
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci., 15 March 2023
Sec. Global Change and the Future Ocean
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1101855
This article is part of the Research TopicEcological Consequences of Climate Change in Boreal Marginal SeasView all 11 articles
The Atlantic herring and Atlantic cod are two marine fish species that have successfully adapted to the brackish Baltic Sea, and the former is able to spawn in near-freshwater conditions in the inner Gulf of Bothnia. Here, we review the state of current knowledge concerning ecological adaptation in the two species and make an attempt to predict how they will be able to cope with future climate change. Previous whole genome sequencing studies in Atlantic herring have revealed hundreds of genetic loci underlying ecological adaptation, including several loci that show very strong associations to variation in salinity and temperature. These results suggest the existence of standing genetic variation available for adaptation to a changing environment. However, although Atlantic herring probably has the genetic potential to adapt, its future status also depends on how climate change will affect plankton production and competing species, such as sprat and three-spined stickleback. In cod, the situation is challenging, as there is only one true Baltic population, spawning east of Bornholm and then dispersing towards the east and north. This Baltic cod population is threatened by overfishing, low oxygen levels in benthic waters and generally bad physiological condition of individual fish, in addition to being completely isolated from gene flow from nearby cod populations at the entrance of the Baltic Sea.
The brackish Baltic Sea is a challenging environment for marine fish, and few marine species have been able to successfully colonize this body of water. Surface salinity drops from 35‰ in the Atlantic Ocean to about 8‰ in the Southern Baltic Sea, and to as low as 2-3‰ in the inner Gulf of Bothnia. Furthermore, the amplitude in temperature variation over the year is higher than in the Atlantic Ocean (Snoeijs-Leijonmalm et al., 2017). Major anthropogenic impacts on the Baltic Sea ecosystem are eutrophication, leading to algal blooms and hypoxia in certain areas, pollution (dioxin, dioxin-like compounds, and other persistent organic pollutant), and climate change (Snoeijs-Leijonmalm et al., 2017). Climate predictions indicate drastic changes during this century with increasing temperature, lower salinity, and less ice coverage in the winter (Reusch et al., 2018).
The Atlantic herring (Clupea harengus) and Atlantic cod (Gadus morhua) are two of the most important marine fish that have been able to adapt to the brackish environment in the Baltic Sea (Ojaver et al., 2010). Linnaeus classified the Baltic herring as a subspecies of the Atlantic herring based on its distinct phenotype (small size, reduced fat content compared with Atlantic herring) and named it Clupea harengus membras (Linnaeus, 1761). Also, the Atlantic cod present in the Baltic Sea may be considered a distinct subspecies with specific adaptation to the Baltic Sea (Linnaeus, 1761). Both species have been subjected to detailed genetic studies in recent years, which have revealed many of the genes and genomic regions that have contributed to their genetic adaptation. The aim of this review is to summarize the current knowledge of genetic adaptation to the environmental conditions in the Baltic Sea as a basis to speculate how climate change may affect these species.
The Atlantic herring has a key ecological role in the North Atlantic Ocean and adjacent waters. It feeds on plankton and thereby constitutes a link between the primary production in the ocean and other fish, including cod, sea birds, and marine mammals that feed on herring (Ojaver et al., 2010). Herring has also been a critical food resource for humans for at least a thousand years (Atmore et al., 2022). It is a benthic spawner that utilizes fully marine environments (35-36‰) as well as brackish environments, including the entire Baltic Sea. Early genetic studies using a handful of biochemical polymorphisms revealed no significant genetic differentiation among populations even between population samples from full marine environments to the inner Gulf of Bothnia (Ryman et al., 1984). This surprising finding was resolved when it became possible to screen large numbers of genetic markers including the sequencing of the entire genome (Lamichhaney et al., 2012; Martinez Barrio et al., 2016; Pettersson et al., 2019; Han et al., 2020). These later studies revealed that there is almost no genetic differentiation at the great majority (>90%) of single nucleotide polymorphisms (SNPs) but highly significant differentiation, often approaching fixation of alternative alleles, between subpopulations for a few percent of the polymorphic sites. The very low genetic differentiation at selectively neutral loci can be explained by the very large population size and some gene flow between subpopulations, resulting in negligible genetic drift. In contrast, natural selection, fueled by the same large population size, is causing striking genetic differentiation at loci underlying ecological adaptation. Thus, whole genome sequencing reveals a considerable degree of local adaptation (Han et al., 2020), in total contrast to the previous results based on a handful of selectively neutral markers. The Atlantic herring data demonstrate that for marine species with large breeding populations, it is essential to study the entire genome to get full insight about population structure and to reveal the genetic markers that confer the most power to distinguish different subpopulations.
Whole genome sequencing has revealed about ten major groups of Atlantic herring that constitute ecotypes genetically adapted to different environmental conditions (Han et al., 2020) (Figure 1). The three major factors distinguishing these ecotypes are (i) spawning time (primarily spring or autumn); (ii) salinity at spawning locations; (iii) water temperature at spawning locations. However, it is clear that there is further genetic differentiation within the major groups. For instance, the spring-spawning herring from the region between the North Sea and the Baltic Sea proper – i.e., the transition zone with a steep salinity gradient – includes genetically differentiated subpopulations. The exact number of subpopulations within the major groups is unknown and it is likewise not known to which extent local populations are isolated from other local populations within the same major group.
