
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 22 July 2022
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.889840
This article is part of the Research TopicThe Impacts of Anthropogenic Activity and Climate Change on the Formation of Harmful Algal Blooms (HABs) and Its Ecological ConsequenceView all 16 articles
In 2016-17, shellfish harvesting closed for the first time in Narragansett Bay, Rhode Island, USA, from domoic acid (DA), a neurotoxin produced by diatoms of the Pseudo-nitzschia genus. Pseudo-nitzschia have occurred frequently for over 60 years in Narragansett Bay’s Long-Term Plankton Time Series (NBPTS), therefore it is surprising that the first closure only recently occurred. Pseudo-nitzschia species are known to vary in their toxin production, thus species identification is critical for understanding the underlying ecological causes of these harmful algal blooms (HABs). DNA in plankton biomass can be preserved for many years, so molecular barcoding of archived samples is useful for delineation of taxa over time. This study used amplification of the Pseudo-nitzschia-specific 18S-5.8S rDNA internal transcribed spacer region 1 (ITS1) in plankton samples and high throughput sequencing to characterize Pseudo-nitzschia species composition over a decade in Narragansett Bay, including eight years before the 2016-17 closures and two years following. This metabarcoding method can discriminate nearly all known Pseudo-nitzschia species. Several species recur as year-round residents in Narragansett Bay (P. pungens var. pungens, P. americana, P. multiseries, and P. calliantha). Various other species increased in frequency after 2015, and some appeared for the first time during the closure period. Notably, P. australis, a species prevalent in US West Coast HABs and known for high DA production, was not observed in Narragansett Bay until the 2017 closure but has been present in several years after the closures. Annual differences in Pseudo-nitzschia composition were correlated with physical and chemical conditions, predominantly water temperature. The long-term composition trends of Pseudo-nitzschia in Narragansett Bay serve as a baseline for identifying the introduction of new species, understanding shifting assemblages that contributed to the 2016-17 closures, and monitoring species that may be cause for future concern.
Pseudo-nitzschia, a cosmopolitan genus of diatom, causes harmful algal blooms (HABs) through the production of the neurotoxin domoic acid (DA), which bioaccumulates in primary and secondary consumers and causes the potentially fatal illness Amnesic Shellfish Poisoning in humans (Bates et al., 1989). Pseudo-nitzschia HABs are frequent on the US Gulf and Pacific coasts (Del Rio et al., 2010; McCabe et al., 2016), though the Northeast US had not experienced levels of DA high enough to prompt shellfish harvest closures until 2016, followed by additional closures in 2017 (Clark et al., 2019; Sterling et al., in press). This included Narragansett Bay, Rhode Island, where for the first time a closure in RI was triggered by DA in shellfish meat exceeding National Shellfish Sanitation Program limits (reviewed in Bates et al., 2018; NSSP, 2019; Sterling et al., in press). This recent emergence of blooms was unexpected, as the RI Department of Environmental Management (RI DEM) has monitored Pseudo-nitzschia HABs in Narragansett Bay since the 1990s without a closure incident (Pers. comm. David Borkman, RI DEM), and Pseudo-nitzschia have been recorded for over 60 years at the site of the Narragansett Bay Long-Term Plankton Time Series (NBPTS) (Smayda, 1959-1997; https://web.uri.edu/gso/research/plankton/, 1999-2022).
Only half of the known Pseudo-nitzschia species are confirmed toxin producers, which makes identification of species important for monitoring toxic events (reviewed in Bates et al., 2018). Additionally, many Pseudo-nitzschia species are morphologically cryptic under light microscopy (Amato & Montresor, 2008; Lundholm et al., 2012). High throughput sequencing techniques and genus-specific amplicon metabarcoding have made it possible to accurately and cost-effectively identify species at high taxonomic resolutions (Canesi & Rynearson, 2016; Lopes dos Santos et al., 2022). Furthermore, this method can be applied to previously archived biomass samples, including those of the NBPTS. Thus, amplicon sequencing is an effective way to analyze the role of species composition in the development of HABs over long periods of time (Lopes dos Santos et al., 2022). For example, a previous study that used Pseudo-nitzschia-specific metabarcoding to distinguish species in Narragansett Bay found that the high toxin-producing species P. australis likely contributed to the 2017 shellfish harvest closure and several Pseudo-nitzschia species more commonly observed at the NBPTS contributed to the precautionary closure in 2016 (Sterling et al., in press). From 2017 – 2019 in Narragansett Bay, low levels of plankton-associated DA were observed with fall and summer maxima, indicating that toxic species of Pseudo-nitzschia remained present in seasonally distinct species assemblages (Sterling et al., in press).
Similar to Pseudo-nitzschia HABs recently appearing in new locations like Narragansett Bay, they are also increasing in frequency and intensity in many regions of the ocean as climate change increases sea surface temperatures and impacts the phenology of biogeochemical cycling (reviewed in Wells et al., 2015; Bates et al., 2018; Testa et al., 2018). One example of this was in 2015 on the US West Coast, when a large bloom of P. australis led to record levels of DA, and an anomalously warm water mass was implicated in bloom formation (McCabe et al., 2016). Narragansett Bay has also been impacted by climate change, with surface water temperatures that increased by 0.23± 0.1°C per decade from 1984 – 2020 and more pronounced winter warming than other seasons (Fulweiler et al., 2015; Benoit and Fox-Kemper, 2021). Additionally, this location has experienced climate-driven nutrient cycle changes as well as a reduction in nutrient inputs due to recent management changes of sewage treatment (Oviatt et al., 2017). Examining whether the long-term patterns in Pseudo-nitzschia species composition correlate with these shifting environmental conditions is necessary for understanding the emergent DA events in Narragansett Bay and predicting future HABs.
In this study, we investigated the following questions: (1) Have particular species of Pseudo-nitzschia increased in prevalence during the closures and subsequent years? (2) Is P. australis a new species in Narragansett Bay? and (3) How have changing environmental conditions influenced Pseudo-nitzschia species composition? To address these questions, we examined more than a decade of archived phytoplankton biomass samples collected weekly since 2008 by the NBPTS for DNA analysis, with corresponding chemical and physical measurements. We used metabarcoding of the ITS1 region to identify nearly all known Pseudo-nitzschia species during the timeframe prior to closures (2008 - 2015), the years in which closures occurred (2016 and 2017), and the subsequent years (2018 - 2019). Characterizing these long-term patterns in species assemblages and environmental conditions provides a baseline for understanding the changes in closure years and following, informing how future Pseudo-nitzschia HABs will be monitored in Narragansett Bay.
Narragansett Bay, a temperate estuary on the northeast continental shelf of the United States, receives riverine freshwater inputs from the north and saline tidal flow from the Atlantic Ocean in the south (Raposa, 2009). Weekly surface water samples from December 2008 to November 2019 were collected at the NBPTS (Figure 1), a mid-bay site located in the West Passage (41° 34.2’ N, 71° 23.4’ W). In this study, two sample sets were combined to analyze over a decade of data: (1) December 2008 - August 2017 samples collected by the NBPTS and (2) September 2017 - November 2019 samples collected at the same location and processed by Sterling et al., in press. The data from Sterling et al., in press are publicly available online through the National Science Foundation Biological and Chemical Oceanography and Data Management Office (BCO-DMO; Jenkins & Bertin, 2021a; Jenkins & Bertin, 2021b). For a comparison of sample set metadata and methods, refer to Table S2.
Figure 1 Map of sampling location in Narragansett Bay, Rhode Island, USA. (A) Location of Narragansett Bay in Rhode Island (light grey) within the US northeast coast. (B) The Narragansett Bay Long Term Plankton Time Series (NBPTS) site is located mid-bay in the West Passage.
Biomass from the seawater samples collected by the NBPTS was filtered onto 25 mm 0.22 μm pore size ExpressPlus filters (MilliporeSigma, Burlington, MA, USA) and stored at -80°C until DNA extraction. The volume filtered for biomass capture varied and was based on the observed Secchi depth to normalize biomass collected, with 100 mL of seawater filtered per 1 m of Secchi depth. The volume filtered for NBPTS samples averaged 300 mL and ranged 100 – 600 mL. The Sterling et al., in pressbiomass samples were passed over 25 mm 5.0 μm pore size polyester membrane filters (Sterlitech, Kent, WA, USA), with an average of 240 mL and range of 75 – 430 mL filtered. Filters were then flash frozen in liquid nitrogen and stored at -80°C until DNA extraction. To ensure the comparability of sample sets using different pore sizes, we performed a comparison of species richness captured on each pore size that is outlined in the supplemental materials Figure S1. Sea surface temperature, sea surface salinity, chlorophyll a concentrations, Pseudo-nitzschia spp. cell counts, and nutrient measurements (nitrate, nitrite, phosphate) were obtained from the NBPTS prior to August 2017 and from the BCO-DMO dataset after September 2017 (NBPTS; Jenkins and Bertin, 2021a; Sterling et al., in press).
