Abstract
Sponge-associated bacteria are a well-known source of unique and diverse natural products with potential biological activities. In this study, a rarely occurring sponge-associated bacterium from the genus Alcanivorax was isolated from the sponge Siphonochalina siphonella and subjected to extraction of metabolites. Metabolites were extracted from the culture supernatant and biomass of the bacterium using the solvent extraction method. Extracts were evaluated for antibacterial and antibiofilm activities against five different biofilm-forming bacteria isolated from the microfouling assemblage. The crude extracts were subjected to chemical profiling to identify their composition using gas chromatography-coupled mass spectrometry (GC-MS). The results obtained show mild to strong antibacterial activity of the extracts against the biofilm-forming bacteria with the minimum inhibitory concentration (MIC) of 3.125 mg ml-1. The percentage inhibition of biofilm formation ranged from 46% to 71%. Compounds detected in GC-MS analysis are mostly fatty acids and their derivatives that include compounds with known antibiofilm activity such as tetradecanoic acid, dodecanoic acid, and hexadecanol from the culture supernatant extract. Eicosane; hexadecanoic acid, methyl ester; and hexadecanoic acid, ethyl ester were detected from the biomass extract. Molecular docking analysis of the compounds from the bacterial extracts confirms their potential antibiofilm properties. The result indicated that Alcanivorax sp. associated with the sponge possess relevant antibiofilm activity and may serve as a novel source of secondary metabolites with the application as antibiofilm agents.
Introduction
The attachment of microbial communities and the formation of a structured assemblage embedded in the extracellular matrix are called biofilms (Karygianni et al., 2020). In the marine environment, biofilm formation on artificial materials is a cause of concern due to its economic significance. Bacterial biofilm is the fundamental cause of biofouling development in the marine environment on artificial and natural surfaces (Unabia and Hadfield, 1999). When an object is submerged in the sea, the biofouling process is initiated by the attachment of biofilm bacteria that then attract larger organisms (Dobretsov and Rittschof, 2020). When this occurs on marine infrastructure and installations, it will have negative environmental and economic impacts on marine transport, aquaculture, fisheries, tourism, and mariculture (Bannister et al., 2019; Cruz, 2020). Biofouling results in an increase in frictional resistance of ship hulls and subsequent increase in fuel consumption. There is also an increase in biodiversity threat via the transfer of invasive microorganisms (Molnar et al., 2008).
Many antibiofilm and antifouling strategies are used to control biofouling on artificial materials (Satheesh et al., 2016). One of the most common methods to get rid of biofilm in water is the use of biocides (Bhoj et al., 2021). Biocides are expensive and have toxicity toward non-target organisms (Schulte et al., 2005). The biofilm bacteria could also develop resistance to the biocides, making them ineffective (Kocot and Olszewska, 2017). Sustainable antibiofilm strategies are required to control biofilms on marine installations and structures. Secondary metabolites from marine bacteria have been found to be sources of novel and unique antimicrobial compounds that could overcome the problem of antimicrobial resistance (Proksch et al., 2002; Kijjoa and Sawangwong, 2004; Vignesh et al., 2011; Mondol et al., 2013; Kumar et al., 2014; Santos-Gandelman et al., 2014; Malve, 2016; Mayer et al., 2021). The natural compounds from marine bacteria and other organisms are considered potential antifouling compounds (Satheesh et al., 2016).
The marine environment has rich biodiversity and covers a major portion of the biosphere, making it one of the largest unexplored sources of bioactive compounds (Malve, 2016). Among marine organisms, sponges are one of the most important organisms due to its worldwide distribution and bioprospecting potentials. Bioactive compounds from sponges are enormous compared with other organisms and represent about 30% of marine natural products (MNPs) reported between the years 2001 and 2010 (Mehbub et al., 2014). The sponge harbors diverse microorganisms that are living in symbiotic association with the sponge host. Microbes make up 25% of its biomass and contribute to its survival (Fahmy and Abdel-Tawab, 2021). Through the association, most of the sponge MNPs are produced as by-products of metabolism (Santos-Gandelman et al., 2014). The MNPs are produced by the sponge and its symbionts and have antagonistic activity against other microbes (Srinivasan et al., 2021). The microbes produce compounds as secondary metabolites that enable them to provide some form of chemical defense to the host through chemical interactions, while the host provides the microbes with nutrients necessary for their growth (Zhang et al., 2022). Some of the bacterial symbionts are unique because they have not been described previously elsewhere, making them potential sources of bioactive compounds (Wang, 2006; Caruso et al., 2018; Rizzo et al., 2020). As sources of diverse and new compounds, bacteria associated with sponges have been explored for many activities including as antifouling compounds (Satheesh et al., 2016; Wang et al., 2017; Liu et al., 2020). Compounds from the sponge and its associated bacteria are chemically diverse, covering a range of chemical classes such as fatty acids, polyketides, alkaloids, benzenoids, amino acids, steroids, and many others (Fusetani, 2004). Antibiofilm compounds reported from these organisms with promising activity include polyethers (Dash et al., 2011), butenolide (Yin et al., 2019), phorbaketal B, and phorbaketal C (Kim et al., 2021).
The Red Sea is home to diverse marine organisms; many of them are endemic to the environment (Berumen et al., 2019). The organisms are the repository of diverse bioactive secondary metabolites, with over 700 compounds reported in the last decade alone (El-Hossary et al., 2020). It is also a source of novel marine microbial secondary metabolites due to its harsh environmental conditions, making it a model in the search for microbial species that can produce promising antibiofilm compounds (Behzad et al., 2016). However, to date, only a few antibiofilm metabolites have been reported from the Red Sea, making it one of the underexplored environments. Hence, in this study, the antibiofilm activity of the bacteria associated with the sponge species Siphonochalina siphonella collected from the Red Sea was assessed using laboratory assays and in silico methods against the biofilm-associated protein (BAP). The results of this study will be useful for the development of novel natural product antibiofilm compounds from marine microbes.
Methodology
Isolation and identification of sponge-associated bacteria
The sponge-associated bacterium (strain AA8) was isolated from a marine sponge. The sponge was collected from the Central Red Sea coast in Jeddah (N21°42.562′ E39°05.764′) and identified as S. siphonella using the method described previously (Lieske et al., 2004). The isolation of bacteria associated with the sponge was done by washing the organism (sponge) with 0.22-µm filter-sterilized seawater (FSW), cut into pieces, and then immersed in an FSW-containing flask. The flask was kept in a rotary shaker for 4 h, and the contents were serially diluted (10-fold serial dilution). An aliquot of 100 µl of the serially diluted sample was inoculated in Zobell marine agar (ZMA) (Himedia, India) plates using the spread plate method and incubated at 28°C for 72 h or until visible colonies appeared on the plates. Individual colonies were selected following subcultures to obtain pure culture. It was identified by the partial 16S rRNA method as reported previously (Abo-State et al., 2018). The PCR product of the 16S rRNA gene was commercially sequenced at Macrogen Inc., Korea. The gene sequence of the selected organism was deposited in the NCBI database (accession number: ON514280). This was followed by phylogenetic tree construction using MEGAX neighbor joining method to determine the phylogenetic position of the strain and its evolutionary relatedness (Kumar et al., 2018).
Extraction of metabolites from the culture supernatant and biomass extract
The solvent extraction method was applied for the extraction of secondary metabolites from the sponge-associated bacterium (strain AA8). The growth of the organism was optimized for conditions such as temperature and incubation time required for reaching the exponential phase. The pH of the media was maintained as 7.4 throughout the experiment. The temperature was varied from 20°C to 35°C (i.e., 20°C, 25°C, 30°C, and 35°C) to select the best growth temperature by measuring the optical density (OD) at 600 nm (OD600) using a spectrophotometer (Shimadzu, Japan) according to an earlier reported method (Selvin et al., 2009). The best growth was observed at 30°C, which has the highest OD600 value. The time required to reach the exponential phase was determined using a previously described method with modifications (Ortega-Morales et al., 2008). The incubation time was varied from 12 h to 24 h at a temperature of 30°C with the observation of the OD at hourly intervals until the exponential phase was attained. The time taken to reach the exponential phase was found to be at the 18th hour. To extract the metabolites, the organism was inoculated in 10 ml of Zobell Marine Broth (ZMB) (HiMedia, India) overnight at 30°C until it reaches the exponential phase. From the overnight culture, 5 ml was transferred into a 500-ml-capacity Erlenmeyer flask containing 250 ml of ZMB. It was incubated with constant shaking (180 rpm) at 30°C for 7 days. After that, centrifugation was carried out at 8,000 × g for 20 min at 4°C. Following that, the supernatant and culture biomass (pellet) were collected separately for the extraction of metabolites. For the cell-free culture supernatant, ethyl acetate was added in equal volume and agitated for 24 h. For the biomass, the cells recovered were washed in phosphate-buffered saline (PBS). Metabolites were extracted from the biomass using an ice-cold solvent mixture of acetonitrile and methanol in a ratio of 1:1. This was followed by shaking, filtration, and evaporation of the solvents as described previously (Nadeem et al., 2020). Each of the crude extracts obtained was weighed and solubilized in dimethyl sulfoxide (DMSO) to a concentration of 50 mg ml-1. The extracts were stored at refrigerated temperature until required for further use.
