- 1CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3CAS Engineering Laboratory for Marine Ranching, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 4University of Chinese Academy of Sciences, Beijing, China
- 5Shandong Province Key Laboratory of Experimental Marine Biology, Qingdao, China
- 6Department of Fish Production, Faculty of Agriculture, Al-Azhar University, Cairo, Egypt
- 7College of Food Science and Engineering, Ocean University of China, Qingdao, China
- 8Processes Design and Development Department, Egyptian Petroleum Research Institute, Cairo, Egypt
- 9The Innovation of Seed Design, Chinese Academy of Sciences, Wuhan, China
Microorganisms colonize plastics in the aquatic environment but their composition on plastics used in aquaculture remains poorly studied. Microorganisms play a significant role in aquaculture in terms of water quality and the health of cultivated species. In the current study, we explored the composition of microorganisms on floating plastics and their surrounding water collected from ponds and open aquaculture areas. Using scanning electron microscopy, the diversity of microbial communities, primarily diatoms, and bacteria were identified on the plastic surfaces. Additionally, epifluorescence microscopy revealed that prokaryotes were colonized on all plastic samples from 0.1 to 29.27×103 cells/cm2, with a high abundance found in open aquaculture areas compared to ponds. Bacterial communities were characterized by 16S rRNA sequencing which showed that bacterial communities on plastics were dominated by Proteobacteria, Cyanobacteria, Bacteroidetes, and Actinobacteria. The level of these microbial communities on the plastics differed from those found in the surrounding seawater samples and the abundance of potentially pathogenic bacteria was higher in plastics than in seawater samples. Moreover, hydrocarbon-degrading bacteria were more abundant in the investigated plastic samples than in the water samples. This study contributes to the knowledge regarding the plastisphere community in aquaculture.
Introduction
Plastics are semisynthetic or synthetic materials made of natural products, such as crude oil, natural gas, cellulose, and coal. Because of their flexibility, plastics have been used in many applications, especially packaging (Lambert and Wagner, 2018). Plastic production has grown rapidly since the 1950s compared to other human-made materials and has even replaced metals and wood (Geyer et al., 2017). Thus, plastic production has increased 189 times to reach 330 million metric tons in 2016 (Lebreton and Andrady, 2019). Nevertheless, plastic production may reach 20% of the petroleum used worldwide and 15% of the annual carbon emission budget (Lebreton and Andrady, 2019). With inadequate waste management, human activities have led to the accumulation of a considerable volume of plastics in the marine environment. In 2010, it was estimated that between 4.8 and 12.7 million tons of plastic litter entered the ocean from 192 coastal cities, accounting for 1.8–4.7 % of the plastic generated globally that year (Jambeck et al., 2015) and this number is predicted to increase in the following decade (Wu et al., 2017).
Plastics can last for a long time in the marine environment and thus present an artificial substrate for microbial colonialization. Microorganisms, such as bacteria, algae, and fungi aggregate on plastics through producing polymeric substances to adhere to each other and the surface forming a biofilm, known as the plastisphere (Amaral-Zettler et al., 2020). It was reported that groups of Rhodobacteraceae Flavobacteriaceae, Alteromonadaceae, and Cyclobacteriaceae are highly abundant on plastics collected from the marine ecosystem (Zettler et al., 2013; De Tender et al., 2015; Oberbeckmann et al., 2016; Xu et al., 2019; Vaksmaa et al., 2021). Microorganisms grown on plastics vary from those in the surrounding water, sediment, and organic particles (Zettler et al., 2013; De Tender et al., 2015). Nevertheless, microorganisms grown on plastics are influenced by different geographic areas (Oberbeckmann et al., 2016) and, to some extent, by polymer type (Basili et al., 2020). Furthermore, plastics in the ocean carry harmful microorganisms, including members of the genus Vibrio and other potentially pathogenic microorganisms such as members of Camplylobacteraceae, Enterobacteriaceae, Pseudomonadaceae, and Shewanellaceae (Zettler et al., 2013; Amaral-Zettler et al., 2020; Zhang et al., 2021). However, the majority of research on the plastisphere that has employed high-throughput DNA sequencing has focused on plastic samples from Europe, with a few focusing on samples from Asia and Africa (Amaral-Zettler et al., 2020) as well as aquaculture (Wen et al., 2020).
Aquaculture, which is one of the fastest expanding segments of the food industry (FAO, 2020), makes extensive use of plastics due to their positive application in management and packaging (Mahapatra et al., 2011). Aquaculture systems vary greatly worldwide depending on species and region. In mariculture systems, plastics are used to keep the structures floating and are fixed in a place using ropes. For cages, plastics are used from small to high-scale facilities for ropes, nets, and buoys. In ponds, plastics are used in pond linings, ropes, floats, and fish feeders. Furthermore, plastics are generally used in the aquaculture process for packaging, feed, transportation, and in the daily life of farmers, such as cups, bags, and bottles (Lusher et al., 2017). Plastic materials from aquaculture facilities may be discarded, lost, washed ashore, or accumulated on the seafloor posing hazards for animals, fishers, and boat traffic (Andréfouët et al., 2014; Bendell, 2015). Additionally, the breakdown of these materials can lead to the formation of microplastics, which could have a further impact on the marine ecosystem. Furthermore, these plastics present a substrate for microorganism colonization as a result of nutrient accumulation and waste (Cole et al., 2009; He et al., 2022), which may increase their longevity (Carson et al., 2013; Viršek et al., 2017), affect their buoyancy (Lobelle and Cunliffe, 2011), potentially degrade them (Oberbeckmann et al., 2016), and host potential pathogens (Radisic et al., 2020; Bhagwat et al., 2021). For instance, fish pathogens (Aeromonas salmonicida) were found to be attached in higher numbers to plastics than to stainless steel used in aquaculture (Carballo et al., 2000). Additionally, nylon and copper nets employed in aquaculture contain potential pathogens belonging to the Winogradskyella and Tenacibaculum taxa (Canada et al., 2020). Conversely, some bacteria found in aquaculture facilities’ biofilms play a critical role in the elimination of toxic metabolic wastes (Moriarty, 1997; King et al., 2004). Therefore, given the extensive use of plastics in aquaculture that can host microbial communities, which could play a critical role in aquaculture ecosystem, the plastisphere in aquaculture needs extensive investigation to determine their ecological effect on cultured species and ecosystem.
In the current study, we compared the microbial communities grown on plastics in two mariculture systems, ponds and marine ranching, in order to examine potential factors affecting the growth of the bacterial community in aquaculture, compare the levels of bacterial diversity, and identify potential pathogens and plastic-degrading bacteria. We employed high throughput 16S rRNA sequencing to identify bacterial populations growing on several kinds of plastic obtained from aquaculture systems.
Materials and Methods
Sampling Locations
Samples were collected during July 2021 from four aquaculture sites surrounding Shandong Province in the Yellow Sea and Bohai Sea of China (Figure S1). We sampled plastics from two aquaculture systems (ponds and marine ranching) located in different locations. Sites located in Laizhou (S2) and Weihai (S3) were closed aquaculture ponds, whereas sites located in Qingdao (S1) and Haiyang (S4) were open mariculture areas. These sites were used for farming sea cucumbers, mussels, and seaweed. Plastics associated with the aquaculture processes (i.e., ropes, raft balls, bottles, and bags) and surrounding water were collected from each site. At every site, six plastic items that differed in texture and color were collected. Plastics were floated in the water and exposed to sun. Using sterilized scissors, blades, and tweezers, submerged plastic parts were cut into small pieces (approximately 5–10 cm), washed with sterilized seawater, placed in 50 mL sterilized tubes, and preserved in an ice box containing dry ice (approximately -87°C) until reaching to the laboratory. Thereafter, plastics were preserved at -20°C and the water samples were preserved at 4°C until analysis.
Water Nutrients
Seawater samples were collected from each site for analysis. Salinity and temperature were measured using an YSI instrument. Additionally, nitrate (NO3-N), phosphate (PO4-P), nitrogen dioxide (NO2-N), and ammonia (NH3-N) were measured through colorimetric analysis using a QuaAAtro autoanalyzer (Seal Analytical, Norderstedt, Germany). Furthermore, nutrients of potassium (k), calcium (Ca), sodium (Na), magnesium (Mg), and strontium (Sr) were determined in the water using inductively coupled plasma–mass spectrometry (ICP–MS).
FT-IR Spectroscopy
Every plastic particle was analyzed using Fourier transform infrared (FT-IR) spectroscopy (Nicolet iS50 FT-IR). To provide knowledge on the chemical structure of the samples, the spectrum was compared with several libraries in the OMNIC software (Thermo Fisher Scientific, USA).
Scanning Electron Microscopy
Each plastic sample was immediately placed in an electron microscopy fixative solution and preserved at 4°C until analysis via scanning electron microscopy (SEM). The fixed samples were washed three times for a total of 15 minutes each time with 0.1 M phosphate buffer (pH 7.4). Thereafter, postfixation with 0.1 M (pH: 7.4) phosphate buffer (1% osmium acid) at room temperature was followed by three 15-minute rinses with 0.1 M phosphate buffer. Following that, the samples were dehydrated in a graded sequence of ethanol concentrations of 30%–50%–70%–80%–90%–95%–100%–100% for 15 minutes each time, and isoamyl acetate for 15 minutes. A critical point dryer was used to dry the samples after they were collected. Finally, SEM images were captured after samples were adhered to metallic stubs using carbon stickers and sputter-coated with gold for 30 s.
