- 1Key Laboratory for Water Quality and Conservation of the Pearl River Delta, Institute of Environmental Research at Greater Bay Area, Ministry of Education, Guangzhou University, Guangzhou, China
- 2State Key Laboratory of Tropical Oceanography (LTO), South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 3University of Chinese Academy of Sciences, Beijing, China
Denitrification is an important pathway for nitrogen sink and N2O emissions, but little is known about the ecological distribution of key functional genes of denitrification and their potential N2O emissions in marine sediments. In this study, we analyzed the abundance, ecological distribution, and diversity of key functional genes (nir and nosZ) for denitrification in the northern South China Sea (SCS) surface sediments. Our results showed that the gene abundances varied from 105 to 108 and from 106 to 107 copies·g-1 for the nirS and nirK, respectively. The nosZ II/nosZ I gene abundance ratios were 1.28–9.88 in shallow-sea and deep-sea sediments, suggesting that the nosZ II gene should play a dominant role in N2O reduction in the northern SCS sediments. Moreover, the significantly higher abundance ratios of nir/nosZ in deep-sea surface sediments implied that there might be stronger N2O emissions potential in deep-sea sediments than in shallow-sea sediments. The ecological distribution profiles of the nirS, nosZ I, and nosZ II gene communities varied with water depth, and denitrification genes in shallow-sea and deep-sea sediments differed in their sensitivity to environmental factors. Water temperature was the major factor affecting both the abundance and the community distribution of the nirS gene in deep-sea sediments. Nitrate was the major factor shaping the community of nosZ I and nosZ II genes in shallow-sea sediments. Our study provides a pattern of ecological distribution and diversity for the nir and nosZ genes and emphasizes the role of these key functional genes in potential N2O emissions of the northern SCS surface sediments.
Introduction
Nitrogen is essential for all creatures and is a major nutrient supporting marine primary production (Wang et al., 2019a). With the rapid development of industry, increased amounts of reactive nitrogen generated by anthropic activities have flooded into estuaries, coastal bays, and oceans (Seitzinger et al., 2010). Fortunately, 40–50% of the external sources of dissolved inorganic nitrogen (DIN) input can be removed by denitrification and released into the ambient environment as N2 to maintain the nitrogen cycle balance in marine sediments (Ward, 2013). Meanwhile, N2O, a powerful and long-lived greenhouse gas with 300 times higher global warming potential than CO2, may also form during denitrification (Makowski, 2019).
The denitrification process includes four stepwise reduction steps: reduction, reduction, NO reduction, and N2O reduction (Knowles, 1982). Among them, reduction and N2O reduction are two key steps. Two enzymes with different structures but similar functions catalyze reduction; the nirS gene encodes the copper-containing reductases and the nirK gene encodes the cytochrome cd1-containing reductases (Zumft, 1997). In most cases, the nirS gene or nirK gene occurs in bacteria as a single copy (Jones et al., 2008; Graf et al., 2014); the two genes cannot co-exist in microorganisms (Coyne et al., 1989). However, many research on reductase has only focused on the nirS gene (Smith et al., 2007; Christopher et al., 2013). The nirK gene was detected in only 30% of denitrifying microorganisms but with a broader range of taxa (Coyne et al., 1989). The norB gene encodes the NO reductase (responsible for the reduction of NO to N2O), but as compared with the nirS or nirK genes, research on the potential of the norB gene as a molecular marker started relatively late (Guo et al., 2011). N2O reductase (Nos), which catalyzes N2O reduction, is encoded by the nosZ gene. The nosZ gene generally occurs as a single copy, except in Pseudomonas raps and Thiomonas denitrificans with the form of a double copy (Sanford et al., 2012). A significant deviation between the predicted N2O emissions based on the gene abundance and expression of the nosZ and the actual N2O emissions detected (Henderson et al., 2010; Morales et al., 2010), indicating that a class of previously undiscovered functional N2O reductase genes might exist. Sanford et al. (2012) compared the amino acid sequences of nosZ genes in public databases and found that, in addition to the known typical nosZ I gene, there was also an emerging atypical nosZ II gene. Emerging studies about nosZ II have focused on the soil as a significant source of N2O, such as arable soil (Graf et al., 2016), grassland soil (Di et al., 2014), and salt marsh ecosystems (Dini-Andreote et al., 2016). The nosZ II gene has been found to have a particularly significant effect on N2O reduction in soils (Hallin et al., 2018). However, there is little research on the nosZ II gene in oceans whose surfaces cover over 70% of the Earth. The abundance and community diversity of the nosZ II gene and its relative importance on N2O reduction in oceans are poorly understood, as well as how these four genes (nirS, nirK, nosZ I, and nosZ II) collectively affect N2O emissions by denitrification in oceans.
The SCS is a largest marginal sea in China and an ideal region for research on the nitrogen cycle (Su, 2004; Yang et al., 2021). The SCS was reported to be a contributor of atmospheric N2O, with about 132% N2O saturation in its waters (Tseng et al., 2016; Ren et al., 2017). The atmospheric contribution of N2O from the SCS has been estimated to be 0.15-0.24 Tg·a-1, which was much larger than that of the open ocean (Zheng et al., 2009; Ren et al., 2017). Moreover, the N2O concentration in coastal waters of northern SCS increased gradually from the surface to the bottom (Xu et al., 2006; Han et al., 2013), suggesting that a source of N2O might exist in deep waters or sediments. The present research on N2O in the SCS mainly focuses on the distribution of N2O in waters and its correlation with apparent oxygen utilization (AOU) (Xu et al., 2006; Zheng et al., 2009; Han et al., 2013; Tseng et al., 2016; Ren et al., 2017). As a hot spot for denitrification, sediment is generally overlooked for N2O emissions in the SCS. The balance between nir (nirS and nirK) and nosZ (nosZ I and nosZ II) genes should determine N2O emissions to some extent (Domeignoz-Horta et al., 2015). Combining the abundances and communities of denitrification-related functional genes will provide valuable information to explain potential N2O emissions driven by the denitrification process.
In this study, we analyzed the distribution and relative importance of key functional genes (nir and nosZ) in N2O emissions during denitrification by using qPCR and high-throughput sequencing analysis. We aimed to provide valuable insight into the implications of key functional genes on N2O emissions by denitrification in marine sediments.
Materials and Methods
Sample Collection and Environmental Parameters Measurement
The South China Sea (SCS) lies in the Northwest Pacific Ocean, expanding from the equator to latitude 23°N and from 99°E to 121°E (Su, 2005). It covers an area of about 3.50 × 106 km2, combined with an average water depth of 1000 m (Chen et al., 2001). A total of 17 surface sediments (0–5 cm) were obtained during 3 cruises of R/V Shiyan 3 (in May 2014, October 2015, and October 2016). The sampling map is shown in Figure 1. Site description, sampling details, and methods of analysis are provided by Wu et al. (2019). Water depth and water temperature was continuously recorded, and the salinity and DIN (including , , and ) of porewater was examined as described by Wu et al. (2016) and Guan et al. (2017).
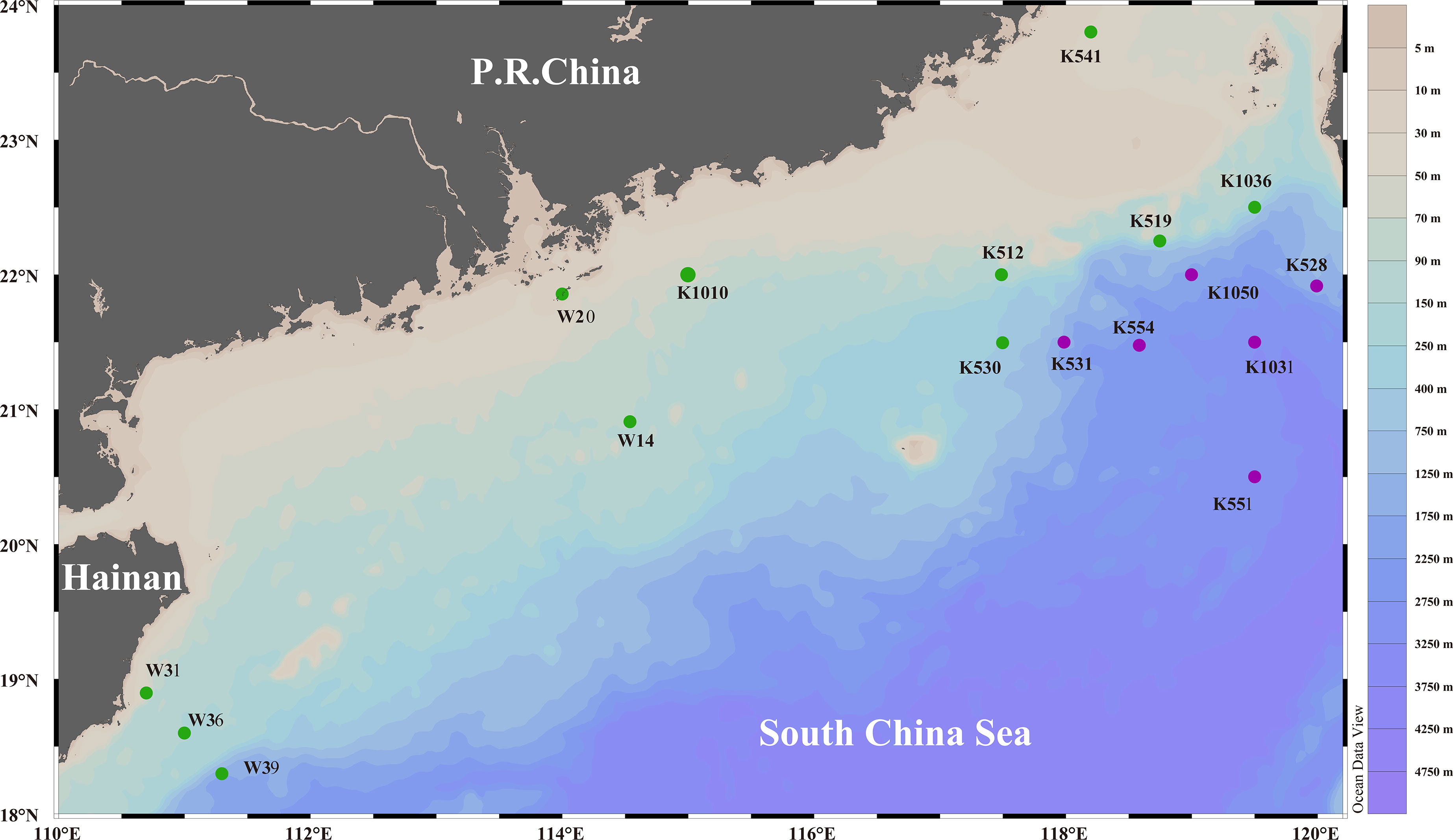
Figure 1 The map of sampling sites. The dots denoted sampling stations (green dots represent shallow-sea stations and purple dots represent deep-sea stations), and the scale bar denoted water depth.
