
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 07 July 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.938742
 Junlin Wang1,2†
Junlin Wang1,2† Fuyuan Zeng1,2†
Fuyuan Zeng1,2† Haiyun Feng1,2
Haiyun Feng1,2 Liangchuan Chen1,2
Liangchuan Chen1,2 Weijie Zhang1,2
Weijie Zhang1,2 Xing Xiao1,2
Xing Xiao1,2 Sean J. Monaghan3
Sean J. Monaghan3 Jichang Jian1,2
Jichang Jian1,2 Huanying Pang1,2*
Huanying Pang1,2* Na Wang4
Na Wang4Vibrio alginolyticus, a zoonotic bacterial pathogen, expresses a type III secretion system (T3SS) that is critical for pathogen virulence and disease development. In this study, the mutant HY9901 ΔaraC was obtained from the laboratory and its biological characteristics were analyzed. The swimming ability of ΔaraC decreased and exhibited a 2,600 times reduction in virulence to zebrafish. However, ΔaraC showed no difference in growth and extracellular protease activity compared to wild type. Biofilm-forming ability was improved at 24 h, but no difference was observed at other time points. The results of drug sensitivity testing showed that compared with the wild-type HY9901 strain, ΔaraC was sensitive to amikacin, tetracycline, neomycin, minocycline, and gentamicin. The transcription levels of T3SS effector proteins HopPmaJ, VopS, VcrV, and VopN were analyzed by qRT-PCR. The results showed that ΔaraC had significantly upregulated the mRNA expression of VopS, VcrV, and VopN, but significantly downregulated the mRNA expression of HopPmaJ at each stage compared with HY9901. Western blotting and the β-galactosidase reporter gene experiment also showed that the deletion of araC gene significantly downregulated the expression of HopPmaJ. Finally, the relative percent survival (RPS) rate of grouper inoculated by intramuscular (IM) injection of HY9901 ΔaraC was 61.3% after being challenged with HY9901. Real-time qPCR analysis showed that vaccination of HY9901 ΔaraC could enhance the expression of immune-related genes, including gata-1, il6, IgM, il-1β, and lyz in liver and spleen, indicating that ΔaraC applied as a live-attenuated vaccine effectively induced an immune response in the zebrafish. This study provides evidence for the subsequent development of an effective live-attenuated V. alginolyticus vaccine.
Vibrio alginolyticus exhibits characteristics of a halophilic and mesophilic Gram-negative bacterium (Rameshkumar et al., 2014; Yang et al., 2021). It is commonly present in the microfauna of the marine environment and a variety of marine animals. It is one of the main pathogens of fish, shrimp, shellfish, and other mariculture animals (Bunpa et al., 2020). In addition to V. alginolyticus representing a conditional pathogen of fish, it poses a serious threat to human public health and threatens the Chinese aquaculture industry, with reports of it causing human food poisoning, otitis media, septicemia, and other diseases (Li et al., 2009; Bunpa et al., 2018). Due to the large-scale outbreak of V. alginolyticus and the long-term overuse and abuse of a large number of antibiotics and other chemical drugs, drug resistance of Vibrio is increasing daily with major implications to the aquaculture industry (Shahimi et al., 2021).
The mechanism of bacterial drug resistance is complex (Stewart, 2001), and there are limited reports on the pathogenic mechanisms of V. alginolyticus. So far, researchers have not solved this problem. Prophylactic prevention of disease outbreaks caused by V. alginolyticus through vaccination is a key objective for marine aquaculture, to minimize impact to the water environment and human health while improving the survival rate of aquatic animals (Aly et al., 2021). Therefore, understanding key molecules with roles in virulence can enable identification of potential vaccine candidates and can also avoid the bacterial drug resistance problem, which continues to plague the aquaculture industry.
Research on the pathogenic mechanisms of different bacteria have revealed that many Gram-negative bacteria contain many broadly conserved genes, which encode a type III protein secretion system (Hueck, 1998). When V. alginolyticus contacts and infects the host, the Type III secretion system (T3SS) operates through the injection of bacterial effector proteins into the host cells, resulting in the death of those cells (Zhou et al., 2013). In recent years, research relating to the V. alginolyticus T3SS has elucidated the role of T3SS apparatus proteins (Zhou et al., 2013; Pang et al., 2022), chaperone proteins (Wu et al., 2020), effector proteins (Pang et al., 2018), and regulatory proteins (Liu et al., 2016; Zhou et al., 2020). From current knowledge on Vibrio parahaemolyticus and Pseudomonas aeruginosa, the transcription of V. alginolyticus T3SS genes is regulated by ExsA of the AraC/XyIS family of transcription regulators (Yahr and Wolfgang, 2006; Liu and Thomas, 2015; Liu et al., 2016).In addition to activating the expression of virulence-related genes, AraC family members can also activate the expression of carbon metabolism and stress response genes (Yang et al., 2013). AraC is an AraC family transcriptional regulator, However, the function of V. alginolyticus AraC is poorly understood. In Escherichia coli, AraC is active in the form of a dimer and reacts with small effector molecules bound to the N-terminal domain of protein, so as to regulate carbon metabolism (Lowden et al., 2010). Gene araC has the ability to positively regulate L-arabinose metabolism because it can promote the expression of L-arabinose structural genes araA, araB, and araD, and araBAD encodes three metabolic enzymes required for L-arabinose metabolism (Sheppard and Englesberg, 1967; Singer and Englesberg, 1971). Moreover, at the molecular level, the details of AraC protein response to arabinose have been clarified (Schleif, 2000; Yang et al., 2011). HopPmaJ is a known effector protein of V. alginolyticus eliciting pathogenic effect on orange-spotted grouper (Epinephelus coioides). Diseased fish showed symptoms of ulcer, liver, and kidney bleeding and swelling (Pang et al., 2018). Moreover, regulatory protein TyeA can upregulate the expression of HopPmaJ (Wu et al., 2020); thus, in order to further understand the function of AraC in T3SS, we cloned the V. alginolyticus araC gene and analyzed it bioinformatically (MO et al., 2021). We obtained the mutant strain HY9901 ΔaraC and studied its biological characteristics and pathogenicity, and showed that AraC regulates the transcription of T3SS effector proteins. In addition, here we report on the potential of HY9901 ΔaraC mutant as a candidate live-attenuated vaccine against V. alginolyticus in zebrafish and the associated proinflammatory and immunoglobulin-related immune response induced.
The bacterial strains, plasmids, and zebrafish used in this study are listed in Table 1. V. alginolyticus HY9901 was isolated from diseased red snapper (Lutjanus sanguineus) (Cai et al., 2007) in Zhanjiang Port, Guangdong Province. Healthy zebrafish (Danio rerio) were purchased from Zhanjiang Aquatic Market. The average length and weight were 3 cm and 0.3 g, respectively. The zebrafish were tested by bacteriological recovery tests and kept in seawater in a circulation system at 28°C for 2 weeks prior to the experiment. The primers were synthesized by Sangon Biotech Co., Ltd.
In-frame deletion was conducted according to the method used previously (Liu et al., 2016). The upstream and downstream homologous arms of araC coding sequence were amplified. Except for BglII and SalI restriction site sequences, the two amplified fragments contained 10-bp overlapping sequences. These products were used as templates for PCR, and the fusion fragments were amplified by primers at both ends of the upstream and downstream homologous arms. The whole fragment was digested with BglII and SalI and connected to the suicide vector pDM4 (enzyme digestion). The obtained plasmid was electroporated into E. coli S17-1 and transferred into V. alginolyticus HY9901 by electroporation. Ampicillin and chloramphenicol were used to screen the monoclonal strains successfully transferred into plasmids on TSA plates. Several insertional mutant single colonies were taken, scribed on an LB plate (4 g/L L-arabinose), and cultured at 28°C for 18 h. Colonies on the plate were selected. Single colonies with the primer araC-up/araC-down were detected using the wild-type V. alginolyticus as the control. After single-colony purification, amplification verification was carried out again, and the sequencing results were verified to determine whether the mutant strain ΔaraC was successfully constructed.
The insertion mutant ΔaraC was continuously subcultured 30 times on TSB medium. The primers araC-F/araC-R were used to detect the genetic stability of the mutant at the 30th generation.
Bacterial growth curves were performed according to Shi et al. (2021). Wild strain HY9901 and mutant strain ΔaraC (OD600 = 0.5) were inoculated at a ratio of 1:100 in fresh TSB medium and cultured at 28°C; OD600 was measured every 2 h including 3 replicates per group. After taking the average value, a growth curve was drawn.
Bacterial swarming motility was performed according to the method of Pearson (Pearson, 2019). A single colony of HY9901 and the mutant strain ΔaraC were inserted vertically into the LBS (0.3% Agar and 2% NaCl added to the LB medium). The diameter of the swimming circle was measured after 12 h during incubation at 28°C.
Extracellular protease activity was assessed according to the method of Zhang et al. (2022) with modifications. HY9901 and mutant strain ΔaraC were coated on TSA plates with sterile cellophane, cultured at 28°C for 24 h, washed with PBS, and centrifuged at low temperature for 30 min; 0.22-μm porous membranes were used to filter the supernatant. Protease activity of the supernatant was measured using azocasein trichloroacetic acid colorimetric determination of solution at OD442.
The crystal violet staining method was performed according to O’Toole (2011). HY9901 and mutant strain ΔaraC were cultured to OD600 = 0.5; 100 μl/well was inoculated onto sterile 96-well plates and cultured at 28°C for 6 h, 12 h, 24 h, 48 h, and 72 h. After 72 h, the bacterial solution was washed with PBS and dried; 150 μl of methanol was added for fixation and dried; 150 μl/well crystal violet ammonia oxalate solution was added, rinsed with water, and then dried upside down before the addition of 150 μl of ethanol. The absorbance was read at OD570. The experiment was repeated three times.
Biofilm formation was investigated using a laser scanning confocal microscope (LSCM) as described by Miquel Guennoc et al. (2017). HY9901 and ΔaraC (OD600 = 0.5) were diluted 50-fold, added to glass-bottom culture dishes (spec: type 28.2 mm, class diameter: 20 mm) (Wuxi NEST, Wuxi, China), and statically cultured in a 28°C biochemical incubator for 24 h. Both strains were gently washed three times with PBS, and then combined and incubated with 10% SYTO9 green in the dark for 20 min, washed three times with saline, mounted in 40% saline-glycerol, and observed by confocal microscopy (Zeiss, LSM710, Germany). The excitation wavelength was 488 nm, scanned from the bottom to the top of the biofilm, Z-section was 1 μm apart, and biofilm parameters—biomass and maximum thickness—were determined. Three samples were assessed for each strain and the average readout was calculated.
