- 1Department of Marine Microbiology and Biogeochemistry, NIOZ Royal Netherlands Institute for Sea Research, Texel, Netherlands
- 2Department of Energy, Environment and Climate Change, Asian Institute of Technology, Pathum Thani, Thailand
- 3Centre for Rural Development and Technology, Indian Institute of Technology, Delhi, India
- 4Biological Oceanography Group, University of Vigo, Vigo, Spain
- 5Department of Earth Sciences, Faculty of Geosciences, Utrecht University, Utrecht, Netherlands
Advancements in chemical, medical, cosmetic, and plastic producing industries have improved agricultural yields, health and human life in general. As a negative consequence, a plethora of chemicals are intentionally and unintentionally released to terrestrial and aquatic environments with sometimes devastating effects for entire ecosystems. One mitigation strategy to counteract this pollution is bioremediation. Bioremediation is an umbrella term for biologically mediated processes during which an undesired compound is transformed, degraded, sequestered and/or entirely removed from the ecosystem. Organisms across all domains of life may mediate bioremediation; yet, fungi are particularly promising candidates. They possess metabolic capabilities to break down complex molecules which make fungi the ultimate degraders of recalcitrant organic matter in nature. Bioremediation by fungi, also termed mycoremediation, has been more frequently investigated in terrestrial than aquatic ecosystems, although fungi also thrive in lacustrine and marine environments. Here, we focus on mycoremediation of emerging pollutants in aquatic environments. In this context, we draw parallels between terrestrial and aquatic fungal taxa, and their role in mycoremediation. We discuss the ability of fungi to break-down (i) pesticides, (ii) pharmaceuticals and personal care products, (iii) plastics, both conventional types and (iv) bioplastics, and fungal role, (v) mitigation of heavy metal pollution. Furthermore, we (vi) discuss possible mycoremediation strategies in applied settings and highlight novel enzyme based mycoremediation strategies.
1 Introduction
Emerging pollutants stem from large-scale manufacturing, use and application of pesticides, pharmaceuticals and personal care products (PPCPs), plastic polymers, and heavy metals. These compounds are designed to satisfy the ever-growing need for a better life, health and ways to ensure food and material supply for a growing human population. However, their production and application are often accompanied by substantial waste generation. This is not paralleled by mitigation strategies. Through transport in groundwater, rivers, air and ocean circulation, these often recalcitrant products/compounds have reached and contaminated virtually all ecosystems on Earth (Lebreton et al., 2017; Pujari and Kapoor, 2021; Roy et al., 2022). For example, organic pesticides have been detected in deep sea sediments of the Pacific Ocean (Ge et al., 2021). Pharmaceuticals and personal care products are frequently detected in waste water treatment systems (Lishman et al., 2006; Oulton et al., 2010) as well as in marine ecosystems (Bayen et al., 2016; Ojemaye and Petrik, 2022). Microplastics have been found in remote places such as the sea ice of Antarctica (Kelly et al., 2020) and the deepest parts of the marine realm, the Mariana Trench (Peng et al., 2020) and nanoplastics are seemingly widespread, too (Ter Halle et al., 2017; Materić et al., 2022).
In nature, these emerging pollutants cause adverse effects, ranging from impacts on the organism to the ecosystem level. Well-known examples include bioaccumulation of methylmercury in shellfish, causing poisoning in humans upon consumption (Sunderland, 2007), ingestion and entanglement of marine organisms in plastic litter (Gregory, 2009; Kurtela and Antolović, 2019), increase of antimicrobial resistance genes due to excess use of antibiotics and their inadequate disposal (Allen et al., 2010) and, finally, adverse ecological consequences due to the large scale use of the pesticide dichlorodiphenyltrichloroethane (DDT) before its ban in many countries (Mansouri et al., 2017). In order to mitigate the increasing amounts and effects of these contaminants, biological processes can be utilized to break down, transform and remove hazardous pollutants from the environment - these processes have been coined ‘bioremediation’. A bioremediation strategy was firstly applied by George M. Robinson in the 1960s by using microbes to mitigate the effects of oil spills. Bioremediation mostly relies on plants (phytoremediation) or microorganisms (microremediation) to degrade pollutants. Prominent examples for bioremediation in aquatic environments are the use of microalgae that accumulate heavy metals (Aksu, 1998; Zhang et al., 2019; Leong and Chang, 2020) or pharmaceuticals (Silva et al., 2019; Chandel et al., 2022). Microbial bioremediation efforts benefit from the large natural diversity of microorganisms featuring a broad spectrum of pathways to metabolise or co-metabolise a wide range of compounds. Furthermore, the adaptation of microorganisms to novel compounds is comparably high: the typically short generation time of microbes is accompanied by high rates of evolutionary adaptation. Microbes may even utilize otherwise hazardous compounds for energy gain or as cellular building blocks. Furthermore, with the aid of genetic manipulation, microbial metabolisms can be “improved” or “designed” to target specific pollutants. With the aid of synthetic biology tools, microbial communities can even be assembled to target specific pollution scenarios (Jaiswal et al., 2019; Borchert et al., 2021; Xiang et al., 2021). Bioremediation strategies are either in-situ, i.e., pollution is treated at the location where it occurs or ex-situ, where the polluted matrix is removed and treatment is carried out elsewhere. Most common examples of microbial bioremediation are cleaning up industrial spills, such as oil spills (Biswas et al., 2022). For oil spills, both bioaugmentation (i.e., the application of microbes to the pollution site) and biostimulation (i.e., stimulation of the natural microbial community to perform faster and more efficiently) are used (Sharma et al., 2020). Further examples include treatment of wastewater effluents in wastewater treatment plants; e.g., to clean up textile dye industry waste (Ihsanullah et al., 2020), or cleaning up environmental metal, such as cadmium (Cd), mercury (Hg) and arsenic (As) pollution (Verma and Kuila, 2019).
2 Fungi as bioremediating agents
Fungi are organisms gaining increasing attention because of their broad potential for bioremediation applications (Supplementary Figure 1). Fungi are a diverse kingdom in the tree of life, belonging to the domain eukarya. Nevertheless, fungal taxonomy is a matter of debate and discussed controversially. The most recent taxonomical update describes nineteen major phyla: Aphelidiomycota, Ascomycota, Basidiobolomycota, Basidiomycota, Blastocladiomycota, Calcarisporiellomycota, Caulochytriomycota, Chytridiomycota, Entomophthoromycota, Entorrhizomycota, Glomeromycota, Kickxellomycota, Monoblepharomycota, Mortierellomycota, Mucoromycota, Neocallimastigomycota, Olpidiomycota, Rozellomycota, and Zoopagomycota (Wijayawardene et al., 2020). The number of described species is only ∼1 million (Wu et al., 2019), but the total number of fungal species is estimated to range between 2 to 4 million (Hawksworth and Lücking, 2017). Fungi are ubiquitous chemoheterotrophic organisms, prevalent throughout terrestrial and aquatic environments. Nevertheless fungi have been studied in terrestrial environments more extensively, while they have gained less attention in aquatic, and particularly in marine environments (Zeghal et al., 2021). The vast majority of known fungi are aerobic and thus inhabit oxic environments; yet anaerobic fungi have been found in ocean oxygen minimum zones or digestive tracts of animals (Stief et al., 2014; Mura et al., 2018; Peng and Valentine, 2021). Fungi are known as the ultimate degraders of complex organic matter, involved in decay processes and known to degrade wood including lignin and cellulose and other plant-based materials, which are common waste products in agriculture (Dinis et al., 2009; Janusz et al., 2017; Goodell et al., 2020). Due to their intra and extracellular enzymatic machinery and their ability to excrete acids, fungi are able to attack and metabolize a wide range of compound classes comprising inorganic and organic pollutants (Dashtban et al., 2010; Harms et al., 2011; Deshmukh et al., 2016). Fungal enzymes are often characterised by a low substrate specificity (El-Gendi et al., 2021). Counterintuitively, this provides an advantage, because the non-specific enzymes catalyse a broad range of reactions enabling fungi to use also a wide range of compounds as carbon and energy source (Harms et al., 2011). The biodegradation capacities of fungi are almost universally linked to oxidative enzymatic reactions mediated by a diverse set of oxidases and peroxidases (Hofrichter, 2002). Hence, fungi typically require an oxic environment for their function.
The enzymatic capabilities of fungi are well classified into intra- and extra-cellular mechanisms (El-Gendi et al., 2021). The most widely studied extracellular enzymes from fungi are different oxidoreductases thus remove enzymes are after the word different, specifically laccases, which use oxygen and peroxidases (manganese peroxidase, lignin peroxidase), which use H2O2 as terminal electron acceptor (Baldrian, 2006; Hofrichter et al., 2010; Rodgers et al., 2010; El-Gendi et al., 2021; Urlacher and Koschorreck, 2021). Among the most recognized pollutant degrading extracellular reactions are oxidations with hydroxyl radicals cleaving double bonds in cyclic or aliphatic structures (Hofrichter et al., 2010). The hydroxyl radicals stem from quinone cycling after the action of laccases (Gómez-Toribio et al., 2009), or peroxide-dependent hydroxylations. Both processes are exergonic in nature and these reactions facilitate the degradation of complex pollutant structures into more easily degradable metabolite intermediates. Intracellularly, cytochromes catalyze a broad variety of oxidation reactions inside the cells (Figure 1 and Table 1), with some species such as Phanerochaete chrysosporium containing 150 Cytochrome P450 genes in its genome (Ning et al., 2010).
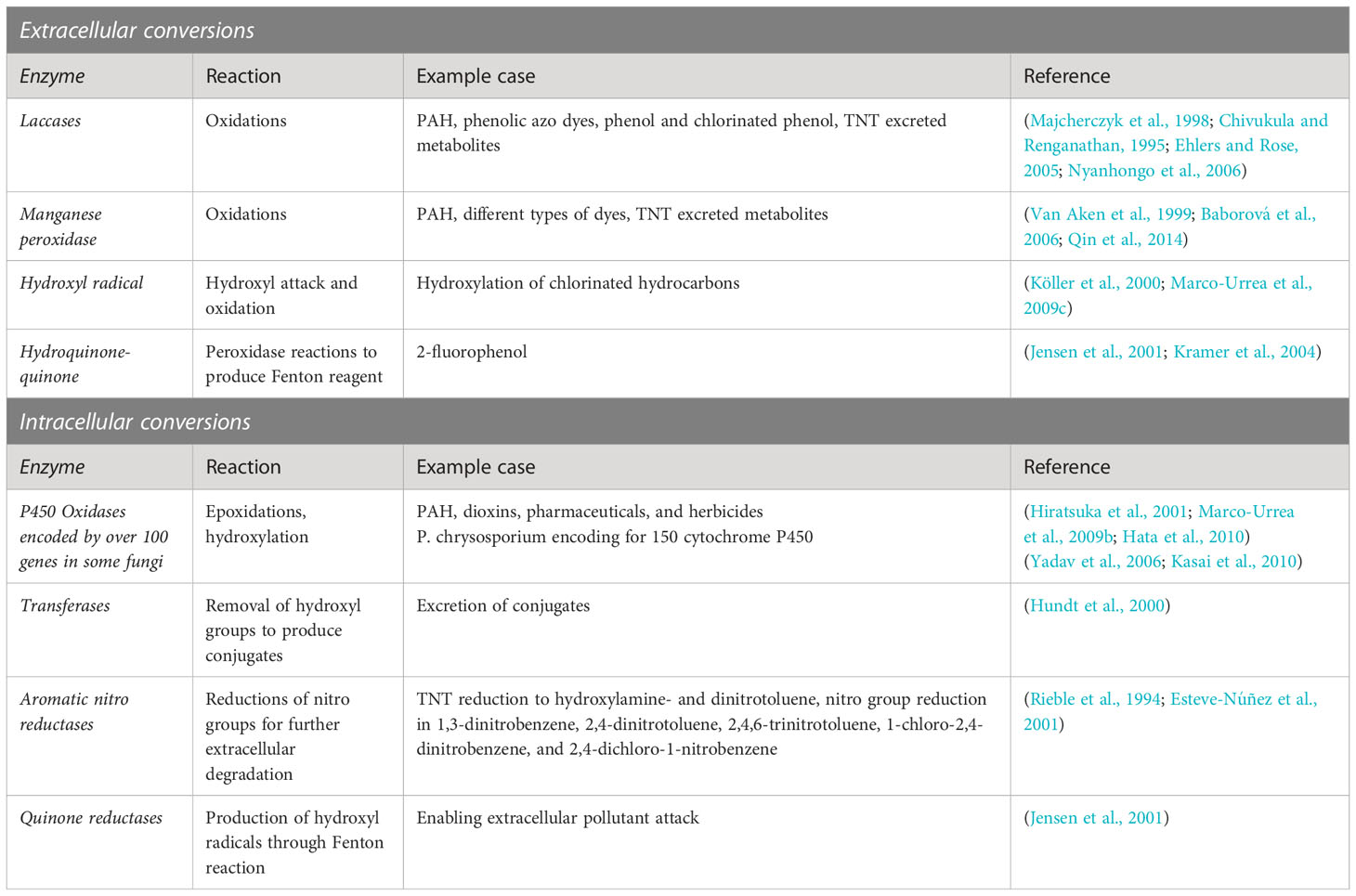
Table 1 Summary of extracellular and intracellular mechanisms, only most general mechanisms are exemplified with the most common reactions and case studies.
The potential of fungi in bioremediation processes is for example highlighted by their ability to degrade petroleum hydrocarbons, comprising alkanes, aromatic and nitrogen-sulfur-oxygen-containing compounds (Adenipekun and Lawal, 2012; Al-Hawash et al., 2018; Miri et al., 2019; Mahmud et al., 2022). Fungi have also been described as suitable bioremediation agents to counteract environmental pollution of toxic metals (Li et al., 2020). Thus, fungi are consequently attractive candidates for biotechnological purposes and industrial bioremediation efforts (Hyde et al., 2019) and have recently been gaining attention as suitable agents for wastewater treatment (Shahid et al., 2021; González-González et al., 2022; Morin-Crini et al., 2022).
This review aims at highlighting the potential of fungi in bioremediation processes. In the following sections, we discuss several emerging pollutant classes and provide an overview of the current literature on the potential of fungi in mitigating the adverse effects these pollutants pose in the environment.
3 Degradation of pesticides
Increased global food demand and the need to prevent global crop losses due to pests have resulted in an extensive annual utilization of about 2 millions tonnes of pesticides worldwide (Sharma et al., 2019; Lykogianni et al., 2021). Pesticides are deliberately applied to kill unwanted plants (weeds), insects, rodents and other living organisms threatening the cultured crops (Matthews, 2015). However, it is postulated that approximately 90% of agricultural pesticides do not reach the intended target organism but instead disperse in the environment (Ortiz-Hernández et al., 2011). Indeed, pesticides have been detected in the atmosphere, soil systems, groundwater, and the ocean (Ernst, 1980; White et al., 2006; Schipper et al., 2008; Wołejko et al., 2020) as a result of high population densities and intensive modern farming practices.
Pesticides such as organochlorines, organophosphates, pyrethroids, and carbamates are associated with detrimental effects including high environmental persistence, bioaccumulation, long-range transmission, and adverse effects to non-target organisms (Kumari et al., 2014; Kumar et al., 2019). Exposure to pesticides can for example cause different types of human cancer, teratogenic and genotoxic defects as well as endocrine and nerve dysfunction (Kim et al., 2017; Shah and Parveen, 2021). Pesticides may cause acute lethal effects, however, some seem to cause more long term health effects and environmental concerns (Pretty and Hine, 2012). For example, the overuse of the insecticide dichlorodiphenyltrichloroethane (DDT) since the 1940s, resulted in adverse ecological consequences, which in some cases only became apparent with a time delay of many years after application (Turusov et al., 2002; Mansouri et al., 2017). Even though the first countries banned DDT in the 1970s, it is still in use in many countries and its derivatives persist in nature for decades (Loganathan and Kannan, 1991). Fungi have been reported to degrade a wide range of different pesticides, including organochlorines, organophosphorus compounds, pyrethroids, and carbamates (Maqbool et al., 2016; Bokade et al., 2021; Bose et al., 2021; Kumar et al., 2021); for example Lindane (Dritsa et al., 2009), Endosulfan (Bhalerao and Puranik, 2007), DDT (Purnomo et al., 2010), Atrazine (Bastos and Magan, 2009; Gonçalves et al., 2012), Dieldrin and Aldrin (Xiao et al., 2011), among others. In some cases, specific fungi are even suggested as candidates to degrade fungicides, such as broad-spectrum pyrazole-carboxamide fluxapyroxad (Podbielska et al., 2020), which is often applied to counteract fungal diseases of apple trees (He et al., 2016). Just as the term ‘pesticide’ encompasses a wider range of different compounds, degradation of these is not confined to one fungal taxon. Known pesticide-degrading fungi belong to the genera Trametes (Bastos and Magan, 2009), Ganoderma (Dritsa et al., 2009), Aspergillus (Bhalerao and Puranik, 2007), Fusarium (Guillén-Jiménez et al., 2012), Pleurotus (Purnomo et al., 2010), Cladosporium, Rhizopus and Penicillium (Gonçalves et al., 2012), Phlebia (Xiao et al., 2011), and Mortiella (Badawi et al., 2009), among many others. These genera belong to different fungal groups such as white rot fungi (WRF), brown rot fungi, filamentous fungi, and yeasts and representative species of all groups have been investigated for the degradation of pesticides.
WRF have been studied extensively due to their ability to degrade wide range of pesticides and structurally highly variable organic pollutants with their non-specific enzymes (Ghosh et al., 2014). WRF possess a variety of oxidative and extracellular ligninolytic enzymes such as lignin peroxidase (LiP), manganese peroxidase (MnP), versatile peroxidase (VP), and laccase (Lac) making them one of the most studied candidates for pesticide bioremediation purposes (Zhuo and Fan, 2021). For example, Trametes versicolor was utilized for the degradation of three different hydrophobic pesticides: chlorpyriphos, dicofol and cypermethrin. These are an organophosphorus, an organochlorine and a synthetic pyrethroid, respectively. Their removal potential at 25°C was 95% (chlorpyriphos), 88% (dicofol) and 93% (cypermethrin) over a time period of 14 days. For all tested pesticides, an initial fast adsorption to fungal pellets was monitored, followed by biodegradation. Based on the identified metabolites, hydrolyzation, dichlorination and oxidation were proposed to be important degradation mechanisms, depending on the pesticide (Hu et al., 2020). T. versicolor has also been shown to remove the herbicides Diuron and Bentazon from agricultural wastewater (Beltrán-Flores et al., 2021). Diuron degradation was also shown for another WRF, Ganoderma lucidum, with Diuron increasing laccase activity (Coelho-Moreira et al., 2017).
In addition to extracellular ligninolytic enzymes, which give fungi access to water-insoluble contaminants, intracellular enzymes such the cytochrome P450 are important in the degradation of pesticides, too (Magan et al., 2010) (Figure 1). T. versicolor degrades also Fipronil, an insecticide with low aqueous solubility belonging to the chemical class of phenylpyrazoles. This was metabolized intracellularly by cytochrome P450 as confirmed by the production of hydroxylated and glycosylated transformation products (Wolfand et al., 2016). This enzyme was also shown to degrade the highly polar pesticides Acetamiprid and Imidacloprid (Hu et al., 2022). P. chrysosporium, with about 150 P450 monooxygenase genes in its genome (Doddapaneni et al., 2005), has been shown to utilize two cytochrome P450 isozymes to degrade 4 neonicotinoids: Acetamiprid, Clothianidin, Imidacloprid, and Thiacloprid (Mori et al., 2021). Furthermore, the degradation of Endosulfan, a chlorinated pesticide, may proceed either via a hydrolytic or oxidative pathway in P. chrysosporium (Kullman and Matsumura, 1996). By using a cytochrome P450 inhibitor, the oxidative metabolism was partially suppressed and rather proceeded via a hydrolytic pathway. The involvement of the P450 system was also shown for the degradation of Lindane (Mougin et al., 1996). For an overview on the application of fungi for pesticide degradation see Table 2, and the reviews of Magan et al. (2010) and Maqbool et al. (2016).