DNA from 65 previously extracted NBPTS samples from December 2008 to April 2017 was used in this study (Canesi & Rynearson, 2016; Rynearson et al., 2020; Sterling et al., in press). An additional 76 NBPTS biomass samples from March 2009 to August 2017 were extracted. NBPTS samples with the highest corresponding Pseudo-nitzschia spp. cell counts under light microscopy for each month were selected. If all cell counts during a month were zero, samples were selected at random. The following frequency of samples was chosen when available: one sample per month from winter (December - February) and spring (March-May), and two samples per month from summer (June-August) and fall (September-November) due to the more frequent occurrence of high Pseudo-nitzschia abundance during these months (Sterling et al., in press). Additionally, 70 publicly available sequenced samples collected at the NBPTS site from September 2017 - November 2019 were used (Jenkins and Bertin, 2021b; Sterling et al., in press). Because only a partial time series exists for 2012, samples from that year were not analyzed as part of this study. In total, the dataset contained 211 samples from the NBPTS site during December 2008 - November 2019.
For all NBPTS samples, including those extracted prior to this study, DNA was extracted from biomass filters using a modified version of the DNeasy Blood & Tissue DNA extraction kit (Qiagen, Germantown, MD, USA) with the addition of 4 μL RNase, a 1 min bead beating step (0.1 mm and 0.5 mm Zirconia/Silica beads, BioSpec Products, Bartlesville, OK, USA), and elution into a total volume of 100 μL Buffer AE. DNA was amplified using a eukaryotic ITS1 forward primer 5’ TCCGTAGGTGAACCTGCGG 3’ (White et al., 1990) and 5.8S reverse primer 5’ CATCCACCGCTGAAAGTTGTAA 3’ (Sterling et al., in press), with MiSeq adapters added to the 5’ ends of each primer for high throughput sequencing: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG - forward primer; 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGAC AG - reverse primer. This reverse primer was designed by Sterling et al., in press using a curated database of Pseudo-nitzschia sequences representing 41 species, thus the primer set can discriminate nearly all known Pseudo-nitzschia species with high specificity, though it is also expected to amplify some other diatom and dinoflagellate genera. The ITS region (both ITS1 and ITS2) have been used to distinguish Pseudo-nitzschia species in several studies (Lundholm et al., 2003; Amato et al., 2007; Casteleyn et al., 2008; Kaczmarska et al., 2008). The ITS1 region alone has been demonstrated as an effective marker to distinguish intra- and interspecific variation of Pseudo-nitzschia species (Hubbard et al., 2008; Hubbard et al., 2014). In Pseudo-nitzschia, the ITS1 locus is naturally variable in length, so the expected PCR product length ranged from 235 to 370 base pairs (White et al., 1990; Sterling et al., in press).
For PCR amplification, the following reagents were used in 25 μL reactions: Phusion Hot Start High-Fidelity Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA), HPLC-purified forward and reverse primers at 0.5 µM concentration (Integrated DNA Technologies, Coralville, IA, USA), and 2 μL of DNA template. A stepwise thermocycle protocol was used to amplify samples: 30 s denaturation at 98°C, 15 cycles of 98°C (10 s), 64.1°C (30 s), 72°C (30 s), 15 cycles of 98°C (10 s), 72°C (30 s), 72°C (30 s), and 10 min final extension at 72°C (Sterling et al., in press). Positive and negative sequencing controls were used as reported in Sterling et al., in press. PCR products were submitted to the RI Genomics and Sequencing Center (Kingston, RI, USA) for sequencing library preparation and high throughput sequencing. There, ITS1 PCR products were cleaned with KAPA pure beads (KAPA Biosystems, Woburn, MA, USA), sequencing indices and adapters were attached using PCR (50 ng template DNA, 8 cycles) and the Illumina Nextera XT Index Kit (Illumina, San Diego, CA, USA) with Phusion High Fidelity Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA), and PCR products were cleaned again with KAPA pure beads before being visualized by agarose gel electrophoresis. Selected samples were run on a Bioanalyzer DNA1000 chip (Agilent, Santa Clara, CA, USA). All samples were quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) prior to pooling, and the final pooled library was quantified with qPCR in a LightCycler480 (Roche, Pleasanton, CA, USA) with the KAPA Biosystems Illumina Kit (KAPA Biosystems, Woburn, MA, USA). Samples were analyzed using v3 chemistry, 600 cycles, and 2x250 bp paired-end sequencing on an Illumina MiSeq (Illumina, Inc., San Diego, CA, USA). These sequencing methods were exactly the same as those in Sterling et al., in press.
Illumina Miseq adapters and primers were trimmed using Cutadapt (v3.2; Martin, 2011) and sequences were quality checked before and after trimming using MultiQC (v1.9; Ewels et al., 2016). Amplicon sequence variants (ASVs) were delineated in DADA2 in R (v1.18.0; Callahan et al., 2016). One sample contained no reads following analysis with DADA2 and was removed. All ASVs, including those sequenced for this study and those from Sterling et al., in press, were assigned taxonomy using the scikit-learn naïve Bayes machine learning classifier in QIIME2 with the default confidence threshold of 0.7 (v2021.4.0; Bolyen et al., 2019). For taxonomic assignment, the curated reference database used in Sterling et al., in press was updated with an additional 170 unique Pseudo-nitzschia National Center for Biotechnology Information (NCBI) GenBank sequences, for a total of 302 sequences representing 51 species (retrieved June 1, 2021) (Table S1). To maximize the number of ASVs classified to the species level, additional ASVs were assigned by manual inspection of a megablast search that reported the top hit of each ASV from the BLAST nt database (retrieved June 24, 2021). Additional ASVs classified using the megablast search required >98% identity and >98% query cover to an NCBI Pseudo-nitzschia species, otherwise classifications were discarded. From this additional classification, we recovered only species already represented in our custom database from the pool of ASVs. All ASVs of the same species were agglomerated in R for downstream analyses.
Data were analyzed and visualized in R (v4.0.2; R Core Team, 2017) within RStudio (v1.3.1056; RStudio Team, 2020) using the following packages: phyloseq (v1.34.0; McMurdie & Holmes, 2013) for ASV dataset manipulation and transformations; ggplot2 (v3.3.3; Wickham, 2016) for scatterplot, heatmap, bar plot, and sample frequency plots; vegan (v2.5.7; Dixon, 2003) for dispersion tests and ANOSIM; indicspecies (v1.7.9; De Cáceres & Legendre, 2009) for indicator species analysis; raster (v3.5.15; Hijmans et al., 2015) for the sampling site map; and viridis (v0.6.2; Garnier et al., 2021) for colorblind-friendly figure color palettes. Relative abundances were calculated as the proportion of a species out of the total Pseudo-nitzschia sequencing reads for each sample, and these were only used in one visualization (Figure 3). Presence-absence Jaccard distances of composition data generated in phyloseq were used in all statistical analyses to avoid distorted relative abundance metrics that may arise from eukaryotic gene copy number variation (reviewed in Canesi & Rynearson, 2016; Gloor et al., 2017). This is a better approach for metabarcoding data than using relative or absolute abundance of sequencing reads (Zaiko et al., 2015; Canesi & Rynearson, 2016; Rynearson et al., 2020).