The injection concentrations of wild-type strain HY9901 and ΔaraC were 104, 105, 106, 107, and 108 CFU/ml. The water temperature is controlled to 28°C. In the experimental group, each fish was injected with 5 μl of bacterial solution by intramuscular injection. The control group was treated with 5 μl of PBS (Table 3). The fish were observed for 14 consecutive days until the mortality was stabilized.
Using the paper diffusion method (Cai et al., 2016), HY9901 and ΔaraC (OD600 = 0.5) were coated on TSA plates, pasted with 30 kinds of antibiotic-sensitive paper, and cultured at 28°C for 12 h, and the sensitivity to the antibiotic was measured and evaluated with vernier caliper.
HY9901 and ΔaraC were cultured in DMEM medium at 28°C until OD600 = 0.5, reinoculated into DMEM medium and cultured at 28°C for 12 h. The bacterial solution was centrifuged, and the precipitated cells were collected and dissolved in Trizol (Takara bio, Inc.). Novogene Co., Ltd. conducted a series of experiments, including mRNA extraction, RNA fragmentation, cDNA synthesis, and RNA SEQ library construction.
RNA of HY9901 and ΔaraC was provided by Novogene Co., Ltd. The T3SS effector genes in this study were hop (Pang et al., 2018), vopN (Tam et al., 2010), vopS (Osorio, 2018), and vcrV (Kinoshita et al., 2020). 16S rRNA is used as an internal reference. The primers are shown in Table 2. Based on the method of Li et al. (2016b), cDNA was synthesized and real-time PCR was used to analyze the expression of T3SS-related genes.
In order to detect the effect of AraC deletion on the secretion of effector protein HopPmaJ, we cultured wild strain HY9901 and deletion strain ΔaraC for 12 h with DMEM. The culture was harvested, washed, and mixed evenly with PBS. A sample combination of 5:1 sample to 6× Protein Loading Buffer (#DL101-02, purchased from Trans Gen Biotechnology Co., Ltd.) was boiled and centrifuged to obtain the whole bacterial protein. After SDS-PAGE, proteins were electrophoretically transferred to a PVDF membrane. The membrane was blocked with TBST solution containing 5% skimmed milk and probed with HopPmaJ serum antibody (the prokaryotic expression vector pET28a-hop was constructed and HopPmaJ serum antibody was prepared by Sangon Biotech Co., Ltd., 1:5,000 diluted in TBST solution). The second antibody was HRP-labeled Goat Anti-Rabbit IgG (#A0208, purchased from Shanghai Beyotime Biotechnology Co., Ltd.) (1:8,000 diluted in TBST solution) and colored by ClarityTM Western ECL substrate (#1705061, purchased from Bio-Rad Co., Ltd.). The automatic chemiluminescence image analysis system (TAN 5200) was used to take photos and record the experimental result.
The recombinant plasmid of the pME6522-hop promoter region was constructed. A 363-bp DNA fragment was amplified upstream of hop by primer pME-hop-F/pME-hop-R. PCR products were digested with PstI and EcoRI and cloned into plasmid pME6522 to construct pME6522-hop-lacZ. The plasmid was transformed into E. coli S17 to obtain strain S17-hop-lacZ, respectively, and integrated into V. alginolyticus HY9901 and mutant strain ΔaraC through conjugation to obtain lacZ transcription fusion strains hop-lacZ::HY9901 and hop-lacZ::ΔaraC. hop-lacZ::HY9901 and hop-lacZ::ΔaraC (OD600 = 0.5) were inoculated into fresh TSB at a ratio of 1:100 and cultured for 18 h. After collection, the strains were applied separately to β-galactosidase detection kits (#RG0036, purchased from Shanghai Biyuntian Biotechnology Co., Ltd.). This allowed assessment of β-Galactosidase activity to judge the regulatory relationship between AraC and effector protein HopPmaJ.
LD50 experiment confirmed that 105 CFU/ml ΔaraC was not lethal to zebrafish (Table 3). The mutant ΔaraC was cultured in TSB, and the concentration was adjusted to 105 CFU/ml with PBS and the water temperature was maintained at 28°C. Two groups (30 tails/group) were injected intramuscularly with 5 μl of ΔaraC (105 CFU ml−1), and PBS was used as negative control and injected in the same volume. Liver and spleen were sampled continuously for 7 days. The ground zebrafish viscera were diluted with PBS and coated on a TCBS plate. They were cultured at 28°C for 18 h and counted.
Vaccination was undertaken according to Ding et al. (2019) with modifications. The water temperature was kept at 28°C. Two groups (80 tails/group) were injected intramuscularly with 5 μl of ΔaraC (105 CFU ml−1), and each fish in the control group was injected with 5 μl of PBS. Four weeks post vaccination, each group (n = 30) was randomly assigned and challenged by intramuscular injection of 5 μl of V. alginolyticus HY9901 (1 × 108 CFU ml−1). Fish were monitored for 14 days. RBS was calculated in the light of the formula (RPS (%) = (1 − immunized group mortality/control group mortality) × 100%).
Histological analysis was conducted to evaluate safety of HY9901ΔaraC as an attenuated vaccine. After 28 days post-immunization, three zebrafish in the control group and the immunized group were randomly selected for sampling, and the spleen and liver tissues were dissected for histopathological observation. The negative control was a PBS-injected control group, and the positive control was the wild strain HY9901-injected group. Organ samples were fixed in 4% paraformaldehyde for 24 h and transferred to 70% ethanol. The samples were processed through an ethanol gradient, transferred to xylene, embedded in paraffin, and sectioned at 5 μm. The sections were sealed after hematoxylin–eosin (H.E.) staining, and observed and photographed under an optical microscope.
After 4 weeks of immunization, the liver and spleen of zebrafish were collected, added with transzol, and homogenized. According to Li et al. (2016b), total RNA was prepared, and reverse transcription, real-time qPCR detected immune-related genes’ expression levels. Primers for il-6, il-6r, il-1β, il-8, igm, gata-1, lyz, rag-1, tlr5, and c/ebpβ are shown in Table 3, and β-actin was used as internal reference.
All animal experiments were conducted strictly based on the recommendations in the Guide for the Care and Use of Laboratory Animals set by the National Institutes of Health. The animal experiments were approved by the Animal Ethics Committee of Guangdong Ocean University (Zhanjiang, China).
The bacteria protocols were approved by the Biosecurity Committee of Guangdong Ocean University (Zhanjiang, China).
The experimental data were analyzed by single-factor analysis of variance (ANOVA) with SPSS19.0 software. ** indicates highly significant difference compared with the control group (p < 0.01). * indicates significant difference compared with the control group (p < 0. 05).
The 1,109-bp fragment was amplified by the correct deletion mutation clone, and the length of the HY9901 amplified fragment was 1,827 bp. After cloning and purification, it was amplified and verified again, and the PCR products were submitted for sequencing. The sequencing results confirmed that the mutant strain ΔaraC was successfully constructed Figure 1.

Figure 1 Construction and confirmation of the knockout mutant strain HY9901ΔaraC. M: DL2000 marker; Lane 1, the 1,827-bp fragment amplified from genomic DNAs of the strain HY9901; Lane 2, the 1,107-bp fragment amplified from genomic DNAs of ΔaraC using primer pairs of araC-up/araC-down.
After the mutant ΔaraC was continuously subcultured for 30 generations, primers araC-F/araC-R were used in PCR to amplify mutant strain ΔaraC and the wild strain. The amplified products were detected by 1.5% agarose gel electrophoresis. The results showed that for the wild strain HY9901, a fragment of 711 bp was obtained. The mutant strain ΔaraC was negative for this fragment (Figure 2A), indicating that the deletion strain ΔaraC can be inherited stably when the gene araC is deleted.
