- 1Department of Life Sciences, Texas A&M University-Corpus Christi, Corpus Christi, TX, United States
- 2Department of Physical and Environmental Sciences, Texas A&M University-Corpus Christi, Corpus Christi, TX, United States
- 3Center for Coastal Studies, Texas A&M University-Corpus Christi, Corpus Christi, TX, United States
Marine environments are sinks for many contaminants, including petroleum-based plastic waste. Bioplastics, or biodegradable plastics derived from renewable resources, are considered promising alternatives as numerous studies have demonstrated their degradation in marine environments. However, their rates of degradation vary and microbial consortia responsible for its degradation are not well characterized. Previous research by our group has shown that polyhydroxyalkanoate (PHA) stimulates sulfate reducing microorganisms (SRM), enriches sulfate reduction gene pools, and accumulates antibiotic and metal resistance genes. Here, we quantify the degradation rate of PHA pellets in marine sediment and present the long-term temporal changes in PHA-associated SRM communities over 424 days. For comparative purposes, polyethylene terephthalate (PET) and ceramic served as biofilm controls and the free-living microorganisms in the overlying water column served as a non-biofilm control. PHA experienced a 51% mass loss after 424 days and a generalized additive mixed model predicted that 100% mass loss would require 909 days. Throughout the course of the 424-day exposure, PHA was colonized by a distinct microbial community while PET and ceramic were colonized by similarly structured communities. SRM comprised a larger proportion of the overall community (25 – 40%) in PHA-associated biofilms as compared to PET and ceramic controls across all timepoints. Further, the diversity of SRM was greater within PHA biofilms than PET and ceramic biofilms. This study shows that PHA degrades relatively slowly and promotes a long-term shift in microbial community structure toward sulfate reduction, demonstrating the ability of this manufactured polymer to alter its environment via the disruption of biogeochemical cycling, indicating that PHA rises to the level of pollutant in benthic marine systems.
Introduction
Humans have significantly altered natural environments through the addition of 5.5 billion tons of petroleum-based plastic (henceforth referred to as plastic) waste (Geyer et al., 2017). Aquatic ecosystems in particular are sinks for plastic waste and it was estimated that between 19 and 23 million metric tons of plastic waste entered aquatic ecosystems in 2016 alone (Borrelle et al., 2020). To combat the crisis of plastic pollution, more than 60 countries have approved plastic bans or levies for the purpose of reducing single-use plastics as well as promoting the manufacture and use of bio-based and/or biodegradable (henceforth referred to as bioplastics) alternatives (E.U, 2018; U.N.E.P, 2018). Earlier this year, the United Nations Environment Assembly (UNEA-5), with the support of delegates from 175 member nations, announced a historic resolution to establish the first global treaty to address plastic pollution by 2024 (UNEA, 2022). As a result of these legislative actions and increased public awareness, the global production of bioplastics has increased and is forecasted to grow substantially in the near future. For example, the production capacity of polyhydroxyalkanoate (PHA) is predicted to more than triple between 2021 and 2026 (European Bioplastics, 2021). Although trading petroleum-based plastic for biodegradable plastic may appear an environmentally sound decision, the effects of bioplastic loading in natural environments are largely unknown.
PHAs are a diverse group of linear polyesters composed of ester bonds between the carboxyl group of one monomer and the hydroxyl group of neighboring monomers (Muneer et al., 2020). A diverse set of bacteria – including but not limited to members of Pseudomonas, Bacillus, Burkholderia, Aeromonas, Halomonas, Rhizobium, and Serratia – synthesize PHA as intracellular carbon and energy storage compounds during periods of excess carbon coupled with nutrient limitation (Samrot et al., 2021; Suzuki et al., 2021). Manufactured PHA is typically the product of microbial synthesis under fermentation conditions (Koller, 2018) and it is increasingly regarded as a promising alternative to petroleum-based plastics given its high degradation potential (Nehra et al., 2017). Further, innovative physical blends and chemical modifications are predicted to expand PHA’s functionality (Muneer et al., 2020). Their physical properties are often compared to polyethylene and polypropylene and not surprisingly, industrial news is promoting PHA as an innovative polymer ‘whose time has finally come” (Tullo, 2019).
Microorganisms capable of biodegrading PHA are abundant, phylogenetically diverse, and widely distributed (Emadian et al., 2017). The degradation of PHA under anaerobic conditions is an important process for efficient waste management in waste treatment plants and landfills as well as natural sediments (Al-Khattaf et al., 2022). Previous work by our group has shown that PHA accumulates sulfate-reducing microorganisms (SRMs) and dissimilatory sulfate-reduction genes in marine sediments (Pinnell and Turner, 2019; Pinnell and Turner, 2020). Seeing that SRM play a predominant role in organic matter mineralization in marine sediments (Jørgensen et al., 2019), increased abundances of SRM and sulfate-reduction genes within these sediments could have substantial impacts on biogeochemical cycling. Culture-based work has shown that SRM contribute to PHA degradation under anaerobic laboratory conditions (Çetin, 2009) while in situ microcosm studies have shown that SRM contribute to PHA degradation in anoxic lake sediments (Mas-Castellà et al., 1995; Urmeneta et al., 1995). Within aquatic environments, members of a diverse set of microbial taxa (e.g., Bacillaceae, Burkholderiaceae, Comamonadaceae, Alteromonadaceae, Pseudoalteromonadaceae, and Alcaligenaceae) are capable of biodegrading PHA (Yamada et al., 2018; Kato et al., 2019; Cho et al., 2021; Park et al., 2021; Suzuki et al., 2021; Cho et al., 2022). A recent synthesis of published PHA degradation data, estimated the average rate of degradation in the marine environment was 0.04-0.09 mg·day-1·cm-2 (Dilkes-Hoffman et al., 2019).
What remains unclear is the temporal variability of the microbial consortia responsible for its degradation and SRM specifically. To address this knowledge gap, we used 16S rRNA gene sequencing to monitor microbial succession during the degradation of PHA in coastal marine sediment over 424-days. Polyethylene terephthalate (PET) and ceramic were included as biofilm controls, and the free-living microbial community in the overlying water was included as a non-biofilm control. We hypothesized that 1) PHA-associated microbial communities would be distinct in comparison to the biofilm controls (PET and ceramic) and 2) the diversity and relative abundance of SRM would increase within PHA-associated microbial communities over time.
Materials and methods
Microcosms
The data analyzed in this study were the product of in situ microcosm experiments conducted by our laboratory. Microcosms utilized a 315 micron mesh to contain the pellets and permit the exchange of water, nutrients, microorganisms, and some grazers but limit the entry of large organisms. Please refer to Pinnell and Turner (2020) for a map of the study site, a detailed description of the randomized experiment design, and laboratory procedures. Briefly, each microcosm contained either 3.0 g of PET pellets (M&G Chemicals, Ettelbruck, Luxembourg), 3.0 g of PHA pellets (polyhydroxybutyrate, specifically) (Doctors Foster and Smith, Rhinelander, WI, USA) or 6.0 g of ceramic pellets (Lyman Products, Middletown, CT, USA) that were deployed in quadruplicate at the sediment water interface (~1 m depth) for up to 424 days (subsampled approximately every 28 days) at the Laguna Madre Field Station located at 27°32’39.0”N and 97°17’07.7”W in the northwestern Gulf of Mexico (GOM). The Laguna Madre is a coastal lagoon and the largest estuarine system in the northwestern GOM. Environmental parameters (i.e., temperature, salinity, DO, and pH) were measured using a 6920 V2-2 Multi-Parameter Water Quality Sonde (YSI, Yellow Springs, OH, USA; Table S1) at the deployment date and each of the 15 collection timepoints.
