- Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, Key Laboratory of Open-Sea Fishery Development, Ministry of Agriculture and Rural Affairs of China, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Science, Guangzhou, China
Microalgae blooms are a frequent occurrence in coastal waters worldwide. It is reasonable to assume that these blooms have various influences on bacterial communities, which in turn may affect the development and dissipation of the bloom. However, the bacterial community characteristics, particularly of attached bacteria, associated with microalgae blooms remain unclear. In this study, we investigated the community profiles of bacteria using high-throughput sequencing during a Phaeocystis globosa bloom in Mirs Bay, southern China, in January 2021. Bacteria living in three habitats, i.e., bacterioplankton, particle-attached bacteria, and colony-attached bacteria, were studied from the exponential growth phase to the decline growth phase of the bloom. Distinct variations in bacterial community composition existed among the three habitats. Bacteroidota, Proteobacteria, and Cyanobacteria were the dominant phyla of bacterioplankton, particle–attached bacteria, and colony-attached bacteria, respectively. Richness and diversity were significantly highest (p < 0.01) in particle-attached bacteria, followed by bacterioplankton, and lowest in colony-attached bacteria. The community diversities of bacterioplankton and particle-attached bacteria decreased significantly (p < 0.05) as the bloom shifted from the exponential to the decline phase. During the decline growth phase of the bloom, Bacteroidota and Verrucomicrobiota were the dominant remarkably abundant bacteria in the bacterioplankton community, whereas Verrucomicrobiota was dominant in the particle-attached bacteria community. No significant difference was observed in the colony-attached bacterial community between the exponential and decline phases of the P. globosa bloom owing to their complex network. The results of this study suggest that P. globose bloom has a profound impact on marine bacteria, particularly species that can decompose organic matter, which could play a crucial role in the dissipation of algal blooms.
Introduction
The formation and dissipation of algal blooms, which are caused by the rapid growth of one or more phytoplankton species, are known to be affected by various environmental factors, such as temperature, salinity, eutrophic conditions, and light intensity (Higashi et al., 2016). The interactions between algae and other organisms, such as marine bacteria that rely on the organic matter secreted by algae, are crucial for algal growth (Anderson et al., 2002; Higashi et al., 2016). The community compositions of bacteria, which play critical roles in the marine ecosystem by participating in biochemical reactions and associate with chemical transformations of almost all elements, are sensitive to environmental changes (Arrigo, 2004; Comte and Del Giorgio, 2010; Ruiz-Gonzalez et al., 2013; Sun et al., 2017; Xia et al., 2020). Previous studies have suggested that bacterial communities can be significantly influenced by algal blooms, such as diatom blooms (Liu et al., 2019; Sun et al., 2020), dinoflagellate Karenia brevis blooms (Meyer et al., 2014), and dinoflagellate Noctiluca scintillans blooms in marine waters (Xia et al., 2020). Therefore, it is hypothesized that the occurrence of Phaeocystis globosa bloom in this study is likely to have a substantial impact on bacteria.
A wide range of interactions between bacteria and phytoplankton determine the composition of bacterial communities (Cole, 1982; Sher et al., 2011; Delmont et al., 2014). Most marine bacteria in seawater are free-living (bacterioplankton), while some are attached bacteria with sophisticated interactions with their hosts (Worden et al., 2015). Bacteria can decompose organic matter such as microalgae cells and use algal polysaccharides as a carbon source (Teeling et al., 2016), making attached bacteria more important in the dissipation of bloom due to their interaction with their attached host. However, most research has focused on bacterioplankton and its interaction with algae, neglecting information on community structures of attached bacteria and their temporal variations with bloom development, which is essential for understanding the complex interactions between microalgae and bacteria, quite limited (Meyer et al., 2014; Li et al., 2020).
In January 2021, a bloom of P. globosa, which alternates between solitary cells (3–10 μm in diameter) and gelatinous colonies (3 cm in maximal diameter), occurred for the first time in Mirs Bay, southern China (Wang et al., 2021a; Wang et al., 2021b). Of the seven identified colony-forming Phaeocystis species, P. globosa is the most widespread, and its bloom could affect the bacterioplankton community by environmental filtering of specific taxa (Li et al., 2020). The gelatinous colonies of P. globosa may also favor the attachment of specific bacteria, further affecting the blooming process. To understand the effects of P. globosa bloom, it is necessary to consider not only bacterioplankton but also attached bacteria, may play an important role in the outbreak and dissipation of algal blooms.
In this study, we investigated the community profiles of bacteria during a P. globosa bloom in Mirs Bay, southern China. Bacteria living in three habitats, i.e., bacterioplankton, particle-attached bacteria, and colony-attached bacteria, were studied from the exponential growth phase to the decline growth phase of the bloom. This study aimed to investigate the differences in bacterial communities among three habitats and their temporal variations during blooming, as well as the interactions between microalgae and bacteria.
Materials and methods
Study sites and sampling protocols
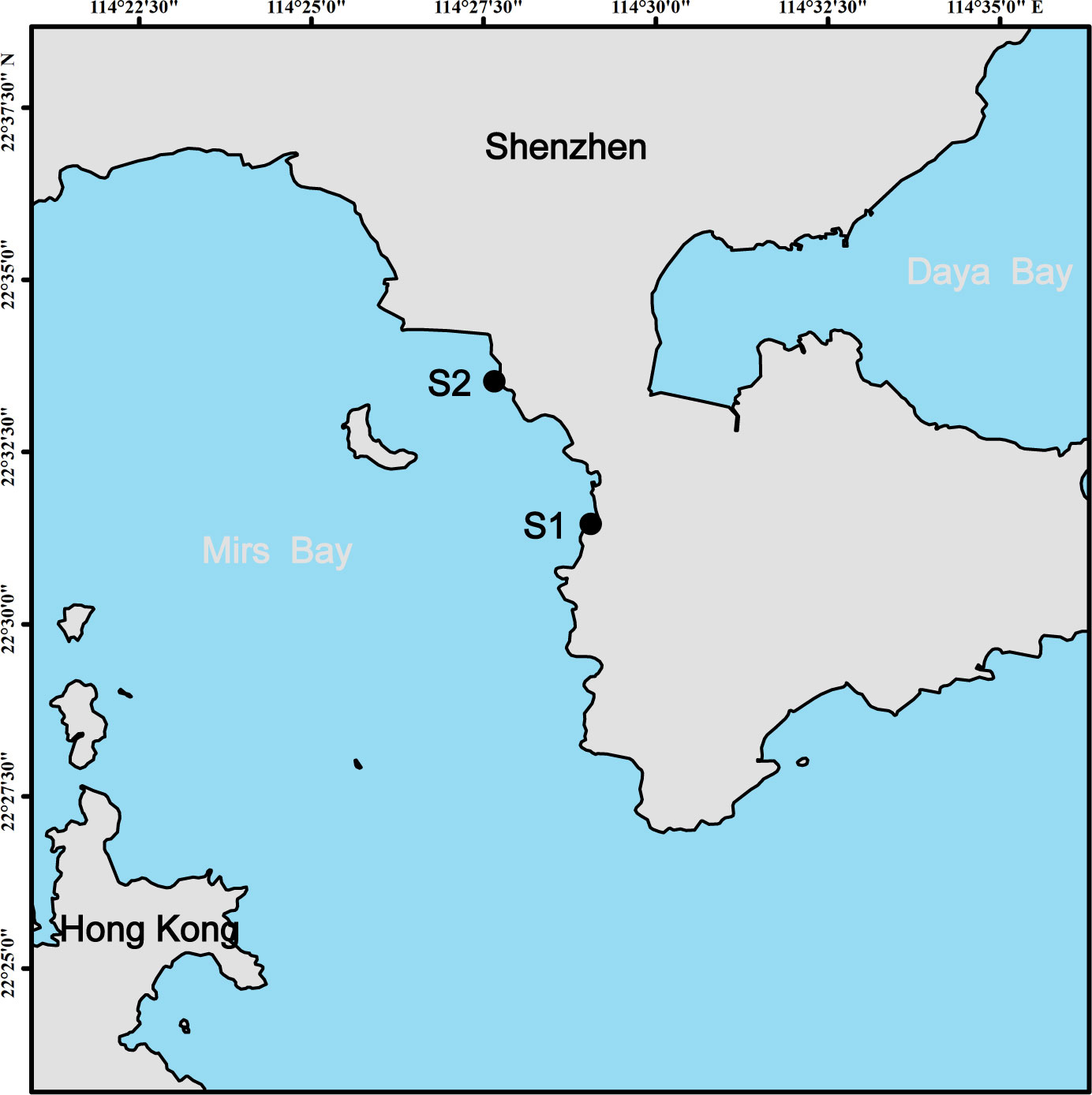
Mirs Bay, located between Dapeng Peninsula in Shenzhen and Kowloon Peninsula in Hong Kong, is a semi-enclosed subtropical bay with a high frequency of HABs due to the rapid development of marine aquaculture, tourism, and commercial shipping (Chan et al., 2008). In 2021, a P. globosa bloom occurred in Mirs Bay along the Shenzhen coast from 23 January to 3 February. During the bloom, triplicate surface water samples were collected daily from January 22 to 28, and every second day from January 30 to February 3 at stations S1 and S2 (Figure 1).
