
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 26 October 2022
Sec. Marine Pollution
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.986493
This article is part of the Research TopicCoastal and Marine Environmental Quality AssessmentsView all 28 articles
 Gajendra Joshi1*
Gajendra Joshi1* Pankaj Verma2
Pankaj Verma2 Balakrishnan Meena1
Balakrishnan Meena1 Prasun Goswami1†
Prasun Goswami1† D Magesh Peter2
D Magesh Peter2 Dilip Kumar Jha2
Dilip Kumar Jha2 Nambali Valsalan Vinithkumar1
Nambali Valsalan Vinithkumar1 Gopal Dharani2
Gopal Dharani2The marine environment is most vital and flexible with continual variations in salinity, temperature, and pressure. As a result, bacteria living in such an environment maintain the adaption mechanisms that are inherent in unstable environmental conditions. The harboring of metal-resistant genes in marine bacteria contributes to their effectiveness in metal remediation relative to their terrestrial counterparts. A total of four mercury-resistant bacteria (MRB) i.e. NIOT-EQR_J7 (Alcanivorax xenomutans); NIOT-EQR_J248 and NIOT-EQR_J251 (Halomonas sp.); and NIOT-EQR_J258 (Marinobacter hydrocarbonoclasticus) were isolated from the equatorial region of the Indian Ocean (ERIO) and identified by analyzing the 16S rDNA sequence. The MRBs can reduce up to 70% of Hg(II). The mercuric reductase (merA) gene was amplified and the mercury (Hg) volatilization was confirmed by the X-ray film method. The outcomes obtained from ICP-MS validated that the Halomonas sp. NIOT-EQR_J251 was more proficient in removing the Hg from culture media than other isolates. Fourier transform infrared (FT-IR) spectroscopy results revealed alteration in several functional groups attributing to the Hg tolerance and reduction. The Gas Chromatography-Mass Spectrometry (GC-MS) analysis confirmed that strain Halomonas sp. (NIOT-EQR_J248 and NIOT-EQR_J251) released Isooctyl thioglycolate (IOTG) compound under mercury stress. The molecular docking results suggested that IOTG can efficiently bind with the glutathione S-transferase (GST) enzyme. A pathway has been hypothesized based on the GC-MS metabolic profile and molecular docking results, suggesting that the compound IOTG may mediate mercuric reduction via merA-GST related detoxification pathway.
Mercury (Hg) pollution poses a global threat to human and environmental health due to its noxiousness, mobility, and lengthy residence duration in the atmosphere (Raphael et al., 2011). Toxic metals rapidly accumulate in the food chain, impacting higher trophic levels, hence these are of principal concern nowadays (Raphael et al., 2011). According to recent research, oceanic release and biomass burning (organic compounds) account for the majority of worldwide Hg emissions, with anthropogenic activities accounting for the remaining significant percentage (Pirrone et al., 2010; Nelson et al., 2012; Serrano et al., 2013). Hg exists in elemental, inorganic, and organic forms in both land and water systems, depending on oxidation-reduction conditions. In the atmosphere, Hg with valence +2 is more extensively spread (Wang et al., 2004). In the biogeochemical cycle, a considerable part of Hg accumulates in seas and oceans by its atmospheric deposition (Bindler, 2003; Wang et al., 2004). The effects of human-induced sources on the Hg contents and its forms are extensively larger in the photic layers of oceans (Strode et al., 2007).
Numerous physico-chemical and biological methodologies have been implemented by industries and regulatory agencies to remediate Hg from polluted sites (Mahbub et al., 2017). Many studies suggested the use of marine bacteria for the bioremediation of harmful metals as they have an immense ability to conquer the continuous fluctuating pattern of pH, temperature, salinity, and other variable factors (Chikere et al., 2012; Naik et al., 2012; Dash et al., 2013). The population of Mercury-resistant bacteria (MRB) belongs to gram-positive as well as gram-negative groups that can easily grow in the toxic environment containing Hg and can convert the toxic form to a relatively nontoxic form (Dash and Das, 2012). The most commonly reported mechanism to develop resistance towards Hg in bacteria is mer operon mediated. It consists of various functional genes like merT, merP, merD, merA, merB, merG, merC along with the promoter and operator (Giri et al., 2014). Many MRB were previously isolated from Indian coastal areas which can thrive at 25 mg/L or greater levels of Hg (Dash and Das, 2014; De et al., 2014). These bacteria eliminate Hg through reductive volatilization, which means Hg2+ gets reduced to elemental Hg (Hg0) through the catalytic activity of mercuric reductase enzyme encoded by merA gene and successfully detoxifies Hg polluted waters (De and Ramaiah, 2007; De et al., 2008).
In our recent study (Joshi et al., 2021), we isolated and characterized several MRBs from the deeper depth of the Central Indian Ocean and evaluated their ability to remove Hg from the culture media. However, no bacterial strain was observed from the surface seawater samples that were resistant to Hg up to 100 mg/L. Though various bacterial strains have been isolated from the coastal and oceanic region, so far. MRB from the equatorial region of the Indian Ocean (ERIO) is not studied. It is hypothesized that the marine bacteria from ERIO could be a potential resource for the reduction of Hg2+ as ERIO is highly dynamic in nature due to high current and intense climatological precipitation (Annamalai, 2010).
The present study was carried out to isolate the culturable MRBs (resistant up to 100 mg/L of Hg) from the surface seawater of the ERIO. To understand the effect of Hg ions on microbes and their mechanism to transform Hg2+ to Hg0, various techniques such as PCR, ICP-MS, FT-IR, SEM, GC-MS, molecular docking etc. were used. The Hg tolerance, genotyping of merA gene, Hg reduction potential, Hg(II) depletion, alteration in functional groups, metabolites formed during Hg2+ reduction were determined for the isolates. This study explores the bacteria from the ERIO for Hg bioremediation potential which may be used in environmental pollution management.
The two sites of ERIO were selected for seawater samples collection. The GPS locations of selected sites were 00° 00′ 559″ N & 80° 37′ 919″ E and 00° 28′ 649″ N & 87° 50′ 546″ E. The samples were collected from the euphotic zone (5m depth) in sterile 100 mL glass bottles with the help of a Niskin water sampler fitted with a CTD rosette sampling device (Seabird Electronics), onboard ORV Sagar Nidhi (Cruise No. SN134) in October, 2018. Seawater samples were analyzed for temperature and salinity using a CTD profiler (Model 911 plus) and were processed immediately for microbial analysis. The CTD profiler was calibrated before use. The details of water sampling sites are given in supplementary information (SI) Table S1 and Figure 1.
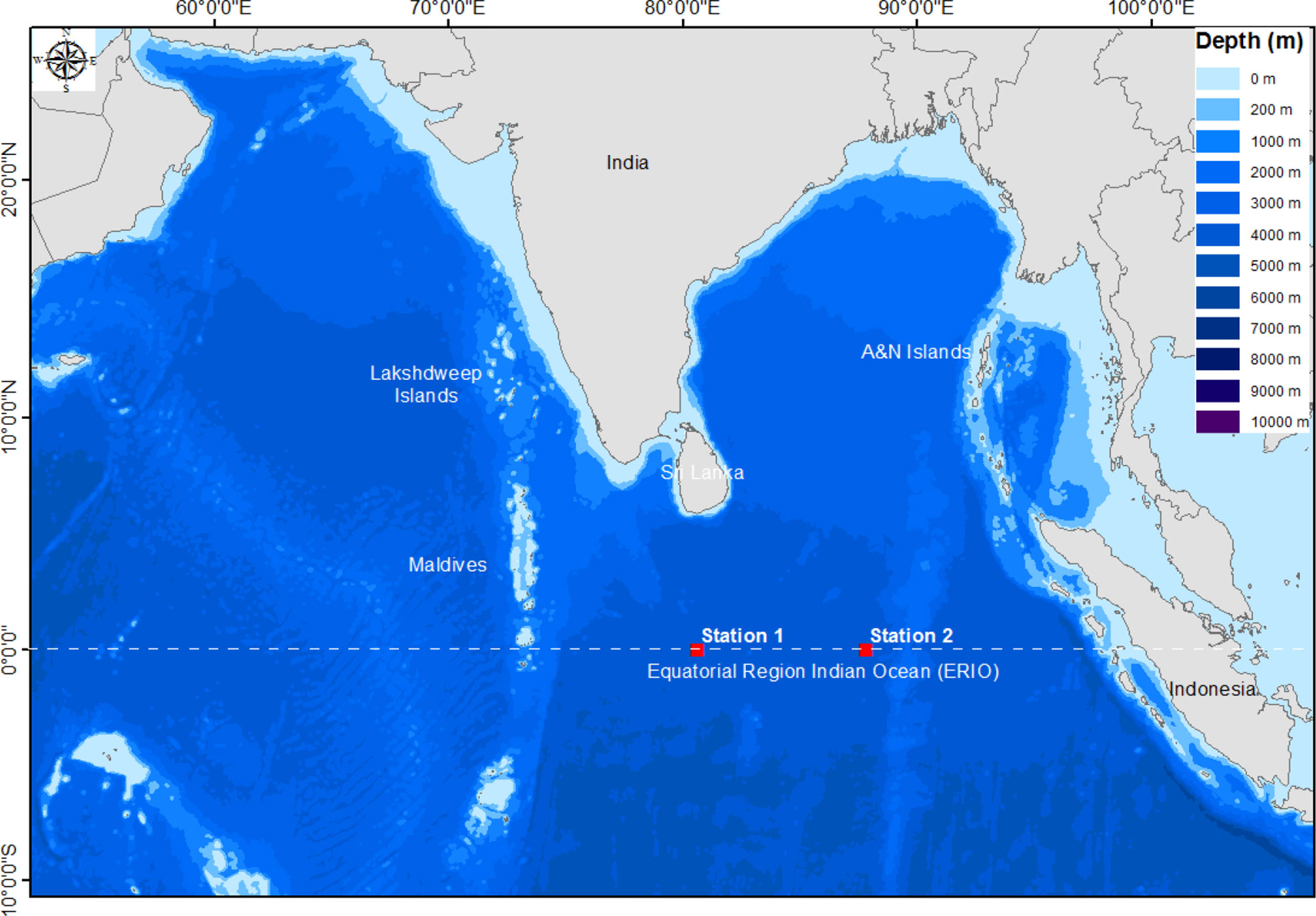
Figure 1 Study area map showing selected sites of ERIO for sample collection. The map was prepared using the Arc GIS software (Version 10.7.1).
To isolate and enumerate total culturable bacteria (TCB) and culturable mercury-resistant bacteria (CMRB) under aerobic conditions, the previously described serial dilution method was adopted (Joshi et al., 2021). In brief, the stock solution (1 g/L) of mercuric chloride (HgCl2, Merck) was prepared and the stability of the Hg stock solution was ensured by reducing the pH level below 2.0 using Suprapur elemental grade nitric acid (HNO3, Merck, India). The detailed procedure is elaborated in the SI. TCB and CMRB were counted using the standard formula (CFU/mL). The percentage (%) of CMRB was calculated using the equation (1) as under:
Hg tolerance of CMRBs was calculated by the broth dilution method (Konopka and Zakharova, 1999; Santos-Gandelman et al., 2014) with some modifications. CMRB cultures grown in Zobell Marine Broth medium (ZMB, Himedia, Mumbai) without Hg supplementation were used as a positive control, whereas ZMB treated with Hg (without bacteria) was used as a negative control in this study. To ensure the uniform bioavailability of Hg to the bacteria, uniform culture conditions were maintained across different experimental treatments. Based on the growth behavior of cultures in the presence of a higher concentration of HgCl2, the selected isolates were classified into two different categories i.e. MRB and moderate MRB (MMRB). The detailed procedure is elaborated in the SI. To understand the effects of different Hg concentrations on bacterial growth, two qualitative analyses were carried out, i.e. monitoring the optical density (OD) at 600 nm and dry biomass.