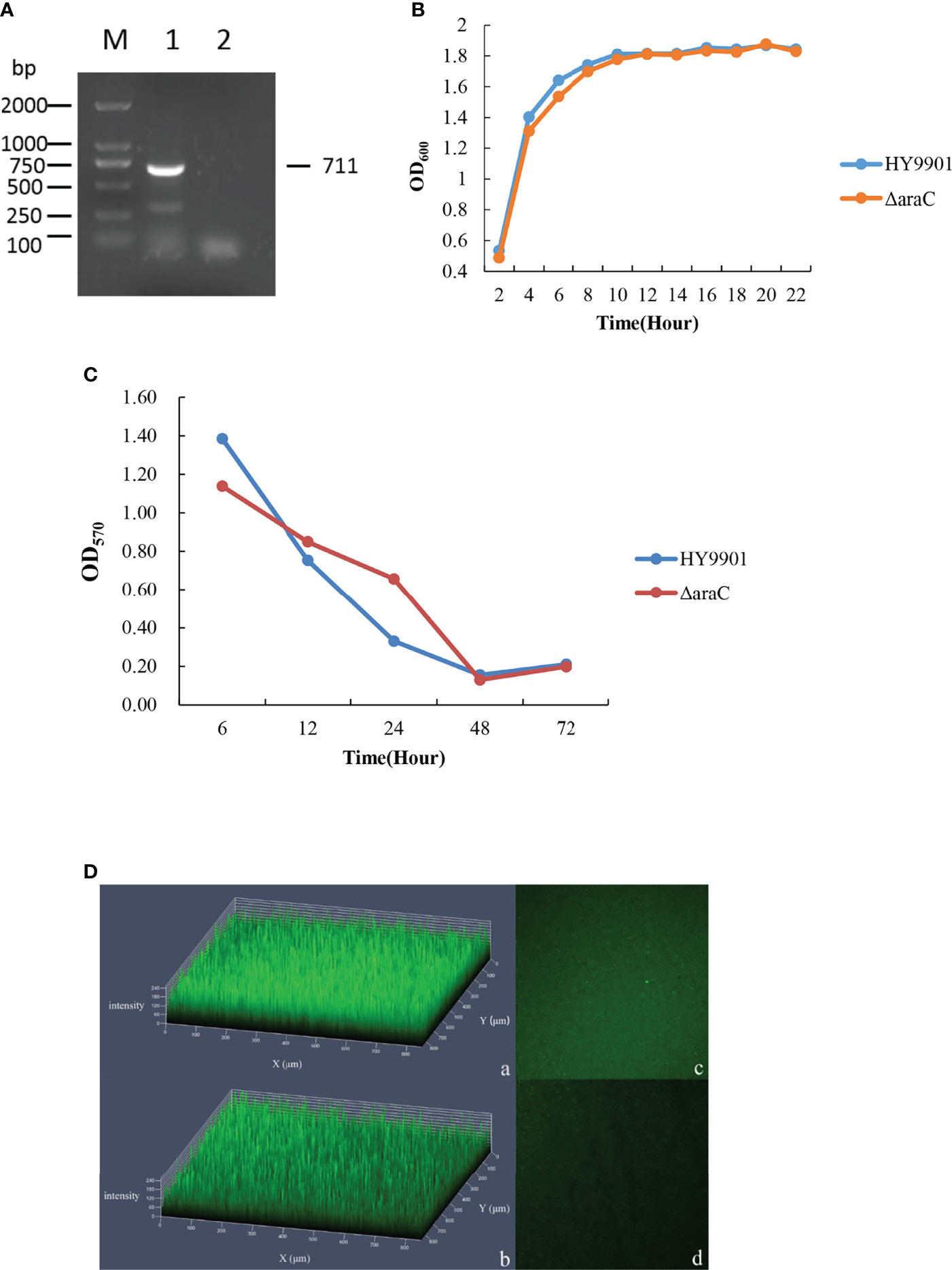
Figure 2 Characterization of the ΔaraC. (A) Genetic stability detection of deletion mutant HY9901ΔaraC. M: DL2000 marker; Lane 1, a fragment of 711 bp is obtained for HY9901 using primer pairs of araC-F/araC-R. Lane 2, the result was negative, and is obtained for HY9901ΔaraC using primer pairs of araC-F/araC-R. (B) Growth rates of HY9901ΔaraC and HY9901. Aliquots of cell culture were taken at various time points and measured for cell density at OD600. (C) Measurement of biofilm by crystal violet ammonium oxalate. (D) Measurement of biofilm by LSCM. a: HY9901ΔaraC 2.5d diagram; b: HY9901 2.5d diagram; c: HY9901ΔaraC 2d diagram; d: HY9901 2d diagram; HY9901 Biofilm thickness: 120 ± 20 μm; HY9901ΔaraC Biofilm thickness: 160 ± 20 μm.
The growth rate of HY9901 and ΔaraC was similar, and the deletion of araC gene has no effect on the growth of V. alginolyticus (p > 0.05) (Figure 2B). The exponential growth phase of the two bacterial strains was from 0 to 6 h, with growth stationary at 18 h, OD600 ≈ 1.8.
HY9901 and ΔaraC were inoculated on the LBS, and the results were as follows: the swarming circle of HY9901 was 39.7 ± 0.4 mm, and that of ΔaraC was 30.8 ± 0.5 mm (Table 4). The swarming circle diameter of HY9901ΔaraC was significantly smaller than that of HY9901 (p < 0.01), indicating that the swarming ability of HY9901 ΔaraC was significantly weakened.
Compared with wild strain HY9901, there was no significant difference in the extracellular protease activity of ΔaraC (p > 0.05) (Table 4).
The biofilm thickness was detected by crystal violet staining (Figure 2C) and confocal scanning (Figure 2D and Table 4). Crystal violet results showed that there were no significant differences in the ability of wild strain HY9901 and mutant strain ΔaraC to form biofilms before 24 h (p > 0.05), The ability of ΔaraC to form a biofilm was stronger than that of wild strain (p < 0.01). There was little difference after 48 h. Confocal electron microscopy (24 h) (Figure 2D) 2.5D results showed that the biofilm of ΔaraC was significantly thicker than that of the wild strain HY9901 (p < 0.01).
LD50 results showed that the 50% lethal dose of HY9901ΔaraC was 3,620 times higher than that of the wild strain (Tables 2, 4). The symptoms of body surface hyperemia, abdominal redness and swelling, and slow swimming were observed in the diseased fish. The results showed that the virulence of HY9901 ΔaraC was significantly decreased when compared with the wild strain (p < 0.01).
According to CLSI standards, the susceptibility of the mutant strain ΔaraC and wild strain HY9901 to 30 kinds of antibiotics was determined by disk diffusion method. Compared with the wild strain, the sensitivity of the ΔaraC strain to amikacin, tetracycline, and neomycin changed from resistant to moderate sensitivity; the sensitivity of the ΔaraC strain to minocycline and gentamicin changed from drug resistant to high sensitivity; and the sensitivity of the ΔaraC strain to chloramphenicol changed from moderate sensitivity to high sensitivity (Table 5).
Through the transcriptome sequencing analysis of HY9901 and mutant strain ΔaraC, a total of 1,376 differentially expressed genes were screened. Compared with HY9901, 635 genes were upregulated and 741 genes were downregulated in ΔaraC (Table S1). A total of 429 differentially expressed genes were enriched by GO function. The results showed that the differentially expressed genes of mutant strain ΔaraC and HY9901 were mainly related to locomotion, cilium, or flagellum-dependent cell motility and bacterial-type flagellum (Figure 3).
qRT-PCR was employed to analyze the transcription levels of T3SS-related effector genes at each time point (6, 12, 24, 48, and 72 h). The results showed that compared with wild strain HY9901, ΔaraC significantly decreased the expression of hopPmaJ (hop) (6, 12, 24, 48, and 72 h) (p < 0.01), but increased the expression of vopN, vopS, and vcrV (6, 12, 24, 48, and 72 h) (p < 0.01) (Figure 4).
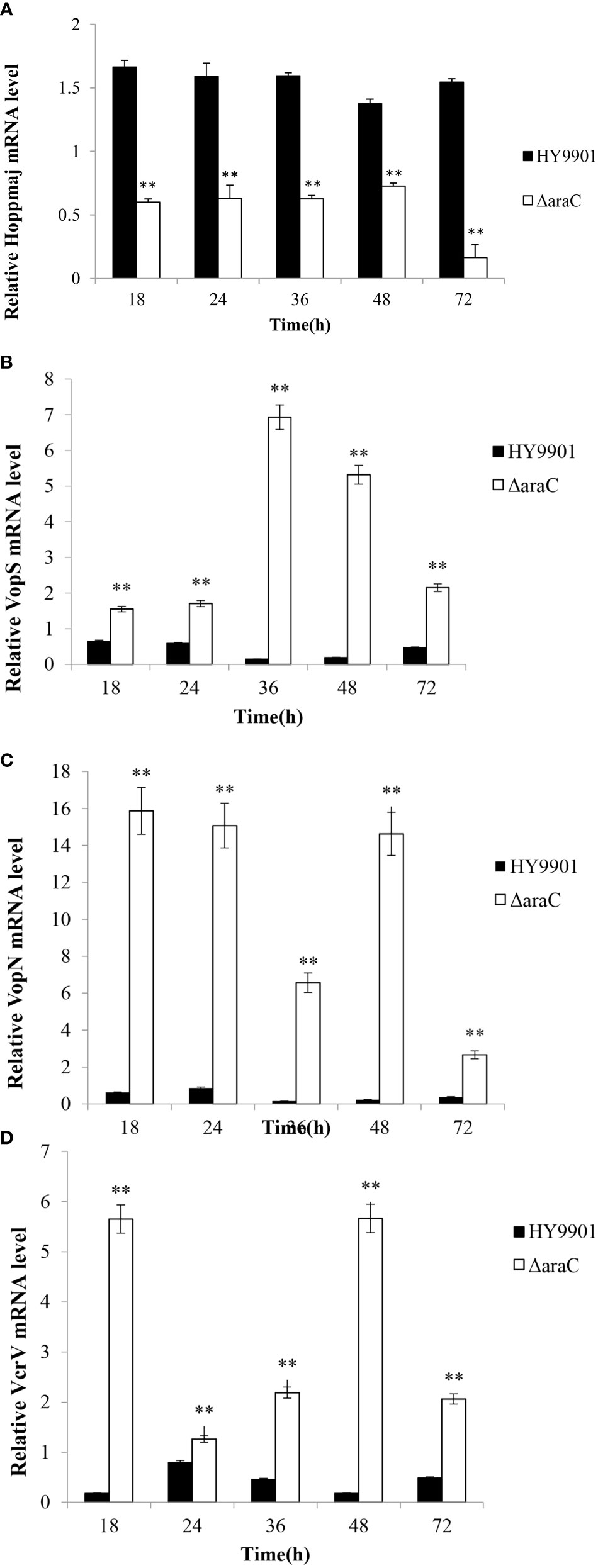
Figure 4 Expression of HY9901 and HY9901ΔaraC T3SS-related genes induced by DMEM. (A) Relative HopPmaJ mRNA level. (B) Relative VopS mRNA level. (C) Relative VopN mRNA level. (D) Relative VcrV mRNA level. *P < 0.05, **P < 0.01.
The mouse anti-serum to recombinant HopPmaJ rabbit antiserum of V. alginolyticus HY9901 strain was analyzed by Western blot with the whole bacterial protein extracted from HY9901 and ΔaraC. The results showed that the protein recognition and the deletion of araC gene significantly reduced the secretion of HopPmaJ protein (12.78 kDa) (Figure 5A).
