- 1Sars-Fang Centre and Ministry of Education (MOE) Key Laboratory of Marine Genetics and Breeding, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Laboratory for Marine Fisheries Science and Food Production Processes, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Key Laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
Cilia are organelles located on the surface of eukaryotic cells and play important roles in numerous physiological and developmental processes. Cilia- and flagella-associated proteins (CFAP family) are well known due to their close relation to the assembly and functionalization of both cilia and flagella. In vertebrates, Cfap206 is reported to function during motile cilia assembly, and its abolished expression could lead to abnormal ciliary beating and decreased sperm motility. However, there is scarcely any information on the function of Cfap206 in marine invertebrates. Here, through using an ideal shellfish research model, the dwarf surf calm Mulinia lateralis, we explored the role of Cfap206 during embryonic ciliogenesis as well as during male gametogenesis. In trochophore larvae, effective knocking down or knocking out of Cfap206 was detected respectively through RNAi and CRISPR/Cas9 technology, showing an obviously decreased number of cilia. During the period of gonadal maturation, we managed to deliver the dsRNA of Cfap206 to male individuals for 14 days through carrier vector feeding. Significant suppression of Cfap206 was observed, together with impaired sperm motility and aberrant sperm tail assembly. The present study provided valuable information on gene function exploration methods in M. lateralis and further enriched the understanding of Cfap206 on ciliogenesis in marine invertebrates. Furthermore, by revealing the relationship between Cfap206 and spermatozoa flagellum, it is possible to generate male sterile populations in hermaphroditic marine organisms to improve crossbreeding efficiency.
Introduction
Cilia are membranous organelles that protrude from the surface of most eukaryotic cells, contributing to multiple functions such as movement, sensation, and signal transmission (Pazour and Witman, 2003; Jakobsen et al., 2006; Verhey et al., 2011; Bangs and Anderson, 2017). The basic structure of cilia is highly conserved, including four parts: matrix, transition zone, axoneme, and ciliary membrane (Elliott and Brugmann, 2019). According to the differential axoneme microtubule composition, cilia could be divided into motile cilia and nonmotile cilia (known as primary cilia) (Verhey et al., 2011). The axoneme of motile cilia usually possesses nine doublet microtubules surrounding a central pair of singlet microtubules (Satir and Christensen, 2007). The microtubule doublets are interconnected with ciliary dynein motors, the activity of which is regulated through radial spokes (RS1, RS2, and RS3) to produce fluid and pulsatile motion. Thus, motile cilia could mediate the movement of cells and extracellular fluid, such as the movement of eggs and sperm, as well as the removal of foreign bodies in the respiratory tract (Afzelius and Eliasson, 1983; Lyons et al., 2006; Orhon et al., 2015). The axoneme of primary cilia lack the central pair of singlet microtubules, and there are a number of surrounding sensory membrane proteins that can assist in performing sensory, signal transduction, and coordination functions (Hua and Ferland, 2018; Tao et al., 2020). Primary cilia are closely related to the transduction of Hedgehog (Hh) pathway (Danwei et al., 2003), Wnt pathway (Ross et al., 2005), platelet-derived growth factor (PDGF) pathway, and Ca2+ signaling cascade (Orhon et al., 2015).
The cilia- and flagella-associated protein (CFAP) family is proven to be associated with the normal assembly of cilia or flagella (Tang et al., 2017). Cfap39 and Cfap65, for example, are required for the assembly of the dynein regulatory complex in the generation of motile cilia (Lin et al., 2012; Beckers et al., 2020) and deletion of Cfap43, Cfap44, Cfap65, Cfap69, and Cfap251 will lead to abnormal sperm flagellum assembly in mice or humans, resulting in male infertility (Tang et al., 2017; Dong et al., 2018; He et al., 2020; Li et al., 2020). In particular, Cfap206 has recently been identified as a downstream target gene of Foxj1, a key transcription factor that has been proven to be closely related to the occurrence of mobile cilia in vertebrates (Stubbs et al., 2008; Thomas et al., 2010; Alten et al., 2012; Vij et al., 2012). Any mutation or deletion of them may lead to severe cilia diseases, such as primary ciliary dysmotility (PCD), hydrocephalus, retinitis pigmentosa, polycystic kidney disease (PKD), and multiple morphological abnormalities of the sperm flagella (MMAF) (Pazour and Rosenbaum, 2002; Satir and Christensen, 2007; Sironen et al., 2020). Moreover, CFAP206 protein was found to be located in cilia, which was required for establishing the dynamin motor RS2 (Beckers et al., 2020). It was reported that the stable assembly of calmodulin- and spoke-associated complex (CSC), which is considered to mediate the specific connection of radial spokes with adjacent structures in cilia, was regulated by Cfap206 in a dependent manner (Heuser et al., 2012; Vasudevan et al., 2015; Toure et al., 2021). Although Cfap206 is a key gene for ciliogenesis, it has only been studied in several model organisms, such as Tetrahymena and Mus musculus. For example, when Cfap206 was knocked out, the cilia of Tetraphymena cells were found to produce abnormal waveforms due to the abnormal RS2 assembly and showed reduced bending amplitude and weakened intertemporal coordination, but there was no significant difference in cilia number and length (Vasudevan et al., 2015). Interestingly, the loss of Cfap206 also leads to abnormal patterns of bending or curling of sperm flagellum in mice; however, the Cfap206-KO mice did show sperm flagella shortening or loss (Shen et al., 2021). Bivalve mollusc is an ancient Lophotrochozoa group of marine invertebrates that possesses rich species diversity. Cilia play critical roles in important physiological activities of bivalve species, such as filter feeding, respiration, reproduction, as well as larval movement. However, knowledge of molecular modulation of ciliogenesis is quite limited in bivalves, and to the best of our knowledge, this is the first study on Cfap206 to point to the regulation of both cilia and flagella. Notably, many bivalves, such as Argopecten irradians (Wei et al., 2021), are hermaphroditic, and a better understanding of the regulation of Cfap206 on sperm flagellum may help to produce a male sterile population, thus improving artificially crossbreeding efficiency.
The dwarf surf clam (Mulinia lateralis) is a small, buried bivalve mollusc belonging to the family Mactridae, which is naturally distributed in estuaries and mudflats along the Atlantic coast from Canada to the Caribbean Sea (Walker and Tenore, 1984). They possess many appealing features for becoming a promising bivalve model, such as small adult size (15–20 mm), rapid development and early transparent shell, short generation time (~2 months), and convenient artificial cultivation (no attachment base) (Calabrese, 1970; Santos and Simon, 1980). In addition, M. lateralis represents the first bivalve with successful demonstration of transgene technology (Lu et al., 1996). At present, our team has established a standard laboratory artificial breeding system for M. lateralis, and we have successfully achieved gene knockdown through RNAi as well as gene knockout through CRISPR/Cas9 microinjection. Thus, we chose M. lateralis to explore the participation of Cfap206 in both ciliogenesis and spermatogenesis in this study. Our study provides the functional characterization of bivalve Cfap206 and assists in a better understanding of cilia/flagella regulation in marine invertebrates. Further investigation of the relationship between Cfap206 and sperm flagellum formation may provide a possible method for the generation of male sterile individuals to improve the crossbreeding efficiency for hermaphroditic marine organisms.
Materials and Methods
Identification, Phylogenetic and Spatiotemporal Expression Analysis of Cfap206 in M. lateralis
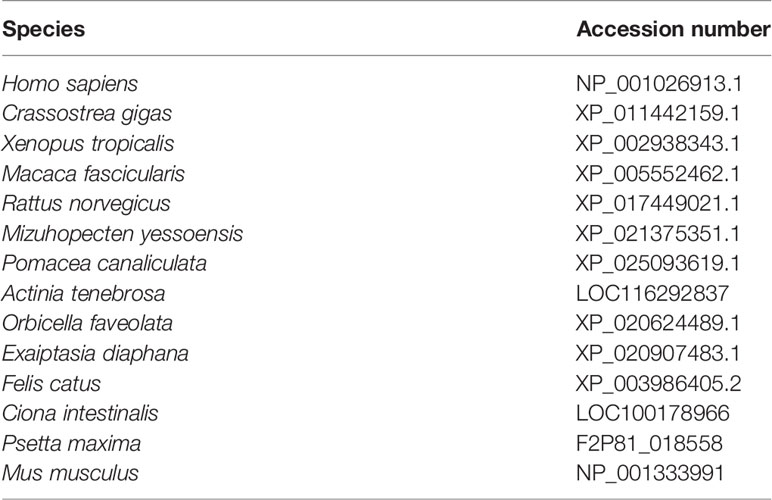
The available CFAP206 protein sequence of 14 representative species were downloaded from NCBI and Uniprot databases (Table 1 shows the accession numbers). These orthologous sequences were used as queries for whole-genome blast with the E-value threshold of 1E−05 in the M. lateralis proteome and genome database. To ensure the integrity and accuracy of Cfap206, its nucleic acid sequence was predicted using ORF Finder, and the amino acid sequence was submitted to the SMART database to verify the presence of the CFAP domain. The molecular weights and isoelectric points were calculated by using Compute PL/Mw tools. The genetic structure of Cfap206 was mapped using the GSDS2.0 website. The Geneious Prime software and PHYRE2.0 website were used to predict the secondary structure and tertiary structure of CFAP206 protein.