The selected isolates were further analyzed by subjecting them to 16S rDNA sequencing. In brief, DNA was isolated and the polymerase chain reaction (PCR) amplification was executed with the universal 16S rDNA primers of 27F (5’-AGAGTTTGATCMTGGCTCAG-3’) and 1495R (5’-GGHTACCTTGTTACGACTT-3’) as described by Dash and Das (2014). The amplified PCR products were sequenced and submitted to NCBI, as described by Kumar et al. (2018). The detailed procedure is elaborated in the SI.
A conserved region of merA gene encoding bacterial mercuric reductase enzyme was detected using the above isolated genomic DNA (Joshi et al., 2021). The PCR reactions were carried out using the primer set F1merA (5’-TCGTGATGTTCGACCGCT-3’) and F2merA (5’-TACTCCCGCCGTTTCCAAT-3’) containing 1 U/μl Taq polymerase (Sigma-Aldrich), 1 × Enzyme buffer, 200 μM of each dNTP (Sigma-Aldrich), 1.25 mM MgCl2, and 0.5 μM of each primer and 50 ng template DNA in a thermal cycler (Applied Biosystems). PCR reaction was carried out under the following amplification conditions: pre-denaturation step at 94°C for 2 min followed by 30 cycles of 94°C for 1 min, 52°C for 1 min and an extension step at 72°C for 1 min and final extension at 72°C for 7 min as described by Dash and Das, (2014). For the isolates that showed negative PCR reactions with F1merA and F2merA primer set, a re-attempt has been made for amplifying the merA gene by gradient PCR method (annealing at 54 ± 5°C) and by using another set of primer A1F (5′-ACCATCGGCGGCACCTGCGT-3’) and A5R (5′-ACCATCGTCAGGTAGGGGACCAA3-′) as described by De et al. (2008). Amplification of the merA gene was confirmed by visualizing it under the Gel Doc system (BioRad).
A simplified X-ray film method was adopted to look into the Hg reduction by bacterial volatilization in the presence of 50 and 100 mg/L of Hg as HgCl2 (Nakamura and Nakahara, 1988; Joshi et al., 2021). Briefly, the bacterial cells were collected by centrifugation at 5,000 rpm for 10 min. and washed with 0.07 M phosphate buffer (0.5 mM EDTA, 0.2 mM magnesium acetate, 5 mM sodium thioglycolate; pH 7.0) and transferred to the microplate. The cells were suspended into 50 µL of 0.07 M phosphate buffer containing 50 and 100 mg/L of Hg as HgCl2 in a microplate. The phosphate buffers (pH 7.0) with 50 and 100 mg/L of HgCl2 (without bacterial cells) were used as a negative control. The plate was covered with X-ray film and incubated in the dark for 2 h at 35°C.
The cultures were grown in a ZMB medium supplemented with 50 mg/L of HgCl2 at 30°C and 150 rpm for 48 h. To collect the Hg associated with the bacterial cells, the cultures were centrifuged at 12,000 rpm for 10 min. To analyse the Hg concentration, the samples were processed as described by (Joshi et al. 2021). The detailed procedure for Hg estimation is elaborated in the SI. The concentration of Hg was analyzed by inductively coupled plasma mass spectrometry (ICP-MS, Agilent -7500) as described in the U.S. EPA (1994). The % of Hg elimination was calculated using equation (2).
To check the accuracy and precision of analysis, the nominal Hg concentration was compared with the replicates of analytical blank, working solution of HgCl2, and control samples (without bacteria) which were analyzed by ICP-MS. The details of Hg recovery are shown in SI Table S2.
The modification in functional groups present in the culture pellets was identified by measuring the spectra in the range of 400 to 4000 cm-1 using Fourier transform infrared (FT-IR) spectroscopy (IR Affinity-I spectrometer, Shimadzu, Japan), as described by Joshi et al. (2021). In brief, 48 h grown cultures in ZMB medium without Hg supplementation were used as a control, whereas cultures with 50 mg/L of Hg supplementation were used as Hg treated. The mixtures (lyophilized cells and 2% KBr) were fixed in the FT-IR spectrometer after compressing them into translucent sample discs, followed by analyzing in ATR-FT-IR mode by following the manufacturer’s protocol.
To inspect the consequence of Hg2+ concentration on the morphology, the bacterial cells were freshly grown in the presence (50 and 100 mg/L - test samples) and absence (control) of Hg2+ as HgCl2; and cell pellets were harvested by centrifugation (5000 rpm at 4°C for 5 min) after 48 h. SEM analysis has been carried out, as reported by Joshi et al. (2021). In brief, the bacterial cells were fixed with 2.5% glutaraldehyde followed by the post-fixing in 1% osmium tetroxide and dehydration of the cells with graded ethanol series (25%, 50%, 80%, 90%, and absolute). The processed samples were scanned using SEM (JEOL-JSM-IT500).
GC-MS analysis was performed to characterize the bacterial response and the metabolomic changes leading to Hg tolerance. The solvent extraction method was used to extract the bacterial metabolites. In brief, the freshly inoculated and exponentially grown MRB and MMRB bacterial cells in ZMB medium (non-exposed and exposed to 50 mg/L of Hg) were freeze-dried using BENCHTOP lyophilizer (VIRTIS Instrument, Gardiner, NY). For extraction of compounds, 50 mg of lyophilized bacterial cells were suspended in ethyl acetate and chloroform (1:1; v/v) and homogenized. After homogenization, the solution (crude extract) containing the metabolites was transferred to the clean glass vial by pipetting. These steps were repeated two-three times to obtain a pure and ample amount of sample. The separated organic fractions (crude extract) were treated with anhydrous NaSO4 (Sigma-Aldrich) to remove moisture, which was again concentrated on the rotary evaporator (BUCHI Rotavapor R-215/V advanced, Switzerland) at RT and stored at -80°C until further analysis. The concentrated crude extract was re-suspended in 1 mL of Dichloromethane (DCM) and 5 µL of the sample was injected into the GC-MS analyzer (Agilent Technologies Instrument 7890A GC System, 240 Ion Trap GC/MS, USA). The GC-MS analysis was carried out under external ionization mode using a fused silica column HP 5 MS column (30 m × 0.320 mm × 0.25 µm). High purity helium was used as a carrier gas at a constant flow rate of 1 mL/min. For analysis, the chromatographic conditions i.e. initial injector and detector temperature, were set at 250°C and 330°C, respectively. The temperature of the column was programmed from 50°C (hold for 2 min) to 320°C (2 min hold), with a constant 5°C increment per minute and 1 min hold at 330°C. A metabolic library of all the separated compounds found via GC-MS analysis of bacterial extract was created and identified using NIST mass spectral library match. The PubChem CID, structures, names, and molecular weight of those bioactive compounds were obtained from the PubChem database.
The compounds obtained in GC-MS analysis were evaluated for bioactivity using the Swiss-ADME server, as described by Daina et al. (2017). The compounds, which target the proteins/enzymes/molecules possessing the detoxification property were selected for molecular docking studies. The 3D structure of normalized GST and IOTG compound (in PDB file format) was used for further docking studies. Molecular docking was performed using AutoDock Vina (ADV) software as described by Trott and Olson (2010) and BIOVA Discovery Studio Visualizer Tool 21.1.0.0 was used to perceive the interactions between receptor and ligand. The detailed procedure is elaborated in the SI.
Using molecular docking results, reference pathways derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) website (http://www.kegg.jp/or http://www.genome.jp/kegg/) and based on the prior reports, a metabolomic pathway was hypothesized for detoxification of Hg in marine isolates.
Three experimental replicates were determined for each analysis and the statistical significance of the results was analyzed by one way ANOVA test. All Statistical analyses were carried out using SPSS (version 17) and GraphPad Prism 7 software. Differences were considered significant at p< 0.05 and values are reported as mean ± standard deviation (SD).
Some microbes hold a vital role in the process of bio-remediation and bioprospecting in which halophilic or halotolerant microbes are the key players (Ventosa et al., 1998; Fukushima et al., 2005). Culture that can grow in heavy metals, organic solvents and produced antibiotics may be used for bioremediation process (De Souza et al., 2006). In the present study, four bacteria of three different species, Alcanivorax xenomutans, Halomonas sp., and Marinobacter hydrocarbonoclasticus were isolated from the ERIO and shown to be resistant to Hg. The physico-chemical characteristics of seawater samples at both sites of the ERIO are presented in SI Table S1.
The total bacterial colonies that appeared on the ZMA medium were counted and documented. The statistical analysis found no significant difference (p > 0.05) between the TCB of both sites. The TCB count at Site 1 and Site 2 was 176.66 ± 47.71 and 131 ± 37.04 (× 10² CFU/mL), respectively (Table S1). De and Ramaiah (2006) reported that the mean total viable count (TVC) on the surface in coastal and oceanic water of the Bay of Bengal (BOB) was 632.38 ± 551.31 and 61.90 ± 80.32 CFU/mL, respectively. The diversity of culturable bacterial counts at the photic zone of the New Britain Trench was 102 to 104 bacterial cells/mL (Liu et al., 2018). The total retrievable bacterial count in the water samples collected from the two different sites of the central region of the Indian Ocean (CRIO) was 286 ± 39.58 and 177 ± 30.51 (× 10² CFU/mL) (Joshi et al., 2021). Our results suggested that the TCB counts observed from different sites of the ERIO in this study are similar to those reported from the CRIO and New Britain Trench, whereas much higher than BOB. In the water column, the distribution of bacteria depends on the availability of utilizable organic carbon (Calleja et al., 2019). Consequently, a relatively large bacterial population is found in the euphotic layer.
The total mercury (THg) concentrations in marine waters in the different parts of the world are in the range of 0.5 – 27,060 ng/L (Gworek et al., 2016). As a consequence of the excessive use of Hg for industrial and agriculture activities and the ultimate disposal of this heavy metal effluent into marine zones, the Hg concentration in the marine environment, including the Indian Ocean is continuously increasing (Sarin, 1991). The Hg vapor released by resistant biota in an open system like the ocean might be the possible reason for re-polluting the environment which had become a part of the local Hg cycle (Lacerda and Pfeiffer, 1992). Among the total aerobic heterotrophic bacteria, the existence of 1-10% of MRB is quite normal in a particular environment (Muller et al., 2001). The MRB population ratio increases with the increasing level of Hg contamination. However, higher MRB had also existed in a comparatively Hg-free environment (Ramaiah and De, 2003). In the current study, the % of CMRB counts was found 16.20% (26.66 ± 10.21 × 10² CFU/mL) at Site 1 whereas 6.84% (9 ± 4 × 10² CFU/mL) at Site 2 (Table S1). There was a significant difference (p< 0.05) among the TCB and CMRB counts on both the sites, as a limited number of bacterial populations can survive in 10 mg/L of Hg as HgCl2. The % frequency of MRB found in this work is lower than that of the other natural environment. In coastal and oceanic water of the BOB, around 97% of MRB were resistant to 10 mg/L of Hg. The authors concluded that MRB contributed to over 20% of the TVC in surface (1-10 m) samples. In the case of oceanic water, the % of MRB at two different sites of the BOB was 92% and 53% of total CFU (Ramaiah and De, 2003; De and Ramaiah, 2006). In our previous study, we reported the occurrence of 17.95% and 27.58% retrievable MRB at two different sites of CRIO (Joshi et al., 2021). Though so far, there are no reports about Hg pollution at the ERIO, the detection of bacterial population resistance to Hg in this study and other related studies suggest that marine bacteria resistant to Hg are widely distributed in the biosphere. Since there are many natural and anthropogenic sources of Hg in the biosphere that makes Hg a global pollutant.
