
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci. , 20 July 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.924130
This article is part of the Research Topic Genetic Adaption and Metabolic Response of Aquatic Animals to Diverse Water Environment Parameters View all 13 articles
 Yongxiang Yu1,2†
Yongxiang Yu1,2† Xiao Liu3†
Xiao Liu3† Yingeng Wang1,2*
Yingeng Wang1,2* Meijie Liao1,2
Meijie Liao1,2 Miaomiao Tang1
Miaomiao Tang1 Xiaojun Rong1,2
Xiaojun Rong1,2 Chunyuan Wang1
Chunyuan Wang1 Bin Li1,2
Bin Li1,2 Zheng Zhang1,2*
Zheng Zhang1,2*As an intestinal organism settled long-term within the gut of marine fish, Vibrio scophthalmi is a potential object for the bacterium genetic variation and adaptation research. The genetic diversity, antimicrobial resistance phenotype, and genotype of 33 V. scophthalmi isolated from diseased marine fish intestines between 2002 and 2020 were evaluated. The results showed that all isolates were frequently resistant to penicillins, cephalosporins, aminoglycosides, and macrolides and displayed multidrug-resistant (MDR) phenotype in vitro. Thirty percent of isolates were resistant to more than 20 different drugs. The average insensitive (resistant and intermediate) rate of V. scophthalmi isolates was 49.5%~81.8% between 2002 and 2020, but the t-test revealed that there was no significant difference in the drug-resistance rate of V. scophthalmi isolates with typical interannual variability. Eleven antimicrobial resistance genes (strB, strA, ant(3 ˝)-I, mphA, blaPSE, qnrS, tetC, tetE, tetM, tetS, and int1) were detected in these isolates, but the antimicrobial resistance phenotypes and genotypes of these isolates were not consistent. Enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) analysis indicated that 33 isolates could be divided into two clusters (G1 and G3) and two single isolates (G2 and G4), and the G2 cluster was isolated from South Sea C. undulates with typical geographical species differences. There was no significant correlation between the drug susceptibility and the genetic types of V. scophthalmi isolates. The results reveal the mismatch phenomenon between antimicrobial resistance and genotype of inherent V. scophthalmi in the marine fish intestines, and the antimicrobial susceptibility isolates might be a potential risk source for storage and transmission of resistance genes.
Vibrio scophthalmi was first isolated from the intestines of juvenile turbot and identified as a species of Vibrio genus based on phenotypic traits, G+C content, DNA-DNA hybridization, and 16S rDNA gene sequence in 1997 (Cerdag-cuellar et al., 1997). Previous studies have indicated that V. scophthalmi has pathogenicity to aquatic animals such as Paralichthys olivaceus (Qiao et al., 2012), Dentex dentex L. (Sitjà-bobadilla et al., 2010), and Thunnus maccoyii (Valdenegro-vega et al., 2013). Furthermore, V. scophthalmi was also the abundant Vibrio species in the healthy turbot intestinal microbiota and serves as an intestinal organism settled long term within the gut of marine fish (Cerdag-cuellar and Blanch, 2010).
As an intestinal bacterium in marine fish, the physiological and metabolic phenotype of V. scophthalmi was highly related to the breeding environment and food foundation of fish. Thus, V. scophthalmi could be a potential object for the bacterium genetic variation and adaptation research in fish culture. Identifying the types of microbial pathogens through various methods such as phenotyping and molecular typing to investigate the prevalence of hospitalized infections is a necessity (Healy et al., 2005). Antimicrobial resistance is an important concern in the public health systems that due to the rise in the resistant strains of bacteria (Nouri et al., 2020). And the enterobacterial repetitive intergenic consensus sequences polymerase chain reaction (ERIC-PCR) is a simple, fast, and affordable PCR-based methods for molecular typing analysis of different isolated bacteria, which were less dependent on effective and variable factors on bacterial growth (Sedighi et al., 2020).
In this study, we evaluated the antimicrobial resistance and genotype characteristics of V. scophthalmi from diseased fish intestines (85% strains isolated from diseased flatfish) from 2002–2020 and gained a better understanding of the relationship between antimicrobial-resistant phenotypes and resistance genotypes of those isolates. The results could provide an overview of the status of bacterial genetic diversity and antimicrobial sensitivity in intensive cultivation of fish and provide guidance on aquatic disease treatment.
From Nov 2002 to Oct 2020, epidemiological and etiological investigations were performed which primarily aims at monitoring the prevalence of bacterial diseases among cultured marine fish in China. During this time, a total of 33 isolates were preliminarily identified to be V. scophthalmi with typical timeline differences. The isolates in our study were isolates from fish intestines with typical intestinal diseases. The detailed information of 33 V. scophthalmi isolates was shown in Supplementary Table 1. The isolate resources were resuspended in 20% glycerol and stored at −80°C in our laboratory for long-term preservation. For all the experiments, the V. scophthalmi isolates were grown in tryptic soy agar medium and incubated aerobically at 28°C for 24 h.
