- 1Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Scientific Observation and Research Field Station of Pearl River Estuary Ecosystem, Guangdong Province, South China Sea Fisheries Research Institute, Guangzhou, China
- 4Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, South China Sea Fisheries Research Institute, Guangzhou, China
Herbivorous fishes play an important role in preventing the overabundance of macroalgae on coral reefs. Understanding the feeding selectivity and consumption of macroalgae by herbivorous fishes can be challenging in studies of their ecological role in the preservation and recovery of coral reefs. Coral reef decline, macroalgal overgrowth and overfishing are clearly visible in the Xisha Islands, China. However, there are seldom studies on the feeding behaviors of herbivorous fishes in this area. We used microscopy, 18S rRNA high-throughput sequencing, and stable isotope analyses to comprehensively examine the diet of eight herbivorous reef fish species common in the Xisha Islands, including one parrotfish, two chub, two unicorn fish, and three rabbitfish. Based on microscopic examination, Siganus argenteus fed on the highest number of macroalgae species (five species), followed by Naso unicornis (four species). Kyphosus cinerascens, K. vaigiensis, N. unicornis and S. punctatissimus fed on the entire macroalgal thallus, indicating their greater ecological importance compared with species that only consume algal fronds. According to the 18S rRNA high-throughput sequencing of fish intestinal contents, cluster analysis revealed that consumed macroalgae composition from the eight fishes always grouped together based on the fish species, but with low similarity. K. vaigiensis removed the highest diversity of macroalgae species as well as the greatest quantity of macroalgae. Calotomus carolinus can consume the red alga Pneophyllum conicum, which is widely distributed on Indo-Pacific coral reefs and can overgrow and kill live corals. N. unicornis was found to occupy the lowest trophic position based on stable isotope analysis. Multi-technique analyses revealed that K. vaigiensis, N. unicornis and S. argenteus showed a high consumption potential of macroalgae, suggesting that they are the key browsers that should receive priority protection in the Xisha Islands. A diverse herbivorous fish fauna is also very important in the Xisha coral reefs. These results not only demonstrated the various functions of different herbivorous fish species in macroalgal removal, but also provided insights into the management of herbivorous fishes on the coral reefs of the South China Sea.
Introduction
Coral reefs are often called the tropical rainforests of the ocean and are the most productive and biologically diverse ecosystem, supporting more than 25% of all known marine species (Moberg and Folke, 1999; Walker and Wood, 2005). However, the world’s coral reefs are declining due to numerous anthropogenic activities and global climate disturbances, such as coastal development, land-based and marine pollution, overfishing, tropical storms and thermal stress (Burke et al., 2011; Liu et al., 2021; Souter et al., 2021). There was a massive loss of almost 14% of the coral from the world’s coral reefs between 2009 and 2018 (Souter et al., 2021). The decline of coral reefs can involve a shift from coral towards macroalgae as the dominant feature (Cheal et al., 2010; Kopp et al., 2010; Bruno et al., 2019). Macroalgae growth can hinder the settlement, survival, and growth of corals, so that a healthy coral reef ecosystem is difficult to reestablish (Puk et al., 2016; Dell et al., 2020). Phase shifts in the balance of coral/macroalgal species have been attributed to insufficient herbivore activity, eutrophication, environmental disturbances (e.g. typhoons, ocean warming and coral bleaching, etc.), or a combination of these factors (Cheal et al., 2010; Russ et al., 2015; Neilson et al., 2018; Bruno et al., 2019; Adam et al., 2021).
Herbivorous reef fishes are the major consumers of macroalgae on coral reefs (Cheal et al., 2010). Chub (Kyphosus spp.) (Dell et al., 2020), unicorn fish (Naso spp.) (Lefevre and Bellwood, 2011; Streit et al., 2015) and rabbitfish (Siganus spp.) (Bennett and Bellwood, 2011; Streit et al., 2015) are recognized as the important macroalgal grazers on coral reefs. Parrotfish are usually as microphages that target microscopic photoautotrophs, and macroalgae is a low component of diet of Parrotfish (Clements et al., 2017; Nicholson and Clements, 2020). The feeding preferences of herbivorous reef fishes differ at the species level (Hoey and Bellwood, 2009; Duran et al., 2019; Dell et al., 2020). Naso spp. and Kyphosus spp. generally target brown macroalgae (Puk et al., 2016). Of the over 50 herbivorous fish species identified off Lizard Island, northern Great Barrier Reef (GBR), only one, Naso unicornis, fed on the erect brown macroalga Sargassum across all habitats (Hoey and Bellwood, 2009). A study of the feeding behaviors of four rabbitfish species in the Seychelles of the Indian Ocean, indicated that Siganus argenteus and S. sutor were generalist herbivores, foraging on turf algae, macroalgae, seagrass and epiphytic algae, while S. corallinus and S. stellatus were specialist herbivores foraging primarily on turf algae growing on the reef substrate (Ebrahim et al., 2020). Herbivore species richness appears to be critical in generally lowering macroalgal abundance because of the complementary feeding habits of a diverse assemblage of herbivores (Burkepile and Hay, 2008). Therefore, it is essential to understand the dietary spectra of the various herbivorous reef fishes to understand their cumulative effect on coral reef health. However, very little is currently known about the macroalgal feeding preferences of the herbivorous reef fishes on the coral reefs off the Xisha Islands, and this limits our understanding of their different functional roles in controlling the spread of macroalgae.
The feeding ecology of herbivorous fishes on coral reefs is usually determined by behavioral observations, counting the number of bites taken by fish feeding in the field (Mantyka and Bellwood, 2007; Dell et al., 2020; Ebrahim et al., 2020), or by microscopic examination of intestinal contents (Choat et al., 2002; Dromard et al., 2015). These methods are convenient and provide detailed information on the dietary spectrum of different species (de Carvalho et al., 2019). However, there are some practical problems; small, fragile, or morphologically indistinct organisms are difficult to identify in intestinal samples, especially after digestion (Dromard et al., 2015; Kume et al., 2021). Stable isotope analysis is a powerful tool which can reveal feeding behavior over an extended period, although with some taxonomic limitations (Rodriguez-Barreras et al., 2020; Kume et al., 2021). High-throughput sequencing is emerging as a molecular method to estimate fish dietary composition by identifying the taxa eaten from genomic DNA recovered from fish intestines (Corse et al., 2010; Leray et al., 2015; Lin et al., 2021; Nalley et al., 2022). This molecular method is more efficient for diet evaluation with enrichment of the reference sequence databases such as GenBank (Devloo-Delva et al., 2019). Nalley et al. (2021) also demonstrated that reference sequences were important for metabarcoding sequencing. However, DNA metabarcoding lacks the biomass information of the diet (Lamb et al., 2019). Different approaches on examining the diets of herbivorous fishes may influence the interpretation of diet and function (Nalley et al., 2022). Therefore, using a combination of these three methods allows comprehensive descriptions of the dietary composition of various fish species.
Coral cover off the Xisha Islands has declined dramatically over the past 15 years (Li et al., 2018) and macroalgae have become dominant in many of their coral reef areas (Chen et al., 2019). Macroalgae cover was higher than coral cover in about 30% of the sites (70 sites) in Xisha coral reef island, and macroalgae cover in Qilianyu Islets and Cays can reach to 20% (Chen et al., 2019). Over 100 fish species have been recorded off the Qilianyu Islets and Cays, with the dominant species belonging to the families Pomacentridae and Labridae (Li et al., 2017). Overfishing has become a serious threat for these reefs (Zhao et al., 2016). However, the diet composition of fishes in this area was seldom investigated. Here, we explored the dietary differences of eight different herbivorous reef fishes common around the Xisha Islands, based on microscopic examination, 18S rRNA high-throughput sequencing and stable isotope analyses. We focused on comparisons of the macroalgal feeding selectivity of herbivores, including one parrotfish (Calotomus carolinus), two chub (Kyphosus cinerascens, and K. vaigiensis), two unicorn fish (Naso brevirostris, and N. unicornis) and three rabbitfish (S. argenteus, S. puellus, and S. punctatissimus). These fishes cover the herbivorous fishes belonging to different Family in Xisha Islands. Our previous study showed that K. vaigiensis, N. brevirostris and N. unicornis were the abundant herbivorous species in this region, while the others were common species (Wang et al., 2022). In addition, chubs, unicorn fish and rabbitfish were always considered as important herbivores in the Indo-Pacific (Dell et al., 2020). The results provided key information on the ecological function of these herbivorous fishes in removing macroalgae from coral reefs, and gave insights into the most effective overfishing prevention measures to keep the coral reef ecosystem at its most healthy.
Materials and Methods
Study Site and Fish Sample Collection
The Xisha Islands, in the central South China Sea, are derived from coral reefs and comprise over 40 islands, reefs and cays, including the Dongdao Atolls, the Huaguang Atolls, the Xuande Atolls, the Yongle Atolls, and some smaller islands (Zhao et al., 2017; Ding et al., 2019; Zhao et al., 2019). The Qilianyu Islets and Cays (16°59′ N, 112°18′ E) of the Xisha Islands comprise an arced reef flat, which extends in a NW-SE direction and curves to the NNE (Shen et al., 2017). Eight herbivorous reef fish species common in the Qilianyu Islets and Cays were collected by SCUBA in June 2020 (Figure S1). Fishes were captured by driver with spear guns between the hours of 18:00 and 20:00. All fishes were collected along the shallow waters around the coral reef regions (water depth below 15 m). The details of the fishes sampled in this study, along with their phylogenetic classification and feeding strategies, are shown in Table 1. The feeding strategy of each species was determined based on the previous published studies. All fishes collected were kept at -4°C and transported to the laboratory, where they were dissected using sterile scissors. In the dissecting process, all fishes were made sure to have food item in the intestines. The anterior intestinal contents were separated and divided into two subsamples. One subsample was used for microscopic examination, and the another was used for 18S rRNA high-throughput sequencing analysis. A small piece of white muscle tissue near the dorsal fin was sampled from each fish and used for stable isotope analysis.
