
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 23 June 2022
Sec. Marine Pollution
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.920293
This article is part of the Research Topic Alternatives to petroleum-based plastics as a potential solution to the global plastic pollution crisis in marine environments: Do they provide sustainable solutions? View all 6 articles
The progressive substitution of petroleum-based polymers, such as polyethylene, polyvinylchloride, or polyethylene terephtalate, by so-called bioplastics facilitated the development and production of many new materials. The continuously refined properties of bioplastic compounds and their blends enable various applications. With growing production and utilization of bioplastic products, these materials are increasingly discarded into the environment. Although many of these materials are labeled biodegradable, there is limited information about their degradability under environmental conditions. We tested the enzymatic degradability of five bioplastic compounds with the rapid pH-Stat titration assay at environmentally relevant seawater temperatures between 5 and 30°C and pH 8.2. These plastics, issued from the European Horizon 2020 Project ´Bioplastics Europe´, are based on polylactic acid (PLA), polybutylene succinate (PBS), and poly(hydroxybutyrate-co-hydroxyvalerate) (PHBV). Suspensions of microparticles (< 200 µm) were incubated with each of the three hydrolytic enzymes, protease, lipase, and esterase. A PLA-based compound blended with polybutylene adipate terephthalate (PBAT) showed the highest hydrolysis rate of 30 nmol·min-1 when incubated with lipase at 30°C. All other materials showed low hydrolysis rates of less than 10 nmol·min-1. Below 20°C, hydrolysis almost ceased. Plate clearing assays with the same enzymes at 37°C and pH 5 and pH 8, respectively, largely confirmed the results of the pH-Stat titration assays. Our findings indicate that there is a potential degradation of most of the materials with at least one of these hydrolytic enzymes. Nonetheless, the rate of enzymatic degradation under environmentally relevant conditions is low, which indicates only a marginal degradability of bioplastics in the marine environment.
Petroleum-derived plastic materials (petro-plastics) shape the daily life of humans and have become indispensable in modern industrialized societies. For decades, the use of plastics is continuously increasing, with an annual global production of about 360 million tons by 2018 (PlasticsEurope, 2019). Excessive use and poorly controlled disposal caused a massive discard of persistent petro-plastics into the environment and a pollution of terrestrial and aquatic ecosystems worldwide (Borrelle et al., 2020). Annually, an estimated amount of about 4-12 million tons of plastic debris ends up in the oceans (Jambeck et al., 2015), where it may remain for centuries before it degrades (Worm et al., 2017). Marine plastic debris covers a wide size range from large abandoned fishing nets to the smallest micro- and nanoparticles (Lebreton et al., 2018). Plastic debris may interfere with marine biota, often causing deleterious effects on organisms and ecosystems (do Sul and Costa, 2014; Gall and Thompson, 2015; Kühn et al., 2015).
To reduce the consumption of fossil resources and to promote the degradability of plastics, extensive research effort is being made to replace conventional petroleum-based polymers with so-called bioplastics. The term bioplastics refers to synthetic polymers derived from renewable biomass sources (Vert et al., 2012), but is also frequently used for biodegradable polymers and materials that combine both of these properties, therefore covering a wide spectrum of different polymers (e.g., Dilshad et al., 2021; Muneer et al., 2021). The majority of bioplastics are aliphatic polyesters composed of monomers linked by ester bonds. Common polymers used as the basis for bioplastics are polylactic acid (PLA), polybutylene succinate (PBS), and poly(hydroxybutyrate-co-hydroxyvalerate) (PHBV) (Garlotta, 2001; Luo and Netravali, 2003; Xu and Guo, 2010; Ferreira et al., 2019; RameshKumar et al., 2020).
Depending on their application, biodegradable plastics have to combine a wide range of properties with the requirements of biodegradability standards. The chemical and mechanical characteristics of such materials can be substantially improved or adjusted by blending with additives and fillers (Angelini et al., 2015; Gug and Sobkowicz, 2016; Shafqat et al., 2021). Currently, bioplastics represent only one percent of the global annual plastic production. However, the demand for bio-based and biodegradable plastics is rapidly growing. Due to the continuous improvement of the compounds and the diversification of applications, the annual bioplastic production will prospectively grow from 2.1 million tons in 2020 to 2.9 million tons in 2025 (European Bioplastics e.V., 2021).
Synthetic biopolymers are degraded by hydrolytic cleavage of their ester bonds, catalyzed by microbial enzymes (Singh and Sharma, 2008). However, only 60% of the produced bioplastics are currently considered biodegradable (European Bioplastics e.V, 2021). Moreover, many of these biodegradable materials are sufficiently degraded only under specific conditions, such as high temperatures and in industrial composting facilities (e.g., Bátori et al., 2018; Ruggero et al., 2019; Folino et al., 2020). In the environment, the degradation rate of most biodegradable plastics seems to be low (Wang et al., 2021). Accordingly, improved biodegradability of customized bioplastics in the environment demands a sound understanding of their enzymatic degradation under realistic conditions.
A variety of hydrolytic enzymes from microbes or other organisms are capable of degrading bioplastics (Meereboer et al., 2020; Ali et al., 2021; Polman et al., 2021). Such enzymes, comprising various peptidases and esterases, are present in the environment, including seawater and marine sediments (Arnosti et al., 2014; Liu and Liu, 2018; Patel et al., 2019). Hydrolases, namely proteinase K, protease, esterase, and lipase, have been used in in vitro assays on the enzymatic degradability of bioplastics (Miksch et al., 2021; Richert and Dąbrowska, 2021). The pH-Stat titration is a rapid assay, which is based on maintaining the pH during a pH-affecting reaction. The hydrolysis of polyesters forms carboxyl end groups, which reduce the pH in the surrounding medium. Accordingly, the pH-Stat detects the drop in pH and adds an alkaline solution. Based on the amount of the added solution over a certain period of time, the degradation rate can be calculated. According to previous studies, we chose for the present investigation a proteolytic enzyme (protease), capable of hydrolyzing peptide and ester bonds, as well as an esterase and a lipase, which hydrolyze short-chain and long-chain esters, respectively. Although the selected enzymes do not derive from a marine microorganism, they represent important enzymes in the class of hydrolases, which are prevalent in almost all organisms and possess a rather broad substrate specificity. Moreover, the chosen enzymes are readily available commercially, and their specificity is equivalent to enzymes synthesized by marine fungi and bacteria.
In the frame of the EU Horizon 2020 project Bioplastics Europe (BPE) (Bioplastics Europe, 2022), we investigated the enzymatic degradability of selected customized bioplastics in seawater by rapid in vitro methods. Five bioplastic compounds, which were designed for different applications, were provided by project partners. These materials were tested for degradation at environmentally relevant seawater temperatures and their hydrolysis rates were compared with the conventional petroleum-based polymer poly(methyl methacrylate) (PMMA) and a biogenic structural protein (collagen).
NaOH standard solution (cat. no. 5564732) was purchased from Omnilab (Bremen, Germany). Agarose (cat. no. 16500) was purchased from Thermo Fisher Scientific (Waltham, Massachusetts, USA). All other chemicals and enzymes (Table 1) were purchased from Sigma-Aldrich (Taufkirchen, Germany).
Bioplastic compounds, in the following referred to as BPE-materials, were provided by Arctic Biomaterials OY (ABM, Tampere, Finland) and NaturePlast SAS (Ifs, France). The materials were supplied as granules (3 x 3 mm, 25 mg). The designations of the materials are composed of the abbreviations of the project, the intended use of the application, and the predominant base polymer (Table 2A). Petroleum-based PMMA was manufactured by Kunststoff- und Farben-Gesellschaft mbH (KFG, Biebesheim, Germany) and supplied as powder with particles < 200 µm (Table 2B). The structural protein collagen was purchased from Sigma (Table 2B). The material was cryo-milled as described under 2.3. and the size fraction < 200 µm was analyzed.
Granules of the bioplastic compounds were ground in a cryogenic mill (SPEX SamplePrep, 6775 Freezer/Mill). The grinding program included 15 min of pre-cooling before the initiation of the grinding process. Grinding was performed in four to eight cycles, with 1 to 2 min of agitation and 1 to 2 min of cooling within each cycle. Grinding was performed with 15 impacts per second (cps). Pre-cooling was essential to achieve sufficient brittleness of the material to avoid agglutination of melting particles. One gram of material was processed per grinding operation. The ground material was sieved for fractions smaller than 200 µm. The yield of the < 200 µm fraction varied depending on the polymer type from about 20% to 60% of the total ground material.
The composition of the bioplastic compounds was verified by NMR spectroscopy. Prior to the NMR analysis, the materials were solubilized in deuterated chloroform (CDCl3). The NMR spectra were recorded on a Bruker Neo spectrometer (equipped with a 1.7 mm cryo probe) at 300 K operating at 600 MHz (1H) and 125 MHz (13C). For each sample, a one-dimensional (1D) 1H-NMR spectrum and a two-dimensional (2D) 1H,13C-HSQC spectrum were acquired. The 1D 1H-NMR spectra were acquired with a 30° pulse, 128 scans, and 65536 data points (acquisition time of 3.6 s, relaxation delay of 1.0 s). The 2D 1H,13C-HSQC spectra were run with 16 scans and 2048 data points in F2 and 256 data points in F1 (acquisition time of 113 ms, relaxation delay of 1.5 s).
