- 1Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2Hainan Provincial Key Laboratory of Tropical Marine Biology Technology, Sanya Institute of Ocean Eco-Environmental Engineering, Sanya, China
- 3Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Department of Science and Technology of Guangdong Province, Guangzhou, China
- 4Innovation Academy of South China Sea Ecology an Environmental Engineering, Chinese Academy of Sciences, Guangzhou, China
- 5Guangdong Key Laboratory of Animal Conservation and Resource Utilization, Guangdong Public Laboratory of Wild Animal Conservation and Utilization, Institute of Zoology, Guangdong Academy of Sciences, Guangzhou, China
Giant clams are conspicuous bivalves that inhabit in coral reefs. Among the giant clams, eight species of subfamily Tridacninae are the most common in the Asia-Pacific. However, very little is known about their evolutionary history. Here, we determined the complete mitochondria genome of Hippopus porcellanus, which was 29,434 bp in size and contained 13 protein-coding genes, 2 rRNAs and 23 tRNAs. The A+T composition of protein-coding regions was 57.99%, and the AT composition of the 3rd codon position was 59.33%, of which in agreement with the invertebrate bias favoring codons ending in A or T. Analysis of phylogenetic relationships according to the concatenated nucleotide data set containing 18S rRNA gene and 13 protein-coding genes, the phylogenetic relationship was analyzed by Maximum likelihood and Bayesian inference methods. The results showed that T. maxima was placed with the clade comprising T. noae, T. squamosa, and T. crocea, in which T. squamosa was highly similar to T. crocea and is consistent with the results of the previous studies using15 mitochondrial markers and nuclear 18S rRNA. Moreover, the inferred divergence time of Tridacnidae species is generally consistent with the fossil record of Tridacnidae. The divergence time of H. porcellanus and H. hippopus was about 10.64 Mya, this result is in agreement with the speculation that H. porcellanus also originated in Miocene. The availability of molecular phylogeny and divergence time estimation provides information genetic relationship of Tridacninae, which could be helpful to the ecological research and conservation of giant clams.
Introduction
Molecular phylogenetic analysis is an ideal method to study the relationship among species (Sigwart and Sutton, 2007; Smith et al., 2011; Kocot et al., 2011). Mitochondrial genomes are characterized by high evolution rate, conserved gene components (Curole and Kocher, 1999), rare recombination (Boore, 1999), and maternal inheritance (Barr et al., 2005). Now mitochondrial genomes have been extensively used for studying phylogenetic relationships at various taxonomic levels (Yamanoue et al., 2007; Yokobori et al., 2007).
In the animal kingdom, mollusca is the second most species-rich phylum, and there are great variations in their mitochondrial genomes. Six bivalve families (Veneridae, Mytilidae, Donacidae, Unionidae, Hyriidae and Margaritiferidae) are known to possess an unusual mtDNA inheritance mode (doubly uniparental inheritance) (Theologidis et al., 2008). Similar to mitochondrial tRNAs of nematode, tRNAs of some pulmonate gastropods also lacks T-stem or D-stem (Yamazaki et al., 1997). Unlike metazoans, the mitochondrial genomes of most molluscs, especially Scaphopoda and Bivalves, contain a large number of rearrangements (Serb and Lydeard, 2003). Several specific mitochondrial genomic features have also been found in the published mitochondria genomes of Tridacninae species. The mitogenome gene order of Tridacna squamosa is different from Fulvia mutica and Acanthocardia tuberculate, the other species of the Cardioidea (Gan et al., 2016). It is worth noting that the mitochondrial gene order of Tridacna crocea and Tridacna squamosa is different from that of other Tridacninae species (Tan et al., 2021).
Giant clams (Cardiidae: Tridacninae) are conspicuous bivalves that inhabit in coral reefs (Yonge, 1975). The adults of the smallest giant clam species, Tridacna crocea are about 15 cm long, while the largest giant clam species, Tridacna gigas, can grow longer than 1 m and weigh more than 300 kg (Rosewater, 1965). Giant clams not only play an important ecological role in coral reef ecosystems, but also become an important food source for coastal communities in Asia and the South Pacific, and make a significantly contribution to shell and aquarium trade (Keys and Healy, 1999). Overfishing is a major threat to giant clams and caused the extinction of a few larger giant clam species (Juinio et al., 1989). Giant clams are currently listed as vulnerable in the International Union for Conservation of Nature (IUCN) list. According to Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (Wells et al., 1983; Tisdell and Menz, 1992; Tisdell et al., 1994), the trade of giant clams is strictly regulated, in which only the trade of certified aquaculture products is allowed, and the trade of wildlife products is not allowed (Bell et al., 1997).
All eight species of Tridacninae giant clams [two Hippopus (H. porcellanus and H. hippopus) and six Tridacna (T. crocea, T. derasa, T. maxima, T. gigas, T. noae and T. squamosa)] are distributed along the Asia-Pacific coast (Neo et al., 2017; Zhang et al., 2020). These giant clams can be classified according to their adult size: large clams (T. derasa and T. gigas), medium clams (H. porcellanus, H. hippopus, and T. squamosa) and small clams (T. crocea, T. noae and T. maxima) (Zhou et al., 2020). Due to morphological similarity, T. noae was previously misidentified as a variant of T. maxima (McLean, 1947; Rosewater, 1965). However, recent genetic analysis found that T. maxima and T. noae are belong to different species (Su et al., 2014). Therefore, identification based on morphological characteristics sometimes caused misidentification. Recently, although molecular methods have resolved some confusion in the identification of giant clams (Schneider and Foighil, 1999; Su et al., 2014), very little is known about the evolutionary history of the Tridacninae species. Moreover, most available phylogenetic analyses were based on short DNA fragments or different genetic markers, yielding different results (Benzie and Willianm, 1998; Schneider and Foighil, 1999; Nuryanto et al., 2007; Herrera et al., 2015). Recently, although Tan et al. (2021) performed a comprehensive phylogenetic analysis of 12 giant clam species using mitochondrial genome and nuclear 18S rRNA data, this study did not deeply analyze the divergence time between Tridacninae species and lacked the mitochondrial genome of H. porcellanus.
In this study, we characterized the complete mitochondrial genome of H. porcellanus, including its structural characteristics and nucleotide composition. We also studied the gene order of giant clam mitochondrial and speculated the scenario of gene order rearrangement. Based on complete mitochondrial genomes, the divergence time among Tridacninae species was estimated using a relaxed molecular clock calibrated with fossil evidence. Moreover, the molecular phylogeny of giant clams in the Asia-Pacific was reconstructed by using mitochondrial genome and nuclear 18S rRNA gene.
Materials and methods
Sample collection and DNA extraction
Individual of H. porcellanus was sampled from the South China Sea. The total genomic DNA was extracted following a modified CTAB DNA extraction protocol (Attitalla, 2011), and then stored at -80°C.
Mitochondrial DNA sequencing and genome assembly
A total amount of 1μg DNA per sample was used as input material for the DNA library preparations. Sequencing library was generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer’s recommendations and index codes were added to the sample. Briefly, genomic DNA sample was fragmented by sonication to a size of 350 bp. The short fragments were then sent to the Total Genomics Solution (TGS) Institute in Shenzhen, China, and sequenced using the Illumina Novaseq 6000 sequencing system (Borgström et al., 2011).
