- 1Fishery College, Zhejiang Ocean University, Zhoushan, China
- 2School of Ocean, Yantai University, Yantai, China
The intertidal salinity fluctuations provide pose a significant physical challenge for the crustaceans that live here. In the present study, Charybdis japonica was selected as the research species to explore the genetic regulatory mechanisms of intertidal crustaceans responding to salinity fluctuations. We cultured C. japonica s with three salinity gradients (the salinity of 15‰, 25‰, and 35‰) and 68.90 Gb clean transcriptome reads were obtained by RNA-seq. All clean reads were then de novo assembled to 41,058 unigene with a mean length of 1,179.39 bp and an N50 length of 2,033 bp. Furthermore, a total of 18,100 (24.74%) unigenes were successfully matched with the sequences from at least one database. We further analyzed the transcriptome structure of C. japonica and a total of 26,853 CDSs and 12,190 SSRs were predicted. The gene expression levels of C. japonica at 25‰ were used as control, and 204 and 5,392 DEGs were found at the salinity of 35‰ and 15‰, respectively. Based on the annotated information of DEGs, we speculated that C. japonicas were subjected to greater stress under the salinity of 35‰ and isotonic intracellular regulation was activated. Furthermore, neuronal activity, oxidation response, intracellular regulatory activity and osmotic regulation-related genes were speculated to be critical genes for C. japonica to cope with salinity fluctuations. The present results will provide fundamental information for revealing the salinity fluctuation-related genetic regulatory mechanisms of C. japonica and other intertidal crustaceans, and then help us predict how future salinity changes will affect the survival of crustaceans.
Introduction
The rapid climate change in the last decades has caused particularly alarming damage to marine ecosystems (Poloczanska et al., 2013; Molinos et al., 2016). Based on previous studies, seawater warming has been predicted to fluctuate dramatically and continuously due to the large amount of carbon dioxide (CO2) absorbed by seawater (Wiltshire et al., 2010). In fact, seawater warming is expected to increase the evaporation and air humidity, and further drive the salinity of ocean water toward more extremes, signifying lower salinity in low-salt areas and higher salinity in high-salt areas (Held and Soden, 2006). It is worth noting that salinity fluctuation may exceed the practical and potential microenvironment of some species and even threaten their survival (Nielsen and Brock, 2009; Waterkeyn et al., 2011).
Crustaceans are one of the most species-abundant taxa in the ocean and are widely distributed (Greenaway, 2003; Bar-On et al., 2018). Although the high global invasive potential of many crustaceans may mean that they are able to adapt to changing salinity, salinity still affects many physiological processes and natural behaviors of crustaceans (Whiteley et al., 2018; Young and Elliott, 2020). This is the case because there is lack effective salinity regulation mechanisms in crustaceans and thus their biological processes such as hormone release, metabolism and immunity will be hindered by osmoregulation disturbance caused by salinity changes (Huni and Aravindan, 1985). In fact, crustaceans also exhibit a variety of behavioral and physiological responses that allow them to relieve salinity changes (Whiteley and Taylor, 2015). Such as crustaceans move away from their original shelter where salinity changes, but may be exposed to new biological or abiotic stresses (Stein and Harzsch, 2021). Meanwhile, crabs such as Carcinus maenas has also been shown to respond to different salinities by modulating hemolymph (Winkler et al., 1988). Additionally, changes in the abundance of some neuroendocrine factors (i.e. neuropeptides) are also thought to involved in osmoregulation during salinity fluctuations by altering the neurobehavioral and immune behaviors of crustaceans (Delorenzi et al., 2000; Zhang et al., 2015). It is worth explaining that the difference in osmoregulation ability also leads to the salinity tolerance-specificity of crustaceans (Rubio et al., 2005; Ye et al., 2009). Specifically, euryhaline crustaceans appear to show greater responsiveness to salinity fluctuations because they are always exposed to continuous and significant salinity changes (Jung et al., 2012). In conclusion, there is no denying that some macroscopical effects of salinity fluctuations on crustaceans have been identified, but we know very little about the internal molecular regulatory mechanisms. Consequently, it is necessary to explore the regulatory mechanisms of crustaceans under salinity fluctuations at the molecular level.
Intertidal ecosystems show particularly pronounced and rapid salinity changes than open subtidal ecosystems, and such changes seem to more easily threaten the crustaceans’ survival compared with slow fluctuations in salinity averages (Somero, 2012; Stein and Harzsch, 2021). With this in mind, intertidal crustaceans may be more tolerant to salinity fluctuations, but the intertidal salinity fluctuations also provide pose a significant physical challenge for the crustaceans. Therefore, intertidal crustaceans can be used as excellent models to explore the responses of marine crustaceans to salinity fluctuations. As an intertidal crustacean, the Charybdis japonica is a euryhaline marine crab and predominantly distributed in the China, Japan, the Korean Peninsula and Southeast Asia countries (Yu et al., 2004). This species can tolerate a wide range of salinity from 10‰ to 35‰, and its optimum survival salinity ranges from 15‰ to 27‰. Therefore, the C. japonica could be chosen as proxies to explore the effects of salinity fluctuations on intertidal crustaceans. The acquisition of transcriptome will help to explore the gene-level regulation mechanisms in C. japonica under salinity fluctuation (Wang et al., 2009), but relevant studies have yet to be developed. At present, RNA sequencing (RNA-seq) based on high-throughput sequencing has been successfully used to identify gene expression patterns in a variety of crustaceans under salinity stresses, such as Oratosquilla oratoria (Lou et al., 2019), Scylla paramamosain (Wang et al., 2018) and others. Meanwhile, salinity fluctuations have also been shown to activate some gene (i.e. CPT-1, LRP1, etc) expression, enzyme (i.e. Na+/K+-ATPase, etc) activity, and metabolic pathways (i.e. glucocorticoid receptor signaling, protein kinase A signaling, etc) in crabs, and ultimately improve their fitness (Chen et al., 2015; Wang et al., 2015; Wang et al., 2018). Therefore, there is every reason to believe that RNA-seq can reveal the genetic modulation of C. japonica under salinity stresses.
Here, RNA-seq was applied to determine the transcriptome of C. japonicas under three salinity gradients and we then identified differentially expressed genes (DEGs) in C. japonicas under different salinities based on comparative transcriptome. We hope to discover the salinity regulation-related genes and relevant regulation mechanisms of C. japonica. Furthermore, the aim was to provide a theoretical basis for predicting the effects of salinity fluctuations caused by climate change on C. japonica and crustaceans.
Methods
Ethics Approval
We have read the animal experiments-related policies and we have guaranteed that our research is in compliance. Meanwhile, C. japonica is not protected animal in China or any other countries. In conclusion, the present research did not require specific authorization. In fact, frost anesthesia was also used to reduce the suffering of all C. japonica individuals during dissection.
C. japonica Maintenance and Salinity Fluctuation Schemes
We have collected 30 healthy female C. japonicas (mean carapace length: 50.43 mm; mean carapace width: 72.33 mm; mean weight: 72.85 g) from the coastal water of Zhoushan (Temperature: 19.89°C; Salinity: 33.13 ‰), China. All individuals were equally and randomly distributed into 3 plastic aquariums (temperature: 20°C, salinity: 25‰, length × width × height: 100 cm × 70 cm × 40 cm) and then kept temporarily for 48 hours. During the temporary rearing process, the sea water in each aquarium is continuously oxygenated and circulated.
As a euryhaline crustacean, C. japonica can survive at salinities ranging from 10‰ to 35‰. Therefore, we designed three salinity gradients (15‰, 25‰ and 35‰). After acclimatization, we have increased the salinity of one aquarium from 25‰ to 35‰, decreased the salinity of another aquarium from 25‰ to 15‰, and kept the salinity of the remaining an aquarium at 25‰. The salinity increase and decrease processes were completed within 1 hour (salinity fluctuation rate: 10‰/hour). C. japonicas remain in each salinity for 12 hours and thus reducing the impact of circadian rhythms on their regulatory mechanisms. In brief, three salinity gradients (15‰, 25‰, and 35‰) were performed in the present study, and 25‰, 15‰, and 35‰ was as control salinity, low- and high- salinity stresses, respectively. It is worth noting that all factors except salinity are similar in the three aquariums.
Sample Collection and RNA Extraction
Three C. japonicas were randomly selected from each salinity group. Considered that the gill tissue is critical organs for respiration and osmotic regulation, and then gill tissues from nine individuals was immediately harvested after frost anesthesia. All obtained gill tissues were separately snap-frozen in liquid nitrogen and then stored at -80°C for the subsequent RNA extraction. We have isolated the total RNA of each gill tissue using a standard Trizol Reagent Kit (Huayueyang Biotech Co., Ltd., Beijing, China) and following the manufacturer’s protocol and all total RNAs was then quantified based on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). We further used RNA Purification Beads (Illumina, San Diego, CA, USA) to delete rRNA from each total RNA to obtain mRNA for subsequent RNA-seq, and purified all the mRNAs for three times with Beads Binding Buffer (Illumina, San Diego, CA, USA).
