
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 06 February 2023
Sec. Marine Megafauna
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1088598
This article is part of the Research Topic Risks, Threats, and Conservation Status of Cetaceans in the Mediterranean and Black Seas View all 15 articles
 Rachele Antonacci1†
Rachele Antonacci1† Giovanna Linguiti1†
Giovanna Linguiti1† Federica Paradiso1
Federica Paradiso1 Chiara Scalone2
Chiara Scalone2 Carmelo Fanizza3
Carmelo Fanizza3 Elena Ciani1
Elena Ciani1 Giulia Cipriano1
Giulia Cipriano1 Salvatrice Ciccarese1
Salvatrice Ciccarese1 Roberto Carlucci1*
Roberto Carlucci1*In the framework of global and EU policies focused on stopping the loss of biodiversity process, deepening the genetic variability, especially of populations species identified as threatened, is crucial for defining conservation units and developing appropriate conservation strategies. This is more urgent for cetacean species in the Mediterranean because they assume a key ecological role in the marine food web and are severely affected by numerous and different anthropogenic pressures. This study aims to increase information on the genetic variability of striped dolphin in the Northern Ionian Sea by investigating the population structure, phylogenetic relationships and phylogeographic patterns using two mtDNA markers. From October 2020 to August 2021, a total of 88 skin tissue samples were collected from free-ranging dolphins in the Gulf of Taranto by applying the non-invasive technique of skin swabbing. An acceptable amount of DNA was extracted from 86 samples and used for subsequent genetic analysis conducted on the partial sequences of 421 and 704 bp in length of the cytb gene and D-loop control region, respectively. In addition, the sequences of the two mtDNA markers were joined together to compose a mtDNA concatenated sequence of 1125 bp for each sampled dolphin in order to investigate the genetic variability of the species population in the study area. Genetic analysis highlighted a low nucleotide diversity and high haplotypic diversity of the striped dolphin of the Gulf of Taranto, suggesting a population in rapid expansion after a period of reduction in size and diversity of the initial population. The phylogenetic analyses revealed the presence of at least two different lineages of Stenella coeruleoalba in the Mediterranean Sea, one specific to the Northern Ionian Sea and one shared with the Mediterranean population, confirming results already obtained for the local unit in the Gulf of Taranto. The results point out a potential problem of hybridization between striped and common dolphins which needs to be further investigated. Therefore, increasing the analysis of several markers may increase understanding of the genetic diversity of the population in the Ionian Sea and represent a useful tool to support the implementation of future effective conservation measures.
Biological diversity and richness are decreasing globally because of the massive impact of anthropogenic activities responsible for habitat destruction and fragmentation, pollution, exotic invasions, and climate change (Féral, 2002; Duffy and Stachowicz, 2006; Storch et al., 2022). This trend, if not quickly stopped or slowed down, will continue to negatively affect biodiversity which is strongly linked to human well-being as highlighted by the New Global Framework for Managing Nature Through 2030 from the Convention on Biological Diversity (CBD) (https://www.cbd.int/article/draft-1-global-biodiversity-framework), the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) with the Global assessment report on biodiversity and ecosystem services (IPBES, 2019) and the United Nations Sustainable Development Goals (Stange et al., 2021). Therefore, a quantitative evaluation of the status and trends of intraspecific and interspecific genetic diversity and that at the community level is necessary to point out possible adaptations to environmental changes in ecosystem services, resilience capacity and stability of functions (Prieto et al., 2015; Hunter et al., 2018; Hoban et al., 2020). This type of investigation can be carried out thanks to novel molecular and advanced genomic tools developed within conservation genetics, a discipline that helps us to resolve taxonomic uncertainties, to define evolutionarily diverged units such as subpopulations within the same species (de los Angeles Bayas-Rea et al., 2018) and to obtain information important for species conservation (sex, population structure, gene flow, inbreeding and outbreeding rates). It also allows us to integrate genetics with demographic and environmental variables to predict extinction risks and find proper conservation measures in order to slow biodiversity erosion (Allendorf et al., 2007; Frankham, 2010; Coker, 2017). More specifically, analyzing genetic variation over time [genetic diversity, differentiation, and distance (Taylor et al., 2010)], differentiations in specific local populations with a defined population genetic structure could be revealed as a result of low gene flow due, for example, to a recent geographic isolation or a divergence (Gaspari et al., 2019). This phenomenon is highly challenging for biodiversity conservation and mostly for that of highly mobile species such as cetaceans assuming a key ecological role in the marine food web (Roman and McCarthy, 2010; Ricci et al., 2019; Carlucci et al., 2021a) even as sentinel species for human and ecosystem health (Bossart, 2011) and a broad geographic range of distribution. Moreover, cetacean species in the Mediterranean Sea are subject to multiple stressors such as habitat fragmentation and loss, alterations to distribution and availability of resources, climate change, chemical and noise pollution as well as several other threats (Coll et al., 2012; Pace et al., 2015; Notarbartolo di Sciara, 2016).
Although genetic variability in some cetacean species has already been investigated, such as for the killer whale in the Northern Pacific Ocean (Barrett-Lennard and Ellis, 2001), the blue whale in southern Australian waters (Attard et al., 2015), the common bottlenose dolphin in the southern Pacific Ocean (de los Angeles Bayas-Rea et al., 2018), the striped dolphin (Bourret et al., 2007; Gkafas et al., 2017; Ciccarese et al., 2019; Gaspari et al., 2019) and the short-backed common dolphin in different areas of the Mediterranean Sea (Santostasi et al., 2021), increasing these studies in the Mediterranean Sea is very urgent given that genetic diversity generally underpins population resilience and persistence, thus determining the success and long term survival rate of a species in threatened and changing environments (Pace et al., 2015). Indeed, in-depth investigations on genetic variability of cetacean species might help recognize subpopulations or metapopulations, clarify possible connectivity between putative populations and prevent the decline of a species due to environmental and genetic threats such as inbreeding or hybridization as demonstrated for the striped dolphin and common dolphin in the Greek seas (Antoniou et al., 2018; Johnson et al., 2022). Moreover, further studies on this theme could provide more reliable information for the assessment of the extinction risk of species listed in the IUCN Red List even if, genetic information is currently used indirectly to assess the conservation status of species according to different criteria considered (IUCN, 2012; Garner et al., 2020).
In the Mediterranean Sea, studies on genetic variability of cetacean species have demonstrated, for all species regularly occurring in the basin, that the Mediterranean populations are differentiated from the Atlantic ones (ACCOBAMS, 2021). Within the Mediterranean, there is currently only evidence of genetic differences between groups living in the same area for the striped dolphin, Stenella coeruleoalba. In the Ligurian Sea, little differentiation is detected between offshore and inshore groups probably linked to different environmental factors and/or prey resources that reflect on group behaviour (Gaspari et al., 2007). In the Gulf of Taranto, the existence of an independent lineage was suggested by studies carried out on genetic variability of the cytb gene sequence (Ciccarese et al., 2019; Linguiti et al., 2021). However, a recent study comparing genetic information from both nuclear and mitochondrial samples from Gibraltar to Israel has revealed that the population of striped dolphins in the Mediterranean is structured with low levels of gene flow across the region (Gaspari et al., 2019).
This state of knowledge and the recent change in the conservation status of the species from Vulnerable to Least Concern (ACCOBAMS, 2021; Lauriano, 2021) in any case do not solve the question about the possible occurrence of subpopulations, as recently identified for the Gulf of Corinth (Bearzi et al., 2022), or metapopulations across the Mediterranean regions. Thus, further genetic investigations are necessary. Therefore, this study aimed to deepen understanding of the genetic variability of individuals of striped dolphin in the Gulf of Taranto through investigations of mtDNA sequences of two markers, the cytochrome b (cytb) gene and the D-loop control region (CR). The cytb gene was chosen as a coding marker of the mtDNA to compare the results with those of a previous study conducted in the same study area (Ciccarese et al., 2019). Moreover, improvements in the sampling activity and in laboratory protocols allowed us to analyze also the D-loop as mtDNA not coding region. This choice was also guided by the large number of cytb and D-loop sequences, both of the striped dolphin and of other Delphinidae species, available in the GenBank database.
The Gulf of Taranto is in the northernmost part of the Northern Ionian Sea (Central Mediterranean Sea) encompassing an area of about 14000 km2 from Santa Maria di Leuca to Punta Alice (Figure 1). It is characterized by a narrow continental shelf with a steep slope and several channels in the western sector and by descending terraces toward the submarine canyon known as the “Taranto Valley” in the eastern one. The complex morphology of the area together with the circulation of water masses involve the occurrence of seasonal and decadal upwelling currents (Civitarese et al., 2010; Matarrese et al., 2011; Carlucci et al., 2014; Pinardi et al., 2016) playing a significant role in sustaining productivity (Capezzuto et al., 2010; Maiorano et al., 2010; Carlucci et al., 2018; Ricci et al., 2019) and favouring the occurrence of valuable habitats from the conservation perspective such as the Santa Maria di Leuca cold-water coral province (D’Onghia et al., 2016; Vassallo et al., 2017) and those inhabited by several species of cetaceans (Bellomo et al., 2019; 2020c; Carlucci et al., 2018b; Carlucci et al., 2018c; Carlucci et al., 2020a; Carlucci et al., 2020b; 2021a; Santacesaria et al., 2019; Cipriano et al., 2022). Unfortunately, the basin is potentially affected by several human pressures or threats resulting in possible direct and indirect impacts on cetaceans (Carlucci et al., 2021; Ricci et al., 2021).
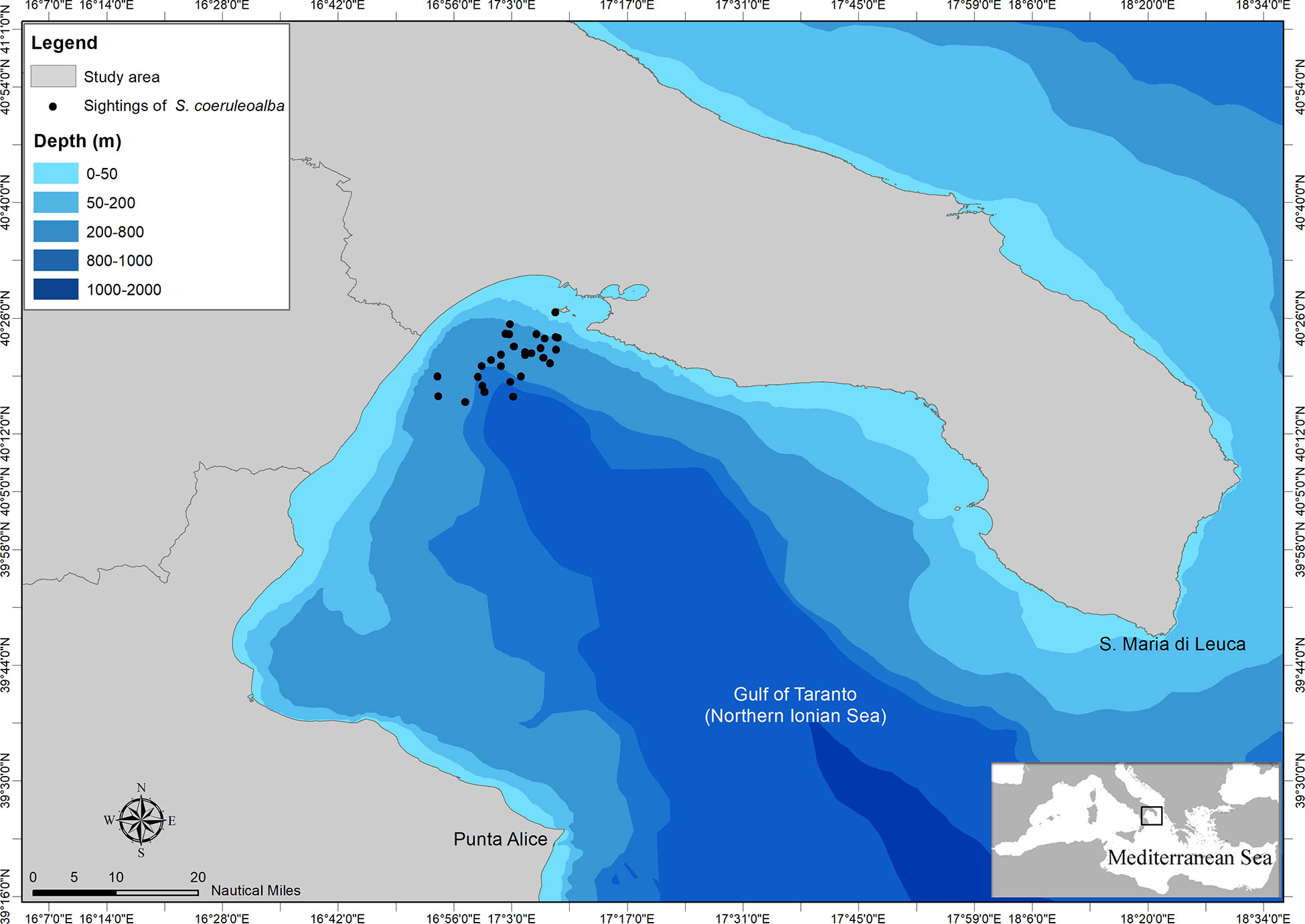
Figure 1 Map of the study area with indication of sightings of striped dolphin from which genetic samples were collected.
