- 1Department of Marine Science and Technology, Faculty of Fisheries and Marine Sciences, IPB University, Bogor, Indonesia
- 2Oceanogen, Bogor, Indonesia
- 3Asosiasi Pengelolaan Rajungan Indonesia (APRI), Surabaya, Indonesia
- 4School of Business, IPB University, Bogor, Indonesia
- 5Department of Aquaculture, Faculty of Marine and Fisheries, Universitas Syiah Kuala, Banda Aceh, Indonesia
- 6Department of Marine Science, Faculty of Marine and Fisheries, Universitas Syiah Kuala, Banda Aceh, Indonesia
- 7Center for Transdisciplinary and Sustainability Science, IPB University, Bogor, Indonesia
- 8Faculty of Fisheries and Marine Sciences, Universitas Khairun, Ternate, Indonesia
- 9Department of Aquatic Resources Management, Faculty of Agriculture, Universitas Sumatera Utara, Medan, Indonesia
- 10Faculty of Fisheries and Marine Science, Universitas Brawijaya, Malang, Indonesia
- 11Department of Fisheries, Faculty of Agriculture, Universitas Gorontalo, Limboto, Indonesia
- 12Department of Fisheries and Marine, Politeknik Negeri Nusa Utara, Tahuna, Indonesia
- 13STP Hatta-Sjahrir Banda Naira, Banda Naira, Indonesia
- 14Politeknik Perikanan Negeri Tual, Maluku Tenggara, Indonesia
- 15Department of Fisheries, Universitas Gadjah Mada, Yogyakarta, Indonesia
- 16Aquatic Resources Management Study Program, Universitas Hasanuddin, Makassar, Indonesia
- 17Marine Science Department, Faculty of Marine Science and Fisheries, Universitas Hasanuddin, Makassar, Indonesia
- 18Fisheries and Marine Department, Faculty of Animal Husbandry and Fisheries, Universitas Tadulako, Palu, Indonesia
- 19Politeknik Negeri Sambas, Sambas, Indonesia
- 20Faculty of Animal Husbandry, Marine and Fisheries, Universitas Nusa Cendana, Kota Kupang, Indonesia
- 21Faculty of Fisheries and Marine Sciences, Artha Wacana Christian University, Kota Kupang, Indonesia
- 22Graduate School, Universitas Hasanuddin, Makassar, Indonesia
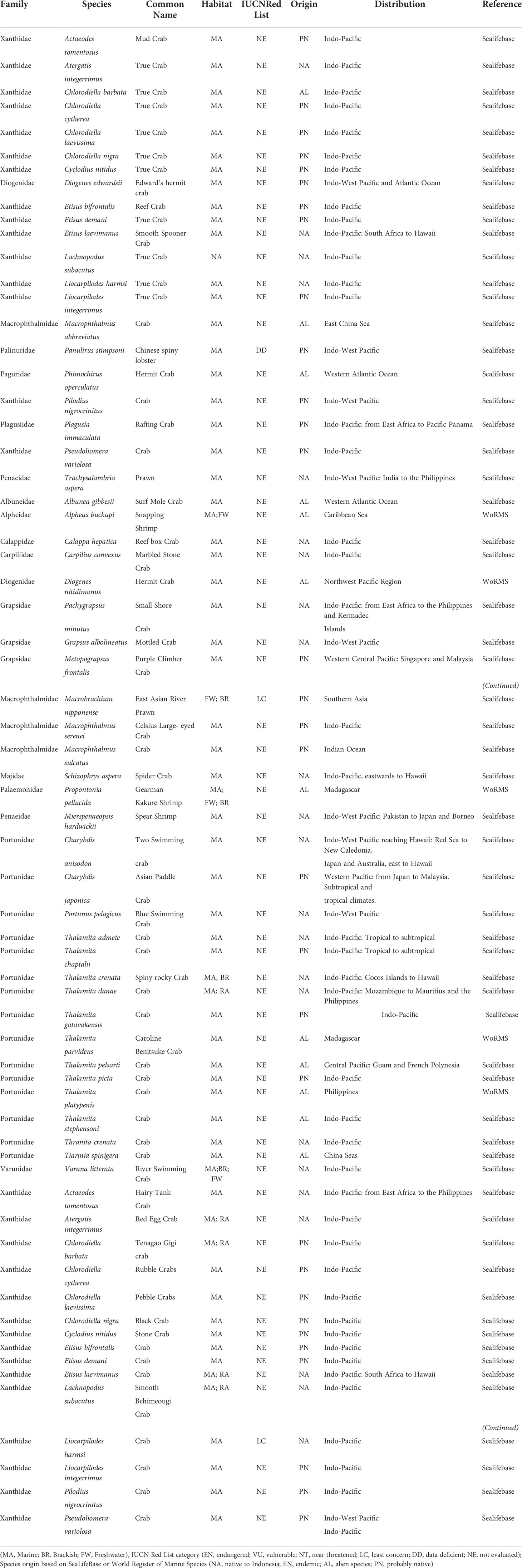
Environmental DNA (eDNA) methods are increasingly viewed as alternate or complementary approaches to conventional capture-based surveys for marine conservation and fisheries management purposes, especially at large spatial scales in mega-biodiversity regions such as Indonesia. Decapod crustacean distribution and diversity across Indonesia are still poorly known, even for economically important fisheries commodities. This study assessed coral reef associated decapod diversity and distribution by sampling 40 sites in three regions (West, Central, East), representing 17 provinces and 10 Fisheries Management Areas (FMAs) across Indonesia, with a special focus on the blue swimming crab Portunus pelagicus. DNA sequencing (Illumina iSeq100) data were analysed in mBRAVE (Multiplex Barcode Research And Visualization Environment) yielded 406 OTUs belonging to 32 families, with 47 genera and 51 species identified. The number of families identified was highest in the Central region (25), while the most genera (31) and species (36) were identified in the West region. Alpha diversity did not differ significantly between regions or provinces, while Beta diversity differed significantly between provinces but not between regions. Our results also showed 31 species are possibility native based on the distribution meanwhile 12 species do not appear to have been recorded based of SeaLifeBase or WorMS. While providing a reference for further exploration of Indonesian coastal and small island decapod biodiversity, the high proportion of unidentified taxa calls for concerted efforts to develop and maintain reference specimen and sequence repositories and expand species conservation status assessments. The economically important decapod crustaceans identified in this study included three crabs (Charybdis anisodon, Charybdis japonica, Portunus pelagicus), a freshwater prawn (Macrobrachium nipponense), a lobster (Panulirus stimpsoni) and two penaeid shrimps (Mierspenaeopsis hardwickii and Trachysalambria aspera). For most decapod taxa, observed patterns indicate management under existing provincial and/or FMA level management structures is appropriate. Furthermore, the data can inform science-based fisheries management strategies, in particular for P. pelagicus.
Introduction
Decapod crustaceans are a highly diverse taxonomic group, distinguished from other Crustacea by having ten pairs of legs (Martin & Davis, 2001; Watling & Thiel, 2013; Gökoğlu, 2021). These crustaceans include crabs, shrimps and prawns, lobsters, slipper lobsters and crayfish. Common in freshwater and marine tropical environments, including coral reef ecosystems, decapods play important ecological roles in many benthic communities (Thiel & Watling, 2015; Dev Roy and Nandi, 2017; Wolfe et al., 2019; West et al., 2020). Furthermore many are important as fisheries species for human consumption (Chan, 2010; Bondad-Reantaso et al., 2012; Gökoğlu, 2021) or the growing marine ornamental trade (Calado et al., 2003; Yuliana et al., 2021). In Indonesia, spiny lobsters (Panulirus spp.), mud crabs (Scylla spp.), and blue swimming crabs (Portunus pelagicus) are internationally traded fisheries commodities that contribute to the national economy and foreign exchange balance (Madduppa et al., 2016; Saputra, 2020).
The number of extant taxa in the speciose order Decapoda is unknown; however, in 2009 there were 14,335 accepted extant decapod species worldwide, comprised of two suborders: Dendrobranchiata (540 extant species) and Pleocyemata (13,795 extant species), the latter divided into ten infraorders (De Grave et al., 2009). A decade later, over 15,000 extant decapod species were recognised (Wolfe et al., 2019). The Brachyura, comprising the majority of crab species, are a particularly complex infraorder taxonomically, and are thought to comprise between 6,700 and 10,000 species (De Grave et al., 2009; Chakravarty et al., 2016), with at least 6,793 species reported from marine ecosystems (Ng et al., 2008; Kumaralingam et al., 2013). Indonesian marine decapod crustaceans have been studied at various scales (Moosa, 1980; Moosa & Aswandy, 1994; Aswandy, 2008; Pratiwi, 2010; Pratiwi, 2012; Pratiwi & Astuti, 2012; Pratiwi & Widyastuti, 2013; Pratiwi & Wijaya, 2013; Anggorowati, 2014; Mashar et al., 2014; Mashar et al., 2015; Anggraeni et al., 2015; Ardika et al., 2015). Hutomo & Moosa (2005) listed 1,502 crustaceans reported from Indonesian marine waters but noted that many Indonesian decapod crustaceans are still poorly documented.
Species distributions may depend on the influence of contemporary factors as well as processes at evolutionary timescales and global to micro spatial scales (Wolfe et al., 2019; Lagos et al., 2021). Within a given latitudinal range, local variations in species richness and community composition may depend on past and present seabed composition and topography, food availability, tidal and sea level patterns, prey-predator relationships, interactions among species, reproduction strategies, climatic variations, ontogenetic factors, etc. (Castilho et al., 2007; Lui et al., 2007; Ndoro et al., 2014; Lagos et al., 2021). Temporal and/or spatial variations in inter and intra-species diversity have been reported in taxonomic groups including decapods (Lui et al., 2007; Andrade et al., 2015; Madduppa et al., 2020a; Madduppa et al., 2021b). Biodiversity and distribution studies are increasingly turning to environmental DNA (eDNA) as a complement to traditional taxonomic surveys as an effective and cost-effective means of obtaining baseline biodiversity data and monitoring various taxa, including crustacea (Thomsen & Willerslev, 2015; West et al., 2020; Madduppa et al., 2021b, Gelis et al., 2021; Gilbey et al., 2021)
Arguably, one of the greatest threats facing marine biodiversity is anthropogenic habitat degradation and resultant species extirpation or loss (Jackson, 2008; Pimm et al., 2015). The rising level of global (e.g. climate change) and local (e.g. pollution, coastal development, overfishing) threats (Roberts & Hawkins, 1999; Jackson, 2008; Cheung et al., 2009; Burke et al., 2012; WWF, 2016; Buckley et al., 2019) is mirrored in the increasing number of taxa listed in the at risk categories in the Red List of Threatened Taxa produced by the International Union for Conservation of Nature (IUCN Red List) (IUCN, 2021). The conservation status evaluations based on IUCN Red List criteria (IUCN, 2012; IUCN,2019) are important for policy implementation and decision making at the global, regional, national, and sub-national levels (Hayward, 2011; Campbell, 2012; Bennun et al., 2018; Betts et al., 2020). Red List assessments can lead to the prioritization of species for practical conservation action such as recovery plans for threatened species, which may include fisheries management measures for directly exploited species (Hayward, 2011; Campbell, 2012; Hornborg et al., 2013). Although the majority of global and regional studies on extinction risk have focused on birds, mammals and amphibians, there are a growing number of IUCN Red List assessments for marine and freshwater vertebrates and invertebrates (IUCN, 2021).
The larvae of some commercially valuable fisheries target species and many other decapod taxa play important roles in marine food chains (Bowser et al., 2013; Mablouké et al., 2013; Pombo et al., 2013; Kwak et al., 2015; Park et al., 2020), Decapods therefore contribute to marine biodiversity and support the fisheries sector both directly and indirectly. In addition to evaluating biodiversity, eDNA studies can help detect the presence of decapods that are of conservation concern, such as species included in the IUCN Red List and those protected under national or international legislation, as well as those targeted by large and/or small-scale fisheries, at all stages of their life-cycle. Therefore, the study of decapod eDNA is important for both fisheries and conservation. Despite the recognition of Indonesia as a mega biodiversity country, with rich marine ecosystems including over half of coral reefs in the Coral Triangle global biodiversity hotspot (Hoegh-Guldberg et al., 2009; Barber et al., 2011; Burke et al., 2012), the distribution and diversity of coral reef associated decapod crustaceans are still poorly known. Studies on or including reef associated decapod crustaceans have focused on limited taxa, spatial patterns of abundance at high taxonomic levels or specific sites (e.g. Prabowo et al., 2021; Madduppa et al., 2021a). There are many knowledge gaps, even for taxa such as the blue swimming crab Portunus pelagicus which are economically important and have become prime fisheries commodities. Therefore, the aim of this study was to apply eDNA methods to assess the species diversity and distribution of decapods found in coral reefs across Indonesia, exploring patterns of diversity at various scales and implications for policy and action. In addition, economically important decapod species were assessed by Fisheries Management Area (FMA) with a special focus on the prime fisheries commodity Portunus pelagicus.
Material and methods
eDNA seawater sample collection
The eDNA seawater samples were collected from 40 sampling sites across Indonesia (Figure 1). Seawater samples were collected from the surface at a depth of about 1 meter. A 3L volume of each seawater sample was then filtered through a 0.45 µM Pall Corporation sterilized filter paper using a vacuum pump to draw the water through the filter. After the filtering process was complete, the filter paper was then cut into two halves using sterilised scissors. Each half was placed into a 2 mL cryotube filled with 1 mL DNA shield (ZymoBIOMICS DNA/RNA shield). Contamination was prevented through the strict sterilisation of all the sampling equipment used at each stage of the sampling procedure with a 30% solution of commercial bleach. On each sampling site, a negative control using sterilized ddh2o was used to filtering at the end of sampling section to monitor any contamination according to (West et al., 2020; West et al., 2022).
