
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 26 May 2022
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.884834
Armophorean ciliates constitute an important component of microeukaryotic community in anaerobic or hypoxic environments. Yet, their diversity remains poorly known due to under-sampling or the scarcity of knowledge. In this study, three metopid ciliates, i.e., Metopus paraes sp. n., Metopus spiculatus sp. n., and Metopus parapellitus sp. n., collected from coastal sediments in Qingdao and Rizhao, China, were investigated using live observation, protargol staining, and molecular phylogenetic methods. M. paraes sp. n. can be distinguished by its long caudal cilia. M. spiculatus sp. n. resembles M. vestitus in many ways, but differs mainly in having a beak-like preoral dome end and a conspicuous tail. The most remarkable features of M. parapellitus sp. n. include an ovate body shape, caudal cilia located at the rear end and right posterior body, and an adoral zone that never extends onto the dorsal surface. Sequence divergences supported the species identification of these three species. Phylogenetic analyses confirmed that the Metopus is not monophyletic, and first revealed that all marine species of Metopus form a well-supported clade. The clustering of these marine forms with M. es (type species) is not rejected by the AU test, which infers that the marine clade represents the genus Metopus together with M. es.
Ciliates inhabiting anaerobic or hypoxic environments have received increased attention recently, not only due to discoveries of new species but also due to studies on the evolution of mitochondria-related organelles and prokaryote–eukaryote symbioses (e.g., Orsi et al., 2012; Fernandes et al., 2018; Lewis et al., 2018; Bourland et al., 2020; Campello-Nunes et al., 2020; Lewis et al., 2020; Rotterová et al., 2020; Li et al., 2021a; Li et al., 2021b; Zhuang et al., 2021). As far as we know, more than half of known anaerobic ciliates belong to class Armophorea Lynn, 2004.
Metopus Claparède and Lachmann, 1858 is the most species-rich anaerobic genus within this class, and includes more than 70 nominal species so far, which makes it morphologically heterogeneous and suggests the need for a splitting (Esteban et al., 1995; Bourland et al., 2014; Bourland et al., 2017a; Bourland et al., 2017b; Omar et al., 2017; Vďačný and Foissner, 2017a; Vďačný and Foissner, 2017b; Bourland et al., 2018a; Bourland et al., 2018b; Rotterová et al., 2018; Vďačný and Foissner, 2019; Bourland et al., 2020; Li et al., 2021). In the last few years, a few new taxa, such as Apometopidae Foissner, 2016, Tropidoatractidae Rotterová et al., 2018, and Heterometopus Foissner, 2016, were established based on the unique morphological features of some former Metopus species. Continual discovery of new species infers that the species diversity is still underestimated within this genus. Additionally, considering that most species of Metopus have not been described using modern methods, and less than 20 species are available of ciliature and molecular data, intensive and extensive survey from under-sampled areas are urgently required to unveil the morphological and genetic biodiversity of this group of ciliates. The taxonomy and phylogeny of this group are still contentious. Metopus species are clustered into two main clades in the phylogeny trees, while most species clustered away from the type species. Interrelationship of the genus is chaotic due to species of Metopus intersecting with substantial genera. Morphology and molecular data from more species are needed for further understanding and clarification of the genus.
The aim of the present study is to reveal the species diversity and elucidate the interspecific relationship within the genus Metopus. During a faunistic survey on ciliates along the coast area of Qingdao, we isolated three marine forms and found that they are new to science: M. paraes sp. n., M. spiculatus sp. n., and M. parapellitus sp. n. Meanwhile, SSU rRNA gene sequencing data were used to investigate their systematic positions. We also discussed the interrelationship between type species and the marine clade.
Metopus paraes sp. n. and M. spiculatus sp. n. were found in black, sulfide-rich sediments of intertidal zones in Bainidi, Qingdao (E120°22′, N36°12′) in June 2020 and in Liujiawan, Rizhao (119°26′, N35°17′) in March 2021, China, respectively. The salinity of both environments was 30‰. M. parapellitus sp. n. was collected from sulfide-rich marine sands near a sewage outfall in Zhanqiao Pier, Qingdao (E120°19′, N36°04′), China, in March 2021; the salinity was 27‰ (Figure 1). Sediments and water samples were collected using plastic bottles (500 ml) and taken back to the laboratory. The samples were kept in anaerobic jars with oxygen-scavenging chemicals (Thermo Scientific Oxoid AnaeroGen) at room temperature (about 25°C). Five autoclaved wheat grains were added to each bottle to promote the growth of bacteria, and the anaerobic ciliates were observed after 2 weeks.
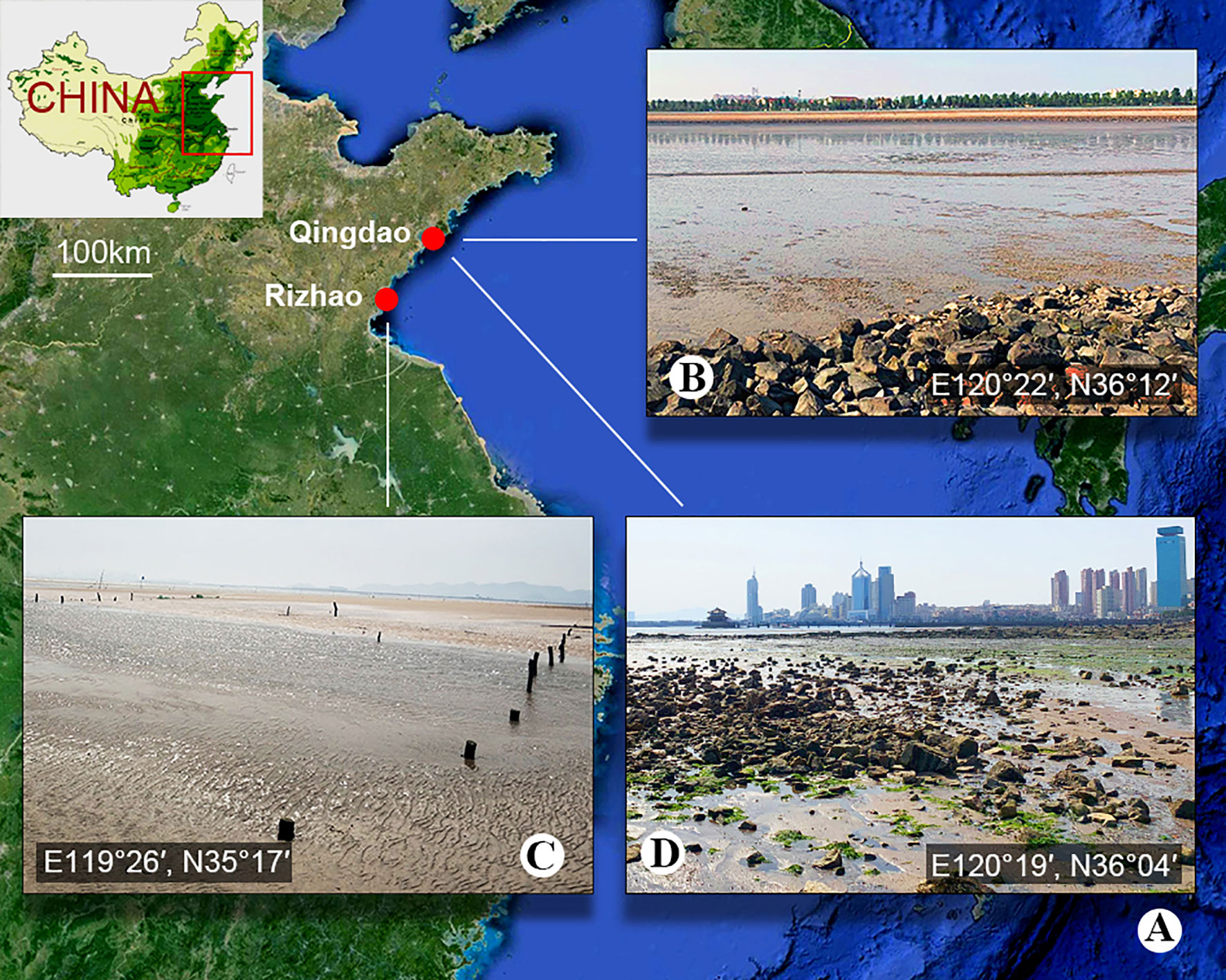
Figure 1 Sample location. (A) Map showing the location of Qingdao and Rizhao, China. (B) The sludge-rich beach of Bainidi in Qingdao. (C) The beach of Liujiawan in Rizhao. (D) The sewage outfall of Zhanqiao Pier in Qingdao.
Living organisms were isolated with a micropipette and observed at 100–1,000× magnification with bright-field and differential interference contrast microscopy using a Zeiss AXIO Imager D2 microscope (Liu et al., 2021). Live cells and DAPI-stained cells were observed under UV light to reveal the autofluorescence of the prokaryotic symbionts and nucleus apparatus (Wu et al., 2020). Protargol staining was performed following the method described in Wilbert (1975) to reveal the ciliature and nucleus apparatus. Protargol powder was prepared according to the protocol described by Pan et al. (2013). Measurements and counts were performed at a magnification of 1,000×. A drawing device was used to draw the specimens.
Three cells of each species were removed using autoclaved micropipettes and washed five times in sterile marine water. Each cell was placed in a 1.5-ml microcentrifuge tube. DNA extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen, Germantown, MD) in accordance with the manufacturer’s instructions. The SSU rRNA gene was amplified using Q5 Hot Start high-fidelity DNA polymerase (NEB, Ipswich, MA). The primers 18S-11F-Karyo (5’-GCCAGTAGTSATATGCTTGTCT-3’) and 5.8SR (5’-TACTGATATGCTTAAGTTCAGCGG-3’) were used for Metopus paraes sp. n. and M. spiculatus sp. n.; 82F (5’-GAAACTGCGAATGGCTC-3’) (Jerome et al., 1996) and 18SR (5’-TGATCCTTCTGCAGGTTCACCTAC-3’) (Medlin et al., 1988) were used as primers for M. parapellitus sp. n. The PCR products were purified using a FastPure Gel DNA Extraction Mini Kit (Nanjing Vazyme Biotech, China) and then cloned using a 5-min TA/Blunt–Zero Cloning Kit (Nanjing Vazyme Biotech, China). Four clones were randomly selected and cultured in a Lysogeny broth medium for 12 h and then sequenced. Sequencing was performed bidirectionally by the Tsingke Biological Technology Company (Beijing, China).
We created a dataset of 73 SSU rDNA sequences, including three newly obtained sequences. These sequences were aligned using the MUSCLE algorithm as described on the EMBL-EBI website (https://www.ebi.ac.uk/Tools/msa/muscle/) (Madeira et al., 2019). The alignments were manually edited by trimming both ends using Bioedit 7.2.3 (Hall, 1999). The final dataset of unambiguously aligned characters consisted of 1,654 positions. Phylogenetic trees were constructed using the maximum likelihood (ML) and Bayesian (BI) methods. ML analysis was performed in RA×ML 8.2.12 (Stamatakis, 2014) according to the GTRGAMMA model. Node support was assessed by 1,000 bootstrap pseudoreplicates. BI analysis was performed using the GTR + I + G model with MrBayes 3.2.6 (Ronquist et al., 2012). Markov Chain Monte Carlo simulations were run for 10,000,000 generations with a sampling frequency of 100 generations; the first 25% of trees were removed as burn-in.