Skin tissue samples were collected during standardized vessel-based surveys carried out while investigating an area of 960 km2 in the northernmost portion of the Northern Ionian Sea from October 2020 to August 2021. Surveys were carried out only in favourable sea-weather conditions (Douglas scale ≤ 3 and Beaufort scale ≤ 4) applying an effort of approximately 5 h per day along 35 nautical miles and adopting a zig-zag line transect sampling (Buckland et al., 2004; Thomas et al., 2010). Genetic sampling was carried out under authorizations provided by Ministry of the Environment and the Protection of the Territory and Sea, under Authorization 367-REG-1570798753503, Prot. n. 28525, applying the non-invasive technique of skin swabbing (Harlin et al., 1999; Cosentino et al., 2015; Ciccarese et al., 2019; Linguiti et al., 2021). This method consists of applying moderate friction using a 4 x 4 cm synthetic fibre scrub pad on the dorsal-lateral region of an individual to collect the superficial layer of the skin, taking advantage of the moment at which, it approaches the boat during sightings and then came to the surface to breath. The scrub pad was attached with plastic fasteners to the tip of a 130 cm long telescopic aluminium stick covered with a soft sponge-like tissue to prevent hurting the animals. Although striped dolphins can react to the skin swab by swimming, jumping, or diving sometimes they came back close to the boat confirming that the sampling method did not cause any damage or irreversible stress. Individuals sampled are always recognizable by the light skin scratches caused by the scrub pad, preventing unwanted resampling of the same dolphin.
The skin tissue samples collected were then immediately removed with sterilized forceps from the scrub pad and transferred to a flask containing a 70% alcohol solution, labelled, and preserved at 4°C. At the same time as the genetic samples were taken, information about sighting date, geographic coordinates, depth (m), group size (number of individuals) and predominant activity state was collected. In particular, the collection of data concerning the predominant activity states of groups of striped dolphins encountered was carried out applying the focal-group protocol with instantaneous scan sampling (Mann, 1999; Neumann, 2001).
The sampling activities were performed with complete respect for the animals, respecting their space and trying not to interfere with their natural behaviour or their activities (also taking into account their attitude towards the research boats and the researchers on board).
Together with the skin tissue samples collected from live individuals, two samples were collected from two stranded striped dolphins found on 2nd March 2021 (on the Marina di Ginosa coast, west of Taranto) and on 27th April 2021 (on the Leporano Marina coast, east off Taranto), respectively. Sampling was carried out by directly taking the skin from the bodies using a sterile dermal biopsy punch curette and preserving the samples at 4°C in labelled flasks containing 70% alcohol (ACCOBAMS-MOP7/2019/Doc 33, 2019).
Total genomic DNA from skin samples was extracted using a Chelex-100 (Sigma) resin suspension in Tris-EDTA (10 mM Tris HCl, 1 mM EDTA, pH 8.0). Chelex solution (500μL at 15%) was added to each tube containing the sloughed skin sample previously preserved at 4°C. Due to the fast sedimentation of the Chelex solution, it was crucial for it to be frequently stirred prior to pipetting and putting it in the tube. The tubes were vortexed and incubated at 100˚C for 20 minutes, and then placed on ice for 2 minutes. The skin samples were then centrifuged at 13000 rpm for 5 minutes, and the supernatant was transferred to new tubes. The DNA was purified using the standard phenol/chloroform method, quantified, and the quality was checked with a Nanodrop 1000 spectrophotometer from Thermo Scientific.
A protocol for sex determination was applied as an additional tool, to photography of sampled individuals, to ensure the uniqueness of the samples from individuals sharing the same haplotype within the same sampling group.
The sex of free-ranging and stranded dolphins was identified with a duplex PCR amplification of the striped dolphin ZFX/ZFY and SRY gene fragments.
A set of three oligonucleotide primers for multiplex PCR amplification of the ZFX and ZFY partial sequences was designed: a forward-orientated oligonucleotide primer designed to anneal to the ZFY, as well as the ZFX sequence (ZFYX0582F, 5’-ATAGGTCTGCAGACTCTTCTA-3’), and two reverse-orientated oligonucleotide primers placed within a polymorphic position between the ZFX (ZFX0923R 5’-AGAATATGGCGACTTAGAACG-3’) and ZFY sequences (ZFY00767R 5’-TTTGTGTGAACTGAAATTACA-3’). PCR amplification reactions were carried out in a 25ml reaction mixture containing 500 ng of sample DNA, 5X PCR buffer (with 15 mM MgCl2 and 5 mM dNTPs), 10 μM of each primer, and 1 U of Taq enzyme – yourSIALc HiFi Polymerase (S.I.A.L.). PCR thermo-cycling conditions consisted of an initial denaturation step at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 60 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s with a final extension step at 72°C for 5 min.
Gender was determined by the banding pattern on a 2% agarose gel, stained with 0.5 μg/ml ethidium bromide, and visualized under ultraviolet light. The expected product size was a 382 bp single band, as determined by electrophoresis, for females and two bands (382 bp and 226 bp long) for males.
To verify the male gender, another primer set (SRYF 5’-GAGAATCCCCAAATGCAAAACTCAGA-3’, SRYR 5’-GGAATTGAGTTGCAAATGGCAGCAA-3’) was used to amplify a 418 bp fragment of the SRY gene. The PCR conditions were those described above.
2-mtDNA regions were amplified for all samples: the cytochrome b (cytb) gene and the mtDNA CR (D-loop), designed based on the striped dolphin mitochondrial genome.
A fragment of the cytb gene was amplified by a PCR reaction, in a volume of 50 μl, using Taq polymerase - Platinum (Life Technology). The primer set (F1cytb 5’-TAACAGTCATGGCCACTGCATT-3’ and R2cytb 5’-TGGTTTGATGTGTGCAGGGGTG-3’) was used under the following conditions: 500 ng of each DNA sample, 10 mM dNTP, 50 mM MgCl2, 1 U Taq, 10 μM of each primer, and 10X PCR buffer. PCR thermo-cycling conditions consisted of an initial denaturation step at 94°C for 4 min, followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, with a final extension step at 72°C for 5 min.
The PCR products were purified and fully-sequenced, in both directions, by a sequencing commercial service, using another forward primer (F2cytb 5’-CCAACCTCTTATCAGCAATC-3’) along with another reverse primer (R1cytb 5’-AGGGTGGAATGGAATTATGTCT-3’). The forward primer (F2cytb) and the reverse one (R1cytb) were drawn respectively downstream of the forward and reverse primers used in the PCR reaction. The sequences acquired were used to assemble and edit the sequence of 421 bp of the cytb fragment for each sample.
A different primer set was used to amplify a fragment of the mtDNA CR (D-loop). A forward primer dLp1.5L (5’-CACCCAAAGCTGRARTTCTA-3’) and a reverse primer dLp8scR (5’-TAGGGACGAAGCACTGTAGG-3’) were used under the same PCR conditions as above. The PCR program consisted of an initial denaturation step at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 60 s, and extension at 72°C for 60 s with a final extension step at 72°C for 10 min. The PCR products were purified and fully-sequenced, in both directions, using a forward primer (dLpF2 5’-CAACATCACAGTACTACGTC-3’) along with another reverse primer (dLp5H 5’-CCATCGWGATGTCTTATTTAAGRGGAA-3’), designed downstream of the primers used in the PCR reaction. The sequences acquired were used to assemble and edit the sequence of 704 bp of the mtDNA CR (D-loop) fragment for each sample.
All cytb and D-loop haplotype sequences of individuals of striped dolphin sampled are available from GenBank database (Accession numbers ON959814 - ON959831).
The cytb and D-loop mtDNA sequences obtained from striped dolphins sampled in the Gulf of Taranto were aligned using Clustal Omega software (EBI). The genetic analysis was conducted on the partial sequences of 421 and 704 bp in length of the cytb gene and D-loop, respectively. The number of haplotypes and polymorphic sites as well as the type (single variable and parsimony information sites; transitions and transversions; synonymous and replacement changes, only for cytb coding sequence) of single nucleotide polymorphisms were assessed using Arlequin v. 3.5.2.2 (Excoffier and Lischer, 2010). Moreover, considering all samples from the Gulf of Taranto as incorporated into a single unit, the sample genetic diversity was estimated by calculating the haplotype diversity (h) and nucleotide diversity (π) with the same software package.
The Fu’s FS (Fu, 1996) and Tajima’s D neutrality (Tajima, 1989) tests, implemented in the program Arlequin v. 3.5.2.2 (Excoffier and Lischer, 2010), was performed in order to test past population expansion. A negative value of FS and Tajima’s D is considered evidence for excess of rare haplotypes over what would be expected under neutrality, as it would be expected from a recent population expansion or from genetic hitchhiking.
Pair-wise FST values among the considered taxonomic groups were also calculated.
The evolutionary relationships was investigated by building up phylogenetic trees based on the cytb and D-loop nucleotide sequences including samples of striped dolphin from the Gulf of Taranto (obtained in this work plus those reported in Ciccarese et al., 2019) and samples, retrieved from GenBank, of the same species from other geographic areas and of other phylogenetically comparable species such as Stenella clymene, S. longirostris, S. frontalis, S. attenuata, Delphinus delphis, Tursiops truncatus and T. aduncus. The corresponding cytb (GenBank ID: LC630882) and D-loop (GenBank ID: NC_012062) sequences from Grampus griseus were used as outgroup. Multiple alignments were carried out with the MUSCLE program (Edgar, 2004) both for the cytb and D-loop sequences. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000), and the units are in the number of base differences per site. Phylogenetic trees were built using the neighbor-joining (NJ) (Saitou and Nei, 1987) method implemented in MEGA X (Kumar et al., 2018; Stecher et al., 2020) and the grouping in the tree was supported by high bootstrap probability values. To support the phylogenetic inference additional phylogenetic analyses such as maximum likelihood (ML) and Bayesian Inference (BI) based on JModelTest (Posada, 2008) were carried out.
Phylogeographic networks were constructed using the sequences of cytb and D-loop of S. coeruleoalba. Networks were constructed adopting the median-joining algorithm implemented in the package Network 10.2.0.0 (Copyright 2004-2022 Fluxus Technology Ltd.). Haplotype frequency distribution in the considered major geographic areas were visualized through pie charts using different color codes. For better visualization of the network topology, branch lengths were not maintained proportional to the number of mutations.
Finally, to deepen knowledge of the genetic structure of the population of striped dolphin in the Gulf of Taranto the sequences of the two mitochondrial DNA-fragments, cytb and D-loop, were joined together to compose a mtDNA concatenated sequence of 1125 bp for each sampled dolphin. This allowed identification of the haplotypes and investigation of their variability.
During the sampling period from October 2020 to August 2021, the number of individuals sampled during each survey varied between 1 and 9 for a total of 88 striped dolphins sampled. An acceptable amount of DNA for subsequent analysis, ranging from 18 to 120 μg, was only extracted from 86 samples from free-ranging individuals (Supplementary Table 1). Sex was determined for 56 individuals, showing a sampling bias in favor of males (42) over females (14) (sex ratio 3:1).