A total of 33 agents were chosen for antimicrobials susceptibility test in vitro for scientific research. Some of the drugs were allowed in veterinary practices at different periods, and others were chosen for the drug resistance census in this study only. The types and concentrations of antimicrobials were listed in Supplementary Table 2. The testing was performed using the disc diffusion method as described in the Clinical and Laboratory Standards Institute M07 (CLSI-M07). Escherichia coli ATCC 25922 was used as the reference strain. The bacterial suspension was prepared and adjusted to approximately 1 × 108 cfu/ml. The disc contains antimicrobial agents that were affixed to the agar plate coated with 100 µl of bacterial suspension. Culture media were maintained in a normal atmosphere incubator for 24 h at 28°C. Each group was replicated twice, and the mean of the inhibition zone was used as the result. The bacterial sensitivity was calculated by referring to the aquatic bacterial susceptibility test standard issued by the CLSI.
To determine the presence of antimicrobial resistance genes (ARGs), PCR detection assays were used to examine the presence or absence of 20 ARGs belonging to six categories (Versalovic et al., 1991; Yamamoto and Harayama, 1995; Byers et al., 1998; Speldooren et al., 1998; Jun et al., 2004; Kim et al., 2004; Chuanchuen and Padungtod, 2009; Nguyen et al., 2009; Colomer-lluch et al., 2011; Liu et al., 2013; Deng et al., 2014; Qiao et al., 2017; Moffat et al., 2020; Yuan et al., 2021; Yu et al., 2021). Primers of all target ARGs were based on the published literature and are shown in Supplementary Table 3. The PCR method employed to determine the presence of ARGs was based on the genomic DNA of V. scophthalmi isolates. The gene analyses used specific primers and PCR conditions modified according to primer Tm values. The PCR fragments were sequenced for both strands, and the sequence identities were conformed using the BLAST database.
The optimized ERIC-PCR condition was employed for the genetic diversity analysis of V. scophthalmi isolates as Versalovic et al. (Versalovic et al., 1991) described with minor modification. The final optimized amplification conditions consisted of initial denaturation at 94°C for 5 min, followed by 30 cycles each of denaturation at 94°C for 30 s, annealing at 55°C for 1 min, extension at 72°C for 5 min, and an additional 10-min extension at 72°C. The amplification was repeated three times to confirm the reproducibility of the ERIC-PCR method. The zero-one manual method was used to analyze the patterns, and the dendrogram was drawn according to the clusters (Movahedi et al., 2021).
All isolates V. scophthalmi were tested for antimicrobial susceptibilities to 33 selected antibiotics in vitro, and the antimicrobial resistance phenotypes were evaluated and summarized in Figure 1. Among the V. scophthalmi isolates, the resistance frequency to penicillins and aminoglycosides reached up to 48.48%–100% and 57.58%–93.94%, respectively. In contrast, there was a low resistance frequency to phenicols (9.09% for florfenicol and 15.15% for chloroamphenicol). When it comes to cephalosporins, tetracyclines, macrolides, and fluoroquinolones, there were significant differences in the antimicrobial resistance rates of V. scophthalmi isolates in different agents that belong to the same category. For cephalosporins, isolates were frequently resistant to cefradine and cefalexin with 75.76% and 72.73%, moderately resistant to cefazolin, ceftriaxone, ceftizoxime, ceftazidime, and cefotaxime with 33.33%–57.58%, and seldom resistant to sulbactam (18.18%). For tetracyclines, isolates were frequently resistant to tetracycline (36.36%) and doxycycline (30.30%) and infrequently resistant to minocycline (3.03%). For macrolides, isolates were frequently resistant to acetylspiramycin (90.91%) and moderately resistant to clarithromycin, erythromycin, and azithromycin (36.36%–42.42%). For fluoroquinolones, isolates were frequently resistant to pipemidic (90.91%), moderately resistant to lomefloxacin (42.42%), and infrequently resistant to ciprofloxacin, ofloxacin, fleroxacin, enrofloxacin, norfloxacin, and nalidixic with 3.03%–15.15%. Incidence of resistance to oxacillin (100%), neomycin (93.94%), acetylspiramycin (90.91%), and pipemidic acid (90.91%) was the highest among individual antimicrobials. Incidence of resistance to minocycline (3.03%), ciprofloxacin (3.03%), and norfloxacin (3.03%) was the least observed among the individual antimicrobials. Thirty percent of isolates were resistant to more than 20 different drugs. The most resistant isolate was VS11, which was resistant to 27 agents, and the least resistant isolate was VS19, which was resistant to 5 agents.
