
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 20 April 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.861691
This article is part of the Research Topic Animal Welfare, Volume I: Animal Welfare in Aquaculture - Physiological Basis and Recent Findings View all 14 articles
Current BLASTP search analysis results suggested that the lobster (Homarus americanus) HaCHH-A and HaCHH-B may be derived from two different four-exon genes. Repeated tissue expression studies have revealed much different expression patterns of these two genes from those reported in the past. With RT-PCR, rapid amplification of complementary DNA (cDNA) ends (RACE), and genomic DNA cloning, we confirmed that the HaCHH-A and HaCHH-B transcripts were derived from two different four-exon CHH genes. By an alternative splicing mechanism, each gene can produce different but larger transcript variants (i.e., sHaCHH-A and sHaCHH-B) mainly in different non-eyestalk tissues of the females. The larger and unspliced transcripts can be detected in the hepatopancreas, gill, heart, nerve cord, brain, ovary, and thoracic ganglion of the reproductive females. The expression patterns of sHaCHH-A and sHaCHH-B in other non-eyestalk tissues suggest that these transcripts have a wide spectrum of expressions during the female reproductive cycle. An in vitro organ explant culture system was developed to investigate the reproductive function of these cDNAs. The results showed that the recombinant proteins for sHaCHH-A and sHaCHH-B inhibited the gene expression of vitellogenin, whereas the double-stranded RNA (dsRNA) for sHaCHH-A and sHaCHH-B stimulated the expression of the vitellogenin gene in vitro. The results of the study may provide insights for the development of techniques to induce gonad development without using eyestalk ablation operation. This is the first in-depth report of the characterization of two four-exon CHH genes in a crustacean.
1. Multiple CHH-A and CHH-B genes exist in the lobster Homarus americanus, and these genes consist of four exons and are interrupted by three introns.
2. The previously reported lobster CHH-A and CHH-B cDNAs were derived from alternative splicing of the third intron, and the newly identified sCHH-A and sCHH-B transcripts were products of four-exon transcripts that exist mainly in non-eyestalk neuronal tissues.
3. The expression patterns of the newly reported sHaCHH-A and sHaCHH-B suggested that they may have a reproductive stage-related function in the biannual reproductive cycle of females.
4. Recombinant proteins for sHaCHH-A and sHaCHH-B inhibited the expression of vitellogenin, and gene knockdown of sHaCHH-A and/or sHaCHH-B caused a significant increase in the gene expression of vitellogenin in the hepatopancreas and ovary.
Reproduction is an important physiological process and requires a large amount of energy in crustaceans. Crustacean reproduction is tightly regulated by the interactions of hormones (i.e., peptides, neuropeptides, juvenoids, and steroids) (Fingerman, 1987; Huberman, 2000; Diwan, 2005). The neuropeptides most popularly studied in crustacean endocrine research are the crustacean hyperglycemic hormone (CHH), molt-inhibiting hormone (MIH), and gonad-inhibiting hormone (GIH). Together, these hormones belong to a large group of neuropeptides of the CHH/MIH/GIH family (Keller, 1992). These hormones are synthesized in the X-organ sinus gland complex located in the optical ganglia of the eyestalk. The eyestalk sinus gland is a neurohemal organ consisting of clustered axon endings of the neurosecretory cell somata (Chan et al., 2003; Chung et al, 2020; Keller, 1992; Montagné et al., 2008; Tensen et al., 1991; Webster et al., 2012). Many studies have shown that the removal of eyestalks could induce gonad maturation in decapods (Panouse, 1943; Primavera, 1978; Okumura uand Aida, 2001; Uawisetwathana, et al., 2011; Magaña-Gallegos et al., 2021). Removal of the eyestalk could remove the source of GIH and result in rapid ovary development. However, the use of this technique has raised serious animal welfare concerns in recent years. In lobster (Homarus americanus), two highly similar HaCHH isoforms (i.e., HaCHH-A and HaCHH-B) have been characterized (Tensen et al., 1991; De Kleijn et al., 1994; De Kleijn and Van Herp, 1995; Chang et al., 1999). They are known to play a role in the reproduction of the female lobster. The HaCHH-A and HaCHH-B isoforms share a high overall amino acid sequence identity and differ only by eight amino acid residues for the deduced pro-hormone. Despite the high sequence homology, each CHH has been reported to regulate reproduction during the biannual reproductive cycle of adult females (De Kleijin et al., 1995). Many CHH family neuropeptides from other crustacean species were discovered in the eyestalk from 1990 to 2000; subsequent studies indicated that the transcripts of these neuropeptides can be found in other non-eyestalk neuronal tissues (Chan et al., 2003; Diwan, 2005; Huberman, 2000; Keller, 1992; Spanings-Pierrot et al., 2000; Webster et al., 2012; Loredo-Ranjel et al., 2017) and even in many non-neuronal tissues (Chen et al., 2004; Shi et al., 2018). Although the discovery of the two lobster CHH isoforms has been reported for 25 years, little information has been acquired since their first discovery. At the time when the complementary DNAs (cDNAs) of HaCHH-A and HaCHH-B were cloned, only a few CHH family neuropeptide sequences have been reported in the GenBank. With the exponential increase in the total number of CHH family neuropeptides reported in recent years, it is important to obtain additional specific information on the gonad maturation of the lobster.
In recent years, China has imported large quantities of H. americanus as high-priced seafood from the US and Canada. Because of the increased need for this species as food, growing interest to develop a large-scale commercial complete aquaculture of H. americanus in China has arisen. One major constraint for the successful aquaculture of this species is the lack of information on the endocrine control of reproduction. Knowledge on its reproduction, larval rearing, and broodstock management is inadequate. For this reason, investigations into the involvement of CHHs in the reproductive cycle of the female lobster are urgently needed. In this study, we further characterized HaCHH-A and HaCHH-B from the study of gene structure and expression. We report new findings for two additional CHH-related peptides and confirm that they are derived from two different four-exon CHH genes that also gave rise to the originally discovered HaCHH-A and HaCHH-B cDNAs. We also demonstrate the potential functions of sHaCHHs during the reproductive cycle of the female lobster.
Female lobsters (average carapace length = 70 ± 3 cm, weight = 500 ± 50 g) were purchased from the local seafood market. These animals were imported from the US or Canada to China. They were brought back to the Marine Research Station of the Guangdong Ocean University, Zhanjiang, China. The lobsters were acclimatized (>1 week) in 1-mt circular fiberglass tanks equipped with running seawater (salinity = 32 ± 5‰), temperature of 20 ± 3°C, and an aeration system under conditions of natural photoperiod. They were fed twice daily with commercial pellet diets and fresh oysters. The molt stages of the lobsters were determined by setogenesis specifically for lobster (Helluy and Beltz, 1991). Except for the tissue extraction studies, females were used in all cloning and expression experiments. Before dissection, the lobsters were anesthetized in ice for 30 min. All experiments were conducted in compliance with the guidelines of the Animal Care and Use Committee of Guangdong Ocean University.
To avoid cloning of multiple highly homologous CHH isoforms and highly polymorphic gene transcripts from different individuals, genomic DNA from a single animal was used as the template. For genomic DNA extraction, tissues from the hepatopancreas (Hp) or testis were dissected from the animal and extracted in an extraction buffer. Purified high-quality genomic DNA was used as the template for PCR. Gene-specific primers were designed to specifically amplify either the HaCHH-A or the HaCHH-B gene, and these primers were located in the exon of the published CHH gene from other decapods (Table 1).
Total RNA was extracted from various tissues using TransZol Up Plus RNA spin column-based kit (TransGen, Beijing, China). The RNA concentration was determined using a NanoDrop spectrophotometer, and 1 μg of total RNA was reverse transcribed using the SMART RACE cDNA Amplification Kit (TaKaRa, Shiga, Japan). PCR amplification of RACE (rapid amplification of cDNA ends) products was performed using a 5′- and 3′-RACE Kit (TaKaRa, Shiga, Japan). The forward primers CHH-F1A: 5′-CGTCATGTTCGCCTGC AGAACTCTG-3′ and CHH-F1B: TCATGATGGCCTGCAGAGCGCTGT and the common reverse primer CHH-RP: 5′-TTACTTGCCGACCATCTGGACG-3′ were used. The internal primers used to generate HaCHH-A and HaCHH-B isoform-specific RT-PCR products were as follows: full-length HaCHH-A and HaCHH-B were generated using the forward primer 5′-CCCATTTGCAAAGCGATGAGTTCG-3′ and the reverse primer 5′-TGTGCCAA ATGGCTCACAGAA-3′; the amplified fragment was cloned into the pMD19 TA cloning vector (Sangon Biotech, Shanghai, China). All the clones were confirmed by nucleotide sequencing analyses.
To determine the messenger RNA (mRNA) expression profiles of sHaCHH-A and sHaCHH-B in the biannual ovarian development cycle, females at different reproductive stages were sacrificed for total RNA extraction. The reproductive stages of these animals were determined based on the coloration of the ovary and the Gonadosomatic Index (GSI) as previously reported (Tiu et al., 2009).
The amino acid sequences of the positive clones were analyzed using ORF Finder (NCBI; http://www.ncbi.nlm.nih.gov/projects/gorf/orfig.cgi). Sequence identities were verified using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was performed using the amino acid sequence, and the online Multiple Sequence Alignment software ClustalW (genome.jp) was employed for the various CHH sequences chosen to represent the major subgroups of the type I CHH genes, using only the most highly conserved orthologous amino acids within the CHH/MIH/GIH from representative decapods. The amino acid sequences used in our phylogenetic analysis are listed in Supplemental Data File 1. The amino acid sequences of HaCHH-A and HaCHH-B were compared to those of other decapods to confirm the identification of each family member.
The transcript levels of HaCHH-A, sHaCHH-A, HaCHH-B, and sHaCHH-B were determined by both semi-quantitative PCR and real-time quantitative PCR (RT-qPCR). For the semi-quantitative PCR, each PCR was carried out in a final volume of 10 μl containing 5 μl of 2× PCR Master Mix (ABI, Richmond Hill, Canada), 0.3 μl of forward and reverse primers, 2.4 μl of nuclease-free water, and 2 μl of cDNA template. For the internal control, we have used the elongation factor 1 (EF1α) and the β-actin gene of the lobster. The PCR conditions were as follows: 94°C for 5 min, 34 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, and 72°C for 3 min. For RT-qPCR, the CFX96 Real Time System (Bio-Rad, Hercules, CA, USA) and qPCR Master Mix (TaKaRa) were used. Total RNA was extracted from the different tissues of mature lobsters. Total RNA from each tissue (1 μg) was treated with gDNA Eraser at 37°C for 15 min to avoid contamination with genomic DNA. Total RNAs were used for cDNA synthesis using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa). The cDNAs from females with vitellogenic oocytes were used for tissue distribution. The relative mRNA abundance of each gene was calculated using the ΔΔCt method, then normalized to that of the Ef1α and β-actin for each sample. Measurements of the expression levels were performed in triplicate or duplicate.