The mtDNA cytb gene fragment of 421 bp was successfully amplified and sequenced in 85 out of the 86 samples extracted (99%) (Table 1). The sequences aligned were compared with those obtained in the previous work of Ciccarese et al. (2019) to verifying the possible match with haplotypes already identified. New haplotypes have been labelled with the abbreviation “Hap” and a progressive number. From this study, six cytb haplotypes were obtained (Table 1). Hap10 was identified in 80 of the 85 striped dolphin individuals (94%), confirming it to be the most common in the Gulf of Taranto as assessed in the previous work (Ciccarese et al., 2019). Haplotypes Hap-3, Hap-14, Hap-29, Hap-31 and Hap-32 were found in single individuals. While the Hap-3 had already been found in the Gulf of Taranto (Ciccarese et al., 2019), the other haplotypes were new findings in the area. In addition, Hap-31 and Hap-32 are new haplotypes also at the global level as they are not present in any database.
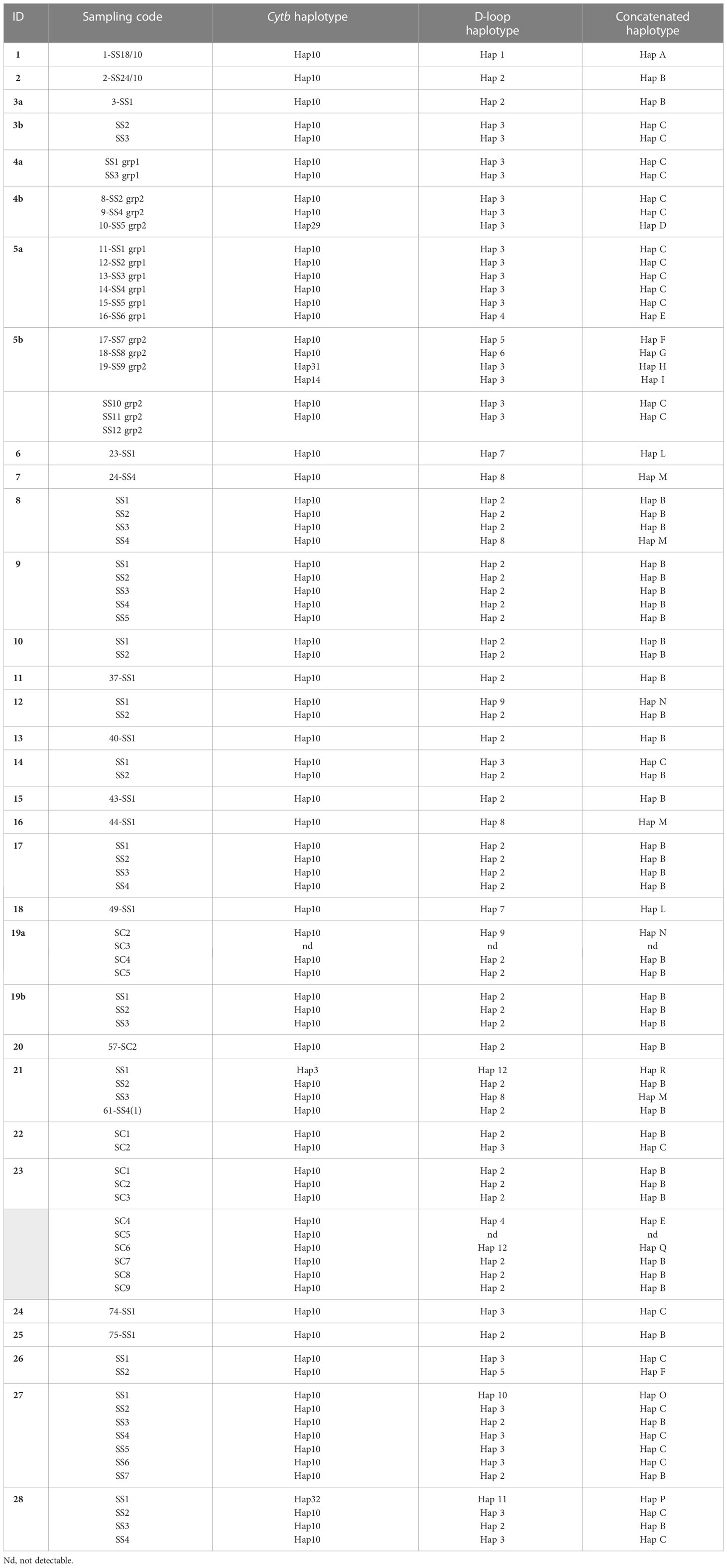
Table 1 Sampling code and haplotypes of cytb, D-loop and concatenated identified from individuals of S. coeruleoalba sampled in the Gulf of Taranto.
The haplotype analysis indicated 23 polymorphic loci, including 6 single variable sites (34, 280, 286, 295, 388 397 bp) and 17 parsimony information sites (91, 94, 109, 130, 178, 181, 184, 217, 244, 250, 259, 304, 310, 329, 341, 350, 394 bp) (Tables 2, 3). Within the observed substitutions, 11 are transition changes and only one is a transversion (ratio 11:1) (Tables 2, 3). The ratio between transition and transversion is in line with ratios observed in mammalian mtDNA ranging from 10:1 to 20:1 (or more) (Irvine et al., 1981) and with previous data on the same species (Ciccarese et al., 2019). Substitutions are observed chiefly at third codon positions except for the transitions at the first codon position of 329, 341 and 350 polymorphic sites. All changes represent silent substitutions (Table 2). The analysis aimed to identify the genetic diversity of the Gulf of Taranto dolphin population showed a mean value of nucleotide diversity (π) equal to 0.000885 ± 0.000946 and a mean value of haplotype diversity (h) equal to 0.1148 ± 0.0475, which are both lower than the values observed in the previous study (Ciccarese et al., 2019). The combined group, i.e. haplotypes found in this (group 1) plus those identified in the previous study by Ciccarese et al. (2019), (group 2), containing all the samples of S. coeruleoalba from the Gulf of Taranto still gives low levels of both nucleotide and haplotype diversity (Table 3).
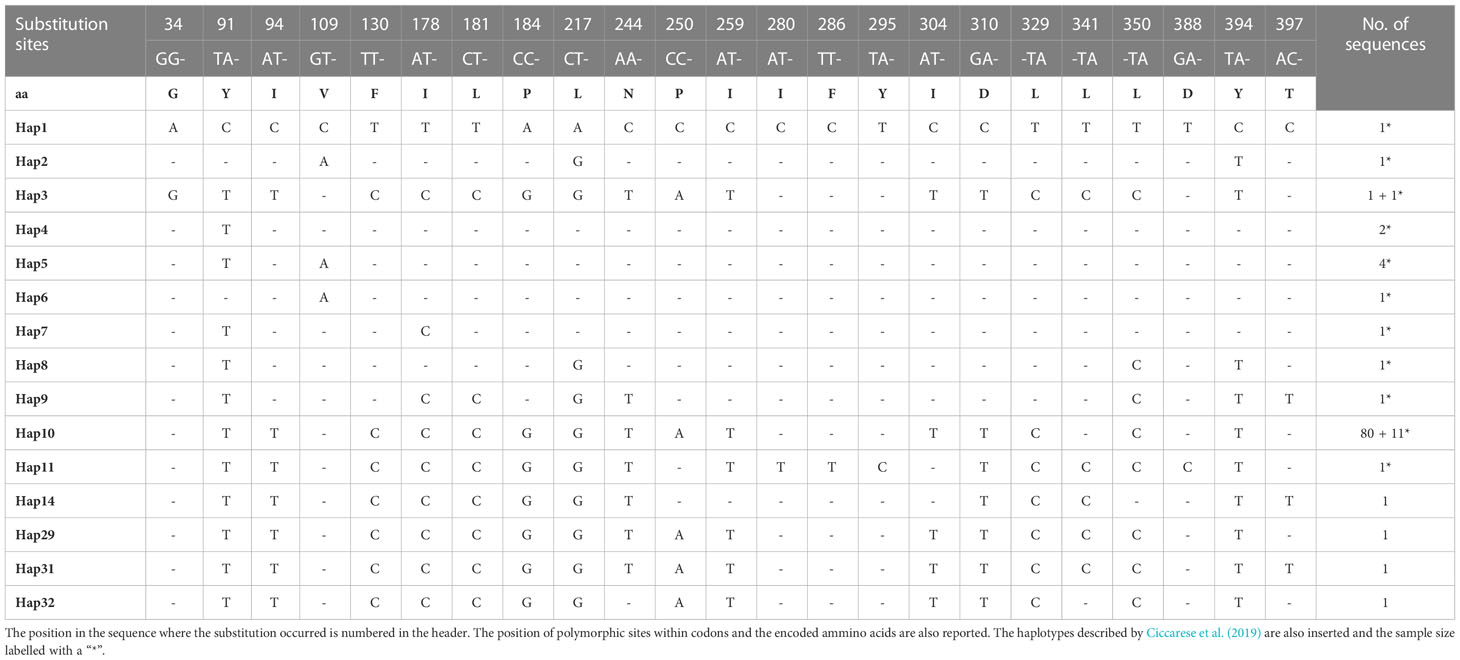
Table 2 Haplotypes identified in the 421bp mitochondrial cytb gene sequences, along with sample size.
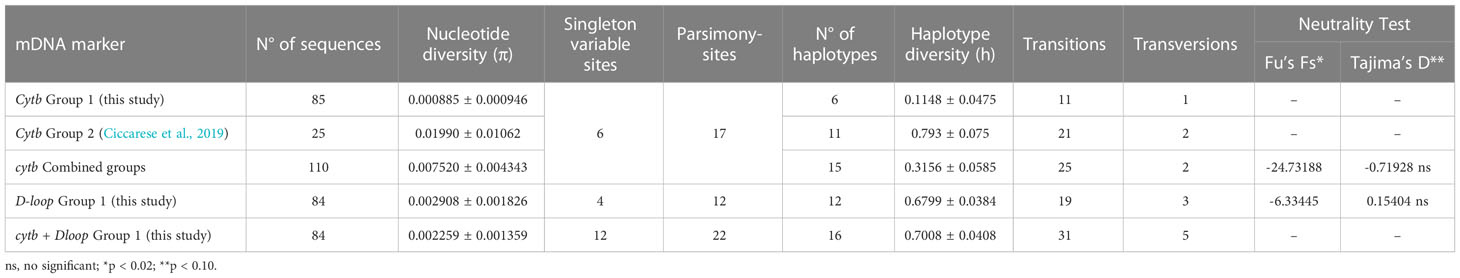
Table 3 Nucleotide and haplotype diversity in mtDNA cytb and D-loop sequences of individuals of S. coeruleoalba sampled in the Gulf of Taranto.
The evolutionary relationship of the cytb haplotypes was investigated by comparing the sequences of S. coeruleoalba from the Gulf of Taranto with sequences, retrieved from the GenBank dataset, of the same species from other geographic areas, and of other phylogenetically comparable species, i.e. S. clymene, S. longirostris, S. frontalis, S. attenuata, D. delphis, T. truncatus and T. aduncus (Supplementary Table 2). The corresponding cytb sequence from G. griseus was used as an outgroup. In particular, the following selection criterion was adopted. Only one gene sequence for each haplotype of each species was included in the analysis. All sequences were combined in the same alignment to build a phylogenetic tree using the NJ method (Figure 2). Since the different methods applied gave overlapping results (data not shown for ML and BI), the NJ tree has been preferred in order to be in line with previous evolutionary analysis performed by Ciccarese et al. (2019), of which this represents an updated version. The grouping in the tree was supported by high bootstrap probability values. The addition of new haplotype sequences reinforces without modifying the paraphyletic distribution of S. coeruleoalba. As a matter of a fact, it is possible to recognize five different groups (Figures 2A–E) divided into two principal branches as indicated by an arrow. In the upper branch, group A is the largest and most represented, with five haplotypes (Hap-3, Hap-10, Hap-29, Hap-31 and Hap-32) identified in the Gulf of Taranto intermingled with sequences derived from other geographic areas such as the Mediterranean Sea, Northeast Atlantic, Pacific Ocean, Indian Ocean, Eastern and Northern Pacific. Among these, Hap-10 and Hap-29 are the only ones to be shared between the dolphins of the Gulf of Taranto and those of the other seas. As previously described (Ciccarese et al., 2019), other haplotypes exclusive to the Gulf of Taranto, Hap-1, 2, 4, 5, 6, 7, 8, and 9, form a separate group in the tree (Figure 2E), suggesting the possible existence of a distinct Ionian (sub)population. Moreover, these haplotypes are closer to Tursiops truncatus sequences than to other sequences of congeneric species. Instead, S. coeruleoalba haplotypes distributed in the other clades are closer to D. delphis haplotypes, with Hap-12 (Figure 2C) clustering with D. delphis haplotypes, and Hap14 apart (Figure 2B) and tightly related to S. clymene as Hap-11 and Hap-13 are (Figure 2D). Moreover, the results of the analysis show the expected species-specific clustering of the cytb gene sequences of S. longirostris, S. frontalis, S. attenuata, D. delphis, T. aduncus and T. truncatus, together with the polyphyletic distribution of the S. clymene, already described (Amaral et al., 2014). S. clymene seems to be the result of an ancient speciation by a natural hybridization between two other species of dolphin closely related to each other, S. coeruleoalba and S. longirostris. This conclusion is in line with our findings. In fact, the sequences of the haplotypes of S. clymene in the tree form monophyletic groups, with the corresponding sequences of S. coeruleoalba and S. longirostris.
