- 1College of Information and Network Engineering, Anhui Science and Technology University, Bengbu, China
- 2Mechanical and Electrical Engineering College, Hainan University, Haikou, China
Microalgae are essential parts of marine ecology, and they play a key role in species balance. Microalgae also have significant economic value. However, microalgae are too tiny, and there are many different kinds of microalgae in a single drop of seawater. It is challenging to identify microalgae species and monitor microalgae changes. Machine learning techniques have achieved massive success in object recognition and classification, and have attracted a wide range of attention. Many researchers have introduced machine learning algorithms into microalgae applications, and similarly significant effects are gained. The paper summarizes recent advances based on various machine learning algorithms in microalgae applications, such as microalgae classification, bioenergy generation from microalgae, environment purification with microalgae, and microalgae growth monitor. Finally, we prospect development of machine learning algorithms in microalgae treatment in the future.
Introduction
Microalgae in the ocean are usually single-celled organisms that play a crucial part in marine ecology (Chew et al., 2017). Microalgae are primary organic matter producers in the sea. Microalgae absorb carbon dioxide and convert it into organic matter, while releasing oxygen through photosynthesis (Chakdar et al., 2021). As a result, microalgae are crucial food sources for organisms in ocean, and they could reduce the greenhouse effect (Mochdia and Tamaki, 2021). In addition, microalgae have considerable social and commercial value. Microalgae are capable of purifying sewage, because they can absorb nitrogen and phosphorus. The high content of oil and fat in microalgae makes them an ideal raw material for biodiesel product (Adamczak et al., 2009; Chowdhury and Loganathan, 2019; Mofijur et al., 2019).
Microalgae species recognition and growth monitor are crucial steps in actual applications (Gomez-Espinoza et al., 2018). Microalgae are commonly microscopic, and there are usually many different kinds of microalgae species in a single sample (Ferro et al., 2018) (Figure 1). These characteristics make the identification, classification, and analysis of microalgae a very challenging task (Andersen and Kawachi, 2005). Traditional manual methods are not only time-consuming, they also require much skill and experience for the operators (Peniuk et al., 2016; Saputro et al., 2019). As a result, the efficiency and scope of microalgae applications are greatly limited. Faster and more efficient methods for the classification, identification, and analysis of microalgae are needed. (Sá et al., 2013; Wei et al., 2017).
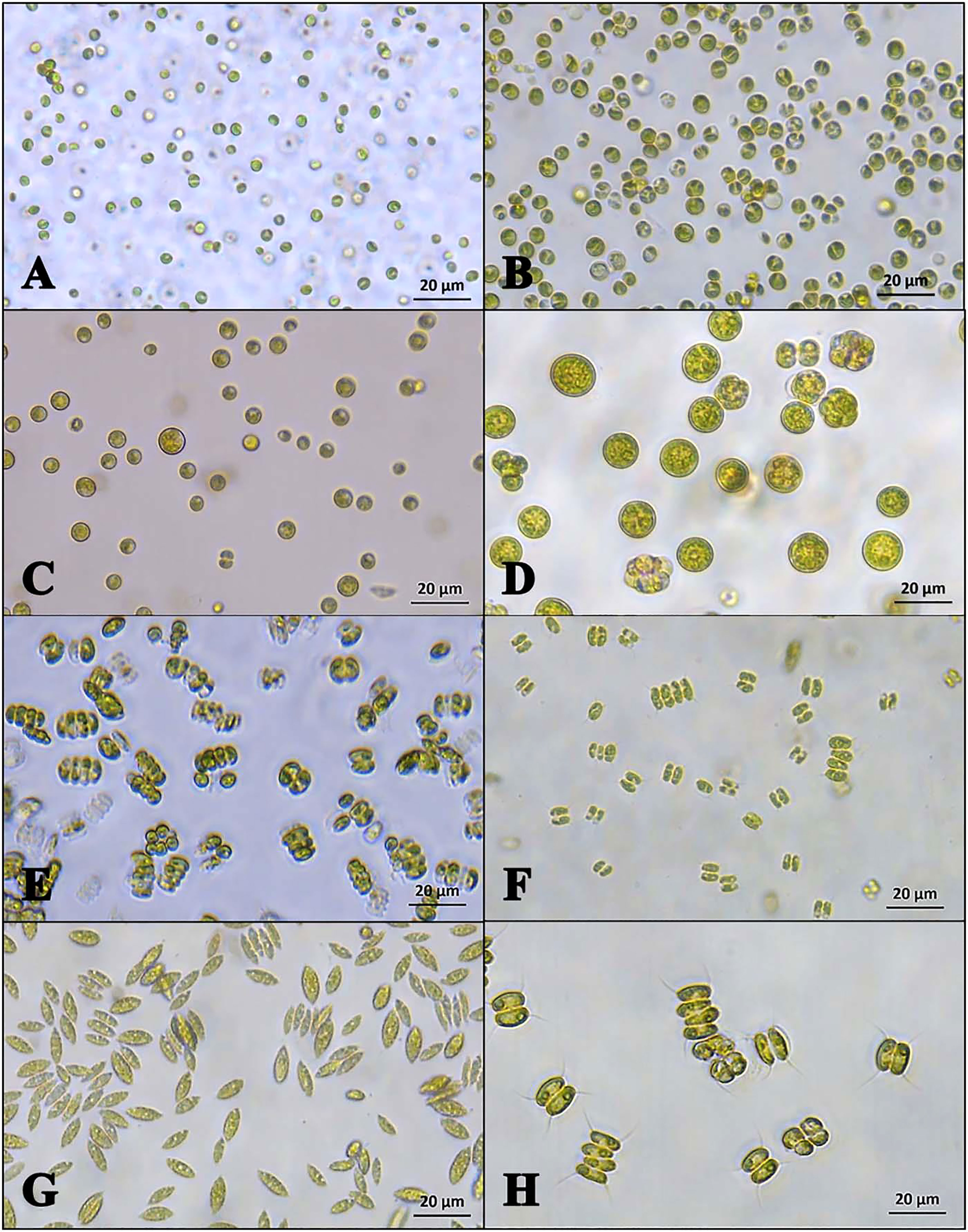
Figure 1 Microscopic images of microalgae: (A) Glycophilic Chlorella or Chlorella saccharophilus; (B) Chlorellasorokiniana; (C) Chlorella vulgaris; (D) Coelastrella; (E) Desmodesmus; (F) Desmodesmus; (G) Scenedesmus obliquus; (H) Scenedesmus. (reproduced with permission from Ferro et al., 2018).
Machine learning is a collection of data-driven algorithms in essential (Rosenblatt, 1958; Rumelhart et al., 1986). In recent years, data resource and computer computing power have enhanced significantly. Machine learning has achieved great success and is applied widely in many fields (El Naqa and Murphy, 2015; Jordan and Mitchell, 2015; Liakos et al., 2018). In particular, machine learning has greatly facilitated the development of digital image processing and speech recognition (McCulloch and Pitts, 1990; He et al., 2015). Many researchers have introduced machine learning techniques into the field of microalgae process to identify the species of microalgae, and monitor the growth process of microalgae with outstanding results as well (Carleo et al., 2019).
This paper summarizes the state of machine learning algorithms used in microalgae treatment, with a focus on summing up the advances made in recent years. Firstly, the article explains the basic principle of machine learning algorithms such as support vector machine, decision tree, random forest, and neural network. The development of microalgae classification, the conversion from microalgae to bioenergy, microalgae for environmental protection, and the monitoring of microalgae growth stage with machine learning algorithms are then explained in detail. With all the summaries, we list machine learning methods different from traditional manual operation in microalgae treatment. This is a pretty reference for the following researchers and workers in the field.
Basic principles of several machine learning algorithms
Artificial intelligence is the theory and method that allows computers to reason and simulate human thinking based on previous perceptions or experiences (Sain, 1996). Computers with artificial intelligence is able to do more complicated work that needs logical ability. As an implementation of artificial intelligence, machine learning has not only become increasingly mature in its theoretical basis, but has achieved great success in practical applications (Dietterich, 1997). Machine learning is a multi-disciplinary interdisciplinary discipline that integrates statistics, data mining, probability theory, information theory, algorithmic analysis, and other fields (Vapnik, 1999).
The basic machine learning process is the analysis and learning of data with algorithms, and subsequent judgment and prediction about the actual situations are made automatically (Wei et al., 2019)(Figure 2A). A framework with many parameters is first built, and then the prepared data is fed into the model. The parameters are continuously adjusted until they match or close to the correct result (Bishop, 2013). Machine learning contains supervised learning, unsupervised learning, semi-supervised learning, and reinforcement learning, depending on the training model. Many different models can be used for machine learning training, and a comprehensive description of centralized representative models are in the following (Mahesh, 2020).
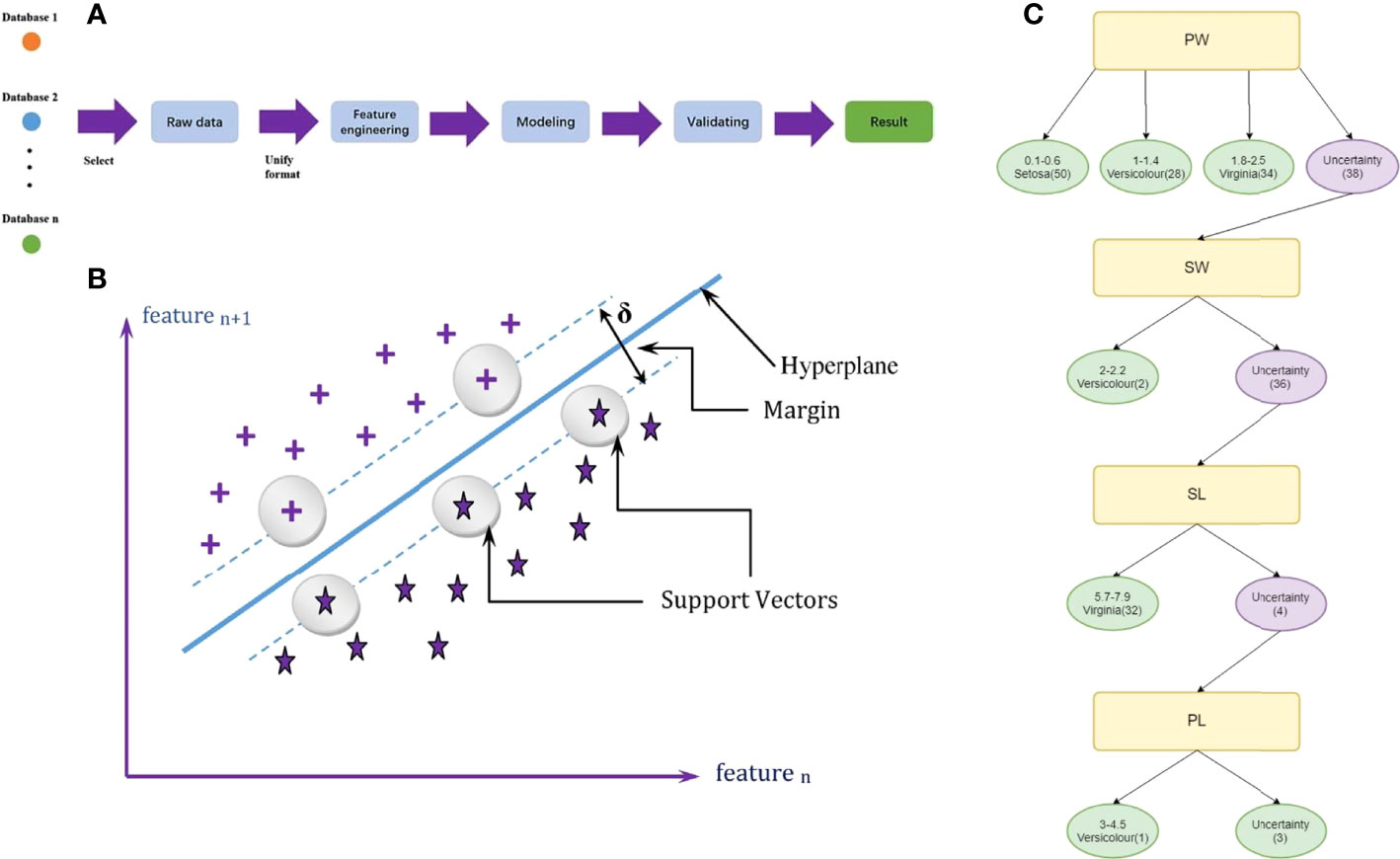
Figure 2 (A) Flowchart for machine learning. (reproduced from an open access article). (B) Schematic diagram of SVM. (reproduced with permission from Deka, 2014). (C) A decision tree for identification based on iris. (reproduced from an open access article).
Support vector machine
Support vector machine(SVM) is a supervised learning framework mainly utilized in classification and regression calculations (Boser et al., 1992). The SVM constructs a hyperplane in the existing data to differentiate the known data according to the specific needs. When the SVM processes the new data, it is continuously optimized according to the output results (Hearst et al., 1998; Suykens and Vandewalle, 1999).
Assume that the data will be processed by the SVM is a sample set: D = (xi, yi), i = 1, 2,…,nx is the input data, y is the output result, and n is the total amount in the case.
Linear separable model
If the data is linearly separable, data types can be classified by a hyperplane (Chen et al., 2005). The optimal plane with the farthest distance from the two types of data and the plane can be represented as:
ω means the coefficient vector that judges the hyperplane direction, and b represents the bias vector which describes the distance between the hyperplane and the data sample set (Deka, 2014) (Figure 2B).
The closest hyperplane between the positive and the negative samples can then be expressed as:
The hyperplanes between different sample data can then be uniformly expressed as:
The correctness of the sample classification is converted to the interval distance between the sample data to the hyperplane, . The coefficients ω and b for optimal hyperplane can be found by searching the maximum value of the equation, . The equation is equivalent to: .
This is a programming problem of convex quadratic, the solution of which should be obtained by introducing the Lagrange multiplier αi, is the solution to the pairwise optimization issue, and the subscript j satisfies .