Intestinal Contents for Microscopical Analysis Examination
Intestinal contents of each fish species were collected for dietary assessment. Intestinal contents of individual fish were spread on glass culture dish and put under a microscope (Zeiss SteREO Discovery.V20, Germany). Photographs of the food items were taken using the microscope. Macrophytes species (Macroalgae and seagrasses) were only observed here, however, quantity analysis of the macrophytes composition wasn’t concerned. Macrophytes species were identified according to their morphological traits (Titlyanov et al., 2017; Huang, 2018).
Composition of Intestinal Contents Measured by 18S rRNA High-Throughput Sequencing Analysis
The total DNA of intestinal contents (0.2 g sample) of individual fish was extracted using a QIAamp® Fast DNA Stool Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s protocols. The total DNA recovered from the same fish species was pooled together for PCR amplification which targeted the V4 region of the eukaryotic 18S rRNA gene using primers 528F (5’-GCGGTAATTCCAGCTCCAA-3’) and 706R (5’-AATCCRAGAATTTCACCTCT-3’), where the barcodes were an eight-base sequence unique to each sample (Cheung et al., 2010). Amplicons were extracted from 2% agarose gels and purified using the AMPure XP Beads (Beckman, Agencourt, USA). These purified amplicons were pooled in equimolar paired-end sequences (2 × 250) on an Illumina platform (Gene Denovo Co., Guangzhou, China). Raw reads for each fish species were deposited into the NCBI Sequence Read Archive (SRA) database with the accession number PRJNA742779. The representative sequences affiliated to the phyla Chlorophyta, Ochrophyta and Rhodophyta obtained in this study were deposited in the GenBank database under accession numbers MZ481947-MZ481964, MZ481932-MZ481946 and MZ481965-MZ482014, respectively.
Raw reads were filtered using FASTP (V0.18.0) to obtain high quality clean reads according to the following rules: (1) remove reads containing more than 10% of unknown nucleotides and (2) remove reads containing less than 50% of bases with a quality (Q-value)>20 (Chen et al., 2018). Paired-end clean reads were merged as raw tags using FLSAH (V1.2.11) with a minimum overlap of 10 bp and mismatch error rates of 2% or less (Magoc and Salzberg, 2011). Noisy sequences of raw tags were then filtered using QIIME (V1.9.1) (Caporaso et al., 2010) under specific filtering conditions (Bokulich et al., 2013) to obtain high quality clean tags. Clean tags were searched against the reference database (http://drive5.com/uchime/uchime_download.html) to perform reference-based chimera checks, using the UCHIME algorithm (Edgar et al., 2011). All chimeric tags were removed to finally obtain effective tags for further analysis.
The effective tags were clustered into operational taxonomic units (OTUs) of ≥ 97% similarity using UPARSE (V9.2.64) (Edgar, 2013). A dominant sequence was selected within each cluster as a representative sequence. The representative sequences were then picked-out to annotate taxonomic assignments using the RDP classifier (V2.2) (Wang et al., 2007) based on the SILVA database (V132) (Pruesse et al., 2007). The OTU sequences belonging to class Actinopterygii were discarded in further analysis. Moreover, if an OTU sequence could not be assigned to any phylum in the SILVA database, it was subjected to a further BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The taxonomic classification was then finally confirmed by its similarity to a BLAST result.
Stable Isotope Analysis
Fish muscle tissue were dried at 60°C to a constant weight. Samples were then ground to a fine, homogeneous powder using an automatic sample grinder (Jxfstprp-24, Jingxin Co., Shanghai, China). Samples were taken for carbon and nitrogen stable isotope analysis using a continuous-flow isotope ratio mass spectrometer (Finnigan MAT 253, Thermo Scientific, USA) coupled to an elemental analyzer (Flash EA 1112, Thermo Scientific, USA). The C and N isotope ratios were determined as δ13C and δ15N, respectively, according to the following formula:
where R is the corresponding ratio 13C/12C or 15N/14N, Rsample is measured for fish, and Rstandard is an international standard (Pee Dee Belemnite for C isotopes and atmospheric N2 for N isotopes).
Data Analysis
Bar charts of relative abundances of eukaryote sequences found in eight fish intestines and a biplot of stable isotope signatures (δ13C and δ15N) of fish muscles were constructed using Origin 2018 software (OriginLab Co., Northampton, MA, USA). A phylogenetic tree of 18S rRNA sequences related to the phyla Chlorophyta, Ochrophyta and Rhodophyta was constructed using MEGA X and the neighbor-joining algorithm, and the maximum composite likelihood method with bootstrap analyses for 1,000 replicates (Kumar et al., 2018). A heat map of the relative abundance of the 18S rRNA sequences belonging to the phyla Chlorophyta, Ochrophyta and Rhodophyta was individually generated using the OmicShare tools (http://www.omicshare.com/tools). Dominant OTUs from the heat maps were selected to analyze taxonomic assignments using BLAST research. Based on the relative abundance of macroalgae sequences in eight fish intestines, data were transformed as square root with Bray-Curtis similarity resemblance. Then cluster analysis of the Bray-Curtis similarity was done with the Primer 5 software. Statistical differences about the stable isotope values of δ13C and δ15N among the different eight fishes were analyzed, with p<0.05 considered significant.
Results
Microscopic Observations of Fish Intestinal Contents
Intestinal contents of the eight fish species collected were analyzed by microscopy. After thorough washing, anterior intestinal contents of fishes were examined and a high proportion of flocculent detritus was found. Coral sand was only detected in the intestinal contents of C. carolinus and S. argenteus. Importantly, macrophytes (macroalgae or seagrasses) were easily observed in fish intestines (Table 2). The number of macrophyte species consumed by the eight herbivorous reef fishes varied. S. argenteus consumed the highest number of macrophyte species (six species), followed by N. unicornis (four species). However, only a few macrophyte species could be identified morphologically, while other fragments belonging to Chlorophyta, Ochrophyta or Rhodophyta were too small and were difficult to identify. The green algae Valonia ventricosa and Halimeda sp. were detected in the fish intestines of N. unicornis and S. argenteus, respectively. The brown alga Turbinaria ornata and the red alga Acanthophora spp. were observed in the intestinal contents of N. unicornis and S. punctatissimus, respectively. In addition, the seagrass Halophila ovalis was identified as a food source of S. argenteus.

Table 2 Macrophyte fragments observed by microscopic examination in the intestinal contents of fishes from the Xisha coral reefs.
Molecular Detection of Macrophyte Species Composition in Fish Intestines
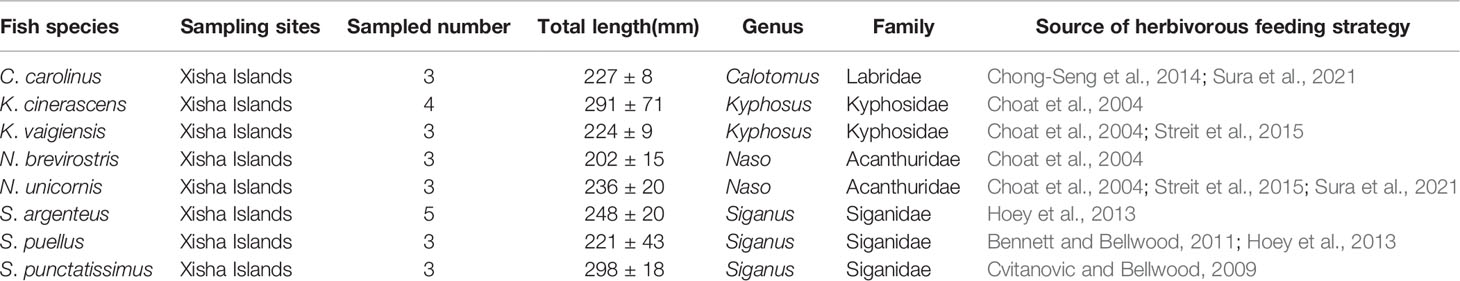
The dietary spectra of the eight fishes were analyzed with 18S rRNA gene high-throughput sequencing at the phylum and class levels (Figure 1). The 15 most abundant phyla accounted for 88.2%–99.3% of the total sequences found in the eight fish species’ intestines (Figure 1A). There were marked differences in the relative abundance of the dominant phyla in each species sample. Porifera was the most abundant taxon in C. carolinus, which was further identified as Demospongiae (sponges) (Figure 1). The dominant phyla in K. cinerascens were Bacillariophyta and Porifera. Ochrophyta, Streptophyta and Bigyra were significantly more abundant in K. vaigiensis, S. argenteus and S. punctatissimus, respectively. However, Cnidaria, mainly Anthozoa (corals), were much higher in S. puellus, N. brevirostris, N. unicornis and C. carolinus (Figure 1). Sequences belonging to Streptophyta were identified as H. ovalis, a seagrass species which occurred in the intestines of C. carolinus, N. brevirostris, S. argenteus and S. punctatissimus.
