- 1College of Agriculture, Ludong University, Yantai, China
- 2Institute of Quality Standards and Testing Technology for Agricultural Products, Chinese Academy of Agricultural Sciences, Beijing, China
Abalone (Haliotis discus hannai Ino) is one of the most vital economic shellfish cultured in China. However, bacterial and viral infection often leads to its high mortality and serious economic losses. In this study, transcriptome sequencing was performed on gill of abalone challenged by Vibrio parahaemolyticus for 0 h (control group, A0h), 8 h (A8h), and 48 h (A48h), respectively. Each library has an average of 53,361,675 clean reads. Approximately 85.71% reads were mapped to the reference genome of Haliotis discus hannai Ino. In comparison with A0h group, 7,656 and 2,789 genes were differentially expressed in the A8h and A48h groups, respectively. The differentially expressed genes (DEGs) were mostly enriched in oxidation–reduction process and oxidoreductase activity from GO analysis as well as immunity and metabolism-related pathways from Kyoto Encyclopedia of Genes and Genomes analysis. Cluster analysis demonstrated that all DEGs were categorized into eight subclusters by adopting the gene expression model. Specifically, a total of 1,070 significantly downregulated genes from two clusters were related to some metabolic pathways, ABC transporters, and cell adhesion. Moreover, 212 upregulated genes in one subcluster were enriched in several immune-related pathways, including nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway and apoptosis. Some members of the pattern recognition receptors family, which can initiate inflammatory signaling and induce apoptosis, were stimulated to upregulate in gill after challenge including C1q, perlucin, and low-density lipoprotein receptor–related protein. In addition, caspase-dependent apoptosis pathway was inactive, whereas caspase-independent apoptosis pathway was stimulating in response to Vibrio challenge. To eliminate invading pathogen, the immune effectors such as interleukin-17, matrix metalloproteinase, lysozyme, superoxide, and ferritin were also stimulated in gill. Moreover, this study also provides more information for comprehending the immunity and defense mechanisms against V. parahaemolyticus infection in abalone.
Introduction
Haliotis discus hannai Ino is a precious marine shellfish with high nutritional and economic value in China. According to the statistical analysis from China Fisheries Yearbook 2020, the yield of abalone in mariculture was 180,267 tons in 2019. However, abalone is susceptible to bacteria (Cai et al., 2008), virus, and parasites (Chang et al., 2005), especially under stressed conditions. This will lead to the high mortality and generate the serious economic loss. Among the conditioned pathogens, Vibrio parahaemolyticus is one of the main bacteria causing diseases for mollusks (Yu et al., 2013; Mok et al., 2019). Therefore, understanding the immune response of abalone against V. parahaemolyticus stress has great significance for its aquaculture production.
Similar to most invertebrates, abalone mainly depends on innate immunity to defense the infection of bacteria or virus due to the lack of adaptive immunity (Nam et al., 2016). Innate immune system consists of cellular and humoral immunity in organism. The first step to stimulate the immune system is to recognize the pathogen-associated molecular patterns (PAMPs) of pathogens via pattern recognition receptors (PRRs) of immune cells. Then, PRRs trigger a series of extracellular cascades, thereby stimulating signal transduction pathways and ultimately activating the effector response system (Janeway and Medzhitov, 2002). Recently, some PRRs have been identified in abalone including C-type lectins (CTLs) (Zhang et al., 2014), galectin (Maldonado-Aguayo et al., 2014), perlucin (Weiss et al., 2000), toll-like receptor (TLR) (Priyathilaka et al., 2019), and peptidogly (Premachandra et al., 2014).
In recent years, transcriptome analysis has become an efficient method to investigate the underlining mechanism of the immunity activation stimulated by pathogens challenge in aquatic species. Some results have been reported in mollusks including Pacific oyster Crassostrea gigas (de Lorgeril et al., 2011), pearl oyster Pinctada martensii (Wang et al., 2016), razor clam Sinonovacula constricta (Zhao et al., 2017), Manila clam Ruditapes philippinarum (Ren et al., 2021), surf clam Mactra veneriformis (Fang et al., 2013), hard clam Meretrix petechialis (Yu et al., 2019), Yesso scallop Patinopecten yessoensis (Jiang et al., 2018), and soft-shell clam Mya arenaria (Araya et al., 2010). Moreover, there are also some research studies concerning the response of abalone against Vibrio challenge (Silva-Aciares et al., 2013; Nam et al., 2016). However, there is no transcriptome analysis about the immunity response against V. parahaemolyticus challenge in gill of abalone. Gill is the first line of defense for substance exchange between internal and external environment, which is of great importance for abalone immunity. Moreover, the results in this study can help us to well comprehend the underlining mechanism about gill immune defense against V. parahaemolyticus invasion in abalone.
Materials and methods
Abalone culture and bacterial challenge
Juvenile abalones (Haliotis discus hannai Ino) were purchased from HaiYi hatchery Co. (Yantai, China) and fed with the formulated diet in a closed circulating water system within 24-h dark photoperiod for 100 days. The production of the manufactured feed (formula in Supplementary Table 1) and challenge method were previously described (Meng et al., 2019). Briefly, 150 abalones with the average shell length of 42.40 mm and the average weight of 5.80 g were averagely divided into three aquariums (15 L) with 50 individuals in each. Before challenge, pooled gills from five abalones in each aquarium were collected and immediately frozen in liquid nitrogen (recorded as A0h). Then, the rest of abalones were Vibrio challenged by both muscle injection (100 μl of bacterial suspension at the final concentration of 4 × 107 CFU/ml) and bath treatment (3 ml of bacterial suspension in each aquarium). After exposure for 8 and 48 h, pooled gill samples were collected from each aquarium (recorded as A8h and A48h) and frozen immediately. All gill samples were stored at −80°C for RNA extraction and transcriptome sequencing.
Library construction and sequencing
Total RNA was extracted from pooled gills of control group (A0h) and challenged groups (A8h and A48h), respectively, using TRIzol reagent. The RNA quality and purity were checked by agarose gel electrophoresis and Nano Photometer® spectrophotometer (IMPLEN, USA). Then, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and subsequently broken into short fragments with fragmentation buffer. The RNA fragments purified with AMPure XP system (Beckman Coulter, Beverly, USA) were reversely transcribed to produce double-strand cDNA using random hexamer primers, which were then ligated to an adapter. The ligation products were amplified by PCR with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer to construct the cDNA sequence library. Then, the library quality was assessed on the Agilent Bio analyzer 2100 system before it was sequenced on an Illumina Hiseq platform.
Transcriptome data analysis
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. The filtering criteria are presented as follows: (1) reads with adapters were removed; (2) reads containing N (N means that the base information could not be determined) were removed; and (3) low-quality reads were removed. Meanwhile, Q20, Q30, and guanine and cytosine (GC) content of the clean data were calculated. All the downstream analyses based on the clean data with high quality. All clean reads were mapped to the abalone reference genome (GenBank accession: PRJNA317403) using HISAT2 v2.0.4 with default parameters (Kim et al., 2015). H. discus hannai reference genome and gene model annotation files were downloaded from http://gigadb.org/dataset/100281.
HTSeq v0.9.1 was applied to count the reads numbers mapped to each gene. Fragments per kilo base of transcript per million fragments mapped (FPKM) value was used to represent expression abundance of the genes as it simultaneously considered the effect of sequencing depth and gene length for the reads count (Trapnell et al., 2010).
Identification and analysis of differentially expressed genes
Differential expression analysis of two groups (A8h vs. A0h and A48h vs. A0h) with three biological replicates per group was performed using the DESeqR package (1.18.0). Genes with an adjusted P-value (Q-value) < 0.05 and log2 (fold change) > 1 were assigned as differentially expressed (Wang et al., 2010).
To further understand the differentially expressed genes (DEGs) and affected pathways responsive to V. parahaemolyticus challenge, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed by adopting the GOseq R package and KOBAS software, respectively (Mao et al., 2005). GO enrichment analysis is conducive to describing gene product attributes in all organisms by mapping unigenes to functional categories, whereas the KEGG enrichment analysis is useful to map unigenes onto known signaling pathways. GO terms and KEGG pathways with corrected P < 0.05 were considered significantly enriched by DEGs.
Moreover, cluster analysis is used to judge the expression patterns of DEGs under different experimental conditions using the Novomagic, which is an online platform for data analysis (https://magic.novogene.com). Then, KEGG enrichment analysis was further performed on the basis of the clustered genes to reveal the possible functional pathway of DEGs with similar expression model.
Validation by quantitative real-time PCR
To confirm the RNA sequencing (RNA-seq) data, eight immune-related DEGs were selected for quantitative real-time PCR (qRT-PCR). The forward and reverse primers of the selected genes were shown in Table 1. Ribosomal Protein S9 (RPS9, EU247757.1) was selected as the internal reference gene. RNA was extracted from polled gill samples of abalone and then reversely transcribed into cDNA template. qRT-PCR with 20-µl volume was performed using the SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biology, China) in ABI 7500 Real-Time PCR System (USA) in line with the introduction. qRT-PCR procedure was set as 95°C for 30 s, 40 cycles of 95°C for 5 s and 55°C for 30 s, followed by the melting curve determination. All samples were performed in triplicates. The relative expressions of immune related genes were calculated by adopting the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Results
Illumina sequencing and data filtering
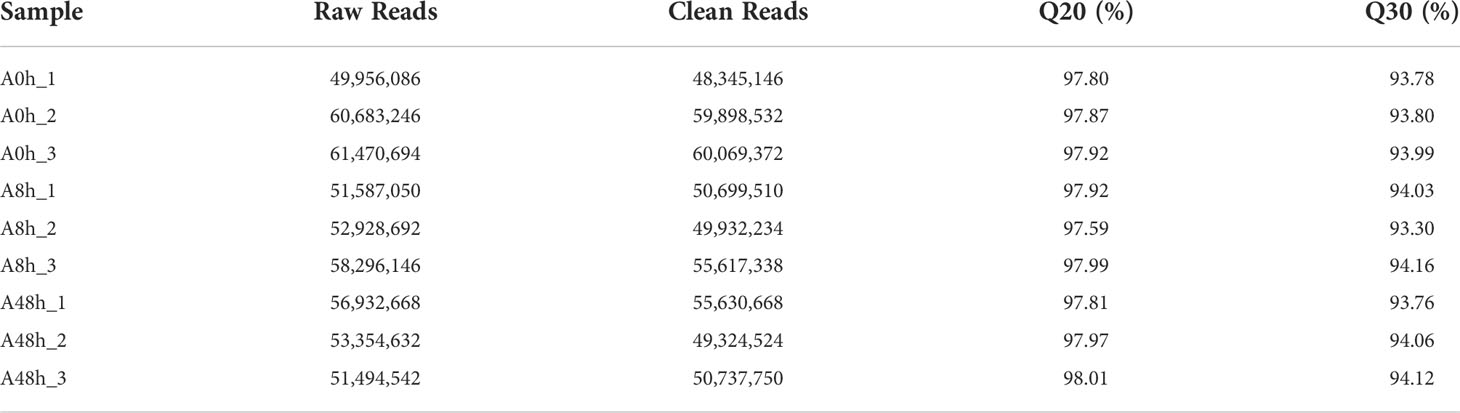
RNA-seq was performed on polled gill samples of A0h, A8h, and A48h groups with three replicates. A total of 57,370,009, 54,270,629, and 53,927,281 raw reads (average) in gill were obtained, respectively. The raw data were uploaded to Sequence Read Archive (SRA) of NCBI with the accession number PRJNA743940. After filtering the low-quality raw reads, the libraries produced 56,104,350, 52,083,027, and 51,897,647 clean reads on average, respectively. Q20 and Q30 of all samples were above 97% and 93%, respectively. The transcriptome data analysis was summarized in Table 2.