Degradation rate
The substrate-associated biofilms (i.e., ceramic, PET, and PHA) were removed during the DNA isolation procedure as described previously (Pinnell and Turner, 2020). Briefly, DNA was isolated using a modified version of a high-salt and sodium dodecyl sulfate-based method (Zhou et al., 1996). DNA was quantified (ng/μL) and assayed for quality (A260/A280) using a BioPhotometer D30 (Eppendorf, Hamburg, Germany). Following biofilm removal, the substrates were rinsed twice with 50 mL of ultrapure water (Milli-Q; MilliporeSigma, Burlington, MA, USA) and incubated in 50 mL of ultrapure water for 12-15 hours at room temperature. The ultrapure water was decanted and the substrates were again rinsed twice with 50 mL of ultrapure water. Substrates were transferred to glass Petri dishes, dried for 24 hours at 60 °C and their aggregate mass was determined to 1.0 μg using a Sartorius Cubis Microbalance (Sartorius AG, Göttigen, Germany). The percent mass loss (X, %) was calculated as a ratio of the final mass (X2) to the initial mass (X1) multiplied by 100 with the following equation:
Virgin substrates that were not exposed to marine sediment were processed identically as controls. To estimate how long it would take for the pellets to completely degrade, a generalized additive mixed model, which allows for random effects in a generalized additive model, was fitted to the data using R version 3.5.2 and the package mgcv. Due to heteroscedasticty in the data, an exponential variance function was added to the model using the R package nlme. The best fit model was chosen based on corrected Akaike Information Criterion (AICc), which estimates prediction error and compares the quality of each model and reduces overfitting on data with smaller sample sizes.
Visualization of biofilms and degradation
The fourth replicate collected at each of the 15 sample collections was stored in the dark at -80 °C, and used for visualizing biofilms and degradation with scanning electron microscopy (SEM). Substrates were thawed, freeze-dried, mounted to stubs using double-stick tape and imaged using a Jeol JCM-6000 Neoscope SEM (JEOL Inc., Peabody, MA, USA). Virgin substrates subjected to the DNA isolation procedure were used as controls.
16S rRNA library preparation, sequencing, and bioinformatics
Amplicon libraries were prepared by amplifying the V4 region of the 16S rRNA gene with an iTaq DNA Polymerase kit (Bio-Rad, Hercules, CA) and the improved 515f/805r primer pair (Walters et al., 2016). Amplification conditions were 95°C for 3 minutes, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 50°C, and 1 minute at 72°C. Final elongation occurred at 72°C for 5 minutes. Amplicons were purified using the EXOSAP-IT Express PCR Cleanup kit (Thermo Fisher Scientific, Waltham, MA). Clean amplicons were pooled in equal proportion and purified using Ampure XP beads (Beckman Coulter, Indianapolis, IN) and the pooled library was sequenced on an Illumina MiSeq instrument using paired-end chemistry (2 x 250 bp) at Molecular Research LP (Shallowater, TX). Raw sequence reads were processed with QIIME2 and phyloseq. DADA2 (Callahan et al., 2016) was used to generated amplicon sequence variants (ASVs) with truncate lengths of 242 and 233 bp on the forward and reverse reads, respectively. Taxonomy was assigned using a Naïve Bayes classifier trained on the SILVA 132 99% OTU database, where sequences had been trimmed to include only those base pairs bound by the 515f/806r primer pair. Reads mapping to chloroplast and mitochondrial 16S sequences were removed, and a mid-point rooted phylogenetic tree was calculated with the ‘q2-phylogeny’ pipeline under default settings. Data was then imported in phyloseq (Callahan et al., 2016) and ASV counts were normalized using cumulative sum scaling (Paulson et al., 2013).
To facilitate temporal comparisons, communities were divided into five stages (i.e., fall 2016, winter 2016, spring 2017, summer 2017, and fall 2017), as described previously (Pinnell and Turner, 2020). These five seasonal stages correspond to 0 – 56, 57 – 140, 141 – 224, 225 – 337, and 338 – 424 days of exposure, respectively. Beta-diversity was analyzed using generalized UniFrac distances (Chen et al., 2012). From these distances, a principal co-ordinates analysis was performed and plotted and a permutational multivariate analysis of variance (PERMANOVA) was used to test for significant differences in community structure using the vegan (Oksanen et al., 2019) and pairwiseAdonis (Arbizu, 2017) packages. To ensure significant differences were not the result of unequal dispersions of variability, permutational analyses of dispersion (PERMDISP) were conducted for all significant PERMANOVA outcomes using the vegan package. Further, the relative abundance of ASVs within each sample were calculated and a heatmap was plotted.
SRM abundance and temporal patterns
To compare relative abundances of SRM, amplified sequence variants (ASVs) mapped to known SRM (67 genera representing 17 families, 11 orders, and 5 phyla; Table S2) were filtered using phyloseq’s ‘subset_taxa’ function. The relative abundances of SRM were then calculated for each substrate type and compared.
Statistical analyses
Unless specified otherwise, R version 3.5.2 (R Core Team, 2017) was used for statistical analysis of data. Pairwise Wilcoxon rank-sum test were performed with a Bonferroni (PW+B) correction for multiple comparisons. Differences in beta diversity were tested using pairwise PERMANOVA with a Bonferroni correction for multiple comparisons and 9999 permutations. Additionally, pairwise PERMDISPs were carried out for all PERMANOVAs using 9999 permutations to test for differences in dispersions.
Data availability
All sequence reads were made available through the BioProject PRJNA551219 at the NCBI’s Sequence Read Archive.
Results
Biodegradation rates
To determine the rate of PHA degradation, the mass loss was calculated as a ratio of the final mass to the initial mass. Following 424 days, the mass of PHA pellets decreased by an average of 1,512 mg, representing a loss of 51% (Figure 1). Generalized additive mixed models were fitted to the PHA mass loss data using all combinations of environmental variables collected during this study (i.e., temperature, salinity, DO, and pH; Table S1) but the rate of degradation was not correlated with environmental variability. The best fit model based on AICc scores used only the length of exposure to account for the loss in mass. The adjusted r-squared value for this model was 0.97 and it predicted that 100% mass loss would occur after 909 days or approximately 2.5 years. Mass loss measurements showed no evidence of PET degradation over the course of this 424-day study (Figure 1).
