
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 13 July 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.942074
Intertidal ecosystems are affected by severe nitrogen (N) pollution as a result of anthropogenic activities, and it is unclear how this may affect intertidal microbial communities, which play critical roles in regulating biogeochemical cycles. To address this gap, we conducted a two-month mesocosm experiment using six targeted concentrations of total N. The findings indicated that N entering seawaters has direct negative effects on the bacterial diversity. Dose dependence was found for the effects of N on bacterial diversity in sediment: low N addition increased the bacterial diversity, but a reduction in bacterial diversity occurred when N exceeded a certain value (≥ 3 mg L−1). Additionally, N enrichment caused clear shifts in bacterial community composition with increases in the relative abundance of Balneola (organic-degrading), Phalacroma mitra (carbohydrate-fermenting), and Bacteroides (phosphorus (P)-solubilizing), and decreases in Leptolyngbya_PPC_6406 (N2-fixing). The increased abundance in P-solubilizing and organic-degrading bacteria and decrease in N-fixing bacteria, combined with the upregulated activity of alkaline phosphatase and downregulation of urease activity, implied that the bacterial assemblage tended to be more effective in P and carbon acquisition but reduced N acquisition. Further path analysis suggested that N had direct effects on bacteria and contributed 50%–100% to the variations in bacterial diversity, whereas environmental changes such as dissolved oxygen and pH played minor roles. Overall, bacteria occurring in sediment were likely more stress-resistant to high N exposure than those occurring in seawater, possibly due to the high buffering capacity of sediment and growth tolerances of bacteria in the sediment. These findings point to the vulnerability of microbes in water systems to increasing global N loading, and that N reduction is needed to combat the loss of microbial diversity.
Intertidal zones, located at the interface between terrestrial and marine ecosystems, are pivotal for maintaining coastal habitat heterogeneity, biodiversity, and ecosystem functions and services (Niu et al., 2021). This region is susceptible to human activities and receives increasing amounts of reactive nitrogen (N) through fertilizer runoff and atmospheric deposition (Ma et al., 2021a). For instance, total N deposition into the oceans, particularly in coastal zones that are considered as hotspots for N pollution, has more than tripled from 20 Tg y−1 to 67 Tg y−1 over several decades (Luo et al., 2017). In addition to its well-known effects in augmenting coastal eutrophication (Deegan et al., 2012), widespread hypoxia (Watson, 2016), and ocean acidification (Kessouria et al., 2021), excessive N input may also alter diversity and composition of microbes, which play critical roles in regulating biogeochemical cycles (Zeng et al., 2016; Wang et al., 2018; Wu et al., 2019). However, the current understanding of the diversity, composition, and potential metabolic function of the bacterial community in response to increasing N loading has mainly been limited to terrestrial systems (e.g., croplands, grasslands, and forests) (Contosta et al., 2015; Wang et al., 2018; Dai et al., 2018). For example, N has been generally reported to decrease microbial diversity through a variety of mechanisms, such as altering the N availability, pH, and redox potential of soils (Contosta et al., 2015; Wang et al., 2018). Coastal microorganisms may also be sensitive to N pollution (Bowen et al., 2020). However, studies of N-driven changes in microbial diversity focusing on coastal ecosystems remain limited, restricting our ability to predict the stress resistance and resilience of marine ecosystems to global environmental changes.
Based on homeostatic mechanisms, when N enters aquatic ecosystems, cellular adjustments in acquisition efficiency will lead to altered emergent properties such as enzyme activities, gene regulation, and cellular elemental composition (Sterner and Elser, 2002). These properties have implications for competition and species success, eventually leading to changes in species dominance and biodiversity (Glibert, 2012). Although the effects of N inputs on microbial biomass and associated activities have been sporadically reported in marine ecosystems, the results generated to date remain preliminary, and there is no consensus on the conclusions. Microbial composition and diversity in marine ecosystems have been found to increase (Nogales et al., 2011), decrease (Schwermer et al., 2008; Dong et al., 2017; Niu et al., 2021), or even remain unaffected (Carrino-Kyker et al., 2012) after N addition. For instance, Nogales et al. (2011) found that N enrichment led to a significant increase in diversity, mainly by increasing cell numbers, whereas no notable effect of nitrate pulse (0.2 g kg−1 dry sediment) on microbial community diversity was observed by Carrino-Kyker et al. (2012). In contrast, Dong et al. (2017) reported that reductions in coastal microbial diversity and shifts in community composition began to manifest following nitrate input (1 mg L−1). Niu et al. (2021) proposed that low ammonium input (≤ 2.4 mg kg−1) could increase the diversity of microorganisms, but excess ammonium input possibly reduces microbial diversity in coastal sediment, suggesting a dose-dependent effect of N enrichment on bacterial diversity. Regarding the mechanisms underlying the effects of N on microbial diversity, some studies have suggested that N is the primary factor, whereas others have indicated that changes in environmental factors (e.g., pH or salinity) are more critical (Zeng et al., 2016; Niu et al., 2021).
Overall, the above-mentioned conflicting results may be attributable to: i) the lack of quantitative relationships between N and microbial diversity, and 2) the unclear relative contributions of N and environmental factors to microbial alterations. These results indicate that consistent and general conclusions regarding marine microbial feedback in response to global N loading are still lacking and require further in-depth studies. Xiangshan Bay (XSB) in China is known as the national “large fishing pond” and receive high N pollutant of up to 2692 t y-1, mainly from extensive mariculture and soil erosion. The total N concentration (TN) in XSB was 1.84−48.90 mg L-1 (average of 4.67−7.48 mg L-1) in 2016, nearly ten folds higher than other bays like Jiaozhou Bay (Li et al., 2018; Wang et al., 2022). Thus, it is an ideal area for this study. To better understand how N affects microbial diversity and communities and further determine their quantitative relationships in XSB, we conducted a two-month experiment in 18 aquaria (60 L) with six targeted N levels.
Xiangshan Bay (XSB) is located on the coastline of northern Zhejiang Province of China in the East China Sea. This region is a shallow, long, and semi-enclosed estuarine basin with broad intertidal zones. Tides in XSB are semi-diurnal, with an average tidal range of 3.3 m. To explore the effects of high N on microbial diversity, we built a mesocosm ecosystem using 18 aquaria (60 L) harboring coastal sediment and seawater. Sediment (with total carbon, nitrogen, and phosphorus in 9.0, 0.7, and 0.6 mg g−1 DW) from the surficial 10 cm was collected from XSB (29.3528° N, 121.5454° W) (Figure S1) and subsequently mixed and added to the aquaria to obtain a sediment layer of 10 cm. The remaining 35 cm was filled with well-mixed water from the middle water depth in XSB. Circulating pumps in the tidal mode (Guangzhou Fort Fisherman Co., Ltd., China) were fixed in each aquarium to simulate tide-induced vertical mixing and sediment resuspension. The experimental system set-up was completed on August 7, 2020, then left stationary for a week to balance sediment resuspension and settlement before running for the N addition experiment (August 14 − October 8).
A gradient of six TN concentrations with three replicates was established: control without N addition (N0), 2 mg L−1 (N2), 5 mg L−1 (N5), 10 mg L−1 (N10), 15 mg L−1 (N15), and 20 mg L−1 (N20) (N added in the form of urea). N0 refers to the background TN concentration; N2, TN concentration of 2 mg L-1, refers to water quality Class V as defined in the environmental quality standards for surface water in China (GB 3838-2002); N5, total ammonium concentration of 5 mg L-1, refers to water quality Class II in the standard for groundwater quality in China (GBT-14848-2017); N10, total nitrate concentration of 10 mg L-1, refers to the China standards for drinking water quality (SAC, 2006) as well as the USA federal maximum level for drinking water (Camargo et al., 2005); N15, TN concentration of 15 mg L-1, refers to Primary A in the discharge standard of pollutants for municipal wastewater treatment plants in China (GB-18918-2002); N20, TN concentration of 20 mg L-1, refers to Primary B in the discharge standard of pollutants for municipal wastewater treatment plants in China (GB-18918-2002). Urea (the most common N fertilizer used in aquaculture) was dissolved in seawater before being introduced into the experimental aquaria (at weekly intervals). Total dosage of 0 g, 0.3 g, 0.8 g, 1.5 g, 2.5 g, and 3 g urea fertilizer were applied in N0, N2, N5, N10, N15, and N20, respectively.
