
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 27 September 2022
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.950716
 Przemysław Dąbek1*†
Przemysław Dąbek1*† Romain Gastineau1†
Romain Gastineau1† Thomas G. Bornman2,3†
Thomas G. Bornman2,3† Claude Lemieux4†
Claude Lemieux4† Monique Turmel4†
Monique Turmel4† Gustaaf Hallegraeff5†
Gustaaf Hallegraeff5† Jean-Luc Mouget6†
Jean-Luc Mouget6† Andrzej Witkowski1†
Andrzej Witkowski1†Haslea ostrearia represents the model species of blue diatoms, a cluster of benthic marine species all belonging to the genus Haslea, noticeable for producing a blue pigment called marennine famous for its greening activity on the gills of bivalves but also for its potential in biotechnology. The exact distribution of H. ostrearia is unknown. It has been long considered a cosmopolitan diatom, but recent studies provided evidence for cryptic diversity and the existence of several other blue species, some of them inhabiting places where diatoms described as H. ostrearia had previously been observed. Recently, a marine diatom with blue tips was isolated into clonal culture from a plankton net sample from Kei Mouth on the Indian Ocean coast of South Africa. It was identified as H. ostrearia through a combination of LM/SEM microscopy and molecular analysis. This constitutes the first established record of this species from South Africa and the Indian Ocean and the second record for the southern hemisphere. Molecular barcoding clearly discriminated the South African strain from an Australian strain and cox1 based molecular phylogeny associated it instead with strains from the French Atlantic Coast, raising questions about the dispersal of this species. The complete mitochondrial and plastid genomes were compared to those of Haslea nusantara and Haslea silbo. Multigene phylogenies performed with all protein-coding genes of the plastome and the mitogenome associated H. ostrearia with H. silbo. In addition, complete sequences of circular plasmids were obtained and one of them showed an important conservation with a plasmid found in H. silbo.
The South African coasts, bounded by the Indian and Atlantic Ocean water masses, are among the most productive coastlines in the world and considered a ‘hot-spot’ of global marine biodiversity (Griffiths et al., 2010; Teske et al., 2011). South Africa has three relatively distinct regions with respect to inshore marine biogeography, i.e., West, South and East coast marine provinces (Stephenson, 1948; Emanuel et al., 1992; Bolton and Anderson, 1997). Lombard et al. (2004) further divided the southern African coastline into five bioregions, i.e., the Namib and the Namaqua Bioregion on the cool temperate West Coast, the South-western Cape Bioregion overlap zone, the warm-temperate Agulhas Bioregion on the south coast and the subtropical Natal Bioregion and the tropical Delagoa Bioregion on the East Coast. The divide between the Agulhas and Natal Bioregions occurs at the Mbashe Estuary, approximately 68 km north of the Kei River mouth (Lombard et al., 2004). Each sector is influenced by the characteristics of the dominant ocean currents in terms of distinct differences in temperature and nutrient concentrations (Smit et al., 2013; Toonen et al., 2016; Smit et al., 2017)
The diversity of marine diatom communities found along the South African near-shore coastal environment appears to be underestimated in comparison with other marine organisms in the area, despite a long and extensive history of marine research in South Africa. The first comprehensive account of benthic marine diatoms of South Africa was produced on the marine littoral species of Simon’s Bay (Heiden and Kolbe, 1928). Early research on the offshore phytoplankton has received attention in the past because of the importance attributed to diatom blooms in the food-web (Hendey, 1937; Boden and Day, 1949; Taylor, 1967). Marine littoral diatoms were again briefly studied many years later on the coast near Knysna, Gordon’s Bay and other sites on the eastern shores of False Bay (Cholnoky, 1963). The ‘Golden era’ in South African marine diatomology was associated with the research of Professor Malcolm Giffen (1902 – 1986), who produced numerous papers on marine and estuarine benthic diatoms of the Indian Ocean, south coast in the Eastern Cape Province (Giffen, 1963; Giffen, 1966; Giffen, 1970a) and the Atlantic Ocean, west coast between Gordon’s Bay and Namaqualand (Giffen, 1970b; Giffen, 1971; Giffen, 1973; Giffen, 1975; Giffen, 1976; Giffen, 1984). More recently, marine benthic diatom research was focused on taxonomy and phylogeny (e.g., Dąbek et al., 2013; Dąbek et al., 2014; Dąbek et al., 2019), local diversity and ecology (Cotiyane-Pondo et al., 2020; Cotiyane-Pondo and Bornman, 2021a; Cotiyane-Pondo et al., 2021b) or epiphytic, epibiontic host-diatoms relationships (Majewska et al., 2019a; Majewska et al., 2019b; Majewska, 2020; Mayombo et al., 2020; Majewska et al., 2022).
Since 2012, authors of the current article (PD, TB and AW) have been conducting an extensive sampling effort in the ‘footstep of Giffen’ aiming to establish a ‘baseline’ diversity of the marine littoral diatoms from the South African coasts. During the austral-spring of 2018, a blue-tipped diatom, identified as Haslea ostrearia (Gaillon) Simonsen (Simonsen, 1974; Poulin et al., 2019), was isolated from the plankton net sample originating from the Kei Mouth rock pools. The blue diatom H. ostrearia is a marine species famous for synthesizing marennine, a blue pigment displaying several biological activities and whose exact chemical structure remains unknown (Pouvreau et al., 2006; Pouvreau et al., 2008; Gastineau et al., 2012a; Gastineau et al., 2014b; Prasetiya et al., 2015; Gastineau et al., 2018; Prasetiya et al., 2019a; Prasetiya et al., 2020; Prasetiya et al., 2021a; Prasetiya et al., 2021b; Francezon et al., 2021). Haslea ostrearia is a benthic/tychopelagic species famous for the ‘greening’ effect of its blue pigment on the natural benthic habitat (e.g., macroalgae or sand, rocks etc.) but especially on oyster gills. This latter phenomenon can be observed in oyster farming facilities from Europe (France, Denmark, Great Britain) and North America (USA) - see summary in Gastineau et al. (2014a) – and in Australia and Tasmania (GH – personal observations). Although other species able to produce blue pigments have recently been described, namely Haslea karadagensis Davidovich, Gastineau & Mouget, 2012, Haslea provincialis Gastineau, Hansen & Mouget 2016, Haslea nusantara Mouget, Gastineau & Syakti 2019 and Haslea silbo Gastineau, Hansen & Mouget 2021 (Gastineau et al., 2012b; Gastineau et al., 2016; Prasetiya et al., 2019b; Gastineau et al., 2021a), the presence of different, genetically distinguishable populations of H. ostrearia in distant areas has also previously been reported (Gastineau et al., 2013; Gastineau et al., 2014a).
Gastineau et al. (2012b; 2014a) previously wrote that the presence of the blue-tipped H. ostrearia in the Indian Ocean had been reported by Simonsen (1974) in the course of his exploration on the ‘Meteor’. But this assertion might originate from an over-interpretation of Simonsen’s work. Indeed, when he erected the genus Haslea with H. ostrearia as the generitype, Simonsen described only the new combination of this diatom without any indication of the precise distribution or origin of the material he was using. Instead, he referred to a work published one year before by Robert (1973), which contained SEM micrographs of H. ostrearia, still labelled Navicula ostrearia at that time. These pictures were obtained from material originating from the Bay of Bourgneuf in Western France, a place of intense oyster farming, where the greening mentioned above regularly occurs.
In the third tome of Malcolm Giffen’s account on the biodiversity of the diatoms from South Africa (Giffen, 1966), he mentioned the presence of a ‘blue’ diatom and ascribed it to N. ostrearia based solely on light microscopy, but with doubts regarding its true identity: ‘The striae are excessively faint and scarcely visible’ and also ‘The species in the group nearest to the above characters is N. ostrearia’. This took place 8 years before Simonsen erected the genus Haslea (Simonen, 1974), and 46 years before the existence of more than one species of blue diatoms was demonstrated (Gastineau et al., 2012b). Since 1966, no record of blue diatoms in South Africa had been reported, and the identity of the population identified remained uncertain. In the current study, the presence of H. ostrearia from the Indian Ocean and from the South African coast is assessed for the first time, by means of LM/SEM and molecular data. Haslea ostrearia is common on the Northern Atlantic coasts (both European and North American) (Gastineau et al., 2014a). Our research also presents the second established record from the southern hemisphere to date, with previous observations restricted to Australia (Tasmania, West and South Australia, New South Wales, and Queensland - Fraser Island) in the Pacific Ocean (Saunders et al., 2010; John, 2016; John, 2018 and GH – unpublished observations).
The aims of this study were: 1) to document morphologically and genetically the South African strain of H. ostrearia; 2) to sequence and characterize its mitochondrial and plastid genomes; and 3) to compare the resulting morphological and molecular data (cox1, mitochondrial and plastidic coding genes) with those previously reported for different populations of H. ostrearia and other blue diatom species from which these genomes are available, namely Haslea nusantara Mouget, Gastineau & Syakti (Prasetiya et al., 2019b) and Haslea silbo Gastineau, Hansen and Mouget (Gastineau et al., 2021a).
The strain of H. ostrearia analyzed in the current study originates from Kei Mouth on the Indian Ocean coast (Eastern Cape Province, South Africa; 32°41’4.72’’ S, 28°22’50.20’’ E). Samples were collected on 11 November 2018 from tidal rockpools during an incoming high tide using a phytoplankton net (Hydro-Bios Apstein net – 438001: 25 cm diameter, 55 µm mesh). Approximately 3 mL of f/2 medium (Guillard, 1975) were added to the 25 ml collection tube to facilitate the growth and survival of diatoms. No ‘greening effect’ on macroalgae nor on other benthic microhabitats was observed during the time of sampling at this location. The sample was transferred to and processed at the Szczecin Diatom Culture Collection, Institute of Marine and Environmental Sciences, University of Szczecin, Poland (SZCZ). Here, a clonal culture of H. ostrearia (strain SZCZP1883) was obtained using the micropipette method and inverted light microscope as described in Andersen & Kawachi (2005). After isolation, the clonal culture was maintained in a plastic Petri dish in a plant growth chamber at 20°C and illuminated at intensity of ca. 100 μmol photons m2 x s-1 (12:12h light:dark) for ca. 10 days. A subsample of the clonal culture biomass was then cleaned and boiled with 37% hydrogen peroxide and microscopic slides were prepared as described in Dąbek et al., 2017; Dąbek et al., 2019). Cleaned material of H. ostrearia (voucher strain no. SZCZP1883) is stored at SZCZ, as the strain is no longer alive.
LM micrographs of cleaned material and living cells were taken at the University of Szczecin using a Zeiss Axio Scope A1 light microscope (Carl Zeiss, Jena, Germany), with a 100x PlanApochromatic oil immersion objective (n.a. = 1.40) equipped with a Canon EOS 500D camera. The preparation methods are described in Li et al. (2015). SEM micrographs were taken using a Hitachi SU8020 microscope (Hitachi, Tokyo, Japan) at the Faculty of Chemical Technology and Engineering, Western Pomeranian University of Technology in Szczecin (Poland). Morphometric measurements of the valves, based on LM and SEM, were performed using ImageJ 1x (Schneider et al., 2012). LM/SEM figures were prepared using Adobe® Photoshop® CS6 (ver. 13). All microscopic examinations (LM and SEM) were performed on the voucher strain SZCZP1883. The culture was maintained in the exponential growth phase and the diatoms were motile.
A 250 mL sample of Haslea ostrearia clonal culture (strain SZCZP1883) was harvested by centrifugation during the exponential growth phase and DNA was extracted following Doyle and Doyle (1990). Total DNA was sent to BGI Shenzhen, where sequencing was performed on a DNBSEQ platform. A total of ca. 40 million clean 100 base pair (bp) paired-end reads was obtained. Reads were assembled with SPAdes 3.14.0 (Bankevich et al., 2012), with a k-mer of 85. Contigs of interest were selected by customized command-line blastn analyses, using sequences from Haslea nusantara (GenBank: MH681881 and MH681882) as query (Prasetiya et al., 2019b). Some contigs of the chloroplast genome were joined using Consed (Gordon and Green, 2013). Protein-coding genes and various open reading frames (ORFs) in both the mitochondrial and plastid genomes were identified using the findORF tools developed at Laval University (Gagnon, 2004). tRNA-scan and Rfam were used for identification of the tRNA and rRNA genes, respectively (Chan and Lowe, 2019; Kalvari et al., 2020). Maps of the organellar genomes were obtained with OGDRAW (Lohse et al., 2013). Genome alignments were performed with progressiveMauve (Darling et al., 2010). Prior to aligning plastomes with progressiveMauve, the second copy of the inverted repeat was removed.
