
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 03 August 2022
Sec. Marine Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.962534
 Umar Farouk Mustapha
Umar Farouk Mustapha You-Xing Peng
You-Xing Peng Yuan-Qing Huang
Yuan-Qing Huang Daniel Assan
Daniel Assan Fei Zhi
Fei Zhi Gang Shi
Gang Shi Yang Huang
Yang Huang Guang-Li Li
Guang-Li Li Dong-Neng Jiang*
Dong-Neng Jiang*The reproductive-related studies, including genetic and genomic such as gonadal transcriptome analyses, have previously focused on the adult spotted scat, with little information on juvenile fish. Transcriptomics is a powerful tool that allows for massive parallel analysis to identify differential expression and the patterns of gene expression holistically at a particular stage in a cell or tissue development. This study presents the first report on gonadal transcriptome analysis of the differentiating (juvenile; 4 months after hatch; stage I ovary and testis) spotted scat gonads. The study revealed potential reproduction and gonadal development-related genes. A total of 25936 genes were identified, of which 16248 were co-expressed, 17893 and 17258 expressed in males and females, respectively, from sequence data of testis I (n = 3) and ovary I (n = 2). A total of 6549 differentially expressed genes (DEGs) were identified between males and females. Genes attributable to male gonad development pathway such as dmrt1, gsdf, and amh are significantly expressed in differentiating testes, while female-related genes such as foxl2, cyp19a1a, 42sp50 and sox3 were expressed considerably in differentiating ovaries. In addition, dmrt1/dmrt1y was not expressed in the female (FPKM=0.00), while its paralog dmrt1b was expressed in both males and females. In the male pathway, dmrt1y and gsdf are critical for sex determination and maintenance while foxl2/foxl3 and cyp19a1a are critical in the female development pathway. The current studies provide an insight into the expression patterns of sex and gonadal-related genes in differentiating gonads of spotted scat.
The spotted scat Scatophagus argus is an emerging aquaculture species with a known candidate sex determination gene and efficient molecular markers for sex identification (Barry and Fast, 1992; Biswas et al., 2016; Mustapha et al., 2018). Spotted scat exhibit XY sex-determination system (XY; males, XX; females) (Mustapha et al., 2018; Huang et al., 2019). The fish is known for its aesthetic and food value and is mainly distributed in the Indo Pacific and Southeast Asia waters with resistance to broad environmental conditions (Carcasson, 1977; Fast et al., 1988; Gandhi, 2002; Gandhi et al., 2013; Biswas et al., 2016). Additionally, species exhibiting traits such as fast growth, sexual dimorphism, and disease resistance are essential for aquaculturists (Ye et al., 2017). The traits in animals attributable to sexual dimorphism have dominant roles in behavior and evolution. The spotted scat exhibits sexual growth dimorphism, with females growing over 15% larger than males in a year (Mustapha et al., 2021).
Gonadal sex determination, differentiation, and sexual development-related genes have recently gained interest in developmental biology. The mechanisms of sex determination in vertebrates are categorized primarily into two (thus, genetic sex determination (GSD) and environmental sex determination (ESD)) (Marshall Graves, 2008). Unlike ESD, the primary sex in GSD is regulated by genetic heritability and determined during fertilization (Budd et al., 2015). The development process of sex-related traits after sex determination mainly involves much more complex gene interactions, and sex differentiation genes primarily play critical roles in these interactions. Gonadal sex differentiation in vertebrates involves the expression of sexually dimorphic genes, which leads to testicular or ovarian development and maintenance (Ayers et al., 2015). Nevertheless, many key genes related to gonadal differentiation have been identified in different species (Cutting et al., 2013; Wang et al., 2018; Mustapha et al., 2021). Meanwhile, the expression of sex-related genes begins to show differential expression during developmental processes, their expressions are mostly not constant and fluctuate during developmental stages (Jiang et al., 2020; Mustapha et al., 2021). Therefore, identifying novel testis and ovarian differentiation regulators is critical to increasing our understanding of gonad sexual differentiation. Similarly, identifying and understanding genes in the gonadal sex differentiation pathways and their expression pattern during developmental stages will be invaluable in sex differentiation and sex control studies.
In recent years, transcriptomic analysis has proven to be a powerful tool to identify potential genes and their expression patterns in tissues and cells at a specific time point. The transcriptomic analysis is a sensitive, tractable, and reliable “omic” technique and has been helpful in developmental studies and other biological analyses (Shi et al., 2013). Numerous studies have capitalized on this powerful tool to analyze the holistic expression patterns in gonads. For instance, the gonad transcriptome of Nile tilapia, Oreochromis niloticus (Tao et al., 2013; Sun et al., 2018; Teng et al., 2020; Lin et al., 2021a), adult spotted scat (He et al., 2019), Chinese tongue sole, Cynoglossus semilaevis (Wang et al., 2019), silver sillago, Sillago sihama (Tian et al., 2019), Hong Kong catfish, Clarias fuscus (Lin et al., 2021b), spot-fin porcupine fish, Diodon hystrix (Chen et al., 2021) and others (Santos et al., 2007; Díaz et al., 2014; Chen et al., 2021; Kohli et al., 2021; Yao et al., 2021; Zhu et al., 2022), revealed numerous differential and male and female pathway genes. As indicated above, previous studies identified several sex-related genes in adult spotted scat by gonadal transcriptomics (He et al., 2019). Meanwhile, a snapshot of gene expression patterns at early gonad development stages remains unclear. This could be due to the difficulty obtaining gonads of juvenile spotted scat and the unavailability of sex-linked markers. Excitedly, recent studies have successfully developed efficient sex-linked markers from Dmrt1 and Dmrt3 (Mustapha et al., 2018; Huang et al., 2022). This allowed for sex typing and distinguishing males from females for subsequent gonad excising.
The current study employed the first transcriptome analysis on the differentiating spotted scat gonads (juvenile fish). Subsequently, comparative transcriptomics revealed the expression patterns of gonadal sex-biased genes and differentially expressed genes (DEGs) between female and male gonads. The Gene Ontology (GO) term and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were functionally analyzed for DEGs and compared with the previous report where necessary. This study provides insight into the expression patterns of sex-related genes during gonad differentiation.
Four months old spotted scat, Scatophagus argus herein referred to as juvenile fish, were used for this study. The spotted scat is a flat fish with extremely small gonads which is difficult to excise at the early stage. However, 3.5 – 4 month is the earliest time we could excise the gonad from spotted which is also at the differentiating stage (Mustapha et al., 2021). The fish were obtained and screened into males and females using sex-linked markers as reported in (Mustapha et al., 2018). The sex-linked markers used were developed on dmrt1 and dmrt3 transcripts. The marker on dmrt3 amplified SNP regions with homozygous CC GG genotypes in females and heterozygous CG GA genotypes in the males. The markers on dmrt1 can be applied in a straightforward manner using DNA extracted with a rapid DNA extraction kit effectively and efficiently (Huang et al., 2022). Gonads were excised for RNA-seq analysis and histology. Similarly, some male and female gonads were selected for histological studies, as detailed previously (Mustapha et al., 2021). All experimental fish used in this study were produced and reared by our research group in Donghai Island experimental base. The experimental procedure was in accordance with the approved protocol of the Animal Research and Ethics Committee of Guangdong Ocean University, China (201903004).
Due to the small nature of the gonads, two to three gonads of same-sex were mixed to obtain enough RNA for subsequent analysis in triplicate. Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer ‘s instructions. The RNA integrity was assessed using RNA Nano 6000 Assay Kit with Bioanalyzer 2100 system (Agilent Technologies, CA, USA). For library preparation, total RNA was used in sample preparations. mRNA purification was carried out using poly-T oligo-attached with magnetic beads from the total RNA. Fragmentation was achieved under elevated temperature using divalent cations in First-Strand Synthesis Reaction Buffer(5X). Subsequently, random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-) were used to synthesize the first-strand cDNA, and DNA Polymerase I and RNase H were used for the second strand cDNA synthesis. The residual overhangs were then converted into blunt ends through exonuclease/polymerase activities. Adenylation of 3’ ends DNA fragments were carried out, and those adapters with hairpin loop structures ligated for hybridization. Furthermore, to obtain cDNA fragments with appropriate length (370 - 420 bp), the AMPure XP system (Beckman Coulter, Beverly, USA) was used for the library fragments purification. PCR was performed, and library quality was assessed. Finally, the clustering of index-coded samples was carried out according to the manufacturer’s instructions (cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia)), following library preparations on Illumina Novaseq platform to generate 150 bp paired-end reads. The transcriptome library dataset was deposited into the NCBI Sequence Read Archive (SRA) database (Accession No. PRJNA810208).
The fastq format raw data/reads were processed via in-house perl scripts to obtain clean data/reads by removing; adapter, poly-N, and low-quality read from the raw data. Subsequently, Q20, Q30, and GC content were calculated from the clean reads, following downstream analysis. The paired-end clean reads obtained were used to align to the reference genome (GWHAOSK00000000.1,https://ngdc.cncb.ac.cn/search/?dbId=gwh&q=GWHAOSK00000000.1&page=1) using Hisat2 v2.0.5. From the data analysis, ovary3 was found to be polluted and hence was removed from subsequent analysis.
The StringTie (v1.3.3b) uses a novel network flow algorithm assemble each sample’s mapped reads in a reference-based approach. The featureCounts v1.5.0-p3 was employed to count the read numbers mapped to each gene. The Fragments Per Kilobase of transcript sequence per Millions (FPKM) of each gene was calculated based on gene length and read count. DESeq2 R package (1.20.0) was used to perform the differential analysis of gene expressions. Genes with an adjusted P-value < 0.05 established by DESeq2 were considered differentially expressed.
Gene Ontology (GO) enrichment analysis of differentially expressed genes was analyzed and the clusterProfiler R package corrected gene length bias. GO terms with corrected P-value ≤ 0.05 were considered significantly enriched DEGs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database helps understand the biological system’s high-level functions and utilities from molecular-level information generated from genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). Similarly, the clusterProfiler R package was used for the statistical enrichment test of DEGs in the KEGG pathways. To screen the target DEGs, genes with P value < 0.05 and (Log2(FC)) >1 were chosen to be highly significant.
Fourteen DEGs were selected, and their expression patterns from the RNA-seq were validated using qPCR. Gonad cDNA of males (N=3) and females (N=3) were used for gene expression profiling. The total RNA (2.0 μg) used for qPCR was isolated from frozen gonads of spotted scat using RNeasy® Mini Kit (50) (163,046,943, QIAGEN, Germany), following the manufacturer’s instructions. The quality and concentration of the RNA were assessed by NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). The total RNA was treated with RNase-free DNase I (Thermo Scientific Corp, Waltham, MA, USA). First-strand cDNA synthesis was performed using the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara) with 1 μg total RNA. The real-time PCR was performed on a Light Cycler 480 (Roche) using SYBR Green Realtime PCR Master Mix (Toyobo, Japan), following manufacturer’s instructions. The real-time PCR conditions used were: denaturation at 95°C, 1 min, followed by 40 cycles of 95-°C, 30 sec, 60°C, 30 sec, 72°C, 30 sec (fluorescent data collection). The β-actin values were used to normalize the expression patterns of genes. The formula; , was used to evaluate the relative abundance of selected genes. The primers used are shown in Table S1.
The size of juvenile spotted scat gonads is too small and cannot be distinguished phenotypically by mere observation. On the other hand, the differentiating gonad can be distinguished by the difference in proliferating germ cells using HE staining (Mustapha et al., 2021). Two markers were used for genotypic sex identification of the four months old spotted scat as detailed in (Mustapha et al., 2018). The average length and weight of the fish sampled were male (6.98 ± 0.273/cm and 11.75 ± 1.258/g) and female (7.13 ± 0.203/cm and 12.27 ± 0.871/g) (Figure 1A). Histological analysis showed that, the gonads of the four months spotted scat were at the differentiating stage or stage I (testisI/testis-I) and (ovaryI/ovary-I) (Figure 1B) similar to previous findings (Mustapha et al., 2021). Few primary oocytes were scattered in the female gonads, while spermatogonia were found in the testes.