CFAP206 protein sequences of 12 species were obtained from the NCBI and Uniprot databases, including Homo sapiens, Mus musculus, Petromyzon marinus, Crassostrea gigas, Mizuhopecten yessoensis, Mulinia lateralis, Capitella teleta, Aplysia californica, Lingula anatina, Ciona intestinalis, Nematostella vectensis, and Lottia gigantea. ClustalW (Larkin et al., 2007) and Genedoc Software (Nicholas, 1997) were used for multiple sequence alignment. The phylogenetic analysis of Cfap206 was performed by MEGA7.0, and the establishment method of the tree was neighbor joining (NJ). The parameter was P-distance, and the value of bootstrap replications was set to 1,000. According to the transcriptome data of M. lateralis obtained in our laboratory, the expression level of Cfap206 in fourteen embryonic stages and seven adult tissues were plotted.
RNAi Based on Electroporation
Based on unpublished genome and transcriptome information from our lab, the exon and intron of Cfap206 gene were annotated by MAKER2 (Version 3.01.03) to obtain cDNA sequence information. Also, the interference sites (110–594 bp) were identified using SiDirect Version 2.0. Cfap206-dsRNA was synthesized by MEGA Script RNAi kit (Thermo Fisher Scientific, America) (RNAi primers were shown in Table 2). Male and female M. lateralis with well-developed gonads were selected, then the fresh sperm and eggs were obtained under standard laboratory conditions. After extrusion of the first polar body (around 15 min after mixing the sperm and eggs), each group collected 6,000–7,000 fertilized eggs for electroporation. Bio-Rad Gene Pulser Xcell (America) was selected as the electroporation instrument, and the electric transfer parameters were set according to Table 3. We chose filtered seawater as the electroporation buffer: blank control group was not processed; electroporation-control group was given electric shock without dsRNA; and experimental group was added with dsRNA (concentration maintained at 250 ng/μl) and was given electric shock. After 30 min on ice, all groups were cultured at room temperature in a 6-well plate to trochophore larvae stage.
Deletion of Cfap206 Gene Using CRISPR-Cas9 System
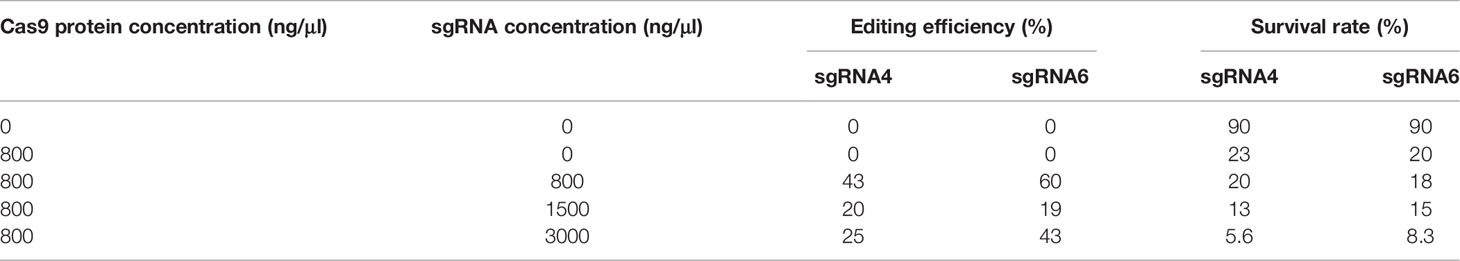
The functional domain of Cfap206 was predicted using the SMART website. Single-guided RNAs (sgRNAs) were designed on the exons of this domain, and sgRNAs containing SNPs were excluded (sgRNA sequences are shown in Table 2). sgRNAs were synthesized using MEGAscript™ T7 Transcription Kit (Thermo Fisher Scientific, America). The editing efficiency of sgRNAs was tested in vitro using the Guide-it sgRNA Screening Kit (Takara, Japan). sgRNAs and Cas9 protein in different concentrations were incubated at 37° C for 15 min (Table 4), and then, microinjection was performed using fertilized eggs of M. lateralis. The control and injected embryos were cultured in filtered seawater at 24°C. The injection volume of the Cas9 protein-sgRNAs mixture is about 0.1 nl. A single embryo was collected approximately 24 h after microinjection, and genomic DNA was extracted by the Chelex®-100 method (Li et al., 2021). The DNA fragment containing the target site was amplified using MightAmp™ DNA Polymerase Ver.3 (Takara, Janpan), and the corresponding verified primers are shown in Table 2. The PCR fragments were sequenced in Sangon Biotech.
Plasmid Construction and Induction of Interfering dsRNA
Following the relevant method developed in the Pacific oyster Crassostrea gigas (Feng et al., 2019), the interference fragment of Cfap206 (110–594 bp) was inserted into the L4440 vector to construct the Cfap206-dsRNA-L4440 plasmid. The Cfap206-dsRNA-L4440 and L4440 plasmids were transfected into competent Escherichia coli cells HT115 for expression. The strain HT115 carrying the Cfap206-dsRNA-L4440 plasmid was inoculated in LB medium containing ampicillin (50 μg/ml) and tetracycline (12.5 μg/ml) at 37°C and 220 rpm to the logarithmic stage (OD: 0.4–0.6). In total, 0.8 Mm IPTG was added to a 40 ml bacterial solution and stirred at 37°C for 4 h to induce dsRNA. The bacteria of the induced group and the noninduced group were centrifuged at 5,500 rpm for 2 min. After the supernatant was removed, the total RNA was extracted by the Trizol method, and the presence of dsRNA was determined by 1.5% agarose gel.
RNAi by Ingested dsRNA-Expressing Bacteria
Three hundred male M. lateralis with healthy growth status, equal size, and immature gonads were selected and divided into three groups (blank-control group, L4440-control group, and Cfap206-dsRNA-L4440 group). Eight liters of filtered seawater was added to the glass tank, and each group was fed with a micropump. The water flow rate was controlled at 12 ml/min, and the water temperature was maintained at about 25°C. Each group was temporarily reared for 5 days without treatment to observe the mortality under natural growth state. During the interference process, Chlorella pyrenoidesa was fed twice a day from days 1 to 7, with 180 μl (1 × 1010/ml) each time. In detail, normal algae were fed at 10 am, and normal algae or algae-bacteria were fed at 10 pm for different groups, and the sea water was changed once a day. From days 8 to 14, to speed up gonadal ripening to maturation, 180 μl of normal C. pyrenoidesa were fed at 10 am, and normal mixed algae (Table 5) or mixed algal-bacteria were fed at 10 pm, and egg yolk was fed continuously during these 2 weeks.
Quantitative RT-PCR
According to the phenol chloroform extraction method described by Hu et al. (2006), the total RNA of embryos was extracted. The Reverse Transcriptase M-MLV (Rnase H-) kit (Takara, Japan) was used to reverse transcription into cDNA, with a concentration of 20 ng/μl after inversion. Fluorescent real-time quantitative PCR was performed using the 2× ChamQ SYBR Color qPCR Master Mix (Vazyme, China). The 2–ΔΔt method is used to quantify the expression of Cfap206. Quantitative RT-PCR primers are shown in Table 2. All qPCR experiments were repeated twice, with two biological replicates and three technical parallels for each sample, using RS23 as an internal control.