The approximately unbiased (AU) test (Shimodaira, 2002) was performed on the SSU rDNA dataset to test the robustness of phylogenetic associations of particular interest (Chi et al., 2021). The constrained ML tree was generated with the enforced hypothetical relationship of respective target taxa and using the same parameter as the unconstrained ML tree. The site-wise likelihoods for the resulting constrained and non-constrained topologies were calculated using PAUP (Swofford, 2002) and then analyzed by CONSEL (Shimodaira and Hasegawa, 2001) to obtain p-values.
Terminology is based on that used by Lynn (2008) and Foissner and Agatha (1999). False kinetids are short, oblique kineties produced by the highly ordered kinetids of the perizonal stripe (Foissner and Agatha, 1999).
Present work urn:lsid:zoobank.org:pub:F3AC3B76-F9D4-4B43-8320-47C38689380E
Metopus paraes sp. n. urn:lsid:zoobank.org:act:AA8E07B1-5AED-48B4-89F4-42D6FB6788EF
Metopus spiculatus sp. n. urn:lsid:zoobank.org:act:BBFC7268-48EA-496E-B295-9F736C987C2E
Metopus parapellitus sp. n. urn:lsid:zoobank.org:act:0FAB167E-D392-4527-AF17-5F1C80D5E65F
Ciliophora Doflein, 1901
Armophorea Lynn, 2004
Metopida Jankowski, 1980
Metopidae Kahl, 1927
Metopus Claparède and Lachmann, 1858
Metopus paraes sp. n. ( Figures 2, 3 and Table 1)
Marine form about 110–125 × 40–45 μm in vivo. Body shape ellipsoidal to obconical, slightly dorsoventrally flattened. Preoral dome convex, about 50% of body length. Cortical granules colorless, ellipsoidal, arranged in lines. Macronucleus elongate, non-extending to preoral dome. On average, 23 somatic kineties including 9 preoral dome kineties. Perizonal stripe rows never forming false kineties. The adoral zone is composed of 42 membranelles on average, occupying about 40% of body length. Paroral membrane stichomonad. Elongate caudal cilia about 25 μm long.
Sludge sediments of intertidal zone in Bainidi, Qingdao (E120°22′, N36°12′), China.
The protargol slide containing the holotype (Figures 2E, F, 3C, D) (registration number: ZWB202006220101) and another slide containing other paratypes (registration number: ZWB202006220102) were deposited in the Laboratory of Protozoology, Ocean University of China, Qingdao, China. The holotype and relevant paratypes were marked by black ink circles on the backs of the slides.
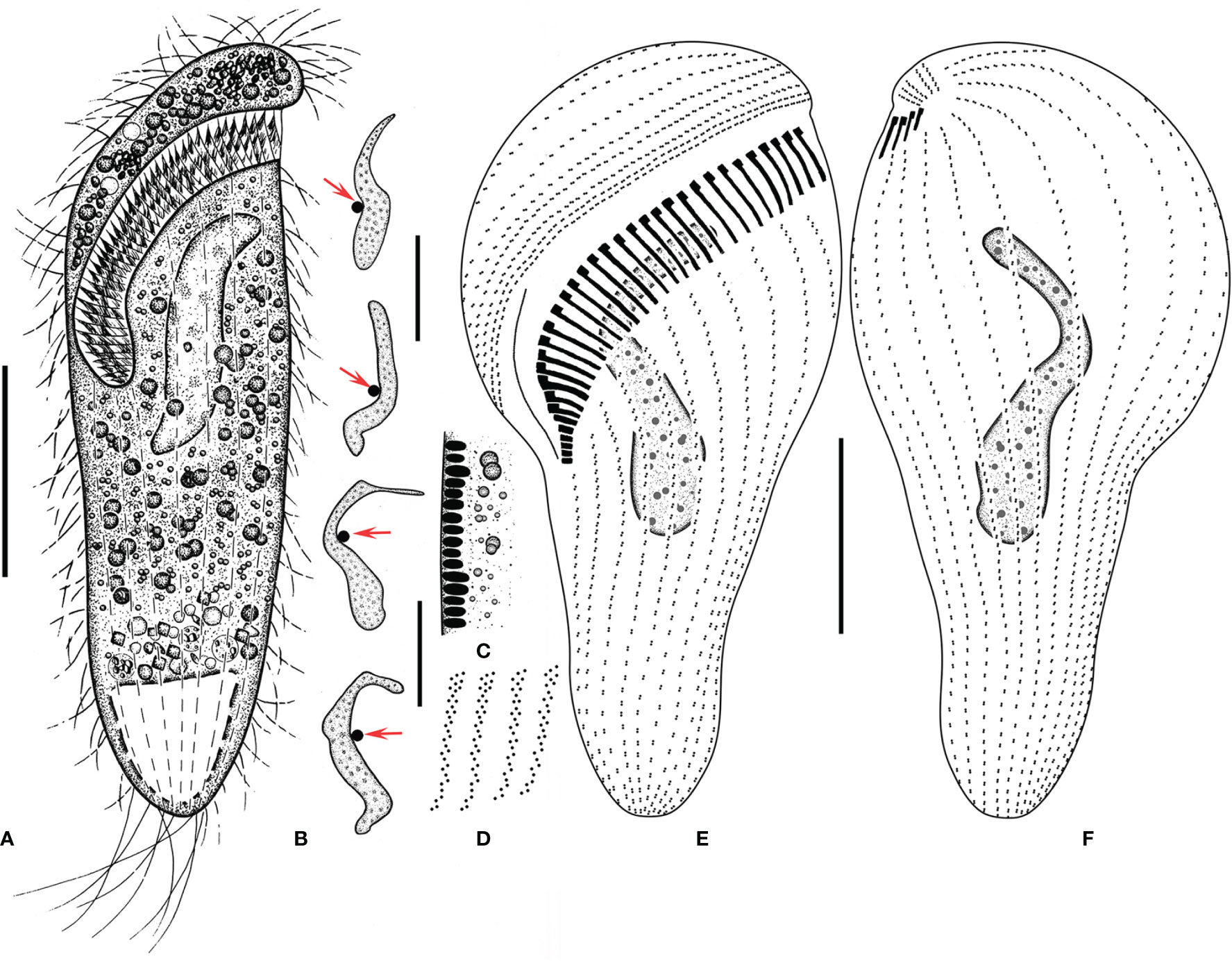
Figure 2 Metopus paraes sp. n. from life (A, C) and after protargol staining (B, D–F). (A) Ventral view of a representative individual, showing body shape. (B) Different shapes of macronucleus, arrows showing micronucleus. (C) Cortical granules from lateral view. (D) Structure of membranelles from the mid-portion of the adoral zone. (E, F) Ventral (E) and dorsal (F) views of the holotype, showing dome kineties, perizonal stripe, adoral membranelles, paroral membrane, and other somatic kineties. Scale bars: 30 μm.
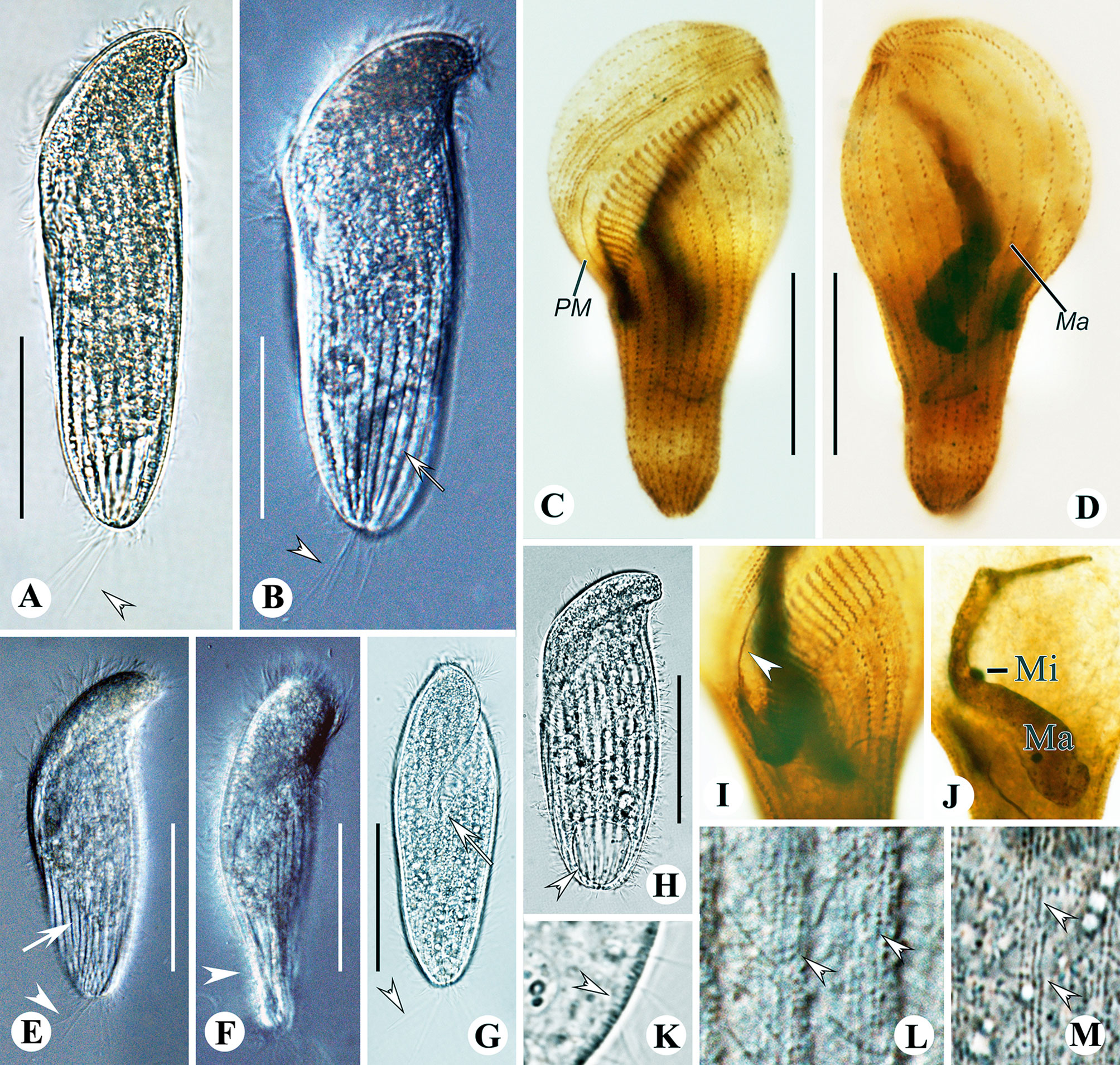
Figure 3 Metopus paraes sp. n. from life with bright-field (A, G, H, K, M), and differential interference contrast (B, E, F, L) illuminations, and after protargol staining (C, D, I, J). (A, B, E) Ventral view showing body shape and caudal cilia (arrowheads) and kinetal furrows (arrows). (C, D) Ventral (C) and dorsal (D) view of the holotype showing ciliature and macronucleus. (F) Right ventral view showing dorsal-ventrally flattened posterior body (arrowhead). (G) Right ventral view showing caudal cilia (arrowhead) and the proximal end of the adoral zone (arrow). (H) Ventral view showing contractile vacuole (arrowhead). (I) Paroral membrane (arrowhead). (J) Macronucleus and micronucleus. (K) Lateral view showing cortical granules (arrowhead). (L) Arrangement of cortical granules (arrowheads). (M) Arrangement and color of cortical granules (arrowheads). Ma, macronucleus; Mi, micronucleus; PM, paroral membrane. Scale bars: 40 μm.