The rate of enzymatic hydrolysis of the bioplastics was measured by pH-Stat titration using a TitroLine® 7000 titrator (SI Analytics GmbH, Mainz, Germany). The titration unit was equipped with a 20-mL exchangeable head and a 1 mm diameter PTFE tube as a titration tip. The unit was connected to a magnetic stirrer (TM 235), a pH-electrode model A 162 2M DIN ID, and a circulation thermostat (Lauda, Lauda-Königshofen, Germany). The reaction vial was a 20-mL glass vial placed in a custom-made thermostat jacket to maintain a constant temperature (Miksch et al., 2021).
pH-Stat titration was performed after Miksch et al. (2021) whilst the rate of acidification due to the hydrolysis of the bioplastic was determined by counter-titration of a base. Briefly, suspensions of bioplastic microparticles (3 mg·ml-1) were prepared in a solution of 3.2% sea salt (Seequasal, Münster, Germany) in deionized water (referred to as artificial seawater, ASW). The suspensions were first stirred in a glass beaker with a magnetic stirring bar at 800 rpm for 16 h before aliquots of 10 mL were subjected to pH-Stat titration. Ten to 100 µL of enzyme solution of commercial lipase, protease, and esterase (Table 2) were added to the reaction vial with a 100-µL micro-syringe (Model 710 N, Hamilton Bonaduz AG, Bonaduz, Switzerland). The pH was kept constant at 8.2 by titration of 10 mmol·L-1 NaOH solution. The addition of NaOH solution after enzyme addition was recorded every minute for 60 min. Each measurement involved an initial recording of the addition of NaOH solution for 60 min without enzyme, to correct for the effects of atmospheric CO2 and polymer autolysis. For each replicate at all temperatures an additional enzyme blank was measured, where the hydrolysis rate of the enzyme in seawater without substrate was determined, to correct for the autocatalytic activity of the enzyme. The hydrolysis of the five bioplastics and the petroleum-based polymer PMMA was assayed at 5, 10, 15, 20, 25, and 30°C. The electrode was calibrated each day before use. Routine measurements were carried out in triplicate.
To compare the enzymatic degradation of the bioplastics with that of a natural polymer, hydrolysis of non-soluble collagen by each of the three enzymes was measured at 20°C using the same methodology.
Plate clearing assays were performed as an alternative measure of enzymatic polymer degradation. Polymer emulsions and polymer agar plates were prepared after Uchida et al. (2000); Urbanek et al. (2020) with slight modifications. Briefly, 0.4 g of plastic particles were dissolved in 10 mL of dichloromethane by stirring at 28°C. After dissolution, 400 µl of Triton-X and 20 mL of distilled water were added. The mixture was sonicated for 2 to 4 min with a Sonopuls ultrasonic homogenizer GM 2070.2 (Bandelin, Berlin, Germany). Next, the dichloromethane was evaporated by stirring under vacuum for 1 h. The remaining emulsion was diluted with 60 mL of distilled water and 800 mg of agarose (Thermo Fisher, cat. no. 16500) was added. After heating the mixture in a microwave oven for 60 s at 600 W, the pH was adjusted to 5 and 8, respectively, by adding 0.1 mol·L-1 NaOH. About 20 ml of this solution was poured into a Petri dish and allowed to cool and polymerize. The enzymes lipase, esterase, and protease (10 µL each) were applied onto the surface of the gel with the help of small ceramic wells (diameter: 10 mm, height: 8 mm) and incubated for 24 h at 37°C. The formation of clear zones (halos) around the wells was interpreted as enzymatic degradation of the polymer. The diameter of the clear zones was used as a measure for the amount of polymer degraded by the enzyme. This plate clearing assay could not be performed on BPE-AMF-PLA, because it was not possible to create an opaque emulsion of this plastic.
Statistical analysis and graphs were done with the program GraphPad Prism version 7.05 for Windows, GraphPad Software, La Jolla, CA, USA, https://www.graphpad.com. The temperature dependency of the hydrolysis rates of the tested polymers by the enzymes was described by non-linear regression models. The temperature dependency of the hydrolysis was described either by a Boltzmann sigmoidal model (Eq. 1) or by an exponential model (Eq. 2):
The 1H-NMR spectra of BPE-C-PLA and BPE-RP-PLA clearly indicate a mixture of PLA (1H signals: CH and CH3) and PBS (1H signals: 3 x CH2) with ratios of about 2.4:1 and 2.6:1, respectively (Figures 1A, B). BPE-AMF-PLA is a mixture of PLA and PBAT (seven 1H signals) in a ratio of about 30:1. (Figure 1C). BPE-SP-PBS shows the indicative 1H signals (three methylene groups) for PBS (Figure 1D). BPE-T-PHBV shows the characteristic signals for poly-(hydroxybutyrate-co-hydroxyvalerate)-copolymer (PHBV) with the HB (three 1H signals) and HV (four 1H signals) subunits and traces of PLA (Figure 1E). Detailed 1H- and 13C-NMR results are listed in the following.
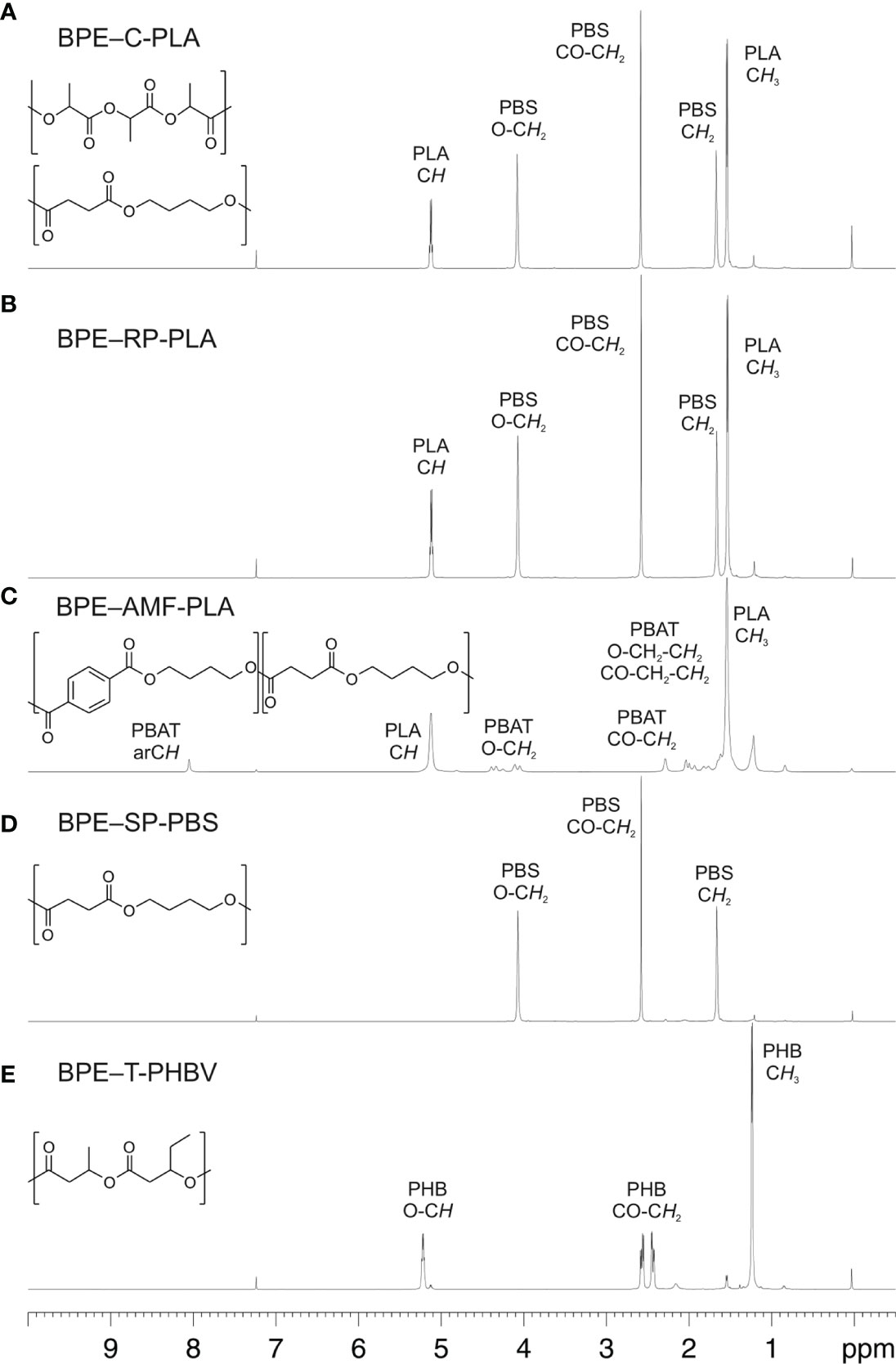
Figure 1 1H-NMR spectra of the investigated BPE materials (structures shown on the left): (A) BPE-C-PLA, (B) BPE-RP-PLA, (C) BPE-AMF-PLA, (D) BPE-SP-PBS, and (E) BPE-T-PHBV. For all spectra the region between 0.5 and 10 ppm is shown. The BPE peaks are assigned.
BPE-C-PLA (Figure 1A): NMR data (600 MHz, CDCl3): 1H, δ5.13 (q, 1H, PLA-CH), 4.08 (br, 4H, PBS-O-CH2), 2.59 (br, 4H, PBS-CO-CH2), 1.67 (br, 4H, PBS-CH2), 1.54 (d, 3H, PLA-CH3); 13C, δ69.1 (PLA-CH), 64.3 (PBS-O-CH2), 29.1 (PBS-CO-CH2), 25.3 (PBS-CH2), 16.6 (PLA-CH3), n.d. (CO).
BPE-RP-PLA (Figure 1B): NMR data (600 MHz, CDCl3): 1H, δ5.12 (q, 1H, PLA-CH), 4.07 (br, 4H, PBS-O-CH2), 2.58 (br, 4H, PBS-CO-CH2), 1.66 (br, 4H, PBS-CH2), 1.54 (d, 3H, PLA-CH3); 13C, δ68.9 (PLA-CH), 64.1 (PBS-O-CH2), 29.0 (PBS-CO-CH2), 25.2 (PBS-CH2), 16.6 (PLA-CH3), n.d. (CO).