For bacterial DNA, three kinds of bacteria (bacterioplankton, particle-attached bacteria, and colony-attached bacteria) were collected. Specifically, 1 L of water was pre-filtered with a 20-µm-pore-size bolting cloth to remove debris and larger organisms and then filtered sequentially through 3-µm-pore-size and 0.2-µm-pore-size membranes (Merck Millipore, Bedford, MA, USA). Bacteria retained on the 3-µm-pore-size membranes and 0.2-µm-pore-size membranes were particle-attached bacteria and bacterioplankton, respectively. Additionally, 40 P. globosa colonies were absorbed by pipette, washed twice with sterile seawater, and filtered with a 3-µm-pore-size polycarbonate membrane. Bacteria retained on the 3-µm-pore-size membranes were colony-attached bacteria. The bacteria retained on the membranes were all stored at −80°C for DNA extraction.
Environmental factor analysis
Water temperature, salinity, dissolved oxygen (DO), and pH were measured using multiprobe sonde (Yellow Springs Instrument Co., Dayton, OH, USA), and the density of P. globosa colony was counted directly in situ to evaluate the different growth stages of P. globosa. The density of Akashiwo sanguinea was measured using FlowCAM8400 (Yokogawa Fluid Imaging Technologies, Inc., Scarborough, Maine, USA). The concentrations of Chl a, , , , , and chemical oxygen demand (COD) were measured according to marine monitoring specifications (GB/T 12763.4-2007, 2007). 0.5 L seawater was filtered through a pre-combusted GF/F membrane, and the concentrations of dissolved inorganic carbon (DIC) and dissolved total carbon (DTC) were measured according to the method of Chen et al. (2020), with dissolved organic carbon (DOC) being the difference value between DTC and DIC.
DNA extraction, sequencing, and analysis
DNA extraction was based on the manufacturer’s instructions of the E.Z.N.A. Water DNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA). The concentration and quality of DNA were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA) and 1% agarose gel electrophoresis, respectively. The PCR amplification primers of bacterial V4-V5 regions were 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) (Parada et al., 2015). PCR reactions, further processing, and sequencing of PCR products were performed following a method described in Shi et al. (2022). In addition, demultiplexed, quality-filtered, and merged methods of raw 16S rRNA gene sequencing reads were described in Shi et al. (2022). The high-quality sequences were then clustered into operational taxonomic units (OTUs) with 97% similarity using UPARSE (Edgar, 2013), with a minimum number of sequences (27106) as the rarefied criterion to standardize uneven sequence depth. The taxonomy of each OTU was analyzed by RDP Classifier based on the 16S rRNA database (Silva v138) with a confidence threshold of 0.7 (Wang et al., 2007).
The calculation and multiple comparisons of α-diversity indices (Chao 1, Pielou’s evenness, and Shannon-Wiener indices) were performed by packages “mother” and “multcomp” of R (Hothorn et al., 2008; Schloss et al., 2009; R Core Team, 2018). The bacterial analysis of similarities (ANOSIM), Non-metric multidimensional scaling (NMDS) based on Bray-Curtis distance and Redundancy analysis (RDA) were conducted in package “vegan” of R (Oksanen et al., 2019) with forward selection used to avoid collinearity of environmental factors and a parsimonious set of significant environmental variables selected using a permutation test. Co-occurrence network and Network topological features (degree and network density) were drawn and calculated using packages “psych” and “igraph” of R (Benjamini et al., 2006; Csárdi and Nepusz, 2006; Ma et al., 2016; Revelle, 2021). The difference significance tests (ANOVA) of bacterial community and adjusted p-values (Benjamini and Hochberg false discovery rate) were calculated using STAMP between the exponential and decline growth phase of P. globosa bloom (Parks et al., 2014).
Results
Density of P. globosa
Changes in P. globosa colony density are shown in Figure 2. Colony density increased from 4.41 to 16.80 colonies L-1 from January 23 to 25 and decreased sharply to 2.53 colonies L-1 on January 27 and fluctuated until February 3. The growth phase of the P. globosa bloom was divided into an exponential growth phase (January 23 to 26) and a decline growth phase (January 27 to February 3) based on colony density. It is worth noting that the concentration of chl a (representing solitary cells) on January 26 was still in high value, and thus the growth of P. globosa bloom on January 26 was classified as in the exponential growth phase, although colony density had begun to decrease.
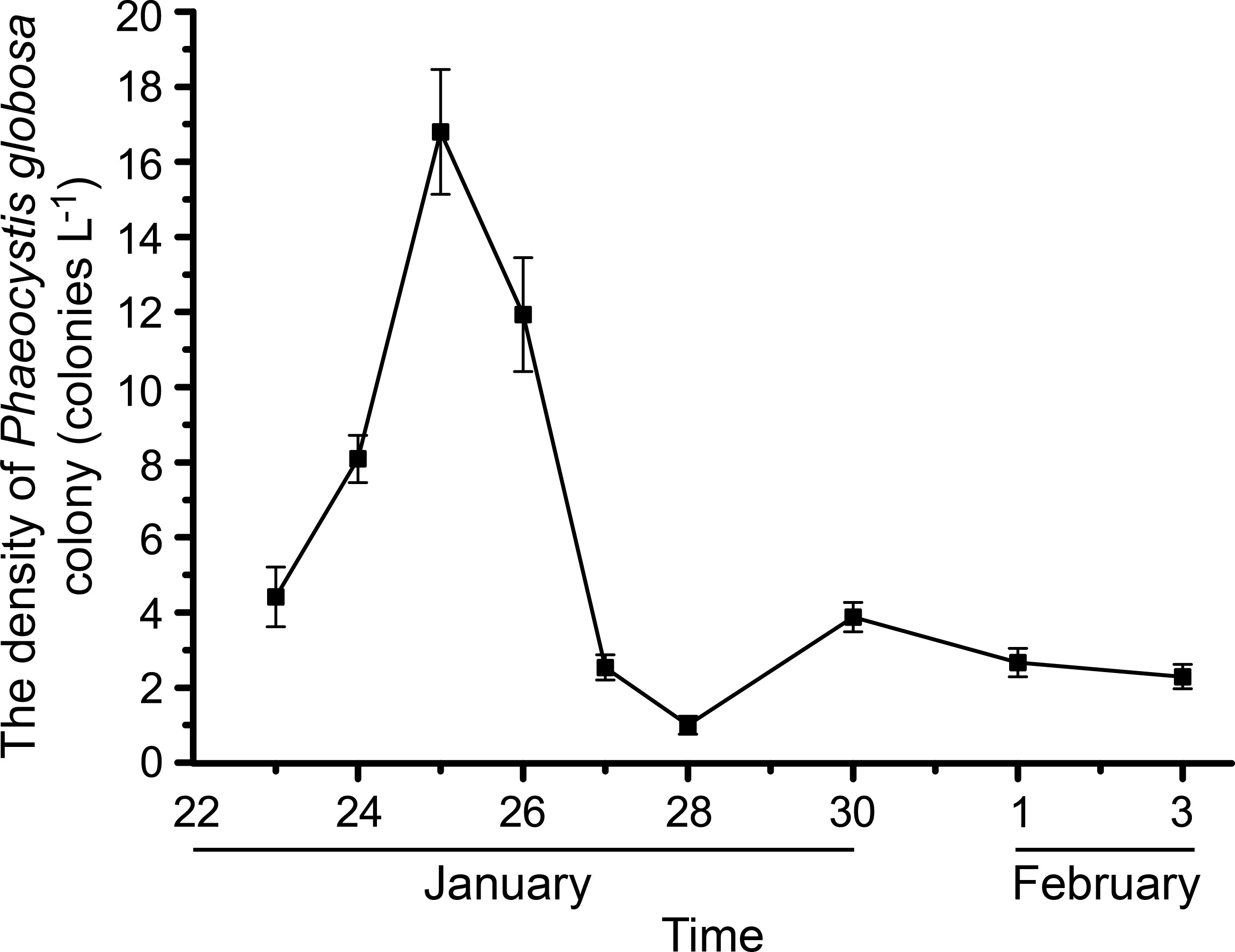
Figure 2 Changes of the phaeocystis globosa colony density (colonies L-1). The error bars represent standard errors of the P. globosa colonies density.
