- 1Fujian Key Laboratory on Conservation and Sustainable Utilization of Marine Biodiversity, Fuzhou Institute of Oceanography, Minjiang University, Fuzhou, China
- 2College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
In this study, a new Cladonema species was identified in a laboratory aquarium in Fuzhou, China, and named Cladonema digitatum sp. nov. based on its morphological characteristics and DNA barcoding. It is distinct from other Cladonema medusae in having a manubrium with finger-like protuberances, radial canals with Y-shaped bifurcations, tentacles with 3–11 adhesive branches, and 3–7 stinging branches growing from the main branch as side branches. The validity of C. digitatum sp. nov. was supported by molecular phylogenetic analyses based on both mitochondrial cytochrome oxidase and mitochondrial 16S rRNA sequences. Similar to other Cladonema medusae, the adhesive and stinging branches of each tentacle, oral tentacle, manubrium, and gonads in C. digitatum displayed considerable phenotypic plasticity, thus making species identification based solely on morphology difficult. Although diagnostic characters such as filiform tentacles and medusa buds of hydroids and nematocysts are also useful for species identification in the genus Cladonema, related information is missing in some Cladonema species. Thus, information on the life cycle and DNA barcoding should be updated to describe new or cryptic species and to improve the taxonomy of the genus Cladonema.
Introduction
Hydromedusae play an important role in marine ecosystems (Colin et al., 2003; Tewksbury et al., 2014). In recent years, increasing jellyfish blooms have exacerbated the adverse ecological effects of medusae as competitors and predators in coastal marine ecosystems (Gravili et al., 2008; Miglietta et al., 2008). Cladonematidae is a family of anthoathecate hydrozoans containing three genera: Cladonema, Eleutheria, and Staurocladia (Ghory et al., 2020). Although Cladonema species are distributed in coastal waters worldwide (Chou and Huang, 1958; Xu, 1993; Schuchert, 2006; Gershwin and Zeidler, 2008; Cedeno-Posso, 2014), they are not frequently caught in plankton nets because they prefer to remain in benthic zones (Schuchert, 2006). Thus, the distribution, abundance, and biodiversity of Cladonema species are likely underestimated (Zhou et al., 2022). Cladonema medusae can also make a difference in local habitats when they gather in Yantai waters during the summer (Chou and Huang, 1958). Furthermore, Cladonema pacificum and Cladonema radiatum are used as model organisms in the fields of developmental (e.g., branching morphogenesis and eye development), regenerative, and evolutionary biology (Graziussi et al., 2012; Fujiki et al., 2019; Fujita et al., 2019; Fujita et al., 2021).
Cladonema medusae can be distinguished from other Cladonematidae by the presence of branched tentacles and two types of tentacle branches: (1) stinging branches, used for predation and self-defense with numerous bulbous nematocyst clusters and (2) adhesive branches, which end with a structure that can adhere to substrates (Schuchert, 2006; Farias et al., 2020). However, species identification is challenging in the genus Cladonema. Zhou et al. (2022) suggested several diagnostic characteristics of the genus, including the number of radial canals and adhesive branches, arrangement of stinging branches and gastric pouches in the manubrium during the medusa stage, existence of filiform tentacles, and the number and arrangement of medusa buds on the hydroids. The diagnostic characteristics often vary among individuals in the same population (Bouillon and Boero, 2000). Several rounds of lumping and splitting among Cladonema species, especially C. radiatum and its subspecies, have occurred because of the phenotypic plasticity of the medusae (Schuchert, 2006; Gershwin and Zeidler, 2008). Currently, the life cycles in only five Cladonema species have been investigated, increasing the difficulties in cryptic species identification in the genus Cladonema (Schuchert, 2006; Gershwin and Zeidler, 2008; Zhou et al., 2022). The use of mitochondrial cytochrome oxidase (COI) and mitochondrial 16S rRNA (16S) in DNA barcoding is an efficient and reliable tool for the identification of hydromedusae (Bucklin et al., 2010a; Bucklin et al., 2010b; Zheng et al., 2014; Schuchert, 2016) and detecting cryptic or new species (Lindner et al., 2011). The use of both morphological descriptions and molecular phylogenetic analyses may provide a more accurate way to describe new or cryptic hydromedusae (Zhou et al., 2013; He et al., 2015).
In this study, Cladonema digitatum sp. nov. was found in an Oryzias melastigma aquarium in our laboratory (Fuzhou, China) and was verified as a new Cladonema species through both morphological and molecular tests. In terms of morphological characteristics and molecular sequences, C. digitatum sp. nov. showed obvious differences from Cladonema multiramosum, another Cladonema species previously found in the same aquarium (Zhou et al., 2022). We further revised the taxonomy within the genus Cladonema and the phylogenetics in Cladonematidae.
Materials and Methods
Sample Collection
Cladonema species were found in an O. melastigma aquarium in Dr. Chen’s laboratory (Fuzhou, China). Polyps were collected, and artificial seawater was prepared to keep the polyps alive for morphology and life cycle observation. Individuals prepared for the molecular tests were purified for at least 72 h in sterilized artificial seawater.
Culturing and Morphological Description
The hydroids of Cladonema were kept in covered glass vessels (diameter: 125 mm) with a polyethylene Petri dish (diameter: 90 mm) placed at the bottom for hydroids to attach to. The culture conditions were maintained at room temperature (20–22°C) and 30–35 ppt salinity. The polyps were fed twice daily with Artemia sp. nauplii, and half of the seawater was changed after feeding to maintain a clean culture environment. Newly released medusae were collected and fed to maturity under the same culture conditions for morphological analysis. Before feeding, the hydroids and medusae were observed under a stereomicroscope (Leica S9D, Leica Microsystems, Wetzlar, Germany) to record their development.
Thirty hydroids and newly released and mature medusae were photographed and measured under a stereomicroscope (Leica M165FC, Leica Microsystems) after an anesthetic treatment with 10% MgSO4. Several hydroids and medusae were used to determine the type, size, and distribution of their nematocysts under a microscope (Leica DM2500, Leica Microsystems), following the methods of Östman (1987).
Photosensitivity Experiment
Newly released (1 to 2 days old), young (7–10 days old), and mature (25–30 days old) medusae were collected to test their reaction to light. The experiments were performed as follows: 50 medusae from each developmental stage were randomly collected in a culture dish. Each group was exposed to strong white light (light phase) for 30 s, then the light source was removed (dark phase) for 5 s, and the group was exposed to light (light phase) again for 5 s. The light-to-dark switch was performed four times, and the number of medusae bouncing up was recorded. For each group, three parallel control groups were used to avoid random errors. The data were analyzed using Prism 6.0. A two-way ANOVA was conducted to test the effect of the developmental stages and light-to-dark switches on the photosensitivity of medusae. A Tukey’s multiple-comparisons test was conducted when the effect was significant.
Molecular Analyses
The DNA was extracted from a pool of five medusae from a single colony using a TIANnamp Marine Animals DNA Kit (TIANGEN, Beijing, China), according to the manufacturer’s instructions. In total, five colonies were sampled. Before DNA extraction, the medusae were examined under a stereoscopic microscope, cultured without food, and cleaned in sterile seawater for at least 72 h. Partial mitochondrial 16S (~0.55 kb) and COI (~0.70 kb) fragments were amplified using 2×Ex Taq Mastermix (Takara Bio, Kyoto, Japan) and the primers which are listed in Table 1. The PCR system and program were as described by Zhou et al. (2022). The 550- and 700-bp amplicons were visualized using 2% agarose gel electrophoresis (130 V, 30 min) and purified with a DNA Purification Kit (Omega, Georgia, USA). The target fragments were ligated into the pMD-19T vector (Takara Bio) and transformed into Escherichia coli JM109 competent cells (Takara Bio). E. coli was uniformly plated on Luria–Bertani agar plates containing 50 µl/ml ampicillin and incubated at 37°C overnight. Colonies containing target DNA fragments were selected by colony PCR using programs and primers as previously described and then sent to Sangon Biotech Co. Ltd. (Shanghai, China) for sequencing using an ABI 3730 automatic DNA sequencer with the universal M13 primer and two sequences per fragment. All samples were sequenced in both directions to ensure sequence accuracy.
The sequences were examined based on chromatogram files. Target sequences were obtained using COI and 16S primers, aligned using the NCBI Nucleotide BLAST program to confirm their accuracy and validity, and deposited in GenBank (Table 1). Pairwise genetic distances between COI and 16S fragments in Cladonematidae species were calculated using MEGA X with the Kimura-2-Parameter (K2P) model. The normality and homogeneity of the data were tested, and the Kruskal–Wallis test was used to assess differences in the genetic distances among the different levels using SPSS 16.0. The sequences of the Cladonematidae species were used to establish the phylogenetic trees using the maximum likelihood (ML, the GTR + I model for COI, and the GTR + I + R model for 16S) and neighbor-joining (NJ, based on the K2P model) method using MEGA X (Tamura et al., 2011) with 1,000 bootstrap replicates. The Corynidae species Coryne eximia (AY512541 and AJ878713) and Coryne pusilla (AY512552 and AY787874) and Sarsia tubulosa (GQ395327 and EU876548) were chosen as outgroups for the phylogenetic tree of 16S (Nawrocki et al., 2010), whereas Coryne eximia (KT981902 and KT981909) was chosen as an outgroup for the phylogenetic tree of COI.
Results
Systematics
Phylum Cnidaria Hatschek, 1888
Class Hydrozoa Owen, 1843
Subclass Hydroidolina Collins, 2000
Order Anthoathecata Cornelius, 1992
Suborder Capitata Kühn, 1913
Family Cladonematidae Gegenbaur, 1857
Genus Cladonema Dujardin, 1843
Cladonema digitatum sp. nov.