The species group name paraes is a composite of the Greek prefix para- (close to, similar to, resembling) and the species group name es, indicating that the new species resembles but differs from the species Metopus es.
Size in vivo 110–125 × 40–45 μm and in protargol preparations 95–130 × 20–50 μm. Shape elongate, ellipsoidal to obconical, shrink and blunt posteriorly (Figures 2A, 3A, B, E, G, H). Widest at preoral dome. Length:width ratio including preoral dome 3.5:1 on average, slightly dorsoventrally flattened, especially at the posterior end, about 30 μm thick at the middle part of the cell while 10 μm at the posterior end (Figure 3F); anterior part and posterior part projecting forward from lateral view, conferring a slightly curved appearance. Preoral dome convex, overhanging left margin, occupying about 50% of body length (Figures 3A, B, E, G, H). Cortex flexible, prominent kinetal furrows at 5 μm intervals (Figure 3B). Cortical granules ellipsoidal and colorless, about 0.5 × 0.2 μm in vivo, arranged in about 4 lines between ciliary rows (Figures 2C, 3K–M). Cytoplasmic granules variable in size, ranging from about 0.5 to 3 μm, packed densely across the cell except for the posterior end and aggregated at the anterior pole (Figures 2A, 3A, B, F). Single macronucleus elongate, shape varying from C-shape to sigmoidal, up to 55 μm long in the middle part of the cell, non-extending to the preoral dome; filled with numerous nucleoli. Single micronucleus, spherical at the middle part of macronucleus, about 4 μm in diameter (Figures 2B, 3J). Contractile vacuole terminally located, rectangular, about 25 μm in width (Figures 2A, 3H). Swimming pace moderate, rotating around the long axis.
Besides perizonal stripe cilia and about 7 caudal cilia up to 15 μm and 25 μm long, respectively, other somatic cilia about 12 μm long in vivo (Figures 2A, 3A, B, E, G). On average, 23 (range: 21–24) somatic kineties and 9 preoral dome kineties (Figures 2E, F, 3C, D; Table 1). The perizonal stripe is invariably composed of dikinetids arranged in five rows, never forming false kineties; rows 1–5 narrowly spaced; rows 1–3 about 1 μm apart; rows 3–5 about 2 μm apart. Row 5 separated from dome kinety 1 by a conspicuous gap (about 3.5 μm wide). Axis of dikinetids in perizonal stripe rows 1 and 2 parallel to kinety axis, while rows 3–5 inclined about 45° to kinety axis (Figures 2E, F, 3C, D). Somatic kineties composed of dikinetids, both basal bodies ciliated at the anterior part while only one basal body ciliated posteriorly. The axis of dikinetids in both ends of each somatic kinety inclined about 45°, but those in the middle part are parallel to kinety axis (Figures 2E, F, 3C, D). The adoral zone consisted of about 42 (35–46) membranelles on average, occupying about 40% of body length, and extending parallel to the dome brim; longest membranelles at mid-portion, about 12 μm wide (Figures 2D–F, 3C). Paroral membrane about 30 μm long, composed of a single file of ciliated basal bodies (Figures 2E, 3C, I).
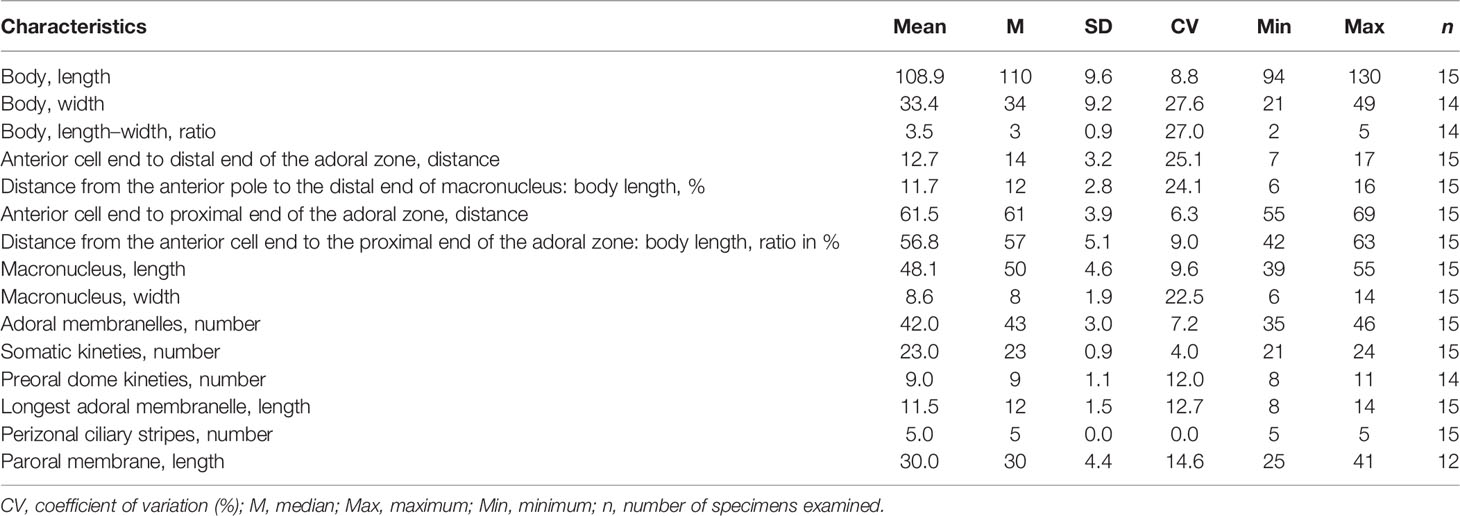
Table 1 Morphometric characteristics of Metopus paraes sp. n. based on protargol-stained specimens. Measurements in μm.
Metopus spiculatus sp. n. ( Figures 4, 5 and Table 2)
Marine form about 55–100 × 25–40 μm in vivo. Body shape oblong to ovoid. Preoral dome extremely compressed, distal end curved and tapered into a conspicuous beak-like structure. Posterior body tapered sharply into an about 25 μm long tail. Cell surface covered by a layer of rod-shaped ectosymbionts arranged perpendicularly. Cytoplasm filled with needle-like intracytoplasmic structures, aggregate in the anterior part. Elongate ellipsoidal macronucleus centrally located below the preoral dome and spherical micronucleus. On average, 21 somatic kineites including 5 preoral dome kineties. Perizonal stripe rows 1 and 2 forming false kineties. The adoral zone consisted of about 20 membranelles, occupying about 30% of body length. Paroral membrane double-rowed with one about twice as long as the other.
Sulfide-rich sediments in an intertidal zone in Liujiawan, Rizhao (119°26′, N35°17′), China.
The protargol slide containing the holotype (Figures 4G, H, 5D, E) (registration number: ZWB202103290301) and two other slides containing paratypes (registration numbers: ZWB202103290302-03) were deposited at the Laboratory of Protozoology, Ocean University of China, Qingdao, China. The holotype and relevant paratypes were marked by black ink circles on the backs of the slides.
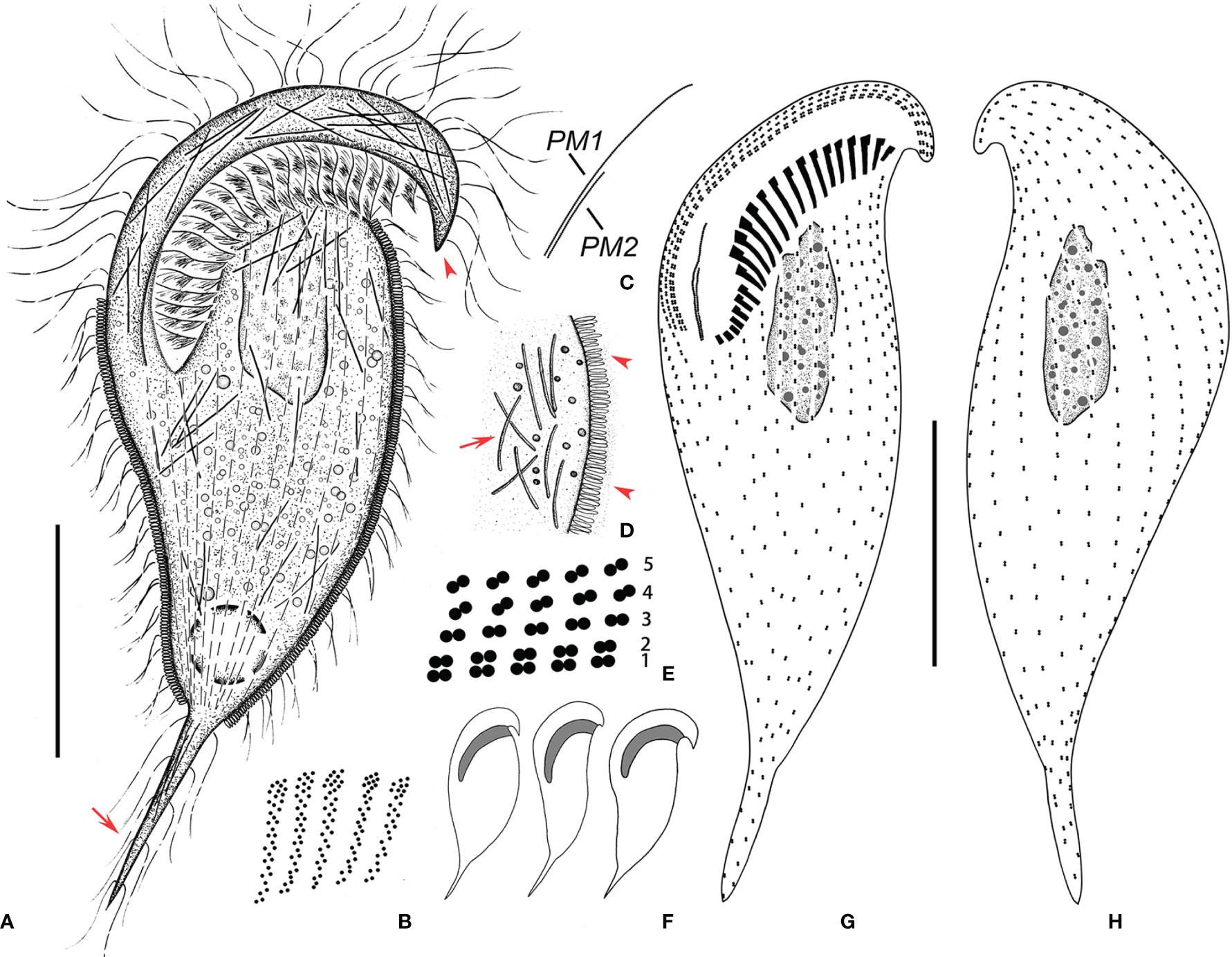
Figure 4 Metopus spiculatus sp. n. from life (A, D, F) and after protargol staining (B, C, E, G, H). (A) Ventral view of a representative specimen, showing body shape, ectosymbionts, contractile vacuole, beak-like preoral dome end (arrowhead), and posterior tail (arrow). (B) Structure of membranelles from the mid-portion of the adoral zone. (C) Two paroral membranes. (D) Ectosymbionts (arrowheads) and needle like structures (arrow). (E) Part of the perizonal stripe and false kineties. (F) Different body shapes. (G, H) Ventral (G) and dorsal (H) view of the holotype showing perizonal stripe, dome kineties, paroral membranes, adoral membranelles, and somatic kineties. PM, paroral membrane. Scale bars, 30 μm.