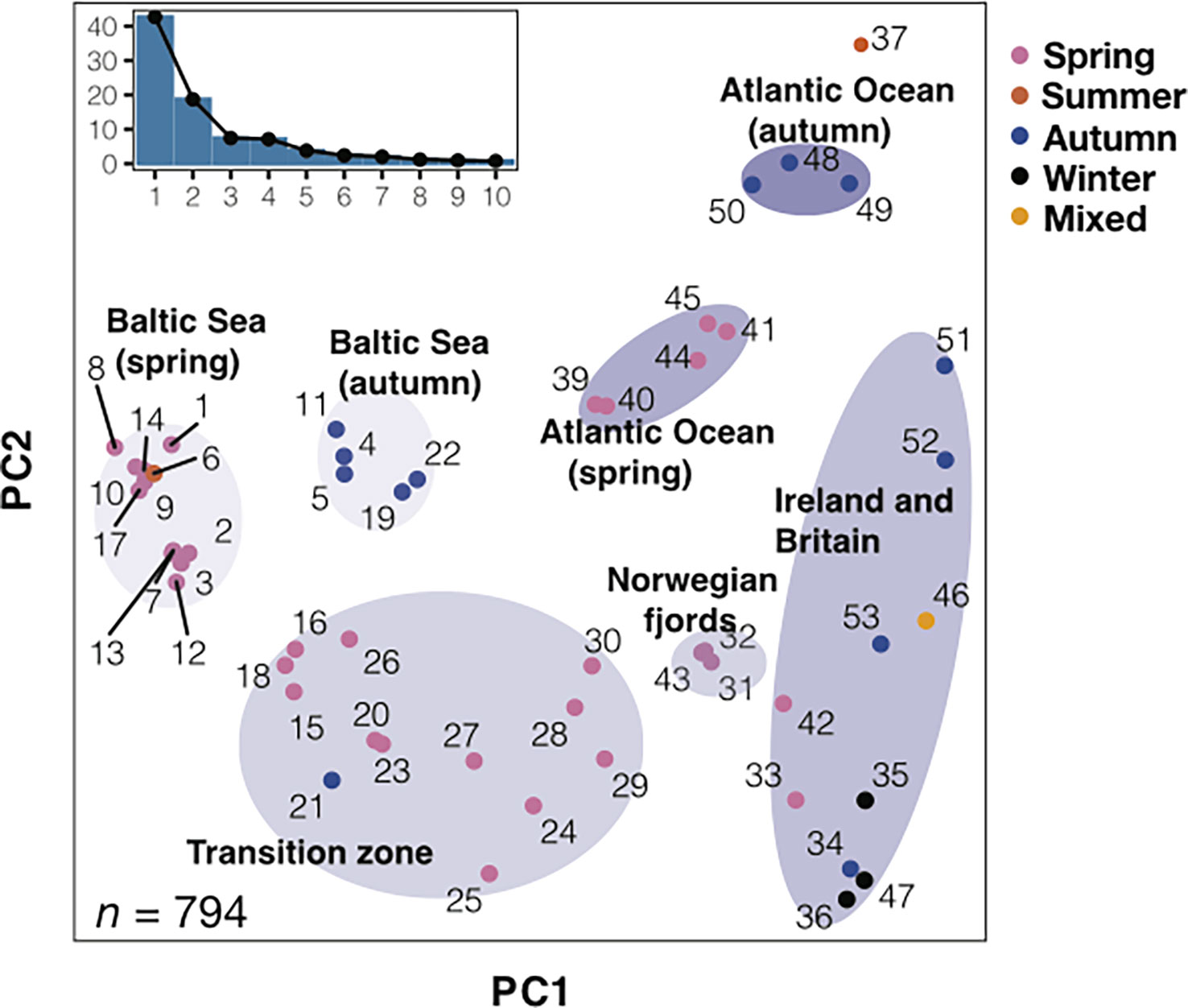
Figure 1 Principal component analysis (PCA) of herring populations. PCA based on 794 highly informative markers representing loci likely under natural selection. Color of each population represents spawning season. Inset bar plot indicates the percentage of explained variance by each principal component. n = number of SNPs used in the analysis. Adapted from Han et al., 2020, Figure 2.
Difference in salinity is a major environmental factor affecting ecological adaptation in herring. Previous data show that it is the salinity at spawning locations that is more important for local adaptation than salinity at feeding (Han et al., 2020). One example of this is a population spawning under brackish conditions in the Ringköbing fjord (Denmark) but which is feeding under fully marine conditions in the North Sea. This population shows allele frequency patterns shared with other populations spawning in brackish waters (Han et al., 2020). Whole genome sequencing has revealed hundreds of gene regions showing strong genetic differentiation between populations from the Atlantic Ocean and the brackish Baltic Sea (Han et al., 2020). Some of these are expected to directly affect osmoregulation and two examples of top candidate genes are LRRC8C and PRLR. LRRC8C encodes a volume-regulated ion channel (VRAC) with an important role in osmoregulation in all vertebrates. Members of this family of ion channels are activated when the cell swells and play a key role in cell volume regulation (Osei-Owusu et al., 2018). The most common LRRC8C allele present in populations from the Atlantic Ocean differs by four amino acid substitutions from the most common allele in the brackish Baltic Sea (Han et al., 2020) (Figure 2). Interestingly, an intermediate form having two of these substitutions is the most common allele in some populations from the transition zone between the Atlantic Ocean and the Baltic Sea (Figure 2), suggesting that this gene shows a step-wise evolutionary adaptation to low salinity.
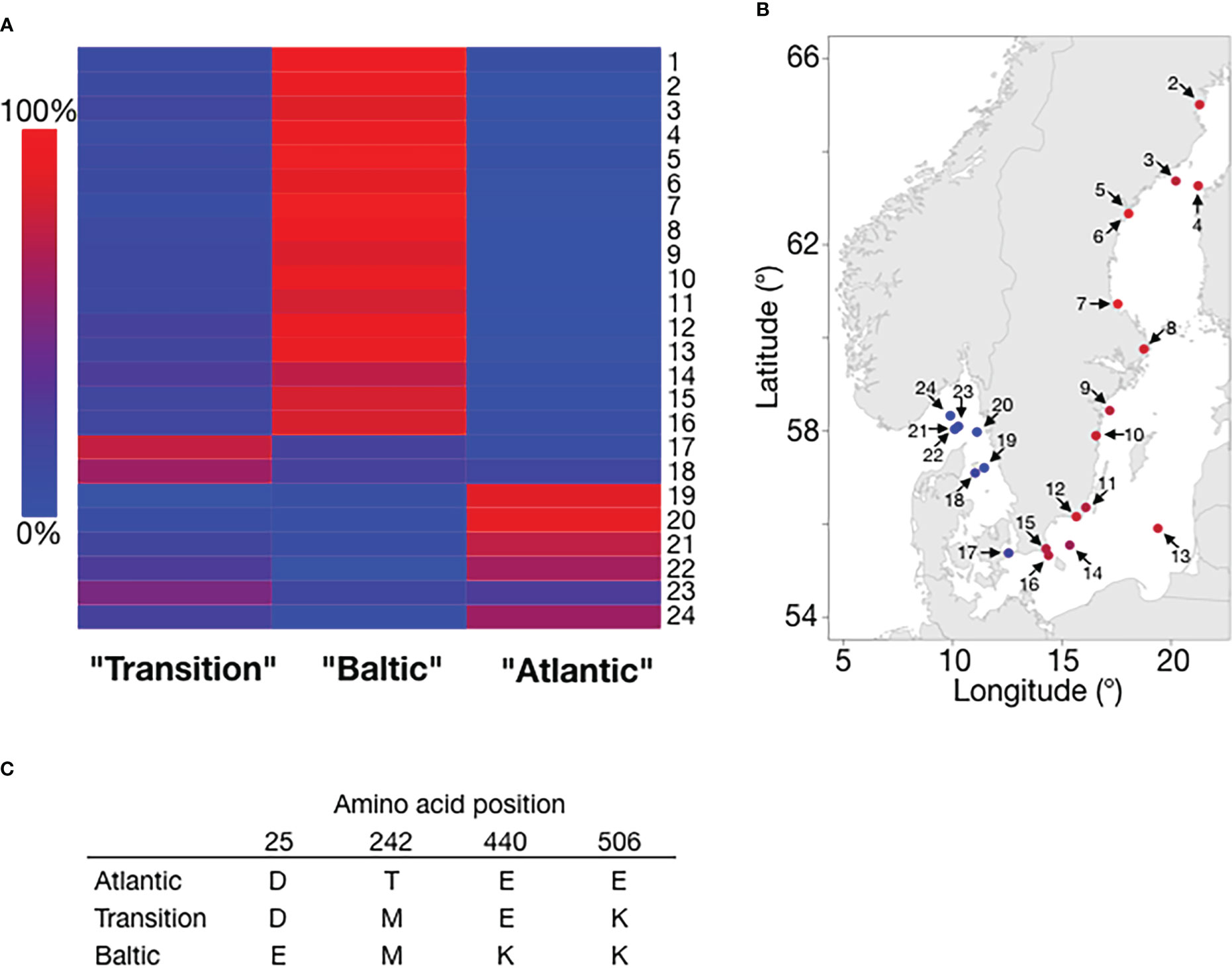
Figure 2 The frequencies of LRRC8C alleles show strong correlation with salinity. (A) Heatmap of the frequencies of the three LRRC8C alleles. Column labels are colloquial names for the common alleles based on their most prevalent location. Samples were collected and genotyped in the collaborative “Forskarhjälpen” project presented in Pettersson et al. (2019). Each row refers to a population sample (numbered 1-24) collected by one participating school and ordered by geographical location (see panel B). Color indicates allele frequency. (B) Geographical presentation of the frequency of the Baltic allele (color code for allele frequency the same as in (A). Sample labels correspond to (A) Sample (1) lacks a reported location, and is omitted. (C) Amino acid residues for the three alleles at the variable positions.
