- 1Shandong Provincial Key Laboratory of Plant Stress, College of Life Sciences, Shandong Normal University, Jinan, China
- 2Key Laboratory of Algal Biology, Institute of Hydrobiology, Chinese Academy of Sciences, Hubei University, Wuhan, China
- 3Laboratory of Bacterial Vaccine, Wuhan Institute of Biological Products Co., Ltd., Wuhan, China
- 4Institute of Biology and Medicine, Wuhan University Science Technology, Wuhan, China
- 5College of Life Science and Healthy, Wuhan University Science Technology, Wuhan, China
Monogalactosyldiacylglycerol (MGDG) is the most abundant polar lipid in thylakoid membrane, wherein it plays critical roles related to thylakoid membrane assembly and function in diatoms. However, diatom MGDG biosynthesis has not been fully characterized. In this study, we investigated the role of a novel MGDG synthase (ptMGD2), which is one of the key enzymes for MGDG biosynthesis, in the model diatom Phaeodactylum tricornutum. An analysis of subcellular localization demonstrated that the ptMGD2 is mainly localized in plastids. Gene disruption by gene editing of ptMGD2 resulted in delayed growth, decrease in oxygen evolution rate, reduced MGDG and digalactosyldiacylglycerol (using MGDG as the substrate) content as well as lipid remodeling. Considered together, these observations provide novel insights into the importance of ptMGD2 for regulating MGDG biosynthesis and its potential roles in biotechnical application of Phaeodactylum.
Introduction
Diatoms, which are one of the major primary producers in oceans (Falkowski et al., 1998; Field et al., 1998), are considered to have originated from red algae and some other eukaryotic host following a series of endosymbiotic events (Moustafa et al., 2009; Sibbald and Archibald, 2020). Although the complicated algal origin of diatoms is still not really understood, whole-genome analysis and phylogeny-based horizontal gene transfer detection have revealed that diverse genes were acquired from other organisms besides through endosymbiosis (Bowler et al., 2008; Lommer et al., 2012; Traller et al., 2016; Mock et al., 2017; Rastogi et al., 2018; Vancaester et al., 2020). Diatoms are economically important species that have commonly been applied as microalgal cell factories for the production of natural and genetically engineered products, including triacylglycerols for biodiesel, polyunsaturated fatty acids with nutraceutical uses, and heterologous recombinant proteins beneficial for human health (Hempel et al., 2017; Vanier et al., 2018).
Phaeodactylum tricornutum is the most thoroughly characterized diatom to date, with an available fully sequenced genome (Bowler et al., 2008). The biochemical and physiological characteristics of P. tricornutum have been studied and large-scale transcriptomic and proteomic analyses for this species have been developed (Chen et al., 2018; Ge et al., 2014; Maheswari et al., 2009; Rastogi et al., 2018). This diatom has been extensively used as a model species for investigating diatom metabolism and evolution. To date, viable methods for genetic manipulations, such as nuclear (Falciatore et al., 1999) and chloroplast (Xie et al., 2014) transformations, have been developed for P. tricornutum. Genetic modifications are becoming routine procedures because of the application of biolistic transformation, electroporation, and conjugation (Falciatore et al., 1999; Miyahara et al., 2013; Karas et al., 2015). Over the past few decades, researchers have developed state-of-the-art molecular tools that have facilitated protein tagging and overexpression (Siaut et al., 2007) and targeted gene mutations in this diatom, such as knockdown (De Riso et al., 2009), transcription activator-like effector nucleases (TALEN) (Daboussi et al., 2014; Weyman et al., 2015), and clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 (Sharma et al., 2018; Slattery et al., 2018; Stukenberg et al., 2018; Moosburner et al., 2020). The use of these tools has provided researchers with valuable information relevant for structural and functional studies of diatoms.
Monogalactosyldiacylglycerol (MGDG) in plant plastids accounts for approximately 50% of the thylakoid membrane lipids. Moreover, MGDG, which is synthesized in the plastid envelope membrane, is a substrate for digalactosyldiacylglycerol (DGDG) synthesis (Block et al., 1983). This biosynthetic reaction is catalyzed by MGDG synthase (MGD), which transfers a galactosyl residue from UDP(uridine diphosphate)-galactose to the sn-3 position of sn-1,2-diacylglycerol (Benning and Ohta, 2005). Therefore, MGD is the principal enzyme for the synthesis of MGDG in phototrophs possessing secondary chloroplast (Dörmann and Benning, 2002; Kalisch et al., 2016). In plants, MGDG biosynthesis may occur via the prokaryotic pathway, omega pathways, and eukaryotic pathway (Petroutsos et al., 2014). However, the MGDG biosynthetic pathway catalyzed by MGD in diatoms remains unclear.Previous research revealed that MGDG is crucial for plant growth and development at least partly because of its involvement in photosynthetic membrane biogenesis (Masuda et al., 2011), photosynthetic reactions (Hölzl and Dörmann, 2007), and tolerance to various adverse environmental conditions (Cook et al., 2021), including phosphorous deficiency (Kobayashi et al., 2009a), salt stress (Wang et al., 2014), submergence (Qi et al., 2004), and wounding (Klecker et al., 2014). The roles of MGD have been extensively investigated in the green sulfur bacterium Chlorobaculum tepidum (Masuda et al., 2011) as well as in the photosynthetic and non-photosynthetic tissues of higher plants, including Arabidopsis (Kobayashi et al., 2009b; Myers et al., 2011) and rice (Basnet et al., 2019). In contrast, little is known about the contribution of such enzymes to diatom MGDG synthesis, including their cellular sublocation, involvement in lipid remodeling, and roles in response to unfavorable environmental conditions. The objective of this study was to characterize one of the MGD-encoding genes in biosynthesis of MGDG in P. tricornutum (ptMGD2) by applying a reverse genetic approach. Specifically, the CRISPR/Cas9 system was used to mutate ptMGD2 and elucidate its importance for MGDG biosynthesis and its effects on the corresponding lipid remodeling. Loss-of-function mutations to this gene had obvious detrimental effects on diatom cells, suggestive of its vital role in the MGDG biosynthetic pathway in marine diatoms.
Materials and Methods
Cell Culture and Growth Conditions
Axenic cells of P. tricornutum Bohlin (CCMP 632 from culture collection of the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, Bigelow Laboratory for Ocean Sciences, USA) were cultured in f/2 medium (Guillard, 1975) at 22 ± 1°C with a 16-h light/8-h dark photoperiod (60 µmol photons m−2 s−1 white light). For the growth experiment, the medium was inoculated with wild-type (Wt) and mutant cells (2 × 105 cells mL−1) from those maintained in enriched seawater at the exponential phase (5 × 106 cells mL−1) and the cultures were bubbled with filtered axenic air. We measured the cell growth rate every 2 days for determination of cell number.
Plasmid Construction
Escherichia coli DH5 alpha cells (Tiangen, Beijing, China) were grown on Luria–Bertani broth or agar supplemented with ampicillin (50 mg L-1), kanamycin (50 mg L-1), or gentamicin (50 mg L-1) as needed. E. coli DH5 alpha cells were used to propagate recombinant plasmids according to the manufacturer’s instructions. The ptMGD2 cDNA sequence was initially cloned into pMD18-T (TaKaRa, Dalian, China) using primers 9619cDNA_F and 9619cDNA_R (Qingke, Beijing, China) before being subcloned into the pPha-T1 vector containing the eGFP (Enhanced Green Fluorescent Protein) sequence. Primers of P9619_GFP_F and P9619_GFP_R were used for the subcellular localization analysis of ptMGD2. We constructed the PtPuc3_diaCas9_sgRNA vector (AddGene, ID: 109219) for the targeted mutagenesis of the ptMGD2 gene. Two different PAM(protospacer adjacent motif)-target sites (ptMGD2 PAM1 and ptMGD2 PAM2) with low homology to other genomic loci were designed targeting the coding sequence of the ptMGD2 gene. Small adapters for the targets of interest were inserted into the sgRNA of PtPuc3_diaCas9_sgRNA vectors according to Nymark et al. (2016). Primers used were: 9619cas_SmaI_F, 9619cas_SmaI_R, 9619cas_AccIII_F, and 9619cas_AccIII_R. All primers used in this study are listed in Supplemental Table S1, and all constructs were confirmed by DNA sequencing.
Quantitative Real-Time PCR
To analyze the effects of high salinity and phosphate deprivation, cells were cultured in f/2 medium supplemented with 0.8 M NaCl or without phosphate. We also analyze the relative abundance of MGDs in both Wt and mutants under normal growth conditions. Total RNA was extracted from P. tricornutum cells collected during the exponential growth phase using the RNAprep Pure kit (BioFlux, Hangzhou, China) and then reverse transcribed to cDNA using the PrimeScript RT Reagent kit (Yeasen, Shanghai, China). Gene expression was examined using the Roche Illuminator system (Roche, Mannheim, Germany) and the qPCR SYBR master mix (Yeasen). The analysis was completed using three biological replicates. Relative expression levels were calculated according to the 2−ΔCt method. qRT-PCR primers used in the study are listed in Supplemental Table S1 (9619rt_F and 9619rt_R for Phatr3_J9619; 14125rt_F and 14125rt_R for Phatr3_J14125; 54168rt_F and 54168rt_R for Phatr3_J54168; H4rt_F and H4rt_R for Phatr3_J26896).
