
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 03 October 2022
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.934307
This article is part of the Research TopicEVOLMAR 2020 - The First Italian Congress on Marine EvolutionView all 7 articles
 Jenny Melo Clavijo1*
Jenny Melo Clavijo1* Corinna Sickinger1
Corinna Sickinger1 Sabrina Bleidißel1
Sabrina Bleidißel1 Gilles Gasparoni2
Gilles Gasparoni2 Sascha Tierling2
Sascha Tierling2 Angelika Preisfeld1
Angelika Preisfeld1 Gregor Christa2*
Gregor Christa2*Photosymbiosis is found in different animal lineages and is best understood in cnidarians. A successful initiation and maintenance of the symbiosis between the animal hosts and the photosymbiotic partners is based on a recognition by specific host receptors. This triggers signaling cascades that promote the photobiont tolerance by the host, including an interpartner nutrient exchange and the ability of the host to cope with increased levels of reactive oxygen species (ROS) generated by the photobiont. Key to the successful symbiosis is the inhibition of the phagosomal maturation resulting in the formation of the symbiosome. In animals other than cnidarians, little is known about the photosymbiosis initiation and maintenance, for instance in sea slugs belonging to the Nudibranchia. Here, we investigated the gene expression profile of Berghia stephanieae, which is able to incorporate Breviolum minutum from its cnidarian prey Exaiptasia diaphana (Rapp, 1829) but is not able to maintain the algae for more than a couple of days during starvation. We show that the recognition of the algae is based on similar mechanisms present in cnidarians, and we identified some additional candidate genes that might be molluscan specific for photobiont recognition. Downstream, B. stephanieae responds to increased levels of ROS but is not able to stop the phagosomal maturation or decrease the immune response against B. minutum, which seem to be the key factors missing in B. stephanieae that accounts for the unstable symbiosis in this slug. Hence, B. stephanieae can be considered a transitional state toward a stable photosymbiosis and can help to elucidate general aspects of the evolutionary processes involved in establishing photosymbioses in animals.
Several animal lineages evolved the ability to engage in a symbiotic relationship—the so-called photosymbiosis—with unicellular algae or Cyanobacteria (Melo Clavijo et al., 2018). This symbiosis is especially beneficial for the animal host in oligotrophic environments (Stanley and Lipps, 2011; Roth, 2014) and is based on a nutritional exchange between the animal host and the photosynthetic symbiont (the photobiont) (Matthews et al., 2017; Rädecker et al., 2021). Most photosymbiotic animals acquire the photobiont from the environment (horizontal transmission), but some marine slugs belonging to the Cladobranchia (Gastropoda, Nudibranchia) evolved a unique strategy. Cladobranchs feed on photosymbiotic cnidarians and specifically incorporate the prey’s symbiotic dinoflagellates (Symbiodiniaceae) into epithelial cells of their own digestive gland system (Rudman, 1987; Burghardt and Wägele, 2004; Burghardt et al., 2005; Burghardt and Wägele, 2006; Burghardt et al., 2008; Wägele et al., 2010). This process makes the slugs the only known secondary host of photobionts in the animal kingdom (Rousseau, 1934; Rousseau, 1935; Wägele and Johnsen, 2001). Depending on the Cladobranchia species, the algae are then directly digested intracellularly (non-symbiotic species) or maintained physiologically active for variable times as photobionts, ranging from a couple of weeks (unstable symbiosis) to several months (stable symbiosis) (reviewed in Rola et al., 2022). In species with unstable symbiosis, the photobionts are often found photosynthetically active in the animal’s feces, suggesting a symbiont expelling mechanism that might be based on vomocytosis (Jacobovitz et al., 2021); in species with stable symbiosis, the healthy photobionts are not expelled and are even able to divide intracellularly (Kempf, 1991; Burghardt and Wägele, 2004; Burghardt and Wägele, 2004; Burghardt et al., 2005; Burghardt and Wägele, 2006; Burghardt et al., 2008; Wägele et al., 2010; Rola et al., 2022).
The evolution of photosymbiosis in Cladobranchia, and hence the mechanisms underpinning the selective photobiont recognition and the subsequent maintenance for the varying time periods, remains unknown. Regardless of the evolutionary distance between the slugs and their cnidarian prey, photobiont recognition and maintenance mechanisms might indeed be similar (Chan et al., 2018; Melo et al., 2020). The winnowing, a series of complex steps that occur to establish a stable symbiosis (Nyholm and McFall-Ngai, 2004), has been well described in cnidarians (Davy et al., 2012) and can serve as a reference to understand if the same mechanisms are at play in Cladobranchia. For instance, in cnidarians the successful initiation of the photosymbiosis is initiated by the interaction of pattern recognition receptors (PRRs) of the host, such as scavenger receptors and lectins, with membrane-associated molecular patterns (MAMPs) of the photobiont like glycans (reviewed in Davy et al., 2012, and Mansfield and Gilmore, 2019). This is followed by an inhibition of the phagosomal maturation, where Rab proteins, V-ATPases, and lysosomes are highly involved (Davy et al., 2012). The resulting membrane surrounding the photobiont intracellularly is known as symbiosome (Neckelmann and Muscatine, 1983; Hinde and Trautman, 2001; Kazandjian et al., 2008) and is highly relevant for interpartner nutritional exchange (Hill and Hill, 2012; Mohamed et al., 2016). A stable photosymbiosis is then based on tolerating the photobiont, involving processes like the quenching of reactive oxygen species (ROS), a constant nutrient exchange (Rädecker et al., 2021), and the decrease of the innate immune response where the complement system and signaling pathways like the transforming growth factor-beta (TGF-β), the Toll-like receptor (TLR), and the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) are major players (Mansfield and Gilmore, 2019). Only recently, the initial steps of the photosymbiosis establishment have been addressed in the context of the functional kleptoplasty of Sacoglossa sea slugs, revealing evidence for a recognition machinery that is in part conserved among evolutionary distant related taxa (Chan et al., 2018; Melo Clavijo et al., 2020). Yet, in Cladobranchia, and other photosymbiotic animals, mechanisms involved in the establishment of photosymbiosis have not yet been studied.
In the present study, we address this knowledge gap by using the cladobranch Berghia stephanieae (Valdés, 2005). B. stephanieae obtains its photobionts by feeding on sea anemones from the genus Exaiptasia (Carroll and Kempf, 1990; Valdés, 2005; Dionísio et al., 2013) and is considered a transitional form toward a stable photosymbiosis: even though the photobionts are kept for a few days to 1 week, their maintenance does not seem to provide any physiological advantages to the slug (Kempf, 1991; Bleidißel, 2010; Mies et al., 2017; Monteiro et al., 2019a, b). Moreover, Exaiptasia diaphana is used as a model organism to study the photosymbiosis in cnidarians (Dungan et al., 2020), and both animals can be cultivated in symbiotic and aposymbiotic (symbiont-free) states under laboratory conditions. Hence, B. stephanieae is an ideal organism for studying the evolution of photosymbiosis in Cladobranchia, because it allows a direct comparison of known mechanisms involved in photobiont recognition and maintenance in taxonomically divergent taxa. Studying the gene expression of B. stephanieae, we found that one major step toward the evolution of a stable photosymbiosis in Cladobranchia might be based on the inhibition of the photobiont digestion/expulsion and the immunosuppression response of the host against the photobiont.
Symbiotic individuals of Exaiptasia diaphana were obtained from a local provider (Seepferdchen24 Meeresaquaristik GmbH, Germany) in February 2019 and were maintained in a 55-l tank (60 cm × 30 cm × 30 cm) filled with circulating artificial seawater (ASW) (AB Reef Salt, Aqua Medic, Germany), keeping the salinity at 33 Practical Salinity Units (PSU), the temperature at 25°C, pH at 8, and light intensity at 30 µmol photons m-2 s-1 provided by RGB top light for reef tanks (Daylight Sunrise 520, Sera), on a 12-h light/12-h dark cycle. To ensure a stable microbiome in the tank (Godoy-Olmos et al., 2019; Xiao et al., 2019), 2 BactoBalls (Fauna Marin GmbH, Germany) were added and replaced every 2 weeks. In parallel, brine shrimp Artemia were cultivated using the Artemio Set and 16 g of ArtemioMix eggs + salt (JBL, Germany). The anemones were fed with freshly hatched brine shrimp nauplii Artemia two to three times per week.
In order to produce aposymbiotic individuals of E. diaphana, 40 animals were placed in a separate 10-l tank with circulating artificial seawater and menthol (20% w/v in ethanol; Carl Roth, Germany) at a final concentration of 0.19 mM as recommended by Matthews et al. (2016). Minor changes to this procedure were done and are described as follows: to induce bleaching without causing mortality, the anemones were placed in the menthol tank for 6 h at light intensity of 30 µmol photons m-2 s-1 provided by RGB top light for reef tanks and later incubated in another 55-l tank with ASW and 0.21 M monolinuron (Algol, JBL, Germany) for 18 h in total darkness. At the beginning of this 18-h incubation period, the anemones were fed with freshly hatched nauplii of Artemia three times per week. Monolinuron acts as a photosynthesis blocker, thus preventing unwanted algal blooms and reducing the possibility of any symbiont recolonization (Arnaud et al., 1994). This process of menthol + monolinuron incubation was done for 4 consecutive days, with a 3-day pause, during which the anemones were kept in the monolinuron tank in total darkness. The bleaching procedure was performed for 1 month at 23°C, with pH 8, and the salinity was kept at 33 PSU, compensating any loss due to evaporation with newly fresh water. The bottom of all tanks was cleaned, and 30% of the water was changed weekly. To confirm the aposymbiotic status, individuals were visually inspected with the stereo microscope (SteREO Discovery V.8, Zeiss, Germany), light microscope (Imager A2, Zeiss, Germany), and fluorescence microscope (Biozero, Keyence, Japan).
Six breeding pairs of B. stephanieae were obtained from a local provider (Seepferdchen24 Meeresaquaristik GmbH, Posthausen) in February 2019 and cultivated in the lab at 23°C, at a day/night cycle of 12 h/12 h. Each pair was kept in a 50-ml plastic container with a lid (75 mm, FAUST, Germany) in 35 ml of ASW. Water changes (50%) were made three times per week, with freshly prepared ASW with a salinity of 33 PSU, pH of 8.0, and temperature of 23°C. The slugs were fed with small symbiotic E. diaphana anemones (7-mm foot and 4-mm oral disk or 4-mm foot and 3-mm oral disk) three times per week. Once the breeding pairs spawned, the egg masses were collected and placed individually in 25-ml plastic containers with lids (55 mm, FAUST, Germany) with ASW and maintained as stated above.
The embryological development was monitored, and as soon as the animals hatched they were separated into two groups: symbiotic and aposymbiotic. Depending on the group, the animals were given several cut tentacles as food, symbiotic tentacles for the symbiotic group and aposymbiotic tentacles, or small aposymbiotic anemone pieces for the aposymbiotic group, three times a week. When the animals had developed cerata and were around 1 cm in length, they were fed with small symbiotic or aposymbiotic anemones (7-mm foot and 4-mm oral disk or 4-mm foot and 3-mm oral disk), according to their feeding group.
In order to identify the Symbiodiniaceae genotype present in B. stephanieae and in its anemone prey E. diaphana, total DNA was extracted from three anemones and three slugs from our cultures using the E.Z.N.A® Mollusc DNA Kit, Omega (Georgia, USA). Then, in a total volume of 20 µl, 5 µl of the total DNA was used as a PCR template using 10 µl of 2× concentrated GoTaq® Green Master Mix (Promega, USA) and 10 mM of forward and reverse primers for the Internal Transcriber Space 2 (ITS2) gene attached to Illumina adapter sequences (ITS2 forward: TCT TTC CCT ACA CGA CGC TCT TCC GAT CT GTG AAT TGC AGA ACT CCG TG (Pochon et al. (2001)); ITS2 reverse: GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T CCT CCG CTT ACT TAT ATG CTT (Stat et al. (2009)); Illumina adapter sequence underlined). A three-step PCR was done using a Gradient Mastercycler® (Eppendorf, Germany), with a 15-min initial denaturation at 95°C, followed by 30 cycles of 1-min denaturation at 94°C, 1 min of primer annealing at 54°C, and 1 min of elongation at 70°C, and ending in a 10-min final elongation at 70°C. The PCR products were visualized with an electrophoresis gel and subsequently purified with 0.7 volume of AMPure XP beads (Beckman Coulter, USA). A second PCR was performed with 10 µl of 2× Q5® High-Fidelity Master Mix (New England Biolabs, UK), 10 mM of Illumina sequencing adapters containing TruSeq indexes, and 5 µl of purified PCR products as template in a 20-µl total volume. The amplification protocol included a 5-min initial denaturation at 95°C, 9 cycles of 1 min at 94°C denaturation, a 1-min annealing step at 52°C, and 1 min of elongation at 70°C, ending with a 10-min elongation at 70°C. The PCR products were purified with 0.7 volume of AMPure XP beads, quantified with a Qubit fluorometer (Thermo Fisher, USA), and concentrated equimolarly to 5 nM. The samples were sequenced on a MiSeq platform (Illumina, Germany) at the Saarland University using the MiSeq Reagent Nano Kit v2 (500-cycle) chemistry (Illumina, Germany). The quality of raw reads was inspected using FastQC v0.11.8 Andrews (2010), adapters were removed, and sequences were quality trimmed using cutadapt v1.15 Martin (2011) with -m 10 -O 17 -e 0 -q 20,20 parameters. Subsequently, the sequences were imported into QIIME 2 v2021.4 (Bolyen et al., 2019) and were denoised with a maximum error rate of 2 using the dada2 plugin (Callahan et al., 2016). For Symbiodiniaceae assignment, we first trained an ITS2 classifier using the Symbiodatabacea database published by Fujise et al. (2021) and then followed the qiime2 pipeline to obtain Symbiodiniaceae annotations for our samples. Annotations were only considered valid if at least 150 sequences could be assigned to the according annotation for each sample (Supplementary Table 1, Supplementary Figure 1; BioProject ID: PRJNA812737; BioSample IDs: SAMN29176735-SAMN29176740; GenBank SRA: SRR19736587-SRR19736592).