Prolactin is best known for its crucial role for lactation in mammals. However, in fish it is well established that prolactin plays a key role in osmoregulation (Manzon, 2002). The PRLR gene encodes the receptor of prolactin and the locus shows a very strong signature of selection in relation to adaptation to the brackish Baltic Sea (Han et al., 2020). It is not yet known if this is caused by changes in the protein sequence or in gene regulation.
With regard to the ability of the Atlantic herring to adapt to variation in water temperature, insights primarily come from a comparison of allele frequencies in herring spawning in the waters around Great Britain and Ireland with those spawning further north in the Atlantic (Norway, Iceland, Greenland, and Canada) (Han et al., 2020). In this comparison, there are essentially no differences in salinity between regions, but a clear difference in water temperatures; the waters around Ireland and Great Britain are among the warmest where herring reproduction takes place. This analysis revealed about ten loci with very strong genetic differentiation between the British Isles and northern Atlantic groups (Han et al., 2020) (Figure 3). Interestingly, four of these represent inversions present on chromosome 6, 12, 17, and 23, and ranging in size from 1.2 Mb to 7.8 Mb. Due to the distribution pattern, the two alternate haplotypes at these four inversions are referred to as “Northern” and Southern” and the relative frequencies of the haplotypes at all four show a strong correlation with water temperature at the spawning location. However, it has not yet been proven whether the genetic adaptations at these loci are directly related to water temperatures or to other environmental factors strongly correlated with temperature. Nevertheless, these are obvious candidate loci that may contribute to genetic adaptation to the ongoing climate change.
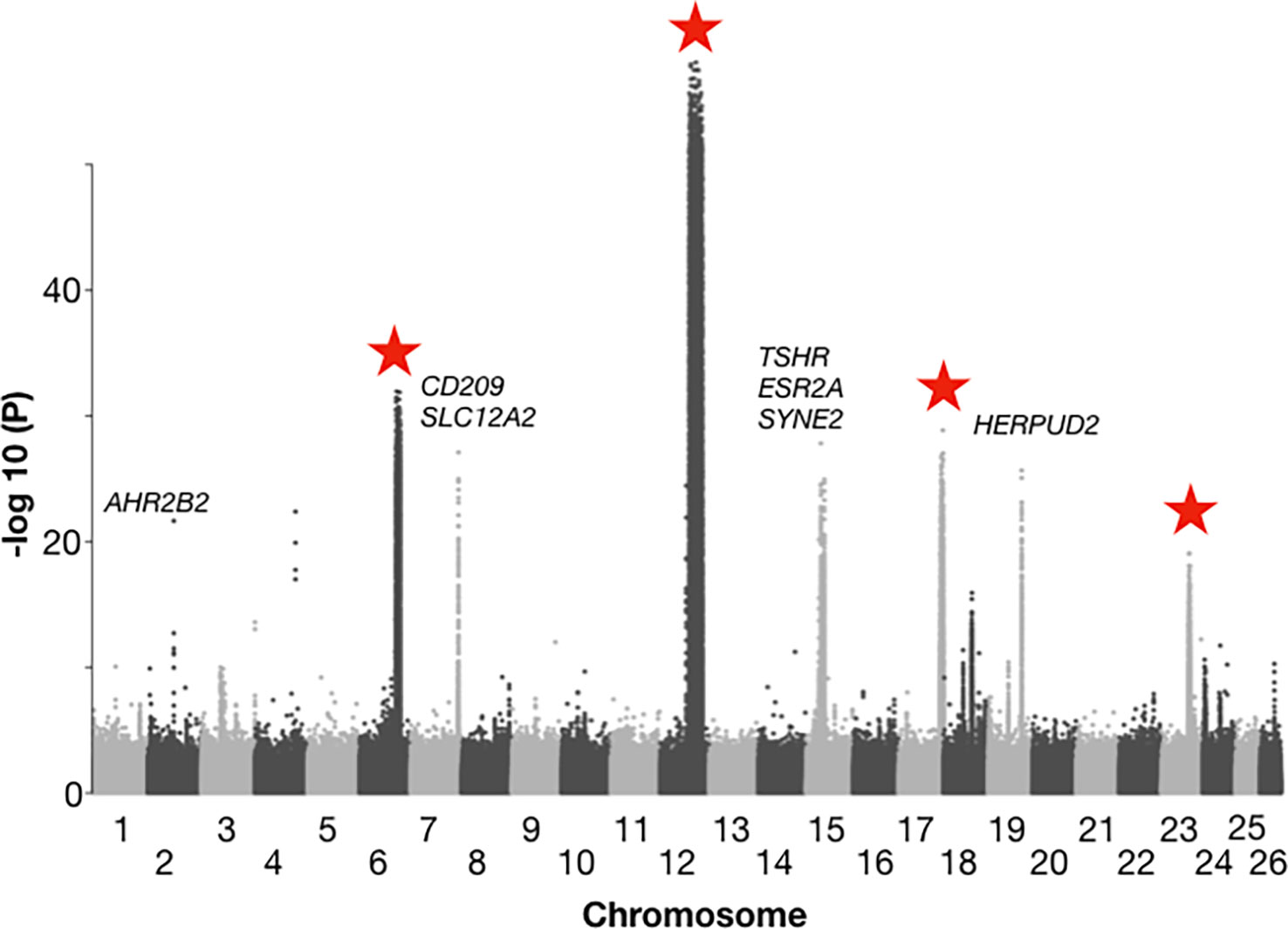
Figure 3 Genetic differentiation between herring populations from Ireland and Britain vs. other populations from the Northeast Atlantic. Inversions are located on chromosomes 6, 12, 17, and 23, and are highlighted with red stars. Strong genetic differentiation is observed in the following intervals: chromosome 6: 22.2 – 24.8 Mb, chromosome 12: 17.8 – 25.6 Mb, chromosome 17: 25.8 – 27.5 Mb, and chromosome 23: 16.3 – 17.5 Mb. The Y-axis represents -log10 of the probability (P) value in a chi square test per SNP testing the null hypothesis of no allele frequency differences between regions. The following genes associated with genetic differentiation are highlighted: AHR2B2, aryl hydrocarbon receptor 2B2; CD209, CD209 antigen-like protein; SLC12A2, solute carrier family 12 member 2; TSHR: thyroid stimulating hormone receptor; ESR2A, estrogen receptor 2A; SYNE2, spectrin repeat containing nuclear envelope protein 2; HERPUD2, HERPUD family member 2. Adapted from Han et al., 2020, Figure 4.