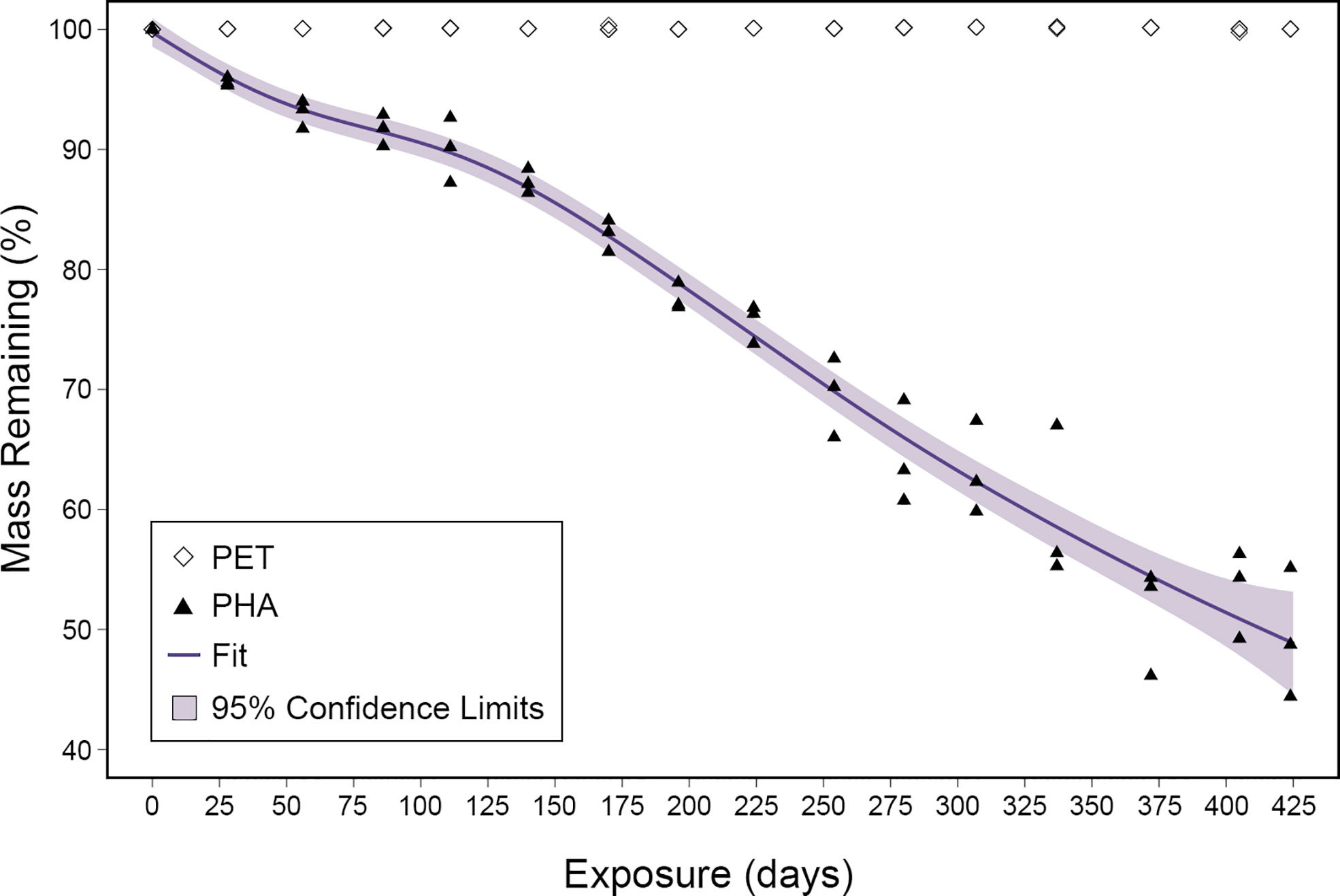
Figure 1 Mass loss ratios plotted for both PHA and PET over the 424-day exposure. The fit of the generalized additive mixed model is displayed as a purple line and the 95% confidence limits are shown as light purple shading.
Visualization of biofilms and signs of degradation
To visualize microbial colonization and degradation, SEM was used to image substrates with and without biofilms. Images revealed that microorganisms formed biofilms on both PHA and PET as early as the first sample collection (28 days exposure; Figure S1). Imaging of substrates after biofilm removal revealed that the PHA experienced significant size reduction and extensive pitting over time (Figure 2). This clear visual size reduction confirms the above mass loss calculation (51% after 424 days; Figure 1). In comparison, PET experienced no size reduction or pitting at any point during the 424 exposure (Figure 2).
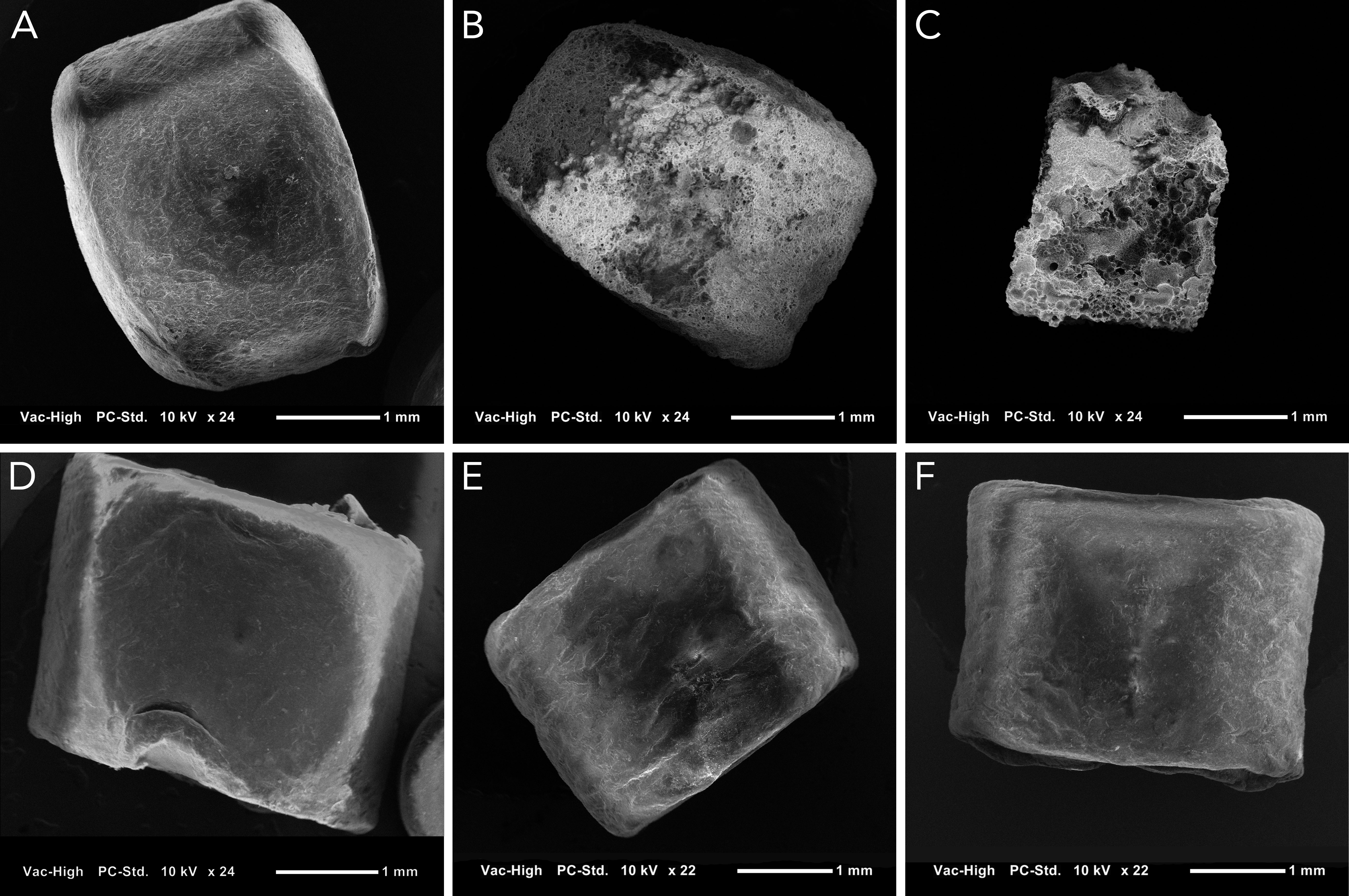
Figure 2 Scanning electron images of PHA and PET pellets without their associated biofilm to display changes to the polymer surface at approximately 25X magnification. (A–C) show PHA pellets after 0, 202, and 424 days of exposure, respectively. (D–F) show PET pellets after 0, 202, and 424 days of exposure, respectively. Extensive pitting is present on PHA pellets, but the surface of PET appears unchanged.