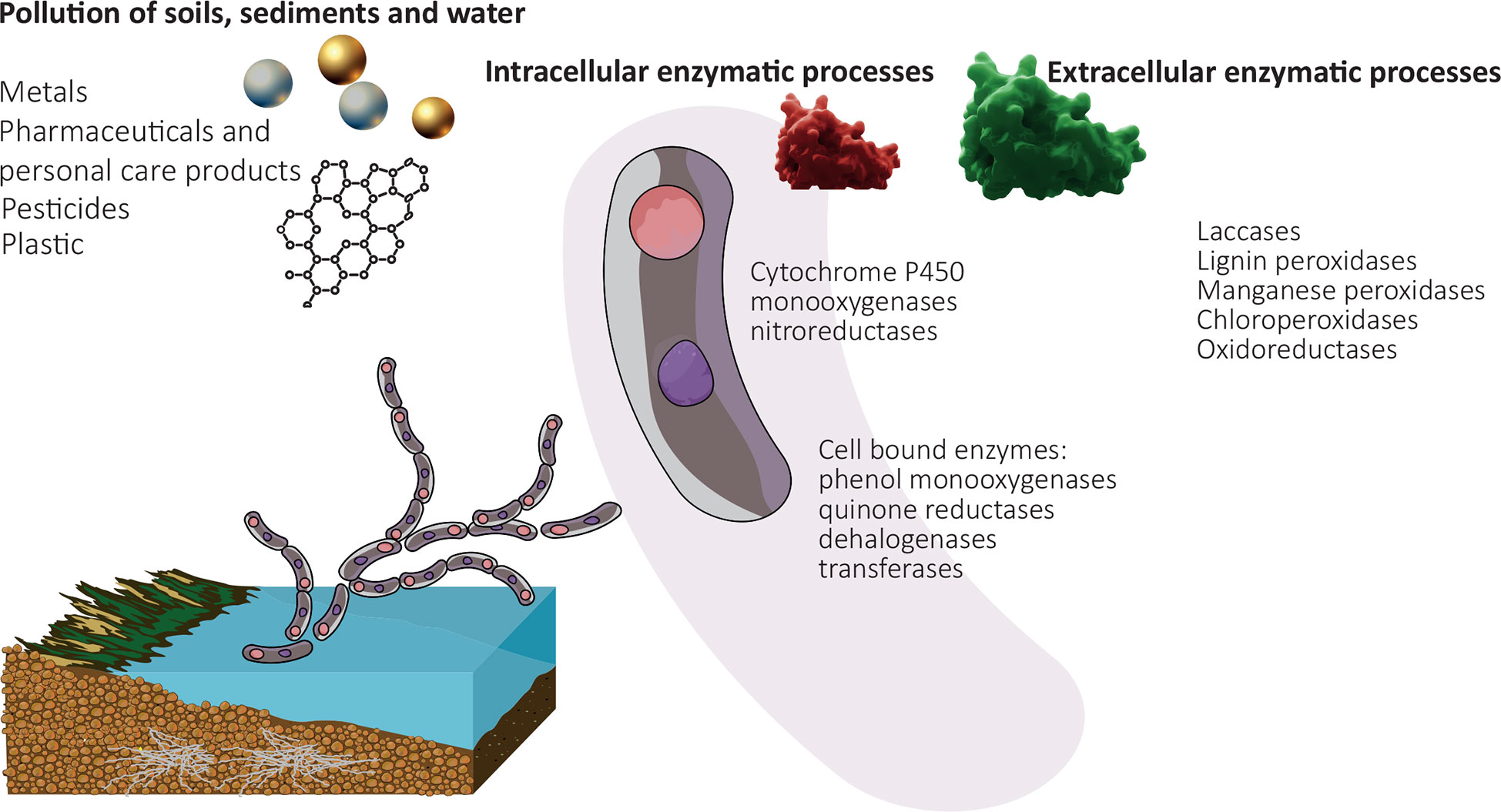
Figure 1 Schematic overview of mycoremediation in the environments. Pollutants and commonly used fungal enzyme classes are highlighted. Pollutants follow intracellular and/or extracellular enzymatic degradation, leading to the production of a diversity of metabolites and/or degradation products.
In addition, it has been shown that a consortium of different fungi, in contrast to single strains, can degrade some pesticides more efficiently. For example, a consortium of 5 fungal isolates, (three from the genus Pleurotus, one clustering with genus Coriolopsis, one unknown) were better suited to degrade Diazinon and methomyl pesticides than single isolates tested (Nyakundi et al., 2011). A similar benefit was found for consortia of fungi (Mortierella LEJ702) and bacteria (Variovorax SRS16 and A. globiformis D47) degrading Diuron (Ellegaard-Jensen et al., 2014). The same Mortierella spp. LEJ702, enhanced the degradation of a herbicide, 2,6-dichlorobenzamide, in a consortium with Aminobacter spp. MSH1 (Knudsen et al., 2013). However, the fungal contribution might not have been to directly degrade the compounds, but that they stimulated bacterial dispersal via hyphae and/or translocated the compound.
The combined application of fungi and certain minerals has in some cases also proven more efficient for pesticide removal than application of fungi or minerals alone. For example, the combined application of the WRF P. chrysosporium together with the boron silicate mineral tourmaline was used for the remediation of agricultural soils contaminated with a variety of organochlorinated pesticides (OCPs) and polycyclic aromatic hydrocarbons (PAHs) (Wang et al., 2014). Synergistic effects amounted to 44% removal of OCPs from the agricultural soils, whereas individual treatment with either P. chrysosporium or tourmaline had lower efficiencies of 34% and 26%, respectively. Tourmaline was suggested to play a role in promoting soil enzyme activities and biodiversity of the active soil microorganisms.
Next to terrestrial fungi, also marine fungi are now slowly gaining attention as potential candidates for the degradation of pesticides. For example, one study investigated seven marine-derived fungal strains of Aspergillus spp. and Penicillium spp. for the removal potential of the insecticide Methyl Parathion (MP). Aspergillus sydowii CBMAI 935 was found to be the most efficient strain removing all MP within 20 days of incubation. The analysis of the metabolites further revealed that the degradation pathway was involved in the formation of the toxic intermediate methyl paraoxon. In a follow-up step, this was fully degraded to p-nitrophenol, reducing toxicity levels by 120-fold (Alvarenga et al., 2014). Furthermore, Aspergillus sydowii CBMAI 935, A. sydowii CBMAI 933, Penicillium miczynskii CBMAI 930 and Trichoderma spp. CBMAI932 have been explored for the degradation of the insecticide Dieldrin (Birolli et al., 2015). P. miczynskii was found to be the most efficient species, degrading 90% of Dieldrin within 14 days. Despite of some of the enzymatic systems being well described, the enzymatic versatility of marine fungi remains vastly unexplored, as recently shown for A. sydowii CBMAI 935 (Soares et al., 2021). This strain expressed new phosphoesterases and methyltransferases suitable for the degradation of chlorpyrifos, methyl parathion and profenofos. These studies are evidence of the potential of marine-derived fungi for bioremediation of pesticides. Harnessing the capabilities of marine fungi could thus further advance mycoremediation strategies.
4 Fungal degradation of pharmaceuticals and personal care products
Pharmaceuticals and personal care products (PPCPs) are an emerging class of contaminants as modern societies are increasingly focused on health, hygiene, and personal care. A substantial fraction of PPCPs ends up in nature, e.g., via sewage streams and wastewater treatment systems (Daughton, 2001; Yang et al., 2017). In fact, there is a lack of regulations regarding use and discharge for many PPCPs (Boxall et al., 2012). This raises concerns in particular related to potentially adverse effects on ecosystems and human health if pharmaceuticals are involved. Pharmaceuticals comprise a vast number of substances and compounds used in healthcare to biochemically or physiologically positively influence the functioning of biological systems, mainly humans and animals. Pharmaceuticals, even though with varying chemical structures are commonly intended to be active and persistent. Pharmaceuticals are transported via hospital and municipal wastewater to wastewater treatment plants (WWTP), which have been identified as the primary source for the increase in antimicrobial resistance genes (Frascaroli et al., 2021). Some of the pharmaceuticals do not alter their chemical composition upon consumption, and not all are removed in WWTPs. In fact, removal efficiency for many of the widely used pharmaceuticals is as low as 10% (Patel et al., 2019). Among the main reasons of concern are their high persistence, water-solubility, bioactivity, bioaccumulation, and toxicity (Brodin et al., 2014; Patel et al., 2019; Kayode-Afolayan et al., 2022). They also trigger antibiotic resistance in bacteria, cause alterations in gene expressions, abnormal protein and enzyme synthesis, and various changes in growth and behaviour of non-target organisms (Brodin et al., 2014; Patel et al., 2019; Kayode-Afolayan et al., 2022). Pharmaceuticals can also be part of personal care products, for example in facial creams or detergents. Furthermore, personal care products may contain a variety of other hazardous compounds. For example, some hair creams contain heavy metals (Ayenimo et al., 2010) (see discussion on heavy metals in section 7) and some scrubs may be based on plastic microbeads (Cheung and Fok, 2017; Lei et al., 2017; Guerranti et al., 2019) (see section 5 and 6 on plastics). Their removal efficiency in communal wastewater is variable (Mohana et al., 2021) as microplastics get adsorbed to sludge which ultimately possess risks upon disposal (Rout et al., 2022; Singh et al., 2022). Even personal care products labelled as ‘natural’ or ‘plant-based’ may contain compounds that are hazardous, which hence does not make them ecologically safe (Klaschka, 2016). Personal care products are widely used, in some western societies by more than >93% of the population (Biesterbos et al., 2013).
Fungi have been shown to break down a range of pharmaceuticals ranging from antibiotics (Rodarte-Morales et al., 2011), anti-inflammatory drugs (Dalecka et al., 2019), anticancer drugs (Jureczko et al., 2021), antidepressants (Rodarte-Morales et al., 2011), diuretics (Al-Aboudi et al., 2017), analgesics (Marco-Urrea et al., 2009b) and beta-blockers (Jaén-Gil et al., 2019). The most studied fungal group able to degrade pharmaceuticals, are WRF. For the degradation of pharmaceuticals, these fungi make often use of the same intracellular and extracellular enzymes that they also use for the degradation of pesticides (see section 3 on pesticide degradation). A variety of fungal strains and their application for the removal of PPCPs are described in detail in Table 3.
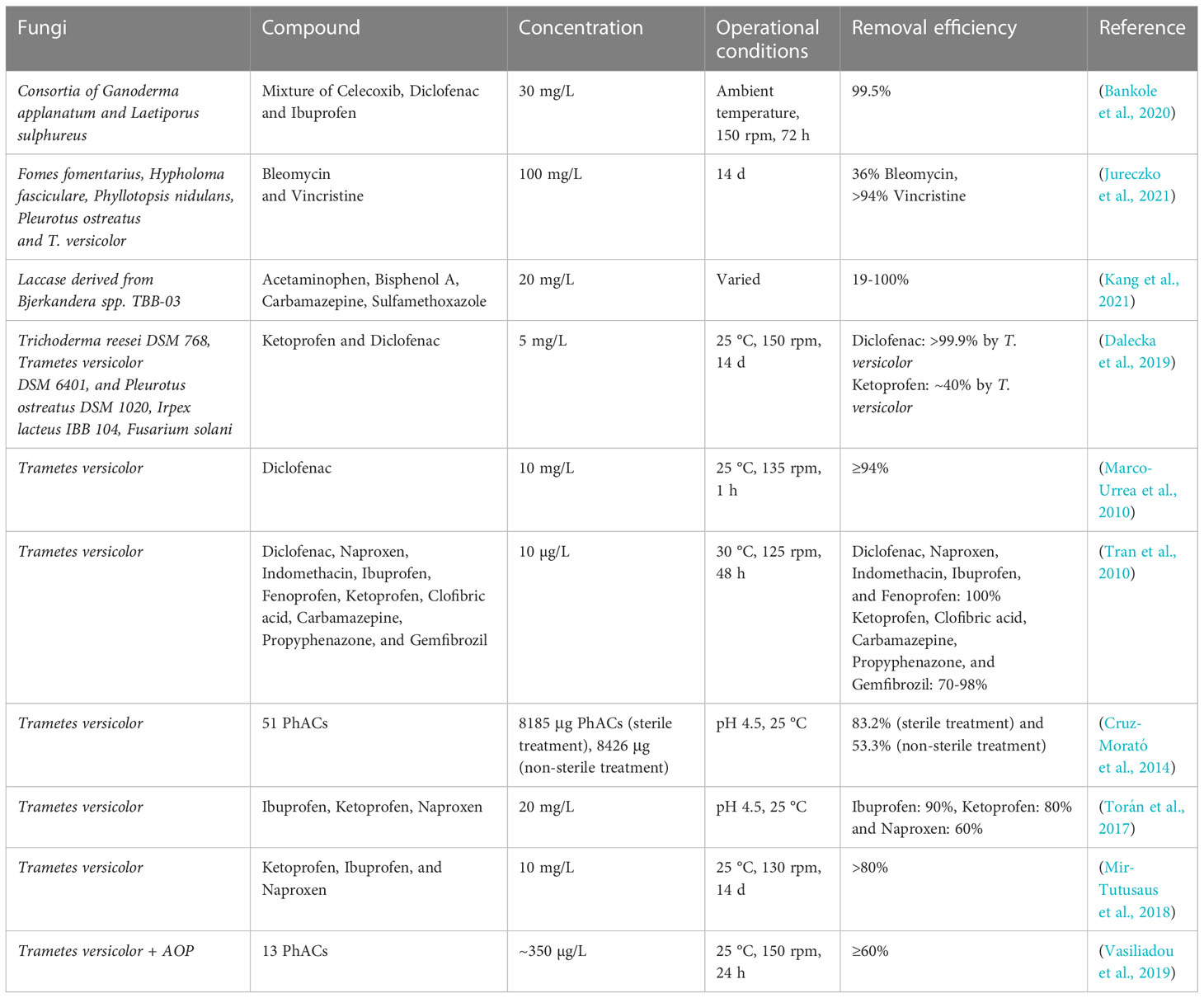
Table 3 Examples of application of fungi for the biodegradation of pharmaceuticals and personal care products.
T. versicolor has been widely investigated for the ability to degrade a variety of pharmaceuticals (Tran et al., 2010). For example, diclofenac, naproxen, indomethacin, ibuprofen, and fenoprofen, while ketoprofen, clofibric acid, propyphenazone, and gemfibrozil were only partially degraded. Lac enzyme was shown to preferentially remove diclofenac, naproxen, indomethacin, and this indicated that intracellular enzymes may be involved in the degradation of other compounds. T. versicolor removed >99.9% and ~40% of diclofenac and ketoprofen, respectively, within 14 days (Dalecka et al., 2019). The removal mechanism was found to be related to the production of laccase along with biosorption. As an interesting finding, a monoculture of T. versicolor was more efficient when compared to mixed cultures containing other fungi, too. This was probably related to competition and growth inhibition. Another study using T. versicolor for diclofenac degradation reported ≥94% removal in just 1 hour (Marco-Urrea et al., 2010). The cytochrome P450 system was found to be the main responsible system in the initial steps of diclofenac degradation and not Lac enzyme, as the time frame of 1 hour would not have enabled measurable laccase activity. Degradation of carbamazepine, ibuprofen and clofibric acid was investigated for T. versicolor, I. lacteus, G. lucidum and P. chrysosporium, showing that all four strains degraded ibuprofen already within 7 days of incubation. However only T. versicolor showed substantial degradation of the more recalcitrant carbamazepine and clofibric acid (Marco-Urrea et al., 2009a).
T. versicolor has been proposed as an efficient fungus for cleaning up hospital wastewater. Hospital wastewater loaded with 51 pharmaceuticals and endocrine disrupting compounds were treated in a bioreactor with complete or partial removal of 46 of these compounds along with significant reduction in toxicity (Cruz-Morató et al., 2014). Analgesic and anti-inflammatory drugs such as ibuprofen, acetaminophen, naproxen, diclofenac, and phenazone present in high concentrations (ranging between 10–100 µg/L) in the hospital effluents were removed by more than 80% within 24 hours. Antibiotic removal rates for sulfamethoxazole, trimethoprim, metronidazole, and its hydroxylated metabolite (dimetridazole), and erythromycin showed large variations ranging from 26-100% compared to the analgesics.
Further examples include the degradation of the anticancer drugs bleomycin and vincristine by Fomes fomentarius, Hypholoma fasciculare and T. versicolor with a high vincristine removal efficiency (>94%) (Jureczko et al., 2021). However, bleomycin was difficult to degrade with only 36% removal by T. versicolor. Probably, bleomycin was toxic at high concentrations. Considering the comparably low concentrations of pharmaceuticals in actual wastewater, WRF could be efficiently applied for their degradation. Bjerkandera spp. TBB-03, has been shown to degrade the pharmaceuticals acetaminophen, carbamazepine, sulfamethoxazole, and the plastic additive bisphenol A (Bilal and Iqbal, 2019). Bisphenol A is a highly estrogenic compound found in high concentrations in WWTP effluents (1000–10,000 μg/L). Oxidative coupling and radical polymerization in acetaminophen and bisphenol A, respectively, were the main degrading mechanisms by Lac enzyme. Complete acetaminophen and bisphenol A removal was reported within 2 hours at 25-40°C and 12 hours at >25°C, respectively. Carbamazepine and sulfamethoxazole were not well degradable, only 22% on sulfamethoxazole removal was observed in the presence of acetaminophen (Kang et al., 2021).
Besides testing only single fungal species also multispecies consortia were tested for pharmaceutical degradation. For example, a consortium of WRF G. applanatum and the edible fungus Laetiporus sulphureus removed 99.5% of all compounds in a mixture of anti-inflammatory drugs (diclofenac, celecoxib, and ibuprofen) within 72 hours (Bankole et al., 2020). In contrast, removal efficiency was much lower (66-92%) when only one strain was present. Significant induction in enzyme production of Lac, LiP and MnP amounted to 201%, 180% and 135%, respectively in the fungal consortia.
5 Conventional plastic polymer degradation by Fungi
Conventional plastics are synthetic polymers, commonly of petrochemical origin (Wayman and Niemann, 2021). Large scale production of plastics begun in the 1950s and exponentially increased until today. Plastics are used in nearly all industry sectors, most importantly packaging, textile, electronics and consumer products (Geyer et al., 2017). Plastic polymer production in 2018 reached 359 Mt, however waste mismanagement and country-specific inadequate policies lead to the disposal of plastics in landfills or direct littering in nature. Of the global plastic production, it is estimated that 1.8-4.1% enters the ocean (Jambeck et al., 2015) via coastal deposition or riverine input (Lebreton et al., 2017; Onink et al., 2021), or through atmospheric deposition (Liss, 2020). Plastic litter in the oceans has raised concern, both in the scientific and public domains and was identified as an environmental threat (Cózar et al., 2014; Jambeck et al., 2015; Geyer et al., 2017; Macleod et al., 2021; Wayman and Niemann, 2021). In the ocean, plastics may float at the surface or sink to the seafloor, depending on the polymer’s density and shape (Wayman and Niemann, 2021). Indeed, it has been suggested that sedimented plastic debris might become a stratigraphic indicator of the Anthropocene epoch (Zalasiewicz et al., 2016). Some plastics are also ingested by marine biota (Giani et al., 2019; Savinelli et al., 2020). Nevertheless, the ultimate fate of marine plastic is still largely unknown. Global calculations of floating marine plastic debris only account for a few percent of the expected amount that has entered the ocean (Thompson et al., 2004; Van Sebille et al., 2015; Koelmans et al., 2017). Several reasons for this “missing plastic paradox” have been discussed, most importantly shear stress and weathering lead to the fragmentation of larger plastic items into ever smaller particles that escape current sampling techniques (Gewert et al., 2015; Romera-Castillo et al., 2018; Chubarenko et al., 2019). Degradation through solar radiation (UV-weathering) and microorganisms likely contribute to plastic disappearance, but specifically the latter is a poorly understood process (Wayman and Niemann, 2021; Zeghal et al., 2021; Delre et al., 2023; Goudriaan et al., 2023). Marine plastic creates a new habitat for eukaryotic and prokaryotic organisms (Scales et al., 2021; Vaksmaa et al., 2021b), which was also termed the plastisphere (Zettler et al., 2013).
A variety of plastic types exists of which the most prevalent ones contain a carbon-carbon backbone, e.g., polyethylene (PE), polypropylene (PP) and polystyrene (PS). However, others, such as polyethylene terephthalate (PET) and nylon, contain also other heteroatoms. Because plastics are rich in chemical energy, it has been suggested that microbes may utilize the plastic as a carbon substrate for energy gain and growth (Yoshida et al., 2016; Auta et al., 2018; Taipale et al., 2019). Nevertheless, the high energy required for the breakdown of the carbon bonds in plastic polymers renders them a potentially problematic substrate for microbial enzymatic degradation. Also, specific metabolic capabilities are likely necessary to account for the variety of plastic formulations. Identification of potential plastic-degrading microbes often includes analysis of microbial biofilm formation and colonization with specific taxa on plastic surfaces (Kettner et al., 2017; Kettner et al., 2019; Lacerda et al., 2020). Several microbes have been isolated from these biofilms, mostly from plastics in terrestrial environments, and tested for their potential to degrade specific polymers in laboratory conditions. These investigations revealed that some microbes, in fact, can degrade certain polymers, typically those containing heteroatoms (Yoshida et al., 2016).
