- 1Ministry of Education (MOE) Key Laboratory of Marine Genetics and Breeding, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
Interferon regulatory factors (IRFs) are a family of transcription factors that control many facets during innate and adaptive immune responses. Vertebrate IRFs play important roles in regulating the expression of interferons (IFNs) and IFN-stimulated genes, while only limited studies were conducted on invertebrate IRFs. In the present study, four IRF family genes (CfIRF1, CfIRF1-like, CfIRF2, and CfIRF8) were identified from Zhikong scallop (Chlamys farreri) through whole-genome scanning. CfIRFs contain a highly conserved N-terminal DNA-binding domain and a variable C-terminal regulatory domain. CfIRFs were constitutively expressed during development as well as in adult tissues, especially in hepatopancreas, hemolymph, gill, and mantle. In hemolymph, qRT-PCR analysis revealed that CfIRF1, CfIRF1-like, and CfIRF2 were significantly upregulated in response to Vibrio anguillarum infection, and their encoding proteins could translocate into nucleus. Dual-luciferase reporter assay on CfIRF1, CfIRF1-like, and CfIRF2 showed that these three proteins were capable to induce a strong activation of ISRE promoters. Notably, in comparison with CfIRF1 and CfIRF1-like, CfIRF2 showed the most sensitive responses in coping with V. anguillarum, and consistently, CfIRF2 exhibited the most significant activation on ISRE. This study would provide valuable information for the innate immune roles of the IRF gene family in bivalve molluscs.
1 Introduction
Interferon regulatory factors (IRFs) are a family of transcription factors that were first identified as regulators of IFN (Type I interferon) and IFN-inducible genes (Miyamoto et al., 1988; Harada et al., 1989), which have been extensively studied in vertebrates, showing diverse functions in regulating immune responses, stress responses, reproduction, development, and carcinogenesis (Tamura et al., 2008; Nehyba et al., 2009; Savitsky et al., 2010). So far, a total of 11 IRF family members (from IRF-1 to IRF-11) have been reported in vertebrates (Inkpen et al., 2019), with nine IRFs identified in mammals and another two members found in several avian and fish species (Nehyba et al., 2002; Huang et al., 2010). All IRF family members possess a highly conserved N-terminal helix-turn-helix DNA-binding domain (DBD). This domain consists of about 120 amino acids, binding to the core IFN-stimulated response element (ISRE) recognition sequence, GAAANNGAAAG/CT/C (Escalante et al., 1998; Marchler-Bauer et al., 2011). As for the C-terminus, most of the IRFs share an IRF-associated domain 1 (IAD1) or a similar IAD2, which mainly mediates the homomeric or heteromeric formation of IRFs as well as the interaction of IRF with non-IRF members, and the resulting protein complex acts as a transcriptional activator or repressor (Ikushima et al., 2013). It was reported that IRF1, IRF3, IRF5, IRF7, and IRF9 are usually functioned as positive mediators of the host’s IFN response, whereas IRF4 usually acts as a repressor. Furthermore, IRF2 and IRF8 can participate in either activating or repressing the target gene transcription, depending on the nature of the pathogen or the signaling pathways that is involved (Taniguchi, 2006; Weiqi et al., 2008; Ikushima et al., 2013). Besides the IAD, the C-terminus is not well conserved, which may confer versatile functions to IRF members (Yanai et al., 2012). For example, besides IFN mediator, IRF activation through the TLRs or other inflammatory cytokines could interfere with NF-κB signaling, which were necessary to maintain the immunity balance (Anda et al., 2012; Cavlar et al., 2012; Xuan et al., 2020).
Previous studies have revealed that IRF genes are present in all principal metazoan groups, and IRF-like genes have been detected in genomic and expressed sequence tag (EST) databases (Davidson et al., 2006; Azumi et al., 2007; Huang et al., 2008). Based on the evolutionary molecular relationships, the IRF proteins could be classified into four subfamilies, namely, IRF-1 group (IRF1, 2, and 11), IRF-3 group (IRF3 and 7), IRF-4 group (IRF4, 8, 9, and 10), and IRF-5 group (IRF5 and 6) (Zhan et al., 2016). For the IRF-1 subfamily, IRF1 and IRF2 were first identified as transcriptional regulators of ISGs and type I IFN, which mainly play important roles in antiviral immunity (Harada et al., 1989). IRF11 has only been identified in teleost fish, and its function study is still in the infancy stage (Huang et al., 2010). For the IRF-3 family, researchers found that phosphorylated IRF7 and IRF3 could jointly regulate the rapid production of IFN initially, and ultimately induce the production of IFN in large quantities through a positive feedback regulatory loop (Marié et al., 1998; Sato et al., 1999). For the IRF-4 subfamily, they showed diverse IFN or NF-κB regulating functions depending on the nature of the binding molecules or the cellular differentiation status, and higher homology was found between IRF4 and IRF8 (Meraro et al., 2002; Lehtonen et al., 2005; Lu, 2008). As for the IRF-5 subfamily, IRF5 is mainly involved in the natural inflammatory response, while IRF6 is mainly involved in the embryonic early development (Hatada et al., 1997; Barnes et al., 2001; Barnes et al., 2004; Green et al., 2015).
Compared with the extensive knowledge of IRFs in vertebrates, studies on IRFs in invertebrates are quite limited. Previously, the interferon response has been thought to be a vertebrate innovation because the genomes of model invertebrates (i.e., Drosophila) do not encode interferon or its major effectors (Green et al., 2015). With the abundance and further analysis of invertebrate genome data, several key molecules in the IFN system have been identified, including IRF, interferon-like protein (IFNLP), interferon receptor (IFNR), and interferon-induced protein (Lelong et al., 2015; Zhang et al., 2015; Huang et al., 2017). More recently, studies have revealed that IRF genes are present in a lot of invertebrate groups, including sea sponges, placozoans, comb jellies, cnidarians, and bivalves, but are not detected in Nematoda and Hexapoda (including insects) (Nehyba et al., 2009; Huang et al., 2010). Although IRFs have been found in various invertebrates, they are different in number and genomic characteristics from the vertebrate IRF family, and there are only few preliminary functional studies on invertebrate IRFs through gene cloning. For example, PfIRF-2 in pearl oyster Pinctada fucata; CgIRF-1, -2, and -8 in pacific oyster Crassostrea gigas; and LvIRF in pacific white shrimp Litopenaeus vannamei were found to participate in the immune response against Gram-negative bacteria (Huang et al., 2013; Li et al., 2015; Huang et al., 2017; Lu et al., 2018). As invertebrates or vertebrates might have experienced a different pressure during evolution, it may in turn lead to the functional differentiation of IRF genes (Huang et al., 2010). Therefore, we need more research into the function of invertebrate IRFs, providing valuable information for the origin of the IRF family as well as the evolution of innate immunity.
Bivalve molluscs belong to the most speciose phylum of marine invertebrates, which could well adapt to the highly diverse and hostile environment with various stressors (bacteria, pollution, etc.). Scallops are highly prized as a food source, while in recent years, their aquaculture industry suffers huge economic loss due to the etiological diversity of pathogens that cause repeated appearance of disease outbreaks (Liu et al., 2004; Teng et al., 2012). Scallops generally lack the adaptive immune system and rely solely on innate immunity mediated by both cellular and humoral components (Loker et al., 2010). Functional studies of IRFs on scallops would be helpful for revealing their immune defense mechanisms and understanding the origin and evolution of bivalve innate immunity. In the present study, we take Chlamys farreri (Zhikong scallop), one of the most important maricultural scallop species in China, as research subject to systematically identify the IRF gene family. Their expression profiles during development and in different healthy adult tissues were analyzed. Meantime, their responses after Vibrio anguillarum challenge in hemocytes were investigated. We further explored their subcellular localization as well as the transcriptional activity using pISRE-Luc reporter plasmids in HEK293T cells, thereby providing insights into the immune function of IRF genes in bivalves.
2 Materials and Methods
2.1 Database Mining, Gene Identification, and Sequence Analysis
To identify IRF genes, the transcriptome and whole genome sequence databases of the C. farreri were searched using the available IRF protein sequences from representative invertebrates and vertebrates, including Homo sapiens, Mus musculus, Gallus gallus, Xenopus tropicalis, Danio rerio, C. gigas, P. fucata, Mytilus galloprovincialis, Hyriopsis cumingii, Pecten maximus, Lottia gigantea, Biomphalaria glabrata, and Elysia chlorotica. These IRF proteins from representative species were retrieved from NCBI (http://www.ncbi.nlm.nih.gov), Ensembl (http://useast.ensembl.org), MolluscDB (http://mgbase.qnlm.ac/), and OysterBase (http://www.oysterdb.com/) databases. TBLASTN was used to obtain the initial pool of IRFs transcriptome sequences from the Zhikong scallop, and then, BLASTN was performed to verify the cDNA sequences by comparing the transcriptome sequences with the whole genome sequences. The candidate CfIRFs sequences were submitted to the ORF Finder program (https://www.ncbi.nlm.nih.gov/orffinder/) to predict the open reading frame (ORF), and the ORFs were translated into amino acid sequences. The translated sequences were submitted to the SMART program (http://smart.embl-heidelberg.de/) for identification of the signal peptide and other conserved domains. The putative isoelectric point (pI) and molecular weight were computed using the Compute pl/Mw (http://web.expasy.org/compute_pi/). The subcellular localization and nuclear localization signals (NLSs) were predicted through the online prediction website (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/, https://www.genscript.com/wolf-psort.html/, https://sunflower.kuicr.kyoto-u.ac.jp/~smatsuda/slplocal.html, http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). The protein structures of all the identified IRF proteins were drawn with IBS1.0.3 software. Multiple alignment analysis of CfIRFs were performed with the ClustalW multiple alignment programs (http://www.ebi.ac.uk/clustalw/).