Physicochemical parameters of water, including dissolved oxygen (DO) and pH, were measured twice before and after N addition using a Horiba multiparameter instrument (U-52, Japan). Water samples (1.5 L) were collected from each aquarium on August 14 and October 8 using a syringe. Of the samples, 0.5 L was used for bacterial alkaline phosphatase activity (APA) (Ma et al., 2018) and chemical parameters such as TN, ammonium (NH4+), nitrate (NO3-), total phosphorus (TP), and total organic carbon (TOC) analyses according to standard methods (AQSIQ, 2007). The remaining water (ca. 1 L) was filtered through a 0.22-μm cellulose acetate membrane for DNA extraction. Simultaneously, sediment pore water samples in each aquarium were collected with a soil moisture sampler (SMS rhizons, Netherlands) and subsequently filtered (0.45 μm) for TN (TNSed), NH4+ (NH4+Sed), and NO3− (NO3−Sed) analysis. Labile phosphorus (Labile-P) in the sediment was measured using diffusive gradients in thin films (Ding et al., 2015). The pH of the top sediment (pHSed) was measured using a soil pH meter (pH 400 and 600, USA). Sediment samples collected with a core sampler (XDB0204, New Landmark, China) were mixed and partitioned into three subsamples; one was stored at −80°C for sediment DNA extractions, the second was stored at 4°C for enzyme analyses within 24 h, including urease activity (UASed) (Mobley et al., 1995) and β-glucosidase activity (GASed) (Dick et al., 2013), and the third was partially air-dried and passed through a 2-mm sieve for total organic carbon (TOCSed) analyses (Lu, 1999).
DNA in the water and sediment was extracted using a FastDNA spin kit (Q-BIOgene, Carlsbad, CA, USA). The genomic DNA concentrations and purity were measured using an Eppendorf Biophotometer Plus (Eppendorf, Germany). Bacterial community composition was assessed by sequencing the V3-V4 region of the 16S rRNA gene using the PCR primers 338F (5’- ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). High-throughput sequencing was performed on an Illumina MiSeq platform (BioMarker Technologies Co. Ltd., China). Trimmomatic (version 0.33) was used to remove low-quality sequences and Cutadapt (version 1.9.1) was used to cut primer sequences (Martin, 2011; Bolger et al., 2014). High-quality sequences were obtained by merging two-paired reads and cleaning the chimeras using USEARCH (version 10) and UCHIME (version 8.1) (Edgar et al., 2011; Edgar, 2013). Operational taxonomic units (OTUs) were defined by clustering qualified sequences at a 97% identity threshold using USEARCH (version 10) (Edgar, 2013). The sequences were taxonomically identified using a BLASTn search of a curated NCBI database. The raw sequences were submitted in the NCBI sequence reading Archive (SRA) with the registration number of PRJNA826550.
QIIME2 (2020.6) was used to annotate species and calculate alpha diversity indices, including the Chao and Shannon indices (Bolyen et al., 2019). One-way analysis of variance (ANOVA) was used to test the differences between the treatments. Redundancy analysis (RDA) was performed using the vegan (v2.3) R package (Dixon, 2003). Bacterial interaction network analysis based on Spearman rank correlation coefficient was conducted using Cytoscape (V3.9.1) with a connection suggesting a strong (Spearman’s |r|>0.4) and significant correlation (p<0.05 after FDR correction). Path analysis was applied to further explore the key factors and paths influencing the N regulation of microbial diversity using AMOS software (IBM SPSS AMOS 26). The best-fit model was developed using maximum likelihood estimation, and the chi-square test (χ2) (p > 0.05) and comparative fit index (CFI) (>0.90) were used to test the overall goodness of the model fit. When conducting the path analyses, multicollinearity among environmental variables was diagnosed, and no multicollinearity was found (variance inflation factor, VIF < 5) (Ma et al., 2018; Ma et al., 2021b).
During the two-month N addition (Table 1), TN in the water showed, as expected, a significant concentration gradient, being significantly higher in N5, N10, N15, and N20 than in N0, whereas no significant difference was found between N2 and N0. There were no differences in the N treatments for TP, TOC, and pH compared with N0. DO was significantly lower in N5, N10, N20 than that in N0, whereas no differences were discerned between N0, N2, and N15. TNSed in sediment pore water was significantly higher in N15 and N20 than in N0, whereas no differences were observed between N0, N2, N5, and N10. No statistical differences were observed among treatments for labile P and TOCSed. The pHSed was significantly lower in the N treatments than in N0. As for the enzyme activities associated with carbon (C), N, and phosphorus (P) cycles, no significant differences in GASed were observed among the treatments, although N10 and N15 tended to be lower than N0. UASed was significantly lower in the high-N treatments (N15 and N20) than in N0, whereas no differences were found among the low-N treatments (N0, N2, N5, and N10). The APA of bacteria was significantly higher in N20 than in N0, whereas no differences were recorded among N15, N10, N5, N2, and N0.
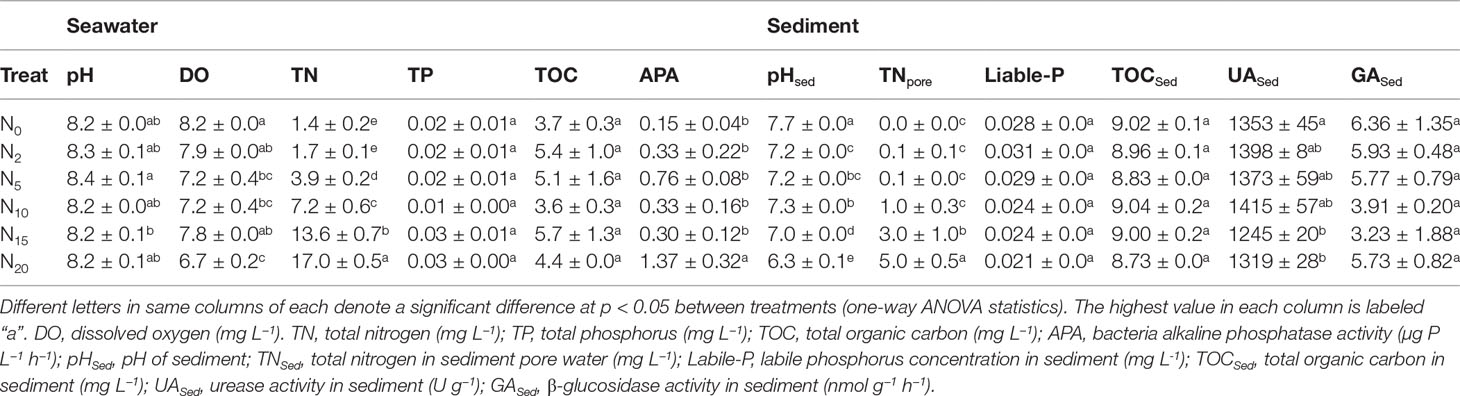
Table 1 Mean (± standard error) measures for seawater and sediment variables across the nitrogen gradient.