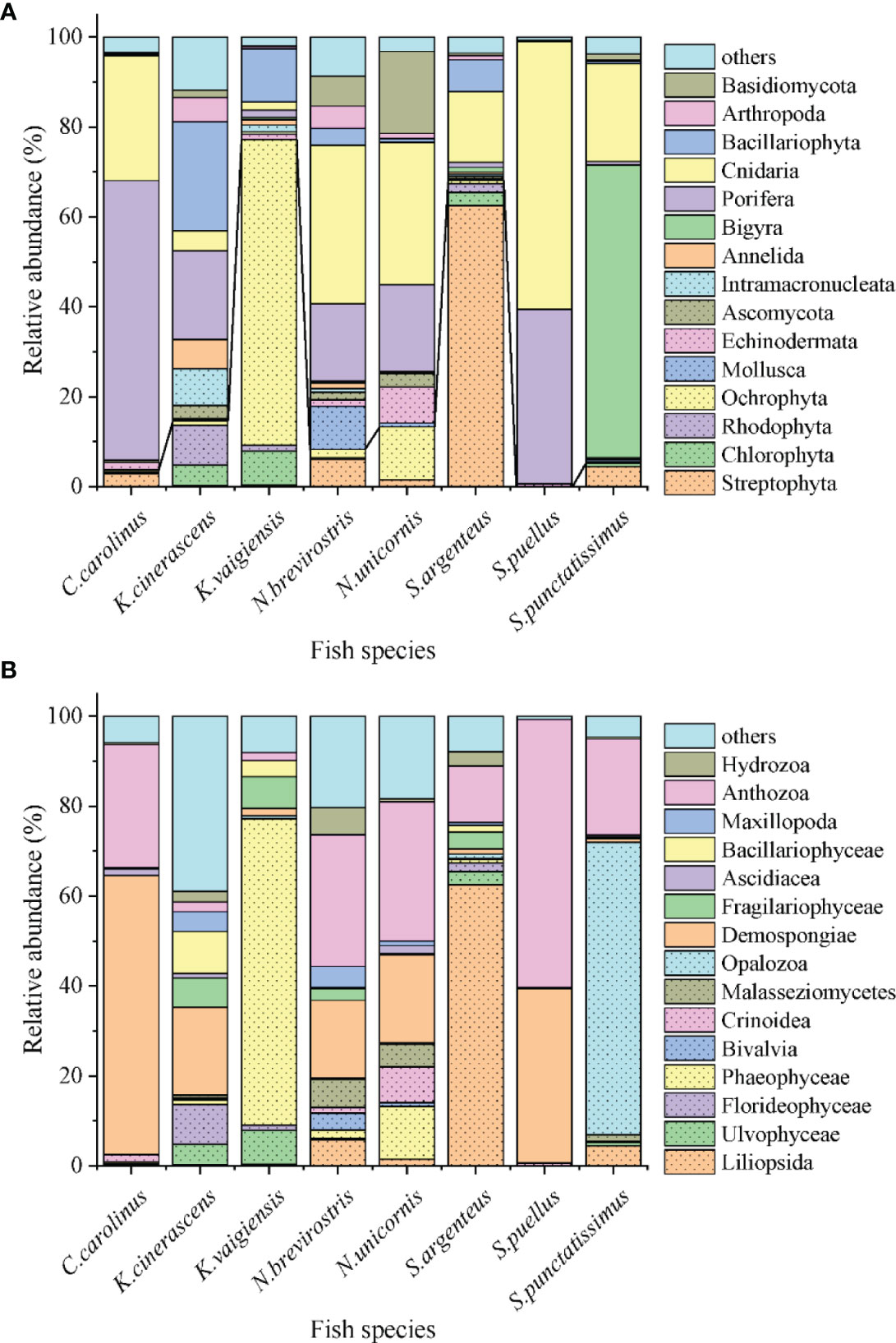
Figure 1 Relative abundances of eukaryote sequences found in fish intestines, determined with 18S rRNA high-throughput sequencing. (A) phylum level. Polylines show the sum of the relative abundance of the phyla Streptophyta, Chlorophyta, Ochrophyta and Rhodophyta. (B) class level.
Sequences referring to the phyla Chlorophyta, Ochrophyta and Rhodophyta found in the eight fishes were further analyzed (Figure 2). A total of 18, 15 and 47 OTUs were observed belonging to Chlorophyta, Ochrophyta and Rhodophyta, respectively (Figure 2A). A phylogenetic tree of the observed OTUs was constructed and is shown in Figure 3. Chlorophyta OTUs were most diverse in K. vaigiensis and S. argenteus (≥10 OTUs). The highest numbers of Rhodophyta OTUs (23 OTUs) were observed in K. cinerascens and K. vaigiensis. Ochrophyta OTUs were most abundant in K. vaigiensis (11 OTUs). The relative abundance of macroalgae found in fish intestines are shown in Figure 2B. K. vaigiensis consumed the greatest abundance of macroalgae (76.9%), followed by K. cinerascens (14.5%). N. unicornis preferred brown algae, while S. punctatissimus consumed more green algae. The diet of S. argenteus contained 2.9% green algae, 0.8% brown algae and 1.9% red algae.
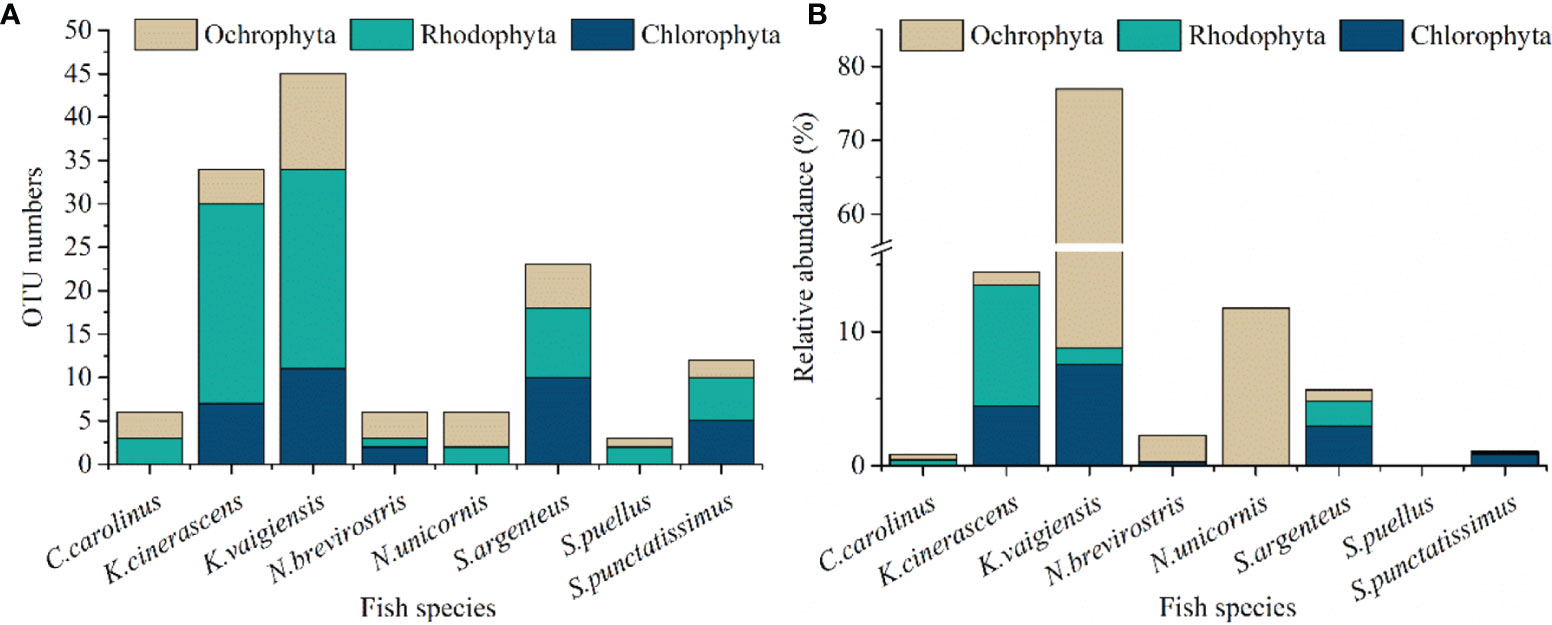
Figure 2 Eukaryote sequences belonging to the phyla Chlorophyta, Ochrophyta and Rhodophyta obtained from intestinal contents of eight coral reef fish species which were collected from the Xisha Islands, China. (A) OTU numbers of taxonomic groups. (B) Relative abundance of taxonomic groups.
Figure 3 Neighbor-joining tree of 18S rRNA sequences affiliated to the phyla Chlorophyta, Ochrophyta and Rhodophyta obtained from the intestinal contents of eight coral reef fish species which were collected from the Xisha Islands, China. The scale bar represents 5% estimated sequence divergence.
