harrison, 2014). In this study, IRI-160AA (designated as the “algicide”) refers to bacterial cell-free exudates containing ammonium and other compounds as active ingredients that act synergistically to affect cell death in dinoflagellates.
The cellular response of dinoflagellates to bacteria (Dungca-Santos et al., 2019) and bacterial cell-free medium (Yang et al., 2014; Zhang et al., 2018a) have been investigated for the interaction of other dinoflagellate-bacteria pairs. However, limited research has focused on the impacts of bacteria or their derivatives on dinoflagellates at the molecular level (e.g., Moustafa et al., 2010; Lei et al., 2015). In this research, transcriptomic analyses were conducted on the harmful dinoflagellate Karlodinium veneficum exposed to IRI-160AA to illustrate the effects of the bacterial derivatives on dinoflagellates at the molecular level. Transcriptomic analyses were also conducted on the same species treated with ammonium to differentiate the impacts of ammonium alone from the algicide.
2 Methods
2.1 Algal stock culture
Non-axenic stock cultures of Karlodinium veneficum (CCMP 2936 [National Center for Marine Algae and Microbiota, https://ncma.bigelow.org/]; dinoflagellate) were maintained in natural seawater with f/2 nutrients (-Si) (Guillard and Ryther, 1962) and a salinity of 20, at 25 ˚C, and with a light intensity of approximately 130 µmol photons m-2 s-1. The cultures were kept under a 12 h: 12 h light: dark cycle and semi-continuously in the exponential growth phase.
2.2 Algicide preparation
IRI-160AA was prepared as in Pokrzywinski et al. (2012) with slight modifications. Briefly, a single colony of Shewanella IRI-160 was transferred to LM medium (Sambrook et al., 2012). The culture was incubated at 25 °C with shaking overnight at 100 rpm. The Shewanella IRI-160 culture was then centrifuged at 6000 rpm for 5 min. The supernatant was discarded, and the cell pellet was resuspended in f/2 medium (-Si) (Guillard and Ryther, 1962). The suspension was centrifuged again as above. The supernatant was discarded, and cell pellets were resuspended in f/2 medium (-Si) (Guillard and Ryther, 1962) and incubated at room temperature for 7 days. To prepare the cell-free algicide, the culture was filtered through a 0.2 µm nylon syringe filter (Corning, Corning, NY, USA). This filtrate is IRI-160AA, containing ammonium and other active compounds.
2.3 Transcriptomic effects of IRI-160AA on Karlodinium veneficum
2.3.1 Culture treatments
Karlodinium veneficum culture was treated with 50 μM NH4Cl (final concentration) in sterile MilliQ water for the ammonium treatment (N=3), or 1.28% (v/v) IRI-160AA algicide (equal to 50 µM final ammonium concentration) for the algicide treatment (N=3). Sterile MilliQ water and f/2 medium were added to the controls (N=3) (Supplementary Table 2). All treatments and controls received the same volumes of MilliQ water and f/2 medium.
In vivo fluorescence (as a proxy for biomass; Pokrzywinski et al., 2012) of the bulk culture was measured at the initial time point (T0). In vivo fluorescence of each treatment and control culture was measured at 1 hour (T1hr), 6 hours (T6hr), and 24 hours (T24hr) after the initial time point. Relative in vivo fluorescence was measured in an AquaFluor handheld fluorometer (Turner, San Jose, CA, USA). During this process, all cultures were in the light phase during the light: dark cycle, with the exception of the cultures at T6hr, which were measured during the dark phase.
At T1hr, immediately before measuring in vivo fluorescence, 40 mL samples were collected for transcriptome analysis by filtering onto 3 µm polycarbonate filters (Millipore, Burlington, MA, USA). The filters were immediately immersed in RLT buffer (RNeasy Mini Kit; Qiagen, Chatswort, CA, USA) on ice and then transferred to -80°C before RNA extraction as described below.
2.3.2 Statistical analysis of in vivo fluorescence measurement
One-way ANOVA was used to test the significant difference (p<0.05) of in vivo fluorescence between the treatments and controls at each time point. If there was a significant difference detected, then Tukey HSD test was conducted to test the significant difference of in vivo fluorescence between all pairs of groups (p<0.05).
2.3.3 Sample preparation for RNA-seq sequencing
RNA was extracted from filtered cells with an RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. RNA concentration was measured using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and the integrity of RNA was evaluated by electrophoresis. Contaminating DNA was digested using a DNA-free™ DNA Removal Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. RNA collected at T1hr was combined into a single composite sample per treatment for sequencing. The quality and integrity of the RNA in the composite samples were assessed using a fragment analyzer (Advanced Analytical Technologies, Inc., Ankeny, IA, USA). Library preparation and sequencing of composite RNA samples were conducted at Delaware Biotechnology Institute (DBI; Newark, DE, USA).
NEXTflex™ Poly(A) Beads (Perkin-Elmer, Waltham, MA, USA) were used to isolate messenger RNA (mRNA) to limit both ribosomal RNA and prokaryotic mRNA contamination in the final sequence library. A pooled library was prepared using the NEXTflex™ Rapid Directional RNA-Seq Kit (Perkin-Elmer) following the manufacturer’s protocol. The library concentration was measured using a Qubit fluorometer (Thermo Fisher Scientific), and the library quality was assessed using a fragment analyzer (Advanced Analytical Technologies, Inc.). Sequencing of 101 base pair single-end reads from the pooled library was performed on a single lane on an Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA).
2.3.4 De novo assembly and gene differential expression analysis
The reference genome of K. veneficum is not available; de novo assembly for the RNA-seq data was applied here. Reads from all samples were assembled into one single assembly first using Trinity (v2.8.5) (Grabherr et al., 2011; Haas et al., 2013). The flags used in the program were –seqType fq (for fastq formats), –single (for single-ended unpaired reads), –SS_lib_type R (for reverse complement reads), and –trimmomatic (to quality trim the reads before the assembly). Other flags used include those to specify the allowed maximal memory (–max_memory 120G) and CPU (–CPU 24), as well as the output directory (with flag –output). To assess the quality of the de novo assembly, Bowtie 2 (v 2.3.4.3) was used to examine the RNA-seq read representation by the assembly (Langmead and Salzberg, 2012). The contig Nx statistic (Haas et al., 2013) and ExN50 statistic (Geniza and Jaiswal, 2017) were estimated using the script bundled with the Trinity toolkit.
2.3.5 Gene transcript abundance estimate and differential expression analysis
Gene transcript abundance in each treatment and control was estimated using Trinity (Grabherr et al., 2011) with Salmon (Patro et al., 2017) as the estimate method. Downstream analysis was followed using all sequences assembled without filtering, as suggested by the Trinity manual (RNA-Seq De novo Assembly Using Trinity; https://github.com/trinityrnaseq/trinityrnaseq/wiki; assessed on 10/04/22). Differential expression (DE) analysis was conducted at the gene level using edgeR (Robinson et al., 2009) bundled in Trinity. DE genes were clustered according to their DE patterns across the samples using the analyze_diff_expr.pl tool in Trinity (Haas et al., 2013). Volcano and MA plots (log-fold-change between experimental groups [M] against the average expression across all the samples [A] for each gene) were also generated to examine the gene expression patterns using the Trinity toolkit (Haas et al., 2013). The differential expression analysis of RNA-Seq data was validated using reverse transcription-quantitative PCR (RT-qPCR) (Supplementary Material).
2.3.6 Gene annotation
Coding regions were identified from the assembly using TransDecoder (v 5.5.0) (Haas et al., 2013). The long open reading frames were extracted first, and then the reading frames were used to search against Uniprot/Swiss-Prot (Bateman, 2019) and protein family (Pfam) (Finn et al., 2014) databases using BLASTP (v 2.9.0) (Camacho et al., 2009) and HMMER (v 3.2.1) (Finn et al., 2015), respectively. Finally, the searching results were integrated to predict the coding regions using TransDecoder (Haas et al., 2013). Translated protein sequences were also generated with TransDecoder (Haas et al., 2013).
The assembled unigenes from Trinity (Grabherr et al., 2011; Haas et al., 2013) and protein sequences from TransDecoder (Haas et al., 2013) were searched against a variety of databases. The BLASTN program (v 2.9.0) (Boratyn et al., 2019) was used to search against the NCBI non-redundant nucleotide (NT) database (Brown et al., 2015), the BLASTP program (v 2.9.0) (Camacho et al., 2009) was used to search against the NCBI non-redundant protein sequences (NR) (Brown et al., 2015). Both BLASTX (v2.9.0) and BLASTP programs (Camacho et al., 2009) were used to search against the Uniprot/Swiss-Prot database (Bateman, 2019). The BLATX program was also used to search against the Eukaryotic Orthologous Groups database (KOG) (Tatusov et al., 2003). All BLAST searches had an e-value cut-off of 0.001, and the number of sequences obtained with each search was limited to 5. Additionally, the predicted protein sequences were searched against the Pfam database (Finn et al., 2014) using the HMMER program (Finn et al., 2015). Finally, the annotation results were loaded into an SQLite database built by the Trinotate program (v 3.2.1) (Bryant et al., 2017). Kyoto Encyclopedia of Genes and Genomes Ortholog database (KEGG), Gene Ontology (GO), and evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG) (Huerta-Cepas et al., 2019) terms were assigned by Trinotate (Bryant et al., 2017). A report was generated using Trinotate with a cut-off p-value less than 0.001 for all annotations. Ribosomal RNA and spliced leader genes were searched in the annotation report to avoid contamination by these genes for further analysis; no such genes were found.
Genes that were highly differentially expressed between control and the treatments (fold-change > 4, FDR < 0.001) were used to construct subsets of annotation reports and used for further analyses below.
Gene ontology enrichment was analyzed using Database for Annotation, Visualization and Integrated Discovery (DAVID; v 6.8) (Huang et al., 2009a; Huang et al., 2009b) for each subset of data comparing genes that were up- or down-regulated by ammonium or the algicide compared to the control. Only the GO terms with a modified Fisher exact p-value < 0.05 were treated as enriched. The annotation of the whole transcriptome was used as the background. The GO enrichment analysis was conducted using all species in the DAVID database. GO terms in the biological process category at the direct level in which only the GO terms directly associated with the genes, and not including their parent terms, were included in the analysis. These GO terms were used to conduct a Venn diagram analysis using InteractiVenn (Heberle et al., 2015) (http://www.interactivenn.net; accessed on 11/29/2022) and a network analysis using EnrichmentMap (Merico et al., 2010) embedded in the Cytoscape software (v 3.8.0) (Shannon et al., 2003).
In addition, to compare with previously reported physiological response of K. veneficum to IRI-160AA (Tilney et al., 2014; Pokrzywinski et al., 2017a; Pokrzywinski et al., 2017b), the algicide-regulated DEGs that were involved in reactive oxygen species (ROS) and stress response, DNA damage response (DDR), cell cycle arrest, programmed cell death (PCD), and photobiology were searched in the annotation reports generated above. Key DEGs related to these processes were identified by searching the literature.
3 Results
3.1 Effects of algicide IRI-160AA and ammonium on K. veneficum
No significant differences of in vivo fluorescence were detected between treatments and the control at T1hr or T6hr (p>0.05; Figure 1). At T24hr, in vivo fluorescence of the control was slightly but significantly higher than the algicide treatment by 1.12 times (p<0.05), while no significant difference was observed between the control and the ammonium treatment or between the ammonium and the algicide treatment (p>0.05).
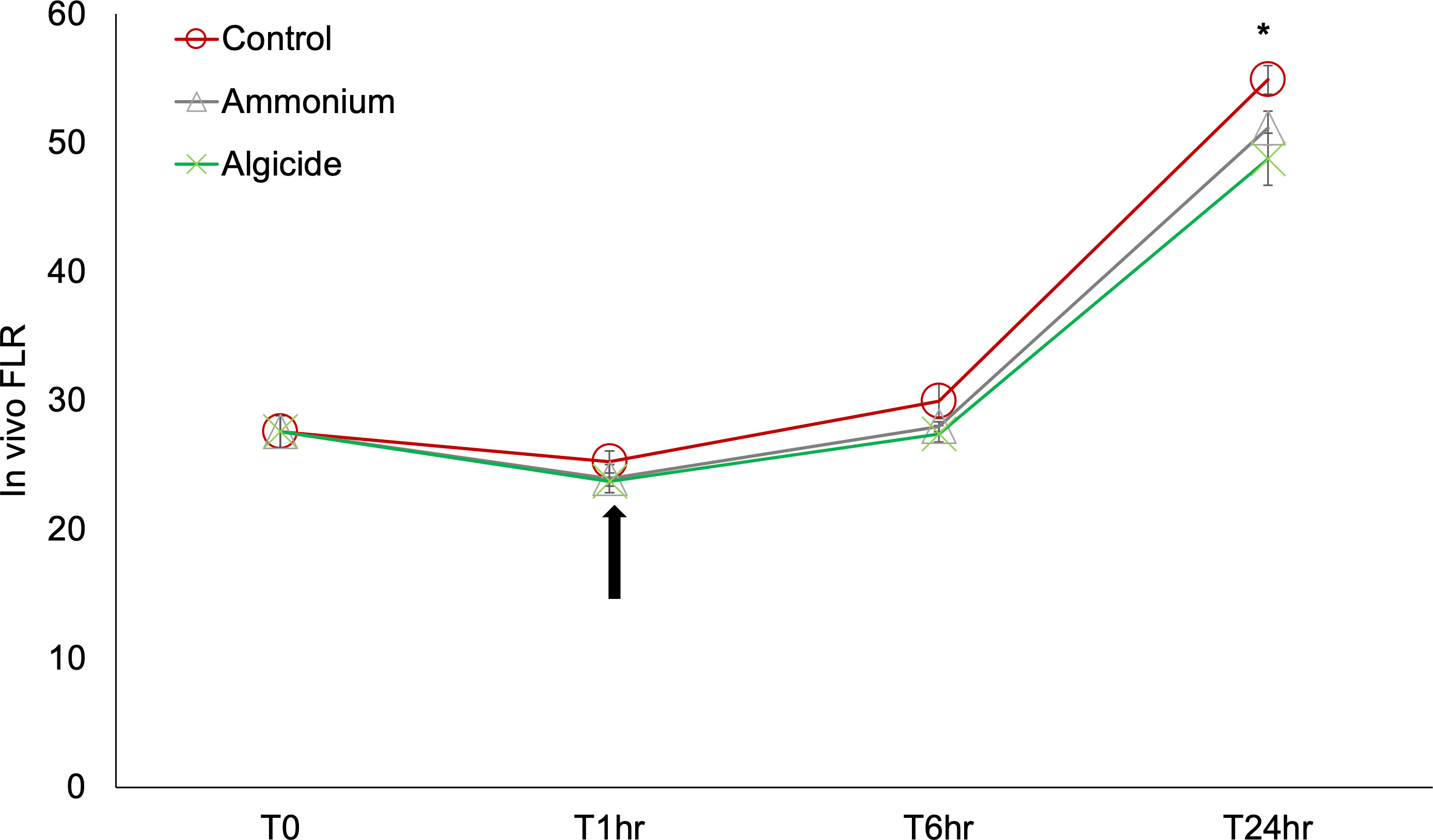
Figure 1 In vivo fluorescence (proxy of biomass) of Karlodinium veneficum treated with ammonium or the algicide IRI-160AA for 24 hours. Error bars indicate standard deviations of three replicates. Asterisk “*” indicates a significant difference between the in vivo fluorescence at the indicated time point (p<0.05). The arrow indicates 1 hour after the treatment, when the samples for RNA-seq analysis were collected.