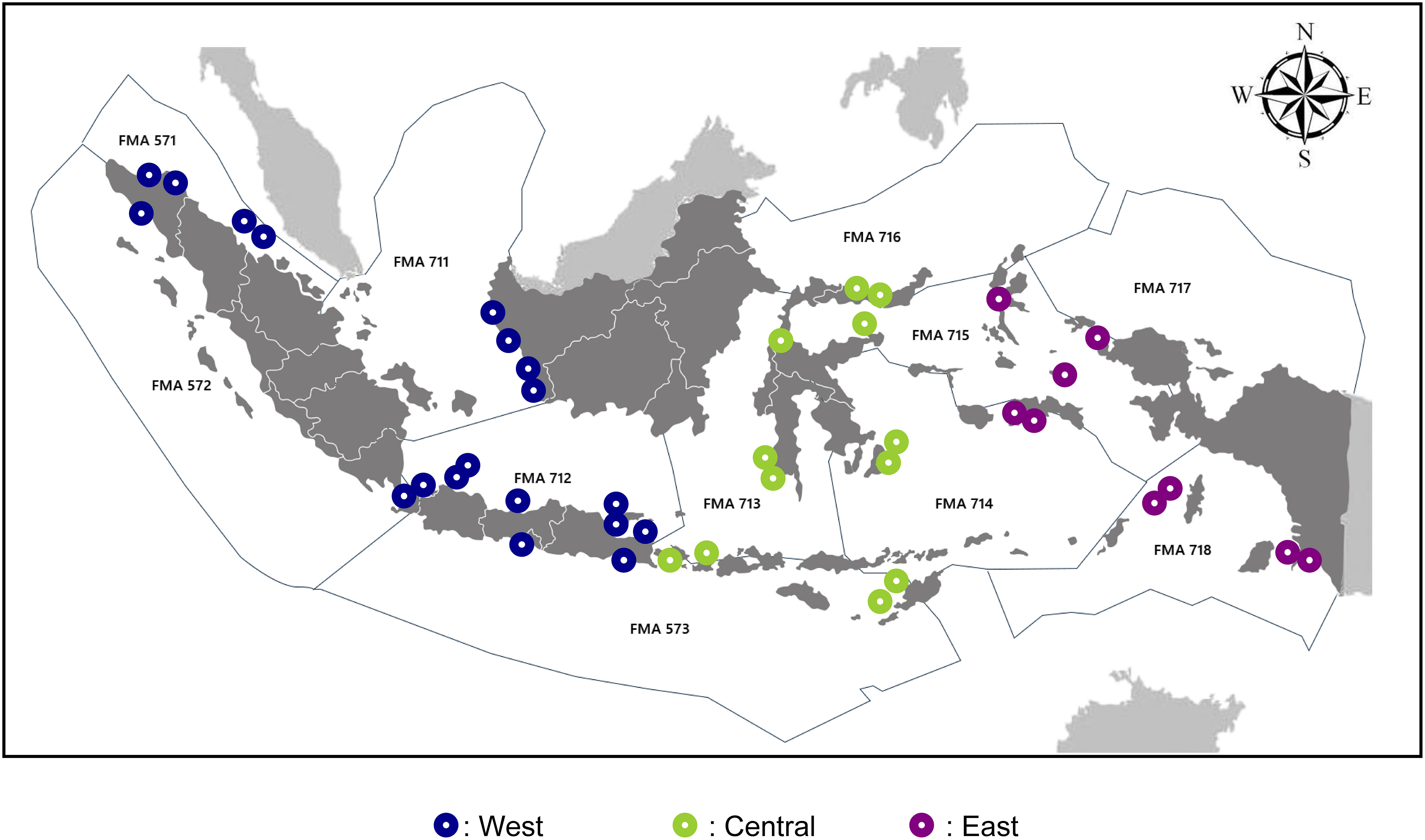
Figure 1 eDNA seawater sampling sites in coral reefs across Indonesia by region (West n = 19, Central n = 12 and East n = 9) and fisheries management area (FMA).
eDNA laboratory analysis, library preparation and next generation sequencing
The eDNA retained in the filter papers was extracted using Geneaid gSYNC™ DNA Extraction Kits following the manufacturer’s protocol. The first PCR amplified the target region using 16S rRNA MiDeca Primers (Forward and Reverse) (Komai et al., 2019) with connecting adapters: 5’-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG (forward sequence adapter) and 5’-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA G (reverse sequence adapter). The primers target a hypervariable region of the 16S rRNA gene (154 – 189 bp) which contains sufficient information to identify Decapods to taxonomic family, genus and species (Komai et al., 2019). To date, primer selection to identify organism based on eDNA metabarcoding by using the hypervariable regions are suitable targets given their sequence variation enables for strong taxonomic resolution in macroeukaryotes (Miya et al., 2015; Berry et al., 2017; Stat et al., 2017; Jeunen et al., 2019). The MiDeca itself is universal primer to amplify from 56 families, 126 genera, and 207 species (Komai et al., 2019). The first PCR reaction volume was 25 μl, consisting of 13 µL KAPA Hifi Hotstart Readymix, 1 µL each of 1 nM primers (Forward and Reverse), 4 µL ddH2O, and 7 µL DNA Template. The DNA amplification PCR profile stages included: (1) pre-denaturation of the template DNA at 95°C for 5 minutes; (2) denaturation of the template DNA at 98°C for 10 seconds; (3) annealing at 60° C for 10 seconds; (4) primary extension at 72°C for 10 seconds and (5) final extension (post extension) at 72°C for 5 minutes with 35 cycles of stages (2)-(4). Two negative controls (i.e. blank template) were used when running the 96 Universal peqStAR PCR machine (Peqlab Ltd, USA) in order to check for contamination. The PCR product quality was visualised through electrophoresis on 2% agarose gel (100 mL 1X TAE buffer and 2 g agarose) run at 100 Volts for 38 minutes. The results were visualized using UV fluorescence via an Alphaimager Mini Gel Documentation System (ProteinSimple Ltd, California, USA)
All PCR products which passed the electrophoresis quality control underwent a second PCR for indexing purposes. The IDT double index and Illumina sequencing adapter for Illumina - Nextera DNA Unique Dual Index, Set A (catalogue number 20027213) (Illumina, San Diego, USA) were added to the target amplicon in the second PCR, using 12.5 μl of Kapa HotStart HiFi 2 × ReadyMix DNA polymerase (Kapa Biosystems Ltd., London, UK) and 2 μl of PCR product. The PCR cycle comprised an initial denaturation at 95°C (3 minutes), then 9 cycles of 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. The first PCR and second PCR products were purified using AMPure XP beads (Beckman Coulter, Inc) before proceeding to the next step.
DNA sequencing was performed on an Illumina iSeq 100 using the standard reagent kit and cycles following the modified protocol of Illumina MiSeq 16S metagenomic sequencing library protocol. The concentration of each amplicon barcode library was assayed using a Qubit fluorometer and diluted to 10 nM before the libraries were pooled. The pooled library was diluted and denatured according to the Illumina MiSeq library preparation guide. Aliquots of 16 µL of the 40 pM amplicon library and 4 µL of the 60 pM PhiX Illumina version 3 control library were pooled as the final product. The Illumina iSeq v.2 Reagent kit for 2×150 bp PE was used with a run-time of about 18 hours and produced a Fastq file. The specific barcode index of the IDT double index and the Illumina sequencing adapter for Illumina - Nextera DNA Unique Dual Index were excluded during the process.
Bioinformatics and data preparation
All eDNA Fastq files were analysed using mBRAVE (Multiplex Barcode Research And Visualization Environment) (Ratnasingham, 2019). Every parameter described hereafter was retrieved from the mBrave platform and was available to the user (as last accessed in May 2021). For each mBRAVE run, the paired end merging of iSeq reads required a minimum 20bp overlap between the forward and reverse reads, while allowing up to 5 nucleotide substitutions. Primer sequences were removed from those merged reads, 20 bp was trimmed from the front of each read meanwhile 19 bp was trimmed from the end of each read, to ensure only the appropriate length of each sequence (~165 bp). Next, the data were filtered to remove sequences of lower average QV value than 20 and sequences shorter than 100bp. This filtering step allowed for a maximum of 2% nucleotides with >20 QV value and a maximum of 1% nucleotides with >10 QV value (Supplementary Figure 1).
Sequences fulfilling these criteria were dereplicated and clustered as Operational Taxonomic Units (OTUs) using a 2.5% similarity threshold. OTUs were taxonomically assigned using an initial 2% ID distance threshold to reference sequences of customized library databases from BOLD Systems. The publicly available BOLD (Barcode of Life Database) reference libraries for Decapoda, as well as a standard contamination reference database, were compared to all OTUs. The Project Analytical Parameters, as outlined above, are provided in Supplementary Figure 1. After selecting parameter values, mBRAVE automatically applied the same parameters to every run in the dataset. Each run generated a run summary in mBRAVE, which was checked to ensure it aligned with expectations for the dataset. These summaries (TSV file) included sequence length distribution, sequence reads, dereplicated sequences, GC composition distribution, run QV score distribution and BIN count vs. OTU count. All singleton reads were removed prior to analysis. To further assign all decapods in the dataset, we examined OTUs in the NCBI (National Centre for Biotechnology Information) GenBank database with high sequence similarity. For highly conservative taxonomic assignment, all decapod OTUs were identified as follows: similarity ≥80% identified to family; similarity 90≥-97% identified to genus; similarity ≥97% identified to species.
Data analysis
Prior to downstream analysis, the taxonomy and read table file were translated to the TaXonTableTools format using a custom python script (https://github.com/TillMacher/xml2 to TTT) (Macher et al., 2021). Species accumulation curves were used to compare the diversity of community data sets using rarefaction (Supplementary Figure 2). This method estimates the expected species richness (mean and standard deviation) by sampling site. The distribution of the number of reads, OTUs, and OTUs identified at species level by sampling site were visualised using bar charts. Venn diagrams showing the taxonomic overlap between sites across Indonesia were produced using an online program (http://bioinformatics.psb.ugent.be/webtools/venn/).
Boxplot were produced to represent alpha diversity (Shannon-Wiener Diversity Index, calculated manually) based on the number of OTUs identified to species level by region and province. Beta diversity, used to evaluate between site differences in read sequence composition, was visualised as a heatmap based on Jaccard-Distances using TaxonTableTools v1.3.0. Species occurrence across regions and sites was examined using ANOSIM. The distribution and relative abundance of economic species by Fisheries Management Area (FMA) was conducted using the ParCat plot routine. These analyses were conducted using TaxonTableTools (TTT) v1.3.0 through GitHub (https://github.com/TillMacher/TaxonTableTools) and using the Python package index (https://pypi.org/project/taxontabletools/) (Macher et al., 2021).
Results
Species distribution
A total of 3,401,773 paired-end reads were generated from the 16S rRNA amplicons obtained from the 40 samples collected from 40 coral reef sampling sites across Indonesia. The species accumulation curve (SAC) shows a linear increase in species richness between stations (Supplementary Figure 2). mBRAVE analysis yielded 406 OTUs across all sampling locations based on a 3% similarity threshold (Figure 2). Taxonomic assignment identified 51 species belonging to 47 genera and 32 families (Figure 3 and Table 1). Based on region, the highest number of families (25) was identified from the Central Indonesia region, while the highest number of genera (31) and species (36) were identified in the West Indonesian region. The East Indonesia region had the lowest taxonomic richness based on the families, genera, and species identified in this study. Figure 4 shows the decapod community composition by family and genus (based on read abundance) varied between sampling sites (aggregated by province). The Portunidae family contributed the highest percentage of read abundance at all sites, followed by Palaemonidae and the unassigned OTUs. At the genus level, Thalamita, Portunus, and Macrobrachium contributed the highest read abundance percentage, while a large proportion of OTUs were unassigned at the genus level, ranking fourth highest in terms of relative abundance. A comparison between the percentage of assigned and unassigned OTUs for all sampling sites in each province and region is shown in Figure 5. Overall, a high percentage of OTUs was unassigned to species level at all sites. The lowest percentages of unidentified OTUs were from the easternmost and westernmost sites, with very high unassigned OTU percentages at some sites in all three regions. The site with the highest percentage of unassigned OTUs was the Wakatobi, Southeast Sulawesi (Central region), followed by Raja Ampat (East region), and Banten (West region).
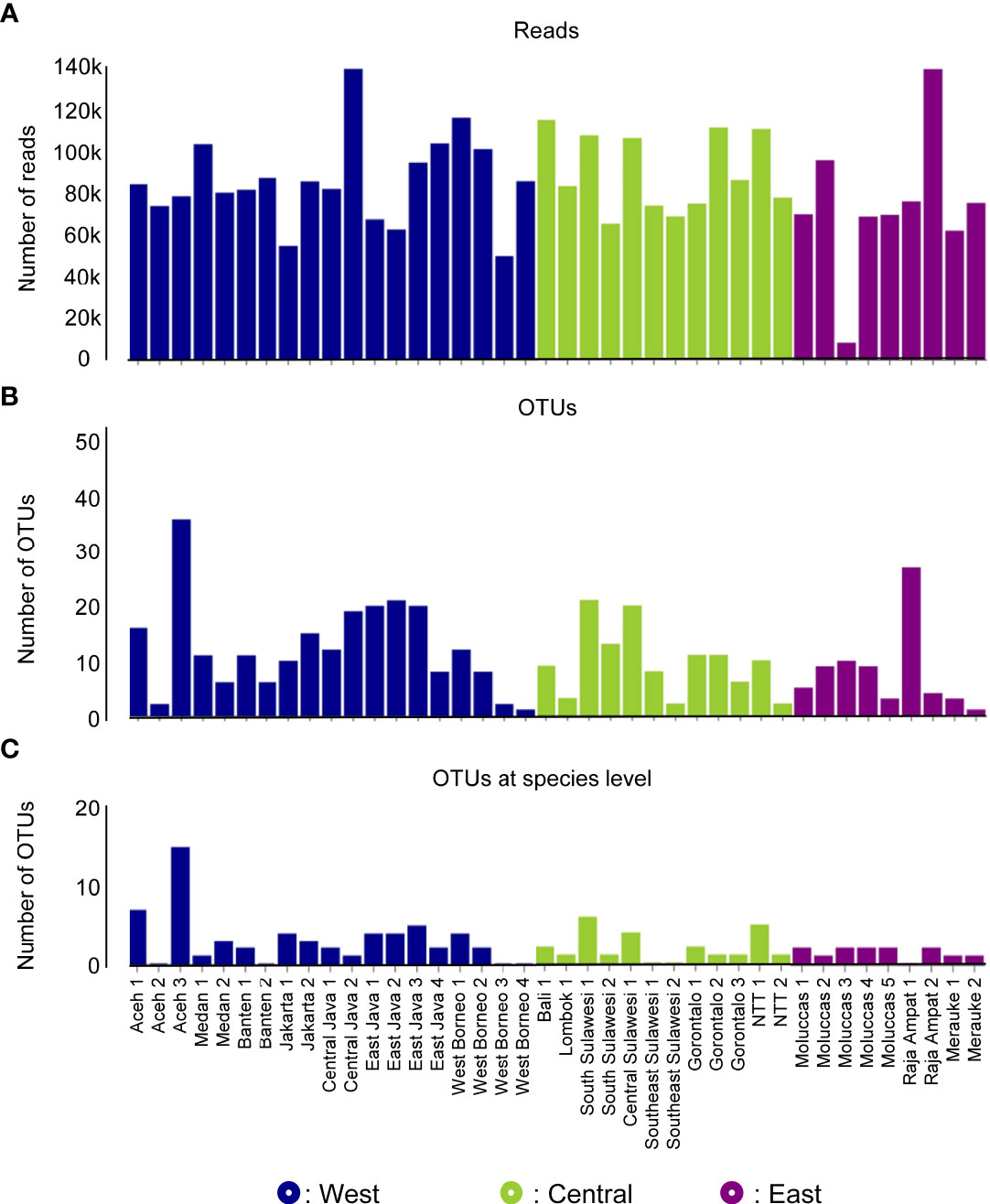
Figure 2 eDNA sequence data obtained by sampling site and region: number of reads (A), number of Operational Taxonomic Units (OTUs) (B), and OTUs identified to species level (C).
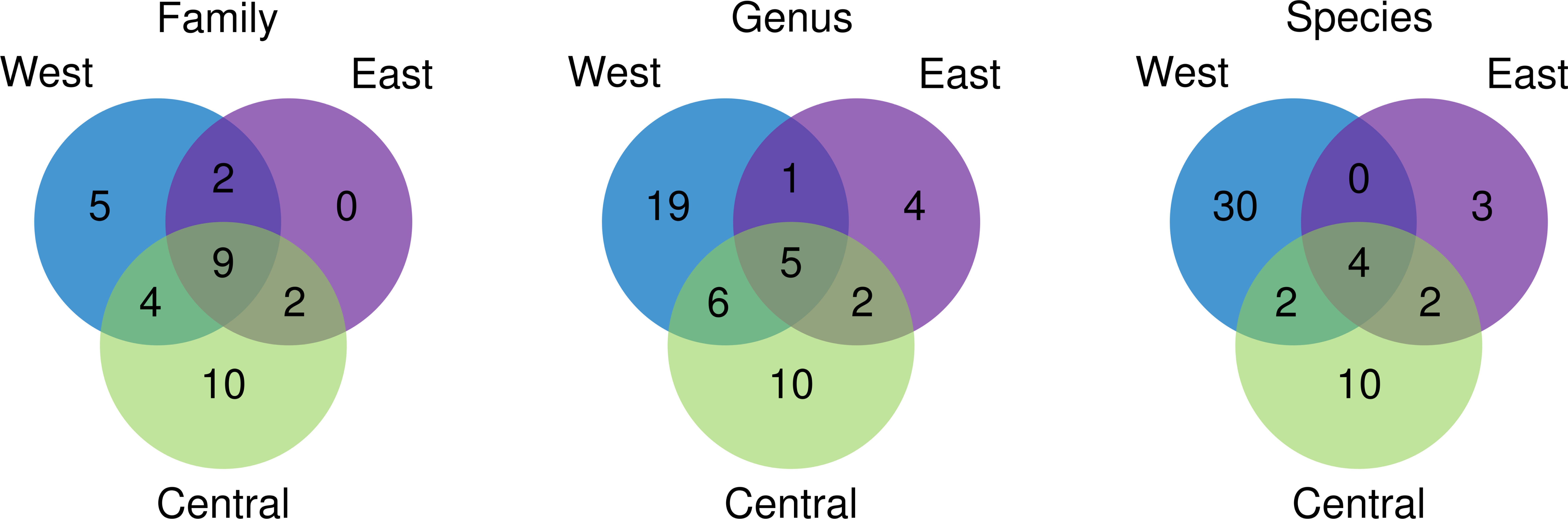
Figure 3 Venn diagram showing the taxonomic overlap of decapods identified from sites in West, Central, and East Indonesian regions at Family, Genus, and Species levels.