RNA-Seq Library Preparation and Sequencing
The purified mRNAs was incubated at 94°C for 8 min to complete fragmentation. The segmented mRNAs was then combined with 8 μl of First Strand Synthesis Act D Mix (Illumina, San Diego, CA, USA) and SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and incubated at 25°C for 50 min to synthesize the first-strand cDNA. Subsequently, we added 5 μl of End Repair Control (Illumina, San Diego, CA, USA) and 20 μl of Second Strand Marking Master Mix (Illumina, San Diego, CA, USA) to the first-strand cDNA, and the formed conjugated product was incubated at 16°C for 60 min to synthesize the second-strand cDNA. Each synthesized double-stranded cDNA was purified using 90 μl of AMPure XP beads (Beckman Coulter, Beverly, USA) and then added with A-tailing and adapter ligation using A-Tailing and Ligation Control. Furthermore, 5 μl of PCR Primer Cocktail (Beckman Coulter, Beverly, USA) and 25 μl of PCR Master Mix (Beckman Coulter, Beverly, USA) were added to the all double-stranded cDNAs mentioned above and the compound was then amplified using the following enrich reaction system: the initial denaturing at 98°C for 30 sec, the 15 cycles of denaturing at 98°C for 10 sec, the annealing at 60°C for 30 sec, and the extension at 72°C for 30 sec, the final extension at 72°C for 5 min. Dilute all cDNA libraries to 10 pM. The quantified cDNA libraries were finally performed on an Illumina HiSeq 2000 platform across one lane with 150 bp paired-end.
Transcriptome Sequencing Reads Processing
The low-quality reads (including reads containing sequencing adaptors, the proportion of unknown nucleotides greater than 10%, and the quality score less than 20) in the raw transcriptomic reads generated by the Illumina sequencing platform were filtered out to obtain the high-quality clean transcriptomic reads. We have combined the clean reads of 9 C. japonicas into a multifasta file and Trinity software (version 2.4.0; Grabherr et al., 2011) was applied to de novo assembly to obtain the transcripts. The parameter of Trinity software was as follows: min_kmer_cov 3. Corset software (version 1.05; Nadia and Alicia, 2014) was applied to map all reads to transcripts to hierarchically cluster the sequences, and ultimately spliced the unigenes for subsequent analyses. The default parameters were used for Corset software. To explore the functional information of these unigenes, we compared these unigenes to the National Center for Biotechnology Information (NCBI) Non-redundant (NR), Swiss-Prot, Gene Ontology (GO), Eukaryotic Othologous Groups (KOG), eggNOG and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLAST software (Altschul, 1990) and the parameter as follows: E < 1e−5.
Transcriptome Structure Analyses
The transcriptome structure (including coding sequences [CDSs] and simple sequence repeats [SSRs]) of C. japonicas were further analyzed. Firstly, we use TransDecoder software to predict the CDSs of unigenes, and software parameters are the default. Subsequently, SSRs of unigenes were excavated based on MISA (version 1.0), and the software parameters (SSR type [including mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, hexanucleotide] - repetition number) as follows: 1-10, 2-6, 3-5, 4-5, 5-5, 6-5.
Salinity Fluctuation – Related Gene Regulation
It is considered that RNA extraction, library construction and sequencing may increase noise in samples and lead to outliers of gene expression, principal component analysis (PCA) was performed based on the expression levels of all genes to demonstrate the clustering of samples. To explore how C. japonicas adapt to fluctuating salinity through genetic regulation, we analyzed the differentially expressed genes (DEGs) and their biological functions in C. japonicas with different salinity gradients. Firstly, all unigenes of individual C. japonica were aligned to the multifasta file mentioned above using the “bwa-mem” algorithm in the BWA (version 0.7.12; Li and Durbin, 2009), the obtained expression level of each unigene of individual was standardized and then calculated the fragments per kilobase of exon model per million mapped fragments (FPKM) using DESeq2 (Leng et al., 2013), the software parameters are the default. Furthermore, we used the gene expression levels of C. japonicas at 25‰ as the control and then quantified the DEGs according to the gene expression levels of C. japonicas at 15‰ and 35‰. The false discovery rate (FDR) ≤ 0.01 and |log2 fold change (FC)| ≥ 2 were used as the screening thresholds of DEGs. The number of DEGs in C. japonicas at 15‰ and 35‰ were visualized using Venn, volcano and MA diagrams. We used Blast2GO software (E < 1e−5; Conesa et al., 2005) to predict the function of DEGs to explore the GO terms and pathways of C. japonica response to salinity fluctuations. Furthermore, we calculated the P-value of each GO term and conducted Bonferroni correction for all P-values. The GO terms with corrected P-values lower than 0.05 were considered to be significantly enriched. Meanwhile, enrichment factors were used to analyze enrichment degree of KEGG pathways, and qvalue after Fisher’s exact test represented enrichment significance.
Accuracy Validation of RNA-Seq Data Based on Quantitative Reverse Transcription PCR (qRT-PCR)
In the present study, qRT-PCR experiments were performed to verify the accuracy of RNA-seq data. Eight genes (including FAXC, Cystatin, ADAMTS1, NKCC, TauD, SNTG1, LIMD, and PTP3) that might contribute to the C. japonica cope with salinity fluctuations were chosen for qRT-PCR experiments and Primer Premier 6.0 software was used to design these genes-specific primers (Table 1). Meanwhile, β-actin (ACTB) gene and 18s gene were selected as the reference genes for the internal standardization of the present qRT-PCR results. We used the nuclease-free water to dilute 9 cDNA samples 20-fold according to the standard curve to obtain cDNA templates for qRT-PCR experiments. A 25 μL qRT-PCR reaction system was designed following the manufacturer’s instructions for SYBR® Premix Ex TaqTM (Tli RNaseH Plus) RR420A kit and the reaction system consisted of 2.0 μl of cDNA template, 12.5 μl of SYBR Premix Ex Taq (2×), 0.5 μl of each of the forward and reverse primers and 9.5 μl of nuclease-free water. Then, the constructed qRT-PCR reaction system was performed in an ABI PRISM 7300 Real-time PCR System and the amplification processes consisted of a holding stage of 30 sec at 95°C, followed by 40 cycles of 5 sec at 95°C and 30 sec at 54°C. Each qRT-PCR reaction was designed to be repeated three times to increase the accuracy of the ultimate results. Finally, 2−ΔΔCT (ΔCT = CTsalinity fluctuations response-related genes − CTACTBand18S, ΔΔCT = ΔCT15‰ or 35‰ – ΔCT25‰) method was applied to calculate the relative expression levels of eight salinity fluctuations response-related genes.
Results
Transcriptome Sequencing Results of Nine C. japonicas
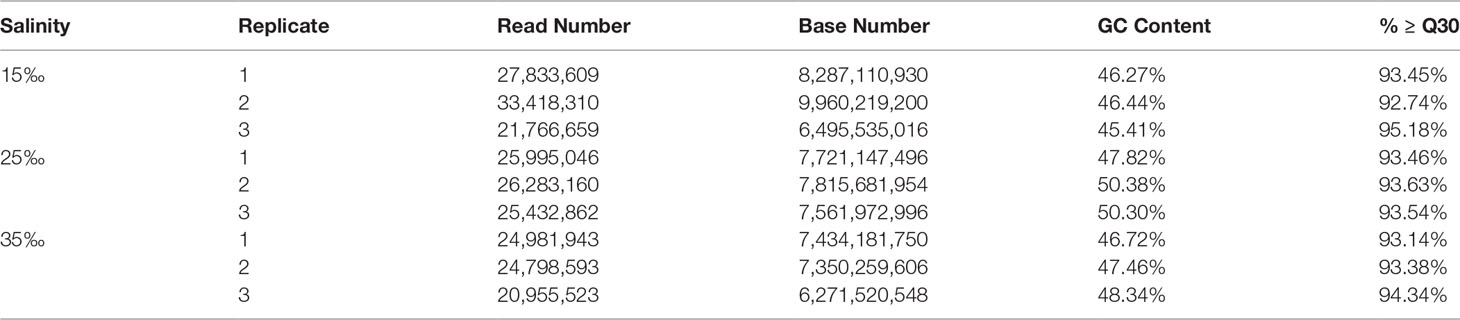
The raw RNA-seq reads of nine C. japonicas sequenced using the Illumina HiSeq 2000 platform were placed in the NCBI Sequence Read Archive (SRA) database under the accession number of SRR17422011, SRR17422010, SRR17422009, SRR17422008, SRR17422007, SRR17422006, SRR17422005, SRR17422004, and SRR17421997 under BioProject PRJNA793994. A total of 68.90 Gb clean reads were obtained after removing the low-quality reads of raw RNA-seq reads and the statistical results of all clean reads are shown in Table 2. The clean reads obtained in each C. japonica were exceeded 6.27 Gb. Meanwhile, the Q30 of all clean reads were exceeded 90%.
C. japonica Transcriptome De Novo Assembling, Annotation and Structure Analyses
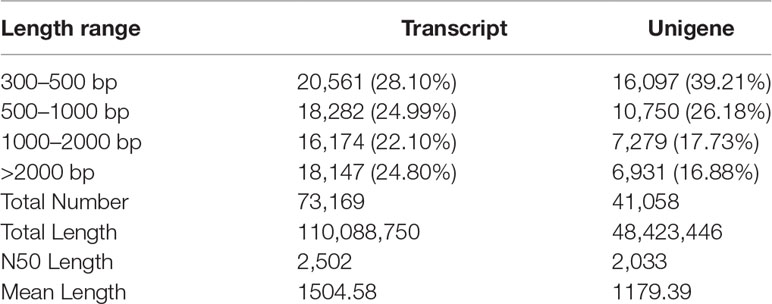
All clean reads of nine C. japonica individuals were applied to perform de novo assembly and 73,169 transcripts were generated. The N50 and mean length of transcripts was 2,502 bp and 1,504.58 bp, respectively. Furthermore, a total of 41,058 unigenes were spliced after redundant transcripts were removed and the N50 and mean length of unigenes was 2,033 bp and 1,179.39 bp, respectively (Table 3). We further compared all unigenes with the sequences in the protein database mentioned above, and the comparison results showed that 16,262, 9,901, 7,834, 11,626, 16,189 and 8,453 unigenes were successfully matched with the sequences in NR, Swiss-Prot, GO, KOG, eggNOG and KEGG database, respectively. Meanwhile, a total of 18,100 (24.74%) unigenes were successfully matched with the sequences from at least one database. We further analyzed the transcriptome structure of C. japonica and a total of 26,853 CDSs and 12,190 SSRs were predicted. Meanwhile, we also found a negative correlation between the number of SSR and base repetition, and the number of mononucleotides, dinucleotides, trinucleotides, tetranucleotides, pentanucleotides and hexanucleotides were 4,124, 4,261, 2,767, 162, 7 and 4, respectively.
