- 1State Key Laboratory of Biocontrol, School of Marine Sciences, Sun Yat-sen University, Guangzhou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
- 3Guangdong Provincial Key Laboratory of Marine Resources and Coastal Engineering/Guangdong Provincial Key Laboratory for Aquatic Economic Animals, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 4Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, China
Rap2a, a member of Ras family small GTPases, plays a key role in growth and development. So far, no Rap2a homolog has been discovered in crustaceans. Herein, a Rap2a homolog from shrimp Litopenaeus vannamei, named LvRap2a, was cloned and identified. LvRap2a showed a high level of sequence conservation to other Rap2a homologs from different species. LvRap2a was found to be abundantly expressed in hemocytes, muscle, hepatopancreas and intestine tissues. Importantly, Rap2a knockdown effectively suppressed the length and weight of shrimp in a growth experiment. Mechanistically, we found that Rap2a could induce the expression of several Wnts, which are the positive regulators in cellular growth, differentiation, and tissue homeostasis. Moreover, LvRap2a expression was observed to be negatively regulated by LvDorsal (NF-κB), a transcription factor of innate immune signaling pathway. These results indicated that LvRap2a contributed to shrimp growth probably by activating the Wnt signaling pathway, and that LvRap2a could be the crosstalk between innate immunity and growth, as well as that LvRap2a could serve as a potential target for shrimp growth promotion.
Introduction
Shrimp aquaculture has grown rapidly in recent decades around the world, and it has become one of the most representative aquaculture activities, contributing to the growing demand for high-quality animal proteins. In 2019, the aquaculture production of shrimps and prawns is approximately 6,555,000 tons, with a value of nearly 41 billion dollars (Fao, 2021). Litopenaeus vannamei has a rapid growth rate and a high valuation in business, and thus has been the most popular cultured shrimp species, accounting for roughly 70% of the world’s cultured shrimp (Fao, 2021). In artificial culture, L. vannamei has a short production cycle that individual can reach a weight of 15 g (usually body weight in the market) after about 80 days of farming from the post larva.
A faster growth rate in commercial aquaculture indicates a shorter culture cycle for farm-raised animals, which can save food consumption and production costs. As a result, it’s critical to figure out how to improve aquatic animal growth/development rates. Environmental conditions and nutritional supplementation have been proven to alter the growth traits of shrimp L. vannamei in several investigations. For instance, L. vannamei cultured at salinity of 30‰ grows faster than those cultured at salinity of 3‰ (Huang et al., 2019), and dietary supplementation with polypeptides or Buton forest onion extracts can improve growth performance (Liao et al., 2019), whereas acidified seawater can inhibit the growth of L. vannamei (Munaeni et al., 2019; Muralisankar et al., 2021). The genetic mechanism of growth traits has also been explored, and a few genes have been identified to be associated with growth traits (Yu et al., 2019; Huang et al., 2020). Monocyte to macrophage differentiation factor 2-like (MMD2) is significantly associated with body weight, especially the nonsynonymous mutation named MMD5 that contributes the most to the trait (Wang et al., 2020). Rap2a has been found to be highly associated with L. vannamei growth performance, particularly body weight, using genome-wide association studies (GWAS) (Yu et al., 2019). Later, the L. vannamei DEAD-box helicase 43 (DDX43) was discovered as a growth-related gene in a quantitative trait locus (QTL) mapping analysis (Huang et al., 2020). However, how the mechanisms underlying these identified genes affect growth traits in shrimp have not been investigated.
Ras proteins, which belong to the small guanosine triphosphatase (GTPase) family, are essential components of many signal transduction pathways and play crucial roles in growth and development (Karnoub and Weinberg, 2008). Rap2a, a member of the Ras family of small GTP-binding proteins, has been shown to regulate growth and development via its ability to modulate the activity of Wnt signaling pathway in two ways. On the one hand, Rap2a activates the Wnt signaling pathway by promoting the autophosphorylation of TRAF2/Nck-interacting kinase (TNIK) by binding to its C-terminating citron homology (CNH) domain (Taira et al., 2004; Mahmoudi et al., 2009). On the other hand, Rap2 can directly interact with low-density lipoprotein receptor-related protein 6 (LRP6), an upstream receptor of Wnt signaling pathway, and maintain LRP6 stability (Park et al., 2013). Thus, Rap2a can maintain the activation level of Wnt pathway, which results in the expression of growth-related genes that stimulate cell proliferation and body expansion (Park et al., 2013). Previous study points that a sequence encoding a potential LvRap2a could be a growth regulated gene via a GWAS analysis (Yu et al., 2019), but the exact function of LvRap2a in regulating shrimp growth is not confirmed. In this study, we de novo cloned and identified the Rap2a homolog from L. vannamei (named as LvRap2a). Our results demonstrated that there is a reliable link between LvRap2a and shrimp growth. LvRap2a was able to positively regulate the expression of several LvWnts, while LvRap2a itself was negatively regulated by LvDorsal, a transcription factor of Toll pathway. These results might provide some information for the possible use of Rap2a expression as a growth trait parameter for selective breeding.
Materials and Methods
LvRap2a Sequence and Phylogenetic Analysis
NCBI was used to retrieve probable LvRap2a sequences (GenBank No. QCYY01001773.1). To confirm the sequences, Rap2a-F/Rap2a-R primers were developed to amplify the nucleotide sequences from L. vannamei cDNA library. The PCR fragments were cloned onto a pEASY-T1 Cloning Vector (TransGen Biotech, China) and sequenced to determine their identity.
BLAST searches were used to find protein sequences of Rap2a homologs from different species in GenBank. We used the Clustal X v2.0 tool (Larkin et al., 2007) to align LvRap2a and its homologs, and the identities between LvRap2a and others were examined using GeneDoc software and manually labeled. Using MEGA 5.2 software (Tamura et al., 2011) and the neighbor-joining (NJ) method, a phylogenetic tree was built based on the full-length amino acid sequences of Rap2a proteins.
Plasmids and dsRNA Construction
Based on the results of the nucleotide sequence, a reporter gene plasmid containing the 5’ flanking promoter region of LvRap2a was constructed into the pGL3 vector and the expression plasmid containing the ORF of LvDorsal was constructed into the pAc5.1A-HA vector with corresponding primers (Table 1).
In vitro transcription of LvDorsal dsRNA, LvRap2a dsRNA, and GFP dsRNA was performed with the T7 RiboMAXTM Express RNAi System kit (Promega) and the primers listed in Table 1.
Expression Pattern of LvRap2a in Different Tissues
The tissue distribution templates were obtained from a previous study (Wang et al., 2015). In brief, the shrimp tissues including scape, gill, stomach, heart, nerve, intestine, hepatopancreas, muscle, and hemocyte were sampled from 15 shrimps. The expression levels of LvRap2a in various tissues of healthy shrimp were determined using quantitative PCR (qPCR) and the primers listed in Table 1.
Dual-Luciferase Reporter Assay
A dual-luciferase reporter assay was used to detect the induction of LvRap2a by LvDorsal overexpression. In particular, Drosophila S2 cells were seeded in a 12-well plate. The cells in each well were then transfected with 0.5 μg of pGL3-LvRap2a, 0.1 μg of pRL-TK renilla luciferase plasmid (internal control; Promega), and 0.5 μg pAc-LvDorsal-HA or 0.5 μg pAc5.1-HA plasmid (as controls). The cells were harvested 48 hours after the transfection. Half of the cells were used to measure the induction of LvRap2a with Dual-Glo Luciferase Assay System (Promega, USA). Half of the cells were used to perform western blotting with rabbit anti-HA antibody (Sigma) and mouse anti-actin clone C4 antibody (Merck Millipore).