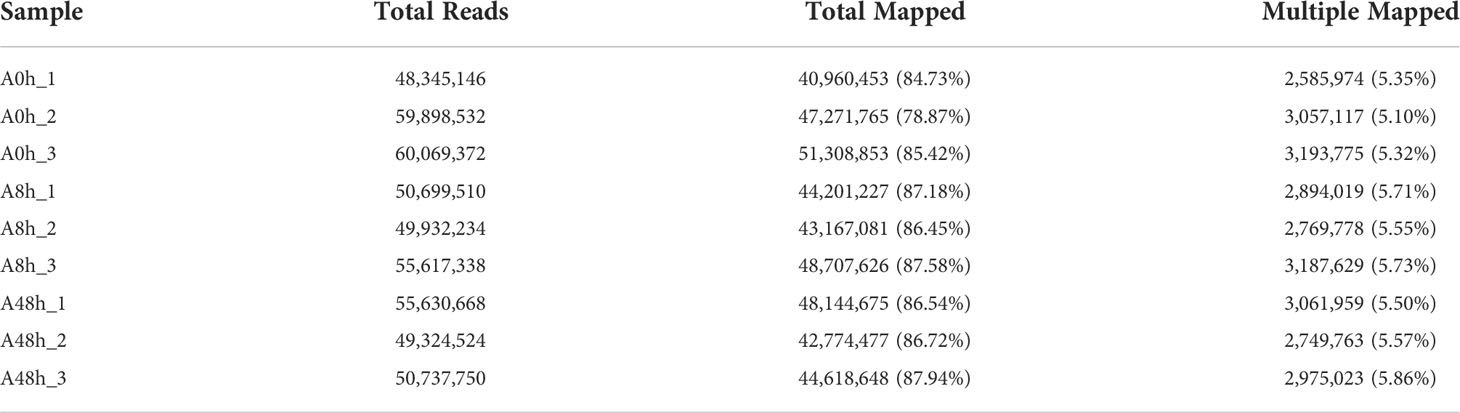
HISAT software was selected to perform genomic localization analysis of the spliced reads (Kim et al., 2015). If the reference genome is selected appropriately and the experiment is free of contamination, then the percentage of reads generated by the experiment that are localized (total mapped reads or fragments) will normally be higher than 70%, and reads with multiple localizations (multiple mapped reads or fragments) are usually no more than 10%. As shown in the results, a total of 411.12 M reads were mapped to the H. discus hannai reference genome (GenBank accession: PRJNA317403). Total mapping rates were ranging from 78.87% to 87.94%, and the multiple mapping rates were 5.1% to 5.86%. The detailed information was summarized in Table 3.
Analysis of DEGs
The transcriptome results were applied as a reference to explore the overall gene expression in gill of abalone challenged by V. parahaemolyticus at 8 and 48 h in comparison with unchallenged group. Altogether, 7,656 genes were differently expressed in the comparison of A8h vs. A0h, including 3,453 upregulated genes and 4,203 downregulated genes (Figure 1A). In addition, 2,789 DEGs were identified in the comparison of A48h vs. A0h containing 1,096 upregulated genes and 1,693 downregulated genes (Figure 1B). The Venn diagram showed that a total of 8,322 genes were significantly differentially expressed in the gill of abalone after V. parahaemolyticus challenge. Among them, 2,123 genes were significantly expressed both at 8- and 48-h treatments (Figure 1C). Some DEGs were involved in various processes of immune defense against pathogens and were classified into various functions, such as PRRs, apoptosis, cytokines and cytokine receptors, signaling transduction, and oxidative stress (Supplementary Table 2).
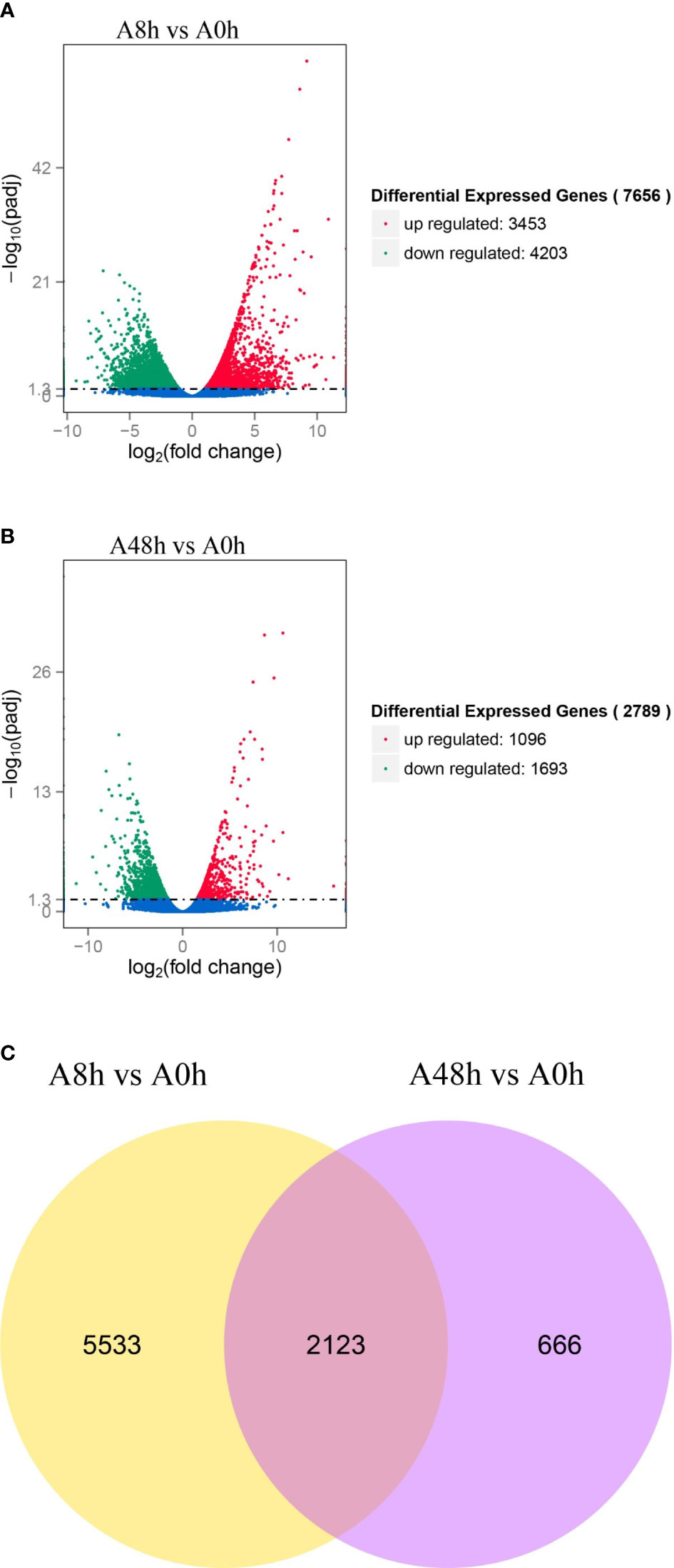
Figure 1 (A) Volcano plot of differentially expressed genes (DEGs) distribution trends between A8h vs. A0h sample. (B) Volcano plot of DEGs distribution trends between A48h vs. A0h sample. Each dot in the volcano plot represents a detected gene. Genes with significant differential expression are denoted by red dots (upregulated) and green dots (downregulated). Genes without significant differential expression are indicated by blue dots. Horizontal ordinate signifies the fold change of DEGs, whereas vertical ordinate stands for statistical significance of DEGs between A8h vs. A0h and A48h vs. A0h groups. (C) Venn diagrams of DEGs. There are 7,656 and 2,789 DEGs between A8h vs. A0h and A48h vs. A0h, respectively. Among all DEGs, a total of 2,123 differential genes were overlapped expressed both in A48h and A8h groups compared with unchallenged group.
GO and KEGG analysis of DEGs
To further understand the function of DEGs, GO and KEGG enrichment were analyzed on the basis of 8,399 DEGs. In terms of the GO analysis, all DEGs are classified into three GO categories: biological process, molecular function, and cellular component, respectively. The 30 most enriched terms under bacterial challenge were shown in Figure 2A. Most DEGs under these terms were associated with the oxidation–reduction process or other oxidative stress processes. The redox of cell membranes and intracellular molecules leads to structural or conformational changes and exerts an important regulatory function in signal transduction and gene transcription, which makes a significant impact on cellular life activities.