Nonlinear model
SVM handles nonlinear data by introducing kernel function to enhance the dimensionality of the feature space (Pradhan, 2012). Suppose that the kernel function ϕ(x) is employed to represent the feature vector after the map of the sample set, the hyperplane representation can be denoted as:
After introducing the Lagrangian operator αi and using the equation κ(xi,xj)=〈ϕ(xi)ϕ(xj)〉 to represent the inner product ϕ(xi)Tϕ(xj) , the solution of the hyperplane equation can be obtained:
The type of the kernel function denotes the changed distribution of the original sample in one-higher dimensional space (Widodo and Yang, 2007). The function is the most significant variable for a nonlinear support vector machine framework. Much research reveals that the efficiency of the framework relays greatly on the kernel function (Meyer et al., 2003; Shahid et al., 2015). The common kernel functions are linear kernel function, polynomial kernel function, radial basis kernel function, and Sigmoid function (Wang et al., 2008).
Decision tree
The decision tree algorithm is also a classification and regression method that belongs to unsupervised learning (Quinlan, 1986). The internal node in a decision tree means an attribute, a branch is a chosen path to obtain the final result, and each leaf node indicates a species (Li et al., 2019) (Figure 2C). To construct a decision tree model, a training dataset is essential.
Decision tree learning essentially generalizes features in the training dataset and gains the rules to partition final sample data into smaller ones. Based on the different partition ways, many decision trees can be obtained through the same training dataset (Myles et al., 2004). A decision tree with excellent performance depends less on the training dataset, and it means the tree owns perfect generalization ability. The most commonly used probability fits the training dataset well, and predicts the following unknown data perfectly. The process of decision tree construction has three steps: feature selection, decision tree generation, and decision tree pruning.
Feature selection
Feature selection refers to the choice of features in the training dataset suitable for the current dataset to be divided into many parts, and one part means a leaf in the decision tree (Pal and Mather, 2003). The dataset will be divided recursively until the sample points can be classified into their respective categories, and the complete tree is constructed. Many prediction criteria can be used to choose features, and each choice leads to a different decision tree algorithm. The standard commonly employed construction tree algorithms are the ID3 algorithm, the C4.5 algorithm, and the CART algorithm. The algorithms utilize information gain, information gain rate, and Gini index respectively, to determine the features that divide the dataset (Patel and Prajapati, 2018).
The essence of the ID3 algorithm is the attributes chosen with the information gain benchmark, and the attribute with the maximum information gain will be utilized to divide the dataset recursively (Wellner et al., 2017). The less the expected information is, the greater the information gain will be, and the dataset owns higher purity. To describe the information gain clearly, the entropy and conditional entropy need to be explained first.
The entropy of the random variable X is expressed as:
n represents the n different discrete values of X, p(xi) represents the probability that X takes the value i.
To describe the non-determinacy of a random variable Y in the situation that the variable X is known, the conditional entropy is introduced:
The ID3 algorithm evaluates the information gain of feature A in the sample set D and the prediction is computed as:
After the information gains of all features are calculated, the feature with the largest information gain will divide the sample set D.
The disadvantage of information gain is that features could have a bias toward characteristics that have many taken values. If the count of different values taken by a feature is greater, the more likely the feature will be used as a split point. The most extreme case is that each result of the feature refers to a different outcome of the feature, then the information entropy is found to be 0, and the information gain is maximized. After improving the defect of the ID3 algorithm, the C4.5 algorithm is derived.
The C4.5 algorithm utilizes the information gain rate to measure the ability of features in ensemble classification (Elomaa, 1994). The information gain rate is described in the following:
Gain (A) represents the information gain generated by dividing the dataset using feature A, and H (A) is the information entropy of feature A. The C4.5 algorithm selects the property with the maximum information gain rate as the division attribute to partition the dataset (Sharma et al., 2013; Nugraha et al., 2020).
The CART algorithm uses the Gini index, which reflects the mixture of the framework as the splitting criterion (Ayyagari, 2020). The smaller the Gini index is, the lower the mixture will be, and the selected feature is better. The Gini index is defined as:
Generation of decision tree
The decision tree generation process grows from the root node and generates sub-nodes recursively top and down according to the chosen feature classification until the dataset is indistinguishable (Zhou and Chen, 2002). Based on various algorithms to generate a decision tree, we traverse the entire data sample from the root node downward to search for the most influential node in the current feature vector as the child node of the layer. Then, we continue to traverse downward and take the child node just obtained as the new parent node, and keep the recursion until the traversal stops at the leaf node (Swain and Hauska, 1977).
Pruning of decision tree
Decision trees are prone to overfitting and often require pruning to minimize the degree of the tree, alleviating overfitting by actively removing some branches and reducing the risk of overfitting. Pruning is one of the methods used to break decision tree branching. There are two pruning ways: pre-pruning and post-pruning. Pre-pruning sets a metric during tree growth and stops growing when that metric is reached. During post-pruning, the tree grows fully until minimum impurity values for all leaf nodes. Post-pruning is often more computationally costly than the pre-pruning manner, particularly in the enormous dataset. But the post-pruning method is still superior to the pre-pruning method in a small sample dataset (Friedl and Brodley, 1997).
Random forest
Random forest is an integrated learning approach, and the decision tree is the primary component unit of the random forest algorithm (Breiman, 2001). Since a single decision tree has the problem of low accuracy and overfitting, it overcomes the limitations by bringing numerous decision trees together. Compared to the decision tree algorithm, the random forest algorithm has better classification and regression performance. Compared with other machine learning algorithms such as SVM and deep learning algorithms, as convolution neural network an example, the random forest algorithm has quicker prediction speed and superior accuracy with relatively lower computing power.
The random forest algorithm is a unite of the Bagging algorithm and decision tree algorithm, which commonly utilizes a decision tree as a basis for classification training. Finally, it makes accurate classification for samples with unknown outcomes by a voting method (Belgiu and Drăguţ, 2016) (Figure 3).
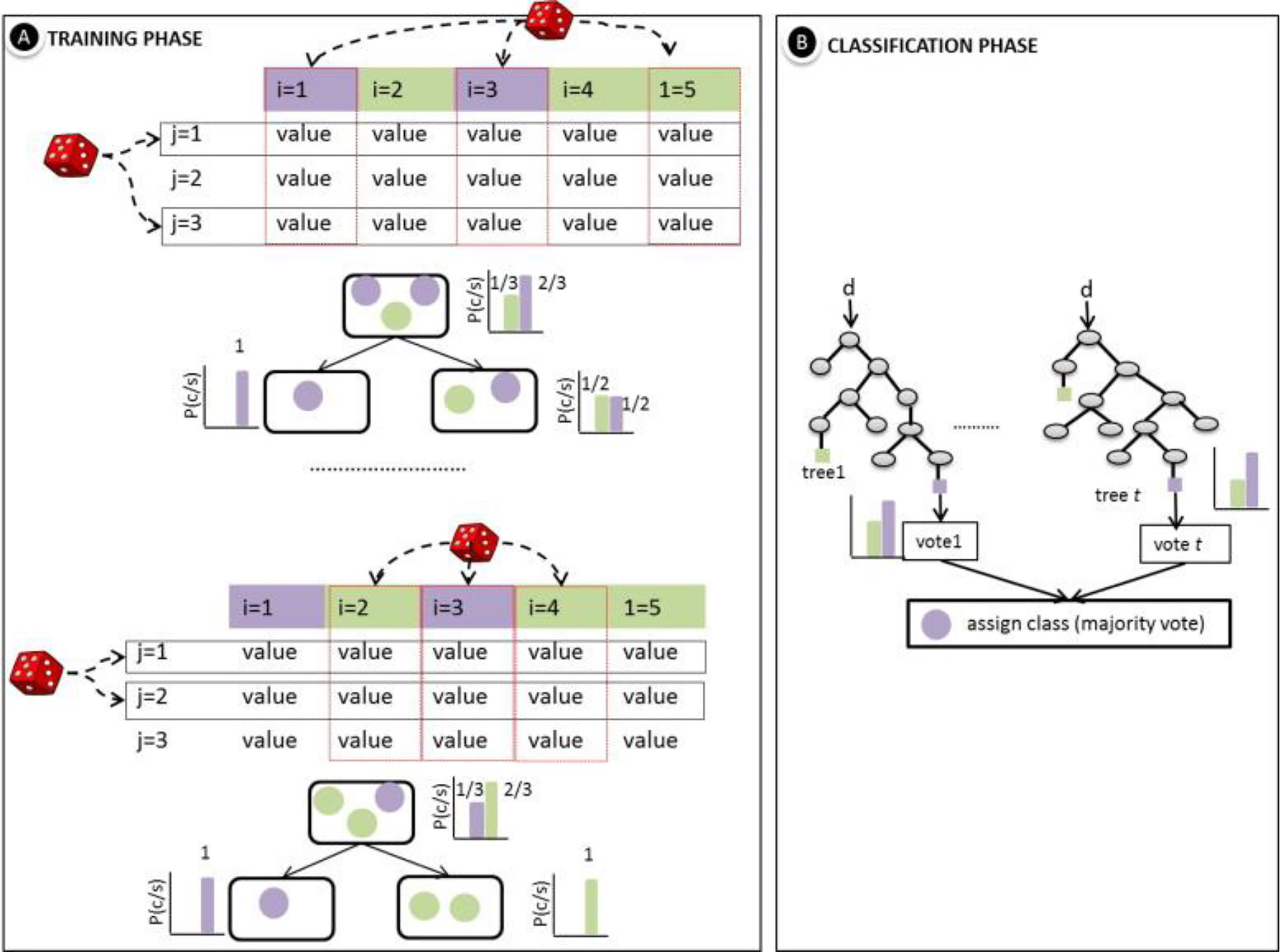
Figure 3 (A) Training steps of random forest. (B) Classification application of random forest. (reproduced with permission from Belgiu and Drăguţ, 2016).
Sample to generate train sets
Each tree in a random forest is different, and different datasets are needed to generate other trees. So different datasets are extracted from the original dataset to form different sub-datasets, which are used to train different decision trees (Paul et al., 2018). A standard method for extracting subsets of data is the Bagging method, which ensures that each tree is unrelated to the other, and thus reduces the risk of overfitting. The Bagging algorithm is a typical parallelized integrated learning method with no strong dependencies between the individual learners.
The Bagging method randomly draws a dataset from the original dataset and then puts the extracted data back into the original dataset before the next random draw. Thus, the dataset would be divided into many training subsets, which are used to construct different decision trees (Shi and Horvath, 2006).
Construct of decision trees
Once the training set for each tree is determined, it is time to construct the decision trees (Qi, 2012). During the construct process of each decision tree for the random forest, some features from the feature set of the sub-dataset are randomly selected to participate in the node split selection calculation as nodes to build the decision tree, and no pruning is done for each generated decision tree. The detailed decision tree construction process can be found in the section 2.2.
Result confirm by vote
The process of sections 2.3.1 and 2.3.2 above is repeated continuously, and will not stop until the number of trees reaches the required quantity. In this way, many different decision trees are built, and these trees are combined (Bonissone et al., 2010; Boulesteix et al., 2012). The classification result of each tree is voted on according to specific rules. The final random forest algorithm classification result is the decision tree result that gains the most votes. In the final vote, there are generally three methods: absolute majority vote, relative majority vote, and weighted vote.
The principle of absolute majority vote indicates that only more than half of the entire votes are cast for an option, and the option is chosen as the predicted outcome (Farnaaz and Jabbar, 2016). The relative majority vote is that the result with the most votes is selected as the expected outcome, and if there is more than one vote owns the most count. The final result is chosen randomly (Speiser et al., 2019). The weighted vote method means that all results are given a weight, which is equivalent to the weighted average process. The classification results of each decision tree are multiplied by the weight, and the weighted choices for each group are added. The category with the maximum number will be considered as the final result (Rodriguez-Galiano et al., 2012).
Neural network
The neural network technique is one of the machine learning ways, and it is skilled in dealing with non-linear data. A neural network is a simulation of the nervous system in the human brain, and the basic building blocks are neurons. The different arrangements of neurons divide neural networks into many types, for example, convolutional neural network, which is ideal for processing image and waveform data (McCulloch and Pitts, 1943).
Neuron
Neurons are the essential components of various neural networks and are the mathematical models of biological perceptual machines (Mohammed et al., 1995; Bakirtzis et al., 1996). By feeding training data into the neuron, a corresponding output can be obtained by some mathematical calculations on the neuron. A neuron is called a perceptron as well.
