- 1Key Laboratory of Applied Marine Biotechnology, School of Marine Sciences, Ningbo University, Ningbo, China
- 2National Engineering Research Center for Facilitated Marine Aquaculture, Marine Science College, Zhejiang Ocean University, Zhoushan, China
- 3Mariculture Research Department, Marine Fisheries Research Institute of Zhejiang Province, Zhoushan, China
Sepia pharaonis is an excellent candidate for aquaculture in China. However, the low survival rate during early feed transition is a bottleneck restricting industrial development. Understanding the changes in digestive physiology and intestinal microflora during feed transition should enable us to meet their nutritional needs to improve production. In this study, we investigate the digestive enzyme of S. pharaonis and undertake histological observations of the digestive gland and intestine. The intestinal microflora 16S rRNA genes were also analyzed using high-throughput sequencing of the pre, mid, and post-feed transition stages (20, 40, and 60 days post-hatching (DPH), respectively). The digestive enzymes from the digestive gland (trypsin and chymotrypsin) rapidly decrease at 40 DPH when compared to their levels at 20 DPH, but mostly recovered by 60 DPH. The alkaline phosphatase and lipase increased sharply by 40 DPH, then peaked at 60 DPH. The intestinal digestive enzymes followed similar trends during feed transition, except for lipase activity, which decreased after 20 DPH and remained low, even at 60 DPH. Feed transition affects the morphogenesis of the digestive tract and feed transition stress leads to the impairment of the digestive gland and intestinal morphology, which reduces the digestive capacity, but almost totally recovers by 60 DPH. Moreover, the comparison of the intestinal microbial composition during feed transition revealed that the dominant phylum Bacteroidetes gradually increased to a peak at 40 DPH and then decreased until 60 DPH. The microbial composition changed with the most abundant genus Pseudomonas being replaced by Acinetobacter. The phylum and family level investigation suggested the microbiota in the rearing water had limited influence on the intestinal microbiota. The intestinal microbiota diversity increased during feed transition. This study improves our understanding of changes and adaptations in cuttlefish during feed transition.
1. Introduction
In recent decades, cephalopods have become candidate species for aquaculture diversification as reported by several researchers (Lee, 1994; Sykes et al., 2014). Cuttlefish, including Sepia officinalis and Sepia pharaonis, are farmed using alternative diets throughout their life cycle (Forsythe et al., 1994; Minton et al., 2001; Domingues et al., 2003; Jiang et al., 2018). However, bottlenecks in the life cycle prevent their transition from pilot-scale to full-scale aquaculture production. The main issue is the limited existing knowledge on cephalopod digestive physiology and nutrition (Navarro et al., 2014; Gallardo et al., 2017). Feed transition is a key step in juvenile rearing, and an important part of large-scale production. The successful transition from live feed to frozen food depends on the quality of the supplied food (i.e., palatability, digestibility) and the species itself (i.e., animal size and digestive system developmental phase). Likewise, it is well known that cuttlefish must be fed live prey as they develop after hatching (Sykes et al., 2013). Cuttlefish accept frozen food (shrimp, fish, or crabs) after the initial hatchling stage (DeRusha et al., 1989; Forsythe et al., 1994; Jiang et al., 2018). Our latest research on optimal weaning protocols for cuttlefish juveniles has validated this on a commercial scale and represents a major step towards improving rearing techniques for S. pharaonis (Jiang et al., 2021). However, the dynamic changes in the digestive gland and the adaptability of the intestine from pre-feed transition to post-feed transition remain largely unexplored. Additionally, the digestive gland is complex, with unsynchronized maturation of the different functions throughout the gland (Boucaud-Camou and Boucher-Rodoni, 1983; Pereda et al., 2009).
The digestive system of cephalopods is highly developed, resembling vertebrates with complete digestive functions (Boucher-Rodoni and Boucaud-Camou, 1987; Safi et al., 2018). Cephalopod digestion begins in the buccal mass, where prey is mechanically broken down and the chyme passes through the esophagus to the stomach, cecum, and intestine, and is excreted through the anus (Martínez et al., 2012). The chyme moves into the stomach and then into the cecum, where enzymes secreted by both tissues further digest the chyme. Subsequently, the degraded chyme (which is now in a liquid form, containing fine suspended particles) resulting from the stomach digestion passes through the vestibule and enters the cecum, while the larger undigested pieces move to the intestine (Morishita, 1972; Boucaud-Camou & Boucher-Rodoni, 1983). The digestive gland is a key component of the cephalopod digestive system and performs many functions, such as synthesis and secretion of digestive enzymes, re-absorption of nutrients, and synthesis and storage of glycogen (Rosa et al., 2005, Martínez et al., 2011). During the digestive process, undigested pieces combine with acidic enzymes secreted by the digestive gland to form chyme, which initiates extracellular digestion (Boucaud-Camou et al., 1976; Pereda et al., 2009; Safi et al., 2018). Investigating the activities of the digestive enzymes allows us to understand the development and maturation of the digestive system during hatchling ontogeny. This knowledge should allow us to replace live prey with frozen food at the correct stage to improve survival in large-scale production of weaned juveniles (Pereda et al., 2009). Cephalopods are carnivorous species; therefore, the dominant enzymes should be a class of proteases, with non-specific proteolytic, alkaline, and acid phosphatase activities (Perrin et al., 2004; Safi et al., 2018). Moreover, neither the histological changes in the digestive gland of feeding cuttlefish nor the cytology of the gland during the digestive process has been quantified. Especially in the feed transition process, revealing these structural changes is essential for understanding digestive cell functions and the exact role of the gland in digestion. Detailed knowledge of the development of digestive enzyme activities will help facilitate a successful feed transition process.
The intestine of the cuttlefish is simple and straight, with the crude residue of the prey entering the intestine from the stomach and cecum (Lobo-da-Cunha, 2019). Few studies have analyzed the microbial diversity in Octopus species (De la Cruz-Leyva et al., 2011; Iehata et al., 2015; Roura et al., 2017), and no information is available on cuttlefish intestinal communities. Intestinal communities are closely linked with the development of the intestinal tract, nutrition, and immune responses of the host animal (Rawls et al., 2004; Sabour et al., 2019). The intestinal microbiota of new hatchlings and juveniles are susceptible to being affected when feeding as the digestive tract develops (Wang et al., 2020; Ivarsson et al., 2022). Therefore, a healthy and resilient intestinal microbiota is essential for maintaining health in captivity.
To date, detailed studies on S. pharaonis digestive physiology have not been undertaken. Previous studies involving S. officinalis and Octopus maya revealed a transition from intracellular to extracellular digestion in digestive gland proteases activities (acid and alkaline), which was related to individual size and food intake (Perrin et al., 2004; Martínez et al., 2012). Thus, the objectives of the present study were (1) to study the changes in the digestive system, digestive enzyme activity and morphology during feed transition; and to (2) investigate the characteristics of intestinal community dynamics during early development in cuttlefish.
2. Materials and methods
2.1 Animals and housing
The current study occurred in Lai Fa Aquaculture Co. Ltd (29°59’ N, 121°99’ E), which specializes in aquatic technological research and application development. S. pharaonis eggs were obtained by artificial breeding. In brief, brood-stock individuals with a male:female ratio close to 1:3 was placed in square concrete tanks (7.8 m × 3.8 m × 1.6 m, length × width × depth; area: 30 m2), at a culture density of 8–10 ind/m2 during the spawning season from February to April in 2021 (Jiang et al., 2022). These cuttlefish were fed with frozen fishes (Larimichthys polyactis and Pampus argenteus) and white shrimp (Penaeus vannamei) twice a day (fed ad libitum), with a feeding rate of 3–5% body weight (bw) d-1 (Jiang et al., 2018). The fertilized eggs were incubated in hatching tanks (8.0 m length × 4.0 m width × 1.6 m depth, area: 32 m2) supplied with sand-filtered and ultraviolet disinfected seawater in a natural photoperiod cycle. The incubation conditions were as follows: salinity of 28 ± 0.8 ‰, water temperature was 24.8 ± 0.7 °C, light intensity was 558.2 ± 69.3 lx, and dissolved oxygen was 6.34 ± 0.28 mg/L. The water quality parameters were measured daily with a YSI Pro DSS instrument, and 30% of the seawater was renewed daily under the same conditions.
Two hundred and ten newly hatched (1-day post hatching, DPH) cuttlefish were randomly collected from the hatching tanks and carefully translocated into fiberglass cylindrical tanks (diameter: 1 m, water depth: 80 cm, volume: 500 L) as described previously in Jiang et al. (2020b). The experimental design included three replicate tanks with 70 individuals assigned to each replicate. Newly hatched juveniles were reared following the methods of Jiang et al. (2020b). Briefly, hatchlings were fed enriched live Artemia (Artemia nauplii) during the first 3 days post-hatching, and then fed live mysids (Neomysis orientalis) twice a day (08:00 and 16:00) until feed transition began. Each tank was gently aerated by an air stone and one airlift. The uneaten food and dead animals were siphoned from the bottom of the tanks daily. Natural seawater was filtered through a filter bed then sterilized using UV light sterilizers before being pumped into the tank. The water parameters were maintained as follows: salinity at 28.5 ± 0.5‰, the temperature at 25.3 ± 1.2 °C, light intensity at 403.8 ± 55.6 lx, and dissolved oxygen at 6.42 ± 0.37 mg/L.
Frozen food was provided to juveniles from 30 DPH, following previously prescribed weaning protocols (Jiang et al., 2021). We followed the optimal weaning protocols, with the ideal prepared food of frozen shrimp (Leptochela gracilis), the feeding frequency was 4 meals d-1, the food ratio was 35% b.w.day-1, and the culture density was 70 cuttlefish m-2. Cephalopods are sensitive to starvation because of their high metabolic and growth rates, which relates to their “live fast, die young” life history. Juvenile cuttlefish can fully adapt to frozen food at 14 days after weaning, but the transition to digestive enzyme activity may not be completely developed at this stage (Jiang et al., 2021). To ascertain the changes in digestive enzyme activity and digestive physiology of cuttlefish during feed transition, the experimental process was divided into three stages, the pre-feed transition period (mysid feeding period, MF), the feed transition period (FT), and the post-feed transition period (PFT). The mysid feeding period (MF) was defined as the newly hatched juveniles from 1-day post-hatching (DPH) to 30 DPH, during which they were fed with live mysids. The feed transition period (FT) was determined as the juvenile growth from 31 DPH to 50 DPH, during which they were fed with frozen shrimp. The post-feeding transition period (PFT), at which point the cuttlefish are defined as juveniles, continued rearing to 60 DPH after the feed transition, during which they were fed with frozen shrimp. The rearing method followed the previous report outlined in Jiang et al. (2021).
2.2 Sample collection
To explore the changes in the digestive physiology from pre-feed transition to post-feed transition, samples were collected at 20 DPH (mantle length: 20.3 ± 1.4 mm, weight: 2.12 ± 0.17 g), 40 DPH (mantle length: 39.4 ± 1.2 mm, weight: 11.42 ± 0.13 g) and 60 DPH (mantle length: 56.1 ± 2.2 mm, weight: 22.84 ± 0.91 g). Six cuttlefish were sampled and all animals were directly immersed in seawater with 3–5% ethanol for 30-50 s before dissection. During the dissection, the digestive gland and intestine were removed and the samples were divided in two: one was immediately frozen with liquid nitrogen and stored at -80 °C for enzymatic analysis, and the other was fixed in 4% formalin buffer for histological analysis. The intestine was immediately frozen in liquid nitrogen and stored at -80°C for microbiome analysis. Concurrently, rearing water at each stage was sampled (200 mL/sample) and then filtered using a 0.22 μm polycarbonate membrane (Millipore, USA). Three replicate water samples were collected for each tank and stored at -80°C until required for the DNA analysis.
2.3 Digestive enzyme analyses
To prepare the enzyme extracts, digestive gland and intestinal tissue samples were manually homogenized (30:1, v/w) on ice (0-4°C) in Tris-mannitol buffer (50 mM Mannitol, 2 mM Tris-HCl, pH 7.5) for 60 s. The homogenate was centrifuged for 10 min at 11,000 rpm, at 4°C to remove the non-homogenized tissue debris. Then, the supernatant extract was used for the analysis of trypsin, chymotrypsin, alkaline phosphatase and lipase activity. All samples were kept in ice during the process described above in order to avoid enzymes denaturation and/or damage. Enzyme extracts were kept at -20°C until analysis. The total protein content in crude extracts was determined at 25°C using bovine serum albumin as a standard based on the method of Bradford (1976). Samples were assayed in triplicate in 96-flat bottom micro-plates and read at 595 nm. The activity of enzymes was measured with commercial assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the protocol described by the manufacturer.
Trypsin (TRYP) activity was determined using Nα-Benzoyl-L-Arginine ethyl ester Hydrochloride as a substrate to a concentration of 20 μM. For analysis, 5 μL of this substrate, 190 μL of 50 mM Tris +10 mM CaCl2 buffer (pH 8.5), and 15 μL of the tissue homogenate were added to the microplate (Kakade et al., 1969). Samples were incubated at 37°C (20 min) and read at 253 nm, the enzyme activities were expressed in terms of unit per mg of total protein (specific activity).
Chymotrypsin activity (CHT) was assayed following the method described by Applebaum et al. (2001), using 0.56 mM of BTEE (N-Benzoyl-L-tyrosine ethyl ester, Sigma 13,110-F) as substrate in 100 mM Tris-HCl buffer, 25 mM CaCl2, pH 7.8 and methanol 2.5% (v/v) at 37°C. The absorbance of the solution was monitored at 540 nm. The reaction was recorded every minute for 30 min in a 96-well plate, and the enzyme activity was expressed as unit per mg of total protein (specific activity).
Alkaline phosphatase (ALP) activity was measured using a modified version of the method described by Nigam (2007). In brief, using p-nitrophenyl-phosphate 2% as substrate in a 1M Tris buffer at pH 10 for ALP, 10 μL of supernatant was added to 10 μL of the substrate in a 96-well flat-bottom plate, and the mixture was incubated at 37°C for 15 min. The absorbance was assessed at 405 nm, the enzyme activity was expressed as unit per mg of total protein (specific activity).