Material Examined
Holotype: MJU-HYD-6, male medusa, diameter: 2.92 mm, height: 2.56 mm.wParatypes: MJU-HYD-6, 30 mature medusae (19 male and 11 female individuals), diameter: 2.22–3.75 mm, height: 1.62–3.17 mm; MJU-HYD-7, 30 newly released medusae, with a diameter of 0.67–1.08 mm and a height of 0.60–0.98 mm; MJU-HYD-8, polyps with trophosomes; MJU-HYD-9, polyps with immature medusa buds.
All specimens were sampled from individuals kept in the laboratory in October 2021 and were deposited at the Institute of Oceanography, College of Geography and Oceanography, Minjiang University, Fuzhou, China.
Diagnosis
Polyp: Hydranth with one oral whorl of 3–6 capitate tentacles and one aboral whorl of 2–4 filiform tentacles; medusa buds naked, one to two per polyp, born on the hydranth body between capitate and filiform tentacles
Adult medusa: umbrella bell-shaped with an apical projection on top; manubrium with 4–7 finger-like protuberances and able to extend beyond the umbrella margin; with gonads around the upper manubrium; mouth with 3–6 bulbous nematocyst clusters; radial canals, simply straight or some with a Y-shaped bifurcation; tentacles 7–9, each bearing 3–7 stinging branches and 3–11 adhesive branches; stinging branches growing from the main branch as side branches. The tentacle bases were bent and cylindrical, each with a black abaxial ocellus.
Description
Hydroid
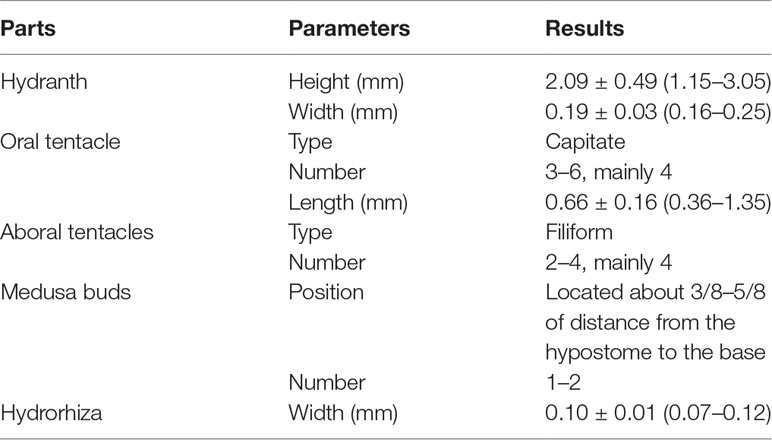
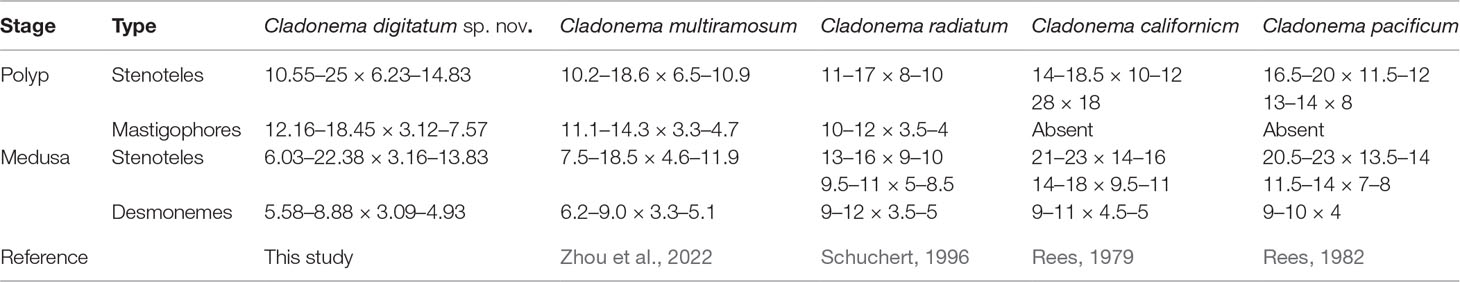
The morphological characteristics of the hydroids are shown in Figure 1 and Table 2. Hydroids are stolonal, are rarely branched (Figure 1D), and originate from ramified stolons. The hydranth was naked and fluctuant in size (1.15–3.05 mm in height and 0.16–0.25 mm in width). The hydrocaulus and stolons were covered with a smooth peri-arc. A hydranth with one oral whorl had 3–6 capitate tentacles (mainly four, 0.36–1.35 mm in length under anesthesia), with bulbous nematocyst clusters at the end. One aboral whorl of 2–4 (mainly four) slender filiform tentacles was located approximately 1/2–5/7 of the distance from the hyposome to the base. The medusa buds were bare, with 1 to 2 buds at different developmental stages per hydranth (up to three), located approximately 3/8–5/8 of the distance from the hypostome to the base (Figures 1B, C). The nematocysts can be described as follows:
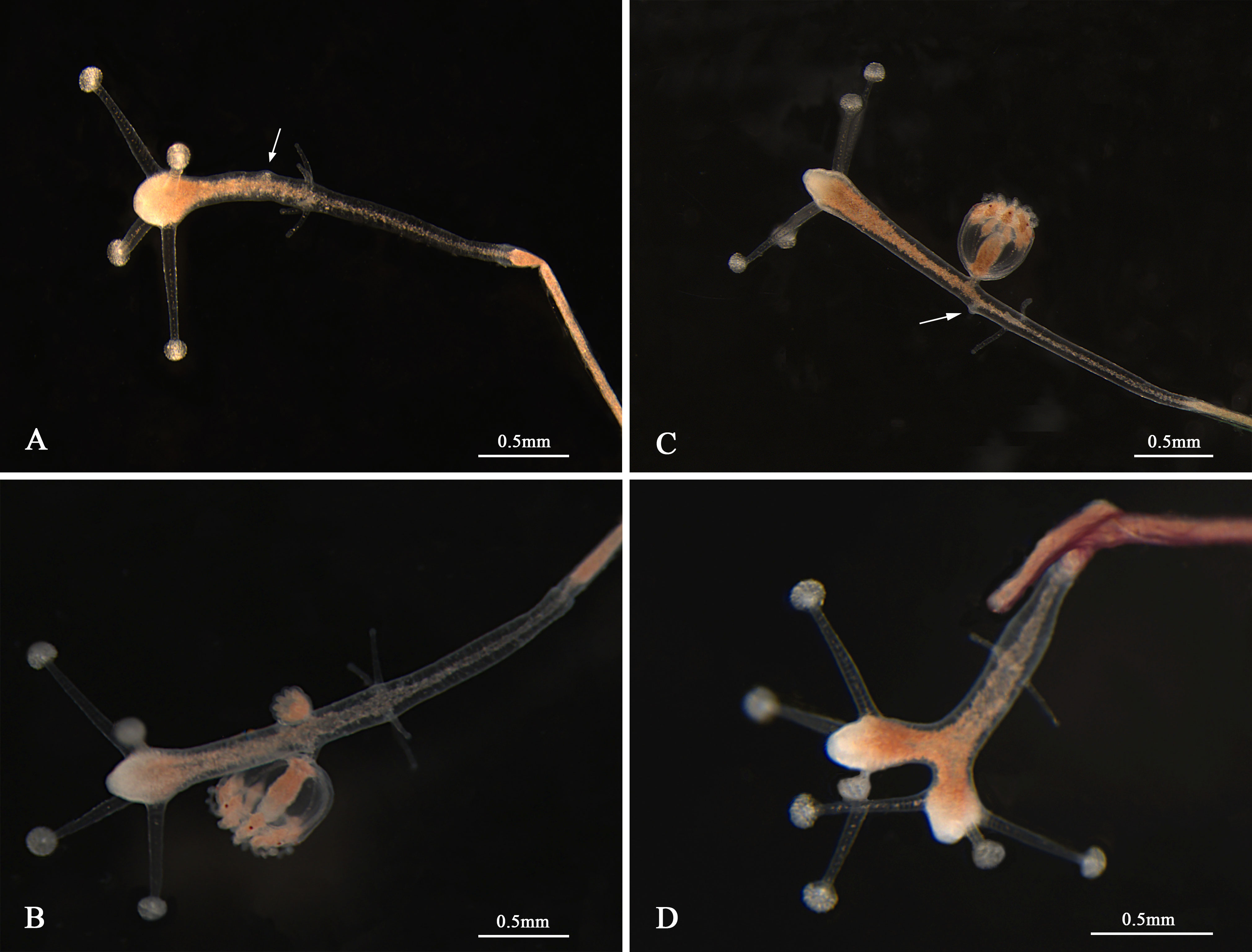
Figure 1 Hydroids of Cladonema digitatum sp. nov. (A) Hydranth. (B, C) Polyp with medusa buds. (D) Branches of a polyp. The arrows marked the early stage of medusa bud.
