
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 16 May 2022
Sec. Global Change and the Future Ocean
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.900200
This article is part of the Research Topic The Adaptation and Response of Aquatic Animals in the Context of Global Climate Change View all 18 articles
 Xiaohui Li1,2
Xiaohui Li1,2 Chen Ling1
Chen Ling1 Qiaoxin Wang1
Qiaoxin Wang1 Cui Feng1
Cui Feng1 Xiangzhong Luo1
Xiangzhong Luo1 Hang Sha1
Hang Sha1 Guoyu He1
Guoyu He1 Guiwei Zou1*
Guiwei Zou1* Hongwei Liang1,3*
Hongwei Liang1,3*The silver carp (Hypophthalmichthys molitrix) is an economically, as well as environmentally, important fish that harbors low environmental hypoxia tolerance and frequently contributes to a loss of aquaculture productivity. The gill is the first tissue attacked by hypoxia; however, the response of the gills of H. molitrix to hypoxia stress at the tissue, cellular, and molecular levels has not been clearly established. The influence of hypoxia on histological features along with gene expression in silver carp gills were explored in this research. The hematoxylin and eosin-stained sections and electron microscopy examinations of gills indicated that the gill lamellae were seriously twisted, gill filaments were dehisced, and the swelling and shedding of epithelial cell layer in the gill tissue were intensified along with the degree of hypoxia. In the hypoxia, semi-asphyxia, and asphyxia groups, the gill transcriptomic assessment of shifts in key genes, as well as modulatory networks in response to hypoxic conditions revealed 587, 725, and 748 differentially expressed genes, respectively. These genes are abundant in immune response signaling cascades (e.g., complement and coagulation cascades, Nucleotide-binding and oligomerization domain (NOD)-like receptor signaling cascade, and differentiation of Th1 along with Th2 cells) and oxygen transport [e.g., MAPK, PI3K-Akt, and hypoxia-inducible factor 1 (HIF-1) signaling cascades]. Genes linked to immune response (e.g., c2, c3, c6, klf4, cxcr4, cd45, and cd40) and oxygen transport (e.g., egln1, egln3, epo, ldh, and vegfa) were additionally identified. According to our findings, the silver carp may be using “HIF-1” to obtain additional oxygen during hypoxia. These findings illustrate that hypoxia stress might damage gill tissue, trigger an immunological response, and activate HIF-1 signaling to increase oxygen availability under hypoxic situations. The findings of this work will help scientists better understand the molecular mechanisms driving hypoxia responses in hypoxia-sensitive fish and speed up the development of hypoxia-resistant varieties.
Most of aerobic life on the planet need molecular oxygen to survive (Van der Meer et al., 2005). Fish interact directly with the aquatic environment, exhibiting significantly greater temporal, as well as spatial, fluctuations in oxygen contents relative to the terrestrial habitat (Zhu et al., 2013). Hypoxia is regarded as a low level of dissolved oxygen (DO) that adversely impacts fish behavioral, biochemical, and physiological activities consisting of reproduction, movement, respiration, development, and metabolism coupled with immunity (Jiang et al., 2017; Abdel-Tawwab et al., 2019). Conditions, for instance, high stocking density, organic matter buildup from unconsumed food, and feces accompanied with the blooms of algae in aquaculture habitats all contribute to the severity of the hypoxic situation (Gallage et al., 2016; Li et al., 2017). As a result, hypoxia occurs often in aquaculture, and the molecular responses of fish to hypoxic stress have generated considerable interest in recent years (Chen et al., 2017; Xia et al., 2018; Mu et al., 2020).
The silver carp is among the four main Chinese carp species that was responsible for 14.7% of total cultured freshwater fish production in 2020 alone (Yearbook, 2020). Silver carp feed on phytoplankton without additional feeding due to normal filter feeding; hence, silver carp aquaculture saves feeding costs. The silver carp has been introduced to or expanded across over 88 countries globally, not only to improve breeding benefits but also to prevent algal bloom and enhance water quality (Li et al., 2020). Nevertheless, H. molitrix harbors a low tolerance for hypoxia in the environment, which leads to higher pond turnover in high-density pond aquaculture (Li et al., 2021). As a result, studying the adaptive approaches of silver carp under hypoxia would be more beneficial.
Hypoxia causes fish to respond to stress in a chronic, as well as acute, manner, leading to diverse molecular, behavioral, morphological, and physiological alterations (Abdel-Tawwab et al., 2019). The principal site of gaseous exchange and the initial target in a hypoxic episode is the fish gill (Tiedke et al., 2015). The crucian carp (Carassius carassius) decreases the size of their inter-lamellar cell mass and exposed lamella, as well as increases the functional surface area of their gills in response to hypoxia (Sollid et al., 2003). When the fish are returned to a normoxic state, the gill structure caused by hypoxia would be changed (Sollid and Nilsson, 2006). As a result, the fish gill plays an indispensable role in the aquatic exchange of gases and may undergo dramatic remodeling as a response to variations in DO contents (Wu et al., 2017). Furthermore, the fish gill is an immune-competent organ with vast mucosal surfaces, referred to as gill-associated lymphoid tissue (Rebl et al., 2014). It is unclear how the gill of H. molitrix responds to hypoxic stress at the tissue, cellular, and molecular levels since it is the first tissue to be affected by hypoxia.
With its high-throughput precision coupled with reproducibility, transcriptome sequencing technology is a useful tool for studying aquatic animal growth along with development, disease-resistant immunological systems, stress physiology, and other functional processes (Zhang et al., 2009; Li and Dewey, 2011). The environmental stress adaption of various aquatic species, for instance, Cyprinus carpio (Xing et al., 2019), Scylla paramamosain (Cheng et al., 2019), Rana chensinensis (Ma et al., 2018), Procambarus clarkia (Zhang et al., 2022), and Gambusia affinis (Hou et al., 2019), has been studied via high-throughput transcriptome sequencing technology. To help comprehend the intrinsic molecular process in the response to hypoxia, the transcriptome alterations in the gill of H. molitrix were determined using the Illumina HiSeq 2500 sequencing technology, and the corresponding histomorphology alterations were also investigated in the current work. The findings of this research will be critical in furthering our understanding of the hypoxia adaptation mechanisms of aquatic species and in speeding up the breeding of hypoxia-resistant types. It also serves as a source of information and data for relevant studies.
Healthy silver carp with a body weight of 52.86 ± 5.64 g and body length of 15.06 ± 1.23 cm were selected as the experimental fish. They were acquired from the Genetics and Breeding Center of Silver Carp, Ministry of Agriculture and Rural Affairs in Jingzhou city, Hubei province, China. The fish were introduced in a re-circulating freshwater system (with heating features and a capacity of 400 L) and given 2 weeks to acclimate before the experiment began. Throughout acclimation, the water was kept at the following parameters: 25.0 ± 0.5°C; pH, 7.5 ± 0.2; and DO, 7.0± 0.5 mg/L. Throughout the study, we maintained a natural photoperiod. The DO level was assessed via a DO meter (HACH, Loveland, CO, USA).
Following acclimation, fish were relocated to indoor rectangular glass tanks measuring 46 cm by length, 33 cm by width, and 28 cm by height, with ten fish in every tank) and fasted for 24 h prior to the hypoxia test (Figure 1A). When the hypoxic stress testing started, 12 glass tanks were employed. Three tanks of fish (n = 10) were used as the control group (CK) and were kept in normoxic conditions. The fish in the remaining nine tanks (n = 10) were deemed as the test groups (T1, T2, and T3). Hypoxia experiments were done by enclosing the water-filled tanks with plastic film, and the fish gradually consumed the oxygen in the aerated water via natural respiration (Chen et al., 2017; Jiang et al., 2020; Li et al., 2021) until the majority of fish attempted to breathe directly via their mouth (three tanks, designated as the hypoxia or T1 group, DO = 0.75 ± 0.04 mg/L), half of the fish lost their balance (three tanks, designated as semi-asphyxia or T2 group, DO = 0.58 ± 0.06 mg/L), and the other half of the fish sunk without the rhythmical opening and closing of the gill flaps (three tanks, designated as asphyxia or T3 group, DO = 0.27 ± 0.06 mg/L), similar to Feng et al. (2022).

Figure 1 Hematoxylin and eosin (H&E) staining of gill sections under (A) normoxia; (B) hypoxia; (C) demi-asphyxia; and (D) asphyxia stress in silver carp. GL, branch leaf; BC, blood cells; PVC, pavement cells; MRC, mitochondria-rich cells.
The fixed gill was dehydrated using a graded ethanol–xylol series and vacuum-embedded in paraffin. The sections (5~6 μm) of the specimens were cut and placed on polylysine-coated slides. hematoxylin and eosin (H&E) staining was done on the slices, and they were photographed under an Olympus microscope (Olympus CH2, Olympus, Japan).
Gill tissues were washed in PBS and preserved in 2.5% glutaraldehyde overnight. After being washed with PBS, the gill tissues were dehydrated using gradient alcohol (70%, 80%, 90%, 95%, and 100%) and then treated with isoamyl acetate for standard critical drying. Then, using a JEOL JSM 7800E field emission scanning electron microscopy (JEOL, Tokyo, Japan), a vacuum ion coating was applied to view and capture pictures.
The isolation of total RNA from the gills of CK, T1, T2, and T3 groups was done with the TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). The processing of the complementary DNA (cDNA) along with the RNA sequencing was done at Frasergen Bioinformatics Co., Ltd (Wuhan, China), with sequencing run on the Illumina HiSeq™ 4000 system to generate 150 bp paired-end reads.
The analyses of the RNA-seq data were done as documented previously (Li et al., 2021). FASTQC (v0.11.5) was employed to verify the quality of the fastq output files. Trimgalore (v0.4.3) was adopted to eliminate adapter sequences. The reads were mapped to the silver carp reference genome (unpublished data). HTSeq (v0.6.1) was utilized to produce raw counts (Anders et al., 2015). To identify DEGs (differentially expressed genes), a fold change (FC) > 2.0 along with a P-adjusted value < 0.05 cut-off (FDR) served as the threshold (Benjamini and Yekutieli, 2001). The R ‘heatmap’ package was adopted to conduct hierarchical DEG clustering. The Gene Ontology (GO) enrichment analysis along with the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses for DEGs were performed via the KOBAS program (v2.1.1), with a P-value of 0.05 signifying significant enrichment.