DNA Extraction and PCR Amplification
Details of DNA extraction and quality determination processes were described by Wu et al. (2019). The primer pairs cd3aF/R3cd (Throback et al., 2004; Yergeau et al., 2007), nosZ 2F/nosZ 2R (Henry et al., 2006), nosZ IIF/nosZ IIR (Jones et al., 2013) were used to amplify the genes of the nirS, nosZ I, and nosZ II, and the products’ lengths were 426, 267, 749 bp, respectively. Detailed protocols about PCR amplification are listed in Supplementary Table 1. PCR products recovered from gel electrophoresis were purified with an agarose gel DNA extraction kit (TaKaRa Bio, Japan).
High-Throughput Sequencing Analysis
The purified products were sequenced on an Illumina HiSeq platform (Genewiz Corporation, Suzhou, China) and then analyzed using Mothur (version 1.39.5), according to high-throughput sequencing standard procedures reported by Schloss et al. (2009). The nosZ I and nosZ II gene sequences were downloaded in Fungene (http://fungene.cme.msu.edu/) as reference sequences, and the reference sequences of nirS gene were derived from Fungene and NCBI (https://www.ncbi.nlm.nih.gov/) as described by Liu et al. (2020).
Briefly, valid raw data were derived after the removal of corresponding primers. The trimmed sequences were generated after the processes of binning, denoising, and trimming, and then they were aligned with prepared reference databases. To ensure the quality of the sequences, screen.seqs and chimera.uchime commands were employed to filter out poor-quality reads and to remove chimeric sequences. Finally, the classifying.seqs command was employed and the level of confidence was set at 85% for the nirS and nosZ genes (Wu et al., 2021a). A 97% sequence identity was used to define the representative sequence of each operational taxon unit (OTU) for these genes for further analysis (Wu et al., 2021a; Wu et al., 2021b). Rarefaction curves, Shannon curves, ranking abundance, and the alpha-diversity index were analyzed by Mothur based on observed OTUs (Schloss et al., 2009) and visualized by Origin 2019b. Origin 2019b was employed to describe the composition of microbial community. The neighbor-joining phylogenetic tree was constructed and visualized using MEGA7.0 (https://www.megasoftware.net/) and EvolView (He et al., 2016). A heat map was plotted by Microsoft Excel (Microsoft Excel 2016).
qPCR Analysis of Key Functional Genes for Denitrification
The abundances of nir and nosZ genes were detected using a LightCycler ® R480 II Real-Time PCR (Roche, Switzerland), and the primers and detailed protocols are listed in Supplementary Table 1. In addition to the primer pairs mentioned above, the nirK gene was amplified and quantified by primer sets F1aCu/R3Cu (Hallin and Lindgern, 1999). Each sample was quantified in triplicate, and negative controls were performed simultaneously. A standard curve was constructed with a plasmid DNA of the target gene with known concentration and copy number. The specificity of PCR products was examined by both the melting curve and gel electrophoresis. High amplification efficiencies (90–110%) and correlation coefficients (R2 > 0.97) were shown for the qPCR results.
Statistical Analysis
The map of the sampling area was drawn using Ocean Data View (http://odv.awi.de). Both the bacterial communities and their correlations with environmental variables were explored by canonical correspondence analysis (CCA), redundancy analysis (RDA) and principal coordinates analysis (PCoA) with Canoco 5 (ter Braak and Šmilauer, 2012). The Pearson correlation analysis of abundance and diversity with environmental variables was constructed and visualized using IBM SPSS statistics 19 and Microsoft Excel (Microsoft Excel 2016). Non-parametric test statistics (Mann-Whitney test) was used to examine the differences of gene abundance ratios in shallow-sea and deep-sea sediments as the data did not conform to a normal distribution.
Nucleotide Sequence Accession Numbers
Raw gene sequences of nirS, nosZ I, and nosZ II were deposited in NCBI under the submission numbers PRJNA792035, PRJNA792063, and PRJNA792068, respectively.
Results
Environmental Characteristics of Surface Sediments
The environmental characteristics of surface sediments were described in (Wu et al., 2019). The water depth at the sediments can reach 2,980 m, with water temperatures varying from 1.58 to 25.36°C (Supplementary Figure 1). Moreover, the water temperature showed a general drop with water depth (Supplementary Table 2), and the average water temperature at depths over 2000 m water was as low as 1.72 ± 0.12°C. Little change was observed in the salinity and pH of porewater, with an average of 35.55 ± 1.65 and 8.31 ± 0.23, respectively. DIN concentrations varied at sampling sites, and mainly existed in the form of , with concentration ranging from 1.8 to 46.43 μM. The concentration of remained at a low level (<1.11 μM) in most samples. The concentration of the W31 sample was as high as 10.61 μM but consistently low (<1.11 μM) in other samples. In addition, DIN concentrations were positively correlated with the concentrations (r = 0.983 and p < 0.01) (Supplementary Table 2).
Abundance and Ratios of Key Functional Genes and Their Relationships With Environmental Factors
The nir and nosZ genes were all detected in sediments (Figure 2 and Supplementary Table 3). An obvious variation was observed in the gene abundance of the nirS, ranging from (6.04 ± 2.78) × 105 to (1.52 ± 0.05) × 108 copies·g-1; maximum and minimum values of nirS gene abundance were reached in the W20 and K554 samples, respectively. The nirS gene abundance inversely correlated with water depth (r = -0.486 and p < 0.05) and correlated favorably with water temperature (r = 0.488 and p < 0.05) (Supplementary Table 2). Moreover, the nirS gene abundance showed a favorable correlation with the nosZ I gene (r = 0.768 and p < 0.01) and the nosZ II gene (r = 0.933 and p < 0.01) (Supplementary Table 2). The nirK gene abundance fluctuated between (1.23 ± 0.22) × 106 and (6.25 ± 1.60) × 107 copies·g-1, and it was strongly correlated with the concentration of (r = 0.752 and p < 0.01) (Supplementary Table 2). The nirK/nirS ratios were <1 in shallow-sea surface sediments (except for the W31 and W14 samples) and >1 in deep-sea surface sediments (Figure 2B and Supplementary Table 3).
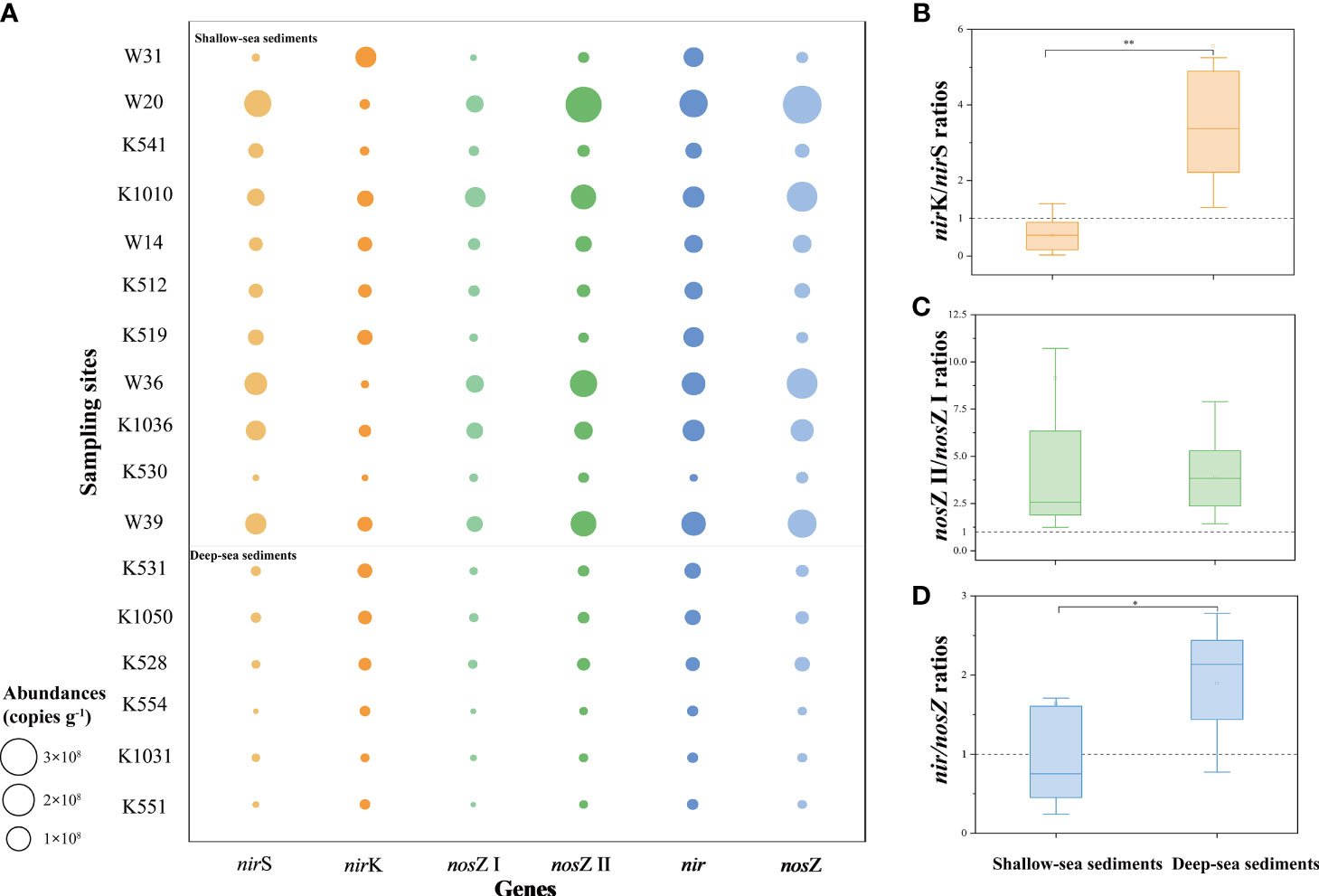
Figure 2 Abundances and ratios of key functional genes for denitrification in surface sediments of SCS. The gene abundances include the nir (nirS and nirK) and nosZ (nosZ I and nosZ II) genes (A), gene ratios include the nirK/nirS gene abundance ratios (B), the nosZ II/nosZ I gene abundance ratios (C) and the nir/nosZ gene abundance ratios (D). The asterisks denoted statistically differences by Mann-Whitney test, two asterisks (**) denotes p < 0.01 and one asterisk (*) denotes p < 0.05.
