
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 13 February 2023
Sec. Aquatic Physiology
Volume 10 - 2023 | https://doi.org/10.3389/fmars.2023.1073322
This article is part of the Research TopicThe defense responses of aquatic animals to the environmentView all 7 articles
To produce cultured pearls, a mantle graft with a nucleus is transplanted into a host pearl oyster, this process is called “transplantation”. The immune response of pearl oyster after transplantation is a major factor that leads to nucleus rejection and death. Butyrate is a histone deacetylase (HDAC) inhibitor which can inhibit the deacetylation process of histones and effectively reduce the inflammatory response. To clarify the function of histone acetylation in immune response after transplantation, butyrate (10 mmol/L) was used for the treatment of pearl oysters before transplantation. Results showed that the proportion of histone H3 acetylation of the hemocytes was significantly increased after butyrate treatment before transplantation (BH group) compared with the control group at 6–24 h. Transcriptome analysis showed that butyrate treatment activated the “lysosome”, inhibited cell migration and cell proliferation at 6 and 12 h, respectively, and activated the intracellular immune recognition response of pearl oyster at 24 h after transplantation. The apoptosis detection revealed no significant difference in the proportion of apoptotic cells between the control and BH group. Moreover, butyrate treatment increased the activity of some immune-related enzymes in the serum of pearl oyster after transplantation.
Pearl oyster Pinctada fucata martensii is one of the most important species for pearl production in southern China. During pearl production, the donor pearl oyster is required for the preparation of mantle graft, which is implanted together with one nucleus into the host pearl oyster (Zhao et al., 2012; Wang et al., 2019). This process, which is called transplantation surgery, can induce the immune response in the host oyster, and overwhelming immune response will result in the rejection of the nucleus or failure development of pearl sac or death of host oyster (Wang et al., 2019). Different omics have been used for the analysis of the immune response of hemocyte, and many crucial pathways and genes have been found after transplantation. A large number of immune-related pathways were enriched, and immune-related genes were upregulated at 6–48 h after transplantation, suggesting that the 6–48 h after transplantation may be a critical period for immune rejection in pearl oysters (Jiao et al., 2019; Cao et al., 2021). How to reduce the excessive immune response of pearl oyster and increase its immune tolerance in the early stage of transplantation remain unclear.
Histone acetylation is one of the most important histone modifications and is an important regulatory modality that regulates the expression of inflammation-related genes and affects the onset and development of inflammation. Studies have shown that acetylation modifications can occur on four histones (H2A, H2B, H3, and H4) (Wang et al., 2012), with acetylation of histone H3 and H4 being the most common (Turner, 1991). Many sites on histone H3 can be modified by acetylation, such as K9, K14, K18, K23 and K27, acetylation of different sites can have different impacts. For example, the localization of nucleosomes is importantly associated with acetylation of the H3K9 site. In addition, acetylation of most loci is involved in gene transcription (Roth and Allis, 1996; Zhang et al., 1998). It was found that acetylation modifications of histone H3 can participate in the inflammatory response by regulating ATP-dependent chromatin remodeling (Chatterjee et al., 2011). Histone H4 acetylation is involved in chromatin transcription and chromatin replication, and the four acetylated lysine of histone H4 are specific for chromatin activity. H4 acetylated on lysine 8 or lysine 16 is preferentially recovered in the “free” histone pool, while acetylation on lysine 5 or lysine 12 is stably bound to chromatin (Mocquard-Bucher et al., 2014).
Histone acetylation and deacetylation are catalyzed by histone acetyltransferase (HAT) and histone deacetylase (HDAC), respectively, and regulate a broad and complex array of physiological processes, such as cell differentiation, proliferation, metabolism, senescence, and immune and apoptosis (Bai et al., 2008; Villagra et al., 2008; Wang et al., 2009). Sirt1 is a NAD+-dependent histone deacetylase, and Sirt1 deacetylates and inhibits FoxO1-dependent apoptosis (Motta et al., 2004). Sirt1 can control the self-renewal and proliferation of mouse embryonic stem cells by regulating the deacetylation of p53 (Han et al., 2005). HDAC4 interacts with the nucleotide excision repair factor XPC, which plays an important role in effectively removing the ultraviolet B-induced DNA damage (Li et al., 2020). HDAC6 plays a crucial role in the innate immune response to intracellular bacterial infection through Toll-like receptor-mediated signaling (Ran and Zhou, 2019). HDAC is associated with the pathogenesis of multiple inflammatory diseases, and HDAC inhibitors (HDI) is a potential therapeutic strategy (Adcock, 2007; De Zoeten et al., 2011).
HDI are a class of compounds, which can interfere with the function of HDAC. It has the characteristics of low toxicity and high specificity (Chuang et al., 2013), improves cell survival and increases the anti-inflammatory response (Kim et al., 2007). HDI is widely used in clinical treatment by increasing the acetylation level of histone and non-histone (Zhang et al., 2015). Butyrate, which is a commonly used HDI, is an important component of the intestinal environment and an apoptosis inducer (Pant et al., 2016), can inhibit HDAC activity, its mode of action is random and non-specific, and can lead to histone H3 and H4 hyperacetylation (Archer and Hodin, 1999). Aside from providing nutrients for intestinal epithelial cells as an energy substance, butyrate is also the main metabolite of intestinal bacteria and has a direct immune regulation impact (Semann et al., 2000). Low concentrations of butyrate can activate the BAX/BCL2 pathway, stimulate tumor cell apoptosis, and exert an anti-tumor proliferation impact (Encarncao et al., 2018). In vertebrates, butyrate affects cell growth through cell cycle arrest (Waby et al., 2010), inhibit the production of pro-inflammatory cytokines (IL-6, IL-12) (Weber and Kerr, 2006), promote the production of anti-inflammatory cytokines (IL-10) (Lee et al., 2017), and regulate the immune system to the colon mucosa maintained in a non-inflammatory state (Nancy et al., 2002). Butyrate also regulates the formation of rat beta-defensin by inhibiting TLR expression, thereby suppressing pro-inflammation upon LPS challenge (Dou et al., 2022). The function of histone acetylation was not clear in the molluscan. In the present study, butyrate was used to treat the pearl oysters before transplantation, and hemocytes were collected at 6, 12, 24 and 48 h after transplantation to construct transcriptome. The impact of butyrate on the immune response of pearl oyster after transplantation was analyzed in detail. This study aimed to determine the mechanism of histone acetylation in the immune regulation of the pearl oyster P. f. martensii and to provide insight into the mechanism of immune regulation during pearl production.
The Pearl Oyster P. f. martensii (approximately 1.5 years old) used in this study was obtained from the sea area of Dajing village, Xuwen County, Guangdong Province. 80 pearl oyster was randomly divided into two groups - one group (BH group) was injected with 100 μL of butyrate (Sangon Biotech, China, 10 mmol × L-1) (Chakravortty et al., 2000), while the other group (PH group) was injected with 100 μL of PBS (Sangon Biotech, China). All pearl oyster were injected through the adductor muscles and were transplanted immediately after the injection treatment. After transplantation, the pearl oysters were cultured in a tank containing filtered seawater. The hemolymphs of pearl oysters from the BH and PH group were collected from adductor muscles at different times (6, 12, 24, and 48 h) by using 2-mL syringes, the hemolymph was centrifuged to obtain hemocytes and serum (4°C, 3,000 rpm, 5 min), and some hemocytes were used to extract the total histone. TRIzol reagent (Invitrogen, USA) was added to the hemocytes, and then stored at −80°C until use. Some hemocytes were fixed with paraformaldehyde (Sangon Biotech, China), and serums were stored by liquid nitrogen quick freezing.
Total histone was extracted from P. f. martensii of the hemocytes and EpiQuik™ total histone extraction kit (Epigentek, USA), and protein concentration was analyzed using a modified BCA protein assay kit (Sangon, China). Each group collected three hemocytes for the acetylation levels of H3 and H4 at different times after injection and transplantation, and acetylation levels were calculated based on the manual of EpiQuik™ Total Histone H3 Acetylation Detection Fast Kit and EpiQuik™ Total Histone H4 Acetylation Detection Fast Kit (Colorimetric, Epigentek, USA).
Total RNA was extracted from the hemocytes of P. f. martensii at 6–48 h after transplantation with TRIzol reagent (Invitrogen, USA), five hemocytes in each group were collected and validated as described by Yang et al. (2018).