Lipase activity (LPS) was determined as described by Versaw et al. (1989). The reaction mixture consisted of 50 mM Tris-HCl buffer (pH 7.2), 100 mM sodium taurocholate, 200 mM b-naphthyl caprylate, and enzyme suspension. The mixture was then incubated at 25°C for 30 min, and 100 mM fast blue was added to produce a chromogenic reaction. The reaction was terminated with the addition of 5% TCA, and the solution was clarified by adding ethanol/ethyl acetate (1:1 v/v). The absorbance of the solution was read at 540 nm, the enzyme activity was expressed as unit per mg of total protein (specific activity).
2.4 Histological analysis
The digestive gland and intestinal tissue samples were consistently removed from the same anatomical site (referring to the same tissue site at different time sampling points) to compare tissue variations during feed transition. The tissue histology was performed following the methods described by Banchroft et al. (1996). The standard microstructures were stained with hematoxylin and eosin (H&E) dye. The sections were prepared on glass slides, dehydrated by immersion in serial dilutions of ethyl alcohol-water mixture, cleaned in xylene, and embedded in paraffin wax following standard procedures. Sections (3–5 μm) were cut using a rotatory microtome and placed on gelatin-coated slides. Histological images were obtained using a light microscopy Olympus Bx53 (Olympus Corporation Tokyo Japan) and analyzed using a digital image analysis software package (Soft Imaging Systems GmbH, Münster, Germany). For each section identification, we analyzed three to five image fields from each of the five specimens in each replicate.
2.5 Intestinal microbiota community analysis
The intestine of two cuttlefish were pooled in one sample, and a total three samples were collected at each stage for microbial community analysis. Total intestinal and water samples bacterial DNA was isolated using the E.Z.N.A.™ Stool DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s protocols. The 16S sequencing of the intestinal community was performed according to our previous studies (Sheng et al., 2021). Briefly, after total bacterial community DNA was extracted and their quality and quantity were checked and estimated, the V3 + V4 region of the 16S rRNA was amplified by PCR with universal prokaryotic primers (341F: CCTAYGGGRBGCASCAG, 806R: GGACTACNNGGGTATCTAAT) and sequenced using a HiSeq 2500 sequencing platform (Illumina, USA) at Biomarker Technologies Corporation (Beijing, China). All PCR reactions were carried out in 30 μL reaction buffer with 15 μL of Phusion R High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers, and about 10 ng template. The PCR program for each sample was as follows: 95°C for 4 min, followed by 27 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C. for 45 s, and a single final extension step of 72°C for 10 min. PCR products were mixed and extracted from 2% agarose gels and then purified using the GeneJET™ Gel Extraction kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions. The library templates were enriched by PCR amplification and the library was sequenced on the HiSeq platform (Illumina, USA).
To obtain high-quality clean reads, the raw tags (paired-end sequences) were filtered by Trimmomatic to obtain clean reads (Bolger et al., 2014). Subsequently, the qualified double-ended raw data were spliced to obtain paired end sequences with a maximum overlap of 200 bp using Flash (Magoc and Salzberg, 2011). Clean tag sequence was then obtained by using the split libraries software in QIIME (Caporaso et al., 2010) to remove sequences containing N bases in the paired-end sequences, single base repeat sequences greater than six, and sequences with a length less than 200 bp. Then, the USEARCH (Edgar et al., 2011) software was used to filter the raw reads based on quality, removing chimeric sequences and non-bacterial sequences, cluster reads with similarities ≥ 97% were used as operational taxonomic units (OTUs), and to assemble an OTU table. To obtain the species classification information corresponding to each OTU, QIIME (http://qiime.org/scripts/assign_taxonomy.html) and RDP Classifier (Wang et al., 2007) (version 2.2 http://sourceforge.net/projects/rdp-classifier/) were used for taxonomic analysis of representative sequences at the 97% similarity OTU level, with a confidence threshold of 0.7, using Silva database (Quast et al., 2012) (Release 138 http://www.arb-silva.de) and RDP functional gene database collation FGR from Gene Bank (Fish et al., 2013) (Release 7.3 http://fungene.cme.msu.edu/). OTU abundance information was normalized by subsampling to the lower depth (no. of reads you normalize/subsample) to equalize library sizes.
2.6 Statistical analysis
All data are presented as the mean ± standard deviation (SD). One way analysis of variance (ANOVA) for enzyme activity results, diversity indices and OTU richness was performed using SPSS software (SPSS, Chicago, IL, USA). When there were significant differences, the group means could be further compared using Tukey’s multiple range test. P < 0.05 was considered to indicate statistical significance. Alpha diversity analysis was performed in Mothur (version v.1.30.1 http://www.mothur.org/wiki/Download_mothur). The index of community richness was determined using the Chao1 estimator (http://www.mothur.org/wiki/Chao). The indices for calculating community diversity were the Shannon index (http://www.mothur.org/wiki/Shannon) and Simpson index (http://www.mothur.org/wiki/Simpson). The sequencing depth index was calculated as good’s coverage (http://www.mothur.org/wiki/Coverage), which determines the classification richness estimator and community diversity. R (version R 4.1.1) calculates the Bray-Curtis distance matrix, and the R language vegan software package performs non-metric multidimensional scaling (NMDS) analysis and mapping. Permutation multivariate analysis of variance (PERMANOVA) combined with permutation multivariate analysis of dispersion (PERMDISP) was used to assess statistical significance in beta diversity by sample type (water vs intestine) and feed transition period (mysid feeding period to post-feeding transition period).
3. Results
3.1 Digestive enzyme activities
Digestive gland specific trypsin activity was initially detected at 20 DPH (4.55 ± 0.12 U/mg prot), and the activity had rapidly decreased by 40 DPH (2.27 ± 0.18 U/mg prot), followed by a marked increase by 60 DPH (4.65 ± 0.09 U/mg prot) (Figure 1). In general, trypsin activity was significantly lower in the intestine than in the digestive gland, whereas the change in trends was similar during feed transition.
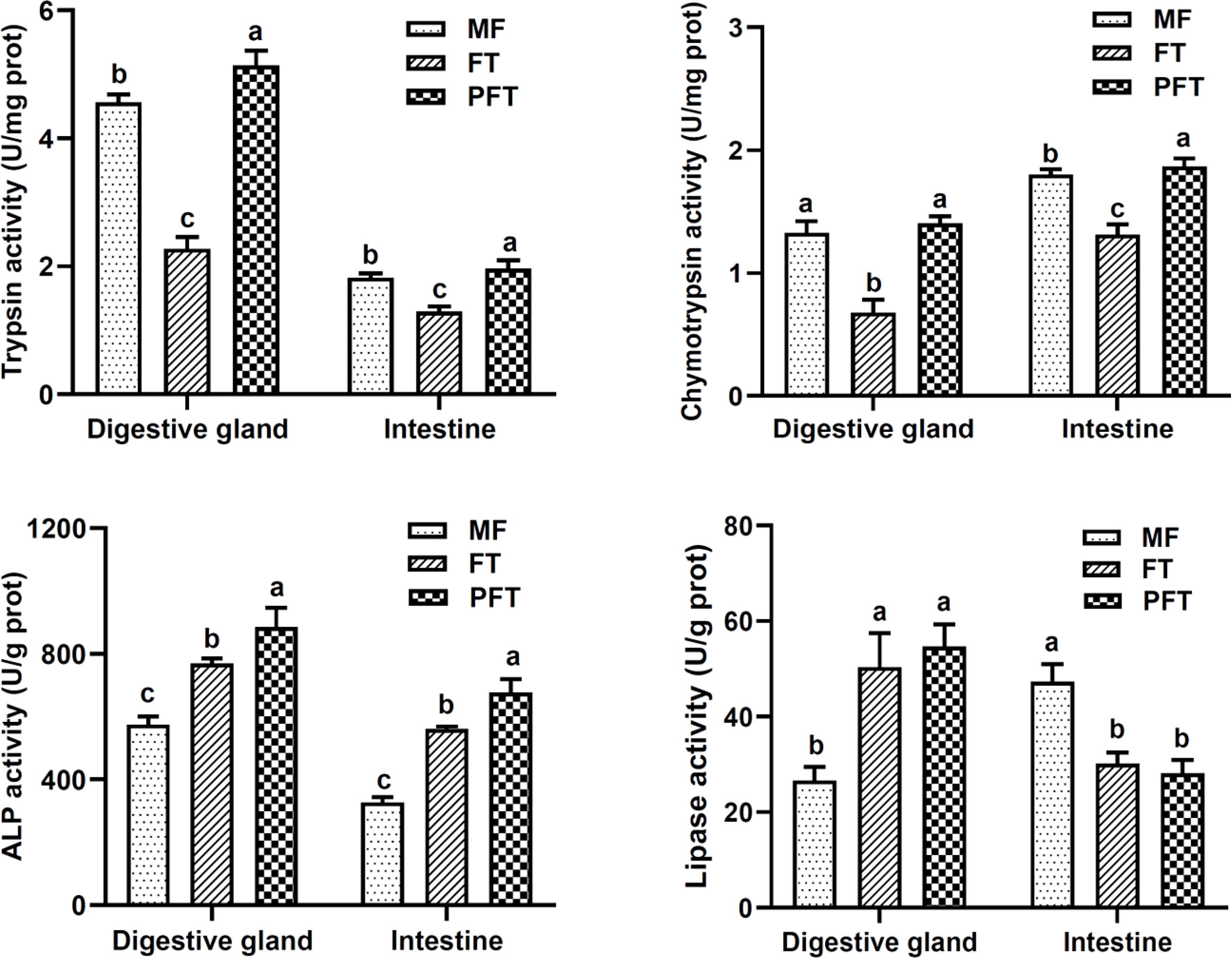
Figure 1 Changes of trypsin, chymotrypsin, lipase and alkaline phosphatase (ALP) in the digestive system from pre-feed transition to post- feed transition. 20 DPH, 40 DPH and 60 DPH in the figure correspond to the mysid feeding period (MF), feed transition period (FT), and the post-feed transition period (PFT), respectively. Different lowercase letters in tissue indicate significant differences among groups.
The digestive gland specific chymotrypsin activity was initially detected at 20 DPH (1.32 ± 0.09 U/mg prot) and exhibited a slight decrease by 40 DPH (0.67 ± 0.11 U/mg prot), and progressively increased until 60 DPH (1.40 ± 0.05 U/mg prot) (Figure 1). Similarly, intestinal specific chymotrypsin activity decreased at 40 DPH and reached its highest observed value the end of the experiment (60 DPH).
The digestive gland specific alkaline phosphatase (ALP) activity was initially detected at 20 DPH (575.10 ± 25.78 U/g prot), then increased sharply by 40 DPH (769.29 ± 15.46 U/g prot), and peaked at 60 DPH (885.37 ± 61.53 U/g prot) (Figure 1). Similarly, intestinal specific alkaline phosphatase (ALP) activity increased sharply after the pre-feed transition stage. The ALP activity continued to increase, and reached its maximum at 60 DPH.
The digestive gland lipase activity was detected at low levels at 20 DPH (26.61 ± 2.84 U/g prot) and had increased sharply at 40 DPH (50.36 ± 7.12 U/g prot), then it had increased slightly at 60 DPH (55.65 ± 4.63 U/g prot) (Figure 1). However, the change in the lipase activities in the intestine showed opposite trend in the digestive gland during the experimental period, it decreased gradually until the end of the experiment (28.09 ± 2.85 U/g prot), with the maximum activity observed at 20 DPH (47.28 ± 3.66 U/g prot) (P < 0.05).
3.2 Histological analyses
The comparative histology of the digestive glands of S. pharaonis at 20 DPH, 40 DPH, and 60 DPH are provided in Figure 2. At 20 DPH, the digestive gland was composed of digestive tubules which were roughly polyhedral, densely packed, and supported by fibrous tissue attached to the inner wall, forming a hive-like arrangement. In addition, the digestive tubules had an irregular lumen with many blood vessels between the tubules. At 40 DPH, the basal lamina boundary between the digestive tubules was unclear, the digestive tubules were filled with “boules”, several basal cells appeared in apoptosis, and the excretory vacuoles were formed. During the post-feed transition period (60 DPH), the digestive tubules were well-structured, digestive gland contained more basal cells, digestive cells, and brownish bodies, and the hemocytes were clearly visible in the inter-tubule connective tissue.