The gene abundances of the nosZ I and nosZ II varied from (5.51 ± 0.39) × 105 to (7.18 ± 0.09) × 107 copies·g-1 and from (3.60 ± 0.17) × 106 to (2.92 ± 0.41) × 108 copies·g-1, respectively (Figure 2A and Supplementary Table 2). The nosZ I gene abundance positively correlated with water temperature (r = 0.520 and p < 0.05) (Supplementary Table 2). Furthermore, the gene abundance of the nosZ II was found to be higher than the nosZ I, with the nosZ II/nosZ I ratio >1 in each sample (Figure 2C). Gene abundances of the nir was calculated as the total gene abundances of the nirS and nirK, and gene abundances of the nosZ as the total gene abundances of the nosZ I and nosZ II. The gene abundances of the nir and nosZ varied from 106 to 108 copies·g-1. The nir/nosZ gene abundance ratios were >1 in deep-sea surface sediments (except for the K528 sample) and <1 in most shallow-sea surface sediments (Figure 2D). Besides, there were significant differences in nirK/nirS ratios (p < 0.01) and nir/nosZ ratios (p < 0.05) between shallow-sea and deep-sea sediments (Figures 2B, D).
Community Composition and Diversity of the nirS Gene
After executing a series of quality control procedures, an average of 8400 ± 380 high-quality reads of nirS gene sequences per sample were screened (Supplementary Table 4). Rarefaction curves (Supplementary Figure 2A), Shannon index (Supplementary Figure 2B), rank abundance (Supplementary Figure 2C) and high coverage (Supplementary Table 4) all suggested that the sequencing results provided adequate bioinformation to study the nirS gene community composition and diversity. Moreover, the community diversity of the nirS gene declined with increasing water depth (OTUs, r = -0.722 and p < 0.01; Ace, r = -0.753 and p < 0.01; Chao1, r = -0.760 and p < 0.01; Shannon, r = -0.535 and p < 0.05), decreasing water temperature (OTUs, r = 0.55 and p < 0.05; Ace r = 0.617 and p < 0.01; Chao1, r = 0.608 and p < 0.05), and decreasing salinity (Shannon, r = 0.496 and p < 0.05; Simpson, r = -0.505 and p < 0.05; Evenness, r = -0.250 and p < 0.05) (Supplementary Table 5).
The dominant OTUs (top 50 OTUs, covering 49.24% of the sequences after removing rare OTUs) of the nirS gene were phylogenetically associated with 10 different clusters (Figures 3A, B and Supplementary Figures 3 and 4). Moreover, only C6 belonged to Alpha-proteobacteria, whereas the other 9 clusters were affiliated to Gamma-proteobacteria (Figure 3B and Supplementary Figure 4). C6 contained only OTU45 and existed exclusively in the K530 sample, closely affiliated to Azospirillum (Figure 3B and Supplementary Figure 4). C1 (29.59% of the total sequences) and C5 (17.15% of the total sequences) showed some similarity with the uncultured bacteria in Changjiang Estuary and the East China Sea (Dang et al., 2008), San Francisco Bay (Lee and Francis, 2017), respectively. Moreover, C1 and C5 were more abundant in deep-sea surface sediments than those sampled in shallow-sea surface sediments (Figure 3A and Supplementary Figure 3). C2 (belonging to Chromohalobacter), C8 (belonging to Pseudomonas), and C9 (belonging to Thermomonas) were found almost exclusively in deep-sea surface sediments (Figures 3A, B). PCA revealed the nirS gene community in deep-sea surface sediments was separated from the shallow-sea surface sediments (accounted for 82.58% of the variance, Supplementary Figure 5). For shallow-sea surface sediments, the environmental variables detected in this study explained 47.49% of the total variance, and nitrate was the major factor shaping the spatial distribution of the nirS gene community (pseudo-F = 1.5 and p > 0.05) (Figure 3C). But for deep-sea surface sediments, the environmental variables detected in this study explained 65.66% of the total variance, and water temperature turned to be the major factor affecting the spatial distribution of the nirS gene community (pseudo-F = 2.1 and p < 0.01) (Figure 3D).
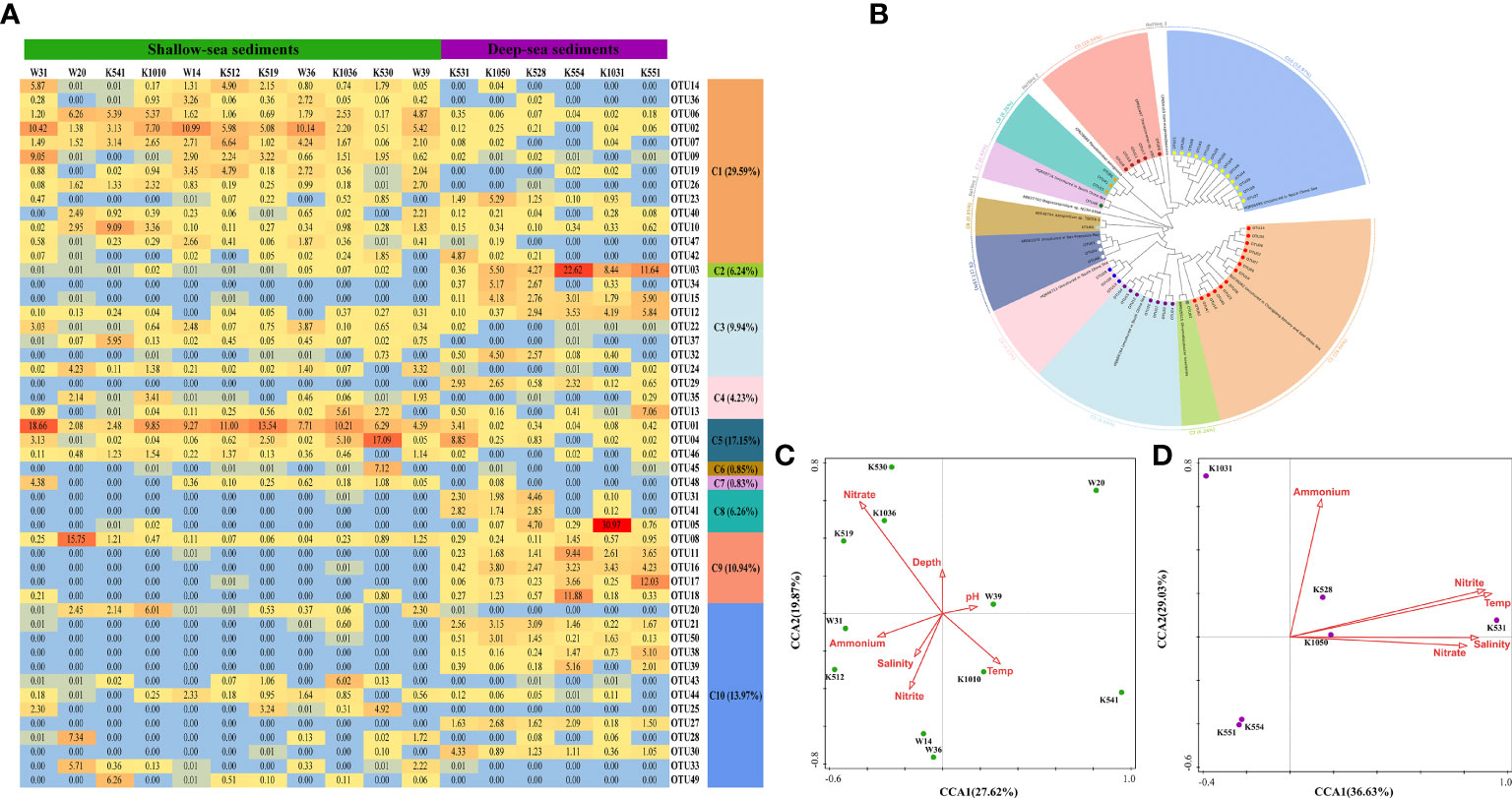
Figure 3 Microbial characteristics of the nirS gene in surface sediments of the northern SCS. (A) The heat map of dominant OTUs for the nirS gene (Top 50 OTUs). (B) Neighbor-joining phylogenetic tree of dominant OTUs for the nirS gene (Top 50 OTUs) and the reference sequences from Genbank (bootstrap 1000). (C) The canonical correspondence analysis (CCA) for the nirS gene derived from shallow-sea surface sediments. (D) The canonical correspondence analysis (CCA) for the nirS gene derived from deep-sea surface sediments.
Community Composition and Diversity of the nosZ Gene
A sum of 0.17 million nosZ I raw sequences for 17 samples and 0.10 million nosZ II raw sequences for 10 samples were detected. Rarefaction curves (Supplementary Figures 6A, 7A), Shannon index (Supplementary Figures 6B, 7B), and rank abundance (Supplementary Figures 6C, 7C) suggested that the sequencing results were sufficient enough to reveal the diversities and community compositions of the nosZ gene. The basic alpha diversities of the nosZ gene are presented in Supplementary Tables 6, 7. Moreover, the coverage of the nosZ gene was above 0.99, indicating that the library OTUs of the nosZ gene had been adequately captured (Supplementary Tables 6, 7). The community diversity of the nosZ I gene was impacted by water depth (OUT, r = -0.605 and p < 0.05), followed by pH (Shannon, r = 0.499 and p < 0.05) (Supplementary Table 8). However, for the nosZ II gene, its diversity was only affected by water depth (Shannon, r = -0.755 and p < 0.05; Simpson, r = 0.694 and p < 0.05) (Supplementary Table 9).