Samples Clustering
PCA was performed on the expression levels of all genes in nine C. japonicas from three salinity gradients (15‰, 25‰ and 35‰). Results showed that C. japonicas from the same salinity had similar gene expression levels and eventually clustered together (Figure 1).
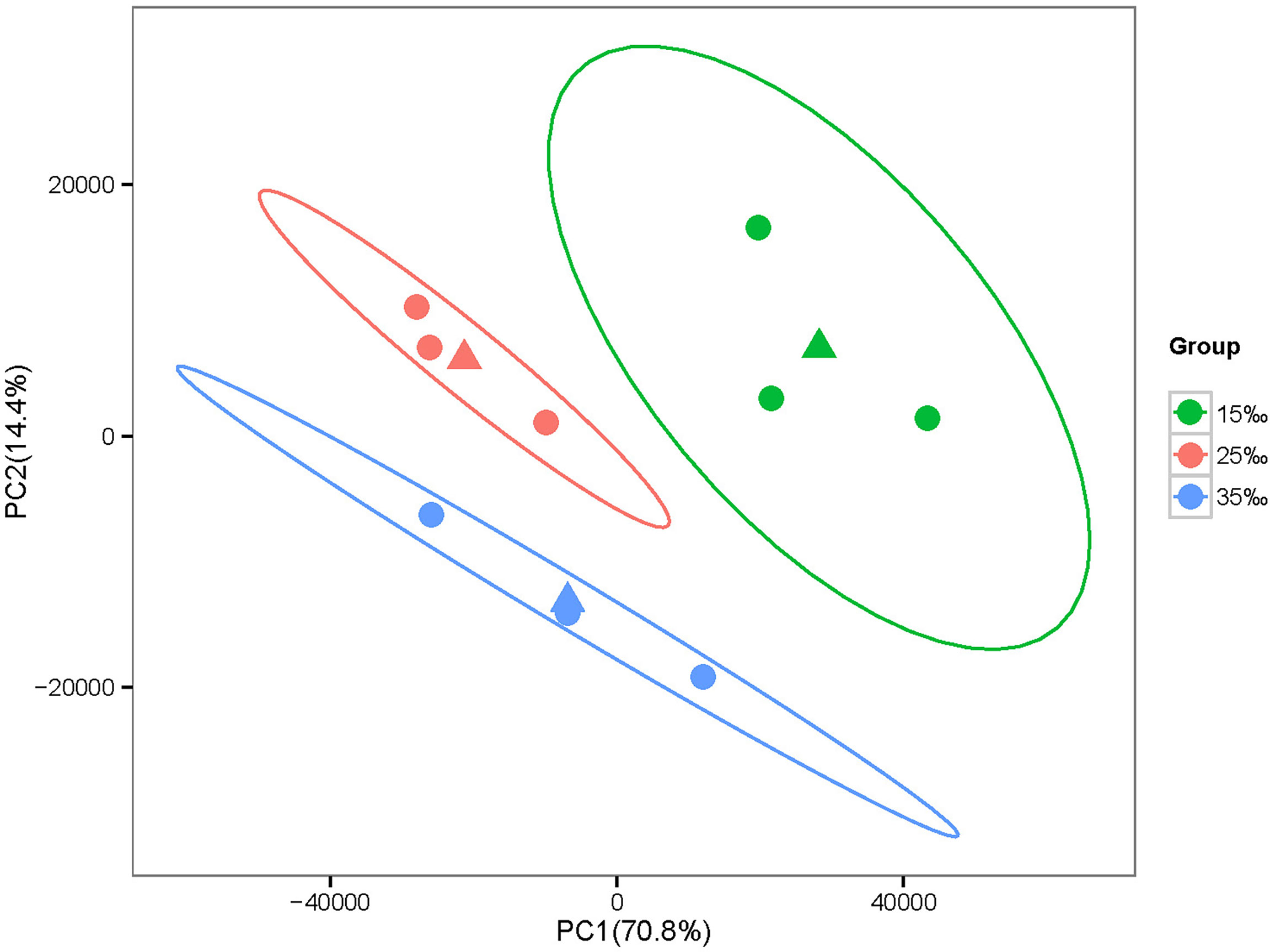
Figure 1 PCA based on the expression levels of all genes. Note: The same color represents three individuals at the same salinity. Meanwhile, the difference between the first principal component (PC1) and the second principal component (PC2) can explain 70.8% and 14.4% of the all analysis results, respectively.
The Number of DEGs That Occur With Salinity Fluctuations
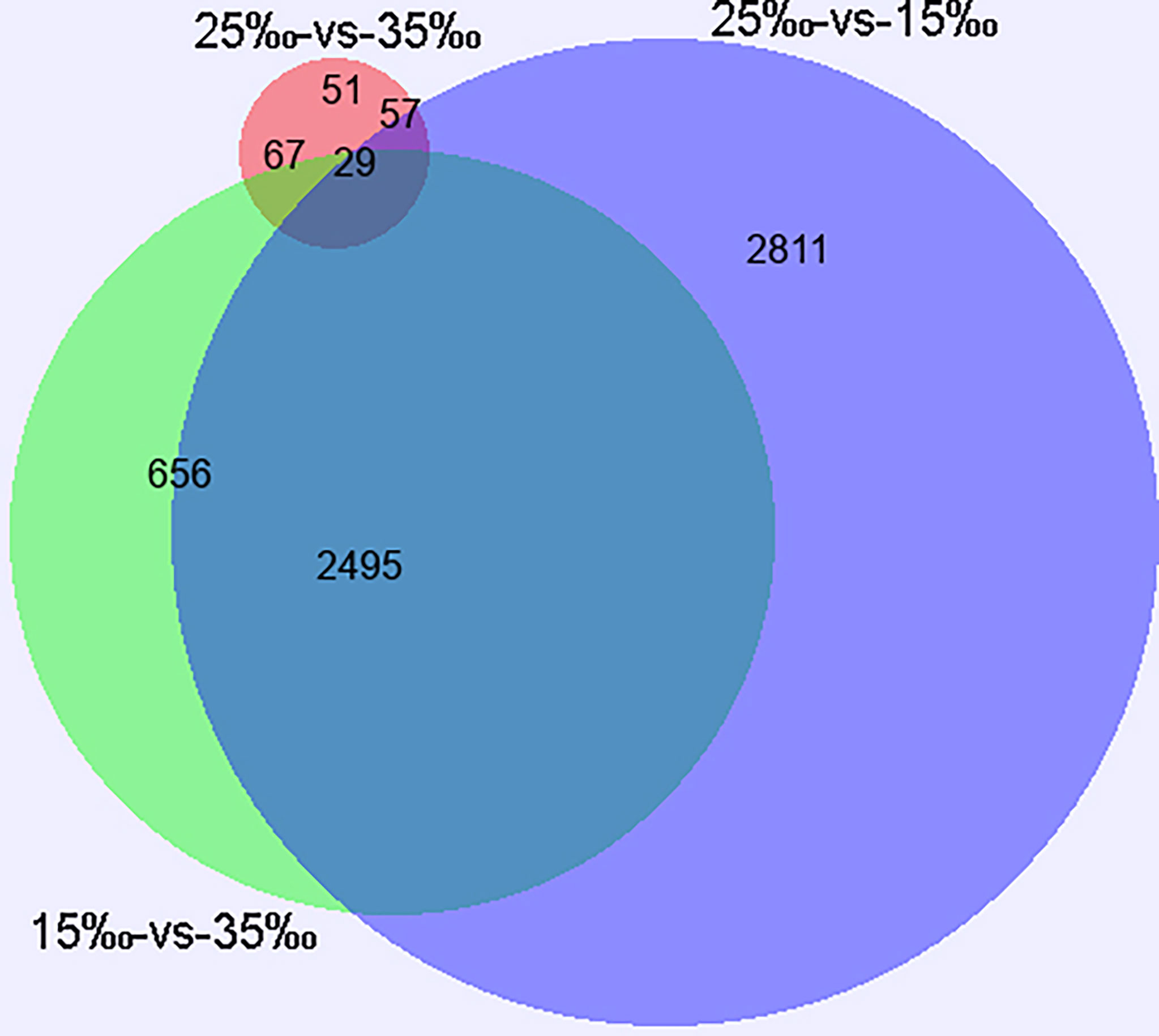
The present study used the gene expression levels of C. japonica at the salinity of 25‰ as controls and then counted the number of genes that expression levels changed in C. japonica at the salinity of 15‰ and 35‰. We compared the number of DEGs of three salinity gradients based on Venn, volcano and MA diagrams. Venn diagram showed that a substantial overlap of DEGs were found at different pairs (15‰-vs-35‰, 25‰-vs-15‰, and 25‰-vs-35‰), although 29 unigenes were differentially expressed in three pairs (Figure 2). Meanwhile, volcano and MA diagrams showed that the expression levels of 204 genes changed significantly (73 up- and 131 down-regulated) when the salinity increased to 35‰ while 5,392 genes were significantly changed (2,362 up- and 3,030 down-regulated) when salinity decreased to 15‰. Furthermore, compared with the salinity of 15‰, the expression levels of 1,681 and 1,566 genes of C. japonica in the salinity of 35‰ were up-regulated and down-regulated, respectively (Figure 3).
