- 1BioSciences at Rice, Rice University, Houston, TX, United States
- 2Department of Biology, Boston University, Boston, MA, United States
- 3Department of Oceanography, Texas A & M University, College Station, TX, United States
Animal waste products are an important component of nutrient cycles and result in the trophic transmission of diverse microorganisms. There is growing recognition that the feces of consumers, such as predators, may impact resource species, their prey, via physical effects and/or microbial activity. We tested the effect of feces from distinct fish trophic groups on coral health and used heat-killed fecal controls to tease apart physical versus microbial effects of contact with fecal material. Fresh grazer/detritivore fish feces caused lesions more frequently on corals, and lesions were 4.2-fold larger than those from sterilized grazer/detritivore feces; in contrast, fresh corallivore feces did not cause more frequent or larger lesions than sterilized corallivore feces. Thus, microbial activity in grazer/detritivore feces, but not corallivore feces, was harmful to corals. Characterization of bacterial diversity in feces of 10 reef fish species, ranging from obligate corallivores to grazer/detritivores, indicated that our experimental findings may be broadly generalizable to consumer guild, since feces of some obligate corallivores contained ~2-fold higher relative abundances of coral mutualist bacteria (e.g., Endozoicomonadaceae), and lower abundances of the coral pathogen, Vibrio coralliilyticus, than feces of some grazer/detritivores. These findings recontextualize the ecological roles of consumers on coral reefs: although grazer/detritivores support coral reef health in various ways (e.g., promoting coral settlement and herbivory through the removal of detritus and sediments from the algal matrix), they also disperse coral pathogens. Corallivore predation can wound corals, yet their feces contain potentially beneficial coral-associated bacteria, supporting the hypothesized role of consumers, and corallivores in particular, in coral symbiont dispersal. Such consumer-mediated microbial dispersal as demonstrated here has broad implications for environmental management.
Introduction
As consumers—including herbivores, predators and parasites—move through the environment and engage in consumer-resource interactions, they transmit microorganisms amongst the animals and plants with which they interact (Barron Pastor and Gordon, 2016; Burns et al., 2017; Ezzat et al., 2020; Vannette, 2020). This trophic transmission likely has important implications for microbiota assembly, as well as for resource species health and environmental acclimatization (Bosch and McFall-Ngai, 2011; McFall-Ngai et al., 2013; Grupstra et al., 2022; Fowler et al., 2023). Tropical coral reefs, for example, harbor diverse fishes that feed on benthic resource species, including stony corals and macroalgae (Hughes, 1994; Hughes et al., 2003; Bellwood et al., 2004). These fish defecate as they swim over the reef, generating a “persistent rain of feces”, containing high densities of live microbiota, that is deposited onto resident corals (Smriga et al., 2010). This frequent deposition of fish feces–along with the microbiota they contain–is likely to affect coral health through several potential mechanisms (Muller Parker, 1984; Ezzat et al., 2019; Ezzat et al., 2020; Umeki et al., 2020; Grupstra et al., 2021).
Corals are filter feeders that can take up nutrients from at least some fish waste products (Coma et al., 2001; Mills and Sebens, 2004; Allgeier et al., 2017); the extent and frequency that they do so from fish fecal material requires exploration. Yet, fish feces also contain particulate matter derived from reef sediments and silts (Krone et al., 2011); when sediments land on a coral colony, smothering and death of coral polyps can ensue, resulting in patches of mortality (lesions). Pathogenic or opportunistic bacteria in fish feces may also have negative effects on coral health (Aeby and Santavy, 2006; Nicolet et al., 2018; Ezzat et al., 2019; Ezzat et al., 2021; Renzi et al., 2022). For example, microbial opportunists in the feces of some grazer/detritivores can trigger lesion formation on corals (Ezzat et al., 2019; Ezzat et al., 2021). This is likely because detritus, turf-, and macroalgae, major food sources for these fishes, can contain diverse coral pathogens (Nugues et al., 2004; Dinsdale et al., 2008; Sweet et al., 2013; Franco et al., 2020) such as members of the genus Vibrio: Vibrio coralliilyticus, for example, can cause lesions or bleaching in corals (Ben-Haim et al., 2003; Vidal-Dupiol et al., 2011; Wilson et al., 2013).
Microbiota in fish feces may not always be detrimental to coral health, however. In fact, feces from corallivores might even support coral health through the delivery of probiotics—live, generally homologous microorganisms that can benefit the recipient animal–on some coral-dominated reefs (Grupstra et al., 2021; Grupstra et al., 2022). We infer this based on increased survival and growth of juvenile corals experimentally inoculated with cultured Symbiodiniaceae cells (Suzuki et al., 2013; McIlroy et al., 2016), as well as increased survival of adult corals under environmental stress that were provided with mixtures of beneficial bacteria or Symbiodiniaceae cells (Peixoto et al., 2017; Morgans et al., 2019; Rosado et al., 2019; Doering et al., 2021; Peixoto et al., 2021; Santoro et al., 2021; Thatcher et al., 2022). The feces of corallivorous fish contain high densities of potential probiotics, including live Symbiodiniaceae cells (Muller Parker, 1984; Castro-Sanguino and Sánchez, 2012; Grupstra et al., 2021) and mutualistic coral bacteria including members of the family Endozoicomonadaceae (Clever et al., 2022). The deposition of feces on corals may thus affect colony health in nuanced ways depending on aspects of fecal composition including diversity of microbiota, nutritional value and sediment content. Studying how feces from coral reef fish in different trophic guilds, whose feces contain different microbial, nutritional and sediment compositions, affect coral health will advance coral reef ecology and inform management and conservation plans.
We tested the effect of feces from distinct consumer guilds on coral health and used heat-killed fecal controls to tease apart physical versus biological effects of contact with fecal material. We then characterized bacterial community assemblages and quantified relative abundances of the coral pathogen V. coralliilyticus in feces of 10 abundant consumers, ranging from obligate corallivores to grazer/detritivores, to determine whether results from the feces addition experiment are generalizable at the guild level. We tested the following hypotheses: 1) microbiota in grazer/detritivore feces, but not corallivore feces, negatively impact coral health through the development of lesions, 2) microbial communities in feces are consumer guild-specific and grazer/detritivore feces contain higher relative abundances of the coral pathogen V. coralliilyticus, whereas corallivore feces contain more coral-associated bacteria that are potentially beneficial.
Methods
Feces addition experimental design
A feces addition experiment was conducted three times (Final N = 11 replicates) using fragments of Pocillopora spp. (Johnston et al., 2018) in Moorea, French Polynesia. An initial experimental iteration was performed in October 2020 (four replicates); two additional iterations took place in June 2021 (5 replicates each). For the initial iteration, four colonies of the cryptic coral genus Pocillopora spp. were collected from the fore reef at ~10 m depth; these colonies were later identified to be members of the species Pocillopora meandrina following methods in Johnston et al. (2018). After 24 h, all corals were cut into five ~10 cm long fragments and left to acclimate for 48 hours, and any macrosymbionts (e.g., Trapezia crabs, shrimp) were removed. All fragments were transferred to glass jars containing 500 ml of 0.2 μm-filtered (sterile) seawater with bubblers and air stones to promote aeration and water circulation, photographed under a dissection microscope with 3.5-180X zoom (AmScope SM-1TSZZ-144S-10M). Initial dark-adapted (20 min) photosynthetic efficiency (Fv/Fm) was then measured using an imaging pulse amplitude modulation (I-PAM) fluorometer (WALZ, Germany) under the default settings.
We then blindly assigned each jar containing a coral fragment to one of the following five treatments (such that one fragment from each coral colony was assigned to each treatment, 4 replicate colonies x 5 treatments = 20 coral fragments in separate jars): fresh feces from a corallivorous butterflyfish (FC); fresh feces from a grazer/detritivore (FG); sterilized feces from a corallivorous butterflyfish (SC); sterilized feces from a grazer/detritivore (SG); no-feces control (C). For the fresh feces treatments (FC, FG), we applied 100 µl of fresh feces isolated from the hindgut of the butterflyfish Chaetodon ornatissimus (FC) or the grazer/detritivore Ctenochaetus striatus (FG) directly onto each coral fragment. Briefly, feces were isolated from the hindgut using sterile tools by making an incision from the anus to the pelvic fin and removing the intestinal tract; feces were then squeezed from the hindgut into sterile collection tubes using sterile tweezers and pipetted onto coral fragments using 1 ml filter tips that were modified to widen the tip opening using a sterilized razor blade. For the sterilized feces treatments (SC, SG), fecal pellets were sterilized in a pressure cooker for 40 minutes at 120 °C and then applied in the same manner as fresh feces. The fresh feces used in the experiment were subsampled for DNA extractions by preservation in DNA/RNA Shield (Zymo Research, CA). No manipulation was conducted on the no-feces control fragments. The experiment ran for ~22 hours; this treatment duration is reflective of time periods over which corals may sometimes be in direct contact with fish feces in situ (up to 48 hours; Ezzat et al., 2019; Ezzat et al., 2021). All fragments were photographed under the dissection microscope before and after the removal of the fecal pellet, and final Fv/Fm measurements were collected from each coral fragment using imaging-PAM fluorometry following a 20-minute dark adaptation.
Minor modifications were incorporated into the design of the second and third experimental iterations: first, all coral colonies used in the experiment were sampled before collection and their species identity was determined as Pocillopora verrucosa using restriction fragment length polymorphism (RFLP) analysis, as outlined in Johnston et al. (2018). For these iterations, five replicate colonies were used instead of four. Additionally, fecal treatments in these iterations were composed of feces from two individuals per fish species that were mixed prior to application (instead of using feces from a single fish individual per treatment as in the first experimental iteration).
The third iteration of the experiment ended after ~15 hours (instead of ~22 hours) because fragments (including control fragments) from three of five colonies were exhibiting signs of tissue loss consistent with stressors outside the scope of our experiment. Tissue loss in some other fragments (that were not selected for use in the experiment) of these coral colonies had started before treatments were applied to experimental fragments. Thus, all data for these three colonies (the entire set of samples—all treatments) were discarded and not included in subsequent analyses. For all subsequent analyses, replicates from all remaining iterations (11 experimental replicates) were analyzed together.
Bacterial culturing
To ensure that the sterilization method for the sterile feces treatments was effective, we streaked agar plates with subsampled material of each fecal treatment in iterations 2 and 3 of the experiment. Briefly, sterile plates (Difco™ Marine Agar 2216, 5.51%) were swabbed with a rice grain-sized mixture of fresh or sterile feces from C. ornatissimus or C. striatus. Culture plates were monitored for bacterial growth for five days and transferred to fresh agar plates. Bacterial growth was observed in ten of ten agar plates with fresh fecal samples and one of eight agar plates with sterilized fecal samples.
Analysis of coral lesions and coral fragment photosynthetic efficiency
To test how microbial communities in fish feces affected coral health, we quantified the frequencies and sizes of coral lesions caused by fecal treatments (in addition to measuring the photosynthetic efficiency of each fragment). In brief, fecal pellets were removed from each fragment and photographs were taken using a dissection microscope. Dissection microscope photographs were analyzed in ImageJ v. 1.53f51. Each fragment was binned to one of three categories: “apparently healthy” (no change in fragment compared to before fecal application), “lesion” (fragment contains a novel patch of bare calcium carbonate, where coral tissue died and sloughed off following fecal application), or “dead” (fragment no longer contains live tissue; all tissue sloughed off following fecal application). The size of each lesion (excluding fragments binned as “apparently healthy” or “dead”) was measured by determining the average polyp size on each fragment in ImageJ by counting the number of polyps in a polygon of standardized size in triplicate. Then, the size of the lesion was measured (in pixels), and the lesion size was expressed as the estimated number of polyps that had died.
The effect of experimental treatment on the frequency of lesions and mortality in coral fragments (categories “healthy”, “lesion”, or “dead”, N =11 per treatment) was tested using an ordinal regression (sensu Gorczynski and Beaudrot, 2021) in the package ordinal (v. 2022.11-16); colony number was included as a separate fixed effect to control for colony-level effects. Differences in lesion sizes (expressed as the estimated number of dead polyps) among treatments were tested using linear models with lmer (package vegan v 1.1-28), with colony number included as a random effect. An ANOVA in the car package (v3.0-12; Anova command) was used to calculate type II p-values. Values were log-transformed to satisfy the assumptions of the linear model. Fragments that had complete mortality (N = 4 total) were excluded from the analysis, as well as one replicate colony (all fragments/treatments) because a lesion in one of the treatments (SG treatment) was not entirely in the image and we were therefore unable to accurately assess its size. Zero values (i.e., fragments that did not have lesions) were not included in the analysis (final N: C, 0; FC, 4; FG, 8; SC, 4; SG, 4). Pairwise comparisons between all treatments were conducted using the package emmeans (v1.7.0) with a Tukey adjustment for multiple comparisons. Model assumptions were visually checked using the check_model function in the package performance v0.9.1.