The structure of a neuron contains many inputs, but only one output (Sagheer et al., 2019) (Figure 4A). x1, x2,…,xn2are the input data of a neuron, and w1,w2…,wn are weights for input data; b is the bias, and f() expresses the activation function. The input parameters are multiplied by the weights and summed. Then biases are added and input into the activation function for processing (Tian and Noore, 2004; Cheng et al., 2015). The result of the activation function is the consequence of this perceptron. The whole process can be represented with the following equation:
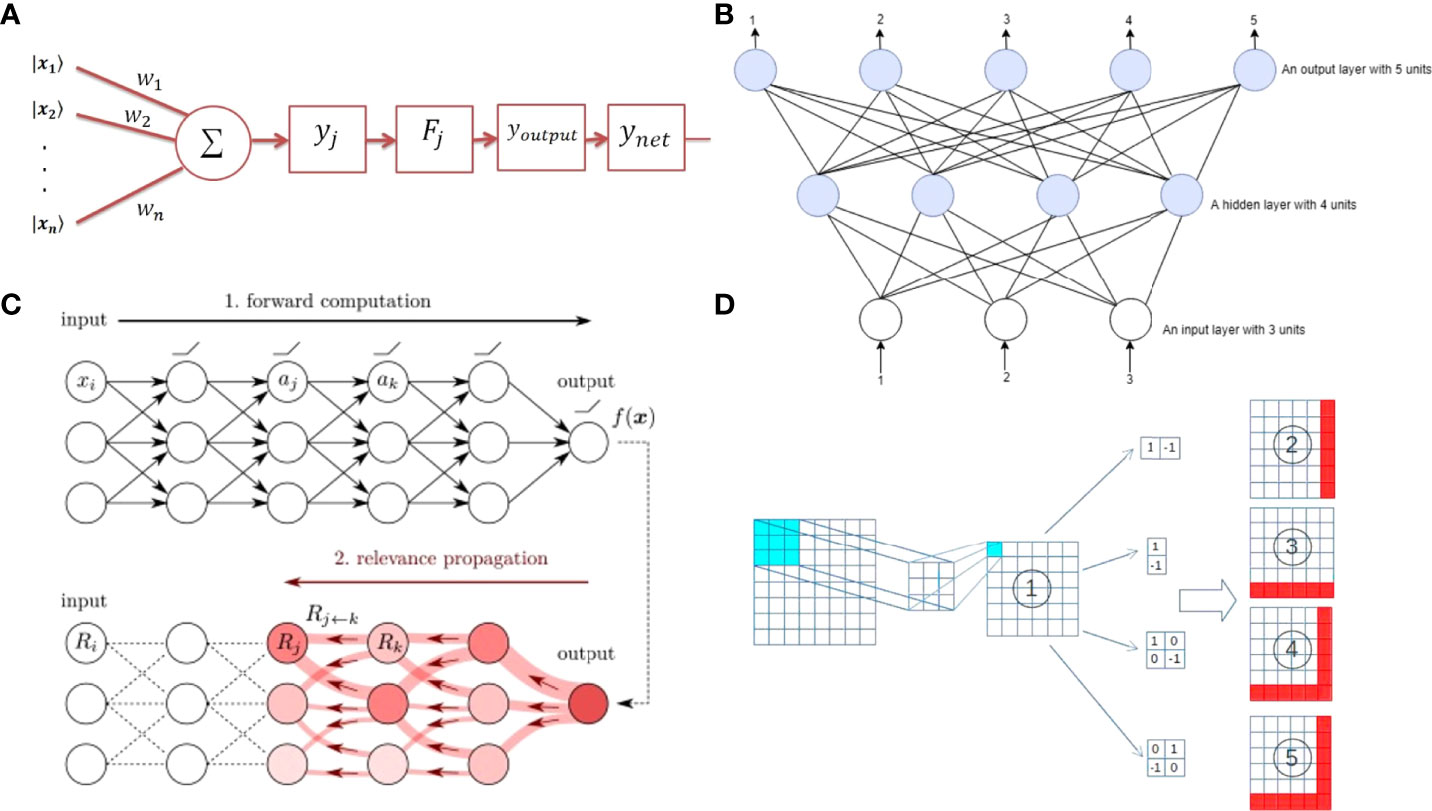
Figure 4 (A) Structure of a perceptron. (reproduced from an open access article). (B) Schematic diagram of an artificial neural network. (reproduced from an open access article). (C) Data forward propagation and error back propagation of a deep neural network. (reproduced from an open access article). (D) Diagram of convolution operation. (reproduced with permission from Sarıgül et al., 2019).
If >x=(x1,x2,…,xn) and ω=(ω1,ω2,…,ωn)T , the above equation could be transferred into y=f(ωx+b)
Common activation functions include ,f(x)=max(0,x ), etc.
Artificial neural network
Neurons have a simple structure and can only deal with linear problems, and neural networks are generally used to handle non-linear problems. Neural networks can solve complex non-linear input-output applications. The network is composed of numerous tiny neurons. In fact, a neural network is a combination of massive neurons according to specific rules. The neural network is called an artificial neural network (Abiodun et al., 2018) (ANN Figure 4B).
The typical ANN has three layers, namely, the input layer, the hidden layer, and the output layer according to their position in the left, middle, and right of the network. The input layer receives input data, the hidden layer is invisible to the outside world and calculates object features, and the output layer gives the final result. Each neuron in the N layer is contacted with all neurons in the N-1 layer, which is also called a fully connected neural network (Hsu et al., 1990).
Deep neural network
If there are more hidden layers in an ANN, there is a more powerful analysis ability, that the neural network owns. If a neural network contains more than two hidden layers, it is called a deep neural network (Montavon et al., 2018) (DNN Figure 4C). In practice, a neural network that includes just one hidden layer can satisfy any requirement, but the hidden layer needs a large number of neurons. A deep network performs the same role with fewer neurons.
DNN determines the relevance of features better through the mapping relations among the input and output data. Any data from the input layer is sent to every neuron in the hidden layer. When the size of the input data is too large, it is easy to over-fit the model with too many parameters. When the input data is unduly small, it is difficult for the model to learn helpful information from the limited data, resulting in underfitting (Cichy and Kaiser, 2019).
Convolutional neural network
Traditional neural network representations are constructed with one-dimensional vectors, which miss the spatial information of the objects. Researchers have devised a convolutional neural network (CNN) by introducing convolutional and pooling operations (LeCun and Bengio, 1995). The CNN can entirely get the local spatial semantic characteristics of an image, and pooling operations are able to extend the perceptual field to obtain more advanced image features for object recognition. Convolutional neural networks are composed of a cascade of a convolutional layer with a local field of perception and pooling layers with a down-sampling effect. The CNN holds the ability to extract hierarchical, multi-scale image features from images. The main application area of the convolutional neural network is image recognition, but it can also be used in video analysis and natural language processing (Albawi et al., 2017).
The convolution layer implements feature detection, extracts crucial information from the input data, and adds non-linear factors to the feature information through the activation function. Convolution is a regional operation in which native information of an image is acquired with a specific size convolution kernel applied to an image (Sarıgül et al., 2019)(Figure 4D). In the convolution layer of a CNN, the convolutional kernel extracts local features by sliding samples over the image matrix. The process is called the convolution operation. The operation could be described with the following formula:
M is the width of the convolution kernel, N is the height of the convolution kernel, W is the weight of the convolution kernel, X is the input data, and b is the bias.
The pooling layer is based on the convolutional layer to extract meaningful information from the image further, reducing the parameters in the network and the amount of computation by reducing the space size. The pooling layer also reduces the overfitting of the model and improves the fault tolerance of the model (Kuo, 2016). There are two main types of pooling, maximum pooling and mean pooling. The maximum pooling is a typical pooling operation that reduces the amount of data through a maximum value. Mean pooling, on the other hand, involves calculating the average value of an convolutional field as the pooling value for that space.
In a CNN, one or more fully connected layers are connected to pooling layers after multiple convolutional layers. Among the layers, each neuron in the N layer connects all neurons in the N-1 layer, but not in the same layer. The fully connected layers play two roles in the overall convolutional neural network. Firstly, it classifies the features based on different details extracted from the convolutional layers. Secondly, it reduces the impact of feature position shifts on the classification to a greater extent. The fully connected layer acts as a classifier (Acharya et al., 2017; Gu et al., 2018).
Microalgae detection and classification with machine learning
As unicellular organisms, microalgae are not only very microscopic, but also do not differ much from one species to another. Combined with the fact that thousands of species of microalgae may be present in a tiny sample, microalgae classification is a very challenging job. Traditional manual classification under a microscope is not only laborious, but also requires a high level of skill and experience for the operators. Therefore, the manual classification method of microalgae is usually inefficient and unsatisfied in terms of accuracy (Barsanti et al., 2021).
Machine learning algorithms based on data-driven models are very advantageous in dealing with different types of unstructured data (Rani et al., 2021). Much progress has been made in introducing machine learning algorithms in microalgae detection and classification work. The scheme allows computers to automatically learn the characteristics of different algae based on existing data and give classification results for new data. The data processed by machine learning algorithms are microalgae images obtained through microscopy, so no-marker and invasion-free data acquisition can be achieved. The operations avoid the tedious process of traditional staining and labeling steps and the damage to the microalgae growth environment (Zheng et al., 2021).
A label-free analysis model was devised by Claire Lifan Chen et al. to perform a rapid classification of microalgae (Chen et al., 2016). High-throughput imaging technology allowed the acquisition of 100,000 images of microalgae per second, while capturing rich information about microalgae and summarizing it into 16 features. They employed multiple machine learning algorithms to classify the unlabeled data. Practical experiments showed that the method was 17% more accurate than the traditional method and was well suited for high-throughput label-free microalgae classification. Çağatay Işıl et al. devised a novel portable cytometer to conduct label-free identification and analysis of microalgae (Işıl et al., 2021b). The device could analyze chemical perturbation in the external environment based on spectral features in microalgae images and classify microalgae based on deep learning technologies. In addition, the device could count the number of microalgae and analyze the interactions between them. The group also utilized a convolutional neural network to analyze the label-free spatial and spectral characteristics of microalgae to analyze the composition and growth status of microalgae (Işıl et al., 2021a). The ultimate goal was to confirm the interactions between microalgae and the response of microalgae to external contamination. The effect of the method was demonstrated by the mixed culture of single microalgae and multiple microalgae in copper-containing solutions. Thanks to the label-free technique, the tested microalgae samples could be directly put back into the original solution without contamination.
Iago Corrêa et al. utilized a convolutional neural network with five convolutional layers and three pooling layers, total eight layers, to classify microalgae (Correa et al., 2017). The dataset consisted of microalgae images and labels. The microalgae images were obtained by the team from South Atlantic seawater using FlowCAM equipment, and the tags were manually classified by multiple experts. The input data of the model could be low-resolution raw microalgae images without feature extraction based on microalgae images. The fully automated microalgae classification accuracy given to the model without image preprocesses and human intervention had reached 88.59%. The performance could be further improved if the data enhancement technique was added. D.P. Yadav et al. improved the traditional ResNeXt convolutional neural network for the recognition and classification of microalgae (Yadav et al., 2020). The dataset was sourced from the Physiology Research Center and the Internet, and the initial dataset contained only 100 images. After data augmentation, the dataset was expanded to 80,000 images, 80% employed for model training and the rest 20% for model validation. The scheme was experimentally validated to achieve 99.97% classification accuracy. P. Otálora et al. presented two frameworks with neural networks to classify microalgae (Otálora et al., 2021). The first framework handled microalgae data from the device FlowCAM, providing 30 features of microalgae. The artificial neural network included 30 neurons in the input layer, and 25 neurons in the hidden layer, and the final output was two types. The second framework processed microalgae images with a convolutional neural network, including 25 layers. The model could achieve 96% accuracy in training and 93.5% accuracy even in actual tests. Mesut Ersin Sonmez et al. used multiple structured convolutional neural networks and a coupled support vector machine algorithm to classify microalgae, all of which yielded excellent results (Sonmez et al., 2022). All microalgae images were obtained from an inverted microscope, with only 20 images of each microalgae in the initial dataset. To ensure the final result, the dataset was extended by applying the data enhancement technique. The final recognition accuracy of the convolutional neural network based on the AlexNet structure with various modifications was as high as 99.66%. In addition, the microalgae features identified by the convolutional neural network were utilized as input parameters for the support vector machine algorithm to improve its recognition accuracy to the same level as the convolutional neural network. Jeffrey Harmon et al. conducted a classification study of spherical microalgae based on a support vector machine approach (Harmon et al., 2020). They first utilized fluorescence imaging technology to obtain trichromatic images of microalgae to quantify the morphological characteristics of microalgae. Then, the morphological features of microalgae were analyzed and finally classified based on the support vector machine algorithm. The accuracy of the method reached 99.8% in experiments, which was higher than the convolutional neural network algorithm in some specific cases. An additional advantage of the model was that it provided morphological information on the microalgal populations. Anaahat Dhindsa et al. designed a new scheme to classify microalgae (Dhindsa et al., 2021). The microalgae images were segmented, and 25 features were extracted by a generalized segmentation algorithm, and then various machine learning algorithms were applied for classification. The classification accuracy increased from 96.1% to 98.2% after the modification of the support vector machine algorithm. The authors mentioned that the introduction of transfer learning into the classification progress was expected to develop the accuracy in the future. Zhanpeng Xu et al. introduced a spectral imager that allowed classification and growth cycle analysis of microalgae (Xu et al., 2020). The device acquired spectral images of microalgae and then analyzed them with a support vector machine algorithm. The last classification accuracy was 94.4%. Based on the random forest algorithm, the growth of microalgae could be predicted from the above data, and the accuracy could reach 98.1%. The accuracy and effectiveness of the model were confirmed after the identification of a mixture of microalgae. Paulo Drews-Jr et al. applied semi-supervised learning in their work on microalgae classification (Drews et al., 2013). The dataset was microalgae data obtained through the FlowCAM device in the Atlantic Ocean. Experiments confirmed that the method could get better results than SVM, and the final recognition accuracy could reach 92% if the active learning algorithm was added. The performance of the method could be further enhanced by improving the dataset and optimizing the image segmentation algorithm.
Sansoen Promdaen et al. performed an in-depth research on the classification of microalgae with unclear boundaries and blurred textures (Promdaen et al., 2014)(Figure 5A). To deal with the issue of vague boundaries, the authors utilized the method of microalgae segmentation based on the image background. To handle the situation of blurred textures, the authors proposed a new texture description method. The dataset with 720 images had multiple sources, including universities, waterworks authorities, networks, etc. The accuracy of the method reached 97.22% in the experiment. Hui Huang et al. employed multiple machine learning algorithms to classify microalgae and microplastics in seawater (Huang et al., 2021). The data processed by the various algorithms were the image data acquired by spectral microscopy. The image stitching technique was introduced to expand the imaging range of images. The effectiveness of each algorithm was verified by testing in a real-world environment. Jhony−Heriberto Giraldo−Zuluaga et al. utilized a digital microscope to take images of the microalgae and obtained the microalgae species through the image process (Giraldo-Zuluaga et al., 2016). Images were characterized by statistical features, which were derived from the calculation and analysis of texture features. The dataset used for training was obtained by processing the original images acquired by the digital microscope. The experiments showed that the effect of the support vector machine algorithm was better than the artificial neural network algorithm, which could reach 98.63%. Zepeng Zhuo et al. constructed a dataset containing 35 species of microalgae specifically for microalgal classification (Zhuo et al., 2022). The content of the dataset was polarized light scattering data of microalgae. They investigated the performance of many machine learning algorithms based on the dataset, and the final result proved that the non-linear support vector machine algorithm could achieve the best performance of 80%. The research work had significant implications for the search for better light polarization.
Figure 5 (A) Images of twelve species of microalgae. (reproduced with permission from Promdaen et al., 2014). (B) Procedure of microalgae classification and recognition based on machine learning. (reproduced with permission from Reimann et al., 2020). (C) Procedure of microalgae classification and recognition based on machine learning. (reproduced with permission from Wang et al., 2021).