Plastic colonizing fungi in terrestrial ecosystems have usually been investigated with isolation-based assays (Taghavi et al., 2021); numerous attempts were made to characterize their plastic degrading potential (Yamada-Onodera et al., 2001; Sowmya et al., 2015; Spina et al., 2021; Khan et al., 2022). As plastic degraders, Aspergillus, Penicillium and Trichoderma are among the most investigated taxa (Sowmya et al., 2015; Ojha et al., 2017; Sáenz et al., 2019; Malachová et al., 2020). For example, Trichoderma viride and Aspergillus nomius were two species isolated from a landfill nearby Medan, Indonesia (Munir et al., 2018). These grew on low density polyethylene (LDPE) causing deformations and reducing the weight and tensile strength of the polymer. Other Aspergillus species isolated from landfills include Aspergillus clavatus (Gajendiran et al., 2016), Aspergillus flavus and Aspergillus terreus (Verma and Gupta, 2019) and with, these were also shown to degrade PE. Fusarium oxysporum, Fusarium falciforme and Purpureocillum lilacinum isolated from an abandoned dumpsite in Northern Italy, showed to cause changes in the PE film morphology (Spina et al., 2021). The anaerobic fungus Paenibacillus spp. (isolated from a landfill in Brazil) was able to degrade PE (Bardají et al., 2019). This fungus was also found to contain the alkB gene in its genome, which encodes the enzyme alkane hydroxylase, which is potentially involved in plastic degradation. Penicillium citrinum isolated from a soils of plastic dump yard in India degraded LDPE (Khan et al., 2022).
In freshwater systems, plastic colonizing fungi or potential degraders have been reported. The fungal strains Cladosporium cladosporioides, Xepiculopsis graminea, and Penicillium griseofulvum and Leptosphaeria spp, isolated from plastic debris from the shoreline of lake Zurich, were tested for their ability to degrade polyurethane (PU) and PE (Brunner et al., 2018). While some degraded PU, none was able to degrade PE. Further incubation experiments in freshwater were conducted with a diversity of plastic debris (PE, PP, PS and polybutylene terephthalate (PBT)) collected from the Urumqi River, (Xue et al., 2021). The fungal communities on plastic differed from the fungal community of the surrounding water and Cladosporium and Alternaria were enriched on plastic debris.
Investigations of plastic-colonizing microbiota in the environment mainly examined bacteria, whereas fungi remain generally an understudied taxa particularly in marine ecosystems (Harrison et al., 2014; Oberbeckmann et al., 2016; Frère et al., 2018; Oberbeckmann et al., 2018; Miao et al., 2019). Furthermore, in contrast to studies from terrestrial environments, many marine studies focused on investigating the natural fungal community on the plastic, while less effort was undertaken to isolate and to characterize single species (Zeghal et al., 2021; Vaksmaa et al., 2021a). Fungi have been found as part of biofilms on plastic surfaces, for example on polyethylene terephthalate (PET) drinking bottles, exposed to the North Sea (Oberbeckmann et al., 2016). Members of the Ascomycota, Basidiomycota and Chytridiomycota together with prokaryotes were identified as colonizers on the PET bottles incubated in-situ for ~6 weeks.
Exposure experiments with PE and PS were carried out in the Baltic Sea, the river Warnow and a wastewater treatment plant with wood as a control surface (Kettner et al., 2017). In total, 81 fungal taxa were identified, including Chytridium, as well as fungi-like Rhinosporideacae, Rhizidiomyces, and Pythium. In contrast to the study with PET in the North Sea (Oberbeckmann et al., 2016), the fungal community on the plastic differed from that on the control surface after ∼2 weeks of incubation. Longer incubations of 44 weeks with PE sheets and dolly ropes (installed at the sea bed), were conducted at a harbor and an offshore location in the North Sea (De Tender et al., 2017). Among the fungal community, Ascomycota were predominantly detected, followed by Basidiomycota, Zygomycota and Lecanoromycetes. Chytridiomycota have been identified as the predominant plastic-colonizing fungal taxon on different synthetic polymers incubated in a laboratory flow through system with North Sea water (Kirstein et al., 2018). Chytridiomycota have been described as a prevalent fungal taxon throughout aquatic environments (Comeau et al., 2016).
Assessment of fungal diversity may depend on a variety of environmental and experimental factors. For example, salinity is a significant factor influencing fungal communities as shown in the Baltic Sea (Rojas-Jimenez et al., 2019). Among experimental factors, the selection of primers, sequencing depth and reference data bases will strongly influence phylogenetic resolution (Zeghal et al., 2021). The potential influence of using different molecular marker genes to analyze fungal communities on plastic was addressed by comparing three genes: ITS2, 18S rRNA V4 and 18S rRNA V9 (Lacerda et al., 2020). These were used for analyzing the fungal community on floating plastics from the Western South Atlantic and the Antarctic Peninsula. The study found that by using 18S v4 primers, only 2.2% (Atlantic) and 4.3% (Antarctica) of the reads could be assigned. By using primers targeting the 18S v9 region, a similarly low percentage was assigned. In stark contrast, primers targeting the ITS2 region worked much better allowing to assign 60% (Atlantic) and 80% of reads (Antarctica). In total, 64 fungal orders were associated with plastics. The fungal community on the different plastics was highly diverse, and no differences were found between polymer types, shapes, size classes or even sampling stations and locations. However, novel fungal phyla of Aphelidomycota, Zoopagomycota, Mucoromycota and Blastocladiomycota were reported for the first time as part of the biofilms on marine plastics.
Fungi are seemingly an important part of microbial biofilms coating plastic fragments in the marine realm. Yet, it remains to be tested in how far fungi on marine plastics are also able to degrade them. In fact, only a few species, such as Zalerion maritimum and Aternaria alternata were shown to degrade PE (Paço et al., 2017; Gao et al., 2022).
6 Fungal degradation of biobased biodegradable plastics
Biodegradable plastics are defined as plastic-like polymers which degrade under standardized conditions (e.g., ISO 17088:2021). However, these mostly include industrial compositing conditions with elevated temperature or moist content, which often do not resemble conditions as found in nature. Thus, even when a certain plastic type is certified as biodegradable plastic, it might only degrade sluggishly or not at all in the environment. On the other hand, plastic types that do not fulfill the biodegradability norm, might, in fact, be degraded by microbes, however, possibly not at the necessary velocity at degradation conditions as determined in the standards. From a purely scientific standpoint, it would nevertheless be considered biodegradable.
Biobased raw materials for bioplastic production are originating from renewable sources such as vegetable oil, corn starch, soybean proteins, sugars, potatoes and even microbes (Rudin and Choi, 2013; Lackner, 2015; Ashter, 2016; Pieja et al., 2017). The common misconception is that biobased plastics are, by default, biodegradable, whereas, in fact, also conventional recalcitrant plastics such as PE can be synthesized from biological source materials. Biodegradable biobased plastics – in the following termed as bioplastics - are intended to (partially) replace conventional petroleum-based plastics. However, at present, they only hold a market share of ~1%. With an increasing demand for ecologically friendly materials and growing restrictions on conventional plastics, the market share is estimated to increase in the future.
One of the most common bioplastics is polylactic acid (PLA), which is an aliphatic polyester, often produced from the fermentation of agricultural products of which the resulting lactic acid monomers can be polymerized (Dubey et al., 2016; Balla et al., 2021). PLA is biodegradable under composting conditions, with high relative humidity at 50°C (Karamanlioglu and Alkan, 2020). Its degradation can be further enhanced by the addition of natural plasticizers, such as acetyl tributyl citrate and CaCO3 (Brdlík et al., 2021). PLA degradation has been reported for several fungal genera such as Aspergillus (Maeda et al., 2005), Fusarium, Penicillium (Torres et al., 1996a; Torres et al., 1996b), Tritirachium (Jarerat and Tokiwa, 2001), Cryptococcus (Masaki et al., 2005), Trichoderma (Lipsa et al., 2016). These genera express protease and cutinase enzymes, described to be responsible for hydrolyzation and depolymerization of PLA. PLA degradation by Tritirachium album ATCC 22563 was stimulated after the addition of 0.1% of gelatin and the responsible enzyme was hypothesized to be a protease (Jarerat and Tokiwa, 2001).
Polyhydroxyalkanoates (PHAs) is a group of biopolymers with over 100 formulations originating from microbial metabolism, such as poly (β-hydroxybutyrate) (PHB), poly (3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV), polyhydroxyvalerate (PHV) etc. PHAs are superior to PLA in biodegradability, as degradation of these occurs in composts as well as in the marine environment, where PLA based plastics may need ~1000 years to degrade (Digregorio, 2009). PHBs are recognized as one of the most common PHAs and often used in investigating fungal degradation potential. PHB degradation has been shown for Penicillium spp., which produced extracellular PHB depolymerase (Zhou et al., 2008). PHBV depolymerase production has been shown for Aspergillus spp. (Nadhman et al., 2012). Aspergillus ustus, was shown to degrade PHB under pressure, similar to deep sea conditions (Gonda et al., 2000). Emericellopsis minima W2, isolated from wastewater was shown to be capable of degrading PHB (Rhee et al., 2002). Fungal strain NKM1712 (Phylum Ascomycota), isolated from soil, stimulated the degradation of poly butylene adipate-co-butylene terephthalate (PBAT) (Kasuya et al., 2009). A PHB depolymerase purified from Penicillium funiculosum (IFO6345), showed a strong hydrolytic activity towards 3-hydroxybutyrate oligomers (Miyazaki et al., 2000).
An interesting finding is that fungi are able to degrade more than one type of bioplastics. A lipase of Cryptococcus spp. strain S-2 was shown to degrade PLA as well as polybutylene succinate (PBS), polycaprolactone (PCL) and PHB (Masaki et al., 2005). Similarly, a cutinase was characterized for Aspergillus oryzae. This showed specific activities of 0.42 U/mg, 11 U/mg and 0.067 U/mg for PBS, poly(butylene succinate-co-adipate) (PBSA) and PLA respectively (Maeda et al., 2005). In addition, Fusarium moniliforme encodes for a cutinase, which allows degradation of polycarpolactone (Murphy et al., 1996).
Environmental exposure studies have shown that fungi colonize and degrade bioplastics in terrestrial, freshwater systems as well as marine environments (Emadian et al., 2017). However more studies can be found that are conducted in soil systems than in marine water or sediments. For example, PLA films were buried to Mediterranean soil for 11 months and showed a low degree of degradation (Rudnik and Briassoulis, 2011). A similar setup was used for tropical soil conditions (Boyandin et al., 2013). PHA pellets were buried in several locations for 10 to 12 months and the daily rate of mass loss ranged from 0.02 to 0.33% across all locations. The fungal species Gongronella butleri, Penicillium spp., Acremonium recifei, Paecilomyces lilacinus, and Trichoderma pseudokoningii were identified as colonizers and potential PHA degraders. Addition of PBSA to the soil was shown to nearly double the fungal biomass (Guliyev et al., 2022). When PBSA was exposed to field soil conditions Tetracaldium spp. doubled in abundance in 328 days (Purahong et al., 2021). Similarly, when PHBV was buried in soil, the latter isolation of potential plastic degraders revealed that fungi Fusarium oxysporium F1–3, Paecilomyces lilacinus F4–5 and Paecilomyces farinosus F4–7 had the highest contribution to the PBHV degradation (Sang et al., 2002).
Recently, a meta-analysis of studies on PHAs degradation in seawater (in-situ and laboratory conditions) found that the average biodegradation rate would range between 0.04 and 0.09 mg per day/cm2 (Dilkes-Hoffman et al., 2019). However, little focus has been on the identification of the total fungal communities colonizing biobased biodegradable polymers in natural environments. For overview of fungi shown to degrade biodegradable plastic polymers, see Supplementary Table 1.
7 Mycoremediation of heavy metals
Metals are generally malleable or ductile elements, but no clear definition exists that well-defines the term heavy metal. Heavy metals may be described as dense metals (≥5 g/cm-3) forming a block of almost all elements in groups 3 to 16 that are in periods 4 and greater of the periodic table of elements (Hawkes, 1997). The block of earth and rare earth elements as well as titanium, aluminum and silicon may then be described as light metals. Almost all metals are scarce and unevenly scattered in the Earth’s crust, but may be concentrated in certain regions as a result of geological or anthropogenic processes (Rankin, 2011). They are also transported by natural phenomena or anthropogenic activities to the atmosphere and aquatic environment, where most heavy metals pose a risk to almost any ecosystem on Earth and human health.
For example, arsenic, nickel, thallium, cadmium, lead and mercury are notably harmful or poisonous (Mathai and Bhanu, 2010; Signes-Pastor et al., 2021). Potential pathways of heavy metal contamination/exposure are manyfold but are often related to erosion of geogenic sources and mining for metal ores combustion of fossil fuels, and disposal of industrial waste (Candelone et al., 1995; Dudka and Adriano, 1997; Lottermoser, 2010; Harms et al., 2011). High levels of heavy metals are also found in certain agricultural products and paints from which they leach into the environment (Ogilo et al., 2017; Karimi et al., 2021). Heavy metals generally persist in nature for long periods of time (Fashola et al., 2016) and may also accumulate in plants or animals, such as arsenic in rice (Rehman et al., 2020) and mercury in fish (Zupo et al., 2019). They are also subjected to trophic transfer (Soliman et al., 2022) and eventually pose a considerable threat to human health (Noman et al., 2022). Heavy metal toxicity has been characterized for plants, animals, humans and microbes, where the metals can cause DNA damage, denature proteins, inhibit enzyme activities, inhibit cell division and disrupt cellular membranes (Rajendran et al., 2003).
Heavy metals, in contrast to organic pollutants, cannot be degraded but their oxidation state may be changed, which can change their toxicity and mobilize or precipitate them. Thus, bioremediation of heavy metals relies on bacteria, microalgae and fungi that have tolerance to heavy metals (Leung et al., 2001; Iyer et al., 2005; Ledrich et al., 2005; Hassan et al., 2009; Marques et al., 2009). Among these, organisms that are able to assimilate heavy metals and thereby alleviate the pollution in nature, are preferentially chosen for bioremediation applications. Fungi possess extra and intracellular biochemical and molecular mechanisms which rely on, firstly binding of the metal to the cell surface via an ion exchange reaction, surface binding, and complexion with functional groups – (known as biosorption) and secondly, cellular uptake and compartmentalization (Goutam et al., 2021).
Extracellular mechanisms intend to preclude the entrance of the toxic metal (Bellion et al., 2006). Thus, binding or biosorption is considered one of the first extracellular barriers to preclude metal toxicity in fungi. For instance, the proportion of Cd binding to cell walls of Paxillus involutus was similar to that found intracellularly (Blaudez et al., 2000). Extracellular release of suppressor enzymes or pollutant chelating agents, as well as suppression of toxicant influx transporters, are other mechanisms to prevent the entrance of metals into the cells (Meharg, 2003).
Intracellular mechanisms and high capacity for metal uptake has been shown for multiple genera of filamentous fungi, including Trichoderma, Penicillium, and Aspergillus species (Dusengemungu et al., 2020), among others. After being incorporated in the cell, intracellular mechanisms tend to reduce metal accumulation, toxicity, or concentration. The active efflux by enhancing cell wall transporters, biochemical transformation into less harmful species and antioxidant cellular processes such as binding to nonprotein thiols, accumulation in vesicles or mobilization and translocation, are the main known mechanisms (Meharg, 2003; Harms et al., 2011).
The ability of fungi to mobilize and translocate molecules and chemical compounds, including toxic metals, between different parts of their mycelium or between their mycelium and plant symbionts makes particularly filamentous fungi interesting organisms for bioremediation applications (Lindahl et al., 2002; Allen et al., 2003; Allen, 2007; Harms et al., 2011). Furthermore, the microtubules system and secretory vesicles serve as paths and media for long-distance transport (Hyde et al., 1999; Horio and Oakley, 2005; Mouriño-Pérez et al., 2006; Uchida et al., 2008). Previous studies in arable lands and tropical forest soils have reported total length estimates of fungal hyphae of 19 to 292 m/g soil, 2 to 34 m/g soil, respectively (Frey et al., 1999; Camenzind and Rillig, 2013). Earlier studies report on fungal hyphae lengths reaching even 10000 m/g soil (Kjøller and Struwe, 1982), attesting to the substantial dimension of networks of filamentous fungi. Despite the fact that translocation of toxic metals has not been documented yet, an increase of Cd in both the cytosol and secretory vesicles of Paxillus involutus soon after exposure (Blaudez et al., 2000), suggests its availability for long-distance transport and detoxification.
Bioremediation of metals (including heavy metals) through fungi have more frequently been addressed in terrestrial ecosystems than in aquatic ecosystems (Chaturvedi et al., 2015). Reduction of toxic Cr (VI) into non-toxic Cr (III) has gained interest for bioremediation. The brown-rot fungus, Gloeophyllum sepiarium, removed 94% of Cr (VI) in chromium contaminated soil within 6 months (Achal et al., 2011). Neocosmospora spp., Aspergillus spp., Penicillium spp. and Rhizopus spp. isolated from arsenic contaminated soils, were shown to survive sodium arsenate concentrations of 10 g/L (Srivastava et al., 2011). In another study Trichoderma spp., and Aspergillus spp. survived up to 10 g/L of arsenate and members of the genera Chaetomium, Myrothecium, Stachybotrys, Rhizomucor, Fusarium, Rhizopus, Microdochium, also showed tolerance up to 10 g/L of arsenate (Singh et al., 2015). Trichoderma asperellum and F. oxysporum and Penicillium janthinellum were shown to bioaccumulate arsenic in the cells (Su et al., 2010).
In aquatic environments, magnetic nanoparticles coated with Aspergillus fumigatus and Aspergillus niger were used as a bio-sorbent to remove Cr(VI) (Saravanan et al., 2021). With a removal efficiency of 249.9 mg/g magnetic nanoparticles could be a suitable removal material of Cr(VI) in aquatic environments. The removal was affected by the presence of chitin or glucan polysaccharides. These have been shown to promote potential binding sites for ion exchange and metal chelation by the presence of ionizable functional groups (e.g., carboxyl, sulfate or phosphate). Several works suggest that biosorption capacity of metals is species-dependent (Dusengemungu et al., 2020). For instance, it has been reported that the EPS of Laccaria bicolor bound less than 30% of the added cadmium and that copper binding could not be detected (Chai et al., 2019).
On the other hand metal sorption experiments with Cu, Pb and Cd and A. fumigatus showed a selective metal binding affinity in the order of Cu(II) > Pb(II) > Cd(II) (Yin et al., 2011). Furthermore, the majority of the fungal isolates (Aspergillus, Penicillium, Alternaria, Geotrichum and Fusarium genera) from water and sediment samples from five contaminated sites in the Moghogha river (Tangier, Morocco) showed tolerance to Pb, Cr, Cu and Zn (Ezzouhri et al., 2009).
Reports on marine fungi as agents for potential heavy metal bioremediation are scarce, though the available results seem promising. For instance, Yarrowia lipolytica is a potential candidate for several biotechnological applications, e.g., treatment of palm oil mill effluents, crude oil as well as 2,4,6-trinitrotoluene (TNT) (Oswal et al., 2002; Zinjarde and Pant, 2002; Jain et al., 2004). Two marine strains of Yarrowia spp. (Idd1 and Idd2) showed high removal efficiencies of Hg. Yarrowia spp. removed more than 97% of Hg from a medium containing 16 μg/mL Hg+2 (Oyetibo et al., 2016). Absorption to the cell wall was the most prominent removal mechanism and responsible for 49-83% of the removal. However, bioaccumulation and volatilization were also important pathways in removing Hg (Oyetibo et al., 2015). Furthermore, Yarrowia lipolytica (NCIM 3589 and 3590) showed a high tolerance to various heavy metals; yet the heavy metal removal capacity was not studied (Bankar et al., 2018). Aspergillus candidus, isolated from waters of Bhavnagar coast was found to tolerate arsenic (Vala, 2009). Higher removal capacities were observed in the treatments exposed to a high As concentration (50 mg/L), and pentavalent As was removed more efficiently than trivalent As (16 mg/g for As (V) and 8.5 mg/g for As (III), respectively).