2.2 Phylogenetic Analysis
The IRF proteins from other vertebrates and invertebrates listed in the Section 2.1 were used for phylogenetic analysis together with the Zhikong scallop IRFs. The amino acid sequences of IRF proteins from these species were retrieved from the NCBI and Ensembl Genome Browser. Protein sequences were aligned using the ClustalW method in the MEGA-X software (Sudhir et al., 2018). Phylogenetic relationships of IRF amino acid sequences were estimated using maximum-likelihood (ML) analyses with FastTree 2.0.0. FastTree accounts for variable rates of evolution across sites by assigning each site to one of 20 categories, with the rates geometrically spaced from 0.05 to 20. FastTree sets each site to its most likely category by using a Bayesian approach with a gamma prior. Branch supports evaluated 10,000 pseudo-replicates of the ultrafast bootstrap procedure (Thi et al., 2017). Whole amino acid sequences were used in the phylogenetic analyses. This analysis involved a total of 59 amino acids across 13 species. The accession numbers of 59 IRFs are listed in Supplementary Table 1.
2.3 Expression Analysis
For expressional analysis, the RPKM (reads per kilo per million reads) value of each IRF gene was retrieved from the published RNASeq datasets of various developmental stages and adult tissues of Zhikong scallop (Li et al., 2017). To visualize the expression patterns of IRF genes in Zhikong scallop, the expressional heatmaps were shown via heatmap package under the R environment and the statistical analysis of the data was performed with edgeR package under R environment using the F-test. Differences were considered significant at p < 0.05. For examining the correlation relationship of IRF genes, a regression analysis was performed.
2.4 Sample Collection and Bacteria Treatment
Two-year-old healthy Zhikong scallops were collected from artificial scallop-rearing substrates installed in Xunshan Fishery Group Co., Rongcheng (Shandong Province, China). All the procedures involved in the handling and the treatment of scallops during this study were approved by the Ocean University of China Institutional Animal Care and Use Committee (OUC-IACUC) prior to the initiation of the study. The scallops were acclimated in the laboratory at ambient seawater temperature for 1 week prior to the experiments, which is within the optimum temperature range for their survival.
Gram-negative (V. anguillarum) bacteria were used to challenge scallops in our study (Zhi et al., 2011). V. anguillarum was cultured in liquid 2216E broth (5 g/L of Tryptone, 1 g/L of yeast extract, and 0.1 g/L of C6H5Fe·5H2O, pH = 7.6) at 28°C and harvested by centrifugation at 2000×g for 5 min, as described by Kong et al. The pellet was suspended in filtered seawater and was adjusted to 1×107 CFU/ml in seawater, respectively (Zhi et al., 2011; Ragab et al., 2014).
A total of 75 individuals were randomly and equally divided into five groups. At 0 h, 5 h, 24 h, 48 h, and 72 h post-infection, 5 individuals were randomly collected from each group. The 0 h group was employed as the control group, and other groups were used as experimental groups. The hemolymph samples were collected from adductor muscles using a syringe and were immediately centrifuged at 800×g, 4°C for 10 min to harvest the hemocytes (Gao et al., 2007). The extracted hemocyte sample was immediately frozen in liquid nitrogen and then subsequently frozen at −80°C before processing.
2.5 RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA was isolated following the method described by Hu et al. (2010), and then was digested with DNase I (TaKaRa, Shiga, Japan). A Nanovue Plus spectrophotometer (GE Healthcare, NJ, USA) was used to assess the concentration and purity of RNA; RNA integrity was determined by agarose gel electrophoresis. First-strand cDNA was synthesized using Moloney murine leukemia virus (MMLV) reverse transcriptase (Thermo, USA) following the manufacturer’s protocol. All of the cDNA products were diluted to 5 ng/ml for use as the template in real-time PCR.
Real-time PCR was conducted using the SsoFast™ EvaGreen® Supermix on a Light Cycler 480 Real-time PCR System (Roche Di-agnostics, Mannheim, Germany). The running program was as follows: 50°C for 2 min, 94°C for 10 min, and 40 cycles at 94°C for 15 s and at 62°C for 1 min. Cytochrome B (CB), DEAD-βox RNA helicase (HELI), and EF1-A gene were designated as internal reference genes for the normalization of gene expression in healthy adults and test subjects during the real-time PCR experiment, respectively (Li et al., 2010; Feng et al., 2013). All the primers used in the real-time PCR were designed using Primer Premier 5.0 and are listed in Table 1.
Data from the real-time PCR were analyzed using the Relative Expression Software Tool (REST) version 2009 (Pfaffl et al., 2002); gene expression is shown as the fold change. For the experimental groups, the control group (0 h) was used for normalization. The statistical analysis of the data was performed with SPSS (version 16.0) software using the independent t-test. Differences were considered to be significant at p < 0.05.
2.6 Subcellular Localizations
The full length of ORFs of three CfIRF genes was amplified from C. farreri cDNA, using primers listed in Table 1. For CfIRF1 and CfIRF2, the PCR products were ligated and subcloned into pEGFP-N1 (Clontech, USA) by way of overlap extension PCR to construct recombinant plasmids pEGFP-CfIRF1 and pEGFP-CfIRF2, while for the CfIRF1-like, PCR products were digested with KpnI and SmaI, and ligated and subcloned into pEGFP-N1 vector digested by the corresponding restriction enzymes to construct recombinant plasmids pEGFP-CfIRF1-like. The constructed recombinant plasmids were subsequently verified by DNA sequencing.
In scallops, as well as in other marine bivalves, there are no mature cell lines. Thus, we choose HEK293T cells to perform our experiment. HEK293 cells were maintained in Modified Eagle Medium (MEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, USA) and antibiotics (100 mg/L streptomycin and 105 U/L penicillin, Gibco) at 37°C in a humidified incubator under 5% CO2. For DNA transfection, cells were seeded and allowed to grow to more than 70% confluence, and then plasmids were transfected by using the Lipofectamine 3000 Reagent (Invitrogen, USA) following the manufacturer’s recommendations. HEK293T cells were transiently co-transfected with 0.8 μg of expression plasmid and 1 μl of Lipofectamine 3000 in each well in a 24-well plate. All assays were performed with three independent transfections.
At 48 h post-transfection, HEK293 cells were washed with PBS twice and fixed with paraformaldehyde for 15 min. Then, the cells were washed three times with PBS and were stained with DAPI (Sigma, USA) to mark the nucleus followed by washing three times. Immunofluorescence was visualized and captured with confocal microscopy (Nikon, Japan).
2.7 Dual-Luciferase Reporter Assays
The full length of ORFs of three CfIRF genes was amplified from C. farreri cDNA, using primers listed in Table 1. For CfIRF1 and CfIRF2, the PCR products were ligated and subcloned into pcDNA3.1 V5/H vector (Invitrogen, USA) by way of overlap extension PCR to construct recombinant plasmids pcDNA3.1-IRF1 and pcDNA3.1-IRF2, while for the CfIRF1-like, the pcDNA3.1-IRF1-like was constructed using the same method as mentioned above but digested with NotI and KpnI. The constructed recombinant plasmids were subsequently verified by DNA sequencing. For reporter plasmids, pISRE-Luc (ClonTech, USA) was used, and pRL-TK renilla luciferase plasmid (Progema, USA) and pGL3-basic vector (Progema, USA) were used as an internal control and blank group, respectively. EndoFree Plasmid MiKit (OMEGA, USA) was used for the transfection of the plasmids according to the manufacturer’s instruction.
The cell culture assays were performed according to Section 2.6. For dual-luciferase reporter assays, HEK293T cells were transiently co-transfected with 0.2 mg of expression plasmid, 0.5 mg of reporter gene plasmid, 0.01 mg of pRL-TK renilla luciferase plasmid, and 0.1 μl of Lipofectamine 3000 in each well in a 24-well plate. The luciferase reporter vector pGL3-basic was used as a blank group. All assays were performed with three independent transfections. At 48 h post-transfection, HEK293 cells were washed with PBS twice and lysed. Firefly and renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instruction. Cell lysate (20 μl) was transferred to a 1.5-ml EP tube and 100 μl of luciferase assay reagent II and 100 μl of Stop & Glo® Reagent were added in sequence, then firefly and renilla luciferase activities were measured, respectively.
3 Results
3.1 Sequence Identification and Analysis
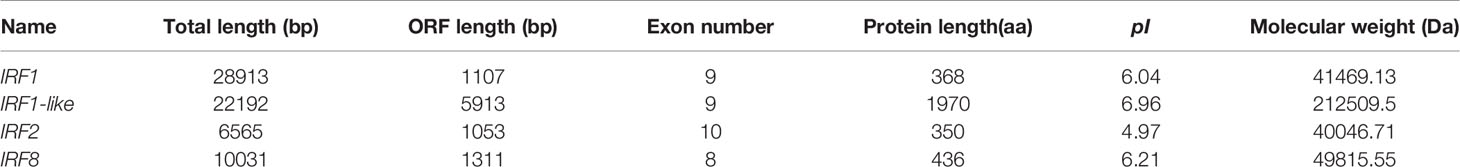
Four IRF genes, CfIRF1, CfIRF1-like, CfIRF2, and CfIRF8, were identified from the genome of Zhikong scallop. The basic information (total length, ORF length, number of exons, amino acids length, theoretical pI, and weight of protein) of these IRF members were summarized in Table 2. The ORFs of CfIRF1, CfIRF1-like, CfIRF2, and CfIRF8 were respectively 1,107, 5,913, 1,053, and 1,311 bp, encoding 368, 1,970, 350, and 436 amino acids. The predicted molecular weights of these four genes ranged from 40.05 to 212.51 KD, with the predicted pI values from 4.97 to 6.96 (Table 2). The genomic structural analysis showed that the length as well as the exon–intron pattern of these four CfIRF genes varied greatly. The longest CfIRF1 gene was 28,913 bp with 9 exons, and the shortest CfIRF2 gene was 6,565 bp with 10 exons. Moreover, CfIRF1-like and CfIRF8 had 9 and 8 exons, respectively (Figure 1).