The oligonucleotide sequences of the primers are given in Table 1. The genes utilized to validate the RNA-seq data were the members of overlapping DEGs from CK versus T1, CK versus T2, and CK versus T3, and their primers were built using Primer 6. B-actin served as the control gene (Li et al., 2013; Li et al., 2016). qPCR was performed on samples that were not related to those utilized for RNA-seq. Following total RNA isolation, cDNA was synthesized using the FastKing RT Kit (TianGen, Beijing, China) and qPCR was done with a 2×SYBR Green MasterMix reagent (Takara, Kyoto, Japan) as previously documented (Zhang et al., 2022). The determination of gene expression was done via the 2-△△Ct method (Livak and Schmittgen, 2001).
All values were presented as SD ± mean. SPSS v15.0 (IBM Corp., USA) was used for data analyses. Before performing ANOVA, the Shapiro–Wilk test was performed to determine the normality of the data and the Levene test was used to determine the homoscedasticity of the data. When the data were homogeneous and met the normal distribution, a one-way ANOVA was used to determine the statistical differences between multiple testing, and P < 0.05 signified statistical significance.
The sections stained with H&E exhibited that the both sides of gill lamellae in the normoxia group were symmetrical and orderly arranged, the epithelial cells were flat and regularly arranged, the red blood cells (RBCs) were evenly distributed in the sinus, and the mitochondria-rich cells were elliptically distributed at the base of the lamellae (Figure 1A). Compared with the CK (normoxia) group, the gill tissue structure of T1 (hypoxia), T2 (semi-asphyxia), and T3 (asphyxia) groups began to be damaged in gradually lower DO. As shown in the T1 group (Figure 1B), the volume of cells with rich mitochondria became larger, the distribution of red blood cells was uneven, and the gill lamellae were gradually curved, thus increasing the gill respiratory area. At the T2 group (Figure 1C), most of the gill lamellae showed a disorder and an “S”-shape distortion, some gill filaments were dehisced in the middle, the RBCs in the blood sinuses were not uniformly distributed and piled up, and the phenomenon of cavitation was common. At the T3 group (Figure 1D), the volume and number of mitochondria-rich cells were increased and the whole gill lamellae were swollen and seriously curved, which deviated from the normal shape. In addition, the statistical analysis showed that the gill filament cracking proportion was higher in hypoxia groups (Figure S1). These results indicated that the severe hypoxia stress caused serious damage to the gill tissue.
The scanning electron microscope section showed that the gill tissue structure was complete and the gill lamellae were arranged neatly without any damage at normoxia (Figure 2A). As shown in the T1 group (Figure 2B), the matrix of the gill lamellae became thin, some gill lamellae were damaged, and the surface of gill arch began to appear as a microridge. At the T2 group (Figure 2C), the arrangement of gill lamellae was disordered and the damage of the microridge on the surface of gill arch began to be obvious. At the T3 group (Figure 2D), the gill lamellae were seriously twisted, the matrix was thinned, and the number of microridge on the surface of the gill arch was greatly expanded.

Figure 2 Electron microscopy examinations of gill sections under (A) normoxia, (B) hypoxia, (C) semi-asphyxia, and (D) asphyxia stress in silver carp. SF: branch leaf.
We constructed a total of 12 cDNA libraries in this research. RNA-seq generated 21,228,388–29,007,113 million high-quality clean reads (Table 2). We mapped 16,801,436–22,573,384 reads (70.50%–80.27% of clean reads) to the genome of silver carp (unpublished), illustrating that the majority of clean reads generated by high-throughput sequencing were mapped (Table 2). Genes with count >1 were considered as highly expressed genes if they accounted for at least 75% of the 12 samples and were utilized for further differential expression assessment, giving a total of 25,319 genes in this research.
To unveil the mechanism via which the silver carp responds to hypoxia, differential gene expression analyses were carried out in the CK gill relative to the T1, T2, and T3 gills (P < 0.05). In comparison to the normoxia group, the hypoxia group harbored 587 DEGs, consisting of 312 upregulated along with 275 downregulated DEGs (Figure 3A). A total of 725 DEGs were found in the semi-asphyxia group, consisting of 426 upregulated coupled with 299 downregulated genes (Figure 3B). In the asphyxia group, a total of 748 DEGs were identified, 499 of which were upregulated with 249 being downregulated (Figure 3C). A Venn diagram exhibited the common and specific DEGs obtained from differential gene expression analyses described above (Figure 4A), and Figure 4B depicted the 224 co-expressed genes’ distinct expression trends in these four groups.
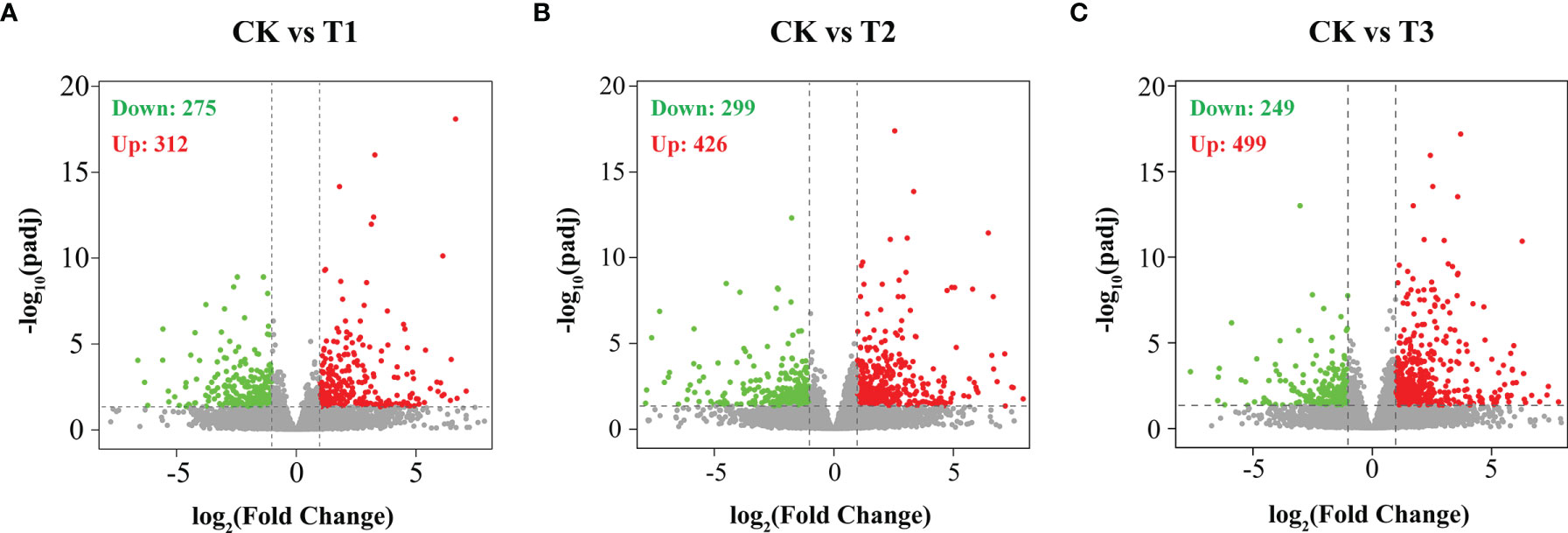
Figure 3 Differentially expressed genes (DEGs) were identified by hypoxia in the gills of silver carp. Volcano plots of DEGs in (A) CK vs. T1, (B) CK vs. T2, and (C) CK vs. T3. Red dots designate upregulated genes, and green dots designate downregulated genes. CK: normoxia, T1: hypoxia, T2: semi-asphyxia, T3: asphyxia.

Figure 4 Clustering analysis of DEGs between the three comparisons. (A) The DEGs in the three comparisons are exhibited in a Venn diagram. (B) The heatmap exhibits the variance in the expression of overlapping DEGs on the basis of the fragments per kilobase of transcript per million mapped read (FPKM) values. Genes with contents with greater than the mean are highlighted in red, while those with expression levels below the mean are highlighted in blue.
The DEGs were analyzed using the GO term analyses to determine considerably (P < 0.05) enriched BPs (biological processes), CCs (cellular components), and MFs (molecular functions). GO analyses exhibited that DEGs in the T1 group were enriched for BPs containing the immune system process, antigen processing and presentation, immune system development, and oxidation–reduction process; for CCs containing MHC protein complex and extracellular region; for MF-containing oxidoreductase activity, iron ion binding, and endopeptidase inhibitor activity (Figure 5A). In the T2 group, the immune system process, the modulation of the immune system process, antigen processing along with presentation, and the response to lipid were enriched in BPs; the MHC protein complex and MHC class II protein complex were enriched in CCs; the oxidoreductase activity, iron ion binding, and MAP kinase phosphatase activity were enriched in MFs (Figure 5B). In the T3 group, immune system process, the response to oxygen-containing compound, the cellular response to chemical stimulus, and the modulation of immune system process were enriched in BPs; the MHC protein complex and MHC class II protein complex were enriched in CCs; and the oxidoreductase activity, iron ion binding, and phospholipase C activity were enriched in MFs (Figure 5C). For better clarity and concise presentation, we analyzed the intersection of significantly enriched GO terms among three comparisons. Genes induced by hypoxia were consistently enriched in GO terms including the immune system process, oxidoreductase activity, the response to endogenous stimulus, iron ion binding, and MHC protein complex (Figure 5D). It is worth mentioning that the numbers of the associated genes and the significance of the GO enrichments roughly increased with the degree of hypoxia stress (hypoxia, semi-asphyxia, and asphyxia).