Overall community structure
The effect of sample type (seawater, ceramic, PET, and PHA) on community structure was analyzed by PCoA, and PERMANOVA and PERMDISP were used to test for significant differences between all communities. The PCoA analysis illustrated that 1) the communities associated with substrates (i.e., biofilms) were clearly different from the free-living communities in the overlying seawater, 2) the communities fouling ceramic and PET were similar when compared across all seasons, and 3) the communities fouling PHA were distinct in comparison to the communities fouling PET and ceramic (Figure 3). Across all timepoints, the compositions of all four communities were significantly different (PERMANOVA; p< 0.05, n= 39-45) but a significant PERMDISP test suggests that unequal dispersions of variation between the four groups may be responsible for the significant difference. However, the distinct clustering of PHA communities clearly illustrates that the composition of PHA-associated communities was significantly different from all other sample types in addition to the unequal dispersion of variation.
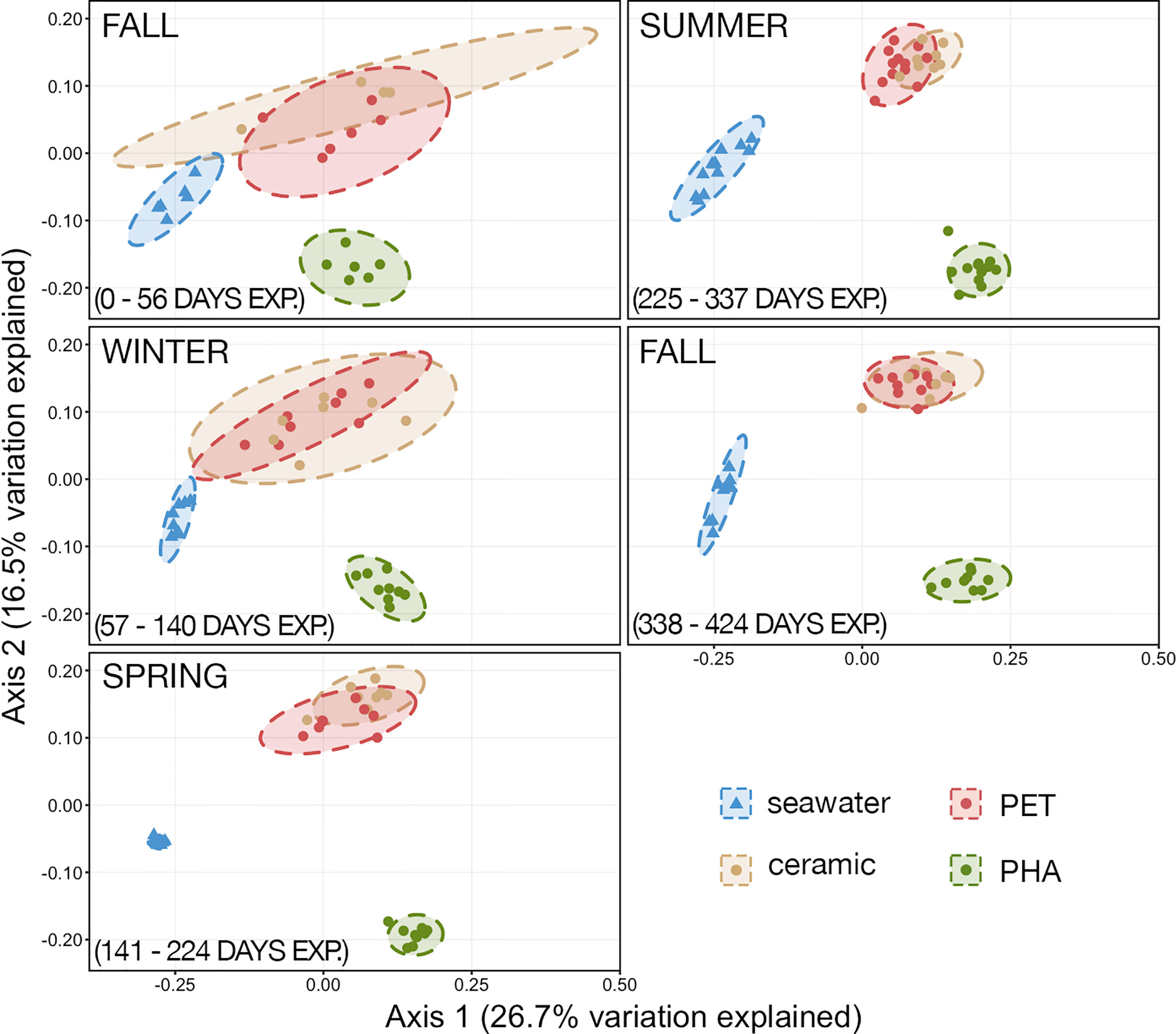
Figure 3 Principal coordinates analysis (PCoA) of generalized Unifrac distances illustrating the variation in microbial community structure. The PCoA demonstrates the clustering of 16S rRNA gene sequences from seawater (n=45), ceramic (n=40), PET (n=40), and PHA (n=45). Dashed lines represent the 95% confidence ellipses for each community type.
Relative abundances from each sample were calculated to identify dominant community members across all time points in each sample type. At the class level, ASVs assigned to δ-proteobacteria were the most abundant among all three biofilm communities especially within PHA (39.2% ± 1.13 SEM) versus PET (25.6% ± 1.13 SEM) and ceramic (25.3% ± 1.00 SEM). Contrastingly, α-proteobacteria was the most abundant class in seawater (47.3% ± 0.91 SEM) and among the top five most abundant classes in PET and ceramic biofilms (8.3% ± 0.84 SEM and 7.0% ± 1.08 SEM, respectively) but it was not abundant in PHA biofilms. While Bacteroidia was among the top five in all four communities, it made up a considerably larger proportion in PET (16.3% ± 0.54 SEM), ceramic (12.3 ± 0.70 SEM), and seawater (16.1% ± 0.54 SEM) compared to PHA (6.4% ± 0.50 SEM). A detailed report of taxonomic classes based on relative abundance across all timepoints is provided in Table S3.
The relative abundances of the top 50 most abundant families were visualized using a heatmap, and it is evident that members of α-proteobacteria, γ-proteobacteria, and Bacteroidetes were far more abundant in seawater, PET, and ceramic (Figure 4). Members of α-proteobacteria, specifically SAR11 (Clade I and Clade III), SAR86, SAR116, were very abundant in seawater, while Rhodobacteraceae was also very abundant in PET and ceramic communities (Figure 4). Members of γ-proteobacteria (i.e., γ-proteobacteria incertae sedis, Halieaceae, Litoricolaceae, Thiotrichaceae, and Chromatiaceae) were abundant in seawater and PET and ceramic. Families more abundant within PHA communities than PET and ceramic communities were largely the sulfur-related taxa Sedimenticolaceae (purple sulfur bacteria), Desulfobulbaceae (SRM), Desulfovibrionaceae (SRM), and Desulfarculaceae (SRM). Desulfobacteraceae (SRM) was abundant across all three substrate-attached communities (PET, ceramic, PHA).