Based on phenotype characteristics, a total of 162 bacterial colonies grown on the ZMA media supplemented with 10 mg/L of Hg as HgCl2 were selected for further analysis. In the presence of 25 mg/L of Hg2+, only 63 isolates out of 162 showed resistance. Further, these 63 isolates were characterized based on their growth pattern in the presence of more than 25 mg/L i.e. 50, 75, and 100 mg/L of Hg2+ concentration. Among the 63 isolates, 21 isolates grew in the presence of 50 mg/L, followed by 9 and 4 isolates at 75 and 100 mg/L of HgCl2, respectively. The growth pattern of 4 isolates in the presence of HgCl2 is shown in Figure S1.
In terms of mean OD, all the 4 isolates i.e. NIOT-EQR_J7, NIOT-EQR_J248, NIOT-EQR_J251, and NIOT-EQR_J258 showed equivalent growth (OD600nm) i.e. 1.77, 1.657, 1.752, and 1.342 in the presence of 50 mg/L of Hg2+ compared to the positive control 1.827, 1.717, 1.82 and 1.396, respectively. Out of 4 isolates, 3 isolates (NIOT-EQR_J7, NIOT-EQR_J248, and NIOT-EQR_J251) showed more than 60% of growth in the presence of 100 mg/L Hg2+ i.e. 1.273, 1.169, and 1.125 as compared to the positive control and were considered as MRB whereas NIOT-EQR_J258 growth was found to be less than 50% in the presence of 100 mg/L of Hg2+ (OD positive control-1.396 and OD with 100 mg/L of Hg2+ - 0.628) considered as MMRB. The statistical analysis revealed a non-significant difference (p > 0.05) among the four isolates in the presence of 25 and 50 mg/L of Hg2+, whereas a significant difference (p< 0.05) in the presence of 75 and 100 mg/L of Hg as HgCl2 when compared to positive control. Hence, we maintained the same Hg concentration (i.e. 50 mg/L) for all analyses. All the isolates showed less than 50% of mean dry biomass in the presence of 100 mg/L Hg2+ compared to the positive control which was outstandingly higher (p< 0.05). The isolate NIOT-EQR_J258 and NIOT-EQR_J251 showed dry biomass of 0.33 g/L and 0.36 g/L, respectively in the presence of 100 mg/L Hg2+. The isolates showed good growth in the presence of 25 and 50 mg/L HgCl2; however, low growth in higher concentrations of Hg2+ (100 mg/L) was observed. The qualitative analysis showed that the positive controls attained an exponential growth phase quickly as compared to the Hg-treated cultures.
Many strains of genera such as Halomonas, Haloferax, Halobacterium, Halococcus, Alcanivorax and Marinobacter could tolerate high concentrations of HgCl2 i.e. from 15 to 400 mg/L (Sorkhoh et al., 2010; Al-Mailem et al., 2011; Dziewit et al., 2013; Abdel-Razik et al., 2020). Several scientific reports are available, suggesting Hg resistant bacterium like Bacillus sp. (100, 62.5, 12.8, 20, and 50 mg/L), P. putida (56 mg/L), Citrobacter (0.2 mg/L), Corynebacterium (6.4 mg/L), Proteus (0.2 mg/L), Pseudomonas (0.2 mg/L), Staphylococcus (12.8 mg/L) and E.Coli (55 mg/L) from different environmental conditions (Zeyaullah et al., 2010; Keramati et al., 2011; Zhang et al., 2012; De et al., 2014; Giri et al., 2014; Santos-Gandelman et al., 2014). Though a small amount of Hg is toxic to the bacterial cells, in this study, Hg tolerance is observed in reported isolates beyond the toxic level. The Hg resistance Gram-positive and Gram-negative isolates may probably be used to detoxify Hg species (Zeyaullah et al., 2010; Dash et al., 2013). This suggests that the isolates from the present study might be potential candidates for effective Hg bioremediation from contaminated sites.
In the current study, the 16S rDNA was successfully amplified (Figure S2). The NIOT-EQR_J7 and NIOT-EQR_J258 were identified as Alcanivorax xenomutans and Marinobacter hydrocarbonoclasticus, whereas NIOT-EQR_J248 and NIOT-EQR_J251 belong to the Halomonas sp. The obtained 16S rDNA sequences were submitted to NCBI under accession numbers MT825200, MT825208, MT825209, and MT825210. The point of divergence and evolutionary closeness is showed by the formation of numerous external nodes and the presence of some clusters (Figure 2). A similar mechanism of Hg tolerance and its remediation may be possessed by evolutionarily related bacteria. Two organisms i.e. NIOT-EQR_J248 and NIOT-EQR_J251, are included in the domain Bacteria, Phylum Proteobacteria, Class Gammaproteobacteria, order Oceanospirillales, and family of Halomonadaceae. One isolates i.e. NIOT-EQR_J7 belongs to the family of Alcanivoracaceae, whereas NIOT-EQR_J258 is a member of the Alteromonadales order and Alteromonadaceae family.
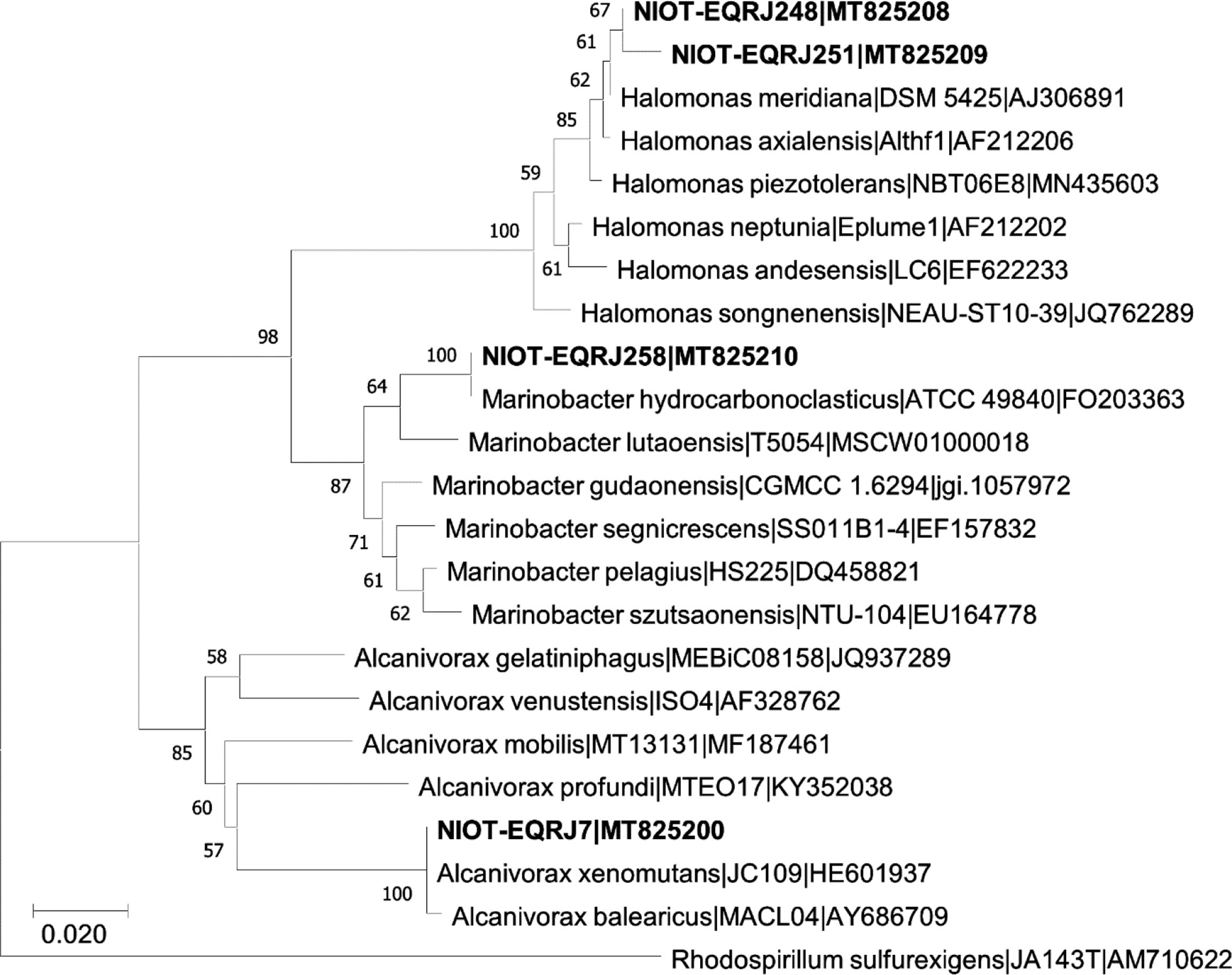
Figure 2 Phylogenetic analysis of Alcanivorax xenomutans strain NIOT-EQR_J7, Halomonas sp. NIOT-EQRJ_248 and NIOT-EQRJ_251; and Marinobacter hydrocarbonoclasticus strain NIOT-EQRJ_258.
Genus Halomonas is reported from most of the saline environments regardless of their geographical location, including the marine environment, salterns, saline lakes, and soil (Llamas et al., 2006). The MRB isolated from the coastal areas of Kuwait were identified as Alcanivorax borkumensis, Marinobacter hydrocarbonoclasticus, and Halomonas taeheungii (Sorkhoh et al., 2010). The 16S rDNA analysis and phenotypic characteristics revealed that the heavy metal-resistant halophilic bacteria WQL9 belong to the genus Halomonas sp (Abdel-Razik et al., 2020). The Marinobacter genus is comprised of widespread marine bacteria found exceptionally in marine or terrestrial environments rich in sodium salts, in the deep sea, coastal seawater, sediment, oceanic basalt, etc. (Handley and Lloyd, 2013). Alcanivorax xenomutans are also rich in the saline environment as they are halophilic and favorable to living in the marine environment. A comparative analysis of the partial sequence of the 16S rDNA of the four strains revealed a high level of similarity to the corresponding sequence of environmental isolates.
In the present study, the presence of a distinct 431-bp size band (using F1merA and F2merA primer set) in two MRB isolates (NIOT-EQR_J248 and NIOT-EQR_J251) confirms that these two isolates harbor the merA gene, which codes for mercuric ion reductase for the detoxification of inorganic Hg (Figure 3). The other isolates NIOT-EQR_J7 and NIOT-EQR_J258 did not showed any amplification, although both the primer sets were used and a gradient method was attempted. However, phenotypically they all were resistant to inorganic Hg and reduced Hg2+ to Hg0. This may suggest the presence of other non-mer mediated Hg resistance mechanisms in NIOT-EQR_J7 and NIOT-EQR_J258 or the presence of a diverse merA gene, which could not be detected by the employed PCR primer sets.