BPE-AMF-PLA (Figure 1C): NMR data (600 MHz, CDCl3): 1H, δ8.05 (s, 4H, PBAT-arCH), 5.13 (q, 1H, PLA-CH), 4.38 (br, 4H, PBAT-O-CH2), 4.08 (br, 4H, PBAT-O-CH2), 2.29 (br, 4H, PBAT-CO-CH2), 1.93 (br, 4H, PBAT-O-CH2-CH2), 1.79 (br, 4H, PBAT-CO-CH2-CH2), 1.63 (br, 4H, PBAT-CO-CH2-CH2), 1.54 (d, 3H, PLA-CH3); 13C, δ68.9 (PLA-CH), 64.8 (PBAT-O-CH2), 63.8 (PBAT-O-CH2), 33.8 (PBAT-CO-CH2), 25.4 (PBAT-O-CH2-CH2), 25.3 (PBAT-O-CH2-CH2), 24.3 (PBAT-CO-CH2-CH2), 16.6 (PLA-CH3), n.d. (CO).
BPE-SP-PBS (Figure 1D): NMR data (600 MHz, CDCl3): 1H, δ4.07 (br, 4H, O-CH2), 2.58 (br, 4H, CO-CH2), 1.66 (br, 4H, CH2); 13C, δ64.1 (O-CH2), 29.0 (CO-CH2), 25.2 (CH2), n.d. (CO).
BPE-T-PHBV (Figure 1E): NMR data (600 MHz, CDCl3): 1H, δ5.22 (h, 2 x 1H, HB-CH+HV-CH), 2.58/2.43 (2 x d, 2 x 2H, HB-CH2), 2.56/2.45 (2 x d, 2 x 2H, HV-CH2), 1.24 (d, 3H, HB-CH3), 1.22 (m, 2H, HV-EtCH2), 0.85 (q, 3H, HV-EtCH3); 13C, δ67.7 (HB-CH+HV-CH), 40.9 (HB-CH2), 40.6 (HV-CH2), 29.6 (HV-EtCH2), 19.7 (HB-CH3) n.d. (CO).
The hydrolysis rates of BPE materials varied with the applied enzymes and temperature.
Hydrolysis of BPE-C-PLA incubated with protease increased exponentially with a temperature to 9.0 ± 2.2 nmol·min-1 at 30°C (Figure 2A). Hydrolysis by lipase and esterase was only detectable at 30°C at rates of 2.6 ± 0.4 nmol·min-1 and 2.4 ± 1.2 nmol·min-1, respectively.
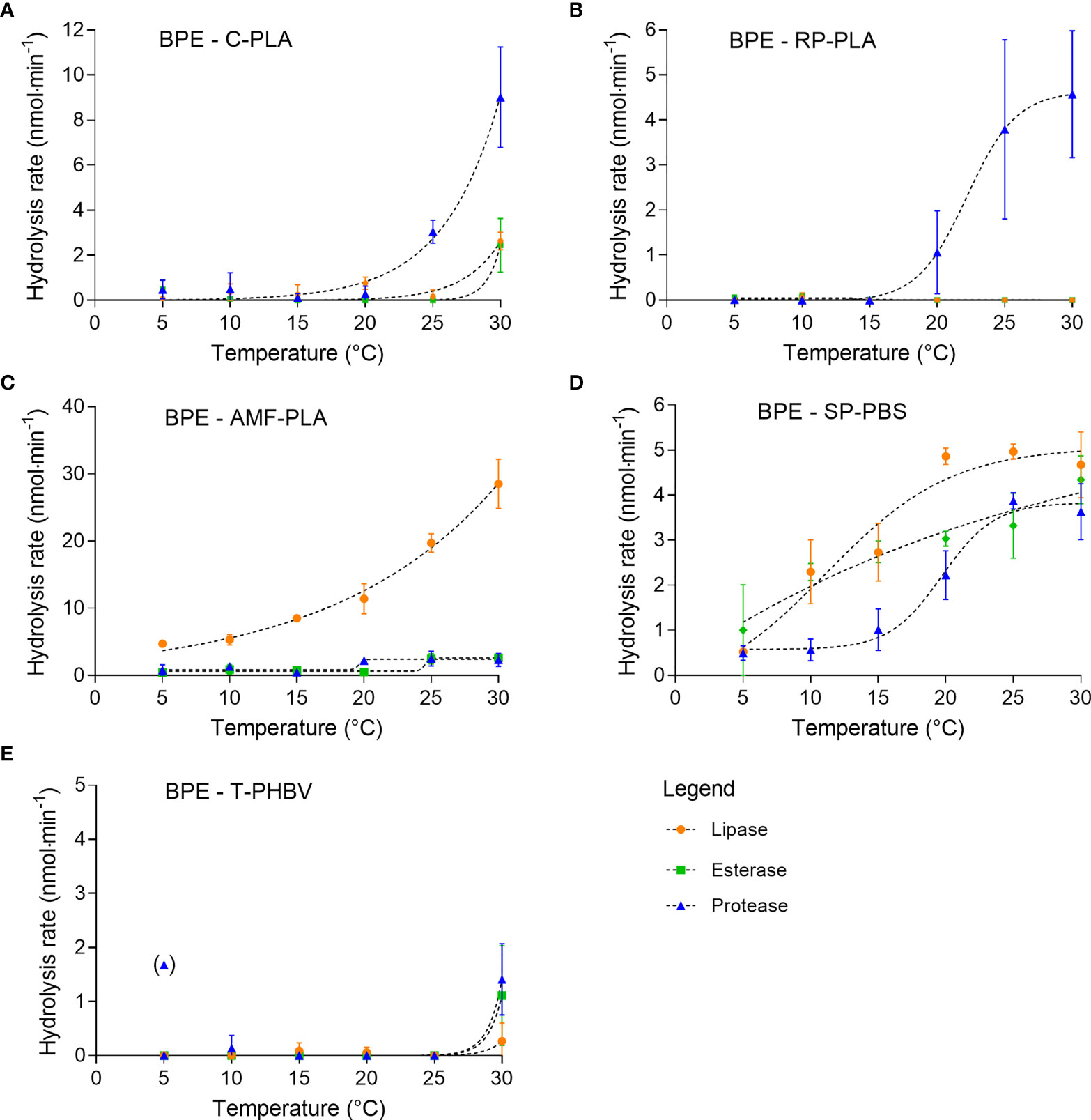
Figure 2 Hydrolytic degradation of the five BPE materials by lipase, esterase, and protease (A–E) measured by pH-stat degradation at temperatures between 5°C and 30°C (means ± SD, n = 3).
BPE-RP-PLA displayed a sigmoidal increase of hydrolysis rate with temperature when incubated with protease (Figure 2B). The highest rate was 4.6 ± 1.4 nmol·min-1 at 30°C. No hydrolysis (< 0.1 nmol·min-1) of this polymer was observed with lipase and esterase at either temperature.
BPE-AMF-PLA showed the highest hydrolysis rate when incubated with lipase (Figure 2C). The hydrolysis rate increased exponentially with temperature from 4.7 ± 0.5 nmol·min-1 at 5°C to 28.5 ± 3.7 nmol·min-1 at 30°C. The same polymer was only poorly hydrolyzed by protease and esterase. Almost no hydrolytic activity was observed with these enzymes below 20°C. Above 20°C, the hydrolysis rate was still low with maximum activities of 2.5 ± 1.1 nmol·min-1 for protease and 2.6 ± 0.5 nmol·min-1 for esterase.
BPE-SP-PBS was hydrolyzed by all three enzymes (Figure 2D). The hydrolytic activity was low and showed a sigmoidal increase with temperature. Accordingly, the highest activities were measured at 30°C at rates of 3.9 ± 0.2 nmol·min-1 for protease, 5.0 ± 0.2 nmol·min-1 for lipase, and 4.3 ± 0.5 nmol·min-1 for esterase.
BPE-T-PHBV showed very low or no hydrolysis when incubated with the three enzymes (Figure 2E). The hydrolytic activity slightly increased at 30°C to 1.4 ± 0.7 nmol·min-1 for protease, 0.3 ± 0.3 nmol·min-1 for lipase, and 1.1 ± 0.9 nmol·min-1 for esterase.
The petroleum-based PMMA was hydrolyzed by lipase at a very low rate, which varied only slightly with temperature (Figure 3). The highest hydrolysis rate of 0.7 ± 0.6 nmol·min-1 was measured at 20°C. Hydrolysis by protease and esterase occurred only at 20°C at very low rates of 0.2 ± 0.4 nmol·min-1 and < 0.01 nmol·min-1, respectively.
