
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 22 August 2022
Sec. Marine Ecosystem Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.982234
This article is part of the Research Topic Degradation, Ecological Restoration and Adaptive Management of Estuarine Wetlands under Intensifying Global Changes View all 20 articles
There are different environmental pressures in coastal and inland wetlands resulting in phenotypic variation among plant populations, which might be related to epigenetic modifications. Phragmites australis is a widespread plant in coastal and inland wetlands, and the phenotype of the population is selected by salinity, but the reasons for the population differences in salt tolerance and phenotype are unclear. We investigated coastal and inland P. australis populations grown under two salinities and two manipulated DNA methylation levels in common gardens. The plants were sprayed with 5-azacytidine (DNA demethylation agent) regularly, and the physiological and morphological traits of reeds were measured. Plant height, density, and basal stem of reeds from different sources were significantly different and correlated with soil conductivity of sampling sites (P < 0.05). Salinity significantly decreased the biomass (37.04%, P < 0.05) and plant height (24.68%, P < 0.05) of inland reeds but had no significant effect on coastal populations (13.48%). P. australis responds to salt stress through phenotypic plasticity, and inland wetland populations exhibit local adaptation to freshwater. Increased salt tolerance in inland populations following DNA demethylation, particularly biomass, plant height, and basal stems (increased 23.62%; 13.08%; 5.35%, respectively), could provide more opportunities in adverse environments. This study will provide important insights into the highly adaptive mechanisms of the large non-model plant.
Coastal wetlands face increasing salinization, but this is not happening in inland wetlands (Jacqueline et al., 2008; Zhou et al., 2022). Plants growing in coastal and inland wetlands experience different environmental stresses. Plants respond to heterogeneous environments through phenotypic variation resulting from genetic variation or epigenetic modification, since the dispersal ability of most plants is limited (Nicotra et al., 2010). Spontaneous new mutations in epigenetic variation, such as methylomes, are more likely to occur than in genetic variation (i.e., DNA sequences) (Herrera and Bazaga, 2010; Becker et al., 2011). DNA methylation (the best-studied epigenetic mechanism in plants) has been widely proved to be strongly associated with adaptive phenotypic traits in natural plant populations (Platt et al., 2015; Liu et al., 2018). Phenotypic variation related to fitness should be subject to natural selection, which leads to population differentiation among different ecological environments. Epigenetic modifications, very similar to the DNA sequence, are usually divergent in other plant species and populations. Methylation alterations of populations associated with morphological changes and adaptive genetic divergence were detected in different habitats (Gao et al., 2010; Herrera and Bazaga, 2010). Thus, DNA methylation variety might be responsible for the phenotypic differentiation in natural populations together with genetic variation.
Environmental stimulation could change DNA methylation patterns in many species (Zhang et al., 2013; Thiebaut et al., 2019). In different conditions, plant populations have specific epigenetic (DNA methylation) loci which were inferred to have a functional role in population differentiation. The adaptive mechanisms of DNA methylation in stress responses are reasonably well understood for model plants Arabidopsis thaliana (Dowen et al., 2012; Schmid et al., 2018); for example, regulated gene expression and DNA methylation in the promoter play an important role in salt tolerance (Baek et al., 2011). However, the relationship between epigenetics and the environment might vary from species to species in non-model species. In the wild plant populations, the 5-methylcytosine of Spartina alterniflora and Borrichia frutescens was significantly correlated with salinity gradients (Foust et al., 2016). The comparison of mangroves (Laguncularia racemosa) in salt marsh and riverside showed morphological dissimilarities and DNA methylation levels of polymorphism (Medeiros et al., 2010). This relationship is consistent in environmental variation and epigenetics related to water availability and temperature adaptation in Allopolyploid orchids (Paun et al., 2010). Methylation and direct environmental relations in stress resistance, geographical distribution, and species differentiation are still the focus of research.
Common reed (Phragmites australis (Cav.) Trin. ex Steud.) is a cosmopolitan species with high phenotypic plasticity (Eller et al., 2017; Packer et al., 2017). Salinity is a significant selective factor in P. australis populations. With the salinity gradient, morphological, physiological, or genetically different, heterogeneous, various ecotypes were formed in the coastal wetland and inland wetland (Zhao et al., 1999; Zhang et al., 2003; Zhuang et al., 2010). Many studies have found that salt tolerance in P. australis was related to phenotypic plasticity and genetic differentiation (Achenbach et al., 2013; Song et al., 2021). It has also shown that methylation patterns of different ecotypes of reed vary in saline-alkali meadow habitats in China, and phenotypic traits were more closely related to epigenetic diversity than genetic diversity (Liu et al., 2020; Qiu et al., 2021). However, it is still unclear whether the differences in phenotypic plasticity of different populations of reeds in coastal habitats are caused by DNA methylation.
The modified DNA methylation triggers phenotypic plasticity and promotes rapid adaptation and transgenerational inheritance (Gonzalez et al., 2016; Puy et al., 2021). In recent years, artificial demethylation methods have been used to study the significant effects of epigenetic modification on functional traits, fitness, and ecological interactions. DNA demethylating agent 5-azacytidine (5-azaC) was a direct inhibition of methylation by inhibiting transcription and indirect elimination of methyltransferase (Gurminder et al., 2000; Bossdorf et al., 2010). Seed soaking methods in previous studies have inside effects like lower germination and reduced vigor and survival (Bossdorf et al., 2010). The novel approach of spraying demethylation has shown a good demethylation effect with less harm to plants (Puy et al., 2018; Münzbergová et al., 2019). DNA methylation is unstable, and environmentally sensitive, high-salinity environments induce genome-wide DNA hypermethylation, indicating robust epigenome regulation in the presence of a stressor factor (Miryeganeh et al., 2021).
In this study, we compared the salt tolerance of coastal and inland populations of P. australis. We quantify trait variation of two populations transplanted to two salinity conditions with specific demethylation treatment. In our study, we aimed to investigate (1) whether the P. australis growing in inland and coastal areas are ecophysiological traits different in response to salinity and (2) whether DNA methylation variation plays a role in population ecophysiological traits variation in salt adaptation.
We collected rhizomes of P. australis from coastal wetland Yellow River Delta and inland wetland Mata Lake (Figure 1). The Yellow River Delta and Mata Lake Wetlands are located in eastern China. Yellow River Delta is the mouth of the Yellow River and is the youngest estuarine wetland in China. Seawater erosion makes the soil and groundwater contain more soluble salt, and the soil salinization is serious (Valladares et al., 2014; Guan et al., 2016). Mata Lake is formed by the convergence of seven rivers and is known as a typical inland shallow lake wetland in northern China. Mata lake is low-lying, with 1,334 ha of reed fields, and has good natural ecological resources (Li et al., 2007). Rhizomes were collected in each wetland as experimental materials. We also collected the soil samples with a five-point sampling method for pH, soil conductivity, and nitrogen and phosphorus content measurement at each site. Soil conductivity was measured by a conductivity meter (FE38, Mettler-Toledo, China). Soil pH was measured by a pH meter (FE28, Mettler-Toledo, China). The total soil N content (%) was measured by Kjeldahl determination (K9860, Hanon, China). The full P content (%) was measured with the molybdenum bismuth photometer and UV spectrophotometer (UV-2550, Shimadzu, Japan), and the leaf ratio of nitrogen to phosphorus was calculated.