Immunofluorescence
Embryos were washed twice with 1×PBS to remove seawater and fixed overnight with 4%PFA at 4°C. The next day, after being centrifuged at 300 rcf at room temperature for 30 s, the embryos were washed with 1×PBS 3 times. After washing, the sample was dehydrated with 80% methanol and 20% DMSO at 4°C overnight. After washing with PBST (1×PBS+0.5%Triton X-100) 3 times, the sample was then added with 500 μl trypsin and incubated for 5 min. After repeated washing with PBST, the sample was incubated with 10% goat serum (NGS) at room temperature for 2 h. Rabbit antiacetylated tubulin antibody was then added to 10%NGS in a ratio of 1:1,000 and the samples were incubated overnight at 4°C. After washing with PBST three times, goat anti-rabbit FITC was added to 2%NGS/PBST at the ratio of 1:200, and the sample was incubated at room temperature for 2 h. Following washing with PBST, the sample was added with phalloidin and DAPI in a ratio of 1:500 and stained for 30 min at room temperature. After being washed with PBS three times, the sample was mounted and the confocal images were taken.
Sperm Motility Test
After 14 days of feeding, sexually mature males were selected for artificial sperm stimulation for each group. Sperm motility was tracked using the CASAS-QH-III automatic sperm analyzer (Tsinghua Tongfang, China). The process was completed by the National Laboratory (Qingdao).
Microscopy
For confocal observation, the cellular morphology was observed with the A1R Ti-E confocal microscopy (Nikon, Japan). For SEM observation, the critical point-dried sample was coated in gold and the sample surface was observed through a Tescan™ Vega3 scanning electron microscope (Czechia).
Results and Discussion
Identification of Cfap206 in M. lateralis and Phylogenetic Analysis
We identified one Cfap206 (MlCfap206) gene through whole-genome searching of M. lateralis, and its genomic structure showed 16 exons, 15 introns, a 5′UTR, and a 3′UTR (Figure 1A). The ORF length and protein length of MlCfap206 were 1875bp and 624 amino acids, respectively. A similar protein structure was revealed for CFAP206 from various species. For example, in mice, Cfap206 encodes 620-amino acids and possesses the conserved FAP206 domain (Shen et al., 2021) (Figure 1B). This may suggest the conservative function of MlCfap206 related to cilia and flagella. The secondary structure of CFAP206 protein is shown in Figure 1C, which contained 27 α-helixes, 25 β-folds, 36 β-bends, and 35 random coils. Based on template d1mhyd (confidence: 87.5), the predicted tertiary ferritin-like structure within the CFAP206 domain (420aa-467aa) is shown in Figure 1D.
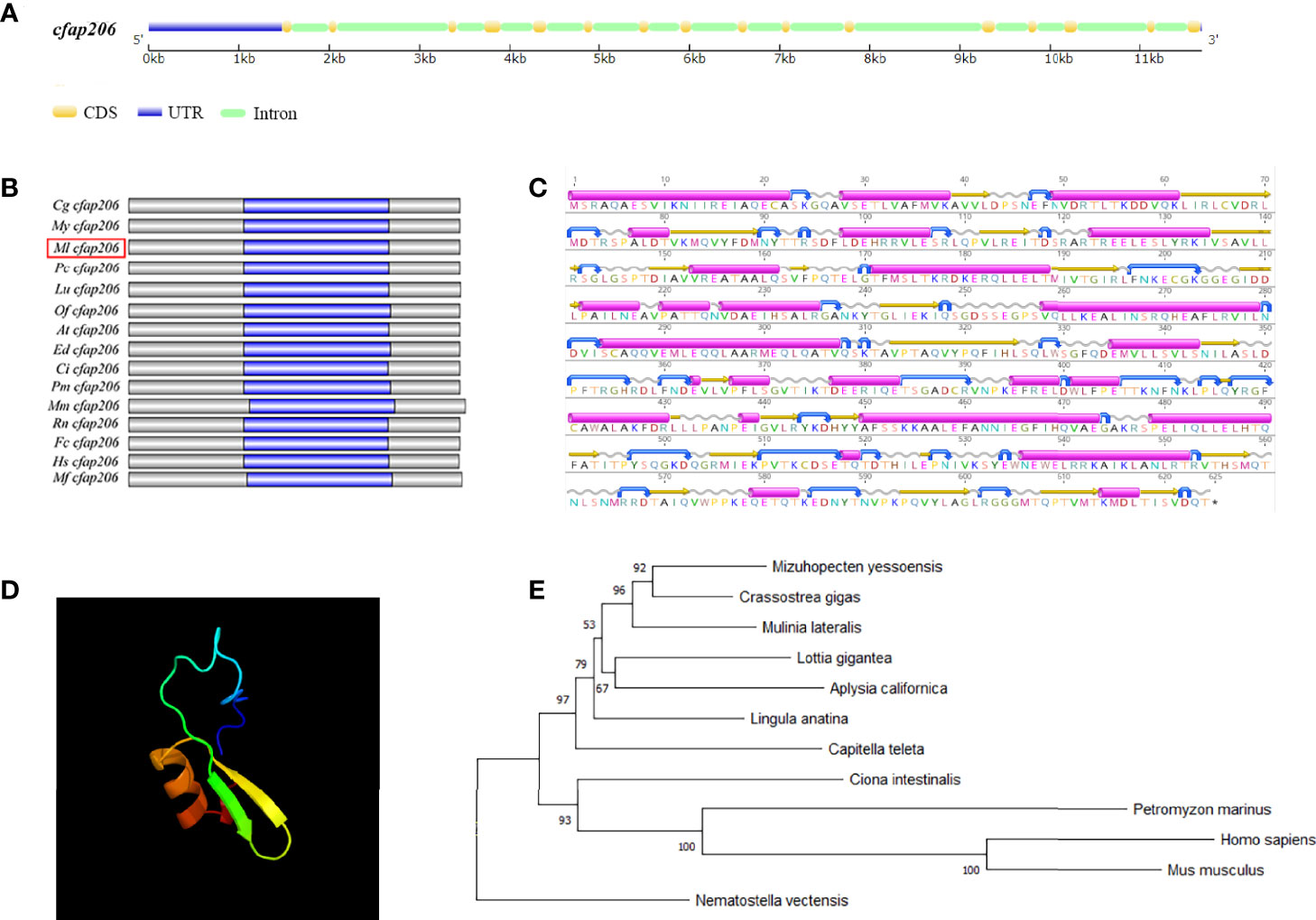
Figure 1 Characterization of Cfap206 in M. lateralis and phylogenetic analysis. (A) Genetic structure of Cfap206. CDS, coding sequence; UTR, untranslated region. (B) Protein structures of CFAP206s of fifteen representative species. Grey represents the whole protein region, and the magenta region represents the CFAP206 domain of each species. (C) Prediction of the secondary structure of CFAP206 protein. Magenta represents α-helixes, yellow represents β-folds, blue represents β-bends, and gray represents random coils. (D) Prediction of the tertiary ferritin-like structure within the CFAP206 domain (420aa-467aa). (E) Phylogenetic analysis of CFAP206 proteins.
Phylogenetic analysis of CFAP206 proteins was performed using the NJ method with 1,000 bootstrap pseudoreplicates (Figure 1E), from 12 selected animals spanning Cnidaria (including N. vectensis), Brachiopoda (L. anatina), Mollusca (including C. gigas, M. yessoensis, M. lateralis, A. californica, and L.gigantea), Annelida (including C. teleta), and Chordata (including H. sapiens, M. musculus, C. intestinalis, and P. marinus). Obviously, MlCFAP206 was first grouped together with CFAP206 proteins from molluscs (such as M. yessoensis and C. gigas). These mollusc CFAP206s were then sequentially clustered with CFAP206s from brachiopods, annelids, and chordates. The clustering relationship of CFAP206 is consistent with the species’ evolutionary relationship.
Spatiotemporal Expression Analysis
Spatiotemporal expression analysis of MlCfap206 in the present study was performed based on our unpublished transcriptomic data. Transcripts per million (TPM) of MlCfap206 in fourteen embryonic stages as well as in seven adult tissues was used to perform the expression level analysis (Figure 2). During development (Figure 2A; Supplementary Table S1), the high expression level of Cfap206 is mainly observed in blastula, gastrula, and trochophore larvae, three key periods for embryonic motile cilia formation and development. A similar pattern could be observed in M. yessoensis based on the published transcriptome data (Hou et al., 2011), which suggests that Cfap206 may be required for embryonic ciliogenesis regulation. Among the seven adult tissues of M. lateralis, the highest expression of Cfap206 was found in the male gonad, followed by foot and gill (Figure 2B; Supplementary Table S2). Functionalization of these three organs needs specialized cilia for sperm tail formation, byssus attachment, as well as for respiration, food intake, or waste removal through gill filaments (Ward et al., 1993; Andrade et al., 2015; Shen et al., 2021). Lines of evidence showed that Cfap206 plays important roles in ensuring the stability of RS and CSC, participating in cilia and flagella formation regulation (Dubruille et al., 2002; Vasudevan et al., 2015). Similar to our findings, previous studies have found that Cfap206 is massively overexpressed in human testicles, and the sperm of mice without the Cfap206 gene shows abnormalities in the morphology, number, and length of flagella (Shen et al., 2021). To date, cilia regulation of gill has only been reported in marine invertebrates including Ciona intestinalis and C. gigas, mainly focusing on calaxin, Meichroacidin, Tektin A1, and Tektin B1 (Shimazaki et al., 2006; Wang et al., 2015), and the regulatory genes of foot cilia have not been reported yet. Our results suggested that Cfap206 may be involved in these organs’ cilia regulation in marine bivalves.