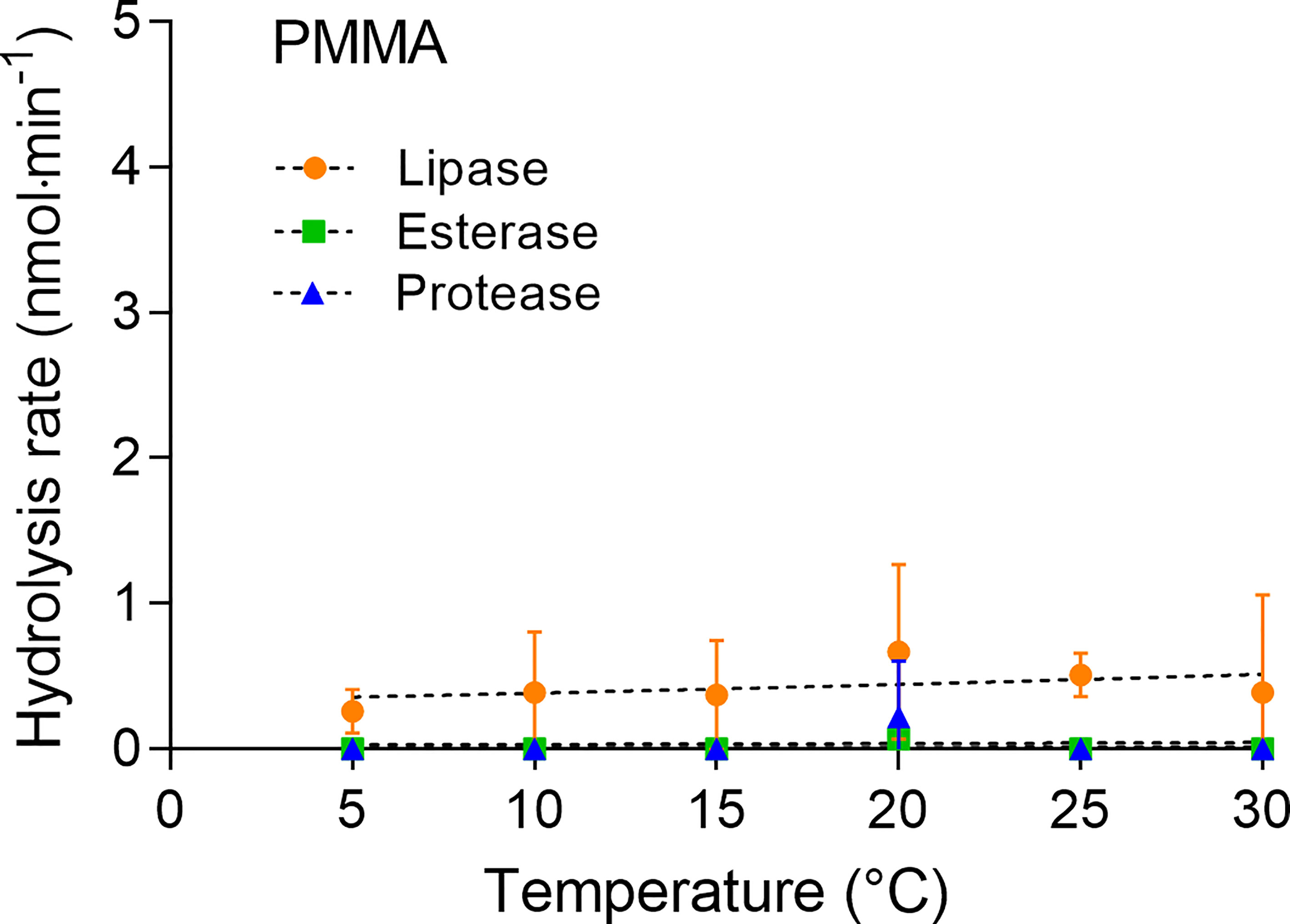
Figure 3 Hydrolytic degradation of PMMA measured by pH-stat titration at temperatures between 5°C and 30°C (means ± SD, n = 3).
Hydrolysis of non-soluble collagen was highest when incubated with protease (Figure 4). At 20°C the hydrolysis rate was 67.7 ± 5.7 nmol·min-1, which was about six times higher than the highest hydrolysis rate of BPE-AMF-PLA at the same temperature (i.e., 11.4 ± 2.2 nmol·min-1, Figure 2C). Hydrolysis of collagen by lipase was very low at 0.7 ± 0.1 nmol·min-1. Almost no hydrolytic activity (< 0.1 nmol·min-1) was measured with esterase (Figure 4).
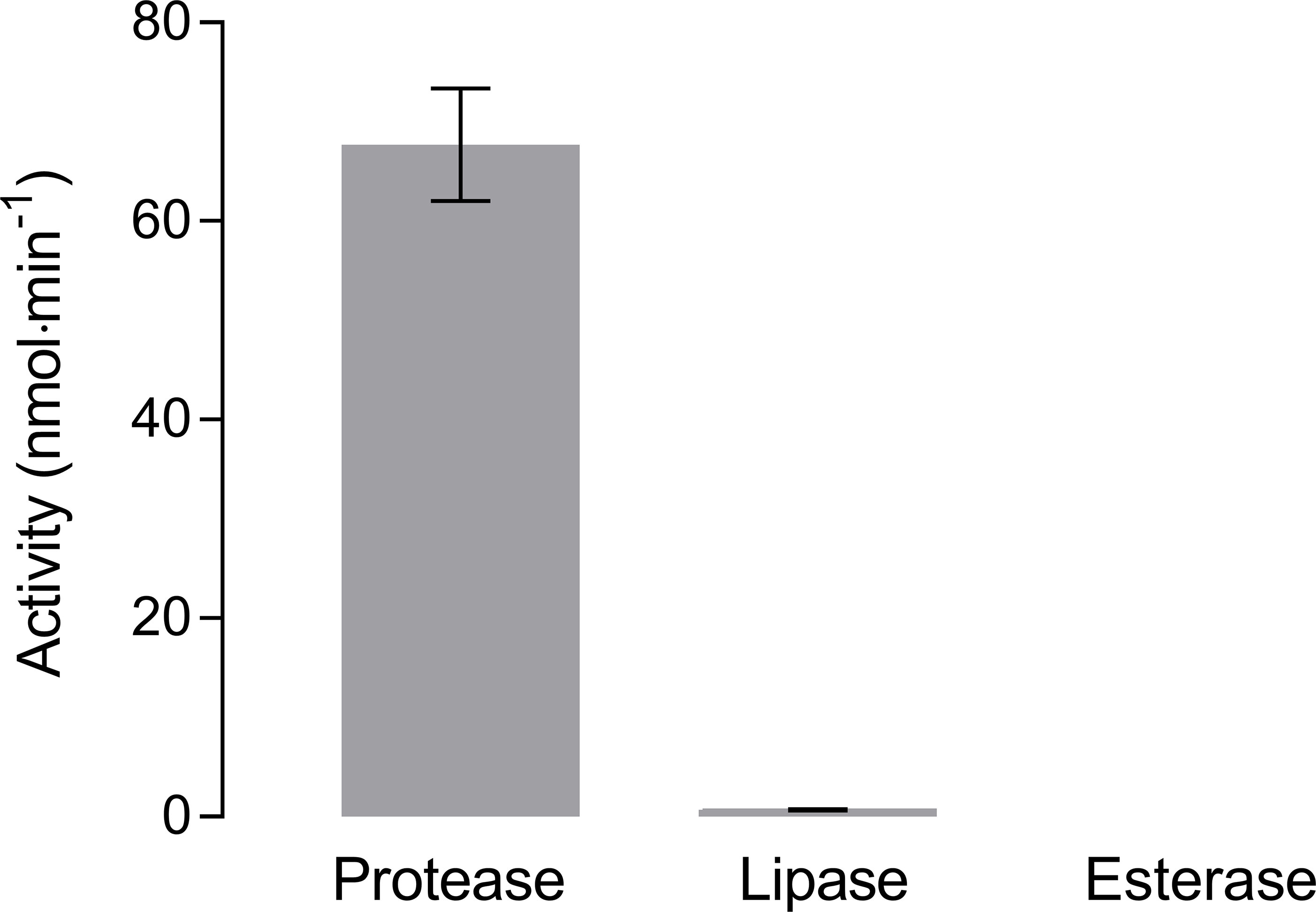
Figure 4 Hydrolysis of non-soluble collagen by protease, lipase, and esterase measured by pH-stat titration at 20°C (means ± SD, n = 3).
Meaningful plate clearing assays were performed with BPE-C-PLA, BPE-RP-PLA, and BPE-SP-PBS. No opaque suspension could be prepared with BPE-AMF-PLA, thus obstructing the plate clearing assays. The control (H2O) showed no clear zones (Table 3 and Figure 5).
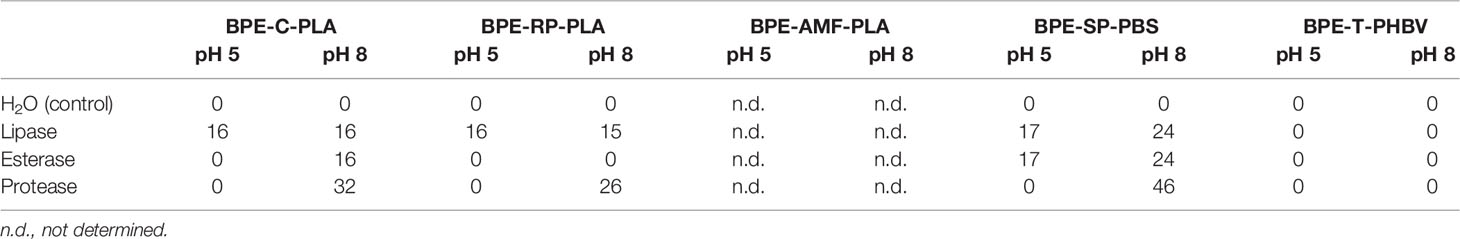
Table 3 Diameter (mm) of clear zones formed around the area of enzyme application for lipase, esterase, and protease on plates of the BPE-polymers at pH 5 and pH 8.
BPE-C-PLA: all three enzymes, protease, lipase, and esterase developed clear zones on plates of BPE-C-PLA at pH 8 (Table 3). The diameter of the clear zone was the largest for protease. At pH 5, a clear zone developed only for esterase.
BPE-RP-PLA: protease and lipase formed clear zones at pH 8 with the largest zone with protease, suggesting the highest hydrolytic activity for this enzyme (Figure 5 and Table 3). At pH 5, a clear zone was observed only for lipase with the same diameter as at pH 8.