Recombinant protein and RNAi technologies were used to study the function of sHaCHH-A and sHaCHH-B in lobster gonad vitellogenesis. For the detailed protocol of recombinant protein production, see Liang et al. (2019). Briefly, the restriction enzyme site sequence-linked primers for sHaCHH-A (expHaCHH-AF: GGATCCCGGTCTGTAGAAGGAGCATC; expHaCHH-AR: GAGCTCCTACCCCTTGATCATATCC) and sHaCHH-B (sHaCHH-B; rHaCHH-B: GGATCCCGGTCAGTAGAAGGAGCGGATATGATCAAGGGGTAGGAGCTC) (Table 1) were used to amplify the mature peptide of sHaCHH-A or HaCHH-B. The cDNA template was derived from the cDNA clones from the above cDNA cloning procedure. The PCR-amplified cDNA fragments were digested with the same enzyme and ligated to the pET32A vector plasmid digested previously with the same restriction enzymes (BamHI and SacI). After bacterial transformation, positive clones were screened by PCR using the T7 promoter and T-terminator primers. The positive clones were verified by PCR with a gene-specific primer for either the HaCHH-A or HaCHH-B gene and DNA sequence determination. The recombinant proteins were produced using Escherichia coli DE21 strain and subsequently purified. The purified and re-natured recombinant proteins for sHaCHH-A and sHaCHH-B were used for in vitro bioassay to study their potential functions.
RNA interference approaches were used to knock down the functions of sHaCHH-A or sHaCHH-B. For double-stranded RNA (dsRNA) synthesis, primers amplifying exon 3 of sHaCHH-A and sHaCHH-B were designed (Table 1). The templates were the cDNAs for sHaCHH-A and sHaCHH-B from the cDNA cloning described above. DsRNAs were synthesized using the T7 RNA Transcription Kit (Vazyme, Nanjing, China). For control, the dsRNA for the tiger frog virus ATPase gene was used (Tiu et al., 2008). The dsRNA template for each gene was amplified (PCR amplification procedure: 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 30 s, then 72°C for 10 min) with the above primers and purified using the FastPure Gel DNA Extraction Mini Kit (Vazyme). After detection of the concentrations of the DNA templates (NanoDrop 2000; Thermo Fisher Scientific, Inc., Waltham, MA, USA), dsRNA was produced with DNA templates using the T7 RNA Transcription Kit (Vazyme). The final dsRNA was diluted to an appropriate concentration (1 µg/µl) with phosphate-buffered saline (PBS). In vitro RNA interference experiments (n = 8) were then performed.
The lobsters were dissected and the ovary developmental stage was determined. Individuals with ovary at the early (yellow) and middle (light green) stages were dissected for the Hp and ovary (i.e., 50–80 mm3). The tissues were first rinsed with ice-cold PBS and then placed into the wells of a sterile culture plate containing 1.5 ml nutrient medium (M199; Sigma, St. Louis, MO, USA). Individual wells were treated with either the tested recombinant proteins, dsRNAs, or the control (Tiu et al., 2008). The culture plate was then shaken on the platform of an orbital shaker at 24–26°C for 3 h. At the end of the culture period, the tissues were extracted for total RNA and for cDNA synthesis. The expression of the lobster vitellogenin gene was monitored by RT-PCR, as previously described. The HaVg1 primers were derived from the lobster vitellogenin cDNA (GenBank no. EF422415.1).
Current GenBank BLASTP sequence homology search results for HaCHH-A (GenBank no. P19806.3) and HaCHH-B (GenBank no. 2105187B) indicated that they are most similar to the prepro-CHH isoform A of the lobster Nephrops norvegicus (GenBank no. AAQ22391.1) and the CHH isoform B (GenBank no. AAQ22392.1). Compared to the crayfish, HaCHH-A and HaCHH-B are most similar to the CHH of Procambarus clarkii (GenBank no. Q25683.3). Compared to the CHH of shrimps, HaCHH-A and HaCHH-B are most similar to the CHH-like of Litopenaeus vannamei (GenBank no. AAN86055.1) and Penaeus monodon (GenBank no. XP037787977), sharing 68%–69% sequence identity in the mature peptide region. Moreover, the HaCHH-A and Ha-CHH-B sequences are more similar to the CHH of the freshwater shrimp Macrobrachium rosenbergii (GenBank no. AAF29534.1). Compared to the crab CHH, the lobster sequences shared the highest sequence identity with the CmCHH (GenBank no. P14944) of Carcinus maenas. Further analysis of these decapod CHH sequences revealed that they were derived from four-exon genes. Alternative splicing of the pre-mRNA for these CHHs gave rise to the transcripts being analyzed. Therefore, the results suggested that the lobster HaCHH-A and HaCHH-B may also be derived from different four-exon genes. Unlike a previous report (DeKleijn et al., 1995), the current RT-PCR results showed that two transcripts can be detected in the eyestalk cDNA (Figure 1A). The small cDNA fragment was about 150 bp, and a larger and more abundant DNA fragment of 310 bp was also amplified. Similar results were observed for the HaCHH-B gene when primers HaCHH1b and HaCHH-RP were used in PCR (Figure 1A). These two DNA fragments were subcloned into pMD19 in E. coli for DNA sequencing. The results revealed that the smaller cDNAs carried coding sequences for the matured peptide of the previously reported HaCHH-A (GenBank no. P198063) and HaCHH-B (GenBank no. 2105187B). The 5′ end of the larger cDNA carried a coding sequence identical to the corresponding HaCHH-A or HaCHH-B, but the 3′ end carried a peptide sequence different from the original HaCHH-A and HaCHH-B cDNA (see below).
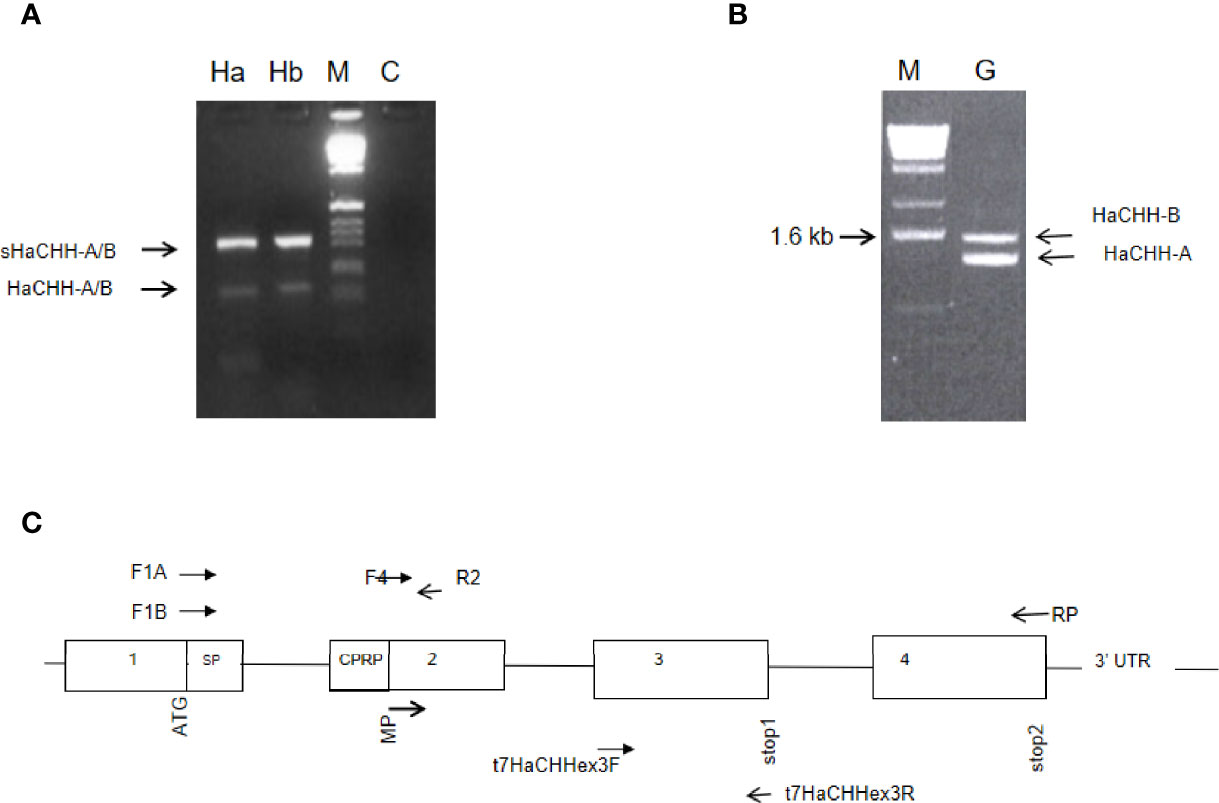
Figure 1 (A) Identification of transcript variants for the HaCHH-A and HaCHH-B genes. Gene-specific forward primers for HaCHH-A (F1A) and HaCHH-B (F1B) were paired with common reverse primer (R2) in the RT-PCR amplification of cDNA from eyestalk (Es). Two major fragments were amplified by each pair of primers. The larger fragment (upper arrow) is the sHaCHH-A or the sHaCHH-B transcript variant; the smaller band (lower arrow) is for transcripts HaCHH-A and HaCHH-B. (B) Gene from eyestalk cDNA of female lobster. Lane Ha: PCR results using primer pair F1A and RP; lane Hb: results for primer F1B/RP; M: DNA size marker; and C: negative, no template control. (B) Genomic PCR detection of multiple HaCHH-A and HaCHH-B genes. The primer pair F4/RP (Figure 2 and Table 1) was used and two distinctive DNA fragments (i.e., 1.4 and 1.6 kb) were amplified. (C) Schematic diagram showing the locations of the primers and the proposed gene structure of the HaCHH genes used in this study.
To study the HaCHH-A and CHH-B genes, PCR amplification of genomic DNA was performed with the DNA templates and cDNAs from a single individual (Figures 1B, C and Table 1). Two major DNA bands with sizes of 1,600 and 1,450 bp were amplified. Because the intensity of the smaller band in the agarose gel was stronger, more copy numbers for the smaller genomic DNA were expected. Sequence determination of the bands revealed that each DNA fragment carries the coding and non-coding sequences for the HaCHH-A (1.4 kb) or the HaCHH-B (1.6 kb) gene (Figures 1B, C). These fragments carry coding sequences for partial exon 2, exon 3, and exon 4 and the interrupting non-coding introns 2 and 3. We employed genomic PCR to attempt to recover the partial exon 1 coding sequence and non-coding intron 1 of both genes, without success. As a comparative study of these CHH genes, genomic information of the four-exon CHH genes from other decapods was collected (Figure 2). Except for the complete CHH gene structure of L. vannamei (from the L. vannamei genome project), the other CHH gene structures were incomplete. Most of these genes lacked information on the first intron. In the L. vannamei CHH gene, intron 1 consisted of several highly repeated microsatellite sequences [i.e., (AT)n, (CT)n, and (GT)n] along the span of a 3.34 intronic region (L. vannamei genomic database, GenBank no. LOC113815764).
