- Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Japan
Dimethlysulfoniopropionate (DMSP) lyase is an enzyme that mediates cleavage of DMSP into dimethyl sulfide (DMS) and acrylate. DMS is an aerosol substance that may affect cloud formation, solar radiation and ocean temperatures. DMSP lyases in marine organisms, such as marine bacteria, release DMS, which might contribute to atmosphere-ocean feedback. Although DMSP lyases were first identified in marine bacteria, eukaryotic DMSP lyases or genes similar to DMSP lyase, DMSP lyase-like (DL-L) genes have been found not only in coccolithophores (Emiliania huxleyi) and symbiotic algae of the Family Symbiodiniaceae, but also in animals, including scleractinian corals (Cnidaria: Anthozoa: Hexacorallia). Comparative genomic analysis showed that gene expansion events of DL-L genes have occurred specifically in the scleractinian genus, Acropora. In the present study, we performed molecular identification of DL-L genes in Acropora digitifera. Thirteen full-length Open Reading Frames were isolated, confirming that these duplicated DL-L genes are likely expressed. A comprehensive survey of available transcriptomic databases revealed that DL-L genes have been identified not only in scleractinians (Hexacorallia), but also Octocorallia (Anthozoa) and even in a jellyfish (Cnidaria: Hydrozoa). Molecular phylogenetic analyses showed that although some sequences from cnidarian transcriptomic databases apparently originated with their symbiotic algae, cnidarian sequences from Anthozoa and Hydrozoa clustered together, indicating that these evolved from a gene in the last common ancestor of Cnidaria, dating to the Precambrian. Interestingly, cnidarian species possessing DL-L genes apparently occur only in coral reefs or shallow, warmer environments, suggesting that these genes may be essential for animals to survive in such environments. Acropora-specific duplicated DL-L genes, which originated during the past warm geological periods, may enable them to adapt to environmental changes.
Introduction
Coral reefs in tropical and subtropical waters harbor about 30% of all marine life, making them the most biodiverse habitats in marine ecosystems (Knowlton et al., 2010). In addition, coral reefs provide many fishery resources, protect coastlines, and fix carbon and nitrogen. Scleractinian or stony corals (Anthozoa, Cnidaria) are the main builders of coral reefs, forming massive calcium carbonate skeletons. Corals are metaorganisms or “holobionts” associated with a variety of microorganisms, such as dinoflagellates of the family Symbiodiniaceae and diverse communities of bacteria, archaea, fungi, viruses, and protists (Rohwer et al., 2002; Ainsworth et al., 2010). They rely mainly on symbiotic algae (Symbiodiniaceae) to trap solar energy for survival, growth, and calcification (Goreau and Goreau, 1959; Muscatine, 1990; Davy et al., 2012). In stony corals, dinoflagellate symbionts use the substrates (nitrogen and phosphorus) produced by their hosts for photosynthesis (Muscatine and Porter, 1977). Prokaryotic microorganisms provide sources of nitrogen, sulfur, and other elements to corals or release them into the water column (Ainsworth et al., 2010; Rädecker et al., 2015). These allow coral reefs to remain highly productive in oligotrophic waters, while also facilitating global biogeochemical cycles.
The sulfur cycle is one of the most important of these cycles, providing biosynthetic proteins and cofactors. Dimethylsulfoniopropionate (DMSP) is an organic sulfur compound that is abundant in marine surface waters. The majority of it is produced by phytoplankton and algae, but it has also been reported in some angiosperms and bacteria (Keller et al., 1989; Hanson et al., 1994; Paquet et al., 1994; Gage et al., 1997; Kocsis et al., 1998; Lyon et al., 2011; Kettles et al., 2014; Curson et al., 2017). Although functions of DMSP are not entirely understood, several physiological functions, such as osmoregulation, oxidative stress protection, and cryoprotection, have been demonstrated in phytoplankton and green algae (Kirst et al., 1991; Karsten et al., 1996; Sunda et al., 2002; Lesser, 2006; Husband et al., 2012; Curson et al., 2018). Biological functions such as chemoattraction, predator deterrence, and mediation of bacterial virulence have been identified (Wolfe and Steinke, 1996; Miller and Belas, 2004; Barak-Gavish et al., 2018). The most notable and important aspect of DMSP is that it is a precursor of dimethyl sulfide (DMS) (van Boeckel and Stefels, 1993), which is a major source of sulfur in the atmosphere, promoting cloud formation and combating greenhouse gasses. DMSP is cleaved by an enzyme called DMSP lyase (EC 4.4.1.3), which converts DMSP to DMS. Recently, studies reported that DMSP lyase products, DMS and acrylate, may have ecological roles such as chemical defenses against predation (Strom et al., 2003; Teng et al., 2021) and enhancing predation by grazers (Shemi et al., 2021). When DMS enters the atmosphere, it is oxidized into aerosol particles that can induce cloud formation and increase reflectivity. This may reduce light levels and water temperatures in marine environments, thus contributing to local climate regulation (Figure 1) (Ayers and Gras, 1991; Vallina and Simo, 2007), although some studies suggest that sea-air DMS flux is low, and that the role of DMS in climate regulation is likely insignificant (Woodhouse et al., 2010; Quinn and Bates, 2011).
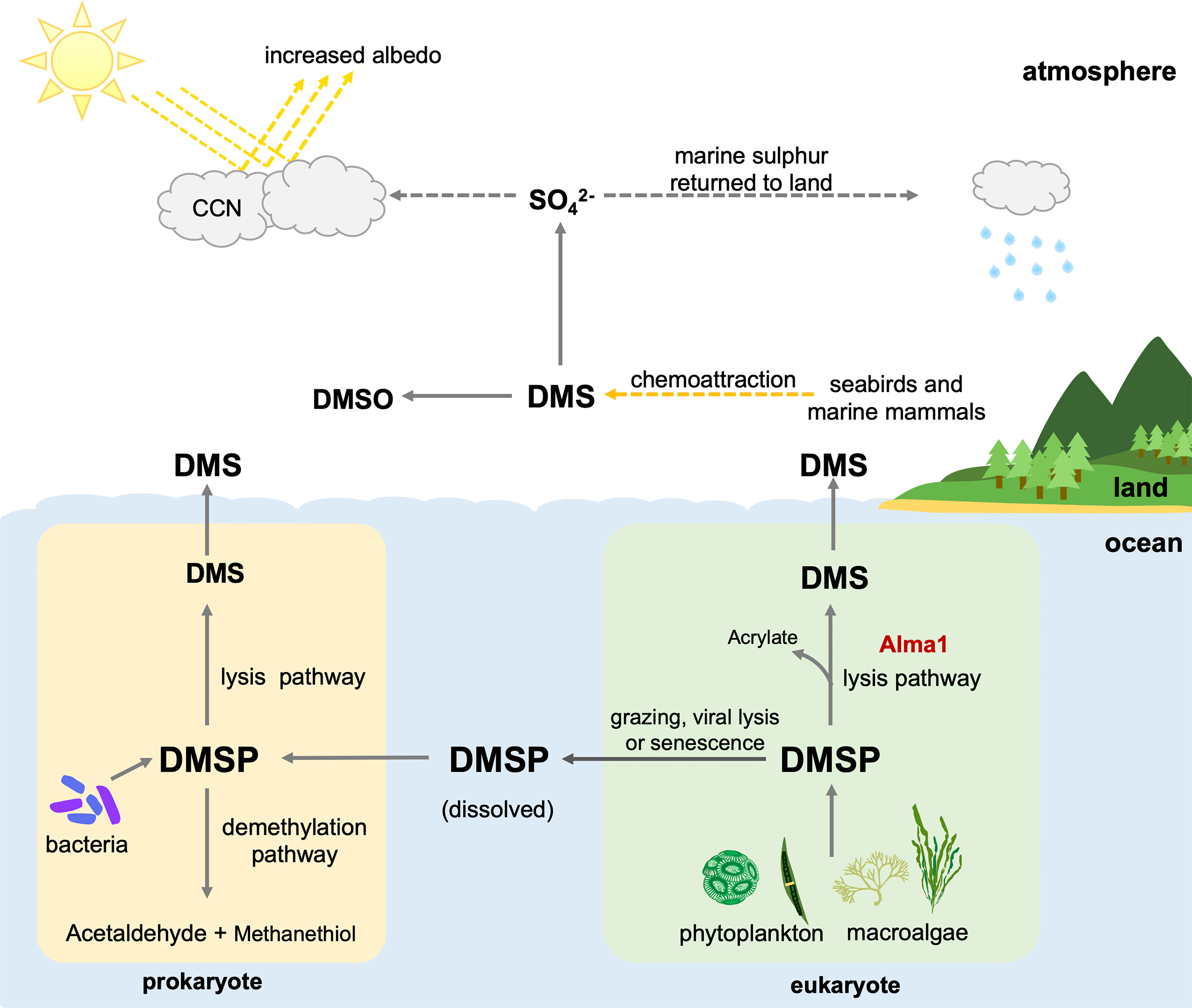
Figure 1 Contributions of prokaryotes and eukaryotes to the biogeochemical cycle and possible functions of dimethylsulfoniopropionic acid (DMSP) and dimethyl sulfide (DMS). CCN, Cloud condensation nuclei; DMSO, dimethyl sulfoxide.
Marine phytoplankton and macroalgae are considered the primary producers of DMSP, while marine bacteria are the primary degraders (Stefels et al., 2007; Reisch et al., 2011). DMSP is released from marine phytoplankton and macroalgae upon cellular lysis caused by zooplankton grazing, viral infection, and senescence (Figure 1) (van Boekel and Stefels, 1993; Wolfe et al., 1997; Hill et al., 1998). Bacteria acquire DMSP mainly from the ocean and demethylate it into methanethiol and acetaldehyde, turning DMSP into DMS via the lysis pathway (Figure 1). DMSP lyase was first identified in bacteria, and at least eight prokaryotic DMSP lyase genes have been identified to date (Li et al., 2021).
Some studies have reported that eukaryotic phytoplankton, including dinoflagellates and algae, possess DMSP lyase activity (Stefels et al., 1995; Steinke and Kirst, 1996; Steinke et al., 1998; Niki et al., 2000; Yoch, 2002), but the responsible gene was difficult to isolate. In 2015, a gene, Alma l, was identified from the eukaryotic coccolithophore, Emiliania huxleyi, and was shown to have high DMSP lyase activity (Alcolombri et al., 2015). It has been suggested that Alma 1 can directly lyse algal DMSP, releasing acrylate and DMS (Figure 1). Interestingly, sequences similar to Alma 1, DMSP lyase-like (DL-L) genes were also found in scleractinian corals and Symbiodiniaceae (Alcolombri et al., 2015). Furthermore, recent comparative genomic analysis of scleractinian and cnidarian genomes showed that numbers of DL-L genes in genomes of the scleractinian coral genus, Acropora, are significantly larger than in other cnidarian genomes and that gene expansion events specifically occurred in Acropora (Shinzato et al., 2021a).