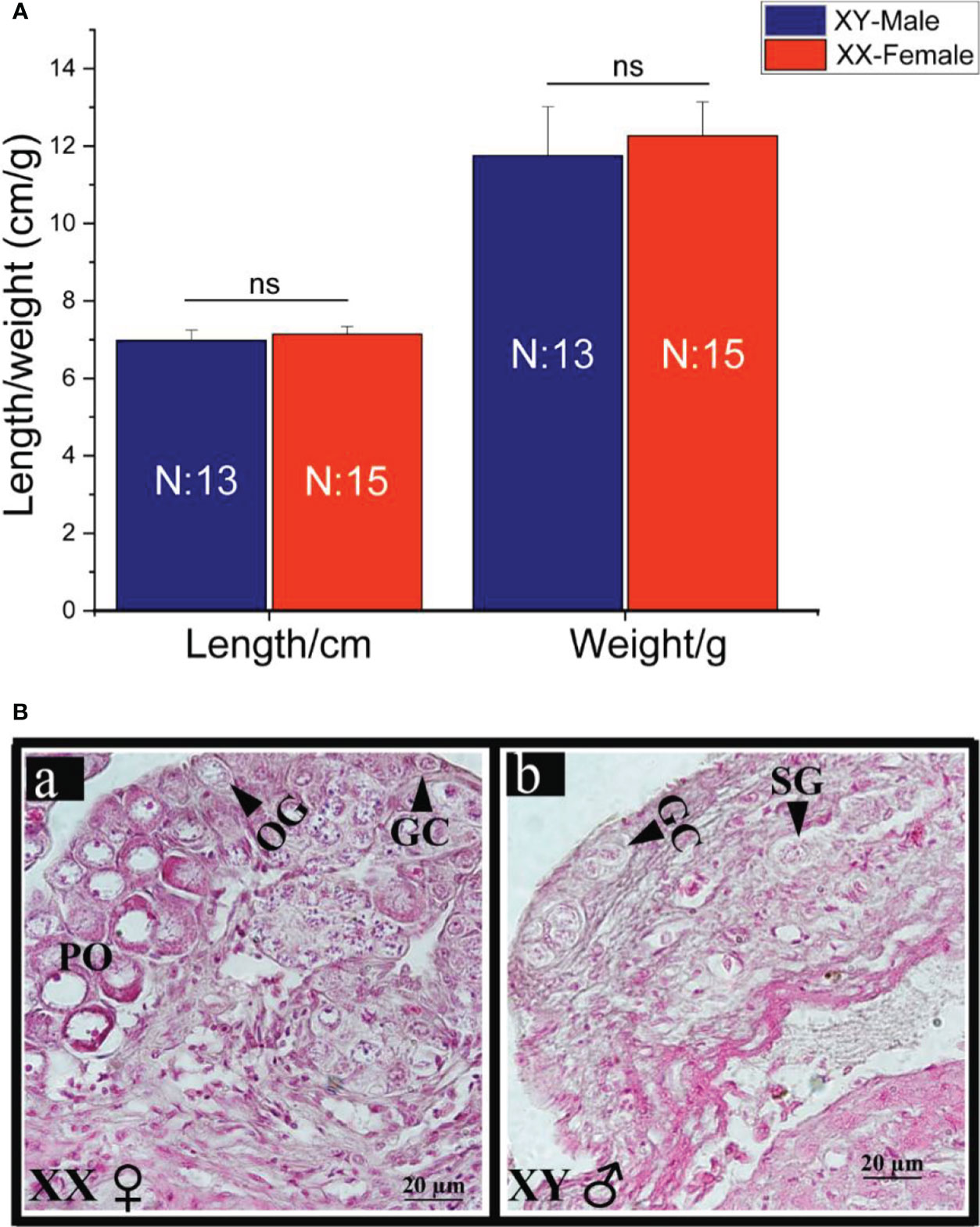
Figure 1 Length and weight data (A) and gonadal histology (B) of juvenile (4mah) spotted scat. Ns; no significant difference (p>0.05), OG, oogonia; PO, primary oocytes; GC, germ cell; SG, spermatogonia; XY, male; XX, female. The gonads (a and b) are both at stage I.
cDNA libraries were prepared from pooled samples of three testes (testis1, testis2, and testis3) and two ovaries (ovary1, ovary2) as detailed in Figure 2, following Illumina sequencing. The total raw and clean reads obtained from each library is shown in Tables 1, 2. After raw data filtering, sequencing error rates and GC content distribution were analyzed, and clean reads were obtained for subsequent analysis. The data summary is shown in Table 1. A sum of six cDNA libraries was constructed in triplicates from testes and ovaries. The total raw reads in each library ranged from 39905724 to 44986798. After filtering adapters, poly-N, and low-quality reads, the total clean reads ranged from 38484126 to 44255056. Also, the clean bases ranged from 5.77G to 6.64G. Both the highest and the lowest raw, clean reads and bases were obtained in the library of the male group (Testis1 and 3, respectively). The percentage of G and C content in the clean reads ranged from 46.55 to 49.82, with a low overall data sequencing error rate ≤ 0.03. In addition, the percentage of bases with Phred values greater than 20 (Q20) and 30 (Q30) in total bases are ≥ 97.52 and ≥ 93.5, respectively.
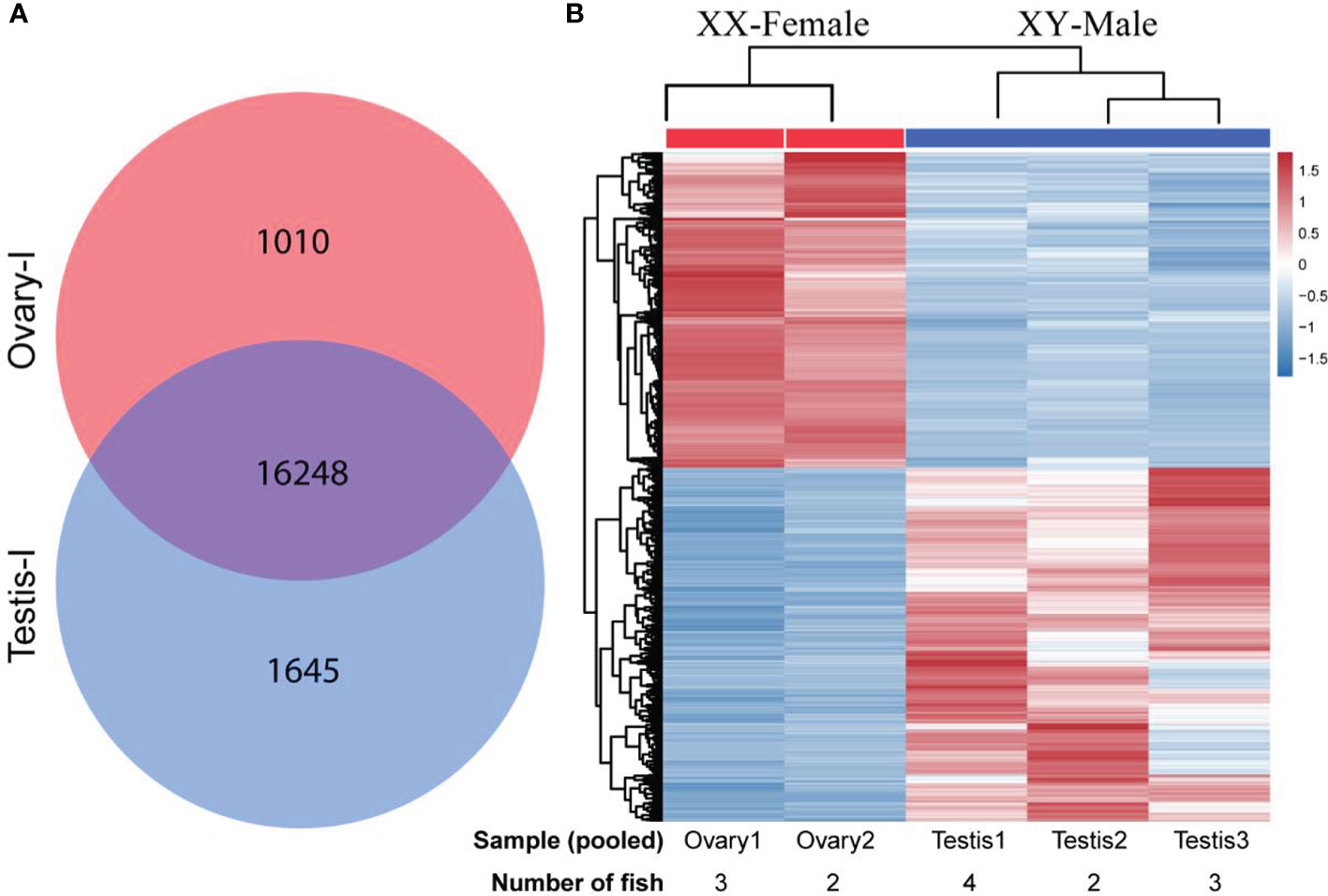
Figure 2 Differentially expressed genes DEGs obtained from gonadal RNA-seq data of XY-male (testisI) and XX-female (ovaryI) spotted scat. (A) Venn diagram depicting the total number of genes expressed in XY-male (17893) and XX-female (17258) gonads of four months old spotted scat. A total of 16248 genes were co-expressed in male and female gonads, while 1645 and 1010 genes were specifically expressed in male and female gonads, respectively. (B) A heatmap depicting hierarchical clustering of 6549 differentially expressed genes (DEGs) in male and female juvenile gonads of spotted scat. The pooled samples, Ovary1, Ovary2, Testis1, Testis2, and Testis3 contained gonads from 3, 2, 4, 2, and 3 fish, respectively.
In addition, the reproducibility, reliability, and gene expression differences between and within biological groups were accessed from all samples’ gene expression values (FPKM) by employing correlation (Figure S1), and principal component analysis (PCA) (Figure S2). These results indicate a high sequencing data quality, hence suitable for subsequent analysis.
In order to obtain the respective mapping rates, the clean reads from each sample were compared with the reference genome using (GWHAOSK00000000.1, https://ngdc.cncb.ac.cn/search/?dbId=gwh&q=GWHAOSK00000000.1&page=1. A total map of >88% and a unique map of > 85% were obtained (Table 2). Unfortunately, ovary3 was found to be polluted and hence were removed from subsequent analysis.
A total of 25936 genes were identified, with 17893 and 17258 expressed in males and females, respectively. Additionally, a total of 16248 were co-expressed in both males and females. In addition, 1645 and 1010 genes were explicitly expressed in male and female gonads, respectively (Figure 2A). A heatmap showing a cluster of differentially expressed genes (DEGs) is indicated in Figure 2B. A total of 6549 DEGs were identified between male and female juvenile gonads (Figure 3). Comparatively, 3463 and 3088 DEGs were detected in females and in males, respectively, respectively. Genes showing higher expression in males, showed lower expression in females and vice versa.
