
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 08 June 2022
Sec. Deep-Sea Environments and Ecology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.873445
Wood arrives on the seabed from natural and anthropogenic sources (e.g., wood falls and wooden shipwrecks, respectively) and creates seafloor habitats for macro-, meio- and microbiota. The way these habitats shape microbial communities and their biogeographic patterns in the deep sea requires study. The objective of this work was to investigate how historic wooden-hulled shipwrecks impact the dispersal of wood-colonizing microbial biofilms. The study addressed how proximity to wooden shipwrecks shapes diversity, richness, and community composition in the surrounding environment. Study sites included two historic shipwrecks in the northern Gulf of Mexico identified as wooden-hulled sailing vessels dating to the late 19th century. Two experimental microbial recruitment arrays containing pine and oak samples were deployed by remotely operated vehicle proximate (0–200 m) to each shipwreck and used to establish new wooden habitat features to be colonized by biofilms. The experiments remained in place for approximately 4 months, were subsequently recovered, and biofilms were analyzed using 16S rRNA gene amplification and sequencing for bacteria and archaea and ITS2 region amplification and sequencing for fungi to determine alpha diversity metrics and community composition. The work examined the influence of wood type, proximity to shipwrecks, and environmental context on the biofilms formed on the surfaces. Wood type was the most significant feature shaping bacterial composition, but not archaeal or fungal composition. Proximity to shipwrecks was also a significant influence on bacterial and archaeal composition and alpha diversity, but not on fungal communities. In all 3 domains, a peak in alpha diversity and richness was observed on pine and oak samples placed ~125 m from the shipwrecks. This peak may be evidence of an ecotone, or convergence zone, between the shipwreck influenced seabed and the surrounding seafloor. This study provides evidence that historic wooden shipwrecks influence microbial biofilm dispersal in the deep sea.
Study of the microbial biogeography of deep ocean habitats has gained attention in recent years (Hamdan et al., 2013; Wilkins et al., 2013; Salazar et al., 2016; Hanson et al., 2019) with focus on how naturally occurring habitat features shape biogeographic patterns of sediment and water column communities (Hanson et al., 2012). While the importance of habitat features on biogeographic patterns is recognized (Hanson et al., 2012; Hanson et al., 2019), until recently, the significance of built habitats on microbial biogeography on the seafloor has been unexplored (Hamdan et al., 2018; Hamdan et al., 2021). Historic constraints (e.g., dispersal limitation) and contemporary features (e.g., habitat specificity) are the main drivers on microbial biogeographic patterns (Martiny et al., 2006; Hanson et al., 2019) in environmental settings. Both constraints have been studied in naturally occurring habitat features in the marine environment including hydrothermal vents, methane seeps, seamounts, and organic falls (Huber et al., 2007; Dick et al., 2013; Anderson et al., 2015). Anthropogenic structures in the marine environment include shipwrecks and energy infrastructure and are encompassed in the term “built environment” (Bugnot et al., 2020). Wood of terrestrial origin on the seafloor presents a habitat feature that may be derived from natural (wood falls) and anthropogenic (historic shipwrecks) sources. Wood delivered in the form of shipwrecks could play a role in deep-sea microbial biogeography.
The Theory of Island Biogeography (MacArthur and Wilson, 1967) explained three main considerations for island habitats: island size, distance from a mainland, and immigration and extinction rates. The Theory of Island Biogeography introduces a biotic equilibrium model in which richness reaches steady state when immigration equals extinction. At equilibrium, species richness and diversity decrease with distance from an island and has been used as indicators of “island-like” seabed habitats (Smith and Baco, 2003; Goffredi and Orphan, 2010; Hamdan et al., 2021). Natural marine habitat features exhibit island-like properties, including whale falls which evidence a distance effect with species richness being higher in proximity to fall sites than in the surrounding seafloor (Smith and Baco, 2003; Goffredi and Orphan, 2010). Anthropogenic structures, specifically, historic shipwrecks, have been recently considered island-like systems, as sediment microbiome diversity and richness declined as a function of distance from a historic metal-hulled shipwreck in the Gulf of Mexico (Hamdan et al., 2021).
While the factors that shape microbial biogeography for sediment and pelagic microorganisms associated with naturally occurring habitat features have been investigated, such studies are generally lacking for marine biofilms. Biofilms form on all submerged surfaces, and settlement of early colonizers and production of extracellular polymers leads to recruitment of microorganisms on hard substrates (Mugge et al., 2019; Mugge et al., 2021). Biofilm formation promotes settlement of macro-fauna (Hadfield, 2011; Dobretsov and Rittschof, 2020) on both natural and built habitat features, and occurs in three steps: adhesion, accumulation, and dispersal of macromolecules and biofilm microfauna (Garrett et al., 2008). Ultimately, dispersal of biofilms defines their ability to reach other habitats and is controlled by both historic constraints and contemporary features.
Considering the estimated 3 million shipwrecks in the world’s oceans, with the majority constructed of wood, shipwrecks may be a more significant source of wood to the seabed than recognized (UNESCO, 2017). Naturally derived wood is delivered to the deep-sea through riverine transport or storm events. While wood falls are most common along coastlines, they have also been observed at deeper water depths (Wolff, 1979; Bienhold et al., 2013). Wooden shipwrecks are found at all explored depths where past shipping lanes occurred. Experimental studies show wood falls are colonized by microorganisms and create hotspots of diversity (Palacios et al., 2009; Fagervold et al., 2012; Bienhold et al., 2013; Kalenitchenko et al., 2015). These studies show geographic location and water depth are important factors in structuring bacterial communities on submerged wood (Palacios et al., 2009; Ristova et al., 2017). Different experimental constraints, including wood type and immersion time also dictate the composition of wood-colonizing microbiomes (Palacios et al., 2009; Fagervold et al., 2012; Kalenitchenko et al., 2015).
The convergence of biofilm formation, maturation, and dispersal on submerged wood provides opportunity to understand how wood habitat features influence biogeography processes on the seabed.
Two historic shipwrecks in the northern Gulf of Mexico investigated for the first time during this study were the setting for this work. Both sites are wooden-hulled, copper-sheathed sailing ships dating to the late 19th century, as previously described (Hampel et al., 2022). Briefly, Site 15711 rests at ~525 m water depth in the Viosca Knoll lease area (Figure 1) and is 20 m long, 7 m wide, with a vertical relief of ~2.5 to 3 m. Debris extends several meters off the bow and the starboard side of the shipwreck. The bow of the vessel is mostly intact with visible copper sheathing, but overall, the hull is not well preserved. Site 15470 rests at ~1800 m water depth in the Mississippi Canyon lease area (Figure 1). It is 32 m long, 12.5 m wide, with a vertical relief of approximately 0.8-1.3 m. A large artifact cluster is located on the starboard side of the vessel. Both sites are estimated to be buried no more than 3-4 ft deep into the seabed.