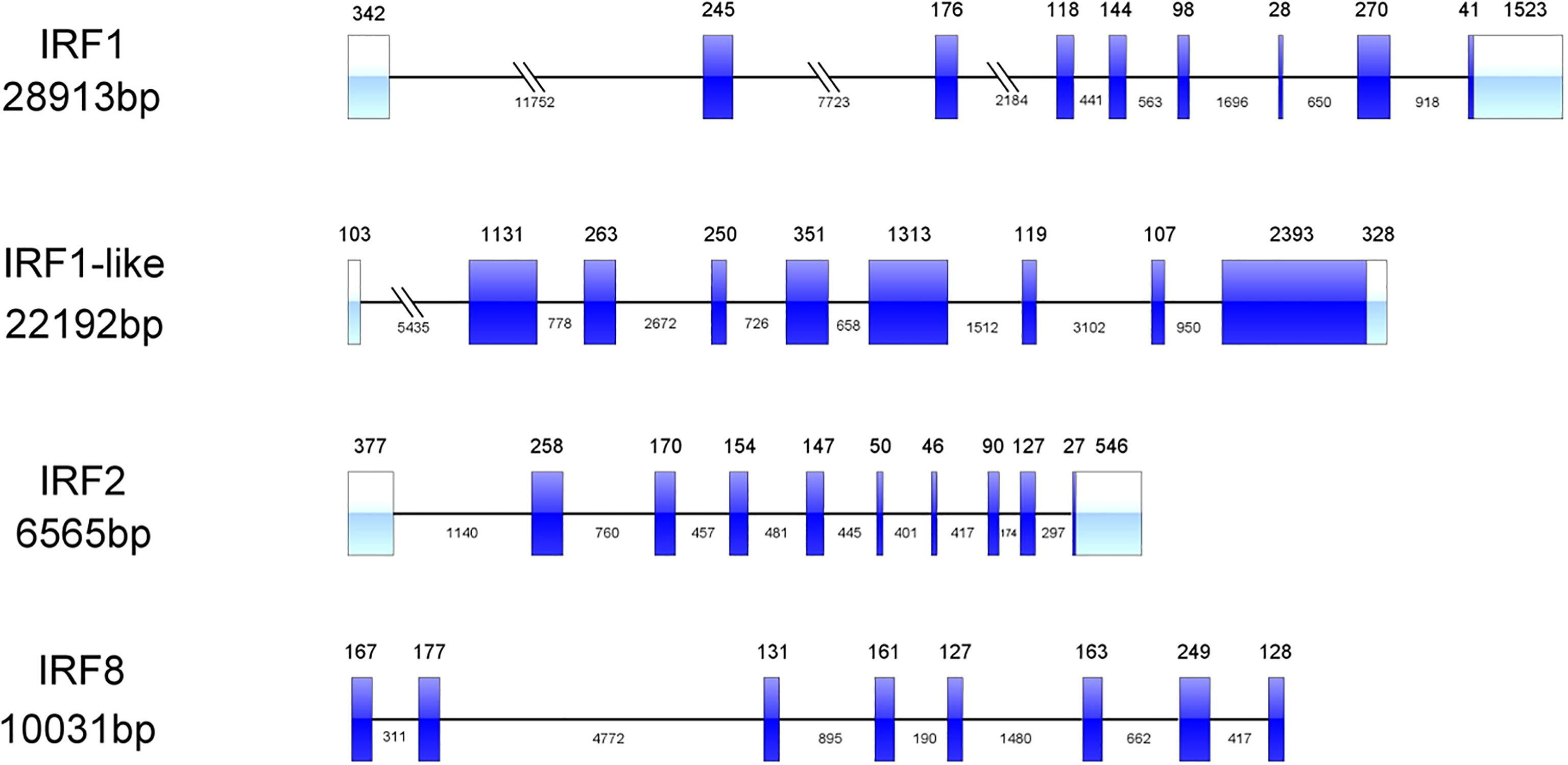
Figure 1 Genetic structure of the CfIRF genes. The light blue boxes indicate the 3’ UTRs and the 5’ UTRs. The dark blue boxes indicate the exons. The horizontal line with sporadic double slash indicates the introns.
All four CfIRF proteins had a single well-conserved DBD domain (113 aa in length), which was found to be helix-turn-helix at the N-terminal (Figure 2). Furthermore, CfIRF8 was predicted to contain an IRF-associated domain (IAD) at the C-terminus, and CfIRF1-like possessed seven C2H2-type (classical) zinc fingers (ZnF_C2H2) at the C-terminal (Figure 2). Multiple sequence alignments of CfIRFs showed that they share high similarity within the DBD domain (Figure 3). Moreover, five conserved tryptophan (W) residues were revealed, in which the W71 residue was found mutated as Y71 in CfIRF1 protein.
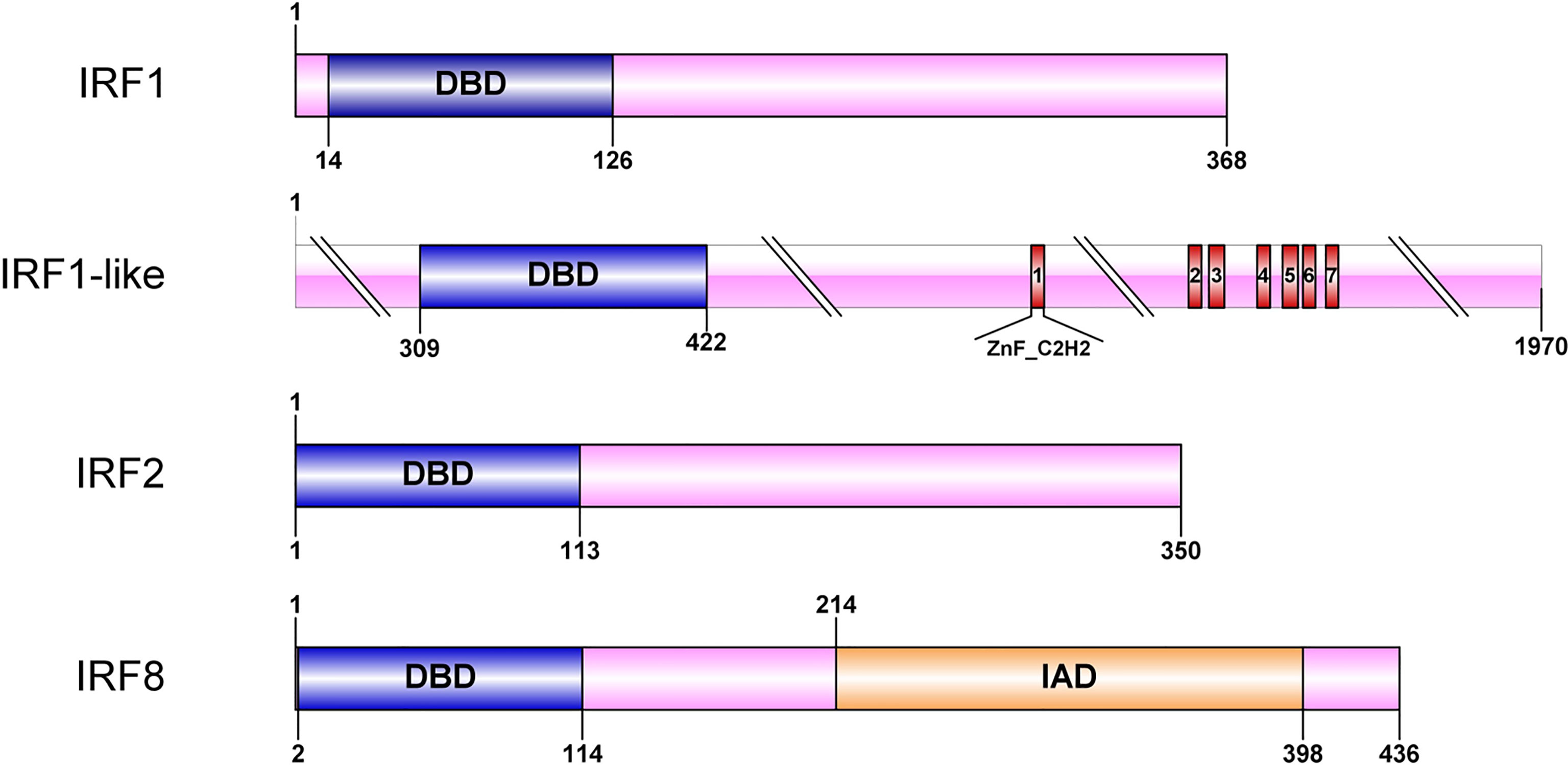
Figure 2 Protein structure of the CfIRF genes. The blue boxes indicate the DBD domain. The orange boxes indicate the IAD domain. The red boxes indicate the ZnF_C2H2 domain.

Figure 3 Alignment of DBD domains of the four CfIRF proteins. Alignment was performed using ClustalW2. Identical residues are indicated in black, and similar residues are in light gray. Dashes indicate gaps. Five conserved tryptophan (W) residues are marked with red boxes.