This project used the DNBSEQ platform for sequencing and performs quality control on the raw reads data obtained. After the quality control was qualified, the data were compared with the reference genome of P. f. martensii using Bowtie2, the gene expression levels of each sample were also calculated using RSEM, and the genes were quantified and subjected to principal component, GO function, KEGG pathway significant enrichment, and other analyses.
The DEG-Seq method allows differentially expressed genes (DEGs) to be detected. The threshold values for significant differential expression were FDR ≤ 0.001 and [log2 Ratio (BP/SP)] > 1. GO function and KEGG pathway analyses were carried out based on DEGs, and enrichment analysis was performed using the phyper function in R software to calculate the p-value, which was usually considered significantly enriched with Q value < 0.05. DEGs were analyzed using GO function and KEGG pathway (Q value<0.05).
Each group involves the collection of three serums at different times after injection and transplantation. Leucine arylamidase (LAP), catalase (CAT), phenol oxidase (PO), lysozyme (LYS), alkaline phosphatase (ALP), malondialdehyde (MDA), superoxide dismutase (SOD), and glutathione peroxidase (GSH-Px) were measured according to the instruction using kits (Beijing Dongge Weiye Technology Co., Ltd., Beijing, China). Meanwhile, we also analyzed the expression of all CAT, ALP, LYS, GSH-Px in the transcriptome at different time after injection and transplantation and used the mean values to represent the expression trend.
The hemocyte was fixed on polylysine-treated glass slides, fixed with 4% paraformaldehyde for 1 h, and then rinsed with PBS. Blocking solution was added, and the sample was incubated for 10 min at 37°C. Permeabilizer solution was added, and the solution was incubated for 2 min at 4°C. Approximately 50 μL of labeling solution (without end transferase) was added to the cells after fixation and permeabilization as a negative control. Fixed and permeabilized cells were treated with DNase I, and 50 μL of TUNEL reaction solution was added as a positive control. The slides were incubated at 37°C for 1 h in the dark, rinsed thrice with PBS, and examined by microscopy. Five different fields of view were randomly selected for photographic counting under white light to detect apoptotic staining in different groups.
All data analyses were performed using SPSS for Windows, and P<0.05 was considered a significant difference. Differences in the levels of histone H3 and H4 acetylation, cell apoptosis and enzyme activities in different groups at the same time point were determined using t-test.
As shown in Figures 1A, B, the acetylation levels of H3 and H4 in the PH and BH group exhibited an increase at 6–24 h and a decrease at 48 h. In comparison with the PH group, the acetylation level of H3 in the BH group was significantly increased at 6–24 h, whereas no significant impact was found in the acetylation level of H4 at 6–48 h.
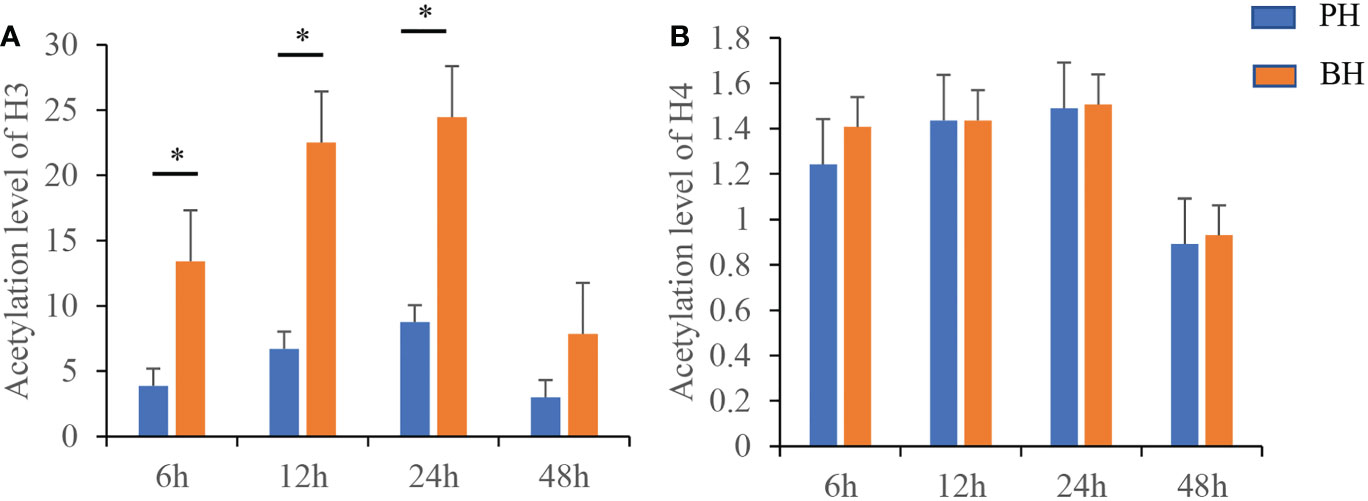
Figure 1 Changes of histone H3 and H4 acetylation at different time points after butyrate stimulation. (A) the level of Histone H3 acetylation in the PH/BH group. (B) the level of Histone H4 acetylation in the PH/BH group. Blue is the PH group and red represents the BH group. Values were shown as mean ± SD (N = 3), *P < 0.05.
At different times (6–48 h) after injection and transplantation, eight transcriptomes from two groups were successfully constructed and sequenced. Basic analysis data for each group of samples are shown in Table 1. Each sample produced an average of 21.34 M data. The comparison rates of the gene and genome were 32.3%–36.78% and 70.06%–71.45%, respectively. The quality assessment of sequencing data resulted in Q20 of 98.19%–99.02% and Q30 of 93.90%–96.49% for each sample, suggesting that the sequencing quality can be effectively used for further analysis (Table 1).
In comparison with the PH group, 3,160 DEGs (FDR<0.001, fold change (FC)≥2) were obtained in the BH group at 6 h, among which 1,454 genes were significantly highly expressed, and 1,706 genes were significantly downregulated in the BH group (Figure 2A). These DEGs were mainly enriched in 61 signaling pathways by KEGG analysis (Q value<0.05), such as “Lysosome”, “Adherents junction”, “Regulation of actin cytoskeleton”, and “Focal adhesion” (Figure 2B). In the lysosome, most protease-related genes such as cathepsins (cathepsin A, C, F, L, X) were upregulated in the BH group. Thus, the function of protein degradation in lysosomes was upregulated after the butyrate treatment and transplantation. Some sugar and lipid metabolism-related genes, such as N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (NAGPA), arylsulfatase B (ARSB), arylsulfatase I/J (ARSI/J), and alpha-N-acetylgalactosaminidase (NAGA) were upregulated in the BH group. This result indicates that butyrate treatment activates the lysosome signaling pathway (Figure 2D). Five β-actin and four α-integrin were found in the “Adherents junction”, “Regulation of actin cytoskeleton”, and “Focal adhesion” (Figures 2E–G), all of which were significantly downregulated in the BH group, indicating that butyrate treatment inhibited cell migration. A total of 163 items were significantly enriched by GO analysis (Q value< 0.05), such as “serine-type carboxypeptidase activity” and “catalytic activity” (Figure 2C), which were related to the regulation of enzyme activity and corresponded to the enrichment of KEGG functions.