A model for automatic classification of live and dead cells in Chlorella was proposed by Ronny Reimann et al (Reimann et al., 2020) (Figure 5B). Microalgae images were acquired by fluorescence microscopy, and features were extracted. Multiple machine learning algorithms were used for the classification prediction of live and dead cells of microalgae, and the random forest algorithm gave the best result with a precision of 96.6%. The model could classify not only individual microalgae, but also the whole microalgae population in terms of live and dead cells with an accuracy of 82%. The dead microalgal cells were significantly larger in diameter and area than the live microalgal system. Yanyan Wang et al. utilized machine learning algorithms to classify live and dead microalgae in the ocean as well (Wang et al., 2021) (Figure 5C). The microalgae features used for machine learning algorithm analysis were all acquired through digital holographic microscopy, which eliminated the need for tedious staining and labeling processes on the microalgae. The method also ensured that there was no impact on the growth environment of the microalgae. The framework achieved an accuracy of 94.8% in the laboratory and reached the same accuracy as the conventional staining method even when validated in practice. B.M. Franco et al. classified a variety of microalgae simultaneously based on an artificial neural network (Franco et al., 2019). The input data for the mode were spectral features of microalgae, and the model was trained using 550 sample data. In the experiment, the model achieved 98% accuracy in the identification of single microalgae. Even for mixtures of multiple microalgae, the model could identify the species of microalgae and analyze the proportion of the total.
The YOLOv3 network was applied in the detection of microalgae by Jungsu Park et al (Park et al., 2021). A dataset of 1114 microalgae images collected using microscopy was composed. Depending on the quantity of extracted microalgae attributes, the dataset was divided into four parts. After the YOLOv3 network was trained on these four datasets, the measured recognition accuracy reached more than 80%. The result fully proved the effectiveness of the approach in recognizing microalgae. Further research by the group showed that the accuracy of recognition could be further improved by replacing the images in the dataset and recognizing objects with color ones. An improved framework based on the YOLOv3 network was proposed by Mengying Cao et al. to identify microalgae (Cao et al., 2021). Features were extracted by the MobileNet network, and the elements could be fused in later operations of this model. The dataset was generated manually by the team through a camera, with a total of 10,000 images after data enhancement. The experiments showed that the correctness of the model for the microalgae identification was improved by 8.59% over the original model, reaching 98.90%. Daniele Gaetano Sirico et al. reported a novel scheme to detect the movement of microalgae in 3D space (Sirico et al., 2022). Mechanical scanning microscope was often challenging to obtain the complete data of microalgae movement, so a digital holographic microscope was employed in the framework to track the trajectory of microalgae movement. Computer software and digital image process algorithms synthesized 3D images of microalgae movements and finalized the tracking of their trajectories. Finally, the model visualized the activity of the microalgae.
Many machine learning algorithms have been widely used in the detection and classification of microalgae, such as support vector machine, random forest, and neural network (Table 1). In particular, deep learning technology represented by convolutional neural network is most widely utilized. The datasets applied in deep learning are essentially acquired by the device FlowCAM. The YOLOv3 network is a kind of deep learning model, which is widely used in microalgae detection due to its perfect recognition effect on small targets.
Conversion from microalgae to energy
Fossil fuels such as oil have insurmountable problems: the non-renewable issue and environmental pollution (Brennan and Owende, 2010). Renewable biofuels are an excellent solution to these problems. Microalgae can be used to make biofuels, and the technology has long been used in reality. But the transfer from microalgae to biofuels has faced issues such as the complexity of the microalgae culture and the uncertainty of the conversation (Enamala et al., 2018; Aghbashlo et al., 2021). Machine learning algorithms can play an essential role in microalgae culture and conversion to biofuels to solve the above problems (Georgianna and Mayfield, 2012). By analysing the existing data, the machine learning algorithms can estimate the optimal environmental and light conditions in the microalgae culture process, predict the biofuel output rate, verify the quality of biofuels, etc. Machine learning algorithms can make the conversion of energy more efficient and the quality of biofuels more assured (Rock et al., 2021; Wang et al., 2022).
Unlike previous methods of population lipid content analysis of microalgae, Baoshan Guo et al. conducted a study of lipid content analysis of individual microalgae by combining optofluidic microscopy images with machine learning (Guo et al., 2017). The method allowed to obtain analytical results in a non-invasive way, without destroying the microalgal structure. The authors demonstrated the effectiveness of the approach through practical experiments with slender-eyed worms and E. coli. They predicted that better results could be achieved if deep learning or unsupervised learning techniques could be introduced. Ahmet Coşguna et al. used a machine learning approach to explore the optimal growth conditions and lipid production factors for microalgae to generate biofuels (Coşgun et al., 2021)(Figure 6). The dataset and potential influence factors were derived from a summary of 102 scientific studies. Through the analysis of the decision tree algorithm, they found 11 combinations of influence conditions for high microalgal production and 13 incorporations of influence factors that could lead to increased lipid content. Rakesh Chandra Joshi et al. reported a new way to estimate the oil content of microalgae with a machine learning method (Joshi et al., 2021). They first obtained images of the microalgae through microscopy. Then, the oil-containing particles in the microalgae images were segmented and analysed for lipid content. A comparison of the results between the traditional method with the model confirmed that the model significantly reduced the computation time, and the predictions were more accurate. Ehecatl Antonio del Rio Chanona et al. devised a novel framework combined with deep learning technology to investigate optimal conditions for microalgae growth and the conversion from microalgae to biofuels (del Rio-Chanona et al., 2019). The behavior of the underlying organisms was studied by coupling hydrodynamic and biodynamic techniques together, and the dataset for deep learning was constructed. The framework reduced the calculation time from months to days and predicted the more appropriate light conditions for microalgae growth and the configuration requirements for the conversion to biofuels. In order to obtain low-cost bio-oil, Bin Long et al. applied machine learning algorithms in the cultivation process of microalgae, hoping to get cheaper microalgae (Long et al., 2022). Factors such as algal density and light condition were thoroughly analysed to provide optimal conditions for the growth of microalgae. A better culture environment and minimal light shading were also considered. The results of the study were equally applicable to the calculation of conditions for the growth of microalgae in large-scale cultures of algae plants in industry and other types of installations.
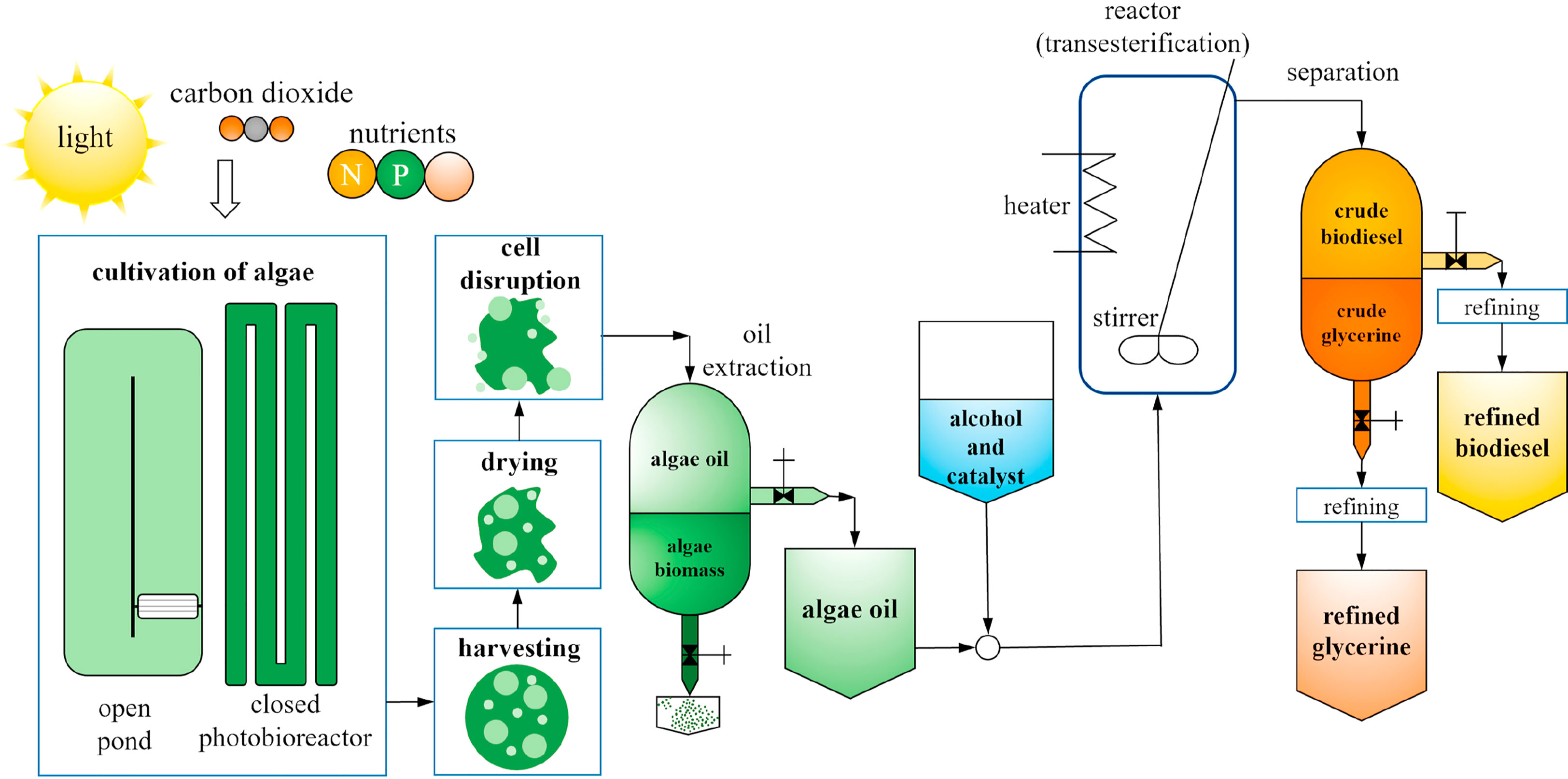
Figure 6 The flowchart of production from microalgae to biodiesel. (reproduced with permission from (Coşgun et al., 2021).
Due to the high price of biodiesel produced from microalgae, some people mixed cheap cooking oil such as canola oil into the biodiesel. Mahdi Rashvand et al. introduced SVM and ANN algorithms in biodiesel quality identification (Rashvand et al., 2019). The SVM algorithm was used to analyse the biodiesel phase shift coefficient and voltage coefficient obtained through the capacitive sensor. In contrast, the ANN was used to analyse the image characteristics of the biodiesel. The experimental data showed that the combination of the two methods together led to optimal identification results. Hossein Moayedi et al. compared several machine learning algorithms that could evaluate the purity of biodiesel obtained from microalgae conversion (Moayedi et al., 2020). The training sample data were obtained from existing biodiesel research results, and eight factors including reaction temperature and catalyst type were used as input parameters for the analysis. The performance of the alternating model tree algorithm was the best after the available metrics argument. A numerical model containing ANN utilized to evaluate the behaviour of biodiesel combustion, emission, etc., produced by microalgae was designed by Satishchandra Salam et al (Salam and Verma, 2019). ANN was trained through the data obtained from a software called Diesel-RK. The model accurately predicted the combustion and emission factors of the internal combustion engine under different response conditions. The redundancy of part system parameters indicated that the model had the potential for further optimization. Hao Chen et al. researched the viscosity of microalgae slurry used in the biofuel manufacturing process with ANN (Chen et al., 2021). The dataset was derived from 1691 experimental data, and the considered parameters included temperature, microalgal mass fraction, shear rate, etc. Experiments demonstrated that this method had better prediction and outperformed the already widely used curve-fitting method. Abhijeet Pathy et al. carried out the prediction of the yield and biochar composition from microalgae to biochar based on machine learning algorithms (Pathy et al., 2020). After drilling the model with the training data, the model was further refined by comparing experimental data on 13 different parameter combinations. The analysis results revealed that temperature played a dominant role in the final yield of biochar. Fangwei Cheng et al. assessed the energy productivity and carbon capture capacity of microalgae through hydrothermal reaction based on machine learning algorithms (Cheng et al., 2020). The dataset contained 800 items, all extracted from the existing literature. Numerous experiments had confirmed that the random forest algorithm was better in the task than the multiple linear regression algorithm and the regression tree model. Hydrothermal reaction methods typically had higher energy production efficiency and carbon capture capacity than conventional methods.
Jie Li et al. introduced the machine learning algorithm to the hydrothermal liquefaction process of converting microalgae into bio-oil to produce high quality, low nitrogen bio-oil (Li et al., 2021). Experiments confirmed that the random forest algorithm was the optimal choice for this multi-task prediction process. Both predicted results and experimental data showed that the lipid content in microalgae and temperature had the most significant effect on oil production. The nitrogen content of microalgae and temperature played a decisive role in the nitrogen content of the final bio-oil. Weijin Zhang et al. researched optimal conditions used to produce bio-fuel for different types of microalgae with machine learning methods in the hydrothermal liquefaction process (Zhang et al., 2021). The hyperparameters included the composition content of microalgae and the primary conditions of hydrothermal liquefaction. After several validations, it was finally shown that the gradient propelled regression algorithm was better than the random forest algorithm for both single-task and multi-task estimation. So far, the whole process has a lot of potential for improvement. The adaptive neuro-fuzzy inference system is a new scheme inference that organically connects fuzzy logic and neuron network. The system employs an integrated algorithm containing the back propagation technique and the least squares method to modulate the model parameters. The algorithm was introduced into the conversion of microalgae to biodiesel production by Momir Milić et al. The training data were obtained from experimental data in the published literature (Milić et al., 2021). The method was a fundamental guide for the conversion of microalgae to biodiesel. Sashi Sonkar et al. utilized machine learning algorithms to study the drying of microalgae pulp for biodiesel production (Sonkar and Mallick, 2020). The collected experimental data were trained by a logistic regression algorithm with regularization to build the binary classification model. Factors such as blower speed and temperature were used as input parameters to evaluate the residual water content and lipid yield of the microalgal slurry. The predictions obtained by the method fundamentally improved the drying speed of microalgae pulp. Nahid Sultana et al. proposed a new model with ANN and a support vector regression algorithm to predict biodiesel produced from microalgae (Sultana et al., 2022). Parameters such as catalyst dosage, reaction time, and reaction temperature were used as input hyperparameters, and the hyperparameters were automatically adjusted by combining the Bayesian algorithm with ANN. The model was validated using a lot of published data, which proved its effectiveness. These numerical simulations used to estimate the yield of microalgae to biodiesel were not only more accurate but also more time and cost efficient.