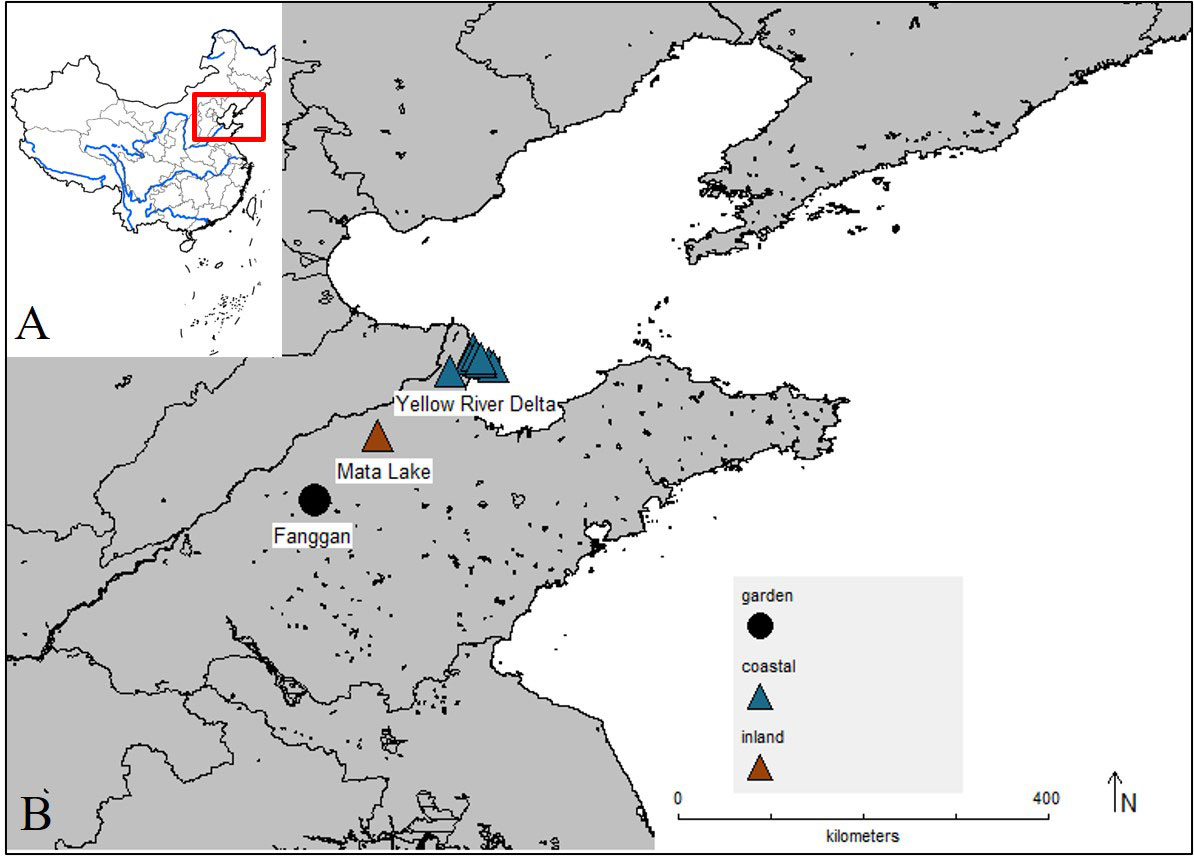
Figure 1 Map of China (A) highlighting the location of the inland and coastal wetland populations of Phragmites australis (B) collected and used in our experiment and the location of common garden (Fanggan).
The rhizomes were taken back to the greenhouse at Shandong University Fanggan experimental station (36°26′N, 117°27′E). They were propagated in shallow sand for 4 weeks to preculture and initiate adventitious shoot at the stem nodes. The adventitious shoots were planted with rhizome when 15–20 cm high into 16-l pots (20 cm in diameter and 25 cm in height) filled with local river sand and 10 g of slow-release fertilizer per pot (Peters Professional NPK 20-20-20 + Fe, The Scotts Company, USA). The seedlings were sufficiently watered with tap water twice a week in the first month and then once a day. The average temperature of the greenhouse was 25.75°C, and humidity was 73.23% during the experiment. The pots were placed in random order to minimize undesired position effects.
The experiment was a 2 × 2 × 2 factorial design with the factors being “origin” (coastal, inland), “demethylation” (0, 100 μmol), and “salt” (0‰, 20‰). There were five samples from inland wetland and 10 samples from coastal wetland, a total of 60 pots. The collected plants were preadapted for 1 month with daily cultivation in the greenhouse. The experiment ran for 106 days after that, from 1 June 2019 to 30 September 2019. Each pot reproduced asexually multiple plants from the first reed seedlings and rhizomes during the experiment.
The DNA demethylation process started on 30 June 2019, and the plants were divided into two chambers, separated by plastic sheets. The plants in one chamber were sprayed with 100 μmol 5-azacytidine solution of DNA demethylation agent and mixed with 2 ml of surfactant (silicone additive), and the other plants in another chamber were sprayed with water and 2 ml surfactant as a control group (Puy et al., 2018; Herrera et al., 2019; Münzbergová et al., 2019). The treatment was carried out in the later afternoon every 2 days from June 30 to August 10. The plants were sprayed around to ensure that each leaf was sprayed.
The salt treatment was added with 40 g salt (NaCl) on July 30 to half pots of each clone both in the demethylation group and in the control group. The salt treatment continued until the experimental harvest time, when additional salt was added and watered normally.
The highest six plant heights in each pot were measured every month. We measured five shoots’ base diameters for mean value and counted the shoot number in each pot at the end of the experiment. The flowering rate and aphid-infected rate were determined as cumulative flowering number and aphid-infected shoot number divided by the total shoots in each pot, respectively. Finally, all pots were harvested and dried at 85°C to constant weight with an oven for dry stems, leaves, flowers, and belowground biomass.
Three fully developed leaves were chosen in each pot before harvest for specific leaf area (SLA) analysis. The leaves were scanned with a scanner (Canon E560) and dried to constant weight. The leaf area was determined by ImageJ ver. 1.52. The specific leaf area was estimated based on the ratio of leaf area to leaf dry mass.
Leaf chlorophyll parameters were measured at the end of experiment. Five circle pieces were sampled with a hole puncher in fully expanded and healthy leaves and measured for fresh weight. After extraction by 95% ethanol (v/v), the concentrations of leaf chlorophyll a and b and total chlorophyll (mass-based) were calculated as below (Lichtenthaler, 1987).
Chla = 13.95 A665 - 6.88 A649
Chlb = 24.96 A649 - 7.32 A665
Chl = Chla+Chlb
To assess the differences in water availability of the leaf, the relative water content (RWC) was measured. Three pieces of leaves were cut out in each pot, and the fresh weight was weighed as Wf with the analytical balance. The leaves were placed in a sealed ziplock bag and soaked for 24 h in tap water, and the leaves were gently blotted and quickly weighed as the saturated water content (Ws). The leaves were dried at 85°C to constant weight with an oven (48 h) for the dry weight (Wd).