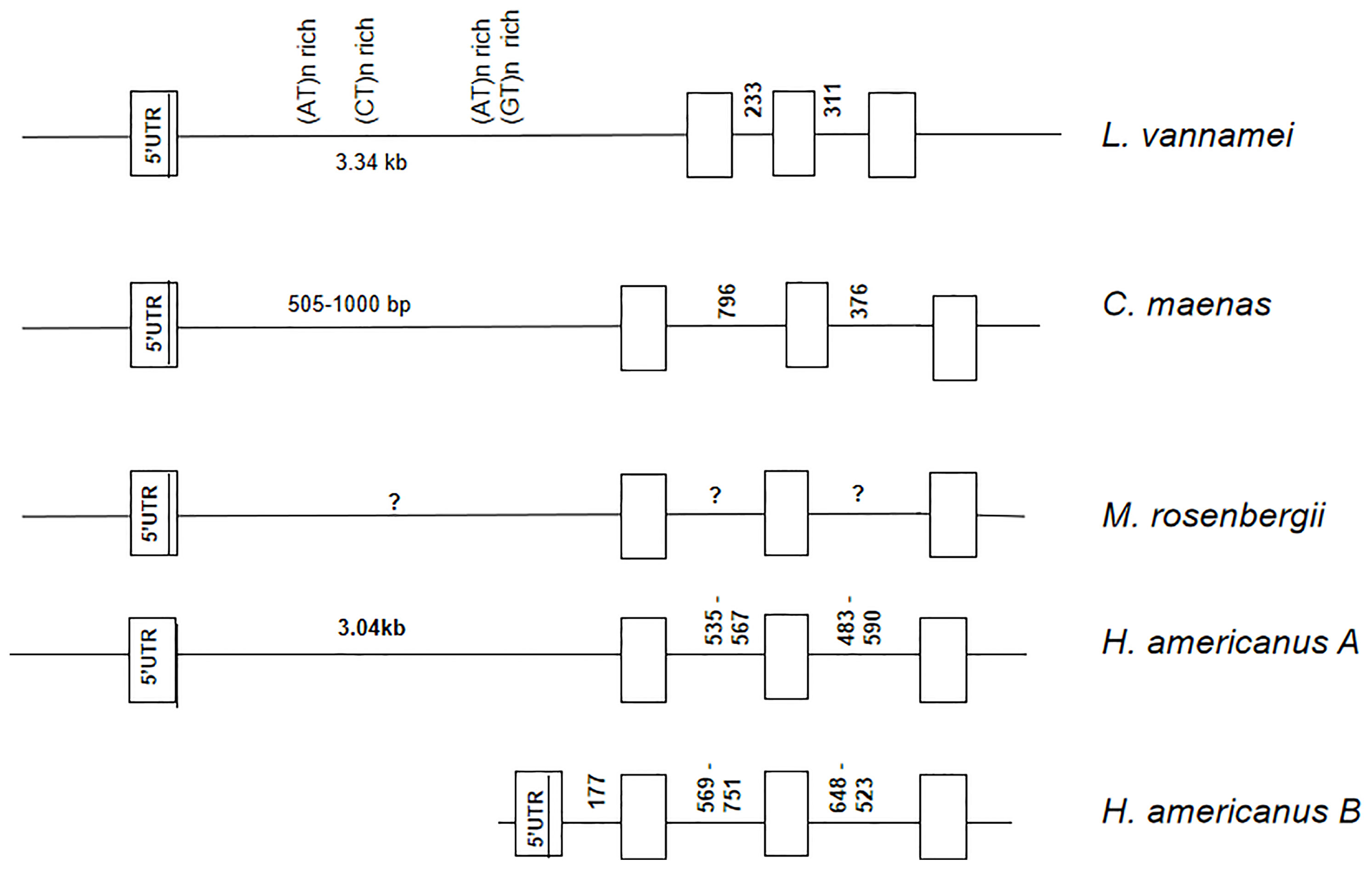
Figure 2 Comparison of the four-exon CHH genes from the lobster Homarus americanus, crab Carcinus maenas, and shrimp Litopenaeus vannamei and Macrobrachium rosenbergii. Exons are indicated by the open boxes and introns are indicated by the horizontal line in between the exons. The first exon consists of the 5′-ÚTR and the ATG start codon. Unknown intron sizes are indicated by an a question mark. Highly repetitive sequence (satellite DNA) clusters were identified in the promoter region of L. vannamei CHH-like gene.
The RT-PCR and 5′ and 3′RACE cloning approaches were used to clone the large and small fragments, as described above. The four cDNA sequences were cloned and the sequences determined. The amino acid sequences of two smaller cDNAs were identical to the previously reported HaCHH-A and HaCHH-B. The two larger cDNAs (i.e., sHaCHH-A and sHaCHH-B) represented the newly discovered transcript variants (Figure 3). As expected, HaCHH-A and sHaCHH-A shared identical amino acids in the N-terminal end, but major amino acid sequence variations occurred in the C-terminal end. Similar results were found for HaCHH-B and sHaCHH-B. For the multiple alignment and phylogenetic study, other CHH subtype I sequences and MIH/GIH subtype II group neuropeptides were included in the ClustalW analysis together with the four HaCHH variants. Within the same color block, a much higher degree of sequence identity was observed in the signal mature peptide region (Figure 4). For the mature peptide, the degree of homology was much higher in the coding sequence of exon 2 and was much lower in the C-terminal end of the protein encoded by exon 3. The amino acid similarity at the N-terminal end of the mature peptide for HaCHH-A and HaCHH-B was much higher, i.e., >97% amino acid identity, but the homology decreased to only 63.5% at the C-terminal end. Despite the similar molecular characteristics of these CHHs, the pI values (ExPASy Compute pI/Mw tool) of these isoforms were quite different. For example, the estimated pI value of sHaCHH-A (i.e., pI = 5.27) was the smallest compared to those of the rest.
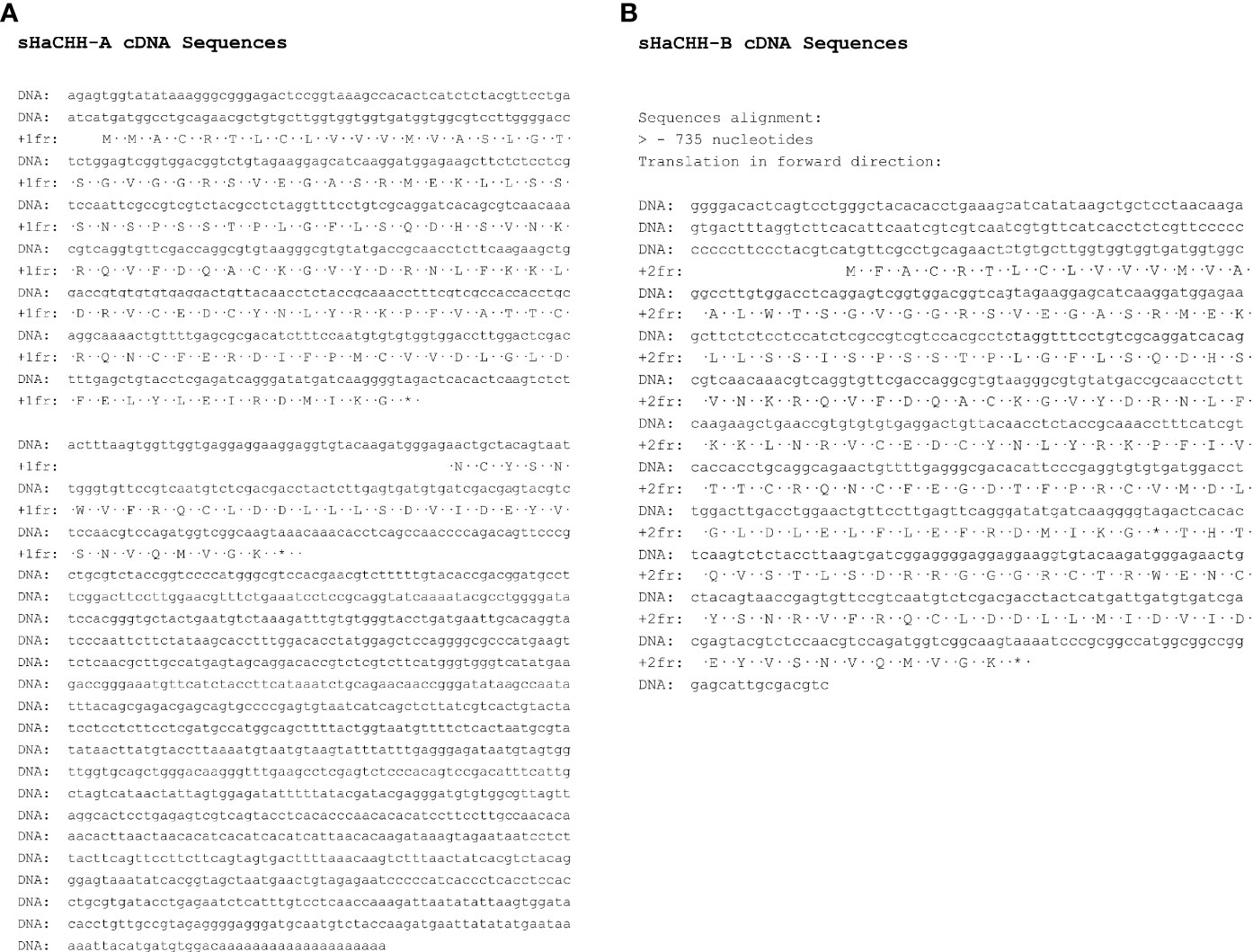
Figure 3 Nucleotide and deduced amino acid sequences of the computed cDNA for sHaCHH-A (A) and sHaCHH-B (B). A one-letter amino acid code was used to indicate the amino acid residues.

Figure 4 Alignment of the lobster four-exon CHH genes with that of other decapods, including lobster (yellow: HaCHH-A and HaCHH-B; NnCHH-1 and NnCHH-2), crab (gray: CmCHH-1 and CmCHH2), shrimp (green: Fm306-c1 and Fm306-c2) (Shi et al., 2018), freshwater shrimp (pink: MrCHH1 and MrCHH2), and crayfish (blue: PcCHH1, PcCHH2). The same block color indicates the same groups of decapods. Signal peptides and mature peptides are indicated. Gaps are introduced in the boundary of the exons in lobster, shrimp, and crab CHH-like genes. A question mark is inserted in the locations of potential intron insertion sites in the CHH-like genes from other decapods. In addition to the conserved cysteine residues (highlighted in red), several conserved amino acids (highlighted in gray) may be important for motif structure formation of the peptide.
The phylogenetic tree results revealed that the crustacean CHH can be divided into three different subgroups: CHH group, CHH-like group, and CHH-L/S group. The CHH group is the three-exon CHH gene consisting of the typical signal peptide–CPRP–mature peptide structure. The CHH-Like group is the more recently discovered CHHs having the signal peptide–mature peptide structure. The CHH-L/S group comprise the four-exon CHH genes that produce alternative transcripts, as described in this study. The lobster sHaCHH-A and sHaCHH-B were closely related to the crayfish CHH, followed by those of the shrimp and crab (Figure 5). As a reference, the MIH/GIH subtype evolved from a common ancestor, giving rise to the three-exon CHH and the four-exon CHH-L/S. It was observed that the four-exon CHH-L/S gene cluster represented a small number relative to the CHH group since only a few (i.e., LvCHH2 and MeCHH-B) of the CHH type were selected for phylogenetic tree construction (Figure 5).