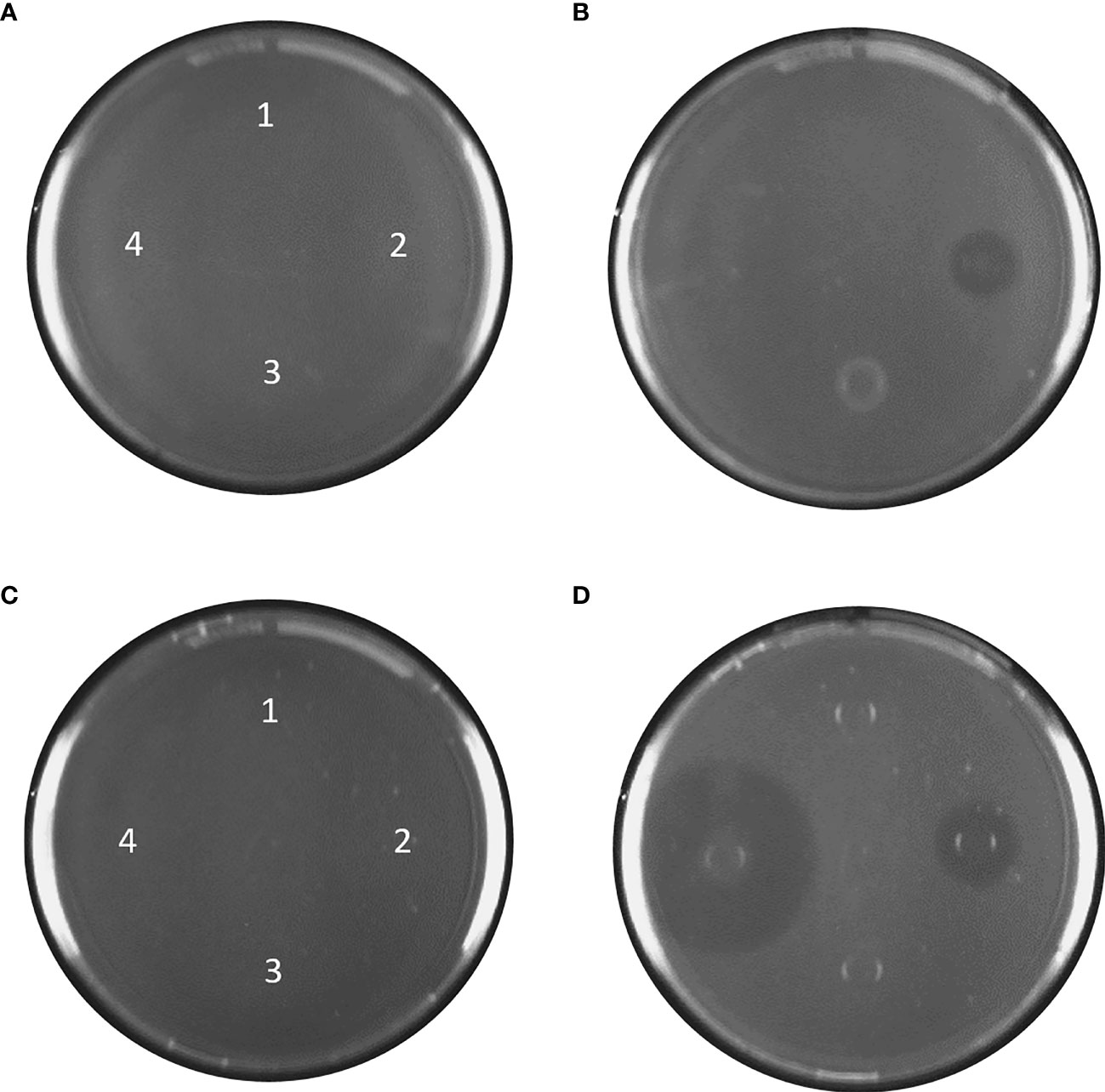
Figure 5 Agar plates with BPE-RP-PLA at pH 5, (A) before incubation and (B) after 24 h at 37°C. BPE-RP-PLA plates at pH 8, (C) before incubation and (D) after 24 h at 37°C. Blank and samples applied at the indicated spots: 1) Deionized water, 2) lipase, 3) esterase, and 4) protease. After 24 h at 37°C, clear zones are visible for lipase at pH 5 (B) and for lipase and protease at pH 8 (D). The contrast of the images was increased by 25% to emphasize the clear zones.
BPE-SP-PBS: large clear zones were formed by all three enzymes at pH 8. At pH 5, clear zones were visible only for lipase and esterase but not for protease (Figure 5 and Table 3). The diameters of the clear zones were substantially smaller at pH 5 than at pH 8.
BPE-T-PHBV: no indication of enzymatic activity was detected at pH 8 and pH 5.
The five bioplastics consisted mainly of PLA, PBS, PBAT, and PHBV, as confirmed by NMR spectroscopy. The bioplastic materials showed different compositions and ratios of these basic materials, which, apparently, affected their degradability. The enzymatic degradability was demonstrated with rapid in vitro assays. pH-Stat titration and plate clearing assays showed mostly low or no degradation at environmental water temperatures. Our results indicate a poor biodegradability of these bioplastics in marine environments.
Hydrolysis rates were highest for BPE-AMF-PLA, BPE-C-PLA, and BPE-RP-PLA, although with different enzymes. These materials are based on PLA, which is the major component, accounting for at least 50% of each blend. Pure PLA is efficiently degraded by protease from Bacillus licheniformis as measured by pH-stat titration (Miksch et al., 2021). However, of the three materials containing PLA as base polymer, only BPE-C-PLA and BPE-RP-PLA showed a noticeable degradability with protease.
BPE-C-PLA and BPE-RP-PLA are blends of PLA and PBS with ratios of 2.5:1 and 2.6:1, respectively. PLA is well degradable by protease from B. licheniformis and other serine proteases (Oda et al., 2000; Lim et al., 2005), whereas PBS polymers are preferably hydrolyzed by lipase (Rizzarelli et al., 2004; Ding et al., 2012). Accordingly, considerable hydrolytic activity was present when these materials were incubated with protease, while the substantially lower hydrolysis rate when incubated with lipase may indicate the degradation of the PBS fraction of the material.
BPE-AMF-PLA is a blend of PLA and PBAT. It was most efficiently hydrolyzed by lipase from Candida antarctica. PLA cannot be hydrolyzed by lipase (Jarerat and Tokiwa, 2003) whereas PBAT is well degraded by lipase from, e.g., Pseudomonas cepacia, Candida cylindracea (Herrera et al., 2002), and Pelosinus fermentans (Biundo et al., 2016). Therefore, the hydrolytic activity of the lipase on BPE-AMF-PLA likely displays the degradation of the PBAT component of the material rather than PLA. Surprisingly, BPE-AMF-PLA was not degraded by protease although the material is composed of PLA by 70 to 80%, according to producer information. Apparently, the PLA was not accessible for the protease or the enzyme may have been inhibited by compounds of the bioplastic blend.
PBS polymers are hydrolyzed by lipase (Rizzarelli et al., 2004; Ding et al., 2012). Among the three PBS-containing materials, only BPE-SP-PBS was significantly degraded by lipase, but also by protease and esterase. Materials sharing the same base polymer may differ in their degradability depending on the associated copolymers or additives. Differences in physical properties, such as molecular weight, crystallinity, or hydrophilic/hydrophobic properties may affect the enzymatic degradation (Singh and Sharma, 2008; Tokiwa et al., 2009) and, thus, determine the degradability of bioplastics.
BPE-T-PHBV was the least degradable of all bioplastic polymers tested herein. A slight increase in hydrolysis was observed with all enzymes though at very low rates and only at the highest temperature of 30°C. According to the course of the degradation curve, an exponential increase may occur above 30°C. Degradation of PHBV in seawater has been observed previously (Rutkowska et al., 2008). However, the catalytic properties were attributed to specific PHBV-depolymerases (Li et al., 2007). Our results indicate that peptidases and esterases play little to no role in the degradation of PHBV, but their contribution to degradation might increase at higher temperatures. However, their relevance in the degradation of PHBV in natural aquatic environments remains to be quantified.
Depending on the composition of the material, pH-Stat titration may underestimate the degradability of the synthetic polymer. During hydrolysis, alkaline degradation products may be formed, which can affect the pH. This would counteract the formation of degradation products with carboxyl end groups, where the acidifying effect is used as a measure of degradability. Alkaline leachates would increase the pH during the blank measurement of the material (Miksch et al., 2021). This effect was observed for BPE-C-PLA and BPE-RP-PLA but not for the other bio-based polymers (data not shown). The use of alkaline additives was confirmed by the manufacturer. The unspecific shift in pH was quantified by the blank measurement without enzyme and can be subtracted from the total rate. However, an increasing alkalization during the progressive enzymatic degradation of the material cannot be excluded.
Enzymatic hydrolysis of the bioplastics increased with temperature. Hydrolysis rates were low below 20°C and highest at 25°C and 30°C. The thermal profiles of BPE-C-PLA and BPE-AMF-PLA showed an exponential increase whereas the profiles of BPE-RP-PLA and BPE-SP-PBS were best described by a sigmoidal regression model. The exponential increase displays the progressive rise in enzyme activity with temperature, which may continue up to the optimum of the enzymes at 40 to 60°C (Distributor Information, Sigma Aldrich). At higher temperatures, enzyme activity will cease due to progressive thermal denaturation of the enzymes. Consequently, the curve will flatten and then follow a sigmoidal course as well with an inflection point well above 30°C. Additionally, hydrolysis is also favored when the incubation temperature approaches the melting point of the material due to the higher accessibility of the substrate to the enzyme (He et al., 2001; Marten et al., 2005; Herzog et al., 2006).
The sigmoidal course of the thermal profiles of BPE-RP-PLA and BPE-SP-PBS at temperatures clearly below the thermal optimum of the enzymes must be attributed to the properties of the material rather than the catalytic properties of the enzymes. Depending on the composition of the material, only a limited fraction of the degradable component may be accessible to the enzyme. Hydrolysis may be limited by the surface availability of the degradable component so that increasing temperature cannot further accelerate the hydrolysis rate. This may particularly affect the hydrolysis of blends and of polymers with amorphous and crystalline fractions, as the amorphous fraction is degraded faster than the crystalline fraction (Göpferich, 1996).