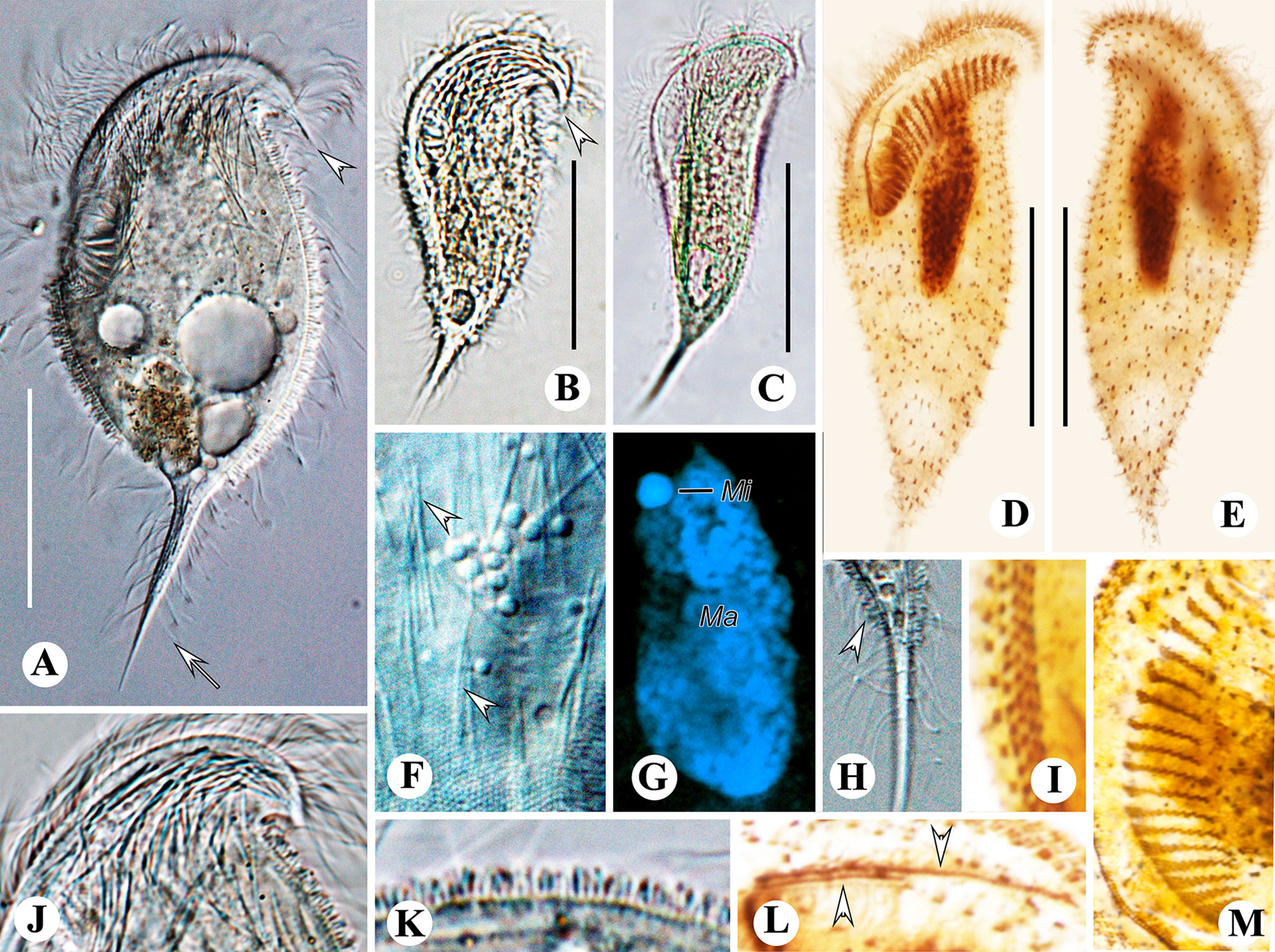
Figure 5 Metopus spiculatus sp. n. from life with differential interference contrast (A, F, H) illuminations, bright-field (B, C, J, K), and after DAPI staining (G) and after protargol staining (D, E, I, L, M). (A–C) Ventral view showing different body shapes, arrowheads showing spine of dome, and arrow showing posterior spine. (D, E) Ventral (D) and dorsal (E) views of the holotype showing ciliature and macronucleus. (F) Unidentified filiform intracytoplasmic structures (arrows). (G) Macronucleus and micronucleus. (H) Posterior spine and ectosymbionts (arrowhead). (I) Perizonal stripe and false kineties. (J) Cortical granules on preoral dome. (K) Lateral view showing ectosymbionts. (L) Two rows of paroral membrane (arrowheads). (M) Adoral membranelles. Mi, micronucleus; Ma, macronucleus. Scale bars: 35 μm (A, G) and 30 μm (B–E).
The Latin adjective “spiculatus” refers to the distinctive, beak-like anterior protrusion.
Size about 55–100 × 25–40 μm. Cell gray and opaque. Shape oblong to ovoid, posterior part tapered acutely into an about 25 μm long tail (Figures 4A, F, 5A–C). Length:width ratio including the preoral dome 2.4:1 on average (n = 15). Preoral dome extremely compressed and slightly twisted, overhanging left margin, tapered into a beak-like structure terminally, occupying about 30% of body length (Figures 4A, F, 5A, B). Body posterior to the preoral dome ellipsoidal in cross-section. Cortex flexible, cortical granules globular, about 0.5 μm in diameter, colorless, loosely arranged in rows, distinguishable only at the preoral dome and at the cell margin due to the coverage of ectosymbionts elsewhere. Cell entirely covered by a layer of rod-shaped, 1.5 × 0.5 μm ectosymbiotic prokaryotes arranged perpendicularly, except for most of the preoral dome and tail zone (Figures 4A, D, 5A–C, F, H, J, K). Cytoplasm colorless, cytoplasmic granules variable in size, ranging from about 0.5 μm to 2 μm. Needle-shaped unidentified intracytoplasmic structures (about 9 μm long) densely packed at the anterior pole and sparsely distributed throughout other portions of the cell, often observed in the tail (Figures 4A–D, 5A, F). Macronucleus about 30 μm long in vivo and about 25 μm long in protargol preparations; elongate ellipsoidal, at the middle-upper part of the cell, not extending beyond the adoral zone into the preoral dome. Micronucleus spherical, about 3 μm across, in depression in the right margin, anterior portion of macronucleus (Figures 4A, 5D, E, G). Contractile vacuole terminal, slightly rightwards, spherical, 10 μm across (Figures 4A, 5B, C). Swimming pace moderate; rotating on the long axis.
Besides perizonal stripe cilia up to 25 μm long, other somatic cilia including cilia on tail about 10 μm in length in vivo (Figures 4A, 5A–C, H). An average of 21 (range: 19–25) somatic kineties, about 3.5 μm apart, extending to posterior tail, including five (4–6) dome kineties; composed of dikinetids (Figures 4G, H, 5D, E; Table 2). Perizonal stripe invariably consists of dikinetids arranged in five rows; rows 1–5 closely spaced at intervals of less than 0.5 μm. Rows 1 and 2 more densely spaced, forming false kineties; no distinct gap between row 5 and dome kinety 1. Dikinetids axis of perizonal stripe parallel to kinety axis in rows 1–2, slightly inclined in row 3, inclined about 45° to kinety axis in rows 4–5 (Figures 4E, G, H, 5D, E, I). The adoral zone is composed of about 20 (17–22) membranelles, occupying about 30% of body length; shorter than the dome brim and perizonal stripe at the anterior part; the longest membranelles at mid-portion, about 6.5 μm long (Figures 4G, 5D, M). The paroral membrane is composed of two parallel rows of basal bodies; one about 19 μm long and the other about 10 μm long (Figures 4C, G, 5D, L).
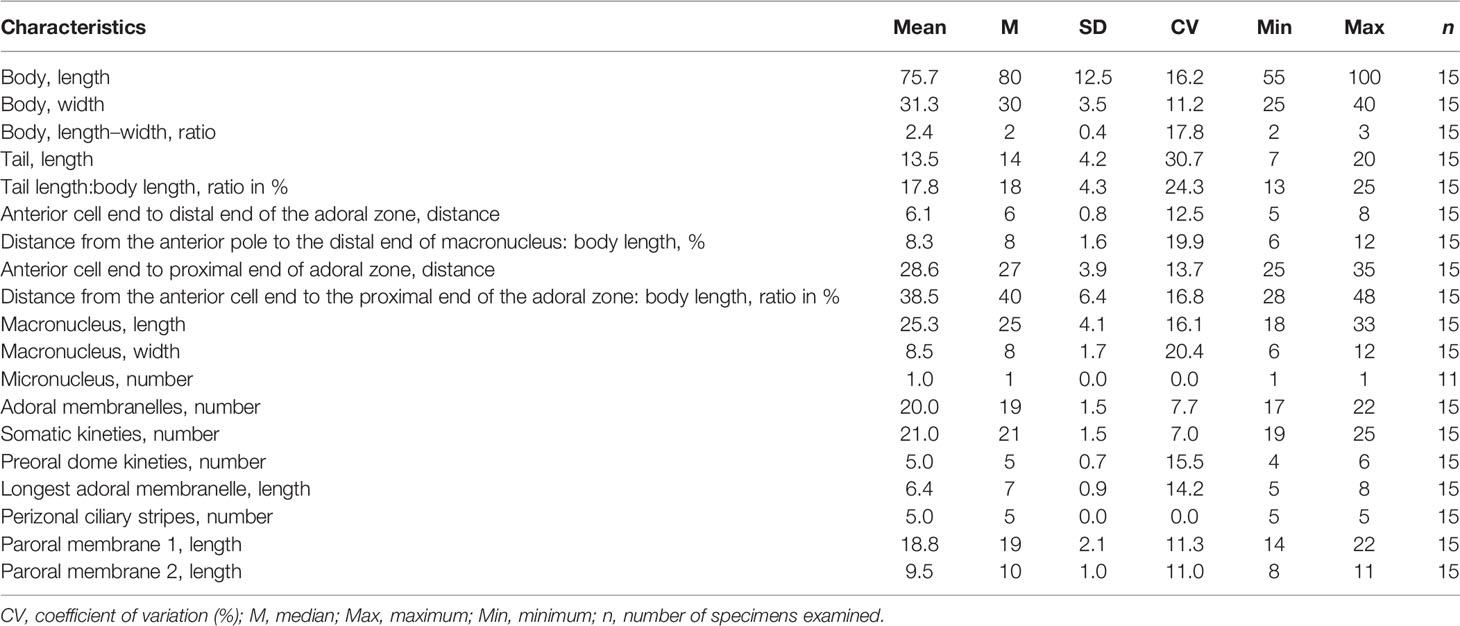
Table 2 Morphometric characteristics of Metopus spiculatus sp. n. based on protargol-stained specimens. Measurements in μm.