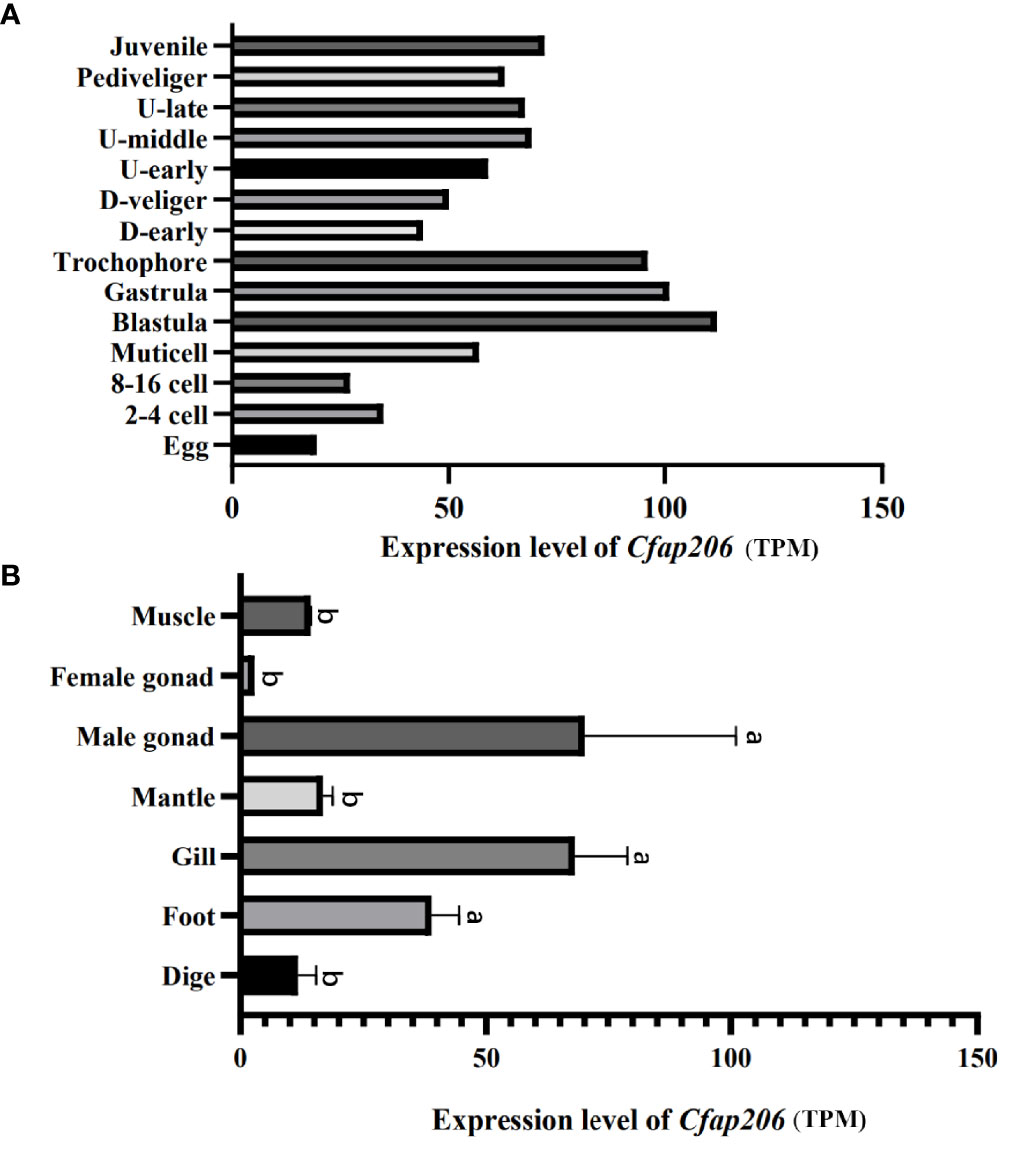
Figure 2 Spatiotemporal expression of Cfap206 in M. lateralis. (A) The expression of Cfap206 in fourteen embryonic stages. (B) The expression of Cfap206 in seven tissues. SPSS28.0 was used for one-way ANOVA analysis of the data, and Duncan method was used to test the significance of the difference between the values of each group.
RNAi to Suppress Cfap206 Expression Affected Embryonic Ciliogenesis
The trochophore larvae possess apical tuft and cilia band which are the typical ciliated features and are responsible for sensation, movement, and feeding (Arenas-Mena, 2010). Moreover, the ciliated trochophore larvae are considered to be the most conserved phylogenetic stage of marine Lophotrochozoans, and several pieces of evidence show they share an ancient and common origin (Paps et al., 2015; Xu et al., 2016; Wang et al., 2020). Thus, we chose trochophore larvae to explore whether or not Cfap206 participate in bivalve embryonic ciliogenesis. Firstly, we generated the Cfap206-dsRNA through in vitro transcription and the target bands were correctly located (Figure 3A). With optimized electroporation conditions and reasonable dose of Cfap206-dsRNA (~250 ng/μl), we detected significant suppression of Cfap206 (p < 0.01) in M. lateralis trochophore larvae by qPCR (Figure 3B). Compared with the control group, the electroporation-blank group showed normal gene expression level, excluding that there might be side effects of electroporation itself on gene expression. Morphological observations of each group of larvae were then made by confocal microscopy and scanning electron microscopy (Figures 3C, D). Similar size as well as spindle-shaped body were observed for the larvae from three groups, indicating the normal speed of embryo development. However, obvious cilia reduction/abolishment showed up in the larvae when Cfap206 was significantly suppressed, demonstrating the irreplaceable requirement of Cfap206 during embryonic ciliogenesis of M. lateralis. Previous studies in model animals such as Xenopus laevis and Tetrahymena revealed that Cfap206 is mainly involved with the regulation of cilia motility (Vasudevan et al., 2015). In X. laevis, moderate ciliary defect occurred during larva development when Cfap206 gene was knocked out, leading to abnormal ciliary movement waveform and decreased motor ability. Similarly, the axoneme of the cilia of Tetrahymena with Cfap206 knocked out were compressed, which swam at only 30% of the speed of the wild type (Vasudevan et al., 2015). However, the relationship between Cfap206 and ciliogenesis is not clear in bivalves. According to our results, it was obvious that the downregulation of Cfap206 expression in M. lateralis led to a significant decrease in the number of cilia. This phenomenon reminded us that there might be function divergence of Cfap206 on cilia regulation among different species, or Cfap206 is effective at different hierarchies during signal transduction which is worthy of further in-depth study.