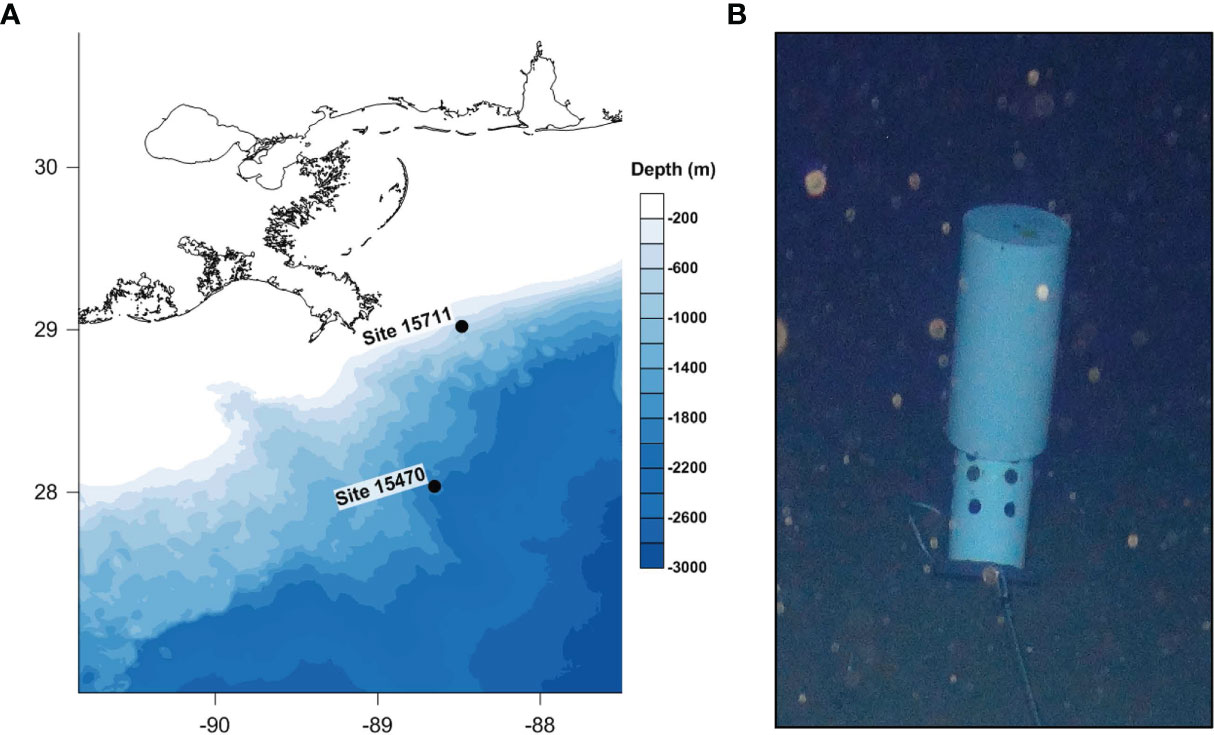
Figure 1 (A) Map depicting the location of historic wooden shipwreck sites in the northern Gulf of Mexico. (B) Image of submerged MRE on the seafloor with the cover floating and coupons exposed.
Microbial Recruitment Experiment (MRE) arrays were deployed in June-July 2019, from Research Vessel Point Sur and recovered in November 2019. MREs were constructed of polyvinyl chloride with a floating cover and body to hold pine and oak surfaces for biofilm recruitment. The cover contained flotation allowing the MRE to remain open when submerged. A line connected the cover to the body to enable the cover to close on recovery. MREs held pine and oak surfaces (called coupons from this point forward) with 4 replicates of each wood type secured at the base (away from the experimental surface) with 100% silicone waterproof sealant. Coupons had a surface area of 1.23 square inches that was cleaned with 70% ethanol immediately before deployment. Five MREs were arranged in an array connected by 25 m lengths of line. The first MRE in the array was connected to a lander, described below, by a 182 m leader line which provided physical separation between the lander and the array, to not interfere with the experiment. For deployment, the array, leader line, and 25 m connecting lines were placed in a deployment cradle on Remotely Operated Vehicle (ROV) Odysseus (Pelagic Research Services) immediately aft of the scientific skid. The leader line was spooled around the cradle with the end secured to a lander comprised of a frame connecting three 17-in glass spheres with up to 50 lbs flotation each. A Teledyne-Benthos R2K acoustic release was enclosed in the top sphere, and the lander was attached to ~200 lbs of weight. When the ROV was in position to deploy the experiment, as determined using an ultra-short baseline acoustic positioning beacon on the ROV and an onboard sonar to determine the distance to the shipwreck, the lander was released from a hydraulic clamp. With the lander and weights on bottom, the ROV flew forward along predetermined flight paths located north and east of the sites towards the shipwrecks. The leader line unspooled as the ROV approached the shipwreck, and MREs were sequentially pulled free from their cradle. The sample holders were negatively buoyant, pulling the holder and flotation to the seafloor, and the cover opened under flotation once free from the ROV (Figure 1). Two MRE arrays were deployed at each site. At 15470, the first array was placed in a northeastern direction with the nearest MRE 100 m from the shipwreck. The second array was placed in a northeastern direction from the shipwreck’s stern with the nearest MRE 22 m away. At 15711, the first array was placed east of the center of the shipwreck with the nearest MRE 12 m away, and the second was placed in a northeastern direction from the stern with the nearest MRE 40 m away.
The arrays were on site for 136 days at 15470 and 138 days at 15711. For recovery, the acoustic release on the lander was triggered to drop the weights. As the lander became buoyant, the leader line raised each MRE off the seabed in sequence. The connecting lines between each MRE engaged the covers closed when under tension, providing protection to biofilms formed on the wood coupons during ascent. Immediately upon recovery, coupons were scraped aseptically with a flame-sterilized scoopula to collect biofilms and placed in lysing matrix E tubes containing sodium phosphate and MT buffers (MP Biomedicals). Samples were stored at -80°C shipboard and at the laboratory.
Genomic DNA was extracted with FastDNA™ SPIN Kit (MP Biomedicals) as previously described (Hamdan et al., 2018), quantified by Qubit 2.0 (Invitrogen) and checked for purity with a NanoDrop (ThermoFisher). For bacterial and archaeal analyses, the 16S rRNA gene was amplified and sequenced at the Integrated Microbiome Resource facility at Dalhousie University (Halifax, Nova Scotia, Canada), as described previously (Hamdan et al., 2021) using bacterial primers B969F/BA1406R and archaeal primers A956F/A1401R to target the V6-V8 variable regions. For fungal analysis, amplification and sequencing of the internal transcribed spacer region 2 (ITS2) was performed at the Integrated Microbiome Resource facility with primers ITS86F/ITS4, which have been shown to be suitable for studying diversity and community structures of fungi (Op de Beeck et al., 2014). Sequencing blanks were included with each individual MiSeq sequencing run to help address background contamination.
Bioinformatics analysis was performed using Quantitative Insights into Microbial Ecology 2 (QIIME2) (Bolyen et al., 2019). The DADA2 plugin was used for sequence quality control, chimera removal, and production of an amplicon sequence variant (ASV) table with quality-based sequence truncation (Callahan et al., 2016). A phylogenetic tree was built using FastTree with a mid-point root and masked to remove highly variable positions using mafft, a denovo multiple sequence alignment. Taxonomy was assigned with the VSEARCH plugin at 99% similarity using the SILVA database (v. 132) (Quast et al., 2013) to classify bacteria and archaea and the UNITE database (v. 8.3) (Nilsson et al., 2018) to classify fungi.