Immunoprecipitation Analysis
Total protein extract (500 μg, in immunoprecipitation (IP) lysis buffer [25mM Tris, 150mM NaCl, 1mM EDTA, 1% NP40, 5% glycerol, pH 7.4]) for IP analysis was carried out according to Pierce Classic IP Kit’s manufacturer’s instructions (Thermo Fisher Scientific, Waltham, USA). Briefly, we first had anti-ptMGD2 antibody (ABclonal, Wuhan, China) bind to magnetic beads using 0.25 mM DSS (disuccinimidyl suberate) in coupling buffer (10 mM Na3PO4, 150 mM NaCl; pH 7.2). Then, we incubated cell lysate with antibody-crosslinked beads overnight at 4°C in IP lysis buffer. Finally, the target antigens were eluted and magnetically separate from the beads by elution buffer (pH 2.0) and the eluent was further identified with tandem mass spectroscopy by ABclonal technology (Wuhan, China). All acquired raw data were processed with pFind (V3.1.6) software (Chi et al., 2018). The peak lists were searched against the protein database from http://protists.ensembl.org/Phaeodactylum_tricornutum/Info/Index. Four missed cleavages were allowed for trypsin. The precursor and fragment ion mass tolerances were 20 ppm and 20 ppm, respectively. Open-search algorithm in pFind was used and acetylation (lysine) was set as variable modifications. Minimum peptide length was set at 6 while the estimated false discovery rate threshold for peptide and protein were specified at maximum 1%. For the other parameters in pFind, we used the algorithm defaults.
Targeted Mutagenesis of the ptMGD2 Using the CRISPR/Cas9 System
The pPtPuc3m diaCas9_sgRNA plasmid was used bacterial conjugation for the delivery of the CRISPR/Cas9 plasmid to P. tricornutum cells as previously described (Sharma et al., 2018; Slattery et al., 2018). The P. tricornutum mutant was first grown in f/2 medium supplemented with Zeocin (50 µg mL-1). Genetically transformed P. tricornutum cells were screened for targeted DNA mutations by PCR and enzymatic analyses as well as by the Sanger sequencing of the regions spanning the ptMGD2 PAM1 and PAM2 target sites. The primers for the PCR amplification of genomic DNA included 9619kn_F/9619kn_R to detect ptMGD2 mutations. Single colony with mutations at the target gene sites containing the conjugative plasmid were then diluted during the two weeks in f/2 liquid medium without antibiotics. Next, cells were diluted spread onto non-selective 50% artificial sea water f/2, 1% agar plates for three weeks. Several colonies randomly picked were selected on 50% artificial sea water f/2, 1% agar plates with and without zeocin (50 μg ml-1). The colonies without conjugative vector failed to grow on the agar plate with zeocin. Finally, such transgene-free colonies were verified by performing PCR using vector specific primers Ble_F/Ble_R to screen for the loss of resistance gene in transgene-free strains and DNA sequencing. All primers used in this study are listed in Supplemental Table S1. To expel the off-target activity of the selected CRISPR sequence, we also performed BLASTN against P. tricornutum genomic sequence (ASM15095v2) and use the CRISPR-offinder (Zhao et al., 2017) as the searching tool for the off-targets of the Phatr3_J9619 throughout the genome (ASM15095v2) (Supplemental Table S3). All constructs were confirmed by DNA sequencing.
Subcellular Localization and Transmission Electron Microscopy
Both Wt and transformants at the exponential growth phase with eGFP were examined using the Zeiss AxioScope A1 microscope (Carl Zeiss, Oberkochen, Germany) (FITC filter; excitation and emission wavelengths of 488 nm and 500–550 nm, respectively, for eGFP; excitation and emission wavelengths of 488 nm and 650–750 nm, respectively, for chlorophyll). Cells at the exponential growth phase were collected to examine the plastid ultrastructure, then processed for transmission electron microscopy imaging (Hitachi, Tokyo, Japan) as previously described (Chen et al., 2018).
Oxygen Evolution Rate Detection
The Wt and mutant P. tricornutum cells (5 × 106 cells ml-1) at the exponential phase were collected to calculate the oxygen evolution rate. The chlorophyll a content was determined as described (Jeffrey and Humphrey, 1975) and the oxygen evolution rate of intact cells in a f/2 medium containing 10 mM NaHCO3 was measured using a Clark-type oxygen electrode (Hanstech Instruments, Ltd., Norfolk, England).
Lipid Analysis
The Wt and mutant P. tricornutum cells (approx. 2 × 108 cells) were harvested by centrifugation at 3000 × g for 10 min, stored at −80°C, and lyophilized for the subsequent lipid extraction and analysis. Total lipids were extracted from 200 mg lyophilized cells (Bligh and Dyer, 1959) and total fatty acids were quantified by gas–liquid chromatography as previously described (Hao et al., 2018). Fatty acids of the total lipids from the Wt and mutants were quantified by gas–liquid chromatography as previously described (Hao et al., 2018). Total lipids were also separated into neutral lipid (NL), glycolipid (GL), and phospholipid (PL) by solid-phase extraction according to Liu et al. (2011) using a 500 mg Sep-Pak™ silica gel cartridge (Waters, Milford, USA).
Global Lipidomics Analysis by Liquid Chromatography With Tandem Mass Spectrometry (LC-MS/MS)
The Wt and mutant P. tricornutum lipids were extracted according to the method developed by Bligh and Dyer (1959). Samples were analyzed using the Waters 2D UPLC system (Waters, Milford, USA) coupled to the Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, USA) with a heated electrospray ionization source. The analysis was controlled using the Xcalibur 2.3 program (Thermo Fisher Scientific). Chromatographic separation was performed on a Waters ACQUITY UPLC CSH C18 column (1.7 μm, 2.1 mm × 100 mm, Waters), and the column temperature was maintained at 55°C. The mobile phase consisted of acetonitrile/water (60:40, v:v), mixed with 10 mM ammonium formate and 0.1% formic acid (A) and isopropanol/acetonitrile (90:10, v:v), mixed with 10 mM ammonium formate and 0.1% formic acid (B) in the positive mode, and in the negative mode, acetonitrile/water (60:40, v:v), mixed with 10 mM ammonium formate (A) and isopropanol/acetonitrile (90:10, v:v), mixed with 10 mM ammonium formate (B). The gradient conditions were as follows: 0-2 min, 40% to 43% B; 2-2.1 min, 43% to 50% B; 2.1-7 min, 50% to 54% B; 7-7.1 min, 54% to 70% B; 7.1 to 13 min,70% to 99% B, 13 to 13.1 min, 99% to 40% B and 13.1-15 min, 40% B. The flow rate was 0.35 mL/min and the injection volume was 5 μL. The mass spectrometric settings for positive/negative ionization modes were as follows: spray voltage, 3.2-3.8 kV; sheath gas flow rate, 40 arbitrary units (arb); aux gas flow rate, 10 arb; aux gas heater temperature, 350°C; capillary temperature, 320°C. The full scan range was 200–2000 m/z with a resolution of 70000, and the automatic gain control (AGC) target for MS acquisitions was set to 3e6 with a maximum ion injection time of 100 ms. Top 3 precursors were selected for subsequent MS/MS fragmentation with a maximum ion injection time of 50 ms and resolution of 17500, the AGC was 1e5. The stepped normalized collision energy was set to 15, 30 and 45 eV. Finally, LC-MS/MS raw data were analyzed and the putative identification of the different lipid species were processed and validation with LipidSearch v.4.1 (Thermo Fisher Scientific). Relative lipid levels were normalized by probabilistic quotient normalization. Student’s t-test was used to determine statistically significant differences of the fatty acid distribution among the MGDG lipid forms. Significance was determined at p < 0.05.
Results and Discussion
Characterization of MGDG Synthase Genes in P. tricornutum
Three homologous genes (Phatr3_J54168, Phatr3_J9619, and Phatr3_J14125) encoding MGDG synthase (ptMGD) were annotated in the P. tricornutum genome. On the basis of recent findings (Dolch et al., 2017), we designated these three MGD genes as ptMGD1, ptMGD2, and ptMGD3, respectively. A phylogenetic analysis revealed all three ptMGD genes are closely related to the corresponding genes in higher plants, but they belong to a separate clade (Supplementary Figure S1A). The MGDs in P. tricornutum diverged from the related enzymes in other photosynthetic organisms, but they share several amino acids, including P189, W287, and C291, that are highly conserved in plant MGDs in the N-domain and play a specific role in lipid binding (Botté et al., 2005; Dubots et al., 2010) (Supplementary Figure S1B). Hence, ptMGDs may have functions in MGDG biosynthesis and other processes that are similar to those of plant MGDs (Jarvis et al., 2000; Awai et al., 2001; Dubots et al., 2010; Kobayashi et al., 2014).
MGDG has been found to play vital roles in photosynthesis (Kalisch et al., 2016) as well as in response to several adverse environmental conditions (Qi et al., 2004), including phosphorous deficiency (Kobayashi et al., 2009a) and salt stress (Wang et al., 2014). We first detected the relative transcriptional levels of the three ptMGD genes in this study and found that ptMGD2 (Phatr3_J9619, which is also subsequently shortened to 9619) was most expressed during logarithmic growth phase (Figure 1A). Next, we also detected that ptMGD2 is salt-stress and phosphate-deprivation inducible (Figures 1B, C), implying ptMGD2 catalyzes MGDG synthesis during an exposure to various stresses. This is consistent with the salinity-induced expression patterns in plants (Qi et al., 2004). It has been reported that ptMGD3 (Phatr3_J14125) has no expression (Dolch et al., 2017) and ptMGD2 (the predicted MGDG synthases designated as MGD3 in the original paper) was up-regulated under phosphorus limitation (Huang et al., 2019). These results above suggest that ptMGD2 may contribute to the maintenance of cellular homeostasis and adaptations to the changing environments in P. tricornutum. Therefore, we mainly focused on ptMGD2 biological functions and its underlying mechanisms in this research.