Procaryotic Cell Abundance on Plastics
To quantify the total prokaryotic abundance on plastic fragments by epifluorescence microscopy, we applied the acridine orange staining protocol as described previously, with a few adaptations (Luna et al., 2002). Acridine orange is a cell-permeable and cationic dye that intercalates with nucleic acids through electrostatic interactions. Each fragment was placed into a sterile 50 mL conical centrifuge tube and covered with 30 mL of filtered (0.2-µm pore size) 2% formalin solution buffered at a pH of 8.5 with a borate buffer and immediately fixed overnight at 4°С. Then, the samples were sonicated three times for 2 min each to release the bacterial cells from plastic samples. The suspension of the plastic fragments was diluted 100 times in prefiltered seawater. Thereafter, each sample was supplemented with acridine stock solution at a final concentration of 0.01% and incubated in the dark for 30 minutes at room temperature. The stained solution was washed 3 times with PBS buffer (pH 7.4) to remove the excess dye. Aliquots were filtered onto a black nucleopore polycarbonate (0.2 µm-pore-size). Finally, 10 µL of each filter was added to microscope slides and examined by epifluorescence microscopy. For each slide, at least 5 randomly selected microscope fields were examined, and the bacterial cells were enumerated and calculated as the mean value of cells abundance per each field.
High-Throughput Sequencing for Biodiversity and Community Composition
DNA Extraction and Illumina Sequencing
Following the manufacturer’s instructions, HiPure Soil DNA Kits (Guangzhou, China) were used to extract DNA from the samples. By utilizing the particular primer pairs of 341F (5’-CCTACGGGNGGCWGCAG-3’ and 806R (5’- GGACTACHVGTTTAAT -3’), the V3–V4 region of the 16S rRNA was amplified by PCR, resulting in a product length of ~466. The PCR amplifications were carried out with three replicates of a 50 μL of mixture containing 10 μL of 5 × Q5 reaction Buffer, 1.5 μL of 2.5 mM of dNTPs, 1.5 μL of each primer (10 μM), 0.2 μL of High-Fidelity DNA Polymerase, and 50 ng of DNA template (Biolabs, New England, USA). A two-minute denaturation step at 95°C was followed by 27 cycles of 98°C for 10 s, 62°C for 30 s, and 68°C for 30 s, with an elongation step of 10 min in the final PCR conditions.
An AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States) was used to extract and purify the amplified products from agarose gels (2%) and an ABI StepOnePlus Real-Time PCR System was used for quantification (Life Technologies, Foster City, USA). Equimolar purified amplicons were pooled and paired end sequenced (PE250) by Guangzhou Genedenovo Biotechnology Co., Ltd (Guangzhou, China) using the Illumina HiSeq 2500 platform (Illumina, Inc., San Diego, CA, USA). All of the raw reads were deposited into the NCBI Sequence Read Archive (SRA) database, bioproject PRJNA815345.
Quality Control and Clustering
FASTP (0.18.0) was used to eliminate reads containing more than 10% unknown nucleotides or fewer than 50% of bases with a quality (Q-value) > 20 from the raw data. FLASH (1.2.11) was then used to combine the pair-ended clean reads with a minimum overlap of 10 bp and error rates of 2%. To obtain clean tags of high quality, paired end clean readings were filtered according to the following conditions: 1) when the number of bases in a continuous poor-quality value (the default quality threshold is ≤3) surpasses the specified length (the default length is 3 bp), raw tags were separated from the first low quality base site; and 2) then, tags with a base length of less than 75% of the tag length were excluded. Next, UPARSE pipeline software was used to perform clustering based on the clean tags into operational taxonomic units (OTUs) of 97 % (9.2.64). The UCHIME method was used to eliminate all chimeric tags, resulting in effective tags. Within each cluster, the tag sequence with the greatest abundance was chosen as the representative sequence. A naive Bayesian model was used to classify typical OTU sequences into organisms, using the RDP classifier (version 2.2) and the SILVA database (version 132), with a confidence threshold of 0.8.
Data Analysis
Community distribution and environmental characteristics relationship was studied using R software and the Vegan package version 2.5.3 using redundancy analysis (RDA) (R core Team, 2020). Additionally, Welch’s t-test and Wilcoxon rank test were used to compare species between groups in the R project Vegan package (version 2.5.3). Furthermore, biomarker characteristics in each group were screened using LEfSe software (version 1.0) and R software (labdsv package version 2.0.1, pROC package version 1.10.0, and random forest package version 4.6.12). Alpha diversity analysis was conducted through Chao1, Shannon, and Simpson indexes, which were calculated in QIIME version 1.9.1. Alpha index comparisons between groups were calculated by Welch’s t-test and Wilcoxon rank test in the R project Vegan package (version 2.5.3). Analysis of the KEGG pathways of the OTUs was done using PICRUSt (version 2.1.4). BugBase was used to classify bacterial phenotypes in the microbiome. Welch’s t-test, Wilcoxon rank test, Kruskal-Wallis H test, and Tukey’s HSD were used in R project Vegan package to analyze function differences between groups (version 2.5.3).Where necessary, one-way ANOVA was employed to determine significant differences between samples.
Results
Water Nutrients
The association between the microbial community and environmental conditions was examined using RDA, including salinity; temperature; and the nutrients PO4-P, NO3-N, NO2-N, and NH3-H. Analysis showed that most phyla were located near salinity and phosphate except of Cyanobacteria, which was located near nitrate and nitrite vectors (Figure 1). The results suggest that salinity plays a key role in the abundance of the microbial community rather than other environmental factors. The deviation explained by RDA was 86.02% in composition at the phylum level. Marine ranching sites had significantly higher abundance of K, but ponds had higher concentrations of NO3 and NO2. Also, nutrients of Mg, and Na were significantly lower in the pond at S2 than at the other sites (Table S1).
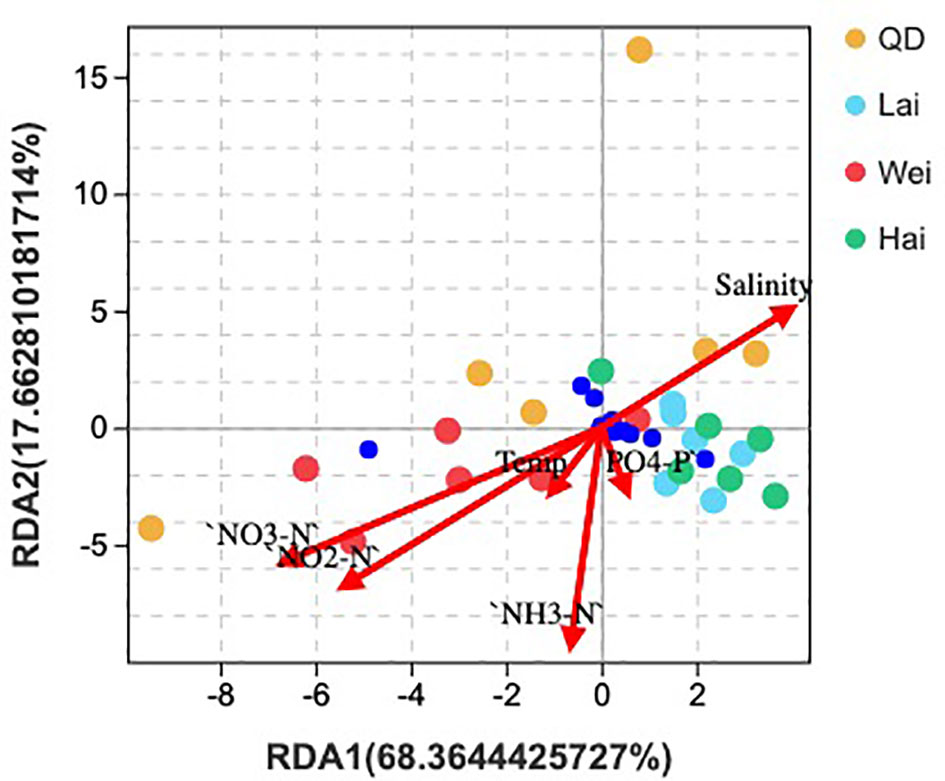
Figure 1 Redundancy analysis on the correlation between relative abundance of microbial community at phylum level and water quality parameters, including phosphate, nitrate, nitrite, ammonia, temperature, and salinity. QD, Qingdao; Lai, Laizhou; Wei, Weihai; Hai, Haiyang.