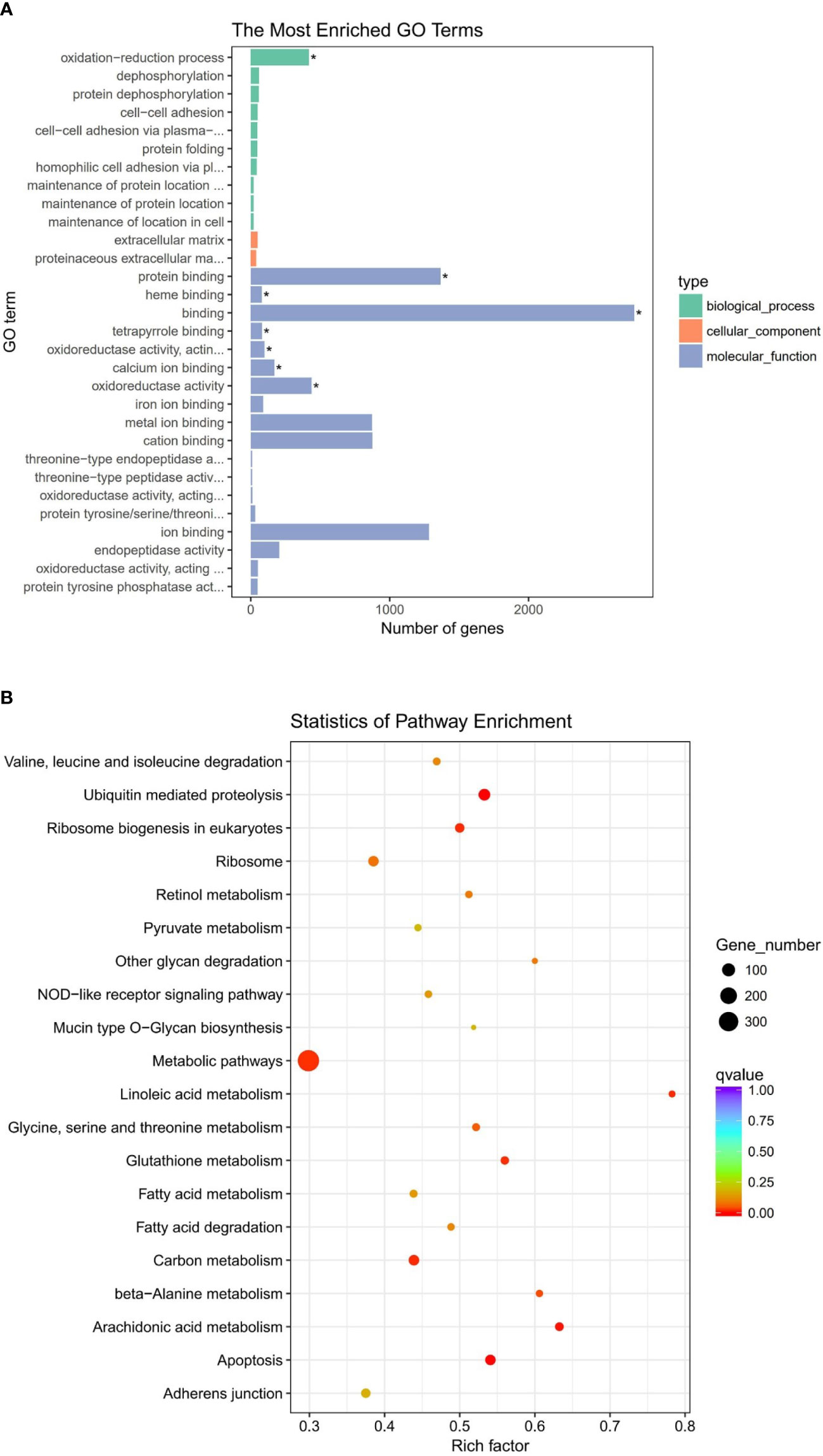
Figure 2 (A) Gene ontology (GO) enrichment analysis of differentially expressed genes (DEGs). Significantly enriched GO term was highlighted with star (*) inside the graph (adjusted P < 0.05). (B) Scatter plot of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs. The vertical axis represents the pathway terms, whereas the horizontal axis signifies the rich factor. The size of the dot represents the number of DEGs in this pathway, and the color of the dot is consistent with different Q-value ranges.
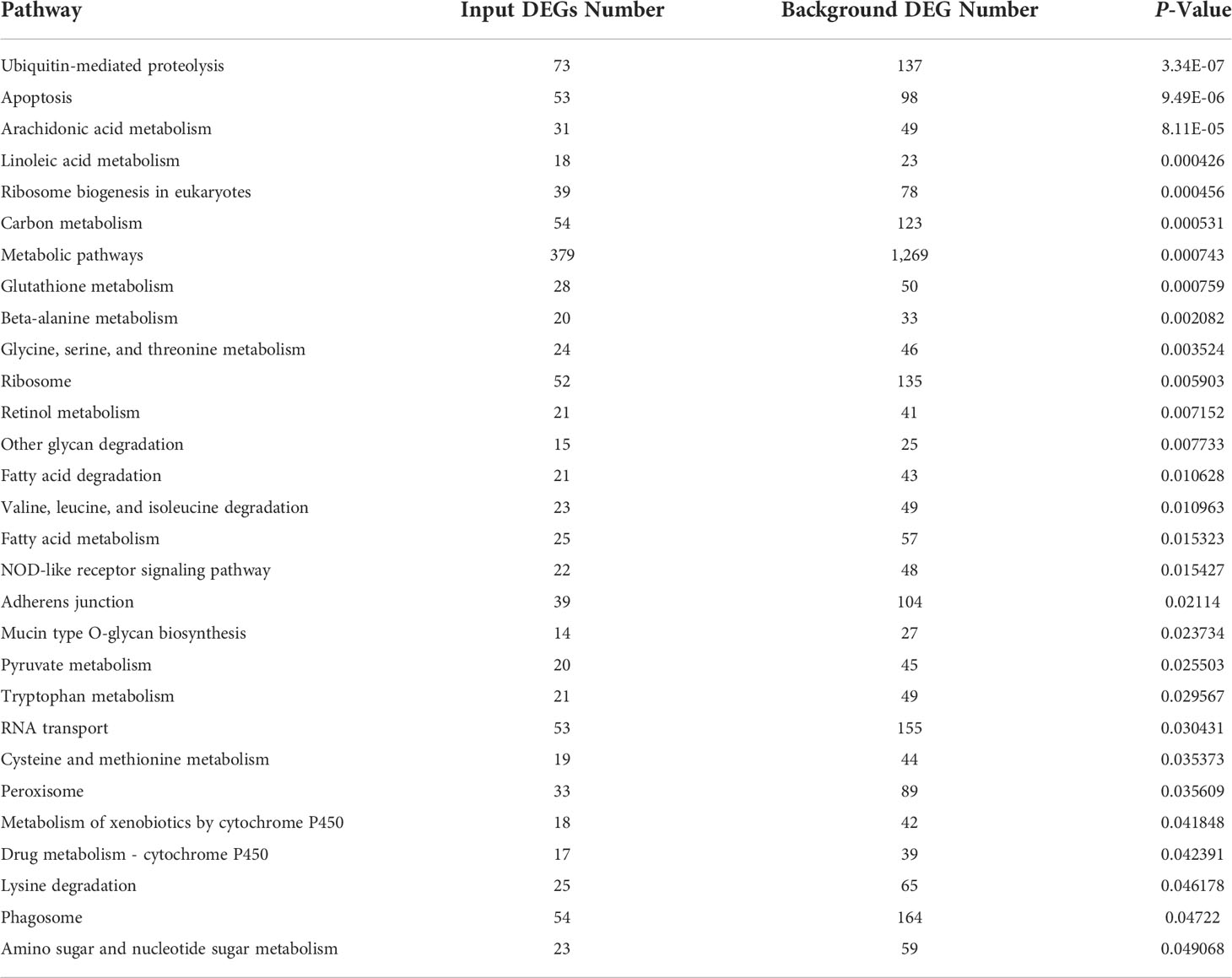
In KEGG pathway analysis, a total of 8,399 DEGs were mapped to 152 KEGG pathways. The scatter plots of top 20 KEGG enrichment pathways were performed in Figure 2B. Among all pathways, metabolism was the predominantly enriched group. Afterward, some immune-related pathways, such as apoptosis, NOD-like receptor signaling pathway, and phagosomes, are significantly enriched in gill of abalone after Vibrio challenge (P < 0.05) (Table 4). The GO and KEGG analysis can make us comprehensively understand the immune response of abalone gill facing with V. parahaemolyticus challenge.
Cluster analysis of DEGs
The clustering of genes with the same or similar expression patterns into classes can be applied to infer the functions of genes. Different clustering algorithms divided the differential genes into several clusters. The genes in the same cluster had similar expression trends under different conditions. The DEGs of abalone after Vibrio challenge were selected and explored. The results showed that the expression profiles of all the DEGs could be divided into eight subclusters (Figure 3), displaying different kinds of expression pattern. Moreover, 1,070 downregulated genes in subcluster_1 and 8 (Figure 4A) and 212 upregulated gens in subcluster_7 were separately enriched to different pathways (Figure 4B). Generally, some metabolic pathways, ABC transporters, and cell adhesion were inhibited after Vibrio infection, whereas several immune-related pathways, containing NOD-like receptor signaling pathway and apoptosis, were stimulated upregulated in gill of abalone for immune defense of Vibrio invasion.
