- 1Key Laboratory of Mariculture, Ministry of Education, Ocean University of China, Qingdao, China
- 2Function Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Zhejiang Province Key Laboratory of Mariculture and Enhancement, Zhejiang Marine Fisheries Research Institute, Zhoushan, China
In order to investigate the effects of razor clams (Sinonovacula constricta) on the food composition and isotopic niches of swimming crabs (Portunus trituberculatus) and kuruma shrimp (Marsupenaeus japonicus) in polyculture systems, this study analyzed 60 P. trituberculatus, 60 M. japonicus and 30 S. constricta to quantify the food sources, food source contributions, and isotopic niches of cultured organisms using 18S rDNA barcoding and stable isotope techniques. The results were as follows: (1) In the P. trituberculatus-M. japonicus (PM) polyculture system, the Sobs and Shannon-Wiener indices of the stomach contents of P. trituberculatus and M. japonicus were not significantly different (P > 0.05). In the P. trituberculatus-M. japonicus-S. constricta polyculture (PMS) system, the Sobs and Shannon-Wiener indices of the stomach contents of P. trituberculatus and M. japonicus were also not significantly different (P > 0.05), but the Sobs indices of P. trituberculatus in the PMS system were significantly higher than those in the PM system (P < 0.05), M. japonicus shows a similar pattern. (2) 18S rDNA barcoding analysis showed the dominant taxa in the stomach contents of both P. trituberculatus and M. japonicus in the PM system were Trebouxiophyceae, Embryophyta and Rotifera, and the food overlap between them was 0.8992, which was significant (Q > 0.6). In the PMS system, the dominant taxa in the stomach contents of P. trituberculatus were Chrysophyceae, Intramacronucleata, and Embryophyta, and in M. japonicus were Chrysophyceae, Embryophyta, and Bacillariophyceae, in this system the food overlap was 0.2061, which was not significant (Q < 0.6). (3) Stable isotope analysis suggested, in both systems, the main food sources of P. trituberculatus and M. japonicus were iced trash fish, zooplankton, phytoplankton, and organic particulate matter (POM). Iced trash fish accounted 77.67% of food sources for P. trituberculatus and 69.42% for M. japonicus in the PM system, and 60.82% and 57.60% in the PMS system. (4) The isotopic niche overlap between P. trituberculatus and M. japonicus was 5.69% in the PM system and 1.21% in the PMS system. These results suggested food competition between P. trituberculatus and M. japonicus, and S. constricta can reduce the competition and isotopic niche overlap, improve the contribution of food sources such as phytoplankton. Razor clams also serve to purify the water and improve the utilization of iced trash fish by filtering phytoplankton (51.10%), POM (32.25%), SOM (7.47%), and iced trash fish (9.18%). Thus, P. trituberculatus-M. japonicus-S. constricta is a healthy and sustainable culture model.
Introduction
Portunus trituberculatus, commonly known as the swimming crab, belongs to Crustacea, Decapoda, and Portunidae, and is widely distributed in the Yellow and Bohai Seas and the East China Sea (Dai et al., 1986). Farm production of P. trituberculatus in China reached 113,800 t in 2019, accounting for 41.5% of the farm production of marine crabs (The Ministry of Agriculture Fishery and Fishery Administration, 2021). P. trituberculatus is an important seawater pond cultured species, and is usually polycultured with Pacific white shrimp (Litopenaeus Vannamei), kuruma shrimp (Marsupenaeus japonicus), razor clams (Sinonovacula constricta), Manila clams (Ruditapes philippinarum), and redlip mullet (Liza haematocheila) (Wang et al., 2009; Zhou et al., 2010; Wang, 2011). P. trituberculatus and M. japonicus can co-exist well in a system due to their different physiological characteristics and food processing methods (Dai et al., 1986; Pérez-Farfante and Kensley, 1997; Buck et al., 2003; Wang et al., 2009; Wang, 2011), improving space utilization and production, iced trash fish are fed in production. Most of the studies about polyculturing P. trituberculatus and M. japonicus have focused on the effects of environmental factors or microbial communities on the ecosystem (Dong, 2013; Ban, 2015). The food habits and trophic niches of P. trituberculatus and M. japonicus are similar (Brzeski and Newkirk, 1997; Yang, 2001; Gao et al., 2020; Tao et al., 2020), but there is a lack of quantitative food source analysis between them in polyculture systems.
Filter-feeding bivalves have become the main species in polyculture seawater ponds due to their ability to improve water quality (Dong et al., 1999), increase nutrient utilization efficiency (Vaughn and Hakenkamp, 2010; Guo et al, 2017), improved survival rate of co-cultured animals (Mckindsey et al., 2007; Vaughn and Hakenkamp, 2010), and enhance the stability of culture systems (Wang, 2011). The effects of filter-feeding bivalves on plankton community structure in pond water and nitrogen and phosphorus budgets in culture systems have been reported (Yang, 1998; Dong et al., 1999; Zhang et al., 2015; Guo et al., 2017), but their effects on food resources of cultured organisms in polyculture systems have rarely been addressed.
In this study, we combined 18S rDNA barcoding and stable isotope techniques with samples collected at the middle of the culture period (October) as experimental material to quantify the food sources, contributions, and isotopic niches of cultured organisms in a P. trituberculatus-M. japonicu (PM) system and a P. trituberculatus-M. japonicu-S. constricta (PMS) system, investigate the feasibility of polyculturing P. trituberculatus with M. japonicu and the effect of S. constricta on their food sources and isotopic niches. Our objectives were to (1) describe food competition between P. trituberculatus and M. japonicu, and (2) determine whether polyculturing S. constricta affected food competition between P. trituberculatus and M. japonicu. The results of this study can provide a scientific basis for rational combination of cultured organisms in integrated ponds with P. trituberculatus.
Materials and Methods
Pond Management
The experiment was conducted on Changbai Island (30°11′15.22″N, 122°2′40.95″E), Zhoushan, Zhejiang Province, China, which has a subtropical monsoon climate with an annual average temperature of 16.1 ~ 16.4 °C and an average precipitation of 1442.9 mm. The experimental pond covered an area of 1.33 ha with an average water depth of 1.2 m. The culture models are P. trituberculatus-M. japonicu (PM) and P. trituberculatus-M. japonicu-S. constricta (PMS). In July 2020, 50 kg of healthy S. constricta (shell length: SL = 1.85 ± 0.14 cm, mean ± SD, n = 53) with uniform size were stocked, 10 kg of vigorous juvenile P. trituberculatus (carapace width: CW = 0.41 ± 0.02cm, mean ± SD, n = 82) and 26 kg of energetic M. japonicus (body length: BL = 1.02 ± 0.07cm, mean ± SD, n = 64) with sound appendages were stocked after half a month (Dong et al., 2021). 40kg of iced trash fish, including fish (Nibea albiflora and Trachinotus blochii), shrimp (Oratosquilla oratoria and Solenocera crassicornis), and crab (Portunus pelagicus) were provided daily at 17:00. Water was changed 1-2 times per month, 30% each time. Sediment consists mainly of mud, with very low levels of benthic microalgae and benthos. The experimental period was from July 2020 to January 2021, and the ranges of water temperature, salinity, dissolved oxygen and transparency during the period were 6.5 ~ 34.0°C, 14.5 ~ 19.0, 6.0 ~ 12.3 mg/L and 40~100 cm, respectively.