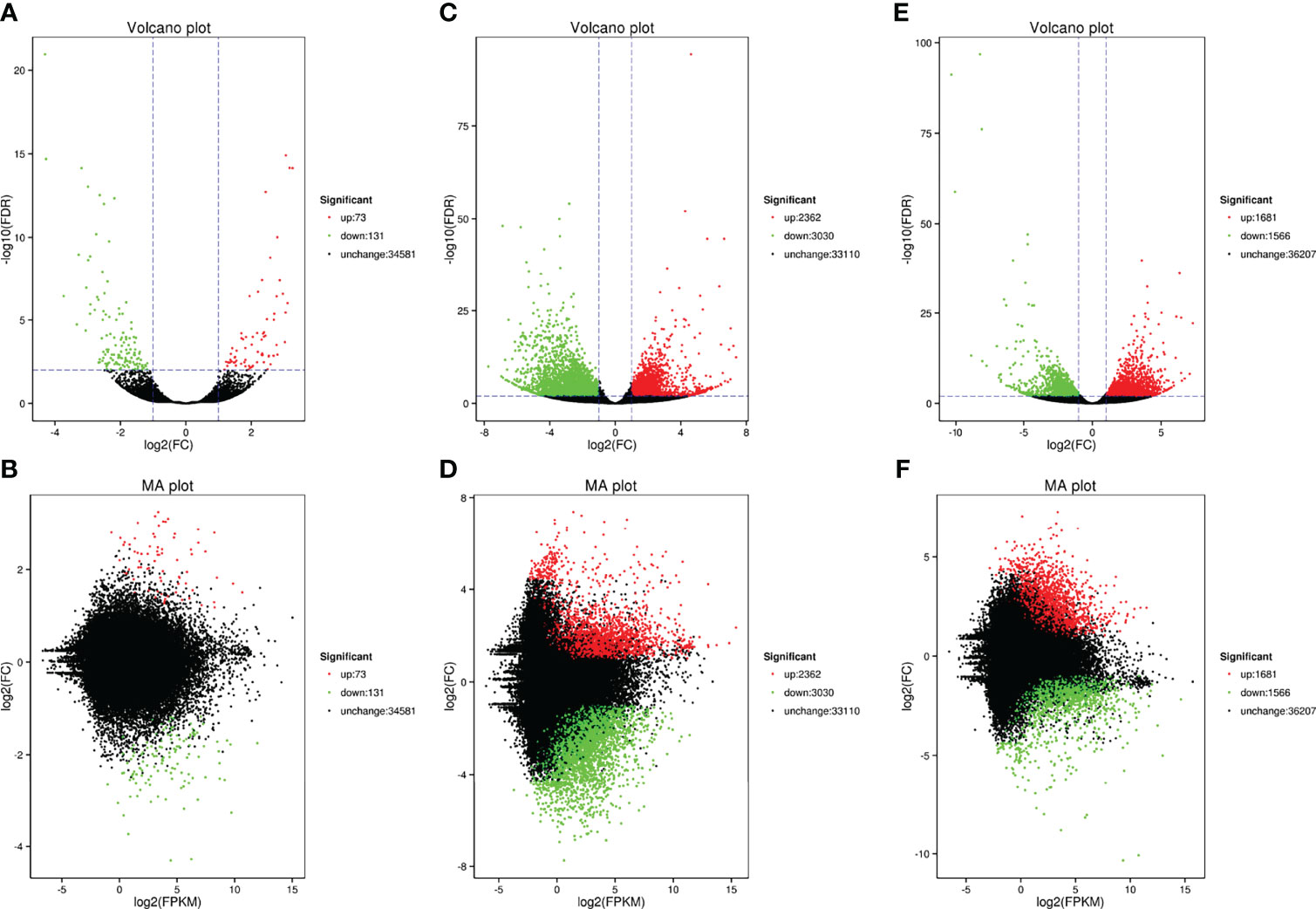
Figure 3 The number of DEGs of C. japonica among three salinity gradients. (A, B)represents the DEGs between the salinity of 25‰ and 35‰; (C, D) represents the DEGs between the salinity of 15‰ and 25‰; (E, F) represents the DEGs between the salinity of 15‰ and 35‰.
The GO Enrichment and KEGG Pathway of DEGs That Occur With Salinity Fluctuations.
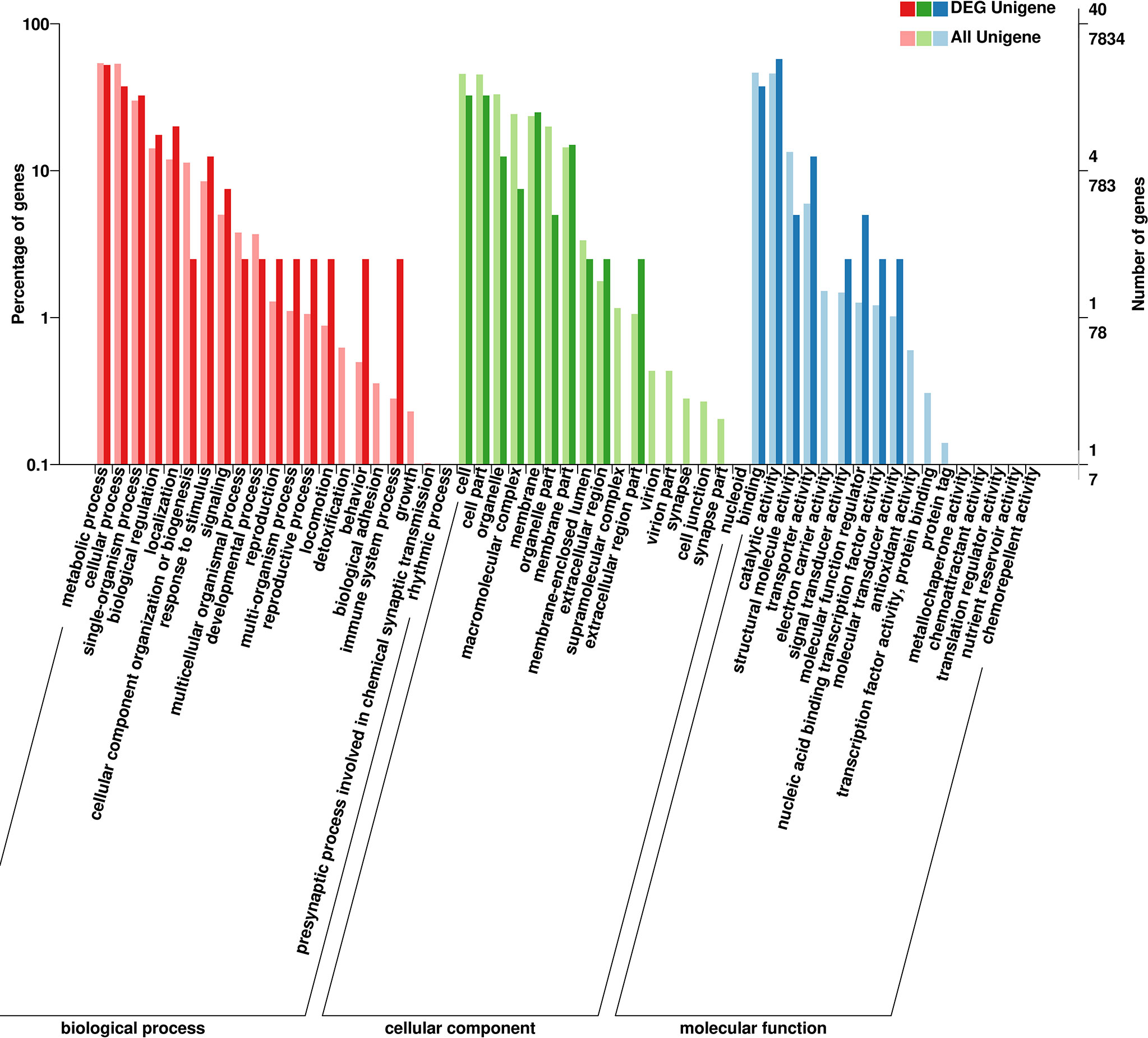
To explore how the above-mentioned DEGs might potentially regulate C. japonica responses to salinity fluctuations, we conducted GO enrichment (Figure 4) and KEGG pathway (Figure 5) analyses for these DEGs.