Acropora is currently the most studied coral genus in the world (Maor-Landaw and Levy, 2016), as it includes the most widespread, abundant, and diverse scleractinian corals on Earth (Wallace and Rosen, 2006). Approximately 180 species have been recorded from the Red Sea to the Indo-Pacific Ocean and the Caribbean Ocean (Wallace, 1999; Veron, 2000; van Oppen et al., 2001). With the increase of research data, more genetic databases (genome and transcriptome databases) and molecular techniques (CRISPR/Cas9 and gene knockdown) have been developed (Yasuoka et al., 2016; Cleves et al., 2018; Ying et al., 2019; Shinzato et al., 2021a). Genetic analyses suggest that Acropora has been evolutionarily very successful and may have become a dominant genus through acquisition and expansion of gene families (Shinzato et al., 2011; Shinzato et al., 2021a). Examples of Acropora-specific gene duplication and expansion events have been reported (Hislop et al., 2005; Shinzato et al., 2021a). Symbiotic relationship with Symbiodiniaceae may also have contributed to rapid adaptation to changing environments (Qin et al., 2019). Interestingly, Shinzato et al. (2021a) showed that DL-L genes are the most diversified gene family in Acropora genomes and that possible coral- or cnidarian-specific stress response genes, including Caspase-X and SCRiPs, have also been tandemly duplicated in Acropora genomes. In this study, to better understand evolution of DL-L genes in animals and their possible involvement in Acropora evolution, we first confirmed that DL-L genes are indeed expressed in A. digitifera as possible functional mRNAs without in-frame stop codons. Then, we performed a comprehensive database survey and molecular phylogenetic analyses to investigate the existence of DL-L genes in various eukaryotes, including scleractinian corals, cnidarians, and other animals, to gain insight into the evolutionary origin of DL-L genes in the animal kingdom.
Materials and Methods
Cloning of DL-L Genes From Acropora digitifera
The same RNA samples used in previous studies including different developmental stages of A. digitifera (eggs, blastulae, gastrulae, planula larvae, early polyps, and adult branches) were used for cDNA synthesis (Shinzato et al., 2011; Shinzato et al., 2021a; Yoshioka et al., 2021). Samples were homogenized in TRIzol reagent (Invitrogen) on ice, and total RNA was extracted following the manufacturer's instruction and Chiu et al. (2020). Extracted total RNA was treated with recombinant DNase I (Roche) to remove contaminating genomic DNA. First-strand cDNA was synthesized from 5 μg of DNase-treated RNA using SuperScriptTM III reserve transcriptase (Thermo Fisher Scientific). Possible DL-L genes of A. digitifera were derived from predicted genes of A. digitifera (Shinzato et al., 2021a), and primers amplifying full length open reading frames (ORFs) were designed for each gene (Supplementary Table 1). PCR products were cloned into the pGEM-T Easy cloning vector (Promega), transformed into competent cells (Escherichia coli, DH5α strain), and plasmids were sequenced at a commercial laboratory (Eurofin Genomics, Tokyo, Japan).
Gene Expression Patterns of Acropora digitifera DL-L Genes Under Different Developmental Stages and Increased Sea Water Temperature
To investigate gene expression patterns of 18 DL-L genes predicted from the A. digitifera genome assembly (Shinzato et al., 2021a), we used previously reported RNA-Seq transcriptome data of A. digitifera from different developmental stages (egg, blastula, gastrula, planula, polyp, Shinzato et al., 2021a), adult stages (Yoshioka et al., 2021), and increased seawater temperature treatments (25°C to 30°C, Shinzato et al., 2021b). Briefly, Illumina adaptor sequences and low-quality reads (Quality score < 20, length < 25 bp) in the RNA-Seq data were trimmed with CUTADAPT v1.16 (Martin, 2011), and cleaned reads were mapped to A. digitifera gene models (Shinzato et al., 2021a) using KALLISTO v0.44.0 (Bray et al., 2016) or MiniMap v2.9 (Li, 2018) with default settings. Mapping counts were normalized using the trimmed mean of M values (TMM) method, and then converted to counts per million (CPM) using EdgeR v3.28.1 (Robinson et al., 2010; McCarthy et al., 2012) in R v3.6.3 (team 2015).
Identification of DL-L Genes From Publicly Available Eukaryote and Cnidarian Transcriptomic Databases
Amino acid sequences were deduced from cloned cDNA sequences using the ExPASy translate tool (https://web.expasy.org/translate/; Gasteiger et al., 2003). To search for genes similar to DMSP lyase gene from transcriptomic databases, the deduced amino acid sequences of cloned A. digitifera DL-L genes, Emiliania huxleyi Alma1 (KR703620.1), and Symbiodinium A1 DMSP lyase (P0DN22) were used as query sequences in homology searches against the Transcriptome Shotgun Assembly (TSA) database at the National Center for Biotechnology Information (NCBI) using tBlastn (A. digitifera DL-L genes: e-value cutoff 1e-50, query coverage >80%, Emiliania huxleyi Alma1 and Symbiodinium A1 DMSP lyase: e-value cutoff 1e-5, query coverage >70%). Sequences satisfying these criteria were retrieved, and amino acid sequences were prepared using the ExPASy translate tool.
Sequence Alignment and Molecular Phylogenetic Analyses
Sequences downloaded from NCBI TSA databases, including cnidarians, animals, phytoplankton and macroalgae were used for phylogenetic analyses, with bacterial DMSP lyase genes (WP028324949.1, WP012448288.1, WP034745099.1, WP020674609.1, and WP006965703.1) as an outgroup. First, all amino acid sequences were aligned using MAFFT v7.310 (Katoh et al., 2002; Katoh and Standley, 2013), and poorly aligned sequences were removed manually. Gaps in alignments were removed using trimAl v1.2 (Capella-Gutiérrez et al., 2009) with the “gappyout” option. After removing gaps, maximum likelihood analyses were performed using RAxML v8.2.10 (Stamatakis, 2014) with the “bootstrap 100” and “protgammaauto” options.
Results
DL-L Genes Isolated From Acropora digitifera RNAs and Their Gene Expression Patterns
Among 18 DL-L genes predicted from the A. digitifera genome (Shinzato at el., 2021a), we successfully obtained 13 DL-L gene sequences with full-length mRNA coding regions from cDNA (Figure 2; Supplementary Figure 1), demonstrating that multiple DL-L genes exist in the A. digitifera genome and that these are actually expressed as possible functional mRNAs without in-frame stop codons. The resultant 13 amino acid sequences had the same Asp/Glu/hydantoin racemase superfamily conserved domain as in Alma1 of E. huxleyi (Figure 2; Supplementary Figure 1) (Alcolombri et al., 2015). These 18 genes showed different levels and patterns of gene expression in different developmental stages (Supplementary Table 2) as shown by previous studies (Shinzato et al., 2021a; Shinzato et al., 2021b; Yoshioka et al., 2021). Gene expression of five genes was significantly changed (one upregulated and four downregulated) by increased sea water temperature (25°C to 30°C, q < 0.05, Supplementary Table 2).
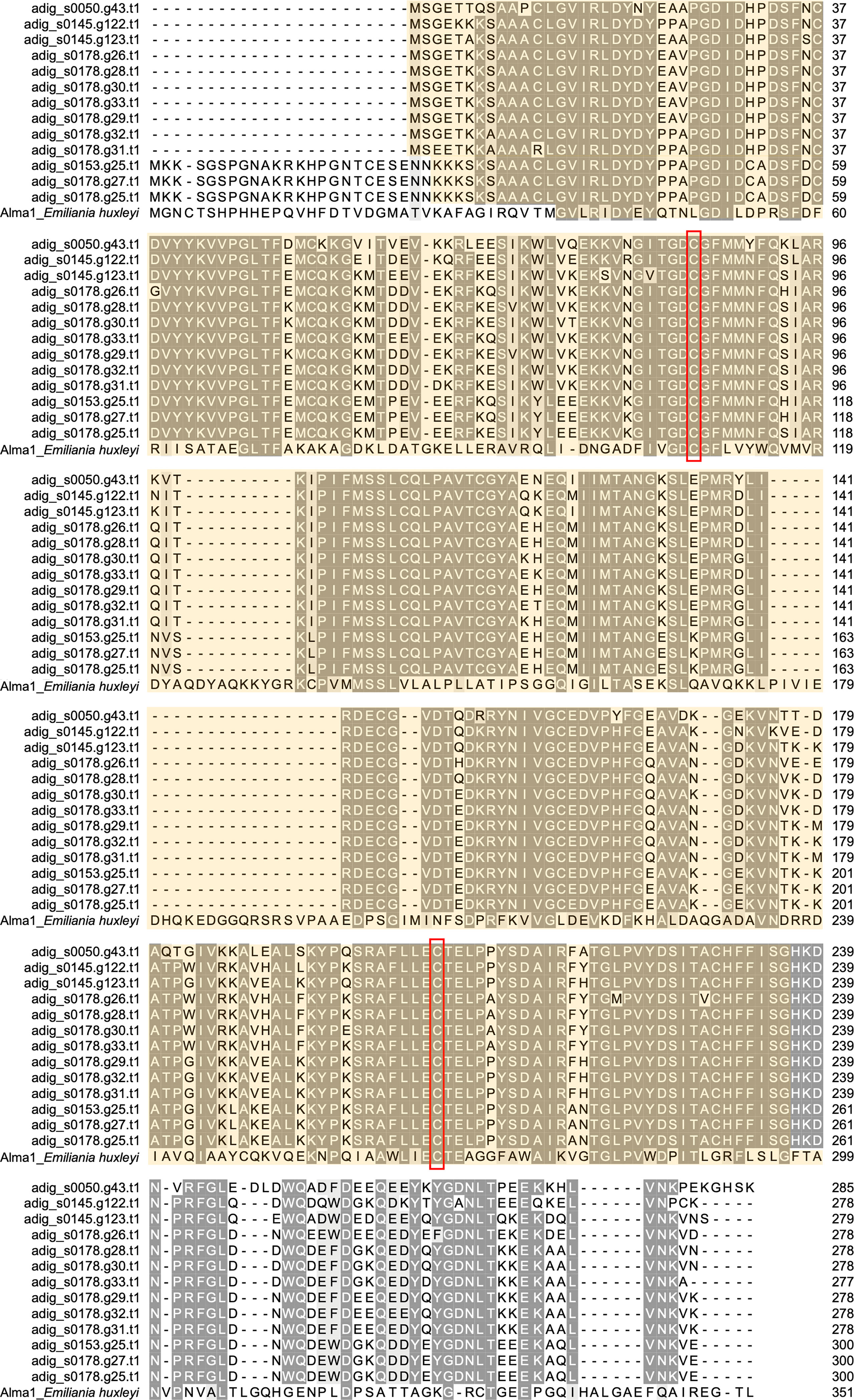
Figure 2 Molecular characterization of DL-L genes in Acropora digitifera. Multiple sequence alignment of deduced amino acid sequences from 13 DL-L genes of A. digitifera and Emiliania huxleyi, Alma1 (KR703620.1). Identical residues are indicated in the same color. Lengths of amino acid sequences are shown at the right and positions of the Asp/Glu/hydantoin racemase superfamily conserved domain are shown in light yellow and the two putative active site residues are shown in red boxes.