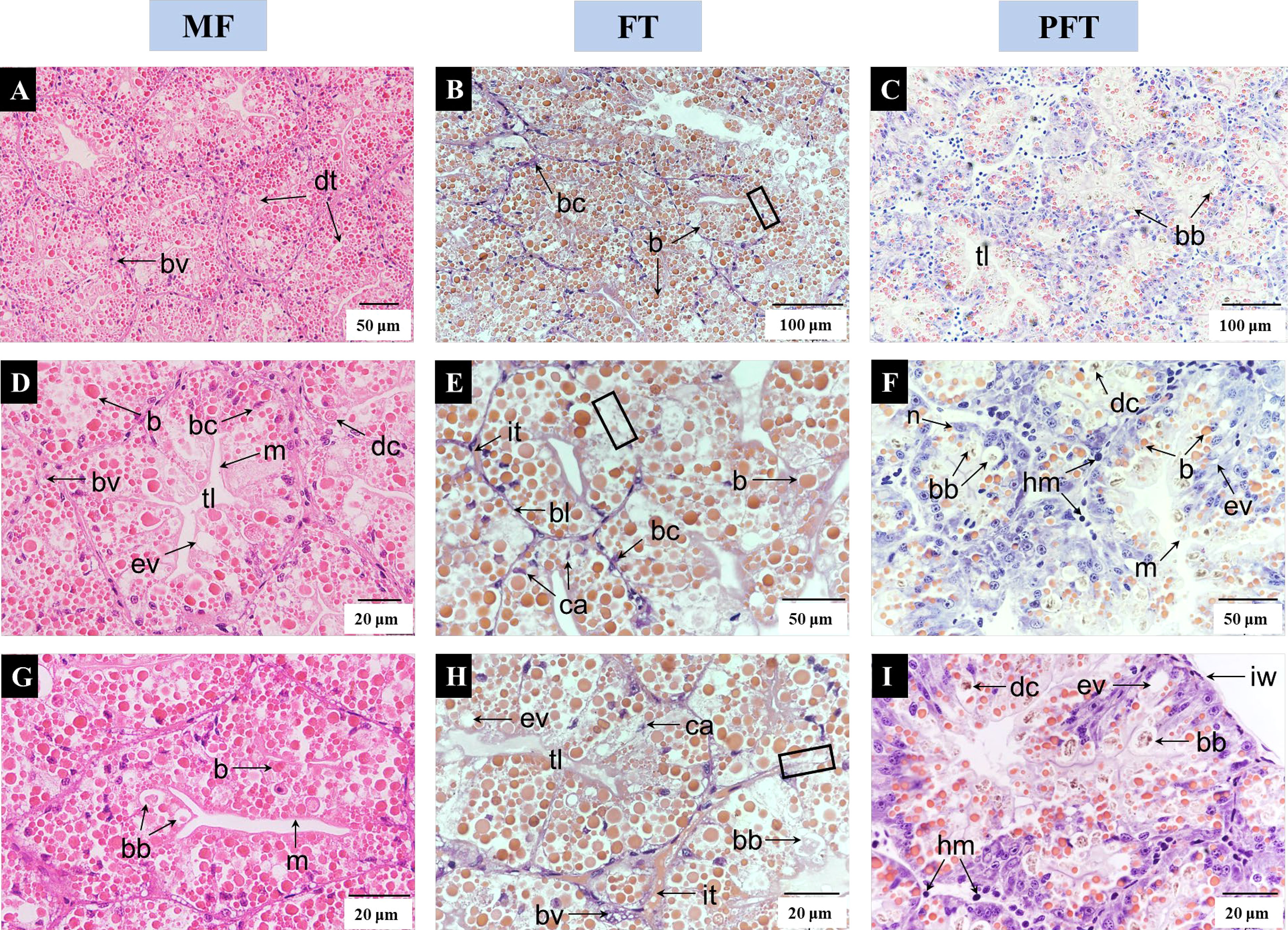
Figure 2 Comparative histology of the S. pharaonis digestive gland stained with hematoxylin and eosin (H&E). 20 DPH (A, D, G), 40 DPH (B, E, H), and 60 DPH (C, F, I) represent individual digestive tubules at the mysid feeding period (MF), feed transition period (FT), and the post-feed transition period (PFT), respectively. Three distinct cell types are found in the digestive tubule, namely basal cells (bc), secretory cells (the secretions are “boules” and “brownish bodies”) and digestive cells (dc). The black border of the digestive gland at 40 DPH indicates that the basal lamina boundary between the digestive tubules was unclear. b “boules” (proteinaceous inclusions characteristic of most cephalopods); bb, brownish bodies; bc, basal cells; bl, basal lamina; bv, blood vessel; ca, cell apoptosis; dc, digestive cells; dt, digestive tubules; ev, excretory vacuole; tl, tubule lumen; hm, haemocytes; it, inter-tubular connective tissue; iw, inner wall; m, microvillus; n, nucleus.
The panoramic view of the intestine showed the serosa located in the epidermis, and closely attached circular muscles and longitudinal muscles, with typhlosoles and longitudinal folds extending to the cavity (Figure 3). Initially, at 20 DPH, the single columnar epithelial cells were closely arranged, the mucous cells were scattered among the epithelial cells and cilia covered the epithelium. Subsequently, at 40 DPH, the single columnar epithelium had a disordered arrangement and cells were vacuolated, with some covered cilium shedding. However, at 60 DPH there was a well-developed arrangement of mucus cells, the massive aggregation of goblet cells was accompanied by the formation of immature mucous cells, and the cilium height had increased significantly when compared to 40 DPH.
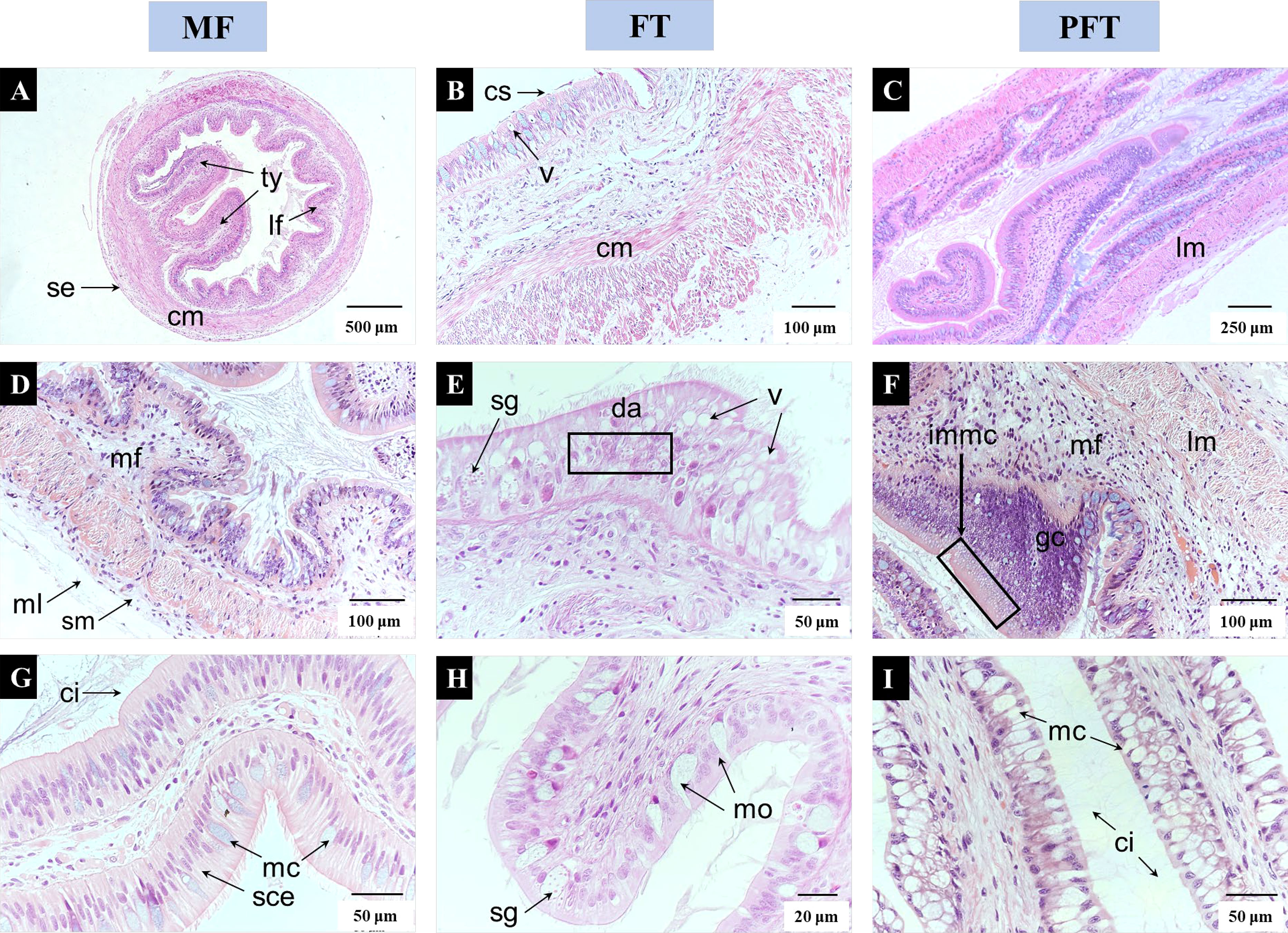
Figure 3 Light microscopic images depicting morphological changes in intestine associated to feed transition with 20 DPH (A, D, G), 40 DPH (B, E, H), and 60 DPH (C, F, I). Panoramic view of the intestine at 20 DPH, showing the serosa (se) located in the epidermis, circular muscle (cm) and longitudinal muscle (lm) are closely attached, typhlosoles (ty) and longitudinal folds (lf) extend to the cavity. A transverse section of the intestine at 40 DPH, showing some cilia appeared to shedding (cs), columnar epithelium formed vacuoles (v) and epithelium cells were disordered arrangement (da). A transverse section of the intestine at 60 DPH, showing numerous goblet cells were accumulated and accompanied by many immature mucous cells (immc), columnar epithelial mucous cells are increased, and cilium are markedly height. ci, cilia; cm, cilia shedding; cs, cilia shedding; da, disordered arrangement; gc, goblets cells; immc, immature mucous cells; lf, longitudinal folds; lm, longitudinal muscle; mc, mucous cell; ml, mucous layer; mo, mucous outflow; sce, single columnar epithelium; se, serosa; sg, secretory granules; sm, submucosa; ty, typhlosoles; v, vacuole.
3.3 Intestinal microbiota community analysis
3.3.1 Abundance and diversity analysis of the intestinal microbiota
The OTUs and alpha diversity statistics are presented in Table 1. The sequences with ≥ 97% similarity were assigned to the same OTUs. A total of 1144.67, 1161.66, 1582.67, 168, 195, and 219 OTUs were generated in mf-W, mw-W, pw-W, mf-I, mw-I, and pw-I groups, respectively. As the number of sequences increased, the Shannon–Wiener curve became flat (Figure 4), indicating that a sufficient sampling depth had been achieved for each sample. The results revealed the bacterial sequences in the water samples were significantly higher than those in the intestine. The number of bacterial sequences in the evaluated samples fluctuated from 99410 to 142,007, and 4652 to 18716, respectively (Table 1). We calculated the OTUs richness, coverage, and diversity richness of each data set. The alpha diversity indexes in the pw-W group were significantly higher than the other groups (P < 0.05). There were no significant differences observed among the intestinal sample groups (P > 0.05), except for the richness index, which was lower in the mf-I group. The Good’s coverage of each sample was > 99%, indicating the identified sequences represented most of the bacteria in each sample.
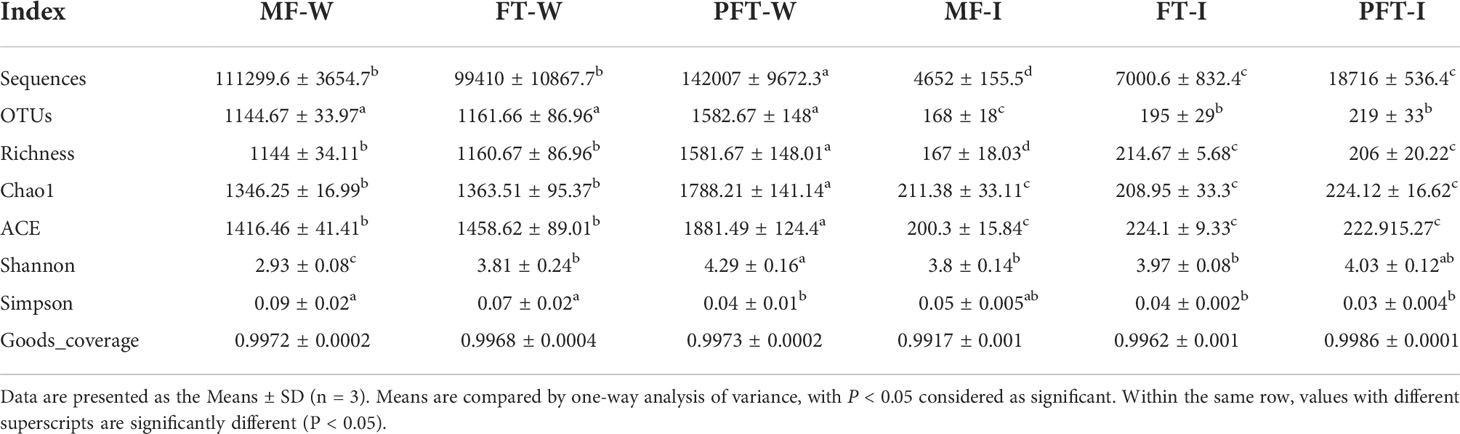
Table 1 Diversity indices and OTU richness for the intestinal bacterial diversity analysis of S. pharaonis during the feed transition.
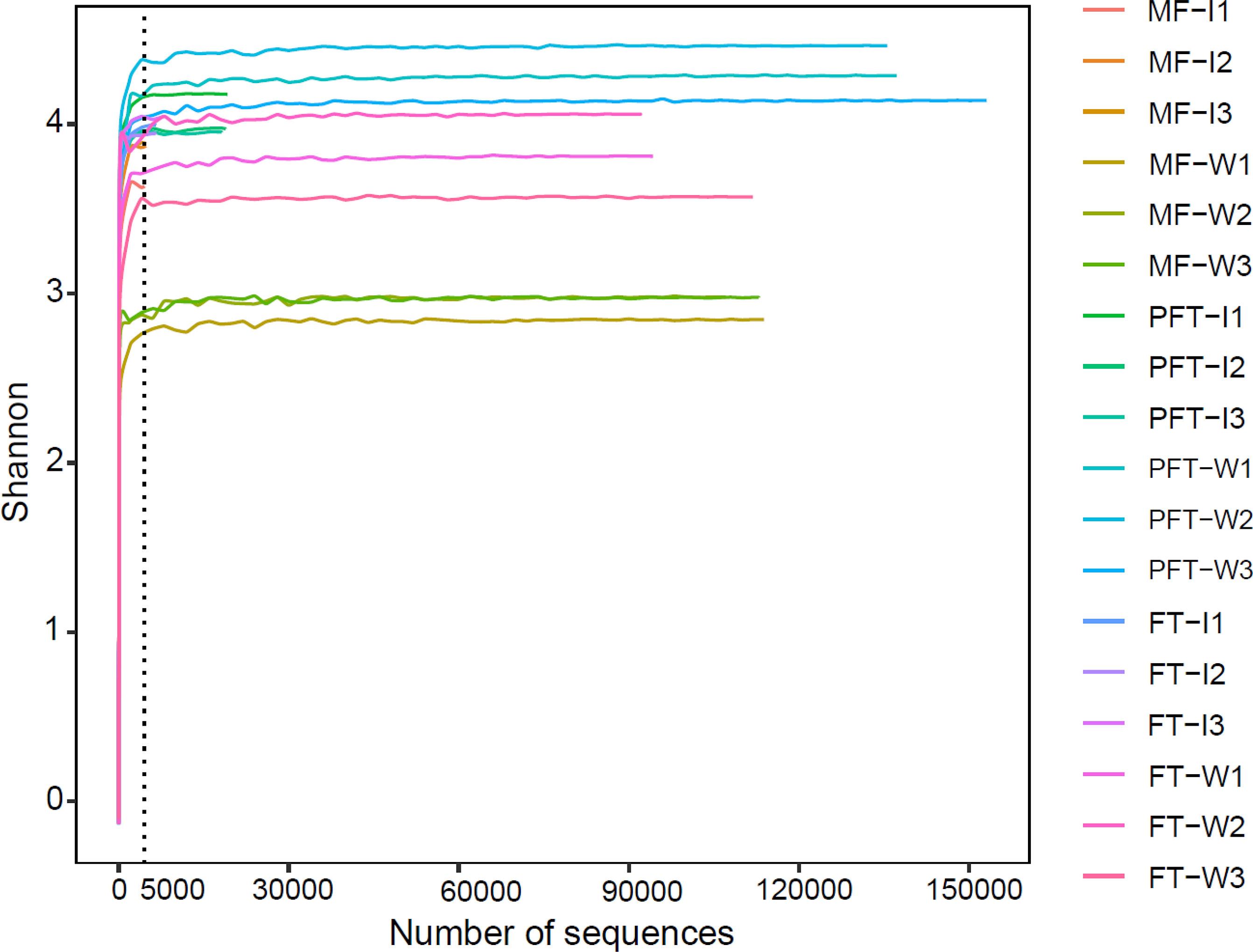
Figure 4 Shannon Wiener curves of the sample bacterial community of S. pharaonis from the mysid feeding period (MF), feed transition period (FT), and the post-feed transition period (PFT). W and I were surrounding water and intestinal samples, respectively. Operational taxonomic units (OTUs) were clustered based on 97% sequence similarity. The number of reads sampled indicates the number of sequencing reads.
