- Department of Ecology, Jinan University, Guangzhou, China
Levanderina fissa (formerly Gyrodinium instriatum) frequently causes blooms in the Pearl River Estuary and has few advantages in interspecific competition with other bloom-forming algal species. Phycosphere bacteria, which closely interact with algal cells, may play an ecologically functional role in the population dynamics and bloom occurrence. To test this hypothesis, we isolated and identified cultivable bacteria coexisting in different growth stages of L. fissa by the gradient dilution method and investigated the characteristics of the bacterial interactions with three diatom species (Chaetoceros curvisetus, Skeletonema dohrnii, and Phaeodactylum tricornutum) and three dinoflagellate species (Scrippsiella acuminata, Karenia mikimotoi, and the host algae) after screening for functional bacteria. One of the isolated bacterial strains, Lf7, which was phylogenetically identified as an Alteromonas species, showed significant inhibitory effects on different algal species except its host. Moreover, all algal species, especially their hosts, showed significant stimulatory effects on bacterial Lf7 growth. These results indicate that the phycosphere bacterium Lf7 may play some ecological roles in the competition between its host alga L. fissa and other phytoplankton. The study also highlights the complicated interactions between phycosphere bacteria and host algae.
1 Introduction
The phycosphere is the planktonic analogue of the rhizosphere in plants (Seymour et al., 2017). Phytoplankton–bacteria relationships in this microenvironment can span mutualism, antagonism, parasitism, and competition, indicating a multifarious and highly sophisticated ecological function of phycosphere bacteria (Cole, 1982; Findlay and Patil, 1984; Amin et al., 2012; Seymour et al., 2017; Höger et al., 2021). Competitive or antagonistic relationships of phycosphere bacteria and phytoplankton have been mostly extensively studied, including their competition for inorganic nutrients (Bratbak and Tingstad, 1985), algicidal abilities of bacteria (Mayali and Azam, 2004; Umetsu et al., 2019), and defense mechanisms of phytoplankton (Teplitski et al., 2004; Rajamani et al., 2011; Stien et al., 2016). Their mutualism associations were recently revealed and proposed to be more prevalent than antagonism (Xie et al., 2013; Buchan et al., 2014; Calatrava et al., 2018).
Some studies have described the phycosphere bacteria of certain algae as friends with benefits (Amin et al., 2009; Kim et al., 2014; Johansson et al., 2019). For example, several clades of Marinobacter ubiquitously found in close association with dinoflagellates and coccolithophores produce an unusually lower affinity dicitrate siderophore, vibrioferrin, which increased algal iron uptake of a representative dinoflagellate partner, Scrippsiella trochoidea, >20-fold (Amin et al., 2009). When Rhizobium sp., the most prevalent and dominant bacterium isolated from Chlorella vulgaris, was cocultured with green algae, it increased the algal cell count by approximately 72% (Kim et al., 2014). Several phycosphere bacterial strains (Arenibacter algicola strain SMS7, Marinobacter salarius strain SMR5, Sphingorhabdus flavimaris strain SMR4y, Sulfitobacter pseudonitzschiae strain SMR1, Yoonia vestfoldensis strain SMR4r and Roseovarius mucosus strain SMR3) stimulate the growth of the diatom partner Skeletonema marinoi under different environmental conditions, for example, low iron concentrations and high and low temperatures (Johansson et al., 2019). Symbiotic bacteria may also provide the host with vitamins and organic acids, including amino acids and siderophores, and are essential to the growth of their host (Croft et al., 2005; Amin et al., 2009; Sandhya and Vijayan, 2019). Moreover, these kinds of phycosphere bacteria are well adapted to a narrow range of compounds secreted by their algal host, forming an exclusively specific association with the host algae (Bell, 1984; Schäfer et al., 2002; Sapp et al., 2007; Sison-Mangus et al., 2014; Palacios et al., 2022). This may not be the only way phycosphere bacteria benefit their algal hosts. Our laboratory isolated several phycosphere bacteria that showed inhibitory activity against algal species other than their host (Wang et al., 2021).
Levanderina fissa (formerly Gyrodinium instriatum) has frequently caused blooms in the Pearl River Estuary in the past two decades, for example, blooms in Shenzhen Bay in 1998, 2003, and 2007 (Wang et al., 2001; Zhu et al., 2004; SOA, 2008); Lingding Bay in 2002 (Wang et al., 2003); and Zhuhai coastal waters in 2009 (Wang et al., 2011). However, blooms caused by this species elsewhere were rare and were observed only in the Gulf of Guayaquil in Ecuador (Jimenéz, 1993); Hakozaki Fishing Port of Japan (Nagasoe et al., 2006; Nagasoe et al., 2010); and Bahia de Acapulco, Mexico (Gárate-Lizárraga et al., 2013). Our previous study showed that L. fissa had few advantages in interspecific competition with three other algal bloom-forming species, namely, Skeletonema dohrnii (Bacillariophyceae), Prorocentrum micans (Dinophyceae), and Chattonella marina (Raphidophyceae), using pure, sterilized cultures (Wang et al., 2021). Thus, we suspect that phycospheric bacteria may play an ecological function role by promoting the growth or competition of L. fissa. To test this hypothesis, we isolated and identified cultivable bacteria coexisting in different growth stages of L. fissa by the gradient dilution method and obtained an isolate with an inhibitory effect on algal growth by screening experiments with C. curvisetus. We also cocultured the isolate Lf7 at different inoculation doses with three diatom species, Chaetoceros curvisetus, Skeletonema dohrnii, and Phaeodactylum tricornutum, as well as three dinoflagellate species, Scrippsiella acuminata, Karenia mikimotoi, and L. fissa, to determine their effects on these commonly dominant phytoplankton species in the Pearl River Estuary. Our research will provide insight into the ecological function of phycosphere bacteria on algal growth, interspecies competition, and bloom formation.
2 Materials and Methods
2.1 Isolation of Bacterial Strains from the Phycosphere of Levanderina fissa
2.1.1 Microalgal Culture
Levanderina fissa (GenBank accession no. ITS: KF435124) was isolated in October 2009 during a bloom in the Pearl River Estuary, South China Sea. The cultures were maintained in autoclaved (121°C, 20 min) f/2 medium (Guillard, 1975). Algal cells at the exponential phase were incubated in 250 mL Erlenmeyer flasks containing 100 mL of sterilized f/2 medium. The medium was made with artificial sea salt (Red Coral Sea, nutrient-free formula) with a salinity of 30-31 and pH of 7.9 ± 0.1. Microalgal cultures were maintained in an incubator at 20 ± 1°C and illuminated with 100 μmol photon/m2·s cool-white fluorescent illumination with a dark:light cycle of 12:12 h.
2.1.2 Isolation of Cultivable Bacteria in the Phycosphere
Samples were aseptically collected at five growth phases of L. fissa, that is, the lag growth phase, the logarithmic growth phase, the stationary phase, and the early and late decline phases. One milliliter of each culture was suspended in 9 mL of Zobell 2216E marine medium (Su et al., 2007) to a dilution of 10-1. Zobell 2216E marine medium was prepared using artificial seawater. These suspensions were further diluted in the same way to dilutions of 10-2-10-6. A 0.1 mL aliquot of the dilutions of 10-3-10-6 was spread onto Zobell 2216E agar plates, and the plates were incubated at 28°C for 5-7 days. Visually distinct bacterial colonies were subcultured on fresh Zobell 2216E agar plates and incubated for 5-7 days. The step was repeated three to four times to obtain purified bacterial colonies.
There were 32 bacterial isolates collected from cultures of L. fissa. The bacterial isolates were then identified based on sequencing of the V3 region of the 16S rRNA gene. Twelve different phylotypes were obtained after removing the duplicate rRNA gene sequences. The isolated bacteria were labelled Lf1 to Lf12. The sequences were submitted to the International Nucleotide Sequence Database Collaboration at NCBI (GenBank accession nos. KF444158-KF444169). The bacterial phylotypes belonged to Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Actinobacteria (Table 1 and Figure 1)
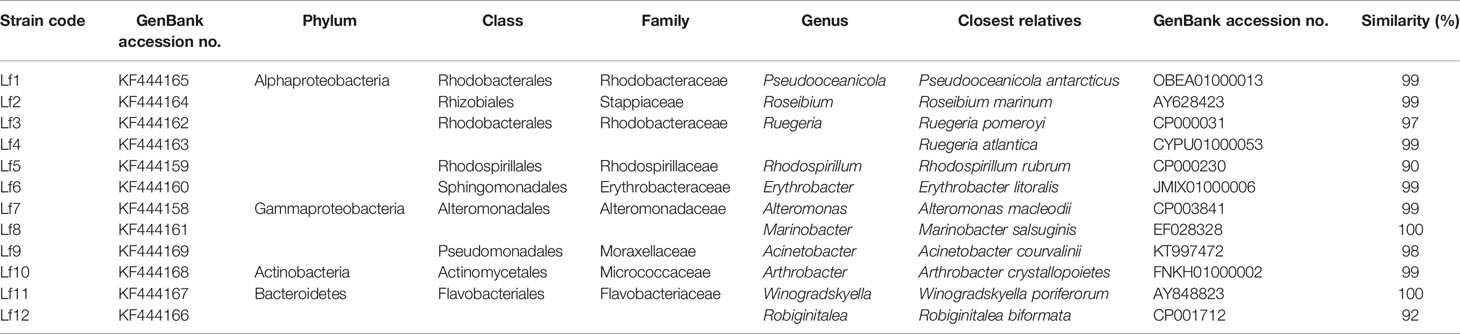
Table 1 The identities of bacteria isolated from phycosphere of Levanderina fissa and their phylogenetically most related sequences from EzTaxon database based on partial 16S rRNA gene sequences.
