
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 02 March 2023
Sec. Aquatic Microbiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.876830
This article is part of the Research Topic The Role of Micronutrients in Mediating Microbial Community Interactions in the Surface Ocean View all 5 articles
 Alexa R. Sterling1
Alexa R. Sterling1 Laura Z. Holland1
Laura Z. Holland1 Randelle M. Bundy2
Randelle M. Bundy2 Shannon M. Burns3
Shannon M. Burns3 Kristen N. Buck3,4
Kristen N. Buck3,4 P. Dreux Chappell5
P. Dreux Chappell5 Bethany D. Jenkins1,6*
Bethany D. Jenkins1,6*The growth of diatoms in the Southern Ocean, especially the region surrounding the West Antarctic Peninsula, is frequently constrained by low dissolved iron and other trace metal concentrations. This challenge may be overcome by mutualisms between diatoms and co-occurring associated bacteria, in which diatoms produce organic carbon as a substrate for bacterial growth, and bacteria produce siderophores, metal-binding ligands that can supply diatoms with metals upon uptake as well as other useful secondary compounds for diatom growth like vitamins. To examine the relationships between diatoms and bacteria in the plankton (diatom) size class (> 3 µm), we sampled both bacterial and diatom community composition with accompanying environmental metadata across a naturally occurring concentration gradient of macronutrients, trace metals and siderophores at 21 stations near the West Antarctic Peninsula (WAP). Offshore Drake Passage stations had low dissolved iron (0.33 ± 0.15 nM), while the stations closer to the continental margin had higher dissolved iron (5.05 ± 1.83 nM). A similar geographic pattern was observed for macronutrients and most other trace metals measured, but there was not a clear inshore-offshore gradient in siderophore concentrations. The diatom and bacteria assemblages, determined using 18S and 16S rDNA sequencing respectively, were similar by location sampled, and variance in both assemblages was driven in part by concentrations of soluble reactive phosphorous, dissolved manganese, and dissolved copper, which were all higher near the continent. Some of the most common diatom sequence types observed were Thalassiosira and Fragilariopsis, and bacteria in the plankton size fraction were most commonly Bacteroidetes and Gammaproteobacteria. Network analysis showed positive associations between diatoms and bacteria, indicating possible in situ mutualisms through strategies such as siderophore and vitamin biosynthesis and exchange. This work furthers the understanding of how naturally occurring gradients of metals and nutrients influence diatom-bacteria interactions. Our data suggest that distinct groups of diatoms and associated bacteria are interacting under different trace metal regimes in the WAP, and that diatoms with different bacterial partners may have different modes of biologically supplied trace metals.
The West Antarctic Peninsula (WAP) region of the Southern Ocean (SO) is impacted disproportionally by climate change. Higher than average air and water temperature increases have led to ice cover retreat (Stammerjohn et al., 2008; Somero, 2012), changing food webs (Atkinson et al., 2004; Ducklow et al., 2007), and increased supply of trace metals to shelf waters (Forsch et al., 2021). Trace metals, such as iron (Fe), are important micronutrients to phytoplankton. Low Fe concentrations in the SO waters adjacent to the WAP have been observed to significantly impact the growth of phytoplankton along with co-limiting silicic acid concentrations and light availability (Martin et al., 1990; Boyd et al., 2000; Gervais et al., 2002; van Oijen et al., 2004; Leblanc et al., 2005). Diatoms, which can bloom to high levels and dominate the phytoplankton community in the area, are some of the most important components of the Antarctic food web (Garibotti et al., 2005; Ducklow et al., 2007; Montes-Hugo et al., 2008). The SO is responsible for 1 Pg carbon export per year, making it 40% of the global ocean carbon sink, and diatoms can be responsible for 97% of the chlorophyll increase during local blooms in the SO (Smetacek et al., 2012) and significantly contribute to carbon fluxes in the region (Mikaloff Fletcher et al., 2006; Frölicher et al., 2015; Le Quéré et al., 2016).
Carbon fixation and ultimately carbon export by diatoms, and other phytoplankton, can be limited by the bioavailability of Fe (Martin et al., 1990) as well as other trace metals. Numerous studies from the SO have documented increased diatom growth with the addition of Fe, including large-scale Fe fertilization studies (e.g. de Baar et al., 1990) and shipboard incubation experiments (Hopkinson et al., 2007; Buck et al., 2010; Browning et al., 2021; Burns et al., 2023). Natural sources of Fe include those from shelf sediments, glacial or sea ice inputs, upwelled Fe from winter mixing (Lannuzel et al., 2010; de Jong et al., 2012; Hatta et al., 2013; Annett et al., 2015) and biological fertilization from whales and krill (Nicol et al., 2010). Additionally, manganese (Mn), required for photosynthesis (Morel and Price, 2003), is also found in very low concentrations in the SO. Mn is derived from many of the same sources as Fe (Martin et al., 1990; Middag et al., 2013; Hulten et al., 2017) and may limit phytoplankton growth near the Drake Passage (Browning et al., 2021; Burns et al., 2023). Cobalt (Co), nickel (Ni), copper (Cu), cadmium (Cd), and zinc (Zn) are other essential trace metals required by phytoplankton (Brand et al., 1983; Sunda, 1989; Bruland et al., 1991), but overall Fe is most often the growth-limiting trace metal to phytoplankton because the Fe requirements to support cell growth are higher than for other trace metals (Sunda, 2012; Twining and Baines, 2013).
Most trace metals in the ocean are bound to organic ligands. In the case of Fe, organic complexation increases the concentration of Fe that can remain dissolved in seawater before precipitation but also decreases the bioavailability of that dissolved Fe (dFe) to phytoplankton (Shaked et al., 2020). Notably, nearly all dFe is bound to organic ligands (Gledhill and van den Berg, 1994; Rue and Bruland, 1995; Buck and Bruland, 2007; Gledhill and Buck, 2012). Siderophores are high affinity extracellular dFe organic ligands produced by bacteria and fungi; however, it is generally thought that marine diatoms do not synthesize these secondary metabolites. Recently diatoms have been shown to possess import mechanisms for metal-bound siderophores (Coale et al., 2019). Bacterially-produced siderophores have been observed in the marine environment under both high (e.g., > 2.9 nM) and low (e.g., < 1.5 nM) dFe conditions (Gledhill et al., 2004; Mawji et al., 2008; Boiteau et al., 2016; Bundy et al., 2018). In addition to siderophore production benefiting heterotrophic bacterial growth, siderophores can stabilize dFe pools that in turn benefit the growth of some phytoplankton, including diatoms (Maldonado and Price, 2001; Gerringa et al., 2006; Rijkenberg et al., 2008; Amin et al., 2009; Kazamia et al., 2018; Coale et al., 2019). In addition to Fe, bacterially-produced siderophores can also bind other trace metals including Zn, Mn, Cu and Co (reviewed in Johnstone and Nolan, 2015). Thus, in the trace-metal-limited SO, siderophores could play an important role of establishing cross-kingdom partnerships between bacteria and diatoms, which may assist in surviving the heterogeneous and dilute nutrient environment in the ocean (Amin et al., 2012; Stocker, 2012).
The immediate area surrounding a diatom cell is known as the phycosphere where carbon exudates and oxygen diffuse from high to low gradients near the cell (Bell and Mitchell, 1972). Heterotrophic bacteria are attracted to this area where they can take advantage of the high carbon concentration by living in close association with diatoms (reviewed in Amin et al., 2012). Bacterial taxa commonly associated with diatoms are Proteobacteria and Bacteroidetes (reviewed in Amin et al., 2012; reviewed in Seymour et al., 2017). These taxa are frequently observed dominating the bacterial community during natural diatom blooms (Pinhassi et al., 2004; Teeling et al., 2012). These associations are also species-specific (Ukeles and Bishop, 1975; Kerkhof et al., 1999; Schäfer et al., 2002) and appear to be maintained by the diatom host through production of specific secondary metabolites (Shibl et al., 2020). Commensalisms may occur in which only the bacteria benefits from a diatom-derived food source (Droop, 2007) or antagonisms may occur when bacteria produce algicides (van Tol et al., 2017) or diatoms produce antibiotics (Kellam and Walker, 1989; Grossart, 1999; Desbois et al., 2008). There could also be mutualistic relationships in which bacteria, while growing on organic exudates from diatoms, produce siderophores, increasing the bioavailability of dFe and other trace metal to the diatoms (Amin et al., 2009; Kazamia et al., 2018; Coale et al., 2019). Together, these different interactions have far-reaching consequences for diatom bloom cycles and global carbon flux.
This study examines potential relationships between diatom taxa and bacteria in the diatom size class (> 3 µm) that were present in the WAP along a natural gradient of dFe, macronutrients, and trace metals in the austral spring (September-October). The austral spring in the WAP is a transition time from sea ice retreat to summer phytoplankton bloom and is undersampled with regard to microbial community composition. Previous studies have shown spring phytoplankton communities in the WAP are dominated by diatoms and haptophytes (Arrigo et al., 2017), but potential microbial interactions as a function of biogeochemical gradients have not been previously investigated. Using high-throughput sequencing, diatoms and associated bacterial communities were characterized and network analysis was used to identify positive correlations between diatoms and associated bacteria. This revealed possible mutualisms that provide insights into potential cross-kingdom interactions in this ecosystem with a naturally occurring gradient of trace metals and macronutrients. Temperature, salinity, macronutrients, trace metals, and siderophores were examined as possible environmental drivers of the bacterial and diatom community compositions. We hypothesized that trace metal concentrations, especially dFe, would influence both diatom community composition as well as the composition of the associated bacterial communities.
Sampling occurred in the SO around the WAP and offshore Drake Passage waters. Samples were collected from 21 stations onboard the R/V/I/B Nathaniel B. Palmer in austral spring from September – October 2016 on the cruise NBP16-08 (Figure 1A). Stations 1 and 13 were the same location, sampled 14-days apart. There were six stations in the Drake Passage, two in Palmer Arch, two in the Antarctic Sound, five in the Bransfield Strait, and six in the Gerlache Strait. Stations covered by ice included the two Antarctic Sound stations (Stations 10, 11), Stations 2 and 17 in the Gerlache Strait, and Station 6 in the Bransfield Strait. Water was collected from Niskin bottles on the standard CTD rosette (SBE 32 Carousel Water Sampler, SeaBird Scientific, Bellevue, WA, USA). For samples analyzed for diatom and bacterial community composition, most stations were sampled at 5 m, except for Station 4 which was sampled at 7 m and Stations 10 and 11 which were sampled at 25 m. Temperature was measured with an SBE 3plus 6800M sensor (SeaBird Scientific, Bellevue, WA, USA), and conductivity was measured with an SBE 4 – 02/O (SeaBird Scientific, Bellevue, WA, USA) and converted to salinity. Temperature and salinity at corresponding depths to DNA samples were used as metadata for the sequencing data, and CTD downcast observations binned to 1 m were used for the depth profiles of the stations (Figure S1). Some CTD observations were not recorded: 6 – 24 m from Station 1, 6 – 9 m from Station 17, and 6 – 7 m from Station 20.