Prior to assembly, raw reads were filtered by using NGS QC Tool Kit v2.3.3 (Patel and Jain, 2012) and 9.29GB clean data were obtained. The GC content of the clean data, Q20-value and Q30 value were 45.36%, 97.55%, and 92.95%, respectively, resulting in 232-fold depth of coverage of the mitochondrial genome, indicating that the quality of the mitochondrial genome sequencing and assembly results was very high. Complete circular assembly graph was checked and further extracted by visualization (e.g., Bandage) of the GFA graph files that were assembled from SPAdes v3.11.0 (Bankevich et al., 2012).
Genome annotation and sequence analysis
Based on the annotated H. hippopus mitochondrial genome with minor revisions, the exact boundary of each gene was determined. The annotation of protein-coding genes (PCGs) was conducted using MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) with invertebrate mitochondrial genetic code. tRNA scan-SE 1.21 was used to identify tRNA genes (Lowe and Eddy, 1997), employing cove only search mode and invertebrate mitochondrial genetic code. The graphical map of H. porcellanus mitogenome was drawn using the online software of mitochondrial visualization tool OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). The mitogenome sequences of H. porcellanus were submitted to GenBank (ON009026).
Ka_Ks calculator was used to estimate the ratio of nonsynonymous to synonymous substitution rates (Ka/Ks) of all 13 protein genes in eight giant clams (subfamily Tridacninae).
Phylogenetic analysis
Eight Tridacninae species mitochondrial genomes (including one obtained in this study) and ribosomal gene sequences were used for phylogenetic analysis (Table 1). Three Cardiidae species were used as outgroups: Acanthocardia tuberculata (NC_008452.1), Cerastoderma edule (NC_035728.1), Fulvia mutica (NC_022194.1). The aligned gene matrices were concatenated for phylogenetic reconstruction using Maximum Likelihood (ML) and Bayesian analysis.
Table 1 Overview of the Tridacninae species, including three outgroup species, examined in this study.
MAFFT algorithm was used for codon-based nucleotide alignment (Katoh and Standley, 2013). Then, under the GTR+R6+F nucleotide substitution model, the maximum likelihood (ML) method in IQ-TREE v1.6.12 was used to reconstruct the phylogenetic tree (Nguyen et al., 2015). A bootstrapping of 1000 replicates was used to support inferred ML tree. ML analyses of nuclear 18S rRNA genes and mitochondrial PCGs were performed separately using the same parameters as above to compare the concatenated-data phylogenetic tree with mitochondrial and nuclear trees. For Bayesian analysis, based on the Bayesian information criterion (BIC), jModelTest v2.1.10 was used to select the most appropriate alternative model for each gene partition (Darriba et al., 2012). A posterior support for phylogenetic relationships among taxa was obtained using MrBayes v3.2.6 (Ronquist et al., 2012). Subsequently, two independent Markov chain Monte Carlo (MCMC) runs were performed using two cold chains and two heated chains, with a total of 10,000,000 generations. Each run initiated with random tree, default priors and sampling tree every 1000 generations, and the first 25% was discarded. FigTree v1.4.2 was used to view the resulting phylogenetic trees (Rambaut, 2014).
Estimate of divergence time