RWC (%) = (Wf - Wd)/(Ws - Wd) ×100%
Photosynthesis was measured on the third youngest fully expanded leaf with LI-6800 (Portable Photosynthesis System, LICOR, USA) in each pot, including net photosynthetic rate (Pn), respiratory rate (E), and stomach conductance (gs). It was conducted on the morning of a sunny day at a 500-ml s−1 flow rate with 70% influx air relative humidity, 400 ppm CO2 concentration, and 1,800 mol m−2 s−1 PAR. We measured the gas exchange parameters four times on July 30 (at the demethylation for 1 month), August 1 (on the first day after salinity treatment), August 8 (at the end of demethylation and continuing salinity treatment), September 17 (before the final harvest), separately.
The phenotypic plasticity indexes (PPI) of traits were calculated, and the calculation formula was as follows: PPI = (Vmax - Vmin)/Vmax, where Vmax represented the maximum value of the trait, and Vmin represented the minimum value of the trait. PPI values range from 0 to 1, where 0 means no plasticity and 1 means maximum plasticity.
We tested the effect of origin, salinity, demethylation, and all their interactions on all the variables with three-way ANOVA. Multiple levels of main effects were compared by Duncan’s multiple-range tests at the 5% significance level. The effects of 5-azaC, salinity, and their interaction were tested as three variance sources, using the tray mean squares as error term. The relationship between soil characteristics and plant traits was analyzed by simple linear analysis. Data analyses were performed using the SPSS 22.0 software package (SPSS Inc., Chicago, IL). Figures were drawn using Origin 9.0 software (OriginLab Co., Northampton, MA).
Salinity significantly decreased total biomass (inland 37.04%; coastal 13.48%) and aboveground biomass (Table 1; Figure 2A). The biomass of the stem, leaf, and flower decreased significantly with the increase in salinity, and the ratio of root to shoot was significantly increased (Table 1; Figure 2B). Different origins mainly produced significant differences in underground biomass and root-to-root ratio. The underground biomass of inland is greater than that of coastal. DNA demethylation increased the biomass of inland and coastal populations, but it was not statistically significant. There is no interaction with biomass between salt, source, and demethylation (Table 1).
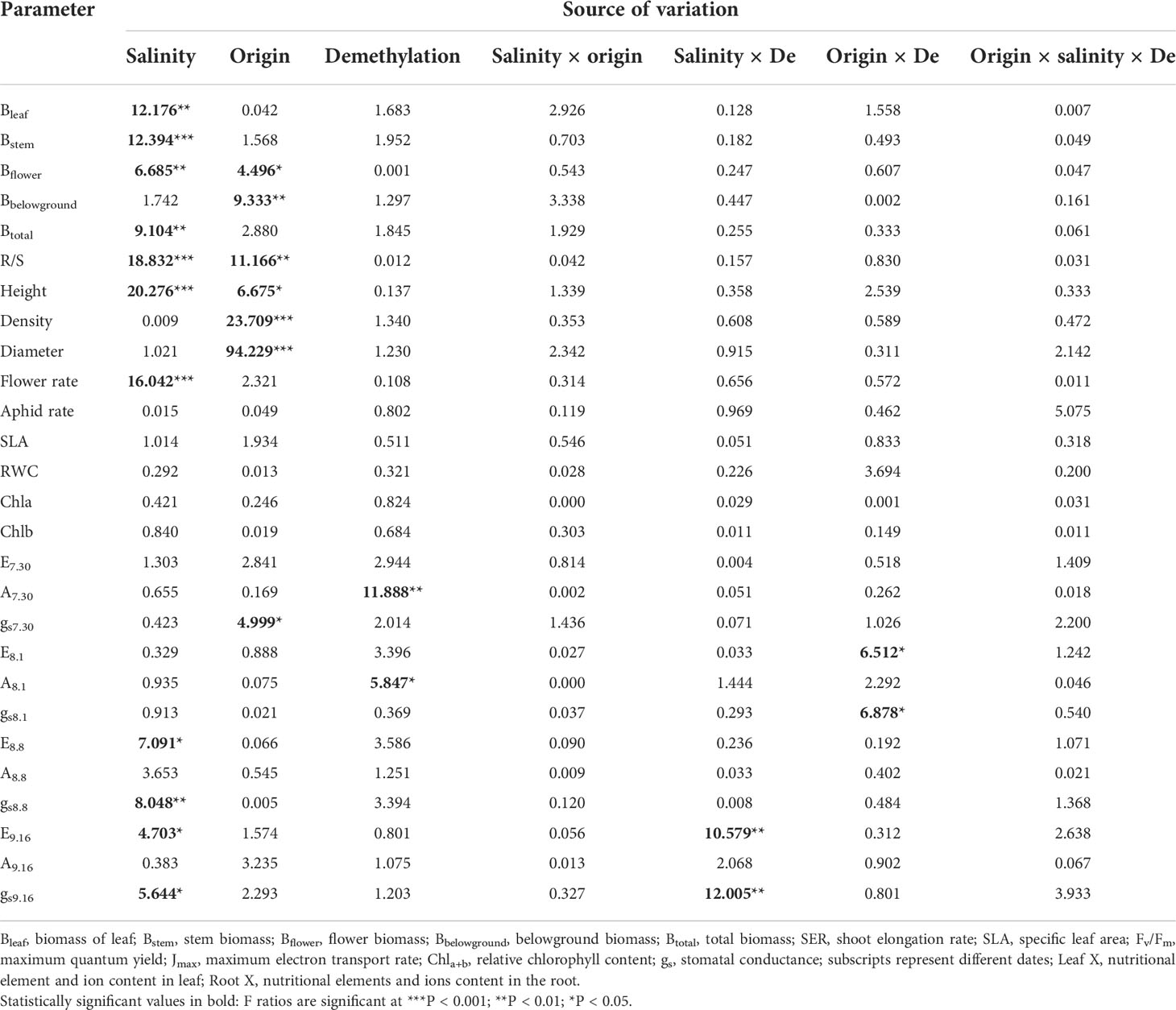
Table 1 Three-factor analysis of variance for phenotypic variance of different Phragmites australis origins with salinity and demethylation treatments.