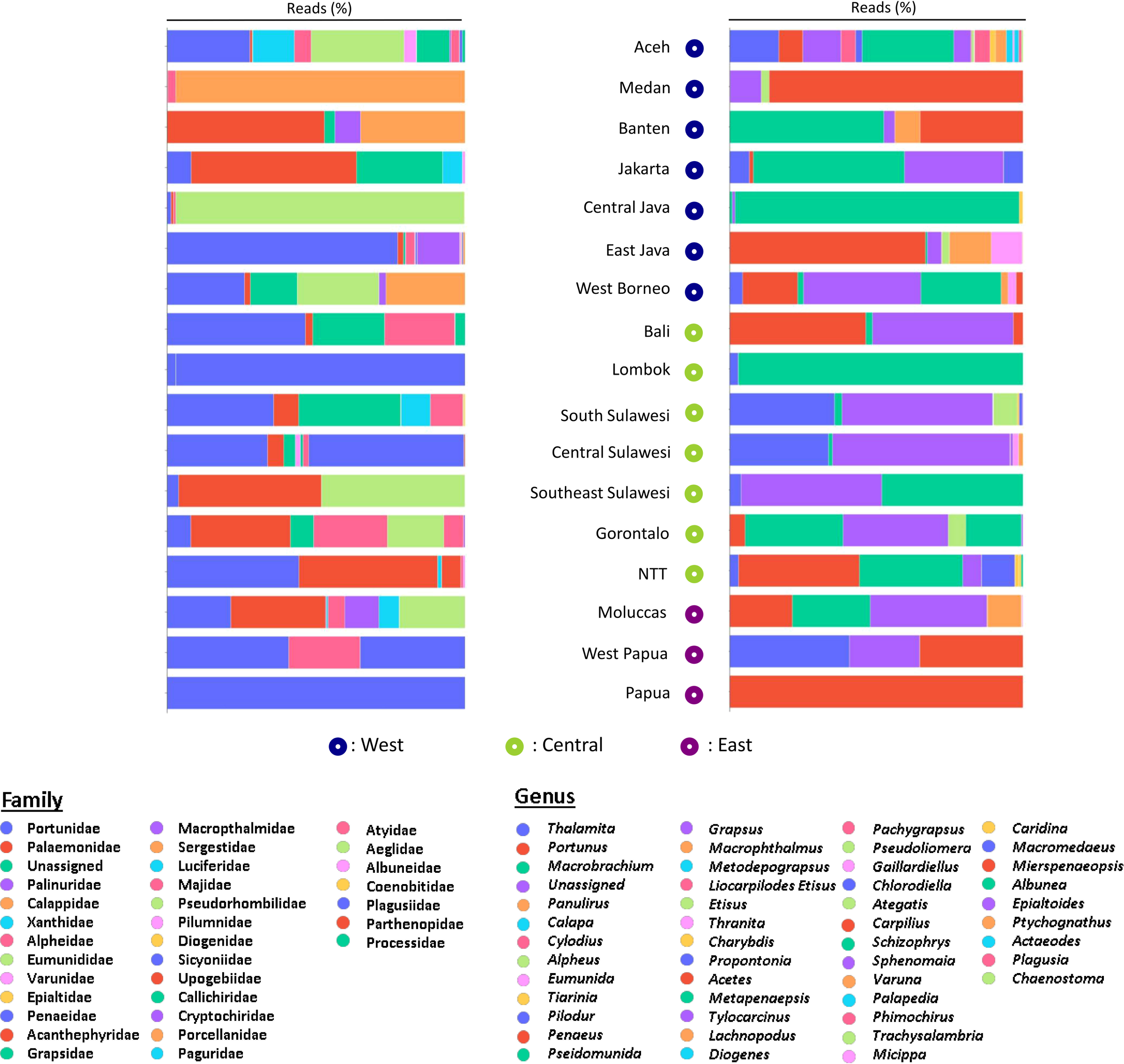
Figure 4 Relative abundance (based on percentage of reads) of decapod Families and Genera at sites in provinces in West, Central and East Indonesia (legends sorted from highest to lowest read percentage).
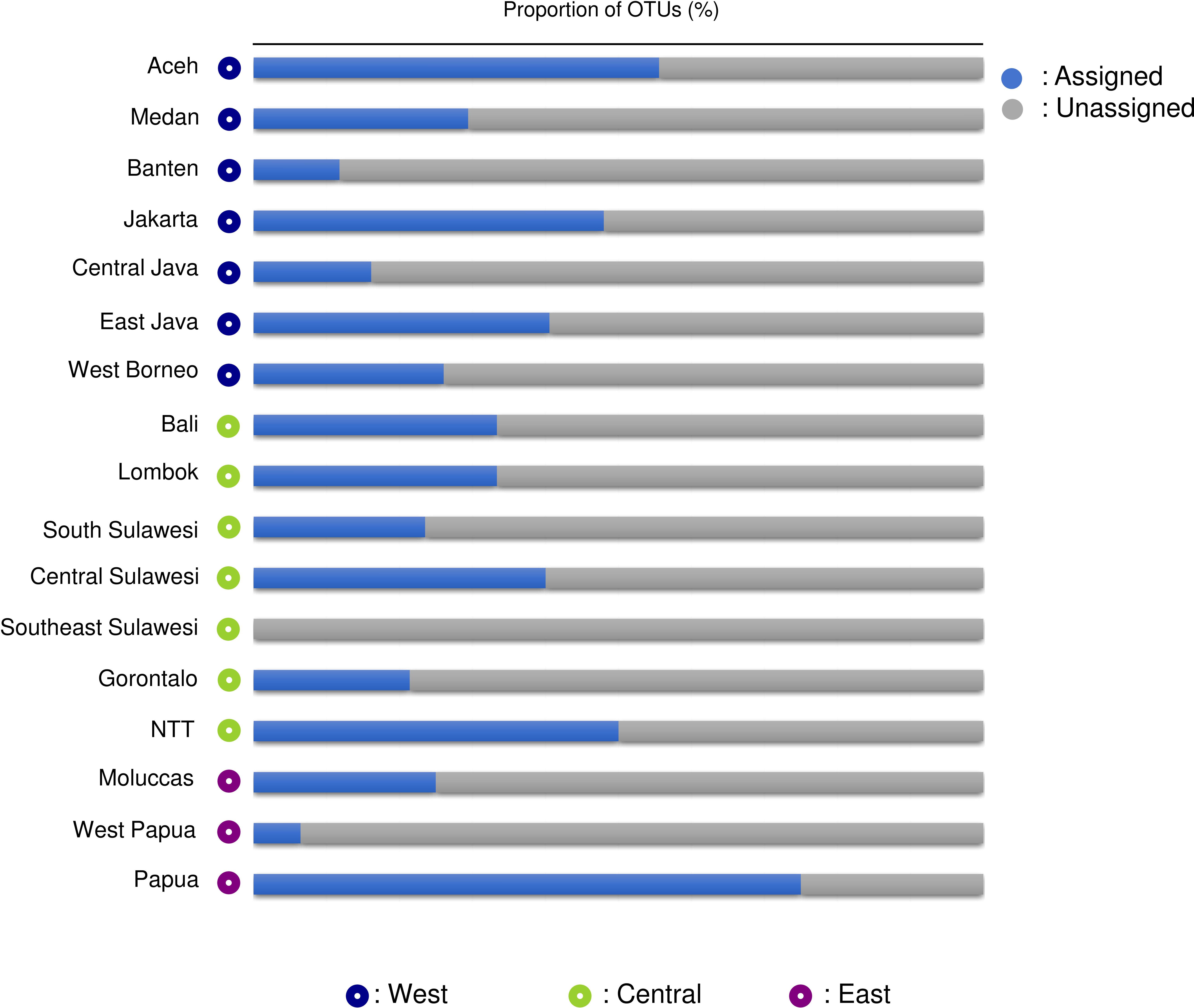
Figure 5 Bar chart showing the percentage of Operational Taxonomic Units (OTUs) assigned and unassigned to species level by province and by region (West, Central, and East Indonesia).
Species diversity
Species richness was used to calculate the alpha diversity of decapods by region and by sites aggregated by province (Figure 6). Species richness was highest in the West region followed by the Central and East regions (Figure 6A). The Shannon-Wiener Diversity index showed similar patterns of species richness (Figure 6B). Alpha diversity did not differ significantly between sites (ANOVA: p = 0.324) or regions (ANOVA: p = 0.406). There was also no significant difference in the Shannon-Wiener diversity index between regions (ANOVA: p = 0.336) or sites (ANOVA: p = 0.624).
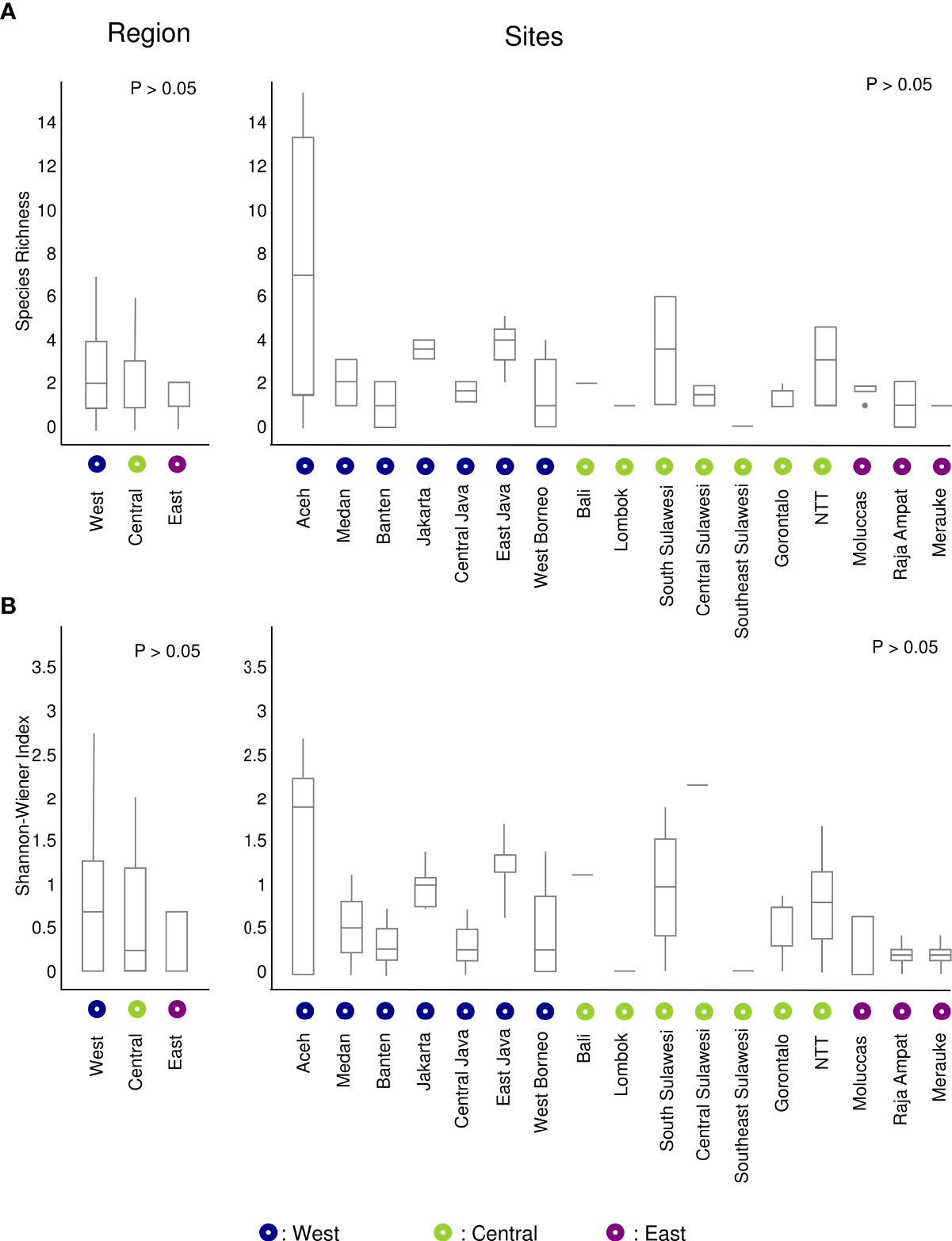
Figure 6 Boxplot representing alpha diversity based on the number of OTUs identified at species level: (A) Species richness, and (B) Shannon-Wiener Index.
Beta diversity was calculated as Jaccard-distances, shown as a heatmap (Figure 7). Jaccard distances close to 1 (yellow) indicate a high dissimilarity between sampling sites. The heatmap is dominated by yellow, with relatively few site pairs having lower dissimilarity (indicated by green and dark blue colours). The ANOSIM analysis found no significant difference in species occurrence between regions (R=0.10524, p = 0.308) but a significant difference between provinces (R=0.00644, p = 0.025).