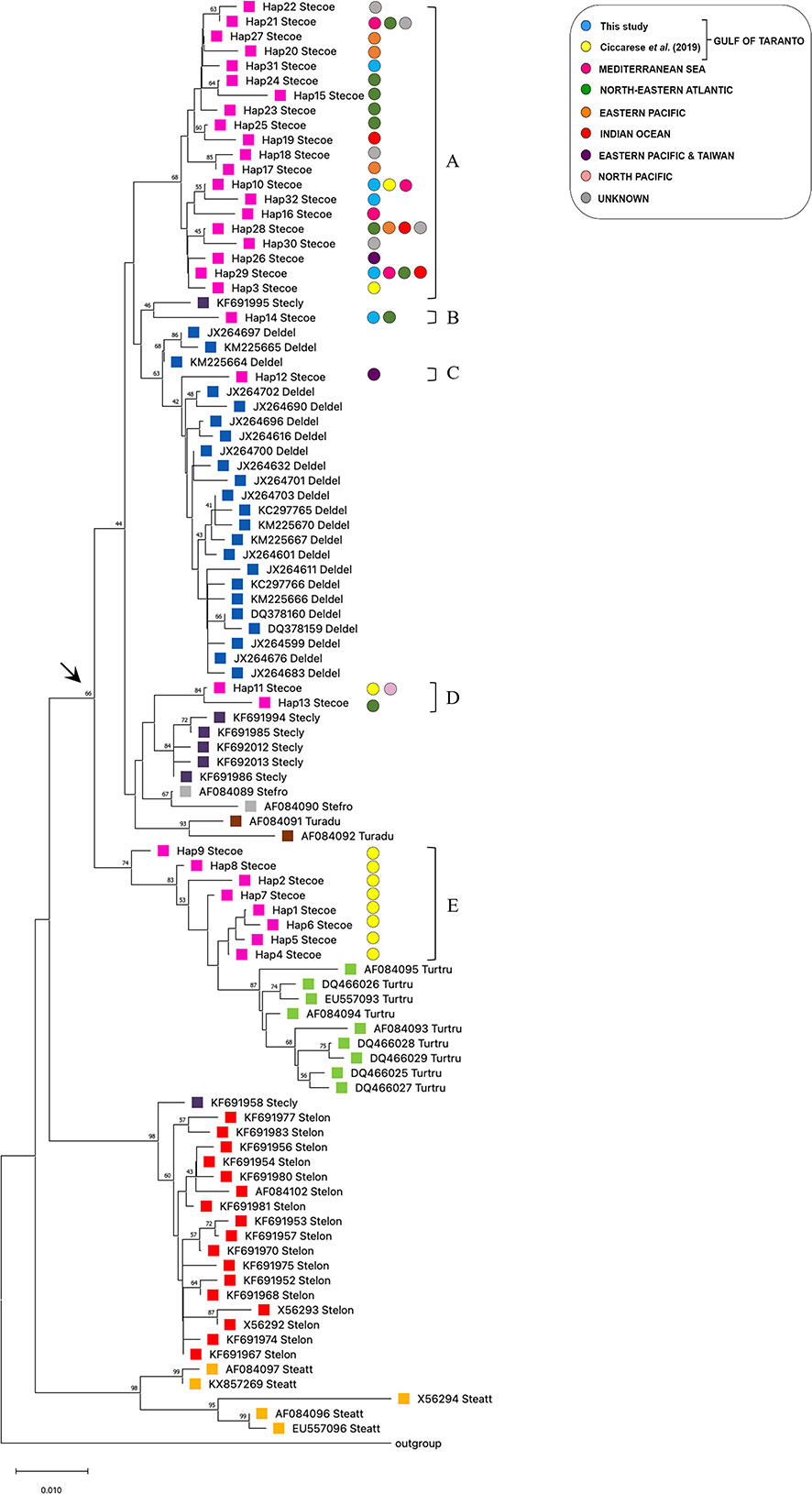
Figure 2 NJ tree inferred from Delphinidae cytb sequences. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). The optimal tree, with the sum of branch length = 0.52128464 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000) and are in the units of the number of base differences per site. This analysis involved 98 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair. There was a total of 421 positions in the final dataset. The colored circles indicate the geographic origin of the samples of the haplotypes of S. coeruleoalba. Every species is highlighted with a different colored square to enhance the distribution on the phylogenetic tree. The arrow indicates the paraphyletic branches containing the Stenella coeruleoalba haplotypes. Letters from (A–E) indicate striped dolphin clades.
The cytb haplotypes were further investigated to evaluate the genetic distances between the two paraphyletic groups of S. coeruleoalba, one exclusive of the Gulf of Taranto (Figure 2E), and one shared with other marine sites. Pair-wise FST comparisons confirmed significant differences between the two groups of S. coeruleoalba individuals. The observed FST value (0.67, Supplementary Table 3) was lower than most of the interspecific comparisons, while being higher than the pair-wise FST distances observed between T. aduncus and S. attenuata, S. frontalis and S. attenuata, as well as between T. aduncus and S. frontalis.
The mtDNA D-loop fragment of 704 bp was successfully amplified and sequenced in 84 out of the 86 samples extracted (approximately 98%). The sequence analysis revealed 12 distinct haplotypes from the study area (Table 1). Hap-2 and Hap-3 were the most frequent, being represented in 40 (48%) and 26 (31%) of samples respectively, followed by Hap-8 identified in 4 samples (5%). Hap-4, Hap-5, Hap-7, Hap-9 and Hap-12 were each found in two individuals and the remaining four haplotypes (Hap-1, Hap-6, Hap-10, Hap-11) were found in single individuals.
The haplotype analysis indicated 21 polymorphic loci, including 3 single variable sites (positions 50, 54 and 215) and 18 parsimony information sites (positions 81, 95, 101, 133, 245, 262, 280, 289, 299, 366, 387, 390, 447, 453, 500, 521, 552 and 600) (Tables 3, 4). The frequency of substitution sites along the D-loop is lower (21 sites/704 bp, one polymorphism every 33 nucleotides) than the cytb (23 sites/421 bp, a polymorphism every 18.3 nucleotides) sequence portion. Within the observed substitutions, 19 are transition changes and three are transversions.
The overall nucleotide diversity and haplotype diversity were 0.002908 ± 0.001826 and 0.6799 ± 0.0384, respectively, for the 84 D-loop sequences analyzed.
The evolutionary relationship of D-loop haplotypes occurring between samples of striped dolphin collected in the Gulf of Taranto and those of other geographic areas is shown in the phylogenetic tree reported in Figure 3. The accession numbers of all D-loop sequences used for this phylogenetic analysis are listed in Supplementary Table 4. This dataset was created by querying the GenBank database. Only one sequence for each haplotype was included in the analysis. Since most of the sequences available in the database covered 88% of the D-loop sequence used for the query, it was decided to shorten all sequences by 78 bp in order to compare them. It should be emphasized that the 78 nucleotides cut at 5’ of each sequence did not contain any polymorphic sites and therefore were not informative for the analysis. Thus, the D-loop region used in the phylogenetic analysis was 626 bp long instead of 704.
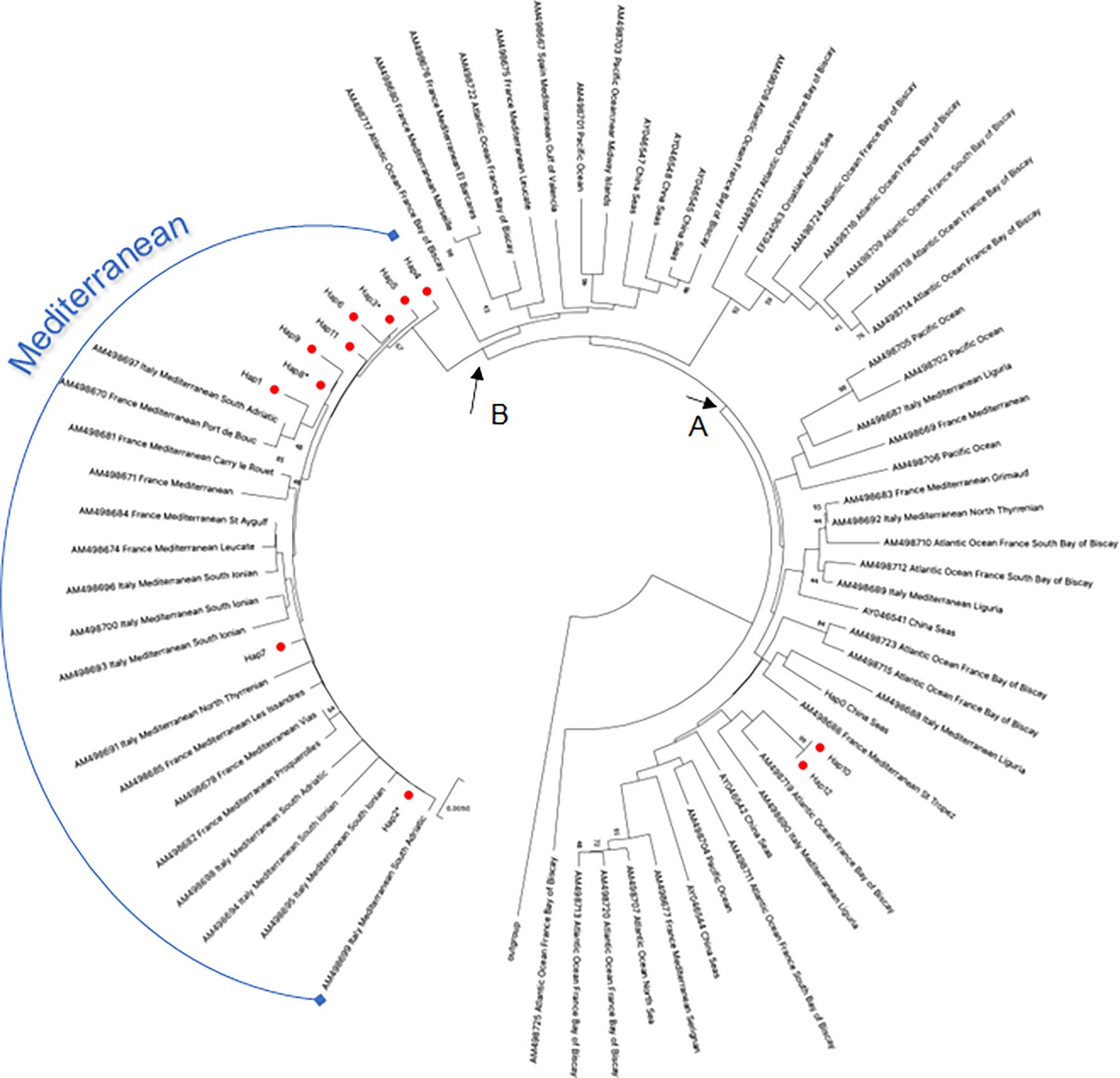
Figure 3 NJ tree inferred from S. coeruleoalba D-loop sequences. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2020). The optimal tree, with the sum of branch length = 1.27736443 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000) and are in the units of the number of base differences per site. This analysis involved 281 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 638 positions in the final dataset. The red circles indicated the haplotypes of S. coeruleoalba identified in the study area. An asterisk indicates haplotypes in the Gulf of Taranto also identified in other geographic areas (Supplementary Table 2). (A, B) nodes indicate the distribution of the striped dolphin haplotypes from the study area as discussed in the text.