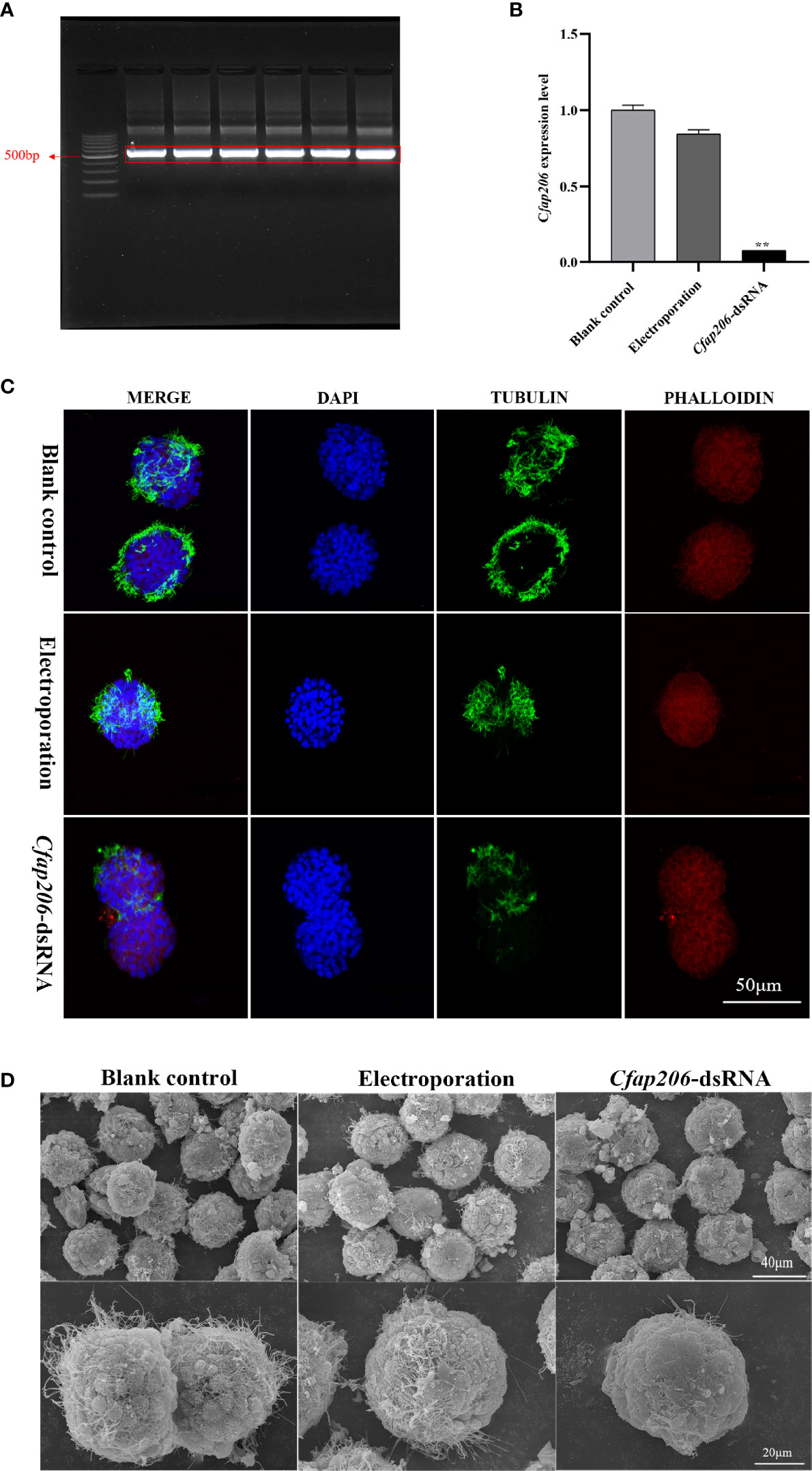
Figure 3 Expression analysis of Cfap206 and morphological observation of trochophore larvae after RNAi. (A) Electrophoresis result of Cfap206 dsRNA. Lane 1, 100 bp DNA ladder. Lanes 2–7, bands of Cfap206-dsRNA (500 bp). (B) The expression of Cfap206 after RNAi (detected by qPCR). **p < 0.01. (C) Confocal observation of trochophore larvae after RNAi. Cytoskeleton was stained red by phalloidin, nucleus was stained blue by DAPI, and cilia were stained green by tubulin antibody. (D) SEM observation of trochophore larvae after RNAi.
CRISPR/Cas9 Knockout of Cfap206 Affected Embryonic Ciliogenesis
The efficiency of sgRNAs plays a key role in the editing effect (Li et al., 2021). Thus, to generate Cfap206-KO larvae of M. lateralis, we first performed the Cfap206-sgRNA screening to avoid the influence of SNP and to perform in vitro efficiency test. Six sgRNAs (sgRNA1-6) were chosen (Figure 4A). Among them, it was found that Cfap206-sgRNA-2, Cfap206-sgRNA-4, and Cfap206-sgRNA-6 could work and cut the target DNA fragment into two pieces in vitro, and Cfap206-sgRNA-4 seems to be the most effective one (Figure 4B). Next, through microinjection together with Cas9 protein (800 ng/µl), we performed the in vivo editing efficiency test of these three effective Cfap206-sgRNAs. A previous study revealed that the concentration ratio of sgRNA-Cas9 was a key factor in inducing effective gene mutation (Li et al., 2021). In the present study, three different concentrations of Cfap206-sgRNA (800/1,500/3,000 ng/µl) were tested and showed a different editing efficiency as well as survival rate. Unfortunately, after Cfap206-sgRNA-2/Cas9 microinjection, no positive editing results turned back. However, Cfap206-sgRNA-4 and Cfap206-sgRNA-6 still worked in vivo and 800 ng/µl was the optimal concentration for microinjection (Table 4). In comparison, under optimal concentration, the in vivo editing efficiency of Cfap206-sgRNA-6 (60%) was higher than Cfap206-sgRNA-4 (43%), and the corresponding positive editing results are shown in Figure 4C. In M. lateralis, the target gene mutations in vivo which were induced by Cfap206-sgRNA/Cas9 mainly included insertion or deletion mutations within 11 bp, similar to the editing results of other invertebrates (Lin and Su, 2016; Hiruta et al., 2018; Li et al., 2021). In addition, we noticed that the editing efficiency of different sgRNAs might be different in vitro or in vivo. This may be due to the influence of complex physicochemical factors in the internal environment on the sensitivity of sgRNA, leading to the difference in the efficiency of sgRNA in vivo and in vitro. Due to the limited number of positive-edited trochophore larvae, we performed bright field morphological observation (about 20–30 larvae were observed) which showed an obvious reduced cilia number after Cfap206 knocked out (Figure 4D), consisting with the Cfap206-RNAi phenotype. It reinforced that Cfap206 played a key role during embryonic ciliogenesis regulation in M. lateralis. In addition, CRISPR/Cas9 system has been widely used as a powerful gene editing tool in agriculture and animal breeding, so the CRISPR/Cas9 system we established in M. lateralis for mediating gene knockout may be beneficial for trait improvement in marine bivalves.
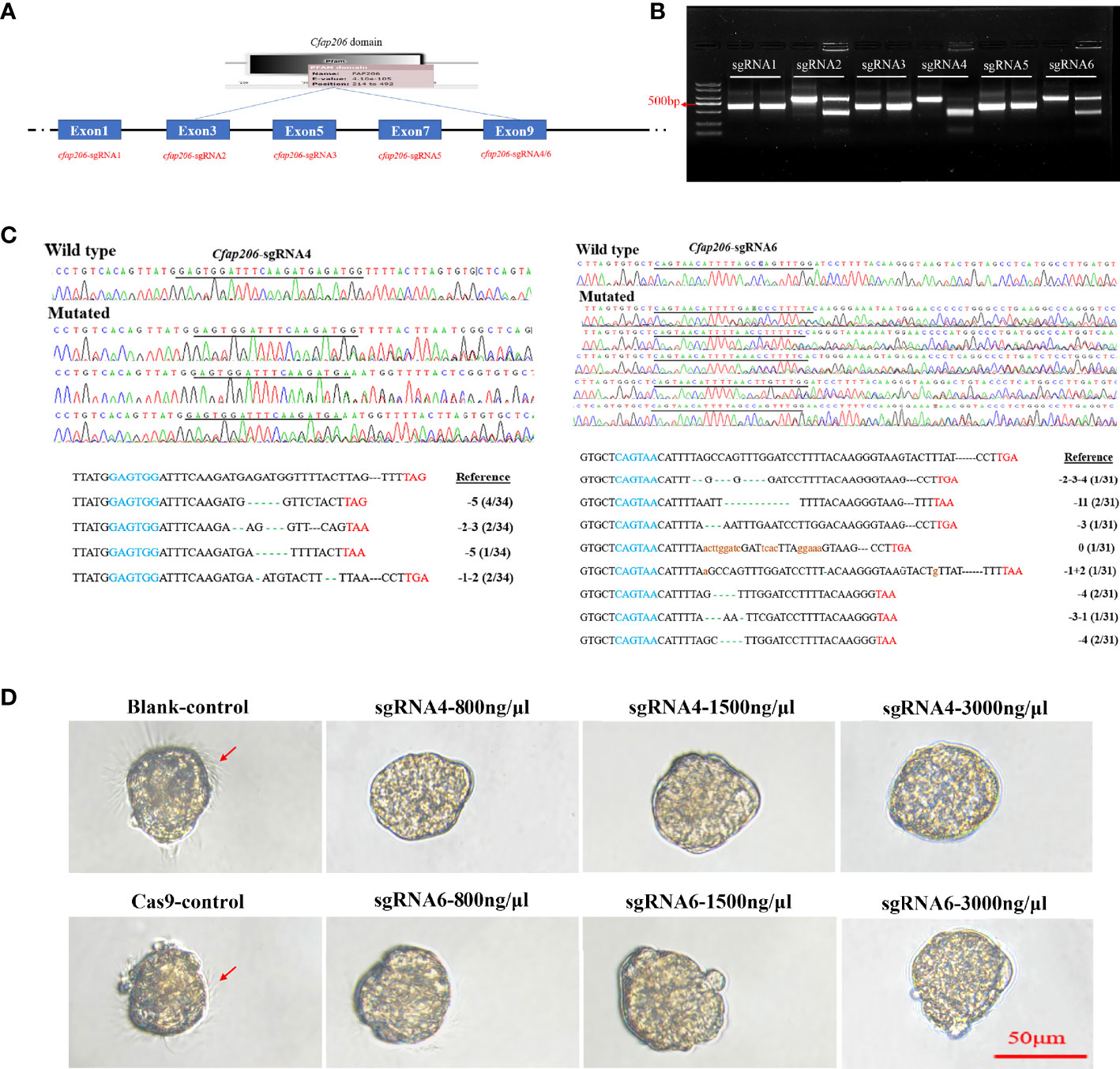
Figure 4 In vitro and in vivo efficiency detection of Cfap206-sgRNAs. (A) sgRNA screening for MlCfap206. (B) In vitro cutting efficiency detection of sgRNA1-6. For each sgRNA, the left lane was the control group and the right lane was the experimental group. (C) Sanger sequencing of PCR products from injected embryos and the identification of mutations. In the left and right bottom panels, the blue sequence marks the start sequence of Cfap206-sgRNA and the red sequence is the stop codon. (D) Light microscope observation of trochophore larvae after Cfap206 knockout.