In the Baltic Sea, the Northern haplotypes are the most common variants at all four inversions, but the Southern haplotypes are also present (Han et al., 2020). This is illustrated for the chromosome 12 inversion in Figure 4. For instance, the Northern haplotype at this locus had a frequency of 94% in a sample from Kalix (BK in Figure 4) while the frequency is about 70% in the Southern Baltic Sea. An interesting finding is that a population sampled from Gamlebyviken (BG in Figure 4) collected on August 20 1979 by one of the authors (LA) had the lowest frequency of the Northern haplotype among all samples from the Baltic Sea. Gamlebyviken is a shallow bay on the east coast of Sweden and it was sampled because it was claimed that it holds a unique local population of Baltic herring. The unusually high frequency of the Southern haplotype at the chromosome 12 inversion supports this assumption, and highlights the importance of local adaptation of the Atlantic herring in general. It is reasonable to assume that the water in this shallow bay warms up faster in the spring, which is favorable for the fitness of herring carrying the Southern haplotype, a haplotype strongly associated with warm water temperatures at spawning.
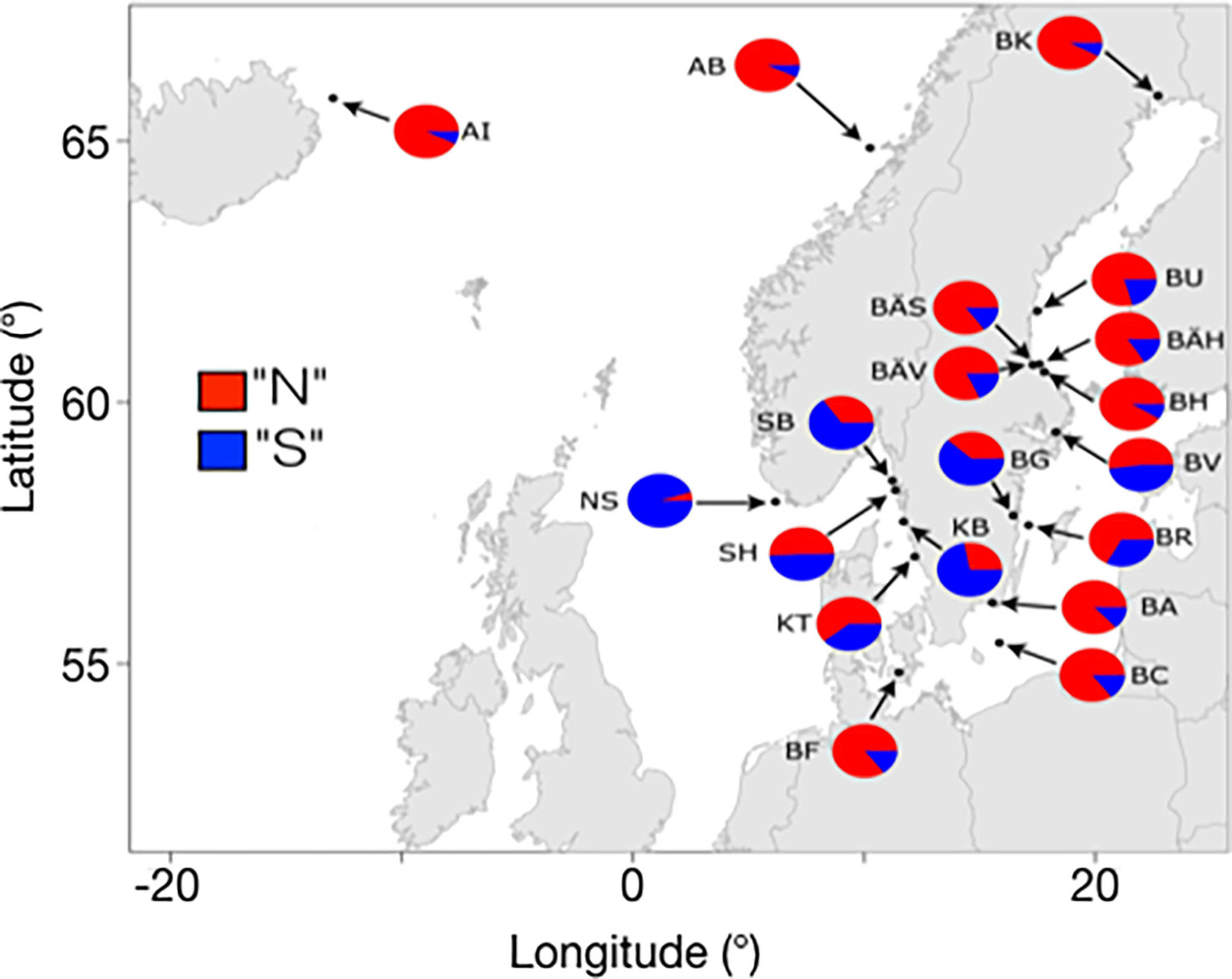
Figure 4 Estimated frequencies for the “Northern” and “Southern” haplotypes for the inversion on chromosome 12 in pooled population samples in the Baltic Sea and East Atlantic. Adapted from Pettersson et al., 2019, Figure 6.
Another gene underlying adaptation to the Baltic Sea is RHO encoding rhodopsin, one of the light receptors in the retina of the eye. Hill et al. (2019) demonstrated that a missense mutation (Phe261Tyr) in this gene is absent from Atlantic herring from fully marine environments with a salinity of 35-36‰ but occurs at high frequency throughout the Baltic Sea and is fixed in the inner Baltic Sea. The mutation is expected to cause a red shift in light absorption providing better vision in the brackish Baltic Sea. The light conditions in the Baltic Sea are red-shifted because blue light tends to be absorbed by dissolved organic carbon in the water (Kratzer and Moore, 2018). Interestingly, exactly the same mutation is present in one third of all fish species living in freshwater or brackish waters (Hill et al., 2019), providing a textbook case of convergent evolution.