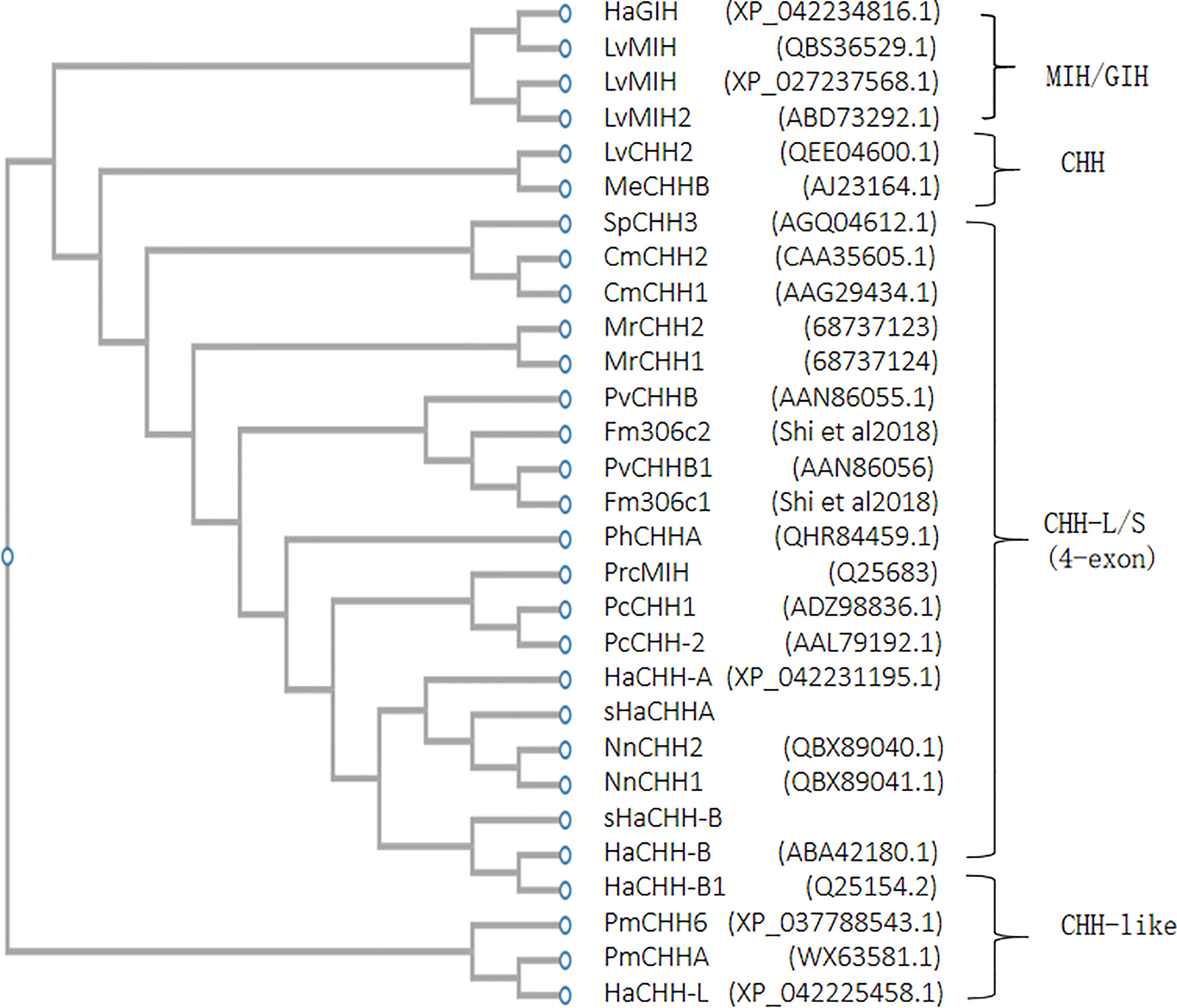
Figure 5 Phylogenetic tree analysis of lobster HaCHH-A and HaCHH-B cDNA within the major subfamilies of decapod CHH/MIH/GIH. The sequences included the lobster (Homarus americanus, Ha) PhCHH-A (QHR84459.1), HaCHH-L (XP_042225458.1), HaGIH (XP_042234816.1), HaCHH-A (XP_042231195.1), sHaCHH-A, NnCHH2 (QBX89040.1), NnCHH1 (QBX89041.1), sHaCHH-B, (ABA42180.1), and HaCHH-B1 (Q25154.2); crab (Carcinus maenas, Cm) CmCHH2 (CAA35605.1), CmCHH1 (AAG29434.1), and SpCHH3 (AGQ04612.1); shrimp (Litopenaeus vannamei, Lv) LvMIH (QBS36529.1), LvMIH (XP_027237568.1), LvMIH2 (ABD73292.1), LvCHH2 (QEE04600.1), PvCHHB (AAN86055.1), Fm306c2 (Shi et al., 2018), PvCHHB1 (AAN86056), Fm306c1 (Shi et al., 2018), PmCHH6 (XP_037788543.1) PmCHHA (WX63581.1), and MeCHHB (AJ23164.1); freshwater shrimp (Macrobrachium rosenbergii, Mr) MrCHH2 (68737123) and MrCHH1(68737124); and crayfish (Procambarus clarkii, Prc) PrcMIH (Q25683), PcCHH1 (ADZ98836.1), and PcCHH-2 (AAL79192.1). GenBank accession numbers are shown in parentheses.
Both semi-quantitative RT-PCR and RT-qPCR were used to study the expression of the HaCHH gene. For semi-quantitative PCR, multiple DNA fragments were amplified by F1A/R4 and F1B/R4 primers from different tissues. The smaller transcripts (i.e., 158 bp) represent the originally reported HaCHH-A or HaCHH-B cDNA, and they can be detected in the eyestalks of non-reproductive females. No specific pattern of these short transcripts was observed during the different stages of the reproductive cycle. The larger transcripts (i.e., 317 bp, which produced sHaCHH-A and sHaCHH-B) can be detected mainly in the Hp and thoracic ganglion. However, DNA fragments of different sizes were also amplified (i.e., especially in the thoracic ganglion) (Figure 6A). Further study by qPCR was performed using females in the early reproductive stages. The results confirmed that HaCHH-A and HaCHH-B were mainly expressed in the eyestalk. Also, sHaCHH-A and sHaCHH-B transcripts were detected in the non-eyestalk neuronal and in non-neuronal tissues, including the Hp, ovary, brain, and thoracic ganglion. However, the transcripts were not detected in the Hp (Figure 6B). The 158- and 317-bp PCR products were also subcloned and DNA sequencing was performed, which confirmed that they were specific for the HaCHH-A and HaCHH-B and the sHaCHH-A and sHaCHH-B isoform sequences, respectively. Nucleotide sequence analysis of 12 clones generated from the PCR products revealed five HaCHH-A and two sHaCHH-A clones and two HaCHH-B and one sHaCHH-B clone, confirming that the large sized clones were from the sHaCHH-A and/or sHaCHH-B and the small sized clones were from the HaCHH-A and/or HaCHH-B.
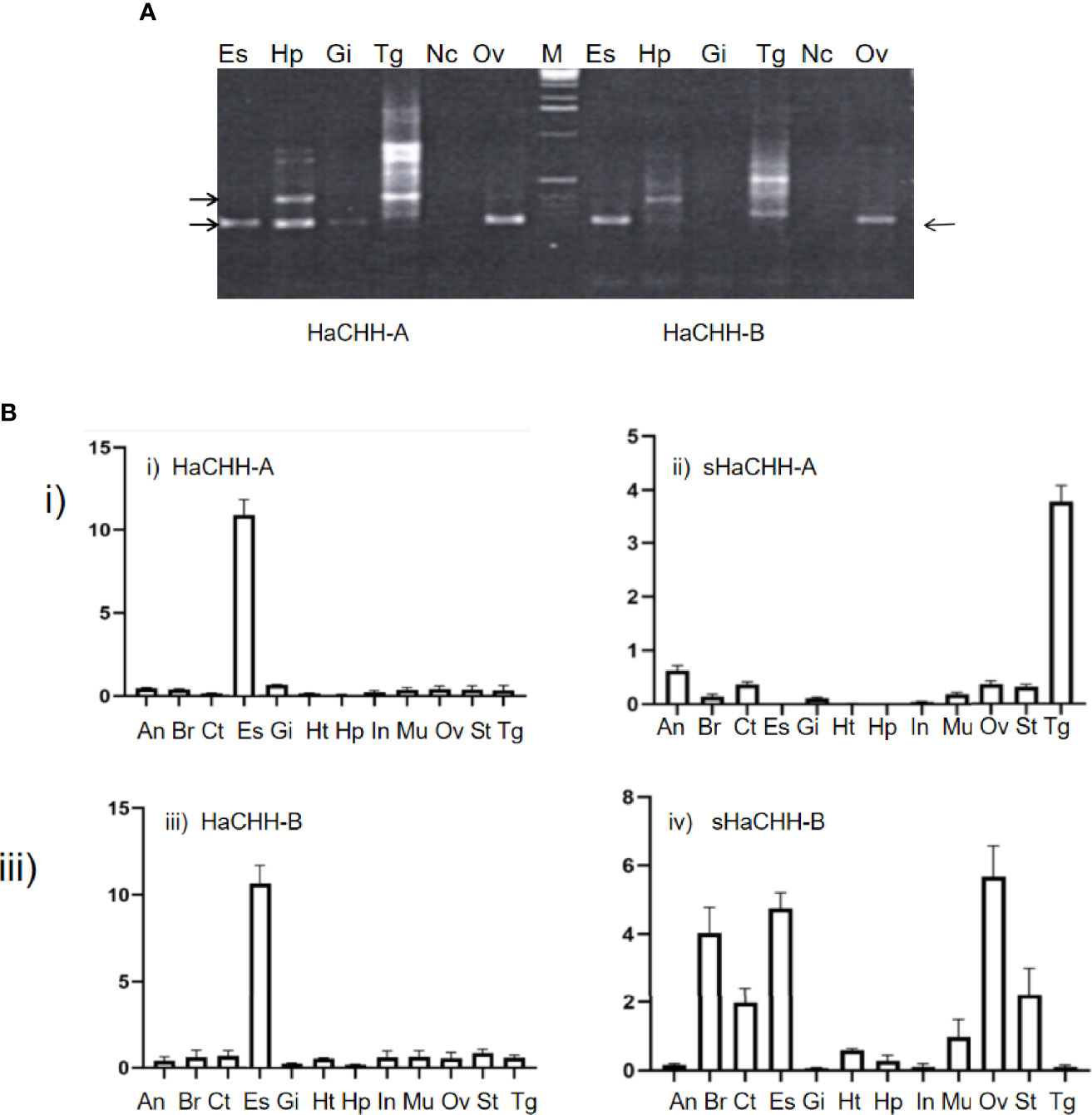
Figure 6 (A) RT-PCR expression study of the HaCHH-A, sHaCHH-A, HaCHH-B, and sHaCHH-B genes in different tissues of the lobster female. The primers F1A and RP were used to amplify HaCHH-A and sHaCHH-A, while primers F1B and RP were used to detect the transcripts of HaCHH-B and sHaCHH-B in the different tissues [eyestalk (Es), hepatopancreas (Hp), gill, (Gi), thoracic ganglion (Tg), nerve cord (Nc), and ovary (Ov)] of lobster during the early reproductive stage (stage I). (A) Representative agarose gels showing the detection of HaCHH-A. HaCHH-B, sHaCHH-A, and sHaCHH-B. The larger (317 bp) fragment is the sHaCHH-A or sHaCHH-B and the smaller (158 bp) fragment is the amplified partial HaCHH-A and HaCHH-B cDNA. (B) qPCR expression study of the i) HaCHH-A, ii) sHaCHH-A, iii) HaCHH-B, and iv) sHaCHH-B genes in different tissues of the female lobster. Primers HaCHH-AF/HaCHH-AR and sHaCHHAF/sHaCHHAR were used to amplify HaCHH-A and sHaCHH-A. Primers HaCHHAF/HaCHHAR and sHaCHH-BF/sHaCHH-BR were used to detect the transcripts of HaCHH-B and sHaCHH-B in the different tissues [abdominal nerve (An), brain (Br), cuticle (Ct), eyestalk (Es), hepatopancreas (Hp), heart (Ht), gill (Gi), intestine (In), muscle (Mu), stomach (St), thoracic ganglion (Tg), nerve cord (Nc), and ovary (Ov)] of lobster during the middle stage of reproduction (II).