Shrimp Culture System and Samples Collection
In an indoor culturing pond (70 cm in length, 43 cm in width, and 41 cm in height), healthy cultured shrimp (supplied by Guangdong Hisenor Group Co., Ltd, Guangzhou, China) were cultured. The average initial weight and length of shrimp are 7.17 ± 0.87 g and 8.48 ± 0.46 cm, respectively. During the experiment period, the water temperature was 25°C, and the salinity of the water was 30‰. Every day, shrimp were fed four times with commercial feeds at 7:00, 11:00, 15:00, and 20:00. To investigate the roles of LvRap2a on shrimp growth and LvWnt induction, 30 shrimp were marked with eyestalk rings and randomly divided into two groups. Shrimp in the treatment group received dsRNA-LvRap2a (14 μg dsRNA in 50 µl PBS) injection, while shrimp in the control group were injected with dsRNA-GFP (14 μg dsRNA in 50 µl PBS). The weight and length from each shrimp was measured on day 0, day 5 and day 10. We were able to track the growth traits of individual shrimp because each eyestalk ring was printed with a unique number. In the meantime, a parallel experiment with 60 shrimp (30 shrimp in each group) was carried out to assess the efficacy of dsRNA-LvRap2a and the activity of the Wnt pathway. On day 0, day 2 and day 10 post dsRNA injection, hepatopancreases and muscle of each group (3 shrimp per sample, 3 samples) were collected for total RNA extraction and further qPCR detection. The experiments were performed twice. Primer sequences were listed in Table 1.
Knockdown of LvDorsal by dsRNA-Mediated RNA Interference
Shrimp were divided into two groups. Shrimp in the experimental group received dsRNA-LvDorsal injection, while shrimp in the control group received dsRNA-GFP injection (each shrimp was injected with 14 μg dsRNA in 50 µl PBS). The hepatopancreas of each group (three shrimp per sample, three samples) were collected 48 hours after dsRNA injection. Total RNA was extracted with RNeasy Mini Kit (Qiagen), and reverse transcribed to cDNA with TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech) following the manufacturer’s instructions. qPCR was used to detect relative expressions of LvDorsal or LvRap2a. Quantitative PCR primers for LvDorsal or LvRap2a were each listed in Table 1.
Quantitative PCR
qPCR was performed via the LightCycler 480 System (Roche, Basel, Germany) in a final reaction volume of 10 μl, which was comprised of 1 μl of 1:10 cDNA diluted with ddH2O, 5 μl of GoTaq qPCR Master Mix (Promega, Madison, WI, USA; cat. no. A6002), and 250 nM of specific primers (Table 1). The cycling program was as follows: 1 cycle of 95°C for 2 min, 40 cycles of 95°C for 15 s, 62°C for 1 min, and 70°C for 1 s. Cycling ended at 95°C with a 5°C/s calefactive velocity to create the melting curve. The expression level of each gene was calculated by using the Livak (2-∆∆CT) method after normalization to LvEF-1α (GenBank accession No. GU136229). Primers were listed in Table 1.
Statistical Analysis
All data were presented as mean ± SD. Student’s t test was used to calculate the comparisons between groups of numerical data. The following p values were considered statistically significant: *p < 0.05 and **p < 0.01.
Results
Sequence Confirmation of the ORF of LvRap2a
The results of nucleotide sequencing were not entirely consistent with the predicted LvRap2a sequences in NCBI (GenBank No. QCYY01001773.1), After PCR, plasmid construction and sequencing, we found that compared with the predicted sequence, there were two “T” missing at 401-402 bp, resulting in subsequent sequence frameshift and advance of termination codon. Therefore, the ORF of LvRap2a identified in this study was 402 bp in length and encoded a putative protein of 133 amino acids with a calculated molecular weight of ~15 kDa (GenBank No. OM972663).
Phylogenetic Analysis of Rap2a Proteins
Rap2a homologs have been discovered in a variety of species, ranging from invertebrates to vertebrates. Rap2a proteins from different species were conserved in the characteristic Ras domains, which were boxed with red lines (Figure 1A). In the effector domain, there were two characteristic amino acids, namely Phe residue at position 39 (marked with black triangle) and Thr residue at position 61 (marked with red triangle), suggesting that LvRap2a may belong to Rap2 family (Figure 1A). Besides, a neighbor-joining (NJ) phylogenetic tree revealed that Rap2a proteins from vertebrates were clustered as a group, while Rap2a proteins from invertebrates and cephalochordate formed a separate group that contained the LvRap2a (Figure 1B). These results suggested that LvRap2a was conserved in protein sequences and could be classified as a new member of Rap2a family.
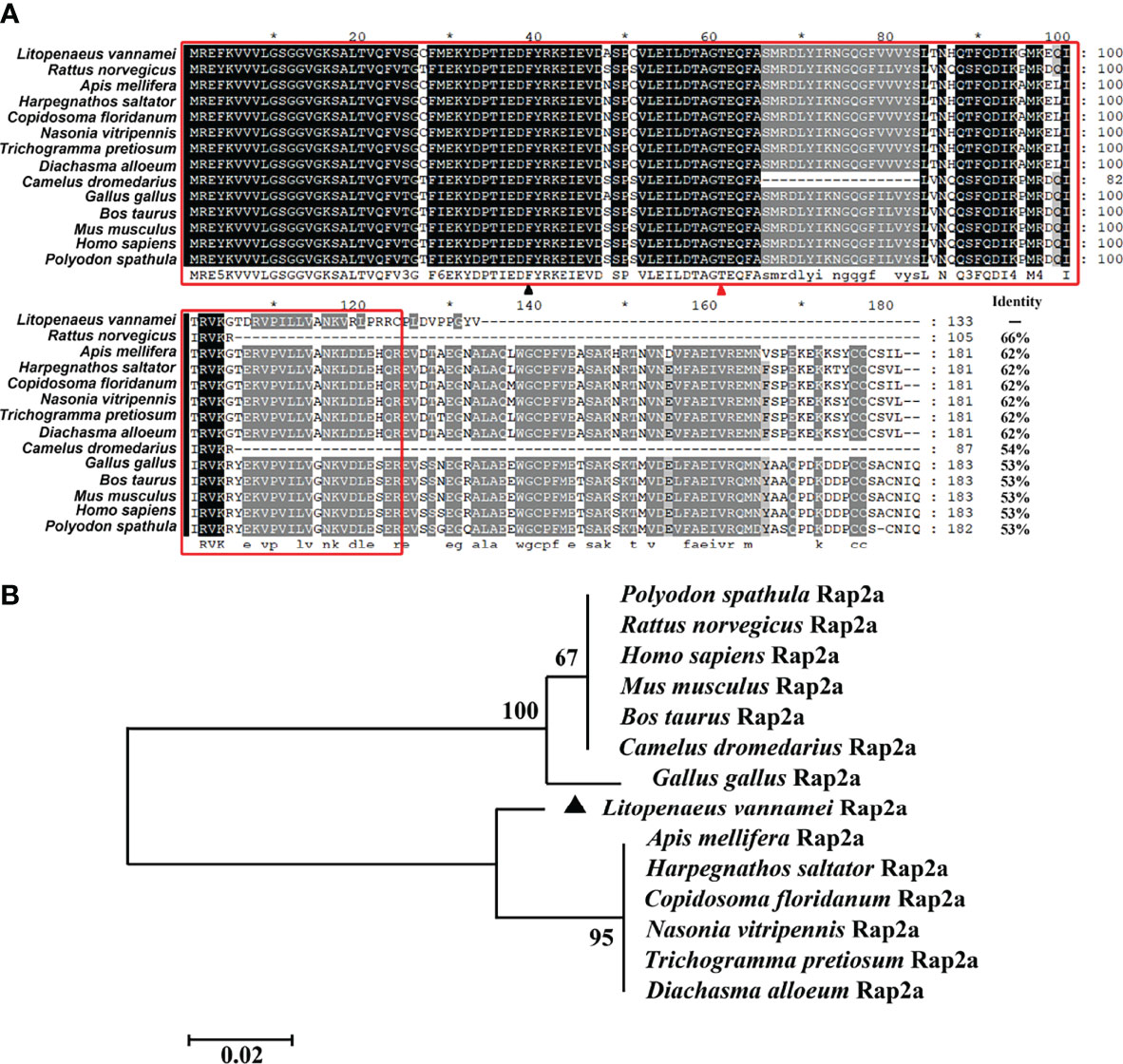
Figure 1 Multiple sequence alignment and phylogenetic tree analysis of Rap2a proteins. (A) Multiple sequence alignment of Rap2a proteins. The conserved Ras domain was boxed with red lines. The Rap characteristic Thr residue at position 61 was marked with red triangle and Rap2 characteristic Phe residue at position 39 was marked with black triangle. (B) Neighbor-joining method was used to construct the phylogenetic tree of Rap2a proteins.