The marine fungi Corollospora lacera and Monodictys pelagica have also been shown to sequester Cd and Pb. Although both strains showed contrasting bioaccumulation patterns. C. lacera was extremely efficient in bioaccumulating Pb but not Cd (up to 250 mg/g and over 7 mg/g of mycelium, respectively), while M. pelagica efficiently bioaccumulated cadmium but not lead (over 60 mg/g and over 6 mg/g of mycelium, respectively) (Taboski et al., 2005). Heavily contaminated coastal sediments from the Mediterranean Sea have been tested for microbial and fungal biodegradation potential, subject to historical deposition of mining waste. Aspergillus niger and Trichoderma spp. Showed 8-fold higher As removal than conventional chemical treatments and higher removal rates than bacteria-mediated remediation. Non-mobile Zn and Cd were removed by fungi-induced bioleaching (Dell'anno et al., 2022). This was more efficient than bacteria augmented treatments as well, likely because of a fungi-mediated pH decrease, which enhanced mobilisation of these metals. Two other Aspergillus species, A. flavus and A. niger, associated with a marine seaweed, tolerated hexavalent chromium (Cr(VI)) at different concentrations. Both strains accumulated similar amounts of Cr(VI) linearly correlating with the levels of Cr(VI) in the experiment, i.e., 4.4, 9.4 and 22.3 mg/g and 3.5, 7.8 and 18.1 mg/g dry weight of fungal biomass in treatments containing 25, 50 and 100 ppm of Cr(VI), respectively (Lotlikar et al., 2018). Both isolates were able to remove up to 25% of the supplied chromium in 15 days. Further species of Aspergillus, such as A. sydowii achieved 26% Cr(VI) removal through exopolysaccharide mediated mechanisms as well as intracellular deposition of Cr2O3. Finally, one member of the Kalmusia genera, Kalmusia italica, isolated from marine sediments has shown tolerance to Ni, Cr, Pb and Zn although the mechanisms for immobilization are yet to be characterized (Sumathi et al., 2020). For a recent review on aquatic fungi and heavy metal accumulation, see (Sharif et al., 2022).
8 Application potential of mycoremediation
Various strategies have been suggested for bioremediation of pollutants in soil, sediment and aqueous environments. The application of mycoremediation can be conducted on site, referred to as in-situ mycoremediation or by excavating or removing the contaminant or contaminated soil, sediment or water matrix to a different location where the mycoremediation is carried out ex-situ (Figure 2). Most common in-situ bioremediation methods are bioaugmentation, bioventing, biosparging and natural attenuation (Azubuike et al., 2016). Ex-situ bioremediation methods depend on the matrix; biopiling, composting, land farming, applied for solid matrixes and bioreactors and water treatment facilities can be used for liquid media such as slurries and water (Kumar et al., 2011). The feasibility of a certain remediation strategies depends on environmental conditions, nature and extensiveness of the pollution, associated costs and the availability of suitable fungal candidates for remediation.
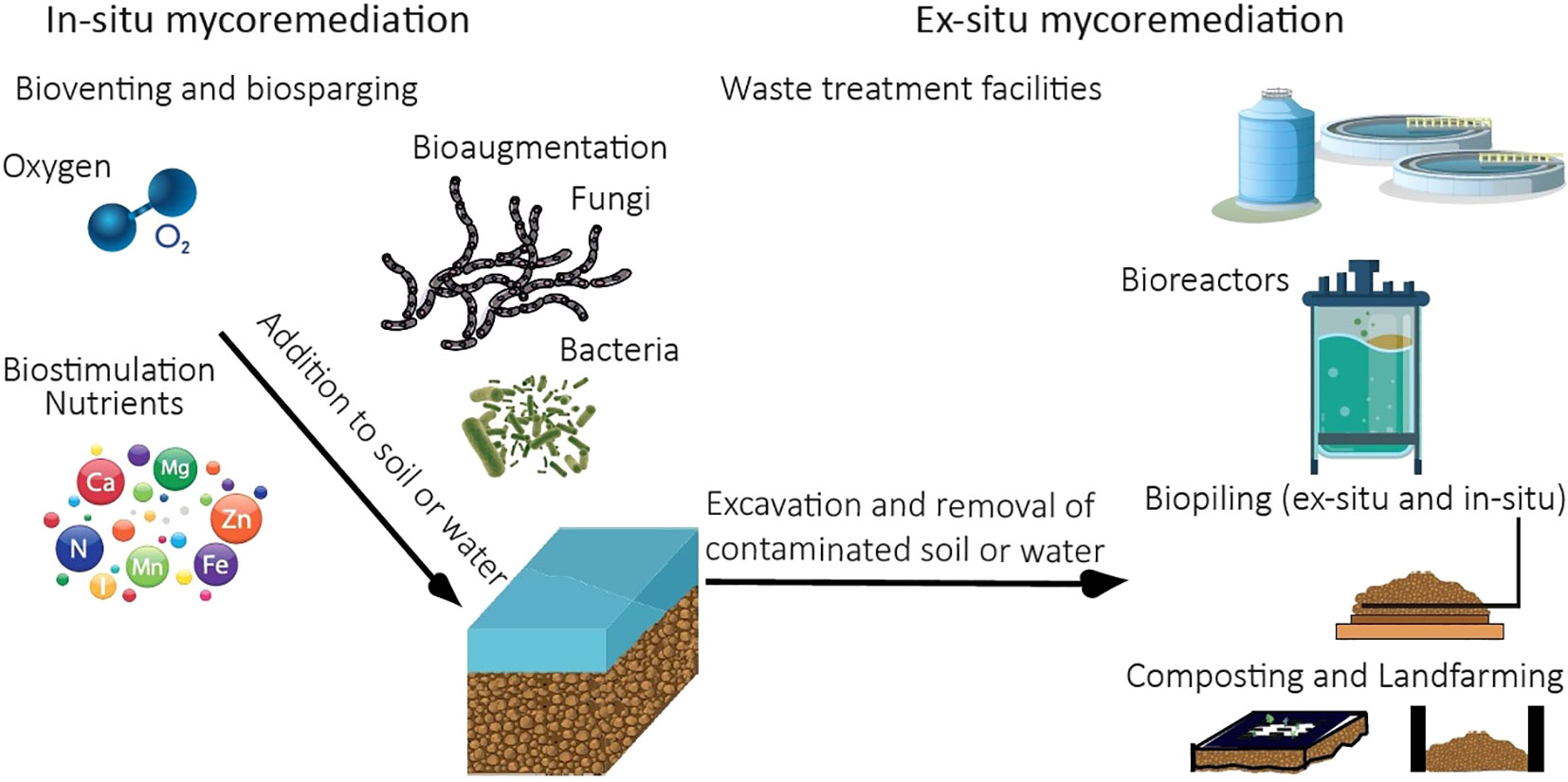
Figure 2 Illustration of most commonly applied in-situ and ex-situ mycoremediation strategies. Bioventing and biosparging - addition of oxygen to promote activity of aerobic microbes. Biostimulation – Addition of nutrients to promote microbes remediating the pollutant. Bioaugmentation – addition of microbes to the site of pollution. Ex-situ strategies – Bioreactors for treatment of pollutants in aqueous matrix. Composting – treatment of polluted matrix (often soil) in confined space (often also includes thermal treatment). Landfarming is based on regular tilling of the soil, collected on a designated bed. Biopiling is a system, which includes irrigation, aeration systems and collection of leachates. In biopiles, the moisture, oxygen, pH and nutrients are controlled.
8.1 In-situ mycoremediation
Implementation of in-situ mycoremediation is unavoidably dependent on the matrix, as it is carried out on site. This requires preliminary investigation and characterization of the polluted site prior to the implementation of a fungi-based bioremediation technique (Winardi et al., 2019). The mycoremediation on site is a multi-factor system, where the pollutant input has already caused changes to the chemical, physical and biological native system. Addition of a live bioremediation component will influence not only the pollutant, but potentially higher organisms and microbial communities as well as physicochemical conditions at the site. In addition, subsequent chemical transformations after application of fungi might be influenced by or influence the microbial communities on the site in positive or negative ways. This is compound specific and depends on native microbial communities, their metabolic capabilities and ability to survive.
In-situ application of fungi is advantageous due to the limited disturbance that this approach induces to the polluted site (in contrast to e.g., soil excavation). As a bioremediation strategy, this has been applied successfully already by using bacteria to treat sites contaminated with hydrocarbons, dyes, heavy metals, and chlorinated solvents. Dispersal of fungi in soils is not dependent on water(saturated)-soil phases (in contrast to bacteria), because fungal hyphae can grow in air-soil interfaces and penetrate both soil and rock matrices. Furthermore, on site treatment minimizes exposure of fungi to shear forces, which would occur for example in bioreactors via stirring. This allows for mycelia to develop and penetrate soils without alterations (Harms et al., 2011).
One of the major limitations of in-situ bioremediation is dilution of the contaminant, leading to lowering of the contaminant concentration. Filamentous fungi are advantageous in situations of low concentration and large dispersion, due to their translocation capabilities inherent to their mycelium growth. Filamentous fungi can extend their hyphae networks in the magnitude of kilometres of soil and are suited for heterogenous environmental conditions (Ingham et al., 1991). Due to their penetration capabilities, fungi can also act as an adjuvant to bacterial degradation by breaking physical barriers in air-soil interfaces where water transport is limited (Kohlmeier et al., 2005; Wick et al., 2007) or even act as fungal highways, allowing bacteria to disperse along fungal mycelium (Junier et al., 2021). This has been demonstrated for hydrocarbon degrading bacteria using common soil fungi such as F. oxysporum or Rhexocercosporidium spp. to mobilize (Kohlmeier et al., 2005). Also, Pythium ultimum was shown to facilitate the chemostatic dispersal of PAH degrading Pseudomonas spp. along its mycelia (Furuno et al., 2022).
In-situ large scale bioremediation applications in soils are, however, not carried out extensively due to some biological, operational/economic challenges. In some cases, additional treatments of the contaminated soil/site are necessary, such as tillage, aeration and water additions (Wick et al., 2007). These may be cost intensive and might make in-situ applications economically unfeasible (Cristorean et al., 2016). In addition, transplanting fungi to polluted sites may result in loss of function. This have been observed for ligninolytic basidiomycetes where mechanical fragmentation of fungi, leads to detrimental effects in their capabilities (Nielsen and Krabben, 1995; Li et al., 2000). Furthermore, one major disadvantage of fungi in in-situ application is competition of the bioremediatory fungus with indigenous microbial communities in particular bacteria, which renders the prospects of applications limited (Baldrian, 2008). Nonetheless, fungi may serve as potential agents at decontamination sites that are physically challenging for bacteria to access or where extreme environmental conditions favour fungal growth. Finally, in-situ mycoremediation can be applied more easily to terrestrial settings, while a successful fungal application to aquatic, specifically marine environments is usually complicated because of dilution and dispersal of the pollutant and the fungus.
8.2 Ex-situ mycoremediation
Implementation of ex-situ mycoremediation can be feasible but heavily depends on whether the pollutant can be collected and transported to facilities for remediation, e.g., designated landfills, large collection tanks and bioreactors or a combination of existing solid waste and wastewater treatment facilities with designated sections. Ex-situ mycoremediation enables better control over the process as it typically allows better monitoring and control of environmental parameters, growth and performance of the fungi. For example, environmental conditions such as oxygen and pH influence the fungal degradation efficiency of certain pesticides (Castillo and Torstensson, 2007), which can be adjusted in bioreactors. Also, the high requirement of most fungi for oxygen and the costs and practicalities associated with facilitating this in-situ makes ex-situ mycoremediation often more feasible. Furthermore, elevated temperatures as can be achieved in ex-situ facilities, typically result in accelerated and improved remediation (Dhiman et al., 2020).
In aqueous media, advances in integrating biological treatment methods using fungi (T. versicolor) (Mir-Tutusaus et al., 2018) or consortia of fungi and bacteria (Del Álamo et al., 2022) to clean up hospital wastewater are extensively investigated. In bioreactor settings, initial coagulation-flocculation pre-treatment steps were shown to improve the viability of fungi within the reactor (Mir Tutusaus et al., 2016). Additionally, the advantage of coupling advanced oxidation processes (AOP) with biological treatment has been investigated utilizing T. versicolor for the removal of pharmaceuticals (Vasiliadou et al., 2019). Addition of the redox mediators, such as quinones, has shown to increase the removal of pharmaceuticals. This appears to be species specific, as the most efficient redox mediators for T. versicolor and G. lucidum were 2,6-dimethoxy-1,4-benzoquinone (DMBQ) and gallic acid, respectively. Furthermore, the combination of DMBQ and T. versicolor resulted in the removal of all 13 pharmaceuticals (≥60%) within 24 hours.
In ex-situ setting, bioremediation can be enhanced in consortia of fungi and other microbes and/or higher organisms or when applied in tandem with other physicochemical remediation approaches. Fungi growing in symbiosis with plants (maize) have been shown to enhance degradation of a range of compounds, including the herbicide Atrazine (Huang et al., 2007). The approach of employing microbial consortia, fungal cultures and their enzymes seems promising for removing PPCPs in conventional waste water treatment plants (Bilal et al., 2019).
Bioremediation of plastic polymers in wastewater treatment plants have been addressed as a potential mitigation strategy for removal of microplastics. At present, the bioremediation has focused on higher aquatic plants and animals as agents, that can physically remove plastic particles from the aqueous phase, e.g., retaining particles on their surface (e.g. seaweeds) or filter particles from the water (e.g., bivalves) (Masiá et al., 2020). Fungi, on the other hand, are able to biochemically degrade plastics in terrestrial and aquatic environments. Thus, their potential as plastic degraders and applicability needs further investigations.
Ex-situ mycoremediation could also benefit from using genetically modified organisms (GMOs) (Jaiswal and Shukla, 2020), specifically designed to target a pollutant or a variety of pollutants. Using GMOs is only feasible in closed systems, where they can be neutralized after treating the polluted matrices and ensuring to ensure no viable GMOs are released to the environment.
8.3 Mycoremediation strategies based on enzyme expression and immobilization
Ascomycete fungi are candidates for genetic manipulation, and expression of enzymes for the upscaling of remediation capabilities (Halaouli et al., 2006; Rodgers et al., 2010). Some candidate enzymes are laccases and tyrosinases with a large range of applications. Peroxidases from basidiomycetes have already been used for commercial applications, with enhanced resistance to peroxide and a wider redox spectrum for degradation through modifications (Ruiz-Dueñas et al., 2009).
Recent studies have been focusing on the exploration and expression of enzymes such as laccase-mediator systems (LMS) (Bilal et al., 2019). However, the major disadvantage of these processes is the high cost of the synthetic mediators. This can be averted by using mediators that have a natural origin such as lignin-derived phenolics (Grijalva-Bustamante et al., 2016). Removal of different pesticides (Carbofuran, Diuron, Bentazone, Tebuconazole, Pyraclostrobin, Clomazone) from aqueous samples using an optimized laccase-mediator system has been investigated. Screening of a variety of mediators including caffeic acid, p-coumaric acid, vanillin, gallic acid, chlorogenic acid, protocatechuic acid, ferulic acid and 2,2′-azino-bis-(3-ethylbenzothiazoline- 6-sulfonate) has shown that the laccase-vanillin system was the most efficient resulting in 77% removal of pesticides (Kupski et al., 2019). Biodegradation of Isoproturon, a widely used herbicide known to produce potentially carcinogenic intermediates, was studied using T. versicolor derived laccase and 1-Hydroxybenzotriazole (HBT) as a redox mediator for enhanced removal in aqueous systems (Zeng et al., 2017). In the absence of HBT, there was negligible degradation of Isoproturon whereas LMS in conjugation with HBT resulted in its complete degradation within 24 hours
Next to that, there have also been advancements towards immobilizing laccases on novel support materials for efficient pesticide degradation (Bilal and Iqbal, 2019). The immobilized laccase from Coriollopsis gallica on mesoporous nanostructured silicon foam (MSU-F), was found to efficiently oxidize dichlorophen pesticide and reduced the associated apoptotic and genotoxic effects (Vidal-Limon et al., 2018). Laccase from Myceliophthora thermophile (MtL) was fixed onto microspheres of poly (glycidyl methacrylate) (PGMA), and used in a biocatalytic system for degradation of azinphos-methyl (AZPM), an organophosphate pesticide (Vera et al., 2018). Not only was this hybrid system found to be effective in a broad range of pH and temperature, had a better stability (thermal, storage and operational), but it also promoted a rapid biodegradation rate for AZPM with ABTS as a mediator. The immobilization of enzymes in nano materials has emerged, too, as biocatalysis strategy to treat pharmaceuticals. Another type of successfully used carrier materials are hollow mesoporous carbon spheres (HMCs), with improved stability (pH, temperature), longer storage life and increased reusability (Shao et al., 2019). In comparison to free enzymes, the immobilized Lac showed good enzymatic activity up to 8 cycles with efficient removal of the antibiotics tetracycline hydrochloride (TCH) and ciprofloxacin hydrochloride (CPH). In both cases, the syringaldehyde mediator SA (3 mmol/L) was found to increase removal efficiencies and reusability of the carrying material. These examples represent the future landscape in mycoremediation technologies based on enzymatic potential without direct reliance on an organism. Despite the effort to express exogenous enzymes from fungi and with a wide range of available successes, the main challenge remains to upscaling the expression of these enzymes for concrete applications.
9 Conclusion and future perspective
This review provides an overview on fungi with a particular focus on their ability to degrade recalcitrant and toxic compounds in aquatic environments. The known role of fungi in bioremediation of different classes of emerging pollutants accumulating in natural systems as a result of anthropogenic activities is highlighted. Fungi with their intra- and extracellular enzymatic machineries have been shown to mitigate the effects of these pollutants, breaking them down or demobilising them and thus act as natural bioremediating agents. In bioremediation applications, the same set of fungal taxa or enzymes are often used to substitute conventional pollution mitigation strategies. Yet, the wealth of fungal taxa and useful enzymes seems not to be explored fully. Specifically in aquatic environments, fungi are understudied. We therefore suggest that future research efforts should aim at unravelling the extent of (aquatic) fungal species that are suitable and effective bioremediation agents and to decipher their metabolic pathways in detail. For technical applications, future bioremediation strategies could (i) exploit the symbiotic action of fungi with bacteria and/or plants or design synthetic communities, (ii) enhance enzyme production via genetic engineering and apply enzymes to different carrying materials for mycoremediation in nature, and (iii) extract compounds such as heavy metals after mycoremediation, and thereby advance the use of natural resources which are exhaustible in nature and/or use the bioremediation process for production of value-added products.
Author contributions
AV, SGC, PG and HN designed the project. All authors, AV, SGC, PG, EZ, VHM, HN contributed to writing the manuscript and approved the submitted version. HN supervised the project.
Funding
This study was financed through the European Research Council (ERC-CoG Grant Nr 772923, project VORTEX to HN). AV was financed through Dutch Research Council (VENI Grant VI.Veni.212.029 and NWO XS Grant OCENW.XS21.4.079).
Acknowledgments
We thank Nelleke Krijgsman for the help with Figures 1 and 2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1070905/full#supplementary-material
References
Achal V., Kumari D., Pan X. (2011). Bioremediation of chromium contaminated soil by a brown-rot fungus, gloeophyllum sepiarium. Res. J. Microbiol. 6, 166–171. doi: 10.3923/jm.2011.166.171
Adenipekun C., Lawal R. (2012). Uses of mushrooms in bioremediation: A review. Biotechnol. Mol. Biol. Rev. 7, 62–68. doi: 10.5897/BMBR12.006
Aksu Z. (1998). “Biosorption of heavy metals by microalgae in batch and continuous systems,” in Wastewater treatment with algae (Springer), 37–53.
Al-Aboudi A., Kana'an B. M., Zarga M. A., Bano S., Atia Tul W., Javed K., et al. (2017). Fungal biotransformation of diuretic and antihypertensive drug spironolactone with gibberella fujikuroi, curvularia lunata, fusarium lini, and aspergillus alliaceus. Steroids 128, 15–22. doi: 10.1016/j.steroids.2017.10.003
Al-Hawash A. B., Dragh M. A., Li S., Alhujaily A., Abbood H. A., Zhang X., et al. (2018). Principles of microbial degradation of petroleum hydrocarbons in the environment. Egyptian J. Aquat. Res. 44, 71–76. doi: 10.1016/j.ejar.2018.06.001
Allen M. F. (2007). Mycorrhizal fungi: Highways for water and nutrients in arid soils. Vadose Zone J. 6, 291–297. doi: 10.2136/vzj2006.0068
Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259. doi: 10.1038/nrmicro2312
Allen M. F., Swenson W., Querejeta J. I., Egerton-Warburton L. M., Treseder K. K. (2003). Ecology of mycorrhizae: A conceptual framework for complex interactions among plants and fungi. Annu. Rev. Phytopathol. 41, 271–303. doi: 10.1146/annurev.phyto.41.052002.095518
Alvarenga N., Birolli W. G., Seleghim M. H., Porto A. L. (2014). Biodegradation of methyl parathion by whole cells of marine-derived fungi aspergillus sydowii and penicillium decaturense. Chemosphere 117, 47–52. doi: 10.1016/j.chemosphere.2014.05.069
Ashter S. A. (2016). “10 - new developments,” in Introduction to bioplastics engineering. Ed. Ashter S. A. (Oxford: William Andrew Publishing), 251–274.