A Bayesian analysis based on concatenation data (13 PCGs) was used to estimate the divergence time in Tridacninae using software BEAST v 1.7.5 (Drummond and Rambaut, 2007). We selected one node as the calibration point: 20.4-23.0 Mya was used as prior divergence time for Hippopus and Tridacna based on the fossil record, with a normal prior distribution, mean 21.7 and standard deviation 0.5 (Schneider and Ó Foighil, 1999). The general time-reversible model with discrete gamma distribution and invariant sites (GTR+G+I) was selected as the best-fit model of nucleotide substitution. The Bayesian analyses were performed using a GTR model with four gamma categories, a Yule process of speciation, and a strict clock model of rate as the tree priors, as well as other default parameters. The Markov Chain Monte Carlo (MCMC), sampling and effective sample size of each parameter were 100 million gene rations, every 10,000 generations and above 200, respectively. Posterior distributions for parameter estimates and likelihood scores to approximate convergence were visualized with the Tracer v1.7.1 (Rambaut et al., 2018). Visual inspection of traces within and across runs, as well as the effective sample sizes (EES) of each parameter (>200), allowed us to confirm that the analyses were adequately sampled. A maximum clade credibility (MCC) tree was chosen by TreeAnnotator v1.7.5 from the output of the MCMC run using the LogCombiner program after the removal of the initial trees (25%) as burn-in (Drummond et al., 2012). The MCC tree was visualized with the program FigTree v1.4.2.
Results and discussion
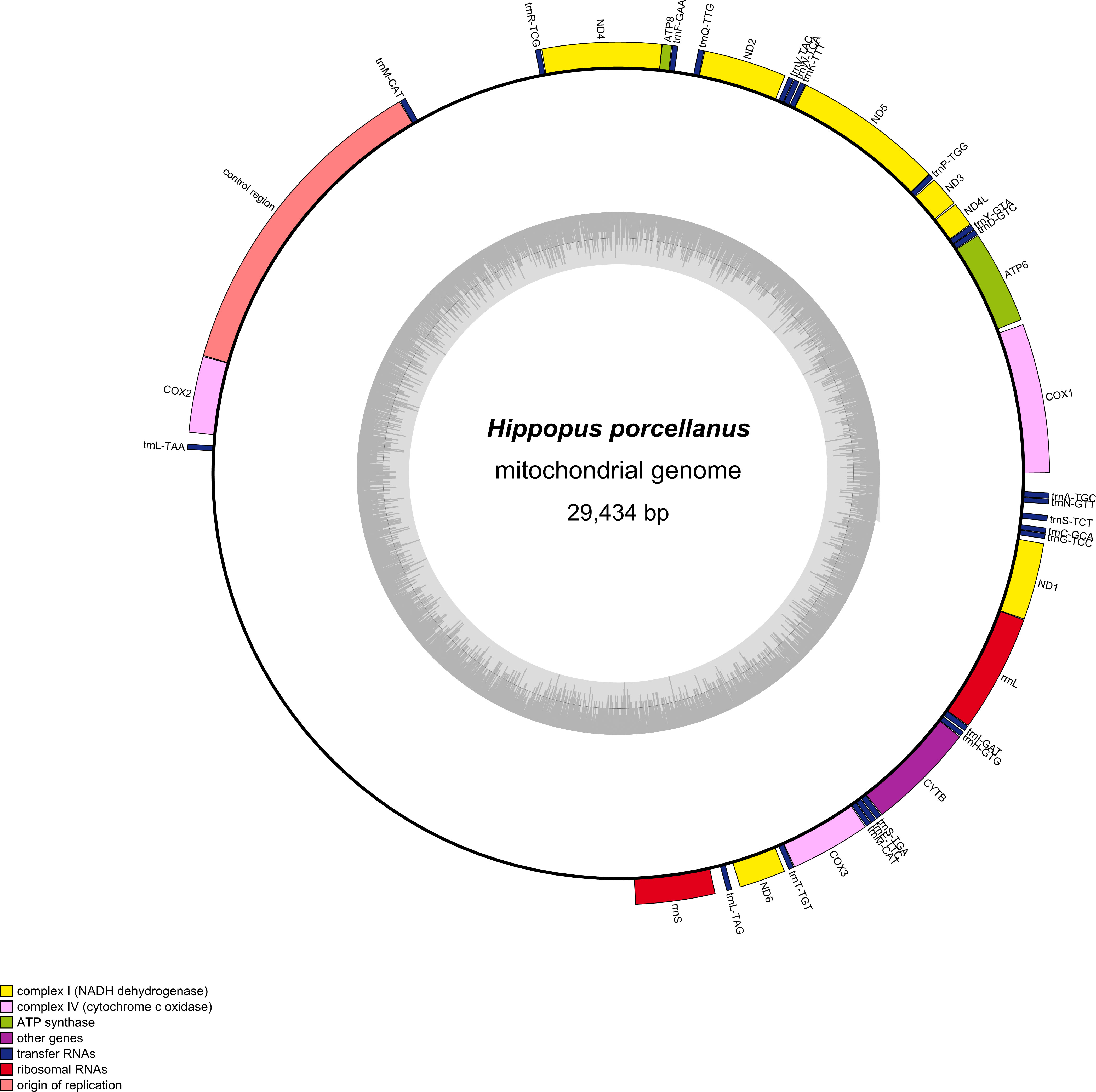
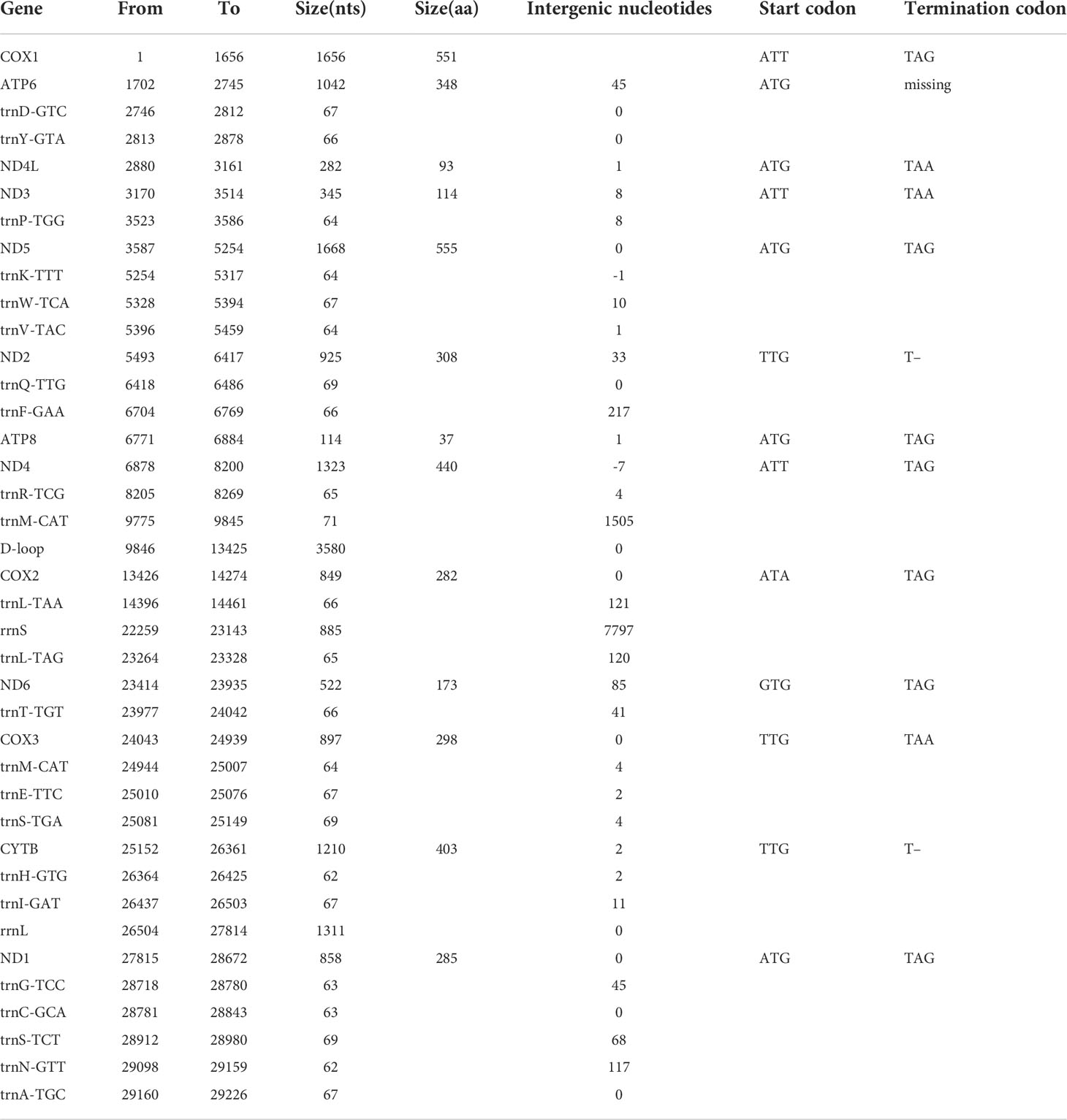
The complete mitochondrial genome size of H. porcellanus was 29,434 bp. The gene content of H. porcellanus was similar to that of other Tridacninae mitogenomes, including two mitochondrial rRNA genes (rrnL and rrnS), 13 PCGs (cox1-3, nad1-6, nad4L, cob, atp6 and atp8), 23 tRNA genes, and a major non-coding region called Control region (Figure 1). The lengths of genes (including rRNAs, tRNAs and PCGs) and intergenic nucleotides of H. porcellanus were 15,392 bp and 14,042 bp, respectively (Table 2). There were 3,887 codons excluding stop codons in the protein-coding genes of H. porcellanus. The length of the D-loop of the mitochondrial genome was 3580 bp. This region was difficult to assemble and annotate, and thus may not be precise. Moreover, we also found two long unannotated sections of the mitochondrial genome between trnM-CAT and trnR-TCG, trnL-TAA and rrnS, and their specific functions need further study. The A+T composition of the protein-coding region was 57.99%, and the AT composition of the third codon position elevated by 59.33%, which is in agreement with the tendency of typical invertebrates to prefer codons ending in A or T (Brown, 1985).
The genomes of Molluscs have unexpected/highly gene rearrangement (Ren et al., 2010). The gene order of Tridacninae species is presented and compared with that of three cardiidae species in Figure 2. Compared with cardiidae species, the mitochondrial genome of Tridacninae species has a higher rate of gene rearrangement. Gene rearrangements have also occurred within Tridacninae species. The gene order of genus Tridacna was different from that of Hippopus in the translocation of rrnS and trna L2, as well as some tRNA. Comparing the gene arrangement within the genus Tridacna, only T. gigas and T. derasa were completely identical to T. maxima. The gene arrangement of T. squamosa was similar to that of T. crocea, but their gene order was completely different from that of the other four giant clams by the translocation of atp8 and nad4, which is in agreement with the previous studies (Tan et al., 2021).