Metopus parapellitus sp. n. ( Figures 6, 7 and Table 3)
Marine form with a size of about 50–100 × 25–40 μm in vivo. Shape ovate to oblong. Preoral dome slightly overhanging left margin and occupying about 50% of body length. Rod-shaped ectosymbionts distributed throughout the cell surface perpendicularly to the cell surface. Macronucleus broadly ellipsoidal, never extending to the preoral dome. On average, 32 somatic kineties including 15 preoral dome kineties. Perizonal stripe rows never forming false kineties. The adoral zone is composed of 21 membranelles on average, occupying about 50% of body length, never extending onto the dorsal surface. Paroral membrane stichomonad. Long caudal cilia about 30 μm long, located at the rear end and right posterior part.
Sulfide-rich marine sands near a sewage outfall in Zhanqiao Pier, Qingdao (E120°19′, N36°04′), China.
The protargol slide containing the holotype (Figures 6H, I, 7D, E) (registration number: ZWB202103170101) and two paratypes slides (registration number: ZWB202103170102, ZWB202103170103) were deposited at the Laboratory of Protozoology, Ocean University of China, Qingdao, China. The holotype and relevant paratypes were marked by black ink circles on the backs of the slides.
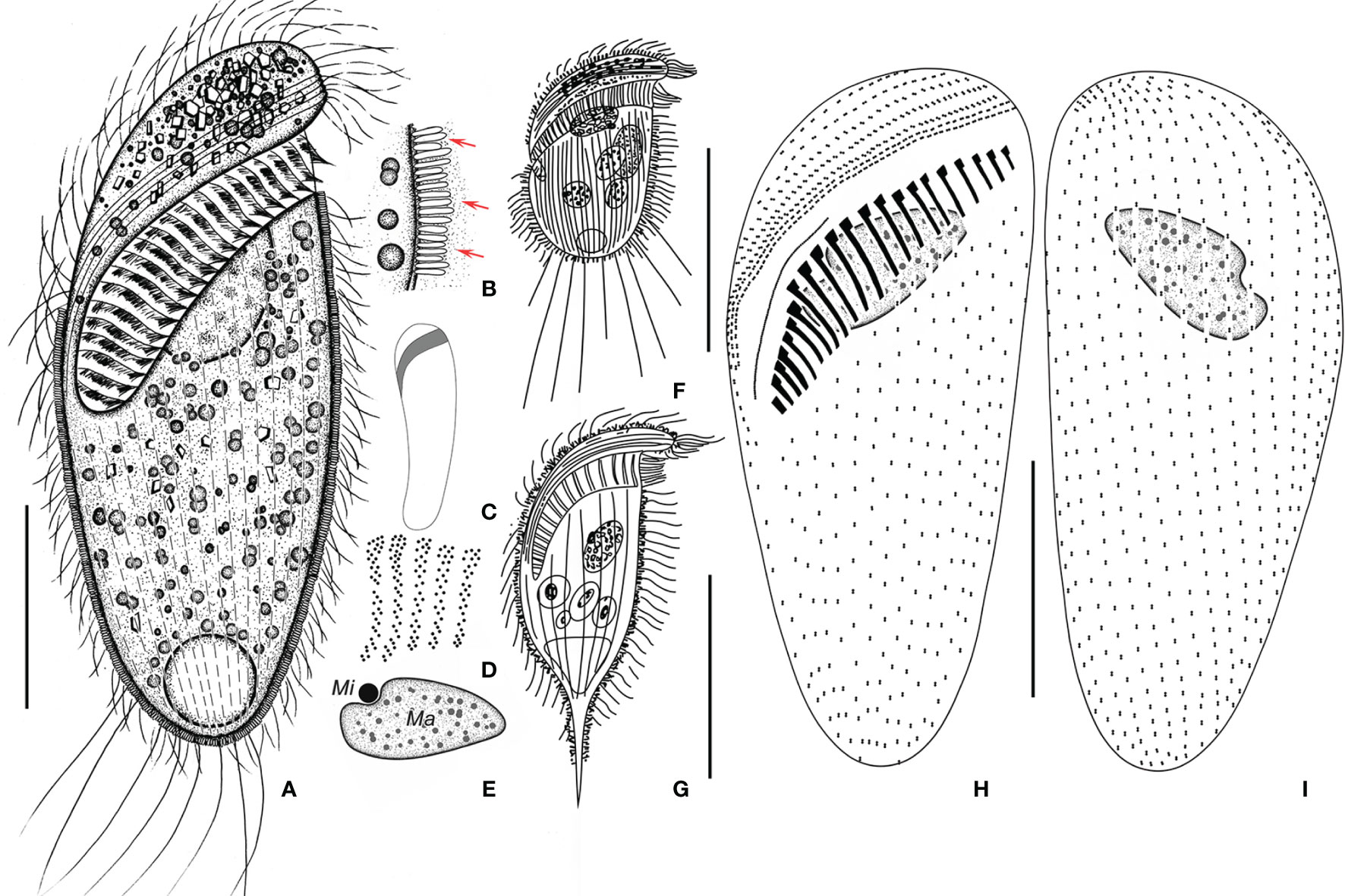
Figure 6 Metopus parapellitus sp. n. from life (A–C, E) and after protargol staining (D, H, I), and M. vestitus (E) and M. contortus pellitus (F) after Kahl, 1932. (A) Ventral view of a representative specimen, showing body shape, ectosymbionts, contractile vacuole, and long caudal cilia. (B) Ectosymbionts (arrows). (C) Left view showing body shape. (D) Structure of membranelles from the mid-portion of the adoral zone. (E) Micronucleus and macronucleus. (F) Metopus vestitus after Kahl (1932). (G) M. contortus pellitus after Kahl, 1932. (H, I) Ventral (H) and dorsal (I) view of the holotype, showing ciliature and macronucleus. Ma, macronucleus; Mi, micronucleus. Scale bars: 30 μm (A, G), 50 μm (G), and 25 μm (H, I).
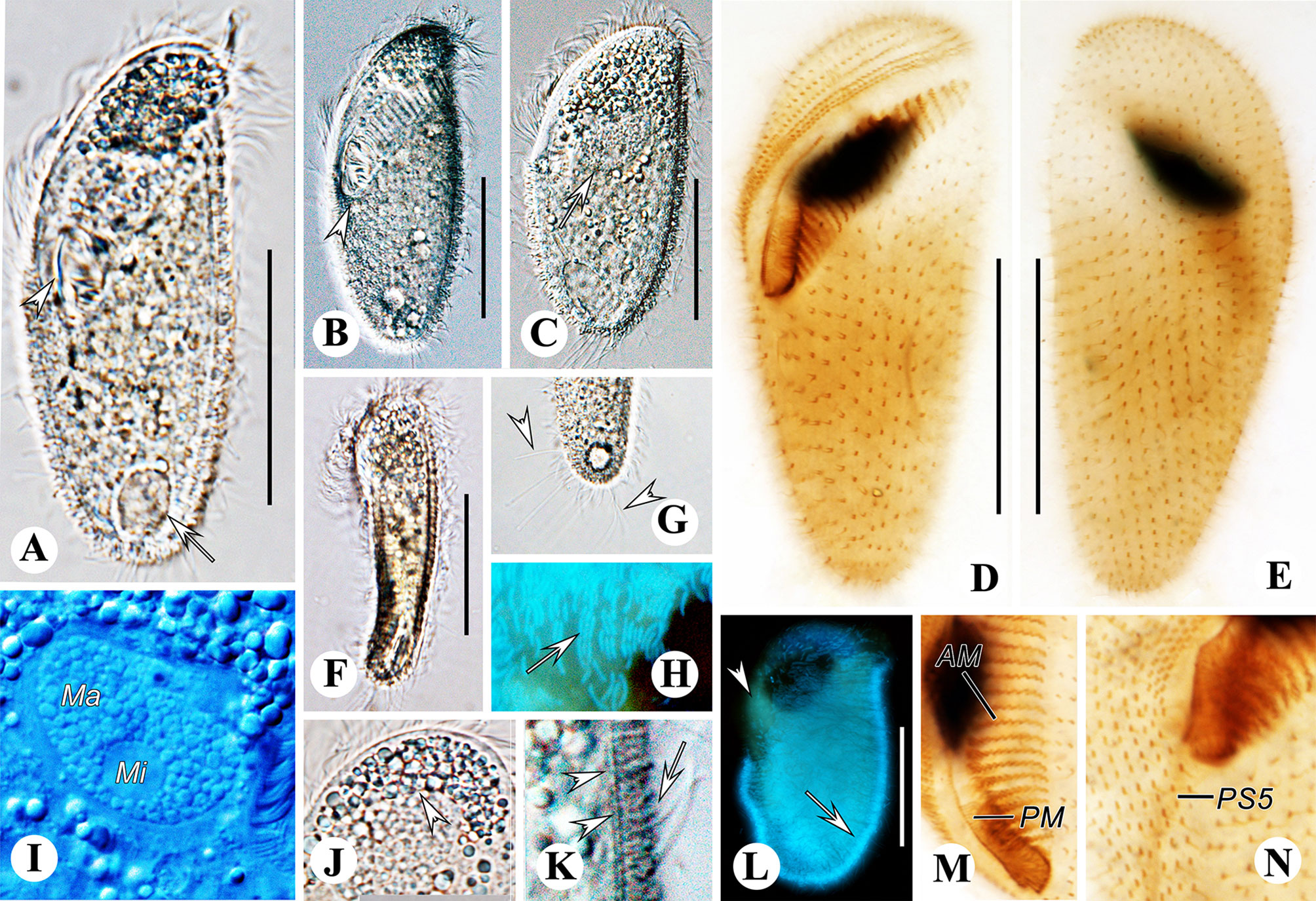
Figure 7 Metopus parapellitus sp. n. from life with bright-field (A, F, G, J), differential interference (B, C, I, K) illuminations and UV light (H, L), and after protargol staining (D, E, M, N). (A) Ventral view showing body shape, paroral membrane (arrowhead), and contractile vacuole (arrow). (B) Ventral view showing body shape and the proximal end of the adoral zone (arrowhead). (C) Dorsal view showing body shape and macronucleus (arrow). (D, E) Ventral (D) and dorsal (E) view of the holotype, showing ciliature and macronucleus. (F) Left view showing body dorsoventrally flattened body. (G) Ventral view showing caudal cilia (arrowheads). (H, L) Autofluorescence of ectosymbionts (arrows) and zone lacking ectosymbionts (arrowhead). (I) Micronucleus and macronucleus. (J) Cytoplasmic granule aggregation at the anterior pole of the cell (arrowhead). (K) Lateral view of ectosymbionts (arrow) and the layer of mucus (arrowheads). (M) Adoral membranelles and paroral membrane. (N) Right lateral view showing the distal end of perizonal stripe row 5. AM, adoral membranelles; Ma, macronucleus; Mi, micronucleus; PM, paroral membrane. Scale bars: 35 μm.