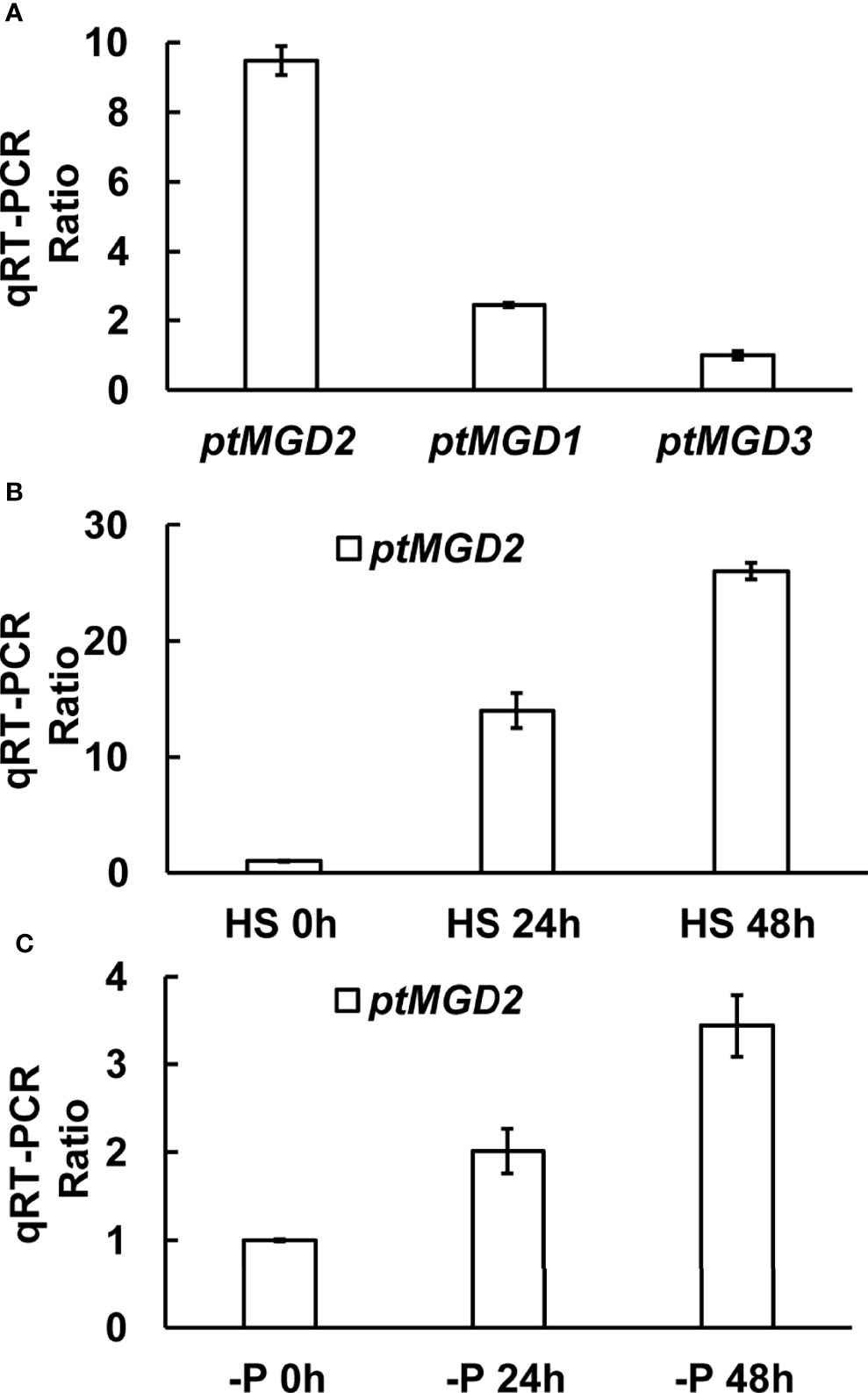
Figure 1 The expression profile of ptMGD transcripts. (A) The expression levels of ptMGD genes under normal growth condition. (B, C) qRT-PCR was performed detect the ptMGD2 expression under high salt stress and phosphate-deprivation, respectively. HS indicates high salinity and -P represents phosphate-deprivation. All values represent means standard deviations of three biological repeats.
Subcellular Localization of ptMGD2
As there is no obvious N-terminal signal peptide predicted by SignalP 5.0 (Almagro Armenteros et al., 2019) in ptMGD2, we examined the subcellular localization of ptMGD2 by constructing a fusion protein containing eGFP. Similar to the MGDs in other photosynthetic organisms (Awai et al., 2001; Basnet et al., 2019), we detected substantial amounts of ptMGD2 in plastids, reflecting this protein’s participation in the synthesis of the MGDG required for photosynthesis (Figure 2A). Furthermore, our analysis of protein–protein interactions by immunoprecipitation indicated that most of the candidate proteins that interact with ptMGD2 are related to photosynthetic processes (e.g., chlorophyll a/b-binding proteins and protein fucoxanthin chlorophyll a/c protein) (Supplementary Table S2). This provides additional evidence of the plastid localization of ptMGD2, which is very likely in the thylakoid membrane. Notably, in a few cells lacking intact plastids (<0.1%), ptMGD2 was detected in the cytoplasm under normal growth conditions (Figure 2B). Compared to the negtive control (Figure 2C). This finding indicates that the ptMGD2 located in cytoplasm may play roles in response of certain stimuli in P. tricornutum.
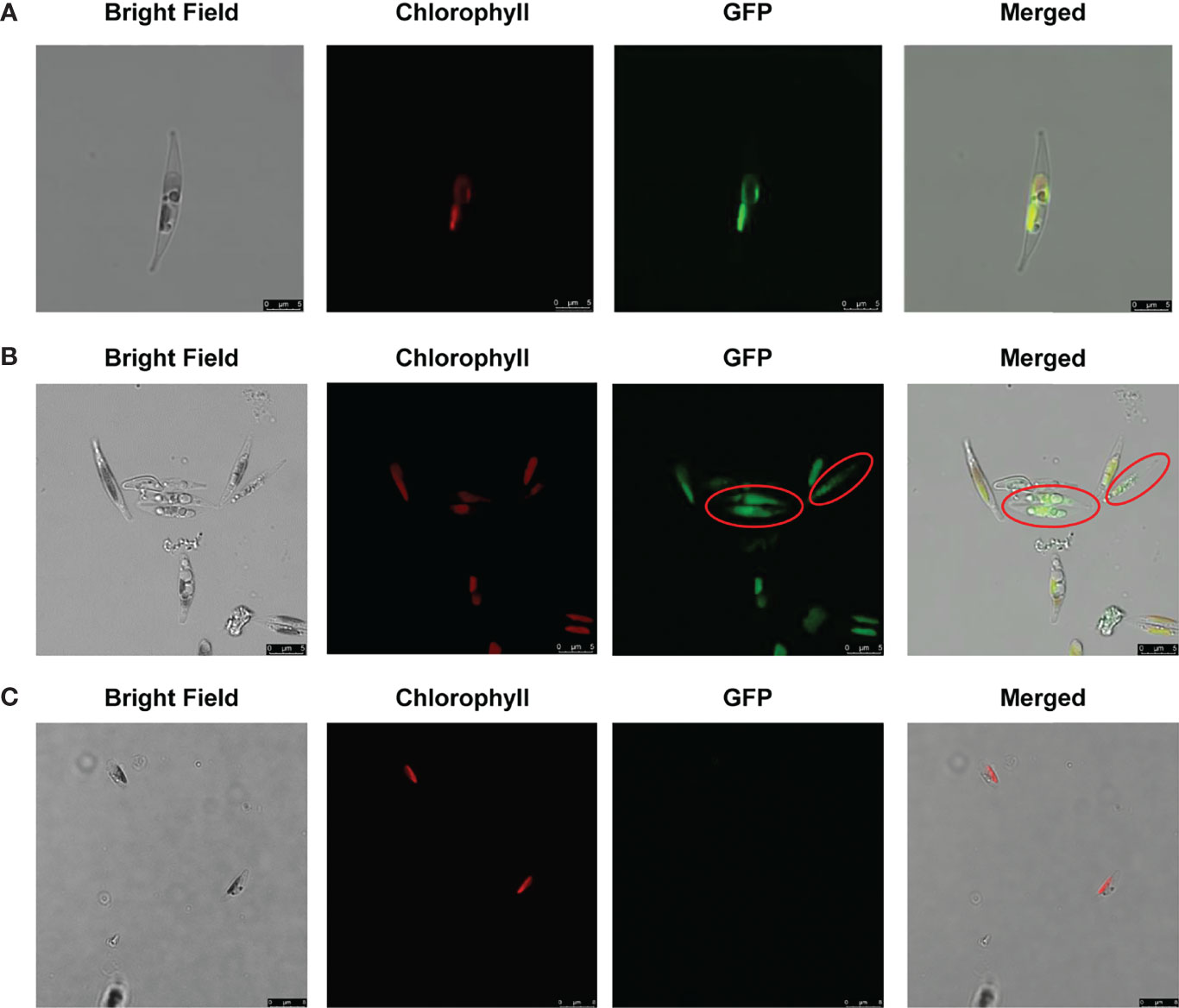
Figure 2 Localization of full-length ptMGD2: eGFP fusion protein expressed in plastids (A) and cytoplasm (B) in P. tricornutum using wild type (Wt) as the negative control (C). Bright field, light microscopical images; chlorophyll, chlorophyll auto-fluorescence; eGFP, GFP fluorescence; merged, merged channel. The scale bar represents 5 μm. Red circles indicate the cytoplasmic subcellular location.