Newly Released Medusa
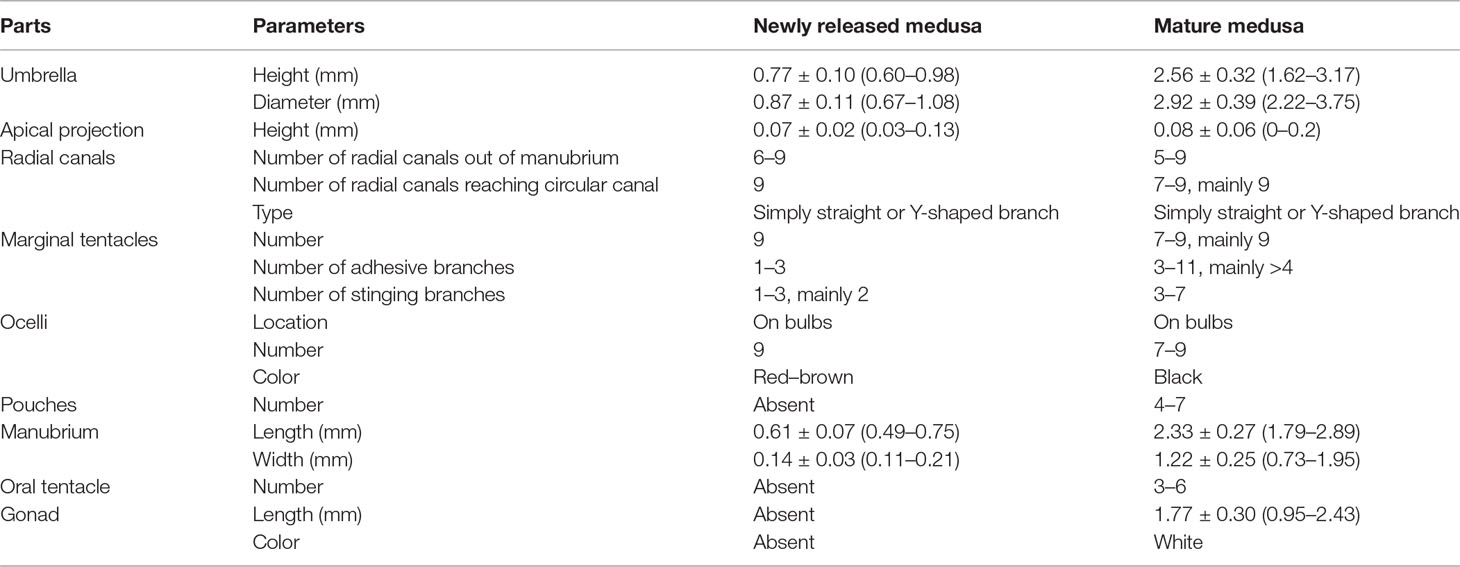
The morphological characteristics of the medusae are shown in Figure 2 and Table 3. The newly released medusa has a bell-shaped umbrella, not fully extended, 0.67–1.08 mm in width, and 0.60–0.98 mm in height. The umbrella had a smooth surface, jelly-thin with an apical projection, and the velum was broad. The manubrium was columnar or spindle-shaped without pouches (0.11–0.21 mm in width and 0.49–0.75 mm in length, within the bell cavity). There was an immature mouth, without bulbous nematocyst clusters. Six to nine radial canals are issued directly from the manubrium, while nine radial canals reached the circular canal, simply straight or some with 1–3 Y-shaped bifurcations. The tentacle bulbs were mainly nine, one opposite each radial canal reaching the circular canal, hourglass-shaped, and with an abaxial red–brown ocellus on the tentacle base. One to three adhesive branches and stinging branches on each tentacle bulb were observed, respectively.
Figure 2 Medusae of Cladonema digitatum sp. nov. (A) Mature male medusa. (B) Young medusa. (C) Newly released medusa. (D) Planiform of medusa. (E) Y-shaped bifurcation. (F) Branches of marginal bulbs.
Adult Medusa
Bell-shaped umbrella, 2.22–3.75 mm in width and 1.62–3.17 mm in height, exumbrella surface smooth, jelly moderately thin with a weak apical projection (Figure 2B), velum broad; manubrium columnar or spindle-shaped, 0.73–1.95 mm in width and 1.79–2.89 mm in length, within bell cavity; manubrium with 4–7 (mainly six) finger-like protuberances in its middle region, with opalescent gonads around the upper 1/2–8/9 (mean 3/4). The mature female gonads were granular, whereas the male gonads were smooth; mouth with 3–6 bulbous nematocyst clusters, mainly six. Five to nine radial canals were issued directly from the manubrium, and seven to nine (mainly nine) canals reached the circular canal. The radial canals are simply straight or with 1–4 Y-shaped bifurcations of mostly two or three. The tentacle bases (7–9) are cylindrical and have one opposite radial canal. One black ocellus per bulb was positioned abaxially at the top of the tentacle base. The adhesive branches were 3–11 (mainly > 4) and the stinging branches 3–7 within each tentacle. The tentacle bases extended with an increase in the adhesive tentacles; stomach and tentacle- base orange. The medusae can swim freely but prefer to stick to bases using adhesive tentacles. The nematocysts can be described as follows:
a) stenoteles (6.03–22.38 × 3.16–13.83 μm)
b) desmonemes (5.58–8.88 × 3.09–4.93 μm)
Distribution
This species was found only in O. melastigma aquaria in our laboratory in Fuzhou, China. C. digitatum may have originated from Hong Kong through the introduction of O. melastigma from the City University of Hong Kong or from a salt lake in Tibet along with Artemia sp. nauplii. Further regional distributions and seasonal variations remain unresolved.
Etymology
This species is named after the Latin term digitatum because of the finger-like protuberances along its manubrium, which are more obvious than those of other species in the genus Cladonema.
Biological Notes
The hydroids of C. digitatum can be easily maintained at temperatures of 20–22°C and salinity of 30–35 ppt. Sometimes the hydranth fractured and separated from the stolon or hydrocaulus. The hydranth part can reattach to a new substrate and develop into a new colony under good conditions. When fed twice a day, the hydroids grew quickly and produced abundant medusa buds within 1 month. The medusa buds took 5 or 6 days to complete their development and eventual release. The medusae matured after 20 days in the same breeding environment as the hydroids. During the developmental period of the medusae, both the stinging and adhesive branches increased with the elongation of the tentacle bases. The pouches on the manubrium gradually became apparent as the medusae developed. The adhesive branches were reduced, and the color of the manubrium became dim with the aging of the medusae.
Photosensitivity
The responses of C. digitatum medusae to light are shown in the Supplementary Video, and the number of medusae bouncing up during each dark phase is shown in Figure 3. Most medusae preferred to stay at the bottom during the first 30 s of the light phase. However, some medusae bounced up when the light was shifted away (1st dark phase), and some medusae chased the moving light. The photosensitivity of medusae was influenced by developmental stages and the light-to-dark switch as well as the interaction between these two factors (Table 4). The newly released medusae were highly sensitive to photostimulation, and half of them bounced up when the light was removed. For the young and mature medusae, however, photosensitivity was significantly reduced from the fourth and second light-to-dark switch, respectively (Figure 3B; two-way ANOVA, F = 23.02, P < 0.01). In the first dark phase, the medusae of the three different stages showed similar responses to light changes (Tukey’s multiple-comparisons test, P < 0.01). However, as the experiment progressed, the number of young and mature medusae bouncing up became less than that of the newly released medusae. In summary, C. digitatum medusae had reduced photosensitivity as they grew up, and they became less sensitive to the light-to-dark switch as the number of trials increased.
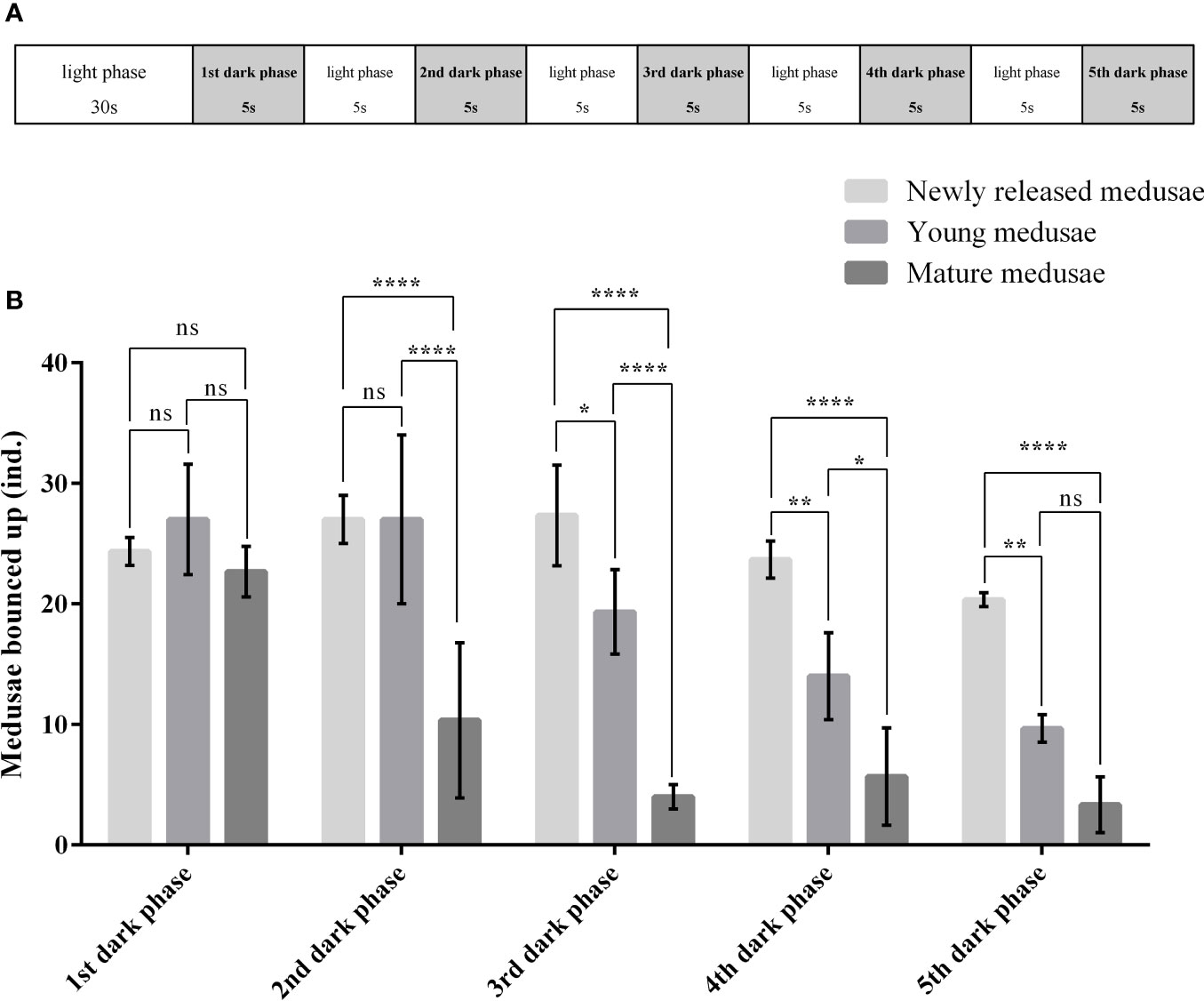
Figure 3 Photosensitivity of Cladonema digitatum medusae at different development stages. (A) Design of the photosensitivity experiment. (B) Number of medusae bouncing up during each dark phase. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
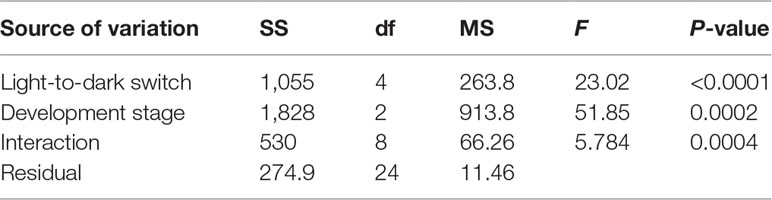
Table 4 Two-way ANOVA for different light to dark switches and development stages on the photosensitivity of medusae.