Sample Collection and Treatment
Samples of cultured organisms (P. trituberculatus, M. japonicus and S. constricta) and potential food sources (iced trash fish, zooplankton, phytoplankton, POM and SOM) were collected on October 13, 2020. 30 each of P.trituberculatus (carapace width: CW = 14.76 ± 1.74 cm, mean ± SD, n = 60) and M. japonicus (body length: BL = 11.02 ± 1.13 cm, mean ± SD, n = 60) in the PM system, and 30 each of P.trituberculatus (carapace width: CW = 20.07 ± 3.55 cm, mean ± SD, n = 60), M. japonicus (body length: BL = 12.05 ± 1.78 cm, mean ± SD, n = 60) and S. constricta (shell length: SL = 8.94 ± 1.44 cm, mean ± SD, n = 90) were collected. The pond is divided into 5 points according to the diagonal line to collect mixed water samples of 10 L each, zooplankton and phytoplankton were collected by filtering 50 L water through No. 13 (20 cm mouth diameter, 112 μm mesh size) and No. 25 (20 cm mouth diameter, 64 μm mesh size) plankton nets, respectively, and the filtrate was extracted onto pre-cauterized (500°C, 5 h) Whatman GF/F membranes for POM. SOM were collected from 1–2 cm of the sediment surface with a column collector. All samples were stored on dry ice for rapid transport back to the laboratory.
Stomach contents were collected in 5 mL lyophilized tubes and transferred to -80°C storage for subsequent 18S rDNA analysis. The foot muscles of S. constricta, cheliped muscles of P. trituberculatus, abdomen muscles of M. japonicus (Hill and Mcquaid, 2009), and all muscles of iced trash fish were treated with 1 mol/L hydrochloric acid and then rinsed with distilled water. Muscles and filter membranes containing samples were dried in a 60 °C oven (DGG-9140A) to constant weight. Muscle samples were ground into powder and collected in 1.5 mL centrifuge tubes, and then stored in a desiccator for subsequent stable isotope analysis.
18S rDNA Barcoding Analysis
Genomic DNA extraction from stomach contents was performed using the E.Z.N.A.® soil DNA kit, the quality of extraction was detected using 1% agarose gel electrophoresis, and DNA concentration and purity were evaluated using NanoDrop2000. PCR amplification of the variable V4 region of the 18S rDNA gene was performed using the universal primers TAREF (5′-CCAGCASCYGCGGTAATTCC-3′) and TARER (5′-ACTTTCGTTCTTGATYRA-3′) with the following amplification procedure: 95°C pre-denaturation for 3 min, 27 cycles (95°C denaturation for 30 s, 55°C). The PCR reaction system was as follows: 5 × TransStart FastPfu buffer 4 μL, 2.5 mM dNTPs 2 μL, upstream primer (5 uM) 0.8 μL, downstream primer (5 uM) 0.8 μL, TransStart FastPfu DNA polymerase 0.4 μL, template DNA 10 ng, made up to 20 μL for three replicates per sample. The PCR products were mixed and recovered on 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen BioPMiences, Union City, CA, USA), and detected using 2% agarose gel electrophoresis. The recovered products were quantified using a Quantus™ Fluorometer (Promega, USA). Libraries were built using NEXTFLEX Rapid DNA-Seq Kit and sequenced using Illumina’s Miseq PE300 platform.
Stable Isotope Analysis
All samples were wrapped in aluminum foil and sent to the stable isotope ratio mass spectrometer (Delta V Advantage, Thermo Fisher PMientific, Inc.) for analysis. The carbon and nitrogen stable isotope values were based on the international reference materials PDB (Pee Dee Belemnite) and atmospheric N2, respectively. Stable isotope abundances were expressed in delta (δ) notation as the deviation from the standards in parts per thousand according to the following equation:
Calculation of the Competition
The competition between cultured organisms can be expressed by food overlap with the following equation:
The competition between cultured organisms can also be expressed by the overlap of isotopic niches with the following equation:
Data Analysis
Owing to the presence of interference data, the original data were spliced and filtered to generate more accurate and reliable data for analysis. Sequences were classified as operational taxonomic units (OTUs) at 97% sequence similarity level by UPASE (version 7.1). The taxonomy of each OTU representative sequence was analyzed using RDP Classifier against the Silva database with a confidence threshold of 0.7.
An SIAR (stable isotope analysis in R) linear mixed model was used to analyze the contribution of different food sources to consumers, and benthos were not considered in the calculation due to their low abundance in the sediment. Aquatic omnivores took muscle tissue, and Δ13C was taken as 1.3‰ ± 0.3‰ without lipid removal. The value of Δ13C was taken as 1.5‰ and Δ15N as 2.5‰ for S. constricta (Mccutchan et al., 2003). The stable isotope Bayesian ellipses in R (SIBER) were used to calculate the isotopic niche (standard elliptical area, SEA; convex hull area, TA) of cultured organisms and their overlapping area (OA) (Jackson et al., 2011). In this paper, we calculate the proportion of the overlap to the niche, and to keep the sample size consistent, the niche is calculated using the standard ellipse area (SEA). The data obtained were analyzed using the MATLAB software. The anova1 function was used for one-way ANOVA to check whether the stable isotope values had the same mean values at a significance level of P < 0.05. The plotting software was ORIGIN2020.
Results
α-Diversity of the Stomach Contents of Cultured Organisms
The Sobs index and Shannon-Wiener index reflect the abundance and diversity of species communities, respectively (Beck, 2010). In the PM system, the Sobs indices of the stomach contents of P. trituberculatus and M. japonicus were 30.5 and 31.0 (Figure 1A), and the Shannon-Wiener indices were 2.09 and 1.95 (Figure 1B), respectively, none of these differences were statistically significant (P > 0.05). In the PMS system, the Sobs indices of the stomach contents of P. trituberculatus and M. japonicus were 51.6 and 50, and the Shannon-Wiener indices were 2.04 and 1.87, respectively. Likewise, none of these differences were statistically significant (P > 0.05). There were, however, significant differences between the Sobs indices of the stomach contents of both P. trituberculatus and M. japonicus in the two systems (P < 0.05). There was no significant difference between the Shannon-Wiener indices (P > 0.05). The Sobs index of the stomach contents of S. constricta was 48.7 and the Shannon-Wiener index was 0.80.
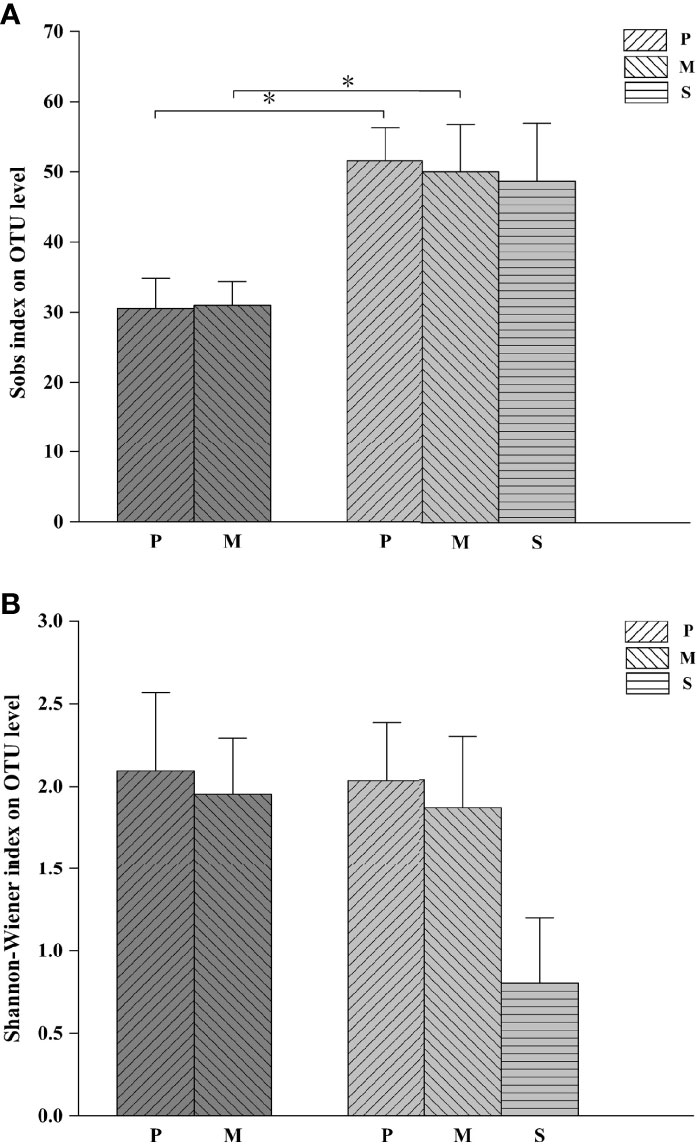
Figure 1 Sobs (A) and Shannon-Wiener (B) indices of stomach contents of cultured organisms. Dark Gray represents the PM system and light gray represents the PMS system; P represents P. trituberculatus; M represents M. japonicus; S represents S. constricta; * indicates significant difference among groups (P < 0.05).