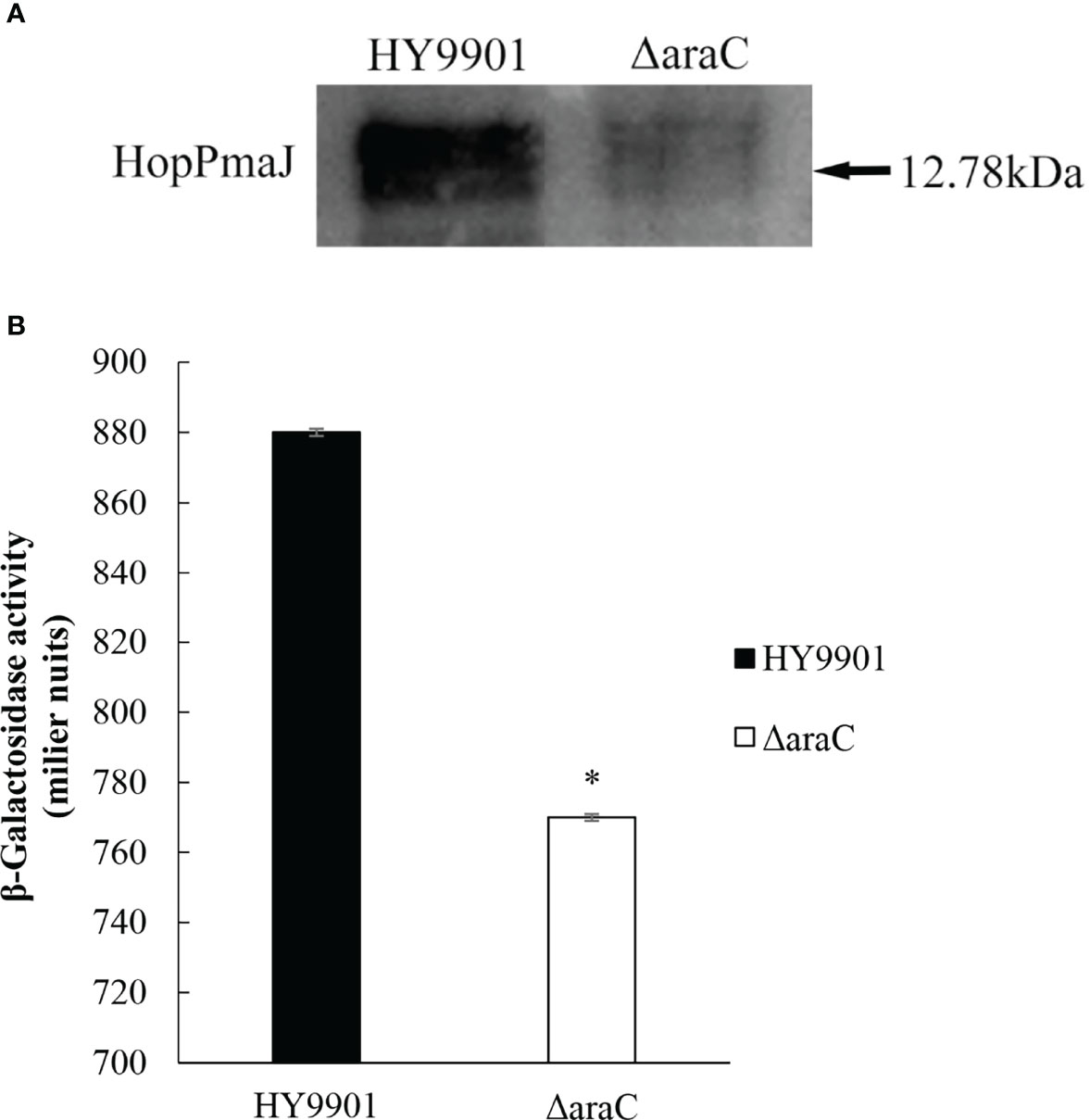
Figure 5 (A) Western blot showing the specificity for the polyclonal antibody against HopPmaJ (12.78 kDa) for the ΔaraC strain (right lane) and wild strain HY9901 (left lane). (B) β-Galactosidase activity from the hop promoter. Promoter activity of hop in HY9901 and ΔaraC. Bacteria were grown in TSB, and levels of β-galactosidase activity were measured (y-axis). The values are the average of three separate experiments, and the standard deviation is indicated by error bars. Asterisks represent statistical difference (p < 0.05).
In this experiment, the recombinant plasmid pME6522-hop-lacZ was introduced into HY9901 and ΔaraC. The lacZ reporter gene fusion experiment was used to study the regulatory relationship between the regulatory factor AraC and HopPmaJ protein (Figure 5B), which can be seen from the results detected in HY9901. β-Galactosidase activity was statistically significantly higher than ΔaraC and has a significant difference (p < 0.05), indicating that AraC can activate the expression of hop gene.
The mutant strain ΔaraC can survive in zebrafish. After 2 days of infection, ΔaraC reaches the maximum concentrations in the liver and spleen, then gradually decreases and disappears on the 7th day (Figure 6), indicating that the live-attenuated vaccine is safe for zebrafish.
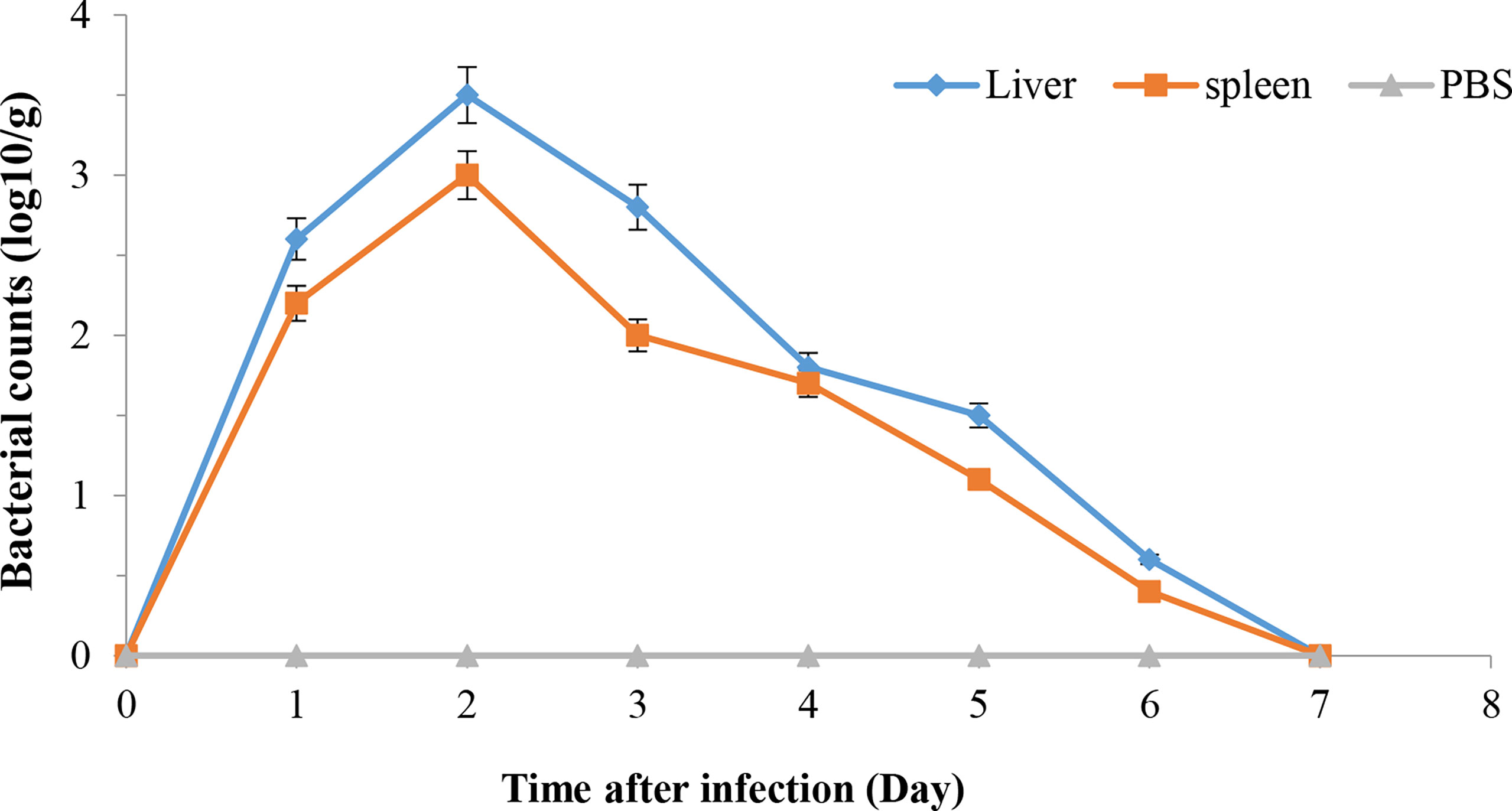
Figure 6 Propagation of HY9901ΔaraC in zebrafish liver and spleen following i.m. injection with 5 μl of 1 × 105 cfu ml−1 ΔaraC. Control fish were i.m. injection with 5 µl of sterile PBS. The number of viable bacteria was shown as the mean ± standard of three samples.
Fish vaccinated with ΔaraC were challenged 4 weeks post vaccination with HY9901. Within 14 days, the mortality rate in the control group injected with sterile PBS was 90%, the mortality rate in the injection immunization group was 35%, and the relative percentage survival was 61.3% (Figure 7).