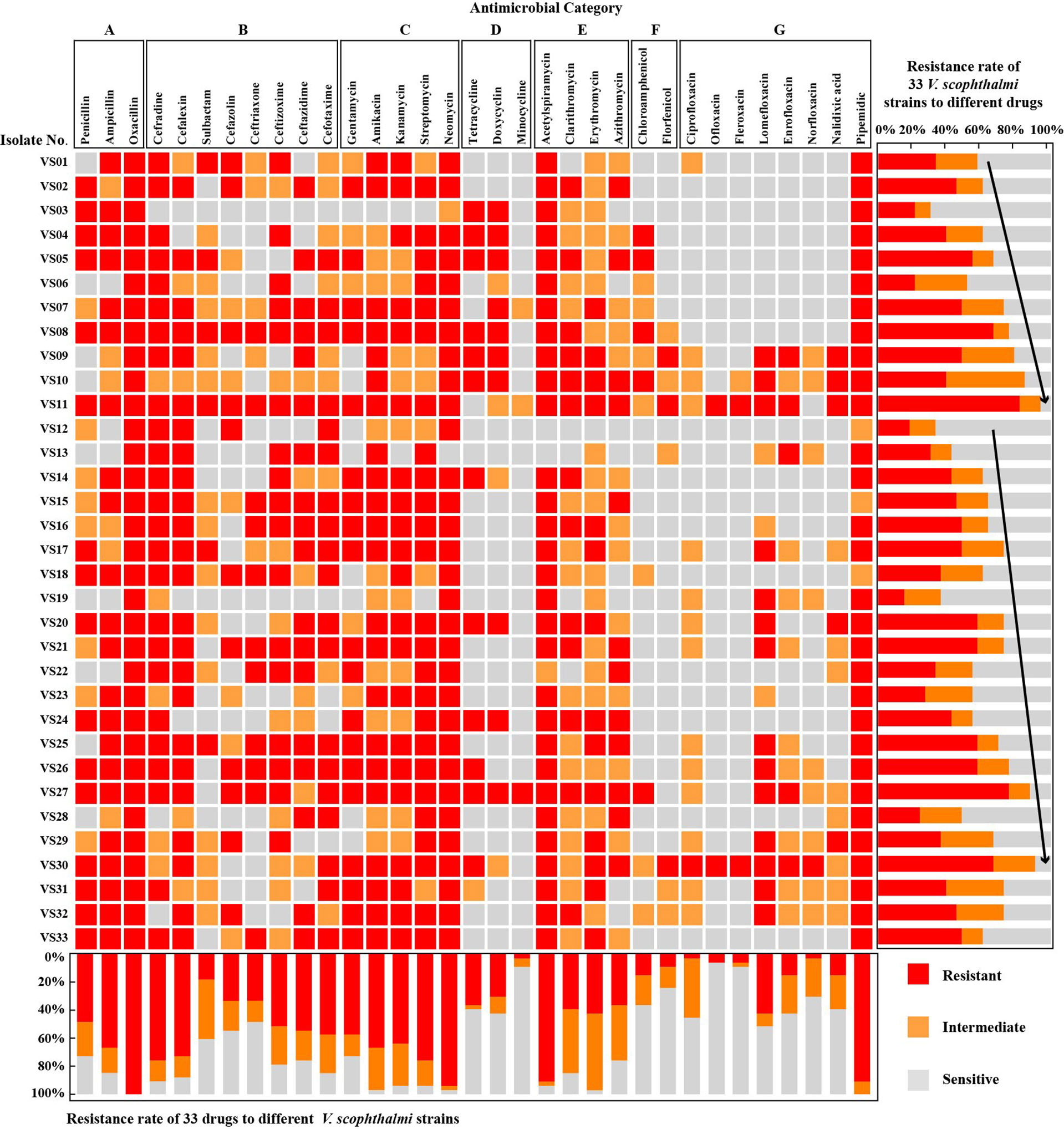
Figure 1 Antimicrobial susceptibility tests of 33 V. scophthalmi isolates to different kinds of antimicrobials.
The proportion of drug-sensitive V. scophthalmi isolated reached the lowest in 2006 with 6.1% and the highest value in 2002 with 69.7%. Combining the resistance rate of V. scophthalmi with the isolation background indicated that there was no significant correlation between the resistance rate of V. scophthalmi and the host. Furthermore, the average insensitive (resistant and intermediate) rate was 49.5%~81.8% between 2002 and 2020, the insensitive rate was lowest in 2002, and the isolates in 2005 and 2019 recorded more than 75% insensitive rates, respectively. But based on the t-test statistical analysis, there was no significant difference in the drug-resistance rate of V. scophthalmi isolates with typical interannual variability.
Isolates that were simultaneously non-susceptible to at least one agent in three or more antimicrobial categories were considered multidrug-resistant (MDR) (Magiorakos et al., 2012). As shown in Figure 2, there were six MDR profiles based on the in vitro susceptibility of 33 V. scophthalmi isolates to seven antimicrobial categories. All isolates were non-susceptible to at least one agent in penicillins (A), aminoglycosides (C), macrolides (E), and fluoroquinolones (G). This suggested that all V. scophthalmi displayed MDR in vitro. Furthermore, the MDR phenotype of V. scophthalmi isolates from 2002 to 2006 also with high growth rate equally. There was no significant correlation between the MDR phenotype of V. scophthalmi and the host.