3.2 De novo assembly and gene annotations
A total of 160,576,929 bases were assembled to 160,206 genes in this research, and 75,297 (47%) genes were annotated with an annotation cut-off of p<0.001. The RNA-seq data was validated by conducting RT-qPCR on 8 genes plus a reference gene across the individual control and treatment cultures (Supplementary Figure 1). The results demonstrated a significant correlation between gene expression fold-change generated from the RNA-seq and RT-qPCR methods (p=0.00018, R=0.9). Within the assembled genes, 7,886 genes were highly differentially expressed across samples (DEGs; FDR < 0.001, fold-change > 4; 4.92% of total genes; Table 1, Figure 2 and Supplementary Figure 2). DEGs were further clustered into 6 subgroups according to their expression patterns (Supplementary Figure 2).
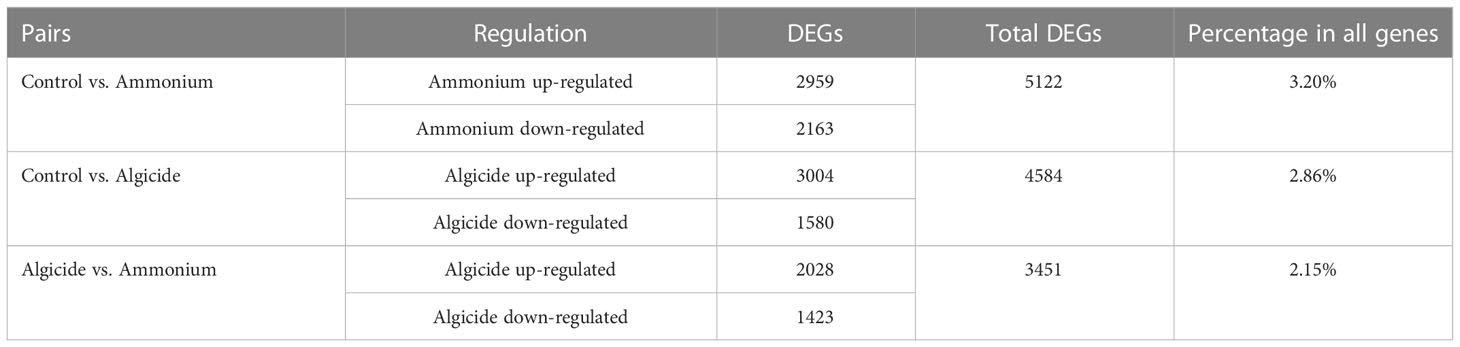
Table 1 Numbers of differentially expressed genes (DEGs) comparing control vs. the ammonium treatment, control vs. the algicide treatment, as well as the ammonium vs. the algicide treatment.
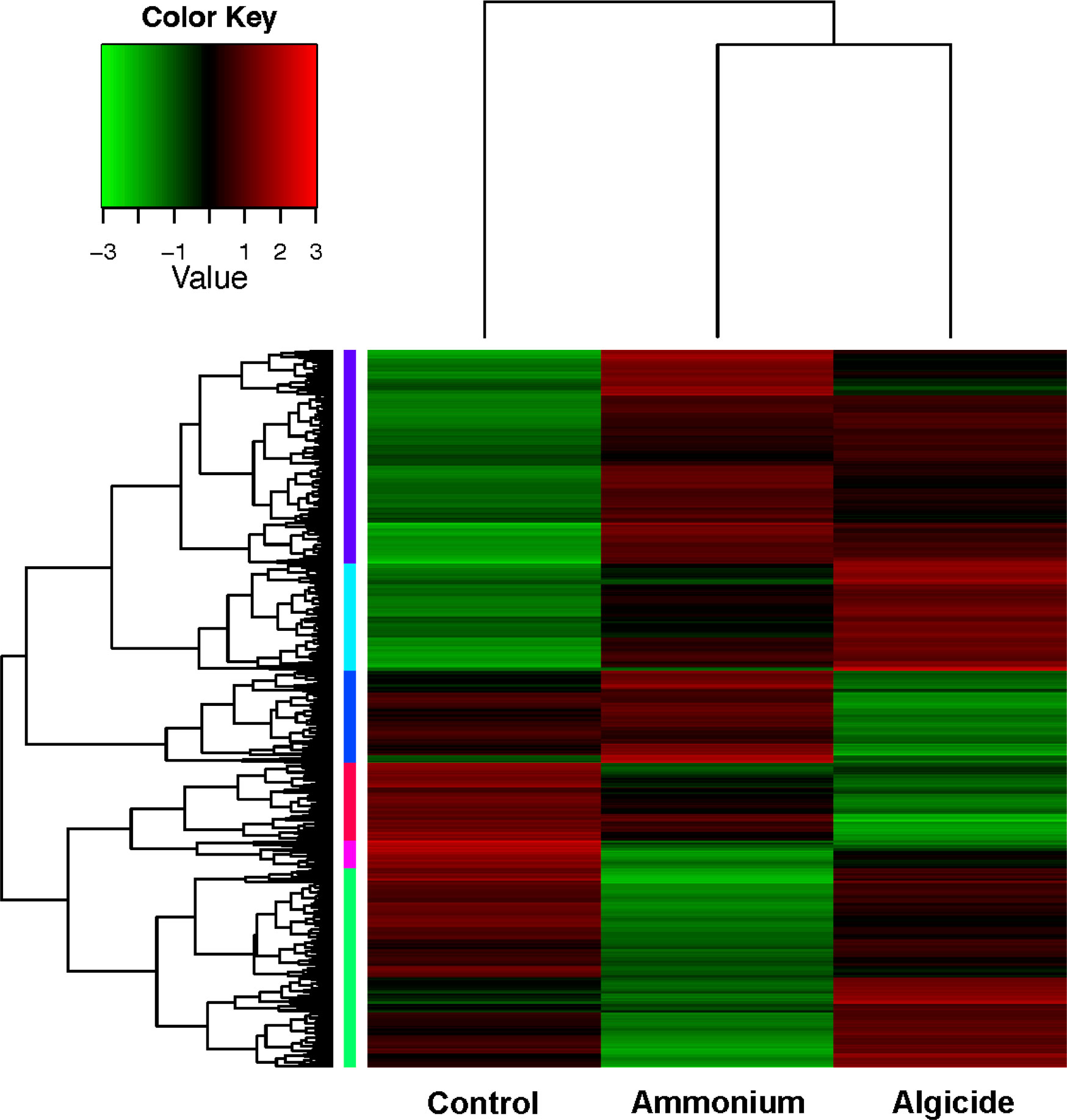
Figure 2 Heatmap of 7,886 highly differentially expressed genes (DEGs; FDR < 0.001 and fold-change > 4). DEGs were clustered into 6 subgroups by their expression patterns (color-coded at the left of the tree; expression patterns of subgroups are shown in Supplementary Figure 2).
Among these DEGs, 2,959 were up-regulated and 2,163 genes down-regulated in the ammonium treatment compared to the control (5,122 DEGs in total; 3.20% of all assembled genes; Table 1), while 3,004 were up-regulated and 1,580 genes were down-regulated in the algicide treatment compared to the control (4,584 DEGs in total; 2.86% of all assembled genes).
3.3 Biological processes enriched by DEGs
Eighteen biological processes were enriched by DEGs that were up- or down-regulated by ammonium and/or the algicide compared to the control (Figure 3). Fifteen processes (83%) were enriched by the DEGs that were regulated either by ammonium (11 processes) or algicide exposure (4 processes) alone, while 3 processes were enriched by DEGs in both ammonium and algicide treatments. These shared processes were (i) regulation of membrane potential, (ii) transmembrane receptor protein tyrosine kinase signaling pathway, and (iii) protein phosphorylation.
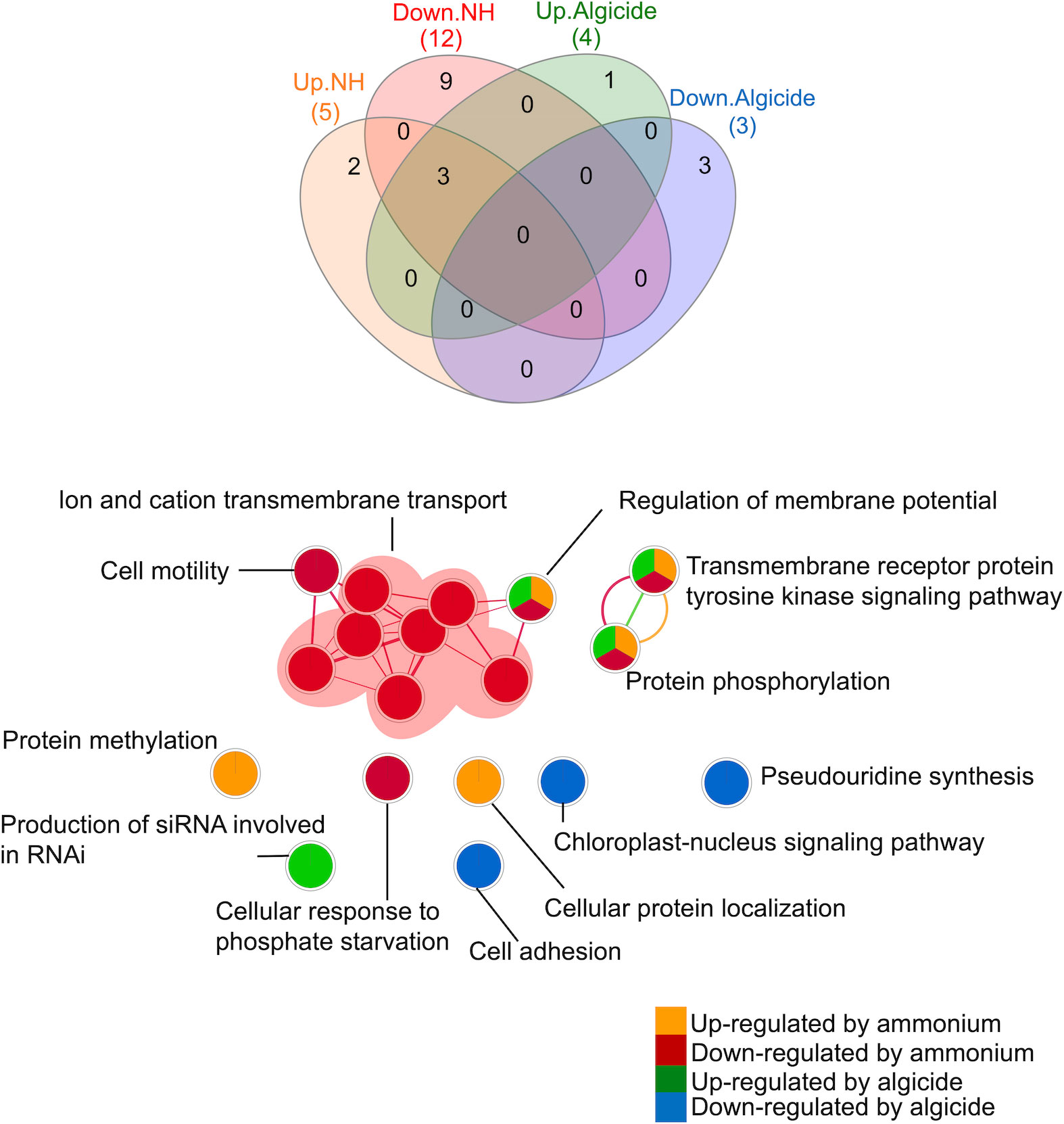
Figure 3 Venn diagram and network analysis of gene ontology (GO) terms in the biological process category enriched by DEGs that were up- or down-regulated by ammonium or the algicide. Up/Down.NH: processes enriched by the DEGs that were up- or down-regulated by ammonium; Up/Down.Algicide: processes enriched by the DEGs that were up- or down-regulated by the algicide. Nodes in the network analysis represent enriched processes (Merico et al., 2010). Edges connect nodes with shared genes, and their thickness is proportional to the overlap between the gene sets represented by the respective nodes (Merico et al., 2010). Venn diagram analysis was done using InteractiVenn (Heberle et al., 2015). Network analysis was done using the Enrichment Map plugin (Merico et al., 2010) in the Cytoscape software (v 3.8.0) (Shannon et al., 2003). siRNA, small interfering RNA; RNAi, RNA interference.
Among the 11 processes (61%) that were enriched by the DEGs that were only up- or down-regulated by ammonium, 2 were enriched by DEGs that were uniquely up-regulated in the ammonium treatment (Figure 3): (i) protein methylation and (ii) cellular protein localization. Nine processes were enriched by DEGs that were uniquely down-regulated in the ammonium treatment, including processes involved in ion and cation transmembrane transport and cell motility, as well as cellular response to phosphate starvation.
A total of 4 biological processes (22% of all enriched processes) were enriched by the DEGs that were only regulated by the algicide but not ammonium treatment (Figure 3). Production of small interfering RNA (siRNA) involved in RNA interference (RNAi) was enriched by DEGs that were uniquely up-regulated in the algicide treatment. Processes that were uniquely enriched by down-regulated DEGs in the algicide treatment included pseudouridine synthesis, cell adhesion, and chloroplast-nucleus signaling pathway.
3.4 Key genes involved in ROS and stress response, DNA damage, cell cycle arrest, PCD, and photobiology in the algicide-regulated DEGs
Four DEGs regulated by the algicide were identified as key genes involved in reactive oxygen species (ROS) and stress response (Table 2). These included genes encoding a peptidyl-prolyl cis-trans isomerase D (aka cyclophilin 40 protein, CyP40), a putative pentatricopeptide repeat-containing protein (PPR40), a multidrug resistance-associated protein (MRP), and a hydroxyacid oxidase (HAO; aka glycolate oxidase, GOX). Among these genes, CyP40 was up-regulated by both ammonium and the algicide, while the others were only down-regulated by the algicide.