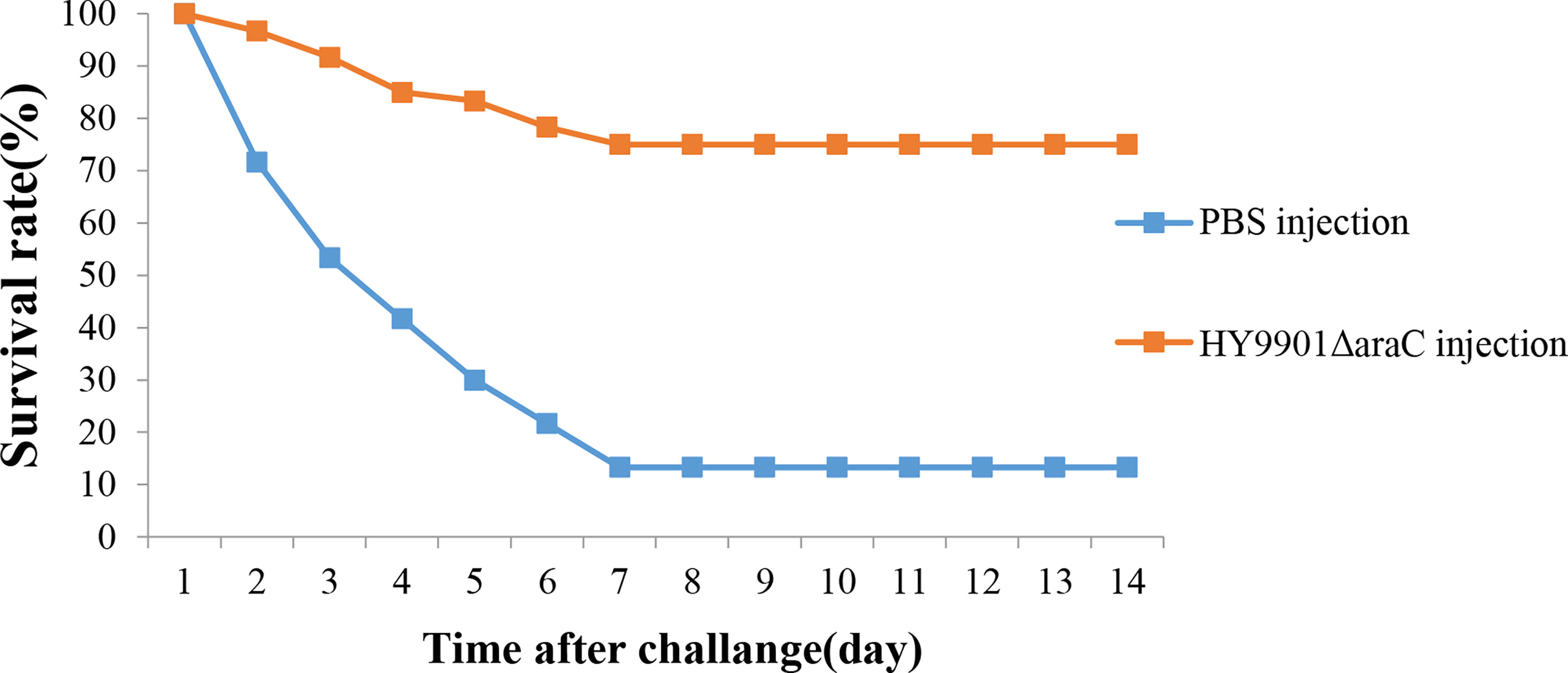
Figure 7 Survival in groups vaccinated with HY9901ΔaraC and PBS following challenge with Vibrio alginolyticus HY9901.
In the tissue sections, the fish infected with wild strain HY9901 showed symptoms such as hyperemia and blurred lymphocyte boundaries, and a small amount of hyperemia in the liver tissue of the immune group, but not in the negative control group (Figure 8).
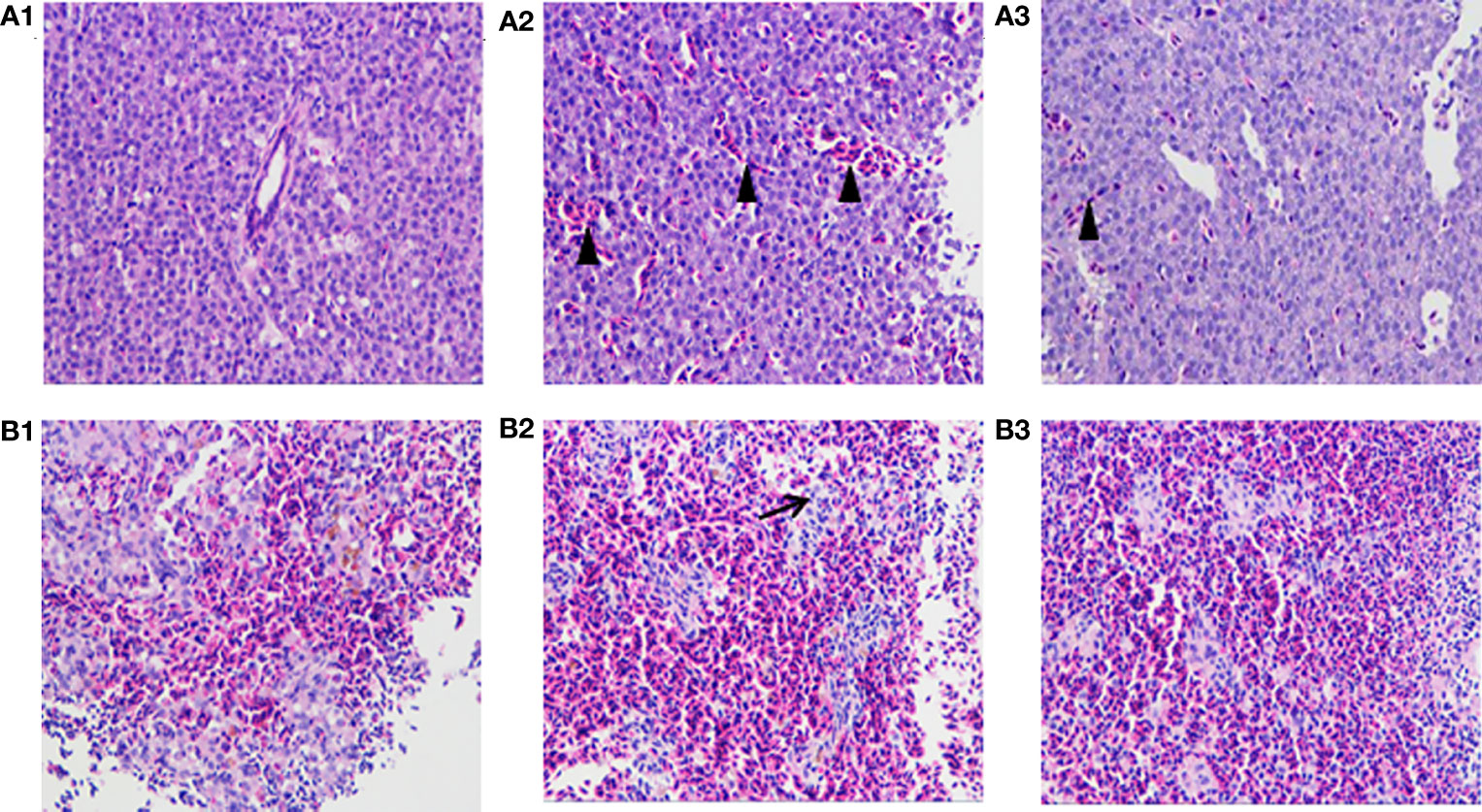
Figure 8 Histological sections of spleen and liver of zebrafish. Zebrafish liver tissue section (A1: PBS liver tissue, A2: HY9901 liver tissue, A3: ΔaraC liver tissue); zebrafish spleen tissue section (B1: PBS spleen tissue, B2: HY9901 spleen tissue, B3:ΔaraC spleen tissue). The triangle (▲) represents hyperemia, and the arrow (↗) represents the blurred boundary of lymphocytes in the figure.
Detection of immune gene expression in zebrafish immunized with HY9901ΔaraC live-attenuated vaccine was assessed by qPCR to analyze the transcriptional levels of pro-inflammatory and immunoglobulin-related immune genes. The results showed that the group vaccinated with HY9901ΔaraC had a significantly increased expression of il-6, il-6R, il-1β, il-8, igm, gata-1, lyz, rag-1, tlr5, and c/ebpβ genes in liver and il-6, il-6R, il-1β, igm, gata-1, lyz, rag-1, tlr5, and c/ebpβ genes in spleen compared to control fish injected with PBS (p < 0.01) (Figure 9).
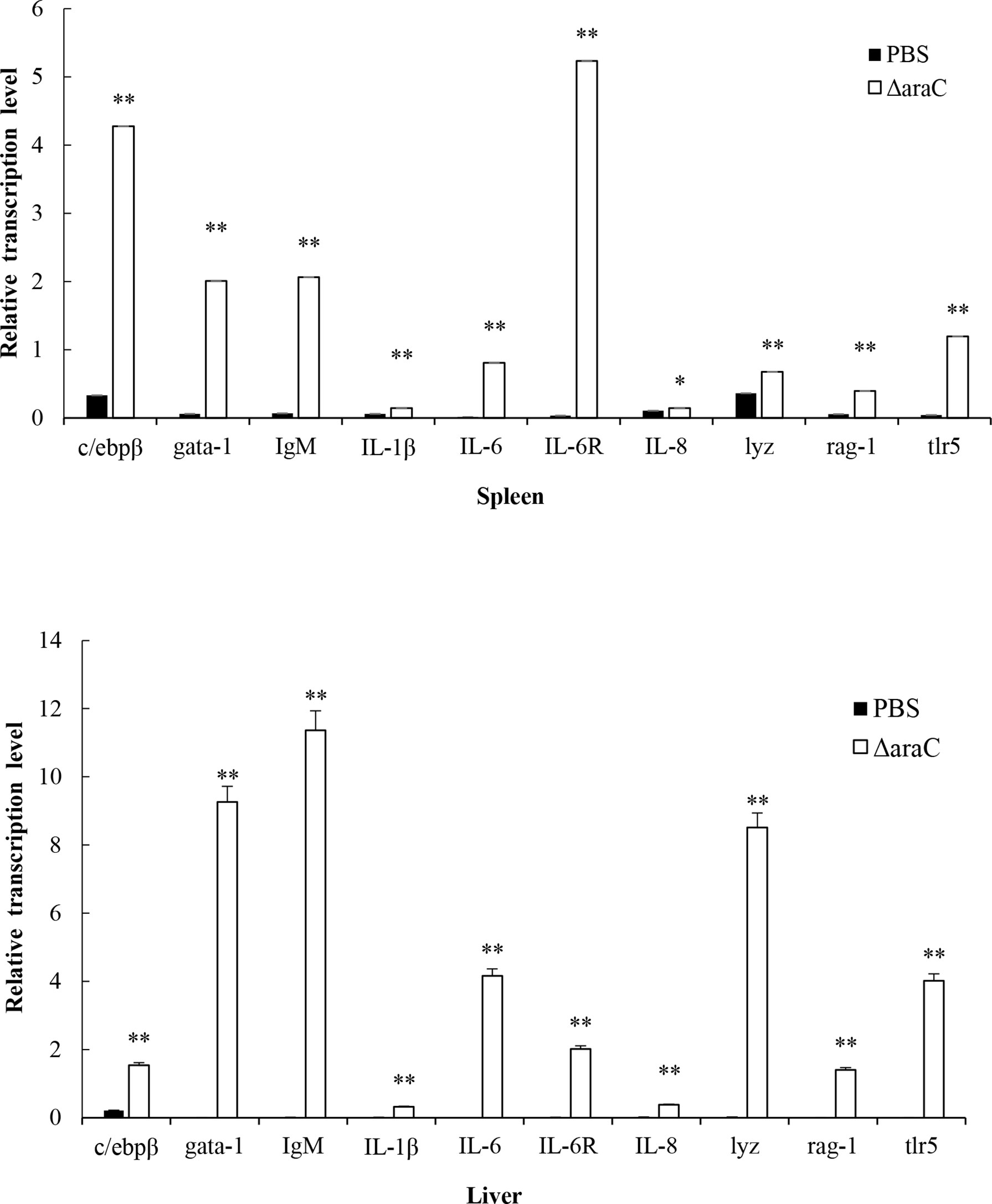
Figure 9 Comparative analysis of the expression of immune-related genes in liver and spleen of zebrafish given the live-attenuated vaccine and unvaccinated zebrafish. The head, kidney, and spleen of grouper were sampled at 1 day before challenge, and the mRNA level of each immune-related gene was normalized to that of β-actin. Bars represent the mean relative expression of three biological replicates and error bars represent standard deviation. *P < 0.05, **P < 0.01.
