
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 04 July 2022
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.863310
This article is part of the Research Topic Omics-based identification, characterization, and validation of marine bioactive peptides View all 6 articles
 Yu Huang1†
Yu Huang1† Xiyang Chen1†
Xiyang Chen1† Haoyue Shu2
Haoyue Shu2 Panpan Xiao1
Panpan Xiao1 Xueqiang Lin1
Xueqiang Lin1 Junmin Xu1
Junmin Xu1 Chao Bian1,3
Chao Bian1,3 Xinxin You1,3†
Xinxin You1,3† Jian Yang2*
Jian Yang2* Qiong Shi1,3,4*
Qiong Shi1,3,4*Recently, the prevalence of hypertension has become a global challenge. Therefore, tremendous efforts have been made to identify and purify antihypertensive peptides (AHTPs) from food-derived proteins to aid the discovery of new hypotensive drugs with fewer side effects. In this study, we performed high-throughput prediction of AHTPs based on multi-omics data, providing an overview of AHTPs in the lined seahorse proteins and suggesting their potential application as bioactive agents to lower blood pressure. We identified 14,695 AHTP-derived genes in the lined seahorse, and most of them were supported by transcriptomic evidence, whereas only 495 genes were further detected by proteome sequencing. Among these predicted AHTP-derived genes, the longest titin had the most hits with 104 AHTPs, some of which were clustered in exon 158, 194, and 204. Another AHTP-rich group was in the collagen family, and four AHTP-rich collagens exhibited much higher transcription in the pouch than in other examined tissues, including brain, testis, and embryos. Additionally, antihypertensive triplets, comprised of the permutations of Gly, Pro, and Lys, prevailed in all collagen sequences due to the representative XaaYaaGly repeat units. In summary, our present findings provide a solid basis for understanding the abundance of various AHTPs in the lined seahorse as well as shed light on the development of antihypertensive products and drugs using seahorses as an important resource.
Seahorses (Hippocampinae), a group of small marine fishes, are famous for their unique body plans (Porter et al., 2013) and male pregnancy (Lin et al., 2016). The lined seahorse (Hippocampus erectus) is an Atlantic species with a main residence in shallow sea beds or coral reefs. The lined seahorse is listed as “vulnerable (VU)” in the International Union for Conservation of Nature (IUCN) assessment (Cardoso et al., 2016), it has been cultured recently for species conservation and commercial significance. Many studies have reported that ethanolic extracts or dried powders of the lined seahorse have anti-fatigue (Kang et al., 2017), antimicrobial (Chen et al., 2020), antioxidant (Ryu et al., 2010; Kim et al., 2016), and immune-modulating (Zhang et al., 2017) activities.
Hypertension (high blood pressure) is a serious physiological condition that increases the risk of heart, brain, and other diseases (Meissner, 2016). The worldwide prevalence of hypertension has become a global challenge, and a dramatic rise in young people makes the situation much worse (Battistoni et al., 2015). Widely used synthetic drugs are often associated with side effects, triggering the development of alternative therapies using natural products, especially food-derived antihypertensive peptides (AHTPs; Abachi et al., 2019; Abdelhedi and Nasri, 2019). AHTPs, which usually consist of 2–15 amino acids, are predicted to be easily absorbed but with fewer undesirable side effects (Abdelhedi and Nasri, 2019). AHTPs can exert an antihypertensive activity mainly through renin and/or angiotensin-converting enzyme (ACE) inhibitor routes, but also employ other ways such as angiotensin II (ATII) receptor blocking, Ca2+ channel blocking, and antioxidant pathways (Majumder and Wu, 2014; Udenigwe and Mohan, 2014; Manzanares et al., 2015).
The first AHTP was purified from snake venom in the early 1970s (Ferreira et al., 1970; Ondetti et al., 1971). Afterwards, a great number of studies have focused on AHTP identification from various plants (Rosales-Mendoza et al., 2013; Aluko, 2015) and animals (Bhat et al., 2017; Lee and Hur, 2017), where marine organisms represent an attractive resource of AHTPs with high activity (Kim et al., 2012; Lee and Hur, 2017; Abdelhedi and Nasri, 2019). For example, an AHTP with strong ACE-inhibitory activity has been purified from rainbow trout (O. mykiss), exhibiting an IC50 of 0.0036 mg/ml (Ketnawa et al., 2019). Salmon (Salmo salar) protein hydrolysates, digested with a mixture of enzymes, exhibit an obvious blood pressure-lowering effect in spontaneously hypertensive rats (Girgih et al., 2016). ACE and renin-inhibitory peptides have been extracted from Atlantic cod (Godus morhua) muscle, showing antihypertensive activity both in vitro and in vivo (Girgih et al., 2015). A recent review has summarized and listed more than 20 marine species, among which numerous AHTPs have been identified (Abdelhedi and Nasri, 2019).
Seahorses are a good material for AHTP isolation as well. In 2015, hydrolysates of three-spot seahorse (H. trimaculatus) proteins, digested with alkaline protease, exerted strong ACE-inhibitory activity (Xu et al., 2015). Recently, another ACE-inhibiting peptide composed of 8 amino acids (PAGPRGPA) was purified from the same species, showing an IC50 of 7.90 µM (Shi et al., 2020). Meanwhile, another team reported that low-molecular weight peptides isolated from big-belly seahorse (H. abdominalis) can improve vasodilation via inhibition of ACE in vivo and in vitro (Je et al., 2020), and the hydrolysate derived from this species has already been proven with antihypertensive potential (Kim et al., 2016).
Although a few attempts have been made to isolate highly active AHTPs from seahorses, the knowledge about seahorse-derived AHTPs is still limited due to a lack of sequence information and a shortage of practical purification methods. Additionally, few studies have focused on the discovery of seahorse proteins that can be hydrolyzed into large amounts of AHTPs. In this study, with the availability of whole genome (Lin et al., 2017), transcriptome (Lin et al., 2016), and proteome (Chen et al., 2020) sequences of the lined seahorse, we can predict AHTPs from the previously published gene sets, and more importantly, to provide guidance on efficient purification of AHTPs from seahorse proteins. We aimed to provide an in-depth exploration of seahorse-derived AHTPs for the potential development of antihypertensive products and drugs.
The genome, transcriptome, and proteome datasets were cited from previous studies (Lin et al., 2016; Lin et al., 2017; Chen et al., 2020). GigaDB (PRJNA347499) was used to download the genome assembly, gene set, and annotation. Transcriptome raw data (SRA392578) were downloaded from the National Center for Biotechnology Information (NCBI). An in-house AHTP database was retrieved from previous reports (Yi et al., 2018; Chen et al., 2020).
For the transcriptome analysis, there were 19 samples, including three embryos (1-day, 3-day, or 10-day post-fertilization), one juvenile body (1-day post-birth), and 15 samples of different tissues (including brain, testis, and brood pouch) from multiple developing stages (i.e., juvenile, rudimentary, pre-pregnancy, pregnancy, and post-pregnancy). For the proteome analysis, the abdominal tissues (both skin and muscle) from the female and male individuals were used.
For transcriptome analysis, the downloaded raw reads were filtered by SOAPnuke (version 1.5.6; Chen et al., 2018), and then mapped to the reference genome assembly by HISAT2 (version 2.0.4; Kim et al., 2019a). Subsequently, we used RSEM (version 1.2.12; Allen et al., 2015) to calculate the transcripts per million (TPM) values of target genes to qualify their transcription levels. Low-expressed transcripts with TPM <1 were discarded before performing any comparison analysis. A heatmap was plotted using the heatmap.2 function in the gplots R package (R core Team, 2021).
For mass spectrometry (MS)-based proteome analysis, both data-independent acquisition (DIA) and data-dependent acquisition (DDA) methods were used. A DDA-based spectrum library was first established using mixed female and male abdomen homogenates (Chen et al., 2020). DIA data were then analyzed by Spectronaut 12 (Bruderer et al., 2015) based on this library. Differentially expressed proteins (DEPs) between female and male-lined seahorses were identified using the MSstats R package (Choi et al., 2014). To obtain non-redundant peptide sequences, we removed duplicates and short peptides that were covered by long ones.
Functional annotation of the gene sets was performed with BLAST searches against public databases including Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa, 2000), swiss-prot, and TrEmbl (Boeckmann et al., 2003). Gene Ontology (GO) enrichment analysis was conducted using the topGO R package (Alexa et al., 2006) with an adjusted p-value of 0.05 as the threshold.
An in-house database with 963 AHTPs was established on the basis of both the public AHTPDB (Kumar et al., 2015) and sequences from public literature (Yi et al., 2018). To identify potential AHTPs in fish explicitly and efficiently, we chose the top 50 AHTPs using the following criteria: First, the top 20 sequences with the highest inhibitory activity were selected, no matter what species they were from. Second, the top 30 fish-derived AHTPs were included, and they are denoted as “Fish” in the “Source” column of Supplementary Table 1. The short sequences of these selected AHTPs were searched against the genome-deduced proteins, transcriptomic sequences, and non-redundant proteomic peptides of the lined seahorse, respectively. Any gene encoding a protein with at least one of the AHTP sequence(s) was considered an AHTP-derived gene. The number of AHTP hits in each protein was counted. To eliminate the effect of protein length, we also calculated the ratio of total AHTP length in a specific protein to the length of this protein, simplified as “AHTP/protein ratio.” The distribution of AHTPs in a target protein was plotted using the SVG module in Perl.