To confirm the identification of the CfIRFs, a phylogenetic tree was constructed through the ML method using 59 IRF proteins across 13 species. Four IRF subfamilies (IRF-1, -3, -4, and -5) were identified (Supplementary Figure 1) and all CfIRFs were clustered into its own clade. The result showed that CfIRF1, CfIRF1-like, and CfIRF2 were clustered into the IRF-1 subfamily. CfIRF8 was firstly grouped together with IRF8-like from Peten maximus and IRF8 from C. gigas, and they were clustered into the IRF-4 subfamily.
3.2 Spatiotemporal Expressions of CfIRFs
The RPKM data were used to analyze the expression patterns of four CfIRF genes during eleven developmental stages (Figure 4A, Supplementary Figure 2A). Their expression during early development (from zygote to trochophore larvae) was quite low (<10 RPKM). After D-shaped larvae formation, CfIRF1-like and CfIRF8 remained at a low expression (<5 RPKM) while expression of CfIRF1 and CfIRF2 was obviously elevated. Although CfIRF2 showed higher expression level than CfIRF1 (>3-fold), their expression pattern was similar, both of which showing the highest expression in creeping larvae and juvenile. In adult tissues, we found that expression of CfIRF1 and CfIRF2 was obviously higher than CfIRF1-like and CfIRF8 (Figure 4C, Supplementary Figure 2B), consistent with their expression tendency during development. In general, CfIRF1-like and CfIRF8 remained low expression in all the tissues (<25 RPKM), while CfIRF1 was dominantly expressed in hepatopancreas and hemolymph (>160 RPKM), and the highest expression of CfIRF2 was observed in gill and hepatopancreas (>220 RPKM). To illustrate the role of CfIRF1 and CfIRF2 in development stages and adult tissues, we performed correlation analysis between the expression of CfIRF1 and CfIRF2 with E2/T ratio. According to the results, CfIRF1 has a significantly positive correlation with CfIRF2 expression both in different developmental stages (r = 0.86, p < 0.001) (Figure 4B) and in adult tissues (r = 0.65, p < 0.05) (Figure 4D).
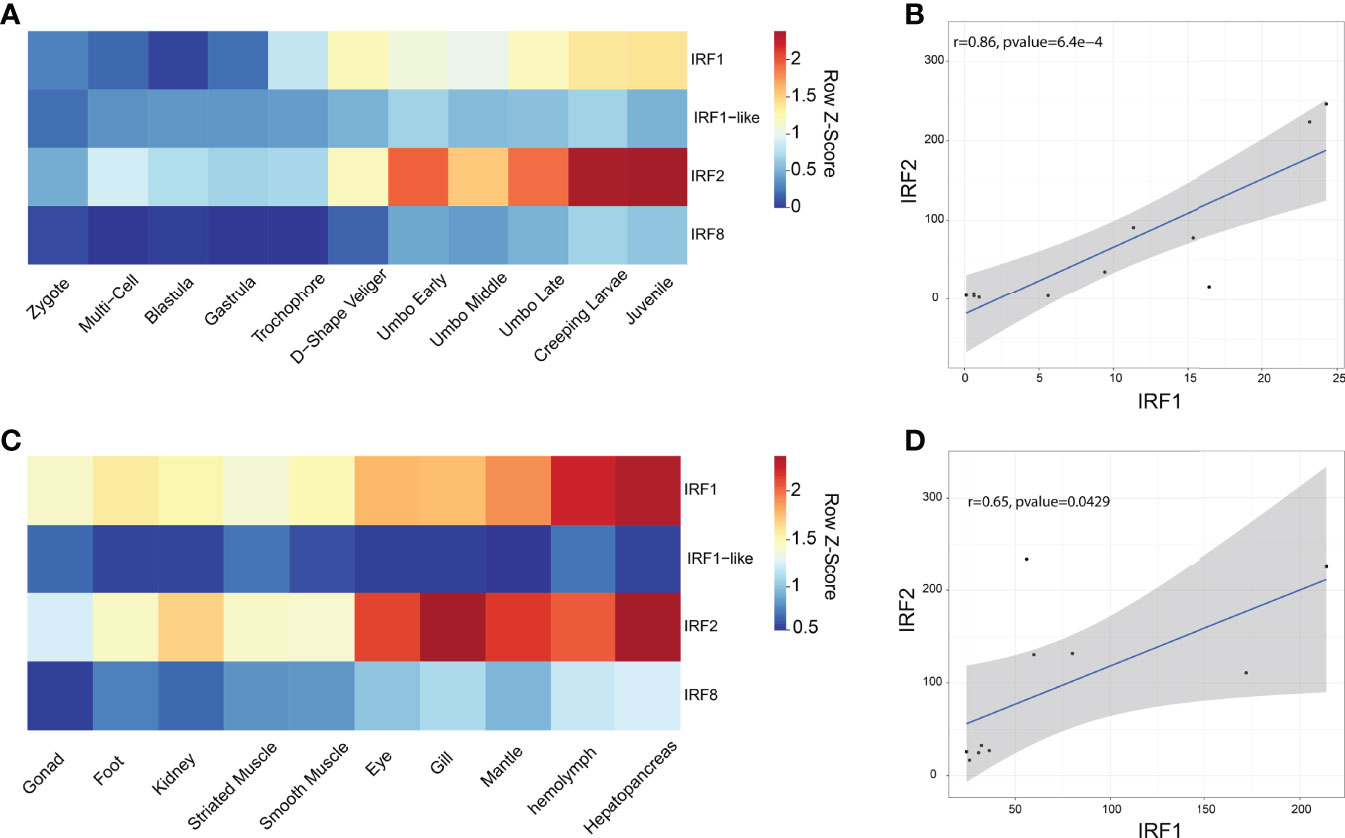
Figure 4 Expression profiles of CfIRFs in different developmental stages and adult tissues. (A) Heatmap of CfIRFs expression profile in different embryonic and larval stages. (B) The positive correlation between CfIRF1 and CfIRF2 in different developmental stages. (C) Expression profiles of CfIRFs in adult tissues. (D) The positive correlation between CfIRF1 and CfIRF2 in adult tissues.
3.3 Temporal Responses of CfIRFs in Coping With Bacterial Infection
To examine the immune responses of CfIRFs to V. anguillarum challenge (Qiu et al., 2007; Costa et al., 2009), their expression level was investigated at four time points (5 h, 24 h, 48 h, and 72 h) after infection (Figure 5). Overall, three CfIRFs from the IRF-1 subfamily (CfIRF1, CfIRF1-like, and CfIRF2) were significantly upregulated. Notably, CfIRF2 showed the most sensitive responses, which showed significant upregulation after 24-h infection (>13-fold, p < 0.05) and sustained at a significantly higher expression level till 72 h. Moreover, the expressions of CfIRF1 and CfIRF1-like were significantly upregulated at 72 h post-infection (>12-fold, p < 0.05), while CfIRF8 only showed mild upregulated tendency after 24-h infection (4- to 8-fold).
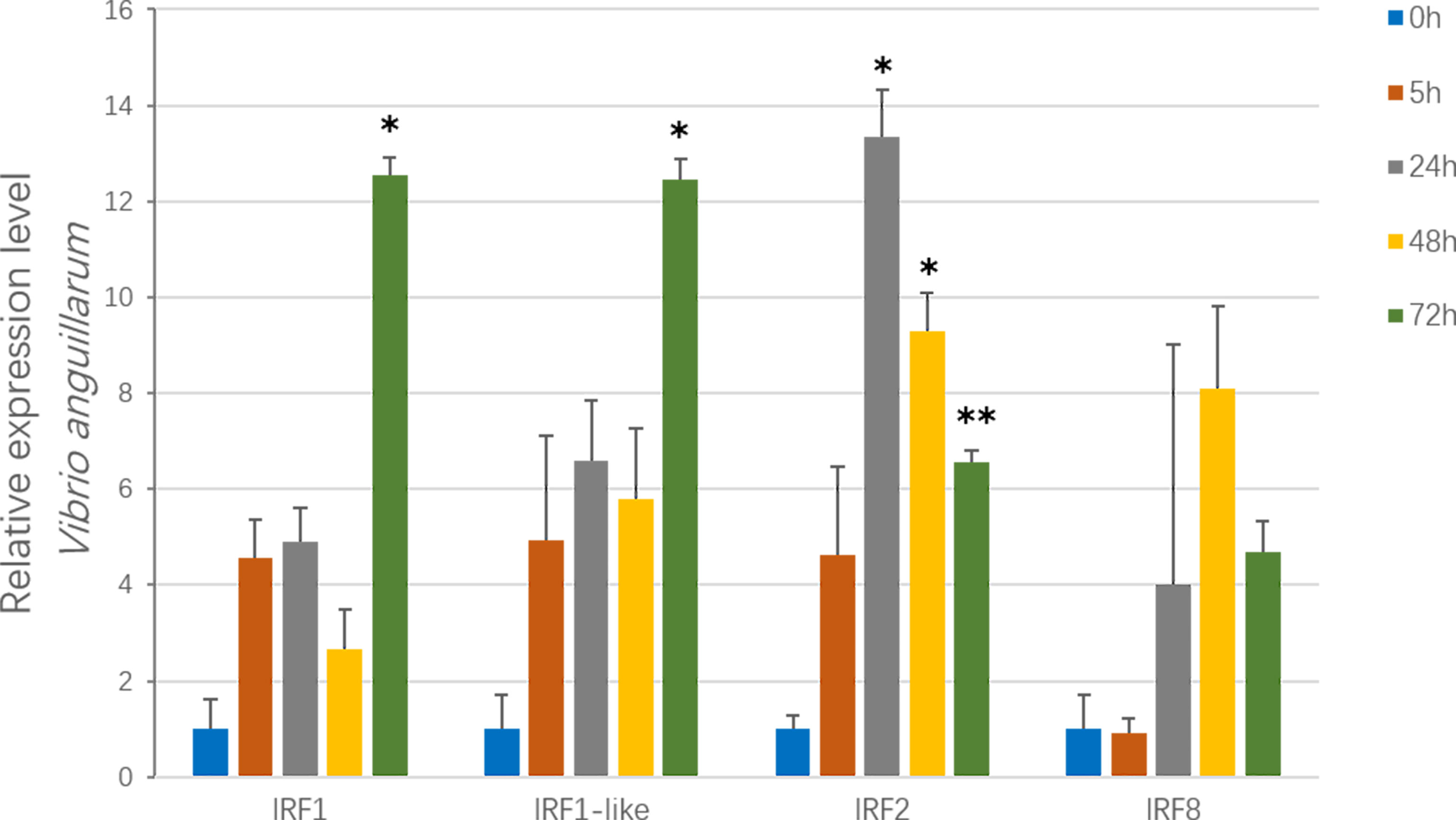
Figure 5 Expression of CfIRF genes after challenge with the V. anguillarum. Vertical bars represent the mean ± S.E. (N = 10). The asterisks indicate significant differences (*p < 0.05, **p < 0.01).