Additionally, the enzymatic activity may be inhibited by chemical additives, which may leach from the hydrolyzed material (Stloukal et al., 2015). To improve the properties and facilitate the processing of plastics, substances such as plasticizers, impact modifiers, or fillers are added during the production process (Jacobsen and Fritz, 1999; Liu et al., 2013; Wiesinger et al., 2021). The broad and diverse application of plastics requires a wide spectrum of additives. Unfortunately, the additives in the bioplastics used in this study were not specified by the manufacturer. The NMR spectra show various small signals in addition to the signals of the base polymers, indicating the presence of additives.
To better estimate the enzymatic degradability of the bioplastics, we compared their hydrolysis rates with those of the conventional petroleum-based polymer PMMA and the biogenic polymer collagen. PMMA is a synthetic polymer primarily made from crude oil. Due to its favorable properties, such as low weight and high mechanical strength and toughness, PMMA is used in a wide range of applications. It is a very stable and persistent polymer, with no susceptibility to hydrolytic degradation (Boudaoud et al., 2018) and almost no bio-degradation in the environment (Kaplan et al., 1979; Smith et al., 1987). Accordingly, PMMA showed among the lowest enzymatic hydrolysis of all materials tested in our experiment. However, it was in a similar range as the hydrolysis of BPE-RP-PLA by lipase and esterase and BPE-T-PHBV by all three enzymes. These results indicate that below 20°C the enzymatic degradation of most bioplastics is as inefficient as the degradation of conventional plastic such as PMMA.
Collagens are the main structural proteins in skin, bones, tendons, and cartilage of vertebrates and invertebrates (Prockop and Kivirikko, 1995) and, thus, highly abundant in the marine environment (Silva et al., 2014). Collagens are virtually insoluble in seawater (Wolf et al., 2006) and very stable due to their dense triple-helix protein structure (Shoulders and Raines, 2009). Although much is known about the rapid degradation of collagen in tissue (McAnulty and Laurent, 1987; Liao and Cui, 2004), studies about the degradation rates of collagens in seawater are rare. Collagens are hydrolyzed by collagenolytic proteases (Ran et al., 2013). Only a few studies addressed collagenolytic enzymes in marine bacteria (Merkel et al., 1975; Kurata et al., 2007; Zhao et al., 2008). Our experiment showed a substantial degradation of collagen by the protease but not by the esterase and the lipase. The hydrolysis rate of collagen was six to 14 times higher than the highest rates for the bioplastic compounds (BPE-AMF-PLA and BPE-SP-PBS both hydrolyzed by lipase at 20°C), and more than 60 times higher than the rates for all other bioplastic compounds. Accordingly, the enzymatic degradation of bioplastics is low compared to the degradation of natural biogenic collagen.
The plate clearing assays largely confirmed the results of pH-Stat titration, albeit with some deviations. Significant hydrolysis rates as detected by pH-Stat titration were confirmed by clear zones on the agar plates at pH 8. Clear zones were larger at pH 8 than at pH 5 for almost all enzymes, confirming favorable conditions for the chosen enzymes at pH values of natural seawater. Lipase from Candida antarctica and esterase from Bacillus subtilis have a pH optima in the neutral to lower alkaline range (Kaiser et al., 2006; Eom et al., 2013), and the serine protease from Bacillus licheniformis in the alkaline range (Sareen and Mishra, 2008). The diameter of the clear zones did not consistently correlate with the hydrolysis rates measured by pH-Stat titration. The diameter of the clear zones ranged from 13 to 46 mm indicating considerable degradation, whereas the corresponding pH-Stat hydrolysis rates were mostly low. Additionally, the plate clearing assay indicated a considerable degradation of BPE-RP-PLA by lipase, which was not detected by the pH-Stat titration. The differences may be explained by the higher temperatures and the longer incubation time of the plate clearing assays. For example, no enzymatic degradation of BPE-T-PHBV was observed up to 25°C but distinctly increased at 30°C. A further exponential increase at higher temperatures is likely. Additionally, the dissolution and subsequent re-crystallization of the plastics in the plate clearing assay may have changed the structural properties of the polymer thereby potentially affecting the degradability of the material. Further studies are needed to confirm the suitability of plate clearing assay for the verification of pH-Stat titration results.
Degradation of bioplastics in the environment requires the presence of the appropriate enzymes or mixtures thereof produced by microorganisms. Besides temperature, salinity, pH, and UV radiation (Voinova et al., 2008), also depth, oxygen, and organic substrate availability determine the microbial community in marine habitats and their catalytic potential (Chen et al., 2019; Tobias-Hünefeldt et al., 2019). Our in vitro experiments proved the general enzymatic degradability of the tested materials by single enzymes, but we did not test the combined effects of enzymes co-occurring in the environment. Mixtures of several enzymes showed synergistic effects on the degradation of natural materials (Spagnuolo et al., 1997; Murashima et al., 2003). A combination of various enzyme classes may show different effects on the hydrolysis of the materials as well. However, the presence of highly active proteases in such mixtures may be adverse as they may degrade other enzymes like lipases and esterase (Drouault et al., 2000) and reduce their catalytic potential.
Three of the tested materials are labeled as industrially compostable (Table 2) at the specific conditions in industrial composting plants of 50 - 60°C [European Bioplastics e.V. (EUBP), 2015]. However, under natural seawater conditions below 20°C, the enzymatic degradation is slow and elevated temperatures of up to 30°C occur only in surface waters of tropical regions (Deser et al., 2010). Plastic debris is not restricted to the surface, where the temperature is usually the highest, but also disperses throughout the water column and down to the seafloor (Kershaw et al., 2015) where temperatures are low. Since the density of the five tested bioplastics is between 1.2 and 1.5 g·cm-3, (Manufacturer Information) these materials likely sink to the ocean floor and are exposed to low temperatures, where they degrade only slowly or not at all.
Degradation rates of the tested bioplastics were low at environmentally relevant temperatures. Accordingly, uncontrolled release of these materials into the environment would inevitably lead to an accumulation in sensitive ecosystems similar to conventional, petroleum-based plastics. To counteract the pollution of the marine environment with putative biodegradable plastics, more effort in research and development is needed to design polymers that are easier to degrade at low environmental temperatures of the oceans.
The datasets presented in this study can be found in the online repository PANGAEA (https://doi.pangaea.de/10.1594/PANGAEA.943844).
LM: Conceptualization, Investigation, Methodology, Writing – original draft. MK: NMR methodology, Writing – Review and editing. LG: Conceptualization, Funding acquisition, Project administration, Writing – Review and editing. RS: Conceptualization, Funding acquisition, Supervision, Writing – Review and editing. All authors contributed to the article and approved the submitted version.
This research has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 860407 (BIO-PLASTICS EUROPE).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors acknowledge the Alfred Wegener Institute, Helmholtz Centre for Polar and Marine Research for supporting this work.
Ali S. S., Elsamahy T., Al-Tohamy R., Zhu D., Mahmoud Y. A. G., Koutra E., et al. (2021). Plastic Wastes Biodegradation: Mechanisms, Challenges and Future Prospects. Sci. Tot. Environ. 780, 146590. doi: 10.1016/j.scitotenv.2021.146590
Angelini S., Cerruti P., Immirzi B., Poskovic M., Santagata G., Scarinzi G., et al. (2015). From Microbial Biopolymers to Bioplastics: Sustainable Additives for PHB Processing and Stabilization. In. Microbial. Factor. (New Delhi) pp, 139–160). doi: 10.1007/978-81-322-2595-9
Arnosti C., Bell C., Moorhead D. L., Sinsabaugh R. L., Steen A. D., Stromberger M., et al. (2014). Extracellular Enzymes in Terrestrial, Freshwater, and Marine Environments: Perspectives on System Variability and Common Research Needs. Biogeochemistry 117 (1), 5–21. doi: 10.1007/s10533-013-9906-5
(2021)Petrochemicals Europe An Industry Sector of Cefic. Available at: https://www.petrochemistry.eu/sector-group/methacrylates/ (Accessed November 28, 2021).
Bátori V., Åkesson D., Zamani A., Taherzadeh M. J., Horváth I. S. (2018). Anaerobic Degradation of Bioplastics: A Review. Waste. Manage. 80, 406–413. doi: 10.1016/j.wasman.2018.09.40
Bioplastics Europe (2022) Sustainable Solutions for Bio-Based Plastics on Land and Sea. Available at: https://bioplasticseurope.eu/ (Accessed January 20, 2022).
Biundo A., Hromic A., Pavkov-Keller T., Gruber K., Quartinello F., Haernvall K., et al. (2016). Characterization of a Poly (Butylene Adipate-Co-Terephthalate)-Hydrolyzing Lipase From Pelosinus Fermentans. Appl. Microbiol. Biotechnol. 100 (4), 1753–1764. doi: 10.1007/s00253-015-7031-1
Borrelle S. B., Ringma J., Law K. L., Monnahan C. C., Lebreton L., McGivern A., et al. (2020). Predicted Growth in Plastic Waste Exceeds Efforts to Mitigate Plastic Pollution. Science 369 (6510), 1515–1518. doi: 10.1126/science.aba3656
Boudaoud N., Benali S., Mincheva R., Satha H., Raquez J. M., Dubois P. (2018). Hydrolytic Degradation of Poly (L-Lactic Acid)/Poly (Methyl Methacrylate) Blends. Polymer. Int. 67 (10), 1393–1400. doi: 10.1002/pi.5659
Chen L., Tsui M. M., Lam J. C., Hu C., Wang Q., Zhou B., et al. (2019). Variation in Microbial Community Structure in Surface Seawater From Pearl River Delta: Discerning the Influencing Factors. Sci. Tot. Environ. 660, 136–144. doi: 10.1016/j.scitotenv.2018.12.480
Deser C., Alexander M. A., Xie S. P., Phillips A. S. (2010). Sea Surface Temperature Variability: Patterns and Mechanisms. Annu. Rev. Mar. Sci. 2, 115–143. doi: 10.1146/annurev-marine-120408-151453
Dilshad E., Waheed H., Ali U., Amin A., Ahmed I. (2021). “General Structure and Classification of Bioplastics and Biodegradable Plastics,” in Bioplastics for Sustainable Development (Singapore: Springer), 61–82. doi: 10.1007/978-981-16-1823-9_2
Ding M., Zhang M., Yang J., Qiu J. H. (2012). Study on the Enzymatic Degradation of Aliphatic Polyester–PBS and its Copolymers. J. Appl. Polymer. Sci. 124 (4), 2902–2907. doi: 10.1002/app.35347
do Sul J. A. I., Costa M. F. (2014). The Present and Future of Microplastic Pollution in the Marine Environment. Environ. Pollution. 185, 352–364. doi: 10.1016/j.envpol.2013.10.036
Drouault S., Corthier G., Ehrlich S. D., Renault P. (2000). Expression of the Staphylococcus Hyicus Lipase in Lactococcus Lactis. Appl. Environ. Microbiol. 66 (2), 588–598. doi: 10.1128/AEM.66.2.588-598.2000
Eom G. T., Lee S. H., Song B. K., Chung K. W., Kim Y. W., Song J. K. (2013). High-Level Extracellular Production and Characterization of Candida Antarctica Lipase B in Pichia Pastoris. J. Biosci. Bioengineer. 116 (2), 165–170. doi: 10.1016/j.biosc.2013.02.016
European Bioplastics e.V. (EUBP) (2015). Available at: https://docs.european-bioplastics.org/publications/bp/EUBP_BP_En_13432 (Accessed January 20, 2022). EN 13432 Certified Bioplastics Performance in Industrial Composting, Fact Sheet.