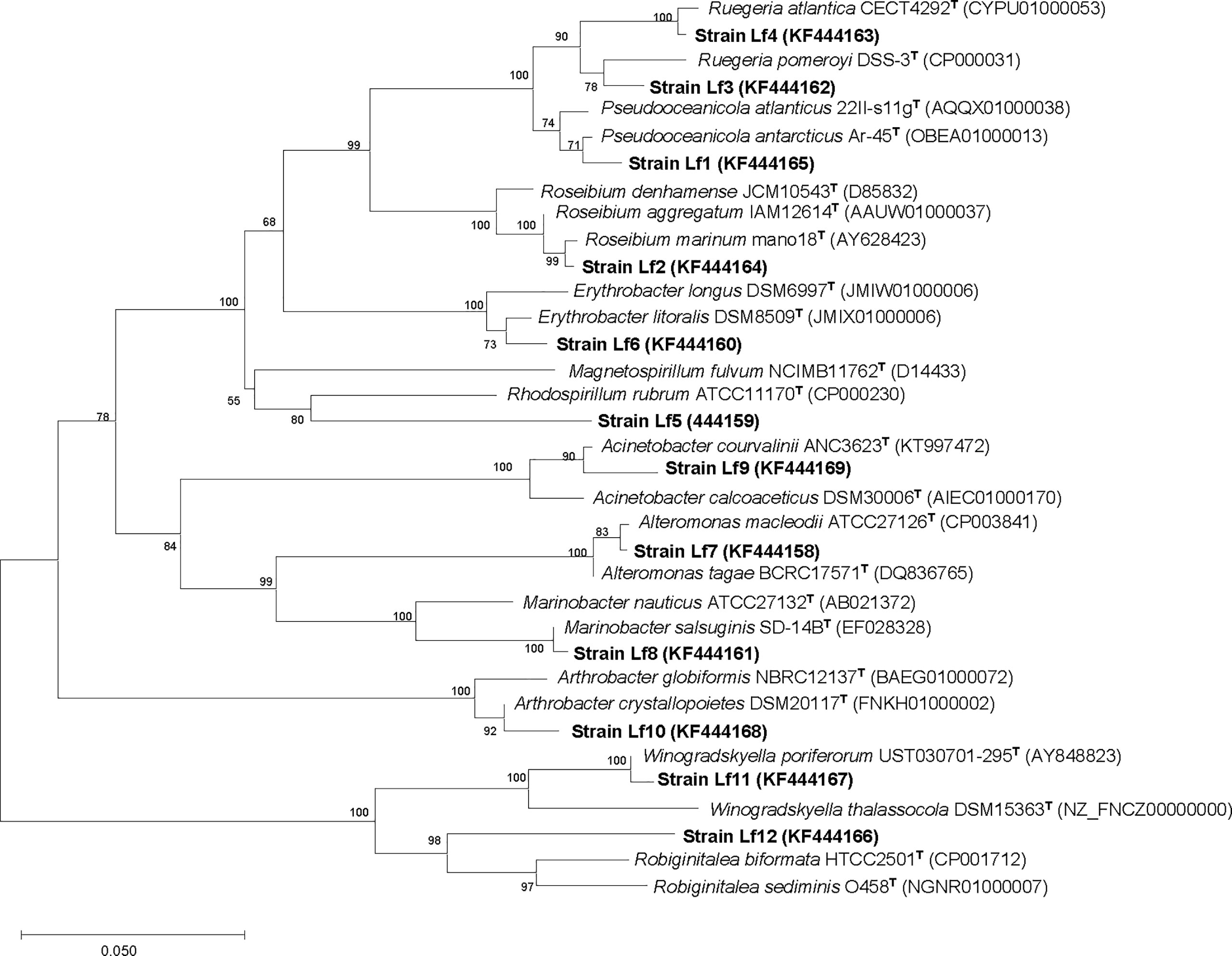
Figure 1 Neighbor-joining tree, showing the phylogenetic position of bacterial strains isolated from phycosphere of Levanderina fissa and type strains of most phylogenetically related taxa (type species included) based on partial 16S rRNA gene sequences. GenBank accession numbers are given. Bootstrap analysis was made with 1000 resamplings; percentages of support are shown at nodes, Bar, 5% sequence divergence.
2.2 Effects of Bacteria on the Growth of Different Microalgal Taxa
2.2.1 Antibacterial Treatment of Microalgal Cultures
The clonal strains of the six phytoplankton species, including three diatom species, C. curvisetus (GenBank accession no.: MW793400), S. dohrnii (GenBank accession no.: MW795694), and P. tricornutum (GenBank accession no.: MW793395), and three dinoflagellate species, K. mikimotoi (GenBank accession no.: MW793400), S. acuminata (GenBank accession no.: MW793402), and L. fissa, were isolated between 2005 and 2009 from southern Chinese coastal waters and maintained in f/2 media (for diatoms) or in f/2 media without silicon (for dinoflagellates).
The stock cultures were treated with a mixture of antibiotics to destroy the external bacteria before the experiments. First, 10 μg/mL penicillin (final concentration) was added to 100 mL of C. curvisetus culture in the exponential phase and incubated for 24 h. Then, 10 mL of the penicillin-treated culture was inoculated into 100 mL of fresh f/2 medium with 10 μg/mL streptomycin sulfate and incubated for another 24 h, followed by treatment with 10 μg/mL kanamycin sulfate. The antibacterial treatment cultures were maintained in sterilized f/2 medium for the experiments.
2.2.2 Bacterial Isolation and Counting
Bacterial isolates were maintained in a Zobell 2216 marine agar plate after removing the repeated isolates by cross-comparison with the results of the rRNA gene sequences. Before the experiment, bacterial colonies were transferred to liquid Zobell 2216 medium, cultivated on a 28°C, 200 r/min shaking table for 8-10 h, and then gradually acclimated to 20°C within 2 h.
The bacterial concentration was determined by measuring the colony-forming unit (CFU) number on Zobell 2216E agar medium. Each distinct bacterial colony was counted in the plate of suitable colonies (30-300), and the concentration of each bacterium (CFU/mL) was obtained according to the dilution factor.
2.2.3 Screening of the Bacterial Strains
The effects of the 12 bacterial isolates on the growth of different microalgal taxa were screened by C. curvisetus. Liquid bacterial cultures of the 12 isolates in exponential phase were added to the exponential C. curvisetus cultures. The inoculation rate was 0.1% (v/v), and the initial bacterial concentration was 7.5×107 CFU/mL in the cocultures. Cultures in f/2 medium (f/2) and with the same volume of liquid Zobell 2216E medium (BC) were used as the negative controls. Algal cultures were cultivated under the same conditions as outlined in Section 2.1.1. All experiments were carried out in sterile 24-well tissue culture plates. The experiment was conducted for 5 days covering the whole exponential phase and reaching the stationary phase for C. curvisetus, and cell numbers were counted every day. The algal cell numbers were counted daily during the incubation period. The cell counts were performed in a cell counting chamber by placing 0.05-0.1 mL of culture into the chamber, fixing with a drop of Lugol’s fixative, and observing under an inverted microscope (Leica DMIRB) at a magnification of 200×. Each sample was counted more than three times until the differences in cell counts were less than 10%. When C. curvisetus always showed negative growth with lower cell densities than initial cell densities, the maximum cell density was recorded as 0. All experiments were conducted in triplicate.
2.2.4 Effects of Bacterial Strain Lf7 on the Growth of Microalgal Taxa
The results of the screening test showed that the bacterial strain Lf7 had the strongest inhibitory effect on the growth of C. curvisetus. Therefore, Lf7 was selected as the bacterial strain to study its effects on the growth of the six different phytoplankton taxa. Four bacterial concentrations were set in the experiment, which were 7.5×107, 7.5×106, 7.5×105, and 7.5×104 CFU/mL, with an inoculation rate of 0.1% (v/v). The experiment was conducted for 6 days to cover the whole exponential phase and reach the stationary phase for the algae, and algal cell numbers and bacterial concentrations were measured every day. The initial cell densities of different microalgal taxa were set differently according to their cell sizes to obtain similar initial biomasses. The size of the microalgal taxa is described in Supplementary Table S1. The experiment was conducted for 6 days to cover the whole exponential phase and reach the stationary phase for the algae according to our former lab experience, and algal cell numbers and bacterial concentrations were measured every day.
This additional experimental design is the same as that in Section 2.2.3.
2.3 Data and Statistical Analysis
2.3.1 Phylogenetic Analysis
Analysis of the 16S rRNA gene sequence was performed using the software package MEGA, version 3.1 (Kumar et al., 2004). The model of Jukes and Cantor (1969) was used to compute the evolutionary distance, based on which a phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987), with a bootstrap analysis derived from 1000 replications. Sequence similarity was calculated using pairwise alignment obtained from the EzTaxon database (Chun et al., 2007).
2.3.2 Inhibitory Effect
The effects of bacterial strains on the growth of algal cells are expressed as the relative maximum cell number (%), which is defined as the maximum/final cell number in each experimental regime compared to the maximum/final cell number obtained in the f/2 medium (f/2). When cell numbers in the test group were always lower than the initial density, indicating that no active growth occurred, neither the maximum cell number (cells/mL) nor the relative maximum cell number (%) was calculated.