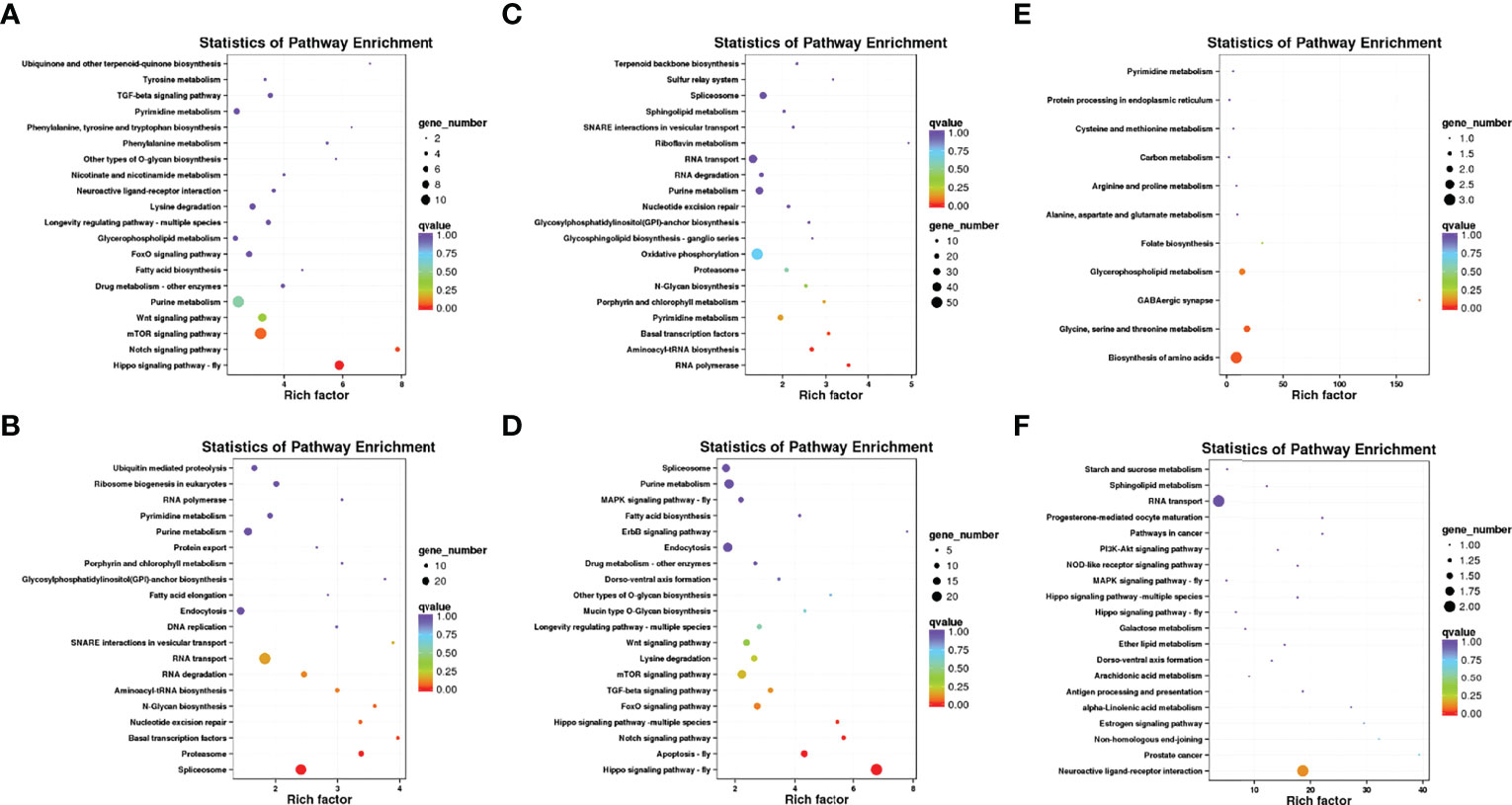
Figure 5 The enrichment analysis of KEGG pathways of DEGs of C. japonica exposed to three salinity gradients. (A) compared with the salinity of 15‰, the KEGG pathways of up-expressed unigenes of C. japonica at the salinity of 35‰ and (B) compared with the salinity of 15‰, the KEGG pathways of down-expressed unigenes of C. japonica at the salinity of 35‰; (C) compared with the salinity of 25‰, the KEGG pathways of up-expressed unigenes of C. japonica at the salinity of 15‰; (D) compared with the salinity of 25‰, the KEGG pathways of down-expressed unigenes of C. japonica at the salinity of 15‰; (E) compared with the salinity of 25‰, the KEGG pathways of up-expressed unigenes of C. japonica at the salinity of 35‰; (F) compared with the salinity of 25‰, the KEGG pathways of down-expressed unigenes of C. japonica at the salinity of 35‰.
GO enrichment showed that the functions of all DEGs mainly includes three categories, including biological processes, cellular components and molecular functions. Results showed that the functions of DEGs and their gene products were remarkably enriched in cell (GO: 0005623) and cell part (GO: 0044464) under cellular component; in catalytic activity (GO: 0003824) and binding (GO: 0005488) under molecular function and in metabolic process (GO: 0009987) and cellular process (GO: 0008152) under biological processes. Meanwhile, whether salinity increases (25 to 35‰ or 15 to 35‰) or decreases (25 to 15‰), the functions of up- or down- regulated genes and their gene products are similar.
KEGG pathway enrichment showed that the up-regulated genes were significantly enriched in the biosynthesis of amino acids pathway (ko01230; P = 0.043468) when the salinity increased to 35‰, but the down-regulated genes were not significantly enriched in any pathway. Meanwhile, although the up-regulated genes were significantly enriched in the RNA polymerase pathway (ko03020; P = 0.005364), basal transcription factors pathway (ko03022; P = 0.031888), and aminoacyl-tRNA biosynthesis pathway (ko00970; P = 0.011846) when the salinity decreased to 15‰, the down-regulated genes were significantly enriched in the hippo signaling pathway (ko04391; P = 9.94E-12), apoptosis pathway (ko04214; P = 0.000653), and notch signaling pathway (ko04330; P = 0.005347). Additionally, compared with the salinity of 15‰, the up-regulated genes of C. japonicas at the salinity of 35‰ were significantly enriched in hippo signaling pathway (ko04391; P = 9.94E-12) and notch signaling pathway (ko04330; P = 0.005347), whereas the down-regulated genes were significantly enriched in the spliceosome pathway (ko00564; P = 0.002237), proteasome (ko03050; P = 0.004687) and basal transcription factors pathway (ko03022; P = 0.031891).
Validation of Gene Expression Level by qRT-PCR
We randomly selected 8 potential salinity regulation-related genes and performed qRT-PCR to obtain gene expression levels and verify the accuracy of transcriptome data. Results showed that the expression trends of the 8 genes were similar, whether based on transcriptome or qRT-PCR (Figure 6). However, it is worth noting that the expression trends of different genes at different salinities are specific. Specifically, the expression levels (relative expression level > 1) of Cystatin gene and SNTG1 gene increased with increasing or decreasing salinity, but the expression levels (relative expression level < 1) of ADAMTS1 gene, NKCC gene, LIMD gene, PTP3 gene decreased with the increase or decrease of salinity. Additionally, the expression level of FAXC gene increased with the decrease of salinity and decreased with the increase of salinity, but the expression trend of TauD gene was opposite.
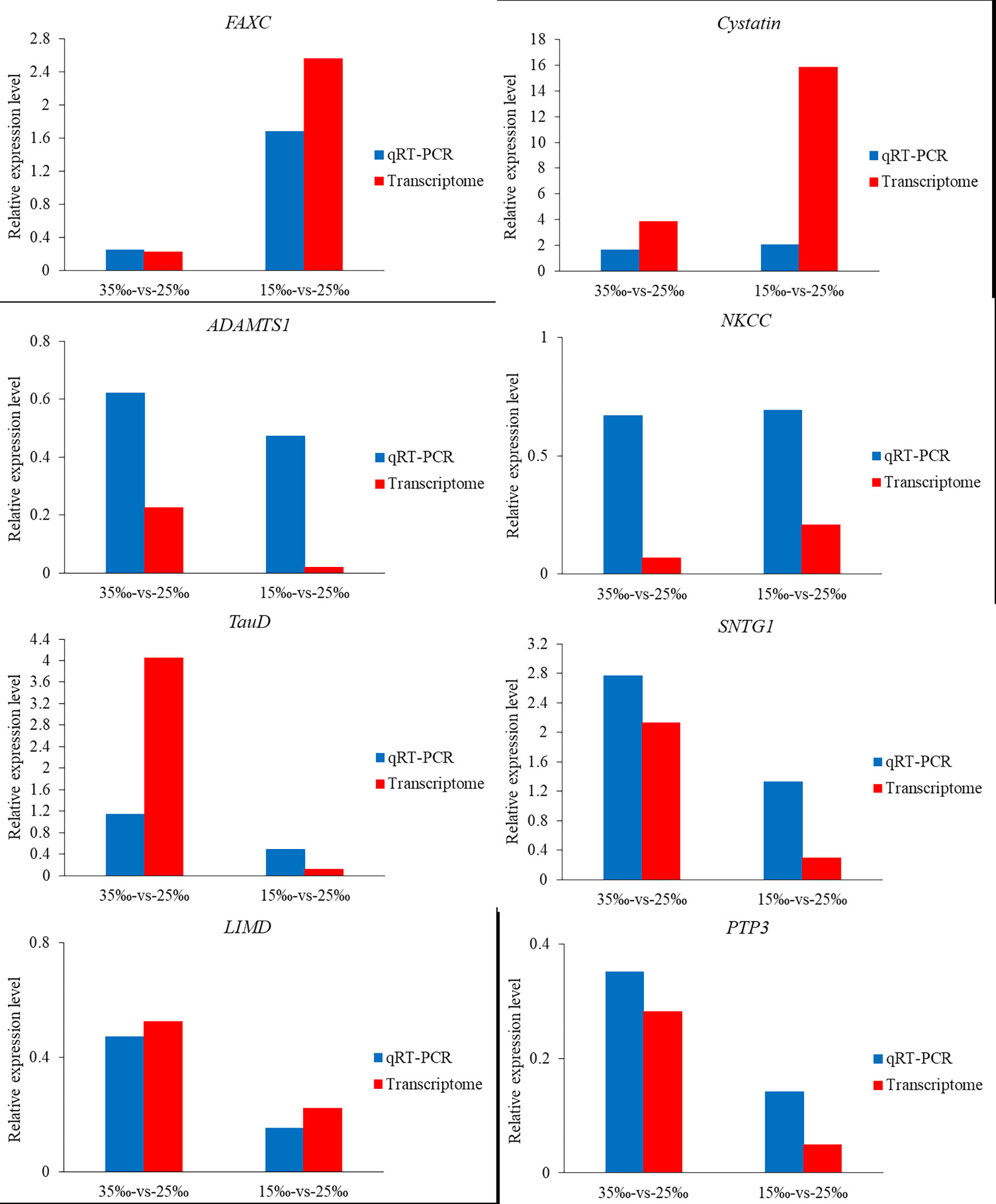
Figure 6 Relative expression level of 8 salinity fluctuation-related genes based on transcriptomes data and qRT-PCR data.