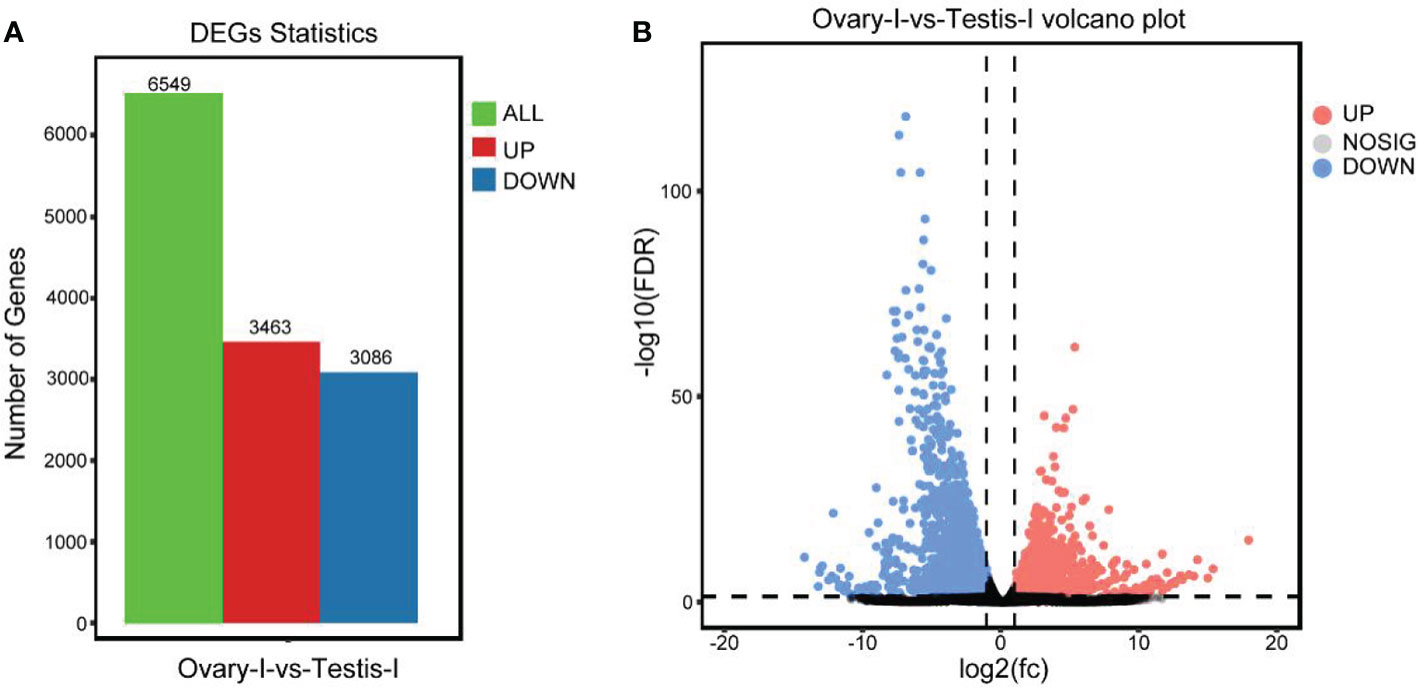
Figure 3 (A) Differentially expressed genes (DEGs) in males (testisI) and females (ovaryI) of spotted scat. The green key represents all 6549 differential express genes. In comparison, red and blue represents up-and down-regulated genes. (B) Volcano plot of DEGs in testisI versus ovaryI. Red and blue keys and dots represent up-regulation (highly expressed) and down-regulated (lowly expressed) genes in females, respectively, and vice versa.
To elucidate the expression pattern of sex differentiation and gonadal development-related genes, we examined over 70 DEGs related to sex differentiation and gonad development and sex steroid synthesis pathway. 43 and 28 DEGs in males and females were examined, respectively (Table 3). The known male-related genes such as dmrt1, gsdf, amh, and cyp17a1 are significantly expressed in males, while female-related genes including foxl2, cyp19a1a, 42sp50, and sox3 are expressed considerably in females.
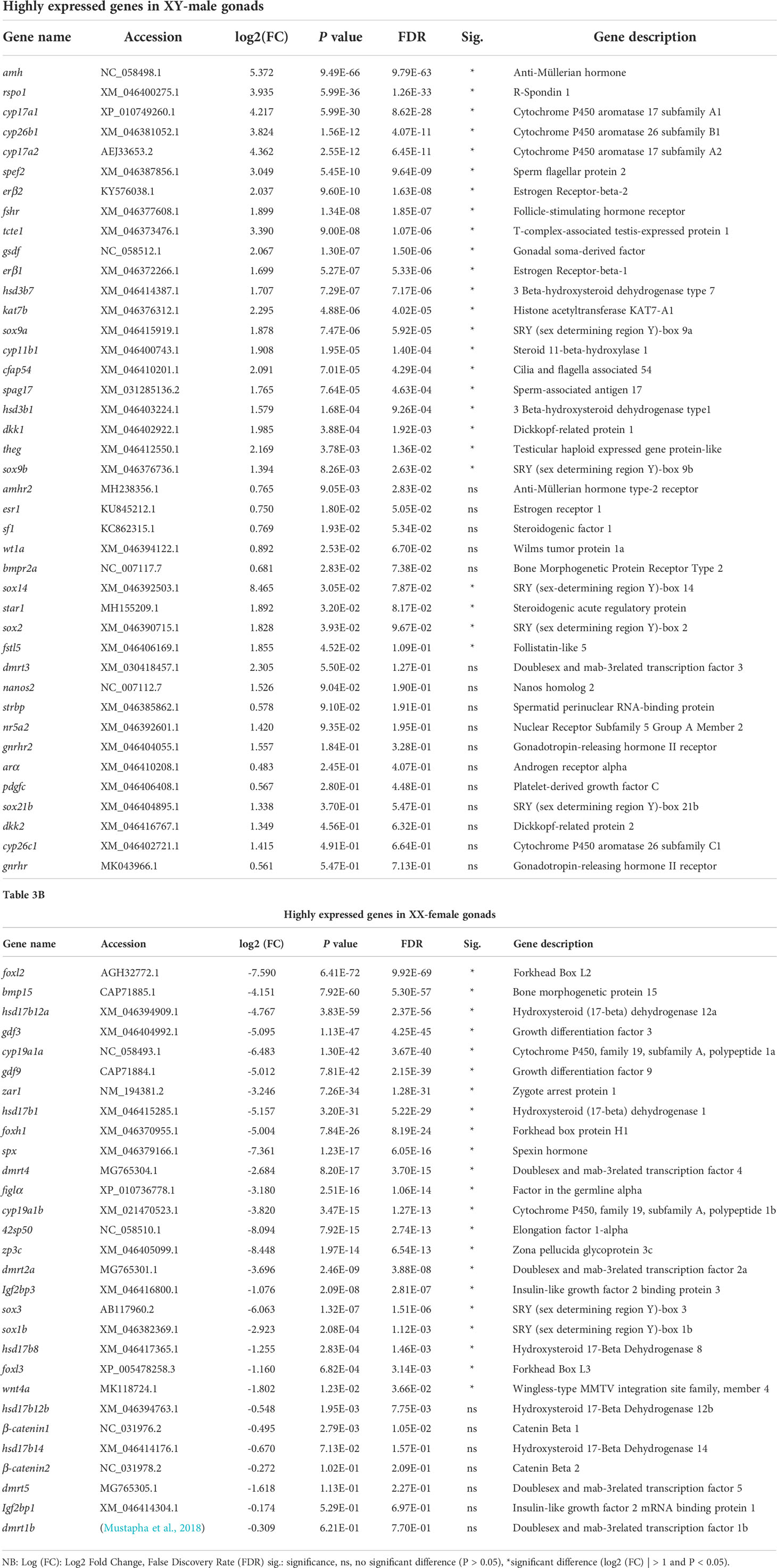
Table 3A The patterns of differentially expressed sex-related and gonadal developmental genes in the differentiating gonads of spotted scat.
All DEGs were enriched and classified into three GO terms, namely biological process (BP), cellular component (CC), and molecular function (MF). Figure 3 shows the 20 most significantly enriched terms in BP, CC, and MF. Amongst them, eight (GO:0071345, GO:0019221, GO:0034097, GO:0001578, GO:0032874, GO:0070304, GO:0070887, and GO:0006415), five (GO:0000313, GO:0019866, GO:0031967, GO:0031975, and GO:0031012), and seven (GO:0004518, GO:0004519, GO:0004536, GO:0004520, GO:0019205, GO:000019206, and GO:0019136) GO terms were highly significant in BP, CC, and MF, respectively. GO term categories in BP, CC, and MF significantly enriched between males and females are shown in (Figure S3).
KEGG analysis revealed eleven significantly enriched pathways. Those significantly enriched KEGG pathways include ribosome biogenesis in eukaryotes, pyrimidine metabolism, ECM-receptor interaction, cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, RNA degradation, DNA replication, drug metabolism – other enzymes, complement and coagulation cascades, mismatch repair and necroptosis (Figure 4). KEGG pathway annotations of genes were mapped to 44 different pathways classified into six main categories. Among the pathways, the “signal transduction” and “biosynthesis of other secondary metabolites” were the most and least predominant pathways, respectively (Figure S4). The top 50 KEGG enrichment is shown in Table S2.
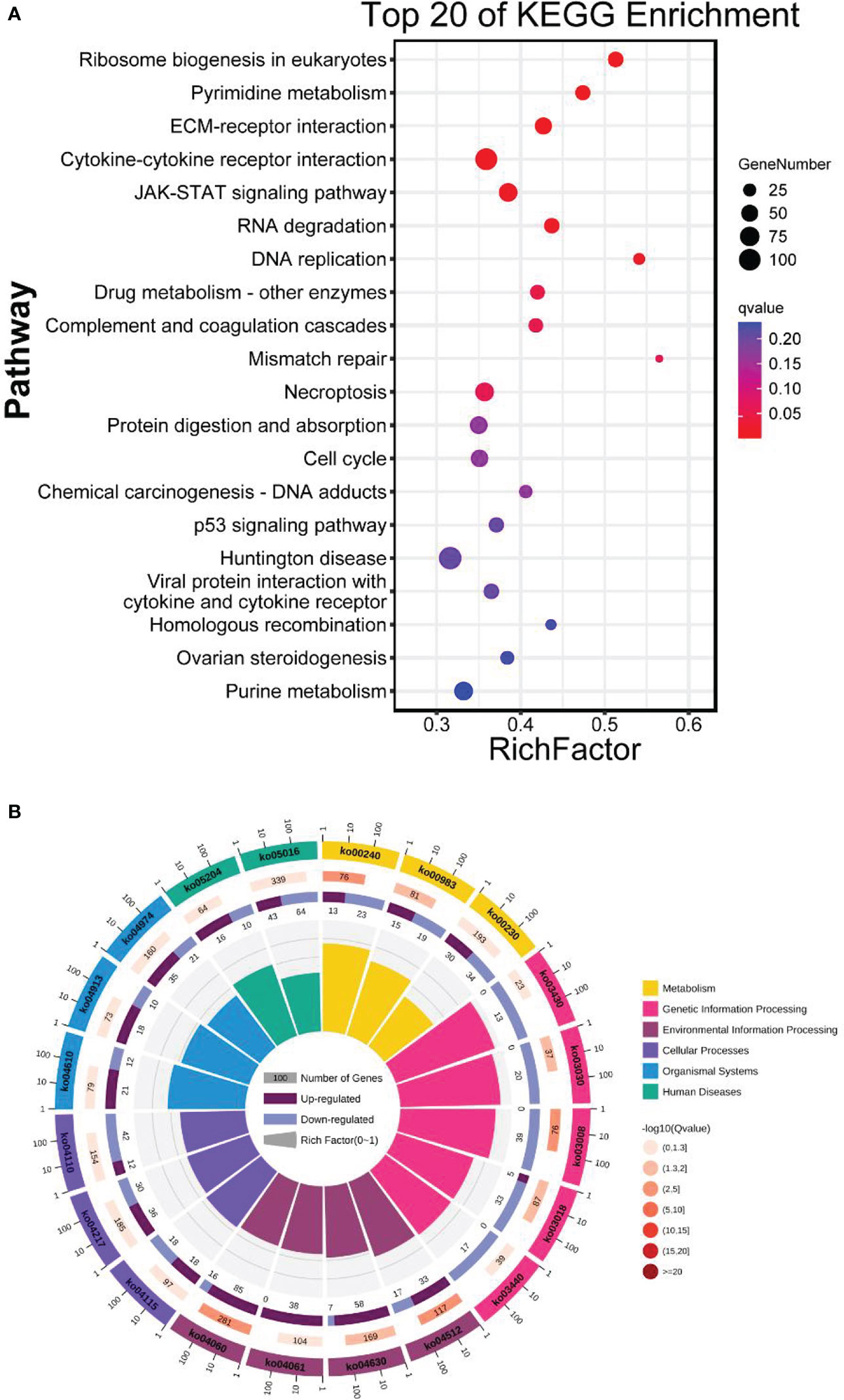
Figure 4 (A) Top 20 KEGG enrichment analysis. The horizontal axis is the ratio of the number of differential genes annotated to the KEGG pathway to the total number of differential genes. The vertical is the enriched KEGG pathways. The size of the dots represents the number of genes annotated on the KEGG pathway term. The colors key, from red to blue, represent significant enrichment. (B) KEGG classification of genes under six main categories; metabolism, genetic information processing, environmental information, and human diseases.