2.3.3 Maximum Specific Growth Rate
The specific growth rate (μ, divisions/d) of algal cells was calculated every day during the experimental period using the following equation:
where N2 and N1 are cell density values at times t2 and t1, respectively. The maximum specific growth rate (μmax) is defined as the maximum value of μ during the experiment.
2.3.4 Statistics
The mean and standard deviation (SD) were calculated for each treatment from three independent replicate cultures. The means and standard deviations of all data were calculated and graphed. Repeated measures ANOVA was used to compare growth curves of different treatments. One-way ANOVA with post hoc (Tukey test) analysis was conducted for multiple comparisons of the maximum specific growth rate, relative maximum cell density, and maximum bacterial concentration. Statistical analyses were performed using SPSS 25.0, and differences were considered significant when p < 0.05. Detailed statistical information is shown in the Supplementary Tables.
3 Results
3.1 Bacteria Isolation from the Phycosphere of Levanderina fissa
The growth of C. curvisetus in the coculture of the 12 bacterial strains isolated from the phycosphere of L. fissa is shown in Figure 2. The cell density increased gradually in the f/2 medium and reached a maximum of 1.71×105 cells/mL at Day 5. The cell density in the BC control, which was supplemented with bacterial medium, increased and peaked at Day 2, with a maximum of 1.67×105 cells/mL. As shown in Figure 2A, most bacteria had no significant inhibition of the growth of C. curvisetus, and only two strains (Lf4 and Lf7) showed significant inhibitory effects (p < 0.01). The cell density of C. curvisetus decreased rapidly after the addition of the bacterial strain Lf7 and dropped to 2.2×104 cells/mL at Day 5, which was only 16.9% of the inoculation density. The growth of algal cells was significantly inhibited in the first 3 days after the addition of the bacterial strain Lf 4 (p < 0.01), and then the cell density increased at Day 4 and Day 5; however, it was still lower than the inoculation density. The cell densities of C. curvisetus in cocultures with Lf7 and Lf4 were always lower than the inoculation densities. Moreover, the maximum cell densities in the cocultures with the other 10 bacterial strains were significantly higher than those of the f/2 and BC media (p < 0.05, Figure 2B). Moreover, only C. curvisetus inoculated with strain Lf7 showed a negative maximum specific growth rate (Figure 2C). The results of the screening experiment suggested that the bacterial strain Lf7 had the most obvious growth inhibition on C. curvisetus. Therefore, Lf7, identified as the Gammaproteobacteria Alteromonas species based on a partial sequence of the 16S rRNA gene (Table 1 and Figure 1), was selected for further experiments.
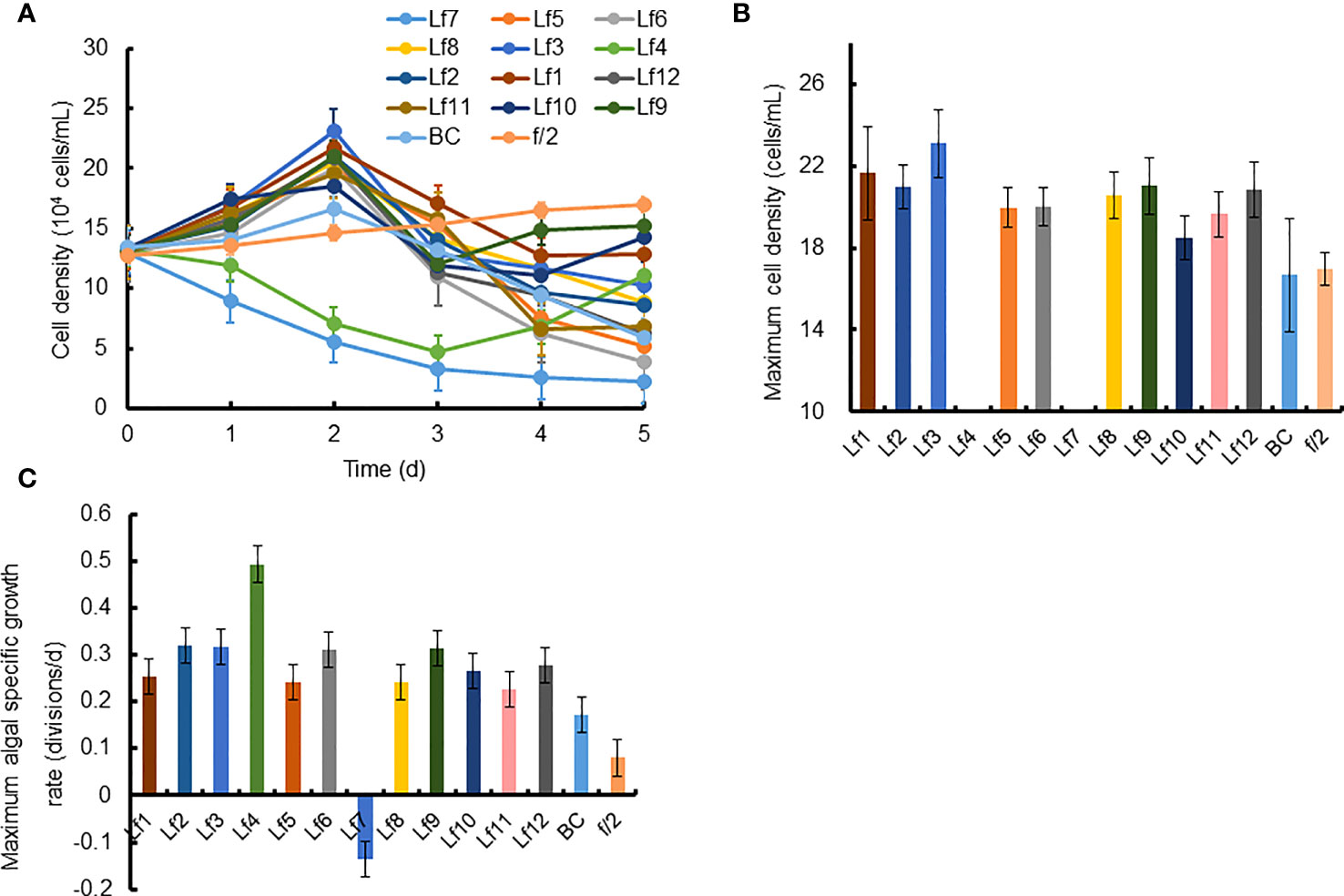
Figure 2 The growth of Chaetoceros curvisetus in the co-culture of bacterial strains isolated from the phycosphere of Levanderina fissa including (A) Growth curve, (B) maximum cell density, and (C) maximum specific growth rate. The maximum cell densities were not shown in Lf4 and Lf7, whose cell numbers were always lower than the initial cell numbers during the experiment. Lf1-Lf12: Cultures addition of the 12 bacterial strains Lf1-Lf12, BC: Culture addition of the liquid Zobell 2216E medium, f/2: Growth in f/2 medium.
3.2 Effects of Bacterial Strain Lf7 on the Growth of Chaetoceros curvisetus
The growth curves of C. curvisetus in different experimental treatments were similar, except for the highest bacterial inoculation group (7.5×107 CFU/mL). The curves initiated at a 1–2-day lag period, then a 2–3-day rapid growth period, followed by a slow increase or a slight decrease in cell density (Figure 3A). Significant growth inhibition occurred only in culture with 7.5×107 CFU/mL bacterium Lf7, in which cell numbers deceased steadily and were always lower than the incubation number (Figure 3A). As shown in Figure 4A, the maximum cell density in the BC culture was 95.7% compared to that in the f/2 medium, while the relative maximum cell densities were over 100% (106-133%) in cultures with bacterial addition at concentrations of 7.5×104-7.5×106 CFU/mL. The algal cells showed no effective growth after adding bacteria at a concentration of 7.5×107 CFU/mL, and the maximum cell density was the inoculation density. The maximum specific growth rate of C. curvisetus showed a negative value when inoculated with 7.5×107 CFU/mL bacterium Lf7, indicating negative growth throughout the whole experimental period (Figure 4B).