DL-L Genes in Eukaryotes
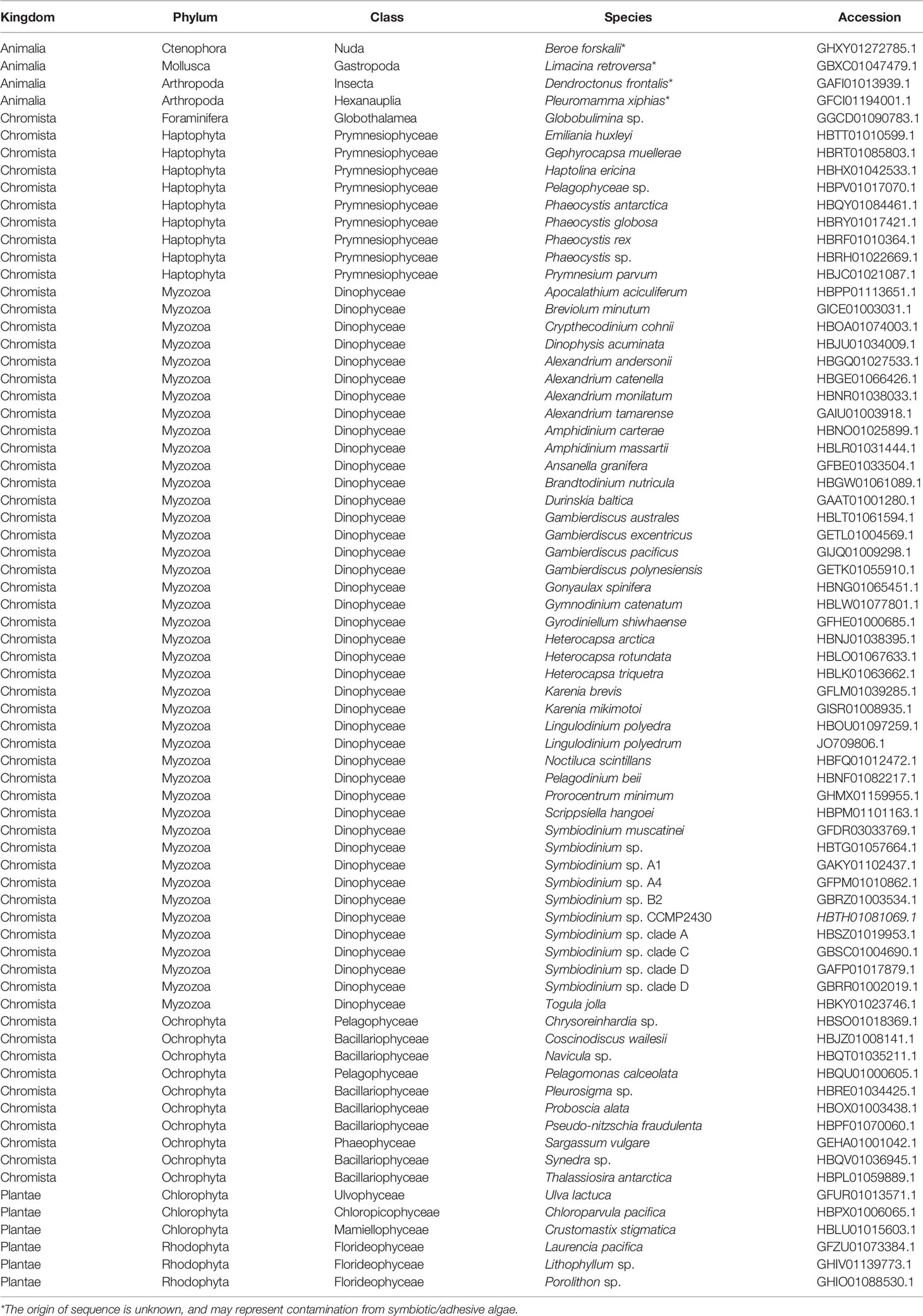
Among the 6,258 eukaryotic TSA databases deposited in NCBI, we identified 123 gene sequences from 72 species that showed similarity to A. digitifera DL-L genes and satisfied the above criteria (tBlastn e-value cutoff 1e-50, query coverage >80%). Most sequences were identified from the Cnidaria (stony corals, sea anemones, jellyfish and hydra), phytoplankton (Phyla Haptophyta, Myzozoa, Ochrophyta) and macroalgae (Phyla Chlorophyta and Rhodophyta) (Table 1). Unexpectedly, four sequences were also identified from other animals, including a ctenophore (Beroe forskalii), a mollusk (Limacina retroversa) and arthropods (Dendroctonus frontalis and Pleuromamma xiphias) (Table 1). We further investigated the amino acid sequence similarity of these sequences with the only known DMSP lyases from eukaryotes to date (Alcolombri et al., 2015), Emiliania huxleyi Alma1 and Symbiodinium A1 DMSP lyase (Supplementary Table 3). Most eukaryote sequences similar to A. digitifera DL-L genes (Table 1) also showed sequence similarities to both DMSP lyases, but percent identities were lower than those to A. digitifera DL-L genes (Supplementary Table 3).
In order to infer evolutionary relationships of the identified sequences from eukaryotes and the four sequences from non-cnidarian animals, we performed molecular phylogenetic analyses with amino acid sequences of the 13 A. digitifera DL-L genes as representatives of Cnidaria and bacterial DMSP lyase genes from prokaryotes as an outgroup. Sequences from prokaryotes and eukaryotes separated into two distinct clusters, indicating that all of the sequences obtained from eukaryote TSA databases originated from eukaryotes, and were not contaminants from symbiotic bacteria (Figure 3; Supplementary Figure 2). All sequences from A. digitifera clustered, demonstrating that they originated from the host coral (Figure 3; Supplementary Figure 2). None of the sequences from non-cnidarian animals clustered with those from A. digitifera. Clustering of the D. frontalis sequence with dinoflagellate sequences displayed 100% bootstrap support, indicating that this sequence was derived from symbiotic or adhesive algae, not from the animal itself. However, the other three sequences from B. forskalii, P. xiphias and L. retroversa did not cluster with those of A. digitifera and other eukaryotes (Figure 3; Supplementary Figure 2); thus, the evolutionary origin (animals or other eukaryotes) are unclear.
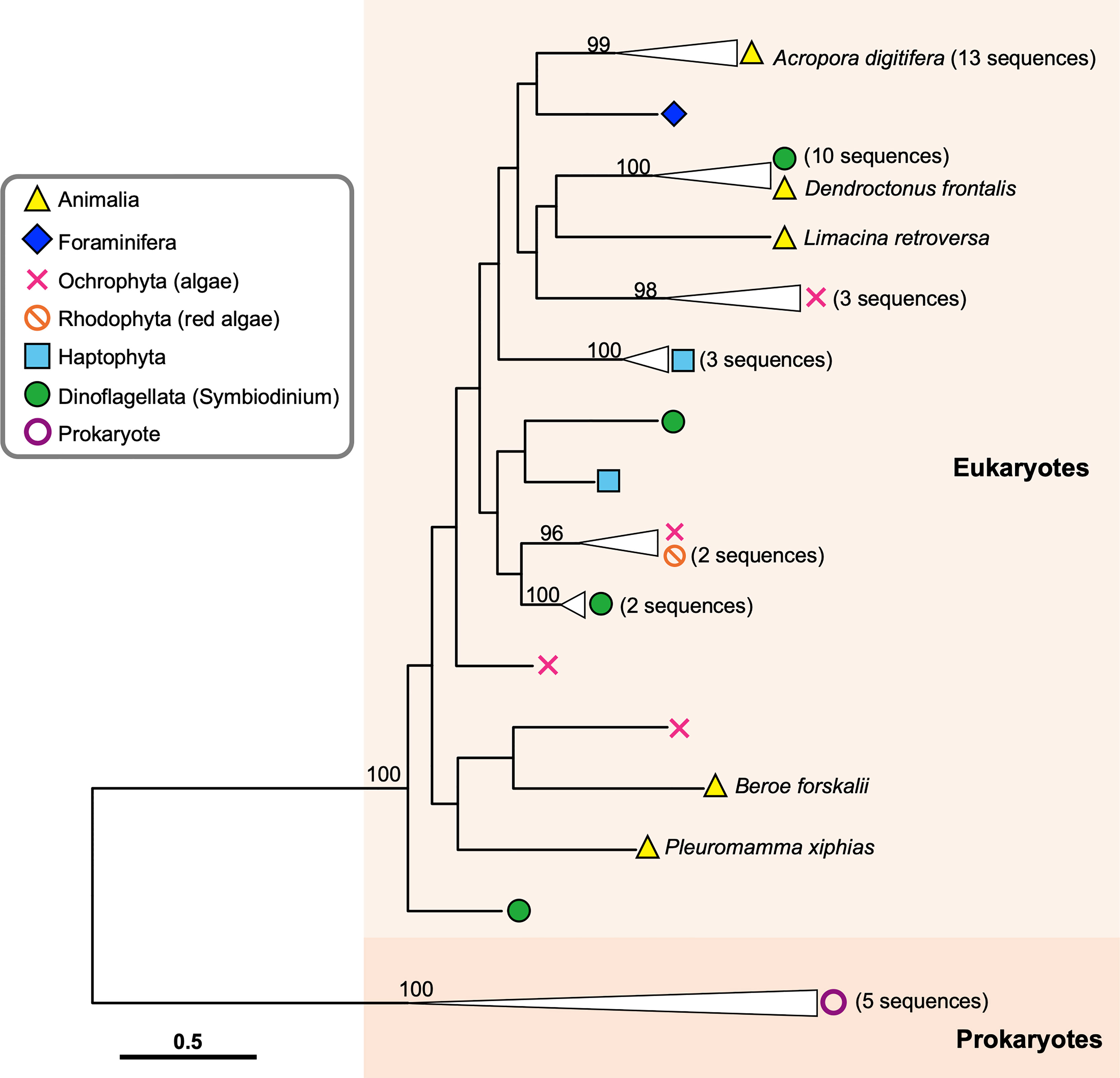
Figure 3 Molecular phylogenetic analysis of DL-L genes identified from transcriptomic databases of eukaryotes. Possible DL-L genes from eukaryotic NCBI TSA databases were analyzed using the maximum likelihood method. 337 gap-trimmed aligned amino acids were used for phylogenetic analysis. Bootstrap support for representative nodes is shown. Eukaryotic and prokaryotic DMSP lyase genes clades are highlighted in different colors. A. digitifera DL-L genes cloned from cDNA are represented as animal genes in the analysis. DL-L genes from the Animalia, Foraminifera, Ochrophyta, Rhodophyta, Haptophyta, Dinoflagellata, and Prokaryota are colored in yellow (triangle), indigo (diamond), pink (cross), orange (prohibited), blue (square), green (circle), and purple (white circle), respectively. The bar indicates 0.5 substitutions per site in aligned regions. The complete phylogenetic tree is shown in Supplementary Figure 2.