Determination of the antibacterial activity of the extracts
The well diffusion method was applied to test the antimicrobial susceptibility of the extracts as described previously by Al-Daghistani et al. (2021). The antibacterial activity of the crude extracts obtained from the sponge-associated bacterium was evaluated against selected biofilm bacteria. The target biofilm-forming bacteria were obtained from the culture collections maintained in our laboratory (Faculty of Marine Sciences, King Abdulaziz University). The biofilm bacteria selected for this study were Pseudoalteromonas sp. IMB1 (NCBI: ON003955), Halomonas sp. IMB2 (NCBI: ON415519), Vibrio alginolyticus IMB11 (NCBI: ON003958), Pseudoalteromonas gelatinilytica IMB14 (NCBI: ON003961), and P. gelatinilytica IMB15 (NCBI: ON003962). These biofilm bacteria were isolated from the microfouling assemblage developed on artificial materials submerged in the Obhur Creek, Jeddah coast (Abdulrahman et al., 2021). For the biofilm bacteria, colonies from the bacterial culture were inoculated in 10 ml of ZMB and incubated overnight. This was followed by centrifugation at 5,000 × g for 5 min. The supernatant obtained after centrifugation was discarded, and the pellets were washed twice with PBS to remove any leftover supernatant. PBS was added to the biomass, and the suspension was adjusted to 0.5 McFarland standard (1.0 × 106 CFU/ml) using a spectrophotometer at OD600. The surface of freshly prepared ZMA plates was uniformly spread with the suspension with a sterile cotton swab, and wells were perforated with a sterile cork borer that was 6 mm in diameter each. The extract (50 µl) was added to each well, and 100% DMSO was used as control. The plates were incubated at 28°C for 24 h. Antibacterial activity was evaluated by measuring the diameter of the zone of inhibition (ZI) formed around the well that indicates the absence of growth. All assays were repeated in triplicate, and the average was recorded. The result was categorized as strong (ZI ≥20 mm), moderate (ZI = 14–19 mm), weak (ZI ≤13 mm), and resistant (when there is no zone of inhibition).
Determination of minimum inhibitory concentration
The minimum inhibitory concentration (MIC) for each extract against the target biofilm bacteria was determined with the help of resazurin dye according to a previously described broth microdilution method with modifications (Elshikh et al., 2016). The MIC was determined in a 96-well microtiter plate. For each well, 100 μl of ZMB was added. In the first column, 100 μl of the stock solution of the extract was added to the first well in the row containing 100 μl of ZMB and mixed with a pipette tip. A 100-μl aliquot from the well was pipetted to the next well in the column. Serial dilution was carried out until the 10th well in the column. Negative and positive controls were used per experiment. Negative control contains only media without extract and culture in column 11, and positive control contains 200 μl of bacterial suspension without extract in the 12th column. The first column has the highest concentration of 25 mg/ml. The standardized culture of each bacterium was prepared and adjusted to a final concentration of 1 × 106 CFU/ml. A 100-μl standardized culture of the tested strain was suspended in each of the wells except for column 11 (media control). Each experiment was repeated three times, and the plates were incubated at 28°C for 24 h under static conditions (Sarker et al., 2007). After incubation, the plates were observed and 30 µl of resazurin solution (0.02%) was added to each well and then incubated statically at 28°C for 2–3 h. Color change was observed visually after the incubation in the wells, and MIC values were recorded as the lowest concentration where the color changes from the resazurin color (purple) to red. Wells that were observed without any color change were regarded as above MIC values. A color change to pink in the growth control (positive) well was the indication of proper growth of the bacteria, while no change in color of the negative control well was considered the absence of contamination (Teanpaisan et al., 2017).
Antibiofilm activity of the extracts
The crude extract was evaluated for antibiofilm activity against the five biofilm-forming bacteria. Each of the tested organisms was prepared for the assay, and the inoculum was standardized using the procedure described by Skogman et al. (2016) with some modifications. Briefly, the selected biofilm organism undergoes preculture by culturing in tubes containing modified nutrient broth (MNB) that was composed of nutrient broth (HiMedia, India) and filtered seawater (0.22 µm) and incubated for 16–18 h at 28°C at 180 rpm in a shaker incubator. The biofilm bacteria culture was diluted 100 times in Luria-Bertani (LB) broth and incubated using the same conditions as above until it reaches exponential growth (OD range: 0.2–0.3 at OD595). The biofilm bacteria culture was further diluted 100 times with LB to arrive at 106 CFU ml-1. Microtiter biofilm inhibition assay was carried out to test the antibiofilm activity. The assay was done in a round-bottom 96-well microtiter plate to determine the ability of each of the extracts to reduce or inhibit biofilm formation using a previously described method (Nunes et al., 2021). The plates were first moistened with sterile filtered seawater (SFSW) for conditioning. The diluted biofilm bacterial culture was added (175 µl) to the microtiter well of 96-well plates. This was followed by the addition of 25 µl of the extract. Each extract was evaluated against all of the biofilm-forming bacteria in replicate (n = 4). For the control, 200 µl of the diluted bacterial culture only was added to the well without extract. Inoculated plates were incubated statically and aerobically at 28°C for 48 h. The whole unattached cells were removed carefully using a multichannel pipette after incubation, and the wells were washed with PBS (three times). The bacteria that attached on the wells were stained by adding 200 µl of 0.2% crystal violet (CV) into the wells and allowed to stay statically at ambient temperature for 15 min. After that, the CV stain was removed from the wells carefully by inverting the plates and washing thrice in PBS. The plate was allowed to air-dry, and 200 µl of 96% ethanol was added to the wells for the solubilization dye from the attached bacterial cells. The plates were incubated with shaking for 20 min at 28°C in a shaker incubator after which the content was carefully transferred to fresh 96-well plates. The OD was measured at 570 nm (OD570) with a multiplate reader (Synergy, Biotek). The experiment was repeated independently in duplicate, and the average for six values with the standard deviation (SD) was considered. The biofilm inhibition was calculated as a percentage using the following formula:
BI = Biofilm Inhibition
OD control = Absorbance of control at 570 nm
OD sample = Absorbance of test sample at 570 nm
Gas chromatography-coupled mass spectrometry analysis of the extracts
Gas chromatography-coupled mass spectrometry (GC-MS) using GCMS-QP2010 system (Shimadzu, Japan) was used to analyze the chemical composition of the crude extracts. The sample was dissolved in isooctane (≥99%, Sigma–Aldrich, Germany) and filtered through a syringe filter prior to injection. The column was programmed, and the analysis was carried out as previously described (Pugazhendi et al., 2017). Identification of volatile compounds from the extracts was done by comparing the retention time and mass spectra with those of mass spectral library search of the National Institute of Standards and Technology library using the similarity index (<92%). Also, the GC-MS data were compared with those of the antibiofilm agent resource database “aBiofilm“ (Rajput et al., 2017) and other relevant literature to identify compounds with known antibiofilm activity.
Collection and preparation of test and reference ligands
The compounds identified from the extract of the strain AA8 through GC-MS were tested for antibiofilm activity against the BAP using an in silico approach. For that, the molecular target BAP (PDB ID-7C7U) was downloaded from the protein databank. Following this, the water molecules and hetero-atom (hetatm) associated with the BAP were removed using the software Biovia Discovery Studio Visualizer (BDVS) (Figure 1). Also, the structures of the identified compounds and the reference ligand were downloaded from PubChem. The details of the identified compounds and the reference are given in Table 1 and in Supplementary Figures S1–S3.
Figure 1
The structure of the molecular target, the biofilm-associated protein (BAP) after the removal of water molecules and HETATM. The BAP was downloaded from RCSB protein data bank (PDB ID: 7C7U, Ma et al. 2021a; Ma et al. 2021b) and modified structure was created using Biovia Discovery Studio Visualizer.
Table 1
| S. No. | Ligands | Molecular weight | Molecular formula | PubChem ID |
|---|---|---|---|---|
| 1 | Butenolide | 84.07 | C4H4O2 | 10341 |
| 2 | Heptane, 2,5-dimethyl- | 128.25 | C9H20 | 16662 |
| 3 | Dodecanoic acid | 200.32 | C12H24O2 | 3893 |
| 4 | Cyclopropaneoctanoic acid, 2-octyl-, methyl ester | 310.5 | C20H38O2 | 543406 |
| 5 | 1-Hexadecanol | 242.44 | C16H34O | 2682 |
| 6 | Octadecanoic acid, 3-hydroxypropyl ester | 342.6 | C21H42O3 | 168316 |
| 7 | Tetradecanoic acid | 228.37 | C14H28O2 | 11005 |
| 8 | Cis-11-Eicosenamide | 309.5 | C20H39NO | 5365374 |
| 9 | Hexadecanoic acid, ethyl ester | 284.5 | C18H36O2 | 12366 |
| 10 | Eicosane | 282.5 | C20H42 | 8222 |
| 11 | n-Pentadecanol | 228.41 | C15H32O | 12397 |
| 12 | Pentadecanoic acid, methyl ester | 256.42 | C16H32O2 | 23518 |
| 13 | Hexadecanoic acid, methyl ester | 270.5 | C17H34O2 | 8181 |
| 14 | l-(+)-Ascorbic acid 2,6-dihexadecanoate | 652.9 | C38H68O8 | 54722209 |
| 15 | Heptadecanoic acid, methyl ester | 284.5 | C18H36O2 | 15609 |
Details of the ligands (reference compound and the compounds identified from the extract of the bacterium Alcanivorax sp. AA8) downloaded from PubChem.
Docking analysis
The docking software PyRx was used to analyze the antibiofilm property of the compounds identified in the bacterial extract against the BAP. In brief, the target protein and the test ligands were carried over to the docking (PyRx) software and converted into PDBQT files. Thereafter, the ligand-binding locations on the receptor protein were marked with the grid box, and the docking was run. Once the docking was over, the docking scores were noted, and the necessary files (receptor and ligands) were carried over to BDSV to visualize the interaction of the ligands with the receptors.
Results
Identification of sponge-associated bacterium
The sponge-associated bacterium isolated was described as a Gram-negative rod and motile. The colonies appear as non-pigmented white colonies that are transparent, circular, and raised with an entire margin (Supplementary Figure S4). The biochemical reaction of the organism to oxidase was negative while that to catalase reagent was positive. Analysis of the 16S rRNA gene sequence with BLAST (restricted to sequence from type material) revealed close similarity with members of the Alcanivorax genus. It shows close identity similarity with Alcanivorax diesosolei (99.33%), A. xenomutans (99.54%), and A. balearicus (98.45), and the next nine members of the genus Alcanivorax have identity similarities between (93.33%–94.88%). The phylogenetic tree of the organism (Alcanivorax sp. AA8) was constructed with other similar species (Figure 2).
Figure 2

Phylogenetic tree constructed using MEGAX. The organism isolated in this study was labeled as AA8 with the full name of the organism in brackets.