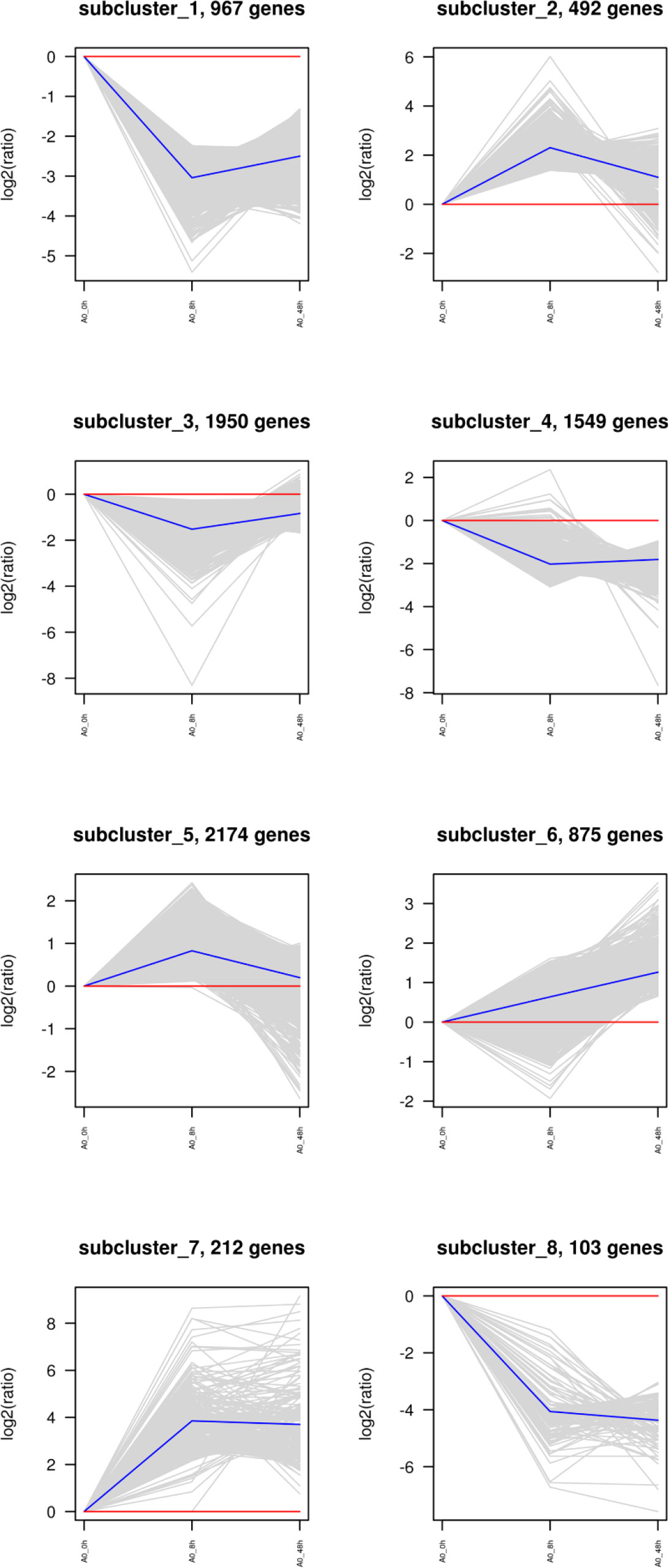
Figure 3 Cluster analysis of all differentially expressed genes (DEGs) between challenged groups (8 h and 48 h) and control group (0 h).
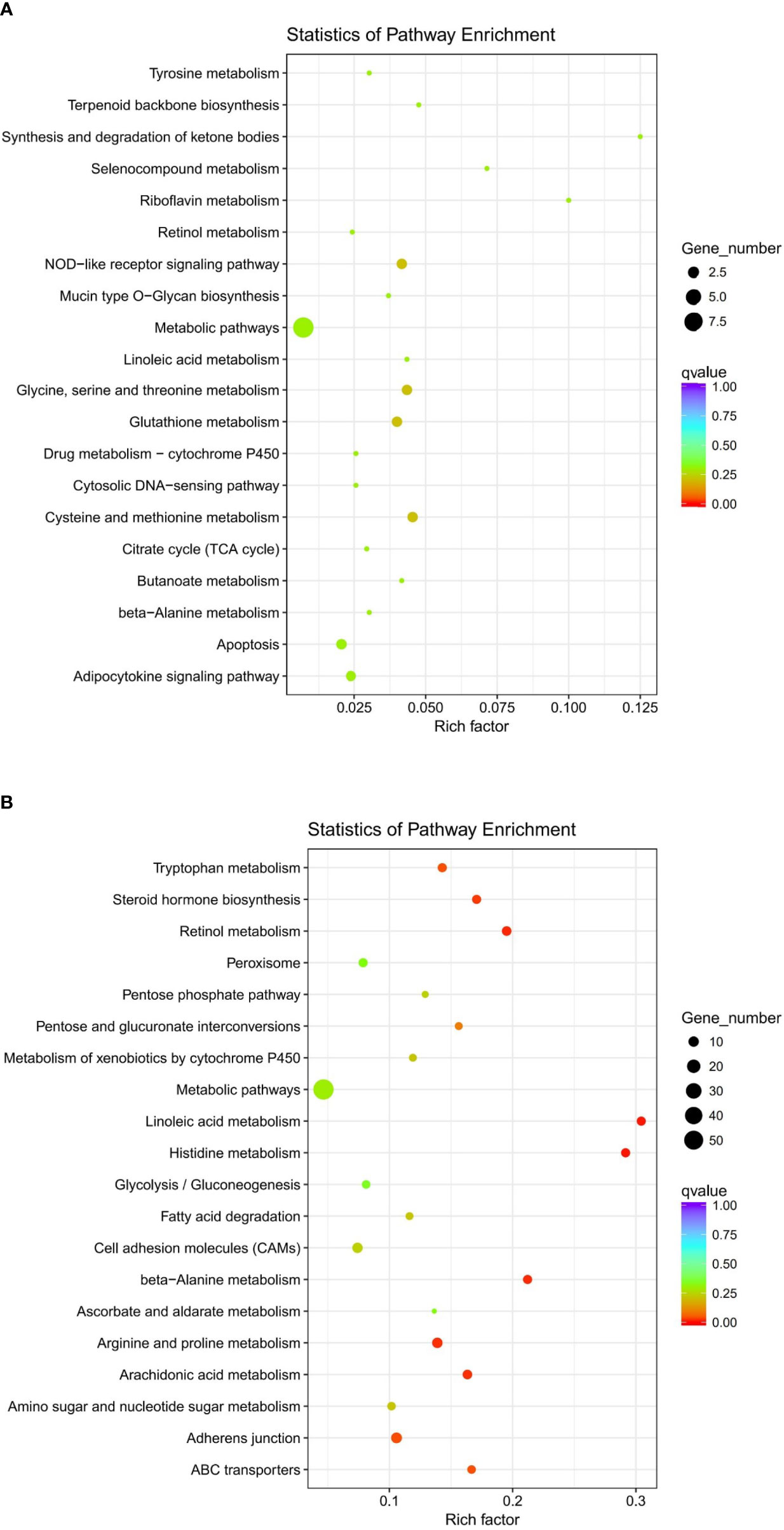
Figure 4 Scatter plot of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of differentially expressed genes (DEGs) by performing cluster analysis. (A) Enriched KEGG pathways of DEGs from subcluster_7. (B) Enriched KEGG pathways of DEGs from subcluster_1 and 8.
Validation of DEGs by qRT-PCR
To verify the expression of genes in transcriptome data, eight immune-related DEGs were selected for qRT-PCR, including matrix metalloproteinase 1 (MMP1), inhibitor of NF-κBα (NFκBIα), caspase 8, ferritin, toll-like receptor 6 (TLR6), glutamine synthetase, tumor necrosis factor α (TNFα), and interferon regulatory factor 1 (IRF1). As displayed in Figure 5, the expressions of MMP1, NFκBIα, glutamine synthetase, and ferritin were upregulated, whereas the expressions of caspase 8, TNFα, TLR6, and IRF1 were downregulated. The gene expression result obtained by qRT-PCR basically conformed to transcriptome data at each challenge time points.
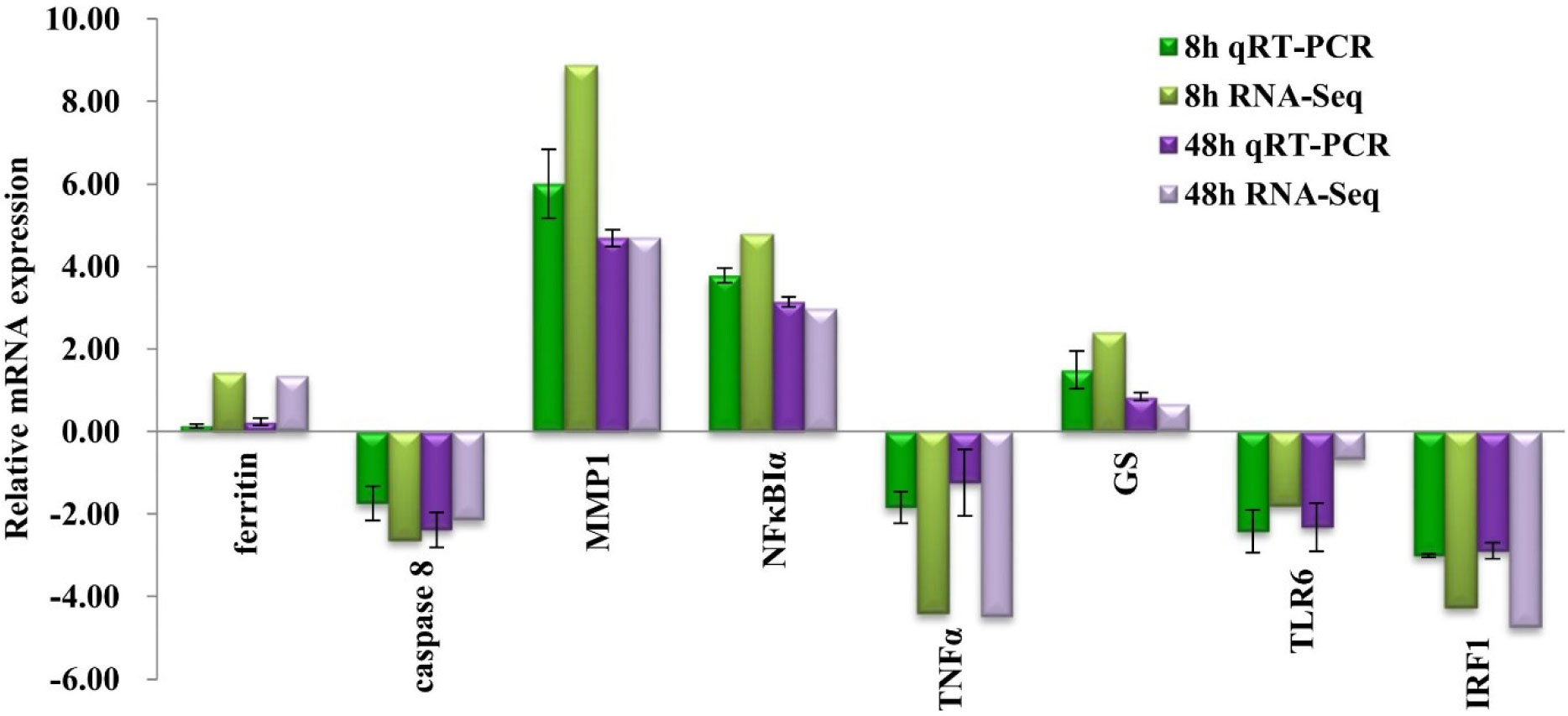
Figure 5 Expression patterns of selected differentially expressed genes (DEGs) at challenge time of 8 and 48 h compared with control group (A0h) with triplicates by qRT-PCR and RNA-seq analysis. The bars above the horizontal axis show that genes whose expression levels have increased after challenge, whereas those below the axis indicate the genes with decreased expression after Vibrio treatment.