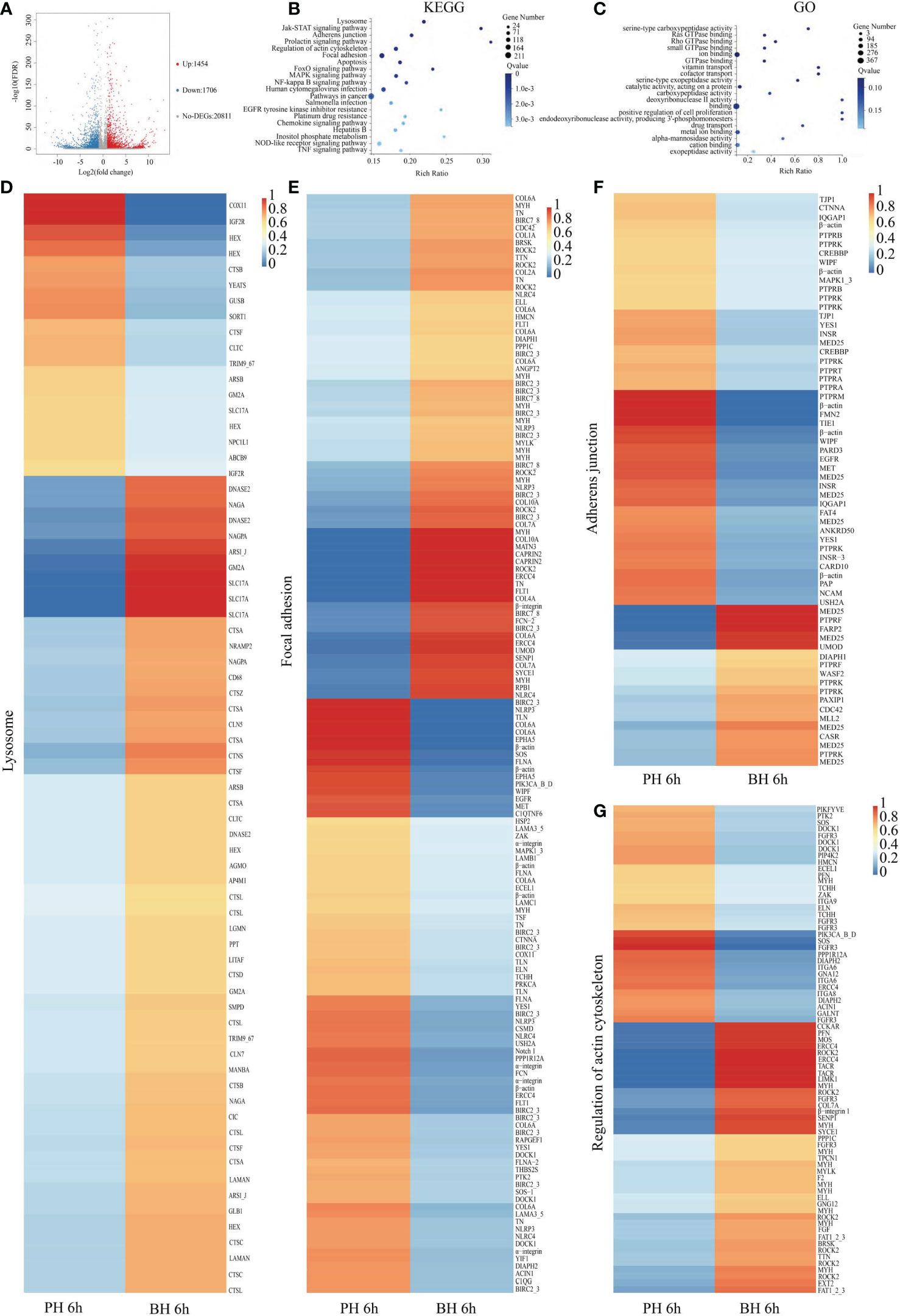
Figure 2 Analysis of DEGs between BH and PH group after transplantation 6 (h) (A). Volcano map of DEGs between BH and PH groups after transplantation 6 (h) Impacted pathway (B) and GO (C) of butyrate on the genes regulated by transplantation. Expression of the related genes in the Lysosome (D), Focal adhesion (E), Adherens junction (F), Regulation of actin cytoskeleton (G) in PH and BH groups after transplantation 6 h.
In comparison with the PH group, 2,671 DEGs (FDR <0.001, fold change (FC) ≥2) were acquired in the BH group at 12 h, of which 899 genes were significantly highly expressed, and 1,772 genes were significantly downregulated in the BH group (Figure 3A). These DEGs were mainly enriched in 19 signaling pathways by KEGG analysis (Q value<0.05), such as “Base excision repair”, “DNA replication”, “Mismatch repair”, “Cell cycle”, and “Nucleotide excision repair” (Figure 3B). These pathways were associated with cell proliferation and mismatch repair, and most of the related genes in the pathway were significantly downregulated in the BH group, indicating that butyrate treatment inhibited cell proliferation (Figures 3D–H). As shown in Figure 3C, 85 items were significantly enriched by GO analysis (Q value< 0.05), such as “DNA replication” and “Cell cycle”, which correspond to the enrichment of KEGG functions.
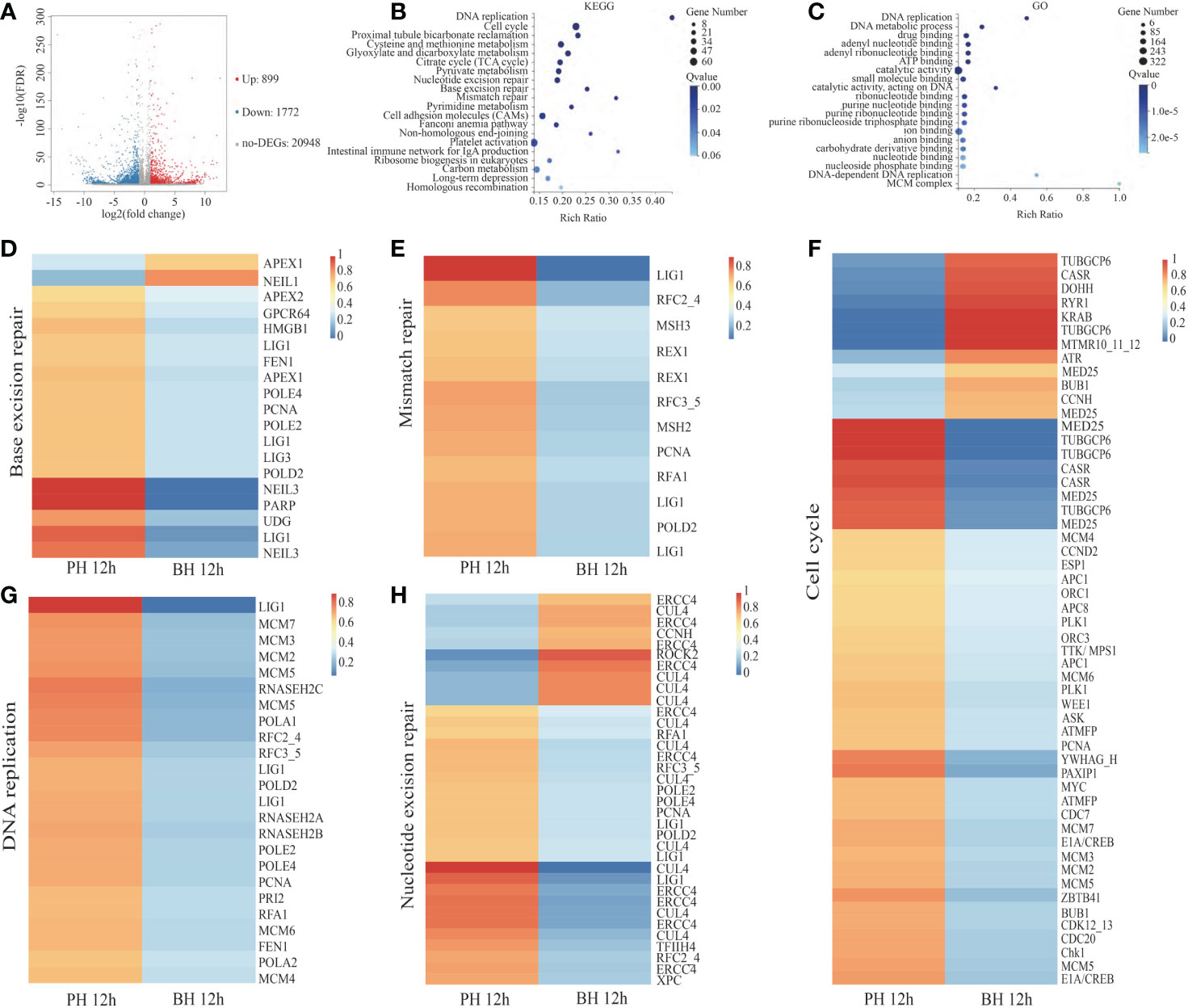
Figure 3 Analysis of DEGs between PH and BH group after transplantation 12 (h) (A). Volcano map of DEGs between PH and BH groups after transplantation 12 (h) Impacted pathway (B) and GO (C) of butyrate on the genes regulated by transplantation. Expression of the related genes in the Base excision repair (D), Mismatch repair (E), Cell cycle (F), DNA replication (G), Nucleotide excision repair (H) in PH and BH groups after transplantation 12 h.