3.3.2 The integral structure of the intestinal bacterial community
The results of beta diversity analysis using NMDS ordination biplot revealed a clear clustering of bacterial communities based on the Bray–Curtis distance of the OTUs detected in the samples (Figure 5). In general, the NMDS 1 axis contained clearly separated clusters of the water samples and intestinal samples. The NMDS 2 axis revealed the same relationship between the clusters. There were clearly separated clusters of the pre-feed transition and post-feed transition results. Two-way ANOVA observed statistical significance (F=16.325, R2 = 0.872, P<0.001) in sample type (water/intestine) and feed transition period (mysid feeding period to post-feeding transition period). The results of the PERMDISP test showed that there was no dispersion effect (F=1.768, P=0.184) in the water and intestinal microbial communities during the feed transition period. Therefore, the bacterial communities from pre-feed transition and post-feed transition were significantly different in both the water samples and intestinal samples.
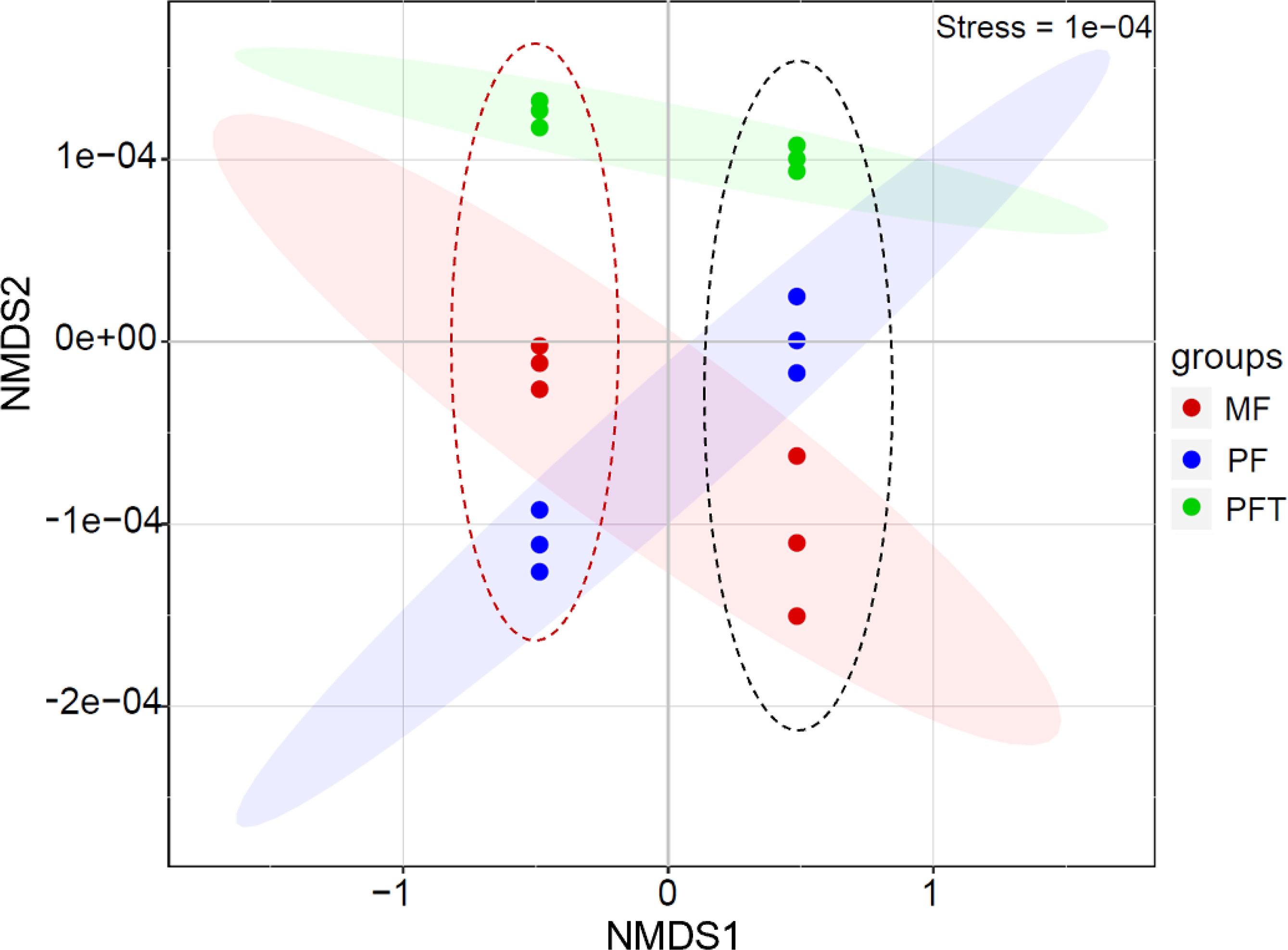
Figure 5 Non-metric multidimensional scaling (NMDS) based on Beta diversity dissimilarity among bacterial communities of the water sample (red dotted line on the left) and intestine (black dotted line on the right) of S. pharaonis from mysids feeding period (MF, red dots), the mid-feed transition period (FT, green dots), and the post-feed transition period (PFT, blue dots).
The composition and overlap of the OTUs in the water samples revealed that 82 OTUs were shared by the three groups, whereas 187, 229, and 263 OTUs were found only in groups mf-W, mw-W, pw-W, respectively (Figure 6A). The Venn diagram of the intestinal samples revealed that 39 OTUs were shared by the three groups, while 83, 100, and 141 OTUs were found only in groups mf-I, mw-I, and pw-I, respectively (Figure 6B).
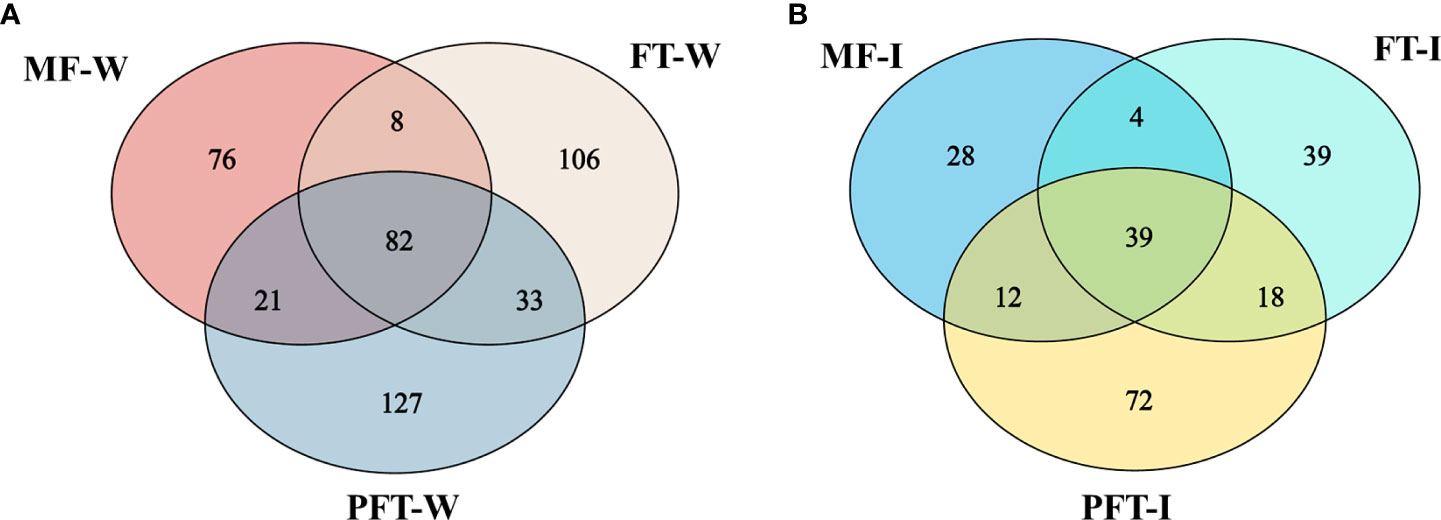
Figure 6 Venn diagram showing the unique and shared OTUs in the water (A) and intestine (B) of S. pharaonis from the pre-feed transition period (mysid feeding period, MF), the feed transition period (FT), and the post-feed transition period (PFT).
3.3.3 Microbiota composition analysis
We identified a total of 25 phyla, the top five phyla were Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes, and Tenericutes (abundance > 1%) (Figure 7). At the phylum level, the intestinal bacterial profiles of S. pharaonis were dominated by Proteobacteria (73.2 ± 0.03%) and Bacteroidetes (15.6 ± 0.02%), with the two phyla accounting for approximately 90% of the total microbiota. Moreover, the intestinal proteobacteria had the highest abundance with no significant variations among the development stages. Bacteroidetes in intestinal samples increased sharply during the mid-feed transition period and decreased in the post-feed transition period. It was notable that the abundance of Firmicutes in intestinal samples gradually decreased and Tenericutes gradually increased during feed transition. Firmicutes increased dramatically in water samples during the post-feed transition period. The relative abundance of the intestinal Actinobacteria did not change significantly, while the relative abundance of Actinobacteria in the rearing water increased sharply (Figure 7A).
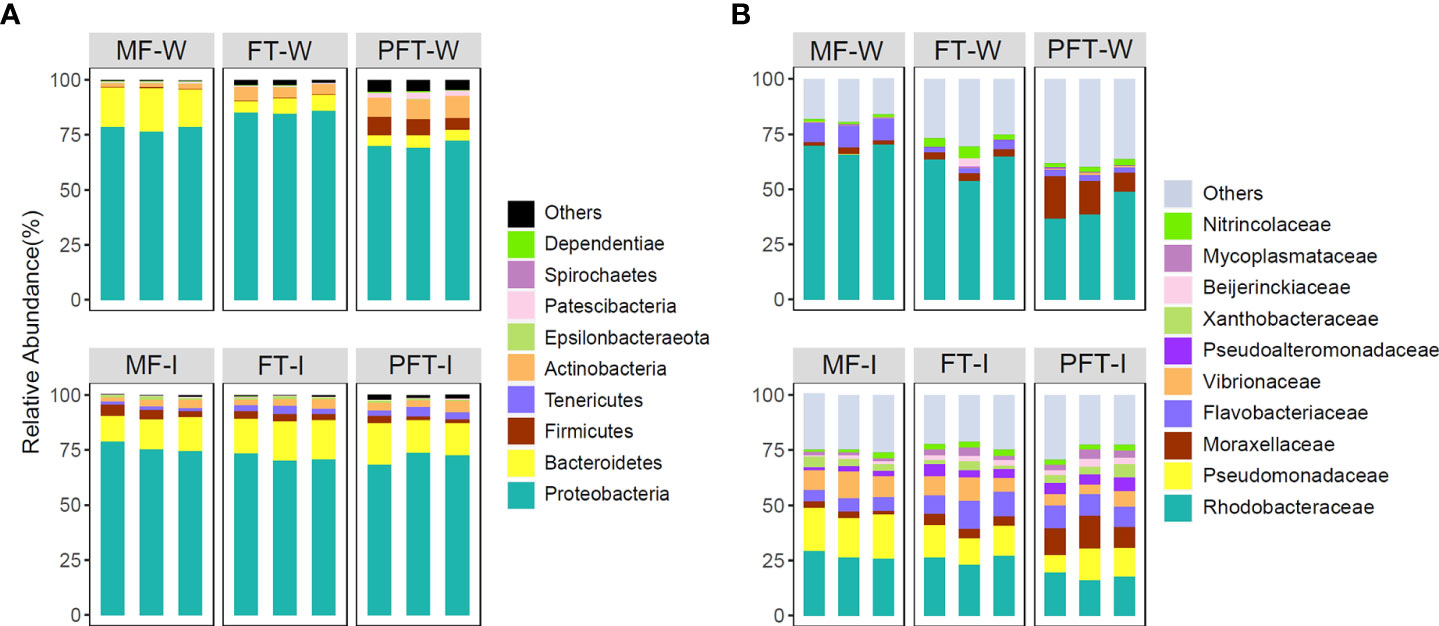
Figure 7 Bacterial composition of different communities. (A) bacterial community bar plot at the phylum level with the relative abundance (top 10); (B) bacterial community bar plot at the family level with the relative abundance (top 10).
A total of 78 family were identified in intestinal bacterial community of S. pharaonis. At the family level, the predominant families were: Rhodobacteraceae (17.8 ~ 27.2%), Pseudomonadaceae (11.6 ~ 19.1%), Flavobacteriaceae (5.7 ~ 10.8%), Vibrionaceae (5.6 ~ 10.1%), Moraxellaceae (2.5 ~ 12.3%), Pseudoalteromonadaceae (2.1 ~ 5.2%), Chitinophagaceae (0.4 ~ 3.3%), Beijerinckiaceae (1.1 ~ 2.9%), Nitrincolaceae (1.5 ~ 2.4%) and Microbacteriaceae (0.5 ~ 1.7%) (Figure 7B). Among them, the abundance of Rhodobacteraceae, Pseudomonadaceae, and Vibrionaceae gradually decreased during the feed transition stages, while the abundance of Flavobacteriaceae, Moraxellaceae, and Pseudoalteromonadaceae gradually increased. In the rearing water samples, the dominant family in each subgroup were all Rhodobacteraceae, followed by Flavobacteriaceae, Moraxellaceae, Nitrincolaceae, and Vibrionaceae. The relative abundance of Rhodobacteraceae, Flavobacteriaceae, and Moraxellaceae in the rearing water accounted for 75% of the total relative abundance, while the abundance of each family in the intestinal bacterial communities had a more diverse distribution.