The differences in macroalgal composition between the intestinal contents of the eight fish species were analyzed (Figure 4 and Table 3). The species composition of green algae or red algae in the eight herbivorous fishes diets was different (Figures 4A, C). Regarding the green algae, Otu000030, identified as Dictyosphaeria cavernosa, was abundant in K. vaigiensis. Otu000055, which was very similar to Ulvella leptochaete, was a dominant component in S. argenteus and K. cinerascens. Both Otu000067 and Otu000102 were affiliated to the genus Cladophora. The occurrence of Otu000067 was higher in K. cinerascens, while Otu000102 dominated in N. brevirostris and S. argenteus. Regarding the red algae (Figure 4C), Otu000056 dominated in S. argenteus and showed a high similarity to Spyridia filamentosa. Otu000077 was related to Centroceras hyalacanthum, and was a major component in S. argenteus and S. punctatissimus. Otu000094 matched with Ceramium sinicola and was relatively abundant in K. cinerascens. Otu000200 and Otu000260 were relatively abundant in K. vaigiensis and were similar to Peyssonnelia rumoiana and P. armorica, respectively. Otu000535 was identified as the red alga P. rosenvingei and was very abundant in C. carolinus. The intestinal contents of C. carolinus also contained a dominant OTU (Otu000554), which was matched with Pneophyllum conicum. The dominant OTUs in N. unicornis included Otu000309 and Otu000874, which were similar to Chondrophycus cf. undulates and C. sinicola, respectively. Regarding the brown algae (Figure 4B), Otu000009, related to Lobophora variegate, was dominant in the intestinal contents of all the fishes, except S. argenteus. Otu000014 was also abundant in most fishes, especially S. argenteus in which it was most abundant. Cluster analysis indicated that macroalgae consumption from different fish of the same species always grouped together with each other. However, there was a low similarity among the fishes (<52%), only N. brevirostris and N. unicornis group at a high similarity (50.8%) (Figure 5).
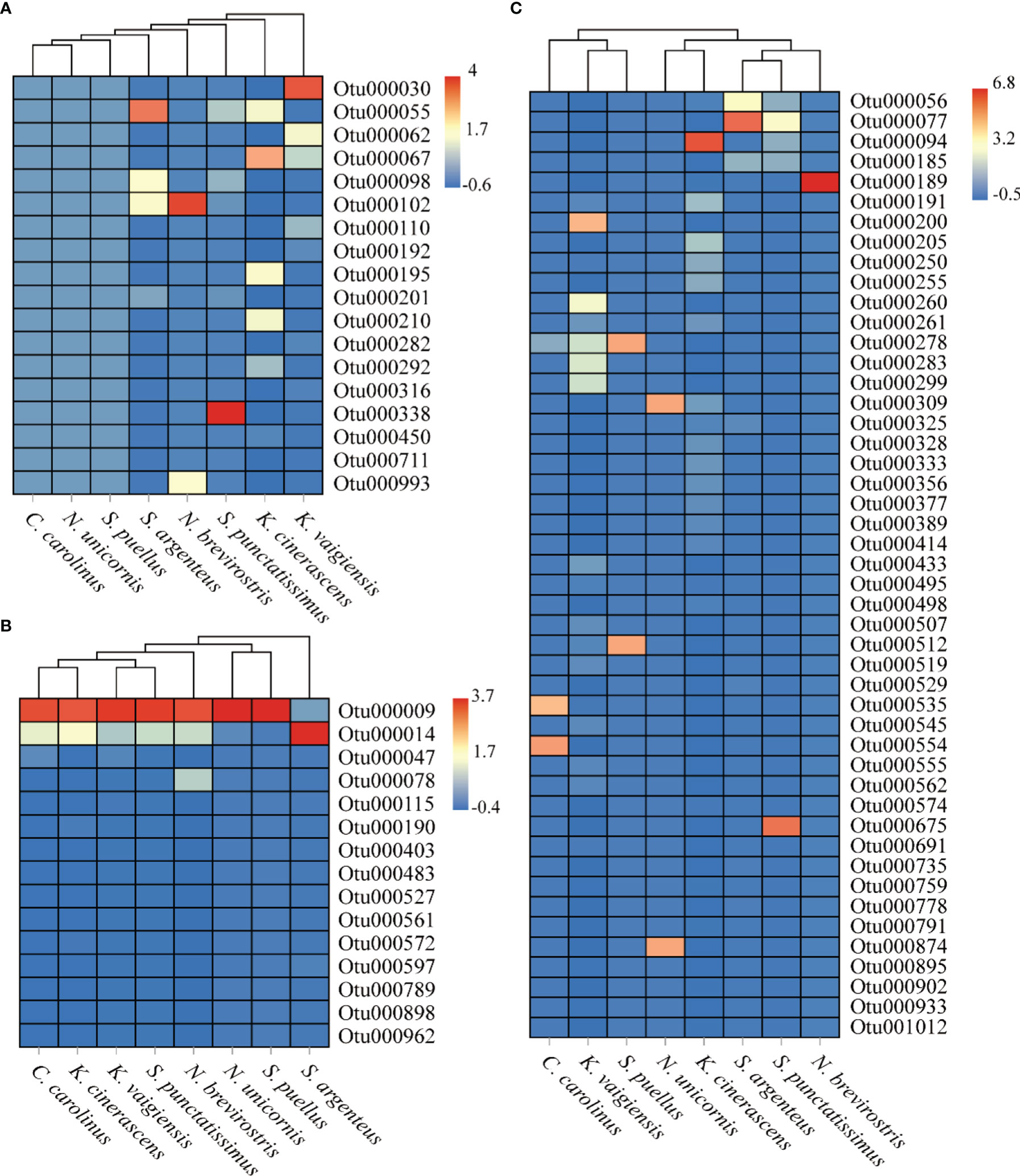
Figure 4 Heat map of the relative abundance of 18S rRNA sequences affiliated with different phyla found in the intestinal contents of eight coral reef fish species which were collected from the Xisha Islands, China. (A) Chlorophyta, (B) Ochrophyta, (C) Rhodophyta.
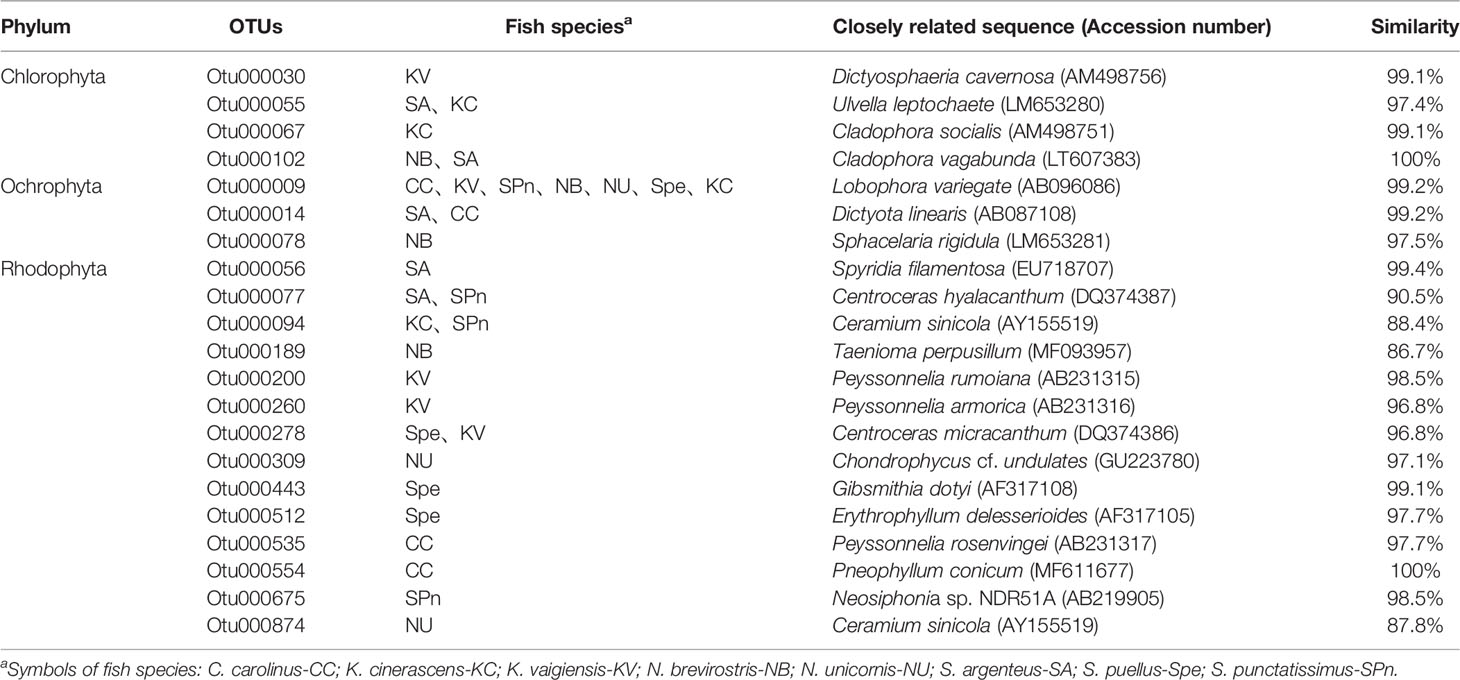
Table 3 Taxonomic assignment of the dominant OTUs affiliated to the phyla Chlorophyta, Ochrophyta and Rhodophyta, found in fish intestines using BLAST methodologies.