Three different phylogenies were conducted. The first one was based on the partial gene of the first subunit of the cytochrome c oxidase gene (cox1) of Haslea spp., rooted with Navicula veneta Kützing (GenBank: MT383644). For this dataset, sequences were aligned with MEGAX (Kumar et al., 2018) and manually trimmed to a final length of 618 bp. For the multigene phylogenies, the datasets were prepared in a similar way to Górecka et al. (2021), reusing most of these previous datasets by appending them. All protein-coding genes were concatenated by alphabetical order and aligned using MAFFT 7 (Katoh & Standley, 2013) (-auto option) and trimmed by trimAl (Capella-Gutiérrez et al., 2009), (-automated1 option). The alignments after trimming were 83327 bp long for the plastidic genes and 23901 bp long for the mitochondrial genes, and Synedra acus Kützing was used as an outgroup in both cases. The best model of evolution was chosen according to ModelTest-NG (Darriba et al., 2020), which was always the GTR+I+G. Maximum Likelihood phylogenies were conducted with IQ-TREE 2.2.0 (Nguyen et al., 2015), with 1000 bootstrap replications for the multigene datasets and 10000 bootstrap replications for the cox1 derived phylogeny.
Haslea ostrearia (Gaillon) Simonsen (Simonsen, 1974: 47)
In the South African field sample, Haslea ostrearia was not blue and the accumulation of intracellular marennine (IMn), at the tips of the cells (Figures 1A–G), was only observed ca. 5-7 days after isolation and culturing. In the culture, diatoms were solitary, free living and motile (see Supplementary video). After 2-3 days of IMn production, a spontaneous secretion of marennine into the medium (extracellular marennine: EMn; blue water production: BW), in the form of small bubbles ca. 5-15 μm in diameter, was also observed. The IMn and BW were observed following every reinoculation after similar time periods than those described above.
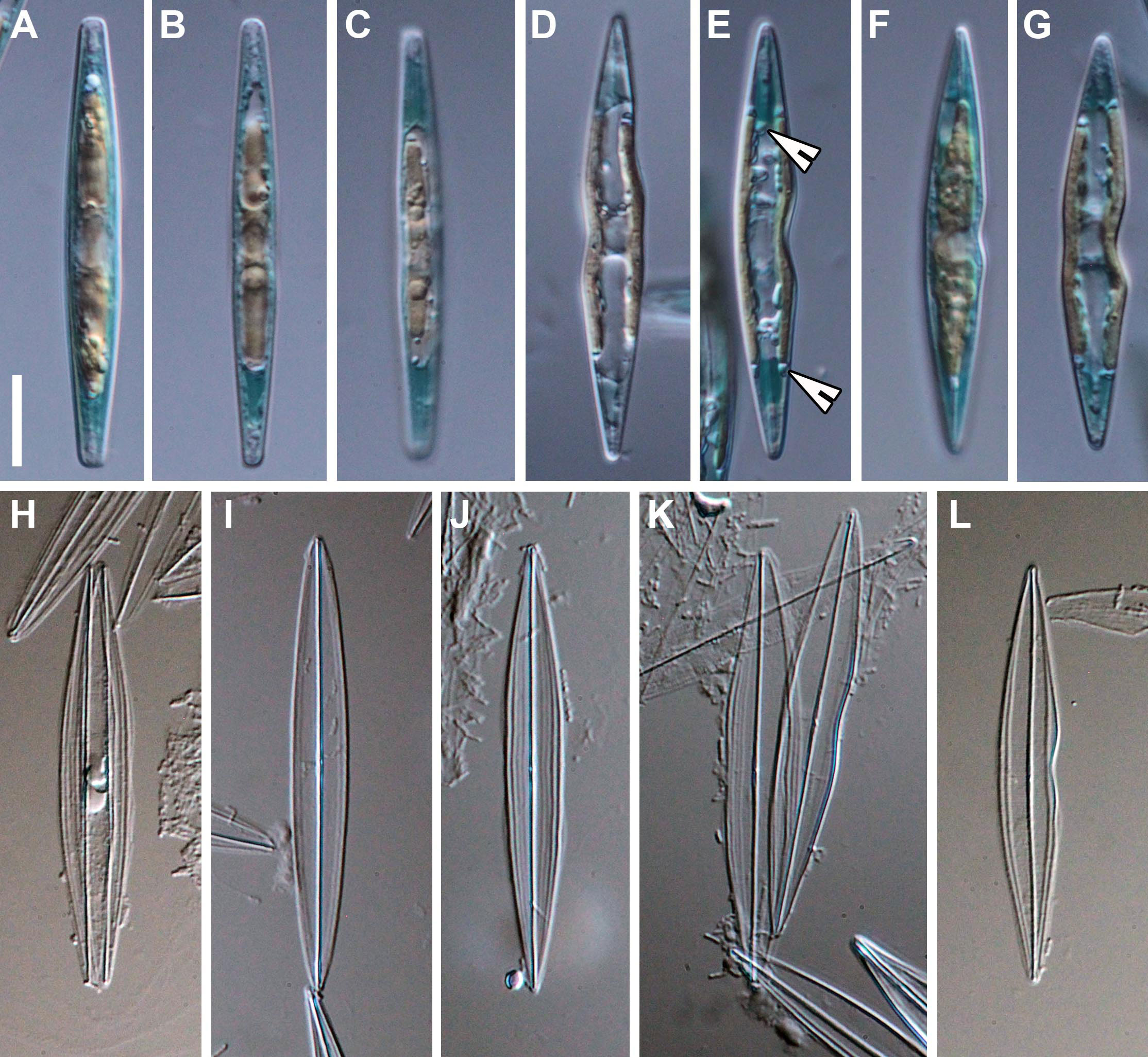
Figure 1 Haslea ostrearia SZCZP1883. (A–G) Live diatoms. Note the chloroplast arrangement and intracellular marennine (IMn) accumulation at the tips of the cells. Note the pyrenoids on 1E (arrowheads). (H–L) Light microscopic images of the cleaned material. Scale bar = 10 μm.
From the valve face view, two chloroplasts were evident per cell with several small pyrenoids (Figure 1E), that are appressed to the sides of the valve without reaching the apical parts (Figures 1C–G). From the girdle view, the chloroplasts were located in the center, extending symmetrically to the apices (Figures 1A–C, F). The nucleus was positioned centrally with small (sometimes numerous) lipid granules also visible (Figures 1C, F). As the size of the cell decreases, the frustules often exhibited shape deformations as shown in Figures 1D–G, K, L.
The valves are lanceolate, spindle-like, with acute endings (Figures 1H–L), ca. 48-50 μm long (= 48.3 μm; n=53) and 5-7 μm wide (= 6.2 μm; n=48) in the central area. The valve outline is sometimes undulated on one or both sides (lips-shaped), usually in the central part (Figures 1D–G, L). In the girdle view, the frustules are rectangular and bulky in the middle (Figures 1A–C). The raphe system is barely visible in LM (Figures 1H–L). The longitudinal striae are curved and parallel to the valve margin (Figures 1H, J) and the transapical striae, barely seen in LM, are straight and perpendicular to the raphe.
The valve is bi-layered (‘sandwich-type’ structure), and the exterior is covered with long, continuous, and apical-oriented siliceous stripes (top layer) and a little relief (Figures 2A–C). The interior is composed of a grate-like layer (basal layer) of small areolae, separated by short bars arranged crosswise (Figures 2A, E–G). Transverse bars of this layer are slightly broader than the longitudinal bars and the areolae are more or less rectangular in shape (0.15 x 0.2 μm). The areolae are occluded externally by hymenes and remnants of this membrane are visible in Figure 2F. Longitudinal striae ca. 48-54 in 10 μm (= 52; n=15), transapical ca. 30-36 in 10 μm (= 34; n=18). The raphe is positioned along the apical axis with a narrow sternum (Figures 2B, C). The central area lacks a lateral extension (Figure 2D), and the central raphe endings are straight, both externally and internally (Figures 2D, F). Internally, the raphe is slightly elevated and straight, with narrow helictoglossae at the poles (Figure 2G). The girdle band is composed of several simple, hyaline copulae, ca. 0.8-1 μm wide throughout (Figures 2A, G).
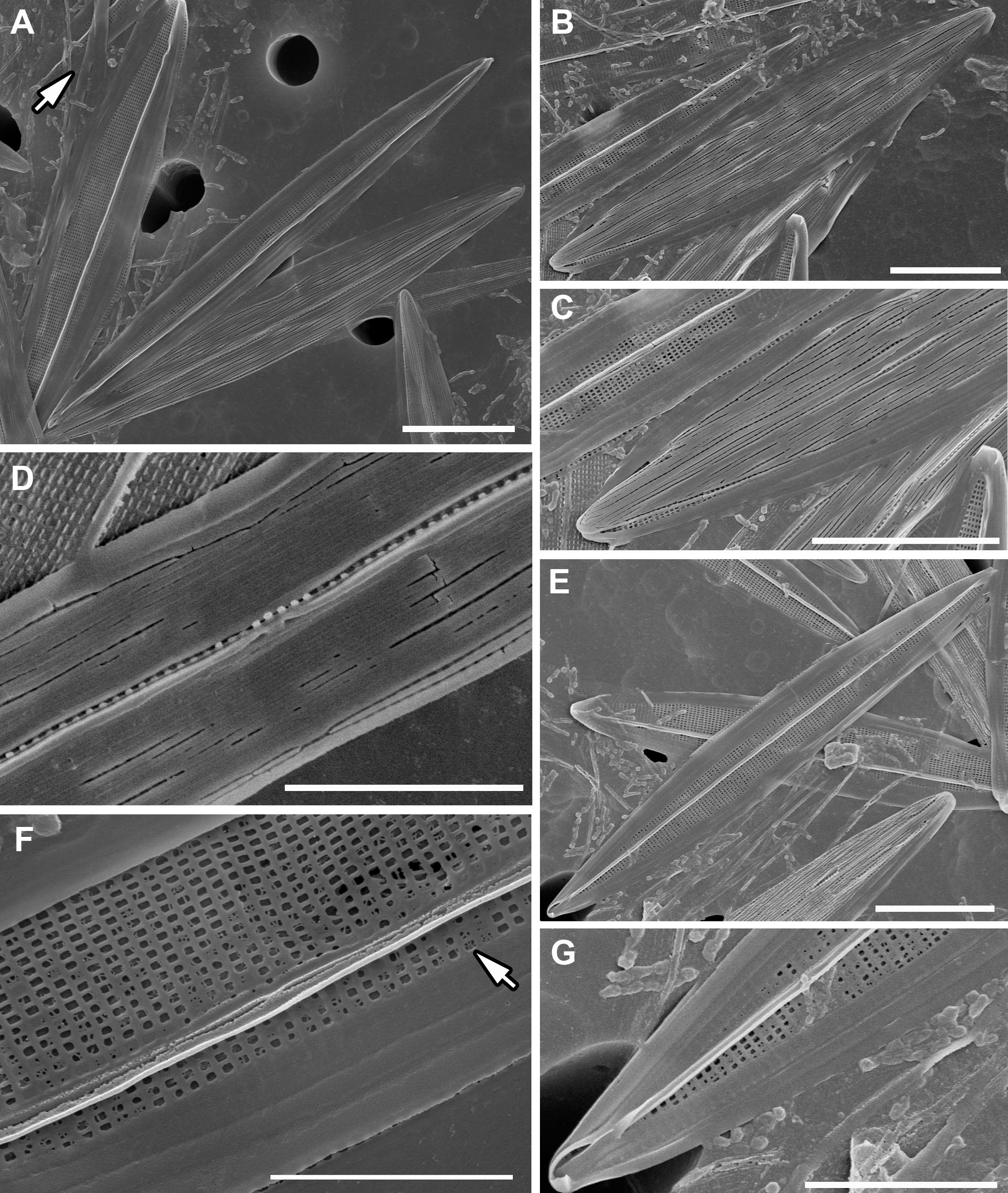
Figure 2 SEM images of H. ostrearia SZCZP1883. (A) General view of the bi-layered (‘sandwich-like’ structure) valves of Haslea ostrearia. Note the hyaline girdle bands (arrow). Scale bar = 10 μm. (B) External valve view with top layer composed of siliceous stripes. Scale bar = 10 μm. (C) Central and apical raphe ending from exterior. Scale bar = 10 μm. (D) Detailed view of a narrow central area of the valve exterior. Note the cracks exposing grate-like basal layer. Scale bar = 5 μm. (E) Internal valve view with basal layer composed of rectangular areolae. Scale bar = 10 μm. (F) Detailed view of central raphe ending and pattern of grate-like layer. Note the remnants of occlusions (arrow). Scale bar = 5 μm. (G) Apical raphe ending with helictoglossa from interior. Scale bar = 5 μm.