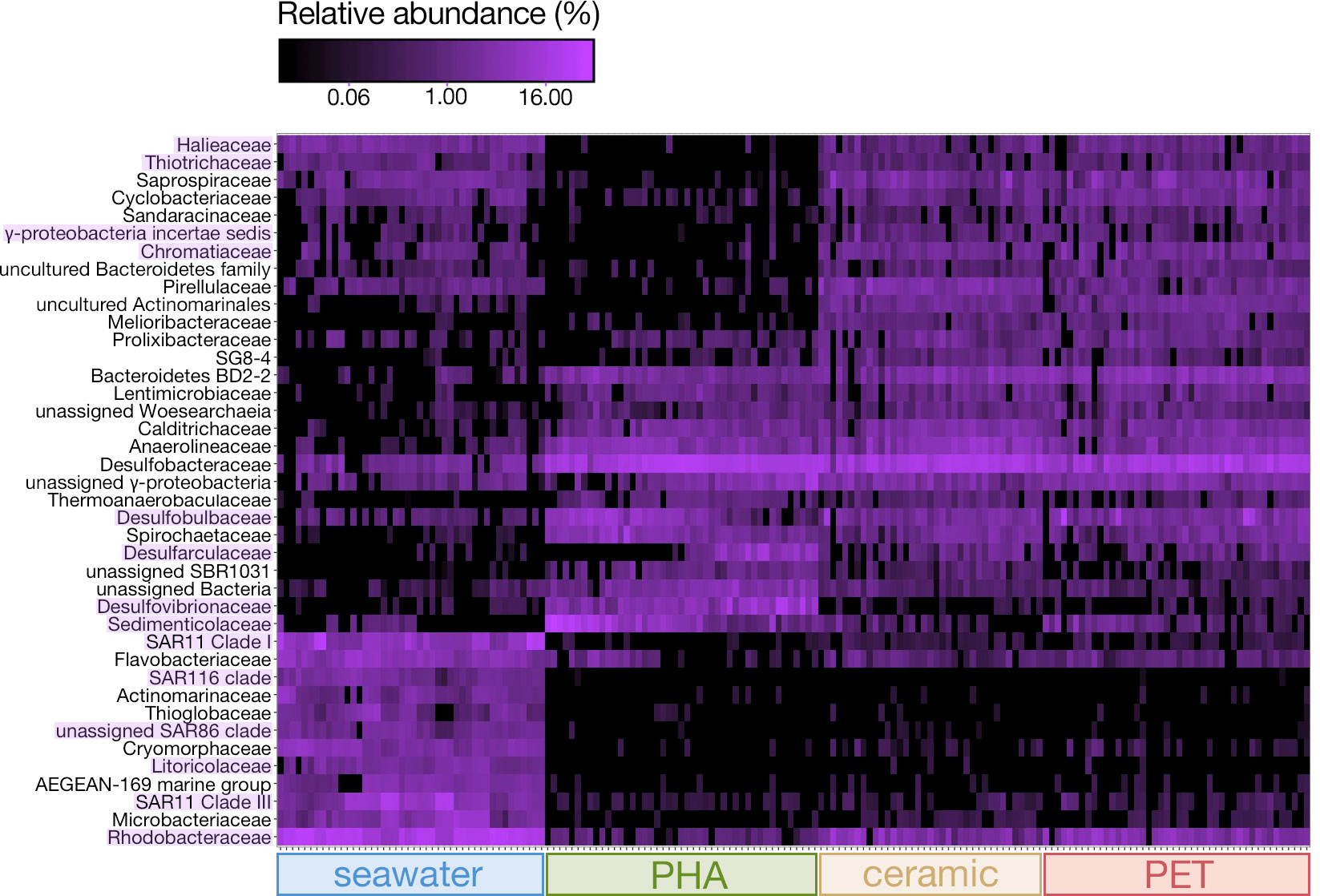
Figure 4 Heatmap based on the relative abundances of the 50 most abundant families across the three substrate types (ceramic, PET, PHA) and seawater and across all timepoints (0 – 424 days exposure). Families of interest with differential abundance between substrates are highlighted in purple.
Temporal changes in SRM
To further explore changes in δ-proteobacteria abundance and diversity, known SRM (Table S1) were filtered from the total microbial community and relative abundances were calculated and compared during each of the five seasons during this study (fall 2016, winter 2016, spring 2017, summer 2017, and fall 2017). Data showed that the overall relative abundance of SRM was higher in PHA biofilms compared to PET and ceramic during all five seasons during the study, and that it was significantly higher during the last four (pairwise Wilcoxon rank-sum; p-value< 0.05; Figure 5). However, the non-significance during the first stage was the result of large variation within PET and ceramic biofilms. In general, SRM abundance within ceramic and PET biofilms increased from 10-15% to 20-25% of the overall microbial community, while SRM abundance within the PHA biofilm increased from 25% to approximately 40%.
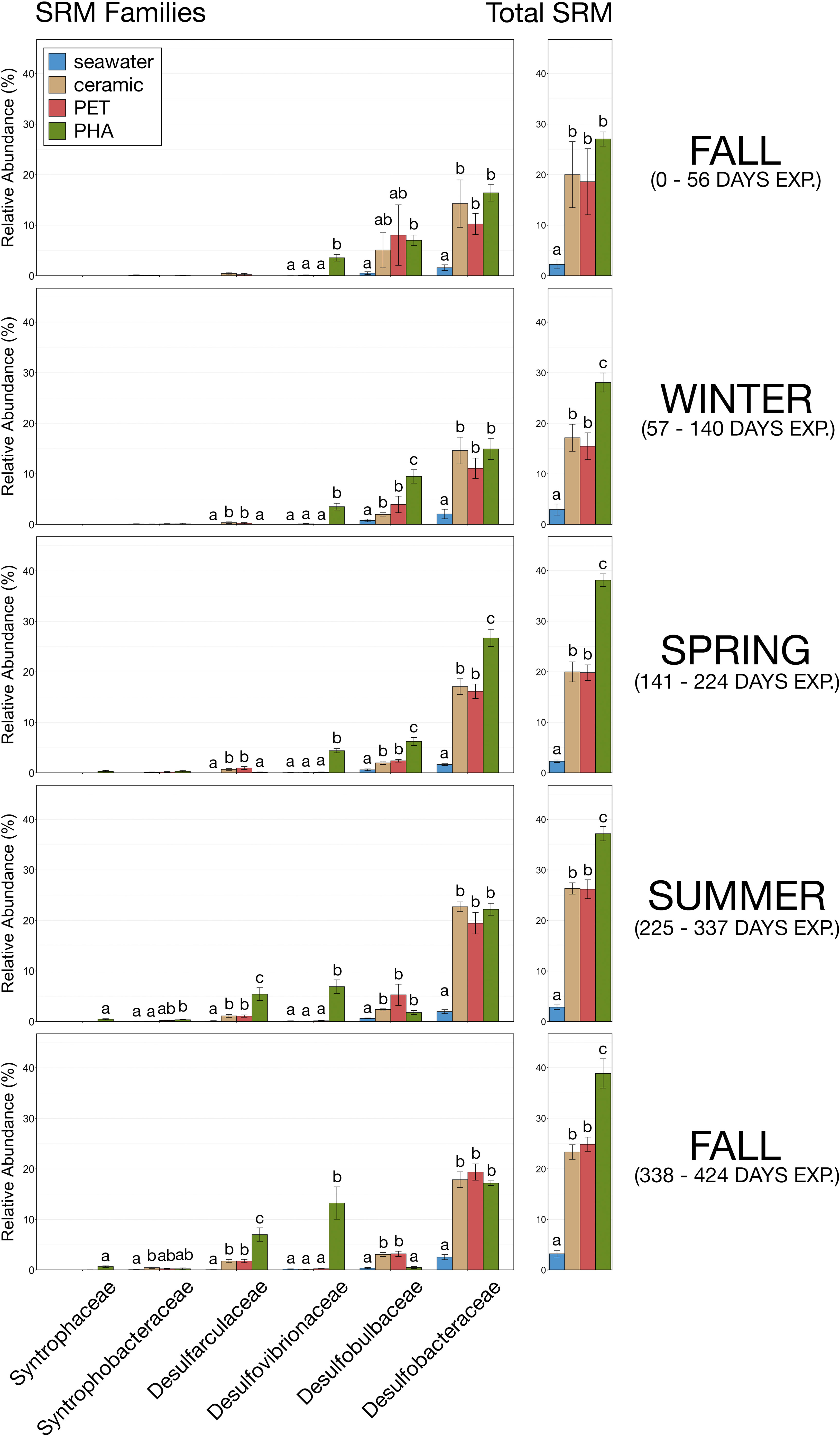
Figure 5 Bar plots demonstrating the relative abundances of the SRM families and total relative abundance of SRM within each type of microbial community (seawater, ceramic, PET, PHA) during each of the five stages: fall 2016 (0 – 56 days exposure), winter 2016 (57– 140 days exposure), spring 2017 (141 – 224 days exposure), summer 2017 (225 – 337 days exposure), and fall 2017 (338 – 424 days exposure). Error bars display standard error of the mean and significant differences in relative abundances are illustrated by different letters (pairwise Wilcoxon rank-sum with Benjamini-Hochberg correction, p-value< 0.05, n=9-12).