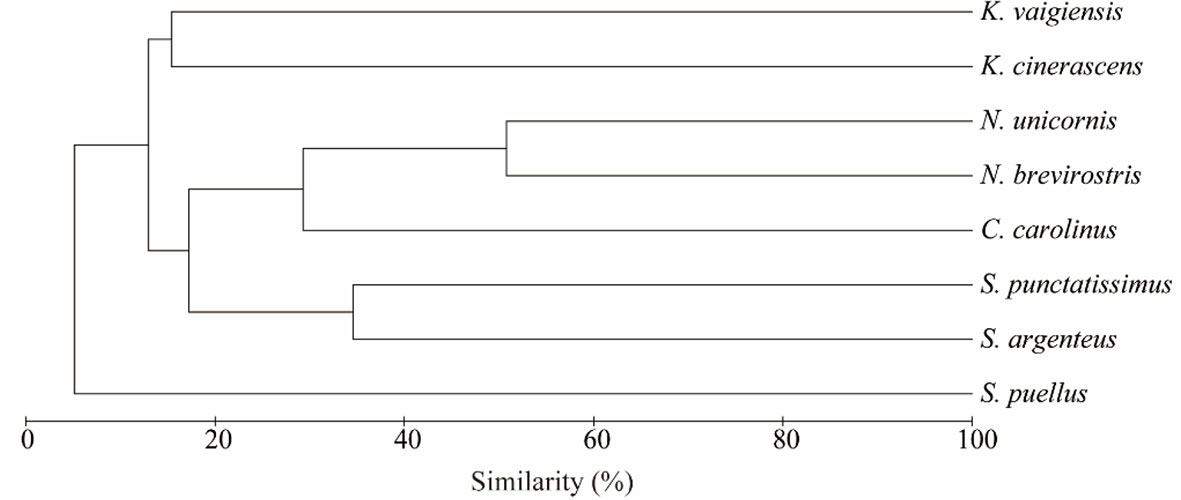
Figure 5 Cluster analysis of macroalgae composition in intestines of eight fish from the Xisha Islands.
Stable Isotope Analysis
Stable isotope values of δ13C and δ15N from the eight herbivorous fish species are shown as a bi-plot (Figure 6). The δ13C values across species ranged from -19.2 to -12.3‰. The δ13C values were more enriched in C. carolinus (-12.3 ± 0.8‰), which was significant different with other five species, except for N. unicornis (-13.4 ± 2.6‰) and K. vaigiensis (-13.8 ± 1.5‰) (Figure S2). However, S. punctatissimus and N. brevirostris had lower enrichment of δ13C values, with -19.2 ± 0.9‰ and -18.0 ± .02‰, respectively. The δ15N values of fishes varied from 5.1 to 8.3‰. K. vaigiensis (8.3 ± 0.5‰) had significantly higher δ15N values than others, except for K. cinerascens (p>0.05). N. unicornis showed the lowest δ15N values (5.1 ± 0.6‰) and exhibited significant different from most species, but not for S. punctatissimus and K. cinerascens. K. cinerascens, S. puellus and S. argenteus had generally similar δ13C and δ15N values with no significant difference. The differences in δ15N observed in the eight fishes indicated that N. unicornis occupied a relatively low trophic position.
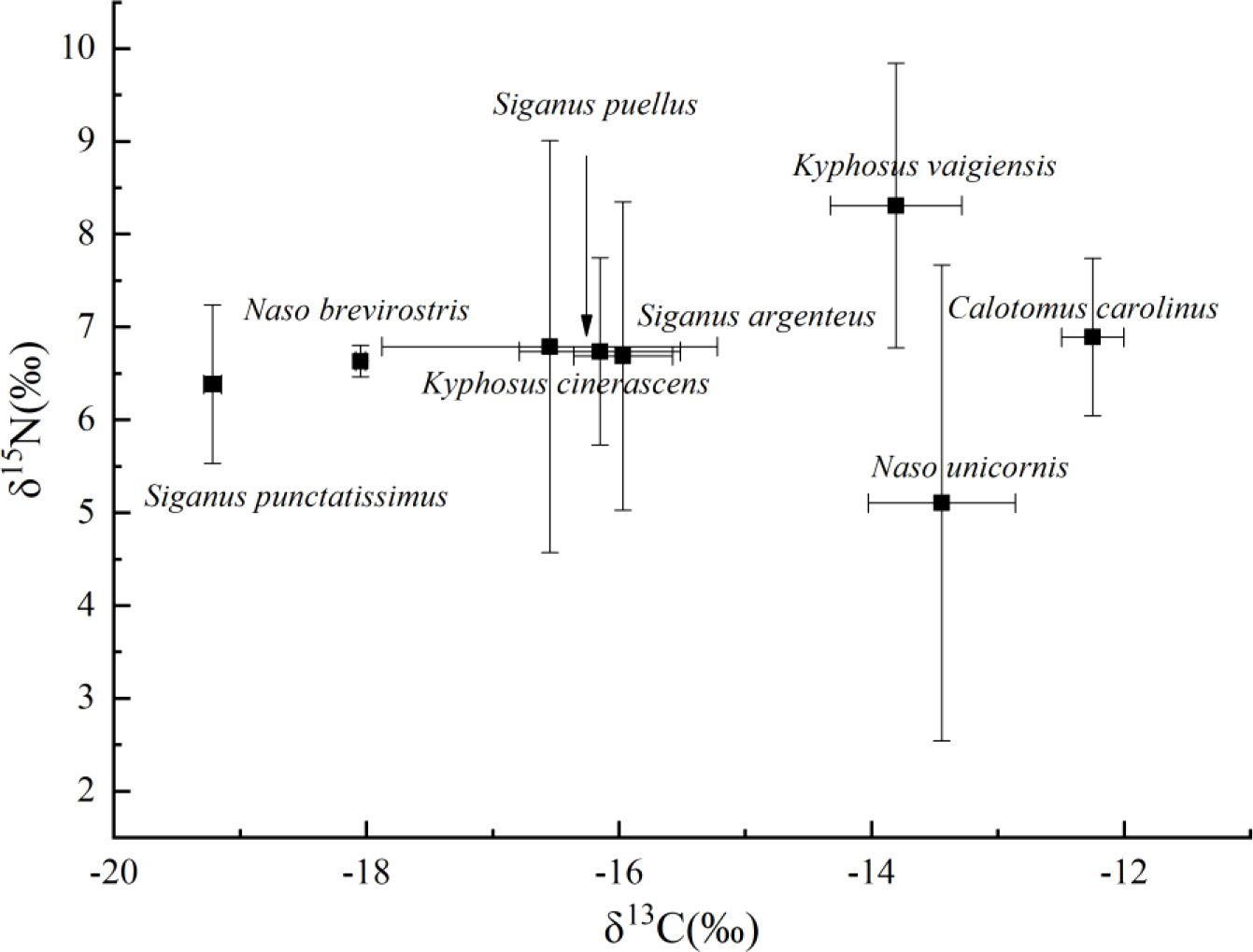
Figure 6 δ13C and δ15N isotope values of eight coral reef fish species from the Xisha Islands. Data points are averages with standard error bars.
Discussion
Microscopic Examination and High-Throughput Sequencing Analyses of Fish Intestinal Contents
Microscopic examination and 18S rRNA high-throughput sequencing analyses were used to investigate the dietary composition of eight herbivorous reef fish species off the Xisha Islands, China. There were some similarities in the results of the two methods. For example, the seagrass H. ovalis was observed in the intestinal contents of S. argenteus based on microscopic examination, and high-throughput sequencing analysis also found that this seagrass (Liliopsida) dominated the intestinal contents of S. argenteus. Microscopic examination revealed coral sand in the intestinal contents of C. carolinus and S. argenteus. High-throughput sequencing analysis also showed a high abundance of the class Anthozoa (corals) in C. carolinus and S. argenteus.
However, diet composition and breadth may vary according to the different approach used (Nalley et al., 2021). In this study, 18S rRNA high-throughput sequencing analysis provided higher taxonomic resolution of dietary composition, compared with microscopic examination. For example, some taxa that were found to dominate the intestinal contents using molecular analysis, such as Porifera, Cnidaria, Bacillariophyta were not detected by microscopic examination. The possible reasons were as follows. First, small organisms or digested organisms are too difficult to identify using a microscope (Dromard et al., 2015; Kume et al., 2021). In this study, microscopy detected a large amount of detritus in the fish intestinal contents. Dromard et al. (2015) using microscopy, also observed a high proportion of detritus in the intestines of Scaridae. Second, high-throughput sequencing is highly sensitive and can detect traces of DNA in mixed samples. This molecular technique can discriminate OTUs with only a single nucleotide variation within the targeted amplified region (Albaina et al., 2016). Third, the number of sampled fishes in this study was small so that some taxa in fish intestine were not observed using the microscope. For example, previous studies have shown that S. puellus can consume sponges (Hoey et al., 2013). While microscopic examination did not find any sponge remains in fish intestinal contents, Demospongiae (sponges) were identified as the main food for S. puellus using high-throughput sequencing. Leray et al. (2015) also showed that pyrosequencing analysis could broaden the recognition of the food webs of coral-dwelling predatory fish and achieved unprecedented taxonomic resolution of their diets. Nalley et al. (2022) reported that the resolution of metabarcding is greater than visual observation; metabarcding can identify specific taxa within the categories of turf algae that were targeted by herbivorous coral reef fishes, while visual inspection was limited to the identification of turf algae.