Scanning Electron Microscopy
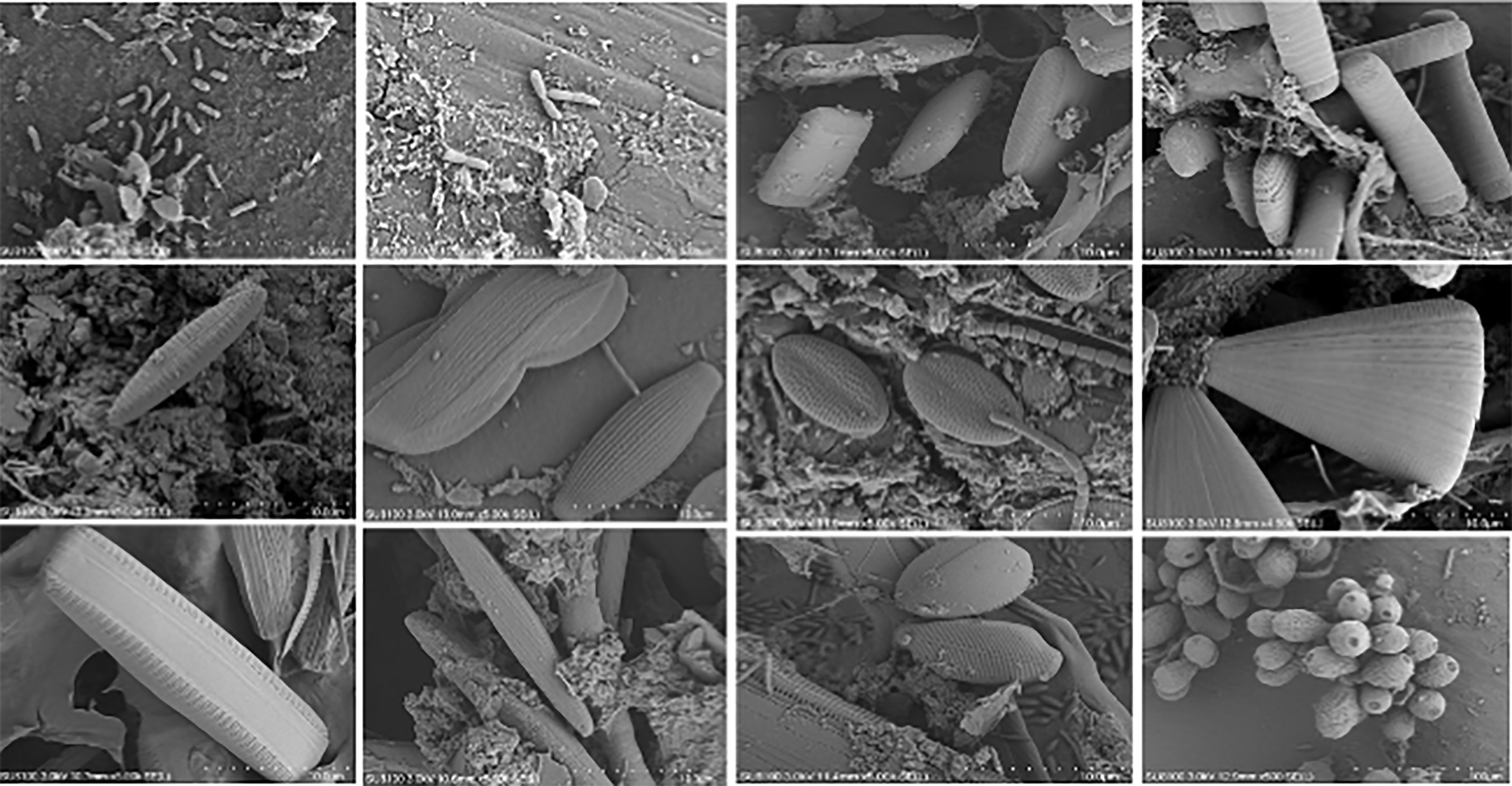
Procaryotic cells were found on plastics mostly with a rod shape. Additionally, pennate diatoms were highly abundant in most samples. Examples of diatom species identified morphologically using SEM include Amphora sp., Nitzschia sp., Navicula sp., Cocconeis sp., and Licmophora sp. (Figure 2). Furthermore, we noticed that the microorganism type on the plastics could be different according to the geographic area. For instance, rod-shapped prokaryotic cells were dominant in plastics collected from S1, whereas diatoms were dominant in S3.
Procaryotic Abundance on Plastics
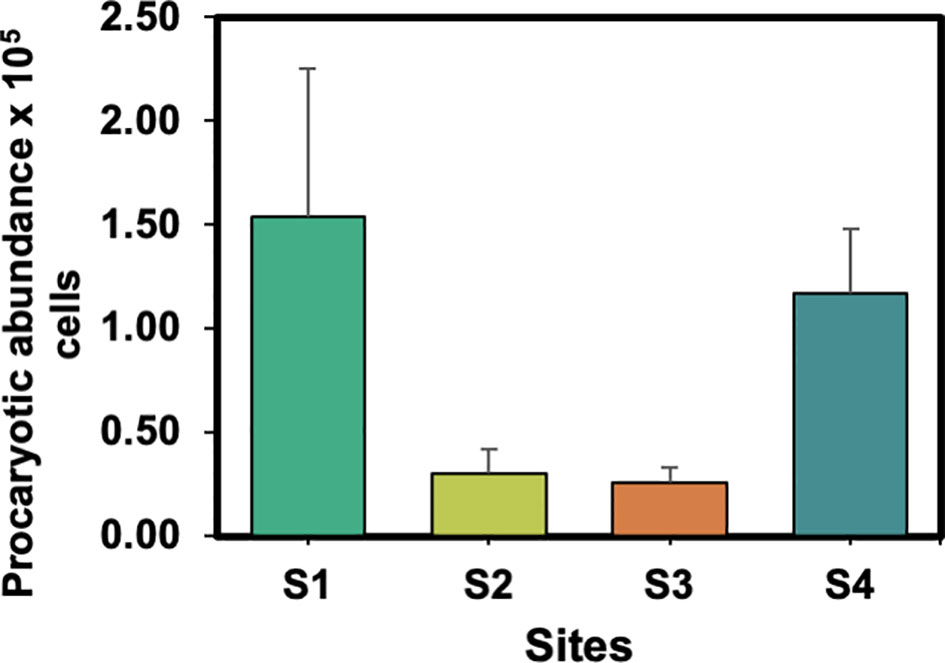
To count the net number of the prokaryotes in the collected samples, we applied the acridine fluorescence dye. The numbers of prokaryotic cells were estimated, assuming a range bacterial number of 0.05–5.07×105 cells/g, with an average of 0.82 ± 1.04 cells/g. When considering the length of the samples, the abundance of the cells was 0.1–29.27×103 cells/cm2, with an average of 2.52 ± 5.37×103 cells/cm2 (Figure 3). The highest abundance of cells was observed in samples of S1, while the lowest abundance of cells was observed in S3. No significant difference was found in the number of cells between sites (p > 0.05). Additionally, similar or different (i.e., PE vs. PVC) polymer type of plastics from different sites showed no significant difference in the number of cells (p > 0.05).
Comparing different site characteristics (i.e., ponds vs. marine ranching), we found a significantly higher number of cells in plastics samples collected from marine ranching (S1 and S4) than samples collected from ponds (S2 and S3; 1.35 ± 1.30×105 and 0.28 ± 0.23×105, respectively; p < 0.05), indicating that plastics in open aquaculture sites have higher abundance of procaryotic cells. Furthermore, we noticed high values of prokaryotic cells on plastics with rough surface and grooves, such as ropes and some aquaculture floats, indicating that the physical characteristics of plastic could determine the number of microorganisms.
Microbial Community Composition
16S high throughput sequencing revealed highly diverse of microbial communities. The effective tags reached a ratio of 85.33–91.96%, with an average of 87.61 ± 1.60% (Figure S2). The annotated OTUs in each sample were 848–2,584, with an average of 1,811 OTUs. There was no significant difference in OTU numbers among sites (p > 0.05). No significant differences in OTU numbers were found between different regions with the same polymer type (p > 0.05). Additionally, there were no significant differences in the OTU number of plastics with different characteristics (p > 0.05). Furthermore, we compared two polymer types (PE vs. PVC) and there were no significant differences between the two polymers in OTU number regardless of the location or plastic characteristics (p > 0.05).
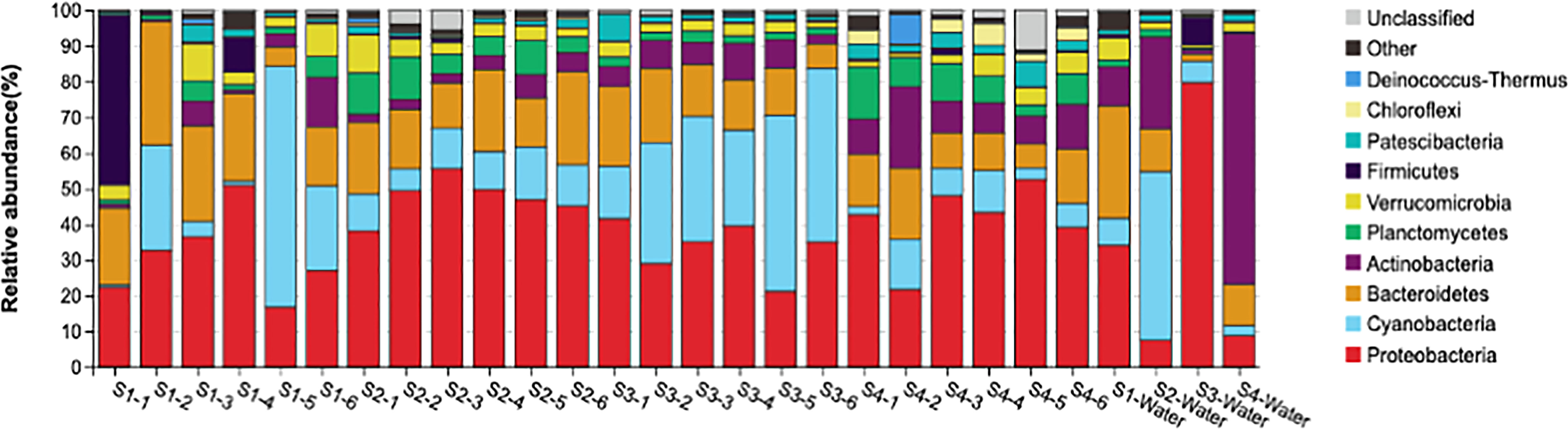
In the water and plastic samples, Proteobacteria had the highest abundance among all samples at the phylum level with relative abundances of 38.29 ± 10.93% and 32.44 ± 33.75%, respectively, which were significantly higher on plastics than seawater (Figure 4 and Figure S3). In the plastic samples, Cyanobacteria, Bacteroidetes, and Actinobacteria exhibited high relative abundances in the descending order of 18.60 ± 17.55%, 17.11 ± 7.14%, and 6.70 ± 5%, respectively, whereas, in water samples Actinobacteria, Cyanobacteria, and Bacteroidetes exhibited relative abundances in the order of 27.01 ± 30.58%, 15.92 ± 20.98%, and 14.31 ± 12.40%, respectively. At the class level, plastic samples had a high abundances of Alphaproteobacteria (31.94 ± 9.88%), Oxyphotobacteria (18.55 ± 17.59%), and Bacteroidia (15.87 ± 6.35%), whereas, Actinobacteria (23.33 ± 31.77%), Gammaproteobacteria (21.07 ± 35.13%), Oxyphotobacteria (15.91 ± 20.98%), and Bacteroidia (13.97 ± 12.43%) had the highest abundance in water samples. The most abundant families in plastic samples were Rhodobacteraceae (20.93 ± 9.50), Flavobacteriaceae (10.19 ± 5.50), and Sphingomonadaceae (4.56 ± 4.92), and the most abundant families in water samples were Microbacteriaceae (22.76 ± 31.78), Cyanobiaceae (12.16 ± 22.20), and Flavobacteriaceae (9.01 ± 11.28).