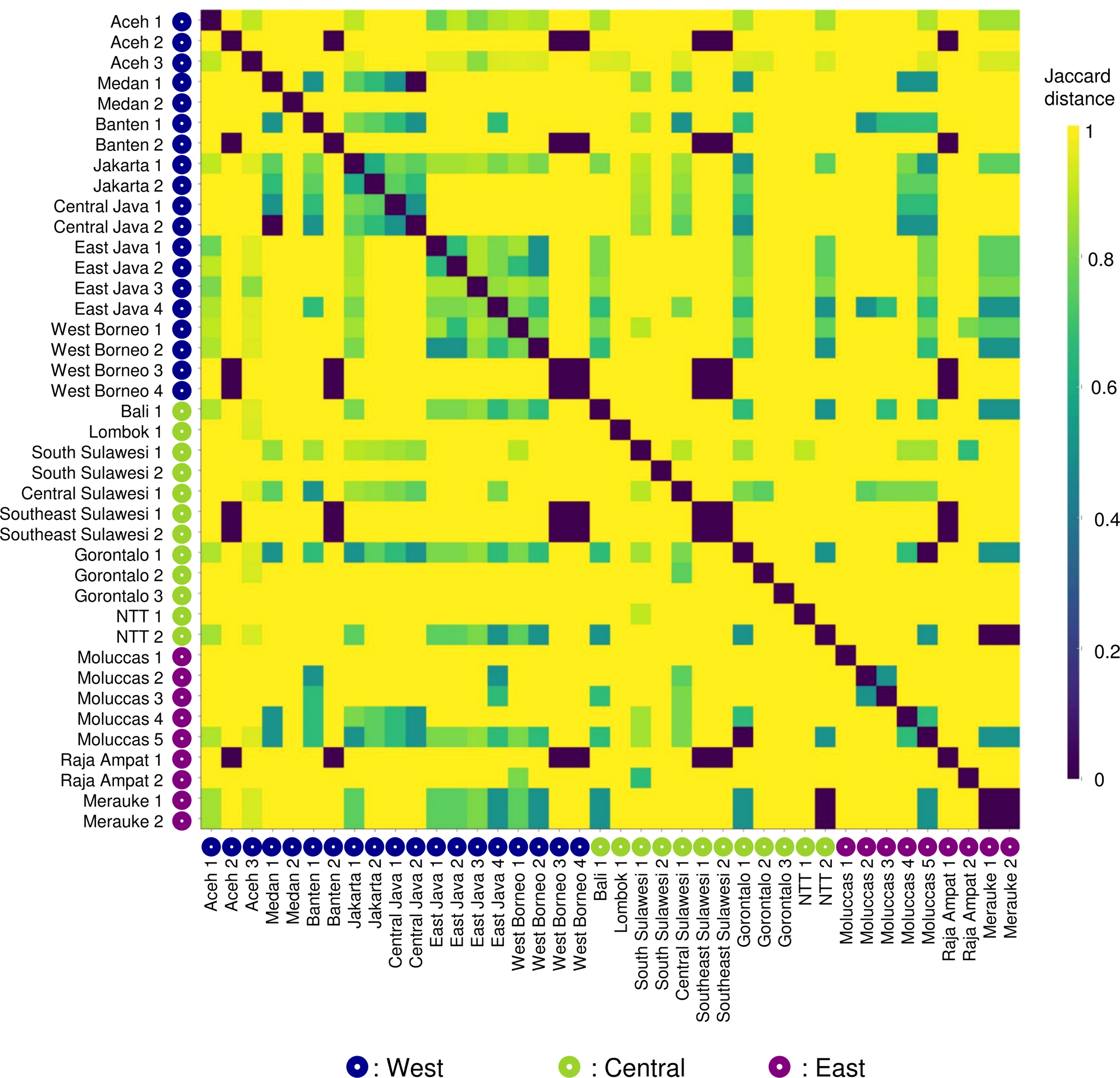
Figure 7 Heatmap analysis of beta diversity calculated as Jaccard distances and examined using ANOSIM analysis reported a non-significant difference in species occurrences between regions (R = 0.01115, p = 0.308) but significant between province differences (R = 0.16915, p = 0.025).
Conservation status
The conservation status for each identified species (Table 1) shows that the majority have not yet been evaluated (NE) based on IUCN Red List criteria, including fisheries target species such as Charybdis anisodon, Charybdis japonica, Macrobrachium nipponense, Mierspenaeopsis hardwickii, Panulirus stimpsoni, Portunus pelagicus, and Trachysalambria aspera. Based on the World Register of Marine Species (WoRMS) (Horton et al., 2017) and SeaLifeBase (Palomares & Pauly, 2021), a total of 23 species were categorised as native (NA), with records from Indonesia. A plurality of 31 species in Table 1 are considered as probably native (PN), based on their known distribution, despite the lack of records from Indonesia in SeaLifeBase or WoRMS databases. The remaining 12 species do not appear to have been recorded for Indonesia and, based on their respective recorded distributions, were classified as non-native or alien (AL).
Economically important decapod crustaceans
This study found seven species of economic importance (fisheries target species) among the 51 decapod crustaceans identified. These were: Charybdis anisodon, Charybdis japonica, Macrobrachium nipponense, Mierspenaeopsis hardwickii, Panulirus stimpsoni, Portunus pelagicus, and Trachysalambria aspera. In terms of distribution by FMA (Figure 8A), C. anisodon and C. japonica were found in FMA 712; M. nipponense in seven FMAs (571; 572; 573; 712; 713; 714; 716); M. hardwickii in FMA 711; and P. stimpsoni in four FMAs (572; 573; 713; 718). P. pelagicus was found in eight FMAs (the exceptions being FMA 715 and FMA 573), and T. aspera in FMA 571. The relative abundance of these fisheries target decapod species (Figure 8B) shows that C. anisodon, C. japonica, M. hardwickii, and T. aspera were dominant in the one FMA in which each species occurred; M. nipponense was most prominent in FMA 714 followed by FMA 712; and P. stimpsoni was dominant in FMA 572. Meanwhile, P. pelagicus was most prominent in FMA 714 followed by FMA 718 and FMA 711.
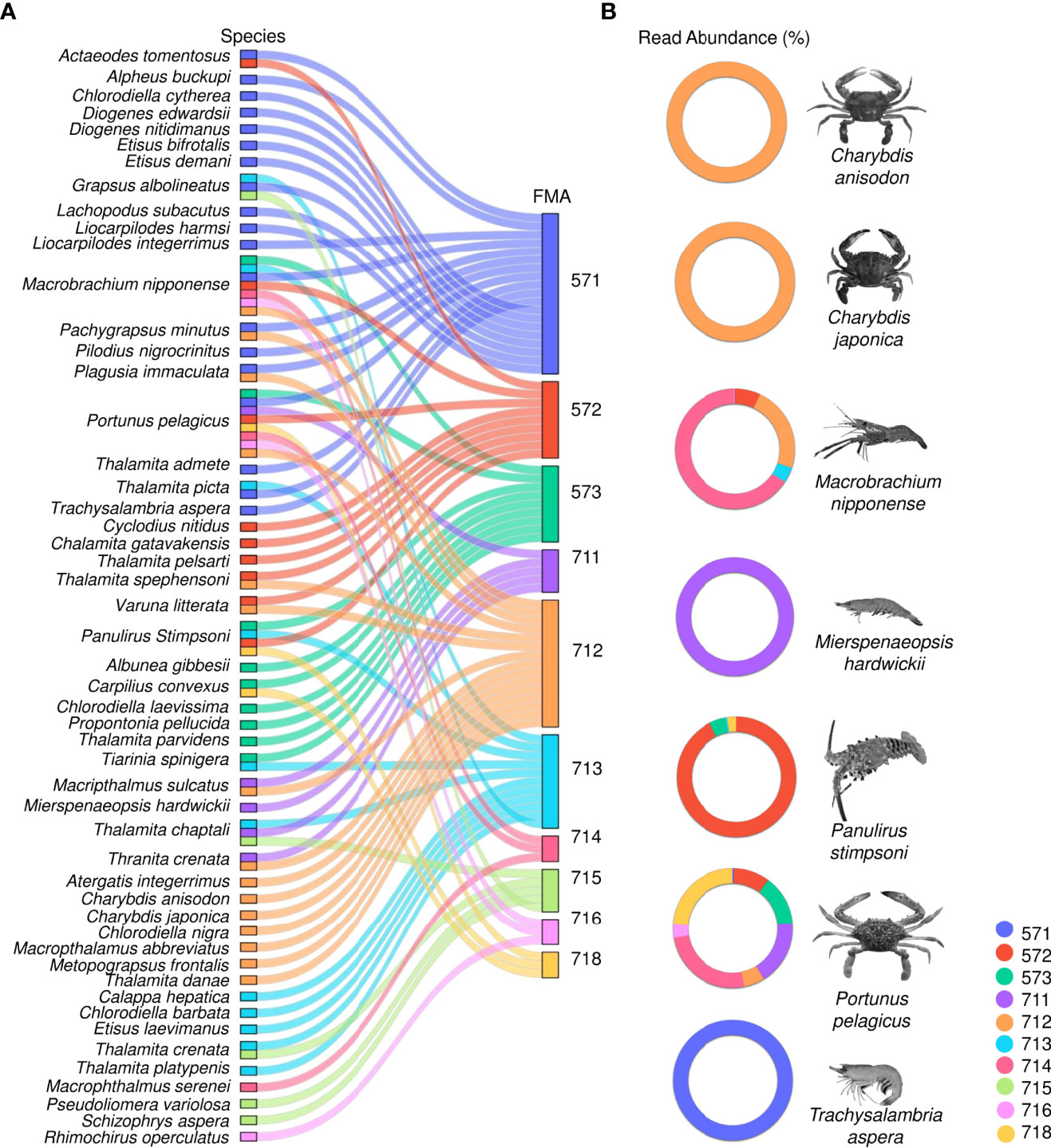
Figure 8 Parcat graphs showing: (A) the taxonomic distribution by Fisheries Management Area (FMA) of all decapod species identified from eDNA; and (B) the relative abundance (read percentage) by Fisheries Management Area (FMA) of the seven economic (fisheries target) decapod species identified.
Discussion
Species distribution and diversity
The current study has successfully mapped hotspots for Decapod crustaceans across Indonesian coral reefs through eDNA metabarcoding using 16s rRNA gene. The 16s rRNA od MiDeca used as selected primer in this study based on universality to amplify wide taxa of Decapods. This result was certainly obtained using universal primers to amplify Decapods with about 20 bp of conservative region from 154-184 bp of target region from 56 families assigned by MiDeca primers (Komai et al., 2019), quite enough interspecific differences for all targeted taxa. The Xanthidae was a dominant family with a total of 30 species followed by Portunidae with 15 species. A similar dominance of these two families was also found in an eDNA metabarcoding study conducted by West et al. (2020) in the eastern Indian Ocean. The Xanthidae crab family consists of gorilla crabs, mud crabs, pebble crabs, and rubble crabs (Integrated Taxonomic Information System) (www.itis.gov). This finding also is also in line with the results of traditional taxonomic surveys, as the Xanthidae is the family with the most species described to date, comprising at least 572 species in 133 genera split into thirteen subfamilies (De Grave et al., 2009). The 15 families identified in all regions (West, Central, East) represent almost half of all families identified in this study. This relatively uniform distribution of many families raises the possibility that for some genera and species (included non-identified taxa) the distribution may also range across all three regions.
In this study, the eastern Indonesia region had the lowest taxonomic richness based on identified families, genera and species, which is contrary to the general perception that this area has the highest marine biodiversity (McKenna et al., 2002; Mangubhai et al., 2012; Purwanto et al., 2021). There could be several reasons for this unexpected finding. One reason is that there were fewer sites in this region compared to the central and western regions. Another is the high number of Decapod sequences unassigned even at the family and genus levels. In general, identification using a genetic approach such as DNA barcoding relies on the species of interest having been sequenced for the genetic marker used and the sequences deposited in an accessible repository with the correct metadata (Hebert & Gregory, 2005; Hubert et al., 2015). In Indonesia, genetic studies to obtain DNA sequences as a means of detecting species and characterizing species have not yet been conducted very often, especially for Decapod crustaceans. The significant finding of a high percentage of unassigned OTUs of Decapod crustaceans in the Indonesia archipelago reflects the results of studies on other taxa. For example, a high proportion of unidentified taxa are reported from eDNA metabarcoding analysis of Indonesian marine bony fish and elasmobranchs as well as invertebrates, in particular molluscs and echinoderms (Andriyono et al., 2019; Juhel et al., 2020; Madduppa et al., 2021; Moore et al., 2021).
The ability of eDNA metabarcoding has been proven to identify such as wide scale of organisms including introduce marine species (Huhn et al., 2020). Environmental DNA (eDNA) methods in detecting the presence of invasive species are currently gaining interest as a comprehensive approach method in ecological investigations (Cristescu & Hebert, 2018). As shown in this current study, eDNA metabarcoding reveals 51 Decapods species that then classified native (NA), endemic (EN), possibility native (PN), and alien (AL) species. The MiDeca primers used in this study have been proven capable of identifying most decapod groups (Komai et al., 2019), and the high number of unidentified or unassigned taxa means that it is likely that sequences for many of the species uncovered in this study are not yet present in worldwide databases (i.e Genbank NCBI). As an example of the paucity of reference sequences, an examination of the coverage for congeners of two OTUs identified as species which are not native to Indonesia is instructive. Purposive or accidental anthropogenic introductions of alien crustacean species are a growing problem around the world (Galil et al., 2011), and some of the 12 species classified as alien (AL) in Table 1 may indeed represent instances of introduced species. Cases of introduced crustacean species have been reported globally (Bezeng and van der Bank, 2019; Bojko et al., 2020; Box et al., 2020) and within Indonesia (Maulina et al., 2020). However, both of the following examples seem more likely to be cases of mistaken identity due to incomplete sequence databases than records of introduced alien species.
The first example, from the Family Albuneidae, was detected at just one site: Rote Island (NTT 1) in Central Indonesia and FMA 753. According to SeaLifeBase (Palomares & Pauly, 2021), the surf mole crab Albunea gibbesii is native to the Atlantic Ocean. Of seven congeners with a distribution known or likely to include Indonesia (Albunea elioti, A holthuisi, A. lucasia, A. microps, A. speciosa, A. symmista, A. thurstoni), only one (A. symmista) has a 16S reference sequence deposited in the NCBI GenBank database. There are therefore at least six closely related species which can be considered a priori quite likely be misidentified as A. gibbesii.
The second example is the Family Alpheidae, with 21 unique OTUs each identified from just one site. These 21 sites represent 9 provinces and 8 FMAs spread across all three regions. Eight of these 21 OTUs were identified to genus level as Alpheus, from four sites in three provinces (East Java, Gorontalo, and Medan) and three FMAs (572, 712, and 716) in West and Central Indonesia. One of these eight OTUs (from Belawan, site Medan 2 in FMA 572, West Indonesia) was identified to species level as the snapping shrimp Alpheus buckupi, a described shrimp (Almeida et al., 2013) native to the Caribbean. The Alpheidae and the genus Alpheus are the second most speciose shrimp family and genus within the Caridea (De Grave & Fransen, 2011). Out of at least 287 Alpheus species (De Grave and Fransen, 2011; Almeida et al., 2013), 187 are described in SeaLifeBase (Palomares & Pauly, 2021), of which 108 are known or likely to be found in Indonesia. However, just 60 species have 16S accessions in the NCBI GenBank, 39 of which are also in SeaLifeBase. Of these, 16 have known distributions which are likely to include Indonesia. Therefore, there are at least 92 (probably substantially more) closely related species (congeners) likely to be found in Indonesia and could reasonably be misidentified as A. buckupi in the absence of conspecific sequence records.
These examples and similar cases for other higher-level taxa identified in this study highlight the extent or scale of the need for barcoding of Indonesian decapods, just to cover currently recognised species. Furthermore, new species are being described across the Indonesian Archipelago and nearby regions (e.g. Ng & Lukhaup, 2015; Spiridonov, 2017), while species ranges are also updated (e.g. Wahyudin et al., 2016), adding to the list of species records for Indonesia. The alpha and beta biodiversity results should be considered tentative and likely an underestimation of true biodiversity, as a high proportion of decapod OTUs were not identified to species, genus or even family level. However, the low overlap in OTUs revealed in the Venn diagram (Figure 3) indicates that the beta diversity analysis (Figure 7) is a valid reflection of relatively fine scale (site, province) differences in species occurrence and decapod community composition, obscuring or precluding a clear pattern at a higher regional scale (i.e., West, Central, and East Indonesia).
The findings of this study highlight the need for scientists and policy makers to work together to improve the genetic biodiversity database for this region, and develop an integrated biodiversity monitoring system as advocated by Kühl et al. (2020). This requires human resources in traditional taxonomy as well as molecular biology disciplines, while long-term safe repositories at the national (and possibly sub-national) level are needed for reference specimens as well as for sequence data and metadata, alongside increased participation in regional and global initiatives such as the Diversity of the Indo-Pacific Network (DIPnet) and Genomic Observatories Metadatabase (GEOME) (Deck et al., 2017; Riginos et al., 2020). The vital importance of these systems and facilities was pointed out by Hebert and Gregory (2005), and the need for such capacity at national and sub-national level, as well as international/global levels, has been highlighted during the current pandemic-induced era of restricted travel as well as increasing restrictions on the movement of biological material such as specimens and samples. Properly curated, eDNA data such as that produced by this study can have value as a historical record, and enable more in-depth studies as reference sequences for unidentified OTUs become available.