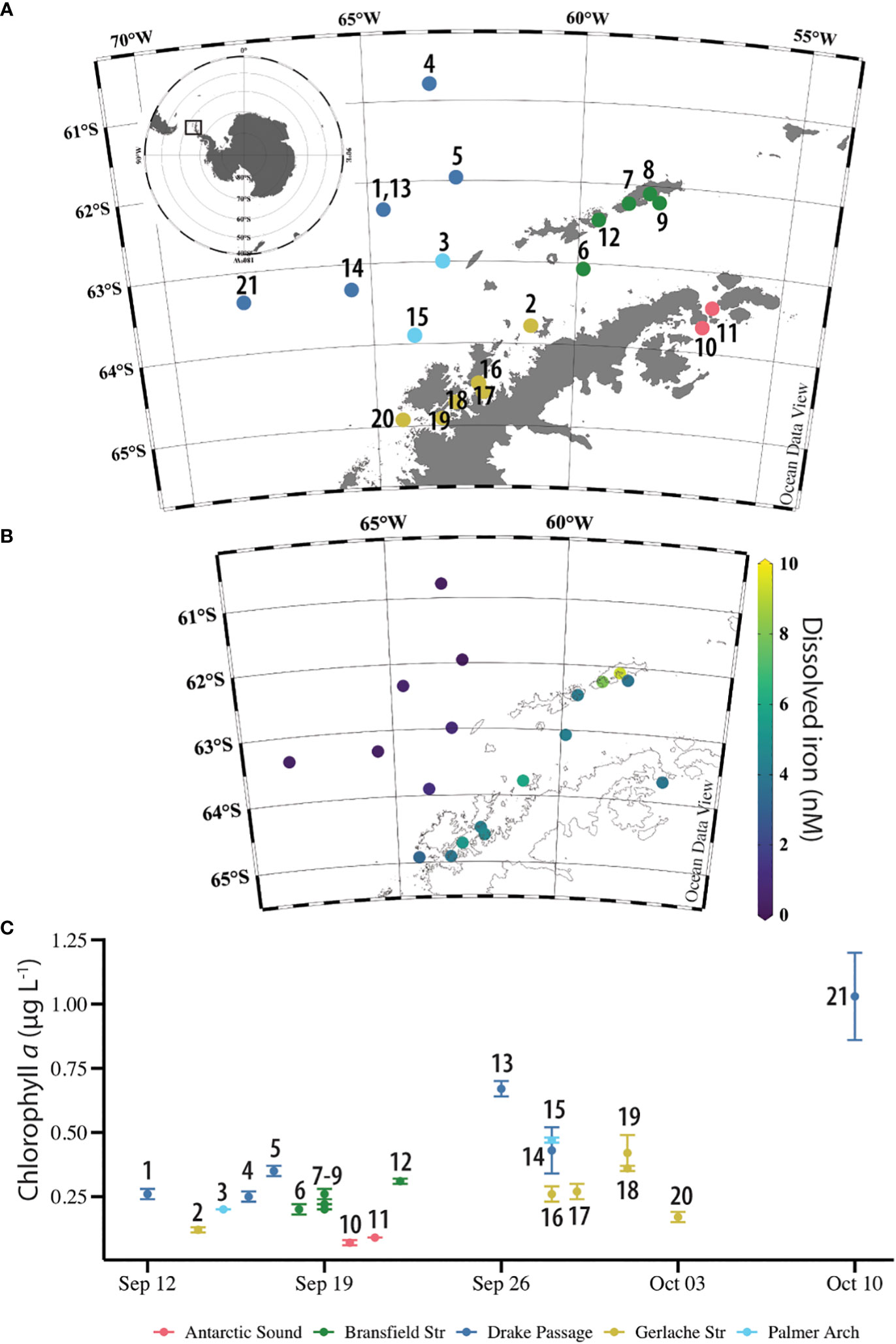
Figure 1 Overview of the sampling location and context of the NBP16-08 stations. (A) Location of 21 stations near the West Antarctic Peninsula. Stations 1 and 13 were the same location, sampled 14-days apart. Stations are labeled with station number and colored by their general location: Drake Passage (dark blue), Palmer Arch (light blue), Bransfield Strait (green), Gerlache Strait (gold), and Antarctic Sound (pink). Plot made in Ocean Data View (Schlitzer 2021). (B) Concentration of dissolved iron (nM) in surface seawater at NBP16-08 stations. There were no surface dissolved iron samples from Station 11. Plot made in Ocean Data View (Schlitzer 2021). (C) Concentration of chlorophyll a (µg L-1) in surface seawater samples at NBP16-08 stations across time and labeled with station number. Stations 10 and 11 were sampled at 25 m. Error bars are the standard deviation of n = 3 replicate averages, except for Station 1 with n = 2. Stations are labeled with station number and colored by their general location: Drake Passage (dark blue), Palmer Arch (light blue), Bransfield Strait (green), Gerlache Strait (gold), and Antarctic Sound (pink).
Macronutrient, trace metal, and siderophore samples were taken from a trace metal clean (TMC) CTD at depths of 20 – 33 m with an average sampling depth of 25 m at all stations for the shallowest surface samples, except for Station 11 which had the shallowest sample at 300 m and was therefore not included in surface sample analysis. The TMC CTD (SBE32 rosette system, Sea-Bird Scientific, Bellevue, WA, USA) was equipped with a Kevlar line and X-Niskin sampler bottles (OceanTestEquipment, Inc., Fort Lauderdale, FL, USA) (Cutter and Bruland, 2012). In the cases where our conventional and TMC CTD sample depths were mismatched, we assumed the concentrations of dissolved trace metals to be uniform in upper 30 m, consistent with more detailed mixed layer studies conducted previously (Figure S1; Hatta et al., 2013; Measures et al., 2013).
Concentrations of chlorophyll a, a proxy for total photosynthetic biomass, were measured by filtering seawater collected from the same depths sampled for DNA sequencing. Seawater (100 mL) was passed through glass-fiber filters (GF/Fs) for 0.6 – 0.8 um particle retention (Cytiva, Marlborough, MA, USA) using a vacuum pump at low pressure of less than -15 in-Hg. Samples were collected in triplicate, except for Station 1 at 5 m which had duplicates. GF/Fs were extracted in 95% ethanol at room temperature in the dark for 12 – 24 hours. Samples were read while on the ship using a 10AU fluorometer (Turner Designs, Inc., San Jose, CA, USA) as both non-acidified and acidified with 10% hydrochloric acid. The 10AU fluorometer was calibrated with a commercially available standard of Anacystis nidulans algae chlorophyll a (Sigma-Aldrich, Inc., St. Louis, MO) Chappell et al. (2022).
For dissolved macronutrient analysis, water samples were collected from the TMC CTD and then filtered through 0.2 µm AcroPak™ membrane filter capsules (Pall Corporation, Port Washington, NY, USA). Filtered water was stored in acid-cleaned polypropylene (Falcon) tubes in the dark at 4°C until analyzed shipboard; a subset of samples was frozen at -20°C for additional shore-based analysis. Samples for nitrate + nitrite, soluble reactive phosphorus (SRP, ‘phosphate’), and dissolved inorganic silicic acid (dSi) were analyzed within 24-hrs of collection shipboard using a Lachat 8500 QuickChem (Hach, Loveland, CO, USA). Upon return from the cruise, nitrate + nitrite, ammonium, SRP, and dSi were analyzed using a Technicon AutoAnalyzer (AA) II (SEAL Analytical Inc., Mequon, WI, USA). Dissolved nitrate + nitrite and SRP were measured with both Lachat 8500 QuickChem and Technicon AAII and averaged. A Lachat 8500 QuickChem was used to measure dSi, with five stations having replicate measurements on the Technicon AAII. Dissolved inorganic nitrogen (DIN) is reported as the summation of nitrite + nitrate and ammonium. Only eight stations had detectable ammonium measurements, contributing 0 – 1% of the total DIN measured.
Trace metal clean techniques were used for the collection, processing, and analysis of all trace metal samples (Cutter et al., 2017). Water samples for dissolved trace metals were collected from the TMC CTD, gravity filtered through 0.2 µm AcroPak™ membrane filter capsules (Pall Corporation, Port Washington, NY, USA) into acid-cleaned low density polyethylene bottles, acidified to pH 1.8 by adding 0.024 M HCl (Optima, Thermo Fisher Scientific, Waltham, MA, USA), and stored at room temperature until analyzed. The following dissolved trace metals were measured: dFe, dissolved Mn (dMn), dissolved Co (dCo), dissolved Ni (dNi), dissolved Cu (dCu), dissolved Cd (dCd), and dissolved Zn (dZn) dissolved Pb (dPb). All sample analyses were supported by analysis of GEOTRACES and SAFe reference samples (Buck et al., 2018; Buck et al., 2019). Extraction and pre-concentration of dissolved metals were done using a seaFAST-pico system (Elemental Scientific, Omaha, NE, USA), and eluents were analyzed on a magnetic sector Element XR High Resolution inductively coupled plasma mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) as detailed previously (Hollister et al., 2020; Burns et al., 2023). Macronutrient and trace metal data used in this analysis and additional methods information is available from NSF Biological & Chemical Oceanography Data Management Office (BCO-DMO) (Buck et al., 2018; Buck et al., 2019; Burns et al., 2023).
Concentrations of solid-phase extractable organic dFe-binding ligands were collected at each station from the TMC rosette, and were concentrated by slowly pumping (18 mL min-1) 10 L of filtered (0.2 µm) seawater onto solid phase extraction columns (Bond Elut ENV, Agilent Technologies, Santa Clara, CA, USA). Solid phase extraction columns were cleaned with HCl-acidified (Optima, Thermo Fisher Scientific, Waltham, MA, USA) MilliQ water (pH 2) prior to use, and then flushed with three column volumes of clean MilliQ before being activated with methanol (Optima, Thermo Fisher Scientific, Waltham, MA, USA) and rinsed again with MilliQ (Bundy et al., 2018). After seawater was pumped onto the columns, each column was flushed with three column volumes of MilliQ before being stored at -20°C until analysis. Prior to analysis, each column was thawed and eluted with 12 - 14 mL of distilled methanol into TMC 15 mL tubes. Methanol eluents were dried down to approximately 500 µL on a Speed-Vac (Thermo Fisher Scientific, Waltham, MA, USA). Samples were then analyzed via liquid chromatography coupled to inductively-coupled plasma mass spectrometry and electrospray ionization mass spectrometry (LC-ICP/ESI-MS) as described in detail elsewhere (Bundy et al., 2018). For the purposes of this work, only data collected from the LC-ICP-MS step have been included, and this data represents the total concentration of solid-phase extractable Fe-binding compounds in each sample. These total concentrations represent concentrations of total siderophores for Fe.
For DNA sequencing of the diatom and bacterial communities, an average of 3.2 L of surface seawater was passed across a 3 µm, 25 mm polyester membrane filter (Sterlitech, Kent, WA, USA) using a peristaltic pump (phytoplankton size fraction, associated with diatoms). The filtered biomass samples were placed in 400 µL of AP1 buffer (DNeasy Plant Mini kit, Qiagen, Germantown, MD, USA) to minimize DNA degradation before extraction, flash frozen in liquid nitrogen, transported on dry ice to the lab, and stored at –80°C until extraction. A modified version of the DNeasy Plant Mini kit protocol (Qiagen, Germantown, MD, USA) was used to extract the DNA of the diatoms and associated bacteria captured on the filters. The modified version included an additional 1-minute bead beating step and use of a QIA-Shredder column (Qiagen, Germantown, MD, USA) with a final elution in 100 µL: RNase free water (Chappell et al., 2019). A Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to assess the extracted DNA for quality and quantity, which determined there was an average of 1.7 ng µL-1. A negative control for sequencing was prepared by extracting a blank 25 mm 3.0 µm polyester membrane filter. Positive controls for bacterial communities were prepared by extracting a commercially available mock community of microbial cells called the ZymoBIOMICS Microbial Community Standard (Zymo Research, Irvine, CA, USA) on a 25 mm 3.0 µm polyester membrane filter as well as using a standard of pre-prepared DNA from microbial cells of the ZymoBIOMICS Microbial Community DNA Standard (Zymo Research, Irvine, CA, USA) for the following PCR steps.
Associated bacteria were examined with primers targeting the V4 region of the 16S SSU rDNA gene using the 515F forward primer: 5’-GTGYCAGCMGCCGCGGTAA-3’ and 806R reverse primer: 5’-GGACTACNVGGGTWTCTAAT-3’ (Apprill et al., 2015; Parada et al., 2016). With the Illumina adapters added, the following full-length primers (Integrated DNA Technologies, Inc., Coralville, Iowa) were used: forward primer 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA-3’ and reverse 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT-3’. Each PCR reaction contained 2 – 8 ng of extracted DNA, 0.2 µM of each primer, 1x Bioline BIO-X-ACT Short Mix (Bioline USA Inc., Taunton, MA, USA), and DEPC-treated water for a total volume of 25 µL. The PCR cycle conditions were based on methods from the Earth Microbiome Project data site (Gilbert et al., 2010), which included an initial denaturation of 3 minutes at 94.0°C, 35 cycles of the following: denaturation for 45 seconds at 94.0°C, annealing for 1 minute at 50.0°C, elongation for 1.5 minutes at 72.0°C, and a final extension of 10 minutes at 72.0°C. The diatom-specific primers targeted the V4 region of the 18S rRNA gene: D512F forward primer: 5’-ATTCCAGCTCCAATAGCG-3’ and D978R reverse primer: 5’-GACTACGATGGTATCTAATC-3’ (Zimmermann et al., 2011). Primers with Illumina adapters were: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGATTCCAGCTCCAATAGCG-3’ and 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACGATGGTATCTAATC-3’ (Chappell et al., 2019). Each PCR reaction contained 2 – 8 ng of extracted DNA, 0.5 µM of forward and reverse primers, 1x Bioline BIO-X-ACT Short Mix, and DEPC-treated water for a total volume of 25 µL. The PCR cycle consisted of an initial denaturation step of 5 minutes at 95.0°C, 32 cycles of the following: denaturation for 1 minute at 95.0°C, annealing for 1 minute at 61.8°C, and extension for 30 seconds at 72.0°C, and a final extension for 10 minutes at 72.0°C. All PCR products were visualized on a 1% agarose gel to confirm amplification of desired sized products. These PCR products were cleaned with KAPA Pure Beads (KAPA Biosystems, Woburn, MA, USA) and visualized again on an agarose gel.
Libraries for both bacterial and diatom amplicons for Illumina sequencing were prepared by the RI Genomics and Sequencing Center (Kingston, RI, USA) by performing a second round of 8-cycle PCR with 50 ng DNA template to attach Nextera XT indices and adapters from the Illumina Nextera XT Index Kit (Illumina, Inc., San Diego, CA, USA) with Phusion High Fidelity PCR Mastermix (Thermo Fisher Scientific, Waltham, MA, USA). Again, PCR products were cleaned with KAPA Pure Beads and visualized on an agarose gel. Quality checks were performed by running certain samples on a BioAnalyzer DNA1000 chip (Agilent Technologies, Inc., Santa Clara, CA, USA). All samples were quantified using a Qubit fluorometer (Invitrogen, Waltham, MA, USA), pooled, and then quantified using qPCR in a LightCycler480 (F. Hoffman -La Roche AG aka Roche, Basel, Switzerland) with the Illumina Kit (KAPA Biosystems, Woburn, MA, USA). Samples were sequenced on the same paired-end 2x250 base pair (bp) MiSeq run using the 500-cycle MiSeq Reagent Kit (Illumina, Inc., San Diego, CA).