Three MREs surfaced open, potentially resulting in the disruption of formed biofilms, and thus were excluded from analysis (Supplementary Table S1). To determine outliers based on non-chimeric reads of individual samples, the median absolute deviation method in RStudio (v. 1.3.959) (Leys et al., 2013) determined lower thresholds using a coefficient of 0.5. The threshold for archaea was 216 non-chimeric reads, which excluded 25 additional samples resulting in 111 archaeal samples retained. The threshold for fungi was 2494 non-chimeric reads, which excluded 32 additional samples resulting in 104 fungal samples retained. For bacteria, all samples excluding those collected from the open MREs were retained, with a minimum of 3366 non-chimeric reads, resulting in 136 samples retained.
Shannon diversity index (Shannon, 1948) and ASV richness were determined in QIIME2 using the core metrics plug-in. A weighted UniFrac analysis (Lozupone and Knight, 2005) was performed in QIIME2 to calculate beta diversity and results were displayed by Principal Coordinates Analysis (PCoA). The weighted UniFrac distance matrix was used in a permutational multivariate analysis of variance (PERMANOVA) and a PERMDISP (Anderson and Walsh, 2013) to identify structuring features on communities. Bubble plots of ASV relative abundance were generated in R-Studio with ggplot2 package (Wickham, 2009) at the highest level of taxonomy possible. A PERMANOVA with transect direction included did not indicate it as a significant factor (p > 0.05), so samples were grouped into 25 m distance categories.
Average Shannon index and ASV richness for replicate samples (n = 4) collected from each MRE in arrays at each site were plotted as a function of distance from the sites (Figure 2). At both sites, bacterial alpha diversity was not statistically impacted by distance from the sites. At 15711, bacterial diversity and richness had a “U” shaped response, and were highest nearest the site, decreased, and then increased again at ~125 m. The same pattern was seen at 15470 for Shannon index, but not ASV richness, which declined consistently with distance. Archaeal alpha diversity was overall lower than bacteria and fungi. Archaeal alpha diversity at 15711 was consistent across distances. At 15470, archaeal richness significantly decreased with distance (p < 0.05), and diversity and richness exhibited the “U” shaped pattern with a primary peak at 25 m and a secondary peak at 125 m. Fungal alpha diversity was not statistically impacted by distance. At 15711, diversity and richness increased with distance. At 15470, diversity and richness exhibited a peak 0-50 m from the shipwreck, a decrease, then a secondary peak from ~125-175 m.
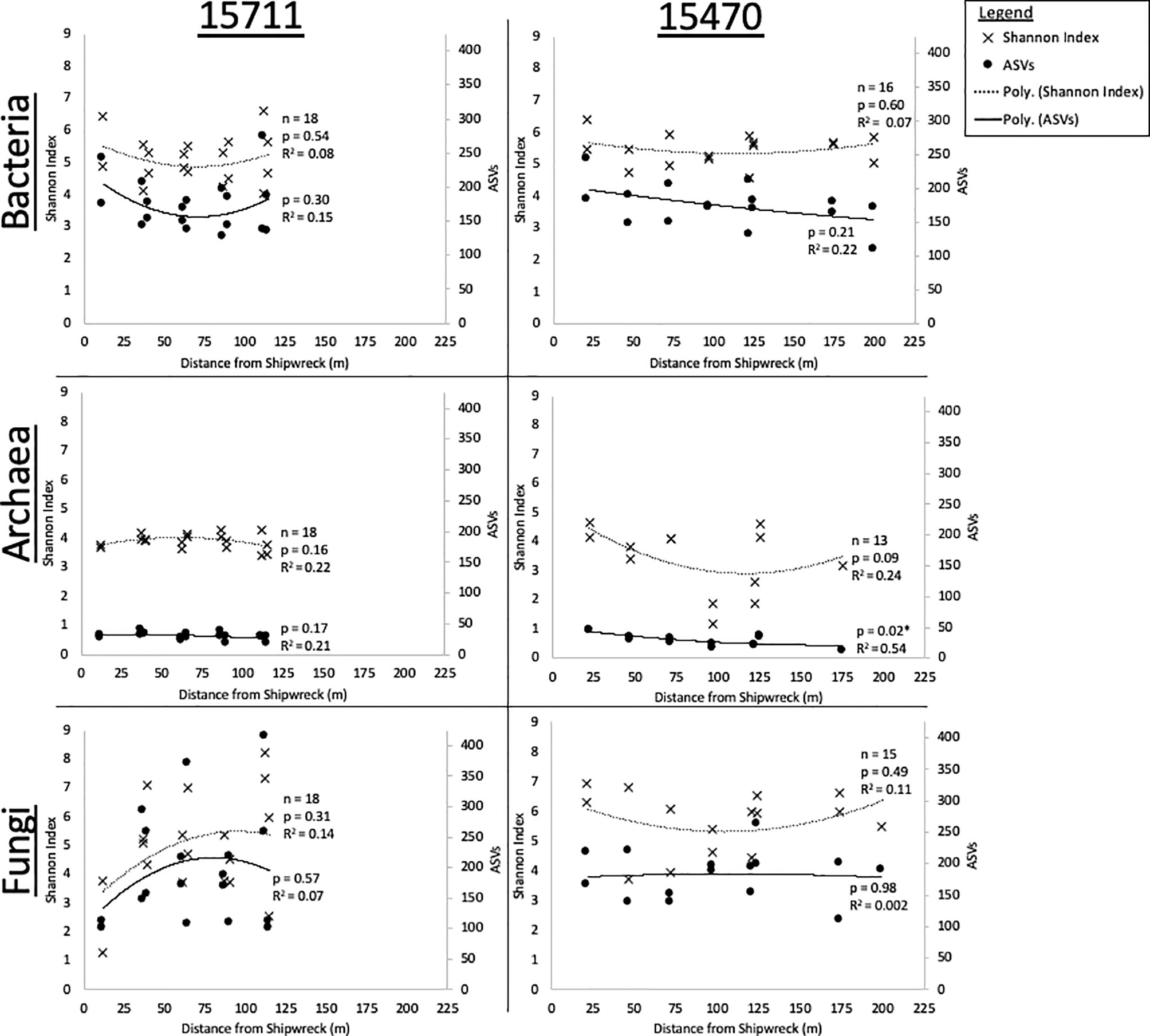
Figure 2 Shannon Diversity and ASV Richness plotted as a function of distance for each site and each domain with replicates averaged. Significance (p < 0.05) of polynomial regression indicated by *.
UniFrac ordinations explained variance in the data driven by site, wood type, and distance from the shipwrecks (Figure 3). PERMANOVA tested statistical significance of these individual and combined factors (Table 1). Wood type was the most significant (p = 0.001) structuring feature on bacteria, followed by site (p = 0.001). While not evident for alpha-diversity metrics, distance was a statistically significant (p = 0.001) structuring feature on bacteria. Site was the strongest structuring feature on archaea (p = 0.001). Distance was also statistically significant (p = 0.003) although the PERMANOVA statistic (Pseudo-F, Table 1) was lower than that for site. Wood archaeal samples grouped at individual sites by distance. Wood type was not a statistically significant structuring feature for archaeal communities (p = 0.08). For fungi, site (p = 0.001) was the strongest structuring influence on communities, followed by wood type (p = 0.001). Unlike the other domains, distance was not a significant structuring factor. PERMDISP tested for significant differences in dispersion and found significant results for bacteria wood type (p = 0.001), archaea site and distance (p = 0.001), and fungi site (p = 0.002) and wood type (p = 0.002) (Supplementary Table S2). Pairwise PERMANOVA and PERMDISP comparisons for distance groups revealed which distance groups were statistically significant (Supplementary Tables S3, S4). Distance groups from 75-124 m were often statistically significant for bacteria and archaea.