The species-group name parapellitus is a composite of the Greek prefix para- (close to, similar to, resembling) and the species-group name pellitus, indicating that the new species resembles but differs from the subspecies Metopus contortus pellitus Kahl, 1932 (originally described as Metopus contortus var. pellitus Kahl, 1932).
Size in vivo 50–100 × 25–40 μm and in protargol preparations 50–95 × 20–40 μm. Length:width ratio including preoral dome 2.7:1 on average. Shape ovate to oblong, dorsoventrally flattened, about 13–20 μm in width and slightly curved from lateral view (Figures 6A, C, 7A–C, F). Cell light gray, dark at anterior pole due to cytoplasmic granule aggregate. Preoral dome convex and slightly overhangs left margin, inclined 45° to long axis; occupying about 50% of body length. Cortex flexible, prominent kinetal furrows at perizonal stripe (Figures 6A, 7A). Cortical granules not observed due to coverage of ectosymbionts. Rod-shaped ectosymbionts about 0.5 × 2.5 μm in vivo, arranged perpendicularly, distributed throughout cell surface but lacking in most part of preoral dome; covered by a layer of thick homogeneous mucus; autofluorescence under UV light, suggesting that the symbionts may belong to methanogens (Figures 6A, B, 7A, H, K, L). Scattered, spherical cytoplasmic globules about 1–2 μm. Anterior pole aggregate composed of densely packed 1–3-μm light gray spherical to rectangular granules (Figures 6A, 7A–C, G). Single macronucleus broadly ellipsoidal, at the anterior part, not extending to the preoral dome but a little higher than the middle adoral zone; composed of numerous nucleoli about 1.5 μm. Single micronucleus 6 μm across, spherical, in top edge depression of macronucleus (Figures 6A, E, 7I). Contractile vacuole terminal, rounded, about 13 μm across (Figures 6A, 7A). Swimming pace moderate; rotating around the long axis.
Perizonal stripe cilia and somatic cilia up to 20 μm and 12 μm long in vivo, respectively. About five caudal cilia 30 μm long in vivo, located at the right rear end of the body (Figures 6A, 7G), On average, 32 (range 28–35) somatic kineties and 15 dome kineties (Figures 6H, I, 7D, E; Table 3). Somatic kineties are composed of dikinetids with both bosal bodies ciliated. The perizonal stripe is invariably composed of dikinetids arranged in five rows, extending slightly onto the dorsal surface; rows 1–3 closely spaced with about 1–1.5 μm intervals, rows 3–5 more loosely spaced with about 2.5–3.5 μm intervals; no conspicuous gap between row 5 and dome kinety 1 (Figures 6H, 7D, N). Dikinetids of perizonal stripe and dome kineties have both basal bodies ciliated; axis of dikinetids in perizonal stripe parallel to kinety axis in rows 1–2; inclined about 45° to kinety axis in rows 3–5, never forming “false kineties”. The adoral zone comprises about 21 membranelles, occupying about 50% of body length, never extends onto dorsal surface, proximal portion enclosed in the buccal cavity, the longest membranelles at mid-portion, about 9 μm wide (Figures 6D, H, 7D, M). The paroral membrane is about 26 μm in length, originating in the buccal cavity at the proximal end of the adoral zone, composed of a single file of ciliated basal bodies (Figures 6B, 7D, M).
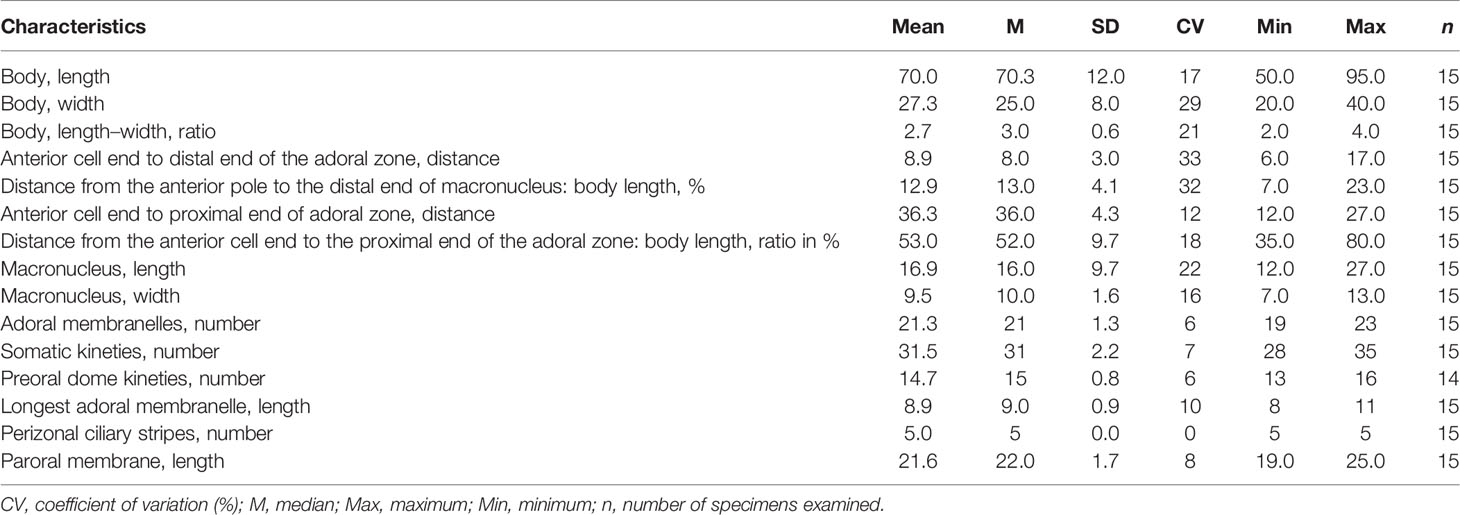
Table 3 Morphometric characteristics of Metopus parapellitus sp. n. based on protargol-stained specimens. Measurements in μm.
The SSU rDNA sequence of Metopus paraes sp. n. is 1,638 bp long, with a GC content of 43.71% and the NCBI accession number OM801553. The sequence of the species is most similar to that of Urostomides bacillatus (KY025569), with an identity of 94.83%. The SSU rDNA sequence of M. parapellitus sp. n. is 1,589 bp long, with a GC content of 44.37%; the NCBI accession number is OM801555. Apart from one unidentified environment sequence, the sequence of M. parapellitus sp. n. is most similar to that of M. contortus (KY432957), with an identity of 98.05%. The SSU rDNA sequence of M. spiculatus sp. n. is 1,650 bp long, with a GC content of 43.88%; the NCBI accession number is OM801554. The sequence of M. spiculatus sp. n. is most similar to that of M. vestitus (MF360251), with an identify of 98.04%.
The topologies of the phylogenetic trees from the ML and BI analyses were identical; therefore, only the ML tree topology is shown; the support values are derived from both algorithms (Figure 8). Armophorea were recovered polyphyletic; they intersected with Litostomatea, Muranotrichea, Parablepharismea, Cariacotrichea, and Odontostomatea and consisted of two unrelated clades, namely, Metopida/Clevelandellida and Armophorida. Apometopidae were confirmed monophyletic and members belonging to the family Tropidoatractidae grouped together. Metopidae and Clevelandellida grouped together with a weak support (38/0.51), while Clevelandellida formed a monophyletic group inside with robust support (99/1.00). The Metopidae/Clevelandellida clade is composed of two main clades. One is a low-support clade (29/0.59) that consists of M. es (Müller, 1776) Lauterborn, 1916, Brachonella contorta (Levander, 1894) Jankowski, 1964, and B. pulchra (Kahl, 1927) Bourland et al., 2018, and six marine species of Metopus. The other has high support (96/1.00) and consists of the remaining metopids and all clevelandellids. Three species from the present study clustered with three marine relatives, viz., M. vestitus Kahl, 1932, M. paravestitus Li et al., 2021, and M. contortus (Quennerstedt, 1867) Kahl, 1932, forming a high-support marine clade (89/1.00), which is parallel to the clade (51/0.69) that consists of M. es and two Brachonella species. Within the marine clade, M. paraes sp. n. branched off early, and the remaining species form a cluster with full support. Metopus spiculatus sp. n. first clusters with M. vestitus with full support, both being sister to M. paravestitus almost with maximum support. Metopus parapellitus sp. n. and M. contortus also constitute a robust clade parallel to the clade containing the above-mentioned marine species.