In comparison with the PH group, 1,912 DEGs (FDR <0.001, fold change (FC) ≥2) were obtained in the BH group at 24 h, of which 1,187 genes were significantly highly expressed, and 725 genes were significantly down-expressed in BH group (Figure 4A). These DEGs were mainly enriched in 108 signaling pathways by KEGG analysis (Q value<0.05), such as “Cytosolic DNA-sensing pathway”, “NF-kappa B signaling pathway”, “Apoptosis”, “NOD-like receptor signaling pathway”, and “TNF signaling pathway” (Figure 4B). The related genes in this pathway were significantly upregulated in the BH group (Figures 4D–F). A total of 34 cyclic GMP-AMP synthase (cGAS) genes in the cytosolic DNA-sensing pathway were upregulated in BH group. Nineteen NOD-like receptors (NLRs) were enriched in the NOD-like receptor signaling pathway, including sixteen NLRP3 and three NLRC4, only three NLRP3 were significantly downregulated, and the remaining genes were significantly upregulated in the BH group. In the apoptosis pathway, 20 apoptosis suppressor genes were enriched, including 19 baculoviral IAP repeat-containing protein 2/3 (BIRC2/3) and 1 Bcl-2 (Bcl-2-like 1), of which only one BIRC2/3 was significantly under-expressed, and the remaining were overexpressed in BH group. Four caspase (Cas) genes were found, of which one Cas gene was significantly overexpressed, and three were under-expressed in the BH group. In the TNF and NF-kappa B signaling pathway, most of DEG genes were apoptosis-suppressing and anti-inflammatory genes. Among these genes, one TNF receptor-associated factor 3 (TRAF3) and two NF-kappa-B inhibitor alpha (NFKBIA) were upregulated in the BH group (Figures 4G–H). Most of interleukin 17 (IL17) genes were downregulated in the BH group (Figures 4I). These results indicate that butyrate treatment activates the intracellular immune recognition response of pearl oyster, activates apoptosis, NF-κB and TNF signaling pathway, but still play a role in inhibiting the inflammatory response.
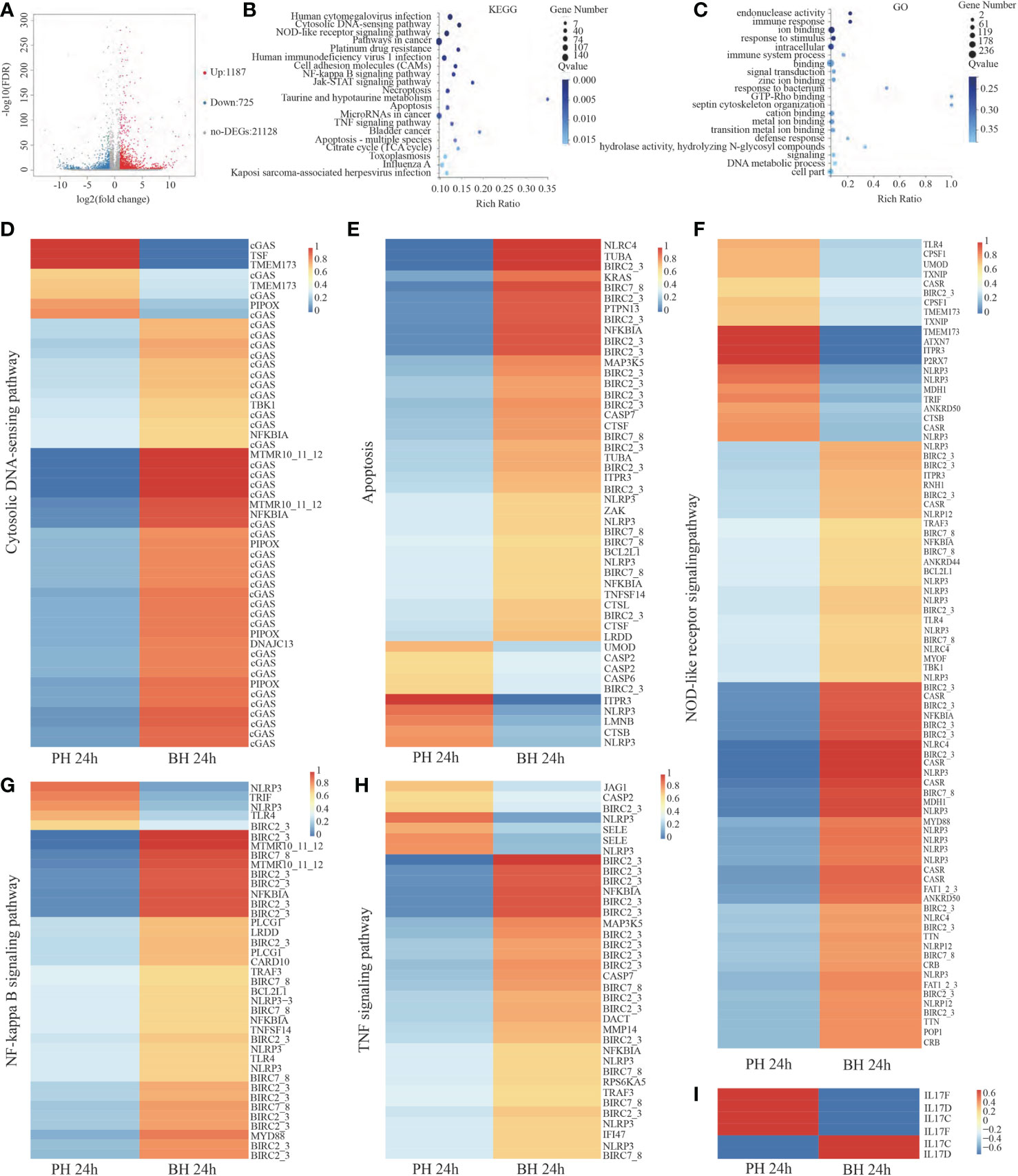
Figure 4 Analysis of DEGs between PH and BH group after transplantation 24 (h) (A). Volcano map of DEGs between PH and BH groups after transplantation 24 (h) Impacted pathway (B) and GO (C) of butyrate on the genes regulated by transplantation. Expression of the related genes in the Cytosolic DNA sensing pathway (D), Apoptosis (E), NOD-like receptor signaling pathway (F), NF-kappa B signaling pathway (G), TNF signaling pathway (H), IL 17 (I) in PH and BH groups after transplantation 24 h.
As shown in Figure 4C, 93 items were significantly enriched by GO analysis (Q value< 0.05), such as “response to stimulus”, “immune response”, and “defense response”, which corresponded to the enrichment of KEGG functions.
The staining of apoptotic cells in the PH and BH group was detected, five different fields of view were randomly selected, and photos were obtained and counted under white light in the same field of view, as shown in Figure 5. The apoptosis in each field of view was calculated. Statistical analysis of the proportion of apoptotic cells and SPSS analysis (P<0.05) showed that the average apoptotic proportion of hemocytes in the BH group at 24 h was 0.2757%, while that in the PH group was 0.2828%. Apoptosis ratio did not significantly differ.