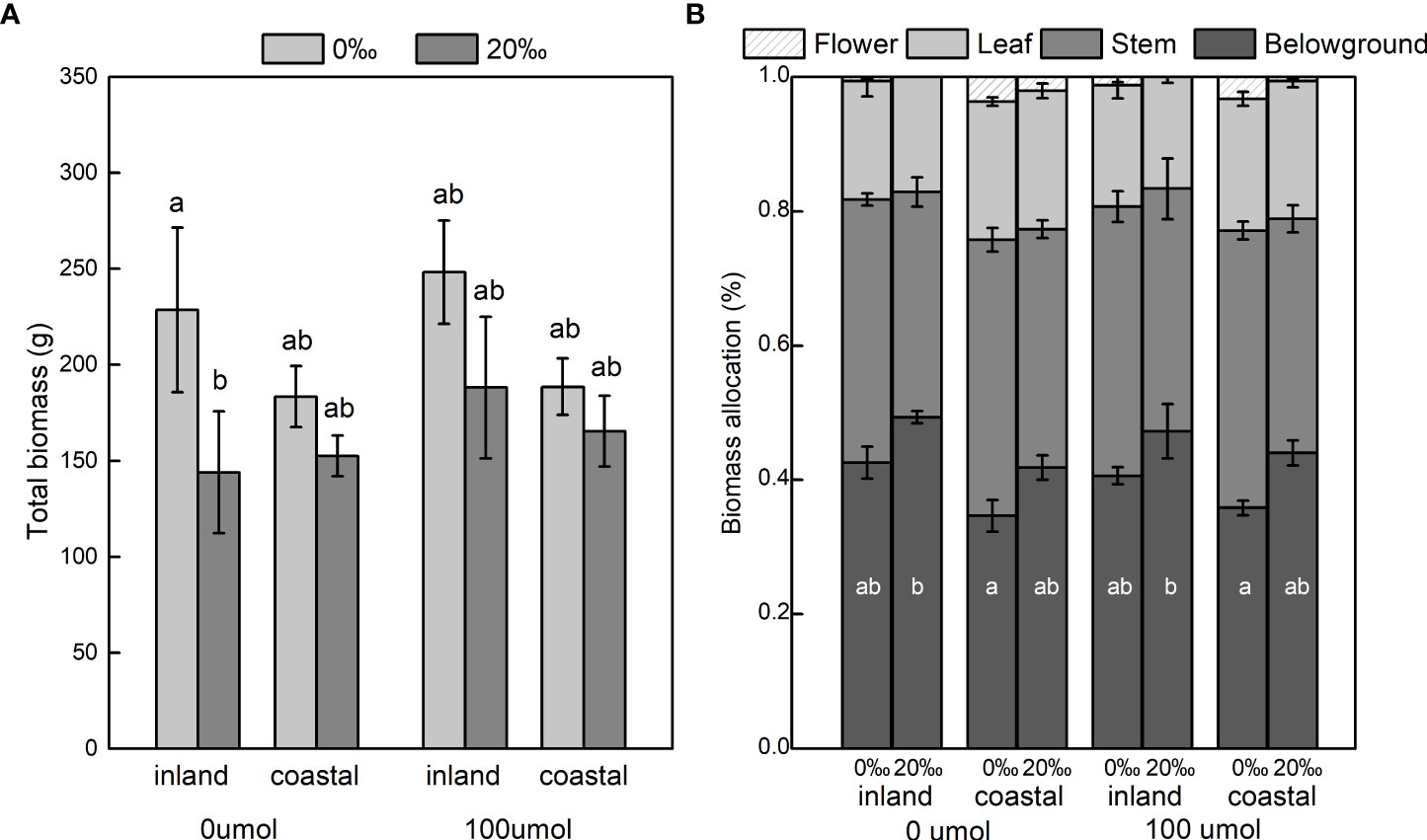
Figure 2 Total dry mass (A) and biomass allocation (B) of Phragmites australis populations grown with salinity (0‰ vs. 20‰) and DNA demethylation (0 vs. 100 μmol) treatment, respectively. Mean ± SE. Letters in a indicate significant differences between all treatments for total biomass, and letters inside bars in b indicate differences between treatments for each plant part. Bars sharing the different letters are different at P < 0.05.
After adding salt, the plant height decreased in the demethylation and no demethylation groups and especially significantly decreased in inland population. There were significant differences in density, diameter, and height between reeds from different origins, but the changing trends of plant height, diameter, and density were different. The inland population had greater plant height and basal diameter than the coastal population (Table 1; Figure 3). The density of reeds was lower in inland populations than coastal populations. Density and basal diameter were significantly negatively correlated Table 2 (r = 0.565, P = 0.000, n = 56). Plant density was positively correlated with soil electrical conductivity Table 2 (Pearson correlation, r = -0.599, P = 0.000, n = 56), while reed base diameter was negatively correlated with soil electrical conductivity at the sampling site Table 2 (Pearson correlation, r = -0.538, P = 0.000, n = 56). There was no significant difference in specific leaf area at the end of the experiment. Except for the flowering rate, the plasticity index of all phenotypic traits of the coastal population was higher than the inland population (Figure 4).
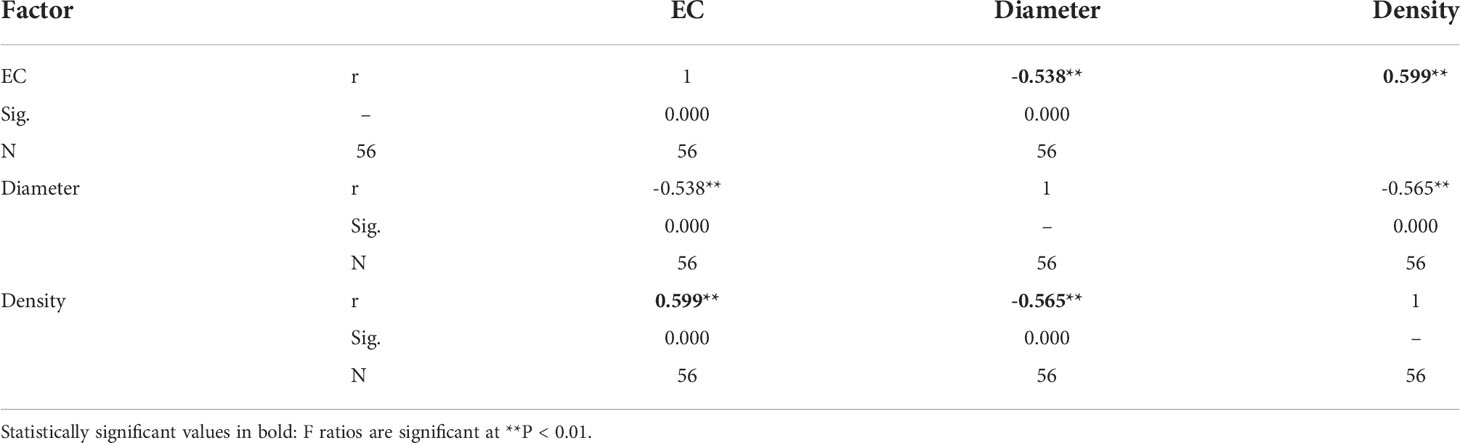
Table 2 Correlation between soil electrical conductivity and Phragmites australis diameter and density.