Antibacterial activity of the extracts
The crude extracts obtained from Alcanivorax sp. AA8 associated with the sponge S. siphonella exhibited antibacterial activity against the tested biofilm bacteria. The crude extracts obtained from Alcanivorax sp. (AA8) were labeled as Alcanivorax extract 1 (AE1) and Alcanivorax extract 2 (AE2) respectively for the extract of the culture supernatant and biomass extract. The results of the antibacterial activity of extracts with their respective MIC against the five biofilm bacteria are presented in Table 2. The results showed that the extracts have varying activities from weak (mild) activity to strong activity against the tested organisms. All of the strains were sensitive to the extracts differently. The extracts recorded 70% moderate activity, 20% mild or weak activity, and 10% strong activity. AE1 recorded moderate activity toward all of the strains except Pseudoalteromonas sp. IMB1 and V. alginolyticus IMB11, while AE2 demonstrated the highest activity with a wider zone of inhibition against all of the tested organisms. Extracts from the biomass culture (AE2) recorded higher activity than extracts from the culture supernatant (AE1). Halomonas sp. IMB2, V. alginolyticus IMB11, P. gelatinilytica IMB14, and P. gelatinilytica IMB15 strains have mild to moderate sensitivity to the extracts, while Pseudoalteromonas sp. IMB1 has mild, moderate, and strong sensitivity to the extracts. Strong activity was recorded only by AE2 against Pseudoalteromonas sp. IMB1. The extracts recorded MIC values of 3.125, 6.25, and 12.5 mg ml-1. The least MIC recorded for AE1 was 6.25, while the least MIC for AE2 was 3.125. The AE2 has better MIC than AE1, maintaining similar trends with the result of antibacterial activity based on the zone of inhibition.
Table 2
| Zone of inhibition (mm) | MIC (mg ml-1) | |||
|---|---|---|---|---|
| Organism | AE1 | AE2 | AE1 | AE2 |
| Pseudoalteromonas sp. IMB1 | 18 ± 2 | 20 ± 1 | 12.5 | 12.5 |
| Halomonas sp. IMB2 | 16 ± 2 | 19 ± 0 | 12.5 | 3.125 |
| Vibrio alginolyticus IMB11 | 8 ± 1 | 14 ± 0 | 6.25 | 6.25 |
| Pseudoalteromonas gelatinilytica IMB14 | 16 ± 0 | 18 ± 1 | 12.5 | 12.5 |
| Pseudoalteromonas gelatinilytica IMB15 | 11 ± 2 | 16 ± 2 | 12.5 | 6.25 |
Zone of inhibition and minimum inhibitory concentration (MIC) of extracts from Alcanivorax sp. AA8 against biofilm bacteria.
Key: IMB1, Pseudoalteromonas sp. IMB1 (ON003955); IMB2, Halomonas sp. IMB2 (ON415519); IMB11, Vibrio alginolyticus IMB11 (ON003958); IMB14, Pseudoalteromonas gelatinilytica IMB14 (ON003961); IMB15, Pseudoalteromonas gelatinilytica IMB15 (ON003962).
AE1, extract of the culture supernatant; AE2, biomass extract.
Biofilm inhibition activity
The biofilm inhibition activity of the extracts shows their ability to reduce/inhibit biofilm development of the target organisms. Extracts of the culture supernatant (AE1) recorded a higher biofilm inhibition activity than the biomass extract (AE2) against all of the strains except P. gelatinilytica IMB15 as shown in Figure 3. The extracts recorded 71% as the highest inhibition activity and 47% as the lowest across the five biofilm-forming strains. The highest was recorded by AE1 against P. gelatinilytica IMB15, while the lowest was recorded against P. gelatinilytica IMB14 by AE1.
Figure 3
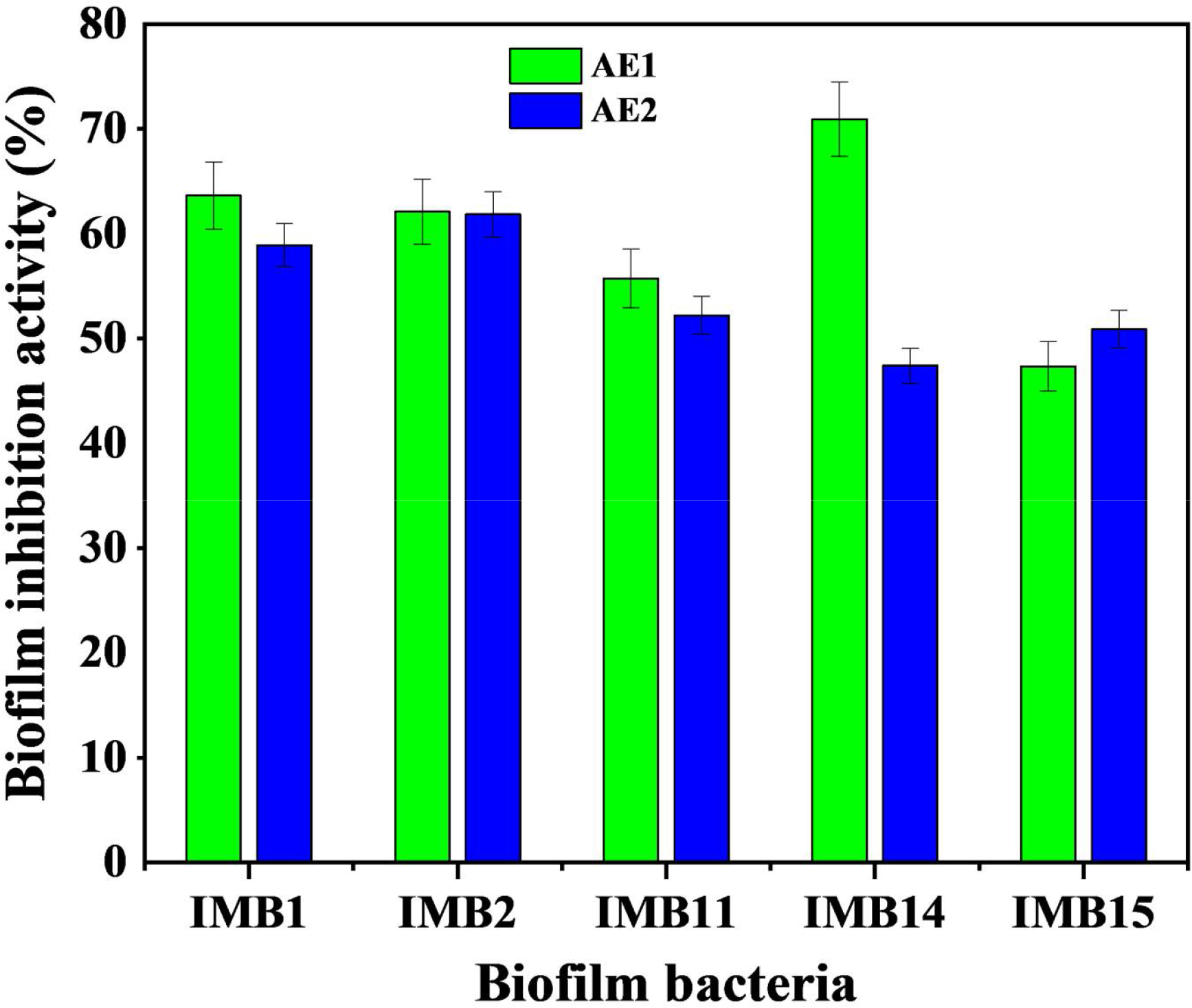
Biofilm inhibition activity of crude extracts from Alcanivorax sp. AA8 bacteria against five biofilm bacteria. Each data point represents an average of eight replicates and SD. Key: IMB1, Pseudoalteromonas sp. IMB1 (ON003955); IMB2, Halomonas sp. IMB2 (ON415519); IMB11, Vibrio alginolyticus IMB11 (ON003958); IMB14, Pseudoalteromonas gelatinilytica IMB14 (ON003961); IMB15, Pseudoalteromonas gelatinilytica IMB15 (ON003962).
Chemical composition of the extracts
GC-MS results (Figure 4) detected several volatile organic compounds from the extracts (Table 1). Some of the detected compounds that have been reported in the literature with antibiofilm properties are presented in Table 3. Compounds detected in the extracts with a known antibiofilm activity include tetradecanoic acid; 1-hexadecanol; dodecanoic acid; and heptadecanoic acid, methyl ester from the culture supernatant extract (AE1) and eicosane and hexadecenoic acid, methyl ester from the biomass extract (AE2).