DNA Barcoding
Among samples of C. digitatum sp. nov., the genetic divergences of both the 16S (558 bp) and COI (709 bp) sequences were 0–0.009 and 0–0.004, respectively. For the 16S rRNA, the pairwise intra-species K2P genetic distances (0–0.053) did not overlap with intra-genus distances in the genera Cladonema (0.087–0.187), Staurocladia (0.108–0.216), and Eleutheria (0.227–0.236), as shown in Table 5, indicating an obvious “barcoding gap” (Table 5; Kruskal–Wallis test, p < 0.01). This result was consistent with that of the phylogenetic tree. In Cladonematidae, C. digitatum sp. nov. displayed the least genetic divergence from Cladonema radiatum [0.133 ± 0.009 (0.117–0.145)], the second closest species was Cladonema sp. [MT709261:0.143 ± 0.002 (0.140–0.147)], the third closest species was C. multiramosum [0.153 ± 0.003 (0.150–0.161)], and the most distant species was Staurocladia sp. [MT709274:0.263 ± 0.002 (0.259–0.268)]. Without Cladonema sp. (AM088484), the intra-species distances [0.003 ± 0.003 (0–0.009)] were smaller than the intra-genus distances [0.145 ± 0.032 (0.087–0.236), Kruskal–Wallis test, p < 0.01], which were also smaller than the intra-family distances [0.212 ± 0.046 (0.093–0.340), Kruskal-Wallis test, p < 0.01] in Cladonematidae. Cladonema sp. (AM088484) showed large genetic divergences with other Cladonematidae species [0.254 ± 0.016 (0.224–0.277)], which were larger than the intra-family distances mentioned above (Kruskal–Wallis test, p < 0.01).
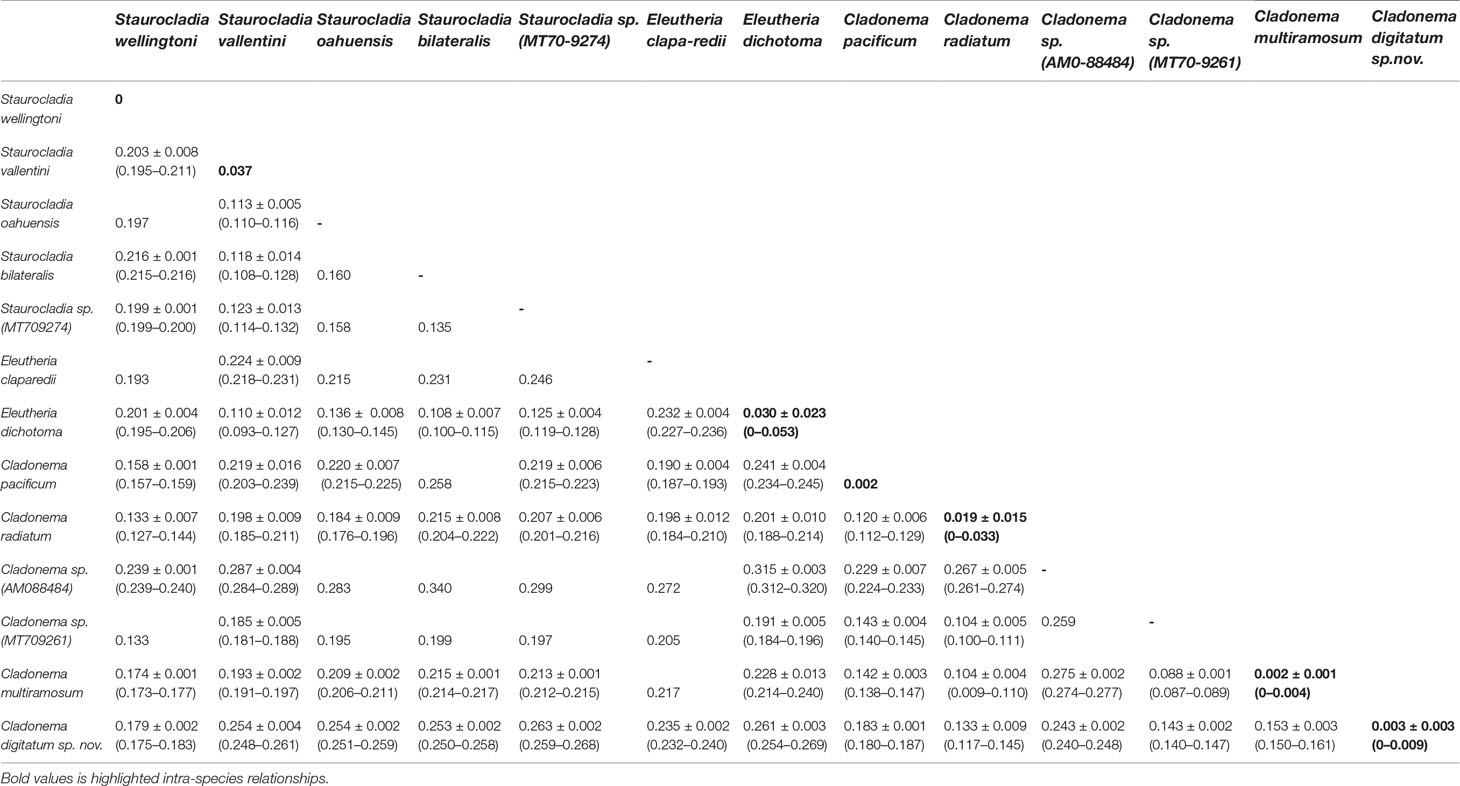
Table 5 Pairwise Kimura 2 parameter genetic distances [mean ± SD (range)] between species in the family Cladonematidae based on mitochondrial 16S rDNA.
The pairwise intraspecies K2P genetic distances for COI are listed in Table 6. C. digitatum sp. nov. showed the closest genetic distance to Cladonema sp. (KC706693, 0.036). Except Cladonema sp. (KC706693), the intra-species K2P genetic distances (0–0.004) did not intersect with the intra-genus distances in the genera Cladonema (0.168–0.241) and Staurocladia (0.128–0.153), suggesting an obvious “barcoding gap”. C. digitatum sp. nov. presented slightly larger distances with other Cladonema species [0.205 ± 0.010 (0.195–0.241)] than those with Staurocladia species [0.183 ± 0.002 (0.180–0.187); Kruskal–Wallis test, p < 0.01]. The intra-genus distances in the genus Cladonema [0.209 ± 0.014 (0.168–0.241)] were also larger than the intra-family distances [0.176 ± 0.017 (0.137–0.211), Kruskal–Wallis test, p < 0.01] in Cladonematidae.