Discussion
Invertebrates mainly rely on innate immunity against pathogen invasion due to the lack of antibodies or specific immune cells. The PRRs on the surface of natural immune cells can recognize and interact with PAMPs of pathogens to stimulate the immune response. In this study, we found that the expressions of some genes related to phagocytosis, apoptosis, and immune effectors were significantly affected by Vibrio challenge, which might indicate their specific function in the immune response of gill in abalone.
Abalone initiates phagocytosis to eliminate invading Vibrio
Phagocytosis is a critical step in the host defense mechanism with the purpose of eliminating pathogens. After phagosomes combined with lysosomes, the pathogens can be degraded by various hydrolases existing in lysosomes. Cathepsin, a kind of lysosomal protease, is a vital regulator in the process of antigen presentation and processing as well as inflammatory response (Turk et al., 2002). The expression of cathepsin L was significantly increased in gill of abalone after Vibrio challenge, which was consistent with the results reported in Pacific abalone (Haliotis discus hannai) (Shen et al., 2015), pearl oyster (Pinctada fucata) (Ma et al., 2010), freshwater mussel (Cristaria plicata) (Hu et al., 2014), and sea cucumber (Apostichopus japonicus) (Yang et al., 2016). Nevertheless, cathepsin B was significantly decreased, which was consistent with the transcriptome result from Nile tilapia (Zhu et al., 2017) but contradictory to the findings from golden pompano (Trachinotus ovatus) (Shen et al., 2021), flounder (Paralichthys olivaceus) (Zhang et al., 2008), and razor clam (Sinonovacula constricta) (Niu et al., 2013). The contradictory results may be associated with the challenge time or species dependence. Moreover, further studies need to be conducted to explain the detailed immune function of cathepsin family in shellfish.
Previous studies have found that C1q is an activation ligand for phagocytosis (Bohlson et al., 2007). C1q belongs to the members of defense collagen family that is characterized by a globular carboxyl terminal portion with the function of recognizing PAMPs of pathogens and the collagen-like N-terminal domain linking the invading organism to phagocyte effector mechanisms of the immune system. Three C1q domain containing (C1qDC) proteins were reported to be strongly induced by both Gram-negative and Gram-positive bacterial in abalone, indicating their involvement in immune response against invading pathogens (Bathige et al., 2016). In addition, C1qDC protein can act as a PRR to mediate phagocytosis against pathogen invasion in Crassostrea gigas (Li et al., 2019) and Pinctada fucata martensii (Liang et al., 2022). According to our results, C1q, C1qDC, C1q, TNF-related protein, and C1q subcomponent subunit B were all upregulated after challenge, indicating that they may promote phagocytosis against Vibrio invasion as a kind of PRR in gill of abalone.
In addition to promoting phagocytosis, C1q also triggers superoxide production following the interaction of C1q collagenous tails with C1q-receptoron the surface of phagocytes to eliminate the invading pathogens (Tenner, 1999). Nicotinamide adenine dinucleotide phosphate (NADPH) oxidase is responsible for the production of superoxide in organisms. The appearance of superoxide will activate antioxidation enzymes including superoxide dismutase (SOD), glutathione peroxidase (GPx), or catalase (CAT), aiming to reduce the damage caused by peroxidation (Bhagat et al., 2016). On the basis of our result, Vibrio challenge significantly increases the expressions of GPx and NADPH oxidase, which may indicate the superoxide production in gill of abalone for bacterial elimination. Briefly, abalone can stimulate the phagocytosis and superoxide productions mediated by C1q and increase the hydrolases activities of phagolysosome to eliminate invading bacteria.
Abalone promote apoptosis and clearance of apoptotic cells to eliminate the invading Vibrio
Apoptosis, called as programmed cell death process, can eliminate injured or infected cells and play a vital role in the stability of intracellular groups, immunity, and morphogenesis (Edinger and Thompson, 2004). TNFα is a cytokine naturally produced by various cells in response to pathogen infection or other stresses (Goetz et al., 2004; De Zoysa et al., 2009). TNFα interacts with the transmembrane receptors on the membrane of infected cell to stimulate the apoptotic signal mediated by TNF receptor-associated death domain (TRADD) and FAS-associated death domain (FADD) and triggers downstream caspase cascade reaction (Sheikh and Huang, 2003). The activated caspase-3 cleaved DNA fragmentation factor 45 (DFF45) into DFF40 that cleaved DNA into fragments (Zhang and Xu, 2000). Moreover, this is the process of exogenous apoptosis inside organisms. Unexpectedly, there are also some factors inhibiting apoptosis such as inhibitor of apoptosis family of proteins (IAP) (Uren et al., 1998). According to the obtained results, Vibrio infection significantly decreased the gene expressions of TNFα, caspase-3, and caspase-7, whereas increased the mRNA level of c-IAP1 in gill of abalone. It seems like that the caspase-dependent apoptosis in gill of abalone tends to be suppressed after Vibrio attack. Studies have demonstrated that lipopolysaccharide, a component of the outer membrane of Gram-negative bacteria, can directly bind and inhibit caspase and eventually inhibit cell apoptosis (Günther et al., 2020). This may be the reason why the expressions of caspases are decreased and caspase-dependent apoptosis is inactive after V. parahaemolyticus challenge in gill of abalone.
Interestingly, caspase-independent apoptosis is found to be involved in the immune defense of abalone as identified in the result. Apoptosis-inducing factor (AIF) is a conservative flavoprotein that exists in the mitochondria intermembrane space. When stimulated by apoptosis, AIF is hydrolyzed into mature proteins with apoptotic potential (Polster et al., 2005). Then, mature AIF proteins translocate into the nucleus, which can lead to chromatin condensation and large-scale DNA fragmentation, resulting in the occurrence of apoptosis (Joza et al., 2001). The involvement of AIF in the innate immune response has already been reported in abalone (Elvitigala et al., 2015) and oyster (Qiao et al., 2021).Voltage-dependent anion channel (VDAC), a kind of porin present in the mitochondrial outer membrane of all eukaryotes, is responsible for transport of low–molecular weight metabolites across the outer membrane of mitochondria (Benz, 1994). Studies have shown that VDAC is a key element in mitochondria-mediated apoptosis and can control the release of several apoptosis-related factors (Shoshan-Barmatz et al., 2010). As shown in our result, the expressions of AIF and VDAC were significantly upregulated in gill of abalone after challenge, suggesting the possibility of AIF release into nucleus to initiate the caspase-independent apoptosis. In general, it is extremely interesting to find that the caspase-dependent apoptosis pathway was passive and inactive, whereas caspase-independent apoptosis pathway was active and stimulating in gill of abalone after Vibrio challenge. Specific experiment is needed for approval in the future.
Many researchers have found that C1q plays a prominent role in the rapid clearance of apoptotic cells (Taylor et al., 2000; Ogden et al., 2001; Vandivier et al., 2002). C1q binds to apoptotic cells and cellular debris through its globular heads recognizing apoptotic cell-associated molecular patterns (ACAMPs) to promote the clearance of apoptotic cells (Gregory, 2000; Navratil et al., 2001). It appears that C1q is a crucial immune element in respond to Vibrio invasion in gill of abalone, which is worthy of investigation in the future.
The immune response of some PRRs and cytokines induced by Vibrio challenge
Perlucin, a typical CTLs protein, was reported to have important function in process of non–self-recognition and disease resistance in abalone (Yao et al., 2021). Recently, Bi et al. reported that perlucin can influence the phagocytosis and the expressions of antimicrobial peptides in white shrimp by binding and agglutinating the bacteria (Bi et al., 2020). As shown in our results, the expressions of perlucins (3 and 5) were significantly upregulated after Vibrio challenge (Supplementary Table 2), verifying its involvement in the immune response in gill of abalone as a kind of PRR.
Low-density lipoprotein receptor–related protein (LRP) is reported to be an indispensable molecule in the recognition of innate immunity. In addition, it can regulate signal transduction, cytokine secretion, phagocytosis, and cell migration of the immune system (Li et al., 2001; Strickland and Ranganathan, 2003; May, 2013). LRP was reported to recognize and eliminate the invading pathogens in immune defense of scallop (Liu et al., 2014) and amphioxus (Gao et al., 2017). According to our result, LRP-1 was significantly upregulated, whereas other types of LRP (4, 6, and 12) showed the downregulated model after bacterial challenge (Supplementary Table 2). This may indicate that LRP-1 acts as a PRR to recognize and eliminate invading pathogens in abalone immune system, providing a new theoretical foundation for the role of LRP in the immunity of aquatic invertebrates.
Interleukin-17 (IL-17) is one of the leading signaling cytokine involved in the immune response of invertebrates and plays an important role in inducing inflammation and clearing extracellular bacteria. The expression of IL-17 was significantly upregulated in gill of abalone after challenge in our study, which was similar with the results from oysters (Roberts et al., 2008), mussels (Saco et al., 2020), and sea urchins (Buckley et al., 2017). Moreover, IL-17 can also stimulate the production of various pro-inflammatory mediators including MMP. MMP is a calcium-dependent endopeptidase, which exerts a vital role in apoptosis and tumor progression (McCawley and Matrisian, 2001). Chen et al. found that MMP-1 was involved in the regulation of phagocytosis, apoptosis and humoral immunity in Haliotis discus hannai and MMP1 silencing could lower the expression of some immune genes (Chen et al., 2019). As displayed in our result, MMP1 was also significantly upregulated after Vibrio challenge, suggesting that it plays a vital role in the innate immunity of abalone against V. parahaemolyticus.