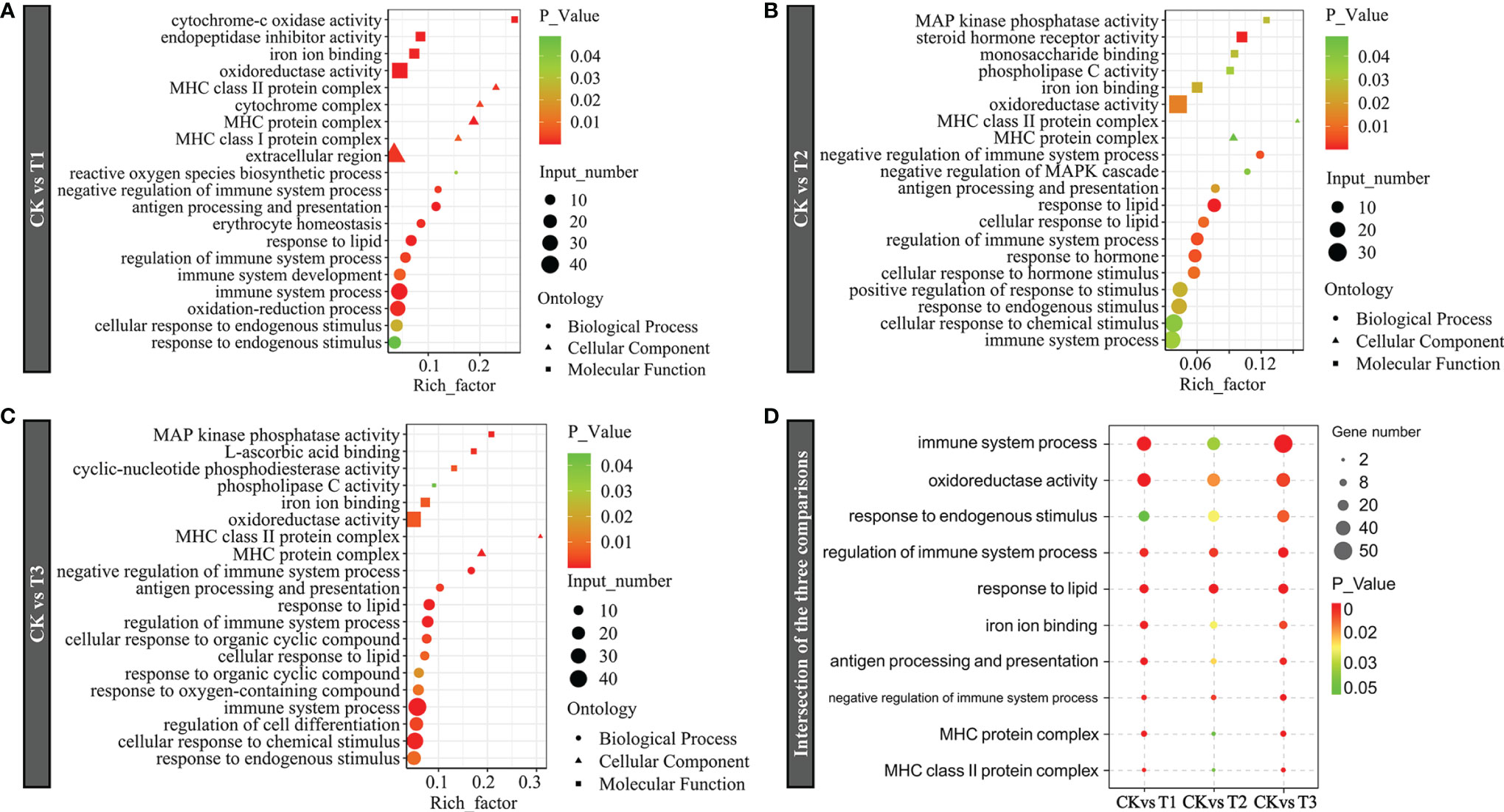
Figure 5 Gene Ontology (GO) term enrichment of dramatically DEGs in (A) CK vs. T1, (B) CK vs. T2, and (C) CK vs. T3. (D) Intersection of the three comparisons. Bubble plots represent remarkably enriched GO terms within the DEGs. The y-axis designates GO terms, and the x-axis exhibits the rich factor. The P-value indicates the enrichment importance of the GO terms, with lower P-values suggesting higher intensity. CK: normoxia, T1: hypoxia, T2: semi-asphyxia, T3: asphyxia.
KEGG enrichment analysis was used to reveal the affected biological pathways. The DEGs induced by hypoxia stress (T1) were enriched in signaling pathways consisting of complement and coagulation, mTOR, Th1 and Th2 cell differentiation, HIF-1, and FoxO (Figure 6A). The genes induced by semi-asphyxia stress (T2) were enriched in signaling pathways including drug metabolism – cytochrome P450, complement and coagulation, and HIF-1 (Figure 6B). The genes induced by asphyxia stress (T3) were enriched in signaling pathways including vitamin B6 metabolism, the immune network for IgA production, antigen processing along with presentation, Th1 and Th2 cell differentiation, HIF-1, FoxO, and MAPK (Figure 6C). Given that the enrichment signaling cascades in T1, T2, and T3 were primarily related to the immune response and oxygen transport, the intersection of enriched KEGG cascades in these two functions was determined for better clarity and concise presentation. As a result, the signaling cascades linked to immune system (complement and coagulation cascades, NOD-like receptor signaling cascade, leukocyte transendothelial migration, and Th1 and Th2 cell differentiation), immune diseases (rheumatoid arthritis, asthma, inflammatory bowel disease, graft-versus-host disease, and systemic lupus erythematosus), as well as oxygen transport (MAPK, FoxO, PI3K-Akt, and HIF-1 signaling cascade) were enriched (Figure 6D). Moreover, the numbers of the associated genes and significance of the KEGG enrichments roughly increased with the degree of hypoxia stress (hypoxia, semi-asphyxia, and asphyxia). Furthermore, the DEGs linked to immune system (Figure 7 and Table 3) as well as oxygen transport (Figure 8) were analyzed.
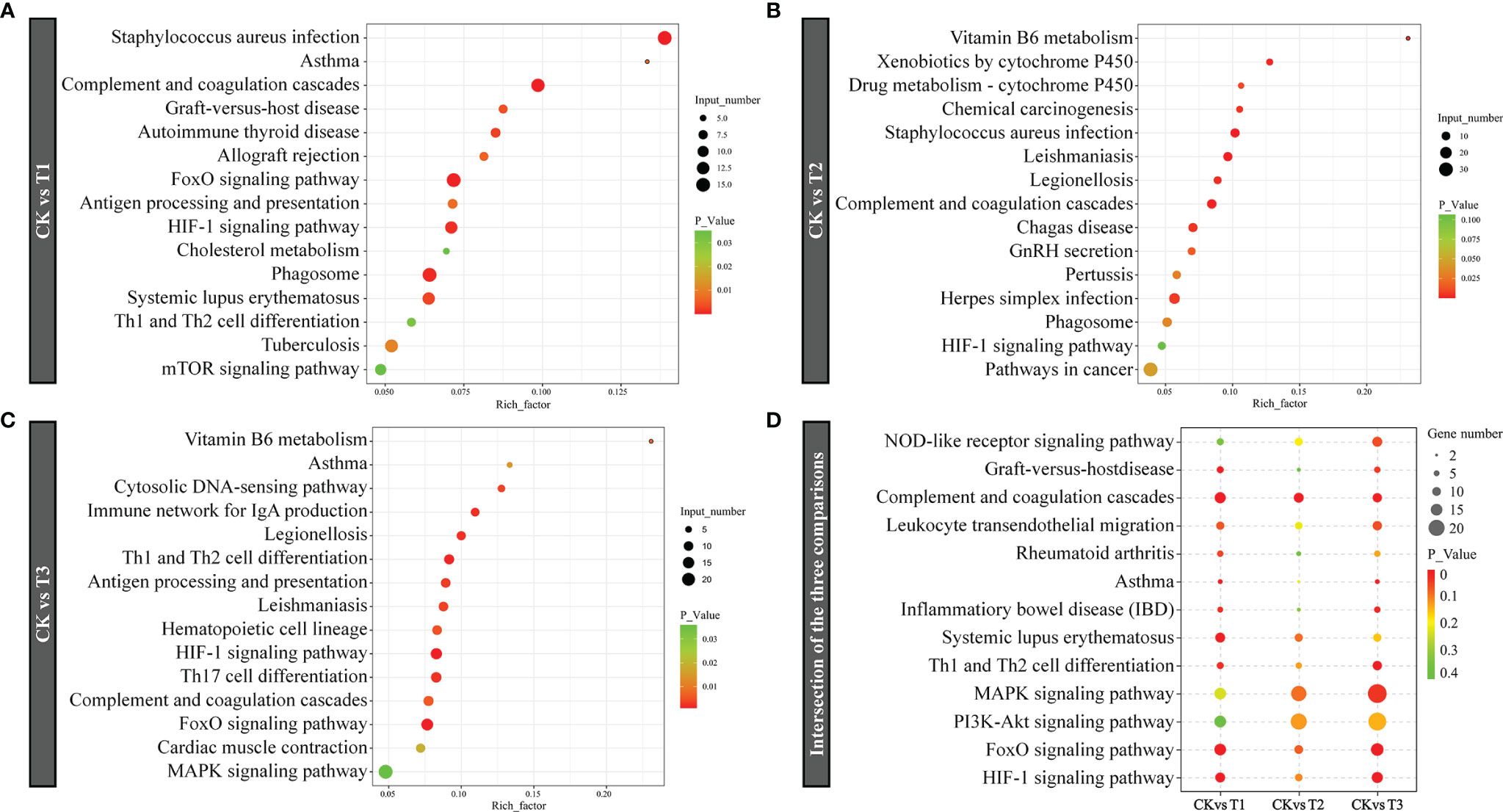
Figure 6 KEGG pathway enrichment of dramatically DEGs in (A) CK vs. T1, (B) CK vs. T2, (C) CK vs. T3, (D) Intersection of the three comparisons in immune response and oxygen transport. The bubble plots highlight KEGG cascades that are considerably enriched in the remarkable DEGs. The y-axis exhibits KEGG cascades. The x-axis denotes the rich factor. The P result signifies the signaling cascades enrichment importance; the lower the P-value, the higher the intensity. CK: normoxia, T1: hypoxia, T2: semi-asphyxia, T3: asphyxia.