Tissue Distribution of LvRap2a in Healthy Shrimp
The mRNA of LvRap2a could be detected in all tested tissues, including scape, gill, stomach, heart, nerve, intestine, hepatopancreas, and hemocyte (Figure 2). In details, the LvRap2a was especially abundant in hemocyte, muscle, hepatopancreas and intestine. The hepatopancreas was the primary organ for nutrient absorption and storage (Cervellione et al., 2017; Vogt, 2019). And muscle was tissue related to growth So hepatopancreas and muscle were chosen to investigate the expression changes of LvRap2a and growth-related genes by subsequent quantitative RT-PCR.
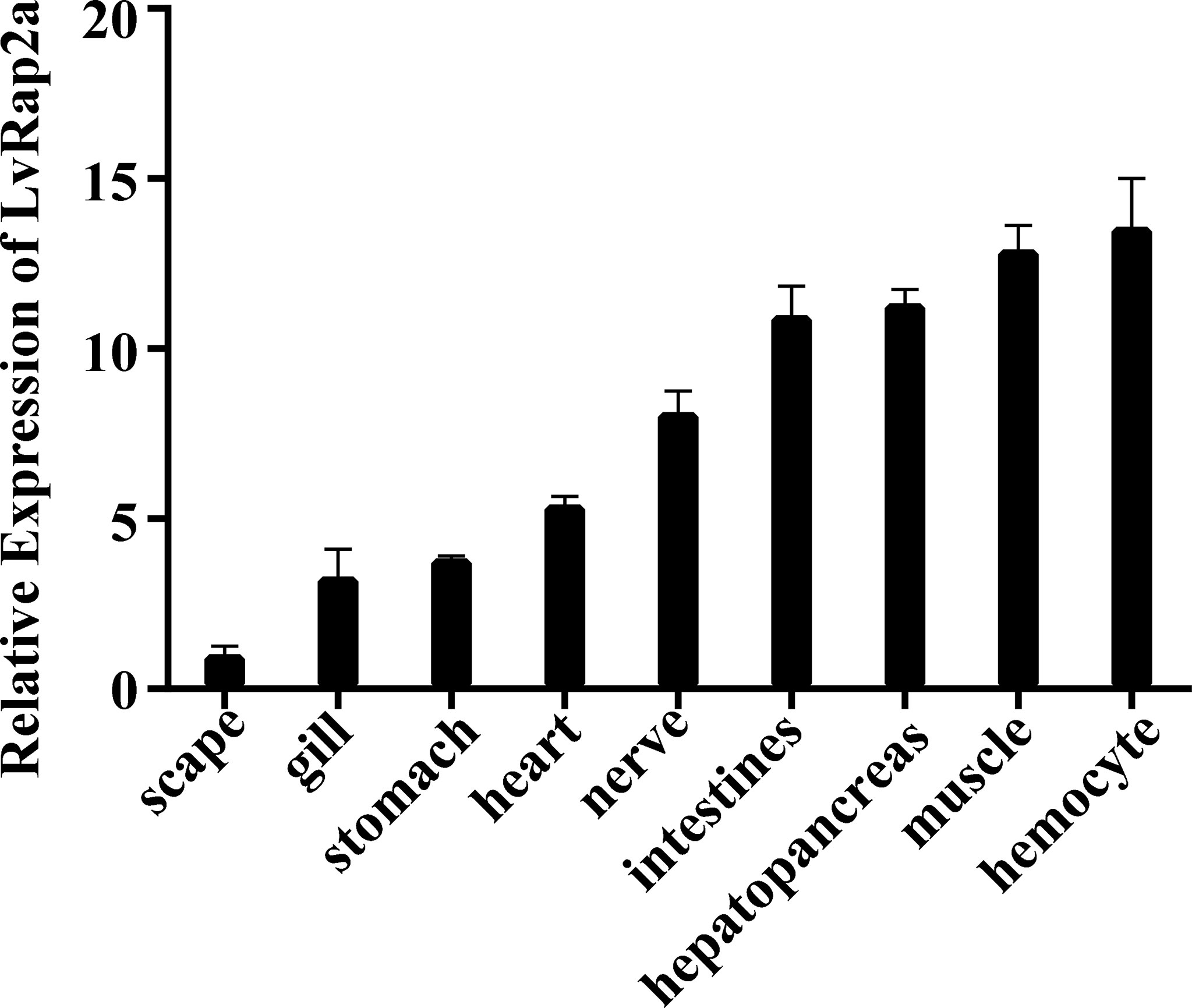
Figure 2 Expression of LvRap2a in healthy shrimp detected by qRT-PCR. Expression of LvRap2a was normalized to that of EF-1α using the Livak (2-ΔΔCT) method and the data were provided as the means ± SD of triplicate assays. Expression level in the scape was set to 1.0.
Roles of LvRap2a in Shrimp Growth Traits
To investigate the role of LvRap2a on shrimp growth, a 10-day indoor culturing experiment was performed. Shrimp labeled with eyestalk rings were divided into two groups and received dsRNA injection (Figure 3A). dsRNA-LvRap2a was utilized to precisely suppress LvRap2a transcription, while dsRNA-GFP was utilized as a control. Every 5 days, each shrimp’s growth-related data (body weight and total length) was measured (Figure 3A). A parallel experiment was also carried out to gain hepatopancreas samples on day 0, day 2 and day 10 for qPCR detection (Figure 3B). The initial weight (p = 0.256) and length (p = 0.246) of shrimp at day 0 from the two experimental groups were almost identical and have no significant difference (Figures 4A, B). At all measure points, shrimp treated with dsRNA-GFP were considerably longer than shrimp treated with dsRNA-LvRap2a in body length (Figure 4A). On day 10, the final shrimp lengths were markedly different (10.73 ± 0.44 cm vs 10.13 ± 0.65 cm) (Figure 4A). Besides, the average weight of LvRap2a silenced shrimp was lower at day 5 (7.53 ± 0.03 g) and day 10 (8.05 ± 1.02 g) than those of the dsRNA-GFP treatments (8.34 ± 1.00 g on day 5, 8.97 ± 0.86 g on day 10) (Figure 4B). The visible growth performances of LvRap2a silenced shrimp and control group at day 10 were shown in Figure 4C. In the parallel experiments, hepatopancreas sampled on day 0 and day 10 were used to determine RNAi efficiency. As shown in Figure 4D, the expression level of LvRap2a was dramaticlly decreased with dsRNA-LvRap2a injection, which was down-regulated to 32.64% of the GFP dsRNA injection groups, indicating that dsRNA-LvRap2a successfully inhibited LvRap2a expression during the 10-day growth experiment.