The expression of eighteen genes related to sex differentiation and gonadal development were analyzed from the RNA-seq data (Figure 5). Male related genes (such as; gsdf, sf1, amh, wt1a, nanos2, cyp11b1 and cyp17a1) and female related genes (such as; foxl2, sox3, cyp19a1a, figla, zar1, bmp15, and 42sp50) were highly expressed in male and female gonads. In the RNA-seq data, dmrt1 FPKM was not detected in the female while dmrt1b was expressed in both male and female (Figure 6A). Subsequently, fifteen genes were selected and validated by qPCR to confirm the accuracy and reliability of the RNA-seq data (Figure 6B). The results showed that male-related genes; gsdf, sf1, amh, wt1a, nanos2, and cyp11b1 were highly expressed in males, while female-related genes; foxl2, sox3, gdf9, cyp19a1a, figla, zar1, bmp15, and 42sp50 were higher in females. All analyzed male and female gene expressions were consistent with the RNA-seq data.
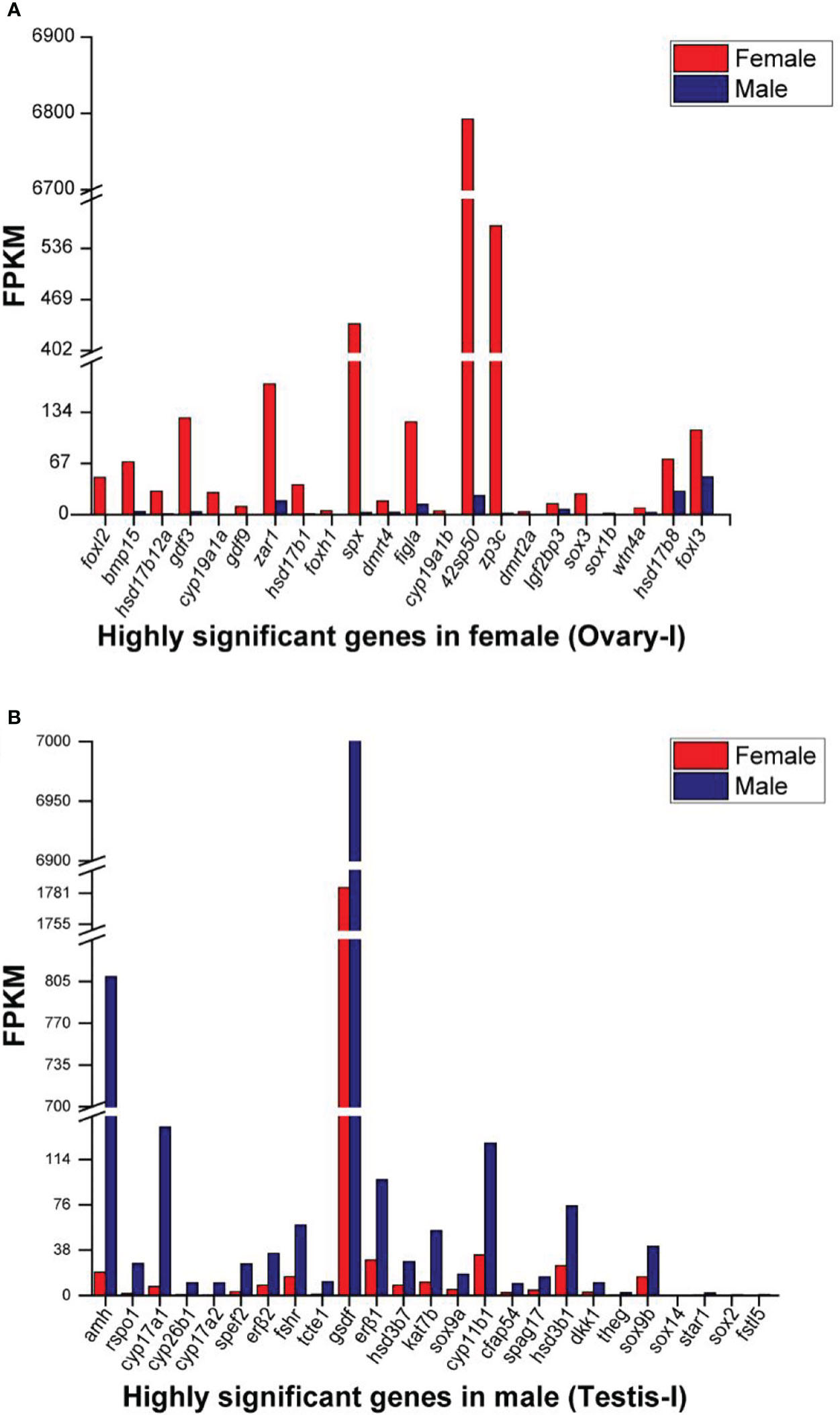
Figure 5 Selected highly differential expressed genes (log2 (FC) >1 and P <0.05) in (A) XX-female (Ovary-I) and (B) XY-male (Testis-I) Spotted scat. The data is based on the mean FPKM values obtained from RNA-seq data.
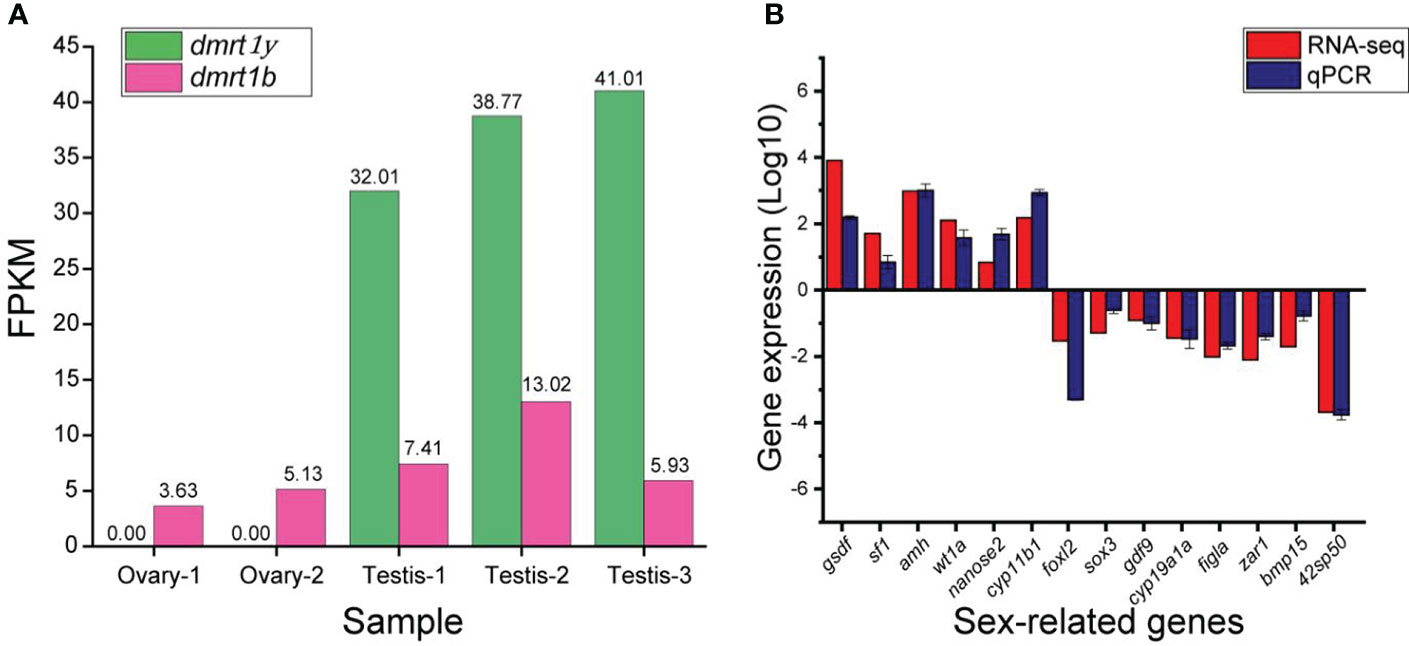
Figure 6 (A) The expression of dmrt1/dmrt1y (on the Y-chromosome) and dmrt1b (on the X-chromosome) in the RNA-seq data of testis and ovary of juvenile spotted scat. Dmrt1y transcript is not found in female, while dmrt1b is expressed in both male and female (B) Comparison of selected sex-related genes’ expression by RNA-seq and qPCR. The male genes are positive while female genes are negative. The positive and negative axis show the gene expression in testis and ovary, respectively.
Knowledge of gonad development is paramount for animal reproduction. During gonad development, the undifferentiated, differentiating, and differentiated stages are key to understanding the underlying gonad development mechanism. Genetically, the underlying mechanism involves complex biological processes, including a set of genes functioning to promote or maintain gonadal development into the testis or ovary. Transcriptomics is an effective way to obtain gene regulatory networks between individuals (male and female). In spotted scat, several transcriptome analyses and comparative studies have been carried out (He et al., 2019). Nonetheless, these studies focused on adult spotted scat (differentiated gonads) with no information on the juveniles (undifferentiated and differentiating gonads). This study conducted transcriptome analysis and comparative studies between four-month-old males and females, differentiating spotted scat gonads. Our study identified several sex-related genes and established the expression patterns of DEGs (differential expression of genes) in the differentiating gonads.
The functional classification of DEGs was carried out using GO enrichment and KEGG pathway analyses. The DEGs were mainly enriched in ribosome biogenesis in eukaryotes, pyrimidine metabolism, ECM-receptor interaction, cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, RNA degradation, DNA replication, drug metabolism – other enzymes, complement and coagulation cascades, mismatch repair and necroptosis (Figure 4). This suggests the involvement of the pathways in spotted scat gonad development. The gonad of female juvenile spotted scat has higher number of germ cells than the male (Mustapha et al., 2021). The KEGG pathways such as RNA degradation, cell cycle, and ovarian steroidogenesis contain substantial amount of DEGs in male and female. The mechanisms accounting for the DEGs and the differences in the genes between male and female will be elucidated in our future study. In addition, the gonadal transcriptome analysis revealed numerous sex-related genes in the differentiating gonads. Based on previous reports, we screened 14 candidate genes related to sex and the gonad development process and verified their expression by qPCR (He et al., 2019; Mustapha et al., 2021). The well-known male pathway genes such as gsdf, amh, and cyp17a1(also essential for female development in common carp (Zhai et al., 2022)) were significantly expressed in male spotted scat (He et al., 2019; Mustapha et al., 2021). Similarly, the expression pattern conforms with that of other fish species (Pfennig et al., 2015; Chi et al., 2017; Wang et al., 2019). On the other hand, female-related genes, including foxl2, cyp19a1a, 42sp50, and sox3, were expressed highly in females in line with other species (Pfennig et al., 2015; Chi et al., 2017; Wang et al., 2019), including adult spotted scat (He et al., 2019; Mustapha et al., 2021). These indicate that these genes might exhibit conserved function in spotted scat. Herein we discuss some important DEGs in the male (dmrt1, gsdf, amh, and cyp17a1) and female (foxl2, cyp19a1a, 42sp50, sox3), sex differentiation pathways.
The functional classification of DEGs was carried out using GO enrichment and KEGG pathway analyses. The transcriptomics revealed numerous genes essential for testis differentiation, among them including dmrt1, gsdf, and amh. The doublesex and mab-3 related transcription factor (Dmrt) family genes are important regulators of sexual fate in teleost (Bellefroid et al., 2013). Dmrt1-6 have been isolated in teleost and consistently exhibited sex-specific or sexual dimorphic expressions in different species (Brunner, 2001; Kondo et al., 2002; Matsuda, 2005; Yamaguchi et al., 2006; Kobayashi et al., 2008; Bewick et al., 2011; Chue and Smith, 2011; Johnsen and Andersen, 2012; Shi et al., 2014; Cui et al., 2017; Lyu et al., 2019). Among them, dmrt1 is critical for testis differentiation in mammals and teleost (Kobayashi et al., 2004; Matson et al., 2011; Minkina et al., 2014). The mammalian Dmrt1 is required to properly differentiate male germ cells and maintain male sexual cell fates (Matson et al., 2011). In teleost, fish such as medaka and zebrafish, dmrt1 mutation led to male-to-female sex reversal (Masuyama et al., 2012; Webster et al., 2017). In spotted scat, dmrt1 is crucial for testis development and is found explicitly on the Y-chromosome in XY males (Mustapha et al., 2018; Huang et al., 2019). The XX-spotted scat is unable to be sex-reversed due to the absence of dmrt1 needed to ensure testis differentiation (Mustapha et al., 2018). In this study, dmrt1 transcript was not found in female differentiating gonads, supporting the identification of dmrt1 as the candidate sex determination gene in spotted scat (Mustapha et al., 2018). Hence, an attempt to induce sex reversal without introducing dmrt1 in female spotted scat might prove futile. This is corroborated by 17α-Methyltestosterone (MT) and Letrozole’s (Le) inability to induce sex-reversal in female spotted scat, while estrogen does in males (Mustapha et al., 2021). Meanwhile, MT and Le could successfully induce sex reversal in many fish species (Megbowon and Mojekwu, 2014; Ranjan et al., 2015; Chen et al., 2016). However, regulatory studies of some genes may be successfully carried out in females spotted scat in vitro but might not be effective in in vivo. For instance, treatment with MT and Le could only up-regulate some male-related genes but could not induce sex reversal in XX females spotted scat (Mustapha et al., 2021). Nonetheless, functional studies are required to establish dmrt1 as the sex determination gene in spotted scat.