Discussion
Although crustaceans appear to cope with salinity fluctuation with a range of natural behavioral (i.e. fluctuation) and physiological (i.e. osmotic regulation) responses, it is undeniable that long-term and constant salinity changes can disrupt the metabolism, immunity, and reproduction of crustaceans (Ruscoe et al., 2004; Rahmi and Zeng, 2007; Whiteley et al., 2018). In fact, intertidal crustaceans are more susceptible to additional salinity changes, because the tide fluctuation often exposes them to extreme salinity (Hofmann et al., 2011; Baumann et al., 2015). Therefore, C. japonica was selected as the research object in this study and the genetic response of intertidal crustaceans to salinity fluctuations was explored.
We explored the gene regulatory mechanisms of C. japonica responding to salinity fluctuations from two aspects of the number and function of DEGs. Although the expression levels of more genes changed significantly as the salinity decreased to 15‰, we cannot simply infer that C. japonicas were under greater stress at this salinity. In fact, we also found that a large number of genes were down-regulated in the C. japonicas of the salinity of 15‰, but it is not clear that the salinity of 15‰ is more suitable for C. japonica survival than 25‰. Future studies still need to set more intensive salinity gradients within the optimum salinity range (15‰ and 27‰) to provide evidence for accurate more optimal salinity of C. japonica from the gene expression level. However, the present results do not prevent us from confirming that C. japonica need to mobilize different numbers of genes in response to different external salinity changes. It is worth noting that C. japonicas with the salinity of 35‰ had a large proportion of up-regulated genes than C. japonicas with the salinity of 15‰, which seems to indicate that C. japonicas are subjected to stress at the salinity of 35‰ and some functional genes come into play.
We further identified the gene functional changes of C. japonica at different salinities and results showed that the function of DEGs were mainly enriched in the cellular structure, membrane, metabolic process, catalytic activity and other GO terms. This means that the DEGs may change the cellular level, enzyme and hormone content of C. japonica, and ultimately help maintain homeostasis in C. japonica during salinity fluctuations (Romano and Zeng, 2012). Previous study has confirmed that the osmotic pressure of crustaceans remains in a steady isotonic state with the surrounding salinity, but this equilibrium is easily upset by salinity fluctuations (Marshall and Grosell, 2006). In order to maintain isotonic state, crustaceans in low-salt environment need to absorb a large number of ions and expel urine constantly, which is mainly to cope with the large amount of water entering the body through gill cells, while crustaceans in high-salt environment need to expel more iron through gill to cope with water loss (Marshall and Grosell, 2006). Except for the osmotic regulation of the cells themselves, the endocrine organs of crustaceans also influence certain critical ion transport enzymes (i.e. Na+-K+-ATPase) through the synthesis and release of hormones, and ultimately promote ion absorption or emission (Pequeux, 1995; Péqueux et al., 2002; Wang et al., 2018). In conclusion, the biological functions (such as material transport, energy conversion and information transfer) related to the osmotic regulation of C. japonica will change with the salinity fluctuation, which will activate or inhibit the ion conversion and hormone release-related critical genes, and ultimately ensure the survival of C. japonica under different salinity.
KEGG pathway enrichment analyses can help us understand the regulatory mechanisms of C. japonica responding to salinity fluctuations in the surrounding environment and the present study also found that DEGs in different salinity were significantly enriched in different pathways. Results showed that the DEGs and related pathways of C. japonica are more pronounced when salinity decreased to 15‰ than when salinity increased to 35‰. It is worth noting that salinity is an important abiotic factor that affect the growth, survival, and molting of crustacean (Liu et al., 2022). Considering that the optimum survival salinity ranges of C. japonica from 15‰ to 27‰, thus we came up with provocative thesis: the salinity of 15‰ is belonging to the optimum survival salinity range, allowing more genes to be mobilized and activated in response to the salinity fluctuations, in fact, heat shock proteins have been shown to be highly expressed at low salt levels to reduce cell damage (Jin et al., 2022); Meanwhile, the salinity of 35‰ reached the survival threshold of crabs, and the functional gene overexpression tended to the survival threshold and ceased to function (Lou et al., 2022). However, future studies are needed to test these hypotheses. The up-regulated genes of C. japonica at the salinity of 35‰ were significantly enriched in the biosynthesis of amino acids pathway. In fact, previous study has suggested that crustaceans generally adapt to salinity fluctuations through both non-isotonic extracellular and isotonic intracellular regulation (Pequeux, 1995; Augusto et al., 2007). Non-isotonic extracellular regulation is the primary osmotic regulation mechanism of crustaceans, and it can play a role in the initial stage of salinity fluctuation through ion transport or hormone regulation. When salinity stress reaches the limit of crustaceans, isotonic intracellular regulation is activated as a compensatory mechanism, and the isotonic equilibrium between the organism and the external salinity is guaranteed by changing the amino acid content in crustaceans (Augusto et al., 2007). Amino acid content has been proved to contribute significantly to the osmotic pressure level of many crustaceans (even up to 70%) (Gerard and Gilles, 1972), and the amino acid content of crustaceans also increases with the increase of ambient salinity (Cobb et al., 1975; Wang et al., 2012; Lu et al., 2015). Therefore, the up-regulated of amino acid synthesis-related genes also means that the salinity of 35‰ has reached the salinity tolerance limit of C. japonica. At this time, C. japonica needs to synthesize amino acids quickly to maintain the isotonic balance between the body and the external environment. The up-regulated genes of C. japonica at the salinity of 15‰ were significantly enriched in the genetic information transmission-related pathways, but the related pathways were significantly enriched by the down-regulated genes of C. japonica at the salinity of 35‰. This may mean that salinity stress may affect the transmission of genetic information, but the exact mechanisms are not clear. Additionally, the down-regulated genes of C. japonica at the salinity of 15‰ were significantly enriched in the hippo and notch signaling pathways, but these two pathways were significantly enriched by the up-regulated genes of C. japonica at the salinity of 35‰. Hippo signaling pathway and notch signaling pathway are widely and evolutionarily highly conserved pathways in invertebrates and vertebrates (Callahan and Egan, 2004; Zhao et al., 2010). It is worth noting that inhibition of hippo signaling pathway might effectively reduce the apoptosis of C. japonica under salinity stress, but inhibition of notch signaling pathway might affect the embryonic development of C. japonica (Callahan and Egan, 2004; Mummery-Widmer et al., 2010; Zhao et al., 2010). This provides evidence for our conjecture above that C. japonica are subjected to greater stress at the salinity of 35‰, which indicates that the salinity of 35‰ may affect cell proliferation or growth of C. japonica, leading to cell apoptosis and ultimately death of C. japonica. Additionally, we also speculate that low salinity within the survival salinities seems to be more likely to limit reproductive behavior of C. japonica, but this hypothesis needs further verification.
Eight genes associated with potential salinity responses were further analyzed in the present study. Salinity stress can affect the neural behavior and thus the maintenance of neuronal activity in C. japonica may help to mitigate the behavioral and endocrine damages caused by environmental changes (Kushinsky et al., 2019). Interestingly, we found that FAXC gene is up-regulated in the salinity of 15‰ and this gene may play an important role in the neuronal axonal development of C. japonica. Salinity fluctuations has been shown to pose a direct challenge to motor axons (especially leg axons), and the membrane potential and volume of axons in some crustaceans also vary with salinity changes (Gleeson et al., 2000; Curtis et al., 2010). Therefore, the up-regulated of FAXC may help promote rapid salinity adaptive behavior in C. japonica under the 15‰. However, the neuronal activity of C. japonica fails at the salinity of 35‰ probably because the C. japonica needed to expend more energy to cope with stress at this salinity (Stein and Harzsch, 2021). At least one study has suggested salinity fluctuations can altered long-term memory of crustaceans (Delorenzi et al., 2000). A gene (SNTG1; Joe et al., 2012) that may alter memory in C. japonica by affecting the balance of water molecules and potassium (K+) outside the cell was found to be up-regulated in the salinity of 15‰ and 35‰. In conclusion, specific neuronal activity adaptations may protect the C. japonica from direct influences of salinity, but further research is needed to determine the mechanisms and extent of these adaptation. The cystatin protein encoded by Cystatin gene is a reversible and competitive inhibitor of cysteine proteases, which plays an important immune-modulatory role in crustaceans (Henskens et al., 1996; Pierre and Mellman, 1998; Li et al., 2010). As an acute-phase reactive negative-related protein, the content of cystatin protein decreases sharply when the organism is subjected to stresses (van-Oss et al., 1975; Lebreton et al., 1979). The Cystatin gene was down-regulated in C. japonica at the salinity of 35‰ but up-regulated in C. japonica at the salinity of 15‰, which seemed to confirm that the C. japonica were under more stress at the salinity of 35‰. Meanwhile, the high expression of oxidative stress-related TauD gene (Yew et al., 2016) also supports that C. japonica were subjected to greater stress at the salinity of 35‰. Additionally, three genes (ADAMTS1, LIMD and PTP3) might involve in intracellular regulatory activity were found to be down-regulated at both the salinity of 15‰ and 35‰, which may indicate that salinity fluctuation changes intracellular activity of C. japonica, but the mechanism is still unclear. Surprisingly, the NKCC gene was also down-regulated under both the salinity of 15‰ and 35‰. In fact, our previous study found that NKCC is a gene that facilitate salt extrusion (Lou et al., 2019). The up-regulation of NKCC in the high-salinity can promoted the transport of sodium (Na+), chlorine (Cl-) and K+ into the cell, which made the crustacean reach the isotonic state with the external salt environment (Hiroi et al., 2008; Velotta et al., 2014). In the present study, the NKCC of C. japonica was down-regulated in the salinity of 35‰, which may be due to the excessive stress suffered by C. japonica at this salinity, the gene potency was thus suppressed. Considering that the salinity of 15‰ is also within the optimum salinity range for C. japonica, further discussion of gene expression in this salinity is not required in this study. In the future, more salinity gradients will be required to accurately explore the expression levels of these functional genes.