Photosynthetic efficiency (Fv/Fm) values were extracted from I-PAM fluorometry scans of each coral fragment post hoc. Measurements were taken following a transect design: one measurement was taken immediately adjacent to the removed fecal pellet location, another measurement was taken halfway between the first measurement and the furthest edge away from the fecal pellet, and the final measurement was taken at the furthest edge of the fragment away from the point where feces were placed. We took the mean of these three values for each fragment. The effect of fecal pellets on untransformed Fv/Fm values were tested in the same way as lesion sizes (including colony as a random effect); p-values were generated using type II Wald chi-square tests.
Field sampling of corallivorous and grazer/detritivorous fishes, and other environmental reservoirs of coral-associated microorganisms
To test the extent to which microbial communities in fish feces—and thereby, their effects on coral health—may be broadly generalizable within fish trophic guilds, we collected additional fecal samples from each of the fish species included in the experiment (the obligate corallivore C. ornatissimus and the grazer/detritivore C. striatus), and other species in the same trophic guilds (grazer/detritivores, obligate corallivores), as well as facultative corallivores. Additionally, samples of corals, algae, sediments, and seawater were collected to test whether bacterial taxa in these samples were also represented in fish feces (N = 5-14 per fish, coral, or algae species/genus, see Table S1). All collections were conducted in October 2020 from the back reef (1-2 m depth) and fore reef (5-10 m depth) in Moorea, between LTER sites 1 and 2 of the Moorea Coral Reef (MCR) Long Term Ecological Research (LTER) site. Obligate corallivores were defined as fish that eat corals nearly exclusively (>90% of stomach content or number of bites; Harmelin-Vivien and Bouchon-Navaro, 1983; Rotjan and Lewis, 2008), and facultative corallivores were defined as fish that were observed to feed on coral for a minor to major part of their diet, while also feeding on algae or other invertebrates (~5-80% of bites from corals; Harmelin-Vivien and Bouchon-Navaro, 1983; Rotjan and Lewis, 2008; Viviani et al., 2019; Ezzat et al., 2020). The selected species included three additional obligate corallivore species (butterflyfishes Chaetodon lunulatus, “CHLU”; Chaetodon reticulatus, “CHRE”; and the filefish Amanses scopas, “AMSC”), three facultative corallivore species (butterflyfishes Chaetodon citrinellus, “CHCI”; and Chaetodon pelewensis, “CHPE”; and the parrotfish Chlorurus spilurus, “CHSP”), and two additional grazer/detritivore species (surgeonfishes Ctenochaetus flavicauda, “CTFL”; and Zebrasoma scopas, “ZESC”). Coral and algal samples were collected from locally abundant coral and algae genera (Pratchett, 2014; Burkepile et al., 2020), including the corals Acropora hyacinthus (“ACR”), Pocillopora species complex (“POC”, Gélin et al., 2017; Johnston et al., 2018), and Porites lobata species complex (“POR”, Forsman et al., 2009; Forsman et al., 2015); and mixed communities of turf algae (“Turf”) as well as macroalgae in the genera Asparagopsis (“Asp”), Dictyota (“Dict”), Lobophora (“Lob”), Sargassum (“Sarg”), and Turbinaria sp. (“Turb”; see Table S1 for replication per species or sample category). Sediment (“SED”; 250 ml) and water (“WAT”; 1.9 L) were collected concomitantly with fish and environmental samples using sterilized containers. Following collection, all samples (fish, coral, algae, sediments and water) were immediately transported on ice to the lab, where they were processed using sterile methods as described in Grupstra et al. (2021) and preserved in DNA/RNA Shield (Zymo Research, CA).
Sequencing and analysis of 16S rRNA gene amplicons from in situ samples
DNA was extracted from all samples using the ZymoBIOMICS DNA/RNA Miniprep kit (Zymo Research, CA) according to the manufacturer’s instructions, but with a 1hr proteinase K incubation (35°C) before the lysis buffer step. Library preparation and sequencing was conducted at the Genomics Core Lab at the Institute of Arctic Biology of the University of Alaska Fairbanks. All samples were sequenced using the 16S rRNA gene V4 primers 515f (GTGYCAGCMGCCGCGGTAA) and 806rB (GGACTACNVGGGTWTCTAAT) on Illumina MiSeq using v3 2x300bp chemistry (Apprill et al., 2015; Parada et al., 2016; Walters et al., 2016). Mock communities (HM-782D, BEI Resources, VA), extraction negatives, and plate negatives were included to facilitate the identification and removal of potential contaminants in silico (Davis et al., 2018).
Bacterial reads were processed in RStudio (version 1.1.456) through the DADA2 pipeline (version 1.11.0, Callahan et al., 2016) and using phyloseq (v1.38). The DADA2 pipeline generated a table of amplicon sequence variants (ASVs), and bacteria taxonomy was assigned using the SILVA rRNA database (version 132, Quast et al., 2012). Non-target (e.g., mitochondrial, chloroplast) reads and singletons were removed. A total of 865 potential contaminant ASVs were identified and removed with the decontam package (v. 1.10.0) using the prevalence method (Davis et al., 2018). A total of 160 samples with >1,000 reads remained after these initial quality control and filtering steps (excluding negatives and mock communities), with a mean of 15,263 reads per sample. All analyses outlined below were conducted based on this dataset, but additional ASV and sample filtering steps were included for several of the approaches (see Table S1 for replication and filtering methods).
To visualize the relationship among samples in an ordination plot, we produced a non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis distance values calculated using metaMDS (K = 3; Vegan v. 2.6-4). Because the dataset was highly diverse (24,095 total ASVs), only common ASVs (>200 reads) were included to facilitate NMDS convergence; samples with <1,000 reads after this additional ASV filtering step were removed and libraries were rarefied to 1,188 reads (NMDS N = 135; see Table S1 for sample sizes per species).
To test for differences in microbial community compositions between sampling groups, PERMANOVA tests were conducted using adonis on Bray-Curtis distances calculated from untransformed counts tables that were rarefied to 1,009 reads per sample (no low abundance ASVs were excluded, in contrast to the NMDS). However, tests of multivariate homogeneity of group dispersion (using betadisper) showed significant heterogeneity, which can increase the probability of Type I errors while using PERMANOVA if the sampling design is unbalanced (Anderson and Walsh, 2013). Because the sampling design was unbalanced—e.g., the obligate corallivore category contained four fish species with 32 samples total, while the facultative corallivore category contained three fish species with 18 samples total (Table S1)—tests among environmental pools (obligate corallivore feces, facultative corallivore feces, grazer/detritivore feces, coral, algae, sediments and water), and among species/sample types (e.g., feces of A. scopas vs. feces of C. striatus) were conducted separately to facilitate subsampling to create a balanced design. Thus, all the categories (environmental pool or type/species) were randomly subsampled (18 random samples per environmental pool or 5 random samples per type/species; see Tables S3, S4 for sample names) prior to conducting PERMANOVA. Pairwise PERMANOVAs were conducted (with a Bonferroni correction for multiple comparisons between environmental pools, or a Benjamini-Hochberg correction for tests between species/sample types) using the package pairwise Adonis v.0.4 on these subsampled datasets.
ASVs shared between the feces of consumers and tissues of resource species (corals and algae), and sediments and water, were identified using the intersect command. To reduce noise and potential “transient” bacterial taxa, low abundance ASVs (<100 reads) were removed; samples with <1,000 reads after this additional ASV filtering step were excluded from the analysis (final N = 149; see Table S1 for replication per species), and samples were rarefied to 1,008 reads.
Lastly, we tested for differences in the relative abundances of members of the family Endozoicomonadaceae, some of which are coral mutualists (Neave et al., 2016; Neave et al., 2017a; Neave et al., 2017b; Tandon et al., 2020), among environmental pools (obligate and facultative corallivore feces, grazer/detritivore feces, corals, algae, sediment and water). All libraries were rarefied to 1,009 reads and normalized to 1. Kruskal-Wallis tests were used to test for differences in relative Endozoicomonadaceae read abundances because the residuals were not normally distributed when using linear models. Pairwise tests between environmental pools were conducted using Dunn tests in the package dunn.test v. 1.3.5. Kruskal-Wallis tests and Dunn tests were also conducted to test for differences between individual sample types/species within each environmental pool (e.g., within algae, Turbinaria sp. vs turf algae).
Quantitative PCR of Vibrio coralliilyticus genes in fish feces
Quantitative PCR (qPCR) was conducted using vcpARTF and vcpARTR qPCR primers developed for the bacterial coral pathogen Vibrio coralliilyticus (Wilson et al., 2013) and results were standardized using primers for general bacteria 967F and 1046R (Sogin et al., 2006; Chen et al., 2011; Shiu et al., 2020). Standard curves (for V. coralliilyticus and general bacteria) were made from a Vibrio coralliilyticus culture (AS008) isolated from corals collected in the Flower Garden Banks National Marine Sanctuary (northwest Gulf of Mexico). Sequencing of the full-length 16S gene region of bacterial DNA with primers 8F and 1513R resulted in 97.5% percent identity with V. coralliilyticus strain U2 (accession MK999891.1) with 100% query cover. The primers vcpARTR and vcpARTF were used to amplify metalloprotease genes and the amplicon was then cleaned using a genejet PCR purification kit (Thermo Fisher, MA); resultant DNA concentrations were acquired using Qubit (Thermo Fisher, MA). A standard curve was made using serial dilutions from 109 to 100 gene copies per µl template; the standard curve for V. coralliiyticus primers vcpARTR and vcpARTF had an efficiency of 106.2%; the standard curve for general bacteria primers 967F and 1046R had an efficiency of 92.3%. Sanger sequencing of gene fragments amplified using vcpARTF and vcpARTR primers from DNA extracted from C. striatus feces resulted in a top hit against a V. coralliilyticus strain P4 metalloprotease gene (accession JQ345042.1) with an e-value of 3x10-12 (query cover 58%, percent identity 85%). Amplification of the V. coralliilyticus target gene at <30 cycles was counted as a positive detection, which roughly corresponded to amplification of the 102 gene copies µl-1 standard (Ct=30.4). Delta cycle threshold (dCT) values were calculated by subtracting the cycle threshold at which the signal from general bacteria primers was detected from the cycle threshold at which V. coralliilyticus was detected. Higher dCT values indicate lower relative abundances of V. coralliilyticus. Differences in dCT values between fecal samples of each trophic guild were tested with an ANOVA, followed by Tukey tests for multiple comparisons (using a Tukey correction). The assumptions for the test were visually assessed.
Results
Microbial activity in grazer/detritivore feces increased coral lesion frequencies and sizes
Fecal treatments resulted in the formation of lesions or complete mortality in some coral fragments, whereas other fragments remained visually healthy (Figures 1A; S1; Supplementary Data File 1; overall ordinal regression results: treatment χ2 = 32.3, p < 0.001; colony χ2 = 6.6, p = 0.761). Fresh grazer/detritivore feces resulted in the formation of lesions or complete mortality in all replicates (N = 9 and 2, respectively) and differed significantly from control (no feces) fragments (estimate = 2.97, p = 0.006). The fresh corallivore feces treatment resulted in lesions or total mortality in only some fragments (N = 4 and 1, respectively) while most fragments remained visually healthy (N = 6) and did not differ significantly from control fragments (estimate = 1.02 p = 0.249). Effects of both sterile feces treatments on fragment health also did not differ from controls (sterile corallivore estimate = 1.31, p = 0.140; sterile grazer estimate = 1.39, p = 0.120) and were comparable to those caused by fresh corallivore feces (i.e., lesions or total mortality in only some fragments; sterile corallivore: N = 4 and 0; sterile grazer: N = 5 and 1). All analyzed control fragments (no feces treatment, N = 11) remained visually healthy during the experiment. Lesion sizes caused by fecal pellets also differed significantly among treatments (Figure 1B; Df = 3, χ2 = 13.49, p = 0.004; Supplementary Data File 2). Pairwise comparisons revealed that mean (± SD) lesion sizes in the fresh grazer feces treatment (41.1 ± 25.8 polyps) were, for example, 4.2 times larger than those in the sterile grazer feces treatments (9.76 ± 4.46 polyps; estimate =1.33; p = 0.049, see Table S2 for all pairwise comparisons). Lesions caused by grazer feces were also three times larger than those caused by fresh corallivore feces (13.9 ± 11.6 polyps), but this pairwise comparison was not significant, potentially due to the low number of lesions caused by fresh corallivore feces (N = 4). Lesion sizes caused by fresh corallivore feces did not differ from sterile corallivore feces (15.3 ± 9.6 polyps). Finally, coral photosynthetic efficiency was not significantly affected by the fecal treatments (Figure S2; lmer results: treatment: Df = 4, χ2 = 9.09, p = 0.059.