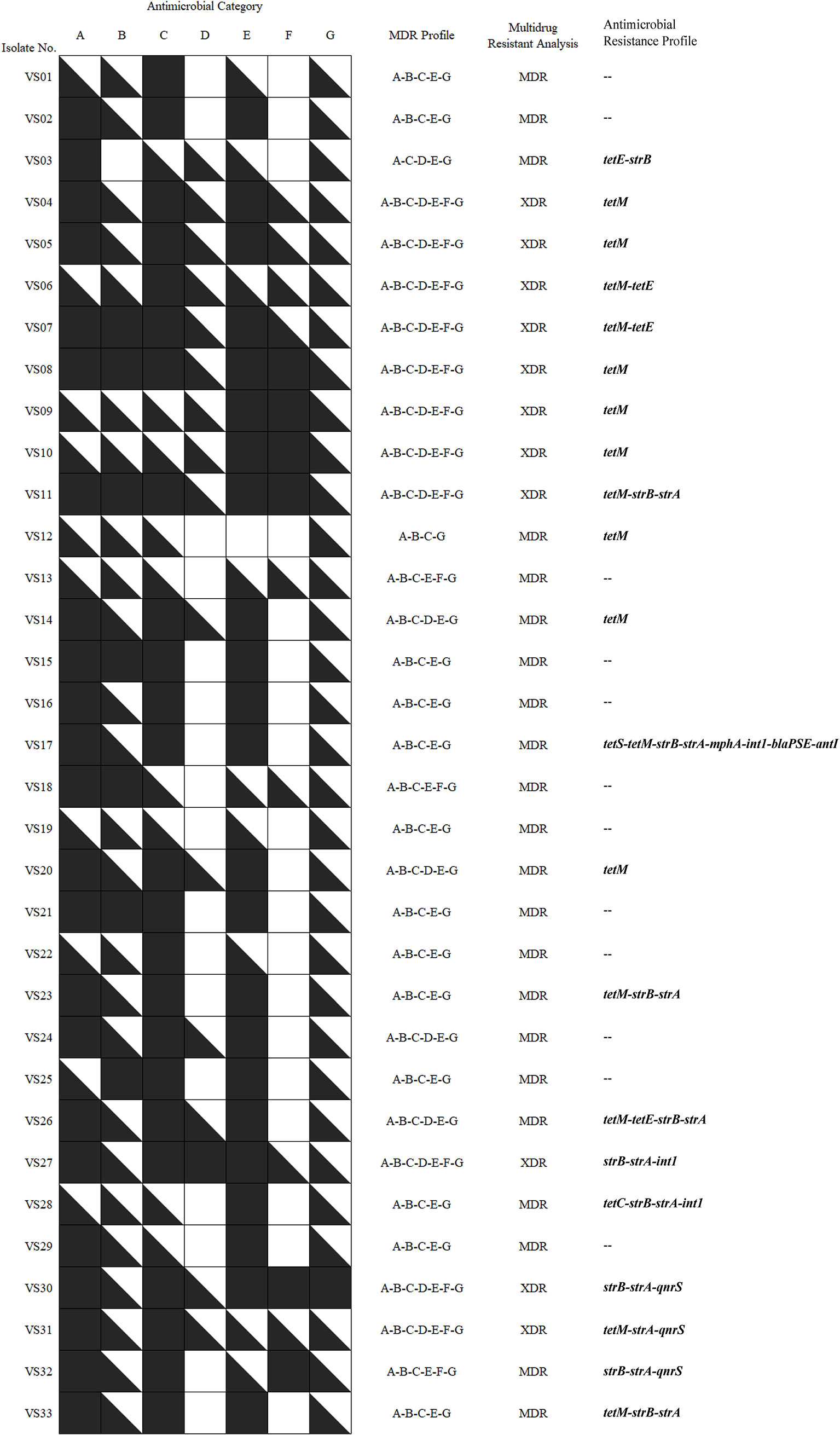
Figure 2 Multidrug-resistant (MDR) profiles of 33 V. scophthalmi isolates to seven different antimicrobial categories.  is the isolate that is non-susceptible to all agents listed in category;
is the isolate that is non-susceptible to all agents listed in category;  is the isolate that is non-susceptible to some but not all agents listed in category; and
is the isolate that is non-susceptible to some but not all agents listed in category; and  is the isolate that is susceptible to all agents listed in the category.
is the isolate that is susceptible to all agents listed in the category.