Nine nudibranchs fed with symbiotic anemones were selected from the culture, and each individual was placed in one 25-ml container with ASW. The animals were separated into three groups, with three animals per group. The first group was fed three times a week with symbiotic anemones (Fed). One hour after feeding, they were snap frozen on dry ice in a -80°C freezer until processing. The second group was fed only once followed by a starvation period of 7 days (Starved). Once the starvation period ended, the animals were snap frozen at -80°C until processing. The third group was fed only once, starved for 7 days, and refed with small symbiotic anemones (Re-fed). One hour after refeeding, they were snap frozen at -80°C until processing.
The same experimental design was applied for other nine nudibranchs fed with aposymbiotic anemones, divided into three groups, with three animals per group. The groups were as follows: Fed with aposymbiotic anemones, Starved, and Re-fed with aposymbiotic anemones after starvation.
During the experiment, the water of all 18 containers was changed (50%) three times a week and they were maintained with the parameters previously described. For each feeding group, samples were collected and stored as stated before.
For the 18 slug samples corresponding to six feeding conditions described before (BioSample IDs: SAMN26426654-SAMN26426659), total RNA was extracted from whole animals using the my-Budget RNA Mini Kit (Bio-Budget Technologies, Germany) following the manufacturer’s instructions. The RNA concentration was quantified with Qubit fluorometric quantification (Thermo Fisher, USA) and NanoDrop™ One/OneC (Thermo Fisher, USA). A total amount of 800 µg RNA per sample was used for sequencing with Novogene (UK). Poly(A) mRNA enrichment, library preparation using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, USA), and 150-bp paired-end sequencing using the Illumina NovaSeq 6000 System were done by Novogene (UK). Samples were run in multiple lanes separating symbiotic from aposymbiotic samples, and all samples were multiplexed using double indices. Batch effects based on using different lanes are excluded based on principal component analysis (PCA) showing that the separation of samples is based on symbiont presence/absence or on feeding state.
A total of 1,210,391,662 reads were obtained (Supplementary Table 2; BioProject ID: PRJNA812737; GenBank SRA: SRR18218271-SRR18218254). Given the presence of the prey and the symbiont genes in the raw reads, a filtering step was performed by mapping the raw reads to the nucleotide sequences of the genome scaffolds of E. diaphana (Baumgarten et al., 2015), Cladocopium sp. C1acro, Fugacium kawagutii (Liu et al., 2018), S. microadriaticum (Aranda et al., 2016), and Breviolum minutum (Shoguchi et al., 2013) using BBSplit as implemented in BBTools v38.90 with qin=33 as input parameter (Bushnell, sourceforge.net/projects/bbmap/). This filtering step resulted in 1,038,020,410 reads that could not be mapped to either the anemone or the dinoflagellates (Supplementary Table 2). An assembly for these unmapped reads was done using Trinity v2.12.0 (Henschel et al., 2012; Haas et al., 2013), which resulted in 1,554,424 transcripts. The assembled transcriptome was then clustered with CD-HIT-EST v4.8.1 with -c 0.9 -n 7 -B 1 -g 1 -s 0.9 parameters (Li and Godzik, 2006; Fu et al., 2012), which reduced the assembly to 1,274,094 transcript clusters (Supplementary Data 1). TransDecoder v5.5.0 (Haas and Papanicolaou, 2016) was then used to translate the transcript clusters into the longest open reading frame (Supplementary Data 2). To assess sequence completeness, the amino acid sequences were compared against the mollusca_odb10 BUSCO database using BUSCO v5.1.3 (Supplementary Table 3). The amino acid dataset was then annotated by a BLASTP search (part of the BLAST+ package v2.9.0) against the UniProt database version 03/2021 (The UniProt Consortium, 2021), the genomes of Fugacium kawagutii, Cladocopium sp. C1acro (Liu et al., 2018), S. microadriaticum (Aranda et al., 2016), and Exaiptasia diaphana (Baumgarten et al., 2015), setting the E-value to 1e-10. Functional annotations of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) identifiers were obtained using eggNOG-mapper v2 (Cantalapiedra et al., 2021). Taxonomic assignment for each protein sequence was done using the UniProt taxonomic database. Sequences were subsequently filtered for metazoan annotations excluding annotations with their best hit to Exaiptasia and annotations of bacteria, fungi, and plants (Supplementary Data 3). The annotations were then screened for innate immune elements involved in symbiont recognition and maintenance such as scavenger receptors, C-type lectins, Toll-like receptors, complement components, the thrombospondin-type-1 repeat (TSR) domain-containing proteins, components of the TGF-ß signaling pathway, genes associated with immune suppression, apoptosis, phagosome maturation, nutrient transporters, and reactive oxygen species (ROS) quenching. An additional search with InterProScan was done in order to identify the transcripts with a characteristic domain architecture correspondent to the innate immune recognition receptors that were not identified based on the annotation, following the classification done in our previous work (Melo Clavijo et al., 2020). Additionally, KEGG pathways of apoptosis, autophagy, Toll-like receptor, complement, and TGF-ß signaling pathways were reconstructed using the online KEGG Mapper tool (https://www.genome.jp/kegg/tool/map_pathway.html).
The transcript abundance of sequences was estimated with kallisto v0.46.1 (Bray et al., 2016) following the Trinity pipeline v2.12.0 (Henschel et al., 2012; Haas et al., 2013). Differential expression analysis was done with DeSeq2 v1.32.0 (Love et al., 2014) implemented in Trinity. Differentially expressed genes (DEGs) were defined with FDR p-values of 0.005 and 0.05. Genes with expression values of more than three TMM in at least two replicates of one condition were considered. The normalized expression data corresponding to all genes found in the transcriptome, the annotated ones, and the DEGs were analyzed via principal components using the prcomp function in R v4.0.2 (R Core Team, 2021). Gene ontology analysis with adaptive clustering were performed using the GOMWU R package (https://github.com/z0on/GO_MWU) to measure whether each GO category was significantly enriched according to the expression value (Wright et al., 2015). EuKaryotic Orthologous Groups (KOG) class enrichment tests were performed using the KOGMWU R Package v1.2 (Dixon et al., 2015) and published gene expression data of animals under different experimental conditions: the coral Stylophora pistillata exposed to heat stress (Meyer et al., 2011; Dixon et al., 2015), different fluorescence morphotypes of the larvae of the coral Acropora millepora (Strader et al., 2016), symbiotic vs. aposymbiotic states of the anemone prey Exaiptasia diaphana (Lehnert et al., 2014), and the dauer and diapause dataset of the nematode Caenorhabditis elegans (Sinha et al., 2012) and the midge Sitodiplosis mosellana (Gong et al., 2013). With the KOGMWU analysis, delta ranks are computed as the difference between the mean rank of genes in a KOG class and the mean rank of all other genes (Dixon et al., 2015; Matz, 2019). These comparisons using KOG classes and delta ranks of different datasets aid to identify a general pattern of expression in our samples, and the effect of the presence of the symbiont and the starvation as a stress factor. Schematic models were created with BioRender.com.
E. diaphana and B. stephanieae both harbored Breviolum minutum as a photobiont. Only in one anemone did we additionally identify Cladocopium and Symbiodinium, but in low frequency (248 and 191 sequences, respectively; Supplementary Figure 1, Supplementary Table 1).
Overall, the gene expression pattern of animals feeding and refeeding with symbiotic prey were distinctly separated in a principal component space from each other and from all other experimental conditions (Figure 1). The samples corresponded to animals feeding and refeeding with aposymbiotic prey, and starved symbiotic and aposymbiotic animals clustered together, independently of the group of genes selected for the analysis (only annotated genes, only differentially expressed genes).
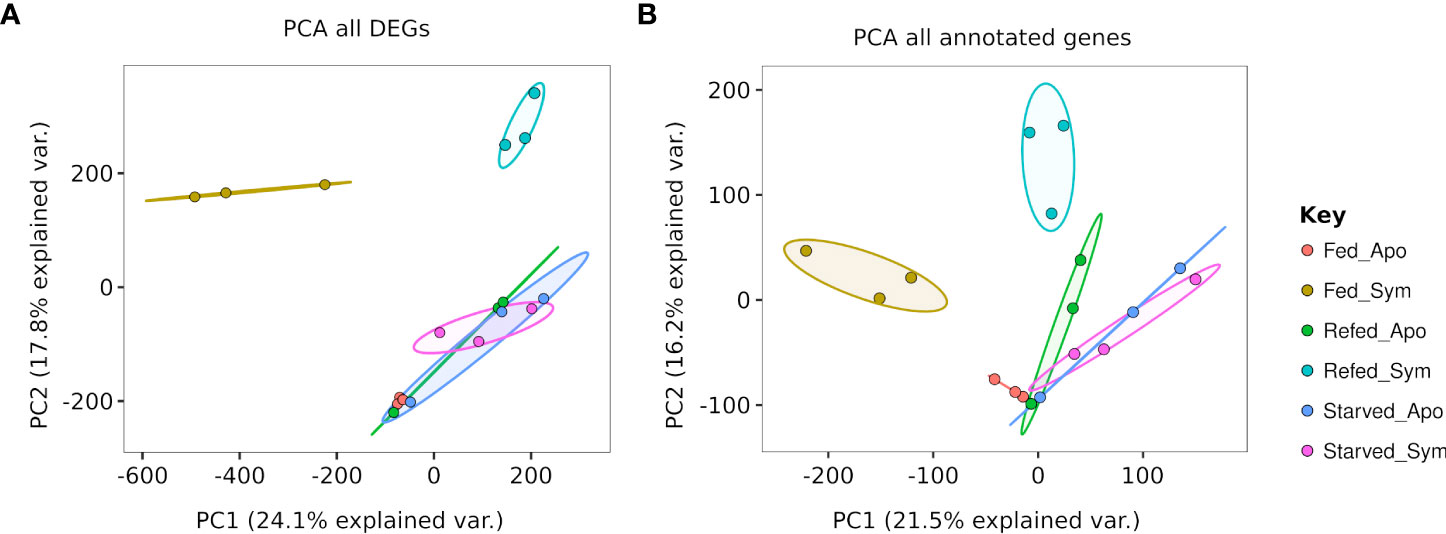
Figure 1 Principal component analysis of rlog-transformed counts for (A) all differentially expressed genes (DEGs) (n = 184,798) and (B) all annotated genes (n = 40,229) identified in the transcriptome of Berghia stephanieae.
For the comparison between symbiotic vs. aposymbiotic fed animals and symbiotic vs. aposymbiotic refed animals, significantly enriched gene ontology (GO) terms were identified in the Biological Processes (BP), Cellular Components (CC), and Molecular Function (MF) categories (Supplementary Figure 2). No significantly enriched GO terms could be found for starved animals.
In symbiotic fed animals, the GO term “antigen processing and presentation of peptide antigen via MHC class II” in the BP category was predominantly upregulated. Processes in the BP category related to protein hydrolysis such as “positive regulation of peptidase activity” and “positive regulation of cysteine-type endopeptidase activity,” “regulation of cell death,” “metabolic and biosynthetic processes,” and “gene expression” were predominantly downregulated. Within the CC category, the GO term “plasmodesma,” important in cellular communication, was enriched exclusively among the upregulated genes. Regarding the MF category, GO terms associated with protein endohydrolysis were strongly upregulated, while signaling receptor activity was downregulated in fed animals.
In refed animals, cellular components and biological processes depending on the microtubule cytoskeleton and related to cellular division were enriched among the upregulated genes. GO terms related to the extracellular region and the nucleoplasm were downregulated in symbiotic refed animals (Supplementary Figure 2).