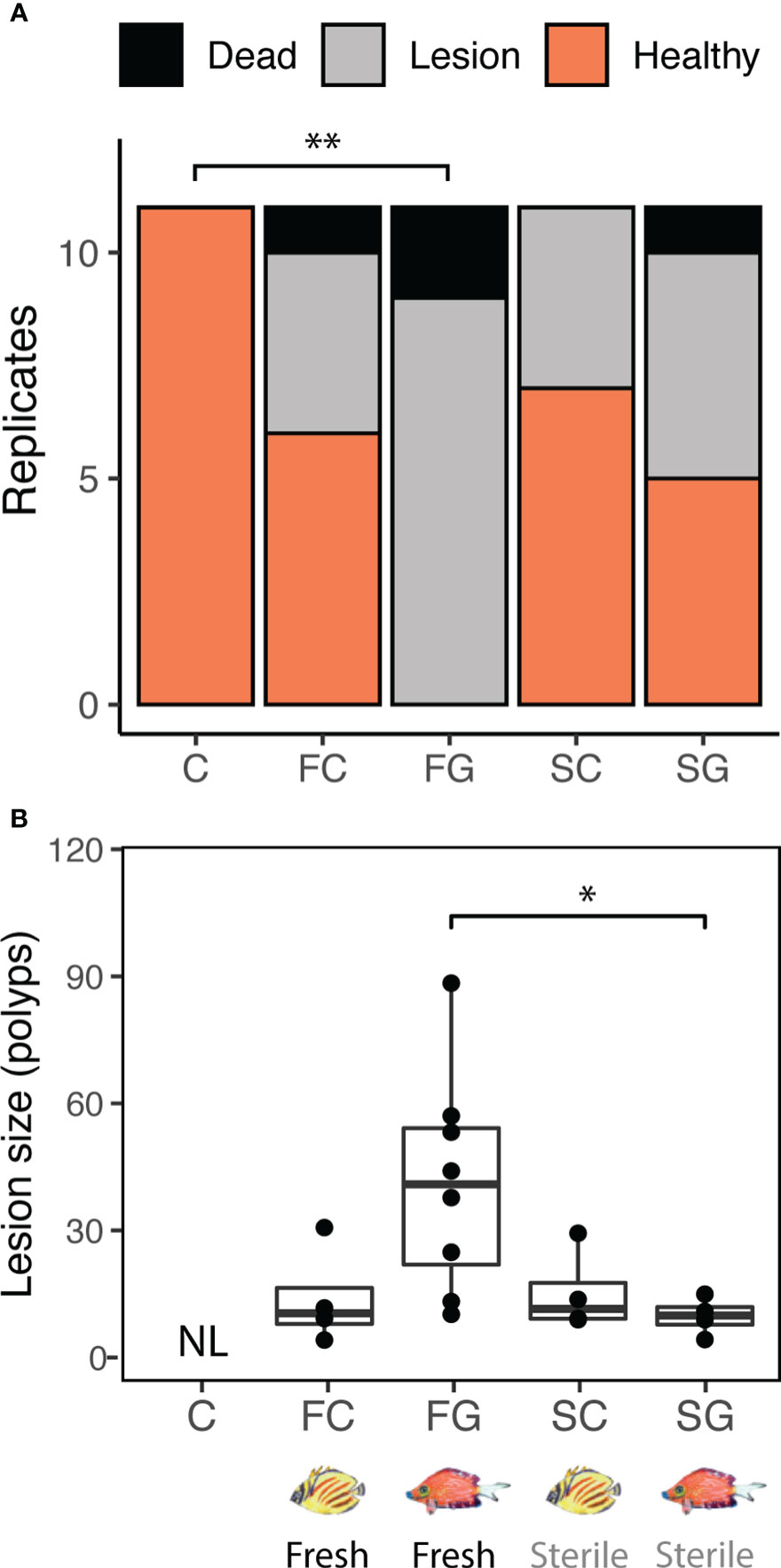
Figure 1 Fish species identity (corallivore or grazer/detritivore) influenced the effect of feces on coral tissue integrity (A) and lesion size (B). (A) Fresh grazer/detritivore feces caused lesions or mortality in all replicates (N = 11/11), whereas fresh corallivore feces only caused lesions or mortality in 45% of replicates (N = 5/11). Sterilized feces similarly caused lesions in 36% (N = 4/11; sterile corallivore feces) to 55% (N = 6/11; sterile grazer feces) of replicates (overall ordinal regression results: treatment χ2 = 32.3, p < 0.001; colony χ2 = 6.6, p = 0.761; **C-FG comparison: estimate = 2.97, p = 0.006). (B) Fresh grazer feces application caused lesions that were, on average, 4.2 times larger than sterile grazer feces (LM results: Treatment x2 = 13.49, Df = 3, p = 0.004; *pairwise comparison FG-SG: estimate = 1.33; adjusted p = 0.049, see Table S2 for individual comparisons). Together, these findings show that grazer feces are more likely to be biologically harmful to corals within 22h of contact than are corallivore feces. “NL”: No lesions observed.
Bacterial communities differed between environmental pools
To determine whether the microbiota in feces of the corallivore and the grazer/detritivore fish species used in the feces addition experiment are representative of microbiota at the guild-level (and may thus have similar effects on coral health), we quantified the bacterial diversity in feces of three fish trophic guilds (four species of obligate corallivore, three species of facultative corallivore, and three species of grazer/detritivore; see Table S1 for replication; Supplementary Data Files 3–5). Bacterial communities in fish feces were also compared to those in corals, algae, sediment and water to characterize the extent to which fish feces contain the microbial taxa associated with their main food sources (all samples combined, N = 160; see Table S1 for replication in each analysis).
Bacterial communities in all environmental pools (i.e., obligate and facultative corallivore feces, grazer/detritivore feces, corals, algae, sediment and water) were significantly different from one another (Figure 2; overall PERMANOVA results: R2 = 0.17, F(5, 107) = 4.1, p < 0.001, pairwise PERMANOVA results: R2 ≥ 0.08, F > 2.6, Bonferroni-adjusted p-values ≤ 0.005; see Table S3). Bacterial communities associated with many of the individual species/sample types also differed from each (e.g., bacteria in feces from C. ornatissimus versus those in Ctenochaetus flavicauda; Figure 2; overall test results R2 = 0.29, F = 3.3, p < 0.05). Out of all pairwise comparisons among species/sample types (Table S4; 210 pairwise comparisons total), most (189, 90%) were significant, including all pairwise comparisons between obligate corallivore species and grazer/detritivore species. Out of the pairwise comparisons that did not differ significantly (21, 10%), most comparisons (14/21) were among different types of algae, and the coral Acropora hyacinthus. Interestingly, several of the obligate and facultative corallivores did not differ significantly from each other (CHLU and CHOR, CHOR and CHRE, CHOR and CHSP; Table S4). Comparisons between two grazers and between a facultative corallivore and a grazer were also insignificant (CTST and ZESC, CHSP and CTST, respectively; Table S4).
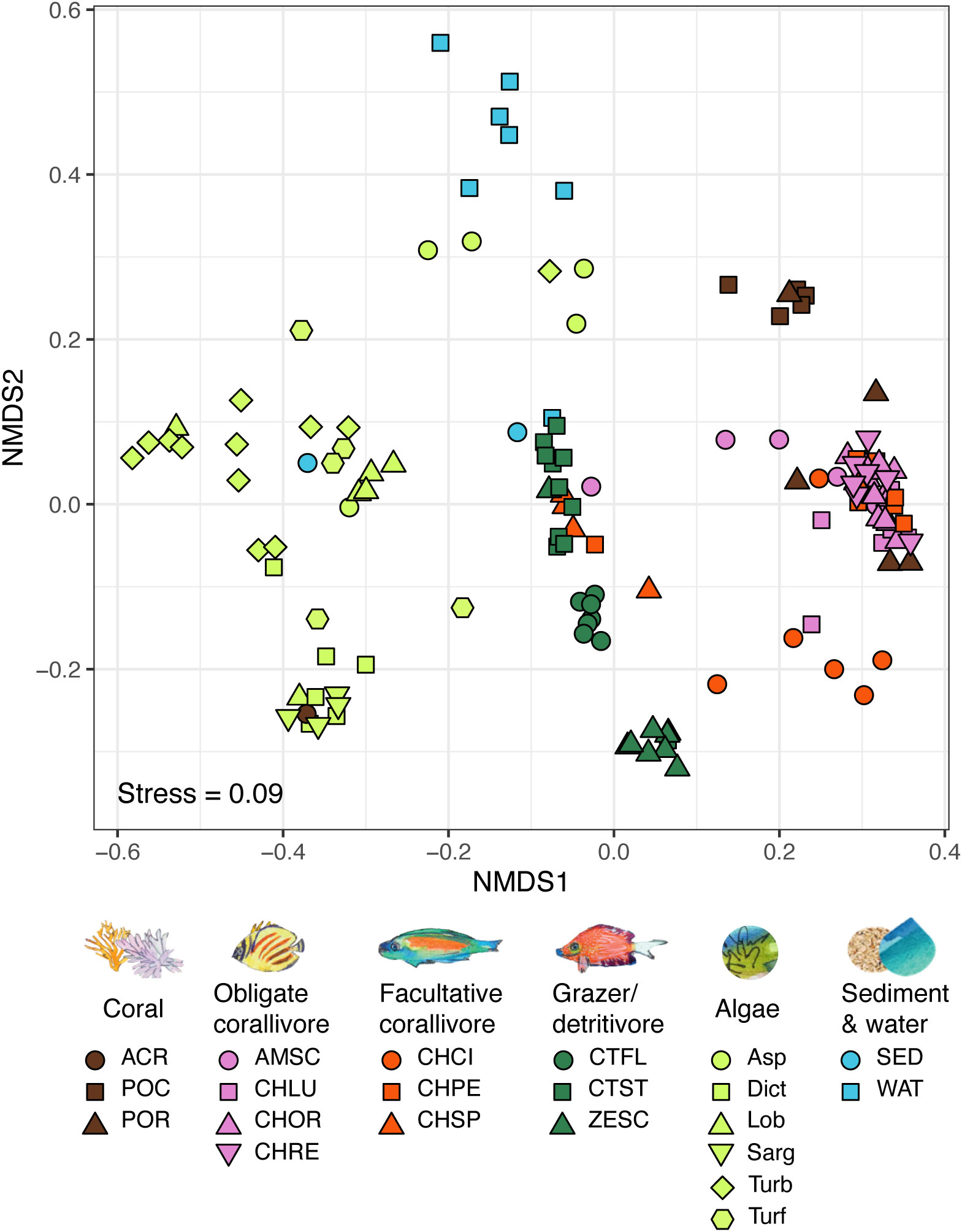
Figure 2 NMDS of bacterial communities based on the 16S rRNA gene in coral reef-associated environmental pools: obligate and facultative corallivore feces, grazer/detritivore feces, corals, algae, sediment and water (see Table S1 for sample sizes). Bacterial communities in all environmental pools differed significantly from each other (overall PERMANOVA results: R2 = 0.17, F(5, 107) = 4.1, p < 0.001, pairwise PERMANOVA results: R2 ≥ 0.08, F > 2.6, Bonferroni-adjusted p-values ≤ 0.005; see Table S3). Bacterial assemblages also significantly differed between most species/sample types (overall PERMANOVA results: R2 = 0.41, F(20,104) = 2.9, p < 0.001; see Table S4). PERMANOVA results are based on Bray-Curtis distances calculated from sequencing libraries rarefied to 1,009 reads per sample. See methods for species abbreviations.
Consumers disperse bacterial taxa associated with the resource that they consume
Feces of individual obligate corallivore species contained up to 92.1% of the distinct ASVs found in Porites lobata species complex (C. reticulatus; 223 of 242 ASVs), and up to 80.4% of ASVs found in Pocillopora species complex (C. reticulatus; 74 of 92 ASVs; Figure 3; Table S5). By comparison, grazer/detritivore feces contained only up to 9.5% of distinct ASVs associated with Porites lobata species complex (C. striatus; 23 of 242) and up to 6.5% of the ASVs identified in Pocillopora species complex (C. striatus; 6 of 92). ASVs associated with A. hyacinthus were much less frequently identified in corallivore feces (up to 2.7%; C. spilurus), but these corals were relatively uncommon at our study site during the sampling period and were not frequently consumed by corallivores (Grupstra et al., 2021).