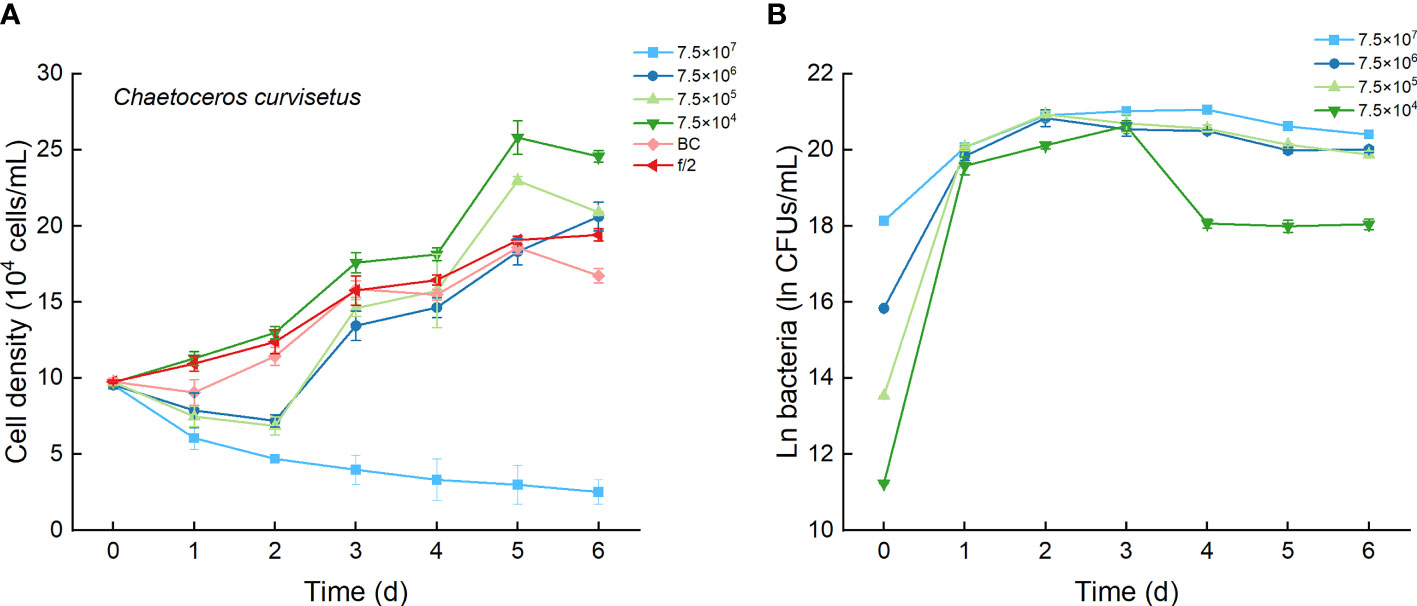
Figure 3 The growth of Chaetoceros curvisetus in the co-cultures with different inoculum dose of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of C. curvisetus in co-cultures with Lf7, (B) Lf7 abundances in the algal cultures.
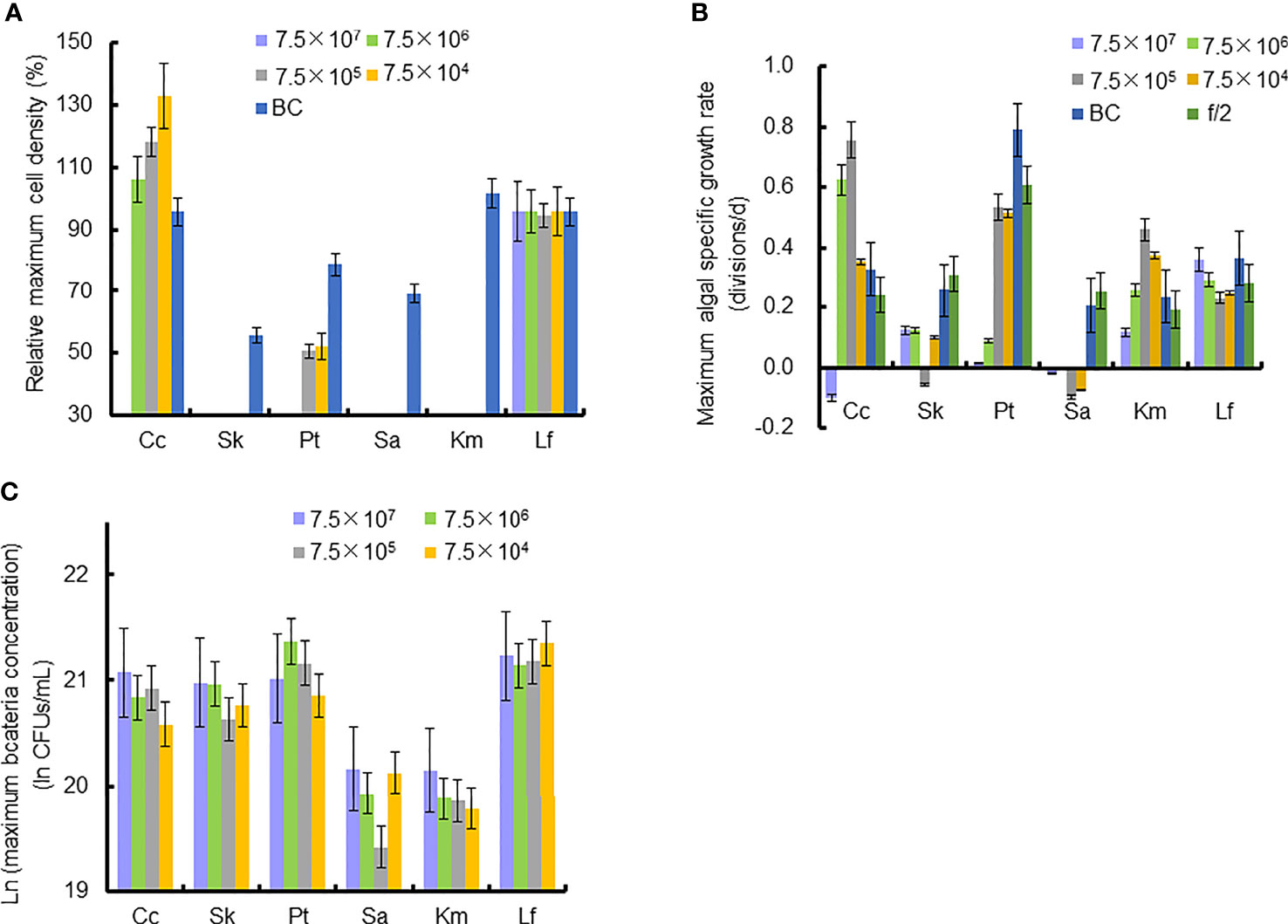
Figure 4 Relative maximum algal cell density, maximum algal specific growth rate, and bacteria concentration in co-cultures of six microalgal taxa with the bacterial strain Lf7 during the six days incubation. (A) Relative maximum algal cell density, (B) maximum algal specific growth rate, (C) maximum Lf7 concentration in the algal cultures. Cc, co-culture of Chaetoceros curvisetus with Lf7; SK, co-culture of Skeletonema dohrnii with Lf7; Pt, co-culture of Phaeodactylum tricornutum with Lf7; Sa, co-culture of Scrippsiella acuminata with Lf7; Km, co-culture of Karenia mikimotoi with Lf7; Lf, co-culture of Levanderina fissa with Lf7.
In the coculture system, bacteria grew well and showed maximum abundances at 2 to 4 days (Figure 3B). The bacterial concentration in the group with the highest bacterial inoculum dose maintained the longest growing period (4 days). In all groups with different bacterial inoculum doses, the maximum bacterial concentration showed no significant difference, ranging from 8.7×108-1.42×109 CFU/mL (Figure 4C).
3.3 Effects of the Bacterial Strain Lf7 on the Growth of Skeletonema dohrnii
The cell densities of S. dohrnii with f/2 medium were significantly higher than those with bacterial medium. The cell densities of S. dohrnii inoculated with strain Lf7 were significantly lower than those inoculated with either f/2 medium or bacterial medium (Figure 5A). The density dropped to 15-24% of the initial inoculation densities, indicating a strong inhibitory effect of the bacterial strain Lf7 on the growth of the algae S. dohrnii. The maximum specific growth rate of S. dohrnii showed a negative value when inoculated with 7.5×105 CFU/mL bacterium Lf7, indicating negative growth throughout the whole experimental period (Figure 4B).
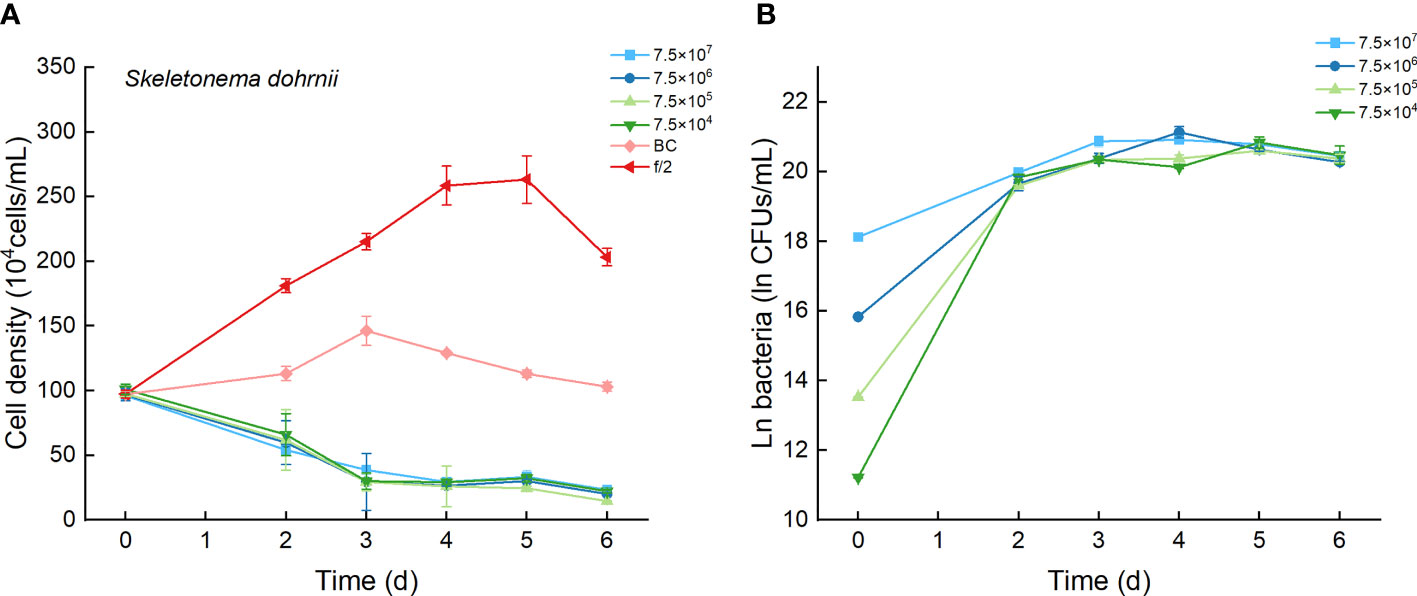
Figure 5 The growth of Skeletonema dohrnii in the co-cultures with different concentrations of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of S. dohrnii in co-cultures with Lf7, (B) Lf7 concentration in the algal cultures.