Figure 4
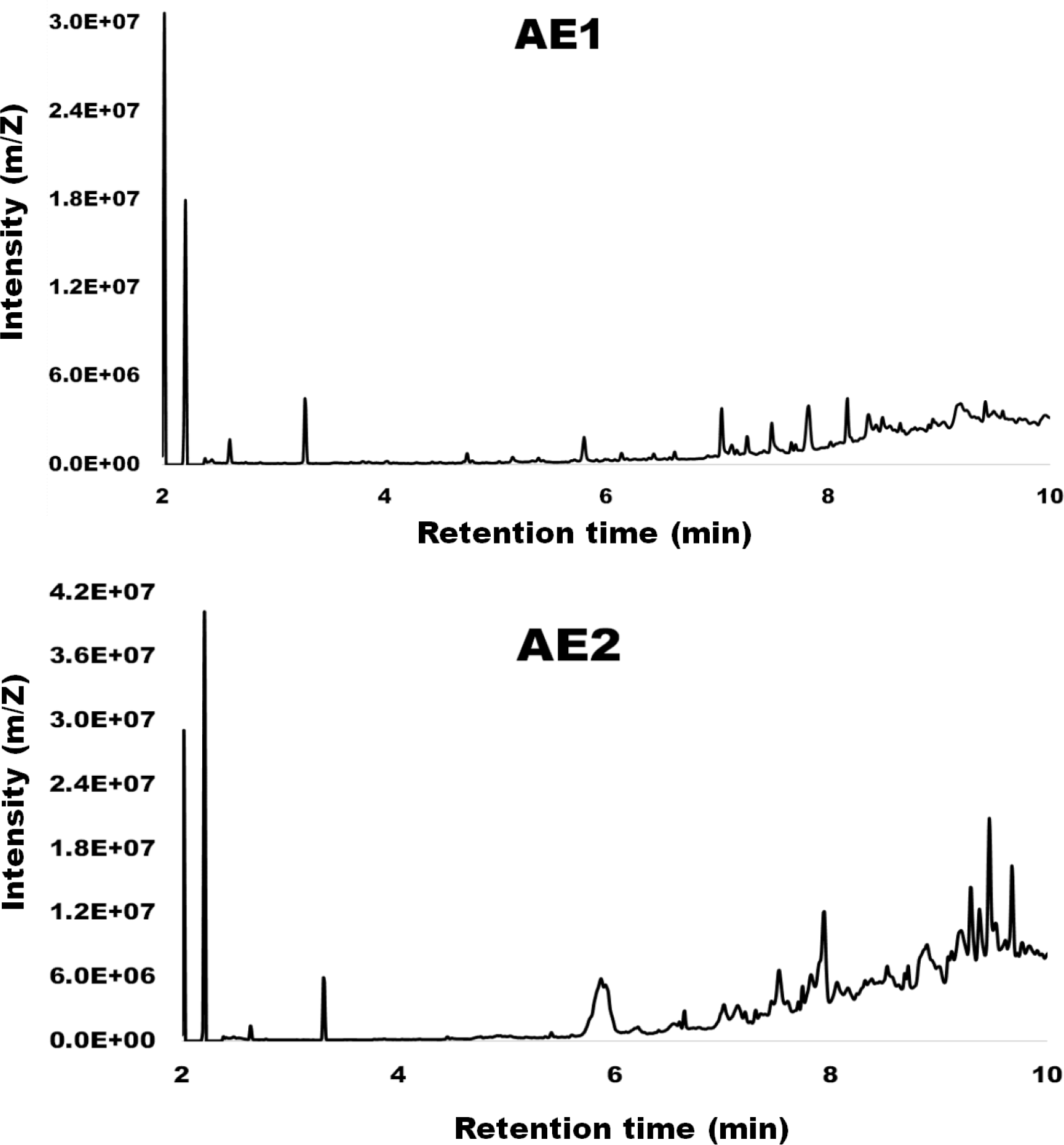
Spectrum of compounds detected by gas chromatography-coupled mass spectrometry (GC-MS) from the culture supernatant extract (AE1) and biomass crude extract (AE2) of Alcanivorax sp. AA8.
Table 3
| Compounds | Molecular formula | Molecular weight | Peak area | Retention time | Type of compound | Activity | Reference |
|---|---|---|---|---|---|---|---|
| Dodecanoic acid1 | C12H24O2 | 200.3178 | 5.29 | 7.492 | Free fatty acid | Antibiofilm | (Jin et al., 2021) |
| Eicosane2 | C20H42 | 282.5475 | 4.24 | 9.575 | Straight-chain alkane | Antibiofilm | (Beema Shafreen et al., 2022) |
| Hexadecanoic acid, methyl ester2 | C17H34O2 | 270.4507 | 13.98 | 9.255 | Fatty acid esters | Antibiofilm | (Dusane et al., 2011) |
| Hexadecanoic acid, ethyl ester2 | C18H36O2 | 284.5 | 4.24 | 9.575 | Fatty acid esters | Antibiofilm | (Wang et al., 2018) |
| 1-Hexadecanol1 | C16H34O | 242.44 | 6.65 | 8.172 | Fatty acid alcohol | Antibiofilm | (Manilal et al., 2020) |
| Tetradecanoic acid1 | C14H28O2 | 228.3709 | 6.68 | 8.49 | Free fatty acid | Antibiofilm | (Rajalaxmi et al., 2016) |
Antibiofilm compounds detected from Alcanivorax sp. AA8 extracts.
Key: 1, extracts from the culture supernatant; 2, biomass extract.
Docking analysis
The docking analysis results of the test ligands (identified compounds) and the reference ligand (butenolide) with the molecular target BAP are given in Tables 4 and 5. The binding affinity of the reference butenolide with the target BAP was -3.9 kcal/mol. Whereas the binding affinities of the compounds identified in the extract of the culture supernatant of the bacterium Alcanivorax sp. AA8 such as heptane, 2,5-dimethyl; dodecanoic acid; cyclopropaneoctanoic acid, 2-octyl-, methyl ester; 1-hexadecanol; octadecanoic acid, 3-hydroxypropyl ester; tetradecanoic acid; cis-11-eicosenamide; and hexadecanoic acid, ethyl ester were -4.7, -5.1, -5, -4.5, -4.7, -5.5, -5.7, and -5.7 kcal/mol, respectively. Similarly, the binding affinity of the compounds identified in the biomass extract of the bacterium Alcanivorax sp. AA8 including eicosane; n-pentadecanol; pentadecanoic acid, methyl ester; hexadecanoic acid, methyl ester; l-(+)-ascorbic acid 2,6-dihexadecanoate; and heptadecanoic acid, methyl ester were -4.1, -4.5, -4.5, -4.3, -5.2, and -4.5 kcal/mol, respectively.
Table 4
| S. No. | Ligands | Binding affinity | Amino acids involved | Interaction Bond |
|---|---|---|---|---|
| 1 | Butenolide | -3.9 | THR722 | H-bond |
| 2 | Heptane, 2,5-dimethyl- | -4.7 | LEU528, LEU720 | Alkyl |
| 3 | Dodecanoic acid | -5.1 | ARG357, ILE419, HIS423, ASP452 | H-bond, Alkyl, Pi-Alkyl |
| 4 | Cyclopropaneoctanoic acid, 2-octyl-, methyl ester | -5 | ALA373, LEU376, TYR400, LEU472, M ET576, PRO660 | H-bond, Alkyl, Pi-Alkyl |
| 5 | 1-Hexadecanol | -4.5 | TYR767, VAL850 | Alkyl, Pi-Alkyl, Pi-Sigma |
| 6 | Octadecanoic acid, 3-hydroxypropyl ester | -4.7 | TYR370, PHE372, ALA373, TYR400, LEU472 | H-bond, Alkyl, Pi-Alkyl |
| 7 | Tetradecanoic acid | -5.5 | PRO356, GLY358, SER359 | H-bond, Alkyl, Pi-Alkyl |
| 8 | Cis-11-Eicosenamide | -5.7 | ALA373, TYR400, LEU472, TYR613 | Alkyl, Pi-Alkyl, Pi-Sigma |
| 9 | Hexadecanoic acid, ethyl ester | -5.7 | LEU364, LYS415, PHE508, ASP701, ALA726 | H-bond, Alkyl, Pi-Alkyl |
The docking result of the compounds identified in the culture supernatant extract of the bacterium Alcanivorax sp. AA8 carried out using the software PyRx.
Table 5
| S. No. | Ligands | Binding affinity | Amino acids involved | Interaction Bond |
|---|---|---|---|---|
| 1 | Butenolide | -3.9 | THR722 | H-bond |
| 2 | Eicosane | -4.1 | ALA373, TYR400, LEU472, TYR613 | Alkyl, Pi-Alkyl |
| 3 | n-Pentadecanol | -4.5 | ALA373, SER374, TYR400, LEU472, THR662 | H-bond, Alkyl, Pi-Alkyl |
| 4 | Pentadecanoic acid, methyl ester | -4.5 | VAL355, GLY358, SER359, PRO418, | H-bond, Alkyl, Pi-Alkyl |
| 5 | Hexadecanoic acid, methyl ester | -4.3 | ALA373, TYR613, ASN656 | H-bond, Alkyl |
| 6 | l-(+)-Ascorbic acid 2,6-dihexadecanoate | -5.2 | LYS775, PRO793, PRO868, VAL877, GLN938, ILE939, THR940 | H-bond, Alkyl |
| 7 | Heptadecanoic acid, methyl ester | -4.5 | LYS502, TYR526, ASP532, GLN596, LYS599, VAL639 | H-bond, Alkyl |
The docking result of the compounds identified in the biomass extract of the bacterium Alcanivorax sp.
The docking result of the compounds identified in the biomass extract of the bacterium Alcanivorax sp. AA8 carried out using the software PyRx.
Visualization of ligand–protein interaction
According to the data obtained from BVDS, the ligands were found to make interactions with various amino acid residues of the BAP with H-bond, Alkyl, Pi-Alkyl, and Pi-Sigma bonding (Tables 4, 5). The reference ligand butenolide interacted with amino acid residues (THR722) of the receptor BAP, whereas the number of amino acid residues that interacted with the test ligands ranged from 2 to 6. Overall, a total of 43 amino acid residues of the receptor protein BAP were found to be involved with the ligands (compounds identified in the bacterial extract) (Figures 5, 6).