European Bioplastics e.V (2021) Bioplastic Market Data. Available at: https://www.european-bioplastics.org/market/ (Accessed November 28, 2021).
Ferreira F. V., Cividanes L. S., Gouveia R. F., Lona L. M. (2019). An Overview on Properties and Applications of Poly (Butylene Adipate-Co-Terephthalate)–PBAT Based Composites. Polymer. Eng. Sci. 59 (s2), E7–E15. doi: 10.1002/pen.24770
Folino A., Karageorgiou A., Calabrò P. S., Komilis D. (2020). Biodegradation of Wasted Bioplastics in Natural and Industrial Environments: A Review. Sustainability 12 (15), 6030. doi: 10.3390/su12156030
Gall S. C., Thompson R. C. (2015). The Impact of Debris on Marine Life. Mar. Pollution. Bull. 92 (1-2), 170–179. doi: 10.1016/j.marpolbul.2014.12.041
Garlotta D. (2001). A Literature Review of Poly (Lactic Acid). J. Polymer. Environ. 9 (2), 63–84. doi: 10.1023/A:1020200822435
Göpferich A. (1996). Mechanisms of Polymer Degradation and Erosion. Biomaterials 17 (2), 103–114. doi: 10.1016/0142-9612(96)85755-3
Gug J., Sobkowicz M. J. (2016). Improvement of the Mechanical Behavior of Bioplastic Poly (Lactic Acid)/Polyamide Blends by Reactive Compatibilization. J. Appl. Polymer. Sci. 133 (45), 43350. doi: 10.1002/app.43350
Herrera R., Franco L., Rodríguez-Galán A., Puiggalí J. (2002). Characterization and Degradation Behavior of Poly (Butylene Adipate-Co-Terephthalate) s. J. Polymer. Sci. Part A.: Polymer. Chem. 40 (23), 4141–4157. doi: 10.1002/pola.10501
Herzog K., Müller R. J., Deckwer W. D. (2006). Mechanism and Kinetics of the Enzymatic Hydrolysis of Polyester Nanoparticles by Lipases. Polymer. Degradation. Stability. 91 (10), 2486–2498. doi: 10.1016/j.polymdegradstab.2006.03.005
He Y., Shuai X., Kasuya K. I., Doi Y., Inoue Y. (2001). Enzymatic Degradation of Atactic Poly (R, S-3-Hydroxybutyrate) Induced by Amorphous Polymers and the Enzymatic Degradation Temperature Window of an Amorphous Polymer System. Biomacromolecules 2 (3), 1045–1051. doi: 10.1021/bm010087w
Jacobsen S., Fritz H. G. (1999). Plasticizing Polylactide—the Effect of Different Plasticizers on the Mechanical Properties. Polymer. Eng. Sci. 39 (7), 1303–1310. doi: 10.1002/pen.11517
Jambeck J. R., Geyer R., Wilcox C., Siegler T. R., Perryman M., Andrady A., et al. (2015). Plastic Waste Inputs From Land Into the Ocean. Science 347 (6223), 768–771. doi: 10.1126/science.1260352
Jarerat A., Tokiwa Y. (2003). Poly (L-Lactide) Degradation by Saccharothrix Waywayandensis. Biotechnol. Lett. 25 (5), 401–404. doi: 10.1023/A:1022450431193
Kaiser P., Raina C., Parshad R., Johri S., Verma V., Andrabi K. I., et al. (2006). A Novel Esterase From Bacillus Subtilis (RRL 1789): Purification and Characterization of the Enzyme. Protein Expression Purificat. 45 (2), 262–268. doi: 10.1016/j.pep.2005.08.030
Kaplan D. L., Hartenstein R., Sutter J. (1979). Biodegradation of Polystyrene, Poly (Metnyl Methacrylate), and Phenol Formaldehyde. Appl. Environ. Microbiol. 38 (3), 551–553. doi: 10.1128/aem.38.3.551-553.1979
Kershaw P. J., Rochman C. M. (2015). Sources, Fate and Effects of Microplastics in the Marine Environment: Part 2 of a Global Assessment. Reports and studies-IMO/FAO/Unesco-IOC/WMO/IAEA/UN/UNEP Joint Group of Experts on the Scientific Aspects of Marine Environmental Protection (GESAMP) eng no. 93, p. 220.
Kühn S., Bravo Rebolledo E. L., van Franeker J. A. (2015). Deleterious Effects of Litter on Marine Life, in Marine Anthropogenic Litter, eds. Bergmann M., Gutow L., Klages M. (Cham, Switzerland: Springer), 75–116. doi: 10.1007/978-3-319-16510-3_4
Kurata A., Miyazaki M., Kobayashi T., Nogi Y., Horikoshi K. (2007). Alkalimonas Collagenimarina Sp. Nov., a Psychrotolerant, Obligate Alkaliphile Isolated From Deep-Sea Sediment. Int. J. Systemat. Evolution. Microbiol. 57 (7), 1549–1553. doi: 10.1099/ijs.0.65084-0
Lebreton L., Slat B., Ferrari F., Sainte-Rose B., Aitken J., Marthouse R., et al. (2018). Evidence That the Great Pacific Garbage Patch is Rapidly Accumulating Plastic. Sci. Rep. 8 (1), 1–15. doi: 10.1038/s41598-018-22939-w
Liao S. S., Cui F. Z. (2004). In Vitro and In Vivo Degradation of Mineralized Collagen-Based Composite Scaffold: Nanohydroxyapatite/Collagen/Poly (L-Lactide). Tissue Eng. 10 (1-2), 73–80. doi: 10.1089/107632704322791718
Li Z., Lin H., Ishii N., Chen G. Q., Inoue Y. (2007). Study of Enzymatic Degradation of Microbial Copolyesters Consisting of 3-Hydroxybutyrate and Medium-Chain-Length 3-Hydroxyalkanoates. Polymer. Degradat. Stability. 92 (9), 1708–1714. doi: 10.1016/j.polymdegradstab.2007.06.001
Lim H. A., Raku T., Tokiwa Y. (2005). Hydrolysis of Polyesters by Serine Proteases. Biotechnol. Lett. 27 (7), 459–464. doi: 10.1007/s10529-005-2217-8
Liu S., Liu Z. (2018). Free Extracellular Enzymes Dominate Initial Peptide Hydrolysis in Coastal Seawater. Mar. Chem. 199, 37–43. doi: 10.1016/j.marchem.2018.01005
Liu W., Li H., Wang X., Du Z., Zhang C. (2013). Effect of Chain Extension on the Rheological Property and Thermal Behaviour of Poly (Lactic Acid) Foams. Cell. Polymer. 32 (6), 343–368. doi: 10.1177/026248931303200602
Luo S., Netravali A. N. (2003). A Study of Physical and Mechanical Properties of Poly (Hydroxybutyrate-Co-Hydroxyvalerate) During Composting. Polymer. Degradation. Stability. 80 (1), 59–66. doi: 10.1016/S0141-3910(02)00383-X
Marten E., Müller R. J., Deckwer W. D. (2005). Studies on the Enzymatic Hydrolysis of Polyesters. II. Aliphatic–aromatic Copolyesters. Polymer. Degradation. Stability. 88 (3), 371–381. doi: 10.1016/S0141-3910(03)00032-6
McAnulty R. J., Laurent G. J. (1987). Collagen Synthesis and Degradation In Vivo. Evidence for Rapid Rates of Collagen Turnover With Extensive Degradation of Newly Synthesized Collagen in Tissues of the Adult Rat. Collagen. Relate. Res. 7 (2), 93–104. doi: 10.1016/S0174-173X(87)80001-8
Meereboer K. W., Misra M., Mohanty A. K. (2020). Review of Recent Advances in the Biodegradability of Polyhydroxyalkanoate (PHA) Bioplastics and Their Composites. Green Chem. 22 (17), 5519–5558. doi: 10.1039/d0gc01647k
Merkel J. R., Dreisbach J. H., Ziegler H. B. (1975). Collagenolytic Activity of Some Marine Bacteria. Appl. Microbiol. 29 (2), 145–151. doi: 10.1128/am.29.2.145-151.1975
Miksch L., Gutow L., Saborowski R. (2021). pH-Stat Titration: A Rapid Assay for Enzymatic Degradability of Bio-Based Polymers. Polymers 13 (6), 860. doi: 10.3390/polym13060860
Muneer F., Nadeem H., Arif A., Zaheer W. (2021). Bioplastics From Biopolymers: An Eco-Friendly and Sustainable Solution of Plastic Pollution. Polymer. Sci. Ser. C. 63 (1), 47–63. doi: 10.1134/S1811238221010057
Murashima K., Kosugi A., Doi R. H. (2003). Synergistic Effects of Cellulosomal Xylanase and Cellulases From Clostridium Cellulovorans on Plant Cell Wall Degradation. J. Bacteriol. 185 (5), 1518–1524. doi: 10.1128/JB.185.5.1518-1524.2003
Oda Y., Yonetsu A., Urakami T., Tonomura K. (2000). Degradation of Polylactide by Commercial Proteases. J. Polymer. Environ. 8 (1), 29–32. doi: 10.1023/A:1010120128048
Patel D., Gismondi R., Alsaffar A., Tiquia-Arashiro S. M. (2019). Applicability of API ZYM to Capture Seasonal and Spatial Variabilities in Lake and River Sediments. Environ. Technol. 40 (24), 3227–3239. doi: 10.1080/09593330.2018.1468492
PlasticsEurope (2019) Market Research Group (PEMRG) and Conversio Market & Strategy GmbH, Plastics – The Facts 2019. Available at: https://plasticseurope.org/wp-content/uploads/2021/10/2019-Plastics-the-facts.pdf (Accessed January 20, 2022). An analysis of European plastics production, demand and waste data.