A contig of 13487 bp containing a cluster of nuclear rRNA genes was retrieved (GenBank: MW561352). The small subunit rRNA gene (SSU) is 1790 bp long, the internal transcribed spacer 1 (ITS1) is 506 bp long, the 5.8S rRNA gene is 155 bp long, the internal transcribed spacer 2 (ITS2) is 358 bp long and the large subunit rRNA gene (LSU) is 2494 bp long. These sequences were submitted to blastn searches. The ITS1-5.8S-ITS2 cluster showed 99.64% and 99.55% identities with its homolog in H. ostrearia NCC 148.75 (GenBank: HE663057) and H. ostrearia NCC 345 (GenBank: HE663059), respectively. The SSU gene showed 99.89% identity its homolog in H. ostrearia (GenBank: AY485523).
From the complete SSU gene of Haslea ostrearia strain SZCZP1883, the variable region V9 was extracted and blasted against the TARA blast server (http://bioinfo.szn.it/tara-blast-server/). The best result retrieved from it was the sequence EUK : MATOU-v1_7499874, with an e-value of 3.22 × 10-24 and 80% of identity. We also extracted from the TARA database W5 (http://taraoceans.sb-roscoff.fr/EukDiv/) all the sequences that were assigned to the genus Haslea, corresponding to the OTUs with the following ‘cid’ identifiers: 121219, 191798, 112785, 186972, 2796140, 627911, 2090776, 267085, 821022, 1912688, 33871, 2825676 (de Vargas et al., 2015). These sequences were aligned with the V9 portion obtained on H. ostrearia SZCZP1883 using clustal omega online (https://www.ebi.ac.uk/Tools/msa/clustalo/). Identities with H. ostrearia ranged between 82.31% for OTU 33871 (21 polymorphisms) and 97.67% for OTU 2796140 (3 polymorphisms).
As a mean to compare, the identity between the V9 regions of H. ostrearia and H. silbo was 98.45% similar, corresponding to 2 polymorphisms. There was no difference for the V9 portion between H. silbo NCC456 (MW679567) and H. silbo SZCZMV2009 (MW679566), nor between H. ostrearia SZCZP1883 (MW561352) and H. ostrearia (AY485523).
The mitogenome is 37444 bp long (GenBank: MW561350) (Figure 3). It contains 33 conserved protein-coding genes, 22 tRNA genes and two rRNA genes. The nad2 and nad6 genes are fused into a single reading frame. There are also four large ORFs: orf162a, orf183a, orf223a and orf252a. Among these, orf162a corresponds to the ubiquitous ORF previously described by Pogoda et al. (2019) as a part of a conserved cluster described as ‘tatC-orf157-rps11’. In two cases, blastp analysis of the putative proteins encoded by these ORFs returned non-conserved ORFs from the mitogenome of H. silbo. More precisely, orf183a returned as best result orf184 (GenBank: QUS63614.1) with an e-value of 1e-92 and 74.32% identity and orf223a returned orf224 (GenBank: QUS63616.1) with an e-value of 2e-121 and 78.12%% of identity. As shown in Figure 4, the H. ostrearia mitogenome is colinear with that of H. nusantara. Both genomes differ from the H. silbo mitogenome by the position of an ORF that corresponds to orf223a in H. ostrearia. The H. ostrearia mitogenome differs from the H. nusantara mitogenome by the presence of an insertion in the 1048 bp intergenic region containing orf252a, which is located between rps14 and cox1. This intergenic region is only 26 bp in H. nusantara. Otherwise, like H. nusantara and unlike H. silbo, no intron was detected in the mitogenome. The cox1 gene of H. ostrearia SZCZP1883 was aligned with the H. ostrearia sequences previously analyzed by Gastineau et al. (2013) as well as with that (GenBank MW595180) obtained from a strain originating from Coffin Bay, South Australia (34°37′S 135°28′E) (Gastineau et al., 2014a). The seven SNPs distinguishing the Atlantic Coast and Kattegat clones of H. ostrearia were found to be completely conserved between the Atlantic and South African clones of H. ostrearia. This is also illustrated below by the cox1 inferred phylogeny. After trimming to identical lengths (721 bp) and alignment with Clustal Omega, the cox1 genes of the SZCZP1883 and Coffin Bay strains revealed 96.95% identity.
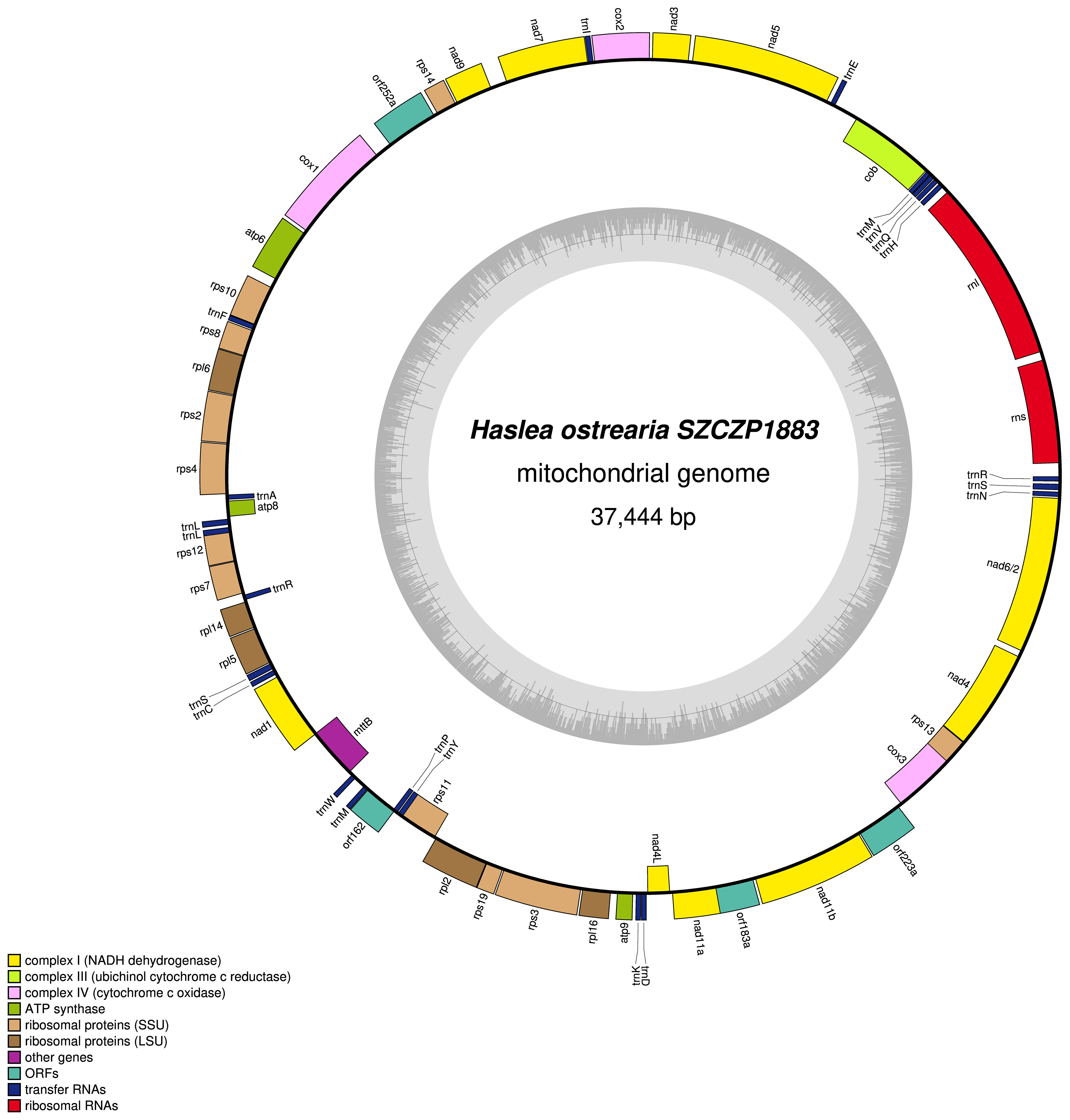
Figure 3 Map of the mitochondrial genome of Haslea ostrearia SZCZP1883. The legend in the lower left part of the graph indicates the type of genes and features represented. Genes displayed on the outside were found on the positive strand, genes displayed on the inside were found on the negative strand. The grey graph indicates the GC content.
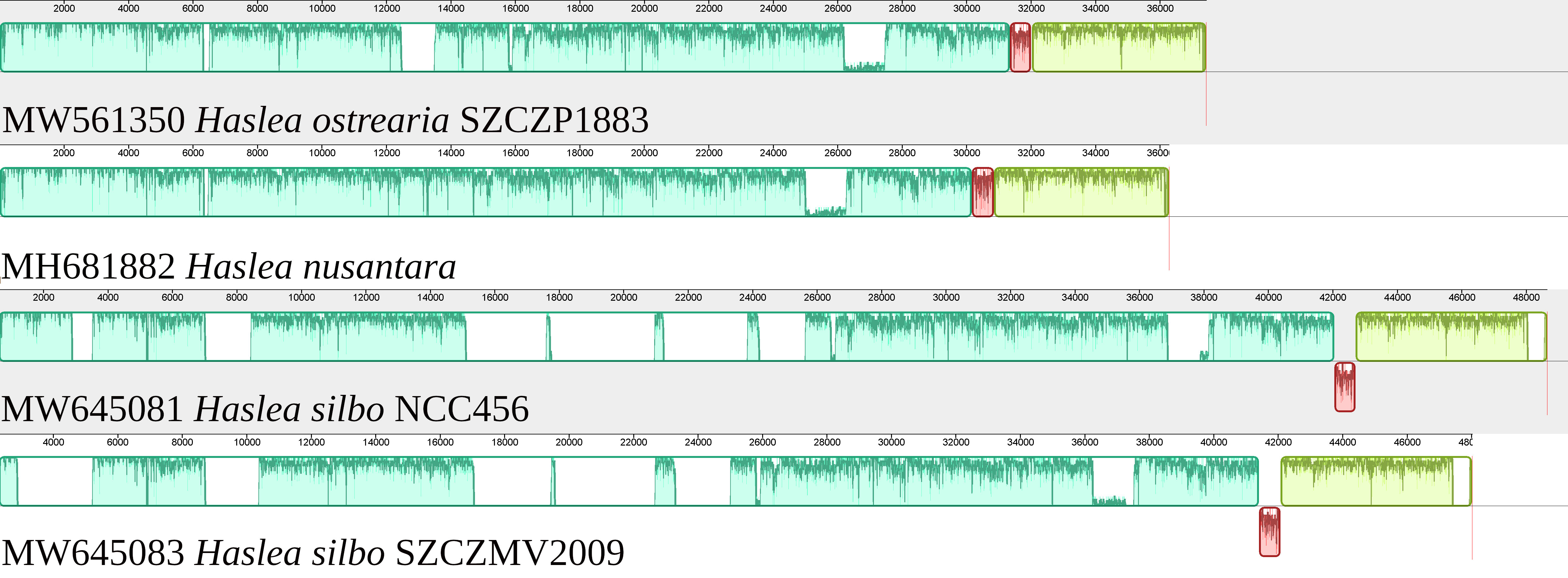
Figure 4 MAUVE alignment of the mitogenomes of Haslea ostrearia SZCZP1883, Haslea silbo and Haslea nusantara. There were 3 blocks of synteny detected by MAUVE. The red block is located on the opposite strand in H. silbo compared to the two other species and corresponds to a non-conserved ORF.