DL-L Genes in Cnidaria
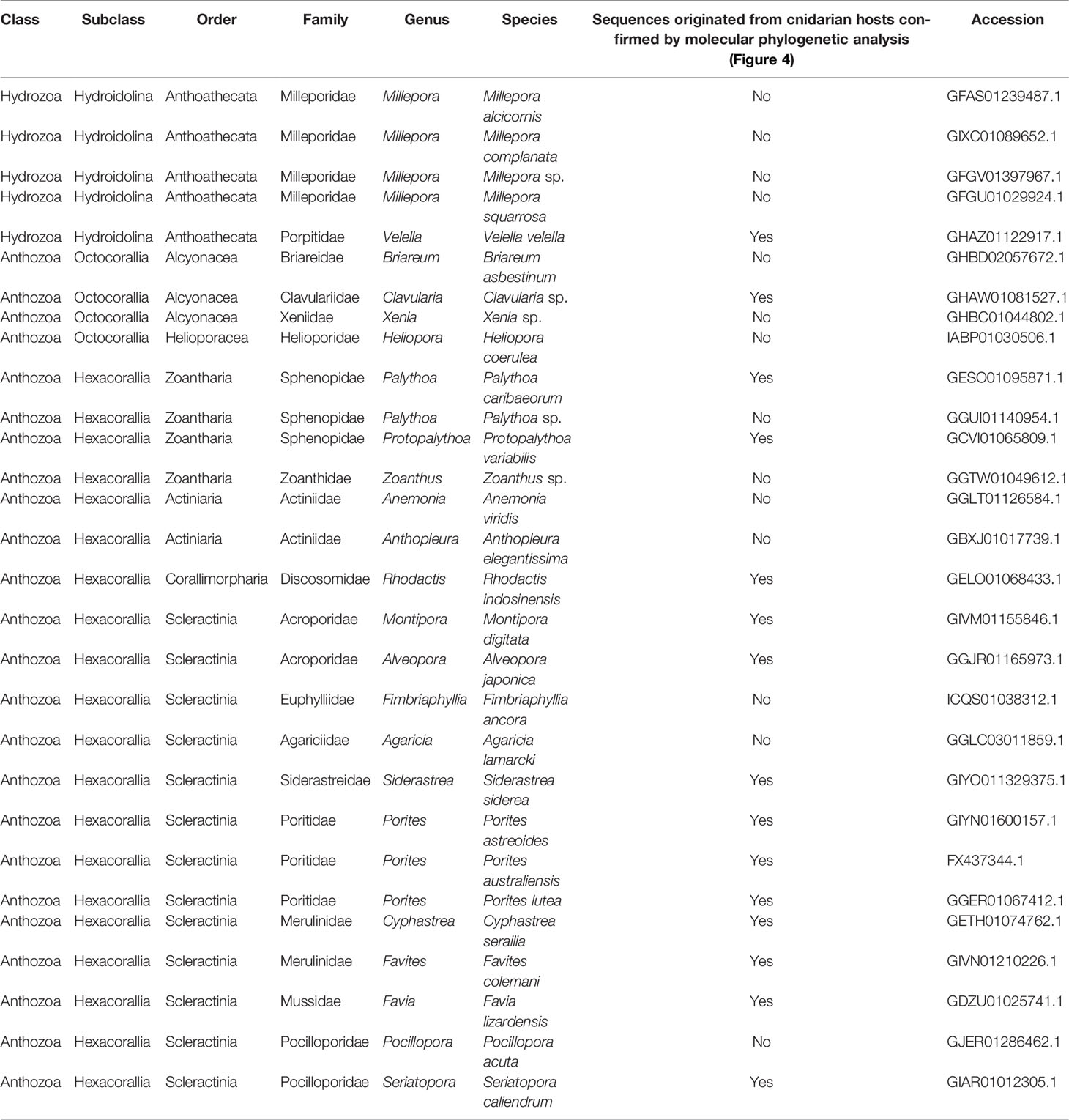
Among 137 Cnidarian TSA databases in NCBI, we found sequences similar to DL-L genes from 35 species (Table 2). Most of these sequences were from Acropora millepora and A. tenuis and had been previously identified (Alcolombri et al., 2015; Shinzato et al., 2021a). Interestingly, possible DL-L genes were found not only in the subclass Hexacorallia, including stony corals, sea anemones, and zoanthids, but also in the Octocorallia, including soft corals and blue corals, and even from Hydroidolina, including jellyfishes and fire corals (Table 2). We also investigated amino acid sequence similarity of these cnidarian sequences with known eukaryote DMSP lyases from E. huxleyi and Symbiodinium A1 (see above), and all sequences except one from Seriatopora caliendrum had significant similarities with these eukaryote DMSP lyases (Supplementary Table 4).
Coral and/or cnidarian transcriptomic databases often contain sequences originating with symbiotic algae, as well as associated microorganisms. In order to identify which sequences were from animal hosts, we further performed molecular phylogenetic analyses of sequences from cnidarian TSA databases together with DL-L genes from the Symbiodiniaceae, coccolithophores and prokaryotes (Figure 4A). Six distinct clades were supported by strong bootstrap probabilities (93 ~ 100%). Most of the sequences belonging to the Hexacorallia clustered together in Clade 1 with A. digitifera DL-L genes, indicating that these originated from hexacorallian hosts. Interestingly, Clavularia sp. of the Octocorallia and Velella velella of the Hydroidolina were also relegated to Clade 1, indicating that these share a common ancestral gene with hexacorallian species. The family Symbiodiniaceae is genetically diverse, and includes multiple genera (LaJeunesse et al., 2018). As expected, some Cnidarian sequences clustered with sequences of symbiotic algae (Symbiodiniaceae) and were assigned to Clades 3-5 (Figure 4A; Supplementary Figure 3), indicating that these were contaminant from symbiotic algae. Among them, a sequence from Pocillopora acuta was clustered into Clade 3, sequences from Anthopleura elegantissima, Millepora squarrosa, Agaricia lamarcki, Heliopora coerulea, Zoanthus sp., Palythoa sp., M. alcicornis, Millepora sp. were clustered in Clade 4, and sequences from Fimbriaphyllia ancora, M. complanata, and Xenia sp. were clustered in Clade 5 (Figure 4A; Supplementary Figure 3; Table 2). Different cnidarian species harbor different types of symbiotic algae; thus, Clades 3-5, containing Symbiodiniaceae could reflect different types of symbiotic algae. All DL-L genes of the coccolithophore E. huxleyi within the Haptophyta clustered in Clade 2, and bacterial DMSP lyase genes clustered in Clade 6. Taken together, all DL-L genes of Cnidaria, including scleractinian corals, soft corals, and jellyfishes, evolved from an ancestral gene that already existed in the last common ancestor of Anthozoa and Hydrozoa, and have completely different evolutionary backgrounds from those of coccolithophores, the Family Symbiodiniaceae, or prokaryotes.