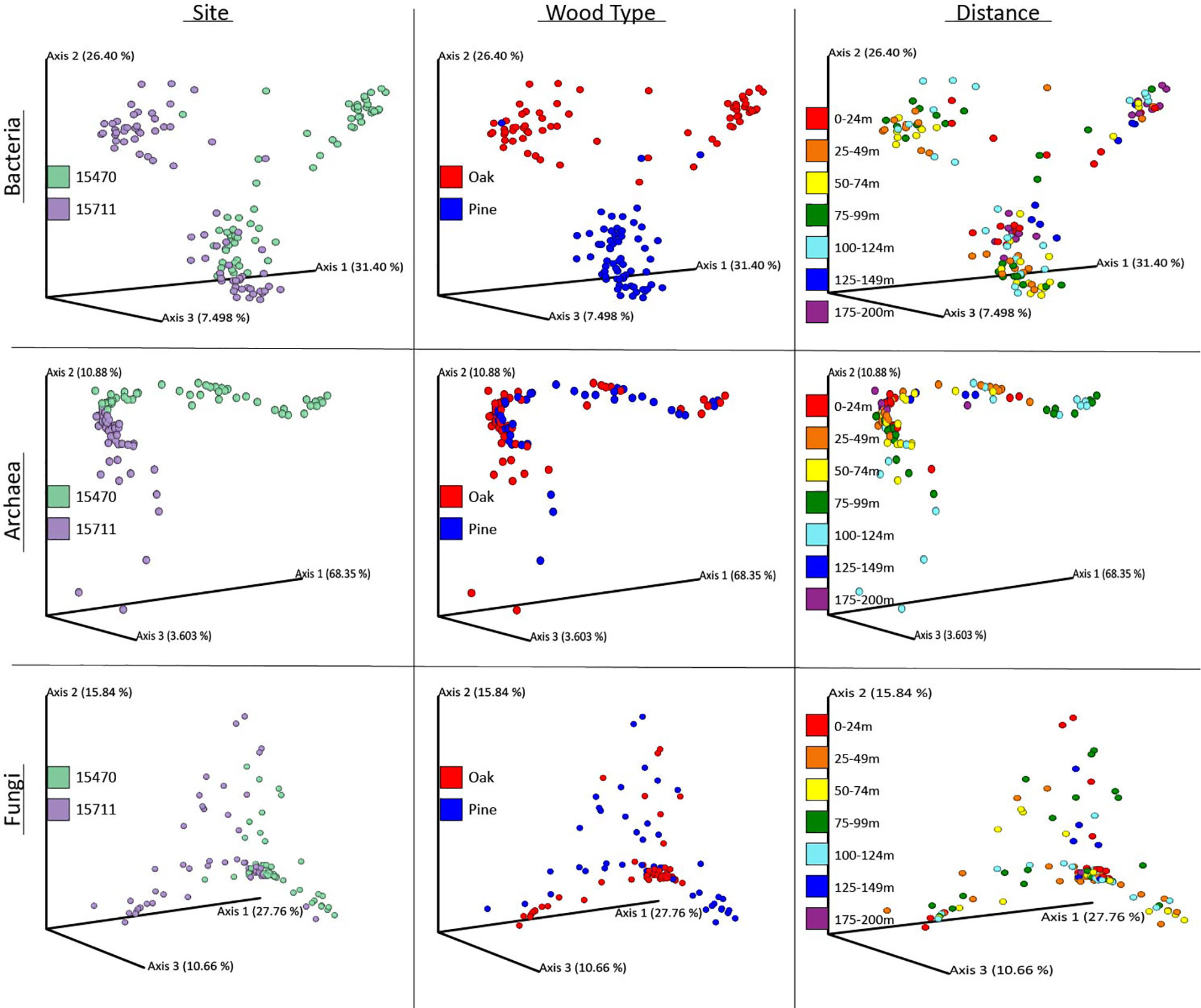
Figure 3 Weighted UniFrac results displayed as PCoA for Site, Wood Type, and Distance for all three domains. Samples were grouped in distance categories spanning 25 m for visualization.
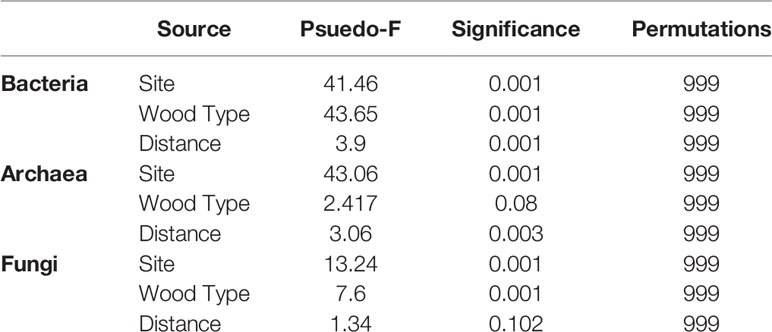
Table 1 PERMANOVA conducted on results from the weighted UniFrac distance matrix to determine differences in microbiome community based on site, wood type, and distance from site.
Bubble plots visualized the composition and relative abundance of the top 30 most abundant ASVs from each domain. Bacterial composition was markedly different on pine samples compared to oak (Figure 4). Myxococcales were highly abundant on pine (15711 = 24%, 15470 = 17%) compared to oak (15711 = 4%, 15470 = 2%) and increased in abundance in the middle of the transects at 15470. Rhodobacteraceae were diverse at both sites, but generally more abundant on oak with different ASVs contributing to the communities. Overall, communities on oak samples were more different in composition across sites compared to pine samples.