We selected the Chao and Shannon indices as our metrics for microbial richness and diversity because they are highly recommended for analyzing microbial α-diversity (Wang et al., 2018). As presented in Table 2, the microbial richness and diversity in both water and sediment were remarkably different among the N treatments. For water, Chao significantly decreased from 1277 in N0 to 678 in N20 and Shannon decreased from 5.0 in N0 to 4.5 in N20 with increasing N concentrations. In sediment, the Chao values were significantly higher in N2 and N15 than in N0, whereas no differences were observed among N5, N10, N20 and N0. For the Shannon index, in addition to N5 which was not different from N0, the other N treatments were significantly higher than that of N0.
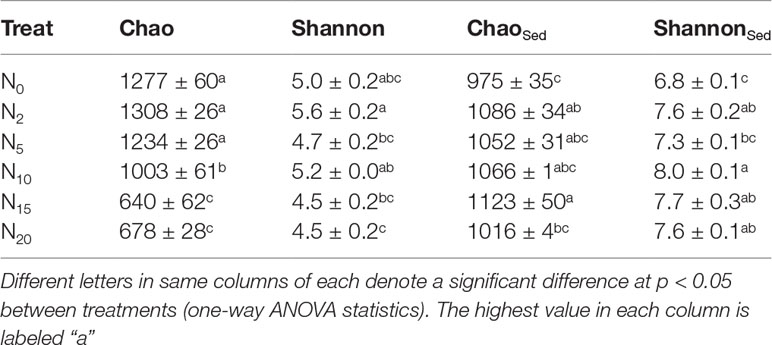
Table 2 Mean (± standard error) measures for microbial richness (Chao) and diversity (Shannon) in water and sediment across the nitrogen gradient.
Further regression analysis demonstrated that the Chao and Shannon indices in water were negatively correlated with TN, suggesting that N addition decreased the α-diversity of bacteria in water (Figures 1A, B). The abundance of bacteria in water decreased with N concentration (Figure S2). Chao and Shannon indices in sediment increased with N addition at low dosages, but opposite trends occurred when N exceeded a certain value (Figures 1C, D). Similar pattern has been found for the abundance of bacteria in sediment (Figure S2).
At the class level, the top ten abundant bacteria in the water belonged to six phyla ranked as follows: Alphaproteobacteria (28%–35%), Oxyphotobacteria (9%–34%), Bacteroidia (5%–23%), Gammaproteobacteria (9%–16%), Acidimicrobiia (4%–15%), Actinobacteria (3%–11%), Rhodothermia (2%–15%), Deltaproteobacteria (0%–9%), Clostridia (0%–1%), and Anaerolineae (0%–1%) (Figure S3A). The most dominant bacteria classes in the sediment belonging to eight phyla were Gammaproteobacteria (13%–28%), Bacteroidia (12%–16%), Actinobacteria (7%–15%), Clostridia (5%–21%), Alphaproteobacteria (7%–12%), Bacilli (3%–5%), Oxyphotobacteria (2%–5%), Deltaproteobacteria (2%–4%), Anaerolineae (1%–5%), and Acidobacteriia (1%–3%) (Figure S3B).
At the genus level, besides a large proportion of unculturable bacteria (17%–28%), the most dominant bacteria in the water were Clade_Ia (2%–15%), Balneola (2%–14%), Phalacroma mitra (0%–18%), Cyanobium_PCC-6307 (0%–14%), Leptolyngbya_PPC_6406 (0%–15%), and Marivita (1%–5%) (Figure 2A). In the sediment, Candidatus sulcia (4%–13%), Candidatus vidania (3%–13%), Bacteroides (1%–4%), Woeseia (0–4%), Faecalibacterium (1%–4%), and Lactobacillus (1%–2%) were the most common bacteria (Figure 2B).
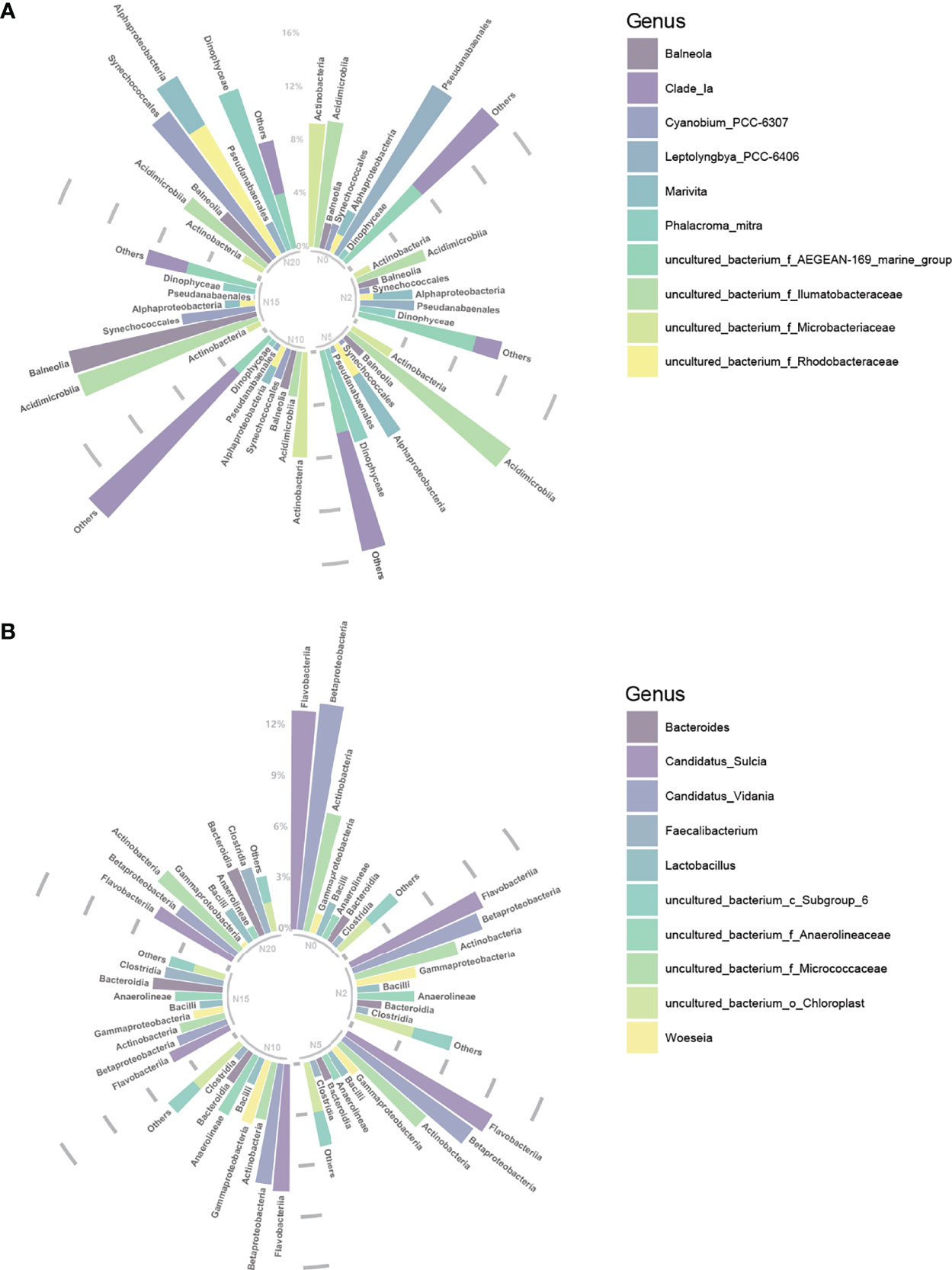
Figure 2 Circular stacked bar-plots demonstrating the top ten abundant bacteria genera in water (A) and sediment (B) under all treatments. Different genera are displayed in different colors.