The Main Eukaryotic Composition of the Stomach Contents of Cultured Organisms
In the PM system, 39 phyla and 68 classes were identified in the stomach contents of P. trituberculatus, and the dominant taxa were Trebouxiophyceae, Embryophyta and Rotifera with relative abundances of 30.88%, 19.46% and 11.95%, respectively (Figure 2A). A total of 25 phyla and 40 classes were identified in the stomach contents of M. japonicus. Trebouxiophyceae, Embryophyta and Rotifera were the dominant taxa, with relative abundances of 47.66%, 18.38% and 13.46%, respectively (Figure 2B). In the PMS system, 32 phyla and 58 classes were identified in the stomach contents of P. trituberculatus, and the dominant taxa were Chrysophyceae, Intramacronucleata and Embryophyta, with relative abundances of 61.37%, 15.70% and 11.81%, respectively (Figure 2C). 36 phyla and 61 classes were identified in the stomach contents of M. japonicus. Chrysophyceae, Embryophyta and Bacillariophyceae were the dominant taxa, with relative abundances of 38.99%, 28.32% and 13.30%, respectively (Figure 2D). 36 phyla and 70 classes were identified in the stomach contents of S. constricta, and the dominant taxa were Dinophyceae, Trebouxiophyceae and Chrysophyceaewith relative abundances of 48.32%, 26.18% and 14.57%, respectively (Figure 2E).
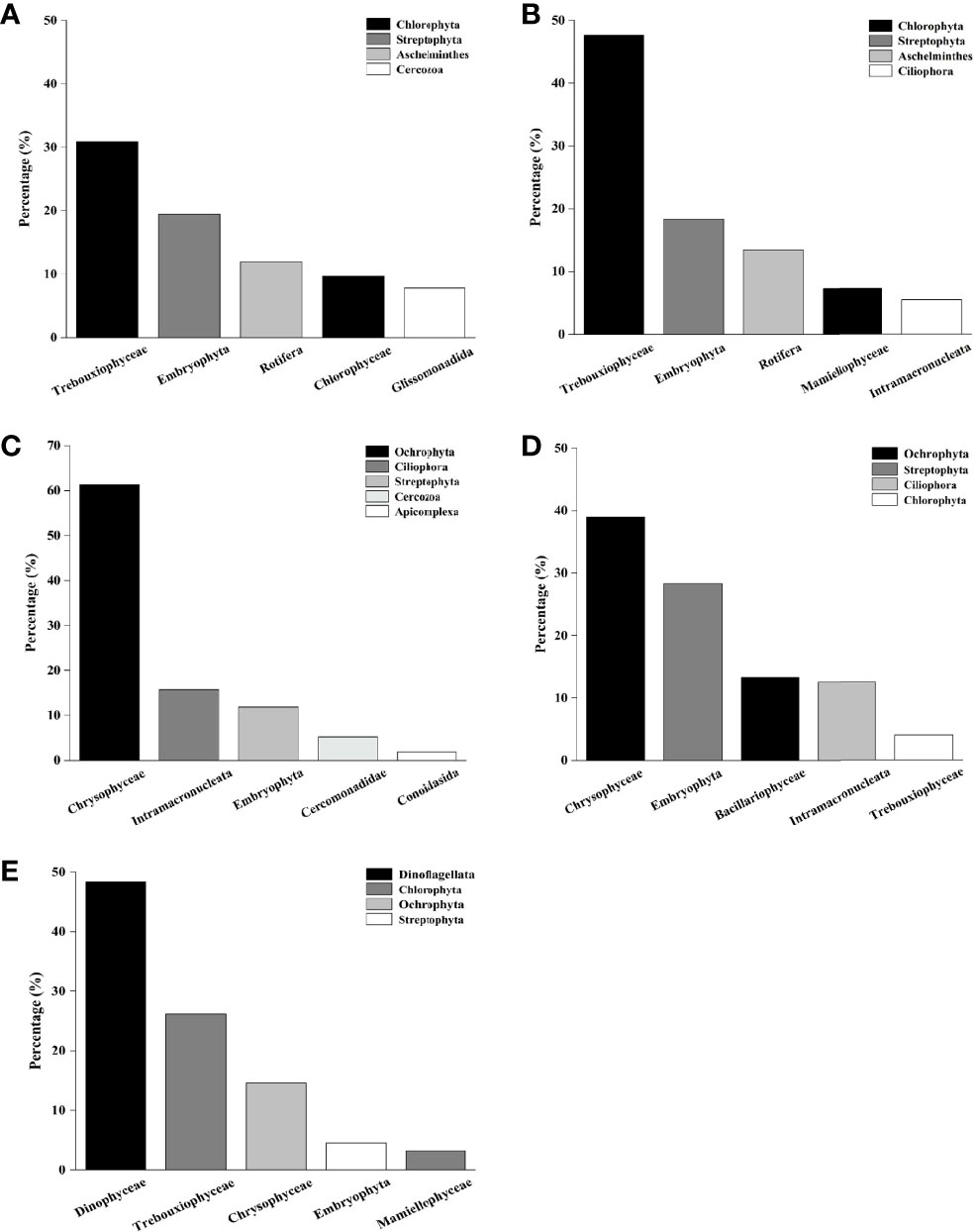
Figure 2 Main eukaryote compositions of stomach contents of cultured organisms. (A) Main eukaryote compositions of stomach contents of P. trituberculatus in the PM system; (B) Main eukaryote compositions of stomach contents of M. japonicus in the PM system; (C) Main eukaryote compositions of stomach contents of P. trituberculatus in the PMS system; (D) Main eukaryote compositions of stomach contents of M. japonicus in the PMS system; (E) Main eukaryote compositions of stomach contents of S. constricta in the PMS system.
In the PM system, the stomach contents of P. trituberculatus and M. japonicus had a total of 199 OTUs and 38 common OTUs (Figure 3), and the food overlap was 0.8992, indicating severe competition (Q > 0.6). In the PMS system, the stomach contents of P. trituberculatus and M. japonicus had a total of 191 OTUs and 19 common OTUs, with a non-significant food overlap of 0.2061 (Q < 0.6).