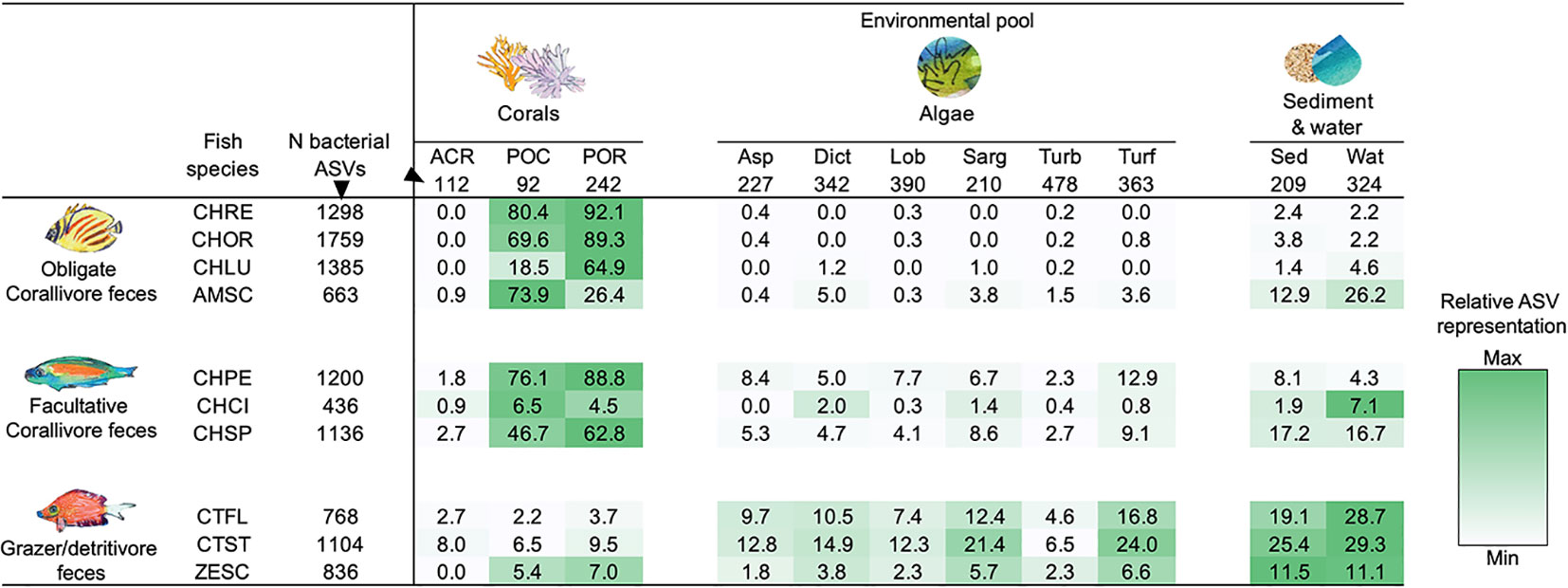
Figure 3 Coral reef fish disperse bacterial taxa associated with the resource that they consume. Listed are the fractions (%) of bacterial ASVs associated with corals, algae, and sediment and water that are also present in the feces of each fish species. Darker shading indicates higher representation of bacterial taxa; shading was scaled for each row (i.e., each consumer species) separately. All taxa with less than 100 reads were removed to exclude “transient” taxa, and all libraries were rarified to 1,008 reads. All samples per species or environmental pool were pooled (see Table S1 for sample sizes). “N bacterial ASVs” indicates the total number of unique ASVs in each environmental pool or fish species after rarefaction. See Table S5 for the number of shared ASVs. See methods text for species abbreviations.
Grazer/detritivore feces contained high representation of ASVs that were associated with different types of algae (Figure 3; Table S5). For example, 24.0% of turf algae ASVs were identified in feces of C. striatus (87 of 363 ASVs), whereas only up to 3.6% of turf algae ASVs were found in obligate corallivore feces (A. scopas; 13 of 363 ASVs). In addition, feces of the grazer/detritivores C. flavicauda and C. striatus contained higher numbers of the ASVs associated with all other categories of algae (up to 21.4%) than did feces of obligate (up to 5.0%) or facultative corallivores (up to 12.9%; Figure 3; Table S5). Feces of these species also contained more of the ASVs found in sediment and water samples (C. striatus; 25.4%, 53 of 209 ASVs and 29.3%, 95 of 324 ASVs), than did other trophic guilds (but note that the obligate corallivore A. scopas and the facultative corallivore C. spilurus also had high representation of ASVs found in sediment and water). Interestingly, while feces from the grazer/detritivore Z. scopas also contained more ASVs associated with algae (1.8-6.6%) than feces of the obligate corallivores (0.0-3.8%), the representation of algal ASVs in Z. scopas feces was noticeably lower than in feces of the other grazer/detritivores (4.6-16.8% for C. flavicauda; 6.5-24.0% for C. striatus). In fact, Z. scopas appeared more similar to two of the sampled facultative corallivores (C. pelewensis: 2.3-12.9% and C. spilurus: 2.7-9.1%; see Figure 3; Table S5) than to grazer/detritivores in terms of the ASVs it shared with algae.
Facultative corallivore feces contained a mix of ASVs associated with corals and algae. For example, C. pelewensis contained 88.8% of the ASVs found in Porites lobata species complex (215 of 242 ASVs; Figure 3; Table S5), as well as up to 12.9% of the ASVs associated with algal samples (47 of 363 ASVs associated with turf algae). Interestingly, feces of the facultative corallivore C. citrinellus contained fewer of the ASVs associated with corals or algae than other members of its guild; fecal samples of this species contained only 0.9-6.5% of ASVs associated with coral samples and 0.0-2.0% of ASVs associated with algae.
Relative abundances of the coral mutualist Endozoicomonadaceae were higher in feces of obligate corallivores than grazer/detritivores
Relative abundances of Endozoicomonadaceae reads in amplicon sequencing libraries significantly differed among environmental pools (Figures 4, S3; Kruskal-Wallis test, df = 5, χ2 = 96.2, p < 0.001). Pairwise tests demonstrated that mean ( ± SE) relative abundances of Endozoicomonadaceae reads were significantly higher (|Z| > 2.8, adjusted p-values < 0.05, see Table S6) in corals (44.3 ± 7.5%), obligate corallivore feces (24.4 ± 1.7%), and facultative corallivore feces (26.8 ± 2.6%) than in grazer feces (11.5 ± 0.9%), algae (3.7 ± 0.4%), or sediment and water (6.0 ± 1.3%). Relative Endozoicomonadaceae read abundances were also higher in feces of the obligate corallivore C. ornatissimus (24.7 ± 1.8) than the grazer/detritivore C. striatus (10.2 ± 1.5), which were the two species used for the feces addition experiment (Kruskal-Wallis test results: χ2 = 15.5, df = 1, p < 0.001). Relative read abundances of Endozoicomonadaceae further differed among species/sample types within environmental pools or trophic guilds (Figure S3; Table S7). For example, relative abundances of Endozoicomonadaceae reads in feces differed among obligate corallivore species; they were lower in feces of A. scopas than in feces of any other obligate corallivores sampled (Kruskal-Wallis test results: χ2 19.26, df = 3, p < 0.001; pairwise Dunn test results |Z| > 2.0, p < 0.05; see Table S7).
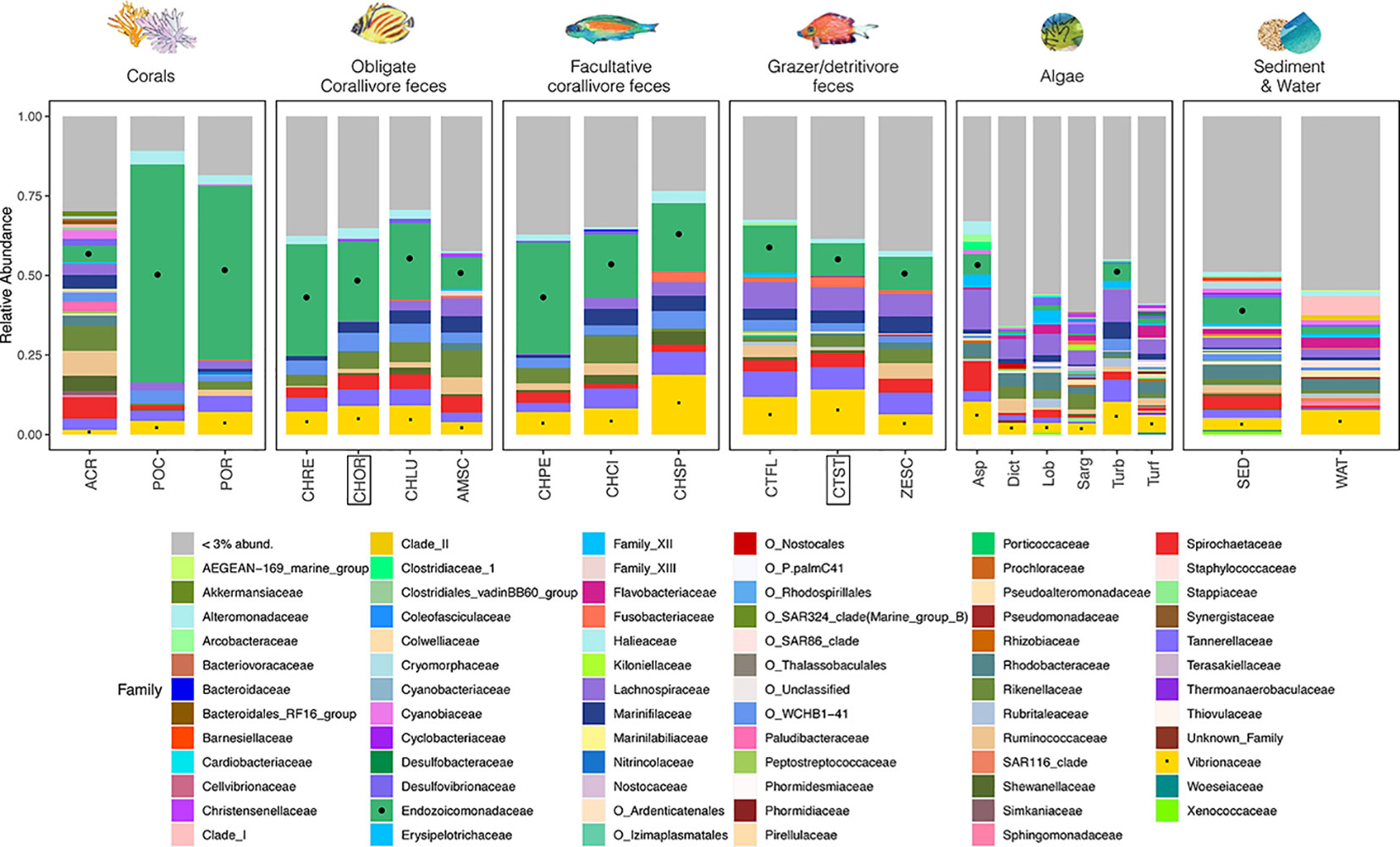
Figure 4 A stacked bar plot illustrates that relative abundances of bacterial families differ among environmental pools, consumer trophic guilds, and species (see Table S1 for sample sizes). Notable families include Endozoicomonadaceae (green, marked with black circles where possible), which are coral mutualists and (in this study) are abundant in the feces of obligate and facultative corallivores (see Figure S3; Tables S6, S7 for pairwise comparisons of relative abundances). The family Vibrionaceae (yellow, marked with black squares), which includes some notable coral pathogens, are present in all sample types. Fish species included in the feces addition experiment (CHOR, CTST; Figure 1) are indicated with boxes. Bacterial families that represent < 3% of total reads were collapsed (“< 3% abund.”). Taxa that are not classified at the family level are listed with their order names (indicated with “O_”).
Vibrio coralliilyticus relative abundances were higher in feces of grazer/detritivores than in corallivore feces
The Vibrio coralliilyticus metalloprotease gene was amplified using qPCR of DNA extracted from feces of each sampled fish species (Figure 5). Using this method, V. coralliiyticus genes were identified (Ct < 30; see Methods) in 54% (N = 20/37) of obligate corallivore feces, 50% of facultative corallivore feces (N = 9/18), and in 65% (N = 20/31) of grazer/detritivore feces. Relative abundances of V. coralliilyticus genes differed between fish guilds (ANOVA: F(2,45) = 16.14, p < 0.001; Supplementary Data File 6). A Tukey post-hoc test revealed that relative abundances of V. coralliilyticus genes were significantly higher in grazer/detritivore feces than in obligate corallivore feces (Figure 5; difference = 4.61, p < 0.001) and facultative corallivore feces (Figure 5; difference = 2.44, p = 0.046). Relative V. coralliilyticus abundances did not significantly differ between obligate and facultative corallivores (difference = 2.18, p = 0.09).