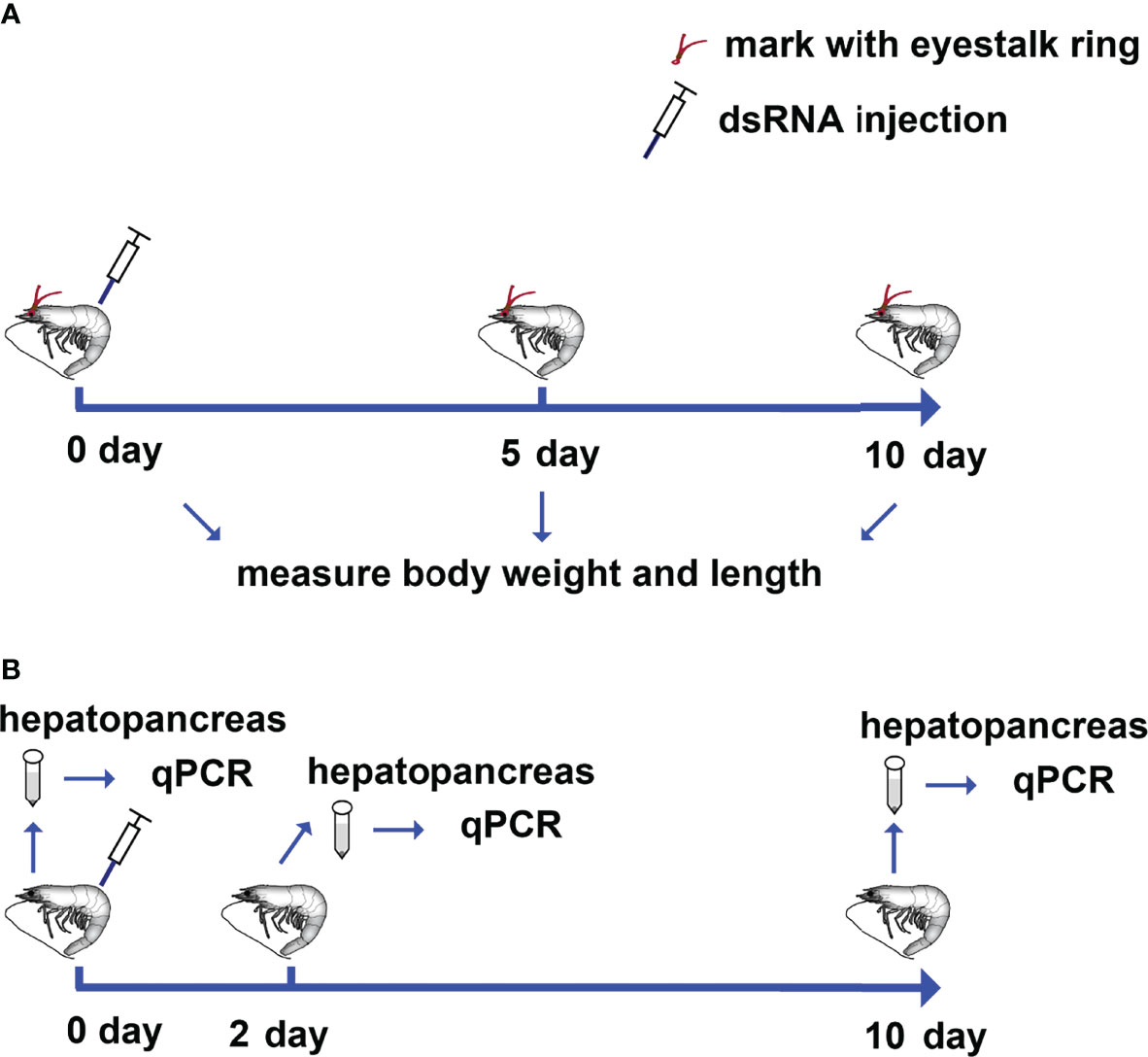
Figure 3 Schematic representation of the procedures for investigating the role of LvRap2a on the growth of shrimp. (A) Each shrimp was marked with a specific eyestalk ring and received dsRNA-LvRap2a or dsRNA-GFP (as control) injections on day 0. Each shrimp’s weight and length were measured on day 0, day 5, and day 10 following dsRNA injection. (B) Shrimp were injected with dsRNA-LvRap2a or dsRNA-GFP (as a control). On day 0, day 2 and day 10 post dsRNA injection, the hepatopancreases of each group (3 shrimp per sample, 3 samples) were collected for total RNA extraction and further qPCR detection.
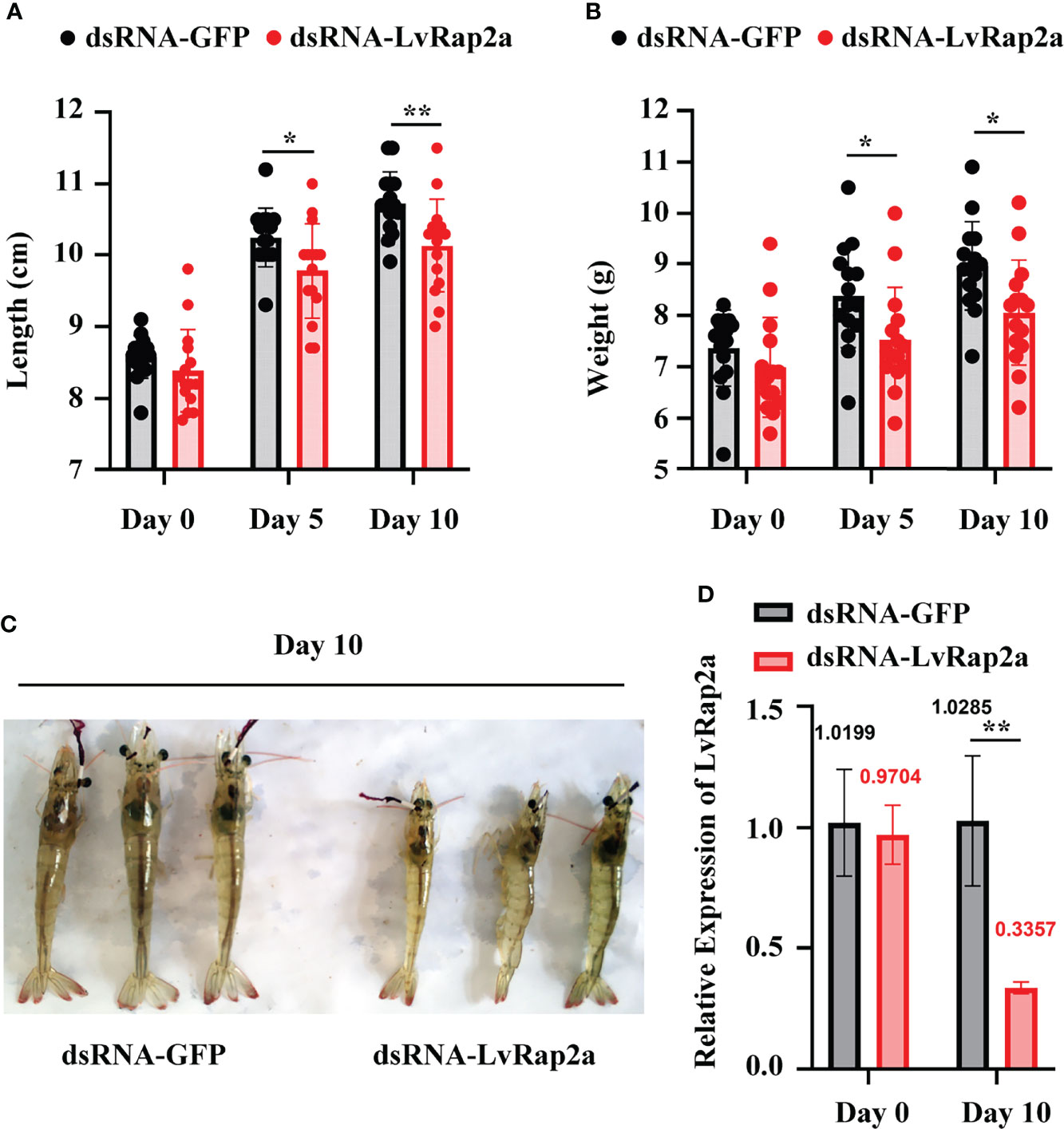
Figure 4 Roles of LvRap2a on the growth traits of shrimp. (A) Length of shrimp on day 0, day 5 and day 10 post dsRNA injection. The data were provided as the means ± SD of 15 shrimp. (B) Weight of shrimp on day 0, day 5 and day 10 post dsRNA injection. The data were provided as the means ± SD of 15 shrimp. (C) Gross signs of shrimp on day 10 post dsRNA treatment. (D) Quantitative PCR analysis of the silencing efficiency of LvRap2a. Asterisks indicate significant differences (*p < 0.05 and **p < 0.01). All the experiments were performed two times with identical results.
LvRap2a Contributes to Shrimp Growth Through Inducing LvWnts
Since the Wnt signaling pathway is highly related to growth, we wondered if LvRap2a contributed to shrimp growth via affecting LvWnts. We detected the expression of 12 known LvWnt in dsRNA-treated shrimp. We found that in both hepatopancreas and muscle, dsRNA-LvRap2a could significantly suppress LvRap2a expression (~0.28-fold in hepatopancreas, and ~0.04-fold in muscle) at 48 hours (2 days) after dsRNA injection (Figure 5). The knockdown of LvRap2a in hepatopancreas could down-regulate the expression of five Wnt members including LvWnt1 (~0.23-fold), LvWnt2 (~0.37-fold), LvWnt9 (~0.07-fold), LvWnt10 (~0.49-fold) and LvWntA (~0.20-fold), implying that LvRap2a may induce those LvWnts to promote shrimp growth (Figure 5A). As for muscle, RNAi of LvRap2a inhibited the expression of LvWnt2, LvWnt5, LvWnt11, LvWnt16 and LvWntA (Figure 5B). However, the expression of LvWnt7 was up-regulated by LvRap2a knocked-down (Figure 5B). Some of the LvWnts were undetectable in the hepatopancreas or muscle because of their low expression in those tissues (Figure 5).