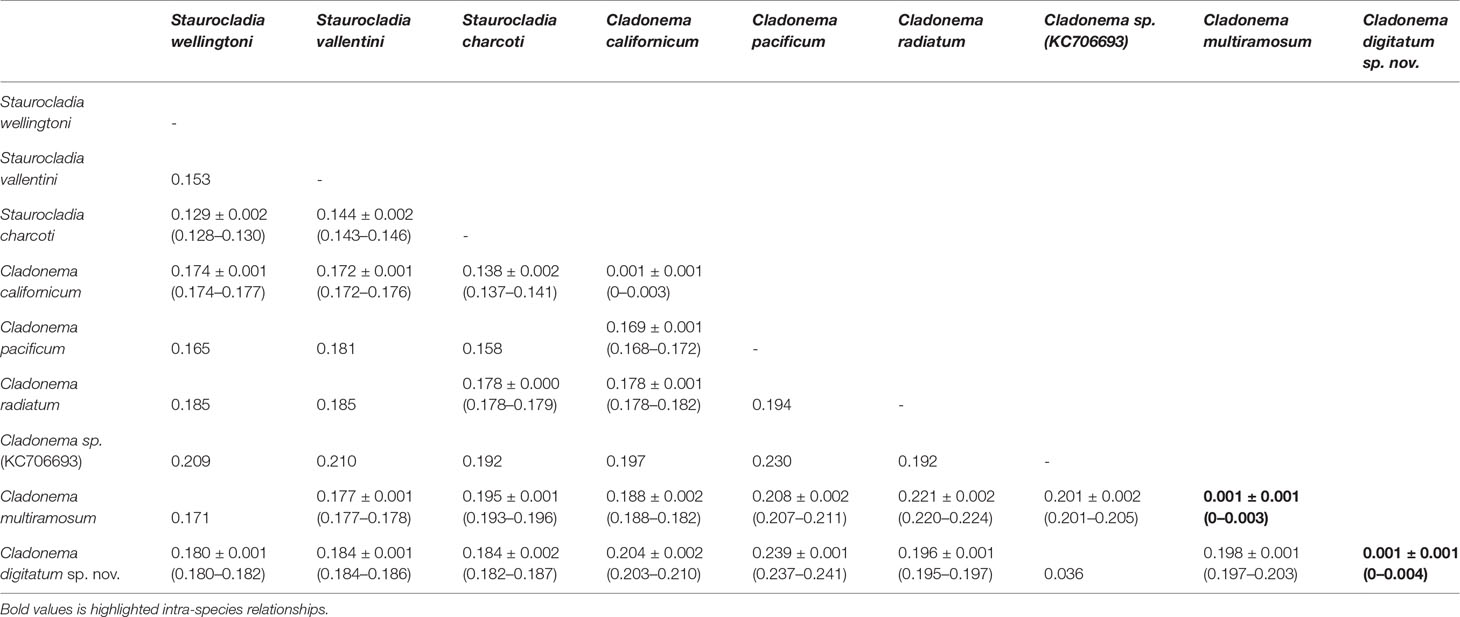
Table 6 Pairwise Kimura 2 parameter genetic distances [mean ± SD (range)] between species in the family Cladonematidae based on mitochondrial cytochrome oxidase.
The NJ and ML topologies of Cladonematidae based on both 16S rRNA and COI revealed an independent clade of C. digitatum sp. nov. with strong support in the genus Cladonema (Figures 4, 5). The genus Cladonema was recovered as monophyletic based on the 16S phylogenetic tree but was polyphyletic based on the COI phylogenetic tree with weak support. In the 16S phylogenetic tree, the genera Eleutheria and Staurocladia were polyphyletic, and the Eleutheria dichotoma clade was nested within the Staurocladia clade with weak support.
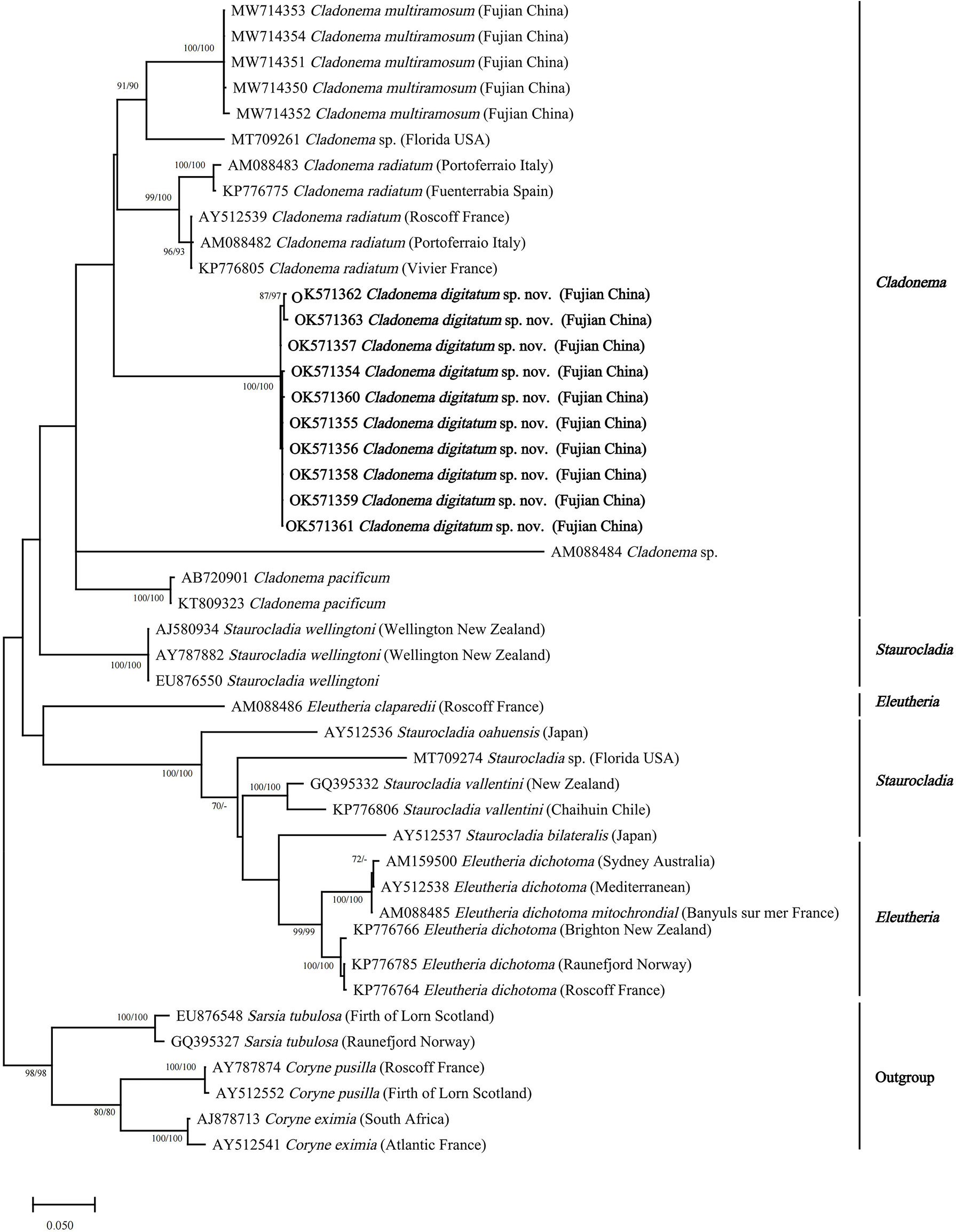
Figure 4 Neighbor-joining clustering of Cladonematidae based on mitochondrial 16S rRNA. Bootstrap values of 1,000 pseudoreplicates higher than 70% were shown above the branches as node-support values. The first number along the branches refers to the maximum likelihood bootstrap values, while the second number refers to the neighbor-joining bootstrap values.
Discussion
The medusa of C. digitatum sp. nov. is distinguishable from other nominal Cladonema species in having a combination of characteristics: a manubrium with finger-like protuberances, radial canals with Y-shaped bifurcations, tentacles with 3–11 adhesive branches, and 3–7 stinging branches growing from the main branch as side branches. The validity of C. digitatum sp. nov. was confirmed by both morphological and molecular analyses.