After sequencing, samples were demultiplexed, and bacteria and diatom reads were separated by primer sequences. Primers were trimmed from both ends of sequences with Cutadapt v1.15 (Martin, 2011). Reads without either primer sequence or less than one bp after trimming were discarded. Trimmed sequences were inputted into DADA2 v1.16 to determine amplicon sequence variants (ASVs; Callahan et al., 2016). For the 16S reads, the truncation length used was 230 bp for forward reads and 160 bp for reverse, decided based on quality scores of the raw reads, with maxN = 0, maxEE = 2 for both directions, truncQ = 2, and rm.phix as true. Only ASVs that were 253 bp in length were retained for analysis, in order to mitigate compounding differences due to read length. ASVs were assigned taxonomy using the naïve Bayes classifier (Wang et al., 2007) within DADA2, using the Silva SSU NR v138 database trimmed to the same 16S V4 region as the primer sequences using Cutadapt (Quast et al., 2013; Yilmaz et al., 2014; Glöckner et al., 2017). A 100% match of an ASV to the Silva database was required to be called a species in the DADA2 species classifier. Matching 100% to an ASV has been shown to be most reliable way of calling species (Edgar, 2018). After assigning taxonomy to the bacterial 16S V4 reads, only reads belonging to Kingdom Bacteria were retained, and any reads identified as chloroplast or mitochondria were removed. The diatom 18S V4 ASVs were created similarly in DADA2 with truncation length of 230 bp for forward reads and 220 bp for reverse reads, and other settings as reported above. Only ASVs that were 390 – 410 bp in length were retained following expectations for that region (Zimmermann et al., 2011). Taxonomy was assigned using the database pr2 v4.12.0 for 18S sequences, which was also trimmed using Cutadapt to the region of interest (Guillou et al., 2013; del Campo et al., 2018). Only ASVs belonging to Class Bacillariophyta were retained. Stations 1, 12, and 13 had triplicate filters sequenced, and a random picker was used to only continue analysis with one of the triplicates to prevent bias towards those three stations. In total, there were 1,027 bacteria ASVs and 124 diatom ASVs used in subsequent analysis.
Statistical analyses were performed in R v4.0.2 (R Core, T 2020) in R Studio v1.1.456 (RStudio, T, 2016) using the vegan v2.5.6 package (Dixon, 2003) for the test for normality, comparison of means, pairwise comparison between groups, tests of group dispersions, and analysis of similarity (ANOSIM) and ggplot2 v3.3.3 for visualizations (Wickham, 2016). Statistical comparisons of means of environmental parameters by station locations (Drake Passage, Palmer Arch, Bransfield Straight, Gerlache Straight, Antarctic Sound) including temperature, salinity, chlorophyll a concentrations, macronutrients, and trace metals were first assessed for normality with the Shapiro-Wilk test using shapiro.test() in R. If the data distribution was normal, an ANOVA was used by aov() in R, followed by a Tukey Test for comparison of groups with TukeyHSD() in R. If the data distribution was not normal, the non-parametric Kruskal-Wallis test was used with Kruskal.test() followed by a Wilcoxon Test for pairwise comparisons of groups with pairwise.wilcox.test() with a Bonferroni correction (p.adjust.method = “BH”). Only comparisons with an adjusted p value of less than 0.05 are shown. For macronutrients and trace metal analysis, Antarctic Sound was excluded since one of the two stations did not have that corresponding data. The same sample grouping by station locations was tested for group dispersions using betadisper() and permutest() in vegan on the Bray-Curtis distance matrix of the relative abundance of the bacteria and diatom ASVs produced by phyloseq using distance(method = “bray”). ANOSIM was performed using anosim() in vegan. Both were run with 999 permutations and an alpha value of 0.05. NMDS plots were made in phyloseq v1.32.0 using default parameters (McMurdie and Holmes, 2013), with stress values equal to or below 0.1 being acceptable for interpretation.
The function bioenv() in vegan was used to determine the best model formula to explain the variation in the Bray-Curtis distance of the relative abundance of the ASVs using the scaled variables, spearman correlations, and Euclidean metric. This analysis uses multivariate statistics to link BIOlogical (i.e. ASVs) to ENVironmental variables, hence it is referred to as BIOENV. Samples from Station 11 in the Antarctic Sound were excluded since they had no corresponding environmental metadata. The following parameters were inputted: temperature, salinity, DIN, SRP, dSi, dFe, dMn, dCo, dNi, dCu, dCd, dZn, and dPb. A second analysis was done including the aforementioned variables along with siderophore concentrations for stations where siderophores were measured (all stations but 7, 11, 14, 17, and 18). For both the diatom and bacteria ASV data sets, the selected BIOENV model parameters were used in the formula for canonical analysis of principal coordinates (CAP) ordination, used with method = “CAP” within the ordinate function of phyloseq, along with the Bray-Curtis distance matrix of the relative abundance of the ASVs (McMurdie and Holmes, 2013).
Relationships between diatom and bacteria ASVs were inferred using sparse inverse covariance estimation for ecological association inference (SPIEC-EASI), which accounts for the compositional nature of amplicon data and the high number of ASVs present in a typically low number of samples (Kurtz et al., 2015). SPIEC-EASI networks were performed within the R package spiec easi v1.1.0 (Kurtz et al., 2015), which performs acceptable data transformation of compositional data sets like ASVs using the centered log-ratio with unit pseudo-count to account for many zero counts. It then estimates relationships between ASVs using neighborhood selection, also known as the MB method (Meinshausen and Bühlmann, 2006), and selects the final network model by using random subsampling by Stability Approach to Regularization Selection or StARS (Liu et al., 2010). The nodes are the ASVs, and connections suggest a linear relationship. A positive linear relationship may indicate a possible mutualism between two ASVs (Steele et al., 2011). Unconnected nodes are likely ASVs which are independent from each other. The predictive power and reproducibility of the network is assessed with a desired stability threshold of 0.05. Networks were run as two groups: 1.) Drake Passage and 2.) Gerlache Strait and Bransfield Strait stations. A subset of the total ASV dataset was selected for a more simple viewing of the networks and to minimize heavy computational load required for more ASVs, a strategy employed by other studies (Kurtz et al., 2015; de Sousa et al., 2019). The top ca. 10% of bacteria ASVs (100 ASVs) and the top ca. 20% of diatom ASVs (25 ASVs) by relative abundance were used. For the Gerlache Strait and Bransfield Strait network, the lambda minimum ratio was set to 1-2, the nlambda was set to 200, and 50 replicates were run. These same settings were used for another network of all 124 diatom ASVs across all stations, and for the network of the 292 Gammaproteobacteria ASVs across all stations the nlambda was lowered to 20. Because the Drake Passage network only had six samples, the parameters were adjusted to a lambda minimum ratio of 1-1, nlambda of 10, and 5 replicates with a subsample ratio of 0.5. The R package igraph v1.2.6 was used to visualize the networks (Csárdi and Nepusz, 2006).
The NBP16-08 cruise sampled a naturally occurring gradient of nutrients and trace metals. Dissolved Fe concentrations near the continent ranged from 3.27 – 9.28 nM and were lower in the offshore Drake Passage waters (0.13 – 0.55 nM) (Figure 1B and Table 1). There was a statistically significant difference between the dFe concentrations in Drake Passage stations and in the Bransfield Strait and Gerlache Strait stations (Table 2). Other dissolved trace metal concentrations were also significantly lower in the Drake Passage compared to the Bransfield Strait and Gerlache Strait regions, including dMn, dCo, dCu, and dZn (Table 2). For dMn, the Palmer Arch stations were more similar to the Drake Passage Stations than the Bransfield Strait stations, with macronutrients (DIN, SRP, and dSi) following the same pattern of lower concentrations offshore (Table 2). The dFe to macronutrient ratios (dFe : DIN, dFe : SRP, dFe: dSi) were also significantly higher in the Bransfield Strait and Gerlache Strait than in the Drake Passage. The Palmer Arch dFe: dSi was significantly different from the Bransfield Strait (Table 2). There was no significant difference in dNi, dPb, or siderophore concentrations between Drake Passage and the more nearshore stations (Table 2). Siderophore concentrations ranged from 0.1 – 7.2 pM, with Drake Passage Station 4 being the highest followed by the Gerlache Strait Station 19 as the second highest (Table 1). Overall, there was no apparent geographic pattern in siderophore concentration distributions. For example, siderophore concentrations did not correlate with dFe or other nutrients, and they did not have any clear onshore to offshore trends. They also varied in concentration with repeat occupations of the same sampling location, such as in the Drake Passage (Table 1).
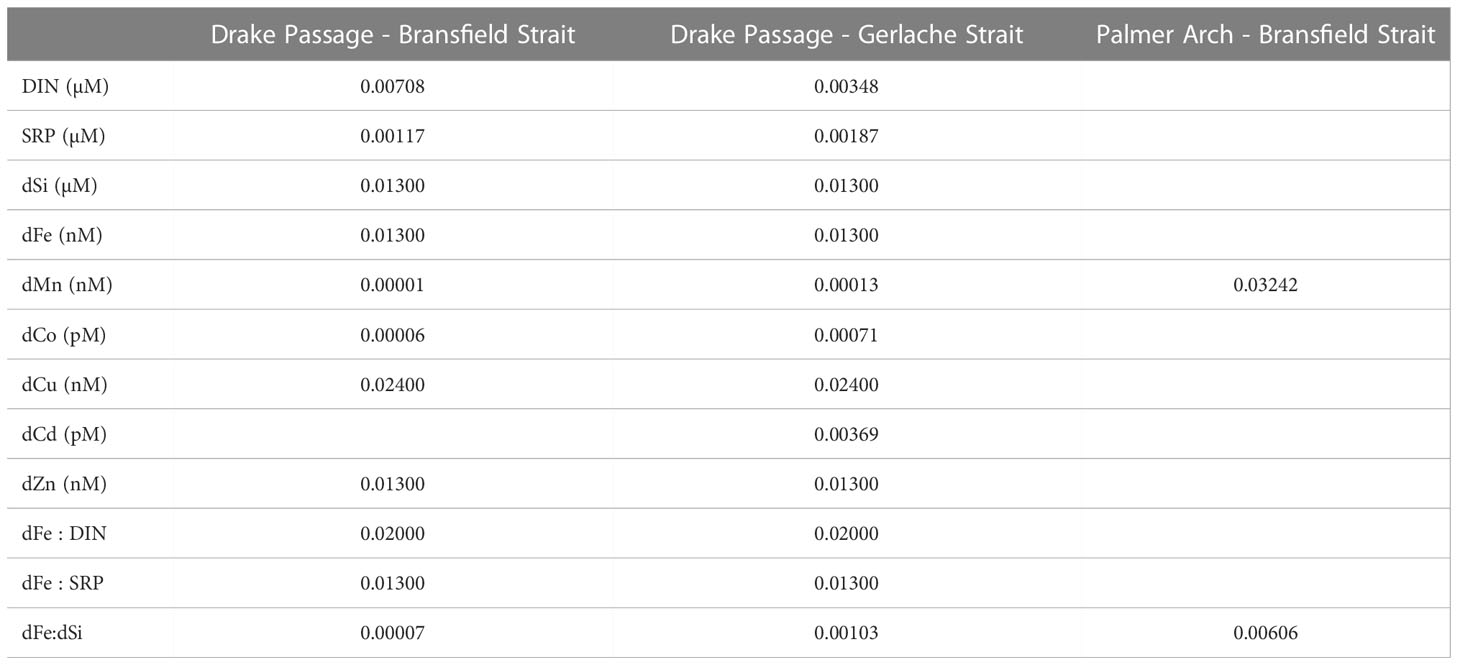
Table 2 Adjusted p-values of pairwise comparisons between groups that were < 0.05 for the environmental parameters including dissolved inorganic nitrogen (DIN), soluble reactive phosphorus (SRP), dissolved silicic acid (dSi), dissolved iron (dFe), dissolved manganese (dMn), dissolved cobalt (dCo), dissolved copper (dCu), dissolved cadmium (dCd), and dissolved zinc (dZn).
Throughout the cruise, low concentrations of chlorophyll a were observed which slightly increased with time over the duration of the cruise (Figure 1C) and did not change with depth (Figure S1A). There was no significant difference in chlorophyll a across sampling locations (Kruskal-Wallis, P = 0.067). The average chlorophyll a concentration across all stations was 0.31 ± 0.21 µg L-1. At the Drake Passage stations, chlorophyll a ranged from 0.25 µg L-1 to 1.03 µg L-1 (Table 1). Surface water temperature stayed within a three-degree range during the cruise from -1.8 to 1.6°C and was not significantly different between station location. The warmest surface temperature (1.6°C) was recorded at 25 m at Station 10 in the Antarctic Sound. From the CTD downcast, it was observed that there was a 3°C increase at 25 m at this station compared to its other depths which was unlike the other Antarctic Sound station (Table 1 and Figure S1B).