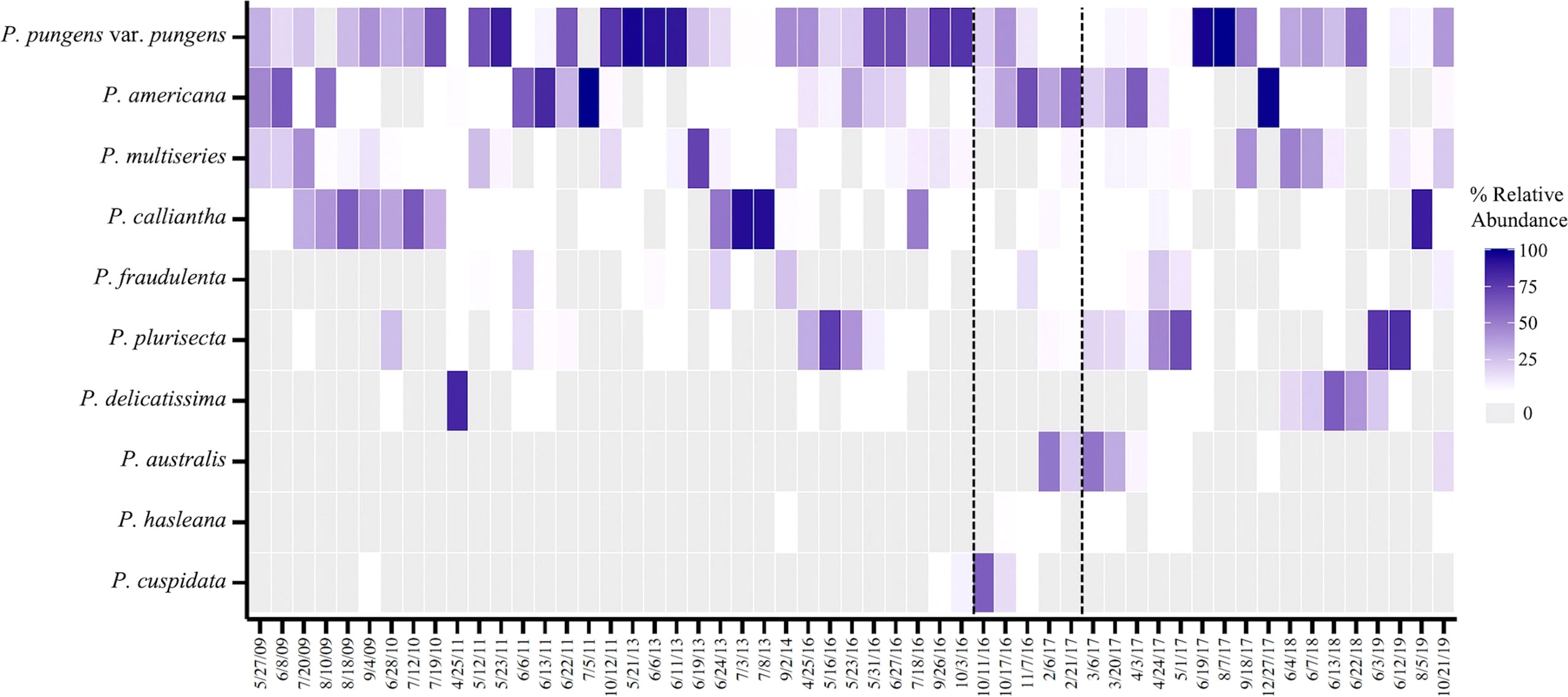
Figure 3 Relative abundance of the top ten species in samples that surpass the RI DEM threshold of 20,000 cells L-1 from Dec 2008 - Nov 2019 (n=55). Dotted lines denote the 2016 precautionary shellfish closure and 2017 closure. Gray boxes represent species that were absent in samples.
Similarities between temporal groupings of species composition were characterized using an analysis of similarity (ANOSIM) on a Jaccard distance matrix. An assumption of ANOSIM is relatively equal variances, or lack of dispersion, between groups being compared. Prior to ANOSIM, dispersion of groups was determined using betadisper() and permutest() (R: vegan) with 999 permutations and a significance level of 0.05. Only groups that did not have significant dispersion were used in ANOSIM, which included year and timeframe, the latter defined as two time periods: before closures (2009 - 2015) and during/after closure years (2016 - 2019). The groupings of samples by both month and season exhibited significant dispersion of groups and thus did not meet the assumptions to test for similarity in ANOSIM (p=0.023; p=0.014). ANOSIM was performed using anosim() (R: vegan) with 999 permutations and a significance level of 0.05. To determine which species preferentially occurred before and during/after closures, an indicator species analysis (ISA) was performed on a Jaccard distance matrix using multipatt() (R: indicspecies), 9999 permutations, and a significance level of 0.05 on p-values adjusted for multiple testing.
A Best Subset of Environmental Variables (BIOENV) multivariate analysis was performed using bioenv() (R: vegan) to correlate environmental conditions with species composition. Environmental variables, including sea surface temperature, sea surface salinity, dissolved inorganic nitrogen (DIN), dissolved inorganic phosphorus (DIP), and chlorophyll a concentration were standardized using log-transformation prior to comparison with a Jaccard distance matrix. The BIOENV analysis was performed for all samples with complete physical and chemical data (n=178), as well as for each timeframe (before: n=81, during/after: n=97).
Within the 141 samples from December 2008 – August 2017 that were sequenced for this study, there were a total of 12.3 x 106 read pairs. Initial reads per sample ranged from 4,821 to 115,087, with an average of 70,104 reads per sample. After DADA2 sequencing error inference, there was an average of 39,673 reads per sample, with a range of 1,335 to 66,903 reads. This is comparable to the average of 35,550 reads per sample reported in the Sterling et al., in press dataset. In total, 5,117 ASVs were recovered in the newly sequenced samples. Taxonomic assignment yielded 57 ASVs at the Pseudo-nitzschia species level (46 QIIME2, 11 megablast). When re-classifying the ASVs from Sterling et al., in press with the updated database, 27 ASVs were assigned to the species level (20 QIIME2, 7 megablast). The number of ASVs per sample ranged from 1 to 15 and an average of 5,773 sequencing reads per sample were assigned to Pseudo-nitzschia spp. Each species was represented by a minimum of 2 and maximum of 17 ASVs, and after aggregating ASVs by species, 17 Pseudo-nitzschia species were characterized in the samples.
During the study period, December 2008 – November 2019, Pseudo-nitzschia species were observed by light microscopy each year, excluding the gap in 2012 (Figure 2). Cell counts at the genus level surpassed the RI DEM HAB abundance threshold of 20,000 cells per liter each year for which there was abundance data except 2015 (Figure 2) (RI DEM, 2021). Specifically, weekly samples surpassed the cell abundance threshold 69 times across all seasons, though more frequently during spring (n=20) and summer (n=33) months (Figure 2). The closure years in 2016 and 2017 alone comprised 38% of these high cell abundance samples: 2016 (n=11) and 2017 (n=17). A subset of our total sequenced samples (n=55) have corresponding cells counts that surpassed the RI DEM HAB threshold and were used to analyze Pseudo-nitzschia species composition during high abundance periods. P. pungens var. pungens occurred most frequently on dates of high cell counts, followed by P. multiseries and P. americana (Figure 3). Only one sample contained just one species (P. pungens var. pungens; Aug 07, 2017), while all other samples contained between 2 and 7 species (Figure 3).