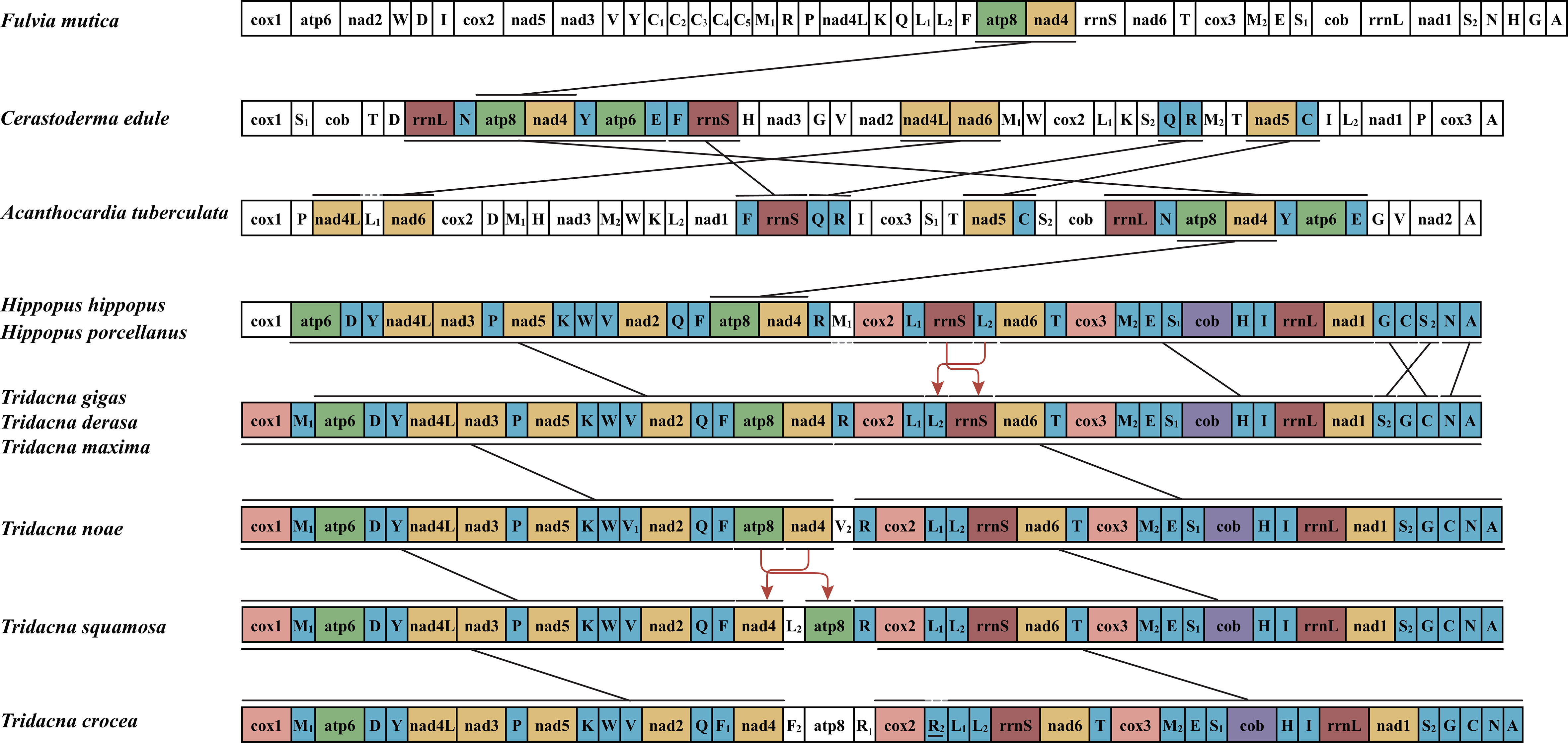
Figure 2 Mitochondrial genome rearrangements in Tridacninae and representatives from Veneroida species (All genes are transcribed from left-to-right. The bars show identical gene blocks. The arrows indicate gene translocation. The non-coding regions are not presented and gene segments are not drawn to scale).
The thirteen PCGs found in the mitochondrial DNA of most other animal (cob, nad1-nad6, cox1-cox3, nad4L, atp6 and atp8) were also determined in all giant clam genomes. All PCGs were encoded on and transcribed from the same strand. Atp8 deletion has been found in the mitochondrial genomes of bivalve molluscs (Gissi et al., 2008), but it was annotated in all giant clams in our study. Thus, the deleted atp8 gene is not unique characteristics of the mitochondrial genome of marine bivalves.
In order to understand the evolutionary dynamics of biological protein-coding sequences, it is important to estimate the nonsynonymous (Ka) and synonymous (Ks) substitution rates of closely related species (Ohta, 1995; Fay and Wu, 2003). The ratio of Ka and Ks is widely accepted as an indicator of the evolution rate of PCG sequence, where Ka/Ks>1, Ka/Ks<1 and Ka/Ks not significantly different from 1indicate positive selection, purifying selection and neutral evolution, respectively (Yang and Bielawski, 2000; Zhang et al., 2006). In this study, the Ka and Ks of eight Tridacninae species, were calculated and illustrated in Figure 3. The Ka/Ks ratios of 13 PCGs ranged from 0.0021 for cox2 in T. derasa _H. hippopus to 0.4101 for atp8 in T. gigas _T.derasa, suggesting the existence of purifying selection among genes. Most amino acid substitutions occur in the atp6, atp8, nad2 and nad6, indicating that the purification selection of these genes is relaxed compared with conservative genes such as cox1 and cob.
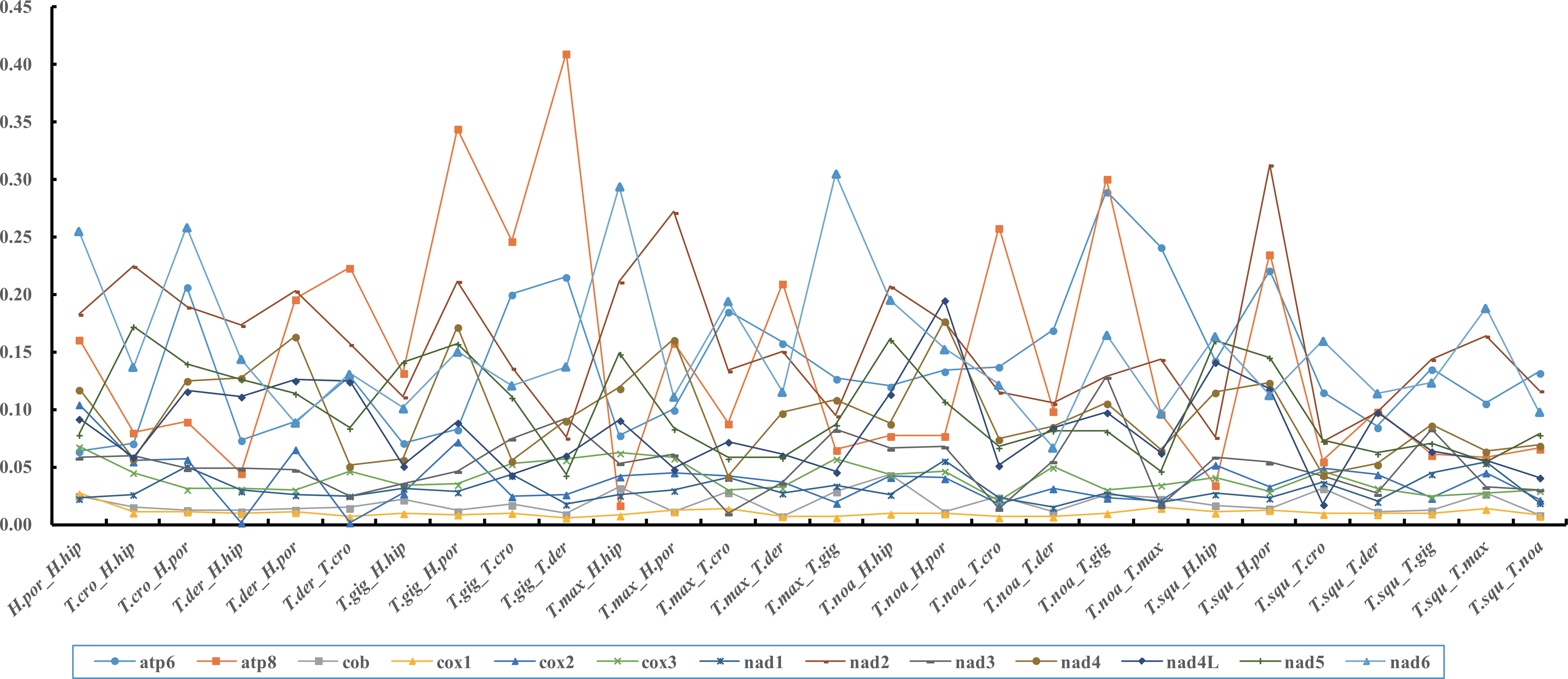
Figure 3 The ratio of nonsynonymous and synonymous substitutions (Ka/Ks) estimated in all 13 protein genes of eight giant clam species.