In addition, traditional machine learning algorithms are often applied in the transformation process from microalgae to bioenergy, while deep learning is rarely applied (Table 2). An important reason for this phenomenon is that there are fewer datasets available for deep learning.
Important role of microalgae in environment protection
Microalgae play an important role in environment protection and pollution prevention. Their absorption of carbon dioxide through photosynthesis can mitigate the global greenhouse effect. (Sundui et al., 2021) In addition, they have an irreplaceable role in wastewater treatment. The addition of machine learning algorithms makes the role of microalgae even more apparent (Cruz et al., 2021) (Figure 7A).
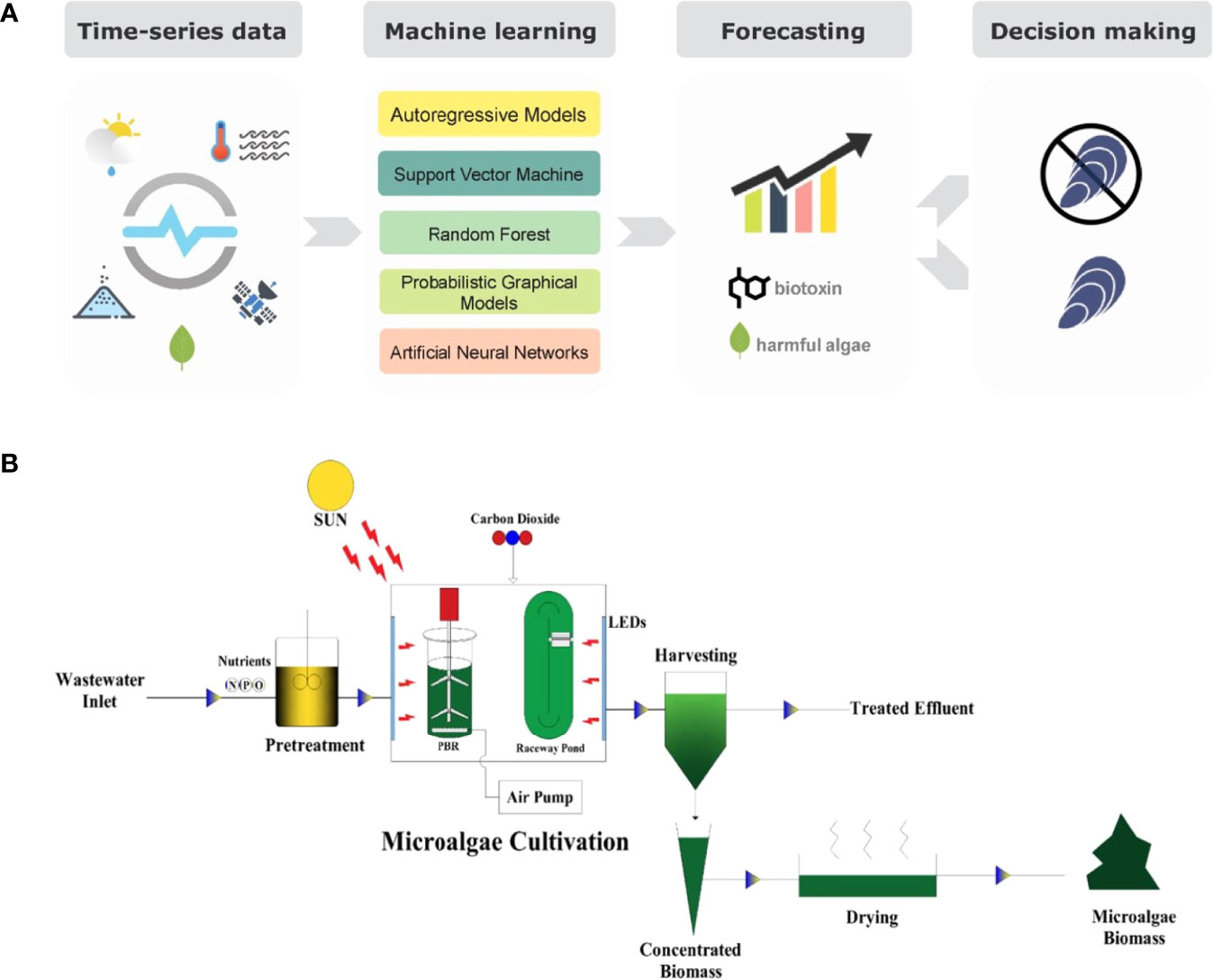
Figure 7 (A) Block diagram of Shellfish contamination prediction based on machine learning. (reproduced with permission from an open access article). (B) Process diagram of wastewater treatment with microalgae. (reproduced with permission from Singh and Mishra, 2021).
Microalgae could be used to treat E. coli in wastewater, and M Žitnik et al. applied a machine learning algorithm to search conditions that worked best for treatment (Žitnik et al., 2019). Parameters such as microalgae concentration, E. coli concentration, pH, and conductivity were analysed by the decision tree algorithm. The results showed that conductivity had the most important effect on the treatment effect of E. coli. Based on the results, targeted optimization of the wastewater treatment system could be carried out. Vishal Singh et al. researched the ways to increase microalgal production and enhance their ability to treat wastewater with machine learning methods as well (Singh and Mishra, 2022). The dataset was derived from publicly available results from recent years and was fully justified by the decision tree algorithm for parameters such as temperature, CO2 content, and pH value. The authors gave different combinations of parameters for improving microalgal production. The way treated wastewater with high nitrogen content and high phosphorus content after experimental validation admirably. They also applied the decision tree algorithm in the prediction of microalgae growth conditions and wastewater treatment conditions (Singh and Mishra, 2021) (Figure 7B). Parameters that were less involved in other algorithms, such as initial inoculum, reactor type, and nutrient concentration in the wastewater, were fully considered in this method. Different combinations of parameters suitable for high yield, high phosphorus removal performance, and high nitrogen removal capability were calculated by this method. The method provided solid theoretical support for the large-scale treatment of wastewater. S. M. Zakir Hossain et al. provided an in-depth analysis of the ability of microalgae to treat municipal sewage (Hossain et al., 2022a). They aimed to use microalgae to remove both nitrogen and phosphorus from sewage. The impact of factors such as temperature, light, and dark cycles on the final results was well demonstrated. The final consequence revealed that the support vector regression algorithm predicted more accurate and efficient results. They also combined the support vector regression algorithm with the crow search approach for single and multi-objective optimization to further improve the effect of microalgae in removing nitrogen and phosphorus from wastewater (Hossain et al., 2022b). Experimental data confirmed the best treatment of wastewater by microalgae at the temperature of 29.3 degrees Celsius, 24 hours of uninterrupted light, and nitrogen to phosphorus ratio of 6:1.
Muzhen Xu et al. investigated the treatment of heavy metals in wastewater by microalgae based on artificial intelligence technology (Xu et al., 2021a). They utilized microscopy to take images of individual microalgae to analyse their behaviour to determine their removal effect on heavy metals. The effect of parameters such as eccentricity and compactness were specifically examined. Copper ion experiments proved that this method had a more effective heavy metal removal efficiency. The team also used machine learning algorithms to study the morphology of microalgae in more depth to obtain the characteristics of microalgae that could efficiently treat heavy metals in wastewater (Xu et al., 2021b). The process used microscopy to acquire images of microalgae, enabling the assessment of the efficiency of heavy metal removal by microalgae in a non-invasive and label-free way. The experimental results showed that the morphology of E. gracilis cells was more conducive to the efficient removal of heavy metals. Microalgae can mitigate the greenhouse effect by absorbing carbon dioxide from the atmosphere through photosynthesis. Domenico D’Alelio et al. studied the impact of microalgae on the global warming issue based on machine learning algorithms (D’Alelio et al., 2020). They trained the model based on known data downloaded from the Web, and then analysed their collection data of 27 years in the North Atlantic. The final analysis showed that as seawater temperature increased, the number of microalgae decreased, and distant marine areas faced nutrient deficiencies in seawater.
At present, there are few machine learning algorithms used to assist microalgae in wastewater treatment and other environmental protection work, mainly support vector machine algorithm and decision tree algorithm (Table 3). The application record of deep learning in this field has not been found yet.
Application of machine learning in the growth phase of microalgae
The yield of microalgae and the ease of harvesting can directly affect their cost. There are many factors influencing the growth and morphology of microalgae, and various investigation has been conducted previously to obtain lower cost microalgae. However, it is either laborious or poorly predicted, making it difficult to provide valuable suggestions for actual production. Numerous factors have been analyzed with machine learning algorithms to predict the growth and final yield of microalgae in recent years.
Susanne Dunker et al. proposed a deep learning scheme for identifying microalgae species and growth cycles (Dunker et al., 2018). 47,000 microscope high-throughput images at 60x magnification were trained on the model. The model achieved 97% accuracy in natural experiments, which was quite good. The framework offered great help for the rapid assessment of water quality. D. M. J. Purnomo et al. studied the growth behavior of microalgae in solutions with different pH values based on an extreme learning machine (Purnomo et al., 2015). The team observed the growth of microalgae for 20 consecutive days and normalized the data to construct the dataset. A cross-validation method was introduced to prevent overfitting problem during model training. Experiments had shown that the method had a high accuracy rate, which could be further improved if used in conjunction with a genetic algorithm. Bi Xiaolin et al. investigated the impact of pH on the growth of microalgae by analyzing hyperspectral images with a machine learning algorithm (Bi et al., 2019). The spectra of all microalgae were represented by 900 pixels, 300 pixels were then randomly selected as the training set, and another 300 pixels were randomly chosen as the validation set. The experimental data revealed that the support vector machine algorithm was the most effective method in identifying microalgae and could reflect their growth conditions. The study provided excellent technical support for monitoring the growth process of microalgae and analyzing their directional movements. Wendie Levasseur et al. studied the effect of light on the growth of microalgae, especially in an environment with alternating light and dark light based on a machine learning algorithm (Levasseur et al., 2022). Medium and high light, and dark light switch frequencies were used as the focus of the analysis. The growth data of low-density green microalgae under different light switch frequencies were compiled and analyzed by inferential statistics. Finally, the authors described different experimental setup to observe the growth of microalgae. Shixuan He et al. analyzed the growth of microalgae based on the support vector machine algorithm to assess the degree of eutrophication in water bodies (He et al., 2018). They first obtained the characteristic of microalgae by Raman spectroscopy to get their growth stages. Then they analyzed the relationship between algal growth and environmental changes. The authors presented a full paper on the effectiveness of the method. A framework that brought together multiple machine learning algorithms was employed to study the growth process of microalgae and the amount of CO2 fixation by S. M. Zakir Hossain et al. Factors such as temperature, nitrogen to phosphorus ratio, and frequency of light and dark cycles were used as input parameters for the whole framework (Hossain et al., 2022c) (Figure 8). All algorithms were utilized together with Bayesian optimization for various predictions. The advantages and disadvantages of each algorithm in prediction were listed in detail in the article.
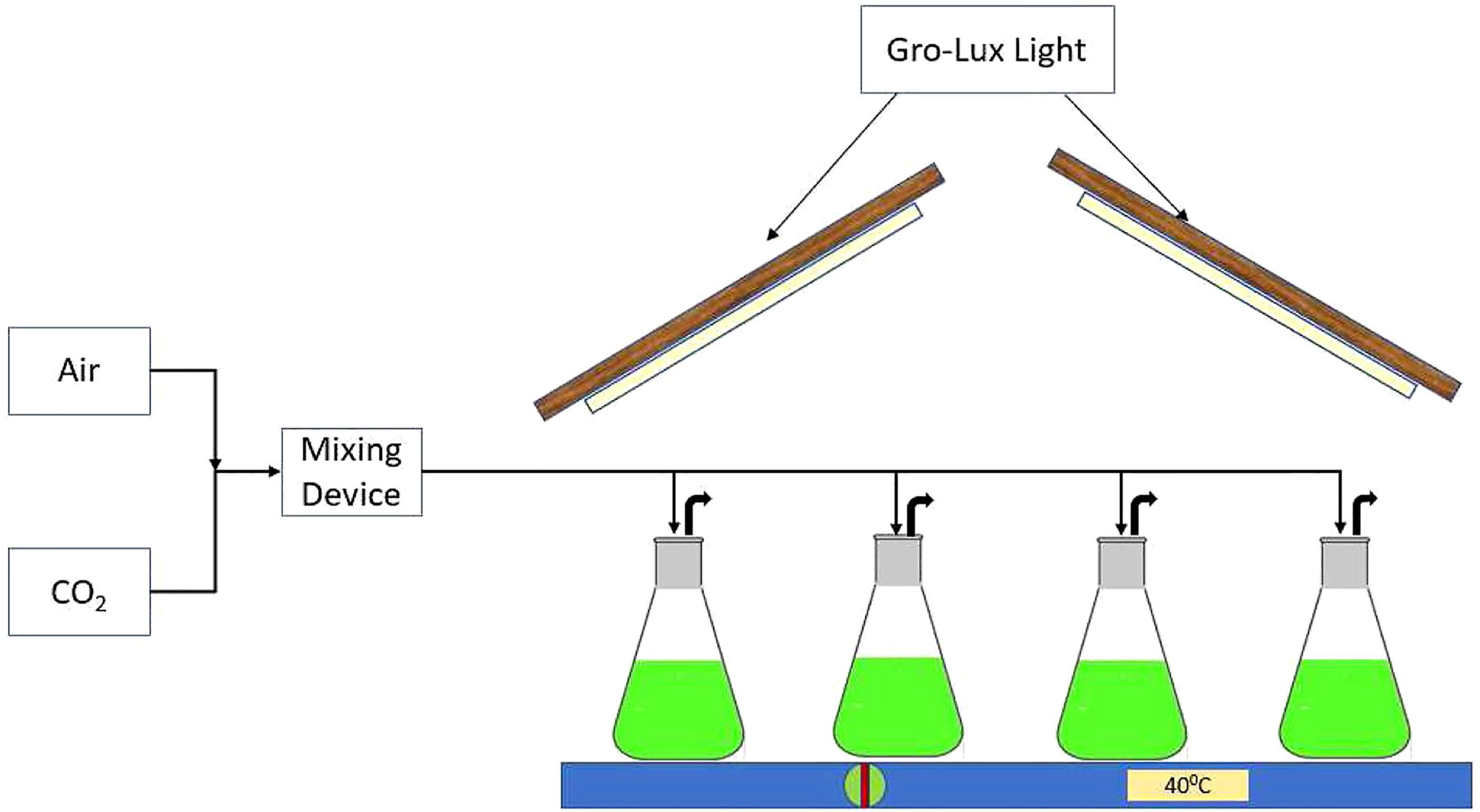
Figure 8 Process diagram of microalgae cultivation environment. (reproduced with permission from Hossain et al., 2022c).