The plastome of Haslea ostrearia SZCZP1883 (129009 bp; GenBank: MW561351) features the typical quadripartite architecture of diatom plastomes (Figure 5). The large single-copy (LSC) region is 64631 bp long and contains 74 conserved protein-coding genes and 28 tRNA genes in addition to the RNA-coding genes ffs and ssrA. The short single-copy (SSC) region is 49916 bp long and contains 52 conserved protein-coding genes and 15 tRNA genes. The SSC also displays between psbA and ycf35 a region with 9 non-conserved ORFs and a protein-coding gene encoding a putative serine recombinase (serC). The inverted repeat (IR) region is 7231 bp long and contains 2 conserved protein-coding genes, 3 rRNA genes and 3 tRNA genes. The MAUVE alignment of the H. ostrearia, H. nusantara and H. silbo plastomes is presented in Figure 6. Due to the presence of many non-conserved ORFs, the SSC regions of the two H. silbo strains are substantially enlarged relative to those of other Haslea species and were previously designated as the LSC region (Gastineau et al., 2021a). In this study, we defined the LSC and SSC regions on the basis of their gene contents, irrespective of their lengths. All LSC regions are colinear in all species, but the SSC regions feature an important rearrangement between ycf35 and clpC, which was previously shown to distinguish H. silbo from H. nusantara (Prasetiya et al., 2019b). As in H. silbo, the H. ostrearia sequence located between ycf35 and psbA contains non-conserved ORFs and a putative serC gene that likely originates from recombination with plasmids, implying that it could be a hotspot for recombination (Hildebrand et al., 1992; Brembu et al., 2014; Ruck et al., 2014; Hamsher et al., 2019; Li et al., 2019; Gastineau et al., 2021a). A blastp search with the protein encoded by the H. ostrearia serC returned as best result (e-value of 2e-107 and 74.64% similarity) the putative serine recombinase encoded in the plastid genome of the diatom Nanofrustulum shiloi (QGW12742.1, in Li et al., 2019).
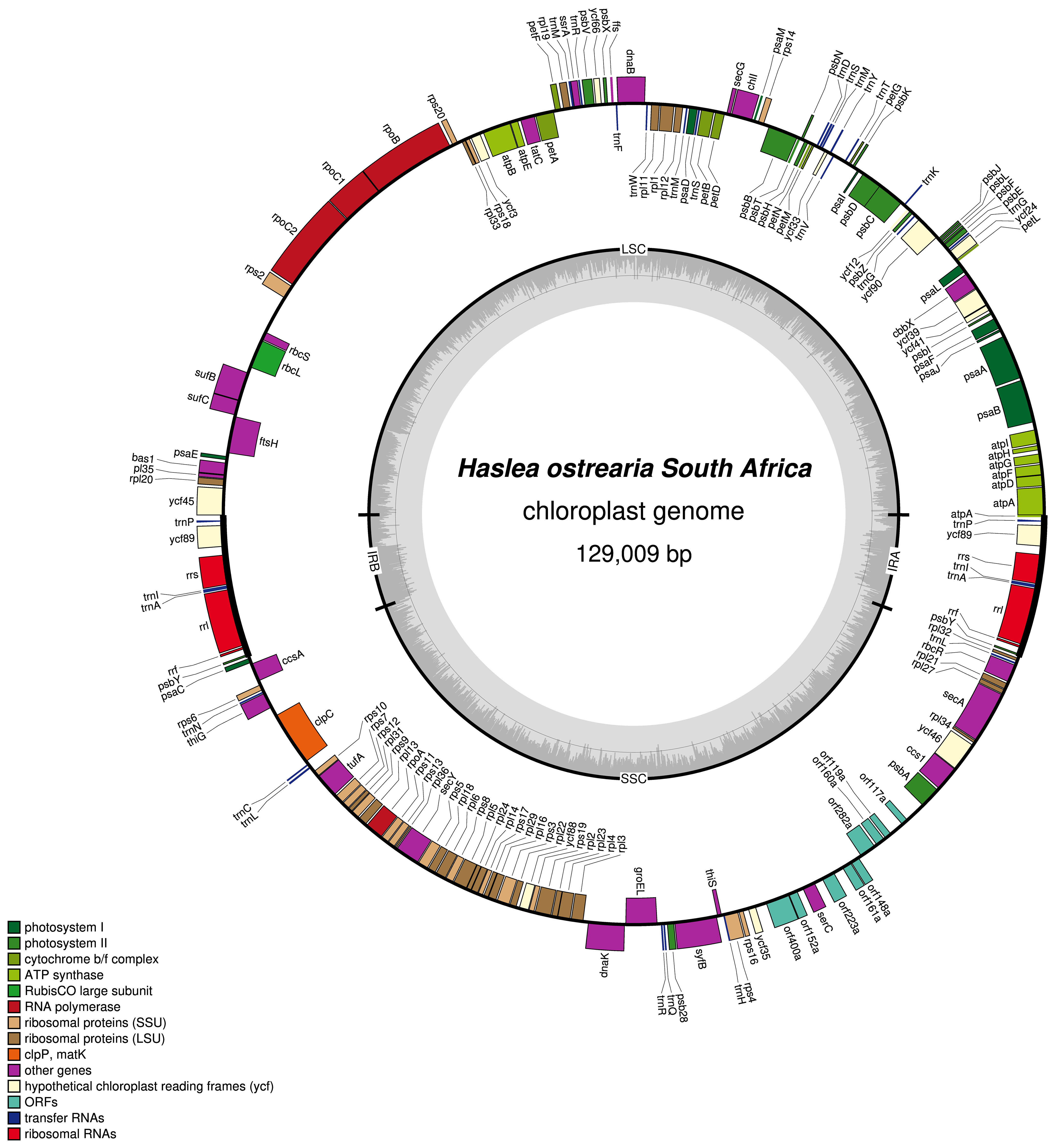
Figure 5 Map of the plastid genome of Haslea ostrearia SZCZP1883. The legend in the lower left part of the graph indicates the type of genes and features represented. Genes displayed on the outside were found on the positive strand, genes displayed on the inside were found on the negative strand. The grey graph indicates the GC content.

Figure 6 MAUVE alignment of the plastomes of Haslea ostrearia SZCZP1883, Haslea silbo and Haslea nusantara. There were 4 major blocks of synteny detected by MAUVE. The large single-copy corresponds to the first pink block, and is colinear among all species. The short single-copy is divided into 3 main blocks of synteny. The red block corresponds to a cluster of genes delimited by ycf35 and clpC, which is on the negative strand for H. ostrearia and H. silbo, and on the positive strand for H. nusantara. The blue block is almost entirely lacking in H. nusantara, and mostly corresponds to DNA of plasmid origin.
T.1wo circular plasmids1 were identified in the H. ostrearia SZCZP1883 assembly. They were recovered as contigs with overlapping ends and are hereafter referred to as pHOZA1 (GenBank: MW561353) and pHOZA2 (GenBank: MW561354), according to the nomenclature used by Hildebrand et al. (1992) and Ruck et al. (2014) and applied to the plasmids observed in H. silbo by Gastineau et al. (2021a). The pHOZA1 and pHOZA2 plasmids are 4343 bp and 3313 bp in size, respectively. pHOZA1 contains a putative serC gene and two non-conserved ORFs (orf389a and orf486a), whereas pHOZA2 contains a gene encoding a putative integrase recombinase and two non-conserved ORFs (orf157a and orf492a). The pHOZA1 and pHOZA2 plasmids were recovered with a coverage of 90X and 180X, respectively; these values are larger than the coverage of 70X that was observed for the plastome and mitogenome. Alignment of the plasmids found among Haslea spp. with the region of the H. ostrearia and H. silbo plastomes encompassing ycf35 and psbA clearly reveals the similarities between plasmid pHOZA1 and the plasmid pHSC2 from H. silbo strain NCC456 (Figure 7). These plasmids are almost identical in size (pHSC2, 4440 bp; pHOZA1, 4343 bp) and they each contain a putative serC and two ORFs. The size of the encoded putative serine recombinase is identical among the two plasmids (222 amino acid residues), but the sizes of the proteins encoded by the two ORFs are more variable (pHSC2, 361 and 513 amino acid residues; pHOZA1 389 and 486 amino acid residues). With regards to pHOZA2, no homologous plasmid was found in the two H. silbo strains, although progressiveMauve identified weak similarity between this plasmid and a small portion of the H. silbo NCC456 plastome. However, these results should be taken with caution as the xerC gene encoded on pHOZA2 is highly divergent from the xerC genes found in the plastomes of H. silbo strains NCC456 and SZCZMV2009.
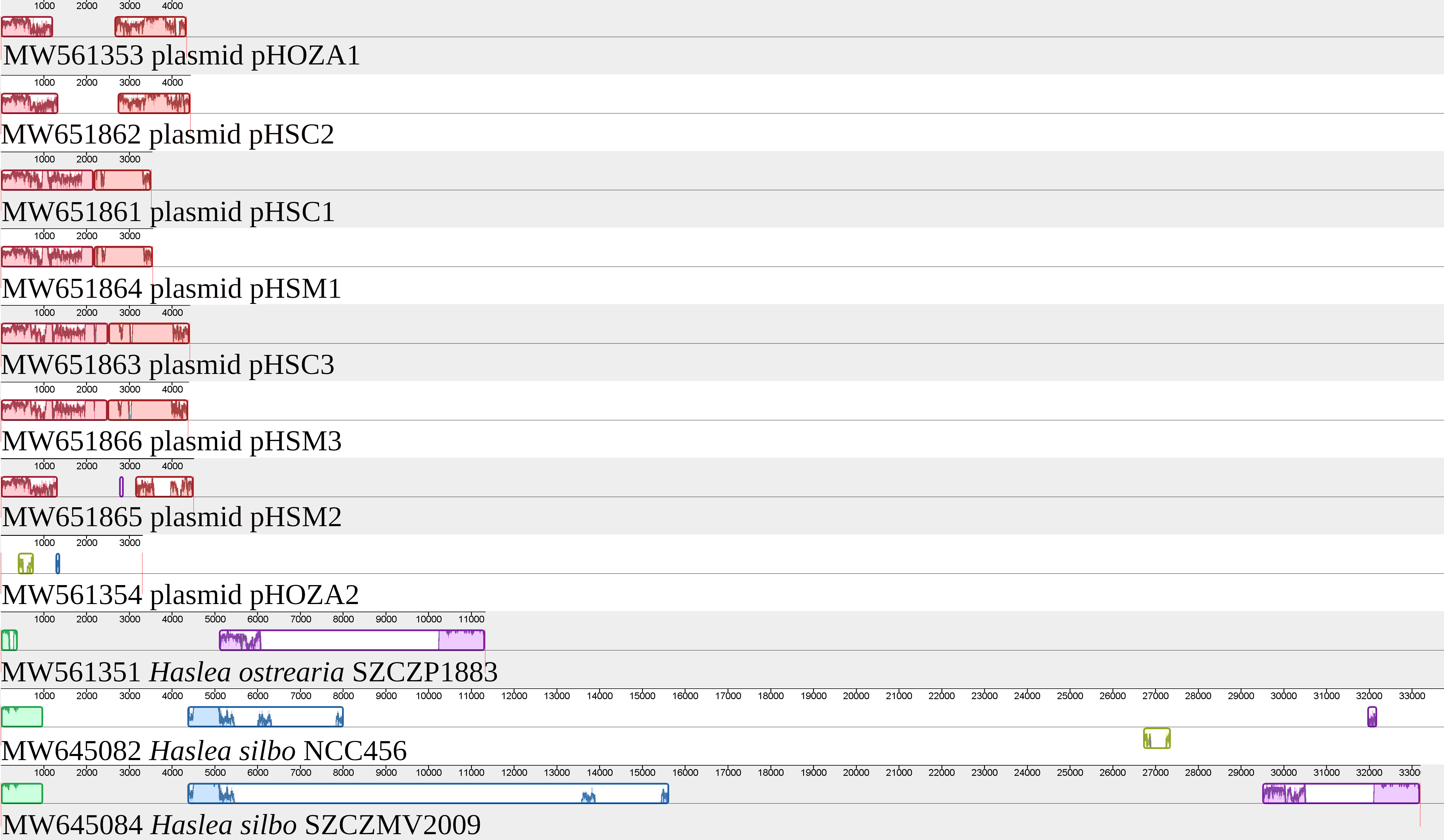
Figure 7 MAUVE alignment of the plasmids and plastomes of Haslea ostrearia SZCZP1883 and Haslea silbo. The alignment illustrates the conservation between some of the plasmids. It shows especially the conservation between the plasmid pHOZA1 from H. ostrearia SZCZP1883 and the plasmid pHSC2 from H. silbo NCC456.
The cox1 inferred phylogeny (Figure 8) clearly discriminated between three different clusters of populations of H. ostrearia, with strong support at the nodes. The clades corresponded to the three different genetic types mentioned above. The first, and most divergent, is represented so far by a single strain from Australia, labelled as OZ-1. A second clade contains strains that originate from the Kattegat Strait (NCC320, NCC321), or that inherited their mitochondrial genomes from these parental strains (NCC390 to NCC398) (Gastineau et al., 2013). Sister to this clade is the third one, which contains strains from the oyster ponds of the French Atlantic Coast, whether it is from near La Rochelle (NCC345) or the Bay of Bourgneuf (NCC 148.78, NCC 158.4), offspring of the strains from Bourgneuf, which inherited their mitochondrial genomes (NCC 384 to NCC 388), and finally strain SZCZP1883 from South Africa.