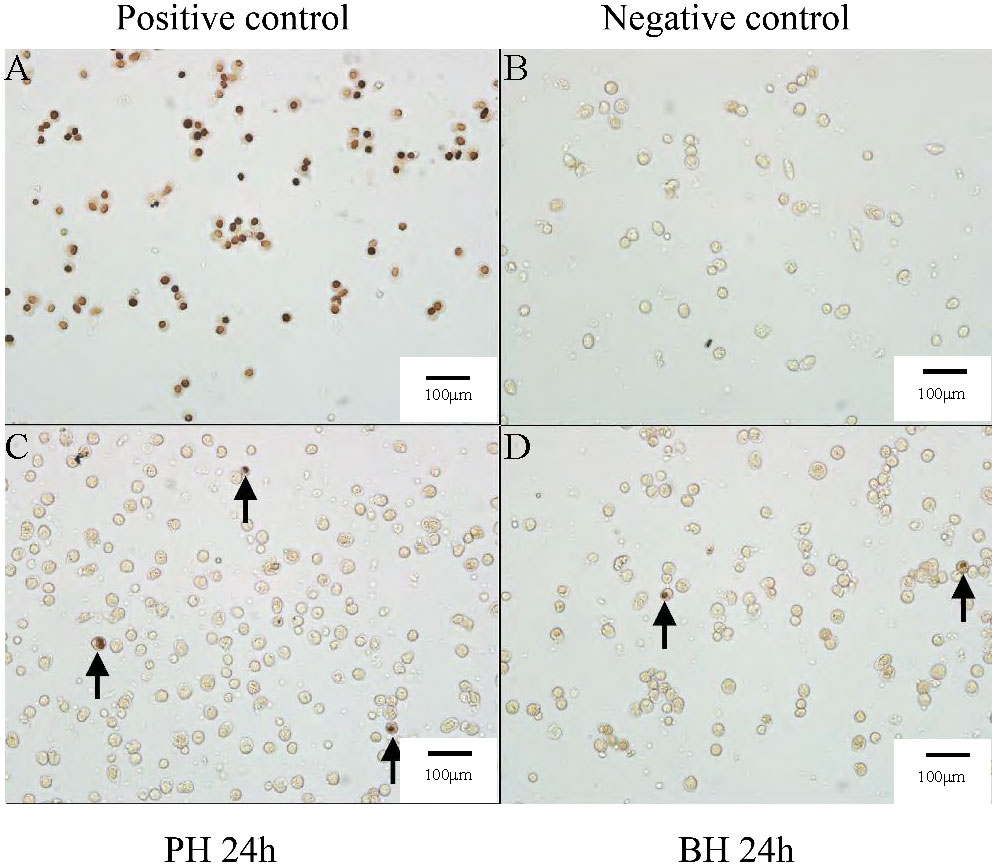
Figure 5 Apoptosis results after butyrate stimulation of transplantation at 24 (h) (A) Apoptosis results in positive control group. (B) Apoptosis results in negative control group. (C) Apoptosis results in PH group after transplantation 24 (h) (D) Apoptosis results in BH group after transplantation 24 h.
In comparison with the PH group, 3,113 DEGs (FDR <0.001, fold change (FC) ≥2) were obtained in the BH group at 48 h, of which 1,595 genes were significantly highly expressed, and 1,518 genes were significantly down-expressed in the BH group (Supplementary Figure 1). These DEGs were mainly enriched in 149 signaling pathways by KEGG analysis (Q value<0.05), such as “Cytosolic DNA-sensing pathway”, “NF-kappa B signaling pathway”, “Apoptosis”, “NOD-like receptor signaling pathway”, and “TNF signaling pathway” (Supplementary Figure 1).
In the cytosolic DNA-sensing pathway, 30 cGAS were upregulated in the BH group, 41 NLRs were enriched in the NOD-like receptor signaling pathway, including 29 NLRP3 and 12NLRC4, nine NLRP3 and two NLRC4 were significantly down-regulated, and the remaining genes were significantly upregulated in the BH group. In the apoptosis pathway, 24 BIRC2/3 were found, of which only 11 BIRC2/3 were significantly under-expressed, and the remaining were over-expressed in the BH group. Six Cas genes were found, of which three Cas genes were significantly over-expressed, and three were under-expressed in the BH group. In the TNF and NF-kappa B signaling pathway, one TRAF3 and one NFKBIA gene were upregulated in the BH group. The results indicate that at 48 h after transplantation, genes associated with intracellular immune recognition responses remain active in the BH group, but the differences in apoptosis and NF-kB signaling pathway between the two groups were narrowed (Supplementary Figure 1). As shown in Supplementary Figure 1, 17 items were significantly enriched by GO analysis (Q value< 0.05), such as “regulation of programmed cell death”, “regulation of cell death”, and “regulation of apoptotic process”, which were consistent to the enrichment of KEGG functions.
A total of one CAT, six ALP, nine GSH-Px, and two LYS genes were identified in the transcriptome and their expression trends were analyzed as the mean of the expression of each gene. Compared with those in the PH group, the expression of CAT and ALP was up-regulated at 6 h and 48 h in the BH group, the expression of LYS was up-regulated at 6 h, 24 h, 48 h, the expression of GSH-PX was up-regulated at 6 - 48 h (Supplementary Figure 2), the activities of LAP, CAT, PO, and SOD remarkably increased at 6 h in the BH group, and the activities of LAP, CAT, PO, ALP, MDA, and SOD were significantly increased at 12 h in the BH group. All eight enzymes were significantly upregulated at 24 h in the BH group. The activities of ALP, MDA, GSH-Px, and SOD were significantly increased at 48 h in the BH group (Figure 6). These results suggest that butyrate stimulation enhances the activity of serum immunity in P. f. martensii.
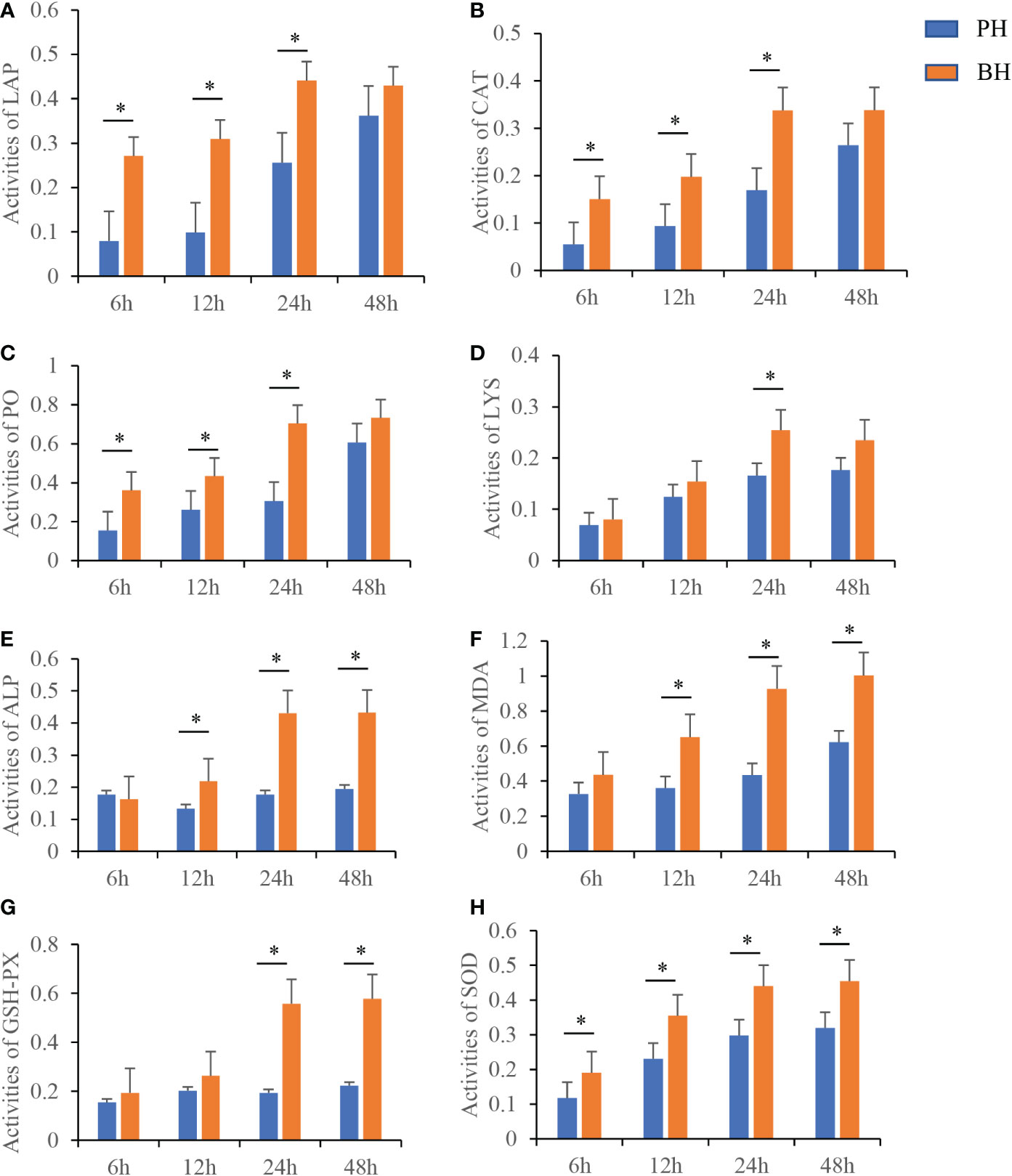
Figure 6 Changes of different activity levels at different time after butyrate stimulation. Activity of LAP (A), CAT (B), PO (C), LYS (D), ALP (E), MDA (F), GSH-Px (G) and SOD (H) in serum at different time points in PH and BH group after transplantation. Values were shown as mean ± SD (N = 3), *P < 0.05.