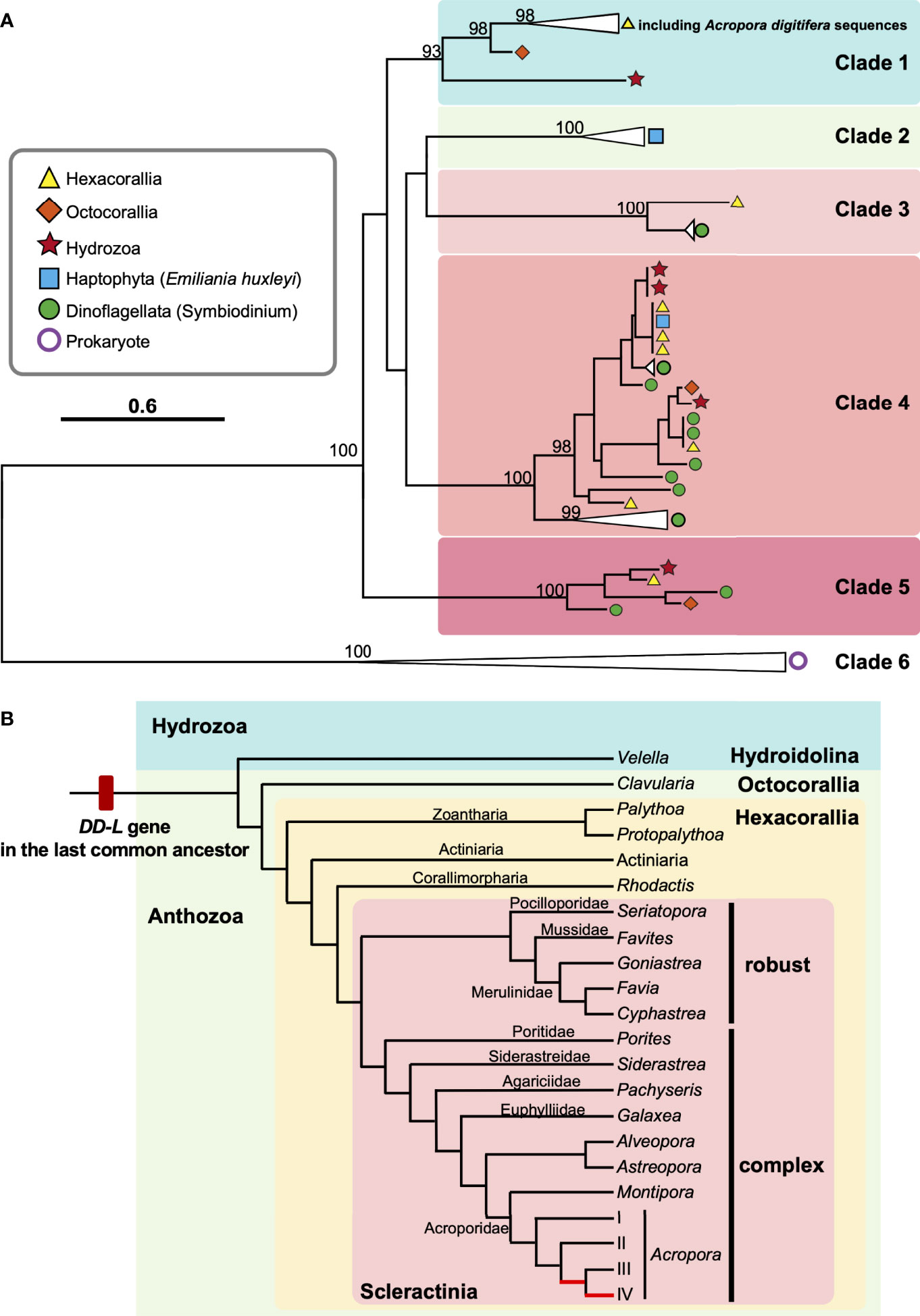
Figure 4 Molecular phylogenetic analysis of DL-L genes identified from transcriptomic databases of Cnidaria. (A) Possible DL-L genes from cnidarian NCBI TSA databases were analyzed using the maximum likelihood method. 667 gap-trimmed aligned amino acids were used for phylogenetic analysis. Bootstrap support for representative nodes is shown. Clades 1-6 of DL-L genes of cnidarians, other eukaryotes and prokaryotes are highlighted in different colors. (A) digitifera DL-L genes identified from cDNA, which were all clustered in Clade 1, are also included. DL-L genes from Hexacorallia, Octocorallia, Hydrozoa, Haptophyta, Dinoflagellata, and Prokaryota are colored in yellow (triangle), orange (diamond), red (start), blue (square), green (circle), and purple (white circle), respectively. The bar indicates 0.6 substitutions per site in aligned regions. The complete phylogenetic tree is shown in Supplementary Figure 3. (B) Updated evolutionary history of DL-L gene in the Phylum Cnidaria. Only species of cnidarians possessing animal-type DL-L genes are shown. Two Acropora-specific gene expansion events proposed by Shinzato et al. (2021a) are shown in red in the phylogenetic tree. Phylogenetic relationships of cnidarians and scleractinian corals are derived from Kayal et al. (2018) and Kitahara et al. (2016).
Discussion
A Variety of Scleractinian Corals Possess DL-L Genes
It has been reported that coral reefs are hotspots for DMSP, which have been attributed to symbiotic algae of corals (Broadbent et al., 2002; Broadbent and Jones, 2004; Broadbent and Jones, 2006; Swan et al., 2012). Recent studies have shown that adult and juvenile A. tenuis and A. millepora without symbiotic algae both produce DMSP (Raina et al., 2013). DL-L genes have also been discovered, not only in Acropora genomes (Alcolombri et al., 2015; Shinzato et al., 2021a), but also in genomes of Montipora, Astreopora, Goniastrea, and two corallimorpharians, Amplexidiscus and Discosoma (Shinzato et al., 2021a).
In this study, we successfully identified full-length ORFs of 13 DL-L genes, which are expressed in A. digitifera, and we identified DL-L genes from a variety of scleractinian lineages (Figures 1, 4A; Supplementary Figure 3). The earliest coral fossil record of a Scleractinian dates to the middle Triassic (240 Ma) (Stanley, 2003). Most extant scleractinians are classified into two major clades, known as the Complexa (complex corals) and Robusta (robust corals), and are assumed to have diverged in the Late Carboniferous (300 Ma) (Romano and Palumbi, 1996; Romano and Cairns, 2000). In genomes of robust corals, a DL-L gene has been detected only in Goniastrea aspera to date (Shinzato et al., 2021a). However, in this study, we identified animal-type DL-L genes from four robust corals, including Cyphastrea serailia, Favites colemani, Favia lizardensis, and Seriatopora caliendrum (Figure 4B; Table 2), indicating that DL-L genes have been preserved in a variety of complex and robust corals, but were lost from some lineages.
Ancient Origin of DL-L Genes in Cnidarians
Although a large proportion of DL-L genes were identified in corals and cnidarians, we also identified similar sequences from non-cnidarian animals, B. forskalii, P. xiphias and L. retroversa. However, none of these share a common ancestry with coral DL-L genes (Figure 3). Although the origins of these sequences are not clear, we suggest that they may have come from symbiotic/adhesive algae, as in the case of a sequence from D. frontalis (Figure 3). Based on our phylogenetic analyses using currently available transcriptomic databases, we conclude that, to date, only cnidarians possess DL-L genes that are clearly of animal origin. Further addition of more phytoplankton DL-L genes may reveal the origins of these from non-cnidarian species.
The Phylum Cnidaria contains three clades (subphyla): Anthozoa (comprising the Octocorallia, Hexacorallia, and Ceriantharia), Endocnidozoa (a clade of parasites) and Medusozoa (consisting of the Cubozoa, Hydrozoa, Scyphozoa, and Staurozoa) (Collins, 2009; Kayal et al., 2018). In this study, we identified eukaryotic DL-L genes, not only in the Scleractinia and Corallimorpharia but also in the Zoantharia (Hexacorallia), Alcyonacea (Octocorallia), and Hydrozoa (Anthoathecata, Hydroidolina) (Figure 4B; Table 2). This indicates that the last common ancestor of Anthozoa and Hydrozoa possessed a DL-L gene and that this gene has been inherited in a wide range of cnidarian species. The earliest cnidarian fossils occur in strata of the Ediacaran (560 Ma, Liu et al., 2014), and together with molecular and paleontological analyses, suggest that the phylum Cnidaria probably originated during the Precambrian Eon from the Cryogenian to the Ediacaran (700-595 Ma, Peterson et al., 2004; Erwin et al., 2011). Divergence of 2 major taxa (Anthozoaria and Medusozoa) may have occurred before the Cambrian (543 Ma) (Cartwright et al., 2007; Park et al., 2012). Consequently, we hypothesize that the DL-L gene may be have originated from an ancient gene that already existed in the last common ancestor of Cnidaria in the pre-Cambrian (Figure 4B).
How did cnidarians, including corals, obtain DL-L genes? Shinzato et al. (2021a) hypothesized that DL-L genes may have been acquired by the common ancestor of scleractinians and corallimorpharians, both of which are hexacorallians (Anthozoa), via horizontal gene transfer from symbiotic Symbiodiniaceae or Emiliania. However, as mentioned above, a variety of anthozoans and hydrozoans possess these genes (Figure 4A). McFadden et al. (2021) suggested that the Anthozoa probably arose in the Cryogenian to Tonian periods (648-894 Ma) and lacked photosymbionts. In the Devonian (383 Ma) from the Palaeozoic Era, anthozoans of the Scleractinia first formed associations with photosymbionts, followed by alcyonacean octocorals (318 Ma) and corallimorpharians (312 Ma), and photosymbioses have been gained and lost repeatedly in all orders through the Jurassic (199-151 Ma, McFadden et al., 2021). In addition, the fossil record shows that scleractinian corals had photosymbionts in most of the upper Triassic since the Mesozoic Era (Stolarski et al., 2011; Frankowiak et al., 2016). Recent molecular dating estimates suggest that the earliest diversification of the Symbiodiniaceae occurred in the Jurassic (∼160 Ma, LaJeunesse et al., 2018) and the first E. huxleyi appeared 270,000 years ago (Thierstein et al., 1977; Paasche, 2001), which were much later than the origin of the Anthozoa. Our phylogenetic analysis (Figure 4) confirms distinct ancestries of DL-L genes in Symbiodiniaceae and cnidarians, indicating that they have completely different evolutionary backgrounds. Therefore, acquisition of DL-L genes via horizontal gene transfer from symbiotic algae (Shinzato et al., 2021a) may not have occurred. Based on the present findings, we propose an updated hypothesis: DL-L genes in cnidarians are “ancient genes” in the animal kingdom, dating back to the pre-Cambrian.
DL-L Genes: Essential for Survival in Coral Reef or Shallow and Warm Water Environments?
As of 10th February 2022, 137 transcriptomic databases from 91 cnidarian species had been registered in NCBI TSA. Interestingly, we realized that all cnidarian species, except V. velella, possessing animal type DL-L genes (15 species) are limited to coral reefs (Table 1). V. velella (Figure 4; Table 2), known as “by-the-wind sailor”, is a cosmopolitan, free-floating, colonial hydrozoan that lives mainly at the water/air interface and floats in temperate and tropical seas (Araya and Aliaga, 2018). This implies that DL-L genes may be essential for animals to survive in coral reef or warm, shallow-water environments, although we acknowledge that publicly available transcriptomic databases of cnidarians to date may be limited to coral reef species.