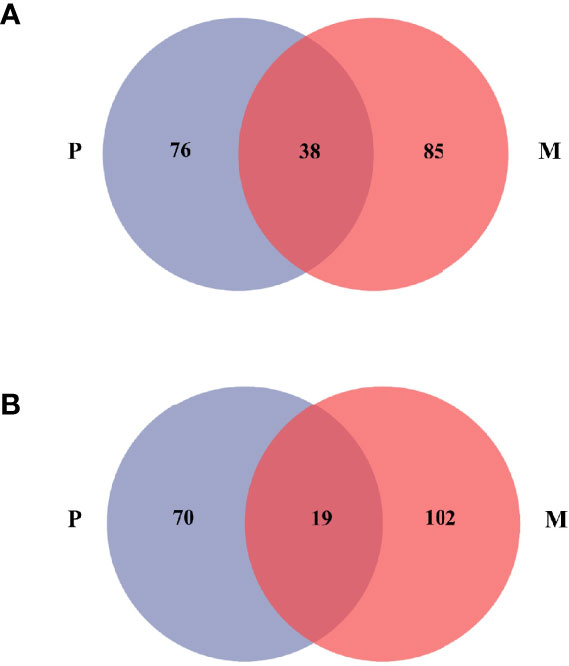
Figure 3 Distribution of OTUs of the stomach contents of P.trituberculatus and M.japonicus in the PM (A) and PMS (B) systems. Blue circle represents P.trituberculatus and red circle represents M.japonicus.
Stable Isotope Characteristics of Cultured Organisms
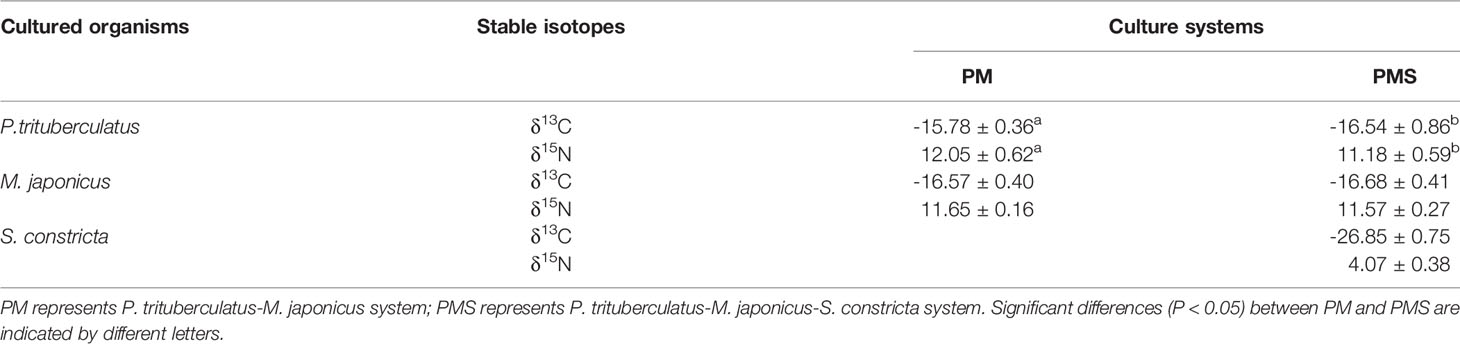
The δ13C and δ15N of P. trituberculatus in the PMS system were -16.54 ± 0.86‰ and 11.18 ± 0.59‰, respectively, and were significantly lower than those of P. trituberculatus in the PM system (δ13C = -15.78 ± 0.36‰ and δ15N = 12.05 ± 0.62‰) (P < 0.05, Table 1). The δ13C and δ15N of M. japonicus in the two systems were not significantly different (P > 0.05), and the δ13C and δ15N of S. constricta were -26.85 ± 0.75‰ and 4.07 ± 0.38‰, respectively.
Contributions of Different Food Sources to Cultured Organisms
An SIAR linear mixed model was used to analyze the contributions of different food sources to the cultured organisms (Figure 4) and showed that the main food sources of P. trituberculatus and M. japonicus included iced trash fish, zooplankton, phytoplankton, and POM. Their contributions to P. trituberculatus in the PM system were 77.67%, 3.78%, 8.22%, and 10.34%, respectively, and their contributions to M. japonicus in the PM system were 69.42%, 1.86%, 6.76%, and 21.97%, respectively. Their contributions to P. trituberculatus in the PMS system were 60.82%, 2.40%, 17.54%, and 19.24%, respectively, and their contributions to M. japonicus in the PMS system were 57.60%, 2.03%, 18.62%, and 21.74%, respectively. The main food sources of S. constricta included phytoplankton (51.10%), POM (32.25%), SOM (7.47%), and iced trash fish (9.18%).
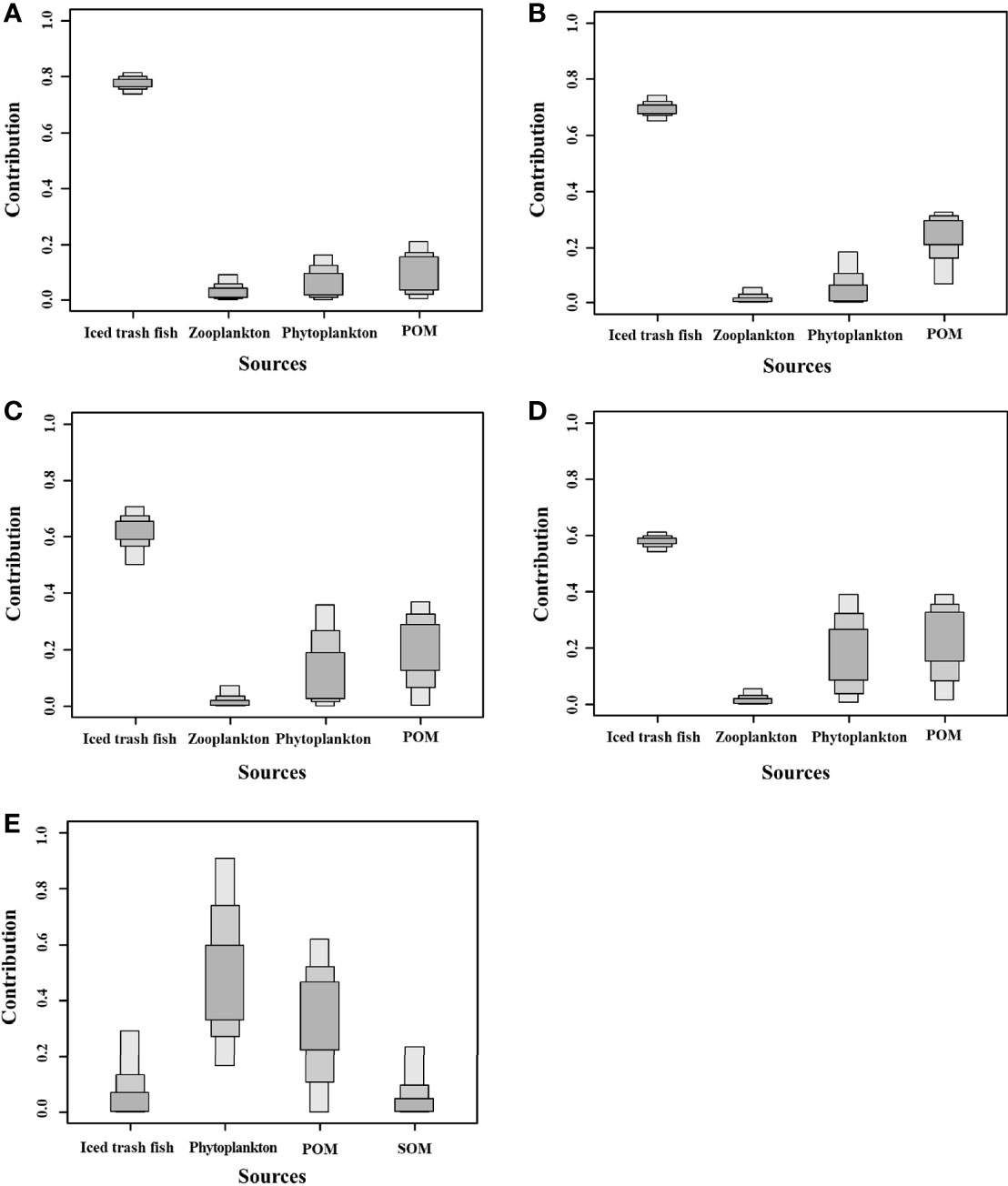
Figure 4 Contributions of different food sources to cultured organisms. (A) Contributions of different food sources to P. trituberculatus in the PM system; (B) Contributions of different food sources to M. japonicus in the PM system; (C) Contributions of different food sources to P. trituberculatus in the PMS system; (D) Contributions of different food sources to M. japonicus in the PMS system; (E) Contributions of different food sources to S. constricta in the PMS system.