We then compared the KOG delta ranks with previously published data on cnidarians that were exposed to heat stress (Meyer et al., 2011; Dixon et al., 2015), on different fluorescence morphotypes of the larvae of the coral Acropora millepora (Strader et al., 2016), on the anemone prey Exaiptasia diaphana, for which a comparison of the gene expression between symbiotic and aposymbiotic animals is available (Lehnert et al., 2014), and with the dauer and diapause dataset of the nematode Caenorhabditis elegans (Sinha et al., 2012) and the midge Sitodiplosis mosellana (Gong et al., 2013), respectively (Figure 2). Two main clusters were identified: one cluster included animals under stress conditions, while the other one consisted of unstressed animals. Comparisons of the gene expression between symbiotic vs. aposymbiotic fed and refed B. stephanieae and its prey E. diaphana clustered within the unstressed group. Starvation in B. stephanieae resulted in a similar gene expression compared to heat-stressed corals, regardless of the symbiotic state. In the stressed cluster, KOG terms belonging to the category “Metabolism” are predominantly downregulated, while KOG terms belonging to “Cellular processes and signaling” and “Information storage and processing” are mainly upregulated. Only the KOG term “Translation, ribosomal structure and biogenesis” was significantly enriched among upregulated genes in symbiotic vs. aposymbiotic fed B. stephanieae, while the terms “Transcription” and “Signal transduction mechanisms” were significantly enriched among the downregulated genes. In aposymbiotic starved vs. fed animals, the term “Cytoskeleton” was significantly enriched among downregulated genes. The term “Posttranslational modification, protein turnover, chaperones” was significantly enriched among upregulated genes.
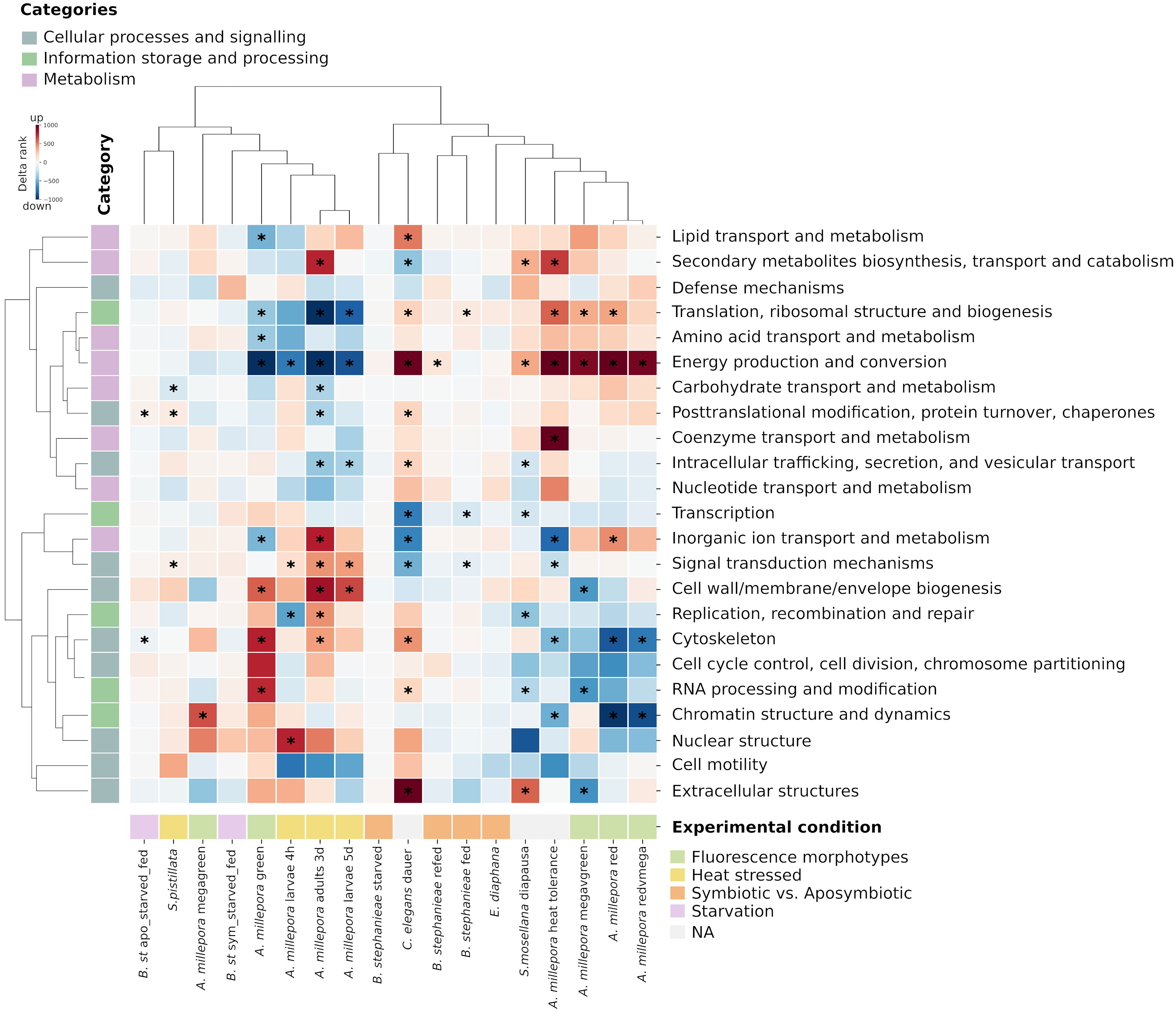
Figure 2 Gene expression profile based on KOG classes. Clustered heatmap of enriched KOG classes in B. stephanieae compared to publicly available datasets of other organisms exposed to stress factors: different fluorescence morphotypes of the larvae of the coral Acropora millepora (Strader et al., 2016), heat-stressed coral Stylophora pistillata (Meyer et al., 2011; Dixon et al., 2015), symbiotic vs. aposymbiotic Exaiptasia diaphana (Lehnert et al., 2014), the dauer state of the nematode Caenorhabditis elegans (Sinha et al., 2012), and the diapause state of the midge Sitodiplosis mosellana (Gong et al., 2013). KOG classes within three main KOG categories are shown. The regulation of the KOG classes is color-coded, where red indicates upregulation and blue downregulation based on the delta rank score. Asterisks show the KOG classes that were significantly enriched (FDR = 0.1).
The symbiont recognition process in E. diaphana is a receptor-mediated contact of the host’s PRRs and the symbiont’s MAMPs. Among the PRRs, the scavenger receptors from class B (SR-B) and E (SR-E), C-type lectins (like collectins), thrombospondin‐type‐1 repeat domain‐containing proteins (TSRs), and the complement system have been previously linked to the symbiont recognition process (Figure 3A) (reviewed in Davy et al., 2012; Mansfield and Gilmore, 2019). The role in the onset of the photosynthetic symbiosis of other PRRs like Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD)-like receptors (NODs), fibrinogen-related proteins (FREPs), and peptidoglycan recognition proteins (PGRPs) is unknown. We found most of these aforementioned receptors in our gene expression analyses of Berghia stephanieae (Table 1).
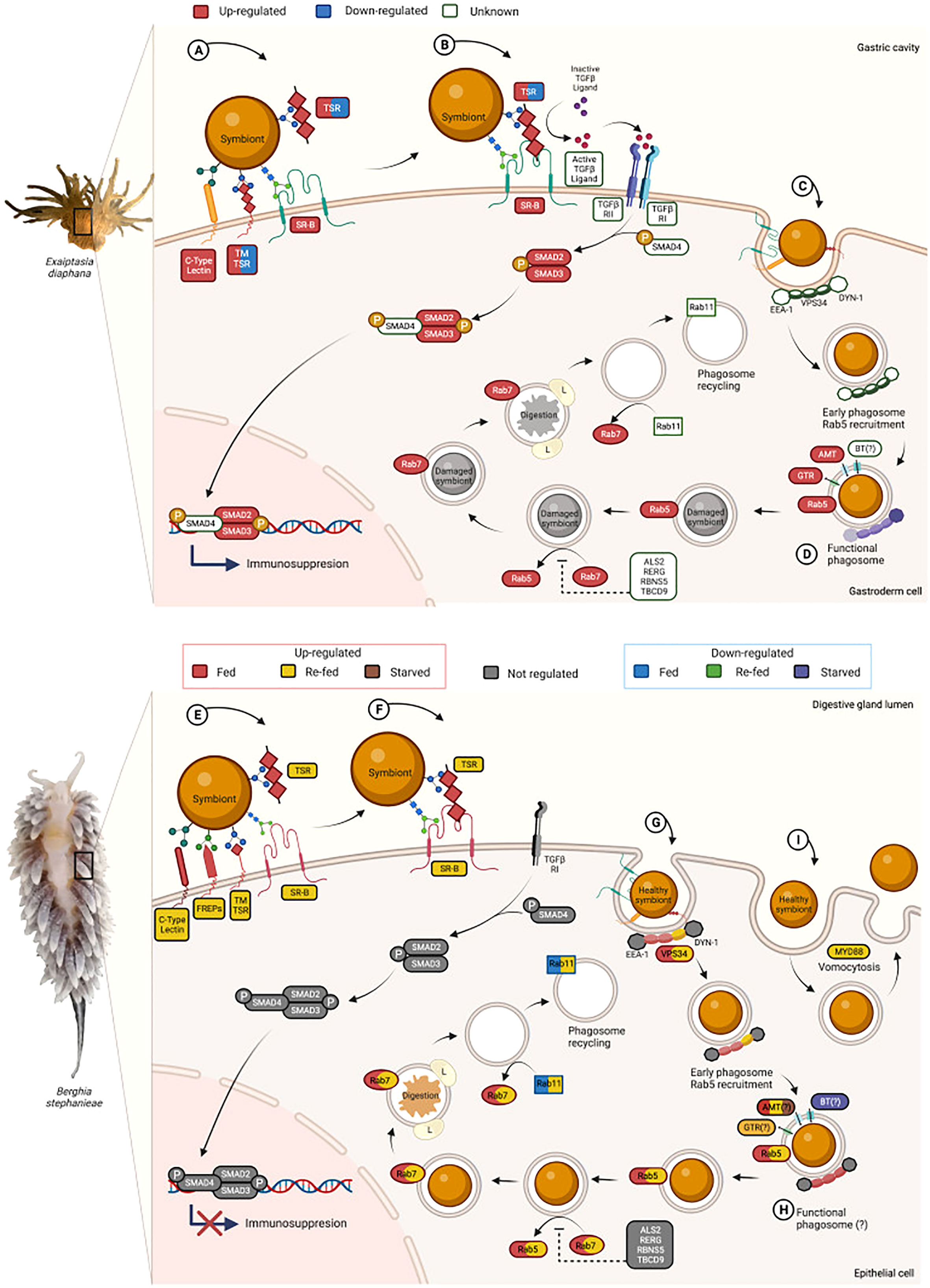
Figure 3 Model of the cellular events involved in the symbiont recognition and maintenance in the sea anemone Exaiptasia diaphana (A-D) based on previously published works (Chen et al., 2003, 2004, 2005, Davy et al., 2012, Detournay et al., 2012, Fransolet et al., 2012, Lehnert et al., 2014, Mohamed et al., 2016, Neubauer et al., 2016, 2017, Ishii et al., 2019) and our results on Berghia stephanieae (E-I). In E. diaphana, the first step of the winnowing is the recognition of the potential symbiont (A) mediated by the host’s pattern recognition receptors (PRRs) such as SR-Bs, SR-E-like, C-type Lectins, and TSRs. (B) The recognition by TSRs and/or SR-Bs seems to trigger the tolerogenic TGF-β pathway, with a high phosphorylation (P) of the transcription factor SMAD2/3 that promotes an immunosuppression response. (C) The phagocytosis of the potential symbiont takes place, where activator proteins like EEA-1, VPS34, and DYN-1 induce the early phagosome formation by the recruitment of Rab5. The phagosomal maturation continues by the replacement of Rab5 with Rab7, which induces the recruitment of lysosome-associated membrane protein (LAMP) and the fusion of the phagosome with lysosomes (L). Once the phagosome content is successfully degraded, the recyclement of the phagosome is triggered by the replacement of Rab7 with Rab11. (D) In stable symbiosis, the phagosomal maturation is inhibited and a functional symbiosome is established, where a nutrient exchange takes place via bicarbonate, glucose, and ammonium transporters, among others. In B. stephanieae, the recognition of the photobiont (E) might be mediated by PRRs such as SR-Bs, SR-E-like, C-type Lectins, TSRs, and/or FREPs. (F) The tolerogenic TGF-β pathway is not regulated, and core components like TGF-β ligand and TGF-βRII are missing. (G) During the early phagosome formation, only the activator protein VPS34 (PIK3C3) is activated and the recruitment of Rab5 is triggered. In contrast with its prey, in B. stephanieae there is no selective symbiont elimination, instead, the phagosomal maturation continues for healthy symbionts, marked by the expression of Rab7. Once the photobiont is successfully digested, the phagosome recycling is activated by the replacement of Rab7 with Rab11. (H) Since the phagosomal maturation is not inhibited, it seems that no functional symbiosome can be established or maintained, so no nutrient exchange takes place. (I) The alternative is that the photobionts cannot escape vomocytosis, given that MyD88 is activated preventing the maintenance of the photobiont in the animal host, and promoting the expulsion of healthy photosynthetically active dinoflagellates.