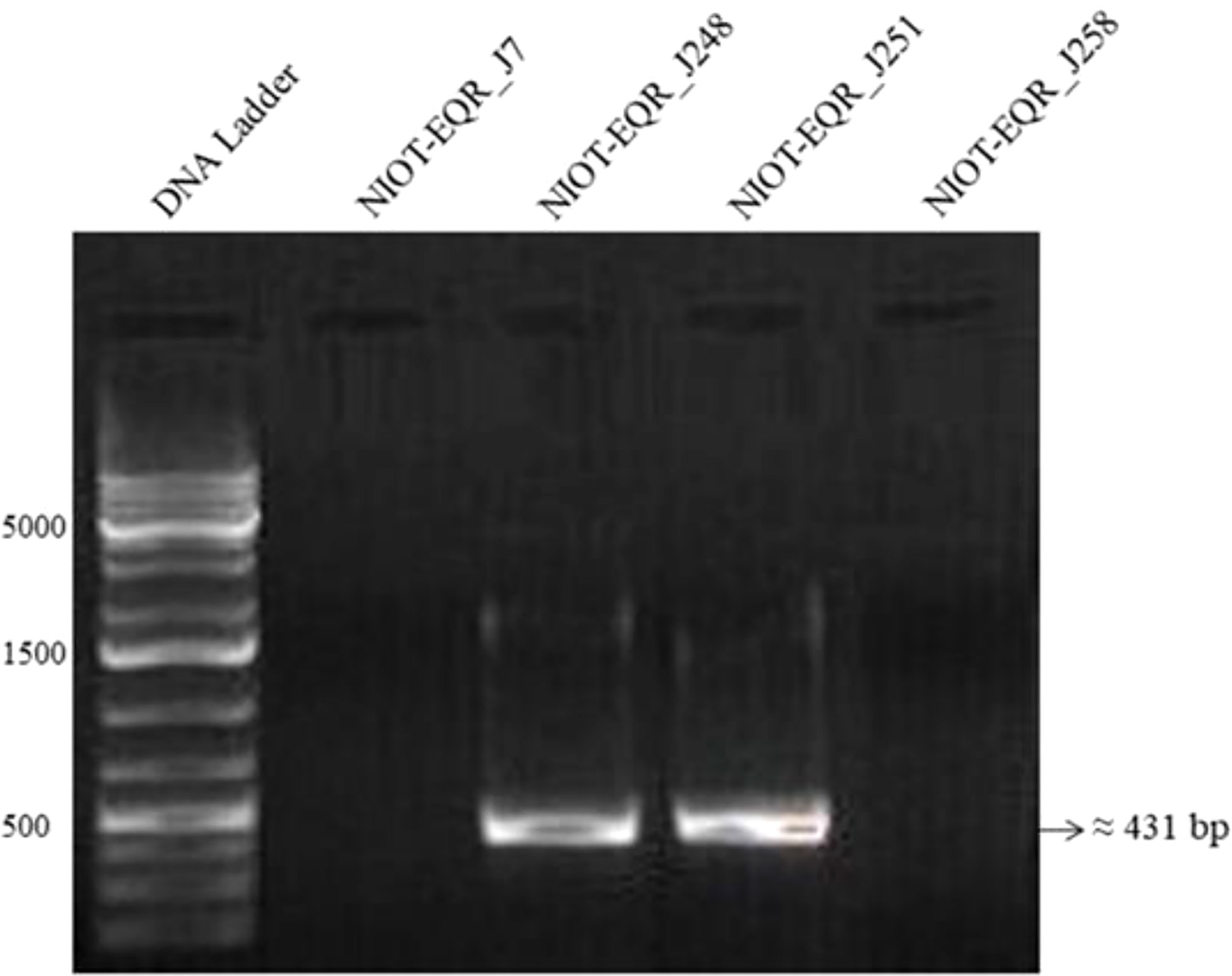
Figure 3 Gel electrophoresis of merA gene amplicons of MRB and MMRB strains using F1 merA and F2 merA primer set. Lane 1: Gene Ruler 1 kb Plus DNA Ladder, Lane 2 and 5: Negative PCR reaction, and Lane 3 and 4: amplified products.
Many studies suggest that mer operon is located on any one of either location such as mobile elements like plasmids, transposons, genomic or chromosomal DNA (Liebert et al., 1999; Schelert et al., 2004; Zeng et al., 2010). The mer operon contains many functional genes along with the operator, promoter, and regulatory elements. All functional genes encrypt for the diverse proteins that contribute to the detection, transportation, and reduction of Hg ions (Barkay et al., 2003; Dash and Das, 2012; Naguib et al., 2018). An extremely noteworthy positive correlation was found among the harboring of mer gene with the phenotypic resistance to Hg and concentration of the Hg in the source environment (Osborn et al., 1997). In this study, we did not find any Hg from the ecosystem (detectable limit - 0.73 µg/L), but the amplification of mer genes and available literature suggested that these are extensively dispersed in a non-highly contaminated or contamination-free environment such as an open ocean, Antarctic sea-ice, high Arctic snow, sea ice brine etc. and plays a key role in the biogeochemical cycle of Hg (Christakis et al., 2021). Thus, the Hg adaptation by these isolates is reasonably accepted.
To confirm the mer gene function in the isolates, a bacterial Hg volatilization study was executed upon the higher concentration of Hg, i.e. 50 and 100 mg/L. It was confirmed after the incubation period of the microtiter plate for 2 h in the dark by developing foggy areas on the X-ray film in the wells region of the microplate where the isolates were present. The results revealed that NIOT-EQR_J7 and NIOT-EQR_J248 showed high fog development on X-ray film in the presence of 50 and 100 mg/L of Hg as compared to other isolates. However, NIOT-EQR_J258 showed a negligible amount of fog in the presence of 50 mg/L and failed to develop the foggy area in the presence of 100 mg/L of Hg. In the case of NIOT-EQR_J251, the development of foggy areas was higher in the presence of 50 mg/L of Hg as compared to 100 mg/L and equivalent to that of NIOT-EQR_J7 (Figure 4A). The un-inoculated phosphate buffers failed to develop any foggy areas confirming the sensitivity pattern for Hg. The bioremediation of Hg is allied with the mer operon, wherein the environmental and genetic factors influence its bioremediation efficiency. These outcomes are in agreement with the results obtained by De et al. (2008) and Joshi et al. (2021). The presence of foggy areas after 2 h on the silver (Ag) contained X-ray film assured Ag reduction by the Hg vapor released by isolates. However, in the negative control, no such signal was detected.
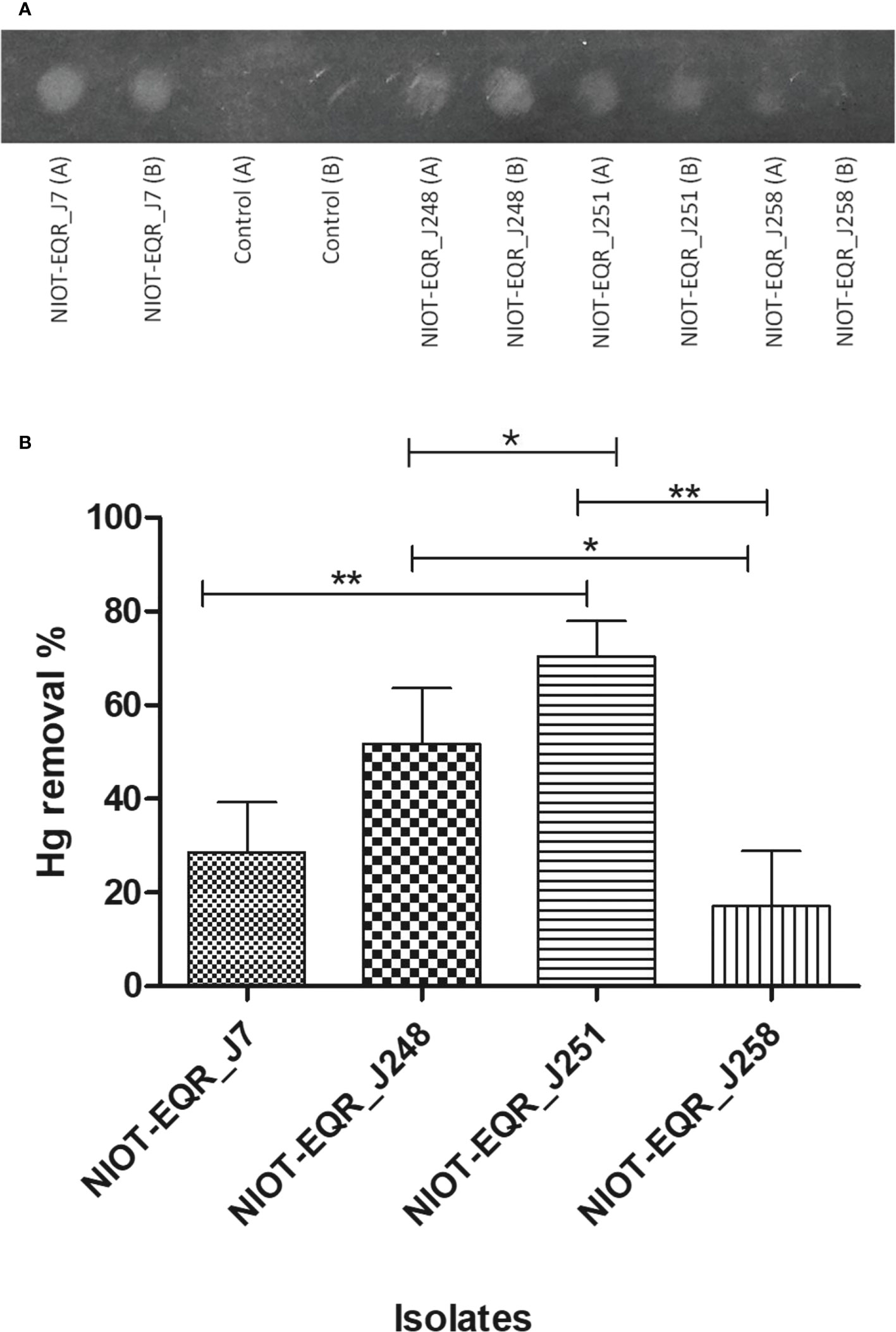
Figure 4 (A) Bacterial volatilization of HgCl2 was confirmed by the X-ray film method. (A) Represents the Hg volatilization in the presence of 50 mg/L whereas (B) corresponds to the 100 mg/L of Hg as HgCl2. (B) Hg depletion potential of the marine cultures in the presence of 50 mg/L of Hg as HgCl2. Bars with an asterisk (* and **) indicate noteworthy differences among the cultures in the level of p< 0.05 and p< 0.01, respectively. Results are represented as mean ± SD (n=3).
Based on the results obtained by the x-ray film method, ICP-MS analysis has been carried out to confirm the Hg removal potential of the isolates in the presence of 50 mg/L of Hg2+. The ICP-MS outcomes showed significant changes in the level of Hg depletion. Among all, the marine culture NIOT-EQR_J251 possessed the highest potential (70.62%) to volatilize Hg. NIOT-EQR_J258 showed the least removal of Hg (17.48%), whereas NIOT-EQR_J7 and NIOT-EQR_J248 were capable to volatilize 29.18% and 52.17% of Hg2+ under similar conditions, respectively. (Figure 4B). NIOT-EQR_J251 could volatilize 32.54 mg of the initial inoculum of 46.07 mg of HgCl2, whereas NIOT-EQR_J258 removes 8.03 mg of the initial supplement of 45.74 mg of Hg2+. In presence of 50 mg/L of Hg2+, NIOT-EQR_J7 and NIOT-EQR_J248 dominantly developed the fog on the x-ray film, whereas, in the ICP-MS analysis, NIOT-EQR_J251 showed the highest Hg removal as compared to the other isolates. This suggests that volatilization was not solely responsible for removing the Hg2+ from the culture media and the contribution of different processes like bioaccumulation and bio-adsorption may be involved.