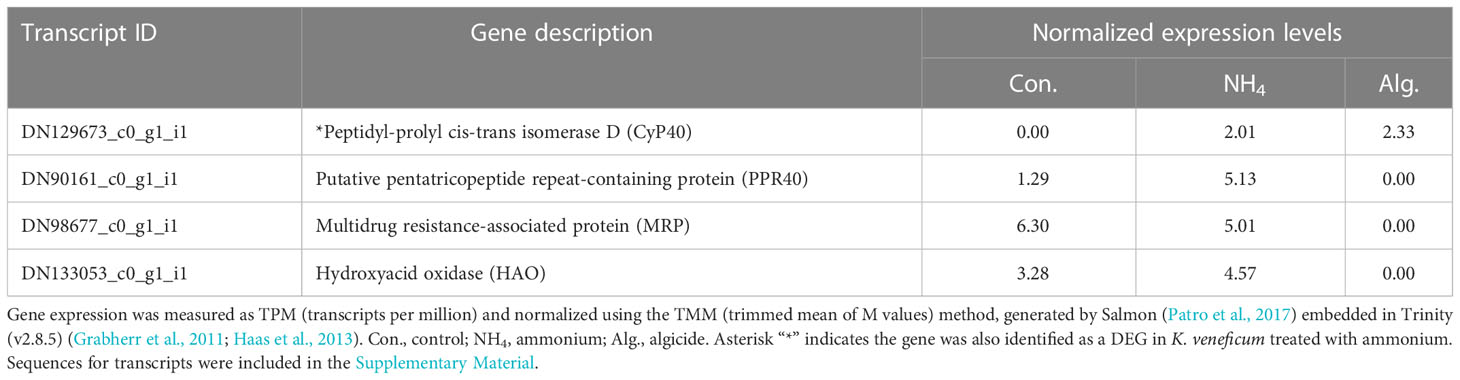
Table 2 Expression of key genes from RNA-seq sequencing involved in oxidative stress response identified in the DEGs of K. veneficum treated with IRI-160AA.
Six key genes in the algicide-regulated DEGs were related to DNA damage, cell cycle arrest, and PCD (Table 3). These genes included those encoding serine/threonine-protein kinases chk1 (CHK1) and chk2 (CHK2), a SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein (SMARCAL1), an E3 ubiquitin-protein ligase 2 (HERC2), a caspase-like enzyme (cathepsin B-like cysteine proteinase; CathB), and a calpain-type cysteine protease (calpain). Among these genes, CHK1, SMARCAL1, HERC2, and CathB were up-regulated by both ammonium and the algicide, while CHK2 and Calpain were only up-regulated by the algicide.
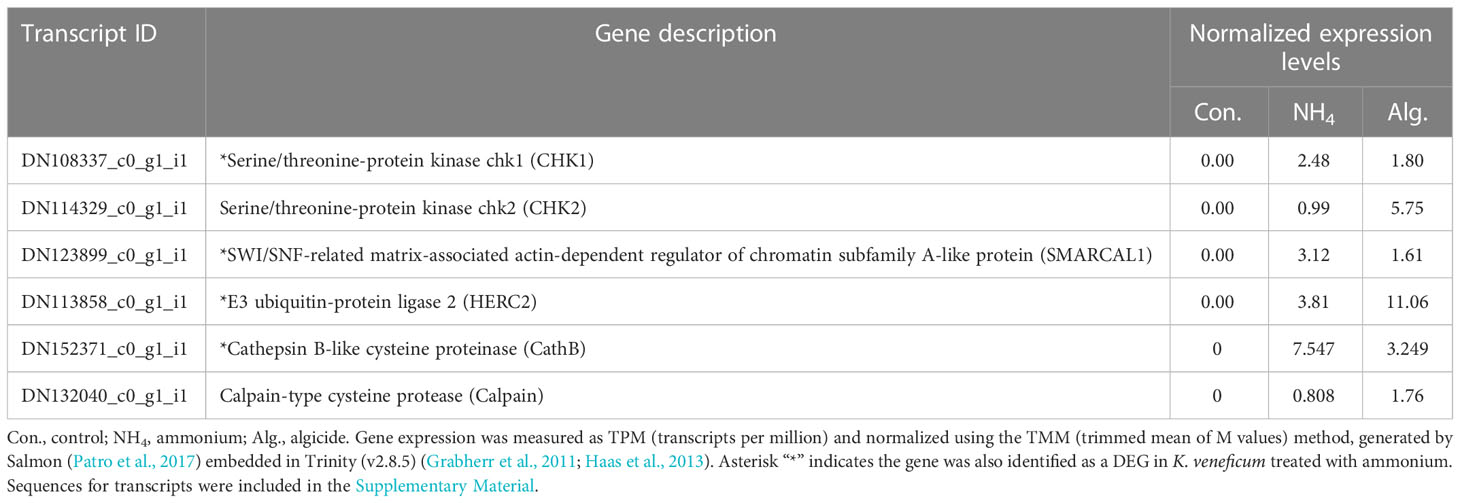
Table 3 Expression of key genes from RNA-seq sequencing related to DNA damage, cell cycle regulation, and programmed cell death (PCD) in the DEGs of K. veneficum treated with IRI-160AA.
Eight genes associated with photosynthesis/photorepair were identified in the algicide-regulated DEGs (Table 4). Among these genes, 3 were involved in chlorophyll biosynthesis (Figure 4), including those encoding a glutamate-1-semialdehyde 2,1-aminomutase (GSAT), an oxygen-dependent coproporphyrinogen-III oxidase (HEMF), and a light-dependent protochlorophyllide reductase (LPOR). LPOR was up-regulated in both ammonium and the algicide treatments, while GSAT was only down-regulated and HEMF was only up-regulated by the algicide. Furthermore, a PETD gene encoding the cytochrome b6-f complex subunit IV was only up-regulated by the algicide. A gene encoding a serine/threonine-protein phosphatase 5 (PAPP5) involved in the chloroplast-nucleus signaling pathway was only down-regulated by the algicide. In addition, 3 genes encoding photorepair proteins were up-regulated by both ammonium and the algicide; one of these genes encoded an ultraviolet-B receptor (UVR8), and two others encoded deoxyribodipyrimidine photolyases.
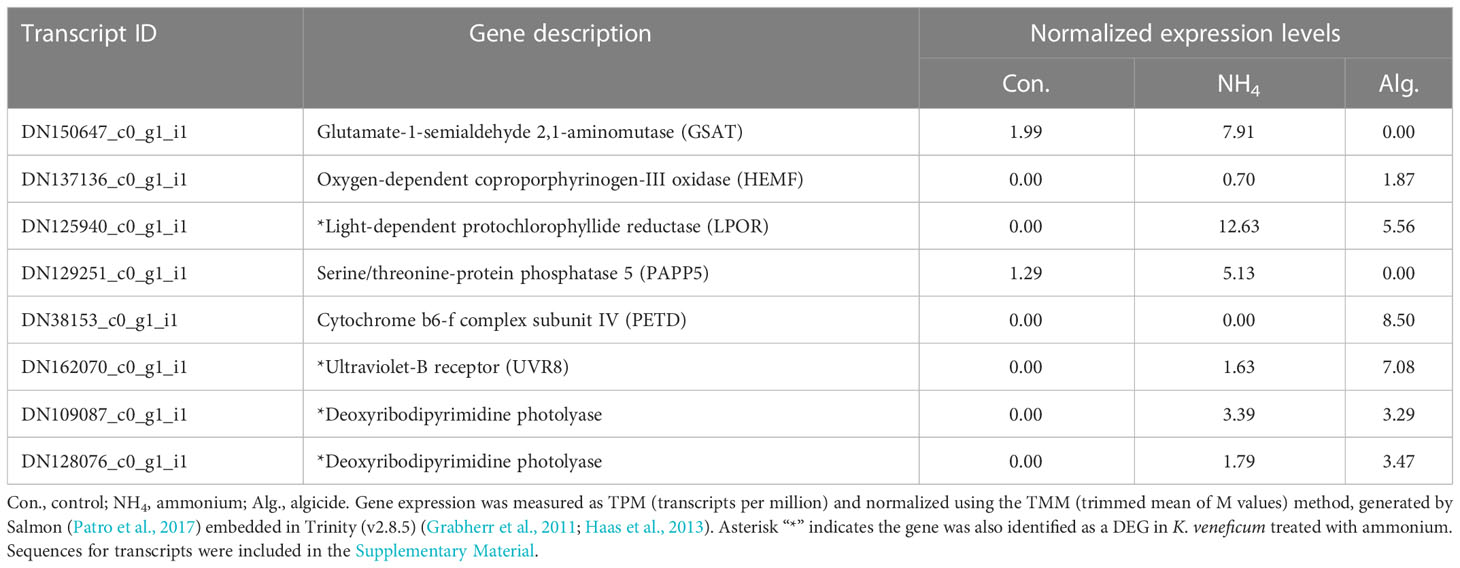
Table 4 Expression of key genes from RNA-seq sequencing related to photosynthesis identified in the DEGs of K. veneficum treated with IRI-160AA.
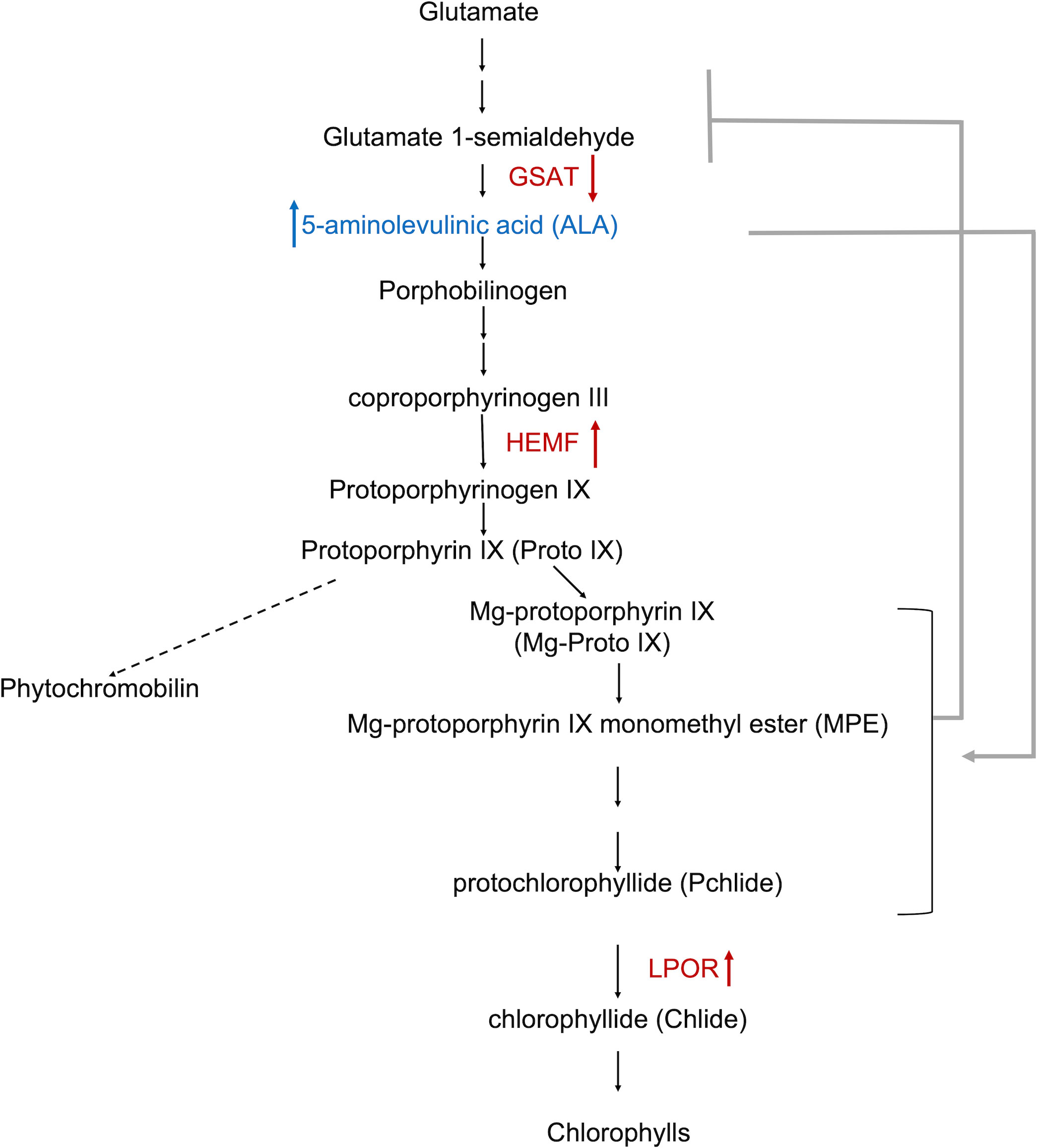
Figure 4 Schematic representation of chlorophyll biosynthesis (Brzezowski et al., 2019). The pathway of phytochromobilin formation is not shown in detail (dashed arrow). The metabolite (5-aminolevulinic acid [ALA]) shown in blue increased in the cell pellets of Karlodinium veneficum treated with IRI-160AA (Wang and Coyne, 2022). DEGs identified in the algicide treatment of K. veneficum were indicated in red. Besides the metabolite and genes, arrows indicate the increase or decrease of the content (of the metabolite) (Wang and Coyne, 2022) or expression (of genes) in K. veneficum treated with IRI-160AA compared to the control. The stimulatory effect of ALA on chlorophyll biosynthesis intermediates (Aarti et al., 2006; Stenbaek et al., 2008; Wu et al., 2018) was indicated by a grey arrow; the feedback inhibitory impact of these intermediates on ALA biosynthesis (Pattanayak and Tripathy, 2011) was indicated by a grey line with a bar. GSAT, glutamate-1-semialdehyde 2,1-aminomutase; HEMF, oxygen-dependent coproporphyrinogen-III oxidase; LPOR, light-dependent protochlorophyllide reductase.
4 Discussion
In this research, a transcriptomic study was conducted on the photosynthetic dinoflagellate K. veneficum exposed to the algicide IRI-160AA to examine the impact of bacterial derivatives on dinoflagellates at the molecular level. This transcriptomic profile was compared with a transcriptomic response of K. veneficum treated with ammonium to differentiate the biological pathways involved in the response to ammonium vs. the algicide.
Here, we provide evidence for a global response in K. veneficum, including changes in gene expression that are related to molecular, physiological and morphological features of the cell (Figure 5). Algicide-regulated DEGs (highly differentially expressed genes) supporting previously reported physiological effects of the algicide were also identified (Pokrzywinski et al., 2017b), including those DEGs involved in stress and ROS response and DNA damage response, as well as DEGs related to PCD. Additionally, results of this analysis suggest the activity of photobiological recovery mechanisms in K. veneficum exposed to IRI-160AA, including an enhancement of chlorophyll biosynthesis and up-regulated photorepair DEGs.