The distribution of ARGs was evaluated and summarized in Figures 2–4. Eleven of 38 resistance genes were detected in at least one isolates. Twenty-one isolates carried one or more ARGs evaluated. Among them, tetM was the most prevalent gene, with the detection frequencies of 48.5%, followed by strA, strB, tetE, qnrS, int1, ant3˝-I, mphA, blaPSE, tetC, and tetS.
None of the isolates carry β-lactam and macrolides resistance genes except isolate VS17. Of all the tetracycline-resistant isolates, tetracycline- and doxycycline-resistant isolates VS24, VS27, and VS30 were found to be negative for any of the tetracycline resistance genes. Comparatively, tetracycline-resistant genes, tetM, tetS, or tetC, were present in isolates VS12, VS17, VS23, VS28, and VS33, which were susceptible to tetracycline antimicrobial agents. Among pipemidic acid–insensitive isolates (n = 33), only three isolates (VS30, VS31, and VS32) carry quinolone resistance genes qnrS. For aminoglycosides resistant genes, strA, strB, and ant3˝-I were detected in 11 aminoglycoside-resistant isolates, but the other aminoglycoside-resistant isolates (n = 22) did not bear any aminoglycoside-resistant genes. Furthermore, integron factors int1 occurred in isolates VS17, VS27, and VS28. The results showed that V. scophthalmi isolates were resistant to some antimicrobial agents but may present negative to relevant resistance genes.
The DNA fingerprints gained by ERIC-PCR consisted of distinct bands ranging in size from 100 to 10,000 bp. The dendrogram was generated by using the software BioNumerics 7.0. All 33 isolates showed 25 ERIC-PCR patterns. As shown in Figure 4, when the relative similarity coefficient is 62%, 33 isolates could be divided into two clusters (G1 and G3) and two single isolates (G2 and G4). G1 and G3 were the dominant groups, in which G1 consisted of 12 (36.4%) isolates, whereas G3 contained 19 isolates (57.6%). Among them, fingerprints of isolate VS15 (G2) and isolates in G1 were quite similar, and the fingerprint of isolate VS04 presented a low similarity with other isolates.
Based on the epidemiological investigation information, VS15 (G2) was isolated from Cheilinus undulates only once, and 11 of 12 hosts in the G1 cluster and 16 of 19 hosts in the G3 cluster belonged to flatfish; in addition to that, VS21 (G1 cluster) was isolated from Epinephelus septemfasciatus and VS13/VS22/VS23 (G2 cluster) were isolated from Sebastes schlegelii and Tetraodontidae, respectively. The G2 and G4 clusters were isolated before 2009, and isolates in G2 and G4 clusters appeared alternately. There was no significant correlation between the ERIC-PCR genetic types, drug-resistant phenotype, and genotypes of V. scophthalmi isolates.
Vibrio spp. was identified as the common and serious pathogen in marine fish and shellfish worldwide, and the use of antimicrobials has greatly contributed to the development and spread of antimicrobial resistance among Vibrio sp. (Loo et al., 2020). To make the aquaculture industry more sustainable, surveillance of bacteria susceptibility and genetic variation was needed. As a native bacterium in the marine fish intestine, a high prevalence of in vitro resistance of V. scophthalmi to penicillins, cephalosporins, aminoglycosides, and macrolides was observed in this study. The ASEAN countries including Malaysia, Myanmar, and the Philippines permit the use of tetracycline and oxytetracyclines in their aquaculture sector (Weese et al., 2015). In European countries, oxytetracycline is approved for use in aquaculture (Rodgers and Furones, 2009). The in vitro resistance rate of V. scophthalmi to doxycycline was 58.33% during 2002–2006. In contrast, the resistance rate of V. scophthalmi to doxycycline was 14.29% during 2007–2020. Martineau et al. found that the reduction of antimicrobial use could reduce the emergence of resistance (Martineau et al., 1996). The significant reduction in the resistance rate of V. scophthalmi to doxycycline, suggests that with the development of a standard and regulatory system of antimicrobials usage, as well as research and development of new techniques such as vaccination, microecologics, and Chinese herbal medicine, the consumption of doxycycline could be gradually reduced in aquaculture in China.
This study is the first large-scale survey on the ARGs of V. scophthalmi isolates. These findings showed that those V. scophthalmi isolates settled long term within the gut of marine fish carried a variety of ARGs, and the V. scophthalmi isolates might be considered as a potential vehicle for the transfer ARGs in species or seafood. Our findings also demonstrated an obvious mismatch between antimicrobial resistance phenotype and genotype in V. scophthalmi isolates, which were also found in other species, such as V. parahaemolyticus, Lactobacillus pentosusand, Leuconostoc pseudomesenteroide, E. coli, and Listeria sp. (Seung et al., 2012; Maria et al., 2014; Lou et al., 2016). The antimicrobial phenotype is mediated by membrane structure, antimicrobial resistance genes, or physiological metabolism. V. scophthalmi isolates in our study were resistant to some antimicrobial agents but may present negative to relevant resistance genes. The mechanism of drug resistance of V. scophthalmi isolates with diverse antimicrobial phenotypes should be investigated further. Furthermore, the antimicrobial resistance phenotype analysis reveals that there was no significant difference in the drug-resistance rate of V. scophthalmi isolated with typical interannual variability. But different kinds of ARGs were detected in V. scophthalmi isolated in recent years. Therefore, there is a great significance of surveillance of antimicrobial susceptibility of V. scophthalmi, which is highly relevant to food safety and public health.