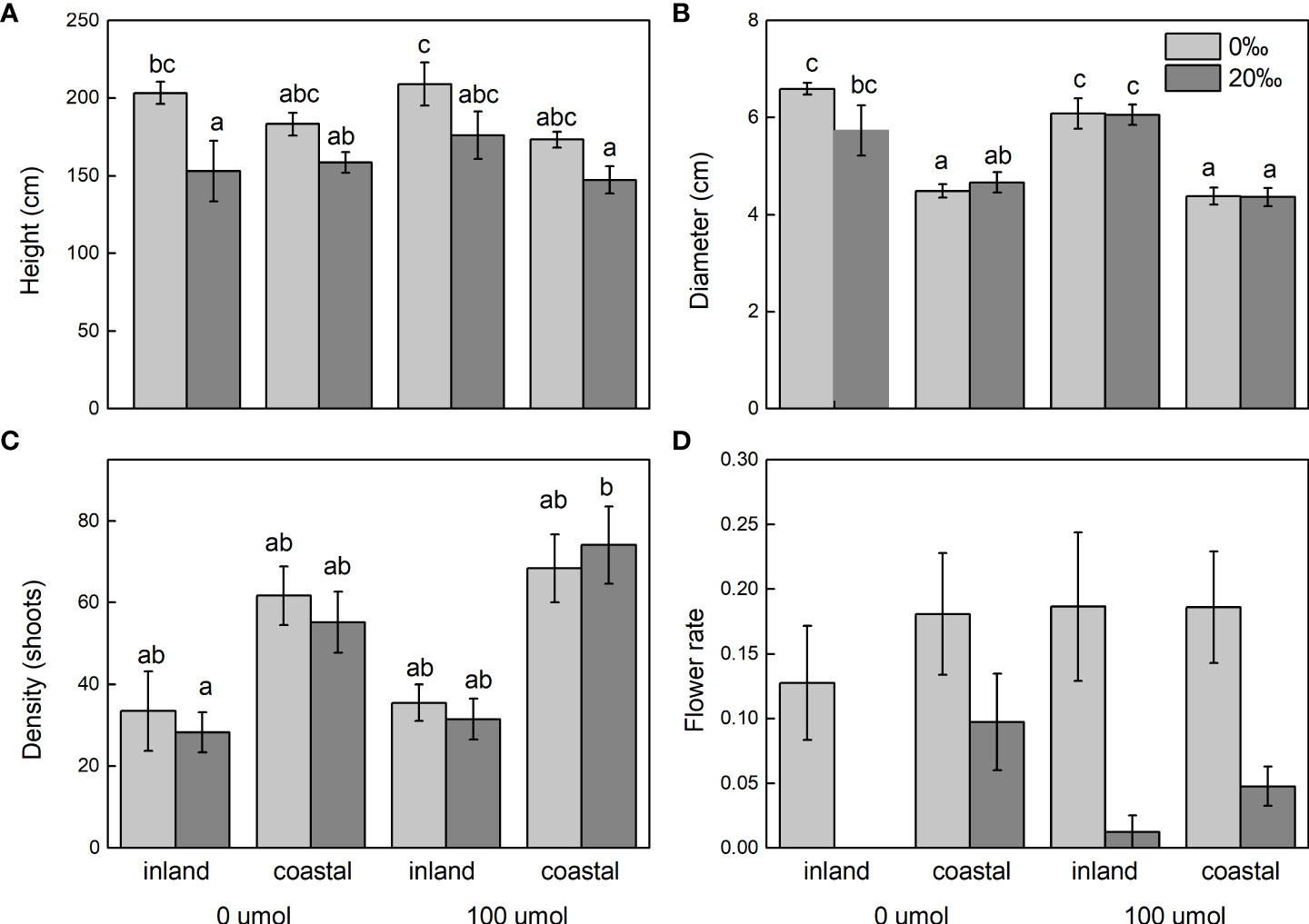
Figure 3 Shoot height (A), diameter (B), density (C), and flower rate (D) of Phragmites australis populations grown with salt (0‰ vs. 20‰) and DNA demethylation (0 vs. 100 μmol) treatment, respectively. Bars sharing the different letters are different at P < 0.05. Mean ± SE.
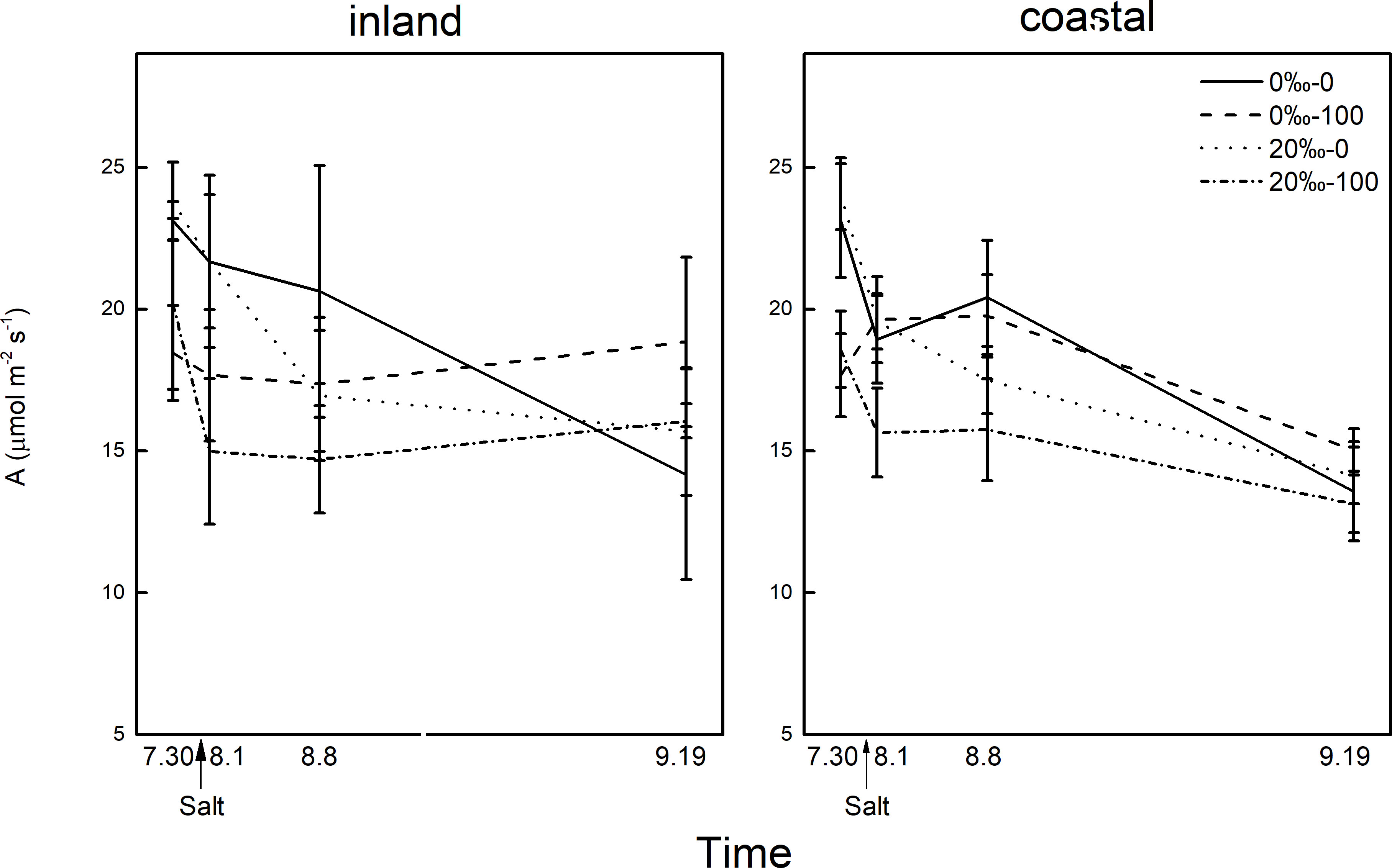
Figure 4 The difference in photosynthetic rate (A)of Phragmites australis populations grown with salt (0‰, 20‰) and DNA demethylation treatment (0, 100 μmol) at different times.
The flower biomass, flowering number, and flowering rate were significantly reduced by the influence of salinity (Table 1). There were also significant differences in flower biomass of reeds from different origins. The flowering number of the coastal population was larger than that of the inland population, and the number of aphids in the coastal population was significantly larger than that of the inland population.
The demethylation treatment significantly reduced the photosynthetic rate, and the transpiration and stomatal conductance of reeds from inland origin was greater than that of coastal reeds (Table 1; Figure 4). The photosynthetic rate decreased the next day after salt addition, and the transpiration rate and stomatal conductance were affected by the interaction between the origin and DNA demethylation. After 1 week of salt added, the photosynthetic transpiration stomatal conductance was significantly reduced, and the stomatal conductance was affected by demethylation. After 45 days of salt treatment, the difference in photosynthesis rate disappeared. Transpiration and stomatal conductance were affected by the interaction of salt and demethylation. At the end of the experiment, there was no significant difference in leaf chlorophyll and relative water content.