In this study, phylogenetic analyses were conducted based on a concatenated nucleotide data set containing 13 PCGs and nuclear 18S rRNA from 8 Tridacninae species and 3 Cardiidae species. The trees inferred from ML and BI methods have same topology, and all nodes have strong support (Figure 4). Based on the previous phylogenetic analysis that using different genetic markers, the relationships among Tridacninae (especially T.squamosa, T. crocea and T. maxima) was uncertain (Benzie and Williams, 1998; Maruyama et al., 1998; Schneider and Ó Foighil, 1999; Nuryanto et al., 2007; Herrera et al., 2015). Among them, the three recent phylogenies based on concatenated multi-gene and mitochondrial genome datasets were the most consistent with the present study (Huelsken et al., 2013; Fauvelot et al., 2020; Tan et al., 2021). In our study, we found that the obvious close relationship between T. squamosa and T. crocea is also agreed with the first tree of Maruyama et al. (1998). Reciprocal hybrids of T. squamosa and T. crocea have been successfully produced in the South China Sea, which also indicated that there is an obvious close relationship between them (Zhou et al., 2020). Based on genetic and morphological description, T. noae was a new species recently rediscovered from T. maxima (Su et al., 2014). This study found that T. maxima was placed with the clade comprising T. noae, T. squamosa, and T. crocea, in which T. squamosa was highly similar to T. crocea. The phylogenetic relationship of four giant clams was 100% supported.
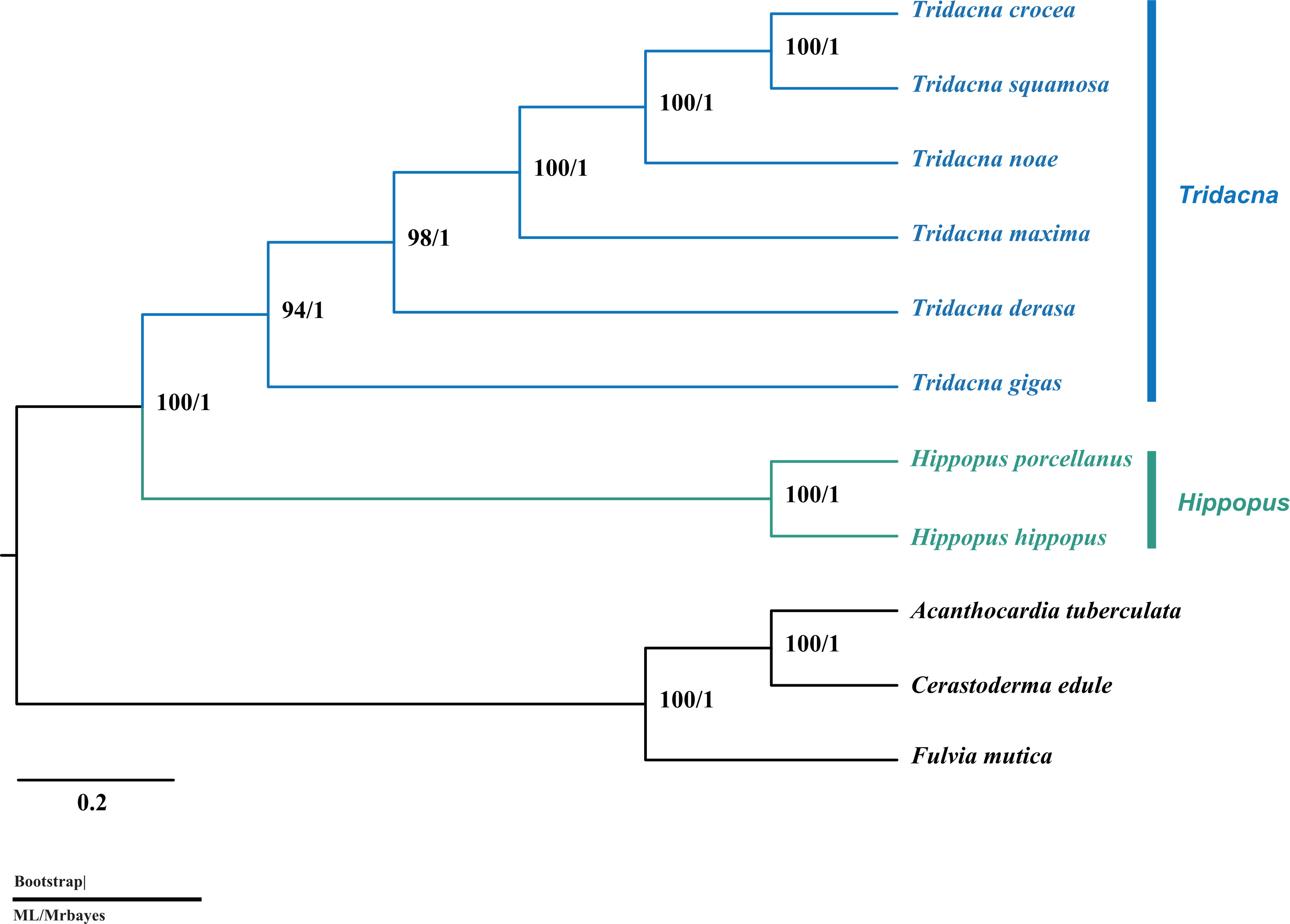
Figure 4 Consensus phylogenetic tree of giant clam species and outgroups from Maximum likelihood and Bayesian inference methods based on a concatenated nucleotide data set containing 13protein-coding genes and 18S rRNA gene.