Previously, we established an in-house fish AHTP database consisting of 963 reliable peptides (Supplementary Table 1; Yi et al., 2018) from the public database AHTPDB (Kumar et al., 2015). In this study, we chose the top 50 AHTP sequences with the highest antihypertensive activity (marked in red in Supplementary Table 1) as queries. To predict AHTP sequences in the lined seahorse, we searched these queries against several omics datasets of this fish, including a genome-based gene set (Lin et al., 2017), 19 transcriptomes (Lin et al., 2016), and two sets of proteomic data (Chen et al., 2020).
Within the genome-based gene set of 22,435 protein-coding genes (Lin et al., 2017), 14,695 (65.50%) of the coded proteins contained 42,347 AHTPs (Table 1), resulting in an average of 2.88 AHTPs per AHTP-containing gene. In the 19 transcriptomes, there were 15,090 transcripts (including 828 novel findings that were not annotated in the genome-based gene set) carrying 44,109 AHTPs, with an average of 2.92 AHTPs per transcript that is comparable to the average number in the genome-based gene set. The total number of AHTPs identified in each transcriptome was similar (around 40,000; Supplementary Table 2). In addition, there were 487 or 248 genes further validated by DDA (Supplementary Table 3) or DIA (Supplementary Tables 4, 5) MS, respectively. In total, 44,776 AHTP hits were identified from 15,523 AHTP-derived genes after integration of multi-omics data followed by removal of redundancies (Table 1).
Among the top 50 AHTPs with the putative highest activity, 32 were identified in the multi-omics data (Figure 1). The frequency distribution of these identified AHTPs shows that Gly-Leu-Pro (GLP), Leu-Gly-Pro (LPG), and Val-Ser-Val (VSV) were the most abundant (Figure 1A). Similarly, GLP, LPG, and VSV were also the major components in the gene sets of various fishes (Yi et al., 2018) and several mammals, including cows, minke whales, and three dolphin species (Jia et al., 2019). The longest AHTP, Leu-Arg-Ile-Pro-Val-Ala (LRIPVA), was only identified in one gene (HE009683, annotated as “dynamin-binding protein”). Presumably, the unidentified 18 AHTP references were too long (more than six amino acids) to be completely matched in the seahorse datasets (highlighted in yellow in Supplementary Table 1).
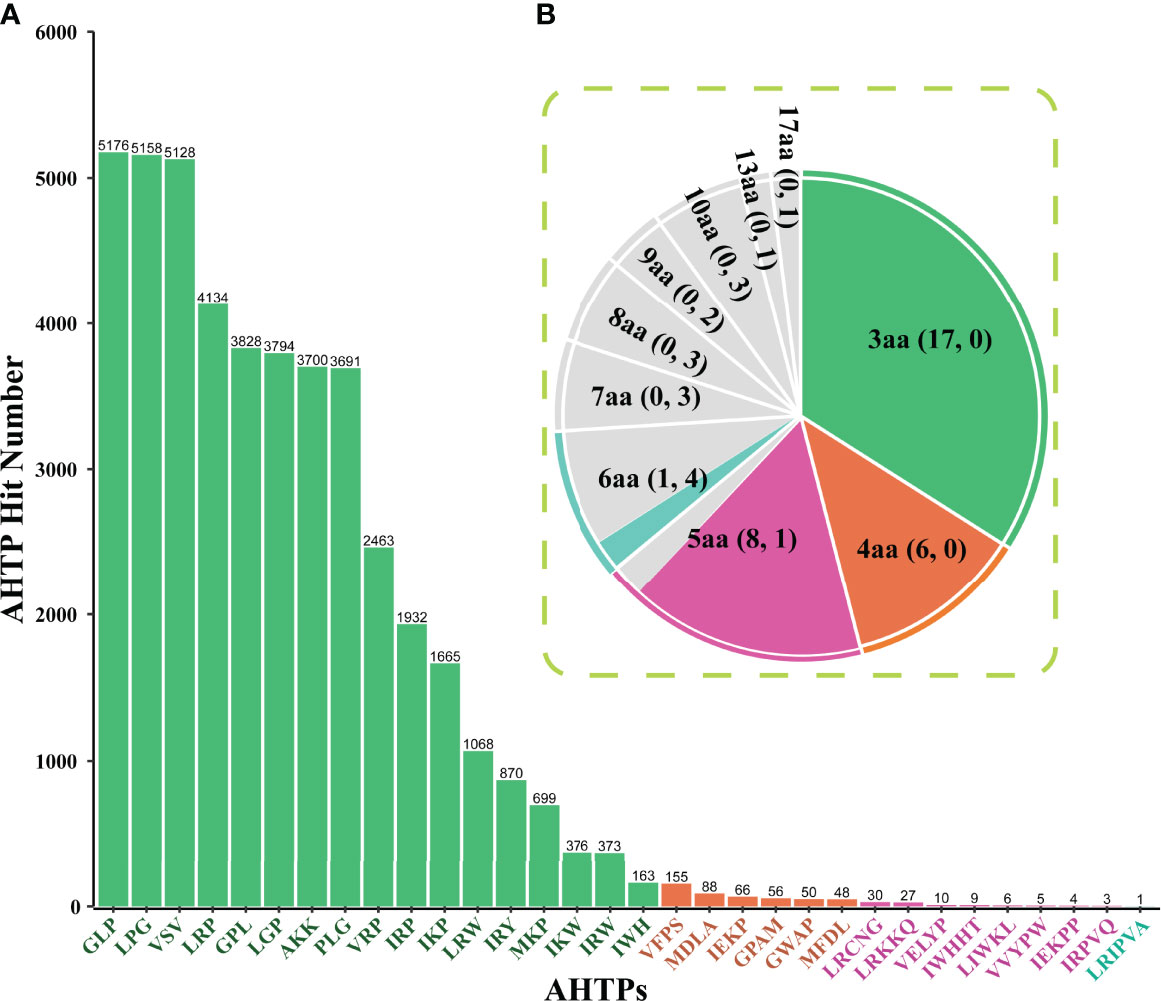
Figure 1 Frequency distribution of AHTPs that were identified from the multi-omics datasets of the lined seahorse. (A) A bar chart for the detailed hit number of each putative AHTP. The x-axis represents the identified 32 AHTPs, and the y-axis stands for the corresponding number of each AHTP. (B) A pie chart for summary of AHTP hits with various length. Green, orange, pink, and aqua refer to AHTP groups consisting of three, four, five, and six amino acids (aa), respectively. Grey means no hits in the lined seahorse genome. The two numbers in every bracket refer to the identified AHTP types and the missing types in each group, respectively.
In order to unravel the potential functions of the 15,523 AHTP-derived genes in lined seahorse, we downloaded functional assignments of the genome-based protein set from a previous work (Lin et al., 2017), and performed a Blast search of the transcriptome-derived novel coding genes against the public RefSeq non-redundant protein database (Acland et al., 2014) and KEGG database (Kanehisa, 2000). Our results show that 93.93% (14,581 out of 15,523) of the AHTP-derived genes were supported by at least one hit in the examined databases (Supplementary Table 6), and these genes were further enriched into 44 functional groups, including “Signal transduction” and “Global and overview maps” (see Supplementary Figure 1 for more details).
AHTP-rich proteins are more likely to be digested into more abundant AHTPs. Here, we defined “AHTP-rich proteins” in two ways. First, they could be proteins simply containing a large number of AHTP sequences. However, longer proteins surely have a higher probability of having more AHTPs, which was supported by a previous report (Yi et al., 2018). To eliminate the protein-length effect, the second way of defining “AHTP-rich proteins” was then to focus on the ratio of the total length of AHTPs in a specific protein to the length of this protein; in other words, the length of total AHTP as a percentage of the protein length (simplified as “AHTP/protein ratio”). Our data demonstrate that each of the above-identified 15,523 AHTP-containing proteins of lined seahorse has 1–104 AHTP(s). If taking the length of AHTPs into consideration, the length of identified AHTPs in each protein ranged from 3 to 315 bp, and the AHTP/protein ratio ranged from 0.09 to 16.98% (Figure 2; Supplementary Table 7).
