- 1Fisheries Research Institute, Wuhan Academy of Agricultural Sciences, Wuhan, China
- 2Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Wuhan, China
- 3Fisheries Research Institute, Wuhan Academy of Agricultural Sciences, Wuhan Xianfeng Aquaculture Technology Co. Ltd, Wuhan, China
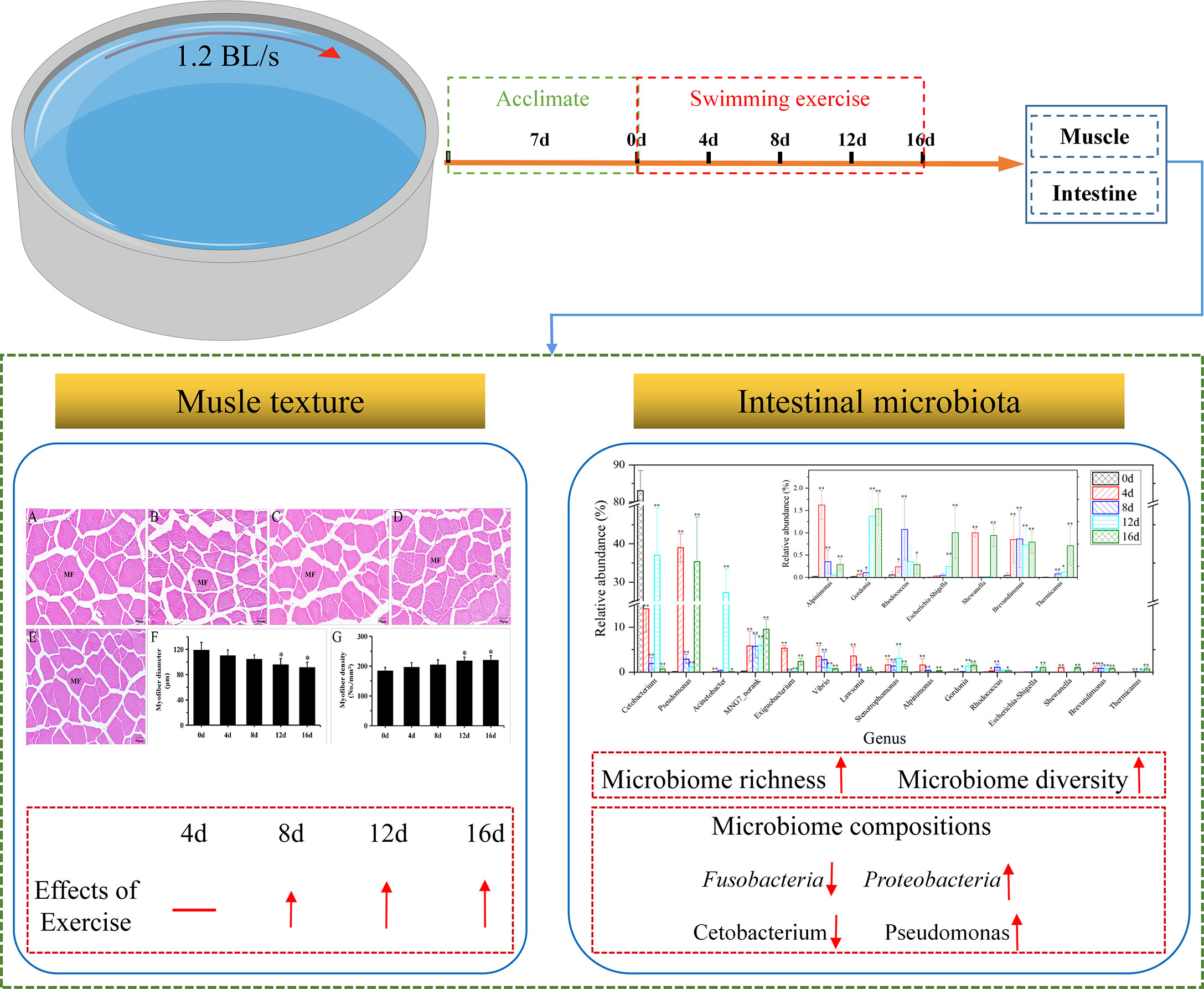
Fillet texture plays a significant role in consumer acceptance, which heavily influenced by exercise according to previous studies. “Xianfeng No. 2”, a novel hybrid of Megalobrama amblycephala (M. amblycephala) (♀) and Ancherythroculter nigrocauda (A. nigrocauda) (♂), with characteristics of quick growth, good disease resistance, and anti-stress capability, is one of an important aquaculture fish species in China, however, the ways to improve fillet quality in this species remains unknown. In this study, a short-term exercise trial (0-16d), hematoxylin and eosin (H&E) staining, and texture analysis were carried out to determine the effects of exercise on muscle cellularity and texture. Furthermore, 16S rRNA analysis was performed to determine the impacts of exercise on the intestinal microbiota. Compared with non-exercised group (0d), exercised groups showed influences in muscle cellularity (decreased muscle fiber diameter and increased muscle fiber density) and improvements in muscle texture (increased hardness, cohesiveness, gumminess, and chewiness). In addition, 16S rRNA sequencing analysis exhibited that short-term exercise could significantly increase the community richness and diversity, as well as alter the composition of gut microflora in this hybrid species. At the level of phylum, the relative richness of Proteobacteria, Firmicutes, Bacteroidetes, and Fusobacteria changed remarkably after short-term exercise. At the level of genus, the relative richness of Pseudomonas, Acinetobacter, Stenotrophomonas, Alpinimonas, Rhodococcus, Shewanella, Brevundimonas changed significantly, suggested that short-term swimming exercise might increase intrinsic antimicrobial resistance and promote denitrification capacity of this hybrid species. Furthermore, a LEfSe analysis of intestinal microflora communities illustrated that Pseudomonas and MNG7 may be utilized to distinguish between exercised and non-exercised fish. Our research, collectively, found an effective way to improve fillet texture by exercise, revealed the impacts of exercise on the intestinal microbiota, further expanded our knowledge of fish under exercise.
Introduction
Fish production has increased dramatically as society has progressed and fisheries have expanded. The current aquaculture sector is particularly interested in the quality of fillets of fish products (Harimana et al., 2019). In addition, fillet texture plays a significant role in consumer acceptance and the suitability for processing (Larsson et al., 2012). Fish meat that is more or less elastic and firm tends to be preferred by consumers (Rasmussen, 2001). The major determinant of muscle texture is skeletal muscle cellularity (i.e., the diameter, density, and number of fibers) (Valente et al., 2013). During postnatal muscle growth, muscle fibers undergo hypertrophy (growing in size) and hyperplasia (growing in number) in fish (Weatherley et al., 1988; Rowlerson and Veggetti, 2001). Genetic characteristics, diet composition, water temperature, as well as light have an impact on the muscle cellularity of fish (Valente et al., 1999; Johnston et al., 2003; Alami-Durante et al., 2010; Martins et al., 2014). The skeletal muscles of fish are the main source of power for swimming exercises (Harimana et al., 2019). Studies have shown that sustained swimming exercise training stimulates muscle hypertrophy and hyperplasia, further affect muscle cellularity and texture (Sänger, 1992; Martin and Johnston, 2005; Ibarz et al., 2011). In Sparus aurata, swimming at a moderate water speed (1-2 body lengths/second [BL/s]) has been documented to influence hypertrophy of white muscle fiber (Ibarz et al., 2011). In Micropterus salmoides, 12 months of moderate exercise may increase muscle cellularity as well as texture (Harimana et al., 2019). In Spinibarbus sinensis, increasing swimming speed or swimming practice time may result in a reduction in the diameter and an increase in the density of muscle fibers (Li et al., 2016). Aquaculture costs would rise substantially if artificial water flow is used on a long-term basis, from an economic point of view, the effect of short-term exercise on fillet quality might be more feasible.
Apart from promoting fish quality, exercise can enhance the immune status (Palstra and Planas, 2011), reduce aggressiveness (Adams et al., 1995), and increase survival (Totland et al., 1987), promote glycogen utilization (Sánchez-Gurmaches et al., 2013), and stimulate fish growth. Sustained exercise-triggered growth has been explored in numerous fish species for instance Seriola lalandi (Brown et al., 2011; Palstra et al., 2015), Danio rerio (Palstra et al., 2010), Spinibarbus sinensis (Li et al., 2013), Piaractus mesopotamicus (Nunes et al., 2013), Polyprion oxygeneios (Khan et al., 2014), Sparus aurata (Ibarz et al., 2011; Sánchez-Gurmaches et al., 2013), and Cyprinus carpio (Shrivastava et al., 2018). To date, nonetheless, there has been little study on the molecular modulatory network in fish in the course of moderate exercise. In previous research, we determined the influence of exercise on growth-linked genes, as well as signaling cascades in Ctenopharyngodon idella and on the synthesis of nutrients-linked genes and signaling cascades in Ictalurus punctatus (Zhang et al., 2021a; Zhang et al., 2021b). Further research is needed to determine the underlying influences of fish under exercise.
The gastrointestinal (GI) tract of vertebrates constitutes a composite bacterial ecosystem containing a complex, as well as dynamic consortium of micro-organisms that are essential to the host nutrition and health (Brugman and Nieuwenhuis, 2010; Cerf-Bensussan and Gaboriau-Routhiau, 2010; Viney and Riley, 2014). In recent decades, the structure and activities of GI microbiota are studied. Numerous allergic, autoimmune, as well as metabolic diseases in modern civilization are linked to alterations in the gut micro-ecosystem (Neish, 2009; Nagalingam and Lynch, 2012). Research on the gut microflora of fish has recently emerged as a cutting-edge area. Fish gut flora is determined by a variety of factors. Water quality directly influences the gut microbiome of fish (Sullam et al., 2012; Wong and Rawls, 2012). The gut microbiome is also strongly influenced by immune status, developmental stage, host genetics, and other host-specific factors (Llewellyn et al., 2014; Ye et al., 2014; Burns et al., 2016; Hennersdorf et al., 2016). Moreover, the diet also shapes the gut microbiome, which displays different compositions depending on the diet (McDonald et al., 2012; Scott et al., 2013; Miyake et al., 2015). In fish, studies on the correlation between exercise and gut microbiota are scarce at present. Our previous research has found that 4-week moderate exercise could increase Proteobacteria and Bacteroidetes in the gut of Ictalurus punctatus (Zhang et al., 2021b). In mammals, there are increasing evidences that exercise can help diversify intestinal microorganisms and restore balance between beneficial and harmful microbes (Matsumoto et al., 2008; Mailing et al., 2019). Compared with sedentary individuals, elite rugby players have greater gut microbiota diversity (Clarke et al., 2014b). In general, greater diversity is associated with better metabolic health (Le Chatelier et al., 2013). However, to our knowledge, research regarding the relationship between short-term exercise and the dynamic changes of gut flora in fish remains unknown, which needs more in-depth investigation. Sequencing bacterial 16S rRNA is a fundamental tool for understanding host gut microflora (Zhang et al., 2021b). As such, analysis of the 16S rRNA can be utilized to shed insights on how the composition of gut flora shifts in response to exercise.
“Xianfeng No. 2” is a genetically stable species of hybrid fish that is bred via crossing M. amblycephala (♀) with A. nigrocauda (♂), which is achieved through continuous selection of four generations of offspring (Zhang et al., 2022). Its mother species, M. amblycephala, is widely regarded as one of the most economically valuable freshwater fishes in China (Zhu et al., 2013). Nonetheless, in recent times, wild M. amblycephala populations have suffered from a decline in germplasm supplies and admixture. At the same time, cultivated M. amblycephala populations have shown signs of growth depression, susceptibility to disease, as well as early sexual maturity (Yi et al., 2013). The aquaculture business is facing an increasing number of pressing challenges, most remarkably the prevention of miniaturization and early puberty in farmed fish and the enhancement of fish resilience to stress (Zhang et al., 2022). The paternal species of “Xianfeng No. 2,” A. nigrocauda, belongs to the culterinae sub-family, which is found in the Yangtze River upper reaches (Liu et al., 2005). It has been employed in artificial hybridization in recent years to produce hybrids with desirable qualities, for instance “Xianfeng No. 1” with oxygen resistance, as well as a gentle nature, which are crucial for fish transportation (Zhang et al., 2020). As a result, the newly produced hybrid “Xianfeng No. 2” has the features of quick growth, good disease resistance, and anti-stress capability, as well as being simpler to net and transport live fish, and is thus considered as a desirable fish with significant economic benefits in pond culture (Zhang et al., 2022).