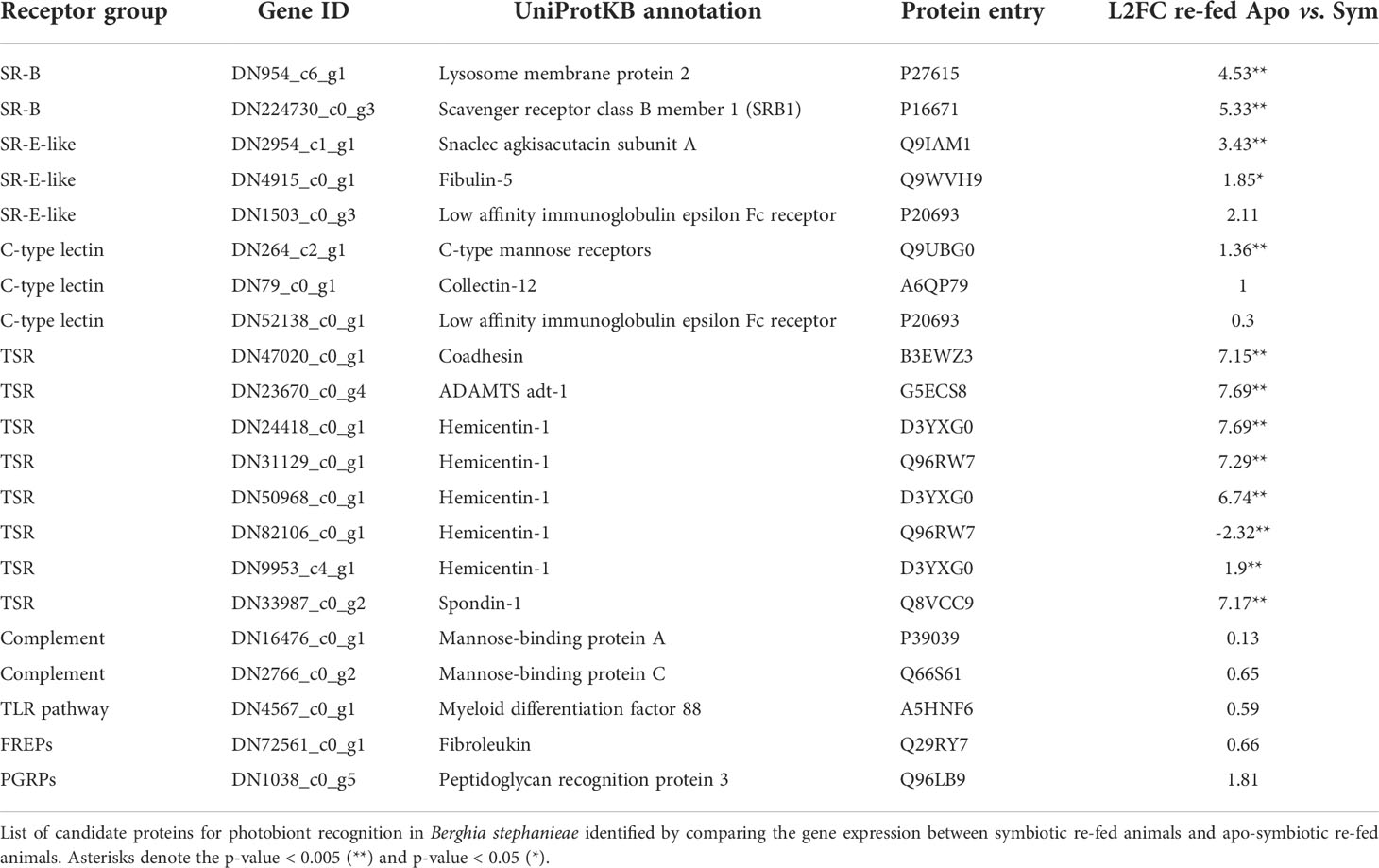
Table 1 Innate immune genes potentially involved in the photosymbiosis initiation in Berghia stephanieae.
Overall, 15 genes were classified as scavenger receptors, from which six belong to class B, three to class E-like, and six to class I. In order to find potential candidates involved in symbiont recognition, we compared the gene expression of these receptors between symbiotic and aposymbiotic refed animals, after a 7-day starvation period. We expected those genes relevant for symbiont recognition to be upregulated in symbiotic refed animals, because our chosen time frame of refeeding of 1 h after a starvation period should induce a photobiont-specific gene expression. In animals regularly fed, this expression pattern might be lost, as observed in Acropora digitifera (Mohamed et al., 2016). Out of the six SR-B genes, two were significantly upregulated: a homologue of the lysosome membrane protein 2 (DN954_c6_g1; L2FC 4.53) was upregulated in symbiotic refed animals, while the other SR-B homologue annotated as SRB1 (DN224730_c0_g3; L2FC 5.33) was upregulated in aposymbiotic refed animals (Supplementary Figure 3). From the three identified SR-E-like receptors, a snaclec agkisacutacin subunit A (p < 0.005: DN2954_c1_g1, L2FC 3.43) and fibulin-5 (p < 0.05: DN4915_c0_g1, L2FC 1.85) were significantly upregulated in symbiotic refed animals (Supplementary Figure 3). None of the SR-I genes were differentially expressed (Supplementary Data 3). Forty-one genes were classified as C-type lectins, and out of them, one C-type mannose receptor was significantly differentially expressed in symbiotic refed animals (p < 0.005: DN264_c2_g1, L2FC 1.36). Additionally, collectin 12 and a low-affinity immunoglobulin epsilon Fc receptor were highly expressed in symbiotic refed animals (Supplementary Figure 3 and Table 1). Nine genes were grouped as SRCR, but none of them was differentially expressed (Supplementary Data 3). Among the TSRs, eight genes were classified as ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs), 15 as semaphorins, three as plexins, 11 as sequences containing only the TSP1 (thrombospondin‐type‐1) domain with transmembrane region (TM), 103 as TSP1 without TM, one astacin metallopeptidase, 2 as F-spondin-like, and 27 as sequences containing TSP1 in combination with other domains (Supplementary Data 3). Out of the 114 TSRs, eight were significantly differentially expressed in symbiotic refed animals (p < 0.005). From those eight TSRs, one was annotated as Coadhesin (DN47020_c0_g1, L2FC 7.15), one as ADAMTS adt-1 (DN23670_c0_g4, L2FC 7.69), five as Hemicentin-1 (DN24418_c0_g1, L2FC 7.69; DN31129_c0_g1, L2FC 7.29; DN50968_c0_g1, L2FC 6.74; DN82106_c0_g1, L2FC -2.32, DN9953_c4_g1, L2FC 1.9), and one as Spondin-1 (DN33987_c0_g2, L2FC 7.17) (Supplementary Figure 3).
Furthermore, 35 activator genes of the complement system were identified (Supplementary Data 3), yet none of them were significantly differentially expressed. Only one mannose-binding protein A and one mannose-binding protein C were highly expressed in symbiotic refed animals compared to aposymbiotic refed ones (Supplementary Figure 4; Table 1).
We identified 40 genes involved in the TLR pathway, out of which 30 were classified as Toll-like receptors (TLRs), five as myeloid differentiation primary response protein (MyD88), and five as the nuclear factor NF-kappa-B (NF-kB). Further, 13 genes were grouped as NOD-like receptors (Supplementary Data 3). None of the TLRs, MyD88, NF-kB, and NODs was significantly differentially expressed, except for one MyD88 that was upregulated in symbiotic refed animals (Supplementary Figure 4; Table 1). Sixty-eight genes were classified as fibrinogen-related proteins (FREPs), and three peptidoglycan recognition proteins (PGRPs) were identified, but none of these genes were differentially expressed in symbiotic refed animals (Supplementary Data 3; Supplementary Figure 4). Only one FREP annotated as fibroleukin (Supplementary Figure 4) and one PGRP annotated as peptidoglycan recognition protein 3 were upregulated in symbiotic refed animals compared to aposymbiotic refed ones (Supplementary Figure 4; Table 1).
Once the symbiont is recognized in cnidarians, signaling cascades like the TGF-β signaling pathway might be triggered, leading to an immunosuppression response (Detournay et al., 2012) (Figure 3B). However, in B. stephanieae, core elements like the activation ligand TGF-β sensu stricto and TGF-β receptor II (TGFβRII) were not present (Figure 3F), and only one bone morphogenetic protein 1 (BMP1) (DN5506_c0_g1, L2FC -2.63) was significantly downregulated in symbiotic refed animals. None of the other TGF-β signaling pathway components were expressed above three TMM in any of the feeding conditions (Supplementary Data 3).
In cnidarians, the potential photobionts are phagocytized, after the successful recognition, into cells of the animal host and are still surrounded by the phagosomal membrane. In order to maintain the photobiont, the phagosomal maturation has to be prevented (Figure 3C) (Hill and Hill, 2012). Usually, newly formed phagosomes fuse with early endosomes and acquire the necessary proteins for maturation including Ras-related protein 5 (Rab5), early endosomal antigen 1 (EEA1), dynamin (DYN-1), and vacuolar sorting protein-34 (VPS34, also known as PIK3C3) (Kinchen and Ravichandran, 2008; Lee et al., 2020). In E. diaphana, Rab5 is only present in phagosomes with healthy photobionts (Chen et al., 2004). The early phagosome matures into late phagosome by the replacement of Rab5 with Rab7, promoting the fusion with lysosomes via lysosome-associated membrane protein (LAMP1 and LAMP2) recruitment, the accumulation of V-ATPases, and the subsequent acidification of the late phagosome (Lee et al., 2020). Once phagosomal degradation is initiated, the endosome is recycled by the replacement of Rab7 and the recruitment of Rab11. In cnidarian photosymbiosis, proteins like Rab22, amyotrophic lateral sclerosis2 (ALS2), TBC1 domain family member 9 (TBCD9), rabenosyn-5 (RBNS5), and Ras-related and estrogen-regulated growth inhibitor (RERG) might block Rab7 by the constant activation of Rab5, preventing the maturation of the phagosome (Mohamed et al., 2016; Lee et al., 2020). In B. stephanieae, Rab5A (DN60084_c0_g1) and Rab7 (DN353992_c0_g1) were significantly upregulated in symbiotic fed and refed animals (p < 0.005: Rab5A L2FC 8.25, L2FC 7.39; Rab7 L2FC 7.71, L2FC 8.09, respectively) (Supplementary Figure 5). Rab7A (DN23467_c1_g2, L2FC 1.90) was significantly upregulated only in symbiotic refed animals (Supplementary Figure 5). None of the activators of Rab5 (EEA1, dynamin) was significantly differentially expressed in any feeding condition; only VPS34/PIK3C3 was upregulated in symbiotic fed and refed animals (Supplementary Figure 5). Rab11A and Rab11B were downregulated in symbiotic fed animals, while they were upregulated in symbiotic refed animals. LAMP1 was significantly differentially expressed in symbiotic refed animals (DN5522_c5_g1, L2FC 1.33, p < 0.05). V-ATPases were significantly upregulated during feeding (p < 0.005: DN35123_c0_g2, L2FC 10.69; p<0.05: DN23173_c0_g1, L2FC 1.62; DN4712_c0_g1, L2FC 1.3) and refeeding with symbiotic prey (p < 0.005: DN35123_c0_g2, L2FC 8.39; p < 0.05: DN10201_c0_g2, L2FC 1.09; DN13065_c0_g2, L2FC 1.06; DN32461_c0_g1, L2FC 2.55; DN4147_c0_g1, L2FC 0.66) (Supplementary Data 3).
In cnidarians, the arrested phagosomes harboring healthy photobionts that evade maturation would communicate with the animal host via membrane transporters to obtain the necessary compounds for photosynthesis (dissolved inorganic carbon in the form of bicarbonate or as CO2), translocate produced organic compounds to the host (e.g., glycerol, glucose, amino acids, lipids), and recycle the host’s waste products like nitrogen (Figure 3D) (Matthews et al., 2017). The metabolic exchange is key for the stability of the symbiosis. In B. stephanieae, we compared symbiotic and aposymbiotic animals for each experimental condition and identified several transporters such as ammonium, bicarbonate, and glucose transporters (Figure 3H; Supplementary Figure 6). Ammonium transporters were significantly upregulated in symbiotic fed animals (p < 0.05: DN2986_c0_g2, L2FC 2.01), symbiotic refed animals (p < 0.005: DN340748_c0_g1, L2FC 8.29), and starved symbiotic animals (p < 0.05: DN8797_c3_g2, L2FC 2.24). Bicarbonate transporters were significantly downregulated in symbiotic starved animals (p < 0.005: DN3291_c0_g2, L2FC -3.58; p < 0.05: DN3291_c0_g1, L2FC -3.41), but not differentially expressed in any other comparison. Once transported over the phagosomal membrane, bicarbonate must be converted to CO2, to make it available for photosynthesis, by carbonic anhydrases (Weis, 1993). Several carbonic anhydrases (Supplementary Figure 6) were significantly upregulated in symbiotic refed (p < 0.005; DN78523_c0_g1, L2FC 1.6; DN72715_c0_g1, L2FC 3.04) and symbiotic starved animals (p < 0.05; DN5108_c0_g1, L2FC -2.46; DN78523_c0_g1, L2FC 1.73; DN87999_c0_g1, L2FC 1.48).
For the animal host, the received glucose is a major nutrient and is essential to assimilating waste products, like ammonium, for the synthesis of amino acids. Hence, we expected that glucose transporters were upregulated in all symbiotic animals, in order to maintain the photobionts. Yet, in B. stephanieae glucose transporters were only significantly upregulated in symbiotic refed animals (p < 0.05: DN1101_c0_g1, L2FC 1.28) (Supplementary Figure 6).