ERIC-PCR is based on the targeting of repeated DNA sequences with oligonucleotide primers which has been broadly employed to perform the epidemiological typing of pathogenic bacteria such as V. parahaemolyticus (Sahilah et al., 2014), Staphylococcus aureus (Akindolire et al., 2018), and Bacillus cereus (Gao et al., 2018). In this study, the ERIC-PCR results suggested a low genetic diversity among V. scophthalmi isolates. In fact, the 33 isolates could only be divided into two clusters (G1 and G3) and two single isolates when the relative similarity coefficient was 62%. V. scophthalmi isolates shared a higher degree of similarity that usually came from close isolation time. What is surprising is that isolates VS30 and VS32, isolated during an outbreak in 2019, shared the same profiles with stain VS33, an isolate obtained in 2020. The same applies to isolates VS24, VS25, and VS26 that were isolated in 2013, and strain VS23 which was isolated in 2012. These findings showed evidence of epidemiological associations among V. scophthalmi isolates isolated at different times. The same ERIC profile was observed in isolates VS23, VS24, VS25, and VS26. However, the four isolates showed different hosts and antimicrobial resistance patterns. The phenomenon indicated that isolates sharing completely the same ERIC profile presented different antimicrobial resistance patterns or hosts, suggesting that the resistant phenotype of V. scophthalmi isolates may be associated with the antimicrobials distributed in the environment and was not associated with the genotype of isolates.
In conclusion, the antimicrobial susceptibility of V. scophthalmi isolated from diseased fish intestines with typical interannual differences in the costal mariculture area of China was highly prevalent and all of them were resistant to multiple antimicrobial agents. The distribution of ARGs reveals the mismatched phenomenon between the antimicrobial resistance phenotype and genotype of V. scophthalmi isolates. Furthermore, the ERIC-PCR analysis showed a low genetic diversity of V. scophthalmi isolates. Further, there was no significant correlation between the genetic types, drug resistance phenotype, and genotypes of V. scophthalmi isolates. The results will provide data support for further understanding the genetic variation of inherent strains in the fish breeding system and protection product development.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YY, YW, and ZZ contributed to conception and design of the research. YY, XL, and MT performed experiment. YY, BL, and CW performed data processing and statistical analysis. YY and XL drafted the manuscript. YW, XR, and ML contributed to revision of manuscript for important intellectual content. YW gave laboratory and project support. All authors read and approved the final manuscript.
This work was funded by the National Key R&D Program of China (2019YFD0900102), Basic Scientific Research Funds for Central Non-profit Institutes, Yellow Sea Fisheries Research Institutes (20603022021013), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2022GH02) and Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD40).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.924130/full#supplementary-material
Akindolire M. A., Kumar A., Ateba C. N. (2018). Genetic Characterization of Antibiotic-Resistant Staphylococcus Aureus From Milk in the North-West Province, South Africa.Saudi J. Biol. Sci. 25, 1348–1355. doi: 10.1016/j.sjbs.2015.10.011
Byers H. K., Stackebrandt E., Hayward C., Blackall L. L. (1998). Molecular Investigation of a Microbial Mat Associated With the Great Artesian Basin. FEMS Microbiol. Ecol. 25, 391–403. doi: 10.1111/j.1574-6941.1998.tb00491.x
Cerdag-cuellar M., Blanch A. R. (2010). Detection and Identification of Vibrio Scophthalmi in the Intestinal Microbiota of Fish and Evaluation of Host Specificity. J. Appl. Microbiol. 93, 261–268. doi: 10.1046/j.1365-2672.2002.01697.x
Cerdag-cuellar M., Rossello-mora R. A., Lalucat J., Jofre J., Blanch A. (1997). Vibrio Scophthalmi Sp. Nov., a New Species From Turbot (Scophthalmus Maximus). Int. J. System. Bacteriol. 47, 58–61. doi: 10.1099/00207713-47-1-58
Chuanchuen R., Padungtod P. (2009). Antimicrobial Resistance Genes in Salmonella Enterica Isolates From Poultry and Swine in Thailand. J. Vet. Med. Sci. 71, 1349–1355. doi: 10.1292/jvms.001349