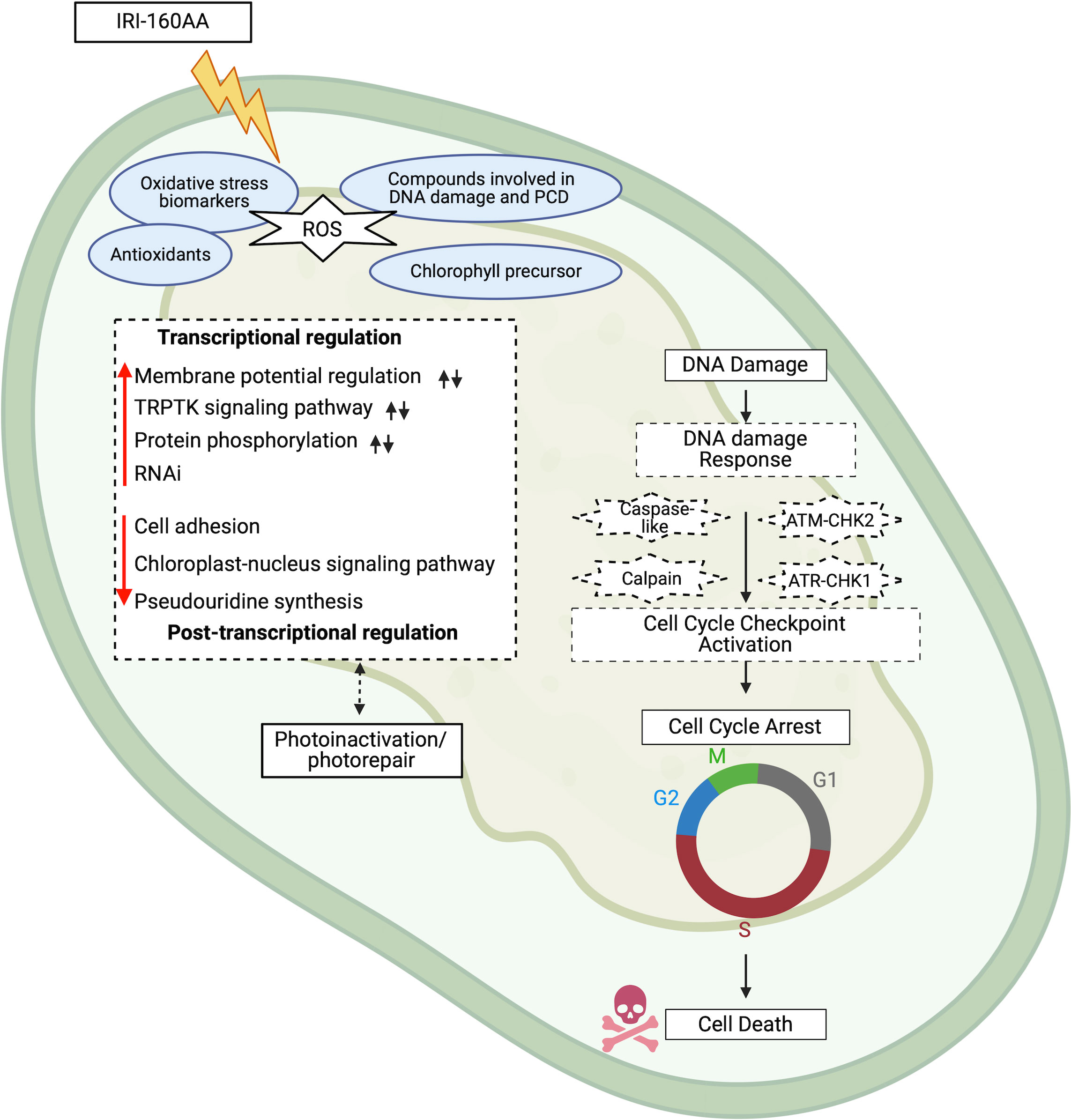
Figure 5 Schematic representation of the cellular impact of IRI-160AA on Karlodinium veneficum. Putative processes revealed by the transcriptome data are portrayed in dashed boxes (Figure 3). Red arrows indicate processes that were stimulated (upward pointing arrows) or suppressed (downward pointing arrows) by the algicide exposure. Black arrows indicate the regulation by ammonium. Processes in solid boxes are those demonstrated by previous physiological data (Tilney et al., 2014; Pokrzywinski et al., 2017a; Pokrzywinski et al., 2017b); genes with putative functions in these processes were also identified and discussed further in the text. Evidence for a global response to the algicide includes impacts on various biological processes at the transcriptional, post-transcriptional, and post-translational levels as supported by the transcriptome data. IRI-160AA stimulated ROS production, inducing the formation of biomarkers for oxidative stress, antioxidants, other compounds involved in DNA damage and programmed cell death, as well as the chlorophyll precursor in the cell pellets of the algicide treatment of K. veneficum (Wang and Coyne, 2022). Results also point to photoinactivation and accompanied photorepair in cells exposed to IRI-160AA. There was also evidence for DNA damage and DNA damage response (DDR), as well as cell cycle checkpoint activation. Support for a genetically programmed pathway to cell death involving the activation of caspase-like and calpain enzymes, as well as the ATM-CHK2 and ATR-CHK1 signaling pathways involved in cell cycle progression is provided in the data. Cell cycles are monitored by cell cycle checkpoints (G1/S, intra-S phase, G2/M, and spindle checkpoints), which can prevent cell cycle progression when DNA damage is detected, leading to DNA repair and resultant cell survival or cell death. ROS, reactive oxygen species; TRPTK, transmembrane receptor protein tyrosine kinase; RNAi, RNA interference. The figure was created with BioRender.com.
4.1 Transcriptomic analysis overview
The number of genes in the de novo assembly of K. veneficum in this research (160,206) was similar to the number of assembled genes from other dinoflagellates, such as Cochlodinium polykrikoides [191,212 genes; (Guo et al., 2016b)] and Karenia mikimotoi [202,600 genes; (Wang et al., 2019)]. Validation of the transcriptome data using RT-qPCR analysis of selected transcripts demonstrated a significant correlation with gene expression (p=0.00018, R=0.9; Supplementary Figure 1). As reviewed by Akbar et al. (2018), a challenge for studies of gene expression in dinoflagellates is the low proportion of genes that can be reliably annotated; for instance, only 28% of all genes were annotated in Alexandrium minutum in a study conducted by Yang et al. (2010). Transcriptome analysis of K. veneficum presented the same challenge, as only 47% of assembled genes were annotated in this study.
The low percentage of DEGs in the assembled genes (4.92%) in this study was also consistent with previous research suggesting that regulation of gene expression may play a reduced role in dinoflagellates, which primarily rely on post-transcriptional regulation (Moustafa et al., 2010; Guo et al., 2016a; Guo et al., 2016b; Riaz et al., 2018; Wang et al., 2019). For instance, Wang et al. (2019) demonstrated only 0.36% of total genes were differentially expressed in K. mikimotoi in responce to UV radiation.
4.2 Gene ontology (GO) enrichment analysis
Among the biological processes affected by ammonium and/or the algicide, only a few (17%) were shared by these treatments, while the majority of these processes (83%) were uniquely regulated by either ammonium treatment (61%) or by the algicide (22%) (Figure 3). The differential effects of IRI-160AA and ammonium on K. veneficum observed in the GO analysis were consistent with the differential impacts revealed by the bioassay (Figure 1), as well as previous research demonstrating the involvement of other compounds in addition to ammonium in the algicidal activity of IRI-160AA (Grasso, 2018; Ternon et al., 2019). For instance, Grasso (2018) demonstrated IRI-160AA had a significant impact on several photochemical parameters, including the maximum quantum yield of PSII (Fv/Fm), photochemical connectivity between PSII reaction centers (ρ), and primary quinone re-oxidation rate (τ) of dinoflagellate species, while these parameters were not significantly affected by ammonium alone. Ternon et al. (2019) demonstrated a synergistic effect of ammonium and n-butylamine, one of the compounds identified in IRI-160AA, that was greater than the effects by each of the individual compounds.
The biological processes enriched by the algicide-regulated DEGs revealed a global response in K. veneficum exposed to IRI-160AA (Figures 2, 3). This global response was evident by transcriptional regulation affecting cellular activities at several levels of organization, from gene expression (e.g., pseudouridine synthesis and RNAi [RNA interfering]), to protein activities (e.g., phosphorylation), to changes affecting morphology (e.g., cell adhesion). These essential biological processes and their implications in K. veneficum’s response to IRI-160AA are discussed below.
The down-regulation of pseudouridine synthesis observed in K. veneficum in response to the exposure to IRI-160AA provides an indication of early impacts on gene expression (Figure 3). Pseudouridine synthesis (pseudouridylation) is an RNA modification (RNA editing) process converting uridine within RNA to pseudouridine (Ψ) (Adachi et al., 2019). Pseudouridylation is among the most abundant RNA modification types (Karijolich et al., 2010; Adachi et al., 2019). Pseudouridylation has been extensively identified on rRNA and spliceosomal snRNAs (small nuclear RNAs); it plays critical roles in cellular processes such as gene expression modulation, protein translation, and mRNA splicing (Adachi et al., 2019). Studies also demonstrated its importance in cell viability and survival (Wu et al., 2016; Antonicka et al., 2017). For instance, inhibition of pseudouridylation on mitochondrial rRNAs led to defects in mitochondrial ribosome assembly, as well as reduced protein synthesis and cell viability in mammalian cell lines (Antonicka et al., 2017); inhibition of pseudouridylation in yeast snRNAs caused a lower pre-mRNA splicing efficiency and led to temperature-sensitive growth inhibition (Wu et al., 2016).
The down-regulated pseudouridine synthesis observed here implies pseudouridylation may play a similar role in regulating gene expression in this species (Figure 3). Spliced leader (SL) RNA trans-splicing is conserved machinery in dinoflagellates, in which a conserved dinoflagellate-specific 22-nucleotide RNA fragment (Dino SL) is spliced onto the 5’ end of mRNA transcripts (Zhang et al., 2007; Song et al., 2018). Though no research has been conducted on the pseudouridylation of Dino SL, studies have revealed pseudouridylation is involved in SL RNA biogenesis in trypanosomes (Michaeli, 2011), where the disruption of pseudouridylation resulted in SL RNA defects and trans-splicing inhibition (Barth et al., 2005). The involvement of pseudouridine synthesis in dinoflagellates, as well as its potential unique role in these species with regard to their conserved Dino SL, remain unknown. Recent research has identified pseudouridine synthases were among the most highly regulated genes in dinoflagellates in response to changes in growth environment, such as glucose supplementation to Symbiodinium (Xiang et al., 2018), as well as life stage transitions, such as a transition from vegetative cells to cysts in Yihiella yeosuensis (Jang et al., 2019).
An RNAi-related process in K. veneficum was also affected by exposure to the algicide (Figure 3). RNAi is an endogenous cellular mechanism widely shared by eukaryotes involving small RNAs, which leads to targeted RNA degradation and transcriptional or post-transcriptional gene silencing (Cerutti et al., 2011; Chang et al., 2012; Heigwer et al., 2018; Rosa et al., 2018). It has been demonstrated that RNAi plays a crucial role in plants’ response to stress and pathogen resistance (Rosa et al., 2018; Zotti et al., 2018). Here, the up-regulated RNAi-related process provides evidence that this mechanism could be involved in the response by K. veneficum to the algicide. This is in line with the presence of the potential RNAi system in dinoflagellates, including K. veneficum, revealed by recent transcriptomics and genomics studies (Zhang and Lin, 2019). The potential role of RNAi machinery in modulating algae-bacteria interactions, including dinoflagellate-bacteria relationships, requires further research.
Processes related to protein phosphorylation were stimulated in the algicide treatment, supporting previous reports that post-transcriptional regulations play key roles in dinoflagellates (Riaz et al., 2018) (Figure 3). Protein phosphorylation is fundamentally important for organisms in response to intra- and extracellular stimuli (Brumbarova and Ivanov, 2016; Böhm et al., 2019; Demongeot et al., 2019); it is involved in a number of critical processes, including signal transduction, DNA damage repair, and cell cycle regulation (Ardito et al., 2017; Li et al., 2019). In dinoflagellates, various conserved kinases have been identified that play key roles in protein translational regulation (Roy et al., 2018), S and M cell cycle progression (Morse et al., 2016), as well as DNA damage response pathways (Morse et al., 2016; Li and Wong, 2019). As dinoflagellates primarily regulate their proteins at the post-transcriptional and translational levels (Riaz et al., 2018; Roy et al., 2018), protein post-translational modifications such as phosphorylation may play an essential role in the response of these species to the algicide exposure.
Cell adhesion plays important roles in stress response, cell communication, signal transduction, cell differentiation, cell migration, and cell cycle progression, as well as programmed cell death in animals (Khalili and Ahmad, 2015; Vlahakis and Debnath, 2017). Reduced cell adhesion has been linked to changes in cell membrane morphology, including plasma membrane blebbing (Fackler and Grosse, 2008). Recent research identified genes encoding cell adhesion proteins in symbiotic dinoflagellates Symbiodinium (Xiang et al., 2015) and Durusdinium (Poquita-Du et al., 2020). These cell adhesion genes were down-regulated by environmental stimuli including light (Xiang et al., 2015), heat (Poquita-Du et al., 2020), and sediment exposure (Poquita-Du et al., 2020). The down-regulation of these genes was correlated with changes to cell surface morphology, accompanied by reduced surface architecture in Symbiodinium (Xiang et al., 2015). While K. veneficum does not form symbiotic relationships, down-regulated genes encoding proteins involved in cell adhesion in this species after exposure to IRI-160AA (Figure 3) may be related to plasma membrane blebbing observed in dinoflagellates exposed to the algicide as previously reported (Pokrzywinski et al., 2017a). In addition, cell adhesion also plays a role in regulating apoptosis in animals (Khalili and Ahmad, 2015; Vlahakis and Debnath, 2017). The suppression of transcripts encoding cell adhesion proteins accompanied by an up-regulation of apoptosis-related genes reported here (Table 3; discussed below), as well as for the dinoflagellate symbiont Durusdinium (Poquita-Du et al., 2020) provides some evidence that this regulatory role is shared with other organisms.
4.3 Linking transcriptomics to previously reported physiological data
Previous research indicated IRI-160AA induced ROS (specifically H2O2) production, DNA damage, and cell cycle arrest, as well as other markers of PCD in dinoflagellates (Pokrzywinski et al., 2017b), accompanied by impaired photosynthetic efficiency and PSII inhibition (Tilney et al., 2014). Recent metabolomics analysis also demonstrated the enrichment of a suite of compounds in K. veneficum by the algicide IRI-160AA, including oxidative stress biomarkers, antioxidants, compounds involved in DNA damage and PCD, as well as the chlorophyll precursor 5-aminolevulinic acid (Figure 5) (Wang and Coyne, 2022). An intertwined relationship between these markers and PCD, as well as photosynthesis, has also been noted in plants (Yoshiyama et al., 2013; Mhamdi and Van Breusegem, 2018) and other algal species (Rezayian et al., 2019; Zhao et al., 2019; Zhao et al., 2020). Below, key DEGs related to these processes are identified (Tables 2–4), and their putative functionality related to previously reported physiological data will be discussed.