Differences in Macroalgal Consumption Among Herbivorous Coral Reef Fishes
Based on microscopy, 18S rRNA high-throughput sequencing and stable isotope analyses, we suggested that K. vaigiensis, N. unicornis and S. argenteus showed a high consumption potential of macroalgae from the coral reef ecosystem. Similarly, previous studies have also shown that K. vaigiensis and N. unicornis were highly effective in consuming macroalgae, especially the brown macroalgae (Lefevre and Bellwood, 2011; Streit et al., 2015; Puk et al., 2016). Similarly, we found that K. vaigiensis and N. unicornis favored brown macroalgae. However, in the northern GBR, Choat et al. (2002) demonstrated the dominant macroalgae genera in the diet of N. unicornis and K. vaigiensis were Turbinara and Dictyota, while Cvitanovic and Bellwood (2009) found that K. vaigiensis in the central region of the GBR preferred the brown macroalgae Sargassum. In this study, K. vaigiensis favored Lobophora and Dictyota, but N. unicornis targeted more on Lobophora and Turbinara. The diet difference of fishes between the GBR and the Xisha Islands may be related with occurrence of the dominant macroalgae in the habitat. Previous study showed that Sargassum was one of the abundant species in nearshore reefs of the GBR (McCook, 1997). However, Sargassum was seldom founded in the Xisha Islands. As for S. argenteus, in the Seychelles of the Indian Ocean, S. argenteus removed a range of substrate algae, including turf algae, macroalgae, seagrass and epiphytic algae (Ebrahim et al., 2020). Here, we also showed S. argenteus can consume macroalgae and seagrass. Furthermore, S. argenteus was supposed to focus more on red and green macroalgae in the northern GBR (Hoey et al., 2013). Here, S. argenteus also was found to favor red and green macroalgae. Of course, macroalgal grazing selectivity of herbivorous fishes can vary with location, season, food availability and according to the other fish species present (Puk et al., 2016).
Ecological Function of Herbivorous Fishes in Removing Macroalgae From Coral Reefs
In addition to identifying the diets of the eight herbivorous reef fishes studied, our results demonstrated the ecological importance of these fishes in removing macroalgae from coral reefs. Herbivorous fishes are broadly classified into four functional groups: excavators, scrapers, grazers, and browsers (Hoey and Bellwood, 2009). Excavators, scrapers and grazers generally consume small macroalgae and algal turfs, while browsers target large, erect macroalgal species and play a critical part in controlling macroalgal spreading (Hoey and Bellwood, 2009; Michael et al., 2013; Ebrahim et al., 2020). Previous studies have shown that C. carolinus, K. cinerascens, K. vaigiensis, N. brevirostris, N. unicornis and S. argenteus in the Moorea, the GBR or other coral reef islands belong to the browser group (Puk et al., 2016; Sura et al., 2021) and this was confirmed in our study. Based on microscopic examination, N. unicornis can consume V. ventricosa and T. ornate, while S. argenteus can consume Halimeda sp. (Table 2). According to the 18S rRNA high-throughput sequencing analysis, we found that C. carolinus, K. cinerascens, K. vaigiensis, N. brevirostris and N. unicornis consumed the brown alga L. variegate, and that K. vaigiensis and N. unicornis foraged especially heavily on this macroalgae. However, S. argenteus fed more on D. linearis. K. vaigiensis played an important role in removing the green alga D. cavernosa, while S. argenteus preferred to feed on the red algae S. filamentosa and C. hyalacanthum. K. vaigiensis also preferred to consume Peyssonnelia spp. The macroalgae mentioned above all belonged to either the upright calcareous or fleshy macroalgae. These two macroalgae groups are problematic as they probably inhibit coral settlement, while the crustose calcareous algae and algal turfs have only minor negative effects on coral settlement (Diaz-Pulido et al., 2010). Overgrowth of Lobophora and Dictyota is widely supposed to reduce coral settlement (Foster et al., 2008; Diaz-Pulido et al., 2010; Evensen et al., 2019; Vieira, 2020) and the spread of Dictyota can even cause coral disease outbreaks (Brandt et al., 2012). Unfortunately, Lobophora and Dictyota, are the dominant species on Indo-Pacific coral reefs (Cardoso et al., 2009; Titlyanov et al., 2017). We found that the nitrogen signature (δ15N) of N. unicornis was significantly lower than most of the other herbivorous fishes, indicating a high consumption potential of macroalgae. In general, we suggested that K. vaigiensis, N. unicornis and S. argenteus were the key browsers removing macroalgae from reefs off the Xisha Islands. This result is similar to that of a previous view, which recommended that K. vaigiensis, N. unicornis and Siganus canaliculatus were the predominant removers of macroalgae, the “true macroalgae browsers” of coral reefs (Puk et al., 2016). K. vaigiensis and N. unicornis were commonly considered as important macroalgal consumers in the coral reefs of the GBR (Verges et al., 2012).
Herbivorous fishes that eat the entire macroalgal thallus have a great functional impact on macroalgae removal from coral reefs than species that only consume algal fronds (Streit et al., 2015). In this study, K. cinerascens, K. vaigiensis, N. unicornis and S. punctatissimus were found to feed on the entire macroalgal thallus by microscopic examination (Table 2). Streit et al. (2015) also showed that K. vaigiensis and N. unicornis bit on the entire macroalgal thallus in almost 90% of bites. Body size, tooth shape, and feeding behavior of different herbivores may result in these functional differences (Streit et al., 2015). S. punctatissimus can fed on Acanthophora sp., while Acanthophora can inhibit the successful settlement of coral planulae (Vermeij et al., 2009). The red alga, P. conicum is widely distributed on Indo-Pacific coral reefs and can overgrow and kill live corals (Antonius, 2001). Coincidentally, we found that C. carolinus consumed P. conicum. In addition, herbivorous fishes can selectively remove some macroalgae including Ulvella, Cladophora, Sphacelaria, Spyridia, Centroceras, Ceramium and Neosiphonia, which often occur in the Xisha Islands (Titlyanov et al., 2017; Huang, 2018). The spread of macroalgae can negatively affect coral reefs through the inhibition of coral fecundity and growth, the reduction of coral larval settlement and recruitment and the increasing prevalence of coral diseases (Dell et al., 2020). Burkepile and Hay (2008) identified herbivore species richness and feeding complementarity as key factors in the effective suppression of the spread of macroalgae in a coral ecosystem. In summary, we recommend reducing fishing mortality for key browser species, such as K. vaigiensis, N. unicornis and S. argenteus, in order to control the spread of macroalgae on the reefs off the Xisha Islands. Gowri et al. (2016) recommended a ban on fishing herbivorous fishes during the times of spawning aggregations, and a ban on the use of small-mesh gill nets in coral management of India. Dell et al. (2020) also encouraged the government to protect and monitor the key browsers in the coral reefs. Some countries, such as Belize, even have implemented bans on fishing herbivorous fishes (Kramer et al., 2017).
Conclusions
In this study, for the first time we compared the capability of macroalgal consumption by eight herbivorous coral reef fishes from the Xisha Islands, China, as determined by microscopy, 18S rRNA high-throughput sequencing and stable isotope analyses. The results of the three methods suggested that K. vaigiensis, N. unicornis and S. argenteus potentially consumed macroalgae more effectively. However, high-throughput sequencing showed that macroalgae consumption of the eight fishes in the same species always clustered together. Moreover, K. vaigiensis and N. unicornis preferred to take brown macroalgae, while S. argenteus targeted more on red and green macroalgae. Meanwhile, a high diversity of herbivorous fishes was very important to the health of the Xisha coral reefs. Because we found herbivorous fishes can selectively remove some macroalgae which showed negative influences on the coral growth and coral larval settlement. For example, C. carolinus can consume a red alga P. conicum, which can directly overgrow and kill live corals. In the future, analysis of feeding behavior of herbivorous fishes should be investigated according to variation of macroalgae composition in Xisha coral reef islands, and macroalgae selectivity for herbivorous fishes should be elaborated in more details.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA742779, MZ481947-MZ481964, MZ481932-MZ481946 and MZ481965-MZ482014].
Ethics Statement
All work undertaken in this study complied with the committee on Laboratory Animal Welfare and Ethics of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (nhdf2022-01).
Author Contributions
Conceptualization, PW, YL and CL; investigation, PW, TW, YX, TH and QQ; methodology, PW, TW and TH; formal analysis, PW and QQ; writing original draft, PW and YX; writing-review & editing, PW, LL, YL, CL, SX and TW; data curation, YL and LL; funding acquisition, CL and PW. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program of China (2018YFD0900803), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0605), Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (No. 2019TS28, No. 2021SD04), Financial Fund of the Ministry of Agriculture and Rural Affairs, P. R. of China (NFZX2021), Key Laboratory of Tropical Marine Biotechnology of Hainan Province (LTMB202203), Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2020TD16), Science and Technology Planning Project of Guangdong Province (2019B121201001) and Fundamental and Applied Fundamental Research Major Program of Guangdong Province (2019B030302004-05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.882196/full#supplementary-material
References
Adam T. C., Burkepile D. E., Holbrook S. J., Carpenter R. C., Claudet J., Loiseau C., et al. (2021). Landscape-Scale Patterns of Nutrient Enrichment in a Coral Reef Ecosystem: Implications for Coral to Algae Phase Shifts. Ecol. Appl. 31, 2227. doi: 10.1002/eap.2227
Albaina A., Aguirre M., Abad D., Santos M., Estonba A. (2016). 18s rRNA V9 Metabarcoding for Diet Characterization: A Critical Evaluation With Two Sympatric Zooplanktivorous Fish Species. Ecol. Evol. 6, 1809–1824. doi: 10.1002/ece3.1986
Antonius A. (2001). Pneophyllum Conicum, a Coralline Red Alga Causing Coral Reef-Death in Mauritius. Coral Reefs 19, 418–418. doi: 10.1007/s003380000126
Bennett S., Bellwood D. R. (2011). Latitudinal Variation in Macroalgal Consumption by Fishes on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 426, 241–252. doi: 10.3354/meps09016
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-Filtering Vastly Improves Diversity Estimates From Illumina Amplicon Sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Brandt M. E., Ruttenberg B. I., Waara R., Miller J., Witcher B., Estep A. J., et al. (2012). Dynamics of an Acute Coral Disease Outbreak Associated With the Macroalgae Dictyota Spp. In Dry Tortugas National Park, Florida, USA. B. Mar. Sci. 88, 1035–1050. doi: 10.5343/bms.2011.1104
Bruno J. F., Cote I. M., Toth L. T. (2019). Climate Change, Coral Loss, and the Curious Case of the Parrotfish Paradigm: Why Don't Marine Protected Areas Improve Reef Resilience? Ann. Rev. Mar. Sci. 11, 307–334. doi: 10.1146/annurev-marine-010318-095300
Burkepile D. E., Hay M. E. (2008). Herbivore Species Richness and Feeding Complementarity Affect Community Structure and Function on a Coral Reef. Proc. Natl. Acad. Sci. U.S.A. 105, 16201–16206. doi: 10.1073/pnas.0801946105
Burke L., Reytar K., Spalding M., Perry A. (2011). Reefs at Risk Revisited (Washington: World Resources Institute).