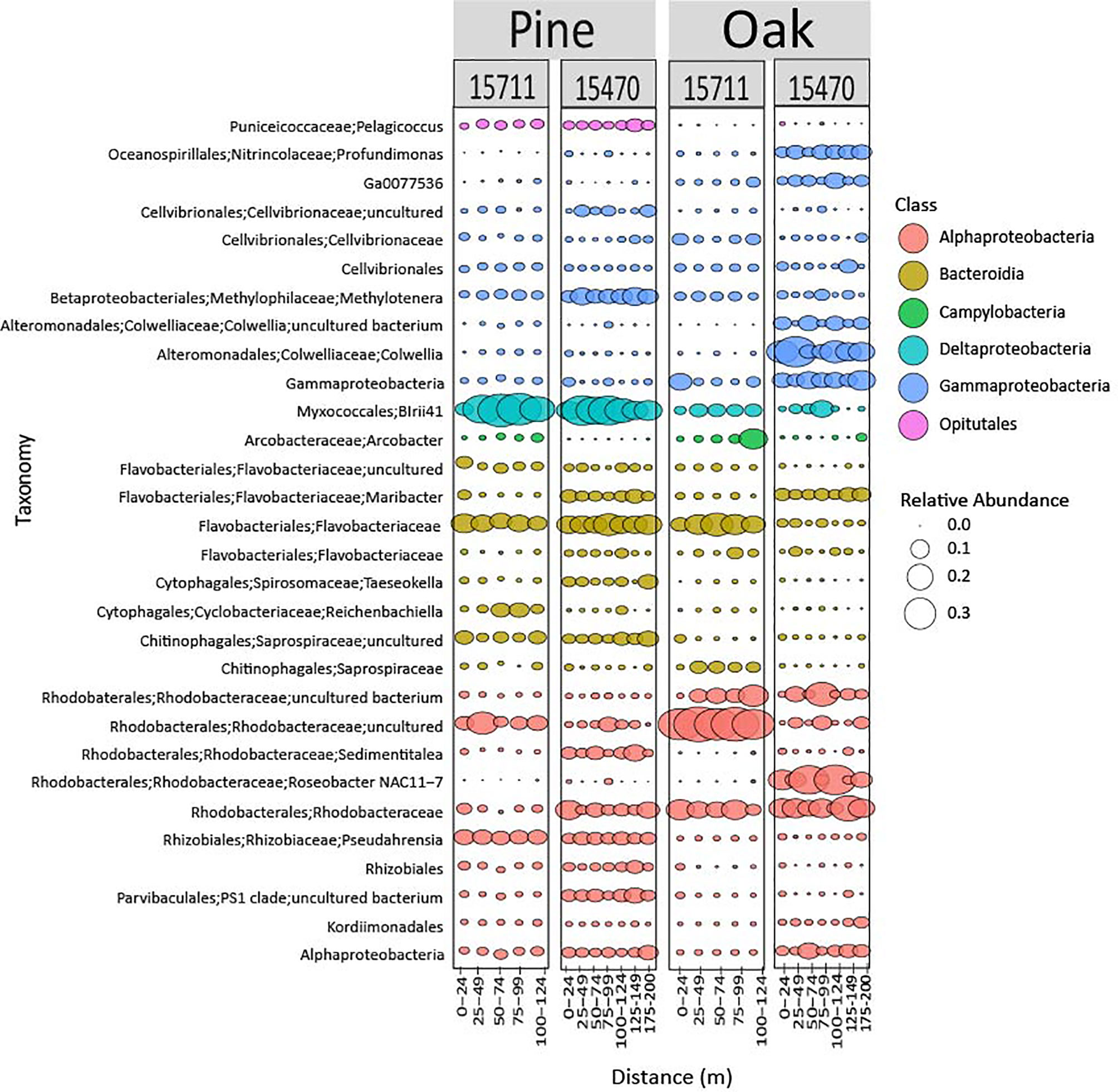
Figure 4 Bubble plot representing the 30 most abundant bacterial taxa from both sites and wood types. Samples are grouped in distance categories spanning 25 m.
Archaeal biofilms were dominated by the phylum Thaumarchaeota, particularly ASVs associated with class Nitrosphaeria, at both sites and on both wood types (Figure 5). One ASV from the genus Nitrosopumilus dominated both sites and wood types but showed some differing abundance across distances. Some subtle differences were observed in lower abundance ASVs across sites on the same wood types.
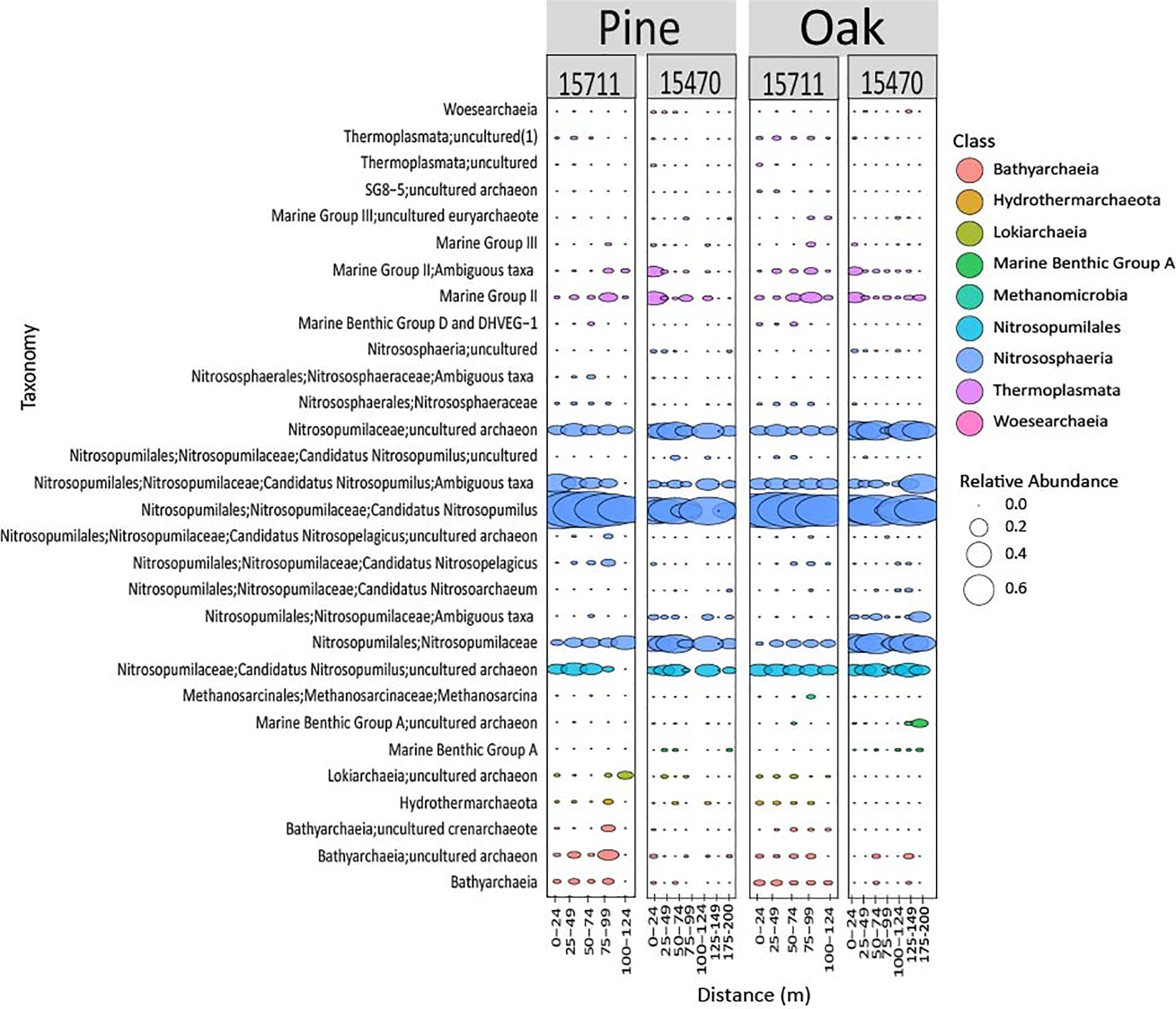
Figure 5 Bubble plot representing the 30 most abundant archaeal taxa from both sites and wood types. Samples are grouped in distance categories spanning 25 m.