Isotopic Niches of Cultured Organisms
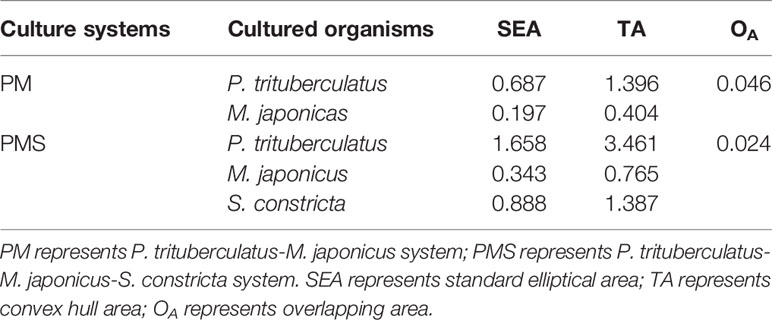
In both culture systems, each cultured organism occupied a unique niche space (Figure 5). In the PM system, SEA and Overlap for P. trituberculatus and M. japonicus were 0.687, 0.197, and 0.046, with a niche overlap of 5.69%. In the PMS system, SEA and Overlap for P. trituberculatus and M. japonicus were 1.658, 0.343, and 0.024, with a niche overlap of 1.21%. The SEA of S. constricta was 0.888, and did not overlap with either P. trituberculatus or M. japonicus. It is noteworthy that the width of the isotopic niches of both P. trituberculatus and M. japonicus in the PMS system expanded relative to the PM system, and the isotopic niche of P. trituberculatus shifted down obviously.
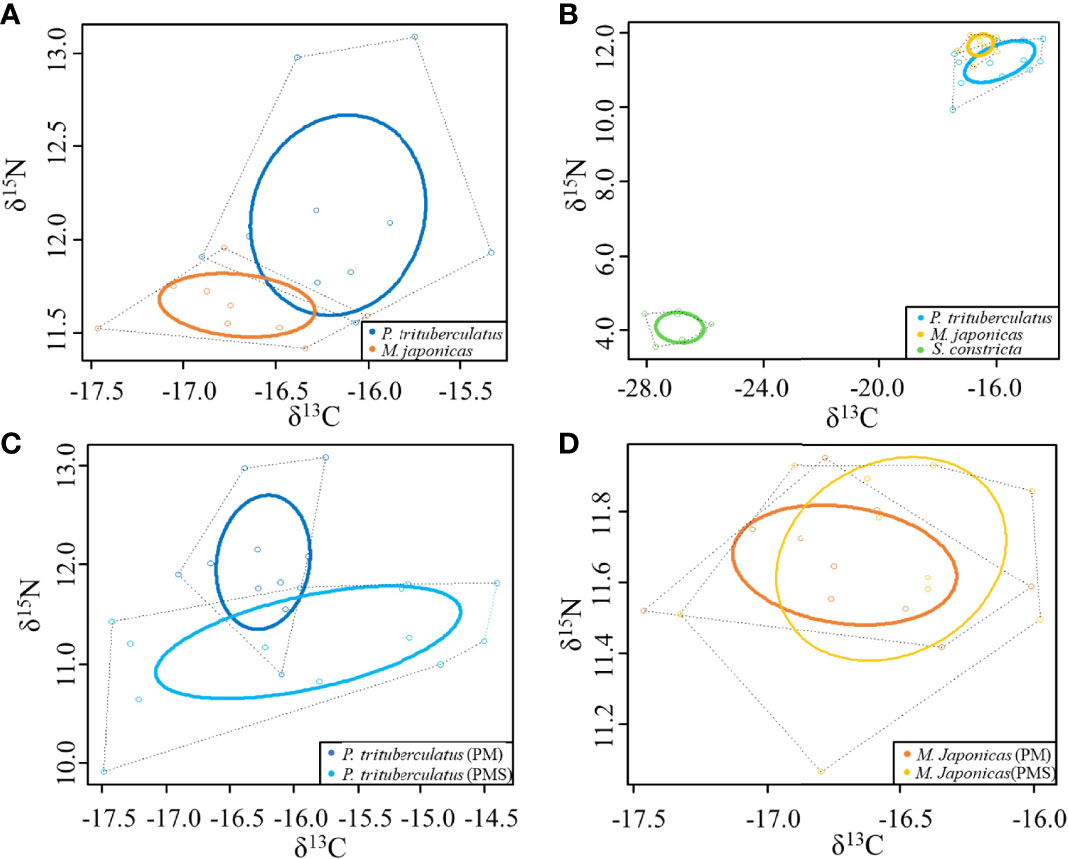
Figure 5 Stable isotopic niches of cultured organisms. (A) Stable isotopic niches of cultured organisms in the PM system; (B) Stable isotopic niches of cultured organisms in the PMS system; (C) Stable isotopic niches of P. trituberculatus in the PM and PMS systems; (D) Stable isotopic niches of M. japonicus in the PM and PMS systems.
Discussion
Food Sources of P. trituberculatus and M. japonicus
Currently, DNA barcoding technologies such as COI (mitochondrial cytochrome oxidase subunit I gene), ITS (internal transcribed spacer region within ribosomal rRNA gene) and 18S rDNA (DNA encoding the small subunit RNA of eukaryotic ribosomes) are widely used to analyze the diets of aquatic animals. 18S rDNA is the most commonly used technology due to its more complete database and better classification ability (Bachy et al., 2013; Leray et al., 2013). 18S rDNA can solve the problem of indistinguishable food fragments from morphological identification, leading to more comprehensive identification of the diets of study subjects (Redmond et al., 2013) and the discovery of easily-overlooked food sources (Rorke et al., 2012), with obvious advantages in terms of data volume, sensitivity, and resolution (Liu et al., 2018). This technique has been applied to food composition studies in sea cucumber (Apostichopus japonicus) and red rock lobster (Jasus edwardsii) (Rorke et al., 2012; Zhang et al., 2016). In this study, we found that the dominant taxa in the stomach contents of both P. trituberculatus and M. japonicus in the PM system included Trebouxiophyceae, Embryophyta, and Rotifera, and their total abundances were 62.29% and 79.50% (Figure 2), respectively. The dominant taxa in the stomach contents of P. trituberculatus and M. japonicus in the PMS system included Chrysophyceae, Intramacronucleata, and Embryophyta, and their total abundances were 88.88% and 79.84%, respectively. That result differed from the stomach contents of shrimp and crabs observed by Yang (2001) may be related to the feeding of iced trash fish and the low abundance of benthos (not collected in this experiment) in this experimental pond. There were no significant differences (P > 0.05) in either the Sobs or Shannon indices of stomach contents of P. trituberculatus or M. japonicus between the two systems (Figure 1), indicating that their food sources are quite similar, likely due to their omnivorous and carnivorous nature (Dai et al., 1986; Pérez-Farfante and Kensley, 1997).