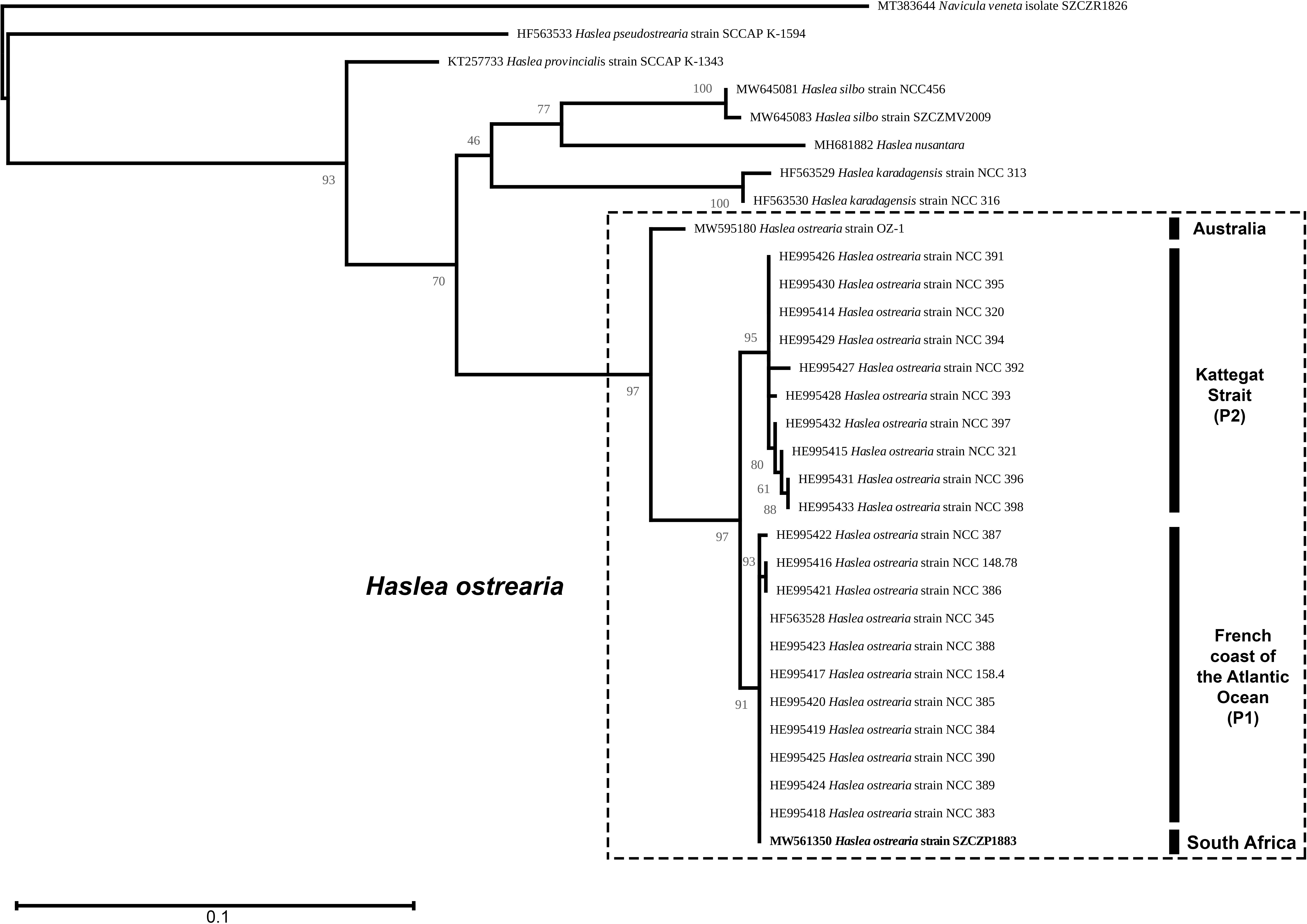
Figure 8 Maximum Likelihood phylogenetic tree inferred from an alignment of the partial cox1 gene of Haslea spp. and rooted with Navicula veneta (Log-likelihood: -2085.461960). The scale indicates the number of substitutions per site. P1 – Progeny of the pair NCC148.78 (Bay of Bourgneuf, France) x NCC320 (Kattegat Strait). P2 – progeny of the pair NCC158.4 (Bay of Bourgneuf, France) x NCC321 (Kattegat Strait), from Gastineau et al., 2013.
The mitochondrial (Figure 9) and plastidic (Figure 10) multigene phylogenies both associate H. ostrearia to H. silbo, with maximum values at the nodes. They also clearly discriminate Haslea spp. from other Naviculaceae, as represented by N. veneta and Seminavis robusta D.B. Danielidis & D.G. Mann in the plastidic phylogeny, or N. veneta and Navicula ramosissima (C. Agardh) Cleve in the mitochondrial phylogeny.
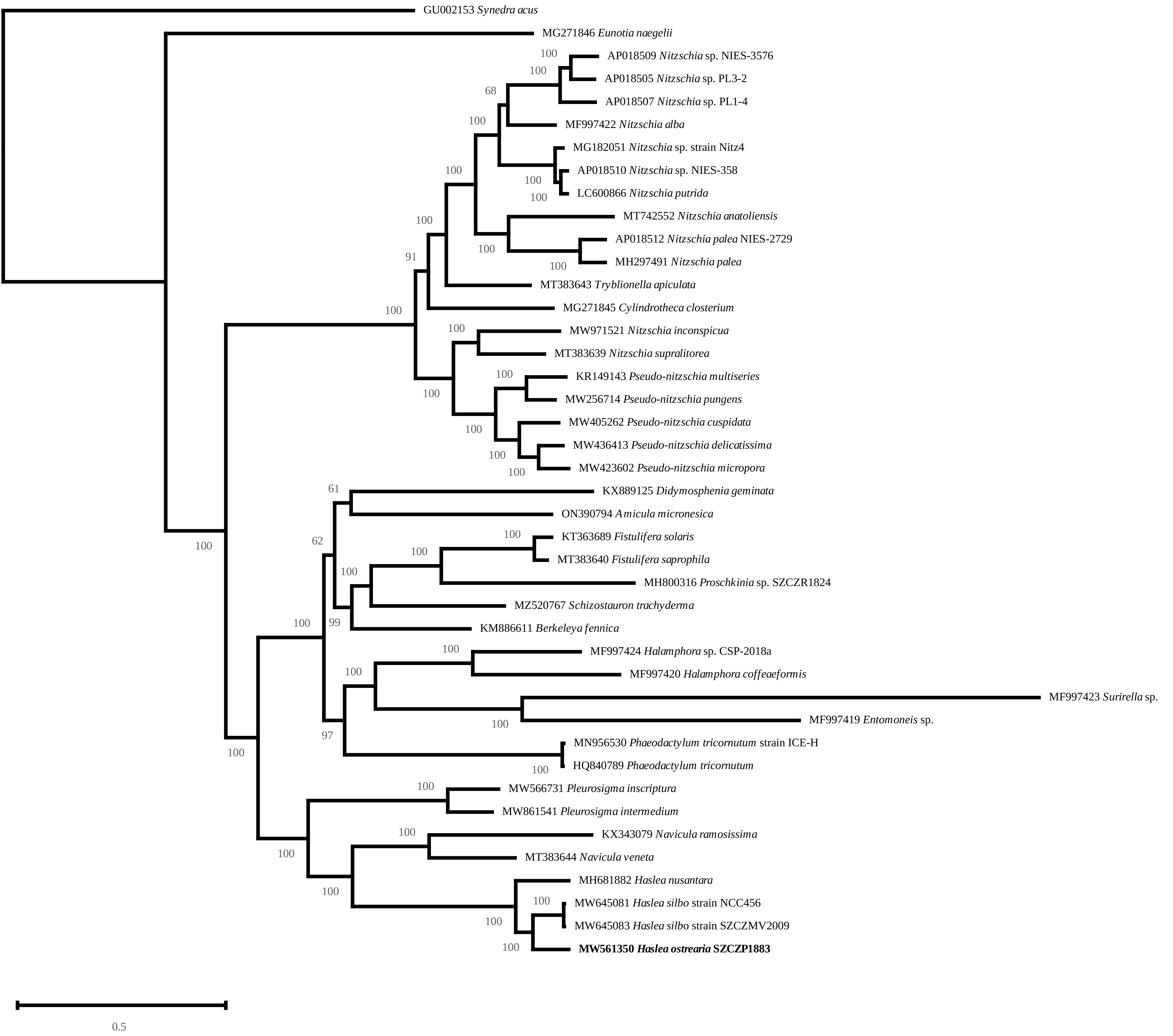
Figure 9 Maximum Likelihood phylogenetic tree inferred from an alignment of all protein coding genes from the mitogenomes of 42 taxa of diatoms and rooted with Synedra acus (Log-likelihood: -504283.7367). The scale indicates the number of substitutions per site.
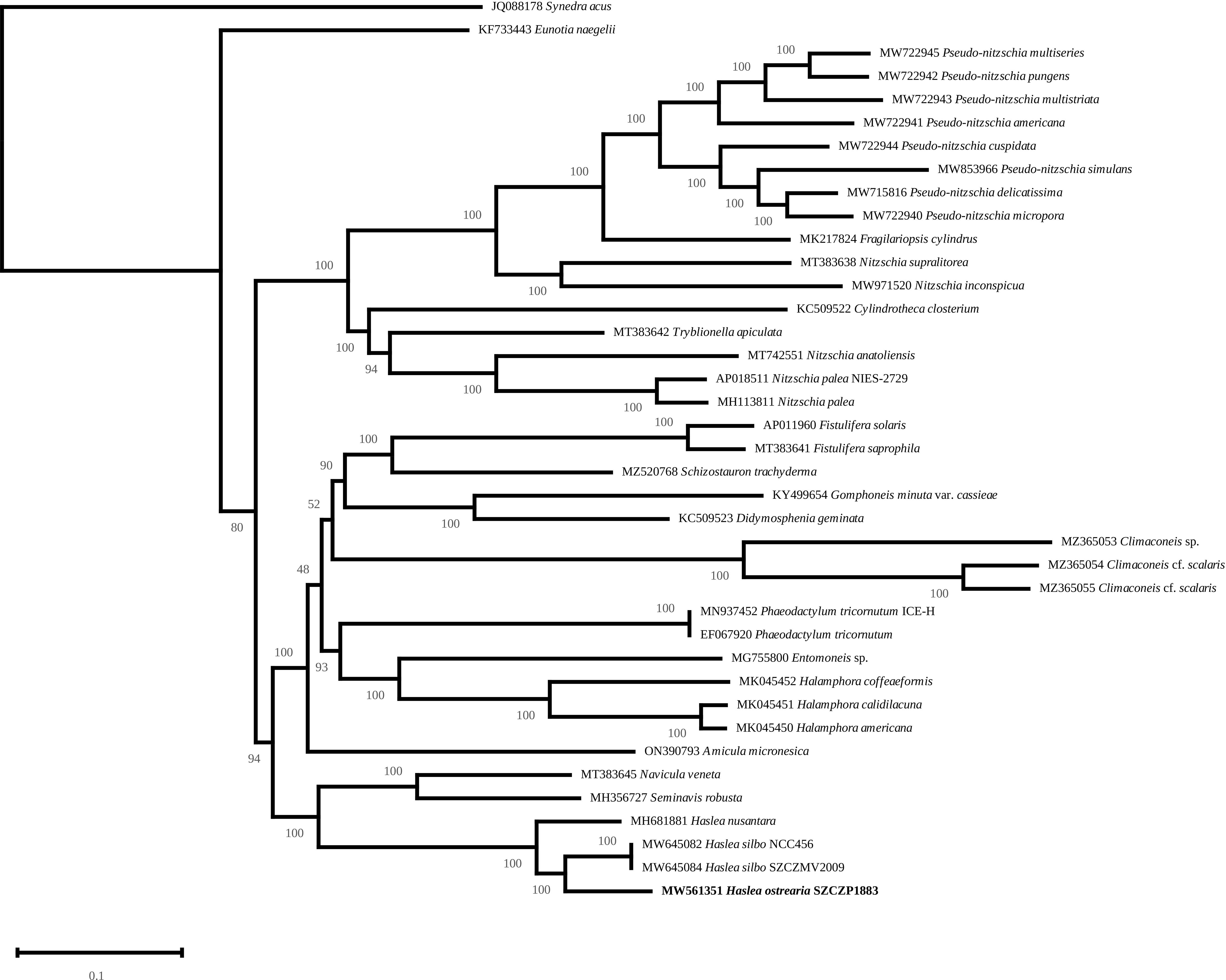
Figure 10 Maximum Likelihood phylogenetic tree inferred from an alignment of all protein coding genes from the plastome of 39 taxa of diatoms and rooted with Synedra acus (Log-likelihood: -1139126.5259). The scale indicates the number of substitutions per site.