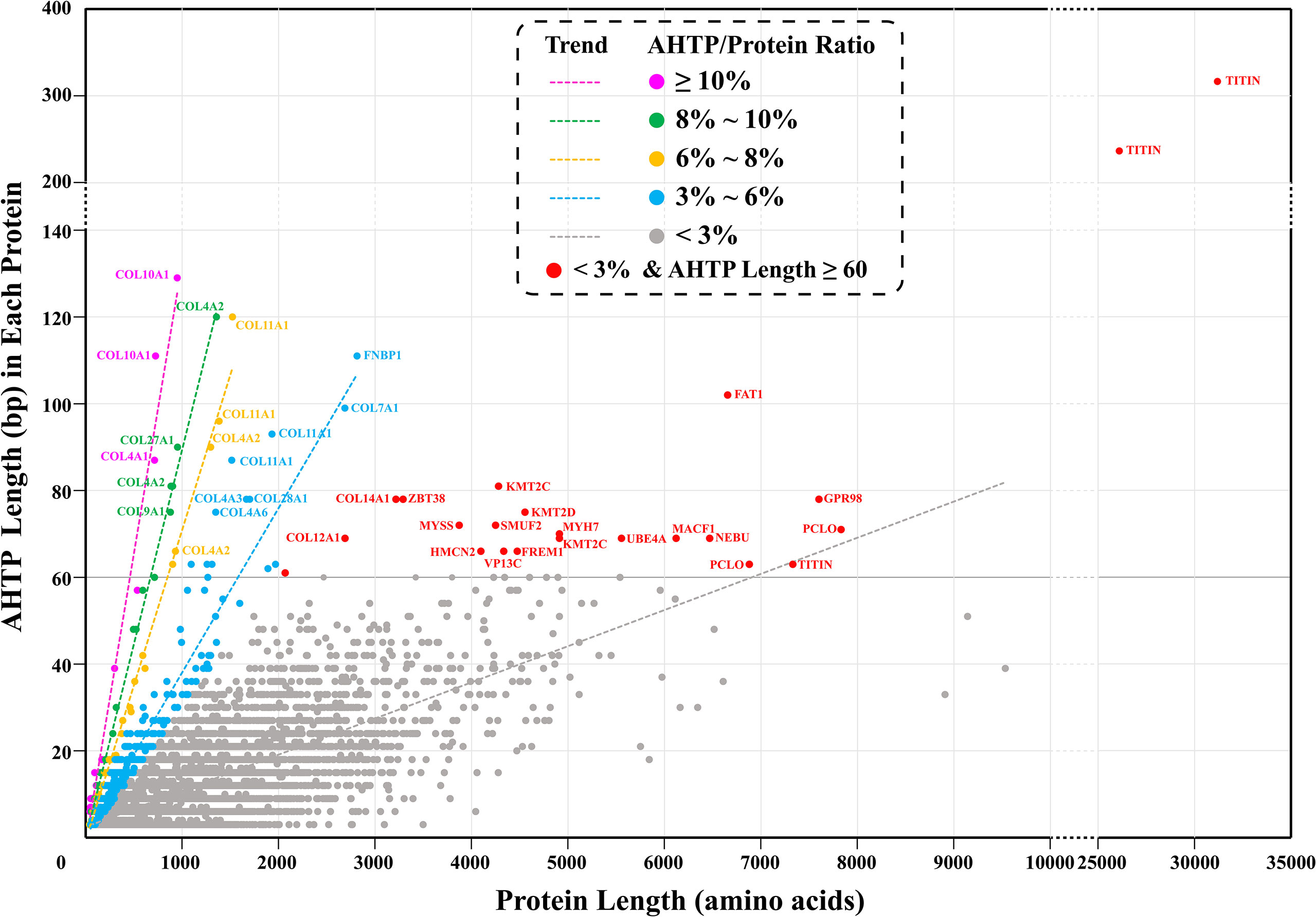
Figure 2 AHTP-rich proteins in the lined seahorse. “AHTP/Protein ratio” means the ratio of the total length of AHTPs in a specific protein to the length of this protein. “Trend” shows the trendline of each group. Each dot represents a protein, and the pink, green, yellow, blue, and gray ones refer to proteins with an AHTP/protein ratio of ≥10, 8–10%, 6–8%, 3–6%, and <3%, respectively. Red dots mark those proteins with an AHTP/protein ratio of less than 3% but containing AHTPs with a total length of more than 60 bp.
The proteins with the largest number of AHTPs were two titin proteins. One (HE002169) contained 104 AHTPs (representing 19 AHTP types; Figure 3), and the other (HE002161) carried 78 AHTPs (belonging to 15 different types; Supplementary Figure 2). Proteins from the collagen family contained the second largest number of AHTPs, but collagens were the biggest group among the AHTP-rich proteins (Figure 4; Supplementary Figure 3). Among the top 49 proteins with the richest AHTPs (>60 bp), 19 of them were collagens, such as COL10A1 (HE002468), COL4A2 (HE013817), and COL11A1 (HE014005), containing 43 (129 bp), 40 (120 bp), and 40 (120 bp) AHTP hits, respectively (Figure 2; Supplementary Table 7). Several other types of collagen (such as COL7A1, COL12A1, COL14A1, and COL19A1) were also identified. Notably, titin and collagen contained the most abundant AHTP hits, which is consistent with previous studies (Yi et al., 2018; Jia et al., 2019).
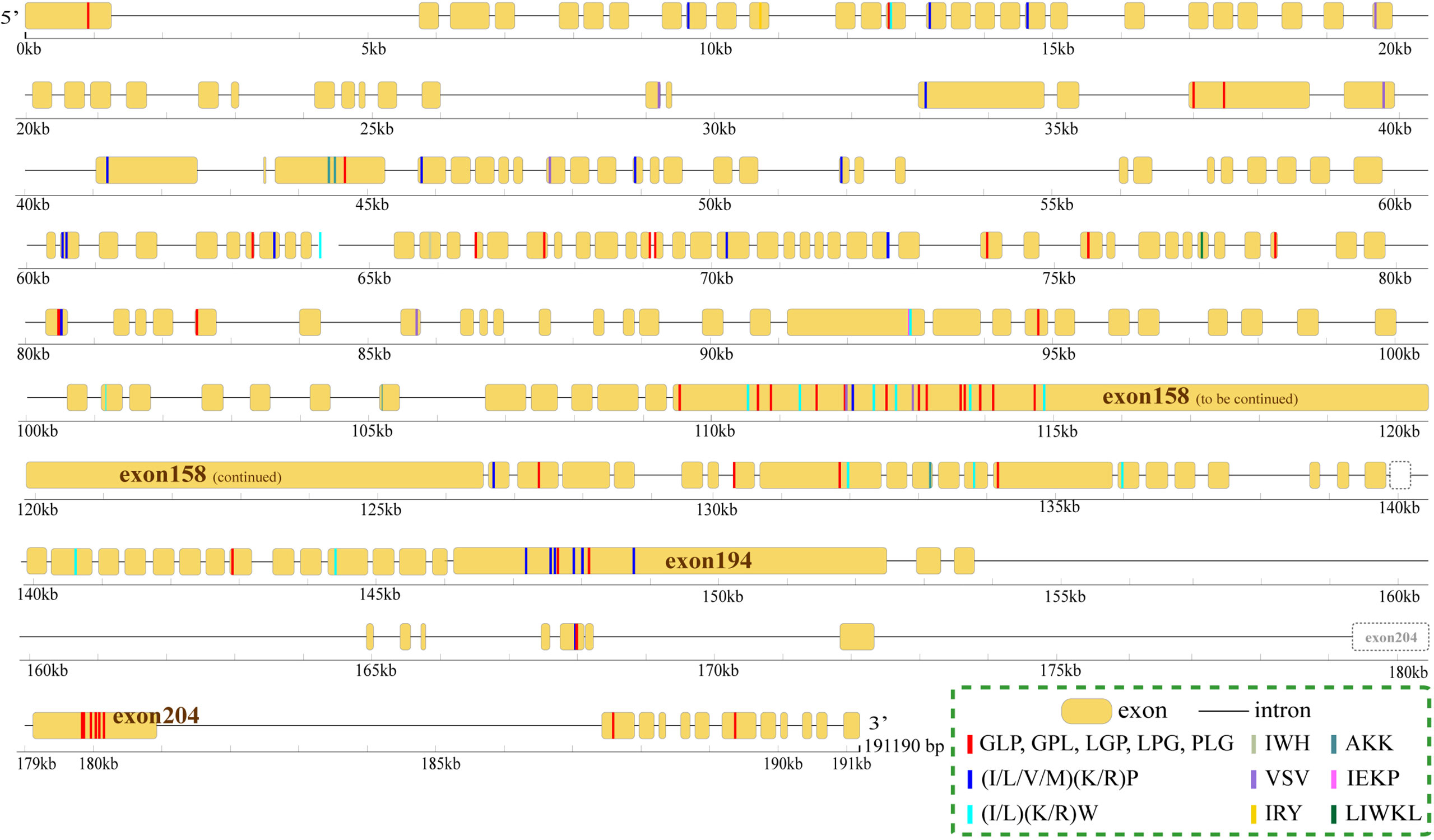
Figure 3 Distribution of AHTPs in the longest titin encoded by the gene HE002169. Yellow bars and the black lines between them represent the exons and introns, respectively. Rulers under them mark their length. The coloured lines inside each exon bar stand for AHTP hits, and each colour represents one AHTP type.

Figure 4 Distribution of AHTPs in collagens. Gray bars indicate the collagen protein sequences. The coloured lines inside each bar stand for AHTP hits, and each color represents one AHTP type. Numbers after each bar refer to the total length of AHTPs in the protein, AHTP/protein ratio, and the number of AHTP hits, respectively. Protein sequences are sorted by the number of AHTP hits (from the highest to the lowest) followed by sequence length (from the shortest to the longest).
In general, longer proteins tend to contain more AHTP sequences and therefore more types of AHTP. However, there were some exceptions. For instance, COL7A1 (HE019378) contained 33 AHTPs, covering 11 AHTP types, while COL10A1 (HE002468 and HE009261) had more AHTPs (43 and 37) but with fewer types (5 and 3, respectively). Another example was COL9A1 (HE020043 and HE020036), carrying 27 AHTPs but representing only two types (Supplementary Table 7). Interestingly, all these collagens with low AHTP diversity were Leu, Pro, and Gly-rich proteins, and peptides with such amino acids have been proven have of high antihypertensive activity (Lee et al., 2014; Jonckheere et al., 2012; Lianliang Liu, 2012).