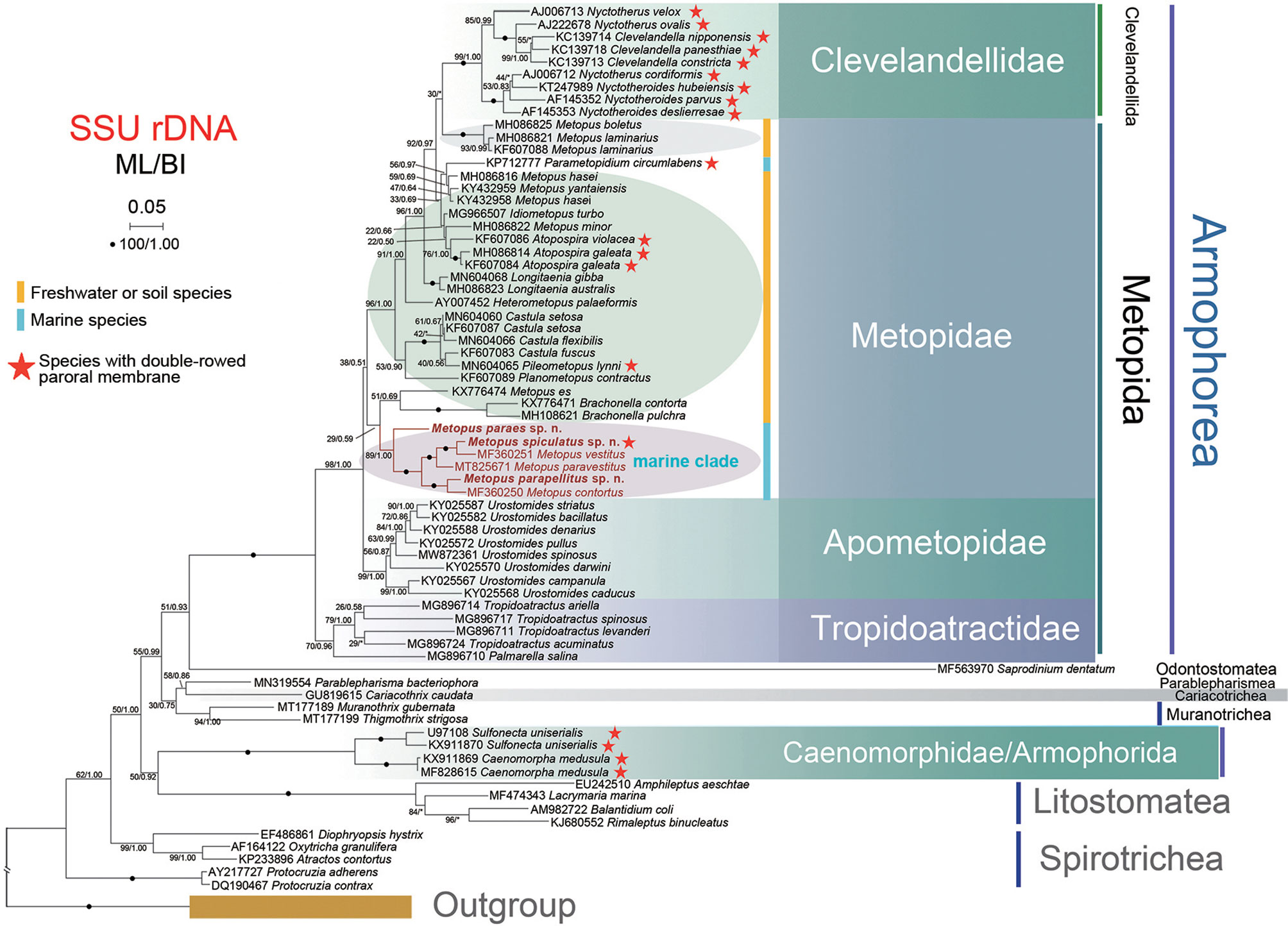
Figure 8 Phylogenetic tree based on SSU rDNA sequences. The tree was constructed by the maximum likelihood method in RA × ML (GTRGAMMAI model). The values at branches represent support in bootstrap values (maximum likelihood method)/posterior probabilities (MrBayes method). New sequences in bold. Yellow bars represent freshwater or soil ciliates in Metopidae. Blue bars represent marine ciliates in Metopidae. Red stars mark ciliates with double-rowed membrane in Armophorea. Asterisks (*) reflect disagreements in topology between BI and ML trees. The scale bar represents 5 changes per 100 positions.
The three species described in the present study are characterized by a five-rowed perizonal stripe, a left torsion in the anterior cell portion, and a frontal lobe that overhangs an obliquely situated adoral zone of membranelles. Therefore, these three species should be assigned to the genus Metopus (Esteban et al., 1995). In addition to traditional taxonomic features, ectosymbiont is also an important feature in identifying these anaerobic species (Esteban et al., 1995; Li et al., 2021a).
Metopus paraes sp. n. is mainly characterized by a combination of the following characteristics: (i) seawater habitat, (ii) an in vivo size of 110–125 × 40–45 μm, (iii) 35–46 adoral membranelles and 21–24 somatic kineties, and (iv) long caudal cilia. It most closely resembles M. es in body size, number of somatic kineties, and adoral membranelles. However, our form can be distinguished from M. es by the long caudal cilia (present vs. absent) and the habitat (marine vs. freshwater). Due to its medium size, elongated body shape, and marine habitat, the new species should also be compared with M. nivaaensis Esteban et al., 1995, M. halophila sensu Esteban et al., 1995, Metopus contortus (Quennerstedt, 1867) Kahl, 1932, and M. paravestitus Li et al., 2021. Our form can be distinguished from M. nivaaensis by the lower number of somatic kineties (21–24 vs. 50) (Esteban et al., 1995). It can be separated from M. halophila sensu Esteban et al., 1995, and M. paravestitus by the ectosymbionts (lacking vs. present). Finally, our form has fewer somatic kineties than M. contortus (21–24 vs. about 40) (Kahl, 1932; Esteban et al., 1995; Bourland et al., 2017a; Li et al., 2021a).
M. spiculatus sp. n. can be distinguished from all other Metopus by the following characteristics: (i) marine habitat, (ii) rod-shaped ectosymbionts, (iii) a beak-like structure at the preoral dome end, (iv) a posterior body that tapers into a tail, (v) an in vivo size of 75–100 × 30–40 μm, (vi) 17–22 adoral membranelles and 19–25 somatic kineties, and (vii) needle-like intracytoplasmic structures. The new species resembles M. vestitus Kahl, 1932 (Figure 6G) in most features except the distinct beak-like preoral dome end (present vs. lacking). Similar to this new species, M. caudatus, Tropidoatractus acuminatus and Tropidoatractus spinosus are medium-sized, and have an oblong body and one acute tail, but all of them lack ectosymbionts, which are considered as an important feature for species identification (Esteban et al., 1995; Rotterová et al., 2018; Li et al., 2021a). In addition, M. rostratus Kahl, 1932 and Tropidoatractus levanderi Rotterová et al., 2018 also possess a beak-like preoral dome end, but both lack a long tail and conspicuous ectosymbionts as shown in our form (Kahl, 1932; Foissner, 2016a; Rotterová et al., 2018).
M. parapellitus sp. n. is mainly characterized by the following characteristics: (i) marine habitat, (ii) rod-shaped ectosymbionts, (iii) long caudal cilia located at the rear end and right posterior body, (iv) an in vivo size of 80–100 × 25–30 μm, and (v) 28–35 somatic kineties and 19–23 adoral membranelles. The species most closely resembles M. contortus pellitus (Figure 6F); these two can be distinguished by body shape (ovate to oblong vs. short ovate), the distribution of caudal cilia (rear end and right posterior body vs. rear end and both sides of the posterior body), and the position of the adoral zone (never extending to the dorsal surface vs. extending to the dorsal surface) (Kahl, 1932). Moreover, both Palmarella salina and Planometopus contractus have an adoral zone located only at the ventral side. However, they possess fewer adoral membranelles than M. parapellitus sp. n. (6–11 vs. 19–23 and 13–15 vs. 19–23) (Rotterová et al., 2018). The species can be easily distinguished from M. contortus by the number of somatic kineties (28–35 vs. 37–47), the number of adoral membranelles (19–23 vs. 33–45), and the ectosymbionts (present vs. lacking) (Esteban et al., 1995). Due to its medium size, ovate body shape, and marine habitat, our species should also be compared with M. halophila sensu Esteban et al., 1995, M. nivaaensis, and M. paravestitus. The species differs from M. halophila sensu Esteban et al., 1995 in the numbers of somatic kineties (28–35 vs. about 20) and of adoral membranelles (19–23 vs. 12–15) (Esteban et al., 1995). M. parapellitus sp. n. also has fewer somatic kineties than M. nivaaensis (28–35 vs. about 50) and fewer adoral membranelles than M. paravestitus (19–23 vs. 27–36) (Li et al., 2021a).
Consistent with previous studies, our analyses showed that the class Armophorea is non-monophyletic (Bourland et al., 2017a; Bourland et al., 2017b; Omar et al., 2017; Rotterová et al., 2018; Li et al., 2021). The obligate anaerobic classes Odontostomatea, Parablepharismea, Cariacotrichea, and Muranothrichea and the partially anaerobic class Litostomatea clustered between the armophorean clades Metopida/Clevelandellida and Armophorida, suggesting possible relationships between these anaerobic groups.
The present study further corroborates the systematic composition of the genus Metopus (Bourland et al., 2017a; Bourland et al., 2017b; Omar et al., 2017; Rotterová et al., 2018; Li et al., 2021). Previous studies suggested that Metopus was heterogeneous and should be split (Bourland et al., 2017a) and the work progresses on (Bourland and Wendell, 2014; Foissner, 2016a; Foissner, 2016b; Bourland et al., 2017a; Bourland et al., 2017b; Vďačný and Foissner, 2017b; Bourland et al., 2018a; Bourland et al., 2018b; Rotterová et al., 2018; Bourland et al., 2020). Perhaps most known species of Metopus from the Metopidae/Clevelandellidae clade will be placed in some other taxa, eventually leaving type species, M. es, and its phylogenetically related species as the “true” Metopus. Brachonella possesses a dominant peroral dome and extreme posteriorization of the cytostome and a highly spiralized adoral zone, whereas Metopus does not, which supported their separation at the genus level (Jankowski, 1964; Bourland et al., 2017a; Bourland et al., 2018b). The six marine species are distantly related with all other species in Metopus, which are either freshwater or soil forms. Moreover, except for Parametopidium circumlabens, an endobiotic species from the sea urchin, the six Metopus species are the only marine representatives in the family Metopidae. These findings raise habitats as a potential genus splitting clue. However, we could not find a synapomorphy to distinguish them from other Metopus species, and the AU test (p = 0.905) does not reject that M. es groups with the marine clade. Any new taxonomic revision is premature for the time being.
Within the marine clade of Metopus, the interspecific grouping might be explained by their morphological similarity. For instance, both M. parapellitus sp. n. and M. contortus have an ovate to elongated body; M. spiculatus sp. n., M. vestitus, and M. paravestitus possess a conspicuous tail, ectosymbionts, and needle-like structures (Kahl, 1932; Esteban et al., 1995; Li et al., 2021).
The datasets presented in this study can be found in online repositories. The names of the repositories and accession number(s) can be found in the GenBank database: OM801553, OM801554, OM801555.
XH conceived and designed the manuscript. WZ conducted the live observation and protargol staining and wrote the manuscript. RL performed the data analyses. XH, XF, and SA-F revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (project no. 41976086) and by the King Saud University, Saudi Arabia (project no. RSP2022R7) as well the China Scholarship Council.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bourland W., Rotterová J., Čepička I. (2017a). Redescription and Molecular Phylogeny of the Type Species for Two Main Metopid Genera, Metopus es (Müller, 1776) Lauterborn, 1916 and Brachonella contorta (Levander, 1894) Jankowski, 1964 (Metopida, Ciliophora), Based on Broad Geographic Sampling. Eur. J. Protistol. 59, 133–154. doi: 10.1016/j.ejop.2016.11.002
Bourland W., Rotterová J., Čepička I. (2017b). Morphologic and Molecular Characterization of Seven Species of the Remarkably Diverse and Widely Distributed Metopid Genus Urostomides Jankowski, 1964 (Armophorea, Ciliophora). Eur. J. Protistol. 61, 194–232. doi: 10.1016/j.ejop.2017.07.003
Bourland W., Rotterová J., Čepička I. (2018a). Morphologic and Molecular Characterization of Brachonella pulchra (Kahl, 1927) Comb. Nov. (Armophorea, Ciliophora) With Comments on Cyst Structure and Formation. Inter. J. Syst. Evol. Microbiol. 68, 3052–3065. doi: 10.1099/ijsem.0.002888
Bourland W., Rotterová J., Čepička I. (2020). Description of Three New Genera of Metopidae (Metopida, Ciliophora): Pileometopus Gen. Nov., Castula Gen. Nov., and Longitaenia Gen. Nov., with Notes on the Phylogeny and Cryptic Diversity of Metopid Ciliates. Protist 171, 125740.