Generation of Two ptMGD2 Mutant Strains
Two different PAM-target sites with low homology to other genomic loci were designed targeting the coding sequence of the ptMGD2 gene in this study. Unfortunately, we failed to obtain knockout strains with transgene-free ptMGD2 mutations for the PAM2 target site. The CRISPR/Cas9-based mutagenesis involving the PAM1 target site in the first exon of ptMGD2 generated two distinct mutations, including 13-bp and 20-bp deletions (Figure 3) (Supplementary Figures S2, S4). Compared with the wild type (Wt), these two mutations resulted in the early termination of protein translation and further led to a lack of glycosyl transferase activity. Moreover, we screened the two CRISPR/Cas9-free homozygous lines by subcloning to assess the possibility of an off-target effect in the following generation as previously described (Sharma et al., 2018). Many off-target proof Cas9 nickase systems have been established in diatoms (Nawaly et al., 2020). To expel the off-target activity of the selected CRISPR sequence, we performed BLASTN against P. tricornutum genomic sequence and found only one match point of Phatr3_J9619. Then, we used the CRISPR-offinder (Zhao et al., 2017) for the off-targets of Phatr3_J9619 throughout the P. tricornutum genome (Supplemental Table S3) and sites of four and five mismatches were confirmed to be exactly the same between Wt and mutants by DNA sequencing (Supplementary Figure S3). These mutants were used for our in-depth investigation of the potential roles for ptMGD2 in galactolipid metabolism and other biological processes.
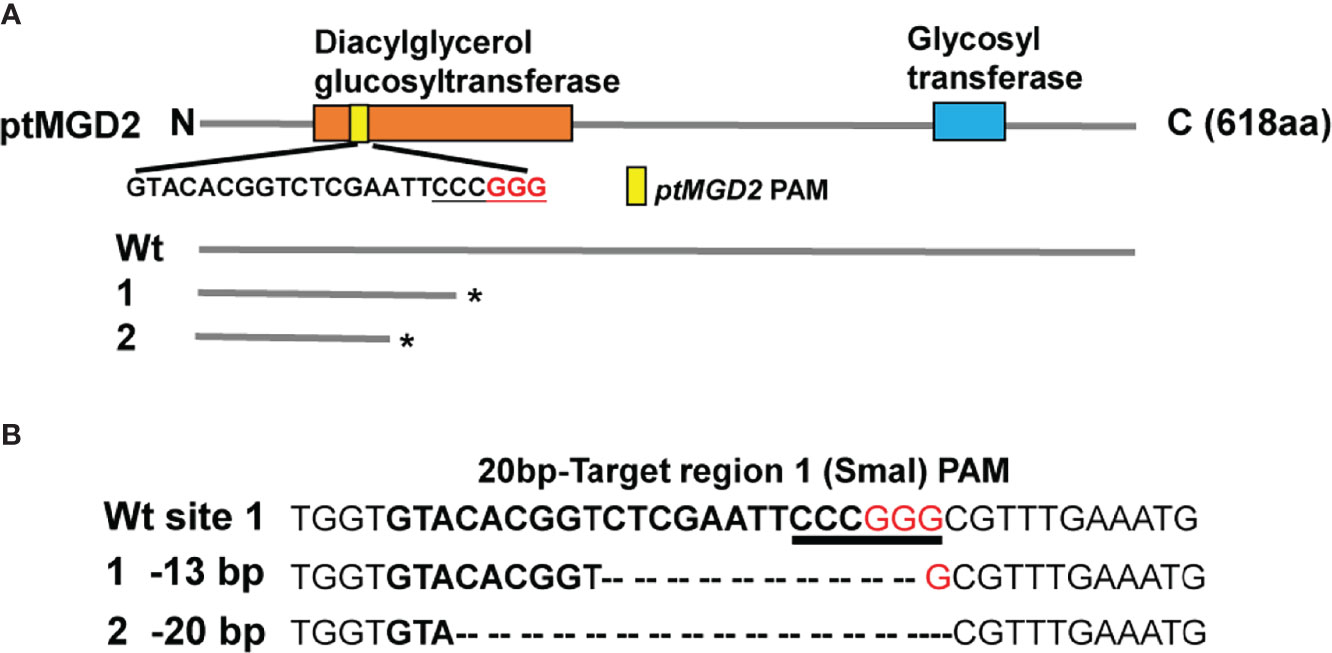
Figure 3 Targeted mutagenesis of ptMGD2 using CRISPR/Cas9 system. (A) Domain architecture of the ptMGD2 protein. The domain analysis was performed with InterProScan and different domains are represented by different colors. The protein lengths are displayed on the right. The nucleotide sequence of the PAM1 target site in the first exon of ptMGD2 is also shown. Compared with wild type (Wt), these two mutations resulted in the early termination of protein translation at Diacylglycerol glucosyltransferase domain. (B) The CRISPR/Cas9-based mutagenesis of ptMGD2 were designed targeting the coding sequence of the ptMGD2 and two mutants were detected by SmaI digestion and DNA sequencing. DNA sequences of the target region with PAM and base pairing sequence of sgRNA are shown in red and bold between Wt and mutants. *The early termination of protein translation.
Effects of the ptMGD2 Mutations on Growth Rate, Oxygen Evolution and Thylakoid Membrane
The Wt control and the ptMGD2 knockout lines were compared regarding their growth rates under normal conditions. The ptMGD2 knockout mutants grew more slowly than the Wt cells (Figure 4A), which is consistent with the effects of MGD deficiency in rice (Basnet et al., 2019). To further explore the potential function of the other two MGDs, we have performed qRT-PCR to measure the mRNA abundance of these MGDs in the ptMGD2 knockout mutants. Upregulation of ptMGD3 in these mutants indicates the ptMGD3 may partially compensate the function of ptMGD2 in diatom (Figure 4B). These findings reflect the importance of galactolipid metabolism mediated by ptMGD2 for the normal growth of this model diatoms. We also examined and compared the growth rate of Wt with mutants under high saline and phosphate deficiency condition (Supplemental Figure S5). These findings revealed that the ptMGD2 mutant cells was more sensitive than Wt cells responding to such conditions, indicating the ptMGD2 gene confers tolerance to environmental stresses.
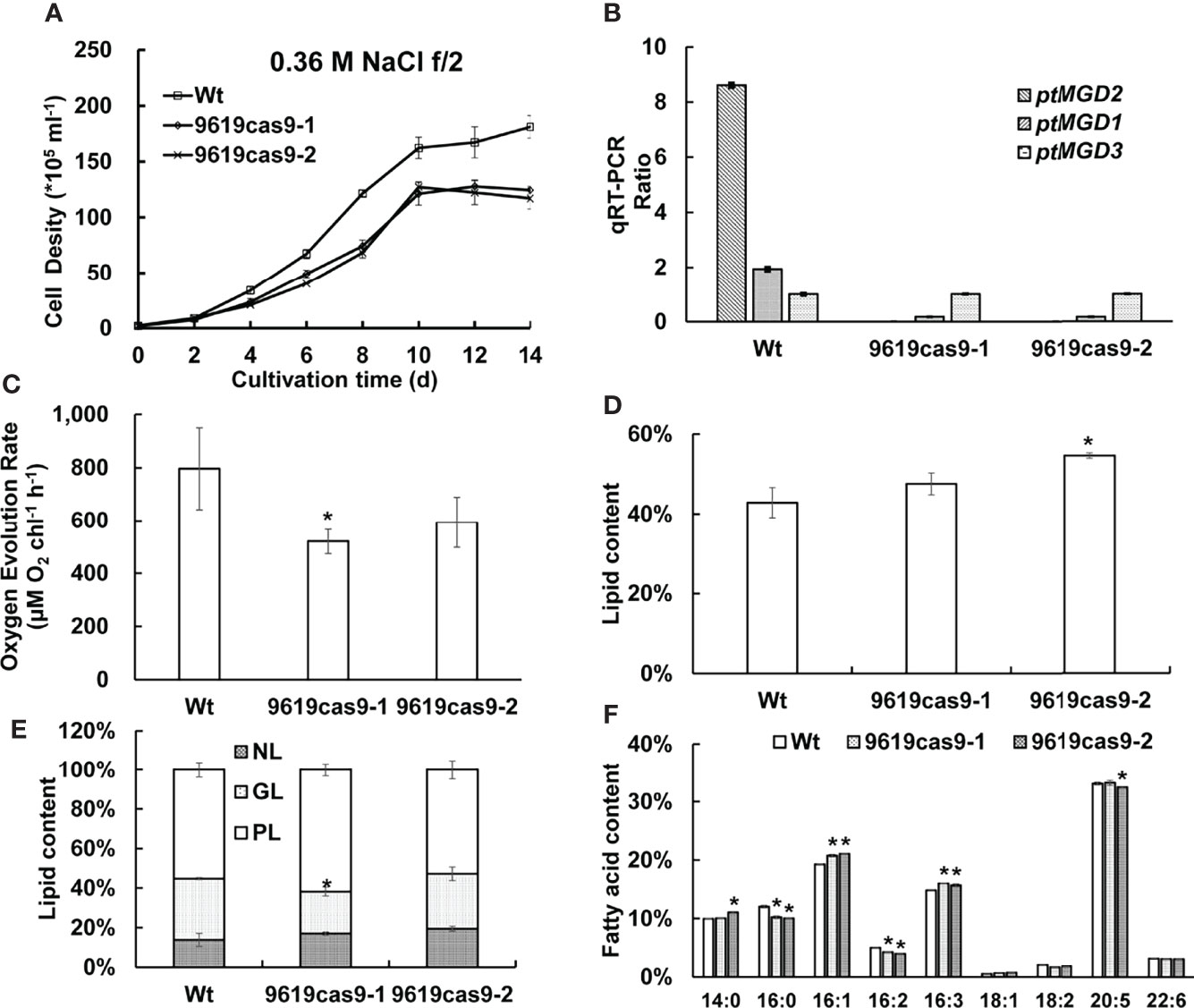
Figure 4 Effect of the ptMGD2 gene inactivation on cell phenotype analyses. The Wt and the knockout lines were compared regarding their growth rates (A) and qRT-PCR ratio of the mRNA abundance of three MGDs in Wt and the ptMGD2 knockout mutants under normal conditions (B). Analysis of oxygen evolution rate (C), total lipid content (D), lipid class ratio (E), fatty acid content (F) from Wt and the ptMGD2 mutants were performed. NL, neutral lipid; GL, glycolipid; PL, phospholipid. All values represent means ± standard deviations of three biological repeats. Asterisk represents statistically significant differences between wild-type and mutants based on Student’s t test (*p < 0.05).