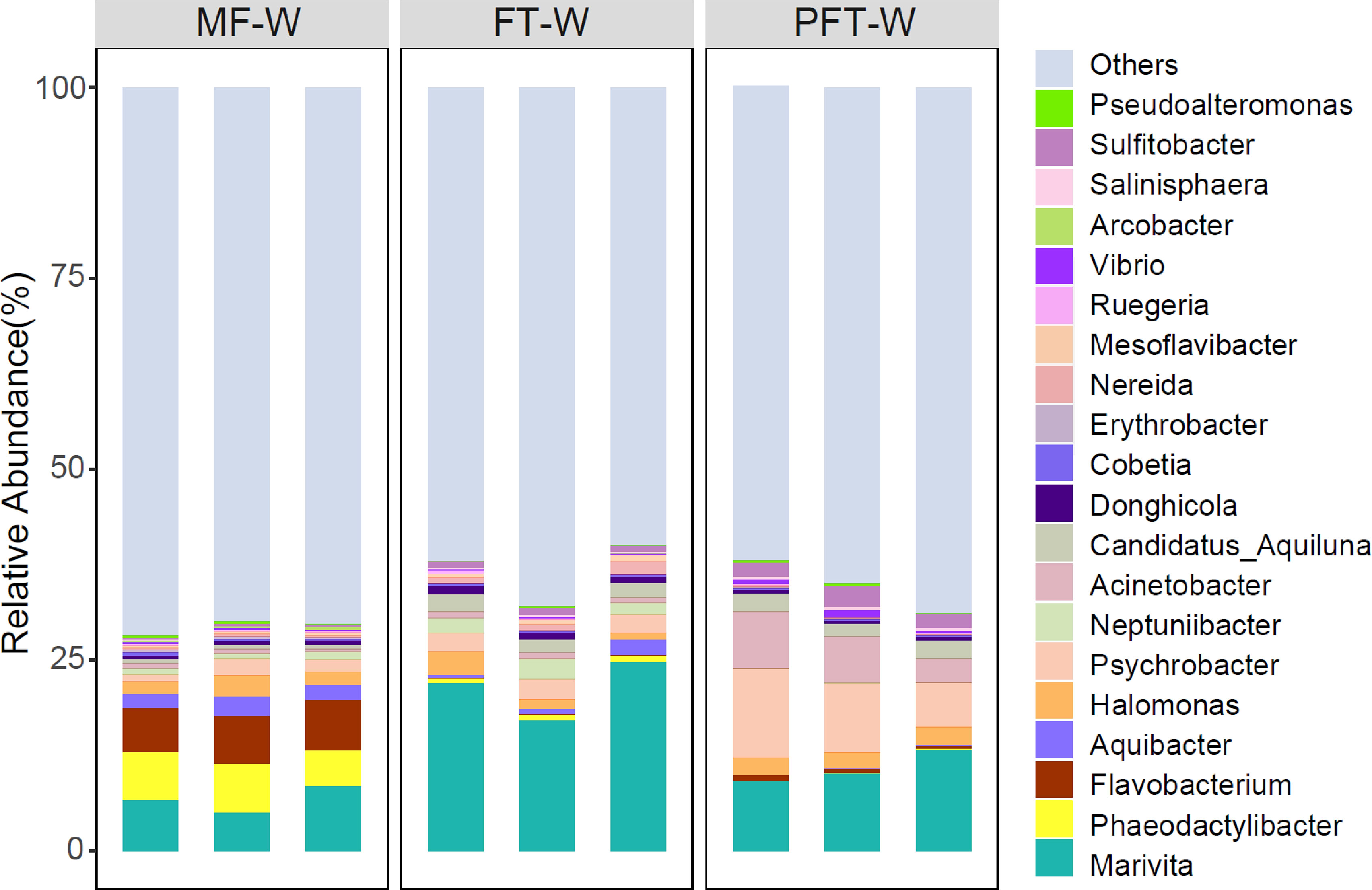
Additionally, the relative abundance of the top twenty genera from water samples are presented in Figure 8. The five most abundant genera were Marivita (6.81%), Flavobacterium (6.22%), Phaeodactylibacter (5.74%), Aquibacter (2.16%), and Halomonas (1.89%), with a combined abundance greater than 22%. The abundance of Marivita increased from 6.81% in the pre-feed transition stage to 21.33% and 10.97% in the mid and post-feed transition stage. While the abundance of Phaeodactylibacter dropped sharply from 6.22% to 0.15% and finally, 0.52%. In contrast, the abundance of Flavobacterium decreased sequentially from 5.74% in the pre-feed transition stage to 0.71% and 0.05% in the mid-and post-feed transition stages, respectively. Likewise, the abundance of Aquibacter decreased from 2.16% to 0.97%, and finally, to 0.03%. In contrast, the abundance of Halomonas remains relatively stable throughout the feed transition period (around 1.8%).
Moreover, the relative abundance of the top twenty genera (relative abundance > 1% in at least one sample) is presented in Figure 9. The top four genera are Acinetobacter (10.4%), Pseudomonas (11.64%), Vibrio (5.49%), and Pseudoalteromonas (5.04%), with a combined abundance greater than 30%. Pseudomonas abundance decreased from 19.1% in the pre-feed transition to 13.4% and 11.6% at mid-feed transition and post-feed transition. Conversely, the abundance of Acinetobacter increased from 1.3% to 2.7% and finally, 10.4%. Similarly, the abundance of Vibrio decreased from 9.9% in the pre-feed transition stage to 8.4% in the mid-feed transition and then 5.4% in the post-feed transition. The abundance of Pseudoalteromonas increased from 1.2% to 3.6% and ended at 5.0%. By comparing the water samples and the intestine of the top twenty genera, only four genera are shared in both groups, including Acinetobacter, Vibrio, Pseudoalteromonas, and Halomonas. The abundance of Acinetobacter in intestinal samples was highest (up to 10.4%) in the intestinal samples, while in the water sample it amounted to only 0.52%. Vibrio abundance ranked 4th in intestinal abundance (5.49%), while in water samples it only ranked 17th (0.19%). Similarly, Pseudoalteromonas abundance was 5.04%, ranking 5th in intestinal samples and 20th in water samples at 0.15%. Interestingly, Halomonas abundance ranked 5th with 1.89% in the water samples, but only 19th with 1.36% in intestinal samples.
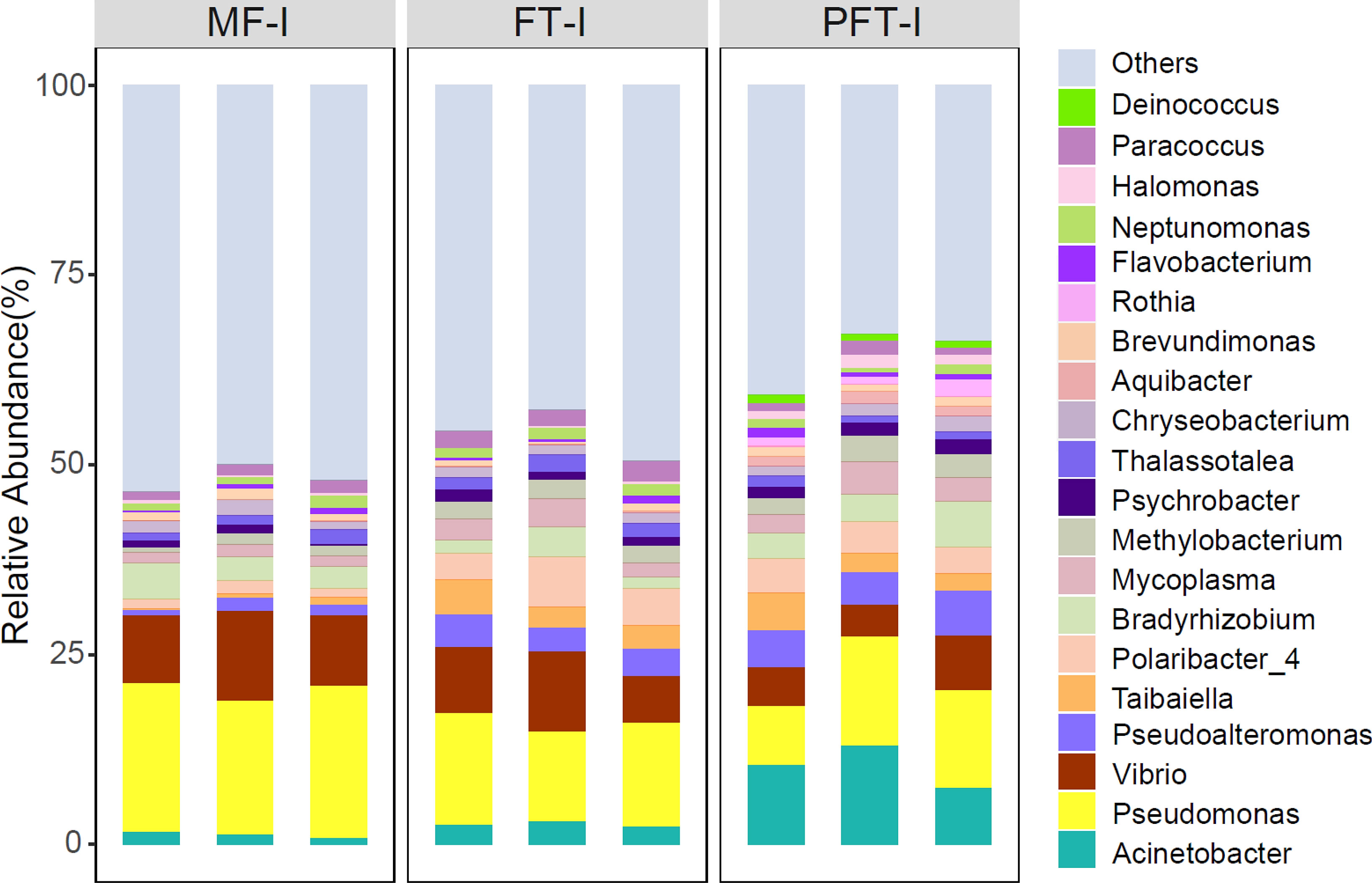
Figure 9 Relative abundances of major bacteria (top 20) in S. pharaonis intestine at the genus level. Microbial community bar plot at the genus level with the relative abundance higher than 1%.
4. Discussion
The present work evaluated the effects of pre-feed transition to post-feed transition on the digestive capacity of the digestive tracts, the progression of the histological changes of the digestive tracts, and the dynamics of intestinal communities in S. pharaonis.
The feed transition process is an extremely important period for large-scale farming, in which losses occur as individuals are stressed by abrupt changes in the quantity and quality of social interactions, nutrition, and environment. Consequently, the weaned cuttlefish must overcome these challenges, which cause imbalances in their intestinal microbiota, damages the morphology of their digestive tracts, and reduces enzyme activity, consequently inhibiting growth (Shang et al., 2020). The development of the digestive system, especially the changes in digestive enzyme activities, provides a base for a successful feed transition strategy that optimizes the digestion and absorption of food by developing correctly formulated feeds (Ma et al., 2021; Pradhan et al., 2014). Although we previously provided an optimal weaning strategy for this species (Jiang et al., 2020a), the changes in the digestive enzyme activities in specific digestive tracts as they develop remain unknown. Previous studies report that Octopus maya, Octopus mimus and Sepia officinalis exhibit unstable changes in the digestive enzymes during the maturation of the digestive glands (Martínez et al., 2012; Gallardo et al., 2017; Safi et al., 2018). In this process, extracellular digestion is performed by trypsin, chymotrypsin, and other digestion enzymes (a transition from predominantly acid intracellular digestion to extracellular alkaline digestion occurs) (Moguel et al., 2010; Safi et al., 2018). Therefore, the changes in the digestive enzyme activities (i.e., trypsin, chymotrypsin, AKP, and lipase) of S. pharaonis juveniles in this study were analyzed as indicators of digestive physiological stress. In the present study, 10 days after initiating feed transition (i.e., 40 DPH), the cuttlefish could not absorb frozen diets in their functional intestine with low trypsin activity. In terms of trypsin activity, the results showed that S. pharaonis juveniles were slightly less able to cope with changes in the nutritional composition and physical properties of the prepared feed. Safi et al. (2018) suggested protein digestion in cuttlefish juveniles mainly occurs by alkaline proteases and alkaline phosphatase. Safi et al. (2018) also suggested extracellular enzymes stabilized after 21 DPH. A stabilization phase in the enzyme activities of cephalopods is predicted to occur when the digestive system matures (Solorzano et al., 2009). In this study, the activity of trypsin decreased in the digestive gland and intestine at 40 DPH. Fasting can lead to a reduction in tissue enzyme activity. The reduction in the tissue enzyme activity could be due to lower protein and carbohydrate digestion, which may result in lower nutrient absorption (Francis et al., 2001; Fuentes-Quesada et al., 2018). During feed transition, intestinal chymotrypsin activity changes like that of the digestive gland, with the intestinal chymotrypsin almost returning to the pre-feed transition level. The results indicated that the low digestive capacity and/or poor nutrient assimilation of the S. pharaonis juvenile intestine is due to the lack of a fully functional digestive system during feed transition. Therefore, it takes time for the intestine to complete the changes, the extended timeframe may be attributed to the properties of chymotrypsin or to the degree of food utilization by the cuttlefish (the digestion and absorption of live prey and frozen food are quite different).