Polman E. M., Gruter G. J. M., Parsons J. R., Tietema A. (2021). Comparison of the Aerobic Biodegradation of Biopolymers and the Corresponding Bioplastics: A Review. Sci. Tot. Environ. 753, 141953. doi: 10.1016/j.scitotenv.2020.141953
Prockop D. J., Kivirikko K. I. (1995). Collagens: Molecular Biology, Diseases, and Potentials for Therapy. Annu. Rev. Biochem. 64 (1), 403–434. doi: 10.1146/annurev.bi.64.070195.002155
RameshKumar S., Shaiju P., O'Connor K. E. (2020). Bio-Based and Biodegradable Polymers-State-Of-the-Art, Challenges and Emerging Trends. Curr. Opin. Green Sustain. Chem. 21, 75–81. doi: 10.1016/j.cogsc.2019.12.005
Ran L. Y., Su H. N., Zhao G. Y., Gao X., Zhou M. Y., Wang P., et al. (2013). Structural and Mechanistic Insights Into Collagen Degradation by a Bacterial Collagenolytic Serine Protease in the Subtilisin Family. Mol. Microbiol. 90 (5), 997–1010. doi: 10.1111/mmi.12412
Richert A., Dąbrowska G. B. (2021). Enzymatic Degradation and Biofilm Formation During Biodegradation of Polylactide and Polycaprolactone Polymers in Various Environments. Int. J. Biol. Macromol. 176, 226–232. doi: 10.1016/j.ijbiomac.2021.01.202
Rizzarelli P., Impallomeni G., Montaudo G. (2004). Evidence for Selective Hydrolysis of Aliphatic Copolyesters Induced by Lipase Catalysis. Biomacromolecules 5 (2), 433–444. doi: 10.1021/bm034230s
Ruggero F., Gori R., Lubello C. (2019). Methodologies to Assess Biodegradation of Bioplastics During Aerobic Composting and Anaerobic Digestion: A Review. Waste. Manage. Res. 37 (10), 959–975. doi: 10.1177/0734242X19854127
Rutkowska M., Krasowska K., Heimowska A., Adamus G., Sobota M., Musioł M., et al. (2008). Environmental Degradation of Blends of Atactic Poly [(R, S)-3-Hydroxybutyrate] With Natural PHBV in Baltic Sea Water and Compost With Activated Sludge. J. Polymer. Environ. 16 (3), 183–191. doi: 10.1007/s10924-008-0100-0
Sareen R., Mishra P. (2008). Purification and Characterization of Organic Solvent Stable Protease From Bacillus Licheniformis RSP-09-37. Appl. Microbiol. Biotechnol. 79 (3), 399–405. doi: 10.1007/s.00253-008-1429-y
Shafqat A., Al-Zaqri N., Tahir A., Alsalme A. (2021). Synthesis and Characterization of Starch Based Bioplatics Using Varying Plant-Based Ingredients, Plasticizers and Natural Fillers. Saudi. J. Biol. Sci. 28 (3), 1739–1749. doi: 10.1016/j.sjbs.2020.12.015
Shoulders M. D., Raines R. T. (2009). Collagen Structure and Stability. Annu. Rev. Biochem. 78, 929–958. doi: 10.1146/annurev.biochem.77.032207.120833
Silva T. H., Moreira-Silva J., Marques A. L., Domingues A., Bayon Y., Reis R. L. (2014). Marine Origin Collagens and its Potential Applications. Mar. Drugs 12 (12), 5881–5901. doi: 10.3390/md12125881
Singh B., Sharma N. (2008). Mechanistic Implications of Plastic Degradation. Polymer. Degradation. Stability. 93 (3), 561–584. doi: 10.1016/j.polymdegradstab.2007.11.008
Smith R., Oliver C., Williams D. F. (1987). The Enzymatic Degradation of Polymers In Vitro. J. Biomed. Mat. Res. 21 (8), 991–1003. doi: 10.1002/jbm.820210805
Spagnuolo M., Crecchio C., Pizzigallo M. D. R., Ruggiero P. (1997). Synergistic Effects of Cellulolytic and Pectinolytic Enzymes in Degrading Sugar Beet Pulp. Biores0 Technol. 60 (3), 215–222. doi: 10.1016/S0960-8524(97)00013-8
Stloukal P., Kalendova A., Mattausch H., Laske S., Holzer C., Koutny M. (2015). The Influence of a Hydrolysis-Inhibiting Additive on the Degradation and Biodegradation of PLA and its Nanocomposites. Polymer. Test. 41, 124–132. doi: 10.1016/j.polymertesting.2014.10.015
Tobias-Hünefeldt S. P., Wing S. R., Espinel-Velasco N., Baltar F., Morales S. E. (2019). Depth and Location Influence Prokaryotic and Eukaryotic Microbial Community Structure in New Zealand Fjords. Sci. Tot. Environ. 693, 133507. doi: 10.1016/j.scitotenv.2019.07.313
Tokiwa Y., Calabia B. P., Ugwu C. U., Aiba S. (2009). Biodegradability of Plastics. Int. J. Mol. Sci. 10 (9), 3722–3742. doi: 10.3390/ijms10093722
Uchida H., Nakajima-Kambe T., Shigeno-Akutsu Y., Nomura N., Tokiwa Y., Nakahara T. (2000). Properties of a Bacterium Which Degrades Solid Poly (Tetramethylene Succinate)-Co-Adipate, a Biodegradable Plastic. FEMS Microbiol. Lett. 189 (1), 25–29. doi: 10.1111/j.1574-6968.2000.tb09201.x
Urbanek A. K., Rybak J., Wróbel M., Leluk K., Mirończuk A. M. (2020). A Comprehensive Assessment of Microbiome Diversity in Tenebrio Molitor Fed With Polystyrene Waste. Environ. Pollut. 262, 114281. doi: 10.1016/j.envpol.2020.114281
Vert M., Doi Y., Hellwich K. H., Hess M., Hodge P., Kubisa P., et al. (2012). Terminology for Biorelated Polymers and Applications (IUPAC Recommendations 2012). Pure. Appl. Chem. 84 (2), 377–410. doi: 10.1351/PAC-REC-10-12-04
Voinova O., Gladyshev M., Volova T. G. (2008). “Comparative Study of PHA Degradation in Natural Reservoirs Having Various Types of Ecosystems,” in Macromolecular Symposia, vol. Vol. 269. (Weinheim: WILEY-VCH Verlag), 34–37. doi: 10.1002/masy.200850906
Wang G. X., Huang D., Ji J. H., Völker C., Wurm F. R. (2021). Seawater-Degradable Polymers—Fighting the Marine Plastic Pollution. Adv. Sci. 8 (1), 2001121. doi: 10.1002/advs.202001121
Wiesinger H., Wang Z., Hellweg S. (2021). Deep Dive Into Plastic Monomers, Additives, and Processing Aids. Environ. Sci. Technol. 55 (13), 9339–9351. doi: 10.1021/acs.est.1c00976
Worm B., Lotze H. K., Jubinville I., Wilcox C., Jambeck J. (2017). Plastic as a Persistent Marine Pollutant. Annu. Rev. Environ. Resour. 42, 1–26. doi: 10.1146/annurev-environ-102016-060700
Wolf K. L., Sobral P. J. A., Telis V. R. N. (2006). Characterizations of collagen fibers for biodegradable films production. Proceedings in 13th world congress of food sciences technology. IUFOST 2006, 929-929. (2006). “Characterizations of Collagen Fibers for Biodegradable Films Production,” in 13th World Congress of Food Science & Technology, 929–929.
Xu J., Guo B. H. (2010). Poly (Butylene Succinate) and its Copolymers: Research, Development and Industrialization. Biotechnol. J. 5 (11), 1149–1163. doi: 10.1023/A:1020200822435
Keywords: enzymatic degradation, hydrolysis, bio-based, polymers, enzymes, plastics
Citation: Miksch L, Köck M, Gutow L and Saborowski R (2022) Bioplastics in the Sea: Rapid In-Vitro Evaluation of Degradability and Persistence at Natural Temperatures. Front. Mar. Sci. 9:920293. doi: 10.3389/fmars.2022.920293
Received: 14 April 2022; Accepted: 16 May 2022;
Published: 23 June 2022.
Edited by:
Tony Robert Walker, Dalhousie University, CanadaReviewed by:
Jingguang Cheng, Tsinghua University, ChinaCopyright © 2022 Miksch, Köck, Gutow and Saborowski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lukas Miksch, bHVrYXMubWlrc2NoQGF3aS5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.