A specific tissue with highly expressed AHTP-rich genes is usually also a good resource for the isolation of abundant AHTPs. Therefore, we calculated the transcription values of target genes as transcripts per million reads (TPM) in the seahorse brain, pouch, and testis at different developmental stages (Supplementary Table 8; Lin et al., 2016; Chen et al., 2020). After discarding those low transcription genes (TPM <1), our results show that samples from the same tissue rather than from the same developmental stage were clustered together, and the transcription pattern of AHTP-derived genes in one-day embryos was remarkably different from those in other samples (Figure 5).
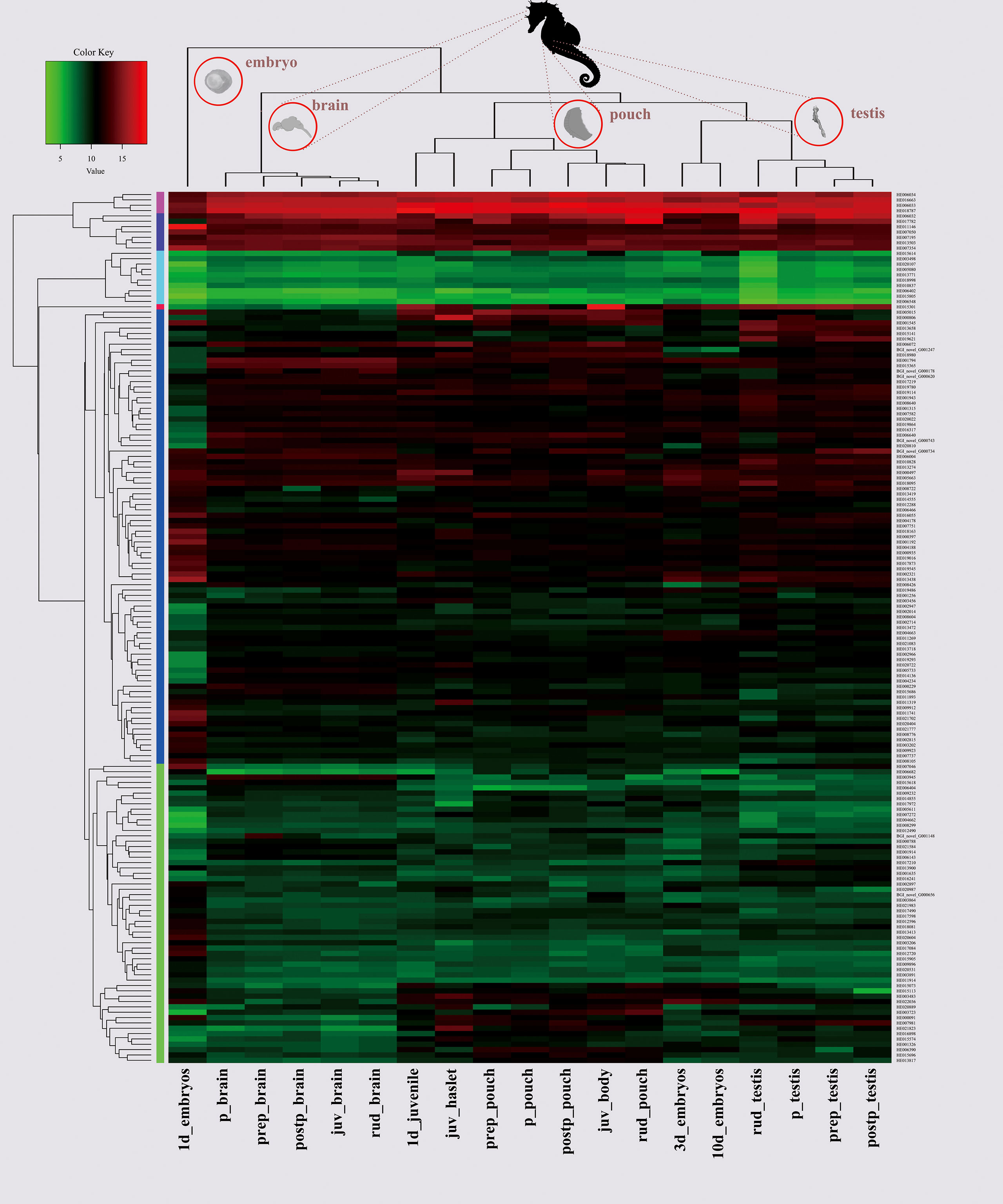
Figure 5 A heatmap of 209 AHTP-rich genes in the seahorse brain, pouch and testis at different developmental stages. X-axis shows clusters of the samples, and Y-axis refers to clusters of the AHTP-derived genes. Color key indicates the intensity associated with normalized transcription values. juv, juvenile; p, pregnancy; prep, pre-pregnancy; postp, post-pregnancy; rud, rudimentary.
In embryos, ATPase6 (HE011100), an essential protein for normal mitochondrial function (Jonckheere et al., 2012) with one IRP peptide, reached an exceedingly high transcription level (with a TPM value of 61,535.34) at the one-day stage, while its transcription went continuously down in 3- to 10-day embryos. Interestingly, it also had the highest transcription in the brain at all developmental stages except pregnancy (still extremely high with a TPM value of 8,688.23). In the brood pouch, where male seahorses nourish and protect embryos, four AHTP-rich collagens (HE016085, HE020257, HE015723, and HE005096) had much higher expression than in other tissues (Supplementary Figure 3; Supplementary Table 8). However, among the five genes encoding AHTP-rich titins (Supplementary Figure 2A), only one (HE002169) had a TPM value of over one thousand in 3 out of the 19 sequenced samples, and there was another gene (HE002168) that had a TPM value of over a hundred in five samples (Supplementary Figure 2B; Supplementary Table 8). Moreover, three actins (HE006032, HE006033, and HE006034), existing in all eukaryotic cells (Pollard, 2016) and each carrying one VFPS and one WHHT, were found to be highly transcribed in all samples (Figure 5). It seems that some proteins with highly or widely transcribed AHTPs could be considered as good markers for AHTP isolation.
In order to validate the AHTP-rich sequences at the level of translation, we employed previously published MS data (Chen et al., 2020) of the lined seahorse to confirm the existence and quantitation of AHTP-containing peptides and proteins. Two 5-month-old lined seahorse individuals (one male and one female) were sampled to construct a 10-fraction DDA library, and the protein intensity in each sample was quantified by the DIA mode.
In total, we detected 23,109 peptides and 4,187 proteins in the DDA spectrum library (Supplementary Table 3), based on which 10,072 and 10,747 peptides from the female and male samples were detected in the DIA mode (Supplementary Tables 4, 5), respectively. Interestingly, the protein numbers identified in both female and male homogenates were the same (1,864).
With expression fold-change ≥2 as the threshold, we determined 231 DEPs, among which 100 were highly expressed in females and the other 131 proteins were highly expressed in males (Figure 6). GO annotation classified these DEPs into 34 functional terms, of which “response to stimulus”, “organelle part,” and “transporter activity” were the top enriched ones exclusively in male (Figure 6B). Interestingly, compared to females, males had more DEPs with clustering into “cellular component” categories, which indicates a sexual difference in protein components.
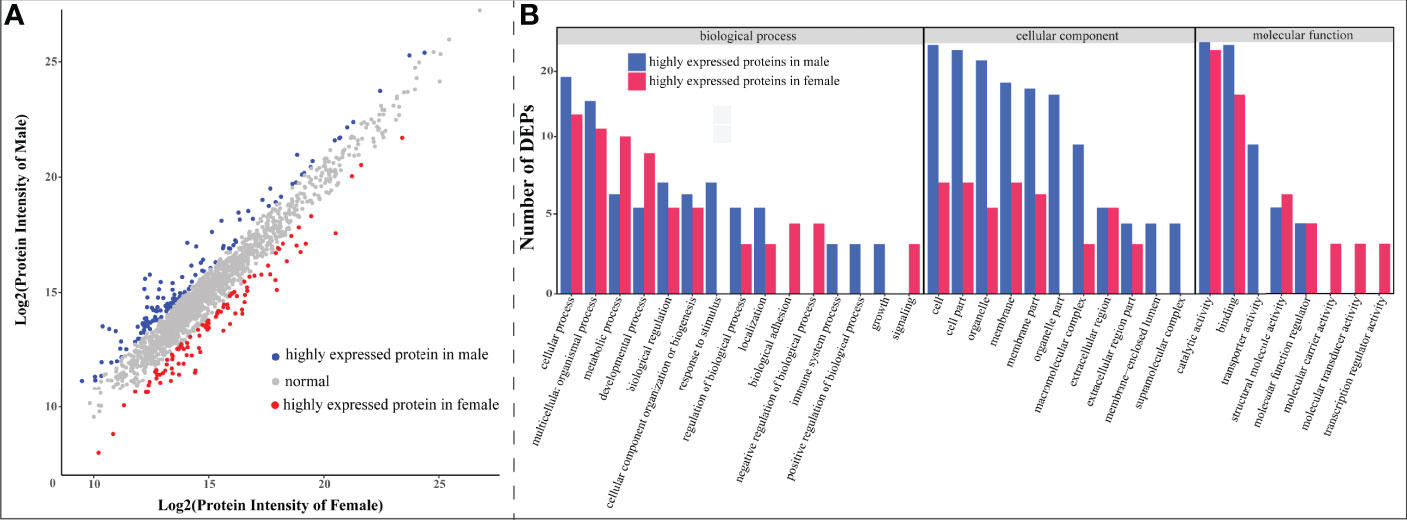
Figure 6 Statistics of differentially expressed proteins (DEPs) between female and male seahorses. (A) A scatter plot of all quantitative proteins. The x-axis and y-axis represent log2(protein intensity) of female and male samples, respectively. The red and blue dots represent DEPs in female and male respectively, while the rest gray dots stand for proteins without sexual difference. (B) Enriched Gene Ontology (GO) functional terms of identified DEPs. The x-axis represents GO term, and the y-axis denotes the number of each DEP. The red and blue bars stand for female and male upregulated proteins, representatively (as shown in A).