Composition of bacterial communities
16S rRNA sequences obtained from samples were clustered into 7235 OTUs, assigned to 54 different phyla. Large variations in bacterial community compositions were observed among the three kinds of bacteria, particularly between colony-attached bacteria and the other two (Figure 3). Specifically, the dominant phyla of bacterioplankton were Bacteroidota (percentages of the total OTUs ranged from 16.80 to 76.30%), Proteobacteria (16.05–58.43%, mainly belonging to Alphaproteobacteria and Gammaproteobacteria) and Cyanobacteria (0.82–31.37%). The dominant phyla of particle-attached bacteria were Proteobacteria (12.97–75.44%, mainly belonging to Alphaproteobacteria and Gammaproteobacteria), Bacteroidota (4.94–48.47%), Verrucomicrobiota (0.91–59.72%) and Cyanobacteria (1.04–55.33%). The community compositions of colony-attached bacteria were simple, with the dominant phyla of Cyanobacteria (26.97–98.90%) and Proteobacteria (0.63–65.91%).
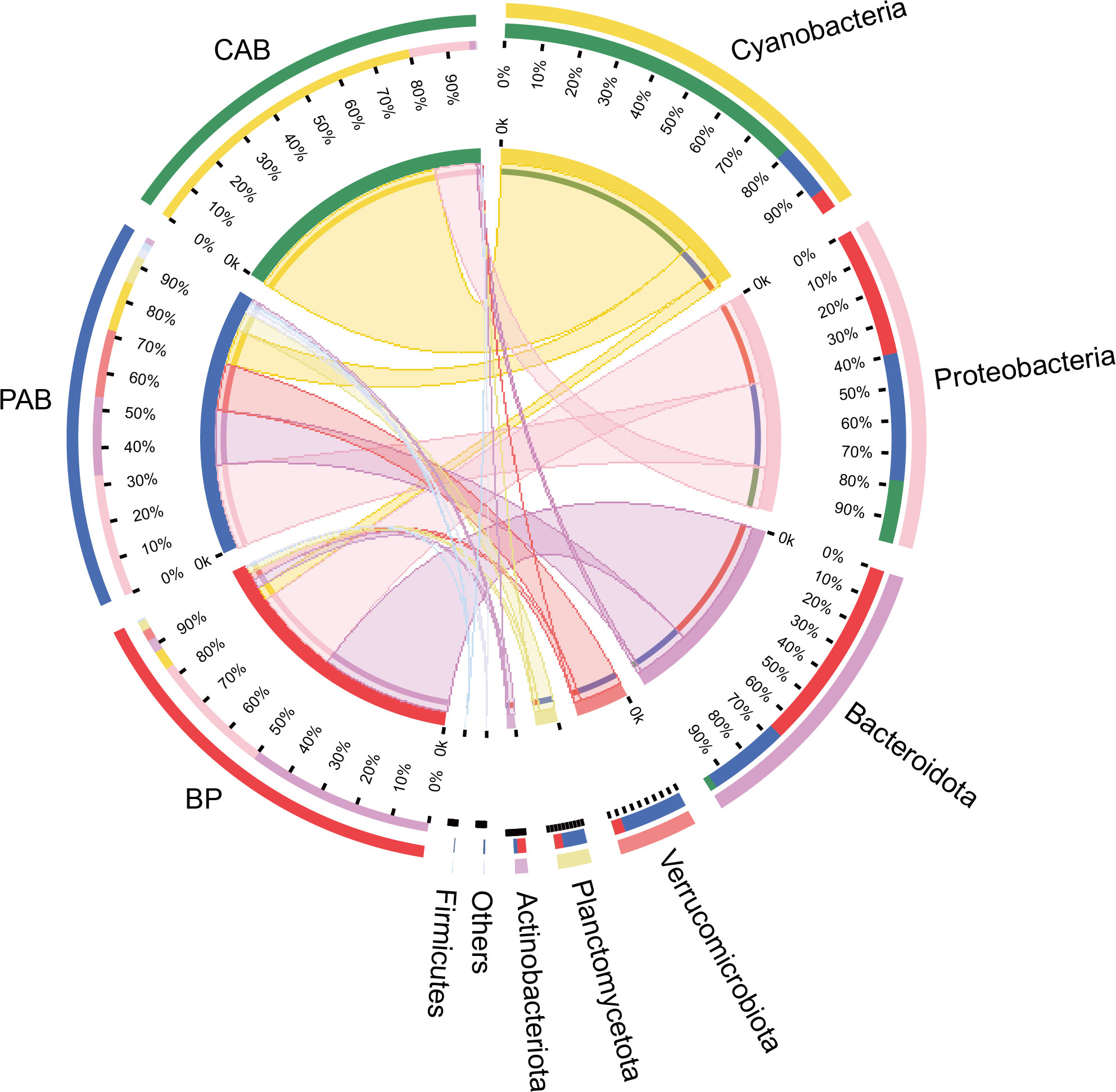
Figure 3 Dominant bacterial lineages (top 7 phyla) of BP (bacterioplankton), PAB (particle-attached bacteria), and CAB (colony-attached bacteria).
α-diversity of bacterial communities
The differences in α-diversity indices among bacterioplankton, particle-attached bacteria and colony-attached bacteria during the P. globosa bloom were extremely significant (p < 0.01). Particle-attached bacteria had the highest α-diversity indices, followed by bacterioplankton, and colony-attached bacteria had the lowest (Figure 4A). In the decline growth phase of the bloom, Shannon-Wiener and Pielou’s evenness indices decreased significantly in bacterioplankton and increased significantly in colony-attached bacteria (p < 0.05), but Chao1 indices showed no significant differences between the exponential and decline growth phases in those two groups (p > 0.05). However, the Chao1 index of particle-attached bacteria increased significantly (p < 0.05), while the Shannon-Wiener and Pielou’s evenness indices decreased extremely significantly (p < 0.01) when the bloom entered the decline growth phase (Figure 4B).