The bacteria with stress-tolerant phenotypes at the genus level are shown in Figure S4. The relative abundance of stress-tolerant bacteria in water decreased with N loading, decreasing from 20% in N0 to 15% in N20. In contrast, stress-tolerant sediment bacteria showed an opposite trend, increasing from 23% in N0 to 39% in N20.Correlation-based network analyses at the bacterial genus level are shown in Figure 3. In seawater, the total number of links for the bacteria-bacteria network was 20, of which 11 links were positive (indicating co-occurrence for taxa) and 9 links were negative (indicating co-exclusion) (Figure 3A). No notable changes have been found in the correlation links along the gradient of N in water (Figure S5). The total number of links in the sediment was 22, of which 10 were positive and 12 were negative. Bacteria belonging to the Bacteroidetes and Proteobacteria phyla displayed the highest number of interactions (Figure 3B). The sum of links increased along with N concentration, with 16 links in low-N treatments while 25 links in high-N treatments (Figure S5). When considering all correlations, the links between bacteria were more complex in sediment than in water, indicating that potential interactions and stability were stronger in bacterial sediment networks (Chen and Wen, 2021).Network analysis between N and bacteria indicated that N correlated positively with the genera Balneola and Phalacroma mitra but negatively with Leptolyngbya_PCC-6406 in water (Figure 3A). N correlated positively with the genera Bacteroides and Faecalibacterium, but negatively with Candidatus sulcia and Candidatus vidania in the sediment (Figure 3B).
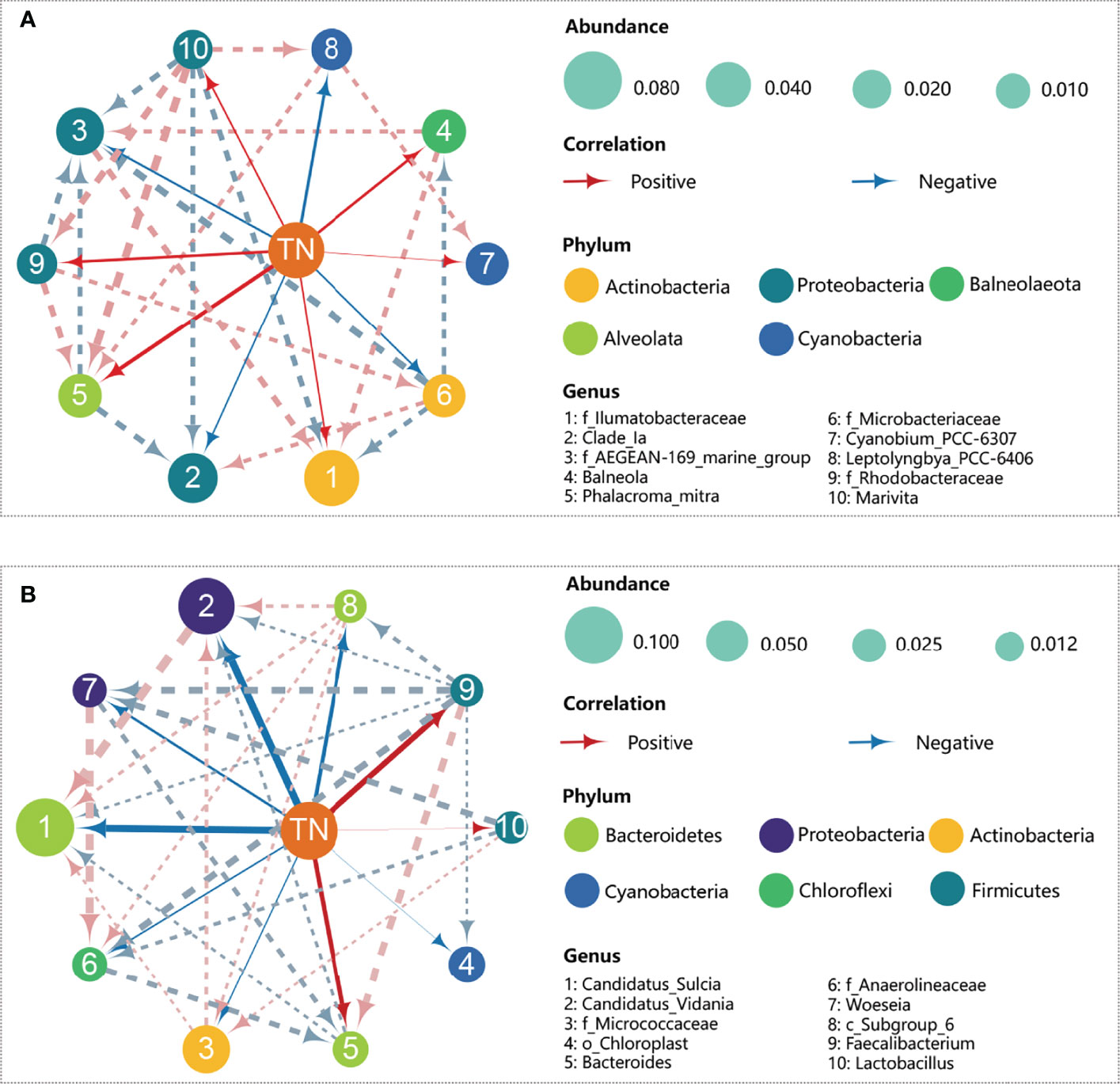
Figure 3 Bacterial interaction networks in water (A) and sediment (B) based on Spearman rank correlation analysis. Top 10 bacterial genera with high absolute abundances were selected for correlation analysis. A connection implies a strong (Spearman’s |r|>0.4) and significant (p<0.05 after FDR correction). The size of each node represents the degree of abundance, the thickness of each line represents the degree of correlation, and the color of each node represents the specified phylum. The dotted and solid lines indicate the bacteria-bacteria and nitrogen-bacteria interactions, respectively. The red and blue lines indicate positive and negative relationships, respectively.
RDA was conducted to explore the distribution patterns of the microbial communities and to further assess the relationships between environmental factors and microbial communities (Figure 4). In seawater, 27.0% of the total variance was explained by the first two constrained axes of the RDA: the first axis explained 19.0%, and the second explained 8.0%. TN, NH4+, and NO3− were the main factors responsible for the distinct structures of the bacterial communities. As for sediment, 20.5% of the total variance was explained by the first two constrained axes of the RDA: the first axis explained 13.1% and the second explained 7.4%. TN, NH4+, NO3−, and pH were significant factors affecting the microbial communities.