T3SS is an important virulence mechanism of Gram-negative bacteria, which has attracted much attention because it participates in the interaction between bacteria and host. The secretion of T3SS virulence proteins is directly or indirectly regulated by the AraC/XylS family of transcription factors in an environment-specific manner (Hueck, 1998; Hu et al., 2017). In Vibrio cholerae, the overall structure of ToxT is similar to that of AraC protein, and it can directly regulate virulence genes (Li et al., 2016a; Narm et al., 2020), but there is no research on the regulatory function of AraC in V. alginolyticus.
In this study, we obtained the mutant strain ΔaraC of V. alginolyticus, explored its biological characteristics and pathogenicity, and evaluated its potential as a live-attenuated vaccine. The extracellular products (ECPs) produced by V. alginolyticus have a variety of extracellular proteases that represent important virulence factors (Hare et al., 1983). Lee et al. (1996) reported that they isolated an alkaline serine protease from the ECP of V. alginolyticus, which can decompose and destroy the host tissue, resulting in fish disease (Lee et al., 1996). (Zhou et al., 2013) reported that there was no significant difference in growth rate, extracellular protease activity, and biofilm-forming ability after the deletion of vscO gene in V. alginolyticus HY9901 (Zhou et al., 2013). The mutant strain ΔsodB of V. alginolyticus also showed no significant difference in growth rate, but its ability to form biofilm was enhanced (Chen et al., 2019). Biofilms, matrix-encapsulated microbial aggregates, attach to biological or abiotic surfaces, and are resistant to antibiotics (Costerton et al., 1999; Hall-Stoodley et al., 2004). Biofilm formation is the function of self-generated EPS matrix wrapping biofilm cells, which are mainly composed of polysaccharides, proteins, lipids, and extracellular DNA (Flemming et al., 2016). Similarly, as found in this study, there was no significant difference in the growth rate and extracellular enzyme activity of HY9901ΔaraC when compared with the wild-type strain HY9901. These results suggested that gene araC may not affect these characteristics of V. alginolyticus. In addition, only at 24 h was the aggregation ability of biofilm significantly enhanced over that of the wild-type strain HY9901, and it was basically the same at other times. During the pathogenic process of bacteria, density sensing systems can regulate the formation of biofilm (Scutera et al., 2014). Therefore, AraC protein significantly affects the biofilm thickness during the stable period of bacterial growth, which may be related to the phenomenon of density sensing, but this needs further experimental verification.
The most important determinants of V. alginolyticus invasion and colonization are biofilms and motility (He et al., 2011). Bacterial motility is achieved via a moving organelle, the flagella (Sourjik and Wingreen, 2012). The flagellum is a spiral filament composed of flagellin (Abram and Koffler, 1964). The synthesis and movement of Vibrio harveyi flagella are controlled by a quorum sensing system, which inhibits the motility and reduces the toxicity to brine shrimp larvae. This highlights that motility is related to the virulence of Vibrio (Echazarreta and Klose, 2019). There are also many studies on the motility and virulence of V. alginolyticus. For example, the deletion of T3SS chaperone proteins vscO gene will weaken the swimming ability and reduce the virulence to fish by 10 times Zhou et al. (2013). The deletion of the effector protein expressed by hop gene also causes an impairment of swimming ability and decreased the virulence to grouper by 2,600 times (Pang et al., 2018). The deletion of the regulatory protein expressed by tyeA gene lessened the swimming ability and attenuated the virulence to zebrafish by nearly 40 times. In addition, qRT-PCR showed that the deletion of apparatus proteins vscQ gene downregulated the expression of flagellin genes flaA and fliG, and the absence of flagellin will affect the normal growth and movement of flagellum (Millikan and Ruby, 2004), which was the reason why it showed a weak population phenotype and reduced its virulence to zebrafish by 4.6 times (Wu et al., 2020). In this study, ΔaraC significantly reduced its swimming ability and toxicity to zebrafish, and transcriptome data showed that araC gene is closely related to movement, flagellum switch, and flagellum composition. The decline of virulence and swimming ability may be related to bacterial flagella. These results suggest that araC gene may affect the swimming ability and virulence of V. alginolyticus by regulating the expression of flagella-related genes, but its specific regulatory mechanism needs to be further studied.
The regulation of T3SS is mainly achieved through the regulation of the expression and secretion of its effector proteins, which have been demonstrated in many independent studies in vitro and in vivo. In P. aeruginosa, ExsA is the central transcriptional regulator of T3SS (Hovey and Frank, 1995), which controls the expression of secretory apparatus translocation mechanism and secreted effector proteins, and ExsD is the anti-activator of ExsA. McCaw et al. (2002) showed that the deletion of exsD gene significantly promoted the secretion of T3SS and the expression of regulatory protein and effector protein through a lacZ reporter gene fusion experiment during which β-galactosidase activity was assayed (McCaw et al., 2002). In V. parahaemolyticus, ExsA and ExsD are also the main positive and negative regulators of T3SS1 (Zhou et al., 2010). The expression and secretion of T3SS1 are inseparable from ExsA, and EMSA experiments showed that the ExsA protein can directly interact with the promoter sequence of effector protein Vp1687. In addition, the destruction of the regulatory protein exsD gene or the overexpression of exsA will also lead to the constructive expression of T3SS1 gene and the secretion of effector protein Vp1656 (Zhou et al., 2008). Liu et al. (2016) found that V. alginolyticus ExsA plays a positive regulatory role on effector proteins Va1686 and Va1687 (Liu et al., 2016). Zhou et al. (2020) showed that TyeA is the regulatory protein of V. alginolyticus T3SS. In the absence of tyeA gene, the mRNA expression of effector proteins VopS, VopN, and HopPmaj was significantly upregulated (Zhou et al., 2020). In this study, the deletion of araC gene also increased the mRNA expression of VopS, VcrV, and VopN, but reduced the mRNA expression of HopPmaJ. Western blotting and β-galactosidase reporter gene experiments had also verified that AraC could activate the expression of HopPmaJ. AraC may regulate the transcription of T3SS-related genes in some way, so as to increase and reduce the expression of effector proteins of V. alginolyticus. However, the specific regulation mechanism needs to be further evaluated. This study laid a foundation for the follow-up studies of T3SS regulation mechanisms.
The mechanism of bacterial drug resistance is complex; in addition to the antagonistic effects of biofilm on antibiotics (Stewart, 2001), the drug resistance of bacteria can also be obtained through the transmission of drug-resistant plasmid conjugation and transduction (Zhou et al., 2018). Furthermore, gene mutation will spontaneously produce a new resistance characteristic (Levy, 1998) or, from other iron carriers, carry drug resistance genes. In addition, V. alginolyticus also carries drug resistance genes. Raissy et al. (2012) tested 20 strains of V. alginolyticus (from seafood including fish, shrimp, lobster, and crab from cages off the Persian Gulf) for drug sensitivity and PCR detection. The results showed that several strains of V. alginolyticus could possess strB, tetS, and ermB genes, which encode streptomycin, tetracycline, and erythromycin resistance, respectively and some strains did not contain tetS gene. However, resistance to tetracycline may be due to the existence of other genes encoding tetracycline resistance, such as tetA, tetB, tetM, and tetK (Raissy et al., 2012). In this study, the results of drug sensitivity tests showed that V. alginolyticus HY9901 was resistant to most antibiotics, while the deleted strain ΔaraC became sensitive to amikacin, tetracycline, neomycin, minocycline, and gentamicin. We speculated that the deletion mutation of araC gene may lead to new drug resistance characteristics, or AraC protein may regulate the expression of drug resistance genes. The specific drug resistance mechanism needs to be further studied.
Drug resistance carried by bacteria will flow from the food chain into human body via aquatic products (Chen et al., 2020). Human infection with drug-resistant bacteria will limit the choice of antibiotics, resulting in difficult treatment, which poses a threat to human health (Shahimi et al., 2021). Therefore, it is necessary to find alternative methods to control V. alginolyticus strains harmful to aquatic animals. Production of a live-attenuated vaccine provides a promising potential prophylactic tool for controlling bacterial fish diseases (Zhang et al., 2013). Vaccines play a major role by reducing or eliminating the risk of infection caused by antibiotic-resistant strains. This can be achieved by antibody-mediated killing of organisms in the blood (Klugman and Black, 2018). In terms of immune protection and production, live-attenuated vaccines have advantages over inactivated vaccines (Chen et al., 2020). Because zebrafish produces fast and many immune-related genes have been found, it is a model suitable for studying immune defense (Randelli et al., 2008). Zhang et al. (2013) found that zebrafish inoculated with live-attenuated Vibrio anguillarum vaccine induced good proinflammatory cytokine IL-1β response but the expression chemokine IL-8 gene transcription decreased compared with non-inoculated fish. However, following challenge, the expression level of adaptive immune-related genes increased, indicating that live-attenuated vaccine triggered protection by inhibiting the expression of inflammatory factors post-challenge and enhanced adaptive immunity (Zhang et al., 2013). In this study, the relative survival rate of zebrafish immunized with vaccine candidate ΔaraC was 61.3%. Evaluation of vaccine safety showed that vaccination of ΔaraC live-attenuated vaccine did not cause obvious pathological changes or clinical symptoms. Compared with the PBS group, HY9901 ΔaraC immunization significantly increased the expression of gata-1, il6, igm, il-1β, lyz, rag-1, tlr5, and c/ebpβ genes in the spleen and liver, possibly relating to protection in zebrafish.