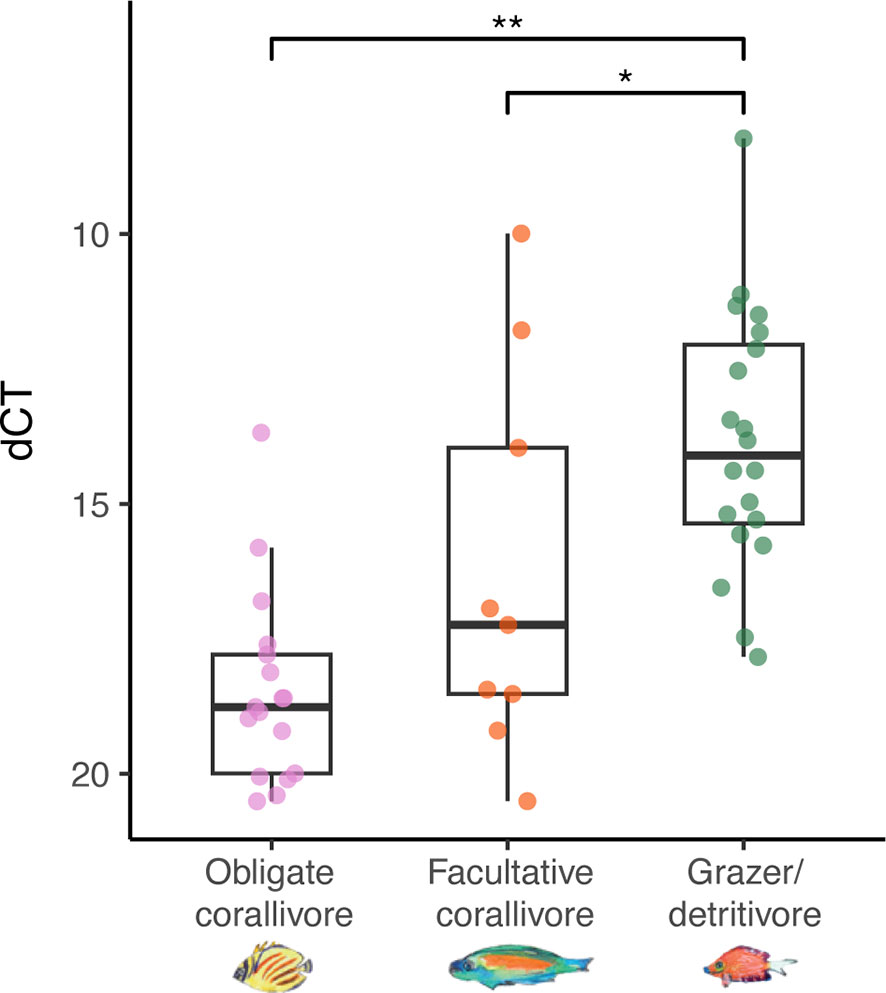
Figure 5 Relative abundances of Vibrio coralliilyticus metalloprotease genes were higher in grazer/detritivore feces than obligate corallivore and facultative corallivore feces (overall ANOVA results: F(2,45) = 16.14, p < 0.001). Delta cycle threshold (dCT) was calculated by subtracting the cycle threshold for the general bacteria assay from the V. coralliilyticus assay; therefore, samples with lower dCT values contained higher relative abundances of V. coralliilyticus. Amplification of the target gene at <30 cycles was counted as a positive detection; negative detections were not included in the analysis. Note that the y-axis has been inverted to make interpretation of the results more intuitive. P-values: * < 0.05; ** < 0.001.
Discussion
The production of waste products is ubiquitous across the animal kingdom; this waste is a key component of nutrient cycles (Roman and McCarthy, 2010; Le Mézo et al., 2022) as well as an important way in which microbiota are transported throughout environments (Grupstra et al., 2022). Animal diets determine the composition of waste products, including their associated microbiota, and thus influence how the waste may affect other organisms in the environment (Barron Pastor and Gordon, 2016; Reese and Dunn, 2018; Clever et al., 2022). Testing how feces derived from animal species in diverse trophic guilds affect cohabitating organisms can inform agriculture, ecosystem management, and restoration. Here, we demonstrate that feces from distinct trophic guilds of coral reef fish—grazer/detritivores and corallivores—affect the health of reef-building corals in distinct ways. We demonstrated that microbiota in grazer/detritivore, but not corallivore, feces are detrimental to coral health. Subsequent characterization of bacterial diversity in grazer/detritivore and corallivore feces indicated that these communities were trophic guild-specific, suggesting that results from the fecal addition experiment presented here are generalizable to trophic guild. Together, these findings highlight an underexplored, indirect result of consumers on resource species, which has potentially important ramifications for ecosystem health and functioning.
Microbiota in feces of grazer/detritivore fish, but not corallivorous fish, are detrimental to coral health
Intact coral reef ecosystems are characterized by high abundances of diverse fishes that release feces, containing up to 2.6×1011 bacterial cells g-1 (dry weight), as they move through their territories (Hughes, 1994; Hughes et al., 2003; Bellwood et al., 2004; Smriga et al., 2010). This frequent deposition of feces is likely to affect coral microbiota assembly, and thereby, colony health. The feces addition experiment presented here demonstrated that fresh feces from the grazer/detritivore C. striatus caused detrimental effects to coral health (higher frequency of lesions and larger lesions) compared to no-feces controls or sterilized feces from the same species (Figures 1B, 2; Table S2); this suggests that microbial activity in fresh grazer/detritivore feces is driving observed detrimental effects to coral health. This may be caused by higher relative abundances of pathogens like V. coralliiyticus in C. striatus feces, or the presence of other potential pathogens that may be associated with the algae that these fish feed on (Figures 3–5; Table S5; Nugues et al., 2004; Dinsdale et al., 2008; Sweet et al., 2013). By comparison, fresh feces from the obligate corallivore C. ornatissimus did not cause significantly larger lesions on corals than sterilized feces from the same species, suggesting that the microbiota in feces of this species were not detrimental to coral health (Figures 1, 2).
To test whether the findings from the feces addition experiment are broadly generalizable, we compared the bacterial assemblages in feces from 10 fish species in three trophic guilds: obligate corallivores, facultative corallivores, and grazer/detritivores. Bacterial communities significantly differed between trophic guilds, as well as between individual species within trophic guilds (Figures 2, 4; Tables S3, S4). While grazer/detritivore feces contained more bacterial taxa associated with algae (Figure 3; Table S5), most corallivore feces contained a higher percentage of coral-associated bacterial ASVs (Figure 3; Table S5) and higher relative abundances of the coral mutualist family Endozoicomonadaceae (Figures 4, S3; Tables S6, S7). Notably, grazer/detritivore feces also contained higher abundances of V. coralliiyticus (Figure 5), which is a known coral pathogen (Ben-Haim et al., 2003). Together, these lines of evidence strongly suggest that bacterial assemblages in fish feces are an important determinant of how feces affect coral health, and that feces from different fish trophic guilds are likely to impact coral health in distinct ways.
Herbivorous fishes support coral dominance through the removal of algal competitors, thereby contributing to coral reef health and resilience (Hughes et al., 2003; Littler et al., 2006; Burkepile and Hay, 2009; Littler et al., 2009). While the grazer/detritivores and facultative corallivores studied here (with the exception of C. spilurus) are not recognized as removing functionally important algae that compete for space and resources with corals, some grazer/detritivores (including C. striatus) graze on early successional turf algae and may thereby promote post-disturbance recovery of reefs (Marshell and Mumby, 2012; Tebbett et al., 2017a; Nalley et al., 2022). Additionally, some grazer/detritivores remove sediments from dense late successional algal turf assemblages (Goatley and Bellwood, 2010; Perry et al., 2022). This process may be vital to reef health because sediments in the algal matrix can inhibit coral recruitment (Birrell et al., 2005; Fabricius, 2005). Removal of sediments from algal turfs also increases the palatability of the algae for other herbivores, increasing top-down control on algal communities (Goatley and Bellwood, 2010; Tebbett et al., 2017a; Tebbett et al., 2017b; Tebbett et al., 2018). While grazer/detritivores and other herbivores are thus functionally important, our research (as well as two previous studies, Ezzat et al., 2019; Ezzat et al., 2021) illuminates an additional dimension of this relationship: grazer/detritivores can disperse pathogens in their feces, which can cause lesions in coral tissues. More work is needed to test the extent to which different (herbivorous) fish species contribute to the dispersal of viable coral pathogens (Grupstra et al., 2022).
Biotic and abiotic factors mediating coral-feces interaction frequencies and outcomes
Corals may acquire nutrients (Coma et al., 2001; Mills and Sebens, 2004; Allgeier et al., 2017; Shantz et al., 2023) and beneficial or harmful microbiota from fish feces (Muller Parker, 1984; Castro-Sanguino and Sánchez, 2012; Ezzat et al., 2019; Umeki et al., 2020; Ezzat et al., 2021; Grupstra et al., 2021; Velásquez-Rodríguez et al., 2021). As shown here (sterile feces treatments in Figure 1), particulate matter in fish feces may also sometimes smother coral polyps, resulting in mortality of the underlying coral tissue (Rogers, 1990; Krone et al., 2011). The frequency and nature of interactions between corals and fish feces are additionally influenced by biotic and abiotic factors. Examples of potentially relevant biotic factors include fish species’ range sizes and spatiotemporal foraging and defecation patterns (Shantz et al., 2015). Relevant abiotic factors include water flow and wave action (Wisnoski and Lennon, 2022), which can move fecal pellets onto or away from corals and may affect the consistency and integrity of fecal pellets.
The distribution of feces by fish varies based on species- and individual-level traits (e.g., territoriality, defecation frequency). Many reef fish are territorial, and the area over which these fish disperse feces is influenced by factors affecting territory size, including species identity, location, depth, and competition with other fish (Wrathall et al., 1992; Cox, 1994; Pratchett et al., 2014). For example, many of the corallivores examined here (obligate corallivores: C. lunulatus, C. ornatissimus, C. reticulatus; facultative corallivores: C. pelewensis, C. citrinellus) are butterflyfishes, many of which live in mating pairs and maintain and defend territories that are often small in size (50 m2 or larger) but can range up to 1,000 m2 depending on the species and location (Wrathall et al., 1992; Cox, 1994; Pratchett et al., 2014). Other butterflyfishes, including C. lunulatus, are less restricted in their movements and may travel between and through territories of other fish (Pratchett, 2005). Mature individuals of the facultative corallivorous parrotfish C. spilurus are solitary and also territorial, but their territories are generally larger (mean territory size ~250m2 in Moorea; Davis et al., 2017). Home ranges for grazer/detritivores vary strongly by species; C. striatus territories can be small (8-16 m2; Krone et al., 2008; Goatley and Bellwood, 2010), whereas territories of some Zebrasoma species can be large (up to 0.6 km2; Claisse et al., 2009; Giffin et al., 2019).
Defecation frequencies and patterns also differ among species. Three of the butterflyfish species in this study were observed to defecate around once per hour (every 45-60 minutes; Grupstra et al., 2021), whereas parrotfishes and some grazer/detritivores may defecate more than four times per hour (Goatley and Bellwood, 2010; Grupstra et al., unpublished data). Latrine use also likely affects the distribution of fish feces (Polunin and Koike, 1987; Nicholson and Sikkel, 2018). For example, C. striatus individuals have been observed to defecate in small sandy areas at the edge of their territories (Krone et al., 2008; Goatley and Bellwood, 2010). They have also been documented to defecate on coral heads at our study site (Ezzat et al., 2019; Grupstra et al., unpublished data), increasing the likelihood of interactions between corals and fish feces. Studies integrating fish species-specific information on fecal qualities and their influence on coral health with spatiotemporal patterns of defecation are needed.
Although it has received relatively little research attention, coprophagy by fish appears common on coral reefs and may reduce the frequency of interactions between corals and fish feces. Reports from the Pacific and Caribbean suggest that 0-92% of the feces released by some abundant reef fish may be eaten by other fish (Bailey and Robertson, 1982; Robertson, 1982; Rempel et al., 2022). The probability that a given fecal pellet will be consumed relates to its nutritional content; protein and lipid-rich feces from some zooplanktivores and other invertebrate feeders (including some obligate corallivores) may be consumed at higher rates than carbohydrate-rich feces from herbivores (Bailey and Robertson, 1982; Rempel et al., 2022). Despite high reported levels of coprophagy from some reef regions, recent observations from Moorea suggest that some (partial) segments of fish feces landed on coral heads in ~91% (20 of 22) of observed defecations (Grupstra et al., 2021). Additional species-specific efforts to quantify coprophagy will improve our understanding of the frequencies at which fish feces come into contact with corals.