Chlorophyll a is the main photosynthetic pigment directly involved in the electron transfer during the light transformation reaction (Yahia et al., 2019). A consequence of a decrease in the galactolipid content is chlorophyll depletion (Kobayashi et al., 2009b). Thus, we compared the chlorophyll a content of the Wt and mutant cells during the exponential growth phase. The data indicated the chlorophyll a level decreased significantly by 20.86% and 24.90% in the ptMGD2 mutants (5.72 and 5.43 vs 7.23 mg mL-1 in Wt), which was similar to the effects of mutations to the corresponding genes in some plant species (Kobayashi et al., 2007; Kobayashi et al., 2014). As with previous results, the loss of MGD has detrimental effects on oxygen evolution and photosynthesis (Basnet et al., 2019). We subsequently examined the differences in the photosynthetic rate between the Wt and mutant cells. Along with the delayed cellular growth, the net photosynthetic oxygen evolution rate of P. tricornutum was 34.61% and 25.47% lower in the two mutants (520.33 and 593.04 μmol O2 chl-1 h-1) than in the Wt (795.71 μmol O2 chl-1 h-1) control during the exponential growth stage as seen in Figure 4C. A similar phenotype was observed in earlier related investigations on plants (Basnet et al., 2019). Crystallographic studies on P. tricornutum revealed that galactolipid molecules functioning as cofactors are present in the PSII–FCPII supercomplex (Pi et al., 2019). Hence, an MGDG deficiency in the PSII reaction center may have led to suppressed oxygen evolution in the two mutants.MGDG is vital for the biogenesis of the photosynthetic membranes and the light reactions of photosynthesis (Myers et al., 2011). To assess the effects of a defective ptMGD2 on plastid development, we compared the plastids of Wt and mutant cells by transmission electron microscopy (Figure 5), which detected differences in the photosynthetic membranes among these cells. More specifically, the Wt cells had normal mature plastids and stacked thylakoid membranes, whereas the plastids in the cells of the two ptMGD2 mutants were underdeveloped with abnormal structures (88.1% and 89.7% in mutants vs 12.5% in Wt), similar to the plastids of plants with mutations to MGD-encoding genes (Jarvis et al., 2000; Fujii et al., 2014). Thus, ptMGD2 is indispensable for thylakoid membrane biogenesis. These results are indicative of the essential role played by ptMGD2 during normal thylakoid membrane biogenesis in P. tricornutum. All these observations provide strong evidence that ptMGD2 contributes to normal growth, plastid biogenesis and photosynthesis.
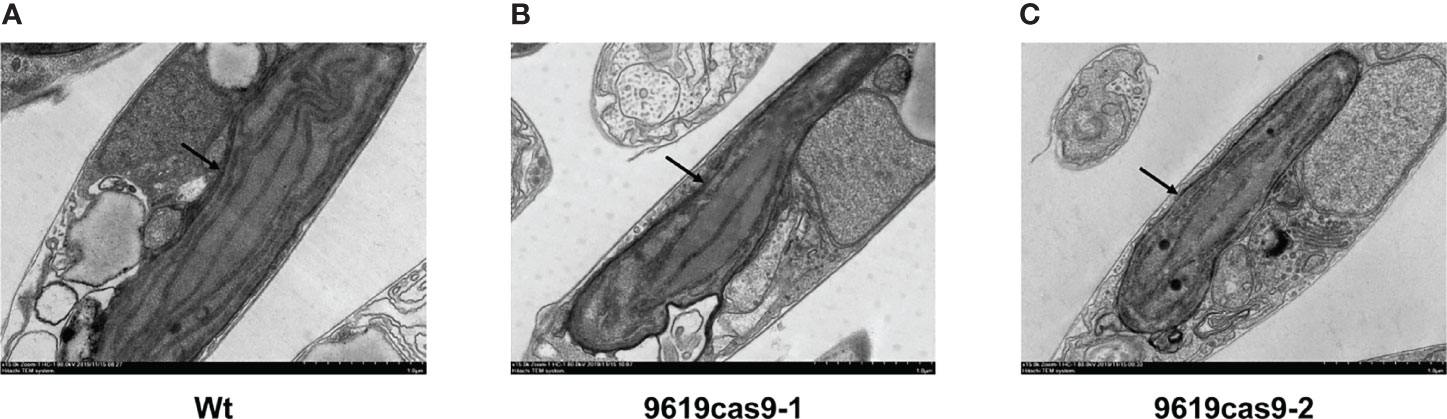
Figure 5 Ultrastructure of thylakoid membranes in plastids from Wt (A) and the ptMGD2 mutants (B, C). The arrows indicate the underdeveloped structures of mutants. Bar = 1.0 μm.
Effects of the ptMGD2 Mutations on Total Lipid Content, Lipid and Fatty Acid Profiles, and MGDG Biosynthesis
Cell division arrest or delay during stress responses in P. tricornutum may be accompanied by the accumulation of lipids (Dolch et al., 2017). To determine the total amount of lipids accumulated in diatom cells, we quantified the lipids abundance. The lipid content per cell was 2.2-fold higher in one of the ptMGD2 mutants (9619cas9-2) than in the Wt control and the other mutant (9619cas9-1) (Figure 4D). To further investigate the differential lipid profiles between the Wt and ptMGD2 mutant cells, we separated the total lipids from the cells collected in the late exponential growth phase into neutral lipid (NL), glycolipid (GL) and phospholipid (PL) (Figure 4E). In the ptMGD2 mutant cells, the photosynthetic membrane lipid (i.e., GL) accounted for only 27.78% and 21.28% of the total lipid content, which was less than the corresponding percentage for the Wt cells (31.03%). Thus, knocking out ptMGD2 (9619cas9-1) clearly decreased the GL content. The storage lipid (i.e., NL) accounted for only 17.02% and 19.44% of the total lipid content in the two mutant strains, which were slightly higher than the corresponding percentage in the Wt cells (13.79%). In contrast, the membrane lipid (i.e., PL) accounted for 61.70% and 52.78% of the total lipid content in the mutant cells, whereas they represented 55.17% of the total lipid content in the Wt cells. Thus, mutant cells exhibited a discrepancy of lipid proportions and GL in these strains may channel carbon for the storage of other lipids. We speculated that the carbon flux may be directed toward other lipid pathways unrelated to triacylglycerol (TAG) accumulation in the 9619cas9-1 mutant.
We also compared the lipid fatty acid profiles among the Wt and mutant cells (Figure 4F). The major fatty acids in P. tricornutum cells are C14, C16, C18, C20, and C22, which is similar to previously published results (Hao et al., 2018). Compared with the Wt control, both mutant strains had less C16:0, C16:2, and C18:2, but more C16:1, C16:3, and C18:1. The differences in the abundance of the other fatty acids between the mutant and Wt cells were insignificant. Considering the decrease in the C16:0 and C16:2 amounts in the two ptMGD2 mutants, we speculated that these fatty acids may serve as important precursors of MGDG in P. tricornutum.
Lipidomic Profiling of the Wt and Mutant Cells
In the current study, we examined the lipidomic profiles of the Wt and mutant cells under normal growth conditions (Supplemental Table S4 Sheet 1) (Figure 6). According to the lipid profiles, the most differentially abundant lipids between the Wt and mutant cells were MGDG and DGDG. Along with the decrease in the MGDG content, we also observed a considerable decrease in the DGDG level in the mutants. More specifically, compared with the Wt control, there was 31.26% and 21.58% less MGDG and 65.71% and 72.89% less DGDG in the two mutants, suggesting ptMGD2 has an essential role related to MGDG anabolism in diatom cells. Accordingly, because it is a substrate for DGDG anabolism, the sharp decline in the MGDG content in the ptMGD2 mutants may result in a drastic decrease in the DGDG level in these cells. Similar concomitant decreasing trends in the MGDG and DGDG contents have been reported for plants (Basnet et al., 2019). Therefore, the decrease in the MGDG level in cells indicated that the inactivation of MGD adversely affects MGDG biosynthesis in P. tricornutum.