This study reports on a blue-tipped marine diatom H. ostrearia from the Indian Ocean and South Africa, encountered for the first time by Giffen (1966) and re-collected in 2018. This is only the second confirmed occurrence in the southern hemisphere after its presence was assessed in Australia and Tasmania (Saunders et al., 2010; John, 2016; John, 2018 and GH & JLM– unpublished observations). H. ostrearia is widely distributed in the northern oceans and seas, such as the Atlantic Ocean, Mediterranean Sea, North Sea and Baltic Sea, and the Pacific Ocean and the coast of Canada and USA (summary and relevant literature in Gastineau et al., 2012b; Gastineau et al., 2014a). The question of the mechanisms driving the dispersal of a benthic, shallow water diatom remains an open one. Ballast waters are known to be an important vector for the introduction of diatoms (e.g., Hallegraeff and Bolch, 1992; Forbes and Hallegraeff, 2002; Klein et al., 2010; Villac et al., 2013). However, this applies mostly to planktonic species, living in the water column, and benthic diatoms may be less represented in ballast water. On the other hand, benthic diatoms are known to form biofilms on migrating animals such as turtles (e.g., Kaleli et al., 2020; Van de Vijver et al., 2020), which includes unidentified species ascribed to the genus Haslea (Majewska et al., 2015). This can be one factor driving the introduction of species into new environments.
In the current case, the possibility of an introduction through aquaculture could not be ruled out. The Kei Mouth area where the South African strain was collected is situated in the warm temperate Agulhas Bioregion, just 68 km south of the border with the subtropical Natal Bioregion and only ca. 100 km north from Kidd’s Beach, the sampling site of Giffen (1966). This entire region is characterized by warm sea temperatures (mean of 18.92°C ± 0.019) with high variability associated with wind driven upwelling (minimum of 11.23°C) and meanders in the warm Agulhas Current (maximum of 27.89°C). This raises the question as to whether the South African strain naturally occurs along this part of the coastline, or whether it was introduced as part of bivalve aquaculture activities with the introduction of oysters from places where greening naturally occurs, e.g., Western France. The coastline is exposed and subject to strong wave action and, as a consequence, marine aquaculture activities are not feasible within the sea itself. Two farms are situated in proximity of the collection site, i.e., Wild Coast Abalone (16 km south of the Kei Mouth), which grows abalone, various microalgae and seaweed and East Coast Rock Lobster in land-based facilities, and a Pacific oyster (Crassostrea gigas Thunberg, also known as Magallana gigas Thunberg) farm located within the Keiskamma Estuary (120 km south of the Kei Mouth). The oyster farm was established after feral Pacific oyster populations were detected in the estuary (Robinson et al., 2005; Hinrichsen, 2019), but has since been closed. As far as we know, no greening of oysters was ever observed at this facility. If the specimen observed by Giffen (1966) was indeed H. ostrearia, and not another blue species of Haslea, it would mean that this introduction occurred before the 1960’s. This is indeed possible, as the Pacific oysters have been actively imported to South Africa for aquaculture since the 1950’s (Robinson et al., 2005). Moreover, as noted in Hinrichsen (2019), there were already failed attempts to cultivate introduced specimens of the European flat oyster Ostrea edulis Linnaeus and the Portuguese oyster Crassostrea angulata Lamarck (also known as Magallana angulata Lamarck) in the 1940’s, both of which were commonly farmed in Western France with an assessed greening effect (e.g., Trochon, 1954), before they both had drastic decreases of population because of epizootic disease in the 60s and 70s, leading to the introduction of C. gigas as a replacement. Therefore, the question about the origin of the South African population of Haslea ostrearia is still open and will requires further molecular and biogeographical studies.
The northern hemisphere population of H. ostrearia is well recognized in terms of morphology (Gastineau et al., 2012b; Poulin et al., 2019). Our observations show that the SZCZP1883 strain, isolated from South Africa, is slightly smaller, on average 48.3 μm long and 6.2 μm wide versus 71.6 μm long and 7.5 μm wide in neotype material as reported by Poulin et al. (2019) – see Table 1. Similar differences are observed when we compare the Pacific populations of the latter species originating from various localities in Australia and Tasmania (Table 1). Our South African clonal population (SZCZP1883) and a wild population observed by Giffen (1966) differ in terms of valve dimension. The length and width of the cells observed by Giffen is almost twice as large as in our strain, and slightly larger than in other populations. However, as reported by Gastineau et al. (2012b), the morphometry of H. ostrearia is relatively diverse and may differ between natural and clonal populations.
The blast analysis of all sequences, from the TARA project W5 dataset (de Vargas et al., 2015), assigned to the genus Haslea provided identities with our strain of H. ostrearia from 82.31 to 97,67%, however, none of them correspond to H. ostrearia specifically. The highest affinity was recovered with OUT 2796140 obtained from station no. 41 in the Arabian Sea. With this ‘low’ percentage of identity in such a conserved gene, it is highly unlikely that this sequence belongs to H. ostrearia. For instance, the V9 region identity of H. ostrearia (SZCZP1883) was higher (98,45%) with the closely related species H. silbo. Moreover, TARA sampling stations near South Africa or in the Indian Ocean were located in the pelagic zone, ca. 100 to 400 km from the coasts, with samples collected from the water column (e.g., station 41, subsurface water 180-2000 m), a habitat where it is unlikely to find a benthic/tychopelagic species like H. ostrearia.
We also report the organellar genomes of H. ostrearia for the first time. While this is the model species of blue diatoms and the first to be described, its organellar genomes were not sequenced up to now. We note that for the mitogenomes of all three blue diatoms analyzed so far, the fusion of nad2 and nad6 into a single reading frame is a constant feature. At the moment, such fusion has not been observed among other diatoms, and it could represent a characteristic feature of the genus Haslea, provided that it is systematically found within all species of Haslea spp. The comparison of the H. ostrearia cox1 gene sequence with those previously published is another interesting result of our study. Based on the comparison of partial cox1 gene sequence, we were able to distinguish three different genetic types of H. ostrearia, representing four populations. Up to now, each of the H. ostrearia populations, on which cox1 molecular barcoding had been performed, was characterized by its own genetic type; this is the first time that the same type is found among two different populations. Interestingly, these populations are geographically distant – NW France (Atlantic Ocean) and the southern coast of South Africa (Indian Ocean). This observation strengthens the idea that mitochondrial genes are efficient markers of populations, keeping in mind that these genes are inherited from one parental clone only (Gastineau et al., 2013). In future studies on H. ostrearia, we plan to repeat the protocol employed here for sequencing the organellar genomes, with the aim of using the complete mitochondrial genome as a marker of population and determining if a biogeographic pattern can be detected.
The region between ycf35 and psbA in the H. ostrearia plastome contains sequences homologous to circular plasmids that has been found in other diatoms, suggesting that this site is a hotspot of recombination with plasmids. Similar observations have previously been reported for H. silbo (Gastineau et al., 2021a) and the distantly related diatom Cylindrotheca fusiformis Reimann & J.C. Lewin (Hildebrand et al., 1992). The plastomes of several diatoms are known to carry recombinase or integrase genes (e.g., Gastineau et al., 2021b; Górecka et al., 2021). However, it is not clear whether they feature circular plasmids that share homology with these genes. Plasmid sequences may be overlooked when processing next generation sequencing data targeted toward organellar genomes. In fact, it was rather easy to retrieve these sequences from the pool of contigs after assembly. We suppose that many other authors working in the field of organellar genomes of diatoms already have such unused data that could become informative, provided that they are duly identified and annotated. Efforts in this direction could provide insights into the diversity of plasmid sequences among diatoms, the origin of these plasmids and the mechanism by which both plastome and plasmid sequences are maintained independently in the same alga. We note that the benefit of such work would concern more than diatoms, as highlighted by recent works on other Stramenopiles which suggested that lateral transfer of plasmids occurs between microalgae (Kim et al., 2022).
This research provides novel data for a well-known blue diatom, Haslea ostrearia, rediscovered from the Indian Ocean coast of South Africa. We see this rediscovery as a tribute to Professor Malcolm H. Giffen, a South African botanist who worked on diatoms after official retirement. In his short diatomological career Professor Giffen made unprecedented progress in research on marine benthic diatoms of South Africa and further afield. He most likely observed the first live cells of the target species and identified it as N. ostrearia (Giffen, 1966).
With new technologies at our disposal 60 years later, we document the latter species by means of morphology (LM and SEM) and molecular (NGS sequencing) methods, as H. ostrearia and confirm its second occurrence in the Southern Hemisphere. Cox1 inferred molecular barcoding validates the Western France and South Africa populations belong to the same distinct genetic type and discriminates the South African strain from the Australia and Kattegat Strait ones. The study presents the first complete mitochondrial and plastid genome of Haslea ostrearia, which has never been published before. Moreover, we compared all three blue diatoms for which the organellar genomes are available, i.e., H. silbo, H. nusantara and now H. ostrearia.
With regards to the origin of the population studied we postulate two potential sources, i.e., 1) introduction through ballast water (a less likely vector for benthic species) and 2) introduction through aquaculture of various oyster species along the South African coast, which predates the observation made by M.H. Giffen.
As the number of complete sequenced genomes of closely related diatoms increases, we believe that the mitochondrial genome could become a reliable marker for tracing biogeographic patterns of diatom populations. With the description of an increasing number of species from the genus Haslea able to produce marennine-like pigments, H. ostrearia has somehow become the model organism for studies on blue diatoms. Beside its known interest in oyster farming, and to a larger extent its potential interest in broader aquaculture and biotechnologies, H. ostrearia has several characteristics that would make it a species of interest. The most obvious is its capacity to produce marennine, with the development and evolution of the metabolic pathway it suggests, but its ability to produce highly branched isoprenoids of the HBI-2 type, which are important proxies in geology, is also to note (Damsté et al., 2004).
The datasets presented in this study can be found in GenBank under the following accession numbers: MW561350, MW561351, MW561352, MW561353, MW561354.
Conceptualization, PD, RG. methodology, PD, RG, TB, CL, MT, GH. Writing—original draft preparation, PD, RG, TB, AW. Supervision, J-LM, AW. Revision, RG, PD, AW, TB. All authors have read and agreed to the published version of the manuscript.
The work has been supported by the GHaNA project: “The Genus Haslea, New marine resources for blue biotechnology and Aquaculture”, grant agreement no. 734708/GHANA/H2020-MSCA-RISE-2016, granted to J-LM. The project was also supported by the 2017–2022 research funds granted for implementation of a co-financed international research project from the Polish Ministry of Science and Higher Education. This work was also supported by the Natural Sciences and Engineering Research Council of Canada under Grant RGPIN-2017-04506. Authors acknowledge the funding from the South Africa-Poland Bilateral Collaboration Project facilitated by the National Research and Development Centre in Warsaw, Poland (NCBiR - project number PL-RPA/TemBioDiaSA/02/2016 granted to AW) and the National Research Foundation of South Africa (NRF - Grant UID 102283 to TB). The project was supported by the Shallow Marine and Coastal Research Infrastructure (SMCRI) platforms of the Department of Science and Innovation (South Africa) hosted by the SAEON Elwandle Coastal Node. The samples were collected with full permission from the South African Department of Agriculture, Forestry and Fisheries and the Department of Environmental Affairs (Permit number: RES2018-83).
Authors are grateful to Marta Krzywda and Ewa Górecka (University of Szczecin) for their fundamental help in developing and maintaining the SZCZ Diatom Culture Collection and to Prof. Rafał Wróbel (Western Pomeranian University of Technology, Szczecin) for his assistance in SEM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.950716/full#supplementary-material
Supplementary Video | Haslea ostrearia SZCZP1883 in the culture. Note the intracellular marennine (IMn) accumulation at the tips of the cells.
Andersen R. A., Kawachi M. (2005). ““Traditional microalgae isolation techniques”,” in Algal culturing techniques. Ed. Andersen R. A. (USA: Elsevier Academic Press), 83–100.