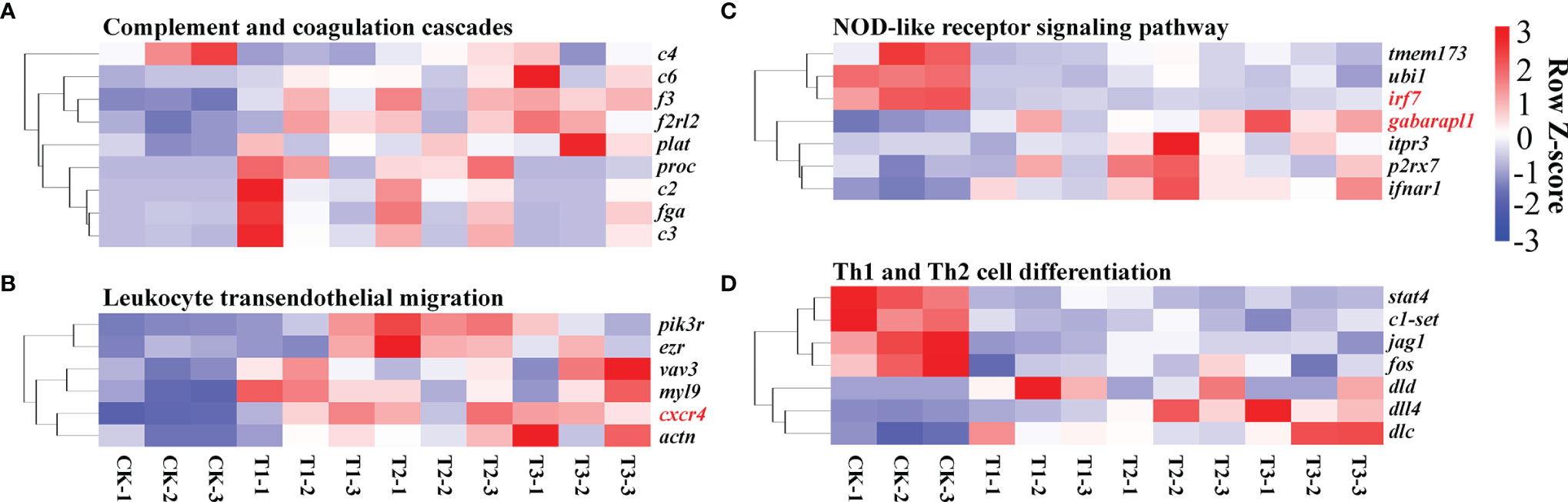
Figure 7 Heat map exhibiting alterations in gene expression in the pathways related to immune response, like complement and coagulation cascades (A), leukocyte transendothelial migration (B), NOD-like receptor signaling cascade (C), and Th1 and Th2 cell differentiation (D). The evaluation criteria used the average RPKM value per gene. Genes with contents greater than the mean are colored red, while those with contents below the mean are colored blue.
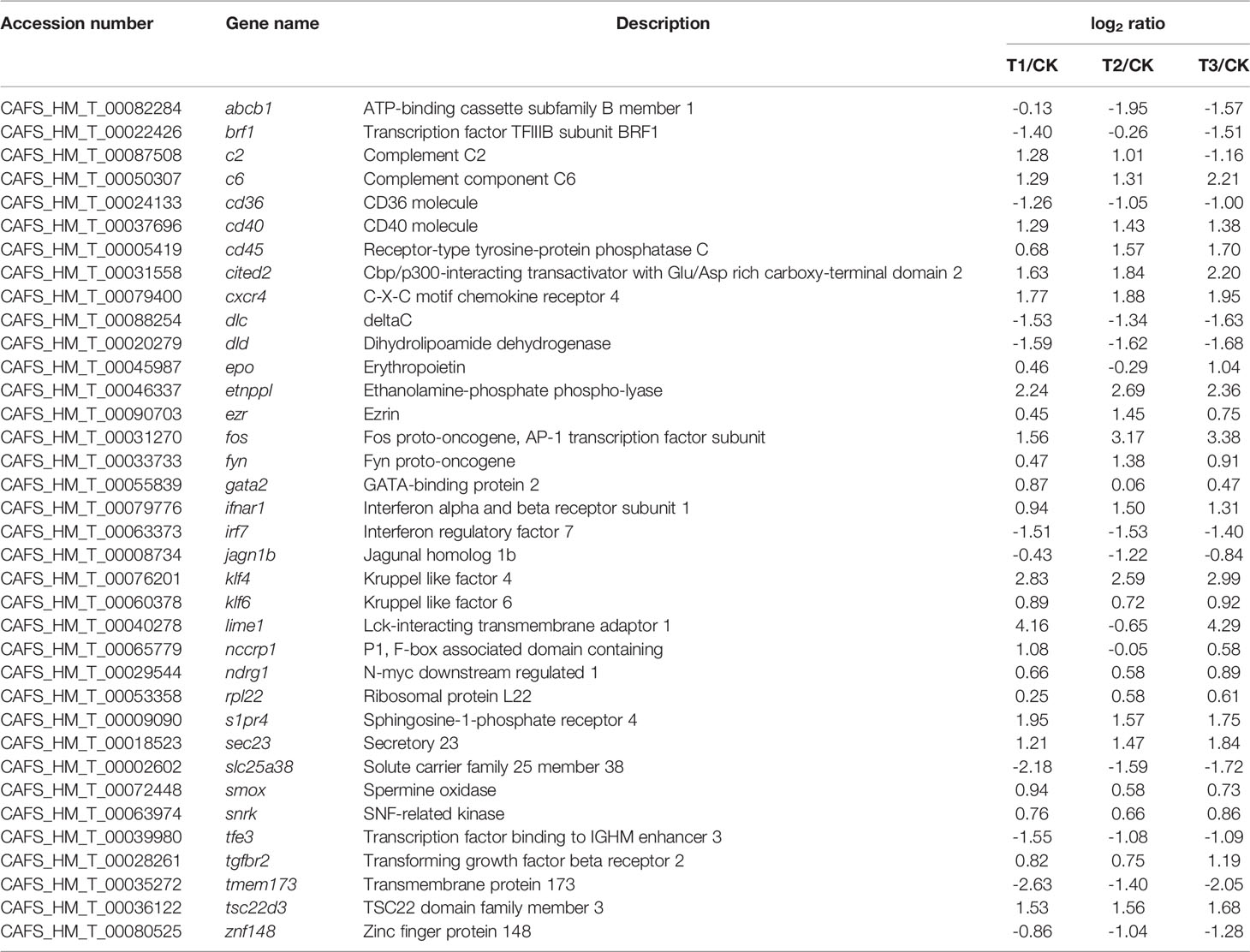
Table 3 The representative immunity-related DEGs in the gills of H. molitrix in varying degrees of hypoxia.
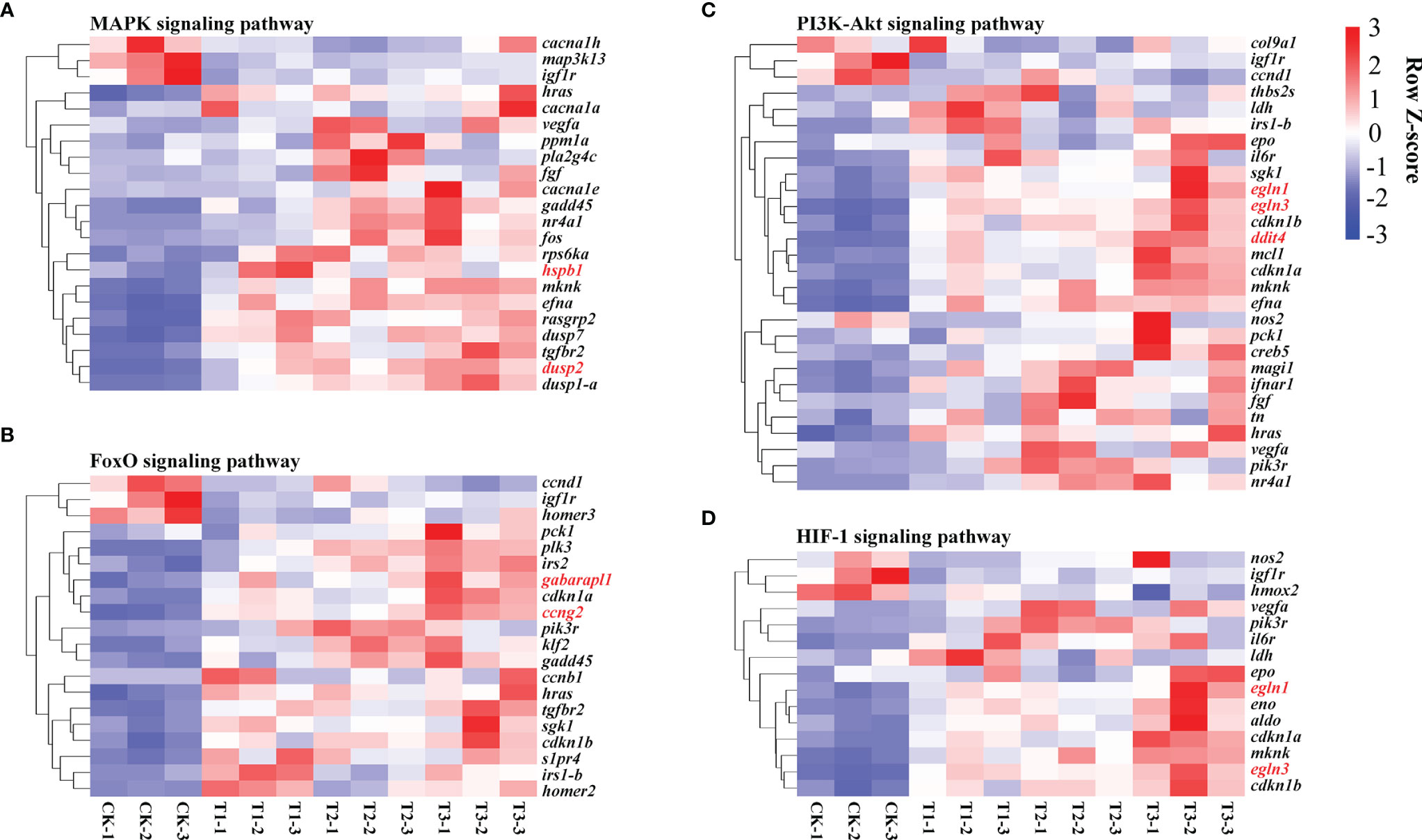
Figure 8 Heat map exhibiting alterations in gene expression in the pathways related to oxygen transport, such as MAPK (A), FoxO (B), PI3K-Akt (C), and HIF-1 signaling cascade (D). The evaluation criteria used the average RPKM value per gene. Genes with contents greater than the mean are colored red, while those with contents below the mean are colored blue.