Many AHTP-derived genes could not be validated by the proteomic data. In particular, in the DDA spectrum library, there were only 620 peptides belonging to 487 AHTP-derived proteins, followed by 346 peptides corresponding to 248 proteins that were identified in the DIA mode. A total of 277 AHTP-containing peptides were shared in both female and male samples, whereas there were 16 female-specific and 53 male-specific peptides, respectively (Supplementary Figure 4). Since these peptides were usually short in length, most of them contained a single AHTP. It seems that AHTP-derived proteins with the highest intensities were consistent between both female and male samples. Noteworthily, the above-mentioned HE002169 (titin) with the biggest number of AHTPs was also among the top lists in both genders (Table 2).
A combination of both DIA data and AHTP screening showed that 95 AHTP-derived DEPs had higher expression in the male sample, while there were 70 DEPs with higher expression in the female sample. Table 3 lists the top five DEPs with the highest fold changes. Interestingly, among the top DEPs in males, there were two toxins, namely, natterin (HE017204) with a VRP peptide that was reported to be of high antihypertensive activity (Fan and Wu, 2021), and neoverrucotoxin (HE016101) carrying “LGP” and “GPL”. In addition, 12-AHTP-containing ryanodine receptor 3 (RYR3; HE015305), a calcium channel probably related to human gender dysphoria (Yang et al., 2017), was also in the top list. In contract, top DEPs with higher expression in female than in male included myosin (HE001010; carrying three AKK peptides) and filensin (HE007393; carrying VSV, AKK, PLG and IKP); both proteins were reported to participate in the formation of cytoskeleton for a complex interacting meshwork (Hohmann and Dehghani, 2019).
Several AHTPs have been purified from different seahorses (Gu and Xu, 2016; Kim et al., 2019b; Je et al., 2020; Shi et al., 2020) already, provoking us to conduct a high-throughput screening to predict potential AHTPs from the lined seahorse at a genomic level. The multi-omics (including genomics, transcriptomics, and proteomics) data enabled us to identify a total of 44,776 AHTPs from 15,523 proteins, accounting for 65.50% of all annotated proteins in the lined seahorse, and this ratio is within the reported range (between 59 and 74%) in various fishes (Yi et al., 2018). It also results in an average of 2.88 AHTPs per protein in the seahorse, and this number ranges from 2.3 to 3.3 in the 18 fishes being studied (Yi et al., 2018). Additionally, most (14,262) of these genes were supported by transcriptomes, but only a few (495) were further detected from the proteome data (Figure 7), suggesting that the coverage of the proteomic data is not deep enough, and some AHTP-derived genes may have very low expression to be detectable in the examined samples.
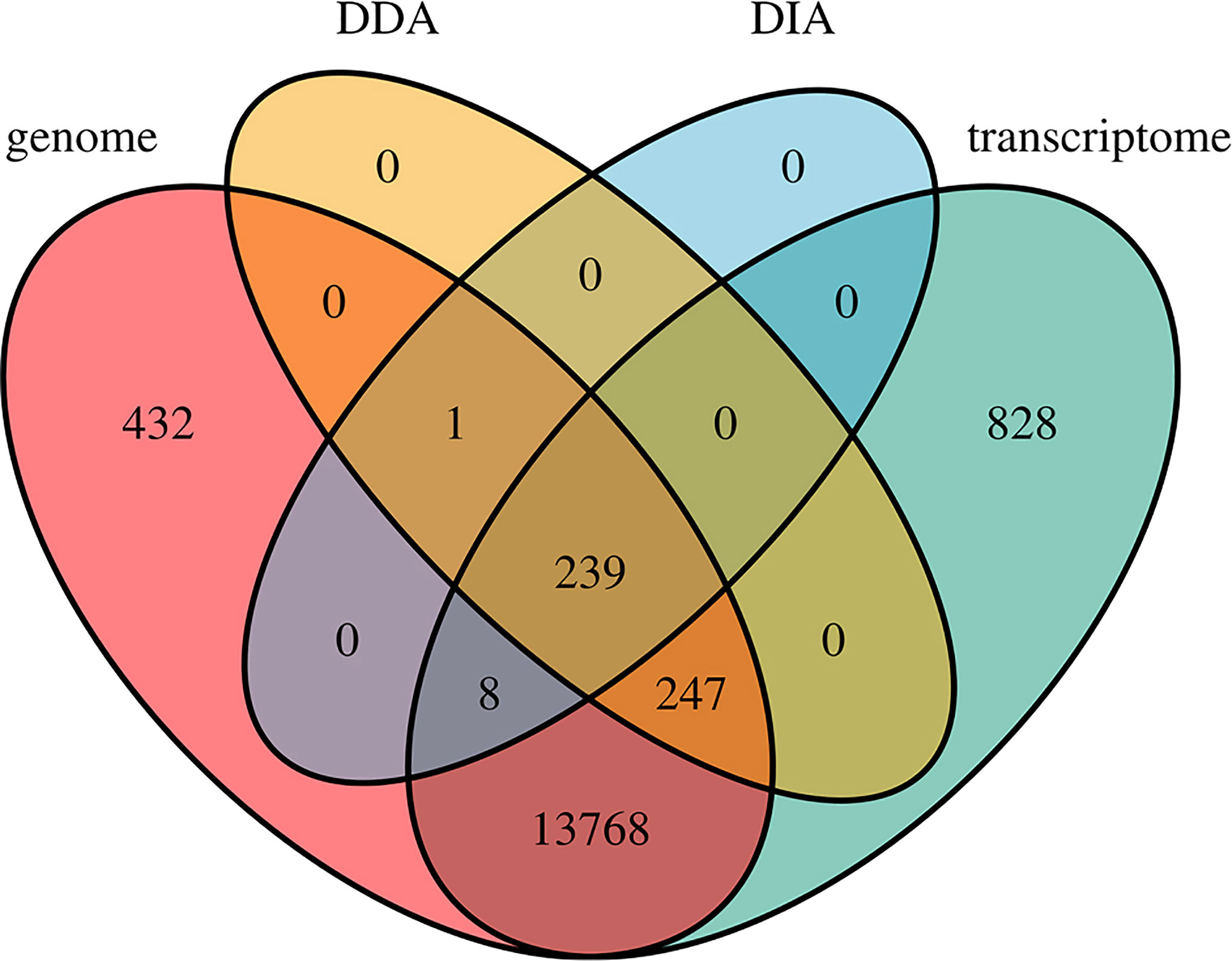
Figure 7 A Venn diagram of AHTP-derived genes from the genome, transcriptome, and proteome (both DIA and DDA modes) datasets.
As reported previously (Yi et al., 2018) in other fishes, including Atlantic cod, three-striped stickleback and grass carp, titins in the lined seahorse were AHTP-rich proteins (Figure 2). However, titins are tremendously large proteins with more than 25,000 amino acids (Krüger and Linke, 2011), which could increase the likelihood of a random match, and such an estimation was supported by the low AHTP/protein ratio of the titins (Figure 2). Among the five titins (including partial sequences) identified in the lined seahorse genome, the longest one (HE002169) contained the most AHTP hits, some of which were clustered in exons 158, 194, and 204 (Figure 3) and had the highest transcription level as well (Supplementary Figure 2). Generally speaking, titins are crucial for the assembly and functioning of sarcomeres (Peng et al., 2007) in fish skeletal and cardiac muscle cells (Seeley et al., 2007). Interestingly, there are certain reports that small peptides from titins of large yellow croaker (Fan et al., 2020), pork (Escudero et al., 2012) and bovine meat (Ferranti et al., 2014) have shown ACE-inhibitory activity. In this study, the abundance of AHTP hits and high expression of titins also support titin proteins as a promising resource for AHTP production.