Colomer-lluch M., Jofre J., Muniesa M. (2011). Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS One 6, el7549. doi: 10.1371/journal.pone.0017549
Deng Y. T., Wu Y. L., Tan A. P., Huang Y. P., Jiang L., Xue H. J., et al. (2014). Analysis of Antimicrobial Resistance Genes in Aeromonas Spp. Isolated From Cultured Freshwater Animals in China. Microbial. Drug Resist. (Larchmont N.Y.) 20, 350–356. doi: 10.1089/mdr.2013.0068
Gao T. T., Ding Y., Wu Q. P., Wang J., Zhang J., Yu S. B., et al. (2018). Prevalence, Virulence Genes, Antimicrobial Susceptibility, and Genetic Diversity of Bacillus Cereus Isolated From Pasteurized Milk in China. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00533
Healy M., Huong J., Bittner T., Lising M., Frye S., Raza S., et al. (2005). Microbial DNA Typing by Automated Repetitive-Sequence-Based PCR. J. Clin. Microbiol. 43, 199–207. doi: 10.1128/JCM.43.1.199-207.2005
Jun L. J., Jeong J. B., Huh M. D., Chung J. K., Choi D. I., Lee C. H., et al. (2004). Detection of Tetracycline–Resistance Determinants by Multiplex Polymerase Chain Reaction in Edwardsiella Tarda Isolated From Fish Farms in Korea. Aquaculture 240, 89–100. doi: 10.1016/j.aquaculture.2004.07.025
Kim S. R., Nonaka L., Suzuki S. (2004). Occurrence of Tetracycline Resistance Genes Tet(M) and Tet(S) in Bacteria From Marine Aquaculture Sites. FEMS Microbiol. Lett. 237, 147–156. doi: 10.1016/j.femsle.2004.06.026
Liu M., Wong M., Chen S. (2013). Mechanisms of Fluoroquinolone Resistance in Vibrio Parahaemolyticus. Int. J. Antimicrob. Agents 42, 187–188. doi: 10.1016/j.ijantimicag.2013.04.024
Loo K. Y., Letchumanan V., Law J. W., Pusparajah P., Goh B. H., Mutalib N. S. A., et al. (2020). Incidence of Antibiotic Resistance in Vibrio Spp. Rev. Aquacult. 12, 2590–2608. doi: 10.1111/raq.12460
Lou Y., Liu H. Q., Zhang Z. H., Pan Y. J., Zhao Y. (2016). Mismatch Between Antimicrobial Resistance Phenotype and Genotype of Pathogenic Vibrio Parahaemolyticus Isolated From Seafood. Food Control. 59, 207–211. doi: 10.1016/j.foodcont.2015.04.039
Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maria D. C. C. M., Nabil B., Leyre L. L., Antonio G., Hikmate A. (2014). Antibiotic Resistance of Lactobacillus Pentosus and Leuconostoc Pseudomesenteroides Isolated From Naturally-Fermented Alorena Table Olives Throughout Fermentation Process. Int. J. Food Microbiol. 172, 110–118. doi: 10.1016/j.ijfoodmicro.2013.11.025
Martineau F., Picard F. J., Roy P. H., Ouellette M., Bergeron M. G. (1996). Species-Specific and Ubiquitous-DNA-Based Assays for Rapid Identification of Staphylococcus Aureus. J. Clin. Microbiol. 34, 93–2888. doi: 10.1128/JCM.34.12.2888-2893.1996
Moffat J., Chalmers G., Reid-Smith R., Mulvey M. R., Boerlin P. (2020). Resistance to Extended-Spectrum Cephalosporins in Escherichia Coli and Other Enterobacterales From Canadian Turkeys. PLoS One 15, e0236442. doi: 10.1371/journal.pone.0236442
Movahedi M., Zarei O., Hazhirkamal M., Karami P., Shokoohizadeh L., Taheri M. (2021). Molecular Typing of Escherichia Coli Strains Isolated From Urinary Tract Infection by ERIC-PCR. Gene Rep. 23, 101058. doi: 10.1016/j.genrep.2021.101058
Nguyen M., Woerther P. L., Bouvet M., Andremont A., Leclercq R., Canu A. (2009). Escherichia Coli as Reservoir for Macrolide Resistance Genes. Emerging. Infect. Dis. 15, 1648–1650. doi: 10.3201/eid1510.090696
Nouri F., Karami P., Zarei O., Kosari F., Alikhani M. Y., Zandkarimi E., et al. (2020). Prevalence of Common Nosocomial Infections and Evaluation of Antibiotic Resistance Patterns in Patients With Secondary Infections in Hamadan, Iran. Infect. Drug Resist. 13, 2365. doi: 10.2147/idr.s259252
Qiao G., Lee D. C., Woo S. H., Li H., Xu D. H., Park S. I. (2012). Microbiological Characteristics of Vibrio Scophthalmi Isolates From Diseased Olive Flounder Paralichthys Olivaceus. Fish. Sci. 78, 853–863. doi: 10.1007/s12562-012-0502-8
Qiao J., Zhang Q., Alali W. Q., Wang J., Meng L., Xiao Y., et al. (2017). Characterization of Extended-Spectrum β-Lactamases (Esbls)-Producing Salmonella in Retail Raw Chicken Carcasses. Int. J. Food Microbiol. 248, 72. doi: 10.1016/j.ijfoodmicro.2017.02.016
Rodgers C. J., Furones M. (2009). “Antimicrobial Agents in Aquaculture: Practice, Needs and Issues,” in The Use of Veterinary Drugs and Vaccines in Mediterranean Aquaculture, Zaragoza: CIHEAM, 41–59.