Ammonium is mainly used in the amino acid synthesis through the glutamine synthetase (GS)— glutamine oxoglutarate aminotransferase (GOGAT). In this GS-GOGAT cycle, glutamate dehydrogenases (GDH) convert ammonium into glutamate and GS transforms ammonium into glutamine. In B. stephanieae, one GDH was significantly downregulated in symbiotic refed animals (p < 0.05: DN134779_c0_g1, L2FC 2.16) (Supplementary Figure 6). One GS homologue was significantly upregulated in symbiotic refed animals (p < 0.005: DN34097_c3_g2, L2FC 2.16) and symbiotic starved animals (p < 0.05: DN34097_c3_g2, L2FC 1.99) (Supplementary Figure 6). No GOGAT homologue could be identified.
The presence of the photobiont may induce additional stress to the animal host by photosynthetic-derived reactive oxygen species (ROS). The maintenance of the photobiont is linked to the ability of the host to cope with this additional stress induced by the photobiont (Richier et al., 2005; 2006; Lesser, 2011). Hence, we expected ROS quenching mechanisms to be active in symbiotic animals. In B. stephanieae, catalases were significantly upregulated in symbiotic refed animals (p < 0.005: DN2499_c9_g2, L2FC 1.83) but also during feeding and refeeding with aposymbiotic prey (p < 0.005: DN5526_c0_g1, L2FC -2.46, -2.63, respectively) (Supplementary Figure 7). Glutathione peroxidases were also significantly upregulated in symbiotic refed animals (p < 0.005: DN11199_c1_g1, L2FC 2.42; p < 0.05: DN39313_c1_g1, L2FC 1.92) but significantly downregulated in aposymbiotic refed animals (DN39313_c1_g2, L2FC -4.59). Glutathione S-transferases were significantly upregulated in symbiotic refed animals (p < 0.005: DN57713_c0_g1, L2FC 2; p < 0.05: DN32195_c4_g1, L2FC 0.89). Likewise, peroxiredoxins and peroxiredoxin-like proteins were significantly differentially expressed during feeding and refeeding with symbiotic prey compared to aposymbiotic fed and refed animals (p < 0.005: DN1905_c0_g2, L2FC 8.75, 9.10; DN40077_c0_g2, L2FC 11.03, 9.63, respectively) (Supplementary Figure 7).
In the present study, we provide the first differential gene expression analysis of a dinoflagellate-bearing sea slug belonging to Cladobranchia. The comparison of symbiotic and aposymbiotic animals either feeding, refeeding, or starving revealed a unique insight into mechanisms involved in the recognition of Symbiodiniaceae obtained from the slugs’ cnidarian prey (Figure 3E). Starvation induced a gene expression profile similar to heat-stressed corals (Meyer et al., 2011; Dixon et al., 2015), including a downregulation of essential metabolic processes (Figure 2). Moreover, in Berghia stephanieae mechanisms to cope with reactive oxygen species are induced in the presence of the photobiont (Supplementary Figure 7). Yet, the immune response against the photobiont is not suppressed and the phagosome maturation is not inhibited. Thus, the slug are probably not able to maintain the photobionts in the long term perhaps due to the inability to initiate the formation of a functional and stable symbiosome that supports a nutrient exchange between host and photobiont.
B. stephanieae and E. diaphana harbored the same symbiodiniaceae genotype (Supplementary Figure 1, Supplementary Table 1), and we found that the recognition of the algae in B. stephanieae (Figure 3E) might depend on the same molecular machinery present in its cnidarian prey E. diaphana, involving SR-B, SR-E-like, C-type lectins, and TSRs (reviewed in Mansfield and Gilmore, 2019) (Figure 3A). Hence, the photobiont recognition seems to be conserved among taxonomically divergent host taxa and independent on the phylogenetic origin of the phototrophic organism, or organelle (Melo Clavijo et al., 2020). Yet, SR-B has multi-ligand recognition and diverse cellular functions. For instance, SR-Bs recognize MAMPs from pathogens like fungi and bacteria, polyanionic ligands such as high-density and low-density lipoproteins (HDL, LDL), and oxidized phospholipids, apoptotic cells, and amyloid proteins (PrabhuDas et al., 2017). Additionally, SR-B is involved in lipid transport and phagocytosis (Silverstein and Febbraio, 2009; Yu et al., 2021). Furthermore, SR-B homologues are also highly expressed in aposymbiotic B. stephanieae. Hence, in the case of SR-B, the determination of a clear involvement in symbiont recognition in the slugs is complicated. This also applies to the other aforementioned receptors, but at least experimental verification has shown the relevance of SR-B, C-type lectins, and TSRs in symbiont recognition in cnidarians (Jimbo et al., 2000; Koike et al., 2004; Neubauer et al., 2016; Neubauer et al., 2017), which is why we consider these receptors as promising candidates for photobiont recognition in B. stephanieae.
Further candidates in photobiont recognition and photosymbiosis initiation in cnidarians belong to the TLRs and NODs, although their involvement is debated (Mansfield and Gilmore, 2019). TLRs and NODs can activate the NF-κB transcription factor, which play a central role in the immune response, inflammatory process, and cytokine release and modulation (Doyle and O’Neill, 2006; Dev et al., 2010). In cnidarian symbiosis, NF-κB is downregulated by Symbiodiniaceae (DeSalvo et al., 2010; Wolfowicz et al., 2016; Mansfield et al., 2017; Mansfield and Gilmore, 2019), yet in B. stephanieae NF-κB is not regulated, nor are TLRs or NODs (Supplementary Figure 4).
A potential new candidate in symbiont recognition in cladobranchs is fibroleukin, which belongs to a highly diversified group of glycoproteins in mollusks known as FREPs. Members of FREPs contain a fibrinogen domain followed by different domains (Romero et al., 2011), can recognize bacteria and fungi (Kenjo et al., 2001; Middha and Wang, 2008; Wu et al., 2011; Xiang et al., 2014; Huang et al., 2015), and are involved in agglutination, clotting, and phagocytosis during the destruction of pathogens (Huang et al., 2015; Buchmann, 2014). There is no evidence for an involvement of fibroleukin in other photosymbioses, but more detailed analyses could illuminate a potential function.
Finally, PGRP 3 was highly expressed in symbiotic B. stephanieae, which belongs to receptors containing a peptidoglycan-binding type 2 amidase (PGRP) domain that binds to peptidoglycan (PG), lipopolysaccharides (LPS), and lipoteichoic acid (Royet et al., 2011). Their main role is in pathogen defense in invertebrates and in the host-microbiota symbiosis establishment, for instance, between the bioluminescent bacteria Vibrio fischeri and the Hawaiian bobtail squid Euprymna scolopes (Dierking and Pita, 2020). LPS and PG are mostly present in the outer cell walls of bacteria (Diks et al., 2001), but so far there is no evidence of their presence in dinoflagellates. Yet, in B. stephanieae PGRP might be involved in bacterial recognition inducing a shift in the bacteriome related to the presence of a photobiont, as previously shown in its prey E. diaphana (Röthig et al., 2016).
After recognizing the algae and a subsequent phagocytosis, B. stephanieae is mainly digesting or excreting the algae and is not establishing a stable mutualistic symbiosis. Well-known photobiont maintenance mechanisms in cnidarians such as the TGF-ß pathway (Detournay et al., 2012; Berthelier et al., 2017), the complement system (Poole et al., 2016), the TLR pathway (Jacobovitz et al., 2021), and downstream NF-κB activation (DeSalvo et al., 2010; Wolfowicz et al., 2016; Mansfield et al., 2017) were not present or regulated in B. stephanieae, which would be essential for symbiont tolerance (reviewed in Mansfield and Gilmore, 2019).
For instance, the recognition of Symbiodiniaceae, possibly via SR-B and/or TSRs, triggers tolerogenic pathways like the TGF-β pathway that leads to the suppression of the host innate immune response in E. diaphana (Detournay et al., 2012) (Figure 3B). Yet, in B. stephanieae, none of the elements of this pathway were regulated. Moreover, core elements of the TGF-ß pathway were missing (e.g., the ligand TGF-β sensu stricto and the TGF-β receptor II); thereupon, there is no immunosuppression via the TGF-β pathway (Figure 3F). Indeed, TGF-β sensu stricto and TGF-β RII are missing in several mollusks (Herpin et al., 2004), which is why alternative pathways could be relevant for immunosuppression, such as the pathways of the complement system.
The complement system is relevant for recognition and promotes phagocytosis (Poole et al., 2016), and its downregulation seems to play a role in the onset and maintenance of the photosymbiosis in cnidarians (Kvennefors et al., 2010; Ganot et al., 2011; Poole et al., 2016; Mansfield and Gilmore, 2019). The activation of the complement system can be achieved through different pathways (Rus et al., 2005; Dunkelberger and Song, 2010; Kemper et al., 2010), resulting in the cleavage of the C3 protein that labels pathogens and recruits the macrophages for phagocytosis (Dunkelberger and Song, 2010). In B. stephanieae, we could only identify a potential triggering of the lectin pathway of the complement system through the mannose binding lectin (MBL2) and/or a possible activation via the alternative pathway through collectin-12 (Ma et al., 2015). Yet, essential ficolins and downstream components for this pathway were not differentially expressed in B. stephanieae (Supplementary Data 3). Thus, the complement pathway might not be relevant for the symbiosis onset in B. stephanieae either.
The transcriptional repression of MyD88 and NF-κB may also promote symbiosis establishment (DeSalvo et al., 2010; Wolfowicz et al., 2016; Mansfield et al., 2017; Mansfield and Gilmore, 2019; Jacobovitz et al., 2021). However, MyD88 is activated and NF-κB is not regulated in the symbiotic B. stephanieae, pointing to an active immune response against the algae, rather than an immunosuppression. Furthermore, it has been shown that the same symbiodiniaceae genotype performs differently depending on the host (Goulet et al., 2019). Thus, even though E. diaphana is in a stable photosymbiosis with Breviolum minutum B1/B2, B. stephanieae might not be able to establish a stable photosymbiosis with the strain due to photobiont intrinsic properties. Further, E. diaphana can be colonized by other less beneficial and productive Symbiodiniaceae such as Symbiodinium microadriaticum and Durusdinium trenchii (Gabay et al., 2018), or the heterologous Cladocopium goreaui (Tortorelli et al., 2020). In previous studies, B. stephanieae fed with anemones harboring S. microadriaticum A1 was able to incorporate these photobionts but only retained them for 10 days maximum (Mies et al., 2017; Monteiro et al., 2019b). Whether B. stephanieae can incorporate and maintain other Symbiodiniaceae genotypes present in its prey remains to be seen. To date, there is no report that identified the native symbiont genotype of this slug in the wild.
Vital for the stability of the symbiosis in intracellular photosymbioses is the inhibition of the phagosome maturation (Hill and Hill, 2012; Mohamed et al., 2016; Mohamed et al., 2020). The high expression of Rab7 in symbiotic B. stephanieae in combination with lysosomes recruiting LAMPs and the V-ATPases rather suggest an ongoing phagosome maturation leading to the photobiont digestion, instead of its maintenance and the establishment of the functional symbiosome (Chen et al., 2003; Chen et al., 2004; Fransolet et al., 2012) (Figure 3G). Yet, V-ATPases can also act as carbon concentration mechanisms by acidifying the phagosome (Kinchen and Ravichandran, 2008; Kinchen et al., 2008). This would then lead to a conversion of bicarbonate to CO2 and a subsequent translocation of CO2 to the photobiont in cnidarians and mollusks (Armstrong et al., 2018; Barott et al., 2022), promoting photosynthesis (Barott et al., 2015). For the system to work properly, carbonic anhydrases and bicarbonate transporters would be needed (Weis et al., 1989; Weis and Levine, 1996; Grasso et al., 2008; Ganot et al., 2011; Meyer and Weis, 2012; Ip et al., 2017; Chew et al., 2019), but carbonic anhydrases were highly expressed only in symbiotic B. stephanieae (Supplementary Figure 6).
Other mollusks use different hosting strategies for harboring Symbiodiniaceae and are able to establish stable symbioses. In clams belonging to the family Cardiidae, Symbiodiniaceae are located in extracellular spaces known as the zooxanthellal tubular system (Norton et al., 1992; Hernawan, 2008). In these clams, a symbiosome per se is absent. Yet, the ectodermal membrane surrounding the zooxanthellal tubular system may act as some sort of symbiosome involved in signaling, acidification, and molecule transport (Armstrong et al., 2018) to establish a stable symbiosis, similar to other extracellular stable photosymbioses such as the acoelomorph Symsagittifera roscoffensis and the green algae Tetraselmis convolutae (Bailly et al., 2014).
In addition, some ammonium and glucose transporters were upregulated in symbiotic B. stephanieae, but starvation had no effect on the expression of these transporters (Supplementary Figure 6), which would be expected in a stable symbiosis. Particularly, ammonium is important for the algae in hospite to ensure a high photosynthetic activity (Taylor, 1978; Koop et al., 2001; Yellowlees et al., 2008), which is actively provided to the algae and is used to control the algal growth (Rädecker et al., 2018; Cui et al., 2019; Xiang et al., 2020). When heat stressed, corals increase the catabolic degradation of amino acids to fuel the GS-GOGAT cycle to generate α-ketoglutarate needed for the tricarboxylic acid (TCA) cycle but simultaneously increase the translocation of ammonium to the algae (Rädecker et al., 2021). As a result, the algae use their photosynthates to grow instead of translocating these to the host (Baker et al., 2018). In the slug’s prey E. diaphana, the GS-GOGAT cycle is highly upregulated (Cui et al., 2019), while in the slug we could only observe an upregulation of GS in symbiotic animals when feeding, but not during starvation (Supplementary Figure 6). Thus, the GS-GOGAT cycle is probably not connected to a symbiotic relationship in B. stephanieae and is likely used to catabolize proteins during starvation to fuel the TCA cycle, similar to heat-stressed corals.