In addition to an increased overall abundance of SRM, PHA biofilms contained more SRM families (Figure 5). During the first fall and winter, SRM within PHA biofilms were largely comprised of three families (Desulfobacteraceae ~15-20%, Desulfobulbaceae ~5-10%, and Desulfovibrionaceae ~3-5%) while SRM in ceramic and PET biofilms were only comprised of two families (Desulfobacteraceae ~10-15%, and Desulfobulbaceae ~2-5%). During the spring, summer, and the second fall Desulfarculaceae began to displace Desulfobulbaceae in all three substrate-associated communities, and Desulfovibrionaceae increased substantially in abundance within the PHA biofilm but remained virtually absent in ceramic and PET biofilms (Figure 5). The most mature biofilms in this study were collected during the second fall when SRM within ceramic and PET biofilms were largely dominated by one family (Desulfobacteraceae; ~20%). Concurrently, Desulfovibrionaceae was nearly as predominant as Desulfobacteraceae in PHA biolfilms. Interestingly, Desulfoarculaceae was significantly more abundant in PHA biofilms than ceramic or PET during the second fall, and Syntrophaceae was only present in PHA biofilms (Figure 5; pairwise Wilcoxon rank-sum, n= 6-12, p< 0.05).
Discussion
Plastic waste in terrestrial and aquatic ecosystems is a crisis that has grown with our society's consumption of these materials. Plastics in the environment are generally considered pollutants (Schmaltz et al., 2020), and research is rapidly expanding in this field (Bucci et al., 2020) to determine effects and important nuances of those effects across species, systems, polymers, and environmental conditions. While petroleum sourced plastic production has increased by ~11 million metric tons annually during the late 2010s (Plastics Europe, 2020), there is interest in polymers that are less persistent, like PHA and are therefore more environmentally friendly (Muneer et al., 2020). While environmental persistence is a vital material property with regard to environmental impacts, others, such as influence to microbial community structure and function, must also be considered.
A microbial consortium rapidly colonized PHA nurdles but it degraded slowly. The lifetime estimate of PHA presented here (909 days) is in agreement with rates reported in a recent meta-study of PHA degradation: 1.5-3.5 years for bottles (Dilkes-Hoffman et al., 2019). The linear shape of the mass loss curve is also in agreement with previous studies of PHA degradation in marine environments (Volova et al., 2010; Dilkes-Hoffman et al., 2019). Biofilms clearly formed on both PHA and PET as early as 28 days exposure, but signs of degradation were only seen on bioplastic. Microorganisms form visible biofilms on pelagic plastic debris in as quickly as two weeks (Zhang et al., 2021) and we have observed diverse microbial communities after 28 days (Pinnell and Turner, 2020). While microbes capable of metabolizing plastic have been reported (Gajendiran et al., 2016; Yoshida et al., 2016), mass loss measurements and SEM imaging showed no evidence of PET degradation in this study. A recent study investigating the prevalence of PET hydrolases within existing metagenomic datasets found that they were very rare enzymes (Danso et al., 2018) and it is likely there were not present in the microorganisms found at the study site. Further, microbes capable of degrading plastic may be largely restricted to highly polluted habitats that select for the growth or evolution of plastic-degraders. For example, polyethylene-biodegrading strains of Rhodococcus ruber and Aspergillus clavatus were isolated from soils with a history of polyethylene contamination (Orr et al., 2004; Gajendiran et al., 2016), a PET-biodegrading strain of Ideonella sakaiensis was isolated from a bottle-recycling facility (Yoshida et al., 2016), and PET-biodegrading consortia containing members of Pseudomonas and Bacillus have recently been isolated from petroleum contaminated soil (León-Zayas et al., 2019; Roberts et al., 2020). By contrast, this study was conducted on a remote dredge material island that is not highly polluted with plastic.
The microbial consortium that colonized PHA was distinct from those that colonized PET and ceramic, and that community remained distinct over time. Members of α-proteobacteria and Bacteroidia are typical marine surface colonizers and would be expected among the most common organisms within biofilm communities in marine environments (Salta et al., 2013; Dang and Lovell, 2016). Their decreased abundance within PHA biofilms compared to PET and ceramic biofilms lends further support to previous work demonstrating that PHA recruited a specific microbial consortia atypical of inert surfaces (Pinnell and Turner, 2019). The decreased abundance of typical biofilm formers on PHA was coupled with a significant increase of sulfate reducers. SRM typically comprise between 5-25% of the total bacterial community in estuarine sediments (Purdy et al., 2002; Bowen et al., 2012; Cheung et al., 2018), and our results show that SRM abundance with PET and ceramic biofilms falls within this typical range, while SRM were considerably more abundant (25-40%) within PHA biofilms.
The microbial consortium in PHA biofilms was increasingly dominated by an increasingly diverse SRM community over time. The most common families of SRM within marine and estuarine sediments are Desulfobacteraceae, Desulfobulbacaea, and Desulfovibrionacaea (Purdy et al., 2002). Throughout the first fall, winter, and spring of this experiment, the biofilms associated with all three substrates were all dominated by these three families, albeit in difference abundances. However, starting in the summer, Desulfobulbaceae was increasingly replaced by Desulfoarculaceae and Desulfovibrionaceae in PHA biofilm communities, but not PET or ceramic. Interestingly, Desulfoarculaceae represents a separate lineage within δ-proteobacteria that are distantly related to most other members and are capable of oxidizing long-chain fatty acids (Sun et al., 2010). Desulfoarculaceae is also underrepresented in the literature, with only five described genera and eight representative genomes in NCBI. Isolates from Desulfarculaceae are strictly anaerobic (Kuever, 2014) and capable of degrading a variety of complex carbon sources including pyruvate (Davidova et al., 2016), benzoate (An and Picardal, 2014), sorbate (Schnell et al., 1991), and 3-hydroxybutyrate (Schnell et al., 1991), the latter of which is the building block of common PHAs. This suggests that members of Desulfarulaceae may be utilizing PHA as a carbon source and their enrichment during the later stages of the experiment could have been facilitated by substrate pitting and the creation of anoxic microenvironments. While studies investigating the interaction between SRM and PHA degradation are scarce and largely limited to older culture-based studies, they demonstrated that PHAs can serve as carbon and electron source for sulfur reduction (Urmeneta et al., 1995) and that SRM are capable of causing pitting to the surface of PHA (Çetin, 2009). Desulfarulaceae may have uniquely evolved to utilize this substrate and they may be a promising source of bioplastic-degrading microbes and enzymes to complement efforts to recycle and valorize bioplastic waste (Jiménez et al., 2022).
Here, we confirmed that PHA biodegrades slowly, on the order of two to three years, in marine sediments. Further, PHA biofilms were unique in their microbial community structure, containing increasingly larger proportions of SRM as compared to PET and ceramic. In addition, we recently reported that PHA biofilms accumulate antibiotic and metal resistance genes in marine sediments and we isolated hemolytic multidrug resistant Bacillus cereus group bacteria from those biofilms (Di Cesare et al., 2021). With the predicted increase in bioplastic production over the next few years, it is crucial to understand how PHA loading impacts natural environments. The stimulation of SRM would enhance rates of sulfate reduction and potentially disrupt the balance between sulfate reducers and methanogens. In a scenario where petroleum-based plastic pollution is traded for bioplastic pollution, this stimulation of SRM may have significant and long-lasting effects on carbon and sulfur sediment biogeochemistry.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA551219.