Herring populations spawn under widely different environmental conditions, with regard to salinity, temperature, water depth, light conditions, and biotic factors (prey and predators). This means that the most sensitive phase of life, embryonic and larval development, takes place under drastically different constraints. This has promoted a homing behavior that is a prerequisite for local genetic adaptation. Indeed, local adaptation in the Baltic Sea is known from a number of other marine fish that also spawn in various coastal or freshwater habitats, such as, cod, plaice, flounder, and stickleback (DeFaveri and Merilä, 2014; Berg et al., 2015; Momigliano et al., 2018; Le Moan et al., 2021). In contrast, the situation is totally different in the European eel, where the entire population spawns in one location, the Sargasso Sea.
Consequently, the eel constitutes a single panmictic population that shows no genetic differentiation between geographic regions (Enbody et al., 2020). This is the situation despite the fact that eels spend most of their adult life under highly variable environmental conditions across Europe and North Africa, but spawning and early development take place under nearly identical conditions in the Sargasso Sea. The homing behavior in the Atlantic herring is not perfect which means that there is some gene flow between subpopulations. Such gene flow combined with the very large size of herring populations is a reasonable explanation for the minute genetic drift in the Atlantic herring and the lack of genetic differentiation at selectively neutral loci. The distribution of FST-values in the Atlantic herring with a sharp contrast between strong genetic differentiation at a few percent of the loci and no genetic differentiation at the great majority of loci constitutes a major deviation from the one expected for selectively neutral alleles under a drift model (Lamichhaney et al., 2017). The most plausible explanation for this deviation is that the strong genetic differentiation at some loci is due to natural selection.
Atlantic cod is, like herring, an important commercial fish species. It is distributed across the North Atlantic and occurs up to the Bothnian Sea in the Baltic. There are at least two genetically distinct populations of cod in the Baltic Sea: one large population east of Bornholm, and one or several smaller populations that spawn in the western Baltic Sea (Hemmer-Hansen et al., 2019; Weist et al., 2019). Currently, it is believed that the eastern Baltic cod only spawn in the Bornholm deep, and that historical spawning areas in the Gotland and Gdansk deep are now too oxygen-depleted for successful spawning (Köster et al., 2017). The eastern Baltic cod population is adapted to the brackish environment and genetically distinct from the adjacent cod populations in the western Baltic, Belt seas, Öresund, and Kattegat. Adaptations include differences in hemoglobin type, osmoregulatory capacity, egg buoyancy, sperm swimming characteristics, and spawning season. All these adaptations to the brackish environment contribute to an effective reproductive barrier between the eastern and western Baltic cod. SNP genotyping (Berg et al., 2015) and whole genome sequencing (Barth et al., 2019) show that the divergence of eastern Baltic cod is genome wide, indicative of genetic drift in a reproductively isolated population, in addition to specific genetic regions under selection (Figure 5). In contrast, a comparison of cod from Kattegat and the North Sea shows a few regions of strong genetic differentiation most likely underlying genetic adaptation (Figure 5). The reproductive isolation of eastern Baltic cod may be regarded as ongoing ecological speciation.
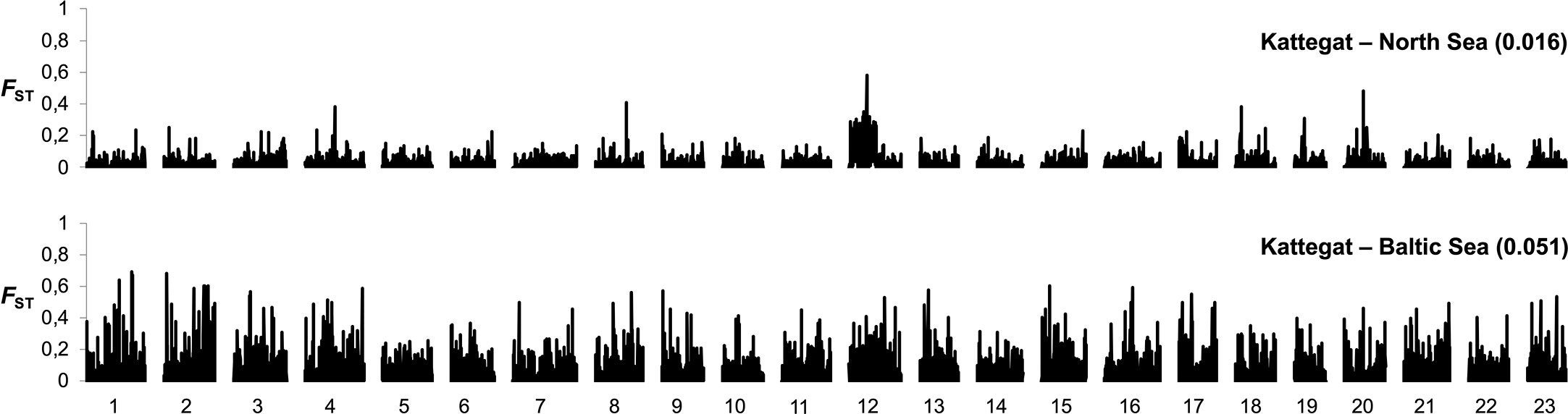
Figure 5 Genome-wide differentiation (FST) in cod based on 8,309 SNPs across all 23 chromosomes in comparisons of cod from Kattegat and North Sea (top) and cod from eastern Baltic Sea and Kattegat (bottom). Median FST estimates in the two pairwise comparisons are denoted in parentheses. Adapted from Berg et al., 2015, Figure 2.
Successful spawning of Baltic cod is restricted to a layer of water in the Bornholm deep, which has high enough salinity to allow the cod eggs to be neutrally buoyant, while still above critical oxygen levels (Nissling et al., 1994). Experimental studies show that a salinity of ≥ 11‰ and oxygen content above 2 ml O2 l-1 are required for successful fertilization of cod eggs, and that the salinity of neutral buoyancy vary between 10-16‰ (Vallin and Nissling, 2000). Eastern Baltic cod produce larger and lighter eggs compared to cod in the western Baltic (Nissling et al., 1994; Thorsen et al., 1996; Nissling and Westin, 1997), that are neutral buoyant at a lower salinity. Baltic cod also produce sperm that are actively swimming at low salinities (Westin and Nissling, 1991).
Eastern Baltic cod spawn later in the season, in May-August, compared to western Baltic cod that spawn in February-April (Poulsen, 1931; Brander, 2005). This temporal difference likely constitutes an effective barrier to reproduction and gene exchange. However, range expansion and mechanical mixing in the Arkona basin occur regularly (Hemmer-Hansen et al., 2019; Schade et al., 2022), while hybridization seems to be rare.