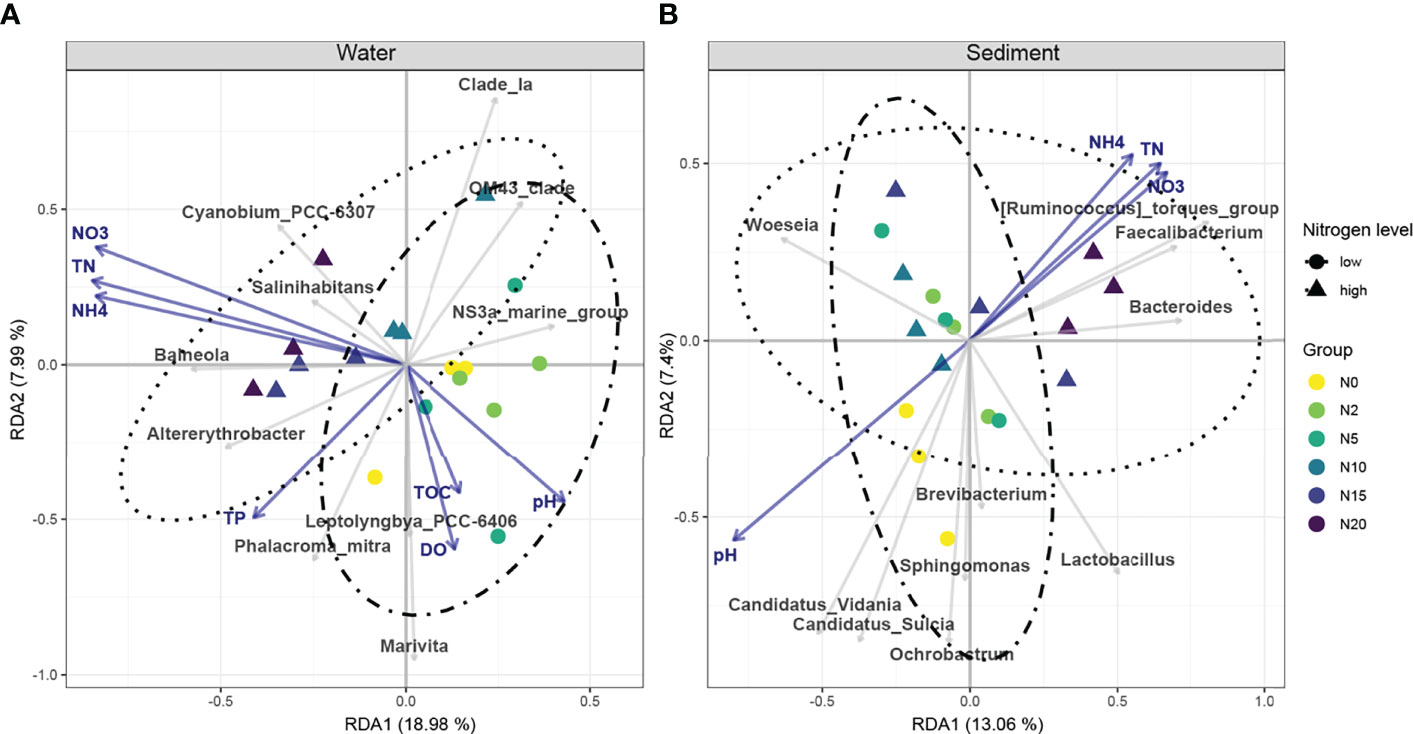
Figure 4 Redundancy analysis (RDA) demonstrating the relative influence of environmental factors on the bacterial communities in water (A) and sediment (B). Only those factors identified to significantly influence bacterial communities are shown in the RDA. Samples were grouped according to nitrogen level. Different groups are displayed in different colors and shapes. Similarity values among the sample groups are represented by the distances between them in each graph. Blue arrows indicate the environmental factors; gray arrows indicate the bacterial genera.
As shown in Figure 5, the models in seawater and sediment were acceptable according to χ2 and CFI. In seawater, the significant path coefficient from TN to diversity was negative, demonstrating the direct negative effect of N input on bacterial diversity. The significant path coefficients from TN to DO and DO to diversity were negative, suggesting a positive effect of N on bacterial diversity through decreasing DO. Overall, 93% of the variation in bacterial diversity was accounted for when TN, DO, and pH were included in the model. In the sediment, dose dependence was observed for the effects of N on bacterial diversity. Low N had positive effects on bacterial diversity, as suggested by the significantly negative coefficients from TN to NO3−Sed and from NO3−Sed to diversity. 73% of the variation in bacterial diversity was accounted for when NO3−Sed and pHSed were included in the model. In contrast, high N had negative effects on bacterial diversity by increasing the NO3−Sed. Overall, 62% of the variation in bacterial diversity was accounted for when NO3−Sed, pHSed, and TOCSed were included in the model.
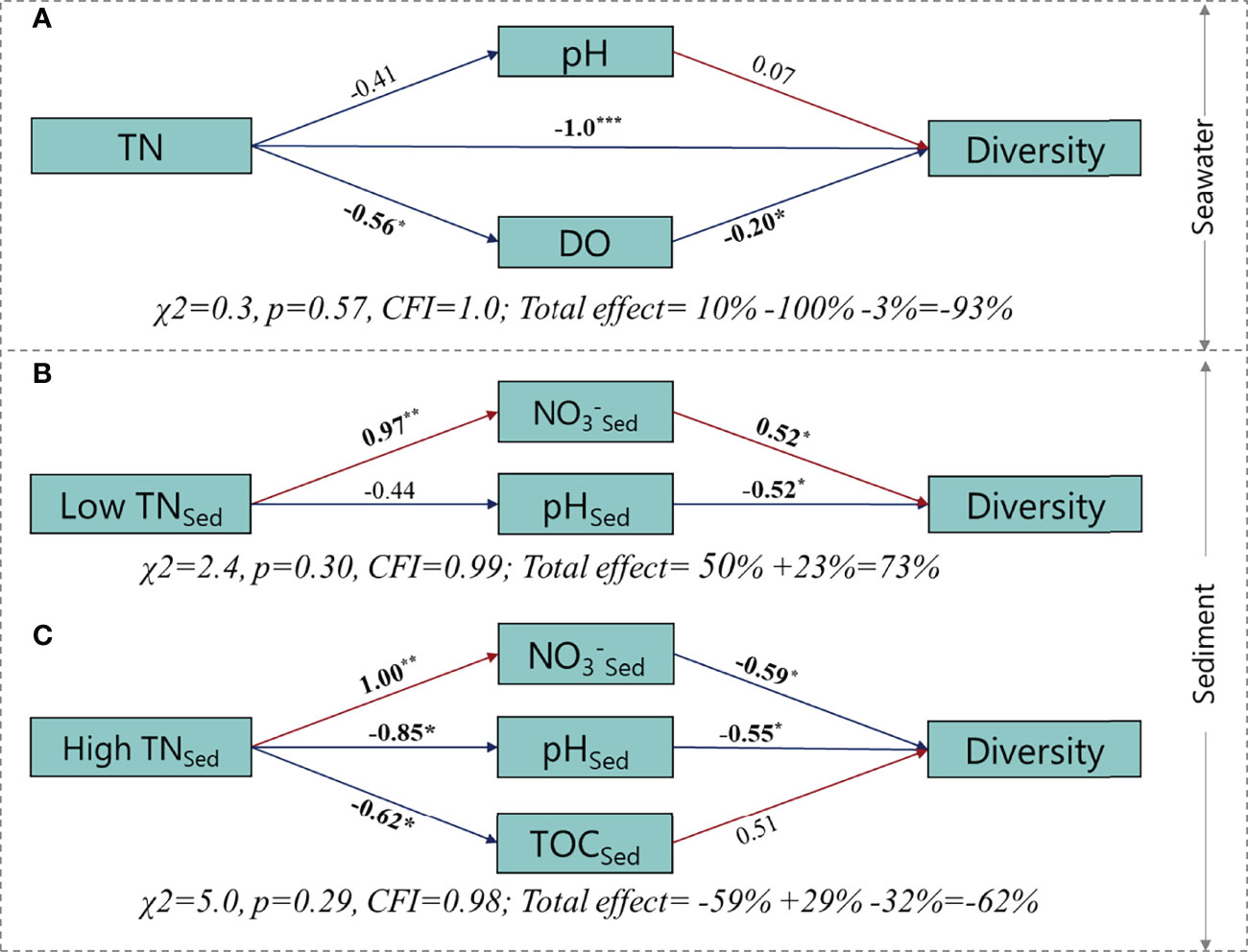
Figure 5 Structural equation modeling depicting the changes of microbial diversity caused by nitrogen loading. Single-headed arrows represent unidirectional causal relationships. The numbers adjacent to the arrows are standardized path coefficients (significant correlations are shown in bold, *p < 0.05, **p< 0.01). The red and blue lines represent significant positive and negative pathways, respectively.