Comparative genomic analysis using reported scleractinian coral genomes showed that DL-L genes are the most diversified gene family among gene families that significantly increased number of genes in the last common ancestor of Acropora (Shinzato et al., 2021a). Why did gene duplication events of DL-L gene specifically occur in the Acropora lineage? The earliest fossil records of Acropora are known from the late Paleocene (65-54 Ma) in Somalia and Austria (Carbone et al., 1993; Baron-Szabo, 2006), indicating that Acropora has existed for more than 50 million years. Acropora was the main reef builder in the Oligocene (28-23 Ma) of Greece and Early Miocene of Egypt (Schuster, 2002), and was widely distributed throughout the Miocene in different regions, including the Indo-Pacific and Caribbean (Budd, 2000). It is suggested that Acropora began to spread throughout the world in the Cenozoic and species diversification occurred in the Eocene and Oligocene (around 25-50 Ma). Molecular dating analysis using whole-genome data indicates that the Acropora ancestor survived warm periods without sea ice from the mid or late Cretaceous to the Early Eocene, when gene expansion of DL-L genes occurred specifically in the Acropora ancestor (Shinzato et al., 2021a). In addition, Acropora species also have high concentrations of DMSP compared to other corals (Broadbent et al., 2002; Guibert et al., 2020), suggesting that Acropora corals actively utilize DMSP and that high concentrations of DMSP may have triggered Acropora-specific duplication of DL-L genes. Several studies have shown that DMSP is involved in a wide range of coral stress responses, including responses to heat, sunlight, air exposure, and hyposalinity (Sunda et al., 2002; Raina et al., 2009; Deschaseaux et al., 2014; Aguilar et al., 2017). Not all A. digitifera DL-L genes (5 out of 18) responded to increased sea water temperature and response patterns of differentially expressed genes vary (1 upregulated and 4 downregulated, Supplementary Table 2), suggesting that the functions of the DL-L genes in A. digitifera have also diverged. Taken together, diversified DL-L genes in Acropora may have acquired new or different functions, not only mediating cleavage of DMSP into DMS, but that they may assist Acropora in adapting to environmental changes, for example from intense heat, light, and salinity (Shinzato et al., 2021a). Eventually Acropora may become the dominant coral genus in extant coral reefs.
The functions of DL-L genes in corals are completely undetermined at this stage. Interestingly, although overexpression of the Alma l genes from E. huxleyi and Symbiodinium A1 in E. coli cells had high DMSP lyase activities, an Alma1 homolog from A. millepora showed almost no DMSP lyase activity (Alcolombri et al., 2015), indicating that not all DL-L genes in eukaryotes retain their DMSP lyase function, and that acquisition of other functions or functional differentiation may have occurred during the process of gene duplication. In particular, duplicated DL-L genes in Acropora will need to be investigated to determine which genes have DMSP lyase activity. We identified expressed sequences of DL-L genes from A. digitifera RNA, and these could be used for molecular and functional characterization in DMSP lyase assays. We also found that expression levels of DL-L genes were indeed diverse in A. digitifera. Therefore, even if some DL-L genes in corals do not function as DMSP lyases, they may have other functions enabling adaptation to coral reef or warm, shallow-water environments. Identifying their actual biological functions in Acropora corals will be important to understand not only adaptation mechanisms to shallow and warmer environments, but also the impact of Acropora corals on the sulfur cycle in the oceans.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CS conceptualized, commenced and supervised the project. Y-LC performed the lab work, database searches and molecular phylogenetic analyses. Y-LC and CS analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported with funding from the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grants (20H03235 and 20K21860) to CS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research was supported by a grant from Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) grants (20H03235 and 20K21860) to CS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.889866/full#supplementary-material
Supplementary Figure 1 | Domain structure of the 13 deduced amino acid sequences of A. digitifera and Emiliania huxleyi, Alma1 (KR703620.1). Lengths of amino acid sequences are shown at the right and positions of the Asp/Glu/hydantoin racemase superfamily conserved domain are shown in gray boxes. Scale bar=20 aa, aa: the number of amino acids.
Supplementary Figure 2 | Maximum likelihood analysis of DL-L genes identified from transcriptomic databases of eukaryotes. A total of 65 DL-L genes from prokaryotes and eukaryotes, including A. digitifera DL-L genes, were aligned using MAFFT v7.310. Then, 337 gap-trimmed aligned amino acid sequences were used for the phylogenetic analysis. Each species name is followed by a different symbol indicating the taxonomy of the species. The bar indicates 0.5 substitutions per site in aligned regions.
Supplementary Figure 3 | Maximum likelihood analysis of DL-L genes identified from transcriptomic databases of cnidarians. A total of 65 DL-L genes from cnidaria and prokaryotes including, A. digitifera DL-L genes, were aligned using MAFFT v7.310. Then, 337 gap-trimmed aligned amino acid sequences were used for the phylogenetic analysis. Each species name is followed by a different symbol indicating the taxonomy of the species. The bar indicates 0.6 substitutions per site in aligned regions.
Supplementary Table 1 | List of the primers for cloning PCR analysis.
Supplementary Table 2 | Gene expression levels (counts per million) of Acropora digitifera DL-L genes in different developmental stages (Shinzato et al., 2021a and Yoshioka et al., 2021) and increased sea water temperature (Shinzato et al., 2021b).
Supplementary Table 3 | DL-L genes in Eukaryotes, excluding cnidarians. Emiliania huxleyi Alma1 (Accession: KR703620.1) and Symbiodinium A1 DMSP lyase (Accession: P0DN22) were used as query sequences in homology searches against the TSA at NCBI using tBlastn (e-value cutoff 1e-5, query range >70%).
Supplementary Table 4 | DL-L genes in cnidarians excluding Acropora. Emiliania huxleyi Alma1 (Accession: KR703620.1) and Symbiodinium A1 DMSP lyase (Accession: P0DN22) were used as query sequences in homology searches against the TSA at NCBI using tBlastn (e-value cutoff 1e-5, query range >70%).
References
Aguilar C., Raina J. B., Motti C. A., Fôret S., Hayward D. C., Lapeyre B., et al. (2017). Transcriptomic Analysis of the Response of Acropora Millepora to Hypo-Osmotic Stress Provides Insights Into DMSP Biosynthesis by Corals. BMC Genomics 18 (1), 612. doi: 10.1186/s12864-017-3959-0
Ainsworth T. D., Thurber R. V., Gates R. D. (2010). The Future of Coral Reefs: A Microbial Perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Alcolombri U., Ben-Dor S., Feldmesser E., Levin Y., Tawfik D. S., Vardi A. (2015). Identification of the Algal Dimethyl Sulfide-Releasing Enzyme: A Missing Link in the Marine Sulfur Cycle. Science 348(6242), 1466–1469. doi: 10.1126/science.aab1586
Araya J., Aliaga J. (2018). El Niño Invaders: The Occurrence of the by-the-Wind Sailor Velella Velella (Linnaeus 1758) in the Southeastern Pacific. Spixiana 14, 132. doi: 10.3800/pbr.14.206
Ayers G. P., Gras J. L. (1991). ). Seasonal Relationship Between Cloud Condensation Nuclei and Aerosol Methanesulphonate in Marine Air. Nature 353, 834–835. doi: 10.1038/353834a0
Barak-Gavish N., Frada M. J., Ku C., Lee P. A., DiTullio G. R., Malitsky S., et al. (2018). Bacterial Virulence Against an Oceanic Bloom-Forming Phytoplankter is Mediated by Algal DMSP. Sci. Adv. 4 (10) eaau5716. doi: 10.1126/sciadv.aau5716
Baron-Szabo R. C. (2006). Corals of the K/T-Boundary: Scleractinian Corals of the Suborders Astrocoeniina, Faviina, Rhipidogyrina and Amphiastraeina. J. Syst. Palaeontol. 4, 1. doi: 10.1017/S1477201905001689
Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 34 (5), 525–527. doi: 10.1038/nbt.3519
Broadbent A. D., Jones G. B. (2004). DMS and DMSP in Mucus Ropes, Coral Mucus, Surface Films and Sediment Pore Waters From Coral Reefs in the Great Barrier Reef. Mar. Freshw. Res. 55, 849–855. doi: 10.1071/MF04114
Broadbent A., Jones G. (2006). Seasonal and Diurnal Cycles of Dimethylsulfide, Dimethylsulfoniopropionate and Dimethylsulfoxide at One Tree Reef Lagoon. Environ. Chem. Lett. 3, 260–267. doi: 10.1071/EN06011
Broadbent A. D., Jones G. B., Jones R. J. (2002). DMSP in Corals and Benthic Algae From the Great Barrier Reef. Estuar. Coast. Shelf. Sci. 55, 547–555. doi: 10.1006/ecss.2002.1021
Budd A. F. (2000). Diversity and Extinction in the Cenozoic History of Caribbean Reefs. Coral. Reefs. 19, 25–35. doi: 10.1007/s003380050222
Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carbone F., Matteucci R., Pignatti J. S., Russo A. (1993). Facies Analysis and Biostratigraphy of the Auradu Limestone Formation in the Berbera-Sheikh Area, Northwestern Somalia. Geol. Rom. 29, 213–235.
Cartwright P., Halgedahl S. L., Hendricks J. R., Jarrard R. D., Marques A. C., Collins A. G., et al. (2007). Exceptionally Preserved Jellyfishes From the Middle Cambrian. PLoS One 2, e1121. doi: 10.1371/journal.pone.0001121
Chiu Y. L., Shikina S., Yoshioka Y., Shinzato C., Chang C. F (2020). CDe Novo Transcriptome Assembly From The Gonads of a Scleractinian Coral, Euphyllia Ancora: Molecular Mechanisms Underlying Scleractinian Gametogenesis. PBMC Genomics 21 (1), 732. doi: 10.1186/s12864-020-07113-9
Cleves P. A., Strader M. E., Bay L. K., Pringle J. R., Matz M. V. (2018). CRISPR/Cas9-Mediated Genome Editing in a Reef-Building Coral. Proc. Natl. Acad. Sci. U. S. A. 115, 5235–5240. doi: 10.1073/pnas.1722151115
Collins A. G. (2009). Recent Insights Into Cnidarian Phylogeny. Smithson. Contrib. Mar. Sci. 38, 139–149.