Genome scan studies have detected several regions in the genome of eastern Baltic cod associated with salinity and oxygen conditions at spawning depth (Berg et al., 2015). Most of these genomic regions also contain outlier loci, within or close to genes, with high divergence between eastern and western Baltic cod. Several of these candidate genes for selection are known to be involved in fish egg development. One notable example is SNP outlier loci located close to the aquaporin gene, AQP1A. The hydration of fish eggs making them larger and lighter, is thought to be governed by water channels or aquaporins (Fabra et al., 2005; Cerdà et al., 2013). Other genes under selection in Baltic cod are members of the solute carrier gene family (SLCs) involved in ion transport, important for egg development. Vitellogenin is involved in hydrolysis of yolk proteins, which produce free amino acids affecting water flux in the egg. Vitellogenin is also important for egg chorion thickness, which is thinner in Baltic cod eggs (Nissling et al., 1994). If a female of western Baltic origin spawns in the Bornholm deep, the eggs will sink down to the anoxic layer and not survive.
Fertilization of eggs is also challenging at low salinity. Baltic cod harbor outlier loci in the vicinity of the zona pellucida glycoprotein-2 gene (ZP2L1) on chromosome 8, possibly involved in sperm binding. Other fish species, such as sand goby, show sperm adaptation to low salinity in Baltic populations (Leder et al., 2021).
Osmoregulation and ion exchange in cod larvae, juveniles and adults are controlled by the expression of different enzymes, such as Na+/K+-ATPase. Both differential gene expression (Larsen et al., 2011) and signals of selection (Berg et al., 2015) on Na+/K+-ATPase genes have been detected in Baltic cod. A SNP located close to the PRL gene, coding for prolactin, another protein important for water and salt balance, also show strong environmental correlation in Baltic cod (Berg et al., 2015; Weist et al., 2019). Interestingly and as mentioned above, the gene (PRLR) encoding the receptor of prolactin shows a very strong genetic differentiation between Atlantic and Baltic herring (Han et al., 2020).
Already in the early 1960ies, Knud Sick reported a two-allele system with three different hemoglobin genotypes for Atlantic cod, where one allele (HbI-2) was almost fixed in Baltic cod (Sick, 1961). The HbI-2 allele is also dominating in cod populations in colder waters in the North Atlantic. Experimental studies have later shown that oxygen uptake is more efficient at low temperature in fish with the HbI-2 allele (reviewed in Andersen, 2012). Molecular and structural studies have revealed that two non-synonymous mutations in the Hemoglobin beta 1 gene are associated with two amino acid substitutions, Met55Val and Lys62Ala, where the Val-Ala variant corresponds to the HbI-2 allele (Andersen et al., 2009). Several population surveys have identified hemoglobin as a strong genetic outlier between eastern and western Baltic cod (e.g., Weist et al., 2019).
Recent studies show that local adaptation to different environments have been facilitated by chromosomal inversions in the Atlantic cod genome (Matschiner et al., 2022; Sodeland et al., 2022). These inverted chromosomal regions act as supergenes, where multiple neighboring genes are inherited together. Four major supergenes have been identified in Atlantic cod, occurring in different frequencies in different cod populations. Genes involved in adaptation to low salinity in the Baltic are located in inversions on chromosome 2 and 12 (Berg et al., 2015; Barth et al., 2019; Matschiner et al., 2022).
As the genetic data for herring and cod clearly show, Baltic populations of these two species are specifically adapted to the Baltic Sea, a marginal marine environment. Increased stress from climate changes will put further pressure on both species. Moreover, the Baltic Sea is an isolated marine basin with narrow openings to the North Sea through the shallow Danish Straits. Consequently, sea warming is more severe and much faster in the Baltic Sea than in the open oceans (Reusch et al., 2018). Currently, the ongoing change in sea surface temperature is about 0.5°C per decade and the average temperature is currently 1.5°C higher than it was 40 years ago (Snoeijs-Leijonmalm et al., 2017). Due to increased precipitation, the salinity is also predicted to decrease in the Baltic Sea (Kniebusch et al., 2019) where most marine species already have adapted to truly marginal levels of salinity (Johannesson and André, 2006; Johannesson et al., 2020). In Atlantic herring, a number of genetic variants show strong correlation with water temperature at spawning (e.g., four inversions) (Han et al., 2020). These will most likely change in frequency in response to increasing temperature. Such a genetic shift might progress smoothly, if the effective sizes of individual subpopulations are large (as is the case in natural populations of herring). However, fishing pressure on the Baltic herring is currently strong and size distributions skewed towards smaller fish (Anonymous, 2021). Furthermore, it is unclear how the current fishing pressure affects local subpopulations, as there is no monitoring at the level of subpopulation. Specific subpopulations, such as the one in Gamlebyviken, may be instrumental to a rapid shift and will be less stressed than other populations by selective mortality as a consequence of increased temperature. From historical samples from the last hundred years, it would be possible to determine the genetic consequences of the current increase of temperature of about 1.5°C.
What appear as additionally problematic for the survival of the Baltic herring is what happens to the ecosystem, not least to the zooplankton communities. For example, in different regions of the Baltic Sea the numbers of three-spined stickleback have increased between 4-fold and 45-fold over the past 35 years, and its predation on fish egg, fish larvae, and zooplankton impacts substantially coastal and open sea food webs (Bergström et al., 2015). Sprat (Sprattus sprattus) largely overlaps with herring in distribution in the Baltic Sea and the two species compete for food by targeting the same species of zooplankton, a competition that has increased in intensity with increasing numbers of Baltic sprat (Möllman et al., 2004; Casini et al., 2010). It is unclear how increasing temperature and possibly lower salinity will affect sprat populations in the Baltic. Finally, bladderwrack (Fucus vesiculosus) and eelgrass (Zostera marina) are important spawning habitats for Baltic herring, and model predictions for the bladderwrack show that it will likely go locally extinct in most of the Baltic Sea over the coming 50-100 years due to increased temperature and reduced salinity (Jonsson et al., 2018; Kotta et al., 2019).
The situation for the isolated eastern cod population is strongly related to the situation of its spawning ground east of the island Bornholm. The semi-pelagic spawning of the eastern cod is depending on correct buoyancy of the eggs that should neither float, nor sink to the bottom. The Baltic cod has eggs with a buoyancy adapted to the lower salinity of the deep Baltic Sea (Nissling and Westin, 1997). It is furthermore, depending on oxygen in the bottom waters, both for the survival of the eggs and for feeding on benthic invertebrates. Over the past century the area of deep benthic habitat that has become hypoxic inside the Baltic Sea has increased multifold (Reusch et al., 2018). However, data indicate that the hydrographic conditions has remained relatively unchanged in the important cod spawning grounds east of Bornholm the last 60 years (Svedäng et al., 2022). Finally, the cod stock has been heavily depleted by a too intense commercial fishing in the past and today it is in a very poor condition, despite low fishing pressure (ICES, 2021). As cod is also a coldwater species (Asgeirsson et al., 1989), increasing temperatures will put further stress and induce selective mortality, and there is a large risk of local extinction of the Baltic cod east of Bornholm in the near future, partly as a consequence of its complete isolation to other cod stocks (Hemmer-Hansen et al., 2019).