N enrichment could directly decrease microbial diversity (indicated by Shannon and Chao index values) in water, consistent with most previous studies showing the N-induced decline in microbial diversity (Aoyagi et al., 2015; Luo et al., 2017; Craig et al., 2021; Niu et al., 2021), possibly due to the high osmotic potential and ion toxicity (Omar and Ismail, 1999). Bacteria in the sediment responded in a different manner: low N addition increased the bacterial diversity, but the diversity decreased when N exceeded a certain value (≥ 3 mg L−1). Further path analysis demonstrated that NO3−Sed (as the end-product of urea, nearly 65% of total N) was the dominant factor influencing bacterial diversity. Low NO3−Sed concentrations had a positive effect on bacterial diversity, likely by providing electron acceptors for bacterial growth during organic matter decomposition, whereas high NO3−Sed concentrations had an opposite effect. Similar patterns have been reported by Niu et al. (2021), in which positive and negative regulation of ammonium on microbial diversity occurred at low and high N concentrations, respectively. Regardless of the form of N applied, the results consistently suggested that the effects of N on sediment bacterial diversity mainly depended on the amount of N added. Such dose-dependent effects—stimulation of bacterial diversity at low N but decrease at high N—might be explained, to some degree, by the C acquisition for bacterial growth that can be stimulated by low NO3− but suppressed at high levels (Wang et al., 2018). This was also corroborated by the decrease in GASed (C acquisition) in the high N treatments in this study.
It is possible that bacteria occurring in water were more sensitive to N enrichment than those in sediment, pointing to the vulnerability of microbes in water systems to increasing global N loading. The sensitivity and vulnerability of bacteria in water systems might be partly explained by the high N concentration in water, in which microbes that are less tolerant to high osmotic potential do not survive, resulting in a sharp decline in microbial biodiversity (Wang et al., 2018). In contrast, the less pronounced response of sediment bacteria to N might be attributed to: i) the stronger resistance of the bacterial community to stresses, as indicated by their more complex interaction networks; ii) the broader growth tolerances of bacteria, as indicated by the high abundance of stress-tolerant bacteria; and iii) sediment harboring higher buffering capacity for reducing the surrounding N concentration through ion adsorption.
Different groups of microorganisms can differ in their ability to process various nutrients (Craig et al., 2021). Urea, potentially serving as both a C and an N source, plays a vital role in driving the changes of bacterial abundance and community composition, likely through altering microbial biomass C and N (Ghosh and leff, 2013; Ma et al., 2017). Moreover, community composition and functional response are highly correlated. In this study, N addition increased the relative abundance of Balneola and Phalacroma mitra in the water. One of the Balneola harboring high environmental tolerances is associated with organic degradation (Kong et al., 2019), whereas the other Phalacroma mitra is a type of acid-producing (okadaic acid) bacteria related to carbohydrate fermentation (Uchida et al., 2018). Cyanobium_PCC-6307 and Leptolyngbya_PPC_6406, as the members of Cyanobacteria, responded differently to N enrichment. N increased the abundance of Cyanobium_PCC-6307, related to N and C fixation (Wei et al., 2022), while decreased the abundance of Leptolyngbya_PPC_6406, a diazotroph adept at N2-fixing (Berman-Frank et al., 2003). Similar patterns have been reported by Berthrong et al. (2014) and Wang et al. (2018), in which N input may reduce the competitive ability of microbes adept at N2-fixing.
In the sediment, N addition decreased the relative abundance of Candidatus _Sulcia and Candidatus_Vidania. Those two bacteria are obligately intracellular and strict aerobic organisms, mainly devoted to essential amino acid synthesis (Bennett and Mao, 2018). In contrast, N addition increased the relative abundance of Faecalibacterium and Bacteroides. Both Faecalibacterium and Bacteroides are associated with acid production (Yang et al., 2020; Ma et al., 2021a), consistent with the significant drop in sediment pH. Therefore, the low pH of the sediment might not only be a cause but also a result of N-induced changes in bacterial composition. Additionally, Bacteroides was identified as a P-solubilizing bacterium, which may increase the availability of microbial P and mitigate P limitation triggered by an imbalance in the N:P ratio (Ma et al., 2021a). Overall, the increase in P-solubilizing and organic degrading bacteria and decrease in N-fixing bacteria following N addition implied that the bacterial assemblage tended to utilize P and C more effectively but reduced N acquisition, which was in line with the niche-based and stoichiometric theory (Sterner and Elser, 2002; Stegen et al., 2012). Shifts in community composition can be accompanied by changes in the hydrolytic ectoenzyme activity (Luo et al., 2017). In principle, N-acquiring enzyme activity might decrease due to the availability of N, whereas C- and P-acquiring enzyme activities should relatively increase with N addition (Luo et al., 2017). However, this was not always the case in this study; a significant increase in APA (P-acquisition) and a minor decrease in UASed (N-acquisition) were observed in the high N treatments, whereas GASed (C acquisition) exhibited a slight reduction in the high N treatments.
N enrichment has been reported to affect bacterial diversity directly by altering N availability and indirectly by changing environmental properties, such as salinity, oxygen conditions, and pH (Luo et al., 2017; Wang et al., 2018; Niu et al., 2021). However, there is currently no consensus on the most dominant mechanism of N in the regulation of bacterial diversity. For instance, Wessén et al. (2010) reported that alterations in bacterial abundance caused by N fertilization were mainly driven by pH. Inconsistent findings reported by Ramirez et al. (2010) suggested that bacterial diversity response to N input primarily resulted from direct effects of elevating N concentration, rather than indirect effects such as pH. Given this, we examined the contribution of each factor to changes in bacterial diversity through path analysis. The results indicated that N was the primary factor affecting bacterial diversity and contributed 50%–100% to the variations in bacterial diversity, whereas environmental changes in DO, sediment pH, and DOC played minor roles, accounting for 3%–32%. We emphasize the need to reduce N loading to enhance the pace and level of recovery of aquatic biodiversity and their resilience to future changes. Furthermore, seasonal variations may be another key driver of the changes in bacteria communities and diversity because the abovementioned factors (e.g., pH and N concentration) were significantly different with seasons (Guo et al., 2021). For example, the N-increased abundance of bacteria was more notable in summer than in other seasons (Ma et al., 2017). Likewise, the relative abundances of some genera, such as Cyanobacterium, increased significantly during the summer season, which may fuel algal blooms (Floreza et al., 2019). Given the gap between artificial and natural coastal ecosystems (such as the different hydrodynamic conditions), our mesocosm study may not completely catch the process of N affecting the diversity and composition of microbes. Thus, further in situ investigations at larger spatial scales are needed.
Overall, bacteria occurring in water and sediment responded differently to N enrichment. N showed direct adverse effects on water bacterial diversity, whereas we discerned dose-dependent effects in sediment: low N addition increased the bacterial diversity, but a reduction in bacterial diversity occurred when N exceeded a certain value (≥ 3 mg L−1). Bacteria in the sediment were more stable and stress-resistant than those in the water to N enrichment, likely due to their stronger stability and growth tolerance. Furthermore, the high buffering capacity of sediment may be another contributor. Bacterial assemblages following N addition tended to utilize P and C more effectively but reduced N acquisition, supported by the following patterns: i) N-driven shifts in bacterial composition with increases in the relative abundance of organic degrading bacteria (Balneola) and P-solubilizing bacteria (Bacteroides) and decreases in N2-fixing bacteria (Leptolyngbya_PPC_6406), and ii) increases in bacterial APA and decrease in UASed. Further path analysis indicated that N was the dominant mechanism driving the decrease in bacterial diversity and composition shift, whereas environmental changes in DO, pH, and DOC played minor roles. The findings of this study indicate that N loading has potential implications for microbial diversity, community composition, and nutrient (C, N, and P) cycling in marine environments, warranting further long-term investigation at larger spatial scales.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA826550.
Y-FX, methodology, software Priya Singh, and data curation. X-MD, investigation and formal analysis. CL, investigation and resources. S-NM, conceptualization, writing- original draft preparation, writing- reviewing and editing, and supervision. J-LX, supervision, conception, revision. Y-DC, revision, project administration. All authors contributed to the article and approved the submitted version.