A short-term (16 day) exercise experiment, H&E staining, and texture analysis were performed to investigate the effects of short-term exercise on muscle cellularity and texture, to improve the quality and consumer acceptability of the hybrid fish. In addition, 16S rRNA analysis was performed to investigate the temporal dynamic changes in important gut microflora underlying the impacts of short-term exercise in this hybrid species to predict the potential effects of exercise and expand our knowledge of fish under exercise.
Materials and methods
Experimental design
All fish were left to acclimatize for two weeks before the experiment in a 1000-liter flow-through tank (pH 7.4 ± 0.3; dissolved oxygen > 7.0 mgL-1; temperature 21.5 ± 1.0°C; natural photoperiod) at the Yangtze River Fisheries Research Institute. The experiment used only male fish with similar sizes to avoid gender-related differences and the influence of female oviposition. Thereafter, 180 hybrid individuals (176.42 ± 16.76 g per tail) were selected at random and distributed equally (N = 30) into 6 swimming circular flumes (Diameter 150 cm; Height 100 cm) where swimming exercise experiments were conducted. Afterward, the fish were acclimatized to their new conditions without directional flow for 7 days. During the exercise training, the six swimming flumes were gradually sped up to the desired speed. Throughout the experiment, the speed was maintained. The nutritional quality of largemouth bass and channel catfish can be improved by raising them at 1.2 BL/s (Harimana et al., 2019; Zhang et al., 2021a), as a result, we chose a speed of swimming of 1.2 BL/s for the experiment. Depending on the exercise time, the fish were divided into one control group (non-exercised or 0d group) and four experimental groups (4d, 8d, 12d, and 16d group). The fish were fed on commercial pelleted feed comprising 30% protein (Haida Co., Ltd. in Guangdong Province, China) during the exercise testing daily once until they appeared to be visual satiation. The diet of this hybrid species remained the same from the adaptive period to the end of the experiment. To ensure fish fully ingested delivered pellets in the feeding period, the water circulation was shut off for two hours every morning. During the exercise experiments, parameters of water quality were monitored once daily for 16d (pH 7.3 ± 0.2, NH3 0.004 - 0.007 mM, nitrate 0.011 - 0.034 mM, nitrite 0.0007 - 0.0015 mM, and dissolved oxygen > 6.8 mgL-1).
Texture analysis
This assay way of texture analysis followed as previously described (Harimana et al., 2019). The TA.XT2i Surface Platform (Stable Miniaturized scale Frameworks, Surrey, United Kingdom) was employed to examine the textural characteristics of a cross section of the trunk muscle adjacent to the head. Six circular flumes were utilized to randomly pick six specimens from every group (0d, 4d, 8d, 12d, and 16d group). Every specimen was split into three (1cm3) sections. The P35 Probe was employed to compress every specimen perpendicular to the muscle fibers at a constant speed of 1 mm s-1 until 75% of the specimen was compressed. Using twofold compression, the Texture Profile Analysis (TPA) of the specimen was created. On the basis of previous descriptions, the resulting values were described as gumminess, springiness, chewiness, hardness, and cohesiveness (Friedman et al., 1963). Data analysis of muscle texture was performed using Excel and SPSS 17.0. Values were expressed as the mean ± SD, and P < 0.05 was considered statistically significant.
Muscle cellularity analysis
A graded series of ethanol was used to dehydrate muscle samples, which were then paraffin-embedded, sectioned, and stained with haematoxylin-eosin. Muscle images (5 images per sample, 3 samples per group) were captured from each slide using via a light microscope (Olympus BX53, Tokyo, Japan). Later, the Image J Launcher was utilized to determine the myofiber properties. A minimum of 150 fibers from five photographs of each sample were examined, and the myofibers diameter was recorded. Myofiber number was counted accurately for detecting myofiber density from five photographs of each sample. The number of fibers per mm2 of muscle cross-sectional area was used to calculate fiber density.
Intestinal sample collection
To assess the effects of moderate exercise on gut microflora, contents of the whole intestinal tract from three fish in every swimming flume was combined into one biological replicate, six biological replicates for every group (0d, 4d, 8d, 12d, and 16d). After rinsing the intestines thrice with ice-cold sterile PBS, the contents were kept at -80°C for DNA isolation. All samples were extracted on a super-clean workbench.
DNA isolation, PCR, and Illumina MiSeq sequencing
Isolation of bacterial DNA was done with the E.Z.N.A.® DNA Kit (Omega Bio-tek, Norcross, United states) from the intestinal contents of fish at various times during the exercise experiment, as described by the manufacturer. Quantitation of DNA was done with the NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, United States), and the DNA quality was assessed using a 1% agarose gel electrophoresis. Amplification of the V3–V4 hypervariable regions of the bacteria 16S rRNA gene was done with the primers (338F 5′-ACTCCTACGGGAGGCAGCA-3′ and 806R 5′-GGACTACHVGGGTWTCTAAT-3′) using a thermocycler PCR platform (GeneAmp 9700, ABI, Foster City, CA, USA). In the PCR reactions, the following protocol was employed: denaturation for 3 minutes at 95°C; 27 cycles of 30 seconds at 95°C, annealing for 30 seconds at 55°C, and extension for 45 seconds at 72°C; and a final extension for 10 minutes at 72°C. In a 20 μL mixture comprising 10 ng of template DNA, 0.4 μL of FastPfu Polymerase, 0.8 μL of each primer (5 μM), 2 μL of 2.5 mM dNTPs, and 4 μL of 5 × FastPfu Buffer. The PCR reactions were done in triplicate. PCR products were extracted using a 2% agarose gel, and the DNA was purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, United States). The samples were quantified using QuantiFluorTM-ST (Promega, Madison, United States) as described by the manifacturer. Using paired-end sequencing (2 × 300), the purified amplicons were sequenced on an Illumina MiSeq (Illumina, San Diego, United States) platform.
Data analysis
Demultiplexing of raw reads was done, and then filtered for quality via Trimmomatic, and merged with FLASH based on the following criteria: (1) All reads at sites with an average quality score of less than 20 across a sliding window of 50 bp were trimmed. (2) Two mismatched nucleotides were permitted in the primers, and ambiguous bases were eliminated from the reads. (3) Sequences larger than 10 bp that overlapped were combined as per their overlap sequence. We used UPARSE (v7.1 http://drive5.com/uparse/), as well as a similarity threshold of 97% to cluster operational taxonomic units (OTUs), and UCHIME was adopted to identify and eliminate chimeric sequences. To examine the taxonomy of each 16S rRNA gene sequence from the Silva database (SSU123), the RDP Classifier algorithm (http://rdp.cme.msu.edu/) was applied to the 16S rRNA sequences. Non-parametric indicators (Chao1, coverage, and Shannon) were utilized to examine the associations between bacterial community coverage levels and diversity features across 0d, 4d, 8d, 12d, and 16d groups. For assessing the sequencing depth of samples, QIIME (v1.8.0) was utilized to generate rarefaction curves. Principal co-ordinates analysis (PCoA) was adopted to investigate and illustrate the similarities between the samples from the five groups on the basis of Bray–Curtis dissimilarity with the Ape software (v5.3). Venn diagrams (Fouts et al., 2012) were generated using R to identify the common and unique OTUs. At the phylum as well as genus levels, abundance and composition diagrams of gut microflora were generated using Origin 8.0 software. Through the Shapiro-Wilk normality test, the normal distribution of all six replicates (mean standard deviation) was determined. A one-way ANOVA was performed to assess for statistically significant changes between experimental groups using the SPSS 17.0 software. Significant differences are deemed to exist when * P< 0.05 and highly significant differences when ** P< 0.01.
Results and discussion
Effects of exercise on the muscle cellularity and texture parameters of hybrid species
Muscle development in fish is characterized by hypertrophy (increase in diameter of muscle fiber) and hyperplasia (increase quantity of muscle fiber), which represent muscle cellularity and may persist into adulthood (Veggetti et al., 1990; Rowlerson et al., 1995). Fish swimming is mostly propelled by skeletal muscle. Multiple studies have documented that long-term swimming affects muscle cellularity dramatically (Martin and Johnston, 2005; Ibarz et al., 2011). In Micropterus salmoides, 1.2 BL/s for 12 months can significantly decrease muscle fiber diameter and increase muscle fiber density (Harimana et al., 2019). In Spinibarbus sinensis, 2 BL/s for 8 weeks can significantly decrease muscle fiber diameter. Nonetheless, if the duration or velocity of swimming exercise increases, the muscle fiber diameter will decrease and the muscle fiber density will rise (Li et al., 2016). Figure 1 illustrates the morphology of myofibers under exercise. In groups 12 d and 16 d, myofibers were arranged more tightly, and the fiber diameters in the exercise groups were shorter than those of the control group (Figures 1A-E). Furthermore, Figures 1F,G exhibits a gradual decrease in myofiber diameters and an increase in myofiber density with longer exercise duration. In addition, exercise for 12 d and 16d significantly decrease muscle fiber diameter and increase muscle fiber density (P < 0.05). These results are in general agreement with above literature. In our study, the muscle fiber diameter decreased by 23.2% and muscle fiber density increased by 19.5% in 1.2 BL/s for 16d to this hybrid species, the change intensity was less than the intensity in 1.2 BL/s for 12 months to Micropterus salmoides (Harimana et al., 2019), these disparities may be due to the difference of species and exercise duration.
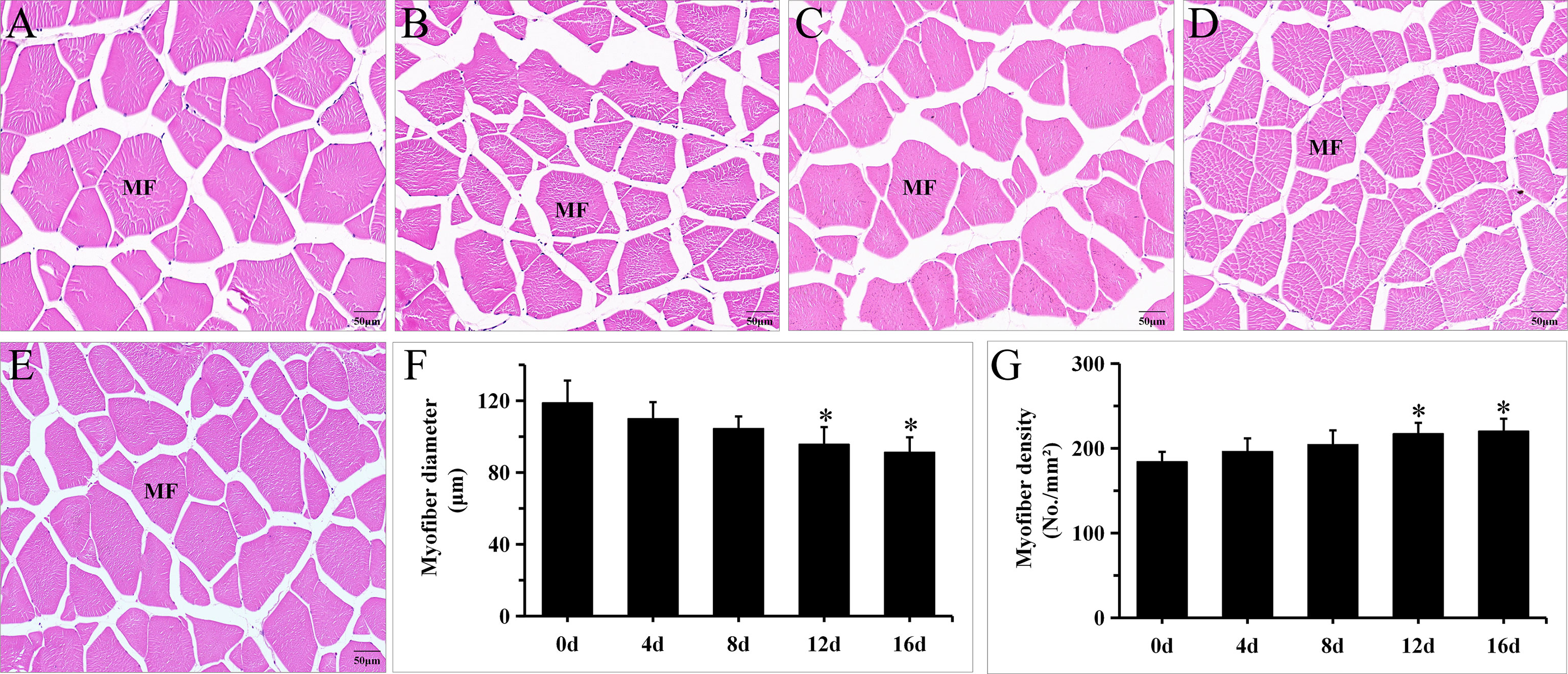
Figure 1 Short-term exercise induced changes in myofiber morphology in hybrid species. Illustrative images of cross-section of muscle in 0d (A), 4d (B), 8d (C), 12d (D) and 16d (E) group of hybrid species. The effects of short-term exercise on the diameter (F) and density (G) of muscle fiber in hybrid species (N = 15, mean ± SD). *P ≤ 0.05. MF, myofiber.