In addition to a nutrient exchange, the swift detoxification of reactive oxygen species (ROS) produced primarily by the photobiont is fundamental in a stable photosymbiosis. In B. stephanieae, ROS quenching mechanisms were predominantly activated in the presence of the algae (Supplementary Figure 7), similar to photosymbiotic cnidarians (Ganot et al., 2011; Meyer and Weis, 2012; Matthews et al., 2017; Yuyama et al., 2018). Starvation-induced stress prior to feeding seemed to enhance ROS quenching, particularly inducing the glutathione peroxidase.
In summary, the main challenges for cladobranchs to establish a stable photosymbiosis are thus the inhibition of the phagosomal maturation, which might be based on the successful nutrient exchange with the algae and suppressing immune responses that promote the expulsion of the photosynthetic partner. In combination with previous observations of both digested and intact photobionts in juveniles and in the feces of B. stephanieae (Figure 4) (Kempf, 1991; Bleidißel, 2010; Mies et al., 2017; Monteiro et al., 2019b), symbiophagy is probably the dominant process that might be even accompanied by vomocytosis as the TLR pathway is not downregulated (Jacobovitz et al., 2021) (Figure 3I). Given that gene regulation can take place at different steps and levels (posttranscriptional, translational, posttranslational, epigenetic), changes in gene expression are not definitive evidence of activation/inhibition of signaling pathways but provide information of the overall regulation of cellular processes at a certain time (Day & Tuite, 1998; Gibney and Nolan, 2010; Zhao et al., 2017).
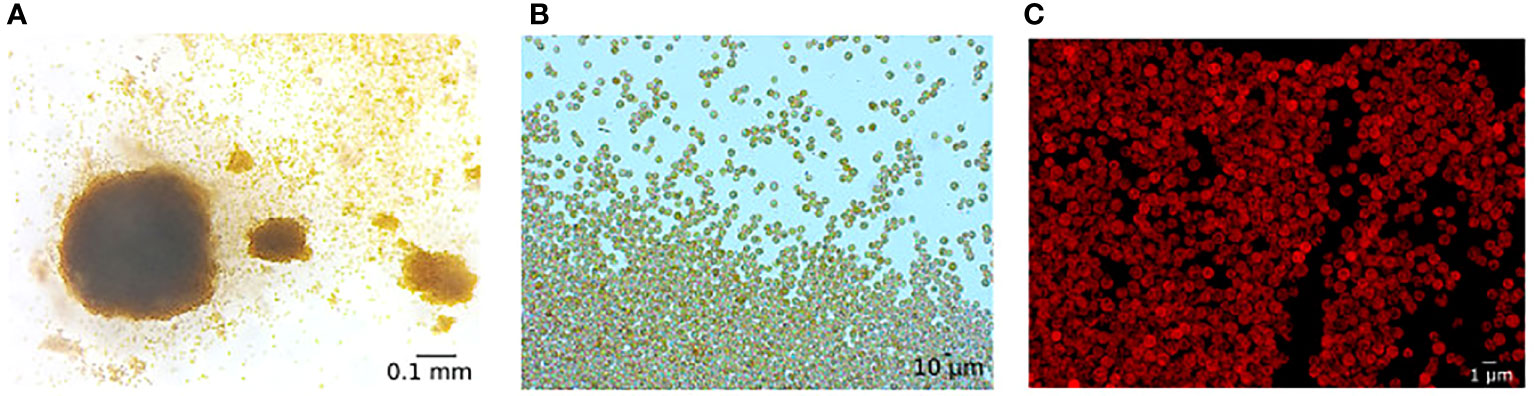
Figure 4 Fecal pellet of B.stephanieae containing intact, photosynthetically active dinoflagellates. Image taken (A) by Corinna Sickinger with a stereo microscope (SteREO Discovery.V8, Zeiss, Germany), 80× magnification, (B) by Jenny Melo with a light microscope (Imager A2, Zeiss, Germany), 200×, and (C) with a fluorescence microscope (BZ-X800E, Keyence, Japan), using the Texas Red Filter and a 400× magnification.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JC and GC sourced, processed the data, performed the data analyses, and edited the figures. GG and ST performed the MiSeq library preparation, sequencing, and data demultiplexing. JC, CS, SB, AP, and GC discussed and interpreted the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Thanks are due to the central research-funding budget at the University of Wuppertal (ZEFFT) for the financial support of our work.
We would like to thank Sabine Stratmann-Lettner and Christina Lo Porto for assistance in the lab. We acknowledge support from the Open Access Publication Fund of the University of Wuppertal, Germany.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.934307/full#supplementary-material
Additional Supplementary Data can be found at online at: 10.6084/m9.figshare.19690843.
Andrews S. (2010) FastQC: A quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Aranda M., Li Y., Liew Y. J., Baumgarten S., Simakov O., Wilson M. C., et al. (2016). Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6 (1), 1–15. doi: 10.1038/srep39734
Armstrong E. J., Roa J. N., Stillman J. H., Tresguerres M. (2018). Symbiont photosynthesis in giant clams is promoted by V-type h+-ATPase from host cells. J. Exp. Biol. 221 (18), p.jeb177220. doi: 10.1242/jeb.177220
Arnaud L., Taillandier G., Kaouadji M., Ravanel P., Tissut M. (1994). Photosynthesis inhibition by phenylureas: A QSAR approach. Ecotoxicol. Environ. Saf. 28 (2), 121–133. doi: 10.1006/eesa.1994.1040
Bailly X., Laguerre L., Correc G., Dupont S., Kurth T., Pfannkuchen A., et al (2014). The chimerical and multifaceted marine acoel Symsagittifera roscoffensis: from photosymbiosis to brain regeneration. Front. Microbiol. 5, 498.
Baker D. M., Freeman C. J., Wong J. C., Fogel M. L., Knowlton N. (2018). Climate change promotes parasitism in a coral symbiosis. ISME J. 12 (3), 921–930. doi: 10.1038/s41396-018-0046-8
Barott K. L., Thies A. B., Tresguerres M. (2022). V-Type h+-ATPase in the symbiosome membrane is a conserved mechanism for host control of photosynthesis in anthozoan photosymbioses. R Soc. Open Sci. 9 (1), 211449. doi: 10.1098/rsos.211449
Barott K. L., Venn A. A., Perez S. O., Tambutté S., Tresguerres M. (2015). Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. PNAS. 112 (2), 607–612. doi: 10.1073/pnas.1413483112
Baumgarten S., Simakov O., Esherick L. Y., Liew Y. J., Lehnert E. M., Michell C. T., et al. (2015). The genome of Aiptasia, a sea anemone model for coral symbiosis. PNAS. 112 (38), 11893–11898. doi: 10.1073/pnas.1513318112
Berthelier J., Schnitzler C. E., Wood-Charlson E. M., Poole A. Z., Weis V. M., Detournay O. (2017). Implication of the host TGFβ pathway in the onset of symbiosis between larvae of the coral Fungia scutaria and the dinoflagellate Symbiodinium sp.(clade C1f). Coral Reefs. 36 (4), 1263–1268. doi: 10.1007/s00338-017-1621-6
Bleidißel S. (2010). Molekulare untersuchungen zur evolution der aeolidida (Mollusca, Gastropoda, nudibranchia, cladobranchia) und zur evolution einer sekundären symbiose mit symbiodinium (Dinoflagellata) in den aeolidida (Bergische Universität Wuppertal, Fakultät für Mathematik und Naturwissenschaften Biologie Dissertationen)
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 (5), 525–527. doi: 10.1038/nbt.3519
Buchmann K. (2014). Evolution of innate immunity: clues from invertebrates via fish to mammals. Front. Immunol. 5, 459. doi: 10.3389/fimmu.2014.00459
Burghardt I., Evertsen J., Johnsen G., Wägele H. (2005). Solar powered sea slugs-mutualistic symbiosis of aeolid nudibranchia (Mollusca, Gastropoda, opisthobranchia) with Symbiodinium. Symbiosi. 38, (227–250).
Burghardt I., Stemmer K., Wägele H. (2008). Symbiosis between Symbiodinium (Dinophyceae) and various taxa of nudibranchia (Mollusca: Gastropoda), with analyses of long-term retention. Org. Divers. Evol. 8 (1), 66–76. doi: 10.1016/j.ode.2007.01.001
Burghardt I., Wägele H. (2004). A new solar powered species of the genus Phyllodesmium ehrenber (Mollusca: Nudibranchia: Aeolidoidea) from Indonesia with analysis of its photosynthetic activity and notes on biology. Zootaxa. 596 (1), 1–18. doi: 10.11646/zootaxa.596.1.1
Burghardt I., Wägele H. (2006). Interspecific differences in the efficiency and photosynthetic characteristics of the symbiosis of “solarpowered” nudibranchia (Mollusca: Gastropoda) with zooxanthellae. Rec. West Austr Mus. 69, (1–9). https://d-nb.info/983779732/34#page=88
Bushnell B. BBTools software package, version 33.89. Available at: http://sourceforge.net/projects/bbmap.
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cantalapiedra C. P., Hernández-Plaza A., Letunic I., Bork P., Huerta-Cepas J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38 (12), 5825–5829. doi: 10.1093/molbev/msab293
Carroll D. J., Kempf S. C. (1990). Laboratory culture of the aeolid nudibranch Berghia verrucicornis (Mollusca, opisthobranchia): Some aspects of its development and life history. Biol. Bull. 179 (3), 243–253. doi: 10.2307/1542315
Chan C. X., Vaysberg P., Price D. C., Pelletreau K. N., Rumpho M. E., Bhattacharya D. (2018). Active host response to algal symbionts in the sea slug Elysia chlorotica. Mol Biol. Evol. 35 (7), 1706–1711. doi: 10.1093/molbev/msy061
Chen M. C., Cheng Y. M., Hong M. C., Fang L. S. (2004). Molecular cloning of Rab5 (ApRab5) in Aiptasia pulchella and its retention in phagosomes harboring live zooxanthellae. Biophys. Res. Commun. 324 (3), 1024–1033. doi: 10.1016/j.bbrc.2004.09.151
Chen M. C., Cheng Y. M., Sung P. J., Kuo C. E., Fang L. S. (2003). Molecular identification of Rab7 (ApRab7) in Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae. Biochem. Biophys. Res. Commun. 308 (3), 586–595. doi: 10.1016/S0006-291X(03)01428-1
Chen M. C., Hong M. C., Huang Y. S., Liu M. C., Cheng Y. M., Fang L. S. (2005). ApRab11, a cnidarian homologue of the recycling regulatory protein Rab11, is involved in the establishment and maintenance of the Aiptasia–Symbiodinium endosymbiosis. Bioph Res. Co. 338 (3), 1607–1616. doi: 10.1016/j.bbrc.2005.10.133
Chew S. F., Koh C. Z., Hiong K. C., Choo C. Y., Wong W. P., Neo M. L., et al. (2019). Light-enhanced expression of carbonic anhydrase 4-like supports shell formation in the fluted giant clam Tridacna squamosa. Gene. 683, 101–112. doi: 10.1016/j.gene.2018.10.023
Core Team. R. (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Cui G., Liew Y. J., Li Y., Kharbatia N., Zahran N. I., Emwas A. H., et al. (2019). Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PloS Genet. 15 (6), e1008189. doi: 10.1371/journal.pgen.1008189
Davy S. K., Allemand D., Weis V. M. (2012). Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76 (2), 229–261. doi: 10.1128/MMBR.05014-11
Day D. A., Tuite M. F. (1998). Post-transcriptional gene regulatory mechanisms in eukaryotes: An overview. J. Endocrinol. 157 (3), 361–371. doi: 10.1677/joe.0.1570361
DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113. doi: 10.3354/meps08372
Detournay O., Schnitzler C. E., Poole A., Weis V. M. (2012). Regulation of cnidarian–dinoflagellate mutualisms: evidence that activation of a host TGFβ innate immune pathway promotes tolerance of the symbiont. Dev. Comp. Immunol. 38 (4), 525–537. doi: 10.1016/j.dci.2012.08.008
Dev A., Iyer S., Razani B., Cheng G. (2010). “NF-κB and innate immunity,” in Current topics in microbiology and immunology (Berlin, Heidelberg: Springer), 115–143.