dsRNA-Expressing Bacteria Feeding Interfered With Adult Ciliogenesis
To further explore whether or not Cfap206 also participates in the male gametogenesis, we managed to deliver the dsRNA of Cfap206 to male individuals for 14 days feeding during the gonadal maturation period. We successfully induced Cfap206-dsRNA expression in competent HT115 E. coli cells with Cfap206-dsRNA-L4440 vector (Figure 5A). After feeding Cfap206-dsRNA-L4440 bacteria, the expression level of Cfap206 in each group was detected on days 7 and 14. Cfap206 was markedly suppressed (~70%) in male gonad compared with that of the control group, and no obvious expression change could be detected after L4440-blank feeding (Figure 5B). At each check point, the survival rate and body size were roughly equal between different groups, suggesting negligible side effects of this experimental approach. Thereafter, sexually mature individuals from each group were chosen to perform the sperm viability investigation (Table 6; Supplementary Videos 1–3). According to the sperm motility, the sperm viability could be rated as A–D four grades, respectively as fast forward movement (A), slow forward movement (B), nonforward movement (C), and extremely slow or inactive movement (D). The results showed that more than 80% of sperms from the control group or L4440-blank group could be rated as grade A/B, while more than 90% of sperms from the Cfap206-dsRNA-L4440 group was rated as grade D, demonstrating the severe defect of sperm motility after significant suppression of Cfap206 (Table 6). Further morphological observation showed that although the head of sperm from the Cfap206-dsRNA-L4440 group was still normal shaped, the sperm tail appeared to be disintegrated (Figure 5C), which could be the obvious cause for their abolished motility. Similarly, it was found that sperm cells with short flagella or without flagella were significantly increased in Cfap206-KO mice compared with wild-type mice (Shen et al., 2021). Previous studies have shown that the sperm flagellum without Cfap206 showed severe axoneme disorganization and peri-axonemal defects, resulting in abnormal sperm morphology and decreased motility (Shen et al., 2021). This is because Cfap206 is closely related to the assembly of the radial spoke RS2 in the axoneme, and the absence of Cfap206 would result in the radial spoke RS2 either not being assembled or being assembled without the front fork (Shen et al., 2021). Cfap206 is also a condition for the stable assembly of CSC elements, and CSC connects three major axonemal complexes involved in the dynein regulation of RS2 (Vasudevan et al., 2015). Taken together, our findings showed that Cfap206 plays critical roles during spermatogenesis, and its participation in the sperm flagella assembly is presented not only in vertebrates but also in marine bivalves. It is worth mentioning that the sexual system of bivalves is diversified, including hermaphrodite, simultaneous hermaphrodite, and sequential hermaphrodite (Collin, 2013). Before the gametes are expelled from the body, hermaphrodite species cannot determine the sex of their gametes. Considering the important role of Cfap206 in spermatogenesis, it may be possible to use gene editing technology to knockout Cfap206 gene to obtain male sterile population, to effectively improve the crossbreeding efficiency of hermaphrodite bivalves.
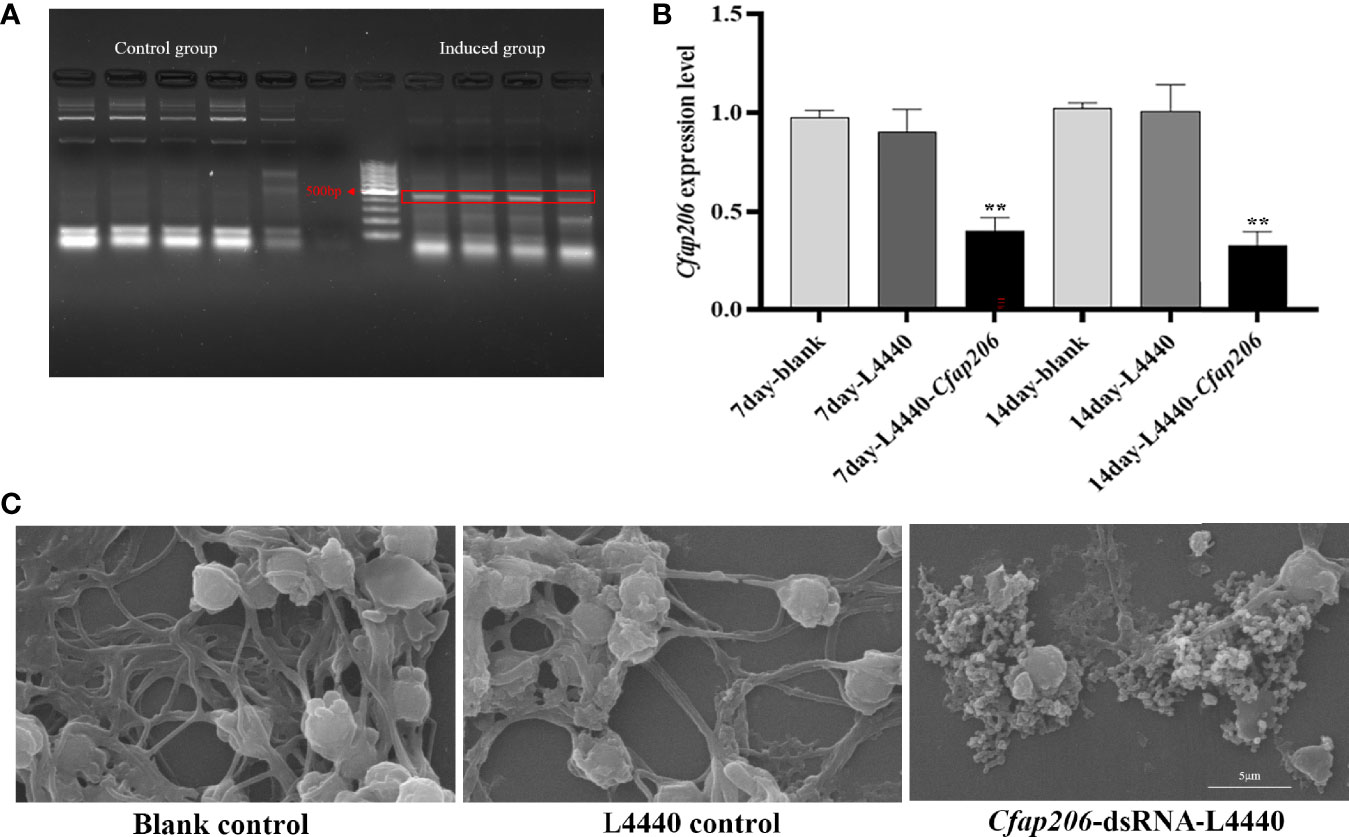
Figure 5 Expression analysis of Cfap206 and morphological observation of sperm after RNAi. (A) Electrophoresis results of dsRNA induced by L4440-Cfap206. Lanes 1–5, control group; lanes 8–11, induced group; and lane 7, 100 bp DNA ladder. (B) The expression of Cfap206 on days 7 and 14. **p < 0.01. (C) Sperm morphology observation using SEM.