Because previous tissue expression results suggested that neuronal tissues express relatively large amounts of sHaCHH-A and sHaCHH-B, we studied their expressions at different gonad maturation stages of the female lobster. The RT-PCR results indicated that they are the predominant transcripts during the different stages of maturation (i.e., stages I to IV). Although both sHaCHH-A and sHaCHH-B were expressed, the transcript level for sHaCHH-B was much lower than that of the original HaCHH-A and HaCHH-B transcripts. In contrast, the other non-eyestalk neuronal tissues (i.e., brain, thoracic ganglion, and nerve cord) expressed mainly the larger transcripts consisting of exons 1–4 (Figure 7A). In the thoracic ganglion and the nerve cord, the expression level of sHaCHH-A was consistently low. The expression level of HaCHH-B appeared higher during the early and active stages of vitellogenesis (Figure 7B). However, no specific expression pattern for sHaCHH-A/sHaCHH-B was recognized in the neuronal tissues. In summary, the expression study indicated that sHaCHH-A and sHaCHH-B are both expressed in non-eyestalk neuronal tissues, including the brain, thoracic ganglion, and ventral nerve cord. The expressions of sHaCHH-A and sHaCHH-B in other non-neuronal tissues, such as the Hp and ovary, during ovarian maturation suggested that these proteins are important in ovary maturation. Moreover, the expressions of these genes are stage-specific as lower transcript levels were detected during the immature stage, but much higher levels of expression in lobster undergo reproduction.
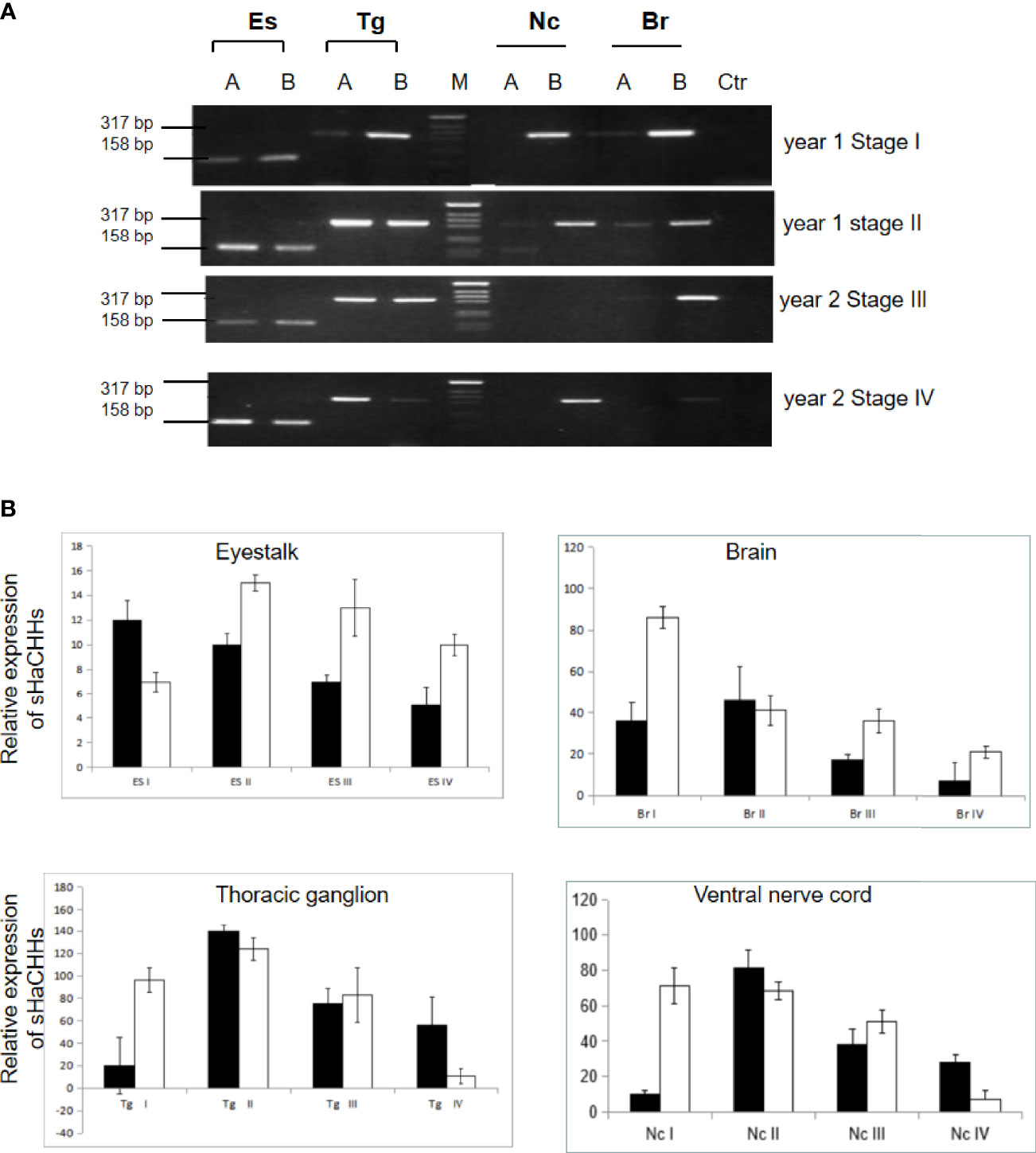
Figure 7 Expression study of sHaCHH-A and sHaCHH-B by Semi-quantitative (A) and (B) qPCR. (A) Agarose gel analysis of the PCR products and detection of the sHaCHH-A, HaCHH-A sHaCHH-B, HaCHH-B. The lanes A used primers for HaCHH-A gene and lane B used primers for HaCHH-B forcDNA from eyestalk (Es), thoracic ganglion (Tg), ventral nerve cord (Nc) and brain (Br).
Purified recombinant protein and dsRNA were added to the nutrient medium in the tissue explant bioassay experiments (Figure 8). In stage I Hp, recombinant sHaCHH-A (i.e., 2 μg) caused a decrease in the transcript level of HaVg1. Despite the lower expression of HaVg1 in the ovary compared to that in the hepatopacreas, an increase in the concentration of rsHaCHH-A caused a decrease in the expression of HaVg1 in a dose-dependent manner (p < 0.05). In stage I Hp and ovary, the dsRNAs for sHaCHH-A caused a significant increase in the expression of HaVg1 (p < 0.05) (Figure 9A). In stage III Hp, rsHaCHH-A caused a decrease of HaVg1 in a dose-dependent manner (p < 0.05). However, the expression level of HaVg1 in the ovary was not affected. When dssHaCHH-A was added to the medium, the expression levels of HaVg1 in the Hp and ovary increased (Figure 9B).
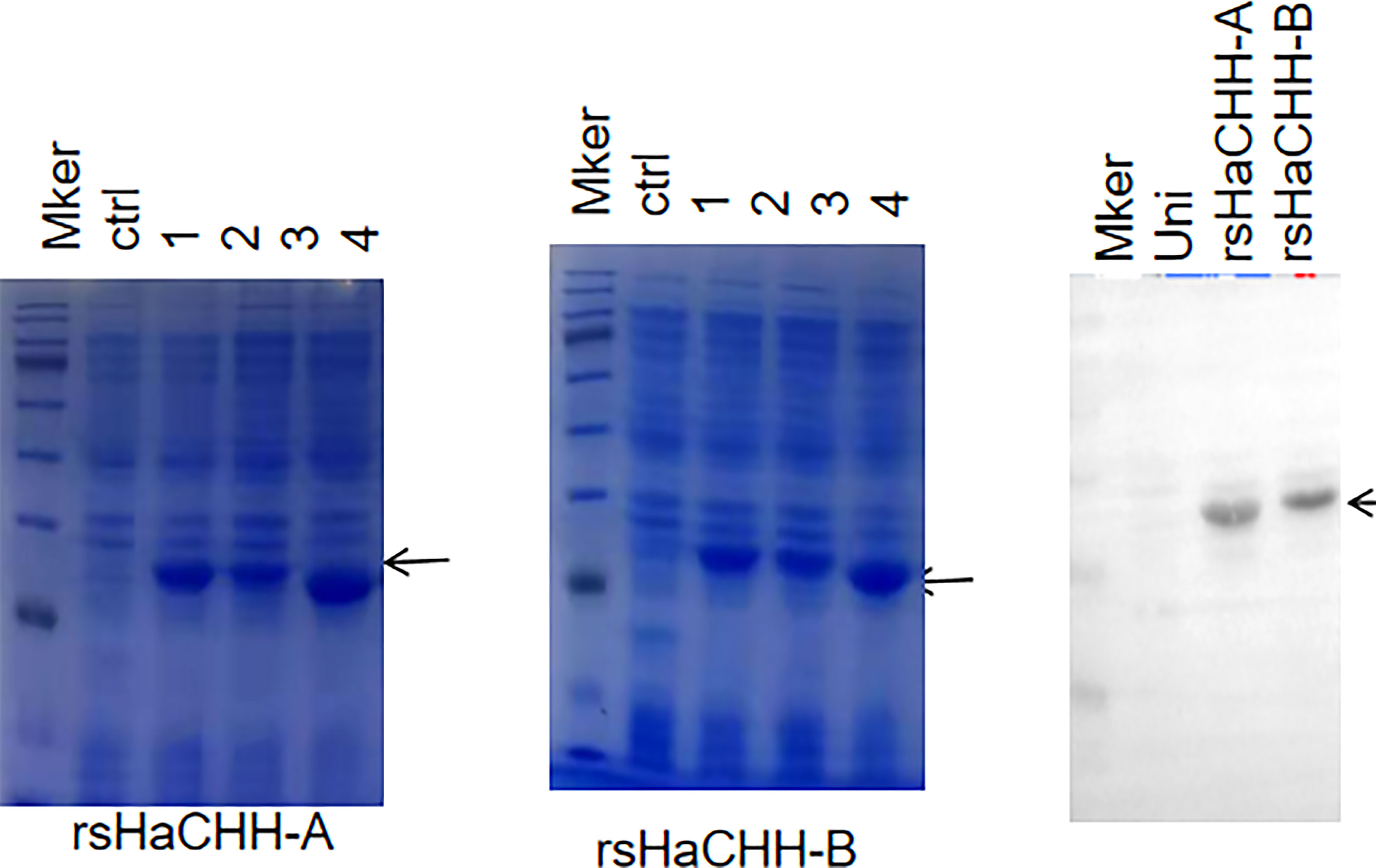
Figure 8 SDS-PAGE (12.5%) analysis of the prokaryotic (Escherichia coli) expression of the recombinant proteins rsHaCHH-A (left) and rsHaCHH-B (middle) after induction by IPTG. Ctrl: uninduced (uni); lanes 1–4: induced and sampled at 2, 4, 6, and 8 h, respectively. Western blot detection of the poly-His-tagged recombinant proteins sHaCHH-A and sHaCHH-B (arrow). Right: The antibody used is the anti-polyhistidine peroxidase conjugate antibody (Sangon Biotech, Shanghai, China).