Fungal community composition was different across sites and wood types (Figure 6). ASVs associated with Sordariomycetes (Phylum Ascomycota) were especially abundant on pine samples at 15711, and two ASVs, Sordariales and Trichoderma, exhibit a pattern of higher abundances in the middle of the transect.
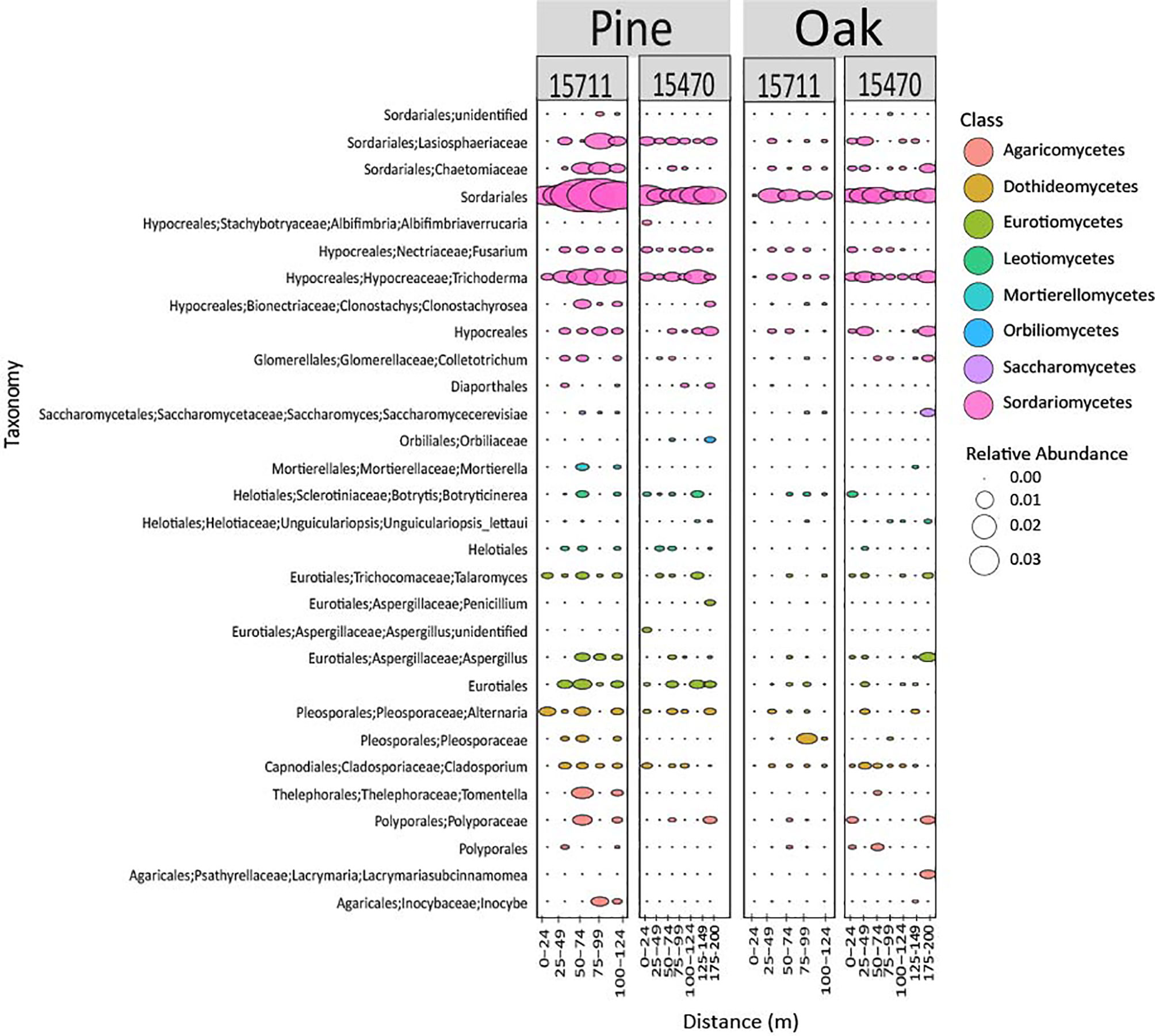
Figure 6 Bubble plot representing the 30 most abundant fungal taxa from both sites and wood types. Samples are grouped in distance categories spanning 25 m.
The goal of this work was to determine how proximity to historic, wooden shipwrecks influenced diversity, richness, and community composition of microbial biofilms on two types of wood. This information may support understanding of how submerged wooden habitats influence biofilm dispersal in the marine environment. PERMANOVA revealed distance from shipwreck sites was a key structuring factor on phylogenetic composition for bacteria and archaea, but not for fungi. Alpha diversity extinction plots did not provide uniform evidence that shipwrecks function as island-like systems for biofilms in the surrounding environment given that a significant relationship was not generally discovered for alpha diversity. Bacterial alpha diversity at both sites and archaeal alpha diversity at Site 15470, however, were highest in samples collected nearest to the shipwrecks (Figure 2) with a statistically significant relationship for archaea at Site 15470. A related study of sediment surrounding these sites also revealed archaeal diversity and taxa richness declined as a function of distance from site 15470 but not 15711 (Hampel et al., 2022). In that work, local environmental conditions, specifically differing organic matter supply between the sites was cited as a reason that alpha diversity decreased as a function of distance from 15470 which rests in a deeper, more oligotrophic setting compared to 15711. The two sites in this study are separated by 111 km of seafloor and 1275 m of vertical depth. Previous studies have shown that location and water depth are important factors driving marine sediment bacterial and archaeal community composition (Kallmeyer et al., 2012; Hoshino et al., 2020), and these factors may also influence biofilm composition. Site 15711 rests in shallower water near the coast (76 km from the Mississippi River delta), and thus receives more nutrient and carbon deposition relative to Site 15470. Discharge from the Mississippi River is also known to influence microbial community composition (Mason et al., 2016; Sánchez-Soto Jiménez et al., 2018). It is, therefore, likely that along with being a more eutrophic setting, 15711 has higher rates of immigration of new taxa from terresterial sources. In the context of The Theory of Island Biogeography, the site has greater proximity to a “mainland” source for immigration, resulting in an unequal extinction rate at the two sites. The dispersal of biofilms from the shipwreck would thus not be as evident at 15711 compared to 15470, the latter further from this “mainland” source and potentially at equilibrium. While the diversity and richness curves do not provide clear evidence for the shipwrecks as island-like systems, there is evidence that island-like functions relating to geographic location and taxa and nutrient supply impact biofilm dispersal around a shipwreck.
Across the two sites, archaeal and bacterial diversity and richness were generally highest nearest the shipwrecks, with secondary peaks near the ends of the transects, with the exception of archaea at Site 15711. The secondary peaks may be indicative of an ecotone, defined as a convergence zone between two ecosystems connecting along an environmental gradient (Levin et al., 2016). Ecotones function as diversity hotspots providing for co-location of taxa from adjacent ecosystems. If the shipwreck ecosystem is converging with the soft sediment dominated ecosystem, an ecotone may appear in water, sediment, or biofilm communities in between and would contain higher alpha diversity on par with end-member environments along the gradient. A recent study presented evidence of an ecotone in sediment microbiomes surrounding a metal-hulled shipwreck (Hamdan et al., 2021). The presence of an ecotone in this study indicates wooden shipwrecks harbor unique communities different from the background habitat of the surrounding seafloor and may be functioning as an island-like system. As noted above, however, the extent to which a shipwreck may function as an island-like system is influenced by geographic location in proximity to a source of taxa and nutrients.