In the fossil record, the divergence time of Hippopus and Tridacna is approximately 20.4-23.0 Mya ago (Schneider and Ó Foighil, 1999). The divergence times of Tridacnidae species estimated in this study is summarized in Figure 5. The results showed the same topology as the phylogenetic tree. Our molecular estimation showed that the genus Hippopus and the genus Tridacna diverged about 21.67 Mya (95% HPD: 20.71-22.69). The six Tridacna giant clams started their divergence about 18.08 Mya (95% HPD: 16.87-19.28) with the separation of T. gigas from other species first. T. derasa diverged from T. maxima, T. noae, T. squamosa and T. crocea 15.65 Mya (95% HPD: 14.77-16.54). T.maxima diverged from T. noae, T. squamosa and T. crocea 10.64 Mya (95% HPD: 9.89-11.47). T. noae diverged from T. crocea and T. squamosa 7.96 Mya (95% HPD: 7.43-8.57). T. crocea and T. squamosa have the closest relationship in our phylogenetic analysis, and the first split of their most recent common ancestors happened at about 5.01 Mya (95% HPD: 4.64-5.42). In the other clade, H. porcellanus and H. hippopus diverged about 10.64 Mya (95% HPD: 10.04-11.40).
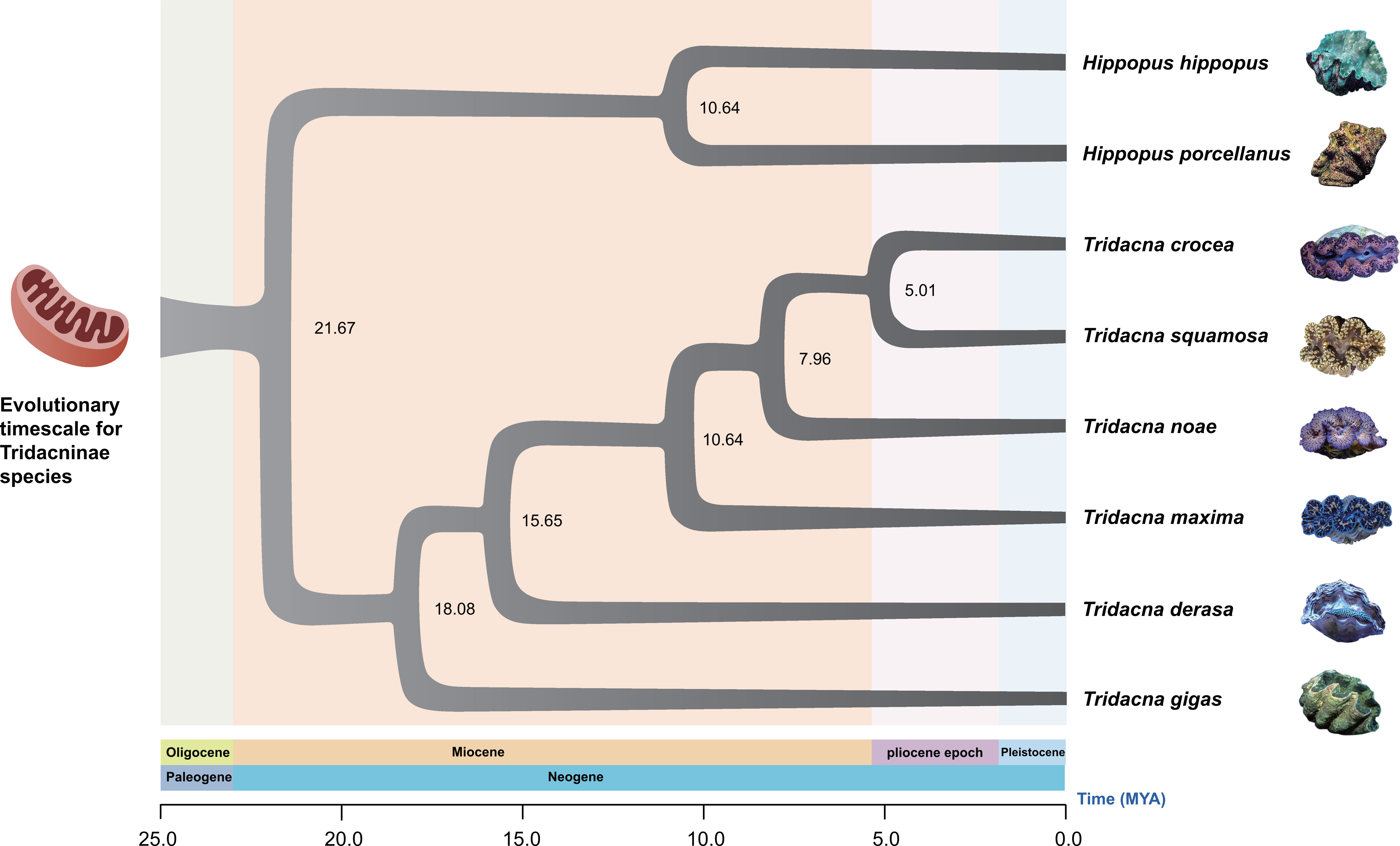
Figure 5 Evolutionary timescale for eight giant clam species inferred from a mitochondrial data set comprising 13 protein codons by mtDNA. Numbers at nodes indicate the mean estimated divergence times (in mya).
In this study, the inferred divergence time of Tridacnidae species is generally consistent with the fossil record of Tridacnidae (Harland et al., 1990; Schneider and Ó Foighil, 1999). Genus Hippopus and Tridacna are believed to evolve independently from Byssocardium-like ancestor in the early Miocene (Stasek, 1962), and Hippopus and Tridacna fossil record was first discovered in the Miocene (Stasek, 1962). The fossil records of T. maxima, T. derasa and T. gigas are in the Late Miocene (about 14 million years later) (Martin, 1880; Beets, 1986). The earliest fossil record of T. squamosa was in the Late Pliocene or Early Pleistocene (Nomura and Zinbo, 1935), while the earliest fossil record of T. crocea was in the Late Pleistocene or Holocene age (Rosewater, 1965). As for H. hippopus, the oldest known occurrence is in the Early Miocene (Cloud et al., 1956), but H. porcellanus lacks known fossil records. Since H. hippopus occurred in the Early Miocene (Cloud et al., 1956), it is reasonable to speculate that H. porcellanus also originated in the Miocene using genetic distance based on partial 16S rDNA gene sequences (Schneider and Foighil, 1999). The results of this study are in agreement with this speculation. In future studies, in order to better understand the phylogenetic relationship and replacement events, a more representative of Tridacnidae mitochondrial genomes should be used in the phylogenetic analysis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
YuZ and ZY conceived the study. HM and DY carried out the field and laboratory work, participated in the data analysis, and drafted the manuscript. JL and YQ collected the giant clam specimens. YaZ and ZX contributed to the phylogenetic analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Hainan Provincial Key R&D Programme (ZDYF2021XDNY183, ZDYF2021XDNY135), National Science Foundation of China (31702340), Guangdong Academy of Sciences (GDAS) Special Project of Science and Technology Development (2018GDASCX-0107), Guangdong Natural Science Foundation (2017A030310442), Science and Technology Planning Project of Guangdong Province, China (2017B030314052), National Marine Genetic Resource Center; China Agriculture Research System of MOF and MARA (CARS-49), and Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences (No.ISEE2018PY01; No.ISEE2018ZD02; ISEE2018PY03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Attitalla I. H. (2011). Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J. Biol. Sci. 14, 998–999. doi: 10.3923/pjbs.2011.998.999
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A.S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barr C. M., Neiman M., Taylor D. R. (2005). Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168, 39–50. doi: 10.1111/j.1469-8137.2005.01492.x
Beets C. (1986). Molluscan fauna of the lower gelingseh beds s.str., sangkulirang area, kalimantan timur (East Borneo). Scripta Geol. 82, 1–82.