Morphological Differences Among Cladonema Species
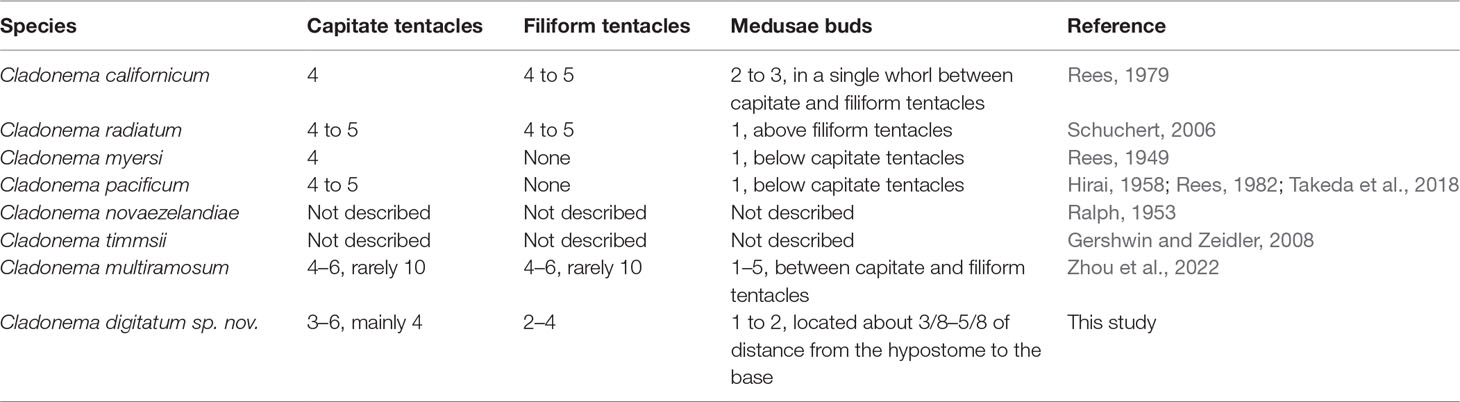
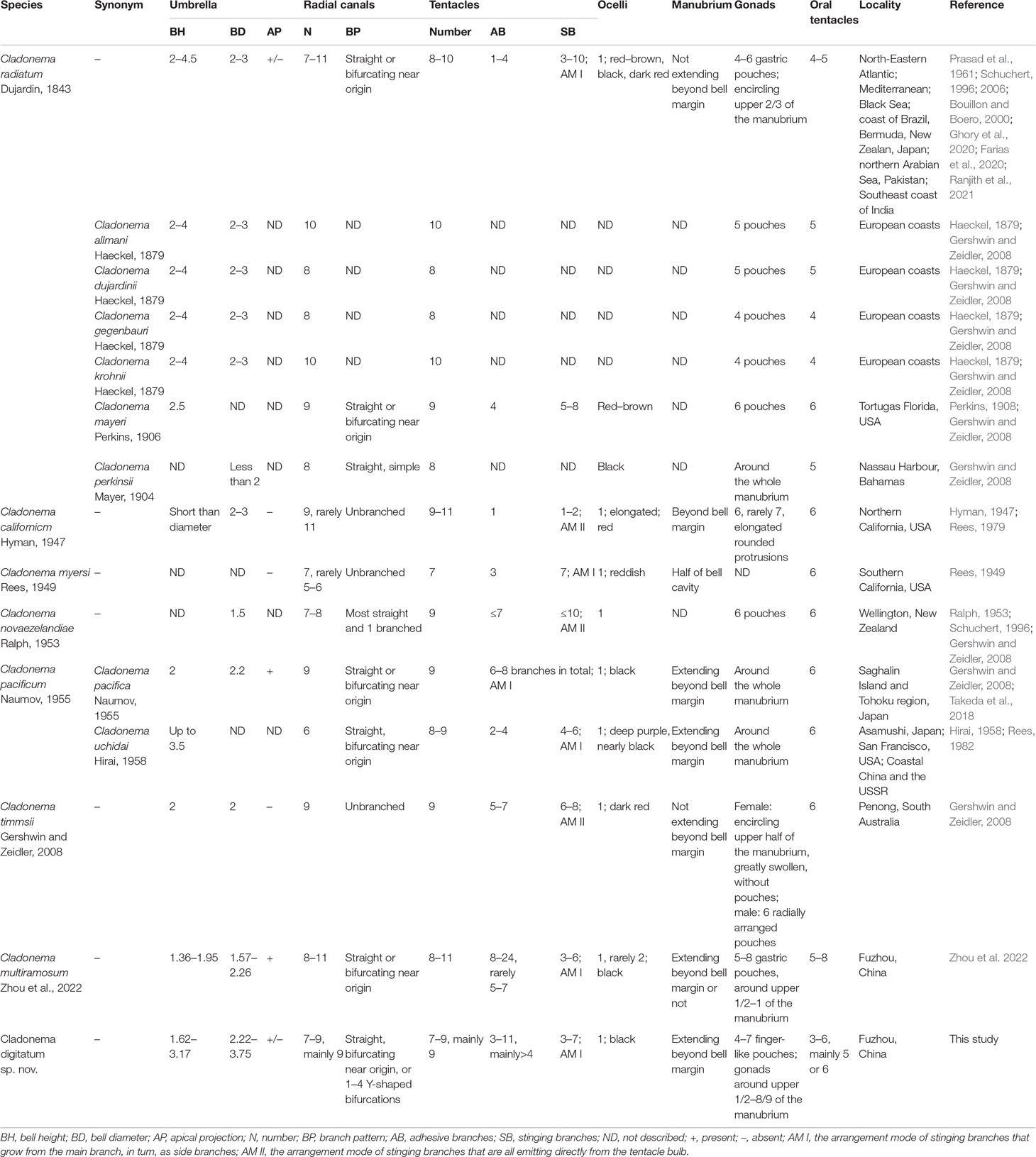
The morphological characteristics of C. digitatum sp. nov. were compared to those of other nominal Cladonema species (Tables 7–9). The hydroid of C. digitatum sp. nov. differs from those of C. pacificum and C. myersi, which lack filiform tentacles (Rees, 1949; Hirai, 1958; Takeda et al., 2018), whereas it is similar to those of C. californicum, C. radiatum, and C. multiramosum, which possess an oral whorl of filiform tentacles (Rees, 1979; Schuchert, 2006; Zhou et al., 2022). C. digitatum sp. nov. has fewer medusa buds per polyp (1 to 2, mainly one) than C. californicum (2 to 3) and C. multiramosum (1–5) (Rees, 1979; Zhou et al., 2022). In addition, the medusa buds of C. digitatum sp. nov. appeared dispersed on the hydranth, whereas those of C. californicum were arranged in a single whorl (Rees, 1979). These findings confirm the validity of the diagnostic characteristics of Cladonema polyps, such as filiform tentacles, and the number and arrangement of medusa buds (Zhou et al., 2022).
Compared with other Cladonema medusae, the main distinct features of C. digitatum sp. nov. are Y-shaped bifurcations in the upper radial canals, stinging branches growing from the main branch as side branches, and finger-like protuberances along the middle region of the manubrium (Table 8). Most C. digitatum sp. nov. have two or three Y-shaped bifurcations, more than those of C. novaezelandiae medusae, another species with a Y-shaped bifurcation in the genus Cladonema (Ralph, 1953). The other distinctive feature of C. digitatum sp. nov. and C. novaezelandiae is the arrangement of their stinging branches, where the stinging branches of C. digitatum sp. nov. grow from the main branch as side branches (Figure 2F), whereas those of C. novaezelandiae all emit directly from the tentacle bulb (Ralph, 1953). This is also the case for C. californicum (Hyman, 1947) and C. timmsii (Gershwin and Zeidler, 2008). Except for C. pacificum, female C. timmsii, and C. myersi, the other Cladonema medusae have gastric pouches (Table 8), and C. digitatum sp. nov. medusae possess the most obvious finger-like protuberances.
Most morphological differences among nominal Cladonema medusae vary (Schuchert, 2006; Zhou et al., 2022). Owing to the overlap in the numbers of radial canals, tentacles, adhesive and stinging branches per tentacle, pouches in the manubrium, and oral tentacles (Table 8), C. digitatum sp. nov. medusae could not be easily distinguished from other Cladonema medusae, except C. californicum, which has only one adhesive and one to two stinging branches per tentacle (Hirai, 1958; Rees, 1979). C. digitatum sp. nov. medusae have more adhesive branches (3–11, mainly > 4) than most Cladonema medusae, next only to C. multiramosum (Zhou et al., 2022). In addition, C. digitatum sp. nov. medusae have a flexible manubrium, and some lack an apical projection, reflecting the morphological plasticity of the medusae. Thus, the diagnostic criteria for mature Cladonema medusae should be a combination of characteristics, including the number of adhesive branches, stinging branches, tentacles, radial canals, the branch patterns of stinging branches and radial canals, and the presence of manubrium pouches.
However, the newly released medusae of C. digitatum sp. nov. differ from those of C. pacificum (Hirai, 1958), C. californicum (Rees, 1979), C. radiatum (Schuchert, 1996), C. myersi (Rees, 1949), and C. multiramosum (Zhou et al., 2022) in two stinging branches and Y-shaped bifurcations in the upper radial canals. The number of adhesive branches (1–3) in C. digitatum sp. nov. covers those of other Cladonema species (one or two). Moreover, with the development of C. digitatum sp. nov. medusae, finger-like protuberances formed on the manubrium and became increasingly obvious. Meanwhile, the number of stinging and adhesive branches increases during development, similar to other Cladonema medusae (Zhou et al., 2022).
As for the nematocysts, the medusae and polyps of C. digitatum sp. nov, C. multiramosum, C. radiatum, C. californicum, and C. pacificum all have stenoteles in variable sizes. Mastigophores only appeared in the stolons of the polyps of C. digitatum sp. nov., C. multiramosum, and C. radiatum and were absent in C. californicum and C. pacificum (Table 9). Desmonemes were only found in medusae, but in small numbers. Although the morphological features of nematocysts are taxonomically useful for many groups of hydromedusae (Östman, 1979; Östman, 1982; Östman, 1987), they are not recommended for species identification in the genus Cladonema because of the polytropic size of nematocysts and time-consuming observation work (Rees, 1979; Rees, 1982; Zhou et al., 2022).
Except for two species without a description of the hydroid stage, Cladonema species could be divided into two groups based on the presence of filiform tentacles in hydroids and gastric pouches in medusae: C. radiatum-like medusae, including C. radiatum, C. californicum, C. multiramosum, and C. digitatum sp. nov. and C. pacificum-like medusae, including C. myersi and C. pacificum. Based on the version of Zhou et al. (2022), we added information on C. digitatum sp. nov. and updated the taxonomy of Cladonema based on the morphological characteristics of the mature medusae.
Key to the accepted nominal species of the genusCladonema:
1a With 7, rarely 5–6 marginal tentacles………………………C. myersi
1b With 8–11 marginal tentacles………………………………………………2
2a Stinging branches all emitting directly from the tentacle bulb………………………………………………………………………3
2b Stinging branches growing from the main branch, in turn, as side branches…………………………………………………………5
3a Most radial canals straight and one-branched………………………………………………C. novaezelandiae
3b Radial canals straight…………………………………………………………4
4a Tentacles with one adhesive branch and 1-2 stinging branches……………………………………C. californicum
4b Tentacles with 5–7 adhesive branches and 6–8 stinging branches…………………………………………C. timmsii
5a Manubrium without pouches……………………………C. pacificum
5b Manubrium with pouches…………………………………………………6
6a Tentacles with 1–4 adhesive branches………………C. radiatum
6b Tentacles with over four adhesive branches………………………7
7a Tentacles with 8–24 adhesive branches (rarely 5–7), radial canals without Y-shaped bifurcation on the upper part……………………C. multiramosum
7b Tentacles with 3–11 adhesive branches (mainly >4), radial canals with 1–4 Y-shaped bifurcations in the upper part……………………C. digitatum sp. nov.