It is considered that swimming activities have a remarkable effect on muscle texture, and that muscle texture correlates dramatically with muscle cellularity as well as collagen content (Periago et al., 2005; Tang et al., 2012). Consequently, the increased density and smaller diameter of muscle fibers result in a higher contents of collagen and hydroxyproline, which increases fish fillet firmness (Bugeon et al., 2003; Rasmussen et al., 2011; Harimana et al., 2019). The fact that swimming exercise significantly improves the texture properties of fish fillet has been reported in Mylopharyngodon piceus (0.9–1.5 BL/s for 12 months) (Harimana et al., 2018), Micropterus salmoides (1.2 BL/s for 12 months) (Harimana et al., 2019), and Spinibarbus sinensis (1 or 2 or 4 BL/s for 8 weeks) (Li et al., 2016). Although studies have shown that short-term exercise decrease the body weight of Cyprinus carpio (2.4–2.6 BL/s for 4 weeks) (Martin and Johnston, 2006) and Carassius auratus (4.5 BL/s for 4 weeks) (Davison and Goldspink, 1978), little is known about the relationship between short-term exercise and muscle texture. In our study, as illustrated in Table 1, excepted for the springiness index, other texture parameters were increased in exercised groups, suggested that short-term exercise could also improve muscle texture. In addition, Hardness, Cohesiveness, Gumminess, and Chewiness were significantly higher in exercised for 12d and 16d group compared with non-exercised group, which suggested that short-term exercise for 12d-16d could effective improve the fillets of this hybrid species. The muscle texture is one of the criteria of flesh quality (Huang et al., 2021). Consumers prefer fish meat that is more or less elastic and firm (Rasmussen, 2001). It is anticipated that this effective method of improving muscle texture will improve consumer acceptability and enhance the economic value of this hybrid species.
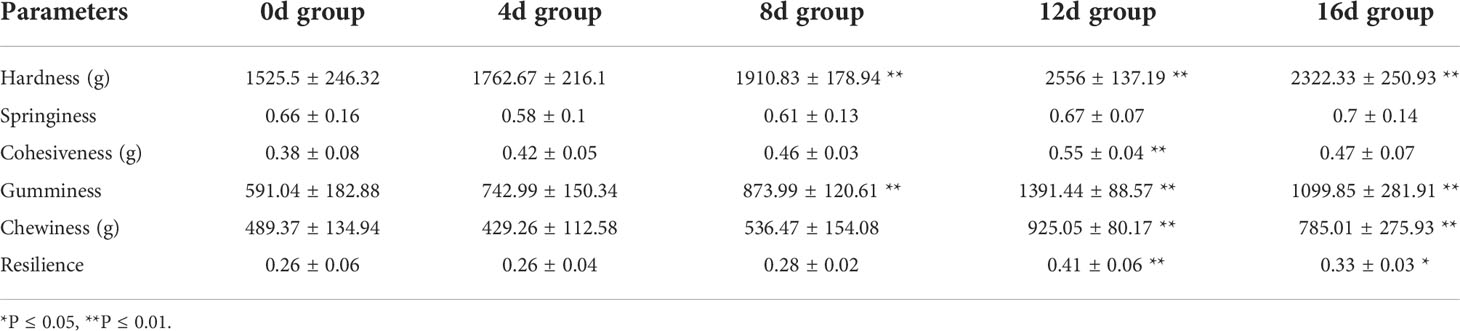
Table 1 The effects of short-term swimming exercise on the muscle texture parameters of this hybrid species (N = 18, mean ± SD).
Sequencing analysis
All the samples yielded a total of 1,304,792 sequences of high quality. The remarkable degree of sequence coverage was exhibited by good coverage (99%). As illustrated in Figure S1, the curve of species accumulation rose rapidly at first and then plateaued as the number of samples multiplied, indicating that there were sufficient samples in this experiment to accurately represent community richness. Besides, the curves of rarefaction (Figure S2) demonstrated that every sample had a high sampling coverage (>99%) and adequate depth of sequencing.
Alpha-diversity indices and beta-diversity analyses
Two measures were employed to quantify microbial diversity: alpha-diversity and beta-diversity. α-diversity represents the number (abundance) and distribution (uniformity) of taxa in a particular sample (Morgan and Huttenhower, 2012). The Shannon index, Simpson index, and Chao 1 index are frequently employed to characterize alpha-diversity (Morgan and Huttenhower, 2012). The Shannon diversity index is the metric that is utilized the most frequently for comparing the diversity in different habitats (Clarke et al., 2014a). When compared, a lower Simpson index and a higher Shannon index are indicative of a more diverse bacterial population (Zhang et al., 2019). Additionally, the higher the value of the Chao 1 index, the greater the richness of species present (Yao et al., 2019).
The indices of alpha-diversity of gut microflora of hybrid fish in different durations of swimming exercise were exhibited in Figure 2. Chao1 index, Shannon index, and Observed OTUs increased remarkably in exercised groups (4d, 8d, 12d, and 16d), while Simpson index decreased significantly in exercised groups compared with that in non-exercised group (0d). As suggested by the above literature, our data suggesting that short-term swimming exercise could markedly improve the community richness, as well as the diversity of gut microbiota in hybrid fish. It has been reported that 4-week moderate exercise increased richness and diversity of gut microbiota in Ictalurus punctatus (Zhang et al., 2021b). Exercise has also been reported to increase diversify intestinal microorganisms in mammals (Matsumoto et al., 2008; Mailing et al., 2019). The data from these literatures support our findings regarding the effects of exercise on richness and diversity of gut microbiota.
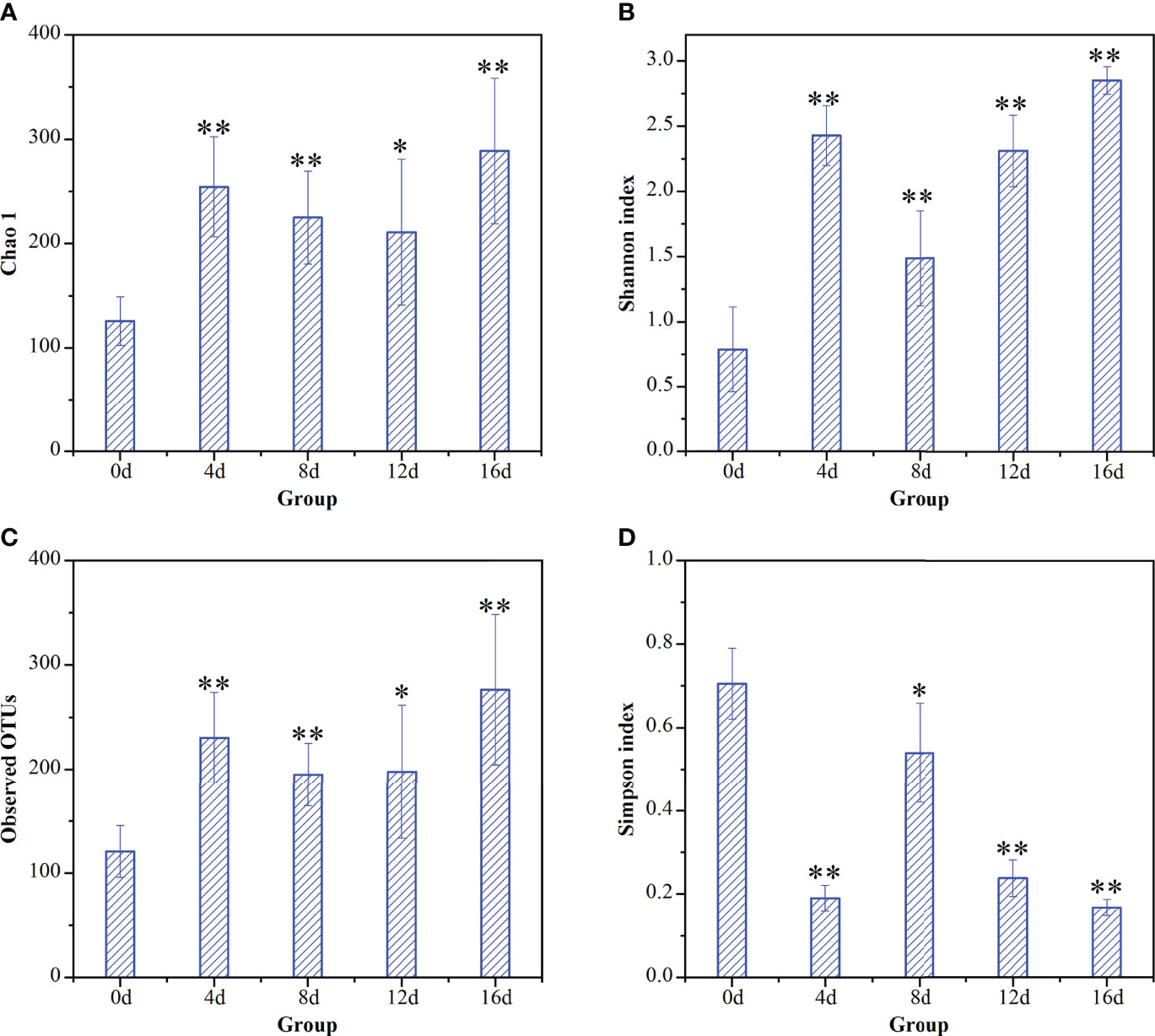
Figure 2 Temporal dynamic changes of short-term exercise on alpha-diversity of gut microflora communities of this hybrid species. Effects of exercise on (A) Chao1 index, (B) Shannon index for species diversity, (C) observed OTUs, and (D) Simpson index in intestinal microbial of hybrid fish. *P ≤ 0.05, **P ≤ 0.01. Data were provided as mean ± SD.
Beta-diversity quantifies the number of taxa shared by two or more samples and can be interpreted as a measure of (dis)similarity (Morgan and Huttenhower, 2012). In general, a healthy condition is defined by a species-rich, diversified, and stable microflora that performs several, as well as complex metabolic functions (Lozupone et al., 2012). Based on unweighted UniFrac distance PCoA analysis, samples from five groups (0d, 4d, 8d, 12d, and 16d) were separated into five distinct portions (Figure 3A). Moreover, the hierarchical clustering tree (Figure 3B) revealed that the non-exercised group (0d) and the 12d-exercised group (12d) were clustered into one branch, whereas the 4d, 8d, and 16d groups were grouped into another branch. The data revealed that short-term swimming activity might have a remarkable effect on the intestinal microflora structure of this hybrid species, making it potentially healthier than the non-exercising fish.