Gsdf (Gonadal soma-derived factor) belongs to the transforming growth factor-β (TGF-β) family and is a key gene in teleost testicular differentiation. The expression pattern of gsdf is highly conserved in fish gonad developmental pathways and is highly expressed in males (Gautier et al., 2011; Imai et al., 2015; Jiang et al., 2016). In Nile tilapia and medaka, knockout and mutation of gsdf induced sex reversal in XY individuals (Imai et al., 2015; Jiang et al., 2016). Additionally, gsdf knockdown in the Chinese tongue sole influenced several sex-related genes (Zhu et al., 2018). In this study, gsdf is highly expressed in the testes similar to the results in the adult spotted scat (He et al., 2019; Jiang et al., 2019; Mustapha et al., 2021). In Nile tilapia, Yeast-two-hybrid assays showed that amhr2 and bmpr2a might be the type II receptors of gsdf (Jiang et al., 2022). Both amhr2 and bmpr2a are expressed at similar levels between the ovary and testis in spotted scat. The male sex-biased expression of gsdf might be also critical for testicular differentiation in spotted scat. We previously reported that, gsdf could be regulated by either dmrt1 alone or interaction with sf1 in spotted scat (Jiang et al., 2019). However, the average gsdf FPKM in both differentiating ovary (FPKM: 1785) and testis (FPKM: 7482) are relatively high in spotted scat, which may be the reason for its indifference detection by qPCR in an earlier report (Mustapha et al., 2021). It is reported that over-expression of gsdf resulted in female-to-male sex reversal in XX Nile tilapia (Kaneko et al., 2015). Thus we deduce that there should be some proteins that antagonize the Gsdf function in the ovary of spotted scat, or gsdf expression in the ovary may be inconsequential. Alternatively, in the XX spotted scat, despite the high level of gsdf expression, it could not induce the testicular differentiation for lack of intact dmrt1 (Mustapha et al., 2018). The present results support our previous conclusion that gsdf acts as a downstream gene of dmrt1 in testicular differentiation in fish (Jiang et al., 2016; Jiang et al., 2019; Jiang et al., 2022) (Figure 7). The spotted scat with male-specific dmrt1 as the candidate sex determination gene is a special model to study genes involved in sex differentiation.
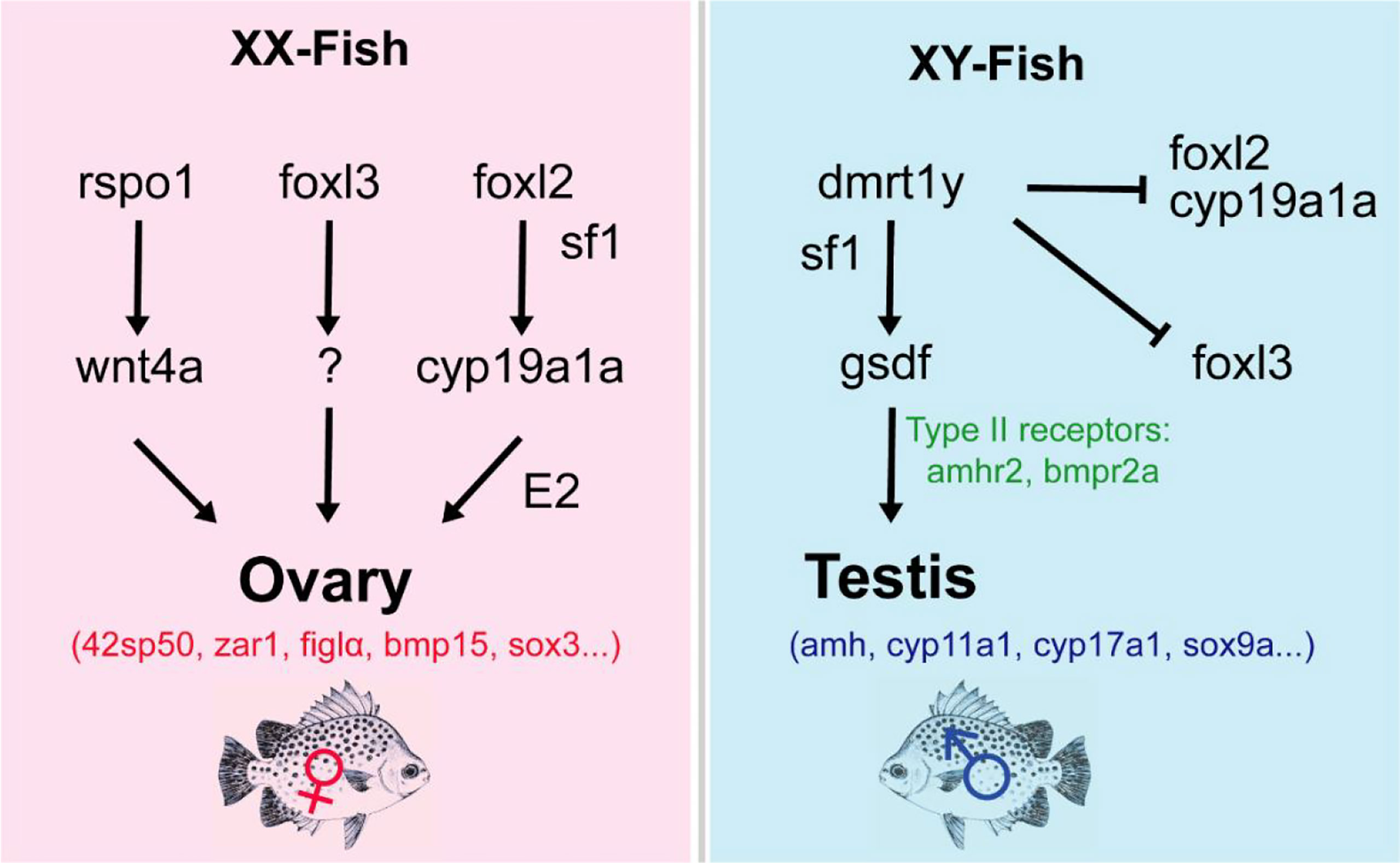
Figure 7 Predicted schematic overview of the sex development gene regulatory network in spotted scat. The Y-chromosome dmrt1 (dmrt1y) is a key regulator of male sex determination, which could act with sf1 to regulate gsdf expression in the male pathway. The absence of dmrt1y in XY will activate foxl2/cyp19a1a to induce female development. Also, foxl2 or together with sf1 can regulate cyp19a1a to stimulate E2 production to induce ovary development. Similarly, rspo1 may regulate wnt4a expression in the ovary, whiles foxl3 may also act with other regulators to ensure female gonad development. ⊥: Inhibition, ↓: stimulation. Female and male DEGs are in red and blue respectively.
Anti-Müllerian hormone (amh) is also a member of the transforming growth factor-β (TGF-β) family genes. Amh expression has been reported in over 20 fish species with possible regulatory interactions with factors like sex steroids and gonadotropic hormones. Amh inhibits proliferation and differentiation of germ cells as well as steroidogenesis in both sexes (Pfennig et al., 2015). Amh-type II receptor gene (amhr2) is involved in germ cell regulation of fish species. In the Japanese flounder undifferentiated gonads, the amh transcript was undetectable (Yoshinaga et al., 2004; Yamaguchi and Kitano, 2012). In species such as Nile tilapia, medaka, zebrafish, and rainbow trout, amh expresses at similar levels in the undifferentiated gonads (Klüver et al., 2007; Schulz et al., 2007; Vizziano et al., 2008; Poonlaphdecha et al., 2013; Eshel et al., 2014) and becomes highly expressed in the male differentiated testis except for medaka (Klüver et al., 2007; Hattori et al., 2009; Yamaguchi et al., 2010; Haugen et al., 2012; Mankiewicz et al., 2013; Poonlaphdecha et al., 2013). However, amhr2 was highly expressed in juvenile ovaries, whereas it was profound in the adult testes in spotted scat (He et al., 2019). Studies indicate that amhr2 is expressed in both male and female gonads, but it is more strongly expressed in males after gonadal sex differentiation in chicken (Cutting et al., 2014). In medaka, amhr2 did not show sexually dimorphic expression during sex differentiation similar to this study (Klüver et al., 2007). In addition, amhr2 was suggested to play an important role in gonadal development, particularly in testicular differentiation and development, including late sex change in the protandrous black porgy (Wu et al., 2010). Amhr2 mutants exhibited male to female sex reversal and displayed excessive germ cell proliferation in medaka (Nakamura et al., 2012), whiles higher expression of amhr2 coincides with increasing germ cells in females (Mustapha et al., 2021). Therefore, amhr2 in spotted scat could be developmentally regulated during gonad differentiation and maturation.
Cytochrome P450 family 17 subfamily A member 1 (cyp17a1) is found to play an essential role in fish steroidogenesis (e.g., production of sex steroids), which are involved in sex determination, differentiation, and fertility. The knockout of cyp17a1 in Nile tilapia, zebrafish, and common carp resulted in female-to-male sex-reversal indicating that cypa17a1 is essential for ovarian differentiation in fish (Zhai et al., 2018; Yang et al., 2021; Zhai et al., 2022). Insufficient estrogen at the critical window of sex differentiation should be the reason for the sex reversal in the XX cyp17a1-/- Nile tilapia and zebrafish as the sex reversal could be rescued and the ovarian characteristics could be maintained after treatment with exogenous E2 or its precursor testosterone (Zhu et al., 2019). Despite the significant-high expressions of cyp17a1 (FPKM: 140.9) and cyp17a2 (FPKM: 10.7) in the testis than in the ovary in spotted scat, they are also expressed in the ovary (FPKM: 7.58 and 0.5, respectively) indicating that they may be essential for the ovary differentiation via inducing E2 synthesis.
Forkhead Box L2 (foxl2) is involved in ovarian differentiation and oogenesis in vertebrates (Bertho et al., 2016). The gene showed sexually dimorphic expression in the differentiating and adult gonads favoring ovaries in almost all fish species studied (Baron et al., 2004; Wang et al., 2004; Nakamoto et al., 2006; Vizziano et al., 2007; Yarmohammadi et al., 2017; Zhang et al., 2017). In Nile tilapia, foxl2 mutants resulted in female-to-male sex reversal (Zhang et al., 2017). Foxl3 is also an important family member gene that is highly expressed in ovaries compared with testis and was found to initiate oogenesis in medaka (Kikuchi et al., 2019; Kikuchi et al., 2020). In XX Japanese medaka and Nile tilapia, foxl3 knockout resulted in the gonads developing an ovary-like testis which could produce fertile sperms indicating that foxl3 is critical to guarantee the differentiation direction of the germ cell in the early fish ovary (Nishimura and Tanaka, 2016; Dai et al., 2021). Additionally, the double knockout of dmrt1 and foxl3 rescued the XX Nile tilapia to develop as female fish indicating that dmrt1 and foxl3 antagonize each other to determine the germ cell fate. Also, the antagonistic effect of dmrt1 and foxl2/foxl3 was reported to determine germline sexual fate (Dai et al., 2021). In spotted scat foxl2 and foxl3 are also highly expressed in the ovary than that in the testis. However, the XX spotted scat lacks dmrt1, hence we deduce that the two genes may still have some other critical functions to guarantee germ cell differentiation as females besides repressing the dmrt1.