Conservation status and management of decapod crustaceans
The conservation status data in Table 1 show that only one of the decapods identified from sites across Indonesia has been evaluated under the IUCN Red List criteria (IUCN, 2012; IUCN, 2019). The Chinese spiny lobster Panulirus stimpsoni is listed as Data Deficient (DD) (Cockcroft et al., 2011) with a distribution which does not include Indonesia. As the valuable rock lobster genus Panulirus is well represented in the NCBI GenBank, in particular in terms of 16S sequences, this may well be a valid first record for Indonesia, although the possibility of confusion with a congener cannot be discounted. Ardura (2018) compared the availability of all genes in the NCBI GenBank then found that 16S rRNA accessions compromised 10.20% of all gene sequences deposited. Conducting studies using eDNA metabarcoding tools with the 16S rRNA gene could support the development of decapod databases, especially in mega biodiversity regions such as Indonesia. The 16S rRNA gene has been recommended as the molecular marker of choice for Decapod biomonitoring with eDNA metabarcoding (Komai et al., 2019; West et al., 2020). However, with respect to conservation status, a search of the IUCN Red List portal (IUCN, 2021) reveals that only a small percentage of Indonesian decapods have been evaluated to date. Just 13.6% of all crustaceans evaluated are marine, of which at most 33% (135 species, 4.5% overall) might be present in Indonesia, not all of which are decapods.
It is increasingly recognised that biodiversity is essential for sustainable development and human well-being, as illustrated by the attention paid to biodiversity in the context of the Sustainable Development Goals (Diz et al., 2018; Friedman et al., 2018; Recuero Virto, 2018; Rees et al., 2018) and the inclusion of Biodiversity as one of nine planetary boundaries for sustainable development (Rockström et al., 2009; Steffen et al., 2015). In this current study, the alpha and beta diversity calculated are based on known indified taxa in addition to notable record of decapod diversity in several location across Indonesia seas. Alpha diversity and Shannon-Wiener diversity showed no significant difference between provinces and regions. However, the ANOSIM did reveal significant between province differences in beta diversity. This is reflected in Figure 8A, which illustrates the geographical spread at FMA level of each of the species identified. These differences have a clear implication for conservation and biodiversity management as well as fisheries. Under the current regional autonomy paradigm (Act 23/2014), the coastal waters from 0-12NM offshore are predominantly managed by provincial governments, with the exception of some strategic national interests (Ambo-Rappe & Moore, 2019). This includes marine conservation area management as well as many aspects of fisheries and marine resource management more generally. The specificity of crustacean decapod communities indicates that province or site (e.g. MPA) based management can be appropriate for many but not all taxa. Similarly, for some taxa management at the larger FMA spatial level appears appropriate, in particular the blue swimming crab Portunus pelagicus. It should be noted that some provinces divided between several FMAs and all FMAs comprising waters under several provincial as well as national jurisdictions (Figure 1, Table 2). Meanwhile under the Ecosystems Approach to Fisheries Management (EAFM) paradigm adopted by Indonesia, conservation areas are considered important, especially with respect to the ecosystem and habitat domain (Nadiarti et al., 2021). However, both provincial level and FMA level management systems and implementation are mostly in development. Biodiversity data on taxa of economic and/or ecological importance such as decapod crustaceans could and should inform this development. In particular, baseline data can provide a basis for monitoring and evaluation of management success.
Economically important decapod crustaceans
eDNA metabarcoding has been utilized as a biomonitoring method to find marine creatures as well as commercially important species. Economic or commercially fish several times unravel in eDNA samples found in marine habitat (Miya et al., 2015; Closek et al., 2019; Madduppa et al., 2021; Gelis et al., 2021). Not only have fish creatures been detected, but other commercial species have also been detected in the marine Decapods group in eDNA samples as shown by West et al. (2020). Meanwhile, this current study was successful in demonstrating the detection of economic species of Decapods in eDNA samples collected. The economically important decapod crustaceans identified in this study included three crabs (Charybdis anisodon, Charybdis japonica, Portunus pelagicus), a freshwater prawn (Macrobrachium nipponense), a lobster (Panulirus stimpsoni) and two penaeid shrimps (Mierspenaeopsis hardwickii and Trachysalambria aspera). The fishing of crabs in Indonesia is mostly carried out by small-scale fishermen using boats less than 10 GT, ranging from small canoes to vessels with inboard engines; in addition, crabs can be retained bycatch in other fisheries (Madduppa et al., 2016). Crabs and other crustacea can also be collected in multi-species gleaning fisheries as well as targeted fisheries (Blankenhorn, 2008). While some blue swimming crabs Portunus pelagicus go directly to small processing plants (Madduppa et al., 2016), many of the crabs caught by small-scale fishermen are landed at sites scattered along the coasts of the Indonesian Archipelago and are generally purchased from the fishermen by small-scale crab or general fisheries produce collectors/traders. Crabs are mostly caught with nets and traps in many areas across Indonesia and various types of trawl gear can pose a threat to non-target crustaceans (including crabs) as well as target stocks (generally shrimp or swimming crabs) and ecosystems (Hamid et al., 2020; Suherman et al., 2020). In most areas of the country trawls are forbidden, but the rules are often controversial, not always enforced, as illustrated by a case study in West Kalimantan (Nadiarti et al., 2021). In East Java, at the time of writing, the ban had been suspended for a type of trawl called cantrang.
Four of these species were identified from just one FMA, indicating that FMA-based management might be appropriate. The crabs Charybdis anisodon and C. japonica were only found in FMA 712, each in one province in West Indonesia, although they co-occurred in this FMA with the widespread blue swimming crab P. pelagicus and the oriental river prawn Macrobrachium nipponense. According to SeaLifeBase (Palomares & Pauly, 2021), these two swimming crabs have wide distributions, with C. anisodon reported from Indonesia and C. japonica reported from several countries in Southeast Asia including Malaysia. This distribution makes it likely that C. japonica is native to the Java Sea, including Probolinggo, the site in East Java where this species was identified. As a commercial fisheries species, C. anisodon is considered of lower economic value than P. pelagicus (Hamid & Wardiatno, 2018) but is frequently caught in fisheries targeting P. pelagicus in areas as far apart as Southeast Sulawesi, Indonesia (Hamid et al., 2020) and Tanzania (Chande & Mgaya, 2003).
Widespread globally and within Indonesia, predominantly freshwater decapods of the genus Macrobrachium include several obligate or landlocked species as well as amphidromous species with a marine larval stage (Wowor et al., 2009) and some fully marine species (Liu et al., 2007). An amphidromous life history can be conducive to dispersal between freshwater catchments and even landmasses (McDowall, 2007), and hence extensive distribution ranges such as that of M. rosenbergii, a valuable fisheries commodity found across the Indo-Pacific (Palomares & Pauly, 2021). The amphidromous life history of many species indicates the possibility of introductions via ship ballast water, where subsequent spread of any introduced populations could occur through amphidromy and/or ballast water. The Sibolga Expedition found or described 30 species of Macrobrachium from Indonesia (Holthuis, 1950) while a recent study on Macrobrachium phylogeny included 55 species (Jose & Harikrishnan, 2019). There are currently 111 Macrobrachium nominal species with 16S sequence accessions in the NCBI GenBank repository (accessed on 4 September 2021). Macrobrachium reported from Indonesia include species considered native and introduced, with 16S sequence accessions for at least 12 species (Wowor et al., 2009; Aprila et al., 2020; Jurniati, 2021; Maulina et al, 2020; Nursyahran et al., 2021). However, the evolutionary history and taxonomy of this genus are complex (Wowor et al., 2009; Siriwut et al., 2021), and here is some evidence for cryptic species and a lack of power in distinguishing taxa at the species level, especially based on single molecular markers (Siriwut et al., 2021).
The oriental river prawn Macrobrachium nipponense is a predominantly amphidromous (Liu et al., 2007; Wowor et al., 2009) Asian prawn (Jose & Harikrishnan, 2019). According to SeaLifeBase (Palomares & Pauly, 2021), M. nipponense has a reported native distribution of Japan and Malaysia, while countries listed as having introduced populations include the Philippines and Singapore. Assuming a correct species level identification, it seems likely this species is native to Indonesia, inter alia due to the widespread occurrence across all three regions, 7 FMAs and 8 provinces. In addition to this one identified species, the vast majority of OTUs assigned to the Family Palaemonidae were also assigned to the genus Macrobrachium with OTUs unassigned at genus but not species level present in all three regions, all 11 FMAs and 14 out of 17 provinces. One species from one other genus was identified (Propontonia pellucida), with OTUs assigned to family level only in all three regions, 9 FMAs and 13 provinces. These results indicate a need for taxonomic research on the Palaemonidae, especially the genus Macrobrachium, in Indonesian waters. In terms of conservation management, the amphidromous lifestyle typical of this genus reinforces the importance of maintaining upstream-downstream connectivity, for example in the context of dams for irrigation, water supplies and electricity generation (Jarvis & Closs, 2019).
Spiny or rock lobsters of the genus Panulirus are valuable fisheries commodities wherever they occur around the world, including Indonesia (Milton et al., 2014; Wahyudin et al., 2016; Teteleptal et al., 2017; Priyambodo et al., 2020). In this study, only one species, the Chinese spiny lobster Panulirus stimpsoni, was identified to species level, with no other OTUs assigned to the Family Palinuridae. It is unclear whether this species is in fact native (indicating an extension to the known range) or introduced. One reason for this doubt is that juvenile Panulirus sp. are (or have been) widely traded for grow-out, often at the puerulus stage (Priyambodo et al., 2020), where species identification may be problematic. Panulirus stimpsoni was identified in West, Central and East Indonesia, from four provinces: Banten, FMA 572; East Java, FMA 573; Central Sulawesi, FMA 713; and the Moluccas, FMA 718. The SeaLifeBase distribution is limited to four countries/territories: China, Hong Kong, Taiwan and Thailand (Palomares & Pauly, 2021). At least eight species are reported from Indonesia: Panulirus homarus, P. ornatus, P. penicillatus, P. longiceps, P. polyphagus, P. versicolor, P. daypus, and P. femoristriga (Wahyudin et al., 2016; Teteleptal et al., 2017; Setyanto et al., 2019). Reports of declining stocks (Teteleptal et al., 2017) indicate a need for sustainable fisheries management of this genus. As pointed out by Setyanto et al. (2019), the first step is to identify the species present and their respective distributions. In this context, this study indicates a ninth spiny lobster species may be widespread across Indonesia.
Two penaeid shrimps species were identified in this study, each from one site/province in West Indonesia: Mierspenaeopsis hardwickii from West Kalimantan, FMS 711 and Trachysalambria aspera from Aceh, FMA 571. Both species have a wide Indo-Pacific distribution including Indonesia, with Mierspenaeopsis hardwickii listed under the superseded synonym of Parapenaeopsis hardwickii in SeaLifeBase (Palomares & Pauly, 2021). In addition, OTUs assigned to the Family Penaeidae, most of which were assigned to the genus Penaeus, were found in all three regions, 10 provinces and 8 FMAs. These data indicate that this shrimp family is commonly found in or associated with coral reef ecosystems across Indonesia. Penaeid shrimps are a major fisheries commodity in Indonesia and heavily fished using legal and illegal gears (Suherman et al., 2020). However, despite their economic and ecological importance, this study reveals a lack of reference sequences as a basis for taxonomic identification based on molecular biology, in particular eDNA methods.
Implications for crustacean fisheries sustainability: Case study on the blue swimming crab Portunus pelagicus
The blue swimming crab (Portunus pelagicus), the most widespread fisheries species identified in this study, is an important Indonesian marine fisheries commodity with great social and economic significance. With an estimated export value of more than 300 Million USD, the majority of crab landings in Indonesia are processed domestically and exported to international markets, with over 80% of the production shipped to the United States (APRI, 2020). The fishery employs around 90-100 thousand fishermen and around 180 thousand workers (mainly women) in processing plants (APRI, 2020, Madduppa et al., 2021a). The third most valuable fishery in Indonesia, blue swimming crab products are exported to countries that demand sustainability, while international consumer demand for seafood products that adhere to the principles of ethical and good fishing practices should also encourage crab conservation. As an example, In the United States of America (USA), the Food Safety Modernization Act of 2011 allows the Food and Drug Administration (FDA) to require a food product traceability system. The increasing demand and high level of exploitation make it vital to manage the blue swimming crab stocks effectively, ethically and sustainably; however signs of overfishing and serial depletion have been reported (Madduppa et al., 2016). Efforts to date include the development of systems and capacity to meet and comply with the sustainability and traceability standards used by the destination countries (Madduppa et al., 2016; APRI, 2020) as well as the delineation and mapping of stocks in six of the eleven Fisheries Management Areas (FMAs) of Indonesia (Madduppa et al., 2021a).
Regulations on allowable catch size and berried females have been issued for several crustacean species, including mudcrabs (genus Scylla), lobsters (Palinuridae) and the blue swimming crab (BSC) Portunus pelagicus (Saputra, 2020). On 19 January 2015, the Ministry of Marine Affairs and Fisheries (MMAF) announced two regulations relevant to BSC fisheries. Ministerial Decree 1/2015 concerning Catching Spiny Lobster (Panulirus spp.), Mud Crab (Scylla spp.), and Blue Swimming Crab (Portunus pelagicus spp.) set a minimum harvest size of 10 cm carapace width for BSC and mandated that egg bearing (berried) female crabs be released and returned alive to the sea. This regulation was replaced by Ministerial Decree 56/2016 on the Prohibition of Taking and/or Exporting Lobsters (Panulirus spp.), Mud crabs (Scylla spp.), and Blue Swimming Crabs (Portunus pelagicus) from Indonesian waters, with similar provisions: crabs may not be retained and/or exported if they are carrying eggs and/or the carapace width is less than 10 cm, and/or or the body weight is less than 60 g. These regulations were revised in 2020 (12/PERMEN-KP/2020) and 2021 (17/PERMEN-KP/2021); however, the rules applying to the BSC remain similar to the previous regulations. Meanwhile, Ministerial Decree 2/KepMen-KP/2015 on the Prohibition of Using Trawls and Seine Nets in Indonesian Fisheries Management Areas affected some fishers but should have reduced by-catch of BSC and related species as well as avoiding habitat damage, and Ministerial Decree 18/KepMen-KP/2021 provides further regulations on the use of fishing gears and fishermen/fishing vessels operating outside their home province or FMA. For blue swimming crab (BSC) fisheries in Indonesia, the Indonesian government established a species-based Fisheries Management Plan (RPP) through Decree of the Minister of Marine Affairs and Fisheries of the Republic of Indonesia Number 70/KepMen-KP/2016 concerning the Management Plan for Crab Fisheries in Indonesian Fisheries Management Areas (MMAF, 2016).
While ministerial regulations are a positive step to support the sustainability of BSC, their impact depends on effective implementation in the field. Despite these advances at the policy level, challenges are still faced in the management of Indonesian blue swimming crab resources at the upstream and downstream levels of the crab industry. Lack of law enforcement for illegal fishing activities, unreliable data on the condition of crab stocks, socio-economic conditions of fishermen which result in low participation in sustainable fisheries practices, and limited attention from the industry to addressing the problems are all issues requiring urgent attention (Saputra, 2020). Furthermore, as in many other developing countries, the challenges also include a lack reliable data, a lack of control over fishery access/community resource management rights, a lack of effective organization of small crab fishermen, a lack of government capacity to support the “social health” of fishing communities, and limited understanding of these issues within the industry and among other key stakeholders. In order to address these issues, the management of Indonesian crab fisheries needs to involve all stakeholders from the central government, provincial governments, fishermen, industry and non-governmental organizations. Based on the Regional Autonomy Act 23/2014, regulation of vessels with a gross tonnage of less than 30 GT is the responsibility of the provincial government. This means that management of the BSC fishery is largely the responsibility of the provincial governments; however, as mandated by the BSC Fisheries Management Plan (FMP), each provincial government needs to develop and implement its own action plan based on and guided by the national plan.