The bacterial concentration increased significantly to 9.1×108-1.29×109 CFU/mL, showing no significant differences between groups with different bacterial inoculum doses (Figure 5B).
3.4 Effects of Bacterial Strain Lf7 on the Growth of Phaeodactylum tricornutum
As the control, P. tricornutum with f/2 experienced a lag phase within two days, reached maximum cell densities on the fourth day, and maintained a high cell density from Day 5 to Day 6 (Figure 6A). At a high inoculation dose of bacteria (7.5×107 and 7.5×106 CFU/mL), the growth of P. tricornutum was strongly inhibited, with a decline of 90% cell density within 3 days. Afterward, the cell density of this algae decreases slowly and remains low in such a coculture condition. Whether at a low inoculation dose of bacteria (7.5×104 and 7.5×105 CFU/mL) or under additions of bacterial medium, the growth of P. tricornutum showed a similar growth pattern: cell densities decreased within 2 days and gradually increased to maximum values on Day 4. The cell densities of P. tricornutum with high inoculation doses of strain Lf7 were significantly lower than those with either low inoculation doses of strain Lf7 or bacterial medium. The maximum specific growth rate of P. tricornutum showed a value close to zero when inoculated with 7.5×104 CFU/mL bacterium Lf7, indicating almost no growth throughout the whole experimental period (Figure 4B).
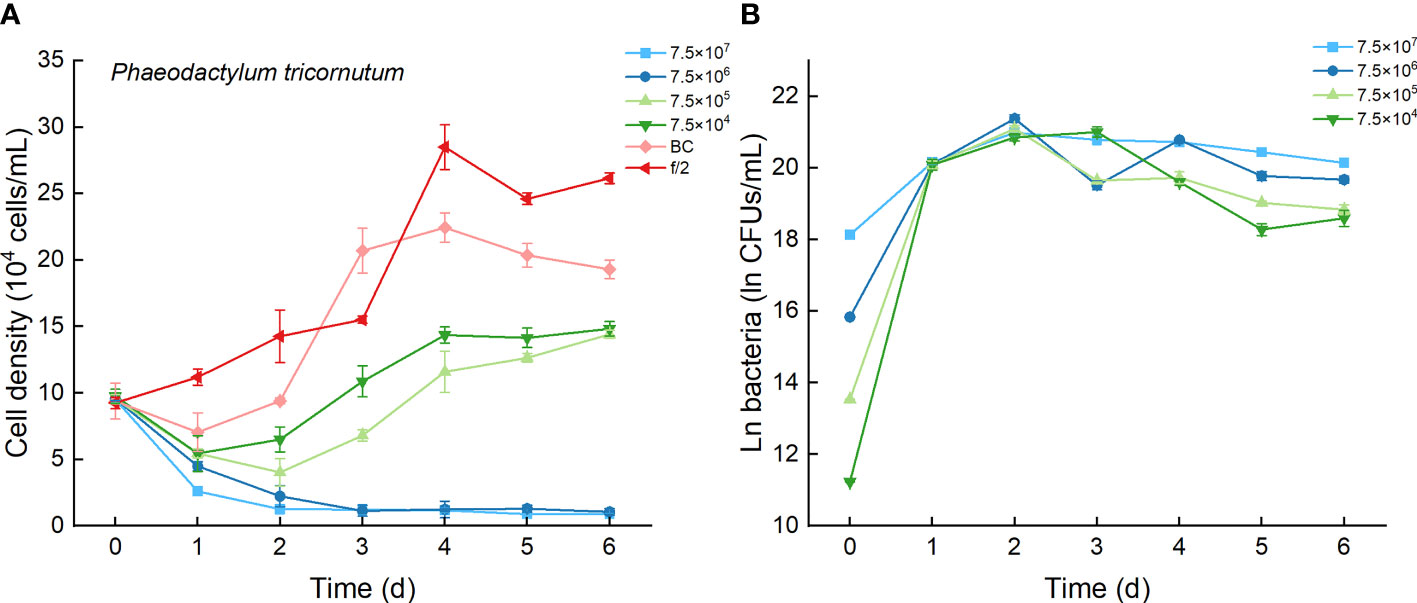
Figure 6 The growth of Phaeodactylum tricornutum in the co-cultures with different concentrations of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of P. tricornutum in co-cultures with Lf7, (B) Lf7 concentration in the algal cultures.
When cocultured with P. tricornutum, CFUs of strain Lf7 increased significantly (Figure 6B). The highest bacterial concentration occurred on Day 2 when the growth of this algae was strongly suppressed. The bacterial concentration increased to 1.14-1.9×109 CFU/mL by the end of the experiment, showing no significant differences between groups with different bacterial inoculum doses (p > 0.05).
3.5 Effects of Bacterial Strain Lf7 on the Growth of Scrippsiella acuminata
As the control, the cell densities of S. acuminata with additions of f/2 medium and bacterial medium showed no significant differences. The cell densities of S. acuminata at all inoculation doses of Lf7 declined dramatically (Figure 7A), indicating a strong inhibitory effect of Lf7 on S. acuminata. Moreover, the concentration of strain Lf7 increased under the coculture conditions, with maximum values within 2.7-5.7×108 CFU/mL from Day 3 to Day 4 (Figure 7B). The maximum specific growth rate of S. acuminata showed a negative value at all inoculation doses of the bacterium Lf7, indicating negative growth throughout the whole experimental period for all the coculture treatments (Figure 4B). Among the three diatoms used in the coculture experiments, Lf7 showed the lowest maximum concentration when cocultured with S. acuminata (Figure 4C).
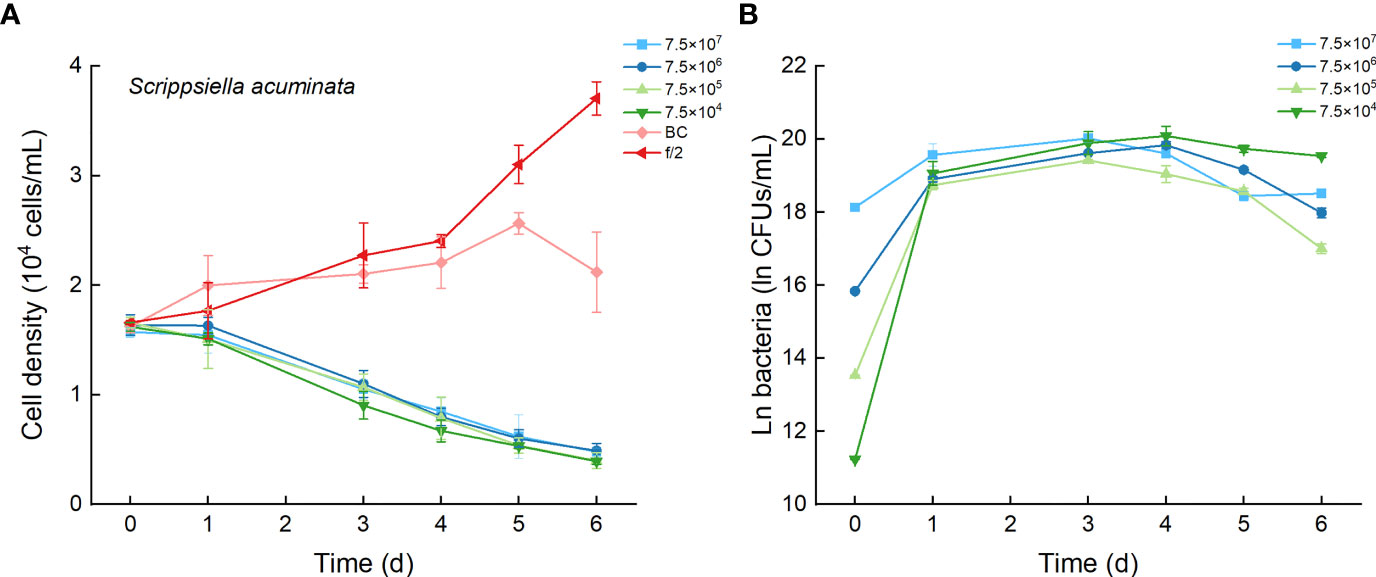
Figure 7 The growth of Scrippsiella acuminata in the co-cultures with different concentrations of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of S. acuminata in co-cultures with Lf7, (B) Lf7 concentration in the algal cultures.