The alkaline phosphatase (ALP) activity revealed an ascending pattern in the digestive gland and intestine along with a considerable surge at 60 DPH. A cephalopod digestive gland is a multi-task annex, involved in the secretion of digestive enzymes, extra-and intracellular digestion, substance storage, and excretion (Pereda et al., 2009; Moguel et al., 2010; Ibarra-García et al., 2018; Safi et al., 2018). Previous studies have confirmed the cephalopod digestive system is highly efficient and mainly attributed this efficiency to the digestive gland, which produce extracellular enzymes that flow to the intestine to accelerate simultaneous digestion and absorption (Swift et al., 2005; Rodrigo and Costa, 2017; Lobo-da-Cunha, 2019). Cephalopods possess a protein-based metabolism, while cephalopod lipid metabolism has not been as extensively studied (Lee, 1994). However, lipids are also an important energy source for the body during food deprivation. In adverse environment, lipids can provide the necessary energy for the organism, and supply essential precursors for biofilm protection (Ishibashi et al., 2013; Bravo-Ruiz et al., 2021). In the present study, we observed an increase in lipase activity in the digestive gland during feed transition, with the activity peaking at 60 DPH. These results may be interpreted as follows: previous studies have reported that the digestive gland is the largest organ in cuttlefish visceral mass and have a lipid content as high as 10–12% (dry weight), suggesting that the digestive gland is the main site of lipid metabolism (Shyla et al., 2010; Jiang et al., 2019). Changes in lipase activity may be attributed to changes in food quality and tissue specificity, since the production and secretion of digestive enzymes are modulated by the digestibility of the food components and tissue metabolism levels (Zambonino-Infante et al., 2009). Considering the digestive gland is the main digestive organ, which produces extracellular enzymes that flow to the digestive tracts, future studies should use enzyme-linked immunization and specific staining marker analyses to clarify the digestive capacity of tissue-specific enzymes and the role and function of the digestive gland.
Ontogenesis histological studies are necessary to optimize rearing techniques and feeding conditions for different species. Ontogenesis studies are useful for establishing appropriate feed transition protocols to ensure healthy growth and increased production of cultured animals during the early life stages in aquaculture (Padros et al., 2011; Ghasemi et al., 2020). Numerous authors have investigated the digestive gland of cephalopods (Swift et al., 2005; Rodrigo and Costa, 2017). Most of these studies have focused on the structural composition of the digestive cells and the potential functions of cells during the digestive process (Semmens, 2002; Lobo et al., 2010; Martínez et al., 2011). We classified the cells in the S. pharaonis digestive gland into three distinct types (basal, excretory, and digestive), which is consistent with previous descriptions of the digestive glands of other mollusks (Gros et al., 2009; Costa et al., 2013). Costa et al. (2014) reported that the digestive gland of S. officinalis displays multiple functions, including digestion (intracellular and extracellular), energy storage (carbohydrate and lipid) and detoxification (accumulation of heavy metals). According to these authors, there are three types of histological cell structures, these are secretory cells with cytoplasmic droplets or “boules”, absorption cells with brown material (brownish bodies), and excretory cells with a single large vacuole (also confirmed in this study). Although the morphology of these cells is well understood, the exact biochemical processes and specific functions of each cell remain unknown. Moreover, at 40 DPH S. pharaonis possessed a blurred boundary of basal lamina between the digestive tubules, apoptosis of the basal cells appeared, and the volume of excretory vacuoles increased. The analysis of the abnormal structure of the digestive tubules and the changes in digestive enzyme levels suggests the activity of digestive enzymes could be improved by increasing the excretory vacuoles and “boules” secretory cells. The results revealed the basal cells decreased and “boules” increased during feed transition, indicating that the digestive gland undergo cellular transformation due to changes in enzymatic activity. These erythrotropic “boules” contain digestive enzymes, digesting particles, and digested material (Costa et al., 2014). In addition, at 60 DPH, the structure of the digestive gland returned to normal, basal cells increased, and hemocytes were clearly visible in the inter-tubule connective tissue, indicating an enhanced metabolic cycle (Pila et al., 2016).
The animal intestine plays a crucial role in nutrient absorption and metabolism, and represents the first line of defense against pathogens and environmental stresses (Sonnenburg and Bäckhed, 2016). Several studies have shown that changes in the feed may significantly affect the composition of the intestine microbiota and digestive enzyme capabilities (Zhao et al., 2012; Iehata et al., 2015; Zhao et al., 2020). Animals possess indigenous intestinal microbiota and stable digestive enzymes play an important role in food digestion, nutrient absorption, and the health of the host (Nayak, 2010; Ray et al., 2012). Moreover, Kar and Ghosh (2008) reported that the composition of digestive enzyme-producing microbiota may be related to their feeding habits. In this study, four important digestive enzymes were examined to assess intestinal digestive capacities during feed transition. We observed significant decreases in chymotrypsin and lipase enzymes at 40 DPH. Similar to largemouth bass, the digestive enzymes and digestibility of commercial feeds decreased during feed transition (Zhao et al., 2020). Conversely, pike perch showed no difference in the digestibility of live prey with compound feed (Hamza et al., 2007), which may correlate with the digestive capacity of different species. Evidence suggests early feed transition can lead to significant deterioration of the intestinal morphology (including villus height and number) causing reduced nutrition absorption and poor physiological performance (Li et al., 2017; Shaw et al., 2012; Braga et al., 2022). As a component of intestinal mucosal layer, intestinal villus plays an important role in the efficient function and absorption of the intestine (Jha and Berrocoso, 2015; Zhu et al., 2020). In present study, feed transition stress also led to: impaired intestinal morphology, an obvious decrease in villus height, vacuolated epithelium cells, and minor covered cilium shedding at 40 DHP. A decrease in the digestive enzymes confirmed feed transition stress. Villus height is an indirect indicator of intestinal epithelium cell function, as longer villi provide a larger nutrient absorptive area (Zhu et al., 2020). We also used optimal feed transition strategies, as earlier feed transition is associated with higher mortality, severe malnutrition, and reduced antioxidant capacity. The antioxidant capacity of an organism usually represents the host’s response to endogenous oxidative damage and improving the antioxidant capacity is beneficial for relieving feed transition stress (Wang et al., 2018c).
In recent years, many farmed animal studies have used high-throughput sequencing to identify the relationships between intestinal microbiota and individual health (Cornejo-Granados et al., 2017; Pinoargote et al., 2018). Intestinal bacteria and bacterial metabolites directly influence the physiology of both invertebrates and vertebrates, including the host metabolism, intestinal barrier integrity, immune regulation, and disease occurrence (Ivana et al., 2012; Destanie et al., 2018; De Vadder and Mithieux, 2018). The intestinal microbiota may come from a variety of sources, and in farmed animals the bacterial colonization in early developing larvae is complex and depend on the microbiota in the: eggs, rearing water, and feed (Bolnick et al., 2014; Piccolo et al., 2017; Wang et al., 2018a). Among the many influencing factors, diet is regarded to have the strongest influence on intestinal microbes (Wang et al., 2018a). In this study, the dominant phyla of intestinal microbes were Proteobacteria (73%) and Bacteroidetes (15%). Proteobacteria did not change significantly during the feed transition period, while Bacteroidetes gradually increased in the mid-feed transition period and decreased in the post-feed transition period. Previous research suggests the relative abundance of Proteobacteria reflects the health status of breeding aquatic animals (Blandford et al., 2018), and most Proteobacteria are heterotrophic, reducing the need for organic input (Bryant and Frigaard, 2006). Interestingly, the levels of both phyla were inconsistent, Firmicutes gradually decreased and Tenericutes gradually increased when fed frozen food during feed transition. Bacteroidetes are closely related to Firmicutes, with both affecting fatty acid absorption and lipid metabolism in zebrafish (Semova et al., 2012). In addition, in aquatic animals, Firmicutes in the intestine promotes nutrient absorption, while Bacteroidetes improves metabolic active and health (Kumar et al., 2019). Research investigating Octopus, showed Proteobacteria was the dominant phyla in wild and cultured animals, as well as in other species (De la Cruz-Leyva et al., 2011; Iehata et al., 2015; Zhu et al., 2020). It is widely believed that a considerable portion of the intestinal microbiota is directly affected by the water (Han et al., 2010). In this study, Proteobacteria had the highest abundance in the cultured water and intestinal system. Bacteroides and Firmicutes were the next most common in the microbiota and displayed opposite trends in the rearing water and intestinal during feed transition. Therefore, the microbiota variations between the rearing water and intestinal system may be due to the feed provided, and not all the microbiota in the water is absorbed and colonized in the intestinal system (Hou et al., 2018; Sun et al., 2018).
During the feed transition period, the intestinal microbiota showed slight dysbiosis, including the most abundant Rhodobacteraceae and Pseudomonadaceae families, which gradually decreased, while Flavobacteriaceae and Moraxellaceae gradually increased. Rhodobacteraceae has been reported to persist throughout shrimp growth stages and becomes the core microbiota in the gut (Yao et al., 2018). Moreover, Rhodobacteraceae may be a great probiotic source in shrimp culture, and various strains (Roseobacter clade) have already been successfully used in aquaculture (Balcázar et al., 2006). Meanwhile, different Flavobacteriaceae members are also known to produce high levels of carotenoids (Sowmya and Sachindra, 2015). Schleder et al. (2020) reported regulating Flavobacteriaceae abundance could be beneficial to shrimp gut health. Dominant Proteobacteria phyla, such as the Pseudomonadaceae family, were positively correlated with most pathways involved in xenobiotic biodegradation and metabolism, amino acid metabolism, and vitamins, but were significantly and negatively correlated with most carbohydrate metabolism pathways (Yao et al., 2019). To further understand the connection between changes in the intestinal microbes and host metabolism, the amino acid and lipid metabolic pathways in the metabolome were further analyzed. A correlation analysis involving the top five microbial families, revealed the changes of several major microbes are strongly correlated with the host amino acid and lipid metabolic pathways (Zhao et al., 2020). Thus, the feed transition effects on cuttlefish should be further investigated at the transcriptomic and metabolomic levels.
The beneficial effect of intestinal ecological function is dependent on the enrichment or presence of certain beneficial genera or species in the bacterial community. In this study, the most dominant genus of the intestinal microbes gradually changed from Pseudomonas to Acinetobacter with the intake of frozen food during the feed transition. Notably, Pseudomonas is the causative agent of visceral white spots disease in large yellow croaker (Zhang et al., 2014; Hu et al., 2014. Acinetobacter can biodegrade typical pharmaceutical compounds and may inhibit pathogens (Wang et al., 2018b). Moreover, Acinetobacter is required for the successful development of Stomoxys calcitrans fly larvae (Lysyk et al., 1999). Therefore, an increased Acinetobacter abundance in intestinal microbes of cuttlefish juveniles may improve immunity and antibacterial activity during the post-feed transition period. However, Acinetobacter is used as a conditional pathogen in the human intestine, and the intake of Acinetobacter may change the intestinal microbial community (Gao et al., 2020). The tolerance to and mechanisms performed by Acinetobacter may vary between organisms. The dominant genus Vibrio decreased in this study, which indicates some pathogenic bacteria reduced and intestinal immunity improved after feed transition. Several studies have reported Vibrio-induced mortalities as it compromises immunity (Hsieh et al., 2008) and causes hepatopancreatic damage (Lavilla Pitogo et al., 1998). Our results in the feed transition phase of cuttlefish showed the Vibrio microbe abundance was significantly different from another dominant genus, Pseudoalteromonas richness dramatically increased as feed transition progressed. Recent studies report that marine Pseudoalteromonas species possess a wide range of bioactivity associated with the secretion of extracellular compounds (Sivasubramanian et al., 2011). The genus Pseudoalteromonas successfully colonized the intestinal system and benefited the host. Moreover, only four dominant genera were shared by water and intestinal samples, but these dominant genera showed different trends, indicating that the microbial community in the intestine of S. pharaonis was less affected by the water environment. Therefore, we suggest the intestinal bacteria may develop an “adult” microbiota before the immune system is mature, to eliminate potential invaders and maintain health. Overall, the abundance of adverse communities decreased at both the family and genus levels, while the beneficial communities increased during the feed transition period. Therefore, intestinal microflora succession patterns should be systematically characterized at different growth stages to determine the core healthy microflora. Moreover, the dominant families and genera vary between different farmed species, and their functions in the intestine of cephalopods require further research.
In conclusion, the present study provides a comprehensive understanding of the changing dynamics in the digestive gland, the intestinal physiology characteristics, and the intestinal microbial community of S. pharaonis during feed transition. Our results show the major changes in the structure of the digestive gland and intestinal system as well as the overall digestive enzyme activity during the feed transition period (from 20 DPH to 40 DPH). In addition, the activity levels and changes in trypsin suggest it is responsive to starvation stress and may be used as an indicator of physiological digestive capacity. The histomorphology results showed the digestive gland and intestine were inordinately damaged during the mid-feed transition period. The comparison of the tissue structures was well synchronized with the digestive enzyme changes. Moreover, they improved during the post-feed transition period, indicating that the juvenile digestive physiological processes are prolonged. The phylum and family level analysis revealed the microbiota in the rearing water had a limited influence on the intestinal microbiota. Conversely, the feeding and/or feed transition physiology affected the intestinal microbiota. During the feed transition period, the juvenile intestine showed an ascension in bacterial diversity, the abundance of adverse communities decreased, while beneficial communities increased. These results lay the foundations for a deeper understanding of cuttlefish feed transition for future research to develop optimal feed transition strategies in aquaculture.
Data availability statement
The data presented in the study are deposited in the Mendeley Data repository, accession number 10.17632/5vwv6d45f8.1.
Ethics statement
Animal handling complied with the Guideline for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006), and all experimental protocols were approved by the Animal Research Ethics Committee of the Chinese Academy of Fishery Sciences. Authors are aware of the general principles stated by the Directive for the use of live cephalopod mollusks in scientific research as pointed out in several studies (Smith et al., 2013; Fiorito et al., 2015).
Author contributions
MJ: Conceptualization, Methodology, Data curation, Writing-Original draft. LX, WX, JY: Animal management, Data curation. RP, HS: Visualization, Investigation. QH, ZL: Writing-Reviewing, Editing. XJ: Provided funding, Supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Ningbo Science and Technology Bureau (2021Z007) and National Natural Science Foundation of China (32102756).
Acknowledgments
The authors are grateful to Lai Fa Aquaculture Co. Ltd. (Zhejiang Province, China) for supplying the experimental S. pharaonis cuttlefish hatchlings and providing logistical support throughout the experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Applebaum S. L., Perez R., Lazo J. P., Holt G. J. (2001). Characterization of chymotrypsin activity during early ontogeny of larval red drum (Sciaenops ocellatus). Fish Physiol. Biochem. 25, 291–300. doi: 10.1023/A:1023202219919
Balcázar J. L., de Blas I., Ruiz-Zarzuela I., Cunningham D., Vendrell D., Muzquiz J. L. (2006). The role of probiotics in aquaculture. Veterinary Microbiol. 114, 173–186. doi: 10.1016/j.vetmic.2006.01.009
Banchroft J. D., Stevens A., Turner D. R. (1996). Theory and practice of histological techniques, 4th Ed (New York, London, San Francisco, Tokyo: Churchil Livingstone).