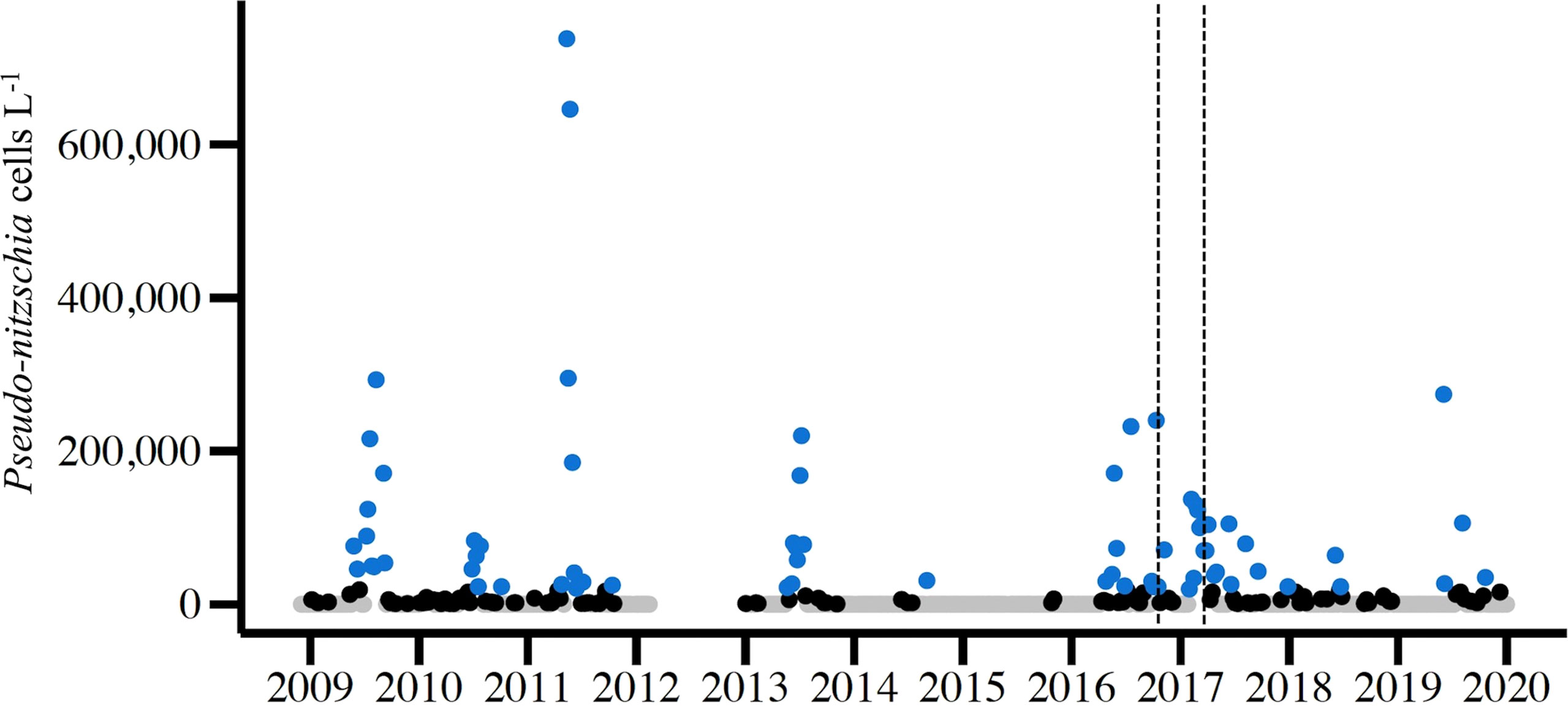
Figure 2 Abundance of live Pseudo-nitzschia spp. cells from light microscopy counts of weekly NBPTS surface seawater samples spanning Dec 2008 – Dec 2019 (n=523). The NBPTS paused during Mar 2012 – Dec 2012, so no cell counts were collected. Blue points represent Pseudo-nitzschia abundance that surpassed the RI Department of Environmental Management (DEM) HAB abundance threshold of 20,000 cells per liter, while black points are non-zero abundances below the threshold and gray points are zero counts. Dotted lines denote the 2016 precautionary shellfish closure (Oct 7–30) and 2017 closure (Mar 1–24).
The 17 species of Pseudo-nitzschia identified in the total dataset exhibited varying annual patterns of occurrence (Figure 4). Four species were observed most frequently in over 40% of samples - P. pungens var. pungens (66%), P. americana (66%), P. multiseries (55%), and P. calliantha (44%) - and were found in samples from each year of the dataset (Figure 4A). Several species appeared for the first time or increased in prevalence during closure and subsequent years. Most notably, P. australis, a well characterized toxin producer, was not present in any of the sequenced samples prior to 2017 and appeared for the first time on February 6, 2017, several weeks before the closure (Figure 4B). But from 2017 – 2019, P. australis continued to be observed, appearing in 23% of sequenced samples. Similarly, the toxin-capable species P. hasleana and P. subpacifica appeared relatively infrequently in the years prior to the shellfish closures, however, each became more prevalent in frequency and were found in nearly each year from 2015-2019 (Figure 4B). Two additional toxin-capable species P. fraudulenta and P. plurisecta were present in low proportions of samples (0 - 32%) in the years prior to closures, but both were observed in higher proportions of samples (25 - 74%) from 2016-2019 (Figure 4B). Several rare Pseudo-nitzschia species that occurred in less than 10% of samples from the entire dataset were more prevalent during the closure years than any other timeframe, including P. hasleana in 2016 and 2017 and P. cuspidata in 2016 (Figure 4B).
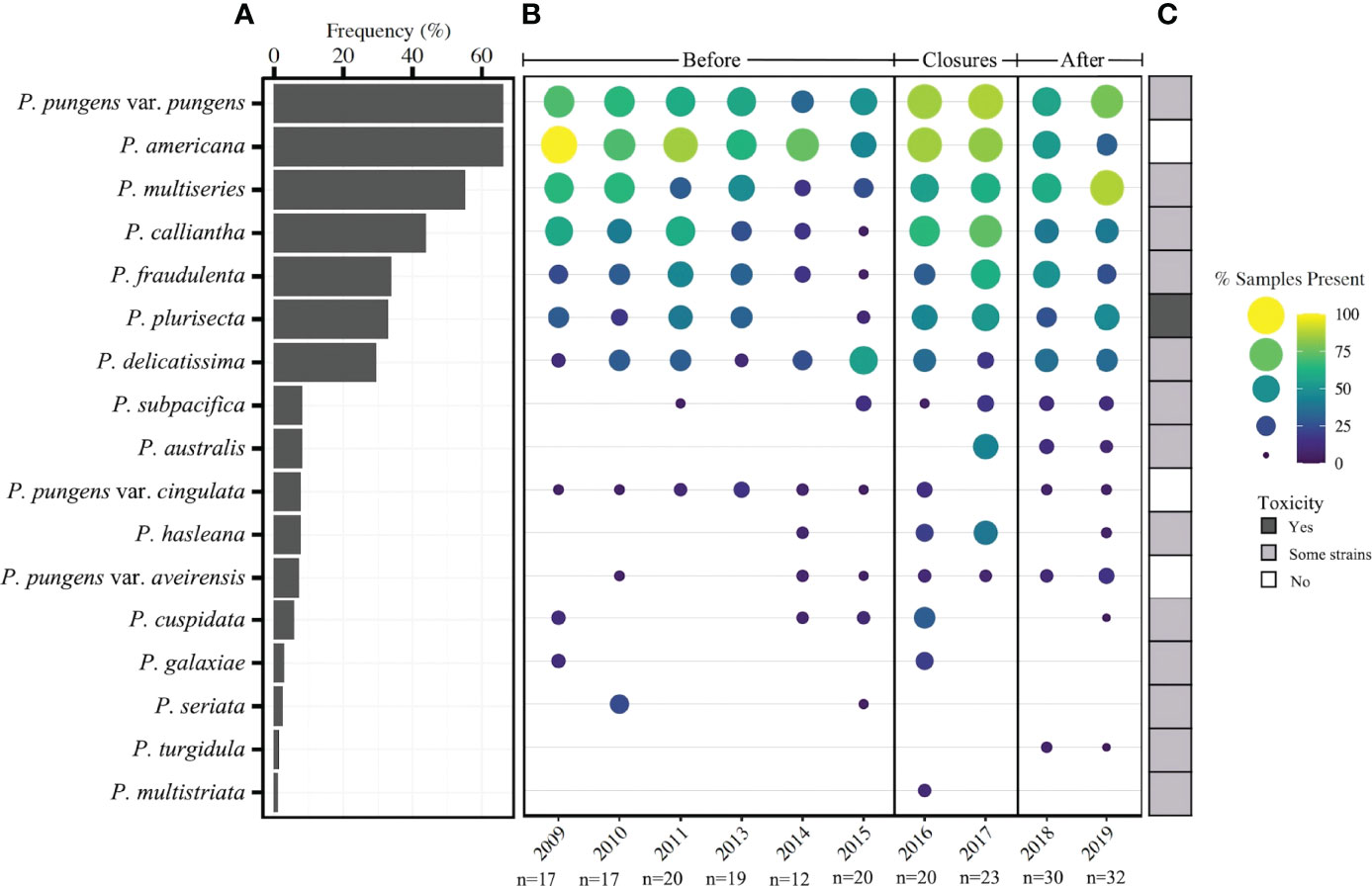
Figure 4 (A) Frequency of occurrence of 17 species of Pseudo-nitzschia from 2008 – 2019 (n=214). (B) Presence of Pseudo-nitzschia species by year for three timeframes: prior to closures, during closure years, and following closures. The size and color of each circle represents the percent of samples in a given year in which Pseudo-nitzschia species occurred based on a presence/absence matrix. (C) Observed toxicity of each species as reviewed in Bates et al., 2018, with “some strains” meaning that some have been reported to produce toxin while others have not.