Lysozyme is extensively regarded as a kind of antimicrobial peptide that takes part in the innate immune response against pathogen infection in animals. Lysozyme was reported to have antibacterial activity against Gram-negative or Gram-positive bacteria in White Shrimp (de-la-Re-Vega et al., 2006) and Pacific oyster (Itoh et al., 2010). The expression of lysozyme was significantly upregulated in abalone after infection by V. harveyi (Yao et al., 2021). In our result, the expression of G-type lysozyme was dramatically increased, whereas C-type lysozyme showed a decreased expression model in gill after challenge. The expression difference of the two types of lysozyme needs to be further investigated in future.
Ferritin is a soluble histone that stores iron in the body. The involvement of ferritin in the immune defense against pathogens was reported in some aquatic species, containing sea cucumber (Ren et al., 2014), Japanese scallop (Zhang et al., 2013), tiger prawn (Maiti et al., 2010), soft-shell clam (Araya et al., 2010), and abalone (He et al., 2016; Qiu et al., 2016). Moreover, this is consistent with our result that the expression of ferritin was also upregulated after challenge.
ATP-binding cassette transporter (ABC transporter) is a transporter on the cytoplasmic membrane, which can control the pumping and output of external substances and nutrients (Pradel et al., 2009). It was approved to be the first defensive barrier against pathogens in bivalves (Luckenbach and Epel, 2008). After Vibrio challenge, the expression of most ABC transporters in the gill of razor clam (Sinonovacula constricta) was downregulated. This reduced the import and export of substances and also contributed to the negative regulation of immune response (Zhao et al., 2017). In our result, the mRNA level of ABCA3 was also downregulated after V. parahaemolyticus challenge. Currently, the research studies of ABC transporter in shellfish are extremely limited. Therefore, the mechanism of ABC transporter in the immune defense of abalone remains to be explored in near future.
Conclusion
To conclude, this study confirmed that gill is not only a filtering tissue but also a sort of immune recognition tissue. Various PRRs have been demonstrated to be sensitive to V. parahaemolyticus invasion in our study including C1q, perlucin, and LRP. It is worth mentioning that C1q may play a predominant role in recognition, triggering and effector functions in gill of abalone. The defensive response against V. parahaemolyticus challenge in gill of abalone contains promoting the recognition of PAMPs by PRRs, initiating apoptosis and clearance of apoptotic cells, activating phagocytosis, as well as releasing some cytokines to comprehensively eliminate the invading pathogen (Figure 6). The obtained findings underscore the importance of gill as the main defensive tissue besides hemocytes, improving our knowledge of the efficient and complex innate immune system of abalone.
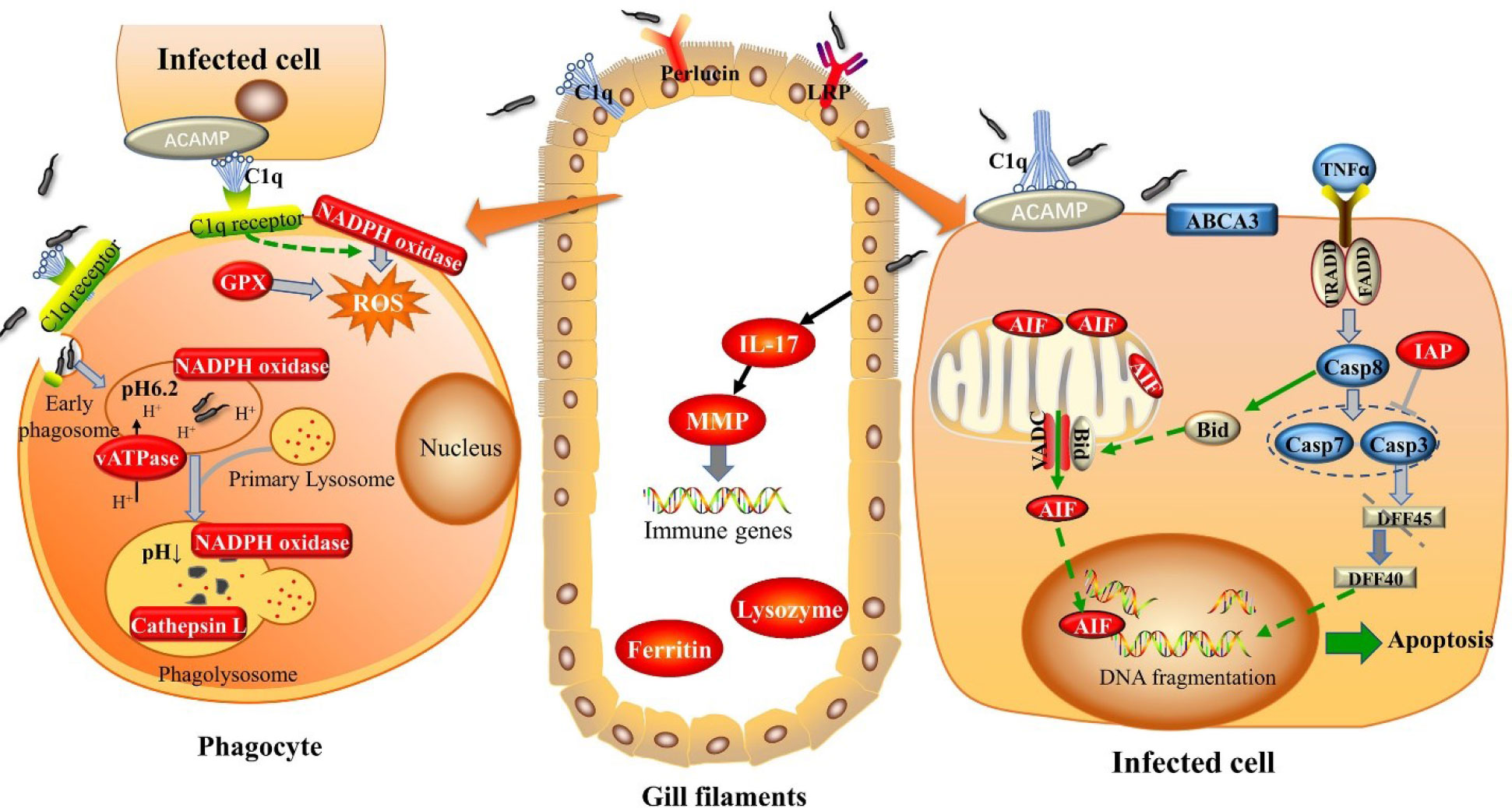
Figure 6 Graphic representation of the immune response in gill of abalone against Vibrio Parahemolyticus invasion.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/743940, PRJNA743940.
Ethics Statement
The animal study was reviewed and approved by Affidavit of Approval of the Institutional Review Board of Ludong University.
Author Contributions
YF: formal analysis, writing—review and editing, and funding acquisition; XY: formal analysis, writing—original draft, and sample analysis; SZ: visualization and methodology; XC: validation; GL: feeding trial and sample analysis; YZ: resources; MW: resources; ML: conceptualization, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The current work was supported by the National Key Research and Development Program of China, “Blue Granary Science And Technology Innovation” special project (2019YFD0902102); National Natural Science Foundation of China (No. 31502173); the Key Laboratory of Mariculture of Ministry of Education, Ocean University of China (No. KLM2018007); and Research Foundation of LuDong University (No. LY2015037).