Conclusion
The habitats of intertidal crustaceans are more likely to approach the salinity tolerance limit and therefore more sensitive to possible future salinity changes. To investigate the genetic response of intertidal crustaceans to salinity fluctuations, C. japonica was selected as subject and transcriptome differences were then compared at three temperature gradients. The present study confirm that C. japonica show specific regulatory mechanisms in response to high- and low-salinity. Additionally, salinity fluctuation is also speculated to affect the physiological processes of C. japonica, such as neuronal activity, oxidation response, intracellular regulatory activity and osmotic regulation. Our present study provides some insights into the genetic regulatory mechanisms of C. japonica and other intertidal crustaceans under further salinity fluctuation caused by climate change.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, accession numbers SRR17422011, SRR17422010, SRR17422009, SRR17422008, SRR17422007, SRR17422006, SRR17422005, SRR17422004 and SRR17421997.
Author Contributions
FL and ZH conceptualized this research conducted the analyses. FL and ZH have conducted all analyses of the present study. BS, YW, and FL have written this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFA0604904), the Zhejiang Provincial Natural Science Foundation of China (LR21D060003) and the National Natural Science Foundation of China (32070513).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Augusto A., Greene L. J., Laure H. J., Mcnamara J. C. (2007). Adaptive Shifts in Osmoregulatory Strategy and the Invasion of Freshwater by Brachyuran Crabs: Evidence From Dilocarcinus Pagei (Trichodactylidae). J. Exp. Zool. Part A. 307, 688–698.doi: 10.1002/jez.a.422
Bar-On Y. M., Phillips R., Milo R. (2018). The Biomass Distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511. doi: 10.1073/pnas.1711842115
Baumann H., Wallace R. B., Tagliaferri T., Gobler C. J. (2015). Large Natural Ph, CO2 and O2 Fluctuations in a Temperate Tidal Salt Marsh on Diel, Seasonal, and Interannual Time Scales. Estuar. Coast. 38, 220–231. doi: 10.1007/s12237-014-9800-y
Callahan R., Egan S. E. (2004). Notch Signaling in Mammary Development and Oncogenesis. J. Mammary. Gland. Biol. 9, 145–163. doi: 10.1023/B:JOMG.0000037159.63644.81
Chen K., Li E. C., Li T. Y., Xu C., Wang X. D., Lin H. Z., et al. (2015). Transcriptome and Molecular Pathway Analysis of the Hepatopancreas in the Pacific White Shrimp Litopenaeus Vannamei Under Chronic Low-Salinity Stress. PLoS One 10, e0131503. doi: 10.1371/journal.pone.0131503
Cobb B. F., Conte F. S., Edwards M. A. (1975). Free Amino Acids and Osmoregulation in Penaeid Shrimp. J. Agr. Food Chem. 23, 1172–1174. doi: 10.1021/jf60202a015
Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Curtis D. L., Vanier C. H., Mcgaw I. J. (2010). The Effects of Starvation and Acute Low Salinity Exposure on Food Intake in the Dungeness Crab, Cancer Magister. Mar. Biol. 157, 603–612. doi: 10.1007/s00227-009-1345-4
Delorenzi A., Dimant B., Frenkel L., Nahmod V. E., Nassel D. R., Maldonado H. (2000). High Environmental Salinity Induces Memory Enhancement and Increases Levels of Brain Angiotensin-Like Peptides in the Crab Chasmagnathus Granulatus. J. Exp. Biol. 203, 3369–3379. doi: 10.1046/j.1420-9101.2000.00229.x
Gerard J. F., Gilles R. (1972). The Free Amino-Acid Pool in Callinectes Sapidus (Rathbun) Tissues and its Role in the Osmotic Intracellular Regulation. J. Exp. Mar. Biol. Ecol. 10, 125–136. doi: 10.1016/0022-0981(72)90098-6
Gleeson R. A., Mcdowell L. M., Aldrich H. C., Hammar K., Smith P. J. (2000). Sustaining Olfaction at Low Salinities: Evidence for a Paracellular Route of Ion Movement From the Hemolymph to the Sensillar Lymph in the Olfactory Sensilla of the Blue Crab Callinectes Sapidus. Cell Tissue Res. 301, 423–431. doi: 10.1007/s004410000246
Grabherr M., Haas B., Yassour M., Levin J., Thompson D., Amit I., et al. (2011). Full-Length Transcriptome Assembly From RNA-Seq Data Without a Reference Genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Greenaway P. (2003). Terrestrial Adaptations in the Anomura (Crustacea: Decapoda). Memoirs. Museum. Victoria. 60, 13–26. doi: 10.24199/j.mmv.2003.60.3
Held I. M., Soden B. J. (2006). Robust Responses of the Hydrological Cycle to Global Warming. J. Clim. 19, 5686–5699. doi: 10.1175/JCLI3990.1
Henskens Y. M., Veerman E. C., Nieuw Amerongen A. V. (1996). Cystatins in Health and Diseases. Biol. Chem. HoppeSeyler. 377, 71–86. doi: 10.1007/s10989-008-9160-1
Hiroi J., Yasumasu S., McCormick S. D., Hwang P. P., Kaneko T. (2008). Evidence for an Apical Na-Cl Cotransporter Involved in Ion Uptake in Teleost Fish. J. Exp. Biol. 211, 2584–2599. doi: 10.1242/jeb.018663
Hofmann G. E., Smith J. E., Johnson K. S., Send U., Levin L. A., Micheli F., et al. (2011). High-Frequency Dynamics of Ocean Ph: A Multi-Ecosystem Comparison. PLoS One 6, e28983. doi: 10.1371/journal.pone.0028983
Huni A. A. D., Aravindan C. M. (1985). The Effect of Salinity on the Oxygen Consumption of Two Intertidal Crustaceans. Comp. Biochem. Physiol. 81, 869–871. doi: 10.1016/0300-9629(85)90921-1
Jin S. H, Deng Z. C, Xu S. Y, Zhang H. B, Han Z. Q. (2022). Genome-Wide Identification and Low-Salinity Stress Analysis of the Hsp70 Gene Family in Swimming Crab (Portunus Trituberculatus). Int. J. Biol. Macromol. 208, 126–135. doi: 10.1016/j.ijbiomac.2022.03.055
Joe M. K., Kee C., Tomarev S. I. (2012). Myocilin Interacts With Syntrophins and is Member of Dystrophin-Associated Protein Complex. J. Biol. Chem. 287, 13216–13227. doi: 10.1074/jbc.M111.224063
Jung D., Sato J. D., Shaw J. R., Stanton B. A. (2012). Expression of Aquaporin 3 in Gills of the Atlantic Killifish (Fundulus Heteroclitus): Effects of Seawater Acclimation. Comp. Biochem. Phys. A. Mol. Integr. Physiol. 161, 320–326. doi: 10.1016/j.cbpa.2011.11.014
Kushinsky D., Morozova E. O., Marder E. (2019). In Vivo Effects of Temperature on the Heart and Pyloric Rhythms in the Crab, Cancer Borealis. J. Exp. Biol. 222, jeb199190. doi: 10.1242/jeb.199190
Lebreton J. P., Joisel F., Raoult J. P., Lannuzel B., Rogez J. P., Humbert G. (1979). Serum Concentration of Human Alpha 2 HS Glycoprotein During the Inflammatory Process: Evidence That Alpha 2 HS Glycoprotein is a Negative Acute-Phase Reactant. J. Clin. Invest. 64, 1118–1129. doi: 10.1172/JCI109551
Leng N., Dawson J. A., Thomson J. A., Ruotti V., Rissman A. I., Smits B. M. G., et al. (2013). EBSeq: An Empirical Bayes Hierarchical Model for Inference in RNA-Seq Experiments. Bioinformatics 29, 1035–1043. doi: 10.1093/bioinformatics/btt087
Li H., Durbin R. (2009). Fast and Accurate Short Read Alignment With Burrows-Wheeler Transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li F., Gai X., Wang L., Song L., Zhang H., Qiu L., et al. (2010). Identification and Characterization of a Cystatin Gene From Chinese Mitten Crab Eriocheir Sinensis. Fish. Shellfish. Immun. 29, 521–529. doi: 10.1016/j.fsi.2010.05.015
Liu X. Y., Han X. L., Han Z. Q. (2022). Effects of Climate Change on the Potential Habitat Distribution of Swimming Crab Portunus Trituberculatus Under the Species Distribution Model. J. Oceanol. Limnol. doi: 10.1007/s00343-021-1082-1
Lou F. R., Gao T. X., Han Z. Q. (2019). Effect of Salinity Fluctuation on the Transcriptome of the Japanese Mantis Shrimp Oratosquilla Oratoria. Int. J. Biol. Macromol. 140, 1202–1213. doi: 10.1016/j.ijbiomac.2019.08.223
Lou F. R., Wang Y. J., Han X. Q., Shui B. N. (2022). Comparative Transcriptome Reveals the Molecular Regulation Mechanism of Charybdis Japonica to High- and Low-Temperature Stresses. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.849485
Lu J. Y., Shu M. A., Xu B. P., Liu G. X., Ma Y. Z., Guo X. L., et al. (2015). Mud Crab Scylla Paramamosain Glutamate Dehydrogenase: Molecular Cloning, Tissue Expression and Response to Hyposmotic Stress. Fisheries. Sci. 81, 175–186. doi: 10.1007/s12562-014-0828-5
Marshall W. S., Grosell M. (2006). Ion Transport, Osmoregulation and Acid-Base Balance. Physiol. Fishes., 177–230.