Another group of AHTP-rich proteins were collagens. There were 81 proteins annotated as “collagen” in the lined seahorse genome. Each of them had at least one AHTP, and the highest number of AHTP hits in a collagen protein (such as HE002468 and COL8A1) could reach as high as 43, covering 5 AHTP types (Figure 4). However, they are usually transcribed at very low levels. The highest transcription was found in a 3-day embryo (TPM = 33.71), while in most samples (14 out of 19) the calculated TPM values were only a few (less than 4). Nevertheless, the four highly expressed collagens, HE005096 (COL1A1), HE016085 (COL1A1), HE020257 (COL1A2), and HE015723 (COL2A1), carried 7, 7, 3, and 1 AHTP(s), respectively (Figure 6; Supplementary Figure 3). Collagens are well known for their abundance in Gly, Pro, and Hyp (hydroxylated Pro) and XaaYaaGly repeats, where Xaa and Yaa can be any amino acid (Gauza-Włodarczyk et al., 2017; Li and Wu, 2018). Likewise, our data show that the most common AHTP type in seahorse collagens was tripeptides comprised of the permutations of Gly, Pro, and Lys (red lines inside the bars in Figure 4), covering 87.31% of all the identified AHTPs in collagens. Such a biased composition suggests that these amino acids may contribute largely to the antihypertensive activity of collagen hydrolysates.
Many peptides digested from collagens exhibit ACE-inhibitory activity (Saiga et al., 2008; Ichimura et al., 2009; Liu et al., 2011; Zhuang et al., 2012), and even some collagen peptide supplements have already been on sale for many years due to their multiple benefits for human health, including lowering blood pressure. Although our findings provide molecular evidence for such potential function to some extent, more investigations are still required for an in-depth understanding of these products’ pros and cons. In addition to their antihypertensive role, fish collagens have other biomedical applications, such as wound healing (Zhou et al., 2016), bone regeneration (Elango et al., 2016), and antimicrobials (Sahiner et al., 2014). However, instead of muscle, many other tissues (including bones, scales, and skin) are the major resources of fish collagen (Jafari et al., 2020), which poses a great challenge for the industrial preparation of collagen-derived peptides.
Some of the previously reported AHTPs have been purified from seahorses and their relatives, pipefishes. Recently, in the alcalase hydrolysate of the big-belly seahorse (H. abdominalis), three peptides (GIIGPSGSP, IGTGIPGIW, and QIGFIW) showed strong ACE inhibitory activity (Kim et al., 2019b). Later on, three more low-molecular peptides (APTL, CNVPLSP, and PWTPL) were proved to improve vasodilation via inhibition of ACE in vivo and in vitro (Je et al., 2020). A Chinese team hydrolyzed three-spot seahorse (H. trimaculatus) protein and identified an ACE-inhibiting peptide, PAGPRGPA (Shi et al., 2020). Two AHTPs (TFPHGP and HWTTQR) have been purified from muscle protein hydrolysates of seaweed pipefish (Syngnathus schlegeli; Wijesekara et al., 2011).
Among these (9) validated AHTPs, three of them were identified in seahorses, namely APTL and PWTPL from the big-belly seahorse, and PAGPRGPA from the three-spot seahorse (Supplementary Figure 5). As the shortest seahorse-derived AHTP to date, APTL had 281 hits spread over 270 lined seahorse proteins (without any dense distribution on specific sequences). Meanwhile, PWTPL and PAGPRGPA had only two and one hit(s) respectively, possibly due to their longer chain. Almost half of these AHTP-derived genes were transcribed (TMP ≥1, Supplementary Figure 5). Interestingly, the peptide PAGPRGPA was hydrolyzed from a collagen protein (COL1A1; encoded by gene HE005096), which was highly expressed in the pouch as mentioned above (Supplementary Figure 5). Unfortunately, the other six reported AHTPs could not be identified in the lined seahorse proteome data, indicating that although being members of the same family (Syngnathidae) or even within the same genus, different species do not share many AHTP types, mainly due to protein sequence divergence.
Advanced omics along with bioinformatic tools are efficient approaches for high-throughput prediction of bioactive peptides such as AHTPs, toxins (Gao et al., 2018; Peng et al., 2021), antimicrobial peptides (Chen et al., 2020) and anticancer peptides (Zhong et al., 2020). In this study, we predicted some ATHP-rich proteins in the lined seahorse, especially those with high expression by integration of genome, transcriptome, and proteome data. However, the reliability of our results might be limited by the quality of the omics data. For example, the number of the annotated collagens and titins from the lined seahorse genome was larger than expected due to fragment assembly. Collagens are encoded by 44 genes in humans and by 59 genes in the diploidy zebrafish (Nauroy et al., 2018; Bretaud et al., 2019), but there are 81 collagen genes predicted in the lined seahorse. Similarly, only two titin genes exist in zebrafish (Huttner et al., 2018), whereas the lined seahorse had five titins (although three were partial; see Supplementary Figure 2). Fragmentary sequences increased the number but shortened the length of predicted genes, and thus influenced the subsequent AHTP prediction, which could be improved by the generation of a high-quality assembly and performing additional cloning validation.
Using the multi-omics-based strategy, we performed high-throughput screening and identification of AHTPs from the lined seahorse. Our comprehensive data indicate that most of the AHTP-derived genes were supported by transcriptomic evidence, whereas only a few were further detected by proteome. Several groups of proteins, especially titins and collagens, contained the most AHTP hits, of which four AHTP-rich collagens had much higher transcription in the pouch than in other tissues. Tripeptides comprising the permutations of Gly, Pro, and Lys were the most abundant AHTP types in collagens. Additionally, three AHTPs that were previously purified from other seahorse species were also identified in the lined seahorse. Although omics-based AHTP predictions rely heavily on the quality of the used data, such a limitation can be overcome by in-depth experimental validation. In summary, our current findings provide a solid basis for understanding the abundance of various AHTPs in the lined seahorse, offering instructions for the choice of suitable samples in practical cases to purify AHTPs, and shed light on the development of antihypertensive products and drugs using seahorses as an important resource.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI [accession: PRJNA347499, SRA392578].
The animal study was reviewed and approved by the Institutional Review Board on Bioethics and Biosafety of BGI, China (No. FT 18134).
Conceptualization, QS and YH. Methodology, YH and PX. Software, XC and CB. Data curation, XC, PX, HS, and XL. Writing—original draft preparation, YH and XC. Writing—review and editing, QS, XY, JX, and JY. Visualization, YH. Supervision, QS. Funding acquisition, QS, XY, and JY. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This research was financially supported by the Shenzhen Science and Technology Program for International Cooperation (no. GJHZ20190819152407214 to QS), the Special Funds for Science Technology Innovation and Industrial Development of Dapeng New District (no. KJYF202101-01 to XY), and the Science and Technology Project of Shenzhen (no. JCYJ20170413155047512 to JY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Dr. Yuhai Yi from the Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, for her instructive advice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.863310/full#supplementary-material.
Abachi S., Bazinet L., Beaulieu L. (2019). Antihypertensive and Angiotensin-I-Converting Enzyme (ACE)-Inhibitory Peptides From Fish as Potential Cardioprotective Compounds. Mar. Drugs 17, 613 doi: 10.3390/md17110613
Abdelhedi O., Nasri M. (2019). Basic and Recent Advances in Marine Antihypertensive Peptides: Production, Structure-Activity Relationship and Bioavailability. Trends Food Sci. Technol. 88, 543–557. doi: 10.1016/j.tifs.2019.04.002