Cyp19a1a is a gonadal aromatase gene involved in ovarian differentiation and has received considerable attention in recent years. There have been quite consistent results from functional studies of cyp19a1a in fish. In the Nile tilapia, cyp19a1a-/- XX fish displayed female-to-male sex reversal (Zhang et al., 2017). The knockout and targeted disruption of cyp19a1a (and, double mutants of cyp19a1a and cyp19a1b) in zebrafish led to failed ovarian differentiation and induced sex skewness towards all-male individuals (Lau et al., 2016; Yin et al., 2017). It is evidenced that, cyp19a1a could be regulated by foxl2 directly or indirectly via interacting with sf1 to produce E2 for ovarian differentiation (Wang et al., 2007; Yamaguchi et al., 2007). In foxl2-/- and the cyp19a1a-/- XX gonads Nile tilapia, dmrt1, gsdf, sf1, and cyp11b2 were upregulated. In spotted scat differentiating gonads, cyp19a1a and cyp19a1b are significantly expressed in the ovary, while sf1 (insignificant) and cyp11b2 (significant) are expressed highly in the testis similar to our previous results (Mustapha et al., 2021). Interestingly, sf1 was found to be functionally important in testis and ovary development. In Nile tilapia, sf1 is highly expressed in the testis, and the down-regulation displayed abnormal gonads in both males and females, while the deficiency caused female to male sex reversal. Additionally, its inactivation led to the death of embryos (Xie et al., 2016; Cao et al., 2022). We, therefore, infer that foxl2, cyp19a1a, and sf1 might essentially interact toward ovarian differentiation, and overexpression (of foxl2 and cyp19a1a) may induce ovarian differentiation by repressing male pathway genes as predicted in Figure 7.
R-spondin1 (rspo1) activates the Wnt/β-catenin signaling pathway involved in female sex differentiation in mammals (Binnerts et al., 2007; Lacour et al., 2017). Overexpression of the rspo1 in XY medaka resulted in upregulation of wnt4b and beta-catenin transcription and decreasing of male pathway genes (dmy, gsdf, sox9a2 and dmrt1) indicating that rspo1 acts as well as female differentiation pathway gene (Wu et al., 2018). Contrarily, rspo1 was expressed higher in the testis than that of the ovary of differentiating spotted scat, while wnt4a is expressed higher in the ovary than that of the testis. β-catenin1 and β-catenin2 were expressed higher (insignificantly) in the ovary than in the testis. Thus, it is still unclear whether the rspo1/Wnt/beta-catenin pathway is important for ovary differentiation in spotted scat (Figure 7).
Eukaryotic translation elongation factor 1A (eEF1A) is one of the most abundant protein synthesis factors in eukaryotic cells. Five eEF1A genes, including eEF1A1-4 and 42sp50, have been isolated and characterized (Gao et al., 1997; Nowell et al., 2000; Infante et al., 2008). Amongst them, 42sp50 is mainly detected in the oocytes of many species including Xenopus laevis, Nile tilapia, and medaka (Abdallah et al., 1991; Deschamps et al., 1991; Kinoshita et al., 2009; Loeber et al., 2010; Chen, 2018; Murakami et al., 2019), and vitally associated with ovarian development (Horiguchi et al., 2018). In the adult medaka, Nile tilapia, and spotted scat (juvenile and adult) gonads, 42sp50 was more profoundly expressed in the ovaries than in testes (Kinoshita et al., 2009; Chen et al., 2017; Mustapha et al., 2021). Similarly, in this study, the expression was found to be significant in the differentiating ovaries. Despite the sexually dimorphic expression, its role in spotted scat might mainly be linked to normal ovarian development and gene knockout or mutation will not induce sex reversal for lack of dmrt1. Even though, male to female sex reversal can influence 42sp50 expression in the gonads of spotted scat (Mustapha et al., 2021). Similarly, the loss of dmrt1 in XY Nile tilapia which led to all-female sex reversal resulted and up-regulation of 42sp50 and cyp19a1a, and down-regulation of gsdf and amh (Dai et al., 2021). However, the knockout of 42sp50 rather impeded oocyte development and caused abnormal proliferation and differentiation of follicular cells at the primary growth stage in female Nile tilapia (Chen, 2018). This suggests that 42sp50 is instead crucial for normal ovarian development.
The sex-related and gonadal development gene expression profile in the differentiating spotted scat gonads was examined using RNA-seq. A total of 6549 differential expressed genes (DEGs) were identified between males (testis stage I) and females (ovary stage I), of which 3463 and 3086 were highly expressed in females (upregulated) and males (downregulated), respectively. The conserved expression pattern of the major sex-related genes indicates their conserved function in sexual differentiation and gonadal development. The candidate sex determination gene on the Y chromosome (dmrt1/dmrt1y) was not found in females while dmrt1b was expressed in both males and females. Genes such as foxl2 and cyp19a1a are key in females while dmrt1 and gsdf are key in male development pathways. Additionally, 42sp50 and amh might ensure the proper functioning of gonads and knockout might not induce sex reversal in spotted scat. The study provides a valuable data source to ascertain the expression pattern of sex-related and gonadal developmental genes in spotted scat. In addition, the study provides a developmentally relevant resource useful in genomic and genetic studies and the establishment of sex control strategies in spotted scat.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA810208.
The animal study was reviewed and approved by Animal Research and Ethics Committee of Guangdong Ocean University, China (201903004). Written informed consent was obtained from the owners for the participation of their animals in this study.
Conceptualization, D-NJ, UFM and G-LL; methodology, UFM and D-NJ.; investigation, UFM, Y-XP, DA, Y-QH, FZ, GS and YH; writing—original draft preparation, UFM and D-NJ; writing—review and editing, UFM and D-NJ; funding acquisition, D-NJ and G-LL. All authors have read and agreed to the published version of the manuscript.
This study was supported by grants from the National Natural Science Foundation of China (Nos. 32172971); Guangdong Basic and Applied Basic Research Foundation (2021A1515010430); the Key Project of “Blue Granary Science and Technology Innovation” of the Ministry of Science and Technology (2018YFD0901203).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.962534/full#supplementary-material
Abdallah B., Hourdry J., Krieg P. A., Denis H., Mazabraud A. (1991). Germ cell-specific expression of a gene encoding eukaryotic translation elongation factor 1 alpha (eEF-1 alpha) and generation of eEF-1 alpha retropseudogenes in xenopus laevis. Proc. Natl. Acad. Sci. U. S. A. 88, 9277–9281. doi: 10.1073/PNAS.88.20.9277
Ayers K. L., Lambeth L. S., Davidson N. M., Sinclair A. H., Oshlack A., Smith C. A. (2015). Identification of candidate gonadal sex differentiation genes in the chicken embryo using RNA-seq. BMC Genomics 16, 704. doi: 10.1186/s12864-015-1886-5
Baron D., Baron D., Cocquet J., Xia X., Fellous M., Guiguen Y., et al. (2004). An evolutionary and functional analysis of FoxL2 in rainbow trout gonad differentiation. J. Mol. Endocrinol. 33, 705–715. doi: 10.1677/jme.1.01566
Barry T. P., Fast A. W. (1992). Biology of the spotted scat (Scatophagus argus) in the Philippines. Asian Fish Sci. 5, 163–179.
Bellefroid E. J., Leclère L., Saulnier A., Keruzore M., Sirakov M., Vervoort M., et al. (2013). Expanding roles for the evolutionarily conserved dmrt sex transcriptional regulators during embryogenesis. Cell. Mol. Life Sci. 70, 3829–3845. doi: 10.1007/S00018-013-1288-2/FIGURES/4
Bertho S., Pasquier J., Pan Q., Le Trionnaire G., Bobe J., Postlethwait J. H., et al. (2016). Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex Dev. 10, 111–129. doi: 10.1159/000447611
Bewick A. J., Anderson D. W., Evans B. J. (2011). Evolution of the closely related, sex-related genes dm-w and dmrt1 in african clawed frogs (xenopus). Evol. (N. Y). 65, 698–712. doi: 10.1111/j.1558-5646.2010.01163.x
Binnerts M. E., Kim K. A., Bright J. M., Patel S. M., Tran K., Zhou M., et al. (2007). R-Spondin1 regulates wnt signaling by inhibiting internalization of LRP6. Proc. Natl. Acad. Sci. U. S. A. 104, 14700–14705. doi: 10.1073/PNAS.0702305104
Biswas G., Sundaray J. K., Bhattacharyya S. B., Kailasam M. (2016). Evaluation of growth performance and survival of wild collected spotted scat , Scatophagus argus ( Linnaeus , 1766 ) during rearing of fry to marketable size juveniles for aquarium trade at varied stocking densities. J. Indian Soc Coast. Agric. Res. 34, 120–126.
Brunner B. (2001). Genomic organization and expression of the doublesex- related gene cluster in vertebrates and detection of putative regulatory regions for DMRT1. Genomics 77, 8–17. doi: 10.1006/geno.2001.6615
Budd A., Banh Q., Domingos J., Jerry D. (2015). Sex control in fish: approaches, challenges and opportunities for aquaculture. J. Mar. Sci. Eng. 3, 329–355. doi: 10.3390/jmse3020329
Cao Z. M., Qiang J., Zhu J. H., Li H. X., Tao Y. F., He J., et al. (2022). Transcriptional inhibition of steroidogenic factor 1 in vivo in Oreochromis niloticus increased weight and suppressed gonad development. Gene 809, 146023. doi: 10.1016/J.GENE.2021.146023
Carcasson R. H. (1977). A field guide to the coral reef fishes of the Indian and West pacific oceans (London: Collins).
Chen J. L. (2018). The role of elongation factor eEF1A1b and 42Sp50 in gametogenesis in Nile tilapia (China: Southwest University School of Life Science).
Chen L., Jiang X., Feng H., Shi H., Sun L., Tao W., et al. (2016). Simultaneous exposure to estrogen and androgen resulted in feminization and endocrine disruption. J. Endocrinol. 228, 205–218. doi: 10.1530/JOE-15-0432
Chen J., Jiang D., Tan D., Fan Z., Wei Y., Li M., et al. (2017). Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, oreochromis niloticus. Sci. Rep. 2017 71 7, 1–15. doi: 10.1038/srep43733
Chen H., Li Z., Wang Y., Huang H., Yang X., Li S., et al. (2021). Comparison of gonadal transcriptomes uncovers reproduction-related genes with sexually dimorphic expression patterns in diodon hystrix. Animals 11, 1042. doi: 10.3390/ani11041042
Chi W., Gao Y., Hu Q., Guo W., Li D. (2017). Genome-wide analysis of brain and gonad transcripts reveals changes of key sex reversal-related genes expression and signaling pathways in three stages of Monopterus albus. PLoS One 12, e0173974. doi: 10.1371/JOURNAL.PONE.0173974
Chue J., Smith C. A. (2011). Sex determination and sexual differentiation in the avian model. FEBS J. 278, 1027–1034. doi: 10.1111/j.1742-4658.2011.08032.x
Cui Z., Liu Y., Wang W., Wang Q., Zhang N., Lin F., et al. (2017). Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 7, 1–10. doi: 10.1038/srep42213
Cutting A. D., Ayers K., Davidson N., Oshlack A., Doran T., Sinclair A. H., et al. (2014). Identification, expression, and regulation of anti-müllerian hormone type-II rreceptor in the embryonic chicken gonad. Biol. Reprod. 90, 1–12. doi: 10.1095/biolreprod.113.116491
Cutting A., Chue J., Smith C. A. (2013). Just how conserved is vertebrate sex determination? Dev. Dyn. 242, 380–387. doi: 10.1002/dvdy.23944
Dai S., Qi S., Wei X., Liu X., Li Y., Zhou X., et al. (2021). Germline sexual fate is determined by the antagonistic action of dmrt1 and foxl3/foxl2 in tilapia. Development 148 dev199380. doi: 10.1242/dev.199380
Deschamps S., Morales J., Mazabraud A., Le Maire M., Denis H., Brown D. D. (1991). Two forms of elongation factor 1 alpha (EF-1 alpha O and 42Sp50), present in oocytes, but absent in somatic cells of xenopus laevis. J. Cell Biol. 114, 1109–1111. doi: 10.1083/JCB.114.6.1109
Díaz N., Ribas L., Piferrer F. (2014). Effects of changes in food supply at the time of sex differentiation on the gonadal transcriptome of juvenile fish. implications for natural and farmed populations. PLoS One 9. e111304. doi: 10.1371/journal.pone.0111304
Eshel O., Shirak A., Dor L., Band M., Zak T., Markovich-Gordon M., et al. (2014). Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15, 774. doi: 10.1186/1471-2164-15-774
Fast A., W., of Hawaii at Manoa. Mariculture Research, U., Center, T., of Marine Biology, H. I., and of Fisheries, I. S. C (1988). Spawning induction and pond culture of the spotted scat (Scatophagus argus Linnaeus) in the Philippines (University of Hawaii at Manoa: Mariculture Research and Training Center, Hawaii Institute of Marine Biology). Available at: https://books.google.com/books?id=t8GtHAAACAAJ.