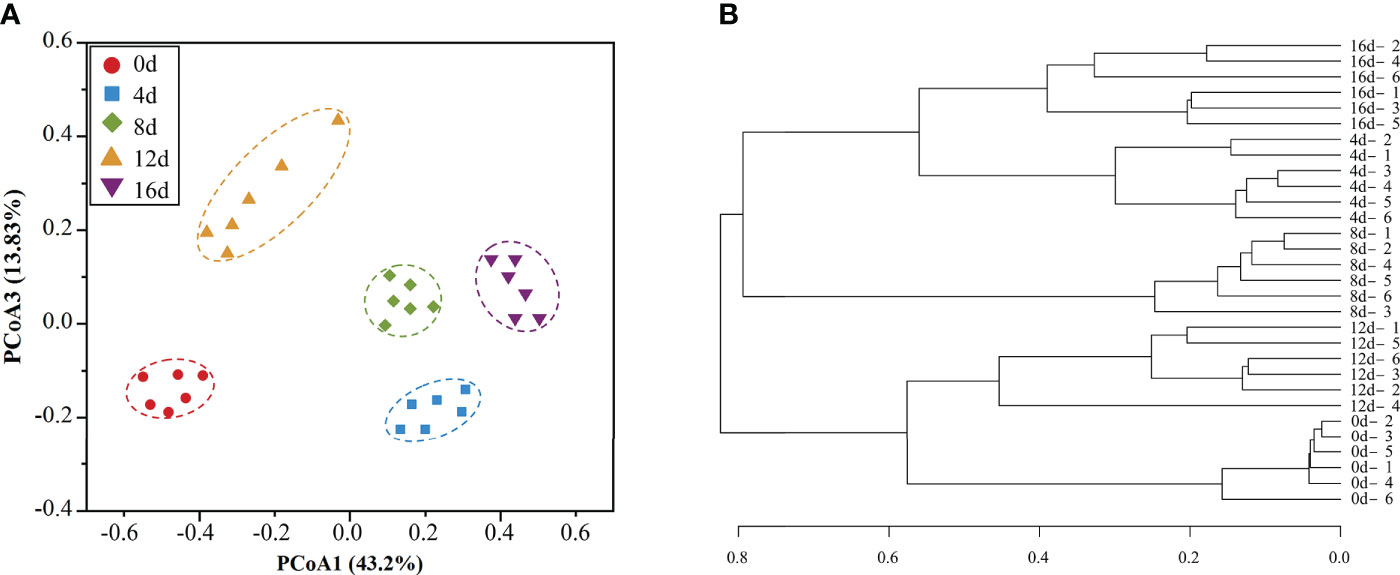
Figure 3 PCoA analysis (A) and Similarity tree (B) of intestinal microbial community of this hybrid species.
OTU distribution at non-exercised and exercised groups
Five groups shared 125 OTUs contained Acidobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Cyanobacteria, Firmicutes, Fusobacteria, Gemmatimonadetes, Planctomycetes, Proteobacteria, and Saccharibacteria at phylum lever according to venn diagrams (Figure 4), demonstrating that these OTUs may be insensitive to swimming activity. In addition, 86 OTUs contained Chlorobi and Synergistetes at phylum lever were unique to the control group. We determined these certain OTUs were susceptible to physical exercise. Additionally, 62 OTUs contained Nitrospirae at phylum lever shared by the four exercised groups but absent from the non-exercised group might be used as biomarkers to differentiate non-exercised and exercised fish. In comparison, 160, 68, 149, and 291 OTUs were only found in the 4d, 8d, 12d, and 16d exercised groups. Furthermore, 395 OTUs contained Armatimonadetes, Nitrospirae, and Parcubacteria at phylum lever were present in 12d group but not in control group, 600 OTUs contained Armatimonadetes, Nitrospirae, and BRC1 at phylum lever were present in 16d group but not in control group. Notably, similar with the Chao index finding, the 16d exercised group had the most OTUs, further demonstrating that this group had a more diverse community. Consequently, we concluded that short-term swimming might considerably affect the gut microflora of this hybrid species.
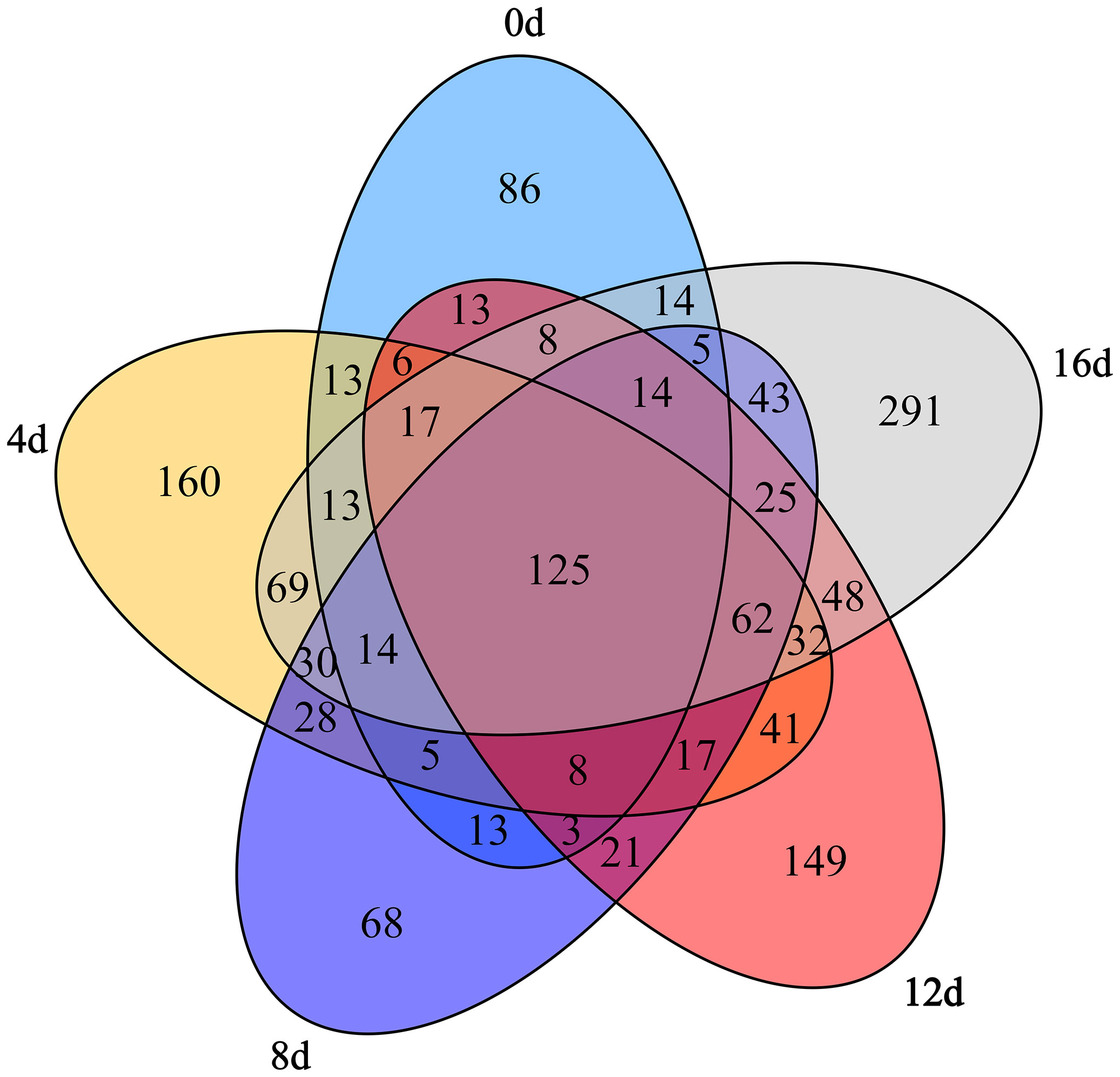
Figure 4 Venn diagrams depicting the number of unique and shared OTUs within the gut microbiota of this hybrid species during different swimming exercise durations.
Phylum-level composition of gut microbiota
As illustrated in Figure 5A, the most predominant phyla were Fusobacteria (71.41%–88.01%), Proteobacteria (11.39%–16.67%), Actinobacteria (0.26%–6.85%), Firmicutes (0.15%–2.09%), and Bacteroidetes (0.02%–0.71%) in the gut bacteria of hybrid fish. Fusobacteria are capable of producing high amounts of butyric acid, which can regulate transepithelial transport, enhance the mucosal barrier, improve the oxidative, as well as inflammatory states of the intestinal mucosa, and suppress colon cancer (Sossai, 2012). Proteobacteria is one of the most prevalent taxa in activated sludge, and it may play a vital role in decomposing complex carbon and eliminating nitrogen (Shu et al., 2015). Additionally, being one of the most prevalent bacteria in fishponds, it participates in several bio-geochemical processes, such as the cycling of carbon, nitrogen, as well as sulfur (Klase et al., 2019). It is established that an increase in Firmicutes might increase the number of lipid droplets and hence enhance fatty acid absorption proportionally (Semova et al., 2012). Oligotrophic actinobacteria play a pivotal role in the degradation of refractory substances including xylan and cellulose (Rawat et al., 2012; Kim et al., 2014). According to reports, Bacteroidetes contributed in the cycling of protein-rich compounds and complex carbon compounds (Shi et al., 2015). Herein, the relative richness of Fusobacteria, Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes accounted for more than 97% of total phyla in the gut of hybrid fish, suggesting that these phyla played key roles in the digestive, absorption, and immune functions of this hybrid species.
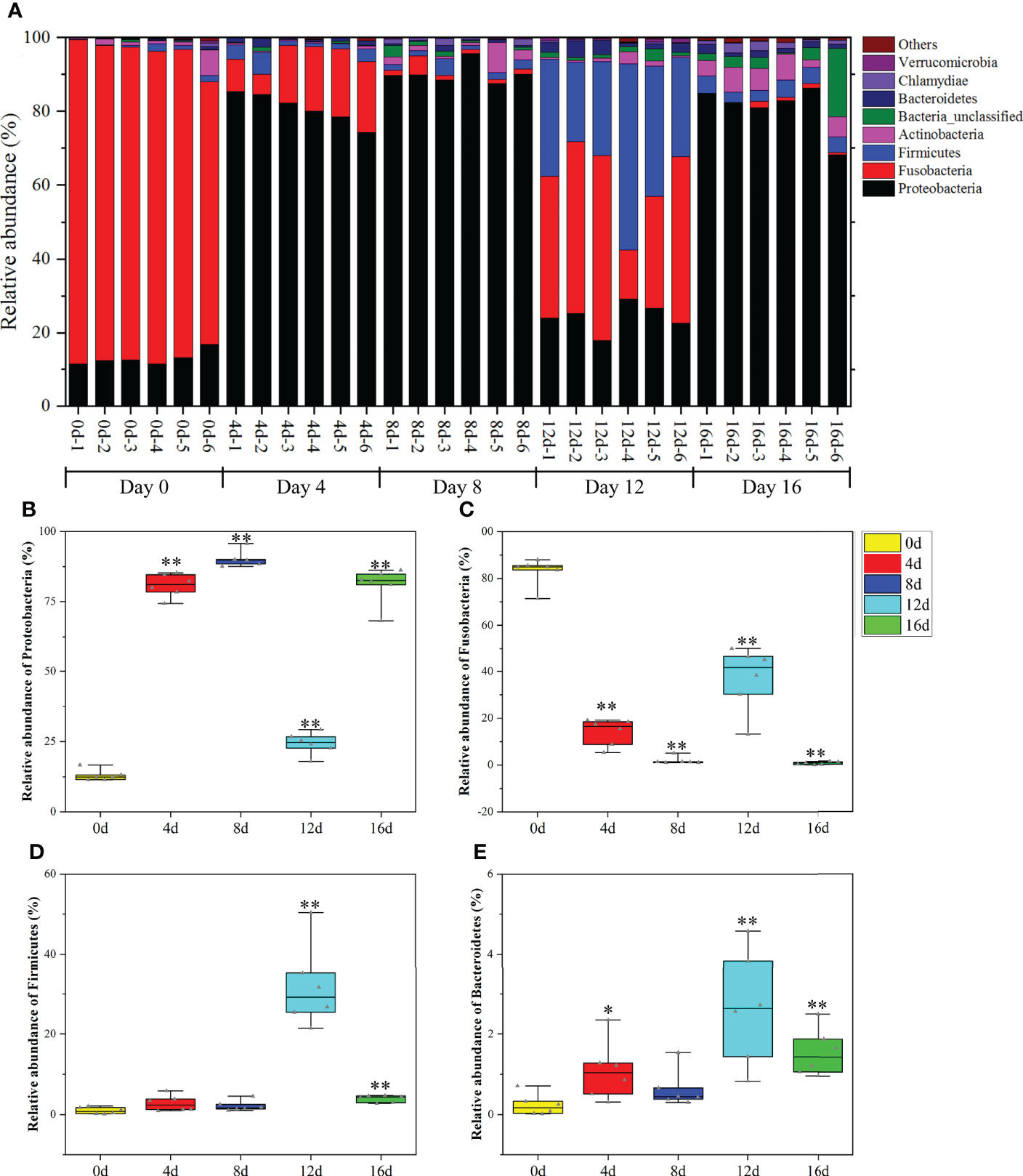
Figure 5 The influences of short-term exercise on microbial phyla in this hybrid species. Relative richness of dominant microbial phyla in the intestine of this hybrid species under different durations of swimming exercise (A). Abundance differences in Proteobacteria (B), Fusobacteria (C), Firmicutes (D), and Bacteroidetes (E) among different durations of swimming exercise groups at the phylum level. *P ≤ 0.05, **P ≤ 0.01. Data were provided as mean ± SD.