4.3.1 Stress and ROS responsive genes
Cyclophilin proteins (CyPs) are conserved proteins among both prokaryotic and eukaryotic organisms (Jandova et al., 2013; Ponmani et al., 2015). They are essential proteins responding to environmental stresses (Jandova et al., 2013; Jia et al., 2013; Ponmani et al., 2015; Yau et al., 2016; Abassi et al., 2017) and stress-induced ROS in animals, plants, and algae (Wu et al., 2009; Jandova et al., 2013; Dos Santos and Park, 2019), including the dinoflagellate Prorocentrum minimum (Ponmani et al., 2015). Here, the up-regulation of the CyP gene encoding the cyclophilin 40 protein (CyP40; peptidylprolyl isomerase D; Table 2) in algicide-treated K. veneficum was consistent with previous observations of ROS production in this and other dinoflagellates exposed to IRI-160AA (Pokrzywinski et al., 2017b). Jandova et al. (2013) demonstrated the cytosolic CyP40 may play a role in regulating the mitochondrial membrane potential (MMP), the mitochondrial permeability transition pore (MPTP), as well as key proteins involved in MPTP regulation, including the mitochondrial CyP protein (CyPD). The transient opening of MPTP can trigger the release of cytochrome c and subsequent apoptosis (Jandova et al., 2013; Wan et al., 2019). The knockdown of the CyP40 gene in UVA-stressed human cells led to apoptosis prevention, accompanied by reduced MMP dissipation and MPTP opening; the mitochondrial superoxide levels were also lower in the CyP40 knockdown cells (Jandova et al., 2013).
While CyP40 was up-regulated after exposure to both ammonium and the algicide, genes that encode vital protective proteins against oxidative stress, including PPR40 (mitochondrial pentatricopeptide repeat [PPR] domain protein) and MRP (multidrug resistance-associated protein), were suppressed only in the algicide treatment (Table 2). PPR domain proteins play an essential role in the stress tolerance of plants (Zsigmond et al., 2008). For instance, Arabidopsis with an inactivated PPR40 protein exhibited a semi-dwarf growth phenotype and a sensitivity to stresses, including salt, plant hormone, and oxidative stress, accompanied by cellular ROS accumulation (Zsigmond et al., 2008). MRPs are membrane proteins in the family of ATP-binding cassette (ABC) transporters that play important protective roles in stress tolerance in animals (Takahashi et al., 2012; Granitzer et al., 2020), plants (Park et al., 2012), and algae (Hou et al., 2016; Gu et al., 2019). Overall, suppression of genes for proteins that offer protection against oxidative stress (PPR40 and MRP) provide evidence for underlying mechanisms that may be involved in increased ROS reported for dinoflagellate cultures exposed to IRI-160AA (Pokrzywinski et al., 2017b; Figure 5).
Exposure to the algicide also resulted in a decrease in the expression of hydroxyacid oxidases (HAOs; aka glycolate oxidase, GOXs); this gene was not differentially regulated in the ammonium treatment (Table 2). HAOs, located in peroxisomes, are key enzymes involved in photorespiration in plants and are a part of the plants’ defensive system against biotic and abiotic stresses (Dellero et al., 2016). HAOs are essential for plants’ survival and photosynthesis even under normal growth conditions (Xu et al., 2009; Zelitch et al., 2009; Rojas and Mysore, 2012; Rojas et al., 2012; Jacobo-Velázquez et al., 2015; Cui et al., 2016; Xu et al., 2018). Rice with suppressed HAO genes exhibited impaired growth and net photosynthetic rates, accompanied by PSII inactivation (Xu et al., 2009). Chern et al. (2013) also reported resultant PCD in rice with silenced HAOs.
Despite their roles in improving photooxidative stress tolerance in plants (Cui et al., 2016), HAOs are also major ROS producers, in that H2O2 is one of the products during the catalytic process (Cui et al., 2016; Dellero et al., 2016; Rademacher et al., 2016). Recalcati et al. (2003) demonstrated a decrease in HAO gene expression in rats under oxidative stress. They noted this could be a mechanism for the organism to prevent the formation of H2O2 and its further cell damage impact. The down-regulation of the HAO gene in K. veneficum responding to IRI-160AA could imply a similar mechanism; it may play a role in the decreased photosynthetic rates and PSII inactivation observed in Tilney et al. (2014), as well as markers associated with PCD demonstrated in Pokrzywinski et al. (2017b) for dinoflagellates exposed to the algicide.
4.3.2 Genes involved in DNA damage response
DNA damage response (DDR; Figure 5) involves complex signaling pathways (Zannini et al., 2014; Francis et al., 2020) that lead to appropriate responses by the cells, including DNA repair, cell cycle checkpoint activation, cell cycle regulation, and cell death (Patil et al., 2013). In eukaryotic cells, the cell cycle consists of a series of sequential phases, including gap phase 1 (G1), S-phase, gap phase 2 (G2), and mitosis (M) (El-Aouar Filho et al., 2017). Cells only enter the next phase when the previous phase is completed (Patil et al., 2013). The cell cycle progression is monitored by a surveillance mechanism, the cell cycle checkpoints, to ensure faithful cell cycle progression and cell division (Patil et al., 2013). The G1/S and G2/M checkpoints inhibit the entry to the S and M phases of the cells, respectively (Patil et al., 2013). The intra-S phase checkpoint monitors the DNA replication initiation and fork stability; it inhibits the cell cycle progression with arrested forks (Willis and Rhind, 2009; Patil et al., 2013). The spindle checkpoint checks for the alignment and segregation of the chromosomes (Patil et al., 2013).
Double- (DSBs) and single-strand breaks (SSBs) are among the most common DNA damage types; the most notable pathways that sense and repair these damages are the ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3 related) signaling pathways, involving the CHK2 and CHK1 checkpoint kinases, respectively (Patil et al., 2013; Francis et al., 2020). The ATM/ATR pathways activate the cell cycle checkpoints, leading to cell cycle arrest, DNA repair, and/or apoptosis (Patil et al., 2013). The up-regulation of both CHK1 and CHK2 genes in K. veneficum exposed to IRI-160AA (Table 3, Figure 5) was consistent with previous observations of DNA damage, cell cycle arrest, and markers associated with PCD in dinoflagellates exposed to the algicide (Pokrzywinski et al., 2017a; Pokrzywinski et al., 2017b). Treatment with ammonium alone resulted in an up-regulation of CHK1 but not CHK2, suggesting the involvement of other compounds present in IRI-160AA in the regulation of checkpoint kinases. Other key genes involved in these pathways have also been identified in K. veneficum in this study (e.g., histone H2AX, replication protein A [RPA], ATM, ATR, and RAD family proteins; not shown) but were not regulated transcriptionally. These genes have also been identified in other dinoflagellates (Li and Wong, 2019), providing evidence for a regulatory role of these pathways in the DDR of dinoflagellates. In addition, the up-regulation of genes encoding fork-repair and checkpoint interacting proteins involved in DNA damage response, including the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein (SMARCAL1) (Bansbach et al., 2009; Postow et al., 2009; Couch et al., 2013; Keka et al., 2015; Poole et al., 2015) and E3 ubiquitin-protein ligase 2 (HERC2) (Izawa et al., 2011; Mohiuddin et al., 2016) observed here was also consistent with the previously indicated DNA damage and S-phase cell cycle arrest reported for dinoflagellates exposed to IRI-160AA (Pokrzywinski et al., 2017a; Pokrzywinski et al., 2017b). These genes were also up-regulated in the ammonium treatment, consistent with the contribution of ammonium in the algicidal effect of IRI-160AA.
4.3.3 Genes encoding caspase and non-caspase proteases involved in PCD
Caspase and/or caspase-like proteases have been used as prominent biomarkers for PCD diagnosis in both prokaryotic and eukaryotic organisms, including bacteria (Lee and Lee, 2018), fungi (Wilkinson and Ramsdale, 2011), animals (Abu-Qare and Abou-Donia, 2001; Rodriguez-Ruiz et al., 2019), plants (Reape and McCabe, 2010; Kabbage et al., 2017), and algae (Johnson et al., 2014; Bidle, 2016; Sun et al., 2020), including dinoflagellates (Johnson et al., 2014; Pokrzywinski et al., 2017b). For instance, Johnson et al. (2014) demonstrated the involvement of a PCD-like pathway in the ROS-mediated cell death of dinoflagellate Karenia brevis, accompanied by induced caspase-like activities and DNA damage. Cathepsin B proteases are caspase-3-like enzymes that initiate and regulate apoptosis in animals (de Castro et al., 2016; Sendler et al., 2016). Cathepsin B-like cysteine proteinases (CathBs) were also identified in plants; their role in PCD induction has been demonstrated (McLellan et al., 2009; Ge et al., 2016; Cai et al., 2018). The up-regulation of the CathB gene in K. veneficum was consistent with the increased caspase-like activity reported for dinoflagellates exposed to IRI-160AA (Table 3; Figure 5) (Pokrzywinski et al., 2017b), as well as their PCD-inducing roles in other organisms mentioned above. CathB was also up-regulated in the ammonium treatment, indicating the participation of ammonium in the induction of a PCD-like pathway by the algicide.
Calpains are non-caspase proteases that play a role in apoptosis execution in animals (Momeni, 2011). Genes encoding calpains have also been identified in K. veneficum (Lin, 2011) and other dinoflagellates (Johnson et al., 2012; Parkinson et al., 2016; Jang et al., 2019) in previous studies. The induction of the calpain gene after exposure to IRI-160AA, but not in the ammonium treatment, provides additional evidence that it plays a role in dinoflagellate PCD-like processes (Table 3; Figure 5).
4.4 Photobiology
As demonstrated previously, cell death in dinoflagellates exposed to IRI-160AA was accompanied by a change in chloroplast morphology (Pokrzywinski et al., 2017a), as well as inactivation of photosystem II, and a disruption of the electron transport chain in dinoflagellates (Tilney et al., 2014). In photosynthetic organisms, rapid and proper assembly of the photosynthetic apparatus is essential for the repair and recovery from damages (Komenda et al., 2012; Lu, 2016), and chlorophyll biosynthesis is critical in this process (Masuda et al., 2002; Sobotka et al., 2005; Komenda et al., 2012). For instance, chlorophyll biosynthesis was required for the assembly of the light-harvesting complex II (LHC-II) and PSI in the green alga Dunaliella salina during recovery from photodamage (Masuda et al., 2002) and was essential for the PSII assembly in cyanobacteria (Sobotka et al., 2005; Komenda et al., 2012). As discussed below, algicide-regulated DEGs involved in the chlorophyll biosynthesis reveal an up-regulation of this process in K. veneficum exposed to IRI-160AA (Table 4; Figure 4). DEGs indicating an impact on the photosystem of K. veneficum by IRI-160AA and accompanied photorepair will also be discussed.
4.4.1 Chlorophyll biosynthesis
The plant hormone 5-aminolevulinic acid (ALA) is an essential growth regulator that can enhance plants’ tolerance to environmental stresses (Brzezowski et al., 2019; Wu et al., 2019). ALA is also the precursor of all tetrapyrroles, including chlorophylls, in plants and algae (Brzezowski et al., 2019; Wu et al., 2019). Chlorophyll biosynthesis is part of a branched, multienzyme pathway shared by hemes and bile pigments (Figure 4) (Brzezowski et al., 2019). ALA is synthesized by a series of sequential steps from glutamate, where glutamate-1-semialdehyde 2,1-aminomutase (GSAT) catalyzes the last step of ALA biosynthesis (Brzezowski et al., 2019). ALA is then converted to downstream metabolites, catalyzed by multiple enzymes, including the oxygen-dependent coproporphyrinogen-III oxidase (HEMF) (Wu et al., 2019). HEMF converts coproporphyrinogen III to protoporphyrinogen IX, which is then converted to protoporphyrin IX (Proto IX) (Wu et al., 2019). Proto IX is the branching point leading to heme and chlorophyll biosynthesis (Brzezowski et al., 2019). In the chlorophyll biosynthesis branch, Proto IX is converted to chlorophylls through a series of sequential enzymatic steps and their respective intermediates; the light-dependent protochlorophyllide reductase (LPOR) catalyzes one of the rate-limiting steps in this process, converting protochlorophyllide (Pchlide) to chlorophyllide (Chlide), which is subsequently converted to chlorophylls (reviewed by Zhang et al., 2018b).
Previous metabolomics study (Wang and Coyne, 2022) revealed that ALA increased in the cell pellets and decreased in the cell filtrate of K. veneficum exposed to the algicide IRI-160AA compared to control cultures, suggesting that IRI-160AA may have stimulated the uptake of ALA from surrounding medium (Figure 4). This was consistent with the stimulating effects of the exogenous application of ALA on its intracellular accumulation observed in plants (Liu et al., 2016). The increase of ALA in K. veneficum cell pellets likely induced an up-regulation of the subsequent enzymatic processes, which was evident by an up-regulation of the HEMF and LPOR genes in the algicide treated K. veneficum, as shown in the transcriptomics data (Table 4; Figure 4). This was supported by other studies that demonstrated the exogenous treatment of ALA enhanced the chlorophyll biosynthesis pathway and increased concentrations of intermediate products in this pathway (Aarti et al., 2006; Stenbaek et al., 2008; Wu et al., 2018). The stimulated ALA uptake (Wang and Coyne, 2022) and subsequent chlorophyll biosynthesis (Figure 4) could be a defense response for K. veneficum to recover from the disruptions of PSII and the electron transport chain due to the exposure to IRI-160AA (Tilney et al., 2014). Additionally, while LPOR was up-regulated by both algicide and ammonium, HEMF was only up-regulated in the algicide treatment, indicating ammonium may have played a role in the chlorophyll biosynthesis involved defense response of K. veneficum, but in a limited manner. This was also evident by the down-regulation of the PAPP5 gene encoding the serine/threonine-protein phosphatase 5 observed here only in the algicide treatment (Table 4). PAPP5 has been demonstrated as a negative regulator of chlorophyll biosynthesis and was involved in the signaling transduction from chloroplast to nucleus (Barajas-López et al., 2013).
Additionally, the down-regulation of the GSAT gene in the algicide treatment of K. veneficum observed in this study could imply a feedback inhibition by ALA on GSAT biosynthesis and reflect a mechanism to avoid the accumulation of downstream intermediates (Table 4; Figure 4). This gene was differentially regulated by the algicide but not in the ammonium treatment, consistent with the limited contribution of ammonium to the algicide-stimulated chlorophyll biosynthesis pathway as discussed above. Chlorophyll biosynthesis intermediates, including Pchlide and its tetrapyrrole precursors (e.g., Proto IX, Mg-protoporphyrin IX [Mg-Proto IX], and Mg-protoporphyrin IX monomethyl ester [MPE]), are potently phototoxic (Stenbaek et al., 2008; Gabruk and Mysliwa-Kurdziel, 2015); the feedback inhibition of chlorophyll biosynthesis intermediates on ALA biosynthesis has also been demonstrated by other studies (Stenbaek et al., 2008; Gabruk and Mysliwa-Kurdziel, 2015).