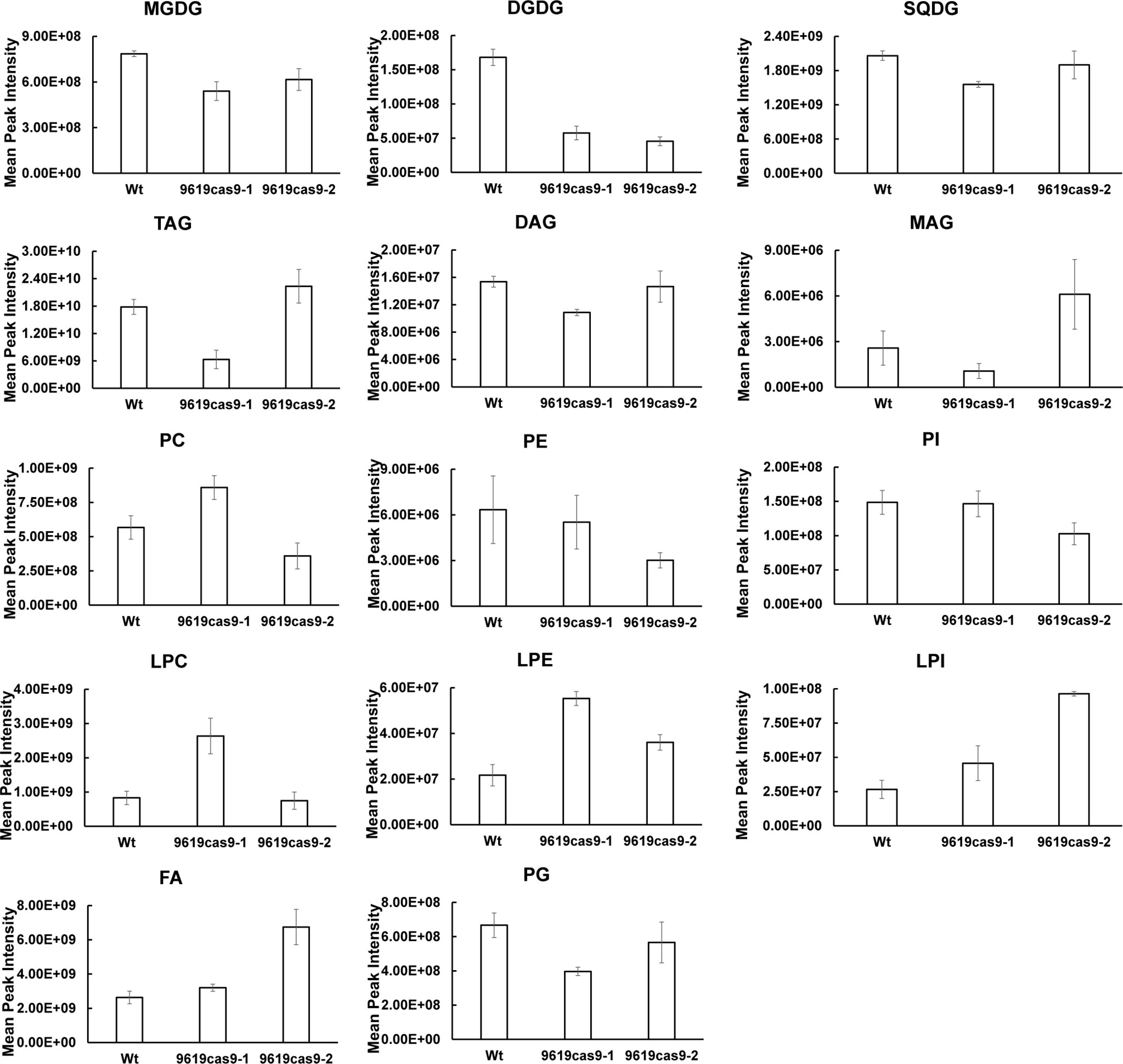
Figure 6 Major Lipidomic Profiling of Wt and the ptMGD2 mutants. MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; TAG, triacylglycerol; DAG, diacylglycerol; MAG, monoacylglycerols; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; FA, free fatty acids; PG, phosphatidylglycerol.
The decreased abundance of MGDG and DGDG in the mutants was accompanied by content changes to other major lipids. Consistent with the total lipid contents, the 9619cas9-2 mutant produced more TAG (1.25-fold) than the Wt control. The large amounts of monoacylglycerols (MAG) and fatty acids (FA) in the 9619cas9-2 mutant may be key metabolites for TAG biosynthesis. The 9619cas9-1 mutant accumulated lysophosphatidylethanolamine (LPE), lysophosphatidylcholine (LPC), and phosphatidylcholine (PC), reflecting a carbon flow from GL to PC and its precursor. Earlier research revealed the increase in phosphatidylethanolamine and PC quantities in the shoots of the Arabidopsis mgd2 mgd3 double mutant in response to phosphorus starvation, but a lack of changes to phosphatidylglycerol and phosphatidylinositol contents (relative to the Wt levels) (Kobayashi et al., 2009a). Additionally, the abundance of nonplastidial lipids reportedly increases following the complementation of an Arabidopsis MGD mutant (mgd1-2) by C. tepidum MgdA (Masuda et al., 2011). Together with the increased lipid content, our data suggest that the 9619cas9-1 mutant cells may be more useful as a feedstock for biodiesel production than the 9619cas9-2 mutant cells (Supplemental Figure S6).In this study, we also characterized the MGDG fatty acid profiles. The fatty acid distribution among the MGDG lipid forms differed from that determined by previous research (Abida et al., 2015). This may be associated with the diversity in the nutrient ratios and sampling time-points (i.e., cultivation stages) between studies. Of these fatty acids (Supplemental Table S4 Sheet 2), the C16:1 and C16:0 contents decreased in the mutant cells (Figure 7A). To more precisely elucidate the MGDG fatty acid composition, we analyzed the MGDG lipid forms in the Wt and mutant cells (Supplemental Table S4 Sheet 3) (Figure 7B). The differences in the MGDG lipid forms between the mutants and the Wt control included the following: MGDG (14:0/16:1), MGDG (16:1/16:1), MGDG (16:0/16:1), MGDG (16:3/16:3), and MGDG (14:0/16:3). Previous studies proved that plastid-derived DAG contains C16 fatty acids at the sn-2 position. Combined with our findings, we demonstrated that loss of ptMGD2 primarily affects MGDG (14:0/16:1), MGDG (16:1/16:1), and MGDG (16:0/16:1), which may be generated within plastids. As one of the major components of galactolipids, polyunsaturated fatty acids influence chloroplast functions to maintain photosynthetic activity (Mcconn and Browse 1998; Selstam, 1998). The results of this study indicate C16 fatty acids are crucial for normal plastid functions. Future biochemical investigations should further analyze the related endogenous enzymatic reactions.
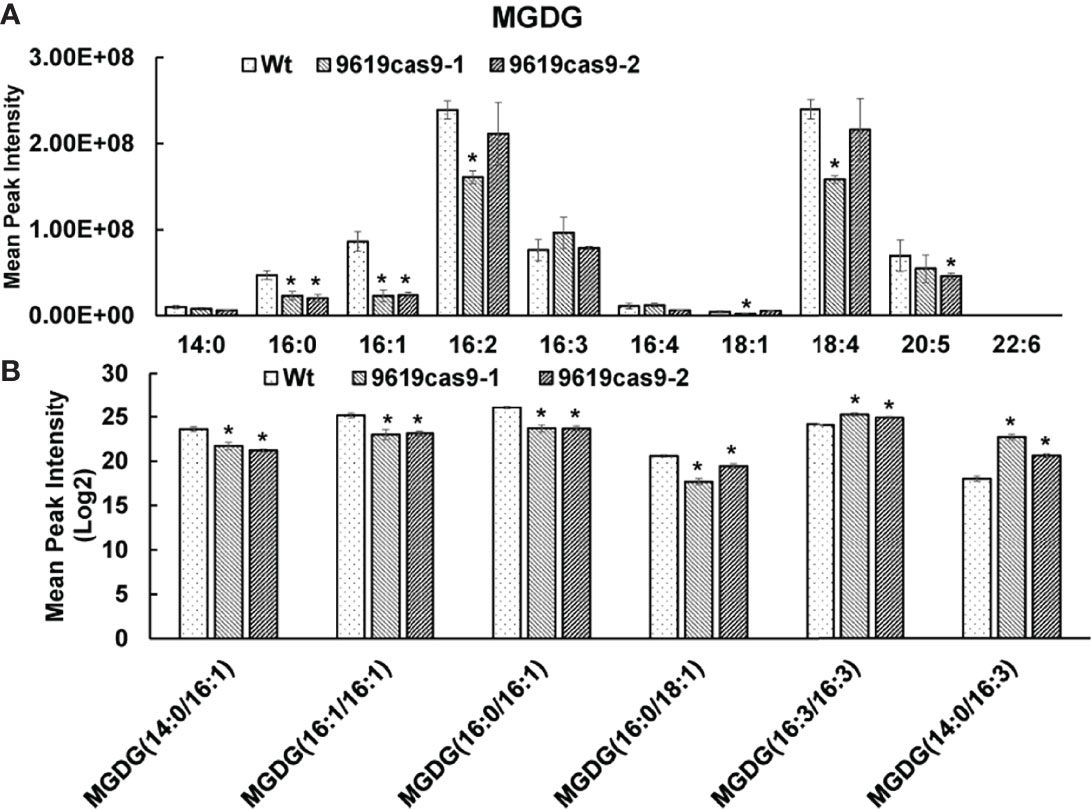
Figure 7 Analysis of the distribution of fatty acids (A) and the fatty acid profiles in the MGDG lipid classes among Wt and mutants (B). All values represent means ± standard deviations of four biological repeats. Asterisk represents statistically significant differences between wild-type and mutants based on Student’s t test (*p < 0.05).
Conclusion
Taken together, by combining genetic and phenotypic analyses of Wt and mutant cells, we elucidated the multifunctional role of ptMGD2 in P. tricornutum. Disruption of ptMGD2 employing the CRISPR/Cas9 system resulted in retarded growth, damaged thylakoid membranes, inhibited oxygen evolution, decreased MGDG biosynthesis, and lipid remodeling. Interestingly, compared with the Wt control, one mutant had a higher TAG content, whereas another mutant contained more PL. The former with the increased TAG abundance may be a genetically engineered strain useful for biotechnology-based production of biofuel. Also, ptMGD2 contributes to the diatom adaptations to adverse environments, such as high saline and phosphate deficiency. Future studies may focus on clarifying the molecular mechanism underlying the stress response mediated by ptMGD2.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
ZC designed research and wrote the manuscript; ZC, SS, RL, LL, XL, and SZ performed research; YZ, PZ, and BW made revisions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31970367, 31600286, 31500622, 31800304), The Key Laboratory of Algal Biology of Chinese Academy of Sciences, the program of Wuhan University Science and technology (No. 25011401).