Author contributions
JT and LP participated in and provided oversight and participated in the experimental design, laboratory procedures, report preparation, and securing funding. LP performed the data analysis, and LP, JC, and JT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Texas General Land Office Coastal Management Program (GLO-CMP), the Texas Research and Development Fund (TRDF), Texas Sea Grant (TSG), and the National Sciences and Engineering Research Council of Canada (NSERC). The majority of computational data analysis was performed on TAMU-CC’s high-performance computing cluster, which is funded in part by the National Science Foundation’s CNS MRI Grant (No. 1429518).
Acknowledgments
This study would not have been possible without the support of our friends and colleagues. Specifically, we thank Thomas Merrick for his assistance with data analysis, Robert “Bobby” Duke and the entire Center for Coastal Studies staff for assistance in the field.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.945822/full#supplementary-material
Supplementary Figure 1 | Scanning electron images of PHA and PET pellets with their associated biofilms at approximately 25X magnification. (A–C) show PHA pellets after 0, 202, and 424 days of exposure, respectively. (D–F) show PET pellets after 0, 202, and 424 days of exposure, respectively.
References
Al-Khattaf F. S., Al-Ansari M. M., Maruthamuthu M. K., Dyona L., Agastian P. (2022). Polyhydroxybutyrate degradation by biocatalyst of municipal sludge water and degradation efficacy in sequencing batch biofilm reactor. Environ. Res. 204, 112336. doi: 10.1016/j.envres.2021.112336
An T. T., Picardal F. W. (2014). Desulfocarbo indianensis gen. nov., sp. nov., a benzoate-oxidizing, sulfate-reducing bacterium isolated from water extracted from a coal bed. Int. J. Systematic Evolutionary Microbiol. 64, 2907–2914. doi: 10.1099/ijs.0.064873-0
Arbizu P. M. (2017) pairwiseAdonis: pairwise multilevel comparison using adonis. Available at: https://github.com/pmartinezarbizu/pairwiseAdonis.
Borrelle S. B., Ringma J., Law K. L., Monnahan C. C., Lebreton L., McGivern A., et al. (2020). Predicted growth in plastic waste exceeds efforts to mitigate plastic pollution. Science 369, 1515–1518. doi: 10.1126/science.aba3656
Bowen J. L., Morrison H. G., Hobbie J. E., Sogin M. L. (2012). Salt marsh sediment diversity: a test of the variability of the rare biosphere among environmental replicates. ISME J. 6, 2014–2023. doi: 10.1038/ismej.2012.47
Bucci K., Tulio M., Rochman C. M. (2020). What is known and unknown about the effects of plastic pollution: A meta-analysis and systematic review. Ecol. Appl. 30, e02044. doi: 10.1002/eap.2044
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Çetin D. (2009). Anaerobic biodegradation of poly-3-hydroxybutyrate (PHB) by sulfate reducing bacterium desulfotomaculum sp. Soil Sediment Contam 18, 345–353. doi: 10.1080/15320380902772653
Chen J., Bittinger K., Charlson E. S., Hoffmann C., Lewis J., Wu G. D., et al. (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 28, 2106–2113. doi: 10.1093/bioinformatics/bts342
Cheung M. K., Wong C. K., Chu K. H., Kwan H. S. (2018). Community structure, dynamics and interactions of bacteria, archaea and fungi in subtropical coastal wetland sediments. Sci. Rep. 8, 14397. doi: 10.1038/s41598-018-32529-5
Cho J. Y., Lee Park S., Lee H.-J., Kim S. H., Suh M. J., Ham S., et al. (2021). Polyhydroxyalkanoates (PHAs) degradation by the newly isolated marine bacillus sp. JY14. Chemosphere. 283, 131172. doi: 10.1016/j.chemosphere.2021.131172
Cho J. Y., Park S. L., Kim S. H., Jung H. J., Cho D. H., Kim B. C., et al. (2022). Novel poly(butylene adipate-co-terephthalate)-degrading bacillus sp. JY35 from wastewater sludge and its broad degradation of various bioplastics. Waste Manage 144, 1–10. doi: 10.1016/j.wasman.2022.03.003
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., Leggewie C., et al. (2018). New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 84, e02773–e02717. doi: 10.1128/AEM.02773-17
Davidova I. A., Wawrik B., Callaghan A. V., Duncan K., Marks C. R., Suflita J. M. (2016). Dethiosulfatarculus sandiegensis gen. nov., sp. nov., isolated from a methanogenic paraffin-degrading enrichment culture and emended description of the family desulfarculaceae. Int. J. Systematic Evolutionary Microbiol. 66, 1242–1248. doi: 10.1099/ijsem.0.000864
Di Cesare A., Pinnell L. J., Brambilla D., Elli G., Sabatino R., Sathicq M. B., et al. (2021). Bioplastic accumulates antibiotic and metal resistance genes in coastal marine sediments. Environ. Pollut. 291, 118161. doi: 10.1016/j.envpol.2021.118161
Dilkes-Hoffman L. S., Lant P. A., Laycock B., Pratt S. (2019). The rate of biodegradation of PHA bioplastics in the marine environment: A meta-study. Mar. Pollut. Bull. 142, 15–24. doi: 10.1016/j.marpolbul.2019.03.020
Emadian S. M., Onay T. T., Demirel B. (2017). Biodegradation of bioplastics in natural environments. Waste Manage 59, 526–536. doi: 10.1016/j.wasman.2016.10.006
E.U (2018). Directive of the European parliament and of the council on the reduction of the impact of certain plastic products on the environment (Brussels, Belgium: E Commission).
European Bioplastics (2021). Bioplastics - facts and figures (Berlin, Germany:European Bioplastics e.V.).
Gajendiran A., Krishnamoorthy S., Abraham J. (2016). Microbial degradation of low-density polyethylene (LDPE) by aspergillus clavatus strain JASK1 isolated from landfill soil. 3 Biotech. 6, 52. doi: 10.1007/s13205-016-0394-x
Geyer R., Jambeck J. R., Law K. L. (2017). Production, use, and fate of all plastics ever made. Sci. Adv. 3, e1700782. doi: 10.1126/sciadv.1700782
Jørgensen B. B., Findlay A. J., Pellerin A. (2019). The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10. doi: 10.1111/1462-2920.14721
Jiménez D. J., Öztürk B., Wei R., Bugg T. D., Gomez C. V. A., Galan F. S., et al. (2022). Merging plastics, microbes, and enzymes: Highlights from an international workshop. Appl. Environ. Microbiol. 88, e00721–e00722. doi: 10.1128/aem.00721-22
Kato C., Honma A., Sato S., Okura T., Fukuda R., Nogi Y. (2019). Poly 3-hydroxybutyrate-co-3-hydroxyhexanoate films can be degraded by the deep-sea microbes at high pressure and low temperature conditions. High Pressure Res. 39, 248–257. doi: 10.1080/08957959.2019.1584196
Koller M. (2018). A review on established and emerging fermentation schemes for microbial production of polyhydroxyalkanoate (PHA) biopolyesters. Fermentation 4, 30. doi: 10.3390/fermentation4020030
Kuever J. (2014). “The family desulfarculaceae,” in The prokaryotes (Berlin, Germany: Springer), 41–44.