Auta H. S., Emenike C. U., Jayanthi B., Fauziah S. H. (2018). Growth kinetics and biodeterioration of polypropylene microplastics by bacillus sp. and rhodococcus sp. isolated from mangrove sediment. Mar. pollut. Bull. 127, 15–21. doi: 10.1016/j.marpolbul.2017.11.036
Ayenimo J., Yusuf A., Adekunle A., Makinde O. (2010). Heavy metal exposure from personal care products. Bull. Environ. contamination Toxicol. 84, 8–14. doi: 10.1007/s00128-009-9867-5
Azubuike C. C., Chikere C. B., Okpokwasili G. C. (2016). Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 32, 180. doi: 10.1007/s11274-016-2137-x
Baborová P., Möder M., Baldrian P., Cajthamlová K., Cajthaml T. (2006). Purification of a new manganese peroxidase of the white-rot fungus irpex lacteus, and degradation of polycyclic aromatic hydrocarbons by the enzyme. Res. Microbiol. 157, 248–253. doi: 10.1016/j.resmic.2005.09.001
Badawi N., Rønhede S., Olsson S., Kragelund B. B., Johnsen A. H., Jacobsen O. S., et al. (2009). Metabolites of the phenylurea herbicides chlorotoluron, diuron, isoproturon and linuron produced by the soil fungus mortierella sp. Environ. pollut. 157, 2806–2812. doi: 10.1016/j.envpol.2009.04.019
Baldrian P. (2006). Fungal laccases - occurrence and properties. FEMS Microbiol. Rev. 30, 215–242. doi: 10.1111/j.1574-4976.2005.00010.x
Baldrian P. (2008). Wood-inhabiting ligninolytic basidiomycetes in soils: Ecology and constraints for applicability in bioremediation. Fungal Ecol. 1, 4–12. doi: 10.1016/j.funeco.2008.02.001
Balla E., Daniilidis V., Karlioti G., Kalamas T., Stefanidou M., Bikiaris N. D., et al. (2021). Poly(lactic acid): A versatile biobased polymer for the future with multifunctional properties–from monomer synthesis, polymerization techniques and molecular weight increase to PLA applications. Polymers 13, 1822. doi: 10.3390/polym13111822
Bankar A., Zinjarde S., Telmore A., Walke A., Ravikumar A. (2018). Morphological response of yarrowia lipolytica under stress of heavy metals. Can. J. Microbiol. 64, 559–566. doi: 10.1139/cjm-2018-0050
Bankole P. O., Adekunle A. A., Jeon B.-H., Govindwar S. P. (2020). Novel cobiomass degradation of NSAIDs by two wood rot fungi, ganoderma applanatum and laetiporus sulphureus: Ligninolytic enzymes induction, isotherm and kinetic studies. Ecotoxicology Environ. Saf. 203, 110997. doi: 10.1016/j.ecoenv.2020.110997
Bardají D. K. R., Furlan J. P. R., Stehling E. G. (2019). Isolation of a polyethylene degrading paenibacillus sp. from a landfill in Brazil. Arch. Microbiol. 201, 699–704. doi: 10.1007/s00203-019-01637-9
Bastos A. C., Magan N. (2009). Trametes versicolor: Potential for atrazine bioremediation in calcareous clay soil, under low water availability conditions. Int. Biodeterioration Biodegradation 63, 389–394. doi: 10.1016/j.ibiod.2008.09.010
Bayen S., Estrada E. S., Juhel G., Kit L. W., Kelly B. C. (2016). Pharmaceutically active compounds and endocrine disrupting chemicals in water, sediments and mollusks in mangrove ecosystems from Singapore. Mar. pollut. Bull. 109, 716–722. doi: 10.1016/j.marpolbul.2016.06.105
Bellion M., Courbot M., Jacob C., Blaudez D., Chalot M. (2006). Extracellular and cellular mechanisms sustaining metal tolerance in ectomycorrhizal fungi. FEMS Microbiol. Lett. 254, 173–181. doi: 10.1111/j.1574-6968.2005.00044.x
Beltrán-Flores E., Sarrà M., Blánquez P. (2021). Pesticide bioremediation by trametes versicolor: Application in a fixed-bed reactor, sorption contribution and bioregeneration. Sci. Total Environ. 794, 148386. doi: 10.1016/j.scitotenv.2021.148386
Bhalerao T. S., Puranik P. R. (2007). Biodegradation of organochlorine pesticide, endosulfan, by a fungal soil isolate, aspergillus niger. Int. Biodeterioration Biodegradation 59, 315–321. doi: 10.1016/j.ibiod.2006.09.002
Biesterbos J. W. H., Dudzina T., Delmaar C. J. E., Bakker M. I., Russel F. G. M., Von Goetz N., et al. (2013). Usage patterns of personal care products: Important factors for exposure assessment. Food Chem. Toxicol. 55, 8–17. doi: 10.1016/j.fct.2012.11.014
Bilal M., Iqbal H. M. N. (2019). Persistence and impact of steroidal estrogens on the environment and their laccase-assisted removal. Sci. Total Environ. 690, 447–459. doi: 10.1016/j.scitotenv.2019.07.025
Bilal M., Iqbal H. M. N., Barceló D. (2019). Persistence of pesticides-based contaminants in the environment and their effective degradation using laccase-assisted biocatalytic systems. Sci. Total Environ. 695, 133896. doi: 10.1016/j.scitotenv.2019.133896
Birolli W., Vacondio B., Da Silva N., Seleghim M., Porto A. (2018). Enantioselective biodegradation of the pyrethroid (±)-lambda-cyhalothrin by marine-derived fungi. Chemosphere 197, 651–660. doi: 10.1016/j.chemosphere.2018.01.054
Birolli W. G., Yamamoto K. Y., De Oliveira J. R., Nitschke M., Seleghim M. H., Porto A. L. (2015). Biotransformation of dieldrin by the marine fungus penicillium miczynskii CBMAI 930. Biocatalysis Agric. Biotechnol. 4, 39–43. doi: 10.1016/j.bcab.2014.06.002
Biswas J. K., Banerjee A., Biswas S. (2022). “Chapter 22 - microbes and marine oil spills: oil-eating bugs can cure oily sea sickness,” in Advances in oil-water separation. Eds. Das P., Manna S., Pandey J. K. (Elsevier), 393–422.
Blaudez D., Botton B., Chalot M. (2000). Cadmium uptake and subcellular compartmentation in the ectomycorrhizal fungus paxillus involutus. Microbiol. (Reading) 146 (Pt 5), 1109–1117. doi: 10.1099/00221287-146-5-1109
Bokade P., Purohit H. J., Bajaj A. (2021). Myco-remediation of chlorinated pesticides: Insights into fungal metabolic system. Indian J. Microbiol. 61, 237–249. doi: 10.1007/s12088-021-00940-8
Borchert E., Hammerschmidt K., Hentschel U., Deines P. (2021). Enhancing microbial pollutant degradation by integrating eco-evolutionary principles with environmental biotechnology. Trends Microbiol. 29, 908–918. doi: 10.1016/j.tim.2021.03.002
Bose S., Kumar P. S., Vo D.-V. N., Rajamohan N., Saravanan R. (2021). Microbial degradation of recalcitrant pesticides: A review. Environ. Chem. Lett. 19, 3209–3228. doi: 10.1007/s10311-021-01236-5
Boxall A. B., Rudd M. A., Brooks B. W., Caldwell D. J., Choi K., Hickmann S., et al. (2012). Pharmaceuticals and personal care products in the environment: What are the big questions? Environ. Health Perspect. 120, 1221–1229. doi: 10.1289/ehp.1104477
Boyandin A. N., Prudnikova S. V., Karpov V. A., Ivonin V. N., Đỗ N. L., Nguyễn T. H., et al. (2013). Microbial degradation of polyhydroxyalkanoates in tropical soils. Int. Biodeterioration Biodegradation 83, 77–84. doi: 10.1016/j.ibiod.2013.04.014
Brdlík P., Boruvka M., Běhálek L., Lenfeld P. (2021). Biodegradation of Poly(Lactic acid) biocomposites under controlled composting conditions and freshwater biotope. Polymers 13, 594. doi: 10.3390/polym13040594
Brodin T., Piovano S., Fick J., Klaminder J., Heynen M., Jonsson M. (2014). Ecological effects of pharmaceuticals in aquatic systems–impacts through behavioural alterations. Philos. Trans. R Soc. Lond B Biol. Sci. 369 (1656), 20130580. doi: 10.1098/rstb.2013.0580
Brunner I., Fischer M., Rüthi J., Stierli B., Frey B. (2018). Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PloS One 13, e0202047. doi: 10.1371/journal.pone.0202047
Camenzind T., Rillig M. C. (2013). Extraradical arbuscular mycorrhizal fungal hyphae in an organic tropical montane forest soil. Soil Biol. Biochem. 64, 96–102. doi: 10.1016/j.soilbio.2013.04.011
Candelone J.-P., Hong S., Pellone C., Boutron C. (1995). Post industrial revolution changes in large-scale atmospheric pollution of the northern hemisphere by heavy metals documented in central Greenland snow and ice. J. Geophysical Res. 1001, 16605–16616. doi: 10.1029/95JD00989
Castillo M. D. P., Torstensson L. (2007). Effect of biobed composition, moisture, and temperature on the degradation of pesticides. J. Agric. Food Chem. 55, 5725–5733. doi: 10.1021/jf0707637
Chai L., Huang M., Cao X., Liu M., Huang Y. (2019). Potential metal-binding ability of proteins in the extracellular slime of laccaria bicolor exposed to excessive Cu and cd. Environ. Sci. pollut. Res. Int. 26, 20418–20427. doi: 10.1007/s11356-019-05201-2
Chandel N., Ahuja V., Gurav R., Kumar V., Tyagi V. K., Pugazhendhi A., et al. (2022). Progress in microalgal mediated bioremediation systems for the removal of antibiotics and pharmaceuticals from wastewater. Sci. Total Environ. 825, 153895. doi: 10.1016/j.scitotenv.2022.153895
Chaturvedi A. D., Pal D., Penta S., Kumar A. (2015). Ecotoxic heavy metals transformation by bacteria and fungi in aquatic ecosystem. World J. Microbiol. Biotechnol. 31, 1595–1603. doi: 10.1007/s11274-015-1911-5
Cheung P. K., Fok L. (2017). Characterisation of plastic microbeads in facial scrubs and their estimated emissions in mainland China. Water Res. 122, 53–61. doi: 10.1016/j.watres.2017.05.053
Chivukula M., Renganathan V. (1995). Phenolic azo dye oxidation by laccase from pyricularia oryzae. Appl. Environ. Microbiol. 61, 4374–4377. doi: 10.1128/aem.61.12.4374-4377.1995
Chubarenko I., Efimova I., Bagaeva M., Bagaev A., Isachenko I. (2019). On mechanical fragmentation of single-use plastics in the sea swash zone with different types of bottom sediments: Insights from laboratory experiments. Mar. pollut. Bull. 150, 110726. doi: 10.1016/j.marpolbul.2019.110726
Coelho-Moreira J., Brugnari T., Sá-Nakanishi A., Castoldi R., Souza C., Bracht A., et al. (2017). Evaluation of diuron tolerance and biotransformation by the white-rot fungus ganoderma lucidum. Fungal Biol. 122 (6), 471–478. doi: 10.1016/j.funbio.2017.10.008
Comeau A. M., Vincent W. F., Bernier L., Lovejoy C. (2016). Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 6, 30120. doi: 10.1038/srep30120
Cózar A., Echevarría F., González-Gordillo J. I., Irigoien X., Ubeda B., Hernández-León S., et al. (2014). Plastic debris in the open ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 10239–10244. doi: 10.1073/pnas.1314705111
Cristorean C., Micle V., Sur I. (2016). A critical analysis of ex-situ bioremediation technologies of hydrocarbon polluted soils. ECOTERRA - J. Environ. Res. Protection.
Cruz-Morató C., Lucas D., Llorca M., Rodriguez-Mozaz S., Gorga M., Petrovic M., et al. (2014). Hospital wastewater treatment by fungal bioreactor: Removal efficiency for pharmaceuticals and endocrine disruptor compounds. Sci. Total Environ. 493, 365–376. doi: 10.1016/j.scitotenv.2014.05.117
Dalecka B., Juhna T., Rajarao G. (2019). Constructive use of filamentous fungi to remove pharmaceutical substances from wastewater. J. Water Process Eng. 33, 100992. doi: 10.1016/j.jhazmat.2016.07.036
Dashtban M., Schraft H., Syed T. A., Qin W. (2010). Fungal biodegradation and enzymatic modification of lignin. Int. J. Biochem. Mol. Biol. 1, 36–50.
Da Silva Coelho-Moreira J., Brugnari T., Sá-Nakanishi A. B., Castoldi R., De Souza C. G., Bracht A., et al. (2018). Evaluation of diuron tolerance and biotransformation by the white-rot fungus ganoderma lucidum. Fungal Biol. 122, 471–478. doi: 10.1016/j.funbio.2017.10.008
Daughton C. G. (2001). “Pharmaceuticals and personal care products in the environment: Overarching issues and overview,” in Pharmaceuticals and care products in the environment (American Chemical Society), 2–38.
Del Álamo A. C., Pariente M. I., Molina R., Martínez F. (2022). Advanced bio-oxidation of fungal mixed cultures immobilized on rotating biological contactors for the removal of pharmaceutical micropollutants in a real hospital wastewater. J. Hazardous Materials 425, 128002. doi: 10.1016/j.jhazmat.2021.128002
Dell'anno F., Rastelli E., Buschi E., Barone G., Beolchini F., Dell'anno A. (2022). Fungi can be more effective than bacteria for the bioremediation of marine sediments highly contaminated with heavy metals. Microorganisms 10 (5), 993. doi: 10.3390/microorganisms10050993
Delre A., Goudriaan M., Morales V. H., Vaksmaa A., Ndhlovu R. T., Baas M., et al (2023). Plastic photodegradation under simulated marine conditions. Marine Pollution Bulletin 187, 114544. doi: 10.1016/j.marpolbul.2022.114544
Deshmukh R., Khardenavis A. A., Purohit H. J. (2016). Diverse metabolic capacities of fungi for bioremediation. Indian J. Microbiol. 56, 247–264. doi: 10.1007/s12088-016-0584-6
De Tender C., Devriese L. I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., et al. (2017). Temporal dynamics of bacterial and fungal colonization on plastic debris in the north Sea. Environ. Sci. Technol. 51, 7350–7360. doi: 10.1021/acs.est.7b00697
Dhiman N., Jasrotia T., Sharma P., Negi S., Chaudhary S., Kumar R., et al. (2020). Immobilization interaction between xenobiotic and bjerkandera adusta for the biodegradation of atrazine. Chemosphere 257, 127060. doi: 10.1016/j.chemosphere.2020.127060
Digregorio B. (2009). Biobased performance bioplastic: Mirel. Chem. Biol. 16, 1–2. doi: 10.1016/j.chembiol.2009.01.001
Dilkes-Hoffman L., Ashworth P., Laycock B., Pratt S., Lant P. (2019). Public attitudes towards bioplastics – knowledge, perception and end-of-life management. Resources Conserv. Recycling 151, 104479. doi: 10.1016/j.resconrec.2019.104479
Dinis M. J., Bezerra R. M., Nunes F., Dias A. A., Guedes C. V., Ferreira L. M., et al. (2009). Modification of wheat straw lignin by solid state fermentation with white-rot fungi. Bioresource Technol. 100, 4829–4835. doi: 10.1016/j.biortech.2009.04.036
Doddapaneni H., Chakraborty R., Yadav J. S. (2005). Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus phanerochaete chrysosporium: Evidence for gene duplications and extensive gene clustering. BMC Genomics 6, 92. doi: 10.1186/1471-2164-6-92
Dritsa V., Rigas F., Doulia D., Avramides E., Hatzianestis I. (2009). Optimization of culture conditions for the biodegradation of lindane by the polypore fungus ganoderma australe. Water air Soil pollut. 204, 19–27. doi: 10.1007/s11270-009-0022-z
Dubey S., Abhyankar H., Marchante V., Brighton J., Blackburn K. (2016). Chronological review of the catalytic progress of polylactic acid formation through ring opening polymerization. Int. Res. J. Pure Appl. Chem. 12, 1–20. doi: 10.9734/IRJPAC/2016/27469
Dudka S., Adriano D. C. (1997). Environmental impacts of metal ore mining and processing: A review. J. Environ. Qual. 26, 590–602. doi: 10.2134/jeq1997.00472425002600030003x
Dusengemungu L., Kasali G., Gwanama C., Ouma K. O. (2020). Recent advances in biosorption of copper and cobalt by filamentous fungi. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.582016
Ehlers G. A., Rose P. D. (2005). Immobilized white-rot fungal biodegradation of phenol and chlorinated phenol in trickling packed-bed reactors by employing sequencing batch operation. Bioresource Technol. 96, 1264–1275. doi: 10.1016/j.biortech.2004.10.015
El-Gendi H., Saleh A. K., Badierah R., Redwan E. M., El-Maradny Y. A., El-Fakharany E. M. (2021). A comprehensive insight into fungal enzymes: Structure, classification, and their role in mankind's challenges. J. Fungi (Basel) 8 (1), 23. doi: 10.3390/jof8010023
Ellegaard-Jensen L., Knudsen B. E., Johansen A., Albers C. N., Aamand J., Rosendahl S. (2014). Fungal-bacterial consortia increase diuron degradation in water-unsaturated systems. Sci. Total Environ. 466-467, 699–705. doi: 10.1016/j.scitotenv.2013.07.095
Emadian S. M., Onay T. T., Demirel B. (2017). Biodegradation of bioplastics in natural environments. Waste Manage. 59, 526–536. doi: 10.1016/j.wasman.2016.10.006
Ernst W. (1980). Effects of pesticides and related organic compounds in the sea. Helgoländer Meeresuntersuchungen 33, 301. doi: 10.1007/BF02414756
Esteve-Núñez A., Caballero A., Ramos J. L. (2001). Biological degradation of 2,4,6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 65, 335–352. doi: 10.1128/MMBR.65.3.335-352.2001
Ezzouhri L., Castro E., Moya M., Espínola F., Lairini K. (2009). Heavy metal tolerance of filamentous fungi isolated from polluted sites in tangier, Morocco. Afr. J. Microbiol. Res. 3, 35–48.
Fashola M. O., Ngole-Jeme V. M., Babalola O. O. (2016). Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health 13, 1047. doi: 10.3390/ijerph13111047
Frascaroli G., Reid D., Hunter C., Roberts J., Helwig K., Spencer J., et al. (2021). Pharmaceuticals in wastewater treatment plants: A systematic review on the substances of greatest concern responsible for the development of antimicrobial resistance. Applied Sciences 11 (15), 6670. doi: 10.20944/preprints202106.0672.v1
Frère L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. (2018). Microplastic bacterial communities in the bay of Brest: Influence of polymer type and size. Environ. pollut. 242, 614–625. doi: 10.1016/j.envpol.2018.07.023
Frey S. D., Elliott E. T., Paustian K. (1999). Bacterial and fungal abundance and biomass in conventional and no-tillage agroecosystems along two climatic gradients. Soil Biol. Biochem. 31, 573–585. doi: 10.1016/S0038-0717(98)00161-8
Furuno S., Remer R., Chatzinotas A., Harms H., Wick L. (2012). Use of mycelia as paths for the isolation of contaminant‐degrading bacteria from soil. Microbial biotechnology 5 (1), 142–148.