There was a significant difference in bacterial community composition by sampling location (Antarctic Sound, Bransfield Straight, Gerlache Straight, Drake Passage, and Palmer Arch; p = 0.001) as indicated by ANOSIM and visualized by the NMDS plot (Figure S2; NMDS stress = 0.1053). For the bacterial communities, there were no significant differences in group dispersions for sampling location (p = 0.19, df = 19), indicating results from ANOSIM are reliable. Similar to the geographic separation in bacterial community composition data, ANOSIM also indicated a significant difference in diatom community composition by station location (p = 0.001) as displayed by the NMDS (Figure S2B; NMDS stress = 0.0931). For the diatom communities, there were also no significant differences in the group dispersions for sampling location (p = 0.56, df = 19). Interestingly, the diatom assemblage at Station 2 in the Gerlache Strait was most similar to the offshore assemblages (Figure S2B). This was unlike the bacterial community at Station 2 which was most similar to the other Gerlache Strait samples (Figure S2A).
The significant separation in bacterial and diatom communities by sampling location indicated potential underlying environmental differences in these locations, which were further explored in the BIOENV and CAP analyses. For the diatom ASVs, BIOENV determined that temperature, SRP, dSi, dMn, dCu, dZn, and dPb, with a correlation of 0.7591, were appropriate to use for subsequent analysis. When BIOENV was re-run to include siderophore concentrations in the parameters assessed, siderophores did not influence the model. Thus, we chose to use the model that included 20 stations in further analysis of the diatom communities. The bacteria ASVs were analyzed separately, and it was determined that SRP, dMn, dCo, and dCu, with a correlation of 0.7644, were appropriate to use. For the same approach to diatom analysis, the model that included 20 stations of bacteria ASVs was used in subsequent analysis.
The environmental variables resulting from BIOENV analysis, which were used in the CAP analysis, are included on the CAP plot to visualize the major contributing variables to each dimension (Figure 2). For the bacterial communities, 37.9% of variation was accounted for by Dimensions 1 and 2, and samples grouped by location (Figure 2A). For the diatom communities, Dimensions 1 and 2 combined on the CAP plot accounted for 57.9% of variation, and samples also grouped by location (Figure 2B). As observed in the NMDS analysis, the diatom assemblage from the Gerlache Strait grouped closer with the Drake Passage samples (Figure 2B). The diatom community at Station 10 in the Antarctic Sound was influenced by temperature and was separate from the other stations (Figure 2B). For both diatom and bacterial communities, changes in dMn, dCu, and SRP were important contributors to Dimension 1, driving the distinct grouping of the near continent stations (Figure 2). For diatoms, Dimension 1 was also driven by dPb changes, especially in the offshore stations (Figure 2B), in contrast to bacterial communities where BIOENV analysis deemed dPb unimportant. Additionally, dSi and dZn contributed to the variance in the diatom community analysis, especially for samples from the Bransfield and Gerlache Straits (Figure 2B). The dCo changes contributed to Dimension 1 for the bacterial communities, especially in the variance of the Gerlache Strait samples, but did not contribute to the diatom community analysis (Figure 2).
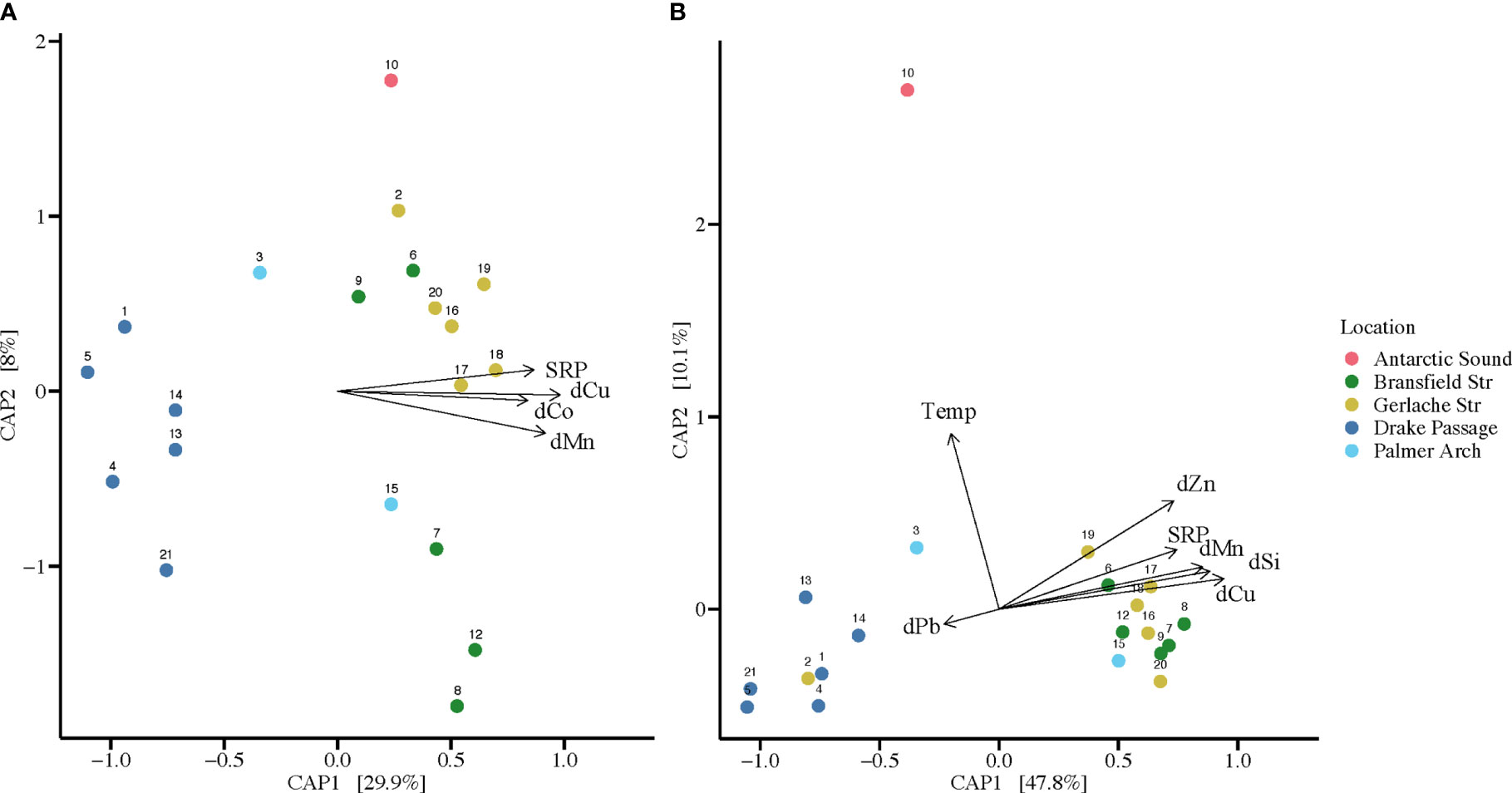
Figure 2 Canonical analysis of principal coordinates (CAP) using significant environmental parameters from the BIOENV test compared to taxonomic composition of communities per location from high-throughput sequencing. Environmental parameters from surface seawater shown include: soluble reactive phosphorus (SRP), dissolved silicic acid (dSi), dissolved manganese (dMn), dissolved cobalt (dCo), dissolved copper (dCu), dissolved zinc (dZn), dissolved lead (dPb), and water temperature (Temp). Colors of the points correspond to the sampling locations (n = 20). Station 11 is not included since it did not have corresponding environmental parameters taken. (A) CAP plot of bacterial communities at each location sampled with vectors of environmental parameters. Dimensions 1 and 2 account for 37.9% of the variation. (B) CAP plot of diatom communities at each sampling location with vectors of environmental parameters. Dimensions 1 and 2 account for 57.9% of the variation.
The average number of sequences initially yielded from the sequencer was similar between the environmental samples (n = 27; 52381 ± 10706) and mock community samples (n = 2; 51011 ± 3154) for the bacteria 16S sequencing with the negative controls having fewer reads (n = 2; 1930 ± 886). After the quality control steps in DADA2 (filtering, denoising, and removing chimeras), there was an average of 42556 ± 8608 reads in the environmental samples (n = 27), 27749 ± 867 reads in mock community samples (n = 2), and 1564 ± 655 reads in the negative controls (n = 2). After processing into ASVs, the mock community samples had far fewer average sequences (n = 2; 27749 ± 867) than the environmental samples (n = 27; 41499 ± 8326) while the negative controls had the least number of sequences similar to their starting average (n = 2; 1531 ± 643). In the bacterial positive controls, the ASVs recovered matched the genera present in the mock communities. There were a smaller number of ASVs present in the positive controls which did not match the starting genera and instead matched a Gammaproteobacterium and chloroplast origin. From the environmental samples amplified with the diatom 18S primers, there were about half as many as raw sequences (n = 27; 24642 ± 5512) resulting in 19521 ± 4605 reads after quality control in DADA2. After ASV processing, there was an average of 19378 ± 4575 sequences per sample (n = 27).
Diatoms were the prevalent phytoplankton taxa across all stations as visually inspected by light microscopy and FlowCam fluid imaging (Figure S3). Diatom ASVs recovered from high-throughput sequencing were grouped together by genus (Figure 3). There were some diatom genera that were found at most stations and considered cosmopolitan taxa within our sampling region and some genera that were only found in certain locations (Figure 3). Diatoms observed included all major classifications of raphid pennate, araphid pennate, radial centric and multipolar centric diatoms. Centric diatoms in the Thalassiosira genus and the raphid pennate diatoms in the Fragilariopsis genus were the most well-represented diatoms by the relative abundance of 18S rDNA at every station (Figure 3). Fragilariopsis was best represented by the relative abundance of the 18S rDNA in the nearshore stations of Bransfield and Gerlache Straits and one Palmer Arch station (Figure 3). Other diatom genera present in the 18S rDNA data at every station included Porosira, Stellarima, Chaetoceros and Corethron (Figure 3). Shionodiscus, Asteroplanus, and Actinocyclus were present in nearly every station as well (Figure 3). The Drake Passage stations were represented by communities that included Thalassiothrix, Pseudo-nitzschia, Hemiaulus, Rhizosolenia, Minidiscus, and Asteromphalus (Figure 3). Many of these taxa in the Drake Passage stations were also present in stations in the Gerlache Strait. Notably, there were diatom ASVs which could not be identified at the genus level, especially one ASV that likely belongs to a raphid pennate diatom and represents a portion of the % relative abundance ranging from 1.2 – 12.8% with an average of 6.9% at the offshore stations (“Raphid pennate ASV2” in Figure 3).