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Boden B. P., Day J. H. (1949). Some marine plankton diatoms from the west coast of South Africa. Trans. R. Soc S. Afr. 32 (4), 321–434. doi: 10.1080/00359194909519870
Bolton J. J., Anderson R. J. (1997). ““Marine vegetation”,” in Vegetation of southern Africa. Eds. Cowling R. M., Richardson D. M., Pierce S. M. (Cambridge, UK: Cambridge University Press), 348–370.
Brembu T., Winge P., Tooming-Klunderud A., Nederbragt A. J., Jakobsen K. S., Bones A. M. (2014). The chloroplast genome of the diatom Seminavis robusta: New features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genom. 16, 17–27. doi: 10.1016/j.margen.2013.12.002
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinform. 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chan P. P., Lowe T. M. (2019). tRNAscan-SE: Searching for tRNA genes in genomic sequences. Meth. Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Cholnoky B. J. (1963). Beiträge zur kenntnis des marinen litorals von südafrika Bot. Mar., 38–83. 5(2/3).
Cotiyane-Pondo P., Bornman T. G. (2021a). Environmental heterogeneity determines diatom egionation on artificial substrata: implications for biomonitoring in coastal marine waters. Front. Ecol. Evol. 9. doi: 10.3389/fevo.2021.767960
Cotiyane-Pondo P., Bornman T. G., Dąbek P. (2021b). Insight into nearshore diatom assemblages from an island ecosystem: composition, spatial variation, and benthic contribution to the pelagic community (Bird Island, South Africa). Reg. Stud. Mar. Sci. 44, 101762. doi: 10.1016/j.rsma.2021.101762
Cotiyane-Pondo P., Bornman T. G., Dąbek P., Witkowski A., Smit A. J. (2020). Austral winter marine epilithic diatoms: Community composition and distribution on rocky substrate around the coast of South Africa. Estuar. Coast. Shelf Sci. 242, 106837. doi: 10.1016/j.ecss.2020.106837
Dąbek P., Ashworth M., Górecka E., Krzywda M., Bornman T. G., Sato S., et al. (2019). Toward a multigene phylogeny of the Cymatosiraceae (Bacillariophyta, Mediophyceae) II: morphological and molecular insights into the taxonomy of the forgotten species Campylosira africana and of Extubocellulus, with a description of two new taxa. J. Phycol. 55, 425–441. doi: 10.1111/jpy.12831
Dąbek P., Ashworth M., Witkowski A., Li C., Bornman T. G., Gonçalves V., et al. (2017). Towards a multigene phylogeny of the Cymatosiraceae (Bacillariophyta, Mediophyceae) I: novel taxa within the subfamily Cymatosiroideae based on molecular and morphological data. J. Phycol. 53, 342–360. doi: 10.1111/jpy.12501
Dąbek P., Sabbe K., Witkowski A., Archibald C., Kurzydłowski K. J., Zgłobicka I. (2013). Cymatosirella Dąbek, Witkowski & Sabbe gen. nov., a new marine benthic diatom genus (Bacillariophyta) belonging to the family Cymatosiraceae. Phytotaxa 121, 42–56. doi: 10.11646/phytotaxa.121.1.2
Dąbek P., Witkowski A., Archibald C. (2014). Minutocellus africana Dąbek & Witkowski sp. nov.: new marine benthic diatom (Bacillariophyta, Cymatosiraceae) from Lamberts Bay, Western Cape Province, South Africa. Nova Hedwigia 99, 223–232. doi: 10.1127/0029-5035/2014/0191
Damsté J. S., Muyzer G., Abbas B., Rampen S. W., Massé G., Allard W. G., et al. (2004). The rise of the rhizosolenid diatoms. Science 304 (5670), 584–587. doi: 10.1126/science.1096806
Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PloS One 5 (6), e11147. doi: 10.1371/journal.pone.0011147
Darriba D., Posada D., Kozlov A. M., Stamatakis A., Morel B., Flouri T. (2020). ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37 (1), 291–294. doi: 10.1093/molbev/msz189
de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., et al. (2015). Ocean plankton. eukaryotic plankton diversity in the sunlit ocean. Science 348 (6237), 1261605. doi: 10.1126/science.1261605
Emanuel B. P., Bustamante R. H., Branch G. M., Eekhout S., Odendaal F. J. (1992). A zoogeographic and functional approach to the selection of marine reserves on the west coast of South Africa. S. Afr. J. Mar. Sci. 12 (1), 341–354. doi: 10.2989/02577619209504710
Forbes E., Hallegraeff G. M. (2002). “Transport of potentially toxic pseudo-nitzschia diatom species via ballast water,” in Proceedings of the 15th international diatom symposium 1998. Ed. Gantner J. J. (A.R.G. Gantner, Ruggell (Liechtenstein): Koenigstein), pp 509–pp 520.
Francezon N., Herbaut M., Bardeau J.-F., Cougnon C., Bélanger W., Tremblay R., et al. (2021). Electrochromic properties and electrochemical behavior of marennine, a bioactive blue-green pigment produced by the marine diatom Haslea ostrearia. Mar. Drugs 19 (4), 231. doi: 10.3390/md19040231
Gagnon J. (2004). Création d’outils pour l’automatisation d’analyses phylogénétiques de génomes d’organites [Development of bioinformatic tools for the phylogenetic analyzes of organellar genomes] [M.Sc. dissertation] (Québec city: University of Laval).
Gastineau R., Davidovich N. A., Bardeau J. F., Caruso A., Leignel V., Hardivillier Y., et al. (2012b). Haslea karadagensis (Bacillariophyta): a second blue diatom, recorded from the Black Sea and producing a novel blue pigment. Eur. J. Phycol. 47, 469–479. doi: 10.1080/09670262.2012.741713
Gastineau R., Davidovich N. A., Davidovich O. I., Lemieux C., Turmel M., Wróbel R. J., et al. (2021b). Extreme enlargement of the inverted repeat region in the plastid genomes of diatoms from the genus Climaconeis. Int. J. Mol. Sci. 22 (13), 7155. doi: 10.3390/ijms22137155
Gastineau R., Davidovich N. A., Hansen G., Rines J., Wulff A., Kaczmarska I., et al. (2014a). Haslea ostrearia-like diatoms: biodiversity out of the blue. Adv. Bot. Res. 71, 441–446. doi: 10.1016/B978-0-12-408062-1.00015-9
Gastineau R., Hansen G., Davidovich N. A., Davidovich O., Bardeau J. F., Kaczmarska I., et al. (2016). A new blue-pigmented hasleoid diatom, Haslea provincialis, from the Mediterranean Sea. Eur. J. Phycol. 51 (2), 156–170. doi: 10.1080/09670262.2015.1110861
Gastineau R., Hansen G., Poulin M., Lemieux C., Turmel M., Bardeau J.-F., et al. (2021a). Haslea silbo, a novel cosmopolitan species of blue diatoms. Biology 10 (4), 328. doi: 10.3390/biology10040328
Gastineau R., Leignel V., Jacquette B., Hardivillier Y., Wulff A., Gaudin P., et al. (2013). Inheritance of mitochondrial DNA in the pennate diatom Haslea ostrearia (Naviculaceae) during auxosporulation suggests a uniparental transmission. Protist 164, 340–351. doi: 10.1016/j.protis.2013.01.001
Gastineau R., Pouvreau J. B., Hellio C., Morançais M., Fleurence J., Gaudin P., et al. (2012a). Biological activities of purified marennine, the blue pigment responsible for the greening of oysters. J. Agric. Food Chem. 60 (14), 3599–3605. doi: 10.1021/jf205004x
Gastineau R., Prasetiya F. S., Falaise C., Cognie B., Decottignies P., Morançais M., et al (2018). “Marennine-like pigments: Blue diatom or green oyster cult?,” in Blue biotechnology: Production and use of marine molecules, vol. vol. 1 . Eds. La Barre S., Bates S. S. Weinheim, Germany, Wiley‐VCH Verlag GmbH & Co. KGaA. doi: 10.1002/9783527801718.ch16
Gastineau R., Turcotte F., Pouvreau J.-B., Morançais M., Fleurence J., Windarto E., et al. (2014b). Marennine, promising blue pigments from a widespread Haslea diatom species complex. Mar. Drugs 12, 3161–3189. doi: 10.3390/md12063161
Giffen M. H. (1963). Contributions to the diatom flora of South Africa I. diatoms of the estuaries of the Eastern Cape Province. Hydrobiologia 21 (3-4), 201–265. doi: 10.1007/BF01025943
Giffen M. H. (1966). Contribution to the diatom flora of South Africa. III. diatoms of the marine littoral regions at Kidd’s Beach near East London, Cape Province, South Africa. Nova Hedwigia 13, 245–292.
Giffen M. H. (1970a). ““Contributions to the diatom flora of South Africa IV. the marine littoral diatoms of the estuary of the Kowie River, Port Alfred, Cape Province”,” in Diatomaceae II, vol. 31 . Eds. Gerloff J., Cholnoky J. B. (Berlin, Stuttgart, Germany: Nova Hedwigia Beih.), 259–312.
Giffen M. H. (1970b). New and interesting marine and littoral diatoms from Sea Point, near Cape Town, South Africa. Bot. Mar. 13, 89–99. doi: 10.1515/botm.1970.13.2.81
Giffen M. H. (1971). Marine littoral diatoms from the Gordon’s Bay, region of False Bay Cape Province, South Africa. Bot. Mar. 14, 1–16. doi: 10.1515/botm.1971.14.s1.1
Giffen M. H. (1973). Diatoms of the marine littoral of Steenberg’s Cove in st. Helena bay, Cape Province, South Africa. Bot. Mar. 16, 32–48. doi: 10.1515/botm.1973.16.1.32
Giffen M. H. (1975). An account of the littoral diatoms from Langebaan, Saldanha Bay, Cape Province, South Africa. Bot. Mar. 18, 71–95. doi: 10.1515/botm.1975.18.2.71
Giffen M. H. (1976). A further account of the marine littoral diatoms of the Saldanha Bay lagoon, Cape Province, South Africa. Bot. Mar. 19, 379–394. doi: 10.1515/botm.1976.19.6.379
Giffen M. H. (1984). A checklist of marine littoral diatoms from Namaqualand, South Africa. Bacillaria 7, 179–200.
Gordon D., Green P. (2013). Consed: a graphical editor for next-generation sequencing. Bioinformatics 29, 2936–2937. doi: 10.1093/bioinformatics/btt515
Górecka E., Gastineau R., Davidovich N. A., Davidovich O. I., Ashworth M. P., Sabir J. S. M., et al. (2021). Mitochondrial and plastid genomes of the monoraphid diatom Schizostauron trachyderma. Int. J. Mol. Sci. 22 (20), 11139. doi: 10.3390/ijms222011139
Griffiths C. L., Robinson T. B., Lange L., Mead A. (2010). Marine biodiversity in South Africa: An evaluation of current states of knowledge. PloS One 5 (8), e12008. doi: 10.1371/journal.pone.0012008
Guillard R. R. L. (1975). ““Culture of phytoplankton for feeding marine invertebrates”,” in Culture of marine invertebrate animals. Eds. Smith W. L., Chanley M. H. (Boston, MA: Springer), 29–60. doi: 10.1007/978-1-4615-8714-9_3
Hallegraeff G. M., Bolch C. J. (1992). Transport of diatom and dinoflagellate resting spores in ships’ ballast water: implications for plankton biogeography and aquaculture. J. Plankton Res. 14 (8), 1067–1084. doi: 10.1093/plankt/14.8.1067
Hamsher S. E., Keepers K. G., Pogoda C. S., Stepanek J. G., Kane N. C., Kociolek J. P. (2019). Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PloS One 14 (7), e0217824. doi: 10.1371/journal.pone.0217824
Heiden H., Kolbe R. W. (1928). ““Die marinen diatomeen der deutschen südpolar-expedition 1901-1903”,” in Deutsche sudpolar-expedition 1901-1903, vol. Vol. VIII. (Berlin und Leipzig: Walter de Gruyter & Co.). herausgegeben von Erich von DrygalskiBotanik (no. 5).
Hendey N. I. (1937). “The plankton diatoms of the southern seas,” in Discovery reports 16 (Cambridge: University Press), 151–364.
Hildebrand M., Hasegawa P., Ord R. W., Thorpe V. S., Glass C., Volcani B. E. (1992). Nucleotide sequence of diatom plasmids: identification of open reading frames with similarity to site-specific recombinases. Plant Mol. Biol. 19 (5), 759–770. doi: 10.1007/BF00027072
Hinrichsen E. (2019) Biodiversity risk and benefit assessment for Pacific oyster (Crassostrea gigas) in South Africa. Available at: https://aquasea.csir.co.za/wp-content/uploads/2019/12/Appendix-C-1.7_BRBA-Pacific-Oyster.pdf.