Gajendiran A., Krishnamoorthy S., Abraham J. (2016). Microbial degradation of low-density polyethylene (LDPE) by aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech. 6, 52–52. doi: 10.1007/s13205-016-0394-x
Gao R., Liu R., Sun C. (2022). A marine fungus alternaria alternata FB1 efficiently degrades polyethylene. J. Hazardous Materials 431, 128617. doi: 10.1016/j.jhazmat.2022.128617
Ge M., Wang X., Yang G., Wang Z., Li Z., Zhang X., et al. (2021). Persistent organic pollutants (POPs) in deep-sea sediments of the tropical western pacific ocean. Chemosphere 277, 130267. doi: 10.1016/j.chemosphere.2021.130267
Gewert B., Plassmann M. M., Macleod M. (2015). Pathways for degradation of plastic polymers floating in the marine environment. Environ. Science: Processes Impacts 17, 1513–1521. doi: 10.1039/C5EM00207A
Geyer R., Jambeck J. R., Law K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3, e1700782–e1700782. doi: 10.1126/sciadv.1700782
Ghosh P., Swati, Thakur I. S. (2014). Enhanced removal of COD and color from landfill leachate in a sequential bioreactor. Bioresour Technol. 170, 10–19. doi: 10.1016/j.biortech.2014.07.079
Giani D., Baini M., Galli M., Casini S., Fossi M. C. (2019). Microplastics occurrence in edible fish species (Mullus barbatus and merluccius merluccius) collected in three different geographical sub-areas of the Mediterranean Sea. Mar. pollut. Bull. 140, 129–137. doi: 10.1016/j.marpolbul.2019.01.005
Gómez-Toribio V., García-Martín A. B., Martínez M. J., Martínez A. T., Guillén F. (2009). Enhancing the production of hydroxyl radicals by pleurotus eryngii via quinone redox cycling for pollutant removal. Appl. Environ. Microbiol. 75, 3954–3962. doi: 10.1128/AEM.02138-08
Gonçalves M. S., Sampaio S. C., Sene L., Suszek F. L., Coelho S. R., Bravo C. E. (2012). Isolation of filamentous fungi present in swine wastewater that are resistant and with the ability to remove atrazine. Afr. J. Biotechnol. 11, 11074–11077. doi: 10.5897/AJB11.4018
Gonda K., Jendrossek D., Molitoris H.-P. (2000). “Fungal degradation of the thermoplastic polymer poly-ß-hydroxybutyric acid (PHB) under simulated deep sea pressure,” in Life at interfaces and under extreme conditions (Springer), 173–183.
González-González R. B., Flores-Contreras E. A., Parra-Saldívar R., Iqbal H. M. N. (2022). Bio-removal of emerging pollutants by advanced bioremediation techniques. Environ. Res. 214, 113936. doi: 10.1016/j.envres.2022.113936
Goodell B., Winandy J. E., Morrell J. J. (2020). Fungal degradation of wood: Emerging data, new insights and changing perceptions. Coatings 10, 1210. doi: 10.3390/coatings10121210
Goudriaan M., Morales V. H., van der Meer M. T. J., Mets A., Ndhlovu R. T., van Heerwaarden J., et al (2023). A stable isotope assay with 13C-labeled polyethylene to investigate plastic mineralization mediated by Rhodococcus ruber. Marine Pollution Bulletin 186, 114369. doi: 10.1016/j.marpolbul.2022.114369
Goutam J., Sharma J., Singh R., Sharma D. (2021). “Fungal-mediated bioremediation of heavy metal–polluted environment,” in Microbial rejuvenation of polluted environment: Volume 2. Eds. Panpatte D. G., Jhala Y. K. (Singapore: Springer Singapore), 51–76.
Gregory M. (2009). Environmental implications of plastic debris in marine settings–entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 364, 2013–2025. doi: 10.1098/rstb.2008.0265
Grijalva-Bustamante G. A., Evans-Villegas A. G., Del Castillo-Castro T., Castillo-Ortega M. M., Cruz-Silva R., Huerta F., et al. (2016). Enzyme mediated synthesis of polypyrrole in the presence of chondroitin sulfate and redox mediators of natural origin. Mater Sci. Eng. C Mater Biol. Appl. 63, 650–656. doi: 10.1016/j.msec.2016.03.042
Guerranti C., Martellini T., Perra G., Scopetani C., Cincinelli A. (2019). Microplastics in cosmetics: Environmental issues and needs for global bans. Environ. Toxicol. Pharmacol. 68, 75–79. doi: 10.1016/j.etap.2019.03.007
Guillén-Jiménez F. D. M., Cristiani-Urbina E., Cancino-Díaz J. C., Flores-Moreno J. L., Barragán-Huerta B. E. (2012). Lindane biodegradation by the fusarium verticillioides AT-100 strain, isolated from agave tequilana leaves: Kinetic study and identification of metabolites. Int. Biodeterioration Biodegradation 74, 36–47. doi: 10.1016/j.ibiod.2012.04.020
Guliyev V., Tanunchai B., Noll M., Buscot F., Purahong W., Blagodatskaya E. (2022). Links among microbial communities, soil properties and functions: Are fungi the sole players in decomposition of bio-based and biodegradable plastic? Polymers 14, 2801. doi: 10.3390/polym14142801
Halaouli S., Asther M., Sigoillot J. C., Hamdi M., Lomascolo A. (2006). Fungal tyrosinases: New prospects in molecular characteristics, bioengineering and biotechnological applications. J. Appl. Microbiol. 100, 219–232. doi: 10.1111/j.1365-2672.2006.02866.x
Harms H., Schlosser D., Wick L. Y. (2011). Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 9, 177–192. doi: 10.1038/nrmicro2519
Harrison J. P., Schratzberger M., Sapp M., Osborn A. M. (2014). Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 14, 232. doi: 10.1186/s12866-014-0232-4
Hassan S. H., Kim S. J., Jung A. Y., Joo J. H., Eun Oh S., Yang J. E. (2009). Biosorptive capacity of Cd(II) and Cu(II) by lyophilized cells of pseudomonas stutzeri. J. Gen. Appl. Microbiol. 55, 27–34. doi: 10.2323/jgam.55.27
Hata T., Kawai S., Okamura H., Nishida T. (2010). Removal of diclofenac and mefenamic acid by the white rot fungus phanerochaete sordida YK-624 and identification of their metabolites after fungal transformation. Biodegradation 21, 681–689. doi: 10.1007/s10532-010-9334-3
Hawksworth D. L., Lücking R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5 (4). doi: 10.1128/microbiolspec
He M., Jia C., Zhao E., Chen L., Yu P., Jing J., et al. (2016). Concentrations and dissipation of difenoconazole and fluxapyroxad residues in apples and soil, determined by ultrahigh-performance liquid chromatography electrospray ionization tandem mass spectrometry. Environ. Sci. pollut. Res. Int. 23, 5618–5626. doi: 10.1007/s11356-015-5750-6
Hiratsuka N., Wariishi H., Tanaka H. (2001). Degradation of diphenyl ether herbicides by the lignin-degrading basidiomycete coriolus versicolor. Appl. Microbiol. Biotechnol. 57, 563–571. doi: 10.1007/s002530100789
Hofrichter M. (2002). Review: Lignin conversion by manganese peroxidase (MnP). Enzyme Microbial Technol. 30, 454–466. doi: 10.1016/S0141-0229(01)00528-2
Hofrichter M., Ullrich R., Pecyna M. J., Liers C., Lundell T. (2010). New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87, 871–897. doi: 10.1007/s00253-010-2633-0
Horio T., Oakley B. R. (2005). The role of microtubules in rapid hyphal tip growth of aspergillus nidulans. Mol. Biol. Cell 16, 918–926. doi: 10.1091/mbc.e04-09-0798
Hu K., Barbieri M. V., López-García E., Postigo C., López De Alda M., Caminal G., et al. (2022). Fungal degradation of selected medium to highly polar pesticides by trametes versicolor: Kinetics, biodegradation pathways, and ecotoxicity of treated waters. Analytical Bioanalytical Chem. 414, 439–449. doi: 10.1007/s00216-021-03267-x
Hu K., Peris A., Torán J., Eljarrat E., Sarrà M., Blánquez P., et al. (2020). Exploring the degradation capability of trametes versicolor on selected hydrophobic pesticides through setting sights simultaneously on culture broth and biological matrix. Chemosphere 250, 126293. doi: 10.1016/j.chemosphere.2020.126293
Huang H., Zhang S., Shan X.-Q., Chen B.-D., Zhu Y.-G., Bell J. N. B. (2007). Effect of arbuscular mycorrhizal fungus (Glomus caledonium) on the accumulation and metabolism of atrazine in maize (Zea mays l.) and atrazine dissipation in soil. Environ. pollut. 146, 452–457. doi: 10.1016/j.envpol.2006.07.001
Hundt K., Martin D., Hammer E., Jonas U., Kindermann M. K., Schauer F. (2000). Transformation of triclosan by trametes versicolor and pycnoporus cinnabarinus. Appl. Environ. Microbiol. 66, 4157–4160. doi: 10.1128/AEM.66.9.4157-4160.2000
Hyde G. J., Davies D., Perasso L., Cole L., Ashford A. E. (1999). Microtubules, but not actin microfilaments, regulate vacuole motility and morphology in hyphae of pisolithus tinctorius. Cell Motil. Cytoskeleton 42, 114–124. doi: 10.1002/(SICI)1097-0169(1999)42:2<114::AID-CM3>3.0.CO;2-N
Hyde K. D., Xu J., Rapior S., Jeewon R., Lumyong S., Niego A. G. T., et al. (2019). The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Diversity 97, 1–136. doi: 10.1007/s13225-019-00430-9
Ihsanullah I., Jamal A., Ilyas M., Zubair M., Khan G., Atieh M. A. (2020). Bioremediation of dyes: Current status and prospects. J. Water Process Eng. 38, 101680. doi: 10.1016/j.jwpe.2020.101680
Ingham E., Griffiths R., Cromack K., Entry J. (1991). Comparison of direct vs fumigation incubation microbial biomass estimates from ectomycorrhizal mat and non-mat soils. Soil Biol. Biochem. 23, 465–471. doi: 10.1016/0038-0717(91)90011-8
Iyer A., Mody K., Jha B. (2005). Biosorption of heavy metals by a marine bacterium. Mar. pollut. Bull. 50, 340–343. doi: 10.1016/j.marpolbul.2004.11.012
Jaén-Gil A., Castellet-Rovira F., Llorca M., Villagrasa M., Sarrà M., Rodríguez-Mozaz S., et al. (2019). Fungal treatment of metoprolol and its recalcitrant metabolite metoprolol acid in hospital wastewater: Biotransformation, sorption and ecotoxicological impact. Water Res. 152, 171–180. doi: 10.1016/j.watres.2018.12.054
Jain M. R., Zinjarde S. S., Deobagkar D. D., Deobagkar D. N. (2004). 2,4,6-trinitrotoluene transformation by a tropical marine yeast, yarrowia lipolytica NCIM 3589. Mar. pollut. Bull. 49, 783–788. doi: 10.1016/j.marpolbul.2004.06.007
Jaiswal S., Shukla P. (2020). Alternative strategies for microbial remediation of pollutants via synthetic biology. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00808
Jaiswal S., Singh D. K., Shukla P. (2019). Gene editing and systems biology tools for pesticide bioremediation: A review. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00087
Jambeck J. R., Geyer R., Wilcox C., Siegler T. R., Perryman M., Andrady A., et al. (2015). Marine pollution. plastic waste inputs from land into the ocean. Science 347, 768–771. doi: 10.1126/science.1260352
Janusz G., Pawlik A., Sulej J., Świderska-Burek U., Jarosz-Wilkołazka A., Paszczyński A. (2017). Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41, 941–962. doi: 10.1093/femsre/fux049
Jarerat A., Tokiwa Y. (2001). Degradation of Poly(L-lactide) by a fungus. Macromol. Bioscience 1, 136–140. doi: 10.1002/1616-5195(20010601)1:4<136::AID-MABI136>3.0.CO;2-3
Jensen K. A. Jr., Houtman C. J., Ryan Z. C., Hammel K. E. (2001). Pathways for extracellular fenton chemistry in the brown rot basidiomycete gloeophyllum trabeum. Appl. Environ. Microbiol. 67, 2705–2711. doi: 10.1128/AEM.67.6.2705-2711.2001
Junier P., Cailleau G., Palmieri I., Vallotton C., Trautschold O. C., Junier T., et al. (2021). Democratization of fungal highway columns as a tool to investigate bacteria associated with soil fungi. FEMS Microbiol. Ecol. 97 (2), fiab003. doi: 10.1093/femsec/fiab003
Jureczko M., Przystaś W., Krawczyk T., Gonciarz W., Rudnicka K. (2021). White-rot fungi-mediated biodegradation of cytostatic drugs - bleomycin and vincristine. J. Hazard Mater 407, 124632. doi: 10.1016/j.jhazmat.2020.124632
Kang B. R., Kim S. Y., Kang M., Lee T. K. (2021). Removal of pharmaceuticals and personal care products using native fungal enzymes extracted during the ligninolytic process. Environ. Res. 195, 110878. doi: 10.1016/j.envres.2021.110878
Karamanlioglu M., Alkan Ü. (2020). Influence of degradation of PLA with high degree of crystallinity on fungal community structure in compost. Compost Sci. Utilization 28, 169–178. doi: 10.1080/1065657X.2020.1864514
Karimi F., Shariatifar N., Rezaei M., Alikord M., Arabameri M. (2021). Quantitative measurement of toxic metals and assessment of health risk in agricultural products food from markazi province of Iran. Int. J. Food Contamination 8, 2. doi: 10.1186/s40550-021-00083-0
Kasai N., Ikushiro S., Shinkyo R., Yasuda K., Hirosue S., Arisawa A., et al. (2010). Metabolism of mono- and dichloro-dibenzo-p-dioxins by phanerochaete chrysosporium cytochromes P450. Appl. Microbiol. Biotechnol. 86, 773–780. doi: 10.1007/s00253-009-2413-x
Kasuya K.-I., Ishii N., Inoue Y., Yazawa K., Tagaya T., Yotsumoto T., et al. (2009). Characterization of a mesophilic aliphatic–aromatic copolyester-degrading fungus. Polymer degradation stability 94, 1190–1196. doi: 10.1016/j.polymdegradstab.2009.04.013
Kayode-Afolayan S. D., Ahuekwe E. F., Nwinyi O. C. (2022). Impacts of pharmaceutical effluents on aquatic ecosystems. Sci. Afr. 17, e01288. doi: 10.1016/j.sciaf.2022.e01288
Kelly A., Lannuzel D., Rodemann T., Meiners K. M., Auman H. J. (2020). Microplastic contamination in east Antarctic sea ice. Mar. pollut. Bull. 154, 111130. doi: 10.1016/j.marpolbul.2020.111130
Kettner M. T., Oberbeckmann S., Labrenz M., Grossart H.-P. (2019). The eukaryotic life on microplastics in brackish ecosystems. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00538
Kettner M. T., Rojas-Jimenez K., Oberbeckmann S., Labrenz M., Grossart H.-P. (2017). Microplastics alter composition of fungal communities in aquatic ecosystems. Environ. Microbiol. 19, 4447–4459. doi: 10.1111/1462-2920.13891
Khan S., Ali S. A., Ali A. S. (2022). Biodegradation of low density polyethylene (LDPE) by mesophilic fungus ‘Penicillium citrinum’ isolated from soils of plastic waste dump yard, Bhopal, India. Environ. Technol. 1–15. doi: 10.1080/09593330.2022.2027025
Kim K.-H., Kabir E., Jahan S. A. (2017). Exposure to pesticides and the associated human health effects. Sci. Total Environ. 575, 525–535. doi: 10.1016/j.scitotenv.2016.09.009
Kirstein I. V., Wichels A., Krohne G., Gerdts G. (2018). Mature biofilm communities on synthetic polymers in seawater - specific or general? Mar. Environ. Res. 142, 147–154. doi: 10.1016/j.marenvres.2018.09.028
Kjøller A., Struwe S. (1982). Microfungi in ecosystems: Fungal occurrence and activity in litter and soil. Oikos 39 (3), 391–422. doi: 10.2307/3544690
Klaschka U. (2016). Natural personal care products–analysis of ingredient lists and legal situation. Environ. Sci. Europe 28, 8. doi: 10.1186/s12302-016-0076-7
Knudsen B. E., Ellegaard-Jensen L., Albers C. N., Rosendahl S., Aamand J. (2013). Fungal hyphae stimulate bacterial degradation of 2,6-dichlorobenzamide (BAM). Environ. pollut. 181, 122–127. doi: 10.1016/j.envpol.2013.06.013
Koelmans A. A., Kooi M., Law K. L., Van Sebille E. (2017). All is not lost: deriving a top-down mass budget of plastic at sea. Environ. Res. Lett. 12, 114028. doi: 10.1088/1748-9326/aa9500
Kohlmeier S., Smits T. H. M., Ford R. M., Keel C., Harms H., Wick L. Y. (2005). Taking the fungal highway: Mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39, 4640–4646. doi: 10.1021/es047979z
Köller G., Möder M., Czihal K. (2000). Peroxidative degradation of selected PCB: a mechanistic study. Chemosphere 41, 1827–1834. doi: 10.1016/S0045-6535(00)00132-6
Kramer C., Kreisel G., Fahr K., Kässbohrer J., Schlosser D. (2004). Degradation of 2-fluorophenol by the brown-rot fungus gloeophyllum striatum: Evidence for the involvement of extracellular fenton chemistry. Appl. Microbiol. Biotechnol. 64, 387–395. doi: 10.1007/s00253-003-1445-x
Kullman S. W., Matsumura F. (1996). Metabolic pathways utilized by phanerochaete chrysosporium for degradation of the cyclodiene pesticide endosulfan. Appl. Environ. Microbiol. 62, 593–600. doi: 10.1128/aem.62.2.593-600.1996
Kumar A., Bisht B. S., Joshi V. D., Dhewa T. (2011). Review on bioremediation of polluted environment: A management tool. International journal of environmental sciences 1 (6), 1079–1093. doi: 10.1080/10590501.2019.1654809
Kumar S. S., Ghosh P., Malyan S. K., Sharma J., Kumar V. (2019). A comprehensive review on enzymatic degradation of the organophosphate pesticide malathion in the environment. J. Environ. Sci. Health C Environ. Carcinog Ecotoxicol Rev. 37, 288–329. doi: 10.1080/10590501.2019.1654809
Kumar M., Yadav A. N., Saxena R., Paul D., Tomar R. S. (2021). Biodiversity of pesticides degrading microbial communities and their environmental impact. Biocatalysis Agric. Biotechnol. 31, 101883. doi: 10.1016/j.bcab.2020.101883
Kumari M., Ghosh P., Joshi S., Thakur I. (2014). Microcosmic study of endosulfan degradation by paenibacillus sp. ISTP10 and its toxicological evaluation using mammalian cell line. Int. Biodeterioration Biodegradation 96, 33–40. doi: 10.1016/j.ibiod.2014.08.003
Kupski L., Salcedo G. M., Caldas S. S., De Souza T. D., Furlong E. B., Primel E. G. (2019). Optimization of a laccase-mediator system with natural redox-mediating compounds for pesticide removal. Environ. Sci. pollut. Res. Int. 26, 5131–5139. doi: 10.1007/s11356-018-4010-y
Kurtela A., Antolović N. (2019). The problem of plastic waste and microplastics in the seas and oceans: impact on marine organisms. Croatian J. Fisheries 77, 51–56. doi: 10.2478/cjf-2019-0005
Lacerda A. L. D. F., Proietti M. C., Secchi E. R., Taylor J. D. (2020). Diverse groups of fungi are associated with plastics in the surface waters of the Western south Atlantic and the Antarctic peninsula. Mol. Ecol. 29, 1903–1918. doi: 10.1111/mec.15444
Lebreton L. C. M., van der Zwet J., Damsteeg J.-W., Slat B., Andrady A., Reisser J. (2017). River plastic emissions to the world’s oceans. Nat. Commun. 8, 15611. doi: 10.1038/ncomms15611
Ledrich M. L., Stemmler S., Laval-Gilly P., Foucaud L., Falla J. (2005). Precipitation of silver-thiosulfate complex and immobilization of silver by cupriavidus metallidurans CH34. Biometals 18, 643–650. doi: 10.1007/s10534-005-3858-8
Lei K., Qiao F., Liu Q., Wei Z., Qi H., Cui S., et al. (2017). Microplastics releasing from personal care and cosmetic products in China. Mar. pollut. Bull. 123 (1-2), 122–126. doi: 10.1016/j.marpolbul.2017.09.016
Leong Y. K., Chang J.-S. (2020). Bioremediation of heavy metals using microalgae: Recent advances and mechanisms. Bioresource Technol. 303, 122886. doi: 10.1016/j.biortech.2020.122886
Leung W. C., Chua H., Lo W. (2001). Biosorption of heavy metals by bacteria isolated from activated sludge. Appl. Biochem. Biotechnol. 91-93, 171–184. doi: 10.1385/ABAB:91-93:1-9:171
Li Q., Liu J., Gadd G. M. (2020). Fungal bioremediation of soil co-contaminated with petroleum hydrocarbons and toxic metals. Appl. Microbiol. Biotechnol. 104, 8999–9008. doi: 10.1007/s00253-020-10854-y
Li Z. J., Shukla V., Fordyce A. P., Pedersen A. G., Wenger K. S., Marten M. R. (2000). Fungal morphology and fragmentation behavior in a fed-batch aspergillus oryzae fermentation at the production scale. Biotechnol. Bioeng 70, 300–312. doi: 10.1002/1097-0290(20001105)70:3<300::AID-BIT7>3.0.CO;2-3
Lindahl B., Finlay R., Olsson S. (2002). Simultaneous, bidirectional translocation of 32P and 33P between wood blocks connected by mycelial cords of hypholoma fasciculare. New Phytol. 150, 189–194. doi: 10.1046/j.1469-8137.2001.00074.x
Lipsa R., Tudorachi N., Darie-Nita R. N., Oprică L., Vasile C., Chiriac A. (2016). Biodegradation of poly (lactic acid) and some of its based systems with trichoderma viride. Int. J. Biol. macromolecules 88, 515–526. doi: 10.1016/j.ijbiomac.2016.04.017
Lishman L., Smyth S. A., Sarafin K., Kleywegt S., Toito J., Peart T., et al. (2006). Occurrence and reductions of pharmaceuticals and personal care products and estrogens by municipal wastewater treatment plants in Ontario, Canada. Sci. Total Environ. 367, 544–558. doi: 10.1016/j.scitotenv.2006.03.021
Loganathan B. G., Kannan K. (1991). Time perspectives of organochlorine contamination in the global environment. Mar. pollut. Bull. 22, 582–584. doi: 10.1016/0025-326X(91)90244-M
Lotlikar N. P., Damare S. R., Meena R. M., Linsy P., Mascarenhas B. (2018). Potential of marine-derived fungi to remove hexavalent chromium pollutant from culture broth. Indian J. Microbiol. 58, 182–192. doi: 10.1007/s12088-018-0719-z
Lottermoser B. (2010). Mine wastes: Characterization, treatment and environmental impacts. Environmental Science.