Genetic studies in Atlantic herring and cod have revealed many genes and genomic regions associated with ecological adaptation. It is now straightforward to set up diagnostic tests to monitor genetic changes at these loci. This can for instance be carried out using so called SNP chips that is a cost-effective way to generate genotypes for many individuals for thousands of SNPs. A fish multi-species SNP chip providing data on thousands of SNPs per species has recently become commercially available (https://www.identigen.com/DnaTraceback/Seafood; accessed November 3, 2022). SNPs for the following species are available on this chip: Atlantic herring, Atlantic horse mackerel, Brown trout, Atlantic cod, Perch, Salmon, and Sprat. This is a valuable resource for many types of population studies, including to monitor stock development as a basis for maintaining genetic diversity and promote sustainable fishery. Careful fishery management is expected to be even more important in the future to avoid population collapse due to the combined effects of climate change and intensive commercial fishing.
The already established genetic markers associated with temperature adaptation, such as the chromosome 12 inversion in the Atlantic herring, that is likely to contribute to the response to future climate changes, should be included in such genetic monitoring. However, it will also be wise to explore genetic changes across the entire genome at regular intervals, on the order once per decade, because new loci may come under selection as the environmental conditions become more extreme. Furthermore, new mutations contributing to genetic adaptation may emerge and need to be included in updated panels of diagnostic genetic markers used for monitoring.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
LA’s research group is funded by Forskningsrådet (2017-02907) and Knut and Alice Wallenberg Foundation (KAW 2016.0361).
CA and KJ acknowledge support from the Linnaeus Centre for Marine Evolutionary Biology, CeMEB.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Andersen Ø. (2012). Hemoglobin polymorphism in atlantic cod – a review of 50 years of study. Mar. Genomics 8, 59–65.
Andersen O., Wetten O. F., De Rosa M. C., Andre C., Carelli Alinovi C., Colafranceschi M., et al. (2009). Haemoglobin polymorphisms affect the oxygen-binding properties in atlantic cod populations. Proc. Biol. Sci. 276, 833–841.
Anonymous (2021). “Fisk- och skaldjursbestånd i hav och sötvatten 2020. havs och vattenmyndigheten,” in Swedish Agency for marine and water management report, vol. 2021. (Swedish), 6.
Asgeirsson B., Fox J. W., Bjarnason J. B. (1989). Purification and characterization of trypsin from the poikilotherm Gadus morhua. Eur. J. Biochem. 180, 85–94.
Atmore L. M., Martinez-Garcia L., Makowiecki D., André C., Lougas L., Barrett J. H., et al. (2022). Population dynamics of Baltic herring since the Viking age revealed by ancient DNA and genomics. Proc. Natl. Acad. Sci. U.S.A. 119, e2208703119.
Barth J. M., Villegas-Ríos D., Freitas C., Moland E., Star B., André C., et al. (2019). Disentangling structural genomic and behavioural barriers in a sea of connectivity. Mol. Ecol. 28, 1394–1411.
Berg P. R., Jentoft S., Star B., Ring K. H., Knutsen H., Lien S., et al. (2015). Adaptation to low salinity promotes genomic divergence in Atlantic cod (Gadus morhua l.). Genome Biol. Evol. 7, 1644–1663.
Bergström U., Olsson J., Casini M., Eriksson B. K., Fredriksson R., Wennhage H., et al. (2015). Stickleback increase in the Baltic Sea – a thorny issue for coastal predatory fish. Estuarine Coast. shelf Sci. 163, 134–142.
Brander K. (2005). Spawning and life history information for north Atlantic cod stocks. ICES Coop Res. Rep. doi: 10.17895/ices.pub.5478
Casini M., Bartolino V., Molinero J. C., Kornilovs G. (2010). Linking fisheries, trophic interactions and climate: Threshold dynamics drive herring clupea harengus growth in the central Baltic Sea. Mar. Ecol. Prog. Ser. 413, 241–252.
Cerdà J., Zapater C., Chauvigné F., Finn R. N. (2013). Water homeostasis in the fish oocyte: New insights into the role and molecular regulation of a teleost-specific aquaporin. Fish Physiol. Biochem. 39, 19–27.
DeFaveri J., Merilä J. (2014). Local adaptation to salinity in the three-spined stickleback? J. Evolutionary Biol. 27, 290–302.
Enbody E. D., Pettersson M. E., Sprehn C. G., Palm S., Wickström H., Andersson L. (2020). Ecological adaptation in European eels is based on phenotypic plasticity. Proc. Natl. Acad. Sci. U.S.A., 118, e2022620118.
Fabra M., Raldúa D., Power D. M., Deen P. M. T., Cerdà J. (2005). Marine fish egg hydration is aquaporin-mediated. Science 307, 545.
Han F., Jamsandekar M., Pettersson M. E., Su L., Fuentes-Pardo A. P., Davis B. W., et al. (2020). Ecological adaptation in Atlantic herring is associated with large shifts in allele frequencies at hundreds of loci. eLife 9, e61076.
Hemmer-Hansen J., Hüssy K., Baktoft H., Huwer B., Bekkevold D., Haslob H., et al. (2019). Genetic analyses reveal complex dynamics within a marine fish management area. Evolutionary Appl. 12, 830–844.
Hill J., Enbody E. D., Pettersson M. E., Sprehn C. G., Bekkevold D., Folkvord A., et al. (2019). Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 116, 18473–18478.
ICES (2021). Baltic Fisheries assesment working group (WGBFAS) (Copenhagen: ICES Scientific Reports).
Johannesson K., André C. (2006). Life on the margin: genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol. Ecol. 15, 2013–2029.
Johannesson K., Le Moan A., Perini S., André C. (2020). A Darwinian laboratory of multiple contact zones. Trends Ecol. Evol. 35, 1021–1036.
Jonsson P. R., Kotta J., Andersson H. C., Herkul K., Virtanen E., Nyström Sandman A., et al. (2018). High climate velocity and population fragmentation may constrain climate-driven range shift of the key habitat former Fucus vesiculosus in the Baltic Sea. Diversity Distribution 24, 892–905.
Kniebusch M., Meier H. E. M., Radtke H. (2019). Changing salinity gradients in the Baltic Sea as a consequence of altered freshwater budgets. Geophysical Res. Lett. 46, 9739–9747.
Köster F. W., Huwer B., Hinrichsen Hans-H., Neumann V., Makarchouk A., Eero M., et al. (2017). Eastern baltic cod recruitment revisited—dynamics and impacting factors. ICES Journal of Marine Science 74, 3–19. doi: 10.1093/icesjms/fsw172
Kotta J., Vanhatalo J., Jänes H., Orav-Kotta H., Rugiu L., Jormalainen V., et al. (2019). Integrating experimental and distribution data to predict future species patterns. Sci. Rep. 9, 1821.