John J. (2016). “Diatom flora of australia. vol. 1: Diatoms from Stradbroke and Fraser Island, Australia,” in Taxonomy and biogeography (Oberreifenberg, Germany: Koeltz Botanical Books).
John J. (2018). “The diatom flora of Australia, vol. 2: Diatoms from Tasmania,” in Taxonomy and biogeography (Oberreifenberg, Germany: Koeltz Botanical Books).
Kaleli A., Car A., Witkowski A., Krzywda M., Riaux-Gobin C., Solak C. N., et al. (2020). Biodiversity of carapace epibiont diatoms in loggerhead sea turtles (Caretta caretta Linnaeus 1758) in the Aegean Sea Turkish coast. PeerJ 8, e9406. doi: 10.7717/peerj.9406
Kalvari I., Nawrocki E. P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., et al. (2020). Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 49 (D1), D192–D200. doi: 10.1093/nar/gkaa1047
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim J. I., Jo B. Y., Park M. G., Yoo Y. D., Shin W., Archibald J. M. (2022). Evolutionary dynamics and lateral gene transfer in Raphidophyceae plastid genomes. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.896138
Klein G., MacIntosh K., Kaczmarska I., Ehrman J. M. (2010). Diatom survivorship in ballast water during trans-pacific crossings. Biol. Invasions 12, 1031–1044. doi: 10.1007/s10530-009-9520-6
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li C., Ashworth M., Witkowski A., Dąbek P., Medlin L., Kooistra W., et al. (2015). New insights into Plagiogrammaceae (Bacillariophyta) based on multigene phylogenies and morphological characteristics with the description of a new genus and three new species. PloS One 10, e0139300, p.1–26. doi: 10.1371/journal.pone.0139300
Li C., Gastineau R., Turmel M., Witkowski A., Otis C., Car A., et al. (2019). Complete chloroplast genome of the tiny marine diatom Nanofrustulum shiloi (Bacillariophyta) from the Adriatic Sea. Mitochondrial DNA. Part B Resour. 4 (2), 3374–3376. doi: 10.1080/23802359.2019.1673245
Lohse M., Drechsel O., Kahlau S., Bock R. (2013). Organellar genome DRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581. doi: 10.1093/nar/gkt289
Lombard A. T., Strauss T., Harris J., Sink K., Attwood C., Hutchings L. (2004). South African national spatial biodiversity assessment 2004 technical report (Pretoria: South African National Biodiversity Institute), 1–96.
Majewska R. (2020). Tursiocola neliana sp. nov (Bacillariophyceae) epizoic on South African leatherback sea turtles (Dermochelys coriacea) and new observations on the genus Tursiocola. Phytotaxa 453, 1–15. doi: 10.11646/phytotaxa.453.1.1
Majewska R., Bosak S., Frankovich T. A., Ashworth M. P., Sullivan M. J., Robinson N. J., et al. (2019a). Six new epibiotic Proschkinia (Bacillariophyta) species and new insights into the genus phylogeny. Eur. J. Phycol. 54 (4), 1–23. doi: 10.1080/09670262.2019.1628307
Majewska R., Mayombo N. A. S., Smit A. J., Goosen W. (2022). New observations on the diatom genus Druehlago (Bacillariophyta) and description of two new species from South Africa. Phycol. 61, 75–88. doi: 10.1080/00318884.2021.2012037
Majewska R., Robert K., Van de Vijver B., Nel R. (2019b). A new species of Lucanicum (Cyclophorales, Bacillariophyta) associated with loggerhead sea turtles from South Africa. Bot. Lett. 167, 1–8. doi: 10.1080/23818107.2019.1691648
Majewska R., Santoro M., Bolaños F., Chaves G., De Stefano M. (2015). Diatoms and other epibionts associated with olive ridley (Lepidochelys olivacea) Sea turtles from the Pacific coast of Costa Rica. PloS One 10 (6), e0130351. doi: 10.1371/journal.pone.0130351
Mayombo N. A. S., Majewska R., Smit A. J. (2020). An assessment of the influence of host species, age, and thallus part on kelp-associated diatoms. Diversity 12 (10), 385. doi: 10.3390/d12100385
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Pogoda C. S., Keepers K. G., Hamsher S. E., Stepanek J. G., Kane N. C., Kociolek J. P. (2019). Comparative analysis of the mitochondrial genomes of six newly sequenced diatoms reveals group II introns in the barcoding region of cox1. Mitochondrial DNA Part A 30, 43–51. doi: 10.1080/24701394.2018.1450397
Poulin M., Méléder V., Mouget J.-L. (2019). Typification of the first recognized blue pigmented diatom, Haslea ostrearia (Bacillariophyceae). Plant Ecol. Evol. 152 (2), 402–408. doi: 10.5091/plecevo.2019.1622
Pouvreau J. B., Morançais M., Masse G., Rosa P., Robert J. M., Fleurence J., et al. (2006). Purification of the blue-green pigment “marennine” from the marine tychopelagic diatom Haslea ostrearia (Gaillon/Bory) Simonsen. J. Appl. Phycol. 18, 769–781. doi: 10.1007/s10811-006-9088-9
Pouvreau J. B., Morançais M., Taran F., Rosa P., Dufossé L., Guérard F., et al. (2008). Antioxidant and free radical scavenging properties of marennine, a blue-green polyphenolic pigment from the diatom Haslea ostrearia (Gaillon/Bory) simonsen responsible for the natural greening of cultured oysters. J. Agric. Food Chem. 56 (15), 6278–6286. doi: 10.1021/jf073187n
Prasetiya F. S., Decottignies P., Tremblay R., Mouget J.-L., Cognie B. (2019a). Does culture supernatant of Haslea ostrearia containing marennine affect short-term physiological traits in the adult blue mussel Mytilus edulis? Aquac. Rep. 15, 100288, p. 1–6. doi: 10.1016/j.aqrep.2019.100228
Prasetiya F. S., Decottignies P., Tremblay R., Mouget J.-L., Sunarto S., Iskandar I., et al. (2020). Not only greening: The effects of marennine produced by Haslea ostrearia on physiological traits of three bivalve species. Aquac. Rep. 18, 100546. doi: 10.1016/j.aqrep.2020.100546
Prasetiya F. S., Gastineau R., Poulin M., Hardivillier Y., Syakti A. D., Lemieux C., et al. (2019b). Haslea nusantara, a new blue diatom from the Java Sea, Indonesia: Morphology, biometry and molecular characterizations. Plant Ecol. Evol. 152 (2), 188–202. doi: 10.5091/plecevo.2019.1623
Prasetiya F. S., Ramdhani D. S., Mulyani Y., Agung M. U. K., Arsad S., Mouget J.-L. (2021b). Antioxidant activities of culture supernatant Haslea ostrearia adapted in Indonesia. AACL Bioflux 14 (1), 111–119.
Prasetiya F. S., Safitri I., Widowati I., Cognie B., Decottignies P., Gastineau R., et al. (2015). Does allelopathy affect co-culturing Haslea ostrearia with other microalgae relevant to aquaculture? J. Appl. Phycol. 28, 2241–2254. doi: 10.1007/s10811-015-0779-y
Prasetiya F. S., Sunarto A. S., Astuty S., Agung M. U. K., Stavrakakis C., Mouget J.-L. (2021a). Effect of culture supernatant from Haslea ostrearia on the clearance rate and survival rate of adult Pacific oyster Crassostrea gigas infected with Vibrio aestuarianus. AACL Bioflux 14 (4), 2050–2060.
Robert J. M. (1973). La diatomée Navicula ostrearia Bory en baie de Bourgneuf. Rev. Des. Travaux l’Institut Des. Peˆches Maritimes (ISTPM) 37, 363–368. Available at: https://archimer.ifremer.fr/doc/1973/publication-2148.pdf
Robinson T. B., Griffiths C. L., Tonin A., Bloomer P., Hare M. P. (2005). Naturalized populations of oysters, Crassostrea gigas, along the South African coast: distribution, abundance and population structure. J. Shellfish Res. 24, 443–450. doi: 10.2983/0730-8000(2005)24[443:NPOOCG]2.0.CO;2
Ruck E. C., Nakov T., Jansen R. K., Theriot E. C., Alverson A. J. (2014). Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 6 (3), 644–654. doi: 10.1093/gbe/evu039
Saunders K., Lane C., Cook S., McMinn A., Hallegraef G. M.. (2010). “Benthic diatoms,” in Algae of Australia: Phytoplankton of temperate coastal waters. Ed. Hallegraeff G. M. (Clayton, Australia: CSIRO Publishing), 83–144.
Schneider C., Rasband W., Eliceiri K. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 9, 671–675. doi: 10.1038/nmeth.2089
Simonsen R. (1974). The diatom plankton of the Indian Ocean expedition of R/V Meteor 1964-5, “Meteor”. Forschungsergebnisse. Reihe D: Biologie 19, 1–107.
Smit A. J., Bolton J. J., Anderson R. J. (2017). Seaweeds in two oceans: Beta-diversity. Front. Mar. Sci. 4. doi: 10.3389/fmars.2017.00404
Smit A. J., Roberts M., Anderson R. J., Dufois F., Dudley S. F. J., Bornman T. G., et al. (2013). A coastal seawater temperature dataset for biogeographical studies: large biases between in situ and remotely-sensed data sets around the coast of South Africa. PloS One 8 (12), e81944. doi: 10.1371/journal.pone.0081944
Stephenson T. A. (1948). The constitution of the intertidal fauna and flora of South Africa, part 3. Ann. Natal Mus. 11, 207–324.
Taylor F. J. R. (1967). Phytoplankton of the south Western Indian Ocean. Nova Hedwigia 12 (3-4), 433–476.
Teske P., von der Heyden S., Mcquaid C., Barker N. (2011). A review of marine phylogeography in southern Africa. S. Afr. J. Sci. 107, 514, 1–11. doi: 10.4102/sajs.v107i5/6.514
Toonen R. J., Bowen B. W., Iacchei M., Briggs J. C. (2016). ““Biogeography, marine”,” in Encyclopedia of evolutionary biology, vol. vol. 1 . Ed. Kliman R. M. (Oxford, UK: Oxford Academic Press), 166–178. doi: 10.1016/B978-0-12-800049-6.00120-7
Trochon P. (1954). Etude sur la reproduction de l’huître plate Ostrea edulis dans les claires de la egion de Marennes-Oléron. special scientific meeting “Oyster and mussel culture”. Int. Council Explor. Sea 21, 1–4. https://archimer.ifremer.fr/doc/00004/11500/8070.pdf
Van de Vijver B., Robert K., Majewska R., Frankovich T. A., Panagopoulou A., Bosak S., et al. (2020). Geographical variation in the diatom communities associated with loggerhead sea turtles (Caretta caretta). PloS One 15 (7), e0247350, 1–20. doi: 10.1371/journal.pone.0236513
Keywords: Bacillariophyceae, marennine, Kei Mouth, mitochondrial genome, plastid genome, cox1 phylogeny
Citation: Dąbek P, Gastineau R, Bornman TG, Lemieux C, Turmel M, Hallegraeff G, Mouget J-L and Witkowski A (2022) The blue diatom Haslea ostrearia from the Indian Ocean coast of South Africa, with comparative analysis of Haslea organellar genomes. Front. Mar. Sci. 9:950716. doi: 10.3389/fmars.2022.950716
Received: 23 May 2022; Accepted: 31 August 2022;
Published: 27 September 2022.
Edited by:
Peter Von Dassow, Pontificia Universidad Católica de Chile, ChileReviewed by:
Daniele De Luca, University of Naples Federico II, ItalyCopyright © 2022 Dąbek, Gastineau, Bornman, Lemieux, Turmel, Hallegraeff, Mouget and Witkowski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Przemysław Dąbek, cGRhYmVrQHVzei5lZHUucGw=
†ORCID: Przemysław Dąbek, orcid.org/0000-0002-3736-3011
Romain Gastineau, orcid.org/0000-0001-8661-5118
Thomas G. Bornman, orcid.org/0000-0003-1868-479X
Claude Lemieux, orcid.org/0000-0001-9580-8042
Monique Turmel, orcid.org/0000-0001-7060-035X
Gustaaf Hallegraeff, orcid.org/0000-0001-8464-7343
Jean-Luc Mouget, orcid.org/0000-0003-0154-7289
Andrzej Witkowski, orcid.org/0000-0003-1714-218X
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.