Lykogianni M., Bempelou E., Karamaouna F., Aliferis K. A. (2021). Do pesticides promote or hinder sustainability in agriculture? The challenge of sustainable use of pesticides in modern agriculture. Sci. Total Environ. 795, 148625. doi: 10.1016/j.scitotenv.2021.148625
Macleod M., Arp H. P. H., Tekman M. B., Jahnke A. (2021). The global threat from plastic pollution. Science 373, 61–65. doi: 10.1126/science.abg5433
Maeda H., Yamagata Y., Abe K., Hasegawa F., Machida M., Ishioka R., et al. (2005). Purification and characterization of a biodegradable plastic-degrading enzyme from aspergillus oryzae. Appl. Microbiol. Biotechnol. 67, 778–788. doi: 10.1007/s00253-004-1853-6
Magan N., Fragoeiro S., Bastos C. (2010). Environmental factors and bioremediation of xenobiotics using white rot fungi. Mycobiology 38, 238–248. doi: 10.4489/MYCO.2010.38.4.238
Mahmud T., Sabo I. A., Lambu Z. N., Danlami D., Shehu A. A. (2022). Hydrocarbon degradation potentials of fungi: A review. J. Environ. Bioremediation Toxicol. 5, 50–56. doi: 10.54987/jebat.v5i1.681
Majcherczyk A., Johannes C., Hüttermann A. (1998). Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of trametes versicolor. Enzyme Microbial Technol. 22, 335–341. doi: 10.1016/S0141-0229(97)00199-3
Malachová K., Novotný Č., Adamus G., Lotti N., Rybková Z., Soccio M., et al. (2020). Ability of trichoderma hamatum isolated from plastics-polluted environments to attack petroleum-based, synthetic polymer films. Processes 8, 467. doi: 10.3390/pr8040467
Mansouri A., Cregut M., Abbes C., Durand M.-J., Landoulsi A., Thouand G. (2017). The environmental issues of DDT pollution and bioremediation: A multidisciplinary review. Appl. Biochem. Biotechnol. 181, 309–339. doi: 10.1007/s12010-016-2214-5
Maqbool Z., Hussain S., Imran M., Mahmood F., Shahzad T., Ahmed Z., et al. (2016). Perspectives of using fungi as bioresource for bioremediation of pesticides in the environment: a critical review. Environ. Sci. pollut. Res. Int. 23, 16904–16925. doi: 10.1007/s11356-016-7003-8
Marco-Urrea E., Aranda E., Caminal G., Guillén F. (2009c). Induction of hydroxyl radical production in trametes versicolor to degrade recalcitrant chlorinated hydrocarbons. Bioresour Technol. 100, 5757–5762. doi: 10.1016/j.biortech.2009.06.078
Marco-Urrea E., PéRez-Trujillo M. R., Cruz-Morató C., Caminal G., Vicent T. (2009b). White-rot fungus-mediated degradation of the analgesic ketoprofen and identification of intermediates by HPLC–DAD–MS and NMR. Chemosphere 78, 474–481. doi: 10.1016/j.chemosphere.2009.10.009
Marco-Urrea E., Pérez-Trujillo M., Cruz-Morató C., Caminal G., Vicent T. (2010). Degradation of the drug sodium diclofenac by trametes versicolor pellets and identification of some intermediates by NMR. J. Hazard Mater 176, 836–842. doi: 10.1016/j.jhazmat.2009.11.112
Marco-Urrea E., Pérez-Trujillo M., Vicent T., Caminal G. (2009a). Ability of white-rot fungi to remove selected pharmaceuticals and identification of degradation products of ibuprofen by trametes versicolor. Chemosphere 74, 765–772. doi: 10.1016/j.chemosphere.2008.10.040
Marques A. P., Rangel A. O., Castro P. M. (2009). Remediation of heavy metal contaminated soils: Phytoremediation as a potentially promising clean-up technology. Crit. Rev. Environ. Sci. Technol. 39, 622–654. doi: 10.1080/10643380701798272
Masaki K., Kamini N. R., Ikeda H., Iefuji H. (2005). Cutinase-like enzyme from the yeast cryptococcus sp. strain s-2 hydrolyzes polylactic acid and other biodegradable plastics. Appl. Environ. Microbiol. 71, 7548–7550. doi: 10.1128/AEM.71.11.7548-7550.2005
Masiá P., Sol D., Ardura A., Laca A., Borrell Y. J., Dopico E., et al. (2020). Bioremediation as a promising strategy for microplastics removal in wastewater treatment plants. Mar. pollut. Bull. 156, 111252. doi: 10.1016/j.marpolbul.2020.111252
Materić D., Holzinger R., Niemann H. (2022). Nanoplastics and ultrafine microplastic in the Dutch wadden Sea – the hidden plastics debris? Sci. Total Environ. 846, 157371. doi: 10.1016/j.scitotenv.2022.157371
Mathai A., Bhanu M. S. (2010). Acute aluminium phosphide poisoning: Can we predict mortality? Indian J. Anaesth 54, 302–307. doi: 10.4103/0019-5049.68372
Meharg A. A. (2003). The mechanistic basis of interactions between mycorrhizal associations and toxic metal cations. Mycol Res. 107, 1253–1265. doi: 10.1017/S0953756203008608
Miao L., Wang P., Hou J., Yao Y., Liu Z., Liu S., et al. (2019). Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 650, 2395–2402. doi: 10.1016/j.scitotenv.2018.09.378
Miri S., Naghdi M., Rouissi T., Kaur Brar S., Martel R. (2019). Recent biotechnological advances in petroleum hydrocarbons degradation under cold climate conditions: A review. Crit. Rev. Environ. Sci. Technol. 49, 553–586. doi: 10.1080/10643389.2018.1552070
Mir-Tutusaus J. A., Caminal G., Sarrà M. (2018). Influence of process variables in a continuous treatment of non-sterile hospital wastewater by trametes versicolor and novel method for inoculum production. J. Environ. Manage 212, 415–423. doi: 10.1016/j.jenvman.2018.02.018
Mir Tutusaus J. A., Sarrà M., Caminal G. (2016). Continuous treatment of non-sterile hospital wastewater by trametes versicolor: How to increase fungal viability by means of operational strategies and pretreatments. J. Hazardous Materials 318, 561–570.
Miyazaki S., Takahashi K., Shiraki M., Saito T., Tezuka Y., Kasuya K.-I. (2000). Properties of a Poly(3-hydroxybutyrate) depolymerase from penicillium funiculosum. J. Polymers Environ. 8, 175–182. doi: 10.1023/A:1015245710406
Mohana A. A., Farhad S. M., Haque N., Pramanik B. K. (2021). Understanding the fate of nano-plastics in wastewater treatment plants and their removal using membrane processes. Chemosphere 284, 131430. doi: 10.1016/j.chemosphere.2021.131430
Mori T., Ohno H., Ichinose H., Kawagishi H., Hirai H. (2021). White-rot fungus phanerochaete chrysosporium metabolizes chloropyridinyl-type neonicotinoid insecticides by an n-dealkylation reaction catalyzed by two cytochrome P450s. J. Hazardous Materials 402, 123831. doi: 10.1016/j.jhazmat.2020.123831
Morin-Crini N., Lichtfouse E., Fourmentin M., Ribeiro A. R. L., Noutsopoulos C., Mapelli F., et al. (2022). Removal of emerging contaminants from wastewater using advanced treatments. a review. Environ. Chem. Lett. 20, 1333–1375. doi: 10.1007/s10311-021-01379-5
Mougin C., Pericaud C., Malosse C., Laugero C., Asther M. (1996). Biotransformation of the insecticide lindane by the white rot basidiomycete phanerochaete chrysosporium. Pesticide Sci. 47, 51–59. doi: 10.1002/(SICI)1096-9063(199605)47:1<51::AID-PS391>3.0.CO;2-V
Mouriño-Pérez R. R., Roberson R. W., Bartnicki-García S. (2006). Microtubule dynamics and organization during hyphal growth and branching in neurospora crassa. Fungal Genet. Biol. 43, 389–400. doi: 10.1016/j.fgb.2005.10.007
Munir E., Harefa R. S. M., Priyani N., Suryanto D. (2018). “Plastic degrading fungi trichoderma viride and aspergillus nomius isolated from local landfill soil in medan,” in IOP Conference Series: Earth and Environmental Science (IOP Publishing), Vol. 126 (1), 012145.
Mura E., Edwards J., Kittelmann S., Kaerger K., Voigt K., Mrázek J., et al. (2018). Anaerobic fungal communities differ along the horse digestive tract. Fungal Biol. 123 (3), 240–246.
Murphy C. A., Cameron J. A., Huang S. J., Vinopal R. T. (1996). Fusarium polycaprolactone depolymerase is cutinase. Appl. Environ. Microbiol. 62, 456. doi: 10.1128/aem.62.2.456-460.1996
Nadhman A., Hasan F., Shah Z., Hameed A., Shah A. (2012). Production of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) depolymerase from aspergillus sp. NA-25. Appl. Biochem. Microbiol. 48, 482–487. doi: 10.1134/S0003683812050080
Nielsen J., Krabben P. (1995). Hyphal growth and fragmentation of penicillium chrysogenum in submerged cultures. Biotechnol. Bioeng 46, 588–598. doi: 10.1002/bit.260460612
Ning D., Wang H., Zhuang Y. (2010). Induction of functional cytochrome P450 and its involvement in degradation of benzoic acid by phanerochaete chrysosporium. Biodegradation 21, 297–308. doi: 10.1007/s10532-009-9301-z
Noman M. A., Feng W., Zhu G., Hossain M. B., Chen Y., Zhang H., et al. (2022). Bioaccumulation and potential human health risks of metals in commercially important fishes and shellfishes from hangzhou bay, China. Sci. Rep. 12,4634. doi: 10.1038/s41598-022-08471-y
Nyakundi W., Magoma G., Ochora J., Nyende A. (2011). Biodegradation of diazinon and methomyl pesticides by white rot fungi from selected horticultural farms in rift valley and central Kenya. J Appl Technol Environ Sanit 1 (2), 107–124.
Nyanhongo G. S., Couto S. R., Guebitz G. M. (2006). Coupling of 2,4,6-trinitrotoluene (TNT) metabolites onto humic monomers by a new laccase from trametes modesta. Chemosphere 64, 359–370. doi: 10.1016/j.chemosphere.2005.12.034
Oberbeckmann S., Kreikemeyer B., Labrenz M. (2018). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02709
Oberbeckmann S., Osborn A. M., Duhaime M. B. (2016). Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PloS One 11, e0159289. doi: 10.1371/journal.pone.0159289
Ogilo J., Anam O., Salim A., Yusuf A. (2017). Assessment of levels of heavy metals in paints from interior walls and indoor dust from residential houses in Nairobi city county, Kenya. Chem. Sci. Int. J. 21, 1–7. doi: 10.9734/CSJI/2017/37392
Ojemaye C. Y., Petrik L. (2022). Pharmaceuticals and personal care products in the marine environment around false bay, cape town, south Africa: Occurrence and risk-assessment study. Environ. Toxicol. Chem. 41, 614–634. doi: 10.1002/etc.5053
Ojha N., Pradhan N., Singh S., Barla A., Shrivastava A., Khatua P., et al. (2017). Evaluation of HDPE and LDPE degradation by fungus, implemented by statistical optimization. Sci. Rep. 7, 39515. doi: 10.1038/srep39515
Onink V., Jongedijk C. E., Hoffman M. J., Van Sebille E., Laufkötter C. (2021). Global simulations of marine plastic transport show plastic trapping in coastal zones. Environ. Res. Lett. 16, 064053. doi: 10.1088/1748-9326/abecbd
Ortiz-Hernández M. L., Sánchez-Salinas E., Olvera-Velona A., Folch-Mallol J. L. (2011). Pesticides in the environment: Impacts and their biodegradation as a strategy for residues treatment. Pesticides - Formulations, Effects, Fate.
Oswal N., Sarma P. M., Zinjarde S. S., Pant A. (2002). Palm oil mill effluent treatment by a tropical marine yeast. Bioresource Technol. 85, 35–37. doi: 10.1016/S0960-8524(02)00063-9
Oulton R. L., Kohn T., Cwiertny D. M. (2010). Pharmaceuticals and personal care products in effluent matrices: A survey of transformation and removal during wastewater treatment and implications for wastewater management. J. Environ. Monit 12, 1956–1978. doi: 10.1039/c0em00068j
Oyetibo G. O., Ishola S. T., Ikeda-Ohtsubo W., Miyauchi K., Ilori M. O., Endo G. (2015). Mercury bioremoval by yarrowia strains isolated from sediments of mercury-polluted estuarine water. Appl. Microbiol. Biotechnol. 99, 3651–3657. doi: 10.1007/s00253-014-6279-1
Oyetibo G. O., Miyauchi K., Suzuki H., Endo G. (2016). Mercury removal during growth of mercury tolerant and self-aggregating yarrowia spp. AMB Express 6, 99. doi: 10.1186/s13568-016-0271-3
Paço A., Duarte K., Da Costa J. P., Santos P. S. M., Pereira R., Pereira M. E., et al. (2017). Biodegradation of polyethylene microplastics by the marine fungus zalerion maritimum. Sci. Total Environ. 586, 10–15. doi: 10.1016/j.scitotenv.2017.02.017
Patel M., Kumar R., Kishor K., Mlsna T., Pittman C. U., Mohan D. (2019). Pharmaceuticals of emerging concern in aquatic systems: Chemistry, occurrence, effects, and removal methods. Chem. Rev. 119, 3510–3673. doi: 10.1021/acs.chemrev.8b00299
Peng G., Bellerby R., Zhang F., Sun X., Li D. (2020). The ocean’s ultimate trashcan: Hadal trenches as major depositories for plastic pollution. Water Res. 168, 115121. doi: 10.1016/j.watres.2019.115121
Peng X., Valentine D. L. (2021). Diversity and N(2)O production potential of fungi in an oceanic oxygen minimum zone. J. Fungi (Basel) 7 (3), 218. doi: 10.3390/jof7030218
Pieja A. J., Morse M. C., Cal A. J. (2017). Methane to bioproducts: The future of the bioeconomy? Curr. Opin. Chem. Biol. 41, 123–131. doi: 10.1016/j.cbpa.2017.10.024
Podbielska M., Książek P., Szpyrka E. (2020). Dissipation kinetics and biological degradation by yeast and dietary risk assessment of fluxapyroxad in apples. Sci. Rep. 10, 21212. doi: 10.1038/s41598-020-78177-6
Pretty J., Hine R. (2012). “Pesticide use and the environment,” in The pesticide detox (Routledge), 23–44.
Pujari M., Kapoor D. (2021). “1 - heavy metals in the ecosystem: Sources and their effects,” in Heavy metals in the environment. Eds. Kumar V., Sharma A., Cerdà A. (Elsevier).
Purahong W., Wahdan S. F. M., Heinz D., Jariyavidyanont K., Sungkapreecha C., Tanunchai B., et al. (2021). Back to the future: Decomposability of a biobased and biodegradable plastic in field soil environments and its microbiome under ambient and future climates. Environ. Sci. Technol. 55, 12337–12351. doi: 10.1021/acs.est.1c02695
Purnomo A. S., Mori T., Kamei I., Nishii T., Kondo R. (2010). Application of mushroom waste medium from pleurotus ostreatus for bioremediation of DDT-contaminated soil. Int. Biodeterioration Biodegradation 64, 397–402. doi: 10.1016/j.ibiod.2010.04.007
Qin X., Zhang J., Zhang X., Yang Y. (2014). Induction, purification and characterization of a novel manganese peroxidase from irpex lacteus CD2 and its application in the decolorization of different types of dye. PloS One 9, e113282. doi: 10.1371/journal.pone.0113282
Rajendran P., Muthukrishnan J., Gunasekaran P. (2003). Microbes in heavy metal remediation. Indian J Exp Biol. 41 (9), 935–944.
Rankin W. J (2011). Minerals, metals and sustainability: Meeting future material needs (CSIRO publishing).
Rehman I. U., Ishaq M., Muhammad S., Din I. U., Khan S., Yaseen M. (2020). Evaluation of arsenic contamination and potential risks assessment through water, soil and rice consumption. Environ. Technol. Innovation 20, 101155. doi: 10.1016/j.eti.2020.101155
Rhee Y.-H., Kim D.-Y., Yun J.-H., Kim H.-W., Bae K.-S. (2002). Purification and characterization of poly (3-hydroxybutyrate) depolymerase from a fungal isolate, emericellopsis minima W2. J. Microbiol. 40, 129–133.
Rieble S., Joshi D. K., Gold M. H. (1994). Aromatic nitroreductase from the basidiomycete phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 205, 298–304. doi: 10.1006/bbrc.1994.2664
Rodarte-Morales A. I., Feijoo G., Moreira M. T., Lema J. M. (2011). Degradation of selected pharmaceutical and personal care products (PPCPs) by white-rot fungi. World J. Microbiol. Biotechnol. 27, 1839–1846. doi: 10.1007/s11274-010-0642-x
Rodgers C. J., Blanford C. F., Giddens S. R., Skamnioti P., Armstrong F. A., Gurr S. J. (2010). Designer laccases: A vogue for high-potential fungal enzymes? Trends Biotechnol. 28, 63–72. doi: 10.1016/j.tibtech.2009.11.001
Rodrigues G. N., Alvarenga N., Vacondio B., De Vasconcellos S. P., Passarini M. R. Z., Seleghim M. H. R., et al. (2016). Biotransformation of methyl parathion by marine-derived fungi isolated from ascidian didemnum ligulum. Biocatalysis Agric. Biotechnol. 7, 24–30. doi: 10.1016/j.bcab.2016.05.001
Rojas-Jimenez K., Rieck A., Wurzbacher C., Jürgens K., Labrenz M., Grossart H.-P. (2019). A salinity threshold separating fungal communities in the Baltic Sea. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00680
Romera-Castillo C., Pinto M., Langer T. M., Álvarez-Salgado X. A., Herndl G. J. (2018). Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat. Commun. 9, 1430. doi: 10.1038/s41467-018-03798-5
Rout P. R., Mohanty A., Aastha, Sharma A., Miglani M., Liu D., et al. (2022). Micro- and nanoplastics removal mechanisms in wastewater treatment plants: A review. J. Hazardous Materials Adv. 6, 100070. doi: 10.1016/j.hazadv.2022.100070
Roy P., Mohanty A. K., Misra M. (2022). Microplastics in ecosystems: Their implications and mitigation pathways. Environ. Science: Advances 1. doi: 10.1039/D1VA00012H
Rudin A., Choi P. (2013). “Chapter 13 - biopolymers,” in The elements of polymer science & engineering (Third edition). Eds. Rudin A., Choi P. (Boston: Academic Press), 521–535.