A model used to estimate the daily productivity and final production of microalgae in open ponds was surveyed by Supriyanto et al. Based on an existing dataset, a decision tree method was employed to calculate the effect of temperature, solar radiation, and other condition on the growth and final yield of microalgae (Supriyanto et al., 2018). The efficiency of the model had been validated by practical evaluation. Its performance could be further improved in the future if more parameters were added to the model. The group investigated the production of mixed microalgae in semi-continuous open ponds based on an artificial neural network as well (Supriyanto et al., 2019) (Figure 9). The neural network included a hidden layer and an output layer, and the input layer contained eight parameters such as algae concentration, temperature, solar radiation, and pH value. The network was trained through a mature dataset. The final prediction was the concentration of microalgae. The data showed that the three-layer neural network model worked well for various input parameters. Victor Pozzobon et al. constructed a machine learning scheme to check nitrate and nitrite levels in the microalgae growth environment (Pozzobon et al., 2021). First of all, different concentrations of nitrate and nitrite samples were extracted from the microalgal growth environment and analyzed spectroscopically with a spectrometer. These data were then analyzed based on a least square regression algorithm and ultimately used to predict nitrate and nitrite concentrations. The method not only significantly reduced the time required for detection but also ensured a sufficiently high accuracy. A novel framework coupling support vector machine algorithm and random forest algorithm was reported by Patricio López Expósito et al. to measure microalgae concentration (Expósito et al., 2017). The laser irradiated the suspended particles of microalgae to get reflectance spectra, which could be analyzed to obtain the concentration of microalgae. The team constructed a dataset of 76 vectors through practical experiments to solve the hyperparameters in the model. The results demonstrated that the model could quickly and accurately estimate the concentration of microalgae. The team also analyzed the floc length and geometric shape during the growth of microalgae in a random forest regression model to reduce the cost of microalgae at harvesting (Lopez-Exposito et al., 2019). A set of length collected by computer software generating virtual flocs after focused reflection operations was employed as the data set for the training model. The trained and optimized model achieved very high accuracy in actual tests. An additional advantage of the model was that it could be quickly adapted to the floc structure according to the actual requirements.

Figure 9 Schematic representation of a framework used to estimate microalgae growth with ANN. (reproduced with permission from Supriyanto et al., 2019).
Machine learning algorithms can analyze the effect of periphyton factors such as DNA, in addition to macroscopic factors that affect the growth of microalgae (Teng et al., 2020). Appropriate edit of the microalgae genes could rapidly increase the production and oil content of microalgae. However, the genomes of microalgae were not only long, but also particularly complex. Therefore, they could not be rapidly localized and analyzed by conventional methods. Likai Wang et al. applied a logistic regression algorithm to learn the 32 known characteristic expressions of stress genes to predict the function of the remaining stress genes (Wang et al., 2018). The authors’ study showed that the method had high accuracy. If more feature expressions were learned in known data by deep learning technique, the performance of the framework was promising to be further improved. Supreeta Vijayakumar et al. analyzed the genomes of microalgae based on machine learning algorithms to explore the feedback of microalgae to changes in light and salinity in the environment (Vijayakumar et al., 2020). They firstly collected data on photosynthesis and genome-based energy metabolism of microalgae. These data were then analyzed by methods such as k-means clustering. The team’s further results showed that the combination of machine learning algorithm and genomic model accomplished the work well. Victor Pozzobon et al. analyzed the viability of microalgae by researching the flow cytometer readings through a machine learning algorithm (Pozzobon et al., 2020). The microalgal activity was obtained by studying the integrity of the cell wall of microalgae after double staining. The validity of the model was verified by freezing the microalgae and observing their activity. The results showed that the model predicted data were consistent with those listed in the published literature.
Among all the microalgae treatment modules in this paper, the microalgae growth status detection module uses the most machine learning algorithms (Table 4). Although deep learning has not been widely used in this work, it has achieved very significant results.
Conclusion
This paper provides a detailed summary of machine learning techniques used in microalgae treatment. The overview refers to the classification and identification of microalgae, the conversion of microalgae into bioenergy, the treatment of waste by microalgae, and the growth of microalgae. Microalgae are critical part of the marine ecological cycle and have a very significant economic value. However, the classification, identification, and purification of microalgae have always been a problem for practitioners because they are so small and diverse. Machine learning techniques are good at operations such as classification and regression, and have been highly successful in digital image processing and speech recognition. The introduction of machine learning techniques to microalgae applications has been equally fruitful. This paper illustrates how data-driven machine learning techniques process input data and calculate the output results, with algorithms such as support vector machine, decision tree, random forest, and neural network as examples. How machine learning algorithms have been applied and the results have been achieved in the areas such as microalgae classification, conversion of microalgae into bioenergy, microalgae purification of the environment, and microalgae growth are then summarized. The paper has tremendous implications for future extensions of machine learning in microalgae applications.
Prospect
Many achievements have been made in the application of machine learning in microalgae identification and treatment. However, there are still many aspects that can be further optimized. First of all, there are very few datasets available for machine learning algorithms in microalgae process. Even though some datasets have been widely used in some specific areas, they still face the problem of over-fitting (Rani et al., 2021). Secondly, when the performance of a single machine learning algorithm is limited, multiple algorithms can be coupled to build a hybrid model. The dataset that accompanies the hybrid model also need to be studied in depth (Sundui et al., 2021). Besides, the improvement of the performance of existing machine learning models is also a key work in the future. For example, the current cost of biodiesel converted from microalgae is significantly higher than diesel derived from fossil fuels. Both modification of existing models and construction of new models to assist the conversion of microalgae to biodiesel require a lot of work (Chowdhury and Loganathan, 2019).
Author Contributions
HN collected literatures and wrote the manuscript. RL contributed to manuscript preparation and co-wrote the manuscript. TZ contributed to the conception of the study and manuscript preparation. All authors provided critical feedback and helped shape the manuscript. All authors read and approved the final manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (Grant No. 52075138), the Anhui Provincial Natural Science Foundation (Grant No. 2008085QF329), the Natural Science Foundation for the Higher Education Institutions of Anhui Province (Grant No. KJ2020A0061), and the Natural Science General Project of Anhui Science and Technology University (Grant No. 2021zryb26).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abiodun O. I., Jantan A., Omolara A. E., Dada K. V., Mohamed N. A., Arshad H. (2018). State-of-the-art in artificial neural network applications: A survey. Heliyon 4, e00938. doi: 10.1016/j.heliyon.2018.e00938
Acharya U. R., Oh S. L., Hagiwara Y., Tan J. H., Adam M., Gertych A., et al. (2017). A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 89, 389–396. doi: 10.1016/j.compbiomed.2017.08.022
Adamczak M., Bornscheuer U. T., Bednarski W. (2009). The application of biotechnological methods for the synthesis of biodiesel. Eur. J. Lipid Sci. Technol. 111, 800–813. doi: 10.1002/ejlt.200900078
Aghbashlo M., Peng W., Tabatabaei M., Kalogirou S. A., Soltanian S., Hosseinzadeh-Bandbafha H., et al. (2021). Machine learning technology in biodiesel research: A review. Prog. Energy Combustion Sci. 85, 100904. doi: 10.1016/j.pecs.2021.100904
Albawi S., Mohammed T. A., Al-Zawi S. (2017). Understanding of a convolutional neural network. 2017 International Conference on Engineering and Technology (ICET) (Antalya, 795 Turkey: IEEE(USA)), 2017, 1–6.
Andersen R. A., Kawachi M. (2005). Microalgae isolation techniques. Algal culturing techniques, 83–100. doi: 10.1016/b978-012088426-1/50007-x
Ayyagari M. R. (2020). Classification of imbalanced datasets using one-class SVM, k-nearest neighbors and CART algorithm. Int. J. Advanced Comput. Sci. Appl. 11, 1–5. doi: 10.14569/IJACSA.2020.0111101
Bakirtzis A. G., Petridis V., Kiartzis S. J., Alexiadis M. C., Maissis A. H. (1996). A neural network short term load forecasting model for the Greek power system. IEEE Trans. Power Syst. 11, 858–863. doi: 10.1109/59.496166
Barsanti L., Birindelli L., Gualtieri P. (2021). Water monitoring by means of digital microscopy identification and classification of microalgae. Environ. Science: Processes Impacts 23, 1443–1457. doi: 10.1039/D1EM00258A
Belgiu M., Drăguţ L. (2016). Random forest in remote sensing: A review of applications and future directions. ISPRS J. photogrammetry Remote Sens. 114, 24–31. doi: 10.1016/j.isprsjprs.2016.01.011
Bi X., Lin S., Zhu S., Yin H., Li Z., Chen Z. (2019). Species identification and survival competition analysis of microalgae via hyperspectral microscopic images. Optik 176, 191–197. doi: 10.1016/j.ijleo.2018.09.077
Bishop C. M. (2013). Model-based machine learning. Philos. Trans. R. Soc. A: Mathematical Phys. Eng. Sci. 371, 20120222. doi: 81410.1098/rsta.2012.0222
Bonissone P., Cadenas J. M., Garrido M. C., Díaz-Valladares R. A. (2010). A fuzzy random forest. Int. J. Approximate Reasoning 51, 729–747. doi: 10.1016/j.ijar.2010.02.003
Boser B. E., Guyon I. M., Vapnik V. N. (1992). A training algorithm for optimal margin classifiers. In Proceedings of the 5th Annual Workshop on Computational Learning Theory ACM Press, 144–152. doi: 10.1145/130385.130401
Boulesteix A. L., Janitza S., Kruppa J., König I. R. (2012). Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip. Reviews: Data Min. Knowledge Discovery 2, 493–507. doi: 10.1002/widm.1072
Brennan L., Owende P. (2010). Biofuels from microalgae–a review of technologies for production, processing, and extractions of biofuels and co-products. Renewable Sustain. Energy Rev. 14, 557–577. doi: 10.1016/j.rser.2009.10.009
Cao M., Wang J., Chen Y., Wang Y. (2021). Detection of microalgae objects based on the improved YOLOv3 model. Environ. Science: Processes Impacts 23, 1516–1530. doi: 10.1039/D1EM00159K
Carleo G., Cirac I., Cranmer K., Daudet L., Schuld M., Tishby N., et al. (2019). Machine learning and the physical sciences. Rev. Modern Phys. 91, 045002. doi: 10.1103/RevModPhys.91.045002
Chakdar H., Hasan M., Pabbi S., Nevalainen H., Shukla P. (2021). High-throughput proteomics and metabolomic studies guide re-engineering of metabolic pathways in eukaryotic microalgae: A review. Bioresource Technol. 321, 124495. doi: 10.1016/j.biortech.2020.124495
Chen H., Fu Q., Liao Q., Zhu X., Shah A. (2021). Applying artificial neural network to predict the viscosity of microalgae slurry in hydrothermal hydrolysis process. Energy AI 4, 100053. doi: 10.1016/j.egyai.2021.100053
Cheng C., Cheng X., Dai N., Jiang X., Sun Y., Li W. (2015). Prediction of facial deformation after complete denture prosthesis using BP neural network. Comput. Biol. Med. 66, 103–112. doi: 10.1016/j.compbiomed.2015.08.018
Cheng F., Porter M. D., Colosi L. M. (2020). Is hydrothermal treatment coupled with carbon capture and storage an energy-producing negative emissions technology? Energy Conversion Manage. 203, 112252. doi: 10.1016/j.enconman.2019.112252
Chen P.-H., Lin C.-J., Schölkopf B. (2005). A tutorial on ν-support vector machines. Appl. Stochastic Models Business Industry 21, 111–136. doi: 10.1002/asmb.537
Chen C. L., Mahjoubfar A., Tai L.-C., Blaby I. K., Huang A., Niazi K. R., et al. (2016). Deep learning in label-free cell classification. Sci. Rep. 6, 21471. doi: 10.1038/srep21471
Chew K. W., Yap J. Y., Show P. L., Suan N. H., Juan J. C., Ling T. C., et al. (2017). Microalgae biorefinery: high value products perspectives. Bioresource Technol. 229, 53–62. doi: 10.1016/j.biortech.2017.01.006
Chowdhury H., Loganathan B. (2019). Third-generation biofuels from microalgae: a review. Curr. Opin. Green Sustain. Chem. 20, 39–44. doi: 10.1016/j.cogsc.2019.09.003
Cichy R. M., Kaiser D. (2019). Deep neural networks as scientific models. Trends Cogn. Sci. 23, 305–317. doi: 10.1016/j.tics.2019.01.009
Correa I., Drews P., Botelho S., Souza M. S. D., Tavano V. M. (2017). “Deep learning for microalgae classification,” in 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA): Cancun, Mexico, 18-21 Dec IEEE(USA) Vol. 2017, 20–25.