D-loop haplotype sequences of the Gulf of Taranto dolphins were distributed in two distinct groupings that separate at node A. One group contains only Hap-10 and Hap-12, distributed among haplotypes of striped dolphins sampled in the Mediterranean and in other seas, such as the Atlantic and Pacific Oceans. This confirms the high motility and extensive migratory patterns of this species through different and distant geographical areas between the Mediterranean, the Atlantic Ocean and the Pacific Ocean up to the Sea of China, also passing through the Gulf of Taranto. Node B forms a second monophyletic group containing most of the striped dolphin sequences sampled in the Gulf of Taranto (from Hap-1 to Hap-9 and Hap 11) intermingled with sequences of striped dolphins sampled exclusively in the Mediterranean Sea, confirming the idea of the distinct evolution of the Mediterranean dolphin population.
A second phylogenetic tree was built to identify evolutionary relationship between D-loop haplotypes of striped dolphins occurring in the Gulf of Taranto and those of different species belonging to the Delphinidae family (Figure 4). The D-loop sequences used for the phylogenetic tree retrieved from Genbank are reported in Supplementary Table 5. The phylogenetic distribution of the D-loop sequences between and within the different Delphinidae species is broadly in agreement with the previous cytb evolutionary analysis (Figure 2). The tree shows the predicted species-specific clustering of S. longirostris, S. attenuata, and T. truncatus sequences, along with the only two S. clymene sequences available in the database intermingled between S. coeruleoalba sequences. The sequences of the haplotypes of T. aduncus also are distributed in two different clades and they both form a monophyletic group with the S. coeruleoalba sequences. In fact, molecular evidence supports T. aduncus as a species indistinct from but more closely related to S. coeruleoalba than to T. truncatus (LeDuc et al., 1999; Charlton et al., 2006; Nishida et al., 2007; Möller et al., 2008; Kingston et al., 2009; Xiong et al., 2009; Vilstrup et al., 2011). As in the previous tree, the sequences of the haplotypes of S. coeruleoalba show a paraphyletic grouping that separates sequences of striped dolphins sampled exclusively in the Mediterranean Sea from sequences of individuals also sampled in other sites. In addition, the greater phylogenetic affinity of S. coeruleoalba with D. delphis than other species indicates the possible cross-breeding between these two dolphin species as already reported in Greek seas (Antoniou et al., 2018).
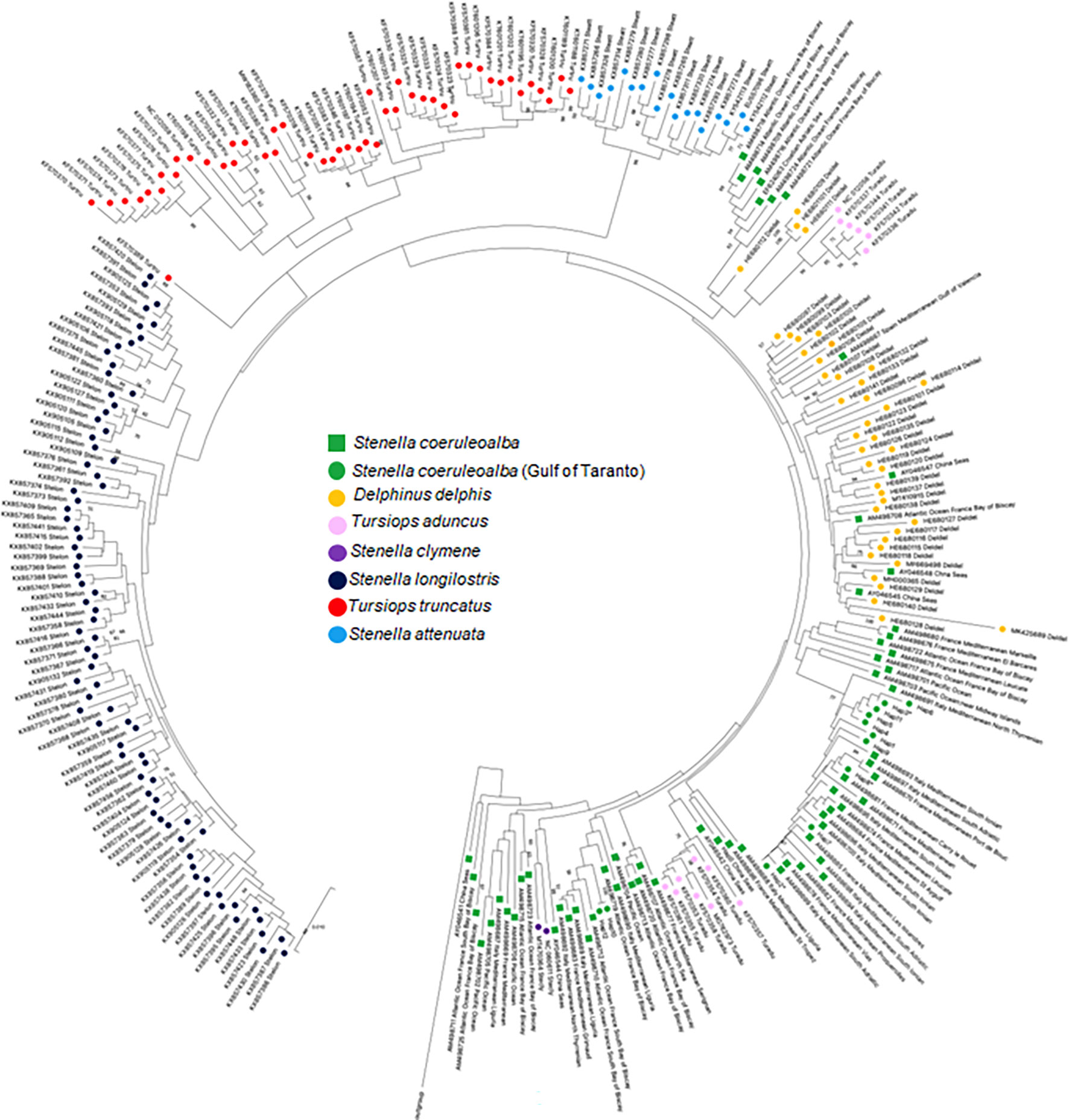
Figure 4 NJ tree inferred from Delphinidae D-loop sequences. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2020). The optimal tree, with the sum of branch length = 0.44828118 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000) and are in the units of the number of base differences per site. This analysis involved 76 nucleotide sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 632 positions in the final dataset. Every species is highlighted with a different colored circle to enhance the distribution on the phylogenetic tree. S. coeruleoalba of the Gulf of Taranto and of other areas were highlighted with different shape but the same color. The haplotype sequences identified in the Gulf of Taranto are highlighted with a red circle. Some of them (Hap-2, Hap-3 e Hap-8) are marked with an asterisk to indicate that their presence has also been documented in other areas.
A total of 84 concatenated sequences were obtained and analyzed to identify two-marker mtDNA haplotypes. The sequence analysis revealed 16 distinct haplotypes named with the acronym “Hap” followed by an alphabetical letter (Table 1 and Supplementary Table 6). The number of concatenated haplotypes is higher than that found for both haplotypes that compose it (6 for cytb and 12 for D-loop), suggesting a remarkable heterogeneity that characterizes the striped dolphins in the study area. Of these, Hap-B (40/84, approximately 48%) and Hap-C (23/84, 27%) are the most represented followed by Hap-M identified in 4 individuals (approximately 4.8%); Hap-E, Hap-F and Hap-L identified in 2 individuals each; and the remaining haplotypes identified in single individuals. Analyzing the combinations in detail, the most frequent cytb haplotype, Hap-10, contributes mainly to the constitution of the two-marker haplotypes (11/16) and it is in combination with all D-loop haplotypes except one (Hap-11). On the other hand, the most represented D-loop haplotype, Hap-2, is always associated with cytb Hap-10 to form Hap-B, which is the most represented concatenated haplotype (Table 1 and Supplementary Table 6).
Hap-3 is also a more represented D-loop haplotype and in most samples (23) it is combined with cytb Hap-10, but it has also been found in association with unique cytb haplotypes (Hap-14, Hap-29 and Hap-31) (Table 1 and Supplementary Table 6). Moreover, the D-loop Hap-12 besides constituting the concatenated Hap-Q haplotype in combination with cytb Hap-10, determines the Hap-R haplotype when associated with cytb Hap-3. Finally, Hap-P is the only combined haplotype determined by the union of two unique haplotypes (cytb Hap-32 and D-loop Hap11) (Table 1 and Supplementary Table 6).
The analysis of concatenated haplotypes indicated 35 polymorphic loci, including 12 single variable sites and 22 parsimony information sites (Table 3). Within the observed substitutions, 31 are transition changes and five are transversions. The nucleotide diversity is 0.002259 ± 0.001359, and the haplotype diversity is 0.7008 ± 0.0408 (Table 3).
The need to increase information about the genetic variability of cetaceans occurring in a semi-closed basin such as the Mediterranean Sea is a crucial point to implement effective measures for the conservation of putative populations or local units in this basin. This study, increasing samples and improving the methodology applied in a previous work (Ciccarese et al., 2019), has allowed us to better understand how wide the genetic variability of this species is at the local and Mediterranean scale. Furthermore, it contributes to broaden the available data source that can be used in the future evaluation phase of the health status of the Mediterranean subpopulation, shedding light on the possible presence of at least two different lineages of S. coeruleoalba in the Mediterranean Sea.
The ability to extract a good quantity (efficiency of extraction of 98%) of high quality of DNA from 88 samples collected during the study period was achieved thanks to improvements in the skill of the on-board team and in the DNA extraction protocol. Great attention was paid to not letting the Chelex solution sediment before being adequately mixed with the skin sample. To prevent fast sedimentation, it was enough to frequently stir the Chelex solution before pipetting and putting it in the sample. Moreover, the time of incubation of the sample with the Chelex resin was prolonged to 20 minutes, 5 minutes longer than the protocol adopted in Ciccarese et al. (2019) because a better yield was observed.
The choice of analyzing the nucleotide sequences of the mitochondrial markers cytb and D-loop was driven by their different evolutionary mutational rate. Cytb changes its amino acid sequence more slowly than any other mitochondrial gene (Simmons & Weller 2001). The protein function limits the nucleotide changes of the gene, as confirmed by our analysis that showed how all nucleotide changes were silent variations. Conversely, D-loop, a non-coding region, tends to be widely used as a marker due to its higher variation than the remaining regions of the mitogenome (Cann et al., 1984; Wang et al., 2019) and thus, has been frequently used for phylogenetic studies of closely related groups, especially for determining intra-specific phylogenies. Both these mitochondrial markers have been widely and successfully used for population differentiation analysis in different species (Imsiridou et al., 2019) as well as in different species among different areas (Giantsis et al., 2014; Turan et al., 2015; Šegvić-Bubić et al., 2016).
Although the π value of D-loop sequences is a little higher (just over three times) than those calculated for the cytb gene sequences of the same samples, both π values of the 2 markers indicate a low level of nucleotide diversity (<0.5%, as suggested by Grant and Bowen, 1998) and, consequently, little genetic divergence of the striped dolphin population in the Ionian Sea. On the contrary, the h value of D-loop sequences is to be considered clearly higher (about six times) than those calculated for the cytb sequences in the same sample group as well as in the combined group. Moreover, the h value calculated for the D-loop marker is >0.5, suggesting a large haplotype diversity (Grant and Bowen, 1998). This condition of high h and low π is attributed to a population in rapid expansion after a period of low effective population size as already suggested by Gaspari et al. (2019). The rapid growth of a population, in fact, enhances the retention of new mutations. The signature of the striped dolphin population expansion in the Ionian Sea was supported by the negative and significant (p<0.02) values of the neutrality Fu’s FS test statistics (Table 3).