Curson A. R., Liu J., Bermejo Martínez A., Green R. T., Chan Y., Carrión O., et al. (2017). Dimethylsulfoniopropionate Biosynthesis in Marine Bacteria and Identification of the Key Gene in This Process. Nat. Microbiol. 2, 17009. doi: 10.1038/nmicrobiol.2017.9
Curson A., Williams B. T., Pinchbeck B. J., Sims L. P., Martínez A. B., Rivera P., et al. (2018). DSYB Catalyses the Key Step of Dimethylsulfoniopropionate Biosynthesis in Many Phytoplankton. Nat. Microbiol. 3, 430–439. doi: 10.1038/s41564-018-0119-5
Davy S. K., Allemand D., Weis V. M. (2012). Cell Biology of Cnidarian-Dinoflagellate Symbiosis. Microbiol. Mol. Biol. Rev. 76 (2), 229–261. doi: 10.1128/MMBR.05014-11
Deschaseaux E. S., Jones G. B., Deseo M. A., Shepherd K. M., Kiene R. P., Swan H. B., et al. (2014). Effects of Environmental Factors on Dimethylated Sulfur Compounds and Their Potential Role in the Antioxidant System of the Coral Holobiont. Limnol. Oceanogr. 59 (3), 758–768. doi: 10.4319/lo.2014.59.3.0758
Erwin D. H., Laflamme M., Tweedt S. M., Sperling E. A., Pisani D., Peterson K. J. (2011). The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals. Science 334, 1091–1097. doi: 10.1126/science.1206375
Frankowiak K., Wang X. T., Sigman D. M., Gothmann A. M., Kitahara M. V., Mazur M., et al. (2016). Photosymbiosis and the Expansion of Shallow-Water Corals. Sci. Adv. 2, e1601122. doi: 10.1126/sciadv.1601122
Gage D. A., Rhodes D., Nolte K. D., Hicks W. A., Leustek T., Cooper A. J., et al. (1997). A New Route for Synthesis of Dimethylsulphoniopropionate in Marine Algae. Nature 387 (6636), 891–894. doi: 10.1038/43160
Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., Bairoch A. (2003). ExPASy: The Proteomics Server for In-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Goreau T. F., Goreau N. I. (1959). The Physiology of Skeleton Formation in Corals. II. Calcium Deposition by Hermatypic Corals Under Various Conditions in the Reef. Biol. Bull. 117, 239–250. doi: 10.2307/1538903
Guibert I., Bourdreux F., Bonnard I., Pochon X., Dubousquet V., Raharivelomanana P., et al. (2020). Dimethylsulfoniopropionate Concentration in Coral Reef Invertebrates Varies According to Species Assemblages. Sci. Rep. 10, 9922. doi: 10.1038/s41598-020-66290-5
Hanson A. D., Rivoal J., Paquet L., Gage D. A. (1994). Biosynthesis of 3-Dimethylsulfoniopropionate in Wollastonia Biflora (L.) DC. (Eevidence That S-Methylmethionine Is an Intermediate). Plant Physiol. 105, 103–110. doi: 10.1104/pp.105.1.103
Hill R. W., White B. A., Cottrell M. T., Dacey J. W. (1998). Virus-Mediated Total Release of Dimethylsulfoniopropionate From Marine Phytoplankton: A Potential Climate Process. Aquat. Microb. Ecol. 14, 1–6. doi: 10.3354/ame014001
Hislop N. R., de Jong D., Hayward D. C., Ball E. E., Miller D. J. (2005). Tandem Organization of Independently Duplicated Homeobox Genes in the Basal Cnidarian Acropora Millepora. Dev. Genes Evol. 215, 268–273. doi: 10.1007/s00427-005-0468-y
Husband J. D., Kiene R. P., Sherman T. D. (2012). Oxidation of Dimethylsulfoniopropionate (DMSP) in Response to Oxidative Stress in Spartina Alterniflora and Protection of a Non-DMSP Producing Grass by Exogenous DMSP+ Acrylate. Environ. Exp. Bot. 79, 44–48. doi: 10.1016/j.envexpbot.2012.01.006
Karsten U., Kück K., Vogt C., Kirst G. O. (1996). “Dimethylsulfoniopropionate Production in Phototrophic Organisms and its Physiological Functions as a Cryoprotectant,” in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds. Eds. Keller M. D., Kiene R. P., Kirst G. O., Visscher P. T. (Boston, MA: Springer), 143–153.
Katoh K., Misawa K., Kuma K. I., Miyata T. (2002). MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Katoh K., Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Kayal E., Bentlage B., Sabrina Pankey M., Ohdera A. H., Medina M., Plachetzki D. C., et al. (2018). Phylogenomics Provides a Robust Topology of the Major Cnidarian Lineages and Insights on the Origins of Key Organismal Traits. BMC Evol. Biol. 18, 1–18. doi: 10.1186/s12862-018-1142-0
Keller M. D., Bellows W. K., Guillard R. R. L. (1989). “Dimethylsulfide Production in Marine Phytoplankton,” in Biogenic Sulfur in the Environment. Eds. Saltzman E. S., Cooper W. J. (Washington, DC: American Chemical Society), 167–182.
Kettles N. L., Kopriva S., Malin G. (2014). Insights Into the Regulation of DMSP Synthesis in the Diatom Thalassiosira Pseudonana Through APR Activity, Proteomics and Gene Expression Analyses on Cells Acclimating to Changes in Salinity, Light and Nitrogen. PLoS One 9, e94795. doi: 10.1371/journal.pone.0094795
Kirst G. O., Thiel C., Wolff H., Nothnagel J., Wanzek M., Ulmke R. (1991). Dimethylsulfoniopropionate (DMSP) in Icealgae and its Possible Biological Role. Mar. Chem. 35 (1-4), 381–388. doi: 10.1016/S0304-4203(09)90030-5
Kitahara M. V., Fukami H., Benzoni F., Huang D. (2016). “The New Systematics of Scleractinia: Integrating Molecular and Morphological Evidence,” in The Cnidaria, Past, Present and Future. Eds. Goffredo S., Dubinsky Z. (Switzerland: Springer), 41–59.
Knowlton N., Brainard R. E., Fisher R., Moews M., Plaisance L., Caley M. J. (2010). Coral Reef Biodiversity. Life World’s Oceans: Diversity Distribution Abundance, editor McIntyre A. (Wiley-Blackwell: Oxford), 65–7465–74. doi: 10.1002/9781444325508.ch4
Kocsis M. G., Nolte K. D., Rhodes D., Shen T. L., Gage D. A., Hanson A. D. (1998). Dimethylsulfoniopropionate Biosynthesis in Spartina Alterniflora1. Evidence That S-Methylmethionine and Dimethylsulfoniopropylamine Are Intermediates. Plant Physiol. 117, 273–281. doi: 10.1104/pp.117.1.273
LaJeunesse T. C., Parkinson J. E., Gabrielson P. W., Jeong H. J., Reimer J. D., Voolstra C. R., et al. (2018). Systematic Revision of Symbiodiniaceae Highlights the Antiquity and Diversity of Coral Endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Lesser M. P. (2006). Oxidative Stress in Marine Environments: Biochemistry and Physiological Ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Li H. (2018). Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 34 (18), 3094–3100. doi: 10.1093/bioinformatics/bty191
Liu A. G., Matthews J. J., Menon L. R., McIlroy D., Brasier M. D. (2014). Haootia Quadriformis N. Gen. N. sp. Interpreted Muscular Cnidarian Impression Late Ediacaran Period (Approx. 560 Ma). Proc. R. Soc. B.: Biol. Sci. 281 (1793), 20141202. doi: 10.1098/rspb.2014.1202
Li C. Y., Wang X. J., Chen X. L., Sheng Q., Zhang S., Wang P., et al. (2021). A Novel ATP Dependent Dimethylsulfoniopropionate Lyase in Bacteria That Releases Dimethyl Sulfide and Acryloyl-CoA. eLife 10, e64045. doi: 10.7554/eLife.64045
Lyon B. R., Lee P. A., Bennett J. M., DiTullio G. R., Janech M. G. (2011). Proteomic Analysis of a Sea-Ice Diatom: Salinity Acclimation Provides New Insight Into the Dimethylsulfoniopropionate Production Pathway. Plant Physiol. 157, 1926–1941. doi: 10.1104/pp.111.185025
Maor-Landaw K., Levy O. (2016). “Survey of Cnidarian Gene Expression Profiles in Response to Environmental Stressors: Summarizing 20 Years of Research, What are We Heading for?,” in The Cnidaria, Past, Present and Future. Eds. Goffredo S., Dubinsky Z. (Switzerland: Springer), 523–543.
Martin M. (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet. J. 17 (1), 10–12. doi: 10.14806/ej.17.1.200
McCarthy D. J., Chen Y., Smyth G. K. (2012). Differential Expression Analysis of Multifactor RNA-Seq Experiments With Respect to Biological Variation. Nucleic Acids Res. 40 (10), 4288–4297. doi: 10.1093/nar/gks042
McFadden C. S., Quattrini A. M., Brugler M. R., Cowman P. F., Dueñas L. F., Kitahara M. V., et al. (2021). Phylogenomics, Origin, and Diversification of Anthozoans (Phylum Cnidaria). Syst. Biol. 70, 635–647. doi: 10.1093/sysbio/syaa103
Miller T. R., Belas R. (2004). Dimethylsulfoniopropionate Metabolism by Pfiesteria-Associated Roseobacter Spp. Appl. Environ. Microbiol. 70, 3383–3391. doi: 10.1128/AEM.70.6.3383-3391.2004
Muscatine L. (1990). The Role of Symbiotic Algae in Carbon and Energy Flux in Reef Corals. Coral. Reefs. 25, 1–29.
Muscatine L. E. O. N. A. R. D., Porter J. W. (1977). Reef Corals: Mutualistic Symbioses Adapted to Nutrient-Poor Environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Niki T., Kunugi M., Otsuki A. (2000). DMSP-Lyase Activity in Five Marine Phytoplankton Species: Its Potential Importance in DMS Production. Mar. Biol. 136, 759–764. doi: 10.1007/s002279900235
Paasche E. (2001). A Review of the Coccolithophorid Emiliania Huxleyi (Prymnesiophyceae), With Particular Reference to Growth, Coccolith Formation, and Calcification-Photosynthesis Interactions. Phycologia 40 (6), 503–529. doi: 10.2216/i0031-8884-40-6-503.1
Paquet L., Rathinasabapathi B., Saini H., Zamir L., Gage D. A., Huang Z. H., et al. (1994). Accumulation of the Compatible Solute 3-Dimethylsulfoniopropionate in Sugarcane and its Relatives, But Not Other Gramineous Crops. Funct. Plant Biol. 21, 37–48. doi: 10.1071/PP9940037
Park E., Hwang D. S., Lee J. S., Song J. I., Seo T. K., Won Y. J. (2012). Estimation of Divergence Times in Cnidarian Evolution Based on Mitochondrial Protein-Coding Genes and the Fossil Record. Mol. Phylogenet. Evol. 62 (1), 329–345. doi: 10.1016/j.ympev.2011.10.008
Peterson K. J., Lyons J. B., Nowak K. S., Takacs C. M., Wargo M. J., McPeek M. A. (2004). Estimating Metazoan Divergence Times With a Molecular Clock. Proc. Natl. Acad. Sci. U. S. A. 101 (17), 6536–6541. doi: 10.1073/pnas.0401670101
Qin Z., Yu K., Chen B., Wang Y., Liang J., Luo W., et al. (2019). Diversity of Symbiodiniaceae in 15 Coral Species From the Southern South China Sea: Potential Relationship With Coral Thermal Adaptability. Front. Microbiol. 10, 2343. doi: 10.3389/fmicb.2019.02343
Quinn P. K., Bates T. S. (2011). The Case Against Climate Regulation via Oceanic Phytoplankton Sulphur Emissions. Nature 480 (7375), 51–56. doi: 10.1038/nature10580
Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C. (2015). Nitrogen Cycling in Corals: The Key to Understanding Holobiont Functioning? Trends Microbiol. 23, 490–497. doi: 10.1016/j.tim.2015.03.008
Raina J. B., Tapiolas D. M., Forêt S., Lutz A., Abrego D., Ceh J., et al. (2013). DMSP Biosynthesis by an Animal and its Role in Coral Thermal Stress Response. Nature 502 (7473), 677–680. doi: 10.1038/nature12677
Raina J. B., Tapiolas D., Willis B. L., Bourne D. G. (2009). Coral-Associated Bacteria and Their Role in the Biogeochemical Cycling of Sulfur. Appl. Environ. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Reisch C. R., Moran M. A., Whitman W. B. (2011). Bacterial Catabolism of Dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2, 172. doi: 10.3389/fmicb.2011.00172
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26 (1), 139–140. doi: 10.1093/bioinformatics/btp616
Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and Distribution of Coral-Associated Bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Romano S. L., Cairns S. D. (2000). Molecular Phylogenetic Hypotheses for the Evolution of Scleractinian Corals. Bull. Mar. Sci. 67 (3), 1043–1068.