The research was supported by the National Natural Science Foundation of China (Grant No. 42107399), Zhejiang Basic Public Welfare Research Program (Grant No. LQ21C030005) and Ningbo Public Welfare Science, Technology Program (Grant No. 2021S060).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Elsevier’s WebShop (https://webshop.elsevier.com/) and Wuhan Science and Technology Plan Project (2020020602012152) for valuable editing of the paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.942074/full#supplementary-material
Aoyagi T., Kimura M., Yamada N., Navarro R.R., Itoh H., Ogata A.(2015). Dynamic Transition of Chemolithotrophic Sulfur-Oxidizing Bacteria in Response to Amendment With Nitrate in Deposited Marine Sediments. Front. Microbiol. 6. 426. doi: 10.3389/fmicb.2015.00426
AQSIQ, P.R.C (2007). The Specification for Marine Monitoring of China-Part 4: Seawater Analysis (GB 17378.4–2007) (General Administration of Quality Supervision, Inspection and Quarantine (Aqsiq) of the People’s Republic of China (in Chinese). http://openstd.samr.gov.cn/bzgk/gb/newGbInfo?hcno=9FB14D0EE23D77A96D54A9BDAAF6EA07
Bennett G. M., Mao M. (2018). Comparative Genomics of a Quadripartite Symbiosis in a Planthopper Host Reveals the Origins and Rearranged Nutritional Responsibilities of Anciently Diverged Bacterial Lineages. Environ. Microbiol. 20 (12), 4461–4472. doi: 10.1111/1462-2920.14367
Berman-Frank I., Lundgren P., Falkowski P. (2003). Nitrogen Fixation and Photosynthetic Oxygen Evolution in Cyanobacteria. Res. Microbiol. 154 (3), 157–164. doi: 10.1016/S0923-2508(03)00029-9
Berthrong S.T., Yeager C.M., Gallegos-Graves L., Steven B., Eichorst S.A., Jackson R.B. (2014). Nitrogen Fertilization has a Stronger Effect on Soil Nitrogen-Fixing Bacterial Communities Than Elevated Atmospheric Co2. Appl. Environ. Microb. 80, 3103–3112. doi: 10.1128/AEM.04034-13
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen E, Rideout J.R., Dillon M.R., Bokulich N., Abnet C.C., Al-Ghalith G.A., et al (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37, 1091–1091. doi: 10.1038/s41587-019-0209-9
Bowen J. L., Giblin A. E., Murphy A. E., Bulseco A.N., Deegan L.A., Johnson D.S. (2020). Not All Nitrogen is Created Equal: Differential Effects of Nitrate and Ammonium Enrichment in Coastal Wetlands. Bioscience 70, 1108–1119. doi: 10.1093/biosci/biaa140
Camargo J. A., Alonso Á., Salamanca A. (2005). Nitrate Toxicity to Aquatic Animals: A Review With New Data for Freshwater Invertebrates. Chemosphere 58, 1255–1267. doi: 10.1016/j.chemosphere.2004.10.044
Carrino-Kyker S. R., Smemo K. A., Burke D. J. (2012). The Effects of pH Change and NO3– Pulse on Microbial Community Structure and Function: A Vernal Pool Microcosm Study. FEMS Microbiol. Ecol. 81, 660–672. doi: 10.1111/j.1574-6941.2012.01397.x
Chen W., Wen D. (2021). Archaeal and Bacterial Communities Assembly and Co-Occurrence Networks in Subtropical Mangrove Sediments Underspartina Alterniflorainvasion. Environ. Microbiome. 16 (1), 10. doi: 10.1186/s40793-021-00377-y
Contosta A. R., Frey S. D., Cooper A. B. (2015). Soil Microbial Communities Vary as Much Over Time as With Chronic Warming and Nitrogen Addition. Soil Biol. Biochem. 88, 19–24. doi: 10.1016/j.soilbio.2015.04.013
Craig H., Antwis R. E., Cordero I., Ashworth D., Robinson C.H., Osborne T.Z., et al. (2021). Nitrogen Addition Alters Composition, Diversity, and Functioning of Microbial Communities in Mangrove Soils: An Incubation Experiment. Soil Biol. Biochem. 153, 108076. doi: 10.1016/j.soilbio.2020.108076
Dai Z. M., Su W. Q., Chen H. H., Barberan A., Zhao H.C., Yu M.J., et al. (2018). Long-Term Nitrogen Fertilization Decreases Bacterial Diversity and Favors the Growth of Actinobacteria and Proteobacteria in Agro-Ecosystems Across the Globe. Global Change Biol. 24 (8), 3452–3461. doi: 10.1111/gcb.14163
Deegan L. A., Johnson D. S., Warren R. S., Peterson B.J., Fleeger J.W., Fagherazzi S., et al. (2012). Coastal Eutrophication as a Driver of Salt Marsh Loss. Nature 490, 388–392. doi: 10.1038/nature11533
Dick W. A., Thavamani B., Conley S., Blaisdell R., Sengupta A., et al. (2013). Prediction of β-Glucosidase and β-Glucosaminidase Activities, Soil Organic C, and Amino Sugar N in a Diverse Population of Soils Using Near Infrared Reflectance Spectroscopy. Soil Biol. Biochem. 56, 99–104. doi: 10.1016/j.soilbio.2012.04.003
Ding S. M., Han C., Wang Y. P., Yao L., Wang Y., Xu D., et al. (2015). In Situ, High-Resolution Imaging of Labile Phosphorus in Sediments of a Large Eutrophic Lake. Water Res. 74, 100–109. doi: 10.1016/j.watres.2015.02.008
Dixon P. (2003). VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 14 (6), 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dong Z. Y., Wang K., Chen X., Zhu J., Hu C., Zhang D., et al. (2017). Temporal Dynamics of Bacterioplankton Communities in Response to Excessive Nitrate Loading in Oligotrophic Coastal Water. Mar. pollut. Bull. 114, 656–663. doi: 10.1016/j.marpolbul.2016.10.041
Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10 (10), 996–1000. doi: 10.1038/NMETH.2604
Edgar R. C., Haas B. J., Clemente J. C., Quince C, Knight R.. and (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27 (16), 2194–2200. doi: 10.1093/bioinformatics/btr381
Floreza J. Z., Camus C., Hengstc M. B., Marchant F., Buschmann A.H. (2019). Structure of the Epiphytic Bacterial Communities of Macrocystis Pyrifera in Localities With Contrasting Nitrogen Concentrations and Temperature. Algal. Res. 44, 101706. doi: 10.1016/j.algal.2019.101706
Ghosh S., Leff G. L. (2013). Impacts of Labile Organic Carbon Concentration on Organic and Inorganic Nitrogen Utilization by a Stream Biofilm Bacterial Community. Appl. Environ. Microb. 79, 7130–7141. doi: 10.1128/AEM.01694-13
Glibert P. M. (2012). Ecological Stoichiometry and its Implications for Aquatic Ecosystem Sustainability. Curr. Opin. Env. Sust. 4 (3), 272–277. doi: 10.1016/j.cosust.2012.05.009
Guo C. C., Zhang X. W., Luan S. M. (2021). Diversity and Structure of Soil Bacterial Community in Intertidal Zone of Daliao River Estuary, Northeast China. Mar. Pollut. Bull. 163, 111965. doi: 10.1016/j.marpolbul.2020.111965
Kessouria F., Mcwilliamsb J.C., Bianchib D., Sutula M., Renault L., Deutsch C. (2021). Coastal Eutrophication Drives Acidification, Oxygen Loss, and Ecosystem Change in a Major Oceanic Upwelling System. PNAS 118 (21), e2018856118. doi: 10.1073/pnas.2018856118
Kong X.K., Li C.H., Wang P., Huang G.X., Li Z.T., Han Z.T. (2019). Soil Pollution Characteristics and Microbial Responses in a Vertical Profile With Long-Term Tannery Sludge Contamination in Hebei, China. Int. J. Environ. Res. Public Health 16, 563. doi: 10.3390/ijerph16040563
Li K.Q., He J., Li J.L., Guo Q., Liang S.K., Li Y.B. (2018). Linking Water Quality With the Total Pollutant Load Control Management for Nitrogen in Jiaozhou Bay, China. Ecol. Indic. 85, 57–66. doi: 10.1016/j.ecolind.2017.10.019
Lu R. K. (1999). Analytical Methods of Soil and Agro-Chemistry (Beijing, China (in Chinese: Agricultural Science and Technology Press).