Dierking K., Pita L. (2020). Receptors mediating host-microbiota communication in the metaorganism: The invertebrate perspective. Front. Immunol. 11, 1251. doi: 10.3389/fimmu.2020.01251
Diks S. H., van Deventer S. J., Peppelenbosch M. P. (2001). Invited review: Lipopolysaccharide recognition, internalisation, signalling and other cellular effects. J. Endotoxin. Res. 7 (5), 335–348. doi: 10.1177/09680519010070050101
Dionísio G., Rosa R., Leal M. C., Cruz S., Brandão C., Calado G., et al. (2013). Beauties and beasts: a portrait of sea slugs aquaculture. Aquac. 408, 1–14. doi: 10.1016/j.aquaculture.2013.04.033
Dixon G. B., Davies S. W., Aglyamova G. V., Meyer E., Bay L. K., Matz M. V. (2015). Genomic determinants of coral heat tolerance across latitudes. Science 348 (6242), 1460–1462. doi: 10.1126/science.1261224
Doyle S. L., O’Neill L. A. (2006). Toll-like receptors: From the discovery of NF-κB to new insights into transcriptional regulations in innate immunity. Biochem. Pharmacol. 72 (9), 1102–1113. doi: 10.1016/j.bcp.2006.07.010
Dungan A. M., Hartman L. M., Tortorelli G., Belderok R., Lamb A. M., Pisan L., et al. (2020). Exaiptasia diaphana from the great barrier reef: A valuable resource for coral symbiosis research. Symbiosis 80 (2), 195–206. doi: 10.1007/s13199-020-00665-0
Dunkelberger J. R., Song W. C. (2010). Complement and its role in innate and adaptive immune responses. Cell Res. 20 (1), 34–50. doi: 10.1038/cr.2009.139
Fransolet D., Roberty S., Plumier J. C. (2012). Establishment of endosymbiosis: the case of cnidarians and Symbiodinium. J. Exp. Mar. Biol. Ecol. 420, 1–7. doi: 10.1016/j.jembe.2012.03.015
Fujise L., Suggett D. J., Stat M., Kahlke T., Bunce M., Gardner S. G., et al. (2021). Unlocking the phylogenetic diversity, primary habitats, and abundances of free-living symbiodiniaceae on a coral reef. Mol. Ecol. 30, 343–360. doi: 10.1111/mec.15719
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28 (23), 3150–3152. doi: 10.1093/bioinformatics/bts565
Gabay Y., Weis V. M., Davy S. K. (2018). Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian-dinoflagellate symbiosis. Biol. Bull. 234 (1), 1–10. doi: 10.1086/696365
Ganot P., Moya A., Magnone V., Allemand D., Furla P., Sabourault C. (2011). Adaptations to endosymbiosis in a cnidarian-dinoflagellate association: Differential gene expression and specific gene duplications. PloS Genet. 7 (7), e1002187. doi: 10.1371/journal.pgen.1002187
Gibney E. R., Nolan C. M. (2010). Epigenetics and gene expression. Heredity 105 (1), 4–13. doi: 10.1038/hdy.2010.54
Godoy-Olmos S., Martinez-Llorens S., Tomas-Vidal A., Monge-Ortiz R., Estruch G., Jover-Cerda M. (2019). Influence of temperature, ammonia load and hydraulic loading on the performance of nitrifying trickling filters for recirculating aquaculture systems. J. Environ. Chem. Eng. 7 (4), 103257. doi: 10.1016/j.jece.2019.103257
Gong Z. J., Wu Y. Q., Miao J., Duan Y., Jiang Y. L., Li T. (2013). Global transcriptome analysis of orange wheat blossom midge, Sitodiplosis mosellana (Gehin)(Diptera: Cecidomyiidae) to identify candidate transcripts regulating diapause. PloS One 8 (8), e71564. doi: 10.1371/journal.pone.0071564
Goulet T. L., Lucas M. Q., Schizas N. V. (2019). Symbiodiniaceae genetic diversity and symbioses with hosts from shallow to mesophotic coral ecosystems. In Mesophotic coral ecosystems (Cham: Springer), pp. 537–51.
Grasso L. C., Maindonald J., Rudd S., Hayward D. C., Saint R., Miller D. J., et al. (2008). Microarray analysis identifies candidate genes for key roles in coral development. BMC Genom. 9 (1), 1–18. doi: 10.1186/1471-2164-9-540
Haas B., Papanicolaou A. J. G. S. (2016) TransDecoder (find coding regions within transcripts). Available at: https://github.com/TransDecoder/TransDecoder.wiki.git.
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8 (8), 1494–1512. doi: 10.1038/nprot.2013.084
Henschel R., Lieber M., Wu L. S., Nista P. M., Haas B. J., LeDuc R. D. (2012). “Trinity RNA-seq assembler performance optimization,” in Proceedings of the 1st conference of the extreme science and engineering discovery environment: Bridging from the eXtreme to the campus and beyond, vol. 45. Association for Computing Machinery, 1–8. doi: 10.1145/2335755.2335842
Hernawan U. E. (2008). Symbiosis between the giant clams (Bivalvia: Cardiidae) and zooxanthellae (Dinophyceae). Biodivers J. 9 (1), 53–58. doi: 10.13057/biodiv/d090112
Herpin A., Lelong C., Favrel P. (2004). Transforming growth factor-β-related proteins: an ancestral and widespread superfamily of cytokines in metazoans. Dev. Comp. Immunol. 28 (5), 461–485. doi: 10.1016/j.dci.2003.09.007
Hill M., Hill A. (2012). The magnesium inhibition and arrested phagosome hypotheses: new perspectives on the evolution and ecology of Symbiodinium symbioses. Biol. Rev. 87 (4), 804–821. doi: 10.1111/j.1469-185X.2012.00223.x
Hinde R., Trautman D. A. (2001). “Symbiosomes,” In: Seckbach J. (eds) Symbiosis. Cellular Origin, Life in Extreme Habitats and Astrobiology, vol 4. Dordrecht (Springer), 207–220. doi: 10.1007/0-306-48173-1_12
Huang B., Zhang L., Li L., Tang X., Zhang G. (2015). Highly diverse fibrinogen-related proteins in the pacific oyster Crassostrea gigas. Fish Shellfish Immunol. 43 (2), 485–490. doi: 10.1016/j.fsi.2015.01.021
Ip Y. K., Koh C. Z., Hiong K. C., Choo C. Y., Boo M. V., Wong W. P., et al. (2017). Carbonic anhydrase 2-like in the giant clam, Tridacna squamosa: Characterization, localization, response to light, and possible role in the transport of inorganic carbon from the host to its symbionts. Physiol. Rep. 5 (23), e13494. doi: 10.14814/phy2.13494
Ishii Y., Maruyama S., Takahashi H., Aihara Y., Yamaguchi T., Yamaguchi K., et al. (2019). Global shifts in gene expression profiles accompanied by environmental changes in cnidarian-dinoflagellate endosymbiosis. G3: Genes Genomes Genet. 9 (7), 2337–2347. doi: 10.1534/g3.118.201012
Jacobovitz M. R., Rupp S., Voss P. A., Maegele I., Gornik S. G., Guse A. (2021). Dinoflagellate symbionts escape vomocytosis by host cell immune suppression. Nat. Microbiol. 6 (6), 769–782. doi: 10.1038/s41564-021-00897-w
Jimbo M., Yanohara T., Koike K., Koike K., Sakai R., Muramoto K., et al. (2000). The d-galactose-binding lectin of the octocoral Sinularia lochmodes: Characterization and possible relationship to the symbiotic dinoflagellates. Comp. Biochem. Physiol. - B Biochem. 125 (2), 227–236. doi: 10.1016/S0305-0491(99)00173-X
Kazandjian A., Shepherd V. A., Rodriguez-Lanetty M., Nordemeier W., Larkum A. W., Quinnell R. G., et al (2008). Isolation of symbiosomes and the symbiosome membrane complex from the zoanthid Zoanthus robustus. Phycologia 47 (3), 294–306. doi: 10.2216/07-23.1
Kemper C., Atkinson J. P., Hourcade D. E. (2010). Properdin: Emerging roles of a pattern-recognition molecule. Annu. Rev. Immunol. 28, 131–155. doi: 10.1146/annurev-immunol-030409-101250
Kempf S. C. (1991). A ‘primitive’symbiosis between the aeolid nudibranch Berghia verrucicornis (A. cost) and a zooxanthella. J. Molluscan Stud. 57 (4), 75–85. doi: 10.1093/mollus/57.Supplement_Part_4.75
Kenjo A., Takahashi M., Matsushita M., Endo Y., Nakata M., Mizuochi T., et al. (2001). Cloning and characterization of novel ficolins from the solitary ascidian, Halocynthia roretzi. J. Biol. Chem. 276 (23), 19959–19965. doi: 10.1074/jbc.M011723200
Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., et al. (2008). A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10 (5), 556–566. doi: 10.1038/ncb1718
Kinchen J. M., Ravichandran K. S. (2008). Phagosome maturation: Going through the acid test. Nat. Rev. Mol. Cell Biol. 9 (10), 781–795. doi: 10.1038/nrm2515
Koike K., Jimbo M., Sakai R., Kaeriyama M., Muramoto K., Ogata T., et al. (2004). Octocoral chemical signaling selects and controls dinoflagellate symbionts. Biol. Bull. 207 (2), 80–86. doi: 10.2307/1543582
Koop K., Booth D., Broadbent A., Brodie J., Bucher D., Capone D., et al. (2001). ENCORE: the effect of nutrient enrichment on coral reefs. Synthesis of results and conclusions. Mar. pollut. Bull. 42 (2), 91–120. doi: 10.1016/S0025-326X(00)00181-8
Kvennefors E. C. E., Leggat W., Kerr C. C., Ainsworth T. D., Hoegh-Guldberg O., Barnes A. C. (2010). Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev. Comp. Immunol. 34, 1219–1229. doi: 10.1016/j.dci.2010.06.016
Lee H. J., Woo Y., Hahn T. W., Jung Y. M., Jung Y. J. (2020). Formation and maturation of the phagosome: A key mechanism in innate immunity against intracellular bacterial infection. Microorganisms 8 (9), 1298. doi: 10.3390/microorganisms8091298
Lehnert E. M., Mouchka M. E., Burriesci M. S., Gallo N. D., Schwarz J. A., Pringle J. R. (2014). Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3-Genes Genom. Genet. 4 (2), 277–295. doi: 10.1534/g3.113.009084
Lesser M. P. (2011). “Coral bleaching: Causes and mechanisms,” in Coral reefs: An ecosystemin transition, vol. pp. (Berlin: Springer), 405–419.