To determine if species assemblages differed temporally, ANOSIM was used on the Jaccard distance matrix of Pseudo-nitzschia species composition. Two groupings were determined to be viable for ANOSIM based on dispersion of group tests (n=199): year (p=0.286), and timeframe (p=0.658), the latter of which was defined as before closures (2009 - 2015) and during/after closures (2016 - 2019). ANOSIM revealed significant differences between species assemblages in each temporal grouping (p=0.001, ANOSIM statistic 0.1082 and 0.0625 respectively) (Figure S2). On average, species richness by year increased over time (Figure S3). An ISA was also performed on a Jaccard matrix to determine which species preferentially occurred in the timeframes before and during/after closures, since the species assemblages between these two timeframes were significantly different (ANOSIM, Figure S2). This analysis revealed that the strongest indicator species of the timeframe prior to closures were P. americana (p=0.003) and P. seriata (p=0.025). Eight species were significant indicators of the timeframe during and after closures, including P. australis (p=0.0001), P. hasleana (p=0.0002), P. multiseries (p=0.0027), P. plurisecta (p=0.017), P. pungens var. aveirensis (p=0.031), P. pungens var. pungens (p=0.033), P. subpacifica (p=0.045), and P. calliantha (p=0.046). All other species included in the analysis did not preferentially occur in either of these two timeframes.
A multivariate correlation analysis (BIOENV) showed that Pseudo-nitzschia species composition correlated with various environmental conditions. The parameters that best correlated with species composition (0.1545) were surface temperature and DIP (Table 1). When the same multivariate correlation analysis was performed on each timeframe (before and during/after closures), the highest correlates prior to closures were temperature and DIP (0.678), while the during/after closure timeframe was most correlated with temperature, DIP, and DIN (0.2725) (Table 1).
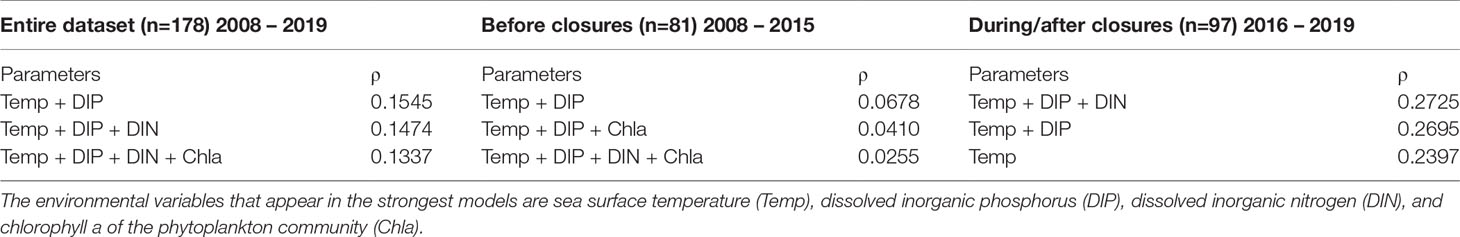
Table 1 BIOENV results of the three models with the highest Spearman rank correlation coefficient (ρ) for three timeframes: 2008 – 2019; 2008 – 2015; 2016 – 2019.
Pseudo-nitzschia species were found at a wide variety of temperatures in Narragansett Bay (Figure 5). During the study period, temperatures ranged from -1.31 – 26.47°C, and Pseudo-nitzschia were present in sequenced samples through nearly this entire range, from -1.31 – 25.51°C. The seven most frequently observed species- P. pungens var. pungens, P. americana, P. multiseries, P. calliantha, P. fraudulenta, P. plurisecta, and P. delicatissima- were present over wide temperature ranges that did not differ greatly in the before closure (2008 - 2015) timeframe and during/after closure (2016 - 2019) timeframe. Several less prevalent species had smaller and more distinct temperature ranges. P. subpacifica (7.6 – 23.3°C), P. pungens var. cingulata (4.48 – 22.18°C), P. pungens var. aveirensis (15.4 – 22.18°C), and P. cuspidata (9.97 – 22.4°C) were observed during relatively higher temperatures, while P. australis (1.6 – 15.4°C) and P. hasleana (2.24 – 22.18°C) tended to occur at relatively lower temperatures (Figure 5).
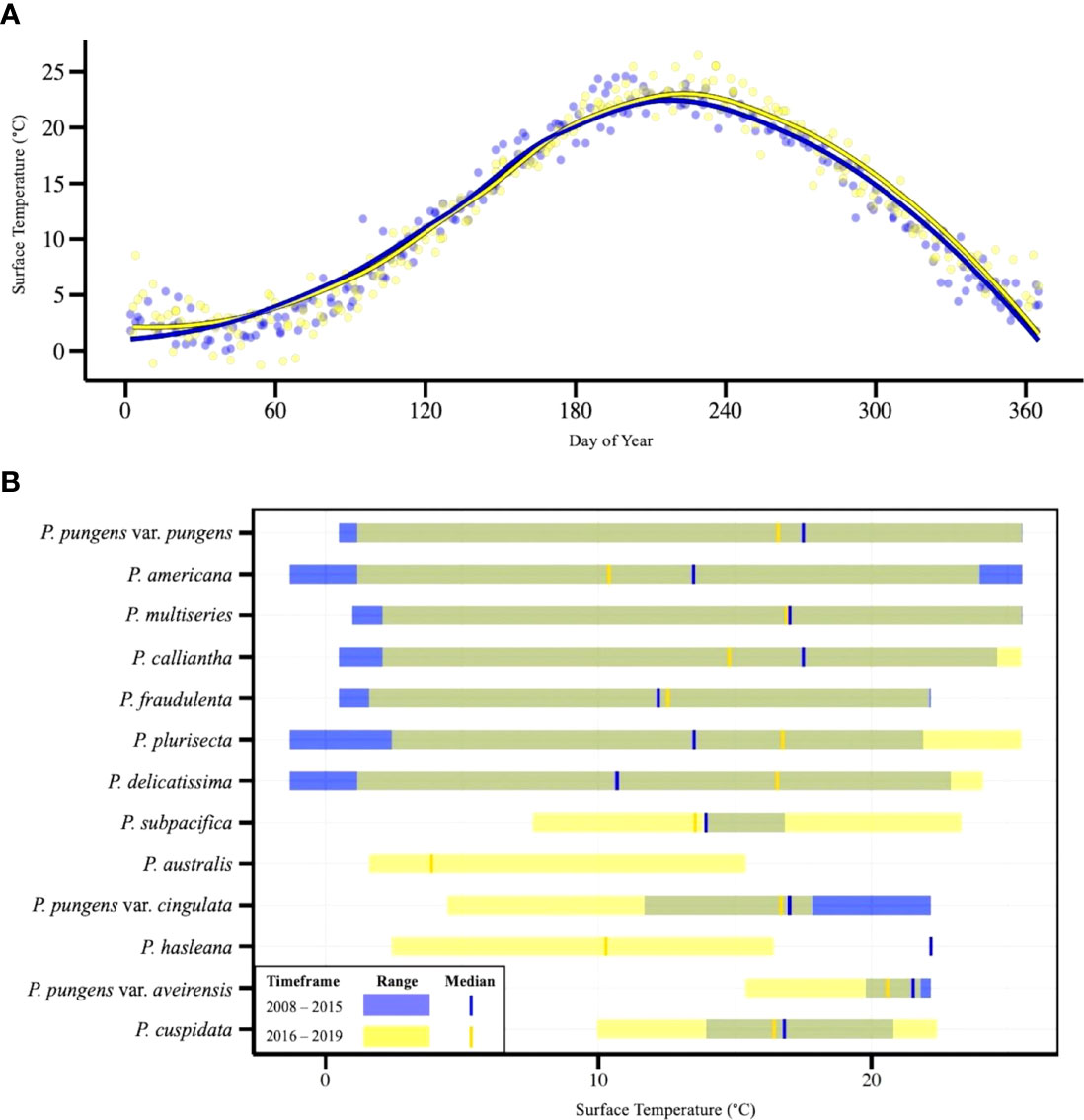
Figure 5 (A) Sea surface temperature at the NBPTS site over an annual cycle with two loess smoother lines representing the timeframes before (2008-2015; blue) and during/after closures (2016-2019; yellow). (B) Thermal ranges of Pseudo-nitzschia species that were found in >5% of samples. Ranges are based on sea surface temperature of presence/absence occurrence during two timeframes. Samples from 2008-2015 are shown in blue; 2016-2019 are shown in yellow. Temperatures for which the temporal thermal ranges overlap appear in green. The median temperature at which species are observed prior to 2015 are represented by vertical blue bars and after 2016 by vertical yellow bars.