Blandford M. I., Taylor-Brown A., Schlacher T. A., Nowak B., Polkinghorne A. (2018). Epitheliocystis in fish: an emerging aquaculture disease with a global impact. Transboundary Emerging Dis. 65 (6), 1436–1446. doi: 10.1111/tbed.12908
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolnick D. I., Snowberg L. K., Hirsch P. E., Lauber C. L., Org E., Parks B. (2014). Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat. Commun. 5 (1), 1–13. doi: 10.1038/ncomms5500
Boucaud-Camou E., Boucher-Rodoni R. (1983). “Feeding and digestion in cephalopods,” in In: The Mollusca. Eds. Saleuddin A. S. M., Wilbur K. M. (NewYork, NY, USA: Academic Press), 149–187.
Boucaud-Camou E., Boucher-Rodoni R., Mangold K. (1976). Digestive absorption in octopus vulgaris (Cephalopoda: Octopoda). J. Zoology 179 (2), 261–271. doi: 10.1111/j.1469-7998.1976.tb02295.x
Boucher-Rodoni R., Boucaud-Camou E. (1987). Fine structure and absorption of ferritin in the digestive organs of Loligo vulgaris and l. forbesi (Cephalopoda, teuthoidea). J. Morphology 193 (2), 173–184.
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Braga R., van der Molen S., Rodriguez Y. E., Fernández-Giménez A. V., Battini N., Rosas C., et al. (2022). Morphophysiological responses of Octopus tehuelchus juveniles during the transition period between endogenous and exogenous feeding. Aquaculture 556, 738269. doi: 10.1016/j.aquaculture.2022.738269
Bravo-Ruiz I., Medina M.Á., Martínez-Poveda B. (2021). From food to genes: Transcriptional regulation of metabolism by lipids and carbohydrates. Nutrients 13 (5), 1513. doi: 10.3390/nu13051513
Bryant D. A., Frigaard N. U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14 (11), 488–496. doi: 10.1016/j.tim.2006.09.001
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Cornejo-Granados F., Lopez-Zavala A. A., Gallardo-Becerra L., Mendoza-Vargas A., Sánchez F., Vichido R., et al. (2017). Microbiome of pacific whiteleg shrimp reveals differential bacterial community composition between wild, aquacultured and AHPND/EMS outbreak conditions. Sci. Rep. 7 (1), 1–15. doi: 10.1038/s41598-017-11805-w
Costa P. M., Carreira S., Costa M. H., Caeiro S. (2013). Development of histopathological indices in a commercial marine bivalve (Ruditapes decussatus) to determine environmental quality. Aquat. Toxicol. 126, 442–453. doi: 10.1016/j.aquatox.2012.08.013
Costa P. M., Rodrigo A. P., Costa M. H. (2014). Microstructural and histochemical advances on the digestive gland of the common cuttlefish, Sepia officinalis l. Zoomorphology 133 (1), 59–69. doi: 10.1007/s00435-013-0201-8
De la Cruz-Leyva M. C., Zamudio-Maya M., Corona-Cruz A. I., González-de la Cruz J. U., Rojas-Herrera R. (2011). A method for isolating RNA from metabolically active bacterial flora associated with octopus. Lett. Appl. Microbiol. 53, 8–13. doi: 10.1111/j.1472-765X.2011.03057.x
DeRusha R. H., Forsythe J. H., DiMarco F. P., Hanlon R. T. (1989). Alternative diets for maintaining and rearing cephalopods in captivity. Lab. Anim. Sci. 4, 306–312.
Destanie R. R., Houa Y., Gloria S., Craig S., Bing M., Milo C. (2018). Differential immune responses and microbiota profiles in children with autism spectrum disorders and co-morbid gastrointestinal symptoms. Brain Behav. Immun. 70, 354–368. doi: 10.1016/j.bbi.2018.03.025
De Vadder F., Mithieux G. (2018). Gut-brain signaling in energy homeostasis: the unexpected role of microbiota-derived succinate. J. Endocrinol. 236, R105–R108. doi: 10.1530/JOE-17-0542
Domingues P., Sykes A., Sommerfield A., Andrade P. (2003). The effects of feeding live or frozen prey on growth, survival and the life cycle of the cuttlefish, Sepia officinalis (Linnaeus 1758). Aquaculture Int. 11, 397–410. doi: 10.1023/B:AQUI.0000004195.92236.3a
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 (16), 2194–2200. doi: 10.1093/bioinformatics/btr381
Fiorito G., Affuso A., Basil J., Cole A., de Girolamo P., D'angelo L., et al. (2015). Guidelines for the care and welfare of cephalopods in research–a consensus based on an initiative by CephRes, FELASA and the Boyd group. Lab. Anim. 49, 1–90. doi: 10.1177/0023677215580006
Fish J. A., Chai B., Wang Q., Sun Y., Brown C. T., Tiedje J. M., et al. (2013). FunGene: the functional gene pipeline and repository. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00291
Forsythe J. W., Derusha R. H., Hanlon R. T. (1994). Growth, reproduction and life span of Sepia officinalis (cephalopoda: mollusca) cultured through seven consecutive generations. J. Zoology 233, 175–192. doi: 10.1111/j.1469-7998.1994.tb08582.x
Francis G., Makkar H. P. S., Becker K. (2001). Antinutritional factors present in plant derived alternate fish feed ingredients and their effects in fish. Aquaculture 199, 197–227. doi: 10.1016/S0044-8486(01)00526-9
Fuentes-Quesada J. P., Viana M. T., Rombenso A. N., Guerrero-Rentería Y., Nomura-Solís M., Gomez-Calle V., et al. (2018). Enteritis induction by soybean meal in Totoaba macdonaldi diets: effects on growth performance, digestive capacity, immune response and distal intestine integrity. Aquaculture 495, 78–89. doi: 10.1016/j.aquaculture.2018.05.025
Gallardo P., Olivares A., Martínez-Yáñez R., Caamal-Monsreal C., Domingues P. M., Mascaró, et al. (2017). Digestive physiology of Octopus maya and o. mimus: temporality of digestion and assimilation processes. Front. Physiol. 8, 355–363. doi: 10.3389/fphys.2017.00355
Gao F. Z., Zou H. Y., Wu D. L., Chen S., He L. Y., Zhang M., et al. (2020). Swine farming elevated the proliferation of acinetobacter with the prevalence of antibiotic resistance genes in the groundwater. Environ. Int. 136, 105484. doi: 10.1016/j.envint.2020.105484
Ghasemi N., Imani A., Noori F., Shahrooz R. (2020). Ontogeny of digestive tract of stellate sturgeon (Acipenser stellatus) from hatching to juvenile stage: Digestive enzymes activity, stomach and proximal intestine. Aquaculture 519, 734751. doi: 10.1016/j.aquaculture.2019.734751
Gros O., Frenkiel L., Aranda D. A. (2009). Structural analysis of the digestive gland of the queen conch Strombus gigas Linnaeus 1758 and its intracellular parasites. J. Molluscan Stud. 75, 59–68. doi: 10.1093/mollus/eyn041
Hamza N., Mhetli M., Kestemont P. (2007). Effects of weaning age and diets on ontogeny of digestive activities and structures of pikeperch (Sander lucioperca) larvae. Fish Physiol. Biochem. 33, 121–133. doi: 10.1007/s10695-006-9123-4
Han S., Liu Y., Zhou Z., He S., Cao Y., Shi P., et al. (2010). Analysis of bacterial diversity in the gut of grass carp (Ctenopharyngodon idellus) based on 16S rDNA gene sequences. Aquaculture Res. 42 (1), 47–56. doi: 10.1111/j.1365-2109.2010.02543.x
Hou D., Huang Z., Zeng S., Liu J., Weng S., He J. (2018). Comparative analysis of the bacterial community compositions of the shrimp gut, surrounding water and sediment. J. Appl. Microbiol. 125 (3), 792–799. doi: 10.1111/jam.13919
Hsieh S. L., Ruan Y. H., Li Y. C., Hsieh P. S., Hu C. H., Kuo C. M. (2008). Immune and physiological responses in pacific white shrimp (Penaeus vannamei) to vibrio alginolyticus. Aquaculture 275, 335–341. doi: 10.1016/j.aquaculture.2007.12.019
Hu J., Zhang F., Xu X., Su Y., Qin Y., Ma Y., et al. (2014). Isolation, identification and virulence of the pathogen of white-spots disease in internal organs of Pseudosciaena crocea. Oceanologia Limnologia Sin. 45 (2), 409–417.
Ibarra-García L. E., Tovar-Ramírez D., Rosas C., Campa-Córdova Á.I., Mazón-Suástegui J. M. (2018). Digestive enzymes of the Californian two-spot octopus, Octopus bimaculoides (Pickford and McConnaughey 1949). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 215, 10–18.
Iehata S., Valenzuela F., Riquelme C. (2015). Analysis of bacterial community and bacterial nutritional enzyme activity associated with the digestive tract of wild Chilean octopus (Octopus mimus Gould 1852). Aquaculture Res. 46, 861–873. doi: 10.1111/are.12240
Ishibashi Y., Kohyama-Koganeya A., Hirabayashi Y. (2013). New insights on glucosylated lipids: Metabolism and functions. BBA-Mol. Cell. Mol. Biol. Lett. 1831 (9), 1475–1485. doi: 10.1016/j.bbalip.2013.06.001
Ivana S., Juliana D. C., Jesse S., Lantz C. M., Rob K., Steven A. F. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Ivarsson E., Wattrang E., Sun L., Cervin G., Pavia H., Wall H. (2022). Evaluation of early feed access and algal extract on growth performance, organ development, gut microbiota and vaccine-induced antibody responses in broiler chickens. Animal 16 (5), 100522. doi: 10.1016/j.animal.2022.100522
Jha R., Berrocoso J. D. (2015). Review: Dietary fiber utilization and its effects on physiological functions and gut health of swine. Animal 9, 1441–1452. doi: 10.1017/S1751731115000919
Jiang M. W., Chen Q., Zhou S. N., Han Q. X., Peng R. B., Jiang X. M. (2021). Optimum weaning method for pharaoh cuttlefish, Sepia pharaonis ehrenberg 1831, in small-and large-scale aquaculture. Aquaculture Res. 52 (3), 1078–1087. doi: 10.1111/are.14963
Jiang M. W., Chen H., Zhou S. N., Han Q. X., Peng R. B., Jiang X. M. (2022). Changes in embryonic development, juvenile growth and physiological adaptation of the cuttlefish Sepia pharaonis in response to photoperiod manipulation. J. Oceanology Limnology 40 (4), 1–16. doi: 10.1007/s00343-021-1243-2
Jiang M. W., Han Z. R., Sheng P., Peng R. B., Han Q. X., Jiang X. M. (2020a). Effects of different weaning protocols on survival, growth and nutritional composition of pharaoh cuttlefish (Sepia pharaonis) juvenile. J. Ocean Univ. China 19 (6), 1421–1429. doi: 10.1007/s11802-020-4479-0
Jiang M. W., Peng R., Peng R. B., Han Q. X., Jiang X. M. (2020b). Effects of size dominance on the survival, growth and physiological activities of juvenile pharaoh cuttlefish (Sepia pharaonis). J. Exp. Mar. Biol. Ecol. 525, 318–327. doi: 10.1016/j.jembe.2020.151318
Jiang M. W., Peng R. B., Wang S. J., Peng R. B., Han Q. X., Jiang X. M. (2018). Growth performance and nutritional composition of Sepia pharaonis under artificial culturing conditions. Aquaculture Res. 49, 2788–2798. doi: 10.1111/are.13741
Jiang M. W., Zhao C. X., Yan R. X., Li J. P., Song W. W., Peng R. B., et al. (2019). Continuous inking affects the biological and biochemical responses of cuttlefish Sepia pharaonis. Front. Physiol. 10, 1429–1441. doi: 10.3389/fphys.2019.01429
Kakade M. L., Simons N., Liener I. E. (1969). An evaluation of natural vs. synthetic substrates for measuring the antitryptic activity of soybean samples. Cereal Chem. 6, 518–526.
Kar N., Ghosh K. (2008). Enzyme producing bacteria in the gastrointestinal tracts of Labeo rohita (Hamilton) and channa punctatus (Bloch). Turkish J. Fisheries Aquat. Sci. 8, 115–120.
Kumar H., Park W., Lim D., Srikanth K., Kim J. M., Jia X., et al. (2019). Whole metagenome sequencing of cecum microbiomes in Ethiopian indigenous chickens from two different altitudes reveals antibiotic resistance genes. Genomics 112 (2), 1988–1999.
Lavilla Pitogo C. R., Leaño E. M., Paner M. G. (1998). Mortalities of pond-cultured juvenile shrimp, Penaeus monodon, associated with dominance of luminescent vibrios in the rearing environment. Aquaculture 164, 337–349. doi: 10.1016/S0044-8486(98)00198-7
Lee P. G. (1994). Nutrition of cephalopods: fueling the system. Mar. Freshw. Behav. Physiol. 25 (3), 35–51.