One-way ANOVA was employed to test the significance of phyla differences (P < 0.05). As exhibited in Figures 5B–E, the richness of Proteobacteria, Firmicutes, and Bacteroidetes remarkably increased in four exercised groups (4d, 8d, 12d, and 16d). On the contrary, the abundance of Fusobacteria decreased remarkably in four exercised groups compared with non-exercised group (0d). Long-term moderate exercise could increase Proteobacteria and Bacteroidetes, as well as decrease Fusobacteria in the gut of channel catfish (Zhang et al., 2021b). Similarly, exercise increased the Proteobacteria and Firmicutes in mice at the phyla level (Choi et al., 2013; Kang et al., 2014). Exercise in normal rats is also associated with higher Bacteroidetes in fecal matter (Evans et al., 2014). The data from these literatures support our findings regarding the effects of exercise on gut microbiota at the phyla level. From a functional point of view, short-term swimming activity appeared to boost glucose as well as nitrogen cycling and metabolism, and the cycling of protein-rich compounds, while suppressing butyric acid production.
Composition of intestinal microbiota at genus level
At the genus level (Figure 6A), the top two genera were Cetobacterium (71.34%– 85.49%) and Aeromonas (10.02%–11.43%) in the gut bacteria of hybrid fish. Cetobacterium was the most common genus of Fusobacteria, and research indicated that Cetobacterium might aid in protein digestion as well as vitamin B12 production (Finegold et al., 2003). In addition, Cetobacterium was detected in the intestines of grass carp (Hao et al., 2017). Aeromonas is common in freshwater organisms. Aeromonads are called facultative pathogens and can infect fish, shrimp, reptiles, amphibians, and other aquatic organisms (Janda and Abbott, 2010). Herein, the relative richness of Cetobacterium and Aeromonas accounted for 82.29 – 98.27% of the total genus in the intestine of hybrid fish, implying that they played key roles in the protein digestion, vitamin production, and immunosuppression activities of hybrid fish intestines.
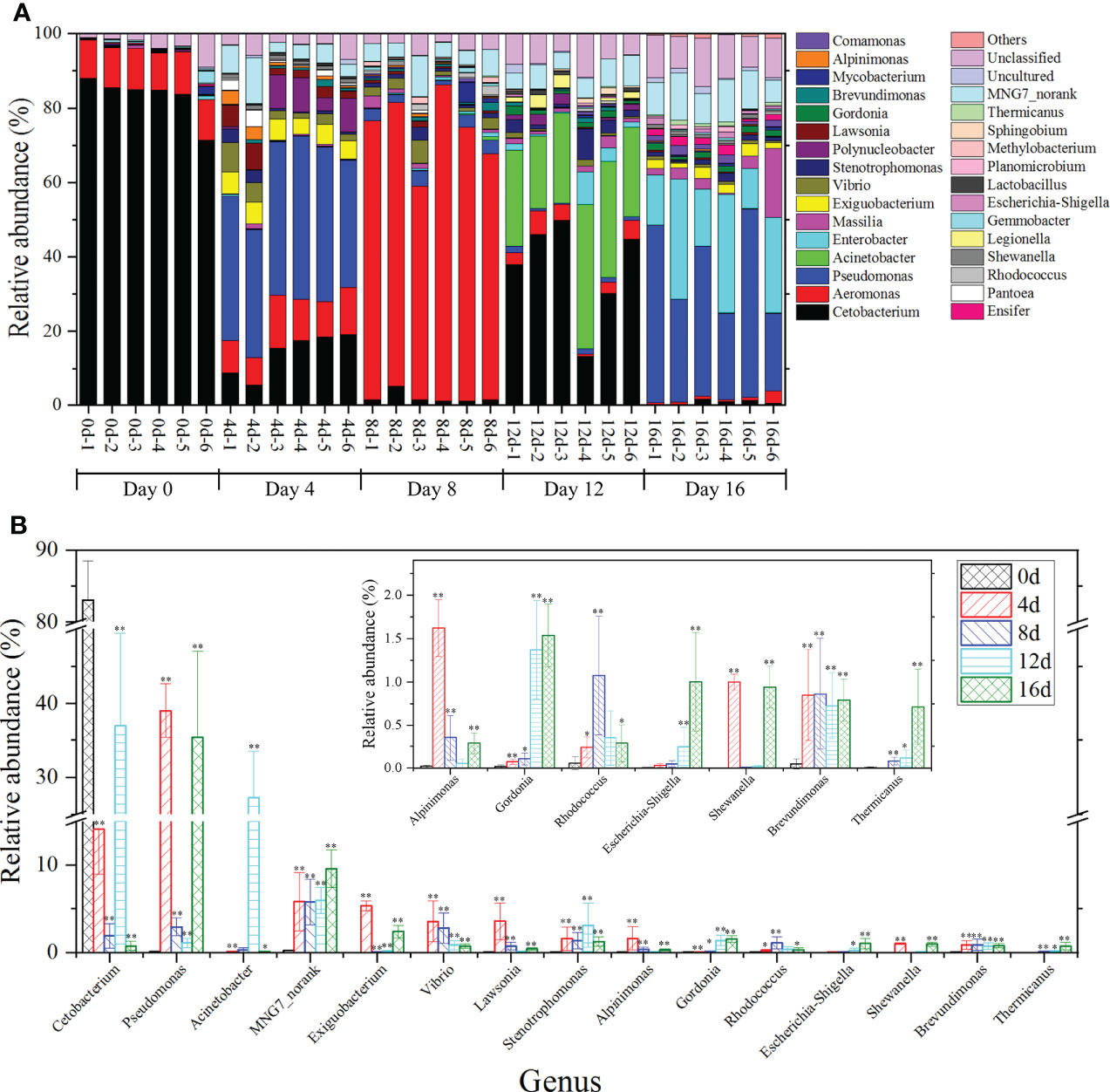
Figure 6 The influences of short-term exercise on microbial genera in this hybrid species. The relative richness of prominent bacterial genera in the gut of this hybrid species during swimming at diverse durations (A). Richness differences in microbial taxa among different durations of swimming exercise groups at the genus level (B). *P ≤ 0.05, **P ≤ 0.01. Data were provided as mean ± SD.
In short-term exercised groups, the most prevalent 15 genera with remarkable differences in relative richness were Cetobacterium, Pseudomonas, Acinetobacter, MNG7_norank, Exiguobacterium, Vibrio, Lawsonia, Stenotrophomonas, Alpinimonas, Gordonia, Rhodococcus, Escherichia-Shigella, Shewanella, Brevundimonas, Thermicanus (Figure 6B). The remarkable decrease of the abundances of Cetobacterium in exercised groups suggested that exercise might weaken protein digestion in the intestine of hybrid fish. Pseudomonas constitutes a metabolically diverse genus that thrives in soil and water, among other habitats (Valenzuela-Heredia et al., 2021). Frequently, members of this genus exhibit innate antimicrobial resistance (Silby et al., 2011). The Acinetobacter genus members are widespread microbes found in a variety of habitats. Certain species are environmental saprophytes and are seldom linked to illness (Turton et al., 2010). Acinetobacter is capable of degrading NAP and PHE (Jiang et al., 2018). Stenotrophomonas bacteria are plant endophytes or rhizosphere- and river sediment-linked microorganisms (Assis et al., 2017). Genus Alpinimonas is a member of the phylum Actinobacteria but its function remained unknown (Zhou et al., 2022). Reportedly, certain Rhodococcus species breakdown chitin (Sun et al., 2014). Shewanella worked as a denitrifier in Pheretima guillemi gut (Zhou et al., 2019). Brevundimonas, a rhizobacterial genus that promotes plant development, increases the yield and growth of plants such as rice, potato, and wheat (Li et al., 2021). The Pseudomonas, Acinetobacter, Stenotrophomonas, Alpinimonas, Rhodococcus, Shewanella, and Brevundimonas were significantly increased in this hybrid species after short-term swimming exercised speculated that exercise could improve intrinsic antimicrobial resistance, accelerate degradation of NAP, PHE, and chitin, as well as promote denitrification capacity. From the perspective of intestinal microbiota, our study provides insight into the underlying mechanisms by which exercise enhances immune function. In addition, the use of short-term exercise in aquaculture could be used to increase immune status and reduce economic losses caused by viral infections. Furthermore, the content of MNG7_norank and Gordonia gradually and significantly increased as the trigger time of swimming exercise extended, which suggested that they could be used as potential marker to differential exercised or non-exercised hybrid fish.
LEfSe analysis based on community abundance
Using the relative richness of OTUs as input data, 1209 bacterial genera could be separated into five different groups with a logarithmic linear discriminant analysis (LDA) score of at least 4.0 (LEfSe use LDA to quantify the impact size of abundance difference between each species). Fusobacteria; Proteobacteria; Proteobacteria and Actinobacteria; Firmicutes and Bacteroidetes; Proteobacteria and Actinobacteria, respectively were the bacterial lineages abundant in the 0d, 4d, 8d, 12d, and 16d groups, as shown in Figure 7.