Acknowledgments
We sincerely show our deepest appreciation to Novogene Co. for technical assistance in bioinformatics analysis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.956317/full#supplementary-material
References
Araya M. T., Markham F., Mateo D. R., McKenna P., Johnson G. R., Berthe F. C. J., et al. (2010). Identification and expression of immune-related genes in hemocytes of soft-shell clams, mya arenaria, challenged with vibrio splendidus. Fish. Shellfish. Immun. 29, 557–564. doi: 10.1016/j.fsi.2010.05.017
Bathige S. D. N. K., Umasuthan N., Jayasinghe J. D. H. E., Godahewa G. I., Park H., Lee J. (2016). Three novel C1q domain containing proteins from the disk abalone haliotis discus discus: Genomic organization and analysis of the transcriptional changes in response to bacterial pathogens. Fish. Shellfish. Immun. 56, 181–187. doi: 10.1016/j.fsi.2016.07.010
Benz R. (1994). Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. B. B. A. 1197, 167–196. doi: 10.1016/0304-4157(94)90004-3
Bhagat J., Ingole B. S., Singh N. (2016). Glutathione s-transferase, catalase, superoxide dismutase, glutathione peroxidase, and lipid peroxidation as biomarkers of oxidative stress in snails: A review. ISJ-Invert. Surviv. J. 13, 336–349. doi: 10.25431/1824-307x/isj.v13i1.336-349
Bi J. X., Ning M. X., Xie X. J., Fan W. F., Huang Y. L., Gu W., et al. (2020). A typical c-type lectin, perlucin-like protein, is involved in the innate immune defense of whiteleg shrimp litopenaeus vannamei. Fish. Shellfish. Immun. 103, 293–301. doi: 10.1016/j.fsi.2020.05.046
Bohlson S. S., Fraser D. A., Tenner A. J. (2007). Complement proteins C1q and MBL are pattern recognition molecules that signal immediate and long-term protective immune functions. Mol. Immunol. 44, 33–43. doi: 10.1016/j.molimm.2006.06.021
Buckley K. M., Ho E., Hibino T., Schrankel C. S., Schuh N. W., Wang G., et al. (2017). IL17 factors are early regulators in the gut epithelium during inflammatory response to vibrio in the sea urchin larva. eLife 6, e23481. doi: 10.7554/elife.23481.024
Cai J. P., Wang Z. X., Cai C. H., Zhou Y. P. (2008). Characterization and identification of virulent klebsiella oxytoca isolated from abalone (Haliotis diversicolor supertexta) postlarvae with mass mortality in fujian, China. J. Invertebr. Pathol. 97, 70–75. doi: 10.1016/j.jip.2007.07.005
Chang P. H., Kuo S. T., Lai S. H., Yang H. S., Ting Y. Y., Hsu C. L., et al. (2005). Herpes-like virus infection causing mortality of cultured abalone haliotis diversicolor supertexta in Taiwan. Dis. Aquat. Organ. 65, 23–27. doi: 10.3354/dao065023
Chen Y. L., Li W. Y., Hu J. J., Li Y., Liu G. M., Jin T. C., et al. (2019). Nucleus-translocated matrix metalloprotease 1 regulates innate immune response in pacific abalone (Haliotis discus hannai). Fish. Shellfish. Immun. 84, 290–298. doi: 10.1016/j.fsi.2018.10.017
de-la-Re-Vega E., García-Galaz A., Díaz-Cinco M. E., Sotelo-Mundo R. R. (2006). White shrimp (Litopenaeus vannamei) recombinant lysozyme has antibacterial activity against gram negative bacteria: Vibrio alginolyticus, vibrio parahemolyticus and vibrio cholera. Fish. Shellfish. Immun. 20, 405–408. doi: 10.1016/j.fsi.2005.06.005
de Lorgeril J., Zenagui R., Rosa R. D., Piquemal D., Bachère E. (2011). Whole transcriptome profiling of successful immune response to vibrio infections in the oyster crassostrea gigas by digital gene expression analysis. PLoS One 6, e23142. doi: 10.1371/journal.pone.0023142
De Zoysa M., Jung S., Lee J. (2009). First mollus can TNF-α homologue of the TNF superfamily in disk abalone: Molecular characterization and expression analysis. Fish. Shellfish. Immun. 2, 625–631. doi: 10.1016/j.fsi.2008.10.004
Edinger D. A. S., Thompson C. B. (2004). Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 16, 663–669. doi: 10.1016/j.ceb.2004.09.011
Elvitigala D. A., Jayasooriya R. G. P. T., Whang I., Lee J. (2015). First report on the gastropod proapoptotic AIF3 counterpart from disk abalone (Haliotis discus discus) deciphering its transcriptional modulation by induced pathogenic stress. Fish. Shellfish. Immun. 47, 697–705. doi: 10.1016/j.fsi.2015.10.006
Fang Y., Yang H. S., Liu B. Z., Zhang L. B. (2013). Transcriptional response of lysozyme, metallothionein, and superoxide dismutase to combined exposure to heavy metals and bacteria in mactra veneriformis. Comp. Biochem. Phys. C 157, 54–62. doi: 10.1016/j.cbpc.2012.10.002
Gao Z., Qu B. Z., Ma Z. Y., Jiao D. Y., Ji G. D., Zhang S. C. (2017). Identification and functional characterization of a novel member of low-density lipoprotein receptor-related protein (LRP)-like family in amphioxus. Gene 618, 42–48. doi: 10.1016/j.gene.2017.04.011
Goetz F. W., Planas J. V., MacKenzie S. (2004). Tumor necrosis factors. Dev. Comp. Immunol. 28, 487–497. doi: 10.1016/j.dci.2003.09.008
Gregory C. D. (2000). CD14-dependent clearance of apoptotic cells: relevance to the immune system. Curr. Opin. Immunol. 12, 27–34. doi: 10.1016/s0952-7915(99)00047-3
Günther S. D., Fritsch M., Seeger J. M., Schiffmann L. M., Snipas S. J., Coutelle M., et al. (2020). Cytosolic gram-negative bacteria prevent apoptosis by inhibition of effector caspases through lipopolysaccharide. Nat. Microbiol. 5, 354–367. doi: 10.1038/s41564-019-0620-5
He J., Jiang J. Z., Gu L., Zhao M. M., Wang R. X., Ye L. T., et al. (2016). Identification and involvement of ferritin in the response to pathogen challenge in the abalone, haliotis diversicolor. Dev. Comp. Immunol. 60, 23–32. doi: 10.1016/j.dci.2016.01.022
Hu X. J., Hu X. P., Hu B. Q., Wen C. G., Xie Y. H., Wu D., et al. (2014). Molecular cloning and characterization of cathepsin l from freshwater mussel, cristaria plicata. Fish. Shellfish. Immun. 40, 446–454. doi: 10.1016/j.fsi.2014.07.005
Itoh N., Okada Y., Takahashi K. G., Osada M. (2010). Presence and characterization of multiple mantle lysozymes in the pacific oyster, crassostrea gigas. Fish. Shellfish. Immun. 29, 126–135. doi: 10.1016/j.fsi.2010.02.027
Janeway C. A., Medzhitov J. R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi: 10.1146/annurev.immunol.20.083001.084359
Jiang W. W., Lin F., Fang J. G., Gao Y. P., Du M. R., Fang J. H., et al. (2018). Transcriptome analysis of the yesso scallop, patinopecten yessoensis gills in response to water temperature fluctuations. Fish. Shellfish. Immun. 80, 133–140. doi: 10.1016/j.fsi.2018.05.038
Joza N., Susin S. A., Daugas E., Stanford W. L., Cho S. K., Li C. Y. J., et al. (2001). Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410, 549–554. doi: 10.1038/35069004
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Liang X. R., Xiong X. W., Cao Y. F., Li Z. X., Chen J. Y., Jiao Y., et al. (2022). Globular C1q domain-containing protein from pinctada fucata martensii participates in the immune defense process. Fish. Shellfish. Immunol. 123, 521–527. doi: 10.1016/j.fsi.2022.02.048
Li H., Kong N., Sun J. J., Wang W. L., Li M. J., Gong C. H., et al. (2019). A C1qDC (CgC1qDC-6) with a collagen-like domain mediates hemocyte phagocytosis and migration in oysters. Dev. Comp. Immunol. 98, 157–165. doi: 10.1016/j.dci.2019.04.010
Liu R., Wang L. L., Sun Y., Wang L. L., Zhang H., Song L. S. (2014). A low-density lipoprotein receptor-related protein (LRP)-like molecule identified from chlamys farreri participated in immune response against bacterial infection. Fish. Shellfish. Immun. 36, 336–343. doi: 10.1016/j.fsi.2013.11.017
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li Y., van Kerkhof P., Marzolo M. P., Strous G. J., Bu G. (2001). Identification of a major cyclic AMP-dependent protein kinase a phosphorylation site within the cytoplasmic tail of the low-density lipoprotein receptor-related protein: implication for receptor-mediated endocytosis. Mol. Cell. Biol. 21, 1185–1195. doi: 10.1128/mcb.21.4.1185-1195.2001
Luckenbach T., Epel D. (2008). ABCB- and ABCC-type transporters confer multixenobiotic resistance and form an environment-tissue barrier in bivalve gills. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1919–R1929. doi: 10.1152/ajpregu.00563.2007
Maiti B., Khushiramani R., Tyagi A., Karunasagar I., Karunasagar I. (2010). Recombinant ferritin protein protects penaeus monodon infected by pathogenic vibrio harveyi. Dis. Aquat. Org. 88, 99–105. doi: 10.3354/dao02163
Maldonado-Aguayo W., Teneb J., Gallardo-Escárate C. (2014). A galectin with quadruple-domain from red abalone haliotis rufescens involved in the immune innate response against to vibrio anguillarum. Fish. Shellfish. Immun. 40, 1–8. doi: 10.1016/j.fsi.2014.06.013
Mao X. Z., Cai T., Olyarchuk J. G., Wei L. P. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
May P. (2013). The low-density lipoprotein receptor-related protein 1 in inflammation. Curr. Opin. Lipidol. 24, 134–137. doi: 10.1097/mol.0b013e32835e809c
Ma J. J., Zhang D. C., Jiang J. J., Cui S. G., Pu H. L., Jiang S. G. (2010). Molecular characterization and expression analysis of cathepsin L1 cysteine protease from pearl oyster pinctada fucata. Fish. Shellfish. Immun. 29, 501–507. doi: 10.1016/j.fsi.2010.05.006
McCawley L. J., Matrisian L. M. (2001). Matrix metalloproteinases: they’re not just for matrix anymore! Curr. Opin. Cell Biol. 13, 534–540. doi: 10.1016/s0955-0674(00)00248-9
Meng X. X., Yang X. Y., Lin G., Fang Y., Ruan Z. L., Liu M. F., et al. (2019). Mannan oligosaccharide increases the growth performance, immunity and resistance capability against vibrio parahemolyticus in juvenile abalone haliotis discus hannai ino. Fish. Shellfish. Immun. 94, 654–660. doi: 10.1016/j.fsi.2019.09.058
Mok J. S., Ryu A., Kwon J. Y., Kim B., Park K. (2019). Distribution of vibrio species isolated from bivalves and bivalve culture environments along the gyeongnam coast in Korea: Virulence and antimicrobial resistance of vibrio parahaemolyticus isolates. Food Control. 106, 106697. doi: 10.1016/j.foodcont.2019.06.023