3.4 Subcellular Localizations of CfIRF1, CfIRF1-Like, and CfIRF2
Subcellular localization prediction showed that CfIRF1, CfIRF1-like, and CfIRF2 all possess nucleus localization (Table 3). Furthermore, 163RSRRRKKPCVKKE175 in CfIRF1 was predicted as a nucleus localization signal with a high score (score: 9.5), further suggesting that CfIRF1 was a nuclear-localized protein. HEK293T cells were transfected with plasmids encoding pEGFP-tagged CfIRF1, CfIRF1-like, or CfIRF2 to investigate their subcellular localization. The immunofluorescence image analysis showed that recombinant CfIRF1-GFP protein was located in the nucleus, consisting of its subcellular localization prediction and the NLS prediction. Unlike CfIRF1, the fluorescent signal of CfIRF1-like and CfIRF2 recombinant proteins was distributed in both nuclei and cytoplasm (Figure 6). Thus, the divergence of CfIRFs by subcellular localization tentatively suggested that these three CfIRFs may be involved with different cellular functions.
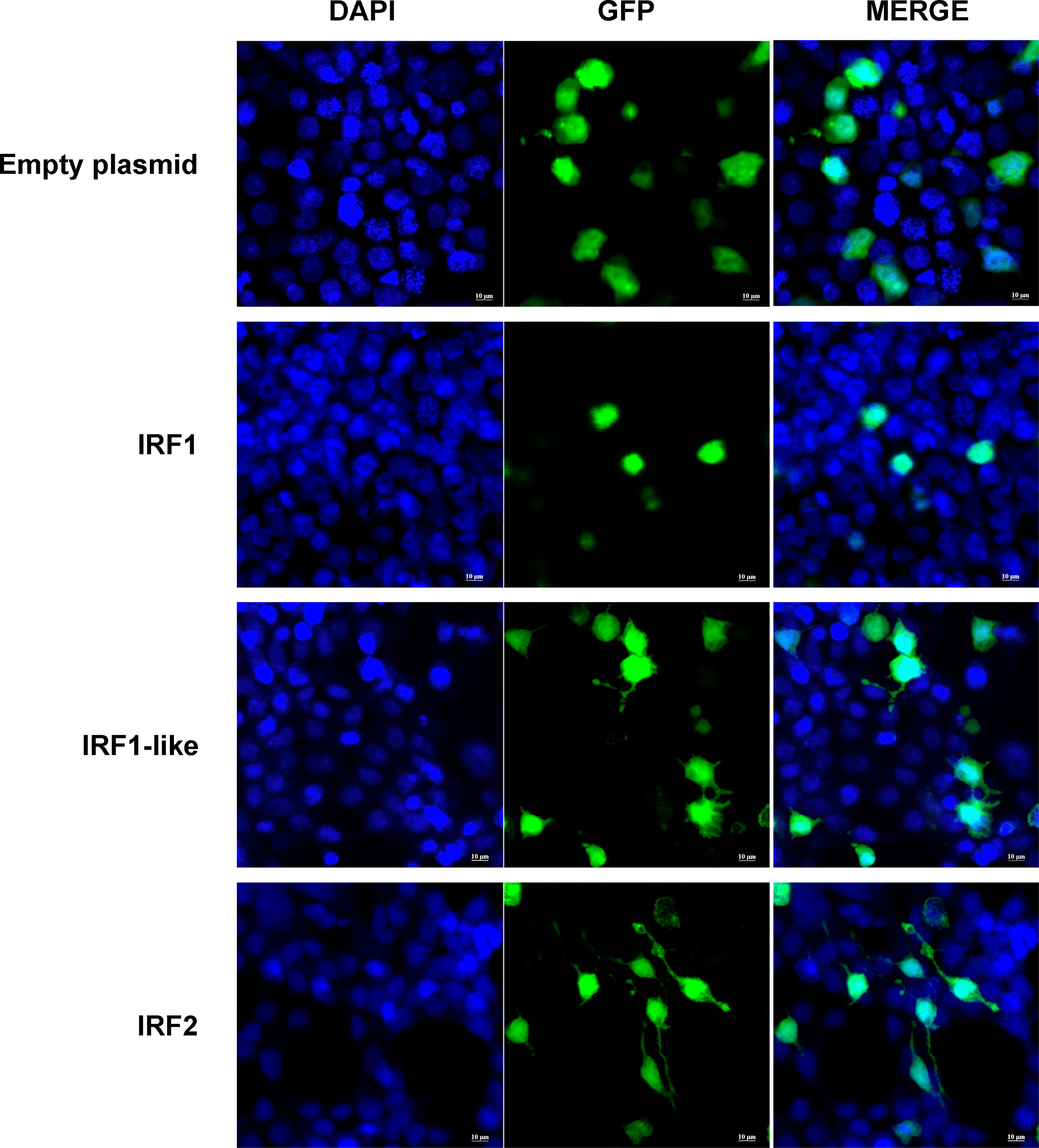
Figure 6 Subcellular localization of CfIRF genes in HEK293T cells. Immunohistochemistry was performed to analyze the expression of CfIRF in hemocytes. The left-hand panels depict DAPI staining, the middle panels depict GFP staining, and the right-hand panels depict merged DAPI/GFP staining. The upper panels depict localization of the GFP negative control, and the lower panels depict localization of the CfIRF-GFP proteins.
Dual-Luciferase Reporter Assays
To analyze the transcription activities of CfIRF1, CfIRF1-like, and CfIRF2, dual-luciferase reporter assays were performed in HEK293T cells. As control, pcDNA3.1-IRFs were co-transfected with pGL3-basic, and it did not show any effect on the ISRE reporter. By using pISRE-Luc, after co-transfection with pcDNA3.1-IRF1, pcDNA3.1-IRF1-like, and pcDNA3.1-IRF2, the luciferase activity of the ISRE reporter was significantly upregulated (Figure 7), suggesting their obvious transcriptional activation on interferon-stimulated response element. These results indicated that these CfIRFs could activate the expression of ISRE luciferase reporter genes, suggesting that scallop IRFs can specifically activate interferon signaling.
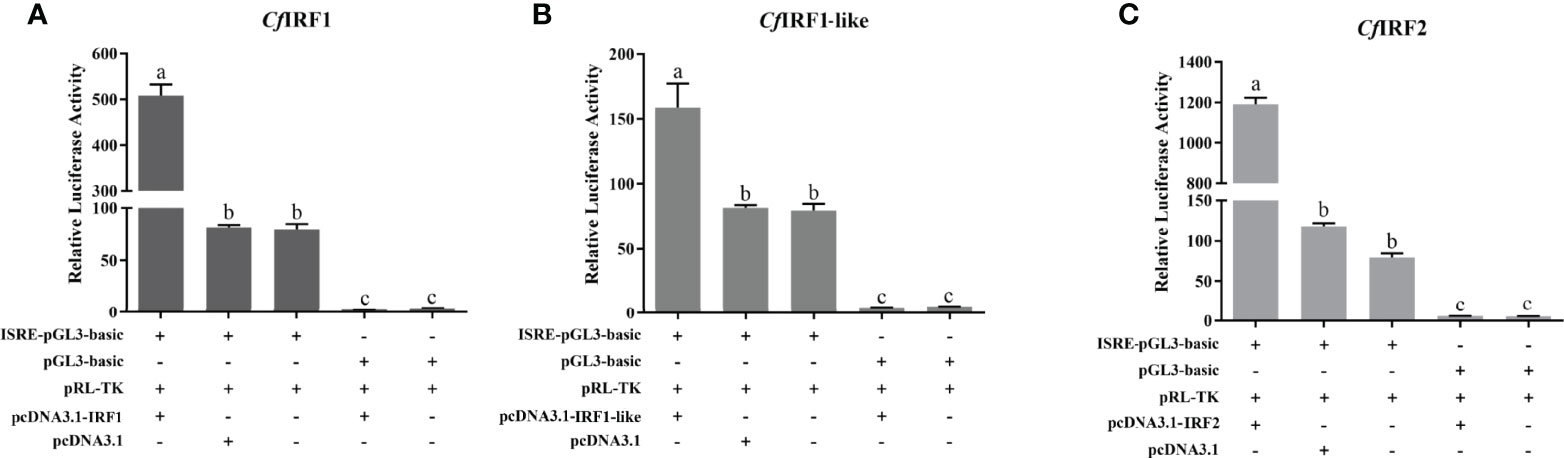
Figure 7 Relative luciferase activity of expression CfIRF plasmids on the luciferase reporter gene pISRE-Luc in HEK293T cells. (A) Relative luciferase activity of expression CfIRF1 plasmids. (B) Relative luciferase activity of expression CfIRF1-like plasmids. (C) Relative luciferase activity of expression CfIRF2 plasmids. All of the groups were co-transfected with pRL-TK as internal reference. Vertical bars represent the mean ± S.E. (N = 3). p-values are calculated by one-way analysis of variance (ANOVA) in SPSS software. Different letters indicate significant differences (p < 0.05).