The phenotypic plasticity index of biomass (0.3–0.8) was higher than other parameters (0.1–0.4; Figure 4) in both inland and coastal origins. The PPI of root biomass, stem root biomass, leaf root biomass, and total biomass, plant height, density, and basal stem in the coastal group were all higher than that in the inland group (Figure 5,).
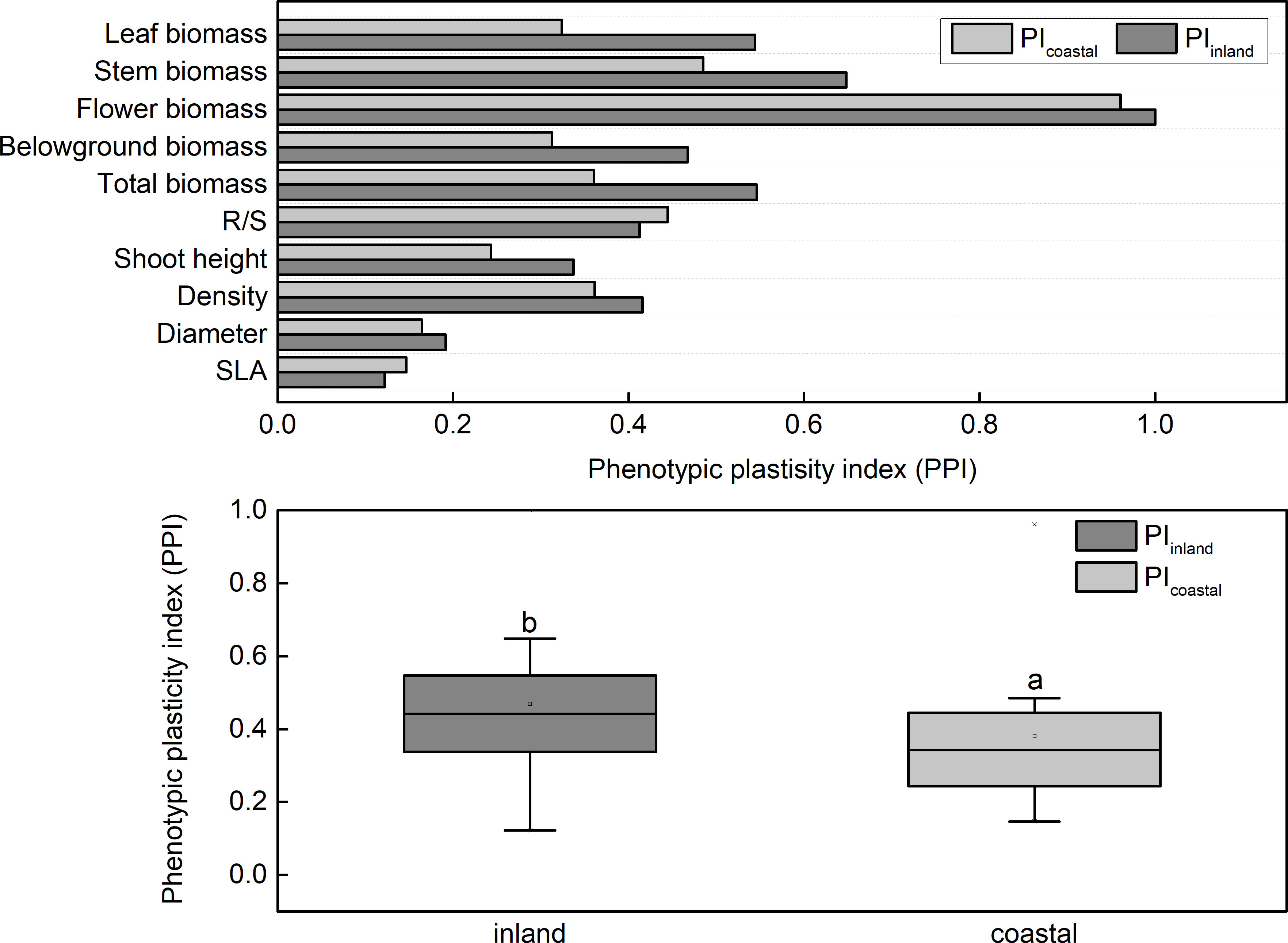
Figure 5 Phenotypic plasticity index (PPI) of Phragmites australis populations grown with salt and DNA demethylation treatment.
In our common garden, we found that the inland population had a higher fitness (biomass-based) in 0‰ salinity; the biomass, shoot height, and diameter of the inland population were negatively affected by salinity. This result showed an advantage of the inland population in freshwater habitat; this advantage means a local adaptation to freshwater. In addition, the inland population of P. australis had higher and thicker shoots than the coastal population, but stem density was much smaller, both in this common garden experiment and in previous research (Hanganu et al., 1999; Song et al., 2021). We determined the relationship between morphology and soil conductivity of the sampling site through correlation analysis. The wild population of P. australis has a phenotypic differentiation between freshwater and saltwater. The long-term environmental differences in the field conditions make the adaptability differences between the populations, which has also been found in other studies (Hanganu et al., 1999; Burdick and Konisky, 2003; Zhang et al., 2003).
There were no significant effects on fitness and traits in the coastal population before and after salt was added, indicating no local adaptation to salt. All populations increased underground biomass to increase water absorption and maintain osmotic pressure under salt stress. Photosynthesis decreased in the short term and maintained a normal level after a long-term response. Reduced flowering rate showed a delayed reproductive period. In our previous studies, it was believed that the salt tolerance of P. australis was mainly affected by phenotypic plasticity (Song et al., 2020; Zhou et al., 2021). In this study, P. australis adjusted the phenotype in the salt environment; however, they can still maintain the physiology without dying, confirming that phenotypic plasticity can increase the ability of the P. australis population to adapt to the salinity environment. Although the importance of phenotypic plasticity and local adaptation differs in populations, the adaptive phenotypic variation resulting from the two strategies together influences the actual performance of the plant.
DNA demethylation resulted in higher biomass and shoot height in the inland population after adding salt. The significant difference caused by salt was weakened after demethylation (Figures 2, 3A), and this alteration was not found in the coastal population. The same trends were found in the diameter, flowering rate, and photosynthetic rate of P. australis. These showed that DNA demethylation enhanced the salt tolerance of the inland population. It may be that the higher level of methylation makes the plant not tolerant to salt. In a recent study, it has been confirmed that hypomethylation leads to more significant activation of salt stress-related genes (jasmonic acid synthesis and signal transduction genes) in tetraploid rice, enhancing its salt tolerance (Wang et al., 2021). Previous studies have found a correlation between the response of epigenetic regulation to specific environmental factors and phenotype through amplified fragment length polymorphism (AFLP) and methylation-sensitive amplification polymorphism (MSAP) (Gurminder et al., 2000; Gao et al., 2010). The correlation between 5-methylcytosine and phenotypic plasticity has also been proved (Fieldes et al., 2005; Gao et al., 2010; Zhang et al., 2013). This experiment further proved that epigenetic variation is reversible; this could adjust the salt adaptation of P. australis. Although the plants were pre-cultured, the maternal effects and genetic diversity might confound the results; more epigenetic modifications cross-generational experiments are still needed.