Sahilah M. A., Laila A. S. R., Azuhairi A., Osman H., Aminah A., Azuhairi A. A. (2014). Antibiotic Resistance and Molecular Typing Among Cockle (Anadara Granosa) Strains of Vibrio Parahaemolyticus by Polymerase Chain Reaction (PCR)-Based Analysis. World J. Microbiol. Biotechnol. 30, 649–659. doi: 10.1007/s11274-013-1494-y
Sedighi P., Zarei O., Karimi K., Taheri M., Karami P., Shokoohizadeh L. (2020). Molecular Typing of Klebsiella Pneumoniae Clinical Isolates by Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction. Int. J. Microbiol. 2020, 1–5. doi: 10.1155/2020/8894727
Seung H. R., Seog G. P., Sung M. C., Young O. H., Hee J. H., Su U. K., et al. (2012). Antimicrobial Resistance and Resistance Genes in Escherichia Coli Strains Isolated From Commercial Fish and Seafood. Int. J. Food Microbiol. 152, 14–18. doi: 10.1016/j.ijfoodmicro.2011.10.003
Sitjà-bobadilla A., Pujalte M. J., Bermejo A., Garay E., Alvarez-Pellitero P., Perez-Sanchez J. (2010). Bacteria Associated With Winter Mortalities in Laboratory-Reared Common Dentex (Dentex Dentex L.). Aquacult. Res. 38, 733–739. doi: 10.1111/j.1365-2109.2007.01719.x
Speldooren V., Heym B., Labia R., Nicolaschanoine M. H. (1998). Discriminatory Detection of Inhibitor-Resistant β-Lactamases in Escherichia Coli by Single-Strand Conformation Polymorphism-Pcr. Antimicrob. Agents Chemother. 42, 879–884. doi: 10.1097/0000113-199804000-00011
Valdenegro-vega V., Naeem S., Carson J., Bowman J. P., Tejedor del Real J. L., Nowak B. (2013). Culturable Microbiota of Ranched Southern Bluefin Tuna (Thunnus Maccoyii Castelnau). J. Appl. Microbiol. 115, 923–932. doi: 10.1111/jam.12286
Versalovic J., Koeuth T., Lupski R. (1991). Distribution of Repetitive DNA Sequences in Eubacteria and Application to Fingerprinting of Bacterial Genomes. Nucleic Acids Res. 19, 6823–6831. doi: 10.1093/nar/19.24.6823
Weese J. S., Giguère S., Guardabassi L., Morley P. S., Papich M., Ricciuto D. R., et al. (2015). Acvim Consensus Statement on Therapeutic Antimicrobial Use in Animals and Antimicrobial Resistance. J. Vet. Internal Med. 29, 487–498. doi: 10.1111/jvim.12562
Yamamoto S., Harayama S. (1995). PCR Amplification and Direct Sequencing of gyrB Genes With Universal Primers and Their Application to the Detection and Taxonomic Analysis of Pseudomonas Putida Strains. Appl. Environ. Microbiol. 61, 9–1104. doi: 10.1128/AEM.61.10.3768-3768.1995
Yuan Y. Z., Zhang Y. G., Qi G. S., Ren H., Gao G. S., Jin X. M., et al. (2021). Isolation, Identification, and Resistance Gene Detection of Vibrio Harveyi From Scophthalmus Maximus. Aquacult. Int. 29, 2357–2368. doi: 10.1007/s10499-021-00752-z
Keywords: Vibrio scophthalmi, antimicrobial susceptibility, antimicrobial resistance genotype, genetic diversity, ERIC-PCR
Citation: Yu Y, Liu X, Wang Y, Liao M, Tang M, Rong X, Wang C, Li B and Zhang Z (2022) Antimicrobial Resistance and Genotype Characteristics of Vibrio scophthalmi Isolated from Diseased Mariculture Fish Intestines With Typical Inter-Annual Variability. Front. Mar. Sci. 9:924130. doi: 10.3389/fmars.2022.924130
Received: 20 April 2022; Accepted: 20 June 2022;
Published: 20 July 2022.
Edited by:
Qingchao Wang, Huazhong Agricultural University, ChinaCopyright © 2022 Yu, Liu, Wang, Liao, Tang, Rong, Wang, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingeng Wang, d2FuZ3lnQHlzZnJpLmFjLmNu; Zheng Zhang, emhhbmd6aGVuZ0B5c2ZyaS5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.