Figure 5
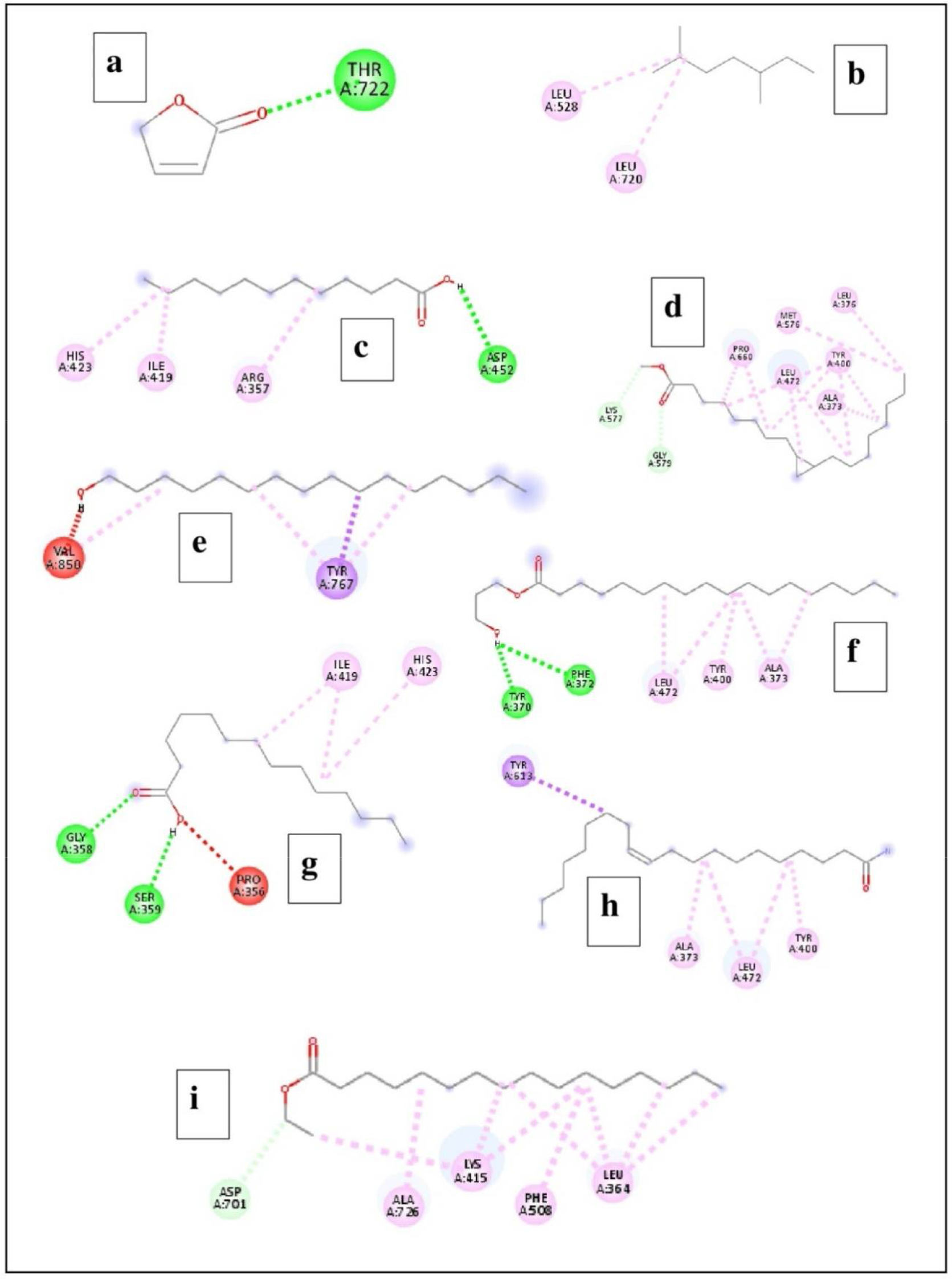
The interaction of compounds identified in the extract from the culture supernatant of Alcanivorax sp. AA8 with the target biofilm-associated protein (BAP). (A) Butenolide; (B) Heptane, 2,5-dimethyl; (C) Dodecanoic acid; (D) Cyclopropaneoctanoic acid, 2-octyl-, methyl ester; (E) 1-Hexadecanol; (F) Octadecanoic acid, 3-hydroxypropyl ester; (G) Tetradecanoic acid; (H) Cis-11-eicosenamide; and (I) Hexadecanoic acid, ethyl ester.
Figure 6
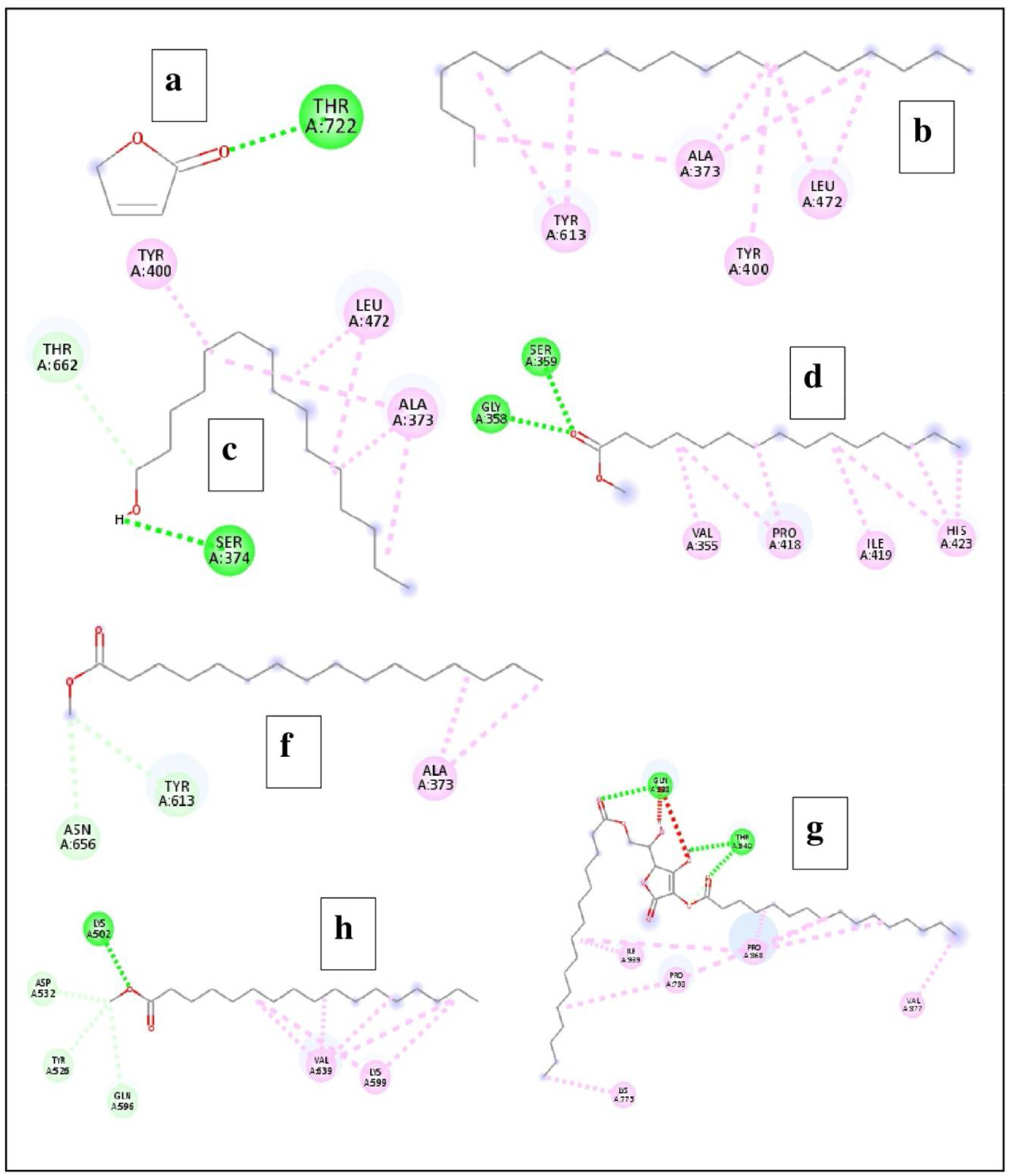
Illustrate the interaction of compounds identified in the biomass extract of the bacterium Alcanivorax sp. AA8 with the target biofilm-associated protein (BAP). (A) Butenolide; (B) Eicosane; (C) n-Pentadecanol; (D) Pentadecanoic acid, methyl ester; (E) Hexadecanoic acid, methyl ester; (F) l-(+)-Ascorbic acid 2,6-dihexadecanoate; and (G) Heptadecanoic acid, methyl ester.
Discussion
Isolation of Alcanivorax sp. from sponge host
Alcanivorax sp. was isolated from the sponge S. siphonella collected from the Red Sea. The bacteria Alcanivorax is widely distributed in marine environments occupying different depths and habitats as free-living or in association with other marine organisms. It is found on the surface water and surface sediments, deep seawater, and deep-sea sediments (Lai et al., 2013; Barbato et al., 2015; Lai et al., 2016; Karthick and Mohanraju, 2018; Song et al., 2018; Liu J. et al., 2019; Liao et al., 2020). It has been reported to be living in a symbiotic relationship with several organisms in the marine environment such as seaweeds and microalgae (Karthick and Mohanraju, 2018; Chernikova et al., 2020). Only rare reports exist on the presence of the Alcanivorax genus in association with sponges such as its association with sponges collected from lower mesophotic reefs (Indraningrat et al., 2019).
Bacterial growth inhibition activity of extracts of Alcanivorax sp.
The extracts from the sponge-associated Alcanivorax sp. demonstrated good antibacterial activity against the biofilm bacteria. There is a broad spectrum of activity across the organism due to the large size of the zone of inhibition against the growth of the tested organisms. The strong antibacterial activity of the extract of Alcanivorax sp. isolated in this study could be attributed to the bioactive compounds present in the extract that inhibited the growth of the tested organisms. Antibacterial compounds such as n-tridecan-1-ol were observed in AE1, while n-tridecan-1-ol and n-pentadecanol were detected in AE2. Both compounds are long-chain fatty alcohols with previously reported antibacterial activity (Togashi et al., 2007). The AE2 extract has more antibacterial compounds than AE1, which might have contributed to the higher antibacterial activity it recorded in the assay. Members of the Alcanivorax genus are well-known as hydrocarboclanistic bacteria associated with petroleum hydrocarbon degradation (Qiao and Shao, 2010; Barbato et al., 2015; Chernikova et al., 2020). However, they produced enzymes that extend their activity beyond oil degradation by performing other activities such as polyester degradation (Zadjelovic et al., 2020). Bioactive compounds have been reported from members of the Alcanivorax genus (Debnath et al., 2007). The chemical defense role of marine symbiotic members of the Alcanivorax genus was reported previously with antimicrobial activity against different bacteria (Pereira et al., 2017). A high antimicrobial activity was reported earlier from symbiotic Alcanivorax dieselolei that was found in association with seaweeds (Karthick and Mohanraju, 2018). This is an indication of the role of Alcanivorax associated with sponge as the ideal source of antibacterial compounds.