To confirm the RNA-seq findings, we employed RT-qPCR to verify 18 DEGs from insulin signaling pathways. RT-qPCR data exhibited that cxcr4 and gabarapl1 (immune response related), hspb1, dusp2, ccng2, ddit4, egln1 and egln3 (oxygen transport related) were remarkably and gradually increased, while irf7 (immune response related) were remarkably and gradually decreased in the hypoxia, semi-asphyxia, and asphyxia groups (Figure 9). These data were congruent with RNA-seq data in terms of the variation trend (Table 4), demonstrating the accuracy and reliability of the RNA-seq investigation.
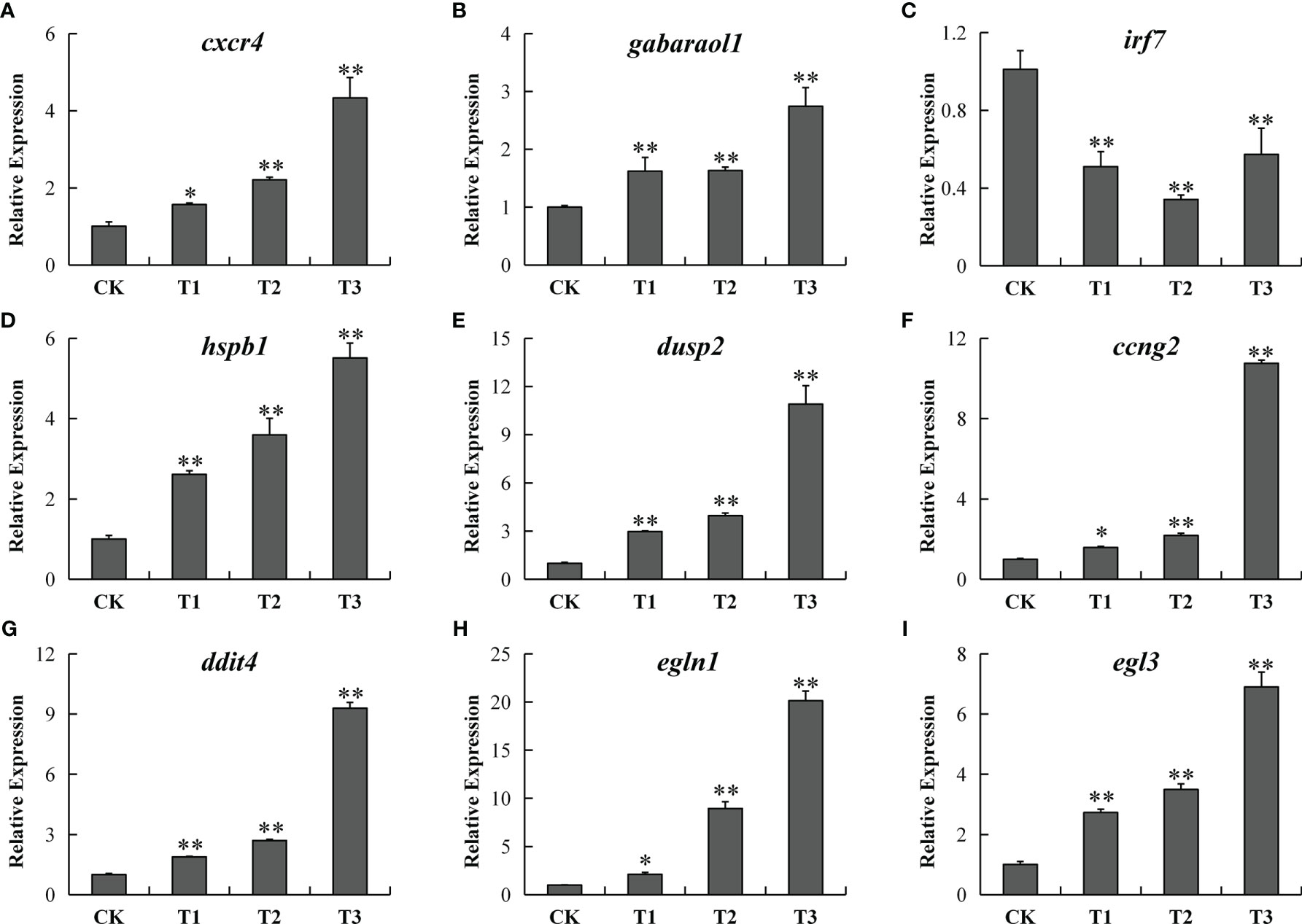
Figure 9 RT-qPCR verification of 9 differentially expressed genes [cxcr4 (A), gabarapl1 (B), irf7 (C), hspb1 (D), dusp2 (E), ccng2 (F), ddit4 (G), egln1 (H), and egln3 (I)] in the CK, T1, T2, and T3 groups. Y-axis exhibits the relative fold change. * designates P < 0.05, ** designates P < 0.01.
Herein, the impacts of varying degrees of hypoxia stress (hypoxia, semi-asphyxia, and asphyxia) were evaluated on the histomorphological in the gill tissue of H. molitrix. Results illustrated that the tissue damage in the gills becomes more and more severe with the increasingly low DO concentration. Using gill transcriptomic analysis, the effects of hypoxia on immune defense and oxygen transport-related genes and signaling cascades in H. molitrix were uncovered. As a result, the classified discussion is detailed below.
Aquatic habitats have a range of oxygen contents, from anoxia to hyperoxia, which results in a corresponding range of tissue and cellular diversity in aquatic animals. Silver carp, being a somewhat hypoxia-sensitive freshwater fish, is susceptible to being harmed by a fall in DO levels caused by weather changes or eutrophication (Li et al., 2021). The gill is the main respiratory organ of fish, and its dominant functions are carrying out gas exchange, promoting metabolism, and regulating osmotic pressure. As the first organ that responds to rapid changes in the DO level of water, the functions of a gill were disrupted by hypoxia, including respiration, nitrogenous waste excretion, and osmoregulation (Khansari et al., 2018). Under hypoxia stress, a Crucian carp could increase its oxygen intake by increasing the number of gill lamellae (Sollid et al., 2003). Studies have also shown that Gymnocypris przewalskii (Matey et al., 2008) and C. carassius (Sollid et al., 2005) can adapt to the hypoxia environment by changing the shape and structure of gills to increase its oxygen intake. In this study, the volume of cells with rich mitochondria became larger and the gill lamellae was gradually curved in the hypoxia condition, thus possibly increasing the gill respiratory area to obtain more oxygen. The results were consistent with the changes of gill tissues of Oncorhynchus mykiss (Matey et al., 2011) and G. przewalskii (Matey et al., 2008) under hypoxia stress. Furthermore, the tissue section and scanning electron microscope section of the gills in silver carp showed that the swelling and shedding of the epithelial cell layer in the gill tissue were intensified, the gill lamellae were seriously twisted, the phenomenon of cavitation became common, and gill filaments were dehisced in the semi-asphyxia and asphyxia conditions. Therefore, the abovementioned results further illustrate that hypoxia would alter the structure of gills to adapt to environmental challenges and severe hypoxia condition could cause a more serious and irreversible tissue damage of the gills in H. molitrix.
The majority of studies on the teleost immune response focused on changes in the expression of genes in the head, kidney, and spleen, which represent the primary hematopoietic along with secondary lymphoid organs of fish, respectively (Ewart et al., 2005; Overturf and LaPatra, 2006; Jørgensen et al., 2011). While the gill is primarily responsible for gas along with electrolyte exchange coupled with excretion (Evans et al., 2005), it has been considered as a key infection site, owing to its continual interaction with the aquatic environment (Dos Santos et al., 2001). Additionally, the fish gills, as a lymphoid tissue connected with the mucosa, contain a variety of leuklarki populations that are responsible for local immune responses, making them an immune-competent organ (Press and Evensen, 1999; Rebl et al., 2014). In this work, immunological cascades (complement and coagulation cascades, leukocyte transendothelial migration, NOD-like receptor signaling cascade, and the differentiation of Th1 and Th2 cells) were enriched in diverse degrees of hypoxic stress. Complement and coagulation systems are two closely linked plasma protein cascades that play critical roles in host defense and hemostasis, respectively (Berends et al., 2014). The complement system, a critical defender against invasive pathogenic microbes, may be triggered by traditional, alternative, or mannan-docking lectin cascades (Kemper et al., 2014). Complement C3 is a critical component of the complement system, and the activation of C3 converges the three complement activation cascades (Meng et al., 2019). C2 and C4 are two effector proteins that have the potential to initiate the classical complement cascade (Petersen et al., 2000). C6 is one of the terminal components of a complement system that, when coupled with other complement components, forms the membrane attack complex, which results in bacterial cell lysis (Shen et al., 2013). The upregulation of complement C2, C6, and C3 in Figure 7A demonstrated that hypoxic stress may trigger the H. molitrix immune defense response.