The dominant OTUs of the nosZ I gene were divided into 12 clusters, all of which belonged to Proteobacteria (Figure 4B and Supplementary Figure 8). Extensive sequences of the 50 OTUs, including C1, C4, C6, and C8, matched with soil or sediment samples. Many cultured pure bacteria have also been detected, such as Aestuarium (C3), Azospirillum (C7), Ruegeria (C9), Polymorphum (C10), Rhodobaca (C11), and Pseudomonas (C12). Among the 12 clusters, only Pseudomonas (C12) belonged to the Gamma-proteobacteria, whereas the other 11 clusters belonged to Alpha-proteobacteria (Figure 4B and Supplementary Figure 8). Moreover, Pseudomonas (C12) was only observed in deep-sea surface sediments. Aestuarium (C3), uncultured pure bacteria (C6), and Ruegeria (C9) were detected to be more abundant in surface sediments of deep sea as compared with those in shallow sea. However, the nosZ I gene in shallow-sea surface sediments was richer in uncultured Proteobacteria (C5) and Azospirillum (C7) than those in deep-sea surface sediments (Figures 4A, B; Supplementary Figure 9).
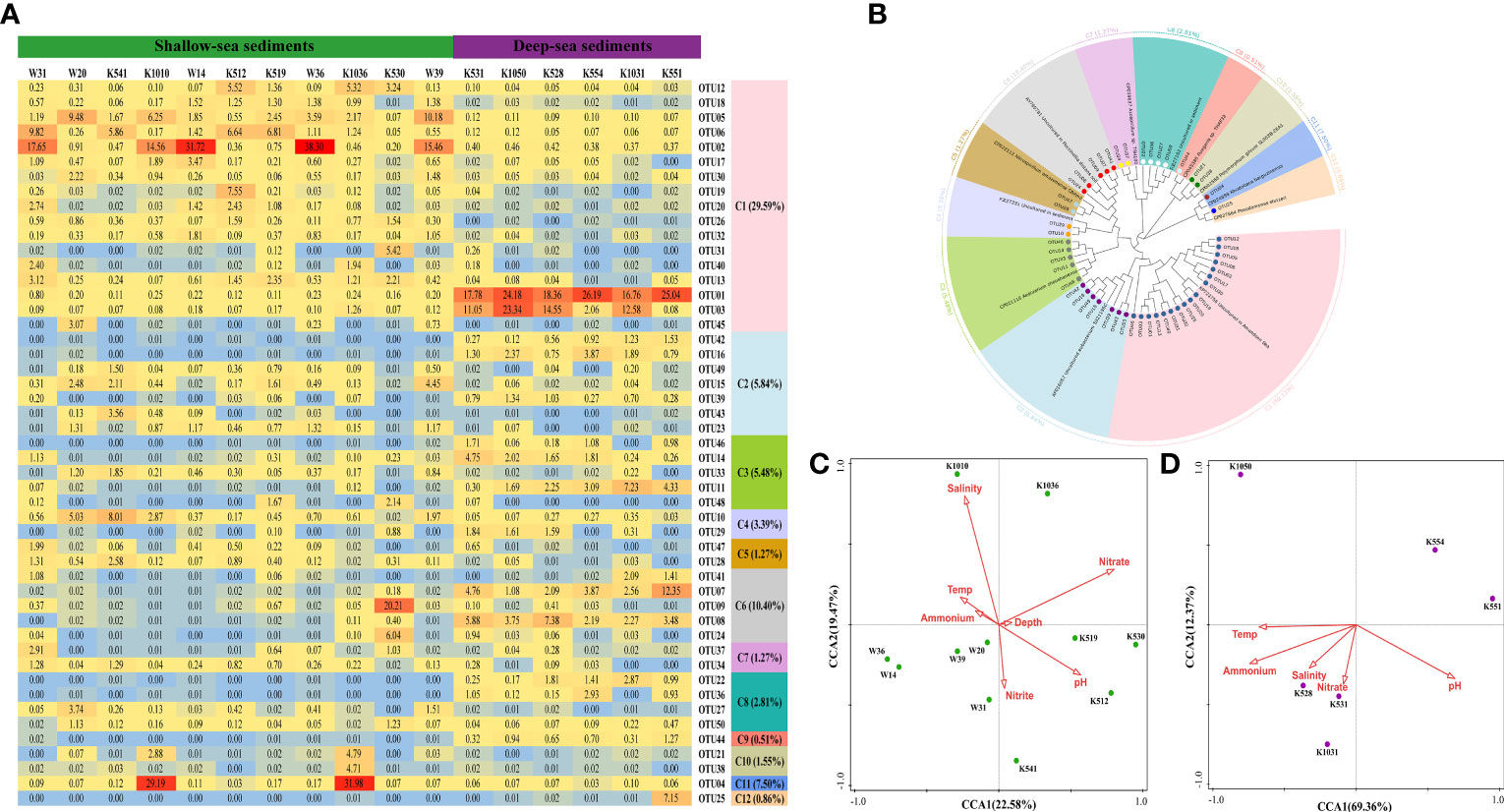
Figure 4 Microbial characteristics of the nosZ I gene in surface sediments of the northern SCS. (A) The heat map of dominant OTUs for the nosZ I gene (Top 50 OTUs). (B) Neighbor-joining phylogenetic tree of dominant OTUs (Top 50 OTUs) and the reference sequences from Genbank (bootstrap 1000). (C) The canonical correspondence analysis (CCA) for the nosZ I gene from shallow-sea surface sediments. (D) The canonical correspondence analysis (CCA) for the nosZ I gene from deep-sea surface sediments.
Dominant OTUs (top 50 OTUs) of nosZ II gene sequences were divided into 10 clusters, including Proteobacteria, Chloroflexi, and Bacteroidetes (Figures 5 A, B and Supplementary Figure 10). C1–C5, containing 69.50% of the total nosZ II gene sequences, belonged to Caldilinea aerophila. C6 (4.37% of the total sequence) was closely related to Magnetospirillum gryphiswaldense. C1, C2, C4, and C6 were richer in shallow-sea surface sediments than deep-sea surface sediments. On the contrary, C3 was only found in deep-sea surface sediments. C7 (9.75% of the total sequence), belonging to Lutibacter profundi, was more abundant in deep-sea surface sediments than shallow-sea surface sediments. C8 (Rhodothermaceae bacterium), C9 (Myxococcales bacterium), and C10 (uncultured bacterium in petrochemical soil and Fyrisan Fen) were just observed in some samples.
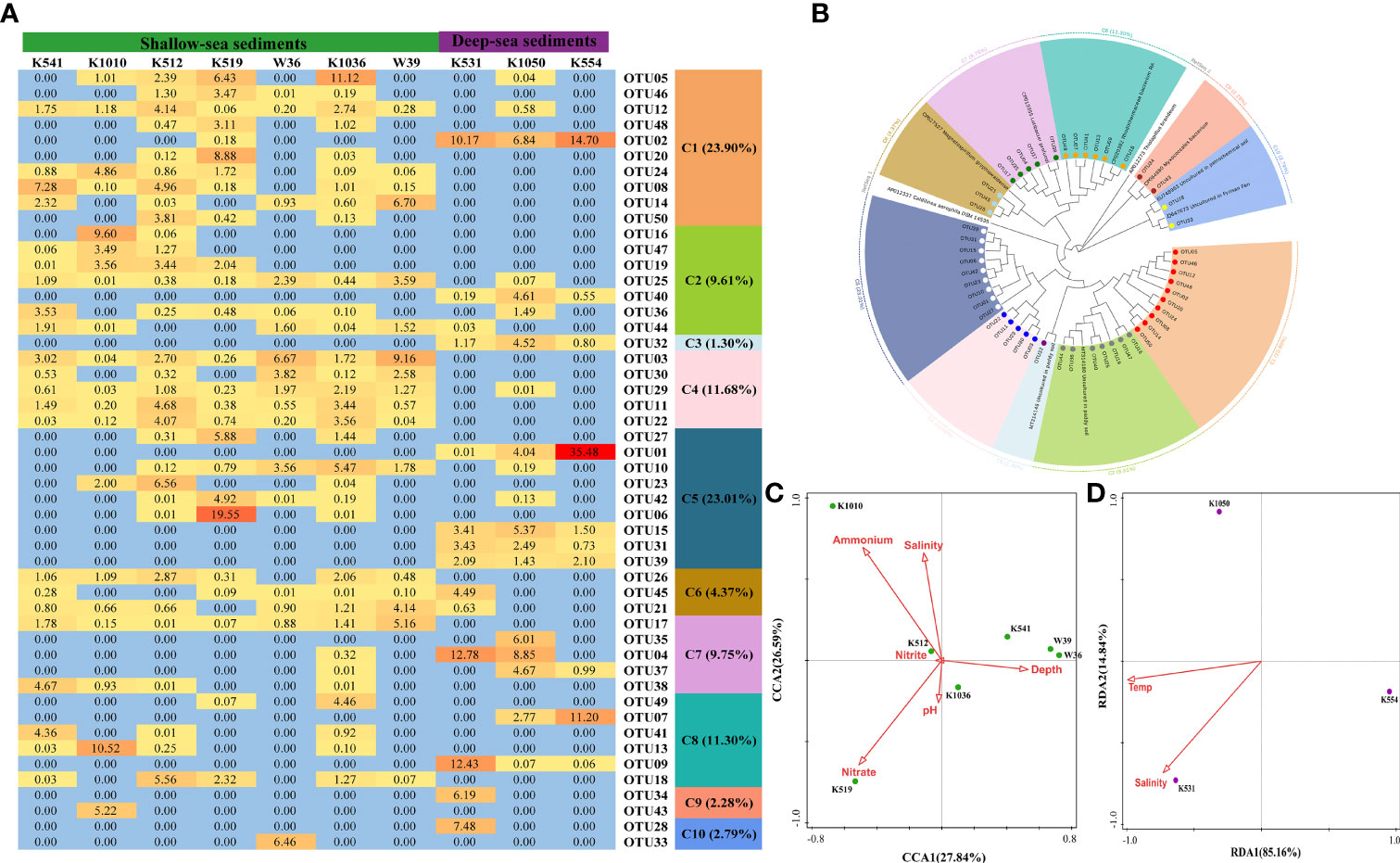
Figure 5 Microbial characteristics of the nosZ II gene in surface sediments of the northern SCS. (A) The heat map of dominant OTUs (Top 50 OTUs). (B) Neighbor-joining phylogenetic tree of dominant OTUs (Top 50 OTUs) and the reference sequences from Genbank (bootstrap 1000). (C) The canonical correspondence analysis (CCA) for the nosZ II gene from shallow-sea surface sediments. (D) The redundancy analysis (RDA) for the nosZ II gene from deep-sea surface sediments.