3.6 Effects of the Bacterial Strain Lf7 on the Growth of Karenia mikimotoi
As the control, the cell densities of K. mikimotoi with additions of f/2 medium and bacterial medium showed no significant differences in their growth curves (p > 0.1, Figure 8A). Only the maximum specific growth rate of K. mikimotoi cocultured with 7.5×106 CFU/mL bacterium Lf7 was significantly lower than that of the control (P. tricornutum cultured with f/2 or BC) (all p <0.05, Figure 4B). However, the cell densities of K. mikimoto at all inoculation doses of Lf7 declined dramatically on the first day and were maintained at a low density, reaching 42-68% of the inoculum dose. Lf7 showed inhibitory effects on the growth of K. mikimotoi under the addition of all bacterial concentrations (Figure 8A).
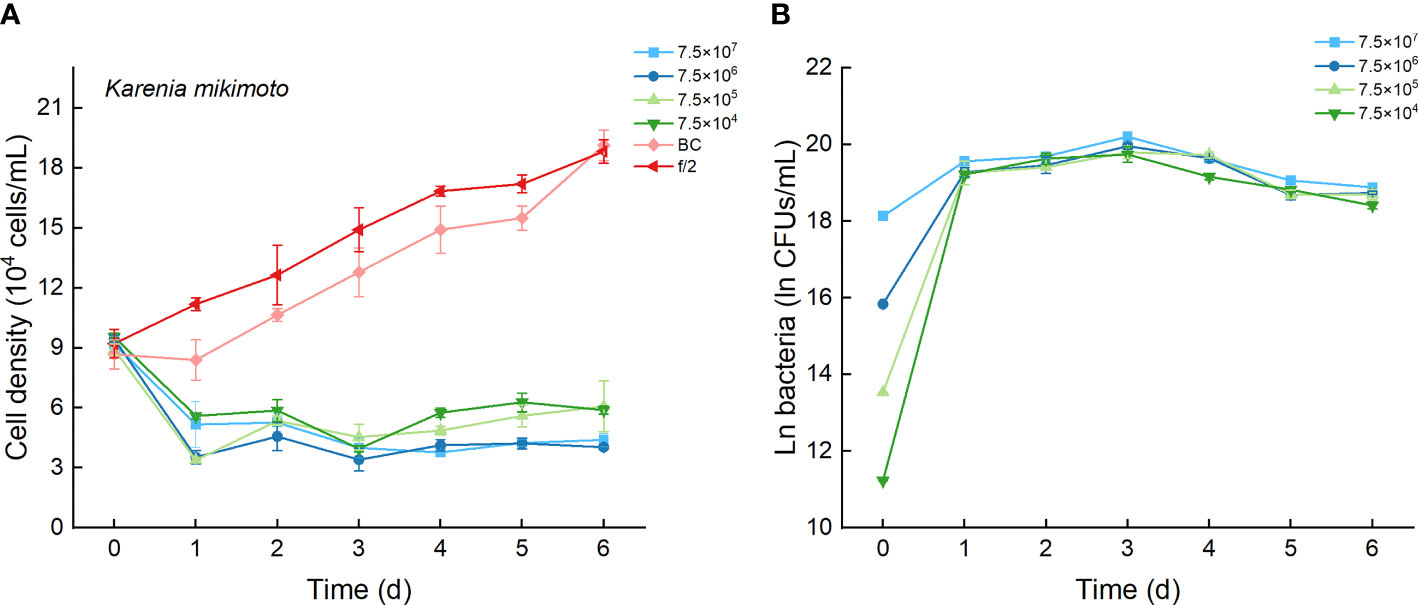
Figure 8 The growth of Karenia mikimotoi in the co-cultures with different concentrations of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of K. mikimotoi in co-cultures with Lf7, (B) Lf7 concentration in the algal cultures.
The concentration of strain Lf7 under coculture conditions with K. mikimotoi was also similar to that with S. acuminata and significantly lower than that with other diatoms, reaching maximum values within 2.7-5.7×108 CFU/mL (p < 0.01, Figures 4C, 8B).
3.7 Effects of Bacterial Strain Lf7 on the Growth of Levanderina fissa
Regardless of treatments (whether with different inoculation doses of bacteria or under additions of different media), the cell densities of L. fissa increased to a maximum value at Day 5 (Figure 9A). Although the daily maximum specific growth rate occurred differently between several of the two treatments (p <0.05, Figure 4B), the growth curves showed no significant differences (p > 0.05), indicating that the growth of L. fissa was not inhibited by Lf7. However, the concentration of strain Lf7 in coculture conditions with L. fissa reached maximum values within 1.52-1.87×109 CFU/mL, with the highest average values among those of strain Lf7 in coculture conditions with all algal species in this experiment (Figures 4C, 9B).
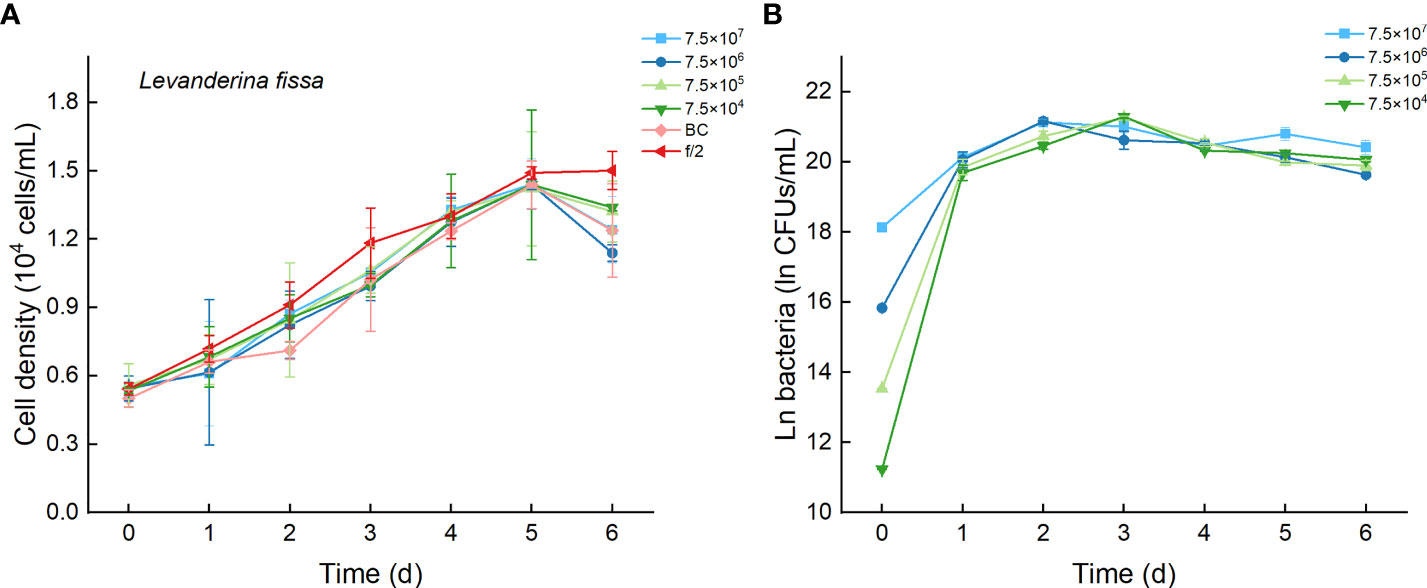
Figure 9 The growth of Levanderina fissa in the co-cultures with different concentrations of bacterial strain Lf7 (7.5×104-7.5×107 CFUs/mL). (A) Growth curves of L. fissa in co-cultures with Lf7, (B) Lf7 concentration in the algal cultures.
4 Discussion
The study isolated and identified a total of 12 strains of cultivable bacteria coexisting in different growth stages of L. fissa by the gradient dilution method. Only strain Lf7, identified phylogenetically as an Alteromonas species using a partial 16S rRNA gene sequence, showed a significant inhibitory effect on different algal species except its host, and its growth was promoted by coculture with the host algae L. fissa.
Alteromonas species are frequently reported as phycosphere bacteria with two opposite interacting actions with their host (Meyer et al., 2017; Cao et al., 2021). These bacteria can directly cause the lysis of their host algae by producing β-glucosidase and chitinase (Imai et al., 1995; Wang et al., 2010; Lin et al., 2013; Umetsu et al., 2019). Shi et al. (2018) isolated an algicidal bacterium (FDHY-03) from a bloom of Prorocentrum donghaiense and found the characteristics of its action against P. donghaiense by digesting cell wall polysaccharides from the megacytic growth zone. 16S rRNA gene sequence analysis placed this strain in the genus Alteromonas. The beneficial effects of Alteromonas species on their hosts have also been reported (Ferrier et al., 2002; Sandhya and Vijayan, 2019). By examining how the entire transcriptome of Isochrysis galbana changed when it was cocultured with A. macleodii isolated from its phycosphere, Cao et al. (2021) revealed transcriptome changes, including notable increases in transcripts related to photosynthesis, carbon fixation, oxidative phosphorylation, ribosomal proteins, biosynthetic enzymes, and transport processes, as well as the depletion of transcripts encoding DNA repair enzymes, superoxide dismutase (SOD), and other stress-response proteins. Taken together with the lab-enhanced growth of I. galbana by coculturing with A. macleodii, the presence of A. macleodii enhanced photosynthesis and biosynthesis of I. galbana and protected it from stress, especially oxidative stress.