4 Discussion
The innate immune system is the first line of defense against the invasion of pathogens. IRFs are key transcription factors involved in type I IFN responses, playing a pivotal role in the regulation of interferon activity (Kimura et al., 1994). In the present study, we successfully identified four IRF genes in a bivalve mollusc C. farreri, namely, CfIRF1, CfIRF1-like, CfIRF2, and CfIRF8. All four CfIRF proteins have a single well-conserved N-terminal helix-turn-helix IRF superfamily domain (also named as DBD). Through the motif of Trp repeats, the conserved DBD domain in vertebrate was proved to recognize and bind DNA sequence containing 5′-GAAA-3′ tetranucleotide as a determinant of interferon regulation (Escalante et al., 1998; Mamane et al., 1999). Consistently, we found well-conserved Trp repeats inside the DBD domain of scallop IRFs, suggesting their similar binding activities with vertebrate IRFs. Moreover, CfIRF8 was predicted to contain an IRF-associated domain (IAD) at the C-terminus, which has been reported to mediate the formation of homologous dimers or the coupling with other transcription factors to form heterodimers (Honda and Taniguchi, 2006; Yanai et al., 2012). According to the phylogenetic analysis, CfIRF1, CfIRF1-like, and CfIRF2 were clustered into the IRF-1 subfamily, and CfIRF8 was clustered into the IRF-4 subfamily.
Spatiotemporal expression levels of CfIRF1 and CfIRF2 were found to be obviously higher than CfIRF1-like and CfIRF8, suggesting the initial requirement for them during development as well as in adult tissues. Correlation analysis showed that CfIRF1 and CfIRF2 have a significantly positive correlation during development as well as in adult tissues, indicating that they may be functionally synergistic. CfIRF1 and CfIRF2 were highly expressed after the D-shape veliger formation, especially in creeping larvae and juvenile, the key stage that multi-organs began to developed. In adult tissues, high expression of both CfIRF1 and CfIRF2 was detected in multiple immune-related tissues, such as hemolymph, hepatopancreas, gill, and mantle (Fan et al., 2018; Lu et al., 2018; Gan et al., 2020), indicating the crucial roles of these genes in the host immune response. Hemocytes, one of the major immune tissues in molluscs, has been reported as the main site where the recognition and elimination of bacterial pathogens occurs (Zhou et al., 2015). Hepatopancreas is the main digestive tissue; thus, it needs to cope with the pathogens incoming with the ingested algae. Furthermore, hepatopancreas was also proved as a toxin-rich tissue, acting as major “centers” for toxin accumulation in C. farreri (Li et al., 2017), which may also induce immune responses. The gill and mantle are constantly in contact with the external environment via water filtering and serve as the front line of the host defense (Lee et al., 2013). The tissue expression patterns of IRF genes have been widely characterized in various species, and high expression has been detected in multiple immune-related tissues. For example, high expression levels of CgIRF-2 were detected in C. gigas hemocytes, hepatopancreas, and mantle, and LcIRF from Larimichthys crocea were highly expressed in hemocytes, gill, and spleen (Lu et al., 2018; Guan et al., 2020). Taken together, the high expression levels of IRFs in these immune-related tissues may suggest their conservative roles in scallop innate immune response.
To provide insights into the functions of CfIRFs during the innate immune response, one of the major bacterial pathogens, V. anguillarum, was employed to perform the infection experiment and the responses of CfIRFs in hemocytes were investigated. Only CfIRFs from the IRF-1 subfamily (CfIRF1, CfIRF1-like, and CfIRF2) were found to be increased significantly post-infection, which confirmed the involvement of these CfIRFs in the innate immune response against bacterial invasion. In comparison with CfIRF1 and CfIRF1-like, CfIRF2 showed the most sensitive response. Subcellular localization prediction showed that CfIRF1 possessed an NLS in its DNA-binding domain, and subcellular localization analysis confirmed that it mainly translocated in the nucleus; meantime, CfIRF1-like and CfIRF2 were expressed in both nucleus and cytoplasm. Similarly, IRFs have shown the divergence in subcellular localization in some other species. For example, in marine bivalves C. gigas and P. fucata, researchers found that CgIRF-1 and CgIRF-2 proteins were both primarily expressed in nucleus and cytoplasm (Huang et al., 2017; Lu et al., 2018), and PfIRF-2 was located in the nucleus (Huang et al., 2013). Previous studies showed that most species only have one IRF1 gene, while in C. gigas, there are two IRF1 members, named CgIRF1a and CgIRF1b (Fan et al., 2018). They found that CgIRF1a significantly activated the ISRE reporter gene, whereas CgIRF1b did not. According to our data, both CfIRF1 and CfIRF1-like showed significant responses against bacterial infection, and they both could activate ISRE significantly; however, they have different functional domains, spatiotemporal expression patterns, and subcellular localizations, suggesting that functional differences may exist for CfIRF1 and CfIRF1-like.
Many studies have shown that IRFs are typical interferon-stimulated genes in mammals, birds, and fish (Andrea et al., 2002; Liu et al., 2018; Zhu et al., 2020). Among IRFs, IRF1 and IRF2 were originally characterized as transcriptional regulators of type I IFNs and IFN-stimulated genes (ISGs), which played an important role in the antiviral immune response (Miyamoto et al., 1988a; Harada et al., 1989b). Our findings from the dual-luciferase reporter gene assays showed that CfIRF1, CfIRF1-like, and CfIRF2 could significantly activate the expression of the IRSE reporter gene, revealing obvious transcriptional activation on interferon-stimulated response element. Similar to their responses against V. anguillarum, in comparison with CfIRF1 and CfIRF1-like, CfIRF2 showed the most significant activation effects, which may contribute to its most sensitive responses to bacterial infection. Similar results have been shown in other bivalves; for example, the recombinant CgIRF-1 or PfIRF-2 exhibited the activity to bind ISRE in vitro. Previous studies in vertebrates have shown that this conserved binding and activation of ISRE is mainly attributable to the DBD domain in IRFs. It was reported that the DBD in vertebrate IRFs could form a helix-turn-helix domain and bind to the core DNA sequence GAAA in the IFN-stimulated response element (ISRE, A/GNGAAANNGAAACT) (Escalante et al., 1998). For instance, IRF-1 from zebrafish (DrIRF-1) could bind to ISRE/IRF-E motifs within the IFN promoters through the DBD helix α3 to induce its transcription (Feng et al., 2015), while compared with the extensive knowledge of IRFs in vertebrates, the possible mechanisms of these bivalve IRFs binding and activating IFN system need further investigation.
5 Conclusion
In conclusion, we identified four IRF genes in Zhikong scallop: CfIRF1, CfIRF1-like CfIRF2, and CfIRF8. CfIRFs contained highly conserved N-terminal DNA-binding domain and variable C-terminal regulatory domain, and were constitutively expressed during development as well as in adult tissues, especially in hepatopancreas, hemolymph, gill, and mantle. Furthermore, we determined that CfIRF1, CfIRF1-like, and CfIRF2 genes played pivotal roles in the innate immune defense against bacterial infection, and their encoding proteins could translocate into nucleus. Functionally, CfIRF1, CfIRF1-like, and CfIRF2 had been proven to induce a strong activation of ISRE promoters. Moreover, in comparison with CfIRF1 and CfIRF1-like, CfIRF2 showed the most sensitive responses in coping with V. anguillarum, and consistently, CfIRF2 exhibited the most significant activation on ISRE. Our data would provide valuable information for further investigations into the evolution and functional characterization of IRFs in bivalve molluscs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
This animal study was reviewed and approved by the Ocean University of China Institutional Animal Care and Use Committee.
Author Contributions
SL and JH conceived and designed the study. NH and XZ performed the experiments. FS and XC participated in data analysis. NH, SL, JH, LZ, SW, and ZB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge the grant support from Project of Sanya Yazhouwan Science and Technology City Management Foundation (SKJC-KJ-2019KY01), Key R&D Project of Shandong Province (2020ZLYS10, 2021ZLGX03), National Key R&D Project (2021YFD1200805) and China Agriculture Research System of MOF and MARA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Materials
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.865707/full#supplementary-material.
Supplementary Figure 1 | The phylogenetic tree was constructed based on the protein sequences of CfIRFs, in addition to those of other species. FastTree 2.0.0 was used to construct the phylogenetic tree by the maximum-likelihood (ML) analyses. Solid circle: vertebrate IRFs, Hollow circle: invertebrate IRFs, Red hollow circle: CfIRFs.
Supplementary Figure 2 | The expression profiles in different developmental stages and adult tissues. (A). Expression levels of CfIRFs in different embryonic and larval stages. (B). Expression levels of CfIRFs in adult tissues. Vertical bars represent the mean ± S.E. (N=3). P values are calculated by one-way analysis of variance (ANOVA) in SPSS software. Different letters indicate significant differences (P < 0.05).