The same trend about nucleotide (<0.5%) and haplotypes (>0.5) diversity values was observed for concatenated haplotypes confirming the hypothesis that striped dolphins in the Gulf of Taranto represent an evolving population. In detail, our results provide evidence of an increase in variability starting from prevalent haplotypes, represented by Hap-10 for cytb (94%) and Hap-2 for the D-loop (48%), along with groups of minor haplotypes that often derive from the founder haplotype after accumulating one or a few mutations.
A phylogeographic analysis reinforced this idea. Two median joining networks were constructed using, respectively, the 32 different haplotypes of striped dolphin (Table 2) based on the 421 bp cytb target region, and the 71 different haplotypes (Supplementary Table 7) based on the 626 bp D-loop target region.
From the point of view of phylogeographic analysis of the cytb sequences, Hap-10 and Hap-29 proved to be central haplotypes closely related to each other as indicated by only one mutational step that separates them (Figure 5). Hap-10, largely consisting of samples from the Gulf of Taranto, represents the Mediterranean haplotype, whereas Hap-29 is shared between individuals from the Mediterranean and other seas. Due to its co-presence in the Atlantic Ocean, Mediterranean Sea and Indian Ocean, Hap-29 could represent a junction point in the evolution between haplotypes of different areas and, due to the geographic continuity between the North-Eastern Atlantic and the Mediterranean Sea, it could have entered the Mediterranean evolving into Hap-10, the most represented haplotype in this area. The identification of Hap-29 in the Gulf of Taranto would confirm this hypothesis. The subsequent evolution of Hap-10 would have generated the grouping of the eight haplotypes (Hap-1, 2, 4, 5, 6, 7, 8 and 9) only observed in the Gulf of Taranto and significantly diverging from all the other haplotypes (Ciccarese et al., 2019). Pair-wise FST comparisons performed with mtDNA cytb sequences seems to support this hypothesis by showing a genetic differentiation between the S. coeruleoalba haplotypes exclusive from the Northern Ionian Sea and those found also in other areas, comparable to interspecific and even intergeneric distances observed in our study.
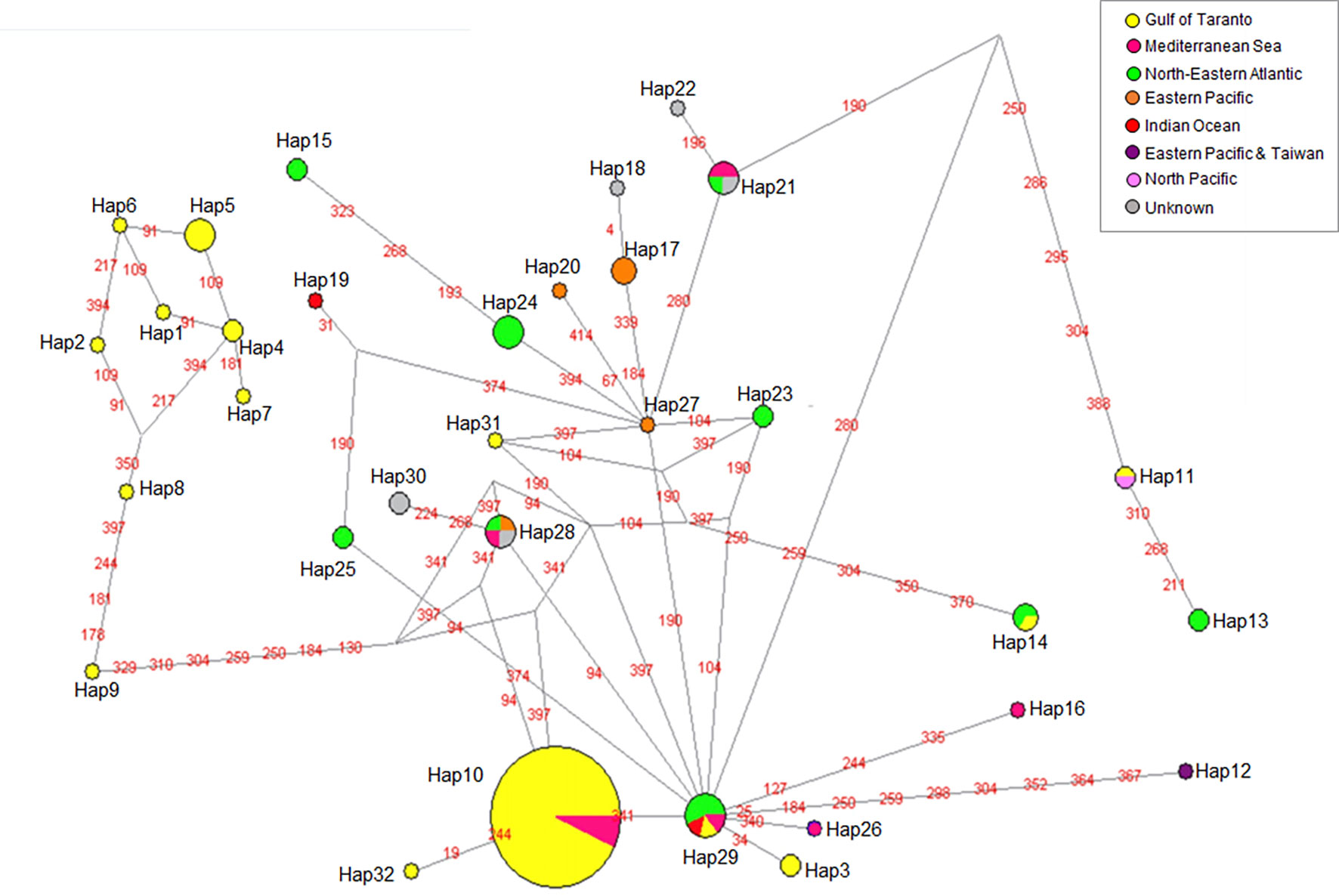
Figure 5 Median-joining network of cytb haplotypes of S. coeruleoalba. Haplotype frequency distribution in the considered geographic areas is visualized through pie charts using different color codes (see the graphic legend). For better visualization of the network topology, branch lengths were not maintained proportional to the number of mutations. The latter are indicated by red labels describing the mutated nucleotide position with respect to the first sequence in the dataset (Hap-1).
The unique Hap-32 may also have been generated directly from Hap-10. In contrast, the other unique haplotype identified in the Gulf of Taranto, Hap-31, is related to Hap-29, as is the case with Hap-3. Whatever the evolutionary steps, our results suggest and confirm the presence of at least two different lineages of S. coeruleoalba in the Mediterranean Sea.
In line with those reported for the cytb gene, the phylogeographic analysis of D-loop haplotypes also revealed evidence of a genetic divergence between the Mediterranean population of striped dolphin and those occurring in other seas (Figure 6). The network topology distinguishes two main haplotype groups. The right group consists of individuals almost exclusively sampled in the Mediterranean Sea, confirming the existence of a Mediterranean lineage; while, the left part of the network, typically reticulated, shows the relationships and connections between different marine sites including the Mediterranean Sea. All haplotypes of the D-loop gene identified in the Gulf of Taranto (this study), except for Hap-10 and Hap-12, are present among Mediterranean haplotypes. Hap-2, which is the most represented among samples, occupies a central position. Its star-like appearance suggests the hypothesis of a Mediterranean population expanding from it, as in the case of Hap-10 of the cytb gene. Specifically, since Hap-2 is always associated with cytb Hap-10, resulting in the concatenated haplotype Hap-B, it might represent the founder of the Mediterranean population as well as of the putative Ionian metapopulation.
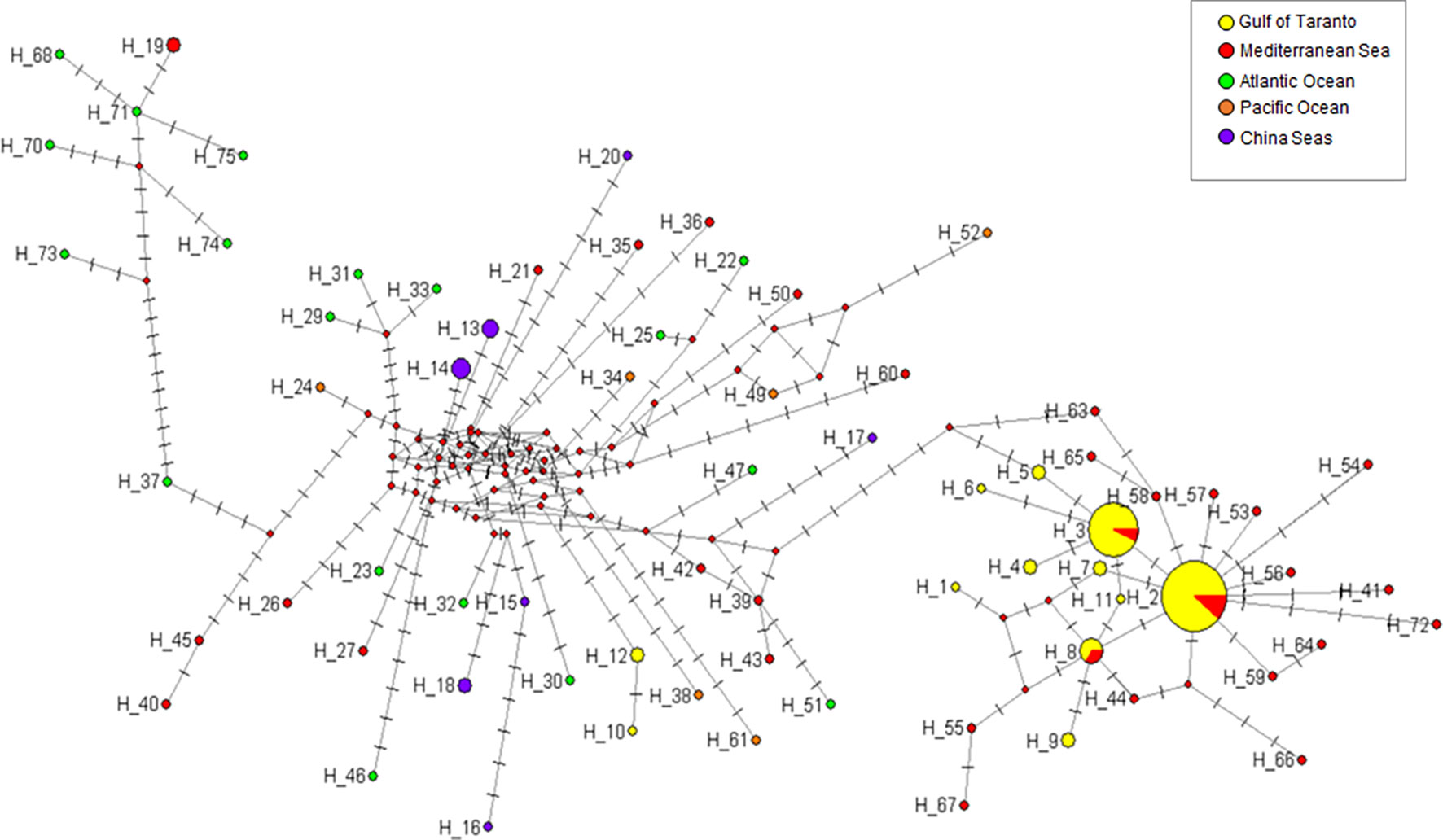
Figure 6 Median-joining network of D-loop haplotypes of S. coeruleoalba. The network was constructed using 71 different sequences (Supplementary Table 7). Haplotype frequency distribution in the considered geographic areas is visualized through pie charts using different color codes. For better visualization of the network topology, branch lengths were not maintained proportional to the number of mutations. The latter are indicated by lines parallel to branch lines, each position line describing a mutated nucleotide position with respect to the first sequence in the dataset (Hap-1). Red circles indicate missing intermediate haplotypes.