León-Zayas R., Roberts C., Vague M., Mellies J. L. (2019). Draft genome sequences of five environmental bacterial isolates that degrade polyethylene terephthalate plastic. Microbiol. Res. Announc 8, e00237–e00219. doi: 10.1128/MRA.00237-19
Mas-Castellà J., Urmeneta J., Lafuente R., Navarrete A., Guerrero R. (1995). Biodegradation of poly-β-hydroxyalkanoates in anaerobic sediments. Int. Biodeterioration Biodegradation 35, 155–174. doi: 10.1016/0964-8305(95)00066-E
Muneer F., Rasul I., Azeem F., Siddique M. H., Zubair M., Nadeem H. (2020). Microbial polyhydroxyalkanoates (PHAs): Efficient replacement of synthetic polymers. J. Polym Environ. 28, 2301–2323. doi: 10.1007/s10924-020-01772-1
Nehra K., Jamdagni P., Lathwal P. (2017). “Bioplastics: A sustainable approach toward healthier environment,” in Plant biotechnology: Recent advancements and developments (Singapore: Springer Singapore), 297–314.
Oksanen J., Blanchet F. G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019) Vegan: Community ecology package. r package version 2.5-5 ed. Available at: https://CRAN.R-project.org/package=vegan.
Orr I. G., Hadar Y., Sivan A. (2004). Colonization, biofilm formation and biodegradation of polyethylene by a strain of rhodococcus ruber. Appl. Microbiol. Biotechnol. 65, 97–104. doi: 10.1007/s00253-004-1584-8
Park S. L., Cho J. Y., Kim S. H., Bhatia S. K., Gurav R., Park S.-H., et al. (2021). Isolation of microbulbifer sp. SOL66 with high polyhydroxyalkanoate-degrading activity from the marine environment. Polymers (Basel) 13, 4257. doi: 10.3390/polym13234257
Paulson J. N., Stine O. C., Bravo H. C., Pop M. (2013). Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 10, 1200–1202. doi: 10.1038/nmeth.2658
Pinnell L. J., Turner J. W. (2019). Shotgun metagenomics reveals the benthic microbial community response to plastic and bioplastic in a coastal marine environment. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01252
Pinnell L. J., Turner J. W. (2020). Temporal changes in water temperature and salinity drive the formation of a reversible plastic-specific community. FEMS Microbiol. Ecol. 12, fiaa230. doi: 10.1093/femsec/fiaa230
Plastics Europe (2020). Plastics–the facts 2020. Brussels, Belgium:Association of Plastics Manufacturers.
Purdy K. J., Embley T. M., Nedwell D. B. (2002). The distribution and activity of sulphate reducing bacteria in estuarine and coastal marine sediments. Antonie Van Leeuwenhoek 81, 181–187. doi: 10.1023/A:1020550215012
R Core Team (2017). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Roberts C., Edwards S., Vague M., León-Zayas R., Scheffer H., Chan G., et al. (2020). Environmental consortium containing pseudomonas and bacillus species synergistically degrades polyethylene terephthalate plastic. mSphere 5, e01151–e01120. doi: 10.1128/mSphere.01151-20
Salta M., Wharton J. A., Blache Y., Stokes K. R., Briand J. F. (2013). Marine biofilms on artificial surfaces: structure and dynamics. Environ. Microbiol. 15, 2879–2893. doi: 10.1111/1462-2920.12186
Samrot A. V., Samanvitha S. K., Shobana N., Renitta E. R., Senthilkumar P., Kumar S. S., et al. (2021). The synthesis, characterization and applications of polyhydroxyalkanoates (PHAs) and PHA-based nanoparticles. Polymers (Basel) 13, 3302. doi: 10.3390/polym13193302
Schmaltz E., Melvin E. C., Diana Z., Gunady E. F., Rittschof D., Somarelli J. A., et al. (2020). Plastic pollution solutions: emerging technologies to prevent and collectmarineplastic pollution. Environ. Int. 144, 106067. doi: 10.1016/j.envint.2020.106067
Schnell S., Wondrak C., Wahl G., Schink B. (1991). Anaerobic degradation of sorbic acid by sulfate-reducing and fermenting bacteria: pentanone-2 and isopentanone-2 as byproducts. Biodegradation 2, 33–41. doi: 10.1007/BF00122423
Sun H., Spring S., Lapidus A., Davenport K., Del Rio T. G., Tice H., et al. (2010). Complete genome sequence of desulfarculus baarsii type strain (2st14). Stand Genomic Sci. 3, 276–284. doi: 10.4056/sigs.1243258
Suzuki M., Tachibana Y., Kasuya K.-i. (2021). Biodegradability of poly(3-hydroxyalkanoate) and poly(ϵ-caprolactone) via biological carbon cycles in marine environments. Polymer J. 53, 47–66. doi: 10.1038/s41428-020-00396-5
Tullo A. H. (2019). PHA: A biopolymer whose time has finally come. chemical and engineering news sect. Section|:Start page| (col. column)|. Washington, DC:Chemical and Engineering news
UNEA (2022). Historic day in the campaign to beat plastic pollution: Nations commit to develop a legally binding agreement (Nairobi, Kenya:United Nations Environmental Assembly).
U.N.E.P (2018). Single-use plastics: A roadmap for sustainability (Nairobi, Kenya:United Nations Environment Programme).
Urmeneta J., Mas-Castella J., Guerrero R. (1995). Biodegradation of poly-(beta)-hydroxyalkanoates in a lake sediment sample increases bacterial sulfate reduction. Appl. Environ. Microbiol. 61, 2046–2048. doi: 10.1128/aem.61.5.2046-2048.1995
Volova T. G., Boyandin A. N., Vasiliev A. D., Karpov V. A., Prudnikova S. V., Mishukova O. V., et al. (2010). Biodegradation of polyhydroxyalkanoates (PHAs) in tropical coastal waters and identification of PHA-degrading bacteria. Polym Degradation Stab 95, 2350–2359. doi: 10.1016/j.polymdegradstab.2010.08.023
Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, e00009–e00015. doi: 10.1128/mSystems.00009-15
Yamada M., Yukita A., Hanazumi Y., Yamahata Y., Moriya H., Miyazaki M., et al. (2018). Poly(3-hydroxybutyrate) production using mannitol as a sole carbon source by burkholderia sp. AIU M5M02 isolated from a marine environment. Fish Sci. 84, 405–412. doi: 10.1007/s12562-017-1164-3
Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 351, 1196–1199. doi: 10.1126/science.aad6359
Zhang B., Yang X., Liu L., Chen L., Teng J., Zhu X., et al. (2021). Spatial and seasonal variations in biofilm formation on microplastics in coastal waters. Sci. Total Environ. 770, 145303. doi: 10.1016/j.scitotenv.2021.145303
Keywords: bioplastic, degradation, marine pollution, sulfate reducing microorganisms, marine pollution, microbial succession
Citation: Pinnell LJ, Conkle JL and Turner JW (2022) Microbial succession during the degradation of bioplastic in coastal marine sediment favors sulfate reducing microorganisms. Front. Mar. Sci. 9:945822. doi: 10.3389/fmars.2022.945822
Received: 17 May 2022; Accepted: 21 July 2022;
Published: 17 August 2022.
Edited by:
Bethanie Carney Almroth, University of Gothenburg, SwedenReviewed by:
Diego Javier Jiménez, University of Los Andes, ColombiaRanjit Gurav, Konkuk University, South Korea
Copyright © 2022 Pinnell, Conkle and Turner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lee J. Pinnell, bGpwaW5uZWxsQGN2bS50YW11LmVkdQ==
†Present address: Lee J. Pinnell, Veterinary Education, Research, and Outreach Program, Texas A&M University, Canyon, TX, United States
 Lee J. Pinnell
Lee J. Pinnell Jeremy L. Conkle
Jeremy L. Conkle Jeffrey W. Turner
Jeffrey W. Turner