Inhibition of biofilm formation
Extracts from the sponge-associated bacterium Alcanivorax sp. recorded a good biofilm inhibition activity against all of the tested biofilm bacteria. Antibiofilm agents from sponge holobionts have proven to be good candidates for tackling biofilm formation (Santos-Gandelman et al., 2014). Many of the sponge-associated bacteria are considered as the source of antibiofilm agents that can inhibit biofilm formation through its multiple targets (Rajput et al., 2017). The good antibiofilm activity recorded in this study is a confirmation of the role of the symbiotic bacteria in protecting the sponge host against invading biofilm bacteria. The promising results obtained from this study are interesting due to the recalcitrant nature of biofilm bacteria and its negative economic impact on the affected industries (Cámara et al., 2022). To overcome the biofilm challenge, inhibition of biofilm on surfaces is a requirement for sustainable growth of the affected industries. More antibiofilm agents were identified through GC-MS analysis from the AE1 extract than AE2, which could be one of the possible reasons for the higher antibiofilm activity recorded by AE1. Alcanivorax sp. is known to produce biosurfactants and many enzymes that have contributed to its ability to produce bioactive compounds (Zadjelovic et al., 2020). Alcanivorax sp. therefore has a great potential application as source of new compounds with multiple biological activities. This suggests that sponges can attract diverse microorganisms with the ability to produce compounds that can protect against predators and colonization.
Chemical compounds from Alcanivorax sp. extracts
From the extracts, several compounds were detected through the GC-MS analysis. The result further confirms the potential of the Alcanivorax sp. to produce secondary metabolites with bioactive properties. The compounds detected in the extracts are mostly fatty acids and their derivatives. Fatty acid-derived compounds are secondary metabolites produced by diverse marine symbiotic organisms with antibacterial, antibiofilm, and antifouling properties (Kaspar et al., 2019; Supardy et al., 2019; Ashitha et al., 2020; Kai, 2020). Fatty acids affect the cell membrane of the bacterial cell and cause disruption by solubilizing the membrane due to their detergent properties (Desbois and Smith, 2010). Fatty acid activity can be either antimicrobial activities in high doses or antibiofilm activities in low doses or synergistic effects with both antimicrobial and antibiofilm activities (Park et al., 2022). Fatty acids are an important component of the bacterial cell that is involved in cell signaling and bacterial adaptation, which is an important requirement in biofilm signal control (De Carvalho and Caramujo, 2018). Fatty acids could be responsible for the inhibition of the biofilm formation under static conditions. Fatty acids can control biofilm development against diverse microbes through many targets, especially by reducing the biomass of the biofilm bacteria, suppressing the cell-to-cell communication expression or quorum quenching (Kumar et al., 2020). Fatty acid compounds have the potential to enable symbiotic bacteria to protect their host against colonization by biofilm bacteria on its surface due to their ability to interfere with the biofilm formation ability (Khadke et al., 2021). Fatty acids have been reported from several marine bacteria with biofilm inhibition activity against biofilm bacteria including Gram-negative bacteria (Rajput et al., 2017). For instance, the biofilm biomass of Vibrio has been reduced significantly by palmitic acid, a free fatty acid, by blocking the formation of biofilm through the suppression of cell-to-cell communication (Santhakumari et al., 2017). Free fatty acids such as tetradecanoic acids and dodecanoic acids that were detected in this study were reported to play key roles in providing protection to hosts against opportunistic organisms (Karimi et al., 2015). Another compound, eicosane, detected in the extract AE2 has been reported with significant antibiofilm activity (Beema Shafreen et al., 2022). Also, hexadecanoic acid, methyl ester and hexadecanoic acid, ethyl ester detected respectively in AE1 and AE2 have antibiofilm potentials and are associated with biosurfactant formation in an endophytic bacterium (Ashitha et al., 2020). Biosurfactant production is an important characteristic of Alcanivorax sp., which is an essential requirement for inhibiting the biofilm formation and biofouling growth (Qiao and Shao, 2010; Alemán-Vega et al., 2020; Mishra et al., 2020). The detection of fatty acids in the extracts along with strong antibiofilm activity in this study indicated the potential of sponge-associated Alcanivorax as a good source of antibiofilm compounds.
In silico analysis
The in silico analysis of the compounds detected from the two extracts of Alcanivorax sp. AA8 showed significant antibiofilm properties. The identified compounds strongly interacted with the molecular target, the BAP. The BAP is a surface protein that plays an important role in primary adhesion, intercellular adhesion, and biofilm formation (De Gregorio et al., 2015). The BAP is generally present in both Gram-positive and Gram-negative bacteria (Cucarella et al., 2004). Therefore, it is assumed that the interaction of the compounds with the target BAP can interfere with the function of the BAP, which has an important role in biofilm formation. Notably, the binding affinity of the identified compounds with the target BAP was better than the binding affinity of the reference ligand butenolide made with the target BAP. Butenolide is a lactone that has broad-spectrum antibiofilm activity (Yin et al., 2019). Comparatively, the compounds present in the bacterial extracts showed better antibiofilm activity than the antibiofilm agent butenolide. Both the in silico and the experimental results demonstrated the ability of the bacterium to produce compounds with antibiofilm properties.
Conclusion
This study revealed the antibiofilm activity of the Alcanivorax sp. AA8 associated with a sponge from the Red Sea. The potential secondary metabolites detected in the extract of the bacterium may serve as promising antibiofilm compounds to tackle the attachment of biofilm bacteria to surfaces. Though further studies are required to identify the compounds responsible for the observed antibiofilm activity, the results demonstrated that bacteria associated with marine sponges could be used as a prospective source for natural antibiofilm and antifouling compounds to control biofouling.
Funding
This research work was funded by Institutional Fund Projects under grant no (IFPRC-086-150-2020). Therefore, authors gratefully acknowledge technical and financial support from the Ministry of Education and King Abdulaziz University, Jeddah, Saudi Arabia.
Acknowledgments
The authors thank Mr. Idris Abdulrahman (Department of Marine Biology, King Abdulaziz University) for his assistance in laboratory experiments. We also acknowledge Dr. Viju Natarajan (Manonmaniam Sundaranar University, India) for his help in molecular docking studies.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Materials.
Author contributions
SS designed the study, analysed the data and conducted in silico analysis, MJ and SS performed the experiments and wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.980418/full#supplementary-material
References
1
AbdulrahmanI.JamalM. T.AlshaeryM.Al-MaaqarS. M.SatheeshS. (2021). Isolation and identification of biofilm bacteria from microfouling assemblage developed on artificial materials submerged in the red sea. JKAU: Mar. Sci.31, 45–54. doi: 10.4197/Mar.31-2.4
2
Abo-StateM. A. M.SalehY. E.-S.GhareebH. M. (2018). Prevalence and sequence of aminoglycosides modifying enzymes genes among e.Coli and klebsiella species isolated from egyptian hospitals. J. Radiat. Res. Appl. Sci.11, 408–415. doi: 10.1016/j.jrras.2018.08.005
3
Al-DaghistaniH. I.Abu-NiaajL. F.BustanjiY.Al-HamaidehK. D.Al-SalamatH.NassarM. N.et al. (2021). Antibacterial and cytotoxicity evaluation of arum hygrophilum bioss. Eur. Rev. Med. Pharmacol. Sci.25, 7306–7316. doi: 10.26355/eurrev_202112_27424
4
Alemán-VegaM.Sánchez-LozanoI.Hernández-GuerreroC. J.HellioC.QuintanaE. T. (2020). Exploring antifouling activity of biosurfactants producing marine bacteria isolated from gulf of California. Int. J. Mol. Sci.21, 6068. doi: 10.3390/ijms21176068
5
AshithaA.RadhakrishnanE. K.MathewJ. (2020). Characterization of biosurfactant produced by the endophyte burkholderia sp. Wyat7 and evaluation of its antibacterial and antibiofilm potentials. J. Biotechnol.313, 1–10. doi: 10.1016/j.jbiotec.2020.03.005
6
BannisterJ.SieversM.BushF.BloecherN. (2019). Biofouling in marine aquaculture: A review of recent research and developments. Biofouling35, 631–648. doi: 10.1080/08927014.2019.1640214
7
BarbatoM.MapelliF.ChouaiaB.CrottiE.DaffonchioD.BorinS. (2015). Draft genome sequence of the hydrocarbon-degrading bacterium alcanivorax dieselolei ks-293 isolated from surface seawater in the eastern mediterranean sea. Genome Announcements3, e01424-15. doi: 10.1128/genomeA.01424-15
8
Beema ShafreenR. M.SeemaS.Alagu LakshmiS.SrivathsanA.TamilmuhilanK.ShresthaA.et al. (2022). In vitro and in vivo antibiofilm potential of eicosane against candida albicans. Appl. Biochem. Biotechnol.194, 4800–4816. doi: 10.1007/s12010-022-03984-8
9
BehzadH.IbarraM. A.MinetaK.GojoboriT. (2016). Metagenomic studies of the red Sea. Gene Part1), 717–723. doi: 10.1016/j.gene.2015.10.034
10
BerumenM. L.VoolstraC. R.DaffonchioD.AgustiS.ArandaM.IrigoienX.et al. (2019). “The red Sea: environmental gradients shape a natural laboratory in a nascent ocean,” in Coral reefs of the red Sea (Cham: Springer), 1–10. doi: 10.1007/978-3-030-05802-9_1
11
BhojY.TharmavaramM.RawtaniD. (2021). A comprehensive approach to antifouling strategies in desalination, marine environment, and wastewater treatment. Chem. Phys. Impact100008. doi: 10.1016/j.chphi.2020.100008
12
CámaraM.GreenW.MacpheeC. E.RakowskaP. D.RavalR.RichardsonM. C.et al. (2022). Economic significance of biofilms: A multidisciplinary and cross-sectoral challenge. NPJ Biofilms Microbiomes8, 42. doi: 10.1038/s41522-022-00306-y
13
CarusoC.RizzoC.ManganoS.PoliA.Di DonatoP.FinoreI.et al. (2018). Production and biotechnological potential of extracellular polymeric substances from sponge-associated Antarctic bacteria. Appl. Environ. Microbiol.84, e01624-17. doi: 10.1128/AEM.01624-17
14
ChernikovaT. N.BargielaR.ToshchakovS. V.ShivaramanV.LunevE. A.YakimovM. M.et al. (2020). Hydrocarbon-degrading bacteria alcanivorax and marinobacter associated with microalgae pavlova lutheri and nannochloropsis oculata. Front. Microbiol.11. doi: 10.3389/fmicb.2020.572931
15
CruzH. (2020). Biofouling challenges and possible solutions, 1.20.003 – final project report (Launceston, Australia: Blue Economy Cooperative Research Centre).