Romano S. L., Palumbi S. R. (1996). Evolution of Scleractinian Corals Inferred From Molecular Systematics. Science 271 (5249), 640–642. doi: 10.1126/science.271.5249.640
Schuster F. (2002). Taxonomy of Oligocene to Early Miocene Scleractinian Corals-From Iran, Egypt, Turkey and Greece. Courier Forschungsinst: Senckenberg. 239, 1–3.
Shemi A., Alcolombri U., Schatz D., Farstey V., Vincent F., Rotkopf R., et al. (2021). Dimethyl Sulfide Mediates Microbial Predator-Prey Interactions Between Zooplankton and Algae in the Ocean. Nat. Microbiol. 6 (11), 1357–1366. doi: 10.1038/s41564-021-00971-3
Shinzato C., Khalturin K., Inoue J., Zayasu Y., Kanda M., Kawamitsu M., et al. (2021a). Eighteen Coral Genomes Reveal the Evolutionary Origin of Acropora Strategies to Accommodate Environmental Changes. Mol. Biol. Evol. 38, 16–30. doi: 10.1093/molbev/msaa216
Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., et al. (2011). Using the Acropora Digitifera Genome to Understand Coral Responses to Environmental Change. Nature 476, 320–323. doi: 10.1038/nature10249
Shinzato C., Takeuchi T., Yoshioka Y., Tada I., Kanda M., Broussard C., et al. (2021b). Whole-Genome Sequencing Highlights Conservative Genomic Strategies of a Stress-Tolerant, Long-Lived Scleractinian Coral, Porites Australiensis Vaughan 1918. Genome Biol. Evol. 13 (12), evab270. doi: 10.1093/gbe/evab270
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stanley G. D. Jr. (2003). The Evolution of Modern Corals and Their Early History. Earth Sci. Rev. 60, 195–225. doi: 10.1016/S0012-8252(02)00104-6
Stefels J., Dijkhuizen L., Gieskes W. W. C. (1995). DMSP-Lyase Activity in a Spring Phytoplankton Bloom Off the Dutch Coast, Related to Phaeocystis Sp. Abundance. Mar. Ecol. Prog. Ser. 123, 235–243. doi: 10.3354/meps123235
Stefels J., Steinke M., Turner S., Malin G., Belviso S. (2007). Environmental Constraints on the Production and Removal of the Climatically Active Gas Dimethylsulphide (DMS) and Implications for Ecosystem Modelling. Biogeochemistry 83 (1), 245–275. doi: 10.1007/s10533-007-9091-5
Steinke M., Kirst G. O. (1996). Enzymatic Cleavage of Dimethylsulfoniopropionate (DMSP) in Cell-Free Extracts of the Marine Macroalga Enteromorpha Clathrata (Roth) Grev. (Ulvales, Chlorophyta). J. Exp. Mar. Biol. Ecol. 201 (1-2), 73–85. doi: 10.1016/0022-0981(95)00207-3
Steinke M., Wolfe G. V., Kirst G. O. (1998). Partial Characterisation of Dimethylsulfoniopropionate (DMSP) Lyase Isozymes in 6 Strains of Emiliania Huxleyi. Mar. Ecol. Prog. Ser. 175, 215–225. doi: 10.3354/meps175215
Stolarski J., Kitahara M. V., Miller D. J., Cairns S. D., Mazur M., Meibom A. (2011). The Ancient Evolutionary Origins of Scleractinia Revealed by Azooxanthellate Corals. BMC Evol. Biol. 11, 1–11. doi: 10.1186/1471-2148-11-316
Strom S., Wolfe G., Holmes J., Stecher H., Shimeneck C., Sarah L. (2003). Chemical Defense in the Microplankton I: Feeding and Growth Rates of Heterotrophic Protists on the DMS-Producing Phytoplankter Emiliania Huxleyi. Limnol. Oceanogr. 48 (1), 217–229. doi: 10.4319/lo.2003.48.1.0217
Sunda W. K. D. J., Kieber D. J., Kiene R. P., Huntsman S. (2002). An Antioxidant Function for DMSP and DMS in Marine Algae. Nature 418, 317–320. doi: 10.1038/nature00851
Swan H. B., Jones G. B., Deschaseaux E. (2012). “Dimethylsulfide, Climate and Coral Reef Ecosystems,” in Proc. 12th Int. Coral Reef Symp, 9–13. Townsville, Qld: James Cook University
Teng Z. J., Wang P., Chen X. L., Guillonneau R., Li C. Y., Zou S. B., et al. (2021). Acrylate Protects a Marine Bacterium From Grazing by a Ciliate Predator. Nat. Microbiol. 6 (11), 1351–1356. doi: 10.1038/s41564-021-00981-1
Thierstein H. R., Geitzenauer K. R., Molfino B., Shackleton N. J. (1977). Global Synchroneity of Late Quaternary Coccolith Datum Levels Validation by Oxygen Isotopes. Geology 5 (7), 400–404. doi: 10.1130/0091-7613(1977)5<400:GSOLQC>2.0.CO;2
Vallina S. M., Simó R. (2007). Strong Relationship Between DMS and the Solar Radiation Dose Over the Global Surface Ocean. Science 315 (5811), 506–508. doi: 10.1126/science.1133680
Van Boekel J. S. W., Stefels W. H. M. (1993). Production of DMS From Dissolved DMSP in Axenic Cultures of the Marine Phytoplankton Species Phaeocystis Sp. Mar. Ecol. Prog. Ser. 97, 11–18. doi: 10.3354/meps097011
van Oppen M. J., McDonald B. J., Willis B., Miller D. J. (2001). The Evolutionary History of the Coral Genus Acropora (Scleractinia, Cnidaria) Based on a Mitochondrial and a Nuclear Marker: Reticulation, Incomplete Lineage Sorting, or Morphological Convergence? Mol. Biol. Evol. 18, 1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916
Veron J. E. N. (2000). Corals of the World. Vol. 1-3. (Townsville Queensland: Australian Institute of Marine Science).
Wallace C. C. (1999). “Staghorn Corals of the World: A Revision of the Coral Genus Acropora (Scleractinia; Astrocoeniina; Acroporidae) Worldwide,” in With Emphasis on Morphology, Phylogeny and Biogeography (Melbourne:CSIRO Publishing).
Wallace C. C., Rosen B. R. (2006). Diverse Staghorn Corals (Acropora) in High-Latitude Eocene Assemblages: Implications for the Evolution of Modern Diversity Patterns of Reef Corals. Proc. Biol. Sci. 273 (1589), 975–982. doi: 10.1098/rspb.2005.3307
Wolfe G. V., Steinke M. (1996). Grazing-Activated Production of Dimethyl Sulfide (DMS) by Two Clones of Emiliania Huxleyi. Limnol. Oceanogr. 41 (6), 1151–1160. doi: 10.4319/lo.1996.41.6.1151
Wolfe G. V., Steinke M., Kirst G. O. (1997). Grazing-Activated Chemical Defence in a Unicellular Marine Alga. Nature 387 (6636), 894–897. doi: 10.1038/43168
Woodhouse M. T., Carslaw K. S., Mann G. W., Vallina S. M., Vogt M., Halloran P. R., et al. (2010). Low Sensitivity of Cloud Condensation Nuclei to Changes in the Sea-Air Flux of Dimethyl-Sulphide. Atmos. Chem. Phys. 10 (16), 7545–7559. doi: 10.5194/acp-10-7545-2010
Yasuoka Y., Shinzato C., Satoh N. (2016). The Mesoderm-Forming Gene Brachyury Regulates Ectoderm-Endoderm Demarcation in the Coral Acropora Digitifera. Curr. Biol. 26 (21), 2885–2892. doi: 10.1016/j.cub.2016.08.011
Ying H., Hayward D. C., Cooke I., Wang W., Moya A., Siemering K. R., et al. (2019). The Whole-Genome Sequence of the Coral Acropora Millepora. Genome Biol. Evol 11 (5), 1374–1379. doi: 10.1093/gbe/evz077
Yoch D. C. (2002). Dimethylsulfoniopropionate: Its Sources, Role in the Marine Food Web, and Biological Degradation to Dimethylsulfide. Appl. Environ. Microbiol. 68 (12), 5804–5815. doi: 10.1128/AEM.68.12.5804-5815.2002
Keywords: DMPS lyase, DMS, Cnidaria, Scleractinia, Acropora, evolution, gene duplication
Citation: Chiu Y-L and Shinzato C (2022) Evolutionary History of DMSP Lyase-Like Genes in Animals and Their Possible Involvement in Evolution of the Scleractinian Coral Genus, Acropora. Front. Mar. Sci. 9:889866. doi: 10.3389/fmars.2022.889866
Received: 04 March 2022; Accepted: 19 May 2022;
Published: 13 June 2022.
Edited by:
Rafel Simó, Institut de Ciències del Mar (ICM-CSIC), SpainReviewed by:
Jean-Baptiste Raina, University of Technology Sydney, AustraliaUria Alcolombri, ETH Zürich, Switzerland
Michael Steinke, University of Essex, United Kingdom
Copyright © 2022 Chiu and Shinzato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuya Shinzato, Yy5zaGluemF0b0Bhb3JpLnUtdG9reW8uYWMuanA=
 Yi-Ling Chiu
Yi-Ling Chiu Chuya Shinzato
Chuya Shinzato