Atherogenesis and other inflammatory vascular disorders are complicated by leukocyte adherence and trans-endothelial migration (Zhang et al., 2011). The NOD-like receptor signaling cascade is responsible for the generation of innate immune responses and plays an indispensable role in inflammation (Viale-Bouroncle et al., 2012). Herein, leukocyte trans-endothelial migration and the NOD-like receptor signaling pathway were enriched in varying groups, indicating that hypoxia might initiate an innate immune response and trigger inflammation processes. The Th1/Th2 balance is thought to be critical for good immunological responses, and an imbalance between the two may result in immune-linked diseases (Ohno et al., 2015). Patients with rheumatoid arthritis, type 1 diabetes, and multiple sclerosis have Th1-dominant immune responses, while those with type 1 hypersensitivity conditions including allergies or asthma exhibit Th2-dominant immune responses (Kidd, 2003). STAT4 is primarily involved in the induction of Th1 responses and suppresses Th2 responses (Chitnis et al., 2004; Cui et al., 2021). STAT4 activation is thought to have an inflammatory impact, and it is involved in the modulation of Th1/Th2 differentiation as well as the autoimmune conditions associated with this dysfunction (Cui et al., 2021). In this research, stat4 was downregulated in hypoxia-treated groups, indicating that Th1 responses were inhibited and Th2 responses were induced in the hypoxia condition in H. molitrix. Furthermore, rheumatoid arthritis, asthma, and Th1 and Th2 cell differentiation were enriched in varying groups in Figure 6D, suggesting that varying degrees of hypoxia would impact Th1/Th2 balance and further cause immune disease including rheumatoid arthritis and asthma in H. molitrix.
In addition to the aforementioned findings, a high number of functional DEGs were identified; and these DEGs have the potential to influence the immune system and various signaling cascades. KLF4 is implicated in the control of endothelial inflammation by dampening the activation of the NF-B cascade (Fang and Davies, 2012). CXCR4 is required for a variety of biological activities, consisting of immune cell trafficking and bone-marrow homeostasis (Lin et al., 2019). IRF7 functions as the pivotal modulator of type 1 IFN-triggered immune responses, which are required for viral immunity (Honda et al., 2005). CD45, the prototypical receptor-like protein tyrosine phosphatase gene, plays an indispensable role in immune cell signal transduction cascades (Ren et al., 2018). CD36 is a membrane-bound glycoprotein that participates in a variety of cellular activities including lipid transport, immunological modulation, coagulation, and atherosclerosis (Endemann et al., 1993). CD40 is a co-stimulatory immune receptor belonging to the TNF receptor superfamily that plays a pivotal role in Th1-cell-triggered immune responses (Cella et al., 1996; Reyes-Moreno et al., 2004). The downregulation of irf7 and cd36 as well as the upregulation of klf4, cxcr4, cd45, and cd40 (Table 3) in hypoxia-treated groups suggested that the immune response ability of the gills in H. molitrix might be weakened by the varying degrees of hypoxia stress (hypoxia, semi-asphyxia, and asphyxia).
The majority of aquatic animals have evolved a variety of molecular approaches for hypoxia modulation, consisting of increased oxygen delivery as well as decreased oxygen consumption (Sun et al., 2016). These processes are controlled by HIF-1 signaling cascade-modulated genes, for instance, erythropoietin, transferrin, transferrin receptor, hexokinase, and lactic dehydrogenase (Zhu et al., 2013). In many species, the HIF-1 signaling cascade induces identical or homologous gene expression, resulting in similar biochemical and physical consequences, such as angiogenesis, oxygen sensing, and erythropoiesis along with oxygen transport (Xiao, 2015).
MAPKs play a role in oxygen sensing as critical modulators of HIF-1 signaling. Hypoxia has been exhibited to change MAPK expression in the zebrafish heart (Marques et al., 2008). As a critical factor in HIF-1 signaling, HIF-1 is activated through the PI3K/Akt signaling (Jiang et al., 2001). The FoxO signaling cascade is remarkably linked to the PI3K/Akt signaling cascade (Farhan et al., 2017), suggesting that it may have an indirect influence on HIF-1 signaling. Furthermore, the HIF-1, MAPK, FoxO, and PI3K-Akt signaling were enriched, and the related genes were remarkably elevated (Figure 8). These findings illustrate that in response to hypoxia, H. molitrix activates these signaling cascades.
Numerous upregulated genes, consisting of nos2, vegfa, igf1r, hmox2, pik3r, il6r, ldh, epo, egln1, eno, aldo, cdkn1a, mknk, egln3, and cdkn1b, which are commonly linked with hypoxia response, were shown to be enriched in HIF-1 signaling. In zebrafish, the egln1 gene, also known as egln1b, produces PHD2, which degrades HIF-1 by prolyl hydroxylation in an aerobic environment (Schofield and Ratcliffe, 2004). Additionally, the EGLN family genes EGLN2 and EGLN3 act as cellular oxygen sensors, catalyzing and hydroxylating HIF alpha proteins (Hu et al., 2019). The EGLN family has been implicated in EPO synthesis and erythropoiesis in murine zygotes (Fisher et al., 2009), and EGLN3 transcriptome and proteomic alterations have been connected to Tibetan pig adaptation to high-altitude hypoxia (Zhang et al., 2017). The induction of EPO at low oxygen levels results in an increase in red blood cell formation, which results in an increase in oxygen delivery in response to ischemia stress (Zhang et al., 2014). HIF activation increases LDH activity, which in turn promotes HIF activation further, implying a positive feedback loop between HIF and LDH (Lu et al., 2002). Hypoxia and a rise in the expression of oxygen-modulated HIF transcription factors are major inducers of VEGFA overexpression (Fang et al., 2019). In our study, the gene expression of egln1, egln3, epo, ldh, and vegfa had an overall increase trend in response to hypoxia, semi-asphyxia, and asphyxia (Figure 8D), indicating that H. molitrix may utilize “HIF-1” to obtain more oxygen to maintain active functions under varying degrees of hypoxia stress.
This study showed that varying degrees of hypoxia induce the tissue damage of gill in silver carp. The H&E sections and electron microscopy examinations of gills indicated that the swelling and shedding of the epithelial cell layer in the gill tissue were intensified, the gill lamellae were seriously twisted, and gill filaments were dehisced with the degree of hypoxia intensified. The present study is the first to use a transcriptome to reveal the response to hypoxia in the gill of H. molitrix. In total, 587, 725, and 748 DEGs were identified in hypoxia, semi-asphyxia, and asphyxia groups, respectively. Further analysis illustrated that c2, c3, c6, klf4, cxcr4, cd45, and cd40 were upregulated as well as complement and coagulation cascades, NOD-like receptor signaling cascade, and Th1 and Th2 cell differentiation were enriched, which involved immune defense in the silver carp exposed to varying degrees of hypoxia. Moreover, egln1, egln3, epo, ldh, and vegfa were upregulated, and HIF-1, MAPK, and PI3K-Akt signaling pathways were enriched, which involved oxygen transport in the response of silver carp to hypoxia. These findings suggest that hypoxia stress may activate the H. molitrix immune defense system and that the HIF-1 signaling pathway is employed to increase oxygen transport in hypoxic situations. The results of this investigation will provide light on the molecular processes driving hypoxia responses in hypoxia-sensitive fish.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA812079.
The animal study was reviewed and approved by the animal care regulations of the Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences.
XHL: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft preparation, Funding acquisition. CL: Investigation, Formal analysis. QW and CF: Investigation, Data curation. XZL: Methodology. HS: Software. GH: Validation. GZ: Project administration, Resources, Funding acquisition. HL: Conceptualization, Supervision, Writing – review and editing, Funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (NO. 2018YFD0900302); the Foundation of Hubei Hongshan Laboratory (NO. 2021hskf014); State Key Laboratory of Developmental Biology of Freshwater Fish (NO. 2021KF005); the Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2020TD33 and NO. YFI202211); and the Earmarked Fund for China Agriculture Research System of MOF and MARA (NO. CARS-45).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.900200/full#supplementary-material
Supplementary Figure 1 | Quantified results of the gill filament cracking proportion in the CK, T1, T2, and T3 groups (mean ± SD, n = 9). *, P < 0.05.