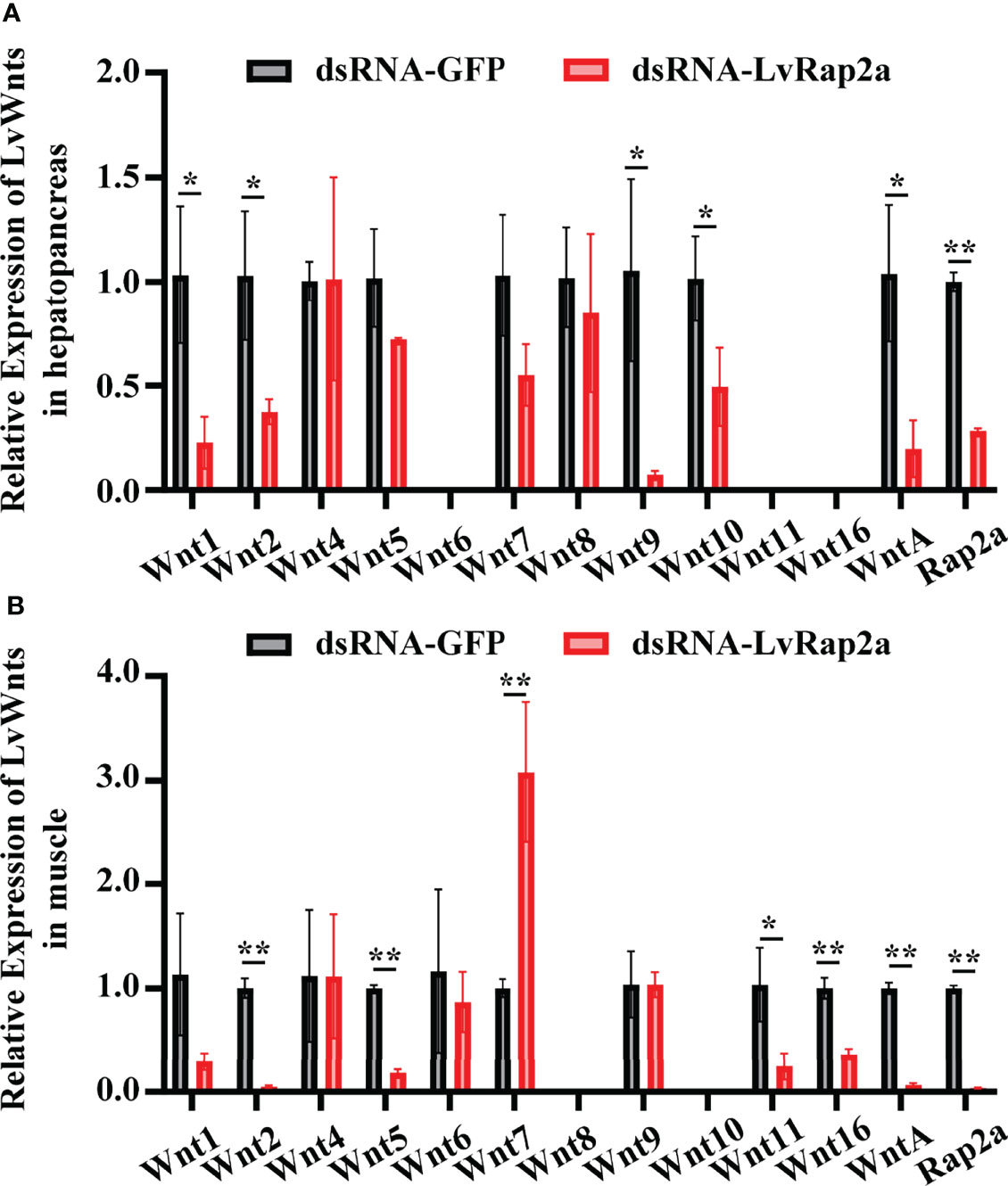
Figure 5 Wnts were regulated by LvRap2a. Relative expression levels of all the twelve Wnts in hepatopancreases (A) and muscle (B) of L. vannamei were detected via qRT-PCR at 48 hours (2-day) post dsRNA injection. The transcription level of each gene in dsRNA-GFP group was normalized to 1.0. Asterisks indicate significant differences (*p < 0.05 and **p < 0.01). All the experiments were performed two times with identical results.
LvRap2a Is Negatively Regulatedby LvDorsal
To explore how LvRap2a was regulated, we analyzed the promoter sequence of LvRap2a. As shown in Figure 6A, two potential Dorsal binding motifs present in the promoter region of LvRap2a, located at -211 to -201 and -174 to -164 from the transcriptional start site (TSS, +1). An in vitro experiment was performed to test whether LvDorsal could regulate the promoter activity of LvRap2a. We constructed a reporter plasmid that contained promoter regions of LvRap2a (Figure 6B). A dual-luciferase reporter assay showed that LvRap2a was unexpectedly down-regulated by ectopic expression of LvDorsal in Drosophila S2 cells (Figure 6C), suggesting that LvDorsal could inhibit LvRap2a induction in vitro. To confirm this, we further explored whether endogenous LvDorsal could be able to inhibit LvRap2a in vivo. LvDorsal expression level was lower in shrimp injected with LvDorsal dsRNA than in those injected with dsRNA GFP (Figure 7A). Injection of dsRNA-LvDorsal significantly increased LvRap2a expression level in vivo (Figure 7B), which were in line with the results in vitro. To sum up, these results strongly indicated that knockdown of LvDorsal increased LvRap2a expression, which in turn triggered LvWnt to promote shrimp growth (Figure 7C).