The Indonesian Blue Swimming Crab Management Association (APRI) (www.apri.or.id) initiated a BSC Fisheries Improvement Program (FIP) at a pilot scale in 2007 and began to work at the national scale in 2014. This program has worked to bring together the stakeholders, in particular from the central government, provincial governments, fishermen, industry and non-governmental organizations. As the blue swimming crab fishers and vessels are largely under the aegis of the provincial government, but fisheries management plans operate at the species and/or FMA level (see Figure 1), coordination between levels is essential. In order to comply with Act 23/2014 and the mandates of the BSC FMP, the Indonesian Blue Swimming Crab Management Association (APRI) program is currently working in three provinces (East Java Province, East Java Province, and Southeast Province) to develop and strengthen the BSC Fishery Management Committee. The duties of the Committee are: (1) Identify and inventory the condition of fisheries management problems in the province; (2) Develop and prepare an action plan for the management of small crab fisheries at the provincial level; (3) Facilitate the implementation of the small crab fishery management action plan at the provincial level; (4) Conduct small crab fishery management activities at the provincial level; (5) Report the implementation results to the Governor. Furthermore, since 2018 the APRI has implemented the Control Document Audit System (CDAS) to meet or anticipate national and international regulatory requirements (APRI, 2019). The CDAS is structured to: a. Promote an ethical culture based on professional internal supervision of Indonesian BSC fisheries, including compliance with Ministerial Regulations through a control and traceability system at all stages from suppliers to buyers; (b) Create a trustworthy, objective, and accountable internal auditing supervisory system with integrity, thereby enabling credible audits to be performed. Specific approaches that have been adopted and implemented by the APRI include the baseline assessments and monitoring of the Length Based Spawning Potential Ratio (LBSPR) and Catch Per Unit Effort (CPUE) for BSC fisheries (Ernawati et al., 2017; Prince et al., 2020). LBSPR assessments could be used to adaptively manage size selectivity within the harvest strategy paradigm now being adopted by Indonesia (Hordyk et al., 2015; Prince et al., 2020; Loneragan et al., 2021) while CPUE is still widely accepted as an indirect measure or index of the relative abundance of target stocks in both fisheries and conservation management (Cheung & Sumaila, 2008; Sagarese et al., 2018). The BSC FIP Program also supports the government plan to develop FMA 2014 as a National fish reservoir (Lumbung Ikan Nasional or LIN). Sometimes referred to as the National Fish Barn, the LIN aims to balance fisheries resources and capture in eastern Indonesia (https://kkp.go.id), with Maluku Province as the pilot region. These efforts need to be continued and scaled up across the distribution of P. pelagicus in Indonesia.
Conclusion
This study reveals patterns in decapod community diversity at relatively small scales, such as at the provincial level and Fisheries Management Areas (FMAs). However, some taxa are widespread across several FMAs, and it is necessary to pay attention to decapod conservation and fisheries management at national or multi-FMA levels. One of the most widespread taxa, and the most economically valuable decapod fisheries commodity, the blue swimming crab Portunus pelagicus has several genetically distinct populations, thus requiring stock-based management which can largely be based on existing FMAs.
The high percentage of unidentified taxa in this study reflects the paucity of reference sequence data for Indonesian marine decapods, although coverage appears slightly higher in West Indonesia compared to Central and East Indonesia. This gap in current database coverage highlights the need for policies to support capacity building and long-term maintenance of systems enabling biodiversity exploration and monitoring, including well-curated and sustainably resourced reference specimen and sequence repositories. Furthermore, the low coverage of Indonesian (and indeed wider Indo-Pacific) marine decapods in the IUCN Red List calls for partnerships to remedy this gap, including concerted efforts to improve distribution data.
Data availability statement
The data presented in the study are deposited in the Figshare repository with the following link https://doi.org/10.6084/m9.figshare.20308905.
Ethics statement
Ethical review and approval was not required for the animal study because we used non-invasive method (eDNA metabarcoding) by collected seawater samples as the samples analyzed in this manuscript.
Author contributions
HM, DB, ZM, and AB contributed on the design of research Idea. HM, LS, BS, DA, VM, AD, MR, ASa, MO, WT, M, JD, ES, NN, JJ, NA, SN, IM, JN, BR, and AM helped on sample collections used in this research. LS analyzed the sample on laboratory. LS analyzed the data. HM and LS contributed to visualize the data. HM, LS, and AM wrote the manuscript and enhanced by KN, NF, NZ, ASu, MI, ESS, NC, DL, PS, and WS. All authors contributed to the article and approved the submitted version.
Funding
The research was funded by the Indonesian Ministry of Education, Culture, Research and Technology under the “World Class Research’’ scheme No: 2346/IT3.L1/PN/2021 awarded to HM for the research project entitled “Redefining hotspots of marine fishes across Indonesian coral reefs with different anthropogenic pressure using environmental DNA metabarcoding for sustainable fisheries”.
Acknowledgments
The authors thank the Ministry of Research and Technology (Ristek/BRIN) for supporting the research entitled “Redefining hotspots of marine fishes across Indonesian coral reefs with different anthropogenic pressure using environmental DNA metabarcoding for sustainable fisheries”; the Institute of Research and Community Service (LPPM) IPB University for undertaking the research administration; the Laboratory of Marine Biodiversity and Biosystematics IPB University, Scientific Diving Laboratory IPB University, and Oceanogen r for the use of their research logistics and facilities; and all contributors and organizations that supported this research. This research article is fully supported by Asosiasi Pengelolaan Rajungan Indonesia (APRI). The authors also thank the Ministry of Finance of the Republic of Indonesia for supporting this research through the funding from LPDP Scholarship. The authors also want to honor the memory of our respected and much-loved co-author and friend Dr. Hawis Madduppa who passed away on 10th May 2022. Hawis Madduppa was an associate professor in marine science and technology at IPB University who also participated actively in several initiatives aimed at improving marine and fisheries research and management in Indonesia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.918295/full#supplementary-material
Supplementary Figure 1 | Overview of mBRAVE Project Analytical Parameter settings used for this study. The 4 steps with analytical parameters included trimming, filtering,clustering, and paired end merging of project MBR-WCRMIDECA.
Supplementary Figure 2 | Decapod species accumulation curve based on species richness from eDNA samples collected from coral reefs across Indonesia.
References
Almeida A. O., Terossi M., Araujo-Silva C. L., Mantelatto F. L. (2013). Description of alpheus buckupi spec. nov., a new amphi-Atlantic snapping shrimp (Caridea: Alpheidae), based on morphological and molecular data. Zootaxa 3652 (4), 437–452. doi: 10.11646/zootaxa.3652.4.3
Ambo-Rappe R., Moore A. M. (2018). Sulawesi seas, Indonesia. World seas: an Environ. Eval., (Elsevier) 559–581. doi: 10.1016/B978-0-08-100853-9.00032-4
Andrade L. S. D., Frameschi I. F., Costa R. C. D., Castilho A. L., Fransozo A. (2015). The assemblage composition and structure of swimming crabs (Portunoidea) in continental shelf waters of southeastern Brazil. Continental shelf research 94, 8–16. doi: 10.1016/j.csr.2014.12.005
Andriyono S., Alam M. J., KIM H. W. (2019). Environmental DNA (eDNA) metabarcoding: Diversity study around the Pondok Dadap fish landing station, Malang, Indonesia. Biodiversitas J. Biol. Diversity 20 (12), 3772–3781. doi: 10.13057/biodiv/d201241
Anggorowati D. A. (2014). Community structure of crustacean fauna at the intertidal zone of West Lombok. Jurnal Zoologi Indonesia 23 (2), 92–100
Anggraeni P., Elfidasari D., Pratiwi R. (2015). Brachyuran crab distribution in Tikus Island, Pari Island, Seribu Islands, in Prosiding seminar nasional masyarakat biodiversitas Indonesia 1 (2), 213–221. doi: 10.13057/psnmbi/m010208
APRI (2019). Summary report of control document audit, Asosiasi Perikanan Rajungan Indonesia (APRI) (Surabaya), 12.
APRI (2020). Annual report of fisheries improvement project, Asosiasi Perikanan Rajungan Indonesia (APRI) (Surabaya).
Aprila L. S., Wowor D., Boer M., Farajallah A. (2020). “Population dynamics of Macrobrachium sintangense and m. lanchesteri in lake lido, West Java,” in IOP conference series: Earth and environmental science, 457 (1), 012008. doi: 10.1088/1755-1315/457/1/012008
Ardika P. U., Farajallah A., Wardiatno Y. (2015). First Record of Hippa adactyla (Fabricius, 1787; Crustacea, Anomura, Hippidae) from Indonesian Waters. Trop. Life Sci. Res. 26 (2), 105–110. Available at: http://www.ncbi.nlm.nih.gov/pubmed/26868713
Ardura A., Zaiko A. (2018). PCR-based assay for Mya arenaria detection from marine environmental samples and tracking its invasion in coastal ecosystems. J. Nat. Conserv. 43, 1–7. doi: 10.1016/j.jnc.2018.02.007
Aswandy I.N.D.R.A. (2008). Struktur komunitas krustasea di estuari cisadane dan parairan laut sekitarnya. Oseanologi dan Limnologi di Indinesia 34 (1), 67–81.
Barber P., Cheng S., Erdmann M., Tenggardjaja K. (2011). “Evolution and conservation of marine biodiversity in the Coral Triangle: insights from stomatopod Crustacea,” in Crustacean Issues 19: Phylogeography and Population Genetics in Crustacea (1st Edition. Eds. Held C., Koenemann S., Schubart C. D. (CRC Press), 129–156. doi: 10.1201/b11113-9
Bennun L., Regan E. C., Bird J., van Bochove J. W., Katariya V., Livingstone S., et al. (2018). The value of the IUCN Red List for business decision-making. Conserv. Lett. 11 (1), e12353. doi: 10.1111/conl.12353
Berry T. E., Osterrieder S. K., Murray D. C., Coghlan M. L., Richardson A. J., Grealy A. K., et al. (2017). DNA Metabarcoding for diet analysis and biodiversity: A case study using the endangered Australian sea lion (Neophoca cinerea). Ecol. Evol. 7 (14), 5435–5453. doi: 10.1002/ece3.3123
Betts J., Young R. P., Hilton-Taylor C., Hoffmann M., Rodríguez J. P., Stuart S. N., et al. (2020). A framework for evaluating the impact of the IUCN Red List of threatened species. Conserv. Biol. 34 (3), 632–643. doi: 10.1111/cobi.13454
Bezeng B. S., van der Bank H. F. (2019). DNA Barcoding of southern African crustaceans reveals a mix of invasive species and potential cryptic diversity. PLoS One 14 (9), e0222047. doi: 10.1371/journal.pone.0222047
Blankenhorn S. U. (2007). Seaweed farming and artisanal fisheries in an Indonesian seagrass bed-complementary or competitive usages? (Universität Bremen). Available at: https://media.suub.uni-bremen.de/bitstream/elib/2462/1/00010932.pdf
Bondad-Reantaso M. G., Subasinghe R. P., Josupeit H., Cai J., Zhou X. (2012). The role of crustacean fisheries and aquaculture in global food security: past, present and future. J. invertebrate Pathol. 110 (2), 158–165. doi: 10.1016/j.jip.2012.03.010
Bowser A. K., Diamond A. W., Addison J. A. (2013). From puffins to plankton: a DNA-based analysis of a seabird food chain in the northern gulf of Maine. PLoS One 8 (12), e83152. doi: 10.1371/journal.pone.0083152
Box A., Colomar V., Sureda A., Tejada S., Nuñez-Reyes V., Cohen-Sanchez A., et al. (2020). Next step of the colonization of the Balearic islands (Spain) by invasive Atlantic blue crab, Callinectes sapidus rathbun 1896 (Crustacea: Decapoda: Portunidae). Bioinvasions Rec 9, 259–265. doi: 10.3391/bir.2020.9.2.11
Buckley S. M., McClanahan T. R., Quintana Morales E. M., Mwakha V., Nyanapah J., Otwoma L. M., et al. (2019). Identifying species threatened with local extinction in tropical reef fisheries using historical reconstruction of species occurrence. PLoS One 14 (2), e0211224. doi: 10.1371/journal.pone.0211224
Burke L., Reytar K., Spalding M., Perry A., Knight M., Kushner B., et al. (2012). Reefs at Risk Revisited in the Coral Triangle (World Resources Institute). Available at: https://files.wri.org/d8/s3fs-public/pdf/reefs_at_risk_revisited_coral_triangle.pdf.
Calado R., Lin J., Rhyne A. L., Araújo R., Narciso L. (2003). Marine ornamental decapods–popular, pricey, and poorly studied. J. crustacean Biol. 23 (4), 963–973. doi: 10.1651/C-2409
Campbell L. M. (2012). Seeing red: inside the science and politics of the IUCN Red List. Conserv. Soc. 10 (4), 367–380. doi: 10.4103/0972-4923.105560
Castilho A. L., Pie M. R., Fransozo A., Pinheiro A. P., Costa R. C. (2007). The relationship between environmental variation and species abundance in shrimp community (Crustacea: Decapoda: Penaeoidea) in south-eastern Brazil. J. Mar. Biol. Assoc. United Kingdom 88 (1), 119–123. doi: 10.1017/S0025315408000313
Chakravarty M. S., Ganesh P. R. C., Amarnath D., Sudha B. S., Vivek V. (2016). Diversity of crabs in Tekkali creek, Srikakulam district, Andhra Pradesh. International journal of fisheries and aquatic studies 4, 414–418. Available at: https://www.fisheriesjournal.com/archives/2016/vol4issue1/PartF/4-1-37.pdf
Chan T. Y. (2010). Annotated checklist of the world’s marine lobsters (Crustacea: Decapoda: Astacidea, Glypheidea, Achelata, Polychelida). Raffles Bull. Zoology 23 (Suppl), 153–181. Available at: https://lkcnhm.nus.edu.sg/wp-content/uploads/sites/10/app/uploads/2017/06/s23rbz153-181.pdf
Chande A. I., Mgaya Y. D. (2003). The fishery of Portunus pelagicus and species diversity of portunid crabs along the coast of Dar es salaam, Tanzania. Western Indian Ocean J. Mar. Sci. 2 (1), 75–84. doi: 10.4314/wiojms.v2i1.28431
Cheung W. W., Sumaila U. R. (2008). Trade-offs between conservation and socio-economic objectives in managing a tropical marine ecosystem. Ecol. economics 66 (1), 193–210. doi: 10.1016/j.ecolecon.2007.09.001
Cheung W. W. L., Lam V. W. Y., Sarmiento J. L., Kearney K., Watson R., Pauly D. (2009). Projecting global marine biodiversity impacts under climate change scenarios. Fish Fisheries 10 (3), 235–251. doi: 10.1111/j.1467-2979.2008.00315.x
Closek C. J., Santora J. A., Starks H. A., Schroeder I. D., Andruszkiewicz E. A., Sakuma K. M., et al. (2019). Marine vertebrate biodiversity and distribution within the central California current using environmental DNA (eDNA) metabarcoding and ecosystem surveys. Front. Mar. Sci. 6 (12), 732. doi: 10.3389/fmars.2019.00732
Cockcroft A., Butler ,. M., MacDiarmid A. (2011)Panulirus stimpsoni. the IUCN Red List of threatened species 2011 (Accessed 30 August 2021).