Conflict of Interest
Author LL was employed by Wuhan Institute of Biological Products Co., Ltd., Wuhan, China.The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.874448/full#supplementary-material
References
Abida H., Dolch L. J., Mei C., Villanova V., Conte M., Block M. A., et al. (2015). Membrane Glycerolipid Remodeling Triggered by Nitrogen and Phosphorus Starvation in Phaeodactylum Tricornutum. Plant Physiol. 167, 118–136. doi: 10.1104/pp.114.252395
Almagro Armenteros J. J., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Awai K., Marechal E., Block M. A., Brun D., Masuda T., Shimada H., et al. (2001). Two Types of MGDG Synthase Genes, Found Widely in Both 16:3 and 18:3 Plants, Differentially Mediate Galactolipid Syntheses in Photosynthetic and Nonphotosynthetic Tissues in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 10960–10965. doi: 10.1073/pnas.181331498
Basnet R., Zhang J., Hussain N., Shu Q. (2019). Characterization and Mutational Analysis of a Monogalactosyldiacylglycerol Synthase Gene Osmgd2 in Rice. Front. Plant Sci. 10, 992. doi: 10.3389/fpls.2019.00992
Benning C., Ohta H. (2005). Three Enzyme Systems for Galactoglycerolipid Biosynthesis are Coordinately Regulated in Plants. J. Biol. Chem. 280, 2397–2400. doi: 10.1074/jbc.R400032200
Bligh E. G., Dyer W. J. (1959). A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Block M. A., Dorne A. J., Joyard J., Douce R. (1983). Preparation and Characterization of Membrane Fractions Enriched in Outer and Inner Envelope Membranes From Spinach Chloroplasts. II. Biochemical Characterization. J. Biol. Chem. 258, 13281–13286. doi: 10.1007/978-94-017-4973-2_5
Botté C., Jeanneau C., Snajdrova L., Bastien O., Imberty A., Breton C., et al. (2005). Molecular Modeling and Site-Directed Mutagenesis of Plant Chloroplast Monogalactosyldiacylglycerol Synthase Reveal Critical Residues for Activity. J. Biol. Chem. 280, 34691–34701. doi: 10.1074/jbc.M505622200
Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., et al. (2008). The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 456, 239–244. doi: 10.1038/nature07410
Chen Z., Luo L., Chen R., Hu H., Pan Y., Jiang H., et al. (2018). Acetylome Profiling Reveals Extensive Lysine Acetylation of the Fatty Acid Metabolism Pathway in the Diatom Phaeodactylum Tricornutum. Mol. Cell. Proteomics 17, 399–412. doi: 10.1074/mcp.RA117.000339
Chi H., Liu C., Yang H., Zeng W. F., Wu L., Zhou W. J., et al. (2018). Open-Pfind Enables Precise, Comprehensive and Rapid Peptide Identification in Shotgun Proteomics. Nature Biotechnol. 36. doi: 10.1038/nbt.4236
Cook R., Lupette J., Benning C. (2021). The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 10, 706. doi: 10.3390/cells10030706
Daboussi F., Leduc S., Marechal A., Dubois G., Guyot V., Perez-Michaut C., et al. (2014). Genome Engineering Empowers the Diatom Phaeodactylum Tricornutum for Biotechnology. Nat. Commun. 5, 3831. doi: 10.1038/ncomms4831
De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene Silencing in the Marine Diatom Phaeodactylum Tricornutum. Nucleic Acids Res. 37, e96. doi: 10.1093/nar/gkp448
Dolch L. J., Lupette J., Tourcier G., Bedhomme M., Collin S., Magneschi L., et al. (2017). Nitric Oxide Mediates Nitrite-Sensing and Acclimation and Triggers a Remodeling of Lipids. Plant Physiol. 175, 1407–1423. doi: 10.1104/pp.17.01042
Dörmann P., Benning C. (2002). Galactolipids Rule in Seed Plants. Trends Plant Sci. 7, 112–118. doi: 10.1016/s1360-1385(01)02216-6
Dubots E., Audry M., Yamaryo Y., Bastien O., Ohta H., Breton C., et al. (2010). Activation of the Chloroplast Monogalactosyldiacylglycerol Synthase MGD1 by Phosphatidic Acid and Phosphatidylglycerol. J. Biol. Chem. 285, 6003–6011. doi: 10.1074/jbc.M109.071928
Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. (1999). Transformation of Nonselectable Reporter Genes in Marine Diatoms. Mar. Biotechnol. 1, 239–251. doi: 10.1007/pl00011773
Falkowski P. G., Barber R. T., Smetacek V. V. (1998). Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science 281, 200–207. doi: 10.1126/science.281.5374.200
Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P. (1998). Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Fujii S., Kobayashi K., Nakamura Y., Wada H. (2014). Inducible Knockdown of MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE1 Reveals Roles of Galactolipids in Organelle Differentiation in Arabidopsis Cotyledons. Plant Physiol. 166, 1436–1449. doi: 10.1104/pp.114.250050
Ge F., Huang W. C., Chen Z., Zhang C. Y., Xiong Q., Bowler C., et al. (2014). Methylcrotonyl-CoA Carboxylase Regulates Triacylglycerol Accumulation in the Model Diatom Phaeodactylum Tricornutum. Plant Cell 26, 1681–1697. doi: 10.1105/tpc.114.124982
Guillard R. R. L. (1975). “Culture of Phytoplankton for Feeding Marine Invertebrates,” in Culture of Marine Invertebrate Animals. Eds. Smith W. L., Canley M. H. (New York: Plenum Press), 29–60.
Hao X., Luo L., Jouhet J., Rebeille F., Marechal E., Hu H., et al. (2018). Enhanced Triacylglycerol Production in the Diatom Phaeodactylum Tricornutum by Inactivation of a Hotdog-Fold Thioesterase Gene Using TALEN-Based Targeted Mutagenesis. Biotechnol. Biofuels 11, 312. doi: 10.1186/s13068-018-1309-3
Hempel F., Maurer M., Brockmann B., Mayer C., Biedenkopf N., Kelterbaum A., et al. (2017). From Hybridomas to a Robust Microalgal-Based Production Platform: Molecular Design of a Diatom Secreting Monoclonal Antibodies Directed Against the Marburg Virus Nucleoprotein. Microb. Cell Fact. 16, 131. doi: 10.1186/s12934-017-0745-2
Hölzl G., Dörmann P. (2007). Structure and Function of Glycoglycerolipids in Plants and Bacteria. Prog. Lipid Res. 46, 225–243. doi: 10.1016/j.plipres.2007.05.001
Huang B., Marchand J., Blanckaert V., Lukomska E., Ulmann L., Wielgosz-Collin G., et al. (2019). Nitrogen and Phosphorus Limitations Induce Carbon Partitioning and Membrane Lipid Remodelling in the Marine Diatom Phaeodactylum Tricornutum. Eur. J. Phycol. 54, 342–358. doi: 10.1080/09670262.2019.1567823
Jarvis P., Dörmann P., Peto C. A., Lutes J., Benning C., Chory J. (2000). Galactolipid Deficiency and Abnormal Chloroplast Development in the Arabidopsis MGD Synthase 1 Mutant. Proc. Natl. Acad. Sci. U.S.A. 97, 8175–8179. doi: 10.1073/pnas.100132197
Jeffrey S. W., Humphrey G. F. (1975). New Spectrophotometric Equations for Determining Chlorophylls a, B, C1, and C2 in Higher Plants, Algae and Natural Phytoplankton. Plant Physiol. Bioch. 8, 53–59. doi: 10.1016/0022-2860(75)85046-0
Kalisch B., Dörmann P., Holzl G. (2016). DGDG and Glycolipids in Plants and Algae. Subcell. Biochem. 86, 51–83. doi: 10.1007/978-3-319-25979-6_3
Karas B. J., Diner R. E., Lefebvre S. C., Mcquaid J., Phillips A. P. R., Noddings C. M., et al. (2015). Designer Diatom Episomes Delivered by Bacterial Conjugation. Nat. Commun. 6, 6925. doi: 10.1038/ncomms7925
Klecker M., Gasch P., Peisker H., Dörmann P., Schlicke H., Grimm B., et al. (2014). A Shoot-Specific Hypoxic Response of Arabidopsis Sheds Light on the Role of the Phosphate-Responsive Transcription Factor PHOSPHATE STARVATION Response1. Plant Physiol. 165, 774–790. doi: 10.1104/pp.114.237990
Kobayashi K., Awai K., Nakamura M., Nagatani A., Masuda T., Ohta H. (2009a). Type-B Monogalactosyldiacylglycerol Synthases are Involved in Phosphate Starvation-Induced Lipid Remodeling, and are Crucial for Low-Phosphate Adaptation. Plant J. 57, 322–331. doi: 10.1111/j.1365-313X.2008.03692.x
Kobayashi K., Fujii S., Sasaki D., Baba S., Ohta H., Masuda T., et al. (2014). Transcriptional Regulation of Thylakoid Galactolipid Biosynthesis Coordinated With Chlorophyll Biosynthesis During the Development of Chloroplasts in Arabidopsis. Front. Plant Sci. 5, 272. doi: 10.3389/fpls.2014.00272
Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H. (2007). Galactolipid Synthesis in Chloroplast Inner Envelope is Essential for Proper Thylakoid Biogenesis, Photosynthesis, and Embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 17216–17221. doi: 10.1073/pnas.0704680104