Conclusion
In the present study, we explore the functional role of Cfap206 during embryonic ciliogenesis as well as during male gametogenesis in a promising bivalve model M. lateralis. Through RNAi and CRISPR/Cas9 technologies, MlCfap206 was effectively knocked down or knocked out in trochophore larvae, and an obvious decreased number of cilia were detected. Through the dsRNA carrier vector feeding, significant suppression of Cfap206 was observed in the male gonad, together with impaired sperm motility and aberrant sperm tail assembly. Our results suggest that Cfap206 is critical for the formation and functionalization of both embryonic cilia and sperm flagellum in M. lateralis, which may facilitate a better understanding of the ciliogenesis in marine invertebrates. Crossbreeding is an effective way to improve the resistance and other production performance of bivalves in shellfish breeding. However, it is quite challenging to obtain pure gametes for hermaphroditic shellfish, making it difficult to perform crossbreeding. Gene editing technology on target genes (such as Cfap206 in the present study) would provide a feasible scheme for producing male sterile population to improve the crossbreeding efficiency of hermaphrodite species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SL, SW, and ZB conceived and designed the study. YW and XZ performed the experiments. YL and NH participated in data analysis. YW, SL, and XH wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
We acknowledge the grant support from the National Natural Science Foundation of China (32130107, 31871499), Major Basic Research Projects of Shandong Natural Science Foundation (ZR2018ZA0748), Project of Sanya Yazhouwan Science and Technology City Management Foundation (SKJC-KJ-2019KY01), and Taishan Scholar Project Fund of Shandong Province of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.864037/full#supplementary-material
Supplementary Video 1 | Sperm motility of control group.
Supplementary Video 2 | Sperm motility of L4440-blank group.
Supplementary Video 3 | Sperm motility of Cfap206-dsRNA-L4440 group.
References
Afzelius B., Eliasson R. (1983). Male and Female Infertility Problems in the Immotile-Cilia Syndrome. Eur. J. Respir. Dis. 127, 144–147.
Alten L., Schuster-Gossler K., Beckers A., Groos S., Ulmer B., Hegermann J., et al. (2012). Differential Regulation of Node Formation, Nodal Ciliogenesis and Cilia Positioning by Noto and Foxj1. Development 139, 1276–1284. doi: 10.1242/dev.072728
Andrade G. R., De Araujo J. L., Nakamura Filho A., Guanabens A. C., Carvalho M. D., Cardoso A. V. (2015). Functional Surface of the Golden Mussel’s Foot: Morphology, Structures and the Role of Cilia on Underwater Adhesion. Mater. Sci. Eng. C Mater. Biol. Appl. 54, 32–42. doi: 10.1016/j.msec.2015.04.032
Arenas-Mena C. (2010). Indirect Development, Transdifferentiation and the Macroregulatory Evolution of Metazoans. Philos. Trans. R Soc. Lond. B Biol. Sci. 365, 653–669. doi: 10.1098/rstb.2009.0253
Bangs F., Anderson K. V. (2017). Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Perspect. Biol. 9 (5), a02817. doi: 10.1101/cshperspect.a028175
Beckers A., Adis C., Schuster-Gossler K., Tveriakhina L., Ott T., Fuhl F., et al. (2020). The FOXJ1 Target Cfap206 Is Required for Sperm Motility, Mucociliary Clearance of the Airways and Brain Development. Development 147 (21), dev188052. doi: 10.1242/dev.188052
Calabrese A. (1970). Reproductive Cycle of the Coot Clam, Mulinia Lateralis (Say), in Long Island Sound. Veliger -Berkeley- 12, 265–269.
Collin R. (2013). Phylogenetic Patterns and Phenotypic Plasticity of Molluscan Sexual Systems. Integr. Comp. Biol. 53, 723–735. doi: 10.1093/icb/ict076
Danwei H., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003). Hedgehog Signalling in the Mouse Requires Intraflagellar Transport Proteins. Nature 426, 83–87. doi: 10.1038/nature02061
Dong F. N., Amiri-Yekta A., Martinez G., Saut A., Tek J., Stouvenel L., et al. (2018). Absence of CFAP69 Causes Male Infertility Due to Multiple Morphological Abnormalities of the Flagella in Human and Mouse. Am. J. Hum. Genet. 102, 636–648. doi: 10.1016/j.ajhg.2018.03.007
Dubruille R., Laurencon A., Vandaele C., Shishido E., Coulon-Bublex M., Swoboda P., et al. (2002). Drosophila Regulatory Factor X Is Necessary for Ciliated Sensory Neuron Differentiation. Development 129, 5487–5498. doi: 10.1242/dev.00148
Elliott K. H., Brugmann S. A. (2019). Sending Mixed Signals: Cilia-Dependent Signaling During Development and Disease. Dev. Biol. 447, 28–41. doi: 10.1016/j.ydbio.2018.03.007
Feng D., Li Q., Yu H. (2019). RNA Interference by Ingested dsRNA-Expressing Bacteria to Study Shell Biosynthesis and Pigmentation in Crassostrea gigas. Mar. Biotechnol. (N. Y.) 21, 526–536. doi: 10.1007/s10126-019-09900-2
He X., Liu C., Yang X., Lv M., Ni X., Li Q., et al. (2020). Bi-Allelic Loss-Of-Function Variants in CFAP58 Cause Flagellar Axoneme and Mitochondrial Sheath Defects and Asthenoteratozoospermia in Humans and Mice. Am. J. Hum. Genet. 107, 514–526. doi: 10.1016/j.ajhg.2020.07.010
Heuser T., Dymek E. E., Lin J., Smith E. F., Nicastro D. (2012). The CSC Connects Three Major Axonemal Complexes Involved in Dynein Regulation. Mol. Biol. Cell 23, 3143–3155. doi: 10.1091/mbc.e12-05-0357
Hiruta C., Kakui K., Tollefsen K. E., Iguchi T. (2018). Targeted Gene Disruption by Use of CRISPR/Cas9 Ribonucleoprotein Complexes in the Water Flea Daphnia pulex. Genes Cells 23, 494–502. doi: 10.1111/gtc.12589
Hou R., Bao Z., Wang S., Su H., Li Y., Du H., et al. (2011). Transcriptome Sequencing and De Novo Analysis for Yesso Scallop (Patinopecten yessoensis) Using 454 GS FLX. PloS One 6, e21560. doi: 10.1371/journal.pone.0021560
Hua K., Ferland R. J. (2018). Primary Cilia Proteins: Ciliary and Extraciliary Sites and Functions. Cell Mol. Life Sci. 75, 1521–1540. doi: 10.1007/s00018-017-2740-5
Hu X., Bao Z., Hu J., Shao M., Zhang L., Bi K., et al. (2006). Cloning and Characterization of Tryptophan 2,3-Dioxygenase Gene of Zhikong Scallop Chlamys farreri (Jones and Preston 1904). Aquaculture Res. 37, 1187–1194. doi: 10.1111/j.1365-2109.2006.01546.x
Jakobsen H. H., Everett L. M., Strom S. L. (2006). Hydromechanical Signaling Between the Ciliate Mesodinium Pulex and Motile Protist Prey. Aquat. Microb. Ecol. 44, 197–206. doi: 10.3354/ame044197
Larkin M. A., Blackshields G., Brown N. P., Chenna R., Mcgettigan P. A., Mcwilliam H., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lin J., Heuser T., Carbajal-Gonzalez B. I., Song K., Nicastro D. (2012). The Structural Heterogeneity of Radial Spokes in Cilia and Flagella Is Conserved. Cytoskeleton (Hoboken) 69, 88–100. doi: 10.1002/cm.21000
Lin C. Y., Su Y. H. (2016). Genome Editing in Sea Urchin Embryos by Using a CRISPR/Cas9 System. Dev. Biol. 409, 420–428. doi: 10.1016/j.ydbio.2015.11.018