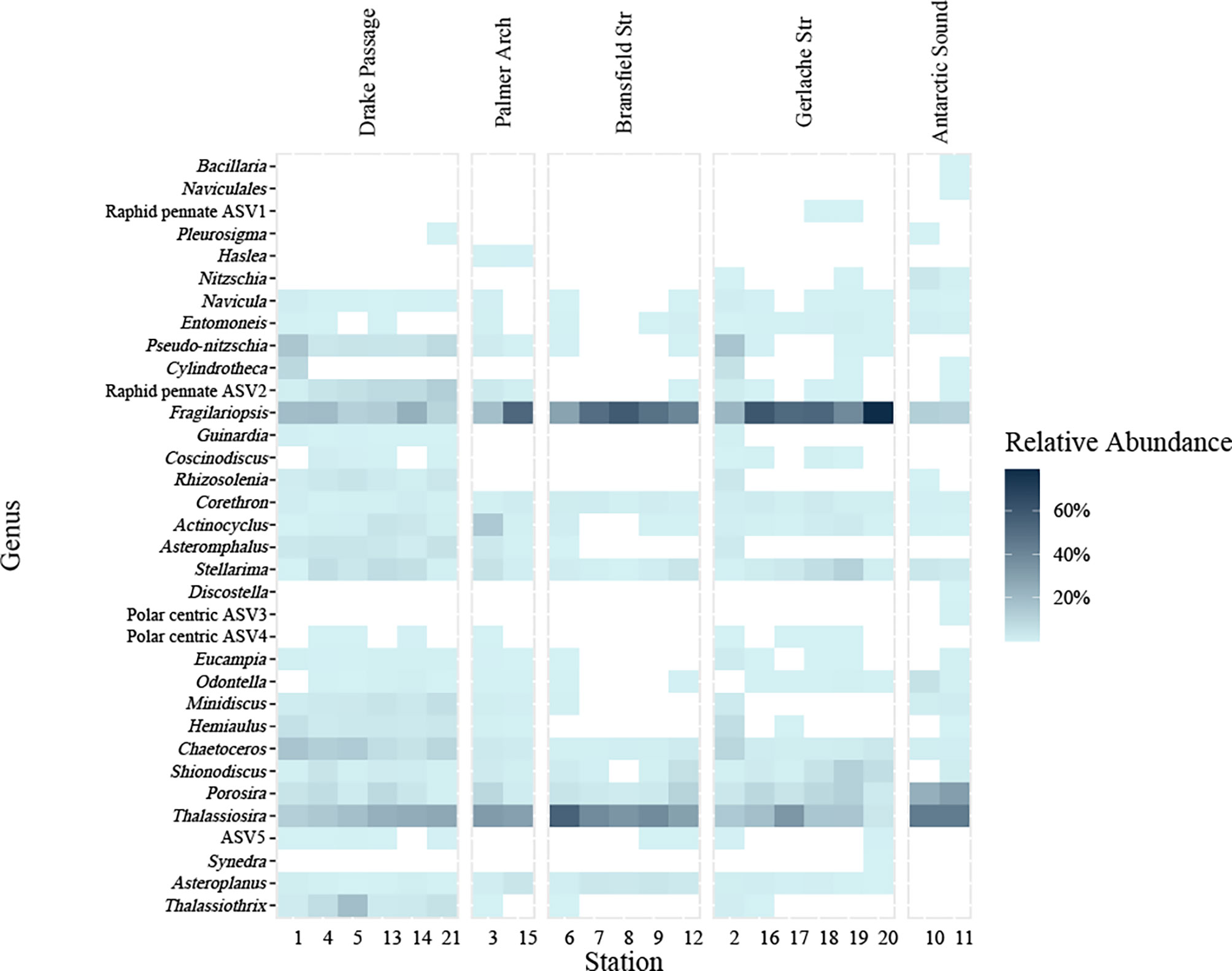
Figure 3 Diatom genera present at stations (bottom) sampled grouped by general location (top) from NBP16-08 from % relative abundance of the sequencing reads of the 18S V4 rDNA barcoding from the total sequencing reads identified as diatoms. A white box denotes that the genus was absent. Amplicon sequence variants (ASVs) that were identified as the same genus are grouped together. Any ASVs not identified to the genus level were retained separately and labeled as their lowest taxonomic assignment. Genera are ordered by family, with raphid pennates at the top from Bacillaria spp. – Fragilariopsis spp. including two unknown genera, radial centric basal Coscinodiscophyceae from Guinardia spp. – Stellarima spp., polar centric Mediophyceae from Discostella spp. – Thalassiosira spp. including two unknown genera, and lastly araphid pennates from Synedra spp. – Thalassiothrix spp. One ASV labeled as ASV5 was not identified at the family level (above Syndedra).
Known groups of diatom-associated bacterial taxa were well represented in terms of relative 16S rDNA abundance in the associated bacterial communities, including Proteobacteria and Bacteroidetes (Amin et al., 2012). Across all stations in the phytoplankton size class, Proteobacteria were the most well-represented by relative abundance of the 16S rDNA gene along with Bacteriodota (Figure 4A). Within the Proteobacteria group, the class Gammaproteobacteria dominated the relative abundance while Alphaproteobacteria were also present (Figure 4B). There were several orders of Gammaproteobacteria that co-occurred at all stations, including Alteromonadales, Cellvibrionales, Oceanospirillales, and Thiomicrospirales (Figure 4C). Also represented were members of Bdellovibrionota, Planctomycetota, and Verrucomicrobiota. ASVs of Bdellovibrionota occurred with more frequency in the samples near landmasses (Figure 4A).
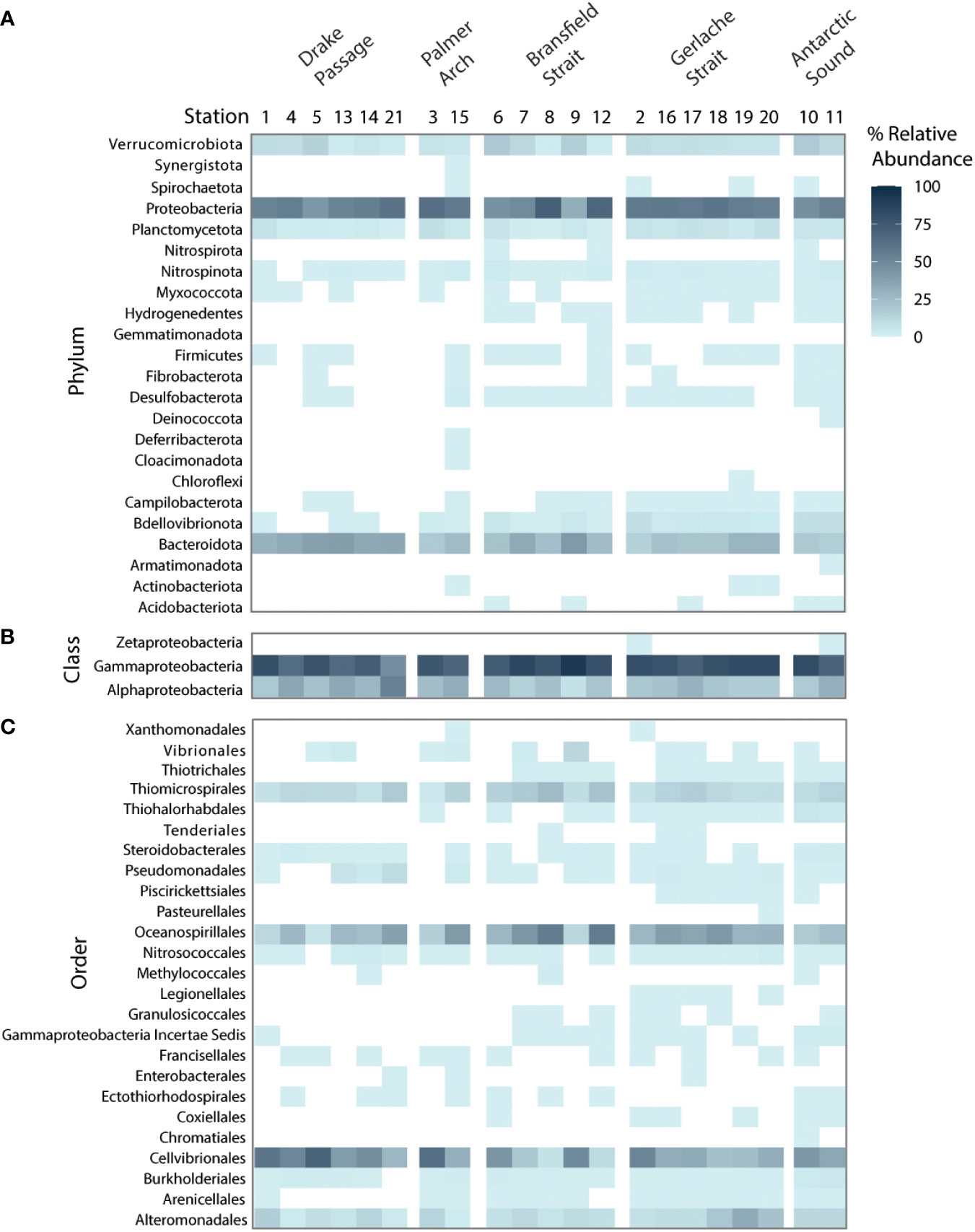
Figure 4 Bacterial taxa recovered from high-throughput sequencing of the V4 16S rDNA at the NBP16-08 stations (n = 21) grouped by sampling location by the % relative abundance of the 16S rDNA gene. A white box denotes absence in a sample. (A) The phyla of all bacterial amplicon sequence variants (ASVs) identified. (B) The classes of Proteobacteria identified. (C) The orders within Gammaproteobacteria identified.
SPIEC-EASI (Kurtz et al., 2015) network analysis of the potential relationships between the diatom taxa and associated bacteria included six stations in the Drake Passage in one network and six stations each from the Gerlache and Bransfield Straits in the other network (Figure 5). In the diatom-bacteria network of the Drake Passage stations, there were 61 positive edge connections (network stability = 0.05) with nine interconnected clusters (subnetworks) that contained both diatoms and bacteria in cross-kingdom connections (Figure 5A, subnetworks #1 - 9). One subnetwork had 14 taxa interconnected but most had six or fewer connected (Figure 5A). In the largest subnetwork (#6) of the 14 connected ASVs, the diatom Chaetoceros debilis acted as a hub connected to members from Bacteroidia NS9, Cryomorphaceae NS10, and Alphaproteobacteria OCS116, while there was a single connection between the diatom Thalassiothrix longissima to a bacteriodetes NS9 marine group member (Figure 5A). In subnetwork #8, Thalassiosira minima was connected to the gammaproteobacterium Paraglaciecola sp. and the Planctomycetes Blastopirellula sp. (Figure 5A). Small subnetwork #9 had Thalassiosira tumida connected to a gammaproteobacterium Pseudohongiella sp. and an alphaproteobacterium Ascidiaceihabitans sp. while also connected to another diatom Shionodiscus ritscheri (Figure 5A). Two very small subnetworks contained Thalassiosira sp., one with it connected to Bacteroidia Lewinella sp. (#7) and another with it connected to NS6 and a member of Nitrincolaceae (#1, Figure 5A). Other diatoms were also connected to members of Bacteroidia, including Hemiaulus sinensis to an NS9 member and Polaribacter sp. (#2, Figure 5A). In subnetwork #5, Chaetoceros neogracilis was connected to Bacteroidia Fluviicola sp. and Cylindrotheca closterium was connected to the gammaproteobacterium Alteromonas australica (Figure 5A). The diatom Asteromphalus sp. was connected to a member of Magnetospiraceae in #4 and in subnetwork #3, the diatom Actinocyclus curvatulus was connected to a member of NS9 and the diatom Porosira glacialis was connected to a Nitrospinia member of LS-NOB (Figure 5A).
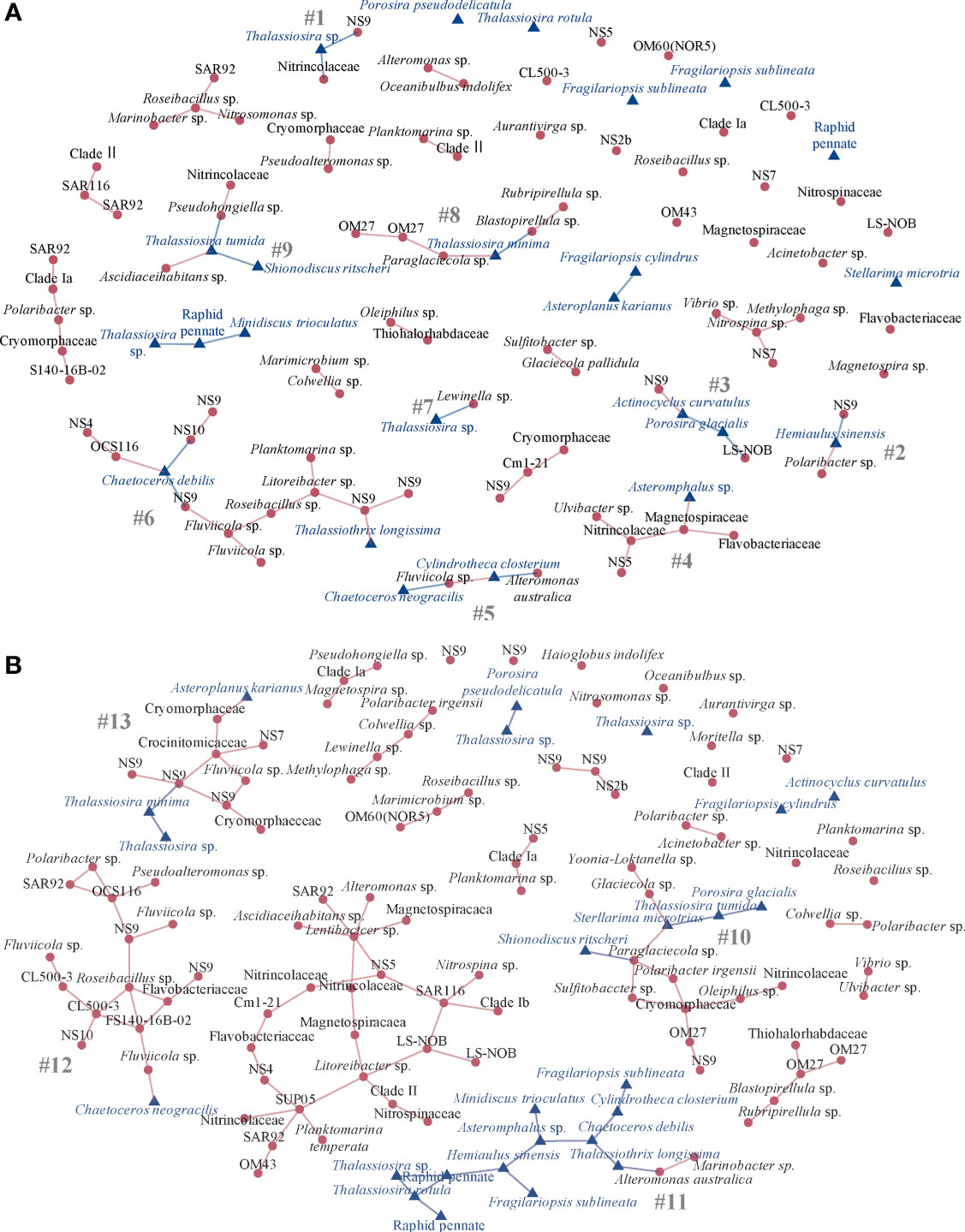
Figure 5 SPIEC-EASI network of the top 100 bacterial amplicon sequence variants (ASVs) and 25 diatom ASVs by relative abundance at the (A) Drake Passage stations (n = 6) with subnetworks #1-9 that contain at least one cross-kingdom connection and (B) Gerlache and Bransfield Strait stations (n = 12) from the NBP16-08 cruise with subnetworks #10-13 that contain at least one cross kingdom connection. The stability of each is 0.05. Only positive edges are shown. Each node is an ASV labeled by lowest taxonomic assignment. Red circle nodes are bacteria, and blue triangle nodes are diatoms.