Gandhi V. (2002). Studies on the food and feeding habits of cultivable butterfish Scatophagus argus (Cuv. and val.). J. Mar. Biol. Ass. India 44, 115–121.
Gandhi V., Venkatesan V., Zacharia P. U. (2013). Biometry analysis, length-weight relationship and sexual dimorphism of the spotted scat, scatophagus argus(Linnaeus 1766)(Perciformes: Scatophagidae) from gulf of mannar, southeast coast of India. J. Mar. Biol. Assoc. India 55, 12–16. doi: 10.6024/jmbai.2013.55.1.01743-02
Gao D., Li Z., Murphy T., Sauerbier W. (1997). Structure and transcription of the gene for translation elongation factor 1 subunit alpha of zebrafish (Danio rerio). Biochim. Biophys. Acta - Gene Struct. Expr. 1350, 1–5. doi: 10.1016/S0167-4781(96)00179-0
Gautier A., Le Gac F., Lareyre J.-J. (2011). The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472, 7–17. doi: 10.1016/J.GENE.2010.10.014
Hattori R. S., Fernandino J. I., Kishii A., Kimura H., Kinno T., Oura M., et al. (2009). Cortisol-induced masculinization: does thermal stress affect gonadal fate in pejerrey, a teleost fish with temperature-dependent sex determination? PLoS One 4, e6548. doi: 10.1371/journal.pone.0006548
Haugen T., Almeida F. F. L., Andersson E., Bogerd J., Male R., Skaar K. S., et al. (2012). Sex differentiation in Atlantic cod (Gadus morhua l.): morphological and gene expression studies. Reprod. Biol. Endocrinol. 10, 47–47. doi: 10.1186/1477-7827-10-47
He F. X., Jiang D. N., Huang Y. Q., Mustapha U. F., Yang W., Cui X. F., et al. (2019). Comparative transcriptome analysis of male and female gonads reveals sex-biased genes in spotted scat (Scatophagus argus). Fish Physiol. Biochem. 45, 1963–1980. doi: 10.1007/s10695-019-00693-8
Horiguchi R., Nozu R., Hirai T., Kobayashi Y., Nakamura M. (2018). Expression patterns of sex differentiation-related genes during gonadal sex change in the protogynous wrasse, halichoeres trimaculatus. Gen. Comp. Endocrinol. 257, 67–73. doi: 10.1016/j.ygcen.2017.06.017
Huang Y., Huang Y., Deng Q., Mustapha U. F., Peng Y., Guangli L., et al. (2022). A rapid method for genetic sex identification in the spotted scat (Scatophagus argus). J. Fish Sci. China 29, 515–524. doi: 10.12264/JFSC2021-0359
Huang Y., Jiang D., Li M., Mustapha U. F., Tian C., Chen H., et al. (2019). Genome survey of male and female spotted scat (Scatophagus argus). Animals 9, 1117. doi: 10.3390/ani9121117
Imai T., Saino K., Matsuda M. (2015). Mutation of gonadal soma-derived factor induces medaka XY gonads to undergo ovarian development. Biochem. Biophys. Res. Commun. 467, 109–114. doi: 10.1016/j.bbrc.2015.09.112
Infante C., Asensio E., Cañavate J., Manchado M. (2008). Molecular characterization and expression analysis of five different elongation factor 1 alpha genes in the flatfish Senegalese sole (Solea senegalensis kaup): differential gene expression and thyroid hormones dependence during metamorphosis. BMC Mol. Biol. 9 19. doi: 10.1186/1471-2199-9-19
Jiang M., Jia S., Chen J., Chen K., Ma W., Wu X., et al. (2020). Timing of gonadal development and dimorphic expression of sex-related genes in gonads during early sex differentiation in the yellow river carp. Aquaculture 518, 734825. doi: 10.1016/j.aquaculture.2019.734825
Jiang D. N., Mustapha U. F., Shi H. J., Huang Y. Q., Si-Tu J. X., Wang M., et al. (2019). Expression and transcriptional regulation of gsdf in spotted scat (Scatophagus argus). Comp. Biochem. Physiol. Part - B Biochem. Mol. Biol. 233, 35–45. doi: 10.1016/j.cbpb.2019.04.002
Jiang D. N., Peng Y. X., Liu X. Y., Mustapha U. F., Huang Y. Q., Shi H. J., et al. (2022). Homozygous mutation of gsdf causes infertility in female Nile tilapia ( Oreochromis niloticus). Front. Endocrinol. (Lausanne) 13. doi: 10.3389/FENDO.2022.813320
Jiang D. N., Yang H. H., Li M. H., Shi H. J., Zhang X. B., Wang D. S. (2016). Gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508. doi: 10.1002/mrd.22642
Johnsen H., Andersen Ø. (2012). Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. the fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505, 221–232. doi: 10.1016/J.GENE.2012.06.021
Kaneko H., Ijiri S., Kobayashi T., Izumi H., Kuramochi Y., Wang D. S., et al. (2015). Gonadal soma-derived factor (gsdf), a TGF-beta superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol. Cell. Endocrinol. 415, 87–99. doi: 10.1016/j.mce.2015.08.008
Kikuchi M., Nishimura T., Ishishita S., Matsuda Y., Tanaka M. (2020). Foxl3, a sexual switch in germ cells, initiates two independent molecular pathways for commitment to oogenesis in medaka. Proc. Natl. Acad. Sci. U. S. A. 117, 12174–12181. doi: 10.1073/PNAS.1918556117/SUPPL_FILE/PNAS.1918556117.SAPP.PDF
Kikuchi M., Nishimura T., Saito D., Shigenobu S., Takada R., Gutierrez-Triana J. A., et al. (2019). Novel components of germline sex determination acting downstream of foxl3 in medaka. Dev. Biol. 445, 80–89.
Kinoshita M., Okamoto G., Hirata T., Shinomiya A., Kobayashi T., Kubo Y., et al. (2009). Transgenic medaka enables easy oocytes detection in live fish. Mol. Reprod. Dev. 76, 202–207. doi: 10.1002/MRD.20942
Klüver N., Pfennig F., Pala I., Storch K., Schlieder M., Froschauer A., et al. (2007). Differential expression of anti-müllerian hormone (amh) and anti-müllerian hormone receptor (amhrII) in the teleost medaka. Dev. Dyn. 236, 271–281. doi: 10.1002/DVDY.20997
Kobayashi T., Kajiura-Kobayashi H., Guan G., Nagahama Y. (2008). Sexual dimorphic expression of DMRT1 and Sox9a during gonadal differentiation and hormone-induced sex reversal in the teleost fish Nile tilapia (Oreochromis niloticus). Dev. Dyn. 237, 297–306. doi: 10.1002/dvdy.21409
Kobayashi T., Matsuda M., Kajiura-Kobayashi H., Suzuki A., Saito N., Nakamoto M., et al. (2004). Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, oryzias latipes. Dev. Dyn. 231, 518–526. doi: 10.1002/dvdy.20158
Kohli S., Gulati P., Narang A., Maini J., Shamsudheen K. V., Pandey R., et al. (2021). Genome and transcriptome analysis of the mealybug Maconellicoccus hirsutus: Correlation with its unique phenotypes. Genomics 113, 2483–2494. doi: 10.1016/J.YGENO.2021.05.014
Kondo M., Froschauer A., Kitano A., Nanda I., Hornung U., Volff J.-N., et al. (2002). Molecular cloning and characterization of DMRT genes from the medaka Oryzias latipes and the platyfish xiphophorus maculatus. Gene 295, 213–222. doi: 10.1016/S0378-1119(02)00692-3
Lacour F., Vezin E., Bentzinger C. F., Sincennes M. C., Giordani L., Ferry A., et al. (2017). R-spondin1 controls muscle cell fusion through dual regulation of antagonistic wnt signaling pathways. Cell Rep. 18, 2320. doi: 10.1016/J.CELREP.2017.02.036
Lau E. S. W., Zhang Z., Qin M., Ge W. (2016). Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci. Rep. 6, 37357. doi: 10.1038/srep37357
Lin X., Tian C., Huang Y., Shi H., Li G. (2021a). Comparative transcriptome analysis identifies candidate genes related to black-spotted pattern formation in spotted scat (Scatophagus argus). Animals 11, 765. doi: 10.3390/ani11030765
Lin X., Zhou D., Zhang X., Li G., Zhang Y., Huang C., et al. (2021b). A first insight into the gonad transcriptome of Hong Kong catfish (Clarias fuscus). Animals 11, 1131. doi: 10.3390/ani11041131
Loeber J., Claußen M., Jahn O., Pieler T. (2010). Interaction of 42Sp50 with the vegetal RNA localization machinery in xenopus laevis oocytes. FEBS J. 277, 4722–4731. doi: 10.1111/J.1742-4658.2010.07878.X
Lyu Q., Hu J., Yang X., Liu X., Chen Y., Xiao L., et al. (2019). Expression profiles of dmrts and foxls during gonadal development and sex reversal induced by 17α-methyltestosterone in the orange-spotted grouper. Gen. Comp. Endocrinol. 274, 26–36. doi: 10.1016/J.YGCEN.2018.12.014
Mankiewicz J. L., Godwin J., Holler B. L., Turner P. M., Murashige R., Shamey R., et al. (2013). Masculinizing effect of background color and cortisol in a flatfish with environmental sex-determination. Integr. Comp. Biol. 53, 755–765. doi: 10.1093/icb/ict093
Marshall Graves J. A. (2008). Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 42, 565–586. doi: 10.1146/ANNUREV.GENET.42.110807.091714
Masuyama H., Yamada M., Kamei Y., Fujiwara-Ishikawa T., Todo T., Nagahama Y., et al. (2012). Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by dmy in the medaka. Chromosom. Res. 20, 163–176. doi: 10.1007/s10577-011-9264-x
Matson C. K., Murphy M. W., Sarver A. L., Griswold M. D., Bardwell V. J., Zarkower D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104. doi: 10.1038/nature10239
Matsuda M. (2005). Sex determination in the teleost medaka, Oryzias latipes.Annual review of genetics. 39, 293–307. doi: 1146/ANNUREV.GENET.39.110304.095800
Megbowon I., Mojekwu T. O. (2014). Tilapia sex reversal using methyl testosterone (MT) and its effect on fish, man and environment. Biotechnology 13, 213–216. doi: 10.3923/BIOTECH.2014.213.216
Minkina A., Matson C. K., Lindeman R. E., Ghyselinck N. B., Bardwell V. J., Zarkower D. (2014). DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev. Cell 29, 511–520. doi: 10.1016/J.DEVCEL.2014.04.017
Murakami Y., Horibe T., Kinoshita M. (2019). Development of an efficient bioreactor system for delivering foreign proteins secreted from liver into eggs with a vitellogenin signal in medaka oryzias latipes. Fish Sci. 85, 677–685. doi: 10.1007/S12562-019-01320-4/FIGURES/3
Mustapha U. F., Huang Y., Huang Y. Q., Assan D., Shi H. J., Jiang M. Y., et al. (2021). Gonadal development and molecular analysis revealed the critical window for sex differentiation, and E2 reversibility of XY-male spotted scat, Scatophagus argus. Aquaculture 544, 737147. doi: 10.1016/J.AQUACULTURE.2021.737147
Mustapha U. F., Jiang D. N., Liang Z. H., Gu H.-T., Yang W., Chen H. P., et al. (2018). Male-Specific Dmrt1 is a candidate sex determination gene in spotted scat (Scatophagus argus). Aquaculture 495, 351–358. doi: 10.1016/j.aquaculture.2018.06.009