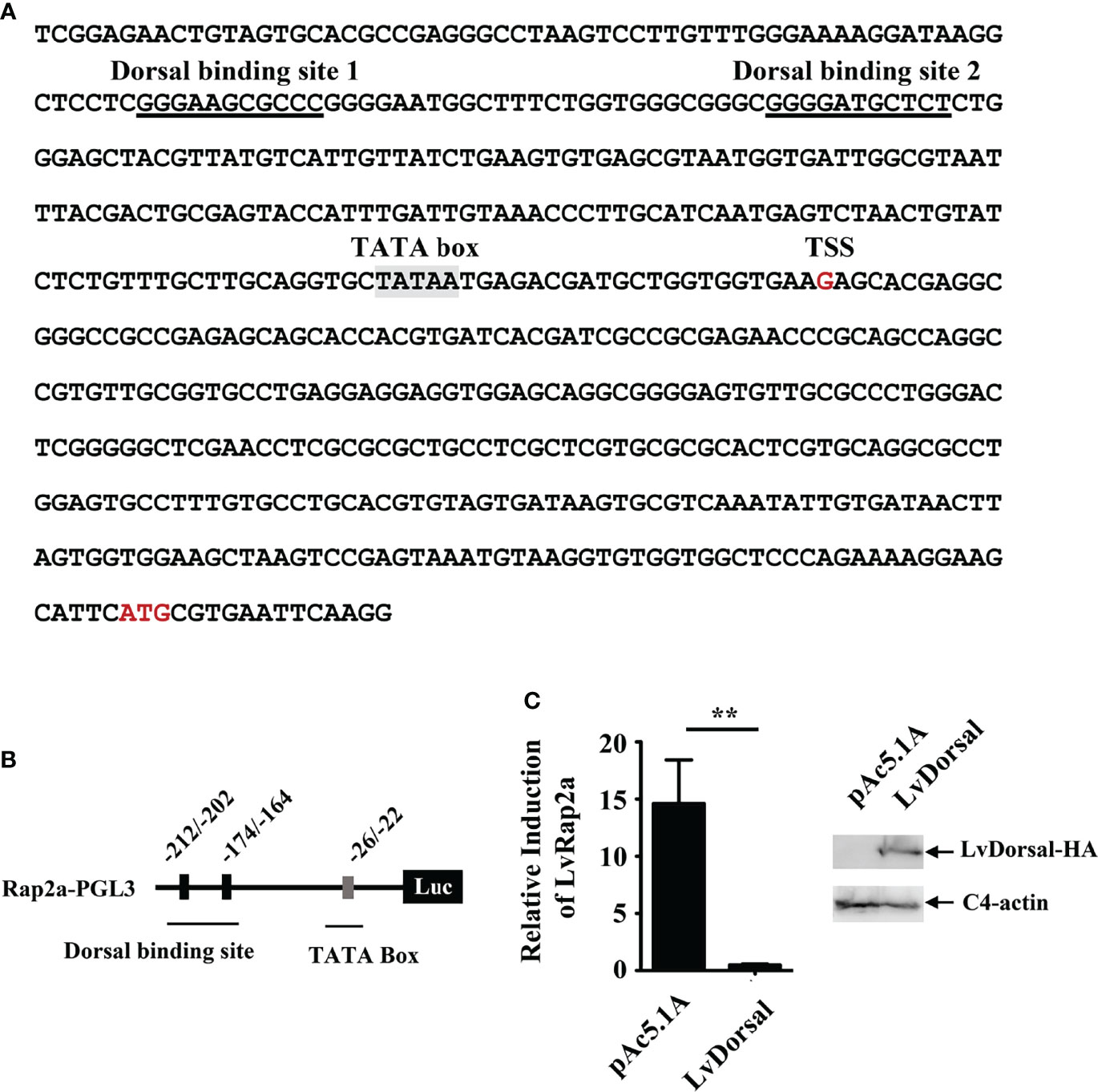
Figure 6 Over-expression of LvDorsal in vitro inhibited LvRap2a induction. (A) The promoter sequences of LvRap2a. The potential Dorsal binding motifs on the promoter of LvRap2a were underlined with black lines and the TATA box was shaded in grey. (B) The structure of the promoter of LvRap2a. There are two putative Dorsal binding motifs located at -212 to -202 and -174 to -164. (C) LvDorsal negatively regulated the promoter activity of LvRap2a in vitro. After 48 h, the cells were harvested for measurement of luciferase activity using the dual-luciferase reporter assay system. Western blotting was performed to confirm LvDorsal expression. C4 actin was used as a protein loading control. Asterisks indicate significant differences (**p < 0.01). All the experiments (C) were performed two times with identical results.
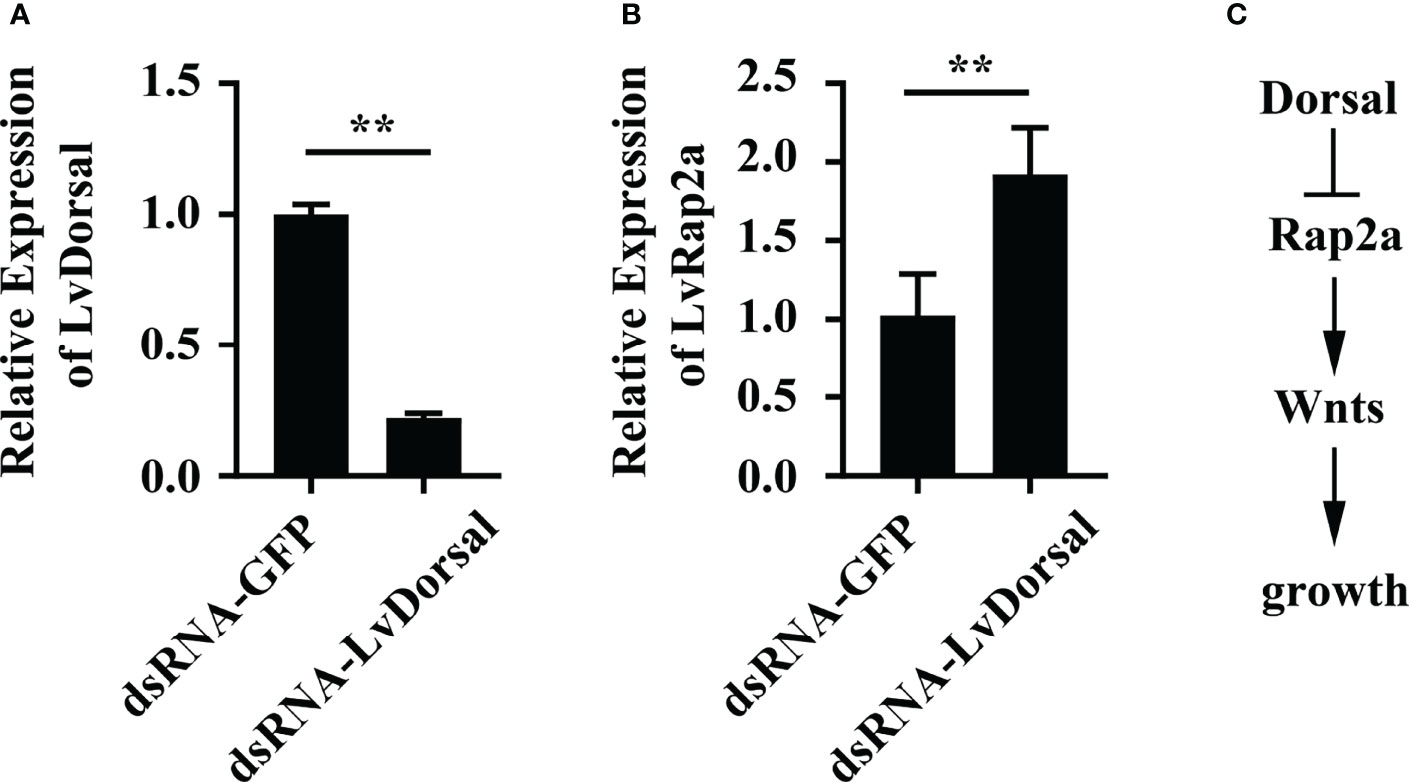
Figure 7 The expression of LvRap2a was negatively regulated by LvDorsal in vivo. (A) The RNAi efficiency of dsRNA-LvDorsal. Samples were taken at 48 h after dsRNA injection. (B) Expression of LvRap2a in LvDorsal silenced shrimp was detected by qRT-PCR analysis. (C) A potential model about the effect of LvDorsal on Rap2a-Wnt cascade mediated shrimp growth. Asterisks indicate significant differences (**p < 0.01). All the experiments (A, B) were performed two times with identical results.
Discussion
The Rap gene family, including five different members of Rap1a, Rap1b, Rap2a, Rap2b, and Rap2c, is widely conserved across eukaryotes (Gloerich and Bos, 2011). The Rap branch of Ras superfamily has a characteristic Thr residue at position 61, whereas most other members of the Ras superfamily of proteins have a glutamine residue in this site (Bokoch, 1993). Rap2 proteins appear be functionally analogous to Rap1 members, but the amino acid 39 (position) in the effector region of Rap2 is Phe that differs from those of Rap1 it is Ser (Taira et al., 2004). In the present study, we observed that LvRap2a contained a conserved Thr residue at position 61 and a Phe residue 39 in its effector domain. Besides, Rap2a proteins from both invertebrates and vertebrates displayed high sequence conservation in the Ras domains. The amino acid sequence conservation and these characteristic amino acids in LvRap2a suggested that LvRap2a could have a similar function to other Rap2a homologs in different species.