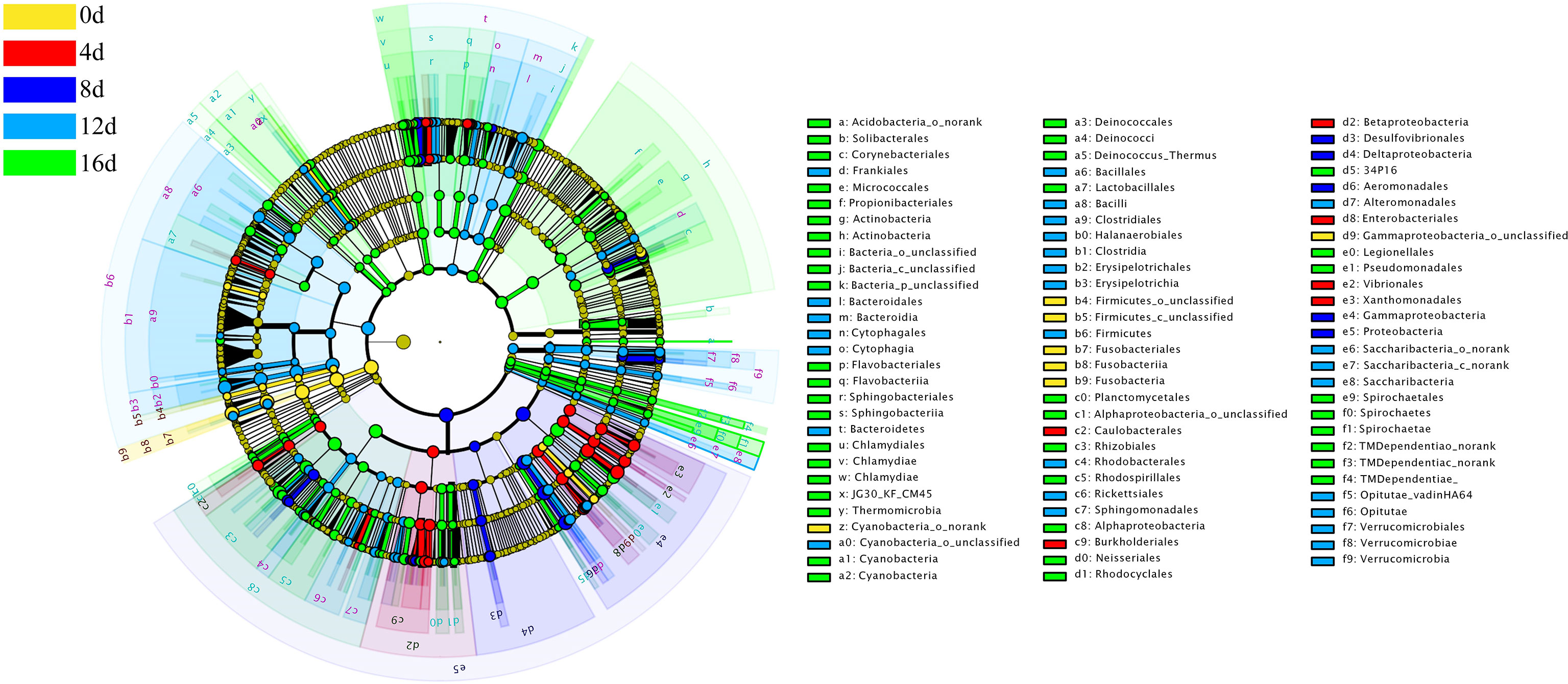
Figure 7 Cladogram illustrating the phylogenetic distribution of microflora communities linked to the different durations of swimming exercise groups. LEfSe determined lineages with linear discriminant analysis (LDA) values of at least 4.0. The color of the class with the highest frequency represents differences. Yellow represents non-exercised group, red represents exercised for 4d group, blue represents exercised for 8d group, blue-green represents exercised for 12d group, and green represents exercised for 16d group.
Among these bacterial communities, only the LDA values for Fusobacteriales, Fusobacteria, Fusobacteriia, Cetobacterium, and Fusobacteriaceae in 0d group were all higher than 4.0 (Figure 8). Besides, the LDA values for Pseudomonadaceae, Pseudomonas, Enterobacteriales, Enterobacteriaceae, Burkholderiales, Betaproteobacteria, Enterobacter, Vibrio, Vibrionaceae, Vibrionales, Xanthomonadaceae, Xanthomonadales, Stenotrophomonas, Oxalobacteraceae, Massilia, and Comamonadaceae in 4d group; Proteobacteria, Gammaproteobacteria, Aeromonadales, Aeromonadaceae, Aeromonas, and Nocardiaceae in 8d group; Firmicutes, Erysipelotrichia, Erysipelotrichales, Erysipelotrichaceae, Peptococcus, Firmicutes, Bacilli, Bacillales, Family_XII, Exiguobacterium, Bacteroidetes, Bacteroidia, Bacteroidales, and Porphyromonadaceae in 12d group; Pseudomonadales, Moraxellaceae, Acinetobacter, Alphaproteobacteria, Rhizobiales, MNG7, Actinobacteria, Actinobacteria, Corynebacteriales, Micrococcales, and Microbacteriaceae in 16d group were all greater than 4.0 (Figure 8). In addition, LDA values of these gut flora greater than 4.0 in exercised groups could be utilized as potential marker to differential exercised or non-exercised fish.
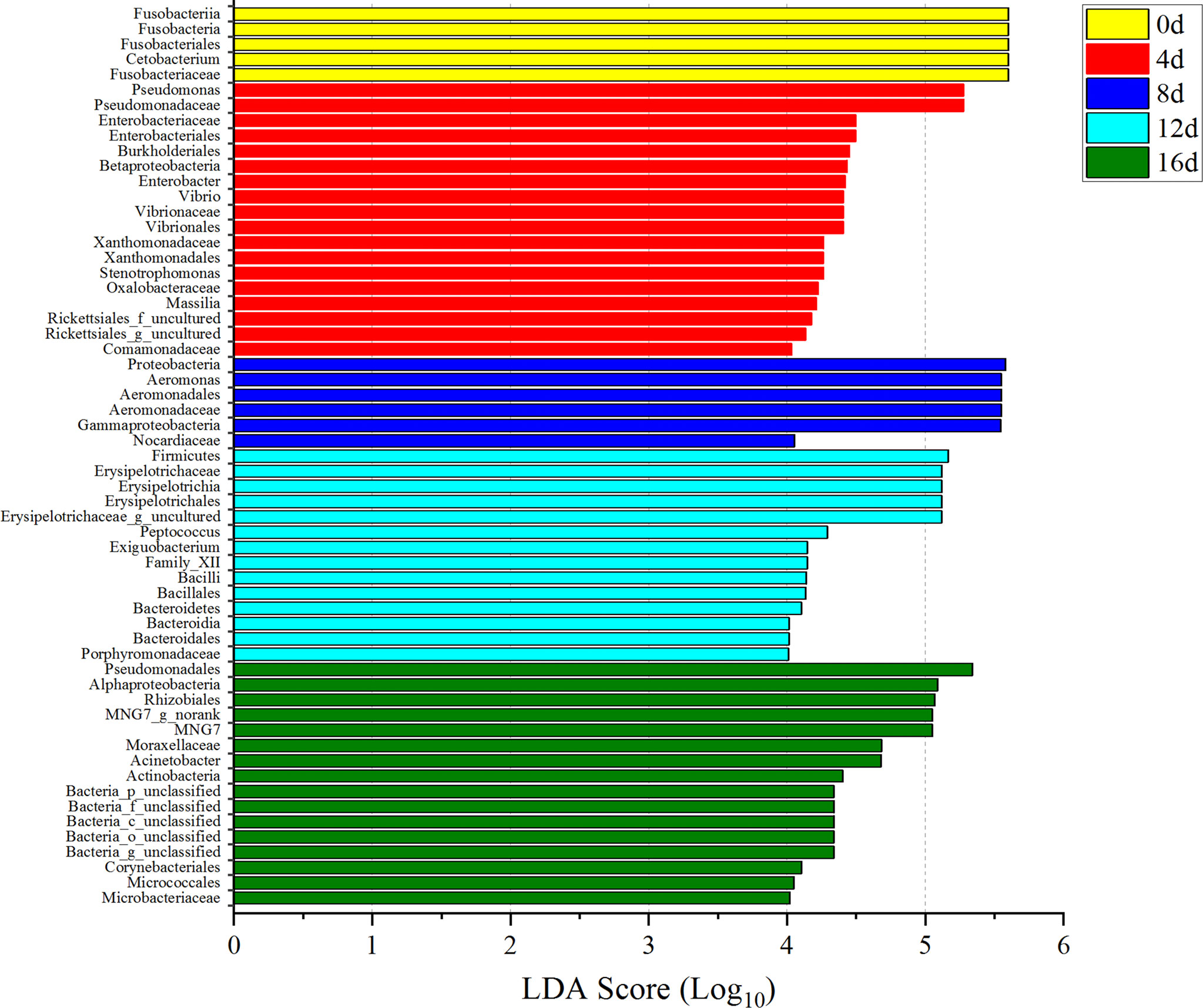
Figure 8 Microbial indicator groups in different durations of swimming exercise with LDA >4.0. Yellow represents non-exercised group, red represents exercised for 4d group, blue represents exercised for 8d group, blue-green represents exercised for 12d group, and green represents exercised for 16d group.
Previous research demonstrated that farmed fish have lower swimming ability when compared to their wild counterparts (Olla and Davis, 1989; Pedersen et al., 2008). The greater swimming performance of wild fish is attributable in part to their cumulative experiences in the outdoors, such as predator avoidance, eating behavior, and adaptation to local environmental changes (Johnsson et al., 1996; Nielsen et al., 2001; Petersson and Järvi, 2006). However, the corresponding molecular marker to identify wild or domestic fish is deficient. From the perspective of swimming exercise, the LDA values of these gut flora greater than 4.0 in exercised groups could also be used as potential marker to differential domestic or wild fish.
Conclusions
The findings are summarized in the Graphical Abstract. In brief, this research work has revealed that short-term culture of this hybrid species in moderate speed water for 12-16d could remarkably decrease muscle fiber diameter and increase muscle fiber density as well as improve its muscle texture (hardness, cohesiveness, gumminess, and chewiness), further improve the taste of fillets. In addition, 16S rRNA sequencing analysis indicated that short-term swimming exercise could significantly increase the community richness and diversity of intestinal microbiota in this hybrid species, therefore, swimming exercise potentially make it be healthier compared with non-exercised fish. Swimming exercise caused remarkable changes of phyla including Proteobacteria, Firmicutes, Bacteroidetes, and Fusobacteria. Besides, short-term swimming exercise might improve intrinsic antimicrobial resistance, accelerate degradation of NAP, PHE, and chitin, as well as promote denitrification capacity by affecting the richness of genera such as Pseudomonas, Acinetobacter, Stenotrophomonas, Alpinimonas, Rhodococcus, Shewanella, Brevundimonas, and so on. The work provides a thorough investigation of the temporal and dynamic impacts of short-term exercise on muscle texture and microflora in this hybrid species and further expanded our understanding of exercise-induced changes in fish.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Wuhan Academy of Agricultural Sciences.
Author contributions
HW: Methodology, Formal analysis, Investigation, Software, Data curation. LZ: Investigation, Data curation, Methodology, Writing – original draft preparation. QL: Investigation, Formal analysis, Data curation. YY: Investigation, Formal analysis, Data curation. YS: Validation. ZS and PL: Investigation, Data curation. TM: Software. JC: Methodology. LH: Resources, Project administration, and funding acquisition. GW: Supervision, Conceptualization, Writing – review and editing, funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Hubei Technology and Innovation (2018ABA105); Special Funds for the Ministry of Science and Technology ‘Science and Technology to help the economy 2020’; and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD74 and No. YFI202213).
Conflict of interest
Authors QL, YS, PL, JC, and GW are employed by Wuhan Xianfeng Aquaculture Technology Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.970858/full#supplementary-material
Supplementary Figure 1 | Species accumulation curves of this hybrid species intestinal microbial samples.
Supplementary Figure 2 | Rarefaction curves of each sample derived from Illumina MiSeq sequencing.
References
Adams C. E., Huntingford F. A., Krpal J., Jobling M., Burnett S. J. (1995). Exercise, agonistic behaviour and food acquisition in Arctic charr, salvelinus alpinus. Environ. Biol. Fishes 43 (2), 213–218. doi: 10.1007/BF00002494
Alami-Durante H., Wrutniak-Cabello C., Kaushik S. J., Médale F. (2010). Skeletal muscle cellularity and expression of myogenic regulatory factors and myosin heavy chains in rainbow trout (Oncorhynchus mykiss): Effects of changes in dietary plant protein sources and amino acid profiles. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 156 (4), 561–568. doi: 10.1016/j.cbpa.2010.04.015
Assis R., Polloni L. C., Patané J. S. L., Thakur S., Felestrino É.B., Diaz-Caballero J., et al. (2017). Identification and analysis of seven effector protein families with different adaptive and evolutionary histories in plant-associated members of the xanthomonadaceae. Sci. Rep. 7 (1), 16133–16133. doi: 10.1038/s41598-017-16325-1
Brown E. J., Bruce M., Pether S., Herbert N. A. (2011). Do swimming fish always grow fast? investigating the magnitude and physiological basis of exercise-induced growth in juvenile new Zealand yellowtail kingfish, seriola lalandi. Fish Physiol. Biochem. 37 (2), 327–336. doi: 10.1007/s10695-011-9500-5
Brugman S., Nieuwenhuis E. E. (2010). Mucosal control of the intestinal microbial community. J. Mol. Med. (Berl) 88 (9), 881–888. doi: 10.1007/s00109-010-0639-9
Bugeon J., Lefevre F., Fauconneau B. (2003). Fillet texture and muscle structure in brown trout (Salmo trutta) subjected to long-term exercise. Aquaculture Res. 34 (14), 1287–1295. doi: 10.1046/j.1365-2109.2003.00938.x
Burns A. R., Stephens W. Z., Stagaman K., Wong S., Rawls J. F., Guillemin K., et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. Isme J. 10 (3), 655–664. doi: 10.1038/ismej.2015.142
Cerf-Bensussan N., Gaboriau-Routhiau V. (2010). The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 10 (10), 735–744. doi: 10.1038/nri2850
Choi J. J., Eum S. Y., Rampersaud E., Daunert S., Abreu M. T., Toborek M. (2013). Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect. 121 (6), 725–730. doi: 10.1289/ehp.1306534
Clarke K. R., Gorley R. N., Somerfield P. J., Warwick R. M. (2014a). Change in marine communities: An approach to statistical analysis and interpretation.