All the samples were identified as plastics, which included polyethylene (PE), polyethylene terephthalate (PET), polyethylene low density LDPE, polyvinyl chloride (PVC), polypropylene (PP), and ethylene vinyl acetate (Table S2). When comparing PVC (aquaculture floats) to PE (bags) samples from all sites, we found Cyanobacteria (33.87 ± 29.16%) in the PVC samples at a higher abundance than in the PE samples (7.03 ± 6.74%). Moreover, Bacteroidetes had a higher abundance on PE (20.45 ± 3.88%) than on PVC (8.68 ± 3.26%). Also, Firmicutes had a higher abundance on PE (12.15 ± 23.64%) than PVC (0.90 ± 0.80%) (Figure S4). According to the Chao1, Simpson and Shannon indexes, different polymer types (i.e., PE and PVC) had no significant difference in alpha diversity (Figure S5).
When considering different sites, 482 OTUs were shared among plastic samples from all sites, which were composed of/or dominated by Proteobacteria and Bacteroidetes phyla (Figure 5). However, there was no significant difference in the number of OTUs between the PE and PVC samples, and 1,186 OTUs were shared among them. Furthermore, PCoA analysis showed that S2 and S4 had distinct and overlapping OTU microbial assemblages from S1 or S3 (Figure 6).
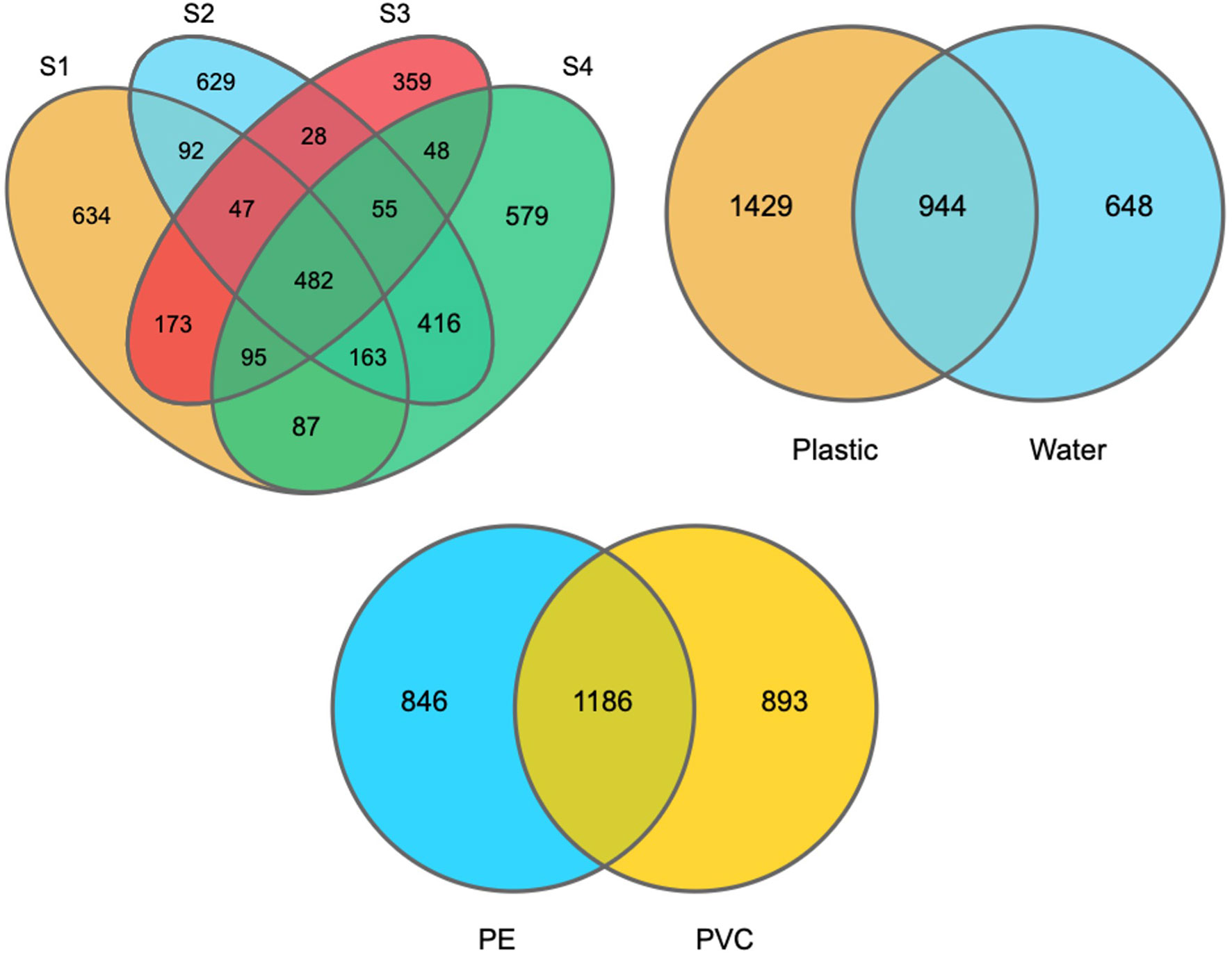
Figure 5 Venn diagram shows the overlapped OTUs between plastic samples from every site (a), plastic vs. water samples, and PE vs. PVC plastic samples.
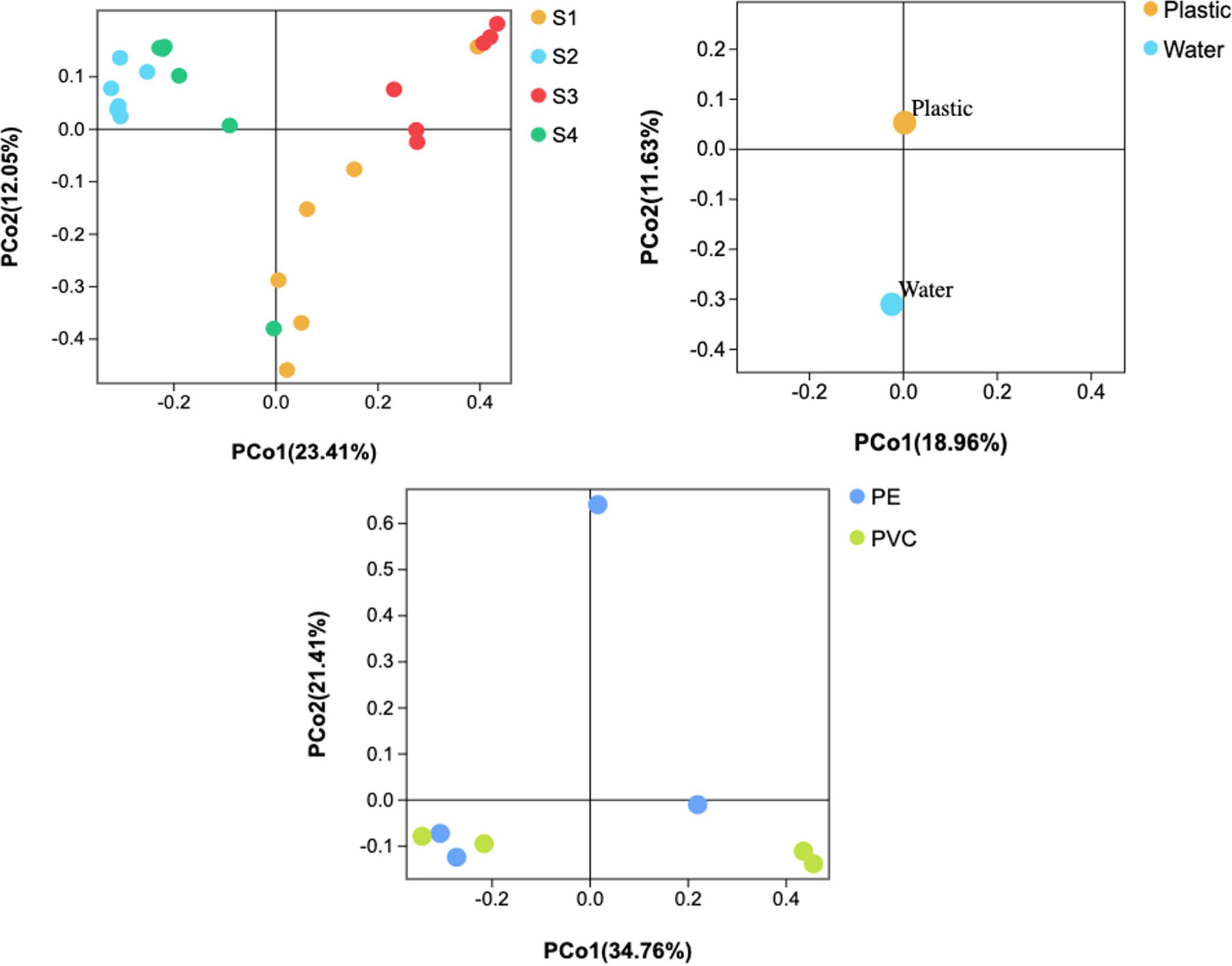
Figure 6 Principal coordinate analysis (PCoA) of microbial communities based on the 16S rRNA sequencing profiles.