Numerous studies have shown that Rap2a is strongly linked to growth and development. In Xenopus laevis, Wnt/beta-catenin signaling during dorsoventral axis specification is thought to be mediated by Rap2a (Choi and Han, 2005). Also, knockdown of Rap2 results in a shortened body axis (Choi and Han, 2005). In mice, Rap2a regulates neurite growth and arborization in mammalian neurons (Kawabe et al., 2010). In Clonorchis sinensis, Rap2 is shown to be involved in normal growth, differentiation, and proliferation during its life cycle, which results in the growth of this parasite (Chen et al., 2011). In this study, our results demonstrated that LvRap2a contributed to shrimp growth from three perspectives. Firstly, LvRap2a was highly expressed in the hepatopancreas and intestine (Figure 2). The hepatopancreas is the main organ of metabolism (Vogt, 2019). R-cells in the hepatopancreas are responsible for nutrient absorption and metabolization, as well as the storage of energy reserves and minerals (Vogt, 2019). Intestine is the main site of the digestive system for nutrient absorption. So, the abundant expression of LvRap2a in these two tissues suggests that LvRap2a may be related to the growth of shrimp. Secondly, the length and weight of shrimp were both significantly lower in LvRap2a silenced group compared with the control group. RNAi is an effective and convenient method for understanding growth (Sánchez Alvarado and Newmark, 1999). For instance, knockdown other growth-related genes, such as MSTN/GDF11 or GSK3, significantly affect the growth of shrimp (Lee et al., 2015; Pang et al., 2021). Finally, we found that LvRap2a could regulate several LvWnts induction. It is widely accepted that Wnt proteins play key roles in cell or individual growth (Pfister and Kühl, 2018). A total of 12 Wnt genes representing 12 Wnt gene subfamilies have been identified in L. vannamei (Du et al., 2018). Screening the expression level of those 12 genes in dsRNA-LvRap2a treated shrimp revealed that LvRap2a promoted the expression of some LvWnts (Figure 5). LvWnt1 is closely related to the development of body segments and appendages in shrimp (Du et al., 2018). In Rex rabbits, Wnt10b promotes hair follicle growth and dermal papilla cell proliferation via the Wnt/beta-Catenin signaling pathway (Wu et al., 2020). LvWntA, LvWnt2, LvWnt5, and LvWnt16 were highly expressed throughout the molting process, which was important in shrimp growth (Du et al., 2018). Wnt11, acted as a directional cue toorient myocyte elongation through the noncanonical Wnt/PCP pathway (Gros et al., 2009). Combining the results above, we believed that LvRap2a might be a key factor in promoting growth via regulating the expression of Wnt members.
In addition to its roles in growth and development, Rap2a is also involved in immune response in mammals. Rap2a inhibited LPS-induced NF-κB activation by interfering with upstream signaling events, leading to decreased p65 serine 536 phosphorylation (Carvalho et al., 2019). In shrimp, Rap homologs have been identified in Fenneropenaeus chinensis and Marsupenaeus japonicus. Studies in both of them are focused on the relationship between Rap and white spot syndrome virus (WSSV) infection (Ren et al., 2012; Huang et al., 2016). As for LvRap2a, it is interesting to find that the expression of LvRap2a was negatively regulated by LvDorsal, the p65 homolog in L. vannamei. LvDorsal is famous for its anti-bacterial and anti-viral roles in shrimp (Li et al., 2018; Li et al., 2019). So, the negative regulatory relationship between Dorsal and Rap2a might raise a potential immune–growth antagonism model in shrimp (Figure 7C). LvDorsal was activated after suffering from pathogenic infection, which inhibited the expression of LvRap2a, and then restrained the activity of Wnt pathway, and finally leading to restrict shrimp growth. Indeed, shrimp growth was slowed by pathogen infections such as hepatopancreatic parvovirus (HPV) (Safeena et al., 2012), Penaeus monodon nudivirus (PmNV) (Yang et al., 2014), infectious hypodermaland hematopoietic necrosis virus (IHHNV) (Chayaburakul et al., 2005) and Enterocytozoon hepatopenaei (EHP) (Chaijarasphong et al., 2021).
In summary, our results revealed that LvRap2a facilitated shrimp growth by positively regulating the expression of several LvWnts, and LvRap2a was negatively regulated by LvDorsal. Our findings in this study might help in development of selective breeding using LvRap2a as the growth trait parameter, and might provide insights into the interaction between innate immunity and growth.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
SW conceived and designed the experiments. SW, MH, QL, HL, and BY performed the experiments and analyzed data. SW, HL, JH and CL acquired funding. SW wrote the draft manuscript. JH oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team. CL responsible for data coordination, and writing, finalizing, and submitting the manuscript. All authors discussed the results and approved the final version. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by National Natural Science Foundation of China (32002416/32022085/31930113), Guangdong Basic and Applied Basic Research Foundation (2019A1515110276), Natural Science Foundation of Guangdong Province (2021A1515010747), Independent Research and Development Projects of Maoming Laboratory (2021ZZ007/2021TDQD004), Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), (SML2021SP301), and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (22lglj05).
Author Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Zhang Lǚping for providing the eyestalk rings (PATENT NO: 201010112910.0).
References
Bokoch G. M. (1993). Biology of the Rap Proteins, Members of the Ras Superfamily of GTP-Binding Proteins. Biochem. J. 289 (Pt 1), 17–24. doi: 10.1042/bj2890017
Carvalho B. C., Oliveira L. C., Rocha C. D., Fernandes H. B., Oliveira I. M., Leão F. B., et al. (2019). Both Knock-Down and Overexpression of Rap2a Small GTPase in Macrophages Result in Impairment of NF-κb Activity and Inflammatory Gene Expression. Mol. Immunol. 109, 27–37. doi: 10.1016/j.molimm.2019.02.015
Cervellione F., Mcgurk C., Berger Eriksen T., Van Den Broeck W. (2017). Effect of Starvation and Refeeding on the Hepatopancreas of Whiteleg Shrimp Penaeus Vannamei (Boone) Using Computer-Assisted Image Analysis. J. fish Dis. 40 (11), 1707–1715. doi: 10.1111/jfd.12639
Chaijarasphong T., Munkongwongsiri N., Stentiford G. D., Aldama-Cano D. J., Thansa K., Flegel T. W., et al. (2021). The Shrimp Microsporidian Enterocytozoon Hepatopenaei (EHP): Biology, Pathology, Diagnostics and Control. J. Invertebr Pathol. 186, 107458. doi: 10.1016/j.jip.2020.107458
Chayaburakul K., Lightner D. V., Sriurairattana S., Nelson K. T., Withyachumnarnkul B. (2005). Different Responses to Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV) in Penaeus Monodon and P. Vannamei. Dis. Aquat. organisms 67 (3), 191–200. doi: 10.3354/dao067191
Chen W., Wang X., Deng C., Lv X., Fan Y., Men J., et al. (2011). Molecular Cloning and Characterization of a Novel Ras-Related Protein (Rap2) From Clonorchis Sinensis. Parasitol. Res. 108 (4), 1021–1026. doi: 10.1007/s00436-010-2147-9
Choi S. C., Han J. K. (2005). Rap2 is Required for Wnt/beta-Catenin Signaling Pathway in Xenopus Early Development. EMBO J. 24 (5), 985–996. doi: 10.1038/sj.emboj.7600571
Du J., Zhang X., Yuan J., Zhang X., Li F., Xiang J. (2018). Wnt Gene Family Members and Their Expression Profiling in Litopenaeus Vannamei. Fish Shellfish Immunol. 77, 233–243. doi: 10.1016/j.fsi.2018.03.034
Gloerich M., Bos J. L. (2011). Regulating Rap Small G-Proteins in Time and Space. Trends Cell Biol. 21 (10), 615–623. doi: 10.1016/j.tcb.2011.07.001
Gros J., Serralbo O., Marcelle C. (2009). WNT11 Acts as a Directional Cue to Organize the Elongation of Early Muscle Fibres. Nature 457 (7229), 589–593. doi: 10.1038/nature07564
Huang W., Cheng C., Liu J., Zhang X., Ren C., Jiang X., et al. (2020). Fine Mapping of the High-pH Tolerance and Growth Trait-Related Quantitative Trait Loci (QTLs) and Identification of the Candidate Genes in Pacific White Shrimp (Litopenaeus Vannamei). Mar. Biotechnol. (New York N.Y.) 22 (1), 1–18. doi: 10.1007/s10126-019-09932-8
Huang M., Dong Y., Zhang Y., Chen Q., Xie J., Xu C., et al. (2019). Growth and Lipidomic Responses of Juvenile Pacific White Shrimp Litopenaeus Vannamei to Low Salinity. Front. Physiol. 10, 1087. doi: 10.3389/fphys.2019.01087
Huang X., Ye T., Jin M., Wang W., Hui K., Ren Q. (2016). Three Members of Ras GTPase Superfamily are Response to White Spot Syndrome Virus Challenge in Marsupenaeus Japonicus. Fish Shellfish Immunol. 55, 623–631. doi: 10.1016/j.fsi.2016.06.038
Karnoub A. E., Weinberg R. A. (2008). Ras Oncogenes: Split Personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531. doi: 10.1038/nrm2438
Kawabe H., Neeb A., Dimova K., Young S. M. Jr., Takeda M., Katsurabayashi S., et al. (2010). Regulation of Rap2A by the Ubiquitin Ligase Nedd4-1 Controls Neurite Development. Neuron 65 (3), 358–372. doi: 10.1016/j.neuron.2010.01.007
Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee J. H., Momani J., Kim Y. M., Kang C. K., Choi J. H., Baek H. J., et al. (2015). Effective RNA-Silencing Strategy of Lv-MSTN/GDF11 Gene and its Effects on the Growth in Shrimp, Litopenaeus Vannamei. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 179, 9–16. doi: 10.1016/j.cbpb.2014.09.005
Liao X. Z., Hu S. K., Wang B., Qin H. P., Zhao J. C., He Z. H., et al. (2019). Dietary Supplementation With Polypeptides Improved Growth Performance, Antibacterial Immune and Intestinal Microbiota Structure of Litopenaeus Vannamei. Fish Shellfish Immunol. 92, 480–488. doi: 10.1016/j.fsi.2019.06.033
Li C., Wang S., He J. (2019). The Two NF-κb Pathways Regulating Bacterial and WSSV Infection of Shrimp. Front. Immunol. 10, 1785. doi: 10.3389/fimmu.2019.01785
Li H., Yin B., Wang S., Fu Q., Xiao B., L, ǚ K., et al. (2018). RNAi Screening Identifies a New Toll From Shrimp Litopenaeus Vannamei That Restricts WSSV Infection Through Activating Dorsal to Induce Antimicrobial Peptides. PLos Pathog. 14 (9), e1007109. doi: 10.1371/journal.ppat.1007109
Mahmoudi T., Li V. S., Ng S. S., Taouatas N., Vries R. G., Mohammed S., et al. (2009). The Kinase TNIK is an Essential Activator of Wnt Target Genes. EMBO J. 28 (21), 3329–3340. doi: 10.1038/emboj.2009.285.