References
Anda S. D., Gómez-Martín D., Díaz-Zamudio M., Alcocer-Varela J. (2012). Interferon Regulatory Factors: Beyond the Antiviral Response and Their Link to the Development of Autoimmune Pathology. Autoimmun. Rev. 11, 98–103. doi: 10.1016/j.autrev.2011.08.006
Andrea K., Mario K., Katharina S., Hansj R. H., Mueller P. P. (2002). Activities of IRF-1. J. Interferon Cytokine Res. 22, 5–14. doi: 10.1089/107999002753452610
Azumi K., Sabau S. V., Fujie M., Usami T., Koyanagi R., Kawashima T., et al. (2007). Gene Expression Profile During the Life Cycle of the Urochordate Ciona Intestinalis. Dev. Biol. 308, 572–582. doi: 10.1016/j.ydbio.2007.05.022
Barnes B. J., Moore P. A., Pitha P. M. (2001). Virus-Specific Activation of a Novel Interferon Regulatory Factor IRF-5, Results in the Induction of Distinct Interferon α Genes. J. Biol. Chem. 276, 23382–23390. doi: 10.1074/jbc.M101216200
Barnes B. J., Richards J., Mancl M., Hanash S., Pitha P. M. (2004). Global and Distinct Targets of IRF-5 and IRF-7 During Innate Response to Viral Infection. J. Biol. Chem. 279, 45194–45207. doi: 10.1074/jbc.M400726200
Cavlar T., Ablasser A., Hornung V. (2012). Induction of Type I IFNs by Intracellular DNA-Sensing Pathways. Immunol. Cell Biol. 90, 474–482. doi: 10.1038/icb.2012.11
Costa M. M., Prado-Alvarez M., Gestal C., Li H., Roch P., Novoa B., et al. (2009). Functional and Molecular Immune Response of Mediterranean Mussel (Mytilus Galloprovincialis) Haemocytes Against Pathogen-Associated Molecular Patterns and Bacteria. Fish Shellfish Immunol. 26, 515–523. doi: 10.1016/j.fsi.2009.02.001
Davidson E. H., Cameron R. A. (2006). Arguments for Sequencing the Genome of the Sea Urchin Strongylocentrotus Purpuratus. Bethesda, MD, USA:National Human Genome Research Institute. Available at: http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/SeaUrchin_Genome.pdf.
Escalante C. R., Yie J., Thanos D., Aggarwal A. K. (1998). Structure of IRF-1 With Bound DNA Reveals Determinants of Interferon Regulation. Nature 391, 103. doi: 10.1038/34224
Fan M., Yue L., Zhou Y., He Z., Li J., Yang Z., et al. (2018). Structural and Functional Analysis of Interferon Regulatory Factors (IRFs) Reveals a Novel Regulatory Model in an Invertebrate, Crassostrea Gigas. Dev. Comp. Immunol. 89, 14–22. doi: 10.1016/j.dci.2018.07.027
Feng L., Yu Q., Xue L., Ning X., Ba O Z. (2013). Identification of Reference Genes for qRT-PCR Analysis in Yesso Scallop Patinopecten Yessoensis. PloS One 8, e75609. doi: 10.1371/journal.pone.0075609
Feng H., Zhang Q.-Y., Yi-Bing G., Jian-Fang Z., Qi- M. (2015). Zebrafish IRF1 Regulates IFN Antiviral Response Through Binding to IFN Phi 1 and IFN Phi 3 Promoters Downstream of MyD88 Signaling. J. Immunol. 194, 1225–1238. doi: 10.4049/jimmunol.1402415
Gan Z., Cheng J., Hou J., Xia L., Lu Y., Nie P. (2020). Molecular and Functional Characterization of Interferon Regulatory Factor 1 (IRF1) in Amphibian Xenopus Tropicalis - ScienceDirect. Int. J. Biol. Macromol. 167, 719–725. doi: 10.1016/j.ijbiomac.2020.11.217
Gao Q., Song L., Ni D., Wu L., Zhang H., Chang Y. (2007). cDNA Cloning and mRNA Expression of Heat Shock Protein 90 Gene in the Haemocytes of Zhikong Scallop Chlamys Farreri. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 147, 704–715. doi: 10.1016/j.cbpb.2007.04.010
Green T. J., Raftos D., Speck P., Montagnani C. (2015). Antiviral Immunity in Marine Molluscs. J. Gen. Virol. 96, 749–759. doi: 10.1099/jgv.0.000244
Guan Y., Chen X., Luo T., Ao J., Chen X. (2020). Molecular Characterization of the Interferon Regulatory Factor (IRF) Family and Functional Analysis of IRF11 in the Large Yellow Croaker (Larimichthys Crocea). Fish Shellfish Immunol. 107, 218–229. doi: 10.1016/j.fsi.2020.10.001
Harada H., Fujita T., Miyamoto M., Kimura Y., Maruyama M., Furia A., et al. (1989). Structurally Similar But Functionally Distinct Factors, IRF-1 and IRF-2, Bind to the Same Regulatory Elements of IFN and IFN-Inducible Genes. Cell 58, 729–739. doi: 10.1016/0092-8674(89)90107-4
Hatada S., Kinoshita M., Takahashi S., Nishihara R., Sakumoto H., Fukui A., et al. (1997). An Interferon Regulatory Factor-Related Gene ( xIRF-6 ) Is Expressed in the Posterior Mesoderm During the Early Development of Xenopus Laevis. Gene 203, 183–188. doi: 10.1016/S0378-1119(97)00512-X
Honda K., Taniguchi T. (2006). IRFs: Master Regulators of Signalling by Toll-Like Receptors and Cytosolic Pattern-Recognition Receptors. Nat. Rev. Immunol. 6, 644–658. doi: 10.1038/nri1900
Huang X. D., Liu W. G., Wang Q., Zhao M., Wu S. Z., Guan Y. Y., et al. (2013). Molecular Characterization of Interferon Regulatory Factor 2 (IRF-2) Homolog in Pearl Oyster Pinctada Fucata. Fish Shellfish Immunol. 34, 1279–1286. doi: 10.1016/j.fsi.2013.02.003
Huang S., Yuan S., Guo L., Yu Y., Li J., Wu T., et al. (2008). Genomic Analysis of the Immune Gene Repertoire of Amphioxus Reveals Extraordinary Innate Complexity and Diversity. Genome Res. 18, 1112–1126. doi: 10.1101/gr.069674.107
Huang B., Zhang L., Du Y., Xu F., Li L., Zhang G. (2017). Characterization of the Mollusc RIG-I/MAVS Pathway Reveals an Archaic Antiviral Signalling Framework in Invertebrates. Sci. Rep. 7, 8217. doi: 10.1038/s41598-017-08566-x
Huang B., Zhi T. Q., Zhen X., Nie P. (2010). Global Characterization of Interferon Regulatory Factor (IRF) Genes in Vertebrates: Glimpse of the Diversification in Evolution. BMC Immunol. 11, 22. doi: 10.1186/1471-2172-11-22
Hu X., Bao Z., Hu J., Shao M., Huang X. (2010). Cloning and Characterization of Tryptophan 2,3-Dioxygenase Gene of Zhikong Scallop Chlamys Farreri (Jones and Preston 1904). Aquac. Res. 37, 1187–1194. doi: 10.1111/j.1365-2109.2006.01546.x
Ikushima H., Negishi H., Taniguchi T. (2013). The IRF Family Transcription Factors at the Interface of Innate and Adaptive Immune Responses. Cold Spring Harb. Symp. Quant. Biol. 78, 105–116. doi: 10.1101/sqb.2013.78.020321
Inkpen S. M., Solbakken M. H., Jentoft S., Eslamloo K., Rise M. L. (2019). Full Characterization and Transcript Expression Profiling of the Interferon Regulatory Factor (IRF) Gene Family in Atlantic Cod (Gadus Morhua). Dev. Comp. Immunol. 98, 166–180. doi: 10.1016/j.dci.2019.03.015
Kimura T., Nakayama K., Penninger J., Kitagawa M., Mak T. W. (1994). Involvement of the IRF-1 Transcription Factor in Antiviral Responses to Interferons. Science 264, 1921–1924. doi: 10.1126/science.8009222
Lee Y., Wickamarachchi W. D. N., Whang I., Oh M., Umasuthan N., De Zoysa M., et al. (2013). Immune Response-Related Gene Expression Profile of a Novel Molluscan Iκb Protein Member From Manila Clam (Ruditapes Philippinarum). Mol. Biol. Rep. 40, 1519–1527. doi: 10.1007/s11033-012-2196-5
Lehtonen A., Veckman V., Nikula T., Lahesmaa R., Kinnunen L., Matikainen S., et al. (2005). Differential Expression of IFN Regulatory Factor 4 Gene in Human Monocyte-Derived Dendritic Cells and Macrophages. J. Immunol. 175, 6570. doi: 10.4049/jimmunol.175.10.6570
Lelong C., Guo X., Sourdaine P., Yan HE, Jouaux A., Mathieu M. (2015). Transcriptome Analysis Reveals Strong and Complex Antiviral Response in a Mollusc. Fish Shellfish Immunol. 46 (1), 131–144.