Although the 18S rDNA barcoding technique has clear advantages in diet analysis of farmed animals, the degree of digestion of food by predators can limit its detection intensity (Albaina et al., 2010; Traugott et al., 2020). Stable isotope techniques can reveal trophic relationships between consumers and prey based on their isotope ratio relationships, and can reveal the diets of organisms over a longer period of time, serving as both a complement and correction to 18S rDNA barcoding (Post, 2002; Boecklen et al., 2011; Nielsen et al., 2018). Nelson et al. (2017) used DNA barcoding and stable isotope techniques to compare the feeding habits of different fish. Georgina et al. (2022) used both techniques, founding that the main contribution to the diet of the green crab (Carcinus maenas) came from phytoplankton. Combining these two methods not only identifies the prey being ingested, but also provides information on what is being absorbed.
In this study, the diets of P. trituberculatus and M. japonicus in both systems consisted primarily of iced trash fish, zooplankton, phytoplankton, and POM, with iced trash fish contributing the most (57.60%-77.67%) (Figure 4). Iced trash fish is nutritious, palatable, and easily available. The crustaceans provide calcium for P. trituberculatus and M. japonicus to form new shells during the molting period (Mykles and Skinner, 1982; Chang et al., 1993), and the fish provide protein and essential micronutrients (Gasco et al., 2018), therefore iced trash fish can meet essential growth needs. POM was the second most important food source, accounting for 10.34%-21.97% of P. trituberculatus and M. japonicus diets. POM is an organic particulate matter formed via microbial fermentation of plankton, feces, and residual bait (Liu, 1999). experimental ponds were likely too large to utilize iced trash fish fully, and POM were formed through biological action and ingested by P. trituberculatus and M. japonicus. In cases of excess animal-based bait, consumers will choose plant-based bait, which is lacking in the main diet, for nutritional supplementation, thus increasing the palatability of food and promoting nutritional balance and growth (Buck et al., 2003). This is likely the reason that phytoplankton acted as the third food source for P. trituberculatus and M. japonicus, accounting for 6.76%-18.62% of their diets. The smallest contribution of food sources in this study was zooplankton, perhaps due to its lower nutritional value relative to iced trash fish and its swimming nature (Genin et al., 2005). Isotopic niche is a means of describing trophic niche that reflects the trophic positions of organisms and the degree of competition for resources among populations (Abrams, 1980; Layman et al., 2007a; Post et al., 2007). In this experiment, the isotopic niches of P. trituberculatus and M. japonicus in both systems overlapped, indicating competition for resource utilization between them (Layman et al., 2007a). Therefore, 18S rDNA and stable isotope techniques analyses revealed competition for food resources between P. trituberculatus and M. japonicus in the ponds. Both P. trituberculatus and M. japonicus are basically living at the bottom of the pond, and they have similar foraging times (Dai et al., 1986; Pérez-Farfante and Kensley, 1997). However, because their feeding methods differ (P. trituberculatus use chelipeds to process food and then ingest it, and some food scraps can be used by M. japonicus) and they use the same resources in different ways (Kassen, 2002), there is less competition for food resources, and the bait utilization rate is improved. Thus, it is feasible to polyculture M. japonicus in P. trituberculatus ponds.
Effect of S. constricta on Food Composition and Isotopic Niche of P. trituberculatus and M. japonicus
In the PM system, the dominant taxa in the stomach contents of P. trituberculatus and M. japonicus were mainly Trebouxiophyceae, Embryophyta, and Rotifera, and the food overlap was obvious (Figure 3, Q > 0.6). After polyculturing with S. constricta, the dominant taxa in the stomach contents of P. trituberculatus were Chrysophyceae, Intramacronucleata, and Embryophyta. The abundance of Chrysophyceae in the stomach contents of M. japonicus increased significantly, and the abundance of Embryophyta and Bacillariophyceae decreased significantly, resulting in a decrease in food overlap (Q < 0.6), and indicating that adding S. constricta in the PM system could change the food composition of P. trituberculatus and M. japonicus and reduce food competition. This change may be related to the growth and metabolism of S. constricta, which changed the phytoplankton community structure in the culture water and increased phytoplankton diversity (Yang, 1998; Dong et al., 1999).
The δ13C and δ15N values of P. trituberculatus were significantly different in the two systems (Table 1, P < 0.05), indicating that P. trituberculatus food sources differed significantly (Deniro and Epstein, 1981). The higher δ15N in the PM system was likely a result of the consumption of more high - δ15N food (iced trash fish), while the lower δ13C in the PMS system was likely a result of the consumption of more low - δ13C food (phytoplankton). This indicates that polyculturing with S. constricta can increase the contribution of phytoplankton to P. trituberculatus’ diet. Salman et al. (2008) found that plant - based sources facilitated the increase of bioflocs in the environment, which mainly consisted of residual bait, zooplankton, phytoplankton, and POM, and this corroborated the explanation for increased overall contributions of zooplankton, phytoplankton, and POM in PMS systems (Li et al., 2018). This may be related to the fact that individuals with higher δ15N tend to be larger within the same consumer population (Wilsona et al., 2009).
Species with wider niches are more adaptable to the environment, and expanding niche width can improve the risk resistance of cultured organisms and make their systems more stable (Layman et al., 2007b; Rossi et al., 2015). In this experiment, the contributions of different food sources to P. trituberculatus in the two systems differed significantly; the contribution of iced trash fish to P. trituberculatus in the PMS system decreased by about 18%, while the total contribution of phytoplankton and POM increased by about 18%, indicating significant reduction and expansion of the isotopic niche of P. trituberculatus. In addition, the isotopic niche overlap of P. trituberculatus and M. japonicus decreased from 5.69% to 1.21% (Figure 5, Table 2), indicating that polyculturing S. constricta in the PM system can weaken food competition. S. constricta not only reduced food competition between P. trituberculatus and M. japonicus, but also filtered a total of 48.90% of POM, SOM, and iced trash fish, which had a significant water purification effect. Therefore, the polyculture of P. trituberculatus, M. japonicus and S. constricta is a healthy and sustainable culture model.
Conclusion
The food composition of P. trituberculatus and M. japonicus in two systems did not differ much, and both main food items were iced trash fish, phytoplankton, zooplankton and POM. Polyculturing S. constricta improved the contribution of plant - based sources, reduced the food competition and isotopic niche overlap between P. trituberculatus and M. japonicus. And S. constricta filtered phytoplankton, POM, SOM and iced trash fish to purify water and improve the utilization of iced trash fish. From the food composition and isotopic niche analysis, P. trituberculatus-M. japonicus-S. constricta integrated culture model is healthier and more feasible.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA809854.
Ethics Statement
All procedures were performed under the Regulations of the Administration of Affairs Concerning Experimental Animals of China, as well as the Regulations of the Administration of Affairs Concerning Experimental Animals of Shandong Province.