Molinos J. G., Halpern B. S., Schoeman D. S., Brown C. J., Kiessling W., Moore P. J., et al. (2016). Climate Velocity and the Future Global Redistribution of Marine Biodiversity. Nat. Clim. Change 6, 83. doi: 10.1038/nclimate2769
Mummery-Widmer J. L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., Chen D., et al. (2010). Genome-Wide Analysis of Notch Signalling in Drosophila by Transgenic RNAi. Nature 458, 987–992. doi: 10.1038/nature07936
Nadia M. D., Alicia O. (2014). Corset: Enabling Differential Gene Expression Analysis for De Novo Assembled Transcriptomes. Genome Biol. 15, 410. doi: 10.1186/s13059-014-0410-6
Nielsen D. L., Brock M. A. (2009). Modified Water Regime and Salinity as a Consequence of Climate Change: Prospects for Wetlands of Southern Australia. Clim. Change 95, 523–533. doi: 10.1007/s10584-009-9564-8
Péqueux A., Le Bras P., Cann-Moisan C., Caroff J., Sébert P. (2002). Polyamines, Indolamines, and Catecholamines in Gills and Haemolymph of the Euryhaline Crab, Eriocheir Sinensis. Effects of High Pressure and Salinity. Crustaceana 75, 567–578. doi: 10.1163/156854002760095606.6842111
Pierre P., Mellman I. (1998). Developmental Regulation of Invariant Chain Proteolysis Controls MHC Class II Trafficking in Mouse Dendritic Cells. Cell 93, 1135–1145. doi: 10.1016/S0092-8674(00)81458-0
Poloczanska E. S., Brown C. J., Sydeman W. J., Kiessling W., Schoeman D. S., Moore P. J., et al. (2013). Global Imprint of Climate Change on Marine Life. Nat. Clim. Change 3, 919. doi: 10.1038/nclimate1958
Rahmi N., Zeng C. (2007). Effects of Temperature and Salinity on the Survival and Development of Mud Crab, Scylla Serrata (Forsskal), Larvae. Aquac. Res. 38, 1529–1538. doi: 10.1111/j.1365-2109.2007.01810.x
Romano N., Zeng C. (2012). Osmoregulation in Decapod Crustaceans: Implications to Aquaculture Productivity, Methods for Potential Improvement and Interactions With Elevated Ammonia Exposure. Aquaculture 334, 12–23. doi: 10.1016/j.aquaculture.2011.12.035
Rubio V., Sánchez-Vázquez F., Madrid J. (2005). Effects of Salinity on Food Intake and Macronutrient Selection in European Sea Bass. Physiol. Behav. 85, 333–339. doi: 10.1016/j.physbeh.2005.04.022
Ruscoe I. M., Shelley C. C., Williams G. R. (2004). The Combined Effects of Temperature and Salinity on Growth and Survival of Juvenile Mud Crabs (Scylla Serrate Forskal). Aquaculture 238, 239–247. doi: 10.1016/j.aquaculture.2004.05.030
Somero G. N. (2012). The Physiology of Global Change: Linking Patterns to Mechanisms. Ann. Rev. Mar. Sci. 4, 39–61. doi: 10.1146/annurev-marine-120710-100935
Stein W., Harzsch S. (2021). The Neurobiology of Ocean Change - Insights From Decapod Crustaceans. Zoology 144, 125887. doi: 10.1016/j.zool.2020.125887
van-Oss C. J., Bronson P. M., Border J. R. (1975). Changes in the Serum Alpha Glycoprotein Distribution in Trauma Patients. J. Trauma 15, 451–455. doi: 10.1097/00005373-197505000-00013
Velotta J. P., McCormick S. D., O’Neill R. J., Schultz E. T. (2014). Relaxed Selection Causes Microevolution of Seawater Osmoregulation and Gene Expression in Landlocked Alewives. Oecologia 175, 1081–1092. doi: 10.1007/s00442-014-2961-3
Wang Z., Gerstein M. M., Snyder M. (2009). RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wang Y., Li E., Yu N., Wang X. D., Cai C. F., Tang B. P., et al. (2012). Characterization and Expression of Glutamate Dehydrogenase in Response to Acute Salinity Stress in the Chinese Mitten Crab, Eriocheir Sinensis. PLoS One 7, e37316–e37316. doi: 10.1371/journal.pone.0037316
Wang H., Tang L., Wei H., Lu J., Mu C., Wang C. (2018). Transcriptomic Analysis of Adaptive Mechanisms in Response to Sudden Salinity Drop in the Mud Crab, Scylla Paramamosain. BMC Genomics 19, 421. doi: 10.1186/s12864-018-4803-x
Wang X., Wang S., Li C., Chen K., Qin J. G., Chen L., et al. (2015). Molecular Pathway and Gene Responses of the Pacific White Shrimp Litopenaeus Vannamei to Acute Low Salinity Stress. J. Shellfish. Res. 34, 1037–1048. doi: 10.2983/035.034.0330
Waterkeyn A., Vanschoenwinkel B., Vercampt H., Grillas P., Brendonck L. (2011). Long-Term Effects of Salinity and Disturbance Regime on Active and Dormant Crustacean Communities. Limnol. Oceanogr. 56, 1008–1022. doi: 10.4319/lo.2011.56.3.1008
Whiteley N. M., Suckling C. C., Ciotti B. J., Brown J., Mccarthy I. D., Gimenez L., et al. (2018). Sensitivity to Near-Future CO2 Conditions in Marine Crabs Depends on Their Compensatory Capacities for Salinity Change. Sci. Rep. 8, 15639. doi: 10.1038/s41598-018-34089-0
Whiteley N. M., Taylor E. (2015). “Responses to Environmental Stresses: Oxygen, Temperature and pH. Physiology,” in The Natural Hstory of the Crustacea (New York: Oxford University Press).
Wiltshire K. H., Kraberg A., Bartsch I., Boersma M., Franke H.-D., Freund J., et al. (2010). Helgoland Roads, North Sea: 45 Years of Change. Estuar. Coast. 33, 295–310. doi: 10.1007/s12237-009-9228-y
Winkler A., Siebers D., Becker W. (1988). Osmotic and Ionic Regulation in Shore Crabs Carcinus Maenas Inhabiting a Tidal Estuary. Helgoländer. Meeresuntersuchungen. 42, 99–111. doi: 10.1007/BF02364206
Ye L., Jiang S., Zhu X. M., Yang Q., Wen W., Wu K. (2009). Effects of Salinity on Growth and Energy Budget of Juvenile Penaeus Monodon. Aquaculture 290, 140–144. doi: 10.1016/j.aquaculture.2009.01.028
Yew S. M., Chan C. L., Kuan C. S., Toh Y. F., Ngeow Y. F., Na S. L., et al. (2016). The Genome of Newly Classified Ochroconis Mirabilis: Insights Into Fungal Adaptation to Different Living Conditions. BMC Genomics 17, 91. doi: 10.1186/s12864-016-2409-8
Young A. M., Elliott J. A. (2020). Life History and Population Dynamics of Green Crabs (Carcinus Maenas). Fishes 5, 4. doi: 10.3390/fishes5010004
Yu C. G., Song H. T., Yao G. Z. (2004). Assessment of the Crab Stock Biomass in the Continental Shelf Waters of the East China Sea. J. Fisheries. China 28, 41–46.
Zhang Y., Buchberger A., Muthuvel G., Li L. (2015). Expression and Distribution of Neuropeptides in the Nervous System of the Crab Carcinus Maenas and Their Roles in Environmental Stress. Proteomics 15, 3969–3979. doi: 10.1002/pmic.201500256
Keywords: Charybdis japonica, intertidal crustaceans, transcriptome, salinity fluctuation regulation, genetic adaption
Citation: Shui B, Wang Y, Lou F and Han Z (2022) Salinity Fluctuation on the Genetic Regulatory Mechanisms of the Crustacean, Charybdis japonica. Front. Mar. Sci. 9:870891. doi: 10.3389/fmars.2022.870891
Received: 07 February 2022; Accepted: 13 June 2022;
Published: 07 July 2022.
Edited by:
Eduardo Almansa, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Xiujuan Shan, Chinese Academy of Fishery Sciences (CAFS), ChinaMelissa Pespeni, University of Vermont, United States
Copyright © 2022 Shui, Wang, Lou and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangrui Lou, bGZyMTk5MjAyQDE2My5jb20=
 Bonian Shui1
Bonian Shui1 Fangrui Lou
Fangrui Lou Zhiqiang Han
Zhiqiang Han