The Gerlache and Bransfield Straits subnetworks were larger, all with 11 – 16 diatom and bacteria taxa connected (Figure 5B). Overall, there were 102 positive edges (network stability = 0.05) with 4 subnetworks of diatom and bacteria connected (Figure 5A). In subnetwork #10, the gammaproteobacterium Paraglaciecola sp. was connected to two diatoms, Stellarima microtrias and Shionodiscus ritscheri, with Stellarima microtrias also connected to other gammaproteobacterium Glaciecola sp. and diatom Thalassiosira tumida (Figure 5B). The majority of subnetwork #11 was diatom taxa, but Thalassiothrix longissima was connected to gammaproteobacterium Alteromonas australica (Figure 5B). In contrast, subnetwork #12 was mostly bacteria but the one diatom Chaetoceros neogracilis was connected to Bacteroidia Fluviicola sp. (Figure 5B). The diatom Asteroplanus karianus was connected to a member of Cryomorphaceae and the diatom Thalassiosira minima was connected to a member of NS9 in subnetwork #13 (Figure 5B).
When the networks were examined by separate Kingdoms, for the Gammaproteobacteria, there were 610 positive connections (stability threshold = 0.05). For the correlation network, there was a core interconnected group of ASVs at the stations with many connections (Figure S4). The diatom community was strikingly less interconnected (Figure S5) and there were only 68 positive connections in the diatom network (stability threshold = 0.05).
The SO surrounding the WAP is a well-known high-nutrient low-chlorophyll area in the global ocean (Moore et al., 2002). Overall, our stations grouped primarily into offshore Drake Passage stations and those corresponding to Bransfield and Gerlache Strait locations, which allowed us to sample across a wide range of macronutrient and especially trace metal concentrations. In particular, dFe concentrations ranged from 0.13 to 9.3 nM across our study region and, based on shipboard incubation experiments conducted in parallel to our field work, dFe was the primary limiting nutrient in the Drake Passage, but was not limiting inshore (Burns et al., 2023). The low dFe and dMn concentrations offshore also suggest a limited influence from shelf sediments at our Drake Passage sites in the spring, as has been observed previously in the austral winter (de Jong et al., 2012; Hatta et al., 2013). In contrast to trace metals and macronutrients, there was no clear geographic pattern with siderophore concentrations. The majority of our siderophore measurements were < 2 pM, with the stations with the highest chlorophyll a with lower (< 1 pM) siderophore concentrations. We sampled very early in the season (austral spring) and photosynthetic biomass was low, perhaps leading to lower in situ siderophore production rates and thus the overall low siderophore concentrations in surface waters. The range of siderophore concentrations we observed is comparable to siderophore concentrations found at sites with comparable dFe : DIN ratios, including in the eastern equatorial Pacific HNLC region (Boiteau et al., 2016) and in the upper mesopelagic at station ALOHA (Bundy et al., 2018). It is also possible that siderophores are cycled rapidly amongst the microbial community, and their concentrations therefore represent a single snapshot in time. Overall, our observed concentration of siderophores was an order of magnitude less than total strong Fe ligand concentrations previously reported from the same region in the SO (Buck et al., 2010; Ardiningsih et al., 2021), which is consistent with previous work showing siderophores comprise less than 10% of the ligand pool that is measured in seawater (Bundy et al., 2018).
With a few exceptions, the diatom and bacteria assemblages generally group by similarity of sampling location: Drake Passage, Palmer Arch, Antarctic Sound, Bransfield Strait, and Gerlache Strait. The Antarctic Sound samples grouped separately from others. Notably, the Antarctic Sound samples were the only ones from 25 m depth instead of 5 m like the other locations, but unlike the other stations, the temperature at Station 10 was warmer at 25 m (1.64°C) as opposed to 5 m water temperatures at all other stations (average -1.56°C). However, at some stations in the Gerlache Strait and Drake Passage, there were also temperatures > 1.00°C between 0 – 25 m (Figure S1B). The diatom assemblage present at Station 2 in the Gerlache Strait was more similar to the assemblages in the Drake Passage, and Station 1 in particular, than to the other Gerlache Strait assemblages. This pattern is different for the bacterial community assemblages in which the Station 2 bacterial community was most similar to the other Gerlache Strait stations. The similarity of the diatom community to those in the Drake passage may be due to the northern location of Gerlache Strait Station 2. This location perhaps received an influx of offshore water with that corresponding diatom assemblage, as it is a mixing spot where the Antarctic Circumpolar Current and Antarctic Peninsula Coastal Current converge (reviewed in Moffat and Meredith, 2018).
The WAP diatom assemblages as assayed by 18S rDNA community composition were comprised of several different genera and included Thalassiosira and Fragilariopsis at all stations. Fragilariopsis spp. have previously been shown to be the dominant diatoms observed in spring WAP assemblages, along with the diatoms Chaetoceros spp. and Navicula spp., as determined from scanning electron microscopy (Pike et al., 2008; Annett et al., 2010). From previous 18S sequencing of diatom communities in the WAP, Fragilariopsis was also found to be the most abundant sequence (Lin et al., 2017). In Marguerite Bay, Antarctica at the long-term site for the Rothera oceanographic and biological Time Series (RaTS), Thalassiosira and Chaetoceros are abundant as observed using DNA sequencing, with fewer sequences belonging to Stellarima microtrias and unknown pennate diatoms in austral summer (Rozema et al., 2017). Chaetoceros spp. and Navicula spp. were also present in our data set although at lower relative 18S rDNA abundance than Thalassiosira and Fragilariopsis. Interestingly, at the offshore stations and Station 2 in the Gerlache Strait there was a portion of ASVs which represented 11% of the ASVs at those stations that could not be taxonomically assigned to the genus level of known diatoms. This may be a limitation of the sequences present in the taxonomic database that might lack representation of diatoms present in the offshore area. Future culturing and sequencing efforts of diatom isolates from the Drake Passage as well as in-situ visual identification through microscopy may be necessary to further understand the potential novel diversity of diatoms in the region.
The members of the diatom and the associated bacterial communities present at these stations were likely driven by the observed gradients in macronutrients and trace metals, especially since SRP, dMn, and dCu seemed to influence both groups based on our multivariate analysis (BIOENV). Low dMn has been shown to co-limit SO diatom growth with dFe (Browning et al., 2021), which results in growth inhibition and decreased carbon production (Pausch et al., 2019). We found that dMn was an important environmental variable explaining the variance in the in situ community composition of diatoms and associated bacteria, providing evidence that dMn may be an important driver of bacteria and/or diatom growth in this region. Dissolved Pb was identified as a variable that explained a significant fraction of the variance in only the diatom community composition. Although not a bioactive element, dPb may adsorb onto phytoplankton biomass (Chuang et al., 2014) and may vary depending on the surface area and morphology of diatoms. Higher dPb concentrations were associated with lower chlorophyll a in samples from the Drake Passage, perhaps reflecting less sorption to cells and the negative correlation of dPb concentrations in the CAP analysis (Figure 2). Finally, dCo was a key driver of the variance in bacteria community composition. Vitamin B12 requires Co (reviewed in Twining and Baines, 2013), and members of Oceanospirillales were commonly observed at stations and have been found to contribute to vitamin B12 synthesis, likely assisting in relieving vitamin stress on the co-occurring diatom communities in the SO (Bertrand et al., 2015). Together, our data suggest that there are distinct diatom communities in the austral spring in the WAP: those who persist under lower trace metals and dSi in some offshore stations and those that require higher concentrations of both trace metals and dSi that are found near the continent. Likewise, there are also distinct associated bacterial communities that may provide specific benefits to the diatoms living in these different nutrient regimes.
When the connectedness of diatom and plankton-associated bacteria communities were visualized, some interesting patterns emerged, suggesting real-time mutualisms that have been observed in other environments (Steele et al., 2011) may also be occurring in this region. Bacteroidetes members were commonly found at every station in the associated assemblage in our study and were connected to diatoms in the network analysis. Genomes of Bacteroidetes members have previously been found to contain siderophore biosynthesis genes indicating this group’s potential for contributing to the overall siderophore pool in the SO (Delmont et al., 2015). The Bacteroidetes Fluviicola sp. was connected to the same diatom, Chaetoceros neogracilis in each network, regardless of station group. It was also connected to the diatom Cylindrotheca closterium in the Drake Passage network. Bacteroidetes can be beneficial to diatom growth (Amin et al., 2012), and this beneficial interaction seems to extend to multiple diatom taxa in the SO. Interestingly, in both networks, there are diatom nodes that are connected to bacterial nodes not identified at the genus level, which may indicate the importance of uncultured bacteria taxa in mutualisms with diatoms. We did not observe other connections to other genera of bacteria that were confirmed siderophore-producers in the subnetworks.
Overall, there was high diversity in the type of bacteria directly connected to diatom nodes in the networks including members of Gammaproteobacteria, Alphaproteobacteria, Bacteriodetes, Planctomycetes, and Nitrospinota. Typically, each pairing between a diatom and bacteria ASV was unique in the networks. For diatom species that are cosmopolitan in the Drake Passage and Gerlache and Bransfield Straits, their interactions with bacteria are distinct, indicating these partnerships may confer unique adaptations to each environment. For example, across both the nearshore and offshore networks, Thalassiosira spp. were connected to different bacteria, implying there are unique diatom-bacteria interactions in the WAP that form dependent on environmental conditions. Since different species or strains within the Thalassiosira genus correlate with different bacteria, this may indicate species-specific interactions even within one cosmopolitan genus in the SO. Interestingly, one strain of Thalassiosira rotula in laboratory experiments predictably recruits similar bacterial symbionts to its surface from a variety of bacterial inoculum from a similar environment (Mönnich et al., 2020). It may be that WAP bacterial seed populations are distinct between offshore and nearshore environments, or it may be that these specific associations formed in response to the environmental changes regardless of seed population. We may be examining a broader view of the diatom-bacteria interactions occurring in situ within the natural laboratory of changing oceanographic properties than has previously been observed in laboratory experiments.
The higher degree interactions betweenthe Gammaproteobacteria may indicate potential mutualisms between bacteria in the environment. Overall, bacteria seem to be interacting with each other more consistently across the stations. For example, Gammaproteobacteria ASVs were connected to other ASVs in one large subnetwork that contains almost every Gammaproteobacteria ASV. These connections may suggest metabolic partnerships occurring between bacteria in the environment. In contrast, the diatoms were less interconnected and thus unlikely to participate in direct mutualisms with other diatoms.
Overall, this study of the SO WAP region gives insights into diatom-bacteria interactions across a gradient of environmental parameters. The rDNA barcoding provides information on the in situ assemblages of diatoms and associated bacteria at each sampling location, and the network analysis of co-occurrence takes this a step further to examine positive correlations between the two groups that may be indicative of mutualisms. Different diatom-bacteria interactions were revealed in the offshore and nearshore station groupings. Although concentrations of dFe were significantly different between Drake Passage and the more nearshore stations, dFe was not a significant contributor to the variance in diatom and associated bacteria communities as we had initially hypothesized. This could be due to the fact that plankton growth in offshore stations is being supported by recycled biogenic dFe (Burns et al., 2023) and the austral spring communities have sufficient dFe. Instead, the concentrations of dMn and dCu, and of SRP, more significantly influenced the variance of both communities.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Sequence data NCBI BioProject ID; PRJNA839781. Macronutrient and trace metal data is available from NSF Biological & Chemical Oceanography Data Management Office (BCO-DMO) (Buck et al., 2018; Buck et al., 2019; Chappell et al., 2022; Burns et al., 2023).
ARS, SMB, RMB, LZH, KNB, PDC and BDJ collected data, BDJ, KNB, and PDC conceived of the study and acquired funding. ARS analyzed sequence data and created figures. ARS. and BDJ drafted the manuscript and all authors contributed to writing. All authors contributed to the article and approved the submitted version.
This research was supported by NSF awards #1443474 and #1756816 to BDJ, NSF award #1443483 to KNB, and NSF award # 1443646 to PDC. Graduate Assistantships to ARS were provided by NSF RI ESPCoR awards OIA #1004057 and #1655221, and a NASA RI Space Grant Fellowship (NNX15AI06H). Work was conducted at a Rhode Island NSF EPSCoR research facilities supported by NSF RI ESPCoR awards OIA #1004057 and #1655221.