Cristescu M. E., Hebert P. D. N. (2018). Uses and Misuses of Environmental DNA in Biodiversity Science and Conservation. Annu. Rev. Ecology Evolution Systematics 49 (1), 209–230. doi: 10.1146/annurev-ecolsys-110617-062306
Deck J., Gaither M. R., Ewing R., Bird C. E., Davies N., Meyer C., et al. (2017). The genomic observatories metadatabase (GeOMe): A new repository for field and sampling event metadata associated with genetic samples. PLoS Biol. 15 (8), e2002925. doi: 10.1371/journal.pbio.2002925
De Grave S., Fransen C. (2011). Carideorum catalogus: The recent species of the dendrobranchiate, stenopodidean, procarididean and caridean shrimps (Crustacea: Decapoda) Zool. Med. Leiden 85 (9), 195–589. Available at: http://www.vliz.be/imisdocs/publications/ocrd/231051.pdf.
De Grave S., Pentcheff N. D., Ahyong S. T., Chan T., Crandall K. A., Dworschak P. C., et al. (2009a). A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zoology 2009 (S21), 1–109. Available at: http://rmbr.nus.edu.sg/rbz/biblio/s21/s21rbz1-109.pdf
De Grave S., Pentcheff N. D., Ahyong S. T., Chan T., Crandall K. A., Dworschak P. C., et al. (2009b). A classification of living and fossil genera of decapod crustaceans. Raffles Bull. Zoology 2009 (S21), 1–109.
Dev Roy M. K., Nandi N. C. (2017). A note on portunid crabs of odisha coast, India. Oceanography Fisheries Open Access J. 1 (3), 66–68. doi: 10.19080/OFOAJ.2017.01.555562
Diz D., Johnson D., Riddell M., Rees S., Battle J., Gjerde K., et al. (2018). Mainstreaming marine biodiversity into the SDGs: the role of other effective area-based conservation measures (SDG 14.5). Mar. Policy 93, 251–261. doi: 10.1016/j.marpol.2017.08.019
Ernawati T. R. I., Sumiono B., Madduppa H. (2017). Reproductive ecology, spawning potential, and breeding season of blue swimming crab (Portunidae: Portunus pelagicus) in Java Sea, Indonesia. Biodiversitas J. Biol. Diversity 18 (4), 1705–1713. doi: 10.13057/biodiv/d180450
Friedman K., Garcia S. M., Rice J. (2018). Mainstreaming biodiversity in fisheries. Marine policy 95, 209–220. doi: 10.1016/j.marpol.2018.03.001
Galil B. S., Clark P. F., Carlton J. T. (Eds.) (2011). “In the Wrong Place - Alien Marine Crustaceans,” in Distribution, Biology and Impacts (Springer Netherlands). Available at: https://doi.org/10.1007/978-94-007-0591-3.
Gelis E. R. E., Kamal M. M., Subhan B., Bachtiar I., Sani L. M. I., Madduppa H. (2021). Environmental biomonitoring of reef fish community structure with eDNA metabarcoding in the Coral Triangle. Environ. Biol. Fishes 104 (8), 887–903. doi: 10.1007/s10641-021-01118-3
Gilbey J., Carvalho G., Castilho R., Coscia I., Coulson M. W., Dahle G., et al. (2021). Life in a drop: Sampling environmental DNA for marine fishery management and ecosystem monitoring. Mar. Policy 124, 104331. doi: 10.1016/j.marpol.2020.104331
Gökoğlu N. (2021). Shellfish processing and preservation (Cham, Switzerland: Springer International Publishing). pp. 7–127 doi: 10.1007/978-3-030-60303-8_2
Hamid A., Kamri S., Irawati N., Wardiatno Y. (2020). Community structure of crustacean bycatch of blue swimming crab (Portunus pelagicus) fisheries in Kendari Bay, Southeast Sulawesi, Indonesia. Aquaculture Aquarium Conserv. Legislation 13 (2), 694–704.
Hamid A., Wardiatno Y. (2018). Biological aspects of Charybdis anisodon (De haa) in lasongko bay, central buton, southeast sulawesi, Indonesia. Biodiversitas J. Biol. Diversity 19 (5), 1755–1762. doi: 10.13057/biodiv/d190523
Hayward M. W. (2011). Using the IUCN Red List to determine effective conservation strategies. Biodiversity Conserv. 20 (12), 2563–2573. doi: 10.1007/s10531-011-0091-3
Hebert P. D., Gregory T. R. (2005). The promise of DNA barcoding for taxonomy. Systematic Biol. 54 (5), 852–859. doi: 10.1080/10635150500354886
Hoegh-Guldberg O., Hoegh-Guldberg H., Veron J. E. N., Green A., Gomez. E., Lough. J., et al. (2009). The Coral Triangle and climate change: Ecosystems, people and societies at risk (Brisbane: WWF Australia)
Holthuis L. B. (1950). The Decapoda of the Sibolga Expedition Part X: The Palaemonidae collected by the Siboga and Snellius expeditions with remarks on other species I. Subfamily Palaemonina (Holand: Leiden), 268.
Hordyk A. R., Loneragan N. R., Prince J. D. (2015). An evaluation of an iterative harvest strategy for data-poor fisheries using the length-based spawning potential ratio assessment methodology. Fisheries Res. 171, 20–32. doi: 10.1016/j.fishres.2014.12.018
Hornborg S., Svensson M., Nilsson P., Ziegler F. (2013). By-catch impacts in fisheries: utilizing the IUCN Red List categories for enhanced product level assessment in seafood LCAS. Environ. Manage. 52 (5), 1239–1248. doi: 10.1007/s00267-013-0096-7
Horton T., Gofas S., Kroh A., Poore G. C., Read G., Rosenberg G., et al. (2017). Improving nomenclatural consistency: a decade of experience in the World Register of Marine Species. Eur. J. Taxonomy 2017 (389), 1–24. doi: 10.5852/ejt.2017.389
Hubert N., Wibowo A., Busson F., Caruso D., Sulandari S., Nafiqoh N., et al. (2015). DNA Barcoding Indonesian freshwater fishes: challenges and prospects. DNA Barcodes 3 (1), 144–169. doi: 10.1515/dna-2015-0018
Huhn M., Madduppa H. H., Khair M., Sabrian A., Irawati Y., Anggraini N. P., et al. (2020). Keeping up with introduced marine species at a remote biodiversity hotspot: awareness, training and collaboration across different sectors is key. Biol. Invasions 22 (2), 749–771. doi: 10.1007/s10530-019-02126-2
Hutomo M., Moosa M. K. (2005). Indonesian Marine and coastal biodiversity: Present status. Indian Journal of Marine Sciences 34 (1), 88–97. Available at: https://www.vliz.be/imisdocs/publications/299010.pdf
IUCN (2012). Guidelines for application of IUCN Red List criteria at regional and national levels version 4.0 (Gland, Switzerland and Cambridge, UK: International Union for Conservation of Nature and Natural Resources), 46.
IUCN (2019). Guidelines for using the IUCN Red List categories and criteria. version 14 (Gland, Switzerland: International Union for Conservation of Nature), 113. IUCN Standards and Petitions Committee.
IUCN (2021). “The IUCN Red List of Threatened Species. Version 2021-1,” in The IUCN Red List of Threatened Species. Available at: https://www.iucnredlist.org.
Jackson J. B. (2008). Ecological extinction and evolution in the brave new ocean. Proc. Natl. Acad. Sci. 105 (Supplement 1), 11458–11465. doi: 10.1073/pnas.0802812105
Jarvis M. G., Closs G. P. (2019). Water infrastructure and the migrations of amphidromous species: impacts and research requirements. J. Ecohydraulics 4 (1), 4–13. doi: 10.1080/24705357.2019.1611390
Jeunen G. J., Knapp M., Spencer H. G., Lamare M. D., Taylor H. R., Stat M., et al. (2019). (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 19 (2), 426–438. doi: 10.1111/1755-0998.12982
Jose D., Harikrishnan M. (2019). Evolutionary history of genus Macrobrachium inferred from mitochondrial markers: a molecular clock approach. Mitochondrial DNA Part A 30 (1), 92–100. doi: 10.1080/24701394.2018.1462347
Juhel J. B., Utama R. S., Marques V., Vimono I. B., Sugeha H. Y., Pouyaud L., et al. (2020). Accumulation curves of environmental DNA sequences predict coastal fish diversity in the Coral Triangle. Proc. R. Soc. B 287 (1930), 20200248. doi: 10.1098/rspb.2020.0248
Jurniati J., Arfiati D., Andriyono S., Hertika A. M. S., Kurniawan A., Tanod W. A. (2021). The morphological characters and DNA barcoding identification of sweet river prawn Macrobrachium esculentum (Thallwitz 1891) from rongkong watershed of South Sulawesi, Indonesia. Biodiversitas J. Biol. Diversity 22 (1), 113–121. doi: 10.13057/biodiv/d220116
Komai T., Gotoh R. O., Sado T., Miya M. (2019). Development of a new set of PCR primers for eDNA metabarcoding decapod crustaceans. Metabarcoding Metagenomics 3, 1–19. doi: 10.3897/mbmg.3.33835
Kühl H. S., Bowler D. E., Bösch L., Bruelheide H., Dauber J., Eichenberg D., et al. (2020). Effective biodiversity monitoring needs a culture of integration. One Earth 3 (4), 462–474. doi: 10.1016/j.oneear.2020.09.010
Kumaralingam S., Sivaperuman C., Raghunathan C. (2013). “Diversity and community structure of brachyuran crabs in north Andaman,” in Ecology and conservation of tropical marine faunal communities (Berlin, Heidelberg: Springer), 171–181. doi: 10.1007/978-3-642-38200-0_11
Kwak S. N., Klumpp D. W., Park J. M. (2015). Feeding relationships among juveniles of abundant fish species inhabiting tropical seagrass beds in cockle bay, north Queensland, Australia. New Z. J. Mar. Freshw. Res. 49 (2), 205–223. doi: 10.1080/00288330.2014.990467
Lagos M. E., Castillo N., Albarrán-Mélzer N., Pinochet J., Gebauer P., Urbina M. A. (2021a). Age dependent physiological tolerances explain population dynamics and distribution in the intertidal zone: A study with porcelain crabs. Marine Environ. Res. 169 (April), 105343. doi: 10.1016/j.marenvres.2021.105343
Liu M. Y., Cai Y. X., Tzeng C. S. (2007). Molecular systematics of the freshwater prawn genus Macrobrachium bate 1868 (Crustacea: Decapoda: Palaemonidae) inferred from mtDNA sequences, with emphasis on East Asian species. ZOOLOGICAL STUDIES-TAIPEI- 46 (3), 272.
Loneragan N. R., Wiryawan B., Hordyk A. R., Halim A., Proctor C., Satria F., et al. (2021). Proceedings from workshops on management strategy evaluation of data-limited fisheries: Towards sustainability–applying the method evaluation and risk assessment tool to seven Indonesian fisheries (Western Australia: Murdoch University).
Lui K. K., Ng J. S., Leung K. M. (2007). Spatio-temporal variations in the diversity and abundance of commercially important decapoda and stomatopoda in subtropical Hong Kong waters. Estuarine Coast. Shelf Sci. 72 (4), 635–647. doi: 10.1016/j.ecss.2006.11.023
Mablouké C., Kolasinski J., Potier M., Cuvillier A., Potin G., Bigot L., et al. (2013). Feeding habits and food partitioning between three commercial fish associated with artificial reefs in a tropical coastal environment. Afr. J. Mar. Sci. 35 (3), 323–334. doi: 10.2989/1814232X.2013.829790
Macher T. H., Beermann A. J., Leese F. (2021). TaxonTableTools: A comprehensive, platform‐independent graphical user interface software to explore and visualise DNA metabarcoding data. Mol. Ecol. Resour. 21 (5), 1705–1714. doi: 10.1111/1755-0998.13358
Madduppa H., Cahyani N. K. D., Anggoro A. W., Subhan B., Jefri E., Sani L. M. I., et al. (2021b). eDNA metabarcoding illuminates species diversity and composition of three phyla (Chordata, Mollusca and Echinodermata) across Indonesian coral reefs. Biodiversity Conserv. 30 (11), 3087–3114. doi: 10.1007/s10531-021-02237-0
Madduppa H., Martaulina R., Zairion Z., Renjani R. M., Kawaroe M., Anggraini N. P., et al. (2021a). Genetic population subdivision of the blue swimming crab (Portunus pelagicus) across Indonesia inferred from mitochondrial DNA: Implication to sustainable fishery. PLoS One 16 (2), e0240951. doi: 10.1371/journal.pone.0240951
Madduppa H., Zairion N. S., Nugroho K., Nugraha B. A. (2016). Setting up traceability tools for the Indonesian blue swimming crab fishery: A case study in southeast sulawesi. Fisheries Aquaculture Modern World 9, 2016. doi: 10.5772/64252
McKenna S. A., Allen G. R., Suryadi S. (Eds.) (2002). A Marine Rapid Assessment of the Raja Ampat Islands, Papua Province, Indonesia (No. 22; RAP Bulletin on Biological Assessment) (Conservation International). Available at: https://indopacificimages.com/wp-content/uploads/2013/07/CI_RAP_Raja-Ampat_2002.pdf.
Mangubhai S., Erdmann M. V., Wilson J. R., Huffard C. L., Ballamu F., Hidayat N. I., et al. (2012). Papuan Bird’s Head Seascape: Emerging threats and challenges in the global center of marine biodiversity. Marine Pollution Bull. 64 (11), 2279–2295. doi: 10.1016/j.marpolbul.2012.07.024
Maulina A., Inocencia A., Putra E. D., Satriani I., Lusiana L. (2020a). The Invasive of Freshwater Prawn Macrobrachium lanchesteri (De Ma) from Watershed in Palangkaraya University, Kalimantan, Indonesia. Natural Science: J. Sci. Technol. 9 (2), 49–53. doi: 10.22487/25411969.2020.v9.i2.15183
Martin J. W., Davis G. E. (2001). An updated classification of the recent Crustacea Vol. 39 (Los Angeles: Natural History Museum of Los Angeles County), 129
Mashar A., Wardiatno Y., Boer M., Butet N. A., Farajallah A. (2014). The diversity and abundance of sand crabs in south coast of central Java. Ilmu Kelautan 19 (4), 226–232. doi: 10.14710/ik.ijms.19.4.226-232
Mashar A., Wardiatno Y., Boer M., Butet N. A., Farajallah A., Ardika P. U. (2015). First record of albunea symmysta (Crustacea: Decapoda: Albuneidae) from Sumatra and Java, Indonesia. AACL Bioflux 8 (4), 611–615. Available at: http://www.bioflux.com.ro/docs/2015.611-615.pdf
McDowall R. M. (2007). On amphidromy, a distinct form of diadromy in aquatic organisms. Fish and Fisheries, 8 (1), 1–13. doi: 10.1111/j.1467-2979.2007.00232.x
Milton D. A., Satria F., Proctor C. H., Prasetyo A. P., Utama A. A., Fauzi M. (2014). Environmental factors influencing the recruitment and catch of tropical Panulirus lobsters in southern Java, Indonesia. Continental Shelf Res. 91, 247–255. doi: 10.1016/j.csr.2014.09.011
Miya M., Sato Y., Fukunaga T., Sado T., Poulsen J. Y., Sato K., et al. (2015). MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2 (7), 150088. doi: 10.1098/rsos.150088
MMAF (2016). Keputusan Menteri Kelautan dan Perikanan Republik Indonesia Nomor 70/KEPMEN-KP/2016, Tentang Rencana Pengelolaan Perikanan Rajungan di Wilayah Pengelolaan Perikanan Negara Republik Indonesia.