DNA sequencing showed that there was a difference between the plastisphere community and surrounding water. For instance, a higher number of unique OTUs were found on plastic samples (1,429 OTUs) than in water samples (648 OTUs), suggesting variance in the number of unique species between plastics and seawater. Additionally, Chao1, Shannon, and Simpson diversity indices showed that higher richness of the microbial community was detected on plastic than in seawater (Figure S5). Furthermore, PCoA analysis showed that water samples had distinct microbial OTU assemblages from most plastic samples (Figure 6).
Bacterial Community Functions
Similar functional types for bacteria were found between plastics and water samples (Figure S7). However, there was a significant difference between all functional composition between plastics and water (Wilcoxon rank test p < 0.05). The highest abundance function for bacteria was membrane transport function and the lowest abundant function was immune disease.
Potential Pathogens
A total of 6,151 OTUs were matched with potentially pathogenic bacteria, with higher abundance on plastics than water (Figure S8). The most abundant phyla on plastics and water were Proteobacteria, Firmicutes, Patescibacteria, Bacteroidetes, and Acidobacteria. For the class of potentially pathogenic bacteria, the most abundant classes in the water were Gammaproteobacteria (21.069 ± 35.13%), Bacilli (1.927 ± 3.62%), Bacteroidia (1.696± 1.32%), and Alphaproteobacteria (1.290 ± 1.18%). In plastic samples, the most abundant classes were Alphaproteobacteria (6.55 ± 4.83%) Gammaproteobacteria (4.66 ± 3.43%), Bacilli (2.14 ± 9.59%), Parcubacteria (1.32 ± 1.77%), and Deltaproteobacteria (1.14 ± 0.76%). The most abundant orders of potentially pathogenic bacteria in the water were Oceanospirillales (14.517 ± 26.96%), Pseudomonadales (4.533 ± 8.78%), and Bacillales (1.927 ± 3.62%). In plastic samples, the abundant orders were Rhodobacterales (3.45 ± 3.56%), Rhizobiales (2.54 ± 1.84%), and Bacillales (2.14 ± 9.58%). Furthermore, the most abundant families in the water samples were Halomonadaceae (13.73 ± 27.45%), Moraxellaceae (4.52 ± 8.77%), and Bacillaceae (1.71 ± 3.39%), whereas, in the plastic samples, the most abundant families were Rhodobacteraceae (3.45 ± 3.58%), Rhizobiaceae (2.07 ± 1.66%), and Planococcaceae (1.95 ± 9.13%). Additionally, the average abundances of Vibrionaceae, Enterobacteriaceae, Pseudomonadaceae, and Shewanellaceae, which are regarded as opportunistic pathogen families, were 0.14 ± 0.25%, 0.31 ± 1.07%, 0.01 ± 0.02% in plastic and 0.01 ± 0.03%, and in water 0.07 ± 0.08%, 0.06 ± 0.05%, 0.02 ± 0.02%, and 0.02 ± 0.03%, respectively. The most abundant genera in the water samples were Cobetia (13.63 ± 23.61%), Psychrobacter (4.42 ± 7.56%), and Bacillus (1.66 ± 2.86%). In plastic samples, the most abundant genera were Ruegeria (1.13 ± 2.39%), Pseudahrensia (0.30 ± 0.30%), Psychrobacter (0.26 ± 1%), and Granulosicoccus (0.26 ± 0.35%). For species abundance, Bacillus hwajinpoensis was the most abundant species in water (1.64 ± 3.26%), while in plastics, Marichromatium sp. (0.23 ± 1.07%) and Marinicella litoralis (0.15 ± 0.16%) were the most abundant species.
Plastic Degrading Bacteria
Various bacterial families that include members known as hydrocarbon degraders were identified, including Rhodobacteraceae, Flavobacteriaceae, Saprospiraceae, Alteromonadaceae, Sphingomonadaceae, Rhodospirillaceae, and Oscillatoriaceae. The most abundant families that include members known as hydrocarbon degraders were Rhodobacteraceae (20.93 ± 9.50%), Flavobacteriaceae (10.19 ± 5.50%), and Saprospiraceae (3.52 ± 4.35%; Figure 7). Among them, the relative abundance of Rhodobacteraceae was higher in plastics (20.93 ± 9.50%) than in the surrounding seawater (7.57 ± 7.10%; p < 0.05; Wilcoxon rank test and LEfSe analysis; Figure S3). Different genera that described previously as hydrocarbon degraders were found on plastics, including Erythrobacter, Lewinella, Winogradskyella, Persicirhabdus, Altererythrobacter, Alcanivorax, Crocinitomix, Alteromonas, Hyphomonas, Oleibacter, Dokdonia, Tenacibaculum, Owenweeksia, and Marinobacter (Figure 7). Among these genera, Erythrobacter was the most abundant genus on plastics (2.10 ± 3.39%; p < 0.05; Wilcoxon rank test), which was higher on plastics than in the surrounding water (0.41 ± 0.32%), constituting 5% of the Proteobacteria phylum, which followed by Lewinella (1.68 ± 4.36%), Winogradskyella (0.83 ± 1.30), Persicirhabdus (0.68 ± 1.13%), and Altererythrobacter (0.52 ± 0.43%).
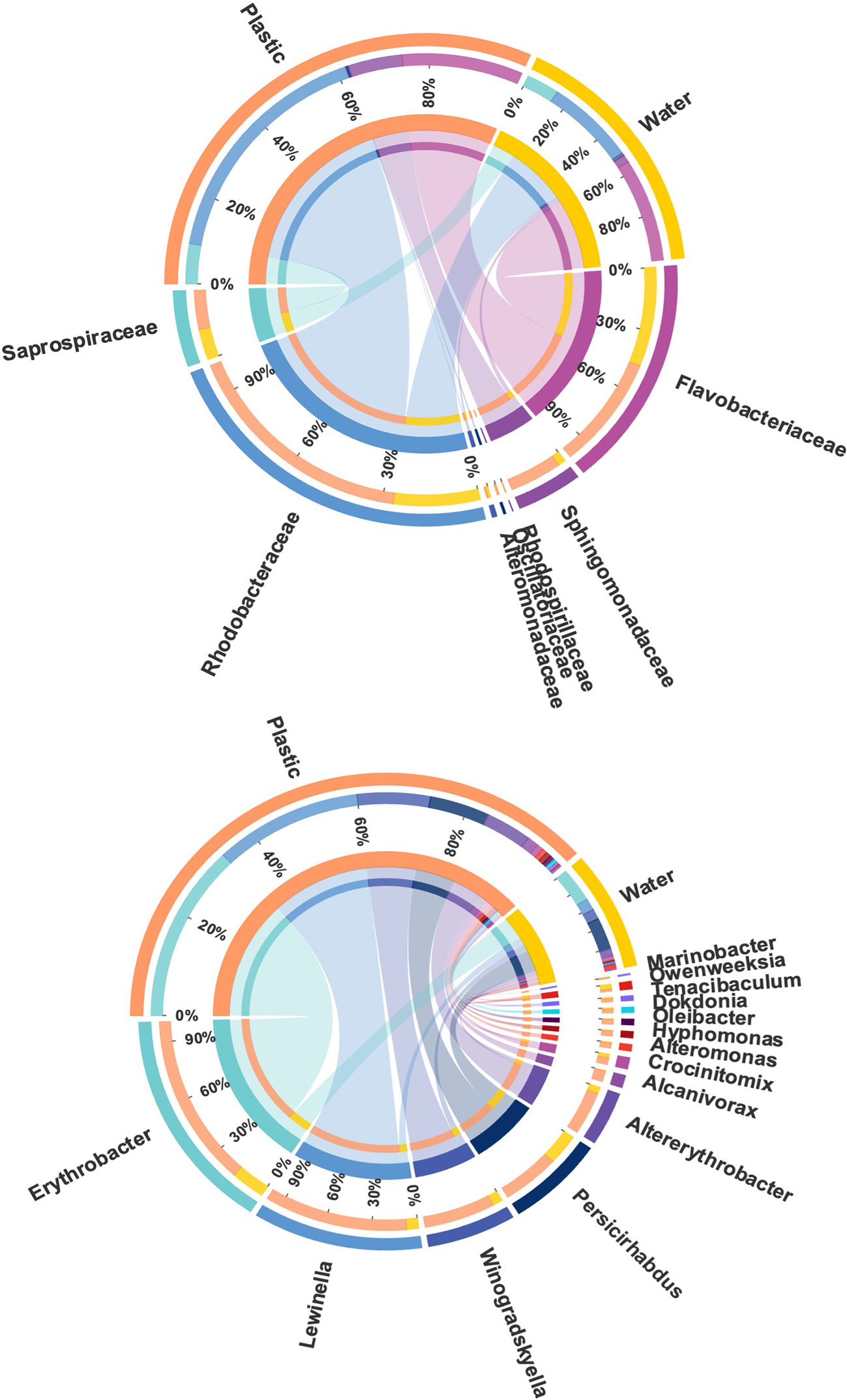
Figure 7 Circos diagram displays the composition relationship between different plastic degrading bacterial families and genera. The lines on both sides indicate the corresponding relationship pair, and the thicker the line, the greater the abundance value.
Discussion
Aquaculture extensively utilizes plastics due to their positive application in management and packaging (Mahapatra et al., 2011). These plastics present a substrate for microorganisms colonization, some of which are considered harmful or pathogenic (Amaral-Zettler et al., 2020). The abundance of organic matter, as well as phosphorus and nitrogenous metabolites, makes the aquaculture field suitable media for microbial growth (Martínez-Porchas and Vargas-Albores, 2017). These microorganisms play a significant role in aquaculture, maintaining the water quality, and the health of cultured species (Moriarty, 1997; Bentzon-Tilia et al., 2016). Therefore, understanding the role of plastics in microbial colonialization in aquaculture systems is critical for understanding the biological impact of plastics in these systems. Herein, we sampled plastics used in two aquaculture systems, i.e., ponds and marine ranching (i.e., open mariculture areas). The sampled plastics originated from aquaculture floats and nets used in suspended culture, as well as bottles and packaging bags.