PCoA discovered that the gene communities of the nosZ I and nosZ II in the deep-sea group were separated from the shallow-sea group (explained by 74.22% and 76.49% of the variance for nosZ I and nosZ II gene, respectively) (Supplementary Figures 12, 13). For shallow-sea surface sediments, nitrate was the major factor shaping the community of nosZ I gene (pseudo-F = 1.9 and p < 0.05) (Figure 4C) and nosZ II gene (pseudo-F = 1.5 and p < 0.05) (Figure 5C). And for deep-sea surface sediments, ammonium and water temperature became the most important factor affecting the community of nosZ I (pseudo-F = 2.6 and p > 0.05) (Figure 4D) and nosZ II gene (pseudo-F = 5.3 and p > 0.05), respectively (Figure 5D).
Discussion
Ecological Distribution and Diversity of the nir Gene
In this study, the gene abundances of the nirS and the nirK was focused on in the northern SCS surface sediments, with nirS gene abundance ranging from 105 to 108 copies·g-1 and nirK gene abundance ranging from 106 to 107 copies·g-1 (Figure 2 and Supplementary Table 3). The gene abundances of the nirS and nirK were similar to the values shown in Fitzroy Estuary (Abell et al., 2009) and Elkhorn Slough (Smith et al., 2014). The abundances of the nirK or nirS genes might have been higher than examined, owning to the primer coverage limitations, since the abundances of the nirK or nirS genes could not be detected with any single universal detection method (Helen et al., 2016). The abundances of nir gene indicate the potential of N2O production in surface sediments of the SCS, as it catalyzes reduction to NO (an essential precursor of N2O). The nosZ gene co-occurred with the nirS gene (nosZ I, r = 0.768 and p < 0.01; nosZ II, r = 0.933 and p < 0.01) at a relatively higher rate than with the nirK gene (nosZ I, r = -0.007 and p > 0.05; nosZ II, r = -0.127 and > 0.05), indicating that the nirS gene might share a common regulatory mechanism that inhibits the loss of the nosZ gene (Supplementary Table 2). However, this mode of genomic linkage was not observed in the nirK gene (Supplementary Table 2). This result was in accordance with a finding that only 10-30% of nirK-harboring bacteria contained the nosZ gene (Clark et al., 2012). Meanwhile, there was also a positive correlation between the gene abundance of the nirK and N2O emissions (Clark et al., 2012). Additionally, Cuhel et al. (2010) observed a significant negative correlation between nirS gene abundance and the N2O/N2 ratio. These results suggested that the nirK gene was more prone to be incomplete denitrifiers, therefore, contributed more to N2O emissions. To some extent, the ratio of nirK/nirS can be seen as an indicator of the percentage of incomplete denitrification, i.e., potential N2O emissions. In this study, we found nirK/nirS >1 in deep-sea surface sediments and nirK/nirS <1 in shallow-sea surface sediments (except for the W31 and W14 samples) (Figure 2B; Supplementary Table 3). Moreover, the ratio of nirK/nirS was around 5.28 in most deep-sea surface sediments (Figure 2B and Supplementary Table 3). These results suggest that the nirK gene cannot be ignored during denitrification, especially in the northern SCS deep-sea surface sediments. More importantly, our study indicated that the northern SCS deep-sea surface sediments had a relatively higher but not strong potential N2O emissions as compared with the northern SCS shallow-water surface sediments (Figure 2 and Supplementary Table 3).
In addition, the abundance of the nirS gene decreased with increasing depth (r = -0.486 and p < 0.05) and decreasing water temperature (r = 0.488 and p < 0.05) (Supplementary Table 2). CCA also showed that water temperature was the major factor affecting the community of nirS gene in deep-sea sediments (Figure 3D). The results might indicate warm environment is more suitable for nirS gene to survive, which was consistent with a previous finding that warm incubation enhanced the growth of nirS-harboring bacteria (Warneke et al., 2011). The lower abundance of the nirS gene in deep-sea sediments also resulted in the significantly higher nirK/nirS ratios (Figure 2B). Some exclusive nirS genes were observed in deep-sea surface sediments, such as those in Chromohalobacter (C2) and Pseudomonas aeruginosa (C8), on average occupying 14.00 ± 9.86% and 17.06 ± 20.30% in the northern SCS deep-sea surface sediments, respectively (Figures 3A, B and Supplementary Figure 3). Similar to the anammox detected in the SCS, the community shift between shallow-sea and deep-sea sediments of the nirS gene might be an adaptation to the low-temperature environment in the deep sea (Oshiki et al., 2016; Wu et al., 2019). Research on surface sediments of San Francisco Bay showed that the environment with higher nitrate and lower water temperature might be more preferred for nirS and nirK genes, respectively (Lee and Francis, 2017). Our study observed that the nirS gene community diversity decreased with increasing salinity (Supplementary Table 5, Shannon, r = 0.496 and p < 0.05; Simpson, r = -0.505 and p < 0.05; Evenness, r = -0.250 and p < 0.05). In addition, concentration was associated with the community composition of the nirK gene (Lee and Francis, 2017). In this study, a positive and stronger correlation between nirK gene abundance and concentration was found (r = 0.752 and p < 0.01) (Supplementary Table S2). Further investigation is needed for a comprehensive understanding between complex environmental characteristics and bacterial community.
Ecological Distribution and Diversity of the nosZ Gene
There are biotic processes (denitrification, nitrification, and the reduction of dissimilatory nitrate to ammonium, i.e., DNRA) and an abiotic process (chemodenitrification) that can be the sources of N2O (Stremińska et al., 2012). However, the only known N2O sink is the N2O reduction to N2 by the N2O reductase encoded by the genes of nosZ I or nosZ II (Sanford et al., 2012; Jones et al., 2013). Although the nosZ II gene has been found to be a different type of N2O reductase gene (Sanford et al., 2012; Yoon et al., 2016), many research on the nosZ gene still ignored the differences between the two distinctive clades, especially in estuaries and oceans (Wang et al., 2019b; Aamer et al., 2020). In this study, the nosZ I and nosZ II genes were both widely distributed in the northern SCS surface sediments, with gene abundances ranging from 105 to 107 copies·g-1 and from 106 to 108 copies·g-1, respectively (Figure 2 and Supplementary Table 3). The gene abundances of the nosZ I were higher than the level reported at Gloucester Point (Wu et al., 2021a) but lower than that measured in the rhizosphere (Zhao et al., 2019). The gene abundances of the nosZ II were similar to the values detected at Gloucester Point (Wu et al., 2021b) and in the riparian zone (Zhao et al., 2018). The gene abundances of the nosZ I were correlated to the nirS gene (r = 0.768 and p < 0.01) rather than the nirK gene (r = -0.007 and p > 0.05), and the gene abundances of the nosZ II were closely associated with the nirS gene (r = 0.933 and p < 0.01) rather than the nirK gene (r = -0.127 and p > 0.05), indicating that the nosZ gene might be closely coupled to the nirS gene rather than the nirK gene during evolution. These results were also observed in Tibetan alpine meadows (Malviya et al., 2016; Samad et al., 2016) and were consistent with a former understanding that nirS-carrying bacteria were more potentially complete denitrifiers (Graf et al., 2014).
Although the higher gene abundances of the nosZ II over the nosZ I at 1.5–10 has been commonly reported in a variety of environments such as soil, sewage treatment plants, wetlands, and rice fields (Tsiknia et al., 2015; Graf et al., 2016), little research has been conducted on the nosZ II gene in marine ecosystems. At Gloucester Point, the gene abundance of the nosZ II was two orders of magnitude larger than the nosZ I based on PCR quantification (Wu et al., 2021a). Another two studies based on metagenomic analysis also supported the dominance of the nosZ II gene in the marine oxygen minimum zone (Bertagnolli et al., 2020) and in salt marshes (Graves et al., 2016). In this study, the nosZ II/nosZ I ratios in the SCS surface sediments were 1.28–9.88, showing that the nosZ II gene was dominant in both shallow- and deep-sea surface sediments (Figure 2C and Supplementary Table 3). The genome of microorganisms can contain different combinations of denitrification genes, and organisms solely equipped with the nosZ gene coding Nos are non-denitrifying N2O microorganisms (Roco et al., 2016; Lycus et al., 2017). Non-denitrifying N2O microorganisms are more likely to be true N2O sinks than denitrifying N2O microorganisms, as they only complete N2O reduction without contributing to N2O production (Hallin et al., 2018). Intergenomic comparisons have shown that 51% of microorganisms with the nosZ II gene, which lacked the nir gene, were non-denitrifying N2O reducers (Graf et al., 2014); only 17% of microorganisms with the nosZ I gene which lacked the nir gene were non-denitrifying N2O reducers (Graf et al., 2014). Pioneering research has also highlighted the role of the nosZ II gene instead of the nosZ I gene acting as an N2O sink in soil (Jones et al., 2014).
Coherent with past research (Jones et al., 2014; Wittorf et al., 2016), the community diversity of the nosZ II gene was higher than the nosZ I gene in the northern SCS surface sediments. At the phylum level, the nosZ II gene was detected in Proteobacteria, Chloroflexi, and Bacteroidetes (Figure 5B and Supplementary Figure 10), and the nosZ I gene was only found in Proteobacteria (Figure 4B and Supplementary Figure 8). A competitive advantage might be provided by higher community diversity of the nosZ II gene in adjusting to different ecology environments. This hypothesis was supported by the Pearson analysis, i.e., no environmental factors except depth were found to be correlated with the community diversity of the nosZ II gene (Shannon, r = -0.755 and p < 0.05; Simpson, r = 0.694 and p < 0.05; Evenness, r = -0.742 and p < 0.05) (Supplementary Table 9). For the nosZ II gene community, Alpha-proteobacteria were more abundant in shallow-sea surface sediments than deep-sea sediments, but the opposite was true for Bacteroides (Figure 5B and Supplementary Figure 11). The high abundance and community diversity of the nosZ II gene indicated that strong N2O reduction potential might exist in the northern SCS surface sediments. For shallow-sea sediments, nitrate was the major factor affecting the community of both nosZ I and nosZ II gene (Figures 4C and 5C), suggesting nitrate may be in short supply in shallow-sea sediments relative to the demand for denitrifying bacteria.