4.4.2 Photosystem impacts and photorepair
In addition to the rapid impacts on photobiology induced by IRI-160AA, Tilney et al. (2014) also observed a light-dependent recovery of the PSII inactivation in K. veneficum exposed to IRI-160AA. The Fv/Fm only recovered in this species under the regular light to dark cycle after the exposure to IRI-160AA but not under continuous dark, implying the involvement of a light-dependent mechanism in the repair of the photosynthetic machinery in this species (Tilney et al., 2014). Concurrent with the recovery of Fv/Fm, however, cell density of this dinoflagellate remained low, implying a defensive response specifically related to photosynthesis function repair in the surviving cells (Tilney et al., 2014).
Although transcript levels in this study reflected short-term impacts on K. veneficum, there was evidence of up-regulation of repair mechanisms for photosynthetic machinery besides the enhanced chlorophyll biosynthesis discussed above. Among those up-regulated in the algicide treatment were a PETD gene encoding the cytochrome b6-f complex subunit IV, as well as UVR8 gene encoding the ultraviolet-B receptor UVR8, and two genes encoding photolyases (Table 4). All of these genes except PETD were also up-regulated in the ammonium treatment, implying the contribution of ammonium to the photosynthetic machinery repair involved in the algicidal effect of IRI-160AA.
PETD is among the proteins with the fastest turn-over rates in plant leaves (Li et al., 2018) and is fundamental for the assembly of the cytochrome b6-f complex (Mirzaei et al., 2018). Li et al. (2017) suggested that PETD proteins could be involved in repairing damage related to electron transport pathways. UVR8, which was differentially expressed in both algicide and ammonium treatments, is involved in the protective responses of plants and plays a role in directly or indirectly maintaining photosynthesis under adverse conditions (Singh et al., 2014). Photolyases, also up-regulated in both treatments, are key enzymes responsible for the light-dependent photoreactivation pathway for DNA damage repair, shown to be a key process to restore the damaged PSII in cyanobacteria (Vass et al., 2013). Recent efforts have identified multiple genes encoding photolyases in dinoflagellate species, implying their potential role in the DNA lesion repair and photosynthesis recovery in these organisms (Shoguchi et al., 2018; Li and Wong, 2019).
Overall, results of this research, together with the metabolomics (Wang and Coyne, 2022) and photobiology (Tilney et al., 2014) data presented previously, demonstrated the photoinhibition effect of IRI-160AA may be accompanied by an activation of defense mechanisms related to photosynthesis impairment, as well as a light-dependent DNA repair mechanism resulting in the eventual restoration of the photosynthetic apparatuses. Results of this study indicate that ammonium may have contributed to this defense mechanism, but in a limited manner.
5 Conclusion
Results of this research demonstrated a distinct impact of ammonium and IRI-160AA on K. veneficum at the transcriptional level. Differentially expressed genes (DEGs) in the ammonium and the algicide treatments only shared 17% enriched biological processes (membrane potential regulation, transmembrane receptor protein tyrosine kinase signaling pathway, and protein phosphorylation), while the vast majority (83%) of the enriched processes were only regulated by ammonium or the algicide alone. Besides those shared with the ammonium treatment, exposure to IRI-160AA also up-regulated a process involved in RNA interference. Processes including pseudouridine synthesis, cell adhesion, and chloroplast-nucleus signaling pathway were down-regulated by the algicide. It is likely that post-transcriptional regulations also participated in the response of K. veneficum exposed to the algicide. DEGs involved in ROS and stress response, DNA damage response (DDR), cell cycle checkpoint activation, and PCD were also identified in K. veneficum treated with the algicide. Results of this research also revealed a photorepair mechanism involving enhanced chlorophyll biosynthesis and light-dependent DNA damage repair in K. veneficum treated with the algicide.
Overall, this research provided insights into the algicidal effect of IRI-160AA on K.veneficum at the transcriptomic level and expanded our knowledge of the molecular response of dinoflagellates to bioactive substances produced by bacteria. Future research may focus on investigating the specific roles of the essential genes identified in this research in dinoflagellate species responding to other biotic and abiotic stresses.
Data availability statement
The data presented in the study are deposited in the NCBI- Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) repository, accession number PRJNA922626.
Author contributions
Conceptualization, YW and KC. Methodology, YW and KC. Software, YW. Validation, YW and KC. Formal analysis, YW. Investigation, YW. Resources, KC. Data curation, YW. Writing—original draft preparation, YW. Writing—review and editing, YW and KC. Visualization, YW. Supervision, KC. Project administration, KC. Funding acquisition, YW and KC. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the National Oceanic and Atmospheric Association (NOAA) Prevention, Control and Mitigation of HABs (PCMHAB) program (Grant # NA15NOS4780176 to K.J. Coyne, M.E. Warner, T. Targett, and J. Cohen; Grant # NA20NOS4780185 to K.L. Pokrzywinski, K.J. Coyne, Y. Wang, and A.J. Kennedy. PCMHAB contribution #65).
Acknowledgments
The content of this manuscript previously appeared online as a Ph.D. dissertation by YW from the University of Delaware (Title: Bacterial algicides: application strategies and cellular response of target organisms).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1112913/full#supplementary-material
References
Aarti P. D., Tanaka R., Tanaka A. (2006). Effects of oxidative stress on chlorophyll biosynthesis in cucumber (Cucumis sativus) cotyledons. Physiol. Plant 128, 186–197. doi: 10.1111/j.1399-3054.2006.00720.x
Abassi S., Wang H., Park B. S., Park J. W., Ki J. S. (2017). A novel cyclophilin b gene in the red tide dinoflagellate Cochlodinium polykrikoides: Molecular characterizations and transcriptional responses to environmental stresses. BioMed. Res. Int. 2017. doi: 10.1155/2017/4101580
Abu-Qare A. W., Abou-Donia M. B. (2001). Biomarkers of apoptosis: Release of cytochrome c, activation of caspase-3, induction of 8-hydroxy-2′-deoxyguanosine, increased 3-nitrotyrosine, and alteration of p53 gene. J. Toxicol. Environ. Health B Crit. Rev. 4, 313–332. doi: 10.1080/109374001301419737
Adachi H., De Zoysa M. D., Yu Y. T. (2019). Post-transcriptional pseudouridylation in mRNA as well as in some major types of noncoding RNAs. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 230–239. doi: 10.1016/j.bbagrm.2018.11.002
Akbar M., Ahmad A., Usup G., Bunawan H. (2018). RNA-Seq as an emerging tool for marine dinoflagellate transcriptome analysis: process and challenges. Processes 6, 5. doi: 10.3390/pr6010005
Anderson D. M., Cembella A. D., Hallegraeff G. M. (2012). Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Ann. Rev. Mar. Sci. 4, 143–176. doi: 10.1146/annurev-marine-120308-081121
Antonicka H., Choquet K., Lin Z., Gingras A., Kleinman C. L., Shoubridge E. A. (2017). A pseudouridine synthase module is essential for mitochondrial protein synthesis and cell viability. EMBO Rep. 18, 28–38. doi: 10.15252/embr.201643391
Ardito F., Giuliani M., Perrone D., Troiano G., Muzio L. L. (2017). The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 40, 271–280. doi: 10.3892/ijmm.2017.3036
Bansbach C. E., Bétous R., Lovejoy C. A., Glick G. G., Cortez D. (2009). The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23, 2405–2414. doi: 10.1101/gad.1839909
Barajas-López J. d. D., Kremnev D., Shaikhali J., Piñas-Fernández A., Strand Å. (2013). PAPP5 is involved in the tetrapyrrole mediated plastid signalling during chloroplast development. PloS One 8:e60305. doi: 10.1371/journal.pone.0060305
Barth S., Hury A., Liang X. H., Michaeli S. (2005). Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 280, 34558–34568. doi: 10.1074/jbc.M503465200
Bateman A. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Bi Y., Wang F., Zhang W. (2019). Omics analysis for dinoflagellates biology research. Microorganisms 7, 288. doi: 10.3390/microorganisms7090288
Bidle K. D. (2016). Programmed cell death in unicellular phytoplankton. Curr. Biol. 26, R594–R607. doi: 10.1016/j.cub.2016.05.056
Böhm M., Boness D., Fantisch E., Erhard H., Frauenholz J., Kowalzyk Z., et al. (2019). Channelrhodopsin-1 phosphorylation changes with phototactic behavior and responds to physiological stimuli in Chlamydomonas. Plant Cell 31, 886–910. doi: 10.1105/tpc.18.00936
Boratyn G. M., Thierry-Mieg J., Thierry-Mieg D., Busby B., Madden T. L. (2019). Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinf. 20, 405. doi: 10.1186/s12859-019-2996-x
Brown G. R., Hem V., Katz K. S., Ovetsky M., Wallin C., Ermolaeva O., et al. (2015). Gene: A gene-centered information resource at NCBI. Nucleic Acids Res. 43, D36–D42. doi: 10.1093/nar/gku1055
Brumbarova T., Ivanov R. (2016). Differential gene expression and protein phosphorylation as factors regulating the state of the Arabidopsis SNX1 protein complexes in response to environmental stimuli. Front. Plant Sci. 7, 1456. doi: 10.3389/fpls.2016.01456
Bryant D. M., Johnson K., DiTommaso T., Tickle T., Couger M. B., Payzin-Dogru D., et al. (2017). A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 18, 762–776. doi: 10.1016/j.celrep.2016.12.063
Brzezowski P., Ksas B., Havaux M., Grimm B., Chazaux M., Peltier G., et al. (2019). The function of PROTOPORPHYRINOGEN IX OXIDASE in chlorophyll biosynthesis requires oxidised plastoquinone in Chlamydomonas reinhardtii. Commun. Biol. 2, 1–9. doi: 10.1038/s42003-019-0395-5
Cai Y.-M. M., Yu J., Ge Y., Mironov A., Gallois P. (2018). Two proteases with caspase-3-like activity, cathepsin b and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis. New Phytol. 218, 1143–1155. doi: 10.1111/nph.14676
Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Cerutti H., Ma X., Msanne J., Repas T. (2011). RNA-Mediated silencing in algae: Biological roles and tools for analysis of gene function. Eukaryot Cell 10, 1164–1172. doi: 10.1128/EC.05106-11
Chang S. S., Zhang Z., Liu Y. (2012). RNA Interference pathways in fungi: Mechanisms and functions. Annu. Rev. Microbiol. 66, 305–323. doi: 10.1146/annurev-micro-092611-150138
Chern M., Bai W., Chen X., Canlas P. E., Ronald P. C. (2013). Reduced expression of glycolate oxidase leads to enhanced disease resistance in rice. PeerJ 2013, e28. doi: 10.7717/peerj.28
Collos Y., Harrison P. J. (2014). Acclimation and toxicity of high ammonium concentrations to unicellular algae. Mar. pollut. Bull. 80, 8–23. doi: 10.1016/j.marpolbul.2014.01.006
Couch F. B., Bansbach C. E., Driscoll R., Luzwick J. W., Glick G. G., Bétous R., et al. (2013). ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623. doi: 10.1101/gad.214080.113
Coyne K. J., Wang Y., Johnson G. (2022). Algicidal bacteria: A review of current knowledge and applications to control harmful algal blooms. Front. Microbiol. 13, 871177. doi: 10.3389/fmicb.2022.871177
Cruz-López R., Maske H. (2016). The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front. Microbiol. 7, 560. doi: 10.3389/fmicb.2016.00560
Cruz-López R., Maske H., Yarimizu K., Holland N. A. (2018). The b-vitamin mutualism between the dinoflagellate Lingulodinium polyedrum and the bacterium Dinoroseobacter shibae. Front. Mar. Sci. 5:274. doi: 10.3389/fmars.2018.00274
Cui L. L., Lu Y. S., Li Y., Yang C., Peng X. X. (2016). Overexpression of glycolate oxidase confers improved photosynthesis under high light and high temperature in rice. Front. Plant Sci. 7:1165. doi: 10.3389/fpls.2016.01165
de Castro M., Bunt G., Wouters F. (2016). Cathepsin b launches an apoptotic exit effort upon cell death-associated disruption of lysosomes. Cell Death Discovery 2, 1–8. doi: 10.1038/cddiscovery.2016.12
Dellero Y., Jossier M., Schmitz J., Maurino V. G., Hodges M. (2016). Photorespiratory glycolate-glyoxylate metabolism. J. Exp. Bot. 67, 3041–3052. doi: 10.1093/jxb/erw090
Demongeot J., Hasgui H., Thellier M. (2019). Memory in plants: Boolean modeling of the learning and store/recall memory functions in response to environmental stimuli. J. Theor. Biol. 467, 123–133. doi: 10.1016/j.jtbi.2019.01.019
Dos Santos I. B., Park S. W. (2019). Versatility of cyclophilins in plant growth and survival: A case study in Arabidopsis. Biomolecules 9, 20. doi: 10.3390/biom9010020
Dungca-Santos J. C. R., Caspe F. J. O., Tablizo F. A., Purganan D. J. E., Azanza R. V., Onda D. F. L. (2019). Algicidal potential of cultivable bacteria from pelagic waters against the toxic dinoflagellate Pyrodinium bahamense (Dinophyceae). J. Appl. Phycol 31, 3721–3735. doi: 10.1007/s10811-019-01839-0
El-Aouar Filho R. A., Nicolas A., De Paula Castro T. L., Deplanche M., De Carvalho Azevedo V. A., Goossens P. L., et al. (2017). Heterogeneous family of cyclomodulins: Smart weapons that allow bacteria to hijack the eukaryotic cell cycle and promote infections. Front. Cell Infect. Microbiol. 7:208. doi: 10.3389/fcimb.2017.00208
Fackler O. T., Grosse R. (2008). Cell motility through plasma membrane blebbing. J. Cell Biol. 181, 879–884. doi: 10.1083/jcb.200802081
Farhat A., Elleuch J., ben Amor F., Barkallah M., Smith K. F., ben Neila I., et al. (2022). A fast and accurate method for specific detection and quantification of the bloom-forming microalgae Karlodinium veneficum in the marine environment. Environ. Sci. pollut. Res. 1, 1–11. doi: 10.1007/S11356-022-21667-Z/TABLES/4
Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42, 222–230. doi: 10.1093/nar/gkt1223
Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38. doi: 10.1093/nar/gkv397
Francis M., Daher A. A., Azzam P., Mroueh M., Zeidan Y. H. (2020). Modulation of DNA damage response by sphingolipid signaling: An interplay that shapes cell fate. Int. J. Mol. Sci. 21, 1–23. doi: 10.3390/ijms21124481
Fukuda Y., Suzaki T. (2015). “Unusual features of dinokaryon, the enigmatic nucleus of dinoflagellates,” in Marine protists: Diversity and dynamics (Tokyo: Springer), 23–45. doi: 10.1007/978-4-431-55130-0_2
Gabruk M., Mysliwa-Kurdziel B. (2015). Light-dependent protochlorophyllide oxidoreductase: Phylogeny, regulation, and catalytic properties. Biochemistry 54, 5255–5262. doi: 10.1021/acs.biochem.5b00704
Ge Y., Cai Y. M., Bonneau L., Rotari V., Danon A., McKenzie E. A., et al. (2016). Inhibition of cathepsin b by caspase-3 inhibitors blocks programmed cell death in Arabidopsis. Cell Death Differ 23, 1493–1501. doi: 10.1038/cdd.2016.34
Geniza M., Jaiswal P. (2017). Tools for building de novo transcriptome assembly. Curr. Plant Biol. 11–12, 41–45. doi: 10.1016/j.cpb.2017.12.004
Gornik S. G., Hu I., Lassadi I., Waller R. F. (2019). The biochemistry and evolution of the dinoflagellate nucleus. Microorganisms 7, 245. doi: 10.3390/microorganisms7080245
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. (Berlin: Springer) 29, 644–652. doi: 10.1038/nbt.1883
Granéli E., Turner J. T. (2006). “An introduction to harmful algae,” in Ecology of harmful algae (Berlin Heidelberg: Springer), 3–7. doi: 10.1007/978-3-540-32210-8_1
Granitzer S., Ellinger I., Khan R., Gelles K., Widhalm R., Hengstschläger M., et al. (2020). In vitro function and in situ localization of multidrug resistance-associated protein (MRP)1 (ABCC1) suggest a protective role against methyl mercury-induced oxidative stress in the human placenta. Arch. Toxicol. 94, 3799–3817. doi: 10.1007/s00204-020-02900-5
Grasso C. R. (2018). Effects of the bacterial algicide IRI-160AA on the microbial community composition of the Delaware inland bays (Master’s thesis, University of Delaware, DE, USA).