Molecular Phylogenetic Analysis
The phylogenetic hypotheses based on 16S rRNA and COI sequences and within-species genetic diversity analyses supported C. digitatum sp. nov. as a distinct species. C. digitatum sp. nov. and Cladonema sp. (KC706693) shared the closest genetic distance (0.036) within the reported intra-species distance range for COI in hydrozoans (0–0.076) (Ortman et al., 2010; Zheng et al., 2014; Liu et al., 2018) and formed a robust clade in the COI phylogenetic tree (Figure 5). A short COI fragment (313 bp) of Cladonema sp. (KC706693) was found in the fish gut content of Moorea Island, French Polynesia (Leray et al., 2013), which might be a geographic population of C. digitatum sp. nov. The genetic distances between C. digitatum sp. nov. and other Cladonema species are almost within the reported intra-genus distances—for example, 0.062–0.236 for 16S and 0.045–0.243 for COI (Zheng et al., 2014).
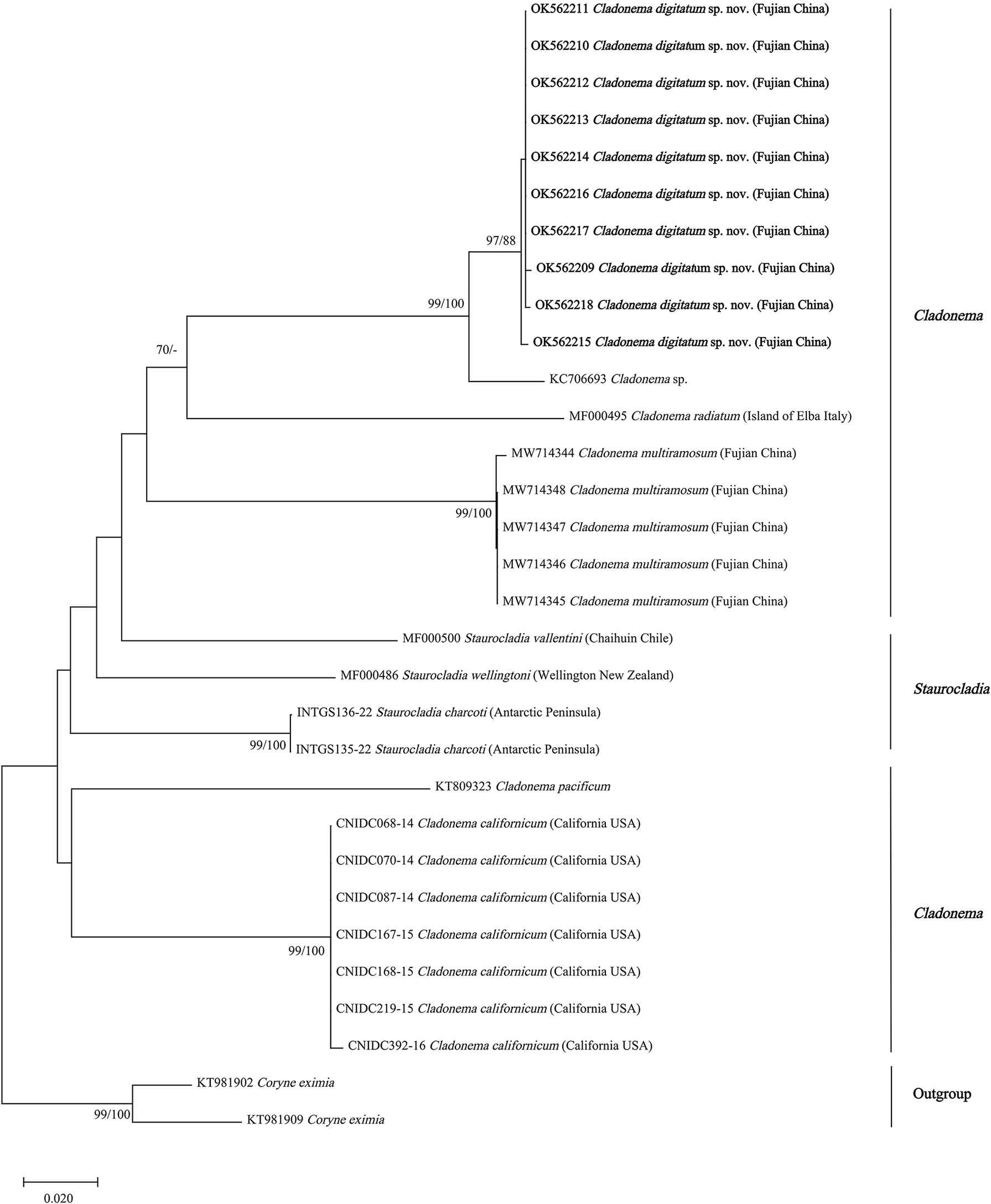
Figure 5 Neighbor-joining clustering of Cladonematidae based on mitochondrial cytochrome oxidase (COI). Bootstrap values of 1,000 pseudoreplicates higher than 70% were shown above the branches as node-support values. The first number along the branches refers to the maximum likelihood bootstrap values, while the second number refers to the neighbor-joining bootstrap values.
With the exception of Cladonema sp., C. radiatum-like species (C. digitatum sp. nov., C. radiatum, and C. multiramosum) clustered together and were presented as a sister group of C. pacificum in both the 16S and COI phylogenetic trees (Figures 4, 5). Unlike the report by Zhou et al. (2022), Cladonema species formed a clade with weak support in the 16S phylogenetic tree of Cladonematidae after adding the data of C. digitatum sp. nov. (Figure 4). However, Cladonema was recovered as polyphyletic based on the COI phylogenetic tree with weak support, which might be attributed to data deficiency. Currently, the 16S sequences of only 10 valid species and the COI sequences of only eight valid species have been reported in Cladonematidae, accounting for approximately half of the valid species in this family (Schuchert, 2022). In addition, the 16s rRNA and COI data of C. myersi, C. novazelandiae, and C. timmisii have not been reported. Although the mature medusae of C. digitatum sp. nov. can be distinguished from C. myersi, C. novazelandiae, and C. timmisii in terms of morphological characteristics, synonyms might still exist among them owing to phenotypic plasticity. Therefore, further sequences are required to revise the phylogenetic tree in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XF, SL, and YZ conducted the experiments. XF and KZ completed the manuscript. KZ, JC and ZW revised the manuscript and provided the funds and resource for this research. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (no. 41806181), the Natural Science Foundation of Fujian Province of China (no. 2021J011042), the Program for Innovative Research Team in Science and Technology in Fujian Province University, and The Talent Introduction Pre-research Project of Minjiang University (no. MJY20017).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the editors and the reviewers for their comments and suggestions on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.891998/full#supplementary-material
References
Bouillon J., Boero F. (2000). Synopsis of the Families and Genera of the Hydromedusae of the World, With a List of the Worldwide Species. Thalassia Salentina. 24, 47–296. doi: 10.1285/i15910725v24p47
Bucklin A., Hopcroft R. R., Kosobokova K. N., Nigro L. M., Ortman B. D., Jennings R. M., et al. (2010a). DNA Barcoding of Arctic Ocean Holozooplankton for Species Identification and Recognition. Deep Sea Res. Part II: Topical Stud. Oceanogr. 57 (1), 40–48. doi: 10.1016/j.dsr2.2009.08.005
Bucklin A., Ortman B. D., Jennings R. M., Wiebe P. H., Nigro, L. M., and (2010b). A “Rosetta Stone” for Metazoan Zooplankton: DNA Barcode Analysis of Species Diversity of the Sargasso Sea (Northwest Atlantic Ocean). Deep Sea Res. Part II: Topical Stud. Oceanogr. 57 (24), 2234–2247. doi: 10.1016/j.dsr2.2010.09.025
Cedeno-Posso C. (2014). First Record of the Genus Cladonema (Medusae and Polyps) in Colombia. Zootaxa. 3793 (5), 597–599. doi: 10.11646/zootaxa.3793.5.8
Chou T.-H., Huang M.-C. (1958). A Study on Hydromedusae of Chefoo. Acta Zoologica Sin. 10 (2), 173–191.
Colin S. P., Costello J. H., Klos E. (2003). In Situ Swimming and Feeding Behavior of Eight Co-Occurring Hydromedusae. Mar. Ecol. Prog. Series. 253, 305–309. doi: 10.3354/meps253305
Ender A., Schierwater B. (2003). Placozoa Are Not Derived Cnidarians: Evidence From Molecular Morphology. Mol. Biol. evolution. 20 (1), 130–134. doi: 10.1093/molbev/msg018
Farias G. B., Leitão S. N., Melo P. A. M. D. C., Júnior M. N., Tosetto E. G. (2020). First in Situ Record of the Medusa Stage of Cladonema Radiatum (Cnidaria: Anthoathecata) in the South Atlantic Ocean. Ocean Coast. Res. 68, e20349. doi: 10.1590/s2675-28242020068349
Folmer O., Black M. B., Hoeh W. R., et al. (1994). DNA Primers for Amplification of Mitochondrial Cytochrome C Oxidase Subunit I From Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 3 (5), 294–299. doi: 10.4028/www.scientific.net/DDF.7.460
Fujiki A., Hou S., Nakamoto A., Kumano G.(2019). Branching Pattern and Morphogenesis of Medusa Tentacles in the Jellyfish Cladonema Pacificum (Hydrozoa, Cnidaria). Zoological Letters. 5 (1), 12. doi: 10.1186/s40851-019-0124-4
Fujita S., Kuranaga E., Nakajima Y.-I. (2019). Cell Proliferation Controls Body Size Growth, Tentacle Morphogenesis, and Regeneration in Hydrozoan Jellyfish. Cladonema pacificum. PeerJ. 7, e7579. doi: 10.7717/peerj.7579
Fujita S., Kuranaga E., Nakajima Y.-I. (2021). Regeneration Potential of Jellyfish: Cellular Mechanisms and Molecular Insights. Genes. 12 (5), 758. doi: 10.3390/genes12050758
Gershwin L.-A., Zeidler W. (2008). Cladonema Timmsii, a New Species of Hydromedusa (Cnidaria: Hydrozoa) From a Salt Lake in South Australia. Zootaxa. 1826 (1), 59–68. doi: 10.11646/zootaxa.1826.1.4
Ghory F. S., Ali Q. M., Ahmed Q. (2020). First Record of Cladonema Radiatum Dujardin 1843 (Hydrozoa: Cladonematidae) From Northern Arabian Sea, Pakistan. Int. J. Fisheries Aquat. Res. 5 (3), 14–16. doi: 15560/11.2.1609
Gravili C., D’ambrosio P., Di Camillo C., Renna G., Bouillon J., Boero F., and (2008). Clytia Hummelincki (Hydroidomedusae: Leptomedusae) in the Mediterranean Sea. J. Mar. Biol. Assoc. United Kingdom. 88 (8), 1547–1553. doi: 10.1017/S0025315408001975
Graziussi D. F., Suga H., Schmid V., Gehring W. J. (2012). The “Eyes Absent” (Eya) Gene in the Eye-Bearing Hydrozoan Jellyfish Cladonema Radiatum: Conservation of the Retinal Determination Network. J. Exp. Zoology Part B: Mol. Dev. Evolution. 318 (4), 257–267. doi: 10.1002/jez.b.22442
Haeckel E. (1879). Das System der Medusen. Erster Teil einer Monographie der Medusen. Denkschriften der Medicinisch-Naturwissenschaftlichen Gesellschaft zu Jena, (1): XX+1–360, 320 plates.