Nam B., Jung M., Subramaniyam S., Yoo S., Markkandan K., Moon J., et al. (2016). Transcriptome analysis revealed changes of multiple genes involved in haliotis discus hannai innate immunity during vibrio parahemolyticus infection. PLoS One 11, e0153474. doi: 10.1371/journal.pone.0153474
Navratil J. S., Watkins S. C., Wisnieski J. J., Ahearn J. M. (2001). The globular heads of C1q specifically recognize surface blebs of apoptotic vascular endothelial cells. J. Immunol. 166, 3231–3239. doi: 10.4049/jimmunol.166.5.3231
Niu D. D., Jin K., Wang L., Sun F. Y., Li J. L. (2013). Identification of cathepsin b in the razor clam sinonovacula constricta and its role in innate immune responses. Dev. Comp. Immunol. 41, 94–99. doi: 10.1016/j.dci.2013.04.014
Ogden C. A., deCathelineau A., Hoffmann P. R., Bratton D., Ghebrehiwet B., Fadok V. A., et al. (2001). C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J. Exp. Med. 194, 781–795. doi: 10.1084/jem.194.6.781
Polster B. M., Basañez G., Etxebarria A., Hardwick J. M., Nicholls D. G. (2005). Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J. Biol. Chem. 280, 6447–6454. doi: 10.1074/jbc.M413269200
Pradel L. C., Mitchell A. J., Zarubica A., Dufort L., Chasson L., Naquet P., et al. (2009). ATP-binding cassette transporter hallmarks tissue macrophages and modulates cytokine-triggered polarization programs. Eur. J. Immunol. 39, 2270–2280. doi: 10.1002/eji.200838867
Premachandra H. K. A., Elvitigala D. A. S., Whang I., Lee J. (2014). Identification of a novel molluscan short-type peptidoglycan recognition protein in disk abalone (Haliotis discus discus) involved in host antibacterial defense. Fish. Shellfish. Immun. 39, 99–107. doi: 10.1016/j.fsi.2014.04.018
Priyathilaka T. T., Bathige S. D. N. K., Lee S., Nam B., Lee J. (2019). Transcriptome-wide identification, functional characterization, and expression analysis of two novel invertebrate-type toll-like receptors from disk abalone (Haliotis discus discus). Fish. Shellfish. Immunol. 84, 802–815. doi: 10.1016/j.fsi.2018.10.062
Qiao X., Hou L. L., Wang J. H., Jin Y. H., Kong N., Li J. L., et al. (2021). Identification and characterization of an apoptosis-inducing factor 1 involved in apoptosis and immune defense of oyster, crassostrea gigas. Fish. Shellfish. Immunol. 119, 173–181. doi: 10.1016/j.fsi.2021.09.016
Qiu R., Kan Y. C., Li D. D. (2016). Ferritin from the pacific abalone haliotis discus hannai: Analysis of cDNA sequence, expression, and activity. Fish. Shellfish. Immunol. 49, 315–323. doi: 10.1016/j.fsi.2015.12.048
Ren C. H., Chen T., Jiang X., Wang Y. J., Hu C. Q. (2014). Identification and functional characterization of a novel ferritin subunit from the tropical sea cucumber, stichopus monotuberculatus. Fish. Shellfish. Immunol. 38, 265–274. doi: 10.1016/j.fsi.2014.03.022
Ren Y. P., Liu H. X., Fu S. Y., Dong W. H., Pan B. P., Bu W. J. (2021). Transcriptome-wide identification and characterization of toll-like receptors response to vibrio anguillarum infection in Manila clam (Ruditapes philippinarum). Fish. Shellfish. Immunol. 111, 49–58. doi: 10.1016/j.fsi.2021.01.007
Roberts S., Gueguen Y., de Lorgeril J., Goetz F. (2008). Rapid accumulation of an interleukin 17 homolog transcript in crassostrea gigas hemocytes following bacterial exposure. Dev. Comp. Immunol. 32, 1099–1104. doi: 10.1016/j.dci.2008.02.006
Saco A., Rey-Campos M., Novoa B., Figueras A. (2020). Transcriptomic response of mussel gills after a vibrio splendidus infection demonstrates their role in the immune response. Front. Immunol. 11. doi: 10.3389/fimmu.2020.615580
Sheikh M. S., Huang Y. (2003). Death receptor activation complexes: it takes two to activate TNF receptor 1. Cell Cycle 2, 559–552. doi: 10.4161/cc.2.6.566
Shen J. D., Cai Q. F., Yan L. J., Du C. H., Liu G. M., Su W. J., et al. (2015). Cathepsin l is an immune-related protein in pacific abalone (Haliotis discus hannai) - purification and characterization. Fish. Shellfish. Immunol. 47, 986–995. doi: 10.1016/j.fsi.2015.11.004
Shen Y., Zhang H., Zhou Y. C., Sun Y., Yang H. R., Cao Z. J., et al. (2021). Functional characterization of cathepsin b and its role in the antimicrobial immune responses in golden pompano (Trachinotus ovatus). Dev. Comp. Immunol. 123, 104128. doi: 10.1016/j.dci.2021.104128
Shoshan-Barmatz V., De Pinto V., Zweckstetter M., Raviv Z., Keinan N., Arbel N. (2010). VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Aspects Med. 31, 227–285. doi: 10.1016/j.mam.2010.03.002
Silva-Aciares F., Moraga D., Auffret M., Tanguy A., Riquelme C. (2013). Transcriptomic and cellular response to bacterial challenge (pathogenic vibrio parahaemolyticus) in farmed juvenile haliotis rufescens fed with or without probiotic diet. J. Invertebr,Pathol. 113, 163–176. doi: 10.1016/j.jip.2013.03.004
Strickland D. K., Ranganathan S. (2003). Diverse role of LDL receptor-related protein in the clearance of proteases and in signaling. J. Thromb. Haemost. 1, 1663–1670. doi: 10.1046/j.1538-7836.2003.00330.x
Taylor P. R., Carugati A., Fadok V. A., Cook H. T., Andrews M., Carroll M. C., et al. (2000). A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 192, 359–366. doi: 10.1084/jem.192.3.359
Tenner A. J. (1999). Membrane receptors for soluble defense collagens. Curr. Opin. Immunol. 11, 34–41. doi: 10.1016/S0952-7915(99)80007-7
Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Turk V., Turk B., Gunčar G., Turk D., Kos J. (2002). Lysosomal cathepsins: structure, role in antigen processing and presentation, and cancer. Adv. Enzyme Regul. 42, 285–303. doi: 10.1016/s0065-2571(01)00034-6
Uren A. G., Coulson E. J., Vaux D. L. (1998). Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem. Sci. 23, 159–162. doi: 10.1016/s0968-0004(98)01198-0
Vandivier R. W., Ogden C. A., Fadok V. A., Hoffmann P. R., Brown K. K., Botto M. (2002). Role of surfactant proteins a, d, and C1q in the clearance of apoptotic cells in vivo and in vitro: calreticulin and CD91 as a common collectin receptor complex. J. Immunol. 169, 3978–3986. doi: 10.4049/jimmunol.169.7.3978
Wang L. K., Feng Z. X., Wang X., Wang X. W., Zhang X. G. (2010). DEGseq: an r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang Z. L., Wang B., Chen G., Jian J. C., Lu Y. S., Xu Y. H., et al. (2016). Transcriptome analysis of the pearl oyster (Pinctada fucata) hemocytes in response to vibrio alginolyticus infection. Gene 575, 421–428. doi: 10.1016/j.gene.2015.09.014
Weiss I. M., Kaufmann S., Mann K., Fritz M. (2000). Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusk haliotis laevigata. Biochem. Bioph. Res. Co. 267, 17–21. doi: 10.1006/bbrc.1999.1907
Yang J. W., Liu H. H., Zheng G., Xiang X. W., Lv Z. M., Wang T. M. (2016). Cathepsin l of the sea cucumber apostichopus japonicus-molecular characterization and transcriptional response to vibrio splendidus infection. Fish. Shellfish. Immunol. 49, 387–395. doi: 10.1016/j.fsi.2016.01.007
Yao T., Lu J., Bai C. M., Xie Z. L., Ye L. T. (2021). The enhanced immune protection in small abalone haliotis diversicolor against a secondary infection with vibrio harveyi. Front. Immunol. 12. doi: 10.3389/fimmu.2021.685896
Yu W. T., Jong K. J., Lin Y. R., Tsai S. E., Tey Y. H., Wong H. C. (2013). Prevalence of vibrio parahaemolyticus in oyster and clam culturing environments in Taiwan. Int. J. Food Microbiol. 160, 185–192. doi: 10.1016/j.ijfoodmicro.2012.11.002
Yu J. J., Wang H. X., Yue X., Liu B. Z. (2019). Dynamic immune and metabolism response of clam meretrix petechialis to vibrio challenge revealed by a time series of transcriptome analysis. Fish. Shellfish. Immunol. 94, 17–26. doi: 10.1016/j.fsi.2019.08.057
Zhang J., Qiu R., Hu Y. H. (2014). HdhCTL1 is a novel c-type lectin of abalone haliotis discus hannai that agglutinates gram-negative bacterial pathogens. Fish. Shellfish. Immunol. 41, 466–472. doi: 10.1016/j.fsi.2014.09.032
Zhang J. H., Xu M. (2000). DNA Fragmentation in apoptosis. Cell Res. 10, 205–211. doi: 10.1038/sj.cr.7290049
Zhang F. T., Zhang Y. B., Chen Y. D., Zhu R., Dong C. W., Li Y. Y., et al. (2008). Expressional induction of paralichthys olivaceus cathepsin b gene in response to virus, poly I:C and lipopolysaccharide. Fish. Shellfish. Immunol. 25, 542–549. doi: 10.1016/j.fsi.2008.07.018
Zhang Y. Y., Zhang R., Zou J. J., Hu X. L., Wang S., Zhang L. L., et al. (2013). Identification and characterization of four ferritin subunits involved in immune defense of the yesso scallop (Patinopecten yessoensis). Fish. Shellfish. Immunol. 34, 1178–1187. doi: 10.1016/j.fsi.2013.01.023
Zhao X. L., Duan X. M., Wang Z. H., Zhang W. W., Li Y., Jin C. H., et al. (2017). Comparative transcriptome analysis of sinonovacula constricta in gills and hepatopancreas in response to vibrio parahaemolyticus infection. Fish. Shellfish. Immunol. 67, 523–535. doi: 10.1016/j.fsi.2017.06.040
Keywords: abalone, Vibrio parahaemolyticus, gill, immune response, transcriptome
Citation: Fang Y, Yang X, Zhang S, Chen X, Lin G, Zhang Y, Wang M and Li M (2022) Transcriptome study on immune response against Vibrio parahaemolyticus challenge in gill of abalone Haliotis discus hannai Ino. Front. Mar. Sci. 9:956317. doi: 10.3389/fmars.2022.956317
Received: 30 May 2022; Accepted: 28 June 2022;
Published: 27 July 2022.
Edited by:
JInghui Fang, Yellow Sea Fisheries Research Institute (CAFS), ChinaReviewed by:
Jianmin Zhao, Yantai Institute of Coastal Zone Research (CAS), ChinaMin Wan, Ocean University of China, China
Copyright © 2022 Fang, Yang, Zhang, Chen, Lin, Zhang, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzhu Li, bGR1bGltaW5nemh1QDE2My5jb20=
†These authors have contributed equally to this work
 Yan Fang
Yan Fang Xiyun Yang
Xiyun Yang Shujuan Zhang
Shujuan Zhang Xuyang Chen
Xuyang Chen Gang Lin
Gang Lin Yuqing Zhang
Yuqing Zhang Mengjiao Wang
Mengjiao Wang Mingzhu Li
Mingzhu Li