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cardoso S. C., Soares M. C., Oxenford H. A., Côté I. M. (2009). Interspecific Differences in Foraging Behaviour and Functional Role of Caribbean Parrotfish. Marine Biodivers. Records 2, 148. doi: 10.1017/S1755267209990662
Cheal A. J., MacNeil M. A., Cripps E., Emslie M. J., Jonker M., Schaffelke B., et al. (2010). Coral-Macroalgal Phase Shifts or Reef Resilience: Links With Diversity and Functional Roles of Herbivorous Fishes on the Great Barrier Reef. Coral Reefs 29, 1005–1015. doi: 10.1007/s00338-010-0661-y
Chen X. Y., Yu K. F., Huang X. Y., Wang Y. H., Liao Z. H., Zhang R. J., et al. (2019). Atmospheric Nitrogen Deposition Increases the Possibility of Macroalgal Dominance on Remote Coral Reefs. J. Geophys. Res-Biogeo 124, 1355–1369. doi: 10.1029/2019JG005074
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: An Ultra-Fast All-In-One FASTQ Preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Cheung M. K., Au C. H., Chu K. H., Kwan H. S., Wong C. K. (2010). Composition and Genetic Diversity of Picoeukaryotes in Subtropical Coastal Waters as Revealed by 454 Pyrosequencing. ISME. J. 4, 1053–1059. doi: 10.1038/ismej.2010.26
Choat J. H., Clements K. D., Robbins W. D. (2002). The Trophic Status of Herbivorous Fishes on Coral Reefs: 1. Dietary Analyses. Mar. Biol. 140, 613–623. doi: 10.1007/s00227-001-0715-3
Choat J. H., Robbins W. D., Clements K. D. (2004). The Trophic Status of Herbivorous Fishes on Coral Reefs: II. Food Processing Modes and Trophodynamics. Mar. Biol. 145, 445–454. doi: 10.1007/s00227-004-1341-7
Chong-Seng K. M., Nash K. L., Bellwood D. R., Graham N. A. J. (2014). Macroalgal Herbivory on Recovering Versus Degrading Coral Reefs. Coral Reefs 33, 409–419. doi: 10.1007/s00338-014-1134-5
Clements K. D., German D. P., Piché J., Tribollet A., Choat J. H. (2017). Integrating Ecological Roles and Trophic Diversification on Coral Reefs: Multiple Lines of Evidence Identify Parrotfishes as Microphages. Biol. J. Linn. Soc. 120, 729–751.doi: 10.1111/bij.12914.
Corse E., Costedoat C., Chappaz R., Pech N., Martin J. F., Gilles A. (2010). A PCR-Based Method for Diet Analysis in Freshwater Organisms Using 18s rDNA Barcoding on Faeces. Mol. Ecol. Resour. 10, 96–108. doi: 10.1111/j.1755-0998.2009.02795.x
Cvitanovic C., Bellwood D. R. (2009). Local Variation in Herbivore Feeding Activity on an Inshore Reef of the Great Barrier Reef. Coral Reefs 28, 127–133. doi: 10.1007/s00338-008-0433-0
de Carvalho D. R., de Castro D. M. P., Callisto M., Chaves A. J. D., Moreira M. Z., Pompeu P. S. (2019). Stable Isotopes and Stomach Content Analyses Indicate Omnivorous Habits and Opportunistic Feeding Behavior of an Invasive Fish. Aquat. Ecol. 53, 365–381. doi: 10.1007/s10452-019-09695-3
Dell C. L. A., Longo G. O., Burkepile D. E., Manfrino C. (2020). Few Herbivore Species Consume Dominant Macroalgae on a Caribbean Coral Reef. Front. Mar. Sci. 7, 676. doi: 10.3389/fmars.2020.00676
Devloo-Delva F., Huerlimann R., Chua G., Matley J. K., Heupel M. R., Simpfendorfer C. A., et al. (2019). How Does Marker Choice Affect Your Diet Analysis: Comparing Genetic Markers and Digestion Levels for Diet Metabarcoding of Tropical-Reef Piscivores. Mar. Freshwater Res. 70, 8–18. doi: 10.1071/MF17209
Diaz-Pulido G., Harii S., McCook L. J., Hoegh-Guldberg O. (2010). The Impact of Benthic Algae on the Settlement of a Reef-Building Coral. Coral Reefs 29, 203–208. doi: 10.1007/s00338-009-0573-x
Ding J. F., Jiang F. H., Li J. X., Wang Z. X., Sun C. J., Wang Z. Y., et al. (2019). Microplastics in the Coral Reef Systems From Xisha Islands of South China Sea. Environ. Sci. Technol. 53, 8036–8046. doi: 10.1021/acs.est.9b01452
Dromard C. R., Bouchon-Navaro Y., Harmelin-Vivien M., Bouchon C. (2015). Diversity of Trophic Niches Among Herbivorous Fishes on a Caribbean Reef (Guadeloupe, Lesser Antilles), Evidenced by Stable Isotope and Gut Content Analyses. J. Sea Res. 95, 124–131. doi: 10.1016/j.seares.2014.07.014
Duran A., Adam T. C., Palma L., Moreno S., Collado-Vides L., Burkepile D. E. (2019). Feeding Behavior in Caribbean Surgeonfishes Varies Across Fish Size, Algal Abundance, and Habitat Characteristics. Mar. Ecol. 00, 12561. doi: 10.1111/maec.12561
Ebrahim A., Martin T. S. H., Mumby P. J., Olds A. D., Tibbetts I. R. (2020). Differences in Diet and Foraging Behaviour of Commercially Important Rabbitfish Species on Coral Reefs in the Indian Ocean. Coral Reefs 39, 977–988. doi: 10.1007/s00338-020-01918-6
Edgar R. C. (2013). UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Evensen N. R., Doropoulos C., Wong K. J., Mumby P. J. (2019). Stage-Specific Effects of Lobophora on the Recruitment Success of a Reef-Building Coral. Coral Reefs. 38, 489–498. doi: 10.1007/s00338-019-01804-w
Foster N. L., Box S. J., Mumby P. J. (2008). Competitive Effects of Macroalgae on the Fecundity of the Reef-Building Coral Montastraea Annularis. Mar. Ecol. Prog. Ser. 367, 143–152. doi: 10.3354/meps07594
Gowri V.S., Pramiladevi R.R.I., Nammalwar P. (2016). Biodiversity of Selected Herbivorous Coral Reef Fishes and Their Role in Coral Management, India. Int. J. Res. Sci. Innov. III, 94–100.
Hoey A. S., Bellwood D. R. (2009). Limited Functional Redundancy in a High Diversity System: Single Species Dominates Key Ecological Process on Coral Reefs. Ecosystems 12, 1316–1328. doi: 10.1007/s10021-009-9291-z
Hoey A. S., Brandl S. J., Bellwood D. R. (2013). Diet and Cross-Shelf Distribution of Rabbitfishes (F. Siganidae) on the Northern Great Barrier Reef: Implications for Ecosystem Function. Coral Reefs. 32, 973–984. doi: 10.1007/s00338-013-1043-z
Kopp D., Bouchon-Navaro Y., Cordonnier S., Haouisee A., Louis M., Bouchon C. (2010). Evaluation of Algal Regulation by Herbivorous Fishes on Caribbean Coral Reefs. Helgoland. Mar. Res. 64, 181–190. doi: 10.1007/s10152-009-0177-4
Kramer P., Kramer P., McField M., Drysdale I., Rueda M., Girà A., et al. (2017). 2015 Report Card for the Mesoamerican Reef (Washington, DC: The Summit Foundation).