Our isolated Lf7 showed neither direct beneficial nor inhibitory effects on its host, as discussed above. However, the strain showed different inhibitory effects on the growth of five other tested algal species, including K. mikimotoi, S. acuminata, P. tricornutum, C. curvisetus, and S. dohrnii. All five algal species occurred in the Pearl River Estuary. Except for the model diatom Phaeodactylum tricornutum, the other four were reported as bloom species elsewhere (Shikata et al., 2008; Begum et al., 2015; Tse and Lo, 2017; Li et al., 2019). L. fissa blooms are frequent in the Pearl River Estuary, while the phenomenon is not common worldwide (Jimenéz, 1993; Nagasoe et al., 2006; Nagasoe et al., 2010; Gárate-Lizárraga et al., 2013). L. fissa showed few advantages in interspecific competition with three other algal bloom-forming species, Skeletonema dohrnii (Bacillariophyceae), Prorocentrum micans (Dinophyceae), and Chattonella marina (Raphidophyceae), using pure, sterilized cultures in previous research (Wang et al., 2021). Thus, the reason L. fissa outcompeted other algae in the Pearl River Estuary and other limited places might be a key factor explaining the frequent blooms. Wang et al. (2017) proposed the assumption that the ability to dissolve organic phosphorus utilization of L. fissa offers this species a competitive advantage in the phytoplankton community. In this study, our results hint at another possible explanation that bacteria associated with L. fissa might have some ecological roles in its competition with other phytoplankton.
Mutualism associations often refer to a serious chemical exchange between phytoplankton and phycosphere bacteria; for example, diatoms secrete the amino acid tryptophan, which is converted by the bacterium into the hormone indole-3-acetic acid (IAA) (increasing bacterial growth simultaneously), which is then transferred from the bacterium back to the diatom to promote its cell division (Amin et al., 2015). Both bacteria and phytoplankton benefited from direct mutualism associations. In our study, the growth of that bacterium benefited most from the original host L. fissa among all tested algae. It is a common case that phycosphere bacteria are more adapted to a narrow range of compounds secreted by their algal host (Bell, 1984; Schäfer et al., 2002; Sapp et al., 2007; Sison-Mangus et al., 2014). Even though strain Lf7 showed no promoting effect on the growth of L. fissa, the inhibitory effect of strain Lf7 on other algae may lead to a competing advantage of its host. This may indicate complicated interactions between phycosphere bacteria and their host algae, which needs further lab studies.
In conclusion, by isolating and identifying cultivable bacteria coexisting in different growth stages of Levanderina fissa and subsequently monitoring the growth status of bacteria and microalgae in cocultural conditions, we obtained the bacterial strain Lf7 (taxonomically belonging to Alteromonas species), which showed inhibitory effects on the growth of five different microalgae (K. mikimotoi, S. acuminata, P. tricornutum, C. curvisetus, and S. dohrnii). We suspect that phycosphere bacteria Lf7 might have some ecological roles in the competition between its host algae L. fissa and other phytoplankton.
Data Availability Statement
The original contributionspresented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
In this paper, ZW and RHvconceived and designed the experiments; CX and XJ contributed to data acquisition and analysis; YT contributed to drafting the manuscript. All authors approved the final submitted manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (No. 42076141 and No. 32071566), Natural Science Foundation of Guangdong Province (No. 2022A1515011074), and Key Program of Marine Economy Development (Six Marine Industries) Special Foundation of Department of Natural Resources of Guangdong Province (No. GDNRC[2021]37).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful for the work of numerous participants who collected and analyzed samples during the periodof experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.908813/full#supplementary-material
References
Amin S. A., Green D. H., Hart M. C., Kuepper F. C., Sunda W. G., Carrano C. J. (2009). Photolysis of Iron–Siderophore Chelates Promotes Bacterial-Algal Mutualism. P. Natl. Acad. Sci. U.S.A. 106, 17071–17076. doi: 10.1073/pnas.0905512106
Amin S. A., Hmelo L. R., Van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and Signalling Between a Cosmopolitan Phytoplankton and Associated Bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions Between Diatoms and Bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Begum M., Sahu B. K., Das A. K., Vinithkumar N. V., Kirubagaran R. (2015). Extensive Chaetoceros Curvisetus Bloom in Relation to Water Quality in Port Blair Bay, Andaman Islands. Environ. Monit. Assess. 187, 1–14. doi: 10.1007/s10661-015-4461-2
Bell W. H. (1984). Bacterial Adaptation to Low-Nutrient Conditions as Studied With Algal Extracellular Products. Microb. Ecol. 10, 217–230. doi: 10.1007/BF02010936
Bratbak G., Tingstad T. F. (1985). Phytoplankton-Bacteria Interactions: An Apparent Paradox? Analysis of a Model System With Both Competition and Commensalism. Mar. Ecol. Progr. Ser. 25, 23–30. doi: 10.1016/0198-0254(86)91170-2
Buchan A., LeCleir G. R., Gulvik C. A., González J. M. (2014). Master Recyclers: Features and Functions of Bacteria Associated With Phytoplankton Blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Calatrava V., Hom E. F., Llamas Á., Fernández E., Galván A. (2018). OK, Thanks! A New Mutualism Between Chlamydomonas and Methylobacteria Facilitates Growth on Amino Acids and Peptides. FEMS Microbiol. Lett. 365, fny021. doi: 10.1093/femsle/fny021
Cao J. Y., Wang Y. Y., Wu M. N., Kong Z. Y., Lin J. H., Ling T., et al. (2021). RNA-Seq Insights Into the Impact of Alteromonas Macleodii on Isochrysis Galbana. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.711998
Chun J., Lee J. H., Jung Y., Kim M., Kim S., Kim B. K., et al. (2007). EzTaxon: A Web-Based Tool for the Identification of Prokaryotes Based on 16S Ribosomal RNA Gene Sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261. doi: 10.1099/ijs.0.64915-0
Cole J. J. (1982). Interactions Between Bacteria and Algae in Aquatic Ecosystems. Annu. Rev. Ecol. Syst. 13, 291–314. doi: 10.2307/2097070
Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. (2005). Algae Acquire Vitamin B12 Through a Symbiotic Relationship With Bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Ferrier M., Martin J. L., Rooney-Varga J. N. (2002). Stimulation of Alexandrium Fundyense Growth by Bacterial Assemblages From the Bay of Fundy. J. Appl. Microbiol. 92, 706–716. doi: 10.1046/j.1365-2672.2002.01576.x
Findlay J. A., Patil A. D. (1984). Antibacterial Constituents of the Diatom Navicula Delognei. J. Nat. Prod. 47, 815–818. doi: 10.1021/np50035a010
Gárate-Lizárraga I., Sevilla-Torres G., Alvarez-Anorve M., Aguirre-Bahena F., Violante-Gonzalez J., Rojas-Herrera A. (2013). First Record of Red Tide Caused by Gyrodinium Instriatum (Dinophyceae: Gymnodiniales) in Bahía De Acapulco, Guerrero. Cicimar. Oceánides 28, 43–47. doi: 10.37543/oceanides.v28i1.120
Guillard R. R. L. (1975). “Culture of Phytoplankton for Feeding Marine Invertebrates,” in Culture of Marine Invertebrate Animals. Eds. Smith W. L., Chanley M. H. (New York: Plenum Press), 29–60. doi: 10.1007/978-1-4615-8714-9_3
Höger A. L., Griehl C., Noll M. (2021). Infection With Intracellular Parasite Amoeboaphelidium Protococcarum Induces Shifts in Associated Bacterial Communities in Microalgae Cultures. J. Appl. Phycol. 33, 2863–2873. doi: 10.1007/s10811-021-02542-9
Imai I., Ishida Y., Sakaguchi K., Hata Y. (1995). Algicidal Marine Bacteria Isolated From Northern Hiroshima Bay, Japan. Fish. Sci. 61, 628–636. doi: 10.1577/1548-8446(1995)020<0016:TPUOFH>2.0.CO;2
Jimenéz R. (1993). “Ecological Factors Related to Gyrodinium Instriatum Bloom in the Inner Estuary of the Gulf of Guayaquil,” in Toxic Phytoplankton Blooms in the Sea. Eds. Smayda T., Shimizu Y.. (Netherlands: Elsevier B.V.) pp. 257–262.
Johansson O. N., Pinder M. I., Ohlsson F., Egardt J., Töpel M., Clarke A. K. (2019). Friends With Benefits: Exploring the Phycosphere of the Marine Diatom Skeletonema Marinoi. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01828
Jukes T. H., Cantor C. R. (1969). “Evolution of Protein Molecules,” in Mammalian Protein Metabolism, vol. 3 . Ed. Munro H. N. (New York: Academic Press), 21–132.