UniFrac PCoAs and PERMANOVA tests showed community composition is influenced by site, wood type, and proximity to a shipwreck. The differences in the environmental setting between sites may explain the statistically significant effect of site in structuring all three domains in this study. Geographic location is known to be important in structuring wood-associated bacterial communities. A study by Ristova et al. (2017) found sunken wood communities were more similar within one geographic location than between different seas. However, wood type has also been shown to be a strong influence on bacterial communities, and more important in bacterial community composition than geographic location or depth (Palacios et al., 2009; Fagervold et al., 2012; Kalenitchenko et al., 2015). The current study agrees with those prior studies where bacteria are concerned, and notably, wood type was also a selective force on phylogenetic composition for fungi, although secondary to site. Although not statistically significant for archaea, wood type could be a selective force to a lesser extent than site and distance. Proximity to historic wooden shipwrecks also shaped community composition of biofilms on wood for bacteria and archaea, while the relationship was not significant for fungi. This is somewhat in conflict with results for alpha diversity where distance was not consistently a significant factor structuring diversity. This may indicate different dispersal mechanisms or different sources of taxa for individual members of the biofilm microbiome.
Previous studies have shown specific phylotypes have an affinity for forming biofilms on different types of sunken wood (Palacios et al., 2009; Fagervold et al., 2012; Kalenitchenko et al., 2015). Myxococcales has been found in association with sunken pine submerged for 20 months in the South Pacific (Fagervold et al., 2012). In the current study, one ASV associated with Myxococcales (Blrii41) was abundant in pine samples, had lower abundances at 25 m from the site, increased in abundance in the middle of the array, and then decreased in abundance at 100-124 m for both sites. This pattern was also seen at 15470 for oak samples. These data provide additional evidence that proximity is a structuring factor on microbiomes but is especially important to wood-associated taxa utilizing wood on the seafloor. Rhodobacteraceae has been found associated with oak in other studies and changes in abundance across distance in this study, exhibiting the same pattern described above for Myxococcales at 15470 (Kalenitchenko et al., 2015). The main archaeal phylotypes observed in this study from Nitrosopumilus are common in the Gulf of Mexico but have also been found on submerged wood (Kalenitchenko et al., 2015; Mason et al., 2016). Nitrosopumilus exhibits changing abundances across distances, although the patterns are not as clear as described for Myxococcales. While information on fungal communities on submerged wood is limited, this study found members of the phylum Ascomycota, which contains wood degraders, in high abundance (Pedersen et al., 2020). This includes those members of Sordariomycetes that exhibited a similar pattern of increasing abundance mid-transects as described for Myxococcales. This is another indication that while proximity overall is not as strong a structuring factor for fungal communities, it may be important to specific wood-associated fungal taxa.
This study reveals that wood-associated biofilm composition is impacted by proximity to wooden shipwrecks. Wood type is a highly selective force on bacterial community composition in deep-sea biofilm communities. Proximity to a shipwreck also shapes bacterial and archaeal community composition although questions remain for fungal communities. This study shows that the presence of wooden shipwrecks and similar anthropogenic structures on the seafloor should be considered in studies of marine microbial biogeography to support understanding of microbial dispersal in the deep-sea.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA802802.
LH designed the study. RDM conducted the data analysis. RDM and LH wrote the manuscript. All authors conducted the laboratory analyses and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
Funding for this study was provided by the National Oceanic and Atmospheric Administration’s (NOAA) Ocean Exploration and Research Program under Cooperative Agreement NA18OAR0110286.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We acknowledge the support of R/V Point Sur’s Captain and Crew, ROV Odysseus (Pelagic Research Services) team, and the members of the shipboard science party that contributed to the study. We thank Maxwell Woolsey and Eric Peterson for contributing to the MRE design.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.873445/full#supplementary-material
Anderson R. E., Sogin M. L., Baross J. A. (2015). Biogeography and Ecology of the Rare and Abundant Microbial Lineages in Deep-Sea Hydrothermal Vents. FEMS Microbiol. Ecol. 91, 1–11. doi: 10.1093/femsec/fiu016
Anderson M. J., Walsh D. C. I. (2013). PERMANOVA, ANOSIM, and the Mantel Test in the Face of Heterogeneous Dispersions: What Null Hypothesis are You Testing? Ecol. Monogr. 83, 557–574. doi: 10.1890/12-2010.1
Bienhold C., Ristova P., Wenzhöfer F., Dittmar T., Boetius A. (2013). How Deep-Sea Wood Falls Sustain Chemosynthetic Life. PloS One 8, 1. doi: 10.1371/journal.pone.0053590
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bugnot A. B., Mayer-Pinto M., Airoldi L., Heery E. C., Johnston E. L., Critchley L. P., et al. (2020). Current and Projected Global Extent of Marine Built Structures. Nat. Sustain. 4, 33–41. doi: 10.1038/s41893-020-00595-1
Callahan B. J., McMurdle P. J., Rosen M. J., Han A. W., Johnson A. J., Holmes S. P. (2016). DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Dick G. J., Anantharaman K., Baker B. J., Li M., Reed D. C., Sheik C. S. (2013). The Microbiology of Deep-Sea Hydrothermal Vent Plumes: Ecological and Biogeographic Linkages to Seafloor and Water Column Habitats. Front. Microbiol. 4, 1–15. doi: 10.3389/fmicb.2013.00124
Dobretsov S., Rittschof D. (2020). Love at First Taste: Induction of Larval Settlement by Marine Microbes. Int. J. Mol. Sci. 21, 731. doi: 10.3390/ijms21030731
Fagervold S. K., Galand P. E., Zbinden M., Giall F., Lebaron P., Palacios C. (2012). Sunken Woods on the Ocean Floor Provide Diverse Specialized Habitats for Microorganisms. FEMS Microbiol. Ecol. 82, 616–628. doi: 10.1111/j.1574-6941.2012.01432.x
Garrett T. R., Bhakoo M., Zhang Z. (2008). Bacterial Adhesion and Biofilms on Surfaces. Pro. Nat. Sci. 18, 1049–1056. doi: 10.1016/j.pnsc.2008.04.001
Goffredi S. K., Orphan V. J. (2010). Bacterial Community Shifts in Taxa and Diversity in Response to Localized Organic Loading in the Deep Sea. Environ. Microbiol. 12, 344–363. doi: 10.1111/j.1462-2920.2009.02072.x