Furthermore, another interesting result is the possibility of highlighting the presence of hybridization phenomena through phylogenetic analyzes. In effect, this type of analysis is useful to highlight phenomena of natural and anthropogenic hybridization (driven by anthropogenic disruption of biological genetic patterns) representing a crucial point for the implementation of effective conservation measures (Faria et al., 2022). In effect, understanding anthropogenic hybridization dynamics can help identify effective and timely management actions for threatened species avoiding genomic extinction potentially led by the presence of admixed individuals and by human disturbances that cause hybridization (Santostasi et al., 2020). In this light, to overcome limits of this study further future analysis on a wider number of mitochondrial and/or nuclear genetic markers could help to better understand and investigate genetic diversity of this species and, consequently, to assist in delineating conservation strategies of local units or putative metapopulations occurring in different regions of the Mediterranean Sea. However, it should be kept in mind that to collect larger amounts of nuclear DNA for genetic analysis it is necessary to apply other sampling methods such as biopsies which require specific permits.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Ministry of the Environment and the Protection of the Territory and Sea.
RC, RA, SC, Conceptualization. RA, SC, GL, CF, RC, methodology and sampling design. RA, GL, FP, CS, EC, formal analysis. RA, RC, GC, GL, writing—original draft preparation. RA, RC, GC, GL, FP, CS, CF, EC, SC, writing—review and editing. RC, RA supervision. All authors contributed to the article and approved the submitted version.
The financial support of the University of Bari is gratefully acknowledged. This research was funded by The National Operational Programme (NOP) on Research and Innovation 2014-2020 promoting actions aimed at fostering mobility and attraction of university researchers – Progetto AIM “Attraction and International Mobility” 1807508_1 – attività 1 – linea 1 (Department of Biology, University of Bari).
We thank Mrs. Angela Pala and Dr. Pietro D’Addabbo for technical assistance in laboratory experiments and with computer software, respectively. The authors would like to acknowledge Dr. Silvia Bruno for her support in computational analyses.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1088598/full#supplementary-material
ACCOBAMS (2021). Conserving whales, dolphins and porpoises in the Mediterranean Sea, black Sea and adjacent areas: an ACCOBAMS status report, (2021). Eds. Notarbartolo di Sciara G., Tonay A. M. (Monaco: ACCOBAMS).
ACCOBAMS-MOP7/2019/Doc 33 (2019). “Best practice on cetacean post mortem investigation and tissue sampling,” in Joint ACCOBAMS and ASCOBANS document(Istanbul).
Allendorf F. W., Luikart G., Aitken S. N. (2007). Conservation and the genetics of populations. Mammalia 2007, 189–197. doi: 10.1515/MAMM.2007.038
Amaral A. R., Lovewell G., Coelho M. M., Amato G., Rosenbaum H. C. (2014). Hybrid speciation in a marine mammal: The clymene dolphin (Stenella clymene). Plos One. 9(1):e83654.
Antoniou A., Frantzis A., Alexiadou P., Paschou N., Poulakakis N. (2018). Evidence of introgressive hybridization between Stenella coeruleoalba and Delphinus delphis in the Greek seas. Mol. Phylogenet. Evol. 129, 325–337. doi: 10.1016/j.ympev.2018.09.007
Attard A., Brander R. W., Shaw W. S. (2015). Rescues conducted by surfers on Australian beaches. Accident Analysis & Prevention. 82, 70–78.
Barrett-Lennard L. G., Ellis G. (2001). Population structure and genetic variability in northeastern pacific killer whales: Towards an assessment of population viability. Anim. Behav. 51, 553–565. doi: 10.1006/anbe.1996.0059
Bearzi G., Bonizzoni S., Santostasi N. L. (2022). Striped dolphin Stenella coeruleoalba (Gulf of Corinth subpopulation). In book: The IUCN Red List of Threatened Species IUCN International Union for Conservation of Nature. doi: 10.2305/IUCN.UK.2022-1.RLTS.T210188066A210188619.en
Bellomo S., Santacesaria F. C., Fanizza C., Cipriano G., Renò V., Carlucci R., et al. (2019). Photo-identification of Physeter macrocephalus in the gulf of Taranto (northern Ionian Sea, central-eastern Mediterranean Sea). In: Proceedings IEEE Metrology for the Sea 2019 IMEKO TC-19. International Workshop on Metrology for the Sea, Genoa, Italy. 2019, pp. 33–37.
Bossart G. D. (2011). Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 48, 676–690, 2011. doi: 10.1177/0300985810388525
Bourret V. J., Macé M. R., Crouau-Roy B. (2007). Genetic variation and population structure of western Mediterranean and northern Atlantic stenella coeruleoalba populations inferred from microsatellite data. J. Mar. Biol. Assoc. United Kingdom 87 (1), 265–269. doi: 10.1017/S0025315407054859
Buckland S. T., Anderson D. R., Burnham K. P., Laake J. L., Borchers D. L., Thomas L., et al (Eds.) (2004). Advanced Distance Sampling: Estimating Abundance of Biological Populations; OUP: Oxford, UK.
Cann R. L., Brown W. M., Wilson A. C. (1984). Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics 106, 479–499. doi: 10.1093/genetics/106.3.479
Capezzuto F., Carlucci R., Maiorano P., Sion L., Battista D., Giove A., et al. (2010). The bathyal benthopelagic fauna in the northwestern Ionian Sea: Structure, patterns and interactions. Chem. Ecol. 26, 199–217. doi: 10.1080/02757541003639188
Carlucci R., Bandelj V., Ricci P., Capezzuto F., Sion L., Maiorano P., et al. (2018a). Exploring spatio-temporal changes in the demersal and benthopelagic assemblages of the northwestern Ionian Sea (Central Mediterranean Sea). Mar. Ecol. Prog. Ser. 598, 1–19. 79. doi: 10.3354/meps12613
Carlucci R., Cipriano G., Santacesaria F. C., Ricci P., Maglietta R., Petrella A., et al. (2020b). Exploring data from an individual stranding of a Cuvier’s beaked whale in the gulf of Taranto (northern Ionian Sea, central-eastern Mediterranean Sea). J. Exp. Mar. Biol. Ecol. 553, 151473. doi: 10.1016/j.jembe.2020.151473.
Carlucci R., Battista D., Capezzuto F., Serena F., Sion L. (2014). Occurrence of the basking shark Cetorhinus maximus (Gunnerus 1765) in the central-Eastern Mediterranean Sea. Ital. J. Zool 81, 280–286. doi: 10.1080/11250003.2014.910275
Carlucci R., Baş A. A., Liebig P., Renò V., Santacesaria F. C., Bellomo S., et al. (2020a). Residency patterns and site fidelity of Grampus griseus (Cuvier, 1812) in the Gulf of Taranto (Northern Ionian Sea, central-eastern Mediterranean Sea). Mammal Res. 65, 445–455.
Carlucci R., Capezzuto F., Cipriano G., D’onghia G., Fanizza C., Libralato S., et al. (2021a). Assessment of cetacean–fishery interactions in the marine food web of the gulf of taranto (Northern Ionian Sea, central Mediterranean Sea). Rev. Fish Biol. Fish 31, 135–156. doi: 10.1007/s11160-020-09623-x
Carlucci R., Cipriano G., Paoli C., Ricci P., Fanizza C., Capezzuto F.. (2018c). Random Forest population modelling of striped and common-bottlenose dolphins in the Gulf of Taranto (Northern Ionian Sea, Central-eastern Mediterranean Sea). Estuar. Coast. Shelf Sci. 204, 177–192.
Carlucci R., Fanizza C., Cipriano G., Paoli C., Russo T., Vassallo P. (2016). Modelling the spatial distribution of the striped dolphin (Stenella coeruleoalba) and common bottlenose dolphin (Tursiops truncatus) in the gulf of taranto (Northern Ionian Sea, central-eastern Mediterranean Sea). Ecol. Indic. 69, 707–721. doi: 10.1016/j.ecolind.2016.05.035
Carlucci R., Manea E., Ricci P., Cipriano G., Fanizza C., Maglietta R., et al. (2021). Managing multiple pressures for cetaceans’ conservation with an ecosystem-based marine spatial planning approach. J. Environ. Manag. 287, 112240. doi: 10.1016/j.jenvman.2021.112240
Carlucci R., Ricci P., Cipriano G., Fanizza C. (2018b). Abundance, activity and critical habitat of the striped dolphin stenella coeruleoalba in the gulf of taranto (Northern Ionian Sea, central Mediterranean Sea). Aquat. Conservation: Mar. Freshw. Ecosyst. 28 (2), 324–336. doi: 10.1002/aqc.2867
Charlton K., Taylor A. C., McKechnie S. W. (2006). A note on divergent mtDNA lineages of bottlenose dolphins from coastal waters of southern Australia. Journal of Cetacean Research & Management 8:173–179.
Ciccarese S., Carlucci R., Ciani E., Corcella E., Cosentino A., Fanizza C., et al. (2019). Cytochrome b marker reveals an independent lineage of Stenella coeruleoalba in the gulf of taranto. PloS One 14 (3), 0213826. doi: 10.1371/journal.pone.0213826
Cipriano G., Carlucci R., Bellomo S., Santacesaria F. C., Fanizza C., Ricci P., et al. (2022). Behavioral Pattern of Risso’s Dolphin(Grampus griseus) in the Gulf of Taranto (Northern Ionian Sea, Central-Eastern Mediterranean Sea). J. Mar. Sci. Eng. 10, 175.
Civitarese G., Gačić M., Lipizer M., Eusebi Borzelli G. L. (2010). On the impact of the bimodal oscillating system (BiOS) on the biogeochemistry and biology of the Adriatic and Ionian seas (eastern Mediterranean). Biogeosciences 7, 3987–3997. doi: 10.5194/bg-7-3987-2010
Coker O. M. (2017). Importance of genetics in conservation of biodiversity. J. Wildl. Manage 1, 11–18.
Coll M., Piroddi C., Albouy C., Lasram F. B.R., Cheung W. W.L., Christensen V., et al. (2012). The Mediterranean Sea under siege: spatial overlap between marine biodiversity: cumulative threats and marine reserves. Glob. Ecol. Biogeogr 21, 465–480.
Cosentino A., Fanizza C., Corcella M. E., Pala R., Carlucci R., Sartori S. M., et al. (2015). Genetic-molecular protocols for the study of the social structure in Stenella Coeruleoalba in the Gulf of Taranto. Biol Mar Mediterr. 22(1):271–272.
de los Angeles Bayas-Rea R., Félix F., Montufar R. (2018). Genetic divergence and fine scale population structure of the common bottlenose dolphin (Tursiops truncatus, montagu) found in the gulf of guayaquil, Ecuador. PeerJ 6, 4589. doi: 10.7717/peerj.4589
D’Onghia G., Calculli E., Capezzuto F., Carlucci R., Carluccio A., Maiorano P., et al. (2016). New records of cold-water coral sites and fish fauna characterization of a potential network existing in the Mediterranean Sea. Mar. Ecol. 37, 1398–1422. doi: 10.1111/maec.12356
Duffy J. E., Stachowicz J. J. (2006). Why biodiversity is important to oceanography: potential roles of genetic, species, and trophic diversity in pelagic ecosystem processes. Mar. Ecol. Prog. Ser. 311, 179–189. doi: 10.3354/meps311179
Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113. doi: 10.1186/1471-2105-5-113 PMID:15318951 doi: 10.1186/1471-2105-5-113
Excoffier L., Lischer H. E. L. (2010). Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Faria D. M., Steel D., Baker C. S., da Silva J. M., de Meirelles A. C. O., Souto L. R. A., et al. (2022). Mitochondrial diversity and inter-specific phylogeny among dolphins of the genus stenella in the southwest Atlantic ocean. PloS One 17 (7), e0270690. doi: 10.1371/journal.pone.0270690
Féral J. P. (2002). How useful are the genetic markers in attempts to understand and manage marine biodiversity? J. Exp. Mar. Biol. Ecol. 268 (2), 121–145. doi: 10.1016/S0022-0981(01)00382-3
Frankham R. (2010). Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143 (9), 1919–1927. doi: 10.1016/j.biocon.2010.05.011
Fu Y. X. (1996) “New statistical tests of neutrality for DNA samples from a population”. Genetics 143, 557–570. doi: 10.1093/genetics/143.1.557
Garner B. A., Hoban S., Luikart G. (2020). IUCN red list and the value of integrating genetics. Conserv. Genet. 21, 795–801. doi: 10.1007/s10592-020-01301-6
Gaspari S., Azzellino A., Airoldi S., Hoelzel A. R. (2007). Social kin associations and genetic structuring of striped dolphin populations (Stenella coeruleoalba) in the Mediterranean Sea. Mol. Ecol. 16 (14), 2922–2933. doi: 10.1111/j.1365-294X.2007.03295.x
Gaspari S., Marsili L., Natali C., Airoldi S., Lanfredi C., Deeming C., et al. (2019). Spatio-temporal patterns of genetic diversity in the Mediterranean striped dolphin (Stenella coeruleoalba). J. Zoological Systematics Evolutionary Res. 57 (3), 721–734. doi: 10.1111/jzs.12265
Giantsis I. A., Abatzopoulos T. J., Angelidis P., Apostolidis A. P. (2014) “Mitochondrial control region variability in Mytilus galloprovincialis populations from the central-Eastern Mediterranean sea.” Int. J. Mol. Sci. 15, 11614–11625. doi: 10.3390/ijms150711614
Gkafas G. A., Exadactylos A., Rogan E., Raga J. A., Reid R., Hoelzel A. R. (2017). Biogeography and temporal progression during the evolution of striped dolphin population structure in European waters. J. biogeography 44 (12), 2681–2691. doi: 10.1111/jbi.13079
Harlin A. D., Würsig B., Baker C. S., Markowitz T. M. (1999). Skin swabbing for genetic analysis: application to dusky dolphins (Lagenorhynchus obscurus). Marine Mammal Science 15(2), 409–425.