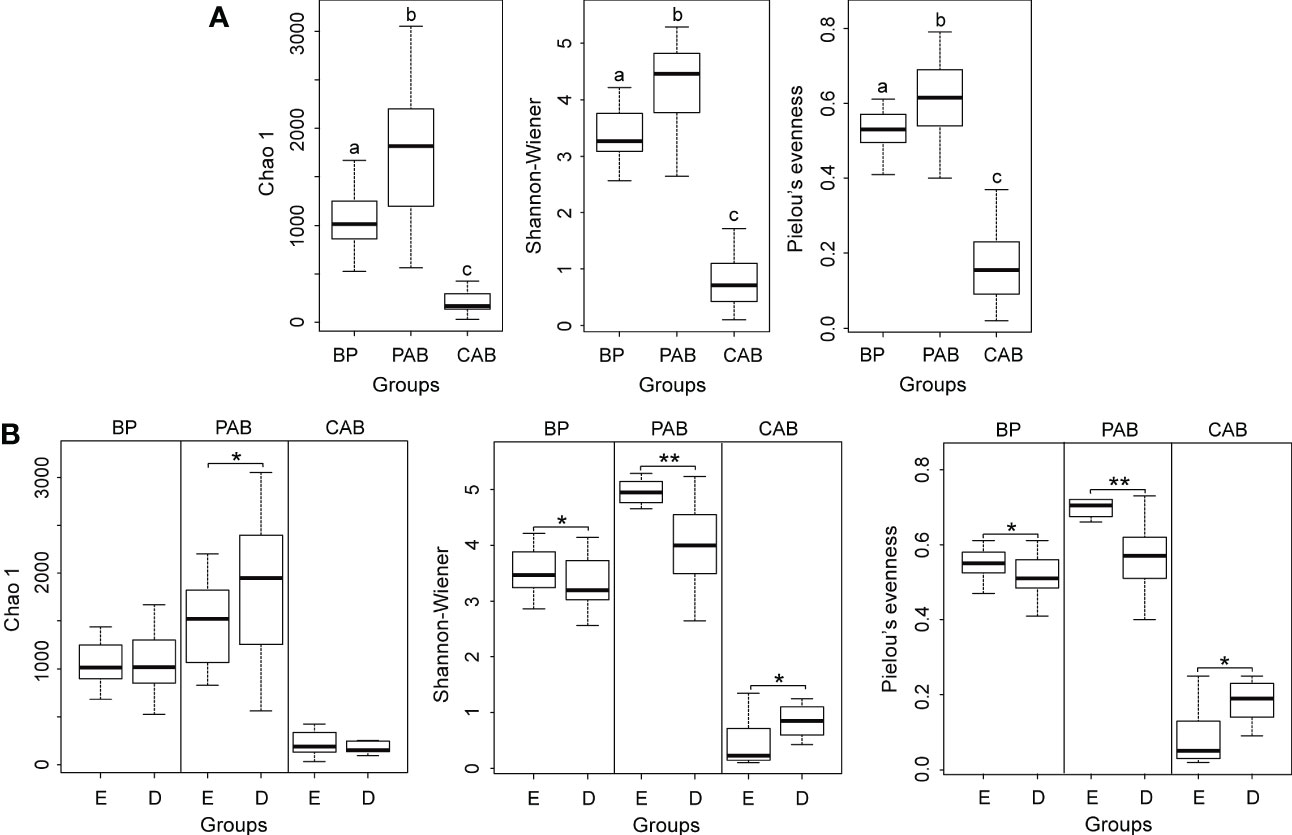
Figure 4 Boxplot of α-diversity indices on genus level among three kinds of bacteria (A) Different lowercase letters represent significant differences, p < 0.01 and between exponential and decline growth phases of Phaeocystis globosa bloom (B). BP, bacterioplankton; PAB, particle-attached bacteria; CAB, colony-attached bacteria; D, exponential growth phase of bloom; E, decline growth phase of bloom. *p< 0.05; **p< 0.01.
Bacterial community structure
During the P. globosa bloom, the bacterial community structures of the three kinds of bacteria were markedly different, and the differences among groups were greater than those within groups (Figures 5A, B). The community compositions of colony-attached bacteria showed smaller variations compared to bacterioplankton and particle-attached bacteria and did not change much throughout the P. globosa bloom (Figure 5C). However, obvious distinctions were observed between the exponential and decline growth phases in bacterioplankton and particle-attached bacteria (Figure 5C). In particular, members of Bacteroidota and Verrucomicrobiota in bacterioplankton and Verrucomicrobiota in particle-attached bacteria were notably higher during the decline growth phase of the P. globosa bloom (Figure 6). Furthermore, the bacterial community structures of bacterioplankton and particle-attached bacteria on January 28 were distinct from the other groups (Figure 5A).
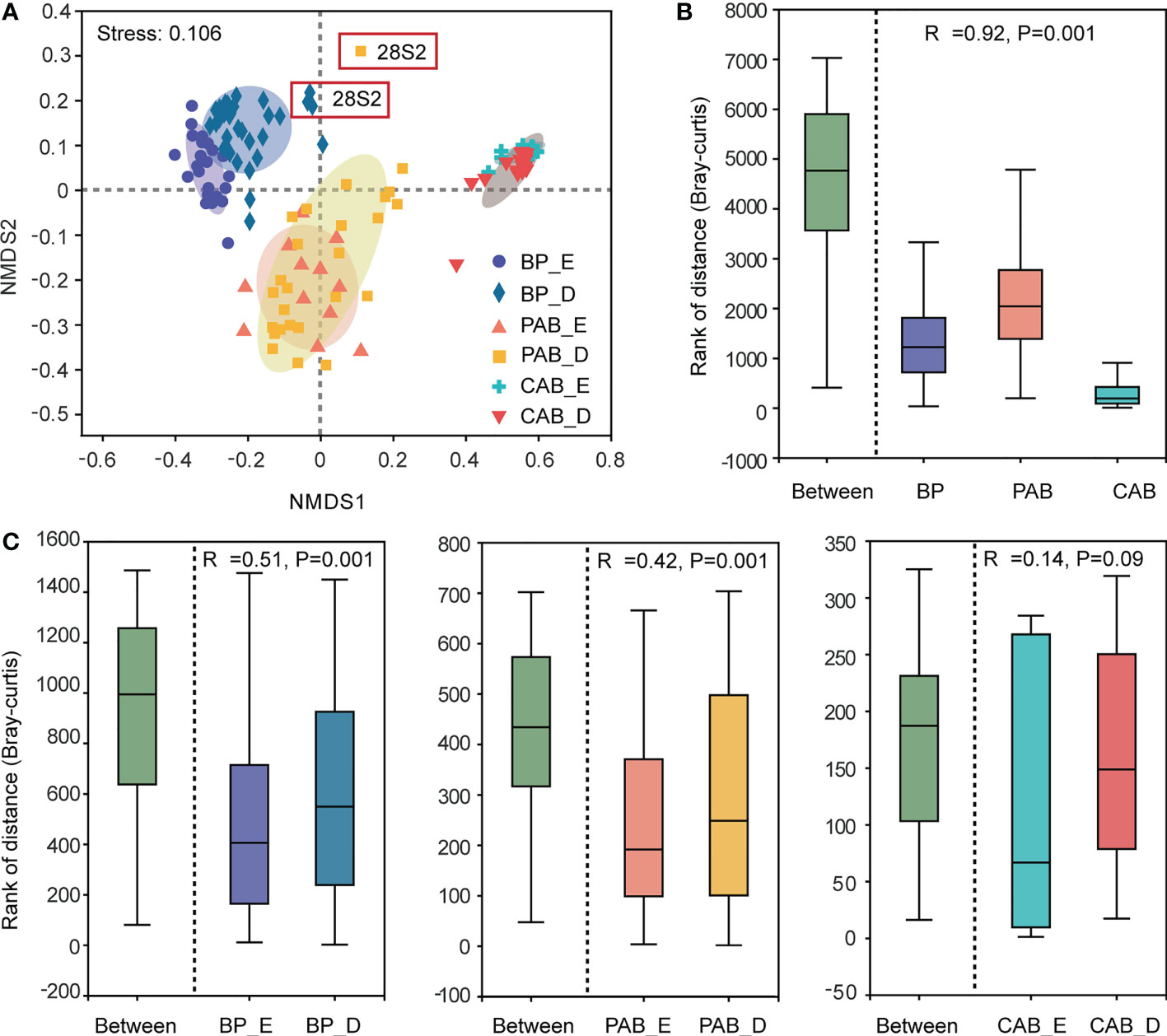
Figure 5 NMDS on genus level (A), distances boxplot among three kinds of bacteria (B) and between exponential and decline growth phase of Phaeocystis globosa bloom (C). BP, bacterioplankton; PAB, particle-attached bacteria; CAB, colony-attached bacteria; E, exponential growth phase of bloom; D, decline growth phase of bloom.