The physical integrity of fecal pellets also likely influences the outcome of interactions between corals and fish feces. In our fecal addition experiment, feces placed on coral fragments mimicked intact feces, yet in situ, fecal pellets sometimes fall apart in the water column (Grupstra et al., 2021), releasing silt to rice grain-sized particles. Such particle sizes may be less likely to smother coral polyps, and might be more readily removed or ingested by corals (Hankins et al., 2018; Grillo et al., 2021); this may be especially the case in areas with high water current or wave action, where fecal pellets are more likely to disintegrate. In our experiment, we did not explicitly examine whether coral polyps consumed parts of fecal pellets. Future studies of coprophagy behavior by Pocillopora spp., and corals in general, are needed to further our understanding of how contact with consumer feces influences coral health.
Finally, many branching corals harbor diverse macrosymbionts including brittle stars, Christmas tree worms, crabs, and small fish (e.g., coral gobies) that can promote coral health (Stier et al., 2012; Doo et al., 2018; Moeller et al., 2023), and these organisms may interact with fish feces that land on coral heads. We observed Trapezia crabs, common macrosymbionts of branching corals, breaking up and consuming corallivore feces applied to coral fragments in situ as well as in the lab (Grupstra et al. personal observation, but note that Trapezia crabs were removed from coral fragments in this experiment). Through such activity, macrosymbionts may in some cases prevent lesions from forming when feces fall directly on coral surfaces. Empirical tests of the extent to which fecal particle size and macrosymbiont activity influence the outcomes of fish fecal contact with corals are needed.
Corallivores promote the dispersal of coral-associated microbiota
Through the dispersal of microbiota, consumer activities affect the assembly of microbiota in resource species (Vašutová et al., 2019; Ezzat et al., 2020; Vannette, 2020; Zhu et al., 2021; Grupstra et al., 2022; Fowler et al., 2023). Here, we show that feces from reef fish contain a high diversity of bacterial taxa (ASVs) that are associated with their food sources. For instance, obligate corallivore feces contained up to 92.1% of the bacterial ASVs found in a locally abundant coral species (Porites lobata species complex; Figure 3; Table S5). We therefore posit that the presence and activity of corallivores promotes the dispersal of bacterial taxa associated with corals. Based on experimental demonstration of the impact of microbial activity on coral health (this study; Ezzat et al., 2019; Ezzat et al., 2021), we infer that at least some bacteria in fish feces are viable. Empirical tests of this (e.g., through culturing, or membrane permeability-based stains and cell sorting or viability PCR; Grupstra et al., 2022) will help reveal the extent to which passage through fish digestive tracts affects the viability of microbiota dispersed in the feces of reef fish.
While some corals provide their offspring with key bacteria (Sharp et al., 2012; Leite et al., 2017; Zhou et al., 2017), most corals acquire at least some, or most, microbial taxa de novo each generation from the environment (Sharp et al., 2010; Apprill et al., 2012; Epstein et al., 2019; Strader et al., 2022). In many coral species, juveniles may take up diverse microbial taxa from environmental pools, followed by a winnowing process in which microbial assemblages are “refined” (Apprill et al., 2009; Littman et al., 2009; Lema et al., 2014; Damjanovic et al., 2019; Epstein et al., 2019; Van Oppen and Blackall, 2019). Corallivore feces may represent hotspots of homologous bacteria and facilitate the acquisition of locally beneficial microbiota by corals and other hosts. While the transfer of beneficial bacteria between coral colonies via fish feces has not been demonstrated, earlier studies have shown that anemones in the genus Aiptasia—which are closely related to corals—can take up Symbiodiniaceae cells from the feces of obligate corallivorous butterflyfishes (Muller Parker, 1984); coral juveniles have also been demonstrated to initiate symbiotic partnerships with Symbiodiniaceae cells from the feces of giant clams (Umeki et al., 2020). Hence, it is likely that juvenile corals can acquire beneficial bacteria from fish feces as well.
We hypothesize that the dispersal of coral-associated bacteria by corallivores is beneficial to stressed adult coral colonies in some cases. Environmental stress can result in disruption of the symbiosis between coral animals and their populations of microorganisms (Ziegler et al., 2016; Grottoli et al., 2018; but see Pogoreutz et al., 2018; Ziegler et al., 2019), yet experimental inoculations with engineered “probiotic” cocktails consisting of cultured coral-derived bacteria and Symbiodiniaceae have increased photosynthetic efficiency and colony health in thermally stressed corals within hours to days after inoculation, potentially by providing nutrition, mitigating toxins and/or deterring pathogens (Peixoto et al., 2017; Morgans et al., 2019; Rosado et al., 2019; Peixoto et al., 2021; Santoro et al., 2021). For example, inoculations with microbiota from heat tolerant coral colonies were shown to reduce bleaching rates in recipient heat-susceptible colonies of the coral species Pocillopora sp. and Porites sp. in Thailand (Doering et al., 2021). Given that corallivore feces contained many of the bacterial ASVs associated with locally abundant corals in this study, and previous work has shown that such feces contain high densities of live coral-associated Symbiodiniaceae cells (Muller Parker, 1984; Castro-Sanguino and Sánchez, 2012; Grupstra et al., 2021), microbiota in corallivore feces may in some cases be expected to have a “stabilizing” effect on the partnership between corals and their microbiomes. This is especially likely given reports that some corallivorous fish preferentially feed on heat tolerant corals during bleaching events (Cole et al., 2009; MacDonald et al., 2021; Rotjan et al., 2022), thereby potentially transferring microbiota associated with stress-resistant colonies to surrounding stress-susceptible colonies. While this experiment did not generate evidence that microbiota in corallivore feces improved coral health (e.g., photosynthetic efficiency was not increased by corallivore feces addition; Figure S2), additional empirical studies are needed to comprehensively test this hypothesis under varied conditions (e.g., size and integrity of fecal pellets, water flow rate, presence/absence of ectosymbionts), as well as with experimentally stressed (e.g., bleaching, diseased) coral colonies.
Conclusions
Our results indicate that microbiota in the feces of grazer/detritivores, but not corallivores, have negative effects on coral health when they come in direct contact with colonies. Feces from grazer/detritivore fish contain higher proportions of bacterial ASVs associated with locally abundant algae, as well as higher relative abundances of some coral pathogens, than feces from corallivores. These findings add a new dimension to our understanding of how fish trophic guilds influence coral health: although grazer/detritivores may support coral dominance in various ways (e.g., promoting coral recruitment, as well as herbivory), they also disperse pathogens that can harm coral health. Corallivore predation can wound corals, yet corallivores also disperse bacteria in their feces that may be beneficial to corals. Future studies are needed to test how consumers in additional fish trophic guilds affect coral health, as well as to disentangle how global stressors impact the outcomes of interactions between corals and fish feces. More broadly, studying how different trophic guilds of consumers contribute to the dispersal of microbiota can aid ecosystem management and conservation, for example through the identification of consumer trophic guilds that promote or harm resource species in different environmental contexts.
Data availability statement
Sequence reads have been uploaded to NCBI’s sequence read archive (SRA) under BioProject PRJNA935035; all other data have been included as supplementary data files. R code to replicate the analyses has been deposited on GitHub: https://github.com/CorreaLab/graco.
Ethics statement
The animal study was reviewed and approved by The institutional animal care and use committee (IACUC) at Rice University [IACUC-21-019].
Author contributions
CG and AC designed the experiment, with input from AV, LH-K, and SC; all authors contributed to data collection; CG and JM processed samples; CG led data analysis, with contributions from all authors; CG wrote the first draft of the manuscript, with contributions by all authors; AC and CG contributed funds for the study. All authors contributed to the article and approved the submitted version.
Funding
This study was made possible through U.S. National Science Foundation awards (OCE #2145472 and #1635798), Rice University start-up funds to AC and fellowships awarded by the Wagoner Foreign Study Scholarship Program to CG, JM, LH-K, and AV. The Kirk W. Dotson Endowed Graduate Fellowship in Ecology and Evolutionary Biology also supported CG, LH-K and AV in completing this work.
Acknowledgments
We thank Dr. Kory Evans, JoJo West, Sean Trainor, Moana Le Rohellec and Dennis Conetta for their help with fish collections and surveys, and the Richard B. Gump Station staff for logistical assistance. We further thank Dr. Amanda Shore for providing the Vibrio coralliilyticus cultures used for the qPCR standard curve, and Dr. Deron Burkepile for providing some materials used in this work. We also thank Julianna Renzi for discussion regarding coral ectosymbionts that informed the interpretation of some of our findings. Additionally, we thank Logan Mullen from the University of Alaska, Fairbanks, for troubleshooting the library preparation and sequencing of the samples included in the manuscript. Lastly, we would like to thank Cyrus Washington, Jake Sperry, and Sara Emami for their contributions to lab work, and Daniel Gorczynski for feedback on the manuscript. This research was funded by a U.S. National Science Foundation Grant OCE 22-24354 (and earlier awards) to the Moorea Coral Reef LTER as well as a generous gift from the Gordon and Betty Moore Foundation. Research was completed under permits issued by the Territorial Government of French Polynesia (Deíleígation à la Recherche) and the Haut-Commissariat de la Reípublique en Polyneísie Francaise (DTRT) (MCR LTER Protocole d’Accueil 2005–2022; Adrienne Correa Protocole d’Accueil 2013-2019), and we thank the Deíleígation à la Recherche and DTRT for their continued support. This work represents a contribution of the Moorea Coral Reef (MCR) LTER Site. All samples were collected under approval of the institutional animal care and use committee (IACUC) at Rice University [IACUC-21-019]. Many thanks to Janavi Mahimtura Folmsbee for designing and creating an artwork used in the manuscript figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1110346/full#supplementary-material
Supplementary Data Files 1 and 2 | Data pertaining to the lesion frequencies and sizes, respectively (Figures 1A, B).
Supplementary Data Files 3–5 | A sample data table, an ASV table and a taxonomy table, respectively, from the 16S V4 rDNA ampli consequencing analysis (Figures 2–4) that can be combined into a phyloseq object. Provided data arepost quality control and filtering of potential contaminant sequences.
Supplementary Data File 6 | qPCR data (Figure 5).
Supplementary Data File 7 | All supplementary tables and figures.
References
Aeby G. S., Santavy D. L. (2006). Factors affecting susceptibility of the coral montastrea faveolata to black-band disease. Mar. Ecol. Prog. Ser. 318, 103–110. doi: 10.3354/meps318103
Allgeier J. E., Burkepile D. E., Layman C. A. (2017). Animal pee in the sea: Consumer-mediated nutrient dynamics in the world’s changing oceans. Global Change Biol. 23, 2166–2178. doi: 10.1111/gcb.13625
Anderson M. J., Walsh D. C. I. (2013). PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 83, 557–574. doi: 10.1890/12-2010.1
Apprill A., Marlow H. Q., Martindale M. Q., Rappé M. S. (2009). The onset of microbial associations in the coral pocillopora meandrina. ISME J. 3, 685–699. doi: 10.1038/ismej.2009.3
Apprill A., Marlow H. Q., Martindale M. Q., Rappé M. S. (2012). Specificity of associations between bacteria and the coral pocillopora meandrina during early development. Appl. Environ. Microbiol. 78, 7467–7475. doi: 10.1128/AEM.01232-12
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Bailey T. G., Robertson D. R. (1982). Organic and caloric levels of fish feces relative to its consumption by coprophagous reef fishes. Mar. Biol. 69, 45–50. doi: 10.1007/BF00396959
Barron Pastor H. J., Gordon D. M. (2016). Effects of dispersal limitation in the face of intense selection via dietary intervention on the faecal microbiota of rats. Environ. Microbiol. Rep. 8, 187–195. doi: 10.1111/1758-2229.12367
Bellwood D. R., Hughes T. P., Folke C., Nyström M. (2004). Confronting the coral reef crisis. Nature 429, 827–833. doi: 10.1038/nature02691
Ben-Haim Y., Zicherman-Keren M., Rosenberg E. (2003). Temperature-regulated bleaching and lysis of the coral pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. doi: 10.1128/AEM.69.7.4236-4242.2003
Birrell C. L., McCook L. J., Willis B. L. (2005). Effects of algal turfs and sediment on coral settlement. Mar. pollut. Bull. 51, 408–414. doi: 10.1016/j.marpolbul.2004.10.022
Bosch T. C. G., McFall-Ngai M. J. (2011). Metaorganisms as the new frontier. Zoology 114, 185–190. doi: 10.1016/j.zool.2011.04.001
Burkepile D. E., Hay M. E. (2009). Nutrient versus herbivore control of macroalgal community development and coral growth on a Caribbean reef. Mar. Ecol. Prog. Ser. 389, 71–84. doi: 10.3354/meps08142
Burkepile D. E., Shantz A. A., Adam T. C., Munsterman K. S., Speare K. E., Ladd M. C., et al. (2020). Nitrogen identity drives differential impacts of nutrients on coral bleaching and mortality. Ecosystems 23, 798–811. doi: 10.1007/s10021-019-00433-2
Burns A. R., Miller E., Agarwal M., Rolig A. S., Milligan-Myhre K., Seredick S., et al. (2017). Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl. Acad. Sci. United States America 114, 11181–11186. doi: 10.1073/pnas.1702511114
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castro-Sanguino C., Sánchez J. A. (2012). Dispersal of Symbiodinium by the stoplight parrotfish Sparisoma viride. Biol. Lett. 8, 282–286. doi: 10.1098/rsbl.2011.0836
Chen C. P., Tseng C. H., Chen C. A., Tang S. L. (2011). The dynamics of microbial partnerships in the coral Isopora palifera. ISME J. 5, 728–740. doi: 10.1038/ismej.2010.151
Claisse J. T., McTee S. A., Parrish J. D. (2009). Effects of age, size, and density on natural survival for an important coral reef fishery species, yellow tang, Zebrasoma flavescens. Coral Reefs 28, 95–105. doi: 10.1007/s00338-008-0447-7
Clever F., Sourisse J. M., Preziosi R. F., Eisen J. A., Guerra E. C. R., Scott J. J., et al. (2022). The gut microbiome variability of a butterflyfish increases on severely degraded Caribbean reefs. Commun. Biol. 5, 770. doi: 10.1038/s42003-022-03679-0
Cole A. J., Pratchett M. S., Jones G. P. (2009). Effects of coral bleaching on the feeding response of two species of coral-feeding fish. J. Exp. Mar. Biol. Ecol. 373, 11–15. doi: 10.1016/j.jembe.2009.02.016
Coma R., Ribes M., Gili J., Hughes R. (2001). The ultimate opportunists: Consumers of seston. Mar. Ecol. Prog. Ser. 219, 305–308. doi: 10.3354/meps219305
Cox E. F. (1994). Resource use by corallivorous butterflyfishes (family chaetodontidae) in Hawaii. Bull. Mar. Sci. 54, 535–545.