Nakamoto M., Matsuda M., Wang D. S., Nagahama Y., Shibata N. (2006). Molecular cloning and analysis of gonadal expression of Foxl2 in the medaka, oryzias latipes. Biochem. Biophys. Res. Commun. 344, 353–361. doi: 10.1016/J.BBRC.2006.03.137
Nakamura S., Watakabe I., Nishimura T., Picard J. Y., Toyoda A., Taniguchi Y., et al. (2012). Hyperproliferation of mitotically active germ cells due to defective anti-müllerian hormone signaling mediates sex reversal in medaka. Development 139, 2283–2287. doi: 10.1242/DEV.076307
Nishimura T., Tanaka M. (2016). The mechanism of germline sex determination in vertebrates. Biol. Reprod. 95, 1–6. doi: 10.1095/BIOLREPROD.115.138271
Nowell M. A., Power D. M., Guerreiro P. M., Llewellyn L., Ramsurn V., Wigham T., et al. (2000). Cloning and expression of an elongation factor-1α in sea bream (Sparus aurata) larvae and adult tissue. Mar. Biotechnol. 2000 2 (2), 173–179. doi: 10.1007/S101269900022
Pfennig F., Standke A., Gutzeit H. O. (2015). The role of amh signaling in teleost fish – multiple functions not restricted to the gonads. Gen. Comp. Endocrinol. 223, 87–107. doi: 10.1016/J.YGCEN.2015.09.025
Poonlaphdecha S., Pepey E., Canonne M., de Verdal H., Baroiller J. F., D’Cotta H. (2013). Temperature induced-masculinisation in the Nile tilapia causes rapid up-regulation of both dmrt1 and amh expressions. Gen. Comp. Endocrinol. 193, 234–242. doi: 10.1016/J.YGCEN.2013.06.007
Ranjan R., Xavier B., Dash B., Edward L. L., Suresh R. D., Kumar P. S. (2015). Efficacy of 17 α-methyl testosterone and letrozole on sex reversal of protogynous grouper, Epinephelus tauvina (Forskal 1775) and spawning performance of sex-reversed males. Aquac. Res. 46, 2065–2072. doi: 10.1111/are.12360
Santos E. M., Paull G. C., Van Look K. J. W., Workman V. L., Holt W. V., van Aerle R., et al. (2007). Gonadal transcriptome responses and physiological consequences of exposure to oestrogen in breeding zebrafish (Danio rerio). Aquat. Toxicol. 83, 134–142. doi: 10.1016/J.AQUATOX.2007.03.019
Schulz R. W., Bogerd J., Male R., Ball J., Fenske M., Olsen L. C., et al. (2007). Estrogen-induced alterations in amh and dmrt1 expression signal for disruption in male sexual development in the zebrafish. Environ. Sci. Technol. 41, 6305–6310. doi: 10.1021/es070785+
Shi S., Chen T., Zhao X. (2013). Comparative transcriptome analysis for metabolic engineering. Methods Mol. Biol. 985, 447–458. doi: 10.1007/978-1-62703-299-5_22
Shi Y., Wang Q., He M. (2014). Molecular identification of dmrt2 and dmrt5 and effect of sex steroids on their expressions in chlamys nobilis. Aquaculture 426–427, 21–30. doi: 10.1016/J.AQUACULTURE.2014.01.021
Sun L., Teng J., Zhao Y., Li N., Wang H., Ji X., et al. (2018). Gonad transcriptome analysis of high-temperature-treated females and high-temperature-induced sex-reversed neomales in Nile tilapia. Int. J. Mol. Sci. 19, 689. doi: 10.3390/ijms19030689
Tao W., Yuan J., Zhou L., Sun L., Sun Y., Yang S., et al. (2013). Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS One 8, 63604. doi: 10.1371/journal.pone.0063604
Teng J., Zhao Y., Chen H. J., Wang H., Ji X. S. (2020). Transcriptome profiling and analysis of genes associated with high temperature–induced masculinization in sex-undifferentiated Nile tilapia gonad. Mar. Biotechnol. 22, 367–379. doi: 10.1007/s10126-020-09956-5
Tian C., Li Z., Dong Z., Huang Y., Du T., Chen H., et al. (2019). Transcriptome analysis of male and female mature gonads of silver sillago (Sillago sihama). Genes (Basel) 10. doi: 10.3390/genes10020129
Vizziano D., Baron D., Randuineau G., Mahè S., Cauty C., Guiguen Y. (2008). Rainbow trout gonadal masculinization induced by inhibition of estrogen synthesis is more physiological than masculinization induced by androgen supplementation. Biol. Reprod. 78, 939–946. doi: 10.1095/BIOLREPROD.107.065961
Vizziano D., Randuineau G., Baron D., Cauty C., Guiguen Y. (2007). Characterization of early molecular sex differentiation in rainbow trout, oncorhynchus mykiss. Dev. Dyn. 236, 2198–2206. doi: 10.1002/dvdy.21212
Wang D., Kobayashi T., Zhou L., Nagahama Y. (2004). Molecular cloning and gene expression of Foxl2 in the Nile tilapia, Oreochromis niloticus. Biochem. Biophys. Res. Commun. 320, 83–89. doi: 10.1016/J.BBRC.2004.05.133
Wang D. S., Kobayashi T., Zhou L. Y., Paul-Prasanth B., Ijiri S., Sakai F., et al. (2007). Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21, 712–725. doi: 10.1210/me.2006-0248
Wang Q., Liu K., Feng B., Zhang Z., Wang R., Tang L., et al. (2019). Transcriptome of gonads from high temperature induced sex reversal during sex determination and differentiation in Chinese tongue sole, Cynoglossus semilaevis. Front. Genet. 10. doi: 10.3389/FGENE.2019.01128
Wang W., Zhu H., Dong Y., Dong T., Tian Z., Hu H. (2018). Identification and dimorphic expression of sex-related genes during gonadal differentiation in sterlet Acipenser ruthenus, a primitive fish species. Aquaculture 500, 178–187. doi: 10.1016/j.aquaculture.2018.10.001
Webster K. A., Schach U., Ordaz A., Steinfeld J. S., Draper B. W., Siegfried K. R. (2017). Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 422, 33–46. doi: 10.1016/j.ydbio.2016.12.008
Wu G. C., Chiu P. C., Lyu Y. S., Chang C. F. (2010). The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Biol. Reprod. 83, 443–453. doi: 10.1095/BIOLREPROD.110.084681
Wu F., Wu L., Wu Q., Zhou L., Li W., Wang D. (2018). Duplication and gene expression patterns of β-catenin in Nile tilapia. Fish Physiol. Biochem. 44, 651–659. doi: 10.1007/S10695-017-0460-2
Xie Q. P., He X., Sui Y. N., Chen L. L., Sun L. N., Wang D. S. (2016). Haploinsufficiency of SF-1 causes female to male sex reversal in Nile tilapia, oreochromis niloticus. Endocrinology 157, 2500–2514. doi: 10.1210/en.2015-2049
Yamaguchi T., Kitano T. (2012). High temperature induces cyp26b1 mRNA expression and delays meiotic initiation of germ cells by increasing cortisol levels during gonadal sex differentiation in Japanese flounder. Biochem. Biophys. Res. Commun. 419, 287–292. doi: 10.1016/J.BBRC.2012.02.012
Yamaguchi A., Lee K. H., Fujimoto H., Kadomura K., Yasumoto S., Matsuyama M. (2006). Expression of the DMRT gene and its roles in early gonadal development of the Japanese pufferfish takifugu rubripes. Comp. Biochem. Physiol. Part D Genomics Proteomics 1, 59–68. doi: 10.1016/J.CBD.2005.08.003
Yamaguchi T., Yamaguchi S., Hirai T., Kitano T. (2007). Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, paralichthys olivaceus. Biochem. Biophys. Res. Commun. 359, 935–940. doi: 10.1016/J.BBRC.2007.05.208
Yamaguchi T., Yoshinaga N., Yazawa T., Gen K., Kitano T. (2010). Cortisol is involved in temperature-dependent sex determination in the Japanese flounder. Endocrinology 151, 3900–3908. doi: 10.1210/EN.2010-0228
Yang L., Zhang X., Liu S., Zhao C., Miao Y., Jin L., et al. (2021). Cyp17a1 is required for female sex determination and Male fertility by regulating sex steroid biosynthesis in fish. Endocrinology 162, bqab205. doi: 10.1210/ENDOCR/BQAB205
Yao Z. L., Chen H. J., Zhao Y., Cao Z. J., Wang H., Ji X. S. (2021). A time course transcriptome analysis of brains from sex-undifferentiated Nile tilapia discloses genes associated with high-temperature-induced masculinization. Aquaculture 530, 735762. doi: 10.1016/J.AQUACULTURE.2020.735762
Yarmohammadi M., Pourkazemi M., Kazemi R. (2017). Differential expression of foxl2 and cyp19a1a mRNA during gonad developmental stages in great sturgeon Huso huso. J. Fish Biol. 90, 1104–1111. doi: 10.1111/JFB.13224
Ye B., Wan Z., Wang L., Pang H., Wen Y., Liu H., et al. (2017). Heritability of growth traits in the Asian seabass (Lates calcarifer). Aquac. Fish 2, 112–118. doi: 10.1016/J.AAF.2017.06.001
Yin Y., Tang H., Liu Y., Chen Y., Li G., Liu X., et al. (2017). Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 158, 3030–3041. doi: 10.1210/en.2016-1865
Yoshinaga N., Shiraishi E., Yamamoto T., Iguchi T., Abe S. I., Kitano T. (2004). Sexually dimorphic expression of a teleost homologue of müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, paralichthys olivaceus. Biochem. Biophys. Res. Commun. 322, 508–513. doi: 10.1016/J.BBRC.2004.07.162
Zhai G., Shu T., Chen K., Lou Q., Jia J., Huang J., et al. (2022). Successful production of an all-female common carp (Cyprinus carpio l.) population using cyp17a1-deficient neomale carp. Engineering 8, 181–189. doi: 10.1016/J.ENG.2021.03.026
Zhai G., Shu T., Xia Y., Lu Y., Shang G., Jin X., et al. (2018). Characterization of sexual trait development in cyp17a1-deficient zebrafish. Endocrinology 159, 3549–3562. doi: 10.1210/EN.2018-00551
Zhang X., Li M., Ma H., Liu X., Shi H., Li M., et al. (2017). Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX nile tilapia. Endocrinology 158, 2634–2647. doi: 10.1210/en.2017-00127
Zhu Y., Cui Z., Yang Y., Xu W., Shao C., Fu X., et al. (2019). Expression analysis and characterization of dmrt2 in Chinese tongue sole (Cynoglossus semilaevis). Theriogenology 138, 1–8. doi: 10.1016/j.theriogenology.2019.06.035
Zhu Q., Han C., Liu S., Ouyang H., Liu D., Zhang Z., et al. (2022). Development and gene expression analysis of gonad during 17α-methyltestosterone-induced sex reversal in mandarin fish (Siniperca chuatsi). Aquac. Rep. 23, 101049. doi: 10.1016/J.AQREP.2022.101049
Keywords: transcriptomics, differentiating gonads, gonadal sex differentiation, gene expression, Scatophagus argus
Citation: Mustapha UF, Peng Y-X, Huang Y-Q, Assan D, Zhi F, Shi G, Huang Y, Li G-L and Jiang D-N (2022) Comparative transcriptome analysis of the differentiating gonads in Scatophagus argus. Front. Mar. Sci. 9:962534. doi: 10.3389/fmars.2022.962534
Received: 06 June 2022; Accepted: 13 July 2022;
Published: 03 August 2022.
Edited by:
Rui Jia, Freshwater Fisheries Research Center (CAFS), ChinaReviewed by:
Hongwei Yan, Dalian Ocean University, ChinaCopyright © 2022 Mustapha, Peng, Huang, Assan, Zhi, Shi, Huang, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong-Neng Jiang, ZG5qaWFuZ0BnZG91LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.