Li C., Li H., Chen Y., Chen Y., Sheng W., Weng S. P., et al. (2015). Activation of Vago by Interferon Regulatory Factor (IRF) Suggests an Interferon System-Like Antiviral Mechanism in Shrimp. Sci. Rep. 5 (1), 1–13. doi: 10.1038/srep15078
Li J., Li L., Zhang S., Zhang G. (2010). Cloning, Genomic Structure, and Expression Analysis of Peroxiredoxin V From Bay Scallop Argopecten Irradians. Fish Shellfish Immunol. 30, 309–316. doi: 10.1016/j.fsi.2010.11.011
Li Y., Sun X., Hu X., Xun X., Zhang J., Guo X., et al. (2017). Scallop Genome Reveals Molecular Adaptations to Semi-Sessile Life and Neurotoxins. Nat. Commun. 8, 1721. doi: 10.1038/s41467-017-01927-0
Liu Y., Cheng Y., Shan W., Ma J., Wang H., Sun J., et al. (2018). Chicken Interferon Regulatory Factor 1 (IRF1) Involved in Antiviral Innate Immunity via Regulating IFN-β Production. Dev. Comp. Immunol. 88, 77–82. doi: 10.1016/j.dci.2018.07.003
Liu S., Jiang X., Hu X., Gong J., Hwang H., Mai K. (2004). Effects of Temperature on non-Specific Immune Parameters in Two Scallop Species: Argopecten Irradians (Lamarck 1819) and Chlamys Farreri (Jones & Preston 1904). Aquac. Res. 35 (7), 678–682. doi: 10.1111/j.1365-2109.2004.01065.x
Loker E. S., Adema C. M., Zhang S. M., Kepler T. B. (2010). Invertebrate Immune Systems–Not Homogeneous, Not Simple, Not Well Understood. Immunol. Rev. 198, 10–24. doi: 10.1111/j.0105-2896.2004.0117.x
Lu R. (2008). Interferon Regulatory Factor 4 and 8 in B-Cell Development. Trends Immunol. 29, 487–492. doi: 10.1016/j.it.2008.07.006
Lu M., Yang C., Li M., Yi Q., Lu G., Wu Y., et al. (2018). A Conserved Interferon Regulation Factor 1 (IRF-1) From Pacific Oyster Crassostrea Gigas Functioned as an Activator of IFN Pathway. Fish Shellfish Immunol. 76, 68–77. doi: 10.1016/j.fsi.2018.02.024
Mamane Y., Heylbroeck C., Génin P., Algarté M., Servant M. J., Lepage C., et al. (1999). Interferon Regulatory Factors: The Next Generation. Gene 237, 1–14. doi: 10.1016/S0378-1119(99)00262-0
Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Bryant S. H. (2011). CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Marié I., Durbin J. E., Levy D. E. (1998). Differential Viral Induction of Distinct Interferon-Alpha Genes by Positive Feedback Through Interferon Regulatory Factor-7. EMBO J. 17 (22), 6660–6669. doi: 10.1093/emboj/17.22.6660
Meraro D., Gleit-Kielmanowicz M., Hauser H., Levi B. Z. (2002). IFN-Stimulated Gene 15 Is Synergistically Activated Through Interactions Between the Myelocyte/Lymphocyte-Specific Transcription Factors, PU.1, IFN Regulatory Factor-8/IFN Consensus Sequence Binding Protein, and IFN Regulatory Factor-4: Characterization of. J. Immunol. 168, 6224–6231. doi: 10.4049/jimmunol.168.12.6224
Miyamoto M., Fujita T., Kimura Y., Maruyama M., Harada H., Sudo Y., et al. (1988). Regulated Expression of a Gene Encoding a Nuclear Factor, IRF-1, That Specifically Binds to IFN-Beta Gene Regulatory Elements. Cell 54, 903–913. doi: 10.1016/S0092-8674(88)91307-4
Nehyba J., Hrdlicková R., Bose H. R. (2009). Dynamic Evolution of Immune System Regulators: The History of the Interferon Regulatory Factor Family. Mol. Biol. Evol. 26, 2539–2550. doi: 10.1093/molbev/msp167
Nehyba J., Hrdlicková R., Burnside J., Bose H. R. (2002). A Novel Interferon Regulatory Factor (IRF), IRF-10, Has a Unique Role in Immune Defense and Is Induced by the V-Rel Oncoprotein. Mol. Cell. Biol. 22, 3942–3957. doi: 10.1128/MCB.22.11.3942-3957.2002
Pfaffl M. W., Horgan G. W., Leo D. (2002). Relative Expression Software Tool (REST©) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 30 (9), e36-e36. doi: 10.1093/nar/30.9.e36
Qiu L., Song L., Xu W., Ni D., Yu Y. (2007). Molecular Cloning and Expression of a Toll Receptor Gene Homologue From Zhikong Scallop, Chlamys Farreri. Fish Shellfish Immunol. 22, 451–466. doi: 10.1016/j.fsi.2006.05.003
Ragab A., Buechling T., Gesellchen V., Spirohn K., Boettcher A. L., Boutros M. (2014). Drosophila Ras/MAPK Signalling Regulates Innate Immune Responses in Immune and Intestinal Stem Cells. EMBO J. 30, 1123–1136. doi: 10.1038/emboj.2011.4
Sato M., Hata N., Asagiri M., Nakaya T., Tanaka N. (1999). Positive Feedback Regulation of Type I IFN Genes by the IFN-Inducible Transcription Factor IRF-7. FEBS Lett. 441, 106–110. doi: 10.1016/S0014-5793(98)01514-2
Savitsky D., Tamura T., Yanai H., Taniguchi T. (2010). Regulation of Immunity and Oncogenesis by the IRF Transcription Factor Family. Cancer Immunol. Immunother. 59, 489–510. doi: 10.1007/s00262-009-0804-6
Sudhir K., Glen S., Li M., Christina K., Koichiro T. (2018). MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547.
Tamura T., Yanai H., Savitsky D., Taniguchi T. (2008). The IRF Family Transcription Factors in Immunity and Oncogenesis. Annu. Rev. Immunol. 26, 535–584. doi: 10.1146/annurev.immunol.26.021607.090400
Taniguchi T. T. (2006). Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immunity 25 (3), 349–360.
Teng W. M., Li W. J., Zhang M., Yu Z. A., Fu C. D. (2012). Isolation, Identification and Pathogenicity of Vibrio Chagasii From Patinopecten Yessoensis. J. Fish. China 36, 937.
Thi H. D., Olga C., Arndt V. H., Quang M. B., Sy V. L. (2017). UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 35 (2), 518–522.
Weiqi H., Chunliu Z., Hao W., Elizabeth H., Elizabeth A. (2008). The Interferon Consensus Sequence-Binding Protein (ICSBP/IRF8) RepressesPTPN13Gene Transcription in Differentiating Myeloid Cells. J. Biol. Chem. 283 (12), 7921–7935.
Xuan M., Yan X., Liu X., Xu T. (2020). IRF1 Negatively Regulates NF-κb Signaling by Targeting MyD88 for Degradation in Teleost Fish. Dev. Comp. Immunol. 110, 103709. doi: 10.1016/j.dci.2020.103709
Yanai H., Negishi H., Taniguchi T. (2012). The IRF Family of Transcription Factors: Inception, Impact and Implications in Oncogenesis. Oncoimmunology 1, 1376. doi: 10.4161/onci.22475
Zhang R., Liu R., Wang W., Xin L., Wang L., Li C., et al. (2015). Identification and Functional Analysis of a Novel IFN-Like Protein (CgIFNLP) in Crassostrea Gigas. Fish Shellfish Immunol. 44 (2), 547–554. doi: 10.1016/j.fsi.2015.03.015
Zhan F. B., Liu H., Lai R. F., Jakovli I., Wang W. M. (2016). Expression and Functional Characterization of Interferon Regulatory Factors (Irf2, Irf7 and Irf9) in the Blunt Snout Bream (Megalobrama Amblycephala). Dev. Comp. Immunol. 67, 239–248. doi: 10.1016/j.dci.2016.09.014
Zhi Z., Wang L., Shi X., Zhang H., Yang G., Wang M., et al. (2011). The Modulation of Catecholamines to the Immune Response Against Bacteria Vibrio Anguillarum Challenge in Scallop Chlamys Farreri. Fish Shellfish Immunol. 31, 1065–1071. doi: 10.1016/j.fsi.2011.09.009
Zhou S.-M., Liu S., Li M., Yang N., Tao Z. (2015). First Description and Expression Analysis of Tumor Necrosis Factor Receptor-Associated Factor 6 (TRAF6) From the Swimming Crab, Portunus Trituberculatus. Fish Shellfish Immunol. 45 (2), 205–210. doi: 10.1016/j.fsi.2015.04.005
Keywords: Chlamys farreri, IRF, transcriptional activation, interferon-stimulated response element, immune response
Citation: Hu N, Lian S, Zhu X, Chen X, Sun F, Zhang L, Wang S, Bao Z and Hu J (2022) Interferon Regulatory Factors Functioned as Activators of the Interferon Pathway in the Scallop Chlamys farreri. Front. Mar. Sci. 9:865707. doi: 10.3389/fmars.2022.865707
Received: 30 January 2022; Accepted: 29 March 2022;
Published: 26 April 2022.
Edited by:
Weiwei You, Xiamen University, ChinaReviewed by:
Lusheng Xin, Chinese Academy of Fishery Sciences (CAFS), ChinaYang Zhang, South China Sea Institute of Oceanology (CAS), China
Copyright © 2022 Hu, Lian, Zhu, Chen, Sun, Zhang, Wang, Bao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Lian, bGlhbnNoYW5zaGFuQG91Yy5lZHUuY24=; Jingjie Hu, aHVqaW5namllQG91Yy5lZHUuY24=
 Naina Hu1
Naina Hu1 Shanshan Lian
Shanshan Lian Lingling Zhang
Lingling Zhang Shi Wang
Shi Wang Zhenmin Bao
Zhenmin Bao