Author Contributions
XX and SD gathered, analyzed and interpreted data, discussed the results and co-wrote the manuscript. DZ, LY, WP and YX done animal collection and maintenance. FW was the major instructor. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Plan Blue Granary Scientific Innovation (No. 2019YFD0900402) and Yellow River Delta Industry Leading Talents Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albaina A., Fox C. J., Taylor N., Hunter E., Maillard M., Taylor M. I. (2010). A TaqMan Real-Time PCR Based Assay Targeting Plaice (Pleuronectes Platessa L.) DNA to Detect Predation by the Brown Shrimp (Crangon Crangon L.) and the Shore Crab (Carcinus Maenas L.)-Assay Development and Validation. J. Exp. Marine Biol. Ecol. 391, 178–189. doi: 10.1016/j.jembe.2010.06.029
Bachy C., Dolan J., López-García P., Deschamps P., Moreira D. (2013). Accuracy of Protist Diversity Assessments: Morphology Compared With Cloning and Direct Pyrosequencing of 18S rRNA Genes and ITS Regions Using the Conspicuous Tintinnid Ciliates as a Case Study. Isme J. 7, 244–255. doi: 10.1038/ismej.2012.106
Ban W. B. (2015). Structure Optimization, Nitrogen and Phosphorus Budget and Emergy Analysis of Polyculture Systems in Different Portunus Trituberculatus Ponds [D] (Qingdao: Ocean University of China (in Chinese).
Beck J. (2010). Comparing Measures of Species Diversity From Incomplete Inventories: An Update. Methods Ecol. Evol. 1 (1), 38–44. doi: 10.1111/j.2041-210X.2009.00003.x
Boecklen W. J., Yarnes C. T., Cook B. A., James A. C. (2011). On the Use of Stable Isotopes in Trophic Ecology. Annu. Rev. Ecol. Evol. Syst. 42, 411–440. doi: 10.1146/annurev-ecolsys-102209-144726
Brzeski V., Newkirk G. (1997). Integrated Coastal Food Production Systems - a Review of Current Literature. Ocean Coastal Manage. 34 (1), 55–71. doi: 10.1016/S0964-5691(97)82690-7
Buck T. L., Breed G. A., Pennings S. C., Chase M. E., Zimmer M., Carefoot T. H. (2003). Diet Choice in an Omnivorous Salt-Marsh Crab: Different Food Types, Body Size, and Habitat Complexity. J. Exp. Marine Biol. Ecol. 292, 103–116. doi: 10.1016/S0022-0981(03)00146-1
Chang E. S., Bruce M. J., Tamone S. L. (1993). Regulation of Crustacean Molting: A Multi-Hormonal System. Am. Zool. 33, 324–329. doi: 10.1093/icb/33.3.324
Crab R., Defoirdt T., Bossier P., Verstraete W. (2012). Biofloc Technology in Aquaculture: Beneficial Effects and Future Challenges. Aquaculture 356–357, 351–357. doi: 10.1016/j.aquaculture.2012.04.046
Deniro M. J., Epstein S. (1981). Influence of Diet on the Distribution of Nitrogen Isotopes in Animals. Geochimica Cosmochim. Acta 45 (3), 341–351. doi: 10.1016/0016-7037(81)90244-1.
Dong J. (2013). Experimental Study on Structure Optimization and Nitrogen and Phosphorus Budget of Polyculture System in Portunus Trituberculatus Ponds [D] (Ocean University of China (in Chinese).
Dong S. L., Wang F., Wang J., Qi Z. X., Lu J. (1999). Effects of Bay Scallop on Plankton and Water Quality of Marineculture Pond. Acta Oceanologica Sinica. 21, 138–144. doi: 10.1017/S0266078400010713
Dong S. P., Wang F., Zhang D. X., Yu L. Y., Pu W. J., Xu X., et al. (2021). Assessment of the Carrying Capacity of Integrated Pond Aquaculture of Portunus Trituberculatus at the Ecosystem Level. Front. Marine Sci. 8, 747891. doi: 10.3389/fmars.2021.747891
Gao S. K., Yu W. W., Zhang S. (2020). The Potential Carbon Source and Trophic Level Analysis of Main Organisms in Coastal Water of Lüsi Fishing Ground, Based on Carbon and Nitrogen Stable Isotope Analysis. Chin. J. Appl. Ecol. 31, 301–308. doi: 10.3969/J.ISSN.2095-4972.2021.03.006
Gasco L., Gai F., Maricchiolo G., Genovese L., Ragonese S., Bottari T., et al. (2018). “Fishmeal Alternative Protein Sources for Aquaculture Feeds,” in Feeds for the Aquaculture Sector. Springer Briefs in Molecular Science (Cham: Springer), 1–28.
Genin A., Jaffe J. S., Reef R., Richter C., Franks P. J. (2005). Swimming Against the Flow: A Mechanism of Zooplankton Aggregation. Science 308, 860–862. doi: 10.1126/science.1107834
Georgina C., Mariana L., Elisabet V., Bettina T., Gregorio B., Dario A.L., et al. (2022). Metabarcoding, Direct Stomach Observation and Stable Isotope Analysis Reveal a Highly Diverse Diet for the Invasive Green Crab in Atlantic Patagonia. Biol. Invasions 24 (2), 505–526. doi: 10.1101/2020.08.13.249896
Guo K., Zhao W., Jiang Z., Dong S. L. (2017). A Study of Organic Carbon, Nitrogen and Phosphorus Budget in Jellyfish-Shellfish-Fish-Prawn Polyculture Ponds. Aquac. Res. 48, 68–76. doi: 10.1111/are.12861
Hill J. M., Mcquaid C. D. (2009). Effects of Food Quality on Tissue-Specific Isotope Ratios in the Mussel Perna Perna. Hydrobiologia 635, 81–94. doi: 10.1007/s10750-009-9865-y
Jackson A. L., Inger R., Parnell A. C., Bearhop S. (2011). Comparing Isotopic Niche Widths Among and Within Communities: SIBER Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 80, 595–602. doi: 10.1111/j.1365-2656.2011.01806.x
Kassen R. (2002). The Experimental Evolution of Specialists, Generalists, and the Maintenance of Diversity. J. Evol. Biol. 15, 173–190. doi: 10.1046/j.1420-9101.2002.00377.x
Layman C. A., Arrington D. A., Montana C. G., Post D. M. (2007a). Can Stable Isotope Ratios Provide for Community-Wide Measures of Trophic Structure? Ecology 88, 42–48. doi: 10.1890/0012-9658(2007)88[42:CSIRPF]2.0.CO;2
Layman C. A., Quattrochi J. P., Peyer C. M., Allgeier J. E. (2007b). Niche Width Collapse in a Resilient Top Predator Following Ecosystem Fragmentation. Ecol. Letters. 10, 937–944. doi: 10.1111/j.1461-0248.2007.01087.x
Leray M., Yang J. Y., Meyer C. P., Mills S. C., Agudelo N., Ranwez V., et al. (2013). A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. Zool. 10, 1–14. doi: 10.1186/1742-9994-10-34
Li C., Cheng Y. X., Guan Q. Z., Xi Y. W., Li J. Y. (2018). Using Stable Isotopes to Estimate the Effects of Different Carbon/Nitrogen Feeds on Feeding Habit of Procambarus Clarkii in the Rice-Crayfish Co-Culture System. J. Fish. China. 42, 1778–1786. doi: 10.11964/jfc.20171211115
Liu G., Ning Y., Xia X. F., Gong M. (2018). The Application of High-Throughput Sequencing Technologies to Wildlife Diet Analysis. Acta Ecologica Sinica. 38, 3347–3356. doi: 10.5846/stxb201706151092
Mccutchan J. H., Lewis W. M., Kendall C., Mcgrath C. C. (2003). Variation in Trophic Shift for Stable Isotope Ratios of Carbon, Nitrogen, and Sulfur. Oikos 102, 378–390. doi: 10.1034/j.1600-0706.2003.12098.x
Mckindsey C. W., Landry T., Obeirn F. X., Davies I. M. (2007). Bivalve Aquaculture and Exotic Species: A Review of Ecological Considerations and Management Issues. J. Shellfish Res. 26 (2), 281–294. doi: 10.2983/0730-8000(2007)26[281:BAAESA]2.0.CO;2
Mykles D. L., Skinner D. M. (1982). Crustacean Muscles: Atrophy and Regeneration During Molting. Soc. Gen. Physiol. 37, 337–357. doi: 10.1371/journal.pone.0181914
Nelson E. J. H., Holden J., Eves R., Tufts B. (2017). Comparison of Diets for Largemouth and Smallmouth Bass in Eastern Lake Ontario Using DNA Barcoding and Stable Isotope Analysis. PloS One 12, e0181914. doi: 10.1371/journal.pone.0181914
Nielsen J. M., Clare E. L., Hayden B., Brett M. T., Kratina P. (2018). Diet Tracing in Ecology: Method Comparison and Selection. Methods Ecol. Evol. 9, 278–291. doi: 10.1111/2041-210X.12869
Ogloff W. R., Yurkowski D. J., Davoren G. K., Ferguson S. H. (2019). Diet and Isotopic Niche Overlap Elucidate Competition Potential Between Seasonally Sympatric Phocids in the Canadian Arctic. Marine Biol. 106, 1–12. doi: 10.1007/s00227-019-3549-6
Pérez-Farfante I., Kensley B. (1997). “Penaeoid and Sergestoid Shrimps and Prawns of the World,” in Keys and Diagnoses for the Families and Genera, vol. 175. , 1–233.