Thank you to the NBP16-08 science party, ASC support staff, and ECO crew. Thank you to J. Atoyan at the RI GSC for sequencing and helpful advice. Sample analyses for macronutrients were performed by William Abbott (USF). We would like to thank our thoughtful reviewers whose input substantially contributed to the final version of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer NC declared a past co-authorship with the author PDC to the handling editor.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.876830/full#supplementary-material
Amin S. A., Green D. H., Hart M. C., Küpper F. C., Sunda W. G., Carrano C. J. (2009). Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proc. Natl. Acad. Sci. 106, 17071–17076. doi: 10.1073/pnas.0905512106
Amin S. A., Parker M. S., Armbrust E. V. (2012). Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 76, 667–684. doi: 10.1128/MMBR.00007-12
Annett A. L., Carson D. S., Crosta X., Clarke A., Ganeshram R. S. (2010). Seasonal progression of diatom assemblages in surface waters of Ryder Bay, Antarctica. Polar Biol. 33, 13–29. doi: 10.1007/s00300-009-0681-7
Annett A. L., Skiba M., Henley S. F., Venables H. J., Meredith M. P., Statham P. J., et al. (2015). Comparative roles of upwelling and glacial iron sources in Ryder Bay, coastal western Antarctic Peninsula. Mar. Chem. 176, 21–33. doi: 10.1016/j.marchem.2015.06.017
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Ardiningsih I., Seyitmuhammedov K., Sander S. G., Stirling C. H., Reichart G.-J., Arrigo K. R., et al. (2021). Fe-binding organic ligands in coastal and frontal regions of the western Antarctic Peninsula. Biogeosciences 18 (15), 4587–4601. doi: 10.5194/bg-18-4587-2021
Arrigo K. R., van Dijken G. L., Alderkamp A.-C., Erickson Z. K., Lewis K. M., Lowry K. E., et al. (2017). Early spring phytoplankton dynamics in the Western Antarctic Peninsula. J. Geophys. Res.Oceans 122 (12), 9350–9369. doi: 10.1002/2017JC013281
Atkinson A., Siegel V., Pakhomov E., Rothery P. (2004). Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature 432, 100–103. doi: 10.1038/nature02950.1
Bell W., Mitchell R. (1972). Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol. Bull. 143, 265–277. doi: 10.2307/1540052
Bertrand E. M., McCrow J. P., Moustafa A., Zheng H., McQuaid J. B., Delmont T. O., et al. (2015). Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. 112, 9938–9943. doi: 10.1073/pnas.1501615112
Boiteau R. M., Mende D. R., Hawco N. J., Mcilvin M. R., Fitzsimmons J. N., Saito M. A., et al. (2016). Siderophore-based microbial adaptations to iron scarcity across the Eastern Pacific Ocean. Proc. Natl. Acad. Sci. 113, 14237–14242. doi: 10.1073/pnas.1608594113
Boyd P. W., Watson A. J., Law C. S., Abraham E. R., Trull T., Murdoch R., et al. (2000). A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature 407, 695–702. doi: 10.1038/35037500
Brand L. E., Sunda W. G., Guillard R. R. L. (1983). Limitation of marine phytoplankton reproductive rates by zinc, manganese, and iron 1. Limnol. Oceanogr. 28, 1182–1198. doi: 10.4319/lo.1983.28.6.1182
Browning T. J., Achterberg E. P., Engel A., Mawji E. (2021). Manganese co-limitation of phytoplankton growth and major nutrient drawdown in the Southern Ocean. Nat. Commun. 12, 1–9. doi: 10.1038/s41467-021-21122-6
Bruland K. W., Donat J. R., Hutchins D. A. (1991). Interactive influences of bioactive trace metals on biological production in Oceanic waters. Limnol. Oceanogr. 36, 1555–1577. doi: 10.4319/lo.1991.36.8.1555
Buck K. N., Bruland K. W. (2007). The physicochemical speciation of dissolved iron in the Bering Sea, Alaska. Limnol. Oceanogr. 52, 1800–1808. doi: 10.4319/lo.2007.52.5.1800
Buck K., Chappell P., Jenkins B. (2018). Dissolved macronutrient concentrations from depth profiles from RVIB Nathanial B. Palmer cruise NBP1608 in the Southern Ocean from September to October 2016. doi: 10.1575/1912/bco-dmo.742819.1
Buck K., Chappell P., Jenkins B. (2019). Dissolved trace metal concentrations from depth profiles during RVIB Nathanial B. Palmer cruise NBP16-08 in the Southern Ocean from 2016-09-11 to 2016-10-10. doi: 10.1575/1912/bco-dmo.781773.1
Buck K. N., Selph K. E., Barbeau K. A. (2010). Iron-binding ligand production and copper speciation in an incubation experiment of Antarctic Peninsula shelf waters from the Bransfield Strait, Southern Ocean. Mar. Chem. 122, 148–159. doi: 10.1016/j.marchem.2010.06.002
Bundy R. M., Boiteau R. M., McLean C., Turk-Kubo K. A., McIlvin M. R., Saito M. A., et al. (2018). Distinct siderophores contribute to iron cycling in the mesopelagic at station ALOHA. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00061
Burns S. M., Bundy R. M., Abbott W., Abdala Z., Sterling A. R., Chappell P. D., et al. (2023). Interactions of bioactive trace metals in shipboard Southern Ocean incubation experiments. Limnol. Oceanography. doi: 10.1002/lno.12290
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1007/s10549-015-3663-1.Progestin
Chappell P. D., Armbrust E. V., Barbeau K. A., Bundy R. M., Moffett J. W., Vedamati J., et al. (2019). Patterns of diatom diversity correlate with dissolved trace metal concentrations and longitudinal position in the northeast Pacific coastal-offshore transition zone. Mar. Ecol. Prog. Ser. 609, 69–86. doi: 10.3354/meps12810
Chappell P. D., Buck K., Jenkins B. D.. (2022). Chlorophyll and phaeopigment concentrations from near-surface profiles collected using the conventional CTD in the Southern Drake Passage. and Antarctic Peninsular region on RVIB Nathaniel B. Palmer cruise NBP 16-08 from September to October 2016. doi: doi:10.26008/1912/bco-dmo.740939.1
Chuang C., Santschi P. H., Jiang Y., Ho Y., Quigg A., Guo L., et al. (2014). Important role of biomolecules from diatoms in the scavenging of particle-reactive radionuclides of thorium, protactinium, lead, polonium, and beryllium in the Ocean: A case study with. Phaeodactylum tricornutum. Limnol. Oceanogr. 56. doi: 10.4319/lo.2014.59.4.1256
Coale T. H., Moosburner M., Horák A., Oborník M., Barbeau K. A., Allen A. E. (2019). Reduction-dependent siderophore assimilation in a model pennate diatom. Proc. Natl. Acad. Sci. 116 (47), 23609–23617. doi: 10.1073/pnas.1907234116
Csárdi G., Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695, 1–9.
Cutter G. A., Bruland K. W. (2012). Rapid and noncontaminating sampling system for trace elements in global Ocean surveys. Limnol. Oceanogr. Methods 10, 425–436. doi: 10.4319/lom.2012.10.425
Cutter G., Casciotti K., Croot P., Geibert W., Heimbürger L.-E., Lohan M., et al. (2017). “Sampling and sample-handling protocols for GEOTRACES cruises, version 3.0 ed,” in GEOTRACES.
de Jong J., Schoemann V., Lannuzel D., Croot P., de Baar H., Tison J. (2012). Natural iron fertilization of the Atlantic sector of the Southern Ocean by continental shelf sources of the Antarctic Peninsula. J. Geophys. Res. Biogeosci. 117. doi: 10.1029/2011JG001679
del Campo J., Kolisko M., Boscaro V., Santoferrara L. F., Nenarokov S., Massana R., et al. (2018). EukRef: Phylogenetic curation of ribosomal RNA to enhance understanding of eukaryotic diversity and distribution. PloS Biol. 16, e2005849. doi: 10.1371/journal.pbio.2005849
Delmont T. O., Eren A. M., Vineis J. H., Post A. F. (2015). Genome reconstructions indicate the partitioning of ecological functions inside a phytoplankton bloom in the Admunsen Sea, Antarctica. Front. Microbiol. 6, 1090. doi: 10.3389/fmicb.2015.01090
Desbois A. P., Lebl T., Yan L., Smith V. J. (2008). Isolation and structural characterisation of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl. Microb. Cell Physiol. 81, 755–764. doi: 10.1007/s00253-008-1714-9
de Sousa A. G. G., Tomasino M. P., Duarte P., Fernández-Méndez M., Assmy P., Ribeiro H., et al. (2019). Diversity and composition of pelagic prokaryotic and protist communities in a thin Arctic sea-ice regime. Microb. Ecol. 78, 388–408. doi: 10.1007/s00248-018-01314-2
Dixon P. (2003). VEGAN, a package of r functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Droop M. R. (2007). Vitamins, phytoplankton and bacteria: Symbiosis or scavenging? J. Plankton Res. 29, 107–113. doi: 10.1093/plankt/fbm009
Ducklow H. W., Baker K., Martinson D. G., Quetin L. B., Ross R. M., Smith R. C., et al. (2007). Marine pelagic ecosystems: The West Antarctic Peninsula. Philos. Trans. R. Soc B Biol. Sci. 362, 67–94. doi: 10.1098/rstb.2006.1955
Edgar R. C. (2018). Updating the 97% identity threshold for 16S ribosomal RNA OTUs. Bioinformatics 34, 2371–2375. doi: 10.1093/bioinformatics/bty113
Forsch K. O., Hahn-Woernle L., Sherrell R. M., Roccanova V. J., Bu K., Burdige D., et al. (2021). Seasonal dispersal of fjord meltwaters as an important source of iron and manganese to coastal Antarctic phytoplankton. Biogeosciences 18, 6349–6375. doi: 10.5194/bg-18-6349-2021
Frölicher T. L., Sarmiento J. L., Paynter D. J., Dunne J. P., Krasting J. P., Winton M. (2015). Dominance of the Southern Ocean in anthropogenic carbon and heat uptake in CMIP5 models. J. Clim. 28, 862–886. doi: 10.1175/JCLI-D-14-00117.1
Garibotti I., Vernet M., Ferrario M. E. (2005). Annually recurrent phytoplanktonic assemblages during summer in the seasonal ice zone west of the Antarctic Peninsula (Southern Ocean). Deep Sea Res. Part I Oceanogr. Res. Pap. 52, 1823–1841. doi: 10.1016/j.dsr.2005.05.003
Gerringa L. J. A., Veldhuis M. J. W., Timmermans K. R., Sarthou G., De Baar H. J. W. (2006). Co-Variance of dissolved fe-binding ligands with phytoplankton characteristics in the canary basin. Mar. Chem. 102, 276–290.