During producing pearls, the transplanted mantle and nucleus could induce the immune response of pearl oyster, and excessive immune response can cause nuclear rejection and death of pearl oysters. Our previous research showed that the H3 acetylation level of the hemocytes was significantly increased after transplantation compared with the un-transplanted pearl oysters, the HAT and HDAC genes were conserved and have a high level of expression in the hemocytes of P. f. martensii, and some HAT and HDAC show dynamic changes after transplantation, indicating that HAT and HDAC were involved in the immune response after transplantation. In vertebrate, butyrate was reported to induce the relaxation of the chromatin structure, provide better access to transcription-related proteins (Jazirehi, 2010), and have an important role in suppressing inflammation (Jiang et al., 2021). In the present study, butyrate was used to treat pearl oysters before transplantation, and hemocytes were collected at 6, 12, 24 and 48 h after transplantation to construct transcriptome. The effect of histone acetylation on the immune response of pearl oyster after transplantation was analyzed in detail.
Our results showed that the level of histone H3 acetylation in the hemocytes of BH group was significantly increased compared with PH group, indicating that the state of histone acetylation modification changed after butyrate stimulation. After transplantation for 6 h, the BH group had 3160 DEGs compared with the PH group. Functional enrichment analysis showed that “Lysosome”, “Adherents junction”, “Regulation of actin cytoskeleton” and “Focal adhesion” were significantly enriched. Lysosome, an intracellular digestive organ, is involved in cellular degradation and waste recycling, and it plays an important role in the phagocytosis and degradation of pathogens. Furthermore, lysosomes participate in some fundamental processes, such as secretion, energy and signaling metabolism, and plasma membrane repair (Settembre et al., 2013). This study showed that lysosomal enzyme-related genes, such as cathepsin and arylsulfatase, were significantly up-regulated in the BH group, indicating that butyrate treatment activates the lysosome signaling pathway, which may help the pearl oyster to deal with the damaged cellular organs or cells. The same conclusion was also reported in HeLa cells, which underwent morphological differentiation after treatment with butyrate, and increased lysosomal enzyme activity (Kelly, 1985). In addition, focal adhesions and adherens junctions are associated with cell migration (Cohen et al., 2013). Inflammatory caspases can regulate cell migration through actin remodeling (Papoff et al., 2018). This study showed thatβ-actin and α-integrins were downregulated in the BH group, integrins can participate in phagocytosis and attachment of hemocytes (Terahara et al., 2006; Lv et al., 2020). Thus, lysosomal signaling pathways, Cell repair and cell migration may be a potential way in which butyrate regulates immunity in pearl oyster after transplantation.
Eukaryotic cells respond to immune stimulation by activating signaling pathways that promote cell cycle arrest and DNA repair. Previous studies have found that the expression of genes related to “cell cycle”, “DNA replication” and “DNA mismatch repair” is up-regulated after transplantation (Jiao et al., 2019). Our data also showed that these DEGs were significantly enriched in cell proliferation-related pathways, while most genes in the pathway were downregulated in the BH group after transplantation 12 h. It was shown that the cell cycle of HEK293, Hela and MCF-7 cell was arrested by butyrate, and that cell cycle arrest may be one of the main reasons for the proliferation-inhibiting impact of butyrate (Li et al., 2015). Therefore, butyrate could suppress inflammatory responses by inhibiting hemocytes proliferation after transplantation.
After transplantation 24 h, 1,912 differential genes were observed between the BH and PH group. The results of functional enrichment analysis illustrated that these DEGs were enriched in cytosolic DNA-sensing, NOD-like receptor signaling, NF-κB signaling, and TNF signaling pathway and apoptosis. cGAS can recognize the cytoplasmic and exogenous DNA fragments (e.g., viruses, bacteria, and parasites) and its own damaged DNA (e.g., damaged mitochondrial and micronuclear DNA (Mackenzie et al., 2017). NLRP3 and NLRC4 in the NOD-like receptor signaling pathway are also intracellular pattern recognition receptors. They can recognize various foreign pathogens (e.g., bacteria, viruses, and fungi) and can be activated by their own “danger signals”. For example, ATP and urate are released when host cells are damaged and necrotic (Jin and Flavell, 2010); In the present study, cGAS, NLRP3, and NLRC4 are upregulated with butyrate treatment before transplantation, suggesting that the activation of these intracellular recognition receptors may be caused by butyrate-induced cell damage. As an anti-inflammatory factor, TRAF3 participates in the process of cellular immune regulation and can inhibit the NF-κB signaling pathway by continuously mediating the degradation of NIK (Huang et al., 2012) and specifically block NF-κB activation by TRAF2/5 (Hauer et al., 2005). NF-κB, as a protein transcription factor, coordinates many inflammatory processes. IκBα, is an important inhibitor of NF-κB activity, which is encoded by NFKBIA (Ali et al., 2013). In the present study, TNF and NF-κB-signaling-pathway-related genes were upregulated with butyrate treatment before transplantation, while most of DEG genes in these pathways were apoptosis-suppressing and anti-inflammatory genes. We analyzed the expression of inflammatory factor IL17 and anti-inflammatory factors TRAF3, NFKBIA and found that the expression of IL17 was mostly decreased and the expression of TRAF3 and NFKBIA were significantly upregulated with butyrate treatment before transplantation, suggesting that butyrate could inhibit the inflammatory response. Chakravortty et al. (2000) found that butyrate prevented NF-κB activation by stabilizing IkappaB-α and IkappaB-β and inhibited the formation of nitric oxide induced by LPS in RAW 264.7 cells. These results support our data about the inhibition of NF-κB signal by butyrate. Moreover, the apoptosis pathway was also significantly enriched in the study, but in which a large number of apoptosis-suppressing genes were induced. And no significant difference was observed in the apoptotic cells between BH and PH group after transplantation for 24 h by apoptosis detection. This phenomenon indicated that butyrate induced the large-scale expansion of apoptosis-suppressing genes in P. f. martensii, thus inhibiting the occurrence of apoptosis. After transplantation for 48 h, the enriched signaling pathways were similar to those at 24 h, but the proportion of induced genes decreased, indicating that the impact of butyrate was gradually decreasing. Therefore, the cytosolic DNA-sensing, NOD-like receptor signaling, NF-κB signaling, and TNF signaling pathway and apoptosis may be the key pathways for the butyrate to regulate the excessive inflammatory response induced by transplantation.
To further explore the function of butyrate, we analyzed the effect of butyrate on the oxidative stress response. Results showed that the content of MDA was significantly increased. It was found that butyrate treatment resulted in an increase in ROS levels, we proposed that the increased MDA levels may be a result of the increasing ROS induced by butyrate. Yuksel et al. (2022) also found an up-regulation of MDA at 48 hours after butyrate treatment. To investigate whether butyrate exerts an antioxidant effect, we analyzed the activities of enzymes such as SOD, GSH-Px and CAT, which are important enzymes that resist intracellular ROS damage, boost the defense system and protect cells from oxidative damage (Jing et al., 2007; Camini et al., 2016). Previous studies have shown that the addition of butyrate to different fish (Abdel-Latif et al., 2020) and Procambarus clarkii (Xiao et al., 2021) diets increased the activity of antioxidant enzymes SOD, CAT and GSH-Px. Li et al. (2021) also found that butyrate increased antioxidant enzyme activities (CAT, SOD and GSH-PX) in the brain. This was consistent with the results of the present study, and the activities of antioxidant and immune-related enzymes were significantly increased after butyrate treatment, suggesting that butyrate may modulate the oxidative stress response of pearl oyster.