16
CucarellaC.TormoM. Á.ÚbedaC.TrotondaM. P.MonzónM.PerisC.et al. (2004). Role of biofilm-associated protein bap in the pathogenesis of bovine staphylococcus aureus. Infect. Immun.72, 2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004
17
DashS.NogataY.ZhouX.ZhangY.XuY.GuoX.et al. (2011). Poly-ethers from winogradskyella poriferorum: Antifouling potential, time-course study of production and natural abundance. Bioresource Technol.102 (16), 7532–7537. doi: 10.1016/j.biortech.2011.05.034
18
DebnathM.PaulA. K.BisenP. S. (2007). Natural bioactive compounds and biotechnological potential of marine bacteria. Curr. Pharm. Biotechnol.8, 253–260. doi: 10.2174/138920107782109976
19
De CarvalhoC. C. C. R.CaramujoM. J. (2018). The various roles of fatty acids. Molecules23 (10), 2583. doi: 10.3390/molecules23102583
20
De GregorioE.Del FrancoM.MartinucciM.RoscettoE.ZarrilliR.Di NoceraP. P. (2015). Biofilm-associated proteins: News from acinetobacter. BMC Genomics16, 933. doi: 10.1186/s12864-015-2136-6
21
DesboisA. P.SmithV. J. (2010). Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol.85 (6), 1629–1642. doi: 10.1007/s00253-009-2355-3
22
DobretsovS.RittschofD. (2020). Love at first taste: Induction of larval settlement by marine microbes. Int. J. Mol. Sci.21 (3), 731. doi: 10.3390/ijms21030731
23
DusaneD. H.PawarV. S.NancharaiahY. V.VenugopalanV. P.KumarA. R.ZinjardeS. S. (2011). Anti-biofilm potential of a glycolipid surfactant produced by a tropical marine strain of serratia marcescens. Biofouling27, 645–654. doi: 10.1080/08927014.2011.594883
24
El-HossaryE. M.Abdel-HalimM.IbrahimE. S.Pimentel-ElardoS. M.NodwellJ. R.HandoussaH.et al. (2020). Natural products repertoire of the red Sea. Mar. Drugs18 (9), 457. doi: 10.3390/md18090457
25
ElshikhM.AhmedS.FunstonS.DunlopP.McgawM.MarchantR.et al. (2016). Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett.38, 1015–1019. doi: 10.1007/s10529-016-2079-2
26
FahmyN. M.Abdel-TawabA. M. (2021). Isolation and characterization of marine sponge-associated streptomyces sp. Nmf6 strain producing secondary metabolite(s) possessing antimicrobial, antioxidant, anticancer, and antiviral activities. J. Genet. Eng. Biotechnol.19, 102–102. doi: 10.1186/s43141-021-00203-5
27
FusetaniN. (2004). Biofouling and antifouling. Natural Prod. Rep.21, 94–104. doi: 10.1039/B302231P
28
IndraningratA. A. G.MichellerS.RunderkampM.SauerlandI.BeckingL. E.SmidtH.et al. (2019). Cultivation of sponge-associated bacteria from agelas sventres and xestospongia muta collected from different depths. Mar. Drugs17, 578. doi: 10.3390/md17100578
29
JinX.ZhouJ.RicheyG.WangM.HongS. M. C.HongS. H. (2021). Undecanoic acid, lauric acid, and n-tridecanoic acid inhibit escherichia coli persistence and biofilm formation. J. Microbiol. Biotechnol.31, 130–136. doi: 10.4014/jmb.2008.08027
30
KaiM. (2020). Diversity and distribution of volatile secondary metabolites throughout bacillus subtilis isolates. Front. Microbiol.11 (559). doi: 10.3389/fmicb.2020.00559
31
KarimiE.JaafarH. Z.GhasemzadehA.EbrahimiM. (2015). Fatty acid composition, antioxidant and antibacterial properties of the microwave aqueous extract of three varieties of labisia pumila benth. Biol. Res.48, 1–6. doi: 10.1186/0717-6287-48-9
32
KarthickP.MohanrajuR. (2018). Antimicrobial potential of epiphytic bacteria associated with seaweeds of little andaman, india. Front. Microbiol.9. doi: 10.3389/fmicb.2018.00611
33
KarygianniL.RenZ.KooH.ThurnheerT. (2020). Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol.28 (8), 668–681. doi: 10.1016/j.tim.2020.03.016
34
KasparF.NeubauerP.GimpelM. (2019). Bioactive secondary metabolites from bacillus subtilis: A comprehensive review. J. Natural Prod.82 (7), 2038–2053. doi: 10.1021/acs.jnatprod.9b00110
35
KhadkeS. K.LeeJ. H.KimY. G.RajV.LeeJ. (2021). Assessment of antibiofilm potencies of nervonic and oleic acid against acinetobacter baumannii using In vitro and computational approaches. Biomedicines9 (9), 1133. doi: 10.3390/biomedicines9091133
36
KijjoaA.SawangwongP. (2004). Drugs and cosmetics from the Sea. Mar. Drugs2 (2), 73–82. doi: 10.3390/md202073
37
KimY. G.LeeJ. H.LeeS.LeeY. K.HwangB. S.LeeJ. (2021). Antibiofilm activity of phorbaketals from the marine sponge phorbas sp. against Staphylococcus aureus. Mar. Drugs19 (6), 301. doi: 10.3390/md19060301
38
KocotA. M.OlszewskaM. A. (2017). Biofilm formation and microscopic analysis of biofilms formed by listeria monocytogenes in a food processing context. LWT84, 47–57. doi: 10.1016/j.lwt.2017.05.042
39
KumarP.LeeJ.-H.BeyenalH.LeeJ. (2020). Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol.28, 753–768. doi: 10.1016/j.tim.2020.03.014
40
KumarS. N.NambisanB.SundaresanA.MohandasC.AntoR. J. (2014). Isolation and identification of antimicrobial secondary metabolites from bacillus cereus associated with a rhabditid entomopathogenic nematode. Ann. Microbiol.64 (1), 209–218. doi: 10.1007/s13213-013-0653-6
41
KumarS.StecherG.LiM.KnyazC.TamuraK. (2018). Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35, 1547–1549. doi: 10.1093/molbev/msy096
42
LaiQ.WangJ.GuL.ZhengT.ShaoZ. (2013). Alcanivorax marinus sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol.63, 4428–4432. doi: 10.1099/ijs.0.049957-0
43
LaiQ.ZhouZ.LiG.LiG.ShaoZ. (2016). Alcanivorax nanhaiticus sp. nov., isolated from deep sea sediment. Int. J. Syst. Evol. Microbiol.66, 3651–3655. doi: 10.1099/ijsem.0.001247
44
LiaoX.LaiQ.YangJ.DongC.LiD.ShaoZ. (2020). Alcanivorax sediminis sp. nov., isolated from deep-sea sediment of the pacific ocean. Int. J. Syst. Evol. Microbiol.70, 4280–4284. doi: 10.1099/ijsem.0.004285
45
LieskeE.FiedlerK. E.MyersR. F. (2004). Coral reef guide: Red sea to gulf of Aden, South Oman (Collins: London: The definitive guide to over 1200 species of underwater life).