Bell J. D., Lane I., Hart A. M. (1997). “Culture, handling and air transport of giant clams from the south pacific,” in Marketing and shipping live aquatic products, northeast region agricultural engineering service. Eds. Paust B., Peters J. B. (Northeast Region Agricultural Engineering Service, New York), 60–66.
Benzie J. A. H., Williams S. T. (1998). Phylogenetic relationships among giant clam species (Mollusca: Tridacnidae) determined by protein electrophoresis. Mar. Biol. 132, 123–133. doi: 10.1007/s002270050378
Boore J. L. (1999). Animal mitochondrial genomes, nucleic. Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Borgström E., Lundin S., Lundeberg J. (2011). Large Scale library generation for high throughput sequencing. PLoS One 6, e19119. doi: 10.1371/journal.pone.0019119
Brown W. M. (1985). “The mitochondrial genome of animals,” in Molecular evolutionary genetics. Ed. Macintyre R. J. (New York: Plenum Press), 95–130.
Cloud P. E. Jr., Schmidt R. G., Burke H. W. (1956). Geology of saipan and mariana islands. part 1. general geology. Geol. Surv. Prof. Paper, 280–A, 1–111. doi: 10.3133/pp280A
Curole J. P., Kocher T. D. (1999). Mitogenomics: digging deeper with complete mitochondrial genomes. Trends. Ecol. Evol. 14, 394–398. doi: 10.1016/S0169-5347(99)01660-2
Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and high-performance computing. Nat. Methods 9, 772. doi: 10.1038/nmeth.2109
Dreyer H., Steiner G. (2006). The complete sequences and geneorganisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica - and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front. Zool. 3, 13. doi: 10.1186/1742-9994-3-13
Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. doi: 10.1186/1471-2148-7-214
Drummond A. J., Suchard M. A., Xie D., Rambaut A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Fauvelot C., Zuccon D., Borsa P., Grulois D., Magalon H., Riquet F., et al. (2020). Phylogeographical patterns and a cryptic species provide new insights into Western Indian ocean giant clams phylogenetic relationships and colonization history. J. Biogeogr. 47, 1086–1105. doi: 10.1111/jbi.13797
Fay J. C., Wu C. I. (2003). Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genomics Hum. Genet. 4, 213–235. doi: 10.1146/annurev.genom.4.020303.162528
Gan H. M., Gan H. Y., Tan M. H., Penny S. S., Willan R. C., Austin C. M. (2016). The complete mitogenome of the giant clam Tridacna squamosa (Heterodonta: Bivalvia: Tridacnidae). Mitochondrial DNA A. 27, 3220–3221. doi: 10.3109/19401736.2015.1007355
Gissi C., Iannelli F., Pesole G. (2008). Evolution of the mitochondrial genome of metazoa as exemplified by comparison of congeneric species. Heredity 101, 301–320. doi: 10.1038/hdy.2008.62
Harland W. B., Armstrong R. L., Cox A. V., Craig L. E., Smith A. G., Smith D. G., et al. (1990). A geologic time scale 1989 (New York: Cambridge Univ. Press).
Herrera N. D., Ter Poorten J. J., Bieler R., Mikkelsen P. M., Strong E. E., Jablonski D., et al. (2015). Molecular phylogenetics and historical biogeography amid shifting continents in the cockles and giant clams (Bivalvia: Cardiidae). Mol. Phylogenet. Evol. 93, 94–106. doi: 10.1016/j.ympev.2015.07.013
Huelsken T., Keyse J., Liggins L., Penny S., Treml E. A., Riginos C., et al. (2013). A novel widespread cryptic species and phylogeographic patterns within several giant clam species (Cardiidae: Tridacna) from the indo-pacific ocean. PloS One 8, e80858. doi: 10.1371/journal.pone.0080858
Imanishi Y., Tanaka M., Fujiwara M. (2013). Complete mitochondrial genome sequence of Japanese cockle Fulvia mutica (Cardiidae). Fish. Sci. 79, 949–957. doi: 10.1007/s12562-013-0662-1
Juinio M. A. R., Meñez L. A. B., Villanoy C. L., Gomez E. D. (1989). Status of giant clam resources of the Philippines. J. mollusc. Stud. 55, 431–440. doi: 10.1093/mollus/55.4.431
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keys J. L., Healy J. M. (1999). Sperm ultrastructure of the giant clam Tridacna maxima (Tridacnidae: Bivalvia: Mollusca) from the great barrier reef. Mar. Biol. 135, 41–46. doi: 10.1007/s002270050599
Kocot K. M., Cannon J. T., Todt C., Citarella M. R., Kohn A. B., Meyer A., et al. (2011). Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456. doi: 10.1038/nature10382
Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic. Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Ma H. T., Zhang Y. H., Xiao S., Chen S. X., Zhang Y., Xiang Z. M., et al. (2019a). The complete mitochondrial genome of giant clam, Hippopus hippopus (Cardiidae: Tridacninae). Conserv. Genet. Resour. 11, 263–266. doi: 10.1007/s12686-018-1003-6
Ma H. T., Zhang Y. H., Zhang Y., Xiao S., Han C. H., Chen S. X., et al. (2019b). The complete mitochondrial genome of the giant clam, Tridacna maxima (Tridacnidae Tridacna). Mitochondrial DNA B. 4, 1051–1052. doi: 10.1080/23802359.2019.1584061
Ma H. T., Xiang Z. M., Zhang Y., Li J., Qin Y. P., Zhang Y. H., et al. (2020). First report of the mitochondrial DNA sequences of the giant clam, Tridacna gigas (Tridacnidae Tridacna). Mitochondrial DNA B. 5, 3614–3615. doi: 10.1080/23802359.2020.1830729
Ma H. T., Lin L., Zhang Y. H., Chen S. X., Shi W., Yu Z. N., et al. (2018). The complete mitochondrial genome sequence of the giant clam Tridacna derasa (Tridacnidae: Tridacna). Mitochondrial DNA B. 3, 913–914. doi: 10.1080/23802359.2018.1501290
Martin K. (1880). Die tertiarschichten auf Java, nach den entdeck-ungen von f. junghuhn (Leiden: Brill).
Maruyama T., Ishikura M., Yamazaki S., Kanai S. (1998). Molecular phylogeny of zooxanthellate bivalves. Biol. Bull. 195, 70–77. doi: 10.2307/1542777
McLean R. A. (1947). “A revision of the pelecypod family tridacnidae,” in Notulae naturae of the academy of natural sciences of philadelphia., 195,1–7.