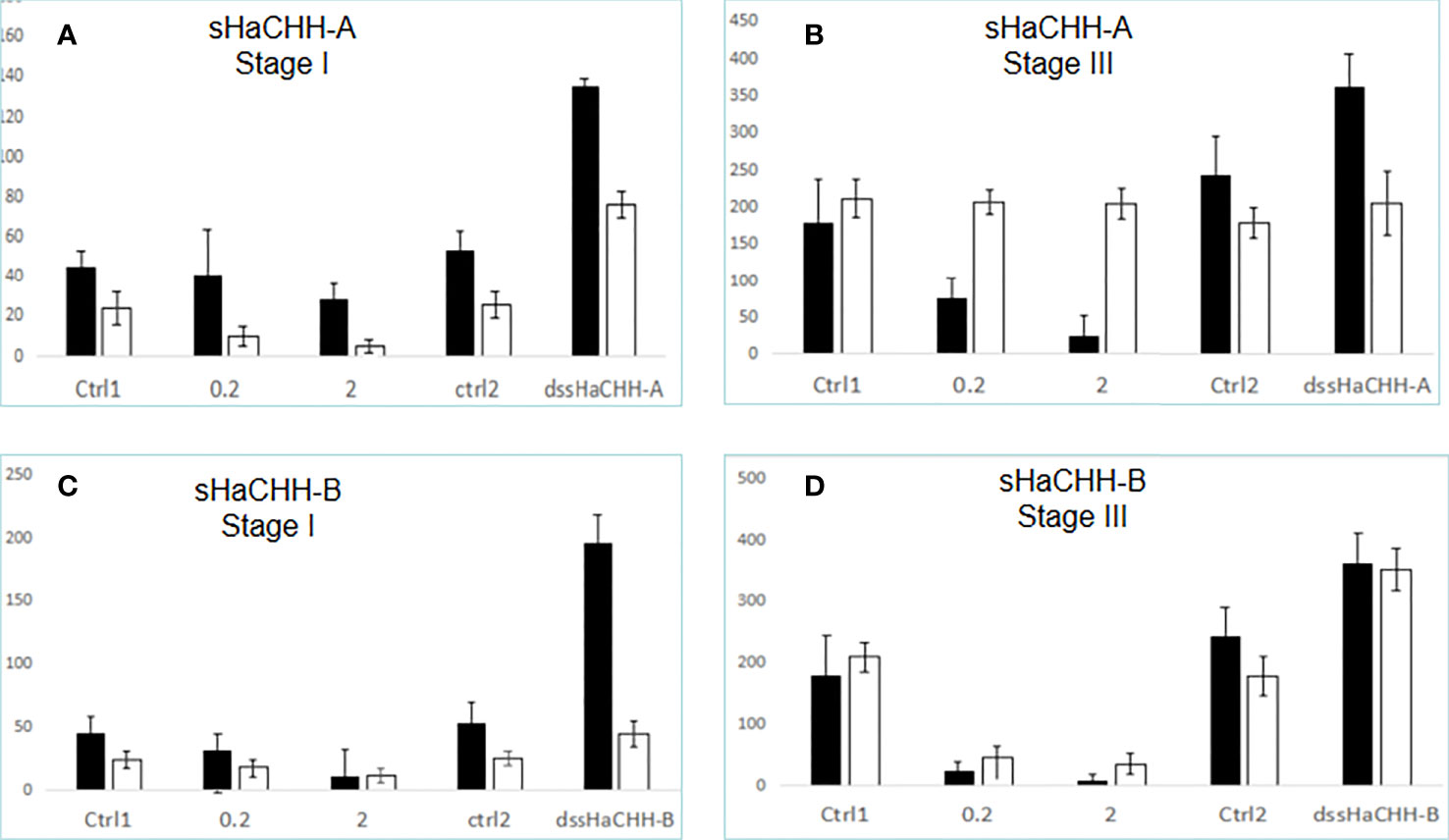
Figure 9 Effect of recombinant proteinand dsRNA for sHaCHH (A, B) and sHaCHH-B (C, D) on HaVg1 gene expression. The Y-axis shows the relative expression level of the HaVg1 gene in the hepatopancreas (black bar) and ovary (open white bar). Triplicate tests (N = 4 samples) were performed for each animal. The lobsters used were in the early stage of vitellogenesis (stage I) and middle–late stage (stage III) of the gonad maturation cycle. Tissue fragments were placed inside the well containing 1.5 ml of the nutrient medium M199 with either the control recombinant protein (0, 0.2, or 2.0 μg/PBS) and ctrl2 dsTFV, 2 μg dssHaCHH-A, or dssHaCHH-B. After incubating for 3 h, the hepatopancreas and ovary fragments were analyzed for the expression of HaVg1. The vitellogenin gene-specific primer was derived from the HaVg1 cDNA, as reported earlier (Tiu et al., 2008). Data are the mean ± SE.
When the recombinant protein for sHaCHH-B was added to the medium, a decrease in the level of HaVg1 was observed in the Hp and ovary from stages I and III (Figures 9C, D). When the dsRNAs for sHaCHH-B were added, the expression levels of HaVg1 also increased in the Hp and ovary fragments. It appears that the level of stimulation was much higher for the dssHaCHH-B-treated Hp. In summary, the results indicated that the recombinant proteins of sHaCHH-A and sHaCHH-B inhibited the expression of HaVg1, but gene knockdown of sHaCHH-A and sHaCHH-B stimulated the expression of HaVg1 in the Hp and ovary fragments (Figure 9).
Major advancement occurred in crustacean endocrinology in the recent application of the next-generation sequencing technique in transcriptome and genome studies. As a result, the number of CHH family genes identified in the same species and the total number of CHH in crustaceans have greatly increased (Havird and Santos, 2016; Chang and Lai, 2018; Oliphant et al., 2018; Hyde et al., 2020; Mykles and Chang, 2020). Therefore, many new CHH sequences have been reported in recent years. However, most CHH sequencing projects mainly focused on economically important species. Therefore, the number of CHH family members identified in lobster is fewer than that of the shrimps and other decapods (Hyde et al., 2020). Based on genomic and transcriptomic sequence data from L. vannamei and P. monodon, it is evident that the CHH family consists of multiple gene members. In L. vannamei, it is estimated that at least >65 CHH family genes are present in the genome (Zhang et al., 2019). However only a small portion of the CHH genes had the four-exon gene organization. Therefore, the four-exon CHH genes from the decapods shown in the phylogenetic tree in Figure 4 represent a very small portion of the same species within the same gene family. Although the number of CHH genes reported in lobster was fewer, it is unlikely that H. americanus consists of only two highly homologous four-exon CHH genes. From the recent lobster genome project (Polinski et al., 2021), we only identified <5 CHH-related genes. Moreover, no positive sequence was returned when we used several lobster genes (from our lobster transcriptome project) as the query sequence to the BLAST search against the lobster genome. Therefore, additional lobster CHH-like sequences may exist, but were missed in the lobster genome project. Despite the ambiguity, the lobster genome should consist of fewer number of CHH/MIH/GIH genes compared to the shrimps. From the expression and multiple alignment results (Figure 5), some generalizations can be proposed. The non-eyestalk expressed transcript is usually the larger CHH variant derived from the four-exon transcript. In addition, due to the broad distribution of the transcripts in the non-eyestalk and non-neuronal tissues, it is suggested that they may have a broad spectrum of functions.
Structurally, the sHaCHH-A and sHaCHH-B identified in this study had the major features of the CHH neuropeptide, and they shared identical lengths with the mature peptides and are conserved in other lobsters. In the lobster N. norvegicus, despite two NnCHH being cloned, the two cDNAs shared >98% amino acid identity in the mature peptide region; a second CHH that shares similar exon 3 has not been reported. In L. vannamei (GenBank no. AAK69346.1) and P. monodon (GenBank no. XP_037787978) CHH-like, they also consisted of identical amino acid residues in the mature peptide region. All the short/smaller CHH-like variants were derived from longer transcripts consisting of exons 1–4. For example, in Fenneropenaeus merguiensis, the gill form CHH-like was derived from a longer transcript (i.e., exons 1–4) and the larger CHH-like was derived from a smaller transcript consisting of exons 1, 2, and 4 (Shi et al., 2018). The mature peptides of sHaCHH-A and sHaCHH-B, and sNnCHH-A and sNnCHH-B consisted of 73 and 74 amino acid sequences, respectively (Mettulio et al., 2004). The phylogenetic tree also revealed that the four-exon CHH-L/S gene (Figure 5) may be derived from the common ancestor gene that gave rise to the more abundant three-exon CHH genes (i.e., LvCHH2; GenBank no. QEE046000.1) and the MIH/GIH subtype. In the lobster H. americanus, a three-exon HaCHH gene was identified from the recent genome sequencing project (GenBank no. XP_042225458.1) and can be grouped into the CHH-like class, as indicated in Figure 5.
Although the four-exon CHH family genes have been reported in several decapods, but only the complete gene structure of L. vannamei is known. For the other decapods—the crab, freshwater shrimp, and crayfish—only the partial gene structure was revealed, and sequence information for the first intron was missing. We speculated that the gene organizations of these four-exon CHHs among decapods are highly similar. The sequence for the first intron of the CHH gene in L. vannamei was obtained from a genome sequencing project. The first intron of the CHH gene is 4.65 kb, and within this region, several highly repetitive simple sequence repeats (SSRs) exist. This highly repetitive sequence would affect the PCR reaction and, therefore, cause failure in genomic PCR (GenBank no. LOC113815764). Therefore, it is likely that the first exon of these decapod CHH genes is also made up of a large intron 1 and may consist of highly repetitive SSRs within the intron that hinder/disrupt the PCR amplification.