Luo L., Meng H., Wu R. N., Gu J.D., et al. (2017). Impact of Nitrogen Pollution/Deposition on Extracellular Enzyme Activity, Microbial Abundance and Carbon Storage in Coastal Mangrove Sediment. Chemosphere 177, 275–283. doi: 10.1016/j.chemosphere.2017.03.027
Ma S. N., Dong X. M., Jeppesen E., Søndergaard M., Cao J.Y., Li Y.Y., et al. (2021). Responses of Coastal Sediment Phosphorus Release to Elevated Urea Loading. Mar. pollut. Bull., 174, 113203. doi: 10.1016/j.marpolbul.2021.113203
Martin M. (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. Embnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Ma S. N., Wang H. J., Wang H. Z., Li Y., Liu M., Liang X.M., et al. (2018). High Ammonium Loading can Increase Alkaline Phosphatase Activity and Promote Sediment Phosphorus Release: A Two-Month Mesocosm Experiment. Water Res. 145, 388–397. doi: 10.1016/j.watres.2018.08.043
Ma S. N., Wang H. J., Wang H. Z., Zhang M, Li Y., Bian S.J., et al. (2021a). Effects of Nitrate on Phosphorus Release From Lake Sediments. Water Res. 194 (2021), 116894. doi: 10.1016/j.watres.2021.116894
Ma Y. X., Wei T., Liu C. F., Liu J., Yang Z.P., Li J., et al. (2017). Response of Microbial Biomass and Bacterial Community Composition to Fertilization in a Salt Marsh in China. Acta Oceanol. Sin. 36, 80–88. doi: 10.1007/s13131-017-1048-5
Mobley H. L., Island M. D., Hausinger R. P. (1995). Molecular Biology of Microbial Ureases. Microbiol. Mole. Biol. R. 59, 451–480. doi: 10.1128/MMBR.59.3.451-480.1995
Niu L. H., Xie X. D., Li Y., Hu Q., Wang C., Zhang W.L., et al. (2021). Effects of Nitrogen on the Longitudinal and Vertical Patterns of the Composition and Potential Function of Bacterial and Archaeal Communities in the Tidal Mudflats. Sci. Total Environ. 806, 151210. doi: 10.1016/j.scitotenv.2021.151210
Nogales B., Lanfranconi M. P., Pia-Villalonga M. J., Bosch R. (2011). Anthropogenic Perturbations in Marine Microbial Communities. FEMS Microbiol. Rev. 35, 275–298. doi: 10.1111/j.1574-6976.2010.00248.x
Omar S. A., Ismail M. A. (1999). Microbial Populations, Ammonification and Nitrification in Soil Treated With Urea and Inorganic Salts. Folia. Microbiol. 44 (2), 205–212. doi: 10.1007/BF02816244
Ramirez K. S., Craine J. M., Fierer N. (2010). Nitrogen Fertilization Inhibits Soil Microbial Respiration Regardless of the Form of Nitrogen Applied. Soil Biol. Biochem. 42, 2336–2338. doi: 10.1016/j.soilbio.2010.08.032
Schwermer C. U., Lavik G., Abed R. M., Dunsmore B., Ferdelman T.G., Stoodley P., et al. (2008). Impact of Nitrate on the Structure and Function of Bacterial Biofilm Communities in Pipelines Used for Injection of Seawater Into Oil Fields. Appl. Environ. Microbiol. 74, 2841–2851. doi: 10.1128/AEM.02027-07
Standardization Administration of the People’s Republic of China (SAC) (2006). Standards for Drinking Water Quality. GB5749–GB2006. http://openstd.samr.gov.cn/bzgk/gb/newGbInfo?hcno=73D81F4F3615DDB2C5B1DD6BFC9DEC86
Stegen J. C., Lin X., Konopka A. E., Fredrickson J.K. (2012). Stochastic and Deterministic Assembly Processes in Subsurface Microbial Communities. Isme J. 6 (9), 1653–1664. doi: 10.1038/ismej.2012.22
Sterner R. W., Elser J. J. (2002). Ecological Stoichiometry: The Biology of Elements From Molecules to the Biosphere (United Kingdom, New Jersey: Princeton University Press), 08540.
Uchida H., Watanabe R., Matsushima R., Oikawa H., Nagai S., Kamiyama T., et al. (2018). Toxin Profiles of Okadaic Acid Analogues and Other Lipophilic Toxins in Dinophysis From Japanese Coastal Waters. Toxins 10, 457. doi: 10.3390/toxins10110457
Wang C., Liu D. W., Bai E. (2018). Decreasing Soil Microbial Diversity is Associated With Decreasing Microbial Biomass Under Nitrogen Addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang X. P., Zhu H. B., Geng Y., Ding K.X., Ye L.N. (2022). Investigation the Effect of the Main Land-Based Pollutants in Xiangshan Bay. Ecol. Chem. Eng. S. 29 (1), 27–38. doi: 10.2478/eces-2022-0004
Watson A. J. (2016). Oceans on the Edge of Anoxia. Science 354 (6319), 1529. doi: 10.1126/science.aaj2321
Wei Y. L., Long Z. J., Ren M. X. (2022). Microbial Community and Functional Prediction During the Processing of Salt Production in a 1000-Year-Old Marine Solar Saltern of South China. Sci. Total Environ. 819, 152014. doi: 10.1016/j.scitotenv.2021.152014
Wessén E., Hallin S., Philippot L. (2010). Differential Responses of Bacterial and Archaeal Groups at High Taxonomical Ranks to Soil Management. Soil Biol. Biochem. 42, 1759–1765.
Wu J., Liu W., Zhang W., Shao Y.H., Duan H.L., Chen B.D., et al. (2019). Long-Term Nitrogen Addition Changes Soil Microbial Community and Litter Decomposition Rate in a Subtropical Forest. Appl. Soil Ecol. 142, 43–51. doi: 10.1016/j.apsoil.2019.05.014
Yang J., Jiang H., Liu W., Huang L.Q., Huang J.R., Wang B.C., et al. (2020). Potential Utilization of Terrestrially Derived Dissolved Organic Matter by Aquatic Microbial Communities in Saline Lakes. ISME J. 14, 2313–2324. doi: 10.1038/s41396-020-0689-0
Keywords: intertidal ecosystem, nitrogen pollution, bacterial community composition, microbial diversity, ecological functions
Citation: Xu Y-F, Dong X-M, Luo C, Ma S-N, Xu J-L and Cui Y-D (2022) Nitrogen Enrichment Reduces the Diversity of Bacteria and Alters Their Nutrient Strategies in Intertidal Zones. Front. Mar. Sci. 9:942074. doi: 10.3389/fmars.2022.942074
Received: 12 May 2022; Accepted: 13 June 2022;
Published: 13 July 2022.
Edited by:
Yuanyuan Feng, Shanghai Jiao Tong University, ChinaReviewed by:
Feng Zhao, Institute of Oceanology (CAS), ChinaCopyright © 2022 Xu, Dong, Luo, Ma, Xu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo-Nan Ma, bWFzaHVvbmFuQG5idS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.