Bourland W., Rotterová J., Luo X., Čepička I. (2018b). The Little–Known Freshwater Metopid Ciliate, Idiometopus Turbo (Dragesco and Dragesco–Kernéis, 1986) Nov. Gen., Nov. Comb., Originally Discovered in Africa, Found on the Micronesian Island of Guam. Protist 169, 494–506. doi: 10.1016/j.protis.2018.05.004
Bourland W. A., Wendell L. (2014). Redescription of Atopospira galeata (Kahl, 1927) Nov. Comb. And A. Violacea (Kahl, 1926) Nov. Comb. with Redefinition of Atopospira Jankowski, 1964 Nov. Stat. and Brachonella Jankowski, 1964 (Ciliophora, Armophorida). Eur. J. Protistol. 50, 356–372. doi: 10.1016/j.ejop.2014.05.004
Bourland W. A., Wendell L., Hampikian G. (2014). Morphologic and Molecular Description of Metopus fuscus Kahl from North America and New rDNA Sequences from Seven Metopids (Armophorea, Metopidae). Eur. J. Protistol. 50, 213–230. doi: 10.1016/j.ejop.2014.01.002
Campello-Nunes P. H., Fernanndes N. M., Franziska S., Fokin S. I., Serra V., Modeo L., et al. (2020). Parablepharisma (Ciliophora) is Not a Heterotrich: A Phylogenetic and Morphological Study with the Proposal of New Taxa. Protist 171, 125716. doi: 10.1016/j.protis.2020.125716
Chi Y., Chen X., Li Y., Wang C., Zhang T., Ayoub A., et al. (2021). New Contributions to the Phylogeny of the Ciliate Class Heterotrichea (Protista, Ciliophora): Analyses at Family-Genus Level and New Evolutionary Hypotheses. Sci. China Life Sci. 64, 606–620. doi: 10.1007/s11427-020-1817-5
Esteban G., Fenchel T., Finlay B. (1995). Diversity of Free–Living Morphospecies in the Ciliate Genus Metopus. Arch. Protistenkd. 146, 137–164. doi: 10.1016/S0003-9365(11)80106-5
Fernandes N. M., Vizzoni V. F., Borges B. D. N., Soares C. A., Silva-Neto I. D., Paiva T. D. S. , et al. (2018). Molecular Phylogeny and Comparative Morphology Indicate That Odontostomatids (Alveolata, Ciliophora) Form a Distinct Class-Level Taxon Related to Armophorea. Mol. Phylogenet. Evol. 126, 382–389. doi: 10.1016/j.ympev.2018.04.026
Foissner W. (2016a). Heterometopus meisterfeldi Nov. Gen., Nov. Spec. (Protozoa, Ciliophora), a New Metopid From Australia. Eur. J. Protistol. 55, 118–127. doi: 10.1016/j.ejop.2015.11.005
Foissner W. (2016b). Terrestrial and Semiterrestrial Ciliates (Protozoa, Ciliophora) From Venezuela and Galápagos. Denisia 35, 1–912.
Foissner W., Agatha S. (1999). Morphology and Morphogenesis of Metopus hasei Sondheim, 1929 and M. inversus (Jankowski 1964) Nov. Comb. (Ciliophora, Metopida). J. Euk. Microbiol. 46, 174–193. doi: 10.1111/j.1550-7408.1999.tb04602.x
Hall T. A. (1999). BioEdit: A User–Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucl. Acid. S 41, 95–98.
Jankowski A. W. (1964). Morphology and Evolution of Ciliophora I. The New System of Sapropelobiotic Heterotrichida. Zool. Zh. 43, 503–517.
Jerome C. A., Simon E. M., Lynn D. H. (1996). Description of Tetrahymena empidokyrea N. Sp., a New Species in the Tetrahymena pyriformis Sibling Species Complex (Ciliophora, Oligohymenophorea), and an Assessment of its Phylogenetic Position Using Small–Subunit rRNA Sequences. Can. J. Zool. 74, 1898–1906. doi: 10.1139/z96-214
Kahl A. (1932). Urtiere Oder Protozoa I: Wimpertiere Oder Ciliata (Infusoria) 3 Spirotricha. Tierwelt Dtl. 25, 399–650.
Lauterborn R. (1916). Die Sapropelische Lebewelt. Ein Beitrag Zur Biologie Des Faulschlammes Naturlicher Gewasser. Verh. Naturkundl. Med. Ver. Heidelb. 13, 395–481.
Lewis W. H., Lind A. E., Sendra K. M., Onsbring H., Williams T. A., Esteban G. F., et al. (2020). Convergent Evolution of Hydrogenosomes from Mitochondria by Gene Transfer and Loss. Mol. Biol. Evol. 37, 524–539. doi: 10.1093/molbev/msz239
Lewis W. H., Sendra K. M., Embley T. M., Esteban G. F. (2018). Morphology and Phylogeny of a New Species of Anaerobic Ciliate, Trimyema finlayi N. Sp., with Endosymbiotic Methanogens. Front. Microbiol. 9, 140. doi: 10.3389/fmicb.2018.00140
Liu W., Shin M. K., Yi Z., Tan Y. (2021). Progress in Studies on the Diversity and Distribution of Planktonic Ciliates (Protista, Ciliophora) in the South China Sea. Mar. Life Sci. Technol. 3, 28–43. doi: 10.1007/s42995-020-00070-y
Li S., Zhuang W., Pérez-Uz B., Zhang Q., Hu X. (2021). Two Anaerobic Ciliates (Ciliophora, Armophorea) from China: Morphology and SSU rDNA Sequence, with Report of a New Species, Metopus paravestitus Nov. Spec. J. Eukaryot. Microbiol. 68, e12822. doi: 10.1111/jeu.12822
Li R., Zhuang W., Wang C., El-Serehy H., Al-Farraj S. A., Warren A., et al. (2021). Redescription and SSU rRNA Gene-Based Phylogeny of an Anaerobic Ciliate, Plagiopyla ovata Kahl, 1931 (Ciliophora, Plagiopylea). Inter. J. Syst. Evol. Microbiol. 71, 004936. doi: 10.1099/ijsem.0.004936
Lynn D. H. (2008). The Ciliated Protozoa: Characterization, Classification and Guide to the Literature. 3rd ed (Dordrecht: Springer).
Madeira F., Park M. Y., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL–EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Medlin L., Elwood H. J., Stickel S., Sogin M. L. (1988). The Characterization of Enzymatically Amplified Eukaryotic 16S–Like rRNA–Coding Regions. Gene 71, 491–499. doi: 10.1016/0378-1119(88)90066-2
Omar A., Zhang Q., Zou S., Gong J. (2017). Morphology and Phylogeny of the Soil Ciliate Metopus yantaiensis N. Sp. (Ciliophora, Metopida), with Identification of the Intracellular Bacteria. J. Eukaryot. Microbiol. 64, 792–6805. doi: 10.1111/jeu.12411
Orsi W., Edgcomb V., Faria J., Foissner W., Fowle H. W., Hohmann T., et al. (2012). Class Cariacotrichea, a Novel Ciliate Taxon from the Anoxic Cariaco Basin, Venezuela. Int. J. Syst. Evol. Micr. 62, 1425–1433. doi: 10.1099/ijs.0.034710-0
Pan X. M., Bourland W., Song W. B. (2013). Protargol Synthesis: An in–House Protocol. J. Eukaryot. Microbiol. 60, 609–614. doi: 10.1111/jeu.12067
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rotterová J., Bourland W., Čepička I. (2018). Tropidoatractidae Fam. Nov., a Deep Branching Lineage of Metopida (Armophorea, Ciliophora) Found in Diverse Habitats and Possessing Prokaryotic Symbionts. Protist 169, 362–405. doi: 10.1016/j.protis.2018.04.003
Rotterová J., Salomaki E., Pánek T., Bourland W., Žihala D., Táborsk P., et al. (2020). Genomics of New Ciliate Lineages Provides Insight into the Evolution of Obligate Anaerobiosis. Curr. Biol. 30, 2037–2050. doi: 10.1016/j.cub.2020.03.064
Shimodaira H. (2002). An Approximately Unbiased Test of Phylogenetic Tree Selection. Systat. Biol. 51, 492–508. doi: 10.1080/10635150290069913
Shimodaira H., Hasegawa M. (2001). CONSEL: For Assessing the Confidence of Phylogenetic Tree Selection. Bioinformatics 17, 1246–1247. doi: 10.1093/bioinformatics/17.12.1246
Stamatakis A. (2014). RAxML Version 8: A Tool for Phylogenetic Analysis and Post–Analysis of Large Phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vďačný P., Foissner W. (2017a). A Huge Diversity of Metopids (Ciliophora, Armophorea) in Soil from the Murray River Floodplain, Australia. I. Description of Five New Species and Redescription of Metopus setosus Kahl, 1927. Eur. J. Protistol. 58, 35–76. doi: 10.1016/j.ejop.2016.12.001
Vďačný P., Foissner W. (2017b). A Huge Diversity of Metopids (Ciliophora, Armophorea) in Soil From the Murray River Floodplain, Australia. II. Morphology and Morphogenesis of Lepidometopus platycephalus Nov. Gen., Nov. Spec. Acta Protozool. 26, 39–57.
Vďačný P., Foissner W. (2019). A Huge Diversity of Metopids (Ciliophora, Armophorea) in Soil from the Murray River Floodplain, Australia. III. Morphology, Ontogenesis and Conjugation of Metopus boletus Nov. Spec., with Implications for the Phylogeny of the SAL Supercluster. Eur. J. Protistol. 69, 117–137. doi: 10.1016/j.ejop.2019.04.002
Wilbert N. (1975). Eine Verbesserte Technik Der Protargolimprägnation Für Ciliaten. Mikrokosmos 64, 171–179.
Wu T., Li Y., Lu B., Shen Z., Song W., Warren A. (2020). Morphology, Taxonomy and Molecular Phylogeny of Three Marine Peritrich Ciliates, Including Two New Species: Zoothamnium apoarbuscula N. Sp. and Z. apohentscheli N. Sp. (Protozoa, Ciliophora, Peritrichia). Mar. Life Sci. Technol. 2, 334–348. doi: 10.1007/s42995-020-00046-y
Keywords: anaerobe, ciliature, metopidae, novel taxa, taxonomy
Citation: Zhuang W, Li R, Feng X, Al-Farraj SA and Hu X (2022) New Contribution to the Diversity of the Anaerobic Genus Metopus (Ciliophora, Armophorea), With Descriptions of Three New Marine Species. Front. Mar. Sci. 9:884834. doi: 10.3389/fmars.2022.884834
Received: 27 February 2022; Accepted: 15 April 2022;
Published: 26 May 2022.
Edited by:
Wen-Jun Li, Sun Yat-sen University, ChinaReviewed by:
Xiangrui Chen, Ningbo University, ChinaCopyright © 2022 Zhuang, Li, Feng, Al-Farraj and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaozhong Hu, eGlhb3pob25naHVAb3VjLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.