Acland A., Agarwala R., Barrett T., Beck J., Benson D. A., Bollin C., et al. (2014). Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 42, 7–17. doi: 10.1093/nar/gkt1146
Alexa A., Rahnenfuhrer J., Lengauer T. (2006). Improved Scoring of Functional Groups From Gene Expression Data by Decorrelating GO Graph Structure. Bioinformatics 22, 1600–1607. doi: 10.1093/bioinformatics/btl140
Allen J. M., Huang D. I., Cronk Q. C., Johnson K. P. (2015). aTRAM - Automated Target Restricted Assembly Method: A Fast Method for Assembling Loci Across Divergent Taxa From Next-Generation Sequencing Data. BMC Bioinf. 16, 98. doi: 10.1186/s12859-015-0515-2
Aluko R. E. (2015). Structure and Function of Plant Protein-Derived Antihypertensive Peptides. Curr. Opin. Food Sci. 4, 44–50. doi: 10.1016/j.cofs.2015.05.002
Battistoni A., Canichella F., Pignatelli G., Ferrucci A., Tocci G., Volpe M. (2015). Hypertension in Young People: Epidemiology, Diagnostic Assessment and Therapeutic Approach. High Blood Press Cardiovasc. Prev. 22, 381–388. doi: 10.1007/s40292-015-0114-3
Bhat Z. F., Kumar S., Bhat H. F. (2017). Antihypertensive Peptides of Animal Origin: A Review. Crit. Rev. Food Sci. Nutr. 57, 566–578. doi: 10.1080/10408398.2014.898241
Boeckmann B., Bairoch A., Apweiler R., Blatter M. C., Estreicher A., Gasteiger E., et al. (2003). The SWISS-PROT Protein Knowledgebase and its Supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370. doi: 10.1093/nar/gkg095
Bretaud S., Nauroy P., Malbouyres M., Ruggiero F. (2019). Fishing for Collagen Function: About Development, Regeneration and Disease. Semin. Cell Dev. Biol. 89, 100–108. doi: 10.1016/j.semcdb.2018.10.002
Bruderer R., Bernhardt O. M., Gandhi T., Miladinović S. M., Cheng L. Y., Messner S., et al. (2015). Extending the Limits of Quantitative Proteome Profiling With Data-Independent Acquisition and Application to Acetaminophen-Treated Three-Dimensional Liver Microtissues. Mol. Cell. Proteomics 14, 1400–1410. doi: 10.1074/mcp.M114.044305
Cardoso P., Stoev P., Georgiev T., Senderov V., Penev L. (2016). Species Conservation Profiles Compliant With the IUCN Red List of Threatened Species. Biodivers. Data J. 4, 2009–2012. doi: 10.3897/BDJ.4.e10356
Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., et al. (2018). SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 7, gix120. doi: 10.1093/gigascience/gix120
Chen X., Yi Y., You X., Liu J., Shi Q. (2020). High-Throughput Identification of Putative Antimicrobial Peptides From Multi-Omics Data of the Lined Seahorse (Hippocampus Erectus). Mar. Drugs 18, 30. doi: 10.3390/md18010030
Choi M., Chang C. Y., Clough T., Broudy D., Killeen T., MacLean B., et al. (2014). MSstats: An R Package for Statistical Analysis of Quantitative Mass Spectrometry-Based Proteomic Experiments. Bioinformatics 30, 2524–2526. doi: 10.1093/bioinformatics/btu305
Elango J., Zhang J., Bao B., Palaniyandi K., Wang S., Wenhui W., et al. (2016). Rheological, Biocompatibility and Osteogenesis Assessment of Fish Collagen Scaffold for Bone Tissue Engineering. Int. J. Biol. Macromol. 91, 51–59. doi: 10.1016/j.ijbiomac.2016.05.067
Escudero E., Toldrá F., Sentandreu M. A., Nishimura H., Arihara K. (2012). Antihypertensive Activity of Peptides Identified in the In Vitro Gastrointestinal Digest of Pork Meat. Meat Sci. 91, 382–384. doi: 10.1016/j.meatsci.2012.02.007
Fan H., Wu J. (2021). Purification and Identification of Novel ACE Inhibitory and ACE2 Upregulating Peptides From Spent Hen Muscle Proteins. Food Chem. 345, 128867. doi: 10.1016/j.foodchem.2020.128867
Fan Y., Yu Z., Zhao W., Ding L., Zheng F., Li J., et al. (2020). Identification and Molecular Mechanism of Angiotensin-Converting Enzyme Inhibitory Peptides From Larimichthys Crocea Titin. Food Sci. Hum. Wellness 9, 257–263. doi: 10.1016/j.fshw.2020.04.001
Ferranti P., Nitride C., Nicolai M. A., Mamone G., Picariello G., Bordoni A., et al. (2014). In Vitro Digestion of Bresaola Proteins and Release of Potential Bioactive Peptides. Food Res. Int. 63, 157–169. doi: 10.1016/j.foodres.2014.02.008
Ferreira S. H., Bartelt D. C., Greene L. J. (1970). Isolation of Bradykinin-Potentiating Peptides From Bothrops Jararaca Venom. Biochemistry 9, 2583–2593. doi: 10.1021/bi00815a005
Gao B., Peng C., Zhu Y., Sun Y., Zhao T., Huang Y., et al. (2018). High Throughput Identification of Novel Conotoxins From the Vermivorous Oak Cone Snail (Conus Quercinus) by Transcriptome Sequencing. Int. J. Mol. Sci. 19, 3901. doi: 10.3390/ijms19123901
Gauza-Włodarczyk M., Kubisz L., Włodarczyk D. (2017). Amino Acid Composition in Determination of Collagen Origin and Assessment of Physical Factors Effects. Int. J. Biol. Macromol. 104, 987–991. doi: 10.1016/j.ijbiomac.2017.07.013
Girgih A. T., Nwachukwu I. D., Hasan F., Fagbemi T. N., Gill T., Aluko R. (2015). Kinetics of the Inhibition of Renin and Angiotensin I-Converting Enzyme by Cod (Gadus Morhua) Protein Hydrolysates and Their Antihypertensive Effects in Spontaneously Hypertensive Rats. E. Food Nutr. Res. 59, 1–9. doi: 10.3402/fnr.v59.29788
Girgih A. T., Nwachukwu I. D., Hasan F. M., Fagbemi T. N., Malomo S. A., Gill T. A., et al. (2016). Kinetics of In Vitro Enzyme Inhibition and Blood Pressure-Lowering Effects of Salmon (Salmo Salar) Protein Hydrolysates in Spontaneously Hypertensive Rats. J. Funct. Foods 20, 43–53. doi: 10.1016/j.jff.2015.10.018
Gu W., Xu Y. (2016). Preparation of Hippocampus ACE Inhibitory Peptide and Determination of Antioxidant Capacity. Sci. Technol. Food Ind. 05, 201–206. doi: 10.13386/j.issn1002-0306.2016.05.031
Hohmann T., Dehghani F. (2019). The Cytoskeleton—A Complex Interacting Meshwork. Cells 8, 362. doi: 10.3390/cells8040362
Huttner I. G., Wang L. W., Santiago C. F., Horvat C., Johnson R., Cheng D., et al. (2018). A-Band Titin Truncation in Zebrafish Causes Dilated Cardiomyopathy and Hemodynamic Stress Intolerance. Circ. Genom. Precis. Med. 11, e002135. doi: 10.1161/CIRCGEN.118.002135
Ichimura T., Yamanaka A., Otsuka T., Yamashita E., Maruyama S. (2009). Antihypertensive Effect of Enzymatic Hydrolysate of Collagen and Gly-Pro in Spontaneously Hypertensive Rats. Biosci. Biotechnol. Biochem. 73, 2317–2319. doi: 10.1271/bbb.90197
Jafari H., Lista A., Siekapen M. M., Ghaffari-Bohlouli P., Nie L., Alimoradi H., et al. (2020). Fish Collagen: Extraction, Characterization, and Applications for Biomaterials Engineering. Polymers 12, 1–37. doi: 10.3390/polym12102230
Je J. G., Kim H. S., Lee H. G., Oh J. Y., Lu Y. A., Wang L., et al. (2020). Low-Molecular Weight Peptides Isolated From Seahorse (Hippocampus Abdominalis) Improve Vasodilation via Inhibition of Angiotensin-Converting Enzyme In Vivo and In Vitro. Process Biochem. 95, 30–35. doi: 10.1016/j.procbio.2020.04.016
Jia K., Bian C., Yi Y., Li Y., Jia P., Gui D., et al. (2019). Whole Genome Sequencing of Chinese White Dolphin (Sousa Chinensis) for High-Throughput Screening of Antihypertensive Peptides. Mar. Drugs 17, 504. doi: 10.3390/md17090504
Jonckheere A. I., Smeitink J. A. M., Rodenburg R. J. T. (2012). Mitochondrial ATP Synthase: Architecture, Function and Pathology. J. Inherit. Metab. Dis. 35, 211–225. doi: 10.1007/s10545-011-9382-9
Kanehisa M. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kang N., Kim S. Y., Rho S., Ko J. Y., Jeon Y. J. (2017). Anti-Fatigue Activity of a Mixture of Seahorse (Hippocampus Abdominalis) Hydrolysate and Red Ginseng. Fish. Aquat. Sci. 20, 1–8. doi: 10.1186/s41240-017-0048-x
Ketnawa S., Suwal S., Huang J. Y., Liceaga A. M. (2019). Selective Separation and Characterisation of Dual ACE and DPP-IV Inhibitory Peptides From Rainbow Trout (Oncorhynchus Mykiss) Protein Hydrolysates. Int. J. Food Sci. Technol. 54, 1062–1073. doi: 10.1111/ijfs.13939
Kim H. S., Je J. G., Ryu B., Kang N., Shanura Fernando I. P., Jayawardena T. U., et al. (2019b). Antioxidant and Angiotensin-I Converting Enzyme Inhibitory Peptides From Hippocampus Abdominalis. Eur. Food Res. Technol. 245, 479–487. doi: 10.1007/s00217-018-3179-0
Kim S. K., Ngo D. H., Vo T. S. (2012). Marine Fish-Derived Bioactive Peptides as Potential Antihypertensive Agents. Avd. Food Nutr. Res. 65, 249-260. doi: 10.1016/B978-0-12-416003-3.00016-0
Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019a). Graph-Based Genome Alignment and Genotyping With HISAT2 and HISAT-Genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim H.-S., Shin B.-O., Kim S.-Y., Wang L., Lee W., Kim Y. T., et al. (2016). Antioxidant Activity of Pepsin Hydrolysate Derived From Edible Hippocampus Abdominalis In Vitro and in Zebrafish Models. Korean J. Fish. Aquat. Sci. 49, 445–453. doi: 10.5657/kfas.2016.0445
Krüger M., Linke W. A. (2011). The Giant Protein Titin: A Regulatory Node That Integrates Myocyte Signaling Pathways. J. Biol. Chem. 286, 9905–9912. doi: 10.1074/jbc.R110.173260
Kumar R., Chaudhary K., Sharma M., Nagpal G., Chauhan J. S., Singh S., et al. (2015). AHTPDB: A Comprehensive Platform for Analysis and Presentation of Antihypertensive Peptides. Nucleic Acids Res. 43, D956–D962. doi: 10.1093/nar/gku1141
Lee S. Y., Hur S. J. (2017). Antihypertensive Peptides From Animal Products, Marine Organisms, and Plants. Food Chem. 228, 506–517. doi: 10.1016/j.foodchem.2017.02.039
Lee J. K., Jeon J. K., Byun H. G. (2014). Antihypertensive Effect of Novel Angiotensin I Converting Enzyme Inhibitory Peptide From Chum Salmon (Oncorhynchus Keta) Skin in Spontaneously Hypertensive Rats. J. Funct. Foods 7, 381–389. doi: 10.1016/j.jff.2014.01.021
Lin Q., Fan S., Zhang Y., Xu M., Zhang H., Yang Y., et al. (2016). The Seahorse Genome and the Evolution of its Specialized Morphology. Nature 540, 395–399. doi: 10.1038/nature20595
Lin Q., Qiu Y., Gu R., Xu M., Li J., Bian C., et al. (2017). Draft Genome of the Lined Seahorse, Hippocampus Erectus. Gigascience 6, gix030. doi: 10.1093/gigascience/gix030
Liu L. (2012). Studies on Bioactive Peptide From Chinese Soft-Shelled Turtle (Pelodiscus Sinensis) With Functionalities of ACE Inhibition and Antioxidation. Afr. J. Biotechnol. 11, 6723–6729. doi: 10.5897/ajb11.3537
Liu Z.-Y., Chen D., Su Y.-C., Zeng M.-Y. (2011). Optimization of Hydrolysis Conditions for the Production of the Angiotensin-I Converting Enzyme Inhibitory Peptides From Sea Cucumber Collagen Hydrolysates. J. Aquat. Food Prod. Technol. 20, 222–232. doi: 10.1080/10498850.2011.560364
Li P., Wu G. (2018). Roles of Dietary Glycine, Proline, and Hydroxyproline in Collagen Synthesis and Animal Growth. Amino Acids 50, 29–38. doi: 10.1007/s00726-017-2490-6
Majumder K., Wu J. (2014). Molecular Targets of Antihypertensive Peptides: Understanding the Mechanisms of Action Based on the Pathophysiology of Hypertension. Int. J. Mol. Sci. 16, 256–283. doi: 10.3390/ijms16010256
Manzanares P., Salom J. B., García-Tejedor A., Fernández-Musoles R., Ruiz-Giménez P., Gimeno-Alcañíz J. V. (2015). Unraveling the Mechanisms of Action of Lactoferrin-Derived Antihypertensive Peptides: ACE Inhibition and Beyond. Food Funct. 6, 2440–2452. doi: 10.1039/c5fo00580a
Meissner A. (2016). Hypertension and the Brain: A Risk Factor for More Than Heart Disease. Cerebrovasc. Dis. 42, 255–262. doi: 10.1159/000446082
Nauroy P., Hughes S., Naba A., Ruggiero F. (2018). The in-Silico Zebrafish Matrisome: A New Tool to Study Extracellular Matrix Gene and Protein Functions. Matrix Biol. 65, 5–13. doi: 10.1016/j.matbio.2017.07.001
Ondetti M. A., Williams N. J., Sabo E., Pluscec J., Weaver E. R., Kocy O. (1971). Angiotensin-Converting Enzyme Inhibitors From the Venom of Bothrops Jararaca. Isolation, Elucidation of Structure, and Synthesis. Biochemistry 10, 4033–4039. doi: 10.1021/bi00798a004
Peng C., Huang Y., Bian C., Li J., Liu J., Zhang K., et al. (2021). The First Conus Genome Assembly Reveals a Primary Genetic Central Dogma of Conopeptides in C. Betulinus. Cell Discov. 7, 11. doi: 10.1038/s41421-021-00244-7
Peng J., Raddatz K., Molkentin J. D., Wu Y., Labeit S., Granzier H., et al. (2007). Cardiac Hypertrophy and Reduced Contractility in Hearts Deficient in the Titin Kinase Region. Circulation 115, 743–751. doi: 10.1161/CIRCULATIONAHA.106.645499
Pollard T. D. (2016). Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 8, a018226. doi: 10.1101/cshperspect.a018226
Porter M. M., Novitskaya E., Castro-Ceseña A. B., Meyers M. A., McKittrick J. (2013). Highly Deformable Bones: Unusual Deformation Mechanisms of Seahorse Armor. Acta Biomater. 9, 6763–6770. doi: 10.1016/j.actbio.2013.02.045
R Core Team (2021). R: A Language and Environment for Statistical Computing(Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.r-project.org/.
Rosales-Mendoza S., Paz-Maldonado L. M. T., Govea-Alonso D. O., Korban S. S. (2013). Engineering Production of Antihypertensive Peptides in Plants. Plant Cell. Tissue Organ Cult. 112, 159–169. doi: 10.1007/s11240-012-0231-9
Ryu B. M., Qian Z. J., Kim S. K. (2010). SHP-1, a Novel Peptide Isolated From Seahorse Inhibits Collagen Release Through the Suppression of Collagenases 1 and 3, Nitric Oxide Products Regulated by NF-κb/P38 Kinase. Peptides 31, 79–87. doi: 10.1016/j.peptides.2009.10.019
Sahiner M., Alpaslan D., Bitlisli B. O. (2014). Collagen-Based Hydrogel Films as Drug-Delivery Devices With Antimicrobial Properties. Polym. Bull. 71, 3017–3033. doi: 10.1007/s00289-014-1235-x
Saiga A., Iwai K., Hayakawa T., Takahata Y., Kitamura S., Nishimura T., et al. (2008). Angiotensin I-Converting Enzyme-Inhibitory Peptides Obtained From Chicken Collagen Hydrolysate. J. Agric. Food Chem. 56, 9586–9591. doi: 10.1021/jf072669w
Seeley M., Huang W., Chen Z., Wolff W. O., Lin X., Xu X. (2007). Depletion of Zebrafish Titin Reduces Cardiac Contractility by Disrupting the Assembly of Z-Discs and A-Bands. Circ. Res. 100, 238–245. doi: 10.1161/01.RES.0000255758.69821.b5
Shi J., Su R., ting W., Chen J. (2020). Purification and the Secondary Structure of a Novel Angiotensin I-Converting Enzyme (ACE) Inhibitory Peptide From the Alcalase Hydrolysate of Seahorse Protein. J. Food Sci. Technol. 57, 3927–3934. doi: 10.1007/s13197-020-04427-0
Udenigwe C. C., Mohan A. (2014). Mechanisms of Food Protein-Derived Antihypertensive Peptides Other Than ACE Inhibition. J. Funct. Foods 8, 45–52. doi: 10.1016/j.jff.2014.03.002
Wijesekara I., Qian Z. J., Ryu B. M., Ngo D. H., Kim S. K. (2011). Purification and Identification of Antihypertensive Peptides From Seaweed Pipefish (Syngnathus Schlegeli) Muscle Protein Hydrolysate. Food Res. Int. 44, 703–707. doi: 10.1016/j.foodres.2010.12.022
Xu K., Shen X., Chen G. (2015). Angiotensin I-Converting Enzyme (ACE) Inhibitory Activity of Enzymatic Hydrolysate From Three-Spot Hippocampus. Sci. Technol. Food Ind. 36, 96–99. doi: 10.13386/j.issn1002-0306.2015.15.012
Yang F., Zhu X. H., Zhang Q., Sun N. X., Ji Y. X., Ma J. Z., et al. (2017). Genomic Characteristics of Gender Dysphoria Patients and Identification of Rare Mutations in RYR3 Gene. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-08655-x
Yi Y., Lv Y., Zhang L., Yang J., Shi Q. (2018). High Throughput Identification of Antihypertensive Peptides From Fish Proteome Datasets. Mar. Drugs 16, 365. doi: 10.3390/md16100365
Zhang Y. Y., Ryu B. M., Qian Z. J. (2017). A Review - Biology, Aquaculture and Medical Use of Seahorse, Hippocampus Spp. Annu. Res. Rev. Biol. 14, 1–12. doi: 10.9734/ARRB/2017/34152
Zhong W., Zhong B., Zhang H., Chen Z., Chen Y. (2020). Identification of Anti-Cancer Peptides Based on Multi-Classifier System. Comb. Chem. High Throughput Screen 22, 694–704. doi: 10.2174/1386207322666191203141102
Zhou T., Wang N., Xue Y., Ding T., Liu X., Mo X., et al. (2016). Electrospun Tilapia Collagen Nanofibers Accelerating Wound Healing via Inducing Keratinocytes Proliferation and Differentiation. Colloids Surf. B. Biointerf. 143, 415–422. doi: 10.1016/j.colsurfb.2016.03.052
Keywords: the lined seahorse, antihypertensive peptide, multi-omics data, high-throughput prediction, titin, collagen
Citation: Huang Y, Chen X, Shu H, Xiao P, Lin X, Xu J, Bian C, You X, Yang J and Shi Q (2022) High-Throughput Identification of Antihypertensive Peptides (AHTPs) and Characterization of AHTP-Derived Genes in the Lined Seahorse (Hippocampus erectus). Front. Mar. Sci. 9:863310. doi: 10.3389/fmars.2022.863310
Received: 27 January 2022; Accepted: 30 May 2022;
Published: 04 July 2022.
Edited by:
Rafik Balti, University of Jendouba, TunisiaReviewed by:
Huixian Zhang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2022 Huang, Chen, Shu, Xiao, Lin, Xu, Bian, You, Yang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Shi, c2hpcWlvbmdAZ2Vub21pY3MuY24=; Jian Yang, amlhbnlAc3pwdC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.