The PE polymer was found to be the most prevalent component of the plastics tested. PE is the most common plastic type utilized in daily life and is primarly used in packaging, pipes, and containers. Additionally plastics, such as LDPE and high-density polyethylene (HDPE) are manufactured in vast quantities using PE (Lusher et al., 2017). Furthermore, PE is widely used in aquaculture in ropes and floats due to its low density (Andrady, 2011; Wu et al., 2020). In the current study, all the sampled plastics were floated on the surface of seawater which could explain the high abundance of PE.
SEM analysis showed that procaryotic cells in rod shape and pennate diatoms were dominantly abundant in the plastic samples. Diatoms play a key role in biofilm formation on plastics as they are the first colonizers of the surface (Eich et al., 2015; Zhao et al., 2021). Studies on the biofilm composition of plastics have showed that diatoms, such as coccolithophores and cyanobacteria, were the most abundant microorganisms on the floating plastics (Casabianca et al., 2019). Additionally, SEM analysis showed that diatoms were dominant in S3, whereas rod-shaped bacteria were dominant in S1. This could indicate that the characteristics of the area affect the type of microorganisms grown on plastics in aquaculture areas. Similarly, the microorganism communities differed significantly among locations (Oberbeckmann et al., 2016).
The average abundance of prokaryotic cells on plastics in the aquaculture areas was lower than procaryotic abundance on those collected from coastal areas impacted with anthropogenic pollution (Basili et al., 2020), but higher than that found on plastic particles collected from the Mediterranean Sea (Dussud et al., 2018). Considering different polymer types, there was no significant difference in the prokaryotic abundance between different polymers among sites. Similar results were observed in plastics collected from the Mediterranean Sea (Basili et al., 2020). However, the abundance of prokaryotic cells was significantly higher in marine ranching sites than in ponds. This might be because of nutrient differences between ponds and marine ranching sites. Marine ranching had a significantly higher abundances of K, Na, and Mg than ponds. A nutrient increase positively correlates with the amount of bacteria attached (Cowan et al., 1991; Donlan, 2002). Similarly, nutrients and salinity affect the growth rate of biofilm (Li et al., 2019; He et al., 2022). Furthermore, plastics with special physical characteristics, such as grooves and rough surface, were found to contain a high number of prokaryotic bacteria. Similarly, physical and chemical features of the substrate shape the microorganism community (Kirstein et al., 2018).
In the water samples, the microbial community was dominated by Proteobacteria, which was followed by Actinobacteria, Cyanobacteria, and Bacteroidetes. Similarly, microbial communities in the marine ranching area of Laoshan Bay was dominated by Proteobacteria, which was followed by Cyanobacteria, and Actinobacteriota (Fang et al., 2021). In aquaculture ponds of shrimp Litopenaeus vannamei in Zhongshan, China, Proteobacteria was dominant, followed by Tenericutes, and Bacteroidetes (Zhang et al., 2019). In another shrimp pond in Dongying, China, Bacteroidetes were the dominant in the water, followed by Proteobacteria, Actinobacteria, and Cyanobacteria (Huang et al., 2018).
The composition of plastics was dominated by Proteobacteria followed by Cyanobacteria, Bacteroidetes, and Actinobacteria. Similarly, Proteobacteria was the most abundant phylum identified on microplastics placed in aquaculture ponds for shrimp, followed by Bacteroidetes, Planctomycetes, and Cyanobacteria (Deng et al., 2021). Additionally, Proteobacteria was the dominant phylum identified on plastics collected from beaches in the Mediterranean Sea (Basili et al., 2020; Vaksmaa et al., 2021). Furthermore, Bacteroidetes, Actinobacteria, and Cyanobacteria were highly abundant among biofilm communities on plastics collected from beaches in the Mediterranean (Basili et al., 2020). In the Mediterranean Sea, Proteobacteria was followed by Bacteroidetes and Cyanobacteria (Vaksmaa et al., 2021). Moreover, plastic marine debris in the Mediterranean Sea was dominated by Cyanobacteria and Alphaproteobacteria (Dussud et al., 2018).
16S rRNA sequencing showed that there was a difference between the plastisphere community and that in the surrounding water. For instance, a higher number of unique OTUs was found on plastic samples (1,429 OTUs) than in water samples (648 OTUs), suggesting variance in the number of unique species between plastics and seawater (Figure 5). Additionally, we observed differences between the microbial community of plastics and surrounding water at the phylum, order, class, family, and genus levels (shown above). Furthermore, the Chao1 and Shannon diversity indexes showed that higher richness of the microbial community on plastic than in seawater (Figure S5). Moreover, PCoA analysis showed that water samples had distinct microbial OTU assemblages from most plastic samples (Figure 6). This indicates that plastic samples in aquaculture areas have a variable abundance of communities compared to seawater. This observation is in agreement with observations of biofilm on microplastics in aquaculture (Deng et al., 2021) and previous studies in the open ocean (De Tender et al., 2015; Dussud et al., 2018; Vaksmaa et al., 2021).
When comparing different polymer types (i.e., PVC vs. PE), we found Cyanobacteria had a higher abundance on PVC samples than PE samples, whereas Bacteroidetes and Firmicutes had a higher abundance on PE samples than PVC samples. This difference might not only be because of different polymer types but also because of the surface characteristics, such as polymer type and surface characteristics influence the type of microorganisms that attached (Amaral-Zettler et al., 2020). Alpha diversity indexes showed that there was no significant difference between the two polymer types and both shared high numbers of OTUs. Collectively, polymer type might influence the abundance of microbial groups on plastics (i.e., specific groups of microbes might prefer one polymer over another). This observation is in agreement with previous studies that concluded polymer type affects the bacterial community attached to plastics; there was a significant difference in the microbial community on PE compared to PP or PS (Vaksmaa et al., 2021).
When considering different sites, S2 and S4 had distinct and overlapping OTUs from S1 and S3. Additionally, only 482 OTUs were shared among sites. Furthermore, our SEM observation showed distinct microorganisms on plastics in S1 and S3 from S2 and S4. Collectively, this observation is in agreement with previous studies concluding that the microbial community structure differs according to the geographic area (Amaral-Zettler et al., 2015; Oberbeckmann et al., 2016; Basili et al., 2020). Furthermore, the membrane transport function was highly represented by the biofilm community on plastics compared to other predicted functions. This observation was reported previously from plastics marine debris collected from the western Mediterranean Sea (Dussud et al., 2018) and submerged plastic pellets in Australia (Bhagwat et al., 2021), which is an essential function for biofilm formation (Dussud et al., 2018).
It has been reported that plastic particles can accumulate pathogenic bacteria and harmful microalgae, which indicates that plastic particles may act as carriers of pathogenic bacteria, resulting in the spread of diseases (Amaral-Zettler et al., 2020; Meng et al., 2021). According to the finding of the current study, bacterial families identified include well-known fish and shellfish potential pathogenic strains, which support the potential for plastics to serve as vectors for possible pathogenic microbes. This might pose a threat to aquaculture profitability and cultured species. For instance, Rhodobacteraceae was the most abundant family on plastic samples, and is widely regarded as potential pathogenic bacteria (Meng et al., 2021). Some members of this family may contribute to shrimp, sea cucumber, and coral disease (Soffer et al., 2015; Zhang et al., 2019; Deng et al., 2021). Additionally, bacterial families commonly regarded as potential pathogens, such as Vibrionaceae, Enterobacteriaceae, Pseudomonadaceae, and Shewanellacea were higher on plastics than in water samples. The abundance of these families was lower than or comparable to those detected on plastics employed in the Yellow Sea (Zhang et al., 2021). Also, the family Vibrionaceae has been widely confirmed on marine plastics and is dominated by the Vibrio genus and a large number of potential pathogens (Amaral-Zettler et al., 2020). The Vibrio genus includes animal and human pathogens that have been responsible for catastrophic pandemics and innumerable epidemics around the world and could result in significant financial loss for aquaculture farms (Laverty et al., 2020). Taken together, our findings support the hypothesis that plastic acts as a vector for potential pathogens.
Conversely, some microbes were reported to have a positive impact on aquaculture management. For instance, some members of the family Rhodobacteraceae produce antibacterial compounds that can inhibit fish pathogens (Henriksen et al., 2022). Also, members of the family Rhizobiaceae were identified among the denitrifying bacteria in the recirculation aquaculture system (Chen et al., 2021). Additionally, the genus Ruegeria had the highest abundance on plastic samples, which is a common bacteria in the aquaculture system and was previously detected on microplastic particles (Zhang et al., 2020). This genus includes members that have probiotic potential due to inhibition of fish pathogens (Sonnenschein et al., 2017). Furthermore, Bacillus hwajinpoensis, which was the most abundant species in water samples, has been reported as a dominant species in aquaculture water and is regarded as probiotic to improve the water quality and inhabit pathogenic bacteria (Wei et al., 2021). Additionally, Marichromatium sp. was the most abundant species identified on plastics, which was reported to improve water quality (Zhu et al., 2019). We anticipate that the extent of the presence of harmful or beneficial bacteria in the aquaculture system may depend on the effective management of the water quality in the farm.
Microorganisms maybe able to provide solutions for plastic pollution through biodegradation (Amaral-Zettler et al., 2020). The Rhodobacteraceae family was found to be the most abundant family on the plastic samples and higher than of seawater. Members of this family are known as hydrocarbon degraders of which Rhodococcus ruber has been shown to degrade PE (Gilan et al., 2004; Dubinsky et al., 2013). Several studies have reported a high abundance of the family Rhodobacteraceae on plastic samples (Bryant et al., 2016; Dussud et al., 2018). Furthermore, we found 14 genera that were previously reported to include hydrocarbon degrading bacteria, of which Erythrobacter had the highest abundance, and these were previously detected on PE plastic samples (Vaksmaa et al., 2021).