Gu S., Xiao S. W., Zheng J. W., Li H. Y., Liu J. S., Yang W. D. (2019). ABC Transporters in Prorocentrum lima and their expression under different environmental conditions including okadaic acid production. Mar. Drugs 17, 259. doi: 10.3390/md17050259
Guillard R. R. L., Ryther J. H. (1962). Studies of marine planktonic diatoms i. cyclotella nana hustedt, and Detonula confervasea (Cleve). Can. J. Microbiol. 8, 229–239. doi: 10.1139/m62-029
Guo R., Lim W. A., Ki J. S. (2016a). Genome-wide analysis of transcription and photosynthesis inhibition in the harmful dinoflagellate prorocentrum minimum in response to the biocide copper sulfate. Harmful Algae 57, 27–38. doi: 10.1016/j.hal.2016.05.004
Guo R., Wang H., Suh Y. S., Ki J. S. (2016b). Transcriptomic profiles reveal the genome-wide responses of the harmful dinoflagellate cochlodinium polykrikoides when exposed to the algicide copper sulfate. BMC Genomics 17, 29. doi: 10.1186/s12864-015-2341-3
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Hackett J. D., Anderson D. M., Erdner D. L., Bhattacharya D. (2004). Dinoflagellates: A remarkable evolutionary experiment. American Journal of Botany (Wiley-Blackwell), 91(10), 1523–1534. doi: 10.3732/ajb.91.10.1523
Hare C. E., Demir E., Coyne K. J., Craig Cary S., Kirchman D. L., Hutchins D. A. (2005). A bacterium that inhibits the growth of Pfiesteria piscicida and other dinoflagellates. Harmful Algae 4, 221–234. doi: 10.1016/j.hal.2004.03.001
Heberle H., Meirelles V. G., da Silva F. R., Telles G. P., Minghim R. (2015). InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 16, 169. doi: 10.1186/s12859-015-0611-3
Heigwer F., Port F., Boutros M. (2018). RNA Interference (RNAi) screening in Drosophila. Genetics 208, 853–874. doi: 10.1534/genetics.117.300077
Hou D.-y., Liang J.-j., Zou C., Li H.-y., Liu J.-s., Yang W.-d. (2016). MRP functional activity and character in the dinoflagellate Prorocentrum lima. J. Appl. Phycol 28, 1667–1676. doi: 10.1007/s10811-015-0679-1
Huang D. W., Sherman B. T., Lempicki R. A. (2009a). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang D. W., Sherman B. T., Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., et al. (2019). EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Imai I., Kimura S. (2008). Resistance of the fish-killing dinoflagellate Cochlodinium polykrikoides against algicidal bacteria isolated from the coastal sea of Japan. Harmful Algae 7, 360–367. doi: 10.1016/j.hal.2007.12.010
Izawa N., Wu W., Sato K., Nishikawa H., Kato A., Boku N., et al. (2011). HERC2 interacts with claspin and regulates DNA origin firing and replication fork progression. Cancer Res. 71, 5621–5625. doi: 10.1158/0008-5472.CAN-11-0385
Jacobo-Velázquez D. A., González-Aguëro M., Cisneros-Zevallos L. (2015). Cross-talk between signaling pathways: The link between plant secondary metabolite production and wounding stress response. Sci. Rep. 5, 1–10. doi: 10.1038/srep08608
Jandova J., Janda J., Sligh J. E. (2013). Cyclophilin 40 alters UVA-induced apoptosis and mitochondrial ROS generation in keratinocytes. Exp. Cell Res. 319, 750–760. doi: 10.1016/j.yexcr.2012.11.016
Jang S. H., Jeong H. J., Chon J. K. (2019). De novo transcriptome of the newly described phototrophic dinoflagellate Yihiella yeosuensis: comparison between vegetative cells and cysts. Mar. Biol. 166, 104. doi: 10.1007/s00227-019-3554-9
Jia Z., Niu J., Huan L., Wu X., Wang G., Hou Z. (2013). Cyclophilin participates in responding to stress situations in Porphyra haitanensis (Bangiales, rhodophyta). J. Phycol 49, 194–201. doi: 10.1111/j.1529-8817.2012.01234.x
Johnson J. G., Janech M. G., Van Dolah F. M. (2014). Caspase-like activity during aging and cell death in the toxic dinoflagellate Karenia brevis. Harmful Algae 31, 41–53. doi: 10.1016/j.hal.2013.08.005
Johnson J. G., Morey J. S., Neely M. G., Ryan J. C., van Dolah F. M. (2012). Transcriptome remodeling associated with chronological aging in the dinoflagellate, Karenia brevis. Mar. Genomics 5, 15–25. doi: 10.1016/j.margen.2011.08.005
Kabbage M., Kessens R., Bartholomay L. C., Williams B. (2017). The life and death of a plant cell programmed cell death (PCD): a genetically regulated program of cellular suicide employed by both unicellular and multicellular organisms. Annu. Rev. Plant Biol. 68, 375–404. doi: 10.1146/annurev-arplant-043015
Karijolich J., Kantartzis A., Yu Y. T. (2010). RNA Modifications: a mechanism that modulates gene expression. Methods Mol. Biol. 629, 1–19. doi: 10.1007/978-1-60761-657-3_1
Keka I. S., Mohiuddin, Maede Y., Rahman M. M., Sakuma T., Honma M., et al. (2015). SMARCAL1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res. 43, 6359–6372. doi: 10.1093/nar/gkv621
Khalili A. A., Ahmad M. R. (2015). A review of cell adhesion studies for biomedical and biological applications. Int. J. Mol. Sci. 16, 18149–18184. doi: 10.3390/ijms160818149
Komenda J., Sobotka R., Nixon P. J. (2012). Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251. doi: 10.1016/j.pbi.2012.01.017
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee B., Lee D. G. (2018). Depletion of reactive oxygen species induced by chlorogenic acid triggers apoptosis-like death in Escherichia coli. Free Radic. Res. 52, 605–615. doi: 10.1080/10715762.2018.1456658
Lei X., Li D., Li Y., Chen Z., Chen Y., Cai G., et al. (2015). Comprehensive insights into the response of Alexandrium tamarense to algicidal component secreted by a marine bacterium. Front. Microbiol. 6:7. doi: 10.3389/fmicb.2015.00007
Li C., Wong J. T. Y. (2019). DNA Damage response pathways in dinoflagellates. Microorganisms 7, 1–40. doi: 10.3390/microorganisms7070191
Li L., Aro E. M., Millar A. H. (2018). Mechanisms of photodamage and protein turnover in photoinhibition. Trends Plant Sci. 23, 667–676. doi: 10.1016/j.tplants.2018.05.004
Li L., Nelson C. J., Trösch J., Castleden I., Huang S., Millar A. H. (2017). Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29, 207–228. doi: 10.1105/tpc.16.00768
Li W., Wang H. Y., Zhao X., Duan H., Cheng B., Liu Y., et al. (2019). A methylation-phosphorylation switch determines Plk1 kinase activity and function in DNA damage repair. Sci. Adv. 5, eaau7566. doi: 10.1126/sciadv.aau7566
Li Y., Zhu H., Lei X., Zhang H., Guan C., Chen Z., et al. (2015). The first evidence of deinoxanthin from deinococcus sp. Y35 with strong algicidal effect on the toxic dinoflagellate Alexandrium tamarense. J. Hazard Mater 290, 87–95. doi: 10.1016/j.jhazmat.2015.02.070
Lin S. (2011). Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569. doi: 10.1016/j.resmic.2011.04.006
Liu D., Kong D. D., Fu X. K., Ali B., Xu L., Zhou W. J. (2016). Influence of exogenous 5-aminolevulinic acid on chlorophyll synthesis and related gene expression in oilseed rape de-etiolated cotyledons under water-deficit stress. Photosynthetica 54, 468–474. doi: 10.1007/s11099-016-0197-7
Lu Y. (2016). Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front. Plant Sci. 7:168. doi: 10.3389/fpls.2016.00168
Masuda T., Polle J. E. W., Melis A. (2002). Biosynthesis and distribution of chlorophyll among the photosystems during recovery of the green alga Dunaliella salina from irradiance stress. Plant Physiol. 128, 603–614. doi: 10.1104/pp.010595
Matthews J. L., Raina J. B., Kahlke T., Seymour J. R., van Oppen M. J. H., Suggett D. J. (2020). Symbiodiniaceae-bacteria interactions: rethinking metabolite exchange in reef-building corals as multi-partner metabolic networks. Environ. Microbiol. 22, 1675–1687. doi: 10.1111/1462-2920.14918
McLellan H., Gilroy E. M., Yun B.-W. W., Birch P. R. J. J., Loake G. J. (2009). Functional redundancy in the Arabidopsis cathepsin b gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 183, 408–418. doi: 10.1111/j.1469-8137.2009.02865.x
Merico D., Isserlin R., Stueker O., Emili A., Bader G. D. (2010). Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PloS One 5, e13984. doi: 10.1371/journal.pone.0013984
Meyer N., Bigalke A., Kaulfuß A., Pohnert G. (2017). Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 41, 880–899. doi: 10.1093/femsre/fux029
Mhamdi A., Van Breusegem F. (2018). Reactive oxygen species in plant development. Development, 145(15), dev164376.
Michaeli S. (2011). Trans-splicing in trypanosomes: Machinery and its impact on the parasite transcriptome. Future Microbiol. 6, 459–474. doi: 10.2217/fmb.11.20
Mirzaei S., Mansouri M., Mohammadi-Nejad G., Sablok G. (2018). Comparative assessment of chloroplast transcriptional responses highlights conserved and unique patterns across triticeae members under salt stress. Photosynth Res. 136, 357–369. doi: 10.1007/s11120-017-0469-5
Mohiuddin, Kobayashi S., Keka I. S., Guilbaud G., Sale J., Narita T., et al. (2016). The role of HERC2 and RNF8 ubiquitin E3 ligases in the promotion of translesion DNA synthesis in the chicken DT40 cell line. DNA Repair (Amst) 40, 67–76. doi: 10.1016/j.dnarep.2016.02.002
Morse D., Daoust P., Benribague S. (2016). A transcriptome-based perspective of cell cycle regulation in dinoflagellates. Protist 167, 610–621. doi: 10.1016/j.protis.2016.10.002
Moustafa A., Evans A. N., Kulis D. M., Hackett J. D., Erdner D. L., Anderson D. M., et al. (2010). Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PloS One 5, e9688. doi: 10.1371/journal.pone.0009688
Nielsen L. T., Kiørboe T. (2015). Feeding currents facilitate a mixotrophic way of life. ISME J. 9, 2117–2127. doi: 10.1038/ismej.2015.27
Park B. S., Guo R., Lim W.-A. A., Ki J.-S. S. (2018). Importance of free-living and particle-associated bacteria for the growth of the harmful dinoflagellate Prorocentrum minimum: Evidence in culture stages. Mar. Freshw. Res. 69, 290–299. doi: 10.1071/MF17102
Park J., Song W. Y., Ko D., Eom Y., Hansen T. H., Schiller M., et al. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69, 278–288. doi: 10.1111/j.1365-313X.2011.04789.x
Parkinson J. E., Baumgarten S., Michell C. T., Baums I. B., LaJeunesse T. C., Voolstra C. R. (2016). Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 8, 665–680. doi: 10.1093/gbe/evw019
Patil M., Pabla N., Dong Z. (2013). Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell. Mol. Life Sci. 70, 4009–4021. doi: 10.1007/s00018-013-1307-3
Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14(4), 417–419.