Multiple species were present during Pseudo-nitzschia blooms in Narragansett Bay, defined here as periods where Pseudo-nitzschia cells surpass the RI DEM cell abundance threshold of 20,000 cells per liter (Figure 2) (RI DEM, 2021). Exceeding this threshold prompts DA Scotia testing of phytoplankton tow net samples followed by DA screening of shellfish meat, which can trigger shellfish harvest closures (RI DEM, 2021). More weekly light microscopy samples surpassed the cell abundance threshold in 2016 and 2017 than any other year of the study period (Figure 2). From 2016 - 2017, P. pungens var. pungens. P. fraudulenta, and P. americana were present in each of the bloom samples, with the first appearance of P. australis occurring in 2017 (Figure 3). Though P. calliantha was the fourth-most prevalent species across the entire study period, it occurred in less than half of the bloom samples (Figures 3, 4A). Most samples that surpassed the abundance threshold contained diverse multi-species assemblages containing as many as seven of the top ten most abundant Pseudo-nitzschia species, with the exception of one bloom sample in which just P. pungens var. pungens occurred (Figure 3).
From 2008 – 2019, 17 species of Pseudo-nitzschia were detected in Narragansett Bay (Figure 4). All species were previously observed except P. seriata, which was recorded annually from 2012 to 2016 in the nearby Gulf of Maine and has only ever been observed in the North Atlantic (Hasle, 2002; Clark et al., 2019; Sterling et al., in press). Some patterns in species composition differ in the years prior to shellfish harvest closures, while others remain consistent. The four resident species (P. pungens var. pungens, P. americana, P. multiseries, and P. calliantha) that occurred in each year of the dataset followed similar patterns of prevalence before, during, and after the closure periods (Figure 4). Furthermore, these resident species occurred across the widest ranges of temperatures and were present through nearly the entire temperature range of Narragansett Bay (Figure 5). This prevalence across temperature ranges indicates a plasticity of these species to a range of environmental conditions. Consistent with our observations, P. pungens var. pungens and P. multiseries have been previously characterized as cosmopolitan, occurring in both equatorial and cold-water regions (Hasle, 2002). Additionally, P. americana has been observed in a wide range of environments, including the Gulf of Maine, Malaysia, Mexico, and Namibia (reviewed in Bates et al., 2018). P. calliantha, is also a confirmed cosmopolite with occurrences in warm and cold environments (Stonik et al., 2011).
Species assemblages varied significantly between years and timeframes (before and during/after closures) (Figure S2). Though prior work in Narragansett Bay observed distinct species assemblages by season (Sterling et al., in press), the seasonal groupings in this dataset were unable to be tested for similarity because the monthly and seasonal groups displayed significant dispersion. Additionally, distinct species groupings by year and timeframe suggest that composition patterns have shifted over time in Narragansett Bay. The indicator species from each of these timeframes explains some of the temporal shifts. Before the closure periods, P. americana, which is a non-toxigenic year-round resident of Narragansett Bay, was the strongest indicator species. This timeframe was characterized by lower species richness and lower prevalence of toxigenic species (Figure 4; Figure S3). P. multiseries and P. plurisecta were two of the strongest indicator species for the during closure/after timeframe, and both toxigenic species increased in prevalence 2014 – 2019 (Figure 4B). Several other important indicator species of the during closure/after timeframe were P. hasleana, P. subpacifica, and P. pungens var. aveirensis, all of which were present in very few samples prior to closures but persisted in nearly each year of the closures and following (Figure 4B).
Temperature was the only consistent environmental driver that appeared in each of the top three multivariate models for the entire species composition dataset, the timeframe prior to closures, and the timeframe encompassing closures and subsequent years (Table 1). This is supported by previous work in Narragansett Bay that showed temperature also correlated with both Thalassiosira spp. and Skeletonema spp. assemblage composition from 2008 - 2014 (Canesi & Rynearson, 2016; Rynearson et al., 2020). The importance of sea surface temperature to phytoplankton community composition may be partially attributed to the wide range of temperatures that Narragansett Bay experiences throughout the year (Figure 5). As sea surface temperatures in Narragansett Bay continue to increase over time due to climate change (Fulweiler et al., 2015; Benoit & Fox-Kemper, 2021), this driver of Pseudo-nitzschia assemblage composition may lead to further shifts in the prevalence of certain species. This could include continued increases in the frequency of P. australis, P. hasleana, P. galaxiae, and P. subpacifica, all of which became more prevalent in Narragansett Bay in closure and subsequent years.
Additionally, availability of nutrients, including both DIP and DIN, impacted Pseudo-nitzschia species composition. DIP was a driver of species composition in all but one of the top multivariate models (Table 1). Phosphorus is a required macronutrient for phytoplankton growth, though its relationship to Pseudo-nitzschia HAB formation is complex because high DIP increases Pseudo-nitzschia growth rate but low DIP correlates with increased DA toxin production (Pan et al., 1998; Sun et al., 2011; Brunson et al., 2018). Furthermore, DIN appears in the multivariate model with the highest correlation in the during closures/after timeframe (Table 1). The importance of DIN concentration to both Pseudo-nitzschia assemblage composition and toxin production in Narragansett Bay is supported by Sterling et al., in press, who found that nitrate and DIN : DIP correlated with species composition and low DIN correlated with elevated particulate DA. The recently reduced DIN in Narragansett Bay following wastewater treatment changes may play a role in impacting the composition and physiology of these Pseudo-nitzschia assemblages (Oviatt et al., 2017).
P. australis was not observed in Narragansett Bay prior to 2017 (Figure 4) and was the strongest indicator species in the during closure/after timeframe as compared to samples prior to the 2016 – 2017 HABs. It appears to be recently introduced to the North Atlantic coasts of the US and Canada during the record HABs of 2016 (Clark et al., 2019, reviewed in Bates et al., 2018). Notably, it was absent during the RI precautionary closure of 2016 in which DA below the NSSP threshold was observed in shellfish meat (Figure 3; reviewed in Bates et al., 2018; Sterling et al., in press), meaning there were other species responsible for this event. Toxigenic species present during the 2016 closure included P. pungen var. pungens and P. cuspidata. P. australis continued to persist in Narragansett Bay in about one-fifth of the samples from 2017 – 2019, suggesting it remained in the region and may be a cause for concern in future toxic events. However, it was not the only toxic species in Narragansett Bay during the 2016 – 2017 closures, so continued monitoring of Pseudo-nitzschia at the species level is important for understanding which species contribute to producing closure levels of DA.
P. australis has been connected to a large 2015 HAB event on the US west coast where increases in both Pseudo-nitzschia abundance and DA were attributed to anomalously warm water temperatures and nutrient-rich upwelling (McCabe et al., 2016). Sea surface temperatures in the North Pacific were 2.5°C higher than long term means, ranging about 12 – 18°C during the closure period from May – November 2015 (McCabe et al., 2016). Increased DA production in West Coast P. australis strains was also associated with warmer temperatures (Zhu et al., 2017). In contrast to this, we found that P. australis occurred at a lower relative thermal range than any other species and was only observed in Narragansett Bay between 1.6 – 15.4°C, with a median temperature of 3.9°C. During the closure in February – March of 2017, the temperatures at which P. australis was observed ranged 2.44 – 3.26°C, so this particular toxic event was not initiated by a warm water mass. Increased toxin concentration in shellfish meat has been linked to higher growth potential of P. australis (McCabe et al., 2016). Clark et al. (2021) reports that the optimal growth temperature for a strain of P. australis isolated from the nearby Gulf of Maine was about 15°C, while studies with US West Coast isolates have found optimal growth temperatures of 17 – 18°C (Monterey Bay, California 2015 isolate; McCabe et al., 2016) and 23 – 26°C (Southern California isolate; Zhu et al., 2017). The presence of P. australis at much lower temperatures in Narragansett Bay suggests there may be other factors influencing its growth and toxin production, such as nutrient availability, bacterial associations, or zooplankton grazing (Maldonado et al., 2002; reviewed in Lelong et al, 2012; Lundholm et al., 2018).