Implications of Key Functional Genes for Potential N2O Emissions by Denitrification
N2O can be produced through nitrification, denitrification, and DNRA; however, N2O exists as an intermediate product in denitrification and as an end product in nitrification and DNRA processes (Zumft, 1997; Sanford et al., 2012; Harris et al., 2021). These microbial processes involving N2O can happen simultaneously in the marine ecosystems, an N2O pool might be formed before it is reduced to N2 through denitrification. Under such a circumstance, denitrification plays an important role in the conversion of N2O (Wang et al., 2014; Zhao et al., 2021). Different abundances of denitrifying genes were associated with varying denitrification enzyme activities (Holtan-Hartwig et al., 2000; Rich et al., 2003) and indicated potential N2O production or consumption (Bergaust et al., 2010; Morales et al., 2010). The abundance analysis of denitrifying genes might offer a closer connection for potential N2O emissions (Cuhel et al., 2010). The nir and nosZ genes are vital denitrification genes and closely related to N2O emissions. The enzyme encoded by nir gene produce a precursor for N2O synthesis; the enzymes encoded by nosZ gene reduces N2O to N2 (Butterbach-Bahl et al., 2014). The gene abundance ratio of nir/nosZ can be regarded as an indicator of potential N2O emissions. Previous research has shown the balance between nir and nosZ genes can partially determine the level of N2O emissions in soils and lakes (Domeignoz-Horta et al., 2015; Saarenheimo et al., 2015). In this study, the nir/nosZ ratios were calculated in the northern SCS sediments. In the shallow-sea surface sediments, the nir/nosZ ratios ranged from 0.27 ± 0.03 to 5.72 ± 0.51, which was in accordance with the results found in rhizosphere soils (Zhao et al., 2019). Most of the nir/nosZ ratios in the shallow-sea surface sediments were < 1, implying that the N2O produced might be reduced in shallow-sea surface sediments. This implication was supported by the relatively low value of N2O concentration (around 12.5 nmol·L-1) in shallow-sea water (Tseng et al., 2016). However, in deep-sea surface sediments, the nir/nosZ ratios were > 1, with the highest value of 2.55 ± 0.33 (except for the K528 sample). These results suggested that potential N2O production was slightly higher than that of reduction during denitrification, thus, resulting in a small amount of N2O emissions. This implication was supported by a relatively higher value of N2O concentration (around 25 nmol·L-1) in deep-sea waters (Tseng et al., 2016). In this study, the nir/nosZ ratios suggested that deep-sea surface sediments might have stronger potential N2O emissions than shallow-sea sediments as the significantly higher nir/nosZ ratios in deep-sea sediments (Figure 2D). Another study also showed N2O concentrations in bottom waters were about 2.5 times than that of surface water in the northern SCS, indicating that N2O was abundantly accumulated in deep sea (Zheng et al., 2009; Ren et al., 2017). Although most research based on significant correlations among N2O, AOU, and has provided circumstantial evidence that N2O was mainly derived from nitrification (Xu et al., 2006; Tseng et al., 2016; Ren et al., 2017). Marchant et al. (2016) reported N2O was mainly produced by denitrification in sediments of the Wadden Sea. Denitrification process cannot be excluded in an oxygen-depleted deep sea of the SCS (Xu et al., 2006; Zheng et al., 2009; Tseng et al., 2016; Ren et al., 2017). Denitrification is more likely to occur under low oxygen conditions, and N2O emissions can be accelerated due to the inhibitory effect of oxygen on the activity of Nos (Körner and Zumft, 1989; Codispoti, 2010). Tseng et al. (2016) reported N2O maxima peaked at depths where dissolve oxygen was minimal at SCS and the West Philippines Sea. Besides, mineralization of organic matter and the product of nitrate from nitrification in the upper water column may provide substrate for denitrification in deep-sea sediments (Gao et al., 2018), and sediments generally contain minerals which can preserve organic substrates (Kleber et al., 2021). Therefore, the substrate can support the abundance of nirK and nosZ genes in the deep-sea sediments. Additional direct evidence based on SP values measured for N2O has verified that both nitrification process and denitrification process can produce N2O in the SCS waters (Zhang et al., 2019). These results all confirmed our speculation based on nir/nosZ ratios. The gene analysis may provide new insight and act as an indicator of N2O emissions, especially when there are difficulties obtaining in situ N2O measurement.
Conclusion
In this study, we investigated the ecological distribution and diversity of key functional genes (nir and nosZ) for denitrification in the northern SCS shallow-sea and deep-sea surface sediments. The results showed that both nir (nirS and nirK) and nosZ (nosZ I and nosZ II) genes were widely distributed in the northern SCS surface sediments. Denitrification genes in shallow-sea and deep-sea sediments differed in their sensitivity to environmental factors. The community distribution and diversity of these genes were affected by water depth, nitrate and water temperature. Furthermore, the significantly higher abundance ratios of nir/nosZ in deep-sea as compared with shallow-sea sediments suggested that deep-sea sediments might have strong potential N2O emissions as compared with shallow-sea sediments. In summary, this study provides an ecological distribution pattern of functional genes of denitrification and the implication on N2O emissions by denitrification in marine sediments.
Data Availability Statement
Raw gene sequences of nirS, nosZ I, and nosZ II were deposited in NCBI under the submission numbers PRJNA792035, PRJNA792063, and PRJNA792068, respectively.
Author Contributions
HX conducted the experimental operation and wrote the original draft. YH was responsible for the conceptualization. HX, YH, and AL reviewed and edited the manuscript. JW was responsible for field sampling. All authors read and approved the final manuscript.
Funding
The work was funded by the National Natural Science Foundation of China (No. 31870100, No. 91851111 and No. 42006122), Basic and Applied Basic Research Foundation of Guangdong Province (No. 2019B1515120066 and No. 2020A1515110597), Young Talent Research Project of Guangzhou Education Bureau (No. 202032795), Innovation Team Project of Guangdong Provincial Department of Education (2021KCXTD016).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Yaohao Hu for his kind help in high-throughput sequencing analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.912402/full#supplementary-material
References
Aamer M., Shaaban M., Hassan M. U., Huang G. Q., Liu Y., Tang H. Y., et al. (2020). Biochar Mitigates the N2O Emissions From Acidic Soil by Increasing the nosZ and nirK Gene Abundance and Soil pH. J. Environ. Manage. 255, 7. doi: 10.1016/j.jenvman.2019.109891
Abell G. C., Revill A. T., Smith C., Bissett A. P., Volkman J. K., Robert S. S. (2009). Archaeal Ammonia Oxidizers and nirS-Type Denitrifiers Dominate Sediment Nitrifying and Denitrifying Populations in a Subtropical Macrotidal Estuary. ISME J. 4, 286–300. doi: 10.1038/ismej.2009.105
Bergaust L., Mao Y., Bakken L., Frostegard A. (2010). Denitrification Response Patterns During the Transition to Anoxic Respiration and Posttranscriptional Effects of Suboptimal pH on Nitrous Oxide Reductase in Paracoccus Denitrificans. Appl. Environ. Microbiol. 76 (19), 6387–6396. doi: 10.1128/AEM.00608-10
Bertagnolli A. D., Konstantinidis K. T., Stewart F. J. (2020). Non-Denitrifier Nitrous Oxide Reductases Dominate Marine Biomes. Environ. Microbiol. Rep. 12, 681–692. doi: 10.1111/1758-2229.12879
Butterbach-Bahl K., Baggs E. M., Dannenmann M., Kiese R., Zechmeister-Boltenstern S. (2014). Nitrous Oxide Emissions From Soils: How Well Do We Understand the Processes and Their Controls? Philos. Trans. R. Soc Lond. B.: Biol. Sci. 368 (1621), 1–15. doi: 10.1098/rstb.2013.0122
Chen C. T. A., Wang S. L., Wang B. J., Pai S. C. (2001). Nutrient Budgets for the South China Sea Basin. Mar. Chem. 75 (4), 281–300. doi: 10.1016/S0304-4203(01)00041-X
Christopher F., Gregory A. C., Jeffrey C., Bess W. (2013). Transitions in nirS-Type Denitrifier Diversity, Community Composition, and Biogeochemical Activity Along the Chesapeake Bay Estuary. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00237
Clark I. M., Buchkina N., Jhurreea D., Goulding K., Hirsch P. R. (2012). Impacts of Nitrogen Application Rates on the Activity and Diversity of Denitrifying Bacteria in the Broadbalk Wheat Experiment. Philos. Trans. R. Soc B-Biol. Sci. 367 (1593), 1235–1244. doi: 10.1098/rstb.2011.0314
Codispoti L. A. (2010). Interesting Times for Marine N2O. Science 327 (5971), 1339–1340. doi: 10.1126/science.1184945
Coyne M. S., Arunakumari A., Averill B. A., Tiedje J. M. (1989). Immunological Identification and Distribution of Dissimilatory Heme Cd1 and Nonheme Copper Nitrite Reductases in Denitrifying Bacteria. Appl. Environ. Microbiol. 55 (11), 2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989
Cuhel J., Simek M., Laughlin R. J., Bru D., Cheneby D., Watson C. J., et al. (2010). Insights Into the Effect of Soil pH on N2O and N2 Emissions and Denitrifier Community Size and Activity. Appl. Environ. Microbiol. 76 (6), 1870–1878. doi: 10.1128/AEM.02484-09
Dang H., Zhang X., Sun J., Li T., Zhang Z., Yang G. (2008). Diversity and Spatial Distribution of Sediment Ammonia-Oxidizing Crenarchaeota in Response to Estuarine and Environmental Gradients in Changjiang Estuary and East China Sea. Microbiology 154, 2084–2095. doi: 10.1099/mic.0.2007/013581-0
Di H., Cameron K., Podolyan A., Dawson A. (2014). Effect of Soil Moisture Status and a Nitrification Inhibitor, Dicyandiamide, on Ammonia Oxidizer and Denitrifier Growth and Nitrous Oxide Emissions in a Grassland Soil. Soil Biol. Biochem. 73, 59–68. doi: 10.1016/j.soilbio.2014.02.011