Munaeni W., Disnawati, Yuhana M., Setiawati M., Bujang A., Ode Baytul Abidin L., et al. (2019). Buton Forest Onion Extract (Eleutherine Bulbosa (Mill.) Potential on Growth Performance of Vannamei Shrimp (Litopenaeus Vannamei). Pakistan J. Biol. Sci. PJBS 22 (1), 15–20. doi: 10.3923/pjbs.2019.15.20
Muralisankar T., Kalaivani P., Thangal S. H., Santhanam P. (2021). Growth, Biochemical, Antioxidants, Metabolic Enzymes and Hemocytes Population of the Shrimp Litopenaeus Vannamei Exposed to Acidified Seawater. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 239, 108843. doi: 10.1016/j.cbpc.2020.108843
Pang Y., Zhang X., Yuan J., Zhang X., Su M., Li F. (2021). Glycogen Synthase Kinase 3 Gene Is Important in Growth and Molting of the Pacific White Shrimp Litopenaeus Vannamei. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.681966
Park D. S., Seo J. H., Hong M., Choi S. C. (2013). Role of the Rap2/TNIK Kinase Pathway in Regulation of LRP6 Stability for Wnt Signaling. Biochem. Biophys. Res. Commun. 436 (2), 338–343. doi: 10.1016/j.bbrc.2013.05.104
Pfister A. S., Kühl M. (2018). Of Wnts and Ribosomes. Prog. Mol. Biol. Trans. Sci. 153, 131–155. doi: 10.1016/bs.pmbts.2017.11.006
Ren Q., Zhou J., Jia Y. P., Wang X. W., Zhao X. F., Wang J. X. (2012). Cloning and Characterization of Rap GTPase From the Chinese White Shrimp Fenneropenaeus Chinensis. Dev. Comp. Immunol. 36 (1), 247–252. doi: 10.1016/j.dci.2011.07.004
Safeena M. P., Rai P., Karunasagar I. (2012). Molecular Biology and Epidemiology of Hepatopancreatic Parvovirus of Penaeid Shrimp. Indian J. Virol. an Off. Organ Indian Virological Soc. 23 (2), 191–202. doi: 10.1007/s13337-012-0080-5
Sánchez Alvarado A., Newmark P. A. (1999). Double-Stranded RNA Specifically Disrupts Gene Expression During Planarian Regeneration. Proc. Natl. Acad. Sci. U S A 96 (9), 5049–5054. doi: 10.1073/pnas.96.9.5049
Taira K., Umikawa M., Takei K., Myagmar B. E., Shinzato M., Machida N., et al. (2004). The Traf2- and Nck-Interacting Kinase as a Putative Effector of Rap2 to Regulate Actin Cytoskeleton. J. Biol. Chem. 279 (47), 49488–49496. doi: 10.1074/jbc.M406370200
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Vogt G. (2019). Functional Cytology of the Hepatopancreas of Decapod Crustaceans. J. morphology 280 (9), 1405–1444. doi: 10.1002/jmor.21040
Wang S., Li H., Qian Z., Song X., Zhang Z., Zuo H., et al. (2015). Identification and Functional Characterization of the TAB2 Gene From Litopenaeus Vannamei. Fish shellfish Immunol. 46 (2), 206–216. doi: 10.1016/j.fsi.2015.06.024
Wang Q., Yu Y., Zhang Q., Luo Z., Zhang X., Xiang J., et al. (2020). The Polymorphism of LvMMD2 and Its Association With Growth Traits in Litopenaeus Vannamei. Mar. Biotechnol. (New York N.Y.) 22 (4), 564–571. doi: 10.1007/s10126-020-09977-0
Wu Z., Zhu Y., Liu H., Liu G., Li F. (2020). Wnt10b Promotes Hair Follicles Growth and Dermal Papilla Cells Proliferation via Wnt/β-Catenin Signaling Pathway in Rex Rabbits. Bioscience Rep. 40 (2). doi: 10.1042/BSR20191248
Yang Y. T., Lee D. Y., Wang Y., Hu J. M., Li W. H., Leu J. H., et al. (2014). The Genome and Occlusion Bodies of Marine Penaeus Monodon Nudivirus (PmNV, Also Known as MBV and PemoNPV) Suggest That it Should be Assigned to a New Nudivirus Genus That is Distinct From the Terrestrial Nudiviruses. BMC Genomics 15 (1), 628. doi: 10.1186/1471-2164-15-628
Keywords: Rap2a, Wnt, NF-κB, growth, shrimp
Citation: Wang S, He M, Li Q, Li H, Yin B, He J and Li C (2022) Rap2a Is Negatively Regulated by NF-κB and Contributes to Growth via Wnt Pathway in Shrimp. Front. Mar. Sci. 9:942009. doi: 10.3389/fmars.2022.942009
Received: 12 May 2022; Accepted: 10 June 2022;
Published: 15 July 2022.
Edited by:
Marcel Martinez-Porchas, Consejo Nacional de Ciencia y Tecnología (CONACYT), MexicoReviewed by:
Jianbo Yuan, Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaGloria Yepiz-Plascencia, Consejo Nacional de Ciencia y Tecnología (CONACYT), Mexico
Copyright © 2022 Wang, He, Li, Li, Yin, He and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaozheng Li, bGljaGFvemhAbWFpbDIuc3lzdS5lZHUuY24=; Jianguo He, PGEgaHJlZj0ibWFpbHRvOmxzc2hqZ0BtYWlsLnN5c3UuZWR1LmNuIj5sc3NoamdAbWFpbC5zeXN1LmVkdS5jbjwvYT4=
 Sheng Wang
Sheng Wang Mingyu He1,2,3
Mingyu He1,2,3 Haoyang Li
Haoyang Li Jianguo He
Jianguo He Chaozheng Li
Chaozheng Li