Abdel-Tawwab M., Monier M. N., Hoseinifar S. H., Faggio C. (2019). Fish Response to Hypoxia Stress: Growth, Physiological, and Immunological Biomarkers. Fish. Physiol. Biochem. 45 (3), 997–1013. doi: 10.1007/s10695-019-00614-9
Anders S., Pyl P. T., Huber W. (2015). HTSeq—A Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics 31 (2), 166–169. doi: 10.1093/bioinformatics/btu638
Benjamini Y., Yekutieli D. (2001). The Control of the False Discovery Rate in Multiple Testing Under Dependency. Ann. Stat 29 (4), 1165–1188. doi: 10.1214/aos/1013699998
Berends E. T., Kuipers A., Ravesloot M. M., Urbanus R. T., Rooijakkers S. H. (2014). Bacteria Under Stress by Complement and Coagulation. FEMS Microbiol. Rev. 38 (6), 1146–1171. doi: 10.1111/1574-6976.12080
Cella M., Scheidegger D., Palmer-Lehmann K., Lane P., Lanzavecchia A., Alber G. (1996). Ligation of CD40 on Dendritic Cells Triggers Production of High Levels of Interleukin-12 and Enhances T Cell Stimulatory Capacity: TT Help via APC Activation. J. Exp. Med. 184 (2), 747–752. doi: 10.1084/jem.184.2.747
Cheng C.-H., Ma H.-L., Su Y.-L., Deng Y.-Q., Feng J., Xie J.-W., et al. (2019). Ammonia Toxicity in the Mud Crab (Scylla Paramamosain): The Mechanistic Insight From Physiology to Transcriptome Analysis. Ecotoxicol. Environ. Saf. 179, 9–16. doi: 10.1016/j.ecoenv.2019.04.033
Chen B.-X., Yi S.-K., Wang W.-F., He Y., Huang Y., Gao Z.-X., et al. (2017). Transcriptome Comparison Reveals Insights Into Muscle Response to Hypoxia in Blunt Snout Bream (Megalobrama Amblycephala). Gene 624, 6–13. doi: 10.1016/j.gene.2017.04.023
Chitnis T., Salama A. D., Grusby M. J., Sayegh M. H., Khoury S. J. (2004). Defining Th1 and Th2 Immune Responses in a Reciprocal Cytokine Environment In Vivo. J. Immunol. 172 (7), 4260–4265. doi: 10.4049/jimmunol.172.7.4260
Cui J., Tong R., Xu J., Tian Y., Pan J., Wang N., et al. (2021). Association Between STAT4 Gene Polymorphism and Type 2 Diabetes Risk in Chinese Han Population. BMC Med. Genomics 14 (1), 1–16. doi: 10.1186/s12920-021-01000-2
Dos Santos N. M., Taverne-Thiele J., Barnes A. C., van Muiswinkel W. B., Ellis A. E., Rombout J. H. (2001). The Gill Is a Major Organ for Antibody Secreting Cell Production Following Direct Immersion of Sea Bass (Dicentrarchus Labrax, L.) in a Photobacterium Damselae Ssp. Piscicida Bacterin: An Ontogenetic Study. Fish. Shellfish. Immunol. 11 (1), 65–74. doi: 10.1006/fsim.2000.0295
Endemann G., Stanton L. W., Madden K. S., Bryant C. M., White R. T., Protter A. A. (1993). CD36 Is a Receptor for Oxidized Low Density Lipoprotein. J. Biol. Chem. 268 (16), 11811–11816. doi: 10.1016/S0021-9258(19)50272-1
Evans D. H., Piermarini P. M., Choe K. P. (2005). The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol. Rev. 85 (1), 97–177. doi: 10.1152/physrev.00050.2003
Ewart K. V., Belanger J. C., Williams J., Karakach T., Penny S., Tsoi S. C., et al. (2005). Identification of Genes Differentially Expressed in Atlantic Salmon (Salmo Salar) in Response to Infection by Aeromonas Salmonicida Using cDNA Microarray Technology. Dev. Comp. Immunol. 29 (4), 333–347. doi: 10.1016/j.dci.2004.08.004
Fang Y., Davies P. F. (2012). Site-Specific microRNA-92a Regulation of Krüppel-Like Factors 4 and 2 in Atherosusceptible Endothelium. Arteriosclerosis. Thrombosis. Vasc. Biol. 32 (4), 979–987. doi: 10.1161/ATVBAHA.111.244053
Fang P., Zhang L., Zhang X., Yu J., Sun J., Jiang Q.-a., et al. (2019). Apatinib Mesylate in the Treatment of Advanced Progressed Lung Adenocarcinoma Patients With EGFR-TKI Resistance—A Multicenter Randomized Trial. Sci. Rep. 9 (1), 1–8. doi: 10.1038/s41598-019-50350-6
Farhan M., Wang H., Gaur U., Little P. J., Xu J., Zheng W. (2017). FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 13 (7), 815. doi: 10.7150/ijbs.20052
Feng C., Li X., Sha H., Luo X., Zou G., Liang H. (2022). Comparative Transcriptome Analysis Provides Novel Insights Into the Molecular Mechanism of the Silver Carp (Hypophthalmichthys Molitrix) Brain in Response to Hypoxia Stress. Comp. Biochem. Physiol. Part D.: Genomics Proteomics 41, 100951. doi: 10.1016/j.cbd.2021.100951
Fisher T. S., Lira P. D., Stock J. L., Perregaux D. G., Brissette W. H., Ozolinš T. R., et al. (2009). Analysis of the Role of the HIF Hydroxylase Family Members in Erythropoiesis. Biochem. Biophys. Res. Commun. 388 (4), 683–688. doi: 10.1016/j.bbrc.2009.08.058
Gallage S., Katagiri T., Endo M., Futami K., Endo M., Maita M. (2016). Influence of Moderate Hypoxia on Vaccine Efficacy Against Vibrio Anguillarum in Oreochromis Niloticus (Nile Tilapia). Fish. Shellfish. Immunol. 51, 271–281. doi: 10.1016/j.fsi.2016.02.024
Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., et al. (2005). IRF-7 Is the Master Regulator of Type-I Interferon-Dependent Immune Responses. Nature 434 (7034), 772–777. doi: 10.1038/nature03464
Hou L., Chen S., Liu J., Guo J., Chen Z., Zhu Q., et al. (2019). Transcriptomic and Physiological Changes in Western Mosquitofish (Gambusia Affinis) After Exposure to Norgestrel. Ecotoxicol. Environ. Saf. 171, 579–586. doi: 10.1016/j.ecoenv.2018.12.053
Hu X.-J., Yang J., Xie X.-L., Lv F.-H., Cao Y.-H., Li W.-R., et al. (2019). The Genome Landscape of Tibetan Sheep Reveals Adaptive Introgression From Argali and the History of Early Human Settlements on the Qinghai–Tibetan Plateau. Mol. Biol. Evol. 36 (2), 283–303. doi: 10.1093/molbev/msy208
Jørgensen H., Sørensen P., Cooper G., Lorenzen E., Lorenzen N., Hansen M., et al. (2011). General and Family-Specific Gene Expression Responses to Viral Hemorrhagic Septicaemia Virus Infection in Rainbow Trout (Oncorhynchus Mykiss). Mol. Immunol. 48 (8), 1046–1058. doi: 10.1016/j.molimm.2011.01.014
Jiang B.-H., Jiang G., Zheng J. Z., Lu Z., Hunter T., Vogt P. K. (2001). Phosphatidylinositol 3-Kinase Signaling Controls Levels of Hypoxia-Inducible Factor 1. Cell Growth Differ. 12 (7), 363–370.
Jiang J.-L., Mao M.-G., Lü H.-Q., Wen S.-H., Sun M.-L., Liu R.-t., et al. (2017). Digital Gene Expression Analysis of Takifugu Rubripes Brain After Acute Hypoxia Exposure Using Next-Generation Sequencing. Comp. Biochem. Physiol. Part D.: Genomics Proteomics 24, 12–18. doi: 10.1016/j.cbd.2017.05.003
Jiang M., Yang H., Peng R., Han Q., Jiang X. (2020). 1h NMR-Based Metabolomic Analysis of Cuttlefish, SepiaLarkiaC (Ehrenberg 1831) Exposed to Hypoxia Stresses and Post-Anoxia Recovery. Sci. Total. Environ. 726, 138317. doi: 10.1016/j.scitotenv.2020.138317
Kemper C., Pangburn M. K., Fishelson Z. (2014). Complement Nomenclature 2014. Mol. Immunol. 61 (2), 56–58. doi: 10.1016/j.molimm.2014.07.004
Khansari A. R., Balasch J. C., Vallejos-Vidal E., Parra D., Reyes-López F. E., Tort L. (2018). Comparative Immune-and Stress-Related Transcript Response Induced by Air Exposure and Vibrio Anguillarum Bacterin in Rainbow Trout (Oncorhynchus Mykiss) and Gilthead Seabream (Sparus Aurata) Mucosal Surfaces. Front. Immunol. 9, 856. doi: 10.3389/fimmu.2018.00856
Kidd P. (2003). Th1/Th2 Balance: The Hypothesis, Its Limitations, and Implications for Health and Disease. Altern. Med. Rev. 8 (3), 223–246.
Li B., Dewey C. N. (2011). RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf. 12 (1), 1–16. doi: 10.1186/1471-2105-12-323
Li H. L., Lin H. R., Xia J. H. (2017). Differential Gene Expression Profiles and Alternative Isoform Regulations in Gill of Nile Tilapia in Response to Acute Hypoxia. Marine. Biotechnol. 19 (6), 551–562. doi: 10.1007/s10126-017-9774-4
Li X., Li F., Zou G., Feng C., Sha H., Liu S., et al. (2021). Physiological Responses and Molecular Strategies in Heart of Silver Carp (Hypophthalmichthys Molitrix) Under Hypoxia and Reoxygenation. Comp. Biochem. Physiol. Part D.: Genomics Proteomics 40, 100908. doi: 1016/j.cbd.2021.100908
Li X., Ma J., Lei W., Li J., Zhang Y., Li Y. (2013). Cloning of Cytochrome P450 3A137 Complementary DNA in Silver Carp and Expression Induction by Ionic Liquid. Chemosphere 92 (9), 1238–1244. doi: 10.1016/j.chemosphere.2013.04.055
Li X., Ma J., Li Y. (2016). Molecular Cloning and Expression Determination of P38 MAPK From the Liver and Kidney of Silver Carp. J. Biochem. Mol. Toxicol. 30 (5), 224–231. doi: 10.1002/jbt.21781
Li C., Wang J., Chen J., Schneider K., Veettil R. K., Elmer K. R., et al. (2020). Native Bighead Carp Hypophthalmichthys Nobilis and Silver Carp Hypophthalmichthys Molitrix Populations in the Pearl River Are Threatened by Yangtze River Introductions as Revealed by Mitochondrial DNA. J. Fish. Biol. 96 (3), 651–662. doi: 10.1111/jfb.14253
Lin Y., Lin Y., Lin X., Sun X., Luo K. (2019). Combination of PET and CXCR4-Targeted Peptide Molecule Agents for Noninvasive Tumor Monitoring. J. Cancer 10 (15), 3420–3426. doi: 10.7150/jca.31087