Neo M. L., Wabnitz C. C., Braley R. D., Heslinga G. A., Fauvelot C. Wynsberge S.V., et al. (2017). Giant clams (Bivalvia: Cardiidae: Tridacninae): a comprehensive update of species and their distribution, current threats and conservation status. Oceanogr. Mar. Biol. 55, 87–388. doi: 10.1201/b21944-5
Nguyen L.-T., Schmidt H. A., Haeseler A. v., Minh B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nomura S., Zinbo N. (1935). Fossil and recent mollusca from the island of kita-Daito-Zima. Sci. Rep. Tohoku Univ. 18, 41–51.
Nuryanto A., Duryadi D., Soedharma D., Blohm D. (2007). Molecular phylogeny of giant clams based on mitochondrial DNA cytochrome c oxidase I gene. HAYATI J. Biosci. 14, 162–166. doi: 10.4308/hjb.14.4.162
Ohta T. (1995). Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J. Mol. Evol. 40, 56–63. doi: 10.1007/BF00166595
Patel R. K., Jain M. (2012). NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7, e30619. doi: 10.1371/journal.pone.0030619
Rambaut A. (2014) FigTree v1.4.2: Tree figure drawing tool. Available at: http://tree.bio.ed.ac.uk/software/figtree.
Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ren J. F., Liu X., Jiang F., Guo X. M., Liu B. (2010). Unusual conservation of mitochondrial gene order in crassostrea oysters: evidence for recent speciation in Asia. BMC Evol. Biol. 10, 394. doi: 10.1186/1471-2148-10-394
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rosewater J. (1965). The family tridacnidae in the indo-pacific. Indo-Pacific. Mollusca. 1, 347–396.
Schneider J. A., Foighil D.Ó. (1999). Phylogeny of giant clams (Cardiidae: Tridacnidae) based on partial mitochondrial 16S rDNA gene sequences. Mol. Phylogenet. Evol. 13, 59–66. doi: 10.1006/mpev.1999.0636
Serb J. M., Lydeard C. (2003). Complete mtDNA sequence of the north American freshwater mussel, lampsilisornata (Unionidae): An examination of the evolution and phylogenetic utility of mitochondrial genome organization in bivalvia (Mollusca). Mol. Biol. Evol. 20, 1854–1866. doi: 10.1093/molbev/msg218
Sigwart J. D., Sutton M. D. (2007). Deep molluscan phylogeny: synthesis of palaeontological and neontological data. Proc. R. Soc B Biol. Sci. 274, 2413–2419. doi: 10.1098/rspb.2007.0701
Smith S. A., Wilson N. G., Goetz F. E., Feehery C., Andrade S. C.S., Rouse G. W., et al. (2011). Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367. doi: 10.1038/nature10526
Stasek C. R. (1962). The form, growth and evolution of the tridacnidae (gaint clams). Arch. ZoologieExpérimentale Générale. 101, 1–40.
Su Y., Hung J. H., Kubo H., Liu L. L. (2014). Tridacna noae (Röding 1798) – a valid giant clam species separated from t. maxima (Röding 1798) by morphological and genetic data. Raffles Bull. Zool. 62, 124–135.
Tan E. Y. W., Randolph Q. Z. B., Neo M. L., Fauvelot C., Huang D. W. (2021). Genome skimming resolves the giant clam (Bivalvia: Cardiidae: Tridacninae) tree of life. Coral Reefs. 2021, 1–14. doi: 10.1007/s00338-020-02039-w
Theologidis I., Fodelianakis S., Gaspar M. B., Zouros E. (2008). Doubly uniparental inheritance (DUI) of mitochondrial DNA in donaxtrunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in bivalvia. Evolution 62, 959–970. doi: 10.1111/j.1558-5646.2008.00329.x
Tisdell C., Menz K. M. (1992). “Giant clam farming and sustainable development: an overview,” in Giant clams in the sustainable development of the south pacific: Socioeconomic issues in mariculture and conservation, vol. 18 . Ed. Tisdell C. (ACIAR Monograph, Canberra, Australia), 18, 3–14.
Tisdell C., Shang Y. C., Leung P. S., et al. (1994). “Development of giant clam mariculture and the need for a review of commercial prospects,” in Economics of commercial giant clam mariculture, vol. 25 . Eds. Tisdell C., Shang Y. C., Leung P. S. (ACIAR Monograph, Canberra, Australia), 25, 11–17.
Wells S. M., Pyle R. M., Collins N. M. (1983). “Giant clams,” in The IUCN invertebrate red data book (Gland, Switzerland), 97–107.
Yamanoue Y., Miya M., Matsuura K., Yagishita N., Mabuchi K., Sakai H., et al. (2007). Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol. Phylogenet. Evol. 45, 89–101. doi: 10.1016/j.ympev.2007.03.008
Yamazaki N., Ueshima R., Terrett J. A., Yokobori S., Kaifu M., Segawa R., et al. (1997). Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organizations of euhadra, cepaea and albinaria and implications of unusual tRNA secondary structures. Genetics 145, 749–758. doi: 10.1093/genetics/145.3.749
Yang Z. H., Bielawski J. P. (2000). Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503. doi: 10.1016/S0169-5347(00)01994-7
Yokobori S., Lindsay D. J., Yushida M., Tsuchiya K., Yamagishi A., Maruyama T., et al. (2007). Mitochondrial genome structure and evolution in the living fossil vampire squid, Vampyroteuthis infernalis, and extant cephalopods. Mol. Phylogenet. Evol. 44, 898–910. doi: 10.1016/j.ympev.2007.05.009
Zhang Z., Li J., Zhao X. Q., Wang J., Wong G. K., Yu J. (2006). KaKs_Calculator: calculating ka and ks through model selection and model averaging. Genomics Proteomics Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhang Y. H., Zhou Z. H., Qin Y. P., Li X. Y., Ma H. T., Wei J. K., et al. (2020). Phenotypic traits of two boring giant clam (Tridacna crocea) populations and their reciprocal hybrids in the south China Sea. Aquaculture 519, 734890. doi: 10.1016/j.aquaculture.2019.734890
Keywords: Tridacninae, mitochondrial genome, 18S, gene arrangement, phylogeny, molecular clock
Citation: Ma H, Yu D, Li J, Qin Y, Zhang Y, Xiang Z, Zhang Y and Yu Z (2022) Molecular phylogeny and divergence time estimates for native giant clams (Cardiidae: Tridacninae) in the Asia-Pacific: Evidence from mitochondrial genomes and nuclear 18S rRNA genes. Front. Mar. Sci. 9:964202. doi: 10.3389/fmars.2022.964202
Received: 08 June 2022; Accepted: 03 October 2022;
Published: 21 October 2022.
Edited by:
Danwei Huang, National University of Singapore, SingaporeReviewed by:
Yafei Mao, Shanghai Jiao Tong University, ChinaPaolo Marra-Biggs, University of Hawaii at Manoa, United States
Copyright © 2022 Ma, Yu, Li, Qin, Zhang, Xiang, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuehuan Zhang, eWh6aGFuZ0BzY3Npby5hYy5jbg==; Ziniu Yu, Y2FybHp5dUBzY3Npby5hYy5jbg==
†These authors have contributed equally to this work
 Haitao Ma
Haitao Ma Dongmei Yu
Dongmei Yu Jun Li
Jun Li Yanping Qin
Yanping Qin Yang Zhang
Yang Zhang Zhiming Xiang
Zhiming Xiang Yuehuan Zhang
Yuehuan Zhang Ziniu Yu
Ziniu Yu