Damjanovic K., Van Oppen M. J. H., Menéndez P., Blackall L. L. (2019). Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01702
Davis K., Carlson P. M., Bradley D., Warner R. R., Caselle J. E. (2017). Predation risk influences feeding rates but competition structures space use for a common pacific parrotfish. Oecologia 184, 139–149. doi: 10.1007/s00442-017-3857-9
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226. doi: 10.1186/s40168-018-0605-2
Dinsdale E. A., Pantos O., Smriga S., Edwards R. A., Angly F., Wegley L., et al. (2008). Microbial ecology of four coral atolls in the northern line islands. PloS One 3, e1584. doi: 10.1371/journal.pone.0001584
Doering T., Wall M., Putchim L., Rattanawongwan T., Schroeder R., Hentschel U., et al. (2021). Towards enhancing coral heat tolerance: A “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome 9, 102. doi: 10.1186/s40168-021-01053-6
Doo S. S., Carpenter R. C., Edmunds P. J. (2018). Obligate ectosymbionts increase the physiological resilience of a scleractinian coral to high temperature and elevated pCO2. Coral Reefs 37, 997–1001. doi: 10.1007/s00338-018-1731-9
Epstein H. E., Torda G., Munday P. L., van Oppen M. J. H. (2019). Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. ISME J. 13, 1635–1638. doi: 10.1038/s41396-019-0358-3
Ezzat L., Lamy T., Maher R. L., Munsterman K. S., Landfield K., Schmeltzer E. R., et al. (2019). Surgeonfish feces increase microbial opportunism in reef-building corals. Mar. Ecol. Prog. Ser. 631, 81–97. doi: 10.3354/meps13119
Ezzat L., Lamy T., Maher R. L., Munsterman K. S., Landfield K. M., Schmeltzer E. R., et al. (2020). Parrotfish predation drives distinct microbial communities in reef-building corals. Anim. Microbiome 2, 1–15. doi: 10.1186/s42523-020-0024-0
Ezzat L., Merolla S., Clements C. S., Munsterman K. S., Landfield K., Stensrud C., et al. (2021). Thermal stress interacts with surgeonfish feces to increase coral susceptibility to dysbiosis and reduce tissue regeneration. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.620458
Fabricius K. E. (2005). Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar. pollut. Bull. 50, 125–146. doi: 10.1016/j.marpolbul.2004.11.028
Forsman Z. H., Barshis D. J., Hunter C. L., Toonen R. J. (2009). Shape-shifting corals: Molecular markers show morphology is evolutionarily plastic in Porites. BMC Evolutionary Biol. 9, 45. doi: 10.1186/1471-2148-9-45
Forsman Z., Wellington G. M., Fox G. E., Toonen R. J. (2015). Clues to unraveling the coral species problem: Distinguishing species from geographic variation in porites across the pacific with molecular markers and microskeletal traits. PeerJ 3, e751–e751. doi: 10.7717/peerj.751
Fowler J. C., Donald M. L., Bronstein J. L., Miller T. E. X. (2023). The geographic footprint of mutualism: How mutualists influence species’ range limits. Ecological Monographs 93, e1558. doi: 10.1002/ecm.1558
Franco A., Rückert C., Blom J., Busche T., Reichert J., Schubert P., et al. (2020). High diversity of Vibrio spp. associated with different ecological niches in a marine aquaria system and description of Vibrio aquimaris sp. nov. Systematic Appl. Microbiol. 43, 126123. doi: 10.1016/j.syapm.2020.126123
Gélin P., Postaire B., Fauvelot C., Magalon H. (2017). Molecular phylogenetics and evolution reevaluating species number, distribution and endemism of the coral genus Pocillopora lamarck 1816 using species delimitation methods and microsatellites. Mol. Phylogenet. Evol. 109, 430–446. doi: 10.1016/j.ympev.2017.01.018
Giffin A. L., Rueger T., Jones G. P. (2019). Ontogenetic shifts in microhabitat use and coral selectivity in three coral reef fishes. Environ. Biol. Fish 102, 55–67. doi: 10.1007/s10641-019-0842-7
Goatley C., Bellwood D. (2010). Biologically mediated sediment fluxes on coral reefs: Sediment removal and off-reef transportation by the surgeonfish Ctenochaetus striatus. Mar. Ecol. Prog. Ser. 415, 237–245. doi: 10.3354/meps08761
Gorczynski D., Beaudrot L. (2021). Functional diversity and redundancy of tropical forest mammals over time. Biotropica 53, 51–62. doi: 10.1111/btp.12844
Grillo J. F., Sabino M. A., Ramos R. (2021). Short-term ingestion and tissue incorporation of polystyrene microplastic in the scleractinian coral Porites porites. Regional Stud. Mar. Sci. 43, 101697. doi: 10.1016/j.rsma.2021.101697
Grottoli A. G., Dalcin Martins P., Wilkins M. J., Johnston M. D., Warner M. E., Cai W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PloS One 13, e0191156–e0191156. doi: 10.1371/journal.pone.0191156
Grupstra C. G. B., Lemoine N. P., Cook C., Correa A. M. S. (2022). Thank you for biting: Dispersal of beneficial microbiota through “antagonistic” interactions. Trends Microbiol 30 (10), 930–939. doi: 10.1016/j.tim.2022.03.006
Grupstra C. G. B., Rabbitt K. M., Howe-Kerr L. I., Correa A. M. S. (2021). Fish predation on corals promotes the dispersal of coral symbionts. Anim. Microbiome 3, 25. doi: 10.1101/2020.08.10.243857
Hankins C., Duffy A., Drisco K. (2018). Scleractinian coral microplastic ingestion: Potential calcification effects, size limits, and retention. Mar. pollut. Bull. 135, 587–593. doi: 10.1016/j.marpolbul.2018.07.067
Harmelin-Vivien M. L., Bouchon-Navaro Y. (1983). Feeding diets and significance of coral feeding among chaetodontid fishes in moorea (French Polynesia). Coral Reefs: J. Int. Soc. Reef Stud. 2, 119–127. doi: 10.1007/BF02395282
Hughes T. P. (1994). Catastrophes, phase shifts, and Large-scale degradation of a Caribbean coral reef. Science 265, 1547–1551. doi: 10.1126/science.265.5178.1547
Hughes T., Baird A. H., Bellwood D. R., Card M., Connolly S. R., Folke C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Johnston E. C., Forsman Z. H., Toonen R. J. (2018). A simple molecular technique for distinguishing species reveals frequent misidentification of Hawaiian corals in the genus Pocillopora. PeerJ 6, e4355. doi: 10.7717/peerj.4355
Krone R., Bshary R., Paster M., Eisinger M., van Treeck P., Schuhmacher H. (2008). Defecation behaviour of the lined bristletooth surgeonfish Ctenochaetus striatus (Acanthuridae). Coral Reefs 27, 619–622. doi: 10.1007/s00338-008-0365-8
Krone R., Paster M., Schuhmacher H. (2011). Effect of the surgeonfish Ctenochaetus striatus (Acanthuridae) on the processes of sediment transport and deposition on a coral reef in the red Sea. Facies 57, 215–221. doi: 10.1007/s10347-010-0239-8
Leite D. C. A., Leão P., Garrido A. G., Lins U., Santos H. F., Pires D. O., et al. (2017). Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00176
Lema K. A., Bourne D. G., Willis B. L. (2014). Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23, 4682–4695. doi: 10.1111/mec.12899
Le Mézo P., Guiet J., Scherrer K., Bianchi D., Galbraith E. (2022). Global nutrient cycling by commercially targeted marine fish. Biogeosciences 19, 2537–2555. doi: 10.5194/bg-19-2537-2022
Littler M. M., Littler D. S., Brooks B. L. (2006). Harmful algae on tropical coral reefs: Bottom-up eutrophication and top-down herbivory. Harmful Algae 5, 565–585. doi: 10.1016/j.hal.2005.11.003
Littler M. M., Littler D. S., Brooks B. L. (2009). Herbivory, nutrients, stochastic events, and relative dominances of benthic indicator groups on coral reefs: A review and recommendations. Smithsonian Contributions to Mar. Sci. 38, 401–414. doi: 10.5479/si.01960768.38.401
Littman R., Willis B., Bourne D. (2009). Bacterial communities of juvenile corals infected with different symbiodinium (dinoflagellate) clades. Mar. Ecol. Prog. Ser. 389, 45–59. doi: 10.3354/meps08180
MacDonald C., Pinheiro H. T., Shepherd B., Phelps T. A. Y., Rocha L. A. (2021). Disturbance and distribution gradients influence resource availability and feeding behaviours in corallivore fishes following a warm-water anomaly. Sci. Rep. 11, 23656. doi: 10.1038/s41598-021-03061-w
Marshell A., Mumby P. J. (2012). Revisiting the functional roles of the surgeonfish Acanthurus nigrofuscus and Ctenochaetus striatus. Coral Reefs 31, 1093–1101. doi: 10.1007/s00338-012-0931-y
McFall-Ngai M., Hadfield M. G., Bosch T. C. G., Carey H. V., Domazet-Lošo T., Douglas A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. 110, 3229–3236. doi: 10.1073/pnas.1218525110
McIlroy S. E., Gillette P., Cunning R., Klueter A., Capo T., Baker A. C., et al. (2016). The effects of symbiodinium (Pyrrhophyta) identity on growth, survivorship, and thermal tolerance of newly settled coral recruits. J. Phycology 52, 1114–1124. doi: 10.1111/jpy.12471
Mills M. M., Sebens K. P. (2004). Ingestion and assimilation of nitrogen from benthic sediments by three species of coral. Mar. Biol. 145, 1097–1106. doi: 10.1007/s00227-004-1398-3
Moeller H. V., Nisbet R. M., Stier A. C. (2023). Cascading benefits of mutualists’ predators on foundation species: A model inspired by coral reef ecosystems. Ecosphere 14, e4382. doi: 10.1002/ecs2.4382
Morgans C. A., Hung J. Y., Bourne D. G., Quigley K. M. (2019). Symbiodiniaceae probiotics for use in bleaching recovery. Restor. Ecology 28 (2), 282–288. doi: 10.1111/rec.13069
Muller Parker G. (1984). Dispersal of zooxanthellae on coral reefs by predators on cnidarians. Biol. Bull. 167, 159–167.