Coşgun A., Günay M. E., Yıldırım R. (2021). Exploring the critical factors of algal biomass and lipid production for renewable fuel production by machine learning. Renewable Energy 163, 1299–1317. doi: 10.1016/j.renene.2020.09.034
Cruz R. C., Costa P. R., Vinga S., Krippahl L., Lopes M. B. (2021). A review of recent machine learning advances for forecasting harmful algal blooms and shellfish contamination. J. Mar. Sci. Eng. 9, 283–99. doi: 10.3390/jmse9030283
D’Alelio D., Rampone S., Cusano L. M., Morfino V., Russo L., Sanseverino N., et al. (2020). Machine learning identifies a strong association between warming and reduced primary productivity in an oligotrophic ocean gyre. Sci. Rep. 10, 3287. doi: 10.1038/s41598-020-59989-y
Deka P. C. (2014). Support vector machine applications in the field of hydrology: a review. Appl. soft computing 19, 372–386. doi: 10.1016/j.asoc.2014.02.002
del Rio-Chanona E. A., Wagner J. L., Ali H., Fiorelli F., Zhang D., Hellgardt K. (2019). Deep learning-based surrogate modeling and optimization for microalgal biofuel production and photobioreactor design. AIChE J. 65, 915–923. doi: 10.1002/aic.16473
Dhindsa A., Bhatia S., Agrawal S., Sohi B. S. (2021). An improvised machine learning model based on mutual information feature selection approach for microbes classification. Entropy 23, 257–271. doi: 10.3390/e23020257
Dietterich T. G. (1997). Machine-learning research. AI magazine 18, 97–97. doi: 10.1609/aimag.v18i4.1324
Drews P., Colares R. G., Machado P., De Faria M., Detoni A., Tavano V. (2013). Microalgae classification using semi-supervised and active learning based on Gaussian mixture models. J. Braz. Comput. Soc. 19, 411–422. doi: 10.1007/s13173-013-0121-y
Dunker S., Boho D., Wäldchen J., Mäder P. (2018). Combining high-throughput imaging flow cytometry and deep learning for efficient species and life-cycle stage identification of phytoplankton. BMC Ecol. 18, 51. doi: 10.1186/s12898-018-0209-5
El Naqa I., Murphy M. J. (2015). What is machine learning? In: El Naqa I., Murphy M. J.. (eds) Machine Learning in Radiation Oncology: Theory and Applications (Cham: Springer International Publishing(Switzerland)).
Elomaa T. (1994). “In defense of C4. 5: Notes on learning one-level decision trees,” In: Cohen W, Hirsh H. (eds) Machine Learning Proceedings 1994. San Francisco (CA): Morgan Kaufmann.
Enamala M. K., Enamala S., Chavali M., Donepudi J., Yadavalli R., Kolapalli B., et al. (2018). Production of biofuels from microalgae - a review on cultivation, harvesting, lipid extraction, and numerous applications of microalgae. Renewable Sustain. Energy Rev. 94, 49–68. doi: 10.1016/j.rser.2018.05.012
Expósito P. L., Suárez A. B., Álvarez C. N. (2017). Laser reflectance measurement for the online monitoring of chlorella sorokiniana biomass concentration. J. Biotechnol. 243, 10–15. doi: 10.1016/j.jbiotec.2016.12.020
Farnaaz N., Jabbar M. A. (2016). Random forest modeling for network intrusion detection system. Proc. Comput. Sci. 89, 213–217. doi: 10.1016/j.procs.2016.06.047
Ferro L., Gentili F. G., Funk C. (2018). Isolation and characterization of microalgal strains for biomass production and wastewater reclamation in northern Sweden. Algal Res. 32, 44–53. doi: 10.1016/j.algal.2018.03.006
Franco B. M., Navas L. M., Gómez C., Sepúlveda C., Acién F. G. (2019). Monoalgal and mixed algal cultures discrimination by using an artificial neural network. Algal Res. 38, 101419. doi: 10.1016/j.algal.2019.101419
Friedl M. A., Brodley C. E. (1997). Decision tree classification of land cover from remotely sensed data. Remote Sens. Environ. 61, 399–409. doi: 10.1016/S0034-4257(97)00049-7
Georgianna D. R., Mayfield S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335. doi: 10.1038/nature11479
Giraldo-Zuluaga J. H., Diez G., Gomez A., Martinez T., Vasquez M. P., Bonilla J., et al. (2016). Automatic identification of scenedesmus polymorphic microalgae from microscopic images. Pattern Analysis and Applications 21, 601–612 doi: 10.1007/s10044-017-0662-3
Gomez-Espinoza O., Guerrero-Barrantes M., Meneses-Montero K., Núñez-Montero K. (2018). Identification of a microalgae collection isolated from Costa Rica by 18S rDNA sequencing. Acta Biológica Colombiana 23, 199. doi: 10.15446/abc.v23n2.68088
Guo B., Lei C., Kobayashi H., Ito T., Yalikun Y., Jiang Y., et al. (2017). High-throughput, label-free, single-cell, microalgal lipid screening by machine-learning-equipped optofluidic time-stretch quantitative phase microscopy. Cytometry Part A 91, 494–502. doi: 10.1002/cyto.a.23084
Gu J., Wang Z., Kuen J., Ma L., Shahroudy A., Shuai B., et al. (2018). Recent advances in convolutional neural networks. Pattern Recognition 77, 354–377. doi: 10.1016/j.patcog.2017.10.013
Harmon J., Mikami H., Kanno H., Ito T., Goda K. (2020). Accurate classification of microalgae by intelligent frequency-division-multiplexed fluorescence imaging flow cytometry. OSA Continuum 3, 430–440. doi: 10.1364/OSAC.387523
Hearst M. A., Dumais S. T., Osuna E., Platt J., Scholkopf B. (1998). Support vector machines. IEEE Intelligent Syst. their Appl. 13, 18–28. doi: 10.1109/5254.708428
He S., Fang S., Xie W., Zhang P., Li Z., Zhou D., et al. (2018). Assessment of physiological responses and growth phases of different microalgae under environmental changes by raman spectroscopy with chemometrics. Spectrochimica Acta Part A: Mol. Biomolecular Spectrosc. 204, 287–294. doi: 10.1016/j.saa.2018.06.060
He K., Zhang X., Ren S., Sun J. (2015). Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. 2015 IEEE International Conference on Computer 929 Vision (ICCV), 7–13 Dec. IEEE(USA) 2015, 1026–1034. doi: 10.1109/ICCV.2015.123
Hossain S. M. Z., Sultana N., Jassim M. S., Coskuner G., Hazin L. M., Razzak S. A., et al. (2022a). Soft-computing modeling and multiresponse optimization for nutrient removal process from municipal wastewater using microalgae. J. Water Process Eng. 45, 102490. doi: 10.1016/j.jwpe.2021.102490
Hossain S. M. Z., Sultana N., Mohammed M. E., Razzak S. A., Hossain M. M. (2022b). Hybrid support vector regression and crow search algorithm for modeling and multiobjective optimization of microalgae-based wastewater treatment. J. Environ. Manage. 301, 113783. doi: 10.1016/j.jenvman.2021.113783
Hossain S. M. Z., Sultana N., Razzak S. A., Hossain M. M. (2022c). Modeling and multi-objective optimization of microalgae biomass production and CO2 biofixation using hybrid intelligence approaches. Renewable Sustain. Energy Rev. 157, 112016. doi: 10.1016/j.rser.2021.112016
Hsu K. Y., Li H. Y., Psaltis D. (1990). Holographic implementation of a fully connected neural network. Proc. IEEE 78, 1637–1645. doi: 10.1109/5.58357
Huang H., Sun Z., Zhang Z., Chen X., Di Y., Zhu F., et al. (2021). The identification of spherical engineered microplastics and microalgae by micro-hyperspectral imaging. Bull. Environ. Contamination Toxicol. 107, 764–769. doi: 10.1007/s00128-021-03131-9
Işıl Ç., dE Haan K., Göröcs Z., Koydemir H. C., Peterman S., Baum D., et al. (2021b). “Label-free imaging flow cytometry for phenotypic analysis of microalgae populations using deep learning,” In: Mazzali CPTAR, Kaindl R Frontiers in Optics + Laser Science Washington, DC: Optica Publishing Group(America). 2021/11/01 2021b.
Işıl Ç., dE Haan K., Göröcs Z., Koydemir H. C., Peterman S., Baum D., Song F., et al. (2021a). Phenotypic analysis of microalgae populations using label-free imaging flow cytometry and deep learning. ACS Photonics 8, 1232–1242. doi: 10.1021/acsphotonics.1c00220
Jordan M. I., Mitchell T. M. (2015). Machine learning: Trends, perspectives, and prospects. Science 349, 255–260. doi: 10.1126/science.aaa8415
Joshi R. C., Dhup S., Kaushik N., Dutta M. K. (2021). An efficient oil content estimation technique using microscopic microalgae images. Ecol. Inf. 66, 101468. doi: 10.1016/j.ecoinf.2021.101468
Kuo C. C. J. (2016). Understanding convolutional neural networks with a mathematical model. J. Visual Communication Image Representation 41, 406–413. doi: 10.1016/j.jvcir.2016.11.003
Lecun Y., Bengio Y. (1995). “Convolutional networks for images, speech, and time series,” in The handbook of brain theory and neural networks Cambridge, MA, USA: MIT Press, vol. 3361, 255–258.
Levasseur W., Pozzobon V., Perré P. (2022). Green microalgae in intermittent light: a meta-analysis assisted by machine learning. J. Appl. Phycology 34, 135–158. doi: 10.1007/s10811-021-02603-z
Liakos K. G., Busato P., Moshou D., Pearson S., Bochtis D. (2018). Machine learning in agriculture: A review. Sensors 18, 2674. doi: 10.3390/s18082674
Li M., Xu H., Deng Y. (2019). Evidential decision tree based on belief entropy. Entropy 21, 897–910. doi: 10.3390/e21090897
Li J., Zhang W., Liu T., Yang L., Li H., Peng H., et al. (2021). Machine learning aided bio-oil production with high energy recovery and low nitrogen content from hydrothermal liquefaction of biomass with experiment verification. Chem. Eng. J. 425, 130649. doi: 10.1016/j.cej.2021.130649
Long B., Fischer B., Zeng Y., Amerigian Z., Li Q., Bryant H., et al. (2022). Machine learning-informed and synthetic biology-enabled semi-continuous algal cultivation to unleash renewable fuel productivity. Nat. Commun. 13, 541. doi: 10.1038/s41467-021-27665-y
Lopez-Exposito P., Negro C., Blanco A. (2019). Direct estimation of microalgal flocs fractal dimension through laser reflectance and machine learning. Algal Res. 37, 240–247. doi: 10.1016/j.algal.2018.12.007
Mahesh B. (2020). Machine learning algorithms-a review. Int. J. Sci. Res. (IJSR) 9, 381–386. doi: 10.21275/ART20203995
Mcculloch W. S., Pitts W. (1943). A logical calculus of the ideas immanent in nervous activity. Bull. Math. biophysics 5, 115–133. doi: 10.1007/BF02478259
McCulloch W. S., Pitts W. (1990). A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 52, 99–115. doi: 10.1016/S0092-8240(05)80006-0
Meyer D., Leisch F., Hornik K. (2003). The support vector machine under test. Neurocomputing 55, 169–186. doi: 10.1016/S0925-2312(03)00431-4
Milić M., Petković B., Selmi A., Petković D., Jermsittiparsert K., Radivojević A., et al. (2021). Computational evaluation of microalgae biomass conversion to biodiesel. Biomass Conversion Biorefinery 11, 1–8. doi: 10.1007/s13399-021-01314-2
Moayedi H., Aghel B., Foong L. K., Bui D. T. (2020). Feature validity during machine learning paradigms for predicting biodiesel purity. Fuel 262, 116498. doi: 10.1016/j.fuel.2019.116498
Mochdia K., Tamaki S. (2021). Transcription factor-based genetic engineering in microalgae. Plants 10, 1602–09. doi: 10.3390/plants10081602
Mofijur M., Rasul M. G., Hassan N. M. S., Nabi M. N. (2019). Recent development in the production of third generation biodiesel from microalgae. Energy Proc. 156, 53–58. doi: 10.1016/j.egypro.2018.11.088
Mohammed O., Park D., Merchant R., Dinh T., Tong C., Azeem A., et al. (1995). Practical experiences with an adaptive neural network short-term load forecasting system. IEEE Trans. Power Syst. 10, 254–265. doi: 10.1109/59.373948
Montavon G., Samek W., MÜLLER K.-R. (2018). Methods for interpreting and understanding deep neural networks. Digital Signal Process. 73, 1–15. doi: 10.1016/j.dsp.2017.10.011
Myles A. J., Feudale R. N., Liu Y., Woody N. A., Brown S. D. (2004). An introduction to decision tree modeling. J. Chemometrics: A J. Chemometrics Soc. 18, 275–285. doi: 10.1002/cem.873
Nugraha W., Maulana M. S., Sasongko A. (2020). Clustering based undersampling for handling class imbalance in C4. 5 classification algorithm. Journal of Physics: Conference Series (UK: IOP Publishing) 1641, 012014.