We analyzed the biological characteristics and pathogenicity of HY9901 mutant strain ΔaraC. The mutant V. alginolyticus strain ΔaraC did not adversely affect growth and extracellular protease production. Furthermore, the biofilm-forming ability did not change significantly, but weakening of the swimming ability was evident, as well as the subsequent virulence to zebrafish. Wild strains and mutant strains were resistant to most antibiotics. qRT-PCR, Western blotting, and β-galactosidase reporter gene experiments showed that AraC could upregulate the expression of the T3SS effector protein HopPmaJ. As a live-attenuated vaccine, it is protective against V. alginolyticus and can induce protective immune responses in zebrafish. These results provide a reference for the study of the regulatory mechanisms of the regulatory protein on T3SS. This study provides further evidence of immune efficacy and safety of live-attenuated bacterial vaccines for potential use in aquaculture. Such data are imperative for evidence-based support for the technology’s application in the prevention and control of fish diseases caused by V. alginolyticus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Animal Ethics Committee of Guangdong Ocean University (Zhanjiang, China).
All the authors have made extensive contributions to the work in this manuscript. JW and HF designed the experiment; JW, FY and HF generated experimental data and wrote manuscripts. HF, LC, WZ, and XX assisted in completing the experiment. HP, SM, JJ, and NW conceived the work and critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 32073015), the Natural Science Foundation of Guangdong Province (No. 2021A1515011078), the Innovation Program in Postgraduate Education of Guangdong Ocean University (No. 202153), the Undergraduate Innovation Team of Guangdong Ocean University (No. CCTD201802), the Innovation and Entrepreneurship Training Program for College Students (No. CXXL2022005) the Science and Technology Innovation Cultivation Program for College Student (No. pdjh2021b0239), and the Innovation Program in Postgraduate Education of Guangdong Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.938742/full#supplementary-material
Abram D., Koffler H. (1964). In Vitro Formation of Flagella-Like Filaments and Other Structures From Flagellin. J. Mol. Biol. 9, 168–185. doi: 10.1016/s0022-2836(64)80098-x
Aly S. M., Eissa A. E., Elbanna N. I., Albutti A. (2021). Efficiency of Monovalent and Polyvalent Vibrio Alginolyticus and Vibrio Parahaemolyticus Vaccines on the Immune Response and Protection in Gilthead Sea Bream, Sparus Aurata (L.) Against Vibriosis. Fish Shellfish Immunol. 111, 145–151. doi: 10.1016/j.fsi.2020.10.011
Bunpa S., Chaichana N., Teng J. L. L., Lee H. H., Woo P. C. Y., Sermwittayawong D., et al. (2020). Outer Membrane Protein A (OmpA) is a Potential Virulence Factor of Vibrio Alginolyticus Strains Isolated From Diseased Fish. J. Fish Diseases. 43, 275–284. doi: 10.1111/jfd.13120
Bunpa S., Nishibuchi M., Thawonsuwan J., Sermwittayawong N. (2018). Genetic Heterogeneity Among Vibrio Alginolyticus Strains, and Design of a PCR-Based Identification Method Using gyrB Gene Sequence. Can. J. Microbiol. 64, 1–10. doi: 10.1139/cjm-2017-0269
Cai X. H., Peng Y. H., Wang Z. C., Huang T., Xiong X. Y., Huang Y. C., et al. (2016). Characterization and Identification of Streptococci From Golden Pompano in China. Dis. Aquat Organ. 119, 207–217. doi: 10.3354/dao02998
Cai S. H., Wu Z. H., Jian J. C., Lu Y. S. (2007). Cloning and Expression of Gene Encoding the Thermostable Direct Hemolysin From Vibrio Alginolyticus Strain HY9901, the Causative Agent of Vibriosis of Crimson Snapper (Lutjanus Erythopterus). J. Appl. Microbiol. 103, 289–296. doi: 10.1111/j.1365-2672.2006.03250.x
Chen J. M., Sun R. X., Pan C. G., Sun Y., Mai B. X., Li Q. X. (2020). Antibiotics and Food Safety in Aquaculture. J. Agric. Food Chem. 68, 11908–11919. doi: 10.1021/acs.jafc.0c03996
Chen Y., Wu F., Pang H., Tang J., Cai S., Jian J. (2019). Superoxide Dismutase B (Sodb), an Important Virulence Factor of Vibrio Alginolyticus, Contributes to Antioxidative Stress and its Potential Application for Live Attenuated Vaccine. Fish Shellfish Immunol. 89, 354–360. doi: 10.1016/j.fsi.2019.03.061
Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial Biofilms: A Common Cause of Persistent Infections. Sci. (New York N.Y.). 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Ding C. C., Liu Q., Li J., Ma J. F., Wang S. Y., Dong Q. L., et al. (2019). Attenuated Listeria Monocytogenes Protecting Zebrafish (Danio Rerio) Against Vibrio Species Challenge. Microbial Pathogenesis. 132, 38–44. doi: 10.1016/j.micpath.2019.03.040
Echazarreta M. A., Klose K. E. (2019). Vibrio Flagellar Synthesis. Front. Cell. Infection Microbiol. 9. doi: 10.3389/fcimb.2019.00131
Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Guennoc C.M., Rose C., Guinnet F., Miquel I., Labbe J., Deveau A. (2017). A New Method for Qualitative Multi-Scale Analysis of Bacterial Biofilms on Filamentous Fungal Colonies Using Confocal and Electron Microscopy. J. Vis. Exp. 119, 54771. doi: 10.3791/54771
Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Hare P., Scott-Burden T., Woods D. R. (1983). Characterization of Extracellular Alkaline Proteases and Collagenase Induction in Vibrio Alginolyticus. J. Gen. Microbiol. 129, 1141–1147. doi: 10.1099/00221287-129-4-1141
He H. H., Wang Q. Y., Sheng L. L., Liu Q., Zhang Y. X. (2011). Functional Characterization of Vibrio Alginolyticus Twin-Arginine Translocation System: Its Roles in Biofilm Formation, Extracellular Protease Activity, and Virulence Towards Fish. Curr. Microbiol. 62, 1193–1199. doi: 10.1007/s00284-010-9844-6
Hovey A. K., Frank D. W. (1995). Analyses of the DNA-Binding and Transcriptional Activation Properties of ExsA, the Transcriptional Activator of the Pseudomonas Aeruginosa Exoenzyme S Regulon. J. bacteriology. 177, 4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995
Hueck C. J. (1998). Type III Protein Secretion Systems in Bacterial Pathogens of Animals and Plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Hu Y. M., Huang H., Cheng X., Shu X. S., White A. P., Stavrinides J., et al. (2017). A Global Survey of Bacterial Type III Secretion Systems and Their Effectors. Environ. Microbiol. 19, 3879–3895. doi: 10.1111/1462-2920.13755
Kinoshita M., Shimizu M., Akiyama K., Kato H., Moriyama K., Sawa T. (2020). Epidemiological Survey of Serum Titers From Adults Against Various Gram-Negative Bacterial V-Antigens. PloS One 15 e0220924. doi: 10.1371/journal.pone.0220924
Klugman K. P., Black S. (2018). Impact of Existing Vaccines in Reducing Antibiotic Resistance: Primary and Secondary Effects. Proc. Natl. Acad. Sci. U. S. A. 115, 12896–12901. doi: 10.1073/pnas.1721095115
Lee K. K., Yu S. R., Yang T. I., Liu P. C., Chen F. R. (1996). Isolation and Characterization of Vibrio Alginolyticus Isolated From Diseased Kuruma Prawn, Penaeus Japonicus. Lett. Appl. Microbiol. 22, 111–114. doi: 10.1111/j.1472-765x.1996.tb01121.x
Levy S. B. (1998). The Challenge of Antibiotic Resistance. Sci. American. 278, 46–53. doi: 10.1038/scientificamerican0398-46
Liu J. X., Lu S. Y., Orfe L. H., Ren C. H., Hu C. Q., Call D. R., et al. (2016). ExsE Is a Negative Regulator for T3SS Gene Expression in Vibrio Alginolyticus. Front. Cell. Infection Microbiol. 6. doi: 10.3389/fcimb.2016.00177
Liu A. C., Thomas N. A. (2015). Transcriptional Profiling of Vibrio Parahaemolyticus exsA Reveals a Complex Activation Network for Type III Secretion. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01089
Li J. Q., Wehmeyer G., Lovell S., Battaile K. P., Egan S. M. (2016a). 1.65 Angstrom Resolution Structure of the AraC-Family Transcriptional Activator ToxT From Vibrio Cholerae. Acta Crystallographica Section F-Structural Biol. Commun. 72, 726–731. doi: 10.1107/s2053230x1601298x
Li X. C., Xiang Z. Y., Xu X. M., Yan W. H., Ma J. M. (2009). Endophthalmitis Caused by Vibrio Alginolyticus. J. Clin. Microbiol. 47, 3379–3381. doi: 10.1128/jcm.00722-09
Li W., Yao Z., Sun L., Hu W., Cao J., Lin W., et al. (2016b). Proteomics Analysis Reveals a Potential Antibiotic Cocktail Therapy Strategy for Aeromonas Hydrophila Infection in Biofilm. J. Proteome Res. 15, 1810–1820. doi: 10.1021/acs.jproteome.5b01127
Lowden M. J., Skorupski K., Pellegrini M., Chiorazzo M. G., Taylor R. K., Kull F. J. (2010). Structure of Vibrio Cholerae ToxT Reveals a Mechanism for Fatty Acid Regulation of Virulence Genes. Proc. Natl. Acad. Sci. United States America. 107, 2860–2865. doi: 10.1073/pnas.0915021107
McCaw M. L., Lykken G. L., Singh P. K., Yahr T. L. (2002). ExsD is a Negative Regulator of the Pseudomonas Aeruginosa Type III Secretion Regulon. Mol. Microbiol. 46, 1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x
Millikan D. S., Ruby E. G. (2004). Vibrio Fischeri Flagellin A is Essential for Normal Motility and for Symbiotic Competence During Initial Squid Light Organ Colonization. J. Bacteriology. 186, 4315–4325. doi: 10.1128/jb.186.13.4315-4325.2004
MO F., Afriyie G., Hu J., Wang J., Zhou S., Wang Z., et al. (2021). Molecular Cloning and Bioinformatics Analysis of araC Gene of Vibrio Alginolyticus. Asian Agric. Res. 13, 19–23. doi: 10.19601/j.cnki.issn1943-9903.2021.02.005
Narm K. E., Kalafatis M., Slauch J. M. (2020). HilD, HilC, and RtsA Form Homodimers and Heterodimers To Regulate Expression of the Salmonella Pathogenicity Island I Type III Secretion System. J. Bacteriology. 202, e00012-20. doi: 10.1128/jb.00012-20
O’Toole G. A. (2011). Microtiter Dish Biofilm Formation Assay. Jove-Journal Visualized Experiments. doi: 10.3791/2437
Osorio C. R. (2018). T3SS Effectors in Vibrios: Homology in Sequence, Diversity in Biological Functions? Virulence 9, 721–723. doi: 10.1080/21505594.2018.1435965
Pang H. Y., Chang Y. S., Zheng H. W., Tan H. M., Zhou S. H., Zeng F. Y., et al. (2022). A Live Attenuated Strain of HY9901ΔvscB Provides Protection Against Vibrio Alginolyticus in Pearl Gentian Grouper (♀ Epinephelus Fuscoguttatus × ♂Epinephelus Lanceolatu). Aquaculture 546 737353. doi: 10.1016/j.aquaculture.2021.737353
Pang H. Y., Qiu M. S., Zhao J. M., Hoare R., Monaghan S. J., Song D. W., et al. (2018). Construction of a Vibrio Alginolyticus Hoppmaj (Hop) Mutant and Evaluation of its Potential as a Live Attenuated Vaccine in Orange-Spotted Grouper (Epinephelus Coioides). Fish Shellfish Immunol. 76, 93–100. doi: 10.1016/j.fsi.2018.02.012
Pearson M. M. (2019). Methods for Studying Swarming and Swimming Motility. Methods Mol. Biol. (Clifton N.J.). 2021, 15–25. doi: 10.1007/978-1-4939-9601-8_3
Raissy M., Moumeni M., Ansari M., Rahimi E. (2012). Antibiotic Resistance Pattern of Some Vibrio Strains Isolated From Seafood. Iranian J. Fisheries Sci. 11, 618–626. doi: 10.4103/0972-124X.99267
Rameshkumar P., Kalidas C., Tamilmani G., Sakthivel M., Nazar A. K. A., Maharshi V. A., et al. (2014). Microbiological and Histopathological Investigations of Vibrio Alginolyticus Infection in Cobia Rachycentron Canadum (Linnaeus 1766) Cultured in Sea Cage. Indian J. Fisheries. 61, 124–127.