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kume G., Kobari T., Hirai J., Kuroda H., Takeda T., Ichinomiya M., et al. (2021). Diet Niche Segregation of Co-Occurring Larval Stages of Mesopelagic and Commercially Important Fishes in the Osumi Strait Assessed Through Morphological, DNA Metabarcoding, and Stable Isotope Analyses. Mar. Biol. 168, 6. doi: 10.1007/s00227-020-03810-x
Lamb P. D., Hunter E., Pinnegar J. K., Creer S., Davies R. G., Taylor M. I. (2019). How Quantitative Is Metabarcoding: A Meta-Analytical Approach. Mol. Ecol. 28, 420–430. doi: 10.1111/mec.14920
Lefevre C. D., Bellwood D. R. (2011). Temporal Variation in Coral Reef Ecosystem Processes: Herbivory of Macroalgae by Fishes. Mar. Ecol. Prog. Ser. 422, 239–251. doi: 10.3354/meps08916
Leray M., Meyer C. P., Mills S. C. (2015). Metabarcoding Dietary Analysis of Coral Dwelling Predatory Fish Demonstrates the Minor Contribution of Coral Mutualists to Their Highly Partitioned, Generalist Diet. Peer J. 3, 1047. doi: 10.7717/peerj.1047
Li Y. C., Chen S. Q., Zheng X. Q., Cai Z. F., Wu Z. J., Wang D. R., et al. (2018). Analysis of the Change of Hermatypic Corals in Yongxing Island and Qilianyu Island in Nearly a Decade. Haiyang Xuebao 40, 97–109. doi: 10.3969/j.issn.0253-4193.2018.08.010.
Lin X. Z., Hu S. M., Liu Y., Zhang L., Huang H., Liu S. (2021). Disturbance-Mediated Changes in Coral Reef Habitat Provoke a Positive Feeding Response in a Major Coral Reef Detritivore, Ctenochaetus Striatus. Front. Mar. Sci. 8, 682697. doi: 10.3389/fmars.2021.682697
Liu Y., Wu P., Li C. H., Xiao Y. Y., Wang T., Lin L. (2021). The Bacterial Composition Associated With Atriolum Robustum, a Common Ascidian From Xisha Coral Reef, China. Symbiosis 83, 153–161. doi: 10.1007/s13199-020-00742-4
Li Y. C., Wu Z. J., Chen S. Q., Cai Z. F., Lan J. X., Tong Y. H., et al. (2017). Discussion of the Diversity of the Coral Reef Fish in the Shallow Reefs Along the Yongxing and Qilianyu Island. Marine Environ. Sci. 36, 509–516. doi: 10.13634/j.cnki.mes.2017.04.005
Magoc T., Salzberg S. L. (2011). FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mantyka C. S., Bellwood D. R. (2007). Macroalgal Grazing Selectivity Among Herbivorous Coral Reef Fishes. Mar. Ecol. Prog. Ser. 352, 177–185. doi: 10.3354/meps07055
McCook L. J. (1997). Effects of Herbivory on Zonation of Sargassum Spp. Within Fringing Reefs of the Central Great Barrier Reef. Mar. Biol. 129, 713–722. doi: 10.1007/s002270050214
Michael P. J., Hyndes G. A., Vanderklift M. A., Verges A. (2013). Identity and Behaviour of Herbivorous Fish Influence Large-Scale Spatial Patterns of Macroalgal Herbivory in a Coral Reef. Mar. Ecol. Prog. Ser. 482, 227–240. doi: 10.3354/meps10262
Moberg F., Folke C. (1999). Ecological Goods and Services of Coral Reef Ecosystems. Ecol. Econ. 29, 215–233. doi: 10.1016/S0921-8009(99)00009-9
Nalley E. M., Donahue M. J., Heenan A., Toonen R. J. (2022). Quantifying the Diet Diversity of Herbivorous Coral Reef Fishes Using Systematic Review and DNA Metabarcoding. Environ. DNA 4, 191–205. doi: 10.1002/edn3.247
Nalley E. M., Donahue M. J., Toonen R. J. (2021). Metabarcoding as a Tool to Examine Cryptic Algae in the Diets of Two Common Grazing Surgeonfishes, Acanthurus Triostegus and A. Nigrofuscus. Environ. DNA 00, 1–12.doi: 10.1002/edn3.206.
Neilson B. J., Wall C. B., Mancini F. T., Gewecke C. A. (2018). Herbivore Biocontrol and Manual Removal Successfully Reduce Invasive Macroalgae on Coral Reefs. Peer J. 6, 5332. doi: 10.7717/peerj.5332
Nicholson G. M., Clements K. D. (2020). Resolving Resource Partitioning in Parrotfishes (Scarini) Using Microhistology of Feeding Substrata. Coral Reefs 39, 1313–1327. doi: 10.1007/s00338-020-01964-0
Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W. G., Peplies J., et al. (2007). SILVA: A Comprehensive Online Resource for Quality Checked and Aligned Ribosomal RNA Sequence Data Compatible With ARB. Nucleic. Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Puk L. D., Ferse S. C. A., Wild C. (2016). Patterns and Trends in Coral Reef Macroalgae Browsing: A Review of Browsing Herbivorous Fishes of the Indo-Pacific. Rev. Fish Biol. Fisher. 26, 53–70. doi: 10.1007/s11160-015-9412-z
Rodriguez-Barreras R., Godoy-Vitorino F., Praebel K., Wangensteen O. S. (2020). DNA Metabarcoding Unveils Niche Overlapping and Competition Among Caribbean Sea Urchins. Reg. Stud. Mar. Sci. 40, 101537. doi: 10.1016/j.rsma.2020.101537
Russ G. R., Questel S. L., Rizzari J. R., Alcala A. C. (2015). The Parrotfish-Coral Relationship: Refuting the Ubiquity of a Prevailing Paradigm. Mar. Biol. 162, 2029–2045. doi: 10.1007/s00227-015-2728-3
Shen J. W., Wang Y., Zhao N., Yang H. Q., Fu F. X., Jin Y. B. (2017). Carbonate Sedimentary Characteristics of the Beach Rocks Around Qilian Islets and Cays, Xisha Islands: Implication for Coral Reef Development and Decline. Palaeogeogr. Palaeocl. 474, 264–278. doi: 10.1016/j.palaeo.2016.05.005
Souter D., Planes S., Wicquart J., Logan M., Obura D., Staub F. (2021). Status of Coral Reefs of the World: 2020 Executive Summary (Townsville: Global Coral Reef Monitoring Network).
Streit R., Hoey A., Bellwood D. (2015). Feeding Characteristics Reveal Functional Distinctions Among Browsing Herbivorous Fishes on Coral Reefs. Coral Reefs. 34, 1037–1047. doi: 10.1007/s00338-015-1322-y
Sura S. A., Molina N. E., Blumstein D. T., Fong P. (2021). Selective Consumption of Macroalgal Species by Herbivorous Fishes Suggests Reduced Functional Complementarity on a Fringing Reef in Moorea, French Polynesia. J. Exp. Mar. Biol. Ecol. 536, 151508. doi: 10.1016/j.jembe.2020.151508
Titlyanov A. E., Titlyanova V. T., Li X., Huang H. (2017). Coral Reef Marine Plants of Hainan Island (London: Academic Press).
Verges A., Bennett S., Bellwood D. R. (2012). Diversity Among Macroalgae-Consuming Fishes on Coral Reefs: A Transcontinental Comparison. PloS One 7, 45543. doi: 10.1371/journal.pone.0045543
Vermeij M. J. A., Smith J. E., Smith C. M., Thurber R. V., Sandin S. A. (2009). Survival and Settlement Success of Coral Planulae: Independent and Synergistic Effects of Macroalgae and Microbes. Oecologia 159, 325–336. doi: 10.1007/s00442-008-1223-7
Vieira C. (2020). Lobophora-Coral Interactions and Phase Shifts: Summary of Current Knowledge and Future Directions. Aquat. Ecol. 54, 1–20. doi: 10.1007/s10452-019-09723-2
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences Into the New Bacterial Taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang T., Liu Y., Quan Q. M., Xiao Y. Y., Wu P., Li C. H. (2022). Species Composition Characteristics Analysis of Qilianyu Reef Fishes of Xisha Islands. J. Fisher. Sci. China 29, 102–117. doi: 10.12264/JFSC2021-0122.
Zhao N., Shen D. S., Shen J. W. (2019). Formation Mechanism of Beach Rocks and Its Controlling Factors in Coral Reef Area, Qilian Islets and Cays, Xisha Islands, China. J. Earth Sci-China 30, 728–738. doi: 10.1007/s12583-018-0981-3
Zhao M. X., Yu K. F., Shi Q., Yang H. Q., Riegl B., Zhang Q. M., et al. (2016). The Coral Communities of Yongle Atoll: Status, Threats and Conservation Significance for Coral Reefs in South China Sea. Mar. Freshwater Res. 67, 1888–1896. doi: 10.1071/MF15110
Keywords: coral reefs, herbivorous fishes, macroalgal species composition, diet, intestinal contents
Citation: Wu P, Wang T, Liu Y, Li C, Xiao Y, Xu S, Han T, Lin L and Quan Q (2022) Differences of Macroalgal Consumption by Eight Herbivorous Coral Reef Fishes From the Xisha Islands, China. Front. Mar. Sci. 9:882196. doi: 10.3389/fmars.2022.882196
Received: 23 February 2022; Accepted: 17 March 2022;
Published: 12 April 2022.
Edited by:
Meilin WU, South China Sea Institute of Onceanology (CAS), ChinaReviewed by:
Ximei Liang, Jiangxi Agricultural University, ChinaYanjun Shen, Chongqing Normal University, China
Xuefeng Wang, Guangdong Ocean University, China
Copyright © 2022 Wu, Wang, Liu, Li, Xiao, Xu, Han, Lin and Quan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Liu, bGl1eW9uZ0BzY3NmcmkuYWMuY24=
 Peng Wu
Peng Wu Teng Wang
Teng Wang Yong Liu
Yong Liu Chunhou Li
Chunhou Li Yayuan Xiao
Yayuan Xiao Shannan Xu
Shannan Xu Tingting Han
Tingting Han Lin Lin
Lin Lin Qiumei Quan1
Qiumei Quan1