The statistical analysis of Hg removal potential among the different species i.e., Alcanivorax xenomutans, Halomonas sp., Marinobacter hydrocarbonoclasticus and within the Halomonas spp. revealed that the Hg removal by NIOT-EQR_J251 was significantly higher (p< 0.01) than NIOT-EQR_J7 and NIOT-EQR_J258. There was a significant difference (p< 0.05) in the removal of Hg between NIOT-EQR_J248 vs. NIOT-EQR_J258 and NIOT-EQR_J248 vs. NIOT-EQR_J251. Al-Mailem et al. (2011) reported Haloferax sp. (HA1 and HA2), Halobacterium sp. HA3, and Halococcus sp. HA4 effectively volatilized (from 40 to 65%) the available 100 mg/L of Hg after 8 days. Many other isolates such as Bacillus sp., Pseudomonas stutzeri, Pseudomonas putida, Vibrio fluvialis could volatilize 60%-95%, 94%, 100%, 60% of Hg2+, respectively, from culture (Zhang et al., 2012; Dash et al., 2013; Giri et al., 2014; Saranya et al., 2017; Zheng et al., 2018).
Hg volatilization by bacteria involves the reduction of Hg2+ via the mer system to Hg0, which is less toxic. It is appealing from the essential and practical viewpoints that halophilic bacteria can also grow in the presence of a higher amount of Hg and volatilized Hg efficiently similar to the non-halophilic bacteria. The ICP-MS analysis did not detect significant Hg loss in the control sample (without bacteria), which confirms the volatility of Hg being biotic (Table S2). Thus, our results suggested that halophilic bacteria can also remove the Hg from the growth media, indicating that it may be potentially applied in contaminated environments. The Hg comes into the ocean either through atmospheric deposition or other several natural and active or passive processes. The results of this study imply that the two marine strains of Halomonas sp., isolated from ERIO may have the ability to remove Hg2+ from the growth medium.
The cultures isolated in this study have shown significant fog development in the presence of 50 mg/L of Hg. Hence, we maintained the same Hg concentration for further studies. The binding of Hg ions to the bacteria depends mainly on the functional groups present on the active sites of bacterial cells and the physicochemical conditions of the solution. In the current study, ATR-FT-IR spectroscopic analysis of the MRB strains was carried out to understand the functional groups involved in the binding process. The IR spectra showed several peaks originating in the 3234.15, 2800.07-3017.95, 2246.72-2383.09, 1618.15-1736.53, 1358.84-1602.80, 1206.39-1273.25, 918.48-1209.93, and 721.47-864.95 cm-1 regions, that belong to OH (alcohol), alkane C-H, S-H stretch, Amide C=O, N-O, C–N, P=O stretch and alkyl halide groups, respectively. As clearly seen from Figures 5A–D and Table S4, in the presence of Hg2+, there was a reduction in the intensity of the IR peaks bands in the S-H Stretch (NIOT-EQR_J7, NIOT-EQR_J248, and NIOT-EQR_J251), Amide C=O stretch (NIOT-EQR_J7), Amine (C-N) (NIOT-EQR_J7, NIOT-EQR_J251), N-O group (NIOT-EQR_J248), and the phosphate group (NIOT-EQR_J7, NIOT-EQR_J251) regions as compared to the control grown without Hg.
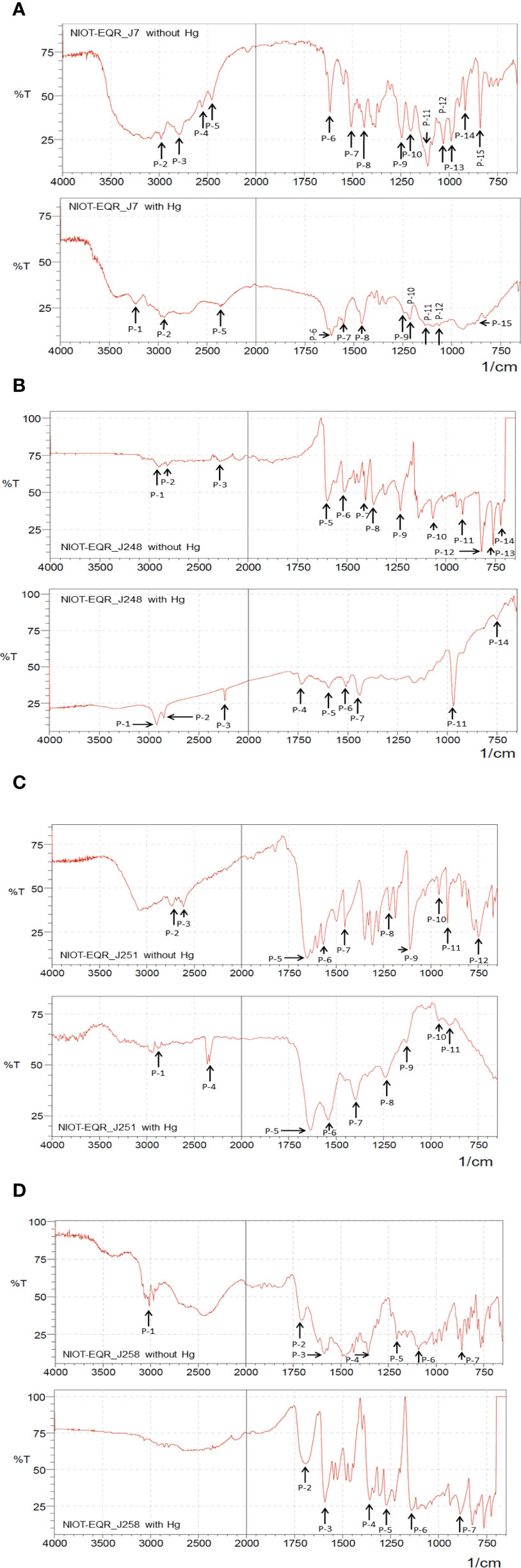
Figure 5 ATR-FT-IR spectra of four isolates in absence and presence of 50 mg/L Hg as HgCl2 (A) NIOT-EQR_J7 (B) NIOT-EQR_J248 (C) NIOT-EQR_J251 and (D) NIOT-EQR_J258.
When the isolate NIOT-EQR_J7 was grown in the presence of Hg2+ supplementation, a slight reduction in the peak intensity attributed to the thiol group (S-H stretch) at 2383.09 cm-1 was observed as compared to the control along with the wavelength that shifted to 2442.42 cm-1. In contrast, the peak intensity corresponding to the thiol group of NIOT-EQR_J248 at 2304.55 cm-1 increased compared to without Hg2+ supplementation and the shift in wavelength to 2246.72 cm-1 was observed. In the case of NIOT-EQR_J251, the peaks corresponding to the S-H group appeared at 2341.34 cm-1 in the presence of Hg2+ whereas, in the absence of Hg2+, the peak was negligible. The peak was absent in both the presence and absence of Hg2+ in the case of isolate NIOT-EQR_J258 (Figures 5A–D). In accord with our findings, previous studies had also reported similar results (Dash and Das, 2014; De et al., 2014; Joshi et al., 2021) and suggested significant changes in the FT-IR spectrum such as a shift in wavenumber in the region of O-H groups, –S-H group, amide C=O, Nitrile C–N stretch, phosphate groups, etc. The changes in peak height and area have also been observed when isolates were grown in the presence of Hg2+ ions compared to the controlled ones. The transmittance of the infrared (IR) in the presence of Hg2+ was lower than in the absence of Hg2+. The changes suggested that the presence of metal leads to a lesser degree of bond stretching, consequently reducing the IR transmittance. The contribution of functional groups in metal binding was legitimized undeniably by the formation of varying spectral bands with and without Hg (Oves et al., 2013).
In context to these four isolates, the shift in wavenumbers, changes in peak height, and the appearance/disappearance of new peaks in the presence of Hg2+; suggest alterations in functional groups (especially -SH group), which might play an important role in the Hg detoxification.
All the isolates were grown in three different concentrations of Hg (0, 50, and 100 mg/L) and were scanned under SEM. The SEM analysis showed that the isolates are small or large rod-shaped (Figure 6). In the presence of the medium concentration of Hg2+ i.e. 50 mg/L, the selected strains were capable to survive and no significant morphological changes were observed. However, the foremost changes like shrinkage or enlargement in bacterial cells, pores formations in the cell wall, and destructions of some of the bacterial cells were noticed in the presence of 100 mg/L of HgCl2.
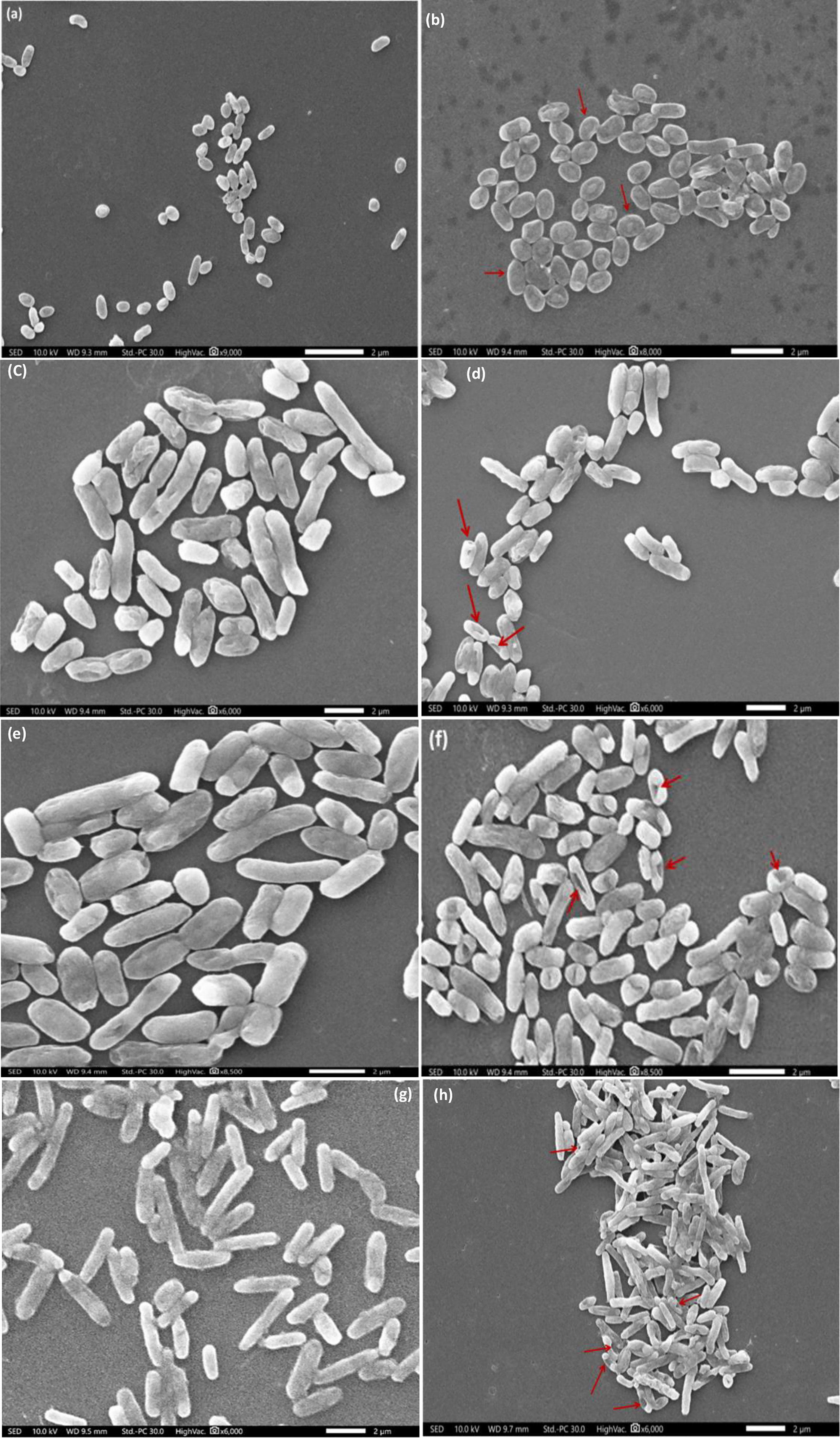
Figure 6 SEM analysis showing Hg effect on bacterial cells: (A, B) NIOT-EQR_J7, (C, D) NIOT-EQR_J248, (E, F) NIOT-EQR_J251 and (G, H) NIOT-EQR_J258. Images were taken in the presence of different concentrations of Hg2+: (A, C, E, G - Control; B, D, F, H - 100 mg/L). The red arrow shows the structural deformities.