Hadfield M. G. (2011). Biofilms and Marine Invertebrate Larvae: What Bacteria Produce That Larvae Use to Choose Settlement Sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hamdan L. J., Coffin R. B., Sikaroodi M., Greinert J., Treude T., Gillevet P. M. (2013). Ocean Currents Shape the Microbiome of Arctic Marine Sediments. ISME J. 7, 685–696. doi: 10.1038/ismej.2012.143
Hamdan L. J., Hampel J. J., Moseley R. D., Mugge R. L., Ray A., Salerno J. L., et al. (2021) . Deep-Sea Shipwrecks Represent Island-Like Ecosystems for Marine Microbiomes. ISME J 15, 2883–2891. doi: 10.1038/s41396-021-00978-y
Hamdan L. J., Salerno J. L., Reed A., Joye S. B., Damour M. (2018). The Impact of the Deepwater Horizon Blowout on Historic Shipwreck-Associated Sediment Microbiomes in the Northern Gulf of Mexico. Sci. Rep. 8, 9057. doi: 10.1038/s41598-018-27350-z
Hampel J. J., Moseley R. D., Mugge R. L., Ray A., Damour M., Jones D., et al. (2022). Deep-Sea Wooden Shipwrecks Influence Sediment Microbiome Diversity. Limnol. Oceanogr 67 (2), 482–497. doi: 10.1002/lno.12008
Hanson C. A., Fuhrman J. A., Horner-Devine M. C., Martiny J. B. H. (2012). Beyond Biogeographic Patterns: Processes Shaping the Microbial Landscape. Nat. Rev. Microbiol. 10, 497–506. doi: 10.1038/nrmicro2795
Hanson C. A., Müller A. L., Loy A., Dona C., Appel R., Jørgensen B. B., et al. (2019). Historical Factors Associated With Past Environments Influence the Biogeography of Thermophilic Endospores in Arctic Marine Sediments. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00245
Hoshino T., Doi H., Uramoto G., Wörmer L., Adhikari R. R., Xiao N., et al. (2020). Global Diversity of Microbial Communities in Marine Sediment. PNAS 117, 44. doi: 10.1073/pnas.1919139117
Huber J. A., Welch D. B. M., Morrison H. G., Huse S. M., Neal P. R., Butterfield D. A., et al. (2007). Microbial Population Structures in the Deep Marine Biosphere. Science 318, 97–100. doi: 10.1126/science.1146689
Kalenitchenko D., Fagervold S. K., Pruski A. M., Vétion G., Yücel M., Le Bris N., et al. (2015). Temporal and Spatial Constraints on Community Assembly During Microbial Colonization of Wood in Seawater. ISME J. 9, 2657–2670. doi: 10.1038/ismej.2015.61
Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D'Hondt S. (2012). Global Distribution of Microbial Abundance and Biomass in Subseafloor Sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Levin L. A., Baco A. R., Bowden D. A., Colaco A., Cordes E. E., Cunha M. R., et al. (2016). Hydrothermal Vents and Methane Seeps: Rethinking the Sphere of Influence. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00072
Leys C., Ley C., Klein O., Bernard P., Licata L. (2013). Detecting Outliers: Do Not Use Standard Deviation Around the Mean, Use Absolute Deviation Around the Mean. J. Exp. Soc Psychol. 49, 764–766. doi: 10.1016/j.jesp.2013.03.013
Lozupone C., Knight R. (2005). UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
MacArthur R. H., Wilson E. O. (1967). The Theory of Island Biogeography (New Jersey: Princeton University Press).
Martiny J. B., Bohannan B. J., Brown J. H., Colwell R. K., Fuhrman J. A., Green J. L., et al. (2006). Microbial Biogeography: Putting Microorganisms on the Map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Mason O. U., Canter E. J., Gillies L. E., Paisie T. K., Roberts B. J. (2016). Mississippi River Plume Enriches Microbial Diversity in the Northern Gulf of Mexico. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01048
Mugge R. L., Brock M. L., Salerno J. L., Damour M., Church R. A., Lee J. S., et al. (2019). Deep-Sea Biofilms, Historic Shipwreck Preservation and the Deepwater Horizon Spill. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00048
Mugge R. L., Salerno J. L., Hamdan L. J. (2021). Microbial Functional Responses in Marine Biofilms Exposed to Deepwater Horizon Spill Contaminants. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.636054
Nilsson R. H., Larsson K. H., Taylor A. F. S., Bengtsson-Palme J., Jeppesen T. S., Schigel D., et al. (2018). The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Op de Beeck M., Lievens B., Busschaert P., Declerck S., Vangronsveld J., Colpaert J. V. (2014). Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PloS One 9, 6. doi: 10.1371/journal.pone.0097629
Palacios C., Zbinden M., Pailleret M., Gaill F., Lebaron P. (2009). Highly Similar Prokaryotic Communities of Sunken Wood at Shallow and Deep-Sea Sites Across the Oceans. Microb. Ecol. 58, 737–752. doi: 10.1007/s00248-009-9538-4
Pedersen N. B., Matthiesen H., Blanchette R. A., Alfredsen G., Held B. W., Westergaard-Nielsen A., et al. (2020). Fungal Attack on Archaeological Wooden Artefacts in the Arctic- Implications in a Changing Climate. Nat. Sci. Rep. 10, 14577. doi: 10.1038/s41598-020-71518-5
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ristova P., Bienhold C., Wenzhöfer F., Rossel P. E., Boetius A. (2017). Temporal and Spatial Variations of Bacterial and Faunal Communities Associated With Deep-Sea Wood Falls. PloS One 12, 1. doi: 10.1371/journal.pone.0169906
Salazar G., Cornejo-Castillo F. M., Benitez-Barrios V., Fraile-Nuez E., Alvarez-Salgado X. A., Duarte C. M., et al. (2016). Global Diversity and Biogeography of Deep-Sea Pelagic Prokaryotes. ISME J. 10, 596–608. doi: 10.1038/ismej.2015.137
Sánchez-Soto Jiménez M. F., Cerqueda-García D., Montero-Muñoz J. L., Aguirre-Macedo M. L., García-Maldonado J. Q. (2018). Assessment of the Bacterial Community Structure in Shallow and Deep Sediments of the Perdido Fold Belt Region in the Gulf of Mexico. Peer J. 6, e5583. doi: 10.7717/peerj.5583
Shannon C. E. (1948). The Mathematical Theory of Communication. Bell Syst. Tech. Jour. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Smith C. R., Baco A. R. (2003). Ecology of Whale Falls at the Deep-Sea Floor. Oceanogr. Mar. Biol. 41, 311–354.
UNESCO. (2017). Wrecks. Available at: http://www.unesco.org/new/en/culture/themes/underwater-cultural-heritage/underwater-cultural-heritage/wrecks/ (Accessed November 15, 2020).
Wilkins D., Van Sebille E., Rintoul S. R., Lauro F. M., Cavicchioli R. (2013). Advection Shapes Southern Ocean Microbial Assemblages Independent of Distance and Environment Effects. Nat. Commun. 4, 2457. doi: 10.1038/ncomms3457
Keywords: biofilm, deep sea, shipwreck, built environment, Gulf of Mexico, dispersal, biogeography, microbiome
Citation: Moseley RD, Hampel JJ, Mugge RL and Hamdan LJ (2022) Historic Wooden Shipwrecks Influence Dispersal of Deep-Sea Biofilms. Front. Mar. Sci. 9:873445. doi: 10.3389/fmars.2022.873445
Received: 10 February 2022; Accepted: 29 April 2022;
Published: 08 June 2022.
Edited by:
Mari Heggernes Eilertsen, University of Bergen, NorwayReviewed by:
Dimitri Kalenitchenko, UiT The Arctic University of Norway, NorwayCopyright © 2022 Moseley, Hampel, Mugge and Hamdan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leila J. Hamdan, bGVpbGEuaGFtZGFuQHVzbS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.