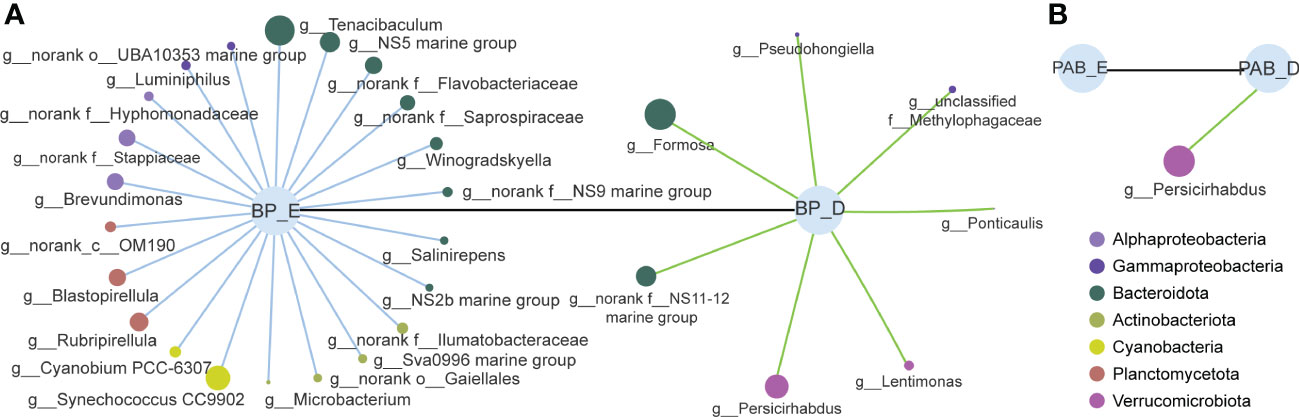
Figure 6 The abundance of bacterioplankton (A) and particle-attached bacteria (B) with significantly differences between exponential and decline growth phase of Phaeocystis globosa bloom. Radiating dots represent bacterial genus, and the dots size and color represent their abundance and phylum they belong to. BP, bacterioplankton; PAB, particle-attached bacteria; E, exponential growth phase of bloom; D, decline growth phase of bloom. (p< 0.01).
Bacterial inter-taxa relationship network
The co-occurrence networks of bacterioplankton, particle-attached bacteria, and colony-attached bacteria had much higher positive correlations than negative correlations (Figure 7). The network of bacterioplankton had 574 nodes and 2603 edges, of which 2565 were positive and 38 were negative correlations. The network of particle-attached bacteria had 893 nodes and 4514 edges, of which 4495 were positive and 19 were negative correlations. The network of colony-attached bacteria had 510 nodes and 6808 edges, of which 6798 were positive and 10 were negative correlations (Figure 7). The network densities of bacterioplankton, particle-attached bacteria, and colony-attached bacteria were 0.016, 0.011 and 0.052, respectively, and their average degrees were 9.07, 10.11 and 26.70, respectively.
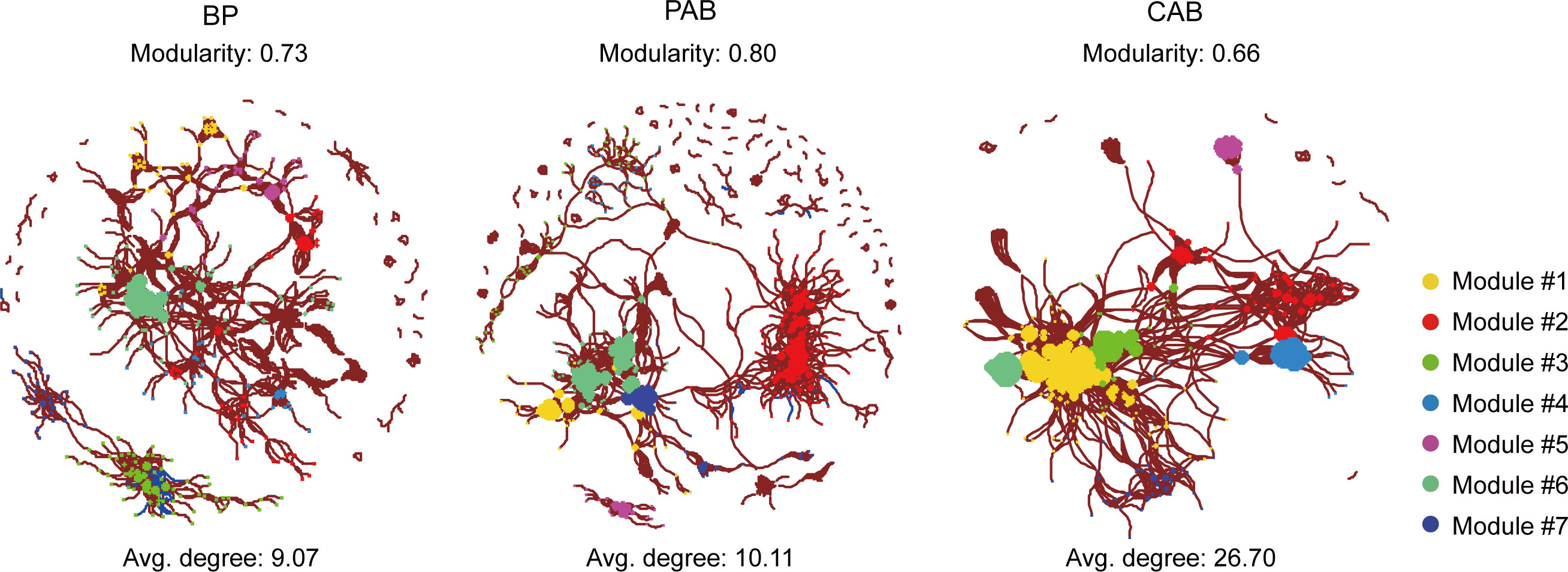
Figure 7 Co-occurrence network of bacterial relationship from BP (bacterioplankton), PAB (particle-attached bacteria), and CAB (colony-attached bacteria) created with a Spearman’s coefficient threshold of ± 0.7 and an adjusted p-value threshold of 0.01. Each node represents bacterial genus and the node size is proportional to the value of degree, lines connecting two nodes indicated the interactions between them. Red lines represent a positive significant correlation and blue lines represent negative significant correlation. Node color represents the bacterial distribution modules.
Relationships between bacteria and environmental factors
As exhibited in Figure 8, the relationships between the community structures of the three kinds of bacteria and the environmental factors varied between the growth phases of the bloom. During the exponential growth phase of the bloom, the community structures of bacterioplankton were positively correlated with COD and concentrations in seawater, whereas, when the bloom shifted to the decline phase, the structures were positively correlated with temperature, salinity, and pH. For particle-attached bacteria, their community structures were positively correlated with DOC, and DO during the exponential growth phase of the bloom, and shifted to salinity during the decline growth phase. Whereas, the community structures of colony-attached bacteria were correlated with seawater temperature, salinity, DO, and COD throughout the bloom, with no obvious bacterial difference observed between the exponential and decline phases.