Dini-Andreote F., Brossi M., van Elsas J., Salles J. (2016). Reconstructing the Geneti Potential of the Microbially-Mediated Nitrogen Cycle in a Salt Marsh Ecosystem. Front. Microbiol. 1 (7). doi: 10.3389/fmicb.2016.00902
Domeignoz-Horta L., Spor A., Bru D., Breuil M., Bizouard F., Leonard J., et al. (2015). The Diversity of the N2O Reducers Matters for the N2O:N2 Denitrification End-Product Ratio Across an Annual and a Perennial Cropping Stystem. Front. Microbiol. 6, 971. doi: 10.3389/fmicb.2015.00971
Gao D., Xiaolong W., Liang H., Wei Q., Dou Y., Li L. (2018). Anaerobic Ammonia Oxidizing Bacteria: Ecological Distribution, Metabolism, and Microbial Interactions. Front. Environ. Sci. Eng. 12 (3), 10. doi: 10.1007/s11783-018-1035-x
Graf D., Jones C., Hallin S. (2014). Intergenomic Comparisons Highlight Modularity of the Denitrification Pathway and Underpin the Importance of Community Structure for N2O Emissions. PLos One 9 (12), e114118. doi: 10.1371/journal.pone.0114118
Graf D. R. H., Zhao M., Jones C. M., Hallin S. (2016). Soil Type Overrides Plant Effect on Genetic and Enzymatic N2O Production Potential in Arable Soils. Soil Biol. Biochem. 100, 125–128. doi: 10.1016/j.soilbio.2016.06.006
Graves C. J., Makrides E. J., Schmidt V. T., Giblin A. E., Cardon Z. G., Rand D. M. (2016). Functional Responses of Salt Marsh Microbial Communities to Long-Term Nnutrient Enrichment. J. Appl. Environ. Microbiol. 82 (9), 2862–2871. doi: 10.1128/AEM.03990-15
Guan F. J., Hong Y. G., Wu J. P., Wang Y., Bing L. Y., Tang B., et al. (2017). A Fast Sodium Hypobromite Oxidation Method for the Sequential Determination of Ammonia Nitrogen in Small Volume. Ecol. Sci. 36 (2), 42–48. doi: 10.14108/j.cnki.1008-8873.2017.02.006
Guo L. Y., Shi F., Yang L. Y. (2011). Advances in Functional Genes and Molecular Ecology in Denitrifiers. Microbiol. China 38 (4), 583–590. doi: 10.3724/SP.J.1011.2011.00462
Hallin S., Lindgern P.-E. (1999). PCR Detection of Genes Encoding Nitrite Reductase in Denitrifying Bacteria. Appl. Environ. Microbiol. 65 (4), 1652–1657. doi: 10.1128/AEM.65.4.1652-1657.1999
Hallin S., Philippot L., Loffler F. E., Sanford R. A., Jones C. M. (2018). Genomics and Ecology of Novel N2O-Reducing Microorganisms. Trends Microbiol. 26 (1), 43–55. doi: 10.1016/j.tim.2017.07.003
Han Y., Zhang G., Zhao Y., Liu S. (2013). Distributions and Sea-to-Air Fluxes of Nitrous Oxide in the Coastal and Shelf Waters of the Northwestern South China Sea. Estuar. Coast. Shelf. Sci. 133, 32–44. doi: 10.1016/j.ecss.2013.08.001
Harris E., Diaz-Pines E., Stoll E., Schloter M., Schulz S., Duffner C., et al. (2021). Denitrifying Pathways Dominate Nitrous Oxide Emissions From Managed Grassland During Drought and Rewetting. Sci. Adv. 7 (6), eabb7118. doi: 10.1126/sciadv.abb7118
Helen D., Heylen K., Tytgat B., Willems A. (2016). Highly Diverse nirK Genes Comprise Two Major Clades That Harbour Ammonium-Producing Denitrifiers. BMC Genomics 17, 155. doi: 10.1186/s12864-016-2465-0
Henderson S. L., Dandie C. E., Patten C. L., Zebarth B. J., Burton D. L., Trevors J. T., et al. (2010). Changes in Denitrifier Abundance, Denitrification Gene mRNA Levels, Nitrous Oxide Emissions, and Denitrification in Anoxic Soil Microcosms Amended With Glucose and Plant Residues. Appl. Environ. Microbiol. 76 (7), 2155–2164. doi: 10.1128/AEM.02993-09
Henry S., Bru D., Stres B., Hallet S., Philippot L. (2006). Quantitative Detection of the nosZ Gene, Encoding Nitrous Oxide Reductase, and Comparison of the Abundances of 16S rRNA, Narg, Nirk, and nosZ Genes in Soils. Appl. Environ. Microbiol. 72 (8), 5181–5189. doi: 10.1128/AEM.00231-06
He Z., Zhang H., Gao S., Lercher M. J., Chen W. H., Hu S. (2016). Evolview V2: An Online Visualization and Management Tool for Customized and Annotated Phylogenetic Trees. Nucleic Acids Res. 44 (W1), W236–W241. doi: 10.1093/nar/gkw370
Holtan-Hartwig L., Dörsch P., Bakken L. (2000). Comparison of Denitrifying Communities in Organic Soils: Kinetics of NO3- and N2O Reduction. Soil Biol. Biochem. 32, 833–843. doi: 10.1016/S0038-0717(99)00213-8
Jones C. M., Graf D. R., Bru D., Philippot L., Hallin S. (2013). The Unaccounted Yet Abundant Nitrous Oxide-Reducing Microbial Community: A Potential Nitrous Oxide Sink. ISME J. 7 (2), 417–426. doi: 10.1038/ismej.2012.125
Jones C. M., Spor A., Brennan F. P., Breuil M.-C., Bru D., Lemanceau P., et al. (2014). Recently Identified Microbial Guild Mediates Soil N2O Sink Capacity. Nat. Clim. Change 4 (9), 801–805. doi: 10.1038/nclimate2301
Jones C., Stres B., Rosenquist M., Hallin S. (2008). Phylogenetic Analysis of Nitrite, Nitric Oxide, and Nitrous Oxide Respiratory Enzymes Reveal a Complex Evolutionary History for Denitrification. Mol. Biol. Evol. 25, 1955–1966. doi: 10.1093/molbev/msn146
Kleber M., Bourg I., Coward E., Hansel C., Myneni S., Nunan N. (2021). Dynamic Interactions at the Mineral–Organic Matter Interface. Nat. Rev. Earth Environ. 2 (6), 402–421. doi: 10.1038/s43017-021-00162-y
Körner H., Zumft W. (1989). Expression of Denitrification Enzymes in Response to the Dissolved Oxygen Level and Respiratory Substrate in Continuous Culture of Pseudomonas Stutzeri. Appl. Environ. Microbiol. 55 (7), 1670–1676. doi: 10.1128/AEM.55.7.1670-1676.1989
Lee J. A., Francis C. A. (2017). Spatiotemporal Characterization of San Francisco Bay Denitrifying Communities: A Comparison of nirK and nirS Diversity and Abundance. Microb. Ecol. 73 (2), 271–284. doi: 10.1007/s00248-016-0865-y
Liu X. H., Wu J. P., Hong Y. G., Jiao L. J., Li Y. B., Wang L. M., et al. (2020). Nitrogen Loss by nirS-Type Denitrifying Bacterial Communities in Eutrophic Coastal Sediments. Int. Biodeterior. Biodegrad. 150, 104955. doi: 10.1016/j.ibiod.2020.104955
Lycus P., Bøthun K. L., Bergaust L., Shapleigh J. P., Bakken L. R., Frostegard A. (2017). Phenotypic and Genotypic Richness of Denitrifiers Revealed by a Novel Isolation Strategy. ISME J. 11 (10), 2219–2232. doi: 10.1038/ismej.2017.82
Makowski D. (2019). N2O Increasing Faster Than Expected. Nat. Clim. Change 9 (12), 909–910. doi: 10.1038/s41558-019-0642-2
Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., et al. (2016). Insights Into Global Diatom Distribution and Diversity in the World's Ocean. Proc. Natl. Acad. Sci. U. S. A. 113 (11), E1516–E1525. doi: 10.1073/pnas.1509523113
Marchant H., Holtappels M., Lavik G., Ahmerkamp S., Winter C., Kuypers M. (2016). Coupled Nitrification-Denitrification Leads to Extensive N Loss in Subtidal Permeable Sediments. Limnol. Oceanogr. 61(3), 1033–1048. doi: 10.1002/lno.10271
Morales S. E., Cosart T., Holben W. E. (2010). Bacterial Gene Abundances as Indicators of Greenhouse Gas Emission in Soils. ISME J. 4 (6), 799–808. doi: 10.1038/ismej.2010.8
Oshiki M., Satoh H., Okabe S. J. E. M. (2016). Ecology and Physiology of Anaerobic Ammonium Oxidizing Bacteria. J. Environ. Microbiol. 18 (9), 2784–2796. doi: 10.1111/1462-2920.13134
Ren Y. Y., Zheng W. J., Ye W.-W., Zhang Y. Y., Zhang G. (2017). Distribution, Formation and Air-Sea Flux of Dissolved Nitrous Oxide in Northern Continental Slope of South China Sea in Summer and Autumn. Oceanol. Limnol. Sin. 48, 29–39. doi: 10.11693/hyhz20160600137
Rich J., Heichen R. S., Bottomley P. J., Cromack K., Myrold D. D. (2003). Community Composition and Functioning of Denitrifying Bacteria From Adjacent Meadow and Forest Soils. Appl. Environ. Microbiol. 69, 5974–5982. doi: 10.1128/AEM.69.10.5974-5982.2003
Roco C. A., Bergaust L. L., Bakken L. R., Yavitt J. B., Shapleigh J. P. (2016). Modularity of Nitrogen-Oxide Reducing Soil Bacteria: Linking Phenotype to Genotype. Environ. Microbiol. 19 (6), 2507–2519. doi: 10.1111/1462-2920.13250
Saarenheimo J., Rissanen A., Arvola L., Nykänen H., Lehmann M., Tiirola M. (2015). Genetic and Environmental Controls on Nitrous Oxide Accumulation in Lakes. PLos One 10, e0121201. doi: 10.1371/journal.pone.0121201
Samad M. S., Biswas A., Bakken L. R., Clough T. J., de Klein C. A., Richards K. G., et al. (2016). Phylogenetic and Functional Potential Links pH and N2O Emissions in Pasture Soils. Sci. Rep. 6, 35990. doi: 10.1038/srep35990
Sanford R. A., Wagner D. D., Wu Q., Chee-Sanford J. C., Thomas S. H., Cruz-Garcia C., et al. (2012). Unexpected Nondenitrifier Nitrous Oxide Reductase Gene Diversity and Abundance in Soils. Proc. Natl. Acad. Sci. U. S. A. 109 (48), 19709–19714. doi: 10.1073/pnas.1211238109
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 75 (23), 7537–7541. doi: 10.1128/AEM.01541-09
Seitzinger S. P., Mayorga E., Bouwman A. F., Kroeze C., Beusen A. H. W., Billen G., et al. (2010). Global River Nutrient Export: A Scenario Analysis of Past and Future Trends. Glob. Biogeochem. Cycle 24 (4), 2621–2628. doi: 10.1029/2009GB003587
Smith J. M., Mosier A. C., Francis C. A. (2014). Spatiotemporal Relationships Between the Abundance, Distribution, and Potential Activities of Ammonia-Oxidizing and Denitrifying Microorganisms in Intertidal Sediments. Microb. Ecol. 69 (1), 13–24. doi: 10.1007/s00248-014-0450-1
Smith C. J., Nedwell D. B., Dong L. F., Osborn A. M. (2007). Diversity and Abundance of Nitrate Reductase Genes (narG and Napa), Nitrite Reductase Genes (nirS and Nrfa), and Their Transcripts in Estuarine Sediments. Appl. Environ. Microbiol. 73 (11), 3612–3622. doi: 10.1128/AEM.02894-06
Stremińska M. A., Felgate H., Rowley G., Richardson D. J., Baggs E. M. (2012). Nitrous Oxide Production in Soil Isolates of Nitrate-Ammonifying Bacteria. Environ. Microbiol. Rep. 4 (1), 66–71. doi: 10.1111/j.1758-2229.2011.00302.x
Su J. L. (2004). Overview of the South China Sea Circulation and Its Influence on the Coastal Physical Oceanography Outside the Pearl River Estuary. Cont. Shelf. Res. 24 (16), 1745–1760. doi: 10.1016/j.csr.2004.06.005
Su J. L. (2005). Overview of the South China Sea Circulation and its Dynamics. J. Acta Oceanol. Sin. 27 (6), 1–8. doi: 10.1111/j.1745-7254.2005.00209.x
ter Braak C., Šmilauer P. (2012). Canoco Reference Manual and User’s Guide: Software of Ordination (Version 5.0) (Ithaca, NY USA: Microcomputer Power).