Clarke S. F., Murphy E. F., O’Sullivan O., Lucey A. J., Humphreys M., Hogan A., et al. (2014b). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63 (12), 1913–1920. doi: 10.1136/gutjnl-2013-306541
Davison W., Goldspink G. (1978). The effect of training on the swimming muscles of the goldfish (Carassius auratus). J. Exp. Biol. 74 (1), 115–122. doi: 10.1242/jeb.74.1.115
Evans C. C., LePard K. J., Kwak J. W., Stancukas M. C., Laskowski S., Dougherty J., et al. (2014). Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One 9 (3), e92193. doi: 10.1371/journal.pone.0092193
Finegold S. M., Vaisanen M. L., Molitoris D. R., Tomzynski T. J., Song Y., Liu C., et al. (2003). Cetobacterium somerae sp. nov. from human feces and emended description of the genus cetobacterium. Syst. Appl. Microbiol. 26 (2), 177–181. doi: 10.1078/072320203322346010
Fouts D. E., Szpakowski S., Purushe J., Torralba M., Waterman R. C., MacNeil M. D., et al. (2012). Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 7 (11), e48289. doi: 10.1371/journal.pone.0048289
Friedman H. H., Whitney J. E., Szczesniak A. S. (1963). The texturometer–a new instrument for objective texture measurement. J. Food Sci. 28 (4), 390–396. doi: 10.1111/j.1365-2621.1963.tb00216.x
Hao Y. T., Wu S. G., Jakovlić I., Zou H., Li W. X., Wang G. T. (2017). Impacts of diet on hindgut microbiota and short-chain fatty acids in grass carp (Ctenopharyngodon idellus). Aquaculture Res. 48 (11), 5595–5605. doi: 10.1111/are.13381
Harimana Y., Tang X., Le G., Xing X., Zhang K., Sun Y., et al. (2018). Quality parameters of black carp (Mylopharyngodon piceus) raised in lotic and lentic freshwater systems. LWT 90, 45–52. doi: 10.1016/j.lwt.2017.11.060
Harimana Y., Tang X., Xu P., Xu G., Karangwa E., Zhang K., et al. (2019). Effect of long-term moderate exercise on muscle cellularity and texture, antioxidant activities, tissue composition, freshness indicators and flavor characteristics in largemouth bass (Micropterus salmoides). Aquaculture 510, 100–108. doi: 10.1016/j.aquaculture.2019.05.051
Hennersdorf P., Kleinertz S., Theisen S., Abdul-Aziz M. A., Mrotzek G., Palm H. W., et al. (2016). Microbial diversity and parasitic load in tropical fish of different environmental conditions. PLoS One 11 (3), e0151594. doi: 10.1371/journal.pone.0151594
Huang X., Hegazy A. M., Zhang X. (2021). Swimming exercise as potential measure to improve flesh quality of cultivable fish: A review. Aquaculture Res. 52 (12), 5978–5989. doi: 10.1111/are.15510
Ibarz A., Felip O., Fernández-Borràs J., Martín-Pérez M., Blasco J., Torrella J. R. (2011). Sustained swimming improves muscle growth and cellularity in gilthead sea bream. J. Comp. Physiol. B 181 (2), 209–217. doi: 10.1007/s00360-010-0516-4
Janda J. M., Abbott S. L. (2010). The genus aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23 (1), 35–73. doi: 10.1128/cmr.00039-09
Jiang Y., Zhang Z., Zhang X. (2018). Co-Biodegradation of pyrene and other PAHs by the bacterium acinetobacter johnsonii. Ecotoxicol Environ. Saf. 163, 465–470. doi: 10.1016/j.ecoenv.2018.07.065
Johnsson J. I., Petersson E., Jönsson E., Björnsson B. T., Järvi T. (1996). Domestication and growth hormone alter antipredator behaviour and growth patterns in juvenile brown trout, salmo trutta. Can. J. Fisheries Aquat. Sci. 53 (7), 1546–1554. doi: 10.1139/f96-090
Johnston I. A., Manthri S., Smart A., Campbell P., Nickell D., Alderson R. (2003). Plasticity of muscle fibre number in seawater stages of Atlantic salmon in response to photoperiod manipulation. J. Exp. Biol. 206 (Pt 19), 3425–3435. doi: 10.1242/jeb.00577
Kang S. S., Jeraldo P. R., Kurti A., Miller M. E., Cook M. D., Whitlock K., et al. (2014). Diet and exercise orthogonally alter the gut microbiome and reveal independent associations with anxiety and cognition. Mol. Neurodegener. 9, 36. doi: 10.1186/1750-1326-9-36
Khan J. R., Trembath C., Pether S., Bruce M., Walker S. P., Herbert N. A. (2014). Accommodating the cost of growth and swimming in fish–the applicability of exercise-induced growth to juvenile hapuku (Polyprion oxygeneios). Front. Physiol. 5. doi: 10.3389/fphys.2014.00448
Kim M., Kim W. S., Tripathi B. M., Adams J. (2014). Distinct bacterial communities dominate tropical and temperate zone leaf litter. Microb. Ecol. 67 (4), 837–848. doi: 10.1007/s00248-014-0380-y
Klase G., Lee S., Liang S., Kim J., Zo Y.-G., Lee J. (2019). The microbiome and antibiotic resistance in integrated fishfarm water: Implications of environmental public health. Sci. Total Environ. 649, 1491–1501. doi: 10.1016/j.scitotenv.2018.08.288
Larsson T., Mørkøre T., Kolstad K., Østbye T. K., Afanasyev S., Krasnov A. (2012). Gene expression profiling of soft and firm Atlantic salmon fillet. PLoS One 7 (6), e39219. doi: 10.1371/journal.pone.0039219
Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500 (7464), 541–546. doi: 10.1038/nature12506
Li Y., Li L., Yu Y., Hu Q., Li X. (2021). Impact of dietary protein content on soil bacterial and fungal communities in a rice-crab Co-culture system. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.696427
Liu H. Z., Zhu Y. R., Wang J. W., Tan D. Q. (2005). Population genetic structure of an endemic cyprinid fish, ancherythroculter nigrocauda, in the upper reaches of the Yangtze river and its implication for conservation. Genes Genomics 27 (4), 361–367. doi: 10.1007/s00109-005-0719-4
Li X.-M., Yuan J.-M., Fu S.-J., Zhang Y.-G. (2016). The effect of sustained swimming exercise on the growth performance, muscle cellularity and flesh quality of juvenile qingbo (Spinibarbus sinensis). Aquaculture 465, 287–295. doi: 10.1016/j.aquaculture.2016.09.021
Li X.-M., Yu L.-J., Wang C., Zeng L.-Q., Cao Z.-D., Fu S.-J., et al. (2013). The effect of aerobic exercise training on growth performance, digestive enzyme activities and postprandial metabolic response in juvenile qingbo (Spinibarbus sinensis). Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 166 (1), 8–16. doi: 10.1016/j.cbpa.2013.04.021
Llewellyn M. S., Boutin S., Hoseinifar S. H., Derome N. (2014). Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00207
Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K., Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489 (7415), 220–230. doi: 10.1038/nature11550
Mailing L. J., Allen J. M., Buford T. W., Fields C. J., Woods J. A. (2019). Exercise and the gut microbiome: A review of the evidence, potential mechanisms, and implications for human health. Exerc Sport Sci. Rev. 47 (2), 75–85. doi: 10.1249/jes.0000000000000183
Martin C., Johnston I. (2005). The role of myostatin and the calcineurin-signalling pathway in regulating muscle mass in response to exercise training in the rainbow trout Oncorhynchus mykiss walbaum. J. Exp. Biol. 208 (11), 2083–2090. doi: 10.1242/jeb.01605
Martin C., Johnston I. (2006). Endurance exercise training in common carp Cyprinus carpio l. induces proliferation of myonuclei in fast muscle fibres and slow muscle fibre hypertrophy. J. Fish Biol. 69 (4), 1221–1227. doi: 10.1111/j.1095-8649.2006.011411.4x
Martins Y. S., Melo R. M., Campos-Junior P. H., Santos J. C., Luz R. K., Rizzo E., et al. (2014). Salinity and temperature variations reflecting on cellular PCNA, IGF-I and II expressions, body growth and muscle cellularity of a freshwater fish larvae. Gen. Comp. Endocrinol. 202, 50–58. doi: 10.1016/j.ygcen.2014.03.047
Matsumoto M., Inoue R., Tsukahara T., Ushida K., Chiji H., Matsubara N., et al. (2008). Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72 (2), 572–576. doi: 10.1271/bbb.70474
McDonald R., Schreier H. J., Watts J. E. (2012). Phylogenetic analysis of microbial communities in different regions of the gastrointestinal tract in panaque nigrolineatus, a wood-eating fish. PLoS One 7 (10), e48018. doi: 10.1371/journal.pone.0048018
Miyake S., Ngugi D. K., Stingl U. (2015). Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 24 (3), 656–672. doi: 10.1111/mec.13050
Morgan X. C., Huttenhower C. (2012). Chapter 12: Human microbiome analysis. PLoS Comput. Biol. 8 (12), e1002808. doi: 10.1371/journal.pcbi.1002808
Nagalingam N. A., Lynch S. V. (2012). Role of the microbiota in inflammatory bowel diseases. Inflammation Bowel Dis. 18 (5), 968–984. doi: 10.1002/ibd.21866
Neish A. S. (2009). Microbes in gastrointestinal health and disease. Gastroenterology 136 (1), 65–80. doi: 10.1053/j.gastro.2008.10.080
Nielsen C., Holdensgaard G., Petersen H. C., Björnsson B. T., Madsen S. (2001). Genetic differences in physiology, growth hormone levels and migratory behaviour of Atlantic salmon smolts. J. Fish Biol. 59 (1), 28–44. doi: 10.1111/j.1095-8649.2001.tb02336.x
Nunes C., Moraes G., Fabrizzi F., Hackbarth A., Arbeláez-Rojas G. A. (2013). Growth and hematology of pacu subjected to sustained swimming and fed different protein levels. Pesquisa Agropecuária Bras. 48 (6), 645–650. doi: 10.1590/S0100-204X2013000600010
Olla B. L., Davis M. W. (1989). The role of learning and stress in predator avoidance of hatchery-reared coho salmon (Oncorhynchus kisutch) juveniles. Aquaculture 76 (3-4), 209–214. doi: 10.1016/0044-8486(89)90075-6
Palstra A. P., Mes D., Kusters K., Roques J. A., Flik G., Kloet K., et al. (2015). Forced sustained swimming exercise at optimal speed enhances growth of juvenile yellowtail kingfish (Seriola lalandi). Front. Physiol. 5. doi: 10.3389/fphys.2014.00506
Palstra A. P., Planas J. V. (2011). Fish under exercise. Fish Physiol. Biochem. 37 (2), 259–272. doi: 10.1007/s10695-011-9505-0
Palstra A. P., Tudorache C., Rovira M., Brittijn S. A., Burgerhout E., Van den Thillart G. E., et al. (2010). Establishing zebrafish as a novel exercise model: Swimming economy, swimming-enhanced growth and muscle growth marker gene expression. PLoS One 5 (12), e14483. doi: 10.1371/journal.pone.0014483
Pedersen L. F., Koed A., Malte H. (2008). Swimming performance of wild and F1-hatchery-reared Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) smolts. Ecol. Freshw. Fish 17 (3), 425–431. doi: 10.1111/j.1600-0633.2008.00293.x
Periago M. J., Ayala M. D., López-Albors O., Abdel I., Martínez C., García-Alcázar A., et al. (2005). Muscle cellularity and flesh quality of wild and farmed sea bass, dicentrarchus labrax l. Aquaculture 249 (1-4), 175–188. doi: 10.1016/j.aquaculture.2005.02.047
Petersson E., Järvi T. (2006). Anti-predator response in wild and sea-ranched brown trout and their crosses. Aquaculture 253 (1-4), 218–228. doi: 10.1016/j.aquaculture.2005.08.012
Rasmussen R. S. (2001). Quality of farmed salmonids with emphasis on proximate composition, yield and sensory characteristics. Aquaculture Res. 32 (10), 767–786. doi: 10.1046/j.1365-2109.2001.00617.x
Rasmussen R. S., Heinrich M. T., Hyldig G., Jacobsen C., Jokumsen A. (2011). Moderate exercise of rainbow trout induces only minor differences in fatty acid profile, texture, white muscle fibres and proximate chemical composition of fillets. Aquaculture 314 (1-4), 159–164. doi: 10.1016/j.aquaculture.2011.02.003
Rawat S. R., Männistö M. K., Bromberg Y., Häggblom M. M. (2012). Comparative genomic and physiological analysis provides insights into the role of acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol. Ecol. 82 (2), 341–355. doi: 10.1111/j.1574-6941.2012.01381.x
Rowlerson A., Mascarello F., Radaelli G., Veggetti A. (1995). Differentiation and growth of muscle in the fish sparus aurata (L): II. hyperplastic and hypertrophic growth of lateral muscle from hatching to adult. J. Muscle Res. Cell Motil. (San Diego: Academic Press) 16 (3), 223–236. doi: 10.1007/BF00121130
Rowlerson A., Veggetti A. (2001). “Cellular mechanisms of post-embryonic muscle growth in aquaculture species,” in Muscle development and growth (San Diego: Academic Press), 103–140.