In this study, we used a DNA metabarcoding approach on more than a decade of plankton samples from Narragansett Bay, RI to characterize Pseudo-nitzschia species diversity before, during, and after two shellfish harvest closures. We found that periods of high Pseudo-nitzschia cell abundances correspond to a wide diversity of species present, supporting the complexity of bloom-forming assemblages. We characterized several species as residents of Narragansett Bay, which occurred in samples in nearly each year of the study period. P. australis, a high toxin producing species, was not present in any of the samples until 2017, and thus is likely to be a newly introduced species. Additionally, species composition was most strongly influenced by water temperature. As water temperatures continue to rise in Narragansett Bay and toxigenic species become more prevalent, it is imperative to continue monitoring Pseudo-nitzschia species composition for increased frequency and potential introduction of toxigenic species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OM672116 - OM672173. https://www.ncbi.nlm.nih.gov/, SAMN25894732 - SAMN25894875.
Conception and design, KR, BJ. Sample collection, TR. Methodology, KR, AS, TR. Formal analysis, KR. Investigation, KR. Resources, BJ. Data curation, KR. Writing—original draft preparation, KR, BJ. Writing—review and editing, all authors. Visualization, KR. Supervision, BJ. Project administration, BJ, TR. Funding acquisition, BJ, MB, TR. All authors contributed to the article and approved the submitted version.
This research was supported by the following awards: MJ and BJ (NA18OAR4170094), TR (NSF1638834), and NSF RI C-AIM EPSCoR Cooperative Agreement (OIA-1655221, OIA-1004057). Sequencing was performed at the Rhode Island RI NSF EPSCoR research facility, the Genomics and Sequencing Center (OIA-1655221). The Narragansett Bay Long-Term Plankton Time Series is supported by the University of Rhode Island and the Rhode Island Department of Environmental Management.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the Plankton Assistants for collecting NBPTS samples and enumerating cell abundances. We thank Z. Pimentel for help with taxonomic assignment, D. Fontaine for helpful suggestions regarding data analysis, and E. Borbee for help with map design. Thank you to G. Armin, S. Song, and V. Sonnet for helpful discussions on clarity of writing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.889840/full#supplementary-material
Amato A., Kooistra W. H. C. F., Levialdi Ghiron J. H., Mann D. G., Pröschold T., Montresor M. (2007). Reproductive Isolation Among Sympatric Cryptic Species in Marine Diatoms. Protist. 158, 193–207. doi: 10.1016/j.protis.2006.10.001
Amato A., Montresor M. (2008). Morphology, Phylogeny, and Sexual Cycle of Pseudo-Nitzschia Mannii Sp. Nov. (Bacillariophyceae): A Pseudo-Cryptic Species Within the P. Pseudodelicatissima Complex. Phycologia. 47 (5), 487–497. doi: 10.2216/07-92.1
Bates S. S., Bird C. J., de Freitas A. S. W., Foxall R., Gilgan M., Hanic L. A., et al. (1989). Pennate Diatom Nitzschia Pungens as the Primary Source of Domoic Acid, a Toxin in Shellfish From Eastern Prince Edward Island, Canada. Can. J. Fisheries. Aquat. Sci. 46 (7), 1203–1215. doi: 10.1139/f89-156
Bates S. S., Hubbard K. A., Lundholm N., Montresor M., Leaw C. P. (2018). Pseudo-Nitzschia, Nitzschia, and Domoic Acid: New Research Since 2011. Harmful. Algae. 79, 3–43. doi: 10.1016/j.hal.2018.06.001
Benoit J., Fox-Kemper B. (2021). Contextualizing Thermal Effluent Impacts in Narragansett Bay Using Landsat-Derived Surface Temperature. Front. Mar. Sci.8, 1247. doi: 10.3389/fmars.2021.705204
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi: 10.1038/s41587-019-0209-9
Brunson J. K., McKinnie S. M., Chekan J. R., McCrow J. P., Miles Z. D., Bertrand E. M., et al. (2018). Biosynthesis of the Neurotoxin Domoic Acid in a Bloom-Forming Diatom. Science. 361 (6409), 1356–1358. doi: 10.1126/science.aau0382
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Canesi K. L., Rynearson T. A. (2016). Temporal Variation of Skeletonema Community Composition From a Long-Term Time Series in Narragansett Bay Identified Using High-Throughput DNA Sequencing. Mar. Ecol. Prog. Ser. 556, 1–16. doi: 10.3354/meps11843
Casteleyn G., Chepurnov V. A., Leliaert F.others (2008). Pseudo-Nitzschia Pungens (Bacillariophyceae): A Cosmopolitan Diatom Species? Harmful. Algae. 7, 241–257. doi: 10.1016/j.hal.2007.08.004
Clark S., Hubbard K. A., Anderson D. M., McGillicuddy D. J. Jr., Ralston D. K., Townsend D. W. (2019). Pseudo-Nitzschia Bloom Dynamics in the Gulf of Maine: 2012-2016. Harmful. Algae. 88, 101656. doi: 10.1016/j.hal.2019.101656
Clark S., Hubbard K. A., McGillicuddy D. J., Ralston D. K., Shankar S. (2021). Investigating Pseudo-Nitzschia Australis Introduction to the Gulf of Maine With Observations and Models. Continental. Shelf. Res. 228, 104493. doi: 10.1016/j.csr.2021.104493
De Cáceres M., Legendre P. (2009). Associations Between Species and Groups of Sites: Indices and Statistical Inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Del Rio R., Bargu S., Baltz D., Fire S., Peterson G., Wang Z. (2010). Gulf Menhaden (Brevoortia Patronus): A Potential Vector of Domoic Acid in Coastal Louisiana Food Webs. Harmful. Algae. 10 (1), 19–29. doi: 10.1016/j.hal.2010.05.006
Dixon P. (2003). VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 14(6), 927-930 doi: 10.1111/j.1654-1103.2003.tb02228.x
Ewels P., Magnusson M., Lundin S., Käller M. (2016). MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 32 (19), 3047–3048. doi: 10.1093/bioinformatics/btw354
Fulweiler R. W., Oczkowski A. J., Miller K. M., Oviatt C. A., Pilson M. E. Q. (2015). Whole Truths vs. Half Truths – And a Search for Clarity in Long-Term Water Temperature Records. Estuarine. Coast. Shelf. Sci. 157, A1–A6. doi: 10.1016/j.ecss.2015.01.021
Garnier S., Ross N., Rudis R., Camargo P. A., Sciaini M., Scherer C. (2021). Viridis—Colorblind-Friendly Color Maps for R. R Package Version 0.6, 2.
Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J. (2017). Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02224
Hasle G. R. (2002). Are Most of the Domoic Acid-Producing Species of the Diatom Genus Pseudo-Nitzschia Cosmopolites? Harmful. Algae. 1 (2), 137–146. doi: 10.1016/S1568-9883(02)00014-8
Hijmans R. J., Van Etten J., Cheng J., Mattiuzzi M., Sumner M., Greenberg J. A., et al. (2015). Package ‘Raster’. R Package. 734.