It can be observed that high Hg concentration causes structural deformities which may be due to its toxic nature. In our previous study, we reported that cultures accustomed to grow in the presence of HgCl2 manifested major morphological abnormalities (Joshi et al., 2021). We also suggested several structural asymmetries allied with the cell wall and cytoplasmic membrane, significantly affecting the cellular mechanism. Resultantly, there is a substantial delay at the beginning of growth and cell division in a higher concentration of Hg2+.
The GC-MS study was included to get insight into the metabolic changes in the MRB isolates under Hg stress. It revealed that the metabolic profiles of MRB isolates are diverse with different biological properties. GC-MS metabolic profiles revealed that the number of compounds increased in the presence of Hg compared to the control isolates, where some compounds were found to be different in Hg-treated samples. A total of 50 metabolites were identified in the absence of Hg, whereas with 50 mg/L of Hg, 64 compounds were identified. The minimum metabolites (6) were identified in NIOT-EQR_J258 without Hg, whereas the maximum metabolites (22) was identified in NIOT-EQR_J251 with Hg2+. The NIOT-EQR_J7 metabolic profile was almost the same in both control and Hg-treated samples. A total of 14 metabolites were present in the control sample, whereas 15 metabolites were identified with Hg. In the case of NIOT-EQR_J248, NIOT-EQR_J251, and NIOT-EQR_J258, a total of 10, 20, and 6 compounds were identified in the control sample, whereas 16, 22, and 11 metabolites were present with Hg, correspondingly.
The GC-MS chromatograms suggested that in the presence of Hg2+, a total of 30 different compounds were detected (3, 8, 13, and 6 nos. in NIOT-EQR_J7, NIOT-EQR_J248, NIOT-EQR_J251, and NIOT-EQR_J258, respectively) compared to control (Table 1 and Figure S3). However, 19 different compounds were observed in the absence of Hg2+ (2, 6, 9, and 2 nos. in NIOT-EQR_J7, NIOT-EQR_J248, NIOT-EQR_J251, and NIOT-EQR_J258, respectively). The new metabolites might be formed due to the bioremediation of Hg by marine strains. An organic compound, Isooctyl thioglycolate (IOTG, molecular formula C10H20O2S and the molecular weight 204.33 g/mol), was obtained from NIOT-EQR_J248 and NIOT-EQR_J251. The compound is also known as Mercaptoacetic acid or thioglycolic acid (http://pubchem.ncbi.nlm.nih.gov/compound/isooctyl-thioglycolate and http://pubchem.ncbi.nlm.nih.gov/compound/Thioglycolic-acid). It contains both thiol and carboxylic acid functional groups. In the marine environment, sulfur-containing amino acids contribute to the range of thiols, the environmental transformation of 3-mercaptopropionic acid as a metabolite of methionine, and the transformation of mercaptopyruvate to mercaptoethanol and mercaptoacetate (Dash and Das, 2014). The higher amount of sulfur-containing amino acids in marine microorganisms bind with Hg ions efficiently and diminishes its noxious effect, resultantly enhancing the bioremediation capability of that organism (Joyner et al., 2003; Dash and Das, 2014). Thus, IOTG may be considered a chemical lead for Hg2+ volatilization by bacteria as sulfhydryl compounds are required to pass off the Hg2+ in a vapor form (Nakamura and Nakahara, 1988). FT-IR analysis also revealed that the Hg reacts with the thiol (-SH) or amine (-NH(1,2,3)) group of proteins before undergoing bioremediation (Deng and Wang, 2012). One of the reasons for the greater expression of the merA gene under similar experimental conditions is the higher existence of sulfur-containing amino acids in marine bacteria, as compared to terrestrial bacteria (Dash and Das, 2014). Based on GC-MS analysis, we report for the first time that in the presence of Hg2+, a compound IOTG is released by MRB strains of Halomonas sp. that may play a key role in Hg bioremediation process. These findings open interesting perspectives on the possibility of using bacterial isolates and/or their derivatives to develop an eco-friendly technique for clean-up of Hg contaminated environment. Secondary metabolites play a vital role in Hg resistance as well as in bioremediation. The link between secondary metabolites and bioremediation needs further exploration.
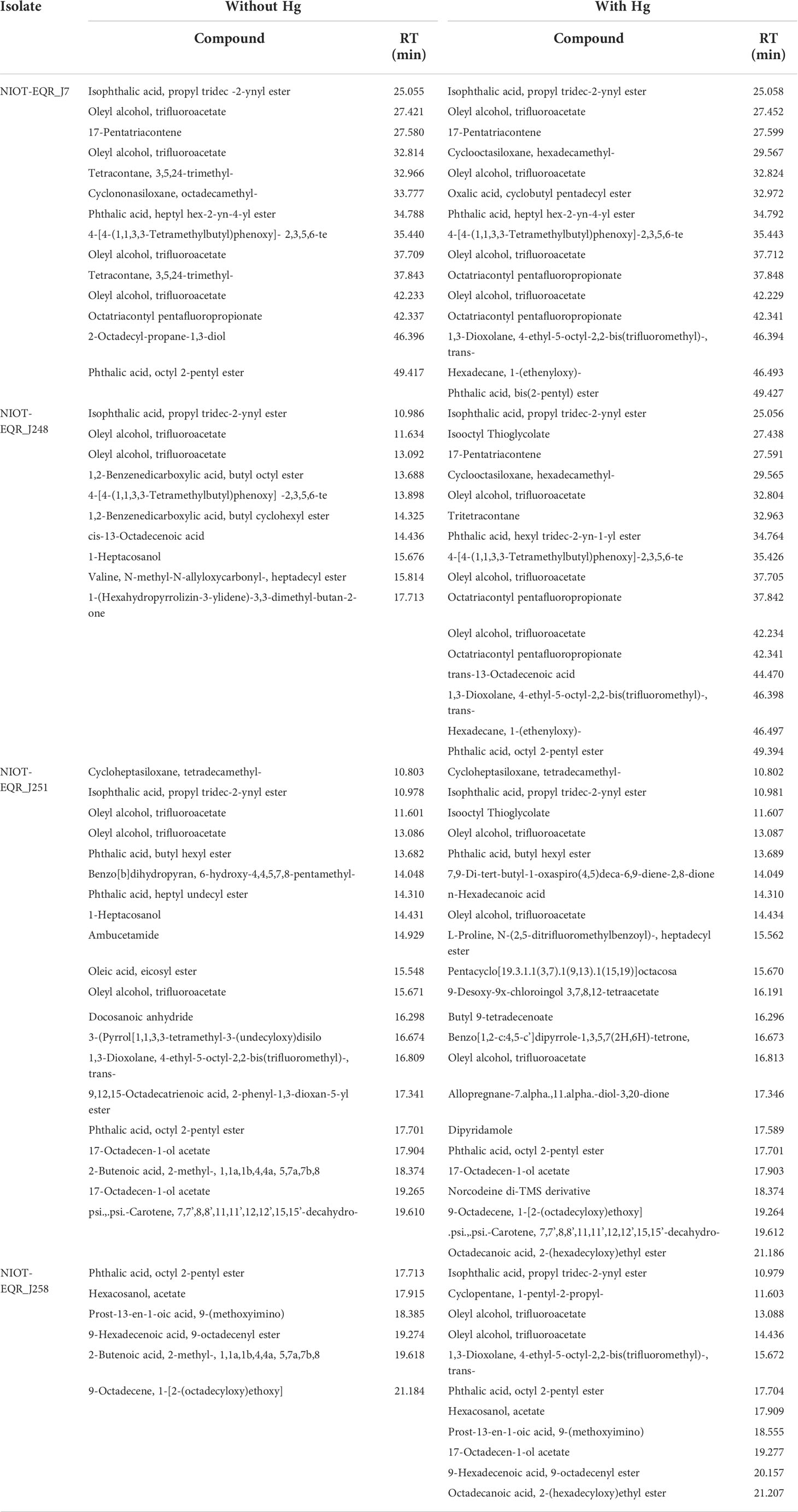
Table 1 Biochemical compounds identified in ethyl acetate and chloroform (1:1; v/v) extract of isolates in the absence and presence of 50 mg/L Hg as HgCl2 by GC-MS.
Based on Swiss-ADME server, among the 30 compounds which can target the number of proteins, one compound i.e. IOTG satisfied all the criteria and showed detoxification properties. The detailed physicochemical properties of ligand (IOTG) are mentioned in SI (Table S5). The SwissTargetPrediction server revealed that various proteins and enzymes may be targeted as receptors including the GST enzyme by the compound IOTG (Figure S4). Its probability to target the GST enzyme was quite higher compared to other GC metabolites. Hence, the compound IOTG was selected for further docking studies to broaden the understanding and obtain insight into the biomolecular interaction with the GST (Figure S5). In docking studies, the highest binding affinity of -15.89 kJ/Mol was observed at the 3rd position. The highest binding affinity of a ligand to a macromolecule refers to the lowest binding energy (Rahman et al., 2021). The molecular interaction results obtained in this study between ligand and protein are found interesting with respect to the GST-GSH interaction. Das et al. (2011) reported that GSH binds with GST dimers and monomers at D2 and M1 (G-site) nominal sites with an energy consumption rate of -39.3 and -32.12 kJ/Mol, respectively. The amino acids such as Glu, Ser, Asn, Thr, Asp, Ile, and Tyr are important for dimer stabilization and identification of GSH binding sites using GST dimer or monomer (Das et al., 2011). In this study, IOTG revealed four hydrogen bonds, four hydrophobic bonds, and five van der Waals interactions along with a best binding affinity with the lowest energy of protein-ligand complexes (Figure S5). The detailed results are mentioned in the SI file. The formation of hydrogen bonds between macromolecule and ligand indicates one of the important interactions and is the result of bioactivity. Therefore, the present docking study suggests that the compound IOTG showed energy-efficient interaction with GST, which may play a significant role in Hg2+ detoxification. The role and importance of specific interacting residues with ligands in the GST active sites could further be analyzed using site-directed mutagenesis.