Conclusion
In the current study, we investigated the microorganisms associated with plastics collected from aquaculture areas (i.e., ponds and marine ranching). Our findings indicated that the amount of bacterial community associated with plastics was significantly different in open aquaculture areas than in closed ponds, regardless of polymers type. Additionally, 16S rRNA gene sequencing and SEM analysis showed that the type of microbial communities differed among the aquaculture areas. Also, our results showed that plastic samples in aquaculture areas had a distinct abundance of the microbial community from seawater samples. Additionally, different polymers may influence the abundance of specific microbial communities. Furthermore, the abundance of potential pathogenic bacteria was higher on plastics than in seawater. However, the dominance of potential probiotic bacteria that have the potential to inhabit pathogens might explain the limited abundance of potential pathogens in the samples collected from aquaculture fields. Moreover, the high abundance of genera including hydrocarbon degrading bacteria indicated that these groups might play a role in plastic degradation. Further research could focus on manipulating and managing beneficial microbes on plastics to enhance the aquaculture management.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA815345].
Author Contributions
MM and CL study design and samples collection. HH samples analysis. MM and AA-Z manuscript writing and review. MM data analysis. HY fund and supervision. All authors revised and/or edited the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (32150410376, 42176106, 42030408), the Major scientific and technological innovation projects in Shandong Province (No. 2019JZZY010812), and the International Partners Program of the Chinese Academy of Sciences (133137KYSB20180069).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank professor Xiangzhao Mao for his guidance in sample analysis, Dr. Chen Tu for his advice in sample collection, and Chenxi Zhang for his help in sample collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.895611/full#supplementary-material
References
Amaral-Zettler L. A., Zettler E. R., Mincer T. J. (2020). Ecology of the Plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Amaral-Zettler L. A., Zettler E. R., Slikas B., Boyd G. D., Melvin D. W., Morrall C. E., et al. (2015). The Biogeography of the Plastisphere: Implications for Policy. Front. Ecol. Environ. 13 (10), 541–546. doi: 10.1890/150017
Andrady A. L. (2011). Microplastics in the Marine Environment. Mar. Pollut. Bull. 62, 1596–1605). doi: 10.1016/j.marpolbul.2011.05.030
Andréfouët S., Thomas Y., Lo C. (2014). Amount and Type of Derelict Gear From the Declining Black Pearl Oyster Aquaculture in Ahe Atoll Lagoon, French Polynesia. Mar. Pollut. Bull. 83 (1), 224–230. doi: 10.1016/j.marpolbul.2014.03.048
Basili M., Quero G. M., Giovannelli D., Manini E., Vignaroli C., Avio C. G., et al. (2020). Major Role of Surrounding Environment in Shaping Biofilm Community Composition on Marine Plastic Debris. Front. Marine Sci. 7. doi: 10.3389/fmars.2020.00262
Bendell L. I. (2015). Favored Use of Anti-Predator Netting (APN) Applied for the Farming of Clams Leads to Little Benefits to Industry While Increasing Nearshore Impacts and Plastics Pollution. Mar. Pollut. Bull. 91 (1), 22–28. doi: 10.1016/j.marpolbul.2014.12.043
Bentzon-Tilia M., Sonnenschein E. C., Gram L. (2016). Monitoring and Managing Microbes in Aquaculture – Towards a Sustainable Industry. Microbial Biotechnol. 9 (5), 576–584. doi: 10.1111/1751-7915.12392
Bhagwat G., Zhu Q., O’Connor W., Subashchandrabose S., Grainge I., Knight R., et al. (2021). Exploring the Composition and Functions of Plastic Microbiome Using Whole-Genome Sequencing. Environ. Sci. Technol. 55 (8), 4899–4913. doi: 10.1021/acs.est.0c07952
Bryant J. A., Clemente T. M., Viviani D. A., Fong A. A., Thomas K. A., Kemp P., et al. (2016). Diversity and Activity of Communities Inhabiting Plastic Debris in the North Pacific Gyre. MSystems 1 (3). doi: 10.1128/msystems.00024-16
Canada P., Pereira A., Nogueira N., Png-Gonzalez L., Andrade C., Xavier R. (2020). Analysis of Bacterial Microbiome Associated With Nylon and Copper Nets in an Aquaculture Context. Aquaculture 516, 734540. doi: 10.1016/j.aquaculture.2019.734540
Carballo J., Seoane R. M., Nieto T. P. (2000). Adhesion of Aeromonas Salmonicida to Materials Used in Aquaculture. Bull. Eur. Assoc. Fish Pathologists 20 (2), 77–82.
Carson H. S., Nerheim M. S., Carroll K. A., Eriksen M. (2013). The Plastic-Associated Microorganisms of the North Pacific Gyre. Mar. Pollut. Bull. 75 (1–2), 126–132. doi: 10.1016/j.marpolbul.2013.07.054
Casabianca S., Capellacci S., Giacobbe M. G., Dell’Aversano C., Tartaglione L., Varriale F., et al. (2019). Plastic-Associated Harmful Microalgal Assemblages in Marine Environment. Environ. Pollution 244, 617–626. doi: 10.1016/j.envpol.2018.09.110
Chen Z., Chang Z., Qiao L., Wang J., Yang L., Liu Y., et al. (2021). Nitrogen Removal Performance and Microbial Diversity of Bioreactor Packed With Cellulosic Carriers in Recirculating Aquaculture System. International Biodeterioration and Biodegradation. 157 105157. doi: 10.1016/j.ibiod.2020.105157
Cole D. W., Cole R., Gaydos S. J., Gray J., Hyland G., Jacques M. L., et al. (2009). Aquaculture: Environmental, Toxicological, and Health Issues. Int. J. Hygiene Environ. Health 212 (4), 369–377. doi: 10.1016/j.ijheh.2008.08.003
Cowan M. M., Warren T. M., Fletcher M. (1991). Mixed-Species Colonization of Solid Surfaces in Laboratory Biofilms. Biofouling 3 (1), 23–34. doi: 10.1080/08927019109378159
Deng H., Fu Q., Li D., Zhang Y., He J., Feng D., et al. (2021). Microplastic-Associated Biofilm in an Intensive Mariculture Pond: Temporal Dynamics of Microbial Communities, Extracellular Polymeric Substances and Impacts on Microplastics Properties. J. Clean. Prod. 319, 128774. doi: 10.1016/j.jclepro.2021.128774
De Tender C. A., Devriese L. I., Haegeman A., Maes S., Ruttink T., Dawyndt P. (2015). Bacterial Community Profiling of Plastic Litter in the Belgian Part of the North Sea. Environ. Sci. Technol. 49 (16), 9629–9638. doi: 10.1021/acs.est.5b01093
Donlan R. M. (2002). Biofilms: Microbial Life on Surfaces. Emerging Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063. Centers for Disease Control and Prevention.
Dubinsky E. A., Conrad M. E., Chakraborty R., Bill M., Borglin S. E., Hollibaugh J. T., et al. (2013). Succession of Hydrocarbon-Degrading Bacteria in the Aftermath of the Deepwater Horizon Oil Spill in the Gulf of Mexico. Environ. Sci. Technol. 47 (19), 10860–10867. doi: 10.1021/es401676y
Dussud C., Meistertzheim A. L., Conan P., Pujo-Pay M., George M., Fabre P., et al. (2018). Evidence of Niche Partitioning Among Bacteria Living on Plastics, Organic Particles and Surrounding Seawaters. Environ. Pollut. 236, 807–816. doi: 10.1016/j.envpol.2017.12.027
Eich A., Mildenberger T., Laforsch C., Weber M. (2015). Biofilm and Diatom Succession on Polyethylene (PE) and Biodegradable Plastic Bags in Two Marine Habitats: Early Signs of Degradation in the Pelagic and Benthic Zone? PLoS One 10 (9), e0137201. doi: 10.1371/journal.pone.0137201
Fang G., Yu H., Sheng H., Tang Y., Liang Z. (2021). Comparative Analysis of Microbial Communities Between Water and Sediment in Laoshan Bay Marine Ranching With Varied Aquaculture Activities. Mar. Pollut. Bull. 173, 112990. doi: 10.1016/j.marpolbul.2021.112990
FAO. (2020). The State of World Fisheries and Aquaculture 2020. Sustainability in action Rome. doi: 10.4060/ca9229en
Geyer R., Jambeck J. R., Law K. L. (2017). Production, Use, and Fate of All Plastics Ever Made. Sci. Adv. 3 (7):e1700782. doi: 10.1126/sciadv.1700782
Gilan I., Hadar Y., Sivan A. (2004). Colonization, Biofilm Formation and Biodegradation of Polyethylene by a Strain of Rhodococcus Ruber. Appl. Microbiol. Biotechnol. 65 (1), 97–104. doi: 10.1007/s00253-004-1584-8
He S., Jia M., Xiang Y., Song B., Xiong W., Cao J., et al. (2022). Biofilm on Microplastics in Aqueous Environment: Physicochemical Properties and Environmental Implications. J. Hazardous Materials 424, 127286. doi: 10.1016/j.jhazmat.2021.127286
Henriksen N. N. S. E., Lindqvist L. L., Wibowo M., Sonnenschein E. C., Bentzon-Tilia M., Gram L. (2022). Role is in the Eye of the Beholder—the Multiple Functions of the Antibacterial Compound Tropodithietic Acid Produced by Marine Rhodobacteraceae. FEMS Microbiol. Rev 46 (3), fuac007. doi: 10.1093/femsre/fuac007
Huang F., Pan L., Song M., Tian C., Gao S. (2018). Microbiota Assemblages of Water, Sediment, and Intestine and Their Associations With Environmental Factors and Shrimp Physiological Health. Appl. Microbiol. Biotechnol. 102 (19), 8585–8598. doi: 10.1007/s00253-018-9229-5
Jambeck J. R., Geyer R., Wilcox C., Siegler T. R., Perryman M., Andrady A., et al. (2015). Plastic Waste Inputs From Land Into the Ocean. Science 347 (6223), 768–771. doi: 10.1126/science.1260352
King R. K., Flick G. J., Pierson M. D., Smith S. A., Boardman G. D., Coale C. W. (2004). Identification of Bacterial Pathogens in Biofilms of Recirculating Aquaculture Systems. J. Aquat. Food Product Technol. 13 (1), 125–133. doi: 10.1300/J030v13n01_11
Kirstein I. V., Wichels A., Krohne G., Gerdts G. (2018). Mature Biofilm Communities on Synthetic Polymers in Seawater - Specific or General? Mar. Environ. Res., 142, 147–154. doi: 10.1016/j.marenvres.2018.09.028
Lambert S., Wagner M. (2018). Microplastics Are Contaminants of Emerging Concern On Freshwater Environments: An overview. Freshwater Microplastics 58, 1–23. doi: 10.1007/978-3-319-61615-5_1.