This study provides an explanation for the molecular mechanism of pearl oyster immune response after butyrate stimulation. Butyrate can increase the acetylation level of H3 and regulate gene expression. The transcriptomic of the hemocytes analysis revealed that butyrate can activate the lysosome pathway and modulate transplantation-induced inflammatory responses by regulating cell proliferation in pearl oysters. “Cytosolic DNA-sensing pathway”, “NOD-like receptor signaling pathway”, “Apoptosis”, “NF-kappa B signaling pathway” and “TNF signaling pathway” may be the key pathways for butyrate to exert its regulation impacts. Moreover, butyrate treatment can enhance the activity of some immune related enzyme in the serum after transplantation. Therefore, butyrate treatment can regulate the immune response of pearl oyster and activate humoral immunity.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: CNGB, CNP0003692.
YJ and YD: Conceptualization, Methodology, Writing-Reviewing and Editing. JY: Data curation, Writing- Original draft preparation. SY and YL: Sample Preparation. All authors contributed to the article and approved the submitted version.
This work was supported by Science and Technology Program of Guangdong Province (Grant No. 2021A1515011199 and 2022A1515010030), and Department of Education of Guangdong Province (Grant No. 2021KTSCX041, 2020ZDZX1045 and 2021KCXTD026), Department of Agriculture and Rurual Affairs of Guangdong Province (Grant No. 2022KJ146), and China Agriculture Research System of MOF and MARA, and Graduate Education Innovation Program of Guangdong Ocean University (Grant no. 202141).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1073322/full#supplementary-material
Supplementary Figure 1 | Analysis of DEGs between PH and BH group after transplantation 48 h. (A). Volcano map of DEGs between PH and BH groups after transplantation 48 h. Impacted pathway (B) and GO (C) of butyrate on the genes regulated by transplantation. Expression of the related genes in the Cytosolic DNA-sensing pathway (D), Apoptosis (E), NOD-like receptor signaling pathway (F), NF-kappa B signaling pathway (G), TNF signaling pathway (H) in PH and BH groups after transplantation 48 h.
Supplementary Figure 2 | Expression of response to stimulus-related genes and antioxidantactivity-related genes at difference times in the transcriptomes of PHand BH group. Expression of CAT (A), ALP (B), LYS (C) and GSHPx (D) at difference times after transplantation. Blue is the PH groupand red represents the BH group.
Abdel-Latif H. M. R., Abdel-Tawwab M., Dawood M. A. O., Menanteau-Ledouble S., El-Matbouli M. (2020). Benefits of dietary butyric acid, sodium butyrate, and their protected forms in aquafeeds: A review. Rev. Fish. Sci. 28 (4), 421–448. doi: 10.1080/23308249.2020.1758899
Adcock I. M. (2007). HDAC inhibitors as anti-inflammatory agents. Br. J. Pharmacol. 150, 829–831. doi: 10.1038/sj.bjp.0707166
Ali S., Hirschfeld A. F., Mayer M. L., Fortuno E. S., Corbett N., Kaplan M., et al. (2013). Functional genetic variation in NFKBIA and susceptibility to childhood asthma, bronchiolitis, and bronchopulmonary dysplasia. J. Immunol. 190 (8), 3949–3958. doi: 10.4049/jimmunol.1201015
Archer S. Y., Hodin R. A. (1999). Histone acetylation and cancer. Curr. Opin. Genet. Dev. 9, 171–174. doi: 10.1016/S0959-437X(99)80026-4
Bai L., Pang W. J., Yang Y. J., Yang G. S. (2008). Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol. Cell. Biochem. 307 (1-2), 129–140. doi: 10.1007/s11010-007-9592-5
Camini F. C., Caetano C. C. D., Almeida L. T., Magalhaes C. L. D. (2016). Implications of oxidative stress on viral pathogenesis. Arch. Virol. 162 (4), 907–917. doi: 10.1007/s00705-016-3187-y
Cao Y. F., Jiao Y., Zheng C., Zhan S. Z., Guo Z. F., Xiong X. W., et al. (2021). Transcriptome analysis provides insights into the function of the cholinergic system in pearl oyster transplantation immunity. Aquaculture 545, 737205. doi: 10.1016/j.aquaculture.2021.737205
Chakravortty D., Koide N., Kato Y., Sugiyama T., Mu M. M., Yoshida T., et al. (2000). The inhibitory action of butyrate on lipopolysaccharide-induced nitric oxide production in raw 264.7 murine macrophage cells. J. Endotoxin Res. 6 (3), 243–247. doi: 10.1179/096805100101532108
Chatterjee N., Sinha D., Lemma-Dechassa M., Tan S., Shogren-Knaak M. A., Bartholomew B. (2011). Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 39 (19), 8378–8391. doi: 10.1093/nar/gkr535
Chuang M. J., Wu S. T., Tang S. H., Lai X. M., Lai H. C., Hsu K. H., et al. (2013). The HDAC inhibitor LBH589 induces ERK-dependent prometaphase arrest in prostate cancer via HDAC6 inactivation and down-regulation. PloS One 8 (9), e73401. doi: 10.1371/journal.pone.0073401
Cohen D. M., Yang M. K., Chen C. S. (2013). Measuring cell–cell tugging forces using bowtie-patterned mPADs (Microarray post detectors). Methods Mol. Biol. 1066, 157–168. doi: 10.1007/978-1-62703-604-7_14
De Zoeten E. F., Wang L. Q., Butler K., Beier U. H., Akimova T., Sai H., et al. (2011). Histone deacetylase 6 and heat shock protein 90 control the functions of Foxp3+ T-regulatory cells. Mol. Cell. Biol. 31 (10), 2066–2078. doi: 10.1128/MCB.05155-11
Dou X. J., Yan D., Ma Z. W., Gao N., Shan A. S. (2022). Sodium butyrate alleviates LPS-induced kidney injury via inhibiting TLR2/4 to regulate rBD2 expression. J. Food. Biochem. 46 (7), e14126. doi: 10.1111/jfbc.14126
Encarncao J. C., Pires A. S., Amaral R. A., Goncalves T. J., Laranjo M., Casalta-Lopes J. E., et al. (2018). Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J. Nutr. Biochem. 56, 183–192. doi: 10.1016/j.jnutbio.2018.02.018
Han M. K., Mantel C., Guo Y., Tao W., Broxtreyer H. E. (2005). Role of SIRT1 in proliferation and self-renewal of mouse embryonic stem cells. Blood 106 (11), 520. doi: 10.1182/blood.V106.11.520.520
Hauer J., Puschner S., Ramakrishnan P., Simon U., Bongers M., Federle C., et al. (2005). TNF receptor (TNFR)-associated factor (TRAF) 3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-κB pathway by TRAF-binding TNFRs. Proc. Natl. Acad. Sci. U.S.A. 102 (8), 2874–2879. doi: 10.1073/pnas.0500187102
Huang X. D., Liu W. G., Guan Y. Y., Shi Y., Wang Q., Zhao M., et al. (2012). Molecular cloning, characterization and expression analysis of tumor necrosis factor receptor-associated factor 3 (TRAF3) from pearl oyster Pinctada fucata. Fish Shellf. Immunol. 33 (3), 652–658. doi: 10.1016/j.fsi.2012.06.026
Jazirehi A. R. (2010). Regulation of apoptosis-associated genes by histone deacetylase inhibitors: Implications in cancer therapy. Anti-Cancer Drugs 21 (9), 805–813. doi: 10.1097/CAD.0b013e32833dad91
Jiang S. S., Ma Y. Y., Li Y. T., Liu R. H., Zeng M. Y. (2021). Mediation of the microbiome-gut axis by oyster (Crassostrea gigas) polysaccharides: A possible protective role in alcoholic liver injury. Int. J. Biol. Macromol. 182, 968–976. doi: 10.1016/j.ijbiomac.2021.04.050
Jiao Y., Yang S., Cao Y. F., Zheng Z., Deng Y. W., Wang Q. H. (2019). Genome and transcriptome analyses providing insight into the immune response of pearl oysters after allograft and xenograft transplantations. Fish Shellf. Immunol. 90, 109–117. doi: 10.1016/j.fsi.2019.04.061
Jin C. C., Flavell R. A. (2010). Molecular mechanism of NLRP3 inflammasome activation. J. Clin. Immunol. 30 (5), 628–631. doi: 10.1007/s10875-010-9440-3