In terms of the evolutionary relationship of the CHH family genes, it was speculated that their original prototype consisted of three exons and two introns. The evolution of the CHH family genes began from gene duplication and mutation (Chan et al., 2003; Montagné et al., 2008; Chung et al., 2010; Chen et al., 2020). Alignment of the four-exon CHH neuropeptide genes revealed both similarities and differences among decapods. The lobster HaCHH-A and HaCHH-B shared >95%–100% amino acid identity with the lobster CHHs. The similarity was much higher (i.e., 99% amino acid identity) in the mature peptide region. In the Norway lobster Nephrops norvegius, two CHHs were cloned (i.e., GenBank nos. AY285782 and AY285783), which differed only in the signal peptide region, but the mature peptide showed 100% sequence identity (Mettulio et al., 2004). HaCHH-A and HaCHH-B shared only 77%–78% amino acid identity with the crayfish P. clarkii CHH-like (78.1%; GenBank no. Q25683) and Astacus leptodactyl (i.e., 77.3%; GenBank no. AAX09331). In earlier studies, the CHH family neuropeptides were reported to be expressed only in the eyestalks. Increasing evidence from recent expression studies has confirmed the expression of CHH family members in other non-eyestalk neuronal and non-neuronal tissues. So far, H. americanus is the only decapod consisting of two 4-exon CHH family genes, and these two genes could produce four different transcripts.
The original HaCHH-A and HaCHH-B transcripts are abundant in the eyestalk; therefore, RT-PCR cannot amplify the larger transcripts in the eyestalk. In early studies, polyclonal antibodies for HaCHH-A and HaCHH-B were produced to determine the titer of CHH during the reproductive cycle of females. For example, in N. norvegicus, CHH antibodies have been generated to localize the neuropeptide by immunocytochemistry (Giulianini et al., 2002). Other antibodies for CHH were produced, and ELISA was performed to measure the concentration of CHH-A in the hemolymph (Chang et al., 1998; Chang et al., 1999). The cross-reactivity of the polyclonal antibody to sHaCHH-A and sHaCHH-B has not been evaluated. Therefore, it is necessary to reexamine the protein level of CHH during the reproductive cycle using more specific antisera against specific isoforms. The role of sHaCHH-A and sHaCHH-B in other neuronal tissues, i.e., brain, thoracic ganglion, and ventral nerve cord, has not been examined. In short, more specific antibody is needed to determine the protein expression patterns of sHaCHH during the lobster reproductive cycle.
The two CHH genes reported in this study can produce a total of four different transcripts (i.e., HaCHH-A, sHaCHH-A, HaCHH-B, and sHaCHH-B). However, the expressions of these transcript variants appeared to be tightly regulated as they may exist in a specific temporal and spatial manner. One of the major controversies in the study of HaCHH-A expression and protein level was the lack of correlation in both the mRNA and peptide levels in the hemolymph during the pre-vitellogenic stage (de Kleijn et al., 1998). For HaCHH-B, the total mRNA and hemolymph CHH peptide levels increased in the mature stage, and there were no differences in peptide storage that may suggest that synthesis is not related to storage (de Kleijn et al., 1998). The interpretation was hampered by the lack of information for these transcripts’ variants. The expression study was focused only on the mRNA in the eyestalk, and the antibody used to detect HaCHH-A and HaCHH-B may not be able to differentiate sHaCHH-A and/or sHaCHH-B. Therefore, a more systematic approach for the study of the titer and expression patterns must be reinvestigated to fully elucidate the roles of these hormones in lobster reproduction control.
Alternative splicing is a method used to create more proteins from the same gene (Baralle and Giudice, 2017; Gallego-Paez et al., 2017; Modrek and Lee, 2002). It is an essential mechanism increasing the complexity of gene expression, and it plays an important role in cellular differentiation and organism development. There are several reports for the alternative splicing of the CHH gene in crustaceans (Dircksen et al., 2003; Chen et al., 2004; Jeon et al., 2012; Shi et al., 2018; Sun et al., 2019). In the Morotoge shrimp Pandalopsis japonica, an eyestalk form and a pericardial organ isoform were identified. Although the reproductive function of the CHH variant was not studied, eyestalk ablation would reduce the expression of Pj-CHH1ES in the brain and Pj-CHH1PO and Pj-CHH2 in the thoracic ganglion (Jeon et al., 2012). The Mar-CHH gene of M. rosenbergii also consists of four exons. The eyestalk transcript (chh) contains exons I, II, and IV, whereas the CHH-l transcript in the heart, gills, antennal glands, and thoracic ganglion contains all four exons. Similarly, the sHaCHH-A and sHaCHH-B transcripts are mainly expressed in non-eyestalk tissues. The production of alternatively spliced variants from a single gene, therefore, would increase the number of proteins in a cell and increase the overall proteome content within the cell. Additionally, these spliced variants may be produced at different stages of the life cycle (Wang et al., 2015). The differential stage-specific expression of the transcripts in lobster might contribute to its regulation during the reproductive cycle in the biannual reproductive pattern of H. americanus. Despite that a similar 3D structure was predicted between HaCHH-A vs. HaCHH-B and sHaCHH-A vs. sHaCHH-B, the pI values (i.e., hydrophobicity) of the corresponding proteins were quite different. The differences in the hydrophobicity may have allowed different isoforms to work optimally in different pH conditions of the Hp and ovary. Therefore, we hypothesized that, with a lower pI value (i.e., 5.27), sHaCHH-A may work optimally under a more acidic condition in the Hp, which has a lower pH for the function of the acidic digestive enzyme.
A major challenge in the research on the CHH/MIH/GIH family is the lack of functional studies for most of the members. Therefore, the bioassay/functional confirmation of most CHHs was not performed. The use of recombinant protein and RNAi technique to study the function of the CHH family genes has been reported for the MIH-like in the sand shrimp and the MIH-like in the banana shrimp F. merguiensis type II neuropeptide (Gu et al., 2000; Tiu et al., 2008; Liang et al., 2019). Opposing results of the dsRNAs and recombinant proteins were demonstrated in the sand shrimp and in the lobster in this study. Since there are multiple numbers of genes in the CHH/MIH/GIH family, the antagonistic results from the recombinant proteins and dsRNAs may provide a tool to investigate the role of this neuropeptide superfamily.
The present study has confirmed that the previously reported HaCHH-A and HaCHH-B cDNAs were derived from alternative splicing of two 4-exon CHH genes. The retention of intron 3 and exon 4 produced the transcript variants sHaCHH-A and sHACHH-B, as reported in this study. Results of the functional study indicated that sHaCHH-A and sHaCHH-B can regulate the gene expression of vitellogenin. The findings in this study have provided new information on the gene structure organization, precise expression patterns for the transcript variants, and the vitellogenin-inhibiting function of sHaCHH-A and sHaCHH-B. The results of this report have also provided a basis for reassessing the expression patterns and hemolymph titers of these transcripts during the reproductive cycle of the female lobster.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
SC: conceptualization, writing—original draft preparation, project administration, and funding acquisition. SC, CGW, and YS: methodology. CGW: software and resources. WW: validation. YS and WW: formal analysis. WG and SC: writing—review and editing. LLS: visualization. YS and SC: supervision. All authors contributed to the article and approved the submitted version.
This study was funded in part by the National Natural Science Foundation of China (#31572606) and Zhanjiang City Fund.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Baralle F. E., Giudice J. (2017). Alternative Splicing as a Regulator of Development and Tissue Identity. Nat. Rev. Mol. Cell Biol. 18, 437–451. doi: 10.1038/nrm
Chang E. S., Chang S. A., Beltz B. S., Kravitz E. A. (1999). Crustacean Hyperglycemic Hormone in the Lobster Nervous System: Localization and Release From Cells in the Subesophageal Ganglion and Thoracic Second Roots. J. Comp. Neurol. 414, 50–56. doi: 10.1002/(SICI)1096-9861(19991108)414:1<50::AID-CNE4>3.0.CO;2-Q
Chang E. S., Keller R., Chang S. A. (1998). Quantification of Crustacean Hyperglycemic Hormone by ELISA in Hemolymph of the Lobster, Homarus Americanus, Following Various Stresses. Gen. Comp. Endocrinol. 111, 359–366. doi: 10.1006/gcen.1998.7120
Chang W. H., Lai A. G. (2018). Comparative Genomic Analysis of Crustacean Hyperglycemic Hormone (CHH) Neuropeptide Genes Across Diverse Crustacean Species. F1000Res 7, 100. doi: 10.12688/f1000research.13732.1
Chan S. M., Gu P. L., Chu K. H. (2003). Crustacean Neuropeptide Genes of the CHH/MIH/GIH Family: Implications From Molecular Studies. Gen. Comp. Endocrinol. 134, 214–219. doi: 10.1016/S0016-6480(03)00263-6
Chen S. H., Lin C. Y., Kuo C. M. (2004). Cloning of Two Crustacean Hyperglycemic Hormone Isoforms in Freshwater Giant Prawn (Macrobrachium Rosenbergii): Evidence of Alternative Splicing. Mar Biotechnol. (NY) 6, 83–94. doi: 10.1007/s10126-003-0014-8
Chen H. Y., Toullec J. Y., Lee C. Y. (2020). The Crustacean Hyperglycemic Hormone Superfamily: Progress Made in the Past Decade. Front. Endocrinol. (Lausanne) 11. doi: 10.3389/fendo.2020.578958
Chung J. S., Christie A., Flynn E. (2020). Molecular Cloning of Crustacean Hyperglycemic Hormone (CHH) Family Members (CHH, Molt-Inhibiting Hormone and Mandibular Organ-Inhibiting Hormone) and Their Expression Levels in the Jonah Crab. Cancer Borealis Gen. Comp. Endocrinol. 1 (295), 113522. doi: 10.1016/j.ygcen.2020.113522
Chung J. S., Zmora N., Katayama H., Tsutsui N. (2010). Review. Crustacean Hyperglycemic Hormone (CHH) Neuropeptides Family: Functions, Titer, and Binding to Target Tissues. Gen. Comp. Endocrinol. 166, 447–454. doi: 10.1016/j.ygcen.2009.12.011
De Kleijn D. P., De Leeuw E. P., Van den Berg M. C., Martens G. J., Van Herp F. (1995). Cloning and Expression of Two mRNAs Encoding Structurally Different Crustacean Hyperglycemic Hormone Precursors in the Lobster. Homarus Americanus Biochim. Biophys. Acta 1260, 62–66.