Randelli E., Buonocore F., Scapigliati G. (2008). Cell Markers and Determinants in Fish Immunology. Fish Shellfish Immunol. 25, 326–340. doi: 10.1016/j.fsi.2008.03.019
Schleif R. (2000). Regulation of the L-Arabinose Operon of Escherichia Coli. Trends Genet. 16, 559–565. doi: 10.1016/s0168-9525(00)02153-3
Scutera S., Zucca M., Savoia D. (2014). Novel Approaches for the Design and Discovery of Quorum-Sensing Inhibitors. Expert Opin. Drug Discovery. 9, 353–366. doi: 10.1517/17460441.2014.894974
Shahimi S., Elias A., Abd Mutalib S., Salami M., Fauzi F., Zaini N. A. M., et al. (2021). Antibiotic Resistance and Determination of Resistant Genes Among Cockle (Anadara Granosa) Isolates of Vibrio Alginolyticus. Environ. Sci. pollut. Res. 28, 44002–44013. doi: 10.1007/s11356-021-13665-4
Sheppard D. E., Englesberg E. (1967). Further Evidence for Positive Control of the L-Arabinose System by Gene araC. J. Mol. Biol. 25, 443–454. doi: 10.1016/0022-2836(67)90197-0
Shi J., Zhao W., Xie J., Zhu Y., Pan Y., Ou J., et al. (2021). Comparison on the Growth Heterogeneity of Vibrio Parahaemolyticus Coupled With Strain Source and Genotype Analyses in Different Oligotrophic Conditions. J. Food protection. 84, 1904–1910. doi: 10.4315/jfp-21-089
Simon R., Priefer U., Pühler A. (1983). A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1, 784–791. doi: 10.1038/nbt1183-784
Singer J., Englesberg E. (1971). Arabinose Transport in araC- Strains of EscherichiA B-R. Biochim. Biophys. Acta 249, 498–505. doi: 10.1016/0005-2736(71)90125-8
Sourjik V., Wingreen N. S. (2012). Responding to Chemical Gradients: Bacterial Chemotaxis. Curr. Opin. Cell Biol. 24, 262–268. doi: 10.1016/j.ceb.2011.11.008
Stewart P. S. (2001). Multicellular Resistance: Biofilms. Trends Microbiol. 9, 204. doi: 10.1016/s0966-842x(01)01983-7
Tam V. C., Suzuki M., Coughlin M., Saslowsky D., Biswas K., Lencer W. I., et al. (2010). Functional Analysis of VopF Activity Required for Colonization in Vibrio Cholerae. Mbio 1 e00289-10. doi: 10.1128/mBio.00289-10
Wu P. W., Zhou S. H., Su Z. H., Liu C., Zeng F. Y., Pang H. Y., et al. (2020). Functional Characterization of T3SS C-Ring Component VscQ and Evaluation of its Mutant as a Live Attenuated Vaccine in Zebrafish (Danio Rerio) Model. Fish Shellfish Immunol. 104, 123–132. doi: 10.1016/j.fsi.2020.05.063
Yahr T. L., Wolfgang M. C. (2006). Transcriptional Regulation of the Pseudomonas Aeruginosa Type III Secretion System. Mol. Microbiol. 62, 631–640. doi: 10.1111/j.1365-2958.2006.05412.x
Yang J., Hocking D. M., Cheng C., Dogovski C., Perugini M. A., Holien J. K., et al. (2013). Disarming Bacterial Virulence Through Chemical Inhibition of the DNA Binding Domain of an AraC-Like Transcriptional Activator Protein. J. Biol. Chem. 288, 31115–31126. doi: 10.1074/jbc.M113.503912
Yang J., Tauschek M., Robins-Browne R. M. (2011). Control of Bacterial Virulence by AraC-Like Regulators That Respond to Chemical Signals. Trends Microbiol. 19, 128–135. doi: 10.1016/j.tim.2010.12.001
Yang B., Zhai S. Y., Li X., Tian J., Li Q., Shan H. W., et al. (2021). Identification of Vibrio Alginolyticus as a Causative Pathogen Associated With Mass Summer Mortality of the Pacific Oyster (Crassostrea Gigas) in China. Aquaculture 535, 736363. doi: 10.1016/j.aquaculture.2021.736363
Zhang Y. L., Tan H. M., Yang S. P., Huang Y. C., Cai S. H., Jian J. C., et al. (2022). The Role of dctP Gene in Regulating Colonization, Adhesion and Pathogenicity of Vibrio Alginolyticus Strain HY9901. J. Fish Diseases. 45, 421–434. doi: 10.1111/jfd.13571
Zhang Z. H., Wu H. Z., Xiao J. F., Wang Q. Y., Liu Q., Zhang Y. X. (2013). Immune Responses Evoked by Infection With Vibrio Anguillarum in Zebrafish Bath-Vaccinated With a Live Attenuated Strain. Veterinary Immunol. Immunopathology. 154, 138–144. doi: 10.1016/j.vetimm.2013.05.012
Zhou X. H., Konkel M. E., Call D. R. (2010). Vp1659 Is a Vibrio Parahaemolyticus Type III Secretion System 1 Protein That Contributes to Translocation of Effector Proteins Needed To Induce Cytolysis, Autophagy, and Disruption of Actin Structure in HeLa Cells. J. Bacteriology. 192, 3491–3502. doi: 10.1128/jb.01493-09
Zhou Z. J., Pang H. Y., Ding Y., Cai J., Huang Y. C., Jian J. C., et al. (2013). VscO, a Putative T3SS Chaperone Escort of Vibrio Alginolyticus, Contributes to Virulence in Fish and Is a Target for Vaccine Development. Fish Shellfish Immunol. 35, 1523–1531. doi: 10.1016/j.fsi.2013.08.017
Zhou X. H., Shah D. H., Konkel M. E., Call D. R. (2008). Type III Secretion System 1 Genes in Vibrio Parahaemolyticus are Positively Regulated by ExsA and Negatively Regulated by ExsD. Mol. Microbiol. 69, 747–764. doi: 10.1111/j.1365-2958.2008.06326.x
Zhou S. H., Tu X. T., Pang H. Y., Hoare R., Monaghan S. J., Luo J. J., et al. (2020). A T3SS Regulator Mutant of Vibrio Alginolyticus Affects Antibiotic Susceptibilities and Provides Significant Protection to Danio Rerio as a Live Attenuated Vaccine. Front. Cell. Infection Microbiol. 10. doi: 10.3389/fcimb.2020.00183
Keywords: Vibrio alginolyticus, type III secretory system, gene araC, characteristics, live-attenuated vaccine
Citation: Wang J, Zeng F, Feng H, Chen L, Zhang W, Xiao X, Monaghan SJ, Jian J, Pang H and Wang N (2022) A T3SS Regulator Mutant HY9901 ΔaraC of Vibrio alginolyticus Decreased the Expression of HopPmaJ and Provided Protection to Danio rerio as a Live-Attenuated Vaccine. Front. Mar. Sci. 9:938742. doi: 10.3389/fmars.2022.938742
Received: 08 May 2022; Accepted: 30 May 2022;
Published: 07 July 2022.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Ming Jiang, Sun Yat-sen University, ChinaCopyright © 2022 Wang, Zeng, Feng, Chen, Zhang, Xiao, Monaghan, Jian, Pang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanying Pang, cGh5aW5nMTIxOEAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.