Moore A. M., Jompa J., Tassakka A. C. M. A., Yasir I., Ndobe S., Umar W., et al. (2021). Sharks and rays (Chondrichthyes) around Banggai Island, Banggai MPA, Indonesia: biodiversity data from an environmental DNA pilot study. Aquaculture Aquarium Conserv. Legislation 14 (2), 725–745. Available at: http://www.bioflux.com.ro/docs/2021.725-745.pdf
Moosa M. K. (1980). Some notes about the crabs from Jakarta Bay and Seribu islands. In: Marine biological resources: overview some research results from Pelita II LON-LIPI, Jakarta. pp 55–79.
Moosa M. K., Aswandy I. (1994). “Crustaceans of seagrass beds in the waters of South Lombok,” in Biological community structure of seagrass on the southern coast of lombok and environmental conditions. Eds. Kiswara W., Moosa M. K., Hutomo M. (Jakarta: Research Center for Oceanography, Indonesian Institute of Sciences), 42–51.
Nadiarti N., Moore A., Abu N., Rahim S. W., Chasanah M. (2021). Ecosystems approach to fisheries management (EAFM) assessment for grouper and snapper fisheries in bontang, East kalimantan, Indonesia. IOP Conf. Series: Earth Environ. Sci. 763, 012031. doi: 10.1088/1755-1315/763/1/012031
Ndoro C. K., Kaunda-Arara B., Ruwa R., Munga C. N. (2014). Influence of seasonality and bathymetry on assemblage structure of decapod crustaceans in the Malindi-Ungwana Bay, Kenya. Western Indian Ocean J. Mar. Sci. 13 (1), 31–46. Available at: https://www.ajol.info/index.php/wiojms/article/download/99087/110343/0
Ng P. K., Guinot D., Davie P. J. (2008). Systema brachyurorum: Part i. an annotated checklist of extant brachyuran crabs of the world. raffles Bull. zoology 17 (1), 1–286.
Ng P. K., Lukhaup C. (2015). Aletheiana tenella, a new genus and new species of freshwater hymenosomatid crab (Crustacea: Decapoda: Brachyura) from Central Sulawesi, Indonesia. Zootaxa 4039 (1), 118–128. doi: 10.11646/zootaxa.4039.1.4
Nursyahran N., Kariyanti K., Jayadi J., Yusuf A., Wahidah S. (2021). Molecular identification of the freshwater prawn Macrobrachium idea in Tempe Lake, South Sulawesi, Indonesia. AACL Bioflux 14, 2174–2180. Available at: http://www.bioflux.com.ro/docs/2021.2174-2180.pdf
Palomares M. L. D., Pauly D. (2021)SeaLifeBase. Available at: www.sealifebase.org.
Park J. M., Kwak S. N., Riedel R. (2020). Crustacean decapod assemblage associated with seagrass (Zostera marina) beds in southern waters of Korea. Diversity 12 (3), 89. doi: 10.3390/d12030089
Pimm S. L., Jenkins C. N., Abell R., Brooks T. M., Gittleman J. L., Joppa L. N., et al. (2014). The biodiversity of species and their rates of extinction, distribution, and protection. science 344 (6187), 1246752. doi: 10.1126/science.1246752
Pombo M., Denadai M. R., Turra A. (2013). Seasonality, dietary overlap and the role of taxonomic resolution in the study of the diet of three congeneric fishes from a tropical bay. PLoS One 8 (2), e56107. doi: 10.1371/journal.pone.0056107
Prabowo B., Idris I., Adiwijaya C., Putri A. R., Sastranegara H., Fahlevy K., et al. (2021). Occurrence distribution of coral-dwelling shrimp among the coral growth forms of the Tidung Island, Jakarta, Indonesia. AACL Bioflux 14 (4), 2027–2039. Available at: http://www.bioflux.com.ro/docs/2021.2027-2039.pdf
Pratiwi R. (2010). Crustaceans association in seagrass ecosystems Lampung Bay waters. Indonesian J. Mar. Sci. 15, 66–76. doi: 10.14710/ik.ijms.15.2.66-76
Pratiwi R. (2012). Species and pattern of distribution of crustaceans fauna in seagrass Tikus Island, Seribu Islands. Oseanologi dan Limnologi di Indonesia 38, 43–55.
Pratiwi R., Astuti O. (2012). Biodiversity of crustacean (Decapod, brachyura, macrura) from kendari waters expedition 2011. Indonesian J. Mar. Sci. 17 (1), 8–14. doi: 10.14710/ik.ijms.17.1.8-14
Pratiwi R., Widyastuti E. (2013). Distributional patterns and zonation of crustaceans mangrove in Lampung Bay. Zoo Indonesia 22 (1), 11–21. doi: 10.52508/zi.v22i1.317
Pratiwi R., Wijaya N. I. (2013). Keanekaragaman komunitas krustasea di kepulauan matasiri kalimantan selatan. Berita Biologi 12 (1), 127–140. doi: 10.14203/beritabiologi.v12i1.525
Prince J., Creech S., Madduppa H., Hordyk A. (2020). Length based assessment of spawning potential ratio in data-poor fisheries for blue swimming crab (Portunus spp.) in Sri Lanka and Indonesia: Implications for sustainable management. Regional Stud. Mar. Sci. 36, 101309. doi: 10.1016/j.rsma.2020.101309
Priyambodo B., Jones C. M., Sammut J. (2020). Assessment of the lobster puerulus (Panulirus homarus and panulirus ornatus, decapoda: Palinuridae) resource of Indonesia and its potential for sustainable harvest for aquaculture. Aquaculture 528, 735563. doi: 10.1016/j.aquaculture.2020.735563
Purwanto Andradi‐Brown D. A., Matualage D., Rumengan I., Awaludinnoer Pada D., Hidayat N. I., Amkieltiela Fox H. E., et al. (2021). The Bird’s Head Seascape Marine Protected Area network—Preventing biodiversity and ecosystem service loss amidst rapid change in Papua, Indonesia. Conserv. Sci. Pract. 3 (6), 1–18. doi: 10.1111/csp2.393
Ratnasingham S. (2019). mBRAVE: The multiplex barcode research and visualization environment. Biodiversity Inf. Sci. Standards 3, e37986. doi: 10.3897/biss.3.37986
Rees S. E., Foster N. L., Langmead O., Pittman S., Johnson D. E. (2018). Defining the qualitative elements of aichi biodiversity target 11 with regard to the marine and coastal environment in order to strengthen global efforts for marine biodiversity conservation outlined in the United Nations sustainable development goal 14. Mar. Policy 93, 241–250. doi: 10.1016/j.marpol.2017.05.016
Riginos C., Crandall E. D., Liggins L., Gaither M. R., Ewing R. B., Meyer C., et al. (2020). Building a global genomics observatory: Using GEOME (the genomic observatories metadatabase) to expedite and improve deposition and retrieval of genetic data and metadata for biodiversity research. Mol. Ecol. Resour. 20 (6), 1458–1469. doi: 10.1111/1755-0998.13269
Roberts C. M., Hawkins J. P. (1999). Extinction risk in the sea. Trends Ecol. Evol. 14 (6), 241–246. doi: 10.1016/S0169-5347(98)01584-5
Rockström J., Steffen W., Noone K., Persson Å., Chapin F. S. III, Lambin E., et al. (2009). Planetary boundaries: exploring the safe operating space for humanity. Ecol. Soc. 14 (2), 32. doi: 10.1038/461472a
Sagarese S. R., Rios A. B., Cass‐Calay S. L., Cummings N. J., Bryan M. D., Stevens M. H., et al. (2018). Working towards a framework for stock evaluations in data‐limited fisheries. North Am. J. Fisheries Manage. 38 (3), 507–537. doi: 10.1002/nafm.10047
Saputra M. A. (2020). Moving within and beyond illegal crustacean fishery: why do Indonesian fishermen not comply with the crustacean catch ban rule? Maritime Stud. 19 (4), 457–473. doi: 10.1007/s40152-020-00194-y
Setyanto A., Wiadnya D. G. R., Prayogo C. (2019). Biodiversity of lobster larvae (Panulirus spp.) from the Indonesian Eastern Indian Ocean. IOP Conf. Series: Earth Environ. Sci. 370, 012046. doi: 10.1088/1755-1315/370/1/012046
Siriwut W., Jeratthitikul E., Panha S., Chanabun R., Ngor P. B., Sutcharit C. (2021). Evidence of cryptic diversity in freshwater Macrobrachium prawns from indochinese riverine systems revealed by DNA barcode, species delimitation and phylogenetic approaches. PLoS One 16 (6), e0252546. doi: 10.1371/journal.pone.0252546
Spiridonov V. A. (2017). Two new species of Thalamita latreille 1829 (Decapoda, portunidae). Crustaceana 90 (7-10), 1211–1233. doi: 10.1163/15685403-00003691
Stat M., Huggett M. J., Bernasconi R., DiBattista J. D., Berry T. E., Newman S. J., et al. (2017). Ecosystem biomonitoring with eDNA: metabarcoding across the tree of life in a tropical marine environment. Sci. Rep. 7 (1), 1–11. doi: 10.1038/s41598-017-12501-5
Steffen W., Richardson K., Rockström J., Cornell S. E., Fetzer I., Bennett E. M., et al. (2015). Planetary boundaries: Guiding human development on a changing planet. Science 347 (6223), 1259855. doi: 10.1126/science.1259855
Suherman A., Santosa M. A., Ihsan Y. N., Wijayanto D., Juwana S. (2020). The eradication of IUU fishing in Indonesia for fisheries resources sustainability by the Task Force 115. Aquaculture Aquarium Conserv. Legislation 13 (5), 2522–2537. Available at: http://bioflux.com.ro/docs/2020.2522-2537.pdf
Teteleptal J. M., Ongkers O. T. S., Pattikawa J. A. (2017). Sustainability status of spiny lobster (Panulirus sp.) fishery in latuhatat waters, Ambon Island, Indonesia. Int. J. Fisheries Aquat. Stud. 5 (6), 205–210. vailable at: https://www.fisheriesjournal.com/archives/2017/vol5issue6/PartC/5-5-60-109.pdf
Thiel M., Watling L. (2015). “The natural history of the Crustacea,” in Lifestyles and feeding biology (New York: Oxford University Press), 593.
Thomsen P. F., Willerslev E. (2015). Environmental DNA–an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Virto L. R. (2018). A preliminary assessment of the indicators for sustainable development goal (SDG) 14 “Conserve and sustainably use the oceans, seas and marine resources for sustainable development”. Mar. Policy 98, 47–57. doi: 10.1016/j.marpol.2018.08.036
Wahyudin R. A., Hakim A. A., Boer M., Farajallah A., Wardiatno Y. (2016). New records of Panulirus femoristriga Von martens 1872 (Crustacea Achelata Palinuridae) from Celebes and Seram Islands, Indonesia. Biodiversity J. 7 (4), 901–906.
Watling L., Thiel M. (2013). The natural history of Crustacea volume 1: Functional morphology and diversity (New York: Oxford University Press), 515.
West K. M., Stat M., Harvey E. S., Skepper C. L., DiBattista J. D., Richards Z. T., et al. (2020). eDNA metabarcoding survey reveals fine‐scale coral reef community variation across a remote, tropical island ecosystem. Mol. Ecol. 29 (6), 1069–1086. doi: 10.1111/mec.15382
West K. M., Adam A. A. S., White N., Barrow D., Lane A., et al. (2022). The applicability of eDNA metabarcoding approaches for sessile benthic surveying in the Kimberley region, north‐western Australia. Environ. DNA 4 (1), 34–49. doi: 10.1002/edn3.184
Wolfe J. M., Breinholt J. W., Crandall K. A., Lemmon A. R., Lemmon E. M., Timm L. E., et al. (2019). A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc. R. Soc. B 286 (1901), 20190079. doi: 10.1098/rspb.2019.0079
Wowor D., Muthu V., Meier R., Balke M., Cai Y., Ng P. K. (2009). Evolution of life history traits in Asian freshwater prawns of the genus Macrobrachium (Crustacea: Decapoda: Palaemonidae) based on multilocus molecular phylogenetic analysis. Mol. Phylogenet. Evol. 52 (2), 340–350. doi: 10.1016/j.ympev.2009.01.002
WWF (2016). Living planet report 2016: Risk and resilience in a new era (Gland, Switzerland: WWF International), 145.
Keywords: megabiodiversity, Coral Triangle, genetic biomonitoring, marine conservation, marine policy, environmental DNA
Citation: Madduppa H, Sani LMI, Nugroho KC, Bengen DG, Muchlisin ZA, Fadli N, Subhan B, Arafat D, Zamani NP, Sunuddin A, Ismet MS, Srimariana ES, Cakasana N, Lestari DF, Santoso P, Setyaningsih WA, Baksir A, Manurung VR, Damora A, Ramadhaniaty M, Sartimbul A, Oli MYUP, Tanod WA, Munira, Dobo J, Setyobudi E, Nadiarti N, Jompa J, Auliyah N, Ndobe S, Mahyudi I, Ninef JSR, Rehatta BM and Moore AM (2022) eDNA metabarcoding of decapod crustaceans across Indonesian seas has implications for biodiversity conservation and fisheries sustainability. Front. Mar. Sci. 9:918295. doi: 10.3389/fmars.2022.918295
Received: 12 April 2022; Accepted: 04 August 2022;
Published: 07 September 2022.
Edited by:
Yaisel Juan Borrell Pichs, University of Oviedo, SpainReviewed by:
Sara Fernández, Galway-Mayo Institute of Technology, IrelandMa. Carmen Anonuevo Ablan Lagman, De La Salle University, Philippines
Alba Ardura, University of Oviedo, Spain
Copyright © 2022 Madduppa, Sani, Nugroho, Bengen, Muchlisin, Fadli, Subhan, Arafat, Zamani, Sunuddin, Ismet, Srimariana, Cakasana, Lestari, Santoso, Setyaningsih, Baksir, Manurung, Damora, Ramadhaniaty, Sartimbul, Oli, Tanod, Munira, Dobo, Setyobudi, Nadiarti, Jompa, Auliyah, Ndobe, Mahyudi, Ninef, Rehatta and Moore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lalu M. Iqbal Sani, aXFiYWxzYW5pQG9jZWFub2dlbi5jb20=; Dietriech G. Bengen, ZGlldGVyQGluZG8ubmV0Lmlk; Neviaty P. Zamani, bmV2aWF0eUBhcHBzLmlwYi5hYy5pZA==
 Hawis Madduppa
Hawis Madduppa Lalu M. Iqbal Sani
Lalu M. Iqbal Sani Kuncoro Catur Nugroho
Kuncoro Catur Nugroho Dietriech G. Bengen1*
Dietriech G. Bengen1* Nur Fadli
Nur Fadli Dondy Arafat
Dondy Arafat Neviaty P. Zamani
Neviaty P. Zamani Adriani Sunuddin
Adriani Sunuddin Meutia Samira Ismet
Meutia Samira Ismet Nadya Cakasana
Nadya Cakasana Vindy Rilani Manurung
Vindy Rilani Manurung Adrian Damora
Adrian Damora Aida Sartimbul
Aida Sartimbul Wendy Alexander Tanod
Wendy Alexander Tanod Eko Setyobudi
Eko Setyobudi Nadiarti Nadiarti
Nadiarti Nadiarti Jamaluddin Jompa
Jamaluddin Jompa Samliok Ndobe
Samliok Ndobe Indra Mahyudi
Indra Mahyudi Jotham S. R. Ninef
Jotham S. R. Ninef Beatrix M. Rehatta
Beatrix M. Rehatta Abigail Mary Moore
Abigail Mary Moore