Nalley E. M., Donahue M. J., Heenan A., Toonen R. J. (2022). Quantifying the diet diversity of herbivorous coral reef fishes using systematic review and DNA metabarcoding. Environ. DNA 4, 191–205. doi: 10.1002/edn3.247
Neave M. J., Apprill A., Ferrier-Pagès C., Voolstra C. R. (2016). Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl. Microbiol. Biotechnol. 100, 8315–8324. doi: 10.1007/s00253-016-7777-0
Neave M. J., Michell C. T., Apprill A., Voolstra C. R. (2017a). Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci. Rep. 7, 40579–40579. doi: 10.1038/srep40579
Neave M. J., Rachmawati R., Xun L., Michell C. T., Bourne D. G., Apprill A., et al. (2017b). Differential specificity between closely related corals and abundant endozoicomonas endosymbionts across global scales. ISME J. 11, 186–200. doi: 10.1038/ismej.2016.95
Nicholson M. D., Sikkel P. C. (2018). Localized defecation in territorial herbivorous fishes. Copeia 106, 532–538. doi: 10.1643/CE-18-007
Nicolet K. J., Chong-Seng K. M., Pratchett M. S., Willis B. L., Hoogenboom M. O. (2018). Predation scars may influence host susceptibility to pathogens: Evaluating the role of corallivores as vectors of coral disease. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-23361-y
Nugues M. M., Smith G. W., Hooidonk R. J. V., Seabra M. I., Bak R. P. M. (2004). Algal contact as a trigger for coral disease. Ecol. Lett. 7, 919–923. doi: 10.1111/j.1461-0248.2004.00651.x
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples: Primers for marine microbiome studies. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Peixoto R., Rosado P., Leite D., Rosado A. S., Bourne D. (2017). Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 8, 265–288. doi: 10.3389/fmicb.2017.00341
Peixoto R. S., Sweet M., Villela H. D. M., Cardoso P., Thomas T., Voolstra C. R., et al. (2021). Coral probiotics: Premise, promise, prospects. Annu. Rev. Anim. Biosci. 9, 265–288. doi: 10.1146/annurev-animal-090120-115444
Perry C. T., Salter M. A., Lange I. D., Kochan D. P., Harborne A. R., Graham N. A. J. (2022). Geo-ecological functions provided by coral reef fishes vary among regions and impact reef carbonate cycling regimes. Ecosphere 13, e4288. doi: 10.1002/ecs2.4288
Pogoreutz C., Rädecker N., Cárdenas A., Gärdes A., Wild C., Voolstra C. R. (2018). Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol. Evol. 8, 2240–2252. doi: 10.1002/ece3.3830
Polunin N. V. C., Koike I. (1987). Temporal focusing of nitrogen release by a periodically feeding herbivorous reef fish. J. Exp. Mar. Biol. Ecol. 111, 285–296. doi: 10.1016/0022-0981(87)90034-7
Pratchett M. S. (2005). Dietary overlap among coral-feeding butterflyfishes (Chaetodontidae) at lizard island, northern great barrier reef. Mar. Biol. 148, 373–382. doi: 10.1007/s00227-005-0084-4
Pratchett M. S. (2014). ““Chapter 6: Feeding preferences and dietary specialisation among obligate coral-feeding butterflyfishes,”,” in Biology of butterflyfishes. Ed. Pratchett M. S. (Boca Raton, Florida: CRC Press), 140–179.
Pratchett M. S., Berumen M. L., Kapoor B. G. Boca Raton, FL: CRC Press (2014). Biology of butterflyfishes.
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Reese A. T., Dunn R. R. (2018). Drivers of microbiome biodiversity: A review of general rules, feces, and ignorance. mBio 9, e01294–e01218. doi: 10.1128/mBio.01294-18
Rempel H. S., Siebert A. K., Van Wert J. C., Bodwin K. N., Ruttenberg B. I. (2022). Feces consumption by nominally herbivorous fishes in the Caribbean: An underappreciated source of nutrients? Coral Reefs 41, 355–367. doi: 10.1007/s00338-022-02228-9
Renzi J. J., Shaver E. C., Burkepile D. E., Silliman B. R. (2022)The role of predators in coral disease dynamics. Coral Reefs 41, 405–422. doi: 10.1007/s00338-022-02219-w
Robertson D. R. (1982). Fish feces as fish food on a pacific coral reef. Mar. Ecol. Prog. Ser. 7, 253–265.
Rogers C. (1990). Responses of coral reefs and reef organisms to sedimentation. Mar. Ecol. Prog. Ser. 62, 185–202. doi: 10.3354/meps062185
Roman J., McCarthy J. J. (2010). The whale pump: Marine mammals enhance primary productivity in a coastal basin. PloS One 5, e13255. doi: 10.1371/journal.pone.0013255
Rosado P. M., Leite D. C. A., Duarte G. A. S., Chaloub R. M., Jospin G., Nunes da Rocha U., et al. (2019). Marine probiotics: Increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Rotjan R. D., Lewis S. M. (2008). Impact of coral predators on tropical reefs. Mar. Ecol. Prog. Ser. 367, 73–91. doi: 10.3354/meps07531
Rotjan R. D., Ray N. E., Cole I., Castro K. G., Kennedy B. R. C., Barbasch T., et al. (2022). Shifts in predator behaviour following climate induced disturbance on coral reefs. Proc. R. Soc B. 289, 20221431. doi: 10.1098/rspb.2022.1431
Santoro E. P., Borges R. M., Espinoza J. L., Freire M., Messias C. S. M. A., Villela H. D. M., et al. (2021). Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci. Adv. 7, eabg3088. doi: 10.1126/sciadv.abg3088
Shantz A. A., Ladd M. C., Ezzat L., Schmitt R. J., Holbrook S. J., Schmeltzer E., et al. (2023). Positive interactions between corals and damselfish increase coral resistance to temperature stress. Global Change Biol. 29, 417–431. doi: 10.1111/gcb.16480
Shantz A. A., Ladd M. C., Schrack E., Burkepile D. E. (2015). Fish-derived nutrient hotspots shape coral reef benthic communities. Ecol. Appl. 25, 2142–2152. doi: 10.1890/14-2209.1
Sharp K. H., Distel D., Paul V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. doi: 10.1038/ismej.2011.144
Sharp K. H., Ritchie K. B., Schupp P. J., Ritson-Williams R., Paul V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. PloS One 5, e10898. doi: 10.1371/journal.pone.0010898
Shiu J., Yu S., Fong C., Ding J., Tan C., Fan T., et al. (2020). Shifting in the dominant bacterial group endozoicomonas is independent of the dissociation with coral symbiont algae. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01791
Smriga S., Sandin S. A., Azam F. (2010). Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol. Ecol. 73, 31–42. doi: 10.1111/j.1574-6941.2010.00879.x
Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., et al(2006). Microbial diversity in the deep Sea and the underexplored “Rare biosphere.”PNAS 103, 1215–12120doi: 10.1002/9781118010549.ch24
Stier A. C., Gil M. A., McKeon C. S., Lemer S., Leray M., Mills S. C., et al. (2012). Housekeeping mutualisms: Do more symbionts facilitate host performance? PloS One 7, e32079. doi: 10.1371/journal.pone.0032079
Strader M. E., Howe-Kerr L. I., Sims J. A., Speare K. E., Shore A. N., Burkepile D. E., et al. (2022). Nitrate enrichment has lineage specific effects on Pocillopora acuta adults, but no transgenerational effects in planulae. Coral Reefs 41, 303–317. doi: 10.1007/s00338-022-02236-9
Suzuki G., Yamashita H., Kai S., Hayashibara T., Suzuki K., Iehisa Y., et al. (2013). Early uptake of specific symbionts enhances the post-settlement survival of Acropora corals. Mar. Ecol. Prog. Ser. 494, 149–158. doi: 10.3354/meps10548
Sweet M. J., Bythell J. C., Nugues M. M. (2013). Algae as reservoirs for coral pathogens. PloS One 8, 69717–69717. doi: 10.1371/journal.pone.0069717
Tandon K., Lu C.-Y., Chiang P.-W., Wada N., Yang S.-H., Chan Y.-F., et al. (2020). Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J. 14, 1290–1303. doi: 10.1038/s41396-020-0610-x
Tebbett S. B., Bellwood D. R., Purcell S. W. (2018). Sediment addition drives declines in algal turf yield to herbivorous coral reef fishes: Implications for reefs and reef fisheries. Coral Reefs 37, 929–937. doi: 10.1007/s00338-018-1718-6
Tebbett S. B., Goatley C. H. R., Bellwood D. R. (2017a). Clarifying functional roles: Algal removal by the surgeonfishes ctenochaetus striatus and Acanthurus nigrofuscus. Coral Reefs 36, 803–813. doi: 10.1007/s00338-017-1571-z
Tebbett S. B., Goatley C. H. R., Bellwood D. R. (2017b). The effects of algal turf sediments and organic loads on feeding by coral reef surgeonfishes. PloS One 12, e0169479. doi: 10.1371/journal.pone.0169479
Thatcher C., Høj L., Bourne D. G. (2022). Probiotics for coral aquaculture: Challenges and considerations. Curr. Opin. Biotechnol. 73, 380–386. doi: 10.1016/j.copbio.2021.09.009
Umeki M., Yamashita H., Suzuki G., Sato T., Ohara S., Koike K. (2020). Fecal pellets of giant clams as a route for transporting symbiodiniaceae to corals. PloS One 15, 1–15. doi: 10.1371/journal.pone.0243087
Vannette R. L. (2020). The floral microbiome: Plant, pollinator, and microbial perspectives. Annu. Rev. Ecology Evolution Systematics 51, 363–386. doi: 10.1146/annurev-ecolsys-011720-013401
Van Oppen M. J. H., Blackall L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vašutová M., Mleczko P., López-García A., Maček I., Boros G., Ševčík J., et al. (2019). Taxi drivers: The role of animals in transporting mycorrhizal fungi. Mycorrhiza 29, 413–434. doi: 10.1007/s00572-019-00906-1
Velásquez-Rodríguez T. M., Zuluaga-Arias C., Montaño-Salazar S. M., González J. M., Sánchez J. A. (2021)The potential of parrotfish faeces in replenishing reefs with coral-associated microbiome. BioRxivdoi: 10.1101/2020.09.15.298737
Vidal-Dupiol J., Ladrière O., Meistertzheim A.-L., Fouré L., Adjeroud M., Mitta G. (2011). Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J. Exp. Biol. 214, 1533–1545. doi: 10.1242/jeb.053165
Viviani J., Lecchini D., Moritz C., Siu G., Galzin R., Viriot L. (2019). Synchrony patterns reveal different degrees of trophic guild vulnerability after disturbances in a coral reef fish community. Diversity Distributions 25, 1210–1221. doi: 10.1111/ddi.12931
Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, e00009–e00015. doi: 10.1128/mSystems.00009-15
Wilson B., Muirhead A., Bazanella M., Huete-Stauffer C., Vezzulli L., Bourne D. G. (2013). An improved detection and quantification method for the coral pathogen vibrio coralliilyticus. PloS One 8, 1–7. doi: 10.1371/journal.pone.0081800
Wisnoski N. I., Lennon J. T. (2022). Scaling up and down: Movement ecology for microorganisms. Trends Microbiol. 31, 242–253. doi: 10.1016/j.tim.2022.09.016
Wrathall T. J., Roberts C. M., Ormond R. F. G. (1992). Territoriality in the butterflyfish Chaetodon austriacus. Environ. Biol. Fish 34, 305–308. doi: 10.1007/BF00004777
Zhou G., Cai L., Yuan T., Tian R., Tong H., Zhang W., et al. (2017). Microbiome dynamics in early life stages of the scleractinian coral Acropora gemmifera in response to elevated pCO2. Environ. Microbiol. 19, 3342–3352. doi: 10.1111/1462-2920.13840
Zhu D., Delgado-Baquerizo M., Ding J., Gillings M. R., Zhu Y.-G. (2021). Trophic level drives the host microbiome of soil invertebrates at a continental scale. Microbiome 9, 189. doi: 10.1186/s40168-021-01144-4
Ziegler M., Grupstra C. G. B., Barreto M. M., Eaton M., Baomar J., Zubier K., et al. (2019). Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 10, 3092. doi: 10.1038/s41467-019-10969-5
Keywords: microbiome, lesion, Vibrio, corallivore, herbivore, grazer, detritivore, reef fish
Citation: Grupstra CGB, Howe-Kerr LI, van der Meulen JA, Veglia AJ, Coy SR and Correa AMS (2023) Consumer feces impact coral health in guild-specific ways. Front. Mar. Sci. 10:1110346. doi: 10.3389/fmars.2023.1110346
Received: 28 November 2022; Accepted: 01 March 2023;
Published: 13 April 2023.
Edited by:
Kshitij Tandon, The University of Melbourne, AustraliaReviewed by:
Andrew A. Shantz, Florida State University, United StatesKimberly B. Ritchie, University of South Carolina Beaufort, United States
Copyright © 2023 Grupstra, Howe-Kerr, van der Meulen, Veglia, Coy and Correa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carsten G. B. Grupstra, cgb.grupstra@gmail.com
 Carsten G. B. Grupstra
Carsten G. B. Grupstra