Pattanayak G. K., Tripathy B. C. (2011). Overexpression of protochlorophyllide oxidoreductase c regulates oxidative stress in Arabidopsis. PloS One 6, 26532. doi: 10.1371/journal.pone.0026532
Peng J., Place A., Yoshida W., Anklin C., Hamann M. T. (2008). Structure of karlotoxin-2, a toxin causing massive fish kills worldwide. Planta Med. 74, P–65. doi: 10.1055/S-2008-1075261
Pokrzywinski K. L., Place A. R., Warner M. E., Coyne K. J. (2012). Investigation of the algicidal exudate produced by Shewanella sp. IRI-160 and its effect on dinoflagellates. Harmful Algae 19, 23–29. doi: 10.1016/j.hal.2012.05.002
Pokrzywinski K. L., Tilney C. L., Modla S., Caplan J. L., Ross J., Warner M. E., et al. (2017a). Effects of the bacterial algicide IRI-160AA on cellular morphology of harmful dinoflagellates. Harmful Algae 62, 127–135. doi: 10.1016/j.hal.2016.12.004
Pokrzywinski K. L., Tilney C. L., Warner M. E., Coyne K. J. (2017b). Cell cycle arrest and biochemical changes accompanying cell death in harmful dinoflagellates following exposure to bacterial algicide IRI-160AA. Sci. Rep. 7, 45102. doi: 10.1038/srep45102
Ponmani T., Guo R., Ki J. S. (2015). A novel cyclophilin gene from the dinoflagellate Prorocentrum minimum and its possible role in the environmental stress response. Chemosphere 139, 260–267. doi: 10.1016/j.chemosphere.2015.06.036
Poole L. A., Zhao R., Glick G. G., Lovejoy C. A., Eischen C. M., Cortez D. (2015). SMARCAL1 maintains telomere integrity during DNA replication. Proc. Natl. Acad. Sci. U.S.A. 112, 14864–14869. doi: 10.1073/pnas.1510750112
Poquita-Du R. C., Huang D., Chou L. M., Todd P. A. (2020). The contribution of stress-tolerant endosymbiotic dinoflagellate Durusdinium to Pocillopora acuta survival in a highly urbanized reef system. Coral Reefs 39, 745–755. doi: 10.1007/s00338-020-01902-0
Postow L., Woo E. M., Chait B. T., Funabiki H. (2009). Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 284, 35951–35961. doi: 10.1074/jbc.M109.048330
Rademacher N., Kern R., Fujiwara T., Mettler-Altmann T., Miyagishima S. Y., Hagemann M., et al. (2016). Photorespiratory glycolate oxidase is essential for the survival of the red alga Cyanidioschyzon merolae under ambient CO2 conditions. J. Exp. Bot. 67, 3165–3175. doi: 10.1093/jxb/erw118
Reape T. J., McCabe P. F. (2010). Apoptotic-like regulation of programmed cell death in plants. Apoptosis 15, 249–256. doi: 10.1007/s10495-009-0447-2
Recalcati S., Tacchini L., Alberghini A., Conte D., Cairo G. (2003). Oxidative stress-mediated down-regulation of rat hydroxyacid oxidase 1, a liver-specific peroxisomal enzyme. Hepatology 38, 1159–1166. doi: 10.1053/jhep.2003.50417
Rezayian M., Niknam V., Ebrahimzadeh H. (2019). Oxidative damage and antioxidative system in algae. Toxicol. Rep. 6, 1309–1313. doi: 10.1016/j.toxrep.2019.10.001
Riaz S., Sui Z., Niaz Z., Khan S., Liu Y., Liu H. (2018). Distinctive nuclear features of dinoflagellates with a particular focus on histone and histone-replacement proteins. Microorganisms 6, 128. doi: 10.3390/microorganisms6040128
Robinson M. D., McCarthy D. J., Smyth G. K. (2009). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodriguez-Ruiz M. E., Buqué A., Hensler M., Chen J., Bloy N., Petroni G., et al. (2019). Apoptotic caspases inhibit abscopal responses to radiation and identify a new prognostic biomarker for breast cancer patients. Oncoimmunology 8, e1655964. doi: 10.1080/2162402X.2019.1655964
Rojas C. M., Mysore K. S. (2012). Glycolate oxidase is an alternative source for H2O2 production during plant defense responses and functions independently from NADPH oxidase. Plant Signal Behav. 7, 752–755. doi: 10.4161/psb.20429
Rojas C. M., Senthil-Kumar M., Wang K., Ryu C. M., Kaundal A., Mysore K. S. (2012). Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and Arabidopsis. Plant Cell 24, 336–352. doi: 10.1105/tpc.111.093245
Rosa C., Kuo Y. W., Wuriyanghan H., Falk B. W. (2018). RNA Interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 56, 581–610. doi: 10.1146/annurev-phyto-080417-050044
Roth P. B., Twiner M. J., Mikulski C. M., Barnhorst A. B., Doucette G. J. (2008). Comparative analysis of two algicidal bacteria active against the red tide dinoflagellate Karenia brevis. Harmful Algae 7, 682–691. doi: 10.1016/j.hal.2008.02.002
Roy S., Jagus R., Morse D., Roy S., Jagus R., Morse D. (2018). Translation and translational control in dinoflagellates. Microorganisms 6, 30. doi: 10.3390/microorganisms6020030
Sambrook J., Fritsch E. F., Maniatis T. (2012). Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 3, 1817.
Sendler M., Maertin S., John D., Persike M., Weiss F. U., Krüger B., et al. (2016). Cathepsin b activity initiates apoptosis via digestive protease activation in pancreatic acinar cells and experimental pancreatitis. J. Biol. Chem. 291, 14717–14731. doi: 10.1074/jbc.M116.718999
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shi X., Liu L., Li Y., Xiao Y., Ding G., Lin S., et al. (2018). Isolation of an algicidal bacterium and its effects against the harmful-algal- bloom dinoflagellate Prorocentrum donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Shoguchi E., Beedessee G., Tada I., Hisata K., Kawashima T., Takeuchi T., et al. (2018). Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics 19, 1–11. doi: 10.1186/s12864-018-4857-9
Singh S., Agrawal S. B., Agrawal M. (2014). UVR8 mediated plant protective responses under low UV-b radiation leading to photosynthetic acclimation. J. Photochem. Photobiol. B 137, 67–76. doi: 10.1016/j.jphotobiol.2014.03.026
Sobotka R., Komenda J., Bumba L., Tichy M. (2005). Photosystem II assembly in CP47 mutant of Synechocystis sp. PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J. Biol. Chem. 280, 31595–31602. doi: 10.1074/jbc.M505976200
Song B., Chen S., Chen W. (2018). Dinoflagellates, a unique lineage for retrogene research. Front. Microbiol. 9, 1556. doi: 10.3389/fmicb.2018.01556
Stenbaek A., Hansson A., Wulff R. P., Hansson M., Dietz K. J., Jensen P. E. (2008). NADPH-dependent thioredoxin reductase and 2-cys peroxiredoxins are needed for the protection of mg-protoporphyrin monomethyl ester cyclase. FEBS Lett. 582, 2773–2778. doi: 10.1016/j.febslet.2008.07.006
Sun Q., Zhou M., Zuo Z. (2020). Toxic mechanism of eucalyptol and β-cyclocitral on Chlamydomonas reinhardtii by inducing programmed cell death. J. Hazard Mater 389, 121910. doi: 10.1016/j.jhazmat.2019.121910
Takahashi K., Tatsunami R., Sato K., Tampo Y. (2012). Multidrug resistance associated protein 1 together with glutathione plays a protective role against 4-hydroxy-2-nonenal-induced oxidative stress in bovine aortic endothelial cells. Biol. Pharm. Bull. 35, 1269–1274. doi: 10.1248/bpb.b12-00069
Tang Y. Z., Koch F., Gobler C. J. (2010). Most harmful algal bloom species are vitamin B1 and b 12 auxotrophs. Proc. Natl. Acad. Sci. U.S.A. 107, 20756–20761. doi: 10.1073/pnas.1009566107
Tarazona-Janampa U. I., Cembella A. D., Pelayo-Zárate M. C., Pajares S., Márquez-Valdelamar L. M., Okolodkov Y. B., et al. (2020). Associated bacteria and their effects on growth and toxigenicity of the dinoflagellate Prorocentrum lima species complex from epibenthic substrates along Mexican coasts. Front. Mar. Sci. 7:569. doi: 10.3389/fmars.2020.00569
Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., et al. (2003). The COG database: An updated vesion includes eukaryotes. BMC Bioinf. 4, 41. doi: 10.1186/1471-2105-4-41
Ternon E., Wang Y., Coyne K. J. (2019). Small polar molecules: A challenge in marine chemical ecology. Molecules 24, 1–17. doi: 10.3390/molecules24010135
Tilney C. L., Pokrzywinski K. L., Coyne K. J., Warner M. E. (2014). Growth, death, and photobiology of dinoflagellates (Dinophyceae) under bacterial-algicide control. J. Appl. Phycol 26, 2117–2127. doi: 10.1007/s10811-014-0248-z
Vass I. Z., Kos P. B., Sass L., Nagy C. I., Vass I. (2013). The ability of cyanobacterial cells to restore UV‐B radiation induced damage to photosystem II is influenced by photolyase dependent DNA repair. Photochemistry and Photobiology 89(2), 384–390.
Vlahakis A., Debnath J. (2017). The interconnections between autophagy and integrin-mediated cell adhesion. J. Mol. Biol. 429, 515–530. doi: 10.1016/j.jmb.2016.11.027
Wan J., Kalpage H. A., Vaishnav A., Liu J., Lee I., Mahapatra G., et al. (2019). Regulation of respiration and apoptosis by cytochrome c threonine 58 phosphorylation. Sci. Rep. 9, 1–16. doi: 10.1038/s41598-019-52101-z
Wang X., Niu X., Chen Y., Sun Z., Han A., Lou X., et al. (2019). Transcriptome sequencing of a toxic dinoflagellate, Karenia mikimotoi subjected to stress from solar ultraviolet radiation. Harmful Algae 88, 101640. doi: 10.1016/j.hal.2019.101640
Wang Y. (2021). Bacterial algicides: application strategies and cellular response of target organisms (Ph.D. dissertation, University of Delaware, DE, USA).
Wang Y., Coyne K. J. (2022). Metabolomic insights of the effects of bacterial algicide IRI-160AA on dinoflagellate Karlodinium veneficum. Metabolites 12, 317. doi: 10.3390/metabo12040317
Wilkinson D., Ramsdale M. (2011). “Proteases and caspase-like activity in the yeast saccharomyces cerevisiae. Biochemical Society Transactions 39(5), 1502–1508. doi: 10.1042/BST0391502
Willis N., Rhind N. (2009). Regulation of DNA replication by the s-phase DNA damage checkpoint. Cell Div 4, 13. doi: 10.1186/1747-1028-4-13
Wu G., Adachi H., Ge J., Stephenson D., Query C. C., Yu Y. (2016). Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J. 35, 654–667. doi: 10.15252/embj.201593113
Wu T. M., Hsu Y. T., Sung M. S., Hsu Y. T., Lee T. M. (2009). Expression of genes involved in redox homeostasis and antioxidant defense in a marine macroalga Ulva fasciata by excess copper. Aquat. Toxicol. 94, 275–285. doi: 10.1016/j.aquatox.2009.07.010
Wu Y., Jin X., Liao W., Hu L., Dawuda M. M., Zhao X., et al. (2018). 5-aminolevulinic acid (ALA) alleviated salinity stress in cucumber seedlings by enhancing chlorophyll synthesis pathway. Front. Plant Sci. 9, 635. doi: 10.3389/fpls.2018.00635
Wu Y., Liao W., Dawuda M. M., Hu L., Yu J. (2019). 5-aminolevulinic acid (ALA) biosynthetic and metabolic pathways and its role in higher plants: a review. Plant Growth Regul. 87, 357–374. doi: 10.1007/s10725-018-0463-8
Xiang T., Jinkerson R. E., Clowez S., Tran C., Krediet C. J., Onishi M., et al. (2018). Glucose-induced trophic shift in an endosymbiont dinoflagellate with physiological and molecular consequences. Plant Physiol. 176, 1793–1807. doi: 10.1104/pp.17.01572
Xiang T., Nelson W., Rodriguez J., Tolleter D., Grossman A. R. (2015). Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 82, 67–80. doi: 10.1111/tpj.12789
Xu H., Zhang J., Zeng J., Jiang L., Liu E., Peng C., et al. (2009). Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J. Exp. Bot. 60, 1799–1809. doi: 10.1093/jxb/erp056
Xu Y. P., Yang J., Cai X. Z. (2018). Glycolate oxidase gene family in Nicotiana benthamiana: Genomewide identification and functional analyses in disease resistance. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-27000-4
Yang I., John U., Beszteri S., Glöckner G., Krock B., Goesmann A., et al. (2010). Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genomics 11, 248. doi: 10.1186/1471-2164-11-248
Yang J., Xie X., Wang X., Dixon R., Wang Y.-P. (2014). Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1–8, E3718–E3725. doi: 10.1073/pnas.1411185111
Yau W. L., Lambertz U., Colineau L., Pescher P., MacDonald A., Zander D., et al. (2016). Phenotypic characterization of a Leishmania donovani cyclophilin 40 null mutant. J. Eukaryotic Microbiol. 63, 823–833. doi: 10.1111/jeu.12329
Yoshiyama K. O., Sakaguchi K., Kimura S. (2013). DNA Damage response in plants: Conserved and variable response compared to animals. Biol. (Basel) 2, 1338–1356. doi: 10.3390/biology2041338
Zannini L., Delia D., Buscemi G. (2014). CHK2 kinase in the DNA damage response and beyond. J. Mol. Cell Biol. 6, 442–457. doi: 10.1093/jmcb/mju045
Zelitch I., Schultes N. P., Peterson R. B., Brown P., Brutnell T. P. (2009). High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 149, 195–204. doi: 10.1104/pp.108.128439
Zhang C., Lin S. (2019). Initial evidence of functional siRNA machinery in dinoflagellates. Harmful Algae 81, 53–58. doi: 10.1016/j.hal.2018.11.014
Zhang F., Ye Q., Chen Q., Yang K., Zhang D., Chen Z., et al. (2018a). Algicidal activity of novel marine bacterium paracoccus sp. strain Y42 against a harmful algal-bloom-causing dinoflagellate, Prorocentrum donghaiense. Appl. Environ. Microbiol. 84, 1015–1033. doi: 10.1128/AEM.01015-18
Zhang H., Hou Y., Miranda L., Campbell D. A., Sturm N. R., Gaasterland T., et al. (2007). Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 104, 4618–4623. doi: 10.1073/pnas.0700258104
Zhang W., Zhong H., Lu H., Zhang Y., Deng X., Huang K., et al. (2018b). Characterization of ferredoxin-dependent biliverdin reductase PCYA1 reveals the dual function in retrograde bilin biosynthesis and interaction with light-dependent protochlorophyllide oxidoreductase LPOR in Chlamydomonas reinhardtii. Front. Plant Sci. 9:676. doi: 10.3389/fpls.2018.00676
Zhao Y., Tang X., Qu F., Lv M., Liu Q., Li J., et al. (2020). ROS-mediated programmed cell death (PCD) of Thalassiosira pseudonana under the stress of BDE-47. Environ. pollut. 262, 114342. doi: 10.1016/j.envpol.2020.114342
Zhao Y., Tang X., Quigg A., Lv M., Zhao Y. (2019). The toxic mechanisms of BDE-47 to the marine diatom Thalassiosira pseudonana-a study based on multiple physiological processes. Aquat. Toxicol. 212, 20–27. doi: 10.1016/j.aquatox.2019.04.010
Zotti M., dos Santos E. A., Cagliari D., Christiaens O., Taning C. N. T., Smagghe G. (2018). RNA Interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag Sci. 74, 1239–1250. doi: 10.1002/ps.4813
Keywords: dinoflagellate, transcriptomics, programmed cell death, algicidal bacteria, Shewanella sp. IRI-160, DNA damage, harmful algal bloom, HAB
Citation: Wang Y and Coyne KJ (2023) Transcriptome profiling reveals a global response in harmful dinoflagellate Karlodinium veneficum to naturally-occurring bacterial algicides. Front. Mar. Sci. 10:1112913. doi: 10.3389/fmars.2023.1112913
Received: 01 December 2022; Accepted: 03 February 2023;
Published: 16 February 2023.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Guanpin Yang, Ocean University of China, ChinaMatthew John Harke, Gloucester Marine Genomics Institute (GMGI), United States
Copyright © 2023 Wang and Coyne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathryn J. Coyne, a2NveW5lQHVkZWwuZWR1
 Yanfei Wang
Yanfei Wang Kathryn J. Coyne
Kathryn J. Coyne