Li S., Wen W., Huang X., Gong X., Feng L., Chen N. (2017). Activities of digestive enzymes and histology of digestive system during larval development of devil stinger (Inimicus japonicus). Aquaculture Res. 48 (10), 5394–5401. doi: 10.1111/are.13353
Lobo J., Costa P. M., Caeiro S., Martins M., Ferreira A. M., Caetano M. (2010). Evaluation of the potential of the common cockle (Cerastoderma edule l.) for the ecological risk assessment of estuarine sediments: Bioaccumulation and biomarkers. Ecotoxicology 19, 1496–1512. doi: 10.1007/s10646-010-0535-7
Lobo-da-Cunha A. (2019). Structure and function of the digestive system in molluscs. Cell Tissue Res. 377 (3), 475–503. doi: 10.1007/s00441-019-03085-9
Lysyk T. J., Kalischuk-Tymensen L., Selinger L. B., Lancaster R. C., Wever L., Cheng K. J. (1999). Rearing stable fly larvae (Diptera: Muscidae) on an egg yolk medium. J. Med. Entomology 36, 382–388. doi: 10.1093/jmedent/36.3.382
Magoc T., Salzberg S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi: 10.1093/bioinformatics/btr507
Ma Y., Han X., Fang J., Jang H. (2021). Role of dietary amino acids and microbial metabolites in the regulation of pig intestinal health. Anim. Nutr. 9, 1–6. doi: 10.1016/j.aninu.2021.10.004
Martínez R., López-Ripoll E., Avila-Poveda O. H., Santos-Ricalde R., Mascaró M., Rosas C. (2011). Cytological ontogeny of the digestive gland in post-hatching Octopus maya, and cytological background of digestion in juveniles. Aquat. Biol. 11 (3), 249–261. doi: 10.3354/ab00305
Martínez R., Santos R., Mascaró M., Canseco L., Caamal-Monsreal C., Rosas C. (2012). Digestive dynamics during chyme formation of Octopus maya (Mollusca, Cephalopoda). Aquaculture Res. 43 (8), 1119–1126. doi: 10.1111/j.1365-2109.2011.02915.x
Minton J. W., Walsh L. S., Lee P. G., Forsythe J. W. (2001). First multi-generation culture of the tropical cuttlefish Sepia pharanois ehrenberg 1831. Aquaculture Internation 9, 379–392. doi: 10.1023/A:1020535609516
Moguel C., Mascaró M., Avila-Poveda O. H., Caamal-Monsreal C., Sanchez A., Pascual C., et al. (2010). Morphological, physiological and behavioral changes during post-hatching development of Octopus maya (Mollusca: Cephalopoda) with special focus on the digestive system. Aquat. Biol. 9 (1), 35–48. doi: 10.3354/ab00234
Morishita T. (1972). Studies on the distribution of proteolytic enzymes in the internal organs of octopus. Bull. Japanese Soc. Sci. Fish 38, 839–843. doi: 10.2331/suisan.38.839
Navarro J. C., Monroig Ó., Sykes A. V. (2014). “Nutrition as a key factor for cephalopod aquaculture,” in Cephalopod culture (Dordrecht: Springer), pp, 77–pp, 95.
Nayak S. K. (2010). Role of gastrointestinal microbiota in fish. Aquaculture Res. 41, 1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x
Nigam A. (2007). Lab Manual in biochemistry, immunology and biotechnology Vol. 110008 (7 West Patel Nagar, New Delhi: Tata McGraw-Hill Publishing Company Limited), pp,198–pp,199.
Padros F., Villalta M., Gisbert E., Estevez A. (2011). Morphological and histological study of larval development of the Senegal sole Solea senegalensis: an integrative study. J. Fish Biol. 79, 3–32. doi: 10.1111/j.1095-8649.2011.02942.x
Pereda S. V., Uriarte I., Cabrera J. C. (2009). Effect of diet and paralarval development on digestive enzyme activity in the cephalopod Robsonella fontaniana. Mar. Biol. 156 (10), 2121–2128. doi: 10.1007/s00227-009-1242-x
Perrin A., Le Bihan E., Koueta N. (2004). Experimental study of enriched frozen diet on digestive enzymes and growth of juvenile cuttlefish Sepia officinalis l. (Mollusca Cephalopoda). J. Exp. Mar. Biol. Ecol. 311, 267–285. doi: 10.1016/j.jembe.2004.05.012
Piccolo B. D., Mercer K. E., Bhattacharyya S., Bowlin A. K., Saraf M. K., Pack L., et al. (2017). Early postnatal diets affect the bioregional small intestine microbiome and ileal metabolome in neonatal pigs. J. Nutr. 147 (8), 1499–1509. doi: 10.3945/jn.117.252767
Pila E. A., Sullivan J. T., Wu X. Z., Fang J., Rudko S. P., Gordy M. A., et al. (2016). Haematopoiesis in molluscs: A review of haemocyte development and function in gastropods, cephalopods and bivalves. Dev. Comp. Immunol. 58, 119–128. doi: 10.1016/j.dci.2015.11.010
Pinoargote G., Flores G., Cooper K., Ravishankar S. (2018). Effects on survival and bacterial community composition of the aquaculture water and gastrointestinal tract of shrimp (Litopenaeus vannamei) exposed to probiotic treatments after an induced infection of acute hepatopancreatic necrosis disease. Aquaculture Res. 49, 3270–3288. doi: 10.1111/are.13791
Pradhan P. K., Jena J., Mitra G., Sood N., Gisbert E. (2014). Effects of different weaning strategies on survival, growth and digestive system development in butter catfish Ompok bimaculatus (Bloch) larvae. Aquaculture 424, 120–130. doi: 10.1016/j.aquaculture.2013.12.041
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rawls J. F., Samuel B. S., Gordon J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. P. Natl. Acad. Sci. U.S.A. 101 (13), 4596–4601. doi: 10.1073/pnas.0400706101
Ray A. K., Ghosh K., Ringø E. (2012). Enzyme-producing bacteria isolated from fish gut: A review. Aquaculture Nutr. 18, 465–492. doi: 10.1111/j.1365-2095.2012.00943.x
Rodrigo A. P., Costa P. M. (2017). The role of the cephalopod digestive gland in the storage and detoxification of marine pollutants. Frontier Physiol. 8, 232–230. doi: 10.3389/fphys.2017.00232
Rosa R, Pereira J, Nunes M.L (2005). Biochemical composition of cephalopods with different life strategies, with special reference to a giant squid, Architeuthis sp. Marine Biology. 146, 739–751. doi: 10.1007/s00227-004-1477-5
Roura Á., Doyle S. R., Nande M., Strugnell J. M. (2017). You are what you eat: A genomic analysis of the gut microbiome of captive and wild Octopus vulgaris paralarvae and their zooplankton prey. Frontier Physiol. 8, 362–375. doi: 10.3389/fphys.2017.00362
Sabour S., Tabeidian S. A., Sadeghi G. (2019). Dietary organic acid and fiber sources affect performance, intestinal morphology, immune responses and gut microflora in broilers. Anim. Nutr. 5 (2), 156–162. doi: 10.1016/j.aninu.2018.07.004
Safi G., Martínez A. S., Le Pabic C., Le Bihan E., Robin J. P., Koueta N. (2018). Digestive enzyme ratios are good indicators of hatchling yolk reserve and digestive gland maturation in early life stages of cuttlefish Sepia officinalis l.: application of these new tools in ecology and aquaculture. J. Comp. Physiol. B 188 (1), 57–76.
Schleder D. D., Blank M., Peruch L. G. B., Poli M. A., Goncalves P., Rosa K. V., et al. (2020). Impact of combinations of brown seaweeds on shrimp gut microbiota and response to thermal shock and white spot disease. Aquaculture 519, 734779. doi: 10.1016/j.aquaculture.2019.734779
Semmens J. M. (2002). Changes in the digestive gland of the loliginid squid Sepioteuthis lessoniana (Lesson 1830) associated with feeding. J. Exp. Mar. Biol. Ecol. 274 (1), 19–39. doi: 10.1016/S0022-0981(02)00165-X
Semova I., Carten J. D., Stombaugh J., Mackey L. C., Knight R., Farber S. A., et al. (2012). Microbiota regulate gut absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12 (3), 277–288. doi: 10.1016/j.chom.2012.08.003
Shang Q., Ma X., Liu H., Liu S., Piao X. (2020). Effect of fiber sources on performance, serum parameters, intestinal morphology, digestive enzyme activities and microbiota in weaned pigs. Arch. Anim. Nutr. 74 (2), 121–137. doi: 10.1080/1745039X.2019.1684148
Shaw D., Gohil K., Basson M. D. (2012). Intestinal mucosal atrophy and adaptation. World J. Gastroenterol. 18, 6357–6375. doi: 10.3748/wjg.v18.i44.6357
Sheng P., Chen Q., Han Q. X., Lu J. Q., Zhang B. L., Jiang M. W., et al. (2021). Bacterial community in gut, shell sediment, and surrounding water of Hyriopsis cumingii fed three different microalgal diets. Aquaculture 540, 1–13. doi: 10.1016/j.aquaculture.2021.736701
Shyla G., Nair C. M., Salin K. R., Sherief P. M., Mukundan M. K. (2010). Liver oil of pharaoh cuttlefish sepia pharaonis ehrenberg 1831 as a lipid source in the feed of giant freshwater prawn, macrobrachium rosenbergii (de man 1879). Aquaculture Nutr. 15 (3), 273–281.
Sivasubramanian K., Ravichandran S., Vijayapriya M. (2011). Antagonistic activity of marine bacteria Pseudoalteromonas tunicata against microbial pathogens. Afr. J. Microbiol. Res. 5, 562–567.
Smith J. A., Andrews P. L., Hawkins P., Louhimies S., Ponte G., Dickel L. (2013). Cephalopod research and EU directive 2010/63/EU: Requirements. impacts and ethical review. J. Exp. Mar. Biol. Ecol. 447, 31–45. doi: 10.1016/j.jembe.2013.02.009
Solorzano Y., Viana M. T., López L. M., Correa J. G., True C. C., Rosas C. (2009). Response of newly hatched Octopus bimaculoides fed enriched Artemia salina: Growth performance, ontogeny of the digestive enzyme and tissue amino acid content. Aquaculture 289, 84–90. doi: 10.1016/j.aquaculture.2008.12.036
Sonnenburg J. L., Bäckhed F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535 (7610), 56–64.
Sowmya R., Sachindra N. M. (2015). Carotenoid production by Formosa sp. kmw, a marine bacteria of flavobacteriaceae family: Influence of culture conditions and nutrient composition. Biocatalysis Agric. Biotechnol. 4, 559–567. doi: 10.1016/j.bcab.2015.08.018
Sun P., Jin M., Ding L., Lu Y., Ma H., Yuan Y., et al. (2018). Dietary lipid levels could improve growth and intestinal microbiota of juvenile swimming crab, Portunus trituberculatus. Aquaculture 490, 208–216. doi: 10.1016/j.aquaculture.2018.02.018
Swift K., Johnston D., Moltschaniwsky N. (2005). The digestive gland of the southern dumpling squid (Eupryma tasmanica): Structure and function. J. Exp. Mar. Biol. Ecol. 315, 177–186. doi: 10.1016/j.jembe.2004.09.017
Sykes A. V., Koueta N., Rosas C. (2014). Historical review of cephalopods culture. Cephalopod Culture, 59–75. doi: 10.1007/978-94-017-8648-5_4
Sykes A. V., Rui A. G., Andrade J. P. (2013). Early weaning of cuttlefish (Sepia officinalis, l.) with frozen grass shrimp (Palaemonetes varians) from the first day after hatching. Aquaculture Res. 44, 1815–1823. doi: 10.1111/j.1365-2109.2012.03186.x
Versaw W. K., Cuppett S. L., Winters D. D., Williams L. E. (1989). An improved colorimetric assay for bacterial lipase in nonfat dry milk. J. Food Sci. Technol. 54, 1557–1558. doi: 10.1111/j.1365-2621.1989.tb05159.x
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267. doi: 10.1128/AEM.00062-07
Wang S., Hu Y., Wang J. (2018b). Biodegradation of typical pharmaceutical compounds by a novel strain acinetobacter sp. J. Environ. Manage. 217, 240–246. doi: 10.1016/j.jenvman.2018.03.096
Wang A. R., Ran C., Ringø E., Zhou Z. G. (2018a). Progress in fish gastrointestinal microbiota research. Rev. Aquaculture 10, 626–640. doi: 10.1111/raq.12191
Wang Y., Wang K., Huang L., Dong P., Wang S., Chen H., et al. (2020). Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle. Microbiome 8 (1), 1–16. doi: 10.1186/s40168-020-00879-w
Wang Y., Xie Q., Sun S., Huang B., Zhang Y., Xu Y., et al. (2018c). Probiotics-fermented Massa Medicata fermentata ameliorates weaning stress in piglets related to improving intestinal homeostasis. Appl. Microbiol. Biotechnol. 102 (24), 10713–10727. doi: 10.1007/s00253-018-9438-y
Yao Z., Yang K., Huang L., Huang X., Wang K., Zhang D. (2018). Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Express 8 (1), 1–10. doi: 10.1186/s13568-018-0644-x
Yao R., Yang Z., Zhang Z., Hu T., Chen H., Huang F., et al. (2019). Are the gut microbial systems of giant pandas unstable? Heliyon 5 (9), e02480. doi: 10.1016/j.heliyon.2019.e02480
Zambonino-Infante J. L., Gisbert E., Sarasquete C., Navarro I., Gutie´rrez J., Cahu C. L. (2009). “Ontogeny and physiology of the digestive system of marine fish larvae,” in Feeding and digestive functions of fish. Eds. Cyrino J. E. O., Bureau D., Kapoor B. G. (Enfield, USA: Science Publishers Inc), 277–348.
Zhang J. T., Zhou S. M., An S. W., Chen L., Wang G. L. (2014). Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, pseudomonas plecoglossicida. J. Fish Dis. 37 (2), 113–121. doi: 10.1111/jfd.12075
Zhao L., He K., Luo J., Sun J., Liao L., Tang X., et al. (2020). Co-Modulation of liver genes and intestinal microbiome of largemouth bass larvae (Micropterus salmoides) during weaning. Front. Microbiol. 11, 1332. doi: 10.3389/fmicb.2020.01332
Zhao J., Shi B., Jiang Q., Ke C. (2012). Changes in gut associated flora and bacterial digestive enzymes during the development stages of abalone (Haliotis diversicolor). Aquaculture 338–341, 147–153. doi: 10.1016/j.aquaculture.2012.01.016
Keywords: digestive tract, digestive enzyme, histology, microbiota, cuttlefish, feed transition
Citation: Jiang M, Xiao W, Ye J, Xu L, Peng R, Han Q, Lü Z, Shi H and Jiang X (2022) Effects of feed transition on digestive tract digestive enzyme, morphology and intestinal community in cuttlefish (Sepia pharaonis). Front. Mar. Sci. 9:941488. doi: 10.3389/fmars.2022.941488
Received: 12 May 2022; Accepted: 11 August 2022;
Published: 06 September 2022.
Edited by:
Eduardo Almansa, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Nicolás Ortiz, Centro Nacional Patagónico, ArgentinaDeiene Rodriguez Barreto, University of La Laguna, Spain
Copyright © 2022 Jiang, Xiao, Ye, Xu, Peng, Han, Lü, Shi and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiamin Jiang, amlhbmd4aWFtaW5AaG90bWFpbC5jb20=
 Maowang Jiang
Maowang Jiang Wencheng Xiao1
Wencheng Xiao1 Xiamin Jiang
Xiamin Jiang