Post D. M. (2002). Using Stable Isotopes to Estimate Trophic Position: Systems, Methods, and Assumptions. Ecology 83, 703–718. doi: 10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2
Post D. M., Layman C. A., Arrington D. A., Takimoto G., Quattrochi J., Montana C. G. (2007). Getting to the Fat of the Matter: Systems, Methods and Assumptions for Dealing With Lipids in Stable Isotope Analyses. Oecologia 152, 179–189. doi: 10.1007/s00442-006-0630-x
Redmond N. E., Morrow C. C., Thacker R. W., Diaz M. C., Esnault N., Boury C. P., et al. (2013). Phylogeny and Systematics of Demospongiae in Light of New Small-Subunit Ribosomal DNA (18S) Sequences. Integr. Comp. Biol. 53, 388–415. doi: 10.1093/icb/ict078
Rorke R. O., Lavery S., Chow S., Takeyama H., Tsai P., Beckley L. E., et al. (2012). Determining the Diet of Larvae of Western Rock Lobster (Panulirus Cygnus) Using High-Throughput DNA Sequencing Techniques. PloS One 42, e42757. doi: 10.1371/journal.pone.0042757
Rossi L., Lascio A. D., Carlino P., Calizza E., Costantini M. L. (2015). Predator and Detritivore Niche Width Helps to Explain Biocomplexity of Experimental Detritus-Based Food Webs in Four Aquatic and Terrestrial Ecosystems. Ecol. Complexity. 23, 14–24. doi: 10.1016/j.ecocom.2015.04.005
Tao L. S. R., Lau D. C. P., Perkins M. J., Hui T. T. Y., Yau J. K. C., Mak Y. K. Y., et al. (2020). Stable-Isotope Based Trophic Metrics Reveal Early Recovery of Tropical Crustacean Assemblages Following a Trawl Ban. Ecol. Indic. 117, 106610. doi: 10.1016/j.ecolind.2020.106610
The Ministry of Agriculture Fishery and Fishery Administration (2021). China Fishery Statistical Yearbook (Beijing: China Agriculture Press).
Traugott M., Thalinger B., Wallinger C., Sint D. (2020). Fish as Predators and Prey: DNAbased Assessment of Their Role in Food Webs. J. Fish Biol. 98, 367–382. doi: 10.1111/jfb.14400
Vaughn C. C., Hakenkamp C. C. (2010). The Functional Role of Burrowing Bivalves in Freshwater Ecosystems. Freshwater Biol. 46, 1431–1446. doi: 10.1046/j.1365-2427.2001.00771.x
Wallace R. (1981). An Assessment of Diet-Overlap Indexes. Trans. Am. Fisheries Soc. 110, 72–76. doi: 10.1577/1548-8659(1981)110<72:AAODI>2.0.CO;2
Wang X. Q., Cao M., Yan B. L., Zhang Q. Q. (2009). Integrated Culture of Swimming Crab Portunus Trituberculatus. Fisheries Sci. 2, 105–108. doi: 10.16378/j.cnki.1003-1111.2009.02.013
Wilsona R. M., Chantona J., Lewisb G., Nowaceka D. (2009). Isotopic Variation (δ15n, δ13c, and δ34s) With Body Size in Post-Larval Estuarine Consumers. Estuarine Coastal Shelf Sci. 83, 307–312. doi: 10.1016/j.ecss.2009.04.006
Yang H. S. (1998). Research Progress on the Environmental Impact of Filter-Feeding Shellfish on Mariculture Area. Marine Sci. 22, 42–44. doi: 10.1017/S0266078400010713
Yang J. M. (2001). A Study on Food and Trophic Levels of Bohai Sea Invertebrates. Modern Fisheries Inf. 16, 8–16.
Zhang K., Tian X. L., Dong S. L., Dong J., Yan F. J. (2015). Nitrogen and Phosphorus Budgets of Polyculture System of Portunus Trituberculatus, Litopenaeus Vannamei and Ruditapes Philippinarum Vol. 45 (Qingdao: Periodical of Ocean University of China, 44–53.
Zhang H., Xu Q., Zhao Y., Yang H. (2016). Sea Cucumber (Apostichopus Japonicus) Eukaryotic Food Source Composition Determined by 18s rDNA Barcoding. Marine Biol. 163, 1–11. doi: 10.1007/s00227-016-2931-x
Keywords: Portunus trituberculatus, integrated pond culture, 18S rDNA barcoding, stable isotope, food composition
Citation: Xu X, Dong S, Zhang D, Yu L, Pu W, Xie Y, Shan H and Wang F (2022) Filter-Feeding Bivalve Weakens Food Competition Between Crustaceans (Portunus trituberculatus, Marsupenaeus japonicus) in Integrated Culture Ponds: Evidence From 18S rDNA Barcoding and Stable Isotope Analysis. Front. Mar. Sci. 9:884968. doi: 10.3389/fmars.2022.884968
Received: 27 February 2022; Accepted: 18 March 2022;
Published: 08 April 2022.
Edited by:
Min Jin, Ningbo University, ChinaReviewed by:
Ping Liu, Chinese Academy of Fishery Sciences (CAFS), ChinaYunfei Sun, Shanghai Ocean University, China
Copyright © 2022 Xu, Dong, Zhang, Yu, Pu, Xie, Shan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Wang, d2FuZ2ZhbmcyNDlAb3VjLmVkdS5jbg==
†These authors have contributed equally to the work
 Xian Xu
Xian Xu Shipeng Dong1,2†
Shipeng Dong1,2† Dongxu Zhang
Dongxu Zhang Hongwei Shan
Hongwei Shan Fang Wang
Fang Wang