Experimental DNA demethylation did not affect the traits of P. australis at the end of experiment, except for short-term photosynthesis damage. Spraying DNA demethylation agent 5-azacytidine changed the salt tolerance among reed populations from different origins but was not harmful to the plant. 5-Azacytidine has a broad cell expression cytotoxicity when less than 100 µmol, has no targeted and whole effect on plant performance, and is expected to increase the gene expression and transposon variation of transposons (Klaas et al., 1989; Griffin et al., 2016). A similar method used found considerable intraspecific heterogeneity in the phenotypic and ecological correlation of plants (Herrera et al., 2019). In other studies, DNA demethylation by seed soaking strongly reduced the growth and adaptability of plants and delayed their flowering (Bossdorf et al., 2010). The toxic effect of 5-azacytidine was excluded from the experiment. Experimental DNA demethylation is a useful and practical method to study the importance of epigenetic modification of the genome for plant response to the environment. For large plants, experimental treatment dose and time should be considered.
Although phenotypic differentiation of populations occurs in heterogeneous habitats and phenotypic changes may be associated with DNA methylation, it has rarely been demonstrated that changes in methylation lead to tolerance to induced stress. Salinity and geographical origin had important effects on the physiology and morphology of P. australis. The variation of shoot height, density, and basal stem diameter of reed plants from the different origins was related to soil conductivity; the population from inland wetland showed a local adaptation to inland. Plant biomass and plant height were affected by salinity, and instantaneous photosynthetic response was also affected, but there is no difference after long-term adaptation. All P. australis responded to the salt environment through phenotypic plasticity. The phenotypic changes observed in 5-azaC-treated plants led to differences in morphological adaptation, which can provide more opportunities for reeds in unfavorable environments. Demethylation did not damage the plant traits. Collaborative investigations of experimental DNA changes and molecular methods should be used in plant experiments to understand the epigenetic variation of plant populations adapting to heterogeneous habitats.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
HS, XG, LL, and WG designed the experiment; HS, ZX, NW, and XL conducted the experiment and collected and analyzed the data; HS wrote the paper, LL, XG, and ND contributed critically to the drafts. All authors made a contribution to this work. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (grant numbers 31970347; 31500264; 32100304) and the Key Research & Development Program of Shandong Province (grant number 2021 CXGC010803).
We thank Dr. Shijie Yi in our laboratory of Shandong University for discussions on their technical assistance and helpful comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.982234/full#supplementary-material
Achenbach L., Eller F., Nguyen L. X., Brix H. (2013). Differences in salinity tolerance of genetically distinct phragmites australis clones. Aob Plant 5, plt019. doi: 10.1093/aobpla/plt019
Baek D., Jiang J., Jung-Sung C., Wang B., Chen J., Xin Z., et al. (2011). Regulated ATHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 52 (1), 149–161. doi: 10.1093/pcp/pcq182
Becker C., Hagmann J., Mueller J., Koenig D., Stegle O., Horgwardr K., et al. (2011). Spontaneous epigenetic variation in the arabidopsis thaliana methylome. Nature 480 (7376), 245–249. doi: 10.1038/nature10555
Bossdorf O., Arcuri D., Richards C. L., Pigliucci M. (2010). Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in arabidopsis thaliana. Evolutionary Ecol. 24 (3), 541–553. doi: 10.1007/s10682-010-9372-7
Burdick D., Konisky R. A. (2003). Determinants of expansion for phragmites australis, common reed, in natural and impacted coastal marshes. Estuaries 26 (2), 407–416. doi: 10.1007/bf02823717
Dowen R. H., Pelizzola M., Schmitz R. J., Lister R., Dowen J. M., Nery J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. United States America 109 (32), E2183–E2191. doi: 10.1073/pnas.1209329109
Eller F., Skalova H., Caplan J. S., Bhattarai G. P., Burger M. K., Cronin J. T., et al. (2017). Cosmopolitan species as models for ecophysiological responses to global change: The common reed phragmites australis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01833
Fieldes M. A., Schaeffer S. M., Krech M. J., Brown J. C. L. (2005). DNA Hypomethylation in 5-azacytidine-induced early-flowering lines of flax. Theor. Appl. Genet. 111 (1), 136–149. doi: 10.1007/s00122-005-2005-9
Foust C. M., Preite V., Schrey A. W., Alvarez M., Robertson M. H., Verhoeven K. J., et al. (2016). Genetic and epigenetic differences associated with environmental gradients in replicate populations of two salt marsh perennials. Mol. Ecol. 25 (8), 1639–1652. doi: 10.1111/mec.13522
Gao L., Geng Y., Li B., Chen J., Yang J. (2010). Genome-wide DNA methylation alterations of alternanthera philoxeroides in natural and manipulated habitats: implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ. 33 (11), 1820–1827. doi: 10.1111/j.1365-3040.2010.02186.x
Gonzalez A. P., Chrtek J., Dobrev P. I., Dumalasova V., Fehrer J., Mraz P., et al. (2016). Stress-induced memory alters growth of clonal offspring of white clover (Trifolium repens). Am. J. Bot. 103 (9), 1567–1574. doi: 10.3732/ajb.1500526
Griffin P.T., Niederhuth C.E., Schmitz R.J.. (2016). A Comparative Analysis of 5-Azacytidine- and Zebularine-Induced DNA Demethylation. G3-Genes Genomes Genetics,6(9) 2773–80. doi: 10.1534/g3.116.030262
Guan B., Yu J., Hou A., Han G., Wang G., Qu F., et al. (2016). The ecological adaptability of phragmites australis to interactive effects of water level and salt stress in the yellow river delta. Aquat. Ecol. 51 (1), 1–10. doi: 10.1007/s10452-016-9602-3
Gurminder S. T., Jacintha M., Chendanda C. C., Reid D. M. (2000). Effect of light quality and 5-azacytidine on genomic methylation and stem elongation in two ecotypes of stellaria longipes. Physiologia Plantarum 109 (3), 313–321. doi: 10.1034/j.1399-3054.2000.100313.x
Hanganu J., Mihail G., Coops H. (1999). Responses of ecotypes of phragmites australis to increased seawater influence: a field study in the Danube delta, Romania. Aquat. Bot. 64 (3-4), 351–358. doi: 10.1016/S0304-3770(99)00062-5
Herrera C. M., Bazaga P. (2010). Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet viola cazorlensis. New Phytol. 187 (3), 867–876. doi: 10.1111/j.1469-8137.2010.03298.x
Herrera C. M., Medrano M., Perez R., Bazaga P., Alonso C. (2019). Within-plant heterogeneity in fecundity and herbivory induced by localized DNA hypomethylation in the perennial herb helleborus foetidus. Am. J. Bot. 106 (6), 798–806. doi: 10.1002/ajb2.1291
Jacqueline S., Kay M., Paul I. B. (2008). Does salinity reduce the tolerance of two contrasting wetland plants, the submerged monocot vallisneria australis and the woody shrub melaleuca ericifolia, to wetting and drying? Mar. Freshw. Res. 59 (4), 291–303. doi: 10.1071/MF07147
Klaas M., John M.c., Crowell D.N., Amasino R.M.. (1989). Rapid induction of genomic demethylation and T-DNA gene expression in plant cells by 5-azacytosine derivatives. Plant Molecular Biology,12(4) 413–23. doi: 10.1007/BF00017581
Lichtenthaler H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymology 148C (1), 350–382. doi: 10.1016/0076-6879(87)48036-1
Li L., Fan X., Liu X. (2007). ASEB grid and its planning applying the demonstrative research on the lake mata in ziho. J. Dezhou Univ. 23 (002), 63–6581.