Gervais F., Riebesell U., Gorbunov M. Y. (2002). Changes in primary productivity and chlorophyll a in response to iron fertilization in the Southern Polar Frontal Zone. Limnol. Oceanogr. 47, 1324–1335. doi: 10.4319/lo.2002.47.5.1324
Gilbert J. A., Meyer F., Antonopoulos D., Balaji P., Brown C. T., Brown C. T., et al. (2010). Meeting report: The terabase metagenomics workshop and the vision of an Earth Microbiome Project. Stand. Genomic Sci. 3, 243–248. doi: 10.4056/sigs.1433550
Gledhill M., Buck K. N. (2012). The organic complexation of iron in the marine environment: A review. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00069
Gledhill M., McCormack P., Ussher S., Achterberg E. P., Mantoura R. F. C., Worsfold P. J. (2004). Production of siderophore type chelates by mixed bacterioplankton populations in nutrient enriched seawater incubations. Mar. Chem. 88, 75–83. doi: 10.1016/j.marchem.2004.03.003
Gledhill M., van den Berg C. M. G. (1994). Determination of complexation of iron (III) with natural organic complexing ligands in seawater using cathodic stripping voltammetry. Mar. Chem. 47, 41–54. doi: 10.1016/0304-4203(94)90012-4
Glöckner F. O., Yilmaz P., Quast C., Gerken J., Beccati A., Ciuprina A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi: 10.1016/j.jbiotec.2017.06.1198
Grossart H.-P. (1999). Interactions between marine bacteria and axenic diatoms (Cylindrotheca fusiformis, Nitzschia laevis, and Thalassiosira weissflogii) incubated under various conditions in the lab. Aquat. Microb. Ecol. 19, 1–11. doi: 10.3354/ame019001
Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., et al. (2013). The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, 597–604. doi: 10.1093/nar/gks1160
Hatta M., Measures C. I., Selph K. E., Zhou M., Hiscock W. T. (2013). Iron fluxes from the shelf regions near the South Shetland Islands in the Drake Passage during the austral-winter 2006. Deep Sea Res. Part II Top. Stud. Oceanogr. 90, 89–101. doi: 10.1016/j.dsr2.2012.11.003
Hollister A. P., Kerr M., Malki K., Muhlbach E., Robert M., Tilney C. L., et al. (2020). Regeneration of macronutrients and trace metals during phytoplankton decay: An experimental study. Limnol. Oceanogr. 65, 1936–1960. doi: 10.1002/lno.11429
Hopkinson B. M., Mitchell B. G., Reynolds R. A., Wang H., Selph K. E., Measures C. I., et al. (2007). Iron limitation across chlorophyll gradients in the southern Drake Passage: Phytoplankton responses to iron addition and photosynthetic indicators of iron stress. Limnol. Oceanogr. 52, 2540–2554. doi: 10.4319/lo.2007.52.6.2540
Hulten M., Middag R., Dutay J.-C., de H., Roy-Barman M., Gehlen M., et al. (2017). Manganese in the west Atlantic Ocean in the context of the first global Ocean circulation model of manganese. Biogeosciences 14, 1123–1152. doi: 10.5194/bg-14-1123-2017
Johnstone T. C., Nolan E. M. (2015). Beyond iron: Non-classical biological functions of bacterial siderophores. Dalt. Trans. 44, 6320–6339. doi: 10.1039/C4DT03559C
Kazamia E., Sutak R., Paz-Yepes J., Dorrell R. G., Rocha Jimenez Vieira F., Mach J., et al. (2018). Endocytosis-mediated siderophore uptake as a strategy for Fe acquisition in diatoms. Sci. Adv. 4, 1–14. doi: 10.1126/sciadv.aar4536
Kellam S. J., Walker J. M. (1989). Antibacterial activity from marine microalgae in laboratory culture. Br. Phycol. J. 24, 191–194. doi: 10.1080/00071618900650181
Kerkhof L. J., Voytek M. A., Sherrell R. M., Millie D., Schofield O. (1999). Variability in bacterial community structure during upwelling in the coastal Ocean. Hydrobiologia 401, 139–148. doi: 10.1023/A:1003734310515
Kurtz Z. D., Müller C. L., Miraldi E. R., Littman D. R., Blaser M. J., Bonneau R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PloS Comput. Biol. 11, e1004226. doi: 10.1371/journal.pcbi.1004226
Lannuzel D., Schoemann V., de Jong J., Pasquer B., van der Merwe P., Masson F., et al. (2010). Distribution of dissolved iron in Antarctic sea ice: Spatial, seasonal, and inter-annual variability. J. Geophys. Res. 115, G03022. doi: 10.1029/2009JG001031
Leblanc K., Hare C. E., Boyd P. W., Bruland K. W., Sohst B., Pickmere S., et al. (2005). Fe and Zn effects on the Si cycle and diatom community structure in two contrasting high and low-silicate HNLC areas. Deep Sea Res. Part I Oceanogr. Res. Pap. 52, 1842–1864. doi: 10.1016/j.dsr.2005.06.005
Le Quéré C., Andrew R. M., Canadell J. G., Sitch S., Korsbakken J. I., Peters G. P., et al. (2016). Global Carbon Budget 2016. Earth Syst. Sci. Data 8.
Lin Y., Cassar N., Marchetti A., Moreno C., Ducklow H., Li Z. (2017). Specific eukaryotic plankton are good predictors of net community production in the Western Antarctic Peninsula. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-14109-1
Liu H., Roeder K., Wasserman L. (2010). Stability approach to regularization selection (stars) for high dimensional graphical models. Adv. Neural Inf. Process. Syst. 24, 1432.
Maldonado M. T., Price N. M. (2001). Reduction and transport of organically bound iron by Thalassiosira Oceanica (Bacillariophyceae). J. Phycol. 37 (2), 298–310.
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Martin J. H., Gordon R. M., Fitzwater S. E. (1990). Iron in Antarctic waters. Nature 345, 156–158. doi: 10.1038/345156a0
Mawji E., Gledhill M., Milton J. A., Tarran G. A., Ussher S., Thompson A., et al. (2008). Hydroxamate siderophores: Occurence and importance in the Atlantic Ocean. Environ. Sci. Technol. 42, 8675–8680. doi: 10.1021/es801884r
McMurdie P. J., Holmes S. (2013). Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8. doi: 10.1371/journal.pone.0061217
Measures C. I., Brown M. T., Selph K. E., Apprill A., Zhou M., Hatta M., et al. (2013). The influence of shelf processes in delivering dissolved iron to the HNLC waters of the Drake Passage, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 90, 77–88. doi: 10.1016/j.dsr2.2012.11.004
Meinshausen N., Bühlmann P. (2006). High-dimensional graphs and variable selection with Lasso. Ann. Stat. 34, 1436–1462. doi: 10.1214/009053606000000281
Middag R., de Baar H. J. W., Klunder M. B., Laan P. (2013). Fluxes of dissolved aluminum and manganese to the weddell Sea and indications for manganese co-limitation. Limnol. Oceanogr. 58, 287–300. doi: 10.4319/lo.2013.58.1.0287
Mikaloff Fletcher S. E., Gruber N., Jacobson A. R., Doney S. C., Dutkiewicz S., Gerber M., et al. (2006). Inverse estimates of anthropogenic CO2 uptake, transport, and storage by the Ocean. Global Biogeochem. Cycles 20. doi: 10.1029/2005GB002530
Moffat C., Meredith M. (2018). Shelf–Ocean exchange and hydrography west of the Antarctic Peninsula: A review. Philos. Trans. R. Soc A Math. Phys. Eng. Sci. 376, 20170164. doi: 10.1098/rsta.2017.0164
Mönnich J., Tebben J., Bergemann J., Case R., Wohlrab S., Harder T. (2020). Niche-based assembly of bacterial consortia on the diatom Thalassiosira rotula is stable and reproducible. ISME J. 14, 1614–1625. doi: 10.1038/s41396-020-0631-5
Montes-Hugo M. A., Vernet M., Martinson D., Smith R., Iannuzzi R. (2008). Variability on phytoplankton size structure in the western Antarctic Peninsula, (1997 – 2006). Deep. Res. II 55, 2106–2117. doi: 10.1016/j.dsr2.2008.04.036
Moore J. K., Doney S. C., Glover D. M., Fung I. Y. (2002). Iron cycling and nutrient-limitation patterns in surface waters of the world Ocean. Deep. Res. II 49, 463–507. doi: 10.1016/S0967-0645(01)00109-6
Moore J. K., Lindsay K., Doney S. C., Long M. C., Misumi K. (2013). Marine ecosystem dynamics and biogeochemical cycling in the community earth system model [CESM1(BGC0]: Comparison of the 1990s with the 2090s under the RCP4.5 and RCP8.5 scenarios. J. Clim. 26, 9291–9312. doi: 10.1016/j.palaeo.2013.06.001
Morel F. M. M., Price N. M. (2003). The biogeochemical cycles of trace metals in the Oceans. Science 300, 944–947. doi: 10.1126/science.1083545
Nicol S., Bowie A., Jarman S., Lannuzel D., Meiners K. M., van der Merwe P. (2010). Southern Ocean iron fertilization by baleen whales and Antarctic krill. Fish. Fish. 11, 203–209. doi: 10.1111/j.1467-2979.2010.00356.x
Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pausch F., Bischof K., Trimborn S. (2019). Iron and manganese co-limit growth of the Southern Ocean diatom Chaetoceros debilis. PloS One 14, 1–16. doi: 10.1371/journal.pone.0221959
Pike J., Allen C. S., Leventer A., Stickley C. E., Pudsey C. J. (2008). Comparison of contemporary and fossil diatom assemblages from the western Antarctic Peninsula shelf. Mar. Micropaleontol. 67, 274–287. doi: 10.1016/j.marmicro.2008.02.001
Pinhassi J., Monteserrat Sala M., Havskum H., Peters F., Guadayol O., Malits A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. doi: 10.1128/AEM.70.11.6753
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Queguiner B. (2013). Iron fertilization and the structure of planktonic communities in high nutrient regions of the Southern Ocean. Deep. Res. II 90, 43–54. doi: 10.1016/j.dsr2.2012.07.024
Rijkenberg M. J. A., Gerringa L. J. A., Timmermans K. R., Fischer A. C., Kroon K. J., Buma A. G. J., et al. (2008). Enhancement of the reactive iron pool by marine diatoms. Mar. Chem. 109, 29–44. doi: 10.1016/j.marchem.2007.12.001
Rozema P. D., Biggs T., Sprong P. A., Burma A. G., Venables H. J., Evans C., et al. (2017). Summer microbial community composition governed by upper-Ocean stratification and nutrient availability in northern Marguerite Bay, Antarctica. Deep Sea Res. Part II Top. Stud. Oceanogr. 139, 151–166. doi: 10.1016/j.dsr2.2016.11.016
Rue E. L., Bruland K. W. (1995). Complexation of iron (III) by natural organic ligands in the Central North Pacific as determined by a new competitive ligand equilibration/adsorptive cathodic stripping voltammetric method. Mar. Chem. 50, 117–138. doi: 10.1016/0304-4203(95)00031-L
Sanudo-Wilhelmy S. A., Olsen K. A., Scelfo J. M., Foster T. D., Flegal A. R. (2002). Trace metal distributions off the Antarctic Peninsula in the weddell Sea. Mar. Chem. 77, 157–170. doi: 10.1016/S0304-4203(01)00084-6
Schäfer H., Abbas B., Witte H., Gerard M. (2002). Genetic diversity of “satellite” bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42, 25–35. doi: 10.1111/j.1574-6941.2002.tb00992.x
Seymour J. R., Amin S. A., Raina J.-B., Stocker R. (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2. doi: 10.1038/nmicrobiol.2017.65
Shaked Y., Buck K. N., Mellett T., Maldonado M. T. (2020). Insights into the bioavailability of Oceanic dissolved Fe from phytoplankton uptake kinetics. ISME J. 14, 1182–1193. doi: 10.1038/s41396-020-0597-3
Shibl A. A., Isaac A., Ochsenkühn M. A., Cárdenas A., Fei C., Behringer G., et al. (2020). Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc. Natl. Acad. Sci. 117, 27445–27455. doi: 10.1073/pnas.2012088117
Smetacek V., Klaas C., Strass V. H., Assmy P., Montresor M., Cisewski B., et al. (2012). Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature 487, 313–319. doi: 10.1038/nature11229
Somero G. N. (2012). The physiology of global change: linking patterns to mechanisms. Annu. Rev. Mar. Sci. 4, 39–61. doi: 10.1146/annurev-marine-120710-100935
Stammerjohn S. E., Martinson D. G., Smith R. C., Yuan X., Rind D. (2008). Trends in Antarctic annual sea ice retreat and advance and their relation to El Niño–Southern Oscillation and Southern Annular Mode variability. J. Geophys. Res. 113, C03S90. doi: 10.1029/2007JC004269
Steele J. A., Countway P. D., Xia L., Vigil P. D., Beman J. M., Kim D. Y., et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425. doi: 10.1038/ismej.2011.24
Stocker R. (2012). Marine microbes see a sea of gradients. Science 338, 628–633. doi: 10.1126/science.1208929
Sunda W. G. (2012). Feedback interactions between trace metal nutrients and phytoplankton in the Ocean. Front. Microbiol. 3, 204. doi: 10.3389/fmicb.2012.00204
Teeling H., Fuchs B. M., Becher D., Klockow C., Gardebrecht A., Bennke C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Twining B. S., Baines S. B. (2013). The trace metal composition of marine phytoplankton. Ann. Rev. Mar. Sci. 5, 191–215. doi: 10.1146/annurev-marine-121211-172322
Ukeles R., Bishop J. (1975). Enhancement of phytoplankton growth by marine bacteria. J. Phycol. 11, 142–149.
van Oijen T., van Leeuwe M. A., Granum E., Weissing F. J., Bellerby R. G. J., Gieskes W. W. C., et al. (2004). Light rather than iron controls photosynthate production and allocation in Southern Ocean phytoplankton populations during austral autumn. J. Plankton Res. 26, 885–900. doi: 10.1093/plankt/fbh088
van Tol H. M., Amin S. A., Armbrust V. E. (2017). Ubiquitous marine bacterium inhibits diatom cell division. ISME J. 11, 31–42. doi: 10.1038/ismej.2016.112
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wickham H. (2016). ggplot2: elegant graphics for data analysis (New York: Springer-Verlag). doi: 10.1007/978-0-387-98141-3
Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Keywords: diatom-bacteria interactions, Southern Ocean, Antarctic Peninsula, phytoplankton microbiome, trace metal limitation
Citation: Sterling AR, Holland LZ, Bundy RM, Burns SM, Buck KN, Chappell PD and Jenkins BD (2023) Potential interactions between diatoms and bacteria are shaped by trace element gradients in the Southern Ocean. Front. Mar. Sci. 9:876830. doi: 10.3389/fmars.2022.876830
Received: 15 February 2022; Accepted: 07 December 2022;
Published: 02 March 2023.
Edited by:
Elena Kazamia, École Normale Supérieure, FranceReviewed by:
Natalie Cohen, University of Georgia, United StatesCopyright © 2023 Sterling, Holland, Bundy, Burns, Buck, Chappell and Jenkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bethany D. Jenkins, YmRqZW5raW5zQHVyaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.