Hubbard K. A., Olson C. E., Armbrust E. V. (2014). Molecular Characterization of Pseudo-Nitzschia Community Structure and Species Ecology in a Hydrographically Complex Estuarine System (Puget Sound, Washington, USA). Mar. Ecol. Prog. Ser. 507, 39–55. doi: 10.3354/meps10820
Hubbard K. A., Rocap G., Armbrust E. V. (2008). Inter- and Intraspecific Community Structure Within the Diatom Genus Pseudo-Nitzschia (Bacillariophyceae). J. Phycol. 44, 637–649. doi: 10.1111/j.1529-8817.2008.00518.x
Jenkins B. D., Bertin M. J. (2021a). “Amplicon Sequence Variants (ASVs) Recovered From Samples and Their Related Identification as Pseudo-Nitzschia Taxa and the Methods Used,” in Biological and Chemical Oceanography Data Management Office (BCO-DMO). (Version 1) Version Date 2021-04-05 [Subset to NBPTS Samples]. doi: 10.26008/1912/bco-dmo.847469.1
Jenkins B. D., Bertin M. J. (2021b). “Pseudo-Nitzschia Spp. Cell Counts, Nutrients Water Temperature and Salinity, and Concentrations of the Toxin Domoic Acid From Weekly Samples and Offshore Cruises With the Northeast U.S. Shelf (NES) Long-Term Ecological Research (LTER),” in Biological and Chemical Oceanography Data Management Office (BCO-DMO). (Version 1) Version Date 2021-04-05 [Subset to NBPTS Samples]. doi: 10.26008/1912/bco-dmo.847448.1
Kaczmarska I., Reid C., Martin J. L., Moniz M. B. J. (2008). Morphological, Biological, and Molecular Characteristics of the Diatom Pseudo-Nitzschia Delicatissima From the Canadian Maritimes. Botany 86, 763–772. doi: 10.1139/B08-046
Lelong A., Hégaret H., Soudant P., Bates S. S. (2012). Pseudo-Nitzschia (Bacillariophyceae) Species, Domoic Acid and Amnesic Shellfish Poisoning: Revisiting Previous Paradigms. Phycologia 51 (2), 168–216. doi: 10.2216/11-37.1
Lopes dos Santos A., Gérikas Ribeiro C., Ong D., Garczarek L., Shi X. L., Nodder S. D., et al. (2022). “Chapter 11 - Phytoplankton Diversity and Ecology Through the Lens of High Throughput Sequencing Technologies,” in Advances in Phytoplankton Ecology. Eds. Clementson L. A., Eriksen R. S., Willis A. ( Elsevier) Amsterdam, Netherlands, 353–413. doi: 10.1016/B978-0-12-822861-6.00020-0
Lundholm N., Bates S. S., Baugh K. A., Bill B. D., Connell L. B., Léger C., et al. (2012). Cryptic and Pseudo-Cryptic Diversity in Diatoms—With Descriptions of Pseudo-Nitzschia Hasleana Sp. Nov. And P. Fryxelliana Sp. Nov. 1. J. Phycol. 48 (2), 436–454. doi: 10.1111/j.1529-8817.2012.01132.x
Lundholm N., Krock B., John U., Skov J., Cheng J., Pančić M., et al. (2018). Induction of Domoic Acid Production in Diatoms—Types of Grazers and Diatoms are Important. Harmful. Algae. 79, 64–73. doi: 10.1016/j.hal.2018.06.005
Lundholm N., Moestrup Ø., Hasle G. R., Hoef-Emden K. (2003). A Study of the Pseudo-Nitzschia Pseudodelicatissima/Cuspidata Complex (Bacillariophyceae): What is P. Pseudodelicatissima? J. Phycol. 39, 797–813. doi: 10.1046/j.1529-8817.2003.02031.x
Maldonado M. T., Hughes M. P., Rue E. L., Wells M. L. (2002). The Effect of Fe and Cu on Growth and Domoic Acid Production by Pseudo-Nitzschia Multiseries and Pseudo-Nitzschia Australis. Limnol. Oceanography. 47 (2), 515–526. doi: 10.4319/lo.2002.47.2.0515
Martin M. (2011). Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet. J. 17 (1), 10–12. doi: 10.14806/ej.17.1.200
McCabe R. M., Hickey B. M., Kudela R. M., Lefebvre K. A., Adams N. G., Bill B. D., et al. (2016). An Unprecedented Coastwide Toxic Algal Bloom Linked to Anomalous Ocean Conditions. Geophysical. Res. Lett. 43 (19), 10366–10376. doi: 10.1002/2016GL070023
McMurdie P. J., Holmes S. (2013). Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 8 (4) e61217. doi: 10.1371/journal.pone.0061217
NSSP (2019) National Shellfish Sanitation Program (NSSP) Guide for the Control of Molluscan Shellfish. Available at: https://www.issc.org/nssp-guide.
Oviatt C., Smith L., Krumholz J., Coupland C., Stoffel H., Keller A., et al. (2017). Managed Nutrient Reduction Impacts on Nutrient Concentrations, Water Clarity, Primary Production, and Hypoxia in a North Temperate Estuary. Estuarine. Coast. Shelf. Sci. 199, 25–34. doi: 10.1016/j.ecss.2017.09.026
Pan Y., Bates S. S., Cembella A. D. (1998). Environmental Stress and Domoic Acid Production by Pseudo-Nitzschia: A Physiological Perspective. Natural Toxins. 6, 127–135. doi: 10.1002/(SICI)1522-7189(199805/08)6:3/4<127::AID-NT9>3.0.CO;2-2
Raposa K. B. (2009). “Ch 7: Ecological Geography of Narragansett Bay,” in An Ecological Profile of the Narragansett Bay National Estuarine Research Reserve. Raposa K.B., Schwartz M.L. (eds.), Rhode Island Sea Grant, Narragansett, R.I.176pp.
R Core Team. (2017). R: A Language and Environment for Statistical Computing ( R Foundation for Statistical Computing). Available at: https://www.R-project.org.
RI DEM (2021). “Rhode Island Department of Environmental Management (RI DEM) and Rhode Island Department of Health (RI DOH),” in Harmful Algal Bloom and Shellfish Biotoxin Monitoring and Contingency Plan. Available at: http://www.dem.ri.gov/programs/benviron/water/shellfsh/pdf/habplan.pdf.
RStudio Team (2020). RStudio: Integrated Development for R (RStudio, PBC). Available at: http://www.rstudio.com.
Rynearson T. A., Flickinger S. A., Fontaine D. N. (2020). Metabarcoding Reveals Temporal Patterns of Community Composition and Realized Thermal Niches of Thalassiosira Spp. (Bacillariophyceae) From the Narragansett Bay Long-Term Plankton Time Series. Biology 9 (1), 19. doi: 10.3390/biology9010019
Sterling A. R., Kirk R. D., Bertin M. J., Rynearson T. A., Borkman D. G., Caponi M. C., et al (in press). Emerging Harmful Algal Blooms Caused by Distinct Seasonal Assemblages of The Toxic Diatom Pseudo-nitzschia. Limnol. Oceanography.
Stonik I. V., Orlova T. Y., Lundholm N. (2011). Diversity of Pseudo-Nitzschia H. Peragallo From the Western North Pacific. Diatom. Res. 26 (1), 121–134. doi: 10.1080/0269249X.2011.573706
Sun J., Hutchins D. A., Feng Y., Seubert E. L., Caron D. A., Fu F.-X. (2011). Effects of Changing Pco2 and Phosphate Availability on Domoic Acid Production and Physiology of the Marine Harmful Bloom Diatom Pseudo-Nitzschia Multiseries. Limnol. Oceanography. 56 (3), 829–840. doi: 10.4319/lo.2011.56.3.0829
Testa J. M., Murphy R. R., Brady D. C., Kemp W. M. (2018). Nutrient- and Climate-Induced Shifts in the Phenology of Linked Biogeochemical Cycles in a Temperate Estuary. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00114
Wells M. L., Trainer V. L., Smayda T. J., Karlson B. S., Trick C. G., Kudela R. M., et al. (2015). Harmful Algal Blooms and Climate Change: Learning From the Past and Present to Forecast the Future. Harmful. Algae. 49, 68–93. doi: 10.1016/j.hal.2015.07.009
White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics,” in PCR Protocols: A Guide to Methods and Applications. Academic Press, Inc., New York, vol. 18. , 315–322.
Wickham H. (2016). Ggplot2. Wiley. Interdiscip. Reviews.: Comput. Stat 3 (2), 180–185. doi: 10.1002/wics.147
Zaiko A., Samuiloviene A., Ardura A., Garcia-Vazquez E. (2015). Metabarcoding Approach for Nonindigenous Species Surveillance in Marine Coastal Waters. Mar. pollut. Bull. 100 (1), 53–59. doi: 10.1016/j.marpolbul.2015.09.030
Keywords: Pseudo-nitzschia, DNA metabarcoding, Narragansett Bay, harmful algal blooms (HAB), long-term trends
Citation: Roche KM, Sterling AR, Rynearson TA, Bertin MJ and Jenkins BD (2022) A Decade of Time Series Sampling Reveals Thermal Variation and Shifts in Pseudo-nitzschia Species Composition That Contribute to Harmful Algal Blooms in an Eastern US Estuary. Front. Mar. Sci. 9:889840. doi: 10.3389/fmars.2022.889840
Received: 04 March 2022; Accepted: 03 June 2022;
Published: 22 July 2022.
Edited by:
Margaret R. Mulholland, Old Dominion University, United StatesReviewed by:
Savvas Genitsaris, National and Kapodistrian University of Athens, GreeceCopyright © 2022 Roche, Sterling, Rynearson, Bertin and Jenkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bethany D. Jenkins, YmRqZW5raW5zQHVyaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.