46
LiuJ.RenQ.ZhangY.LiY.TianX.WuY.et al. (2019). Alcanivorax profundi sp. nov., isolated from deep seawater of the mariana trench. Int. J. Syst. Evol. Microbiol.69, 371–376. doi: 10.1099/ijsem.0.003145
47
LiuL.ZhengY.-Y.ShaoC.-L.WangC.-Y. (2019). Metabolites from marine invertebrates and their symbiotic microorganisms: molecular diversity discovery, mining, and application. Mar. Life Sci. Technol.1 (1), 60–94. doi: 10.1007/s42995-019-00021-2
48
LiuL. L.WuC. H.QianP. Y. (2020). Marine natural products as antifouling molecules - a mini-review (2014-2020). Biofouling36(10, 1210–1226. doi: 1080/08927014.2020.1864343
49
MaJ. F.XuZ. H.ZhangY. K.ChengX.FanS. L.WangJ. W.et al. (2021a). Biofilm associated protein - BSP domain. doi: 10.2210/pdb7C7U/pdb
50
MaJ.ChengX.XuZ.ZhangY.ValleJ.FanS.et al. (2021b). Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch. EMBO. J.40, e107500. doi: 10.15252/embj.2020107500
51
MalveH. (2016). Exploring the ocean for new drug developments: Marine pharmacology. J. Pharm. Bioallied Sci.8 (2), 83–91. doi: 10.4103/0975-7406.171700
52
ManilalA.SabuK. R.ShewangizawM.AkliluA.SeidM.MerdekiosB.et al. (2020). In vitro antibacterial activity of medicinal plants against biofilm-forming methicillin-resistant staphylococcus aureus: Efficacy of moringa stenopetala and rosmarinus officinalis extracts. Heliyon6, e03303. doi: 10.1016/j.heliyon.2020.e03303
53
MayerA. M. S.GuerreroA. J.RodríguezA. D.Taglialatela-ScafatiO.NakamuraF.FusetaniN. (2021). Marine pharmacology in 2016–2017: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs19 (2), 49. doi: 10.3390/md19020049
54
MehbubM. F.LeiJ.FrancoC.ZhangW. (2014). Marine sponge derived natural products between 2001 and 2010: Trends and opportunities for discovery of bioactives. Mar. Drugs12, 4539–4577. doi: 10.3390/md12084539
55
MishraR.PandaA. K.De MandalS.ShakeelM.BishtS. S.KhanJ. (2020). Natural anti-biofilm agents: Strategies to control biofilm-forming pathogens. Front. Microbiol.11. doi: 10.3389/fmicb.2020.566325
56
MolnarJ. L.GamboaR. L.RevengaC.SpaldingM. D. (2008). Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ.6, 485–492. doi: 10.1890/070064
57
MondolM. A. M.ShinH. J.IslamM. T. (2013). Diversity of secondary metabolites from marine bacillus species: chemistry and biological activity. Mar. Drugs11 (8), 2846–2872. doi: 10.3390/md11082846
58
NadeemM. S.RazeethM.ChoudhryH. M. Z.AnwarF.ZamzamiM. A.MurtazaB. N.et al. (2020). Lc-ms/ms-based metabolic profiling of escherichia coli under heterologous gene expression stress. J. Cell. Biochem.121, 125–134. doi: 10.1002/jcb.28962
59
NunesS. D. O.RosaH. D. S.CanellasA. L. B.RomanosM. T. V.Dos SantosK. R. N.MuricyG.et al. (2021). High reduction of staphylococcal biofilm by aqueous extract from marine sponge-isolated enterobacter sp. Res. Microbiol.172, 103787. doi: 10.1016/j.resmic.2020.10.002
60
Ortega-MoralesB. O.Chan-BacabM. J.Miranda-TelloE.FardeauM.-L.CarreroJ. C.SteinT. (2008). Antifouling activity of sessile bacilli derived from marine surfaces. J. Ind. Microbiol. Biotechnol.35, 9–15. doi: 10.1007/s10295-007-0260-2
61
ParkS.LeeJ. H.KimY. G.HuL.LeeJ. (2022). Fatty acids as aminoglycoside antibiotic adjuvants against staphylococcus aureus. Front. Microbiol.876932. doi: 10.3389/fmicb.2022.876932
62
PereiraL. B.PalermoB. R. Z.CarlosC.OttoboniL. M. M. (2017). Diversity and antimicrobial activity of bacteria isolated from different brazilian coral species. FEMS Microbiol. Lett.364 (16), 164. doi: 10.1093/femsle/fnx164
63
ProkschP.EdradaR.EbelR. (2002). Drugs from the seas – current status and microbiological implications. Appl. Microbiol. Biotechnol.59 (2), 125–134. doi: 10.1007/s00253-002-1006-8
64
PugazhendiA.Abbad WazinH.QariH.BasahiJ. M.GodonJ. J.DhavamaniJ. (2017). Biodegradation of low and high molecular weight hydrocarbons in petroleum refinery wastewater by a thermophilic bacterial consortium. Environ. Technol.38, 2381–2391. doi: 10.1080/09593330.2016.1262460
65
QiaoN.ShaoZ. (2010). Isolation and characterization of a novel biosurfactant produced by hydrocarbon-degrading bacterium alcanivorax dieselolei b-5. J. Appl. Microbiol.108, 1207–1216. doi: 10.1111/j.1365-2672.2009.04513.x
66
RajalaxmiM.Beema ShafreenR.IyerP. M.Sahaya VinoR.BalamuruganK.PandianS. K. (2016). An in silico, in vitro and in vivo investigation of indole-3-carboxaldehyde identified from the seawater bacterium marinomonas sp. as an anti-biofilm agent against vibrio cholerae o1. Biofouling32, 1–12. doi: 10.1080/08927014.2016.1154545
67
RajputA.ThakurA.SharmaS.KumarM. (2017). Abiofilm: A resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res.46, D894–D900. doi: 10.1093/nar/gkx1157
68
RizzoC.GugliandoloC.Lo GiudiceA. (2020). Exploring Mediterranean and Arctic environments as a novel source of bacteria producing antibacterial compounds to be applied in aquaculture. Appl. Sci.10, 4006. doi: 10.3390/app10114006
69
SanthakumariS.NilofernishaN. M.PonrajJ. G.PandianS. K.RaviA. V. (2017). In vitro and in vivo exploration of palmitic acid from synechococcus elongatus as an antibiofilm agent on the survival of artemia franciscana against virulent vibrios. J. Invertebrate Pathol.150, 21–31. doi: 10.1016/j.jip.2017.09.001
70
Santos-GandelmanF. J.Giambiagi-DemarvalM.OelemannM. R. W.LaportS. M. (2014). Biotechnological potential of sponge-associated bacteria. Curr. Pharm. Biotechnol.15, 143–155. doi: 10.2174/1389201015666140711115033
71
SarkerS. D.NaharL.KumarasamyY. (2007). Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods42, 321–324. doi: 10.1016/j.ymeth.2007.01.006
72
SatheeshS.Ba-AkdahM. A.Al-SofyaniA. A. (2016). Natural antifouling compound production by microbes associated with marine macroorganisms: A review. Electronic J. Biotechnol.19, 26–35. doi: 10.1016/j.ejbt.02.002
73
SchulteS.WingenderJ.FlemmingH.-C. (2005). Efficacy of biocides against biofilms in, Directory of microbicides for the protection of materials: A handbook. Ed. PaulusW. (Dordrecht: Springer Netherlands), 93–120.
74
SelvinJ.ShanmughapriyaS.GandhimathiR.Seghal KiranG.Rajeetha RavjiT.NatarajaseenivasanK.et al. (2009). Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes nocardiopsis dassonvillei mad08. Appl. Microbiol. Biotechnol.83, 435–445. doi: 10.1007/s00253-009-1878-y
75
SkogmanM. E.VuorelaP. M.FallareroA. (2016). A platform of anti-biofilm assays suited to the exploration of natural compound libraries. J. Vis. Exp.118, 54829. doi: 10.3791/54829
76
SongL.LiuH.CaiS.HuangY.DaiX.ZhouY. (2018). Alcanivorax indicus sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol.68, 3785–3789. doi: 10.1099/ijsem.0.003058
77
SrinivasanR.KannappanA.ShiC.LinX. (2021). Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs19, 530. doi: 10.3390/md19100530
78
SupardyN. A.IbrahimD.Mat NorS. R.NoordinW. N. M. (2019). Bioactive compounds of pseudoalteromonas sp. IBRL PD4.8 inhibit growth of fouling bacteria and attenuate biofilms of vibrio alginolyticus FB3. Polish J. Microbiol.68 (1), 21–33. doi: 10.21307/pjm-2019-003
79
TeanpaisanR.KawsudP.PahumuntoN.PuripattanavongJ. (2017). Screening for antibacterial and antibiofilm activity in thai medicinal plant extracts against oral microorganisms. J. Traditional Complementary Med.7, 172–177. doi: 10.1016/j.jtcme.2016.06.007
80
TogashiN.ShiraishiA.NishizakaM.MatsuokaK.EndoK.HamashimaH.et al. (2007). Antibacterial activity of long-chain fatty alcohols against staphylococcus aureus. Molecules12 (2), 139–148. doi: 10.3390/12020139
81
UnabiaC. R. C.HadfieldM. G. (1999). Role of bacteria in larval settlement and metamorphosis of the polychaete hydroides elegans. Mar. Biol.133 (1), 55–64. doi: 10.1007/s002270050442
82
VigneshS.RajaA.JamesR. A. (2011). Marine drugs: Implication and future studies. Int. J. Pharmacol.7 (1), 22–30. doi: 10.3923/ijp.2011.22.30
83
WangG. (2006). Diversity and biotechnological potential of the sponge-associated microbial consortia. J. Ind. Microbiol. Biotechnol.33 (7), 545–545. doi: 10.1007/s10295-006-0123-2
84
WangK.-L.WuZ.-H.WangY.WangC.-Y.XuY. (2017). Mini-review: Antifouling natural products from marine microorganisms and their synthetic analogs. Mar. Drugs15 (9), 266. doi: 10.3390/md15090266
85
WangW. D.ZhangN. N.ChandaW.LiuM.DinS.DiaoY. P.et al. (2018). Antibacterial and anti-biofilm activity of the lipid extract from Mantidis ootheca on Pseudomonas aeruginosa. Journal of Zhejiang University. ScienceB 19 (5), 364–371. doi: 10.1631/jzus.B1700356
86
YinQ.LiangJ.ZhangW.ZhangL.HuZ.-L.ZhangY.et al. (2019). Butenolide, a marine-derived broad-spectrum antibiofilm agent against both gram-positive and gram-negative pathogenic bacteria. Mar. Biotechnol.21, 88–98. doi: 10.1007/s10126-018-9861-1
87
ZadjelovicV.ChhunA.QuareshyM.SilvanoE.Hernandez-FernaudJ. R.Aguilo-FerretjansM. M.et al. (2020). Beyond oil degradation: Enzymatic potential of alcanivorax to degrade natural and synthetic polyesters. Environ. Microbiol.22, 1356–1369. doi: 10.1111/1462-2920.14947
88
ZhangS.SongW.NothiasL.-F.CouvillionS. P.WebsterN.ThomasT. (2022). Comparative metabolomic analysis reveals shared and unique chemical interactions in sponge holobionts. Microbiome10, 22–22. doi: 10.1186/s40168-021-01220-9
Summary
Keywords
sponge, symbiotic bacteria, biofilm, biofouling, antibiofilm, bioactive compounds, alcanivorax
Citation
Jamal MT and Satheesh S (2022) Antibiofilm activity of secondary metabolites of sponge-associated bacterium Alcanivorax sp. from the Red Sea. Front. Mar. Sci. 9:980418. doi: 10.3389/fmars.2022.980418
Received
28 June 2022
Accepted
26 September 2022
Published
14 October 2022
Volume
9 - 2022
Edited by
Ahmed M. Sayed, AlMaaqal University, Iraq
Reviewed by
Karim Abdelkader, Beni-Suef University, Egypt; Carmen Rizzo, Stazione Zoologica Anton Dohrn Napoli, Italy
Updates
Copyright
© 2022 Jamal and Satheesh.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sathianeson Satheesh, ssathianeson@kau.edu.sa; satheesh_s2005@yahoo.co.in
This article was submitted to Marine Biotechnology and Bioproducts, a section of the journal Frontiers in Marine Science
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.