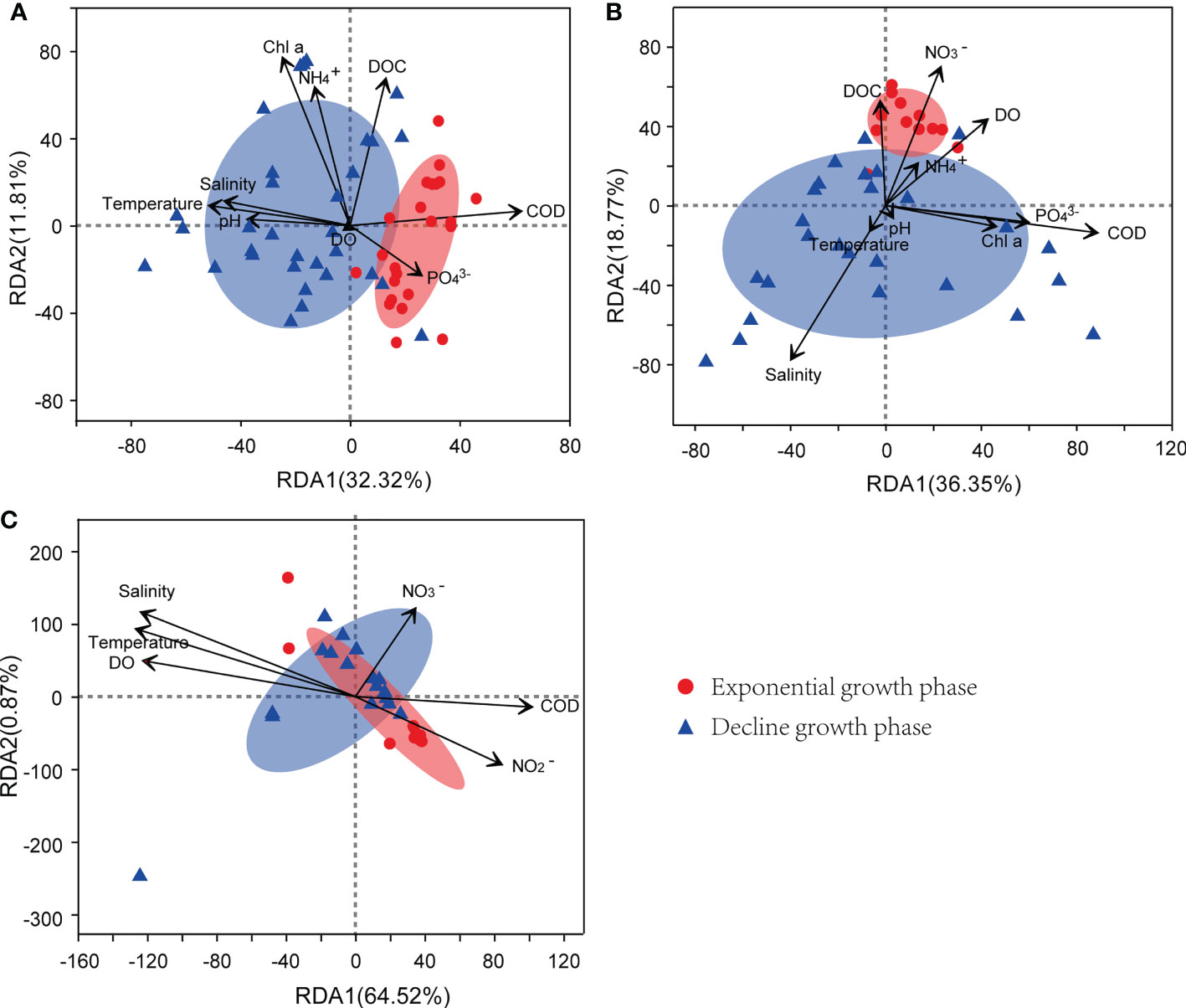
Figure 8 RDA of environmental factors and bacterial community structure from bacterioplankton (A), particle-attached bacteria (B), and colony-attached bacteria (C). E, exponential growth phase of Phaeocystis globosa bloom; D, decline growth phase P. globosa bloom.
Discussion
The bacterial communities harbored in the three habitats were influenced by the inhabited niches and the growth phases of microalgae bloom. We found marked variations in bacterial community structures among the three types of bacteria, potentially due to the diverse lifestyles of bacterioplankton and attached bacteria (Acinas et al., 1999; Fuchsman et al., 2011; Li et al., 2015). The physical and biochemical characteristics of bacterial hosts impact their diversity, with particle-attached bacteria exhibiting greater diversity than bacterioplankton and colony-attached bacteria, consistent with the findings in the Black and Beaufort Seas (Fuchsman et al., 2011; Ortega-Retuerta et al., 2013). In contrast, a lower taxonomic richness of particle-attached bacterial communities compared with bacterioplankton was reported by Li et al. (2015) in the South China Sea, which may be attributed to the P. globosa bloom outbreak in the present study. Further research is required to gain a better understanding of the mechanisms involved.
The present study showed that the characteristics of bacterial communities were related to the growth phases of microalgae bloom, likely due to the varying availability of substrates. Exopolysaccharides were the most available polysaccharides at the beginning of the bloom and were utilized first by most bacteria species. As the bloom progressed, recalcitrant substrates such as sulfated polysaccharides in cellular substrates become dominant, allowing specific bacteria to acquire a dominant position (Teeling et al., 2016). These differences in substrate availabilities at different growth phases of P. globosa bloom likely caused the variations in dominant species and the richness of bacterial communities. Additionally, the change of bloom species also influenced the bacterial community. The A. sanguinea bloom at station S2 on January 28 caused a marked shift in the bacterial communities of bacterioplankton and particle-attached bacteria (Figure S1). For example, Candidatus puniceispirillum, a member of the SAR11 clade of Alphaproteobacteria, increased significantly compared to station S1 on January 28 and station S2 on January 26. C. pelagibacter, another member of the SAR11 clade, is known to be a dominant genus in the bacterial community and plays a crucial role in carbon cycling during A. sanguinea blooms (Yang et al., 2012). Therefore, the explosive growth of C. puniceispirillum during the A. sanguinea bloom in this study may be attributed to their similar abilities to C. pelagibacter.
It has been reported that the dominant phyla of bacterioplankton in the South China Sea are Proteobacteria and Cyanobacteria (Shi et al., 2018; Sun et al., 2021; Ren et al., 2022). However, the present study found that Bacteroidota was the most abundant bacterioplankton, likely due to the high concentrations of nutrients (e.g. ) that favor these organisms during bloom periods, as well as their ability to degrade complex proteins and mucopolysaccharides released by P. globosa (Cottrell and Kirchman, 2000; Janse et al., 2000; Brussaard et al., 2005; Lamy et al., 2009; Almutairi, 2015). Remarkable differences in bacterial community structure and diversity were also observed between the exponential and decline growth phases of the bloom, likely due to the increased proportion of recalcitrant polysaccharides (e.g., fucose-containing sulfated polysaccharides) derived from algal fragmentation, which favored specific bacteria such as Persicirhabdus sp. of Verrucomicrobiota (Martinez-Garcia et al., 2012; Sichert et al., 2020; Orellana et al., 2021). On the other hand, the dominant bacterioplankton that differed significantly between the exponential and the decline growth phases of bloom were all belonged to Bacteroidota, indicating their important role in the degradation of complex organic carbon, particularly in the forms of polysaccharides and hydrocarbons (Bauer et al., 2006; Wolińska et al., 2017). The amounts of organic matter which served as electron donors for organic matter-degrading bacteria such as denitrifying bacteria and sulfate-reducing bacteria can be evaluated by the COD value. In the present study, the community structures of bacterioplankton were positively correlated with COD during the exponential growth phase of the bloom. Notably, the COD value decreased drastically as the bloom developed into the decline phase due to algal cell death or sinking to the bottom, yet the bacterioplankton community did not change accordingly that much. This suggested that the bacterioplankton community was regulated not only by the quantity of organic matter but also by its quality, i.e., biological and chemical composition and form characteristics (Stelzer et al., 2014). Furthermore, environmental factors such as temperature, salinity, and pH were also essential in shaping the community profiles of bacterioplankton.
Particle-attached bacteria possess a critical capability of attaching to particulate matters including Phaeocystis aggregates, providing them many advantages including increased nutrient acquisition, reduced predation risk, and enhanced interactions among populations (Dang et al., 2008). The aggregates of particles or microalgae cells serve as organic-rich microhabitats with abundant substrates, harboring large amounts of bacteria (Alldredge et al., 1986; Li et al., 2015). In the present study, Proteobacteria (Alphaproteobacteria and Gammaproteobacteria) was the dominant phylum of particle-attached bacteria due to their great adhesion ability, as reported by Li et al. (2015). As the bloom shifted from the exponential phase to the decline phase, the homogeneity of their hosts (dominated by algal fragments) caused a significant decrease in the community diversity of particle-attached bacteria (Kiørboe and Jackson, 2001; Azam and Malfatti, 2007; Fang et al., 2015). In addition, the dissolved organic matter released into ambient waters was further utilized by bacterioplankton, such as Persicirhabdus sp. of Verrucomicrobiota, whose abundance significantly increased in the decline growth phase of the bloom, resulting in the complex relationship among organic particles, phytoplankton, particle-attached bacteria, and bacterioplankton, and further studies are needed to explore this.