Kratzer S., Moore G. (2018). Inherent optical properties of the Baltic Sea in comparison to other seas and oceans. Remote Sens. 10, 418.
Lamichhaney S., Barrio A. M., Rafati N., Sundström G., Rubin C.-J., Gilbert E. R., et al. (2012). Population-scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proc. Natl. Acad. Sci. U.S.A. 109, 19345–19350.
Lamichhaney S., Fuentes-Pardo A. P., Rafati N., Ryman N., Mccracken G. R., Bourne C., et al. (2017). Parallel adaptive evolution of geographically distant herring populations on both sides of the north Atlantic ocean. Proc. Natl. Acad. Sci. U.S.A. 114, E3452–E3461.
Larsen P. F., Nielsen E. E., Meier K., Olsvik P. A., Hansen M. M., Loeschcke V. (2011). Differences in salinity tolerance and gene expression between two populations of Atlantic cod (Gadus morhua) in response to salinity stress. Biochem. Genet. 50, 454–466.
Leder E. H., André C., Le Moan A., Töpel M., Blomberg A., Havenhand J. N., et al. (2021). Post-glacial establishment of locally adapted fish populations over a steep salinity gradient. J. Evolutionary Biol. 34, 138–156.
Le Moan A., Bekkevold D., Hemmer-Hansen J. (2021). Evolution at two time frames: ancient structural variants involved in post-glacial divergence of the European plaice (Pleuronectes platessa). Heredity 126, 668–683.
Manzon L. A. (2002). The role of prolactin in fish osmoregulation: A review. Gen. Comp. Endocrinol. 125, 291–310.
Martinez Barrio A., Lamichhaney S., Fan G., Rafati N., Pettersson M., Zhang H., et al. (2016). The genetic basis for ecological adaptation of the Atlantic herring revealed by genome sequencing. eLife 5, e12081.
Matschiner M., Barth J. M. I., Tørresen O. K., Star B., Baalsrud H. T., Brieuc M. S. O., et al. (2022). Supergene origin and maintenance in Atlantic cod. Nat. Ecol. Evol. 6, 469–481.
Möllman C., Kornilovs G., Fetter M., Köster F. W. (2004). Feeding ecology of central Baltic Sea herring and sprat. J. Fish Biol. 65, 1563–1581.
Momigliano P., Jokinen H., Fraimout A., Florin A.-B., Norkko A., Merilä J. (2018). Extraordinarily rapid speciation in a marine fish. Proc. Natl. Acad. Sci. U.S.A. 114, 6074–6079.
Nissling A., Kryvi H., Vallin L. (1994). Variation in egg buoyancy of Baltic cod Gadus morhua and its implications for egg survival in prevailing conditions in the Baltic Sea. Mar. Ecol. Prog. Ser. 110, 67–74.
Nissling A., Westin L. (1997). Salinity requirements for successful spawning of Baltic and belt Sea cod and the potential for cod stock interactions in the Baltic Sea. Mar. Ecol. Prog. Ser. 152, 261–271.
Ojaveer H., Jaanus A., MacKenzie B. R., Martin G., Olenin S., et al. (2010). Status of biodiversity in the Baltic Sea. PLoS ONE 5 (9), e12467. doi: 10.1371/journal.pone.0012467
Osei-Owusu J., Yang J., Vitery M. D. C., Qiu Z. (2018). Molecular biology and physiology of volume-regulated anion channel (VRAC). Curr. Top. Membr 81, 177–203.
Pettersson M. E., Rochus C. M., Han F., Chen J., Hill J., Wallerman O., et al. (2019). A chromosome-level assembly of the Atlantic herring genome–detection of a supergene and other signals of selection. Genome Res. 29, 1919–1928.
Poulsen E. M. (1931). “Biological investigations upon the cod in Danish waters,” in Meddelelser fra kommisionen for danmarks fiskeri-og havundersøgelser, (Copenhagen: C. A. Reitzels publisher) vol. Vol. IX., No 1.
Reusch T. B. H., Dierking J., Andersson H. C., Bonsdorff E., Carstensen J., Casini M., et al. (2018). The Baltic Sea as a time machine for the future coastal ocean. Sci. Adv. 4, eaar8195.
Ryman N., Lagercrantz U., Andersson L., Chakraborty R., Rosenberg R. (1984). Lack of correspondence between genetic and morphological variability patterns in Atlantic herring (Clupea harengus). Heredity 53, 687–704.
Schade F. M., Weist P., Dierking J., Krumme U. (2022). Living apart together: Long-term coexistence of Baltic cod stocks associated with depth-specific habitat use. PloS One 17, e0274476.
Snoeijs-Leijonmalm P., Schubert H., Radziejewska T. (2017). Biological oceanography of the Baltic Sea (Dordrecht: Springer Science+Business Media).
Sodeland M., Jentoft S., Jorde P. E., Mattingsdal M., Albretsen J., Kleiven A. R., et al. (2022). Stabilizing selection on Atlantic cod supergenes through a millennium of extensive exploitation. Proc. Natl. Acad. Sci U.S.A. 119, e2114904119.
Svedäng H., Savchuk O., Villnäs A., Norkko A., Gustafsson B. G., Wikström S. A., et al. (2022). Re-thinking the “ecological envelope” of Eastern Baltic cod (Gadus morhua): conditions for productivity, reproduction, and feeding over time. ICES J. Mar. Sci. 79, 689–708.
Thorsen A., Kjesbu O. S., Fyhn H. J., Solemdal P. (1996). Physiological mechanisms of buoyancy in eggs from brackish water cod. J. Fish Biol. 48, 457–477.
Vallin L., Nissling A. (2000). Maternal effects on egg size and egg buoyancy of Baltic cod, Gadus morhua: implications for stock structure effects on recruitment. Fisheries Res. 49, 21–37.
Weist P., Schade F. M., Damerau M., Barth J. M. B., Dierking J, Andre C., et al. (2019). Assessing SNP-markers to study population mixing and ecological adaptation in Baltic cod. PloS One 14, e0218127.
Keywords: Atlantic herring, Atlantic cod, genetic adaptation, climate change, Baltic Sea
Citation: Andersson L, André C, Johannesson K and Pettersson M (2023) Ecological adaptation in cod and herring and possible consequences of future climate change in the Baltic Sea. Front. Mar. Sci. 10:1101855. doi: 10.3389/fmars.2023.1101855
Received: 18 November 2022; Accepted: 23 February 2023;
Published: 15 March 2023.
Edited by:
Agneta Andersson, Umeå University, SwedenReviewed by:
Juan Andrés López, University of Alaska Fairbanks, United StatesCopyright © 2023 Andersson, André, Johannesson and Pettersson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leif Andersson, bGVpZi5hbmRlcnNzb25AaW1iaW0udXUuc2U=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.