de Kleijn D. P., Janssen K. P., Waddy S. L., Hegeman R., Lai W. Y., Martens G. J., Van Herp F. (1998). Expression of the Crustacean Hyperglycaemic Hormones and the Gonad-Inhibiting Hormone During the Reproductive Cycle of the Female American Lobster Homarus Americanus. J. Endocrinol. 156 (2), 291–298. doi: 10.1677/joe.0.1560291
De Kleijn D. P., Janssen K. P., Martens G. J., Van Herp F. (1994). Cloning and Expression of Two Crustacean Hyperglycemic-Hormone mRNAs in the Eyestalk of the Crayfish. Orconectes Limosus Eur. J. Biochem. 224, 623–629. doi: 10.1111/j.1432-1033.1994.00623.x
De Kleijn D. P., Van Herp F. (1995). Molecular Biology of Neurohormone Precursors in the Eyestalk of. Crustacea. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 112, 573–579. doi: 10.1016/0305-0491(95)00126-3
Dircksen H., Böcking D., Heyn U., Mandel C., Chung J. S, Baggerman G., et al. (2003). Crustacean Hyperglycaemic Hormone (CHH)-Like Peptides and CHH-Precursor-Related Peptides From Pericardial Organ Neurosecretory Cells in the Shore Crab, Carcinus Maenas, are Putatively Spliced and Modified Products of Multiple Genes. Biochem. J. 356, 159–170. doi: 10.1042/bj3560159
Fingerman M. (1987). The Endocrine Mechanisms of Crustaceans. J. Crustacean Biol. 7, 1–24. doi: 10.2307/1548622
Gallego-Paez L. M., Bordone M. C., Leote A. C., Saraiva-Agostinho N., Ascensão-Ferreira M., Barbosa-Morais N. L. (2017). Alternative Splicing: The Pledge, the Turn, and the Prestige: The Key Role of Alternative Splicing in Human Biological Systems. Hum. Genet. 136, 1015–1042. doi: 10.1007/s00439-017-1790-y
Giulianini P. G., Pandolfelli N., Lorenzon S., Ferrero E. A., Edomi P. (2002). An Antibody to Recombinant Crustacean Hyperglycaemic Hormone of Nephrops Norvegicus Cross-Reacts With Neuroendocrine Organs of Several Taxa of Malacostracan Crustacea. Cell Tissue Res. 307, 243–254. doi: 10.1007/s00441-001-0484-9
Gu P. L., Yu K. L., Chan S. M. (2000). Molecular Characterization of an Additional Shrimp Hyperglycemic Hormone: cDNA Cloning, Gene Organization, Expression and Biological Assay of Recombinant Proteins. FEBS Lett. 472, 122–128. doi: 10.1016/S0014-5793(00)01420-4
Havird J. C., Santos S. R. (2016). Here We Are, But Where Do We Go? A Systematic Review of Crustacean Transcriptomic Studies From 2014-2015. Integr. Comp. Biol. 56 (6), 1055–1066. doi: 10.1093/icb/icw061
Helluy S. M., Beltz B. S. (1991). Embryonic Development of the American Lobster (Homarus Americanus): Quantitative Staging and Characterization of an Embryonic Molt Cycle. Biol. Bull. 180, 355–371. doi: 10.2307/1542337
Huberman A. (2000). Shrimp Endocrinology: A Review. Aquaculture 191, 191–208. doi: 10.1016/S0044-8486(00)00428-2
Hyde C. J., Fitzgibbon Q. P., Elizur A., Smith G. G., Ventura T. (2020). CrustyBase: An Interactive Online Database for Crustacean Transcriptomes. BMC Genomics 21, 637. doi: 10.1186/s12864-020-07063-2
Jayasankar V., Tomy S., Wilder M. N. (2020). Insights on Molecular Mechanisms of Ovarian Development in Decapod Crustacea: Focus on Vitellogenesis-Stimulating Factors and Pathways. Front. Endocrinol. (Lausanne) 11. doi: 10.3389/fendo.2020.577925
Jeon J.-M., Kim B.-K., Lee J. H., Kim H. K., Kang C.-K., Mykles D. L., et al. (2012). Two Type I Crustacean Hyperglycemic Hormone (CHH) Genes in Morotoge Shrimp (Pandalopsis Japonica): Cloning and Expression of Eyestalk and Pericardial Organ Isoforms Produced by Alternative Splicing and a Novel Type I CHH With Predicted Structure Shared With Type II CHH Peptides Comp. Biochem. Physiol. B Biochem. Mol. Biol. 162 (4), 88–99. doi: 10.1016/j.cbpb.2012.04.003
Keller R. (1992). Crustacean Neuropeptides: Structures, Functions and Comparative Aspects. Experientia 48, 439–448. doi: 10.1007/BF01928162
Liang H. F., Liu Y., Zhou T. T., Li ,. X. Y., Li B., Chan S.F, et al. (2019). Molecular Characterization, RNA Interference and Recombinant Protein Approach to Study the Function of the Putative Molt Inhibiting Hormone (FmMIH1) Gene From the Shrimp Fenneropenaeus Merguiensis. Peptides 122, 1–12. doi: 10.1016/j.peptides.2017.10.017
Loredo-Ranjel R., Fanjul-Moles M. L., Escamilla-Chimal E. G. (2017). Crustacean Hyperglycemic Hormone Is Synthesized in the Eyestalk and Brain of the Crayfish Procambarus Clarkii. PloS One 12 (4), e0175046. doi: 10.1371/journal.pone.0175046
Magaña-Gallegos E., Arévalo M., Cuzon G., Gaxiola G. (2021). Effects of Using the Biofloc System and Eyestalk Ablation on Reproductive Performance and Egg Quality of Litopenaeus Vannamei (Boon) (Decapoda: Dendrobranchiata: Penaeidae). Anim. Reprod. Sci. 228, 106749. doi: 10.1016/j.anireprosci.2021.106749
Mettulio R., Edomi P., Ferrero E. A., Lorenzon S., Giulianini P. G. (2004). The Crustacean Hyperglycemic Hormone Precursors a and B of the Norway Lobster Differ in the Preprohormone But Not in the Mature Peptide. Peptides 25, 1899–1907. doi: 10.1016/j.peptides.2004.06.013
Modrek B., Lee C. (2002). A Genomic View of Alternative Splicing. Nat. Genet. 30, 13–19. doi: 10.1038/ng0102-13
Montagné N., Soyez D., Céline D., Jean-Yves G., Toullec O. (2008). New Insights Into Evolution of Crustacean Hyperglycaemic Hormone in Decapods–First Characterization in Anomura. FEBS J. 275, 1039–1052. doi: 10.1111/j.1742-4658.2007.06245.x
Mykles D. L., Chang E. S. (2020). Hormonal Control of the Crustacean Molting Gland: Insights From Transcriptomics and Proteomics. Gen. Comp. Endocrinol. 294, 113493. doi: 10.1016/j.ygcen.2020.113493
Okumura T., Aida K. (2001). Effects of Bilateral Eyestalk Ablation on Molting and Ovarian Development in the Giant Freshwater Prawn. Macrobrachium Rosenbergii Fish Sci. 67, 1125–1135. doi: 10.1046/j.1444-2906.2001.00370.x
Oliphant A., Alexander J. L., Swain M. T., Webster S. G., Wilcockson D. C. (2018). Transcriptomic Analysis of Crustacean Neuropeptide Signaling During the Moult Cycle in the Green Shore Crab, Carcinus Maenas. BMC Genomics 19, 711–720. doi: 10.1186/s12864-018-5057-3
Panouse J. B. (1943). Influence De L’ablation Du Pédoncle Oculaire Sur La Croissance De L’ovaire Chez La Crevette Leander Serratus. Comptes Rendus l’Académie Des. Sci. 217, 553–555.
Polinski J. M., Zimin A. V., Clark K. F., Kohn A. B., Sadowski N., Timp W. (2021). The American Lobster Genome Reveals Insights on Longevity, Neural, and Immune Adaptations. Sci. Adv. 7 (26). doi: 10.1126/sciadv.abe8290.
Primavera J. H. (1978). Induced Maturation and Spawning in Five-Month-Old Penaeus Monodon Fabricius by Eyestalk Ablation. Aquacult 13, 355–359. doi: 10.1016/0044-8486(78)90184-9
Reddy P. R., Kishori B. (2017). Mini Review. Crustacean Endocrinology: Intriguing Towards Quality Protein Production. J. Endocrinol. Thyroid Res. 1 (2), 555556. doi: 10.19080/JETR.2017.01.555556
Shi L., Li B., Zhou T. T., Wang W., Chan S. F. (2018). Functional and Evolutionary Implications From the Molecular Characterization of Five Spermatophore CHH/MIH/GIH Genes in the Shrimp. Fenneropenaeus Merguiensis PloS One 19, 13(3):e0193375. doi: 10.1371/journal.pone.0193375
Sun D., Lv J., Gao B., Liu P., Li J. (2019). Crustacean Hyperglycemic Hormone of Portunus Trituberculatus: Evidence of Alternative Splicing and Potential Roles in Osmoregulation. Cell Stress Chaperones 24, 517–525. doi: 10.1007/s12192-019-00980-6
Tensen C. P., Verhoeven A. H., Gaus G., Janssen K. P., Keller R., Van Herp F. (1991). Isolation and Amino Acid Sequence of Crustacean Hyperglycemic Hormone Precursor-Related Peptides. Peptides 12, 673–681. doi: 10.1016/0196-9781(91)90119-A
Tiu S. H. K., Chan S.-M. (2007). The Use of Recombinant Protein and RNA Interference Approaches to Study the Reproductive Functions of a Gonad-Stimulating Hormone From the Shrimp Metapenaeus Ensis. FEBS J. 274, 4385–4395. doi: 10.1111/j.1742-4658.2007.05968.x
Tiu S. H., Hui H. L., Tsukimura B., Tobe S. S., He J. G., Chan S. M. (2009). Cloning and Expression Study of the Lobster (Homarus Americanus) Vitellogenin: Conservation in Gene Structure Among Decapods. Gen. Comp. Endocrinol. 160, 36–46. doi: 10.1016/j.ygcen.2008.10.014
Uawisetwathana U., Leelatanawit R., Klanchui A., Prommoon J., Klinbunga S., Karoonuthaisiri N. (2011). Insights Into Eyestalk Ablation Mechanism to Induce Ovarian Maturation in the Black Tiger Shrimp. PloS One 6, e24427. doi: 10.1371/journal.pone.0024427
Wang Y., Liu J., Huang B. O., Xu Y. M., Li J., Huang L. F., et al. (2015). Mechanism of Alternative Splicing and its Regulation. Biomed. Rep. 3, 152–158. doi: 10.3892/br.2014.407
Webster S. G. (2001). Crustacean Hyperglycaemic Hormone (CHH)-Like Peptides and CHH-Precursor-Related Peptides From Pericardial Organ Neurosecretory Cells in the Shore Crab, Carcinus Maenas, are Putatively Spliced and Modified Products of Multiple Genes. Biochem. J. 356, 159–170. doi: 10.1042/bj3560159
Webster S. G., Keller R., Dircksen H. (2012). The CHH-Superfamily of Multifunctional Peptide Hormones Controlling Crustacean Metabolism, Osmoregulation, Moulting, and Reproduction. Gen. Comp. Endocrinol. 175, 217–233. doi: 10.1016/j.ygcen.2011.11.035
Keywords: shrimp, eyestalk, crustacean hyperglycemic hormone, alternative splicing, vitellogenin
Citation: Wang CG, Wang W, Shi LL, Shen YC and Chan SF (2022) Alternative Splicing of the Lobster (Homarus americanus) Crustacean Hyperglycemic Hormone A and B Genes Produce 2 Protein Variants Involved in Vitellogenin Inhibition. Front. Mar. Sci. 9:861691. doi: 10.3389/fmars.2022.861691
Received: 25 January 2022; Accepted: 11 March 2022;
Published: 20 April 2022.
Edited by:
Benjamin Costas, University of Porto, PortugalReviewed by:
Piero Giulio Giulianini, University of Trieste, ItalyCopyright © 2022 Wang, Wang, Shi, Shen and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Chun Shen, c2hlbnl1Y2h1bkAxNjMuY29t; Siuming F. Chan, c2l1bWluZzU3M0BzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.