He J., Zheng L., Zhang W., Lin Y., Cao W. (2015). Morphology and Molecular Analyses of a New Clytia Species (Cnidaria: Hydrozoa: Campanulariidae) From the East China Sea. J. Mar. Biol. Assoc. United Kingdom. 95 (2), 289–300. doi: 10.1017/S0025315414000836
Hirai E. (1958). “On the Species of Cladonema radiatum var. mayeri Perkins”. in: Bulletin of the Marine Biological Station of Asamushi, ( Tohoku University) 9, 23–25.
Hyman L. H. (1947). Two New Hydromedusae From the California Coast. Trans. Am. Microscopical Society. 66 (3), 262–268. doi: 10.2307/3223391
Leray M., Yang J. Y., Meyer C. P., Mills S. C, Agudelo N., Ranwez V., et al (2013). A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. zoology. 10, 34. doi: 10.1186/1742-9994-10-34
Lindner A., Govindarajan A. F., Migotto A. E. (2011). Cryptic Species, Life Cycles, and the Phylogeny of Clytia (Cnidaria: Hydrozoa: Campanulariidae). Zootaxa. 2980, 23–36. doi: 10.11646/zootaxa.2980.1.2
Liu C., Gu Z., Xing M., Sun Y., Chen S., Chen Z. (2018). Sex Determination and Differentiation in Aurelia Sp.1: The Absence of Temperature Dependence. J. Oceanol. Limnol. 36 (2), 457–464. doi: 10.1007/s00343-017-6170-x
Miglietta M. P., Rossi M., Collin R. (2008). Hydromedusa Blooms and Upwelling Events in the Bay of Panama, Tropical East Pacific. J. Plankton Res. 30 (7), 783–793. doi: 10.1093/plankt/fbn038
Nawrocki A. M., Schuchert P., Cartwright P. (2010). Phylogenetics and Evolution of Capitata (Cnidaria: Hydrozoa), and the Systematics of Corynidae. Zoologica Scripta. 39 (3), 290–304. doi: 10.1111/j.1463-6409.2009.00419.x
Ortman B. D., Bucklin A., Pagès F., Youngbluth M.(2010). DNA Barcoding the Medusozoa Using mtCOI. Deep Sea Res. Part II: Topical Stud. Oceanogr. 57 (24–26), 2148–2156. doi: 10.1016/j.dsr2.2010.09.017
Östman C. (1979). Two Types of Nematocysts in Campanulariidae (Cnidaria, Hydrozoa) Studied by Light and Scanning Electron Microscopy. Zoologica Scripta. 8 (1–4), 5–12. doi: 10.1111/j.1463-6409.1979.tb00614.x
Östman C. (1982). Nematocysts and Taxonomy in Laomedea, Gonothyraea and Obelia (Hydrozoa, Campanulariidae). Zoologica Scripta. 11 (4), 227–241. doi: 10.1111/j.1463-6409.1982.tb00536.x
Östman C. (1987). “New Techniques and Old Problems in Hydrozoan Systematics,” in Modern Trends in the Systematics, Ecology and Evolution of Hydroids and Hydromedusae. Eds. Bouilon J., Boero F., Cicogna F., Cornelius P. F. S. (Oxford: Clarendon Press), 67–82.
Perkins H. F. (1908). Notes on Medusae of the Western Atlantic. ( Carnegie Institution of Washington), 1. 135–149.
Prasad R., Tampi P., Durve V. (1961). Note on the Occurrence of the Anthomedusa Cladonema in the Indian Region. J. Mar. Biol. Assoc. India. 3 (1 & 2), 251–252.
Ralph P. M. (1953). A Guide to the Athecate (Gymnoblastic) Hydroids and Medusae of New Zealand. Tuatara. 5, 59–75.
Ranjith L., Kalidas C., Saravanan R., Gagadis I., Prabu D. L., Kavitha M, et al. (2021). Observation on the Morphological and Gonadal Aspects of Cladonema Radiatum (Class: Hydrozoa) From Tuticorin Bay, Southeast Coast of India. Zootaxa. 4990 (3), 591–595. doi: 10.11646/zootaxa.4990.3.12
Rees W. J. (1949). On Cladonema Myersi, a New Species of Hydroid From the Californian Coast. Proc. Zoological Soc. London. 119 (4), 861–865. doi: 10.1111/j.1096-3642.1950.tb00912.x
Rees J. T. (1979). The Polyp and Immature Medusa Stages of Cladonema Californicum, Hyman 1947 (Anthomedusae: Cladonemidae) With Biological Notes and a Discussion of the Taxonomy of the Genus Cladonema. J. Natural History. 13 (3), 295–302. doi: 10.1080/00222937900770231
Rees J. T. (1982). The Hydrozoan Cladonema in California: A Possible Introduction From East Asia. Pacific Science. 36 (4), 439–444.
Schuchert P. (1996). The Marine Fauna of New Zealand: Athecate Hydroids and Their Medusae (Cnidaria: Hydrozoa). Oceanogr. Lit. Review. 1 (45), 1–159.
Schuchert P. (2006). The European Athecate Hydroids and Their Medusae (Hydrozoa, Cnidaria): Capitata Part 1. Rev. suisse Zoologie. 113 (2), 325–410. doi: 10.5962/bhl.part.80356
Schuchert P. (2016). The Polyps of Oceania Armata Identified by DNA Barcoding (Cnidaria, Hydrozoa). Zootaxa. 4175 (6), 539–555. doi: 10.11646/zootaxa.4175.6.3
Schuchert P. (2022) World Hydrozoa Database. Cladonema Dujardin 1843. Available at: http://marinespecies.org/aphia.php?p=taxdetails&id=117049.
Takeda N., Deguchi R., Itabashi T. (2018). “Reproductive Strategies in Marine Hydrozoan Jellyfish: Sexual Medusae and Asexual Polyps,” in Reproductive and Developmental Strategies ( Springer), 157–174.
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. evolution. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Tewksbury J. J., Anderson J. G. T., Bakker J. D., Billo T. J., Dunwiddie P. W., Groom M. J., et al. (2014). Natural History’s Place in Science and Society. BioScience. 64 (4), 300–310. doi: 10.1093/biosci/biu032
Xu Z. (1993). Revisions of Nominal Species on the Hydromedusae of China Sea Areas. J. Oceanogr. Taiwan Strait 12 (3), 197–204.
Zheng L., He J., Lin Y., Cao W., Zhang W. (2014). 16s rRNA is a Better Choice Than COI for DNA Barcoding Hydrozoans in the Coastal Waters of China. Acta Oceanologica Sinica. 33 (4), 55–76. doi: 10.1007/s13131-014-0415-8
Zhou K., Gu Y., Wang L., Chen J. (2022). Discovery of Cladonema Multiramosum Sp. Nov. (Cnidaria: Hydrozoa: Cladonematidae) Using DNA Barcoding and Life Cycle Analyses. Acta Oceanologica Sin 41, 44–52. doi: 10.1007/s13131-021-1900-5
Keywords: Cladonema, morphology, life cycle, DNA barcoding, 16S rRNA, COI, photosensitivity
Citation: Fang X, Lin S, Zhang Y, Wang Z, Zhou K and Chen J (2022) Identification of a Novel Species, Cladonema digitatum sp. nov. (Cnidaria: Hydrozoa: Cladonematidae), Using DNA Barcoding and Life Cycle Analyses. Front. Mar. Sci. 9:891998. doi: 10.3389/fmars.2022.891998
Received: 08 March 2022; Accepted: 16 June 2022;
Published: 28 July 2022.
Edited by:
Rachel Collin, Smithsonian Tropical Research Institute (SI), United StatesReviewed by:
Maria Pia Miglietta, Texas A&M University at Galveston, United StatesAllen G. Collins, NOAA Fisheries, United States
Copyright © 2022 Fang, Lin, Zhang, Wang, Zhou and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Konglin Zhou, zhoukl@mju.edu.cn; Jianming Chen, chenjm@mju.edu.cn
 Xinyu Fang
Xinyu Fang Shen Lin1
Shen Lin1 Jianming Chen
Jianming Chen