The formation and enlargement of gelatinous colony is a defense strategy for Phaeocystis, allowing them to avoid predation and become cosmopolitan marine phytoplankton (Wang et al., 2015; Wang et al., 2021a). These colonies are capable of maintaining their position in the euphotic zone by regulating their buoyancy based on light intensity and water flow (Wang et al., 2010). Correspondingly, the majority of colony-attached bacteria were species belonging to autotrophic cyanobacteria, which require light absorption for photosynthesis. Intensive host selection pressure (unique host) resulted in the lowest diversity of colony-attached bacteria, but this could also be an advantage in maintaining the stability of symbiotic relationships between colony-associated bacteria and their hosts (Edwards et al., 2015). A more complex microbial ecosystem with greater connectivity is more stable and robust during environmental changes, as indicated by the high network density, average degree, and the number of edges with the least nodes in colony-attached bacteria (Santolini and Barabási, 2018; Huang et al., 2022). This complex network may explain why there was no significant difference in colony-attached bacterial communities between the exponential and decline growth phases of the P. globosa bloom.
Conclusion
Our study reveals the profound impacts of P. globosa bloom on marine bacteria, with the bacterioplankton and particle-attached bacterial communities varying as the bloom progressed. Bacteria, particularly those with organic decomposing abilities (e.g., Verrucomicrobiota and Bacteroidota), could play a key role in the dissipation of P. globosa blooms. Our findings emphasize the importance of studying the intricate connections between bacteria and microalgae blooms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA776989.
Author contributions
RS and HH conceived the original ideas and collected samples during the outbreak of P. globosa bloom for the manuscript. RS developed the outline and analyzed the data, prepared the manuscript with some contributions to drafting from SZ, MD, and TH and critical refinement by ZQ. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31900094, 41976149), Guangdong Basic and Applied Basic Research Foundation (2021A1515011377), Guangzhou Basic and Applied Basic Research Foundation (202201010306), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD15), Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (2021SD03).
Acknowledgments
We are thankful for the comments and suggestions of the reviewers and the editor that helped to improve the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1075059/full#supplementary-material
References
Acinas S. G., Anton J., Rodriguez-Valera F. (1999). Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl. Environ. Microbiol. 65, 514–522. doi: 10.1128/AEM.65.2.514-522.1999
Alldredge A. L., Cole J. J., Caron D. A. (1986). Production of heterotrophic bacteria inhabiting macroscopic organic aggregates (marine snow) from surface waters. Limnol. Oceanogr. 31, 68–78. doi: 10.4319/lo.1986.31.1.0068
Almutairi A. (2015). Spatial–temporal variations and diversity of the bacterioplankton communities in the coastal waters of Kuwait. Mar. pollut. Bull. 100 (2), 699–709. doi: 10.1016/j.marpolbul.2015.09.016
Anderson D. M., Glibert P. M., Burkholder J. M. (2002). Harmful algal blooms and eutrophication: nutrient sources composition, and consequences. Estuaries 25, 704–725. doi: 10.1007/BF02804901
Arrigo K. R. (2004). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Azam F., Malfatti F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Bauer M., Kube M., Teeling H., Richter M., Lombardot T., Allers E., et al. (2006). Whole genome analysis of the marine bacteroidetes ‘Gramellaforsetii’ reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 8, 2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x
Benjamini Y., Krieger A. M., Yekutieli D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. doi: 10.1093/biomet/93.3.491
Brussaard C. P. D., Mari X., Van Bleijswijk J. D. L., Veldhuis M. J. W. (2005). A mesocosm study of Phaeocystis globosa (Prymnesiophyceae) population dynamics. II. significance for the microbial community. Harmful Algae 4 (5), 875–893. doi: 10.1016/j.hal.2004.12.012
Chan K. M., Leung K. M., Cheung K. C., Wong M. H., Qiu J. W. (2008). Seasonal changes in imposex and tissue burden of butyltin compounds in Thais clavigera populations along the coastal area of mirs bay. China. Mar. pollut. Bull. 57 (6–2), 645–651. doi: 10.1016/j.marpolbul.2008.02.040
Chen J., Li H., Zhang Z., He C., Shi Q., Jiao N., et al. (2020). DOC dynamics and bacterial community succession during long-term degradation of ulva prolifera and their implications for the legacy effect of green tides on refractory DOC pool in seawater. Water Res. 185, 116268. doi: 10.1016/j.watres.2020.116268
Cole J. J. (1982). Interactions between bacteria and algae in aquatic cosystems. Annu. Rev. Ecol. Syst. 13, 291–314. doi: 10.1146/annurev.es.13.110182.001451
Comte J., Del Giorgio P. A. (2010). Linking the patterns of change in composition and function in bacterioplankton successions along environmental gradients. Ecology 91, 1466–1476. doi: 10.1890/09-0848.1
Cottrell M., Kirchman D. (2000). Natural assemblages of marine proteobacteria and members of the cytophaga–flavobacter cluster consuming low- and high-molecular weight dissolved organic matter. Appl. Environ. Microbiol. 66, 1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000
Csárdi G., Nepusz T. (2006). The igraph software package for complex network research. Inter. J. Complex Syst. 1695. .Available at: http://igraph.sf.net.
Dang H., Li T., Chen M., Huang G., Dang H. Y. (2008). Cross-ocean distribution of rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/AEM.01400-07
Delmont T. O., Hammar K. M., Ducklow H. W., Yager P. L., Post A. F. (2014). Phaeocystis antarctica bloom strongly influence bacterial community structures in the amundsen Sea polynya. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00646
Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edwards J., Johnson C., Santos-Medellín C., Lurie E., Podishetty N. K., Bhatnager S., et al. (2015). Structure, variation, and assembly of the root- associated microbiomes of rice. P. Natl. Acad. Sci. U.S.A. 112 (8), E911–E920. doi: 10.1073/pnas.1414592112
Fang J., Zhang L., Li J., Kato C., Zhang Y., Tamburini C., et al. (2015). The POM-DOM piezophilic microorganisms continuum (PDPMC)-the role of piezophilic microorganisms in the global ocean carbon cycle. sci. China Earth Sci. 58, 106–115. doi: 10.1007/s11430-014-4985-2
Fuchsman C. A., Kirkpatrick J. B., Brazelton W. J., Murray J. W., Staley J. T. (2011). Metabolic strategies of free-living and aggregate-associated bacterial communities inferred from biologic and chemical profiles in the black Sea suboxic zone. FEMS Microbiol. Ecol. 78 (3), 586–603. doi: 10.1111/j.1574-6941.2011.01189.x
GB/T 12763.4-2007 (2007). Specifications for oceanographia survey–part 4: Survey of chemical parameters in sea water (Beijing: Standards press of china).
Higashi A., Fujitani Y., Nakayama N., Tani A., Ueki S. (2016). Selective growth promotion of bloom-forming raphidophyte Heterosigma akashiwo by a marine bacterial strain. Harmful Algae 60, 150–156. doi: 10.1016/j.hal.2016.11.009
Hothorn T., Bretz F., Westfall P. (2008). Simultaneous inference in general parametric models. Biometrical J. 50 (3), 346–363. doi: 10.1002/bimj.200810425
Huang Y., Liu Y., Geng J., Lü H., Zhao H., Xiang L., et al. (2022). Maize root-associated niches determine the response variation in bacterial community assembly and function to phthalate pollution. J. Hazard. Mater. 429, 128280. doi: 10.1016/j.jhazmat.2022.128280
Janse I., Zwart G., van der Maarel M. J. E. C., Gottschal J. C. (2000). Composition of the bacterial community degrading Phaeocystis mucopolysaccharides in enrichment cultures. Aquat. Microb. Ecol. 22, 119–133. doi: 10.3354/ame022119
Kiørboe T., Jackson G. A. (2001). Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46 (6), 1309–1318. doi: 10.4319/lo.2001.46.6.1309
Lamy D., Obernosterer I., Laghdass M., Artigas L. F., Breton E., Grattepanche J. D., et al. (2009). Temporal changes of major bacterial groups and bacterial heterotrophic activity during a Phaeocystis globosa bloom in the eastern English channel. Aquat. Microb. Ecol. 58, 95–107. doi: 10.3354/ame01359
Li J., Wei B., Wang J., Liu Y., Dasgupta S., Zhang L., et al. (2015). Variation in abundance and community structure of particle-attached and free-living bacteria in the south China Sea. Deep-Sea Res. Pt. II 122, 64–73. doi: 10.1016/j.dsr2.2015.07.006
Li N., Zhao H., Jiang G., Xu Q., Tang J., Li X., et al. (2020). Phylogenetic responses of marine free-living bacterial community to Phaeocystis globosa bloom in beibu gulf, China. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01624
Liu Y., Debeljak P., Rembauville M., Blain S., Obernosterer I. (2019). Diatoms shape the biogeography of heterotrophic prokaryotes in early spring in the southern ocean. Environ. Microbiol. 21, 1452–1465. doi: 10.1111/1462-2920.14579
Ma B., Wang H., Dsouza M., Lou J., He Y., Dai Z., et al. (2016). Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 10 (8), 1891–1901. doi: 10.1038/ismej.2015.261
Martinez-Garcia M., Brazel D. M., Swan B. K., Arnosti C., Chain P. S., Reitenga K. G., et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of verrucomicrobia. PloS One 7, e35314. doi: 10.1371/journal.pone.0035314
Meyer K. A., O’Neil J. M., Hitchcock G. L., Heil C. A. (2014). Microbial production along the West Florida shelf: responses of bacteria and viruses to the presence and phase of Karenia brevis blooms. Harmful Algae 38, 110–118. doi: 10.1016/j.hal.2014.04.015
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019) Vegan: community ecology package. Available at: https://CRAN.R-project.org/package=vegan.