Li W., Godzik A. (2006). Cd-hit: A fast program for clustering and com-paring large sets of protein or nucleotide sequences. Bioinformatics. 22 (13), 1658–1659. doi: 10.1093/bioinformatics/btl158
Liu H., Stephens T. G., González-Pech R. A., Beltran V. H., Lapeyre B., Bongaerts P., et al. (2018). Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1 (1), 1–11. doi: 10.1038/s42003-018-0098-3
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ma Y. J., Hein E., Munthe-Fog L., Skjoedt M. O., Bayarri-Olmos R., Romani L., et al. (2015). Soluble collectin-12 (CL-12) is a pattern recognition molecule initiating complement activation via the alternative pathway. J. Immunol. 195 (7), 3365–3373. doi: 10.4049/jimmunol.1500493
Mansfield K. M., Carter N. M., Nguyen L., Cleves P. A., Alshanbayeva A., Williams L. M., et al. (2017). Transcription factor NF-κB is modulated by symbiotic status in a sea anemone model of cnidarian bleaching. Sci. Rep. 7, 16025. doi: 10.1038/s41598-017-16168-w
Mansfield K. M., Gilmore T. D. (2019). Innate immunity and cnidarian-symbiodiniaceae mutualism. Dev. Comp. Immunol. 90, 199–209. doi: 10.1016/j.dci.2018.09.020
Martin. M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17 (1), 10–12. doi: 10.14806/ej.17.1.200
Matthews J. L., Crowder C. M., Oakley C. A., Lutz A., Roessner U., Meyer E., et al. (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. PNAS 114 (50), 13194–13199. doi: 10.1073/pnas.1710733114
Matthews J. L., Sproles A. E., Oakley C. A., Grossman A. R., Weis V. M., Davy S. K. (2016). Menthol-induced bleaching rapidly and effectively provides experimental aposymbiotic sea anemones (Aiptasia sp.) for symbiosis investigations. J. Exp. Biol. 219 (3), 306–310. doi: 10.1242/jeb.128934
Matz (2019). Package “KOGMWU”. Functional Summary and Meta-Analysis of Gene Expression Data. R documentation. https://cran.r-project.org/web/packages/KOGMWU/KOGMWU.pdf
Melo Clavijo J., Donath A., Serôdio J., Christa G. (2018). Polymorphic adaptations in metazoans to establish and maintain photosymbioses. Biol. Rev. 93 (4), 2006–2020. doi: 10.1111/brv.12430
Melo Clavijo J., Frankenbach S., Fidalgo C., Serôdio J., Donath A., Preisfeld A., et al. (2020). Identification of scavenger receptors and thrombospondin-type-1 repeat proteins potentially relevant for plastid recognition in sacoglossa. Ecol. Evol. 10 (21), 12348–12363. doi: 10.1002/ece3.6865
Meyer E., Aglyamova G. V., Matz M. V. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol. Ecol. 20 (17), 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x
Meyer E., Weis V. M. (2012). Study of cnidarian-algal symbiosis in the “omics” age. Biol. Bull. 223 (1), 44–65. doi: 10.1086/BBLv223n1p44
Middha S., Wang X. (2008). Evolution and potential function of fibrinogen-like domains across twelve Drosophila species. BMC Genom. 9 (1), 1–8. doi: 10.1186/1471-2164-9-260
Mies M., Voolstra C. R., Castro C. B., Pires D. O., Calderon E. N., Sumida P. Y. (2017). Expression of a symbiosis-specific gene in Symbiodinium type A1 associated with coral, nudibranch and giant clam larvae. R Soc. Open Sci. 4 (5), 170253. doi: 10.1098/rsos.170253
Mohamed A. R., Andrade N., Moya A., Chan C. X., Negri A. P., Bourne D. G., et al. (2020). Dual RNA-sequencing analyses of a coral and its native symbiont during the establishment of symbiosis. Mol. Ecol. 29 (20), 3921–3937. doi: 10.1111/mec.15612
Mohamed A. R., Cumbo V., Harii S., Shinzato C., Chan C. X., Ragan M. A., et al. (2016). The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome. Mol. Ecol. 25 (13), 3127–3141. doi: 10.1111/mec.13659
Monteiro E. A., Güth A. Z., Banha T. N., Sumida P. Y., Mies M. (2019a). Implications of feeding frequency, prey size and condition, and intraspecific competition for the commercial aquaculture of the nudibranch Berghia stephanieae. J. World Aquac. Soc 51 (1), 244–254. doi: 10.1111/jwas.12645
Monteiro E. A., Güth A. Z., Banha T. N., Sumida P. Y., Mies M. (2019b). Evidence against mutualism in an aeolid nudibranch associated with symbiodiniaceae dinoflagellates. Symbiosis 79 (2), 183–189. doi: 10.1007/s13199-019-00632-4
Neckelmann N., Muscatine L. (1983). Regulatory mechanisms maintaining the Hydra-Chlorella symbiosis. Proceedings of the Royal society of London. Series B. Biological sciences 219 (1215), 193–210. doi: 10.1098/rspb.1983.0067
Neubauer E. F., Poole A. Z., Detournay O., Weis V. M., Davy S. K. (2016). The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian-dinoflagellate symbiosis. PeerJ 4, e2692. doi: 10.7717/peerj.2692
Neubauer E. F., Poole A. Z., Neubauer P., Detournay O., Tan K., Davy S. K., et al. (2017). A diverse host thrombospondin-type-1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian-dinoflagellate symbiosis. Elife 6, e24494. doi: 10.7554/eLife.24494.049
Norton J. H., Shepherd M. A., Long H. M., Fitt W. K. (1992). The zooxanthellal tubular system in the giant clam. Biol. Bull. 183 (3), 503–506. doi: 10.2307/1542028
Nyholm S. V., McFall-Ngai M. (2004). The winnowing: establishing the squid–vibrio symbiosis. Nat. Rev. Microbiol. 2 (8), 632–642. doi: 10.1038/nrmicro957
Pochon X., Pawlowski J., Zaninetti L., Rowan R. (2001). High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar. Biol. 139 (6), 1069–1078. doi: 10.1007/s00227010067
Poole A. Z., Kitchen S. A., Weis V. M. (2016). The role of complement in cnidarian-dinoflagellate symbiosis and immune challenge in the sea anemone Aiptasia pallida. Front. Microbiol. 7, 519. doi: 10.3389/fmicb.2016.00519
PrabhuDas M. R., Baldwin C. L., Bollyky P. L., Bowdish D. M., Drickamer K., Febbraio M., et al. (2017). A consensus definitive classification of scavenger receptors and their roles in health and disease. J. Immunol. 198 (10), 3775–3789. doi: 10.1007/s002270100674
Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., et al. (2021). Heat stress destabilizes symbiotic nutrient cycling in corals 118, 5. doi: 10.1073/pnas.2022653118
Rädecker N., Raina J. B., Pernice M., Perna G., Guagliardo P., Kilburn M. R., et al. (2018). Using Aiptasia as a model to study metabolic interactions in cnidarian-Symbiodinium symbioses. Front. Physiol. 9, 214. doi: 10.3389/fphys.2018.00214
Rapp W. (1829). Ueber die polypen im allgemeinen und die actinien insbesondere (Weimar: Verlage des Grofsherzogl; Sächs. privileg. Landes-Industrie-Comptoirs).
Richier S., Furla P., Plantivaux A., Merle P. L., Allemand D. (2005). Symbiosis-induced adaptation to oxidative stress. Journal of Experimental Biology 208 (2), 277–85. doi: 10.1111/j.1742-4658.2006.05414.x
Richier S., Sabourault C., Courtiade J., Zucchini N., Allemand D., Furla P. (2006). Oxidative stress and apoptotic events during thermal stress in the symbiotic sea anemone, Anemonia viridis. FEBS J. 273 (18), 4186–4198.
Rola M., Frankenbach S., Bleidissel S., Sickinger C., Donath A., Frommlet J. C., et al. (2022). Cladobranchia (Gastropoda, nudibranchia) as a promising model to understand the molecular evolution of photosymbiosis in animals. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.745644
Romero A., Dios S., Poisa-Beiro L., Costa M. M., Posada D., Figueras A., et al. (2011). Individual sequence variability and functional activities of fibrinogen-related proteins (FREPs) in the Mediterranean mussel (Mytilus galloprovincialis) suggest ancient and complex immune recognition models in invertebrates. Dev. Comp. Immunol. 35 (3), 334–344. doi: 10.1016/j.dci.2010.10.007
Roth M. S. (2014). The engine of the reef: photobiology of the coral–algal symbiosis. Front. Microbiol. 5, 422. doi: 10.3389/fmicb.2014.00422
Röthig T., Costa R. M., Simona F., Baumgarten S., Torres A. F., Radhakrishnan A., et al. (2016). Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Front. Mar. Sci. 3, 234. doi: 10.3389/fmars.2016.00234
Rousseau C. (1934). Sur la estructure de l’epithelium hépatique de eolidiens. C. R. hebd. séances Acad. Sci. 198, 677–679.
Rousseau C. (1935). Histophysiologie du foie des eolidiens Etude de leurs xanthelles. cytologie des cellules nématophages. Arch. Anat. Microsc. 31, 305–395.
Royet J., Gupta D., Dziarski R. (2011). Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat. Rev. Immunol. 11 (12), 837–851. doi: 10.1038/nri3089
Rus H., Cudrici C., Niculescu F. (2005). The role of the complement system in innate immunity. Immunol. Res. 33 (2), 103–112. doi: 10.1385/IR:33:2:103
Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23 (15), 1399–1408. doi: 10.1016/j.cub.2013.05.062
Silverstein R. L., Febbraio M. (2009). CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2 (72), re3–re3. doi: 10.1126/scisignal.272re3
Sinha A., Sommer R. J., Dieterich C. (2012). Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC Genom. 13 (1), 1–17. doi: 10.1186/1471-2164-13-254
Stanley Jr. G.D., Lipps J. H. (2011). Photosymbiosis: the driving force for reef success and failure. Paleontol. Soc. Paper 17, 33–60. doi: 10.1017/S1089332600002436
Stat M., Pochon X., Cowie R. O., Gates R. D. (2009). Specificity in communities of Symbiodinium in corals from Johnston atoll. Mar. Ecol. Prog. Ser. 386, 83–96. doi: 10.3354/meps08080
Strader M. E., Aglyamova G. V., Matz M. V. (2016). Red fluorescence in coral larvae is associated with a diapause-like state. Mol. Ecol. 25 (2), 559–569. doi: 10.1111/mec.13488
Taylor D. L. (1978). Nutrition of algal-invertebrate symbiosis. II. effects of exogenous nitrogen sources on growth, photosynthesis and the rate of excretion by algal symbionts in vivo and in vitro. Proc. R Soc. B: Biol. Sci. 201 (1145), 401–412. doi: 10.1098/rspb.1978.0052
The UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D1. doi: 10.1093/nar/gkaa1100
Tortorelli G., Belderok R., Davy S. K., McFadden G. I., Van Oppen M. J. (2020). Host genotypic effect on algal symbiosis establishment in the coral model, the anemone exaiptasia diaphana, from the great barrier reef. Front. Mar. Sci. 6, 833. doi: 10.3389/fmars.2019.00833
Valdés A. (2005). A new species of aeolidiella berg(Mollusca: Nudibranchia: Aeolidiidae) from the Florida keys, USA. Veliger. 47 (3), 218–223. https://www.biodiversitylibrary.org/partpdf/97944
Wägele M., Johnsen G. (2001). Observations on the histology and photosynthetic performance of “solar-powered” opisthobranchs (Mollusca, Gastropoda, opisthobranchia) containing symbiotic chloroplasts or zooxanthellae. Org. Divers. Evol. 1 (3), 193–210. doi: 10.1078/1439-6092-00016
Wägele H., Raupach M. J., Burghardt I., Grzymbowski Y., Händeler K. (2010). “Solar powered seaslugs (Opisthobranchia, Gastropoda, mollusca): incorporation of photosynthetic units: A key character enhancing radiation?,” in Evolution in action (Berlin, Heidelberg: Springer), 263–282.
Weis V. M. (1993). Effect of dissolved inorganic carbon concentration on the photosynthesis of the symbiotic sea anemone Aiptasia pulchella carlgren: Role of carbonicanhydrase. J. Exp. Mar. Biol. Ecol. 174 (2), 209–225. doi: 10.1016/0022-0981(93)90018-J
Weis V. M., Levine R. P. (1996). Differential protein profiles reflect the different lifestyles of symbiotic and aposymbiotic Anthopleura elegantissima, a sea anemone from temperate waters. J. Exp. Biol. 199 (4), 883–892. doi: 10.1242/jeb.199.4.883
Weis V. M., Smith G. J., Muscatine L. (1989). A “CO2 supply” mechanism in zooxanthellate cnidarians: role of carbonic anhydrase. Mar. Biol. 100 (2), 195–202. doi: 10.1007/BF00391958
Wolfowicz I., Baumgarten S., Voss P. A., Hambleton E. A., Voolstra C. R., Hatta M., et al. (2016). Aiptasia sp larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Sci. Rep. 6, 32366. doi: 10.1038/srep32366
Wright R. M., Aglyamova G. V., Meyer E., Matz M. V. (2015). Gene expression associated with white syndromes in a reef-building coral, Acropora hyacinthus. BMC Genom. 16, 371. doi: 10.1186/s12864-015-1540-2
Wu C., Söderhäll K., Söderhäll I. (2011). Two novel ficolin-like proteins act as pattern recognition receptors for invading pathogens in the freshwater crayfish pacifastacus leniusculus. Proteomics. 11 (11), 2249–2264. doi: 10.1002/pmic.201000728
Xiang T., Lehnert E., Jinkerson R. E., Clowez S., Kim R. G., DeNofrio J. C., et al. (2020). Symbiont population control by host-symbiont metabolic interaction in symbiodiniaceae-cnidarian associations. Nat. Commun. 11 (1), 1–9. doi: 10.1038/s41467-019-13963-z
Xiang Z., Qu F., Wang F., Li J., Zhang Y., Yu Z. (2014). Characteristic and functional analysis of a ficolin-like protein from the oyster Crassostrea hongkongensis. Fish Shellfish Immunol. 40 (2), 514–523. doi: 10.1016/j.fsi.2014.08.006
Xiao R., Wei Y., An D., Li D., Ta X., Wu Y., et al. (2019). A review on the research status and development trend of equipment in water treatment processes of recirculating aquaculture systems. Rev. Aquac. 11 (3), 863–895. doi: 10.1111/raq.12270
Yellowlees D., Rees T. A. V., Leggat W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31 (5), 679–694. doi: 10.1111/j.1365-3040.2008.01802.x
Yu L., Dai Y., Mineo C. (2021). Novel functions of endothelial scavenger receptor class b type I. Curr. Atheroscler. Rep. 23 (2), 1–9. doi: 10.1007/s11883-020-00903-2
Yuyama I., Ishikawa M., Nozawa M., Yoshida M. A., Ikeo K. (2018). Transcriptomic changes with increasing algal symbiont reveal the detailed process underlying establishment of coral-algal symbiosis. Sci. Rep. 8 (1), 1–11. doi: 10.1038/s41598-018-34575-5
Keywords: nudibranchia, photosynthetic symbiosis, onset of symbiosis, de novo transcriptome, symbiont recognition
Citation: Clavijo JM, Sickinger C, Bleidißel S, Gasparoni G, Tierling S, Preisfeld A and Christa G (2022) The nudibranch Berghia stephanieae (Valdés, 2005) is not able to initiate a functional symbiosome-like environment to maintain Breviolum minutum (J.E.Parkinson & LaJeunesse 2018). Front. Mar. Sci. 9:934307. doi: 10.3389/fmars.2022.934307
Received: 02 May 2022; Accepted: 25 August 2022;
Published: 03 October 2022.
Edited by:
Marco Munari, Anton Dohrn Zoological Station, ItalyReviewed by:
Raúl A. González-Pech, The Pennsylvania State University (PSU), United StatesCopyright © 2022 Clavijo, Sickinger, Bleidißel, Gasparoni, Tierling, Preisfeld and Christa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jenny Melo Clavijo, bWVsb2NsYXZpam9AdW5pLXd1cHBlcnRhbC5kZQ==; Gregor Christa, Y2hyaXN0YUB1bmktd3VwcGVydGFsLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.