Hoban S., Bruford M., Jackson J. D. U., Lopes-Fernandes M., Heuertz M., Hohenlohe P. A., et al. (2020). Genetic diversity targets and indicators in the CBD post-2020 global biodiversity framework must be improved. Biol. Conserv. 248, 108654. doi: 10.1016/j.biocon.2020.108654
Hunter M. E., Hoban S. M., Bruford M. W., Segelbacher G., Bernatchez L. (2018) Next-generation conservation genetics and biodiversity monitoring.” Evolutionary Appl. 11 (7), 1029–1034. doi: 10.1111/eva.12661
Irvine A. B., Scott M. D., Wells R. S., Kaufmann J. H. (1981). Movements and activities of the Atlantic bottlenose dolphin, Tursiops truncatus, near Sarasota, Florida. Fish. Bull. 79, 671–688.
Imsiridou A., Papapetrou M., Tilikidis A., Loukovitis D., Minos G., Gouva E., et al. (2019). Can the population structure of three Greek marine species (Sardina pilchardus, Penaeus kerathurus, Mullus barbatus) become a tool for their future characterization as PGI products. J. Nutr. Food Lipid Sci. 2019, 1901–1908.
IPBES (2019). Global assessment report on biodiversity and ecosystem services of the intergovernmental science-policy platform on biodiversity and ecosystem services. Eds. Brondizio E. S., Settele J., Díaz S., Ngo H. T. (Bonn, Germany: IPBES secretariat), 1148 pages.
IUCN. (2012). IUCN Red List Categories and Criteria: Version 3.1; IUCN: Gland, Switzerland; Cambridge, UK.
Johnson C., Reisinger R., Palacios D., Friedlaender A., Zerbini A., Willson A., et al. (2022). Protecting blue corridors, challenges and solutions for migratory whales navigating international and national seas (Switzerland: WWF, Oregon State University, University of California, Santa Cruz, Publisher: WWF International).
Kingston S. E., Adams L. D., Rosel P. E. (2009). Testing mitochondrial sequences and anonymous nuclear markers for phylogeny reconstruction in a rapidly radiating group: molecular systematics of the Delphininae (Cetacea: Odontoceti: Delphinidae). BMC Evol. Biol. 9, 245.
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lauriano G. (2021). Striped dolphin, Stenella coeruleoalba, Mediterranean subpopulation. The IUCN Red List of Threatened Species 2021.
LeDuc R. G., Perrin W. F., Dizon A. E. (1999). Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Marine mammal science 15(3), 619–648.
Linguiti G., Fanizza C., Ciani E., Bellomo S., Cipriano G., Santacesaria F. C., et al. (2021). “October. “Assessment of genetic diversity of the striped dolphin population in the gulf of taranto (Northern Ionian Sea, central Mediterranean Sea),” in 2021 international workshop on metrology for the sea; learning to measure Sea health parameters (MetroSea) (Reggio Calabria: IEEE), 144–147.
Maiorano P., Sion L., Carlucci R., Capezzuto F., Giove A., Costantino G., et al. (2010). The demersal faunal assemblage of the NW Ionian Sea (Central mediterranean): Current knowledge and perspectives. Chem. Ecol. 26, 219–240. doi: 10.1080/02757541003693987
Mann J. (1999). Behavioral sampling methods for cetaceans: A review and critique. Mar. Mamm. Sci. 15, 102–122. doi: 10.1111/j.1748-7692.1999.tb00784.x
Matarrese R., Chiaradia M. T., Tijani K., Morea A., Carlucci R. (2011). Chlorophyll a'multi-temporal analysis in coastal waters with MODIS data. Ital. J. Remote Sens. 43, 39–48. doi: 10.5721/ItJRS20114333
Möller L. M., Bilgmann K., Charlton-Robb K., Beheregaray L. (2008). Multi-gene evidence for a new bottlenose dolphin species in southern Australia. Molecular Phylogenetics and Evolution. 49(2):674–681. doi: 10.1016/j.ympev.2008.08.011
Neumann D. R. (2001). Activity budget of free-ranging common dolphins (Delphinus delphis) in the northwestern bay of plenty, new Zealand. Aquat. Mamm. 27, 121–136.
Nishida S, Goto M, Pastene LA, Kanda N, Koike H (2007). Phylogenetic relationships among cetaceans revealed by Y-chromosome sequences. Zool Sci 7, 723–732.
Notarbartolo di Sciara G. (2016). Chapter One - Marine Mammals in the Mediterranean Sea: An Overview. InAdvances in Marine Biology, Notarbartolo di Sciara G., Podestà M., Curry B. E., Eds. volume 75, 2–248.
Pace D. S., Tizzi R., Mussi B. (2015). Cetaceans value and conservation in the Mediterranean Sea. J. Biodiversity Endangered Species S1:(004). doi: 10.4172/2332-2543.S1.004
Pinardi N., Lyubartsev V., Cardellicchio N., Caporale C., Ciliberti S., Coppini G., et al. (2016). Marine rapid environmental assessment in the gulf of taranto: A multiscale approach. Nat. Hazards Earth Syst. Sci. 16, 2623–2639. doi: 10.5194/nhess-16-2623-2016
Posada D. (2008). JModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 25 (7), 1253–1256. doi: 10.1093/molbev/msn083
Prieto I., Violle C., Barre P., Durand J. L., Ghesquiere M., Litrico I. (2015). Complementary effects of species and genetic diversity on productivity and stability of sown grasslands. Nat. Plants 1 (4), 1–5. doi: 10.1038/nplants.2015.33
Ricci P., Libralato S., Capezzuto F., D’Onghia G., Maiorano P., Sion L., et al. (2019). Ecosystem functioning of two marine food webs in the north-Western Ionian Sea (Central Mediterranean Sea). Ecol. Evol. 9, 10198–10212. doi: 10.1002/ece3.5527
Ricci P., Manea E., Cipriano G., Cascione D., D’Onghia G., Ingrosso M., et al. (2021). Addressing cetacean–fishery interactions to inform a deep-Sea ecosystem-based management in the gulf of taranto (Northern Ionian Sea, central Mediterranean Sea). J. Mar. Sci. Eng. 9, 872. doi: 10.3390/jmse9080872
Roman J., McCarthy J. J. (2010). The whale pump: marine mammals enhance primary productivity in a coastal basin. PloS one 5(10), e13255.
Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Santacesaria F. C., Bellomo S., Fanizza C., Maglietta R., Renò V., Cipriano G., et al. (2019). Long-term residency of Tursiops truncatus in the Gulf of Taranto (Northern Ionian Sea, Central-eastern Mediterranean Sea). In Proceedings of the IMEKO Metrology for the Sea, Genova, Italy, 3–5 October 2019.
Santostasi N. L., Bonizzoni S., Gimenez O., Eddy L., Bearzi G. (2021). Common dolphins in the gulf of Corinth are critically endangered. Aquat. Conservation: Mar. Freshw. Ecosyst. 31, 101–109. doi: 10.1002/aqc.2963
Santostasi N. L., Ciucci P., Bearzi G., Bonizzoni S., Gimenez O. (2020). Assessing the dynamics of hybridization through a matrix modelling approach. Ecological Modelling 431, 109120
Šegvić-Bubić T., Marrone F., Grubišić L., Izquierdo-Gomez D., Katavić I., Arculeo M., et al. (2016). Two seas, two lineages: How genetic diversity is structured in Atlantic and Mediterranean greater amberjack Seriola dumerili riss(Perciformes, carangidae). Fisheries Res. 179, 271–279. doi: 10.1016/j.fishres.2016.03.018
Stange M., Barrett R. D., Hendry A. P. (2021). The importance of genomic variation for biodiversity, ecosystems and people. Nat. Rev. Genet. 22 (2), 89–105. doi: 10.1038/s41576-020-00288-7
Stecher G., Tamura K., Kumar S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. doi: 10.1093/molbev/msz312
Storch D., Šímová I., Smyčka J., Bohdalková E., Toszogyova A., Okie J. G. (2022). Biodiversity dynamics in the anthropocene: how human activities change equilibria of species richness. Ecography 2022 (4). doi: 10.1111/ecog.05778
Tajima F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Taylor B. L., Martien K., Morin P. (2010). “Identifying units to conserve using genetic data,” in Marine mammal ecology and conservation–a handbook of techniques (Oxford: Oxford University Press), 306–344.
Thomas L., Buckland S. T., Rexstad E. A., Laake J. L., Strindberg S., Hedley S. L., et al. Distance software: Design and analysis of distance sampling surveys for estimating population size. J. Appl. Ecol. (2010) 47, 5–14.
Turan C., Gürlek M., Ergüden D., Yağıoğlu D., Reyhaniye A., Özbalcılar B., et al. (2015). “Population genetic analysis of Atlantic bonito sarda sarda (Bloc) using sequence analysis of mtDNA d-loop region,” in Fresenius environmental bulletin, vol. 24. Parlar Scientific Publications. Available at: https://hdl.handle.net/20.500.12462/9277.
Vassallo P., D’Onghia G., Fabiano M., Maiorano P., Lionetti A., Paoli C., et al. (2017). A trophic model of the benthopelagic fauna distributed in the Santa maria di leuca cold-water coral province (Mediterranean Sea). Energy Ecol. Environ. 2, 114–124. doi: 10.1007/s40974-016-0047-2
Vilstrup J. T., Ho S. Y., Foote A. D., Morin P. A., Kreb D., Krützen M., et al. (2011). Mitogenomic phylogenetic analyses of the Delphinidae with an emphasis on the Globicephalinae. BMC Evol Biol. 11, 65. doi: 10.1186/1471-2148-11-65.
Wang C., Chen Y.-s., Han J.-l., Mo D.-l., Li X.-j., Liu X.-h. (2019). Mitochondrial DNA diversity and origin of indigenous pigs in south China and their contribution to Western modern pig breeds. J. Integr. Agric. 18, 2338–2350. doi: 10.1016/S2095-3119(19)62731-0
Keywords: striped dolphin, genetic variability, cytochrome b, D-loop control region, genetic markers
Citation: Antonacci R, Linguiti G, Paradiso F, Scalone C, Fanizza C, Ciani E, Cipriano G, Ciccarese S and Carlucci R (2023) Mitochondrial DNA diversity and genetic structure of striped dolphin Stenella coeruleoalba in the Northern Ionian Sea. Front. Mar. Sci. 10:1088598. doi: 10.3389/fmars.2023.1088598
Received: 03 November 2022; Accepted: 12 January 2023;
Published: 06 February 2023.
Edited by:
Susana Caballero, Faculty of Sciences, University of Los Andes, ColombiaReviewed by:
Ioannis A. Giantsis, University of Western Macedonia, GreeceCopyright © 2023 Antonacci, Linguiti, Paradiso, Scalone, Fanizza, Ciani, Cipriano, Ciccarese and Carlucci. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Carlucci, cm9iZXJ0by5jYXJsdWNjaUB1bmliYS5pdA==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.