Li W., Wu H., Li F., Tian S., Kherraf Z. E., Zhang J., et al. (2020). Biallelic Mutations in CFAP65 Cause Male Infertility With Multiple Morphological Abnormalities of the Sperm Flagella in Humans and Mice. J. Med. Genet. 57, 89–95. doi: 10.1136/jmedgenet-2019-106344
Li H., Yu H., Du S., Li Q. (2021). CRISPR/Cas9 Mediated High Efficiency Knockout of Myosin Essential Light Chain Gene in the Pacific Oyster (Crassostrea gigas). Mar. Biotechnol. (N. Y.) 23, 215–224. doi: 10.1007/s10126-020-10016-1
Lu J.-K., Chen T. T., Allent S. K., Matsubara T., Burns J. C. (1996). Production of Transgenic Dwarf Surfclams, Mulinia lateralis, With Pantropic Retroviral Vectors. Proc. Natl. Acad. Sci. U. S. A. 93, 3482–3486. doi: 10.1073/pnas.93.8.3482
Lyons R. A., Saridogan E., Djahanbakhch O. (2006). The Reproductive Significance of Human Fallopian Tube Cilia. Hum. Reprod. Update 12, 363–372. doi: 10.1093/humupd/dml012
Nicholas K. B. (1997). Genedoc: Analysis and Visualization of Genetic Variation, EMBNEW. Embnew. News 4, 14. doi: 10.11118/actaun201361041061
Orhon I., Dupont N., Pampliega O., Cuervo A. M., Codogno P. (2015). Autophagy and Regulation of Cilia Function and Assembly. Cell Death Differ. 22, 389–397. doi: 10.1038/cdd.2014.171
Paps J., Xu F., Zhang G., Holland P. W. (2015). Reinforcing the Egg-Timer: Recruitment of Novel Lophotrochozoa Homeobox Genes to Early and Late Development in the Pacific Oyster. Genome Biol. Evol. 7, 677–688. doi: 10.1093/gbe/evv018
Pazour G. J., Rosenbaum J. L. (2002). Intraflagellar Transport and Cilia-Dependent Diseases. Trends Cell Biol. 12, 551–555. doi: 10.1016/S0962-8924(02)02410-8
Pazour G. J., Witman G. B. (2003). The Vertebrate Primary Cilium Is a Sensory Organelle. Curr. Opin. Cell Biol. 15, 105–110. doi: 10.1016/S0955-0674(02)00012-1
Ross A. J., May-Simera H., Eichers E. R., Kai M., Hill J., Jagger D. J., et al. (2005). Disruption of Bardet-Biedl Syndrome Ciliary Proteins Perturbs Planar Cell Polarity in Vertebrates. Nat. Genet. 37, 1135–1140. doi: 10.1038/ng1644
Santos S. L., Simon R. J. L. (1980). Response of Soft-Bottom Benthos to Annual Catastrophic Disturbance in a South Florida Estuary. Marine Ecol. Prog. 3, 347–355. doi: 10.3354/meps003347
Satir P., Christensen S. T. (2007). Overview of Structure and Function of Mammalian Cilia. Annu. Rev. Physiol. 69, 377–400. doi: 10.1146/annurev.physiol.69.040705.141236
Shen Q., Martinez G., Liu H., Beurois J., Wu H., Amiri-Yekta A., et al. (2021). Bi-Allelic Truncating Variants in CFAP206 Cause Male Infertility in Human and Mouse. Hum. Genet. 140, 1367–1377. doi: 10.1007/s00439-021-02313-z
Shimazaki A., Sakai A., Ogasawara M. (2006). Gene Expression Profiles in Ciona intestinalis Stigmatal Cells: Insight Into Formation of the Ascidian Branchial Fissures. Dev. Dyn. 235, 562–569. doi: 10.1002/dvdy.20657
Sironen A., Shoemark A., Patel M., Loebinger M. R., Mitchison H. M. (2020). Sperm Defects in Primary Ciliary Dyskinesia and Related Causes of Male Infertility. Cell Mol. Life Sci. 77, 2029–2048. doi: 10.1007/s00018-019-03389-7
Stubbs J. L., Oishi I., Izpisua Belmonte J. C., Kintner C. (2008). The Forkhead Protein Foxj1 Specifies Node-Like Cilia in Xenopus and Zebrafish Embryos. Nat. Genet. 40, 1454–1460. doi: 10.1038/ng.267
Tang S., Wang X., Li W., Yang X., Li Z., Liu W., et al. (2017). Biallelic Mutations in CFAP43 and CFAP44 Cause Male Infertility With Multiple Morphological Abnormalities of the Sperm Flagella. Am. J. Hum. Genet. 100, 854–864. doi: 10.1016/j.ajhg.2017.04.012
Tao F., Jiang T., Tao H., Cao H., Xiang W. (2020). Primary Cilia: Versatile Regulator in Cartilage Development. Cell Prolif. 53, e12765. doi: 10.1111/cpr.12765
Thomas J., Morle L., Soulavie F., Laurencon A., Sagnol S., Durand B. (2010). Transcriptional Control of Genes Involved in Ciliogenesis: A First Step in Making Cilia. Biol. Cell 102, 499–513. doi: 10.1042/BC20100035
Toure A., Martinez G., Kherraf Z. E., Cazin C., Beurois J., Arnoult C., et al. (2021). The Genetic Architecture of Morphological Abnormalities of the Sperm Tail. Hum. Genet. 140, 21–42. doi: 10.1007/s00439-020-02113-x
Vasudevan K. K., Song K., Alford L. M., Sale W. S., Dymek E. E., Smith E. F., et al. (2015). FAP206 Is a Microtubule-Docking Adapter for Ciliary Radial Spoke 2 and Dynein C. Mol. Biol. Cell 26, 696–710. doi: 10.1091/mbc.E14-11-1506
Verhey K. J., Dishinger J., Kee H. L. (2011). Kinesin Motors and Primary Cilia. Biochem. Soc. Trans. 39, 1120–1125. doi: 10.1042/BST0391120
Vij S., Rink J. C., Ho H. K., Babu D., Eitel M., Narasimhan V., et al. (2012). Evolutionarily Ancient Association of the Foxj1 Transcription Factor With the Motile Ciliogenic Program. PloS Genet. 8, e1003019. doi: 10.1371/journal.pgen.1003019
Walker R. L., Tenore K. R. (1984). Growth and Production of the Dwarf Surf Clam Mulinia lateralis (Say 1822) in a Georgia Estuary. Gulf Res. Rep. 7, 357–363. doi: 10.18785/grr.0704.07
Wang X., Liu B., Liu F., Huan P. (2015). A Calaxin Gene in the Pacific Oyster Crassostrea gigas and Its Potential Roles in Cilia. Zoology. Sci. 32, 419–426. doi: 10.2108/zs150009
Wang J., Zhang L., Lian S., Qin Z., Zhu X., Dai X., et al. (2020). Evolutionary Transcriptomics of Metazoan Biphasic Life Cycle Supports a Single Intercalation Origin of Metazoan Larvae. Nat. Ecol. Evol. 4, 725–736. doi: 10.1038/s41559-020-1138-1
Ward J. E., Macdonald L. B. A., Thompson R. J. (1993). Mechanisms of Suspension Feeding in Bivalves Resolution of Current Controversies by Means of Endoscopy. Limnol. Oceanography 38, 466. doi: 10.4319/lo.1993.38.2.0265
Wei H., Li W., Liu T., Li Y., Liu L., Shu Y., et al. (2021). Sexual Development of the Hermaphroditic Scallop Argopecten irradians Revealed by Morphological, Endocrine and Molecular Analysis. Front. Cell Dev. Biol. 9, 646754. doi: 10.3389/fcell.2021.646754
Keywords: Mulinia lateralis, Cfap206, ciliogenesis, RNAi, CRISPR/Cas9, breeding
Citation: Wang Y, Zhu X, Lian S, Li Y, Hu N, Hu X, Bao Z and Wang S (2022) Functional Characterization of Cfap206 for Bivalve Ciliogenesis by RNAi and CRISPR/Cas9 Technologies. Front. Mar. Sci. 9:864037. doi: 10.3389/fmars.2022.864037
Received: 28 January 2022; Accepted: 01 March 2022;
Published: 24 March 2022.
Edited by:
Yuehuan Zhang, South China Sea Institute of Oceanology (CAS), ChinaReviewed by:
Xiaotong Wang, Ludong University, ChinaErnesto Maldonado, National Autonomous University of Mexico, Mexico
Copyright © 2022 Wang, Zhu, Lian, Li, Hu, Hu, Bao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shanshan Lian, bGlhbnNoYW5zaGFuQG91Yy5lZHUuY24=
†These authors share first authorship
 Yinghui Wang
Yinghui Wang Xiaomei Zhu1†
Xiaomei Zhu1† Shanshan Lian
Shanshan Lian Xiaoli Hu
Xiaoli Hu Zhenmin Bao
Zhenmin Bao Shi Wang
Shi Wang