In prior studies, many pathways have been reported for Hg2+ detoxification in microbes using various tools and technologies (Chang et al., 2020; Cursino et al., 2000; Chang et al., 2021). In the present study, a metabolic pathway is hypothesized based on the GC-MS metabolites and protein-ligand interaction (Figure S6) and an attempt has been made to correlate the proposed pathway with the different components of the already existing pathways. In this preliminary study, we found that general resistance/detoxification mechanisms of NIOT-EQR_J248 and NIOT-EQR_J251 in response to inorganic mercury (Hg2+) exposure were a multisystem combined process.
GST is the major detoxifying enzyme catalyzing the conjugation of reduced GSH to different cytotoxic substrates, which is induced when exposed to heavy metals (Goswami et al., 2014; Huang et al., 2019; Chang et al., 2020). The GSH conjugation reaction is the first step of the mercapturic acid pathway, which is one of the most important detoxification processes. Hg2+ has a high affinity for the thiol group, which could reduce and sequester Hg2+ (Zagorchev et al., 2013; Norambuena et al., 2018). Low molecular weight (LMW) thiol compounds act as cellular antioxidants and could contribute toward defending the cells from reactive oxygen species (ROS) produced due to heavy metals, free radicals, lipid peroxides, and peroxides (Forman et al., 2009). The results obtained in this study are in the agreement with previous findings, which suggested that thiol compounds are playing a crucial role in heavy metal resistance (Zagorchev et al., 2013; Norambuena et al., 2018; Chang et al., 2021). In this study, thiol compound namely IOTG was obtained when isolates were grown in presence of Hg2+. Based on the related reactions and literature, the GST enzyme may also catalyse the conjugation of the reduced form of IOTG to xenobiotic substrates for detoxification in Halomonas sp. NIOT-EQR_J248 and NIOT-EQR_J251.
Our preliminary study suggests that the detoxification of Hg2+ is the immediate result of the GST and merA function as shown in Figure S6. The existence of the glutathione reductase gene in some bacterial mer operons also supports the role of LMW thiols in Hg2+ detoxification (Norambuena et al., 2018). The E. coli, together with the integrated merA - GST gene, was able to survive in the high Hg stress environment and transform Hg2+ to Hg0 (Cursino et al., 2000). It is correlated that IOTG may act as Hg(II)-buffering agents and subsequently, Hg2+ is reduced by merA. In the case of merA, mercuric ion was uptaken by the active process i.e. mer mediated transport (merP and merT) and converted to Hg0 form by mercuric reductase. On the other hand, the Hg2+ that was uptaken through the passive process could subsequently be inactivated by GST or conjugated to merA. Thus, an alternative method is proposed via which Hg resistance level may be augmented in bacteria: the sequestering of Hg given a protein-ligand interaction may lead to an improved way for the volatilization process.
The overall results of this study revealed that bacterial isolates Alcanivorax xenomutans, Halomonas sp., and Marinobacter hydrocarbonoclasticus isolated from the ERIO possessed significant tolerance ability to Hg. In presence of 50 mg/L Hg2+, the potent strains Halomonas sp. (NIOT-EQR_J248 and NIOT-EQR_J251) efficiently (upto 70%) convert the Hg2+ to Hg0 (elementary form) under laboratory conditions. These strains could be used in in-situ conditions to significantly avert the adverse environmental conditions arising from the inorganic Hg contamination. Due to metabolic changes taking place under Hg stress conditions, the two Halomonas strains i.e. NIOT-EQR_J248 and NIOT-EQR_J251, released a compound, namely IOTG. Further, the role of IOTG is reported in the bacterial-based Hg detoxification mechanism. In this study, amplification of merA gene and GC-MS-based metabolic profile suggests that Hg detoxification is the result of a simultaneous process of GST and Mer mechanism. Based on the results obtained in this study IOTG may play a crucial role in strains NIOT-EQR_J248 and NIOT-EQR_J251 to reduce the Hg2+ concentration and establish a comparatively steady cellular environment for survival under Hg stress. Further studies are required at the molecular level using gene inactivation or knockouts to reveal the contribution made by different pathways in bacterial Hg reduction. This study will be useful to design effective environmentally friendly technology for bioremediation of Hg.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
GJ: Conceptualization, Methodology, Formal analysis, Investigation, Writing-original draft, Writing-review & editing. PV: Investigation, Formal analysis, Writing-review & editing. BM: Investigation. PG: Formal analysis. DMP: Investigation. DKJ: Writing-review & editing. NVV: Supervision, Project administration, Writing-review & editing. GD: Supervision, Project administration, Writing-review & editing. All authors contributed to the article and approved the submitted version.
The authors thank the Ministry of Earth Sciences, Govt. of India, for the funding support.
The authors gratefully acknowledge the Ministry of Earth Sciences (MoES), Government of India, for the financial support and providing the necessary laboratory facilities to conduct this research. We are thankful to the Director, Dr. G. A. Ramadass and former director Dr. M.A. Atmanand, the National Institute of Ocean Technology (NIOT), for their continuous support and encouragement during this study. The authors are also indebted grateful to Mr. Karuppasamy Mookan of OSTI, NIOT, for his technical assistance in SEM analysis. Authors also wish to put a special word of thanks to Dr. Lawrance Anburajan, Scientist-D, Atal Centre for Ocean Science and Technology for Islands (ACOSTI), NIOT, Port Blair for his valuable suggestions and co-operation. We thank all the scientific staff of ACOSTI-NIOT and OSTI-MBT along with the cruise staff for their support in the onboard and laboratory analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.986493/full#supplementary-material
Abdel-Razik M. A., Azmy A. F., Khairalla A. S., AbdelGhani S. (2020). Metal bioremediation potential of the halophilic bacterium, Halomonas sp. strain WQL9 isolated from lake qarun, Egypt. Egypt. J. Aquat. Res. 46 (1), 19–25. doi: 10.1016/j.ejar.2019.11.009
Al-Mailem D. M., Al-Awadhi H., Sorkhoh N. A., Eliyas M., Radwan S. S. (2011). Mercury resistance and volatilization by oil utilizing Haloarchaea under hypersaline conditions. Extremophiles 15, 39–44. doi: 10.1007/s00792-010-0335-2
Annamalai H. (2010). Moist dynamical linkage between the equatorial Indian ocean and the south Asian monsoon trough. J. Atmos. Sci. 67 (3), 589–610. doi: 10.1175/2009JAS2991.1
Barkay T., Miller S. M., Summers A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi: 10.1016/S0168-6445(03)00046-9
Bindler G. R. (2003). Estimating the natural background atmospheric deposition rate of mercury utilizing ombrotrophic bogs in southern Sweden. Environ. Sci. Technol. 37, 40–46. doi: 10.1021/es020065x
Calleja M. L., Al-Otaibi N., Morán X. (2019). Dissolved organic carbon contribution to oxygen respiration in the central red Sea. Sci. Rep. 9 (1), 4690. doi: 10.1038/s41598-019-40753-w
Chang J., Shi Y., Si G., Yang Q., Dong J., Chen J. (2020). The bioremediation potentials and mercury(II)-resistant mechanisms of a novel fungus penicillium spp. DC-F11 isolated from contaminated soil. J. Hazard. Mater. 396, 122638. doi: 10.1016/j.jhazmat.2020.122638
Chang J., Si G., Dong J., Yang Q., Shi Y., Chen Y., et al. (2021). Transcriptomic analyses reveal the pathways associated with the volatilization and resistance of mercury(II) in the fungus lecythophora sp. DC-F1. Sci. Total. Environ. 752, 142172. doi: 10.1016/j.scitotenv.2020.142172
Chikere C. B., Chikere B. O., Okpokwasili G. C. (2012). Bioreactor-based bioremediation of hydrocarbon-polluted Niger delta marine sediment, Nigeria. 3. Biotech. 2, 53–66. doi: 10.1007/s13205-011-0030-8
Christakis C. A., Barkay T., Boyd E. S. (2021). Expanded diversity and phylogeny of mer genes broadens mercury resistance paradigms and reveals an origin for MerA among thermophilic archaea. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.682605
Cursino L., Mattos S. V., Azevedo V., Galarza F., Bucker D. H., Chartone-Souza E., et al. (2000). Capacity of mercury volatilization by mer (from escherichia coli) and glutathione s-transferase (from schistosoma mansoni) genes cloned in escherichia coli. Sci. Total. Environ. 261, 109–113. doi: 10.1016/S0048-9697(00)00629-X
Daina A., Michielin O., Zoete V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717. doi: 10.1038/srep42717
Das A., Chalil S., Nigam P., Magee P., Janneh O., Owusu-Apenten R. (2011). Glutathione transferase-P1-1 binding with naturally occurring ligands: Assessment by docking simulations. J. Biophys. Chem. 2, 401–407. doi: 10.4236/jbpc.2011.24046
Dash H. R., Das S. (2012). Bioremediation of mercury and the importance of bacterial mer genes. Int. Biodeter. Biodegr. 75, 207–213. doi: 10.1016/j.ibiod.2012.07.023
Dash H. R., Das S. (2014). Bioremediation potential of mercury by bacillus species isolated from marine environment and wastes of steel industry. Bioremediat. J. 3, 204–212. doi: 10.1080/10889868.2014.899555
Dash H. R., Mangwani N., Chakraborty J., Kumari S., Das S. (2013). Marine bacteria: Potential candidates for enhanced bioremediation. Appl. Microbiol. Biotechnol. 97, 561–571. doi: 10.1007/s00253-012-4584-0
De J., Dash H. R., Das S. (2014). Mercury pollution and bioremediation - a case study on biosorption by a mercury-resistant marine bacterium. in: Das s (ed) microbial biodegradation and bioremediation (London: Elsevier), 137–166. doi: 10.1016/B978-0-12-800021-2.00006-6
Deng X., Wang P. (2012). Isolation of marine bacteria highly resistant to mercury and their bioaccumulation process. Bioresour.Technol. 121, 342–347. doi: 10.1016/j.biortech.2012.07.017
De J., Ramaiah N. (2006). Occurrence of large fractions of mercury-resistant bacteria in the bay of Bengal. Curr. Sci. 91 (3), 368–372.