Orellana L. H., Francis T. B., Ferraro M., Hehemann J., Fuchs B. M., Amann R. (2021). Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 16 (3), 630–641. doi: 10.1038/s41396-021-01105-7
Ortega-Retuerta E., Joux F., Jeffrey W. H., Ghiglione J. F. (2013). Spatial variability of particle-attached and free-living bacterial diversity in surface waters from the Mackenzie river to the Beaufort Sea (Canadian Arctic). Biogeosciences 10, 2747–2759. doi: 10.5194/bg-10-2747-2013
Parada A. E., Needham D. M., Fuhrman J. A. (2015). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
R Core Team (2018). R: a language and environment for statistical computing (Vienna, Austria: R foundation for statistical computing). Available at: https://www.R-project.org/.
Ren L., Lu Z., Xia X., Peng Y., Gong S., Song X., et al. (2022). Metagenomics reveals bacterioplankton community adaptation to long-term thermal pollution through the strategy of functional regulation in a subtropical bay. Water Res. 216 (5), 118298. doi: 10.1016/j.watres.2022.118298
Revelle W. (2021). Psych: procedures for personality and psychological research (Evanston, Illinois, USA: Northwestern University). Available at: https://CRAN.R-project.org/package=psych.
Ruiz-Gonzalez C., Proia L., Ferrera I., Gasol J. M., Sabater S. (2013). Effects of large river dam regulation on bacterioplankton community structure. FEMS Microbiol. Ecol. 84, 316–331. doi: 10.1111/1574-6941.12063
Santolini M., Barabási A. L. (2018). Predicting perturbation patterns from the topology of biological networks. P. Natl. Acad. Sci. U.S.A. 115 (27), E6375–E6383. doi: 10.1073/pnas.1720589115
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75 (23), 7537–7541. doi: 10.1128/AEM.01541-09
Sher D., Thompson J. W., Kashtan N., Croal L., Chisholm S. W. (2011). Response of Prochlorococcus ecotypesto co-culture with diverse marine bacteria. ISME J. 5, 1125–1132. doi: 10.1038/ismej.2011.1
Shi R., Huang H., Qi Z., Han T. (2022). The extent and pattern of mariculture impacts on spatial and seasonal variations of sediment bacterial communities among three coastal waters. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.782456
Shi R., Li J., Qi Z., Zhang Z., Liu H., Huang H. (2018). Abundance and community composition of bacterioplankton in the northern south China Sea during winter: geographic position and water layer influences. Biologia 73 (2), 197–206. doi: 10.2478/s11756-018-0023-8
Sichert A., Corzett C. H., Schechter M. S., Unfried F., Markert S., Becher D., et al. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5, 1026–1039. doi: 10.1038/s41564-020-0720-2
Stelzer R. S., Scott J. T., Bartsch L. A., Parr T. B. (2014). Particulate organic matter quality influences nitrate retention and denitrification in stream sediments: evidence from a carbon burial experiment. Biogeochemistry 119, 387–402. doi: 10.1007/s10533-014-9975-0
Sun F., Wang C., Wang Y., Tu K., Zheng Z., Lin X. (2020). Diatom red tide significantly drive the changes of microbiome in mariculture ecosystem. Aquaculture 520, 734742. doi: 10.1016/j.aquaculture.2019.734742
Sun F., Wang Y., Wu M., Sun C., Jiang Z., Cheng H., et al. (2021). Bacterial community variations in the south China Sea driven by different chemical conditions. Ecotoxicology 30, 1808–1815. doi: 10.1007/s10646-021-02455-w
Sun W., Xia C., Xu M., Guo J., Sun G. (2017). Seasonality affects the diversity and composition of bacterioplankton communities in dongjiang river, a drinking water source of Hong Kong. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01644
Teeling H., Fuchs B. M., Bennke C. M., Krüger K., Chafee M., Kappelmann L., et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5, e11888. doi: 10.7554/eLife.11888
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang X., Huo Y., Yang F., Wang Y. (2021b). Induced allelopathic effects of Thalassiosira weissflogii on colony formation in Phaeocystis globosa. Water 13, 581. doi: 10.3390/w13050581www.mdpi
Wang X., Song H., Wang Y., Chen N. (2021a). Research on the biology and ecology of the harmful algal bloom species Phaeocystis globosa in China: progresses in the last 20 years. Harmful Algae 107 (2), 102057. doi: 10.1016/j.hal.2021.102057
Wang X., Tang K. W., Smith W. O. Jr. (2010). Temperature effects on growth, colony development and carbon partitioning of three Phaeocystis species. Aquat. Biol. 9 (3), 239–249. doi: 10.3354/ab00256
Wang X., Wang Y., Ou L., He X., Chen D. (2015). Allocation costs associated with induced defense in Phaeocystis globosa (Prymnesiophyceae): the effects of nutrient availability. Sci. Rep. 5, 10850. doi: 10.1038/srep10850
Wolińska A., Kuźniar A., Zielenkiewicz U., Izak D., Szafranek-Nakonieczna A., Banach A., et al. (2017). Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 119, 128–137. doi: 10.1016/j.apsoil.2017.06.009
Worden A. Z., Follows M. J., Giovannoni S. J., Wilken S., Zimmerman A. E., Keeling P. J. (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594. doi: 10.1126/science.1257594
Xia X., Leung S. K., Cheung S., Zhang S., Liu H. (2020). Rare bacteria in seawater are dominant in the bacterial assemblage associated with the bloom-forming dinoflagellate Noctiluca scintillans. Sci. Total Environ. 711, 135107. doi: 10.1016/j.scitotenv.2019.135107
Keywords: Phaeocystis globosa bloom, bacterioplankton, particle-attached bacteria, colony-attached bacteria, Mirs Bay
Citation: Shi R, Qi Z, Han T, Dai M, Zhang S and Huang H (2023) Responses of bacterioplankton, particle- and colony-attached bacterial communities to Phaeocystis globosa blooms in Mirs Bay, China. Front. Mar. Sci. 10:1075059. doi: 10.3389/fmars.2023.1075059
Received: 20 October 2022; Accepted: 02 March 2023;
Published: 15 March 2023.
Edited by:
Paulo João Vieira Vale, Portuguese Institute for Sea and Atmosphere (IPMA), PortugalReviewed by:
Pengbin Wang, Ministry of Natural Resources, ChinaSun Cuici, South China Sea Institute of Oceanology (CAS), China
Copyright © 2023 Shi, Qi, Han, Dai, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghui Huang, aHVhbmdoaEBzY3NmcmkuYWMuY24=
 Rongjun Shi
Rongjun Shi Zhanhui Qi
Zhanhui Qi Tingting Han
Tingting Han Ming Dai
Ming Dai Shufei Zhang
Shufei Zhang Honghui Huang
Honghui Huang