Kobayashi K., Nakamura Y., Ohta H. (2009b). Type A and Type B Monogalactosyldiacylglycerol Synthases are Spatially and Functionally Separated in the Plastids of Higher Plants. Plant Physiol. Bioch. 47, 518–525. doi: 10.1016/j.plaphy.2008.12.012
Liu J., Huang J. C., Sun Z., Zhong Y. J., Jiang Y., Chen F. (2011). Differential Lipid and Fatty Acid Profiles of Photoautotrophic and Heterotrophic Chlorella Zofingiensis: Assessment of Algal Oils for Biodiesel Production. Bioresource Technol. 102, 106–110. doi: 10.1016/j.biortech.2010.06.017
Lommer M., Specht M., Roy A. S., Kraemer L., Andreson R., Gutowska M. A., et al. (2012). Genome and Low-Iron Response of an Oceanic Diatom Adapted to Chronic Iron Limitation. Genome Biol. 13, R66. doi: 10.1186/gb-2012-13-7-r66
Maheswari U., Mock T., Armbrust E. V., Bowler C. (2009). Update of the Diatom EST Database: A New Tool for Digital Transcriptomics. Nucleic Acids Res. 37, D1001–D1005. doi: 10.1093/nar/gkn905
Masuda S., Harada J., Yokono M., Yuzawa Y., Shimojima M., Murofushi K., et al. (2011). A Monogalactosyldiacylglycerol Synthase Found in the Green Sulfur Bacterium Chlorobaculum Tepidum Reveals Important Roles for Galactolipids in Photosynthesis. Plant Cell 23, 2644–2658. doi: 10.1105/tpc.111.085357
Mcconn M., Browse J. (1998). Polyunsaturated Membranes are Required for Photosynthetic Competence in a Mutant of Arabidopsis. Plant J. 15, 521–530. doi: 10.1046/j.1365-313x.1998.00229.x
Miyahara M., Aoi M., Inoue-Kashino N., Kashino Y., Ifuku K. (2013). Highly Efficient Transformation of the Diatom Phaeodactylum Tricornutum by Multi-Pulse Electroporation. Biosci. Biotechnol. Biochem. 77, 874–876. doi: 10.1271/bbb.120936
Mock T., Otillar R., Strauss J., McMullan M., Paajanen P., Schmutz J., et al. (2017). Evolutionary Genomics of the Coldadapted Diatom Fragilariopsis Cylindrus. Nature 541, 536–540. doi: 10.1038/nature20803
Moosburner M. A., Gholami P., McCarthy J. K., Tan M., Bielinski V. A., Allen A. E. (2020). Multiplexed Knockouts in the Model Diatom Phaeodactylum by Episomal Delivery of a Selectable Cas9. Front. Microbiol. 11, 5. doi: 10.3389/fmicb.2020.00005
Moustafa A., Beszteri B., Maier U. G., Bowler C., Valentin K., Bhattacharya D. (2009). Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms. Science 324, 1724–1726. doi: 10.1126/science.1172983
Myers A. M., James M. G., Lin Q. H., Yi G., Stinard P. S., Hennen-Bierwagen T. A., et al. (2011). Maize Opaque5 Encodes Monogalactosyldiacylglycerol Synthase and Specifically Affects Galactolipids Necessary for Amyloplast and Chloroplast Function. Plant Cell 23, 2331–2347. doi: 10.1105/tpc.111.087205
Nawaly H., Tsuji Y., Matsuda Y. (2020). Rapid and Precise Genome Editing in a Marine Diatom, Thalassiosira Pseudonana by Cas9 Nickase (D10A). Algal Res. 47, 101855. doi: 10.1016/j.algal.2020.101855
Nymark M., Sharma A. K., Sparstad T., Bones A. M., Winge P. (2016). A CRISPR/Cas9 System Adapted for Gene Editing in Marine Algae. Sci. Rep. 6, 24951. doi:10.1038/srep24951
Petroutsos D., Amiar S., Abida H., Dolch L. J., Bastien O., Rebeille F., et al. (2014). Evolution of Galactoglycerolipid Biosynthetic Pathways–From Cyanobacteria to Primary Plastids and From Primary to Secondary Plastids. Prog. Lipid Res. 54, 68–85. doi: 10.1016/j.plipres.2014.02.001
Pi X., Zhao S. H., Wang W. D., Liu D. S., Xu C. Z., Han G. Y., et al. (2019). The Pigment-Protein Network of a Diatom Photosystem II-Light-Harvesting Antenna Supercomplex. Science 365, eaax4406. doi: 10.1126/science.aax4406
Qi Y., Yamauchi Y., Ling J., Kawano N., Li D., Tanaka K. (2004). Cloning of a Putative Monogalactosyldiacylglycerol Synthase Gene From Rice (Oryza Sativa L.) Plants and its Expression in Response to Submergence and Other Stresses. Planta 219, 450–458. doi: 10.1007/s00425-004-1245-2
Rastogi A., Maheswari U., Dorrell R. G., Vieira F. R. J., Maumus F., Kustka A., et al. (2018). Integrative Analysis of Large Scale Transcriptome Data Draws a Comprehensive Landscape of Phaeodactylum Tricornutum Genome and Evolutionary Origin of Diatoms. Sci. Rep. 8, 4834. doi: 10.1038/s41598-018-23106-x
Selstam E. (1998). “Development of Thylakoid Membranes With Respect to Lipids,” in Lipids in Photosynthesis: Structure, Function and Genetics. Advances in Photosynthesis and Respiration, vol. vol 6 . Eds. Paul-André S., Norio M. (Dordrecht: Springer).
Sharma A. K., Nymark M., Sparstad T., Bones A. M., Winge P. (2018). Transgene-Free Genome Editing in Marine Algae by Bacterial Conjugation - Comparison With Biolistic CRISPR/Cas9 Transformation. Sci. Rep. 8, 14401. doi: 10.1038/s41598-018-32342-0
Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., et al. (2007). Molecular Toolbox for Studying Diatom Biology in Phaeodactylum Tricornutum. Gene 406, 23–35. doi: 10.1016/j.gene.2007.05.022
Sibbald S. J., Archibald J. M. (2020). Genomic Insights Into Plastid Evolution. Genome Biol. Evol. 12, 978–990. doi: 10.1093/gbe/evaa096
Slattery S. S., Diamond A., Wang H. L., Therrien J. A., Lant J. T., Jazey T., et al. (2018). An Expanded Plasmid-Based Genetic Toolbox Enables Cas9 Genome Editing and Stable Maintenance of Synthetic Pathways in Phaeodactylum Tricornutum. ACS Synth. Biol. 7, 328–338. doi: 10.1021/acssynbio.7b00191
Stukenberg D., Zauner S., Dell’aquila G., Maier U. G. (2018). Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum Tricornutum. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00740
Traller J. C., Cokus S. J., Lopez D. A., Gaidarenko O., Smith S. R., John P. M., et al. (2016). Genome and Methylome of the Oleaginous Diatom Cyclotella Cryptica Reveal Genetic Flexibility Toward a High Lipid Phenotype. Biotechnol. Biofuels 9, 258. doi: 10.1186/s13068-016-0670-3
Vancaester E., Depuydt T., Osuna-Cruz C. M., Vandepoele K. (2020). Comprehensive and Functional Analysis of Horizontal Gene Transfer Events in Diatoms. Mol. Biol. Evol. 37, 3243–3257. doi: 10.1093/molbev/msaa182
Vanier G., Stelter S., Vanier J., Hempel F., Maier U.G., Lerouge P., et al., (2018). Alga-Made Anti-Hepatitis B.
Wang S., Uddin M. I., Tanaka K., Yin L., Shi Z., Qi Y., et al. (2014). Maintenance of Chloroplast Structure and Function by Overexpression of the Rice MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE Gene Leads to Enhanced Salt Tolerance in Tobacco. Plant Physiol. 165, 1144–1155. doi: 10.1104/pp.114.238899
Weyman P. D., Beeri K., Lefebvre S. C., Rivera J., Mccarthy J. K., Heuberger A. L., et al. (2015). Inactivation of Phaeodactylum Tricornutum Urease Gene Using Transcription Activator-Like Effector Nuclease-Based Targeted Mutagenesis. Plant Biotechnol. J. 13, 460–470. doi: 10.1111/pbi.12254
Xie W. H., Zhu C. C., Zhang N. S., Li D. W., Yang W. D., Liu J. S., et al. (2014). Construction of Novel Chloroplast Expression Vector and Development of an Efficient Transformation System for the Diatom Phaeodactylum Tricornutum. Mar. Biotechnol. 16, 538–546. doi: 10.1007/s10126-014-9570-3
Yahia E. M., Carrillo-López A., Barrera G. M., Suzán-Azpiri H., Bolaños M. Q. (2019). “Chapter 3 - Photosynthesis,” in Postharvest Physiology and Biochemistry of Fruits and Vegetables. Ed. Yahia E. M. (Sawston: Woodhead Publishing).
Keywords: monogalactosyldiacylglycerol, MGDG synthase, MGDG biosynthesis, glycolipids, diatom
Citation: Shang S, Liu R, Luo L, Li X, Zhang S, Zhang Y, Zheng P, Chen Z and Wang B (2022) Functional Characterization of the Monogalactosyldiacylglycerol Synthase Gene ptMGD2 in the Diatom Phaeodactylum tricornutum. Front. Mar. Sci. 9:874448. doi: 10.3389/fmars.2022.874448
Received: 12 February 2022; Accepted: 06 June 2022;
Published: 15 July 2022.
Edited by:
Richard Dorrell, École Normale Supérieure, FranceReviewed by:
Bogumil Karas, Western University, CanadaJeffrey Leblond, Middle Tennessee State University, United States
Yusuke Matsuda, Kwansei Gakuin University, Japan
Copyright © 2022 Shang, Liu, Luo, Li, Zhang, Zhang, Zheng, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuo Chen, Y2hlbnpodW80MzU3QDE2My5jb20=; Peng Zheng, cGVuZ3poMTk4NEB3dXN0LmVkdS5jbg==
 Shuo Shang
Shuo Shang Ruyi Liu1,2
Ruyi Liu1,2 Peng Zheng
Peng Zheng Zhuo Chen
Zhuo Chen Baoshan Wang
Baoshan Wang