Kim B. H., Ramanan R., Cho D. H., Oh H. M., Kim H. S. (2014). Role of Rhizobium, a Plant Growth Promoting Bacterium, in Enhancing Algal Biomass Through Mutualistic Interaction. Biomass Bioenerg. 69, 95–105. doi: 10.1016/j.biombioe.2014.07.015
Kumar S., Tamura K., Nei M. (2004). MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform. 5, 150–163. doi: 10.1093/bib/5.2.150
Lin J., Zheng W., Tian Y., Wang G., Zheng T. (2013). Optimization of Culture Conditions and Medium Composition for the Marine Algicidal Bacterium Alteromonas Sp. DH46 by Uniform Design. J. Ocean. U. China 12, 385–391. doi: 10.1007/s11802-013-2153-5
Li X., Yan T., Yu R., Zhou M. (2019). A Review of Karenia Mikimotoi: Bloom Events, Physiology, Toxicity and Toxic Mechanism. Harmful Algae 90, 101702. doi: 10.1016/j.hal.2019.101702
Mayali X., Azam F. (2004). Algicidal Bacteria in the Sea and Their Impact on Algal Blooms. J. Eukar. Microbiol. 51, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Meyer N., Bigalke A., Kaulfuss A., Pohnert G. (2017). Strategies and Ecological Roles of Algicidal Bacteria. FEMS Microbiol. Rev. 41, 880–899. doi: 10.1093/femsre/fux029
Nagasoe S., Kim D. I., Shimasaki Y., Oshima Y., Yamaguchi M., Honjo T. (2006). Effects of Temperature, Salinity and Irradiance on the Growth of the Red Tide Dinoflagellate Gyrodinium Instriatum Freudenthal Et Lee. Harmful Algae 5, 20–25. doi: 10.1016/j.hal.2005.06.001
Nagasoe S., Shikata T., Yamasaki Y., Matsubara T., Shimasaki Y., Oshima Y., et al. (2010). Effects of Nutrients on Growth of the Red-Tide Dinoflagellate Gyrodinium Instriatum Freudenthal Et Lee and a Possible Link to Blooms of This Species. Hydrobiology 651, 225–238. doi: 10.1007/s10750-010-0301-0
Palacios O. A., López B. R., de-Bashan L. E. (2022). Microalga Growth-Promoting Bacteria (MGPB): A Formal Term Proposed for Beneficial Bacteria Involved in Microalgal-Bacterial Interactions. Algal. Res. 61, 102585. doi: 10.1016/j.algal.2021.102585
Rajamani S., Teplitski M., Kumar A., Krediet C. J., Sayre R. T., Bauer W. D. (2011). N-Acyl Homoseine Lactone Lactonase, AiiA, Inactivation of Quorum-Sensing Agonists Produced by Chlamidomonas Reinhardtii (Chlorophyta) and Characterization of aiiA Transgenic Algae. J. Phycol. 47, 1219–1227. doi: 10.1111/j.1529-8817.2011.01049.x
Saitou N., Nei M. (1987). The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sandhya S. V., Vijayan K. K. (2019). Symbiotic Association Among Marine Microalgae and Bacterial Flora: A Study With Special Reference to Commercially Important Isochrysis Galbana Culture. J. Appl. Phycol. 31, 2259–2266. doi: 10.1007/s10811-019-01772-2
Sapp M., Schwaderer A. S., Wiltshire K. H., Hoppe H. G., Gerdts G., Wichels A. (2007). Species-Specific Bacterial Communities in the Phycosphere of Microalgae? Microb. Ecol. 53, 683–699. doi: 10.1007/s00248-006-9162-5
Schäfer H., Abbas B., Witte H., Muyzer G. (2002). Genetic Diversity of ‘Satellite’bacteria Present in Cultures of Marine Diatoms. FEMS Microbiol. Ecol. 42, 25–35. doi: 10.1016/S0168-6496(02)00298-2
Seymour J. R., Amin S. A., Raina J. B., Stocker R. (2017). Zooming in on the Phycosphere: The Ecological Interface for Phytoplankton–Bacteria Relationships. Nat. Microbiol. 2, 1–12. doi: 10.1038/nmicrobiol.2017.65
Shi X., Liu L., Li Y., Xiao Y., Ding G., Lin S., et al. (2018). Isolation of an Algicidal Bacterium and its Effects Against the Harmful-Algal-Bloom Dinoflagellate Prorocentrum Donghaiense (Dinophyceae). Harmful Algae 80, 72–79. doi: 10.1016/j.hal.2018.09.003
Shikata T., Nagasoe S., Matsubara T., Yoshikawa S., Yamasaki Y., Shimasaki Y., et al. (2008). Factors Influencing the Initiation of Blooms of the Raphidophyte Heterosigma akashiwo and the Ciatom Skeletonema costatum in a Port in Japan. Limnol. Oceanogr. 53, 2503—2518. doi: 10.4319/lo.2008.53.6.2503
Sison-Mangus M. P., Jiang S., Tran K. N., Kudela R. M. (2014). Host-Specific Adaptation Governs the Interaction of the Marine Diatom, Pseudo-Nitzschia and Their Microbiota. ISME J. 8, 63–76. doi: 10.1038/ismej.2013.138
SOA (State Oceanic Administration People’s Republic of China) (2008) Bulletin of Marine Environmental Quality in Shenzhen City in 2007. Available at: http://www.soa.gov.cn/zwgk/hygb/zghyhjzlgb/201212/t20121206_21225.html.
Stien D., Sanchez-Ferandin S., Lami R. (2016). Quorum Sensing and Quorum Quenching in the Phycosphere of Phytoplankton: A Case of Chemical Interactions in Ecology. J. Chem. Ecol. 42, 1201–1211. doi: 10.1007/s10886-016-0791-y
Su J. Q., Yang X. R., Zheng T. L., Tian Y., Jiao N. Z., Cai L. Z., et al. (2007). Isolation and Characterization of a Marine Algicidal Bacterium Against the Toxic Dinoflagellate Alexandrium Tamarense. Harmful Algae 6, 799–810. doi: 10.1016/j.hal.2007.04.004
Teplitski M., Chen H., Rajamani S., Gao M., Merighi M., Sayre R. T., et al. (2004). Chlamydomonas Reinhardtii Secretes Compounds That Mimic Bacterial Signals and Interfere With Quorum Sensing Regulation in Bacteria. Plant Physiol. 134, 137–146. doi: 10.1104/pp.103.029918
Tse S., Lo S. (2017). Comparative Proteomic Studies of a Scrippsiella Acuminata Bloom With its Laboratory-Grown Culture Using a 15N-Metabolic Labeling Approach. Harmful Algae 67, 26–35. doi: 10.1016/j.hal.2017.05.009
Umetsu S., Kanda M., Imai I., Sakai R., Fujita M. J. (2019). Questiomycins, Algicidal Compounds Produced by the Marine Bacterium Alteromonas Sp. And Their Production Cue. Molecules 24, 4522. doi: 10.3390/molecules24244522
Wang Z., Guo X., Qu L., Lin L. (2017). Effects of Nitrogen and Phosphorus on the Growth of Levanderina Fissa: How it Blooms in Pearl River Estuary. J. Ocean U. China 16, 114–120. doi: 10.1007/s11802-017-3080-7
Wang H. K., Huang L. M., Huang X. P., Song X. Y., Wang H. J., Wu N. J., et al. (2003). A Red Tide Caused by Gyrodinium Instriatum and its Environmental Characters in Zhujiang River Estuary. J. Trop. Environ. 22, 55–62. doi: 10.1007/BF02860423
Wang Z. H., Jiang S., Gu Y. G., Kang W., Feng J., Lin L. C., et al. (2011). Effects of Cochlodinium Bloom in Pearl River Estuary in China on the Growth of Other Harmful Algal Bloom Species. J. Shenzhen U. Sci. Engi. 28, 553–558. doi: 10.3969/j.issn.1000-2618.2011.06.015
Wang X., Li Z., Su J., Tian Y., Ning X., Hong H., et al. (2010). Lysis of a Red-Tide Causing Alga, Alexandrium Tamarense, Caused by Bacteria From its Phycosphere. Biol. Control 52, 123–130. doi: 10.1016/j.biocontrol.2009.10.004
Wang Z., Qi Y., Yin Y., Jiang T., Xie L. (2001). Studies on the Cause and the Occurrence Reasons of a Gyrodinium Instriatum Red Tide in Shenzhen Bay in Spring of 1998. Mar. Sci. 25, 47–49. doi: 10.3969/j.issn.1000-3096.2001.05.015
Wang Z., Wang C., Li W., Wang M., Xiao L. (2021). Interspecies Competition Between Scrippsiella Acuminata and Three Marine Diatoms: Growth Inhibition and Allelopathic Effects. Aquat. Toxicol. 237, 195878. doi: 10.1016/j.aquatox.2021.105878
Xie B., Bishop S., Stessman D., Wright D., Spalding M. H., Halverson L. J. (2013). Chlamydomonas Reinhardtii Thermal Tolerance Enhancement Mediated by a Mutualistic Interaction With Vitamin B12-Producing Bacteria. ISME J. 7, 1544–1555. doi: 10.1038/ismej.2013.43
Keywords: algicidal bacteria, phycosphere, harmful algal bloom, growth, algae-bacteria interactions
Citation: Tang Y, Xie C, Jin X, Wang Z and Hu R (2022) Inhibitory Effect of Isolated Bacteria from the Phycosphere of Levanderina fissa on the Growth of Different Microalgae. Front. Mar. Sci. 9:908813. doi: 10.3389/fmars.2022.908813
Received: 31 March 2022; Accepted: 27 April 2022;
Published: 25 May 2022.
Edited by:
Zhangxi Hu, Guangdong Ocean University, ChinaReviewed by:
Xinfeng Dai, Ministry of Natural Resources, ChinaArief Rachman, National Research and Innovation Agency, Indonesia
Copyright © 2022 Tang, Xie, Jin, Wang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaohui Wang, dHd6aEBqbnUuZWR1LmNu; Yali Tang, bGl0YW5neWFsaUAxNjMuY29t; Ren Hu, dGh1cmVuQGpudS5lZHUuY24=
 Yali Tang
Yali Tang Changliang Xie
Changliang Xie Xiaotong Jin
Xiaotong Jin Zhaohui Wang
Zhaohui Wang Ren Hu*
Ren Hu*