Throback I. N., Enwall K., Jarvis A., Hallin S. (2004). Reassessing PCR Primers Targeting Nirs, nirK and nosZ Genes for Community Surveys of Denitrifying Bacteria With DGGE. FEMS Microbiol. Ecol. 49 (3), 401–417. doi: 10.1016/j.femsec.2004.04.011
Tseng H. C., Chen C. T. A., Borges A. V., Delvalls T. A., Lai C. M., Chen T. Y. (2016). Distributions and Sea-to-Air Fluxes of Nitrous Oxide in the South China Sea and the West Philippines Sea. Deep-Sea. Res. Part I-Oceanogr. Res. Pap. 115, 131–144. doi: 10.1016/j.dsr.2016.06.006
Tsiknia M., Paranychianakis N., Varouchakis E. A., Nikolaidis N. P. (2015). Environmental Drivers of the Distribution of Nitrogen Functional Genes at a Watershed Scale. FEMS Microbiol. Ecol. 91 (6), fiv052. doi: 10.1093/femsec/fiv052
Wang Q., Jiang G., Ye L., Pijuan M., Yuan Z. (2014). Heterotrophic Denitrification Plays an Important Role in N2O Production From Nitritation Reactors Treating Anaerobic Sludge Digestion Liquor. Water Res. 62, 202–210. doi: 10.1016/j.watres.2014.06.003
Wang J., Kan J. J., Qian G., Chen J. F., Xia Z. Q., Zhang X. D., et al. (2019b). Denitrification and Anammox: Understanding Nitrogen Loss From Yangtze Estuary to the East China Sea (ECS). Environ. Pollut. 252 (Pt B), 1659–1670. doi: 10.1016/j.envpol.2019.06.025
Wang W. L., Moore J. K., Martiny A. C., Primeau F. W. (2019a). Convergent Estimates of Marine Nitrogen Fixation. Nature 566 (7743), 205–211. doi: 10.1038/s41586-019-0911-2
Warneke S., Schipper L., Matiasek M., Scow K., Cameron S., Bruesewitz D., et al. (2011). Nitrate Removal, Communities of Denitrifiers and Adverse Effects in Different Carbon Substrates for Use in Denitrification Beds. Water Res. 45 (17), 5463–5475. doi: 10.1016/j.watres.2011.08.007
Wittorf L., Bonilla-Rosso G., Jones C. M., Bäckman O., Hulth S., Hallin S. (2016). Habitat Partitioning of Marine Benthic Denitrifier Communities in Response to Oxygen Availability. Environ. Microbiol. Rep. 8 (5), 486–492. doi: 10.1111/1758-2229.12393
Wu J. P., Hong Y. G., Chang X. Y., Jiao L. J., Li Y. B., Liu X. H., et al. (2019). Unexpectedly High Diversity of Anammox Bacteria Detected in Deep-Sea Surface Sediments of the South China Sea. FEMS Microbiol. Ecol. 95 (3), fiz013. doi: 10.1093/femsec/fiz013
Wu J. P., Hong Y. G., Guan F. J., Wang Y., Tan Y. H., Yue W. Z., et al. (2016). A Rapid and High-Throughput Microplate Spectrophotometric Method for Field Measurement of Nitrate in Seawater and Freshwater. Sci. Rep. 6 (1), 20165. doi: 10.1038/srep20165
Wu J. P., Hong Y. G., Liu X. H., Hu Y. H. (2021b). Variations in Nitrogen Removal Rates and Microbial Communities Over Sediment Depth in Daya Bay, China. Environ. Pollut. 286, 117267. doi: 10.1016/j.envpol.2021.117267
Wu J. P., Hong Y. G., Wilson S. J., Song B. (2021a). Microbial Nitrogen Loss by Coupled Nitrification to Denitrification and Anammox in a Permeable Subterranean Estuary at Gloucester Point, Virginia. Mar. Pollut. Bull. 168, 112440. doi: 10.1016/j.marpolbul.2021.112440
Xu J. R., Wang Y. S., Zhang F. Q., Sun C. C., Yin J. P., Wang Q. J., et al. (2006). Studies on Formation Mechanism and Distribution of Nitrous Oxide in Northeastern South China Sea. J. Trop. Oceanogr. 25 (4), 66–74. doi: 10.1111/j.1744-7917.2006.00098.x
Yang J. Y., Tang J. M., Guo X. H., Kao S.-J. (2021). Nitrogen Cycling Processes and its Budget in China Marginal Sea: Case Studies in the South China Sea. Oceanol. Limnol. Sin. 52 (2), 9. doi: 10.11693/hyhz20200800234
Yergeau E., Kang S., He Z., Zhou J., Kowalchuk G. (2007). Functional Microarray Analysis of Nitrogen and Carbon Cycling Genes Across an Antarctic Latitudinal Transect. ISME J. 1 (2), 163–179. doi: 10.1038/ismej.2007.24
Yoon S., Nissen S., Park D., Sanford R. A., Löffler F. E. (2016). Nitrous Oxide Reduction Kinetics Distinguish Bacteria Harboring Clade II NosZ. Appl. Environ. Microbiol. 82 (13), 3793–3800. doi: 10.1128/AEM.00409-16
Zhang G., Liu S., Casciotti K. L., Forbes M. S., Gu X., Ren Y., et al. (2019). Distribution of Concentration and Stable Isotopic Composition of N2O in the Shelf and Slope of the Northern South China Sea: Implications for Production and Emission. J. Geophys. Res.: Ocean. 124 (8), 6218–6234. doi: 10.1029/2019jc014947
Zhao S., Wang X., Pan H., Wang Y., Zhu G. (2021). High N2O Reduction Potential by Denitrification in the Nearshore Site of a Riparian Zone. Sci. Total. Environ. 813, 152458. doi: 10.1016/j.scitotenv.2021.152458
Zhao S., Wang Q., Zhou J., Yuan D., Zhu G. (2018). Linking Abundance and Community of Microbial N2O-Producers and N2O-Reducers With Enzymatic N2O Production Potential in a Riparian Zone. Sci. Total. Environ. 642, 1090–1099. doi: 10.1016/j.scitotenv.2018.06.110
Zhao S., Zhou J., Yuan D., Wang W., Zhou L., Pi Y., et al. (2019). NirS-Type N2O-Producers and nosZ II-Type N2O-Reducers Determine the N2O Emission Potential in Farmland Rhizosphere Soils. J. Soil. Sed. 20, 461–417. doi: 10.1007/s11368-019-02395-3
Zheng L., Zhang G., Xu J., Zhang F., Zhang J. (2009). Distribution and Air-Sea Fluxes of N2O in South China Sea in Spring. Mar. Environ. Sci. 28 (3), 233–237. doi: 10.3969/j.issn.1007-6336.2009.03.001
Keywords: denitrification, functional genes, ecological distribution, potential N2O emissions, South China Sea
Citation: Xiang H, Hong Y, Wu J and Long A (2022) Ecological Distribution and Diversity of Key Functional Genes for Denitrification in Surface Sediments of the Northern South China Sea: Implications for Potential N2O Emissions. Front. Mar. Sci. 9:912402. doi: 10.3389/fmars.2022.912402
Received: 04 April 2022; Accepted: 09 May 2022;
Published: 13 June 2022.
Edited by:
Meilin Wu, South China Sea Institute of Oceanology (CAS), ChinaReviewed by:
Hui Wang, Shantou University, ChinaYanpei Zhuang, Jimei University, China
Hongpo Dong, East China Normal University, China
Copyright © 2022 Xiang, Hong, Wu and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiguo Hong, eWdob25nQGd6aHUuZWR1LmNu; Aimin Long, bG9uZ2FtQHNjc2lvLmFjLmNu
 Hua Xiang1,2,3
Hua Xiang1,2,3 Yiguo Hong
Yiguo Hong