Otálora P., Guzmán J. L., Acién F. G., Berenguel M., Reul A. (2021). Microalgae classification based on machine learning techniques. Algal Res. 55, 102256. doi: 10.1016/j.algal.2021.102256
Pal M., Mather P. M. (2003). An assessment of the effectiveness of decision tree methods for land cover classification. Remote Sens. Environ. 86, 554–565. doi: 10.1016/S0034-4257(03)00132-9
Park J., Baek J., You K., Nam S. W., Kim J. (2021). Microalgae detection using a deep learning object detection algorithm, YOLOv3. J. Korean Soc. Water Environ. 37, 275–285. doi: 10.15681/KSWE.2021.37.4.275
Patel H. H., Prajapati P. (2018). Study and analysis of decision tree based classification algorithms. Int. J. Comput. Sci. Eng. 6, 74–78. doi: 10.26438/ijcse/v6i10.7478
Pathy A., Meher S., Balasubramanian P. (2020). Predicting algal biochar yield using eXtreme gradient boosting (XGB) algorithm of machine learning methods. Algal Res. 50, 102006. doi: 10.1016/j.algal.2020.102006
Paul A., Mukherjee D. P., Das P., Gangopadhyay A., Chintha A. R., Kundu S. (2018). Improved random forest for classification. IEEE Trans. Image Process. 27, 4012–4024. doi: 10.1109/TIP.2018.2834830
Peniuk G. T., Schnurr P. J., Allen D. G. (2016). Identification and quantification of suspended algae and bacteria populations using flow cytometry: applications for algae biofuel and biochemical growth systems. J. Appl. phycology 28, 95–104. doi: 10.1007/s10811-015-0569-6
Pozzobon V., Levasseur W., Guerin C., Perré P. (2021). Nitrate and nitrite as mixed source of nitrogen for chlorella vulgaris: fast nitrogen quantification using spectrophotometer and machine learning. J. Appl. Phycology 33, 1389–1397. doi: 10.1007/s10811-021-02422-2
Pozzobon V., Levasseur W., Viau E., Michiels E., Clément T., Perré P. (2020). Machine learning processing of microalgae flow cytometry readings: illustrated with chlorella vulgaris viability assays. J. Appl. Phycology 32, 2967–2976. doi: 10.1007/s10811-020-02180-7
Pradhan A. (2012). Support vector machine-a survey. Int. J. Emerging Technol. Advanced Eng. 2, 82–85. doi: 10.1007/978-3-662-47926-1_26
Promdaen S., Wattuya P., Sanevas N. (2014). Automated microalgae image classification. Proc. Comput. Sci. 29, 1981–1992. doi: 10.1016/j.procs.2014.05.182
Purnomo D. M. J., Purbarani S. C., Wibisono A., Hendrayanti D., Bowolaksono A., Mursanto P., et al. (2015). Genetic algorithm optimization for extreme learning machine based microalgal growth forecasting of chlamydomonas sp. 2015 International Conference on Advanced Computer Science and Information Systems (ICACSIS), 10-11 Oct. 2015, 243–248. doi: 10.1109/ICACSIS.2015.7415189
Qi Y. (2012). “Random forest for bioinformatics,” In: Zhang C., Ma Y.. (eds.) Ensemble Machine Learning: Methods and Applicationsg (Boston, MA: Springer US).
Rani P., Kotwal S., Manhas J., Sharma V., Sharma S. (2021). Machine learning and deep learning based computational approaches in automatic microorganisms image recognition: Methodologies, challenges, and developments. Arch. Comput. Methods Eng 29, 641–677. doi: 10.1007/s11831-021-09639-x
Rashvand M., Zenouzi A., Abbaszadeh R. (2019). Potential of image processing, dielectric spectroscopy and intelligence methods in order to authentication of microalgae biodiesel. Measurement 148, 106962. doi: 10.1016/j.measurement.2019.106962
Reimann R., Zeng B., Jakopec M., Burdukiewicz M., Petrick I., Schierack P., et al. (2020). Classification of dead and living microalgae chlorella vulgaris by bioimage informatics and machine learning. Algal Res. 48, 101908. doi: 10.1016/j.algal.2020.101908
Rock A., Lucie N., Green D. H. (2021). Synthetic biology is essential to unlock commercial biofuel production through hyper lipid-producing microalgae: a review. J. Appl. Phycology 2, 41–59. doi: 10.1080/26388081.2021.1886872
Rodriguez-Galiano V. F., Ghimire B., Rogan J., Chica-Olmo M., Rigol-Sanchez J. P. (2012). An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. photogrammetry Remote Sens. 67, 93–104. doi: 10.1016/j.isprsjprs.2011.11.002
Rosenblatt F. (1958). The perceptron: a probabilistic model for information storage and organization in the brain. psychol. Rev. 65, 386. doi: 10.1037/h0042519
Rumelhart D. E., Hinton G. E., Williams R. J. (1986). Learning representations by back-propagating errors. nature 323, 533–536. doi: 10.1038/323533a0
Sagheer A., Zidan M., Abdelsamea M. M. (2019). A novel autonomous perceptron model for pattern classification applications. Entropy 21, 763–786. doi: 10.3390/e21080763
Sain S. R. (1996). The nature of statistical learning theory. Technometrics 38, 409–409. doi: 10.1080/00401706.1996.10484565
Salam S., Verma T. N. (2019). Appending empirical modelling to numerical solution for behaviour characterisation of microalgae biodiesel. Energy Conversion Manage. 180, 496–510. doi: 10.1016/j.enconman.2018.11.014
Sá C., Leal M. C., Silva A., Nordez S., André E., Paula J., et al. (2013). Variation of phytoplankton assemblages along the Mozambique coast as revealed by HPLC and microscopy. J. Sea Res. 79, 1–11. doi: 10.1016/j.seares.2013.01.001
Saputro T. B., Purwani K. I., Ermavitalini D., Saifullah A. F. (2019). Isolation of high lipids content microalgae from wonorejo rivers, Surabaya, Indonesia and its identification using rbcL marker gene. Biodiversitas J. Biol. Diversity 20, 1380–1388. doi: 10.13057/biodiv/d200530
Sarıgül M., Ozyildirim B. M., Avci M. (2019). Differential convolutional neural network. Neural Networks 116, 279–287. doi: 10.1016/j.neunet.2019.04.025
Shahid N., Naqvi I. H., Qaisar S. B. (2015). One-class support vector machines: analysis of outlier detection for wireless sensor networks in harsh environments. Artif. Intell. Rev. 43, 515–563. doi: 10.1007/s10462-013-9395-x
Sharma S., Agrawal J., Sharma S. (2013). Classification through machine learning technique: C4. 5 algorithm based on various entropies. Int. J. Comput. Appl. 82, 28–32. doi: 10.5120/14249-2444
Shi T., Horvath S. (2006). Unsupervised learning with random forest predictors. J. Comput. Graphical Stat 15, 118–138. doi: 10.1198/106186006X94072
Singh V., Mishra V. (2021). Exploring the effects of different combinations of predictor variables for the treatment of wastewater by microalgae and biomass production. Biochem. Eng. J. 174, 108129. doi: 10.1016/j.bej.2021.108129
Singh V., Mishra V. (2022). Evaluation of the effects of input variables on the growth of two microalgae classes during wastewater treatment. Water Res. 213, 118165. doi: 10.1016/j.watres.2022.118165
Sirico D. G., Cavalletti E., Miccio L., Bianco V., Memmolo P., Sardo A., et al. (2022). Kinematic analysis and visualization of tetraselmis microalgae 3Dmotility by digital holography. Appl. Optics 61, B331–B338. doi: 10.1364/AO.444976
Sonkar S., Mallick N. (2020). Application of machine learning for development of a drying protocol for microalga chlorella minutissima in a single rotary drum dryer for biodiesel production. Authorea 26, 2020. doi: 10.22541/au.160372833.38766717/v1
Sonmez M. E., Eczacioglu N., Gumuş N. E., Aslan M. F., Sabanci K., Aşikkutlu B. (2022). Convolutional neural network - support vector machine based approach for classification of cyanobacteria and chlorophyta microalgae groups. Algal Res. 61, 102568. doi: 10.1016/j.algal.2021.102568
Speiser J. L., Miller M. E., Tooze J., Ip E. (2019). A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 134, 93–101. doi: 10.1016/j.eswa.2019.05.028
Sultana N., Hossain S. M. Z., Abusaad M., Alanbar N., Senan Y., Razzak S. A. (2022). Prediction of biodiesel production from microalgal oil using Bayesian optimization algorithm-based machine learning approaches. Fuel 309, 122184. doi: 10.1016/j.fuel.2021.122184
Sundui B., Calderon O. A. R., Abdeldayem O. M., Lázaro-Gil J., Rene E. R., Sambuu U. (2021). Applications of machine learning algorithms for biological wastewater treatment: Updates and perspectives. Clean Technol. Environ. Policy 23, 127–143. doi: 10.1007/s10098-020-01993-x
Supriyanto, Noguchi R., Ahamed T., Mikihide D., Watanabe M. M. (2018). “A decision tree approach to estimate the microalgae production in open raceway pond,” in IOP Conference Series: Earth and Environmental Science, 3rd International conference on biomass: Accelerating the technical development and commercialization for sustainable bio-based products and energy, Bogor, Indonesia:IOP Publishing (UK), 209. 012050.
Supriyanto, Noguchi R., Ahamed T., Rani D. S., Sakurai K., Nasution M. A., et al. (2019). Artificial neural networks model for estimating growth of polyculture microalgae in an open raceway pond. Biosyst. Eng. 177, 122–129. doi: 10.1016/j.biosystemseng.2018.10.002
Suykens J. A. K., Vandewalle J. (1999). Least squares support vector machine classifiers. Neural Process. Lett. 9, 293–300. doi: 10.1023/A:1018628609742
Swain P. H., Hauska H. (1977). The decision tree classifier: Design and potential. IEEE Trans. Geosci. Electron. 15, 142–147. doi: 10.1109/TGE.1977.6498972
Teng S. Y., Yew G. Y., Sukačová K., Show P. L., Máša V., Chang J.-S. (2020). Microalgae with artificial intelligence: A digitalized perspective on genetics, systems and products. Biotechnol. Adv. 44, 107631. doi: 10.1016/j.biotechadv.2020.107631
Tian L., Noore A. (2004). Short-term load forecasting using optimized neural network with genetic algorithm. 2004 International Conference on Probabilistic Methods Applied to Power 1140 Systems, 12-16 Sept. 2004, 135–140. doi: 10.1109/PMAPS.2004.243045
Vapnik V. N. (1999). An overview of statistical learning theory. IEEE Trans. Neural Networks 10, 988–999. doi: 10.1109/72.788640
Vijayakumar S., Rahman P. K. S. M., Angione C. (2020). A hybrid flux balance analysis and machine learning pipeline elucidates metabolic adaptation in cyanobacteria. iScience 23, 101818. doi: 10.1016/j.isci.2020.101818
Wang Y., Ju P., Wang S., Su J., Zhai W., Wu C. (2021). Identification of living and dead microalgae cells with digital holography and verified in the East China Sea. Mar. pollut. Bull. 163, 111927. doi: 10.1016/j.marpolbul.2020.111927
Wang W., Men C., Lu W. (2008). Online prediction model based on support vector machine. Neurocomputing 71, 550–558. doi: 10.1016/j.neucom.2007.07.020
Wang Z., Peng X., Xia A., Shah A. A., Huang Y., Zhu X., et al. (2022). The role of machine learning to boost the bioenergy and biofuels conversion. Bioresource Technol. 343, 126099. doi: 10.1016/j.biortech.2021.126099
Wang L., Xi Y., Sung S., Qiao H. (2018). RNA-Seq assistant: machine learning based methods to identify more transcriptional regulated genes. BMC Genomics 19, 546. doi: 10.1186/s12864-018-4932-2
Wei J., Chu X., Sun X.-Y., Xu K., Deng H.-X., Chen J., et al. (2019). Machine learning in materials science. InfoMat 1, 338–358. doi: 10.1002/inf2.12028
Wei L., Su K., Zhu S., Yin H., Li Z., Chen Z., et al. (2017). Identification of microalgae by hyperspectral microscopic imaging system. Spectrosc. Lett. 50, 59–63. doi: 10.1080/00387010.2017.1287094
Wellner B., Grand J., Canzone E., Coarr M., Brady P. W., Simmons J., et al. (2017). Predicting unplanned transfers to the intensive care unit: a machine learning approach leveraging diverse clinical elements. JMIR Med. Inf. 5, e8680. doi: 10.2196/medinform.8680
Widodo A., Yang B.-S. (2007). Support vector machine in machine condition monitoring and fault diagnosis. Mechanical Syst. Signal Process. 21, 2560–2574. doi: 10.1016/j.ymssp.2006.12.007
Xu M., Harmon J., Hasunuma T., Isozaki A., Goda K. (2021a). “Ai on a chip for identifying microalgal cells with high heavy metal removal efficiency,” in 2021 21st International Conference on Solid-State Sensors, Actuators and Microsystems (Transducers), Orlando, FL, USA, 20-24 June 2021. IEEE(USA) 385–388.
Xu M., Harmon J., Yuan D., Yan S., Lei C., Hiramatsu K., et al. (2021b). Morphological indicator for directed evolution of euglena gracilis with a high heavy metal removal efficiency. Environ. Sci. Technol. 55, 7880–7889. doi: 10.1021/acs.est.0c05278
Xu Z., Jiang Y., Ji J., Forsberg E., Li Y., He S. (2020). Classification, identification, and growth stage estimation of microalgae based on transmission hyperspectral microscopic imaging and machine learning. Optics Express 28, 30686–30700. doi: 10.1364/OE.406036
Yadav D. P., Jalal A. S., Garlapati D., Hossain K., Goyal A., Pant G. (2020). Deep learning-based ResNeXt model in phycological studies for future. Algal Res. 50, 102018. doi: 10.1016/j.algal.2020.102018
Zhang W., Li J., Liu T., Leng S., Yang L., Peng H., et al. (2021). Machine learning prediction and optimization of bio-oil production from hydrothermal liquefaction of algae. Bioresource Technol. 342, 126011. doi: 10.1016/j.biortech.2021.126011
Zheng X., Duan X., Tu X., Jiang S., Song C. (2021). The fusion of microfluidics and optics for on-chip detection and characterization of microalgae. Micromachines 12, 1137–1156. doi: 10.3390/mi12101137
Zhou Z.-H., Chen Z.-Q. (2002). Hybrid decision tree. Knowledge-based Syst. 15, 515–528. doi: 10.1016/S0950-7051(02)00038-2
Zhuo Z., Wang H., Liao R., Ma H. (2022). Machine learning powered microalgae classification by use of polarized light scattering data. Appl. Sci. 12, 3422. doi: 10.3390/app12073422
Keywords: microalgae, machine learning, environment protection, biodiesel, convolutional neural network
Citation: Ning H, Li R and Zhou T (2022) Machine learning for microalgae detection and utilization. Front. Mar. Sci. 9:947394. doi: 10.3389/fmars.2022.947394
Received: 18 May 2022; Accepted: 29 June 2022;
Published: 26 July 2022.
Edited by:
Ravi Raghavbhai Sonani, Jagiellonian University, PolandReviewed by:
Datta Madamwar, Charotar University of Science and Technology, IndiaBharati Pandey, Panjab University, India
Copyright © 2022 Ning, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teng Zhou, emhvdXRlbmdAaGFpbmFudS5lZHUuY24=
 Hongwei Ning
Hongwei Ning Rui Li
Rui Li Teng Zhou
Teng Zhou