De J., Ramaiah N. (2007). Characterization of marine bacteria highly resistant to mercury exhibiting multiple resistances to toxic chemicals. Ecol. Ind. 7, 511–520. doi: 10.1016/j.ecolind.2006.05.002
De J., Ramaiah N., Vardanyan L. (2008). Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 10, 471–477. doi: 10.1007/s10126-008-9083-z
De Souza M. J., Nair S., Bharathi P. L., Chandramohan D. (2006). Metal and antibiotic resistance in psychrotrophic bacteria from Antarctic marine waters. Ecotoxicol. 15 (4), 379–3384. doi: 10.1007/s10646-006-0068-2
Dziewit L., Pyzik A., Matlakowska R., Baj J., Szuplewska M., Bartosik D. (2013). Characterization of Halomonas sp. ZM3 isolated from the zelazny most post-flotation waste reservoir, with a special focus on its mobile DNA. BMC Microbiol. 13, 59. doi: 10.1186/1471-2180-13-59
Forman H. J., Zhang H., Rinna A. (2009). Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 30, 1–12. doi: 10.1016/j.mam.2008.08.006
Fukushima T., Mizuki T., Echigo A., Inoue A., Usami R. (2005). Organic solvent tolerance of halophilic α-amylase from a haloarchaeon, haloarcula sp. strain s-1. Extremophiles 9, 85–89. doi: 10.1007/s00792-004-0423-2
Giri S., Dash H. R., Das S. (2014). Mercury resistant bacterial population and characterization of bacillus sp. isolated from sediment of solid waste discharged point of steel industry. Natl. Acad. Sci. Lett. 37 (3), 237–243. doi: 10.1007/s40009-014-0229-4
Goswami P., Hariharan G., Godhantaraman N., Munuswamy N. (2014). An integrated use of multiple biomarkers to investigate the individual and combined effect of copper and cadmium on the marine green mussel (Perna viridis). J. Environ. Sci. Health A. Tox. Hazard. Subst. Environ. Eng. 49 (13), 1564–1577. doi: 10.1080/10934529.2014.938534
Gworek B., Bemowska-Kałabun O., Kije´nska M., Wrzosek-Jakubowska J. (2016). Mercury in marine and oceanic waters–a review. Water Air. Soil pollut. 227 (10), 371. doi: 10.1007/s11270-016-3060-3
Handley K. M., Lloyd J. R. (2013). Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by marinobacter species. Front. Microbiol. 4 (136). doi: 10.3389/fmicb.2013.00136
Huang N., Mao J., Zhao Y., Hu M., Wang X. (2019). Multiple transcriptional mechanisms collectively mediate copper resistance in cupriavidus gilardii CR3. Environ. Sci. Technol. 53, 4609–4618. doi: 10.1021/acs.est.8b06787
Joshi G., Meena B., Verma P., Nayak J., Vinithkumar N. V., Dharani G. (2021). Deep-sea mercury resistant bacteria from the central Indian ocean: A potential candidate for mercury bioremediation. Mar. Pollut. Bull. 169, 112549. doi: 10.1016/j.marpolbul.2021.112549
Joyner J. L., Peyer S. M., Lee R. W. (2003). Possible roles of sulphur containing amino acids in a chemoautotrophic bacterium-mollusc symbiosis. Biol. Bull. 205, 331–338. doi: 10.2307/1543296
Keramati P., Hoodaji M., Tahmourespour A. (2011). Multi-metal resistance study of bacteria highly resistant to mercury isolated from dental clinic effluent. Afr. J. Microbiol. Res. 5, 831–837. doi: 10.5897/AJMR10.860
Konopka A., Zakharova T. (1999). Quantification of bacterial lead resistance via activity assays. J. Microbiol. Methods 37, 17–22. doi: 10.1016/S0167-7012(99)00032-9
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kyoto Encyclopedia of Genes and Genomes (KEGG). Available at: http://www.genome.jp/kegg/.
Lacerda L. D., Pfeiffer W. C. (1992). Mercury from gold mining in Amazon environment - an overview. Quim. Nova. 55, 238–294.
Liebert C. A., Hall R. M., Summers A. O. (1999). Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522. doi: 10.1128/MMBR.63.3.507-522.1999
Liu Q., Fang J., Li J., Zhang L., Xie B.-B., Chen X.-L., et al. (2018). Depth-resolved variations of cultivable bacteria and their extracellular enzymes in the water column of the new Britain trench. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00135
Llamas I., del Moral A., Martínez-Checa F., Arco Y., Arias S., Quesada E. (2006). Halomonas maura is a physiologically versatile bacterium of both ecological and biotechnological interest. Antonie. Van. Leeuwenhoek. 89, 395–403. doi: 10.1007/s10482-005-9043-9
Mahbub K. R., Bahar M. M., Labbate M., Krishnan K., Andrews S., Naidu R., et al. (2017). Bioremediation of mercury: Not properly exploited in contaminated soils! Appl. Microbiol. Biotechnol. 101, 963–976. doi: 10.1007/s00253-016-8079-2
Muller A., Rasmussen L., Sorenson S. (2001). Adaptation of the bacterial community to mercury contamination, FEMS microbial. Lett. 16, 49–53. doi: 10.1111/j.1574-6968.2001.tb10861.x
Naguib M. M., El-Gendy A. O., Khairalla A. S. (2018). Microbial diversity of mer operon genes and their potential rules in mercury bioremediation and resistance. Open Biotechnol. J. 12, 56–77. doi: 10.2174/1874070701812010056
Naik M. M., Pandey A., Dubey S. K. (2012). “Bioremediation of metals mediated by marine bacteria,” in Microorganisms in environmental management. Eds. Satyanarayana T., Johri B., Prakash A. (Dordrecht: Springer), 665–682. doi: 10.1007/978-94-007-2229-3_29
Nakamura K., Nakahara H. (1988). Simplified X-ray film method for detection of bacterial volatilization of mercury chloride by escherichia coli. Appl. Environ. Microbiol. 54, 2871–2873. doi: 10.1128/aem.54.11.2871-2873.1988
National Library of Medicine Compound summary - isooctyl thioglycolate. Available at: http://pubchem.ncbi.nlm.nih.gov/compound/isooctyl-thioglycolate.
National Library of Medicine Compound summary - thioglycolic acid. Available at: http://pubchem.ncbi.nlm.nih.gov/compound/Thioglycolic-acid.
Nelson P. F., Morrison A. L., Malfroy H. J., Cope M., Lee S., Hibberd M. L., et al. (2012). Atmospheric mercury emissions in Australia from anthropogenic, natural and recycled sources. Atmos. Environ. 62, 291–302. doi: 10.1016/j.atmosenv.2012.07.067
Norambuena J., Wang Y., Hanson T., Boyd J. M., Barkay T. (2018). Low-molecular-weight thiols and thioredoxins are important players in Hg(II) resistance in thermus thermophilus HB27. Appl. Environ. Microbiol. 84, e01931–e01917. doi: 10.1128/AEM.01931-17
Osborn A. M., Bruce K. D., Strike P., Ritchie D. A. (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19, 239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x
Oves M., Khan M. S., Zaidi A. (2013). Biosorption of heavy metals by bacillus thuringiensis strain OSM29 originating from industrial effluent contaminated north Indian soil. Saudi. J. Biol. Sci. 20 (2), 121–129. doi: 10.1016/j.sjbs.2012.11.006
Pirrone N., Cinnirella S., Feng X., Finkelman R. B., Friedli H. R., Leaner J., et al. (2010). Global mercury emissions to the atmosphere from anthropogenic and natural sources. Atmos. Chem. Phys. 10, 5951–5964. doi: 10.5194/acp-10-5951-2010
Rahman M. S., Zilani M. N. H., Islam M. A., Hasan M. M., Islam M. M., Yasmin F., et al. (2021). In vivo neuropharmacological potential of gomphandra tetrandra (Wall.) sleumer and in-silico study against ß-amyloid precursor protein. Processes 9, 1449. doi: 10.3390/pr9081449
Ramaiah N., De J. (2003). Unusual rise in mercury resistant bacteria in coastal environments. Microb. Ecol. 45, 444–454. doi: 10.1007/s00248-001-1068-7
Raphael E. C., Augustina O. C., Frank E. O. (2011). Trace metals distribution in fish tissues, bottom sediments and water from okumeshi river in delta state, Nigeria. Environ. Res. J. 5, 6–10. doi: 10.3923/erj.2011.6.10
Santos-Gandelman J. F., Cruz K., Crane S., Muricy G., Giambiagi-deMarval M., Barkay T., et al. (2014). Potential application in mercury bioremediation of a marine sponge-isolated bacillus cereus strain Pj1. Curr. Microbiol. 69, 374–380. doi: 10.1007/s00284-014-0597-5
Saranya K., Sundaramanickam A., Shekhar S., Swaminathan S., Balasubramanian T. (2017). Bioremediation of mercury by vibrio fluvialis screened from industrial effluents. BioMed. Res. Int. 6509648, 6. doi: 10.1155/2017/6509648
Sarin R. (1991). In proceeding of the third indo-Dutch workshop on aquatic toxicology (Chennai: Centre for Environment Studies, Anna University), 1–29.
Schelert J., Dixit V Hoang V., Simbahan J., Drozda M., Blum P. (2004). Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon sulfolobus solfataricus by use of gene disruption. J. Bacteriol. 186, 427–437. doi: 10.1128/JB.186.2.427-437.2004
Serrano O., Martínez-Cortizas A., Mateo M., Biester H., Bindler R. (2013). Millennial scale impact on the marine biogeochemical cycle of mercury from early mining on the Iberian peninsula. Global Biogeochem. Cy. 27, 21–30. doi: 10.1029/2012GB004296
Sorkhoh N. A., Ali N., Dashti N., Al-Mailem D. M., Al-Awadhi H., Eliyas M., et al. (2010). Soil bacteria with the combined potential for oil utilization, nitrogen fixation, and mercury resistance. Int. Biodeterior. Biodegr. 64, 226–231. doi: 10.1016/j.ibiod.2009.10.011
Strode S., Jaegle L., Selin N., Jacob D., Park R., Yantosca R., et al. (2007). Air-sea exchange in the global mercury cycle. Global Biogeochem. Cy. 21, GB1017. doi: 10.1029/2006GB002766
Trott O., Olson A. J. (2010). AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. doi: 10.1002/jcc.21334
U.S. EPA (1994). Method 200.8: Determination of trace elements in waters and wastes by inductively coupled plasma-mass spectrometry,” revision 5.4 (U.S. Environmental Protection Agency: Cincinnati, OH).
Ventosa A., Nieto J. J., Oren A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62, 504–544. doi: 10.1128/MMBR.62.2.504-544.1998
Wang Q., Kim D., Dionysiou D. D., Soriala G. A., Timberlake D. (2004). Sources and remediation for mercury contamination in aquatic systems - a literature review. Environ. pollut. 131, 323–336. doi: 10.1016/j.envpol.2004.01.010
Zagorchev L., Seal C. E., Kranner I., Odjakova M. (2013). A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 14, 7405–7432. doi: 10.3390/ijms14047405
Zeng X., Tagn J., Jiang P., Liu H., Dai Z., Liu X. (2010). Isolation, characterization and extraction of mer gene of Hg2+ resisting strain D2. Trans. Nonferr. Met. Soc China 20, 507–512. doi: 10.1016/S1003-6326(09)60170-9
Zeyaullah M., Islam B., Ali A. (2010). Isolation, identification and PCR amplification of merA gene from highly mercury polluted yamuna river. Afr. J. Biotechnol. 9, 3510–3514. Available at: https://www.ajol.info/index.php/ajb/article/view/82390.
Zhang W., Chen L., Liu D. (2012). Characterization of a marine-isolated mercury-resistant pseudomonas putida strain SP1 and its potential application in marine mercury reduction. Appl. Microbiol. Biotechnol. 93, 1305–1314. doi: 10.1007/s00253-011-3454-5
Keywords: Mercury resistant bacteria, merA gene, FT-IR, GST enzyme, Metabolic pathway, Molecular docking
Citation: Joshi G, Verma P, Meena B, Goswami P, Peter DM, Jha DK, Vinithkumar NV and Dharani G (2022) Unraveling the potential of bacteria isolated from the equatorial region of Indian Ocean in mercury detoxification. Front. Mar. Sci. 9:986493. doi: 10.3389/fmars.2022.986493
Received: 05 July 2022; Accepted: 06 September 2022;
Published: 26 October 2022.
Edited by:
Meilin Wu, South China Sea Institute of Oceanology, (CAS), ChinaReviewed by:
Rajamanickam Krishnamurthy, Arignar Anna Government Arts and Science College Chennai, IndiaCopyright © 2022 Joshi, Verma, Meena, Goswami, Peter, Jha, Vinithkumar and Dharani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gajendra Joshi, Z2FqZW5kcmFqb3NoaTRAZ21haWwuY29t
†Present address: Prasun Goswami, Indian Institute of Science Education and Research, Kolkata, India
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.