Sánchez-Gurmaches J., Cruz-Garcia L., Ibarz A., Fernández-Borrás J., Blasco J., Gutiérrez J., et al. (2013). Insulin, IGF-I, and muscle MAPK pathway responses after sustained exercise and their contribution to growth and lipid metabolism regulation in gilthead sea bream. Domest. Anim. Endocrinol. 45 (3), 145–153. doi: 10.1016/j.domaniend.2013.08.001
Sänger A. M. (1992). Effects of training on axial muscle of two cyprinid species: Chondrostoma nasus (L.) and leuciscus cephalus (L.). J. Fish Biol. 40 (4), 637–646. doi: 10.1111/j.1095-8649.1992.tb02611.x
Scott K. P., Gratz S. W., Sheridan P. O., Flint H. J., Duncan S. H. (2013). The influence of diet on the gut microbiota. Pharmacol. Res. 69 (1), 52–60. doi: 10.1016/j.phrs.2012.10.020
Semova I., Carten J. D., Stombaugh J., Mackey L. C., Knight R., Farber S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12 (3), 277–288. doi: 10.1016/j.chom.2012.08.003
Shi X., Ng K. K., Li X.-R., Ng H. Y. (2015). Investigation of intertidal wetland sediment as a novel inoculation source for anaerobic saline wastewater treatment. Environ. Sci. Technol. 49 (10), 6231–6239. doi: 10.1021/acs.est.5b00546
Shrivastava J., Raskovic B., Blust R., De Boeck G. (2018). Exercise improves growth, alters physiological performance and gene expression in common carp (Cyprinus carpio). Comp. Biochem. Physiol. a-Molecular Integr. Physiol. 226, 38–48. doi: 10.1016/j.cbpa.2018.08.007
Shu D., He Y., Yue H., Wang Q. (2015). Microbial structures and community functions of anaerobic sludge in six full-scale wastewater treatment plants as revealed by 454 high-throughput pyrosequencing. Bioresource Technol. 186, 163–172. doi: 10.1016/j.biortech.2015.03.072
Silby M. W., Winstanley C., Godfrey S. A., Levy S. B., Jackson R. W. (2011). Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 35 (4), 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Sossai P. (2012). Butyric acid: What is the future for this old substance? Swiss Med. Weekly 142, w13596. doi: 10.4414/smw.2012.13596
Sullam K. E., Essinger S. D., Lozupone C. A., O’Connor M. P., Rosen G. L., Knight R., et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 21 (13), 3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x
Sun Y., Zhang J., Wu S., Wang S. (2014). Statistical optimization for production of chitin deacetylase from rhodococcus erythropolis HG05. Carbohydr. Polymers 102, 649–652. doi: 10.1016/j.carbpol.2013.11.010
Tang X., Xu G., Dai H., Xu P., Zhang C., Gu R. (2012). Differences in muscle cellularity and flesh quality between wild and farmed coilia nasus (Engraulidae). J. Sci. Food Agric. 92 (7), 1504–1510. doi: 10.1002/jsfa.4734
Totland G., Kryvi H., Jødestøl K., Christiansen E., Tangerås A., Slinde E. (1987). Growth and composition of the swimming muscle of adult Atlantic salmon (Salmo salar l.) during long-term sustained swimming. Aquaculture 66 (3-4), 299–313. doi: 10.1016/0044-8486(87)90115-3
Turton J. F., Shah J., Ozongwu C., Pike R. (2010). Incidence of acinetobacter species other than a. baumannii among clinical isolates of acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48 (4), 1445–1449. doi: 10.1128/jcm.02467-09
Valente L. M., Moutou K. A., Conceicao L. E., Engrola S., Fernandes J. M., Johnston I. A. (2013). What determines growth potential and juvenile quality of farmed fish species? Rev. Aquaculture 5, S168–S193. doi: 10.1111/raq.12020
Valente L., Rocha E., Gomes E., Silva M., Oliveira M., Monteiro R., et al. (1999). Growth dynamics of white and red muscle fibres in fast-and slow-growing strains of rainbow trout. J. Fish Biol. 55 (4), 675–691. doi: 10.1111/j.1095-8649.1999.tb00710.x
Valenzuela-Heredia D., Henríquez-Castillo C., Donoso R., Lavín P., Ringel M. T., Brüser T., et al. (2021). An unusual overrepresentation of genetic factors related to iron homeostasis in the genome of the fluorescent pseudomonas sp. ABC1. Microbial Biotechnol. 14 (3), 1060–1072. doi: 10.1111/1751-7915.13753
Veggetti A., Mascarello F., Scapolo P. A., Rowlerson A. (1990). Hyperplastic and hypertrophic growth of lateral muscle in dicentrarchus labrax (L.). Anat. Embryol. 182 (1), 1–10. doi: 10.1007/BF00187522
Viney M. E., Riley E. M. (2014). “From immunology to eco-immunology: More than a new name,” in Eco-immunology: Evolutive aspects and future perspectives. Eds. Malagoli D., Ottaviani E. (Dordrecht: Springer Netherlands), 1–19.
Weatherley A., Gill H., Lobo A. (1988). Recruitment and maximal diameter of axial muscle fibres in teleosts and their relationship to somatic growth and ultimate size. J. Fish Biol. 33 (6), 851–859. doi: 10.1111/j.1095-8649.1988.tb05532.x
Wong S., Rawls J. F. (2012). Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 21 (13), 3100–3102. doi: 10.1111/j.1365-294x.2012.05646.x
Yao J., Liu J., Zhang Y., Xu S., Hong Y., Chen Y. (2019). Adding an anaerobic step can rapidly inhibit sludge bulking in SBR reactor. Sci. Rep. 9 (1), 10843–10843. doi: 10.1038/s41598-019-47304-3
Ye L., Amberg J., Chapman D., Gaikowski M., Liu W. T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. Isme J. 8 (3), 541–551. doi: 10.1038/ismej.2013.181
Yi S., Gao Z.-X., Zhao H., Zeng C., Luo W., Chen B., et al. (2013). Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by solexa sequencing. BMC Genomics 14 (1), 1–12. doi: 10.1186/1471-2164-14-754
Zhang L., Jiang X., Liu X., Zhao X., Liu S., Li Y., et al. (2019). Growth, health, rumen fermentation, and bacterial community of Holstein calves fed lactobacillus rhamnosus GG during the preweaning stage1. J. Anim. Sci. 97 (6), 2598–2608. doi: 10.1093/jas/skz126
Zhang L., Li X., Yu Y., Zhang L., Dong L., Gan J., et al. (2021a). Comparative analyses of liver transcriptomes reveal the effect of exercise on growth-, glucose metabolism-, and oxygen transport-related genes and signaling pathways in grass carp (Ctenopharyngodon idella). Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol., 262, 111081. doi: 10.1016/j.cbpa.2021.111081
Zhang S.-Q., Xiong X.-M., Shi R.-H., Luo L.-F., Wang G.-Y., Tang D.-W., et al. (2022). Screening and characterization of sex-specific markers for the hybrid (Megalobrama amblycephala♀× Ancherythroculter nigrocauda♂) based on 2b-RAD and transcriptome sequencing. Aquaculture 548, 737704. doi: 10.1016/j.aquaculture.2021.737704
Zhang H. H., Xu M. R. X., Wang P. L., Zhu Z. G., Nie C. F., Xiong X. M., et al. (2020). High-quality genome assembly and transcriptome of ancherythroculter nigrocauda, an endemic Chinese cyprinid species. Mol. Ecol. Resour. 20 (4), 882–891. doi: 10.1111/1755-0998.13158
Zhang L., Yu Y., Dong L., Gan J., Mao T., Liu T., et al. (2021b). Effects of moderate exercise on hepatic amino acid and fatty acid composition, liver transcriptome, and intestinal microbiota in channel catfish (Ictalurus punctatus). Comp. Biochem. Physiol. Part D: Genomics Proteomics, 40, 100921. doi: 10.1016/j.cbd.2021.100921
Zhou J. S., Cheng J. F., Li X. D., Li Y. H. (2022). Unique bacterial communities associated with components of an artificial aquarium ecosystem and their possible contributions to nutrient cycling in this microecosystem. World J. Microbiol. Biotechnol. 38 (4), 72. doi: 10.1007/s11274-022-03258-9
Zhou G.-W., Yang X.-R., Sun A.-Q., Li H., Lassen S. B., Zheng B.-X., et al. (2019). Mobile incubator for iron (III) reduction in the gut of the soil-feeding earthworm pheretima guillelmi and interaction with denitrification. Environ. Sci. Technol. 53 (8), 4215–4223. doi: 10.1021/acs.est.8b06187
Keywords: hybrid (Megalobrama amblycephala×Ancherythroculter nigrocauda), Xianfeng No. 2, short-term exercise, muscle cellularity and texture, intestinal microbiota, 16S rRNA
Citation: Wei H, Zhang L, Li Q, Yu Y, Sun Y, Song Z, Li P, Mao T, Chen J, He L and Wang G (2022) Dynamic effects of short-term exercise on muscle texture and gut flora in the hybrid (Megalobrama amblycephala♀ × Ancherythroculter nigrocauda♂). Front. Mar. Sci. 9:970858. doi: 10.3389/fmars.2022.970858
Received: 17 June 2022; Accepted: 05 September 2022;
Published: 30 September 2022.
Edited by:
Filomena Fonseca, University of Algarve, PortugalReviewed by:
Yao Zheng, Freshwater Fisheries Research Center (CAFS), ChinaFangzhou Hu, Hunan Normal University, China
Han Liu, Huazhong Agricultural University, China
Copyright © 2022 Wei, Zhang, Li, Yu, Sun, Song, Li, Mao, Chen, He and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li He, aGVsaTI4QHNvaHUuY29t; Guiying Wang, NDY5OTI2NjQ4QHFxLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Huijie Wei1†
Huijie Wei1† Qing Li
Qing Li Li He
Li He