Liu L., Pei C., Liu S., Guo X., Guo W. (2018). Genetic and epigenetic changes during the invasion of a cosmopolitan species (Phragmites australis). Ecol. Evol. 8 (13), 6615–6624. doi: 10.1002/ece3.4144
Liu L., Yin M., Guo X., Yu X., Song H., Eller F., et al. (2020). The river shapes the genetic diversity of common reed in the yellow river delta via hydrochory dispersal and habitat selection. Sci. Total Environ. 764, 144382. doi: 10.1016/j.scitotenv.2020.144382
Medeiros C., Parisod C., Fernandes R. A., Mata C. S., Cardoso M. A., Ferreira P. C. G. (2010). Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One 5 (4), e10326. doi: 10.1371/journal.pone.0010326
Miryeganeh M., Marlétaz F., Gavriouchkina D., Saze H. (2021). De novo genome assembly and in natura epigenomics reveal salinity-induced DNA methylation in the mangrove tree bruguiera gymnorhiza. New Phytol. 233 (5), 2094–2110. doi: 10.1111/nph.17738
Münzbergová Z., Latzel V., Šurinová M., Hadincová V. (2019). DNA Methylation as a possible mechanism affecting ability of natural populations to adapt to changing climate. Oikos 128 (1), 124–134. doi: 10.1111/oik.05591
Nicotra A. B., Atkin O. K., Bonser S. P., Davidson A. M., Finnegan E. J., Mathesius U., et al. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15 (12), 684–692. doi: 10.1016/j.tplants.2010.09.008
Packer J. G., Meyerson L. A., Skálová H., Pyšek P., Kueffer C. (2017). Biological flora of the british isles: Phragmites australis. J. Ecol. 105 (4), 1123–1162. doi: 10.1111/1365-2745.12797
Paun O., Bateman R. M., Fay M. F., Hedren M., Civeyrel L., Chase M. W. (2010). Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol. Biol. Evol. 27 (11), 2465–2473. doi: 10.1093/molbev/msq150
Platt A., Gugger P. F., Pellegrini M., Sork V. L. (2015). Genome-wide signature of local adaptation linked to variable CpG methylation in oak populations. Mol. Ecol. 24 (15), 3823–3830. doi: 10.1111/mec.13230
Puy J., D. H., Carmona C. P., Bello F. D., Hiiesalu I., Latzel. V. (2018). Improved demethylation in ecological epigenetic experiments: Testing a simple and harmless foliar demethylation application. Methods Ecol. Evol. 9 (3), 744–753. doi: 10.1111/2041-210x.12903
Puy J., de Bello F., Dvorakova H., Medina N. G., Latzel V., Carmona C. P. (2021). Competition-induced transgenerational plasticity influences competitive interactions and leaf decomposition of offspring. New Phytol. 229 (6), 3497–3507. doi: 10.1111/nph.17037
Qiu T., Liu Z., Yang Y., Liu B. (2021). Epigenetic variation associated with responses to different habitats in the context of genetic divergence in phragmites australis. Ecol. Evol. 11 (17), 11874–11889. doi: 10.1002/ece3.7954
Schmid M. W., Heichinger C., Coman Schmid D., Guthorl D., Gagliardini V., Bruggmann R., et al. (2018). Contribution of epigenetic variation to adaptation in arabidopsis. Nat. Commun. 9 (1), 4446. doi: 10.1038/s41467-018-06932-5
Song H., Guo X., Yu X., Liu L., Wang N., Eller F., et al. (2021). Is there evidence of local adaptation of phragmites australis to water level gradients and fluctuation frequencies? Sci. theTotal Environ. 756, 144065. doi: 10.1016/j.scitotenv.2020.144065
Song H., Jespersen E., Guo X., Du N., Xie L., Pei L., et al. (2020). Differences in relative air humidity affect responses to soil salinity in freshwater and salt marsh populations of the dominant grass species phragmites australis. Hydrobiologia 848, 3353–3369. doi: 10.1007/s10750-020-04285-z
Thiebaut F., Hemerly A. S., Ferreira P. C. G. (2019). A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00246
Valladares F., Matesanz S., Guilhaumon F., Araujo M. B., Balaguer L., Benito-Garzon M., et al. (2014). The effects of phenotypic plasticity and local adaptation on forecasts of species range shifts under climate change. Ecol. Lett. 17 (11), 1351–1364. doi: 10.1111/ele.12348
Wang L., Cao S., Wang P., Lu K., Chen Z. J. (2021). DNA Hypomethylation in tetraploid rice potentiates stress-responsive gene expression for salt tolerance. Proc. Natl. Acad. Sci. 118 (13), e2023981118. doi: 10.1073/pnas.2023981118
Zhang Y. Y., Fischer M., Colot V., Bossdorf O. (2013). Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 197 (1), 314–322. doi: 10.1111/nph.12010
Zhang S., Wang R., Zhang Z., Guo W., Liu J., Song B. (2003). Study on morphological variation of phragmites australis in the yellow river downstream wetland. Acta Phytoecological Sin. 27 (1), 78–85. doi: 10.1023/A:1022289509702
Zhao K. F., Feng L. T., Zhang S. Q. (1999). Study on the salinity-adaptation physiology in different ecotypes of phragmites australis in the yellow river delta of China: Osmotica and their contribution to the osmotic adjustment. Estuar. Coast. Shelf Sci. 49 (1), 37–42. doi: 10.1016/S0272-7714(99)80006-7
Zhou D., Ni Y., Yu X., Lin K., Guo W. (2021). Trait-based adaptability of phragmites australis to the effects of soil water and salinity in the yellow river delta. Ecol. Evol. 11 (16), 11352–11361. doi: 10.22541/au.161342230.01798188/v1
Zhou Q., Wang S., Liu J., Hu X., Liu Y., He Y., et al. (2022). Geological evolution of offshore pollution and its long-term potential impacts on marine ecosystems. Geosci. Front. 12(5), 101427. doi: 10.1016/j.gsf.2022.101427
Keywords: DNA methylation, Phragmites australis, phenotypic plasticity, physiological traits, salt tolerance
Citation: Song H, Guo X, Liu L, Xu Z, Wang N, Liu X, Du N and Guo W (2022) Role of DNA methylation in ecophysiological responses to salinity in natural populations of Phragmites australis from coastal and inland habitats. Front. Mar. Sci. 9:982234. doi: 10.3389/fmars.2022.982234
Received: 30 June 2022; Accepted: 02 August 2022;
Published: 22 August 2022.
Edited by:
Junhong Bai, Beijing Normal University, ChinaCopyright © 2022 Song, Guo, Liu, Xu, Wang, Liu, Du and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Guo, eGlhb2d1b3llYWhAeWVhaC5uZXQ=; Weihua Guo, d2hndW9fc2R1QDE2My5jb20=
†ORCID: Xiao Guo, orcid.org/0000-0003-2268-5090
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.