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2– ΔΔCT Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu H., Forbes R. A., Verma A. (2002). Hypoxia-Inducible Factor 1 Activation by Aerobic Glycolysis Implicates the Warburg Effect in Carcinogenesis. J. Biol. Chem. 277 (26), 23111–23115. doi: 10.1074/jbc.M202487200
Ma Y., Li B., Ke Y., Zhang Y., Zhang Y. (2018). Transcriptome Analysis of Rana Chensinensis Liver Under Trichlorfon Stress. Ecotoxicol. Environ. Saf. 147, 487–493. doi: 10.1016/j.ecoenv.2017.09.016
Marques I. J., Leito J. T., Spaink H. P., Testerink J., Jaspers R. T., Witte F., et al. (2008). Transcriptome Analysis of the Response to Chronic Constant Hypoxia in Zebrafish Hearts. J. Comp. Physiol. B. 178 (1), 77–92. doi: 10.1007/s00360-007-0201-4
Matey V., Iftikar F. I., De Boeck G., Scott G. R., Sloman K. A., Almeida-Val V. M., et al. (2011). Gill Morphology and Acute Hypoxia: Responses of Mitochondria-Rich, Pavement, and Mucous Cells in the AmazonianLark (Astronotus Ocellatus) and the Rainbow Trout (Oncorhynchus Mykiss), Two Species With Very Different Approaches to the Osmo-Respiratory Compromise. Can. J. Zool. 89 (4), 307–324. doi: 10.1139/z11-002
Matey V., Richards J. G., Wang Y., Wood C. M., Rogers J., Davies R., et al. (2008). The Effect of Hypoxia on Gill Morphology and Ionoregulatory Status in the Lake Qinghai Scaleless Carp, Gymnocypris Przewalskii. J. Exp. Biol. 211 (7), 1063–1074. doi: 10.1242/jeb.010181
Meng X., Shen Y., Wang S., Xu X., Dang Y., Zhang M., et al. (2019). Complement Component 3 (C3): An Important Role in Grass Carp (Ctenopharyngodon 17Larki) Experimentally Exposed to Aeromonas Hydrophila. Fish. Shellfish. Immunol. 88, 189–197. doi: 10.1016/j.fsi.2019.02.061
Mu Y., Li W., Wei Z., He L., Zhang W., Chen X. (2020). Transcriptome Analysis Reveals Molecular Strategies in Gills and Heart of Large Yellow Croaker (Larimichthys Crocea) Under Hypoxia Stress. Fish. Shellfish. Immunol. 104, 304–313. doi: 10.1016/j.fsi.2020.06.028
Ohno K., Sato Y., Ohshima K.-i., Takata K., Miyata-Takata T., Takeuchi M., et al. (2015). A Subset of Ocular Adnexal Marginal Zone Lymphomas may Arise in Association With IgG4-Related Disease. Sci. Rep. 5 (1), 1–10. doi: 10.1038/srep13539
Overturf K., LaPatra S. (2006). Quantitative Expression (Walbaum) of Immunological Factors in Rainbow Trout, Oncorhynchus Mykiss (Walbaum), After Infection With Either Flavobacterium Psychrophilum, Aeromonas Salmonicida, or Infectious Haematopoietic Necrosis Virus. J. Fish. Dis. 29 (4), 215–224. doi: 10.1111/j.1365-2761.2006.00707.x
Petersen S. V., Thiel S., Jensen L., Vorup-Jensen T., Koch C., Jensenius J. C. (2000). Control of the Classical and the MBL Pathway of Complement Activation. Mol. Immunol. 37 (14), 803–811. doi: 10.1016/S0161-5890(01)00004-9
Press C. M., Evensen Ø. (1999). The Morphology of the Immune System in Teleost Fishes. Fish. Shellfish. Immunol. 9 (4), 309–318. doi: 10.1006/fsim.1998.0181
Rebl A., Korytář T., Köbis J. M., Verleih M., Krasnov A., Jaros J., et al. (2014). Transcriptome Profiling Reveals Insight Into Distinct Immune Responses to Aeromonas Salmonicida in Gill of Two Rainbow Trout Strains. Marine. Biotechnol. 16 (3), 333–348. doi: 10.1007/s10126-013-9552-x
Ren X., Deng R., Zhang K., Sun Y., Teng X., Li J. (2018). SpliceRCA: In Situ Single-Cell Analysis of mRNA Splicing Variants. ACS Cent. Sci. 4 (6), 680–687. doi: 10.1021/acscentsci.8b00081
Reyes-Moreno C., Girouard J., Lapointe R., Darveau A., Mourad W. (2004). CD40/CD40 Homodimers Are Required for CD40-Induced Phosphatidylinositol 3-Kinase-Dependent Expression of B7. 2 by Human B Lymphocytes. J. Biol. Chem. 279 (9), 7799–7806. doi: 10.1074/jbc.M313168200
Schofield C. J., Ratcliffe P. J. (2004). Oxygen Sensing by HIF Hydroxylases. Nat. Rev. Mol. Cell Biol. 5 (5), 343–354. doi: 10.1038/nrm1366
Shen Y., Zhang J., Xu X., Fu J., Li J. (2013). A New Haplotype Variability in Complement C6 Is Marginally Associated With Resistance to Aeromonas Hydrophila in Grass Carp. Fish. Shellfish. Immunol. 34 (5), 1360–1365. doi: 10.1016/j.fsi.2013.02.011
Sollid J., De Angelis P., Gundersen K., Nilsson G. (2003). Hypoxia Induces Adaptive and Reversible Gross Morphological Changes in Crucian Carp Gills. J. Exp. Biol. 206 (20), 3667–3673. doi: 10.1242/jeb.00594
Sollid J., Kjernsli A., De Angelis P. M., Røhr A. K., Nilsson G. E. (2005). Cell Proliferation and Gill Morphology in Anoxic Crucian Carp. Am. J. Physiology-Regulatory. Integr. Comp. Physiol. 289 (4), R1196–R1201. doi: 10.1152/ajpregu.00267.2005
Sollid J., Nilsson G. E. (2006). Plasticity of Respiratory Structures—Adaptive Remodeling of Fish Gills Induced by Ambient Oxygen and Temperature. Respir. Physiol. Neurobiol. 154 (1-2), 241–251. doi: 10.1016/j.resp.2006.02.006
Sun S., Xuan F., Fu H., Ge X., Zhu J., Qiao H., et al. (2016). Comparative Proteomic Study of the Response to Hypoxia in the Muscle of Oriental River Prawn (Macrobrachium Nipponense). J. Proteomics 138, 115–123. doi: 10.1016/j.jprot.2016.02.023
Tiedke J., Borner J., Beeck H., Kwiatkowski M., Schmidt H., Thiel R., et al. (2015). Evaluating the Hypoxia Response of Ruffe and Flounder Gills by a Combined Proteome and Transcriptome Approach. PloS One 10 (8), e0135911. doi: 10.1371/journal.pone.0135911
Van der Meer D. L., van den Thillart G. E., Witte F., de Bakker M. A., Besser J., Richardson M. K., et al. (2005). Gene Expression Profiling of the Long-Term Adaptive Response to Hypoxia in the Gills of Adult Zebrafish. Am. J. Physiology-Regulatory. Integr. Comp. Physiol. 289 (5), R1512–R1519. doi: 10.1152/ajpregu.00089.2005
Viale-Bouroncle S., Felthaus O., Schmalz G., Brockhoff G., Reichert T. E., Morsczeck C. (2012). The Transcription Factor DLX3 Regulates the Osteogenic Differentiation of Human Dental Follicle Precursor Cells. Stem Cells Dev. 21 (11), 1936–1947. doi: 10.1089/scd.2011.0422
Wu C.-B., Liu Z.-Y., Li F.-G., Chen J., Jiang X.-Y., Zou S.-M. (2017). Gill Remodeling in Response to Hypoxia and Temperature Occurs in the Hypoxia Sensitive Blunt Snout Bream (Megalobrama Amblycephala). Aquaculture 479, 479–486. doi: 10.1016/j.aquaculture.2017.06.020
Xia J. H., Li H. L., Li B. J., Gu X. H., Lin H. R. (2018). Acute Hypoxia Stress Induced Abundant Differential Expression Genes and Alternative Splicing Events in Heart of Tilapia. Gene 639, 52–61. doi: 10.1016/j.gene.2017.10.002
Xiao W. (2015). The Hypoxia Signaling Pathway and Hypoxic Adaptation in Fishes. Sci. China Life Sci. 58 (2), 148–155. doi: 10.1007/s11427-015-4801-z
Xing H., Chen J., Peng M., Wang Z., Liu F., Li S., et al. (2019). Identification of Signal Pathways for Immunotoxicity in the Spleen of Common Carp Exposed to Chlorpyrifos. Ecotoxicol. Environ. Saf. 182, 109464. doi: 10.1016/j.ecoenv.2019.109464
Yearbook C. F. S. (2020). China Fishery Statistical Yearbook Vol. 2020 (Beijing, China: China Agriculture Press), 24–34.
Zhang J., Alcaide P., Liu L., Sun J., He A., Luscinskas F. W., et al. (2011). Regulation of Endothelial Cell Adhesion Molecule Expression by Mast Cells, Macrophages, and Neutrophils. PloS One 6 (1), e14525. doi: 10.1371/journal.pone.0014525
Zhang B., Chamba Y., Shang P., Wang Z., Ma J., Wang L., et al. (2017). Comparative Transcriptomic and Proteomic Analyses Provide Insights Into the Key Genes Involved in High-Altitude Adaptation in the Tibetan Pig. Sci. Rep. 7 (1), 1–11. doi: 10.1038/s41598-017-03976-3
Zhang Z., Ju Z., Wells M. C., Walter R. B. (2009). Genomic Approaches in the Identification of Hypoxia Biomarkers in Model Fish Species. J. Exp. Marine. Biol. Ecol. 381, S180–S187. doi: 10.1016/j.jembe.2009.07.021
Zhang L., Song Z., Zhong S., Gan J., Liang H., Yu Y., et al. (2022). Acute Hypoxia and Reoxygenation Induces Oxidative Stress, Glycometabolism, and Oxygen Transport Change in Red Swamp Crayfish (ProcambarusLarkia): Application of Transcriptome Profiling in Assessment of Hypoxia. Aquacult. Rep. 23, 101029. doi: 10.1016/j.aqrep.2022.101029
Zhang Y., Wang L., Dey S., Alnaeeli M., Suresh S., Rogers H., et al. (2014). Erythropoietin Action in Stress Response, Tissue Maintenance and Metabolism. Int. J. Mol. Sci. 15 (6), 10296–10333. doi: 10.3390/ijms150610296
Keywords: Hypophthalmichthys molitrix, hypoxia, gill, tissue damage, transcriptome
Citation: Li X, Ling C, Wang Q, Feng C, Luo X, Sha H, He G, Zou G and Liang H (2022) Hypoxia Stress Induces Tissue Damage, Immune Defense, and Oxygen Transport Change in Gill of Silver Carp (Hypophthalmichthys molitrix): Evaluation on Hypoxia by Using Transcriptomics. Front. Mar. Sci. 9:900200. doi: 10.3389/fmars.2022.900200
Received: 20 March 2022; Accepted: 15 April 2022;
Published: 16 May 2022.
Edited by:
Hongsheng Yang, Institute of Oceanology (CAS), ChinaReviewed by:
Shuming Zou, Shanghai Ocean University, ChinaCopyright © 2022 Li, Ling, Wang, Feng, Luo, Sha, He, Zou and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guiwei Zou, em91Z3dAeWZpLmFjLmNu; Hongwei Liang, bGlhbmdod0B5ZmkuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.