Rudnik E., Briassoulis D. (2011). Comparative biodegradation in soil behaviour of two biodegradable polymers based on renewable resources. J. Polymers Environ. 19, 18–39. doi: 10.1007/s10924-010-0243-7
Ruiz-Dueñas F. J., Morales M., García E., Miki Y., Martínez M. J., Martínez A. T. (2009). Substrate oxidation sites in versatile peroxidase and other basidiomycete peroxidases. J. Exp. Bot. 60, 441–452. doi: 10.1093/jxb/ern261
Sáenz M., Borodulina T., Diaz L., Banchon C. (2019). Minimal conditions to degrade low density polyethylene by aspergillus terreus and Niger. J. Ecol. Eng. 20 (6), 44–51. doi: 10.12911/22998993/108699
Sang B. I., Hori K., Tanji Y., Unno H. (2002). Fungal contribution to in situ biodegradation of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) film in soil. Appl. Microbiol. Biotechnol. 58, 241–247. doi: 10.1007/s00253-001-0884-5
Saravanan A., Kumar P. S., Govarthanan M., George C. S., Vaishnavi S., Moulishwaran B., et al. (2021). Adsorption characteristics of magnetic nanoparticles coated mixed fungal biomass for toxic Cr(VI) ions in aquatic environment. Chemosphere 267, 129226. doi: 10.1016/j.chemosphere.2020.129226
Savinelli B., Vega Fernandez T., Galasso N., D'anna G., Pipitone C., Prada F., et al. (2020). Microplastics impair the feeding performance of a Mediterranean habitat-forming coral. Mar. Environ. Res. 155, 104887. doi: 10.1016/j.marenvres.2020.104887
Scales B. S., Cable R. N., Duhaime M. B., Gerdts G., Fischer F., Fischer D., et al. (2021). Cross-hemisphere study reveals geographically ubiquitous, plastic-specific bacteria emerging from the rare and unexplored biosphere. mSphere 6, e00851–e00820. doi: 10.1128/mSphere.00851-20
Schipper P. N. M., Vissers M. J. M., van der Linden A. M. A. (2008). Pesticides in groundwater and drinking water wells: Overview of the situation in the Netherlands. Water Sci. Technol. 57, 1277–1286. doi: 10.2166/wst.2008.255
Shah Z. U., Parveen S. (2021). Pesticides pollution and risk assessment of river ganga: A review. Heliyon 7, e07726. doi: 10.1016/j.heliyon.2021.e07726
Shahid M. K., Kashif A., Fuwad A., Choi Y. (2021). Current advances in treatment technologies for removal of emerging contaminants from water – a critical review. Coordination Chem. Rev. 442, 213993. doi: 10.1016/j.ccr.2021.213993
Shao B., Liu Z., Zeng G., Liu Y., Yang X., Zhou C., et al. (2019). Immobilization of laccase on hollow mesoporous carbon nanospheres: Noteworthy immobilization, excellent stability and efficacious for antibiotic contaminants removal. J. Hazard Mater 362, 318–326. doi: 10.1016/j.jhazmat.2018.08.069
Sharif N., Bibi A., Zubair N., Munir N. (2022). “Chapter 11 - heavy metal accumulation potential of aquatic fungi,” in Freshwater mycology. Eds. Bandh S. A., Shafi S. (Elsevier), 193–208.
Sharma A., Kumar V., Shahzad B., Tanveer M., Sidhu G. P. S., Handa N., et al. (2019). Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci. 1, 1446. doi: 10.1007/s42452-019-1485-1
Sharma R., Singh N. S., Dhingra N., Parween T. (2020). “Bioremediation of oil-spills from ShoreLine environment,” in Modern age waste water problems : Solutions using applied nanotechnology. Eds. Oves M., Ansari M. O., Zain Khan M., Shahadat M., Ismail I. M. I. (Cham: Springer International Publishing), 275–291.
Signes-Pastor A. J., Martinez-Camblor P., Baker E., Madan J., Guill M. F., Karagas M. R. (2021). Prenatal exposure to arsenic and lung function in children from the new Hampshire birth cohort study. Environ. Int. 155, 106673. doi: 10.1016/j.envint.2021.106673
Silva A., Delerue-Matos C., Figueiredo S. A., Freitas O. M. (2019). The use of algae and fungi for removal of pharmaceuticals by bioremediation and biosorption processes: A review. Water 11, 1555. doi: 10.3390/w11081555
Singh S., Kumar Naik T. S. S., Anil A. G., Dhiman J., Kumar V., Dhanjal D. S., et al. (2022). Micro (nano) plastics in wastewater: A critical review on toxicity risk assessment, behaviour, environmental impact and challenges. Chemosphere 290, 133169. doi: 10.1016/j.chemosphere.2021.133169
Singh M., Srivastava P. K., Verma P. C., Kharwar R. N., Singh N., Tripathi R. D. (2015). Soil fungi for mycoremediation of arsenic pollution in agriculture soils. J. Appl. Microbiol. 119, 1278–1290. doi: 10.1111/jam.12948
Soares P. R. S., Birolli W. G., Ferreira I. M., Porto A. L. M. (2021). Biodegradation pathway of the organophosphate pesticides chlorpyrifos, methyl parathion and profenofos by the marine-derived fungus aspergillus sydowii CBMAI 935 and its potential for methylation reactions of phenolic compounds. Mar. pollut. Bull. 166, 112185. doi: 10.1016/j.marpolbul.2021.112185
Soliman M. M., Hesselberg T., Mohamed A. A., Renault D. (2022). Trophic transfer of heavy metals along a pollution gradient in a terrestrial agro-industrial food web. Geoderma 413, 115748. doi: 10.1016/j.geoderma.2022.115748
Sowmya H. V., Ramalingappa, Krishnappa M., Thippeswamy B. (2015). Degradation of polyethylene by penicillium simplicissimum isolated from local dumpsite of shivamogga district. Environment Dev. Sustainability 17, 731–745. doi: 10.1007/s10668-014-9571-4
Spina F., Tummino M. L., Poli A., Prigione V., Ilieva V., Cocconcelli P., et al. (2021). Low density polyethylene degradation by filamentous fungi. Environ. pollut. 274, 116548. doi: 10.1016/j.envpol.2021.116548
Srivastava P. K., Vaish A., Dwivedi S., Chakrabarty D., Singh N., Tripathi R. D. (2011). Biological removal of arsenic pollution by soil fungi. Sci. Total Environ. 409, 2430–2442. doi: 10.1016/j.scitotenv.2011.03.002
Stief P., Fuchs-Ocklenburg S., Kamp A., Manohar C.-S., Houbraken J., Boekhout T., et al. (2014). Dissimilatory nitrate reduction by aspergillus terreus isolated from the seasonal oxygen minimum zone in the Arabian Sea. BMC Microbiol. 14, 35. doi: 10.1186/1471-2180-14-35
Su S., Zeng X., Bai L., Jiang X., Li L. (2010). Bioaccumulation and biovolatilisation of pentavalent arsenic by penicillin janthinellum, fusarium oxysporum and trichoderma asperellum under laboratory conditions. Curr. Microbiol. 61, 261–266. doi: 10.1007/s00284-010-9605-6
Sumathi S., Priyanka V., Krishnapriya V., Suganya K. (2020). Identification of a novel strain of fungus kalmusia italica from untouched marine soil and its heavy metal tolerance activity. Bioremediation J 25 (2), 91–107. doi: 10.1080/10889868.2020.1853029
Sunderland E. M. (2007). Mercury exposure from domestic and imported estuarine and marine fish in the U.S. seafood market. Environ. Health Perspect. 115, 235–242. doi: 10.1289/ehp.9377
Taboski M. A., Rand T. G., Piórko A. (2005). Lead and cadmium uptake in the marine fungi corollospora lacera and monodictys pelagica. FEMS Microbiol. Ecol. 53, 445–453. doi: 10.1016/j.femsec.2005.02.009
Taghavi N., Singhal N., Zhuang W.-Q., Baroutian S. (2021). Degradation of plastic waste using stimulated and naturally occurring microbial strains. Chemosphere 263, 127975. doi: 10.1016/j.chemosphere.2020.127975
Taipale S. J., Peltomaa E., Kukkonen J. V. K., Kainz M. J., Kautonen P., Tiirola M. (2019). Tracing the fate of microplastic carbon in the aquatic food web by compound-specific isotope analysis. Sci. Rep. 9, 19894. doi: 10.1038/s41598-019-55990-2
Ter Halle A., Jeanneau L., Martignac M., Jardé E., Pedrono B., Brach L., et al. (2017). Nanoplastic in the north Atlantic subtropical gyre. Environ. Sci. Technol. 51, 13689–13697. doi: 10.1021/acs.est.7b03667
Thompson R. C., Olsen Y., Mitchell R. P., Davis A., Rowland S. J., John A. W., et al. (2004). Lost at sea: where is all the plastic? Science 304, 838. doi: 10.1126/science.1094559
Torán J., Blánquez P., Caminal G. (2017). Comparison between several reactors with trametes versicolor immobilized on lignocellulosic support for the continuous treatments of hospital wastewater. Bioresour Technol. 243, 966–974. doi: 10.1016/j.biortech.2017.07.055
Torres A., Li S.-M., Roussos S., Vert M. (1996a). Screening of microorganisms for biodegradation of poly(lactic acid) and lactic acid-containing polymers. Appl. Environ. Microbiol. 62, 2393–2397. doi: 10.1128/aem.62.7.2393-2397.1996
Torres A., Li S. M., Roussos S., Vert M. (1996b). Degradation of l- and DL-lactic acid oligomers in the presence ofFusarium moniliforme andPseudomonas putida. J. Environ. polymer degradation 4, 213–223. doi: 10.1007/BF02070690
Tran N., Urase T., Kusakabe O. (2010). Biodegradation characteristics of pharmaceutical substances by whole fungal culture trametes versicolor and its laccase. J. Water Environ. Technol. 8, 125–140. doi: 10.2965/jwet.2010.125
Turusov V., Rakitsky V., Tomatis L. (2002). Dichlorodiphenyltrichloroethane (DDT): ubiquity, persistence, and risks. Environ. Health Perspect. 110, 125–128. doi: 10.1289/ehp.02110125
Uchida M., Mouriño-Pérez R. R., Freitag M., Bartnicki-García S., Roberson R. W. (2008). Microtubule dynamics and the role of molecular motors in neurospora crassa. Fungal Genet. Biol. 45, 683–692. doi: 10.1016/j.fgb.2007.10.013
Urlacher V. B., Koschorreck K. (2021). Pecularities and applications of aryl-alcohol oxidases from fungi. Appl. Microbiol. Biotechnol. 105, 4111–4126. doi: 10.1007/s00253-021-11337-4
Vaksmaa A., Hernando-Morales V., Zeghal E., Niemann H. (2021a). “Microbial degradation of marine plastics: Current state and future prospects,” in Biotechnology for sustainable environment. Eds. Joshi S. J., Deshmukh A., Sarma H. (SingaporeSpringer Singapore), 111–154.
Vaksmaa A., Knittel K., Abdala Asbun A., Goudriaan M., Ellrott A., Witte H. J., et al. (2021b). Microbial communities on plastic polymers in the Mediterranean Sea. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.673553
Vala A. (2009). Tolerance and removal of arsenic by a facultative marine fungus aspergillus candidus. Bioresource Technol. 101, 2565–2567. doi: 10.1016/j.biortech.2009.11.084
Van Aken B., Hofrichter M., Scheibner K., Hatakka A., Naveau H., Agathos S. (1999). Transformation and mineralization of 2,4,6-trinitrotoluene (TNT) by manganese peroxidase from the white-rot basidiomycete phlebia radiata. Biodegradation 10, 83–91. doi: 10.1023/A:1008371209913
Van Sebille E., Wilcox C., Lebreton L., Maximenko N., Hardesty B. D., Van Franeker J. A., et al. (2015). A global inventory of small floating plastic debris. Environ. Res. Lett. 10, 124006. doi: 10.1088/1748-9326/10/12/124006
Vasiliadou I. A., Molina R., Pariente M. I., Christoforidis K. C., Martinez F., Melero J. A. (2019). Understanding the role of mediators in the efficiency of advanced oxidation processes using white-rot fungi. Chem. Eng. J. 359, 1427–1435. doi: 10.1016/j.cej.2018.11.035
Vera M., Nyanhongo G. S., Pellis A., Rivas B., Guebitz G. (2018). Immobilization of myceliophthora thermophila laccase on poly(glycidyl methacrylate) microspheres enhances the degradation of azinphos-methyl. J. Appl. Polymer Sci. 136, 47417. doi: 10.1002/app.47417
Verma N., Gupta S. (2019). Assessment of LDPE degrading potential aspergillus species isolated from municipal landfill sites of Agra. SN Appl. Sci. 1, 701. doi: 10.1007/s42452-019-0746-3
Verma S., Kuila A. (2019). Bioremediation of heavy metals by microbial process. Environ. Technol. Innovation 14, 100369. doi: 10.1016/j.eti.2019.100369
Vidal-Limon A., García Suárez P. C., Arellano-García E., Contreras O. E., Aguila S. A. (2018). Enhanced degradation of pesticide dichlorophen by laccase immobilized on nanoporous materials: A cytotoxic and molecular simulation investigation. Bioconjugate Chem. 29, 1073–1080. doi: 10.1021/acs.bioconjchem.7b00739
Wang C., Yu L., Zhang Z., Wang B., Sun H. (2014). Tourmaline combined with phanerochaete chrysosporium to remediate agricultural soil contaminated with PAHs and OCPs. J. Hazard Mater 264, 439–448. doi: 10.1016/j.jhazmat.2013.10.073
Wayman C., Niemann H. (2021). The fate of plastic in the ocean environment – a minireview. Environ. Science: Processes Impacts 23, 198–212. doi: 10.1039/D0EM00446D
White L. M., Ernst W. R., Julien G., Garron C., Leger M. (2006). Ambient air concentrations of pesticides used in potato cultivation in prince Edward island, Canada. Pest Manage. Science: formerly Pesticide Sci. 62, 126–136. doi: 10.1002/ps.1130
Wick L. Y., Remer R., Würz B., Reichenbach J., Braun S., Schäfer F., et al. (2007). Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 41, 500–505. doi: 10.1021/es061407s
Wijayawardene N., Hyde K., Al-Ani L. K. T., Tedersoo L., Haelewaters D., Becerra A. G., et al. (2020). Outline of fungi and fungus-like taxa. mycosphere 11, 1160–1456. doi: 10.5943/mycosphere/11/1/8
Winardi, Soetarto E. S., Haryono E., Sudrajat (2019). Research roadmap of bioremediation: Review of in situ method on land bioremediation. J. Phys. Conf. Ser. (Online) 1175, 8. doi: 10.1088/1742-6596/1175/1/012130
Wołejko E., Jabłońska-Trypuć A., Wydro U., Butarewicz A., Łozowicka B. (2020). Soil biological activity as an indicator of soil pollution with pesticides – a review. Appl. Soil Ecol. 147, 103356. doi: 10.1016/j.apsoil.2019.09.006
Wolfand J. M., Lefevre G. H., Luthy R. G. (2016). Metabolization and degradation kinetics of the urban-use pesticide fipronil by white rot fungus trametes versicolor. Environ. Science: Processes Impacts 18, 1256–1265. doi: 10.1039/C6EM00344C
Wu B., Hussain M., Zhang W., Stadler M., Liu X., Xiang M. (2019). Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology. doi: 10.1080/21501203.2019.1614106
Xiang L., Li G., Wen L., Su C., Liu Y., Tang H., et al. (2021). Biodegradation of aromatic pollutants meets synthetic biology. Synthetic Syst. Biotechnol. 6, 153–162. doi: 10.1016/j.synbio.2021.06.001
Xiao P., Mori T., Kamei I., Kiyota H., Takagi K., Kondo R. (2011). Novel metabolic pathways of organochlorine pesticides dieldrin and aldrin by the white rot fungi of the genus phlebia. Chemosphere 85, 218–224. doi: 10.1016/j.chemosphere.2011.06.028
Xue N., Fang Q., Pan X., Zhang D. (2021). Distinct fungal plastisphere across different river functional zones: A watershed scale study. Sci. Total Environ. 752, 141879. doi: 10.1016/j.scitotenv.2020.141879
Yadav J. S., Doddapaneni H., Subramanian V. (2006). P450ome of the white rot fungus phanerochaete chrysosporium: Structure, evolution and regulation of expression of genomic P450 clusters. Biochem. Soc. Trans. 34, 1165–1169. doi: 10.1042/BST0341165
Yamada-Onodera K., Mukumoto H., Katsuyaya Y., Saiganji A., Tani Y. (2001). Degradation of polyethylene by a fungus, penicillium simplicissimum YK. Polymer Degradation Stability 72, 323–327. doi: 10.1016/S0141-3910(01)00027-1
Yang Y., Ok Y. S., Kim K.-H., Kwon E. E., Tsang Y. F. (2017). Occurrences and removal of pharmaceuticals and personal care products (PPCPs) in drinking water and water/sewage treatment plants: A review. Sci. Total Environ. 596-597, 303–320. doi: 10.1016/j.scitotenv.2017.04.102
Yin Y., Hu Y., Xiong F. (2011). Sorption of Cu(II) and Cd(II) by extracellular polymeric substances (EPS) from aspergillus fumigatus. Int. Biodeterioration Biodegradation 65, 1012–1018. doi: 10.1016/j.ibiod.2011.08.001
Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199. doi: 10.1126/science.aad6359
Zalasiewicz J., Waters C. N., Ivar Do Sul J. A., Corcoran P. L., Barnosky A. D., Cearreta A., et al. (2016). The geological cycle of plastics and their use as a stratigraphic indicator of the anthropocene. Anthropocene 13, 4–17. doi: 10.1016/j.ancene.2016.01.002
Zeghal E., Vaksmaa A., Vielfaure H., Boekhout T., Niemann H. (2021). The potential role of marine fungi in plastic degradation – a review. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.738877
Zeng S., Qin X., Xia L. (2017). Degradation of the herbicide isoproturon by laccase-mediator systems. Biochem. Eng. J. 119, 92–100. doi: 10.1016/j.bej.2016.12.016
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “Plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang Z., Yan K., Zhang L., Wang Q., Guo R., Yan Z., et al. (2019). A novel cadmium-containing wastewater treatment method: Bio-immobilization by microalgae cell and their mechanism. J. Hazardous Materials 374, 420–427. doi: 10.1016/j.jhazmat.2019.04.072
Zhou H., Wang Z., Chen S., Liu D., Xia H. (2008). Purification and characterization of extracellular poly (β-hydroxybutyrate) depolymerase from penicillium sp. DS9701-D2. Polymer-Plastics Technol. Eng. 48, 58–63. doi: 10.1080/03602550802539627
Zhuo R., Fan F. (2021). A comprehensive insight into the application of white rot fungi and their lignocellulolytic enzymes in the removal of organic pollutants. Sci. Total Environ. 778, 146132. doi: 10.1016/j.scitotenv.2021.146132
Zinjarde S. S., Pant A. (2002). Emulsifier from a tropical marine yeast, yarrowia lipolytica NCIM 3589. J. Basic Microbiol. 42, 67–73. doi: 10.1002/1521-4028(200203)42:1<67::AID-JOBM67>3.0.CO;2-M
Keywords: bioremediation, mycoremediation, fungi, pharmaceuticals, heavy metals, plastics, pollutants
Citation: Vaksmaa A, Guerrero-Cruz S, Ghosh P, Zeghal E, Hernando-Morales V and Niemann H (2023) Role of fungi in bioremediation of emerging pollutants. Front. Mar. Sci. 10:1070905. doi: 10.3389/fmars.2023.1070905
Received: 15 October 2022; Accepted: 30 January 2023;
Published: 06 March 2023.
Edited by:
Renata Denaro, National Research Council (CNR), ItalyReviewed by:
Sandhya Mishra, National Botanical Research Institute (CSIR), IndiaNatalia González-Benítez, Rey Juan Carlos University, Spain
Copyright © 2023 Vaksmaa, Guerrero-Cruz, Ghosh, Zeghal, Hernando-Morales and Niemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annika Vaksmaa, YW5uaWthLnZha3NtYWFAbmlvei5ubA==
 Annika Vaksmaa
Annika Vaksmaa Simon Guerrero-Cruz
Simon Guerrero-Cruz Pooja Ghosh
Pooja Ghosh Emna Zeghal
Emna Zeghal Victor Hernando-Morales4
Victor Hernando-Morales4 Helge Niemann
Helge Niemann