Laverty A. L., Primpke S., Lorenz C., Gerdts G., Dobbs F. C. (2020). Bacterial Biofilms Colonizing Plastics in Estuarine Waters, With an Emphasis on Vibrio Spp. And Their Antibacterial Resistance. PLoS One 15 (8 August), e0237704. doi: 10.1371/journal.pone.0237704
Lebreton L., Andrady A. (2019). Future Scenarios of Global Plastic Waste Generation and Disposal. Palgrave Commun. 5 (1), 1–11. doi: 10.1057/s41599-018-0212-7
Li W., Zhang Y., Wu N., Zhao Z., Xu W., Ma Y., et al. (2019). Colonization Characteristics of Bacterial Communities on Plastic Debris Influenced by Environmental Factors and Polymer Types in the Haihe Estuary of Bohai Bay, China. Environ. Sci. Technol. 53 (18), 10763–10773. doi: 10.1021/acs.est.9b03659
Lobelle D., Cunliffe M. (2011). Early Microbial Biofilm Formation on Marine Plastic Debris. Mar. Pollut. Bull. 62 (1), 197–200. doi: 10.1016/j.marpolbul.2010.10.013
Luna G. M., Manini E., Danovaro R. (2002). Large Fraction of Dead and Inactive Bacteria in Coastal Marine Sediments: Comparison of Protocols for Determination and Ecological Significance. Appl. Environ. Microbiol. 68 (7), 3509–3513. doi: 10.1128/AEM.68.7.3509-3513.2002
Lusher A., Hollman P., Mendoza-Hill J., FAO (2017) Status of Knowledge on Their Occurrence and Implications for Aquatic Organisms and Food Safety. Available at: http://oceanrep.geomar.de/49179/1/Microplastics in fisheries and aquaculture.pdf.
Mahapatra B. C., Sarkar B., Barik N. K., Jayasankar P. (2011) Application Of Plastics In Manufacture. Available at: http://krishi.icar.gov.in/jspui/bitstream/123456789/1840/1/.
Martínez-Porchas M., Vargas-Albores F. (2017). Microbial Metagenomics in Aquaculture: A Potential Tool for a Deeper Insight Into the Activity. Rev. Aquac. 9, 42–56. doi: 10.1111/raq.12102. John Wiley & Sons, Ltd.
Meng J., Zhang Q., Zheng Y., He G., Shi H. (2021). Plastic Waste as the Potential Carriers of Pathogens. Curr. Opin. Food Sci. 41, 224–230. doi: 10.1016/j.cofs.2021.04.016
Moriarty D. J. W. (1997). The Role of Microorganisms in Aquaculture Ponds. Aquaculture 151 (1–4), 333–349. doi: 10.1016/S0044-8486(96)01487-1
Oberbeckmann S., Osborn A. M., Duhaime M. B. (2016). Microbes on a Bottle: Substrate, Season and Geography Influence Community Composition of Microbes Colonizing Marine Plastic Debris. PLoS One 11 (8). doi: 10.1371/journal.pone.0159289
R Core Team. (2020). R: A Language and Environment for Statistical Computing. (Vienna, Austria:R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Radisic V., Nimje P. S., Bienfait A. M., Marathe N. P. (2020). Marine Plastics From Norwegian West Coast Carry Potentially Virulent Fish Pathogens and Opportunistic Human Pathogens Harboring New Variants of Antibiotic Resistance Genes. Microorganisms 8 (8), 1–13. doi: 10.3390/microorganisms8081200
Soffer N., Zaneveld J., Vega Thurber R. (2015). Phage-Bacteria Network Analysis and Its Implication for the Understanding of Coral Disease. Environ. Microbiol. 17 (4), 1203–1218. doi: 10.1111/1462-2920.12553
Sonnenschein E. C., Nielsen K. F., D’Alvise P., Porsby C. H., Melchiorsen J., Heilmann J., et al. (2017). Global Occurrence and Heterogeneity of the Roseobacter-Clade Species Ruegeria Mobilis. ISME J. 11 (2), 569–583. doi: 10.1038/ismej.2016.111
Vaksmaa A., Knittel K., Abdala Asbun A., Goudriaan M., Ellrott A., Witte H. J., et al. (2021). Microbial Communities on Plastic Polymers in the Mediterranean Sea. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.673553
Viršek M. K., Lovšin M. N., Koren Š., Kržan A., Peterlin M. (2017). Microplastics as a Vector for the Transport of the Bacterial Fish Pathogen Species Aeromonas Salmonicida. Mar. Pollut. Bull. 125 (1–2), 301–309. doi: 10.1016/j.marpolbul.2017.08.024
Wei Y., Bu J., Long H., Zhang X., Cai X., Huang A., et al. (2021). Community Structure of Protease-Producing Bacteria Cultivated From Aquaculture Systems: Potential Impact of a Tropical Environment. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.638129
Wen B., Liu J. H., Zhang Y., Zhang H. R., Gao J. Z., Chen Z. Z. (2020). Community Structure and Functional Diversity of the Plastisphere in Aquaculture Waters: Does Plastic Color Matter? Sci. Total Environ. 740, 140082. doi: 10.1016/j.scitotenv.2020.140082
Wu F., Wang Y., Leung J. Y. S., Huang W., Zeng J., Tang Y., et al. (2020). Accumulation of Microplastics in Typical Commercial Aquatic Species: A Case Study at a Productive Aquaculture Site in China. Sci. Total Environ. 708, 135432. doi: 10.1016/j.scitotenv.2019.135432
Wu W. M., Yang J., Criddle C. S. (2017). Microplastics Pollution and Reduction Strategies. Front. Environ. Sci. Eng. 11 (1). doi: 10.1007/s11783-017-0897-7
Xu X., Wang S., Gao F., Li J., Zheng L., Sun C., et al. (2019). Marine Microplastic-Associated Bacterial Community Succession in Response to Geography, Exposure Time, and Plastic Type in China’s Coastal Seawaters. Mar. Pollut. Bull. 145, 278–286. doi: 10.1016/j.marpolbul.2019.05.036
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 47 (13), 7137–7146. doi: 10.1021/es401288x
Zhang Y., Lu J., Wu J., Wang J., Luo Y. (2020). Potential Risks of Microplastics Combined With Superbugs: Enrichment of Antibiotic Resistant Bacteria on the Surface of Microplastics in Mariculture System. Ecotoxicol. Environ. Saf. 187, 109852. doi: 10.1016/j.ecoenv.2019.109852
Zhang M., Pan L., Huang F., Gao S., Su C., Zhang M., et al. (2019). Metagenomic Analysis of Composition, Function and Cycling Processes of Microbial Community in Water, Sediment and Effluent of Litopenaeus Vannamei Farming Environments Under Different Culture Modes. Aquaculture 506, 280–293. doi: 10.1016/j.aquaculture.2019.03.038
Zhang B., Yang X., Liu L., Chen L., Teng J., Zhu X., et al. (2021). Spatial and Seasonal Variations in Biofilm Formation on Microplastics in Coastal Waters. Sci. Total Environ. 770, 145303. doi: 10.1016/j.scitotenv.2021.145303
Zhang Z., Zhang W., Hu Z., Li C., Shao Y., Zhao X., et al. (2019). Environmental Factors Promote Pathogen-Induced Skin Ulceration Syndrome Outbreak by Readjusting the Hindgut Microbiome of Apostichopus Japonicus. Aquaculture 507, 155–163. doi: 10.1016/j.aquaculture.2019.03.054
Zhao S., Zettler E. R., Amaral-Zettler L. A., Mincer T. J. (2021). Microbial Carrying Capacity and Carbon Biomass of Plastic Marine Debris. ISME J. 15 (1), 67–77. doi: 10.1038/s41396-020-00756-2
Keywords: plastisphere, aquaculture plastic, microbial community, pathogens, plastic polymers
Citation: Mohsen M, Lin C, Hamouda HI, Al-Zayat AM and Yang H (2022) Plastic-Associated Microbial Communities in Aquaculture Areas. Front. Mar. Sci. 9:895611. doi: 10.3389/fmars.2022.895611
Received: 14 March 2022; Accepted: 12 May 2022;
Published: 04 July 2022.
Edited by:
Tanveer Adyel, Deakin University, AustraliaReviewed by:
Sedat Gundogdu, Çukurova University, TurkeyJun Sun, China University of Geosciences Wuhan, China
Copyright © 2022 Mohsen, Lin, Hamouda, Al-Zayat and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenggang Lin, bGluY2hlbmdnYW5nQHFkaW8uYWMuY24=
 Mohamed Mohsen
Mohamed Mohsen Chenggang Lin
Chenggang Lin Hamed I. Hamouda
Hamed I. Hamouda Ahmed M. Al-Zayat6
Ahmed M. Al-Zayat6 Hongsheng Yang
Hongsheng Yang