Jing G., Li Y., Xie L., Zhang R. Q. (2007). Different effects of Pb2+ and Cu2+ on immune and antioxidant enzyme activities in the mantle of Pinctada fucata. environ. Toxicol. Pharmacol. 24 (2), 122–128. doi: 10.1016/j.etap.2007.04.002
Kelly R. E. (1985). Sodium butyrate induced alterations in lysosomal enzyme activity. In Vitro Cell. Dev. Biol. 21 (7), 373–381. doi: 10.1007/BF02623468
Kim H. J., Rowe M., Ren M., Hong J. S., Chen P. S., Chuang D. M. (2007). Histone deacetylase inhibitors exhibit anti-inflammatory and neuroprotective effects in a rat permanent ischemic model of stroke: Multiple mechanisms of action. J. Pharmacol. Exp. Ther. 321 (3), 892–901. doi: 10.1124/jpet.107.120188
Lee C., Kim B. G., Kim J. H., Chun J., Im J. P., Kim J. S. (2017). Sodium butyrate inhibits the NF-kappa b signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int. Immunopharmacol. 51, 47–56. doi: 10.1016/j.intimp.2017.07.023
Li D., Bai X., Jiang Y., Cheng Y. (2021). Butyrate alleviates PTZ-induced mitochondrial dysfunction, oxidative stress and neuron apoptosis in mice via Keap1/Nrf2/HO-1 pathway. Brain Res. Bull. 168, 25–35. doi: 10.1016/j.brainresbull.2020.12.009
Li L. P., Sun Y. X., Liu J. Q., Wu X. D., Chen L. J., Ma L., et al. (2015). Histone deacetylase inhibitor sodium butyrate suppresses DNA double strand break repair induced by etoposide more effectively in MCF-7 cells than in HEK293 cells. BMC Biochem. 16. doi: 10.1186/s12858-014-0030-5
Li S. S., Zhou M., Ze K., Sun X. Y., Zhao C. M., Li Z. R., et al. (2020). Protective role of histone deacetylase 4 from ultraviolet radiation-induced DNA lesions. Mol. Carcinog. 59 (11), 1292–1301. doi: 10.1002/mc.23257
Lv Z., Qiu L. M., Wang W. L., Liu Z. Q., Liu Q., Wang L. L., et al. (2020). The members of the highly diverse crassostrea gigas integrin family cooperate for the generation of various immune responses. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01420
Mackenzie K. J., Carroll P., Martin C. A., Murina O., Fluteau A., Impson D. J. S., et al. (2017). cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548 (7668), 461–465. doi: 10.1038/nature23449
Mocquard-Bucher E., Galvani A., Thiriet C. (2014). Histone H4 acetylation links nucleosome turnover and nucleosome assembly: Lessons from the slime mold Physarum polycephalum. front. Life Sci. 7, 12–21. doi: 10.1080/21553769.2013.848241
Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., et al. (2004). Mammalian SIRT1 represses forkhead transcription factors. Cell 116 (4), 551–563. doi: 10.1016/S0092-8674(04)00126-6
Nancy S., Bienvenu J., Coffin B., Andre F., Descos L., Flourie B., et al. (2002). Butyrate strongly inhibits In vitro stimulated release of cytokines in blood. Dig. Dis. Sci. 47 (4), 921–928. doi: 10.1023/A:1014781109498
Pant K., Yadav A. K., Gupta P., Islam R., Jain A., Venugopal S. K. (2016). Butyrate induces ros-mediated apoptosis via modulation of mir-22/sirt-1 pathway in hepatoma cells. Hepatology 64.
Papoff G., Presutti D., Lalli C., Bolasco G., Santini S., Manelfi C., et al. (2018). CASP4 gene silencing in epithelial cancer cells leads to impairment of cell migration, cell-matrix adhesion and tissue invasion. Sci. Rep. 8. doi: 10.1038/s41598-018-35792-8
Ran J., Zhou J. (2019). Targeted inhibition of histone deacetylase 6 in inflammatory diseases. Thorac. Cancer 10 (3), 405–412. doi: 10.1111/1759-7714.12974
Roth S. Y., Allis C. D. (1996). Histone acetylation and chromatin assembly: A single escort, multiple dances? Cell 87 (1), 5–8. doi: 10.1016/S0092-8674(00)81316-1
Semann M. D., Bohmig G. A., Osterreicher C. H., Burtscher H., Parolini O., Diakos C., et al. (2000). Anti-inflammatory effects of sodium butyrate on human monocytes: Potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 14 (15), 2380–2382. doi: 10.1096/fj.00-0359fje
Settembre C., Fraldi A., Medina D. L., Ballabio A. (2013). Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell. Biol. 14 (5), 283–296. doi: 10.1038/nrm3565
Terahara K., Takahashi K. G., Nakamura A., Osada M., Yoda M., Hiroi T., et al. (2006). Differences in integrin-dependent phagocytosis among three hemocyte subpopulations of the pacific oyster “Crassostrea gigas”. DCI 30 (8), 667–683. doi: 10.1016/j.dci.2005.09.009
Turner B. M. (1991). Histone acetylation and control of gene expression. J. Cell Sci. 99 (1), 13–20. doi: 10.1242/jcs.99.1.13
Villagra A., Cheng F., Wang H. W., Suarez I., Glozak M., Maurin M., et al. (2008). The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 10 (1), 92–100. doi: 10.1038/ni.1673
Waby J. S., Chirakkal H., Yu C. W., Griffiths G. J., Benson R. S. P., Bingle C. D., et al. (2010). Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol. Cancer 9 (1), 275. doi: 10.1186/1476-4598-9-275
Wang W., Meng Z. Q., Shi F. W. (2012). Modification and biological role of histone. Hereditas 34, 19–27.
Wang W., Wu Y. Y., Lei Q. N., Liang H. Y., Deng Y. W. (2019). Deep transcriptome profiling sheds light on key players in nucleus implantation induced immune response in the pearl oyster Pinctada martensii. Fish Shellf. Immunol. 69, 67–77. doi: 10.1016/j.fsi.2017.08.011
Wang Z. B., Zang C. Z., Cui K. R., Schones D. E., Barski A., Peng W. Q., et al. (2009). Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138 (5), 1019–1031. doi: 10.1016/j.cell.2009.06.049
Weber T. E., Kerr B. J. (2006). Butyrate differentially regulates cytokines and proliferation in porcine peripheral blood mononuclear cells. Vet. Immunol. Immunopathol. 113 (1-2), 139–147. doi: 10.1016/j.vetimm.2006.04.006
Xiao C. Y., Zhang Y. F., Zhu F. (2021). Effect of dietary sodium butyrate on the innate immune response of Procambarus clarkii and disease resistance against white spot syndrome virus. Aquaculture 541. doi: 10.1016/j.aquaculture.2021.736784
Yang J. M., Luo S. J., Li J. H., Zheng Z., Du X. D., Deng Y. W. (2018). Transcriptome analysis of growth heterosis in pearl oyster Pinctada fucata martensii. FEBS Open Bio. 8 (11), 1794–1803. doi: 10.1002/2211-5463.12502
Yuksel B., Ozkan A. D., Aydn D., Betts Z. (2022). Evaluation of the antioxidative and genotoxic effects of sodium butyrate on breast cancer cells. Saudi J. Biol. Sci. 29 (3), 1394–1401. doi: 10.1016/j.sjbs.2021.12.061
Zhang W., Bone J. R., Edmondson D. G., Turner B. M., Roth S. Y. (1998). Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17 (11), 3155–3167. doi: 10.1093/emboj/17.11.3155
Zhang H., Dai X. F., Qi Y., He Y., Du W., Pang J. J., et al. (2015). Histone deacetylases inhibitors in the treatment of retinal degenerative diseases: Overview and perspectives. J. Ophthalmol. 2015, 1–9. doi: 10.1155/2015/250812
Keywords: Pinctada fucata martensii, histone acetylation, transplantation, inflammatory response, butyrate
Citation: Yang JM, Yang S, Liao YS, Deng YW and Jiao Y (2023) Increased histone H3 acetylation inhibit the inflammatory response and activate the serum immunity of Pearl oyster Pinctada fucata martensii. Front. Mar. Sci. 10:1073322. doi: 10.3389/fmars.2023.1073322
Received: 18 October 2022; Accepted: 31 January 2023;
Published: 13 February 2023.
Edited by:
Daniel Carneiro Moreira, University of Brasilia, BrazilReviewed by:
Ngoc Tuan Tran, Shantou University, ChinaCopyright © 2023 Yang, Yang, Liao, Deng and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Jiao, amlhb3lAZ2RvdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.